-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
Human neutrophil antigen 2 (HNA-2) is coded by CD177 gene that involves in human myeloproliferative disorders. HNA-2 expression varies among humans and about 3–5% people lack HNA-2 expression. HNA-2 deficient people are susceptible to produce HNA-2 alloantibodies, which play a pathological role in various human diseases including transfusion-related acute lung injury, neonatal alloimmune neutropenia, autoimmune neutropenia, drug-induced immune neutropenia, and graft failure following marrow transplantation. The level of HNA-2 expression has also been identified as a prognostic biomarker for the gastric cancer. Although HNA-2 is among the most important clinical antigens, the underlying genetic mechanism of HNA-2 deficiency and expression variations has remained unknown. Here, we demonstrate that HNA-2 deficiency and expression variations are primarily caused by a novel CD177 genetic polymorphism that disrupts HNA-2 expression. The illumination of genetic mechanism for HNA-2 deficiency and expression variations will enable the development of effective HNA-2 genetic tests. Our findings will facilitate prognosis and diagnosis of HNA-2-related human disorders.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005255
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005255Summary
Human neutrophil antigen 2 (HNA-2) is coded by CD177 gene that involves in human myeloproliferative disorders. HNA-2 expression varies among humans and about 3–5% people lack HNA-2 expression. HNA-2 deficient people are susceptible to produce HNA-2 alloantibodies, which play a pathological role in various human diseases including transfusion-related acute lung injury, neonatal alloimmune neutropenia, autoimmune neutropenia, drug-induced immune neutropenia, and graft failure following marrow transplantation. The level of HNA-2 expression has also been identified as a prognostic biomarker for the gastric cancer. Although HNA-2 is among the most important clinical antigens, the underlying genetic mechanism of HNA-2 deficiency and expression variations has remained unknown. Here, we demonstrate that HNA-2 deficiency and expression variations are primarily caused by a novel CD177 genetic polymorphism that disrupts HNA-2 expression. The illumination of genetic mechanism for HNA-2 deficiency and expression variations will enable the development of effective HNA-2 genetic tests. Our findings will facilitate prognosis and diagnosis of HNA-2-related human disorders.
Introduction
Transfusion-related acute lung injury (TRALI) is associated with the transfusion of leukocyte alloantibodies from donors or associated with the presence of alloantibodies in recipients of blood [1,2]. Alloantibodies against human neutrophil alloantigenes (HNAs) are a very strong trigger for the development of TRALI [1,2]. Human neutrophil antigen 2 (HNA-2) alloantibodies have been linked to the induction of TRALI and various pulmonary reactions [3–6] while anti-HNA-3 alloantibodies are frequently implicated in severe and fatal TRALI [7]. Animal models have firmly established a pathological role for HNA-2 alloantibodies in TRALI [8,9]. Furthermore, HNA-2 alloantibodies have been implicated in multiple human disorders such as neonatal alloimmune neutropenia, autoimmune neutropenia, drug-induced immune neutropenia, and graft failure following marrow transplantation [10–13]. Accordingly, HNA-2 is among the most important clinical antigens.
HNA-2 is heterogeneously expressed on subpopulations of neutrophils and approximately 3–5% Americans do not express HNA-2 [14]. HNA-2 deficient subjects are predisposed to the production of HNA-2 alloantibodies when exposed to the HNA-2 antigen during blood transfusion, pregnancy, and bone marrow transplantation. HNA-2 is encoded by the CD177 gene that contains nine exons at Chromosome 19q13.31 region, where a CD177 pseudogene highly homologous to CD177 between exon 4 and 9 is also located (Fig 1A) [15–17]. The genetic studies of CD177 were significantly hampered by the presence of CD177 pseudogene [18,19]. HNA-2 is also known as PRV-1 as CD177 mRNA is over-expressed in polycythemia rubra vera patients [20]. CD177 has an open reading frame of 1311 nucleotides that encode 437 amino acids with a signal peptide of 21 residues. HNA-2 (or CD177) is expressed as a GPI-linked receptor with a mature peptide consisting of residue 22 to 408 [15,21]. HNA-2 plays important roles in neutrophil functions and myeloid cell proliferation. The interaction between HNA-2 and PECAM-1 facilitates neutrophil transendothelial migration [22,23]. In addition, HNA-2 is required for the attachment of proteinase 3 (PR3) to neutrophils [24–27], which plays a pivotal role in PR3-ANCA-mediated neutrophil activation [28]. CD177 mRNA levels are elevated in several conditions associated with increased neutrophil counts [14,29]. Furthermore, elevated levels of neutrophil CD177 mRNA are associated with increased neutrophil production and quantitation of neutrophil CD177 mRNA is a diagnostic tool for polycythemia vera [14]. Moreover, the level of HNA-2 expression has been identified as a prognostic biomarker for gastric cancer [30].
Fig. 1. CD177 gene structure and HNA-2 expression. 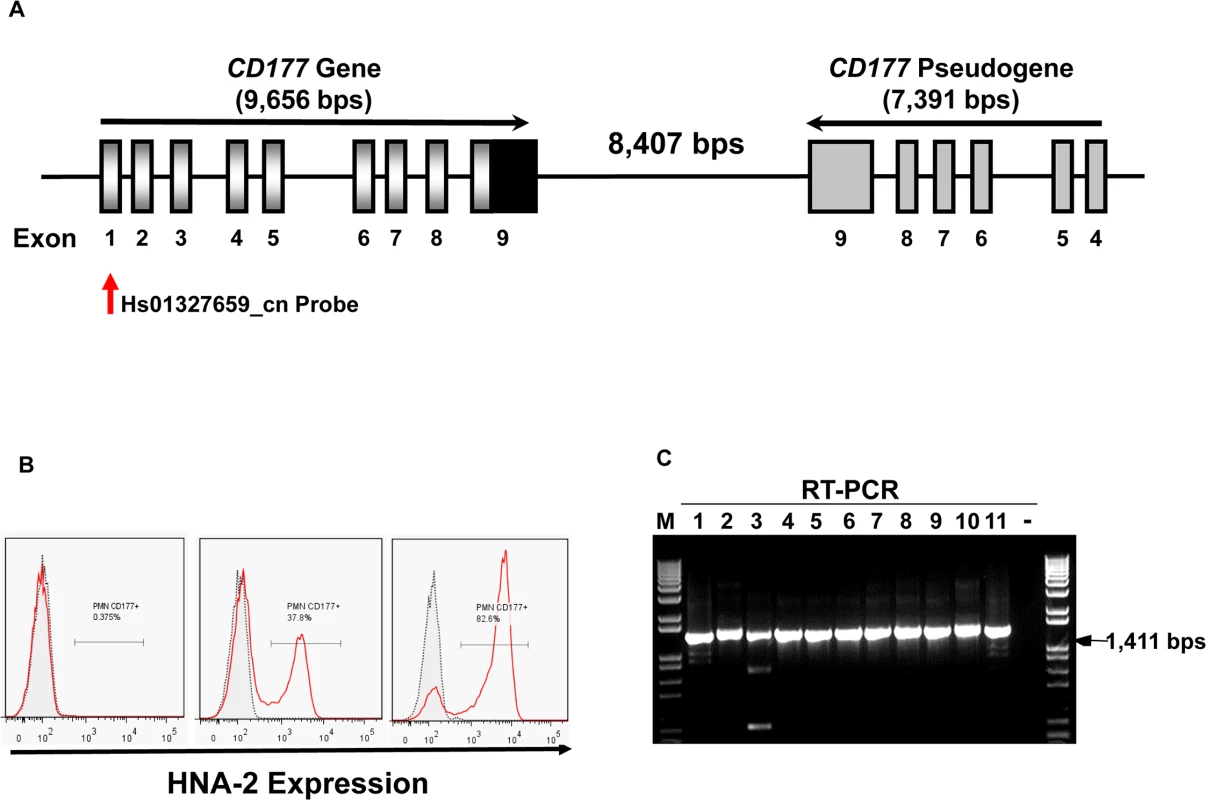
A). Structures of CD177 gene and pseudogene at chromosome 19q13.31 region. CD177 gene with nine exons and its pseudogene with six pseudo-exons are separated by 8,407 nucleotides. CD177 pseudogene is highly homologous to CD177 between exon 4 and 9. Vertical arrow indicates the CNV assay probe location. B). HNA-2 expression varies in different donors. Characteristic light-scatter properties were used to identify neutrophils in flow cytometry. Neutrophils from different donors vary in the percentages of HNA-2 positive cells. Right panel shows the absence of HNA-2 expression on neutrophils from a donor. C). Amplification of full-length CD177 cDNA from HNA-2 deficient donors. All HNA-2 deficiency donors expressed full-length CD177 mRNA the full-length CD177 cDNAs (1,411 bps) were detected in RT-PCR. The CD177 non-synonymous coding SNPs (cSNPs) were reported to associate with HNA-2 expression variations, however, the effect of those non-synonymous CD177 coding SNPs on HNA-2 expression was unknown [18,31,32]. CD177 mRNA splicing variants were found in two HNA-2 deficient donors but it remains inconclusive whether CD177 splicing abnormality was actually responsible for HNA-2 deficiency [33]. Therefore, the underlying genetic mechanism of HNA-2 deficiency has remained elusive since the observation of HNA-2 deficiency four decades ago [10]. Elucidation of the molecular genetics and basis of the HNA-2 deficiency is a prerequisite for the use of effective genetic tests in prognosis and diagnosis of HNA-2-related human diseases. In the current study, we demonstrated that a novel nonsense CD177 coding SNP 829A>T is the primary genetic determinant for HNA-2 deficiency and expression variations in humans.
