-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
The plant hormone auxin plays a critical role in regulating various aspects of plant growth and development, and the spatial accumulation of auxin within organs, which is primarily attributable to local auxin biosynthesis and polar transport, is largely responsible for lateral organ morphogenesis and the establishment of plant architecture. Here, we show that three Arabidopsis INDETERMINATE DOMAIN (IDD) transcription factors, IDD14, IDD15, and IDD16, cooperatively regulate auxin biosynthesis and transport and thus aerial organ morphogenesis and gravitropic responses. Gain-of-function of each IDD gene in Arabidopsis results in small and transversally down-curled leaves, whereas loss-of-function of these IDD genes causes pleiotropic phenotypes in aerial organs and defects in gravitropic responses, including altered leaf shape, flower development, fertility, and plant architecture. Further analyses indicate that these IDD genes regulate spatial auxin accumulation by directly targeting YUCCA5 (YUC5), TRYPTOPHAN AMINOTRANSFERASE of ARABIDOPSIS1 (TAA1), and PIN-FORMED1 (PIN1) to promote auxin biosynthesis and transport. Moreover, mutation or ectopic expression of YUC suppresses the organ morphogenic phenotype and partially restores the gravitropic responses in gain - or loss-of-function idd mutants, respectively. Taken together, our results reveal that a subfamily of IDD transcription factors plays a critical role in the regulation of spatial auxin accumulation, thereby controlling organ morphogenesis and gravitropic responses in plants.
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003759
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003759Summary
The plant hormone auxin plays a critical role in regulating various aspects of plant growth and development, and the spatial accumulation of auxin within organs, which is primarily attributable to local auxin biosynthesis and polar transport, is largely responsible for lateral organ morphogenesis and the establishment of plant architecture. Here, we show that three Arabidopsis INDETERMINATE DOMAIN (IDD) transcription factors, IDD14, IDD15, and IDD16, cooperatively regulate auxin biosynthesis and transport and thus aerial organ morphogenesis and gravitropic responses. Gain-of-function of each IDD gene in Arabidopsis results in small and transversally down-curled leaves, whereas loss-of-function of these IDD genes causes pleiotropic phenotypes in aerial organs and defects in gravitropic responses, including altered leaf shape, flower development, fertility, and plant architecture. Further analyses indicate that these IDD genes regulate spatial auxin accumulation by directly targeting YUCCA5 (YUC5), TRYPTOPHAN AMINOTRANSFERASE of ARABIDOPSIS1 (TAA1), and PIN-FORMED1 (PIN1) to promote auxin biosynthesis and transport. Moreover, mutation or ectopic expression of YUC suppresses the organ morphogenic phenotype and partially restores the gravitropic responses in gain - or loss-of-function idd mutants, respectively. Taken together, our results reveal that a subfamily of IDD transcription factors plays a critical role in the regulation of spatial auxin accumulation, thereby controlling organ morphogenesis and gravitropic responses in plants.
Introduction
Auxin is a key plant hormone that plays critical roles in the regulation of plant growth and development. A combination of physiological, genetic, biochemical, and molecular studies has greatly enriched our understanding of auxin biosynthesis, transport, and signal transduction [1]–[3]. Increasing evidence indicates that auxin is essential for nearly all developmental processes, including gametogenesis, embryogenesis, lateral organ formation and patterning, branching, and tropic responses [4], [5]. It is generally believed that most auxin-mediated developmental events are highly dependent on the differential accumulation of auxin within plant organs (auxin gradients), which are mainly attributable to both local auxin biosynthesis and the intercellular polar transport of auxin [1], [5].
Direct evidence that local auxin biosynthesis is involved in the regulation of plant organogenesis comes from studies of several genes in the auxin biosynthetic pathway of Arabidopsis, including YUCCA (YUC) and TRYPTOPHAN AMINOTRANSFERASE of ARABIDOPSIS (TAA). YUCs encode the flavin monooxygenases that catalyze a key step in converting tryptophan into IAA, a main auxin in plants [6]. Overexpression of YUC genes in Arabidopsis substantially elevates the endogenous IAA level and causes distinct phenotypes such as epinastic cotyledons, elongated hypocotyls, and narrow and curly leaves [6]–[8]. Although a yuc single mutant in Arabidopsis does not show an obvious phenotype, the mutation of multiple YUC genes leads to a diversity of auxin-related phenotypes, including reduced apical dominance, crinkled leaves, simple venation, and abnormal flower development, demonstrating that YUC-modulated local auxin biosynthesis is critical for plant morphogenesis and architecture formation [9], [10]. Consistently, such developmental defects in yuc mutants can be rescued by local expression of iaaM, a bacterial auxin biosynthetic gene, but not by the application of exogenous auxin [9]. The expressions of YUC genes are overlapping and spatiotemporally regulated in various organs [9], [10], suggesting that YUCs function redundantly and cooperatively in different organs. Further studies demonstrate that TAA1 and its homologs function in auxin biosynthesis in response to environmental and developmental signals in Arabidopsis. The taa1 plant has a decreased level of endogenous IAA and displays defects in shade avoidance and root-specific ethylene sensitivity, and the simultaneous mutation of TAA1 and its close homologs (TAR1 and TAR2) results in phenotypes that are obviously auxin-related, such as reduced gravitropic response of roots, shortened hypocotyls, and monopteros-like seedlings with a single cotyledon [11]–[13]. Recently, YUCs and TAAs have been shown to function in a key two-step auxin biosynthetic pathway [14]–[16]. Therefore, the regulation of YUCs and TAAs may represent an important mechanism to alter auxin gradients and thus modify plant morphogenesis and responses to environmental cues. Indeed, several transcription factors have been shown to participate in the regulation of YUC expression. STYLISH1 (STY1), one of the SHORT INTERNODES (SHI) transcription factors and NGATHA3 (NGA3), a member of the B3 transcription factor family, are both proposed to act cooperatively to direct style development, partially through the activation of YUC2- and YUC4-mediated auxin biosynthesis in the apex of the gynoecium [17], [18]. LEAFY COTYLEDON2 (LEC2), a B3 domain transcription factor acting as a central regulator of embryogenesis, has been found to be involved in modulating de novo auxin biosynthesis via the activation of YUC2 and YUC4 [19]. Recent studies have also revealed that PIF4-activated YUC8 expression is required for hypocotyl growth under high temperature conditions [20], and a putative transcription factor SPOROCYTELESS/NOZZLE (SPL/NZZ) has been found to be involved in the regulation of lateral organ morphogenesis by repressing transcription of YUC2 and YUC6 [21].
In addition to local auxin biosynthesis, polar auxin transport also contributes greatly to auxin redistribution. The polar auxin transport is mediated by AUX1/LIKE AUX1 (AUX1/LAX), PIN-FORMED (PIN), and ATP-binding cassette class B (ABCB) transporters [2], [22]–[24]. Mutation in AUX1/LAX, which encode auxin influx carriers, influences lateral root formation, root hair development, phyllotaxy, and tropic responses [22], [24]–[28]. Similarly, single or multiple mutations in auxin efflux carrier genes, PINs and ABCBs, lead to severe defects in multiple developmental processes including embryogenesis, lateral organ formation and patterning, vascular development, and tropic responses [29]–[33], demonstrating that polar auxin transport is also essential for plant morphogenesis and tropic responses.
The INDETERMINATE DOMAIN (IDD) transcription factors belong to a plant-specific transcription factor family that contains a conserved ID domain with four zinc finger motifs [34], [35]. The founding member of this family, maize INDETERMINATE1 (ID1), is a key regulator of flowering transition [36]. The Arabidopsis genome contains 16 IDD members, and the characterization of several IDD members in Arabidopsis has indicated that IDD genes are involved in regulation of multiple developmental processes. For example, IDD8/NUTCRACKER (NUC) is involved in the regulation of flowering time through modulation of sugar metabolism [37], and IDD14 regulates starch metabolism in response to cold stimulus [38]. IDD15/SHOOT GRAVITROPISM 5 (SGR5) is involved in the gravitropic response of inflorescence stems, possibly through alteration of gravity sensing [39]–[41]. IDD8/NUC, IDD3/MAGPIE (MAG), and IDD10/JACKDAW (JKD) regulate root development and patterning [42], [43], while IDD1/ENHYDROUS (ENY) participates in seed maturation and germination [44]. Recent studies have demonstrated that a rice ID1 homolog is required for flowering induction [45]–[47], and that the rice Loose Plant Architecture1 (LPA1), an ortholog of Arabidopsis IDD15, influences shoot gravitropism and architecture [48].
Here, we identified a gain-of-function mutant of Arabidopsis IDD14, idd14-1D, which exhibited small and transversally down-curled leaves. Further characterization of loss - and gain-of-function mutants of IDD14 and its close homologs, IDD15 and IDD16, revealed that these three IDD genes redundantly but differentially regulate multiple aspects of organ development and gravitropic responses. Moreover, we provide evidence that these IDD transcription factors can directly target auxin biosynthetic and transport genes and alter auxin accumulation in multiple organs. These results reveal that IDD-mediated auxin biosynthesis and transport are critical for lateral organ morphogenesis and gravitropic responses in plants.
