-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaChromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
Enhancers and promoters often contain multiple binding sites for the same transcription factor, suggesting that homotypic clustering of binding sites may serve a role in transcription regulation. Here we show that clustering of binding sites for the transcription termination factor TTF-I downstream of the pre-rRNA coding region specifies transcription termination, increases the efficiency of transcription initiation and affects the three-dimensional structure of rRNA genes. On chromatin templates, but not on free rDNA, clustered binding sites promote cooperative binding of TTF-I, loading TTF-I to the downstream terminators before it binds to the rDNA promoter. Interaction of TTF-I with target sites upstream and downstream of the rDNA transcription unit connects these distal DNA elements by forming a chromatin loop between the rDNA promoter and the terminators. The results imply that clustered binding sites increase the binding affinity of transcription factors in chromatin, thus influencing the timing and strength of DNA-dependent processes.
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003786
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003786Summary
Enhancers and promoters often contain multiple binding sites for the same transcription factor, suggesting that homotypic clustering of binding sites may serve a role in transcription regulation. Here we show that clustering of binding sites for the transcription termination factor TTF-I downstream of the pre-rRNA coding region specifies transcription termination, increases the efficiency of transcription initiation and affects the three-dimensional structure of rRNA genes. On chromatin templates, but not on free rDNA, clustered binding sites promote cooperative binding of TTF-I, loading TTF-I to the downstream terminators before it binds to the rDNA promoter. Interaction of TTF-I with target sites upstream and downstream of the rDNA transcription unit connects these distal DNA elements by forming a chromatin loop between the rDNA promoter and the terminators. The results imply that clustered binding sites increase the binding affinity of transcription factors in chromatin, thus influencing the timing and strength of DNA-dependent processes.
Introduction
An intriguing question for understanding protein-DNA recognition is how low-abundant transcription factors recognize their target sites in genomic DNA [1], [2]. Empirical studies revealed that regulatory regions, such as enhancers and promoters, comprise modular units of a few hundred base pairs that harbour multiple binding sites for the same transcription factor. Such ‘homotypic clustering sites’ (HTCs) have been identified in 2% of the human genome, being enriched at promoters and enhancers [3]. HTCs have been shown to play a role in Drosophila development, regulating early patterning genes [4]–[6]. Genome-wide binding analyses in yeast have demonstrated that the occupancy of transcription factors is higher at clustered binding sites compared to single ones [7]. Studies in mammalian cells have shown that clustering of binding sites facilitate the cooperative binding of nuclear receptors to their target sites in vivo, suggesting that HCTs coordinate the recruitment of transcription initiation factors [8]–[10]. Alternatively, cooperative binding could arise through indirect effects, e.g. by changing the accessibility of neighbouring binding sites in chromatin [11].
To assess the functional relevance of homotypic clustering of transcription factor binding sites, we studied the 3′-terminal region of murine rRNA genes (rDNA), which contains ten binding sites (T1–T10) for the transcription termination factor TTF-I. Binding of TTF-I to the terminator elements is required to stop elongating RNA polymerase I (Pol I) and termination of pre-rRNA synthesis occurring predominantly at the first terminator T1 [12]–[15]. In addition to the downstream terminators, there is a single TTF-I binding site, termed T0, located 170 bp upstream of the transcription start site [16]. Binding of TTF-I to this site is required for efficient transcription initiation and for the recruitment of chromatin remodelling complexes that establish distinct epigenetic states of rRNA genes. The interaction of TTF-I with CSB (Cockayne Syndrome protein B), NoRC (Nucleolar Remodeling Complex), or NuRD (Nucleosome Remodeling and Deacetylation complex), respectively, has been shown to recruit histone modifying enzymes which lead to the establishment of a specific epigenetic signature that characterizes active, silent or poised rRNA genes [17]–[20].
TTF-I has been shown to oligomerize in vitro and to link two DNA fragments in trans [21]. These characteristics enable TTF-I bound to the upstream binding site T0 and the downstream terminators T1–T10 to loop out of the pre-rRNA coding region [22], [23]. Formation of a chromatin loop facilitates re-initiation and increases transcription initiation rates at the rRNA gene [22], [24]. TTF-I is a multifunctional protein that is not only essential for transcription termination, but also directs efficient rDNA transcription, mediates replication fork arrest [25], establishes specific epigenetic features and determines the topology of rDNA. The conservation of multiple TTF-I binding sites downstream of the pre-rRNA coding region raises the question whether homotypic clustering of terminator elements is functionally relevant. Here we demonstrate that HTCs serve a chromatin-specific function. Packaging into chromatin increases the binding affinity of TTF-I to clustered terminator elements, augments the efficiency of transcription termination, enhances transcription initiation, and changes the higher-order structure of rRNA genes. The homotypic clusters at the rRNA gene coordinate the timing of molecular events, coordinating transcription termination and initiation and the occurrence of higher-order chromatin domains, suggesting an important chromatin-dependent role for clustered binding sites in the genome.
Results
Multiple termination sites enhance the efficiency of transcription in vitro
The rDNA terminators in human and mouse exhibit an overall similar structure, containing 10 to 11 TTF-I binding sites in close proximity (Fig. S1A). We focused on the murine rDNA terminator, which comprises 10 termination sites (T1–T10) spaced by 18–123 bp, preventing the accommodation of nucleosomes in between the TTF-I binding sites. The consensus sequence of these TTF-I binding sites share almost perfect sequence identity within the core motif GGTCGACCAG, while the surrounding nucleotides vary slightly (Fig. 1A). In electrophoretic mobility shift assays (EMSAs) recombinant TTF-I bound with comparable affinity to all terminators assayed (data not shown). The DNA binding affinity of TTF-I was quantified by microscale thermophoresis, recording changes of nucleoprotein complex mobility in a small temperature gradient [26]. By titrating a wide range of TTF-I∶DNA ratios, the binding constant of TTF-I to free Sal-box DNA was determined to be 0.5 µM (Fig. 1B), a relatively low DNA binding affinity which is one order of magnitude lower than the KD of other transcription factors [27]–[29].
