-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
The detection of causative nonsynonymous single nucleotide variants (SNVs) is essential for the understanding of the pathogenesis of human inherited diseases. In this paper, we propose a statistical method called SPRING (Snv PRioritization via the INtegration of Genomic data) to combine six functional effect scores calculated by existing methods and five association scores derived from multiple genomic data sources to estimate the statistical significance that a nonsynonymous SNV is pathogenic for a query disease. We find that SPRING is effective in identifying disease-causing SNVs for diseases whose genetic bases are either partly known or completely unknown across a variety of inheritance styles. With real exome sequencing data, we show the qualified potential of SPRING in not only the detection of causative SNVs in simulation studies but also the identification of pathogenic de novo mutations for autism, epileptic encephalopathies and intellectual disability.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004237
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004237Summary
The detection of causative nonsynonymous single nucleotide variants (SNVs) is essential for the understanding of the pathogenesis of human inherited diseases. In this paper, we propose a statistical method called SPRING (Snv PRioritization via the INtegration of Genomic data) to combine six functional effect scores calculated by existing methods and five association scores derived from multiple genomic data sources to estimate the statistical significance that a nonsynonymous SNV is pathogenic for a query disease. We find that SPRING is effective in identifying disease-causing SNVs for diseases whose genetic bases are either partly known or completely unknown across a variety of inheritance styles. With real exome sequencing data, we show the qualified potential of SPRING in not only the detection of causative SNVs in simulation studies but also the identification of pathogenic de novo mutations for autism, epileptic encephalopathies and intellectual disability.
Introduction
Pinpointing genetic variants underlying human inherited diseases is the primary step towards the understanding of the pathogenesis of these diseases [1]. With the accelerating advancement of the next generation sequencing technology, it becomes an efficient strategy to selectively sequence coding regions of a genome, resulting in the exome sequencing technique [2]. With the increase of sequencing throughput and the decrease of sequencing costs, exome sequencing has been widely used in not only the detection of pathogenic variants for Mendelian diseases [3]–[5] but also the discovery of susceptible loci for complex diseases [6]–[8].
A majority of genetic variants captured by exome sequencing studies are nonsynonymous single nucleotide variants (SNVs), whose occurrences may change structures of encoded proteins, thereby affecting functions of proteins and further causing diseases [5]. It has been shown that among the large number (typically around 8,000–10,000) of nonsynonymous SNVs sequenced in an exome, a significant fraction occurs with low minor allele frequency (MAF≤1%), belonging to the category of rare genetic variation [9], [10]. Recent studies have also shown that a non-negligible fraction of disease-causing SNVs occur de novo, representing the most extreme form of rare variants [11]–[13]. The existence of such rare or de novo mutations, together with the fact that the number of affected and normal individuals being sequenced is typically quite limited, has been obstructing direct applications of such traditional statistical genetics methods as family-based linkage analysis and population-based association studies to the analysis of exome sequencing data [14]. Indeed, prevalent applications of exome sequencing have been appealing for an effective computational method for the identification of pathogenic variants from a large number of sequenced nonsynonymous SNVs [1], [5].
To meet the requirement in the analysis of exome sequencing data, existing methods for predicting functional implications of nonsynonymous SNVs have been borrowed. These methods, with examples including SIFT [15], PolyPhen2 [16], LRT [17], MutationTaster [18], GERP [19], PhyloP [20], and many others [21]–[25], typically predict damaging effects of a nonsynonymous SNV on the function of its hosting protein based on individual or combined use of such information as sequence properties [15], structure characteristics [16] and database annotations [18]. Genome-scale prediction results of these methods have also been collected in databases such as the dbNSFP [26]. However, for a specific query disease, the alteration of the function of a gene hosting a variant does not necessarily mean that the variant is pathogenic for the query disease. For example, in the UniProtKB/Swiss-Prot database [27] (release 2012_09), there have been 37 SNVs reported in gene ABCB4 (corresponding to multidrug resistance protein 3, MDR3_HUMAN). Among these variants, 11 (e.g., p.Arg150Lys) has been reported to be causative for intrahepatic cholestasis of pregnancy (MIM: 147480) [28], [29], 3 (e.g., p.Gly983Ser) causative for progressive familial intrahepatic cholestasis type 3 (MIM: 602347) [30]–[32], and 6 (e.g., p.Ala934Thr) causative for gallbladder disease type 1 (MIM: 600803) [33], [34]. Therefore, in order to access whether a nonsynonymous SNV is causative for a query disease, it is not enough to only predict the functionally damaging effects of the variant — the association information between the disease and the gene hosting the variant is also important.
With this understanding, we propose in this paper a statistical method called SPRING (Snv PRioritization via the INtegration of Genomic data) for the detection of pathogenic nonsynonymous SNVs for a given query disease in exome sequencing studies. Given a query disease and a set of candidate nonsynonymous SNVs, SPRING calculates a q-value for each candidate variant to indicate the statistical significance that the variant is causative for the query disease and thus provides a means of prioritizing the candidate variants. SPRING achieves this goal by using a rigorous statistical model to integrate six functional effect scores that are calculated by SIFT, PolyPhen2, LRT, MutationTaster, GERP and PhyloP to indicate the functional implication of a nonsynonymous SNV and five association scores that are derived from gene ontology, protein-protein interactions, protein sequences, protein domain annotations and gene pathway annotations to describe the potential association between the variant and the query disease. The integrated p-values are further converted to q-values for addressing the multiple testing correction problem [35], [36]. We perform a series of comprehensive validation experiments to access the effectiveness of SPRING. Results show that our method is valid for diseases whose genetic bases are either partly known or completely unknown, effective for diseases with a variety of inheritance styles, and capable of identifying disease-causing SNVs in whole-exome sequencing studies. We further show the capability of our method in detecting causative de novo mutations for autism, epileptic encephalopathies, and intellectual disability.
Results
Principles of the proposed method
The computational assessment of functional implications of nonsynonymous SNVs has been usually formulated as a task of predicting functionally damaging effects of such SNVs. To this end, existing methods predict the potential impact of a nonsynonymous SNV on the function of its host protein based on such information as sequence properties [15], structure characteristics [16], and database annotations [18]. The principle behind these methods is that a functionally damaging SNV usually raises a significant change on the structure and function of the host protein, and the sequence at the mutation position is more conserved, while a neutral SNV typically results in a minor or negligible change in protein structure and function, and the sequence of the resulting protein is less conserved.
The computational identification of disease genes has been typically modeled from the viewpoint of one-class prioritization. Given a query disease and a list of candidate genes, existing methods rank candidate genes according to their strength of association with the query disease. This is usually done according to the guilt-by-association principle [37], which assumes that genes related to the same disease are correlated in their functions, and such correlation can be calculated from such genomic data as gene sequences [38], gene functional annotations [39], protein-protein interactions [40], etc [41], [42].
However, for a specific query disease, the alteration of the function of a gene hosting an SNV does not necessarily mean the association between the gene and the query disease, as we have analyzed previously that the SNVs occurring in ABCB4 may cause three diseases intrahepatic cholestasis of pregnancy, progressive familial intrahepatic cholestasis type 3 and gallbladder disease type 1. On the other hand, the association between a gene and a query disease does not mean every functional variant in the gene is causative for the query disease. For example, SCN2A (MIM: 182390) has been validated to be causative for autism (MIM: 209850) [43] and early infantile epileptic encephalopathy-11 (EIEE11, MIM: 613721) [44], [45] but benign familial neonatal-infantile seizures-3 (BFIS3, MIM: 607745) [46], [47]. Nevertheless, based on the dbSNP database, among the 37 SNVs found in SCN2A, only 7 are detected to be pathogenic, and the other 30 are not reported to be causative for any disease up to now [48]. Therefore, in order to access whether an SNV is causative for a query disease, one needs to integrate both the functional implication of the variant and the association information between the disease and the gene hosting the variant. Based on this reasoning, we model the identification of causative SNVs for a query disease from a set of candidate SNVs as a prioritization problem and propose a computational approach called SPRING to address this problem.
Specifically, as illustrated in Figure 1, given a query disease and a set of candidate SNVs, SPRING calculates a q-value for each candidate variant to indicate the statistical significance that the variant is causative for the query disease and thus provides a means of ranking the candidate variants. SPRING achieves this goal by using a statistical method called Fisher's combined probability test with dependence correction to integrate six functional effect p-values that characterize the functional implication of a variant and five association p-values that describe the potential association between the variant and the query disease. The six functional effect p-values are derived from existing approaches for prediction functional implications of SNVs, including SIFT [15], PolyPhen2 [16], LRT [17], MutationTaster [18], GERP [19], and PhyloP [20]. The five association p-values are derived from genomic data sources, including gene ontology, protein-protein interactions, protein sequences, domain annotations, and pathway annotations. To address the multiple testing correction problem, p-values resulting from the Fisher's method [49] are further converted to q-values to control the positive false discovery rate (pFDR) [35], [36].