Results
cuf1-D/idd14-1D Displays Pleiotropic Leaf Phenotypes
To gain insight into how lateral organ morphogenesis is controlled in plants, we screened for mutants with altered aerial organ morphology in a transgenic Arabidopsis population harboring a T-DNA activation-tagging plasmid (pSKI015) [49]. A semi-dominant mutant was identified by its smaller and dramatically down-curling leaf phenotype, and thus designated as curlyfolia1-D (cuf1-D) and subsequently as idd14-1D (see below) (Figure 1A). To examine the genetic nature of cuf1-D, we backcrossed cuf1-D with wild type (WT) plants. All F1 plants exhibited an intermediate phenotype as cuf1-D/+, and F2 plants displayed a phenotypic segregation of WT∶cuf1-D/+∶cuf1-D as 1∶2∶1 (82∶173∶87, P = 0.89, X2-test), in which all WT plants were BASTA sensitive. This observation suggests that cuf1-D results from a semi-dominant mutation of a single gene that is likely to co-segregate with a T-DNA insertion event. The most striking phenotype in cuf1-D was the leaf size and shape. Detailed quantification showed that the average areas of fully-expanded leaf blades in heterozygous and homozygous cuf1-D were only about 70% and 47% of that in WT, respectively (Figure 1B). The leaves of cuf1-D were also dramatically curled downward in a transverse direction. The leaf transverse curvature (TC) index in WT was about 0.08, whereas the leaf TC index in heterozygous and homozygous cuf1-D reached about 0.29 and 0.36, respectively (Figure 1C). Moreover, the lamina of a mature WT rosette leaf displayed an elliptical shape with a leaf length/width index at about 1.5, while the heterozygous and homozygous cuf1-D leaves were comparatively narrow and their leaf indices were about 1.9 (Figure 1D). In addition to the morphological changes in leaves, cuf1-D was also late flowering and dwarfed (Figure 1A, Table S1). These observations demonstrate that the dominant mutation in cuf1-D has pleiotropic effects on lateral organ development and plant architecture.
Fig. 1. Phenotype of cuf1-D plants. 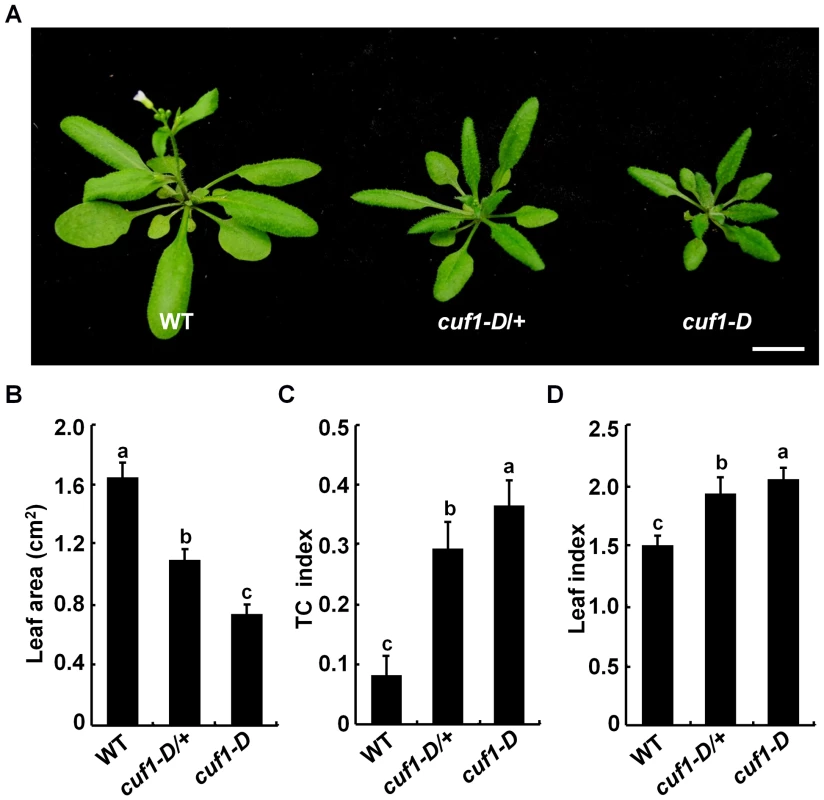
(A) Morphology of 4-week-old wild-type (WT), cuf1-D/+, and cuf1-D plants. The scale bar represents 1 cm. (B) Blade areas of WT, cuf1-D/+, and cuf1-D leaves. (C) Transverse curvature (TC) index of WT, cuf1-D/+, and cuf1-D leaves. (D) Leaf index (ratio of length to width) in WT, cuf1-D/+, and cuf1-D plants. At least 10 sixth leaves from each genotype were used for determination of the leaf area, TC index, and leaf index as described in the methods section, respectively. Data are shown as mean values ± one SD, and the letters (a to c) indicate the significance (P<0.05) among the genotypes according to one-way ANOVA test (SPSS 13.0, Chicago, IL, USA). CUF1 Encodes the INDETERMINATE DOMAIN 14 Transcription Factor
Since cuf1-D is a single gene mutation that is likely to co-segregate with a T-DNA insertion event, we amplified the genomic DNA adjacent to the left border of the T-DNA by thermal asymmetric interlaced PCR (TAIL-PCR). The sequencing analysis indicated that a T-DNA was inserted in the intergenic region between At1g68120 and At1g68130 (Figure 2A). Genotyping analysis showed that the T-DNA insertion co-segregated with the leaf phenotype in heterozygous and homozygous cuf1-D mutants (Figure 2B), suggesting that cuf1-D was caused by the T-DNA insertion event. Because cuf1-D was a dominant mutant, we monitored the transcripts of genes flanking the T-DNA insertion using semi-quantitative reverse transcription PCR (RT-PCR) analysis. Compared to those in the WT, the transcripts of At1g68130 (IDD14) were dramatically elevated, while At1g68140 mRNA levels were slightly decreased in cuf1-D (Figure 2C). To determine whether the elevated levels of IDD14 transcripts are responsible for the cuf1-D phenotype, we introduced a p35S::anti-IDD14 construct into cuf1-D and a p35S::IDD14 construct into WT plants, respectively. Transgenic plants overexpressing IDD14 fully recapitulated the phenotype of cuf1-D, while the expression level of At1g68140 in these plants was comparable with that in WT plants (Figure 2D, S1B). Moreover, introduction of p35S::anti-IDD14 into cuf1-D restored the cuf1-D to WT morphology (Figure 2D). These results demonstrate that cuf1-D results from the ectopic expression of IDD14, and accordingly, the cuf1-D was thus re-designated as idd14-1D.
Fig. 2. Molecular characterization of CUF1/IDD14. 
(A) Scheme of the genomic region flanking T-DNA insertion in cuf1-D. Genes are shown as thick arrows and intergenic regions are shown as lines. The orientation of T-DNA left border (LB), right border (RB), and the four CaMV 35S enhancers (4×35S) are indicated. (B) Linkage analysis of the T-DNA and cuf1-D phenotype. The 954-bp genomic fragments in WT and cuf1-D/+ plants were amplified with primers in the genomic region flanking the T-DNA insertion site, and the 894-bp fragments in cuf1-D/+ and cuf1-D were amplified with an LB primer and a downstream genomic primer. (C) Expression analysis of the genes flanking the T-DNA in WT and cuf1-D plants. ACTIN2 (ACT2) was used as an internal control. Note that transcripts of At1g68130 (IDD14) are highly elevated in cuf1-D. (D) Morphology of 25-days-old WT, cuf1-D carrying p35S::anti-IDD14, and p35S::IDD14 plants. The scale bar represents 2 cm. (E) Alignment of the ID domains in IDD14, IDD15, and IDD16. ZF1-ZF4 represents the four C2H2-type zinc finger motifs. The arrowheads indicate the conserved cysteine and histidine residues. (F) Morphology of 4-week-old transgenic plants overexpressing IDD15 or IDD16. The scale bar represents 1 cm. In Arabidopsis, the IDD14 transcription factor is phylogenetically sub-grouped with two close homologs: IDD15 (SGR5) and IDD16, to form a small subfamily that is distinct from the other IDD family members (Figure S1A) [34], [39]. IDD14, IDD15 and IDD16 share 52%–62% amino acid identity, and their ID domains are highly conserved with 89%–95% amino acid identity (Figure 2E). To investigate the possible redundancy of IDD15 and IDD16 with IDD14, we generated transgenic Arabidopsis plants overexpressing IDD15 or IDD16, respectively. The ectopic expression of either IDD15 or IDD16 resulted in a similar leaf phenotype as observed in idd14-1D (Figure 2F, S1B), suggesting that the three IDD members may have redundant function during plant development.
