-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklama: A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
Cilia are architecturally complex organelles that protrude from the cell membrane and have signalling, sensory and motility functions that are central to normal tissue development and homeostasis. There are two broad categories of cilia; motile and non-motile, or primary, cilia. The central role of primary cilia in health and disease has become prominent in the past decade with the recognition of a number of human syndromes that result from defects in the formation or function of primary cilia. This rapidly growing class of conditions, now known as ciliopathies, impact the development of a diverse range of tissues including the neural axis, craniofacial structures, skeleton, kidneys, eyes and lungs. The broad impact of cilia dysfunction on development reflects the pivotal position of the primary cilia within a signalling nexus involving a growing number of growth factor systems including Hedgehog, Pdgf, Fgf, Hippo, Notch and both canonical Wnt and planar cell polarity. We have identified a novel ENU mutant allele of Ift140, which causes a mid-gestation embryonic lethal phenotype in homozygous mutant mice. Mutant embryos exhibit a range of phenotypes including exencephaly and spina bifida, craniofacial dysmorphism, digit anomalies, cardiac anomalies and somite patterning defects. A number of these phenotypes can be attributed to alterations in Hedgehog signalling, although additional signalling systems are also likely to be involved. We also report the identification of a homozygous recessive mutation in IFT140 in a Jeune syndrome patient. This ENU-induced Jeune syndrome model will be useful in delineating the origins of dysmorphology in human ciliopathies.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003746
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003746Summary
Cilia are architecturally complex organelles that protrude from the cell membrane and have signalling, sensory and motility functions that are central to normal tissue development and homeostasis. There are two broad categories of cilia; motile and non-motile, or primary, cilia. The central role of primary cilia in health and disease has become prominent in the past decade with the recognition of a number of human syndromes that result from defects in the formation or function of primary cilia. This rapidly growing class of conditions, now known as ciliopathies, impact the development of a diverse range of tissues including the neural axis, craniofacial structures, skeleton, kidneys, eyes and lungs. The broad impact of cilia dysfunction on development reflects the pivotal position of the primary cilia within a signalling nexus involving a growing number of growth factor systems including Hedgehog, Pdgf, Fgf, Hippo, Notch and both canonical Wnt and planar cell polarity. We have identified a novel ENU mutant allele of Ift140, which causes a mid-gestation embryonic lethal phenotype in homozygous mutant mice. Mutant embryos exhibit a range of phenotypes including exencephaly and spina bifida, craniofacial dysmorphism, digit anomalies, cardiac anomalies and somite patterning defects. A number of these phenotypes can be attributed to alterations in Hedgehog signalling, although additional signalling systems are also likely to be involved. We also report the identification of a homozygous recessive mutation in IFT140 in a Jeune syndrome patient. This ENU-induced Jeune syndrome model will be useful in delineating the origins of dysmorphology in human ciliopathies.
Introduction
Primary cilia provide highly regulated cellular sensory functions with diverse roles in the development and maintenance of many tissues and organs [1]. Primary cilia protrude from the surface of most eukaryotic cells except fungi and higher plants. The structural support for this arrangement comes from a tubulin-based scaffold known as the axoneme, the assembly and function of which is coordinated through a centriole-derived basal body. The basal body is tethered to the plasma membrane by transition fibres that are believed to form a selective gate or pore which functions to control the movement of proteins between the cytoplasm and cilium [2]. The sensory function of the cilia derives from the deployment of plasma membrane-spanning receptors and channels within the ciliary membrane that provide a link between the extracellular environment and the cytoplasm [1].
Components of the primary cilia have to be transported from their site of synthesis within the cytoplasm to the cilium itself using a microtubule motor-based system known as intraflagellar transport (IFT). The axoneme grows from the distal tip and individual axonemal subunits must be transported along the nascent axoneme by IFT [3]. The IFT complex is simultaneously in close physical association with both the axoneme and the ciliary membrane enabling transport of membrane bound proteins such as growth factor receptors [2]. Given the blind ending structure of the cilia, IFT must proceed in a two-way fashion. The IFT particle is composed of at least two distinct sub-complexes, complex B and complex A, which are specialized for either outbound (anterograde) transport or the return (retrograde) journey respectively [4], [5], [6], [7]. The IFT particles form linear arrays known as IFT trains, which are transported by different motor protein complexes depending on the direction of movement. The anterograde transport complex B uses a kinesin-based motor while the retrograde transport complex A uses a dynein-based motor system [8].
The IFT-A complex consists of at least 6 subunits with a highly stable core sub-complex of three proteins, while the IFT-B complex consists of around 14 subunits with a salt-stable core of 9 [9]. The nature of the interaction between the IFT-A and B complexes and their respective motor protein complexes remains unclear but anterograde transport components become cargo for the retrograde complex and the converse is also true [10], [11]. Thus, mutation of any individual subunit is likely to impact on the function of the entire system. However, each transport complex has specific roles within the cilia, mutations within IFT-B tend to result in short or absent cilia while mutations in IFT-A more typically result in distorted cilia with a distended tip due to accumulation of stranded IFT cargo [12].
Over the past decade our appreciation of the importance of primary cilia function in development and disease has grown rapidly. There are now at least 12 disorders that have been attributed to mutations in cilia proteins and collectively they impact on nearly every organ system in the body [13]. The skeletal ciliopathies are a diverse group of conditions involving congenital malformation of variable skeletal systems including the craniofacial skeleton, ribs and limbs. One example is the short-rib polydactyly (SRP) group which includes at least 6 distinct autosomal recessive conditions including Jeune syndrome [14]. Common features of the SRPs include a small thoracic cage due to a failure of normal rib growth, shortened tubular bones and pelvic defects. In addition, individual conditions present with a diverse range of soft-tissue defects including malformations of the heart, intestine, genitalia, pancreas, liver and polycystic kidneys. There is significant phenotypic overlap between these conditions and recent identification of causative genetic mutations confirms that the difficulty in distinguishing the features of particular SRPs stems from defects common to the structure or function of the primary cilia [15].
The primary cilia are endowed with an array of signal transduction components such as ion channels, receptor tyrosine kinases, Notch receptors and matrix receptors including the Hedgehog signalling components Ptch1, Smo1 and Gli, PDGFRa, IGFR1, EGFR and Tie2 [16]. The function and activity of these sensory components is regulated, at least in part, by controlling their localisation within the cilia and in part by controlling access to downstream signalling components. The co-localisation of multiple signal transduction components within the cilia raises the possibility that the cilia act as a site to coordinate cross talk between signalling systems [16]. Thus, the broad range of phenotypes resulting from mutations in primary cilium-associated proteins is likely due to cell type specific disturbances in a range of different signalling systems.
We carried out an ENU mutagenesis screen to identify genes with critical functions during early embryonic development [17] and have identified a mutant with a strong primary cilia phenotype. Linkage and candidate gene sequencing identified a missense mutation in the core IFT-A gene Ift140. This mutation results in mid-gestation lethality, exencephaly, craniofacial dysmorphism, skeletal malformation and mixed poly - and oligodactyly. A number of these defects can be attributed to disregulated Hedgehog signalling while other defects indicate involvement of additional signalling systems. Based on the identification of an Ift140 mutation in this mouse model we also sequenced the human orthologue in a cohort of SRP and Jeune syndrome patients. We present the identification of a novel IFT140 mutation in a case of Jeune syndrome. IFT140 has recently been implicated in Mainzer-Saldino and Jeune syndromes and conditional deletion in mice causes a polycystic kidney phenotype [18], [19], [20]. Together these data confirm the validity of our mouse model of Jeune syndrome and will underpin further investigations into the mechanisms responsible for this condition.
Results
Cauli is a novel ciliopathy mouse model with a recessive missense mutation in the Ift140 gene
The cauli strain was identified via a comprehensive phenotype-driven ENU screen undertaken to identify novel genes involved in embryogenesis [17]. The vast majority of cauli homozygous embryos survive to approximately embryonic day (E) 13.5, however a small proportion has lived beyond this age (n = 10/130). No live embryos were obtained >E16.5. Fully penetrant phenotypes identified in cauli mutants include exencephaly (n = 130/130), anopthalmia (n = 130/130) and polydactyly of the hindlimbs (Figure 1B; n = 32/32 embryos analysed at >E12.5). Cauli mutants were developmentally delayed compared to wildtype littermates and additional phenotypes observed in these mice include oligodactyly of the forelimbs, gaping mouth, omphaloceole, oedema, curly tail and caudal neural tube closure defects (Figures 1A and B).