Fig. 1. Workflow of SPRING. 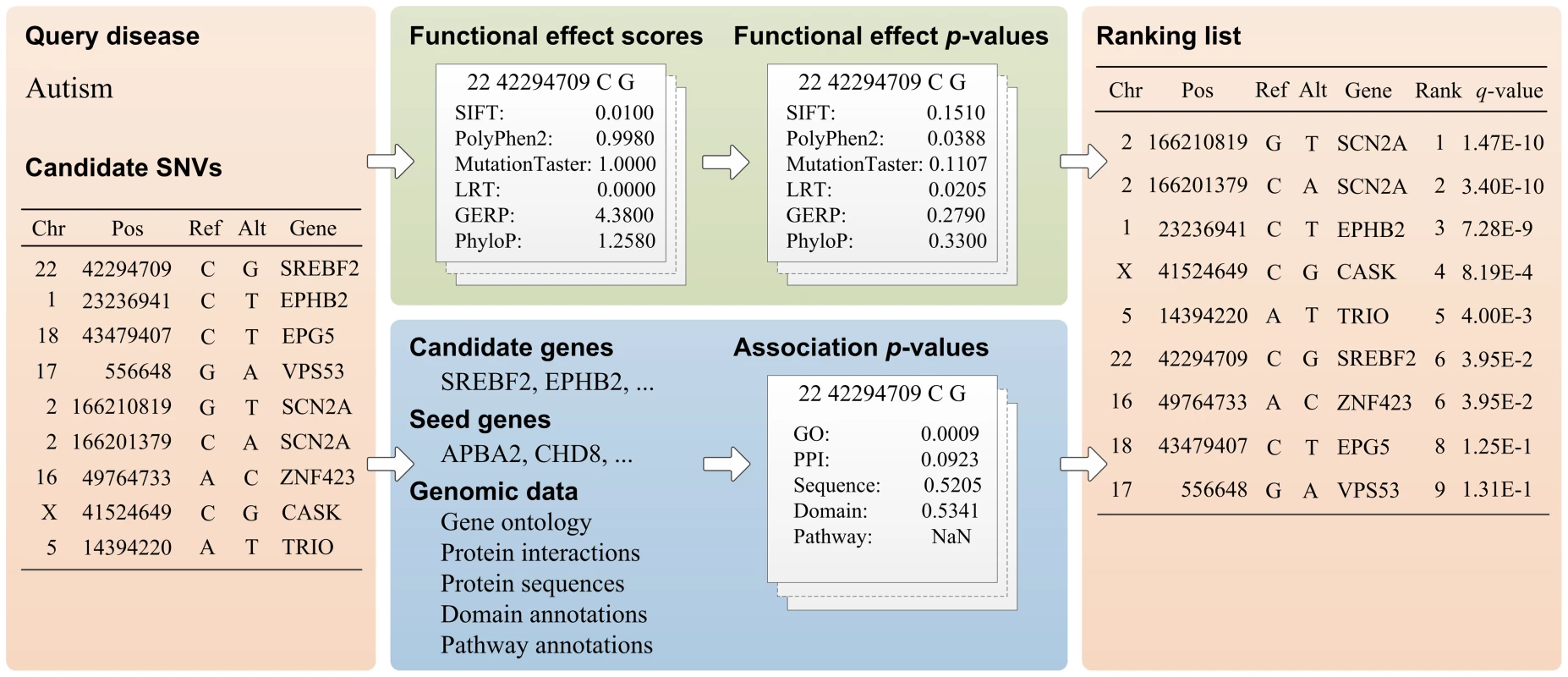
Given a query disease and a set of candidate SNVs as inputs, SPRING calculates a q-value for each candidate and generates a ranking list of the candidates as the output. A q-value is calculated by using Fisher's method with dependence correction to integrate six functional effect p-values and five association p-values. Data sources
We extracted a list of 1,436 diseases from the OMIM database (accessed in November 2012) [50] and downloaded a total of 1,206 genes associated with these diseases using the tool BioMart [51] (Table S1).
We downloaded from the UniProtKB/Swiss-Prot database [27] (release 2012_09) 23,320 disease-causing SNVs annotated as “Disease” and 37,193 neutral SNVs annotated as “Polymorphism.” We downloaded functional effect scores for SNVs (calculated by SIFT, PolyPhen2, LRT, MutationTaster, GERP and PhyloP) from the dbNSFP database [26] (version 2.0b4), which included prediction scores for not only missense mutations but also nonsense mutations. Matching these two data sources, we obtained 12,610 disease-causing SNVs and 23,403 neutral SNVs with at least one functional effect score available. We downloaded exome sequencing data of eight HapMap individuals [52] that represented three populations (Europe, Asia and Africa) and derived a set of SNVs for each individual (Text S1).
We downloaded protein-protein interaction data from the STRING database [53] (Version 9.0). Focusing on high confident interactions (confidence scores greater than or equal to 0.9), we extracted 9,966 proteins and 116,648 interactions between the proteins. We downloaded the gene ontology (GO) and associated annotations (accessed on November 2, 2012). Focusing on the biological process domain, we calculated semantic similarity between 14,283 genes. We downloaded sequences of human proteins from the UniProt database (release 2012_09) and calculated their pairwise similarities using SSEARCH [54]. Focusing on e-values less than or equal to 0.001, we obtained 20,281 proteins and 912,018 similarities between them. We downloaded annotations of protein structural domains from the Pfam database [55] (version 26.0). Focusing on the manually curated part (Pfam-A), we collected 1,066 domains that were contained in at least five human proteins and derived pairwise similarities between 12,713 proteins that contained at least one of these domains. We downloaded annotations of 232 pathways from the KEGG database [56] (release 58.0) and derived pairwise similarities between 5,951 genes accordingly.
Performance on diseases with partly known genetic bases
We validated SPRING for diseases whose genetic bases were partly known (i.e., some genes had been annotated as associated with the diseases). To simulate this situation, we extracted 113 diseases annotated as associated with two or more genes from the OMIM database (Table S1). Taking each of these diseases as the query disease, we collected its causative SNVs from the Swiss-Prot database [27] to obtain a set of test SNVs, and we ranked each of them against three sets of control SNVs, including (1) a neutral control set composed of SNVs annotated as “Polymorphism” in the Swiss-Prot database, except for those selected for estimating functional effect p-values, (2) a disease control set consisting of SNVs annotated as “Disease” in the Swiss-Prot database, except for those in the test set, and (3) a combined control set obtained as the union of the neutral and the disease sets. The disease control set was used to assess the capability of our method in distinguishing variants causative for the query disease from those causative for other diseases but irrelevant to the query one. The combined control set was used to simulate the real situation in which an individual might carry not only neutral variants but also variants responsible for diseases other than the query one. In the calculation of functional effect p-values, we partitioned SNVs annotated as “Polymorphism” in the Swiss-Prot database into two equal parts at random and used one part to estimate the null distribution and the other part as the neutral control set. In the calculation of association p-values, we selected seed genes for the query disease as genes annotated as associated with the disease, except for the one hosting the test SNV.
We summarized ranks of the test SNVs in Figure 2 (A–C). There are a total of 1,501 disease SNVs annotated as causative for the 113 diseases. In the validation against the neutral control set (11,702 SNVs), SPRING ranks 1,161 test SNVs among top 10 and 1,306 among top 50. In contrast, with a random guess procedure, one could only expect 10×1,501/11,702≈1.28 test SNVs enriched among top 10 and 6.41 among top 50. In the validation against the disease control set (12,605 SNVs), SPRING ranks 185 test SNVs among top 10 and 653 among top 50, while random guess can only enrich 1.19 test SNVs among top 10 and 5.95 among top 50. In the validation against the combined control set (24,307 SNVs), SPRING ranks 164 test SNVs among top 10 and 628 among top 50, while random guess can only enrich 0.62 test SNVs among top 10 and 3.09 among top 50. These results suggest the capability of our method in identifying SNVs causative for diseases whose genetic bases are partly known.
Fig. 2. Rank distributions of the test SNVs. 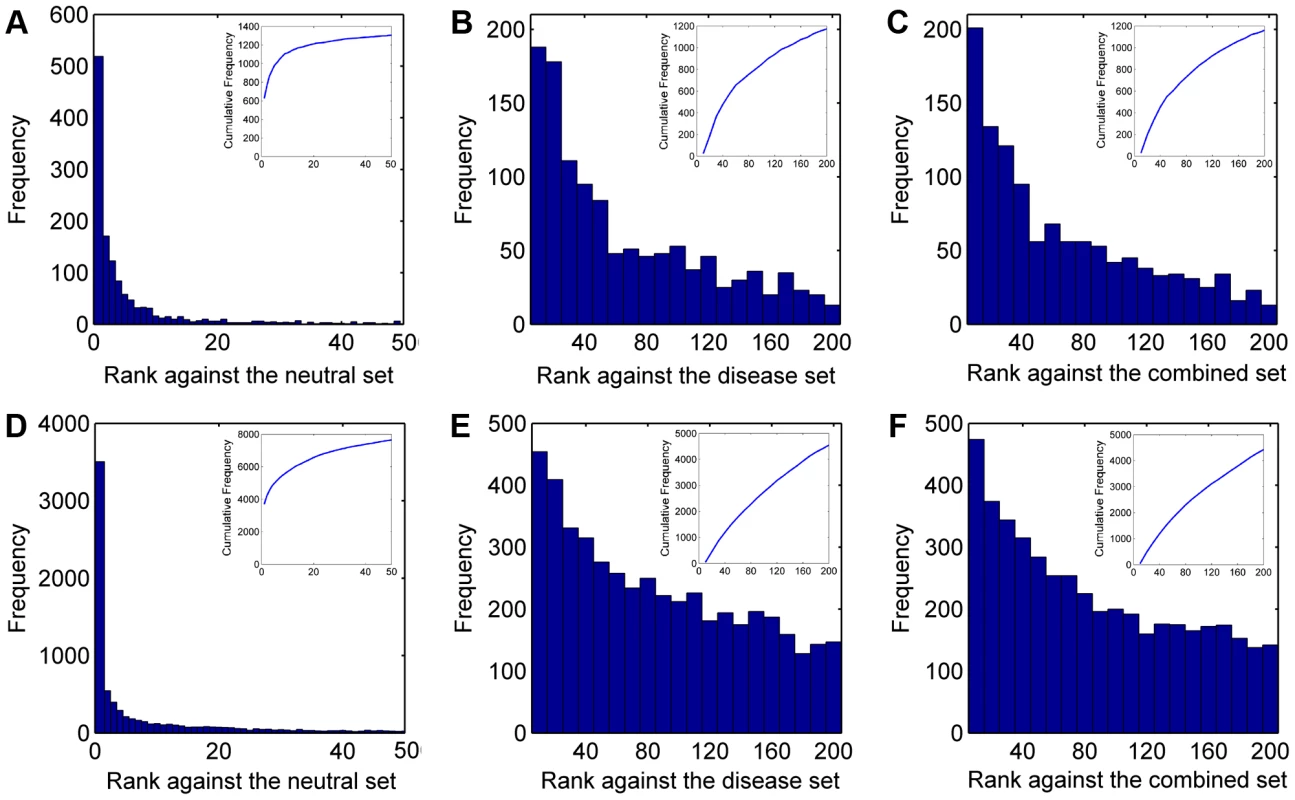
(A–C) Results for diseases with partly known genetic bases when validating against the neutral, disease, and combined control sets, respectively. (D–F) Results for diseases of unknown genetic bases when validating against the neutral, disease, and combined control sets, respectively. We then derived two criteria to quantify the performance of our method. Dividing the rank of a test SNV by the total number of candidates, we obtained the rank ratio of the SNV. Averaging rank ratios of all test SNVs for a query disease, we obtained the first criterion called the Mean Rank Ratio (MRR). At a certain threshold of the rank ratio, we defined the sensitivity as the fraction of test SNVs ranked above the threshold, and specificity as the fraction of control SNVs ranked below the threshold. Varying the threshold, we plotted the rank operating characteristic (ROC) curve (sensitivity versus 1-specificity) and further calculated the area under this curve as the second criterion called the AUC score. As shown in Figure 3 (A and B), the average MRR and AUC for the 113 diseases are 0.0071 and 0.9930 respectively in the validation against the neutral control set, 0.0466 and 0.9535 respectively in the validation against the disease control set, and 0.0275 and 0.9725 respectively in the validation against the combined control set. These results further suggest the effectiveness of our method, considering that random guess can only yield an MRR of 0.5 and an AUC of 0.5.