The Differential and Overlapping Expression of IDD14, IDD15, and IDD16 in Multiple Organs
To explore the functions of this IDD subfamily, we examined the tissue-specific expression patterns of these genes in multiple organs of transgenic plants harboring a pIDD::GUS (β-Glucuronidase) construct. As shown in Figure 3A–3C, IDD14 was mainly expressed in cotyledons and the vasculature of rosette leaves, and a weak level of expression was observed in hypocotyls and floral organs. However, the GUS signal was undetectable in roots and inflorescence stems. IDD15 was highly expressed in petioles, hypocotyls, roots, floral organs, and especially in inflorescence stems. In inflorescence stems, GUS staining was mainly present in the cortex, endodermis and vasculature tissues (Figure 3A–3C). IDD16 was highly expressed in leaves, hypocotyls, roots, vasculature of cotyledons, floral organs, and in the endodermis and vasculature of inflorescence stems (Figure 3A–3C). RNA in situ hybridization assayed in the inflorescence stems validated the expressions of the three IDD genes detected by the GUS reporter (Figure 3D). Additionally, consistent with the previous finding that IDD members act as transcription factors [38], [39], an IDD14-GFP fusion protein in transgenic plants carrying p35S::IDD14-GFP, which recapitulated the phenotype of idd14-1D, was found to be localized in nuclei (Figure S2A, S2B).
Fig. 3. Differential and overlapping expression of IDD14, IDD15, and IDD16. 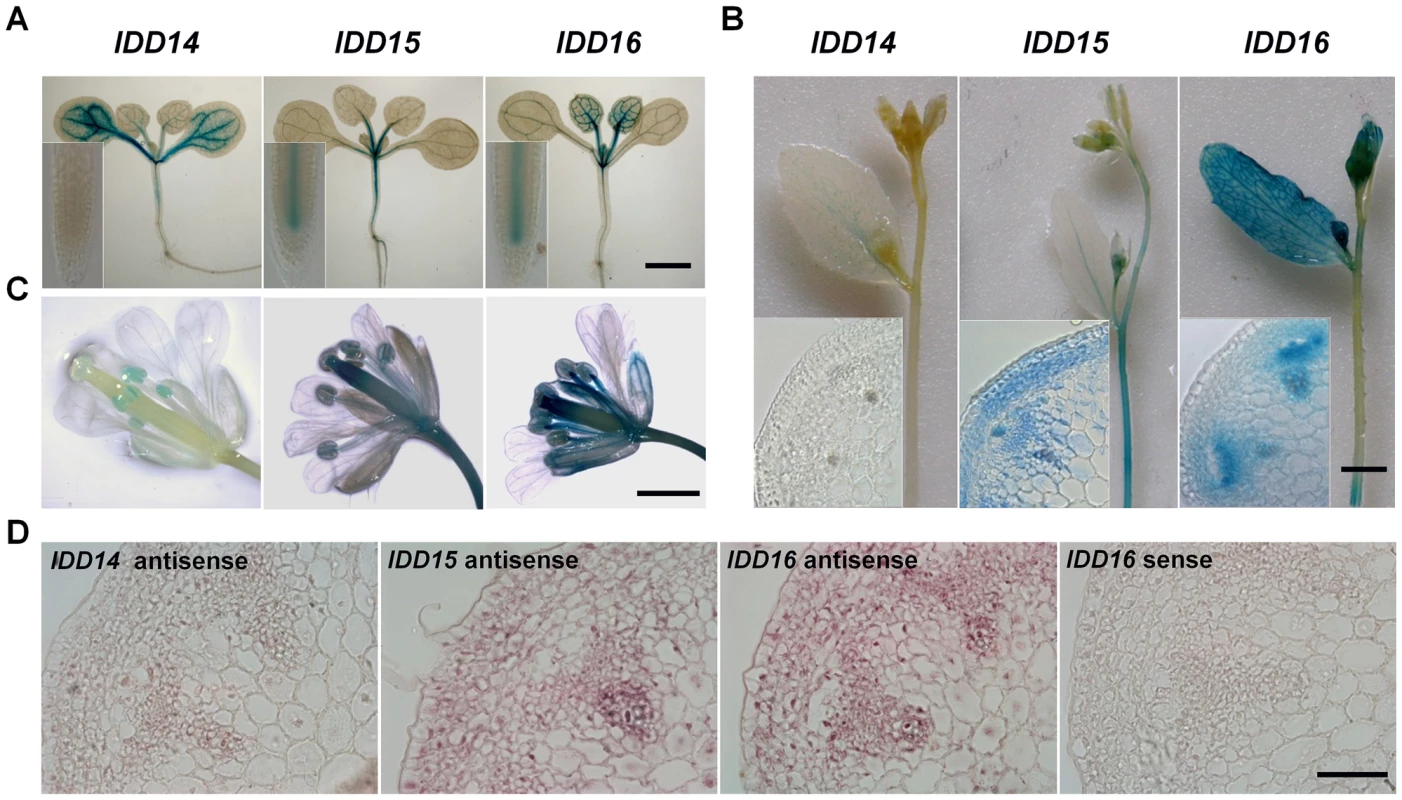
(A) Expression of IDD14, IDD15, and IDD16 in seedlings assayed by GUS staining. The 12-day-old transgenic seedlings carrying pIDD14::GUS, pIDD15::GUS, or pIDD16::GUS constructs were subjected to GUS staining assays. Insets show the primary roots. The scale bar represents 1 mm. (B) GUS staining of the florescence stems of pIDD14::GUS, pIDD15::GUS, and pIDD16::GUS transgenic plants. Insets show the transverse sections of inflorescence stems. The scale bar represents 2 mm. (C) GUS staining of the floral organs of pIDD14::GUS, pIDD15::GUS, and pIDD16::GUS transgenic plants. The scale bar represents 1 mm. (D) Expression of IDD14, IDD15, and IDD16 in inflorescence stems assayed by RNA in situ hybridization. A section hybridized with an IDD16 sense probe is shown as a control. The scale bar represents 50 µm. IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropic Responses
To gain further insight into the functions of this IDD subfamily, we obtained the T-DNA insertion mutant idd14-1 (CS367164) and idd15-5 (Salk_087765) from the Arabidopsis Biological Resource Center (ABRC), in which the T-DNA is inserted in an exon of IDD14 or IDD15. Semi-quantitative RT-PCR analyses indicated that the transcripts of IDD14 or IDD15 were undetectable in idd14-1 or idd15-5, respectively (Figure 4A, 4B). As no idd16 mutant is publically available, we generated IDD16-RNAi transgenic plants with an IDD16-specific cDNA fragment (Figure 4A). Semi-quantitative RT-PCR analysis indicated that IDD16 transcripts were dramatically reduced in transgenic IDD16-RNAi plants (Figure 4B). We then generated double and triple mutants of the three IDD genes, which allowed us to closely examine their differential and redundant functions during plant growth and development.
Fig. 4. Pleiotropic organ phenotypes in loss-of-function idd mutants. 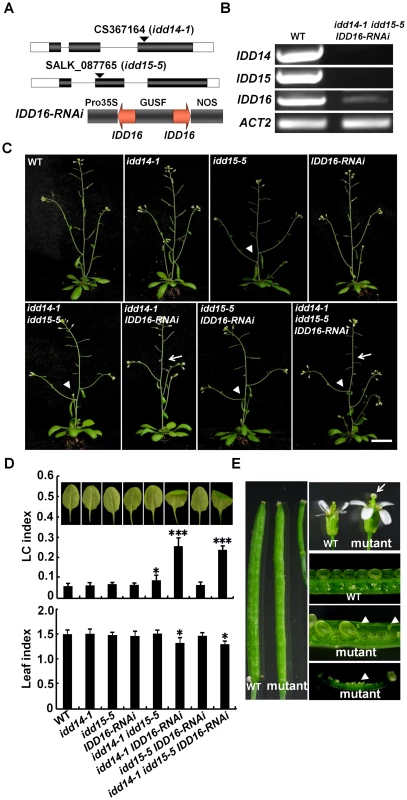
(A) Schematic illustration of the idd14-1, idd15-5, and the IDD16-RNAi construct. A 248-bp specific IDD16 cDNA fragment was used for construction of p35S::IDD16-RNAi. (B) Semi-quantitative RT-PCR analysis of IDD genes in WT and idd mutants. The transcripts of IDD14, IDD15, and IDD16 in the idd triple mutant are shown as representatives. (C) 36-day-old plants of idd single, double, and triple mutants. Arrows indicate the infertile siliques. The arrowheads show the increased angles between the inflorescence stems and branches. The scale bar represents 2 cm. (D) Morphology of sixth leaves in 25-day-old idd mutants. The longitudinal curvature (LC) index and leaf index were determined from at least 10 leaves in each genotype. The data are shown as mean values ± one SD (Student's t-test, *P<0.05 and ***P<0.001). (E) Enlarged floral organs and infertile siliques in the idd triple mutant. The arrow indicates the stigma lacking pollen. The arrowheads show the unfertilized ovules. We first examined the aerial organ morphogenesis in these idd mutants. As shown in Figure 4C and 4D, none of the idd single mutants exhibited any obvious organ phenotype. However, the leaves of idd14-1 IDD16-RNAi and idd triple mutants were not only downward-curled in the longitudinal direction, but were also more rotund when compared to WT leaves. Furthermore, idd14-1 IDD16-RNAi and the idd triple mutant had enlarged floral organs and infertile siliques. Careful examination showed that the infertile siliques resulted from the asynchronous elongation of stamen filaments and styles, and thus had poorly pollinated stigmas (Figure 4C, 4E). Manual pollination of styles in these mutants resulted in the development of normal siliques. In contrast to these dramatic phenotypes, the idd15-5 IDD16-RNAi plants had only slightly curled leaves (Figure 4C, 4D). These observations imply that IDD14 and IDD16 have redundant roles in directing leaf and floral organ morphogenesis.