Fig. 1. An Ift140 mutation is responsible for the ciliopathic phenotype observed in cauli. 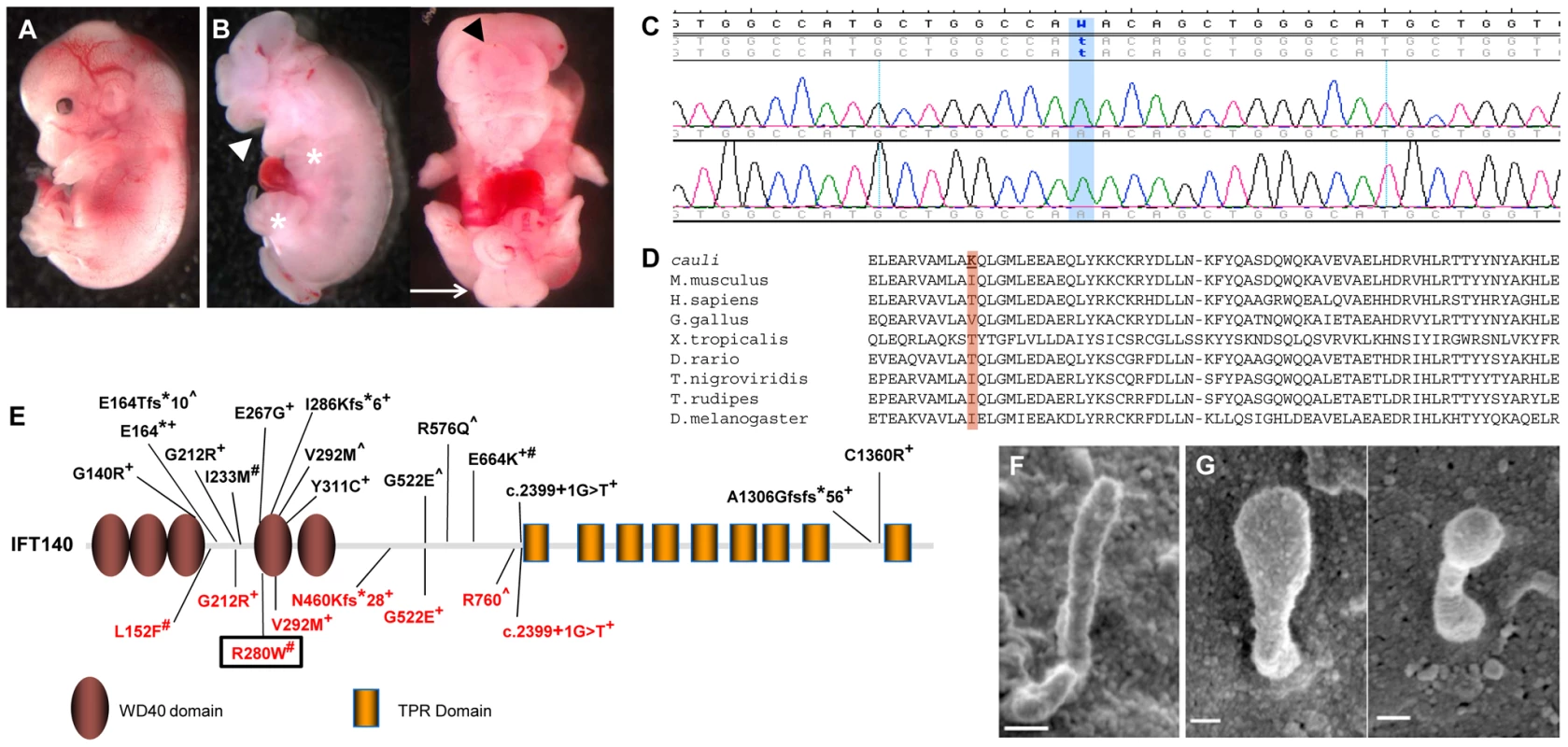
Representative E13.5 wildtype (A) and mutant (B) embryos showing exencephaly (black arrowhead), open mouth (white arrowhead), polydactyly (asterisks) and caudal neural tube closure defects (arrow) in mutants. Chromatogram of cauli mutant showing the homozygous missense mutation (c.2564T>A) in the Intraflagellar Transport Protein 140 (Ift140) gene (C). IFT140 protein alignment showing the isoleucine to lysine substitution at position 855 of the protein in cauli and the corresponding amino acid across several species (D). Schematic of the IFT140 protein detailing protein domains, location of Ift140cauli/cauli mutation and reported human mutations (E). Mainzer-Saldino (black), Jeune asphyxiating thoracic dystrophy (red), +compound heterozygous, #homozygous ∧no second mutation identified. Black box represents mutation reported in this paper. Primary cilia from E10.5 Ift140+/+ (F) and Ift140cauli/cauli (G) limb buds show a severely altered cilia morphology in the mutant. Linkage analysis in cauli identified a 7 Mb locus on chromosome 17 between markers rs3667809 and rs3684506 harbouring 218 genes. The Ift140 gene was prioritised as a strong putative candidate due to the similarity of the cauli phenotype with other ciliopathy mouse models [21], [22], [23]. Direct sequencing of the Ift140 gene (NCBI RefSeq transcript NM_134126.3) identified a T to A substitution in exon 19 at position 2564 (Figure 1C; c.2564T>A), resulting in an isoleucine to lysine amino acid change at position 855 of the protein (p.I855K).
We utilized two algorithms designed to assess the potential effect of the isoleucine to lysine mutation on Ift140 function. The prediction output from PolyPhen-2 for the cauli Ift140 p.I855K mutation indicates a ‘possibly damaging’ effect of the mutation on protein function, with a PSIC score of 0.503. SIFT prediction output denotes that the mutation is ‘not tolerated’, with a probability of 0.00, projected to be deleterious. Alternative amino acids found at this residue in other species (Threonine and Valine) were all predicted to be ‘tolerated’ by this analysis (Figure 1D). This residue resides within a coiled-coil domain immediately upstream of the first tetratricopeptide repeat (TPR) domain of mouse IFT140, as predicted by the algorithms TPRpred and coils/pcoils [24] (http://toolkit.tuebingen.mpg.de/). Interestingly, secondary structure prediction using the PSIPRED algorithm [25] indicates that the I855K mutation will not disrupt the α-helix in which it is embedded and therefore likely has a subtle impact on the overall structure of Ift140. Thus, the implications of the I855K change for mutant Ift140 function are unclear.
Ultrastructural examination of the primary cilia of E10.5 Ift140+/+ and Ift140cauli/cauli limb buds by SEM (Figures 1F and G) revealed, that when present, cilia morphology was severely disrupted in Ift140cauli/cauli mutant embryos. Cilia in these mice were much broader and had a bulbous appearance, consistent with the accumulation of cargo at the tip due to a retrograde transport defect.
Ift140cauli/cauli embryos exhibit multiple developmental defects
Morphological differences between Ift140+/+ and Ift140cauli/cauli embryos were analysed using a number of techniques. Skeletal preparations of surviving E16.5 mutant embryos (n = 2) revealed severe craniofacial defects including failure of calvarial development, cervical vertebral fusion and agenesis or hypoplasia of many facial bones (Figures 2A–F). The exoccipital was fused to the first cervical vertebra. The shape of the exoccipital, bassioccipital and basisphenoid, appeared relatively normal. In contrast, the tympanic ring, alisphenoid and palatine were absent or rudimentary while the maxilla and premaxilla were hypoplastic and malpositioned due to the absence of components with which they would normally articulate. Examination of the thoracic skeleton revealed severe rib defects in Ift140cauli/cauli embryos when compared to the ordered structure seen in Ift140+/+ controls (Figures 2G and H). Although the normal complement of 13 ribs was present, anomalies of the ribs in Ift140cauli/cauli embryos include abnormal costovertebral articulations (the joints between the heads of each rib and the thoracic vertebrae), lateral bifurcation (branching) and thickened ossified protuberances (exostoses, Figure 2H). Due to the gestational lethality observed in Ift140cauli/cauli embryos further skeletal analysis was not possible. To further examine the origins of axial defects observed in skeletal preparations, somite patterning was analysed in E11.5 embryos by in situ hybridisation with the somatic marker myogenin (Figures 2I–L). Consistent with the rib phenotype identified in older embryos, somites in E11.5 Ift140cauli/cauli mutants were extremely disorganised with a bifid appearance, more evident in somites located in the rostral region (Figures 2J and L). This pattern is in stark contrast to the highly ordered somite pattern seen in Ift140+/+ control embryos (Figures 2I and K). Sections of wholemount embryos confirmed the disruption of normal myogenin staining and revealed the accumulation of blood within distended and irregularly located inter-somitic vessels (Figures 2K and L). In addition to the obvious cranial neural tube defects, irregularities in the integrity of the axial neural tube were also revealed by in situ hybridisation analysis. Msx1 marks the dorsal-most neural tube cell population and in E11.5 wildtype controls appears as a continuous line of staining, while in Ift140cauli/cauli mutants staining is irregular and discontinuous (Figures 2M and N). Analysis of older embryos (E12.5) with a Sox9 in situ probe reveals the straight, aligned neural tube of Ift140+/+ embryos (Figure 2O) in comparison to the irregular and extremely convoluted neural tube seen in Ift140cauli/cauli mutants (Figure 2P).
Fig. 2. Ift140cauli/cauli embryos exhibit skeletal, somite and neural tube defects. 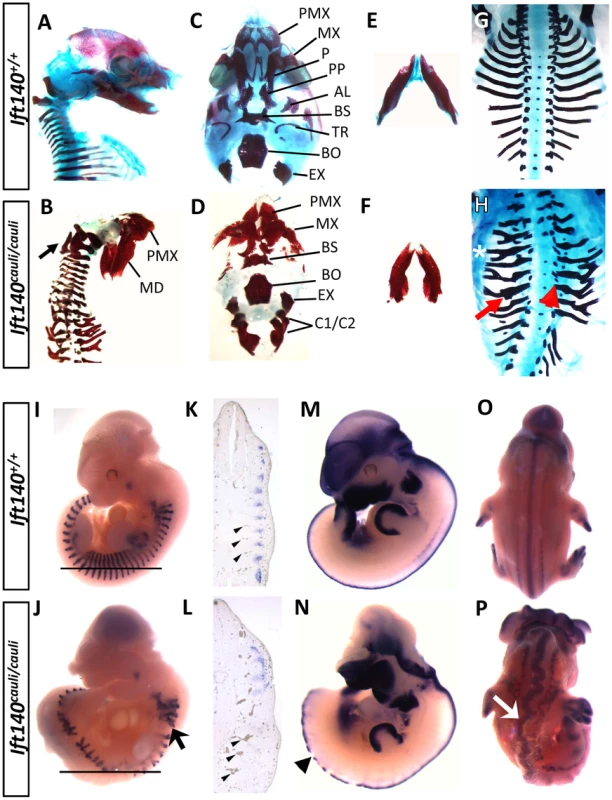
Morphological and expression analysis of Ift140+/+ and Ift140cauli/cauli embryos. Lateral view of E16.5 skull in Ift140+/+ (A) and Ift140cauli/cauli (B) embryos. Ift140cauli/cauli embryos exhibit fusion of the exoccipital bone and C1/C2 vertebrae (arrow in B). Ventral view of skull base in Ift140+/+ (C) and Ift140cauli/cauli (D) embryos. Ift140+/+ (E) and Ift140cauli/cauli (F) mandibles. The normal organisation of the ribs seen in E16.5 Ift140+/+ embryos (G) is severely disrupted in Ift140cauli/cauli (H) with lateral branching (asterisk), thickened ossified nodules (red arrow) and abnormal costovertebral articulations (red arrowhead). (I–P) In situ hybridisation of gene expression patterns of myogenin (I–L), Msx1 (M,N) and Sox9 (O,P). Myogenin staining at E11.5 reveals the highly ordered segmental structure of a Ift140+/+ embryo (I) while in the Ift140cauli/cauli embryo (J) myogenin staining reveals the presence of disorganised and branched somite-derived structures (myotome; arrow). (K,L) Sections of wholemount embryos at the level indicated by the horizontal line in I and J, illustrating the loss of segmental myogenin staining and the accumulation of blood within distorted and irregular intersomitic vessels (arrowheads) in Ift140cauli/cauli embryos. (M,N) Msx1 expression delineates the dorsal margin of the neural tube in an E11.5 Ift140+/+ embryo (M) but highlights the disrupted neural tube structure in an Ift140cauli/cauli embryo (arrowhead in N). In addition, the neural tube is convoluted and irregular in appearance, as shown in E12.5 embryos stained for Sox9 (arrow in P). PMX, premaxilla; MD, mandible; MX, maxilla; P, palatine; PP, palatal process; AL, alisphenoid; BS, basisphenoid; TR, tympanic ring; BO, basioccipital; EX, exoccipital; C1/C2, fused 1st and 2nd cervical vertebrae. Ift140cauli/cauli mutant embryos begin to die at E13.5 during the stage when palatal fusion occurs. It is therefore difficult to determine if older mutants have a specific palate fusion defect or whether palate fusion is impeded due to a global disruption of growth preceding death. To investigate the impact of the cauli mutation on palatal development we performed histological analysis at E13.5 in healthy individuals. Coronal sections through the palatal shelves of Ift140+/+ and Ift140cauli/cauli embryos at E13.5 identified hypoplastic palatal shelves in Ift140cauli/cauli mutants, suggesting that palate fusion would be impeded in older mice (Figures 3A and B). Histological analysis of E13.5 Ift140+/+ and Ift140cauli/cauli embryos also highlighted numerous deformities of the internal organs (Figures 3C–F). Although the kidneys in Ift140cauli/cauli embryos appear grossly normal and comparable to Ift140+/+ kidneys, an unusual cavity is evident around these structures, consistent with an accumulation of fluid (Figures 3C and D). The lungs are severely misshapen in Ift140cauli/cauli embryos with a rounded appearance, unlike the cone-shape lobes clearly evident in Ift140+/+ sections (Figures 3E and F). Analysis of heart morphology indicates an acute heart phenotype in Ift140cauli/cauli embryos when compared to the normal heart structure of Ift140+/+ controls (Figures 3E and F). The unusual accumulation of blood within the atria may be a consequence of the malformed tricuspid and mitral atrioventricular valves and the irregular interventricular septum formation identified in Ift140cauli/cauli embryos (Figure 3F).