Results
Copy number variations (CNVs) of CD177 gene
The percentages of neutrophils expressing HNA-2 were heterogeneous among normal healthy blood donors in flow cytometry analysis (Fig 1B). In 294 normal healthy blood donors, the percentage of HNA-2-positive neutrophils ranged from 0.0% to 97.8%. Among 294 blood donors, we have identified 11 donors (or 3.7%) deficient for HNA-2 and the percentage of HNA-2 deficient blood donors is consistent with those previously reported [6,10,34].
Copy number variations (CNVs) are the primary cause of human neutrophil antigen 1 (HNA-1 or FcγRIIIB) deficiency and expression variations [35–38]. To investigate whether CD177 CNVs are involved in HNA-2 deficiency, we determined CD177 CNVs using TaqMan CNV assay kit Hs01327659_cn with the probe targeting the unique CD177 exon 1 region (Fig 1A). Among 294 human subjects, 95.2% (280/294) of subjects were two-copy CD177 carriers and 4.8% (14/294) were three-copy CD177 carriers. No human subjects had CD177 gene deletions among 294 subjects. Notably, all 11 HNA-2 deficient donors identified in the flow cytometry analysis carried two copies of CD177 gene. In addition, those 11 HNA-2 deficient donors produced full-length CD177 mRNAs as demonstrated by RT-PCR (Fig 1C). Our data clearly demonstrated that CD177 gene deletion (or CNVs) and the lack of mRNA expression are not the cause of HNA-2 deficiency.
Detection of a novel nonsense CD177 coding SNP (cSNP)
We subsequently determined CD177 cDNA sequences of all 11 HNA-2 deficient donors along with 119 HNA-2 positive donors. In addition to CD177 coding SNPs (cSNPs) identified previously, we discovered five novel cSNPs (SNP 824G>C or rs17856827G>C, 828A>C or rs70950396A>C, 829A>T or rs70950396A>T, 832G>A, and 841A>G or rs201266439) (S1 Table), which form two haplotypes (Fig 2). Most importantly, the CD177 SNP 829A>T is a nonsense polymorphism that creates a translation stop codon at amino acid position 263 (Lysine → Stop codon change) in CD177 open reading frame. Consequently, those two haplotypes were designated as the open reading frame haplotype (or ORF allele: 824G/828A/829A/832G/841A) and the stop codon haplotype (or STP allele: 824C/828C/829T/832A/841G) (Fig 2). To determine the origin of the novel CD177 cSNP haplotype, we have also sequenced CD177 genomic DNA PCR products. Based on genomic DNA sequencing analysis, 72.1% (212/294) of donors were homozygous 829AA donors and the homozygous 829TT donors accounted for 3.1% (9/294) in our study population. The minor allele (829T) frequency is 15.5% (S2 Table). The distribution of SNP 829A>T genotypes was consistent with the Hardy-Weinberg equilibrium in 294 blood donors (χ2 = 0.76, P = 0.38) (S2 Table).
Fig. 2. Identification of novel CD177 coding SNPs and SNP haplotypes. 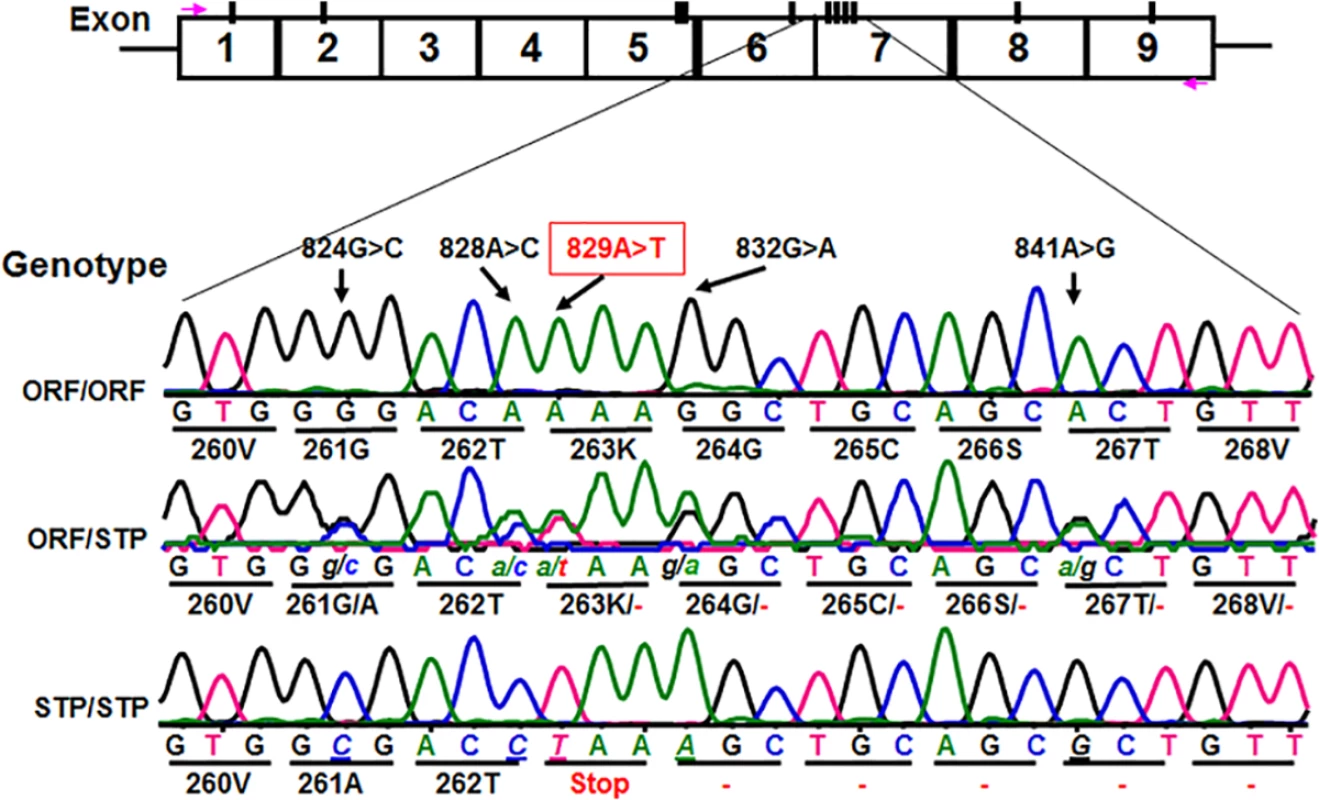
The CD177 cDNAs from 11 HNA-2 deficiency donors contain five novel cSNPs (824G>C, 828A>C, 829A>T, 832G>A, and 841A>G) that form a previously unidentified haplotype (824C/828C/829T/832A/841G). The CD177 cSNP 829A>T is a nonsense polymorphism that creates a translation stop codon at amino acid position 263 (Lysine → Stop codon change) in CD177 open reading frame. The upper row of tracers is the haplotype containing 829A allele (designated as ORF or open reading frame allele) while the lower row tracer shows the haplotype containing 829T allele (designated as STP or stop codon allele). The middle tracer was from a heterozygous donor. Association of the CD177 SNP 829A>T genotypes with HNA-2 deficiency and expression variations
To examine whether the CD177 SNP 829A>T affects HNA-2 expression, the donor genotypes and HNA-2 expressions were statistically analyzed. As shown in Fig 3A, all nine 829TT homozygous donors were negative for HNA-2 expression in flow cytometry analysis. In addition, the percentages of HNA-2 positive neutrophils from 73 heterozygous donors (829AT) were significantly lower than those from 212 homozygous 829AA donors (P < 0.0001). Western blot analyses also confirmed the absence of HNA-2 protein in 829TT homozygous donors and significantly less HNA-2 protein being expressed in the 829AT donors when compared to the 829AA homozygous donors (Fig 3B). Our data strongly support the notion that the SNP 829A>T allele is a crucial determinant for HNA-2 deficiency and expression variations. To verify our findings, we recruited an independent cohort containing 102 blood donors, among whom nine HNA-2 deficient donors were identified (S1 Fig). Similar to those of the first cohort, all nine HNA-2 deficient donors in the replication cohort were SNP 829TT homozygotes as demonstrated by sequencing analysis of genomic DNA and cDNA (S2 Fig). Again, the SNP 829A>T genotypes were significantly associated with the percentages of HNA-2 positive neutrophils (S1 Fig) and the HNA-2 protein expression (S3 Fig). Our data confirmed that the SNP 829A>T is a crucial genetic determinant for HNA-2 deficiency and expression variations.