Consistent with the previous finding that idd15 displays increased angles between inflorescence stems and siliques [40], we noticed that the orientation angles of both branches and siliques were obviously increased in idd15-5 (Figure 4C). This phenotype was further enhanced in the idd15-5 IDD16-RNAi and idd triple mutants, but not in idd14-1 idd15-5 plants (Figure 4C, 5A), indicating that IDD15 and IDD16 act cooperatively to control silique and branch orientation. As idd15 has a reduced gravitropic response in inflorescence stems [39]–[41], it is likely that the altered orientation of branches and siliques is related to the gravitropism defect in the idd mutants. To test this, we investigated the gravitropic responses in gain - and loss-of-function of idd plants. As expected, idd15-5 inflorescence stems exhibited an obviously reduced gravitropic response, and this phenotype was greatly enhanced in idd15-5 IDD16-RNAi and the idd triple mutant (Figure 5B, S3A), demonstrating that IDD15 and IDD16 function coordinately in gravitropic responses. Interestingly, although idd14-1 did not show any defect in gravitropic response, the inflorescence stems of idd14-1D were hypersensitive to gravistimulation (Figure 5B, S3A), indicating that ectopic expression of IDD14 also influences gravitropic responses.
Fig. 5. Silique orientation and gravitropic response in gain- and loss-of-function idd mutants. 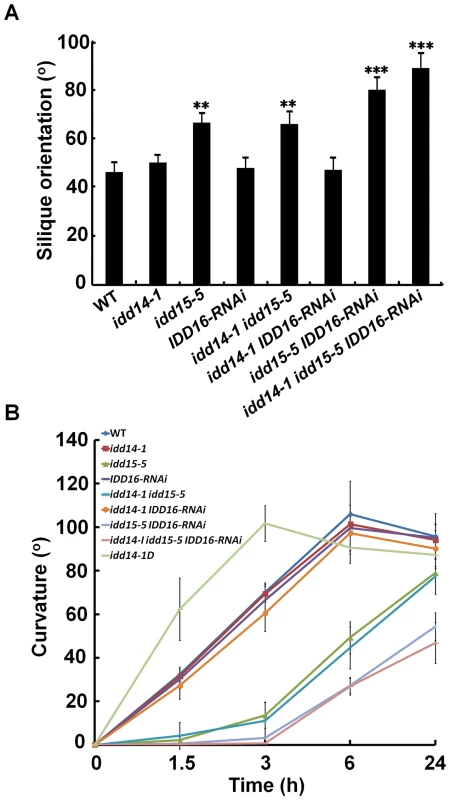
(A) Silique orientation in the idd single, double, and triple mutants. The angles between siliques and inflorescence stems were measured in 45-day-old WT and idd mutant plants. The data are shown as mean values ± one SD (Student's t-test, **P<0.01 and ***P<0.001). (B) Gravitropic responses of inflorescence stems in idd14-1D and loss-of-function idd mutants. The degree of curvature was determined from at least 18 inflorescence stems in each genotype after gravistimulation at the indicated times. The data are shown as mean values ± one SE. Surprisingly, although the IDD genes were found to be expressed in hypocotyls and roots, we did not observe any obvious phenotype in hypocotyls or roots of either idd14-1D or the idd triple mutant, including gravitropic response phenotypes (Figure S3B, S3C). The only aberration we observed was a slightly waved primary root phenotype in the idd triple mutant when grown vertically (Figure S3D). These findings suggest that the three IDDs may primarily function in lateral aerial organs.
IDD Affects Auxin Accumulation by Modulating Auxin Homeostasis and Transport
Previous studies have suggested that IDD14 and IDD15 are involved in the regulation of starch metabolism [38], [40]. However, the narrow, epinastic leaves in gain-of-function IDD mutant or transgenic plants seem to resemble, to some extent, those observed in auxin overproduction mutants or transgenic plants, such as yucD or p35S::YUCs transgenic plants [6], [7], [9]. By contrast, the rotund and curly rosette leaves, abnormal floral phenotype, and gravitropism defect in the loss-of-function idd mutants are also documented in the mutants defective in auxin biosynthesis or transport [2], [9]–[11]. This led us speculate that auxin accumulation or signaling may also be involved in IDD-mediated organ development and/or gravitropic response. To test this, we used the idd14-1D and idd triple mutant plants as representatives of gain - and loss-of-function idd mutants for our further analysis. We first examined auxin accumulation in their leaf, inflorescence stem, and root, by monitoring the expression of DR5∶GUS or DR5∶GFP, a widely used auxin gradient reporter [50]. On day 3 after leaf initiation, an obvious GUS signal was observed in WT leaf distal tips. A stronger and spatially-expanded GUS signal was observed in idd14-1D leaf tips. No obvious GUS signal, and thus no localized auxin maxima, was observed in the leaf tips of idd triple mutant plants (Figure 6A). Similar differential GUS patterns were subsequently observed in 5-day-old expanding and 15-day-old expanded leaves (Figure 6A). In inflorescence stems, the GUS signal was mainly observed in vascular tissues in WT, whereas strong GUS staining was present in the cortex and endodermis tissues in idd14-1D and weak GUS expression without a tissue-specific pattern was observed in the idd triple mutant (Figure 6B). The increased or decreased auxin accumulation was also found in the meristem regions of primary roots in idd14-1D and the idd triple mutant, respectively (Figure 6C). Further quantification of the GUS activity in these organs confirmed the variations of DR5∶GUS signals observed in idd14-1D and the idd triple mutant (Figure 6D). This observation strongly suggests that alteration of IDD expression affects auxin accumulation in multiple organs.