Fig. 3. Ift140cauli/caul mutants show palate defects, hydrops fetalis and malformation of the lungs and heart. 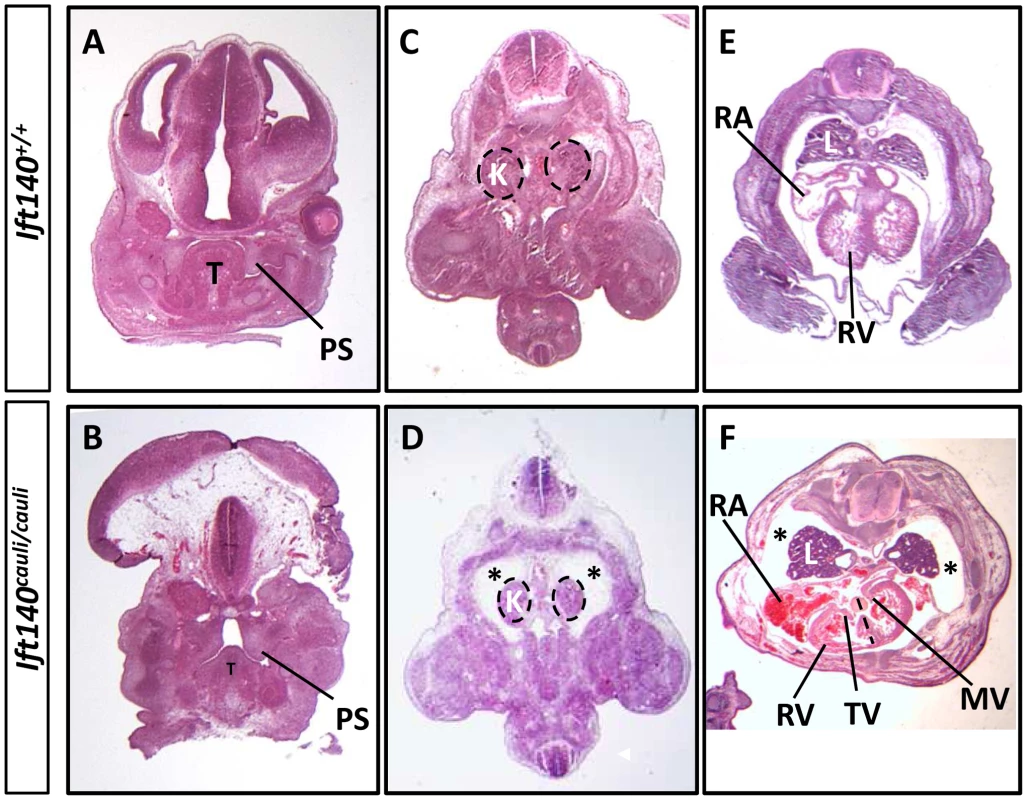
Coronal sections of E13.5 Ift140+/+ (A) and Ift140cauli/cauli (B) palates, highlighting hypoplastic palatal shelves in Ift140cauli/cauli embryos. Transverse sections of E13.5 Ift140+/+ (C and E) and Ift140cauli/cauli (D and F) embryos at the level of the kidneys (C and D) and heart (E and F). Ift140cauli/cauli mutant embryos have grossly normal kidneys but show accumulation of fluid around the kidneys and lungs (asterisks in D and F), which is not evident in Ift140+/+ controls (C and E). The lungs of Ift140cauli/cauli embryos are also abnormal in shape (F), unlike the cone-shaped lobes seen in Ift140+/+ controls (E). The atrioventricular valves of the heart are well formed in Ift140+/+ embryos (E), but both the tricuspid and mitral valves are abnormal in Ift140cauli/cauli mutants (F). Ift140cauli/cauli embryos appear to have ventricular hypotrophy and the interventricular septum is not well formed (depicted by dashed line in F). An irregular accumulation of blood can also be seen in the atria and ventricles of Ift140cauli/cauli mutants (F). T, tongue; PS, palatal shelf; K, kidneys; L, lung; RA, right atrium; RV, right ventricle; TV, tricuspid valve; MV, mitral valve. Molecular signalling is disrupted in Ift140cauli/cauli limb buds
The Shh/Grem1/Fgf loop is a key signalling system responsible for controlling the outgrowth and patterning of the limb [26]. Components of this signalling system were analysed in Ift140cauli/cauli limb buds by in situ hybridisation (Figure 4). Many members of the Shh/Grem1/Fgf signalling loop show disrupted expression between Ift140+/+ control and Ift140cauli/cauli mutant limb buds at E11.5–13.5 days of development (Figure 4). Ectopic Shh expression is evident along the anterior margin of Ift140cauli/cauli limb buds (Figures 4A–D). Expression of Ptch1, the Shh receptor and a Shh-responsive gene, is reduced posteriorly but expanded anteriorly across limb buds of Ift140cauli/cauli mutants consistent with the presence of anterior Shh (Figures 4E–H). The distinct expression pattern of the Bmp antagonist Grem1 seen in Ift140+/+ limb buds is altered in Ift140cauli/cauli mutants (Figures 4I–L, M′ and N′). Grem1 expression appears weaker in mutants compared to controls but is expanded both anteriorly and posteriorly. Gli3, a major mediator of Shh signalling in the limb, is spatially restricted in the anterior portion of the fore - and hindlimb buds of Ift140cauli/cauli embryos (Figures 4M–P). Expression of dHand, which has a major role in limb patterning at least in part through a reciprocal regulatory interaction with Shh, is reduced posteriorly but expanded into the anterior limb bud (Figures 4Q–T). Dusp6 (Mkp3) is a downstream mediator of Fgf signalling. Expression of Dusp6 is elevated across the anterioposterior axis but is anteriorly expanded (Figures 4U–X). Consistent with this, while Fgf8 expression is maintained in a distinct pattern outlining the AER at the distal tip of wildtype and Ift140cauli/cauli limb buds, expression is elevated in mutants (Figures 4Y–B′). In addition, Fgf8 staining also indicates that in Ift140cauli/cauli mutants there is a disruption in the location of the AER at the border between the dorsal and ventral limb bud ectoderm relative to Ift140+/+ controls (Figures 4K′ and L′). Twist1 has been demonstrated to have a central role in patterning the limbs and in particular has been shown to regulate Shh expression. Given the alteration in Shh expression in Ift140cauli/cauli mutants it was important to examine the level and location of Twist1 expression. Interestingly, expression of Twist1 in Ift140cauli/cauli mutants did not appear to undergo any substantial spatial alteration but did appear to be repressed in Ift140cauli/cauli embryos relative to controls (Figures 4C′–F′). Analysis of digit number was often difficult in younger embryos when overt signs of digit outgrowth where not yet apparent. Sox9 expression highlights the presence of digital rays within the autopod revealing oligodactyly, proximal and distal branching of digits and polydactyly in Ift140cauli/cauli embryo limb buds (Figures 4G′–J′).
Fig. 4. Molecular signalling is disturbed in Ift140cauli/cauli embryos. 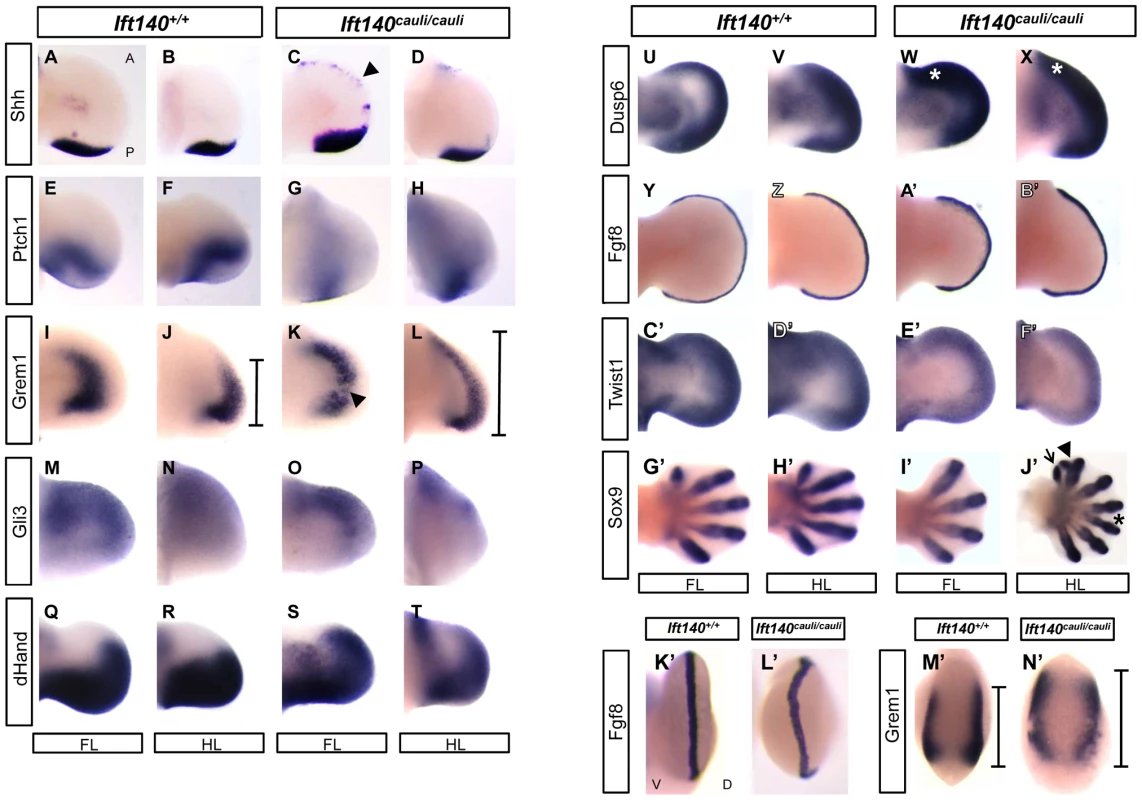
WISH analysis of the forelimbs and hindlimbs of Ift140+/+ and Ift140cauli/cauli embryos. Dorsal view of fore- and hindlimb buds (A–J′), where anterior is always to the top of the image. Distal view of forelimb buds (K′–M′), where dorsal side is facing to the right of the image. Arrowhead in (C) indicates ectopic Shh expression domain. Bars in (J,L and M′,N′) indicate anterior-posterior extent of Grem1 expression. Arrowhead in (K) indicates disruption in Grem1 expression in mutant forelimb. Asterisk in (W and X) indicates elevated anterior Dusp6 expression. Arrow, arrowhead and asterisk in J′ indicate a single ectopic digit, bifid ectopic digit and proximal syndactyly respectively. A, anterior; P, posterior; D, dorsal; V, ventral; FL, forelimb; HL, hindlimb. All embryos are E11.5 except G′–J′ which are E13.5. Abnormal epithelial morphology in Ift140cauli/cauli limb bud ectoderm
Scanning electron microscopy of E10.5 Ift140+/+ control and Ift140cauli/cauli mutant limb buds identified a defect in the epithelial cellular architecture (Figures 5A and B). In Ift140+/+ control limb buds, epidermal ectodermal cells have distinct, clearly defined cellular borders (Figure 5A). In contrast, individual cells in Ift140cauli/cauli mutants have poorly defined cellular borders and individual cells are difficult to identify from surface topology (Figure 5B). In addition, while the primary cilia are easily identified in >90% of wildtype cells, <20% of Ift140cauli/cauli mutant epithelial cells harbour an identifiable cilium in SEM images (Figures 5A, B, E).
