-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
Fusion protein RUNX1-ETO (AML1-ETO, RUNX1-RUNX1T1) is expressed as the result of the 8q22;21q22 translocation [t(8;21)], which is one of the most common chromosomal abnormalities found in acute myeloid leukemia. RUNX1-ETO is thought to promote leukemia development through the aberrant regulation of RUNX1 (AML1) target genes. Repression of these genes occurs via the recruitment of the corepressors N-COR and SMRT due to their interaction with ETO. Mechanisms of RUNX1-ETO target gene upregulation remain less well understood. Here we show that RUNX1-ETO9a, the leukemogenic alternatively spliced transcript expressed from t(8;21), upregulates target gene Alox5, which is a gene critically required for the promotion of chronic myeloid leukemia development by BCR-ABL. Loss of Alox5 expression reduces activity of RUNX1-ETO9a, MLL-AF9 and PML-RARα in vitro. However, Alox5 is not essential for the induction of leukemia by RUNX1-ETO9a in vivo. Finally, we demonstrate that the upregulation of Alox5 by RUNX1-ETO9a occurs via the C2H2 zinc finger transcription factor KLF6, a protein required for early hematopoiesis and yolk sac development. Furthermore, KLF6 is specifically upregulated by RUNX1-ETO in human leukemia cells. This identifies KLF6 as a novel mediator of t(8;21) target gene regulation, providing a new mechanism for RUNX1-ETO transcriptional control.
Published in the journal: . PLoS Genet 9(10): e32767. doi:10.1371/journal.pgen.1003765
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003765Summary
Fusion protein RUNX1-ETO (AML1-ETO, RUNX1-RUNX1T1) is expressed as the result of the 8q22;21q22 translocation [t(8;21)], which is one of the most common chromosomal abnormalities found in acute myeloid leukemia. RUNX1-ETO is thought to promote leukemia development through the aberrant regulation of RUNX1 (AML1) target genes. Repression of these genes occurs via the recruitment of the corepressors N-COR and SMRT due to their interaction with ETO. Mechanisms of RUNX1-ETO target gene upregulation remain less well understood. Here we show that RUNX1-ETO9a, the leukemogenic alternatively spliced transcript expressed from t(8;21), upregulates target gene Alox5, which is a gene critically required for the promotion of chronic myeloid leukemia development by BCR-ABL. Loss of Alox5 expression reduces activity of RUNX1-ETO9a, MLL-AF9 and PML-RARα in vitro. However, Alox5 is not essential for the induction of leukemia by RUNX1-ETO9a in vivo. Finally, we demonstrate that the upregulation of Alox5 by RUNX1-ETO9a occurs via the C2H2 zinc finger transcription factor KLF6, a protein required for early hematopoiesis and yolk sac development. Furthermore, KLF6 is specifically upregulated by RUNX1-ETO in human leukemia cells. This identifies KLF6 as a novel mediator of t(8;21) target gene regulation, providing a new mechanism for RUNX1-ETO transcriptional control.
Introduction
Acute myeloid leukemia (AML) is the most prevalent form of adult leukemia [1]. Chromosomal translocations are found in over 80% of AML, the most common of which is t(8;21), occurring in up to 40% of AML cases categorized within the French-American-British (FAB) subtype M2 [2]–[6]. This translocation results in the expression of fusion protein RUNX1-ETO. Although sufficient for in vitro immortalization, RUNX1-ETO requires additional cooperating mutations to induce leukemia in vivo [7]–[9]. RUNX1-ETO also exists as C-terminally truncated forms due to alternative splicing at exons 9 (RUNX1-ETO9a) and 11 (RUNX1-ETO11a) [10], [11]. Both isoforms lack the NHR4/MYND domain and are expressed in human t(8;21)+ leukemia patient samples, and RUNX1-ETO9a (RE9a) strongly promotes leukemia development in mice [10], [11].
RUNX1-ETO (RE) is known to be a transcriptional repressor through its recruitment of the corepressors N-CoR and SMRT and their associated histone deacetylases [12]–[14]. RE can also activate promoters cell-specifically, however it is unclear whether such gene activation occurs in a direct or indirect fashion [15]. The mechanisms by which RE upregulates its target genes have been less thoroughly investigated, although recent studies have utilized ChIP-chip and ChIP-seq to identify putative RE target genes to examine their regulation and importance in leukemia development [16]–[18]. One interesting recent finding is that RE upregulates at least some of its target genes via its interaction with the histone acetyltransferase p300, and that loss of this interaction significantly delays leukemia onset [19]. Additionally, our group recently reported that RE9a recruits PRMT1 to some RE9a-activated genes, leading to H3K4 methlyation, H3K9/14 acetylation and transcriptional activation [20]. Further mechanisms of gene upregulation remain to be investigated.
One gene strongly upregulated in t(8;21) leukemia is ALOX5, encoding an enzyme required for the synthesis of leukotrienes, which are small, lipid-derived signaling molecules that trigger pathways implicated in both inflammation and cancer, such as proliferation, cell survival and angiogenesis [21]–[24]. In fact, inhibitors of the ALOX5 pathway have shown promise in treating a number of epithelial cancers [25]–[27]. In addition, ALOX5 has previously been shown to function in both normal hematopoiesis and leukemia development. Using a human CD34+ cell model, it has been demonstrated that leukotrienes both increase proliferation and exert an anti-apoptotic effect on human hematopoietic stem cells [28]. More recently, it was further demonstrated that ALOX5 is required for the induction of chronic myeloid leukemia (CML) by BCR-ABL, and a specific ALOX5 inhibitor is able to significantly delay CML onset when used either alone or in combination with the BCR-ABL kinase inhibitor imatinib [29].
Despite significant advances in our molecular understanding of AML, frontline treatment for this disease is still induction and consolidation chemotherapy, similar to the protocol established 30 years ago, and overall survival for older patients has not improved over the same time period [30], [31]. Furthermore, although t(8;21)+ AML is considered to have a favorable prognosis for chemotherapeutic response, the 10-year overall survival for patients with this cytogenetic signature is only 61% [32]. New molecular targets and a better understanding of the mechanisms of RUNX1-ETO-mediated transcriptional changes leading to disease development are needed for safer and more effective treatment of this disease. Here we investigate the role of ALOX5 in AML development, establishing ALOX5 as an upregulated gene in t(8;21) leukemia that is also important in cellular dysregulation by multiple oncogenic fusion proteins. We further discover that RE9a upregulates Alox5 via the C2H2 zinc finger transcription factor Krϋppel-like factor 6 (KLF6), a protein critically required for early hematopoiesis [33]. Finally, KLF6 itself is upregulated by both RE and RE9a, establishing a new mechanism for the upregulation of target genes by t(8;21) fusion proteins and a new pathway to study in AML development.
Results
Upregulation of ALOX5 in t(8;21)-associated acute myeloid leukemia
In order to understand the mechanism by which RUNX1-ETO (RE) contributes to t(8;21) acute myeloid leukemia (AML) development, our group recently conducted gene expression microarray and ChIP-chip analyses to identify potential disease-related RE target genes [18]. One gene confirmed to be highly upregulated and specifically detected by ChIP in the RE9a murine leukemia model is Alox5. The detected ChIP peak is 47 kilobases downstream of the transcription start site in intron 9. ALOX5 is required for CML development due to the depletion of leukemia stem cells in the absence of Alox5 expression [29]. To determine whether ALOX5 is also upregulated in human t(8;21) AML, we analyzed publicly available microarray data of AML M2 patients with or without the 8;21 translocation [34]. As shown in Supporting Figure S1A, ALOX5 expression is approximately 2.3 fold higher in t(8;21)+ patients than in patients not harboring the translocation, and both patient groups had elevated ALOX5 levels relative to normal CD34+ control samples. This upregulation correlates very well to the Alox5 upregulation seen in the RE9a mouse model (Figure 1A).
