-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
Whole genome sequencing is a powerful tool in the discovery of single nucleotide polymorphisms (SNPs) and small insertions/deletions (indels) among mutant strains, which simplifies forward genetics approaches. However, identification of the causative mutation among a large number of non-causative SNPs in a mutant strain remains a big challenge. In the unicellular biflagellate green alga Chlamydomonas reinhardtii, we generated a SNP/indel library that contains over 2 million polymorphisms from four wild-type strains, one highly polymorphic strain that is frequently used in meiotic mapping, ten mutant strains that have flagellar assembly or motility defects, and one mutant strain, imp3, which has a mating defect. A comparison of polymorphisms in the imp3 strain and the other 15 strains allowed us to identify a deletion of the last three amino acids, Y313F314L315, in a protein phosphatase 2A catalytic subunit (PP2A3) in the imp3 strain. Introduction of a wild-type HA-tagged PP2A3 rescues the mutant phenotype, but mutant HA-PP2A3 at Y313 or L315 fail to rescue. Our immunoprecipitation results indicate that the Y313, L315, or YFLΔ mutations do not affect the binding of PP2A3 to the scaffold subunit, PP2A-2r. In contrast, the Y313, L315, or YFLΔ mutations affect both the stability and the localization of PP2A3. The PP2A3 protein is less abundant in these mutants and fails to accumulate in the basal body area as observed in transformants with either wild-type HA-PP2A3 or a HA-PP2A3 with a V310T change. The accumulation of HA-PP2A3 in the basal body region disappears in mated dikaryons, which suggests that the localization of PP2A3 may be essential to the mating process. Overall, our results demonstrate that the terminal YFL tail of PP2A3 is important in the regulation on Chlamydomonas mating.
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003841
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003841Summary
Whole genome sequencing is a powerful tool in the discovery of single nucleotide polymorphisms (SNPs) and small insertions/deletions (indels) among mutant strains, which simplifies forward genetics approaches. However, identification of the causative mutation among a large number of non-causative SNPs in a mutant strain remains a big challenge. In the unicellular biflagellate green alga Chlamydomonas reinhardtii, we generated a SNP/indel library that contains over 2 million polymorphisms from four wild-type strains, one highly polymorphic strain that is frequently used in meiotic mapping, ten mutant strains that have flagellar assembly or motility defects, and one mutant strain, imp3, which has a mating defect. A comparison of polymorphisms in the imp3 strain and the other 15 strains allowed us to identify a deletion of the last three amino acids, Y313F314L315, in a protein phosphatase 2A catalytic subunit (PP2A3) in the imp3 strain. Introduction of a wild-type HA-tagged PP2A3 rescues the mutant phenotype, but mutant HA-PP2A3 at Y313 or L315 fail to rescue. Our immunoprecipitation results indicate that the Y313, L315, or YFLΔ mutations do not affect the binding of PP2A3 to the scaffold subunit, PP2A-2r. In contrast, the Y313, L315, or YFLΔ mutations affect both the stability and the localization of PP2A3. The PP2A3 protein is less abundant in these mutants and fails to accumulate in the basal body area as observed in transformants with either wild-type HA-PP2A3 or a HA-PP2A3 with a V310T change. The accumulation of HA-PP2A3 in the basal body region disappears in mated dikaryons, which suggests that the localization of PP2A3 may be essential to the mating process. Overall, our results demonstrate that the terminal YFL tail of PP2A3 is important in the regulation on Chlamydomonas mating.
Introduction
Forward genetics allows the identification of mutants with phenotypes of interest and the mechanistic understanding of biological processes [1], [2]. While gene lesions generated by insertional mutagenesis can be identified by Southern blot analysis [3]–[6] or PCR-based approaches [7]–[9], identification of mutations induced by radiation or chemical mutagenesis rely on time-consuming meiotic mapping [10]–[14]. Recently, single nucleotide polymorphism (SNP) discovery by whole genome sequencing (WGS) provides a faster and more efficient method to identify causative mutations [15]–[18]. However, in model organisms such as Arabidopsis thaliana, the number of SNPs can vary from 2,000 to 900,000, depending on the strain background [19]. In Caenorhabditis elegans, the number of SNPs between two strains is ∼100,000 [16]. In the unicellular biflagellate green alga Chlamydomonas reinhardtii, ∼38,000 SNPs were identified in individual mutant strains [18]. Therefore, identification of the causative SNP from a large number of SNPs remains a challenge.
In Chlamydomonas, a model organism for the study of flagellar function, photosynthesis, biofuels, and sex determination, the causative genes in several hundred mutant strains generated by radiation or chemical mutagenesis remain unidentified [20]. UV mutagenesis mutant screens generated 12 impotent (imp) mutant strains that have either abolished or reduced mating efficiency [21], [22]. In Chlamydomonas, the sex of a cell is controlled by two alleles, plus or minus, at the mating-type (MT) locus [23]. The differentiation from exponentially growing vegetative cells to gametes is triggered by nitrogen starvation via an unknown mechanism. When gametes of the opposite mating-types are mixed together, they agglutinate via flagellar membrane-associated proteins, agglutinins, to trigger a mating signal transduction pathway. This signal cascade leads to cell fusion and the formation of zygotes. Among the previously characterized imp mutant strains, imp2, imp5, imp6, imp7, and imp9 are allelic and encode SAG1, which is the plus agglutinin [24]. The imp10 and imp12 mutant strains encode SAD1, the minus agglutinin [24]. The imp8 strain is defective in O-glycosylation and is allelic with the GAG1 locus [25]. The imp1 and imp11 mutant strains map within the MT locus [26]–[29] and carry mutations in FUS1 in plus and MID1 in minus cells, respectively. Only imp3 and imp4 remain unidentified among the original collection of impotent mutants. Unlike the other imp strains that abolish mating (<1%), the mating efficiency of imp3 and imp4 strains varies from 10% to 50%, in contrast to >80% in wild-type cells 1 hour after mixing of the gametes. Neither mutation is linked to the MT locus or to the other [21]. Saito et al. [30] suggested that activation of uncharacterized flagellar adenylate cyclases is blocked in imp3 cells, and IMP3 is required in the mating signaling pathway. However, the causative genes in imp3 and imp4 remain unidentified due to their partial, weak mating phenotype and the difficulty to map this phenotype.
Serine/threonine phosphorylation is generated by 300–400 kinases but is reversed by a relative small number of phosphatases. The serine/threonine phosphatase, protein phosphatase 2A (PP2A), plays an important role in signaling pathways. It is a ubiquitous enzyme that is involved in diverse cellular processes; they include cell cycle control, cell growth, microtubule stability, and signaling [31]. The PP2A heterotrimeric holoenzyme contains 3 subunits; they are a catalytic subunit (C subunit), a scaffold subunit (A subunit), and a regulatory subunit (B subunit). The catalytic subunit (PP2Ac) is highly conserved across species and it shares significant sequence similarity to the PP4 and PP6 protein phosphatases [32]. The catalytic activity of PP2Ac can be modulated by post-translational modifications that include phosphorylation/dephosphorylation in the conserved C-terminal motif T304PDY307FL309 on T304 and Y307 and methylation/demethylation of L309 [33]. The scaffold subunit of PP2A contains multiple HEAT repeats that confer conformational flexibility to both the catalytic subunit and the regulatory subunit [32]. The regulatory subunit of PP2A falls into four distinct families, which are known as B (PR55), B′ (B56 or PR61), B″ (PR72), and B′″ (PR93/PR110). It is believed that different families of the B subunit target PP2A to different cellular locations and bind to different substrates [32], [33]. In Chlamydomonas, sequence similarity reveals four catalytic subunits, two scaffold subunits, and five regulatory subunits [34].
In this study, we took advantage of whole genome sequencing of 16 different wild-type and mutant strains to generate a SNP/indel library. SNP/indel comparison, in conjunction with meiotic mapping, allowed the quick identification of the causative mutation in the imp3 mutant strain, which contains a C-terminal three amino acid deletion in the conserved TPDYFL motif of a PP2A catalytic subunit (PP2A3). The deletion of YFL affects not only the stability of PP2A3, but also the accumulation of PP2A3 around the basal body area.
Results
Construction of a Chlamydomonas SNP/indel library
A previous study using 101-bp paired-end Illumina sequencing of an IFT80 mutant strain, NG30/ift80, revealed that over 38,000 SNPs/indels are present compared to the Chlamydomonas reference genome [18]. It was a challenge to identify the causative mutation from such a large number of changes. We reasoned that if changes are found in other unlinked mutant strains or in wild-type strains, they are not causal and could be eliminated from further analysis. Therefore, a collection of changes from multiple strains would be necessary to remove as many non-causative changes as possible to reveal the causative mutation in a given mutant strain.
To build a library of changes, we first sequenced four wild-type strains (CC-124, CC-125, isoloP (CC-4402), and isoloM (CC-4403)). In Chlamydomonas, the major laboratory strains CC-124 and CC-125 were first isolated from a single diploid zygote, 137c, in 1945 [35] and these strains have been used as the parents in many mutant isolations. CC-125 is also the background strain of CC-503, the strain used for the Chlamydomonas reference genome assembly. CC-124 carries a minus mating-type locus (mt−) and CC-125 is mating-type plus (mt+). The other two wild-type strains, isoloP (mt+) and isoloM (mt−), were generated by crossing CC-124 by CC-125 to obtain meiotic progeny. Several progeny from this cross that gave the fastest and highest percentage of mating were backcrossed to CC-124. This procedure was repeated ten times in an attempt to obtain isogenic strains that were named isoloP and isoloM. Sequencing of these wild-type strains identifies 13,000 to over 100,000 changes relative to the reference genome. We also sequenced is a highly polymorphic strain S1C5 (CC-1952) that is frequently used in meiotic mapping with molecular markers. Over 2 million changes are identified from this strain. In addition, since we were interested in identifying the causative mutations from a variety of mutant strains, we also performed whole genome sequencing on ten mutant strains that were generated by either chemical or UV-mutagenesis. Five of them (fla18, fla24, fla9, uni1, ift80) have flagellar assembly defects, four (ida3, pf23, pf7, pf8) have motility defects, and one has a mating defect (imp3). One additional mutant strain, cnk10, which has a flagellar assembly defect, was generated by insertional mutagenesis of the CC-125 strain (Lin and Dutcher, unpublished). The number of changes in individual mutant strains varies from 22,000 to over 150,000 (Table 1). The sequencing coverage of individual strains ranges from 26× to 162× (Table 1). Overall, 2,557,197 changes are included in this SNP/indel library and it is available at http://stormo.wustl.edu/SNPlibrary/.
