-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
Strains of Extraintestinal Pathogenic Escherichia coli (ExPEC) exhibit an array of virulence strategies and are a major cause of urinary tract infections, sepsis and meningitis. Efforts to understand ExPEC pathogenesis are challenged by the high degree of genetic and phenotypic variation that exists among isolates. Determining which virulence traits are widespread and which are strain-specific will greatly benefit the design of more effective therapies. Towards this goal, we utilized a quantitative genetic footprinting technique known as transposon insertion sequencing (Tn-seq) in conjunction with comparative pathogenomics to functionally dissect the genetic repertoire of a reference ExPEC isolate. Using Tn-seq and high-throughput zebrafish infection models, we tracked changes in the abundance of ExPEC variants within saturated transposon mutant libraries following selection within distinct host niches. Nine hundred and seventy bacterial genes (18% of the genome) were found to promote pathogen fitness in either a niche-dependent or independent manner. To identify genes with the highest therapeutic and diagnostic potential, a novel Trait Enrichment Analysis (TEA) algorithm was developed to ascertain the phylogenetic distribution of candidate genes. TEA revealed that a significant portion of the 970 genes identified by Tn-seq have homologues more often contained within the genomes of ExPEC and other known pathogens, which, as suggested by the first axiom of molecular Koch's postulates, is considered to be a key feature of true virulence determinants. Three of these Tn-seq-derived pathogen-associated genes—a transcriptional repressor, a putative metalloendopeptidase toxin and a hypothetical DNA binding protein—were deleted and shown to independently affect ExPEC fitness in zebrafish and mouse models of infection. Together, the approaches and observations reported herein provide a resource for future pathogenomics-based research and highlight the diversity of factors required by a single ExPEC isolate to survive within varying host environments.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003716
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003716Summary
Strains of Extraintestinal Pathogenic Escherichia coli (ExPEC) exhibit an array of virulence strategies and are a major cause of urinary tract infections, sepsis and meningitis. Efforts to understand ExPEC pathogenesis are challenged by the high degree of genetic and phenotypic variation that exists among isolates. Determining which virulence traits are widespread and which are strain-specific will greatly benefit the design of more effective therapies. Towards this goal, we utilized a quantitative genetic footprinting technique known as transposon insertion sequencing (Tn-seq) in conjunction with comparative pathogenomics to functionally dissect the genetic repertoire of a reference ExPEC isolate. Using Tn-seq and high-throughput zebrafish infection models, we tracked changes in the abundance of ExPEC variants within saturated transposon mutant libraries following selection within distinct host niches. Nine hundred and seventy bacterial genes (18% of the genome) were found to promote pathogen fitness in either a niche-dependent or independent manner. To identify genes with the highest therapeutic and diagnostic potential, a novel Trait Enrichment Analysis (TEA) algorithm was developed to ascertain the phylogenetic distribution of candidate genes. TEA revealed that a significant portion of the 970 genes identified by Tn-seq have homologues more often contained within the genomes of ExPEC and other known pathogens, which, as suggested by the first axiom of molecular Koch's postulates, is considered to be a key feature of true virulence determinants. Three of these Tn-seq-derived pathogen-associated genes—a transcriptional repressor, a putative metalloendopeptidase toxin and a hypothetical DNA binding protein—were deleted and shown to independently affect ExPEC fitness in zebrafish and mouse models of infection. Together, the approaches and observations reported herein provide a resource for future pathogenomics-based research and highlight the diversity of factors required by a single ExPEC isolate to survive within varying host environments.
Introduction
Within the Escherichia coli lineage there are several distinct virulent subgroups that are principally classified by an ability to cause a common set of diseases. One specific subgroup, Extraintestinal Pathogenic E. coli (ExPEC), is typically thought to be a benign inhabitant of the lower intestinal tract of warm-blooded vertebrates. However, outside this niche, ExPEC strains have the ability to persist in an array of secondary host-associated habitats where they can cause urinary tract infections (UTIs), meningitis, bacteremia and sepsis in both humans and domesticated animals [1], [2], [3], [4]. The combined medical, agricultural and economic burden of ExPEC-related diseases is likely to increase as antibiotic resistance spreads [1], [5], [6]. Delineation of the genetic elements utilized by ExPEC to infect such a diverse spectrum of host niches and cause disease promises to advance our understanding of pathogen evolution and behavior while also highlighting more effective strategies to combat these pervasive opportunistic pathogens.
Despite colonizing similar ecological niches, ExPEC isolates can differ by 20–30% of their respective gene inventories and to date, a specific and ubiquitous molecular trait or function exclusive to this cohort has not been characterized [7], [8]. Without knowledge of a unifying ExPEC-associated feature, the development of broad-spectrum therapeutic strategies is made exceptionally difficult [9]. Previous work indicates that individual ExPEC strains appear to have evolved distinct genetic repertoires that promote unique and at times subtle fitness advantages during colonization of specific host environments [10], [11]. It is becoming clear that the working definition of ExPEC is more multigenic in nature. Further complicating the search for ExPEC-associated traits is the 30–40% of genes within their genomes that still require functional annotation [7]. This problem will likely persist as genome sequencing continues to outstrip rates of experimental characterization. Consequently, there is a pressing need to develop methods that mesh high-throughput genomics with context-based functional observations—a notion that is becoming increasingly appreciated [12], [13]. To this end, we adapted a previously described transposon mutagenesis technique known as ‘Tn-seq’ to identify genes that promote pathogen fitness during colonization of a vertebrate host [14], [15].
The use of transposons to conduct unbiased forward genetic screens in bacteria has provided many avenues for investigation over the last two decades [16], [17], [18]. However, these approaches are, for the most part, time and labor intensive and often lack reliable quantitative metrics and sampling depth to identify genes of interest for retrospective follow-up. On the other hand, transposon mutagenesis as a tool is quite amenable to innovation [19], [20], [21]. Tn-seq, also known as ‘INSeq’, utilizes deep sequencing to monitor the composition of insertion variants within bulk mutant pools [14], [15]. This is accomplished through use of a modified mariner transposon that contains recognition sites within the two distal inverted repeats that are specific for the restriction enzyme MmeI. Cleavage of DNA by this enzyme occurs 18–20 base pairs from its recognition site, such that excision of the transposon by MmeI captures flanking genomic sequences. These sequences serve as tags that can be used to track the relative abundance of distinct insertion variants within heterogenetic mutant libraries. This technique ultimately allows the fitness of thousands of mutant variants to be quantified simultaneously following exposure to specific selective pressures. By comparing the composition and abundance of mutants before and after passage through selective conditions, such as within a host organism, a genome-wide map of loci that are important for bacterial fitness can be assembled.
The screening approach reported here involved injection of 48 h post-fertilization (hpf) zebrafish embryos with transposon-mutagenized pools of ExPEC derived from a single parent isolate. Embryonic zebrafish are particularly useful for this type of selection screen because their vertebrate physiology closely matches that of humans, and they have proven useful for the identification and characterization of virulence determinants that enable ExPEC to survive within host niches [10], [11], [22], [23]. Of particular importance, zebrafish embryos possess many of the same innate defenses that mammalian hosts rely upon to resist ExPEC (e.g. phagocytes, complement and antimicrobial peptides) [10], [24], [25], [26]. This attribute underscores the ability of the zebrafish host to mirror a set of overlapping selective pressures that ExPEC naturally encounter. Zebrafish embryos can be used to model both localized and systemic infections and the complete recovery and enumeration of bacteria from within the entire host is relatively straightforward [10], [11]. Through three independent biological experiments employing Tn-seq we identified 970 genes (∼18% of gene content) that promote the competitive fitness of ExPEC during either localized or systemic infections. A large number of these genes have unknown functions, while many others have reported roles in iron transport, bacterial secretion, two-component signaling and metabolism. To focus on genes that are likely important to ExPEC pathogenesis specifically, we devised a gene ontology-like method called ‘TEA’ (Trait Enrichment Analysis), that enabled us to organize candidate genes according to their association with certain bacterial lineages or groups of bacteria that are phenotypically similar. The data presented in this report indicate that TEA, in combination with Tn-seq, provides an effective, streamlined approach for identifying biologically relevant fitness and virulence determinants that are employed by ExPEC within various host environments. Datasets generated in this study have been made freely available through a curated and searchable web-based data viewer (http://pathogenomics.path.utah.edu/F11_TnSeq/).
Results
Mutant library construction and screen design
We previously described the use of the embryonic zebrafish as a surrogate host to study genotype-phenotype relationships of a variety of human and non-human ExPEC isolates [10], [11]. During this initial work we observed that the human cystitis (bladder infection) isolate F11 is particularly adept at growing within and eliciting death of zebrafish during either localized or systemic infections. Elucidation of the virulence gene repertoire (i.e. genetic determinants that promote pathogenic behaviors such as colonization or destruction of host tissues) of this ExPEC strain using Tn-seq provides a starting point for future comparative pathogenomics studies. Figure 1 presents an outline detailing the negative selection screen carried out in this study.
Fig. 1. Schematic of the Tn-seq selection screen used to identify genes necessary for fitness in a zebrafish infection model. 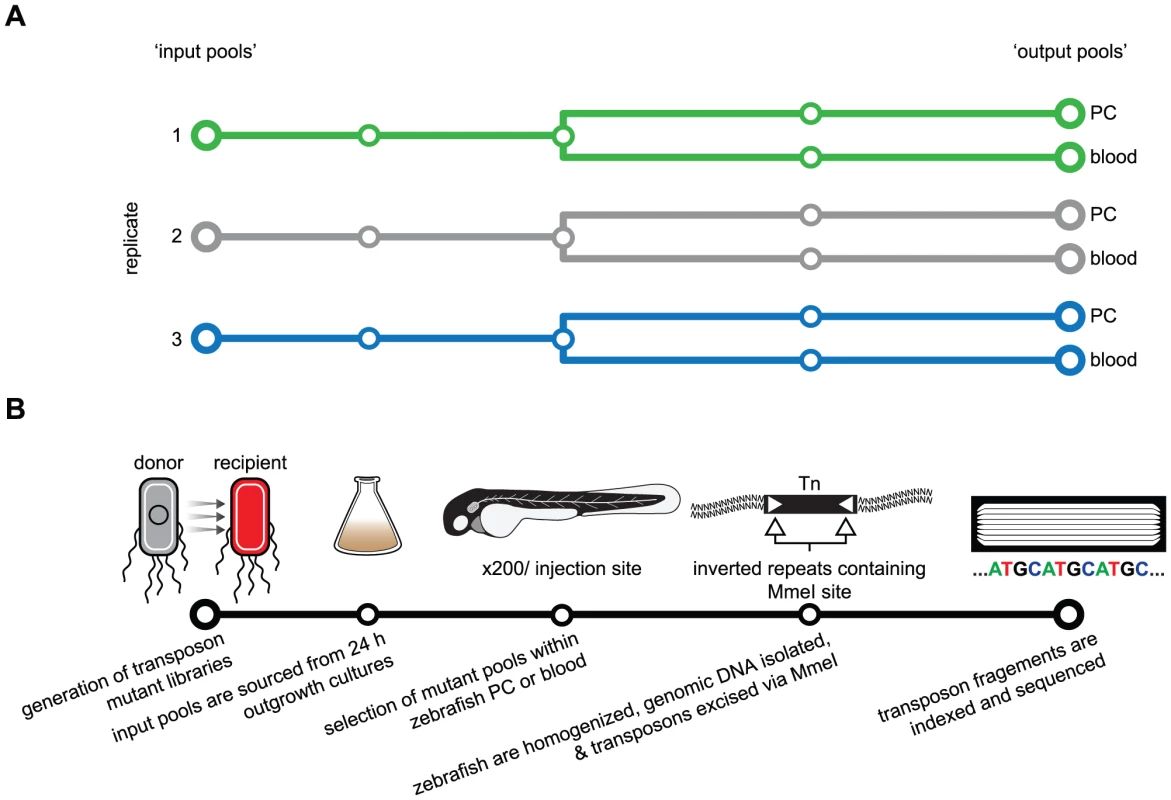
(A) Three selection screens were performed (green, grey and blue tracks) using three ‘input’ pools to generate a total of six ‘output’ pools. (B) For each replicate screen, an independent transposon mutant library was created. Expansion of mutant libraries from frozen stocks was done at 37°C for 24 h to produce input pools and starting inoculum. Approximately 200 zebrafish embryos were injected in the pericardial cavity (PC) or circulation valley (CV, blood) with approximately 3,000 mutant bacteria. After a selection period of 20 h, zebrafish were homogenized to facilitate recovery and extraction of total DNA. The restriction enzyme MmeI was used to excise transposons and flanking F11 genomic DNA for analysis by deep sequencing. Transposon mutagenesis was accomplished by using the previously described pSAM vector and conjugation [15]. This plasmid, which contains a mariner transposon flanked by MmeI modified inverted repeats and the himar1C9 transposase, was retrofitted with an E. coli compatible antibiotic resistance cassette and promoter elements to generate pSAM-Ec (Figure S1). Multiple, independent mating events were performed to assemble three transposon mutant libraries to be used in three separate replicate screens (Figure 1A). It was determined that each pool contained a minimum of 50,000 distinct insertion variants by enumerating selectable, transposon positive colonies immediately following conjugation when mutant siblings are minimal. Successful transposition of single inserts into individual bacterial chromosomes was verified by detection of the transposon by Southern blot analysis (Figure S2). Because the cost of a pre-screen sequence analysis was prohibitive, we utilized a more economical means to confirm the diversity and saturation of mutant pools. The frequency of variants defective for lactose utilization, curli production and glycogen storage was determined using standard colorimetric plating assays (Figure S2), which demonstrated that our transposon mutant libraries were sufficiently saturated and ready for in vivo screening. Post-hoc sequencing later confirmed that our libraries contained 60,000 to 80,000 mutant variants each, corresponding to an insertion event approximately every 75 bp within the F11 chromosome (Table S1).