Fig. 1. Chromatin-specific termination at the homotypic cluster of TTF-I. 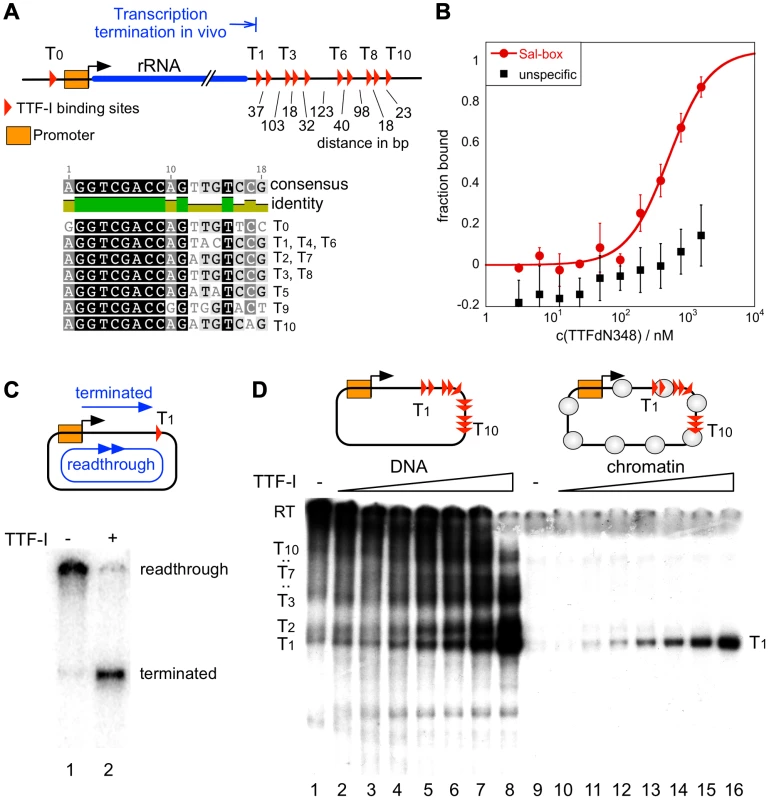
(A) Overview of the murine rRNA gene and the location of the TTF-I binding sites. A homotypic cluster of TTF-I sites is located in the terminator region. The distances between TTF-I binding sites, their orientation and the gene promoter are indicated. A comparison of the TTF-I binding sites T0 and the termination sites T1 to T10 is depicted. (B) Increasing amounts of TTF-IΔN348 were incubated with 50 nM of either a fluorescently labelled 30-mer oligonucleotide containing a Sal-box motif (T2) or a control oligonucleotide of the same length. Protein-DNA interactions are quantified by microscale thermophoresis. Curve fitting with a Hill coefficient of 1 resulted in a KD of 500 nM+/−120 nM for the T2 sequence. (C) Transcription reaction using the circular rDNA minigene plasmid pMr-SB containing a single termination site, a partially purified nuclear extract lacking most of the nuclear TTF-I (DEAE280), performed in the presence or absence of recombinant TTF-I. The positions of the long read-through and the terminated transcripts are indicated. (D) Transcription on free DNA and chromatin, using the pMrWT-T DNA containing the promoter with the TTF-I binding site T0 and the full terminator with the 10 termination sites. DNA (lanes 1–8) and chromatin (lanes 9–16) were incubated with increasing concentrations of TTF-I as indicated and the DEAE280 extract. The position of the long, non-terminated read-through transcript (RT) and the terminated transcripts are indicated. In vitro transcription assays on a circular minigene comprising the rDNA promoter fused to a single termination site (pMrSB) yielded long read-through transcripts in the absence of TTF-I. The addition of recombinant TTF-I led to the synthesis of terminated transcripts whose lengths correspond to the distance from the transcription start site to the terminator T1 (Fig. 1C). If the template contained all ten terminators (pMrT1-T10), both read-through transcripts and a heterogeneous population of transcripts randomly terminated at any of the TTF-I binding sites were synthesized due to sub-saturating TTF-I levels in the extract (Fig. 1D). In the presence of increasing concentrations of recombinant TTF-I the amount of transcripts stopped at terminator T1 progressively increased (Fig. 1D, lanes 1–8 and Fig. S2). Thus, TTF-I binds to all sites with similar affinity and randomly terminates transcription until at saturating concentrations TTF-I occupies all ten terminators.
A strikingly different result was obtained on rDNA templates assembled into chromatin with an extract from Drosophila embryos [30] (Figure S1B). Consistent with Pol I transcription on chromatin requiring binding of TTF-I to the promoter-proximal terminator T0 and ATP-dependent chromatin remodelling [31], [32], transcription was repressed in the absence of TTF-I (Fig. 1D, lane 9). The addition of TTF-I relieved transcriptional repression, yielding only a single RNA species of 686 nt. On chromatin templates, already lowest TTF-I concentrations terminated transcription specifically at T1 (Fig. 1D, lanes 10–16 and Fig. S2). The result suggests that transcription in chromatin is only initiated when the termination sites are set, meaning that the TTF-I binding site at the promoter is only bound after sequestering TTF-I at the terminator. The qualitative difference between transcription on free DNA and chromatin templates indicates that on chromatin templates TTF-I either binds preferentially to T1 or the overall binding affinity of TTF-I to all terminator sites is increased in chromatin.
Clustered termination sites facilitate cooperative binding of TTF-I to chromatin
Next, we performed electrophoretic mobility shift assays (EMSAs) and DNase I footprinting experiments to compare TTF-I binding to free DNA and chromatin. Consistent with the transcription data on free DNA, EMSAs on terminator DNA fragments containing more than one TTF-I binding sites yielded heterogeneous nucleoprotein complexes, reflecting binding to each binding site with similar affinity (Fig. 2A). On chromatin templates, DNase I footprinting experiments demonstrate that TTF-I simultaneously bound to all terminator binding sites (Fig. 2B). Together with the transcription results on chromatin templates, this suggests that homotypic clustering of target sites increases the binding affinity of TTF-I to chromatin.