Fig. 3. Performance of SPRING in the validation experiments. 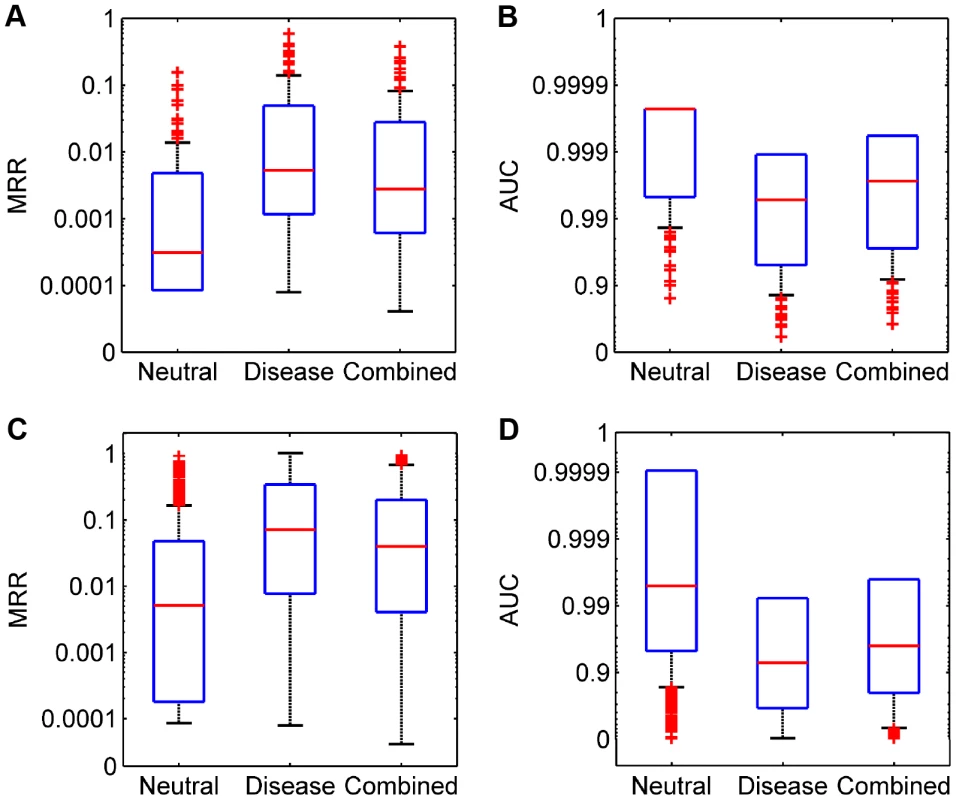
(A) and (B) MRRs and AUCs for diseases with partly known genetic bases, respectively. (C) and (D) MRRs and AUCs for diseases of unknown genetic bases, respectively. We analyzed the statistical significance of candidate SNVs and found that q-values of test SNVs were much smaller than those of control ones. Hence, when ranking a test SNV against control ones according to their q-values, the test SNV was likely to be ranked among top positions (Text S1). We further assessed whether the number of seed genes affected the performance of our method and found this factor having little influence (Text S1).
Performance on diseases of unknown genetic bases
We validated SPRING for diseases whose genetic bases were unknown (i.e., no genes had been annotated as associated with the diseases). To simulate this situation, we extracted a total of 1,436 diseases annotated as associated with at least one gene from the OMIM database (Table S1). Taking each of these diseases as the query disease and pretending that the genetic basis of this disease was unknown, we collected annotated causative SNVs of the disease to obtain test SNVs, and we ranked each of them against the three control sets described previously. Different from the validation for diseases with partly known genetic bases, we identified 10 diseases that had the highest phenotype similarities to the query disease according to pre-calculated pairwise phenotype similarity scores between 5,080 diseases [57] and used genes known as associated with these disease as seed genes for calculating association p-values.
We summarized ranks of the test SNVs in Figure 2 (D–F). There are a total of 12,610 disease SNVs annotated as causative for the 1,436 diseases. In the validation against the neutral control set (11,702 SNVs), SPRING ranks 5,703 test SNVs among top 10 and 7,635 among top 50, while random guess can only rank 10.78 test SNVs among top 10 and 53.88 among top 50. In the validation against the disease control set (12,605 SNVs), SPRING ranks 454 test SNVs among top 10 and 1,785 among top 50, while random guess can only enrich 10.00 test SNVs among top 10 and 50.02 among top 50. In the validation against the combined control set (24,307 SNVs), SPRING ranks 435 test SNVs among top 10 and 1,748 among top 50, while random guess can only enrich 5.19 test SNVs among top 10 and 25.94 among top 50. These results suggest the capability of our method in identifying SNVs causative for diseases whose genetic bases are unknown.
We then summarized MRRs and AUCs for individual diseases in Figure 3 (C and D). The average MRR and AUC for all 1,436 diseases are 0.0440 and 0.9560 respectively in the validation against the neutral control set, 0.2037 and 0.7964 respectively in the validation against the disease control set, and 0.1265 and 0.8735 respectively in the validation against the combined control set. We analyzed the statistical significance of candidate SNVs and found that the q-values of test SNVs were also much smaller than those of control ones (Text S1). We further assessed the influence of the number of neighboring diseases and found that our method was robust to this parameter (Text S1).
These results demonstrate the effectiveness of SPRING in dealing with diseases whose genetic bases have not been deciphered yet and thus have no information about associated genes. On one hand, since phenotypically similar diseases may have genetic overlap [58], it could be a feasible way to borrow genes known as associated with diseases of high phenotype similarities to a query disease to facilitate the inference of SNVs causative for the query disease. On the other hand, we also notice the drop in performance when compared with the results in the previous section. We suppose the reason is that seed genes selected here may not be as reliable as those for diseases with partly known genetic bases.
Prediction power for diseases of different genetic styles
We assessed the prediction power of SPRING for diseases with different genetic styles. We first classified the 1,436 diseases into a group of 1,378 Mendelian diseases and a group of 58 complex diseases according to the Genetic Association database (released in November, 2012) [59] (Table S1). Results show that SPRING can recover disease-causing SNVs for both groups (Figure 4). For example, in the validation against the combined control set, the average MRR and AUC for Mendelian diseases are 0.1272 and 0.8729 respectively, and those for complex diseases are 0.1115 and 0.8886 respectively. The two-sided Wilcoxon rank sum test suggests that the performance of our method on these two categories of disease is not significantly different (p-value = 0.5118).
Fig. 4. Performance of SPRING for diseases of different inheritance styles. 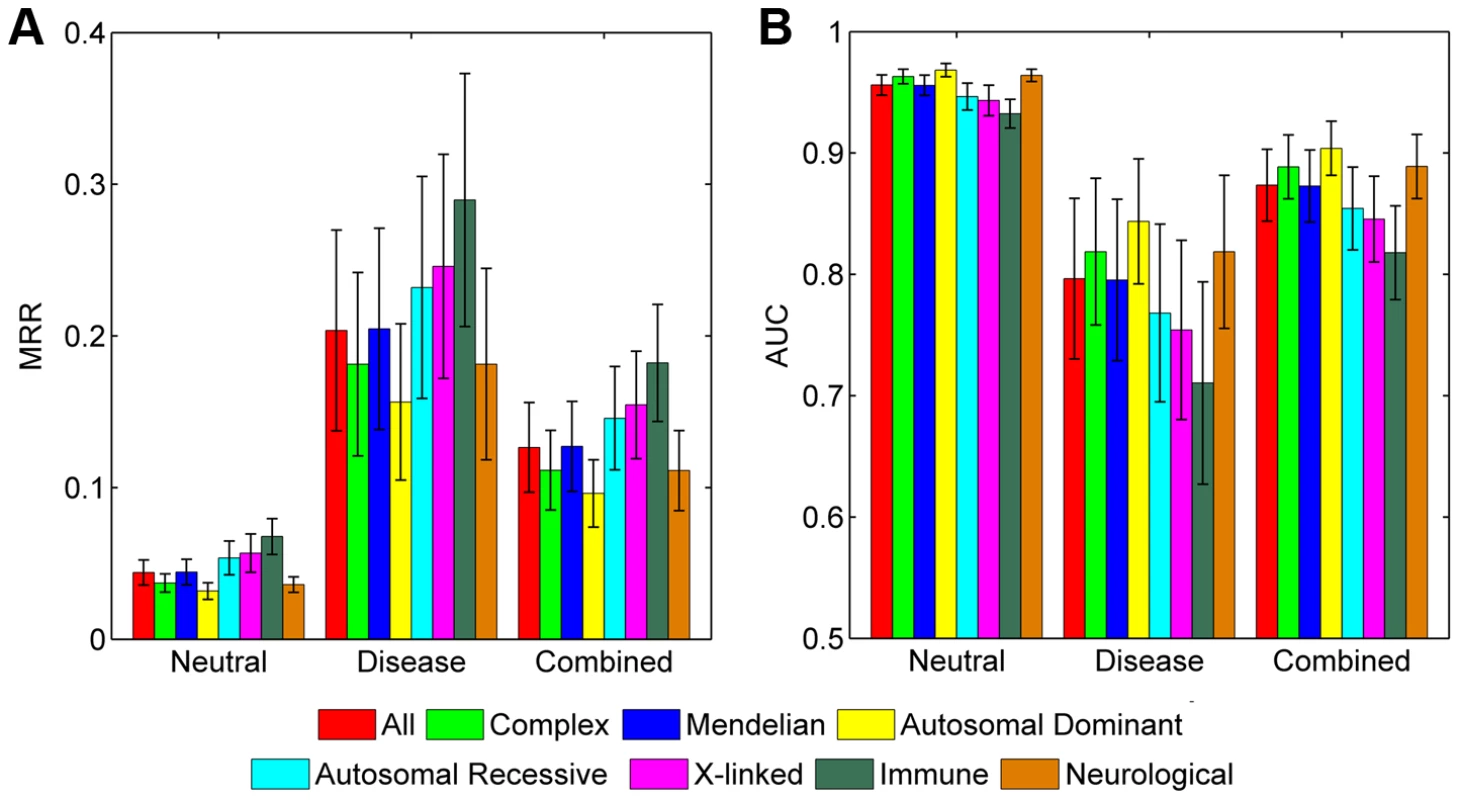
(A) MRRs when validating against the neutral, disease, and combined control sets, respectively. (B) AUCs when validating against the neutral, disease, and combined control sets, respectively. We then classified the 1,378 Mendelian diseases into three categories according to their inheritance patterns, obtaining 396 autosomal dominant diseases (MIM: 1xxxxx), 468 autosomal recessive diseases (MIM: 2xxxxx), and 109 X-linked diseases (MIM: 3xxxxx). The rest of 405 diseases (MIM: 6xxxxx) are not included in the comparison. Results show that SPRING can recover disease-causing SNVs for all these three classes of diseases (Figure 4). For example, in the validation against the combined control set, the average MRR and AUC are 0.09617 and 0.9039 respectively for autosomal dominant diseases, 0.1457 and 0.8543 respectively for autosomal recessive diseases, and 0.1545 and 0.8455 respectively for X-linked diseases. Pairwise two-sided Wilcoxon rank sum tests suggest that the performance on autosomal dominant diseases is significantly different from that on both autosomal recessive diseases (p-value = 3.897×10−9) and X-linked diseases (p-value = 1.643×10−3), while the performance on the latter two classes is not significantly different (p-value = 0.7942).