Fig. 3. Association of CD177 SNP 829A>T with HNA-2 expression. 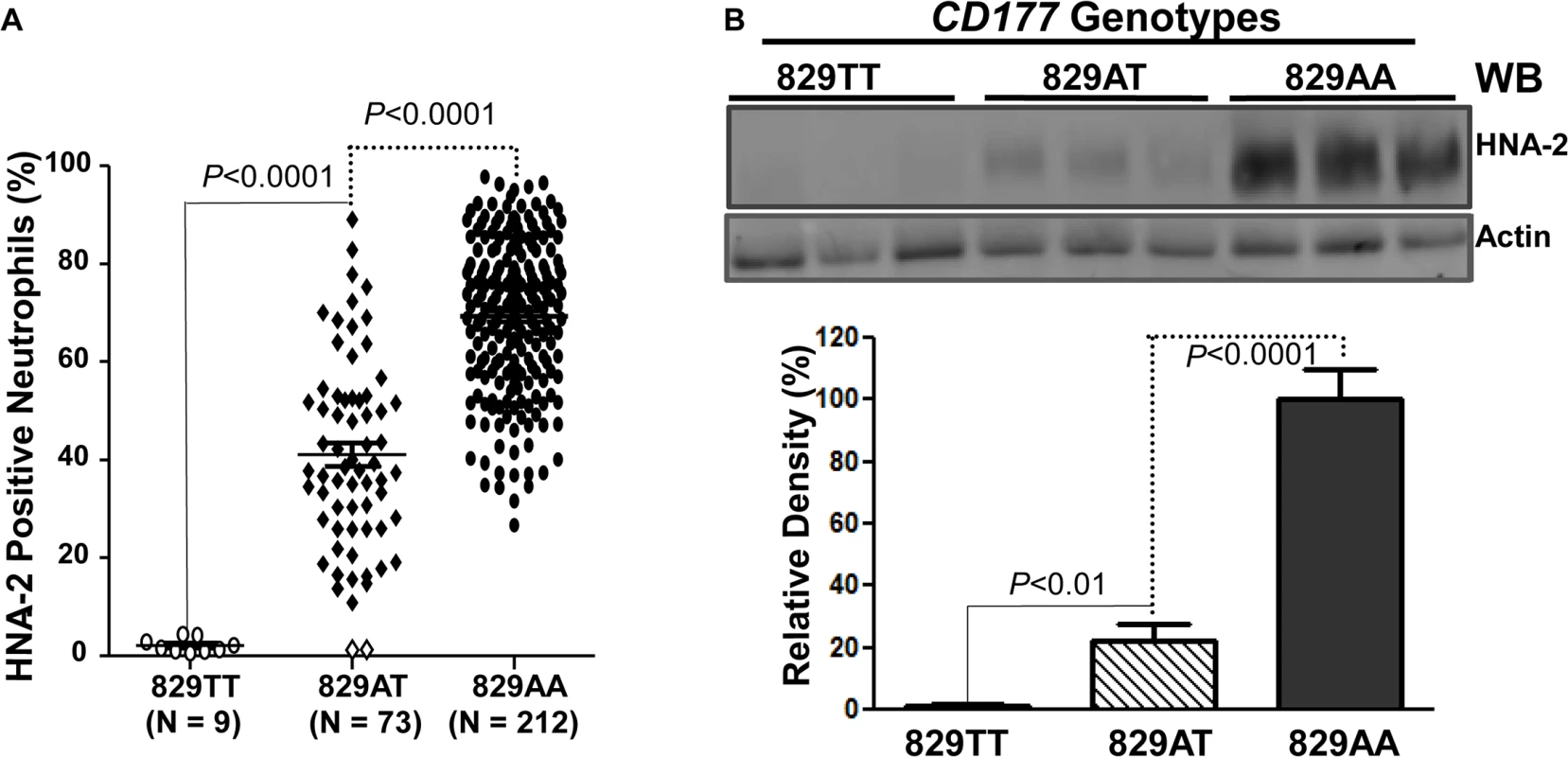
A). CD177 genotypes were determined with genomic DNA sequence analysis. HNA-2 expression was examined with flow cytometry analysis. All 829TT homozygous donors (829TT, N = 9) were negative for HNA-2. The percentages of HNA-2 positive neutrophils from heterozygous donors (829AT, N = 73) were significantly (P < 0.0001) lower than those from 829AA homozygous donors (829AA, N = 212). Two heterozygous 829AT donors indicated by the empty diamonds in the middle column were also negative for HNA-2 expression. B). HNA-2 expression in whole blood leukocytes was assayed in Western blot analyses. Representative blot image is shown (Top panel). No HNA-2 protein could be detected in all 829TT homozygous donors (829TT, N = 9) while heterozygous donors (829AT, N = 9) express significantly less HNA-2 as compared to the 829A homozygous donors (829AA, N = 9) (Lower panel). Identification of a rare mutation in HNA-2 deficiency donors
Similar to all nine homozygous 829TT donors, two 829AT heterozygous donors were also negative for HNA-2 expression (Fig 3A, empty diamonds in the middle column). Analysis of their CD177 cDNA sequences revealed that both HNA-2 deficient donors who were heterozygous for SNP 829A>T also had a heterozygous deletion of the guanidine nucleotide at nucleotide 997 (997G deletion). To determine haplotypes of the SNP 829A>T and the 997G deletion, we cloned and sequenced cDNA from those two HNA-2 deficient donors. As shown in Fig 4A, two species of CD177 mRNAs were found in those two donors. The SNP 829T (STP) allele is in the linkage disequilibrium with the wild-type CD177 997G allele while the 829A (ORF) allele carries the 997G deletion. Genomic DNA sequence analysis confirmed that the guanidine nucleotide deletion occurs at genomic level (Fig 4B). Our data indicate that the presence of the 829T allele in combination with the deletion mutation at nucleotide 997 on another chromosome could also lead to the HNA-2 expression deficiency in an individual. However, we found that only two out of 294 blood donors carried the 997G deletion mutation at one chromosome with genomic sequencing analysis. Therefore, the allele frequency of the 997G deletion mutation is estimated to be 0.0034 in the study population. In those two 829AT heterozygous donors, the 997G deletion allele was coincidentally paired with the 829T allele, which facilitated the discovery of the rare 997G deletion mutation in the study. We failed to identify any donors with the CD177 997G deletion among 102 additional blood donors of the replication cohort, confirming that the 997G deletion is a rare mutation in the population.