Fig. 2. Multiple termination sites enable cooperative binding of TTF-I to chromatin. 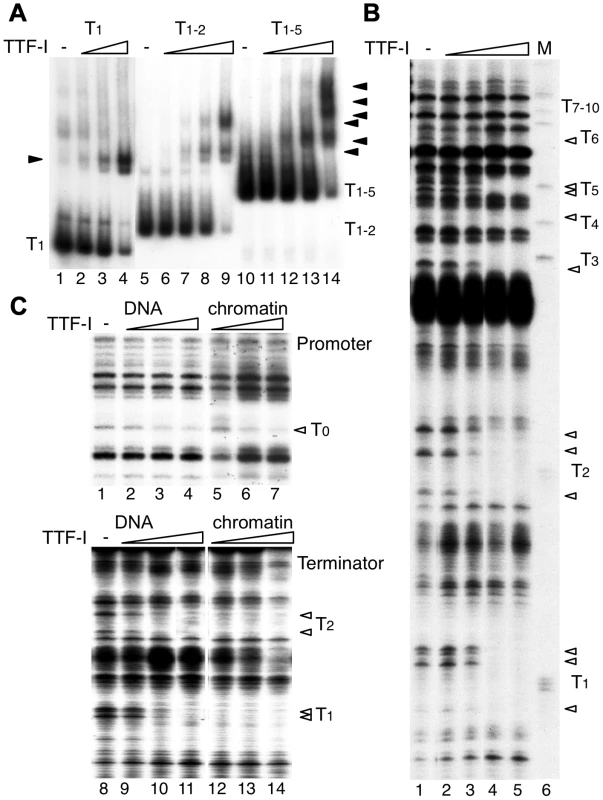
(A) Electrophoretic mobility shift assays (EMSA) were performed with a single TTF-I binding site (T1, lanes 1–4), two binding sites (T1–2, lanes 5–9) and an array of five binding sites (T1–5, lanes 10–14) and increasing concentrations of TTF-I as indicated. Nucleoprotein complexes are resolved on native polyacrylamide gels and detected by autoradiography. The positions of the free DNA molecules and the TTF-I-DNA complexes (triangles) are indicated. (B) Monitoring TTF-I binding to the chromatinized terminator by DNase I footprinting. The pMr-T plasmid containing the full terminator was reconstituted into chromatin with Drosophila embryo extract. Chromatin was incubated with increasing concentrations of TTF-I as indicated and partially digested with DNase I. Footprints were analysed by a primer extension reaction using a radioactively labelled oligonucleotide and resolving the DNA on 6% sequencing gels. The marker was generated by partial digestion of the plasmid with the restriction enzyme SalI and analysed by the same primer extension reaction. The SalI sites (T1 to T10) represent the TTF-I binding sites and the triangles indicate sites of DNase I protection. (C) Comparative footprinting of TTF-I binding to the promoter and terminator of free DNA and chromatin. Identical amounts of pMrWT-T were used as free DNA (lanes 1 to 4 and 8 to 11) or chromatin (lanes 5 to 7 and 12 to 14) and incubated with increasing amounts of TTF-I as indicated. DNA was partially digested with DNase I and the purified DNA was analysed by primer extension reactions, either using a radiolabelled oligonucleotide binding close to the promoter (lanes 1 to 7) or binding close to T1 in the terminator region (lanes 8 to 14). DNA was separated on 8% sequencing gels, dried and analysed after autoradiography. The TTF-I binding sites T1, T2 and T0 and the protected DNase I cutting sites (triangles) are indicated. To compare the binding affinity of TTF-I to free DNA and reconstituted chromatin, we performed DNase I footprinting assays, monitoring DNase I cleavage sites by primer extension which allows simultaneous analysis of TTF-I occupancy at the promoter and terminator(s) (Fig. 2C). TTF-I binding can be observed by the disappearance of a DNase I sensitive site that is apparent within the TTF-I binding sites of free DNA and reconstituted chromatin (Fig. 2B and C). In agreement with the binding studies and the in vitro transcription experiments, TTF-I binds on free DNA to the promoter-proximal terminator T0 and the downstream terminators with similar affinity (Fig. 2C, compare lanes 2–4 and lanes 9–11). On chromatin templates, TTF-I binding to the upstream site T0 is comparable to its binding to free DNA (Fig. 2C, upper panel). However, on chromatin templates TTF-I binds with higher affinity to the clustered sites, fully occupying all terminator sites at low protein concentrations (Fig. 2C, lower panel). Significantly, TTF-I occupied the binding sites at the terminators prior to the promoter-proximal site (compare lanes 5–7 and 12–14), showing a specific role of chromatin and binding site clustering for increasing the binding affinity of TTF-I. The sequential binding of TTF-I, first to the terminators and then to the gene promoter in chromatin was also confirmed using a different method. Affinity purification of either TTF-I bound free DNA or chromatin revealed binding of TTF-I to the gene terminators reconstituted into chromatin already at concentrations one order of magnitude lower than with the gene promoter (Fig. S3). Like in the footprinting assay, this effect was not detectable using free DNA, where both TTF-I binding regions were occupied with similar affinity. Apparently, the clustered arrangement of binding sites increases the affinity of TTF-I, thus promoting the association of TTF-I with the downstream terminators T1–10 prior to the upstream site T0, a process that appears to be essential for both TTF-I dependent transcription activation and transcription termination.
Clustered terminators act as a transcriptional enhancer in vivo
To study the functional relevance of clustered sites in vivo, we transfected CHO cells with reporter plasmids containing the murine Pol I promoter, an internal ribosomal entry site (IRES), Firefly luciferase cDNA and either no terminator (pTΔ) or one (pT1), two (pT2) or ten (pT10) termination sites. As shown in Figure 3A, the presence of one or two terminators (pT1 and pT2) enhanced transcription of the luciferase reporter 8 - to 12-fold compared to the terminator-deficient vector. The presence of ten termination sites (pT10) decreased luciferase activity, presumably due to squelching of endogenous TTF-I. In support of this view, transient overexpression of TTF-I (pTTFΔN348) revealed a linear correlation between the number of terminators and reporter gene activity, showing further transcriptional enhancement by the pT10 construct (Fig. 3B and Fig. S4B). Additional controls revealed that the stimulatory effect depends on the TTF-I binding sites at the promoter and the terminator (Fig. S4A) and co-transfection of pTTFΔN470 revealed that the chromatin-binding domain of TTF-I is required. pTTFΔN470 represents a deletion mutant that is capable of binding to its target sites on free DNA but not on chromatin [31]. Therefore, pTTFΔN470 cannot activate transcription in a chromatin context [31] and transfection of this construct did not further activate transcription of the pT10 construct (Fig. 3C and Fig. S4B). The control shows that chromatin-specific activities of TTF-I are required for efficient transcriptional activation. Notably, there was no luciferase expression using reporters with TTF-I binding site(s) in the reverse orientation (pT1R, pT10R), supporting the importance of the topological arrangement of the HCTs for efficient Pol I transcription.