Fig. 6. Altered auxin accumulation and transport in gain- and loss-of-function idd mutants. 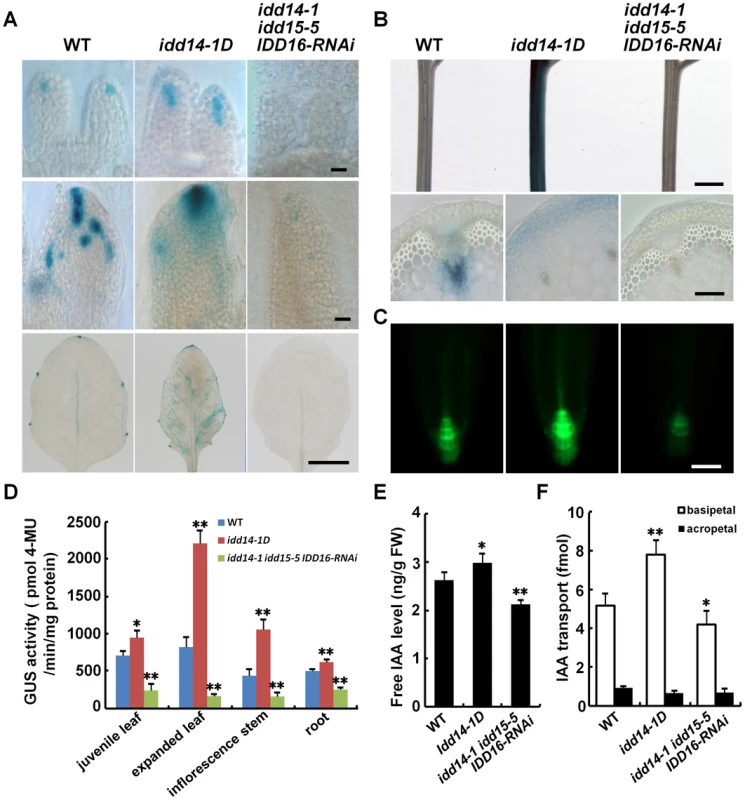
(A) Auxin accumulation assayed with the DR5∶GUS reporter in WT, idd14-1D, and idd triple mutant leaves. The leaves at 3, 5, and 15 days after initiation (from top to bottom panels) were subjected to GUS staining. The scale bars represent 50 µm in the top and middle panels and 5 mm in the bottom panel. (B) Expression of the DR5∶GUS reporter in inflorescence stems of WT, idd14-1D, and idd triple mutant plants. The scale bars represent 2 mm in the top panel and 100 µm in the bottom panel. (C) GFP fluorescence signals in the primary roots of WT, idd14-1D, and idd triple mutant carrying a DR5∶GFP reporter. The scale bar represents 50 µm. (D) Quantification of the GUS activity in the organs of the WT, idd14-1D, and idd triple mutant carrying a DR5∶GUS reporter. Data are from three biological replicates and shown as mean values ± one SD (Student's t-test, *P<0.05 and **P<0.01). (E) Endogenous free IAA levels in WT, idd14-1D, and idd triple mutant plants. Aerial organs of 15-day-old plants were used for measurement of free IAA levels. Data are from four biological replicates and are shown as mean values ± one SD (Student's t-test, *P<0.05 and **P<0.01). (F) Polar auxin transport capability of WT, idd14-1D, and idd triple mutant stems. The inflorescence stem segments of 5-week-old plants were used to determine the basipetal IAA transport efficiency and the background acropetal movement. Data are from four biological replicates and shown as mean values ± one SD (Student's t-test, *P<0.05 and **P<0.01). Auxin homeostasis and transport are key factors that determine the accumulation of auxin in plant organs [1], [2], [4]. To assess the contributions of auxin homeostasis and transport to IDD-mediated auxin accumulation, we first quantified the endogenous free IAA levels in the idd14-1D and idd triple mutants. Consistent with enhanced or decreased expression of DR5∶GUS reporter observed, the endogenous IAA level was increased by about 13% in idd14-1D but decreased by approximately 19% in idd triple mutant plants as compared to that in WT (Figure 6E). This result implied that the IDD genes may affect auxin biosynthesis. We then measured the auxin transport capability of inflorescence stems in idd14-1D and idd triple mutant. As shown in Figure 6F, the basipetal IAA transport efficiency was increased by over 50% in idd14-1D stems but decreased about 18% in idd triple mutant stems when compared with that in WT stem, demonstrating that IDD also modulates the auxin transport process. To further examine whether auxin signaling is affected in gain - and loss-of-function idd mutants, we monitored the expression of the DR5∶GUS reporter, IAA5, and IAA29 in response to exogenous IAA treatment in idd14-1D and the idd triple mutant, and observed that the expression of the DR5∶GUS reporter, IAA5, and IAA29 was normally induced following application of IAA as that in WT plants (Figure S4A, S4B), suggesting that IDD has no effect on auxin perception or signaling. We also observed that the expression of IDD14, IDD15, and IDD16 transcripts was not modulated by auxin treatment (Figure S4C). Taken together, our results strongly suggest that the three IDD genes are involved in the establishment of auxin gradients through the regulation of auxin biosynthesis and transport.
IDD Directly Activates the Expression of YUC5, TAA1, and PIN1
To identify the genes downstream of IDD, we first carried out a real-time quantitative RT-PCR (qRT-PCR) analysis to examine the transcript abundances of genes known to function in auxin biosynthesis and transport in the idd14-D and idd triple mutants. Among the YUC and TAA family genes, the transcription of YUC1, YUC2, YUC3, YUC4, YUC5, YUC8, and TAA1 was found to be elevated in idd14-1D, and the transcription levels of YUC2 and YUC5 were decreased in idd triple mutant, when compared to those in WT plants (Figure 7A). Among 14 genes related to auxin transport, the transcription of AUX1, PIN1, ABCB1, ABCB4 and WAG1 were found to be elevated in idd14-1D but reduced in the idd triple mutant compared to those in WT plants. PIN4 and PINOID (PID) transcripts were only elevated in idd14-1D or deceased in the idd triple mutant (Figure 7A). We further investigated the expression of 11genes, which had the apparently altered expression in idd14-1D or the idd triple mutant (with a significance at P<0.01), in the transgenic plants overexpressing IDD15 or IDD16 and the idd15-5 IDD16-RNAi plants. As expected, their differential expressions in gain - and loss-of-function IDD15 and IDD16 plants were much similar to those observed in idd14-1D and the idd triple mutant (Figure S5A), demonstrating that these genes are also downstream of IDD15 and IDD16.
Fig. 7. IDD Activates the expression of auxin biosynthetic and transport genes. 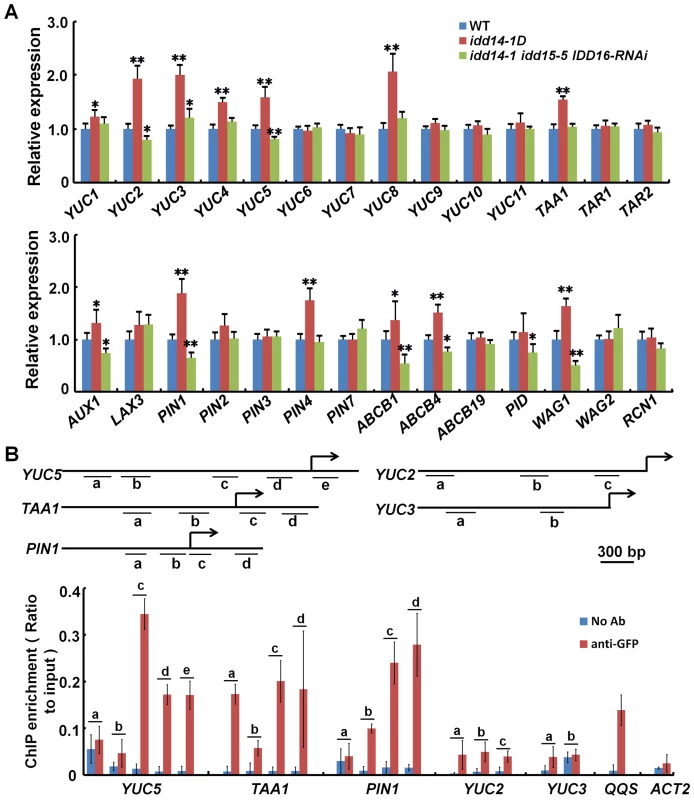
(A) Relative expression levels of the genes involved in auxin biosynthesis (top panel) and transport (bottom panel) in WT, idd14-1D, and idd triple mutant plants. RNAs isolated from aerial organs of 4-week-old plants were subjected to qRT-PCR analysis, and the data are from three biological replicates and shown as mean values ± one SD (Student's t-test, *P<0.05 and **P<0.01). (B) ChIP assay performed with p35S::IDD16-GFP transgenic plants by anti-GFP antibody. The DNA fragments with a putative IDD-binding motif in the promoter and upstream regions of YUC5, TAA1, PIN1, YUC2, and YUC3 are indicated as the letters a–e (top panel), and the enrichment values of their PCR products were quantified by qPCR (bottom panel). A reported IDD-targeted fragment in the QQS promoter and a fragment in the ACT2 promoter were used as the positive and negative controls, respectively. The data are from three biological replicates and shown as mean values ± one SD. As the IDDs are transcription factors, we speculated that some of the genes differentially expressed in gain - and loss-of-function idd mutants might be directly targeted by IDD. We first identified the genes whose expression was rapidly induced by the activation of IDD, using the chemically-inducible IDD14 transgenic plants. After the transgenic seedlings were treated with the inducer, IDD14 was dramatically induced by 0.5 h, and the expression of YUC5, TAA1, and PIN1 was obviously elevated within 2 h (Figure S5B), suggesting that three genes might be direct targets of IDDs. To examine whether IDD can directly bind to the promoter regions of YUC5, TAA1, or PIN1, we performed chromatin immunoprecipitation (ChIP) assays with both p35S::IDD16-GFP and p35S::IDD14-GFP transgenic plants. It has been reported that the maize ID1 and ID domain proteins could bind to a specific 11 bp DNA consensus motif, T-T-T-G-T-C-G/C-T/C-T/a-T/a-T [35]. As such, we targeted similar possible IDD-binding motifs in the promoter and/or upstream coding regions of YUC5, TAA1, PIN1, YUC2, and YUC3, and carried out the ChIP analysis (Figure 7B). As expected, we found that the three fragments containing a putative IDD-binding motif in YUC5, TAA1, or PIN1 were greatly enriched by IDD16 after GFP immunoprecipitation, whereas no binding activity was detected in the promoter regions of YUC2 or YUC3 (Figure 7B). Such enrichment was detected in a control DNA fragment of the QUA-QUINE STARCH (QQS) promoter, which was previously reported to be a target of IDD14 [38], but no enrichment was detectable in a control DNA fragment in the ACTIN2 (ACT2) promoter which lacked the putative IDD-binding motif (Figure 7B). Likewise, similar enrichments of these DNA fragments were further confirmed by ChIP assayed with IDD14 protein (Figure S5C). In addition, we indeed visualized the enhanced or attenuated PIN1 accumulation in the roots of idd14-1D and idd triple mutant carrying a pPIN1::PIN1-GFP construct, respectively (Figure S6). These results illustrate that IDD can directly target YUC5, TAA1, and PIN1, to activate their expression.