We further identified 48 immune diseases and 263 neurological disorders and found that our method was also capable of recovering disease-causing SNVs for both classes of diseases (Figure 4). For example, in the validation against the combined control set, the average MRR and AUC are 0.1822 and 0.8178 respectively for immune diseases, and 0.1112 and 0.8889 respectively for neurological disorders. The two-sided Wilcoxon rank sum test suggests that the performance of our method on these two classes of diseases is of marginal difference (p-value = 0.0159).
Prediction power for rare SNVs
One of the superiorities of exome sequencing is the capability of finding disease-causing rare SNVs for a query disease. To demonstrate the effectiveness of SPRING in identifying causative rare SNVs, we collected 932 causative rare SNVs with minor allele frequency (MAF) less than 0.01 from the dbSNP database [48] and identified a total of 444 diseases annotated as caused by these variants. For each of these rare SNVs, we pretended that the genetic basis of the corresponding disease was unknown, and we ranked the SNV against the three control sets described previously. Results show that the MRR and AUC are 0.0340 and 0.9660 respectively in the validation against neutral controls, 0.1676 and 0.8324 respectively in the validation against disease controls, and 0.1029 and 0.8971 respectively in the validation against combined controls. All these results support the effectiveness of SPRING in the identification of disease-causing rare SNVs. We then compared distributions of functional effect scores for these rare SNVs with those for the same number of SNVs selected at random from a HapMap individual. Results show that the rare SNVs are typically assigned more extreme functional effect scores than SNVs occurring in a normal individual (Text S1). Since the sequence conservation property has been used in the derivation of the functional effect scores, we conjecture that the effectiveness of our method in this validation experiment can be partly attributed to the rarity of such rare mutations in a random human.
Prediction power of individual data sources
We assessed prediction power of individual data sources by repeating the validation experiment for diseases of unknown genetic bases using a single data source. Results, as summarized in Table 1, show that functional effect data sources are effective in the discrimination of disease-causing SNVs against neutral controls but are ineffective in distinguishing such SNVs from disease controls, and thus these data sources show low effectiveness in distinguishing causative SNVs from combined controls. For example, the MRR and AUC for SIFT are 0.1792 and 0.8205 respectively in the validation against neutral controls, 0.5015 and 0.4984 respectively in the validation against disease controls, and 0.3816 and 0.6183 respectively in the validation against combined controls. ROC curves (Figure 5, dotted lines) also support this observation. It is not surprising to see the effectiveness of these data sources in the validation against neutral controls, since the power of the mechanism used for calculating functional effect scores has been verified in numerous studies [15]–[25], and our p-value transformation strategy does not affect the comparison of such scores. The ineffectiveness of these data sources in the validation against disease controls can be attributed to the absence of disease-specific features to identify which exactly disease a variant is associated with. As a result, functional effect scores lack the power of discriminating between variants causing different diseases.
Fig. 5. ROC curves of individual data sources. 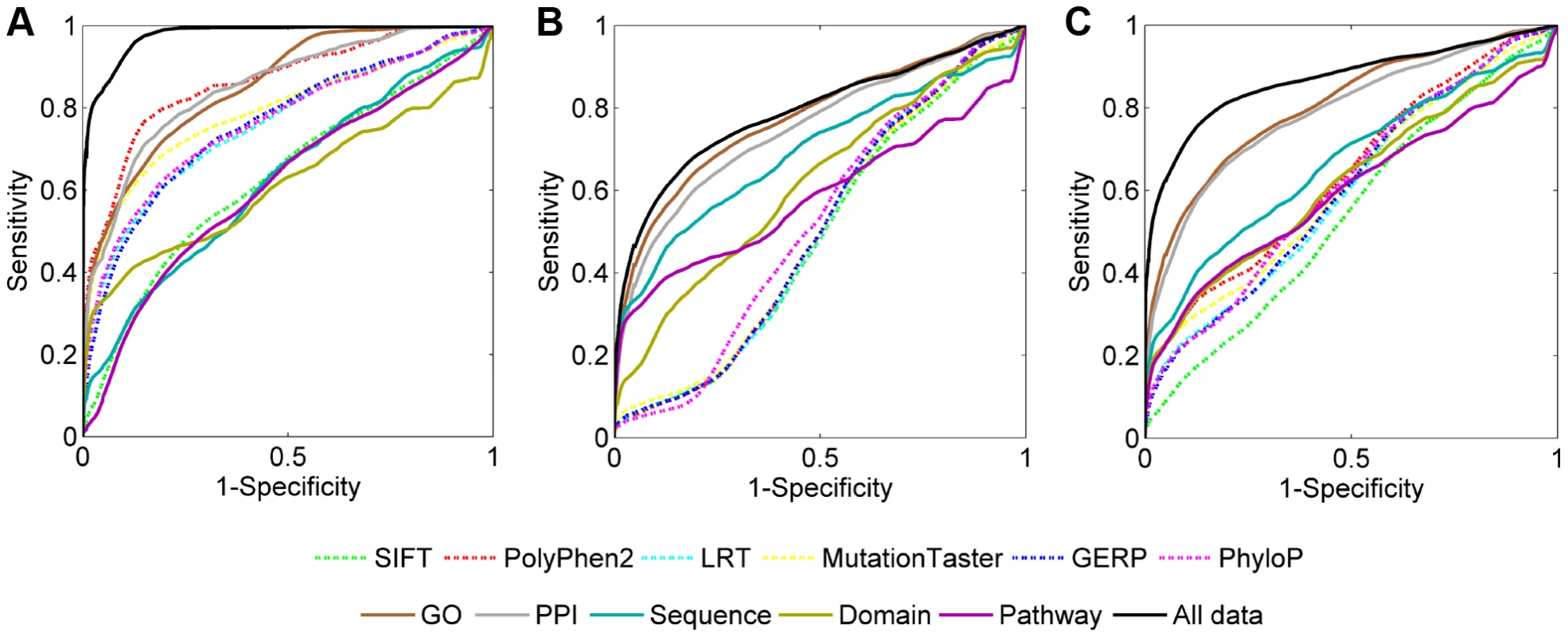
(A) Results when validating against the neutral control set. (B) Results when validating against the disease control set. (C) Results when validating against the combined control set. Tab. 1. Performance of individual data sources. 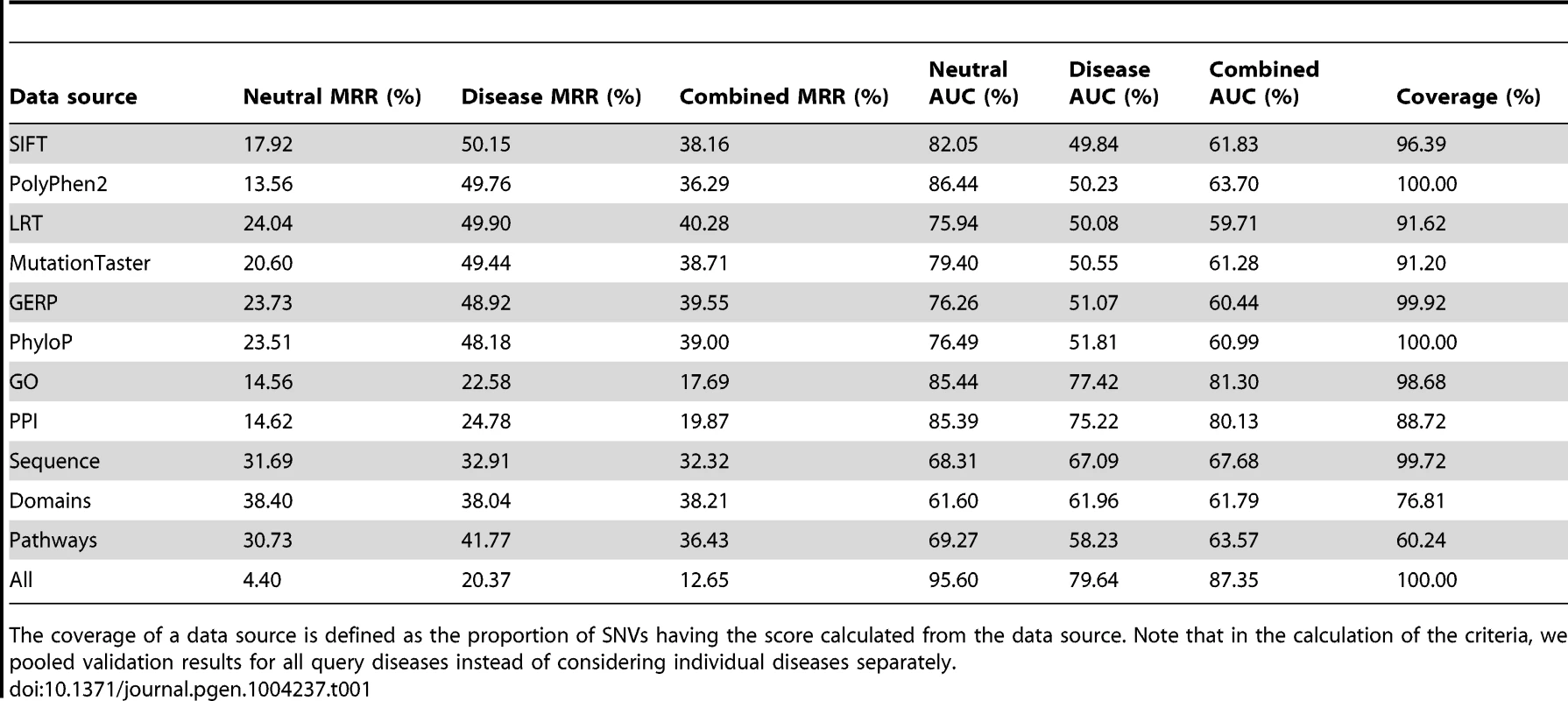
The coverage of a data source is defined as the proportion of SNVs having the score calculated from the data source. Note that in the calculation of the criteria, we pooled validation results for all query diseases instead of considering individual diseases separately. Table 1 also shows that association data sources exhibit medium effectiveness in distinguishing disease-causing SNVs from neutral, disease, and combined controls. For example, the MRR and AUC for GO are 0.1456 and 0.8544 respectively in the validation against neutral controls, 0.2258 and 0.7742 respectively in the validation against disease controls, and 0.1769 and 0.8130 respectively in the validation against combined controls. This observation is also supported by ROC curves (Figure 5, solid lines) and can be explained as follows. The association score assigned to a variant is calculated based on the gene hosting the variant. Therefore, SNVs, regardless of their functional implications, will be assigned identical scores as long as they occur in the same gene. This reasoning, together with the fact that distributions of association scores for neutral and disease controls are not significantly different (Text S1), results in the above observation.