Fig. 3. Multiple termination sites enhance transcription in vivo. 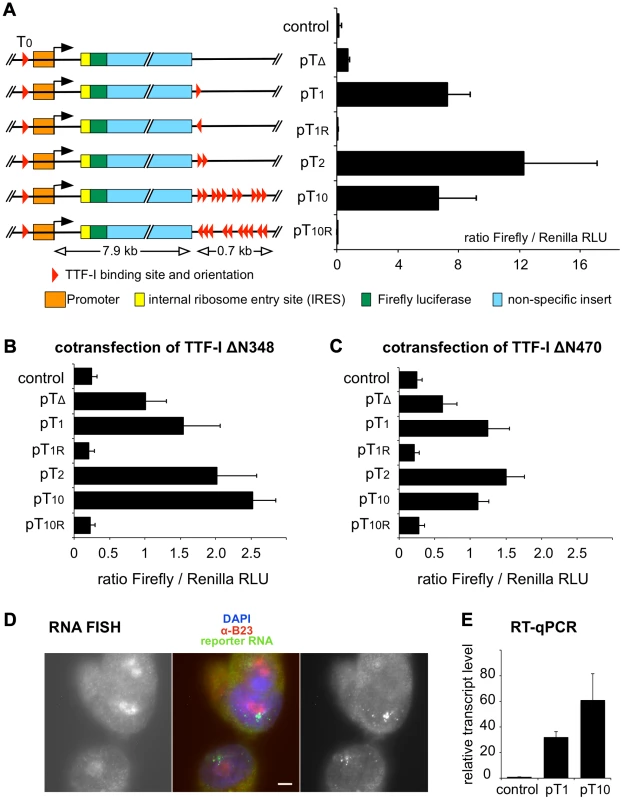
(A) Reporter plasmids containing the rDNA promoter, Firefly luciferase and either no (pTΔ), one (pT1), two (pT2), ten (pT10) termination sites and T1 and T1–10 in reverse orientation (pT1r and pT10r) were co-transfected with a Renilla luciferase encoding plasmid (pRL-TK) into CHO cells. As a control, empty pBluescript vector was co-transfected. Transcriptional activities were analysed using a dual luciferase reporter assay. The ratio of Firefly/Renilla relative light units (RLU) of three independent experiments is given. Error bars indicate standard deviations. The functional elements and the sizes of the reporter plasmids are depicted. (B) Reporter plasmids were co-transfected with a GFP-TTFΔN348 expression vector and analysed as described in (A). (C) Reporter plasmids were co-transfected with a GFP-TTFΔN470 expression vector and analysed as described in (A). (D) RNA FISH using CHO cell lines with stably integrated rDNA minigenes. CHO-pT10 cells containing an rDNA minigene with a full terminator, were stained with DAPI (in blue in the middle panel), with α-B23 antibody staining the nucleoli (left panel; shown in red in the middle panel), and integrated reporter gene transcripts were visualized by FISH (right panel; shown in green in the middle panel). Bar: 5 µm. (E) Transcription levels of genomically inserted pT1 and pT10 constructs were assayed using RT-qPCR. Comparative quantitation was performed and RNA levels of the Firefly luciferase sequence were normalized to β-actin expression. Relative transcript levels of three independent experiments are given in relation to non-transfected CHO Flp-In cells (control), error bars denote standard deviations. To examine whether the number of terminators affects gene activity and/or the spatial organization of rDNA in a genomic context, we generated stable cell lines that harbour a single copy of mouse rRNA minigenes, either containing only T1 (CHO-pT1) or all ten terminators (CHO-pT10) (Fig. S5). Using the Flip-In system we generated comparable rRNA minigene lines, integrated at the same genomic site of CHO cells. This strategy allows us to rule out effects of inefficient chromatin packaging and minigene dosage in transfection experiments. The nuclear localisation of the ectopic rDNA was not affected, as 3D immuno-FISH experiments revealed that comparable number of rDNA was associated with the nucleoli in the stable cell lines (33 of 104 alleles were associated with nucleoli in CHO-pT1 and 50 of 160 alleles in CHO-pT10 cells; Fig. S5A, B). RNA FISH experiments confirmed that all cell lines were transcriptionally active (Fig. 3D). Expression analysis of the rRNA minigene by qRT-PCR and reporter assays revealed that both the level of the ectopic pre-rRNA and the Pol I-driven luciferase activity were increased in CHO-pT10 compared to CHO-pT1 cells (Fig. 3E and Fig. S5C), reinforcing the activating role of clustered termination sites in rDNA transcription.
HTC is required for gene looping and efficient loading of Pol I specific factors
To decipher the molecular mechanism underlying HTC-driven transcriptional activation, we compared transcription factor occupancy within the stable cell lines, containing single rDNA minigenes with either one (CHO-pT1) or ten terminators (CHO-pT10, Fig. 4A). As shown in Figure 4B, binding of Pol I and UBF was enhanced at the promoter, the transcribed region and the terminators of CHO-pT10 compared to CHO-pT1 cells. In addition, we observed increased binding of TBP to the promoter of CHO-pT10 cells, demonstrating that augmented rDNA transcription is a direct consequence of enhanced transcription initiation and polymerase occupancy. Pol I enrichment downstream of the terminator region was reduced in CHO-pT10 cells, consistent with clustered TTF-I binding sites promoting efficient termination. Similar results were obtained with the transient transfection of the constructs (Fig. S6).
Fig. 4. Clustered termination sites enhance transcription and are required for chromatin looping. 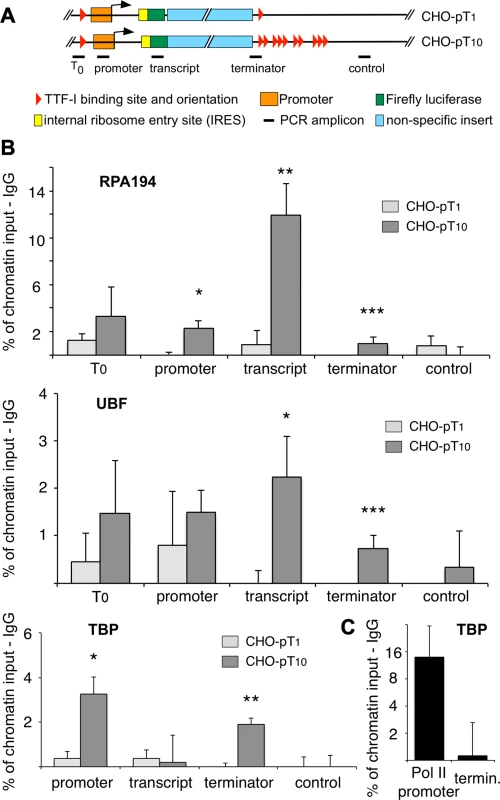
(A) Overview to the stably integrated rDNA minigenes and the locations of the PCR amplicons. (B) Chromatin-immunoprecipitation (ChIP) assays on stably integrated rDNA reporter genes using the indicated antibodies. Occupancies were measured by qPCR, calculated as percentage of input chromatin and background signals as determined from control IPs with unspecific antibodies (α-HA or α-IgG) were subtracted. At least three independent biological replicates were performed. Error bars indicate the standard error of the mean. For statistical analysis, a two-sided, homoscedatic student's t-test was performed, stars denote significances. * p<0.05, ** p<0.01, *** p< = 0.001. (C) ChIP experiment using an rDNA reporter in which the Pol I spacer promoter, core promoter and enhancer regions of a pT10 reporter construct were replaced by a Pol II promoter containing a canonical TBP binding site. The experiment was performed as described in (B). Active rRNA genes are known to form chromatin loops, connecting the promoter with the terminator to promote recycling of Pol I [22], [23], [33]. To examine whether multiple terminators facilitate loop formation, we determined the occupancy of TBP at the terminator in the stable cell lines CHO-pT1 and CHO-pT10 (Fig. 4B lower panel and 4C). The close proximity of a protein to DNA results in crosslinking and co-purification of the DNA, even though the factor does not directly contact the DNA at this site. Such binding events indicate the close spatial proximity of distant DNA sites, comparable to 3C assays [34]. Obviously, TBP was found to be associated with the promoter of CHO-pT1 and CHO-pT10 as part of the initiation complex, while no binding was observed in the transcribed region (Fig. 4B, TBP panel). Strikingly, TBP was also enriched at the terminator of CHO-pT10 but not CHO-pT1 cells, suggesting that clustered TTF-I binding sites are in close proximity with the gene promoter. Consistent with multiple terminators facilitating initiation of transcription, TBP and Pol I occupancy was about 4-fold higher in CHO-pT10 compared to CHO-pT1 (Fig. 4B, TBP panel). To exclude the possibility that clustered TTF-I binding sites on their own recruit TBP to the 3′-end of rRNA genes, we examined TBP occupancy on a reporter plasmid in which the ten terminators were fused to a Pol II promoter. TBP was enriched at the Pol II promoter but close to background at the terminator (Fig. 4C), emphasizing the importance of TTF-I binding sites at both elements, the promoter and the terminators, to form chromatin loops.