Fig. 1. Upregulation of Alox5 in acute myeloid leukemia and by RUNX1-ETO9a. 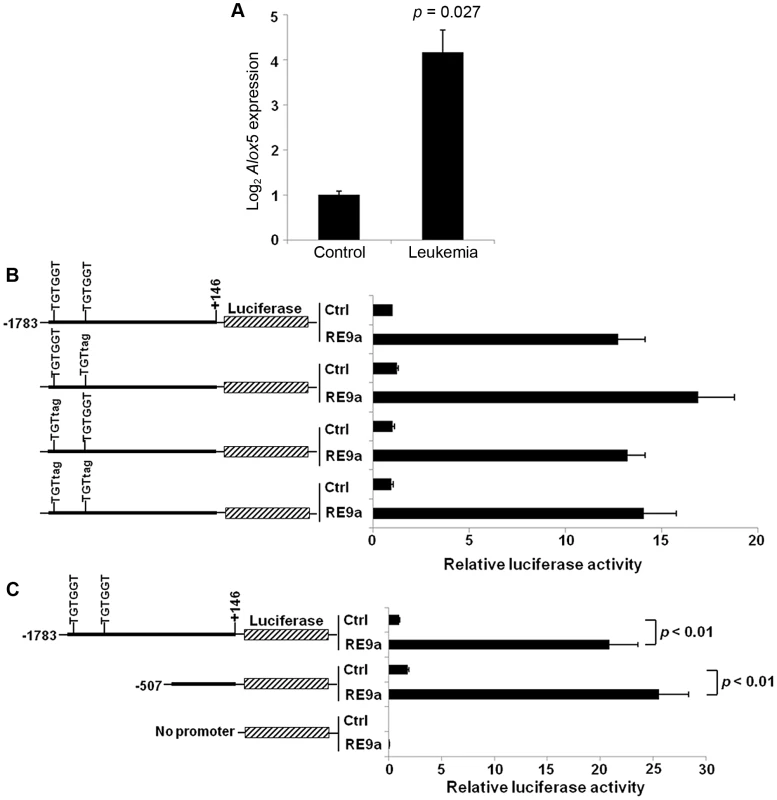
(A) Normalized log2 expression of Alox5 in control or RE9a-leukemic murine lin−c-Kit+ bone marrow cells. mRNA transcript levels were normalized to Gapdh and control was set to 1. Data show averages and standard deviations from 3 independent mice each. (B) RE9a regulation of mouse Alox5 promoter-luciferase reporter. Numbers indicate base pair relative to transcription start site. Two RUNX1 binding sites (TGTGGT) were either wildtype or mutated to TGTtag to abrogate RE9a binding [54]. Indicated promoter-luciferase reporter was co-transfected with control (Ctrl) or RE9a plasmid and expression was normalized to Renilla luciferase. Wildtype promoter+control set to 1. (C) RE9a regulation of truncated mouse Alox5 promoter-luciferase reporter. Luciferase assay performed as described in (B), with −1783 to +146 promoter+control set to 1. As an initial step in determining the molecular mechanism by which Alox5 is upregulated, we cloned a ∼1.9 kb Alox5 promoter fragment (bp −1783 to +146) upstream of a luciferase reporter. The vector backbone of this reporter contained 6 RUNX1 consensus binding sites which were mutated to prevent non-specific regulation by RE9a due to binding at these sites [15]. In promoter-luciferase transactivation studies, the Alox5 promoter is strongly upregulated by RE9a (Figure 1B). This promoter fragment contains two consensus RUNX1 binding sites (TGTGGT), indicating that RE9a may bind directly to the promoter at these sites to affect gene expression. However, when these sites are mutated either individually or together, RE9a is still able to upregulate the reporter (Figure 1B). Furthermore, when the promoter region is truncated to exclude these two RUNX1 binding sites, there is no decrease in the activation of the Alox5 promoter by RE9a (Figure 1C), implying that RE9a either does not upregulate Alox5 directly or functions via binding at a non-canonical RUNX1 binding site.
RE9a cooperates with KLF6 to upregulate Alox5
To determine if RE9a upregulates Alox5 through an imperfect RUNX1 binding site, we examined the truncated promoter from Figure 1C and found 5 motifs that differ from the consensus RUNX1 binding site (TGYGGT) by a single nucleotide (Figure 2A). We then serially truncated this promoter fragment to examine which regions are important for both the basal and RE9a-inducible expression of Alox5. When truncated from −366 to −86, we find a significant increase in the basal activity, indicating the presence of a basal cis repressive element in this region (Figure 2B). When truncated from −55 to −30, there is a large reduction in basal activity, which is unsurprising as this truncation removes an SP1 binding site (GGGCGG) known to be important for Alox5 expression [35]. When examining inducible activity, however, only the shortest truncation shows a significant loss of promoter activation upon addition of RE9a (Figure 2C), implying that sequences between −30 and +57, which is the region typically considered to comprise the core promoter for gene expression [36], play an important role in Alox5 regulation by RE9a.
Fig. 2. Analysis of Alox5 promoter regulation by RE9a. 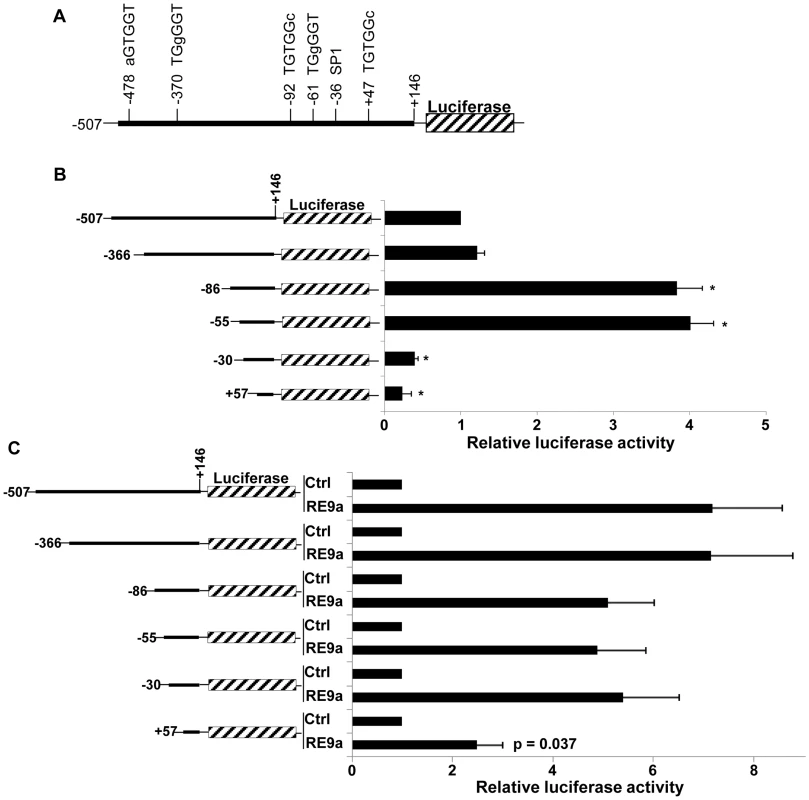
(A) Schematic of Alox5 promoter-luciferase reporter with motifs differing from RUNX1 consensus binding site (TGYGGT) indicated. Bases differing from consensus site labeled in lowercase. Transcription factor SP1 binding site also indicated. Numbers represent base pairs relative to transcription start site. (B) Basal regulation of Alox5 promoter-luciferase truncations. Indicated reporters were transfected in the absence of RE9a and expression was normalized to Renilla luciferase and the −507 to +146 construct was set to 1. * = p<0.01 relative to −507 reporter. (C) Inducible regulation of Alox5 promoter-luciferase by RE9a. Indicated reporters were co-transfected with control or RE9a and expression was normalized to Renilla luciferase. Each control (Ctrl) transfection normalized to 1. p-value relative to −507 reporter +RE9a. Upon closer examination of this region, we identified a GGGTG motif (reverse complement: CACCC) which is known to be a binding site for KLF6, a zinc finger DNA-binding transcription factor that was previously identified as an important cell-specific positive regulator of Leukotriene C4 Synthase (LTC4S) expression, which functions downstream of ALOX5 in the synthesis of certain leukotrienes [37]. Although the CACCC motif is a binding site for most KLF family members [38], since KLF6 is both an important hematopoietic regulator and induces expression of another member of the ALOX5 pathway [33], [37], we decided to focus on KLF6 and hypothesized that RE9a may function through KLF6 to regulate Alox5 expression. Interestingly, we found that KLF6 is both independently capable of activating the Alox5 reporter and, when introduced in combination with RE9a, the activation is greater than the sum of the effect of RE9a and KLF6 alone (Figure 3A). This demonstrates that RE9a can indeed cooperate with KLF6 to upregulate Alox5 expression. To examine whether KLF6 is required for RE9a regulation of Alox5, we utilized two different shRNAs targeting KLF6 that decrease its endogenous expression by approximately 50% (Figure 3B). Both of these shRNAs are able to significantly reduce the ability of RE9a to activate the Alox5 promoter, demonstrating that RE9a does require KLF6 for full regulation of Alox5 (Figure 3C). Possible explanations for an incomplete loss of reporter induction after KLF6 knockdown include an incomplete loss of KLF6 expression (Figure 3B) and that RE9a may function with other factors in addition to KLF6 in the upregulation of Alox5. Furthermore, when the KLF6 binding motif was mutated to a sequence previously shown to disrupt KLF6-mediated gene activation [37], the upregulation of Alox5 by both RE9a and KLF6 was significantly reduced (Figure 3D). Interestingly, although the specific KLF6 binding site GGGTG found in the murine promoter is not conserved in humans, a GC-box (GGGCGGG) is present at this site, which also allows KLF6 binding [39]. Additionally, the human ALOX5 promoter does contain 7 CACCC or GGGTG motifs within 500 bp of the transcription start site, indicating that KLF6 function in human ALOX5 regulation may be conserved through these sites. Supporting this possibility, both exogenous Flag-KLF6 and HA-RE9a display enrichment at the ALOX5 promoter region in K562 cells when examined by ChIP, although endogenous RUNX1 does not (Figure S2). Finally, since RE9a functions with KLF6 in the regulation of gene expression, we were interested to determine whether RE9a and other RUNX1 proteins were capable of interacting with KLF6 in the same complex. Using coimmunoprecipitation, we find that KLF6 is indeed capable of interacting with both RUNX1 and the fusion proteins RUNX1-ETO and RE9a, which is the first report of such interaction (Figure 3E). However, in all conditions tested we can only detect over-expressed, exogenous KLF6 with available antibodies and therefore could not confirm the interactions between KLF6 and these proteins at the endogenous level. We also confirm the previously reported interaction between KLF6 and endogenous SP1 [39]. Given that RUNX1-ETO is also able to interact with SP1 via the Runt domain [40] (which is common to wildtype RUNX1), this raises the possibility that KLF6, SP1 and Runt domain-containing proteins may function together in gene regulation and provides direction for future transcriptional studies.