Tab. 1. Strains used to generate a Chlamydomonas SNP/indel library. 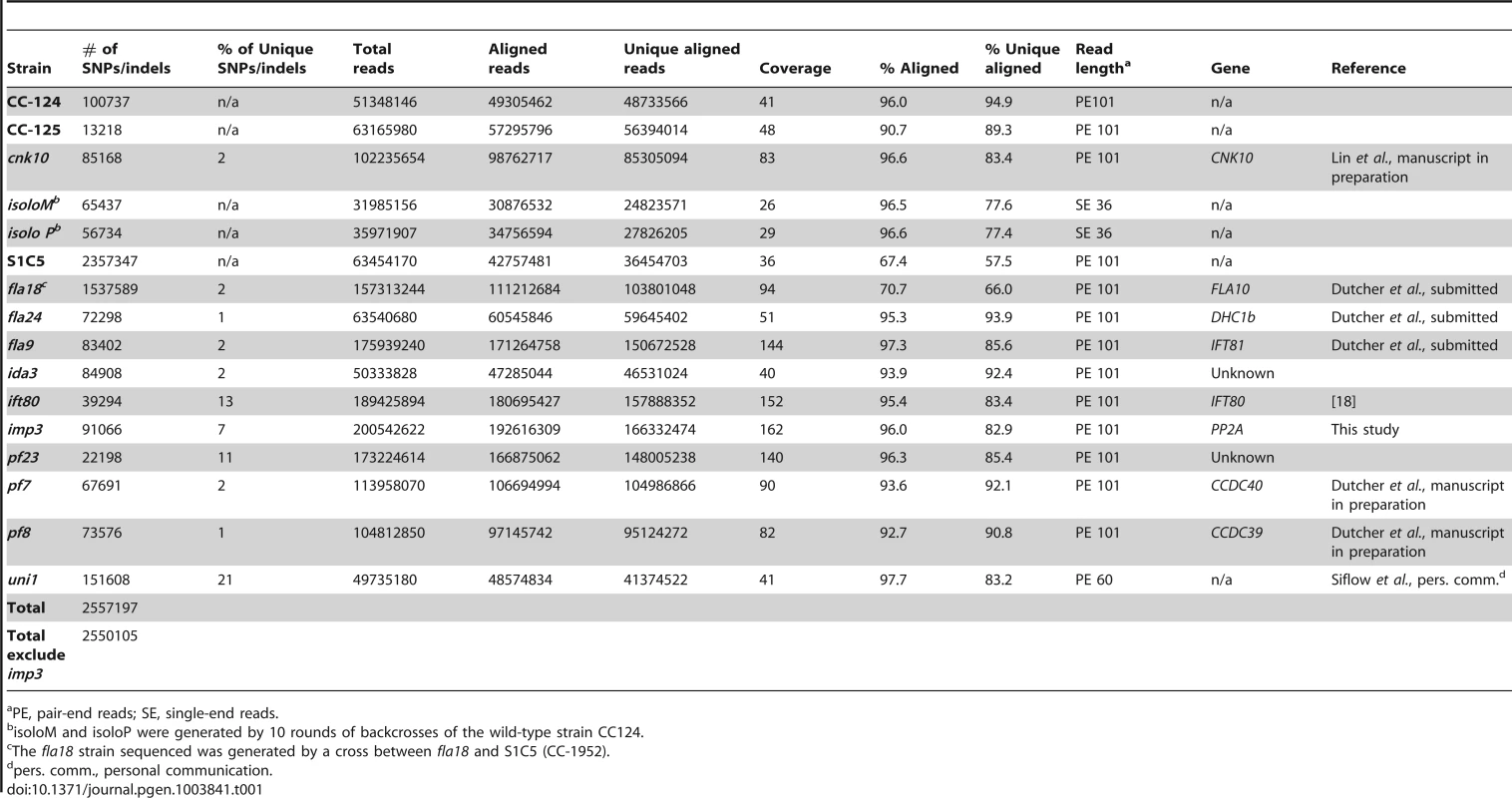
PE, pair-end reads; SE, single-end reads. After collecting the SNPs/indels, we analyzed the distribution of the changes across the 17 chromosomes relative to the reference genome [36] (Figure 1). In the wild-type strain CC-125 (mt+), changes are spread evenly across all chromosomes from the reference genome (Figure 1A, Table 2). The wild-type strain CC-124 (mt−), which came from the same zygote as CC-125, contains 100,737 changes, which is about eight times the number of changes (13,218) found in CC-125. Around 90% of changes found in CC-124 are concentrated on five chromosomes: 3, 6, 12, 16, and 17 (Figure 1A, Table 2). A detailed analysis of numbers of SNPs/indels every 100 kb along the chromosomes in CC-124 reveals that the polymorphisms are not distributed evenly across these five chromosomes (Figure 1B). On chromosome 3, most SNPs/indels are between 8.5 Mb and 9.1 Mb. On chromosome 6, most SNPs/indels are within the first 1.9 Mb, which contains the MT locus. The mating-type locus is known to be polymorphic between the two sexes [37]. CC-124 carries the MT minus locus, which is not shared with the reference strain CC-503. On chromosome 12, most changes are observed between 9.0 Mb to 9.8 Mb. On chromosome 16, three distinct regions of SNPS/indels lie between 0.9–1.0 Mb, 1.5–2.0 Mb, and 6.4–7.8 Mb. A large number of changes within the 0.9–1.0 Mb were observed in a previous study of Chlamydomonas strains, ift80 and ac17 [18]. On chromosome 17, most changes are observed between 0.3 Mb to 1.5 Mb. The other two wild-type strains, isoloP (mt+) and isoloM (mt−), which are meiotic progeny of CC-124 and CC-125 after ten rounds of backcrosses to CC-124, were expected to show difference only on chromosome 6, which contains the MT locus. However, comparison of the sequence of these two strains indicates that they are not isogenic on chromosomes 3 and 17. The isoloM strain maintains large numbers of changes from the CC-124 parent on chromosome 3; isoloP contains large numbers of changes inherited from the CC-124 parent on chromosome 17 (Figure 1A, Table 2).
Fig. 1. Distribution of SNPs/indels along the 17 Chlamydomonas chromosomes compared to the reference genome (v5.3). 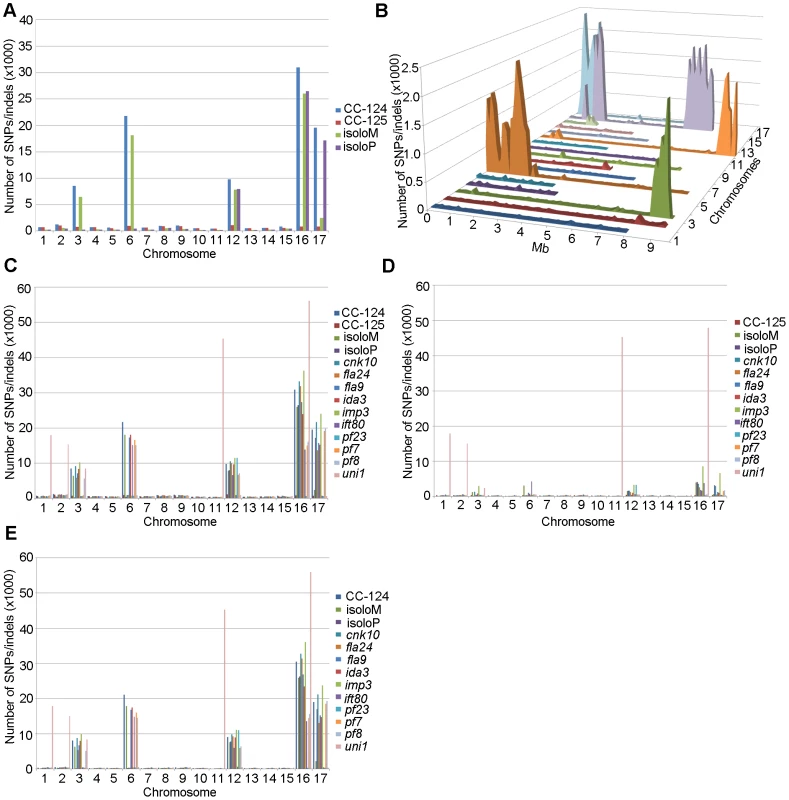
(A) Distribution of SNPs/indels in four wild-type strains (CC-124, CC-125, isoloP, and isoloM). (B) Distribution of SNPS/indels along the length of each chromosome in the CC-124 strain in Mb. The number of SNPs/indels is summed over each 100 kb interval. (C) Distribution in 14 strains used in this study, with the exception of fla18 and S1C5. (D) Distribution in strains in part C after the subtraction of SNPs/indels found in the CC-124 strain. The uni1 strain is the most divergent. (E) Distribution in 14 strains after the subtraction of SNPs/indels found in the CC-125 strain. Tab. 2. Distribution of SNPs on all 17 chromosomes in wild-type strains. 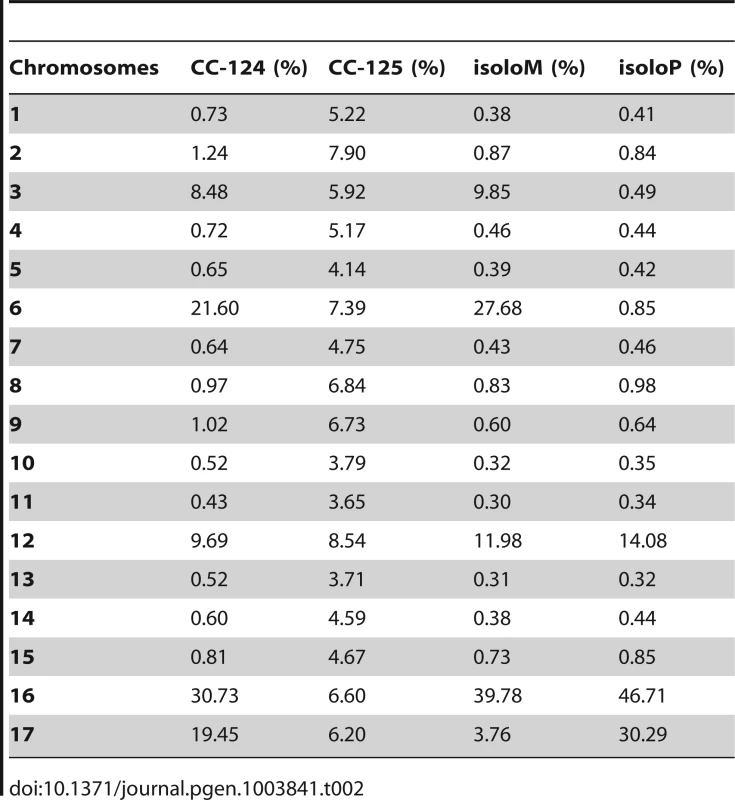
We performed the same analysis on the ten mutant strains, with the exclusion of fla18, because the sequenced strain came from a cross between the fla18 mutant strain and S1C5. Ninety-six percent of fla18 SNPs/indels are found in the S1C5 strain. Prior to Illumina sequencing, the ten strains were crossed to either CC-124 or CC-125 at least once. An accumulation of changes on chromosomes 3, 6, 12, 16, and 17 are observed in most strains (Figure 1C). To ask whether these changes are the same as found in CC-124, we subtracted SNPs/indels found in CC-124 from individual strains. The numbers of changes drop dramatically from 10,000∼35,000 to 1,000∼5,000 in almost all strains (Figure 1D). This suggests that changes in these strains are likely come from the CC-124 parent. In comparison, removal of CC-125 changes from these strains does not have an obvious effect on numbers of changes in these strains (Figure 1E). There is no correlation with the position of the causative mutant and an accumulation of SNPs along the chromosomes. One mutant strain, uni1, has a distinct distribution of changes along all chromosomes when compared to other strains (Figure 1C). Accumulation of changes is found on chromosomes 1, 2, 11, and 16. Removal of CC-124 or CC-125 changes from uni1 has no significant effect on the distribution (Figure 1D and 1E). A comparison between 30,958 changes on chromosome 16 found in CC-124 and 56,192 changes on chromosome 16 found in uni1 indicates that only 8,229 changes are common between these two strains. Thus, the SNPs/indels found in uni1 have significantly different distribution than all other strains we analyzed. This mutant strain was generated from either strain 89 or 90 (CC-1009 or CC-1010) [38]. Pröschold et al. [39] categorized the common used laboratory Chlamydomonas strains into three basic sublines based on several criteria, including their ability to utilize nitrate, the mating-type locus, the number of rDNA repeats, and the presence of cell wall digestion metalloproteases (autolysins). Strains 89 and 90 belong to Subline II and CC-124 and CC-125 belong to Subline III. They have difference in all criteria described above. Thus it is not surprising to observe the difference of SNP/indel accumulations between uni1 and 137c strains.
Identification of the causative mutation in the imp3 mutant strain
The 162× sequencing coverage of imp3 reveal 91,066 changes in this mutant strain, which came from a 137c background. Comparison and subtraction between imp3 and all other 15 strains finds 7,092 (7%) of these changes are unique to imp3 (Table 1). Out of 954 changes in predicted exons, 145 are predicted to be synonymous changes and were excluded from further analysis. Changes vary from 1 to 297 on individual chromosomes (Table 3). In order to identify the causative mutation, we meiotically mapped the imp3 mutant strain.