For each independent biological experiment, a single mutant library was cultivated from a frozen stock in M9 minimal media overnight at 37°C (Figure 1B). During this initial outgrowth period, variants with insertions that disabled genes critical for normal replication within broth culture were likely reduced in abundance or completely eliminated. Prior to injection of 48 hpf zebrafish embryos, a 1 ml aliquot of each overnight culture was pelleted, washed and suspended in phosphate buffered saline (PBS) at the desired density. A second aliquot was pelleted and stored as a reference ‘input’ population for later sequencing and comparative analyses with ‘output’ populations. A dose of approximately 3,000 colony forming units (CFU) from each mutant library was injected into either the pericardial cavity (PC) or into the blood via the circulation valley (CV) of zebrafish embryos (200 embryos per niche per experiment were used to ensure adequate sampling of mutant variants) (Figure S3). Injection of the PC provides a model of localized tissue infection, whereas injection into the CV results in rapid dissemination of ExPEC via the bloodstream, modeling bacteremia or sepsis-like infections [10], [11]. We previously determined that a variety of ExPEC isolates have the capacity to grow unchecked within the PC, while only a subset are able to persist and multiply within the bloodstream. Presumably, the microenvironments encountered by ExPEC following injection into the zebrafish circulatory system are more challenging than the PC with respect to nutrient availability, host defenses or other as-yet-undefined factors [10], [11]. F11 is able to survive and replicate within both the PC and blood. Once injected, selection proceeded for 18 to 20 h—a timeframe in which the mutant F11 population grows to between 104 and 106 CFU/embryo and elicits death in ∼40–60% of the animals (Figure S4). Total genomic DNA of the surviving F11 variants was recovered from batch-homogenized fish (alive and dead) to produce output pools. In parallel, equal amounts of genomic material from input and corresponding output pools were digested using the MmeI restriction enzyme followed by enrichment of transposon-containing fragments with flanking genomic sequences via gel electrophoresis. The fragments were then excised and prepared for indexed sequencing on a single lane of an Illumina HiSeq 2000. General sequence-based features of each mutant pool, including number of detected mutant variants, sequencing depth, saturation and insertional bias, are summarized in Table S1.
Genetic footprinting by Tn-seq reveals distinguishable gene sets that have distinct functional compositions within the ExPEC genome
Gene insertion tolerance after passage in broth culture
Beyond targeting T+A dinucleotides, transposable mariner elements have a relatively low insertional bias (Table S1 and Figure 2: F11 genome) [27]. Using sequence data from the three input samples exclusively, the number of insertion sequences mapped per 100 bp for all annotated regions was calculated to determine an insertion frequency (Table S2, Methods). As anticipated, we observed that transposon insertion rates within our mutant libraries mostly occurred at a steady rate independent of chromosomal region. An approach akin to traditional genetic footprinting was used to determine the specific insertional tolerance of each gene within the F11 genome [28], [29]. Gene insertion rates that were more than 1 standard deviation below the genome-wide mean were considered hypo-tolerant (<2.1 inserts/100 bp, n = 609, Figure 2: hypo-tolerant), those within +/−1 standard deviation were tolerant (2.1 < inserts/100 bp <4.5, n = 4,272) and those that were more than 1 standard deviation above the genome-wide mean were hyper-tolerant (>4.5 inserts/100 bp, n = 431, Figure 2: hyper-tolerant). Genes assigned to the hypo-tolerant gene set presumably provide ‘essential’ or ‘core’ physiological functions. In contrast, genes within the hyper-tolerant gene set are likely not required during growth in broth culture and/or exhibit certain genetic features that support high rates of transposon insertion. Using the Kyoto Encyclopedia of Genes and Genomes (KEGG) functional hierarchies, the hypo-tolerant gene set, as expected, was found to be significantly enriched for informational functions related to transcription and translation and included various noncoding RNAs (ncRNA) and transfer RNAs (tRNA) (Figure 3A and B). The hyper-tolerant gene set contained a significantly elevated proportion of ribosomal RNAs, ncRNAs and genes annotated as ‘hypothetical’ (Figure 3B and F).
Fig. 2. Transposon insertion rates for loci within different gene sets. 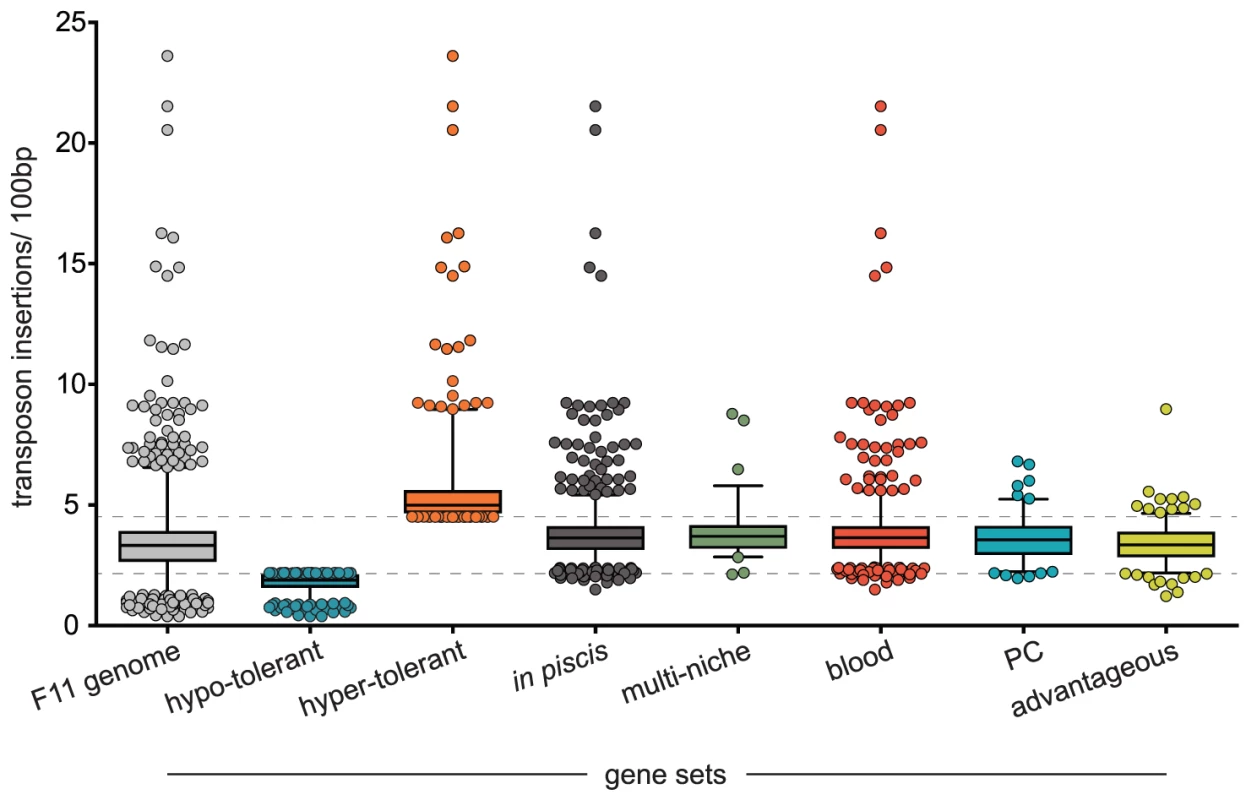
From each input pool (1, 2 and 3) an average number of insertions detected per 100 bp was calculated for each annotated region within the F11 genome. Box plots indicate median and interquartile ranges with whiskers extending to the 1st and 99th percentiles. Dashed lines mark the upper and lower boundaries for hypo and hyper-tolerant gene sets, respectively. Gene set sizes: F11 genome - 5,312; hypo-tolerant - 609; hyper-tolerant - 431; in piscis - 970; multi-niche - 76; blood - 772; PC - 122; advantageous - 227. Note, the insertion rates reported here are expected to be overestimated—please refer to Methods section ‘Candidate Identification’ for a complete description of how the number of transposon insertion events was determined. Fig. 3. Functional KEGG categories enriched within gene sets. 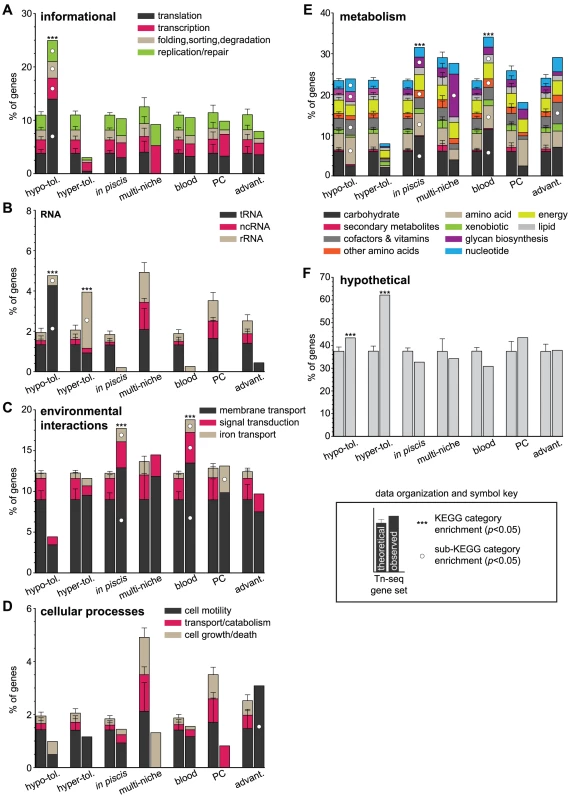
To orient the reader to graph organization and symbols, a key is provided in the lower right corner. The cumulative percent of genes (y-axis) within each Tn-seq-derived gene set (x-axis) belonging to the functional categories (A) informational, (B) RNA (C) environmental interactions, (D) cellular processes, (E) metabolism and (F) hypothetical is depicted. The proportion of genes contributed by specific sub-KEGG categories is represented by stacked elements within each column. Significant enrichments for KEGG and sub-KEGG categories are denoted with asterisks and white circles, respectively. Gene set abbreviations: hypo-tol. = hypo-tolerant; hyper-tol. = hyper-tolerant; advant. = advantageous. Theoretical KEGG category compositions for Tn-seq gene sets and p values were determined using 10,000 Monte Carlo simulations (Methods). Genes required for in vivo fitness
To identify genes necessary for in vivo fitness, a modified genetic footprinting approach was used (Figure S5). For input-output pairs in each experimental replicate (i.e. insertion mutants present in cognate input and output pools), a normalized occurrence frequency reflecting their abundance was calculated. This approach relies on an ability to count the number of specific mutant variants in corresponding input and output pools. However, strong negative selection within the zebrafish host may have completely purged variants with disruptions in genes critical for fitness. Therefore, we established criteria to determine which mutant variants were likely lost due to selection and not by chance through experimental bottlenecks (Figure S5 and Methods). Missing variants that met these criteria were assigned a value equal to the limit of detection. Within each replicate experiment, we employed a Wilcoxon signed-rank test on the collection of inserts for each gene to determine if the distribution of insert occurrence frequencies between input and output samples was significantly different. This is a conservative approach that requires consistency between insertion variants across a given gene for it to be identified as a fitness determinant. Genes with 10 or more measured inserts, a Wilcoxon p value less than 0.05 and an average decrease in variant abundance of at least 2-fold were considered to have a significant influence on in vivo fitness (FDR <10%). Of the 970 genes identified across all replicates and niches, only 21 overlapped with the hypo-tolerant gene set, suggesting that the vast majority of genes identified are specifically required by F11 to survive within the zebrafish embryo (Figure 2: in piscis, http://pathogenomics.path.utah.edu/F11_TnSeq/). This ‘in piscis’ gene set represents ∼18% of the annotated regions within the F11 chromosome and is enriched for genes with functions that fall under the KEGG category ‘environmental interactions’ and the subcategories membrane and iron transport (Figure 3C). Additionally, metabolic functions were also highly represented, encompassing genes involved in glycan biosynthesis and the metabolism of amino acids and carbohydrates (Figure 3E). The proportion of candidate fitness genes that were identified within each of the three mutant libraries is visualized in Figure S6. Genes within the six different in piscis gene sets showed only weak, though still in some cases significant, overlap (denoted in Figure S6), suggesting that the screening of multiple libraries was important for a more thorough enumeration of regions that influence in vivo fitness. Of note, we speculate that we did not fully reach a level of experimental saturation that matched the mutagenic saturation of our libraries. This observation may indicate a point of consideration for future Tn-seq-based screens using high-throughput surrogate hosts such as zebrafish where initial inoculum doses are relatively small and the stochastic nature of infection can influence the resolution of more subtle phenotypes. However, based on the findings presented in the following sections, we conclude that this perceived experimental noise did not compromise our ability to accurately identify and confirm genes of importance to ExPEC fitness and virulence.