Fig. 4. Detection of a rare CD177 nucleotide deletion mutation. 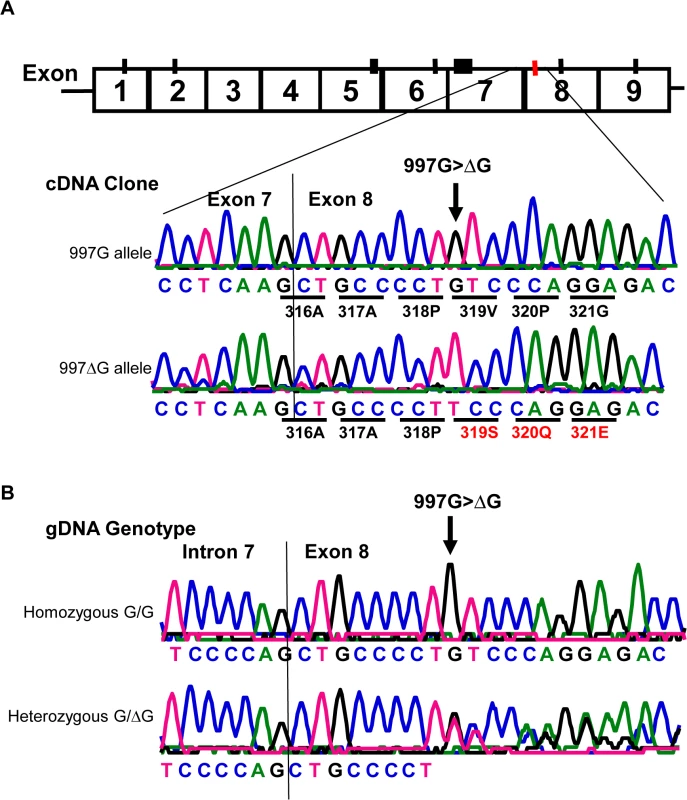
A). Sequencing analysis of cDNA clones from two HNA-2 deficient donors who were heterozygous for the SNP 829A>T. The upper tracer is the wild-type (997G allele) CD177 cDNA clone sequence. The wild-type 997G allele is in linkage disequilibrium with SNP 829T (or STP) allele as demonstrated by the sequence of cDNA clones from two HNA-2 deficient donors. The lower tracer is the sequence of CD177 cDNA clone with 997G deletion (designated as 997ΔG) that is in linkage disequilibrium with SNP 829A allele (or ORF) in CD177 cDNA clones. Those two ORF/STP heterozygous donors manifested as HNA-2 deficient phenotype had the combination of one chromosome carrying 997G deletion (997ΔG) and the other carrying the 829T (STP) allele. B). Genomic DNA sequence analysis confirmed that the guanidine nucleotide deletion occurs at genomic level. The upper tracer shows the wild-type genomic DNA sequence with the 997G. The lower tracer is the CD177 genomic sequence of a heterozygous donor with one chromosome containing 997G deletion and the other containing the wild-type. Effect of CD177 cSNPs on HNA-2 expression and alloantibody binding
Although the genotypes of CD177 non-synonymous SNPs were reportedly associated with HNA-2 expression variations in several genetic analyses [18,31,32], it is unknown whether those CD177 cSPNs directly affect HNA-2 expression. To examine the effect of non-synonymous CD177 cSNPs on HNA-2 expression and on the binding to HNA-2 alloantibodies, we cloned the full-length CD177 cDNA variants containing common non-conservative cSNPs (SNP 134A>T, 652A>G, 656G>T, and 1084G>A) within the coding region for HNA-2 mature peptide (aa22-408). As shown in Fig 5, there were no significant differences in the expression of HNA-2 (Fig 5A) or in the binding to HNA-2 alloantibodies (Fig 5B) among four CD177 variants consisting of four non-conservative amino acid substitutions (His31Leu, Asn204Asp, Arg205Met, and Ala348Thr). Our data support the notion that non-synonymous CD177 cSNPs do not have a direct role in the HNA-2 alloantibody production and expression variations. However, cells transfected with CD177 variants of either STP haplotype (CD177-STP) or 997G deletion (CD177-997ΔG) failed to express HNA-2 on cell surface (Fig 5C) and had no reactivity with HNA-2 alloantibodies (Fig 5D). Our data confirmed that either STP allele or 997G deletion mutation will lead to the HNA-2 expression deficiency. To further confirm that the nonsense SNP 829A>T in the STP haplotype is the key factor for HNA-2 expression, we generated a CD177 expression construct carrying the sole change at SNP 829A>T position. The T substitution at nucleotide position 829 alone led to the absence of HNA-2 expression in transfection experiments (S4 Fig), confirming that the SNP 829A>T is the sole determinant for HNA-2/CD177 expression in the STP haplotype.
Fig. 5. Effect of CD177 SNPs or mutation on HNA-2 expression and HNA-2 alloantibody binding. 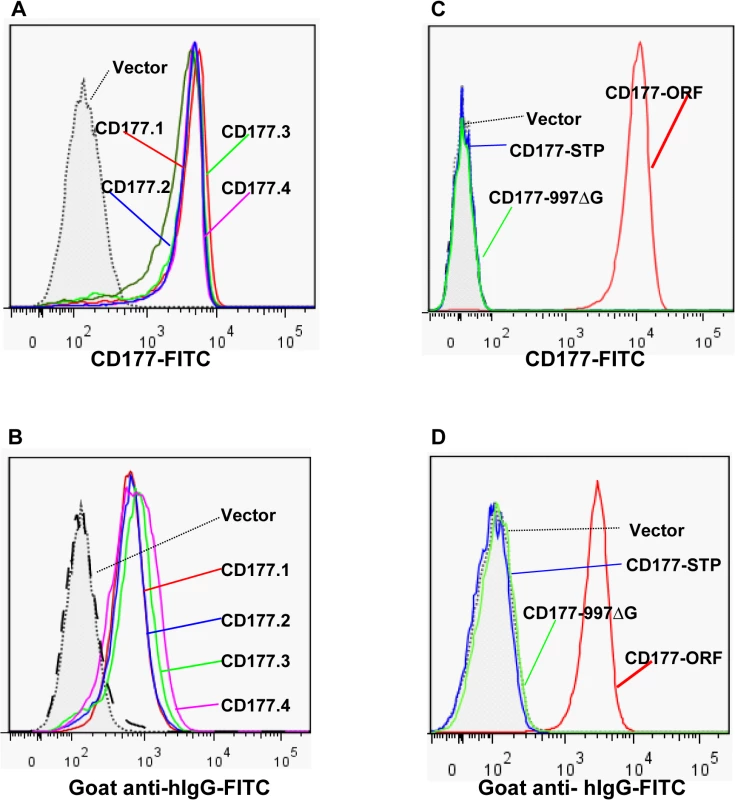
A). Expression of common CD177 SNP haplotypes or variants (CD177.1: 31L-204N-205R-348A. CD177.2: 31H-204N-205R-348A. CD177.3: 31L-204D-205M-348A. CD177.4: 31L-204N-205R-348T) on 293 cells. No significant differences of HNA-2 expressions were observed among four CD177 haplotypes consisting of four common non-synonymous cSNPs (SNP 134A>T, 652A>G, 656G>T, and 1084G>A), which encode the amino acid substitutions for 31His>Leu, 204Asn>Asp, 205Arg>Met, and 348Ala>Thr. B). No significant differences were observed in the HNA-2 alloantibody binding to four common CD177 variants (CD177.1: 31L-204N-205R-348A. CD177.2: 31H-204N-205R-348A. CD177.3: 31L-204D-205M-348A. CD177.4: 31L-204N-205R-348T). C). HNA-2 expression was absent in cells stably transfected with the CD177 expression constructs of the SNP 829T allele (CD177-STP) or 997G deletion mutation (CD177-997ΔG). The SNP 829A allele (or CD177-ORF) serves as the positive control for HNA-2 expression. D). Cells stably transfected with the expression constructs containing either SNP 829T (CD177-STP) allele or 997G deletion (CD177-997ΔG) allele failed to react with HNA-2 alloantibodies. The SNP 829A allele (or CD177-ORF) serves as the positive control for HNA-2/CD177 expression. All experiments were repeated at least three times. Discussion
The phenomenon of HNA-2 deficiency was observed more than four decades ago [10], however, the underlying genetic mechanism of HNA-2 deficiency has remained unknown. In the current study, we identified five common CD177 cSNPs (SNP 824G>C, 828A>C, 829A>T, 832G>A, and 841A>G, minor allele frequency = 0.155) in complete linkage disequilibrium. Among five SNPs, the nonsense SNP 829A>T changes the amino acid codon #263 from lysine to a stop codon, which leads to the HNA-2 expression deficiency. Neutrophils from all 829T allele homozygous donors failed to express HNA-2. In addition, the percentages of HNA-2 positive neutrophils from the SNP 829A>T heterozygous donors (ORF/STP) were significantly lower than those from ORF homozygous donors. In vitro, the T substitution at the nucleotide position 829 alone led to HNA-2 expression deficiency in transfection experiments, confirming that the SNP 829A>T is the sole determinant for HNA-2 expression in the STP haplotype. Our study was the first to unravel the genetic mechanism for HNA-2 deficiency, which plays critical roles in human immunological diseases including TRALI, immune neutropenia, and bone marrow graft failure [3–6,10–13]. The delineation of the HNA-2 genetics undoubtedly will enable the development of effective genetic and clinical diagnosis tools in human medicine.
Intriguingly, similar to neutrophils from all homozygous donors of 829T allele, neutrophils from two 829AT heterozygous donors were also negative for HNA-2 expression (Fig 3A). Analysis of cDNA sequences of those two 829AT heterozygous donors deficient for HNA-2 revealed that the 829A allele (or ORF allele) in those two donors had a guanidine deletion at the nucleotide position 997, which leads to the CD177 reading-frame shift starting from the amino acid codon #319 (Fig 4) and the creation of a stop codon at the amino acid codon #342. The CD177 997G deletion also leads to the early termination of HNA-2 peptide translation, similar to the consequence of the 829T allele. Furthermore, the CD177 variant carrying the nucleotide 997G deletion failed to express HNA-2 on cell surface in the transfection experiments (Fig 5C), confirming the contribution of the 997G deletion mutation to HNA-2 deficiency in those two specific individuals. The CD177 nucleotide 997G deletion mutation was extremely rare (mutant allele frequency = 0.0034) and was absent in the replication cohort of 102 donors. The coincidental appearance of the 997G deletion allele and the 829T allele in the HNA-2 deficient donors facilitated the discovery of the rare 997G deletion mutation in the study. Therefore, at the presence of 829T allele, the rare CD177 997G deletion may also contribute to HNA-2 deficiency. However, the 997G deletion mutation with the allele frequency of 0.0034 will have much less impact on overall HNA-2 deficiency as compared to the SNP 829A>T (the 829T allele frequency was 0.155, S2 Table).
Previous genetic studies suggested that the CD177 non-synonymous SNPs might affect HNA-2 expression [18,31,32], however, the effect of those CD177 cSNPs on HNA-2 expression is unclear. In the current study, we carried out transfection experiments to examine whether common non-conservative cSNPs (SNP 134A>T, 652A>G, 656G>T, and 1084G>A) within the HNA-2 mature peptide (aa22-408) affect the HNA-2 expression and the binding of HNA-2 alloantibodies. We found that the expression of HNA-2 and the binding to HNA-2 alloantibodies were not significantly different among those natural CD177 variants containing non-conservative amino acid substitutions (His31Leu, Asn204Asp, Arg205Met, and Ala348Thr) (Fig 5A and 5B). The expression of HNA-2 in normal neutrophils is also affected by methylations of CD177 promoter and the CD177 SNP 42G>C (rs45441892) at the third codon (Pro3Ala) of the HNA-2 signal peptide was associated with methylation levels of CD177 promoter [39]. However, we found no association between the SNP 42G>C genotypes and the percentages of HNA-2 positive neutrophils in our study (ANOVA, P = 0.1209, S5 Fig). Taken together, those non-synonymous CD177 cSNPs do not seem to have a significant effect on HNA-2 deficiency and expression.
The CD177 mRNA splicing abnormality was previously suggested to be the cause of HNA-2 deficiency as alternatively spliced CD177 mRNA species were detected in two HNA-2 deficient donors [33]. However, no further evidence was provided to support the alternative splicing hypothesis of HNA-2 deficiency in the report [33]. Alternative mRNA splicing is a physiological process and is an essential mechanism to produce different products from a single human gene [40–42]. It seems unlikely that HNA-2 deficient subjects have an abnormal mRNA splicing machinery as HNA-2 deficient donors appear healthy [6]. We have detected full-length CD177 mRNAs in all 11 HNA-2 deficient donors in the main study (Fig 1C) and in all nine HNA-2 deficient donors from the replication study. The combination of the alternative spliced CD177 mRNA isoforms and the regular CD177 mRNA isoform occurred only in two out of nine SNP 829TT homozygous donors in our replication cohort (S2 Fig). Our data refute the notion that the alternative splicing is a major cause of HNA-2 deficiency.
Although gene deletions or copy number variations (CNVs) are the primary cause for HNA-1 (or FcγRIIIB) deficiency [35–38], we did not find any CD177 gene deletion in our blood donors. We found that all HNA-2 deficient donors expressed full-length CD177 mRNAs. We also found that the SNP 829T allele was in complete linkage disequilibrium with SNP 134A, 156G, 593G, 652A, 656G, 671C, 782C, 793C, 824C, 828C, 832A, 841G, 1084G, and 1333G. Our data clearly demonstrated that the gene deletion or the lack of mRNA expression is not responsible for HNA-2 deficiency, in contrast to the HNA-1 deficiency. Interestingly, we found that all heterozygous donors of the SNP 829A>T determined by genomic DNA analysis primarily produced the SNP 829A allele (or ORF allele) mRNA based on their cDNA sequences. The nonsense SNP 829T allele tracer peak barely above the background was typically considered as sequence noise in the cDNA sequence analysis for heterozygous donors. This observation suggests that the CD177 mRNAs containing the nonsense 829T allele are much less stable than the CD177 mRNAs containing the common 829A allele within the same donor. This may explain the observation of associations between expression variations and certain CD177 cSNPs and the inability to discover the SNP 829A>T using the cDNA sequencing strategy in previous studies [31–33].
After transcription, the CD177 mRNA of the nonsense 829T allele may be quickly degraded by the mechanism of nonsense-mediated mRNA decay [43], which will lead to the low abundance of CD177 829T allele mRNA and the dominance of CD177 829A allele mRNA in the heterozygous individual. The nonsense-mediated mRNA decay mechanism per se may contribute to the CD177 mRNA expression deficiency in humans with different diseases, which may explain that the partial HNA-2 peptide was undetectable from those HNA-2 deficient donors in a previous study [44] and in the current study using multiple anti N-terminus of HNA-2 mAbs and HNA-alloantibodies (S6 Fig). Therefore, HNA-2 alloantibodies likely target the whole mature peptide of CD177 in HNA-2 deficient subjects. In heterozygous donors for the SNP 829A>T, only the 829A allele is able to express HNA-2. CD177 promoter DNA methylation regulates HNA-2 expression under physiologic conditions [39]. Non-selective methylation on the 829A allele alone is sufficient to effectively abrogate the HNA-2 expression in a specific cell during granulopoiesis, which may explain that the percentages of HNA-2 positive granulocytes were significantly lower in the 829A>T heterozygous donors than those in the 829A (or ORF) allele homozygous donors (Figs 3 and S1). Therefore, our data strongly support the concept that the SNP 829A>T is also a primary genetic factor for HNA-2 expression variations in humans.
As an important biomarker, HNA-2 (CD177) is over-expressed in neutrophils from patients with myeloproliferative disorders including polycythemia vera, essential thromobocythemia, idiopathic myelofibrocythemia, and hypereosinophilic syndrome [6,14]. HNA-2 was an indicator of increased erythropoietic activity in thalassemia syndromes as HNA-2 expression was significantly elevated in β-thalassemia patients compared to healthy controls [45]. HNA-2 overexpression may also have a direct role in the pathogenesis of myeloproliferative disorders as HNA-2 enhances cell proliferation in vitro [46,47]. Not surprisingly, the low percentage of HNA-2 positive neutrophil is significantly associated with myelodysplastic syndrome and chronic myelogenous leukemia [48,49], suggesting that the reduced levels of membrane-bound HNA-2 may decrease the proliferation and differentiation potentials of myeloid cells. It is possible that the selection pressure to limit the spread of myeloproliferative disorders during evolution may be an important factor in maintaining the CD177 nonsense polymorphism in humans. Therefore, the CD177 SNP 829A>T may be an important genetic risk factor for various myeloproliferative disorders. Approximately 3% of Caucasians, 5% of African Americans, and 1–11% of Japanese are HNA-2 deficient [14]. In the current study, we found that between 3.7% (main cohort) and 8.8% (replication cohort) blood donors (>98% of them were Caucasians from the State of Minnesota) were HNA-2 deficient. Our data indicate that percentages of HNA-2 deficient humans may vary in different regions and be affected by sample sizes.
In summary, the elucidation of the molecular mechanism of HNA-2 deficiency and expression variations fills the critical knowledge gap in the genetics of HNA-2 antigen system. Our findings will enable the development of reliable genetic assays for HNA-2 system and will facilitate the diagnosis and prognosis of HNA-2-associated human disorders.
Materials and Methods
Ethics statement
The human study was approved by the Institutional Review Board for Human Use at the University of Minnesota with Study #1301M26461. Memorial Blood Centers (737 Pelham Boulevard, St. Paul, Minnesota 55114) provided healthy donor blood samples without identifications for research purpose as a service and no consent form was provided per the Memorial Blood Centers policy.
Study subjects
Healthy American blood donors were recruited at the Memorial Blood Center in St. Paul, Minnesota. The age of healthy blood donors ranged from 19 to 84 years-old and >98% of donors in the study were self-declared Caucasians living in the State of Minnesota.
Evaluation of HNA-2 (CD177) expressions on neutrophils
The expression of HNA-2 and the percentage of HNA-2 positive neutrophils were determined with flow cytometry analysis. Leukocytes stained with either FITC-conjugated anti-CD177 mAb (MEM-166, mIgG1, Thermo Scientific) or mIgG1-FITC isotype control were analyzed on a FACS Canto flow cytometer (BD Biosciences). The FlowJo software (Tree Star Inc.) was used to evaluate flow cytometry data. Characteristic light-scatter properties were used to identify neutrophils in flow cytometry. Using the same criteria as in the literature [31], donors had less than 5% of granulocytes positive for MEM-166 staining in flow cytometry analysis were called as HNA-2 deficient.
Western blot analysis of HNA-2 protein
Peripheral blood leukocytes (2 × 107 cells) were lysed in PBS containing 1% NP-40 and 1× protease inhibitor cocktail (Roche, Indianapolis, ID) for 1 hr on ice. The total proteins (50 μg) from each donor were used for Western blotting analysis under non-reducing condition with mouse anti-CD177 mAbs and rabbit anti-actin mAb (LI-COR Biosciences, Lincoln, NE). IRDye 800CW-labeled goat anti-mouse and IRDye 600-labeled goat anti-rabbit antibodies were used for imaging analysis with the instrument software on an Odyssey Infrared Imager according to vendor’s instructions (LI-COR Biosciences).
Nucleic acid isolation
Human genomic DNA was isolated from EDTA anti-coagulated peripheral blood using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN) by following the vendor’s instruction. Total RNA was purified from peripheral blood leukocytes using TRIzol total RNA isolation reagent (Invitrogen, Carlsbad, CA).
Determination of CD177 CNVs
The CNVs of CD177 gene were determined using TaqMan Copy Number Assay kit (the probe location of the assay ID Hs01327659_cn is shown in Fig 1A) (Applied Biosystems, Foster City, CA) and RNase P reference assay (Applied Biosystems, Part# 4403326). Duplex quantitative real-time PCR reactions were carried out on an Applied Biosystems 7500 Real-Time PCR System according to the manufacturer’s instructions. All samples were tested in duplicates, and fluorescence signals were normalized to ROX. TaqMan assay quantitative PCR amplification curves were analyzed using 7500 Software on a plate by plate basis, and the CN was assigned from the raw Cq values using CopyCaller software (version 2.0; Applied Biosystems).
RT-PCR and cDNA sequencing
Five μg of total RNA was used for cDNA synthesis with the SuperScript Preamplification System (Invitrogen). The 1411-bp cDNA fragment covering the entire CD177 coding region was amplified with RT-PCR using the sense primer (5’-CTGAAAAAGCAGAAAGAGATTACCAGCCACAG-3’) and anti-sense primer (5’-GTCCAAGGCCATTAGGTTATGAGGTCAGA-3’). The PCR reaction was performed with 2 μl of cDNA, 200 nM of each primer, 200 μM of dNTPs, 2.0 mM of MgSO4, and 1 U of Platinum Taq DNA polymerase High Fidelity (Invitrogen) in a 25 μl reaction volume. Platinum Taq High Fidelity DNA polymerase was used as it allows the amplification of complex cDNA or DNA templates with high accuracy and yield. The ABI Veriti 96-well Thermal Cycler was used for the PCR reaction starting with 94°C for 3 min, 35 cycles of denaturing at 94°C for 30 s, annealing at 56°C for 45 s, extension at 68°C for 1 min and 30 s with a final extension at 72°C for 7 min. All the PCR products, treated with ExoSAP-IT (Affymetrix, Santa Clara, CA), were assessed by direct Sanger sequencing on an ABI 3730xl DNA Analyzer with BigDye v3.1 Sequencing kit (Applied Biosystems). CD177 cDNA was also directly cloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA). Multiple clones containing CD177 cDNA were selected and subsequently sequenced to confirm CD177 SNPs. Two sense primers and two antisense primers were used to sequence the full-length CD177 cDNA coding region (sequencing primers are listed in S3 Table). The electropherogram data, aligned by the DNASTAR software (DNAStar, Madison, WI) were used for the identification of gene polymorphisms.
Genomic DNA sequence analysis of CD177 gene
Since CD177 and its pseudogene contain a highly homologous region between exon 4 and 9 (Fig 1A) [16,17], we used the long-template PCR strategy to obtain the CD177-specific products for sequence analyses. Long-template PCR was carried out to amplify the CD177 genomic DNA containing all 9 exons using the sense primer (5’-CTGAAAAAGCAGAAAGAGATTACCAGCCACAG-3’) and antisense primer (5’-GTCCAAGGCCATTAGGTTATGAGGTCAGA-3’). The PCR reaction was performed with 200 ng DNA, 200 nM of each primer, 200 μM of dNTPs, 2.0 mM of MgSO4, and 2 U of Platinum Taq DNA polymerase High Fidelity (Invitrogen) in a 25 μl reaction volume. The ABI Veriti 96-well Thermal Cycler was used for the PCR reaction starting with 95°C for 3 min; 10 cycles of denaturing at 94°C for 30 s, annealing at 64°C for 30 s, extension at 68°C for 8 min and 30 s; 30 cycles of denaturing at 94°C for 30 s, annealing at 54°C for 30 s, extension at 68°C for 8 min and 30 s; with a final extension at 68°C for 5 min. The CD177 DNA fragment (8728 base pairs) was sequenced with a primer (5’-TCTTTGCCCCACACTAAACA-3’) on an ABI 3730xl DNA Analyzer with BigDye v3.1 Sequencing kit.