Fig. 5. Epithelial cellular architecture and levels of Ift140 are altered in Ift140cauli/caul mutants. 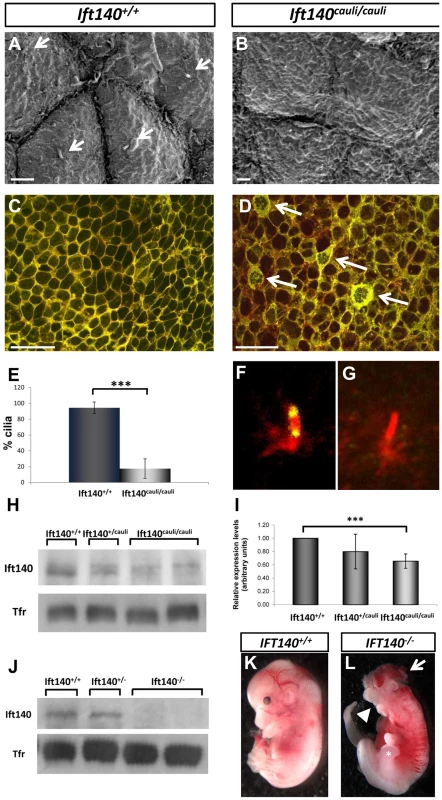
Scanning electron micrographs and immunohistochemistry of epithelia from E10.5 Ift140+/+ (A and C) and Ift140cauli/cauli (B and D) limb buds. The rigid cellular architecture seen in wildtype limbs is highly disrupted in the mutant, as evidenced by the lack of cilia (compare arrows in A), the presence of thick, disorganised cell junctions (arrows in D) and the more diffuse E-cadherin staining in the mutant (D). Overlay of E-cad (green) and phalloidin (red) in Ift140+/+ (C) and Ift140cauli/cauli (D) epithelium. Cilia counts identify a significant decrease in cilia (***p = 2.05×10−7) in limb buds of Ift140cauli/cauli when compared to Ift140+/+ controls (E). Ift140 can be detected at the base and tip of wildtype limb bud cilia (F) but it undetectable in the majority of mutant cilia (G). Western blot analysis shows a reduction of Ift140 protein levels in Ift140cauli/cauli tissue when compared to control and heterozygous samples (H), and a significant reduction of Ift140 transcript levels (***p = 0.0026) in Ift140cauli/cauli mutant embryos (I). Embryos harbouring a homozygous Ift140 null allele show a complete lack of Ift140 protein by western blot (J) and exhibit identical phenotypes to those identified in Ift140cauli/cauli embryos (K,L), including exencephaly (white arrow), open mouth (white arrowhead) and an expanded hindlimb field (asterisk). Scale bar; 2 µM (A and B), 30 µM (C and D). Epithelial cadherin (E-cad) is a component of cell to cell adherens junctions and is required for the maintenance of tight intracellular junctions and oriented alignment of the cytoskeletal networks [27]. Cytoskeletal filamentous actin (F-actin) also localises to these adhesion junctions and is important for regulating tight junction functions within and between cells [28]. In wildtype epithelium, E-cad and F-actin closely co-localise at discrete cellular junctions clearly defining cell borders (Figure 5C). However in Ift140cauli/cauli mutants adherens junctions become diffuse and dispersed in the mutant epithelium and E-cad and F-actin become partially delocalised (Figure 5D). In a proportion of Ift140cauli/cauli epithelial cells, very high levels of E-cad can be seen in a thick band around the cell periphery and a network of E-cad and F-actin appears within the cytoplasm of many of these cells (Figure 5D).
The prediction that the I855K mutation is likely to have a modest impact on structure of Ift140 prompted us to investigate the impact of the cauli missense mutation on Ift140 function by examining the distribution of Ift140 in the primary cilia of limb bud epithelial cells. In wildtype epithelium Ift140 localises to the base and tip of the primary cilium (Figure 5F). In Ift140cauli/cauli mutant epithelium there were undetectable levels of Ift140 in the majority of cilia (Figure 5G) while in rare cases (∼5%) a small amount of Ift140 could be seen in association with a mutant cilium. The lack of detectable Ift140 in mutant cilia prompted an investigation of the levels of Ift140 expressed by mutant cells. Western blot of protein extracted from whole E11.5 embryos indicated a reduction of Ift140 protein to approximately 85% of wildtype levels in Ift140cauli/+embryos and approximately 30% of wildtype levels in Ift140cauli/cauli embryos (Figure 5H). We then examined the level of Ift140 mRNA in Ift140cauli/cauli mutant and wildtype littermates by real-time PCR and observed an approximately 30% decrease in Ift140caul/caulii embryos compared to wildtype controls (Figure 5I). This data suggests that the Ift140cauli/cauli phenotype results from haploinsufficiency. To examine this hypothesis further, we generated Ift140 null embryos. Western blot analysis confirmed the loss of Ift140 in this model (Figure 5J) and analysis of Ift140−/− embryos demonstrated an identical phenotype to that observed in Ift140cauli/cauli embryos (Figures 5K and L) with the exception that Ift140−/− embryos die at around E11.5, two days earlier than Ift140cauli/cauli embryos. The similarity of the phenotypes in the two models supports the theory that the cauli phenotype results from haploinsufficiency.
An IFT140 mutation identified in a human patient with Jeune Syndrome
Patient JS 1-1 was the product of the first pregnancy to healthy, first-cousin Sudanese parents (Figure 6A). There was no extended family history of skeletal disorders. Short limbs (less than 5th centile for gestational age) and a short chest cage were noted on a third trimester ultrasound performed for growth assessment. No other abnormalities were observed at this time. The couple were counselled about a likely poor prognosis given the degree of thoracic hypoplasia. The child was delivered at 37 weeks of gestation with a birth head circumference of 37 cm (>98th centile) and birth length of 48 cm (50th centile). The baby was in poor condition and required high frequency ventilation with high peak pressures (32 mm Mercury) due to pulmonary hypoplasia and hypertension. On examination, the child had short square fingers and hands with five digits each, low nasal bridge, normal ears and palate, and hepatomegaly. A small muscular ventricular septal defect was found on echocardiography, and ultrasound of the enlarged liver showed normal hepatic texture. Normal liver and renal function was documented. The baby deteriorated, despite ventilation, and died aged 7 days. Post mortem radiography was obtained and revealed short long bones, with metaphyseal cupping, bowed femora and humeri, short ribs and thorax, trident appearance to the acetabular roofs, and advanced tarsal bone ossification (Figures 6B–E). These features are considered typical of classic Jeune asphyxiating thoracic dystrophy) syndrome.