Niche-dependent gene requirements
Seventy-six genes (1.4% of total gene content) were found to be necessary for fitness during both localized (PC) and systemic (blood) infections (http://pathogenomics.path.utah.edu/F11_TnSeq/). This ‘multi-niche’ gene set appears to collectively encode functions that directly influence gene expression and membrane architecture, both of which are known to be critical for pathogen fitness. Indeed, a number of the multi-niche genes have already been recognized for their role in bacterial stress resistance, regulation of virulence genes and pathogenesis (Table 1). Transposon inserts within several of the multi-niche genes resulted in some of the most dramatic declines in F11 fitness in vivo (Table 1). Notable candidates include EcF11_3256/bipA, EcF11_1255/lrhA and EcF11_2271/rfaH [30], [31], [32]. This gene set is also remarkably enriched for factors that function in glycan biosynthesis, including several genes involved in the production of capsular polysaccharides, which is a well-documented ExPEC-associated virulence trait (Figure 3E) [33], [34].
Tab. 1. Biological significance of Tn-seq-derived candidate genes. 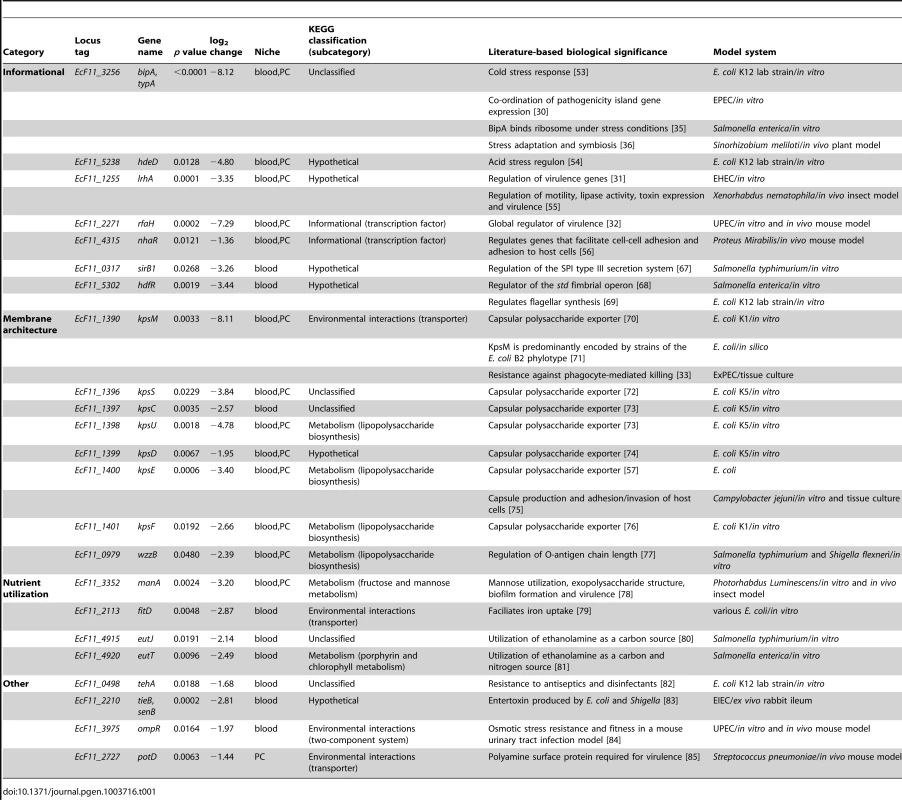
As noted above, the zebrafish PC is more permissive to ExPEC growth than the bloodstream [10], [11]. Corroborating this observation, we found 772 genes to be exclusively required for blood-borne fitness while only 122 genes were exclusive to the PC (http://pathogenomics.path.utah.edu/F11_TnSeq/). This blood-specific gene set was enriched for 7 different KEGG functional categories, whereas the PC-specific gene set was enriched for only 1 (Figure 3, p<0.05). Iron transport, the sole category enriched within the PC gene set, was also enriched within the blood set (Figure 3C). Additional categories that were enriched among blood-borne fitness genes covered signal transduction, membrane transport and the metabolism of amino acids, carbohydrates and lipids (Figure 3C and E). These observations indicate that the ability of ExPEC to survive and grow systemically within the zebrafish is, in part, dictated by a functionally diverse set of genes that facilitate the utilization of nutrients that are likely scavenged from the host.
Retrospective validation of a Tn-seq-derived fitness determinant
We chose the candidate gene EcF11_3256/bipA—which has not been previously reported to play a part in the virulence potential of ExPEC—to experimentally confirm the proficiency of our Tn-seq approach. An insertion map of this gene, depicting the reduction in fitness for each variant across all six pools, is presented in Figure 4A. The average changes in fitness for bipA and neighboring genes were also examined and indicated that the disruption of only bipA correlated with the observed fitness defect (Figure 4B). BipA is conserved across several bacterial phyla and has previously been shown to be a ribosome-associated GTPase that can coordinate the expression of virulence and stress response genes [30], [35], [36]. A targeted deletion of the EcF11_3256/bipA allele was generated and the resulting mutant derivative injected into the zebrafish PC and CV at a one-to-one ratio with wild type F11. F11Δ3256 exhibited a significant reduction in fitness compared to the wild type strain in both environments (Figure 4C). Independent challenges within the PC and bloodstream revealed that F11Δ3256 also exhibits significantly attenuated killing kinetics compared to wild type (Figure 4D). These results confirm that EcF11_3256/bipA is a critical component of F11's genetic repertoire and promotes both the fitness and virulence of this ExPEC strain within distinct host environments. Collectively, the identification of genes with known roles in bacterial pathogenesis (Table 1) and our observations with the Tn-seq-derived in piscis candidate gene EcF11_3256/bipA validate the Tn-seq protocol described here as a viable approach for detecting fitness and virulence determinants of ExPEC.
Fig. 4. Retrospective deletion of the candidate gene EcF11_3256/bipA confirms Tn-seq as a useful tool for the identification of loci required for fitness and virulence. 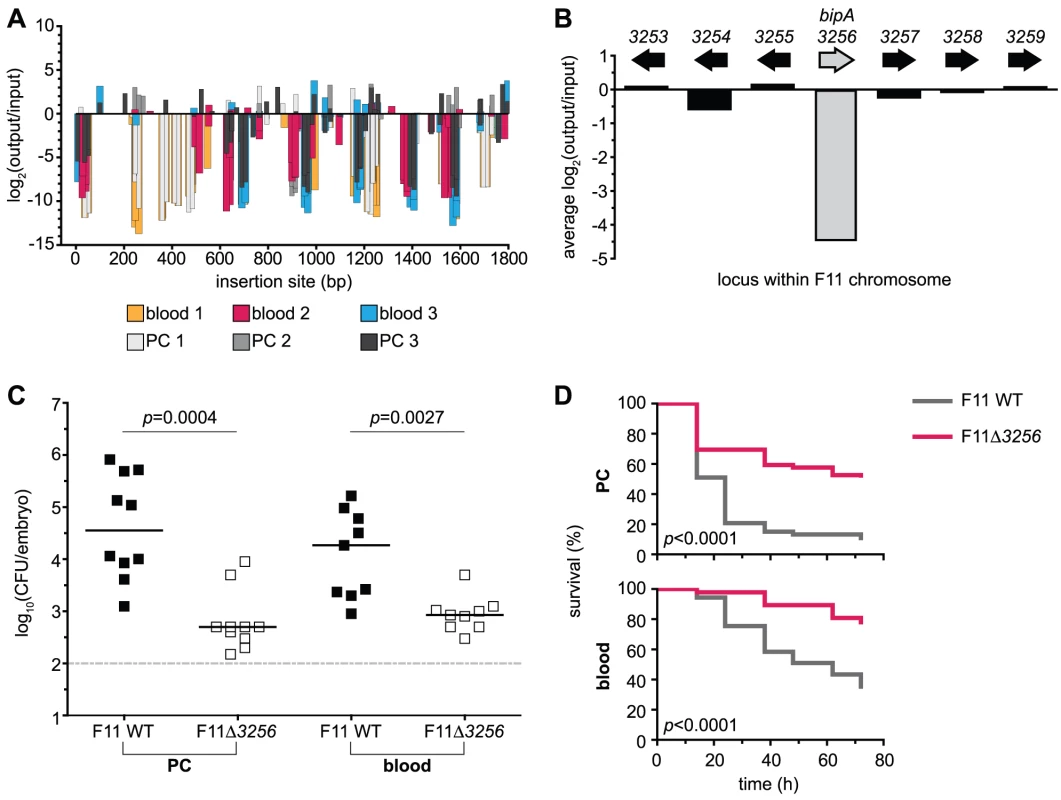
(A) Inserts found within the locus EcF11_3256/bipA are plotted with respect to their position of integration (x-axis). Magnitude and direction of bars represent the change in fitness of F11 (y-axis) that correlated with a given insertion event. Color of bars denotes pool of origin. Changes in fitness are presented as log2 of the ratio of occurrence frequencies observed between output and input pools (output/input). (B) The average of insert-based fitness changes in (A) is plotted on the x-axis (shaded bar, bipA). For comparison, the average alterations in occurrence frequency ratios for proximal genes are also plotted. (C) Equal numbers (1,000–2,000 CFU total) of wild type and F11Δ3256 were inoculated into the PC (left) or bloodstream (right) of zebrafish embryos. Fish were sacrificed and bacterial loads enumerated ∼20 h post-inoculation by differential plating (n = 9 to 10 zebrafish). Dashed line marks the limit of quantification; p values were determined using a paired t-test, bars indicate medians. (D) The pericardial cavity (PC, top) and blood (bottom) of 48 hpf embryos were inoculated with approximately 3,000 CFU. Fish were scored for death at 0, 12, 24, 36, 48, 60 and 72 h post-inoculation (hpi). Data are presented as Kaplan-Meier survival plots and p values were calculated using a log-rank (Mantel-Cox) test (n>45 for each survival curve). Disruptions that confer an in vivo fitness advantage
We identified 227 genes (∼4% of gene content), that when disrupted conferred a fitness advantage to F11 within the zebrafish host (http://pathogenomics.path.utah.edu/F11_TnSeq/). Twenty of these gene disruptions were advantageous in both the PC and blood, 124 were advantageous only in the PC and 84 conveyed significant advantages in only the blood. KEGG functional categories enriched within the ‘advantageous’ gene set include cell motility, nucleotide metabolism and biosynthesis of vitamins and cofactors (Figure 3D and E). This indicates that motility is likely dispensable for survival within zebrafish and agrees with previous work showing that loss of this trait can provide bacteria with a fitness advantage in some host settings [37]. Other factors encoded within the advantageous gene set may control processes that are easily trans-complemented by neighboring bacterial cells through the production of ‘common goods’ or represent negative regulators of virulence determinants that promote competitive growth [38]. Thirteen of the 227 advantageous genes were hyper-tolerant to transposon insertion, suggesting that the inactivation of these genes may have provided a growth advantage prior to in piscis selection (Figure 2: advantageous).