Fig. 3. Alox5 regulation by RE9a and KLF6. 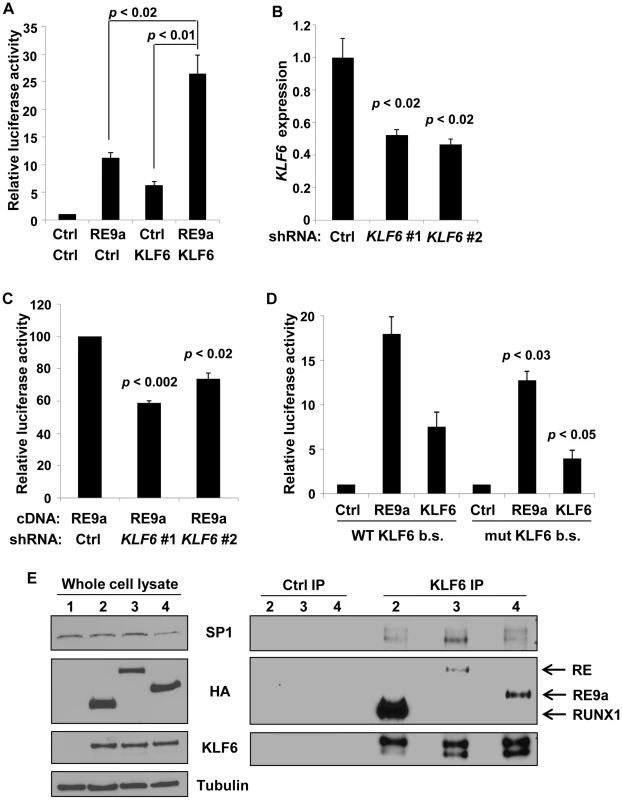
(A) Coregulation of Alox5 reporter by RE9a and KLF6. Alox5 −507 to +146 reporter co-transfected with RE9a, KLF6 or both. Expression was normalized to Renilla luciferase and control (Ctrl) was set to 1. (B) Knockdown of endogenous KLF6 via shRNA. K562 cells were transfected with control or one of two independent shRNAs targeting KLF6 and analyzed by qRT-PCR for KLF6 expression. Expression values were normalized to GAPDH and control transfected value was set to 1. Data show averages with standard deviations of 3 independent transfections. (C) Knockdown of KLF6 impairs ability of RE9a to upregulate Alox5 promoter-luciferase reporter. K562 cells pre-transfected with control or KLF6 shRNA were co-transfected with Alox5 −507 to +146 reporter and RE9a. Expression was normalized to Renilla luciferase and control +RE9a was set to 100. (D) Mutation of KLF6 binding site significantly decreases activation of Alox5 promoter by RE9a and KLF6. Wildtype or KLF6 binding site (b.s.)-mutated (GGGTG to GATCG) Alox5 −30 to +146 reporter co-transfected with RE9a or KLF6. Expression was normalized to Renilla luciferase. Control (Ctrl) was set to 1. p-values are compared to wildtype reporter co-transfected with corresponding transgene. (E) KLF6 can interact with RUNX1, RE and RE9a. KLF6 and RUNX1, RE or RE9a were co-transfected into K562 cells, and lysates were immunoprecipitated with control or KLF6 antibody. Also shown is interaction with endogenous SP1. α-tubulin serves as a loading control for the whole cell lysate. After IP, KLF6 appears as multiple bands likely because KLF6 is expressed endogenously as multiple splicing isoforms which are enriched to more easily detectable levels by immunoprecipitation [55]. KLF6 is specifically upregulated in t(8;21)+ cells
To determine whether KLF6 is also a target of RE gene regulation in human AML, we again examined available patient data and find that KLF6 expression is specifically higher in t(8;21)+ leukemia samples (Figure S1B and [34]). KLF6 is also upregulated in K562 cells 24 hr after transfection with RE9a (Figure 4A), supporting induction of KLF6 as a mechanism of Alox5 upregulation by RE9a in the above luciferase studies. Furthermore, when comparing mRNA levels in AML M2 human leukemia cell lines, the t(8;21)+ cell lines SKNO and Kasumi-1 express significantly higher levels of KLF6 than does the t(8;21) - cell line HL60 (Figure 4B). Finally, when RE or RE9a are introduced retrovirally into HL60 cells, there is a significant and dramatic upregulation of KLF6 (Figure 4C). This upregulation is greater for RE than RE9a, which agrees with previous findings that RE more strongly dysregulates gene expression than its leukemic isoforms [41], [42]. Collectively, these data demonstrate that KLF6 is a target of t(8;21) gene upregulation.