Fig. 6. An IFT140 mutation identified in a Jeune Syndrome patient. 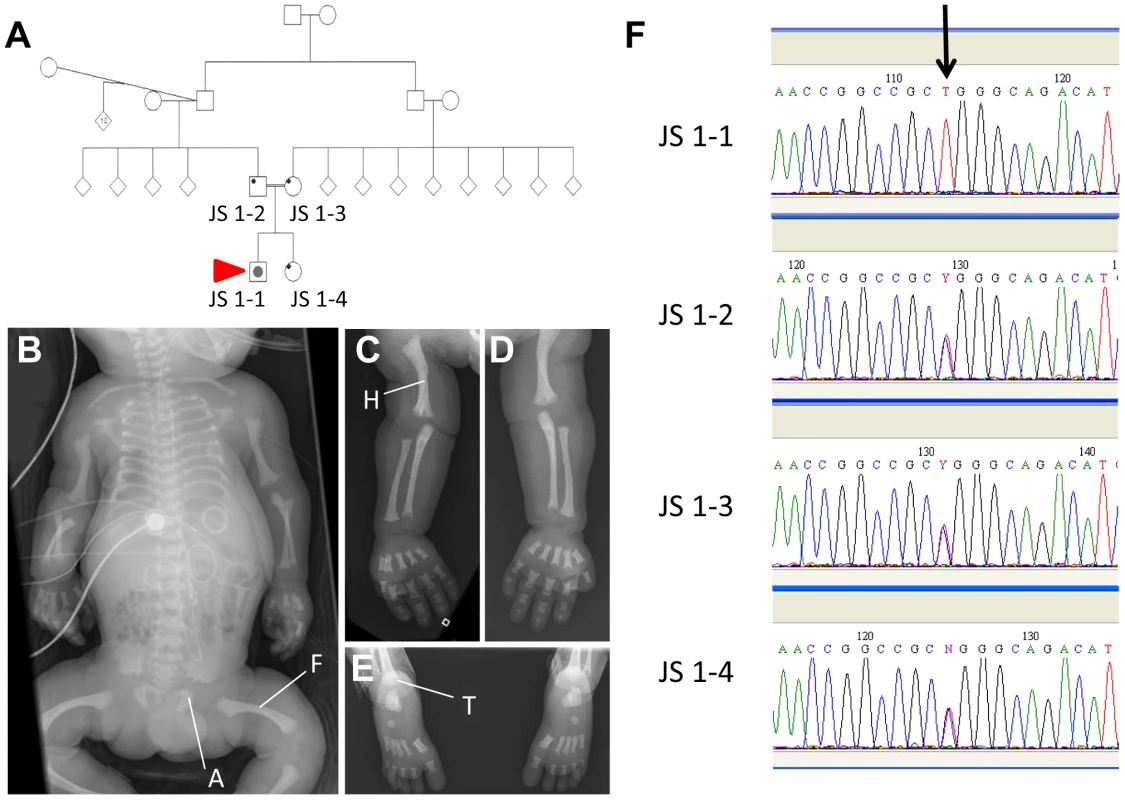
Family pedigree showing the relationship between JS1-1, JS 1-2, JS 1-3 and JS 1-4 (A; red arrow indicates the proband). Whole body (B), right (C) and left (D) upper limb and feet (E) radiographs from JS1-1. Note the short ribs, short long bones with bowed humeri and femora, “trident” appearance of the acetabular roofs and metaphyseal irregularity (B), short long bones with metaphyseal cupping and bowed humerus (C and D), and metaphyseal cupping and advanced tarsal bone ossification for age (E). A, acetabulum; F, femur; H, humerus; T, tarsus. Sequence chromatogram identifying the C>T homozygous mutation in JS1-1 (F; arrow), and heterozygous mutation in immediate family members. Sequencing the IFT140 gene in JS 1-1 identified a novel C>T homozygous mutation at position 838 in this gene that was heterozygous in both parents and the unaffected sibling (Figure 6F), and was not present in a cohort of 90 healthy controls. The Arg to Trp amino acid change at position 280 resulting from this variation attained a PSIC score of 0.972, indicative of a ‘probably damaging’ effect of the mutation on protein function.
Discussion
There are now at least 12 clinical entities defined as ciliopathies [13]. Given the ubiquity of the primary cilia and the number of signalling systems that make use of the cilium as a site in which to deploy receptors and other signalling components, the number of congenital and acquired conditions attributed to defects in cilia function is expected to rise. Mutations in IFT140 have recently been identified in the skeletal ciliopathies Mainzer-Saldino (OMIM 266920) and Jeune (OMIM 208500) syndromes [this paper and 18], [19]. These mutations do not appear to cluster to a particular domain within the IFT140 protein (Figure 1E). While there is significant phenotypic overlap between these two syndromes, including radiological and renal findings, there are also significant differences, such as the existence of skin pigmentation anomalies in Mainzer-Saldino syndrome and polydactyly in Jeune syndrome. These phenotypic differences may be attributable to the impact of specific mutations on the function of IFT140 within the cilium. Mutation of IFT80, a component of the anterograde transport complex B, has also been demonstrated to cause Jeune syndrome [29]. IFT140 resides within the retrograde complex A, thereby suggesting substantially different consequences for the mechanisms of transport within the cilia. Further, mutations in additional IFT components including TTC21B/IFT139, DYNC2H1 and WDR19/IFT144 have been reported in association with Jeune syndrome [30], [31], [32], [33]. Similarly, IFT-A complex mutations have been associated with Sensenbrenner syndrome, a condition with substantial clinical overlap with Jeune syndrome [32]. This suggests that the common phenotypes resulting from each of these mutations are a result of defective cilia function rather than the specific mutation and indicates that this sub-group of skeletal ciliopathies may represent a spectrum of malformations rather than distinct syndromes. There are clear limitations to this view of skeletal ciliopathies since IFT-A complex mutations also lead to clinically distinct entities such as isolated nephronophthisis [32]. These complexities highlight the need to understand the developmental mechanisms underlying specific skeletal ciliopathies through study of appropriate animal models.
The Ift140cauli/cauli mutant mouse phenocopies many of the features of Jeune syndrome although the mouse phenotype is generally more extreme than in humans. The severe dysmorphology seen in Ift140cauli/cauli embryos is incompatible with continued development beyond mid-gestation in the majority of cases. Despite considerable efforts only a very small number of embryos have been obtained beyond E15.5 and in these cases severe skeletal malformation was observed. Skeletal phenotypes include severely disorganised ribs with extensive exostoses, vertebral and palatal defects and agenesis/hypoplasia of the craniofacial skeleton. These phenotypes are broadly consistent with those observed in Jeune syndrome cases including the case presented here. The range of phenotypes observed in Ift140cauli/cauli embryos is also broadly similar to that exhibited by other IFT-A mutant mice [23], [34], [35] consistent with the concept that defective cilia function underpins the dysmorphology observed in each case.
The severity of the Ift140cauli/cauli mutant phenotype may also reflect the nature of the Ift140 mutation in this model. While the cauli strain arose following a missense mutation in Ift140 we have observed a substantial reduction in both protein and RNA levels in mutant tissues. These data together with the identical phenotype observed in Ift140cauli/cauli and Ift140−/− embryos suggests that the cauli phenotype results from loss of Ift140 rather than altered function following amino acid substitution. Ift140 is an essential component of the IFT-A complex and retrograde transport will be severely compromised in its absence. Thus, it is likely that much of the developmentally important signalling normally mediated through the primary cilia is compromised in Ift140cauli/cauli embryos resulting in the devastating phenotype observed.
A missense mutation would normally not be expected to result in RNA degradation and it is not clear why the Ift140cauli/cauli mutation results in loss of one third of the Ift140 RNA population. One possible explanation for this observation is that the c2564T>A mutation, which is 17 bp upstream of the 3′ splice site in exon 19, leads to an abnormal splicing event. It is likely that this would introduce a nonsense mutation downstream of the splicing event and the aberrantly spliced RNA would then be subject to nonsense mediated decay [36]. In support of this, the cauli mutation disrupts predicted overlapping SRSF5 and SRSF6 binding sites raising the possibility that an exonic splice enhancer (ESE) could be impacted [37], [38]. ESEs have been shown to be an integral part of both ubiquitous and alternate splicing mechanisms and exonic mutations disrupting them can result in exon-skipping and the generation of non-sense mutations [39], [40]. There are a growing number of conditions attributable to mutation of an ESE [41], [42], [43], [44], [45], [46].
The somites in Ift140cauli/cauli mutant embryos exhibit a dramatic loss of normal patterning, exhibiting both branching and irregular distribution. There does not appear to be an obvious loss of individual somites suggesting that segmentation of the lateral plate mesoderm proceeds relatively normally. However, irregular morphology of somites suggests that some aspects of somitogenesis, such as the mesenchymal to epithelial transition, may not be regulated appropriately. These somite patterning anomalies are likely responsible for the later rib defects in Ift140cauli/cauli since the ribs are derived from the dermomyotome, a dorsolateral compartment of the somites [47]. In addition, the existence of bony protuberances emanating from many Ift140cauli/cauli mutant ribs suggests a disregulation of osteogenesis. Ift140cauli/cauli mutants exhibit elevated Ptch1 expression within the flank indicating that rib exostoses may result from altered Hh signalling. Mutants are significantly growth retarded relative to littermates, which is most likely related to the mid-gestational demise of most mutants but may also be affected by reduced axial growth. Ift140cauli/cauli mutants exhibit a convoluted neural tube, a phenotype thought to be caused by disruption of coordination between neural tube and axial elongation. Convergent extension defects have been linked to the disruption of primary cilia formation previously [48], [49]. Disrupted convergent extension during neurulation results in failure to narrow the mid-line thereby promoting neural tube closure defects [50]. Ift140cauli/cauli mutants exhibit both cranial and lumbar level neural tube closure defects and it will be interesting in future studies to examine neurulating Ift140cauli/cauli embryos for defects in convergent extension activities.
The polydactylous phenotype observed in Ift140cauli/cauli mutants is similar to that reported in other cilia mutant mouse lines and is highly reminiscent of patterning defects exhibited by a number of Shh signalling mutants [21], [34], [51]. The primary cilia function downstream of Shh and IFT proteins have been demonstrated to be essential for all Shh signalling downstream of Smo [52]. The relative distribution of Gli3 activator and Gli3 repressor across the anterior-posterior axis of the limb bud is a crucial determinant of autopod patterning [53]. Shh signalling inhibits the proteolytic cleavage of full length Gli3 into the shorter Gli3 repressor form which is both dependent on IFT and occurs within the primary cilia [54], [55], [56]. Loss of IFT results in reduced production of Gli3 repressor and consequently an increase in activation of genes such as Grem and dHand, whose expression is linked to Shh and Gli3 action. Our analysis demonstrates that expression of these genes is expanded into the anterior of the autopod in cauli embryos where Gli3 repressor normally predominates, indicating that a loss of Gli3 repression has occurred. The ectopic expression of Ptch1 in the anterior limb bud is consistent with previously described IFT-A mutants [23], and may result from Gli activator effects due to reduced Gli cleavage. An altered ratio of Gli3 repressor to the full-length activator form across the limb paddle results in production of a variable number of additional digits in a number of different mouse models [51], [57], [58] and very likely underlies the polydactyly phenotype in cauli embryos.
The expression of Shh is regulated by a complex network of transcription factors. Ectopic expression of dHand similar to that observed in the anterior limb bud of cauli embryos has been shown to be sufficient to induce ectopic Shh [59]. In addition, Twist1 has been shown to repress Shh expression in the anterior limb bud and Twist1 null mice have polydactyly [60]. We observed a repression of Twist1 in Ift140cauli/cauli mutants and it is therefore possible that the reduction in Twist1 levels is responsible for releasing the repression of Shh in the anterior limb bud. Further experimentation will be required to establish if this is the case and what role these changes have on autopod morphology.
Polydactyly is observed in 100% of Ift140cauli/cauli hindlimbs and 50% of forelimbs. On the other hand, approximately one quarter of Ift140cauli/cauli forelimbs exhibit oligodactyly while it is never seen in the hindlimbs. The variable phenotype between fore - and hindlimbs is difficult to reconcile and suggests that inherent differences in signalling or temporal differences in developmental mechanisms between fore - and hindlimbs result in disparate outcomes. While the co-existence of both oligodactyly and polydactyly within forelimb autopod appears contradictory, especially in light of the highly penetrant polydactyly in the hindlimb autopod, it is important to note that digit number is a continuum and that the final outcome is the result of a complex interplay of transcriptional influences. Mixed forelimb oligo/polydactlyly is also observed in mice with reduced Twist1 levels [61]. Twist1 is at the heart of a transcriptional network involved in the control of Fgf and Shh signalling within the developing limb [60]. Partial loss of Twist1 results in reduced growth of the posterior limb bud and an expansion of the anterior limb bud due to ectopic anterior Shh expression [61]. The ectopic anterior Shh also posteriorises anterior digits and the pattering of the resulting autopod is highly variable. It is unclear why there is incomplete penetrance of each of these outcomes but this may result from stochastic variation in the level of Shh and Fgf signalling. We observe anterior ectopic Shh and it is possible that a similar mechanism operates in the Ift140cauli/cauli forelimb. In particular, given the likelihood that the cauli Ift140 mutation primarily disrupts IFT rather than any specific impact on Hh signalling, differences in the level of transport during development could provide significantly variable signalling outcomes. Defects in retrograde transport have been demonstrated to result in the accumulation of large amounts of IFT cargo along the length of the axoneme [12] which likely blocks even residual levels of transport. However, the primary cilium is disassembled and reformed anew during each cell cycle and it is possible that a higher level of signalling is achieved in the newly formed cilium and that this declines as the amount of stranded cargo increases. Since rapid cell proliferation is a hallmark of embryonic development, the opportunity for differences in the level of Hh signalling is high, especially with respect to generating differential outcomes between rapidly and slowly proliferating populations.
Motile cilia function at the node, the embryonic organiser, and help determine cardiac left-right asymmetry by the generation of directional flow of signalling components [62]. Looping of the heart tube is the first visible manifestation of the consequences of nodal signalling during breaking bilateral symmetry. IFT is required in both primary and motile cilia and as such it is likely that mutation of Ift140 disrupts ciliary transport in the motile cilia of the node. The consequences of disrupted IFT for motility are unclear but 50% of mice with immotile nodal cilia are able to generate normal left-right asymmetry. This suggests a mechanism of signalling in nodal cilia that are normally motile that is independent of the directional flow of signalling components in establishing asymmetry [63]. In addition, cilia have been found in developing heart tissue of E9.5–E12.5 mouse embryos [64]. Shh generated by the pulmonary endoderm has been shown to be important in the specification of progenitors required for atrial and pulmonary trunk septation and disruption of this signalling results in both atrial and atrioventricular septation defects [65]. It is therefore likely that mutation of Ift140 impacts on cardiogenesis both during the looping phase and during later septation events.
The early demise of Ift140cauli/cauli mutant embryos precludes an analysis of a number of issues that are likely to result from Ift140 mutation. A targeted deletion of Ift140 in collecting ducts results in polycystic kidneys beginning around postnatal day 5 [20] consistent with the previous association of IFT140 mutations with a range of kidney pathologies including cysts [19]. Ift140cauli/cauli mutant kidneys appear normal at E13.5 which excludes agenesis but is too early to assess more typically IFT-A associated defects. Similarly, the IFT140 mutation described here in the patient with Jeune syndrome did not result in any obvious kidney dysfunction but this may be due to the early post-natal lethality of the condition in this case. An Ift144 mutation has been shown to result in craniofacial defects such as cleft palate [23] but Ift140cauli/cauli mutants die around the stage at which palate fusion occurs. Since Ift140cauli/cauli mutants are often growth retarded relative to littermates it is likely that palatal shelf elevation and fusion could be delayed making it difficult to determine if Ift140 mutation results in cleft palate. Similarly, mid-gestational lethality of Ift140cauli/cauli mutants makes it difficult or impossible to study the impact of Ift140 mutation on a range of other organ systems. Further analysis of these issues will require the use of conditional Ift140 alleles such as the one presented here, the value of which has already been demonstrated in the study of kidney pathology [20].
The data presented here describes the first Ift140 mouse model of Jeune syndrome. We support this with the identification of a recessively inherited IFT140 mutation in a patient with Jeune syndrome. The difference in severity of the phenotype between mice and humans harbouring IFT140 mutations makes a direct comparison difficult. However, this unique ENU-induced Ift140cauli/cauli mouse model will continue to provide insights into the mechanisms controlling normal limb and skeletal development and the congenital anomalies that arise when these signalling systems are perturbed. These insights will be invaluable in increasing our understanding of the origins of Jeune syndrome and related skeletal ciliopathies and provide a foundation for further analysis of the signalling systems regulated through the primary cilium.
Materials and Methods
Mice
Overt craniofacial and limb phenotypes were identified in IFT140cauli/cauli embryos obtained from a large-scale ENU mutagenesis program at the Australian Phenomics Facility, Canberra. Embryonic stem cell clones and chimeric mice were generated from Ift140-targeted ES cell line EPD0073_5_G01 (www.komp.org) as part of the Australian Phenome Network, “ES Cell to Mouse” initiative. Ift140−/− embryos were generated by crossing mice heterozygous for the floxed Ift140 allele with a CMV-Cre deletor strain and then back crossing Ift140fl/− progeny. All mouse procedures were approved by the Murdoch Children's Research Institute Ethics Committee, MCRI AEC #A647, #A684 and #728. All experiments involved harvest of embryos and mice were culled according to MCRI SOP#21. Embryos were then cooled and placed into fixative.
Mapping and mutation analysis
The cauli line was maintained on an inbred C57BL6/J background and outcrossed to the C3H/HeH mapping strain. Linkage analysis was carried out using 13 F2 progeny using a panel of fluorescent microsatellite markers to localise the mutation to a specific chromosomal interval. Using the UCSC genome browser [66] the linkage interval was examined for putative genes and top candidate genes sequenced. DNA from affected cauli embryos were screened for mutations in Ift140 (NM_134126) by sequencing all exons, intron/exon boundaries and most of the 5′ and 3′ untranslated regions.
PCR, sequencing and genotyping
Gene-specific primers were designed for amplification of all 30 exons of the murine Ift140 gene and DNA amplified with AmpliTaq (Roche) DNA Polymerase using standard PCR cycling conditions with an annealing temperature of 55°C. PCR products were sequenced with a BigDye v3.1 Terminator Cycle Sequencing Kit (Applied Biosystems) and products read using an ABI 3130xl capillary genetic analyser (Applied Biosystems). Sequencing chromatograms were compared to the published gDNA sequence and any differences identified and determined for potential pathogenicity using PolyPhen-2 and SIFT [67], [68]. Conservation of Ift140 protein sequence was analysed using Clustal W [69].
Adult mouse ear clips/tails and embryonic sacs were digested for genotyping in 0.4 mg/ml Proteinase K in Proteinase K buffer (10 mM Tris-HCl, pH 7.5; 10 mM NaCl; 10 mM EDTA, pH 8; 0.5% SDS) overnight at 55°C. DNA was then purified by isopropanol extraction for amplification and High Resolution Melt (HRM) analysis of Ift140 exon 19 using primers Ift140.genF: 5′-CCAGAACTGGAAGCCAGAGT and Ift140.genR: 5′ - CTGGGACTACTCACCAGCAT. For HRM, a PCR reaction was performed in 10 µl containing 2.5 µl of 1/500 diluted gDNA, 5 µl Lightcycler480 High Resolution Melting Master Mix (Roche), 3 mM MgCl2 and 0.2 µM of each primer. DNA was amplified for 45 cycles of 95°C for 10 sec, 62°C for 15 sec, 72°C for 5 sec. HRM melting curve data were obtained by slowly increasing the temperature from 75°C to 90°C at a rate of 0.02°C per second. Results were analysed by comparing each melt curve to control genotype DNA. Tissue from Ift140 null mice was extracted and amplified using a common forward PCR primer and two different allele specific reverse primers to produce unique products for each genotype. Primers are detailed in Table 1.
Tab. 1. Oligonucleotide primers used in PCR. 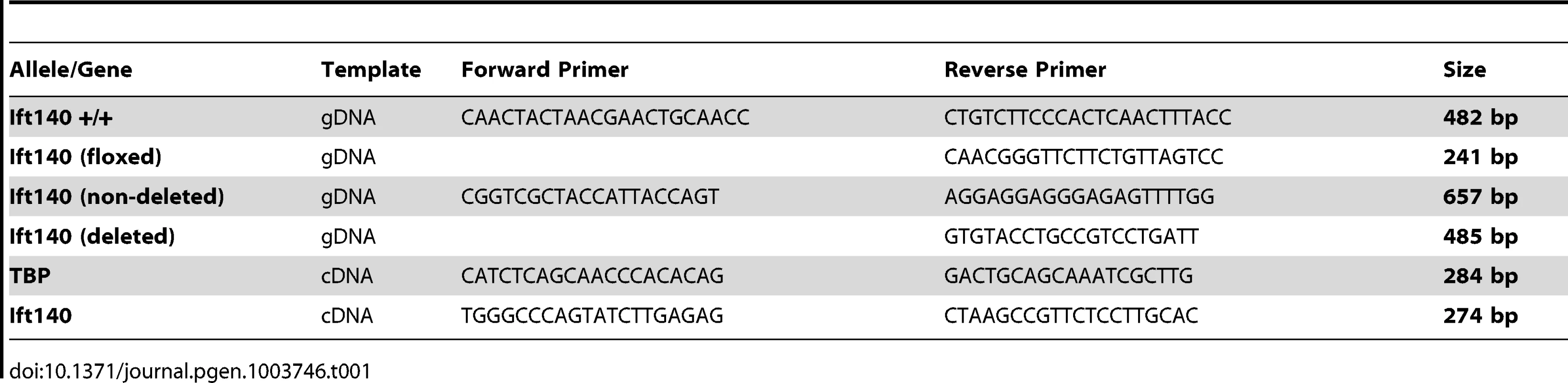
In silico analysis of cauli Ift140 mutation
Pathogenicity of the cauli p.I855K mutation was estimated using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph/) where structural query options were set to default. A PSIC score above 0.85 identifies that the particular mutation was never or almost never observed in that protein family and would be classified as ‘probably-damaging’, scores of 0.15 to 0.85 classified as ‘possibly damaging’, and scores of <0.15 as ‘benign [70]. A second algorithm, SIFT (http://sift.jcvi.org/) was used to support the predicted effect of the cauli p.I855K amino acid substitution on protein function. A SIFT BLink analysis was performed using Ift140 protein ID NP_598887.3. Exonic splice enhancer analysis was performed using ESEfinder v3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home).
Tissue collection
Adult female mice were anaesthetised with isoflurane and culled by cervical dislocation according to the National Health and Medical Research Council Australian code of practice for the care and use of animals for scientific purposes (RCH AEEC approval #A647). Embryos were dissected free from all extra embryonic membranes in Phosphate Buffered Saline (PBS) and processed dependent on the subsequent application. Between 2–6 independent embryos were processed for each analysis. Heterozygous embryos were indistinguishable to wild-type.
Hematoxylin and Eosin (H&E) staining
Embryos were fixed in 4% PFA for up to several weeks then transferred to 70% Ethanol prior to processing through xylene and paraffin using an Excelsior ES tissue processor (Thermo Scientific). Tissues were embedded in the required orientation using a Histocentre 2 Embedding Station (Shandon Life Sciences) and 10 µM sections cut on an MR-2 microtome (RMC products). A standard H&E protocol was followed with 5 min staining in hematoxylin, 1 min staining in eosin and mounted with Entellan (Merck). Images were taken on a Nikon Eclipse 80i microscope (Pathtech).
Skeletal preparations
Embryos >E16.5 were processed for skeletal preparations to visualize bone (Alizarin red) and cartilage (Alcian blue) as previously described [71], with modifications. All steps were carried out at room temperature. Embryos were skinned and eviscerated prior to fixation in 95% (v/v) ethanol. Once required, tissues were incubated in Alcian blue stain (0.06% (w/v) Alcian blue in 80% (v/v) ethanol, 20% (v/v) acetic acid) overnight, re-fixed in 95% ethanol for several hours and cleared in 2% (w/v) KOH×2 hr. Embryos were then incubated with Alizarin red solution (0.03% (w/v) Alizarin red in 1% (w/v) KOH) overnight then immersed in 1% (w/v) KOH/20% (v/v) glycerol solution for further clearing. For long-term storage, embryos were transferred into 50% (v/v) ethanol/50% (v/v) glycerol. Images were taken on a Leica MZ6 microscope using a Leica DFC290 camera (both Leica Microsystems Ltd).
Scanning Electron Microscopy (SEM)
E10.5 limb buds were dissected from wild-type, heterozygous and homozygous embryos, fixed and processed for scanning electron microscopy as previously described [72]. Tissues were viewed using a Philips XL30 FE scanning electron microscope.
In situ probe synthesis and whole mount hybridisation (WISH)
Total RNA was isolated from adult mouse brain tissue using a Qiagen RNeasy Midi Kit (Qiagen) or whole E11.5 wild-type embryos in Trizol (Invitrogen) by phenol/chloroform extraction. cDNA was generated from 1 µg RNA using random hexamers (Sigma) and BioScript reverse transcriptase (Bioline). cDNA fragments specific for the 3′UTR sequences of myogenin, Sox9 and Twist1 were amplified from RNA and ligated into pCRII-TOPO using the TOPO TA cloning kit (Invitrogen). Plasmids containing Msx1, Shh, Ptch1, Grem1, Gli3, dHand, Dusp6 and Fgf8 were obtained from C.Wicking, IMB, University of Queensland. RNA probes were generated by plasmid amplification with M13 primers, synthesised with T7 or SP6 RNA polymerase using a MAXIscript kit (Ambion), integrating dig-UTP (Roche). Probes were treated with Turbo DNase (Ambion) and unincorporated nucleotides removed using NucAway Spin Columns (Ambion). A sense probe was always generated for analysis of first-use probes.
E10.5–E13.5 cauli embryos were processed for whole mount in situ hybridisation as previously described [73], with modifications. After methanol rehydration, embryos were permeabilised by incubation in 10 µg/ml proteinase K for a time optimised for the stage of embryonic development; E10.5 for 45 min, increased per day of development by 10 min increments. Embryos were post-fixed in 0.2% glutaraldehyde/4% paraformaldehyde (PFA), equilibrated in prehybridisation buffer and incubated at 65°C overnight with the in situ riboprobe. After adequate washing to remove unbound probe, embryos were blocked with 10% sheep serum and incubated with an anti-digoxigenin-AP antibody (1∶1000, Roche). After additional washing, colour was developed with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and the reaction stopped with EDTA buffer. The colour was fixed with 4% PFA, background staining reduced by storage in 100% methanol and images taken using a Nikon Eclipse 80i microscope (Pathtech).
Immunohistochemistry
Embryos at E10.5 were processed for immunohistochemistry by fixation in 4% paraformaldehyde (PFA)×30 min, permeabilised/blocked in 0.1% Triton X-100/2% bovine serum albumin (BSA) in PBS, and washed for several hours/overnight in PBS. Embryos were incubated with a rabbit anti-Ift140 polyclonal antibody (1∶50; Proteintech) and mouse acetylated α-tubulin antibody (1∶2000; Sigma) overnight at 4°C, washed for several hrs/overnight in PBS and incubated overnight in the dark with Alexa Fluor 488-conjugated goat anti rabbit IgG and Alexa Fluor 594-conjugated goat anti mouse IgG secondary antibodies (both 1∶1500; Molecular Probes). Following several washes in PBS, embryos were post fixed in 4% PFA×5 min and re-washed in PBS. Embryo limb tips were dissected off and mounted with Vectasheild (Vector Laboratories).
For analysis of cellular architecture, E10.5 embryos were processed as above, incubated with a mouse monoclonal anti-E-cadherin (1∶200; BD Transduction Laboratories), secondary Alexa Fluor 594-conjugated goat anti mouse IgG secondary antibody (1∶1000; Molecular Probes) and incubated with Alexa Fluor 488 phalloidin (1∶1000; Molecular Probes).
Immunofluorescence was analysed using a Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems).
Immunoblotting and quantitative real-time RT-PCR
Independently, whole E11.5 wild-type, heterozygous and homozygous embryos were lysed by sonication (Vibra Cell, Sonics and Materials) in Laemlli SDS Sample Buffer plus 0.1 M DTT. A western blot was performed using standard procedures. Briefly, 100 µg of protein from each genotype was run through a 6 and 10% Tris-Glycine gel and transferred onto PDVF membrane (GE Healthcare) for 1 hour at 100 V. Membranes were preblocked with 5% non-fat milk and incubated with a rabbit anti-Ift140 polyclonal (1∶2000; Proteintech) or mouse anti-Human Transferrin Receptor monoclonal (1∶1000; Invitrogen) antibody. Protein was detected with rabbit (1∶2000; Cell Signaling Technologies) or mouse (1∶10,000; Dako) HRP-linked secondary antibodies, respectively. Blots were processed using the Amersham ELC Prime detection kit (VWR International). Results are representative of at least 3 independent immunoblots, with protein from a minimum of 2 mutant embryos. Protein levels were calculated using ImageQuant TL, v7 software (GE Healthcare Life Sciences).
Whole E13.5 embryos were collected in Trizol reagent (Invitrogen) and sonicated for RNA extraction. Total RNA was extracted from embryos of each genotype using standard chloroform extraction procedures. First-strand cDNA synthesis was performed on 1 µg RNA from each genotype using BioScript reverse transcriptase (BioLine) using random primers. Quantitative real-time PCR reactions were performed in triplicate on a Lightcycler 480 System (Roche) using GoTaq qPCR Master Mix (Promega). Data were normalised to the housekeeping gene TBP (TATA Binding Protein). Primer sequences are detailed in Table 1.
Human patient samples
All human analyses were approved by the Royal Children's Hospital Human Research Ethics Committee project 31190. Genomic DNA was isolated from blood samples received from the proband (JS 1-1), his parents (JS 1-2 and JS 1-3) and saliva from the unaffected sibling (JS 1-4). The candidate gene IFT140 (NM_014714.3) was sequenced, including all 31 exons, intron/exon boundaries and most of the 5′ and 3′ untranslated regions and any variations to the GRCh37 human reference assembly identified. Alterations in the nucleotide sequence of JS 1-1 were analysed using the following criteria: 1) segregation was consistent with recessive inheritance; 2) PCIS score >0.15 by Polyphen-2; 3) SNP allele frequency <0.005 in public databases dbSNP [74] and 1000 genomes [75]; 4) no evidence of homozygous mutant alleles in a cohort of normal healthy controls.
Zdroje
1. BerbariNF, O'ConnorAK, HaycraftCJ, YoderBK (2009) The Primary Cilium as a Complex Signaling Center. Current Biology 19: R526–R535.
2. RosenbaumJL, WitmanGB (2002) Intraflagellar transport. Nat Rev Mol Cell Biol 3 : 813–825.
3. KozminskiKG, BeechPL, RosenbaumJL (1995) The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. The Journal of Cell Biology 131 : 1517–1527.
4. ColeDG, DienerDR, HimelblauAL, BeechPL, FusterJC, et al. (1998) Chlamydomonas Kinesin-II–dependent Intraflagellar Transport (IFT): IFT Particles Contain Proteins Required for Ciliary Assembly in Caenorhabditis elegans Sensory Neurons. The Journal of Cell Biology 141 : 993–1008.
5. PipernoG, SiudaE, HendersonS, SegilM, VaananenH, et al. (1998) Distinct Mutants of Retrograde Intraflagellar Transport (IFT) Share Similar Morphological and Molecular Defects. The Journal of Cell Biology 143 : 1591–1601.
6. PipernoG, MeadK (1997) Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proceedings of the National Academy of Sciences 94 : 4457–4462.
7. KozminskiKG, JohnsonKA, ForscherP, RosenbaumJL (1993) A motility in the eukaryotic flagellum unrelated to flagellar beating. Proceedings of the National Academy of Sciences 90 : 5519–5523.
8. HirokawaN (1998) Kinesin and Dynein Superfamily Proteins and the Mechanism of Organelle Transport. Science 279 : 519–526.
9. TaschnerM, BhogarajuS, LorentzenE (2012) Architecture and function of IFT complex proteins in ciliogenesis. Differentiation 83: S12–S22.
10. PazourGJ, DickertBL, WitmanGB (1999) The DHC1b (DHC2) Isoform of Cytoplasmic Dynein Is Required for Flagellar Assembly. The Journal of Cell Biology 144 : 473–481.
11. PorterME, BowerR, KnottJA, ByrdP, DentlerW (1999) Cytoplasmic Dynein Heavy Chain 1b Is Required for Flagellar Assembly in Chlamydomonas. Molecular Biology of the Cell 10 : 693–712.
12. PiginoG, GeimerS, LanzavecchiaS, PaccagniniE, CanteleF, et al. (2009) Electron-tomographic analysis of intraflagellar transport particle trains in situ. The Journal of Cell Biology 187 : 135–148.
13. NovarinoG, AkizuN, Gleeson JosephG (2011) Modeling Human Disease in Humans: The Ciliopathies. Cell 147 : 70–79.
14. HuberC, Cormier-DaireV (2012) Ciliary disorder of the skeleton. American Journal of Medical Genetics Part C: Seminars in Medical Genetics 160C: 165–174.
15. WatersA, BealesP (2011) Ciliopathies: an expanding disease spectrum. Pediatric Nephrology 26 : 1039–1056.
16. ChristensenST, ClementCA, SatirP, PedersenLB (2012) Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. The Journal of Pathology 226 : 172–184.
17. CaruanaG, FarliePG, HartAH, Bagheri-FamS, WallaceMJ, et al. (2013) Genome-wide ENU mutagenesis in combination with high density SNP analysis and exome sequencing provides a rapid route to the identification of novel mouse models of developmental disease. PLoS ONE 8: e55429.
18. SchmidtsM, FrankV, EisenbergerT, al TurkiS, BizetAA, et al. (2013) Combined NGS Approaches Identify Mutations in the Intraflagellar Transport Gene IFT140 in Skeletal Ciliopathies with Early Progressive Kidney Disease. Human Mutation 34 : 714–724.
19. PerraultI, SaunierS, HaneinS, FilholE, Bizet AlbaneA, et al. (2012) Mainzer-Saldino Syndrome Is a Ciliopathy Caused by IFT140 Mutations. The American Journal of Human Genetics 90 : 864–870.
20. JonassenJA, SanAgustinJ, BakerSP, PazourGJ (2012) Disruption of IFT complex A causes cystic kidneys without mitotic spindle misorientation. J Am Soc Nephrol 23 : 641–651.
21. CameronDA, PennimpedeT, PetkovichM (2009) Tulp3 is a critical repressor of mouse hedgehog signaling. Developmental Dynamics 238 : 1140–1149.
22. PattersonVL, DamrauC, PaudyalA, ReeveB, GrimesDT, et al. (2009) Mouse hitchhiker mutants have spina bifida, dorso-ventral patterning defects and polydactyly: identification of Tulp3 as a novel negative regulator of the Sonic hedgehog pathway. Hum Mol Genet 18 : 1719–1739.
23. AsheA, ButterfieldNC, TownL, CourtneyAD, CooperAN, et al. (2012) Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Human Molecular Genetics 21 : 1808–1823.
24. LupasA, Van DykeM, StockJ (1991) Predicting coiled coils from protein sequences. Science 252 : 1162–1164.
25. BuchanDWA, WardSM, LobleyAE, NugentTCO, BrysonK, et al. (2010) Protein annotation and modelling servers at University College London. Nucleic Acids Research 38: W563–W568.
26. BenazetJ-D, BischofbergerM, TieckeE, GoncalvesA, MartinJF, et al. (2009) A Self-Regulatory System of Interlinked Signaling Feedback Loops Controls Mouse Limb Patterning. Science 323 : 1050–1053.
27. KnustE, BossingerO (2002) Composition and formation of intercellular junctions in epithelial cells. Science 298 : 1955–1959.
28. TsukitaS, FuruseM, ItohM (2001) Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2 : 285–293.
29. BealesPL, BlandE, TobinJL, BacchelliC, TuysuzB, et al. (2007) IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat Genet 39 : 727–729.
30. DavisEE, ZhangQ, LiuQ, DiplasBH, DaveyLM, et al. (2011) TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 43 : 189–196.
31. DagoneauN, GouletM, GenevièveD, SznajerY, MartinovicJ, et al. (2009) DYNC2H1 Mutations Cause Asphyxiating Thoracic Dystrophy and Short Rib-Polydactyly Syndrome, Type III. The American Journal of Human Genetics 84 : 706–711.
32. BredrupC, SaunierS, Oud MachteldM, FiskerstrandT, HoischenA, et al. (2011) Ciliopathies with Skeletal Anomalies and Renal Insufficiency due to Mutations in the IFT-A Gene WDR19. The American Journal of Human Genetics 89 : 634–643.
33. SchmidtsM, ArtsHH, BongersEMHF, YapZ, OudMM, et al. (2013) Exome sequencing identifies DYNC2H1 mutations as a common cause of asphyxiating thoracic dystrophy (Jeune syndrome) without major polydactyly, renal or retinal involvement. Journal of Medical Genetics 50 : 309–323.
34. LiemKF, AsheA, HeM, SatirP, MoranJ, et al. (2012) The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. The Journal of Cell Biology 197 : 789–800.
35. QinJ, LinY, NormanRX, KoHW, EggenschwilerJT (2011) Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proceedings of the National Academy of Sciences 108 : 1456–1461.
36. ChangY-F, ImamJS, WilkinsonMF (2007) The Nonsense-Mediated Decay RNA Surveillance Pathway. Annual Review of Biochemistry 76 : 51–74.
37. WangJ, SmithPJ, KrainerAR, ZhangMQ (2005) Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Research 33 : 5053–5062.
38. LiuH-X, ZhangM, KrainerAR (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & Development 12 : 1998–2012.
39. DietzHC, ValleD, FrancomanoCA, KendziorRJ, PyeritzRE, et al. (1993) The skipping of constitutive exons in vivo induced by nonsense mutations. Science 259 : 680–683.
40. WoolfeA, MullikinJ, ElnitskiL (2010) Genomic features defining exonic variants that modulate splicing. Genome Biology 11: R20.
41. CaputiM, KendziorRJ, BeemonKL (2002) A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes & Development 16 : 1754–1759.
42. ErikssonM, BrownWT, GordonLB, GlynnMW, SingerJ, et al. (2003) Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423 : 293–298.
43. MoseleyCT, MullisPE, PrinceMA, PhillipsJA (2002) An Exon Splice Enhancer Mutation Causes Autosomal Dominant GH Deficiency. Journal of Clinical Endocrinology & Metabolism 87 : 847–852.
44. BoichardA, VenetL, NaasT, BoutronA, ChevretL, et al. (2008) Two silent substitutions in the PDHA1 gene cause exon 5 skipping by disruption of a putative exonic splicing enhancer. Molecular Genetics and Metabolism 93 : 323–330.
45. CartegniL, KrainerAR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30 : 377–384.
46. LiuH-X, CartegniL, ZhangMQ, KrainerAR (2001) A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet 27 : 55–58.
47. KatoN, AoyamaH (1998) Dermomyotomal origin of the ribs as revealed by extirpation and transplantation experiments in chick and quail embryos. Development 125 : 3437–3443.
48. FerranteMI, RomioL, CastroS, CollinsJE, GouldingDA, et al. (2009) Convergent extension movements and ciliary function are mediated by ofd1, a zebrafish orthologue of the human oral-facial-digital type 1 syndrome gene. Human Molecular Genetics 18 : 289–303.
49. ZengH, HooverAN, LiuA (2010) PCP effector gene Inturned is an important regulator of cilia formation and embryonic development in mammals. Developmental Biology 339 : 418–428.
50. WallingfordJB, HarlandRM (2002) Neural tube closure requires Dishevelled-dependent convergent extension of the midline. Development 129 : 5815–5825.
51. HuiC-c, JoynerAL (1993) A mouse model of Greig cephalo-polysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet 3 : 241–246.
52. HuangfuD, AndersonKV (2005) Cilia and Hedgehog responsiveness in the mouse. Proceedings of the National Academy of Sciences of the United States of America 102 : 11325–11330.
53. WangB, FallonJF, BeachyPA (2000) Hedgehog-Regulated Processing of Gli3 Produces an Anterior/Posterior Repressor Gradient in the Developing Vertebrate Limb. Cell 100 : 423–434.
54. MaySR, AshiqueAM, KarlenM, WangB, ShenY, et al. (2005) Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Developmental Biology 287 : 378–389.
55. HaycraftCJ, BanizsB, Aydin-SonY, ZhangQ, MichaudEJ, et al. (2005) Gli2 and Gli3 Localize to Cilia and Require the Intraflagellar Transport Protein Polaris for Processing and Function. PLoS Genet 1: e53.
56. LiuA, WangB, NiswanderLA (2005) Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development 132 : 3103–3111.
57. ShethR, BastidaMF, RosM (2007) Hoxd and Gli3 interactions modulate digit number in the amniote limb. Developmental Biology 310 : 430–441.
58. HillP, GötzK, RütherU (2009) A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Developmental Biology 328 : 506–516.
59. ChariteJ, McFaddenDG, OlsonEN (2000) The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development 127 : 2461–2470.
60. O'RourkeMP, SooK, BehringerRR, HuiCC, TamPP (2002) Twist plays an essential role in FGF and SHH signal transduction during mouse limb development. Dev Biol 248 : 143–156.
61. KrawchukD, WeinerSJ, ChenY-T, LuBC, CostantiniF, et al. (2010) Twist1 activity thresholds define multiple functions in limb development. Developmental Biology 347 : 133–146.
62. SloughJ, CooneyL, BruecknerM (2008) Monocilia in the embryonic mouse heart suggest a direct role for cilia in cardiac morphogenesis. Developmental Dynamics 237 : 2304–2314.
63. TanSY, RosenthalJ, ZhaoX-Q, FrancisRJ, ChatterjeeB, et al. (2007) Heterotaxy and complex structural heart defects in a mutant mouse model of primary ciliary dyskinesia. The Journal of Clinical Investigation 117 : 3742–3752.
64. Ibañez-TallonI, HeintzN, OmranH (2003) To beat or not to beat: roles of cilia in development and disease. Human Molecular Genetics 12: R27–R35.
65. HoffmannAD, PetersonMA, Friedland-LittleJM, AndersonSA, MoskowitzIP (2009) sonic hedgehog is required in pulmonary endoderm for atrial septation. Development 136 : 1761–1770.
66. KentWJ, SugnetCW, FureyTS, RoskinKM, PringleTH, et al. (2002) The human genome browser at UCSC. Genome Res 12 : 996–1006.
67. FrankDU, FotheringhamLK, BrewerJA, MugliaLJ, Tristani-FirouziM, et al. (2002) An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development 129 : 4591–4603.
68. RamenskyV, BorkP, SunyaevS (2002) Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30 : 3894–3900.
69. BorodovskyM, McIninchJ (1993) Recognition of genes in DNA sequence with ambiguities. Biosystems 30 : 161–171.
70. AdzhubeiIA, SchmidtS, PeshkinL, RamenskyVE, GerasimovaA, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7 : 248–249.
71. WallinJ, WiltingJ, KosekiH, FritschR, ChristB, et al. (1994) The role of Pax-1 in axial skeleton development. Development 120 : 1109–1121.
72. ManjiSS, MillerKA, WilliamsLH, AndreasenL, SiboeM, et al. (2011) An ENU-induced mutation of Cdh23 causes congenital hearing loss, but no vestibular dysfunction, in mice. Am J Pathol 179 : 903–914.
73. FowlesLF, BennettsJS, BerkmanJL, WilliamsE, KoopmanP, et al. (2003) Genomic screen for genes involved in mammalian craniofacial development. Genesis 35 : 73–87.
74. SherryST, WardMH, KholodovM, BakerJ, PhanL, et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29 : 308–311.
75. Genomes Project C (2010) A map of human genome variation from population-scale sequencing. Nature 467 : 1061–1073.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Příjem alkoholu a menstruační cyklus
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
- Akutní intermitentní porfyrie
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