Tab. 3. Distribution of changes found in the <i>imp3</i> mutant strain. 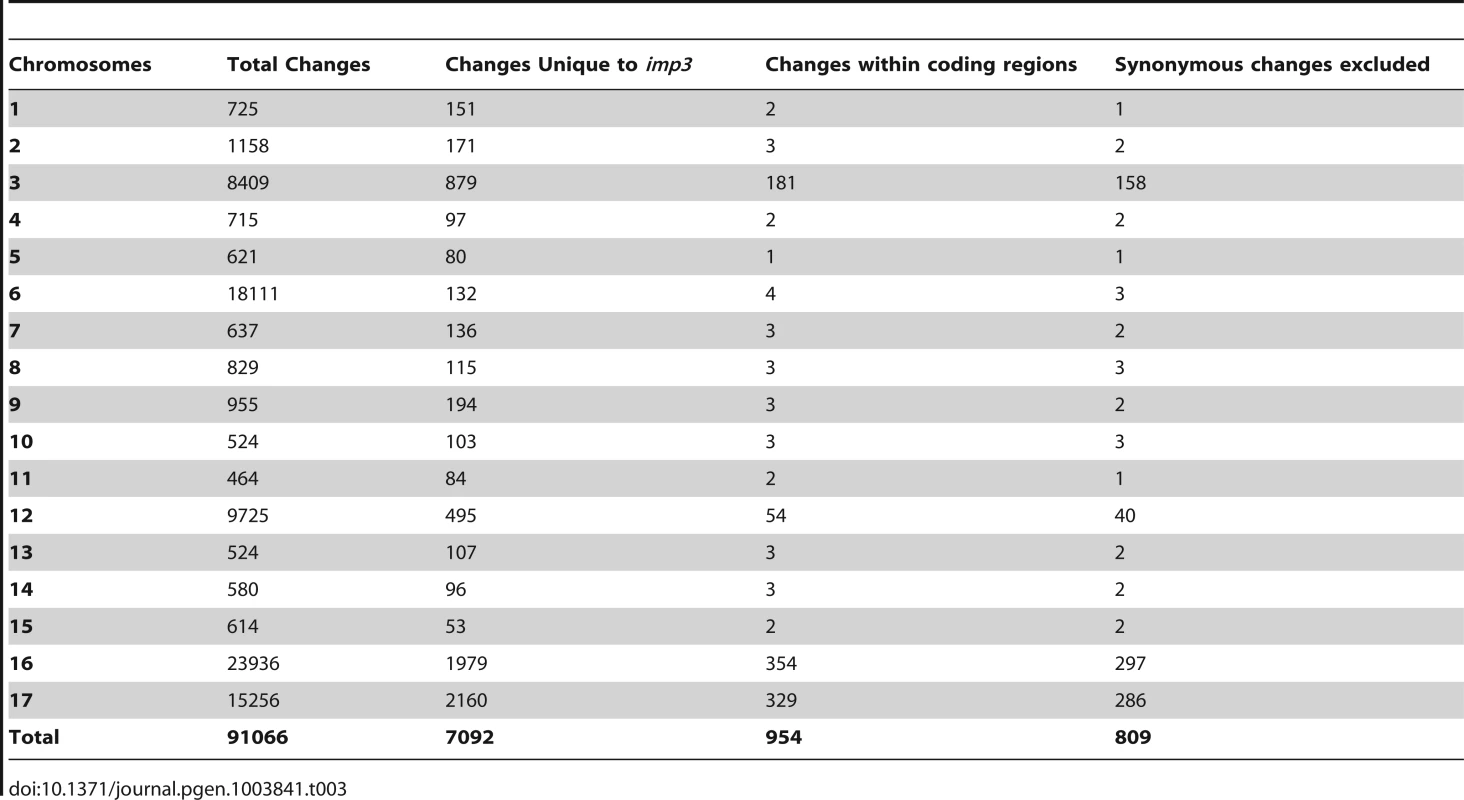
The imp3 phenotype confers reduced mating efficiency, a phenotype that is challenging to analyze quickly. When wild-type gametes mate, the zygotes develop thick cell walls and form a dark green, multi-layered sheet of cells called the pellicle (Figure 2A). The mating between imp3 mt+ and imp3 mt− do not form the thick pellicle sheet observed in wild-type cells or in mating between imp3 and wild-type cells (Figure 2A). Observation under the dissecting microscope reveals dark thick multi-layer pellicle between imp3 and wild-type gametes (Figure 2B). Mating between two imp3 strains produces a single cell-layer pellicle that is light in color (Figure 2C and 2D). Individual progeny from 30 complete tetrads from a cross between imp3 and wild-type cells were tested for this phenotype by mating with imp3 mt+ and imp3 mt− tester strains. All 30 tetrads showed 2∶2 segregation of the single layer pellicle phenotype. This phenotype made it easy to distinguish between imp3 and IMP3 cells; this assay facilitated the molecular mapping of the IMP3 locus.
Fig. 2. Pellicle phenotype after cell mating. 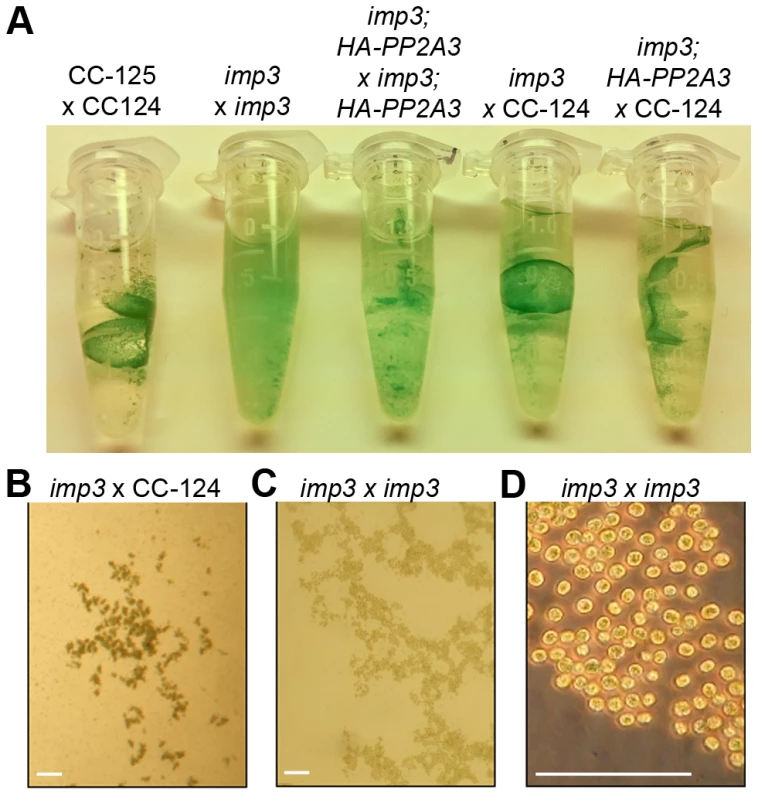
(A) Pellicle formation after overnight mating between plus and minus gametes. Sheets of dark green pellicles are observed in matings between wild-type gametes (CC-125×CC-124), wild-type mated with either the untransformed (imp3×CC-124) or transformed imp3 mutant strain (imp3; HA-PP2A3×CC-124), and between imp3; HA-PP2A3 gametes. These pellicles are resistant to vortex or pipette disruption. In contrast, only small clumps of pellicles are found in the mating between imp3 cells and they are easily disrupted by vortexing or pipetting. (B) Thick, dark green, multilayer pellicle found in mating between wild-type and imp3 gametes. (C) Single layer pellicle that is light in color is found in matings homozygous for the imp3 mutation. (D) Magnification view of cells in (C). Scale bar, 0.1 mm. To map the imp3 mutation, meiotic progeny from imp3 crossed by the highly polymorphic strain S1C5 were obtained. Progeny from twenty tetrads that showed the single layer pellicle phenotype were mapped with dCAPS markers (Table S1). The single layer pellicle phenotype showed very tight linkage (18 parental: 0 recombinant) to the SCA8-2 marker, which maps to ∼6.6 Mb on chromosome 2 in a previous version of the genome assembly (v4) and maps to ∼3.5 Mb on chromosome 9 in the latest version (v5.3) of the genome assembly. To fine map the imp3 mutation, additional progeny from over 100 tetrads were used for mapping. All three markers, 2-98, SCA8-2, and 55-193, which are about 17, 8, and 3 cM away from the imp3 mutant, map to chromosome 2 in v4 genome assembly (Table S1). Both 2-98 and 55-193 are at ∼6.6 Mb and ∼7.1 Mb on chromosome 2 in v5.3 genome assembly, respectively (Table S1). Thus, we believe that genomic DNA with an unknown length around the SCA8-2 marker is misassembled in v5.3 genome assembly to chromosome 9 and the imp3 mutation maps to chromosome 2. However, since we were unclear whether the imp3 mutation is misassembled in v5.3 genome assembly, we examined polymorphisms in predicted exons on both chromosomes 2 and 9. Two 3 nucleotide-insertion changes at ∼2.3 Mb on chromosome 2 in v5.3 genome assembly are found in imp3 (Table 3). Since both changes are over 4 Mb away from the 2-98 and 55-193 markers, they were eliminated from further study. Two changes are found on chromosome 9 in v5.3 genome assembly (Table 3) and they are both within the mapping region of ∼8 cM defined by the SCA8-2 marker. Both changes map to chromosome 2 in the v4 genome assembly. The first SNP change, which maps to position 4,049,338 on chromosome 9, has 5 Illumina sequencing reads and a Phred quality score of 16.9. It is a G to A change that causes a non-synonymous change from R (cGg) to Q (cAg) in a RegA/RlsA-like protein (g9750). The exact same change is observed in an aflagellate mutant cnc1 not included in the SNP/indel library (Dutcher and Nauman, unpublished) and thus is unlikely to be the causative change in the imp3 mutant strain. The second change, which maps to position 3,721,280 on chromosome 9, has 122 reads and a Phred quality score of 214. It is a deletion of 9 nucleotides immediately before the stop codon of a PP2A catalytic protein (PP2A3, g9684) and the deletion is predicted to remove the last 3 amino acids, YFL, which are conserved in almost all type 2A phosphatases (PP2A, PP4, and PP6; Figure 3B, S1, and S2). This change was confirmed by Sanger sequencing.
Fig. 3. Phylogenetic analysis of PP2A, PP4, and PP6. 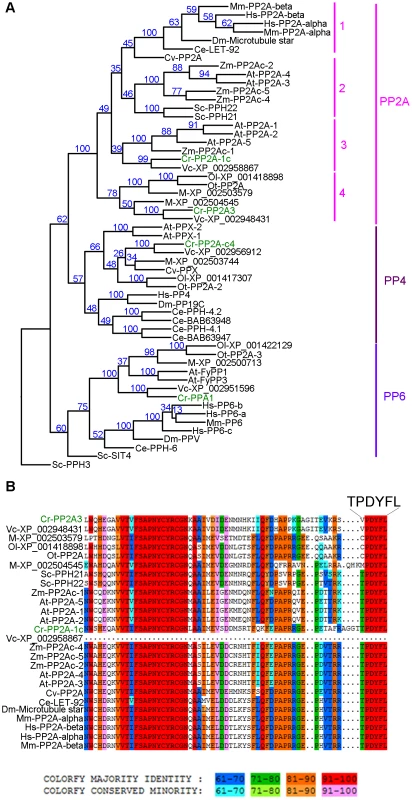
(A) Phylogenetic analysis of 55 protein phosphatases from 13 organisms. Numbers next to each node were obtained from sampling of 100 bootstrap analyses. Chlamydomonas proteins are indicated in green. (B) Sequence alignment of the C-terminal 65 amino acids from all PP2A proteins defined by phylogenetic analysis in (A). The six terminal amino acids are shown in a larger font. The full alignment is shown in Figure S2. Chlamydomonas proteins are indicated in green. Sequence similarity percentage is displayed by colors shown below the alignment. Organism abbreviation: At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Cr, Chlamydomonas reinhardtii; Cv, Chlorella variabilis; Dm, Drosophila melanogaster; Hs, Homo sapiens; M, Micromonas sp. RCC299; Mm, Mus musculus; Ol, Ostreococcus lucimarinus; Ot, Ostreococcus tauri; Sc, Saccharomyces cerevisiae; Vc, Volvox carteri; Zm, Zea mays. The PP2A catalytic subunits in Chlamydomonas
A previous study using sequence similarity indicated that the Chlamydomonas genome contains four potential PP2A catalytic subunits, PP2A-1c (g4366), PP2A3 (g9684), PP2A-c4 (Cre12.g494900), and PPA1 (Cre06.g308350) [34]. Due to the sequence similarities observed among PP2A, PP4, and PP6 in all organisms [32], we asked whether the four Chlamydomonas proteins are PP2A catalytic subunits using phylogenetic analysis. A phylogenetic tree was built based on 55 PP2A, PP4, and PP6 protein sequences from green algae (Chlamydomonas reinhardtii, Chlorella variabilis, Micromonas, Ostreococcus lucimarinus, Ostreococcus tauri, and Volvox carteri), yeast (Saccharomyces cerevisiae), land plants (Arabidopsis thaliana and Zea mays), invertebrates (Caenorhabditis elegans and Drosophila melanogaster), and mammals (Mus musculus and Homo sapiens) (Table S2, Figure 3A and S1). This phylogenetic tree shows that Chlamydomonas PP2A-1c and PP2A3 are PP2A-like proteins. PP2A-c4 is a PP4-like protein and PPA1 belongs to the PP6 family. Within the PP2A family, four subgroups are distinguished using a bootstrap analysis (Figure 3A). PP2A proteins from invertebrates, mammals, and a green alga Chlorella form subgroup 1 in the PP2A family. The subgroup 2 is composed of PP2A proteins from land plants and yeast. PP2A proteins from land plants and green algae are found in subgroup 3. The subgroup 4, which includes PP2A3, is a green algal-specific subgroup. Protein sequences of all 26 proteins from the PP2A family were aligned. The alignment reveals that while subgroups 1, 2, and 3 have the conserved T304PDYFL309 C-terminus, the proteins in the fourth subgroup do not have the conserved T304, instead, it is replaced with V, C, or M (Figure 3B and S2). This suggests that T304 is not conserved in the green algal-specific subgroup.