When compared with a single data source, the integration of all 11 data sources demonstrates much lower MRRs (0.0440, 0.2037 and 0.1265 in the validation against neutral, disease and combined controls, respectively) and much higher AUCs (0.9560, 0.7964 and 0.8735 in the validation against neutral, disease and combined controls, respectively), suggesting the effectiveness of data integration. This conclusion is also supported by Figure 5 in that the ROC curves for integrating all data sources (black solid lines) climb much faster than those for individual data sources towards the top left corner of the plot. Besides, the coverage of our method also benefits from data integration. For example, in the validation experiments, only 88.72% SNVs have PPI information, and the coverage for pathway data is even as low as 60.24%. With the integration of multiple data sources, however, the causative effect of a variant for a query disease can be predicted as long as the variant appears in a data source, and thus the coverage of our method is extended to the union of variants included in individual data sources.
Considering that the data sources are correlated, the prediction power of an individual data source may not reflect its real contribution to the final performance of our method. We therefore evaluated relative contribution of a data source by erasing scores derived from the data source and repeating the validation experiment for diseases of unknown genetic bases against the combined control set. We calculated the proportion of test SNVs whose rank ratio increased after the removal of a data source to measure the relative contribution of the data source. Obviously, the larger the value of this criterion, the higher the relative contribution of a data source. As shown in Table 2, relative contributions of different data sources are quite different. For functional effect data sources, SIFT has the highest contribution, followed by LRT. For association data sources, GO and PPI both have high contributions, followed by pathway information. The results also suggest that diseases in different categories prefer different data sources. For example, GO, PPI, pathway and SIFT show much higher contributions for complex diseases than for Mendelian diseases, while PPI has much lower contribution for immune diseases than for neurological disorders.
Tab. 2. Relative contributions of individual data sources. 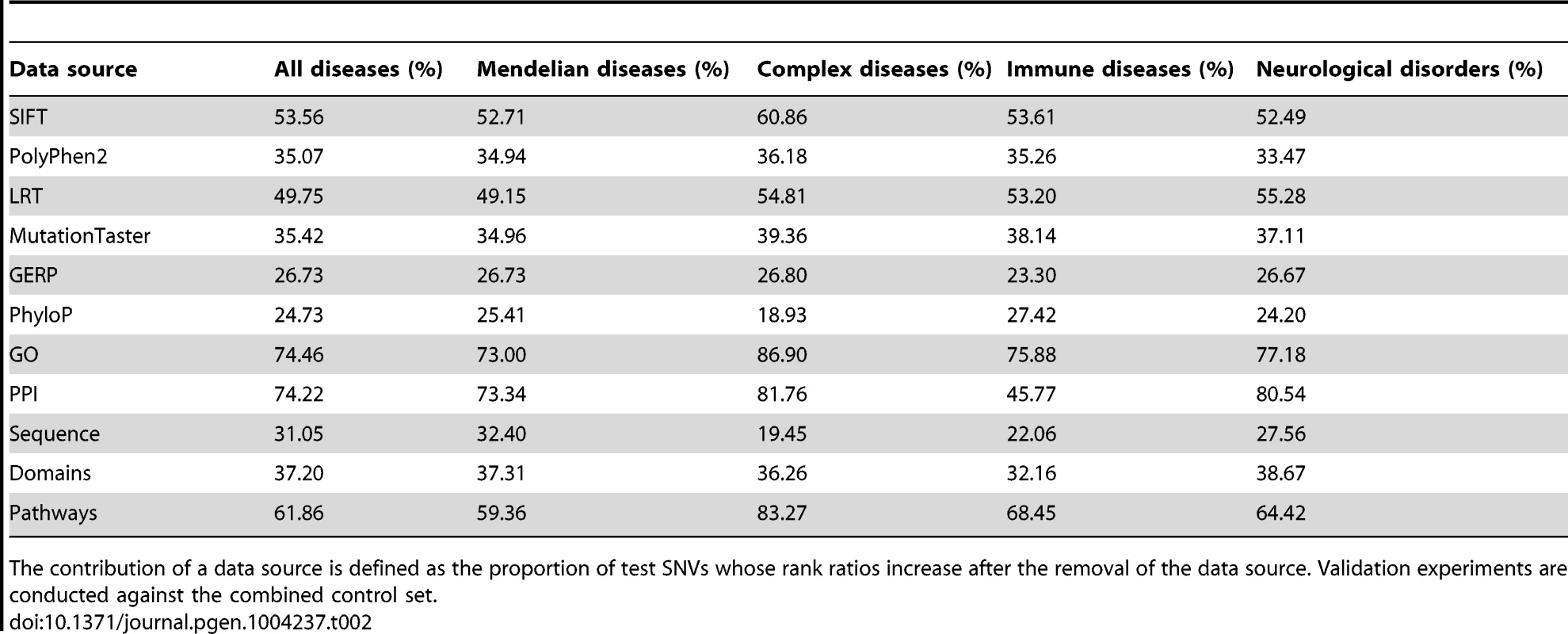
The contribution of a data source is defined as the proportion of test SNVs whose rank ratios increase after the removal of the data source. Validation experiments are conducted against the combined control set. We further adopted a sequential backward selection (SBS) strategy to select a subset of data sources for each of the 1,436 diseases. Results (Text S1 and Table S2) show that the selection procedure can improve the performance of SPRING. However, different diseases show different preferences on the selected data sources, and such preferences are diverse. We therefore suggest either seeking for the simplicity to use all data sources without selection or resorting to a cross-validation experiment to select a subset of data sources for a query disease when there is no strong prior knowledge indicating the preference of the disease to the data sources. In the reset of this paper, we use all data sources without selection by default.
Estimation of the false positive rate and true positive rate
The q-values calculated by SPRING and can be used in two ways. First, for a set of candidate SNVs, their q-values can be used as bases for prioritizing the SNVs. Second, for a single SNV, its q-value can be used to predict whether the SNV is causative for a query disease. We therefore assessed whether the false positive rate (FPR) and true positive rate (TPR) can be controlled at a desired level at a given q-value threshold.
For a HapMap individual, we calculated q-values for SNVs reported in the individual for each of the 1,436 diseases and derived the FPR for a disease as the proportion of SNVs whose q-values are less than or equal to a threshold. Results show that the TFP can be well controlled (Figure 6). For example, at the q-value thresholds 0.1, 0.05, 0.01 and 0.005, the average FPRs for all diseases are 7.16%, 4.94%, 2.01% and 1.34%, respectively. We notice these numbers are greater than those obtained using the neutral control set (average FPRs at the above q-value thresholds are 4.18%, 2.43%, 0.71% and 0.48%, respectively), and we suppose the reason behind this phenomenon is that some variants occurring in these HapMap subjects may actually be related to some diseases [60]. We further performed a simulation study by embedding different proportions of disease SNVs into the neutral control set and found that both FPR and TPR could also be well controlled (Text S1).
Fig. 6. Estimated false positive rates under different <i>q</i>-value cut-offs when using neutral SNVs in the Swiss-Prot database and exomes of the eight HapMap individuals as negative control sets. 
Simulation studies for exome sequencing data
We assessed the effectiveness of SPRING in identifying disease-causing SNVs in real exome sequencing data. For this purpose, we generated a large number of synthesized exomes by inserting each SNV causing one of the 1,436 diseases into the exome of a Hapmap individual, and we applied SPRING to rank the embedded SNV against the other SNVs in each synthesized exome. Results, as summarized in Figure 7, demonstrate the effectiveness of our method in distinguishing disease-causing SNVs from those occurring in normal individuals. For example, According to Figure 7 (A), for the eight individuals, 45.18%–49.19% causative SNVs are ranked among top 10, and 70.89%–74.15% are ranked among top 50. According to Figure 7 (B and C), the average MRRs for the eight individuals range from 0.0225 to 0.0237 (the median MRRs range from 0.0025 to 0.0029), and the average AUCs range from 0.9764 to 0.9775 (the median AUCs range from 0.9972 to 0.9978). These results suggest that our method is effective in finding true disease-causing SNVs in exome sequencing studies.