Trait Enrichment Analysis (TEA) is used to filter Tn-seq-derived candidate genes for putative virulence determinants
To focus the list of Tn-seq-derived candidate genes for further study, we developed a qualitative ranking system using a customized algorithm referred to as Trait Enrichment Analysis (TEA). The underlying premise of TEA was inspired by molecular Koch's postulates, which are a widely acknowledged set of guidelines used by investigators to critically assess the contributions made by certain genetic elements to the virulent nature of pathogenic organisms [39]. In particular, TEA draws on the first axiom of these postulates, which states that, “The phenotype or property under investigation should be associated with pathogenic members of a genus or pathogenic strains of a species.” [40]. Thus, the primary goal of TEA is to identify correlations between bacterial genes and bacterial traits (i.e. genotype-phenotype relationships).
TEA was carried out as outlined in Figure 5. Briefly, protein sequences were acquired from an ecologically diverse, manually curated collection of 165 bacteria representing six phyla and assembled to form the TEA metaproteome database (TEA-MD, Table S3). Each bacterial strain was annotated with ‘trait categories’ defining habitat, niche of isolation, taxonomic lineage and phenotype (i.e. pathogen or non-pathogen). Homologues were retrieved for each of the 5,146 protein-coding genes contained within the F11 chromosome using the Basic Local Alignment Search Tool: BLASTp (Methods) [41]. Because proteins in the TEA-MD are linked to a set of traits defined by their source bacterium, homologue collections associated with F11 proteins can be appraised for the presence of a universal characteristic in a similar fashion to traditional gene ontology and KEGG enrichment analyses. As a result, hypotheses can then be generated regarding a protein's evolutionary origins (i.e. horizontal vs. vertical inheritance or homoplasy) and possible influence on bacterial physiology or behavior based on its combination of trait associations. A complete description of the TEA-MD, including the number of organisms and proteins comprising each trait category and the relative proportions of phyla compared to sequenced isolates in GenBank, is provided as supporting information (Figures S7 and S8).
Fig. 5. Diagrammatic workflow of ‘Trait Enrichment Analysis’ (TEA). 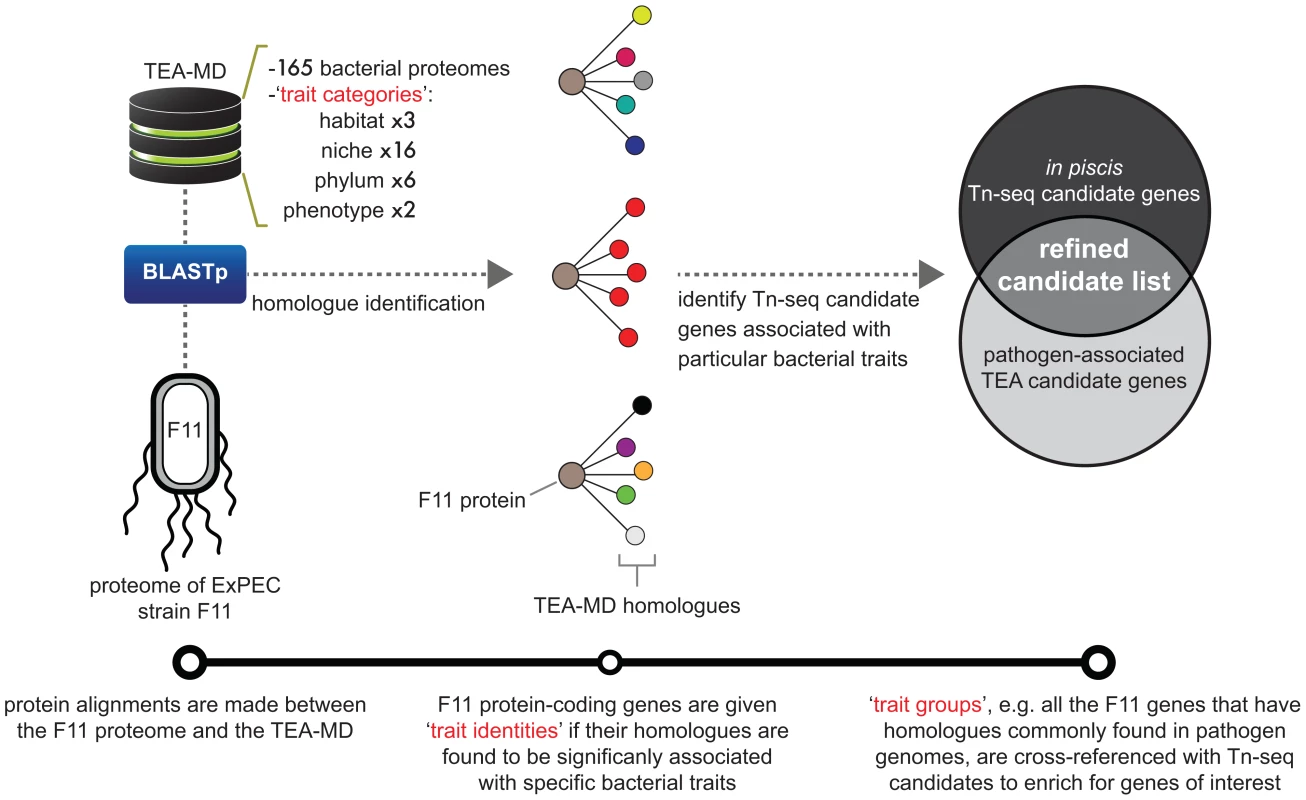
This visualization depicts the TEA method and describes the relationships between various data types. Bottom track: linear representation of steps followed in the execution of TEA. Protein alignments are made between F11 proteins and the TEA-MD to identify homologues. Microbes within the TEA-MD are annotated with four separate traits contained within the trait categories: habitat (1 of 3), niche (1 of 16), phylum (1 of 6) and phenotype (1 of 2). The composition of traits associated with homologue sets for each F11 protein is assessed for enriched traits in order to assign trait identities. Trait groups, which are F11 genes that share a particular trait identity, are used to organize Tn-seq-derived candidate genes. TEA-MD = TEA-metaproteome database; BLASTp = Basic Local Alignment Search Tool used to align protein sequences. Pathogen-associated genes within Tn-seq gene sets
TEA, as it was employed here, aimed to assign ‘trait identities’ to F11 genes, with particular attention being paid to genes that are common to known pathogens and therefore, putative virulence determinants (Figure 5). It was found that approximately 22% of the F11 genome is made up of genes with pathogenic identity—1,166 F11 genes are significantly more associated with pathogen genomes (Methods). Considering Tn-seq-derived candidate genes, it was anticipated that the in piscis gene set be enriched for these types of genes. Indeed, a significant portion of the 970 in piscis genes had pathogenic identity (246 genes, 25% of gene set, p = 0.009) (Table S4 and http://pathogenomics.path.utah.edu/F11_TnSeq/). The enrichment of pathogen-associated genes within the in piscis gene set was largely attributed to the composition of both the PC and blood gene sets, which also had a significant number of genes with pathogenic identity as determined by TEA (PC: 39 genes, ∼32% of gene set, p = 0.008; blood: 192 genes, ∼25% of gene set, p = 0.0485). Interestingly, the multi-niche gene set was not enriched for pathogen-associated genes (15 genes, ∼20% of gene set, p = 0.786).
We conclude from these data that genes required for F11 fitness in multiple niches are likely to encode more basic ‘core’ type functions that may enable both pathogenic and non-pathogenic strains alike to better deal with general stresses encountered within varied host environments, but not necessarily in broth culture. In contrast, the niche-specific genes, which are more likely to be pathogen-associated, may endow bacteria like F11 with salient, pathoadaptive traits that are perhaps contextually regulated. Along these lines, we unexpectedly found that the advantageous gene set was also enriched for pathogen-associated genes (Table S4, 69 genes, ∼30% of gene set, p = 0.004). We hypothesize that the disruption of these particular genes during the Tn-seq selection screen alleviated fitness costs associated with their maintenance and/or expression within the zebrafish host. Future investigation of the advantageous gene set may illuminate evolutionary tradeoffs that are incurred by the acquisition and retention of these loci by pathogens as they traverse different host-associated environments.
Examination of Tn-seq candidates filtered by TEA
To investigate the effectiveness of TEA to help identify previously uncharacterized genes important for ExPEC fitness in vivo, we selected 3 from the subset of 246 pathogen-associated Tn-seq-derived in piscis genes for closer examination. EcF11_3082, EcF11_2628 and EcF11_3933 were individually deleted from the F11 chromosome and tested in competitive infection assays against wild type F11 within the zebrafish PC and blood. These genes were chosen based on their performance in the initial Tn-seq selection screen and their association with known pathogens, as determined by TEA. Trait identities for each gene are illustrated in Figure 6 along with the results from competitive infection assays.
Fig. 6. Targeted deletion of candidate genes with pathogenic identity attenuates the fitness of F11 within zebrafish. 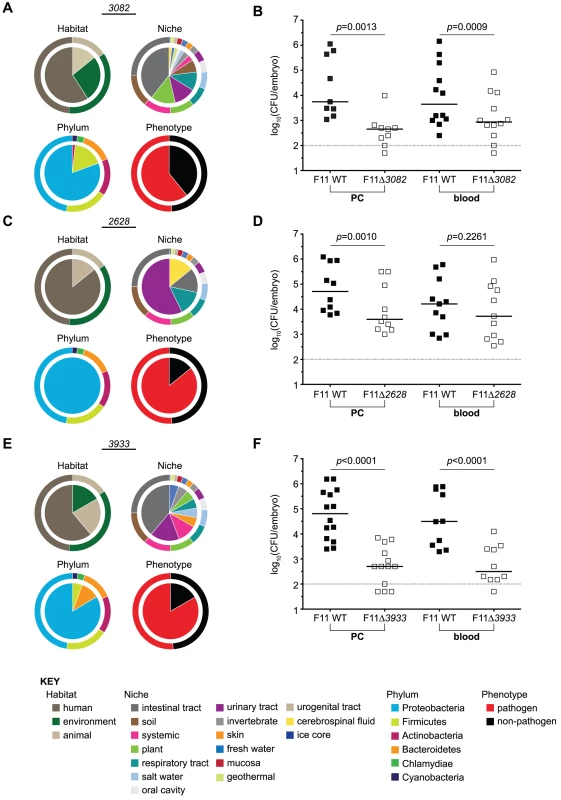
The composition of traits (bottom KEY) represented among bacteria encoding homologues of (A) EcF11_3082, (C) EcF11_2628, or (E) EcF11_3933 are presented for the trait categories habitat of isolation, niche of isolation, phylum and phenotype. For reference, segmented outer rings bordering each pie graph depict the composition of traits of all genes contained within the TEA-MD. Equal numbers (1,000–2,000 CFU total) of wild type and (B) F11Δ3082, (D) F11Δ2628, or (F) F11Δ3933 were inoculated into the PC (left) or bloodstream (right) of embryos. Fish were sacrificed and bacterial counts determined ∼20 h post-inoculation by differential plating (n = 9 to 14 zebrafish). Dashed line marks the limit of quantification; p values were determined using a paired t-test, bars indicate medians. The EcF11_3082 gene product is homologous to the transcriptional repressor MprA/EmrR. In E. coli lab strains, MprA/EmrR has been shown to negatively regulate biosynthesis of the microcin B17 antibiotic and a multidrug efflux pump encoded by emrA and emrB [42], [43]. Fifty-six strains within the TEA-MD harbor an EcF11_3082-like gene, 34 (61%) of which are pathogens (Figure 6A, significance of pathogenic identity: p = 0.034). Tn-seq indicated that EcF11_3082 promotes the fitness of F11 in both the PC and bloodstream in all three replicates. Average fitness deficits observed through Tn-seq for EcF11_3082 insertion mutants ranged from 5 to 21-fold. This was confirmed by one-to-one competitive assays in which survival of the EcF11_3082 deletion mutant was markedly diminished relative to wild type F11 following inoculation of the PC and blood (Figure 6B).
