-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
Elevated homocysteine (tHcy) is strongly associated with risk for common disorders such as stroke, cardiovascular disease and Alzheimer disease. Lowering tHcy levels has proven to have variable success in reducing clinical risk, so the question remains, “Are we correctly targeting these disorders by lowering tHcy?” Understanding folate one-carbon metabolism pathway (FOCM) genetic variation will aid us in developing new targets for therapy. The FOCM is essential in regulation of the epigenome, which controls genes in ways beyond nucleotide sequence. We present data generated from stroke-only and general populations where we identify strong association of genetic risk factors for variation in one-carbon metabolism function, characterized by the post-methionine load test. We show that GNMT harbors genetic and epigenetic differences that influence gene function, which may have downstream effects on the epigenome of the cell, affecting disease risk. We developed a genetic risk score that predicts post-methionine load homocysteine levels that may be useful in clinic. Finally, we identified a novel association between ischemic stroke and ALDH1L1, which emphasizes the clinical importance of this work. Our results highlight the importance of a concerted effort to target the FOCM (beyond tHcy) and parallel pathways in future pharmacogenetic work using the genetic variation we describe here.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004214
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004214Summary
Elevated homocysteine (tHcy) is strongly associated with risk for common disorders such as stroke, cardiovascular disease and Alzheimer disease. Lowering tHcy levels has proven to have variable success in reducing clinical risk, so the question remains, “Are we correctly targeting these disorders by lowering tHcy?” Understanding folate one-carbon metabolism pathway (FOCM) genetic variation will aid us in developing new targets for therapy. The FOCM is essential in regulation of the epigenome, which controls genes in ways beyond nucleotide sequence. We present data generated from stroke-only and general populations where we identify strong association of genetic risk factors for variation in one-carbon metabolism function, characterized by the post-methionine load test. We show that GNMT harbors genetic and epigenetic differences that influence gene function, which may have downstream effects on the epigenome of the cell, affecting disease risk. We developed a genetic risk score that predicts post-methionine load homocysteine levels that may be useful in clinic. Finally, we identified a novel association between ischemic stroke and ALDH1L1, which emphasizes the clinical importance of this work. Our results highlight the importance of a concerted effort to target the FOCM (beyond tHcy) and parallel pathways in future pharmacogenetic work using the genetic variation we describe here.
Introduction
As the fourth leading cause of death and the leading cause of disability in American adults, stroke constitutes a major public health burden. Epidemiological data consistently demonstrate an association between elevated plasma homocysteine (tHcy) and increased risk for stroke [1], cardiovascular disease [2], and dementia [3], but clinical trials of interventions to lower homocysteine have failed to demonstrate global benefit, with B12 supplementation helping to reduce risk only in subsets of the populations studied [4]–[6]. Collectively, these data support a more complicated relationship than simple biomarker and disease risk and indicate the need for new targets for risk reducing therapies. This begs the question, “Have we already missed the target of greatest clinical benefit by the time we lower homocysteine levels?” The folate one-carbon metabolism pathway (FOCM) is not only involved in the regulation of homocysteine, methionine and B-vitamin levels but also the methylation of proteins, histones, DNA and RNA. To this end, the demethylation of S-adenosyl-methionine, which gives rise to S-adenosyl-homocysteine, is the sole source of de novo methyl groups for the cell. Dysregulation of this step in the FOCM could have broad implications on many cellular processes, including risk for stroke and cardiovascular disease.
The post-methionine load test is a more sensitive tool for diagnosing hyperhomocysteinemia than circulating plasma homocysteine levels [7]–[10]. Additionally, “ΔPOST”, or the difference in tHcy levels before and after methionine loading in the clinic, gives a measurement of one's ability to convert methionine to homocysteine in real time and likely reflects methyl group availability in the cell. We utilized this test to analyze genetic determinants of methionine metabolism and how these differences between individuals may be functionally regulated.
Here we present a genomic, genetic and epigenetic investigation into the regulation of methionine metabolism in the Vitamin Intervention for Stroke Prevention Trial (VISP) and the Framingham Heart Study (FHS). We first present genome-wide association (GWAS) data linking five loci to differences in ability to convert methionine to homocysteine. Strikingly, all of the most significant genes identified within these loci are members of the same pathway (FOCM), a feature rarely observed in GWAS studies.
We observed haplotype differences at the GNMT [MIM 606628] locus, our most significant GWAS finding, and devised a scheme to test methionine loading in vitro based on GNMT genotype. Additionally, we have shown epigenetic regulation of the GNMT promoter, based on a CpG-SNP rs11752813, which likely contributes to GNMT transcription and methionine metabolism. These data may one day contribute to identification of new targets for stroke and cardiovascular disease prevention as well as other complex diseases where epigenetics play a role.
Results
Framingham Heart Study (FHS) and Vitamin Intervention for Stroke Prevention Trial (VISP) cohorts
The FHS cohort is a community based longitudinal study to determine the risk for cardiovascular disease and is comprised of randomly recruited participants and their family members in the town of Framingham, Massachusetts (Table 1). VISP was a multi-center, double-blind, randomized, controlled clinical trial designed to determine if vitamin supplementation reduced recurrent cerebral infarction, nonfatal myocardial infarction or mortality and is made up of unrelated individuals. The VISP cohort has a higher proportion of men when compared to the FHS, which is not surprising when considering the VISP participants have all had a stroke (Table 1). Likewise, VISP also has a greater percentage of diabetics and hypertensive individuals (Table 1). The VISP cohort consists of individuals in the top quartile of circulating tHCY levels, which was part of the recruitment requirements; whereas FHS is made up of a normal distribution of tHCY levels (Table 1). FHS participants have higher vitamin B6, B12, and folate levels on average than VISP participants. BMI and smoking status are approximately the same between VISP and FHS.
Tab. 1. Summary statistics for Framingham Heart Study (FHS) and Vitamin Intervention for Stroke Prevention (VISP) subjects. 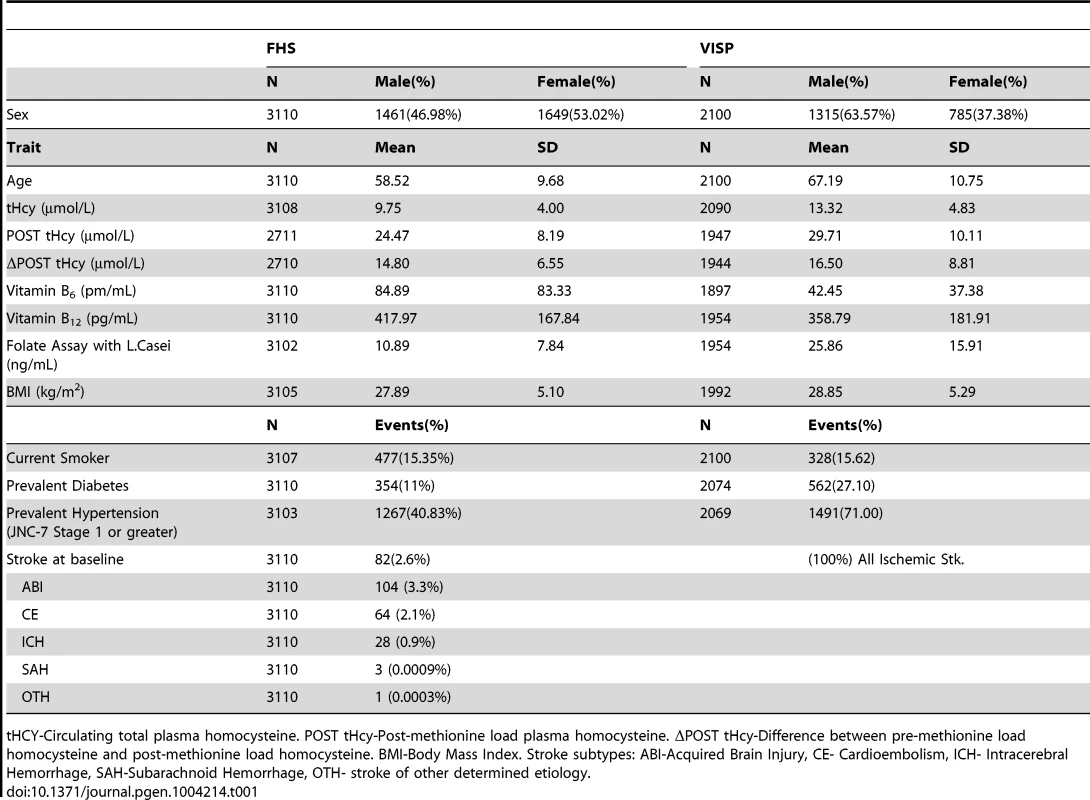
tHCY-Circulating total plasma homocysteine. POST tHcy-Post-methionine load plasma homocysteine. ΔPOST tHcy-Difference between pre-methionine load homocysteine and post-methionine load homocysteine. BMI-Body Mass Index. Stroke subtypes: ABI-Acquired Brain Injury, CE- Cardioembolism, ICH- Intracerebral Hemorrhage, SAH-Subarachnoid Hemorrhage, OTH- stroke of other determined etiology. The VISP study consisted of 1725 (82.1%) individuals of European descent, 258 (12.2%) individuals from African descent and 117 (5.6%) individuals of other ancestral populations. All VISP participants are unrelated. FHS samples are primarily Caucasian. In FHS the 3110 individuals contributing to GWAS belong to 1055 families with extended family size ranging from 1 to 140. In FHS, 1772 individuals have at least one blood relative in the family, 279 individuals have at least one first degree relative, 278 have at least one second degree relative, and 586 have at least one third degree relative.