Fig. 7. Validation results for synthesized exomes. 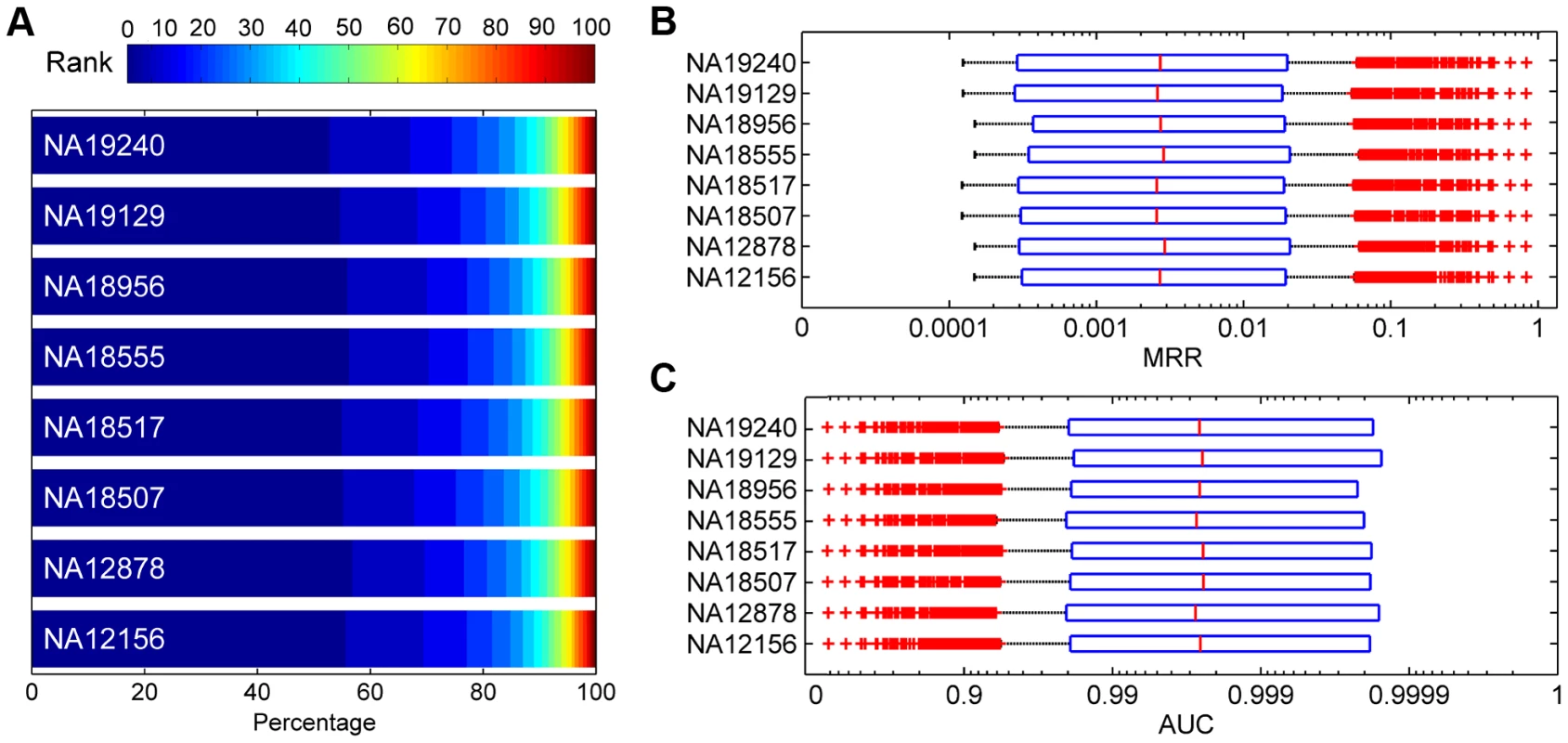
(A) Distributions of ranks for test SNVs. (B) Boxplots of MRRs. (C) Boxplots of AUCs. Detection of causative nonsynonymous de novo mutations for autism, epileptic encephalopathies and intellectual disability
De novo mutations are genetic variants that are not inherited from parents. As the most extreme form of rare genetic variation, de novo mutations have been subjected to less stringent evolutionary selection pressure and thus are usually more functionally damaging than inherited genetic variants [11]. Facilitated by the whole-exome sequencing technique, recent studies have shown that individual de novo germline mutations occurring in single genes could be the major cause of rare Mendalian diseases such as Schinzel–Giedion syndrome [61], Kabuki syndrome [62] and Bohring–Opitz syndrome [63]. Moreover, recent studies have also suggested that the collection of de novo mutations affecting different genes in different individuals might explain a proportion of common complex diseases such as schizophrenia [13], [64], autism [65]–[69], epileptic encephalopathies [70], and intellectual disability [7], [71], [72].
To demonstrate the power of our method in diagnosing disease-causing de novo mutations, we applied SPRING to a whole-exome sequencing data set of autism (PMID 22495306 [65]). From the literature [65], we collected 135 and 87 unique candidate nonsynonymous de novo mutations from the exome sequencing data of probands and siblings respectively, and 214 of these mutations can be mapped to the dbNSFP database. With the criterion that a seed gene should have been reported as associated with autism (MIM: 209850) by independent studies before the publication of this data set, we selected from the OMIM database a total of 34 seed genes and then applied SPRING to prioritize the candidate mutations (Table S3).
In the literature [65], a gene SCN2A was reported as associated with autism, and two probands each carried a nonsense de novo mutation in this gene. SPRING assigned very significant q-values (1.47×10−10 and 3.40×10−10) to these two mutations and ranked them first and second in a list of 214 candidates. Considering that the probability of ranking these two mutations at the top by a random guess procedure is only , the capability of our method in identifying causative de novo mutations in this application is strongly supported.
We noticed that SCN2A had been previously annotated as causative for autism by an independent study [43] and thus was included in seed genes in the above analysis. Although this situation is consistent with the application of SPRING in finding novel causative variants occurring in known causative genes, another important application would require the identification of causative variants occurring in genes not yet studied. To simulate this scenario, we removed SCN2A from the seed genes and prioritized the candidate mutations again. We found that the two mutations were now ranked third and sixth. Considering that the probability of ranking these two mutations among top 6 by chance is only , the capability of our method in detecting novel causative de novo mutations in this application is also supported.
We further extended applications of SPRING to three other whole-exome sequencing data sets of autism [67]–[69], one data set of epileptic encephalopathies [70] and two data sets of intellectual disability [71], [72]. Detailed analyses were given in Text S1 and Table S3.
Large scale prediction of causative nonsynonymous SNVs for 5,080 diseases
We further performed a large scale prediction of causative SNVs for a total of 5,080 diseases in the phenotype similarity matrix [57]. Focusing on SNVs collected in both the dbSNP and dbNSFP databases, we extracted a total of 174,394 SNVs that occurred in genes described by at least one of the five genomic data sources. We then used SPRING to prioritize all these candidate SNVs and distinguished suspicious causative ones for each disease. For a query disease whose genetic basis was partly known, we selected genes annotated as associated with the disease as seed genes. For a query disease whose genetic basis was completely unknown, we selected genes annotated as associated with 10 diseases of the highest phenotype similarities with the query disease as seed genes. Prediction results for the 5,080 diseases, together with an online service and the standalone software of SPRING, are available at http://bioinfo.au.tsinghua.edu.cn/spring.
Discussion
In this paper, we formulate the identification of causative nonsynonymous SNVs for a query disease as a prioritization problem, and we propose a method called SPRING to solve this problem by integrating multiple genomic data sources. We demonstrate the superior performance of our method by conducting a series of validation experiments, showing that our method is valid for diseases whose genetic bases are either partly known or completely unknown, effective for diseases with a variety of inheritance styles, and capable of identifying disease-causing de novo mutations in whole-exome sequencing studies.
The success of our method can be attributed to a combination of several aspects. First, we take the advantage of both functional implications of candidate SNVs and potential associations between a query disease and genes hosting the SNVs. In contrast, existing methods for predicting functional effects of SNVs or prioritizing candidate genes utilize only part of such information and can hardly achieve the goal of identifying causative variants. Second, we ground the inference of causative variants on a rigorous statistical model for integrating multiple data sources, not only explicitly considering correlations between the data sources, but also carefully controlling statistical significance of prediction results. As a result, our method does not rely on a single data source to make inference and is capable of maintaining a relatively low false positive rate at a reasonably high true positive rate.
Certainly, our method can be further improved in the following directions. First, we resort to the phenotype similarity profile to collect seed genes for diseases of unknown genetic bases, and hence the quality and coverage of such a profile directly determine the performance and scope of applications of our method. Following the direction of data integration, it might be helpful to incorporate multiple phenotype similarity profiles such as those calculated based on the human phenotype ontology [73] into our method. Along this direction, how to utilize these phenotype similarity profiles in a more sophisticated way will be a question worth exploring.
Second, our method currently uses five genomic data sources to infer the association relationship between a query disease and a gene hosting a variant. With the development of high-throughput experimental techniques, more and more genome-wide functional genomics data such as transcriptional regulation, microRNA regulation, DNA methylation and histone modification will be available. How to include these data sources in our method will be an important aspect.
Finally, we only focus on SNVs in protein coding regions in this paper. However, genetic variants occurring in splicing sites, promoter regions, introns, and other parts of a genome are also important in the pathogenesis of a disease. How to extend our method to enable the identification of disease-causing variants in these genomic regions will be an important direction.
Methods
Calculation of functional effect p-values
We derive six p-values to assess the statistical significance that a candidate SNV is causative for a query disease from the viewpoint of whether the SNV has damaging effect on the function of the gene containing the SNV. For this purpose, we first extract from the Swiss-Prot database [27] SNVs annotated as “Polymorphism” to obtain a set of neutral SNVs that show no damaging effect on functions of their host genes. Then, for each of the six bioinformatics tools for predicting functional implications of SNVs (SIFT [15], PolyPhen2 [16], LRT [17], MutationTaster [18], GERP [19], and PhyloP [20]), we collect predictive scores for the neutral SNVs from the dbNSFP database [26] and estimate the empirical distribution of the scores. Finally, given a candidate SNV, we extract its predictive score for each of the six tools and calculate a p-value as the probability that a neutral SNV is assigned at least the same extreme score as that of the candidate SNV.