Transposon insertions into the EcF11_2628 locus decreased the fitness of F11 within the blood by an average of 15-fold, as assessed by Tn-seq. The TEA-MD contains 7 homologues of EcF11_2628, 6 of which belong to ExPEC strains that were isolated from a variety of host-associated niches (Figure 6C, significance of pathogenic identity: p = 0.035). EcF11_2628 encodes a hypothetical protein that is predicted by PHYRE (Protein Homology/analogY Recognition Engine) to function as a toxic extracellular metalloendopeptidase [44]. A structurally similar toxin, AsaP1, was recently described as a critical virulence factor encoded by the natural fish pathogen Aeromonas salmonicida subsp. Achromogenes [45]. Deletion of this gene from F11 resulted in decreased fitness within the PC and surprisingly, had no discernible effects in the bloodstream during one-to-one competition assays with the wild type strain (Figure 6D). These results run counter to those obtained with Tn-seq, where disruption of EcF11_2628 reduced the fitness of F11 within the blood. This discrepancy may be attributable to the fact that Tn-seq is in effect a one-to-3,000 competitive assay among transposon insertion mutants within each fish, whereas in our follow-up assays specific gene knockout mutants are tested in one-to-one competition with wild type. Within these different experiments, individual mutants may be subject to very different pressures and frequency-dependent selection events.
Tn-seq results indicated that disruption of a third pathogen-associated gene, EcF11_3933, attenuates the fitness of F11 about 4-fold within the PC. Fifteen of the 18 EcF11_3933 homologues (83%) found in the TEA-MD are encoded by known pathogens, including multiple ExPEC strains, E. coli O157:H7, Neisseria gonorrhoeae, Vibrio cholera, Haemophilus influenza, Photorhabdus luminescens and Erwinia amylovora (Figure 6E, significance of pathogenic identity: p = 0.001). EcF11_3933 is uncharacterized and annotated as a homologue of the broadly conserved DNA protecting protein DprA. A related but distinct protein, DprA/Smf, has been suggested to confer competence for the uptake of foreign DNA by various bacterial species [46], [47], though it is presumed to function differently in lab strains of E. coli [48]. In competitive assays, deletion of EcF11_3933 rendered F11 significantly less fit than the wild type strain in both the PC and bloodstream of zebrafish (Figure 6F).
Tn-seq and TEA-derived in piscis candidate genes contribute to pathogen fitness within mouse models of bacteremia and urinary tract infection
To extend our experimental and sequence-based observations with the candidate genes EcF11_3256/bipA, EcF11_3082, EcF11_2628 and EcF11_3933, we employed a more traditional host organism for investigating ExPEC pathogenicity—the laboratory mouse [49], [50]. Evaluating Tn-seq and TEA-derived in piscis candidate genes using a murine model provided an opportunity to further establish the biological relevance of our multilayered approach. Murine models of bacteremia are an established means to assess the general capacity of ExPEC isolates to replicate and cause disease within a host [50], [51], [52]. One-to-one mixtures of wild type F11 and each mutant strain (108 total CFU) were subcutaneously injected into adult female Swiss Webster mice. Approximately 12 to 15 h later, bacterial titers within the spleen and liver were determined (Figure 7). Mutants lacking EcF11_3256/bipA (Figure 7A), EcF11_3082 (Figure 7B) and EcF11_3933 (Figure 7D) were significantly outcompeted by the wild type F11 parent strain. However, F11Δ3256 exhibited a more modest defect in the spleen (p = 0.0828). In contrast, there was no observable competitive difference between wild type and F11Δ2628 in either organ (Figure 7C).
Fig. 7. Tn-seq and TEA-derived candidate genes promote the fitness of F11 within murine models of bacteremia and urinary tract infection. 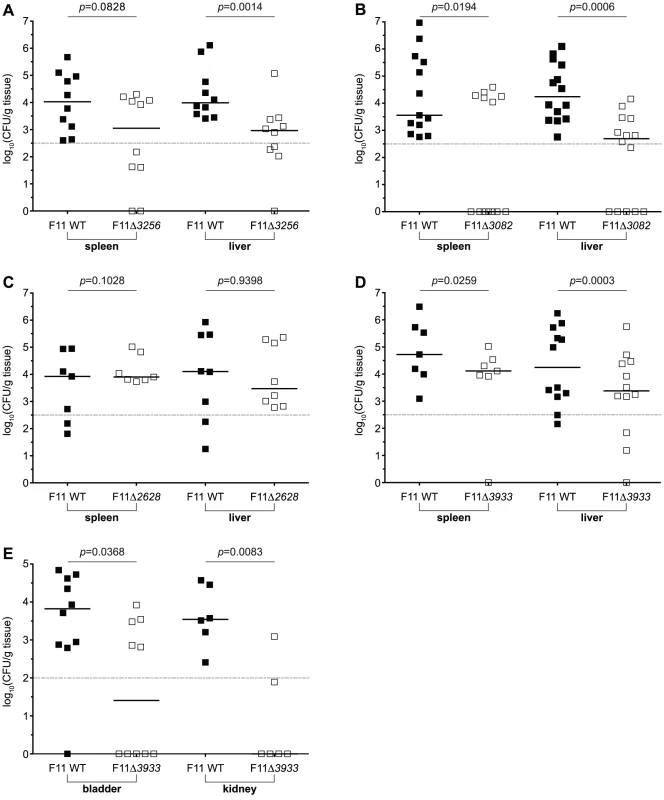
Equal numbers (108 total CFU) of wild type F11 and (A) F11Δ3256, (B) F11Δ3082, (C) F11Δ2628, or (D) F11Δ3933, were subcutaneously injected into the nape of the neck of adult female Swiss Webster mice. Approximately 12 to 15 h later, bacterial titers within the spleen (left) and liver (right) were determined by differential plating. (E) Equal numbers (107 total CFU) of wild type F11 and F11Δ3933 were transurethrally inoculated into the urinary tract of adult female CBA/J mice. Bacterial titers present in the bladder (left) and kidneys (right) were determined 3 d post-inoculation by differential plating. Dashed line marks the limit of quantification. n = 7 to 14 mice; p values were determined using a paired t-test, bars indicate medians. Of note, we found that F11Δ3256 displayed a cold sensitivity phenotype when grown at 20°C in broth culture, which is consistent with reports on BipA function in other bacteria (Figure S9A) [53]. Therefore, we were initially concerned that the growth rate of this mutant at 28.5°C—the temperature used to raise zebrafish embryos—was compromising our ability to distinguish its temperature and host-dependent phenotypes. However, F11Δ3256, like the other mutants, grew similar to the wild type strain at both at 28.5°C and 37°C (Figure S9). We conclude that the attenuated fitness phenotypes observed with F11Δ3256 in competitive assays in both zebrafish and mice are independent of temperature effects.
We next tested the fitness contributions made by each of the four candidate genes above within the murine urinary tract, which is another commonly used model system for understanding ExPEC pathogenicity. Three days after inoculation of one-to-one mixtures of wild type and mutant derivatives (107 total CFU) into the urinary tract of adult female CBA/J mice, kidneys and bladders were harvested and bacterial titers determined. Of the four genes assayed, only EcF11_3933 was critical for the competitive fitness of F11 within the bladder and kidneys (Figure 7E).
Our ability to identify candidate genes necessary for ExPEC fitness within mammalian host niches through initial prescreening in embryonic zebrafish highlight the utility of this teleost host to model overlapping aspects of ExPEC biology. By comparing the specific host and niche-specific phenotypes of various ExPEC isolates and mutant strains in futures investigations, we will be able to gain a deeper understanding of the host-pathogen interactions engaged by ExPEC.
Discussion
Application of Tn-seq to investigate the multigenic basis of ExPEC pathogenicity
The genetic and phenotypic plasticity of ExPEC isolates, which is in part due to horizontal transfer of pathoadpative accessory genes, presumably enables these pathogens to traverse a wide range of environmental and host-associated niches [7], [8], [10], [11]. It will be important to decipher how ExPEC employ their genetic repertoires so that more specific and efficacious therapeutic strategies can be developed. Moreover, knowledge of key events that occur during the evolutionary assembly of pathogen genomes will likely be gleaned as genotype-phenotype relationships are mapped. To this end, we functionally dissected the contents of an ExPEC reference genome using the recently described transposon-based screening technique Tn-seq (Figure 1) [14], [15]. With mutant pools derived from the ExPEC reference isolate F11 and multi-niche zebrafish infection models, we found that ∼18% of the F11 genome significantly affected bacterial fitness under the pathogenic conditions tested. These fitness determinants can be grouped based on the niches where they were required, which in some instances correlated with particular functions like iron transport and amino acid metabolism (Figure 3). Gene set and gene-based contributions to fitness can be explored through an online and curated data viewer (http://pathogenomics.path.utah.edu/F11_TnSeq/). Going forward, it is our intent that this work will expedite future comparative and functional pathogenomic analyses focused on elucidating the genetic basis of pathogen behavior and evolution.
Seventy-six genes (1.4% of genome content) were found to significantly influence the fitness of F11 in both the PC and blood of zebrafish. We suggest that these multi-niche genes largely provide core functions that are critical, but not necessarily specific, to pathogen physiology. It was not surprising that several of these genes were recognizable and annotated as being involved in the regulation of gene expression and membrane architecture—two functions that are important to pathogens and non-pathogens alike (Table 1). In addition to the translational regulator EcF11_3256/BipA, which we validated in follow up experiments using a targeted deletion mutant and multiple in vivo host model systems (Figure 4 and 7A), several other informational genes, including hdeD, lrhA, rfaH and nhaR, were also identified by our Tn-seq screens as being important to the fitness of F11 within multiple niches. These genes have been shown to affect acid stress resistance, toxin expression, adhesion to host tissues and virulence gene regulation in a diverse array of pathogens, including enterohemorrhagic E. coli (O157:H7), Xenorhabdus nematophila, Proteus mirabilis and a sub-group of ExPEC isolates like F11 known as uropathogenic E. coli (UPEC) (Table 1) [31], [32], [54], [55], [56]. Additionally, genes responsible for the production of capsule—which is a well-established virulence feature that serves as a protective polysaccharide barrier surrounding the bacterial cell wall—were also important in both the PC and blood (Table 1) [57]. Further identification and, importantly, functional characterization of multi-niche genes will lead to a deeper understanding of the general fitness and virulence gene networks employed by ExPEC and other virulent bacteria within varying host environments.
Aside from multi-niche genes, we observed a striking numerical imbalance of genes that are exclusively required for either blood-borne or tissue-localized PC fitness (772 and 122 genes, respectively). The large number of genes and variety of enriched KEGG functional categories within the blood-specific gene set corroborated previous experimental observations that indicated the blood of embryonic zebrafish is a hostile environment and highly restrictive to bacterial growth [10], [11]. Specifically, genes involved in nutrient acquisition and utilization made up a significant portion of the blood-specific genes (Figure 3). These results provide evidence that during systemic infections nutrients are more limiting than during localized infection of the PC, requiring ExPEC to employ a greater portion of their genetic repertoire to maintain fitness in the blood. However, additional factors could also account for the discrepancy between the PC and blood-specific gene sets, including instances of trans-complementation. Once injected into the PC, bacteria remain confined until the embryo succumbs to the infection [10]. Because bacteria stay in close proximity to one another within the PC, the chances for trans-complementation and the sharing of common goods are high and may result in relaxed selection. A recent study demonstrated that extensive trans-complementation can occur within mixed mutant populations of Yersinia pestis in a mouse model of pneumonic plague [58]. However, the authors noted that this appears to be a unique characteristic of Y. pestis, as other pathogens with tropism for lung tissue do not exhibit such a high degree of wild type-to-mutant rescue. In our assays, trans-complementation effects within the PC were apparently limited as we were able to identify genes required for ExPEC survival within this niche.
Ultimately, this initial genome-wide analysis of the components necessary for ExPEC fitness within pathogenic environments garnered two key insights: (i) the number of genes that influence fitness in multiple niches is relatively small and (ii) there are distinct subsets of genes employed by ExPEC within different host environments. This suggests that do-it-all genes do not necessarily dictate the generalist and plastic nature of ExPEC. Instead, ExPEC may evolutionarily maintain sets of context-specific genes that can be employed, possibly in different combinations, to respond in a highly specific-manner to a variety of conditions. This characteristic likely contributes to the seemingly redundant composition and versatility of contemporary ExPEC genomes. Further functional dissection of ExPEC genomes by Tn-seq-based approaches will help clarify which genes underlie the salient traits of this lineage and inform the design of broad-spectrum therapies that target only ExPEC while leaving beneficial or benign E. coli strains within the normal microbiota unperturbed.