Introduction of PP2A3 transgene into imp3 rescues the mutant phenotype
In order to demonstrate that the deletion of YFL at the C-terminus of PP2A3 is the causative mutation in imp3, we performed plasmid rescue. The PP2A3 gene contains only a single exon, which is predicted to encode a 315 amino acid protein with a predicted molecular weight of 35,676 daltons. An HA-tagged PP2A3 gene with the epitope tag HA immediately following the start codon to avoid compromising the C-terminus and under the regulation of the 637 bp endogenous PP2A3 promoter was transformed into the imp3 mutant strain, and whole cell extract from 6 putative transformants were screened by immunoblotting with an anti-HA antibody (Table S3). The HA-PP2A3 protein is predicted to have a molecular weight of 36,760 daltons. The anti-HA antibody recognized a ∼37 kD band in one transformant (imp3; HA-PP2A3, Figure 4A). Given the importance of Y313 and L315 (the equivalent of Y307 and L309 in mammalian cells) for the function of the PP2A catalytic subunit [33], we generated various N-terminal HA-tagged mutant forms (Y313Δ, L315A, L315Δ, and Y313F314L315Δ) under the same promoter and transformed them into the imp3 cells individually (Table S3). All mutant forms of the HA-PP2A3 protein are expressed, as detected by the anti-HA antibody but the amount of the protein is significantly less than the transformed wild-type HA-PP2A3 protein (Figure 4A). Additionally, we generated a V310T substitution to investigate whether this change has any effect on rescue of the imp3 mutant strain. The abundance of this protein is about 1.5 fold higher than the wild-type HA-PP2A3 transformant (Figure 4A). An immunoblot with a monoclonal antibody against α-tubulin was used to quantify protein loading (Figure 4A). Two smaller bands (∼27 kD and ∼23 kD) were also detected by the anti-HA antibody and they may correspond to proteolyzed/truncated PP2A3.
Fig. 4. Introduction of HA-tagged PP2A3 transgene rescues the imp3 mutant phenotype. 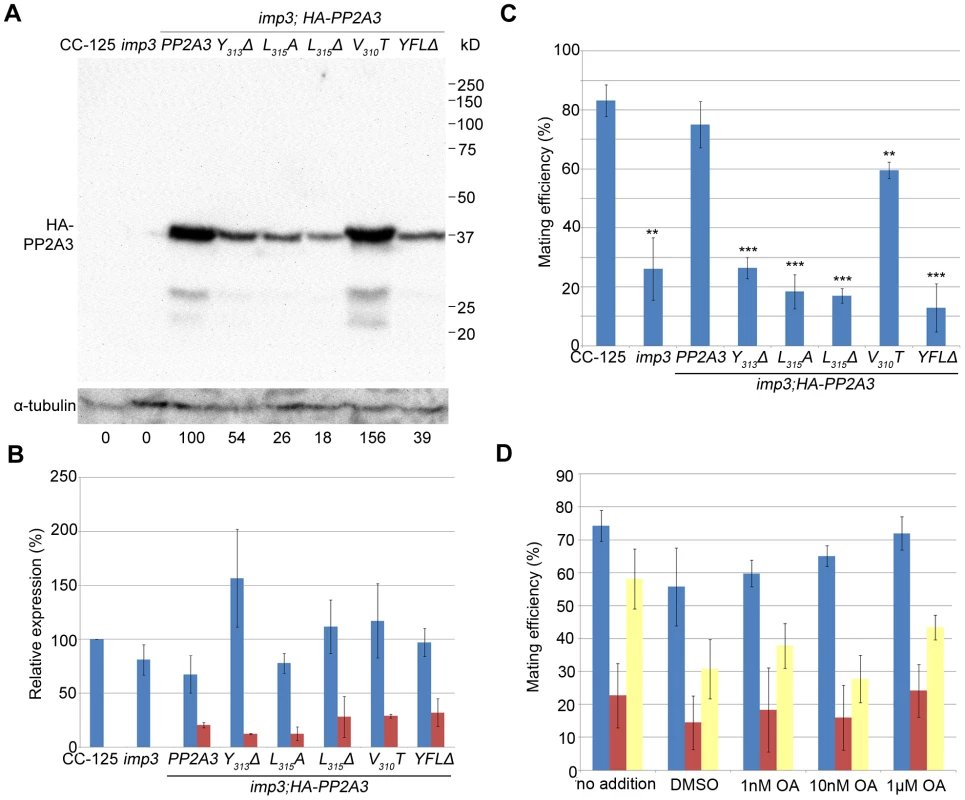
(A) A representative immunoblot with the anti-HA antibody in transformants carrying either the wild-type or mutant HA-PP2A3. The HA-PP2A3 band is observed ∼37 kD and the two small bands (∼27 kD and ∼23 kD) may represent proteolyzed or truncated HA-PP2A3. The same membrane was re-probed with a monoclonal antibody against α-tubulin to serve as a measure of loading. Relative amount of the HA-PP2A3 protein in individual samples were measured by ImageJ and standardized by the amount of α-tubulin in the same sample. (B) Real-time PCR shows relative expression levels of PP2A3 (blue) and HA-PP2A3 (red) with the transcript level in wild-type CC-125 set at 100%. Each column represents three biological replicates and error bars show standard errors. (C) Mating efficiency of individual strains with the wild-type CC-124 after 1 hour of mating. Results represent three biological replicates and error bars show standard errors. **, p<0.01; ***, p<0.001. (D) Effects of okadaic acid on mating in wild-type (blue), imp3 (red), and imp3; HA-PP2A3 (yellow). The addition of DMSO serves as a control. To ask whether the difference in protein abundance observed in the HA-PP2A3 transformants is due to the abundance of the transgenic HA-PP2A3 transcript or due to protein stability, we measured the transcript levels of PP2A3 by real-time PCR. In the wild-type strain CC-125 and imp3, real-time PCR detected only transcript levels of the endogenous PP2A3 transcript (Figure 4B, blue). In all HA-PP2A3 transformants, levels of two transcripts were detected. The first primer set detected the combined transcript levels of endogenous PP2A3 and transgenic HA-PP2A3 (Figure 4B, blue). Overall transcript levels of PP2A3 in all strains tested are comparable. The second primer set detected only the transcript level of transgenic HA-PP2A3 (Figure 4B, red). The transcript levels of transgenic HA-PP2A3 in all transformants are about one-quarter of the total PP2A3 transcript levels. There is no significant difference among different mutant transformants compared to the wild-type HA-PP2A3 transformant (Figure 4B, red). Therefore, we conclude the differences observed in the protein levels of HA-PP2A3 in different mutant transformants are not due to the abundance of the transgenic HA-PP2A3 transcripts, but rather due to the stability of the HA-PP2A3 proteins.
Transformation of wild-type HA-PP2A3 into imp3 cells successfully rescued the mating defect (Figure 4C, HA-PP2A3). Mating between imp3; HA-PP2A3 plus and minus gametes and mating between wild-type and imp3; HA-PP2A3 both produce thick pellicles (Figure 2A). The V310T change, which is found in 5 independent transformants, partially rescues the mating efficiency to about 60% (Figure 4C). None of the changes in the YFL motif rescues the mating phenotype (Figure 4B, Y313Δ, L315A, L315Δ, and YFLΔ), which indicates the importance of the last 3 amino acids in the function of PP2A3 during mating. Thus, we conclude that PP2A3 is encoded by the IMP3 gene and the deletion of nine nucleotides at the C-terminus of this gene causes the defective mating efficiency of imp3 cells.
We further asked whether inhibition of PP2A3 has any effect on mating efficiency. Okadaic acid (OA), a polyether fatty acid, was shown to inhibit the phosphatase activity of PP2A [40], enhance phosphorylation of Y307 [41], and inhibit methylation of L309 [42], [43] in vitro. OA inhibits PP2A at very low concentrations and the dissociation constant (Ki) between OA and PP2A is ∼0.032 nM [44]. From in vivo studies, however, the amount of OA required to inhibit PP2A varies from 10 nM in human lung cancer cells [45] to ∼1 µM in MCF7 breast cancer cells [46]. It is suggested that the entry rate of OA can be affected by pH, temperature, and exposure time to OA [47]. We tested the effect of OA on Chlamydomonas mating at concentrations of 1 nM, 10 nM, and 1 µM. The mating efficiency between wild-type CC-124 (mt−) and CC-125 (mt+) is around 75% (Figure 4D). The addition of DMSO and different concentration of OA, for one hour at room temperature, has no significant effect on the mating efficiency (Figure 4D, blue bars). Similarly, addition of DMSO or OA has no significant effect on the mating efficiency of imp3 mt+×imp3 mt− (Figure 4D, red bars) and imp3; HA-PP2A3 mt+×imp3; HA-PP2A3 mt− (Figure 4D, yellow bars). Pre-treatment of cells with autolysin, an enzyme that removes Chlamydomonas cell walls, before the addition of OA, leads to similar results (data not shown). In a study on phosphoproteome in Chlamydomonas, cells pre-incubated with 1.5 µM OA for 29 hours accumulate 38% more phosphorylated proteins [48]. Therefore, our OA results indicate that either one hour inoculation is not sufficient for OA to enter Chlamydomonas, or the effect of OA on Chlamydomonas is more complicated than simple inhibition of PP2A3.
PP2A3 interacts with a PP2A scaffold protein and localizes to both cell bodies and flagella
To identify interacting proteins of PP2A3 and to investigate whether changes in the YFL motif lead to changes in protein-protein interactions, we performed immunoprecipitation with the anti-HA antibody. Two major bands of ∼65 kD and ∼37 kD are obtained by immunoprecipitation from whole cell extract from imp3 gametes transformed with wild-type HA-PP2A3 but not with untransformed imp3 gametes (Figure 5A). The ∼37 kD band is the HA-PP2A3, indicated by an immunoblot probed with an anti-HA antibody (Figure 5A). The ∼65 kD band was excised and subjected to mass spectrometry. The protein with the most number of peptides (94; 27 are unique) is PP2A-2r (Cre11.g477300) and it has a predicted size of 64,729 daltons. Changes in Y313Δ, L315Δ, V310T, or YFLΔ did not affect the pull-down of PP2A-2r by the HA antibody (Figure 5A and S3). Mass spectrometry of the ∼65 kD band pulled down by HA-PP2A3-V310T and by HA-PP2A3-YFLΔ resulted in 68 (27 unique) and 82 (26 unique) peptides of PP2A-2r, respectively. Thus, the interaction between the catalytic subunit PP2A3 and the scaffold subunit PP2A-2r is not affected by the changes at the C-terminus of PP2A3. PP2A-2r is one of the two PP2A scaffold proteins in the genome and was found in the flagellar proteome [34], [49]. Another scaffold subunit FAP14, which is also found in the flagellar proteome, has a predicted molecular weight of 100,787 daltons. Given that no significant band at ∼100 kD was identified in the immunoprecipitation (Figure 5A and S3), it is unlikely that PP2A3 interacts with FAP14.