Alteration of Auxin Accumulation Suppresses Organ Morphogenesis in idd Mutants
Since idd14-1D contains a high auxin level while the idd triple mutant has low endogenous auxin content, we attempted to genetically modify auxin biosynthesis to examine whether this could suppress or restore the phenotype observed in gain - and loss-of-function idd mutants. We first generated an idd14-1D yuc2 yuc6 triple mutant through genetic crosses, and observed that loss-of-function yuc2 yuc6 completely suppressed the leaf phenotypes of idd14-1D (Figure 8A, S7), and partially attenuated the hypersensitivity of idd14-1D inflorescence stems to gravistimulation (Figure 8B). Further, when we overexpressed YUC2 in the idd triple mutant, the ectopic expression of YUC2 fully rescued the infertile silique defect (Figure 8C), but only partially restored the silique orientation in the idd triple mutant (Figure 8D). These results provide further evidence that auxin biosynthesis is genetically downstream of this IDD subfamily.
Fig. 8. Genetic interaction of auxin biosynthesis and IDD-mediated organ morphogenesis and gravitropism. 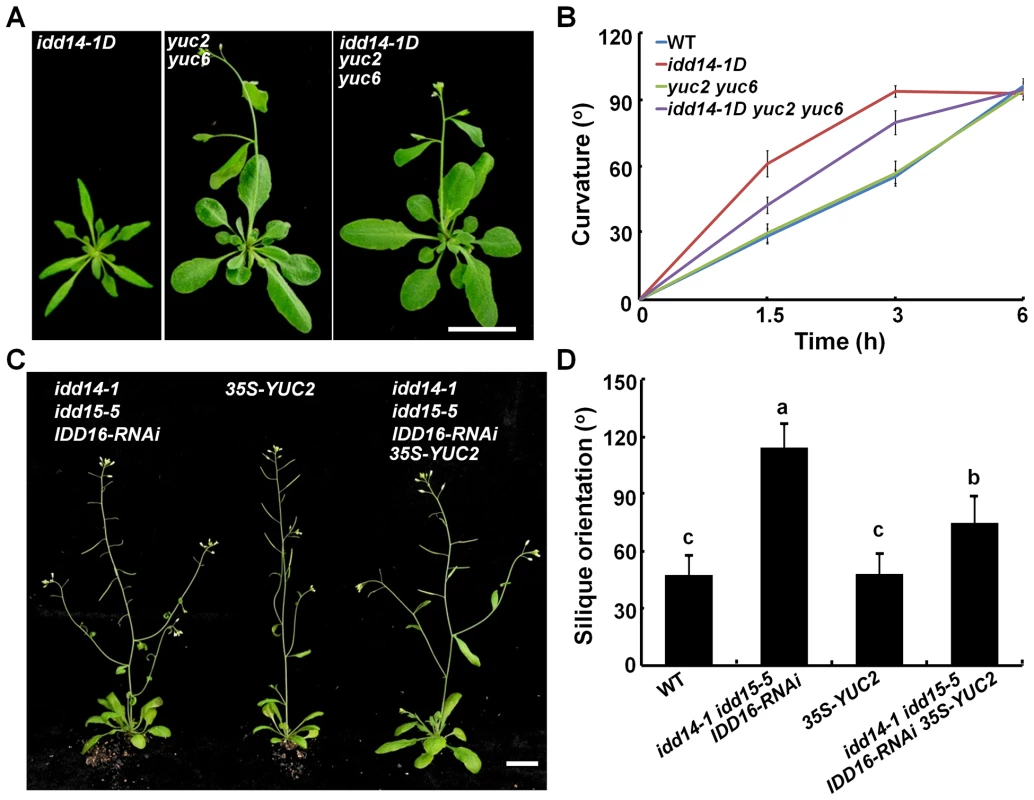
(A) Aerial organ morphology of 25-day-old idd14-1D, yuc2 yuc6, and idd14-1D yuc2 yuc6 plants. The scale bar represents 2 cm. (B) Gravitropic responses of WT, idd14-1D, yuc2 yuc6, and idd14-1D yuc2 yuc6 inflorescence stems. (C) 45-day-old plants of the idd triple mutant, 35S-YUC2, and idd triple mutant carrying a p35S::YUC2 construct. The scale bar represents 2 cm. (D) The silique orientation of WT, idd triple mutant, 35S-YUC2, and idd triple mutant carrying a p35S::YUC2 construct (one-way ANOVA test, P<0.05). As the amyloplast movement in endodermal cells of idd15 stem has been reported to be defective under gravistimulation [39]–[41], we also investigated whether ectopic expression of YUC2 has an effect on amyloplast movement in endodermal cells of the idd triple mutant stems. As shown in Figure S8, the retarded amyloplast movement in endodermal cells of the idd triple mutant stems under gravistimulation was not rescued by ectopically expressed YUC2 (Figure S8), suggesting that altered auxin accumulation does not affect IDD-mediated amyloplast responsiveness to gravistimulation.
Discussion
The Distinct IDD Subfamily Is Critical for Organ Development and Plant Architecture Formation
The IDD family has been defined as a plant-specific transcription factor family [35], [51], and previous characterizations of a few IDD members have indicated that IDDs are involved in the regulation of transition to flowering and starch metabolism [36]–[38]. IDD14, IDD15, and IDD16 belong to a subfamily that is distinct from the other IDD members in Arabidopsis, rice, and maize [34]. The mutation of IDD15 in Arabidopsis and rice reduces gravitropic response in inflorescence stems [39]–[41], [48]. However, because the loss-of-function idd14 mutant does not have obvious organ phenotype, and IDD16 has not yet been characterized, the functions of this subfamily are still largely unknown.
In this study, we characterized both gain - and loss-of-function mutants of this IDD subfamily, and discovered that these three IDD genes function redundantly and cooperatively in regulating organ morphogenesis and gravitropic responses. Gain-of-function of each IDD led to a small, narrow, and down-curled leaf phenotype. Although idd single mutants did not have obvious organ morphological phenotypes (except the reduced gravitropic response observed in idd15), our further characterization of the idd double and triple mutants clearly demonstrates that IDD14 and IDD16 act redundantly to regulate the morphology of aerial organs and affect fertility, while IDD15 and IDD16 cooperatively control the gravitropic responses and plant architecture.
Such redundant and cooperative roles of the three IDD genes in organ morphogenesis and gravitropism are consistent with their differential and overlapping expression patterns in particular organs. IDD14 and IDD16, but not IDD15, were expressed in juvenile leaves, whereas IDD15 and IDD16, but not IDD14, were highly expressed in inflorescence stems. The three IDD genes were all expressed in floral organs, and the abnormal flower phenotype in idd14-1 IDD16-RNAi was enhanced in the idd triple mutant. Interestingly, although the IDD genes were also expressed in hypocotyls and roots, we could not observe obvious changes in their morphology or gravitropic response in either gain - or loss-of-function idd mutants, except the slightly waved roots in idd triple mutant seedlings. This may be attributable to decreased expression of WAG1, a gene that is an indirect target of IDD and has been identified as a suppressor of root waving (Figure 7A) [52].
IDD14, IDD15, and IDD16 Regulate Auxin Gradients by Promoting Auxin Biosynthesis and Transport
Previous studies have shown that IDD15 and IDD14 are involved in the regulation of starch metabolism [38], [40]. Here, with a combination of phenotypic, genetic, and molecular approaches, we demonstrated that IDD14, IDD15, and IDD16 modulate auxin accumulation by affecting auxin biosynthesis and transport, thereby modifying organ morphogenesis and architecture formation. First, the organ phenotype in both gain - and loss-of-function idd mutants appears to be related to altered auxin homeostasis and distribution. For example, the narrow, epinastic leaves in the plants overexpressing IDD are similar to those in auxin overproduction mutants or transgenic plants, such as yucca (yuc1D), yucca6-1D, and p35S::YUCs transgenic plants [6], [7], [9], whereas the rotund and curly rosette leaves and abnormal floral phenotype in the loss-of-function idd mutants are similar to those observed in the mutants defective in auxin biosynthesis or transport [9], [53]. Disruption of IDD genes also influenced the silique and branch angles, root waving, and gravitropism of inflorescence stems, which have been well documented to be related to polar auxin transport [2], [54]–[57]. Second, expression analyses using DR5∶GUS and DR5∶GFP reporter clearly indicated that gain - or loss-of-function of IDDs enhanced or reduced auxin gradients, which was further confirmed by the increased or decreased endogenous auxin content and transport ability. Third, the expression of several genes involved in auxin biosynthesis and transport were altered in both gain - and loss-of-function idd mutants, and the IDD proteins could directly bind to the promoter regions of YUC5, TAA1, and PIN1 to activate their expression. In addition, genetic manipulation of auxin biosynthesis could fully or partially restore the pleiotropic phenotypes in idd14-1D or the idd triple mutant. These results demonstrate that IDD indeed modulates auxin gradients by promoting auxin biosynthesis and transport.
