-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
Meiotic recombination is a mandatory process for sexual reproduction. We identified a protein specifically implicated in meiotic homologous recombination that we named: meiosis specific with OB domain (MEIOB). This protein is conserved among metazoan species and contains single-strand DNA binding sites similar to those of RPA1. Our studies in vitro revealed that both recombinant and endogenous MEIOB can be retained on single-strand DNA. Those in vivo demonstrated the specific expression of Meiob in early meiotic germ cells and the co-localization of MEIOB protein with RPA on chromosome axes. MEIOB localization in Dmc1−/− spermatocytes indicated that it accumulates on resected DNA. Homologous Meiob deletion in mice caused infertility in both sexes, due to a meiotic arrest at a zygotene/pachytene-like stage. DNA double strand break repair and homologous chromosome synapsis were impaired in Meiob−/− meiocytes. Interestingly MEIOB appeared to be dispensable for the initial loading of recombinases but was required to maintain a proper number of RAD51 and DMC1 foci beyond the zygotene stage. In light of these findings, we propose that RPA and this new single-strand DNA binding protein MEIOB, are essential to ensure the proper stabilization of recombinases which is required for successful homology search and meiotic recombination.
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003784
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003784Summary
Meiotic recombination is a mandatory process for sexual reproduction. We identified a protein specifically implicated in meiotic homologous recombination that we named: meiosis specific with OB domain (MEIOB). This protein is conserved among metazoan species and contains single-strand DNA binding sites similar to those of RPA1. Our studies in vitro revealed that both recombinant and endogenous MEIOB can be retained on single-strand DNA. Those in vivo demonstrated the specific expression of Meiob in early meiotic germ cells and the co-localization of MEIOB protein with RPA on chromosome axes. MEIOB localization in Dmc1−/− spermatocytes indicated that it accumulates on resected DNA. Homologous Meiob deletion in mice caused infertility in both sexes, due to a meiotic arrest at a zygotene/pachytene-like stage. DNA double strand break repair and homologous chromosome synapsis were impaired in Meiob−/− meiocytes. Interestingly MEIOB appeared to be dispensable for the initial loading of recombinases but was required to maintain a proper number of RAD51 and DMC1 foci beyond the zygotene stage. In light of these findings, we propose that RPA and this new single-strand DNA binding protein MEIOB, are essential to ensure the proper stabilization of recombinases which is required for successful homology search and meiotic recombination.
Introduction
Meiosis is a central process of sexual reproduction. This specialized cell division program allows halving the genome of diploid germ cells to produce haploid gametes. In order to ensure proper segregation of homologous chromosomes during the first meiotic division, these must become connected through chiasmata [1]. Crucially, formation of chiasmata depends on the occurrence of inter-homolog crossovers (CO) during the first meiotic prophase. COs originate from the recombination mediated-repair of programmed double strand breaks (DSBs) during meiotic prophase I. Meiotic recombination differs from mitotic recombination in that it uses a chromatid from the homolog instead of the sister chromatid as a template for repair [2]. It also favors CO formation and involves specific proteins [3]. In mice, about 250–300 DSBs are generated during the leptotene stage by the catalytic activity of the conserved topoisomerase-like transesterase SPO11 [4]–[6]. DNA ends at DSBs are resected to produce single stranded DNA for homology search [7]. Only a subset of DSBs form COs, the remaining DSBs being repaired without chromosome arm exchanges. The decision to form or not a CO is thought to be made before or during strand invasion of the homologous chromosome [8]. This being the case, the nature and the regulation of proteins loaded at broken ends is likely to be important for the outcome of the DSBs. Single-strand DNA (ssDNA) formed during DNA metabolism is coated by the trimeric replication protein A (RPA) complex (composed of RPA1 70 kDa, RPA2 32 kDa, RPA3 14 kDa) serving to protect from degradation and to prevent secondary structure formation. RPA binds ssDNA with high affinity through oligonucleotide binding (OB) domains [9], [10]. During homologous recombination, RPA has to be removed from the 3′ssDNA of broken ends to allow the formation of a presynaptic nucleofilament. This is essential for homology search and the formation of a physical connection between the invading ssDNA and a homologous duplex DNA template. Whereas mitotic recombination only needs RAD51 to search and invade a homologous sequence, meiotic recombination requires an additional recombinase, the meiosis-specific DMC1 protein [11]. In plants and mammals, the presence of BRCA2, which directly interacts with RAD51 and DMC1, is necessary for their localization, however, the exact mechanisms involved in RAD51 and DMC1 loading during meiosis remain to be determined [12]–[14]. Working models based on data mainly obtained in S. cerevisiae and A. thaliana proposed that Rad51 is essential for proper Dmc1 loading and that Rad51 recombinase activity has to be inhibited to favor the homolog search bias [15]–[17].
An event tightly associated with meiotic recombination is the pairing of homologous chromosomes through the formation of the synaptonemal complex (SC). SC is a tripartite structure comprising two lateral or axial elements (AE) and a central element. Early during the leptotene stage an AE is formed along each chromosome. However, it is only after successful homology search, that complete synapsis between homologs is observed connecting the AEs of homologous chromosomes with transverse filaments at the pachytene stage [18], [19]. Subsequently, proper SC formation is required to the integrity of CO formation [20], [21].
In order to better understand the complex events that occur during meiosis, it is crucial to identify the specific proteins required for meiotic recombination. Hereby, we characterized a conserved and meiosis-specific protein containing ssDNA binding domains homologous to those of RPA1. We named this protein meiosis-specific with OB domains (MEIOB) in vertebrates. In fly, mutation of the likely Meiob gene homolog hold'em (hdm) reduces meiotic crossover formation and sensitizes somatic cells to DNA-damaging agents [22]. Unlike MEIOB, hdm activity is not specific to meiosis. In the present work, we demonstrate that murine Meiob is specifically required after DSB formation during early steps of meiosis to ensure proper DSB repair by homologous recombination, a prerequisite for efficient crossover formation and male and female fertility.
Results
MEIOB is evolutionarily conserved and expressed during meiosis
To identify candidate genes possibly contributing to meiosis, we performed a transcriptome analysis of magnetic-activated cell sorted (SSEA1+) male and female embryonic germ cells at 13.5 days post-coïtum (dpc). At this stage, female but no male germ cells enter meiosis. We identified Meiob (referred to as RIKEN cDNA 4930528F23 gene at the time) among the most differentially expressed genes in female germ cells (complete data set will be published elsewhere). The Meiob murine gene is located on chromosome 17 and is composed of 14 exons (Figure S1A) coding for a predicted protein of 470 amino acids. Amplification of the full length Meiob transcript from 13.5 dpc mouse ovary gave a unique band (Figure S1B), the sequencing of which confirmed the predicted sequence (data not shown).
Search using tBlastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with either the full length sequence or short conserved motifs identified Meiob homologs in the genomes of almost all metazoans (Figure 1A), all except Nematoda. Multiple alignments of full length amino acid sequences indicated a high degree of conservation of the various Meiob homologs in vertebrates (e.g. 91% of similarity and 85% of identity between mouse and human) while invertebrates displayed sequences with a much lower conservation (e.g. 23% of identity between mouse and fly). No ortholog could be retrieved in the genomes of fungi or plants, although a homolog was identified in the single-celled organism, Capsaspora owczarzaki. Interestingly, the closest Meiob paralog identified was the replication protein A large subunit (Rpa1). Consistently, InterProScan Search (http://www.ebi.ac.uk/Tools/pfa/iprscan/) identified three DNA binding domains (dbd), termed oligonucleotide/oligosaccharide binding (OB) folds, which were homologous to those of RPA1 (Figure 1B). However, the orthologs of the Meiob group formed a family distinct from the Rpa1 group (Figure 1A, phylogenetic tree of MEIOB and RPA1 using MULTALIN) [23]. These data suggest that Meiob may have evolved from an ancestral Rpa1 shortly before the appearance of multicellularity.
Fig. 1. MEIOB is evolutionarily conserved and expressed in meiotic cells. 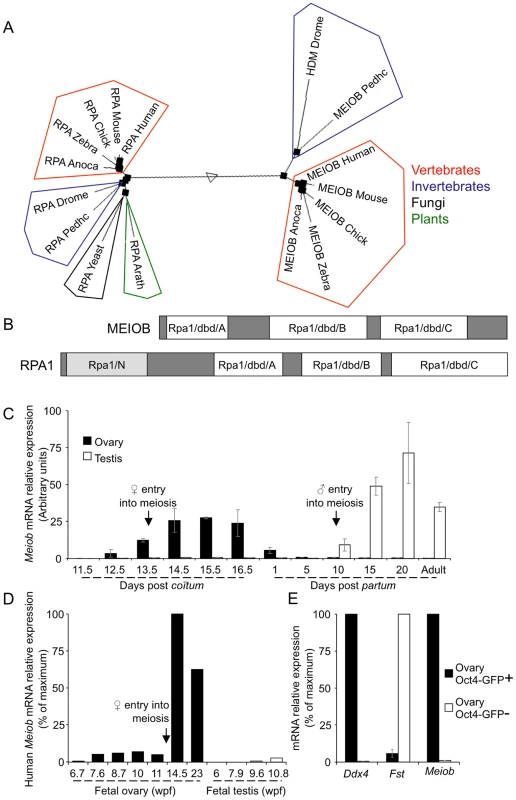
(A) Sequences coding for putative MEIOB orthologs are found present in vertebrates and most invertebrates and are absent in fungi and plants. Multiple alignments of full length MEIOB and RPA1 protein sequences were processed with Multalin Software (http://multalin.toulouse.inra.fr/multalin/). Meiob and the Rpa1 genes form distinct families. Represented species: Anoca, Anolis carolensis; Arath, Arabidopsis thaliana; Chick, Gallus gallus; Drome, Drosophila melanogaster; Human, Homo sapiens; Mouse, Mus musculus; Pedhc, Pediculus humanus corporis; Yeast, Sacharomyces cerevisiae; Zebra, Danio rerio. (B) Schematic representation of RPA1 and MEIOB protein domains. In white, three DNA binding domains (dbd) A, B and C similar to those of RPA1 are found in MEIOB protein. In light grey, RPA1 contains an N-terminal domain (RPA1N) absent in MEIOB protein. (C) Meiob expression pattern in mouse gonads. Meiob mRNA expression was measured using RT-qPCR in whole fetal and post-natal mouse ovaries (black column) and testes (white column). Gonads were harvested at the indicated development stages. Black arrows indicate meiosis initiation. Mean±SEM, n = 3. (D) MEIOB expression pattern in whole fetal human ovaries and testes. MEIOB mRNA expression was measured using RT-qPCR. Gonads were harvested at the indicated developmental stages. Wpf, weeks post fertilization. β-actin mRNA was used as the endogenous reporter. Data are expressed as a percentage of the maximum mRNA expression. (E) Meiob mRNA expression profile in ovary germ and somatic cells. Ddx4, Fst and Meiob expressions were measured using RT-qPCR in purified germ cell fraction (Oct4-GFP+, black column) and somatic cell fraction (Oct4-GFP−, white column) from mouse Oct4-Gfp 13.5 dpc ovaries. Mean±SEM, n = 5. RT-qPCR analysis of Meiob transcription pattern in adult and fetal tissues detected Meiob transcripts exclusively in fetal ovary, postnatal testis and liver (Figures S2A and S2B). In the mouse ovary, Meiob expression started at 12.5 dpc, reached a maximum at 15.5 dpc and decreased to become undetectable in post natal life (Figure 1C). In testes, Meiob expression started at 10 days post partum (dpp), reached a maximum at 20 dpp and was maintained throughout adult life (Figure 1C). This expression profile was in accordance with previously published data screening genes expressed in spermatogenic cells [24]. Additionally, in human fetal gonads, MEIOB expression was only detected in the ovary starting at the 14th weeks post fertilization (Figure 1D). Thus patterns observed in both mouse and human gonads correlate with an expression during the meiotic prophase I suggesting a conserved role within this process.
We next investigated which cell type expressed Meiob in the gonads using fluorescence activated cell sorted (FACS) germ and somatic cells from 13.5 dpc Oct4-GFP ovaries. Meiob expression levels were 112 fold higher in germ cells compared to somatic cells (Figure 1E). This germ cell-specific expression was further confirmed using w/w mouse fetal ovaries devoid of germ cells (Figure S2C). To determine whether the corresponding MEIOB protein was produced in these tissues, we generated two rabbit polyclonal antibodies raised against different MEIOB peptides and confirmed their specificity using a human tagged-MEIOB produced in HEK-293 cells (Figures 2A and S5A). Western blot analysis of various populations of FACS-sorted spermatocytes (Figure S3) detected a 52 kDa band consistent with the predicted MEIOB molecular weight. MEIOB protein was detected solely during early meiosis prophase I (‘early 4N’ fraction containing leptotene, zygotene and few pachytene spermatocytes) and not during later stages of meiosis (Figure 2A). No obvious signal was detected in whole adult testis protein extracts, possibly reflecting the few number of cells expressing MEIOB in this tissue (see ‘early 4N’ in Figure S3) and/or low expression levels.
Fig. 2. MEIOB is localized on the chromosomal axes. 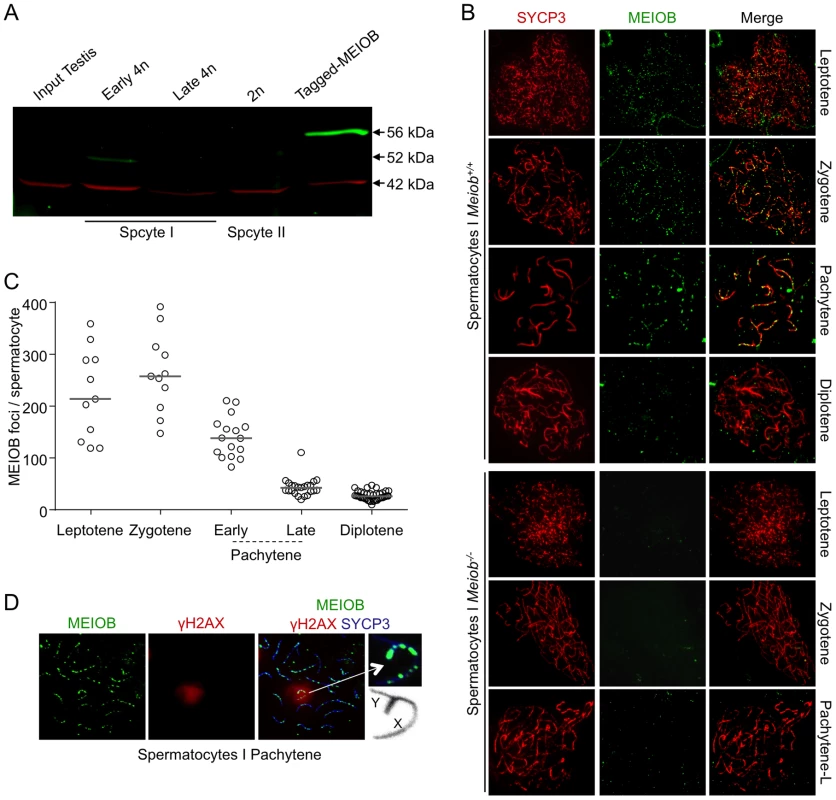
(A) MEIOB protein production in mouse adult testicular cells. MEIOB (green) and β-ACTIN (red) expression were analyzed by western blot using anti-MEIOB and anti-β-ACTIN antibodies in indicated cellular extracts. Testis cells were sorted by FACS after Hoechst 33342 staining (See Materials and Methods section and Figure S3). Early 4n: leptotene, zygotene, pachytene; Late 4n: late pachytene, diplotene; 2n: spermatocytes II; Tagged-MEIOB: HEK-293 cells transfected with tagged-MEIOB cDNA. (B) Representative chromosome spreads stained for SYCP3 (synaptonemal axial element) and MEIOB protein from Meiob+/+ and Meiob−/− spermatocytes. SYCP3 staining was used to visualize the chromosome axes. MEIOB was specifically observed in wild type meiocyte spreads. (C) Quantification of MEIOB foci in Meiob+/+ spermatocytes from adult testes. Leptotene: n = 11; zygotene: n = 11; early pachytene: n = 16; late pachytene: n = 23; diplotene: n = 40; total mice analyzed: n = 3. Median numbers of foci are marked by horizontal lines. (D) MEIOB, SYCP3 and γH2AX were detected in representative chromosome spreads of pachytene wild type spermatocytes. γH2AX stained sex body composed of X and Y chromosomes synapsed by pseudo-autosomal region. MEIOB foci are localized on sex chromosomes. MEIOB is localized on meiotic chromosomes
MEIOB immunostaining performed on chromosomal spreads from adult testes indicated that MEIOB form foci located on chromosome axes stained with SYCP3 in spermatocytes (Figure 2B). These MEIOB foci were visible as early as leptotene stage and persisted throughout the zygotene stage, with about 250 foci per spermatocyte, and decreased drastically during the pachytene stage (Figure 2C). We observed a similar staining in 15.5 dpc oocytes, corresponding to leptotene, zygotene and early pachytene stages (Figure S4). Hardly any signal was observed on chromosome axes in spreads from Meiob−/− mice confirming the specificity of our antibodies. Signal off the axes was considered as non specific as it was retrieved in Meiob−/− spermatocytes and only foci on the axes were repeatedly observed when using chromosome spreads prepared with various protocols (see materials and methods section and Figure S7). At the diplotene stage, about 25 weak MEIOB foci were detected. However we cannot formally exclude that some of these foci are unspecific due to the absence of the equivalent stage in the Meiob−/− mice. During the pachytene stage, γH2AX labels the sex body containing the X and Y chromosomes that can only pair on a limited portion called the pseudo-autosomal region (PAR). Immunostaining for MEIOB and γH2AX indicated that MEIOB was retrieved on chromosomes located in the sex body at this stage and its distribution on the X appeared similar to that observed on autosomes (Figure 2D). Of note, we always retrieved at least one bright focus in the PAR and foci in the PAR were observed until mid/late pachytene stage, when few foci persisted on autosomes.
MEIOB binds single strand DNA
The MEIOB being related to RPA1, a conserved ssDNA binding protein, we hypothesize that MEIOB too may bind ssDNA. To investigate whether MEIOB OB-domains confer any ssDNA binding activity, we performed ssDNA and double-strand DNA (dsDNA) oligonucleotide pull-down assays using recombinant tagged-MEIOB protein produced in a cell-free system. Pull-down assays performed with biotin 60-mer revealed a significantly greater retention of tagged-MEIOB protein on ssDNA than on dsDNA (Figure 3A). A similar experiment carried out with whole testis protein extract indicated that the endogenous protein also preferentially binds ssDNA (Figure 3B). Of note, MEIOB binding efficiency on ssDNA increased with oligonucleotide length as 30 mer ssDNA retained two fold less MEIOB than did 60 mer ssDNA. Extract from HEK-293 cells expressing tagged-MEIOB produced the same results (Figure S5B).
Fig. 3. MEIOB protein binds single strand DNA. 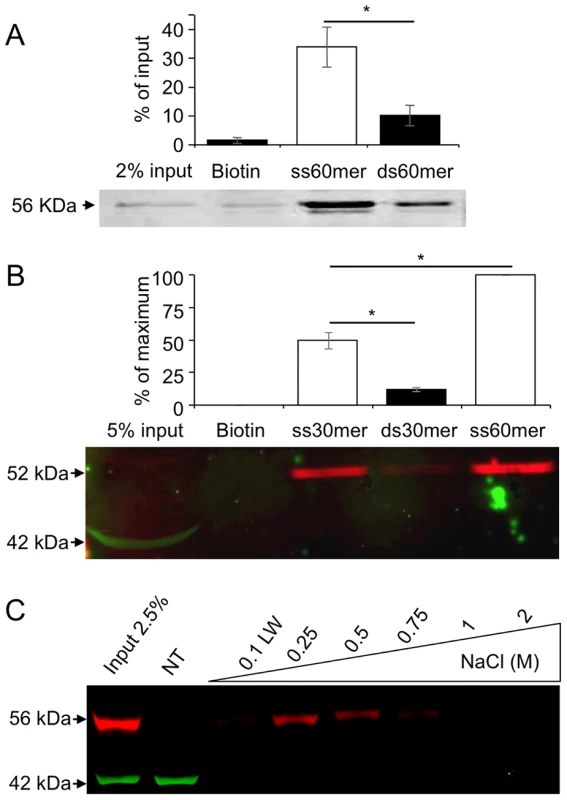
(A) Tagged-MEIOB expressed in a cell-free system or (B) testicular protein extracts were applied to beads coupled with biotin or biotinylated single strand (ss) or double strand (ds) DNA of different lengths (30 mer and 60 mer). Retained proteins were subjected to western blot hybridized with anti-β-ACTIN (green) and anti-c-MYC (A) or anti-MEIOB (B) antibodies (red). Endogenous MEIOB protein (B) and tagged-MEIOB (A) displayed respectively a size of 52 and 56 kDa, corresponding to their theoretical molecular weight. Band intensity quantifications were relative to pull down input protein extract (A) or maximal MEIOB band intensity (B) as the endogenous protein was undetectable in whole testicular protein extract. Tagged-MEIOB protein expressed in cell-free system analyzed: n = 3. Testicular protein extracts analyzed: n = 3. Mean±SEM, *, p<0.05 (paired Student's t-test). (C) ssDNA cellulose affinity chromatography. Whole native protein extract of HEK-293 expressing tagged-MEIOB was applied to ssDNA matrix. Bound proteins were successively eluted with the indicated NaCl solutions. First fractions of each NaCl elution were resolved by western blot hybridized with anti-c-MYC (red) and anti-β-ACTIN (green) antibodies. Controls, last eluted fractions are presented in Figure S5C. NT, non-transfected HEK-293 protein extract; LW, last wash of binding buffer. In order to evaluate the robustness of MEIOB binding to ssDNA, we performed ssDNA affinity chromatography. Protein extracts from HEK-293 cells expressing tagged-MEIOB were loaded on a column containing a ssDNA matrix, and proteins were eluted by progressively increasing salt concentration. Detection of eluted MEIOB by western blot revealed that MEIOB protein was eluted at a concentration of 0.75M NaCl (Figures 3C and S5C), indicating high affinity for ssDNA all be it lower than that of the trimeric RPA (most RPA being eluted at 1M and above, data not shown).
MEIOB is localized at ssDNA sites in vivo
Based on our in vitro observations and protein domain predictions we hypothesized that MEIOB could target ssDNA in vivo. We therefore compared MEIOB localization to that of RPA, RAD51 and DMC1 in spermatocyte chromosome spreads. MEIOB and RPA foci overlapped considerably (Figures 4A and S6A), indicating that MEIOB is present on recombination initiation sites. However, some foci appeared solely stained for MEIOB. Respectively, 71, 76 and 86% of RPA2 foci were stained for MEIOB and 26, 49 and 61% of MEIOB foci were stained for RPA2 at leptotene, zygotene and early pachytene stages. MEIOB staining was retrieved in most RAD51 or DMC1 foci (Figure 4A), particularly in the early pachytene stage when over 80% of RAD51 or DMC1 foci were stained for MEIOB (Figures S6B and S6A). On the other hand, only about half of the MEIOB foci were stained for RAD51 or DMC1. To better characterize MEIOB recruitment we investigated its localization on chromosome spreads from Spo11−/− and Dmc1−/− mice. In the absence of SPO11, no DSBs are formed [4], [5] and we observed no MEIOB foci on the chromosome axes in spermatocytes (Figure 4B). In the absence of the DMC1 recombinase, DSBs cannot be repaired and are believed to accumulate with extensive resection [25]. As a consequence Dmc1−/− spermatocytes failed to complete synapsis and arrested at a stage termed ‘pachytene-like’ equivalent to late zygotene or early pachytene cells in wild type cells based on the stage of AE formation (i.e. with SYCP3 staining) [26], [27]. In Dmc1−/− spermatocytes, MEIOB accumulated on chromosome axes suggesting that MEIOB is recruited to the DSB sites before strand invasion (Figure 4C). Consistent with an expected localization on ssDNA and with presumed longer single stranded DNA ends at DSB sites in Dmc1−/− meiocytes, MEIOB staining was brighter in Dmc1−/− as compared to wild type spermatocytes (Figure 4C). Furthermore, in Dmc1−/− spermatocytes, MEIOB showed near perfect colocalization with almost all RPA2 foci (Figure S7A) and also colocalized with ATR foci (Figure S7B) a protein involved in ssDNA signaling [28].
Fig. 4. MEIOB is located at the DNA double strand breaks. 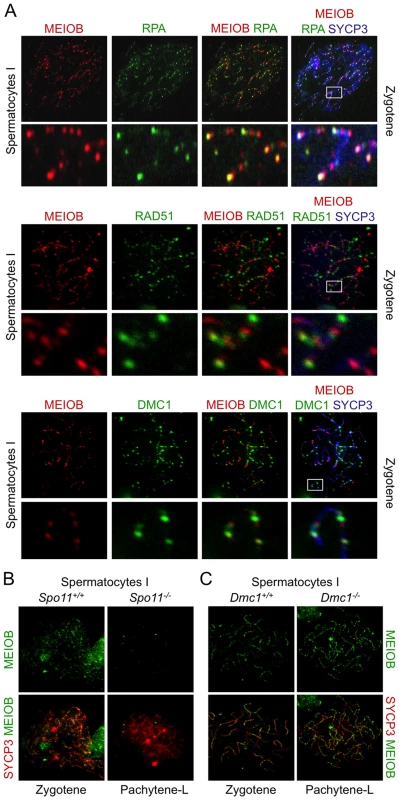
(A) SYCP3, MEIOB and RPA2, RAD51 or DMC1 were detected in chromosome spreads of zygotene wild type spermatocytes. RPA2 and MEIOB were mostly colocalized at these stages. White squares indicate magnified regions. (B) MEIOB staining in Spo11−/− spermatocytes that were defective in DSB formation. Hardly any foci were observed on the chromosome axes in Spo11−/− spermatocytes. (C) MEIOB staining in Dmc1−/− spermatocytes that were arrested due to absence of strand invasion. MEIOB foci in Dmc1−/− appeared brighter in comparison to Dmc1+/+. Meiob is required for fertility and meiosis completion
To investigate the role of MEIOB in vivo, Meiob+/− mice were generated through the deletion of exons 2 to 8 (Figure S1A). Transmission of Meiob mutant allele exhibited the expected Mendelian distribution. RT-qPCR amplifying various parts of the Meiob transcript (including exons 9 and 10 still present in the mutant) confirmed the complete absence of Meiob mRNA in the adult testis of homozygous mutants and confirmed that our genetic model was indeed a null mutant (Figures 5A and S1C). Meiob−/− mice developed and grew normally. Histological analysis revealed no anatomical defect (including in the liver, data not shown). However, Meiob−/− male and female adult mice mated for four months with Meiob+/− or Meiob+/+ counterparts produced no offspring, though vaginal plugs were formed normally (Figure 5B). Meiob thus appears mandatory for both female and male fertility.
Fig. 5. Meiob is required for fertility, germ cell survival and progression through meiosis prophase I. 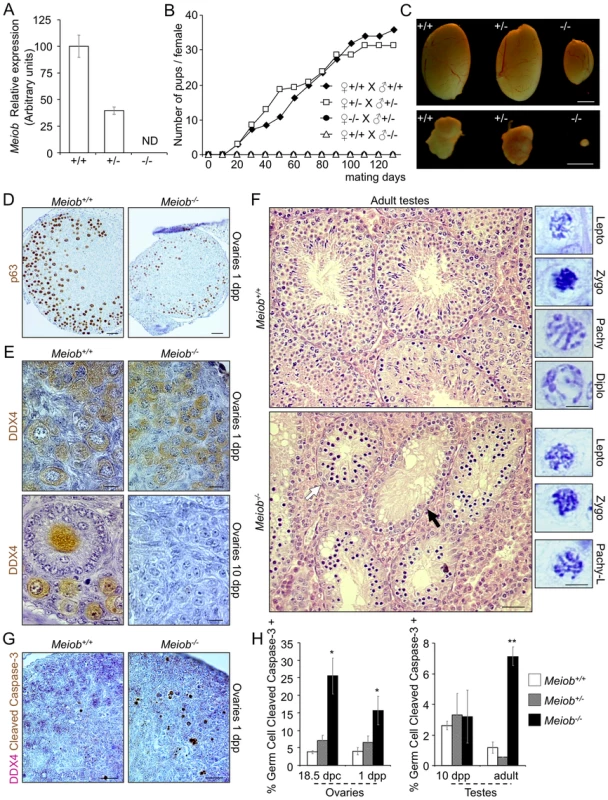
(A) Meiob mRNA is completely absent in adult testis of homozygous mutant mice. Meiob mRNA expression was investigated by RT-qPCR in Meiob+/+, +/− and −/− adult testes. ND, none detected. Mean±SEM; n = 2. (B) Meiob mutant homozygous mice are infertile. Mice of different genotypes were mated over a four month period and births were monitored. Values were expressed in cumulated number of pups per female (♀+/+ x ♂+/+ n = 9; ♀+/− x ♂+/− n = 7; ♀−/− x ♂+/− n = 3; ♀+/+ x ♂−/− n = 4). (C) Adult mice Meiob−/− testes and ovaries are strongly reduced in size by comparison with wild type gonads. Scale bars, 2 mm. (D) P63 immunohistochemistry in 1 dpp Meiob+/+ and −/− ovaries. Scale bars, 40 µm. (E) DDX4 immunohistochemistry in 1 dpp and 10 dpp Meiob+/+ and −/− ovaries. Scale bars, 10 µm. (F) Left panel. Histological sections (H&E staining) of adult Meiob+/+ and −/− testes. White arrow, seminiferous tubule containing abnormal accumulation of spermatocytes; black arrow, seminiferous tubule containing only spermatogonia. Scale bars, 40 µm. Right panel. Spermatocyte chromatin morphology in adult Meiob+/+ and −/− testes. Scale bars, 5 µm. (G) Histological sections of 1 dpp Meiob+/+ and −/− ovaries stained for cleaved caspase-3/DDX4. Scale bar, 20 µm. (H) Quantification of germ cell apoptosis in Meiob+/+, +/− and −/− testes and ovaries, respectively white, striped and black columns, at the indicated stages of development based on cleaved caspase-3/DDX4 immunohistochemistry. Mean±SEM, n = 3; *p<0.05 **p<0.001 (unpaired Student's t-test). Next we analyzed gametogenesis in Meiob−/− mice to identify the defective steps. Meiob−/− adult testes and ovaries presented a strong reduction in size when compared to Meiob+/+ gonads (Figure 5C) (testes being 3.8 times smaller (Figure S8A) and mutant ovaries hard to distinguish). During fetal life, histological analysis revealed no morphological alteration in the Meiob−/− gonads (data not shown). During post-natal life however, in 1 dpp Meiob−/− ovary, despite there being no obvious diplotene stage, the marker of late pachytene/diplotene oocytes, P63-staining, indicated the presence of numerous oocytes similar to those forming primordial follicles (Figure 5D). Germ cell counting based on immuno-detection of the germ cell marker DDX4 revealed a subsequent drop in oocyte number starting at 3 dpp (Figure S9A) until 10 dpp when no more germ cells could be observed in the Meiob−/− ovaries (Figures 5E and S9A). Meiob−/− post-natal testes histology revealed no alteration until 10 dpp. In Meiob−/− adult testes, no stage beyond primary spermatocyte was observed. Some tubules contained only spermatogonia and others contained an accumulation of spermatocytes that were mostly at the leptotene, zygotene and pachytene-like stages based on chromatin compaction and shape (Figure 5F). Furthermore, no spermatozoon was observed in Meiob−/− epididymis (Figure S8B). Cleaved caspase-3 staining indicated a significant increase of apoptotic germ cells starting after meiotic initiation in both sexes: from 18.5 dpc in Meiob−/− ovaries and from 10 dpp in Meiob−/− testes (Figures 5G and 5H). Meiob+/− male or female mice did not show any reproductive alteration when compared to Meiob+/+ mice, including number of pups per litter (Figure 5B), size of adult ovaries and testes (Figures 5C and S8A), follicle population size in adult ovaries (Figure S9B), germ cell apoptosis (Figures 5G and 5H) or seminal vesicle weight (Figure S8C).
The Meiob mutant displays impaired homologous synapsis
As defects in Meiob−/− occur during meiosis prophase I, we first investigated bivalent chromosome formation in wild type and Meiob null meiocytes. Homologous chromosomes pair and become connected along their lengths by synaptonemal complexes (SCs). AE formation is stained by SYCP3 initiated along the shared cores of sister chromatid pairs during leptotene. During zygotene a zipper like connection (stained by SYCP1) between the two AEs is initiated and completed by pachytene to fully connect homologous chromosomes. As illustrated on chromosome spreads in figure 6, the linear co-staining of SYCP3 and SYCP1 reflects fully synapsed bivalents in pachytene wild type spermatocytes. In Meiob−/− spermatocytes, as shown with SYCP3 staining, formation of AE appear to be unaffected as cells progress from leptotene to zygotene stages (Figure 2B). However, no cell with complete synapsis could be observed in adult testes (Figures 6A and 6B). The most advanced stage with regards synapsis presented an abnormal pairing of chromosomes resembling a stage between zygonema and pachynema (here termed ‘pachytene-like’ stage) (Figures 2B and 6). In such pachytene-like Meiob−/− spermatocytes, SYCP1 staining presented considerable cell-to-cell heterogeneity with regards the number of SYCP1 stretches per cell (Figure 6A). Most (∼60%) pachytene-like spermatocytes contained no SYCP1 complex and others presented from one up to nineteen (in very rare cells, less than 1%) SYCP1 stretches (Figure 6B). Similarly, 1 dpp Meiob−/− oocytes presented pachytene-like features with incomplete SYCP3/SYCP1 colocalization indicating a major defect in synapsis (Figure 6C). Both adult testes and newborn ovaries frequently displayed non-homologous pairing in Meiob deficient meiocytes with pairing between chromosomes of different sizes or combinations of more than two chromosomes (Figures 6A and 6C, white square magnifications).
Fig. 6. Meiob is necessary for the complete formation of the synaptonemal complex between homologous chromosomes. 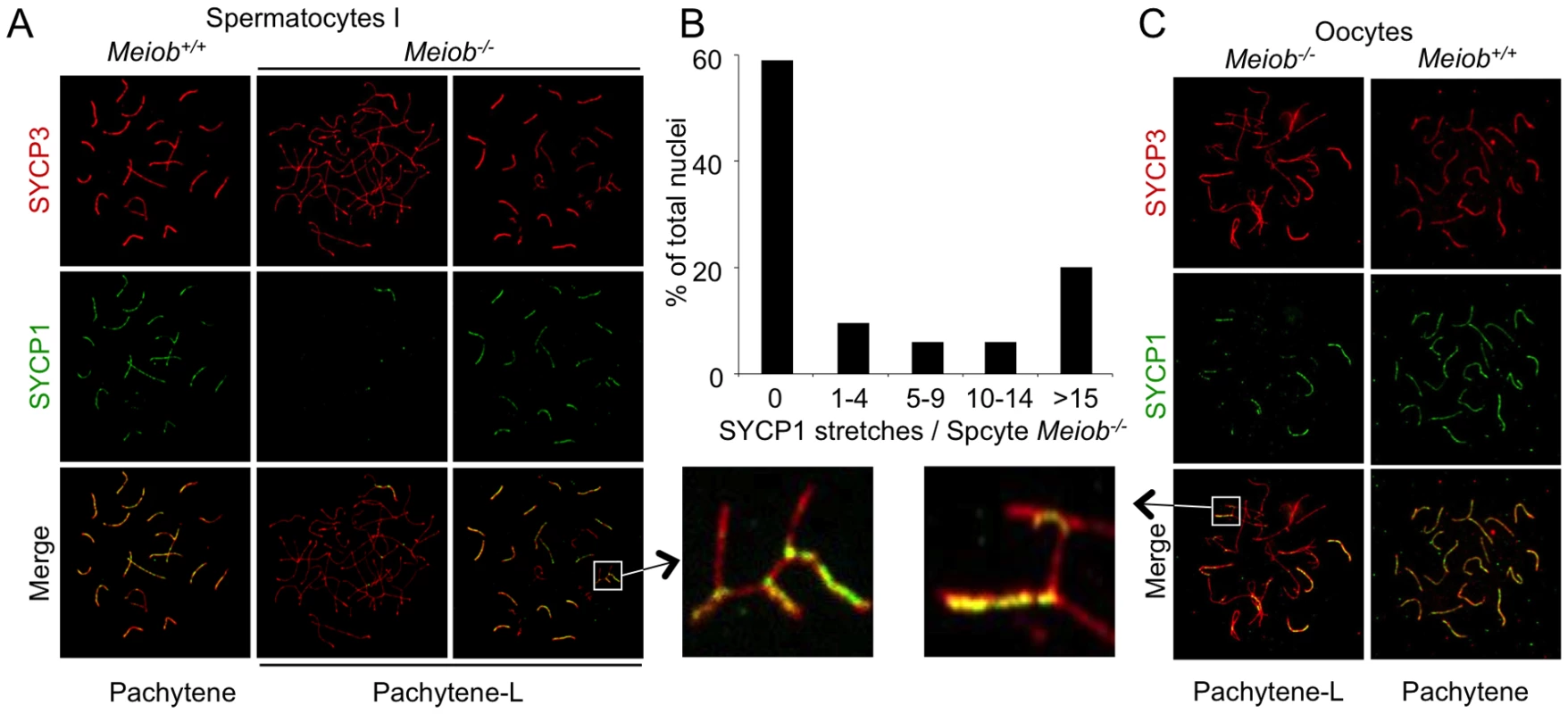
(A) SYCP3 (chromosome axis) and SYCP1 (synaptonemal-complex transverse filament) were detected on chromosome spreads of pachytene and pachytene-like stage spermatocytes of Meiob+/+ and Meiob−/− adult mice, respectively. (B) Quantification of SYCP1 stretches in Meiob−/− spermatocytes at pachytene-like stage. Total cells analyzed: n = 85, mice analyzed: n = 3. (C) Representative Meiob+/+ and Meiob−/− oocyte chromosome spreads at respectively pachytene and pachytene-like stages from newborn (1 dpp) ovaries, stained for SYCP3 and SYCP1. White squares indicate magnified regions that provide evidence for non-homologous synapsis in Meiob−/− meiocytes. DNA DSB repair is impaired during meiosis in Meiob mutants
DSB formation and repair is essential during prophase I of meiosis. We monitored these events using γH2AX staining, γH2AX being a marker of DNA DSB [29], [30]. In sections from Meiob−/− adult testes, all spermatocytes presented a robust staining for γH2AX while such a signal is usually observed in only few cells in wild type testes (Figure 7B). Analysis of chromosome spreads of spermatocytes from wild type and Meiob−/− mice confirmed the presence of γH2AX in leptotene and zygotene cells suggesting that DNA DSBs are normally formed. In all observed chromosome spreads of pachytene-like spermatocytes from Meiob−/− mice, the γH2AX signal was maintained whereas it had disappeared as expected in pachytene spermatocytes from wild type mice (Figure 7A). Additionally, in the Meiob−/− pachytene-like spermatocytes, no structure resembling the sex body was formed (Figure 7A). In 1 dpp ovaries, wild type oocytes are mostly in late pachynema and diplonema and none are stained for γH2AX whereas Meiob−/− oocytes retained a robust γH2AX-staining at this age (Figure S10). Thus, Meiob appears to be required for DNA DSB repair during meiosis. In several meiotic mutants the persistence of γH2AX staining is frequently associated with synapsis defects (i.e. Sycp1−/− [31]). Considering our finding of Meiob deficiency impairing synapsis, we attempted to correlate the extent of synapsis to DNA repair through SYCP1/γH2AX co-immunostaining in Meiob−/− spermatocytes. In these pachytene-like cells, even those with the highest rates of synapsis showed persistence of γH2AX staining (Figure 7C) indicating that the observed γH2AX signal is unlikely the consequence of the synapsis defect and strongly suggests persistence of unrepaired breaks.
Fig. 7. γH2AX is persistent in Meiob−/− spermatocytes. 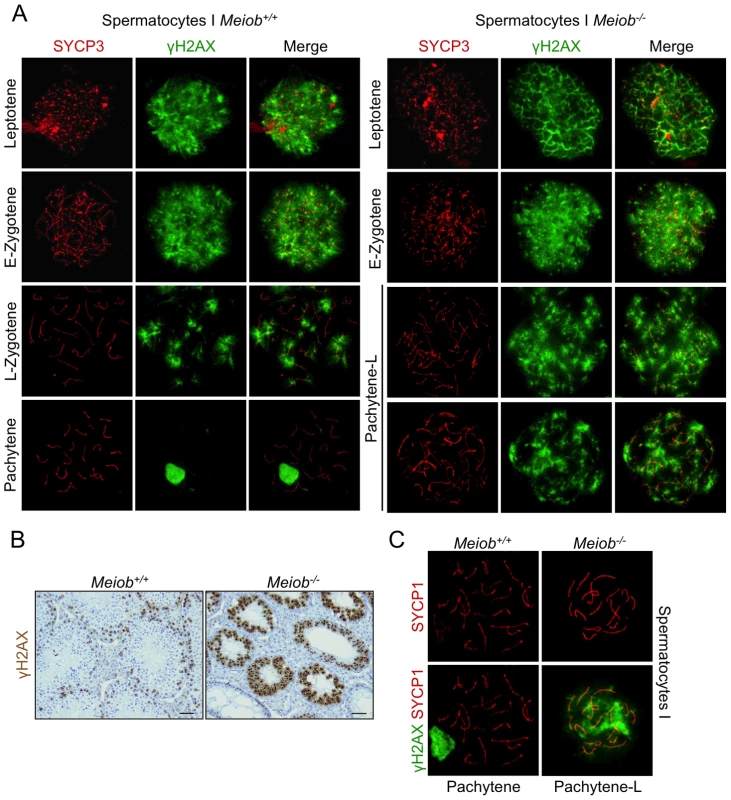
(A) Meiob+/+ and Meiob−/− spermatocyte chromosome spreads at various meiosis prophase I stages, stained for γH2AX, a marker of DNA DSBs, and SYCP3. (B) Histological sections of Meiob+/+ and −/− adult testis stained for γH2AX. Scale bar, 40 µm. (C) γH2AX and SYCP1 are detected in Meiob+/+ and Meiob−/− spermatocyte chromosome spreads at pachytene and pachytene-like stages, respectively. Meiob mutants are defective for meiotic homologous recombination
Taking into account MEIOB spatial and temporal localization and its requirement for proper DSB repair, we investigated homologous recombination in Meiob−/− spermatocytes through immunolocalization of RPA2, RAD51, DMC1 and MLH1. In Meiob−/− leptotene and zygotene/pachytene-like cells, RPA foci were formed on chromosome axes and in normal abundance when compared to wild type equivalent stages (Figures 8A and 8B). This equally confirms that DNA DSBs were produced with no overt defect in the absence of MEIOB. Immunolocalizations of RAD51 and DMC1 were performed with antibodies recognizing specifically RAD51 or DMC1. In Meiob−/− leptotene stage spermatocytes, RAD51 and DMC1 foci were localized on chromosome axes with no overt differences in comparison to wild type counterparts (Figure 9). In wild type zygotene stage these foci were maintained and their numbers decreased over the course of the pachytene stage. However in Meiob−/−, we observed a massive decrease in both RAD51 and DMC1 stainings at zygotene and pachytene-like stages. By comparison with wild type, the number of RAD51 and DMC1 foci respectively decreased by 70% and 69% at mid-zygotene in Meiob−/− (Figure 9B). Measurement of RAD51 and DMC1 foci intensities indicated no significant change in the mean intensity during leptotene and zygotene stages (Figure S11A). Intense (high and medium) foci tended to decrease first in mid-zygotene and all foci intensely and mildly (low) stained considerably decreased in late zygotene/pachytene like stage in Meiob−/− spermatocytes (Figure S11B). These data indicate that while the formation of RAD51 and DMC1 foci is unaltered in Meiob−/− mutants, their stabilization is impaired. The absence of synapsis, the persistence of γH2AX and the reduction of RAD51 and DMC1 foci observed in the absence of MEIOB suggest a strong defect in recombination with consequential impairment of CO formation. To test this hypothesis we investigated CO formation using MLH1 immunostaining. MLH1 is believed to mark future CO sites in mid-to-late pachytene, and is essential at the late stages of recombination in the formation of CO [32]. As expected at least one MLH1 focus was observed per bivalent chromosome in wild type pachytene spermatocytes. In contrast, no MLH1 foc1 were observed in Meiob−/− spermatocytes (Figure 10A). This likely reflects a blockade prior to mid-pachytene preventing thus CO formation. Meiob−/− oocytes are eliminated at a developmental time-point where most wild type oocytes reached a stage beyond pachytene. This allowed us to address whether MEIOB is required or not for the formation of MLH1 foci in cells that likely correspond to pachytene stage based on the developmental time-course of wild type cells. Many oocytes are in pachytene stage, and have MLH1 foci in wild type 17.5 dpc embryos. Interestingly we were unable to observe bright MLH1 foci along the AEs in the oocytes of 17.5 dpc Meiob−/− embryos (Figure 10B). Altogether these data suggest that MEIOB may be involved in both early/homology search –related and possibly also in later/post-homology search steps of meiotic recombination and CO formation.
Fig. 8. Meiob is not necessary for normal RPA foci number. 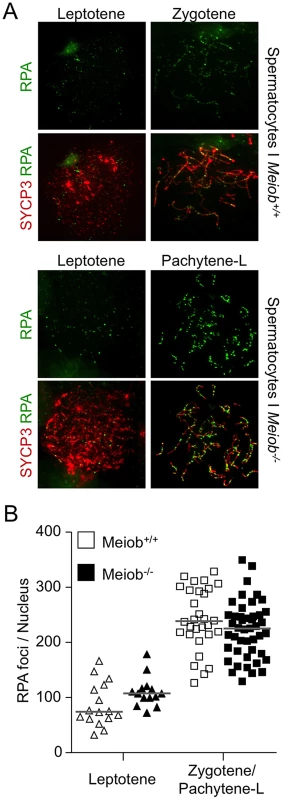
(A) SYCP3 and RPA2 were detected in chromosome spreads of wild type leptotene and zygotene and Meiob−/− leptotene and pachytene-like spermatocytes. (B) Quantification of RPA2 foci in wild type spermatocytes at leptotene (n = 10) and zygotene (n = 24) stages and in mutant spermatocytes at leptotene (n = 8) and zygotene/pachytene-like stages (n = 39). Three mice were analyzed for each genotype. RPA2 foci appeared brighter in Meiob−/− spermatocytes by comparison with wild types. Median numbers of foci are marked by horizontal lines (unpaired Student's t-test). Fig. 9. Meiob is necessary for the maintenance of RAD51/DMC1 recombinase foci during meiotic prophase I. 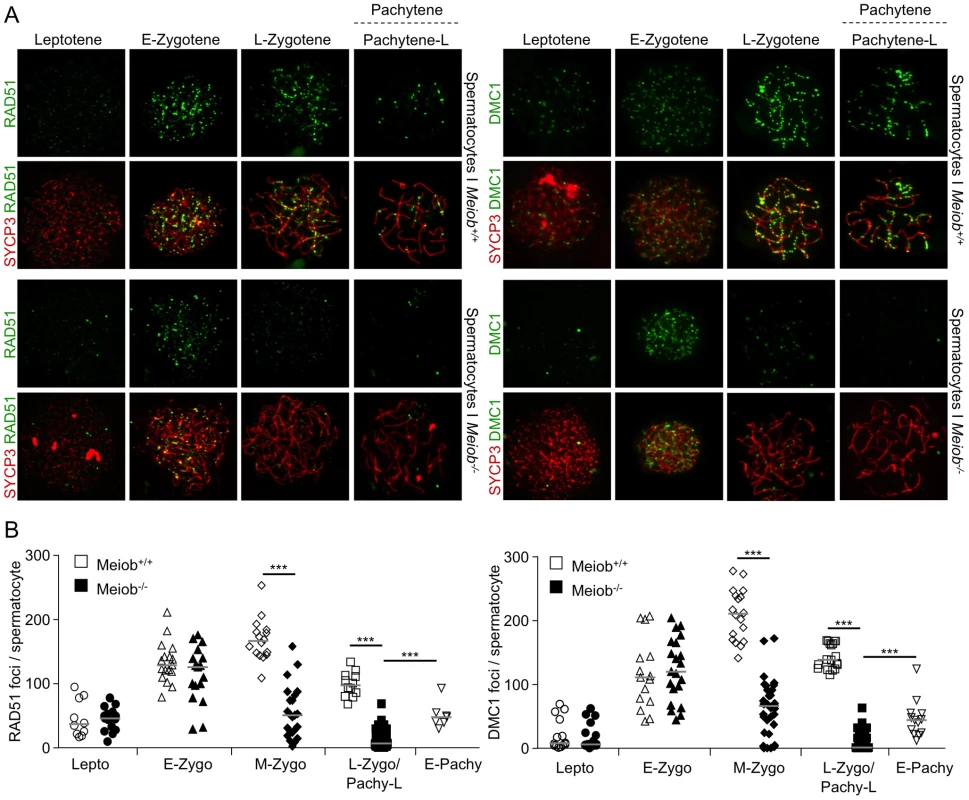
(A) SYCP3 and RAD51 or DMC1 were detected in chromosome spreads of leptotene, early zygotene, late zygotene, and pachytene/pachytene-like spermatocytes from Meiob+/+ or −/− mice using antibodies that recognized specifically RAD51 or DMC1. RAD51 and DMC1 foci appeared at leptotene stage and disappeared prematurely during the zygotene stage in Meiob−/− spermatocytes. (B) Quantification of RAD51 and DMC1 foci in Meiob+/+ spermatocytes at leptotene (Lepto; n = 10 and 14), early-zygotene (E-Zygo; n = 17 and 15), mid-zygotene (M-Zygo; n = 16 and 18), late-zygotene (L-Zygo; n = 12 and 18) and early-pachytene (E-Pachy; n = 7 and 13) stages and in and Meiob−/− spermatocytes at leptotene (n = 15 and 16), early-zygotene (n = 17 and 21), mid-zygotene (n = 23 and 32) and pachytene-like (Pachy-Like; n = 48 and 43) stages. Median numbers of foci are marked by horizontal lines; ***p<0.0001 (unpaired Student's t-test). Fig. 10. Meiob is necessary for CO formation. 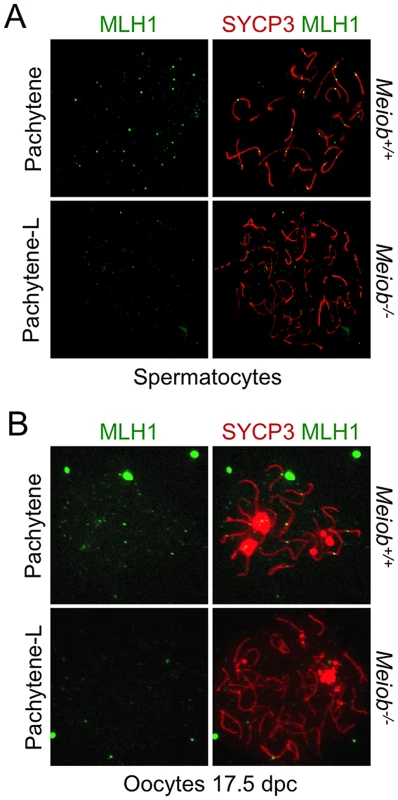
MLH1 and SYCP3 were stained in Meiob+/+ pachytene and Meiob−/− pachytene-like spermatocyte (A) and oocyte (B) nuclear spreads. MLH1 foci were undetectable along axial elements in Meiob−/− meiocytes. Discussion
In this study, we have identified a new major player in mammalian meiosis and in particular have demonstrated for the first time an absolute requirement of MEIOB for meiosis prophase I completion in the mouse. The expression profile and deletion of Meiob indicated a specific role for this protein during meiosis prophase I. Furthermore, our data indicate that MEIOB is mandatory for DSB repair and crossover formation. MEIOB also appears to favor faithful and complete synapsis. As observed with other mutant mice in which these steps are impaired, these defects caused meiotic arrest, meiocyte death by apoptosis and infertility. The complete infertility of both male and female mice indicates that Meiob is one of the core genes required for meiosis, as the deletion of other meiotic genes in mice sometimes only induces male infertility [12], [34]–[35]. In summary, this mammalian meiotic mutant impairs meiotic recombination in both male and female and impairs RAD51 and DMC1 stabilization.
Our data support the idea that MEIOB binds ssDNA. First, protein domain prediction proposed that MEIOB contains three ssDNA binding domains, denoted OB domains, similar to those of RPA1 known to have a high affinity for ssDNA [36]. Second, in vitro DNA binding assays demonstrated that MEIOB is particularly retained by ssDNA. Moreover, MEIOB foci formed along AE and colocalized to a large extent with RPA which marks ssDNA filament [37], [38]. The number of foci corresponded to the expected number of DNA DSBs generated by SPO11 and no MEIOB foci were observed in the absence of SPO11-mediated DSBs. Lastly, when 3′ssDNA accumulated (i.e. in Dmc1−/−, in the absence of strand invasion) MEIOB colocalized almost perfectly with both RPA and ATR, known to sense ssDNA [28], [39]. Altogether these results lead us to propose that the MEIOB protein binds ssDNAs generated during the 5′ to 3′ resection of DNA ends at meiotic DSB sites. We thus propose that MEIOB may be an RPA paralog specifically required for meiotic recombination. Although the data in the present study do not formally prove that the ssDNA binding activity of MEIOB is required for meiosis, this is an appealing hypothesis that needs pursuing in future studies.
Meiob−/− meiocytes are arrested during prophase I of meiosis at a stage resembling late zygotene stage and characterized by persistent DSBs and defects in chromosomal pairing. This incomplete synapsis is likely the consequence of the defective DSB repair as is observed in most mutants with impaired homologous recombination [12], [26]. In Meiob−/− meiocytes the number of DSBs appears unaffected as we observed the expected number of RPA foci [38] and the RAD51 and DMC1 foci were formed but did not persist. These observations point towards the lack of MEIOB primarily causing a defect in the process of recombination. Unfortunately, due to their embryonic lethality, most mutations that impair homologous recombination have not been characterized during mammalian meiosis. Notably, the lethality of Rad51−/− disallows the comparison of Meiob−/− to these murine meiocytes. Of note, we would like to point out that Meiob−/− phenotype is similar to that of Dmc1−/− mice [26], [27]. However in the Dmc1−/− spermatocytes, RAD51 persists in the pachytene-like stage in contrast to that observed in Meiob−/− mice ([26], Figure S12). Interestingly, in Meiob−/− pachytene-like cells, despite the absence of RAD51 and DMC1 foci, we observed abundant and brighter RPA foci and strong γH2AX staining indicating the presence of unrepaired DSBs and of ssDNA. We thus conclude that the transitory loading of RAD51 and DMC1 observed in Meiob−/− is insufficient to allow the completion of homology search, homolog alignment and SC formation. As RAD51 recombinase activity is believed to be inactivated during meiotic recombination, DMC1 is proposed to catalyze homology search and strand exchange of most meiotic recombination events [40]. Thus, in the Meiob mutant, DMC1 would appear to have been prevented from performing its role due to its precocious destabilization. To our knowledge, no murine meiotic mutant has yet presented such a phenotype with a premature disappearance of RAD51 and DMC1 before pachynema.
The formation of RAD51-DNA presynaptic filament is promoted by BRCA2 in mammals to overcome the inhibitory effect on the heterotrimeric RPA [41]. BRCA2 is known to bind both RAD51 and DMC1; Brca2−/−;Tg have an impaired number of RAD51 and DMC1 foci in leptotene and zygotene mouse spermatocytes [12]. Thus the Meiob−/− defect does not seem to involve BRCA2 in a general manner as one would then have also expected an earlier defect in the loading of RAD51 and DMC1. Moreover, the growing percentage of RAD51 and DMC1 foci that are stained for MEIOB at late zygotene and early pachytene stages support a later role of MEIOB on the activity of recombinases. In this line, biochemical findings raise the possibility that the maintenance of RAD51 presynaptic filament in vivo might involve some RAD51 accessory factors such as RAD54, HOP2-MND1 and recently the SWI5-SFR1 complex [42]–[44]. The spermatocyte defects observed in Rad54−/− and Hop2−/− mice differ from those observed in the Meiob mutant. Indeed meiotic recombination is only slightly affected in Rad54−/− mice which in turn show no major fertility defect [45]. In Hop2−/− mice, despite a failure of meiotic recombination, RAD51 and DMC1 foci accumulate and persist through the pachytene-like stage [46]. The role of SWI5-SFR1 has to date only been investigated in mouse embryonic stem cells [47], however genetic studies in yeast have found evidence for the SWI5-SFR1 yeast orthologs Mei5 and Sae3, being involved in meiotic recombination [48], thereby suggesting that the mammalian SWI5-SFR1 may also play a role in meiotic recombination. Interestingly, this complex is devoid of DNA-binding activity in mice [49] whereas their respective orthologs Mei5 and Sae3 in S. cerevisiae possess DNA-binding activity [50]. Furthermore, the expression of Mei5 and Sae3 is restricted to meiosis and mediates Dmc1 activity by enhancing its ability to form nucleofilaments on ssDNA [51]. One may reasonably speculate therefore that MEIOB may influence the stability of the RAD51-DMC1 filament in cooperation with SWI5-SFR1 or a meiotic counterpart. Such a hypothesis will of course require further investigation. Of note, SWI5-SFR1 and Mei5-Sae3 complexes directly interact with RAD51 [49], [50]. We were unable to detect a direct interaction between MEIOB and RAD51 (data not shown). However our experiments were performed in a heterologous system and with an overexpressed protein, thus we cannot formally allow us to exclude a direct interaction between MEIOB and RAD51. Finally, we propose that the presence of two ssDNA binding proteins, namely RPA and MEIOB, confers special properties to resected DNA allowing the proper stabilization of two proteins on the meiotic presynaptic filament: DMC1 and RAD51. Along this line, one may consider MEIOB as a new meiotic-specific mediator for RAD51/DMC1.
While meiosis recombination is a conserved process in eukaryotes, there are clear differences among organisms [52]. Interestingly, yeast, plants and C. elegans do not have a Meiob homolog (Figure 1A). One may therefore consider that in these organisms, either meiosis is achieved via slightly different mechanisms or the function of MEIOB is performed by other proteins. For instance, in plants, there are multiple copies of RPA1 (RPA1-like proteins) [53], [54] some of which may assume the role of MEIOB. In plants, RPA1-like proteins have been proposed to form part of trimeric RPA complexes alongside RPA2 and 3 sub-units [55]; to our knowledge no such additional sub-units are conserved among vertebrates and the analysis of transcriptomic data did not allow us to pin point any meiosis-specific additional RPA2-like protein in mice. We therefore propose that either MEIOB acts with the canonical RPA2 and 3 (thus replacing RPA1) or by its own and possibly with different partners. The only Meiob homolog that has been described previously is hold'em (hdm) in Drosophila [22] that is implicated in crossover formation. However hdm−/− fly and Meiob−/− mice present drastic divergence during meiosis suggesting that their function may have diverged during evolution. Indeed in hdm−/− flies, some crossovers still occur though with lower frequencies compared to wild-type fly and the DSBs are repaired though with a delay. Furthermore, hdm is also involved in DNA repair in somatic cells [22] while Meiob is almost exclusively expressed in meiotic germ cells. Joyce et al. also provided evidence that hdm physically interacts with Ercc1, a member of the exchange class of gene products, and proposed that the Ercc1/Mei9/Mus81/hdm complex functions in the meiotic recombination pathway to resolve DSB-repair intermediates into crossovers [22]. This function appears to be required later by comparison to the here-described recruitment of MEIOB to DSB sites. Furthermore, such an interaction is unlikely to explain the entirety of Meiob−/− phenotype described here. Indeed, in mice, Ercc1 deletion does not prevent synapsis or sex body formation and males are able to produce few spermatozoa albeit with DNA damage [56]. We thus conclude that, in mice, the function of MEIOB is not through an interaction with ERCC1 in late recombination nodules and that additional partners need to be identified. On the other hand, one could also argue that this may be due to the meiotic process itself having slightly different requirements in-between mammals and drosophila. Therefore, we cannot exclude an additional role of MEIOB in CO resolution. Such a role could be responsible of the late staining observed for MEIOB (Figures 2B and 2C). In this line we could speculate that MEIOB might target additional ssDNA sites such as the D-loop intermediate, as it is thought for RPA [57], or the second end of the DSB (i.e. the one not involved in the initial homology search), that are formed during the process of CO. The second end is released to be captured on the recombination intermediate to form a double holiday junction (dHJ). This would fit with our reports of rare cells displaying some amount of pairing, thus having performed the initial homology search (first end), and with the complete lack of MLH1 foci observed in Meiob−/− cells. Such a speculative proposition might also be fueled by the persistent colocalizsation of MEIOB with recombinases and will require additional studies. Putative MEIOB partners are under investigation and their identification should help define the precise function of MEIOB during meiotic recombination and the mechanisms allowing recombinase stabilization in this process.
Materials and Methods
Mice and embryos
All animal studies were conducted in accordance with the guidelines for the care and use of laboratory animals of the French Ministry of Agriculture. Mice were raised and mated, and fetal gonads were isolated as previously described [58], [59]. Mice used in this study were NMRI mice (Naval Maritime Research Institute), Dmc1 and Spo11 mutant mice, w/w mice, Oct4-GFP mice (that have been previously described, respectively [5], [26], [60], [61]) and Meiob mutant mice (see below).
Meiob mutant allele construction
The Meiob null allele was established at the Mouse Clinical Institute/Institut Clinique de la Souris (MCI/ICS), Illkirch, France (http://www.ics-mci.fr/). The targeting vector was constructed as follows: a 4.5 kb fragment encompassing exon 2 and part of the first intron, Red Fluorescent Protein (Rfp) cDNA, followed by a neomycin resistance cassette surrounded by two loxP sites and a 4 kb fragment encompassing exons 9 and 10 (Figure S1A). Meiob endogenous ATG in exon 2 was conserved to ensure Rfp cDNA expression and there was a stop codon at the end of Rfp cDNA (Figure S1A). The linearized construct was electroporated in BD10 C57BL/6J mouse ES cells. After selection, targeted clones were identified by PCR using external primers and further confirmed by Southern blot with a Neo probe, 5′ external probe and 3′ external probe (data not shown). Two positive ES clones were injected into 129/Sv blastocysts and the derived male chimaeras produced germ-line transmission. Meiob+/Rfp-loxPNeoloxP mice were crossed with mice carrying ubiquitous Cre in order to remove the Neo resistance cassette and generate the final Meiob null allele (Figure S1A). Mice were genotyped by PCR of tailed DNA using REDExtract-N-Amp Tissue PCR Kit (Sigma) according to the manufacturer's instructions. Sequences of primers used are shown in Table S2.
Fertility assay
Male and female Meiob+/+, +/ − & −/− mice were mated over the course of four months. Mating partners were inverted every two months. Vaginal plugs were regularly checked to verify that coïtum occurred normally. Births and pups were counted and referenced every day and pups were sacrificed. Results are expressed in cumulative numbers of pups per couple per ten days.
Collection of human fetal gonads
Human fetal material was provided by the Department of Obstetrics and Gynecology at the Antoine Béclère Hospital, (Clamart, France) following legally induced abortions in the first trimester of pregnancy and therapeutical termination of pregnancy in the second trimester. Human fetal gonads were harvested as previously described [62]. Our study was approved by the Biomedicine Agency (reference number PFS12-002), and all women gave their informed consent.
Generation of anti-MEIOB antibody
Rabbit polyclonal anti-MEIOB antibodies were generated and affinity purified by Eurogentec (Angers, France) using double X protocol with ADP TEA SRN LAR QGH T and IRE NKE TNV ADE IDS polypeptides.
Flow cytometric characterization of MEIOB localization
Testicular single-cell suspensions and cell sorting were processed as previously described [63]. Dissociation was performed with two wild type adult testes resulting in the sorting of two hundred million testicular cells. Seven hundred thousand cells from each sorting gate were lysed and submitted to western blot analysis.
Real-time quantitative PCR and PCR
RNA extractions and RT-qPCR were performed as previously described [64]. Sequences of primers are shown in Table S2. β-actin mRNA was used as the endogenous reporter. Data are expressed as a percentage of the maximum mRNA expression unless otherwise stated in which case an external reference (F9 cells) was used to normalize the expression of numerous points.
Cell culture and plasmid transfection
Human Epithelial Kidney cells (HEK-293, ATCC) were cultivated in DMEM High Glucose (Gibco) containing 15% FSB (Gibco). Human MEIOB complete cDNA was inserted into a pCMV6-Entry plasmid (Origene cat# RC228391) containing c-Myc and Flag tags in the C-terminal domain of the protein. This plasmid was transfected into HEK-293 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Tagged-MEIOB was observed in both nuclear and cytoplasmic compartments of the transfected HEK-293 cells (Figure S5A).
Protein extraction and recombinant protein production
Protein extracts were produced from cell lines or tissues under native conditions. HEK-293 cells were harvested, centrifuged 5 min at 500 g and then lysed in Cell Lysis Buffer (Cell signaling, Danvers, USA) complemented with 1 mM 2-mercaptoethanol and Complete protease inhibitor (Roche, Mannheim, Germany). For adult testis, the albuginea was first removed and testis was lysed in the same buffer using ceramic spheres and FastPrep-24 Instrument (MP Biomedicals) with two pulses of 20 seconds. Extracts were then gently sonicated (two pulses of 30 seconds), centrifuged 10 min at 14 000 g and supernatant was immediately used for functional applications. Human recombinant tagged-MEIOB protein was produced in a cell-free system. The TnT T7 Quick Coupled Transcription/Translation System (Promega) was used with 1 µg of the tagged-MEIOB plasmid in a 50 µl reaction according to the manufacturer's instructions.
Western blotting
Prior to gel migration, protein samples were supplemented with Laemmli buffer and resolved by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were electrophoretically transferred to polyvinylidene difluoride membranes (PVDF) (Amersham Biosciences, Buckinghamshire, England) before hybridization with appropriate primary antibodies and fluorescent dye coupled secondary antibodies (See Table S1). Images were acquired using Typhoon 9400 scanner (Amersham Biosciences) and quantified with ImageJ software [65].
Histology, immunohistochemistry and germ cell counting
Histological sections, germ cell counting and immunohistochemical stainings using appropriate antibodies (Table S1) were performed as previously described [59].
Preparation of meiocyte chromosome spreads
Testes used for spermatocyte spread preparation were harvested from mice aged approximately 2 to 6 months. Ovaries used for the preparation of chromosome spreads were harvested in 15.5 and 17.5 dpc embryos and 1 dpp pups. Three types of meiocyte chromosome preparations were used. For γH2AX, SYCP1 and RPA2 stainings, chromosome spreads were prepared according to the drying-down technique [66] with minor, previously described modifications [67]. For RAD51, DMC1 and ATR stainings, chromosome spreads were prepared as previously described [68]. This later protocol was also used for MLH1 staining in oocytes and the former protocol was used for MLH1 staining in spermatocytes. For MEIOB staining, the two above protocols were used and an additional spermatocyte surface-spread technique was applied as follows. Spermatocytes were dispersed in an 80 mM NaCl drop, attached to glass slides, fixed with a 4% paraformaldehyde 0.03% sodium dodecyl sulfate solution and washed in 0.4% Photo-Flo 200 (Sigma). RAD51 and DMC1 antibodies were previously published [69]. The specificity of the DMC1 antibody was further validated using Dmc1−/− spermatocytes and no signal was retrieved (data not shown). Due to the lack of Rad51−/− spermatocytes (i.e. Rad51 deletion is lethal), no such control was performed for the RAD51 antibodies and one cannot formally exclude that these might crossreact.
Immunofluorescence on preparations of meiocyte chromosome spreads
Spermatocyte and oocyte preparations were washed with 0.4% Photoflo 200 (Sigma) and slides were air-dried 15 minutes before permeabilizing and blocking. They were then incubated overnight with the appropriate primary antibodies in blocking solution at room temperature, followed by 1 hour incubation with secondary antibodies at 37°C (see Table S1 for antibodies). Slides were mounted with Vectashield DAPI medium (Vector Laboratories). Imaging was performed using a AX70 epifluorescent microscope (Olympus) equipped with a charge-coupled camera (Roper Scientific) and IPlab software (Scanalytics) or with a DM5500 B epifluorescent microscope (Leica Microsystems) equipped with a CoolSNAP HQ2 camera (Photometrics) and Leica software (Metamorph). Images were processed and specific structures were quantified with ImageJ software (Cell Counter and Foci Picker3D programs). For quantification of foci colocalization, foci overlapping for the most part were considered as colocalized without restriction to foci that share the same centroid (e.g. immediately adjacent foci were not considered as colocalized).
ssDNA/dsDNA pull down assay
HPLH purified biotinylated oligonucleotides were used for the DNA pull down assays: ss30-mer 2 : 5′-GAT CTC AGC GAT TCA CAC GCG TCC TAA CTC G-3′-BiotinTEG, ss60-mer: 5′-GAT CTG CAC GAC GCA CAC CGG ACG TAT CTG CTA TCG CTC ATG TCA ACC GCT CAA GCT GC-3′-Biotin TEG (Eurogentec). Double strand hybridizations were performed in 50 mM NaCl, 25 mM Tris-HCl, pH 7.5 buffer with complementary sequences at molecular equivalence by a denaturing step (3 minutes 94°C) and a slow progressive return to room temperature. 0.2 pMol of DNA was immobilized onto 1 µg Dynabeads M-280 Streptavidin (Dynal) following the protocol supplied by the manufacturer. Protein extracts were pre-incubated on ice for 10 minutes in modified DBB (DBB with 25 mM Tris-HCl, 1 mM EDTA plus 5 mg/ml BSA for the cell-free protein assays or plus 10 µg/ml BSA for assays with total protein extracts) before addition of 500 µg Dynabeads with immobilized ss - or ds-DNA probes. DNA binding assays were conducted either with 3 µl of the tagged-MEIOB cell-free productions, with 600 µg of protein from HEK-293 cells transfected with hMEIOB (Figure S5A) or with 3 mg testis protein extracts. The DNA-protein mixture was incubated for 1 hour at 4°C under gentle agitation. After magnetic separation, the beads were washed three times in 500 µl binding buffer without BSA, before being washed once in 500 µl rinsing buffer (modified DBB with 150 mM NaCl). Elution of DNA binding proteins was performed by resuspending the beads in 20 µl Laemmli buffer and boiling the samples for 5 minutes before magnetic separation of the beads and western blotting of the samples.
ssDNA-cellulose affinity chromatography experiments
HEK-293 cells were harvested 48 h after transfection of the tagged-MEIOB plasmid. Three mg of total protein extract were diluted 1/5 in DNA-Binding Buffer (DBB: 50 mM Tris-HCl, 100 mM NaCl, 10% (w/v) glycerol, Complete Protease Inhibitor, 1 mM 2-mercaptoethanol, pH 7.4), 1 ml of which was loaded in Poly-Prep columns (Biorad), previously loaded with 500 µl of ss-DNA-cellulose (Amersham Biosciences). Protein extract was incubated 90 min at 4°C under gentle agitation. Columns were washed with forty bed volumes of ss-DBB and the last 1 ml of wash was kept to ensure that complete elution occurred (0.1M LW). Columns were then washed with elution buffer containing increasing concentrations of NaCl: 0.25, 0.5, 0.75, 1, 2M. For each step columns were washed with ten bed volumes, the first 1 ml (Figure 3C) and last 1 ml were kept (Figure S5C). Eluted fractions were concentrated using Nanosep 3KDa (Pall) according to the manufacturer's instructions. Salt concentration was then equalized with ss-DBB. Concentrated protein fractions were recovered in Laemmli buffer and subjected to western blot with the appropriate antibodies (Table S1).
Supporting Information
Zdroje
1. Bascom-SlackCA, RossLO, DawsonDS (1997) Chiasmata, crossovers, and meiotic chromosome segregation. Adv Genet 35 : 253–284.
2. SchwachaA, KlecknerN (1994) Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76 : 51–63.
3. AndersenSL, SekelskyJ (2010) Meiotic versus mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32 : 1058–1066.
4. KeeneyS, GirouxCN, KlecknerN (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 : 375–384.
5. BaudatF, ManovaK, YuenJP, JasinM, KeeneyS (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6 : 989–998.
6. RomanienkoPJ, Camerini-OteroRD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6 : 975–987.
7. SunH, TrecoD, SzostakJW (1991) Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64 : 1155–1161.
8. BishopDK, ZicklerD (2004) Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117 : 9–15.
9. WoldMS, KellyT (1988) Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A 85 : 2523–2527.
10. BochkarevaE, KorolevS, Lees-MillerSP, BochkarevA (2002) Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J 21 : 1855–1863.
11. NealeMJ, KeeneyS (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442 : 153–158.
12. SharanSK, PyleA, CoppolaV, BabusJ, SwaminathanS, et al. (2004) BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131 : 131–142.
13. HollomanWK (2011) Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol 18 : 748–754.
14. SiaudN, DrayE, GyI, GerardE, TakvorianN, et al. (2004) Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J 23 : 1392–1401.
15. CloudV, ChanYL, GrubbJ, BudkeB, BishopDK (2012) Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337 : 1222–1225.
16. KurzbauerMT, UanschouC, ChenD, SchlogelhoferP (2012) The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24 : 2058–2070.
17. TsubouchiH, RoederGS (2006) Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev 20 : 1766–1775.
18. ZicklerD, KlecknerN (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33 : 603–754.
19. FrauneJ, SchrammS, AlsheimerM, BenaventeR (2012) The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp Cell Res 318 : 1340–1346.
20. PageSL, HawleyRS (2003) Chromosome choreography: the meiotic ballet. Science 301 : 785–789.
21. KouznetsovaA, BenaventeR, PastinkA, HoogC (2011) Meiosis in mice without a synaptonemal complex. PLoS One 6: e28255.
22. JoyceEF, TannetiSN, McKimKS (2009) Drosophila hold'em is required for a subset of meiotic crossovers and interacts with the dna repair endonuclease complex subunits MEI-9 and ERCC1. Genetics 181 : 335–340.
23. CorpetF (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16 : 10881–10890.
24. KogoH, Kowa-SugiyamaH, YamadaK, BolorH, TsutsumiM, et al. (2010) Screening of genes involved in chromosome segregation during meiosis I: toward the identification of genes responsible for infertility in humans. J Hum Genet 55 : 293–299.
25. BishopDK, ParkD, XuL, KlecknerN (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69 : 439–456.
26. PittmanDL, CobbJ, SchimentiKJ, WilsonLA, CooperDM, et al. (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1 : 697–705.
27. YoshidaK, KondohG, MatsudaY, HabuT, NishimuneY, et al. (1998) The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell 1 : 707–718.
28. CimprichKA, CortezD (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9 : 616–627.
29. RogakouEP, PilchDR, OrrAH, IvanovaVS, BonnerWM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273 : 5858–5868.
30. CelesteA, PetersenS, RomanienkoPJ, Fernandez-CapetilloO, ChenHT, et al. (2002) Genomic instability in mice lacking histone H2AX. Science 296 : 922–927.
31. de VriesFA, de BoerE, van den BoschM, BaarendsWM, OomsM, et al. (2005) Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19 : 1376–1389.
32. GuillonH, BaudatF, GreyC, LiskayRM, de MassyB (2005) Crossover and noncrossover pathways in mouse meiosis. Mol Cell 20 : 563–573.
33. HerranY, Gutierrez-CaballeroC, Sanchez-MartinM, HernandezT, VieraA, et al. (2011) The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J 30 : 3091–3105.
34. HakemR, de la PompaJL, EliaA, PotterJ, MakTW (1997) Partial rescue of Brca1 (5–6) early embryonic lethality by p53 or p21 null mutation. Nat Genet 16 : 298–302.
35. YangF, EckardtS, LeuNA, McLaughlinKJ, WangPJ (2008) Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol 180 : 673–679.
36. FanningE, KlimovichV, NagerAR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34 : 4126–4137.
37. AlaniE, ThresherR, GriffithJD, KolodnerRD (1992) Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol 227 : 54–71.
38. MoensPB, KolasNK, TarsounasM, MarconE, CohenPE, et al. (2002) The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J Cell Sci 115 : 1611–1622.
39. IchijimaY, IchijimaM, LouZ, NussenzweigA, Camerini-OteroRD, et al. (2011) MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev 25 : 959–971.
40. BishopDK (2012) Rad51, the lead in mitotic recombinational DNA repair, plays a supporting role in budding yeast meiosis. Cell Cycle 11 : 4105–06.
41. SiaudN, BarberaMA, EgashiraA, LamI, ChristN, et al. (2011) Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet 7: e1002409.
42. ChiP, San FilippoJ, SehornMG, PetukhovaGV, SungP (2007) Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev 21 : 1747–1757.
43. MazinAV, AlexeevAA, KowalczykowskiSC (2003) A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J Biol Chem 278 : 14029–14036.
44. YuanJ, ChenJ (2011) The role of the human SWI5-MEI5 complex in homologous recombination repair. J Biol Chem 286 : 9888–9893.
45. WesolyJ, AgarwalS, SigurdssonS, BussenW, Van KomenS, et al. (2006) Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol Cell Biol 26 : 976–989.
46. PetukhovaGV, RomanienkoPJ, Camerini-OteroRD (2003) The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell 5 : 927–936.
47. AkamatsuY, JasinM (2010) Role for the mammalian Swi5-Sfr1 complex in DNA strand break repair through homologous recombination. PLoS Genet 6: e1001160.
48. HayaseA, TakagiM, MiyazakiT, OshiumiH, ShinoharaM, et al. (2004) A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell 119 : 927–940.
49. TsaiSP, SuGC, LinSW, ChungCI, XueX, et al. (2012) Rad51 presynaptic filament stabilization function of the mouse Swi5-Sfr1 heterodimeric complex. Nucleic Acids Res 40 : 6558–6569.
50. SayAF, LedfordLL, SharmaD, SinghAK, LeungWK, et al. (2011) The budding yeast Mei5-Sae3 complex interacts with Rad51 and preferentially binds a DNA fork structure. DNA Repair (Amst) 10 : 586–594.
51. FerrariSR, GrubbJ, BishopDK (2009) The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J Biol Chem 284 : 11766–11770.
52. La VolpeA, BarchiM (2012) Meiotic double strand breaks repair in sexually reproducing eukaryotes: we are not all equal. Exp Cell Res 318 : 1333–1339.
53. IshibashiT, KimuraS, FurukawaT, HatanakaM, HashimotoJ, et al. (2001) Two types of replication protein A 70 kDa subunit in rice, Oryza sativa: molecular cloning, characterization, and cellular & tissue distribution. Gene 272 : 335–343.
54. ShultzRW, TatineniVM, Hanley-BowdoinL, ThompsonWF (2007) Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol 144 : 1697–1714.
55. ChangY, GongL, YuanW, LiX, ChenG, et al. (2009) Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol 151 : 2162–2173.
56. HsiaKT, MillarMR, KingS, SelfridgeJ, RedheadNJ, et al. (2003) DNA repair gene Ercc1 is essential for normal spermatogenesis and oogenesis and for functional integrity of germ cell DNA in the mouse. Development 130 : 369–378.
57. BurgoynePS, MahadevaiahSK, TurnerJM (2007) The management of DNA double-strand breaks in mitotic G2, and in mammalian meiosis viewed from a mitotic G2 perspective. Bioessays 29 : 974–986.
58. LiveraG, DelbesG, PairaultC, Rouiller-FabreV, HabertR (2006) Organotypic culture, a powerful model for studying rat and mouse fetal testis development. Cell Tissue Res 324 : 507–521.
59. GuerquinMJ, DuquenneC, LahayeJB, TourpinS, HabertR, et al. (2010) New testicular mechanisms involved in the prevention of fetal meiotic initiation in mice. Dev Biol 346 : 320–330.
60. BernexF, De SepulvedaP, KressC, ElbazC, DelouisC, et al. (1996) Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development 122 : 3023–3033.
61. YoshimizuT, SugiyamaN, De FeliceM, YeomYI, OhboK, et al. (1999) Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev Growth Differ 41 : 675–684.
62. Le BouffantR, SouquetB, DuvalN, DuquenneC, HerveR, et al. (2011) Msx1 and Msx2 promote meiosis initiation. Development 138 : 5393–5402.
63. BastosH, LassalleB, ChicheporticheA, RiouL, TestartJ, et al. (2005) Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65 : 40–49.
64. SouquetB, TourpinS, MessiaenS, MoisonD, HabertR, et al. (2012) Nodal signaling regulates the entry into meiosis in fetal germ cells. Endocrinology 153 : 2466–2473.
65. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 : 671–675.
66. PetersAH, PlugAW, van VugtMJ, de BoerP (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5 : 66–68.
67. ChicheporticheA, Bernardino-SgherriJ, de MassyB, DutrillauxB (2007) Characterization of Spo11-dependent and independent phospho-H2AX foci during meiotic prophase I in the male mouse. J Cell Sci 120 : 1733–1742.
68. WojtaszL, DanielK, RoigI, Bolcun-FilasE, XuH, et al. (2009) Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 5: e1000702.
69. ColeF, KauppiL, LangeJ, RoigI, WangR, et al. (2012) Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 14 : 424–430.
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Akutní intermitentní porfyrie
- Příjem alkoholu a menstruační cyklus
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
- Transfer zmraženého embrya zlepšuje výsledky IVF
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