Integrative analysis of histone marks reveals similarity to classical enhancer elements
Homo - and heterotypic clusters of transcription factor binding sites were shown to mark potential regulatory regions with enhancer function [35]–[39] characterized by eukaryotic histone marks like H3K27ac, which is involved in long-range chromatin interactions [40]. As the repetitive rDNA is left out of standard ChIP-Seq analyses, we artificially added a single mouse rDNA repeat to the current mouse genome version mm9 and mapped ChIP-Seq data of H3K27ac, H3K27me3, H3K4me1, H3K4me2 and H3K4me3 to this expanded reference genome (Fig. 5A). We observed enrichment of H3K27ac and H3K4me2 in the terminator and promoter region of murine rDNA, enforcing our previous results and confirming that the homotypic cluster of TTF-I binding sites represents an active enhancer element. In contrast, H3K27me3, commonly associated to repressed genes, is depleted at the terminator compared to the rDNA gene body. Therefore, the mouse rDNA terminator exhibits a histone modification profile typical for enhancer elements involved in Pol II transcription.
Fig. 5. Distribution of histone modifications at the murine rDNA. 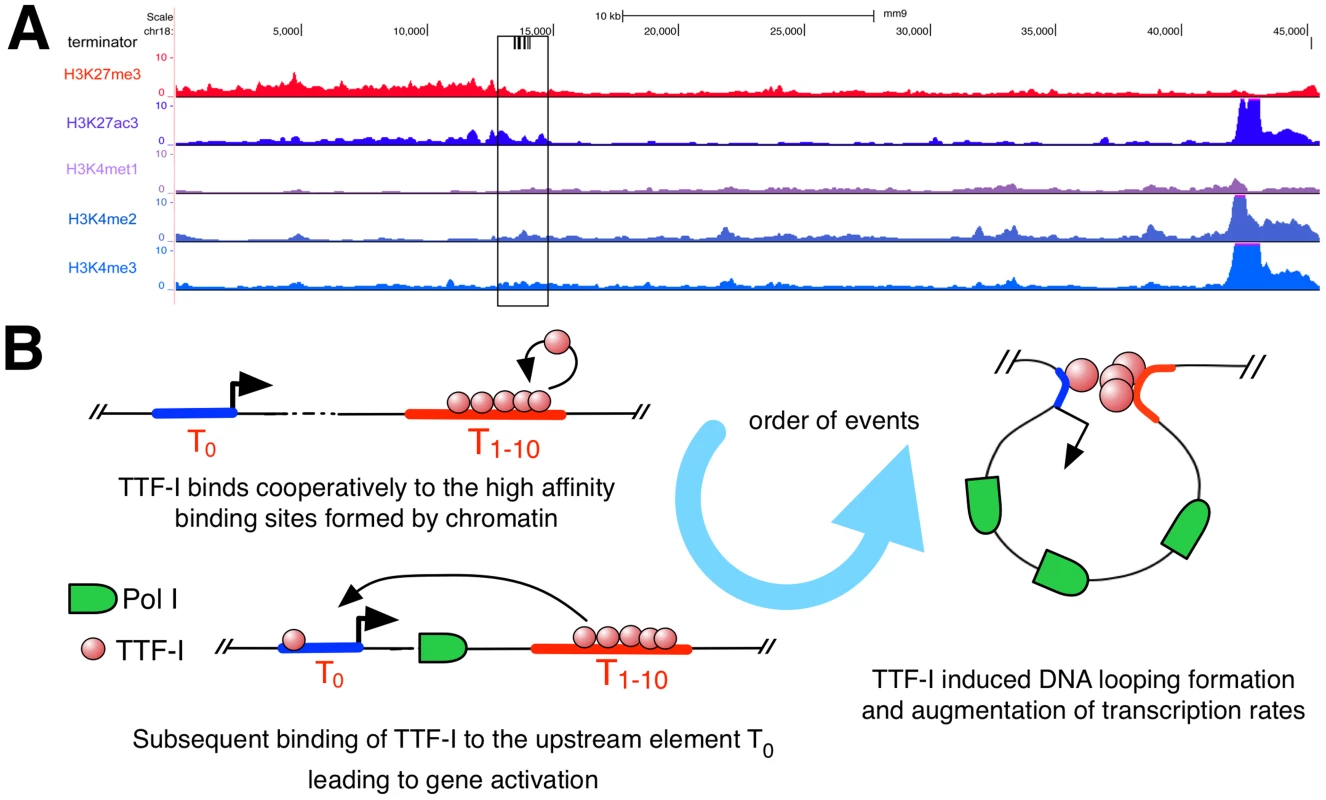
(A) Enrichment of histone modifications at the rDNA locus in 3T3-L1 cells. The whole rDNA repeat is plotted from position +1 (the TSS) to position 45.309. The terminator track indicates TTF-I binding sites by black vertical lines. The black box highlights the clustered terminator elements at the 3′ end of the gene. ChIP-Seq tracks of histone modifications display relative enrichments compared to input. (B) Model depicting the order of binding events at the rRNA gene. The promoter is coloured in blue, a right-headed arrow marks the TSS and the clustered termination sites are depicted in red. Discussion
Clustering of transcription factor binding sites, comprising either multiple binding sites for the same factor (homotypic clustering) or different DNA binding motifs (heterotypic clustering), is an important regulatory feature of eukaryotic gene expression, about 62% of transcription factor genes and 66% of developmentally regulated genes comprising clustered binding sites in vertebrates [3]. Therefore, this feature has been widely used for computational prediction. In Drosophila, predicted HTCs are present in more than 70% of regulatory regions and have been suggested to function as developmental enhancers [6], [41]. Clustered binding sites are suggested to exert a positive effect on transcription by either of the following mechanisms. They could increase the local concentration of transcription factors or facilitate multiple interactions with components of the transcription machinery. Alternatively, they could provide functional redundancy [37], [42], allowing cooperative binding of the factors through interactions among the multiple binding sites or indirectly through multiple interactions with the transcriptional machinery [10], [43]–[46]. Here, we have uncovered a novel chromatin-based mechanism underlying HTC-directed transcriptional activation. We show that packaging into chromatin converts multiple low-affinity terminators downstream of the rDNA transcription unit into a high-affinity binding platform for TTF-I. This preferential binding of TTF-I to the downstream terminators is a prerequisite for TTF-I binding to the promoter-proximal binding site, connecting the promoter with the terminator to allow efficient recycling of Pol I.
Cooperative binding of proteins has been shown to disrupt nucleosomes, thereby increasing the accessibility of transcription factors to regulatory sites [47], [48]. Our data reveal an alternative mechanism that increases the affinity of transcription factors. We show that binding of TTF-I to its target sites in chromatin is higher than to free DNA, suggesting that a specific nucleosomal arrangement or interactions with histones may trigger cooperative binding of TTF-I. Thus, HTCs attract transcription factors to functionally relevant sites, avoiding binding to single target sites in the genome. High-affinity binding of TTF-I to clustered termination sites will ensure loading of the downstream terminators (T1–T10) prior to TTF-I binding to the promoter-proximal binding site T0 in vivo. Sequential binding of TTF-I to the 3′ - and 5′-end of the rDNA transcription unit will ensure that transcription initiation will take place exclusively at rRNA genes that are associated with TTF-I and will be properly terminated. In addition, a direct interaction between the promoter and the terminator is only established when the terminator comprises several TTF-I binding sites. This mode of binding and the formation of an intragenic loop may serve two functions. First, it links the terminator with the respective transcription unit to be activated. Second, it enhances transcription at genes associated with TTF-I by forming a highly active ribomotor structure [22], [49]. Thus, homotypic clustering of TTF-I binding sites coordinates transcription initiation and termination, thereby affecting both the timing and the efficiency of rDNA transcription.
It is well established that gene activation by a distal regulatory element correlates with long-range interactions between enhancer(s) and gene promoters by factor-mediated formation of chromatin loops [50]. With regard to human and rat rRNA genes, previous studies suggested a role for TBP and c-Myc in loop formation at active rRNA genes [23], [33]. However, genome-wide ChIP-Seq data did not reveal significant enrichment of c-Myc - and TBP at the terminator (Fig. S7B). Moreover, murine rRNA genes lack clustered E-boxes (Fig. S7A), and therefore the participation of c-Myc in loop formation is not very likely. Similar loop mechanisms were shown for RNA polymerase II transcribed genes, suggesting a common theme involving the interaction of promoters with transcription termination regions that enhance the transcriptional activity and gene regulation [51].
Active enhancer elements are characterized by eukaryotic histone marks, e.g. H3K27ac or H3K4me1, which are involved in long-range chromatin interactions [40]. Notably, our integrative genomic analysis revealed characteristic enrichment of histone marks at the terminator, which can be observed in human as well [52]. The results support our finding that the homotypic cluster of TTF-I binding sites displays all hallmarks of a functional enhancer, such as distal location, presence of HTCs, regulatory histone marks and the potential to exert gene activation by direct, protein-mediated DNA loops. Chromatin-dependent high-affinity binding of TTF-I to the clustered binding sites adds a further regulatory level on the enhancer function, i.e., coordination of transcription termination and initiation.
Materials and Methods
Protein expression and microscale thermophoresis
Histidine-tagged full-length TTF-I and the deletion mutants TTFΔN210 and TTFΔN348 were purified on a heparin column (Bio-Rad), followed by purification with Ni-NTA agarose according to the manufacturer's instructions (Qiagen). For microscale thermophoresis experiments, 50 nM of fluorescently labelled DNA oligonucleotides were incubated with 5 nM–2.4 µM of protein for 10 min at 30°C in 80 mM Tris-HCl (pH 7.6), 80 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 10% glycerol and 0.05% IGEPAL CA-630. Affinity measurements were carried out in a Monolith NT.015T (NanoTemper Technologies) as described [26].
MNase footprinting and transcription
300 ng of chromatin reconstituted with Drosophila extract was digested with 1.5 U of MNase (Sigma) for 40 s in 10 mM Tris-HCl (pH 7.6), 80 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.5 mM ATP, and 200 ng/µl BSA. Reactions were then stopped by the addition of 0.2 volumes of 4% SDS, 100 mM EDTA, 1 µg of glycogen, 10 µg of proteinase K. Purified DNA was analysed by a single round of PCR (denaturation, 5 min at 95°C; annealing, 2 min at 56°C; extension, 1 min at 72°C) using radioactively labelled oligonucleotides that hybridize to the rDNA promoter or terminator. Primer extension fragments were resolved on 8% sequencing gels and visualized by autoradiography.
Transcription experiments were performed on pMrWT-T, a template comprising the murine rDNA promoter (from −170 to +155 with regard to the transcription start site) fused to a 3.5 kb 3′-terminal rDNA fragment (BamHI/EcoRV Fragment) harbouring all ten terminators. (T1–T10). The promoter and the terminator elements are separated by 686 bp. Transcription reactions were performed as described [53].
Cell culture
CHO and CHO Flp-In cells (Invitrogen) were grown in DMEM (GIBCO) supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. For transient transfections, 200.000 cells were transfected with 1 µg of plasmid DNA. Prior to transfection of the CHO Flp-In cells, 100 µg/ml zeocin (Invitrogen) was added to the medium and for transfection 0.25, 0.5 or 1.0 µg of the rRNA reporter construct and the flipase encoding plasmid pOG44 (Invitrogen) in a ratio of 1∶9 were used. During the selection process, 500 µg/ml of hygromycin (PAA) was added to the medium; afterwards the stable cell lines were passaged with 250 µg/ml of hygromycin.
Constructs and reporter gene assays
Transiently transfected rRNA minigenes [22] contain mouse rDNA (BK000964) sequences from position −1932 to +181, an IRES, the firefly luciferase gene, and rDNA terminator regions from position +13169 to +15278 (T10 constructs) in a pGL3-Basic vector (Promega). Plasmids for genomic integration contain in addition the enhancer/promoter regions from position −2148 to +181 cloned into a pcDNA5-FRT vector (Invitrogen).
Cells were transfected with Pol I driven firefly luciferase reporter constructs and a Pol II renilla luciferase control plasmid, pRL-TK (Promega). TTF-I co-transfections were performed with the expression vectors TTFΔN348-EGFP or TTF-IΔN470-EGFP in a TTF-I∶reporter ratio of 10∶1. Protein expression was monitored by Western Blot analysis. Reporter gene measurements were performed using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions using a single-tube luminometer (Stratec Biomedical Systems).
Isolation of RNA and genomic DNA
RNA isolation was performed with the NucleoSpin RNA II kit (Macherey-Nagel). Purified RNA (500 ng) was used for cDNA preparation with the iScript Select kit (Biorad).
To determine the number of integration sites, genomic DNA was isolated by cells lysis (1% SDS, 50 mM Tris-HCl (pH 8.0), 20 mM EDTA and 250 µg of RNase A), the addition of proteinase K and incubation at 37°C o.n. The supernatant was precipitated with ethanol and ammonium acetate.
qPCR
Quantitative real-time PCR was performed in a Rotor-Gene cycler (Qiagen) using a HotStar master mix containing SYBR green (Qiagen). Primer sequences and annealing temperatures are listed in the in Table S2. Fold inductions were calculated using the comparative quantitation software (Qiagen). Post-PCR melting curves and agarose gels of PCR products (Fig. S3F) were used to assess the quality of primer pairs.
Chromatin immunoprecipitation
Cells were transfected with 10 µg of DNA and cross-linked with 1% formaldehyde for 10 min (α-Pol I and α-UBF ChIPs) or 10 mM DMA for 30 min +1% formaldehyde for 10 min (α-TBP) at RT. The reactions were quenched with 125 mM glycine. Cells were washed twice in ice-cold PBS and the cell pellets were lysed in SDS lysis buffer (1% SDS, 50 mM Tris-HCl pH 8.0, 20 mM EDTA, protease inhibitors). Chromatin was sheared in a Biorupter sonicator (Diagenode) to fragments of 400–1000 bp in length. The samples were diluted in IP dilution buffer (20 mM Tris-HCl, 2 mM EDTA, 1% Triton X-100, 150 mM NaCl, pH 8.0, protease inhibitors). Paf53 antibody for Pol I detection and the pre-serum were obtained from the Grummt lab [54]. Antibodies targeting RPA194 (sc-28714), UBF (sc-9131), TBP (sc-273) and normal rabbit IgG (sc-2027) were purchased from Santa Cruz. Antibodies (5 µg) and chromatin were incubated on a rotating wheel at 4°C o.n. Pre-blocked Protein-G sepharose (500 µg/ml sonicated salmon sperm DNA and 100 µg/ml BSA in IP dilution buffer) was added to isolate the immune-complexes and incubated for 2 h at 4°C. Beads were washed twice with IP dilution buffer, once with high salt buffer (20 mM Tris-HCl, 2 mM EDTA, 1% Triton X-100, 150 mM NaCl, pH 8.0), LiCl buffer (0.25 M LiCl, 1% NP40, 1% Deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0) and twice with TE buffer (10 mM Tris-HCl, 1 mM EDTA pH 8.0). Elution was performed using 250 µl of 1% SDS, 0.1 M NaHCO3. RNase A was added to a concentration of 100 µg/ml and incubated for 2.5 h at 37°C. Following the Proteinase K digestion (100 µg/ml, 2.5 h at 37°C), reverse crosslinking was carried out at 64°C o.n. DNA was isolated by phenol/chloroform/isoamylalcohol extraction and precipitated with ethanol and sodium acetate.
FISH experiments
Fluorescence in situ hybridizations on metaphase chromosome spreads and on interphase nuclei combined with nucleolar immunostaining were performed as described [55]. For RNA FISH, cells grown on coverslips were fixed for 10 min at room temperature with 3.7% formaldehyde/5% acetic acid/0.5% (w/v) NaCl, washed twice with 1× PBS, once in 50 mM NH4Cl/1× PBS pH 7.4, and once in 1× PBS. Coverslips were then transferred to 70% ethanol and incubated o.n. at 4°C. Before hybridization, coverslips were rehydrated in 2× SSC/50% formamide for 15 min at RT. Hybridization mixtures were added for o.n. incubation at 37°C. Post-hybridization washes were carried out as follows: 2×25 min at 37°C in 50% formamide/2× SSC and 2×5 min in 2× SSC at RT. The subsequent immunostaining, DNA staining and mounting was performed as in interphase DNA FISH experiments. Nick-translated, biotin-labeled pcDNA5-FRT-rRNA reporter served as hybridization probe in all experiments.
Visualisation of histone modification data at the mouse rDNA locus
A custom build of the mm9 assembly was generated by replacing unsequenced bases at the 5′-end of chromosome 18 with a murine rDNA repeat (GenBank accession no. BK000964). We used Bowtie [56] to align published ChIP-seq data sets of 3T3L1 and MEL cells (for details see Table S1) to the custom assembly using ‘–best -k 1’ settings. Input-normalized bedGraph files were generated using the makeUCSCfile.pl script contained in the HOMER software suite (http://biowhat.ucsd.edu/homer/, [57]) using standard settings.
Supporting Information
Zdroje
1. BergOG, Hippel vonPH (1987) Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. Journal of Molecular Biology 193 : 723–750.
2. BergOG, Hippel vonPH, Hippel vonPH (1988) Selection of DNA binding sites by regulatory proteins. Trends in Biochemical Sciences 13 : 207–211.
3. GoteaV, ViselA, WestlundJM, NobregaMA, PennacchioLA, et al. (2010) Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Research 20 : 565–577.
4. DavidsonEH (2002) A Genomic Regulatory Network for Development. Science 295 : 1669–1678.
5. SchroederMD, PearceM, FakJ, FanH, UnnerstallU, et al. (2004) Transcriptional Control in the Segmentation Gene Network of Drosophila. PLoS Biol 2 (9) e271.
6. ErivesA, LevineM (2004) Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci USA 101 : 3851–3856.
7. RheeHS, PughBF (2011) Comprehensive Genome-wide Protein-DNA Interactions Detected at Single-Nucleotide Resolution. Cell 147 : 1408–1419.
8. SauerF, HansenSK, TjianR (1995) Multiple TAFIIs directing synergistic activation of transcription. Science 270 : 1783–1788.
9. SauerF, HansenSK, TjianR (1995) DNA template and activator-coactivator requirements for transcriptional synergism by Drosophila bicoid. Science 270 : 1825–1828.
10. HertelKJ, LynchKW, ManiatisT (1997) Common themes in the function of transcription and splicing enhancers. Current Opinion in Cell Biology 9 : 350–357.
11. VasheeS, MelcherK, MelcherK, DingWV, et al. (1998) Evidence for two modes of cooperative DNA binding in vivo that do not involve direct protein-protein interactions. Current biology : CB 8 : 452–458.
12. GrummtI, RosenbauerH, NiedermeyerI, MaierU, OhrleinA (1986) A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell 45 : 837–846.
13. GrummtI, MaierU, OhrleinA, HassounaN, BachellerieJP (1985) Transcription of mouse rDNA terminates downstream of the 3″ end of 28S RNA and involves interaction of factors with repeated sequences in the 3″ spacer. Cell 43 : 801–810.
14. La VolpeA, SimeoneA, SimeoneA, D'EspositoM, et al. (1985) Molecular analysis of the heterogeneity region of the human ribosomal spacer. Journal of Molecular Biology 183 : 213–223.
15. BartschI, SchonebergC, GrummtI (1987) Evolutionary changes of sequences and factors that direct transcription termination of human and mouse ribsomal genes. Molecular and Cellular Biology 7 : 2521–2529.
16. ClosJ, NormannA, OhrleinA, GrummtI (1986) The core promoter of mouse rDNA consists of two functionally distinct domains. Nucleic Acids Research 14 : 7581–7595.
17. StrohnerR, NemethA, NemethA, JansaP, et al. (2001) NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. The EMBO Journal 20 : 4892–4900.
18. SantoroR, LiJ, GrummtI (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nature Genetics 32 : 393–396.
19. YuanX, FengW, ImhofA, GrummtI, ZhouY (2007) Activation of RNA Polymerase I Transcription by Cockayne Syndrome Group B Protein and Histone Methyltransferase G9a. Molecular Cell 27 : 585–595.
20. XieW, LingT, ZhouY, FengW, ZhuQ, et al. (2012) The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proceedings of the National Academy of Sciences 109 : 8161–8166.
21. SanderEE, GrummtI (1997) Oligomerization of the transcription termination factor TTF-I: implications for the structural organization of ribosomal transcription units. Nucleic Acids Research 25 : 1142–1147.
22. NémethA, GuibertS, TiwariVK, OhlssonR, LängstG (2008) Epigenetic regulation of TTF-I-mediated promoter–terminator interactions of rRNA genes. The EMBO Journal 27 : 1255–1265.
23. DenissovS, LessardF, MayerC, StefanovskyV, van DrielM, et al. (2011) A model for the topology of active ribosomal RNA genes. EMBO reports 12 (3) 231–7 doi:10.1038/embor.2011.8
24. NémethA, LängstG (2011) Genome organization in and around the nucleolus. Trends in Genetics 27 : 149–156.
25. GerberJK, GögelE, BergerC, WallischM, MüllerF, et al. (1997) Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 90 : 559–567.
26. ZillnerK, Jerabek-WillemsenM, DuhrS, BraunD, LängstG, et al. (2012) Microscale thermophoresis as a sensitive method to quantify protein: nucleic acid interactions in solution. Methods Mol Biol 815 : 241–252.
27. ColemanRA, PughBF (1995) Evidence for functional binding and stable sliding of the TATA binding protein on nonspecific DNA. J Biol Chem 270 : 13850–13859.
28. MaerklSJ, QuakeSR (2007) A Systems Approach to Measuring the Binding Energy Landscapes of Transcription Factors. Science 315 : 233–237.
29. BaaskeP, WienkenCJ, ReineckP, DuhrS, BraunD (2010) Optical Thermophoresis for Quantifying the Buffer Dependence of Aptamer Binding. Angew Chem Int Ed 49 : 2238–2241.
30. BeckerPB, WuC (1992) Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Molecular and Cellular Biology 12 : 2241–2249.
31. LangstG, BlankA, BeckerPB, GrummtI (1997) RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. The EMBO Journal 16 : 760–768.
32. LangstG, BeckerPB, GrummtI (1998) TTF-I determines the chromatin architecture of the active rDNA promoter. The EMBO Journal 17 : 3135–3145.
33. ShiueC-N, BerksonRG, WrightAPH (2009) c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. 28 : 1833–1842.
34. LingJQ, LiT, HuJF, VuTH, ChenHL, QiuXW, CherryAM, HoffmanAR (2006) CTCF Mediates Interchromosomal Colocalization Between Igf2/H19 and Wsb1/Nf1. Science 312 : 269–272.
35. StanojevicD, SmallS, LevineM (1991) Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science 254 : 1385–1387.
36. ArnoneMI, DavidsonEH (1997) The hardwiring of development: organization and function of genomic regulatory systems. Development 124 : 1851–1864.
37. PapatsenkoDA, MakeevVJ, LifanovAP, RegnierM, NazinaAG, et al. (2002) Extraction of Functional Binding Sites from Unique Regulatory Regions: The Drosophila Early Developmental Enhancers. Genome Research 12 : 470–481.
38. BermanBP, NibuY, PfeifferBD, TomancakP, CelnikerSE, et al. (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci USA 99 : 757–762.
39. HalfonMS, GradY, ChurchGM, MichelsonAM (2002) Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Research 12 : 1019–1028.
40. RyeM, SætromP, HåndstadT, DrabløsF (2011) Clustered ChIP-Seq-defined transcription factor binding sites and histone modifications map distinct classes of regulatory elements. BMC Biology 9 : 80.
41. VavouriT, ElgarG (2005) Prediction of cis-regulatory elements using binding site matrices — the successes, the failures and the reasons for both. Current Opinion in Genetics & Development 15 : 395–402.
42. SommaMP, PisanoC, LaviaP (1991) The housekeeping promoter from the mouse CpG island HTF9 contains multiple protein-binding elements that are functionally redundant. Nucleic Acids Research 19 : 2817–2824.
43. GinigerE, PtashneM (1988) Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci USA 85 : 382–386.
44. LinYS, CareyM, PtashneM, GreenMR (1990) How different eukaryotic transcriptional activators can cooperate promiscuously. Nature 345 : 359–361.
45. AndersonGM, FreytagSO (1991) Synergistic activation of a human promoter in vivo by transcription factor Sp1. Molecular and Cellular Biology 11 : 1935–1943.
46. HeX, SameeMAH, BlattiC, SinhaS (2010) Thermodynamics-Based Models of Transcriptional Regulation by Enhancers: The Roles of Synergistic Activation, Cooperative Binding and Short-Range Repression. PLoS Comput Biol 6: e1000935.
47. VicentGP, ZaurinR, NachtAS, Font-MateuJ, Le DilyF, et al. (2010) Nuclear Factor 1 Synergizes with Progesterone Receptor on the Mouse Mammary Tumor Virus Promoter Wrapped around a Histone H3/H4 Tetramer by Facilitating Access to the Central Hormone-responsive Elements. Journal of Biological Chemistry 285 : 2622–2631.
48. AdamsCC, WorkmanJL (1995) Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Molecular and Cellular Biology 15 : 1405–1421.
49. Kempers-VeenstraAE, OliemansJ, OffenbergH, DekkerAF, PiperPW, et al. (1986) 3′-End formation of transcripts from the yeast rRNA operon. The EMBO Journal 5 : 2703.
50. CookPR (2003) Nongenic transcription, gene regulation and action at a distance. Journal of Cell Science 116 : 4483–4491.
51. Lykke-AndersenS, MapendanoCK, Heick JensenT (2011) An ending is a new beginning: transcription termination supports re-initiation. Cell Cycle 10 : 863–865.
52. ZentnerGE, SaiakhovaA, ManaenkovP, AdamsMD, ScacheriPC (2011) Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res 39 : 4949–4960.
53. NémethA, StrohnerR, GrummtI, LängstG (2004) The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Research 32 : 4091–4099.
54. SeitherP, ZatsepinaO, HoffmannM, et al. (1997) Constitutive and strong association of PAF53 with RNA polymerase I. Chromosoma 106 : 216–225.
55. NémethA, ConesaA, Santoyo-LopezJ, MedinaI, MontanerD, et al. (2010) Initial Genomics of the Human Nucleolus. PLoS Genet 6: e1000889 doi:10.1371/journal.pgen.1000889.g004
56. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10: R25.
57. HeinzS, BennerC, SpannN, BertolinoE, LinYC, et al. (2010) Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell 38 : 576–589.
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Akutní intermitentní porfyrie
- Příjem alkoholu a menstruační cyklus
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
- Transfer zmraženého embrya zlepšuje výsledky IVF
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