Refinement of candidate gene sets by TEA
A longstanding challenge associated with unbiased, high-throughput functional genomics techniques like Tn-seq has been the need to develop methods that provide an effective transition to focused follow-up studies. Much work has been done to quantitatively filter datasets using sophisticated determinations of statistical significance and false discovery control procedures. Analysis of the molecular pathways represented within functional genomics gene sets has also been addressed extensively. However, less attention has been given to systematic methods that exploit the qualitative features of individual genes and how such considerations can streamline candidate gene vetting. To address this issue, we developed a Trait Enrichment Analysis (TEA) tool to organize Tn-seq-derived candidate genes in a biologically meaningful way (Figure 5). With a manually curated set of 165 microbial proteomes, TEA is able to assign meta-features to F11 genes based on their association with specific bacterial traits (i.e. habitat, niche of isolation, phylum and phenotype). In this way, TEA provides a tractable method to categorize Tn-seq candidate genes into a more focused subset—something not easily accomplished using larger microbial sequence databases like GenBank and BioCyc or functional annotation platforms such as DAVID. TEA uncovered several genes with previously unappreciated roles in ExPEC pathogenicity, thus creating new opportunities to further understand the genetic underpinnings of this lineage.
For the purposes of our investigation, we used TEA to identify F11 genes with homologues that are more often associated with the genomes of known pathogens. We describe such genes as having ‘pathogenic identity’ and reason that, because of their affiliation with pathogens, they are more likely to contribute to pathogenic behavior—a principal tenet of molecular Koch's postulates [39], [40]. In considering the overlap between Tn-seq-derived in piscis genes and the TEA-derived gene set made up of F11 genes with pathogenic identity, we were able to identify 246 genes that potentially influence the in vivo fitness capacity of F11 specifically. We selected three of these pathogen-associated candidate genes for closer inspection.
The candidate gene EcF11_3082 was identified by Tn-seq and confirmed during one-to-one co-challenge experiments to be important for the fitness of F11 within the blood and PC of zebrafish and spleen and liver of mice (Figure 6B and 7B). This gene appears to be relatively widespread within the TEA-MD, being found in 56 genomes, 61% of which are classified as pathogens. The range of bacteria encoding homologues of EcF11_3082 includes isolates from 12 (out of 16) different niches and 3 (out of 6) phyla represented in the TEA-MD (Figure 6A). These observations suggest that EcF11_3082 may play a broad role in bacterial fitness within a variety of pathogenic and non-pathogenic contexts. EcF11_3082 is predicted to encode an MprA/EmrR-like transcriptional repressor, which was shown to regulate the expression of a downstream multidrug efflux pump encoded by emrA and emrB. Although our Tn-seq selection screen and co-challenge experiments identified EcF11_3082 as a fitness determinant, disruption of the emrA and emrB genes present in F11 did not appreciably affect fitness within the zebrafish host, as determined by Tn-seq. This suggests that either disruption of EcF11_3082 led to aberrant and deleterious expression of emrAB that resulted in attenuation of the EcF11_3082 mutant, or the particular MprA/EmrR allele harbored by F11 influences other aspects of bacterial physiology not yet defined. Further investigation is required to determine the exact influence of EcF11_3082 on pathogenicity.
EcF11_2628, which encodes a ‘conserved hypothetical protein’ that has some homology to a secreted metalloendopeptidase toxin, was also identified by Tn-seq and confirmed as an important fitness determinant using a targeted knockout strain (Figure 6D). However, we were unable to ascertain if this gene is required during colonization of a mammalian host. The F11Δ2628 mutant variant did not exhibit any statistically significant decline in competitive fitness during systemic or urinary tract infection of mice (Figure 7C and data not shown). Further investigation is required to determine the physiological context in which this putative toxin promotes ExPEC fitness. Nonetheless, homologues of EcF11_2628 were found in only 7 proteobacterial genomes within the TEA-MD (Figure 6C and D). Of these, 5 are ExPEC strains that were isolated from human patients with either meningitis (strain S88) or UTI (strains CFT073, UTI89, 536 and UMN026). Another ExPEC strain (APEC01) harboring an EcF11_2628 homologue came from a case of avian sepsis that occurred subsequent to respiratory tract infection. The sole non-pathogen that carried an EcF11_2628 homologue was ED1a, a commensal isolate from the human gut that is classified as belonging to the B2 phylogenetic group—the E. coli lineage that encompasses the vast majority of human ExPEC strains [7]. This observation is intriguing because the EcF11_2628 allele carried by ED1a could represent either a genetic vestige from a pathogenic past or a new acquisition that is edging ED1a down an evolutionary path towards a more ExPEC-like countenance. Elucidating the function of this hypothetical gene should help explain its seemingly restricted distribution within a phylogenetically narrow range of bacteria.
The final pathogen-associated gene that we examined, EcF11_3933, encodes a variant of the DNA protection protein DprA [46], [47]. However, this particular dprA allele is distinct from those previously characterized in other bacteria. Within the TEA-MD, 83% of the EcF11_3933 homologues were found in the genomes of known pathogens. Interestingly, pathogens harboring an EcF11_3933-like gene included ExPEC isolates as well as other members of Proteobacteria and species within the phyla Bacteroidetes and Firmicutes (Figure 6E). In our Tn-seq selection screens, EcF11_3933 was initially identified because it appeared to affect fitness within the PC, but subsequent analyses revealed that EcF11_3933 could also promote pathogen fitness within the bloodstream of both zebrafish and mice (Figure 6F and 7D). Indeed, there were indications that EcF11_3933 contributed to blood-borne fitness, but it was filtered out of the final Tn-seq-derived blood gene set because it did not meet all of the predetermined cutoffs, though only by a thin margin. These results identify limits to Tn-seq as it is applied here, but also highlight the value of using multiple infection models to discover and assess the roles of fitness and virulence determinants.
Additional work using a murine UTI infection model demonstrated that EcF11_3933 is also important for competitive fitness within the murine bladder and kidneys (Figure 7E). This is in contrast to EcF11_3256, EcF11_3082 and EcF11_2628, which we found to be dispensable for colonization of the urinary tract (data not shown). Interestingly, alleles of c4222, which is a homologue of EcF11_3933 harbored by the UPEC strain CFT073, were shown by microarray to be transcriptionally induced within a panel of UPEC isolates during active infection of the human urinary tract [59]. Together, previous observations and those presented here indicate that EcF11_3933 is of broad importance to the pathogenic potential of F11 and other ExPEC isolates within a variety host-associated niches. Moreover, the distribution of EcF11_3933-like genes among a diverse array of pathogens (e.g. ExPEC, the enteric pathogen E. coli O157:H7, Neisseria gonorrhoeae, Vibrio cholera, Haemophilus influenza, Photorhabdus luminescens and Erwinia amylovora), which can colonize and cause disease within a broad range of animal, insect and plant hosts, suggests that EcF11_3933 also represents a multi-lineage virulence determinant. Further analysis of EcF11_3933 and its pathogen-associated homologues may prove useful in defining common virulence strategies employed by ExPEC as well as other more distantly related bacterial species.
Concluding remarks
Cumulatively, this investigation demonstrates that the application of Tn-seq, coupled with TEA, provides an efficient means to identify novel or previously unappreciated fitness and virulence determinants. The utility of TEA could easily be enhanced through expansion of the TEA-MD by, for example, the addition of metadata obtained from other host-microbe experimental systems. TEA serves as an accelerated annotation layer for bridging high-throughput sequencing experiments and functional characterization. In moving forward, it will be important to continue the careful curation of pertinent biological information associated with the rapidly expanding and often redundant and ill-defined collections of microbial genomics data so that meaningful connections can be mined. Ultimately, the use of Tn-seq and TEA with an expanded number of bacterial isolates and host systems may provide a detailed map of the gene families and signaling cascades employed by ExPEC and other pathogens to colonize host-associated environments.
Methods
Ethics statement
Animals used in this study were handled in accordance with University of Utah approved IACUC protocols that followed the standard guidelines described at www.zfin.org and in the Guide for the Care and Use of Laboratory Animals, 8th Edition.
Bacterial strains and plasmids
All bacterial strains and plasmids used in this study are listed in Table S5. Unless specified otherwise, bacteria were cultured statically at 37°C for 24 h in 20 ml of a defined M9 minimal medium (6 g/L Na2HPO4, 3 g/L KH2PO4, 1 g/L NH4Cl, 0.5 g/L NaCl, 1 mM MgSO4, 0.1 mM CaCl2, 0.1% glucose, 0.0025% nicotinic acid, 0.2% casein amino acids and 16.5 mg/ml thiamine in H2O) prior to injection into zebrafish embryos or mice. Antibiotics (kanamycin or ampicillin) were added to the growth medium when necessary to maintain recombinant plasmids or select for mutants.
Targeted gene knockouts of Tn-seq-derived candidate genes were generated in ExPEC strain F11 using the lambda Red-mediated linear transformation system [60], [61]. Briefly, a kanamycin resistance gene was amplified using polymerase chain reaction (PCR) from the pKD4 plasmid with 40-base pair overhangs specific to the 5′ and 3′ ends of each targeted locus. PCR products were introduced via electroporation into F11 carrying pKM208, which encodes an IPTG (isopropyl-β-D-thiogalactopyranoside)-inducible lambda red recombinase. Knockouts were confirmed by PCR. Primer sets used are listed in Table S6.
Retrofitting of the previously constructed pSAM_Bt vector to be used for the transposon mutagenesis of ExPEC isolate F11 was accomplished using source vectors and primers as listed in Tables S5 and S6, respectively. The Plac promoter from pGFP-Mut3.1 (Clonetech), along with its associated ribosome binding sequence, was amplified via PCR. Engineered 5′ BamHI and 3′ NdeI restriction sites were used to sub-clone the resulting fragment into a BamHI/NdeI (New England Biolabs) double digested pSAM_Bt vector upstream of the himar1C9 transposase gene. The kanamycin resistance gene from pKD4 was amplified and ligated using the restriction sites MfeI and XbaI, replacing the erythromycin resistance gene ermG in pSAM_Bt. The resulting transposon mutagenesis vector, pSAM-Ec, was stored and propagated in the pir+ E. coli strain EcS17.
Zebrafish embryos
*AB wild-type zebrafish embryos were collected from a laboratory-breeding colony that was maintained on a 14 h/10 h light/dark cycle. Embryos were grown at 28.5°C in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.4 mM CaCl2, 0.16 mM MgSO4) containing 0.000016% methylene blue as an anti-fungal agent.
Infection of zebrafish
One ml from a 24-h bacterial culture of each transposon mutant library or isogenic bacterial strain was pelleted, washed once with 1 ml sterile PBS (Hyclone) and re-suspended in approximately 1 ml PBS to obtain appropriate bacterial densities for microinjection. Prior to injection, 48 h post-fertilization (hpf) embryos were manually dechorionated, briefly anesthetized using 0.77 mM ethyl 3-aminobenzoate methanesulfonate salt (tricaine) (Sigma-Aldrich) and embedded in 0.8% low-melt agarose (MO BIO Laboratories) without tricaine. Approximately 1 nl of bacteria was injected directly into the pericardial cavity (PC) or the blood via the circulation valley (CV) located ventral to the yolk sac using a YOU-1 micromanipulator (Narishige), a Narishige IM-200 microinjector and a JUN-AIR model 3-compressor setup. For each experiment, average colony forming units (CFU) introduced per injection were determined by adding 10 nl of the inoculum into 1 ml 0.7% NaCl that was then serially diluted and plated on Luria-Bertani (LB) agar plates. For co-challenge experiments, input doses were plated on LB agar +/ − kanamycin (50 µg/mL) to determine relative numbers of the wild type and mutant strains present. After injection, embryos were carefully extracted from agar and placed in either a 10 cm petri dish containing E3 medium for transposon screening or, individually into wells of a 96-well microtiter plate (Nunc) containing E3 medium for co-challenge experiments. The E3 medium used during this incubation period lacked both tricaine and methylene blue.
To harvest DNA from embryos infected with transposon mutant libraries at the end of the selection period, all infected fish were collected (both alive and dead) in a 1.6 ml tube, being careful to remove as much E3 medium as possible by gently pulsing the tube in a microfuge to facilitate separation. Embryos were suspended in 500 µl of 0.5% Triton X-100 in PBS and homogenized using a mechanical PRO 250 homogenizer (PRO Scientific). Homogenates were then centrifuged at 18,000 g for 5 minutes to sediment bacteria away from host cellular components released during Triton X-100-mediated lysis. The resulting pellet, which appears black because of remaining zebrafish debris, served as the starting material for DNA extraction using the Wizard Genomic DNA Purification Kit (Promega) as per the manufacturer's protocol with a brief modification: the duration of the initial lysis step was extended as needed to dissolve the tough pellet.
To quantify bacterial numbers at the completion of co-challenge experiments, embryos were homogenized at the indicated time points in 500 µL PBS containing 0.5% Triton X-100 using a mechanical PRO 250 homogenizer. Homogenates were then serially diluted and plated on LB agar +/ − kanamycin (50 µg/mL) to determine relative numbers of wild type and mutant bacteria. For co-challenge assays, only embryos where the total bacterial CFU recovered was equal to or greater than the limit of quantification (i.e. at least 20 CFU counted on the lowest serial dilution, which corresponds to 100 CFU per embryo) are reported. For lethality assays, fish were inspected for death at the indicated time points over a 72 h period. Death is defined here as the complete absence of heart rhythm and blood flow. Survival graphs depict total pooled results from at least 3 independent experiments in which groups of approximately 15 to 20 embryos were injected at a time.
Mouse infections
For bacteremic co-challenge experiments, seven to eight-week old female Swiss Webster mice (Charles River) were anesthetized using isoflurane inhalation and injected subcutaneously via the nape of the neck with 200 µl of a 1∶1 wild type to mutant bacterial suspension containing a total of 108 bacteria in sterile PBS. Spleens and livers recovered 12 to 15 h later were weighed and homogenized in 500 µl sterile PBS. Homogenization was done with a Bullet Blender Storm 24 (Next Advance) using three SSB32 3.2 mm stainless steel beads for 5 minutes on power level 6. Homogenates were serially diluted and plated on LB agar +/ − kanamycin (50 µg/mL) to determine the number of both wild type and mutant bacteria. Bacteremic co-challenge experiments were repeated at least twice. For these assays, only organs where the total bacterial CFU recovered was equal to or greater than the limit of quantification (i.e. at least 20 CFU counted on the lowest serial dilution, which corresponds to approximately 200 to 600 CFU per g organ) are reported.
For co-challenge experiments within the murine urinary tract infection, seven - to nine-week old female CBA/J mice (Jackson Labs) mice were anesthetized using isoflurane inhalation and inoculated via transurethral catheterization with 50 µl of a 1∶1 wild type to mutant bacterial suspension containing a total of 107 bacteria suspended in PBS. Bladders and kidneys were recovered 3 days later and each was weighed and homogenized in 1 ml containing 0.025% Triton X-100. Homogenates were serially diluted and plated on LB agar +/ − kanamycin (50 µg/mL) to determine number of both wild type and mutant bacteria.
Statistical analysis of zebrafish and mouse infections
For co-challenge experiments, numbers of wild type and mutant bacteria present in the inoculum and recovered from host tissues were enumerated by differential plating on selective media as described above. To determine if bacterial counts differed between wild type and mutant-infected animals, a paired t-test was performed on log10-transformed values. Graphs and statistics were generated using GraphPad Prism 5.
Transposon library generation
Transposon mutagenesis
A detailed protocol for the transposon mutagenesis of ExPEC strain F11 using the E. coli donor strain EcS17 carrying pSAM-Ec and the methods to verify and sequence mutant libraries is provided in Protocol S1. Briefly, EcS17/pSAM-Ec and F11 were mixed 2∶1 (donor∶recipient), deposited onto nitrocellulose 0.45 µm filter discs (Millipore) and incubated for 5 h on agar plates at 37°C. Because the Plac promoter allows for constitutive low-level expression of the himar1C9 transposase, no induction was required. Bacteria from individual mating discs were recovered in 2 ml of 1× M9 salts by vortexing. A 100 µl aliquot was serially diluted and plated on selective agar plates to determine mutagenesis frequency. The remaining 1.9 ml of mating mixture was added to 20 ml of selective media and allowed to grow shaking at 37°C until an optical density at 600 nm (OD600) of ∼0.5 was reached. One ml aliquots from individual matings were then store at −80°C until used in selection screening. In the event a mutant library did not contain a satisfactory number of mutants, for example, <50,000 mutants, frozen axillary stocks were briefly thawed, combined and refrozen.
Bioinformatic analysis
Sequence alignment
Sequence reads were trimmed to remove transposon sequence and the remaining 12 bp sequences were aligned to the F11 genome consisting of 119 contigs (GenBank: AAJU00000000). The alignment was preformed with Bowtie [62], allowing no mismatches to the trimmed 12-mers. The number of times each 12-mer was observed (abundance) and the insertion position were tracked in a relational database. Because our strategy for determining candidate regions relied on the Wilcoxon signed-rank test, which is relatively resistant to spurious observations, there was no limit to how many times a given 12-mer was allowed to align to the genome.
Candidate identification
To distinguish loci within the F11 genome that were hypo or hyper-tolerant to transposon insertion, the average number of inserts observed within each annotated region, considering input pools 1, 2 and 3 alone, was calculated. A Z-test was performed on the population of 5,312 loci—those that fell below a standard score of −1 were considered hypo-tolerant and those that were above a standard score of +1 were considered hyper-tolerant. Standard scores were used here as an intuitive demarcation within the distribution of insertion tolerances and are not necessarily used to indicate any particular statistical significance. Of note, because we allowed reads to be aligned to the genome multiple times, we acknowledge that the specific values calculated for insertion tolerances for each annotated F11 region is overestimated. Indeed, we found that on average, reads could be aligned approximately twice to the F11 chromosome (distribution median = 2). This accounts for the elevated insertion rates reported in Figure 2 and Table S2, which are approximately two times greater than the inferred genome-wide insertion rate of 1 in 75 bp that is reported in the results section. Although this approach added a source of noise, it also allowed us to quantify and rank regions that would have otherwise been assigned a ‘0’ (i.e. putative essential genes). The design of future Tn-seq experiments should evaluate the pros and cons of such an approach.
To calculate changes in fitness in vivo due to transposon insertion, occurrence frequencies for insertion variants were first determined by normalizing insert abundance. The normalized occurrence rate of each insertion was obtained by dividing the number of times an insert was observed by the sum of insert counts for the insert positions that were detected in both the input and output pools of the replicate screen in question. Next, a Wilcoxon signed-rank test was performed on each F11 locus to compare the distribution of occurrence frequencies of inserts found within both input and respective output pools. Because this basic approach did not immediately account for inserts that were absent from output pools, which possibly indicates a strong negative selection event, we assigned the limit of detection (1/sequencing depth) to lost inserts that were present within a cognate input pool and within the top 1% of occurrence frequencies of all inserts that were lost. By incorporating these lost inserts prior to performing the Wilcoxon signed-rank test, we could more confidently account for inserts that likely represented a true negative selection signal while not considering inserts that were absent due to sampling error or passive loss. Regions having significantly altered frequency distributions (p<0.05) and at least 10 measured inserts were assigned to the in piscis gene set if the average occurrence frequency of inserts within the output samples was reduced by at least 2-fold, or assigned to the advantageous gene set if the average occurrence frequency of inserts within the output samples was greater than that of the input by at least 2-fold. Using these orthogonal vetting parameters we calculated that the false discovery rate (FDR), as defined by Benjamini and Hochberg, 1995, for the collection of observations with a maximal p value of 0.05 to be 0.096 and 0.094 for the in piscis and advantageous gene sets, respectively [63]. Figure S5 provides a diagrammatic summary of the bioinformatic pipeline used above to determine Tn-seq candidate genes and gene sets. Table S7, along with an online data viewer (http://pathogenomics.path.utah.edu/F11_TnSeq/), can be used to anonymously access and browse final metric-based data that were generated using these methods. Data analyses were done using custom software written in Python and the open source math tool SciPy.
KEGG-based functional analysis of gene sets
Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologies (KO) were assigned to genes within the F11 genome using the KEGG automatic annotation server (KAAS) [64], [65], [66]. Genes without KO assignments were considered to have a ‘hypothetical’ function and genes encoding RNAs (e.g. tRNAs, non-coding RNAs and ribosomal RNAs) were manually assembled into a custom ‘RNA’ orthology. To test for enrichment of a particular KEGG functional category within a gene set, Monte Carlo simulations were performed by sampling an equivalent number of genes from the F11 genome at random 10,000 times without replacement. The functional composition of observed gene sets was then compared to theoretical sample sets using a Z-test. If a particular functional category was significantly overrepresented compared to chance (p<0.05) it was considered enriched. Data analyses were done using custom software written in Python and the open source math tool SciPy.
Trait enrichment analysis (TEA)
165 bacterial proteomes were downloaded from the National Center for Biotechnology Information's (NCBI) microbial genome sequence collection and assembled into the TEA metaproteome database (TEA-MD) (Table S3). Each strain included in the TEA-MD was annotated for habitat, niche of isolation, phylum and phenotype. Annotations were retrieved manually by referring to original sequencing project resources. Homologues within the TEA-MD were identified for each protein-coding gene within the F11 genome using the Basic Local Alignment Search Tool BLASTp [41]. Homologue assignment was conferred if bi-directional alignments produced an E value<10−6 with similarity over 50% of query length. The resulting set of TEA-MD microbes harboring homologues of specific F11 proteins were then used to determine particular trait associations. For each F11 protein, 1,000 Monte Carlo simulations were performed by sampling an equivalent number of bacterial genomes from the TEA-MD without replacement. Genes that were found to have significant trait enrichments (p<0.05) were also required to have at least 5 TEA-MD homologues, 60% of which belonged to pathogen genomes, to be further considered and placed into trait groups. TEA was performed in parallel and independent of Tn-seq. Cross-referencing of several well-known bacterial virulence factors, in addition to broadly conserved genes involved in basic bacterial physiology, confirmed that our TEA approach could reliably distinguish pathogenic identity between these two gene classes (Table S8). Data analyses were done using custom software written in Python and the open source math tool SciPy.
Supporting Information
Zdroje
1. EwersC, JanssenT, WielerLH (2003) [Avian pathogenic Escherichia coli (APEC)]. Berliner und Munchener tierarztliche Wochenschrift 116 : 381–395.
2. ShpigelNY, ElazarS, RosenshineI (2008) Mammary pathogenic Escherichia coli. Current opinion in microbiology 11 : 60–65.
3. TanC, XuZ, ZhengH, LiuW, TangX, et al. (2011) Genome sequence of a porcine extraintestinal pathogenic Escherichia coli strain. Journal of bacteriology 193 : 5038.
4. CarvalloFR, DebroyC, BaezaE, HinckleyL, GilbertK, et al. (2010) Necrotizing pneumonia and pleuritis associated with extraintestinal pathogenic Escherichia coli in a tiger (Panthera tigris) cub. Journal of veterinary diagnostic investigation : official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc 22 : 136–140.
5. FoxmanB (2010) The epidemiology of urinary tract infection. Nature reviews Urology 7 : 653–660.
6. JohnsonJR, JohnstonB, ClabotsC, KuskowskiMA, CastanheiraM (2010) Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 51 : 286–294.
7. TouchonM, HoedeC, TenaillonO, BarbeV, BaeriswylS, et al. (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS genetics 5: e1000344.
8. WilesTJ, KulesusRR, MulveyMA (2008) Origins and virulence mechanisms of uropathogenic Escherichia coli. Experimental and molecular pathology 85 : 11–19.
9. MuzziA, MasignaniV, RappuoliR (2007) The pan-genome: towards a knowledge-based discovery of novel targets for vaccines and antibacterials. Drug discovery today 12 : 429–439.
10. WilesTJ, BowerJM, ReddMJ, MulveyMA (2009) Use of zebrafish to probe the divergent virulence potentials and toxin requirements of extraintestinal pathogenic Escherichia coli. PLoS pathogens 5: e1000697.
11. WilesTJ, NortonJP, SmithSN, LewisAJ, MobleyHL, et al. (2013) A phyletically rare gene promotes the niche-specific fitness of an E. coli pathogen during bacteremia. PLoS pathogens 9: e1003175.
12. van OpijnenT, CamilliA (2012) A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome research 22 : 2541–2551.
13. PerezJC, KumamotoCA, JohnsonAD (2013) Candida albicans Commensalism and Pathogenicity Are Intertwined Traits Directed by a Tightly Knit Transcriptional Regulatory Circuit. PLoS biology 11: e1001510.