Generation of HNA-2 expression constructs
The human HNA-2 expression constructs were generated by cloning Hind III/Xba I-flanked RT-PCR products containing full-length CD177 coding region (nucleotide position 25 to 1419, GenBank accession number: NM_020406.2) into the eukaryotic expression vector pcDNA3 (Gibco BRL). The Hind III/Xba I-flanked CD177 cDNA was amplified from the synthesized cDNA of a blood donor using the upper primer 5’-CCCAAGCTTACCAGCCACAGACGGGTCATGAG-3’ and the lower primer 5’-TGCTCTAGAGAGGTCAGAGGGAGGTTGAGTGTG-3’. The changes at nucleotide position 134, 652, 656, 824, 828, 829, 832, 841, 997, and 1084 were generated using QuikChange Site-Directed mutagenesis kit (Stratagene, La Jolla, CA) and primer sets listed in the S3 Table.
Generation of cell lines expressing HNA-2
The 293 cells (human embryonic kidney cell line) from ATCC (ATCC#CRL-1573, Manassas, VA) were maintained in the DMEM medium supplemented with 10% fetal calf serum and L-glutamine (2 mM) in 5% CO2. Transfection reactions were carried out in the 100 mm cell culture dishes with the plasmid DNA (20 μg) purified with OMEGA Plasmid Maxi Kit (Omega Bio-Tek, Norcross, GA) and 40 μl of Lipofectamine 2000 reagent (Invitrogen). Transfected cells were cultured in DMEM medium supplemented with 10% fetal calf serum for two days before harvesting the cells for HNA-2 expression or the selection of stable cell lines with the supplement of G418 (final concentration: 1 mg/ml). The polyclonal cells surviving the G418 selection were sorted with Stemcelll EasySep Cell Sorter for equivalent HNA-2 expression. The expression of HNA-2 on the transfected 293 cell lines was determined with FITC-conjugated anti-CD177 mAb as described previously. In addition, five defined HNA-2 alloantibodies from the American Red Cross Neutrophil Serology Laboratory were used to evaluate the binding of HNA-2 to the cell lines expressing CD177 variants.
Statistical analysis
The ANOVA and the nonparametric t-test (Mann-Whitney test) were used to determine whether HNA-2 positive cell population sizes and the HNA-2 deficiency are statistically associated with the nonsense CD177 cSNPs. The χ2 test was used to determine whether the observed genotype frequencies are consistent with Hardy-Weinberg equilibrium.
Supporting Information
Zdroje
1. Fung YL, Silliman CC (2009) The role of neutrophils in the pathogenesis of transfusion-related acute lung injury. Transfus Med Rev 23 : 266–283. doi: 10.1016/j.tmrv.2009.06.001 19765516
2. Middelburg RA, van Stein D, Briet E, van der Bom JG (2008) The role of donor antibodies in the pathogenesis of transfusion-related acute lung injury: a systematic review. Transfusion 48 : 2167–2176. doi: 10.1111/j.1537-2995.2008.01810.x 18564387
3. Bux J, Becker F, Seeger W, Kilpatrick D, Chapman J, et al. (1996) Transfusion-related acute lung injury due to HLA-A2-specific antibodies in recipient and NB1-specific antibodies in donor blood. Br J Haematol 93 : 707–713. 8652399
4. Fadeyi EA, De Los Angeles Muniz M, Wayne AS, Klein HG, Leitman SF, et al. (2007) The transfusion of neutrophil-specific antibodies causes leukopenia and a broad spectrum of pulmonary reactions. Transfusion 47 : 545–550. 17319838
5. Leger R, Palm S, Wulf H, Vosberg A, Neppert J (1999) Transfusion-related lung injury with leukopenic reaction caused by fresh frozen plasma containing anti-NB1. Anesthesiology 91 : 1529–1532. 10551607
6. Stroncek DF (2007) Neutrophil-specific antigen HNA-2a, NB1 glycoprotein, and CD177. Curr Opin Hematol 14 : 688–693. 17898576
7. Storch EK, Hillyer CD, Shaz BH (2014) Spotlight on pathogenesis of TRALI: HNA-3a (CTL2) antibodies. Blood 124 : 1868–1872. 25006121
8. Sachs UJ, Hattar K, Weissmann N, Bohle RM, Weiss T, et al. (2006) Antibody-induced neutrophil activation as a trigger for transfusion-related acute lung injury in an ex vivo rat lung model. Blood 107 : 1217–1219. 16210340
9. Tamarozzi MB, Soares SG, Sa-Nunes A, Paiva HH, Saggioro FP, et al. (2012) Comparative analysis of the pathological events involved in immune and non-immune TRALI models. Vox Sang 103 : 309–321. doi: 10.1111/j.1423-0410.2012.01613.x 22624696
10. Lalezari P, Murphy GB, Allen FH Jr. (1971) NB1, a new neutrophil-specific antigen involved in the pathogenesis of neonatal neutropenia. J Clin Invest 50 : 1108–1115. 5552408
11. Stroncek DF, Herr GP, Maguire RB, Eiber G, Clement LT (1994) Characterization of the neutrophil molecules identified by quinine-dependent antibodies from two patients. Transfusion 34 : 980–985. 7974707
12. Stroncek DF, Herr GP, Plachta LB (1994) Neutrophil-specific antigen NB1 inhibits neutrophil-endothelial cell interactions. J Lab Clin Med 123 : 247–255. 8301201
13. Stroncek DF, Shapiro RS, Filipovich AH, Plachta LB, Clay ME (1993) Prolonged neutropenia resulting from antibodies to neutrophil-specific antigen NB1 following marrow transplantation. Transfusion 33 : 158–163. 8430456
14. Stroncek DF, Caruccio L, Bettinotti M (2004) CD177: A member of the Ly-6 gene superfamily involved with neutrophil proliferation and polycythemia vera. J Transl Med 2 : 8. 15050027
15. Kissel K, Santoso S, Hofmann C, Stroncek D, Bux J (2001) Molecular basis of the neutrophil glycoprotein NB1 (CD177) involved in the pathogenesis of immune neutropenias and transfusion reactions. Eur J Immunol 31 : 1301–1309. 11465086
16. Bettinotti MP, Olsen A, Stroncek D (2002) The use of bioinformatics to identify the genomic structure of the gene that encodes neutrophil antigen NB1, CD177. Clin Immunol 102 : 138–144. 11846455
17. Caruccio L, Bettinotti M, Director-Myska AE, Arthur DC, Stroncek D (2006) The gene overexpressed in polycythemia rubra vera, PRV-1, and the gene encoding a neutrophil alloantigen, NB1, are alleles of a single gene, CD177, in chromosome band 19q13.31. Transfusion 46 : 441–447. 16533288
18. Caruccio L, Walkovich K, Bettinotti M, Schuller R, Stroncek D (2004) CD177 polymorphisms: correlation between high-frequency single nucleotide polymorphisms and neutrophil surface protein expression. Transfusion 44 : 77–82. 14692971
19. Dittmar K, Lim JB, Caruccio L, Bettinotti M, Stroncek D (2003) Assessment of the relative number of copies of the gene encoding human neutrophil antigen-2a(HNA-2a), CD177, and a homologous pseudogene by quantitative real-time PCR. Immunohematology 19 : 122–126. 15373677
20. Temerinac S, Klippel S, Strunck E, Roder S, Lubbert M, et al. (2000) Cloning of PRV-1, a novel member of the uPAR receptor superfamily, which is overexpressed in polycythemia rubra vera. Blood 95 : 2569–2576. 10753836
21. Skubitz KM, Stroncek DF, Sun B (1991) Neutrophil-specific antigen NB1 is anchored via a glycosyl-phosphatidylinositol linkage. J Leukoc Biol 49 : 163–171. 1825110
22. Bayat B, Werth S, Sachs UJ, Newman DK, Newman PJ, et al. (2010) Neutrophil transmigration mediated by the neutrophil-specific antigen CD177 is influenced by the endothelial S536N dimorphism of platelet endothelial cell adhesion molecule-1. J Immunol 184 : 3889–3896. doi: 10.4049/jimmunol.0903136 20194726
23. Sachs UJ, Andrei-Selmer CL, Maniar A, Weiss T, Paddock C, et al. (2007) The neutrophil-specific antigen CD177 is a counter-receptor for platelet endothelial cell adhesion molecule-1 (CD31). J Biol Chem 282 : 23603–23612. 17580308
24. von Vietinghoff S, Tunnemann G, Eulenberg C, Wellner M, Cristina Cardoso M, et al. (2007) NB1 mediates surface expression of the ANCA antigen proteinase 3 on human neutrophils. Blood 109 : 4487–4493. 17244676
25. Abdgawad M, Gunnarsson L, Bengtsson AA, Geborek P, Nilsson L, et al. (2010) Elevated neutrophil membrane expression of proteinase 3 is dependent upon CD177 expression. Clin Exp Immunol 161 : 89–97. doi: 10.1111/j.1365-2249.2010.04154.x 20491791
26. Bauer S, Abdgawad M, Gunnarsson L, Segelmark M, Tapper H, et al. (2007) Proteinase 3 and CD177 are expressed on the plasma membrane of the same subset of neutrophils. J Leukoc Biol 81 : 458–464. 17077162
27. Hu N, Westra J, Huitema MG, Bijl M, Brouwer E, et al. (2009) Coexpression of CD177 and membrane proteinase 3 on neutrophils in antineutrophil cytoplasmic autoantibody-associated systemic vasculitis: anti-proteinase 3-mediated neutrophil activation is independent of the role of CD177-expressing neutrophils. Arthritis Rheum 60 : 1548–1557. doi: 10.1002/art.24442 19404956
28. Jerke U, Rolle S, Dittmar G, Bayat B, Santoso S, et al. (2011) Complement receptor Mac-1 is an adaptor for NB1 (CD177)-mediated PR3-ANCA neutrophil activation. J Biol Chem 286 : 7070–7081. doi: 10.1074/jbc.M110.171256 21193407
29. Gohring K, Wolff J, Doppl W, Schmidt KL, Fenchel K, et al. (2004) Neutrophil CD177 (NB1 gp, HNA-2a) expression is increased in severe bacterial infections and polycythaemia vera. Br J Haematol 126 : 252–254. 15238147
30. Toyoda T, Tsukamoto T, Yamamoto M, Ban H, Saito N, et al. (2013) Gene expression analysis of a Helicobacter pylori-infected and high-salt diet-treated mouse gastric tumor model: identification of CD177 as a novel prognostic factor in patients with gastric cancer. BMC Gastroenterol 13 : 122. doi: 10.1186/1471-230X-13-122 23899160
31. Moritz E, Chiba AK, Kimura EY, Albuquerque D, Guirao FP, et al. (2010) Molecular studies reveal that A134T, G156A and G1333A SNPs in the CD177 gene are associated with atypical expression of human neutrophil antigen-2. Vox Sang 98 : 160–166. doi: 10.1111/j.1423-0410.2009.01233.x 19695014
32. Wolff J, Brendel C, Fink L, Bohle RM, Kissel K, et al. (2003) Lack of NB1 GP (CD177/HNA-2a) gene transcription in NB1 GP - neutrophils from NB1 GP-expressing individuals and association of low expression with NB1 gene polymorphisms. Blood 102 : 731–733. 12623849
33. Kissel K, Scheffler S, Kerowgan M, Bux J (2002) Molecular basis of NB1 (HNA-2a, CD177) deficiency. Blood 99 : 4231–4233. 12010833
34. Matsuo K, Lin A, Procter JL, Clement L, Stroncek D (2000) Variations in the expression of granulocyte antigen NB1. Transfusion 40 : 654–662. 10864984
35. de Haas M, Kleijer M, van Zwieten R, Roos D, von dem Borne AE (1995) Neutrophil Fc gamma RIIIb deficiency, nature, and clinical consequences: a study of 21 individuals from 14 families. Blood 86 : 2403–2413. 7662988
36. Fromont P, Bettaieb A, Skouri H, Floch C, Poulet E, et al. (1992) Frequency of the polymorphonuclear neutrophil Fc gamma receptor III deficiency in the French population and its involvement in the development of neonatal alloimmune neutropenia. Blood 79 : 2131–2134. 1532916
37. Huizinga TW, Kuijpers RW, Kleijer M, Schulpen TW, Cuypers HT, et al. (1990) Maternal genomic neutrophil FcRIII deficiency leading to neonatal isoimmune neutropenia. Blood 76 : 1927–1932. 1978690
38. Willcocks LC, Lyons PA, Clatworthy MR, Robinson JI, Yang W, et al. (2008) Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med 205 : 1573–1582. doi: 10.1084/jem.20072413 18559452
39. Jelinek J, Li J, Mnjoyan Z, Issa JP, Prchal JT, et al. (2007) Epigenetic control of PRV-1 expression on neutrophils. Exp Hematol 35 : 1677–1683. 17976520
40. Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, et al. (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302 : 2141–2144. 14684825
41. Modrek B, Lee C (2002) A genomic view of alternative splicing. Nat Genet 30 : 13–19. 11753382
42. Xu Q, Modrek B, Lee C (2002) Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res 30 : 3754–3766. 12202761
43. Popp MW, Maquat LE (2013) Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47 : 139–165. doi: 10.1146/annurev-genet-111212-133424 24274751
44. Stroncek DF, Shankar RA, Noren PA, Herr GP, Clement LT (1996) Analysis of the expression of NB1 antigen using two monoclonal antibodies. Transfusion 36 : 168–174. 8614969
45. Montaser LM, El-Rashidi FH, Essa ES, Azab SM (2011) Analysis of CD177 neutrophil expression in beta-thalassemia patients. APMIS 119 : 674–680. doi: 10.1111/j.1600-0463.2011.02755.x 21917004
46. Dillon M, Minear J, Johnson J, Lannutti BJ (2008) Expression of the GPI-anchored receptor Prv-1 enhances thrombopoietin and IL-3-induced proliferation in hematopoietic cell lines. Leuk Res 32 : 811–819. 17980909
47. Mnjoyan Z, Li J, Afshar-Kharghan V (2005) Expression of polycythemia rubra vera-1 decreases the dependency of cells on growth factors for proliferation. Haematologica 90 : 405–406. 15749675
48. Meyerson HJ, Osei E, Schweitzer K, Blidaru G, Edinger A, et al. (2013) CD177 expression on neutrophils: in search of a clonal assay for myeloid neoplasia by flow cytometry. Am J Clin Pathol 140 : 658–669. doi: 10.1309/AJCPDFBEBQZW1OI7 24124144
49. Stroncek DF, Shankar R, Litz C, Clement L (1998) The expression of the NB1 antigen on myeloid precursors and neutrophils from children and umbilical cords. Transfus Med 8 : 119–123. 9675788
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Akutní intermitentní porfyrie
- Primární hyperoxalurie – aktuální možnosti diagnostiky a léčby
- Doc. Eva Vlčková: Transthyretinová amyloidóza již díky moderní terapii nemusí být fatální diagnózou, musíme ji však zachytit a léčit včas
- Spolehlivost LH testů
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



