-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
Eukaryotic DNA replication origins are selected in G1-phase when the origin recognition complex (ORC) binds chromosomal positions and triggers molecular events culminating in the initiation of DNA replication (a.k.a. origin firing) during S-phase. Each chromosome uses multiple origins for its duplication, and each origin fires at a characteristic time during S-phase, creating a cell-type specific genome replication pattern relevant to differentiation and genome stability. It is unclear whether ORC-origin interactions are relevant to origin activation time. We applied a novel genome-wide strategy to classify origins in the model eukaryote Saccharomyces cerevisiae based on the types of molecular interactions used for ORC-origin binding. Specifically, origins were classified as DNA-dependent when the strength of ORC-origin binding in vivo could be explained by the affinity of ORC for origin DNA in vitro, and, conversely, as ‘chromatin-dependent’ when the ORC-DNA interaction in vitro was insufficient to explain the strength of ORC-origin binding in vivo. These two origin classes differed in terms of nucleosome architecture and dependence on origin-flanking sequences in plasmid replication assays, consistent with local features of chromatin promoting ORC binding at ‘chromatin-dependent’ origins. Finally, the ‘chromatin-dependent’ class was enriched for origins that fire early in S-phase, while the DNA-dependent class was enriched for later firing origins. Conversely, the latest firing origins showed a positive association with the ORC-origin DNA paradigm for normal levels of ORC binding, whereas the earliest firing origins did not. These data reveal a novel association between ORC-origin binding mechanisms and the regulation of origin activation time.
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003798
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003798Summary
Eukaryotic DNA replication origins are selected in G1-phase when the origin recognition complex (ORC) binds chromosomal positions and triggers molecular events culminating in the initiation of DNA replication (a.k.a. origin firing) during S-phase. Each chromosome uses multiple origins for its duplication, and each origin fires at a characteristic time during S-phase, creating a cell-type specific genome replication pattern relevant to differentiation and genome stability. It is unclear whether ORC-origin interactions are relevant to origin activation time. We applied a novel genome-wide strategy to classify origins in the model eukaryote Saccharomyces cerevisiae based on the types of molecular interactions used for ORC-origin binding. Specifically, origins were classified as DNA-dependent when the strength of ORC-origin binding in vivo could be explained by the affinity of ORC for origin DNA in vitro, and, conversely, as ‘chromatin-dependent’ when the ORC-DNA interaction in vitro was insufficient to explain the strength of ORC-origin binding in vivo. These two origin classes differed in terms of nucleosome architecture and dependence on origin-flanking sequences in plasmid replication assays, consistent with local features of chromatin promoting ORC binding at ‘chromatin-dependent’ origins. Finally, the ‘chromatin-dependent’ class was enriched for origins that fire early in S-phase, while the DNA-dependent class was enriched for later firing origins. Conversely, the latest firing origins showed a positive association with the ORC-origin DNA paradigm for normal levels of ORC binding, whereas the earliest firing origins did not. These data reveal a novel association between ORC-origin binding mechanisms and the regulation of origin activation time.
Introduction
Eukaryotic DNA replication initiates at specific chromosomal sites called origins. An origin is selected in G1-phase by the origin recognition complex (ORC) that directly binds chromosomal DNA, triggering a series of molecular events that culminate in the loading of an MCM helicase complex onto DNA (reviewed in [1]–[4]). Origin activation (unwinding; firing) occurs only during the subsequent S-phase, when the MCM complex is activated to give two oppositely oriented helicases that will unwind DNA at the bidirectional replication forks (reviewed in [5]–[7]). Temporal separation of the origin selection and activation steps helps ensure a chromosome is replicated only once per cell cycle (reviewed in [8]). However, it is unclear how the specific molecular events essential for the first step, in particular origin binding by ORC in G1 phase, might regulate the second step, origin activation in S-phase.
While the understanding of the roles of origin-binding factors has progressed to a mechanistic level, we have less understanding of how the firing of eukaryotic replication origins is regulated during S-phase. In particular, a eukaryotic chromosome requires the action of multiple origins for its timely and accurate replication [9]–[13]. Individual origins vary in their time of activation during S-phase, creating a distinct spatial and temporal pattern of genome duplication that, in multicellular organisms, shows cell-type specificity and is associated with normal cell differentiation. Indeed, disruption of replication timing contributes to genome instability [14]–[17]. The conservation of replication timing patterns—for example histone genes and centromeres are replicated early during S-phase, while telomeres are replicated late in many organisms—and their strong association with genome stability and differentiation have spurred research to define the mechanisms that control origin activation time [18]–[21]. Despite important advances, including several reported in recent studies, the specific molecular features of DNA replication origins that control their activation time remain incompletely understood [22]–[27].
Many studies examining origin activation time have used the model eukaryote budding yeast S. cerevisiae and dealt with factors with broad effects on DNA replication and other chromosomal processes, such as S-phase kinases, Forkhead family transcription factors, and chromatin structure [22], [28]–[30]. Indeed, early studies in yeast established that chromosomal context could have a substantial impact on an origin's activation time [31], [32]. For example, an origin that normally fires early could be made to fire late by placing it within a region of heterochromatin, while specific modifications associated with actively transcribed chromatin, such as histone acetylation, could advance an origin's replication time in both yeast and flies [33]–[36]. Thus chromatin structure can clearly regulate origin activation time, although in many cases the molecular step affected is unknown.
Although a major focus of the timing studies has been on factors extrinsic to core origin function, a few studies have raised the possibility that origin-binding factors, required for origin activation per se, can influence origin activation time. In particular, recent studies reveal that a collection of origin-activation factors are limiting in S-phase such that their over-expression can advance the replication time of a normally late-firing origin [23], [24]. This observation raises questions about the mechanisms that underlie the differential affinities of origins for these limiting factors. Although specific chromatin structures likely contribute, it is unclear how they affect core factors that establish the origin-protein complex that is recognized during S-phase—such as ORC or the MCM complex [37].
In S. cerevisiae, ORC selects origins in part by interacting in a sequence-specific manner with a conserved DNA element present in all yeast origins [38], [39]. At the simplest level, one might predict that stronger interactions between ORC and its binding site would enhance origin activity and therefore contribute to earlier, more robust origin activation during S-phase. Indeed, a study of S. pombe origin activation time provides evidence in support of this idea [40]. However, a previous study from our group revealed that the relationship between ORC-origin interactions and origin activation time might be more complex [41]. In particular the ORC binding site within a specific origin, ARS317, also known as the HMR-E silencer that controls heterochromatin formation at the silent mating-type locus HMR, binds ORC with a remarkably high affinity in vitro compared to several other replication origins. A noteworthy characteristic of HMR-E, in addition to its silencer function, is that it functions extremely poorly as a replication origin, firing in only a small percentage of cell cycles and then only very late in S-phase (reviewed in [42]). Thus, paradoxically, the high-affinity ORC-origin interaction at ARS317 fails to promote efficient or early origin activation. In fact, mutations that weaken the ORC-DNA interaction enhance the firing efficiency and advance the replication time of ARS317. In addition, several efficient and early activating origins examined in the same study have weak ORC-DNA interactions in vitro. Thus a high-affinity ORC-origin interaction mediated by sequence-specific ORC-DNA contacts is insufficient to promote - and can in fact inhibit - robust, early origin activation. However, we examined only a small number of yeast origins in this study, leaving in question whether this conclusion could be extended to form a general paradigm about the relationship between ORC-origin interactions and origin firing.
An additional complication is revealed by recent studies establishing that, in addition to sequence-specific interactions between ORC and origin DNA, chromatin also contributes to yeast ORC's ability to bind origins, as it does for metazoan ORC [43]–[45]. For example, the bromo adjacent homology (BAH) domain of Orc1, a nucleosome-binding domain within the largest subunit of ORC, contributes to ORC-origin binding in both yeast and mammalian cells [43], [46], [47]. However, individual yeast origins vary substantially in terms of their requirement for the Orc1BAH domain for ORC binding, suggesting that the mechanisms governing ORC-origin binding in budding yeast vary between origins, with some, such as ARS317 using sequence-specific ORC-DNA interactions and others using as yet incompletely defined ORC-chromatin interactions [43]. Thus the stability of an ORC-origin complex in vivo could be achieved by sequence-specific ORC-origin DNA interactions, as at ARS317, or by interactions between ORC and accessory proteins (e.g. chromatin), or by some combination of these mechanisms. It is entirely unknown whether these different mechanisms of ORC-origin binding might ultimately relate to the regulation of origin activation in S-phase.
In this study we addressed these issues by employing a genome-wide approach to determine the relative contribution to ORC-origin binding of ‘intrinsic’ features (i.e. the DNA sequence that comprises the ORC binding site) versus ‘extrinsic’ features (e.g. adjacent ‘chromatin’, including both nucleosomes and non-histone proteins). This approach allowed us to classify origins into several groups based on the type of mechanism that stabilized the ORC-origin complex. We focused further comparative analysis on two distinct groups of origins, each of which bound ORC with similar strengths in vivo. The first group was comprised of DNA-dependent origins, such as ARS317, in which the DNA sequence of the ORC binding site was a primary determinant of ORC-origin affinity. The second group was comprised of ‘chromatin-dependent’ origins in which the ORC-origin DNA interaction was insufficient to explain the ORC binding strength in vivo. As expected from a biologically meaningful classification, DNA-dependent and ‘chromatin-dependent’ origins differed based on several other structural and functional criteria. Significantly, the ‘chromatin-dependent’ group was enriched with origins that fire early in S-phase whereas the DNA-dependent group that included ARS317 was enriched for later firing origins. Moreover, the latest-firing origins in the genome, as a group, showed a positive correlation between in vivo and in vitro ORC-origin binding affinity, indicating that many of these origins followed the DNA sequence-dependent ORC-origin interaction paradigm. In contrast, the earliest-firing origins, as a group, showed no correlation between in vivo and in vitro ORC-origin binding affinity, suggesting that ‘chromatin’ had a larger impact on ORC-origin binding at many of the earliest firing origins. Taken all together these data provided evidence that sequence-specific ORC-DNA interactions that promote ORC-origin binding stability are often associated with the suppression of origin activation, whereas ORC-‘chromatin’ interactions that modulate this stability are often associated with the enhancement of origin activation. We discuss the interesting mechanistic implications of this unanticipated connection between the mode of ORC-origin binding and origin activation time.
Results
Classifying yeast origins based on the contribution of the ORC-origin-DNA interface to the strength of the ORC-origin interaction in vivo
The paradigm for yeast origin selection by ORC is that sequence-specific ORC-origin-DNA (hence referred to as ORC-DNA) interactions drive this process. Specifically, ORC binds to a bipartite ∼35 bp element consisting of a 17-bp EACS-element (extended ARS consensus sequence) and a less conserved B1-element that contains a common 3 bp WTW motif [48]–[50]. However, recent work has shown that local chromatin structure may also contribute to origin selection by ORC [43], [44]. This observation raises the possibility that some yeast origins might rely primarily on ORC-‘chromatin’ interactions for ORC binding while other origins rely on sequence-specific ORC-DNA interactions. If the sequence-specific ORC-DNA interaction controls ORC binding at many yeast origins in vivo, then we would expect a correlation between in vivo and in vitro ORC-origin binding strengths, whereas origins that deviated from this correlation would be putative ‘chromatin-dependent’ origins in which features extrinsic to the ORC binding sequence likely modulate origin-binding by ORC in vivo (Figure 1A). (The term ‘chromatin’ is used here in its broadest sense to include both histone - and non-histone-chromosomal proteins. Therefore at ‘chromatin-dependent’ origins we envision that ORC binds the origin locus through direct contacts with histones or non-histone chromosomal proteins such as transcription factors). Therefore we examined ORC-origin affinity in vivo and in vitro for a large fraction of yeast origins to classify them based on which mechanism (i.e. DNA sequence vs. extrinsic, ‘chromatin’ factors) mediates their binding to ORC in vivo.
Fig. 1. Classifying yeast origins based on the contribution of the ORC-origin-DNA interface to the strength of the ORC-origin interaction in vivo. 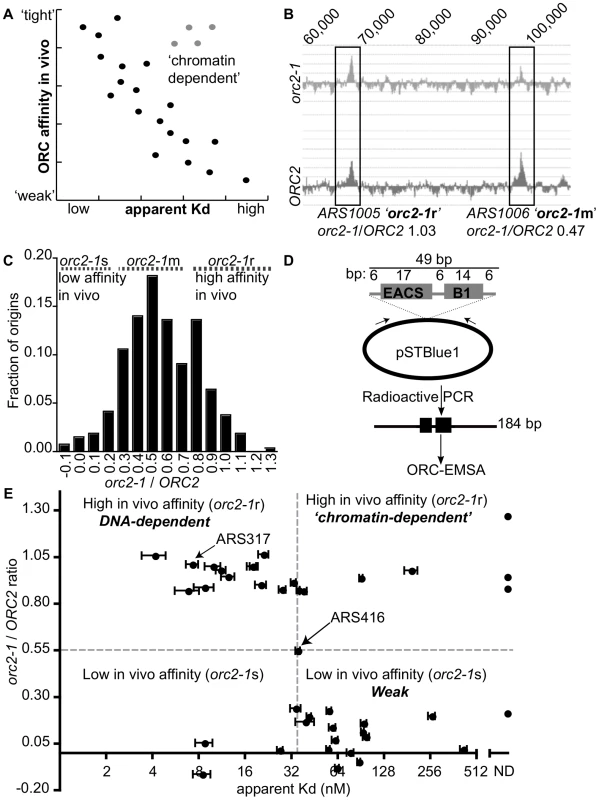
(A) If the strength of ORC-origin interactions in vivo were due to interactions between ORC and the essential ORC binding site within origins, then we would expect a graph in which in vivo ORC-origin binding strengths (x-axis) plotted against in vitro ORC-origin-DNA binding strengths (apparent Kd) to show a correlation (black dots). Such a correlation would allow us to identify ‘exceptions to the rule’ such as those shown as putative ‘chromatin-dependent’ origins (gray dots). (B) ORC ChIP peaks are shown for two adjacent origins on chromosome X, ARS1005, and ARS1006. The relevant chromosomal coordinates are shown at the top of the figure. ARS1005 has a high-affinity for ORC in vivo (orc2-1-resistant (orc2-1r)) relative to ARS1006 that has a moderate affinity for ORC in vivo (orc2-1-moderately sensitive (orc2-1m)). (C) The fraction of the confirmed origins defined in our wild-type (ORC2 ChIP-chip) array (n = 261; y-axis) was plotted against the corresponding orc2-1/ORC2 ratios (x-axis). For a small number of origins the corresponding genomic region in orc2-1 cells showed a slight depletion in ORC binding and hence generated a negative value of −0.1. (D) Schematic of the DNA probes used in EMSAs with ORC. All probes contained the confirmed or predicted ORC binding site in the orientation shown for each origin listed in Table 1. (E) Scatter plot comparing the ORC-origin binding affinities in vivo (orc2-1/ORC2 (y-axis)) and in vitro (apparent Kd (x-axis)). The average apparent Kd and standard error obtained from three independent experiments are shown. ARS317 (HMR-E) and ARS416 (ARS1) known to bind ORC with high- and moderate- affinity in vitro, respectively, are indicated [41]. To measure ORC's affinity for confirmed origins in vivo, we used data from an experiment in which ORC occupancy, as measured by ChIP, was compared genome-wide between wild-type and orc2-1 mutant cells [51]. Orc2-1 is a temperature-sensitive allele that reduces the amount of Orc2, an essential subunit of ORC, by ∼10-fold even at permissive growth temperatures. Orc2-1 cells grow more slowly than wild-type cells and exhibit additional defects even at permissive temperatures [52]. However, simply over-producing orc2-1 is sufficient to rescue these growth defects, including temperature-sensitivity, suggesting that the primary defect is reduction in levels of ORC. Moreover, detailed analyses of ARS317, the HMR-E silencer origin, reveals that mutations in the ORC binding site of this origin that enhance ORC-origin binding affinity in vitro, fully suppress the defects in this origin caused by the orc2-1 allele. Collectively these data provide evidence that the primary defect caused by the orc2-1 mutation is reduced ORC, such that ORC concentration becomes limiting in the nucleus [41], [51]. Thus, as expected, in the orc2-1 mutant, an origin with high in vivo affinity for ORC remains fully occupied by ORC as measured by ChIP, while an origin with a low in vivo affinity shows reduced occupancy (Figure 1B and [51]). Therefore, we used the ratio of the areas of origin-associated ORC binding peaks in orc2-1 mutant cells to that in wild-type cells (hence referred to as orc2-1/ORC2 ratio) as a measure of ORC's ‘affinity’ for that origin in vivo. The orc2-1/ORC2 ratio for every peak identified in the wild-type array that was associated with a confirmed origin in the yeast origin database (261/351 confirmed origins; [53]; http://cerevisiae.oridb.org) is plotted in Figure 1C. The confirmed origins exhibit a range of orc2-1/ORC2 ratios. To aid in further analyses, we arbitrarily divided the origins into three groups: origins with orc2-1/ORC2 ratios</ = 0.3 were termed ‘low in vivo affinity’ and considered orc2-1-sensitive (orc2-1s; n = 35); origins with orc2-1/ORC2 ratios greater than 0.3 but less than 0.8 were termed ‘moderate in vivo affinity’ and considered orc2-1-moderately sensitive (orc2-1m; n = 175); origins with orc2-1/ORC2 ratios of >/ = 0.8 were termed ‘high in vivo affinity’ and considered orc2-1-resistant (orc2-1r; n = 51).
Our goal was to distinguish between origins that used ORC-DNA interactions to achieve normal ORC binding in vivo from origins that used ORC-‘chromatin’ interactions. An expectation was that the established ORC-DNA interaction explained the ORC binding strength for many origins in vivo, as it did for ARS317 and ARS1 (ARS416). Thus if the in vivo-in vitro affinity correlation ‘rule’ for budding yeast origins was followed for some origins, we could be more confident that ‘exceptions to the rule’ would provide useful insights. As a proof-of-principle experiment, we selected 18 origins from among the lowest (orc2-1/ORC2 ratios</ = 0.3) and 20 origins among the highest (orc2-1/ORC2 ratios>/ = 0.8) in vivo affinity groups and determined the strength of the interaction between purified origin-DNA and purified ORC in vitro (Table 1). ARS416 (ARS1) was used to represent the ‘moderate in vivo affinity’ group. The ORC binding site for each of these 39 origins was cloned into a bacterial plasmid, and plasmid-specific PCR primers were used to generate radiolabeled DNA fragments of 184 bp with the ORC binding site centered within the fragment and arranged in the orientation shown (Figure 1D). Data from standard Electrophoretic Mobility Shift Assays (EMSAs) were used to measure ORC binding to these elements and calculate apparent Kds for each ORC-DNA complex. ARS317 whose affinity for ORC in vivo could be accounted for by the strength of its ORC-DNA interaction was used as an internal standard in every EMSA [41], [51].
Tab. 1. ORC binding data for the 39 origins used in ORC binding reactions in Figure 1E. 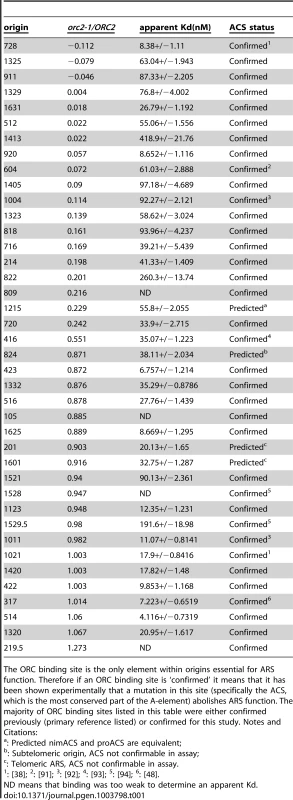
The ORC binding site is the only element within origins essential for ARS function. Therefore if an ORC binding site is ‘confirmed’ it means that it has been shown experimentally that a mutation in this site (specifically the ACS, which is the most conserved part of the A-element) abolishes ARS function. The majority of ORC binding sites listed in this table were either confirmed previously (primary reference listed) or confirmed for this study. Notes and Citations: The orc2-1/ORC2 ratio of each examined origin was plotted against the apparent Kd determined from the EMSA experiments (Figure 1E). Many of these origins followed the yeast ORC-DNA paradigm in that their affinities for ORC in vivo correlated with their ORC-DNA affinities measured in vitro. For example, 11 of the 20 ‘high in vivo affinity’ origins bound ORC with relatively low Kds in vitro, indicating a high-affinity ORC-DNA interaction. We will refer to origins within this class as DNA-dependent because their high affinity for ORC in vivo correlated with a strong ORC-DNA interaction in vitro. Conversely, 14 of the 18 ‘low in vivo affinity’ origins bound ORC with relatively high Kds (apparent Kd>4x the Kd for the ORC-ARS317 complex), indicating a low affinity ORC-DNA interaction. We will refer to origins within this class as Weak because their low affinity for ORC in vivo correlated with their weak ORC-DNA interaction in vitro. ARS1, the representative ‘moderate in vivo affinity’ origin bound ORC with a moderate Kd, as expected from previous work [41], [51].
As described above, for many origins tested, the ORC-DNA interaction was a good predictor of an origin's ‘in vivo affinity’ for ORC. However, there were several exceptions. For example, five ‘high in vivo affinity’ origins showed unexpectedly weak ORC-DNA interactions in vitro. In fact, for three of these (ARS105, ARS219.5 and ARS1528) the confirmed ORC binding site bound ORC so poorly in vitro that an apparent Kd could not be determined. We will refer to origins that behave in this way as ‘chromatin-dependent’ because their high affinity for ORC in vivo did not correlate with their weak ORC-origin DNA interaction measured in vitro suggesting that features extrinsic to the ORC binding site—i.e. ‘chromatin’—were required for normal levels of ORC binding to these origins. The origin classifications defined above—DNA-dependent, ‘chromatin-dependent’ and Weak—will be used throughout this manuscript. Three of the 18 ‘low in vivo affinity’ origins also deviated from the paradigm's prediction, binding ORC more tightly in vitro than predicted, suggesting ‘chromatin’ played a negative role in ORC binding. While origins of this type were not pursued further in this study, they might be a consequence of transcription as reported previously [54], [55]. We note also that for both the ‘high and low in vivo affinity’ origin groups, there is a continuum in apparent Kd values. For example, a few origins in both groups bound ORC with moderate Kds in vitro, similar to that of ARS1, which has a ‘moderate in vivo affinity.’ We referred to these origins as complex to indicate that they use some combination of intrinsic ORC-DNA interactions and extrinsic ORC-‘chromatin’ interactions for normal levels of ORC binding in vivo.
Extending comparisons of in vivo and in vitro ORC-origin interaction strengths genome-wide
To extend this approach to yeast origins on a genome-wide scale, we performed an EMSA using purified ORC (at 0.3, 3.0 and 30 nM) and purified, sheared yeast genomic DNA. We then screened the population of ORC-bound fragments by hybridizing the amplified and labeled DNA pool to tiled arrays (genomic EMSA or gEMSA) (Figure 2A). For each sample, ORC bound fragments were identified as binding peaks with a P-value selected to maximize the number of confirmed origins identified and to capture weaker ORC-DNA interactions (P-value>/ = 10−5). As a first step to determining the effectiveness of this approach, we examined the behavior of those origins we had analyzed with EMSAs (Figure 1E). Specifically, we examined the ORC binding signals generated by the gEMSA at each ORC concentration for each of these origins. Peaks associated with origins ARS422 and ARS822 are shown in Figure 2B, demonstrating how the gEMSA data recapitulated the origin-specific EMSA data for these ARSs.
Fig. 2. Extending comparisons of in vivo and in vitro ORC-origin interaction strengths genome-wide. 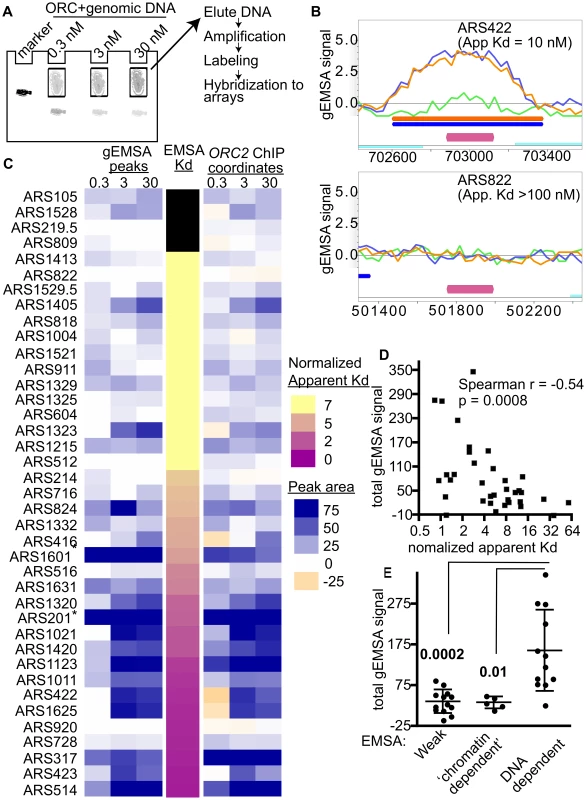
(A) Outline of genomic EMSA: Purified ORC at the indicated concentrations was incubated with 0.5 pM of purified sheared yeast genomic DNA. After the reactions reached equilibrium, ORC-bound and -unbound DNA fragments were separated by native gel electrophoresis. To mark the position of ORC-DNA complexes in the gel, a control reaction was performed with a specific radiolabeled origin-containing fragment (184 bp marker). (B) ORC binding in the gEMSA was visualized in MochiView [88]. Screen shots are shown for two origins that were also examined by origin-specific EMSAs in Figure 1. The signals for each feature (oligo) on the array are indicated on the y-axis and the chromosomal coordinates are indicated on the x-axis. A magenta bar marks the coordinates comprising the oriDB annotated ARS. The signal strength for the 0.3 nM (green), 3 nM (blue) and 30 nM (orange) ORC concentrations are plotted and the coordinates corresponding to the ChIPotle called peaks at each concentration are shown by the correspondingly colored bars. The ORC-ARS422 complex had an apparent Kd of 10 nM by origin-specific EMSAs (Figure 1E and Table 1), while the ORC-ARS822 complex had an apparent Kd estimated to be >100 nM. (C) Heat maps depicting the ORC-origin binding strengths in the gEMSA for the 39 origins examined in Figure 1E or their normalized Kds (narrow map, center) are shown. gEMSA ORC-origin binding strength was defined two different ways, depending on the heat map. The heat map to the left (gEMSA peaks) defined the ORC-origin complex as the area of the peak that was called by the ChIPOTle program analysis of the gEMSA data in each independent array (ORC concentration shown at top of columns). If no binding peak was called, the ORC-origin binding strength was assigned a “0” value and colored white. The heat map on the right (ORC2 ChIP coordinates) defined the ORC-origin complex in the gEMSA as the coordinates that comprised the ChIPOTle called ORC-origin peak for the previously published ORC2 ChIP array [51]. Thus in this map, DNA regions that were depleted in the experimental sample relative to total DNA were assigned a negative value and colored beige. The two different approaches produced similar conclusions, suggesting that for most of these origins the gEMSA peak corresponded well to the in vivo ORC-origin peak. (D) Correlation analysis of gEMSA-derived binding strength versus normalized apparent Kds as determined by origin-specific EMSAs in Figure 1E. The normalized apparent Kd values were calculated by dividing apparent Kds by the apparent Kd for ARS317 (HMR-E). The gEMSA-derived binding strength was a quantitative expression of the data shown in the right map in © as described above. Specifically, the gEMSA signal for each ORC concentration for each origin was summed to give a “Total gEMSA signal.” Thus this value simply expressed a value for total ORC-origin complex formation observed across the ORC titration. Our analysis included 35 origins for which apparent Kd values could be determined. A Spearman correlation coefficient and P-value are indicated. (E) We grouped origins into classes based on their EMSA derived apparent Kd values and their sensitivity to the orc2-1 mutation for ORC binding in vivo. We compared the Total gEMSA signal for each of these three groups: weak orc2-1s (apparent Kd>5x ARS317, n = 13), weak orc2-1r (apparent Kd>10x ARS317, n = 5), and tight orc2-1r (apparent Kd</ = 3x ARS317, n = 12). T tests were used to determine the significance of the difference between the weak or ‘chromatin-dependent’ groups and the DNA-dependent group. P-values are indicated in figure. Two different heat maps were generated to represent the strength of ORC-origin binding produced in the gEMSA for the 39 origins examined by EMSAs (Figure 2C). In one heat map the ORC-origin binding strength was defined as the area of the gEMSA peak called by ChIPOTle that overlapped the annotated ARS (Figure 2C, left panel labeled ‘gEMSA peaks’). The peak area was the sum of the signals for each feature (oligo) on the array that was included in the ChIPOTle-called peak. In the second heat map, ORC-origin binding strength was the sum of the signals of each oligo in the array that corresponded to the coordinates of the in vivo ORC binding peak from the ORC ChIP-chip experiment (Figure 2C, right panel labeled ‘ORC2 ChIP coordinates’) [51]. The two approaches to defining ORC-origin binding strength in vitro produced similar results. Finally, these two representations of the gEMSA data were compared to origin-specific EMSA derived Kds (normalized to the Kd for the ORC-ARS317 interaction; third narrow centrally positioned heat map). The 39 origins were ranked from weakest to strongest for ORC binding based on their normalized apparent Kds.
These analyses revealed that the gEMSA data recapitulated the origin-specific EMSA data well though not perfectly: in general the weakest ORC binding sites tested in vitro by EMSAs were associated with weaker binding signals in the gEMSA and vice versa (Figure 2C). Correlation analysis of the apparent Kds determined by EMSAs and the Total gEMSA signal (i.e. the sum of gEMSA ORC-origin binding strength for each ORC concentration) revealed significant co-variation (Spearman r coefficient = −0.54 and a P-value = 0.0008; Figure 2D). However, eight of the 39 origins examined did not produce gEMSA data that matched the predictions based on the EMSA-derived Kds. Two of these were telomeric ARSs that consistently produced broad peaks at all ORC concentrations tested. We removed core-X telomeric ARSs from further consideration in all subsequent bioinformatics analyses for this and other reasons. Three of these origins produced tighter binding in the gEMSA than predicted by the origin-specific EMSAs (ARS1405, ARS1323 and ARS824). For these origins we noted that the in vivo and in vitro ORC binding peaks were somewhat off set, suggesting that the binding we observed in the gEMSA might involve a site to which ORC may not normally have access in vivo. Conversely, three ARSs bound ORC more weakly than expected in the gEMSA (ARS516, ARS728, ARS920) based on their Kds. It is possible that some DNA fragments do not elute efficiently from the gel matrix or were underrepresented for some other technical reason. Further refinements are ongoing and will address these possibilities. Regardless, overall the gEMSA data recapitulated the origin-specific EMSA derived binding strength for 31 out of 39 origins (79.5%), indicating that the approach could be useful for identifying yeast origins that are ‘chromatin-dependent’ for ORC binding.
Comparison of ORC-DNA affinities from EMSAs with orc2-1/ORC2 ratios of the same origins revealed a cluster of origins that relied on extrinsic (i.e. ‘chromatin’) factors for efficient ORC binding in vivo (Figure 1E). We examined whether the gEMSA recapitulated this property of these origins. The Total gEMSA signal for each origin (y-axis) was plotted for three distinct groups of origins classified by their behavior in the origin-specific EMSAs as Weak, ‘chromatin-dependent’ or DNA-dependent (Figure 2E, see also Figure 1E and associated text for origin classification information). These analyses also revealed that the gEMSA ORC-origin binding strength captured the expected differences between ‘chromatin-dependent’ and DNA-dependent ORC binding mechanisms. In particular, as predicted based on origin-specific EMSAs, putative ‘chromatin-dependent’ and Weak origins bound ORC with similar (low) ‘affinities’ in the gEMSA and were different from the gEMSA ORC-binding strengths observed for DNA-dependent origins. Therefore the gEMSA approach depicted the expected differences in ORC-DNA interactions at many different origins.
The gEMSA showed ORC-DNA selectivity
The gEMSA identified a large number of ORC binding peaks at the selected stringency (P value>/ = 10−5) (Figure 3A and Figure S1 for maps of chromosome III and VI). This fact was not surprising given that no competitor DNA was used in the gEMSA ORC binding reactions and that the genome contains 4300 matches to the ORC binding site compared to only ∼400–700 bound by ORC in vivo. However, it was important to assess the selectivity of the gEMSA for predicted ORC binding sites. Figure 3A shows the overlap between the gEMSA peaks and either all origins (including Likely and Dubious origins) annotated in the OriDB (n = 740) or the total number of annotated yeast genes (n = 6607). A high proportion of annotated origins overlapped with the gEMSA peakes (69%, 72% and 76% for the 0.3 nM, 3 nM and 30 nM ORC arrays, respectively). While this analysis does not account for peak size or degree of overlap between peaks and origins, these data suggest that a majority of annotated origins were bound by ORC. In contrast, only 43%, 41% and 44% of annotated genes overlapped with the gEMSA peaks generated by the 0.3 nM, 3 nM and 30 nM ORC concentrations, respectively. We note that in S. cerevisiae, intergenic regions are small and therefore many origins are annotated as overlapping with genes simply for that reason. Furthermore, there are multiple bona fide ORC-binding sites within yeast genes. Thus some overlap with genes was not unexpected. Next we examined the sequence annotations overlapping the gEMSA peaks in comparison to the genome (Figure 3B). This analysis showed that while both genes and ORC-ORFs (protein coding regions that associate with ORC in vivo [51]) were depleted in the gEMSA relative to the genome, two classes of origins, likely and confirmed, were enriched. Interestingly, confirmed origins showed the greatest level of enrichment (∼4 fold over genome) suggesting that ORC bound more of these loci through specific ORC-DNA interactions compared to origins in either the dubious or likely categories. Together Figures 3A and 3B indicate that the gEMSA captured expected selectivity of yeast ORC-DNA interactions.
Fig. 3. The genomic EMSA showed ORC-DNA selectivity. 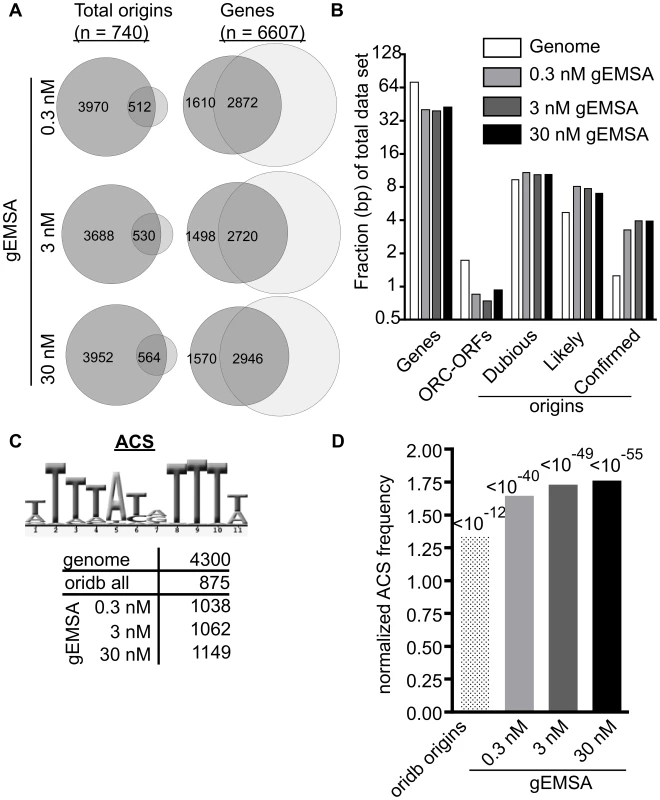
(A) Venn diagrams showing overlap between the complete set of annotated origins from oriDB or the complete set of yeast genes (annotated as of March 2011 on SGD) and the peaks called by ChIPotle (P value = 10−5) for the gEMSA data at 0.3, 3 and 30 nM ORC. (B) The fraction of base pairs in the yeast genome (y-axis) or in the gEMSA data set are shown for the genomic loci indicated on the x-axis. (C) Number of matches to the 11 bp ORC binding site consensus-motif (ACS) that were found in the indicated data sets. (D) The enrichment of an ACS motif (shown in (C)) was determined by plotting the normalized frequency (normalized to the frequency that the same motif is found within the whole genome) that an ACS match meeting a LOD cut-off of 70% was found within all of the base pairs comprising the relevant data set (x-axis). P-values for significance of enrichment indicated above bars. We reasoned that if ORC was showing DNA sequence specificity in the gEMSA data, then an optimal ORC binding site motif should be enriched regardless of whether that motif actually existed within a bona fide chromosomal origin because ORC was free to sample all genomic DNA in these experiments. Therefore we queried the relevant data sets for the 11 bp ACS (ARS consensus site), a conserved motif within the ORC binding site essential for origin function (Figure 3C). We also queried the gEMSA data sets for more stringently defined ORC binding sites as shown and described in Figure S2. The genome (12.1 Mbps) contained 4300 matches to the ACS while the OriDB (1.9 Mbps) contained 875 matches, a ∼1.3 fold increase in motif frequency. The ORC gEMSA datasets had a ∼1.6-fold increase in the 11 bp ACS motif frequency over the genome (1038, 1062, and 1149 matches to this ACS at 0.3 nM (1.8 Mbps), 3 nM (1.8 bps) and 30 nM (1.9 Mbps) ORC concentrations, respectively). Even greater enrichment was seen for more stringently defined matches to the ORC binding site (Figure S2). Thus as predicted if ORC-DNA sequence specificity were a greater driving force behind ORC binding in the gEMSA compared to ORC binding in vivo, DNA sequences preferred by ORC were more enriched in the gEMSA data sets than in the OriDB (Figure 3D). These data provide compelling evidence that chromatin regulates ORC-origin binding both positively and negatively.
We also analyzed the gEMSA data for the presence of motifs representing the binding sites of 89 different sequence-specific DNA binding proteins to test whether other elements were enriched by ORC, either because ORC might be capable of binding these sequences directly or because these motifs are often associated with ORC binding sites (Table S1). Motifs for only five of the 89 proteins met the initial cut-off (LOD>/ = 60%: Mata2, Nhp6a, Nhp6b, Sfl1 and Sum1), and only the Sum1 motif met a P-value cut-off of 10−5 at more stringent LOD scores (>/ = 80% LOD). The Sum1 motif is AT-rich as is the ORC binding site, and, perhaps relevantly, Sum1 has been implicated in origin regulation [56]–[58]. Regardless, the enrichment of origins and sequences preferred by ORC compared to other motifs indicated that the gEMSA was capturing ORC's known sequence specificity. Thus, together, the data in Figures 2 and 3 indicated that the gEMSA captured ORC's affinity and specificity for many origins.
Features extrinsic to the ORC binding site dominated ORC-origin binding at ∼40% of yeast origins that bound ORC with high affinity in vivo
Our gEMSA data provided a measure of the in vitro ORC-DNA bining strength that we could compare to the in vivo ORC-origin binding strength (orc2-1/ORC2 ratio) from the ChIP-chip experiment. In figure 4A origins are ranked from lowest (top) to highest (bottom) in vivo affinity (i.e. orc2-1/ORC2 ratios), and their corresponding in vitro binding strengths (i.e. gEMSA peak signals for each ORC concentration) are shown. Data for ARS1 (a.k.a. ARS416) and the DNA-dependent origin ARS317 (a.k.a. HMR-E silencer) are indicated. As was observed in the analysis of the 39 origins in Figure 2C, in general lower in vivo affinities (yellow; low orc2-1/ORC2 ratios) were generally associated with weaker in vitro binding (low gEMSA signals), while higher in vivo affinities (purple; high orc2-1/ORC2 ratios) were associated with stronger in vitro binding (stronger gEMSA signals). The co-variance of the orc2-1/ORC2 ratios and the gEMSA data had a Pearson r coefficient of 0.20 (P-value = 0.002) indicating a positive relationship, as expected based on the proof-of-principle experiments in Figures 1 and 2 (Figure S3).
Fig. 4. Features extrinsic to the ORC binding site dominated ORC-origin binding at many origins. 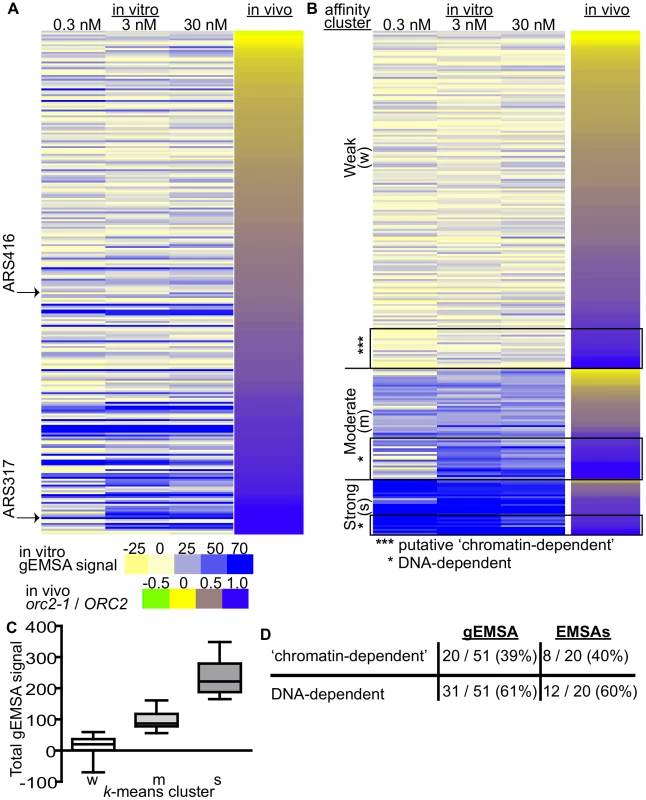
(A) Heat map of peak areas overlapping the 261 confirmed origins identified in the gEMSAs. The origins are ranked by their orc2-1/ORC2 ratios (in vivo affinity, narrow heat map). Positions of ARS1 (ARS416) and ARS317 are indicated. (B) The gEMSA data in (A) were clustered using k-means clustering. Within each of the three clusters, the origins were ranked by their orc2-1/ORC2 ratios. The boxed origins in the weak cluster (W) were called ‘putative chromatin-dependent’ origins. The origins for the DNA-dependent cluster were obtained from the Moderate (M) and Strong (S) clusters (n = 31). Core-X telomeric ARSs within this group were removed prior to all subsequent computational analyses so that for the working set of DNA-dependent origins n = 20. (C) Box-and-whisker plots of the total gEMSA signal for each of the three clusters in (B). (D) Based on analyses of the gEMSA data in (C), 39% of the ‘high in vivo affinity’ origins (orc2-1/ORC2>/ = 0.8) were ‘chromatin-dependent’, and 61% were DNA-dependent. These percentages agreed well with those calculated from the 39 origin-specific EMSA Kds shown in Figure 1E. We clustered the 261 origins based on their in vitro ORC-origin binding strength into three distinct clusters: Weak, Moderate and Strong (Figure 4B and 4C). Interestingly, the majority of confirmed origins (n = 176) were in the Weak cluster. We reasoned that the origins that might rely most substantially on interactions extrinsic to ORC-DNA for ORC binding were those that bound ORC with a high affinity in vivo (purple; orc2-1/ORC2 ratios>/ = 0.8) within the Weak in vitro (gEMSA) cluster (*** in Figure 4B). There are 51 origins with orc2-1/ORC2 ratios>/ = 0.8, and of these 20 were found to have weak (gEMSA) in vitro ORC-DNA affinity. Importantly these twenty origins included the five previously categorized as ‘chromatin-dependent’ for ORC binding (Figure 1E), as well as one defined as ‘complex’ (ARS1332) (Table 2). Only one origin categorized as DNA-dependent, ARS516, (Figure 1E) was also found within this group. ARS516 has a Kd of 28 nm that is close to the arbitrary cut-off used to define ‘chromatin-dependent’ origins. Thus ARS516's appearance in this group was not completely surprising. Importantly, ARS516 was the only origin classified by EMSA as DNA-dependent (Figure 1E) that fell within this putative ‘chromatin-dependent’ origin group based upon the gEMSA data. In addition, not a single origin within the EMSA-defined ‘chromatin-dependent’ group fell within the DNA-dependent group defined by the gEMSA. These observations suggest good agreement between the origin-specific EMSA and gEMSA data, as we would expect based on the analyses in Figure 2. Seven of the newly identified putative ‘chromatin-dependent’ origins had confirmed ORC binding sites, and therefore we measured ORC's interaction with these sites directly by origin-specific EMSAs (Table 2). Each of these seven origins bound ORC with Kds>4x ARS317, validating their placement in this ‘chromatin-dependent’ category. Thus the gEMSA approach successfully identified origins where stable binding of ORC in vivo likely relied on features extrinsic to the paradigmatic ORC-DNA interface. Based on these analyses, ORC used interactions with factors extrinsic to the established ORC binding site for stable association with up to 40% of yeast origins (Figure 4E).
Tab. 2. ‘Chromatin-dependent’ origins identified based on clustering of the gEMSA data. 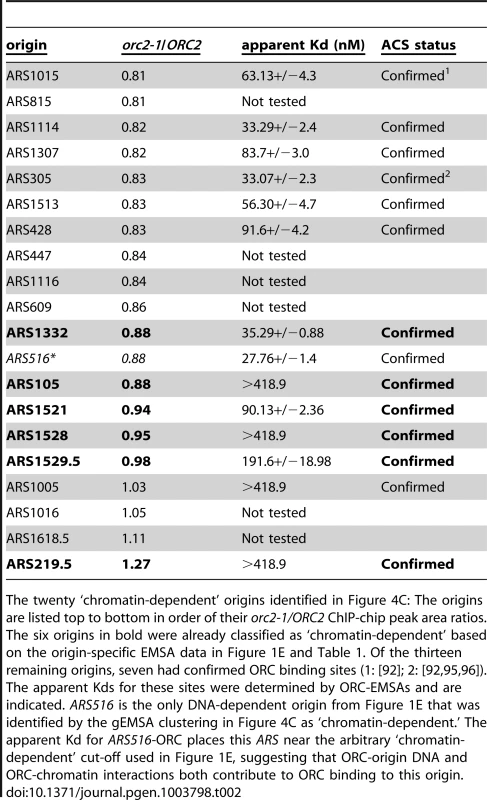
The twenty ‘chromatin-dependent’ origins identified in Figure 4C: The origins are listed top to bottom in order of their orc2-1/ORC2 ChIP-chip peak area ratios. The six origins in bold were already classified as ‘chromatin-dependent’ based on the origin-specific EMSA data in Figure 1E and Table 1. Of the thirteen remaining origins, seven had confirmed ORC binding sites (1: [92]; 2: [92], [95], [96]). The apparent Kds for these sites were determined by ORC-EMSAs and are indicated. ARS516 is the only DNA-dependent origin from Figure 1E that was identified by the gEMSA clustering in Figure 4C as ‘chromatin-dependent.’ The apparent Kd for ARS516-ORC places this ARS near the arbitrary ‘chromatin-dependent’ cut-off used in Figure 1E, suggesting that ORC-origin DNA and ORC-chromatin interactions both contribute to ORC binding to this origin. Local nucleosome architecture and its dependence on ORC around selected groups of origins
The experiments discussed above defined two classes of origins that had similar affinities for ORC in vivo but had different ORC-DNA interaction strengths in vitro. To address whether local chromatin structure might be associated with these effects, we examined the average nucleosome signals relative to the ORC binding site at twenty DNA-dependent, and eighteen ‘chromatin-dependent’ origins (Figure 5A) [59]. For the DNA-dependent group, we excluded the 11 core-X telomere-associated origins for these and all subsequent analyses because they contain repetitive sequences and telomeric chromatin can affect origin function [33]. As controls we also examined 20 randomly selected origins and 20 origins from the Weak category (i.e. bound ORC poorly in vitro and in vivo). These plots revealed that each origin group was similar in having a nucleosome depleted region (NDR) 3′ of the ORC binding site, as expected. For the ‘chromatin-dependent’ origins the NDR was on average slightly larger and the flanking nucleosomes more pronounced (i.e. less ‘fuzzy’ suggesting more stable positioning) than those surrounding the DNA-dependent origins or the control groups. In addition, ‘chromatin-dependent’ and DNA-dependent origins showed differential association in terms of the gene orientation that surrounded them, consistent with local differences in chromatin environments that might result from differences in local transcription associated activities (e.g. transcription termination versus initiation) (Figure S4).
Fig. 5. Local nucleosome architecture and its dependence on ORC around selected groups of origins. 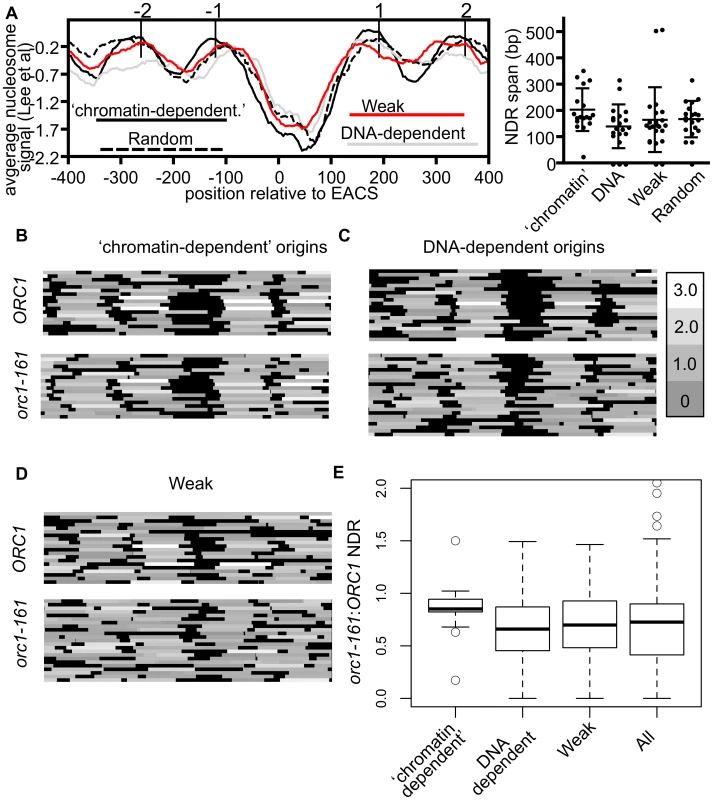
(A) Average nucleosome signals (left panel) and size of NDRs relevant to the indicated origins. Box-and-whisker plots showing median, lower and upper quartiles (box edges) and minimums and maximums excluding outliers for the NDR size of the origins are shown (right panel). These analyses used nucleosome data from [59]. (B–D) Nucleosome heat maps shown around the indicated origins in G1-phase in wild-type cells or orc1-161 cells shifted to the non-permissive temperature for orc1-161 [44] [63]. (B) ‘Chromatin-dependent’, (C) DNA-dependent, (D) and Weak origins. (E) Box-and-whisker plots indicating effects of removing ORC (orc1-161) on size of the NDR for ‘chromatin-dependent’, DNA-dependent, Weak and all 261 origins (All) examined in this study. The difference in means between the ‘chromatin-dependent’ and DNA-dependent origins had a P-value = 0.035. While the nucleosome positioning analyses revealed that local nucleosome positioning was, in general, similar between the origin-groups defined here, they also revealed that ‘chromatin-dependent’ origins were distinct from Orc1BAH-dependent origins defined in a previous study [43]. Specifically, Orc1BAH-dependent origins require the bromo adjacent homology (BAH) domain present on the N-terminus of Orc1 for normal levels of ORC binding in vivo [43]. The Orc1BAH domain is a nucleosome-binding module [60], [61], and origins that require the Orc1BAH domain for ORC binding have a distinctive local nucleosome architecture including a smaller NDR compared to Orc1BAH-independent origins and a striking shift of the −2 and −1 nucleosomes toward the ORC binding site [43]. Thus there were clear differences between the local nucleosome architectures of Orc1BAH-dependent origins and ‘chromatin-dependent’ origins. Consistent with these observations, ‘chromatin-dependent’ and Orc1BAH-dependent origins comprised distinct groups (Figure S5).
Both intrinsic (DNA sequence) and extrinsic factors (e.g. nucleosome remodelers, sequence-specific DNA binding proteins) help define the average nucleosome occupancy profiles that exist in vivo [62]. For example, the sequence of the ORC binding site tends to exclude nucleosomes, whereas other sequences, such as nucleosome positioning elements (NPEs) favor nucleosomes [44]. To ask whether ‘chromatin-dependent’ and DNA-dependent origins might differ in this respect, in vitro and in vivo nucleosome occupancy profiles were assessed at these origin groups and two additional groups, Weak origins and randomly selected origins (Figure S6). For ‘chromatin-dependent’ and Weak origins a difference between in vitro and in vivo nucleosome positioning for the +1 nucleosome was evident compared to DNA-dependent and randomly selected origins, which were more similar to each other. These data suggest that extrinsic factors are more relevant to positioning the +1 nucleosome, in particular, at ‘chromatin-dependent’ and Weak origins relative to DNA-dependent origins.
Experimental evidence from both in vivo and in vitro studies reveal that ORC binding to its sites within replication origins helps position neighboring nucleosomes and maintain an NDR [44]. Thus in the absence of ORC, nucleosomes normally positioned on either side of the origin encroach toward the origin reducing the size of the NDR. To determine whether ‘chromatin-dependent’ and DNA-dependent origins differed in their requirement for ORC to position flanking nucleosomes, we compared nucleosome positioning in wild-type cells to mutant cells where ORC-origin binding was abolished (orc1-161 cells in G1 phase incubated at the non-permissive temperature) [44], [63] (Figure 5B–E). At ‘chromatin-dependent’ origins, loss of ORC binding had, on average, a comparatively small effect on local nucleosome positioning compared to all other origin groups analyzed. In particular, the positions of the −1 and +1 nucleosomes did not change substantially at many of the ‘chromatin-dependent’ origins, resulting in only modest reductions in the average size of the NDR and a tight distribution around the average (Figure 5E). In contrast, at DNA-dependent and Weak origins loss of ORC binding resulted in more dramatic changes in local nucleosome positioning, particularly the positioning of the −1 and +1 nucleosomes, resulting in a larger reduction in the size of the NDR surrounding these origins. Both of these groups behaved more similarly to all origins compared to the ‘chromatin-dependent’ group. These data suggest that ‘chromatin-dependent’ origins do not rely as heavily on ORC binding compared to other origins to establish the normal NDR.
In summary, the local nucleosome architecture of ‘chromatin-dependent’ and DNA-dependent origins, while similar, relied on different mechanisms to establish this architecture. Furthermore ‘chromatin-dependent’ origins were distinct from the previously defined group of Orc1BAH-dependent origins. These data were consistent with the hypotheses that ORC recognized a distinct chromatin-environment at ‘chromatin-dependent’ origins and that more than one type of ORC-chromatin interaction influenced origin selection by ORC in vivo. Plasmid loss assays of selected ‘chromatin-dependent’ and DNA-dependent origins suggested that the former origins were more sensitive to native, local chromatin configurations (Figure S7). This observation is consistent with the idea that local chromatin configurations reflect functional differences between ‘chromatin-dependent’ and DNA dependent origins. Therefore, while an ORC binding site motif can be found in both groups of origins (Figure S8), we found that there were differences in the local chromatin architecture. Collectively these data suggested that differences between the ORC sites and local chromatin structure were relevant to the different modes of ORC-origin binding.
Association between ORC-origin binding mechanisms and the time of origin activation
In a previous study, we showed that the high-affinity ORC-DNA interaction at ARS317 contributed to its late-activation time and inefficiency as origin [41]. To ask whether this observation remains relevant when many origins are examined and to address whether ORC-origin binding mechanisms, as opposed to ORC-origin binding affinity per se, might be relevant to this origin regulation, the mean replication time (Trep values) acquired from a copy-number based analysis of DNA replication, was assessed for several groups of relevant origins [64] (Figure 6A). We observed a difference between the mean Treps for ‘chromatin-dependent’ and DNA-dependent origins. In general, this difference was observed with additional data sets that measured origin replication time directly or other properties linked to origin replication time (Figure S9). Because ‘chromatin-dependent’ ORC binding sites had weak intrinsic ORC-origin DNA interactions, one possibility was that this difference in timing in Figure 6A could be explained by weak ORC-origin binding alone. Therefore we also examined the Trep times for a comparable group of Weak origins. In contrast to ‘chromatin-dependent’ origins, origins within this Weak class were distributed over the timing spectrum and produced a mean Trep time similar to that of the DNA-dependent class of origins. We note that for origins within this Weak group, the sensitivity of our current assays did not allow us to distinguish between DNA-dependent and ‘chromatin-dependent’ ORC binding mechanisms, and thus both types of origins may be present in this Weak class. Regardless, these data indicated that enrichment for early-firing origins was a distinct property of the ‘chromatin-dependent’ group.
Fig. 6. Association between ORC-origin binding mechanisms and the time of origin activation. 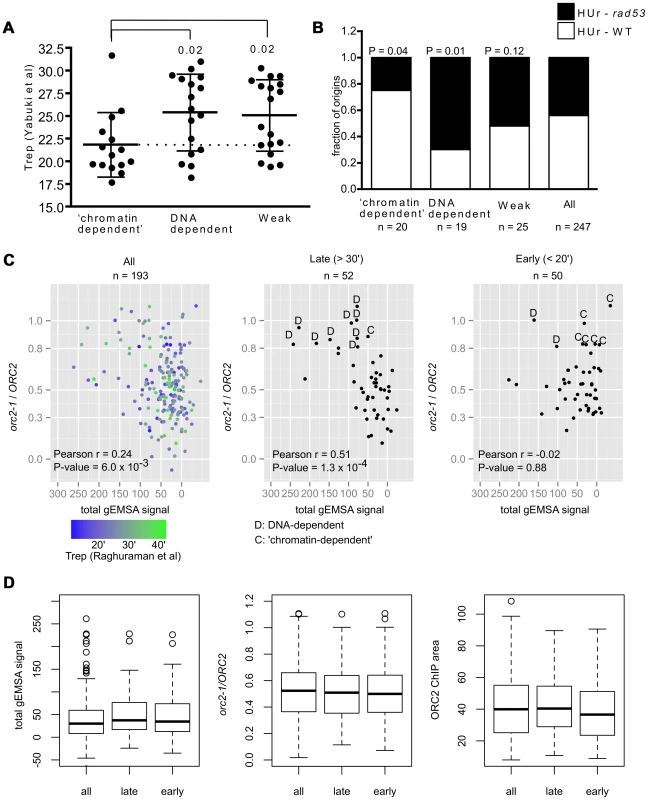
(A) Vertical scatter plots of Trep values for three different classes of origins [64]. Dotted line extends from the mean for ‘chromatin-dependent’ origins. Similar results were observed with other data sets (Figure S9). P-values for significance of differences in the means between DNA-dependent or Weak and ‘chromatin-dependent’ origins are indicated. (B) The ability of origins in each of these groups to fire in the presence of HU in either wild-type cells (HUr-WT, white) or only in rad53 mutant cells (HUr-rad53-1, black) is indicated in bar graphs. Data were from [66], [67]. P-values for the significance of the differences in distributions between selected origin groups and ‘all’ origins are indicated. (C) The orc2-1/ORC2 ratios (y-axis, in vivo binding ORC-origin binding strength) were plotted against the Total gEMSA signal (x-axis, in vitro ORC-origin binding strength) for all origins common to this study's working data set and the Meselson-Stahl timing data set [68], for the 52 latest-firing origins (Trep>30 minutes), and the 50 earliest-firing origins (Trep<20 minutes). (D) Distributions in binding strength (gEMSA signals summed over all three concentrations of ORC), in vivo binding strength (orc2-1/ORC2 ratio), and Wild-type (ORC2) ChIP peak area for all 193 origins in (C), the 52 latest origins in (C) and the 50 earliest origins in (C). A replication origin's sensitivity to the ribonucleotide reductase inhibitor hydroxyurea (HU) correlates strongly with origin activation time and is often used to distinguish between origins that activate early and late in S-phase. In general early-firing origins are resistant to HU, firing efficiently even in its presence, whereas late-firing origins are sensitive to HU, failing to fire in its presence unless the Rad53-dependent checkpoint is inactivated [65]. Thus Rad53 function prevents origins that normally fire later in S-phase from activation in the presence of HU. Two genome-wide studies have measured origin activation in the presence of HU in wild-type and rad53 mutant cells [66], [67]. We used these data to determine the origin activation behavior of our various origin groups in the presence of HU. ‘Chromatin-dependent’ and DNA-dependent origins showed different origin activation behavior in the presence of HU (Figure 6B). Specifically, fifteen of the 20 ‘chromatin-dependent’ origins were activated in the presence of HU in wild-type cells (HUr-WT) while five required inactivation of Rad53 (rad53) for their activation under these conditions (HUr-rad53-1). The DNA-dependent origins behaved in the opposite manner; only four of the twenty activated in the presence of HU in wild-type cells, while fifteen required inactivation of Rad53. The control group of ‘Weak’ origins distributed between the two types of activation in HU, producing a distribution similar to that observed for all origins, suggesting weak ORC-DNA interactions per se could not explain the skew observed for ‘chromatin-dependent’ origins. Thus based on both direct measurements of replication time during S-phase and HU-sensitivity, ‘chromatin-dependent’ origins were functionally different from DNA-dependent origins even though both groups bound ORC with similar strengths in vivo. Thus an origin's high-affinity for ORC per se was not strongly associated with its behaving like ARS317 in terms of origin activation time. Rather, high-affinity binding achieved by sequence-specific ORC-origin DNA interactions was strongly associated with an origin behaving like ARS317.
The association between origin activation time and differences in ORC-origin binding mechanisms was striking. Because we defined in vivo binding affinity using the orc2-1/ORC2 ratio for each origin, one concern was that the origin groups might be differentially affected by signal to noise ratios and thus that the origin activation timing effects we observed resulted from a flaw in the classification method. Therefore we performed additional control analyses (Figures S10 and S11). First, we compared the orc2-1/ORC2 ratios of all origins compared to the ORC2 ChIP-chip peak areas to determine whether origins with small ratios were strongly biased to small ORC2 peak areas indicating that our ‘high-affinity in vivo’ origins might simply result from low signal to noise issues (Figure S10A). This analysis indicated that reduced orc2-1/ORC2 ratios could be observed across the range of peak sizes and that high orc2-1/ORC2 ratios were not clustered among the smallest peaks. Moreover, the ORC2 peak sizes of ‘chromatin-dependent’ and DNA-dependent origins overlapped for the majority of origins in both groups. If anything, the largest skew in ORC2 peak size toward small peaks was for the ‘Weak’ origin control group that showed no obvious skew in terms of origin activation time relative to ‘all’ origins in our data set (Figure S10B and Figures 6A and 6B). We did note however that four ‘chromatin-dependent’ origins generated ORC2 peak areas that were smaller than any DNA-dependent origins (Figure S10A) and therefore we removed these four origins from our origin activation time analyses (Figure S11). Removal of these four smallest peaks did not affect our conclusions. These important controls provided evidence that the orc2-1/ORC2 ratio method for classifying origins based on their relative ‘in vivo affinity’ did not bias the ‘chromatin-dependent’ origins to smaller and thus noisier peaks, suggesting that their enrichment for origins activated in early S-phase was not an artifact of the classification system.
Recent studies reveal that Forkhead transcription factors, Fkh1 and Fkh2 (Fkh1/2) regulate the origin activation time of many origins by an as yet undetermined mechanism that may involve higher-order chromosomal architecture and clustering of origins within the nucleus as well as local mechanisms [22], [26]. In particular, many early-firing origins require Fkh1/2 for their early activation time. Thus we expected that ‘chromatin-dependent’ origins, being enriched for early-firing origins, and Fkh1/2 activated origins would show some relationship, and indeed ‘chromatin-dependent’ origins were enriched for Fkh1/2 activated origins compared to DNA-dependent origins or all origins in the genome (Figure S9D) although there were early-activated origins in both groups that were distinct (Figure S12).
The analyses above revealed a striking association between the mode of ORC binding to an origin in vivo and the timing of origin activation during S-phase. However, these analyses necessarily focused on a relatively small group of origins that had a high-affinity in vivo and that we could therefore assign either DNA-dependent (n = 20) or ‘chromatin-dependent’ (n = 20) mechanisms for ORC binding. Of course many more replication origins fire either very early (Trep<20′; n = 50) or very late (Trep>30′; n = 52) in S-phase that were not represented in our groups that were formed using an arbitrary orc2-1/ORC2 ratio cut-off of 0.8. Therefore we probed this association further by extending our analyses to origins for which Treps were determined in a genome-wide Meselson-Stahl experiment [68]. This data set was used because a larger number of origins were shared between it and the working data set in this study. All origins shared by the two data sets (n = 193), all of the latest firing (Trep>30′, n = 52) and all of the earliest firing (Trep<20′, n = 50) origins were plotted on separate graphs in which in vivo ORC-origin binding strength (orc2-1/ORC2 ratio, y-axis) and in vitro ORC-origin binding strengths (total gEMSA signal, x-axis) were compared (Figure 6C). As expected, when all of the origins were plotted, a positive correlation was observed between in vivo and in vitro ORC-origin binding strengths (Figure 6C, left panel, r = 0.25, P = 0.006). The 52 latest-firing origins (Trep>30 minutes) showed an even greater positive relationship than all origins (r = 0.51, P = 0.0001), indicating that, as a group, these origins followed the ORC-origin DNA paradigm more closely than origins in general. In contrast, the 50 earliest firing origins (Trep<20 minutes) failed to show any relationship between in vivo and in vitro binding strength (r = −0.02, P = 0.88), indicating that, as a group, these early-firing origins did not follow the ORC-origin DNA paradigm at all. We note however that many origins among the latest and earliest firing groups lie in a similar region of the plot, suggesting that they use similar mechanisms for ORC-origin binding in vivo. Indeed, by comparing the average in vivo and in vitro affinities and ORC2 peak sizes for all of the origins examined in Figure 6C, it was evident that the latest and earliest firing origins were quite similar in terms of these values (Figure 6D). However, we note that the current resolution of the ORC-origin affinity measurements in vivo and in vitro limit our ability to definitively assign ORC-origin binding mechanisms to many of the origins that fall within a portion of the graph (orc2-1/ORC2 ∼0.3–0.6 and total gEMSA signals <50). Nevertheless, as separate groups, the latest and earliest firing origins in the genome showed different relationships between their in vivo and in vitro binding affinities as plotted in Figure 6C. All together the data in Figure 6 provided evidence that the paradigmatic ORC-DNA interaction was a more substantial component of ORC-binding to many late-firing origins, whereas early-firing origins showed a greater dependence on ‘chromatin.’ Furthermore, the mode of ORC-origin binding (i.e. DNA-dependent versus ‘chromatin’-dependent) appeared to be more strongly associated with origin activation time than ORC-origin affinity per se.
Discussion
Although the DNA replication origins of the budding yeast Saccharomyces cerevisiae were originally defined in part because they shared a conserved DNA sequence element, the ACS, recent work provides strong evidence that features of chromatin also contribute to defining DNA replication origins in this organism. In this study we established an unbiased, general approach for querying the relative contributions of DNA sequence versus ‘chromatin’ to origin selection by the yeast ORC. Our approach allowed for the comparison of ORC-origin interaction strengths in vivo and in vitro at a genomic scale. We then focused on origins that bound ORC with relatively high affinities in vivo where we could most confidently distinguish between the two basic binding mechanisms—sequence-specific versus ‘chromatin’-based interactions—ORC used in origin-binding. This comparative strategy allowed us to estimate ∼40% of yeast origins that bind ORC with a ‘high-affinity’ in vivo rely on features extrinsic to the canonical ORC-origin DNA interface for normal ORC-origin complex formation in vivo. By definition, these features are exclusive to the chromosomal context that exists in vivo on chromosomes and therefore, by the broadest definition, involve chromatin. This strategy let us examine molecular features and functional properties of ‘chromatin-’ and DNA-dependent origins, revealing unanticipated connections between distinct ORC-origin binding mechanisms and the timing of origin activation during S-phase.
Obtaining genome-wide protein-DNA affinity measurements in vivo and in vitro
DNA sequence alone cannot explain the binding patterns of most sequence-specific DNA binding proteins in eukaryotes. Therefore a simple, quantitative approach to query how ‘chromatin’ might influence these patterns would be useful. Our approach requires that a relevant protein's ‘affinity’ for genomic DNA is measurable both in vivo and in vitro. The basic idea is that target-sites that show large discrepancies between in vitro and in vivo affinities are strong candidates for genomic loci where ‘chromatin’, as opposed to intrinsic sequence-specific protein-DNA interactions, plays a key role in the protein's binding. We exploited the established paradigm of ORC-origin DNA specificity in yeast to test this approach and distinguish between ‘chromatin-dependent’ and DNA-dependent ORC-origin binding mechanisms.
To examine ORC-origin interaction strengths in vivo we used a routine genome-wide chromatin immunoprecipitation (ChIP) approach. Because ChIP directly assesses the efficiency with which a specific DNA locus is precipitated, and many factors other than a protein's affinity for its target site can affect this efficiency, we exploited an ORC mutant (orc2-1) whose primary defect is to substantially reduce the concentration of ORC [52]. By comparing the efficiency of ChIP (i.e. binding signal represented as the area of a peak formed on a Nimblegen array) in orc2-1 mutant to wild-type cells, we were able to obtain a proxy for the in vivo ‘affinity’ of most origins. However, because orc2-1 only allowed a single low concentration of ORC to be assessed, useful information about differences in ORC-origin interaction mechanisms for origins within lower ORC-origin affinity ranges was not obtainable (e.g. the Weak class probably contains a mixture of ‘chromatin-dependent’ and DNA-dependent and complex origins). In addition, because the orc2-1 allele destabilizes the ORC complex, sufficient amounts of the mutant complexes are not obtainable for examination of ORC-DNA interactions [69]. Thus we must also acknowledge that orc2-1-sensitivity of an ORC-origin interaction may reflect features of this protein-DNA complex in addition to affinity. Recent methods for reducing yeast protein concentrations in a gradual, quantized manner should be useful for improving the in vivo step of this approach [70].
To measure ORC-origin affinities in vitro we adapted the traditional gel-shift (EMSA) that is commonly used in the analysis of ORC-origin binding and that works well with many DNA binding proteins. The genomic EMSA (gEMSA) provided a simple and efficient way to isolate ORC-bound fragments away from unbound DNA, and the genomic data could be compared directly to quantitative and highly reproducible ARS-specific EMSAs. While ARS-specific EMSAs routinely include non-specific competitor, the gEMSA used the genomic DNA itself as the common source for both target sites and competitor DNA, which may be one reason a large number of binding peaks were identified. While the gEMSA did not allow us to measure actual Kd values, in general it was effective—many characterized origins with known ORC-origin DNA interaction strengths behaved as predicted in the gEMSA. Direct analyses of our group of 39 selected origins suggested that some discrepancies between in vivo and in vitro binding strengths were probably attributable to technical issues rather than biology. For example for two origins that produced strong ORC-origin binding in vitro we observed no binding in the gEMSA. Such discrepancies might result from DNA-sequence effects on chromosomal DNA shearing or recovery from DNA purification columns or the gel matrix. As in any forward genetic screen, care must be taken in evaluating positive hits. Nevertheless, based on our direct EMSAs of 39 selected origins in our initial proof-of-principle screen and an additional fourteen origins defined as either ‘chromatin-dependent’ (8) or DNA-dependent (6) using the gEMSA, we conclude that the gEMSA successfully estimated the sequence-specific ORC-origin DNA binding behavior for 44 out of 53 origins (success rate of 83%). Moreover, the functional follow-up experiments provided evidence that we had effectively distinguished between distinct classes of origins. Refinements to the approach are expected to improve our ability to classify more origins as either ‘chromatin-’ or DNA-dependent or ‘complex’, which should enable the development of more refined hypotheses about the mechanisms controlling origin selection and function in yeast.
Flexibility in origin selection provided by a multifaceted ORC-origin interface
The Orc1BAH domain, a conserved nucleosome-binding module in Orc1 has substantial effects on ORC-origin binding in yeast and human cells [43], [46], [47]. Thus we expected that the ‘chromatin-dependent’ origins defined in this study would be enriched for origins that required the Orc1BAH domain for stable ORC binding that we defined in our previous study [43]. However, the data defied this expectation: ‘chromatin-dependent’ origins were distinct from Orc1BAH-dependent origins. This result suggests that regions of ORC, in addition to the Orc1BAH domain, contributed to ORC-chromatin interactions at origins. It also suggests that the putative Orc1BAH-nucleosome interaction does not, on its own, provide a particularly high-affinity binding interaction, a defining feature of the ‘chromatin-dependent’ origins identified in this study.
Clearly, ORC-DNA interactions also contribute to ORC-origin binding energy in yeast. Therefore, these data reveal that multiple, independent molecular interfaces contribute to ORC-origin stability. A distinct combination of molecular interfaces may define the stability and dynamics of the ORC-origin complex at any individual origin. This flexibility in the ORC-origin interface allows ORC to select origins within a wide variety of chromosomal environments even while requiring a certain level of specificity (e.g. avoiding transcribed regions). Such adaptability makes biological sense because chromatin varies substantially over a chromosome but origins must be distributed to ensure accurate duplication. While this idea has been discussed extensively with respect to metazoan origin selection, this study indicates that it is also relevant in the model S. cerevisiae where it can be further scrutinized at a detailed mechanistic level [71]–[73].
Is there a conserved role for ORC-origin DNA interactions?
The defined ORC binding site was recognizable even within yeast origins where it did not appear to contribute much to ORC-origin binding energy, suggesting that ORC-origin DNA contacts play a critical role beyond stabilizing ORC's association with an origin. Indeed, several elegant biochemical studies have established that yeast origin DNA is an allosteric regulator of ORC: for example, origin DNA reduces the ATPase activity of ORC, an activity important for the MCM loading reaction in G1-phase, and ORC-origin DNA complexes stimulate Cdc6 ATPase and changes in the ORC-DNA footprint [74]–[76]. Cdc6 directly binds ORC in G1-phase, and the Cdc6 ATPase is necessary for MCM loading [75], [77], [78]. These biochemical data indicate that some features of the yeast ORC binding site likely contribute to ORC's role in loading the replicative MCM helicase complex onto origin DNA. While such a role for DNA in origin function would be expected to be fundamental and therefore conserved in other organisms, sequence-specific ORC-DNA contacts have not been observed in metazoans. It is probable, however, that an allosteric role for DNA, even for particular nucleotides, could function in the absence of any obvious sequence-specific ORC-origin DNA binding. As suggested by others, the constraints on origin function in a single-celled microbe such as yeast compared to metazoans may be an explanation for the differences in sequence-specific ORC-origin DNA interactions observed between yeast and metazoans [4]. In particular, if origin formation and function are favored within intergenic regions, yeast with their gene-rich, compact genomes offer far fewer probable positions than metazoans. This fact may increase the evolutionary pressure on yeast to establish more efficient origin selection mechanisms that include ORC-DNA specificity. In this view, metazoan and yeast origin selection are fundamentally the same; the difference is simply that because metazoan chromosomes offer so much more opportunity, the selection of any single individual origin (or MCM complex loading site) in a given region can be considerably less efficient while still supplying sufficient replicative power.
ORC-origin interaction mechanisms and the regulation of origin activation
A particularly remarkable observation from this study was that ‘chromatin-dependent’ and DNA-dependent origins showed differential associations with origin activation time, with ‘chromatin-dependent’ origins showing greater enrichment of earlier firing, HU-resistant origins compared to DNA-dependent origins. Moreover, as a group, the latest firing origins in the yeast genome showed a positive correlation between in vivo and in vitro ORC binding affinities but the earliest firing origins did not. These data raise the possibility that ORC-origin dynamics are related in some way to mechanisms that modulate the timing of origin activation during S-phase, such as the recruitment of limiting S-phase factors that control the temporal origin activation pattern in yeast [23], [24]. One possibility is that the chromatin structures that promote early origin activation also promote ORC binding, and the weak ORC-DNA interactions that appear more enriched at earlier firing origins are a consequence of that. Alternatively, perhaps weaker ORC-origin DNA interactions associated with ‘chromatin-dependent’ ORC-origin binding have a functional purpose by promoting release of and/or conformational changes in ORC during S-phase that enhance the recruitment and/or effective function of these S-phase factors. In this view, ORC might have either a regulatory or responsive role during the S-phase portion of the ‘origin cycle’ yet to be defined [63]. Another interesting possibility is that the ORC-origin dynamics associated with early-activated origins enhances ORC's established role in loading the MCM complex onto origin DNA during G1-phase. Indeed, previous studies in both yeast and mammalian cells provide evidence that an origin's activation time is pre-determined during G1-phase, the same phase of the cell cycle when ORC and Cdc6 are loading MCM complexes onto origin DNA [79]. In addition, a relatively recent study reveals a causal relationship between ORC occupancy and MCM loading kinetics during M - and G1-phases and earlier origin activation in S. pombe [40], [79]. Finally, elegant biochemical studies provide evidence that cycles of ATP hydrolysis by ORC and Cdc6 that are likely coupled to ORC-origin-DNA-binding-and-release can drive reiterative MCM loading in G1-phase (i.e. multiple MCM loading events) [78], [80]. These data are consistent with a model in which the rate of an MCM loading event could be enhanced by ORC-chromatin interactions because such interactions could enhance the rate of ORC-origin-DNA-binding-and-release. Thus an origin that achieved similar levels of ORC occupancy solely through DNA contacts would be less efficient at such cycles, as ORC-origin DNA release and/or re-association would not be aided by auxiliary contacts from chromatin. This model directly connects chromatin-mediated ORC-origin dynamics to ORC's established biochemical role in loading an MCM complex onto origin DNA—that is, an origin is activated early in S-phase because it possesses a greater number of MCM complexes due to accelerated ORC-Cdc6-dependent MCM complex loading in G1-phase. An attractive feature of this idea, as discussed in the literature, is that it provides a mechanism by which to achieve a defined, quantitative difference between origins that could explain why they have differential affinities for limiting S-phase factors required for origin activation [81]–[83]. However, it is critical to acknowledge that a strong correlation between MCM complex levels and origin activation time has not been reported [22], [82], [83]. Thus while observations about ORC-origin dynamics and origin activation reported here are consistent with an MCM complex effect on origin activation, many additional experiments and multiple technical approaches will be required to address this issue definitively.
Variations in replication timing between different genomic regions are controlled primarily at the level of origin activation time and are often associated with gene expression changes that drive cell differentiation. In addition, replication-timing differences can have significant consequences on other chromosomal processes, e.g. rates of mutation and evolution [84]–[86]. These observations have spurred active investigation into the mechanisms that regulate differences in origin activation times during S-phase. Several regulators of origin firing times in yeast have been identified to date, including both local (e.g. in-cis DNA elements and/or local chromatin structure) and global (higher-order chromosomal architecture) features [22], [25], [27], [28], [35]. Examples of in-cis mechanisms include telomeric heterochromatin and Rpd3-mediated chromatin modifications delaying origin activation time, and centromeres advancing origin activation time. The data presented in this study suggest that the ORC binding site itself may be another in-cis regulator of an origin's activation time and, conversely, raise the possibility that previously defined in-cis regulators of origin activation time may act, at least in part, by modulating ORC-origin interaction dynamics. Recent examples of mechanisms to control origin activation timing at a more global level demonstrate a strong association between higher-order chromosome structure, as measured by chromosome capture methods, and origin activation time [14], [22]. The recent yeast studies further demonstrate a role for forkhead transcription factors (Fkh1/2) in modulating origin activation time by in-cis mechanisms [22], [26]. While it remains possible that these effects of Fkh1/2 perturb ORC-origin dynamics, at least at some origins, it is also possible, based on data presented both here and in the Fkh1/2 studies, that ORC-origin interaction dynamics and Fkh1/2 effects are independent phenomena associated with origin activation time. Indeed, perhaps the most critical issue is that neither of these phenomena—Fkh1/2 regulation or ORC-origin binding mechanisms—is absolute with respect to its association with origin activation time. For example, of the 100 origins that have Treps</ = 25′, 43 are not defined as Fkh1/2-regulated origins. Similarly, while we observed an enrichment of early origins within our ‘chromatin-dependent’ group, several fired later in S-phase, and conversely, a few DNA-dependent origins fired early in S-phase. If differential origin activation time simply reflects origins' adaptations to functioning in diverse chromatin environments to help insure efficient and accurate chromosomal duplication, then it is reasonable that multiple independent molecular contributions, acting in different combinations at different origins, impinge on the probability that a given origin will fire at a particular time in S-phase.
Materials and Methods
Electrophoretic Mobility Shift Assays
ORC was purified as described [41]. For origin-specific EMSAs (Figure 1E, Tables 1, 2 and S2), 49 bp ORC binding sites were synthesized as complementary oligos, annealed, and ligated into the KpnI and NotI sites of the pSTBlue1 vector. All ORC binding sites used in EMSAs were functionally confirmed in ARS assays. To synthesize DNA probes for EMSAs, standard PCR was performed using specific primers that annealed to regions near the multilinker cloning site pSTBlue1 with 40 uCi [32P]dCTP. The radiolabeled 184 bp DNA probe fragments were gel extracted from acrylamide gels before use in ORC binding reactions.
EMSAs were performed in 25 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 150 mM KCl, 40 ug BSA, 5% glycerol, 5 mM DTT, 1 µg calf-thymus DNA. 2 mM ATP was present in all binding reactions except for negative control reactions. Each reaction contained 5 pM of radiolabeled DNA probe and ORC at a final concentration of 0.3, 3, 15 or 30 nM. Reactions were incubated at 25°C for 20 minutes, and ORC-Bound DNA was separated from free DNA probe on 3.5% acrylamide gel run at 200 volts for 2.5 hours at 4°C. A DNA probe with the high-affinity ARS317 ORC binding site (apparent Kd ∼7 nM) served as an internal control in all experiments. Gels were vacuum dried and exposed to phosphor screens for at least 5 hours before imaging. Apparent Kd values were derived by fitting data to a one-site binding hyperbola and constraining the Bmax to 1 in Graphpad Prism 4.0. Each reaction was performed in triplicate and the average value and standard error for the apparent Kd that was determined is reported.
Genomic EMSA
Chromosomal DNA was isolated from 5 ml cultures of yeast grown to saturation. DNA was purified with standard phenol/chloroform/isoamylalcohol extractions, sheared through sonication to an average size of 200 bp, and silicia membrane column-purified after excising from an agarose gel. Binding reactions between ORC and 0.5 pM genomic DNA were performed as in the EMSAs except no competitor was used. Four independent reactions per ORC titration point (0.3, 3, and 30 nM) were performed, and the contents separated by gel electrophoresis as above. Because the genomic DNA was not labeled, one lane of the gel was reserved for the binding reaction between 30 nM ORC and a radiolabeled 184 bp ARS317 EMSA probe to mark the position of ORC-bound genomic fragments. The regions of the gel containing ORC-genomic fragment complexes corresponding in size to the ORC-ARS317 complex were excised. Excised gel fragments from the four replicates were combined and the DNA extracted from fragments with a standard NaOAC crush/soak method to yield bound DNA. Bound DNA and total DNA (unbound) were LM-PCR amplified (Sigma Whole Genome Amplification kit) for a total of 28 cycles per sample. Samples were shipped to Nimblegen for Cy3/Cy5 labeling and hybridization upon tiled microarrays (2006-10-12_Ansari_tiling_51mer) for each ORC titration point. Three total arrays were hybridized corresponding to 0.3, 3, and 30 nM ORC binding reactions. Data from the arrays were processed as described previously [51]. The distributions of log2 ratios of bound (Cy3) to unbound DNA (Cy5) for each ORC titration point were background subtracted so that the distributions centered over 0. The most repetitive probes (1.1%) were removed from analyses. ORC binding regions (peaks) were determined with the program ChIPOTle using a window size of 240 bp, a step size of 60 bp, and a P-value of 10−5 [87]. The raw ORC gEMSA data has been deposited to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accession number GSE48440.
Bioinformatic analyses
The coordinates and sequences for the 740 confirmed, likely, and dubious origins were downloaded from the OriDB (version 2011). Gene annotations and chromosomal coordinates were downloaded from SGD. Venn diagrams were generated using the web based Venn diagram generator from the Whitehead Institute of Biomedical Research (http://jura.wi.mit.edu/bioc/tools/venn.php). We chose a P-value cutoff of 10−5 to maximize the number of confirmed origins identified. 4482, 4218 and 4516 peaks representing 1,847,244, 1,796,733, and 1,913,647 base pairs were called in the 0.3, 3 and 30 nM ORC arrays, respectively. The fraction of base pairs within 0.3, 3, and 30 nM ORC gEMSA peaks that overlapped the chromosomal features in Figure 3B was determined using MochiView [88].
The ACS motif reported in this study (Figure 3C) was generated with a motif finding algorithm in Mochiview. Specifically, an 11 bp motif was generated from the sequences of 67 confirmed ACS elements annotated in the OriDB using a 4th order Markov model of the full genome. To determine the number, frequency, and P-value of enrichment for the ACS elements derived from the various origin groups (yeast genome, all OriDB origins, and the gEMSA) data were scanned with 11 bp windows for sequences meeting or exceeding 70% cut-offs of the likelihood-of-discovery (LOD) value of the ACS. To determine potential enrichment of ACS matches in the gEMSA and OriDB, one-tailed Fisher's Exact tests were performed in Mochiview with frequency of ACS matches in the yeast genome serving as the control.
To convert the gEMSA binding signals into a simple representation of ORC-origin binding strength, we summed the gEMSA ORC-binding strengths obtained for each ORC concentration as described in the Figure 2 legend (this value is referred to as “Total gEMSA signal” in relevant graphs in Figure 2 and Figure 6). The gEMSA ORC-binding strength for each array (i.e. amount of ORC-origin complex formation at each concentration of ORC used) was represented as the area of the peak (peak area) that defined the ORC-origin binding complex. The peak area was defined as the sum of the signals for each feature (oligo) that comprised the defined ORC-origin binding peak, and the signal for each feature was represented as the log2 of the ratio of the experimental (gel-shifted DNA; Cy5) to total (sheared genomic DNA; Cy3) (i.e. signal = log2(Cy5/Cy3). To characterize the strengths of ORC-DNA interactions in the gEMSA, a k-means algorithm in the Gene Cluster 3.0 software was applied to a matrix consisting of the 0.3 nM, 3 nM, and 30 nM gEMSA signals within the coordinates of in vivo ORC2 ChIP binding sites for 261 origins [51]. Clustering was repeated 10,000 times. ORC-DNA binding strengths in the gEMSA best fit into three distinct clusters based on the number of times the clustering was found in the 10,000 repetitions and the ability to separate moderate binding from weak ORC-DNA binding strengths.
For nucleosome mapping, nucleosome intensity signals from independent data sets were used, as indicated in Figure 5. The signals were plotted around the ORC binding sites that were defined by using the ORCACS sequence as determined by [44]. For origins lacking an ORCACS sequence, the proACS sequence, an ORC binding site derived from a phylogenetic study were used [38]. Origins lacking a predicted ACS were omitted from these analyses. All nucleosome traces were centered on the start of the EACS (Expanded ACS) on the T rich strand [49], [50]. For origins containing an ORCACS match, the first nucleotide of the ORCACS on the T rich strand marked the start of the EACS. For origins containing a proACS match, the −1 nucleotide relative to the start of the proACS on the T rich strand marked the beginning of the EACS.
ChIP-chip nucleosome intensities were processed as described previously [43], [59]. For a given origin, the nucleosome intensities were positioned relative to the start of the EACS, as above. Signal intensities with 4 base pair spacing were binned, and intensities were averaged for each bin and plotted relative the start of the EACS. The size in base pairs of the nucleosome depleted region (NDR) was calculated with a Perl script, where the NDR was defined by width in base pairs between the beginning coordinate of the +1 nucleosome the end coordinate of the −1 nucleosome. The genome-wide normalized nucleosome signals mapped in vivo (growth in permissive YPD) and in vitro were positioned relative to the T rich starts of the EACSs, as above [62]. Data were smoothed by binning values into 4 base pair spacing. Bins were averaged and plotted relative to the EACS.
To determine the effect of the orc1-161 mutation on nucleosome configuration around ‘chromatin-dependent’ and DNA-dependent origins, data from an experiment in which nucleosomes were mapped in wild-type and orc1-161 mutant cells arrested in G1 phase at non-permissive temperatures for orc1-161 were used [44]. Nucleosome signals were oriented in relation to the start of the T rich strand of the ORC binding sites, as described above. Nucleosome signals were plotted in heat maps. The NDRs of origins in the orc1-161 and ORC1 backgrounds were calculated with the Perl script as described [43].
To examine the relationship between ORC binding mechanisms and origin activation timing, several independent data sets were used. The Trep values were obtained from published data sets and downloaded from OriDB [64], [68]. T1/2 values were obtained from [30], [83]. To determine whether origin activation in the presence of hydroxyurea required Rad53, genome-wide data from wild-type cells and rad53-1 mutant cells from two independent studies were combined [66], [67]. If an origin was shown to fire in the presence of HU (HU-resistant) in wild-type cells in either of these two studies, we considered it HU-resistant (HUr-WT (Figure 6B)). If it only fired in the presence of HU in rad53 mutant cells in one or both studies, it was termed HU-sensitive (referred to in Figure 6B as HUr-rad53). Data were available for all 20 ‘chromatin-dependent’ origins (n = 20) but only 19 of the DNA-dependent origins (i.e. one DNA-dependent origin, ARS1108, was not detected under either condition in ether data set). Data for all 25 origins in the Weak class were available. To determine the association with CLB5-regulation (Figure S5B) and FKH-mediated regulation (Figure S5C and Figure S6), data from two independent genome-wide studies were used [22], [30].
Plasmid replication assays (ARS assays)
Core origin and expanded origin constructs were PCR-amplified and cloned into the NotI site of the pARS/PM vector [43], [89]. Core origins were comprised of the DNA sequences annotated in oriDB. Expanded origins contained ∼1 kb chromosomal sequence centered surrounding a functionally confirmed ORC binding site (Table S3). The sub-clones of ARS1529.5 in Figure S7 were generated using sequence from the KANMX coding sequence as substitutes for regions of ARS1529.5 as indicated in Figure S7 and Table S3. Plasmids were transformed into wild-type W303 yeast using standard techniques, and ARS assays were performed with four independent cultures per plasmid as in [50].
Supporting Information
Zdroje
1. StillmanB (2005) Origin recognition and the chromosome cycle. FEBS Lett 579 : 877–884.
2. BellSP, DuttaA (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71 : 333–374.
3. SclafaniRA, HolzenTM (2007) Cell cycle regulation of DNA replication. Annu Rev Genet 41 : 237–280.
4. RemusD, DiffleyJF (2009) Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol 21 : 771–777.
5. BoosD, FrigolaJ, DiffleyJF (2012) Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol 24 : 423–430.
6. TakaraTJ, BellSP (2009) Putting two heads together to unwind DNA. Cell 139 : 652–654.
7. LabibK (2011) Building a double hexamer of DNA helicase at eukaryotic replication origins. EMBO J 30 : 4853–4855.
8. DiffleyJF (2011) Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc Lond B Biol Sci 366 : 3545–3553.
9. van BrabantAJ, BuchananCD, CharboneauE, FangmanWL, BrewerBJ (2001) An origin-deficient yeast artificial chromosome triggers a cell cycle checkpoint. Mol Cell 7 : 705–713.
10. TheisJF, IreneC, DershowitzA, BrostRL, TobinML, et al. (2010) The DNA damage response pathway contributes to the stability of chromosome III derivatives lacking efficient replicators. PLoS Genet 6: e1001227.
11. KawabataT, LuebbenSW, YamaguchiS, IlvesI, MatiseI, et al. (2011) Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell 41 : 543–553.
12. IbarraA, SchwobE, MendezJ (2008) Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A 105 : 8956–8961.
13. GeXQ, JacksonDA, BlowJJ (2007) Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev 21 : 3331–3341.
14. RybaT, HirataniI, SasakiT, BattagliaD, KulikM, et al. (2011) Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput Biol 7: e1002225.
15. DebatisseM, Le TallecB, LetessierA, DutrillauxB, BrisonO (2012) Common fragile sites: mechanisms of instability revisited. Trends Genet 28 : 22–32.
16. WatanabeY, MaekawaM (2010) Spatiotemporal regulation of DNA replication in the human genome and its association with genomic instability and disease. Curr Med Chem 17 : 222–233.
17. SmithL, PlugA, ThayerM (2001) Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc Natl Acad Sci U S A 98 : 13300–13305.
18. TiengweC, MarcelloL, FarrH, DickensN, KellyS, et al. (2012) Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep 2 : 185–197.
19. MullerCA, NieduszynskiCA (2012) Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res 22 : 1953–1962.
20. Di RienziSC, LindstromKC, MannT, NobleWS, RaghuramanMK, et al. (2012) Maintaining replication origins in the face of genomic change. Genome Res 22 : 1940–1952.
21. KorenA, TsaiHJ, TiroshI, BurrackLS, BarkaiN, et al. (2010) Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet 6: e1001068.
22. KnottSR, PeaceJM, OstrowAZ, GanY, RexAE, et al. (2012) Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148 : 99–111.
23. MantieroD, MackenzieA, DonaldsonA, ZegermanP (2011) Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30 : 4805–4814.
24. TanakaS, NakatoR, KatouY, ShirahigeK, ArakiH (2011) Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol 21 : 2055–2063.
25. PohlTJ, BrewerBJ, RaghuramanMK (2012) Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae. PLoS Genet 8: e1002677.
26. LookeM, KristjuhanK, VarvS, KristjuhanA (2013) Chromatin-dependent and -independent regulation of DNA replication origin activation in budding yeast. EMBO Rep 14 : 191–198.
27. NatsumeT, MullerCA, KatouY, RetkuteR, GierlinskiM, et al. (2013) Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol Cell 50 : 661–674.
28. KnottSR, ViggianiCJ, TavareS, AparicioOM (2009) Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23 : 1077–1090.
29. DonaldsonAD, RaghuramanMK, FriedmanKL, CrossFR, BrewerBJ, et al. (1998) CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell 2 : 173–182.
30. McCuneHJ, DanielsonLS, AlvinoGM, CollingwoodD, DelrowJJ, et al. (2008) The temporal program of chromosome replication: genomewide replication in clb5{Delta} Saccharomyces cerevisiae. Genetics 180 : 1833–1847.
31. FriedmanKL, DillerJD, FergusonBM, NylandSV, BrewerBJ, et al. (1996) Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev 10 : 1595–1607.
32. FergusonBM, FangmanWL (1992) A position effect on the time of replication origin activation in yeast. Cell 68 : 333–339.
33. StevensonJB, GottschlingDE (1999) Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev 13 : 146–151.
34. LiuJ, McConnellK, DixonM, CalviBR (2012) Analysis of model replication origins in Drosophila reveals new aspects of the chromatin landscape and its relationship to origin activity and the prereplicative complex. Mol Biol Cell 23 : 200–212.
35. VogelauerM, RubbiL, LucasI, BrewerBJ, GrunsteinM (2002) Histone acetylation regulates the time of replication origin firing. Mol Cell 10 : 1223–1233.
36. AggarwalBD, CalviBR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430 : 372–376.
37. DouglasME, DiffleyJF (2012) Replication timing: the early bird catches the worm. Curr Biol 22: R81–82.
38. NieduszynskiCA, KnoxY, DonaldsonAD (2006) Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev 20 : 1874–1879.
39. ChastainPD2nd, BowersJL, LeeDG, BellSP, GriffithJD (2004) Mapping subunit location on the Saccharomyces cerevisiae origin recognition complex free and bound to DNA using a novel nanoscale biopointer. J Biol Chem 279 : 36354–36362.
40. WuPY, NurseP (2009) Establishing the program of origin firing during S phase in fission Yeast. Cell 136 : 852–864.
41. Palacios DeBeerMA, MullerU, FoxCA (2003) Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev 17 : 1817–1822.
42. FoxCA, McConnellKH (2005) Toward biochemical understanding of a transcriptionally silenced chromosomal domain in Saccharomyces cerevisiae. J Biol Chem 280 : 8629–8632.
43. MullerP, ParkS, ShorE, HuebertDJ, WarrenCL, et al. (2010) The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev 24 : 1418–1433.
44. EatonML, GalaniK, KangS, BellSP, MacAlpineDM (2010) Conserved nucleosome positioning defines replication origins. Genes Dev 24 : 748–753.
45. BerbenetzNM, NislowC, BrownGW (2010) Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet 6.
46. NoguchiK, VassilevA, GhoshS, YatesJL, DePamphilisML (2006) The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. Embo J 25 : 5372–5382.
47. KuoAJ, SongJ, CheungP, Ishibe-MurakamiS, YamazoeS, et al. (2012) The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature
48. ChangF, TheisJF, MillerJ, NieduszynskiCA, NewlonCS, et al. (2008) Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW sequence within the origin recognition complex binding site. Mol Cell Biol 28 : 5071–5081.
49. TheisJF, NewlonCS (1997) The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc Natl Acad Sci U S A 94 : 10786–10791.
50. ChangF, MayCD, HoggardT, MillerJ, FoxCA, et al. (2011) High-resolution analysis of four efficient yeast replication origins reveals new insights into the ORC and putative MCM binding elements. Nucleic Acids Res 39 : 6523–6535.
51. ShorE, WarrenCL, TietjenJ, HouZ, MullerU, et al. (2009) The origin recognition complex interacts with a subset of metabolic genes tightly linked to origins of replication. PLoS Genet 5: e1000755.
52. ShimadaK, PaseroP, GasserSM (2002) ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev 16 : 3236–3252.
53. SiowCC, NieduszynskaSR, MullerCA, NieduszynskiCA (2012) OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res 40: D682–686.
54. NieduszynskiCA, BlowJJ, DonaldsonAD (2005) The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic Acids Res 33 : 2410–2420.
55. DonatoJJ, ChungSC, TyeBK (2006) Genome-wide hierarchy of replication origin usage in Saccharomyces cerevisiae. PLoS Genet 2: e141.
56. IrlbacherH, FrankeJ, MankeT, VingronM, Ehrenhofer-MurrayAE (2005) Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev 19 : 1811–1822.
57. WeberJM, IrlbacherH, Ehrenhofer-MurrayAE (2008) Control of replication initiation by the Sum1/Rfm1/Hst1 histone deacetylase. BMC Mol Biol 9 : 100.
58. LynchPJ, FraserHB, SevastopoulosE, RineJ, RuscheLN (2005) Sum1p, the origin recognition complex, and the spreading of a promoter-specific repressor in Saccharomyces cerevisiae. Mol Cell Biol 25 : 5920–5932.
59. LeeW, TilloD, BrayN, MorseRH, DavisRW, et al. (2007) A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39 : 1235–1244.
60. OnishiM, LiouGG, BuchbergerJR, WalzT, MoazedD (2007) Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell 28 : 1015–1028.
61. ArmacheKJ, GarlickJD, CanzioD, NarlikarGJ, KingstonRE Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 334 : 977–982.
62. KaplanN, MooreIK, Fondufe-MittendorfY, GossettAJ, TilloD, et al. (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458 : 362–366.
63. GibsonDG, BellSP, AparicioOM (2006) Cell cycle execution point analysis of ORC function and characterization of the checkpoint response to ORC inactivation in Saccharomyces cerevisiae. Genes Cells 11 : 557–573.
64. YabukiN, TerashimaH, KitadaK (2002) Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7 : 781–789.
65. SantocanaleC, DiffleyJF (1998) A Mec1 - and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395 : 615–618.
66. FengW, CollingwoodD, BoeckME, FoxLA, AlvinoGM, et al. (2006) Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol 8 : 148–155.
67. CrabbeL, ThomasA, PantescoV, De VosJ, PaseroP, et al. (2010) Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol 17 : 1391–1397.
68. RaghuramanMK, WinzelerEA, CollingwoodD, HuntS, WodickaL, et al. (2001) Replication dynamics of the yeast genome. Science 294 : 115–121.
69. BellSP, KobayashiR, StillmanB (1993) Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262 : 1844–1849.
70. Heidinger-PauliJM, MertO, DavenportC, GuacciV, KoshlandD Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol 20 : 957–963.
71. MechaliM (2001) DNA replication origins: from sequence specificity to epigenetics. Nat Rev Genet 2 : 640–645.
72. MechaliM (2010) Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11 : 728–738.
73. AladjemMI (2007) Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet 8 : 588–600.
74. KlemmRD, AustinRJ, BellSP (1997) Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell 88 : 493–502.
75. RandellJC, BowersJL, RodriguezHK, BellSP (2006) Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell 21 : 29–39.
76. SpeckC, ChenZ, LiH, StillmanB (2005) ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol 12 : 965–971.
77. LiangC, WeinreichM, StillmanB (1995) ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81 : 667–676.
78. BowersJL, RandellJC, ChenS, BellSP (2004) ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell 16 : 967–978.
79. RaghuramanMK, BrewerBJ, FangmanWL (1997) Cell cycle-dependent establishment of a late replication program. Science 276 : 806–809.
80. TsakraklidesV, BellSP (2010) Dynamics of pre-replicative complex assembly. J Biol Chem 285 : 9437–9443.
81. MendezJ, Zou-YangXH, KimSY, HidakaM, TanseyWP, et al. (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell 9 : 481–491.
82. RhindN, YangSC, BechhoeferJ (2010) Reconciling stochastic origin firing with defined replication timing. Chromosome Res 18 : 35–43.
83. YangSC, RhindN, BechhoeferJ (2010) Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol Syst Biol 6 : 404.
84. LangGI, MurrayAW (2011) Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol Evol 3 : 799–811.
85. StamatoyannopoulosJA, AdzhubeiI, ThurmanRE, KryukovGV, MirkinSM, et al. (2009) Human mutation rate associated with DNA replication timing. Nat Genet 41 : 393–395.
86. ChenCL, RappaillesA, DuquenneL, HuvetM, GuilbaudG, et al. (2010) Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res 20 : 447–457.
87. BuckMJ, NobelAB, LiebJD (2005) ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol 6: R97.
88. HomannOR, JohnsonAD (2010) MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8 : 49.
89. MarahrensY, StillmanB (1994) Replicator dominance in a eukaryotic chromosome. Embo J 13 : 3395–3400.
90. ZhuC, ByersKJ, McCordRP, ShiZ, BergerMF, et al. (2009) High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res 19 : 556–566.
91. ShirahigeK, IwasakiT, RashidMB, OgasawaraN, YoshikawaH (1993) Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol 13 : 5043–5056.
92. XuW, AparicioJG, AparicioOM, TavareS (2006) Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7 : 276.
93. CelnikerSE, SwederK, SriencF, BaileyJE, CampbellJL (1984) Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol 4 : 2455–2466.
94. BreierAM, ChatterjiS, CozzarelliNR (2004) Prediction of Saccharomyces cerevisiae replication origins. Genome Biol 5: R22.
95. HuangRY, KowalskiD (1993) A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J 12 : 4521–4531.
96. HuangRY, KowalskiD (1996) Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res 24 : 816–823.
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