14. van OpijnenT, BodiKL, CamilliA (2009) Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nature methods 6 : 767–772.
15. GoodmanAL, McNultyNP, ZhaoY, LeipD, MitraRD, et al. (2009) Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell host & microbe 6 : 279–289.
16. JoshiSM, PandeyAK, CapiteN, FortuneSM, RubinEJ, et al. (2006) Characterization of mycobacterial virulence genes through genetic interaction mapping. Proceedings of the National Academy of Sciences of the United States of America 103 : 11760–11765.
17. HenselM, SheaJE, GleesonC, JonesMD, DaltonE, et al. (1995) Simultaneous identification of bacterial virulence genes by negative selection. Science 269 : 400–403.
18. SaenzHL, DehioC (2005) Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Current opinion in microbiology 8 : 612–619.
19. HamerL, DeZwaanTM, Montenegro-ChamorroMV, FrankSA, HamerJE (2001) Recent advances in large-scale transposon mutagenesis. Current opinion in chemical biology 5 : 67–73.
20. LehouxDE, SanschagrinF, LevesqueRC (2001) Discovering essential and infection-related genes. Current opinion in microbiology 4 : 515–519.
21. HayesF (2003) Transposon-based strategies for microbial functional genomics and proteomics. Annual review of genetics 37 : 3–29.
22. TredeNS, LangenauDM, TraverD, LookAT, ZonLI (2004) The use of zebrafish to understand immunity. Immunity 20 : 367–379.
23. JaultC, PichonL, ChlubaJ (2004) Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Molecular immunology 40 : 759–771.
24. WangZ, ZhangS, WangG, AnY (2008) Complement activity in the egg cytosol of zebrafish Danio rerio: evidence for the defense role of maternal complement components. PloS one 3: e1463.
25. LiX, WangS, QiJ, EchtenkampSF, ChatterjeeR, et al. (2007) Zebrafish peptidoglycan recognition proteins are bactericidal amidases essential for defense against bacterial infections. Immunity 27 : 518–529.
26. LieschkeGJ, OatesAC, CrowhurstMO, WardAC, LaytonJE (2001) Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98 : 3087–3096.
27. LampeDJ, ChurchillME, RobertsonHM (1996) A purified mariner transposase is sufficient to mediate transposition in vitro. The EMBO journal 15 : 5470–5479.
28. SmithV, BotsteinD, BrownPO (1995) Genetic footprinting: a genomic strategy for determining a gene's function given its sequence. Proceedings of the National Academy of Sciences of the United States of America 92 : 6479–6483.
29. SmithV, ChouKN, LashkariD, BotsteinD, BrownPO (1996) Functional analysis of the genes of yeast chromosome V by genetic footprinting. Science 274 : 2069–2074.
30. GrantAJ, FarrisM, AlefounderP, WilliamsPH, WoodwardMJ, et al. (2003) Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Molecular microbiology 48 : 507–521.
31. IyodaS, HondaN, SaitohT, ShimutaK, TerajimaJ, et al. (2011) Coordinate control of the locus of enterocyte effacement and enterohemolysin genes by multiple common virulence regulators in enterohemorrhagic Escherichia coli. Infection and immunity 79 : 4628–4637.
32. NagyG, DobrindtU, SchneiderG, KhanAS, HackerJ, et al. (2002) Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infection and immunity 70 : 4406–4413.
33. BurnsSM, HullSI (1999) Loss of resistance to ingestion and phagocytic killing by O(−) and K(−) mutants of a uropathogenic Escherichia coli O75:K5 strain. Infection and immunity 67 : 3757–3762.
34. BucklesEL, WangX, LaneMC, LockatellCV, JohnsonDE, et al. (2009) Role of the K2 capsule in Escherichia coli urinary tract infection and serum resistance. The Journal of infectious diseases 199 : 1689–1697.
35. deLivronMA, RobinsonVL (2008) Salmonella enterica serovar Typhimurium BipA exhibits two distinct ribosome binding modes. Journal of bacteriology 190 : 5944–5952.
36. KissE, HuguetT, PoinsotV, BatutJ (2004) The typA gene is required for stress adaptation as well as for symbiosis of Sinorhizobium meliloti 1021 with certain Medicago truncatula lines. Molecular plant-microbe interactions : MPMI 17 : 235–244.
37. GaugerEJ, LeathamMP, Mercado-LuboR, LauxDC, ConwayT, et al. (2007) Role of motility and the flhDC Operon in Escherichia coli MG1655 colonization of the mouse intestine. Infection and immunity 75 : 3315–3324.
38. MorrisJJ, LenskiRE, ZinserER (2012) The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3: e00036–12.
39. FalkowS (2004) Molecular Koch's postulates applied to bacterial pathogenicity–a personal recollection 15 years later. Nature reviews Microbiology 2 : 67–72.
40. FalkowS (1988) Molecular Koch's postulates applied to microbial pathogenicity. Reviews of infectious diseases 10(Suppl 2): S274–276.
41. AltschulSF, MaddenTL, SchafferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research 25 : 3389–3402.
42. del CastilloI, Gonzalez-PastorJE, San MillanJL, MorenoF (1991) Nucleotide sequence of the Escherichia coli regulatory gene mprA and construction and characterization of mprA-deficient mutants. Journal of bacteriology 173 : 3924–3929.
43. LomovskayaO, LewisK, MatinA (1995) EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. Journal of bacteriology 177 : 2328–2334.
44. KelleyLA, SternbergMJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols 4 : 363–371.
45. ArnadottirH, Hvanndal I andresdottirV, BurrSE, FreyJ, et al. (2009) The AsaP1 peptidase of Aeromonas salmonicida subsp. achromogenes is a highly conserved deuterolysin metalloprotease (family M35) and a major virulence factor. Journal of bacteriology 191 : 403–410.
46. TadesseS, GraumannPL (2007) DprA/Smf protein localizes at the DNA uptake machinery in competent Bacillus subtilis cells. BMC microbiology 7 : 105.
47. KarudapuramS, ZhaoX, BarcakGJ (1995) DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. Journal of bacteriology 177 : 3235–3240.
48. SmeetsLC, BeckerSC, BarcakGJ, Vandenbroucke-GraulsCM, BitterW, et al. (2006) Functional characterization of the competence protein DprA/Smf in Escherichia coli. FEMS Microbiology Letters 263 : 223–228.
49. NortonJP, MulveyMA (2012) Toxin-Antitoxin Systems Are Important for Niche-Specific Colonization and Stress Resistance of Uropathogenic Escherichia coli. PLoS pathogens 8: e1002954.
50. SmithSN, HaganEC, LaneMC, MobleyHL (2010) Dissemination and systemic colonization of uropathogenic Escherichia coli in a murine model of bacteremia. mBio 1: e00262–10.
51. WelchRA, DellingerEP, MinshewB, FalkowS (1981) Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature 294 : 665–667.
52. PicardB, GarciaJS, GouriouS, DuriezP, BrahimiN, et al. (1999) The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infection and immunity 67 : 546–553.
53. PfennigPL, FlowerAM (2001) BipA is required for growth of Escherichia coi K12 at low temperature. Molecular genetics and genomics : MGG 266 : 313–317.
54. KrinE, DanchinA, SoutourinaO (2010) Decrypting the H-NS-dependent regulatory cascade of acid stress resistance in Escherichia coli. BMC microbiology 10 : 273.
55. RichardsGR, HerbertEE, ParkY, Goodrich-BlairH (2008) Xenorhabdus nematophila lrhA is necessary for motility, lipase activity, toxin expression and virulence in Manduca sexta insects. Journal of bacteriology 190 : 4870–4879.
56. HimpslSD, LockatellCV, HebelJR, JohnsonDE, MobleyHL (2008) Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. Journal of medical microbiology 57 : 1068–1078.
57. WhitfieldC (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annual review of biochemistry 75 : 39–68.
58. PricePA, JinJ, GoldmanWE (2012) Pulmonary infection by Yersinia pestis rapidly establishes a permissive environment for microbial proliferation. Proceedings of the National Academy of Sciences of the United States of America 109 : 3083–3088.
59. HaganEC, LloydAL, RaskoDA, FaerberGJ, MobleyHL (2010) Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS pathogens 6: e1001187.
60. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America 97 : 6640–6645.
61. MurphyKC, CampelloneKG (2003) Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC molecular biology 4 : 11.
62. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology 10: R25.
63. BenjaminiY, HochbergY (1995) Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society, Series B 57 : 289–300.
64. KanehisaM, GotoS (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research 28 : 27–30.
65. KanehisaM, GotoS, SatoY, FurumichiM, TanabeM (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids research 40: D109–114.
66. MoriyaY, ItohM, OkudaS, YoshizawaAC, KanehisaM (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic acids research 35: W182–185.
67. RakemanJL, BonifieldHR, MillerSI (1999) A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. Journal of bacteriology 181 : 3096–3104.
68. JakominM, ChessaD, BaumlerAJ, CasadesusJ (2008) Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA and HdfR. Journal of bacteriology 190 : 7406–7413.
69. KoM, ParkC (2000) H-NS-Dependent regulation of flagellar synthesis is mediated by a LysR family protein. Journal of bacteriology 182 : 4670–4672.
70. PavelkaMSJr, WrightLF, SilverRP (1991) Identification of two genes, kpsM and kpsT, in region 3 of the polysialic acid gene cluster of Escherichia coli K1. Journal of bacteriology 173 : 4603–4610.
71. JakobsenL, SpangholmDJ, PedersenK, JensenLB, EmborgHD, et al. (2010) Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. International journal of food microbiology 142 : 264–272.
72. RiggGP, BarrettB, RobertsIS (1998) The localization of KpsC, S and T and KfiA, C and D proteins involved in the biosynthesis of the Escherichia coli K5 capsular polysaccharide: evidence for a membrane-bound complex. Microbiology 144(Pt 10): 2905–2914.
73. BronnerD, SieberthV, PazzaniC, RobertsIS, BoulnoisGJ, et al. (1993) Expression of the capsular K5 polysaccharide of Escherichia coli: biochemical and electron microscopic analyses of mutants with defects in region 1 of the K5 gene cluster. Journal of bacteriology 175 : 5984–5992.
74. McNultyC, ThompsonJ, BarrettB, Lord L andersenC, et al. (2006) The cell surface expression of group 2 capsular polysaccharides in Escherichia coli: the role of KpsD, RhsA and a multi-protein complex at the pole of the cell. Molecular microbiology 59 : 907–922.
75. BachtiarBM, ColoePJ, FryBN (2007) Knockout mutagenesis of the kpsE gene of Campylobacter jejuni 81116 and its involvement in bacterium-host interactions. FEMS immunology and medical microbiology 49 : 149–154.
76. CieslewiczM, VimrE (1997) Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Molecular microbiology 26 : 237–249.
77. KalynychS, RuanX, ValvanoMA, CyglerM (2011) Structure-guided investigation of lipopolysaccharide O-antigen chain length regulators reveals regions critical for modal length control. Journal of bacteriology 193 : 3710–3721.
78. AmosMR, Sanchez-ContrerasM, JacksonRW, Munoz-BerbelX, CicheTA, et al. (2011) Influence of the Photorhabdus luminescens phosphomannose isomerase gene, manA, on mannose utilization, exopolysaccharide structure and biofilm formation. Applied and environmental microbiology 77 : 776–785.
79. OuyangZ, IsaacsonR (2006) Identification and characterization of a novel ABC iron transport system, fit, in Escherichia coli. Infection and immunity 74 : 6949–6956.
80. StojiljkovicI, BaumlerAJ, HeffronF (1995) Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. Journal of bacteriology 177 : 1357–1366.
81. BuanNR, SuhSJ, Escalante-SemerenaJC (2004) The eutT gene of Salmonella enterica Encodes an oxygen-labile, metal-containing ATP:corrinoid adenosyltransferase enzyme. Journal of bacteriology 186 : 5708–5714.
82. TurnerRJ, TaylorDE, WeinerJH (1997) Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrobial agents and chemotherapy 41 : 440–444.
83. NataroJP, SeriwatanaJ, FasanoA, ManevalDR, GuersLD, et al. (1995) Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infection and immunity 63 : 4721–4728.
84. SchwanWR (2009) Survival of uropathogenic Escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology 155 : 1832–1839.
85. WareD, JiangY, LinW, SwiatloE (2006) Involvement of potD in Streptococcus pneumoniae polyamine transport and pathogenesis. Infection and immunity 74 : 352–361.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