Specifically, SIFT relies on multiple sequence alignments generated using PSI-BLAST [74] to calculate predictive scores for SNVs. The smaller a SIFT score, the stronger the evidence that an SNV is functionally damaging. Therefore, for SIFT, we calculate the probability that a neutral SNV is assigned a predictive score smaller than or equal to that of the candidate SNV as the functional effect p-value for the candidate. PolyPhen2 integrates sequence, structure and annotation information to build a classification model that predicts the probability that an SNV is functionally damaging. The greater a PolyPhen2 probability, the stronger the evidence that an SNV is functionally damaging. Therefore, we calculate the probability that a neutral SNV is assigned a predictive probability larger than or equal to that of the candidate SNV as its functional effect p-value. Methods for calculating p-values for LRT is similar to that for SIFT, and the other three tools (MutationTaster, GERP, and PhyloP) are similar to that for PolyPhen2.
Calculation of association p-values
We derive five p-values to assess the statistical significance that a candidate SNV is causative for a query disease from the viewpoint of whether the gene containing the variant is associated with the query disease. For this purpose, we first rely on phenotype similarities between diseases [57] and known associations between diseases and genes to obtain a set of seed genes for the query disease. Meanwhile, we calculate five functional similarity scores between every pair of genes based on a variety of genomic data sources (gene ontology, protein-protein interactions, protein sequences, domain annotations, and pathway annotations). Then, for each functional similarity, we resort to the guilt-by-association principle [23] to calculate a score, indicating the strength of association between the gene hosting the candidate SNV (i.e., candidate gene) and the query disease. Finally, we convert the association score to a p-value by estimating the probability that a non-disease related gene is assigned at least the same extreme score as that of the candidate gene.
In detail, given a candidate SNV, we obtain the candidate gene for the SNV by mapping the SNV back onto the genome and identifying the gene hosting the SNV. Given a query disease, we obtain seed genes for the disease by two means. First, if the genetic basis of the query disease has been studied and hence there exist some genes annotated as associated with the disease, we use these genes related to the disease as seed genes. Second, if the genetic basis of the query disease is unknown and thus these is no gene having been annotated as related to the disease, we sort all diseases except for the query one according to their similarity scores to the query disease in non-increasing order and select genes known as associated with top 10 diseases in the ranking list as seed genes. Obviously, the later strategy can also be used to complement the former when the number of genes annotated as a query disease is limited.
The five pairwise functional similarity scores for genes are calculated as follows. For the network similarity score, we extract 9,966 proteins and 116,648 high confidence interactions (with confidence scores greater than or equal to 0.9) between the proteins from the STRING database [53] (version 9.0) to obtain a protein-protein interaction network. Then, we calculate the diffusion kernel of this network as , where is a free parameter controlling the magnitude of diffusion, D a diagonal matrix containing node degrees, and A the adjacency matrix of the network. In our study, we follow the literature [75] to choose . Finally, we define the network similarity between two genes i and j as the corresponding element kij in the diffusion kernel.
For the semantic similarity score, we focus on 18,850 terms in the biological process domain of the gene ontology (GO, released on November 2, 2012) [76] and 186,080 annotations regarding 14,283 genes to calculate the semantic similarity between every pair of genes using the method of Resnik [77], as detailed in one of our previous studies [39].
For the sequence similarity score, we extract 20,281 protein sequences from the UniProt database (release 2012_09), use the Smith-Waterman algorithm [78] implemented in SSEARCH [54] to perform a local sequence alignment for every pair of protein sequences (with default parameters and the e-value cut-off of to 0.001), and obtain the similarity scores by dividing the negative logarithmic transformed e-values by the maximum of all transformed e-values.
For the domain similarity score, we extract from the Pfam database (version 26.0) [55] 13,672 protein families. Focusing on domains with at least 5 human proteins annotated, we obtain 1,066 domains. Then, we denote a human protein as a 1,066 dimensional binary vector, with a dimension representing the presence or absence of a domain in the protein. Finally, we calculate the similarity measure of two proteins as the cosine of the angle of the corresponding vectors.
For the pathway similarity score, we extract from the KEGG database (release 58.0) [56] 16,662 annotations of 5,951 proteins and 232 pathways. Then, we denote a protein as a 232 dimensional binary vector, with a dimension representing the presence or absence of the protein in a pathway. Finally, we calculate the similarity measure of two proteins as the cosine of the angle of the corresponding vectors.
Given a candidate gene, a set of seed genes and a type of gene functional similarity, we calculate the association score for the candidate gene according to the guilt-by-association principle by summing over similarities between the candidate gene and the seed genes. Furthermore, we convert the association score to a p-value by estimating the probability that a non-disease related gene is assigned at least the same extreme score as that of the candidate gene. Performing these steps for each of the five gene similarities, we obtain five p-values to indicate degrees that the candidate gene containing the candidate SNV is associated with the query disease.
Integration of multiple p-values by Fisher's method with dependence correction
We adopt Fisher's combined probability test [79] to combine the p-values derived above and obtain a single p-value that indicates the statistical significance that a candidate SNV is causative for the query disease. Given k p-values to be combined, denoted as , we calculate a Fisher's combination test statistic T as
It is evident that T has an asymptotic chi-squared distribution with 2k degrees of freedom when all null hypotheses are true. The final combined p-value can then be calculated accordingly.However, the above Fisher's method assumes independence of all p-values to be combined, which is obviously not true in our problem. Therefore, we further apply a dependence correction strategy [49] to adjust the combined p-value. Briefly, Yang et al. assumes that the null distribution of the Fisher's combination test statistic T follows a scaled chi-squared distribution with v degrees of freedom () when the p-values to be combined are not independent and suggests to estimate the parameters r and v using the method of moments [49]. In detail, with definitions and , the population mean and the variance of the T statistic are derived as
respectively, and the corresponding sample mean and variance are calculated as respectively. The covariance can be calculated approximately as,
with , , , , an unbiased estimator of the correlation coefficient between the two test statistics used to derive p-values and , and n the sample size in the calculation of . Furthermore, can be calculated approximately as
with being the sample correlation coefficient between the test statistics [49]. It has been shown that the maximum difference between and the unbiased estimator is less than 0.001 when , and the maximum error between and its approximation given above is no more than 0.00019 [49]. Matching the mean and variance of the population and the sample, r and v can be estimated as
The adjusted p-value can then be calculated according to the scaled chi-squared distribution () [49]. In our studies, we use a set of SNVs annotated as “Polymorphism” in the Swiss-Prot database to estimate the sample correlation coefficients ().It is possible that some data sources are absent for a candidate SNV. To deal with this missing data problem, we ignore the missing data source in the calculation of the Fisher's test statistic and the adjusted p-value. The total number of p-values to be combined will then decrease accordingly.
We further perform multiple testing corrections on the adjusted p-values by calculating q-values for candidate SNVs. Briefly, Storey et al. [35], [36] proposed to control the positive false discovery rate (pFDR, the expected proportion of false positives among all significant hypotheses, given at least one hypothesis having been rejected) in a multiple testing problem and put forward a method to calculate q-values from p-values. Numerical studies have shown the significant improvement of the test power by controlling pFDR using the this method [35], [36] instead of controlling the false discovery rate (FDR) using the traditional step-up procedure of Benjamini–Hochberg [80]. Therefore, in our study, we adopt q-values to measure the statistical significance that an SNV is causative for a query disease.
Supporting Information
Zdroje
1. CooperGM, ShendureJ (2011) Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nature Reviews Genetics 12 : 628–640.
2. ChoiM, SchollUI, JiW, LiuT, TikhonovaIR, et al. (2009) Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences 106 : 19096–19101.
3. NgSB, TurnerEH, RobertsonPD, FlygareSD, BighamAW, et al. (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461 : 272–276.
4. NgSB, BuckinghamKJ, LeeC, BighamAW, TaborHK, et al. (2010) Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 42 : 30–35.
5. BamshadMJ, NgSB, BighamAW, TaborHK, EmondMJ, et al. (2011) Exome sequencing as a tool for Mendelian disease gene discovery. Nature Reviews Genetics 12 : 745–755.
6. VissersLE, de LigtJ, GilissenC, JanssenI, SteehouwerM, et al. (2010) A de novo paradigm for mental retardation. Nature genetics 42 : 1109–1112.
7. O'RoakBJ, DeriziotisP, LeeC, VivesL, SchwartzJJ, et al. (2011) Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics 43 : 585–589.
8. GirardSL, GauthierJ, NoreauA, XiongL, ZhouS, et al. (2011) Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet 43 : 860–863.
9. BodmerW, BonillaC (2008) Common and rare variants in multifactorial susceptibility to common diseases. Nature genetics 40 : 695–701.
10. WuJ, JiangR (2013) Prediction of Deleterious Nonsynonymous Single-Nucleotide Polymorphism for Human Diseases. The Scientific World Journal 2013 Article ID 675851.
11. KongA, FriggeML, MassonG, BesenbacherS, SulemP, et al. (2012) Rate of de novo mutations and the importance of father/'s age to disease risk. Nature 488 : 471–475.
12. RivièreJ-B, van BonBW, HoischenA, KholmanskikhSS, O'RoakBJ, et al. (2012) De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nature genetics 44 : 440–444.
13. XuB, Ionita-LazaI, RoosJL, BooneB, WoodrickS, et al. (2012) De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nature genetics 44 : 1365–1369.
14. LiM-X, KwanJS, BaoS-Y, YangW, HoS-L, et al. (2013) Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies. PLoS genetics 9: e1003143.
15. KumarP, HenikoffS, NgPC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols 4 : 1073–1081.
16. AdzhubeiIA, SchmidtS, PeshkinL, RamenskyVE, GerasimovaA, et al. (2010) A method and server for predicting damaging missense mutations. Nature methods 7 : 248–249.
17. ChunS, FayJC (2009) Identification of deleterious mutations within three human genomes. Genome research 19 : 1553–1561.