Fig. 5. HA-PP2A3 interacts with a scaffold subunit protein and localizes to the flagella. 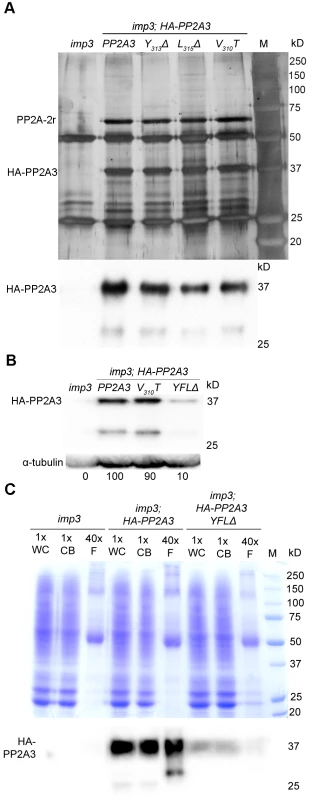
(A) Upper panel, immunoprecipitation of wild-type and mutant HA-PP2A3 proteins from whole cell extracts of gametes by anti-HA-antibody. Proteins were separated on a 10% polyacrylamide gel and visualized by silver staining. M, protein standards. Bottom panel, immunoblot with the anti-HA antibody shows a band with the expected size (∼37 kD) and a faint band with a smaller size (∼29 kD). (B) A representative immunoblot with the anti-HA antibody to detect HA-PP2A3 from flagellar proteins in HA-PP2A3 transformants. The same membrane was re-probed with a monoclonal antibody against α-tubulin to serve as a measure of loading. Relative amount of the HA-PP2A3 protein in individual samples were measured by ImageJ and standardized by the amount of α-tubulin in the same sample. (C) Relative distribution of HA-PP2A3 in whole cells (WC), cell bodies (CB), and flagella (F). Protein from about 1.25×107 whole cells and cell bodies, and flagella isolated from about 5×108 cells were used. Upper panel, Coomassie blue staining of a 10% polyacrylamide gel to visual protein extracts from different compartments of cells. Lower panel, immunoblot with the anti-HA antibody. Since PP2A-2r is present in the flagellar proteome, we asked whether PP2A3 localizes to the flagella. Flagella were isolated from imp3 and the HA-tagged transformants with the wild-type PP2A3, V310T, and YFLΔ genes. The HA-PP2A3 is detected in both wild-type and mutant transformants, but not in the untransformed imp3 flagella with the anti-HA antibody (Figure 5B). The YFLΔ transgene strain contains only ∼10% of HA-PP2A3 of those found in the wild-type HA-PP2A3 and the V310T strain; this is similar to observations in the whole cell extract immunoblots (Figure 4A). A smaller band (∼29 kD), which may represent proteolyzed/truncated HA-PP2A3, is again recognized by the anti-HA antibody (Figure 5B).
To compare the relative distribution of HA-PP2A3 in Chlamydomonas cell bodies and flagella, we perform immunoblots of HA-PP2A3 on the basis of cell equivalents (Figure 5C). Protein from equal numbers of whole cells and cell bodies, and from flagella isolated from about 40 times more cells, were used in the analysis. In both imp3; HA-PP2A3 and imp3; HA-PP2A3 YFLΔ strains, the HA-PP2A3 signal intensity is comparable in all three portions (Figure 5C). While the flagellar proteins represent less than 5% of protein found in whole cell extract [50], we do not find a significant enrichment of HA-PP2A3 in the flagella. In contrast, we observed a significant reduction of the HA-PP2A3 signal in imp3; HA-PP2A3 YFLΔ when compared to imp3; HA-PP2A3. Similar to previous observation (Figure 4A and Figure 5B), we noticed additional smaller bands in whole cells, cell bodies, and flagella. It is intriguing that the smaller bands observed in whole cells/cell bodies and in flagella are different in size and intensity. It is likely that these represent truncated PP2A3 proteins but the functions of these truncated proteins are unknown.
Localization of PP2A3 changes in the imp3 mutant strain and in mating cells
To ask where the HA-PP2A3 protein localize, we performed immunofluorescence with the HA antibody in six transformant strains (Figure 6A). In wild-type (CC-125) and untransformed imp3 gametes, there is some non-specific binding of the antibody in the cell body. In imp3; HA-PP2A3 gametes, robust signals are observed throughout the cells. In addition, in ∼80% of imp3; HA-PP2A3 gametes, accumulation of the signal is observed around the basal body area (Figure 6C, blue bars). The same signal intensity and localization is observed in imp3 cells transformed with HA-PP2A3 carrying a V310T mutation. In comparison, in imp3 cells transformed with the mutant forms of HA-PP2A3 (Y313Δ, L315A, L315Δ, and YFLΔ), the signal intensities of HA are significantly reduced, consistent with what we observed in the immunoblots (Figure 4A). Less than 10% of these cells showed basal body localization (Figure 6C). Therefore, we conclude that mutations of the terminal YFL affect the localization of PP2A3 to the basal body region.
Fig. 6. HA-PP2A3 localizes to the basal body region in gametes. 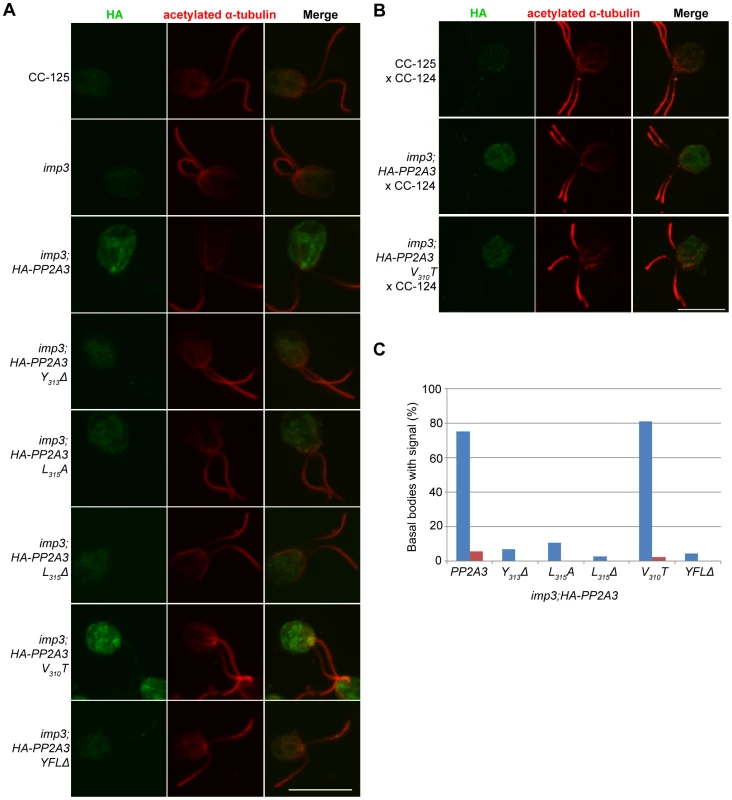
(A) Immunofluorescence with the anti-HA antibody (green, first column) in unmated gametes of wild-type (CC-125), imp3, or imp3 transformed with wild-type and mutant forms of HA-PP2A3. Immunofluorescence with the anti-acetylated α-tubulin (red, second column) identifies the Chlamydomonas flagella. Merged images of both staining are shown in the third column. Scale bar, 10 µm. (B) Immunofluorescence of the HA-PP2A3 location in wild-type dikaryons (CC-125×CC-124), in dikaryons between wild-type and imp3 gametes transformed with wild-type or V310T HA-PP2A3. (C) Percentages of cells in each strain that showed the basal body localization of the HA-PP2A3 signal. Blue columns represent results from the unmated gametes. Red columns represent data from dikaryons identified by their four flagella. For each point, over 50 cells were collected. Given the accumulation of the HA-PP2A3 proteins in wild-type HA-PP2A3 and V310T cells, we asked whether mating of these gametes with wild-type gametes would lead to change of localization of HA-PP2A3 (Figure 6B). When Chlamydomonas cells mate, the flagella adhere to each other, leading to cell fusion to form a single cell with four flagella and two nuclei, which is known as a dikaryon. We examined dikaryons one hour after mixing wild-type gametes (CC-125×CC-124); they show a low background of non-specific staining. In contrast, dikaryons formed between imp3; HA-PP2A3 (wild-type or V310T) and wild-type gametes show strong signals throughout the cells. However, less than 5% of these dikaryons show staining around the basal body area (Figure 6C, red bars). These results indicate that PP2A3 moves out of the basal body region in dikaryons.
Discussion
Whole genome sequencing to generate a Chlamydomonas SNP/indel library
Whole genome sequencing has become an important tool to allow quick identification of causative mutations in Chlamydomonas ([18] and Dutcher et al., submitted), Caenorhabditis elegans [16], Drosophila [51], and humans [52]. In Drosophila, direct comparison of sequences from parental and EMS mutagenized chromosomes leads to the identification of causative SNPs. This removes the need for sequence alignment to the reference genome sequences, which eliminates the natural variation of SNPs within different strains [51]. However, this approach is not feasible to identify mutants whose original strain backgrounds are unavailable. In C. elegans, a cross to a highly polymorphic strain and whole genome sequencing of a pool of 50 F2 progeny eliminates the need for meiotic mapping. The number of SNPs drops significantly within a ∼2 Mb region where the mutation resides. Thus, the number of SNPs of interest is reduced dramatically; it becomes easier to identify the causative mutation [16]. In the studies of human variants, databases such as dbSNP and The 1000 Genomes Project [53] are available to filter non-causative SNPs/indels. The filtering results in a reduction of ∼98% of SNPs/indels in a given individual and thus it becomes feasible to identify causative mutations for rare Mendelian diseases [52]. Similar to the human 1000 Genomes Project, a 1001 Genomes Project on Arabidopsis thaliana was initiated in 2008 [19]. Sequencing of 80 Arabidopsis strains identified ∼5.7 million SNPs/indels [54].
We previously used meiotic mapping to narrow the regions of interest to 269 kb in NG6/fla8-3 and 458 kb in NG30/ift80, respectively. Identification of one and six nonsynonymous changes in these regions eventually led to discovery of the causative mutations in these mutant strains [18]. In an approach similar to that used in C. elegans [16], we combined a pool of 14 progeny from a cross between a pf27 mutant strain and the highly polymorphic S1C5 strain for whole genome sequencing. This approach narrows the region of interest to ∼2 Mb on chromosome 12 ([55] and Alford et al., submitted), and eliminates the need for genome-wide mapping. However, the number of SNPs within 2 Mb remains large and it is hard to identify the causative mutation easily without further fine scale mapping. Therefore, generation of a SNP/indel library similar to the databases generated in other organisms is necessary to further eliminate common SNPs/indels found in Chlamydomonas strains.
While the two major laboratory strains (CC-124 and CC-125) were isolated from a single diploid zygote, 137c, they have >80,000 SNP/indel difference (Table 1). Both strains, and the reference strain used for genome assembly, belong to Subline III, as described by Pröschold et al. [39]. Therefore, strains that belong to Subline I, and strains from Subline II, such as uni1, are expected to contain more changes. The two isolo strains, originated from CC-124 and CC-125, after 10 rounds of backcrosses to CC-124, are expected to contain changes similar to each other and to CC-124. Instead, in addition to the expected difference observed on chromosome 6, the isolo strains show >30,000 polymorphisms between each other on chromosomes 3 and 17 (Figure 1A and Table 2). It is worth noted that the sequencing of isolo strains was performed on the Genome Analyzer IIx platform with 36 base-pair single-end reads and the sequencing of almost all other strains except uni1 was performed on the HiSeq platform with 101 base-pair paired-end reads. Thus, the quality of alignments and coverage on the genome are slightly lower in the isolo strain sequence. As a result, SNP/indel calling in the isolo strains may be less comprehensive than that in other strains (Figure 1A, Table 1, and Table 2). Nevertheless, the accumulation of changes on chromosomes 3, 6, 12, 16, and 17 found in CC-124 is observed in one or both isolo strains. The difference observed between isoloP and isolo M on chromosomes 3 and 17 suggests that in addition to the recombination-suppressed regions found on chromosome 6, there are additional regions on these two chromosomes that may show suppression of meiotic recombination. It is unclear why these regions are recombination-suppressed.
Given that many Chlamydomonas mutants were generated from the 137c background (Table S4), the SNP/indel library that we generated greatly facilitates quick identification of causative mutations in six mutant strains, fla18, fla24, fla9 (Dutcher et al., submitted), imp3 (this study), pf7, and pf8 (Dutcher et al., manuscript in preparation). The percentage of unique SNPs/indels after filtering is about 1∼2% in most strains and 7% in imp3 (Table 1). Even though we cannot eliminate the need for meiotic mapping in this study, we were able to combine this SNP/indel library and transcriptional profiles during flagellar regeneration [56] to identify mutations in flagellar assembly genes without the need of meiotic mapping (Dutcher et al., submitted). Thus, we anticipate that this SNP/indel library will facilitate rapid discovery of more causative mutations in Chlamydomonas mutant strains.
The importance of the terminal YFL in Chlamydomonas PP2A3
In humans, there are two isoforms (α and β) of PP2A catalytic subunits (PP2Ac). They share high sequence identity (97%), but the α isoform is ∼10 times more abundant than the β isoform at both transcript and protein levels [31]. In the invertebrates, Drosophila [57] and C. elegans [58], only one PP2A is present in each genome. In contrast, Arabidopsis has at least five PP2Ac proteins, and they can be divided into two subfamilies based on sequence similarity. The sequence identity between two subfamilies, I and II, is ∼80% [59]. The subfamily I proteins are involved in abscisic acid (ABA) and brassinosteroid signaling [60], [61]. The subfamily II proteins play an important role in auxin distribution and plant development [59]. In green algae, there are two PP2Ac proteins present in the genome. While one Chlamydomonas PP2A protein (PP2A-1c) belongs to the same group as the Arabidopsis PP2A subfamily I, the other Chlamydomonas PP2A protein, PP2A3/IMP3, is found in a green algal-specific cluster (Figure 3A). The sequence identity between PP2A-1c and PP2A3 is only 63%. While our results suggest that PP2A3 functions in flagella, it is likely that PP2A3 and its homologs in the green algae-specific cluster have additional functions since Ostreococcus lacks flagella.
Our study on PP2A3 provides several insights in the importance of the terminal YFL in PP2A. First, it is necessary for PP2A function in Chlamydomonas mating. Transgenes with mutations of individual amino acids or deletion of all three amino acids fail to rescue the mating defect (Figure 4C). Second, mutations or deletion of YFL do not affect the binding of PP2A3 to the scaffold subunit. In a study on the structure of the PP2A holoenzyme in vitro, the removal of the last 15 amino acids at the C-terminus of human PP2A does not affect the formation of the holoenzyme, which includes the catalytic subunit, the scaffold subunit, and the regulatory subunit PR61 γ1 isoform [62]. In cultured mammalian cell lines including COS7, NIH 3T3, and neuro-2a, changes of T304, Y307 and L309 alter the binding between PP2A and different regulatory subunits [63], [64]. Our study provides evidence that YFL is not required to form the core PP2A enzyme, which contains the catalytic C subunit and the scaffold A subunit (Figure 5A and S3), but it is unclear whether it can form the holoenzyme since the regulatory subunit that interacts with PP2A3 has not been identified. Third, loss of YFL affects the protein stability of PP2A3 in Chlamydomonas. It is not clear if this instability is observed in all kinds of cells. In mammalian cell lines, deletion or mutation of T304, Y307 and L309 do not affect the stability of HA-tagged PP2Ac as judged by immunoblots probed with an anti-HA antibody [63], [64]. However in yeast cells, contradictory results were obtained with tagged versus untagged proteins [65], [66]. It suggests that the HA tag might destabilize the PPH21p-L369Δ mutant [66]. In our study, the change or deletion of Y313, L315, and deletion of YFL, but not the V310T change, leads to reduction of PP2A3 protein levels. We think it is unlikely that the HA tag has an effect on the stability of PP2A3 mutants, but to completely rule out the possibility, a PP2A3-specific antibody will be necessary. Fourth, loss of YFL affects the localization of PP2A3 to the basal body region in Chlamydomonas. In C. elegans embryo, LET-92, the catalytic subunit of PP2A, localizes to centrosomes as well as in the cytoplasm. The centrosome localization of LET-92 depends on RSA-1 (Regulator of Spindle Assembly 1), a B″ type PP2A regulatory subunit that also localizes to centrosomes [67]. In monkey kidney CV-1 cells, immunofluorescence using a monoclonal antibody against PP2Ac revealed that PP2Ac localizes to both microtubule and centrosomes [68]. It was shown in HeLa cells both the B′ type PP2A regulatory subunit B56α and the PP2A scaffold subunit localize to the centrosomes and it was proposed that B56α serves as a chaperone that facilitates the translocation and function of the catalytic subunit [69]. In Chlamydomonas, the basal bodies serve as nucleation sites for assembly of flagella during interphase and function as centrioles during mitosis [70]. Our finding that PP2A3 localize to the basal body region is consistent with PP2Ac being found in centrosomes of other organisms. More importantly, our result suggests that mutations in the terminal YFL affect the basal body localization of PP2A3. It is likely that PP2A3 binds to a Chlamydomonas regulatory subunit similar to RSA-1 or B56α in the centrosomes and mutations in PP2A3 YFL attenuate the interaction between PP2A3 and the regulatory subunit.
PP2A3 and Chlamydomonas mating
In the Chlamydomonas mating signaling pathway triggered by agglutination of flagella from plus and minus cells, two proteins have identified. The TRPP2 protein Polycystin-2 (PKD2; Cre17.g715300) localizes to the flagellar membrane as well as to the cell body and is enriched fourfold in flagella during gametogenesis [71]. A cGMP-dependent protein kinase (PKG/CGK2; Cre02.g076900) is also present in both the cell body and flagella. Flagellar agglutination during mating leads to phosphorylation of a tyrosine residue in PKG that activates its protein kinase activity [50]. Knockdown of PKD2 and PKG via RNA interference leads to reduced mating efficiency, which resembles the imp3 phenotype [50], [71]. The signal transduction cascade is postulated to involve at least one additional uncharacterized tyrosine kinase that phosphorylates PKG [50].
Given the localization of PP2A3 to the basal body region as well as to the flagella, we propose that PP2A3 regulates the mating signaling pathway through one or more of the following mechanisms. Similar to other signaling pathways, phosphorylation and dephosphorylation of components in the mating signaling pathway need fine regulation. As a phosphatase, PP2A3 may act as a negative regulator of phosphorylation of PKG. The presence of PP2A3 keeps PKG unphosphorylated in gametes. In the imp3 gametes, since the amount of the PP2A3 protein is significantly reduced, dephosphorylation of PKG is likely to be compromised. Alternatively, PP2A3 could act as a positive regulator to facilitate the movement of signaling proteins as well as agglutinin proteins into the flagella to allow the proper functions of these proteins on the flagellar membrane during mating. It has been shown that the transport of PKD2 to the flagellar membrane requires intraflagellar transport (IFT) [71] but the entry of a truncated SAG1 protein to the flagellar membrane is IFT-independent [72]. It is possible that PP2A3 acts either on these proteins directly or on the transport machineries themselves. Comparison of PKD2 levels in wild-type and imp3 cells, as well as exploration of the relationship between PP2A3 and IFT will provide some answers to the function of PP2A3 in protein trafficking to the flagellar membrane.
The involvement of important signaling proteins such as PKD2 makes the mating process of Chlamydomonas an outstanding model to study ciliary/flagellar signaling. Our finding that PP2A3 involved in this process suggests that this ubiquitous enzyme may play an important role in this signaling. In addition, identification of the causative mutation in the imp4 mutant strain, which shares the same phenotype as imp3 but is unlinked to imp3 [21], may reveal additional players in this intriguing pathway.
Materials and Methods
Chlamydomonas strains
Chlamydomonas reinhardtii strains, CC-124 (mt−), CC-125 (mt+), CC-1952 (S1C5), CC-3864 (fla18), CC-3866 (fla24), CC-1918 (fla9), CC-2668 (ida3), CC-465 (imp3), CC-3660 (pf23), CC-568 (pf7), CC-560 (pf8), CC-1926 (uni1), CC-916 (NG30, ift80), were obtained from the Chlamydomonas Resource Center. Both isoloM (CC-4403, mt−) and isoloP (CC-4402, mt+) were generated in this laboratory and deposited to the Chlamydomonas Resource Center. All mutant strains obtained were backcrossed at least once with either CC-124 or CC-125, and progeny with the mutant phenotype were used in further analysis. The cnk10 mutant strain was generated by insertional mutagenesis in the CC-125 background.
Whole genome sequencing and read alignment
Chlamydomonas genomic DNA was prepared as previously described [18]. Approximately 108 cells were used in DNA preparation. About three µg of genomic DNA from each strain was submitted to Genome Technology Access Core (Department of Genetics, Washington University in St. Louis) for Illumina sequencing. The 36 bp single-ended (SE) sequencing of isoloP and isoloM and the 60 bp pair-ended (PE) sequencing of uni1 were performed with the Genome Analyzer IIx platform. The 101 bp PE sequencing of the other 13 strains was performed with the HiSeq platform. Almost all samples with the exception of ift80 and uni1 were individually tagged with a unique 7-nucleotide index and multiple samples were subjected to one sequencing flow cell lane (Table S4). The sequencing reads of ift80 were obtained from a previously published result [18]. The indexed sequencing reads were de-multiplexed before being subjected to read alignment.
For read alignment, the genome sequence of Chlamydomonas v5.3.1 (Creinhardtii_236.fa.gz) was downloaded from http://www.phytozome.net/chlamy.php [36]. An indexed database was built from 54 FASTA sequences (17 chromosomes+37 scaffolds) by Novoindex (novocraft.com). The SE sequencing reads were aligned to the database by Novoalign (version 2.08.02) with the following options: -o SAM -r random -l 25 -e 100 -a AGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG -H -c 12. The PE sequencing reads were aligned to the database by Novoalign with the following options: -o SAM -r random -l 30 -e 100 -i 230 140 -a AGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTA -H -c 12 -h 90 120. SAMtools (version 0.1.18) [73] was used to convert the resulting SAM (Sequence Alignment/Map) files to BAM (Binary Sequence Alignment/Map) format and sort the BAM files. Duplicated reads were removed by Picard (version 1.46; picard.sourceforge.net) and the resulting BAM files were sorted by SAMtools.
SNP calling and the SNP/indel library
The same genomic DNA sequence (v5.3.1) was indexed by SAMtools and used as a reference file in SAMtools mpileup (options: -u -g), which outputs a BCF (Binary Call Format) file for BCFtools (options: -b -v -c -g; vcfutils.pl varFilter -D 999) to call for SNP/indel variants using Bayesian inference [73]. The numbers of SNPs/indels from individual strains were obtained from the resulting VCF (Variant Call Format) files.
VCF files of all 16 strains sequenced are available on the SNP/indel library webpage (http://stormo.wustl.edu/SNPlibrary/). The webpage is designed to allow direct comparison of SNPs/indels from one strain to one or more strains of interest. It also allows users to upload their own VCF files and compare to SNPs/indels from one or more of the 16 strains we provide. The output is a VCF file which maintains its original format minus the common SNPs/indels. To detect unique changes in the imp3 mutant strain, we compared the SNPs/indels to the SNPs/indels in all other 15 strains. The resulting new VCF file was then subjected to SnpEff (version 3.0j) [74] to identify SNPs/indels within the coding regions or exon/intron boundaries (options: -no-upstream -no-downstream -no-intergenic -no-intron -no-utr -hom -o txt). The SnpEff reference database for was built based on the genome assembly and gene annotation (Creinhardtii_236_gene.gff3.gz) of Chlamydomonas v5.3.1 [36].
Meiotic mapping of imp3
In order to quickly identify the imp3 progeny from crosses between imp3 and wild-type cells as well as between imp3 and S1C5 cells, we developed a low-density mating assay. In this assay, approximately 1o6 cells were subjected to 0.1 ml of nitrogen-free medium (M-N/5) in a well of a 96-well plate for 4 hours at 21°C. Half of the volume of cells was mixed with imp3 mt+ testers and half were mixed with imp3 mt− testers. Starting at 30 minutes after mixing and at 30-minute increments for four hours, the cells were monitored under the dissecting microscope for agglutination and the formation of pellicle. The cells were incubated under constant light overnight at 21°C and then scored again after 18 hours. Mating between IMP3 cells and imp3 testers formed multi-layer, dark green pellicle around the four-hour time point, which was maintained until the 18-hour time point. Mating between imp3 and imp3 testers formed a thin, single-cell layer beginning around the two-hour time point but it never accumulated into the multi-layer pellicle within the next 16 hours. In the cross between imp3 and S1C5, over 100 independent imp3 progeny were picked from over 100 tetrads and used for meiotic mapping.
Crude DNA was obtained from about 106 cells from each progeny. Cells were resuspended in 10 µl 1× Vent Buffer (20 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8) and 1 mg/ml proteinase K (Sigma-Aldrich). The cells were incubated at 58°C for 30 minutes for proteinase K activity and 95°C for 15 minutes to inactivate proteinase K. For PCR, 0.5 µl of crude DNA was used in 20 µl reactions. Each PCR sample contained 1× TAQ buffer (50 mM KCl, 10 mM Tris-HCL, 0.1% Triton X-100, pH 9.0), 1.5 mM MgCl2, 0.4 µM dCAPS primers, 0.4 mM dNTPs, 5% DMSO and 4% TAQ polymerase. The cycling protocol was 95°C for 2 minutes, and 30 cycles of 95°C for 20 seconds, annealing temperature for 20 seconds (Table S1), and 72°C for 1 minute, followed by a final extension time of 5 minutes at 72°C. If digestion of the PCR product was necessary to detect the polymorphism, 10 µL of the PCR product, 1.2 µL of an enzyme's corresponding buffer, 1.2 µL of 10× BSA if necessary, 0.2 µL of the enzyme, and the remaining volume with water to reach a final 15 µL. This was placed at 37°C from 5 minutes to 1 hour depending on the enzyme's efficiency as suggested by the manufacturer (New England Biolabs).
Protein sequence alignment and phylogenetic tree
The protein sequence of PP2A3 was used in BLAST on NCBI and 55 proteins with the expected E-value less than or equal to 1E-100 were collected from green algae (Chlamydomonas reinhardtii, Chlorella variabilis, Micromonas, Ostreococcus lucimarinus, Ostreococcus tauri, and Volvox carteri), yeast (Saccharomyces cerevisiae), land plants (Arabidopsis thaliana and Zea mays), invertebrates (Caenorhabditis elegans and Drosophila melanogaster), and mammals (Mus musculus and Homo sapiens) (Table S2). Sequences alignment was performed with MUSCLE [75] and the output ClustalW (strict) file was subjected to Colorfy [76] for color-coded protein sequence alignment.
From the protein sequence alignment (Figure S1), the low similarity region (∼30 amino acids from the N-terminus) was trimmed from individual protein sequences (Table S2) and multi-sequence alignment was again performed by MUSCLE. The output Phylip interleaved file was subjected to SEQBOOT for 100 bootstraps in the PHYLIP package (3.69; http://evolution.genetics.washington.edu/phylip.html). The 100 sets of samples were subjected to PROTDIST, which uses protein distance matrix to calculate distance. An unrooted tree was generated by a neighbor-joining method implemented by NEIGHBOR for each set of samples. A consensus tree from the 100 unrooted trees was computed by CONSENSE. The output tree was visualized by DRAWGRAM and the bootstrap numbers for each branch were obtained from the outfile file generated by CONSENSE (Figure 3A).
Based on the phylogenetic tree, proteins from the PP2A subfamily were collected and subjected to another round of multi-sequence alignment by MUSCLE and color-coded by Colorfy (Figure S2). Sequence alignment of amino acids close to the C-terminus is shown in Figure 3B.
Plasmid DNA constructs and Chlamydomonas transformation
The full-length PP2A3 genomic DNA, which contains 637 bp upstream of the start codon, the 948 bp coding region, and 424 bp downstream of the stop codon was amplified by primers PP2A-inF2 and PP2A-inR (Table S5) with the Phusion DNA polymerase (New England Biolabs) with the following cycling condition: 98°C for 30 sec, and 30 cycles of 98°C for 10 seconds, 69°C for 10 seconds, and 72°C for 1 minute, followed by a final extension time of 2 minutes at 72°C. The resultant PCR product was gel-purified by UltraClean 15 DNA Purification Kit (MO BIO Laboratories) and cloned into a pBlueScript vector digested with EcoRV using the In-Fusion HD Cloning Kit (Clontech). The resultant PP2A3-pBS plasmid was subjected to Sanger sequencing (GENEWIZ) to ensure the sequence accuracy.
The HA tag was inserted right after the start ATG codon via nested PCR. DNA polymerase and cycling conditions were the same as above with the exception of extension time was reduced to 20 seconds. The primers (PP2A-HA-F, PP2A-3R, Table S5) in the first round of PCR with the PP2A3-pBS plasmid generated above as a template, introduced the HA tag and generated a 361 bp fragment, which was gel purified and used as a template in the second round of PCR. A second forward primer (PP2A-3F-HA), along with the same reverse primer (PP2A-3R), produced a 399 bp fragment. This fragment was then digested with AvrII and AflII, gel purified, and cloned into PP2A3-pBS digested with the same enzymes to generate the HA-PP2A3-pBS plasmid. The plasmid was subject to Sanger sequencing (GENEWIZ) to ensure the sequence accuracy.
The mutations to the C-terminus of the protein were introduced by knitting PCR. DNA polymerase and cycling conditions were the same as above with the exception of extension time was 30 seconds. Using the HA-PP2A3-pBS plasmid as a template, the PP2A3-inf-3F primer was paired with different reverse primers each bearing a mutation (Y313Δ-R; Y315A-R; L315Δ-R; V310T-R; YFLΔ-R, Table S5) to amplify ∼430 bp bands. The PP2A3-inf-3R primer was paired with different forward primers each bearing a mutation (Y313Δ-F; Y315A-F; L315Δ-F; V310T-F; YFLΔ-F, Table S5) to amplify ∼410 bp bands. All bands were gel purified and the corresponding bands (i.e., PP2A3-inf-3F-Y313Δ-R and Y313Δ-F-PP2A3-inf-3R) were used as templates in the second round of PCR, with PP2A3-inf-3F and PP2A3-inf-3R as primers. The resultant ∼830 bp products from the second round of PCR, were gel purified, digested with NcoI and PmlI and cloned into HA-PP2A3-pBS plasmid digested with the same enzymes. All mutant plasmids were subject to Sanger sequencing (GENEWIZ) to ensure they bear the correct mutations.
Chlamydomonas transformation was performed as previously described with modification [76]. One µg of plasmid DNA was used in each transformation into the imp3 cells, using 1 µg of the pSI103 plasmid, which confers resistance to paromomycin [77], for co-transformation. Paromomycin-resistant (paroR) cells were selected on TAP plates with 10 µg/ml paromomycin. In these TAP plates, the concentration of glacial acetic acid was 0.75 ml/L instead of 1 ml/L [35]. The reduction of acetic acid in the medium confers more robust paromomycin resistance. Crude DNA was prepared from paroR colonies and PCR with primers PP2A-HA-short and PP2A-4R (Table S5) was performed to identify HA-positive colonies. The HA-positive colonies were then subjected to immunoblots with anti-HA antibody to confirm the presence of HA-tag in the transformants. The number of paroR colonies, number of HA-positive colonies by PCR and by immunoblot are summarized in Table S3.
Immunoblots and immunofluorescence
The primary antibodies used in this study were anti-HA High Affinity antibody (3F10, Roche, 1∶2000 dilution in immunoblots and 1∶200 dilution in immunofluorescence), anti-α-tubulin antibody (Sigma-Aldrich, T6199, 1∶2000 dilution in immnoblots), and anti-acetylated - α-tubulin antibody (Sigma-Aldrich, T7451, 1∶250 dilution in immunofluorescence). The secondary antibodies used for immunoblots were HRP-conjugated goat anti-mouse antibody (BioRad, 1∶5000), and HRP-conjugated goat anti-rat antibody (Sigma-Aldrich, 1∶5000). Alexa 488-conjugated goat anti-rat antibody (Invitrogen, 1∶500) and Alexa 594-conjugated goat anti-mouse antibody (Invitrogen, 1∶500) were used in immunofluorescence.
Cells used in immunoblots were gametes, which were cultured on R plates for 5 days at 25°C and resuspended in nitrogen-free medium (M-N/5) for 4 hours at room temperature prior to protein extraction. For whole cell extracts, ∼107 cells were resuspended in 30 µl of 20 mM HEPES (pH 7.0) solution with the addition of 1× ProteaseArrest (G-Biosciences) and 10% SDS. The samples were incubated at 37°C for 5 minutes and diluted with 90 µl of 20 mM HEPES (pH 7.0) and 1× ProteaseArrest [78]. For flagellar extracts, ∼5×109 Chlamydomonas cells were resuspended in 4°C HEPES/Sr/DTT buffer (pH 7.1) buffer. Flagella were then detached from cells by pH shock on ice, collected by centrifugation at 4°C [79], and resuspended in 40 µl of 20 mM HEPES (pH 7.0) and 1× ProteaseArrest.
Protein extract from all samples isolated were store at −80°C. The amount of protein in each sample was measured by Bradford assay (Bio-Rad protein assay) before loaded into 10% polyacrylamide gels. The proteins were transferred to PVDF membrane (Millipore) after SDS-PAGE, blocked with 5% nonfat dry milk in PBST (Phosphate Buffered Saline with 0.02% Tween-20) at room temperature for 1 hour, probed with primary antibody in 3% milk in PBST overnight at 4°C, washed with PBST 3 times, 5 minutes each, probed with secondary antibody in 3% nonfat dry milk in PBST 1 hour at room temperature, and washed with PBST 3 times, 5 minutes each. ECL Plus Western Blotting Detection Reagents (GE Healthcare Life Sciences) or SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) was used to expose the signal, which was captured by a FluorChem H2 image (Alpha Innotech). Signal quantification analysis was performed by ImageJ (NIH).
For immunofluorescence, gametes were treated with autolysin for 30 minutes before fixed with 2% paraformaldehyde as previously described [80]. The autolysin treatment was omitted in immunofluorescence of the dikaryons. The images were captured with an UltraVIEW VoX laser scanning disk confocal microscope (PerkinElmer) and acquired by Volocity software (PerkinElmer).
Immunoprecipitation and mass spectrometry
Immunoprecipitation was performed as described by Olson et al. [81] with modifications. About 109 gametes were used and proteins were cross-linked with freshly prepared DSP (Dithiobis-succinimidyl propionate; Pierce) on ice for 30 minutes before the cross-link was stopped by the addition of 100 mM Tris-HCl (pH 7.5). Cells were lysed by sonication and lysates were centrifuged at 20,000 g for 30 minutes at 4°C to remove insoluble materials. Protein concentration from the supernatant was determined by Bradford assay. One microgram of anti-HA high affinity antibody (3G10) was inoculated with 20 µl of protein G magnetic beads (Invitrogen) for 30 minutes at room temperature and cross-linked with 5 mM BS3 (Bis[sulfosuccinimidyl] suberate; Pierce) in conjugation buffer (20 mM sodium phosphate, pH 7.0, 0.15 M NaCl) for 30 minutes at room temperature. The cross-link reaction was stopped by the addition of 50 mM Tris-HCl (pH 7.5) and incubated at room temperature for 15 minutes. The anti-HA-antibody-conjugated protein G magnetic beads were then incubated with cell lysates that contain ∼12 mg of proteins in each sample overnight at 4°C. The beads were washed and the bound proteins were eluted with 1× SDS PAGE loading buffer with the addition of 100 mM DTT at 90°C for 10 minutes. About 1/10 of the elution was used in SDS-PAGE and immunoblots with anti-HA antibody and the rest was separated by a 10% polyacrylamide gels and stained by either silver [82] or 0.1% Coomassie Brilliant blue R-250. The ∼65 kD band stained by Coomassie blue was cut out and sent to the Taplin Mass Spectrometry Facility (Harvard University) for mass spectrometry.
Real-time RT-PCR
Chlamydomonas total RNA extraction and real-time RT-PCR was performed as previously described [76]. Primers used in these reactions were PP2A3-HA-short and PP2A3-4R for HA-PP2A3 transcripts and PP2A3-7F and PP2A3-3R for endogenous+HA-PP2A3 transcripts (Table S5).
Supporting Information
Zdroje
1. Lawson NathanD, Wolfe ScotA (2011) Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Developmental cell 21 : 48–64.
2. KileBT, HiltonDJ (2005) The art and design of genetic screens: mouse. Nature Reviews Genetics 6 : 557–567.
3. PazourGJ, DickertBL, VucicaY, SeeleyES, RosenbaumJL, et al. (2000) Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. The Journal of Cell Biology 151 : 709–718.
4. DutcherSK, TrabucoEC (1998) The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes δ-Tubulin, a new member of the tubulin superfamily. Molecular Biology of the Cell 9 : 1293–1308.
5. TaxFE, VernonDM (2001) T-DNA-associated duplication/translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiology 126 : 1527–1538.
6. AmsterdamA, BurgessS, GollingG, ChenW, SunZ, et al. (1999) A large-scale insertional mutagenesis screen in zebrafish. Genes & Development 13 : 2713–2724.
7. AlonsoJM, StepanovaAN, LeisseTJ, KimCJ, ChenH, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 : 653–657.
8. GollingG, AmsterdamA, SunZ, AntonelliM, MaldonadoE, et al. (2002) Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nature Genetics 31 : 135–140.
9. EsparzaJM, O'TooleE, LiL, GiddingsTHJr, KozakB, et al. (2013) Katanin localization requires triplet microtubules in Chlamydomonas reinhardtii. PLoS ONE 8: e53940.
10. JanderG, NorrisSR, RounsleySD, BushDF, LevinIM, et al. (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiology 129 : 440–450.
11. MutoA, OrgerMB, WehmanAM, SmearMC, KayJN, et al. (2005) Forward genetic analysis of visual behavior in zebrafish. PLoS Genetics 1: e66.
12. WansleebenC, van GurpL, FeitsmaH, KroonC, RieterE, et al. (2011) An ENU-mutagenesis screen in the mouse: identification of novel developmental gene functions. PLoS ONE 6: e19357.
13. DutcherSK, MorrissetteNS, PrebleAM, RackleyC, StangaJ (2002) ε-Tubulin is an essential component of the centriole. Molecular Biology of the Cell 13 : 3859–3869.
14. IominiC, LiL, EsparzaJM, DutcherSK (2009) Retrograde intraflagellar transport mutants identify Complex A proteins With multiple genetic interactions in Chlamydomonas reinhardtii. Genetics 183 : 885–896.
15. GerholdAR, RichterDJ, YuAS, HariharanIK (2011) Identification and characterization of genes required for compensatory growth in Drosophila. Genetics 189 : 1309–1326.
16. DoitsidouM, PooleRJ, SarinS, BigelowH, HobertO (2010) C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE 5: e15435.
17. MinevichG, ParkDS, BlankenbergD, PooleRJ, HobertO (2012) CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192 : 1249–1269.
18. DutcherSK, LiL, LinH, MeyerL, GiddingsTH, et al. (2012) Whole-genome sequencing to identify mutants and polymorphisms in Chlamydomonas reinhardtii. G3: Genes|Genomes|Genetics 2 : 15–22.
19. OssowskiS, SchneebergerK, ClarkRM, LanzC, WarthmannN, et al. (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Research 18 : 2024–2033.
20. SilflowCD, LefebvrePA (2001) Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiology 127 : 1500–1507.
21. GoodenoughUW, HwangC, MartinH (1976) Isolation and genetic analysis of mutant strains of Chlamydomonas reinhardi defective in gametic differentiation. Genetics 82 : 169–186.
22. HwangCJ, MonkBC, GoodenoughUW (1981) Linkage of mutations affecting minus flagellar membrane agglutinability to the mt − mating-type locus of Chlamydomonas. Genetics 99 : 41–47.
23. GoodenoughU, LinH, LeeJ-H (2007) Sex determination in Chlamydomonas. Semin Cell Dev Biol 18 : 350–361.
24. FerrisPJ, WaffenschmidtS, UmenJG, LinH, LeeJ-H, et al. (2005) Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell 17 : 597–615.
25. VallonO, WollmanFA (1995) Mutations affecting O-glycosylation in Chlamydomonas reinhardtii cause delayed cell wall degradation and sex-limited sterility. Plant Physiology 108 : 703–712.
26. FerrisPJ, WoessnerJP, GoodenoughUW (1996) A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Molecular Biology of the Cell 7 : 1235–1248.
27. MisamoreMJ, GuptaS, SnellWJ (2003) The Chlamydomonas Fus1 protein is present on the mating type plus fusion organelle and required for a critical membrane adhesion event during fusion with minus gametes. Molecular Biology of the Cell 14 : 2530–2542.
28. FerrisPJ, GoodenoughUW (1997) Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146 : 859–869.
29. LinH, GoodenoughUW (2007) Gametogenesis in the Chlamydomonas reinhardtii minus mating type is controlled by two genes, MID and MTD1. Genetics 176 : 913–925.
30. SaitoT, SmallL, GoodenoughU (1993) Activation of adenylyl cyclase in Chlamydomonas reinhardtii by adhesion and by heat. The Journal of Cell Biology 122 : 137–147.
31. JanssensV, GorisJ (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353 : 417–439.
32. ShiY (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139 : 468–484.
33. JanssensV, LonginS, GorisJ (2008) PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends in Biochemical Sciences 33 : 113–121.
34. ElamCA, WirschellM, YamamotoR, FoxLA, YorkK, et al. (2011) An axonemal PP2A B-subunit is required for PP2A localization and flagellar motility. Cytoskeleton 68 : 363–372.
35. Harris E, Stern D, Witman G (2009) The Chlamydomonas sourcebook: introduction to Chlamydomonas and its laboratory use: Academic Press.
36. MerchantSS, ProchnikSE, VallonO, HarrisEH, KarpowiczSJ, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 : 245–250.
37. FerrisPJ, ArmbrustEV, GoodenoughUW (2002) Genetic Structure of the Mating-Type Locus of Chlamydomonas reinhardtii. Genetics 160 : 181–200.
38. HuangB, RifkinMR, LuckDJ (1977) Temperature-sensitive mutations affecting flagellar assembly and function in Chlamydomonas reinhardtii. The Journal of Cell Biology 72 : 67–85.
39. PröscholdT, HarrisEH, ColemanAW (2005) Portrait of a Species: Chlamydomonas reinhardtii. Genetics 170 : 1601–1610.
40. CohenP, KlumppS, SchellingDL (1989) An improved procedure for identifying and quantitating protein phosphatases in mammalian tissues. FEBS Letters 250 : 596–600.
41. ChenJ, MartinB, BrautiganD (1992) Regulation of protein serine-threonine phosphatase type-2A by tyrosine phosphorylation. Science 257 : 1261–1264.
42. LiM, DamuniZ (1994) Okadaic acid and microcystin-LR directly inhibit the methylation of protein phosphatase 2A by its specific methyltransferase. Biochemical and Biophysical Research Communications 202 : 1023–1030.
43. FloerM, StockJ (1994) Carboxyl methylation of protein phosphatase 2A from Xenopus eggs is stimulated by cAMP and inhibited by okadaic acid. Biochemical and Biophysical Research Communications 198 : 372–379.
44. TakaiA, SasakiK, NagaiH, MieskesG, IsobeM, et al. (1995) Inhibition of specific binding of okadaic acid to protein phosphatase 2A by microcystin-LR, calyculin-A and tautomycin: method of analysis of interactions of tight-binding ligands with target protein. Biochemical Journal 306 : 657.
45. LiY, WangX, YueP, TaoH, RamalingamSS, et al. (2013) Protein phosphatase 2A and DNA-dependent protein kinase are involved in mediating rapamycin-induced Akt phosphorylation. Journal of Biological Chemistry 288 : 13215–13224.
46. FavreB, TurowskiP, HemmingsBA (1997) Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. Journal of Biological Chemistry 272 : 13856–13863.
47. NamboodiripadAN, JenningsML (1996) Permeability characteristics of erythrocyte membrane to okadaic acid and calyculin A. American Journal of Physiology - Cell Physiology 270: C449–C456.
48. WagnerV, GeßnerG, HeilandI, KaminskiM, HawatS, et al. (2006) Analysis of the phosphoproteome of Chlamydomonas reinhardtii provides new insights into various cellular pathways. Eukaryotic Cell 5 : 457–468.
49. PazourGJ, AgrinN, LeszykJ, WitmanGB (2005) Proteomic analysis of a eukaryotic cilium. The Journal of Cell Biology 170 : 103–113.
50. WangQ, PanJ, SnellWJ (2006) Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell 125 : 549–562.
51. BlumenstielJP, NollAC, GriffithsJA, PereraAG, WaltonKN, et al. (2009) Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182 : 25–32.
52. BamshadMJ, NgSB, BighamAW, TaborHK, EmondMJ, et al. (2011) Exome sequencing as a tool for Mendelian disease gene discovery. Nature Reviews Genetics 12 : 745–755.
53. Genomes Project Consortium (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491 : 56–65.
54. CaoJ, SchneebergerK, OssowskiS, GuntherT, BenderS, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nature Genetics 43 : 956–963.
55. HuangB, PipernoG, RamanisZ, LuckDJ (1981) Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. The Journal of Cell Biology 88 : 80–88.
56. AlbeeAJ, KwanAL, LinH, GranasD, StormoGD, et al. (2013) Identification of cilia genes that affect cell cycle progression using whole genome transcriptome analysis in Chlamydomonas reinhardtti. G3: Genes|Genomes|Genetics 979–91 doi: 10.1534/g3.113.006338
57. OrgadS, BrewisND, AlpheyL, AxtonJM, DudaiY, et al. (1990) The structure of protein phosphatase 2A is as highly conserved as that of protein phosphatase I. FEBS Letters 275 : 44–48.
58. KitagawaD, FlückigerI, PolanowskaJ, KellerD, ReboulJ, et al. (2011) PP2A phosphatase acts upon SAS-5 to ensure centriole formation in C. elegans embryos. Developmental cell 20 : 550–562.
59. BallesterosI, DomínguezT, SauerM, ParedesP, DupratA, et al. (2013) Specialized functions of the PP2A subfamily II catalytic subunits PP2A-C3 and PP2A-C4 in the distribution of auxin fluxes and development in Arabidopsis. The Plant Journal 73 : 862–872.
60. PernasM, García-CasadoG, RojoE, SolanoR, Sánchez-SerranoJJ (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling1. The Plant Journal 51 : 763–778.
61. TangW, YuanM, WangR, YangY, WangC, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nature Cell Biology 13 : 124–131.
62. XuY, XingY, ChenY, ChaoY, LinZ, et al. (2006) Structure of the protein phosphatase 2A holoenzyme. Cell 127 : 1239–1251.
63. LonginS, ZwaenepoelK, LouisJV, DilworthS, GorisJ, et al. (2007) Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic subunit. Journal of Biological Chemistry 282 : 26971–26980.
64. Nunbhakdi-CraigV, SchuechnerS, SontagJ-M, MontgomeryL, PallasDC, et al. (2007) Expression of protein phosphatase 2A mutants and silencing of the regulatory Bα subunit induce a selective loss of acetylated and detyrosinated microtubules. Journal of Neurochemistry 101 : 959–971.
65. WeiH, AshbyDG, MorenoCS, OgrisE, YeongFM, et al. (2001) Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. Journal of Biological Chemistry 276 : 1570–1577.
66. GentryMS, LiY, WeiH, SyedFF, PatelSH, et al. (2005) A novel assay for protein phosphatase 2A (PP2A) complexes in vivo reveals differential effects of covalent modifications on different Saccharomyces cerevisiae PP2A heterotrimers. Eukaryotic Cell 4 : 1029–1040.
67. SchlaitzA-L, SraykoM, DammermannA, QuintinS, WielschN, et al. (2007) The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell 128 : 115–127.
68. SontagE, Nunbhakdi-CraigV, BloomGS, MumbyMC (1995) A novel pool of protein phosphatase 2A is associated with microtubules and is regulated during the cell cycle. The Journal of Cell Biology 128 : 1131–1144.
69. FleggCP, SharmaM, Medina-PalazonC, JamiesonC, GaleaM, et al. (2010) Nuclear export and centrosome targeting of the protein phosphatase 2A subunit B56α: role of B56α in nuclear export of the catalytic subunit. Journal of Biological Chemistry 285 : 18144–18154.
70. DutcherSK (2003) Elucidation of basal body and centriole functions in Chlamydomonas reinhardtii. Traffic 4 : 443–451.
71. HuangK, DienerDR, MitchellA, PazourGJ, WitmanGB, et al. (2007) Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. The Journal of Cell Biology 179 : 501–514.
72. BelzileO, Hernandez-Lara CarmenI, WangQ, Snell WilliamJ (2013) Regulated membrane protein entry into flagella is facilitated by cytoplasmic microtubules and does not require IFT. Current Biology 23 : 1460–1465.
73. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25 : 2078–2079.
74. CingolaniP, PlattsA, WangLL, CoonM, NguyenT, et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome ofDrosophila melanogaster strain w1118; iso-2; iso-3. Fly 6 : 80–92.
75. EdgarR (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 : 1792–1797.
76. LinH, KwanAL, DutcherSK (2010) Synthesizing and salvaging NAD+: lessons learned from Chlamydomonas reinhardtii. PLoS Genetics 6: e1001105.
77. SizovaI, FuhrmannM, HegemannP (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277 : 221–229.
78. PiaseckiBP, LaVoieM, TamL-W, LefebvrePA, SilflowCD (2008) The Uni2 phosphoprotein is a cell cycle–regulated component of the basal body maturation pathway in Chlamydomonas reinhardtii. Molecular Biology of the Cell 19 : 262–273.
79. DutcherSK, HuangB, LuckDJ (1984) Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. The Journal of Cell Biology 98 : 229–236.
80. LeeJ-H, LinH, JooS, GoodenoughU (2008) Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133 : 829–840.
81. OlsonBJSC, OberholzerM, LiY, ZonesJM, KohliHS, et al. (2010) Regulation of the Chlamydomonas cell cycle by a stable, chromatin-associated retinoblastoma tumor suppressor complex. The Plant Cell Online 22 : 3331–3347.
82. MortzE, KroghTN, VorumH, GörgA (2001) Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1 : 1359–1363.
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