Fig. 4. Regulation of KLF6 by RUNX1-ETO and RE9a. 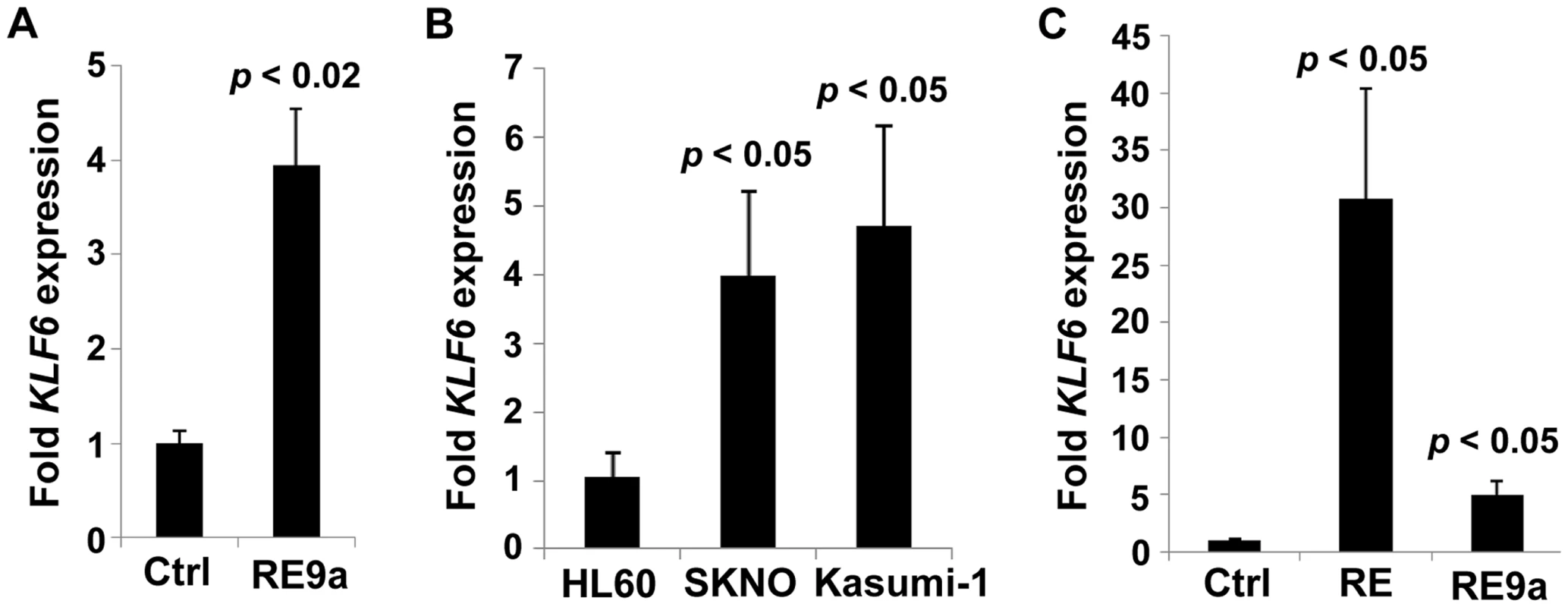
(A) Expression of KLF6 in control- (Ctrl) or RE9a-transfected K562 cells. RE9a or empty vector was co-transfected into K562 cells along with a GFP-expressing vector to determine transfection efficiency. KLF6 mRNA levels were normalized to GAPDH with Ctrl set to 1, and samples were then normalized to account for transfection rate by percent GFP-expressing cells as determined by flow cytometry. Data show averages and standard deviations of three independent transfections. (B) Expression of KLF6 in HL60 [t(8;21)-negative] and SKNO and Kasumi-1 [t(8;21)-positive] cell lines. KLF6 mRNA levels were normalized to GAPDH and HL60 was set to 1. Data show averages and standard deviations of 3 independent RNA isolations. (C) Expression of KLF6 in control-, RUNX1-ETO- or RUNX1-ETO9a-transduced HL60 cells. Following 2 rounds of retroviral transduction, KLF6 levels determined as in (B), with control-transduced cells (Ctrl) set to 1. Data display averages and standard deviations of 3 independent transductions. Lack of Alox5 impairs cellular dysregulation by multiple oncogenic fusion proteins
To examine the potential functional implications of Alox5 in t(8;21)-induced self-renewal, we performed serial replating assays after retroviral transduction of wildtype and Alox5-/ - murine bone marrow cells. In this assay, wildtype cells transduced with retrovirus encoding RE9a-IRES-Puror display increased self-renewal and maintain replating capacity through at least 13 weeks in weekly replating culture, whereas control cells transduced with a vector encoding the puromycin resistance gene alone (MIP) rapidly lose colony forming ability and stop replating after about 3 weeks (Figure 5A). Alox5-/ - cells transduced with MIP alone behave similar to their wildtype counterparts and display limited self-renewal. Interestingly, although RE9a-infected Alox5-/ - cells initially replate longer than MIP control cells, after approximately 5 weeks in culture they begin to produce far fewer colonies than RE9a-infected wildtype cells and eventually lose replating capacity altogether (Figure 5A). These Alox5-/ - cells also form smaller colonies as compared to wildtype (Figure 5B) and stain more positively for the myeloid differentiation marker CD11b when examined by flow cytometry (Figure 5C; Wildtype: 26–36%; Alox5-/-: 56–64%). These data indicate that lack of Alox5 impairs the ability of RE9a to increase murine hematopoietic cell self-renewal, perhaps in part by altering cellular differentiation.
Fig. 5. Alox5 involvement in hematopoietic cell self-renewal. 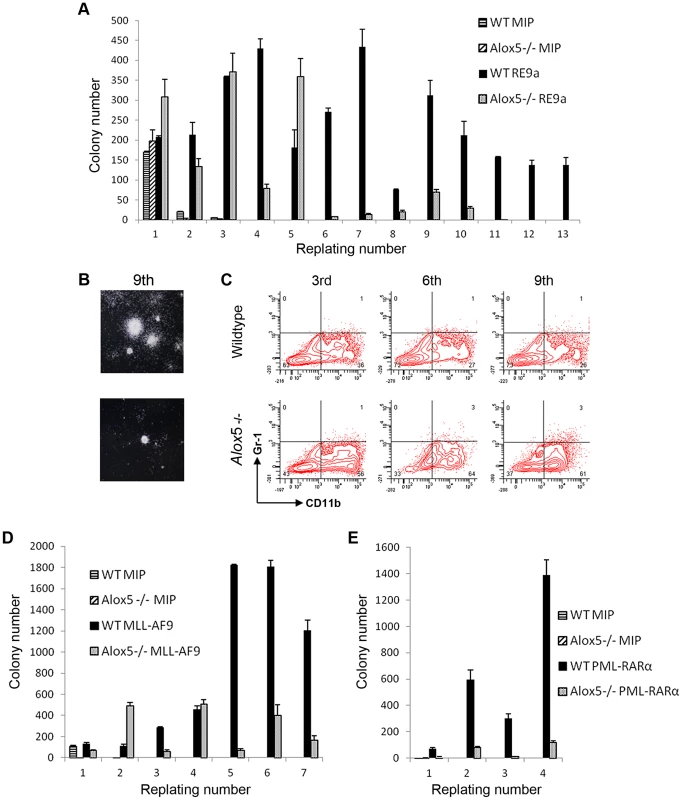
(A) Alox5 required for long-term self-renewal of hematopoietic cells by RE9a. Colony numbers from wildtype or Alox5-/- bone marrow cells transduced with control (MIP) or RE9a retrovirus and serially replated in methylcellulose. Data shown are averages with standard deviations of a representative dataset. Four independent assays were performed. (B) Typical colony images after 9th replating from (A) taken using Nikon Eclipse TS100 microscope with 2×/0.06 objective lens and Nikon DS Camera Control Unit DS-U2 system. (C) Flow cytometric analysis of replated cells from (A). Cells from 3rd, 6th and 9th replatings were stained for myeloid lineage markers Gr-1 and CD11b. Representative data from four independent assays shown. (D) and (E) Lack of Alox5 decreases colony formation potential of hematopoietic cells transduced with MLL-AF9 and PML-RARα. Wildtype or Alox5-/- bone marrow cells were transduced with MIP and MLL-AF9 (D) or PML-RARα (E) retrovirus and serially replated in methylcellusose. Data shown are averages and standard deviations of representative datasets. Three independent assays were performed. To determine whether the self-renewal defects observed in Alox5-/ - cells were specific to RE9a or were more broadly applicable, we performed similar replating assays using the oncogenic fusion proteins MLL-AF9, resulting from t(9;11) and most frequently occurring in acute monoblastic leukemia (AML M5) [43], and PML-RARα, resulting from t(15;17) and observed in acute promyelocytic leukemia (APL/AML M3) [44]. Notably, decreases in colony numbers as compared to wildtype were observed after transduction of Alox5-/ - cells with both MLL-AF9 and PML-RARα (Figure 5D–E), demonstrating that loss of Alox5 impairs the increased self-renewal capability induced by multiple fusion oncoproteins.
It was previously reported that although Alox5 is required for induction of CML by BCR-ABL, lack of Alox5 in hematopoietic stem cells in the absence of BCR-ABL did not result in any significant hematopoietic defects [29]. Having observed that lack of Alox5 also impaired increased self-renewal induced by RE9a, MLL-AF9 and PML-RARα, we next examined whether exogenous expression of Alox5 itself had any effect. To test this, we transduced wildtype bone marrow cells with retrovirus encoding murine Alox5 and performed serial replating assays. As shown in Supporting Figure S3, exogenous Alox5 expression conferred no replating advantage relative to MIP-transduced control cells, demonstrating that while ALOX5 aids in increased self-renewal by multiple oncogenes, it is incapable of inducing this increase on its own.
RE9a-expressing Alox5-/ - cells are capable of leukemia induction
Alox5-/ - hematopoietic cells transduced with multiple fusion oncogenes showed clear defects in self-renewal in vitro (Figure 5). It is important to determine whether these defects are also present in vivo. To investigate this possibility, we harvested wildtype and Alox5-/ - fetal liver cells, retrovirally transduced them with virus encoding either RE9a-IRES-GFP or GFP alone (MigR1) and transplanted them into lethally irradiated recipient mice. As expected, neither wildtype nor Alox5-/ - cells transduced with MigR1 induced leukemia (Figure 6A). In contrast, both wildtype and Alox5-/ - cells expressing RE9a induced leukemia in recipient mice, with a median latency of approximately 30 weeks (Figure 6A). Both wildtype and Alox5-/ - cells were also able to induce secondary and tertiary leukemias in further rounds of transplantations to new recipient mice (data not shown). Additionally, leukemic mice of both genotypes displayed similar blast cells in the peripheral blood, bone marrow and spleen (Figure 6B).
Fig. 6. Loss of Alox5 does not block RE9a leukemia induction in vivo. 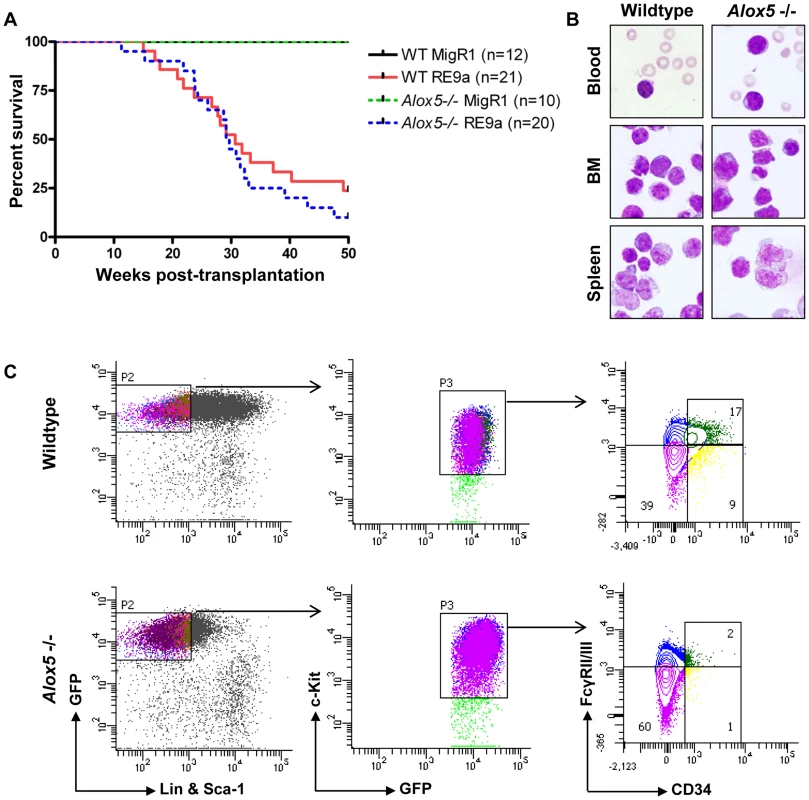
(A) Survival of mice receiving wildtype or Alox5-/- fetal liver cells transduced by control (MigR1) or RE9a retrovirus. Number of mice in each cohort shown at right. WT median survival: 30.71 weeks; Alox5-/- median survival: 29.43 weeks; p = 0.39. (B) Presence of hematopoietic blast cells in tissues of mice transplanted with RE9a-transduced wildtype or Alox5-/- cells. Peripheral blood smears and cytocentrifugation of bone marrow and spleen cells were stained with Wright-Giemsa solutions. (C) Immunophenotype of myeloid progenitor cells in wildtype and Alox5-/- leukemias. Distribution of EGFP+Lin−Sca-1−c-Kit+ leukemic cells harvested from spleen shown based on expression of CD34 and Fcγ receptors II/III (FcγRII/III). At least 4 mice analyzed per genotype, with representative distributions shown. We have previously observed that RE9a-leukemic mice display a phenotype in their myeloid progenitor populations, in which the normal distribution of common myeloid progenitors (CMPs), granulocyte/monocyte progenitors (GMPs) and megakaryocyte/erythroid progenitors (MEPs) is lost, and a single population arises that is similar in immunophenotype to MEPs, but has increased expression of Fcγ receptors II/III (FcγRII/III) by flow cytometry [10]. Similar leukemia cells were also reported in an Inv(16) AML mouse model and termed abnormal myeloid progenitors [45]. To determine whether Alox5-/ - leukemic cells also display this aberrant progenitor profile, we first examined untreated, non-leukemic bone marrow cells from wildtype and Alox5-/ - mice and observed normal distributions of the 3 progenitor populations (Figure S4). We next checked the progenitor populations from leukemic mice. Although there were variations among both wildtype and Alox5-/ - leukemic mice in terms of mean fluorescence intensity of GFP expression and percent of cells that were Sca-1 positive, we found that leukemic mice transplanted with either genotype of RE9a-infected cells displayed a similar abnormal myeloid progenitor immunophenotype (Figure 6C) as compared to controls (Figure S4). Some Alox5-/- leukemic mice displayed smaller GMP compartments than their wildtype counterparts (Figure 6C), but as the differences varied from mouse to mouse and had no effect on leukemia development (Figure 6A), we do not interpret this as being an important functional difference.
These results demonstrate that although loss of Alox5 results in self-renewal defects in vitro, other factors exist in vivo that allow RE9a to overcome these defects and promote leukemia development.
Discussion
In order to design better and more specific treatments for AML, we need a more thorough understanding of the underlying molecular mechanisms of cellular transformation that lead to disease. The 8;21 translocation that causes expression of the RUNX1-ETO DNA-binding fusion proteins is highly associated with AML M2, but its mechanisms of gene dysregulation are not completely understood. This is especially true for upregulated RE target genes. Here, we demonstrate that Alox5 is an upregulated t(8;21) target gene and establish for the first time that KLF6 cooperates in transcriptional dysregulation with leukemia fusion proteins during target gene upregulation. Although the Alox5 promoter is activated only weakly by wildtype RUNX1, both full-length RUNX1-ETO and its splicing isoform RUNX1-ETO9a strongly induce promoter activity, demonstrating that Alox5 is upregulated by multiple t(8;21) fusion proteins (Figure S5).
ALOX5 is a promising molecular target for the treatment of CML, as it has been demonstrated that a small molecule inhibitor of ALOX5 significantly delays leukemia onset in mice [29]. We show here that lack of ALOX5 also leads to in vitro defects in hematopoietic cells transduced by RE9a, MLL-AF9 and PML-RARα, all of which are oncogenes involved in AML development. However, these results did not translate in vivo, as Alox5-/ - cells infected with RE9a are still able to induce leukemia in mice (Figure 6A), and a similar result was obtained in a pilot experiment using an MLL-AF9 model of AML (data not shown). Additionally, no significant differences were observed in the differentiation states of wildtype and Alox5-/ - leukemia cells, either in analysis of their progenitor populations or expression of lineage markers Gr1, CD11b, CD4 and B220 or progenitor markers c-Kit and Sca-1 (Figure 6C and data not shown). It is not entirely clear why the importance of ALOX5 demonstrated in vitro is not observed in vivo, although this difference in phenotype between the two systems has been observed previously. One such example is RUNX1-ETO itself, which induces a significant increase in hematopoietic self-renewal yet requires cooperating mutations to induce leukemia in vivo [46]. Additionally, lack of Stat5 expression blocks the replating potential of the AML-inducing oncogene MOZ-TIF2 but only delays the onset of AML in vivo [47]. We observe a similar discrepancy for the requirement of Alox5 between the in vitro and in vivo systems, for which multiple mechanisms may exist. One possibility arises from the fact that there are a limited number of factors present in the in vitro culture conditions. Additional factors or different concentrations of factors present in vivo may allow for Alox5-/ - cell survival and transformation. In a CML model, BCR-ABL-expressing Alox5-/ - leukemia stem cells (LSCs) display an increased apoptotic rate as compared to wildtype LSCs, indicating the potential importance of the stem cell population for Alox5 and leukemia development [48]. Given this importance, it is possible that interactions between RE9a-infected stem cells and the niche in vivo allow them to survive and self-renew, whereas this interaction is absent in vitro. Another possible cause is cell-specific differences, as total bone marrow cells were used in vitro and fetal liver cells were used for the transplantation experiments. Recent studies indicate that the fetal liver and adult bone marrow contain differing ratios of hematopoietic stem cells (HSCs) with unequal differentiation potential, with the fetal liver specifically enriched for HSCs with long-term myeloid differentiation potential [49]. It is possible that different subtypes of HSCs respond differently to the introduction of RE9a, leading to the observed results. In addition, it is also possible that ALOX5 is required more by RE than RE9a in leukemia development, especially given that RE more strongly upregulates KLF6 expression. Other plausible possibilities also exist. Therefore, the available evidence suggests that ALOX5 is not a suitable molecular target for the treatment of t(8;21) AML alone. Based on our replating data, it appears that ALOX5 does play a role in RE-induced increases in self-renewal. However, as this does not translate to a delay in leukemia onset in vivo, targeting ALOX5 alone is likely insufficient to generate therapeutic benefit, although its inhibition in combination with other treatments may show efficacy and this remains to be examined.
It is well established that RE represses target gene expression via the interaction of the ETO domain with N-CoR and SMRT and their associated histone deacetylases [50]. The mechanism by which RE upregulates gene expression is less well understood, although it was recently reported that the interaction between RE and p300 accounts for increased expression of at least some RE target genes and leukemogenicity [19]. Our current study has identified RE and RE9a as a novel positive transcriptional regulators of Alox5. Interestingly, the data indicate that regulation of Alox5 by RE9a is indirect, as no peak of RE9a binding was observed in the Alox5 promoter region when performing ChIP-chip on RE9a-leukemia cells [18], and no RUNX1 binding motifs are present in the RE9a-responsive region of the Alox5 promoter (Figure 2). We further demonstrate that KLF6 is a critical factor for induction of the Alox5 promoter in cooperation with RE9a. To the best of our knowledge, this is the first report demonstrating both KLF6 regulation of Alox5 expression and the involvement of KLF6 in gene upregulation by t(8;21) fusion proteins. It will be important to examine whether this effect is specific to Alox5 or if KLF6 more broadly participates in t(8;21)-mediated transcriptional alterations. If future work determines that KLF6 does in fact participate in the regulation of disease-related RE and RE9a target genes, KLF6 itself may become an interesting target for future study in the treatment of AML.
The involvement of KLF6 here is especially interesting in light of the fact that KLF6 expression, like ALOX5, is also significantly upregulated in human AML M2 t(8;21)+ patient samples (Figure S1 and [34]). Therefore, KLF6 should be widely available in these leukemia cells to participate with t(8;21) fusion proteins in transcriptional regulation. We further demonstrate that increased KLF6 expression is induced by introduction of both RE and RE9a into the non-t(8;21) AML M2 cell line HL60 (Figure 4). Interestingly, according to published ChIP-seq data, RUNX1-ETO binds multiple sites within and nearby the KLF6 gene, indicating it may be a direct RUNX1-ETO target gene and this should be more closely studied in the future [16], [17].
Finally, we demonstrate for the first time that KLF6 interacts with RUNX1 and the Runt domain-containing t(8;21) fusion proteins. It is intriguing to note the potential implications of this interaction since, as with Runx1, knockout of Klf6 in mice results in embryonic lethality with severe defects in differentiation across all hematopoietic lineages [33]. It will be interesting to examine a potential role of RUNX1 and KLF6 cooperation in early hematopoietic development. For instance, does RUNX1, like RUNX1-ETO9a, also co-regulate gene expression with KLF6 and if so, what are the functions of these genes in early hematopoiesis? Do KLF6 and RUNX1 function partially redundantly in hematopoietic development? Furthermore, KLF6 is only 1 of at least 9 KLF family members with important roles in blood cell function and disease development. KLFs 4, 5 and 10 are important in T-cell activation and trafficking and KLF4 may function as a tumor suppressor in adult T-cell leukemia, KLF2 promotes memory B-cell differentiation and KLF3-deficient mice display a myeloproliferative disorder [38]. It will be interesting then to determine whether RUNX1 or t(8;21) fusion proteins work with any of these other KLF family members in gene expression and normal or disease-related blood cell development.
Materials and Methods
Ethics statement
C57BL/6J wildtype and Alox5-/- mice used in this study were housed in a pathogen-free facility. All procedures were performed in strict accordance with the recommendations of the Institutional Animal Care and Use Committee of the University of California, San Diego, CA, and every effort was made to minimize suffering.
Human and mouse gene expression
Human AML patient data [34] was analyzed using GraphPad Prism4 (GraphPad Software). For human cell line KLF6 expression, total RNA was harvested from 106 untreated HL60, Kasumi-1 and SKNO cells using the RNeasy Mini Kit (Qiagen). Retrovirally transduced HL60 cells were infected twice with virus produced by co-transfection of packaging vector and MSCV-IRES-Puror control or containing RUNX1-ETO in 293T cells. Infected HL60 cells were selected 2 days in 2 µg/ml puromycin to enrich for infected cells. 1 µg of RNA was used to generate cDNA using oligo (dT) and random primers (qScript cDNA SuperMix, Quanta Biosciences), and subject to qPCR on an iCycler (BioRad) using KAPA SYBR FAST Universal 2X qPCR Master Mix (KAPA Biosystems). KLF6 primer sequences: forward: TTCTCGGCGCTGCCGTCTCT, reverse: TCGCCAATGGGGTCGGAGGTA. For mouse Alox5 expression, lin−c-Kit+ hematopoietic cells were enriched from wildtype or leukemic mice using the Lineage Cell Depletion Kit and CD117 MicroBeads (Miltenyi Biotec) RNA extraction, cDNA synthesis and qPCR performed as above. Alox5 primer sequences: forward: CTCTTCCAAGCTCGAAGTGC, reverse: TGATGCTACCGAGTGACGAG.
Luciferase reporter assay
The indicated Alox5 promoter regions were cloned into pGL2 vector (Promega) with the six consensus RUNX1 binding sites in the vector backbone mutated from TGTGGT to TGTtag (pGLX2) to prevent binding of RE9a to the vector [15]; 106 K562 cells were nucleofected (Lonza) with 5 µg promoter-firefly luciferase DNA, 100 ng Renilla control luciferase DNA, and 2–3 µg p3xFlag-CMV-7.1 vector (Sigma-Aldrich) alone or containing RUNX1-ETO, RUNX1-ETO9a or KLF6 cDNA and analyzed 24 hr-post nucleofection using the Dual-Luciferase Reporter Assay System (Promega) on a Monolight 3010 (BD Biosciences). Unless otherwise stated, luciferase data show averages with standard deviations of 3 independent experiments, each performed in duplicate. For knock-down studies, 106 K562 cells were first transfected with 6 µg pSUPER.retro.puro (Oligoengine) containing shRNA, selected 2 days in 2 µg/ml puromycin, and then transfected and analyzed as above. Hairpin sense-strand sequences: shKLF6#1: CGGCTGCAGGAAAGTTTAC; shKLF6#2: GGAGAAAAGCCTTACAGAT. Significance was determined by Student t-test.
Immunoprecipitation and antibodies
K562 cells were nucleofected with 5 µg each p3xFlag-CMV-7.1-KLF6 and pcDNA6-HA-RUNX1, RUNX1-ETO or RUNX1-ETO9a. Pre-cleared lysates were incubated with rotation overnight at 4°C with 1 µg control or KLF6 antibody (Santa Cruz Biotechnology) and washed 5 times with lysis buffer [51] prior to SDS-PAGE. Antibody suppliers: α-tubulin (Covance), HA (Roche), SP1 (Santa Cruz), ALOX5 (Abcam).
Retroviral transduction and replating assay
Alox5-/ - mice[52] were purchased from Jackson Laboratory. Retroviral transduction and replating assays were performed as previously described [20]. Briefly, total bone marrow cells from wildtype or Alox5-/ - mice were transduced with retrovirus MSCV-IRES-Puror (MIP) [53] vector control or MIP containing HA-RUNX1-ETO9a, murine HA-Alox5, MLL-AF9 or PML-RARα cDNAs, as indicated. Infected cells were selected 1 week in 1 µg/ml puromycin in M3434 (STEMCELL Technologies). Ten thousand cells from each transduction were replated in duplicate every 7 days after colony and cell counting.
Fetal liver cell isolation, transduction, transplantation and flow cytometry
These assays were performed as previously described [10]. Briefly, fetal liver cells were harvested from day E13.5–16.5 wildtype or Alox5-/ - mouse embryos and transduced twice with MigR1 or MIG-RUNX1-ETO9a retrovirus. Lethally irradiated (900 rad) wildtype recipient mice were intravenously transplanted with the transduced fetal liver cells. Gr-1, CD11b, Sca-1, c-Kit, CD34 and FcγRII/III fluorescently conjugated antibodies were purchased from eBioscience. Staining and analyses were performed as previously described [10]. Kaplan-Meier survival curves and statistical analyses were performed using GraphPad Prism4.
Chromatin immunoprecipitation
ChIP assay was performed as described previously [20]. Cell lines were generated by retroviral transduction of K562 cells with MIP-HA-RE9a or MIP-Flag-KLF6. Each ChIP reaction contained chromatin from 107 cells and 5 µg antibody. Antibodies used were: HA (Santa Cruz) for RE9a, Flag (Sigma-Aldrich) for KLF6, and N-terminal RUNX1 [10] for endogenous RUNX1 in the absence of exogenous RE9a and KLF6. Following immunoprecipitation, enrichment of regions of the ALOX5 promoter was measured by qPCR using the following primers (numbers indicate nucleotides relative to transcription start site): ALOX5-A (−522 to −234) forward: AGCCTCTGTGCTCCAGAATCCATC, reverse: CGTTCACTCGTTCTCTCCTGAATTG; ALOX5-B (−259 to −79) forward: CAATTCAGGAGAGAACGAGTGAACG, reverse: GCAGTACTTCTCTCCCACTCTTCACG; ALOX5-C (+149 to +596) forward: CACTGACGACTACATCTACCTCAGCCTC, reverse: ATCTTGAAGTGGAGGGGAAACCTTG, and enrichment was normalized to IgG control.
Supporting Information
Zdroje
1. National Cancer Institute (2013) Adult Acute Myeloid Leukemia Treatment. http://www.cancer.gov/cancertopics/pdq/treatment/adultAML Accessed April 3, 2013.
2. RoweD, CotterillSJ, RossFM, BunyanDJ, VickersSJ, et al. (2000) Cytogenetically cryptic AML1-ETO and CBF beta-MYH11 gene rearrangements: incidence in 412 cases of acute myeloid leukaemia. Br J Haematol 111 : 1051–1056.
3. Groupe Francais de Cytogenetique Hematologique (1990) Acute myelogenous leukemia with an 8;21 translocation. A report on 148 cases from the Groupe Francais de Cytogenetique Hematologique. Cancer Genet Cytogenet 44 : 169–179.
4. NuciforaG, RowleyJD (1995) AML1 and the 8;21 and 3;21 translocations in acute and chronic myeloid leukemia. Blood 86 : 1–14.
5. LangabeerSE, WalkerH, RogersJR, BurnettAK, WheatleyK, et al. (1997) Incidence of AML1/ETO fusion transcripts in patients entered into the MRC AML trials. MRC Adult Leukaemia Working Party. Br J Haematol 99 : 925–928.
6. RegeK, SwansburyGJ, AtraAA, HortonC, MinT, et al. (2000) Disease features in acute myeloid leukemia with t(8;21)(q22;q22). Influence of age, secondary karyotype abnormalities, CD19 status, and extramedullary leukemia on survival. Leuk Lymphoma 40 : 67–77.
7. PetersonLF, BoyapatiA, AhnEY, BiggsJR, OkumuraAJ, et al. (2007) Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood 110 : 799–805.
8. MullerAM, DuqueJ, ShizuruJA, LubbertM (2008) Complementing mutations in core binding factor leukemias: from mouse models to clinical applications. Oncogene 27 : 5759–5773.
9. MulloyJC, CammengaJ, BerguidoFJ, WuK, ZhouP, et al. (2003) Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood 102 : 4369–4376.
10. YanM, KanbeE, PetersonLF, BoyapatiA, MiaoY, et al. (2006) A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med 12 : 945–949.
11. KozuT, FukuyamaT, YamamiT, AkagiK, KanekoY (2005) MYND-less splice variants of AML1-MTG8 (RUNX1-CBFA2T1) are expressed in leukemia with t(8;21). Genes Chromosomes Cancer 43 : 45–53.
12. GelmettiV, ZhangJ, FanelliM, MinucciS, PelicciPG, et al. (1998) Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol 18 : 7185–7191.
13. LutterbachB, WestendorfJJ, LinggiB, PattenA, MoniwaM, et al. (1998) ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol 18 : 7176–7184.
14. WangJ, HoshinoT, RednerRL, KajigayaS, LiuJM (1998) ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A 95 : 10860–10865.
15. HinesR, BoyapatiA, ZhangDE (2007) Cell type dependent regulation of multidrug resistance-1 gene expression by AML1-ETO. Blood Cells Mol Dis 39 : 297–306.
16. PtasinskaA, AssiSA, MannariD, JamesSR, WilliamsonD, et al. (2012) Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia 26 : 1829–1841.
17. MartensJH, MandoliA, SimmerF, WierengaBJ, SaeedS, et al. (2012) ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood 120 : 4038–4048.
18. LoMC, PetersonLF, YanM, CongX, JinF, et al. (2012) Combined gene expression and DNA occupancy profiling identifies potential therapeutic targets of t(8;21) AML. Blood 120 : 1473–1484.
19. WangL, GuralA, SunXJ, ZhaoX, PernaF, et al. (2011) The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 333 : 765–769.
20. ShiaWJ, OkumuraAJ, YanM, SarkeshikA, LoMC, et al. (2012) PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood 119 : 4953–4962.
21. TongWG, DingXZ, TalamontiMS, BellRH, AdrianTE (2005) LTB4 stimulates growth of human pancreatic cancer cells via MAPK and PI-3 kinase pathways. Biochem Biophys Res Commun 335 : 949–956.
22. KimEY, SeoJM, ChoKJ, KimJH (2010) Ras-induced invasion and metastasis are regulated by a leukotriene B4 receptor BLT2-linked pathway. Oncogene 29 : 1167–1178.
23. KimGY, LeeJW, ChoSH, SeoJM, KimJH (2009) Role of the low-affinity leukotriene B4 receptor BLT2 in VEGF-induced angiogenesis. Arterioscler Thromb Vasc Biol 29 : 915–920.
24. MezhybovskaM, WikstromK, OhdJF, SjolanderA (2006) The inflammatory mediator leukotriene D4 induces beta-catenin signaling and its association with antiapoptotic Bcl-2 in intestinal epithelial cells. J Biol Chem 281 : 6776–6784.
25. RiouxN, CastonguayA (1998) Inhibitors of lipoxygenase: a new class of cancer chemopreventive agents. Carcinogenesis 19 : 1393–1400.
26. HennigR, VenturaJ, SegersvardR, WardE, DingXZ, et al. (2005) LY293111 improves efficacy of gemcitabine therapy on pancreatic cancer in a fluorescent orthotopic model in athymic mice. Neoplasia 7 : 417–425.
27. PidgeonGP, LysaghtJ, KrishnamoorthyS, ReynoldsJV, O'ByrneK, et al. (2007) Lipoxygenase metabolism: roles in tumor progression and survival. Cancer Metastasis Rev 26 : 503–524.
28. ChungJW, KimGY, MunYC, AhnJY, SeongCM, et al. (2005) Leukotriene B4 pathway regulates the fate of the hematopoietic stem cells. Exp Mol Med 37 : 45–50.
29. ChenY, HuY, ZhangH, PengC, LiS (2009) Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat Genet 41 : 783–792.
30. RaiKR, HollandJF, GlidewellOJ, WeinbergV, BrunnerK, et al. (1981) Treatment of acute myelocytic leukemia: a study by cancer and leukemia group B. Blood 58 : 1203–1212.
31. RobozGJ (2012) Current treatment of acute myeloid leukemia. Curr Opin Oncol 24 : 711–719.
32. GrimwadeD, HillsRK, MoormanAV, WalkerH, ChattersS, et al. (2010) Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 116 : 354–365.
33. MatsumotoN, KuboA, LiuH, AkitaK, LaubF, et al. (2006) Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood 107 : 1357–1365.
34. ValkPJ, VerhaakRG, BeijenMA, ErpelinckCA, van Waalwijk van DoornKhosrovaniBarjesteh, et al. (2004) Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 350 : 1617–1628.
35. SilvermanES, LeL, BaronRM, HallockA, HjobergJ, et al. (2002) Cloning and functional analysis of the mouse 5-lipoxygenase promoter. Am J Respir Cell Mol Biol 26 : 475–483.
36. Juven-GershonT, KadonagaJT (2010) Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339 : 225–229.
37. ZhaoJL, AustenKF, LamBK (2000) Cell-specific transcription of leukotriene C(4) synthase involves a Kruppel-like transcription factor and Sp1. J Biol Chem 275 : 8903–8910.
38. CaoZ, SunX, IcliB, WaraAK, FeinbergMW (2010) Role of Kruppel-like factors in leukocyte development, function, and disease. Blood 116 : 4404–4414.
39. BotellaLM, Sanchez-ElsnerT, Sanz-RodriguezF, KojimaS, ShimadaJ, et al. (2002) Transcriptional activation of endoglin and transforming growth factor-beta signaling components by cooperative interaction between Sp1 and KLF6: their potential role in the response to vascular injury. Blood 100 : 4001–4010.
40. WeiH, LiuX, XiongX, WangY, RaoQ, et al. (2008) AML1-ETO interacts with Sp1 and antagonizes Sp1 transactivity through RUNT domain. FEBS Lett 582 : 2167–2172.
41. LiuY, ChenW, GaudetJ, CheneyMD, RoudaiaL, et al. (2007) Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity. Cancer Cell 11 : 483–497.
42. DekelverRC, YanM, AhnEY, ShiaWJ, SpeckNA, et al. (2013) Attenuation of AML1-ETO cellular dysregulation correlates with increased leukemogenic potential. Blood 121 : 3714–7.
43. SwansburyGJ, SlaterR, BainBJ, MoormanAV, Secker-WalkerLM (1998) Hematological malignancies with t(9;11)(p21-22;q23)–a laboratory and clinical study of 125 cases. European 11q23 Workshop participants. Leukemia 12 : 792–800.
44. GrignaniF, FagioliM, AlcalayM, LongoL, PandolfiPP, et al. (1994) Acute promyelocytic leukemia: from genetics to treatment. Blood 83 : 10–25.
45. KuoYH, LandretteSF, HeilmanSA, PerratPN, GarrettL, et al. (2006) Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell 9 : 57–68.
46. de GuzmanCG, WarrenAJ, ZhangZ, GartlandL, EricksonP, et al. (2002) Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol 22 : 5506–5517.
47. TamWF, HahnelPS, SchulerA, LeeBH, OkabeR, et al. (2013) STAT5 is crucial to maintain leukemic stem cells in acute myelogenous leukemias induced by MOZ-TIF2. Cancer Res 73 : 373–384.
48. PengC, ChenY, ShanY, ZhangH, GuoZ, et al. (2012) LSK derived LSK - cells have a high apoptotic rate related to survival regulation of hematopoietic and leukemic stem cells. PLoS One 7: e38614.
49. BenzC, CopleyMR, KentDG, WohrerS, CortesA, et al. (2012) Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 10 : 273–283.
50. PetersonLF, ZhangDE (2004) The 8;21 translocation in leukemogenesis. Oncogene 23 : 4255–4262.
51. AhnEY, DekelverRC, LoMC, NguyenTA, MatsuuraS, et al. (2011) SON Controls Cell-Cycle Progression by Coordinated Regulation of RNA Splicing. Mol Cell 42 : 185–198.
52. ChenXS, ShellerJR, JohnsonEN, FunkCD (1994) Role of leukotrienes revealed by targeted disruption of the 5-lipoxygenase gene. Nature 372 : 179–182.
53. PetersonLF, WangY, LoMC, YanM, KanbeE, et al. (2007) The multi-functional cellular adhesion molecule CD44 is regulated by the 8;21 chromosomal translocation. Leukemia 21 : 2010–2019.
54. MeyersS, DowningJR, HiebertSW (1993) Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol 13 : 6336–6345.
55. NarlaG, DifeoA, ReevesHL, SchaidDJ, HirshfeldJ, et al. (2005) A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res 65 : 1213–1222.
Štítky
Genetika Reprodukční medicína
Článek Defending Sperm FunctionČlánek How to Choose the Right MateČlánek Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental TransitionČlánek Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in SpermatogenesisČlánek The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate EmbryogenesisČlánek Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time inČlánek Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral MyelinationČlánek A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase inČlánek Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 10- Intrauterinní inseminace a její úspěšnost
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
- Růst a vývoj dětí narozených pomocí IVF
- Akutní intermitentní porfyrie
- Příjem alkoholu a menstruační cyklus
-
Všechny články tohoto čísla
- Defending Sperm Function
- How to Choose the Right Mate
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
- Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental Transition
- A Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
- Tracking Proliferative History in Lymphocyte Development with Cre-Mediated Sister Chromatid Recombination
- Correlated Occurrence and Bypass of Frame-Shifting Insertion-Deletions (InDels) to Give Functional Proteins
- Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
- Loss of miR-10a Activates and Collaborates with Activated Wnt Signaling in Inducing Intestinal Neoplasia in Female Mice
- Both Rare and Copy Number Variants Are Prevalent in Agenesis of the Corpus Callosum but Not in Cerebellar Hypoplasia or Polymicrogyria
- Reverse PCA, a Systematic Approach for Identifying Genes Important for the Physical Interaction between Protein Pairs
- Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice
- Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in Spermatogenesis
- Coordinate Regulation of Mature Dopaminergic Axon Morphology by Macroautophagy and the PTEN Signaling Pathway
- Cooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
- Mobility of the Native Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling
- FliZ Is a Global Regulatory Protein Affecting the Expression of Flagellar and Virulence Genes in Individual Bacterial Cells
- Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing
- Condensin II Subunit dCAP-D3 Restricts Retrotransposon Mobilization in Somatic Cells
- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
- Human Intellectual Disability Genes Form Conserved Functional Modules in
- Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
- ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences
- Different Roles of Eukaryotic MutS and MutL Complexes in Repair of Small Insertion and Deletion Loops in Yeast
- The Spore Differentiation Pathway in the Enteric Pathogen
- Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization
- The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
- Genome Wide Analysis Reveals Zic3 Interaction with Distal Regulatory Elements of Stage Specific Developmental Genes in Zebrafish
- Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State
- The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
- Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways
- The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally
- Direct Monitoring of the Strand Passage Reaction of DNA Topoisomerase II Triggers Checkpoint Activation
- Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in
- A Reversible Histone H3 Acetylation Cooperates with Mismatch Repair and Replicative Polymerases in Maintaining Genome Stability
- ALS-Associated Mutations Result in Compromised Alternative Splicing and Autoregulation
- Robust Demographic Inference from Genomic and SNP Data
- Preferential Binding to Elk-1 by SLE-Associated Risk Allele Upregulates Expression
- Rad52 Sumoylation Prevents the Toxicity of Unproductive Rad51 Filaments Independently of the Anti-Recombinase Srs2
- The Serum Resistome of a Globally Disseminated Multidrug Resistant Uropathogenic Clone
- Identification of 526 Conserved Metazoan Genetic Innovations Exposes a New Role for Cofactor E-like in Neuronal Microtubule Homeostasis
- SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast
- Integrated Enrichment Analysis of Variants and Pathways in Genome-Wide Association Studies Indicates Central Role for IL-2 Signaling Genes in Type 1 Diabetes, and Cytokine Signaling Genes in Crohn's Disease
- Genome-Wide High-Resolution Mapping of UV-Induced Mitotic Recombination Events in
- Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in
- Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral Myelination
- Two Portable Recombination Enhancers Direct Donor Choice in Fission Yeast Heterochromatin
- Mining the Human Phenome Using Allelic Scores That Index Biological Intermediates
- Yeast Tdh3 (Glyceraldehyde 3-Phosphate Dehydrogenase) Is a Sir2-Interacting Factor That Regulates Transcriptional Silencing and rDNA Recombination
- A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
- A Review of Bacteria-Animal Lateral Gene Transfer May Inform Our Understanding of Diseases like Cancer
- High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity
- Partitioning the Heritability of Tourette Syndrome and Obsessive Compulsive Disorder Reveals Differences in Genetic Architecture
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- A GDF5 Point Mutation Strikes Twice - Causing BDA1 and SYNS2
- Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in
- Natural Genetic Variation of Integrin Alpha L () Modulates Ischemic Brain Injury in Stroke
- Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
- Evolutionary Change within a Bipotential Switch Shaped the Sperm/Oocyte Decision in Hermaphroditic Nematodes
- Limiting of the Innate Immune Response by SF3A-Dependent Control of MyD88 Alternative mRNA Splicing
- Multiple Signaling Pathways Coordinate to Induce a Threshold Response in a Chordate Embryo
- Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube
- Genome Wide Analysis of Narcolepsy in China Implicates Novel Immune Loci and Reveals Changes in Association Prior to Versus After the 2009 H1N1 Influenza Pandemic
- Mismatch Repair Genes and Modify CAG Instability in Huntington's Disease Mice: Genome-Wide and Candidate Approaches
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
- Function and Evolution of DNA Methylation in
- Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome
- Transcription Termination and Chimeric RNA Formation Controlled by FPA
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