Association of Folate One-Carbon Metabolism genes with methionine metabolism
In a GWAS of 2,710 persons from the FHS study, five loci (GNMT, CBS [MIM 613381], CPS1 [MIM 608307], ALDH1L1 [MIM 600249] and PSPH [MIM 172480]) reached our pre-determined genome-wide significance threshold of 5×10−8 for the ΔPOST phenotype. These findings were confirmed in the VISP study sample of 2,100 persons for whom two of these loci (GNMT and CBS) independently reached genome wide significance (Figure S1, S2 and Table S1). The results of a sample size-weighted meta-analysis consisting of 4,810 subjects from both FHS and VISP confirm and strengthen the independent GWAS findings (Figure 1 and Table 2). Strikingly, all five loci identified are involved in the FOCM pathway (Figure S3). The most significant association was with GNMT (rs9296404, p = 1.60×10−63) located on 6p21.1, a region associated with large artery atherosclerotic stroke [11], [12].
Fig. 1. Meta-analysis and chromosome 2, 3, 6, 7 and 21 regional association of single nucleotide polymorphisms (SNPs) for ΔPOST in both VISP and FHS cohorts. 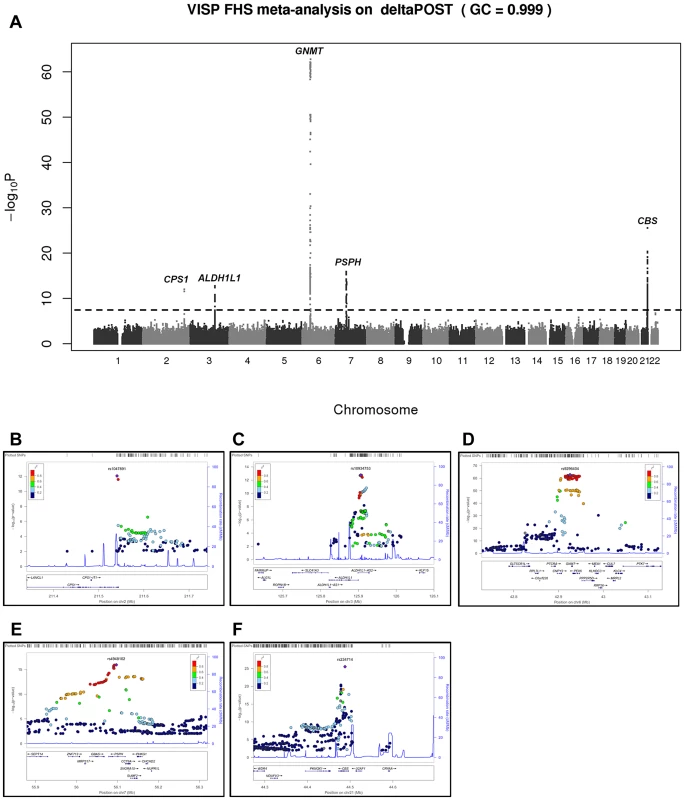
A sample size-weighted meta-analysis was used. (A) Manhattan plot of meta-analysis association results for the combined VISP and FHS samples. Association p-values are noted on the Y axis (-log10 P value), with points above the dashed line indicating SNPs reaching or exceeding genome-wide significance (P≤5×10−8). (B–F) Locus Zoom plots showing the regional association of chromosomes 2, 3, 6, 7 and 21. Y-axis shows –log10 p-value≤0.01. X-axis shows Mb position on each chromosome. Each circle represents an independent SNP and color shading represents r2 values. Tab. 2. Most significant meta-analysis SNPs. 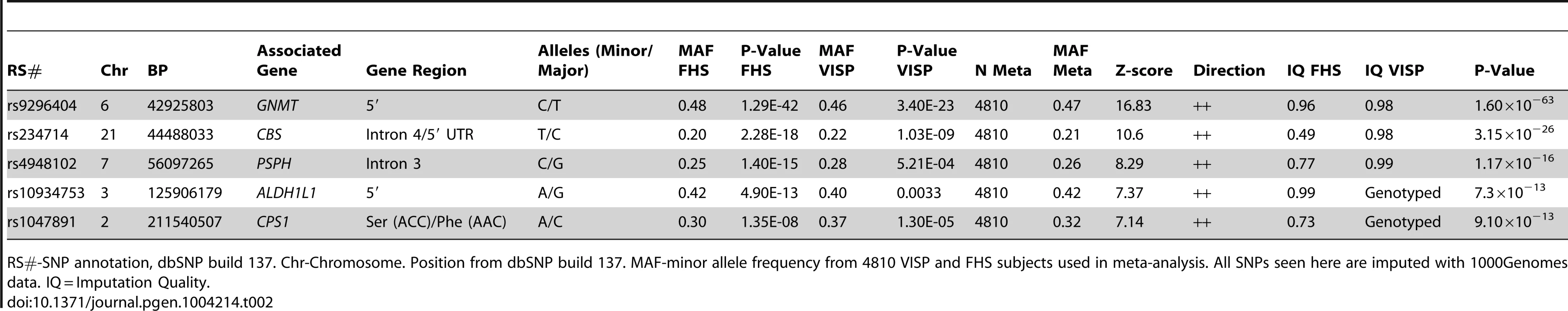
RS#-SNP annotation, dbSNP build 137. Chr-Chromosome. Position from dbSNP build 137. MAF-minor allele frequency from 4810 VISP and FHS subjects used in meta-analysis. All SNPs seen here are imputed with 1000Genomes data. IQ = Imputation Quality. Haplotype analysis of GNMT locus
Using ten genotyped single nucleotide polymorphisms (SNPs) on chromosome 6 in the GNMT region, which were significantly-associated with ΔPOST in the VISP population, (Figure S1A), we conducted a haplotype analyses (Haploview software) [13]. Two major haplotypes emerged, encompassing ∼81% of the individuals in the VISP population (n = 2,100) (Figure 2A,B) and corresponding to a high methionine metabolizing haplotype (ΔPOST = 19.4 µmol/L) and a low methionine metabolizing haplotype (ΔPOST = 14.5 µmol/L) (Figure 2C). One SNP, rs10948059, which is a genotyped and located in the GNMT promoter, captures 100% of alleles with a mean max r2 of 0.722 (range 0.512–0.850). These data suggest functional differences in the GNMT gene impact an individual's ability to metabolize dietary methionine. The lack of a disruptive coding mutation identified by GWAS, or in sequencing of GNMT in 24 high and 24 low methionine metabolizers (data not shown), and the expectation that a higher rate of transcription of the GNMT gene should lead to higher tHcy levels, suggest a regulatory mechanism for the differences in ΔPOST rather than protein dysfunction. This metabolic difference mediated by genetic variation is of functional significance in both the general population (FHS) and a population with tHcy above the 25th percentile as required by the inclusion criteria for the clinical trial (VISP).
Fig. 2. Haplotype analysis of Chr6 SNPs significantly-associated with ΔPOST in the VISP cohort. 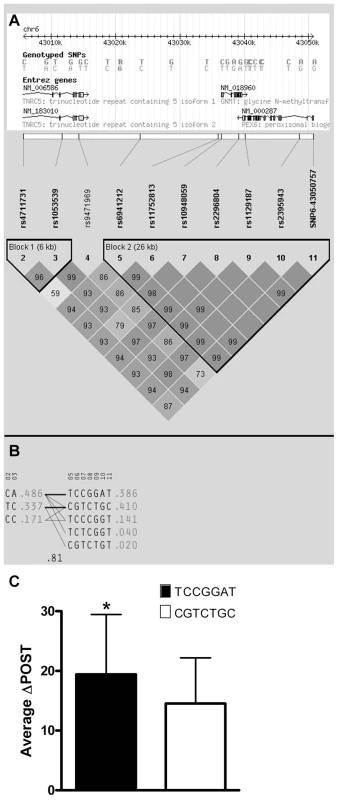
Haplotype analyses of the 10 most-associated SNPs, which are all genome wide significant, from chromosome 6, were performed using Haploview version 4.2 [13]. (A) Shows results for the chromosome 6 genomic haplotype structure in VISP, encompassing the GNMT gene. Genetic coordinates, Entrez gene structure, haplotype blocks and linkage disequilibrium (LD) pattern between SNPs are shown. Within LD pattern, r2 values are shown and are represented by shading. The darker the shading the closer the r2 value is to 1.0. All SNPs assessed were genotyped. (B) Shows haplotypes generated by Haploview for haplotype blocks 1 and 2. Haplotype block 2 is characterized by two major haplotypes, which account for 80% of haplotypes observed. (C) Shows the mean ΔPOST values in the VISP sample for each of the 2 major haplotype block 2 haplotypes. (*) p≤0.001 by student's T-test; error bars represent standard deviation from the mean (SD). Promoter analysis of GNMT
The known GNMT promoter [14] from the high methionine metabolizing haplotype and the low methionine metabolizing haplotype were cloned giving rise to GNMTΔHighLuc and GNMTΔLowLuc constructs (Sequence alignments in Figure S4). GNMT is most highly expressed in the liver; therefore HepG2 cells were used to test promoter activity in the two haplotype groups. There was a ∼30% difference in gene promoter activity between GNMTΔHighLuc and GNMTΔLowLuc constructs (Figure 3A), when cultured with L-methionine, which correlates with the differences seen between the average ΔPOST levels in our haplotype analysis (Figure 2C).
Fig. 3. GNMT promoter analysis of major haplotype groups. 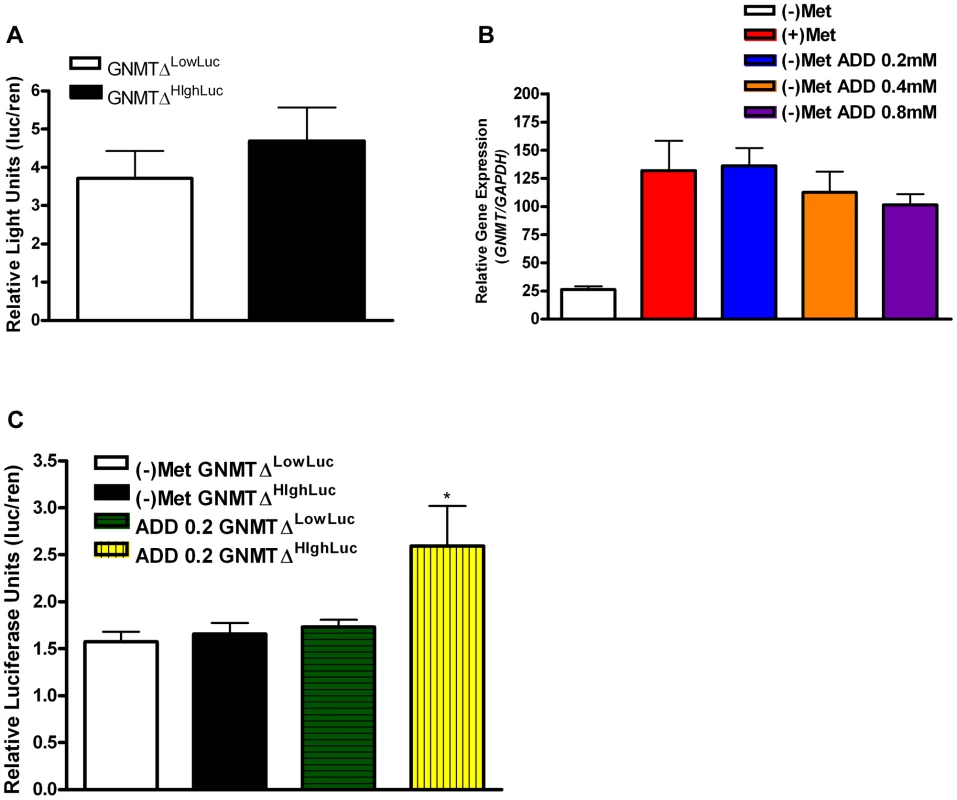
(A) Histogram shows mean luciferase activity of the high-methionine-metabolizing haplotype (TCCGGAT) and the low-methionine-metabolizing haplotype (CGTCTGC) represented by constructs GNMTΔHighLuc and GNMTΔLowLuc cultured in standard DMEM with L-methionine (0.2 mM). (B) Shows GNMT qPCR analysis in HepG2 cells. Culturing conditions are as follows: (+)Met = standard complete DMEM cultured for 48 hours, (−)Met = complete DMEM without L-Methionine culture for 48 hours, (−)Met ADD 0.2 = DMEM without L-Methionine culture for 24 hours, addition of L-Methionine at 0.2 mM for 24 hours, (−)Met ADD 0.4 = DMEM without L-Methionine culture for 24 hours, addition of L-Methionine at 0.4 mM for 24 hours, (−)Met ADD 0.8 = DMEM without L-Methionine culture for 24 hours, addition of L-Methionine at 0.8 mM for 24 hours. (C) (−)Met GNMTΔHighLuc and (−)Met GNMTΔLowLuc represent HepG2 cells transfected with GNMTΔHighLuc and GNMTΔLowLuc constructs and cultured without L-methionine for 48 hours. ADD 0.2 GNMTΔLowLuc and ADD 0.2 GNMTΔHighLuc represent HepG2 cells transfected with GNMTΔHighLuc and GNMTΔLowLuc constructs cultured without L-methionine for 24 hours and with 0.2 mM L-methionine for 24 hours. * P<0.05 by students t-test. N = 3 biological replicates for all analyses. Error bars represent standard deviation from the mean (SD). The quantitative trait, ΔPOST, is dependent on methionine dosing, therefore we starved HepG2 cells of L-methionine for 24 hours and then treated them with L-methionine for 24 hours. As seen in Figure 3C, the GNMTΔHighLuc construct responded to methionine starvation and treatment with ∼2× greater activity than the GNMTΔLowLuc construct. Above standard L-methionine culturing conditions (0.2 mM) a feedback mechanism appears to be induced, which reduces GNMT expression (Figure 3B). These data suggest a mechanism by which elevated levels of tHcy may arise.
Epigenetic analysis of C/G SNP rs11752813
Given the differences in promoter activity seen in Figure 3, we sought to identify functional variants that may play a role transcriptional activity. SNP rs11752813 was significantly associated in our meta-analysis (Figure 1A, p = 7.99×10−32), and either creates or eliminates a CpG site that can be methylated depending on genotype (rs11752813 and flanking sequence: C(C/G)A). The high methionine metabolizing haplotype (Figure 2) harbors the C/C genotype (known as GNMTΔHighLuc in Figure 3) at rs11752813, and the low methionine metabolizing haplotype (Figure 2) harbors the G/G genotype (known as GNMTΔLowLuc in Figure 3) at rs11752813. The presence of a “G” at the rs11752813 locus creates a CpG site while the presence of a “C” eliminates this CpG site. Analysis of ΔPOST values in the VISP (n = 2100) study shows that the individuals that harbor the “G” genotype at rs11752813 have significantly lower ΔPOST on average, indicating a less active GNMT gene (Figure 4A). Bisulfite pyrosequencing of the rs11752813 locus show that the G/G genotype can be methylated whereas the C/C genotype is not methylated (Figure 4B). These results are consistent with the central dogma of DNA methylation that only CpG sites can be methylated. Finally, 23 individuals harboring the G/G genotype at rs11752813 were bisulfite pyrosequenced and percent methylation status was plotted against individual ΔPOST values (Figure 4C). As seen in Figure 4C, even between individuals with the G/G genotype there is a strong correlation between percent methylation and ΔPOST values. These data indicate that the G/G genotype at rs11752813 creates a closed chromatin state that inhibits GNMT transcription and methionine metabolism.
Fig. 4. Epigenetic Evaluation of rs11752813. 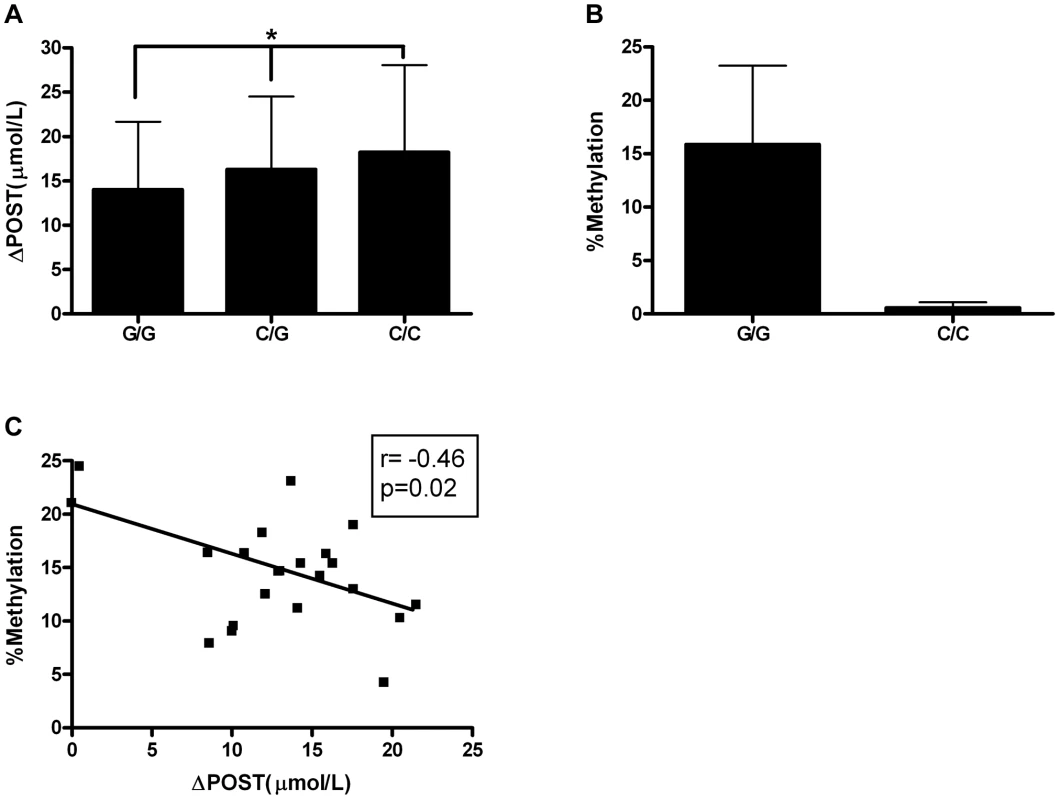
(A) Average ΔPOST (µmol/L) in VISP trial based on rs11752813 genotype. (*) p<0.001 by one-way ANOVA and all genotypes are significantly different by students t-test. (B) Percent methylation analyzed by bisulfite pyrosequencing of n = 23 individuals (G/G) and n = 6 individuals (C/C). (C) Linear regression analysis of percent methylation of rs11752813 based on genotype and ΔPOST (µmol/L). Genetic risk score of most significant ΔPOST variants
We next investigated cumulative effects of ΔPOST risk variants by generating a combined genetic risk score using the most significant SNPs from Table 2. Risk scores are normally distributed in both VISP and FHS (Figure 5A,C). As the risk variant load increases, the average ΔPOST levels in the VISP and FHS samples increases (Figure 5B,D). This score explains 13% of the variability of ΔPOST in FHS and 6.3% of the variability in VISP (Table 3). Because the SNPs used in this analysis are all imputed, for both the VISP and FHS studies, we repeated the analysis utilizing the most significant genotyped SNPs from each locus. This analysis yielded similar results indicating that using imputed SNPs for this risk score does not distort the analysis (Figure S5).
Fig. 5. Risk score in FHS and VISP studies. 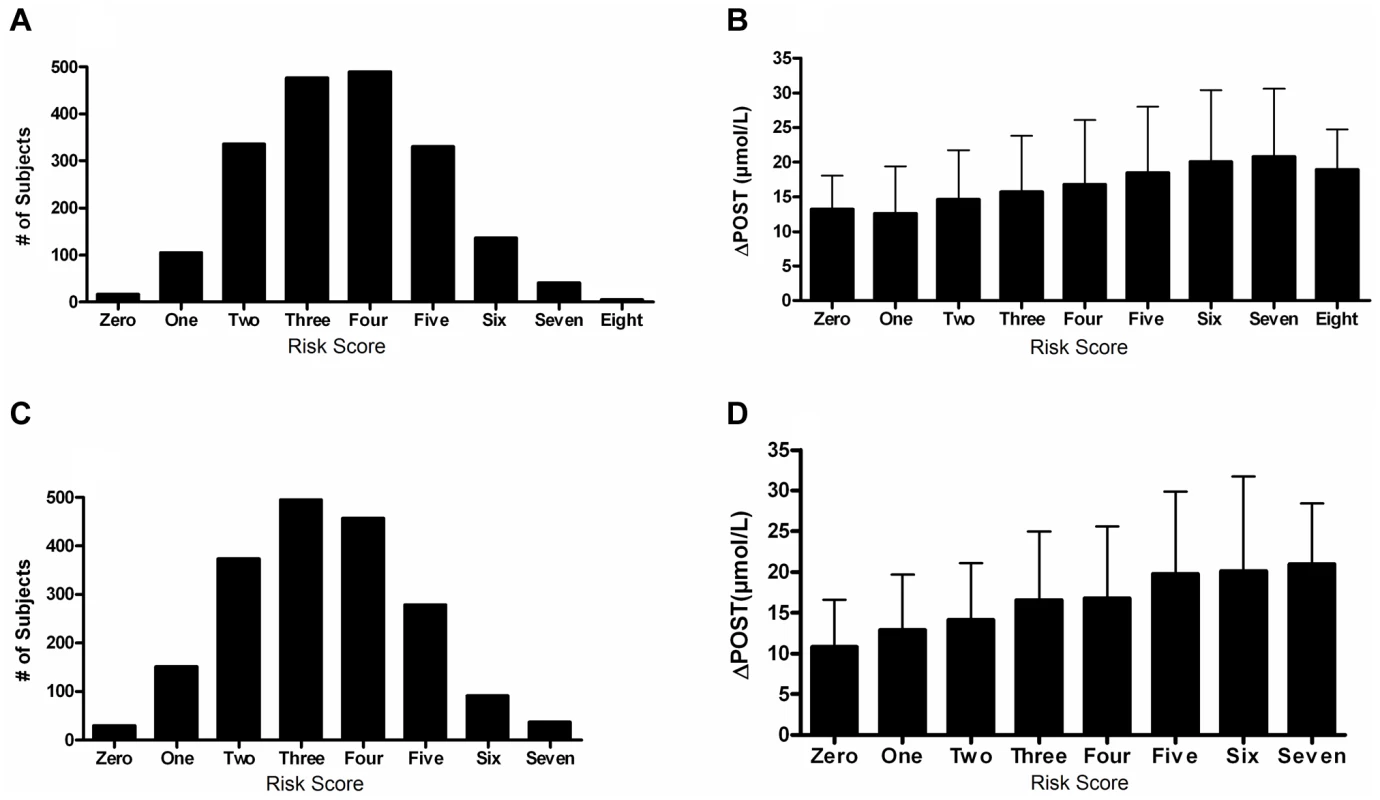
(A) Distribution of risk scores among the FHS sample shows a normal distribution. Y-axis represents the number of individuals who have the given risk score seen on the x-axis. (B) X-axis represents the number of risk variants per subject in FHS. Y-axis represents the average ΔPOST value for each group containing a specific risk score. (C) Distribution of risk scores among the VISP sample shows a normal distribution. Y-axis represents the number of individuals who have the given risk score seen on the x-axis. (D) X-axis represents the number of risk variants per subject in VISP; Y-axis represents the average ΔPOST value for each group containing a specific risk score. Risk variants considered were SNPs at each of the 5 loci most significantly associated with ΔPOST in the meta-analysis. For each SNP a score of 0 was applied for homozygous non-risk variant, 1 for heterozygous at risk variant and 2 for homozygous at risk variant, derived from dosage values of 1000genomes imputation. Tab. 3. Variance and effect size explained by risk score. 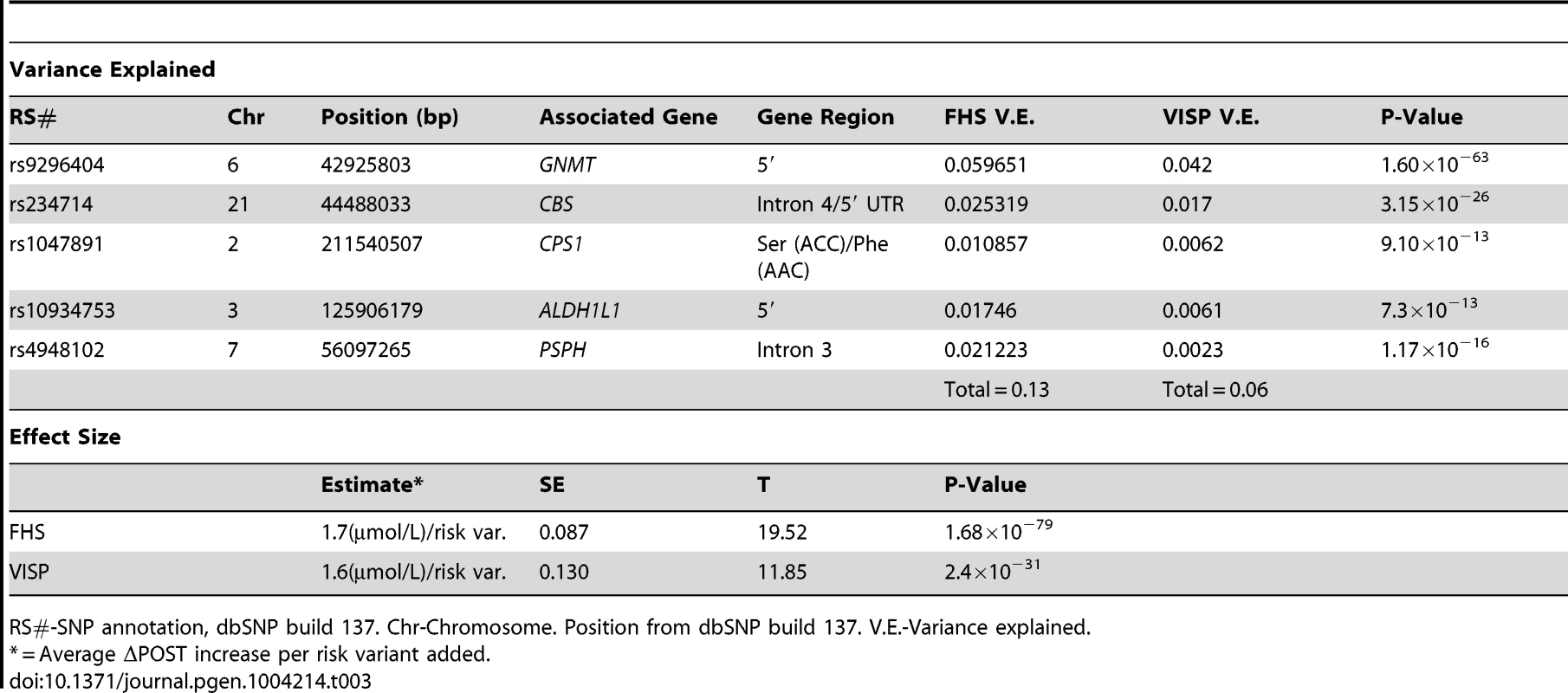
RS#-SNP annotation, dbSNP build 137. Chr-Chromosome. Position from dbSNP build 137. V.E.-Variance explained. ALDH1L1 is associated with ischemic stroke in the Framingham Heart Study
Importantly, when interrogating the most significant five SNPs associated with ΔPOST, we also identified an association between the aldehyde dehydrogenase 1 family member L1 gene (ALDH1L1) and incident ischemic stroke in the FHS cohort (rs10934753, hazard ratio = 1.26, p = 0.015, n = 168 cases 4008 controls, analyses adjusted for age, sex and family relationships). The protein encoded by ALDH1L1 converts 10-formyltetrahydrofolate to tetrahydrofolate and is an essential component of the FOCM pathway. These results provide a new and significant link between the FOCM pathway and risk of initial ischemic stroke. It is important to note that the VISP population consists of exclusively ischemic stroke patients examined for recurrent stroke over a 2 year period; rs10934753 was not associated with recurrent stroke. Additionally, we did not observe an association of genetic variation in GNMT or the other 3 loci with incident ischemic stroke in FHS but our sample has limited power to detect moderate effect size (e.g. power ranges from 20–40% to detect a hazard ratio of 1.20 for a variant with minor allele frequency ranging from 0.1–0.5).
Discussion
Elevated tHcy has long been associated with increased risk for stroke and cardiovascular disease but to date functional evidence for the driving genetic forces behind elevated tHcy levels have only been attributable, in part, to dysfunction in the methylenetetrahydrofolate reductase gene (MTHFR [MIM 607093]) and CBS genes [15]–[18]. MTHFR, which also participates in the FOCM pathway, is tightly related to all the genes products identified in our study and has been implicated in susceptibility to vascular disease, neural tube defects, colon cancer and acute leukemia [19]–[24]. It is interesting to note that a prior GWAS in the individual FHS and VISP cohorts or in a meta-analysis yielded no significant results for baseline tHcy alone. This highlights the usefulness of the post-methionine load test in the diagnosis of hyperhomocysteinemia as well as the fact that we can specifically detect genetic variations that lead to differential methionine metabolism.
In the current study we followed up with a functional evaluation of our GWAS findings. These independent GWAS analyses and a meta-analysis of FHS and VISP find the GNMT locus as the top result. However, there were differences in the GWAS results from FHS and VISP are likely attributable to two factors: one being power (FHS consists of 610 more individuals), and two, VISP is a more homogenous population than FHS lacking a normal distribution of tHcy. Our functional studies start with the GNMT gene as it contributes to the majority of the variance in both VISP and FHS. The GNMT association makes biological sense given that GNMT catalyses the conversion of S-adenosyl-methionine (SAM) to S-adenosyl-homocysteine (SAH), using SAM as the methyl donor [25], and affects global cellular epigenetic status as the sole source of methyl groups for the cell including those used in DNA, histone, protein and RNA modifications. Additionally, it is known that global hypomethylation is seen in atherosclerosis [26], and we suspect that variation in GNMT could affect risk status. Further, hyperhomocysteinemia is a risk factor for stroke and cardiovascular disease, and these data indicate GNMT may represent a new pharmacogenetic target for reducing stroke risk. It is our assertion that targeting the FOCM pathway before methionine is converted to homocysteine may allow us to modulate parallel pathways, such as DNA and histone methylation, which directly impact stroke risk. A recent review by Krishna et al. [27] describes in detail a “tHcy memory effect” that may alter the epigenetic state of the cell and promote deleterious chances after tHcy is lowered. This further strengthens the argument to identify genetic risk, through use of our risk score, and examine parallel targets for therapy.
We did not observe any of the deleterious GNMT mutations associated with Glycine N-Methyltransferase Deficiency [MIM 606664] (characterized by elevated levels of plasma S-adenosylmethionine and normal plasma sarcosine) in sequencing of GNMT in 24 high and 24 low methionine metabolizers. Additionally, a lookup of large GWAS studies of cardiovascular and cerebrovascular disease, identified many of our most significant chromosome 6 findings in a meta-analysis for blood lipids [28], with rs2274517 being the most significant result (p = 1.37×10−4), suggesting that GNMT may play a broader role in risk traits for CVD beyond tHcy measures.
Our functional studies support the role of GNMT in variation of methionine metabolism. The consistent and biologically plausible results from the individual GWAS and meta-analysis emphasize that the other significant associations observed in the FOCM pathway cannot be ignored. The cystathionine-beta-synthase (CBS) gene has been associated with stroke [20] and methionine metabolism [29]. Additionally, CBS mutations are associated with homocystinuria, iridodonesis and agitated motion of the iris [MIM 236200] [30]–[33]. Within the carbamoyl phosphate synthetase one (CPS1) gene, ΔPOST rs1047891 was found to be associated with a missense Ser(ACC)/Phe(AAC) mutation (p = 9.10×10−13). These findings are related to a sex-specific association of CPS1 with tHcy and women, performed in the Woman's Health Genome Study [34] and a Filipino population [35]. We meta-analyzed ΔPOST, rather than tHcy, and included both men (54%) and women (46%). Phosphoserine phosphatase (PSPH) mutations have been associated with phosphoserine phosphatase deficiency [MIM 614023], which results in pre - and postnatal growth retardation, moderate psychomotor retardation, and facial features suggestive of Williams syndrome [36], [37].
Taken together, these data present a new link to the genetics of the FOCM pathway with methionine metabolism both in stroke and non-stroke populations. Because of the impact that the FOCM pathway has on the biology of the cell, including overall epigenetic state and DNA methylation, gluconeogenesis, and DNA repair, understanding how individual genetic composition impacts this pathway is essential. The FOCM has also been implicated in many aspects of human health and the work presented may be relevant to several key biological mechanisms, affecting tumorogenesis [38], B-vitamin utilization [39], as well as cardiovascular and cerebrovascular disease risk. Additionally, it is necessary to repeat these analyses in studies of different ethnicities as both FHS and VISP are comprised of mainly individuals of European descent.
While a direct link between tHcy levels and stroke and cardiovascular disease remains debated, we have shown that understanding sequence variation in the FOCM pathway may provide a link to functional differences in the population, that in turn tie one carbon metabolism to a broad range of disease risk factors. Additionally, because tHcy-lowering therapies have had variable success in reducing stroke risk in subtype populations [6], [40], [41] and have in some cases been harmful [42], we believe that understanding how these genetic variants impact the overall FOCM and related pathways is essential to understanding the pathogenesis of stroke. Methionine metabolism provides a first clue to the impact of this pathway on cell biology.
Materials and Methods
Ethics statement
All human research was approved by the relevant institutional review boards, and conducted according to the Declaration of Helsinki. The Framingham Heart Study protocol was approved by the institutional review board (IRB) of the Boston University School of Medicine and all participants provided written, informed consent. The VISP study protocol was approved by the IRBs of the Wake Forest University School of Medicine (coordinating center) and University of North Carolina Chapel Hill School of Medicine (statistical center). The local IRB for each of the individual recruiting sites approved the VISP protocol and all participants provided written, informed consent. The Genomics and Randomized Trial Network (GARNET) analysis of the VISP data was approved by University of Virginia School of Medicine IRB.
Genome wide analysis of Framingham Heart Study
FHS started in 1948 for evaluation of cardiovascular diseases and risk factors [43]. In 1971, 5124 children of the original cohort, and spouses of these children, referred to as Offspring cohort, were enrolled and have been examined approximately every four years [44]. Genotyping was performed on the Affymetrix 500K mapping array and the Affymetrix 50K supplemental array. Circulating homocysteine levels were measured on 3465 Offspring participants (N = 3464 with tHcy and N = 2999 with POST) during examination cycle 6 (1995–1998). The study sample for GWAS consists of a subset of 3110 individuals with at least one phenotype and GWAS data (N = 3108 with tHCY, N = 2711 with POST, N = 2710 with Δ POST). The sample used to examine the association between the SNPs identified in GWAS of homocysteine phenotypes and incident stroke includes 4176 original cohort and Offspring individuals (N stroke = 200 stroke; N ischemic stroke = 168).
Homocysteine measurement and stroke classification
Plasma tHcy levels were measured using high-performance liquid chromatography with fluorescence detection [45].
Clinical stroke was defined as rapidly developing signs of focal neurologic disturbance of presumed vascular etiology lasting more than 24 hours. Additional details of stroke classification and diagnosis can be found in prior publications [45]–[48].
Imputation and statistical analyses
Imputation of about 11 million 1000 Genomes SNPs (1000G Phase I Integrated Release Version 3 Haplotypes: 2010–11 data freeze, 2012-03-14 haplotypes) was performed using MACH version 1.0.16 (http://www.sph.umich.edu/csg/abecasis/MACH/) based on 412,053 good quality SNPs (excluded SNPs were characterized by call rate <97%, pHWE<1E-6, Mishap p<1e-9, >100 Mendel errors, MAF<1%).
Prior to association analysis, homocysteine phenotypes were normalized by replacing its observed value with the corresponding quantile under normal distribution. For GWAS, linear mixed effects models were fitted with the transformed phenotypes as dependent variables, individual SNP genotype as a fixed effect, and person specific random effects with correlation coefficient between two individuals being twice their kinship coefficient to account for correlation within extended families [49]. FHS GWAS has adjusted for age, sex and first 10 eigenstrat principal components in the linear mixed effects mode (Table S2). Cox proportional hazard model with a robust variance to account for familial relationship was fitted to relate SNPs identified in GWAS with stroke outcomes [50].
VISP Imputation
Imputation was performed using all SNPs and samples passing basic quality filters. In brief, SNPs were selected using the recommended composite quality filter that emerged from the genotype data cleaning process. Samples were selected to have an overall missing call rate <2%, while certain sample-chromosome combinations were also excluded where a gross chromosomal anomaly was detected or when the chromosome-specific missing call rate was >5%. These study data were imputed to a phase 1 interim release from the 1000 Genomes (1000G) Project [51]. Imputation target variants were defined as those with MAF≥0.005 across all 629 1000G samples.
Imputation was carried out using BEAGLE imputation software [52] (v3.3.1) for chromosomes 1–22 and the X chromosome. The imputed dataset contained total 7,500,450 variants; 766,577 of which (10.2%) were observed from the array genotyping. In addition to the primary imputation analysis, additional imputations were run on chromosome 22 and the X chromosome, masking a random 10% of observed SNPs to empirically assess imputation quality. The squared correlation between observed and imputed allelic dosages (dosage r2) was used to summarize the imputation quality. The median dosage r2 was 0.933 for chromosome 22 masked SNPs and 0.930 for X chromosome masked SNPs. The imputed dataset, along with a detailed report on imputation methodology, is available through the authorized access portion of the VISP dbGaP posting.
Genotyping was performed on the Illumina HumanOmni1-Quad-v1 array (Illumina, Inc.) at the Center for Inherited Disease Research, Johns Hopkins University. The genome-wide association analysis was conducted using PLINK v1.0.7. Multivariate linear regression model was used to test correlation of quantitative traits and SNP markers. Using the KING software, the top 10 principle components were derived from genotype data [53] and subsequently used to adjust for population heterogeneity, in addition, age and gender were also included as covariates in the model. The VISP population consists of genetically confirmed unrelated individuals and no adjustments were made to the analysis for relatedness. To normalize phenotypic traits, inverse normal transformation was applied to values of POST and ΔPOST. The same regression model was employed to perform association tests between the phenotype and expected allele counts.
Meta-analysis of Δ post methionine load test homocysteine levels
Meta-analysis of the 2100 VISP and 3110 FHS cohorts was conducted using the METAL software [54]. The sample size of each study was used as weight, and the sign of the beta value of each SNP coded allele was used as the direction for association (Table S3).
Regional association plots
Figure 1 regional association plots were created using the locus zoom “plot your own data” function (https://statgen.sph.umich.edu/locuszoom/genform.php?type=yourdata). Plots were created utilizing the genome build/LD population hg19/1000 Genomes Mar 2012 EUR.
Cell culture
Unless otherwise noted, HepG2 cell lines were cultured in complete media containing high glucose Dulbecco's Modified Eagle's Medium (DMEM) (Invitrogen) with 10% (v/v) FBS, 2 mM L-glutamine supplemented with 1× nonessential amino acids, 1 mM sodium pyruvate (Invitrogen) and 1X antibiotic-antimycotic (Invitrogen). Cells were maintained at 37°C in a 5% CO2 incubator.
In vitro L-methionine treatment
HepG2 cells were culture in complete media as described above for 24 hrs in 25 cm2 tissue culture flasks or 6 well culture plates. For methionine starvation, after 24 hrs media was removed and replaced with either complete media or complete media lacking L-methionine. After 24 hrs, cells starved of methionine were supplemented with L-methionine (Ameresco) at concentrations of (0.2 mM, 0.4 mM, 0.8 mM).
Real time qPCR
RNA isolation was conducted using the Qiagen RNeasy kit according to standard manufacture's protocols. cDNA synthesis was conducted using 1 ug of total RNA and the Verso cDNA Synthesis Kit (Fisher Scientific). A 3∶1 mix (v/v) of random hexamers and anchored oligo-dT was used following standard theromocycling conditions. For GNMT quantitative real-time PCR, Taqman MGB probes and primers were used (Hs002219089) (Applied Biosystems). All samples were run in triplicate in 10 µl reaction volumes. PCR conditions were the default settings of the ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) using the standard curve setting to achieve raw data, which was analyzed in Microsoft Excel. The cycle threshold (Ct) was determined during the geometric phase of the PCR amplification plots as automatically set by the 7900 software. Relative differences in transcript levels were quantified using the ΔΔCt method with GAPDH [MIM 138400] (probe 4333764F) mRNA as an endogenous control.
Creation of luciferase plasmids
GNMT promoters were PCR amplified using primers (Forward: 5′-CGGGGTACCACAGAGCGAGACTGTGTC-3′, Reverse: 5′-GCGAGATCTCCTGCGCCGCGCCTGGCT-3′) as previously described [14]. One VISP case was chosen for TA cloning from the high ΔPOST and low haplotype ΔPOST haplotypes. Promoters were cloned into the StrataClone PCR Cloning vector according to the standard protocols (Agilent Technologies). Promoters were next restriction digested and ligated into the pGL3 luciferase plasmid (Promega) using KpnI and SacI enzymes (New England Biolabs) giving rise to GNMTΔHighLuc and GNMTΔLowLuc.
Haplotype analysis
Haploview version 4.2 [13] was used to analyze haplotype blocks on chromosome 6 from the VISP population. 10 SNPs with p≤5×10−8 were assessed using version 3, release 27, analysis panel CEU+TSI.
Average ΔPOST values were taken from the VISP population, and any individuals with missing data from 1 or more SNPs were excluded from the analysis. The top two haplotypes, encompassing 80% of the total VISP population were assessed.
GNMT promoter analysis
2×104 HepG2 cells were transfected in quadruplicate with GNMTΔHighLuc or GNMTΔLowLuc and pGL4.74[hRluc/TK] (Promega) at a 10∶1 ratio using TransIt-LT1 transfection reagent (Mirus biosciences) in 96 well plates. Luciferase assays were conducted following the Dual-glo kit standard protocol (Promega). Luciferase readings were taken using the Beckman Coulter DTX880 luminometer at a 1 second integration time. Firefly luciferase measures from GNMTΔHighLuc or GNMTΔLowLuc were taken for each well, followed by treatment for renilla luciferase activity and renilla measurement. Relative luciferase activity of each promoter was calculated by dividing the average firefly luciferase counts from GNMTΔHighLuc or GNMTΔLowLuc by pGL4.74[hRluc/TK] for each independent condition. L-Methionine treatment used 0.2 mM reagent. Total relative luciferase activity for each plasmid encompasses the average of 3 biological replicates.
Pyrosequencing
Pyrosequencing was performed as previously described [55]. Primers are as follows, Forward: AGTAGAGAAGTGTTAGTTAGGTTTTAT, Reverse (Biotin labeled): ACCCATACAAAAAAAACAAAAAAAATCTC, Sequencing primer: TTTGGATTAGGTGGATAG.
Risk score analysis
Scores were determined by using imputation dosage measures from VISP and FHS. Alleles were assessed for the average ΔPOST values in FHS and VISP. If a dosage for a homozygous SNP was associated with high homocysteine on average (i.e. close to the value 2) the number was not changed. However, if the homozygous allele was not associated with the risk variant (i.e. close to 0) but was represented as a number above 1.5, 2 was subtracted from that number and made positive. After correction, homozygous risk variants would have a dosage value near 2, heterozygous variants would have a value near 1, and non risk variants would be assigned a number near 0. All imputation dosage values were summed. The sum of each risk value was then taken for each individual to give a score from 0 to a possible 10, and that score was rounded to the nearest integer.
For calculation of the variance explained by the risk score, linear regression was used for VISP, and a linear mixed effects model was used for FHS, as these methods were used in the initial GWAS for each study.
Statistics
All statistics for Figures 2–4 were performed using GraphPad Prism 4. Tests used are indicated in Figure legends. Significance threshold was set at p = 0.05.
Supporting Information
Zdroje
1. FurieKL, KellyPJ (2006) Homocyst(e)ine and stroke. Semin Neurol 26 : 24–32.
2. RefsumH, UelandPM, NygardO, VollsetSE (1998) Homocysteine and cardiovascular disease. Annu Rev Med 49 : 31–62.
3. SeshadriS (2006) Elevated plasma homocysteine levels: risk factor or risk marker for the development of dementia and Alzheimer's disease? J Alzheimers Dis 9 : 393–398.
4. LippiG, PlebaniM (2012) Hyperhomocysteinemia in health and disease: where we are now, and where do we go from here? Clin Chem Lab Med 50 : 2075–80.
5. JiY, TanS, XuY, ChandraA, ShiC, et al. (2013) Vitamin B supplementation, homocysteine levels, and the risk of cerebrovascular disease: A meta-analysis. Neurology 81 : 1298–1307.
6. SpenceJD, StampferMJ (2011) Understanding the complexity of homocysteine lowering with vitamins: the potential role of subgroup analyses. JAMA 306 : 2610–2611.
7. BostomAG, JacquesPF, NadeauMR, WilliamsRR, EllisonRC, et al. (1995) Post-methionine load hyperhomocysteinemia in persons with normal fasting total plasma homocysteine: initial results from the NHLBI Family Heart Study. Atherosclerosis 116 : 147–151.
8. BostomAG, RoubenoffR, DellaripaP, NadeauMR, SutherlandP, et al. (1995) Validation of abbreviated oral methionine-loading test. Clin Chem 41 : 948–949.
9. GalimbertiG, ContiE, ZiniM, PiazzaF, FenaroliF, et al. (2008) Post-methionine load test: A more sensitive tool to reveal hyperhomocysteinemia in Alzheimer patients? Clin Biochem 41 : 914–916.
10. van der GriendR, BiesmaDH, BangaJD (2002) Postmethionine-load homocysteine determination for the diagnosis hyperhomocysteinaemia and efficacy of homocysteine lowering treatment regimens. Vasc Med 7 : 29–33.
11. TraylorM, FarrallM, HollidayEG, SudlowC, HopewellJC, et al. (2012) Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE Collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol 11 : 951–62.
12. HollidayEG, MaguireJM, EvansTJ, KoblarSA, JannesJ, et al. (2012) Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet 44 : 1147–1151.
13. BarrettJC, FryB, MallerJ, DalyMJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21 : 263–265.
14. LeeCM, ShihYP, WuCH, ChenYM (2009) Characterization of the 5′ regulatory region of the human Glycine N-methyltransferase gene. Gene 443 : 151–157.
15. ClarkeR, DalyL, RobinsonK, NaughtenE, CahalaneS, et al. (1991) Hyperhomocysteinemia: an independent risk factor for vascular disease. N Engl J Med 324 : 1149–1155.
16. BoersGH (2000) Mild hyperhomocysteinemia is an independent risk factor of arterial vascular disease. Semin Thromb Hemost 26 : 291–295.
17. TooleJF (2002) Vitamin intervention for stroke prevention. J Neurol Sci 203–204 : 121–124.
18. GuillandJC, FavierA, Potier de CourcyG, GalanP, HercbergS (2003) [Hyperhomocysteinemia: an independent risk factor or a simple marker of vascular disease? 2. Epidemiological data]. Pathol Biol (Paris) 51 : 111–121.
19. StankovicS, Majkic-SinghN (2010) Genetic aspects of ischemic stroke: coagulation, homocysteine, and lipoprotein metabolism as potential risk factors. Crit Rev Clin Lab Sci 47 : 72–123.
20. BersanoA, BallabioE, BresolinN, CandeliseL (2008) Genetic polymorphisms for the study of multifactorial stroke. Hum Mutat 29 : 776–795.
21. ChristensenB, ArbourL, TranP, LeclercD, SabbaghianN, et al. (1999) Genetic polymorphisms in methylenetetrahydrofolate reductase and methionine synthase, folate levels in red blood cells, and risk of neural tube defects. Am J Med Genet 84 : 151–157.
22. AlluriRV, MohanV, KomandurS, ChawdaK, ChaudhuriJR, et al. (2005) MTHFR C677T gene mutation as a risk factor for arterial stroke: a hospital based study. Eur J Neurol 12 : 40–44.
23. TheodoratouE, MontazeriZ, HawkenS, AllumGC, GongJ, et al. (2012) Systematic meta-analyses and field synopsis of genetic association studies in colorectal cancer. J Natl Cancer Inst 104 : 1433–1457.
24. DongLM, PotterJD, WhiteE, UlrichCM, CardonLR, et al. (2008) Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA 299 : 2423–2436.
25. CookRJ, WagnerC (1984) Glycine N-methyltransferase is a folate binding protein of rat liver cytosol. Proc Natl Acad Sci U S A 81 : 3631–3634.
26. BaccarelliA, WrightR, BollatiV, LitonjuaA, ZanobettiA, et al. (2010) Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology 21 : 819–828.
27. KrishnaSM, DearA, CraigJM, NormanPE, GolledgeJ (2013) The potential role of homocysteine mediated DNA methylation and associated epigenetic changes in abdominal aortic aneurysm formation. Atherosclerosis 228 : 295–305.
28. TeslovichTM, MusunuruK, SmithAV, EdmondsonAC, StylianouIM, et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466 : 707–713.
29. GiustiB, SaraciniC, BolliP, MagiA, MartinelliI, et al. (2010) Early-onset ischaemic stroke: analysis of 58 polymorphisms in 17 genes involved in methionine metabolism. Thromb Haemost 104 : 231–242.
30. LefaucheurR, Triquenot-BaganA, QuillardM, GenevoisO, HannequinD (2008) [Stroke and iridodonesis revealing a homocystinuria caused by a compound heterozygous mutation of cystathionine beta-synthase]. Rev Neurol (Paris) 164 : 728–732.
31. MuddSH (2011) Hypermethioninemias of genetic and non-genetic origin: A review. Am J Med Genet C Semin Med Genet 157 : 3–32.
32. KluijtmansLA, BoersGH, KrausJP, van den HeuvelLP, CruysbergJR, et al. (1999) The molecular basis of cystathionine beta-synthase deficiency in Dutch patients with homocystinuria: effect of CBS genotype on biochemical and clinical phenotype and on response to treatment. Am J Hum Genet 65 : 59–67.
33. KrausJP (1994) Komrower Lecture. Molecular basis of phenotype expression in homocystinuria. J Inherit Metab Dis 17 : 383–390.
34. PareG, ChasmanDI, ParkerAN, ZeeRR, MalarstigA, et al. (2009) Novel associations of CPS1, MUT, NOX4, and DPEP1 with plasma homocysteine in a healthy population: a genome-wide evaluation of 13 974 participants in the Women's Genome Health Study. Circ Cardiovasc Genet 2 : 142–150.
35. LangeLA, Croteau-ChonkaDC, MarvelleAF, QinL, GaultonKJ, et al. (2010) Genome-wide association study of homocysteine levels in Filipinos provides evidence for CPS1 in women and a stronger MTHFR effect in young adults. Hum Mol Genet 19 : 2050–2058.
36. CaiuloA, BardoniB, CamerinoG, GuioliS, MinelliA, et al. (1989) Cytogenetic and molecular analysis of an unbalanced translocation (X;7) (q28;p15) in a dysmorphic girl. Hum Genet 84 : 51–54.
37. Veiga-da-CunhaM, ColletJF, PrieurB, JaekenJ, PeeraerY, et al. (2004) Mutations responsible for 3-phosphoserine phosphatase deficiency. Eur J Hum Genet 12 : 163–166.
38. LocasaleJW (2013) Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13 : 572–583.
39. TibbettsAS, ApplingDR (2010) Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr 30 : 57–81.
40. GalanP, BrianconS, BlacherJ, CzernichowS, HercbergS (2008) The SU.FOL.OM3 Study: a secondary prevention trial testing the impact of supplementation with folate and B-vitamins and/or Omega-3 PUFA on fatal and non fatal cardiovascular events, design, methods and participants characteristics. Trials 9 : 35.
41. LonnE, YusufS, ArnoldMJ, SheridanP, PogueJ, et al. (2006) Homocysteine lowering with folic acid and B vitamins in vascular disease. N Engl J Med 354 : 1567–1577.
42. TooleJF, MalinowMR, ChamblessLE, SpenceJD, PettigrewLC, et al. (2004) Lowering homocysteine in patients with ischemic stroke to prevent recurrent stroke, myocardial infarction, and death: the Vitamin Intervention for Stroke Prevention (VISP) randomized controlled trial. JAMA 291 : 565–575.
43. DawberTR, KannelWB, LyellLP (1963) An approach to longitudinal studies in a community: the Framingham Study. Ann N Y Acad Sci 107 : 539–556.
44. FeinleibM, KannelWB, GarrisonRJ, McNamaraPM, CastelliWP (1975) The Framingham Offspring Study. Design and preliminary data. Prev Med 4 : 518–525.
45. ArakiA, SakoY (1987) Determination of free and total homocysteine in human plasma by high-performance liquid chromatography with fluorescence detection. J Chromatogr 422 : 43–52.
46. CarandangR, SeshadriS, BeiserA, Kelly-HayesM, KaseCS, et al. (2006) Trends in incidence, lifetime risk, severity, and 30-day mortality of stroke over the past 50 years. JAMA 296 : 2939–2946.
47. SeshadriS, BeiserA, Kelly-HayesM, KaseCS, AuR, et al. (2006) The lifetime risk of stroke: estimates from the Framingham Study. Stroke 37 : 345–350.
48. WolfPA, KannelWB, DawberTR (1978) Prospective investigations: the Framingham study and the epidemiology of stroke. Adv Neurol 19 : 107–120.
49. ChenMH, YangQ (2010) GWAF: an R package for genome-wide association analyses with family data. Bioinformatics 26 : 580–581.
50. TherneauTM, GrambschPM (2000) Modeling Survival Data: Extending the Cox Model. Springer-Verlag 170–172.
51. AltshulerD, GibbsR, PeltonenL, DermitzakisE, SchaffnerS, et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467 : 52–58.
52. BrowningB, BrowningS (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84 : 210–223.
53. ManichaikulA, MychaleckyjJC, RichSS, DalyK, SaleM, et al. (2010) Robust relationship inference in genome-wide association studies. Bioinformatics 26 : 2867–2873.
54. WillerCJ, LiY, AbecasisGR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26 : 2190–2191.
55. JackA, ConnellyJJ, MorrisJP (2012) DNA methylation of the oxytocin receptor gene predicts neural response to ambiguous social stimuli. Front Hum Neurosci 6 : 280.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3- Délka menstruačního cyklu jako marker ženské plodnosti
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