IDD-Mediated Auxin Gradients and Starch Metabolism May Coordinate Gravitropic Response
The gravitropic response in plants requires a coordination of three sequential processes: gravity perception, signal transduction, and asymmetric growth response [58]. It is widely believed that the starch-filled amyloplasts (statoliths) within specific gravi-sensing cells (statocytes) perceive gravity stimulation [59], [60]. Some other molecules, such as InsP3 and Ca2+ have been found to be involved in gravity signaling [60]. A large body of evidence indicates that auxin plays a key role in gravitropic signaling and asymmetric organ growth, and that it may possibly be involved in gravi-sensing [2], [60]. For example, many mutants related to auxin biosynthesis and especially transport such as taa1, aux1, pin1, and pin2, exhibit a defect in gravitropic responses [2], [11], [61]. Other mutants with altered silique or branch architecture, such as plethora (plt) in Arabidopsis and lazy1 (la1) in rice, also show altered auxin accumulation or transport within their organs [54], [56], [62].
Among the three IDD members we characterized, IDD15 and its rice ortholog, LPA1, have been previously reported to affect the gravitropic response by altering amyloplast sedimentation in the endodermis [39]–[41], [48]. Recently, IDD14 was also found to mediate starch degradation by directly activating the expression of QQS [38], suggesting that IDD14 also participates in the regulation of starch metabolism. Our detailed characterization of gain - and loss-of-function of three IDDs provides substantial evidence that IDD-mediated auxin biosynthesis and transport contribute to the organ morphogenesis and also, to some extent, gravitropic responses, because the genetic manipulation of auxin biosynthesis does not alter the responsiveness of amyloplast to gravistimulation but partially restores the gravity sensitivity or defect in gain - or loss-of-function idd mutants. Therefore, it is likely that IDD-mediated auxin accumulation and starch metabolism coordinately control the gravitropic responses. The IDD-regulated starch metabolism might be primarily involved in gravi-sensing while the IDD-regulated auxin gradient may be primarily involved in signaling and responses.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana accession Col-0 was used in this study. idd14-1D was isolated from a population generated by T-DNA activation-tagging mutagenesis. idd14-1 (CS367164) and idd15-5 (SALK_087765) were obtained from ABRC. All seeds were sterilized and geminated on 1/2 MS medium after vernalization for 2 days at 4°C, and the plants were grown in a culture room or growth chamber at 22±1°C with an illumination intensity of 80–90 µmol m−2 s−1 and a 16-h light/8-h dark photoperiod, as described previously [63].
Leaf Curvature and Gravitropism Assays
Leaf curvature was determined as described previously [64], [65]. Briefly, the transverse curvature (TC) index was defined as TC = 1 - cw/pw, where cw and pw are the curved width and the pressed width of leaves, respectively. The longitudinal curvature (LC) index was defined as LC = 1 - cl/pl, where cl and pl are the curved length and the pressed length of leaves, respectively. To quantify the pressed width or length of leaves, the sixth leaf of 25-day-old plants was dissected transversely or longitudinally on a desk, and then the extreme distances between the margins of the leaf were measured before and after pressing.
To quantify the gravitropic responses of roots and hypocotyls, seedlings were grown vertically for 4 days and then turned horizontally, and curvature angles were measured [58]. For quantification of the gravitropic responses of inflorescence stems, 32-day-old plants with inflorescence stems of approximately 4–8 cm were gravistimulated by rotating them 90° in darkness, and stem curvatures were measured as the angles between the growing direction of apex and horizontal base line [39], [58].
Genotyping of T-DNA Mutants
The T-DNA flanking sequence of idd14-1D was amplified by TAIL-PCR [66]. Three primers, P1, P2, and pSK-LB2 were used for co-segregation analysis. P1 and P2 were located in the Arabidopsis genome flanking the T-DNA insertion site, and pSK-LB2 was a primer corresponding to the left border of the T-DNA sequence. Similarly, idd14-1 and idd15-5 were genotyped with corresponding primers (Table S2), and yuc2 and yuc6 were genotyped according to the methods from a previous study [9].
Plasmid Construction and Plant Transformation
To generate p35S::IDDs and p35S::anti-IDD14 transgenic plants, the IDD coding sequences were amplified by RT-PCR and ligated into the pGEM-T-Easy vector (Promega, USA), and then verified by sequencing. The resulting plasmids were digested with EcoRI and cloned into pVIP96 [67]. The IDD14 cDNA was also cloned into pER8 to generate a chemically inducible IDD14 construct. To investigate the tissue-specific expression of IDD14, IDD15, and IDD16, approximately 2-kb promoter fragments of IDD genes were amplified from genomic DNA and then fused with the β-glucuronidase (GUS) gene into pBI101. To generate the p35S::IDD16-GFP and p35S::IDD14-GFP constructs, an IDD16 or IDD14 coding sequence lacking a stop codon was amplified and then cloned in frame into pMDC83 (Invitrogen, USA). To generate the p35S::IDD16-RNAi construct, a specific IDD16 cDNA fragment was amplified and ligated inversely into pBluescript SK-GUSF [68], and then an XbaI-BglII digested fragment was sub-cloned into pVIP96. A YUC2 cDNA fragment was amplified by RT-PCR and cloned into pVIP96myc to generate the p35S::YUC2 construct. All the primers used are listed in Table S2.
All constructs were introduced into Arabidopsis by Agrobacterium tumefaciens–mediated transformation via the floral dip as described previously [69]. At least 20 independent lines harboring a single T-DNA insertion from each construct were generated, and 4–5 independent lines of T3 homozygous plants were used for detailed characterization.
Gene Expression Analysis
Total RNA was isolated using a guanidine thiocyanate extraction buffer [70]. For semi-quantitative RT-PCR or qRT-PCR analysis, cDNA was synthesized from 1 µg of total RNA using SuperScript III Reverse Transcriptase (Invitrogen, USA). qRT-PCR was performed with a Rotor-Gene 3000 thermocycler (Corbett Research, Australian) with a SYBR Premix Ex Taq II kit (Takara, Japan). The relative expression level for each gene was normalized to the ACTIN2 and the data were collected from three biological replicates, as described previously [49].
The histochemical GUS assay was carried out according to previously described protocol [71]. The GUS activities were quantified by monitoring cleavage of the β-glucuronidase substrate 4-methylumbelliferyl β-D-glucuronide (MUG), as described [71].
For RNA in situ hybridization, a specific cDNA region of IDD14, IDD15, or IDD16 was transcribed in vitro to generate sense and antisense probes using the Digoxigenin RNA labeling kit (Roche, Switzerland). The WT inflorescence stems were fixed and embedded in paraffin (Sigma-Aldrich, USA), and then sectioned to a 10 µm thickness. RNA in situ hybridization was performed according to a previously described method [54].
Free IAA Measurement
Aerial organs from 15-day-old plants of WT, idd14-1D, and idd triple mutant plants were used for measurement of free IAA content. The extraction, purification, and analysis of free IAA by gas chromatography-mass spectrometry was performed according to the methods described by Edlund et al. [72], except that an Agilent GC and a LECO Pegasus TOF mass spectrometer was used, with separation using a DB-5ht column (Agilent, USA) [63].
Auxin Transport Assays
Auxin transport in inflorescence stems was measured according to the methods of a previously published protocol [73]. 25-mm inflorescence segments were cut from 5-week-old plants, and the segments were submerged inversely into an auxin transport buffer (100 nM 3H-IAA, 0.05% MES, pH 5.5–5.7) in a 0.5-ml micro-centrifuge tube. Control experiments were performed by submerging the base of inflorescence stems to measure acropetal IAA movement. After 12 h, 5-mm stem segments were dissected from the non-submerged ends and used to quantify the radiolabeled auxin using a scintillation counter.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed as described previously [74]. Briefly, 2 g of p35S::IDD16-GFP or p35S::IDD14-GFP transgenic plants grown on 1/2 MS plates for 16 days was harvested, and then submerged in 1% formaldehyde to crosslink the DNA with DNA-binding proteins. The chromatin pellets were extracted and sheared by sonication. 5 µl anti-GFP antibodies (Abcam, UK) were used to immunoprecipitate the DNA-IDD16 or DNA-IDD14 complexes. DNA was released with proteinase K and then purified. The enrichment of DNA fragments was determined by quantitative PCR with the primers listed in Table S2.
Amyloplast Staining
Plants were grown in soil until the primary inflorescence stems bolted to a height of 4–9 cm, and then gravistimulated by rotating them upside down [40]. 1-cm-long inflorescence stems below the apex were fixed, embedded in paraffin, and sectioned to a 10-µm thickness. A periodic acid-Schiff kit (Sigma-Aldrich, USA) was used for amyloplast staining, according to the manufacturer's instructions.
Supporting Information
Zdroje
1. ZhaoY (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61 : 49–64.
2. PetrášekJ, FrimlJ (2009) Auxin transport routes in plant development. Development 136 : 2675–2688.
3. HayashiK (2012) The interaction and integration of auxin signaling components. Plant Cell Physiol 53 : 965–975.
4. WoodwardAW, BartelB (2005) Auxin: regulation, action, and interaction. Ann Bot 95 : 707–735.
5. VannesteS, FrimlJ (2009) Auxin: a trigger for change in plant development. Cell 136 : 1005–1016.
6. ZhaoY, ChristensenSK, FankhauserC, CashmanJR, CohenJD, et al. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 : 306–309.
7. KimJI, SharkhuuA, JinJB, LiP, JeongJC, et al. (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145 : 722–735.
8. WoodwardC, BemisSM, HillEJ, SawaS, KoshibaT, et al. (2005) Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol 139 : 192–203.
9. ChengY, DaiX, ZhaoY (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 : 1790–1799.
10. ChengY, DaiX, ZhaoY (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19 : 2430–2439.
11. StepanovaAN, Robertson-HoytJ, YunJ, BenaventeLM, XieDY, et al. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 : 177–191.
12. TaoY, FerrerJL, LjungK, PojerF, HongF, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 : 164–176.
13. YamadaM, GreenhamK, PriggeMJ, JensenPJ, EstelleM (2009) The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151 : 168–179.
14. MashiguchiK, TanakaK, SakaiT, SugawaraS, KawaideH, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108 : 18512–18517.
15. WonC, ShenX, MashiguchiK, ZhengZ, DaiX, et al. (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108 : 18518–18523.
16. ZhaoY (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5 : 334–338.
17. SohlbergJJ, MyrenåsM, KuuskS, LagercrantzU, KowalczykM, et al. (2006) STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J 47 : 112–123.
18. TriguerosM, Navarrete-GómezM, SatoS, ChristensenSK, PelazS, et al. (2009) The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell 21 : 1394–1409.
19. StoneSL, BraybrookSA, PaulaSL, KwongLW, MeuserJ, et al. (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc Natl Acad Sci USA 105 : 3151–3156.
20. SunJ, QiL, LiY, ChuJ, LiC (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594.
21. LiLC, QinGJ, TsugeT, HouXH, DingMY, et al. (2008) SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol 179 : 751–764.
22. BennettMJ, MarchantA, GreenHG, MayST, WardSP, et al. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273 : 948–950.
23. PetrášekJ, MravecJ, BouchardR, BlakesleeJJ, AbasM, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 : 914–918.
24. SwarupK, BenkováE, SwarupR, CasimiroI, PéretB, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10 : 946–954.
25. BainbridgeK, Guyomarc'hS, BayerE, SwarupR, BennettM, et al. (2008) Auxin influx carriers stabilize phyllotactic patterning. Genes Dev 22 : 810–823.
26. JonesAR, KramerEM, KnoxK, SwarupR, BennettMJ, et al. (2009) Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol 11 : 78–84.
27. MarchantA, BhaleraoR, CasimiroI, EklöfJ, CaseroPJ, et al. (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14 : 589–597.
28. StoneBB, Stowe-EvansEL, HarperRM, CelayaRB, LjungK, et al. (2008) Disruptions in AUX1-dependent auxin influx alter hypocotyl phototropism in Arabidopsis. Mol Plant 1 : 129–144.
29. BenkováE, MichniewiczM, SauerM, TeichmannT, SeifertováD, et al. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 : 591–602.
30. ChenR, HilsonP, SedbrookJ, RosenE, CasparT, et al. (1998) The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA 95 : 15112–15117.
31. FrimlJ, VietenA, SauerM, WeijersD, SchwarzH, et al. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 : 147–153.
32. MravecJ, KubešM, BielachA, GaykovaV, PetrášekJ, et al. (2008) Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135 : 3345–3354.
33. ScarpellaE, MarcosD, FrimlJ, BerlethT (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev 20 : 1015–1027.
34. ColasantiJ, TremblayR, WongAY, ConevaV, KozakiA, et al. (2006) The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7 : 158.
35. KozakiA, HakeS, ColasantiJ (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32 : 1710–1720.
36. ColasantiJ, YuanZ, SundaresanV (1998) The INDETERMINATE1 gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93 : 593–603.
37. SeoPJ, RyuJ, KangSK, ParkCM (2011) Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J 65 : 418–429.
38. SeoPJ, KimMJ, RyuJY, JeongEY, ParkCM (2011) Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat Commun 2 : 303.
39. MoritaMT, SakaguchiK, KiyoseS, TairaK, KatoT, et al. (2006) A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant J 47 : 619–628.
40. TanimotoM, TremblayR, ColasantiJ (2008) Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Mol Biol 67 : 57–69.
41. YamauchiY, FukakiH, FujisawaH, TasakaM (1997) Mutations in the SGR4, SGR5 and SGR6 loci of Arabidopsis thaliana alter the shoot gravitropism. Plant Cell Physiol 38 : 530–535.
42. LevesqueMP, VernouxT, BuschW, CuiH, WangJY, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143.
43. WelchD, HassanH, BlilouI, ImminkR, HeidstraR, et al. (2007) Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21 : 2196–2204.
44. FeurtadoJA, HuangD, Wicki-StordeurL, HemstockLE, PotentierMS, et al. (2011) The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23 : 1772–1794.
45. MatsubaraK, YamanouchiU, WangZX, MinobeY, IzawaT, et al. (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 148 : 1425–1435.
46. ParkSJ, KimSL, LeeS, JeBI, PiaoHL, et al. (2008) Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J 56 : 1018–1029.
47. WuC, YouC, LiC, LongT, ChenG, et al. (2008) RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA 105 : 12915–12920.
48. WuX, TangD, LiM, WangK, ChengZ (2013) Loose Plant Architecture 1, an INDETERMINATE domain protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol 161 : 317–329.
49. HuZ, QinZ, WangM, XuC, FengG, et al. (2010) The Arabidopsis SMO2, a homologue of yeast TRM112, modulates progression of cell division during organ growth. Plant J 61 : 600–610.
50. UlmasovT, MurfettJ, HagenG, GuilfoyleTJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 : 1963–1971.
51. EnglbrechtCC, SchoofH, BöhmS (2004) Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5 : 39.
52. SantnerAA, WatsonJC (2006) The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J 45 : 752–764.
53. BlakesleeJJ, BandyopadhyayA, LeeOR, MravecJ, TitapiwatanakunB, et al. (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19 : 131–147.
54. LiP, WangY, QianQ, FuZ, WangM, et al. (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17 : 402–410.
55. YoshiharaT, IinoM (2007) Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol 48 : 678–688.
56. PrasadK, GriggSP, BarkoulasM, YadavRK, Sanchez-PerezGF, et al. (2011) Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr Biol 21 : 1123–1128.
57. OlivaM, DunandC (2007) Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytol 176 : 37–43.
58. FukakiH, FujisawaH, TasakaM (1996) SGR1, SGR2, SGR3: novel genetic loci involved in shoot gravitropism in Arabidopsis thaliana. Plant Physiol 110 : 945–955.
59. HashiguchiY, TasakaM, MoritaMT (2013) Mechanism of higher plant gravity sensing. Am J Bot 100 : 91–100.
60. BaldwinKL, StrohmAK, MassonPH (2013) Gravity sensing and signal transduction in vascular plant primary roots. Am J Bot 100 : 126–142.
61. HagaK, SakaiT (2012) PIN auxin efflux carriers are necessary for pulse-induced but not continuous light-induced phototropism in Arabidopsis. Plant Physiol 160 : 763–776.
62. PinonV, PrasadK, GriggSP, Sanchez-PerezGF, ScheresB (2012) Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci USA 110 : 1107–1112.
63. JingY, CuiD, BaoF, HuZ, QinZ, et al. (2009) Tryptophan deficiency affects organ growth by retarding cell expansion in Arabidopsis. Plant J 57 : 511–521.
64. NathU, CrawfordBC, CarpenterR, CoenE (2003) Genetic control of surface curvature. Science 299 : 1404–1407.
65. WuF, YuL, CaoW, MaoY, LiuZ, et al. (2007) The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19 : 914–925.
66. LiuYG, MitsukawaN, OosumiT, WhittierRF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8 : 457–463.
67. HuY, XieQ, ChuaNH (2003) The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15 : 1951–1961.
68. QinG, GuH, ZhaoY, MaZ, ShiG, et al. (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17 : 2693–2704.
69. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743.
70. HuY, BaoF, LiJ (2000) Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J 24 : 693–701.
71. JeffersonRA, KavanaghTA, BevanMW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 : 3901–3907.
72. EdlundA, EklöfS, SundbergB, MoritzT, SandbergG (1995) A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol 108 : 1043–1047.
73. LewisDR, MudayGK (2009) Measurement of auxin transport in Arabidopsis thaliana. Nat Protoc 4 : 437–451.
74. GendrelAV, LippmanZ, MartienssenR, ColotV (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2 : 213–218.
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Akutní intermitentní porfyrie
- Příjem alkoholu a menstruační cyklus
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
- Transfer zmraženého embrya zlepšuje výsledky IVF
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