18. SchwarzJM, RödelspergerC, SchuelkeM, SeelowD (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods 7 : 575–576.
19. CooperGM, StoneEA, AsimenosG, ProgramNCS, GreenED, et al. (2005) Distribution and intensity of constraint in mammalian genomic sequence. Genome research 15 : 901–913.
20. Siepel A, Pollard KS, Haussler D (2006) New methods for detecting lineage-specific selection. Springer. pp. 190–205.
21. JiangR, YangH, SunF, ChenT (2006) Searching for interpretable rules for disease mutations: a simulated annealing bump hunting strategy. BMC Bioinformatics 7 : 417.
22. YueP, MoultJ (2006) Identification and analysis of deleterious human SNPs. Journal of molecular biology 356 : 1263–1274.
23. JiangR, YangH, ZhouL, KuoC-CJ, SunF, et al. (2007) Sequence-based prioritization of nonsynonymous single-nucleotide polymorphisms for the study of disease mutations. The American Journal of Human Genetics 81 : 346–360.
24. BrombergY, RostB (2007) SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic acids research 35 : 3823–3835.
25. LehmannKV, ChenT (2013) Exploring functional variant discovery in non-coding regions with SInBaD. Nucleic Acids Res 41: e7.
26. LiuX, JianX, BoerwinkleE (2011) dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Human mutation 32 : 894–899.
27. BairochA, ApweilerR, WuCH, BarkerWC, BoeckmannB, et al. (2005) The universal protein resource (UniProt). Nucleic acids research 33: D154–D159.
28. JacqueminE, De VreeJML, CresteilD, SokalEM, SturmE, et al. (2001) The wide spectrum of multidrug resistance 3 deficiency: from neonatal cholestasis to cirrhosis of adulthood. Gastroenterology 120 : 1448–1458.
29. LucenaJ-F, HerreroJI, QuirogaJ, SangroB, Garcia-FoncillasJ, et al. (2003) A multidrug resistance 3 gene mutation causing cholelithiasis, cholestasis of pregnancy, and adulthood biliary cirrhosis. Gastroenterology 124 : 1037–1042.
30. DixonP, WeerasekeraN, LintonK, DonaldsonO, ChambersJ, et al. (2000) Heterozygous MDR3 missense mutation associated with intrahepatic cholestasis of pregnancy: evidence for a defect in protein trafficking. Human molecular genetics 9 : 1209–1217.
31. MüllenbachR, LintonK, WiltshireS, WeerasekeraN, ChambersJ, et al. (2003) ABCB4 gene sequence variation in women with intrahepatic cholestasis of pregnancy. Journal of medical genetics 40: e70–e70.
32. Pauli-MagnusC, LangT, MeierY, Zodan-MarinT, JungD, et al. (2004) Sequence analysis of bile salt export pump (ABCB11) and multidrug resistance p-glycoprotein 3 (ABCB4, MDR3) in patients with intrahepatic cholestasis of pregnancy. Pharmacogenetics and Genomics 14 : 91–102.
33. RosmorducO, HermelinB, BoellePY, ParcR, TabouryJ, et al. (2003) ABCB4 gene mutation—associated cholelithiasis in adults. Gastroenterology 125 : 452–459.
34. RosmorducO, HermelinB, PouponR (2001) MDR3 gene defect in adults with symptomatic intrahepatic and gallbladder cholesterol cholelithiasis. Gastroenterology 120 : 1459–1467.
35. StoreyJD (2002) A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 64 : 479–498.
36. StoreyJD (2003) The positive false discovery rate: A Bayesian interpretation and the q-value. Annals of Statistics 2013–2035.
37. AltshulerD, DalyM, KruglyakL (2000) Guilt by association. Nat Genet 26 : 135–137.
38. GeorgeRA, LiuJY, FengLL, Bryson-RichardsonRJ, FatkinD, et al. (2006) Analysis of protein sequence and interaction data for candidate disease gene prediction. Nucleic acids research 34: e130–e130.
39. JiangR, GanM, HeP (2011) Constructing a gene semantic similarity network for the inference of disease genes. BMC systems biology 5: S2.
40. KöhlerS, BauerS, HornD, RobinsonPN (2008) Walking the interactome for prioritization of candidate disease genes. The American Journal of Human Genetics 82 : 949–958.
41. AertsS, LambrechtsD, MaityS, Van LooP, CoessensB, et al. (2006) Gene prioritization through genomic data fusion. Nature biotechnology 24 : 537–544.
42. ChenY, HaoJ, JiangW, HeT, ZhangX, et al. (2013) Identifying potential cancer driver genes by genomic data integration. Sci Rep 3 : 3538 doi:10.1038/srep03538
43. WeissLA, EscaygA, KearneyJA, TrudeauM, MacDonaldBT, et al. (2003) Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol Psychiatry 8 : 186–194.
44. KamiyaK, KanedaM, SugawaraT, MazakiE, OkamuraN, et al. (2004) A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. The Journal of neuroscience 24 : 2690–2698.
45. LiaoY, AnttonenA-K, LiukkonenE, GailyE, MaljevicS, et al. (2010) SCN2A mutation associated with neonatal epilepsy, late-onset episodic ataxia, myoclonus, and pain. Neurology 75 : 1454–1458.
46. LiaoY, DeprezL, MaljevicS, PitschJ, ClaesL, et al. (2010) Molecular correlates of age-dependent seizures in an inherited neonatal-infantile epilepsy. Brain 133 : 1403–1414.
47. BerkovicSF, HeronSE, GiordanoL, MariniC, GuerriniR, et al. (2004) Benign familial neonatal-infantile seizures: characterization of a new sodium channelopathy. Annals of neurology 55 : 550–557.
48. SherryS, WardM-H, KholodovM, BakerJ, PhanL, et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic acids research 29 : 308–311.
49. YangJJ (2010) Distribution of Fisher's combination statistic when the tests are dependent. Journal of Statistical Computation and Simulation 80 : 1–12.
50. HamoshA, ScottAF, AmbergerJS, BocchiniCA, McKusickVA (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic acids research 33: D514–D517.
51. HaiderS, BallesterB, SmedleyD, ZhangJ, RiceP, et al. (2009) BioMart Central Portal—unified access to biological data. Nucleic acids research 37: W23–W27.
52. GibbsRA, BelmontJW, HardenbolP, WillisTD, YuF, et al. (2003) The international HapMap project. Nature 426 : 789–796.
53. SnelB, LehmannG, BorkP, HuynenMA (2000) STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic acids research 28 : 3442–3444.
54. PearsonWR (1991) Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics 11 : 635–650.
55. BatemanA, CoinL, DurbinR, FinnRD, HollichV, et al. (2004) The Pfam protein families database. Nucleic acids research 32: D138–D141.
56. KanehisaM, GotoS, KawashimaS, OkunoY, HattoriM (2004) The KEGG resource for deciphering the genome. Nucleic acids research 32: D277–D280.
57. van DrielMA, BruggemanJ, VriendG, BrunnerHG, LeunissenJA (2006) A text-mining analysis of the human phenome. European journal of human genetics 14 : 535–542.
58. WuX, LiuQ, JiangR (2009) Align human interactome with phenome to identify causative genes and networks underlying disease families. Bioinformatics 25 : 98–104.
59. BeckerKG, BarnesKC, BrightTJ, WangSA (2004) The genetic association database. Nature genetics 36 : 431–432.
60. XueY, ChenY, AyubQ, HuangN, BallEV, et al. (2012) Deleterious-and Disease-Allele Prevalence in Healthy Individuals: Insights from Current Predictions, Mutation Databases, and Population-Scale Resequencing. The American Journal of Human Genetics 91 : 1022–1032.
61. HoischenA, van BonBW, GilissenC, ArtsP, van LierB, et al. (2010) De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nature genetics 42 : 483–485.
62. LiY, BögershausenN, AlanayY, KiperPÖS, PlumeN, et al. (2011) A mutation screen in patients with Kabuki syndrome. Human genetics 130 : 715–724.
63. HoischenA, van BonBW, Rodríguez-SantiagoB, GilissenC, VissersLE, et al. (2011) De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nature genetics 43 : 729–731.
64. XuB, RoosJL, DexheimerP, BooneB, PlummerB, et al. (2011) Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nature genetics 43 : 864–868.
65. SandersSJ, MurthaMT, GuptaAR, MurdochJD, RaubesonMJ, et al. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485 : 237–241.
66. IossifovI, RonemusM, LevyD, WangZ, HakkerI, et al. (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74 : 285–299.
67. O'RoakBJ, VivesL, GirirajanS, KarakocE, KrummN, et al. (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485 : 246–250.
68. NealeBM, KouY, LiuL, Ma'ayanA, SamochaKE, et al. (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485 : 242–245.
69. IossifovI, RonemusM, LevyD, WangZ, HakkerI, et al. (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74 : 285–299.
70. AllenAS, BerkovicSF, CossetteP, DelantyN, DlugosD, et al. (2013) De novo mutations in epileptic encephalopathies. Nature 501 : 217–221.
71. de LigtJ, WillemsenMH, van BonBW, KleefstraT, YntemaHG, et al. (2012) Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367 : 1921–1929.
72. RauchA, WieczorekD, GrafE, WielandT, EndeleS, et al. (2012) Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380 : 1674–1682.
73. RobinsonPN, KöhlerS, BauerS, SeelowD, HornD, et al. (2008) The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. American journal of human genetics 83 : 610–615.
74. AltschulSF, MaddenTL, SchäfferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research 25 : 3389–3402.
75. KondorRI, LaffertyJ Diffusion kernels on graphs and other discrete input spaces; 2002. 315–322.
76. AshburnerM, BallCA, BlakeJA, BotsteinD, ButlerH, et al. (2000) Gene Ontology: tool for the unification of biology. Nature genetics 25 : 25–29.
77. ResnikP (2011) Semantic similarity in a taxonomy: An information-based measure and its application to problems of ambiguity in natural language. Journal of Artificial Intelligence Research 11 : 95–130.
78. SmithTF, WatermanMS (1981) Identification of common molecular subsequences. Journal of molecular biology 147 : 195–197.
79. FisherRA, GenetikerS, GeneticianS, BritainG, GénéticienS (1970) Statistical methods for research workers: Oliver and Boyd Edinburgh.
80. BenjaminiY, HochbergY (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 57 : 289–300.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání









