-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Divergence of the Yeast Transcription Factor Affects Sulfite Resistance
Changes in gene expression are commonly observed during evolution. However, the phenotypic consequences of expression divergence are frequently unknown and difficult to measure. Transcriptional regulators provide a mechanism by which phenotypic divergence can occur through multiple, coordinated changes in gene expression during development or in response to environmental changes. Yet, some changes in transcriptional regulators may be constrained by their pleiotropic effects on gene expression. Here, we use a genome-wide screen for promoters that are likely to have diverged in function and identify a yeast transcription factor, FZF1, that has evolved substantial differences in its ability to confer resistance to sulfites. Chimeric alleles from four Saccharomyces species show that divergence in FZF1 activity is due to changes in both its coding and upstream noncoding sequence. Between the two closest species, noncoding changes affect the expression of FZF1, whereas coding changes affect the expression of SSU1, a sulfite efflux pump activated by FZF1. Both coding and noncoding changes also affect the expression of many other genes. Our results show how divergence in the coding and promoter region of a transcription factor alters the response to an environmental stress.
Published in the journal: . PLoS Genet 8(6): e32767. doi:10.1371/journal.pgen.1002763
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002763Summary
Changes in gene expression are commonly observed during evolution. However, the phenotypic consequences of expression divergence are frequently unknown and difficult to measure. Transcriptional regulators provide a mechanism by which phenotypic divergence can occur through multiple, coordinated changes in gene expression during development or in response to environmental changes. Yet, some changes in transcriptional regulators may be constrained by their pleiotropic effects on gene expression. Here, we use a genome-wide screen for promoters that are likely to have diverged in function and identify a yeast transcription factor, FZF1, that has evolved substantial differences in its ability to confer resistance to sulfites. Chimeric alleles from four Saccharomyces species show that divergence in FZF1 activity is due to changes in both its coding and upstream noncoding sequence. Between the two closest species, noncoding changes affect the expression of FZF1, whereas coding changes affect the expression of SSU1, a sulfite efflux pump activated by FZF1. Both coding and noncoding changes also affect the expression of many other genes. Our results show how divergence in the coding and promoter region of a transcription factor alters the response to an environmental stress.
Introduction
Transcriptional regulation plays a key role in development and an organism's response to physiological and environmental changes. However, changes in gene regulation that occur over the course of evolution are more difficult to interpret. Genome-wide patterns of gene expression divergence show that while many aspects of regulation are conserved between distantly related species [1]–[3], there is also extensive variation in gene expression levels within and between closely related species [4]. In many, but not all instances, gene expression divergence is consistent with a neutral model of evolutionary change [5]–[8]. Yet, understanding regulatory divergence requires identifying the genetic basis of divergence in gene expression and knowing which changes in gene expression translate into changes in phenotype and fitness.
Substantial progress has been made in understanding the genetic basis of regulatory divergence. Changes in gene expression are influenced by both cis-regulatory sequences and trans-acting factors, with cis-regulatory changes being enriched in interspecific comparisons [9], [10]. Expression changes caused by cis-regulatory elements frequently involve gain or loss of transcription factor binding sites, e.g. [11], [12], although other changes, such as nucleosome position, can also play an important role [13]. Even when changes in gene expression can be attributed to specific cis-regulatory elements, the phenotypic consequences of such changes are hard to know, especially if they depend on the combined effects of many cis-regulatory changes. While changes in trans-acting factors can simultaneously influence the expression of many genes, significant efforts are needed to identify the genetic basis of trans-acting changes in gene expression.
The phenotypic effects of changes in gene expression have in some cases been identified [14]. This has primarily been accomplished by mapping, association and transgenic studies that identify genetic changes underlying a phenotype. While these approaches typically identify changes in protein coding sequences, cis-regulatory changes are more frequently found to underlie interspecific compared to intraspecific differences [14]. Furthermore, changes in protein coding sequences can affect the expression of many genes [15], [16], and in some cases their phenotypic effects depend on multiple differentially expressed genes [17].
What has been more difficult to investigate is the combined influence of multiple regulatory changes. Multiple changes of small effect may frequently go undetected, at least individually, but together could have a substantial impact on divergence [18]. Evidence for adaptive evolution via multiple cis-regulatory changes has been found based on concerted changes in the expression of genes that function in the same pathway or biological process [19]–[21]. Multiple cis-regulatory changes at a single locus have also been found to make substantial contributions to phenotypic divergence between species [22]–[26].
Statistical tests of neutrality are particularly well-suited to identifying multiple adaptive substitutions at a single locus since multiple substitutions are often needed to detect a significant deviation from a neutral pattern of molecular evolution. Rapidly evolving noncoding sequences have been identified in a number of species [27]–[30], and in some instances are known to cause notable changes in gene expression [31], [32]. Although tests of neutrality rely on the concentration of multiple changes at single loci, clustering of changes may occur if there are genetic, developmental or selective constraints at other loci [33].
One mechanism by which multiple, coordinated changes in gene expression may arise is through changes in transcriptional regulators. However, changes in transcription factors can also be constrained by their pleiotropic effects on gene expression. The negative effects of pleiotropy may in some cases be eliminated by altering the regulation of a transcription factor; thereby limiting downstream changes in gene expression to specific times during development, within particular cells or tissues, or to certain environmental conditions [33], [34].
In this study, we investigated changes in gene expression and phenotype caused by a rapidly evolving transcription factor, FZF1. To directly target genes that have potentially accrued multiple cis-regulatory changes, we screened four Saccharomyces genomes for noncoding sequences with non-neutral patterns of divergence. FZF1 was among the genes identified and it also shows a non-neutral pattern of amino acid divergence [35]. To examine the phenotypic consequences of FZF1 divergence we used cross-species complementation assays and found divergence in both its coding and upstream noncoding sequence affect sulfite resistance. Whereas divergence upstream of FZF1 affects its expression in response to sulfites, divergence in the coding region of FZF1 affects the expression of SSU1, an efflux pump that mediates sulfite resistance [36]–[38]. Coincident with their effects on sulfite resistance, both the coding and noncoding regions of FZF1 affect the expression of many other genes. Our results show how divergence in the coding and promoter region of a transcription factor affect the response to an environmental stress.
Results
Patterns of sequence divergence at FZF1
To identify promoter sequences likely to have diverged in function, we screened the noncoding sequences of four Saccharomyces species for accelerated substitution rates. We used a likelihood ratio test to compare a model of sequence evolution where the ratio of the noncoding to synonymous substitution rate, dNC/dS, is constant across lineages versus a model where dNC/dS is free to vary across lineages. Out of 2,539 noncoding regions tested, we identified 145 that showed significant variation in the noncoding substitution rate across species (Likelihood ratio test, P<0.05, Bonferroni corrected, Dataset S1). In these regions, a higher noncoding substitution rate in one or more lineages may be the result of loss of constraint, or in some cases, positive selection.
One of the noncoding regions that we identified lies upstream of the transcription factor FZF1. We selected FZF1 for further analysis because it is known to function in sulfite resistance, a hypothesized adaptation to vineyard environments [39], and its potential role in gene expression divergence. The substitution rate upstream of FZF1 is characterized by an accelerated rate along the lineages leading to Saccharomyces cerevisiae and Saccharomyces paradoxus relative to that along the lineages leading to Saccharomyces mikatae and Saccharomyces bayanus (Figure 1). However, previous studies have shown that signals of selection are highly dependent on the alignment [40], [41]. To determine whether the evidence for rate heterogeneity upstream of FZF1 is dependent on the alignment used, we generated additional alignments using alternative alignment parameters and algorithms, and tested each for substitution rate heterogeneity. Both the alignment parameters and the algorithm affected the evidence for rate heterogeneity, with 9 out of 18 alignments showing evidence of rate heterogeneity (Table S1, Likelihood ratio test, P<0.05, Bonferroni corrected). Although the high substitution rate combined with uncertainty in the placement of insertions or deletions makes it difficult to know the correct alignment, dNC/dS along the S. cerevisiae and S. paradoxus lineage was consistently estimated to be greater than or equal to one (Figure 1).
Fig. 1. Variation in noncoding substitution rates upstream of FZF1. 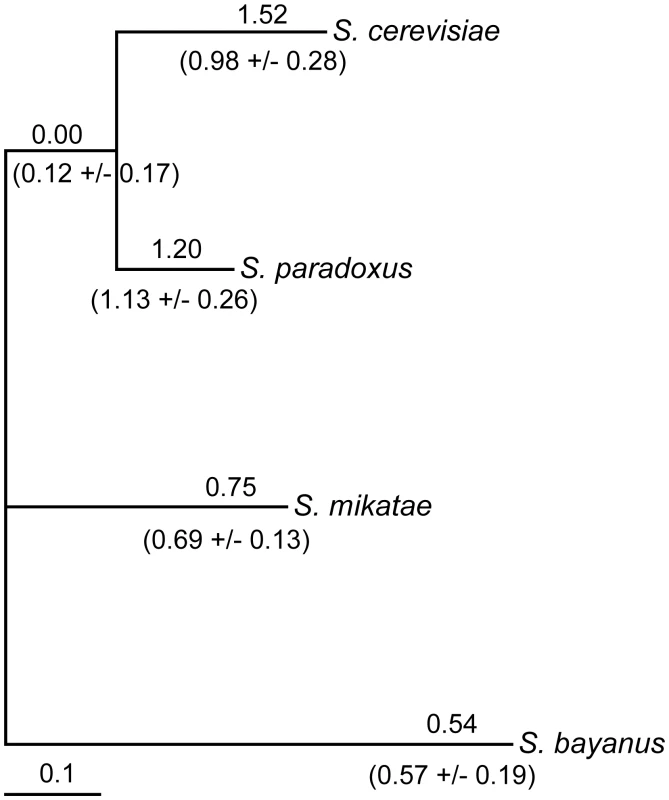
The ratio of the noncoding substitution rate upstream of FZF1 relative to genome-wide substitution rates at fourfold degenerate sites, dNC/dS, is shown above each lineage for the original alignment. The mean and standard deviation of dNC/dS from 18 different alignments (Table S1) is shown below each lineage. dNC/dS was estimated for each lineage using an unconstrained model by maximum likelihood methods implemented in HyPhy. The tree is scaled to the fourfold synonymous substitution rate. The protein coding sequence of FZF1 also shows evidence for non-neutral evolution based on a sliding window analysis of the nonsynonymous to synonymous substitution rate ratio (dN/dS) between S. cerevisiae and S. paradoxus [35]. However, caution should be taken when interpreting the results of the dN/dS test in the context of a sliding window analysis since dS can vary for a number of reasons [42]. Upon re-examination of divergence in FZF1, we found that the window with the signal of positive selection, dN/dS = 1.95, is characterized by a synonymous substitution rate of 0.18, which is lower than the average of 0.46 across the entire gene, and a nonsynonymous substitution rate of 0.34, which is higher than the average of 0.14 across the entire gene. Despite some uncertainty regarding the evidence for non-neutral evolution, we decided that FZF1 was a reasonable candidate to test for functional divergence.
The phenotypic effects of FZF1 divergence
FZF1 encodes a five zinc finger transcription factor that activates the plasma membrane sulfite pump, SSU1 [37]. Gain of function mutations in FZF1 result in hyperactivation of SSU1 and increased sulfite resistance [36], [38]. To determine whether FZF1 has diverged in its ability to confer sulfite resistance, we tested FZF1 alleles from four Saccharomyces species: S. cerevisiae, S. paradoxus, S. mikitae, and S. bayanus, for their ability to complement a deletion of FZF1 in S. cerevisiae. The S. cerevisiae allele of FZF1 showed nearly complete complementation of the FZF1 deletion, as measured by the delay in exponential growth following sulfite treatment (Figure S1). In comparison, FZF1 alleles from the other three species all showed a shorter delay in growth relative to that of S. cerevisiae, indicating that these FZF1 alleles confer greater resistance to sulfites (Figure 2, Kruskal-Wallis test, P = 5.3×10−13).
Fig. 2. FZF1 alleles from different species have diverged in function. 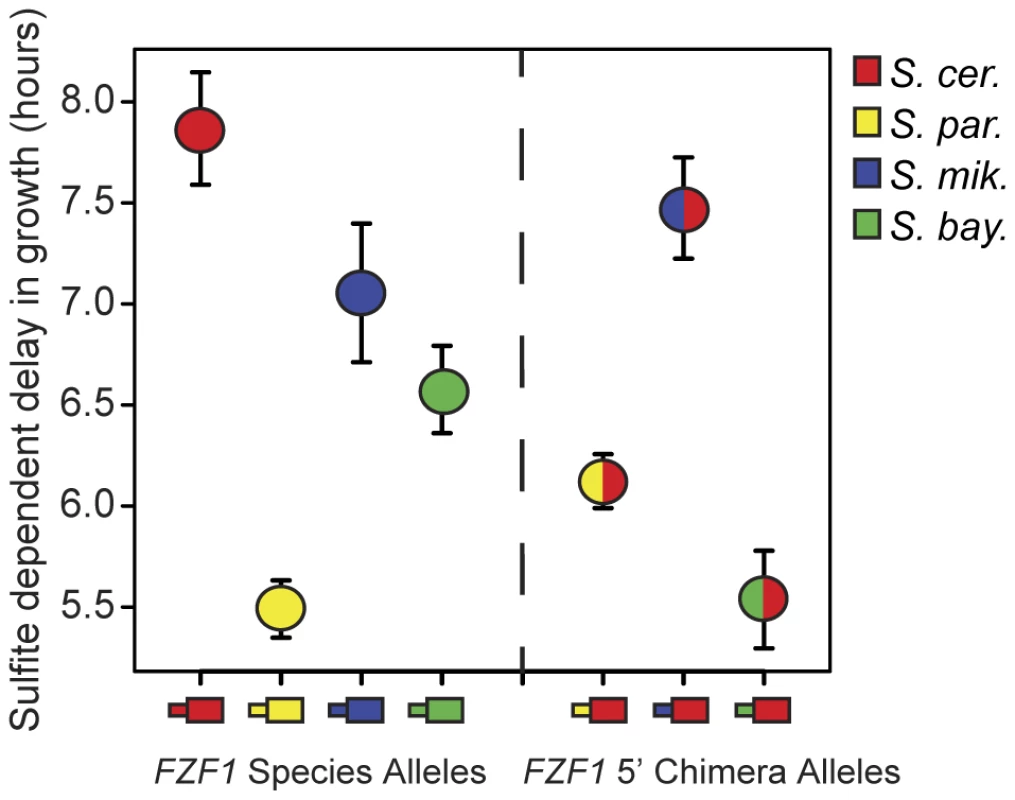
The left side of the figure shows sulfite resistance of FZF1 alleles from four different species: S. cerevisiae (red), S. paradoxus (yellow), S. mikatae (blue), and S. bayanus (green) in an S. cerevisiae strain background. The right side of the figure shows sulfite resistance of chimeric alleles of FZF1 composed of the 5′ noncoding region from S. paradoxus, S. mikatae and S. bayanus combined with the S. cerevisiae coding region. Error bars show the 95% confidence interval of the mean. To determine whether divergence in FZF1 activity resulted from changes in its protein coding sequence or upstream noncoding sequence, we also tested chimeric constructs containing each species' FZF1 upstream noncoding sequence combined with the S. cerevisiae FZF1 coding sequence. These FZF1 5′ noncoding chimeras conferred significant differences in sulfite resistance (Figure 2, Kruskal-Wallis test, P = 2.5×10−18), indicating that the 5′ noncoding region alone makes a significant contribution to FZF1 divergence. Both the S. paradoxus - S. cerevisiae and S. mikatae - S. cerevisiae chimeric alleles showed sulfite resistance intermediate to that of their full length parental alleles, although only the former chimera was significantly different from both parent alleles (Wilcoxon rank sum test, P = 1.9×10−14 for the S. cerevisiae parent and P = 4.2×10−8 for the S. paradoxus parent). In contrast, the S. bayanus 5′ noncoding region upstream of an S. cerevisiae coding sequence conferred greater resistance than either of the two full length parent alleles (Figure 2, Wilcoxon rank sum test, P = 4.6×10−16 for the S. cerevisiae parent and P = 2.4×10−8 for the S. bayanus parent).
Multiple changes are responsible for divergence between S. cerevisiae and S. paradoxus alleles of FZF1
The S. cerevisiae and S. paradoxus alleles of FZF1 confer the largest difference in sulfite resistance. This phenotypic divergence corresponds to the lineages showing the highest noncoding to synonymous substitution rates and the elevated nonsynonymous to synonymous substitution rate within a portion of the coding region. Thus, we further mapped the differences in sulfite resistance between the S. cerevisiae and S. paradoxus FZF1 alleles.
The S. cerevisiae FZF1 protein is 900 amino acids long and has 195 bases in the 5′ noncoding region. Between the S. cerevisiae and S. paradoxus FZF1 alleles there are 67 amino acid differences and 82 differences in the 5′ noncoding region, 31 of which are insertion/deletion differences. To delineate which subset of these differences are responsible for divergence in sulfite resistance, we generated ten sets of reciprocal chimeric constructs between the two species (Figure 3). The FZF1 chimeric breakpoints were located (1) in the middle of the 5′ noncoding region, (2) at the junction between the 5′ noncoding and the coding region, (3) in the coding region between the first zinc finger domain, known to bind DNA [37], and the region under positive selection [35], and (4) at the junction between the coding and 3′ noncoding region. Five sets of chimeric constructs contain a single region in the opposite background and the remaining sets of constructs contain five of the ten possible pairwise combinations of each region.
Fig. 3. FZF1 gene region and chimeric alleles. 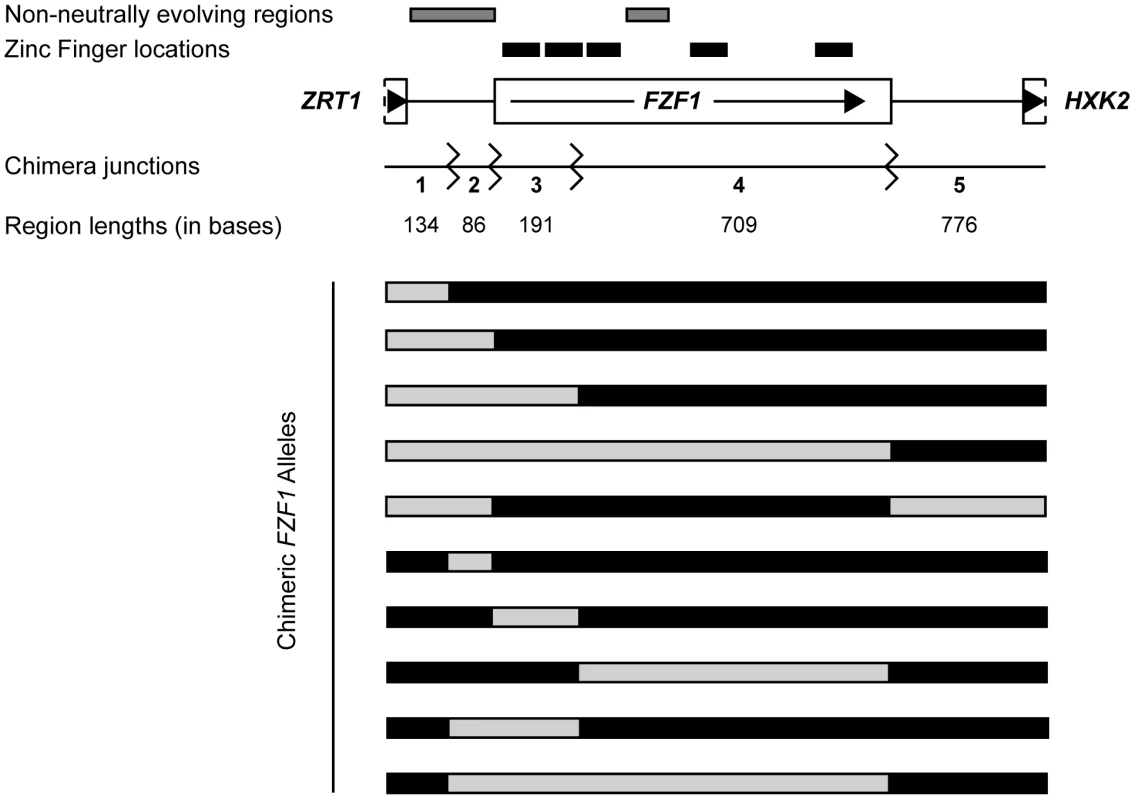
The FZF1 gene region is shown along with the breakpoints used to generate reciprocal chimeric alleles. Regions with non-neutral evolution are indicated by gray boxes and the predicted zinc fingers are indicated by black boxes [73]. Region lengths between chimera junctions are given for S. cerevisiae. One set of chimeric alleles between S. cerevisiae and S. paradoxus are shown below. The reciprocal set is not shown. Including the full length S. cerevisiae and S. paradoxus alleles of FZF1, the 22 constructs show a nearly continuous distribution of sulfite resistance (Figure 4). Using an additive model, the estimated effects of the first three FZF1 regions individually account for 8.2%, 39.0%, and 49.5%, respectively, of the difference in sulfite resistance between the S. cerevisiae and S. paradoxus alleles (Table 1). The latter two regions are not statistically significant. Some of the variation in sulfite resistance can be attributed to non-additive interactions among regions. The additive model explains a total of 66% of the variance among alleles, significantly less than a model that allows for pairwise epistatic interactions, which explains 70% of the variance (Likelihood ratio test, 2Δln(L) = 56.48, 10 d.f., P = 1.7×10−8). However, out of all the pairwise interactions, only the interaction between the two coding regions is individually significant after correcting for multiple tests (Table 1). The interaction indicates that the two coding regions have a smaller effect in combination compared to that expected from each region individually.
Fig. 4. Multiple noncoding and coding changes contribute to sulfite resistance. 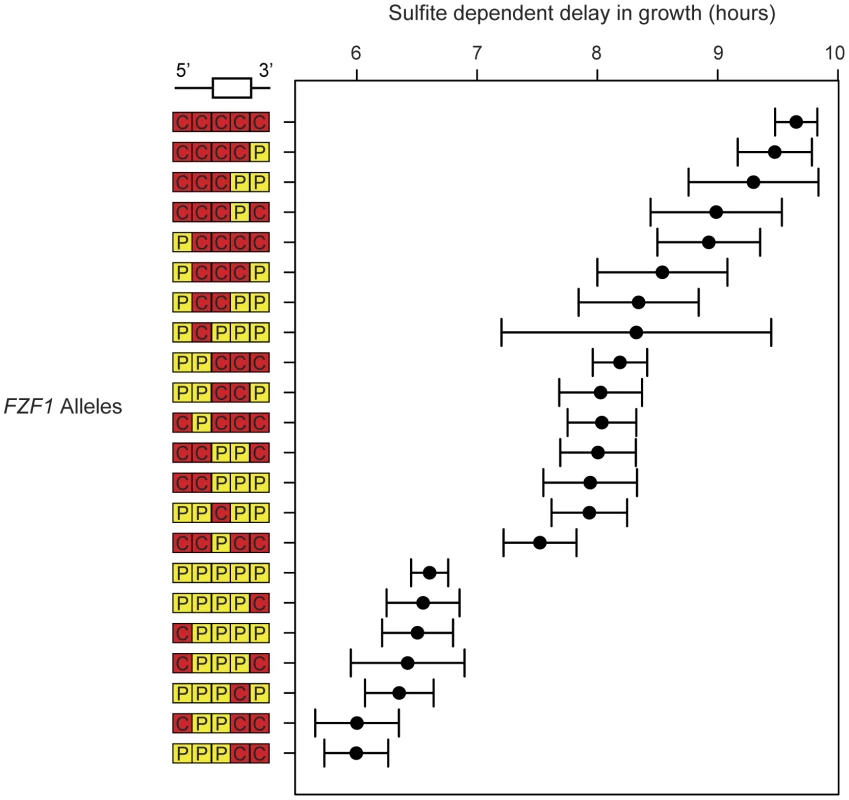
Sulfite resistance is shown for chimeric alleles of FZF1 from S. cerevisiae and S. paradoxus. Chimera breakpoints are shown in Figure 3 and are labeled 5′ to 3′ based on the origin of each region: S. cerevisiae (red, “C”) and S. paradoxus (yellow, “P”). Error bars show the 95% confidence interval of the mean. Tab. 1. Estimated effects of FZF1 divergence on sulfite resistance. 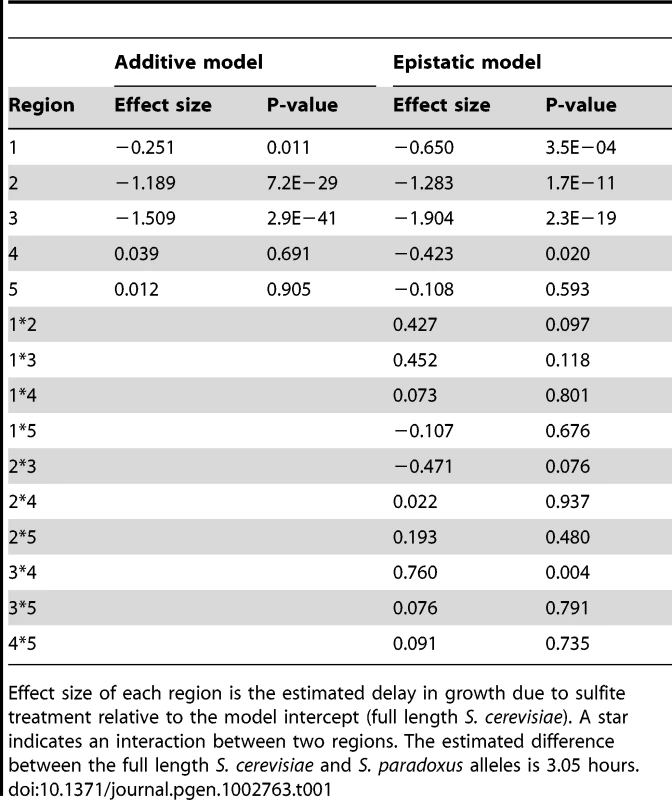
Effect size of each region is the estimated delay in growth due to sulfite treatment relative to the model intercept (full length S. cerevisiae). A star indicates an interaction between two regions. The estimated difference between the full length S. cerevisiae and S. paradoxus alleles is 3.05 hours. Changes in gene expression caused by FZF1 divergence
FZF1-dependent changes in sulfite resistance may be mediated by changes in the expression of FZF1 or the expression of other genes. To characterize changes in gene expression caused by FZF1 divergence, we measured expression of FZF1 and SSU1, a sulfite efflux pump activated by FZF1 [37], [38]. Using quantitative PCR, we measured the expression of both genes before and after sulfite treatment of strains carrying an S. cerevisiae, S. paradoxus, or two reciprocal chimeric FZF1 alleles, which divide the coding and 5′ noncoding regions of the S. cerevisiae and S. paradoxus FZF1 allele.
All of the FZF1 alleles increased in expression following sulfite treatment. However at time-points 15, 30 and 60 minutes after sulfite treatment, the FZF1 alleles with an S. paradoxus promoter were expressed at higher levels than those containing an S. cerevisiae promoter (Wilcoxon rank sum test, P = 6.7×10−9, P = 1.7×10−4, P = 0.008, respectively, Figure 5A). No significant differences were found due to the FZF1 coding region alone from the two species. Yet, 30 minutes after sulfite treatment, the two FZF1 alleles with the S. paradoxus promoter showed significant differences in expression; the allele with an S. cerevisiae coding region remained at a higher level relative to the allele with an S. paradoxus coding region (Wilcoxon rank sum test, P = 0.0012). Similarly, the FZF1 allele with an S. cerevisiae promoter and S. paradoxus coding region showed higher expression at the 30 minute time-point relative to the full length S. cerevisiae allele, although this difference was not significant (Wilcoxon rank sum test, P = 0.15). Differences in gene expression that depend on changes within a coding region have previously been found in yeast [43] and could result from feedback regulation.
Fig. 5. FZF1 alleles affect the expression of both FZF1 and SSU1 subsequent to sulfite treatment. 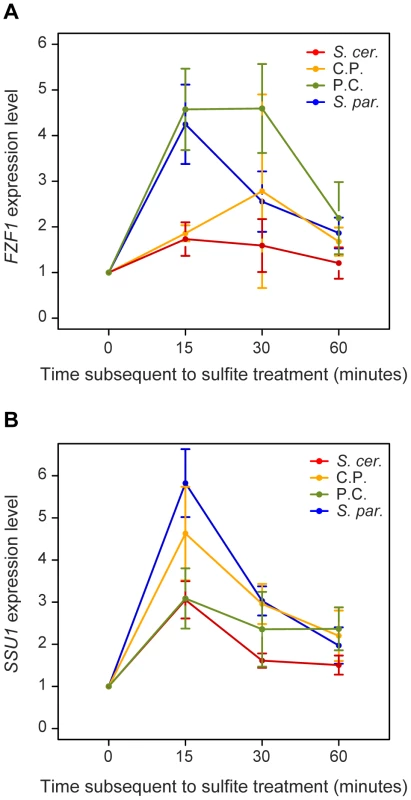
Expression of FZF1 (A) and SSU1 (B) was measured prior to and at three time-points after sulfite treatment for strains carrying four FZF1 alleles: S. cerevisiae (S. cer, red), S. paradoxus (S. par, blue), S. cerevisiae 5′ noncoding with S. paradoxus coding (C.P., gold), S. paradoxus 5′ noncoding and S. cerevisiae coding (P.C., green). Expression levels were normalized to the 0 time-point. Each point is the mean of 3–4 individual observations. Error bars represent the 95% confidence interval of the mean. The FZF1 alleles also caused an increase in SSU1 expression after sulfite treatment (Figure 5B). Unlike FZF1 expression, SSU1 expression primarily depended on the origin of the FZF1 coding region. For both the 15 and 30 minute time-points, FZF1 alleles containing the S. paradoxus coding region caused higher levels of SSU1 expression relative to those containing the S. cerevisiae coding region (Wilcoxon rank sum test, P = 1.15×10−5, P = 8.94×10−6, respectively). No significant differences in SSU1 expression were found as a result of the FZF1 5′ noncoding region alone.
If FZF1-dependent differences in sulfite resistance are mediated by activation of FZF1 and SSU1, they may also be influenced by levels of FZF1 and SSU1 expression prior to sulfite treatment. Immediately prior to sulfite treatment, FZF1 alleles with the S. cerevisiae coding sequences were expressed at 1.5-fold higher levels than those with the S. paradoxus coding sequence (Wilcoxon rank sum test, P = 1.3×10−6). The 5′ noncoding region caused no significant differences in FZF1 expression prior to sulfite treatment. In comparison, expression of SSU1 prior to sulfite treatment was 1.09-fold higher for FZF1 alleles containing the S. cerevisiae coding region and 1.12-fold higher for FZF1 alleles containing the S. cerevisiae 5′ noncoding region relative to the corresponding S. paradoxus regions (Wilcoxon rank sum test, P = 0.011, P = 6.5×10−4, respectively). Because the S. paradoxus allele of FZF1 causes higher levels of sulfite resistance, levels of FZF1 expression prior to sulfite treatment do not appear to be related to sulfite resistance.
The effect of FZF1 divergence on SSU1 expression suggests that FZF1 may also affect the expression of other genes. To examine this possibility, we measured genome-wide changes in expression caused by the S. cerevisiae and S. paradoxus FZF1 alleles and the two reciprocal 5′ noncoding chimeras. Gene expression was measured using microarrays before and 15 minutes after addition of sulfites. Out of 6127 open reading frames queried, 655 showed FZF1-dependent differences in expression across both time-points and 648 showed FZF1-dependent differences in expression that varied by time-point (ANOVA, P<0.01 for both). For both tests, permutation resampling of the data indicated a false discovery rate of 9.8%. Out of the combined set of 1,096 genes that showed FZF1-dependent differences in expression, 87% showed significant changes following sulfite treatment (ANOVA, P<0.01), of which 219 and 271 showed a >2-fold decrease and increase, respectively, in expression following sulfite treatment. Consistent with other studies of the stress response [44], [45], many of the genes that decreased in expression are involved in ribosome biogenesis (64 genes) and many of the genes that increased in expression are involved in oxidation reduction (51 genes) and response to abiotic stimulus (49 genes)(Dataset S2). Overall, strains carrying the S. cerevisiae FZF1 allele showed more pronounced changes in expression than those carrying the S. paradoxus allele (Figure S2), consistent with the possibility that many of the expression differences are not due to direct differential activation or repression by FZF1, but rather a consequence of downstream differences in sulfite resistance initiated by FZF1. A small number of genes, including SSU1, showed a larger increase in expression in strains carrying the S. paradoxus compared to the S. cerevisiae FZF1 allele. Excluding two putative genes, SSU1 showed the largest differences in expression between the S. cerevisiae and S. paradoxus alleles at 15 minutes and was one of the most significant FZF1-dependent differences across both time-points.
FZF1-dependent changes in gene expression may be caused by protein coding changes or by regulatory changes in the FZF1 5′ noncoding region. To distinguish between these possibilities, we classified FZF1-dependent expression changes into those that can be attributed to the 5′ noncoding region, coding region, or an interaction between the two regions. Most of the genes that showed FZF1-dependent differences in gene expression across both time-points were characterized by an interaction between the coding and 5′ noncoding regions (ANOVA, P<0.01, Figure 6). Interestingly, in many cases, the chimeric alleles caused these genes to be expressed at higher or lower levels compared to both of the full length alleles of each species. In contrast, most of the genes showing allele-specific differences in gene expression that varied by time-point were characterized by effects that depended on the coding region of FZF1 (ANOVA, P<0.01, Figure 6). Together, these results suggest that both the FZF1 coding and 5′ noncoding region contribute to downstream changes in gene expression.
Fig. 6. Changes in gene expression caused by FZF1 divergence. 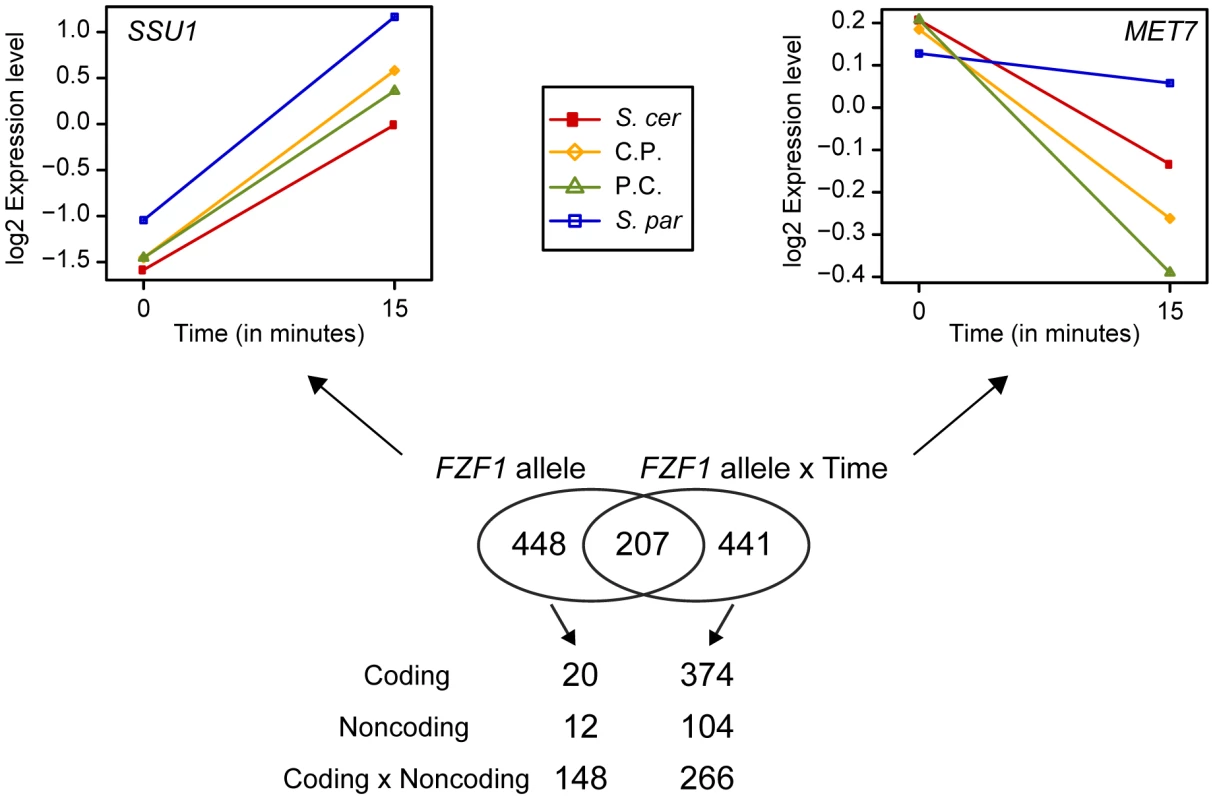
A Venn diagram of the number of genes with expression differences that depended on the FZF1 allele or an interaction between the FZF1 allele and time. An example of a gene with expression differences due to the FZF1 allele alone (SSU1) or an interaction between the FZF1 allele and time (MET7) are shown above the Venn diagram: S. cerevisiae (S. cer, red), S. paradoxus (S. par, blue), S. cerevisiae 5′ noncoding with S. paradoxus coding (C.P., gold), S. paradoxus 5′ noncoding and S. cerevisiae coding (P.C., green). The number of genes with expression differences that can be attributed to the coding region, upstream noncoding region or an interaction between the two regions is shown below the Venn diagram. Discussion
Identification of genes that have diverged in function between species is a key element to understanding species' diversity and evolution. Divergence in transcription factors are of particular interest as they can coordinately regulate the expression of many changes, but by doing so may be limited in how they can evolve. In this study, we used patterns of non-neutral sequence evolution to identify genes likely to have diverged in their regulation. We investigated one candidate, FZF1, by testing species-specific alleles for their ability to complement a deletion of FZF1 in S. cerevisiae. We found that FZF1 has diverged in its ability to confer resistance to sulfites, and used chimeric constructs to show that divergence in sulfite resistance is due to changes in multiple coding and upstream noncoding regions. Finally, we found that divergence at FZF1 affects the expression of FZF1, SSU1 and many other genes. Our results provide insight into how both phenotypic and regulatory divergence is caused by evolution of a transcription factor.
Identification of FZF1 and evidence for non-neutral evolution
We identified FZF1 based on a genome-wide screen for patterns of non-neutral divergence. FZF1 shows evidence of non-neutral divergence in its promoter region based on an accelerated substitution rate in some lineages but not others. In the coding region, evidence of non-neutral divergence is also present and is based on an elevated ratio of nonsynonymous to synonymous substitutions. However, upon closer examination we found a number of uncertainties regarding the evidence for non-neutral patterns of divergence. In the noncoding region, the evidence for substitution rate heterogeneity depends on the alignment. In the coding region, the cause of the elevated nonsynonymous to synonymous substitution rate is ambiguous because the synonymous substitution rate decreases in the same region that the nonsynonymous substitution rate increases. Interestingly, the strongest evidence for non-neutral evolution comes from divergence between the S. cerevisiae and S. paradoxus alleles, which also show the greatest difference in sulfite resistance. Thus, the pattern of divergence for FZF1 is at least consistent with non-neutral evolution. With respect to a potential cause of non-neutral divergence, both positive selection and loss of constraint can result in elevated substitution rates. However, loss of constraint by itself does not provide a good explanation for the loss of sulfite resistance along the S. cerevisiae lineage and the gain of sulfite resistance along the S. paradoxus lineage relative to the intermediate levels of sulfite resistance in S. mikatae and S. bayanus.
While patterns of non-neutral divergence led us to test FZF1 alleles for functional divergence, the value of such an approach remains difficult to assess. First, the evidence for non-neutral evolution is not definitive. Second, we only tested a single candidate. Third, the coding region with the largest effect on sulfite resistance does not include the region with evidence for non-neutral evolution. One factor that may be critical in selecting candidates is whether there is a clearly defined phenotype to test. Many of the other genes that exhibit substitution rate heterogeneity are known to impact a variety of phenotypes, making it difficult to know which one to test. Testing FZF1 was facilitated by its narrowly defined function in sulfite resistance. Thus, while some fascinating examples have emerged, e.g. [46], further work is needed to evaluate whether non-neutral patterns of divergence provide an effective screen for genes that have diverged in function.
Evolution of FZF1 through multiple coding and noncoding changes
Chimeric FZF1 alleles from S. cerevisiae and S. paradoxus indicate that both upstream noncoding regions and the first coding region make additive contributions to divergence in FZF1 activity. A second coding region, including the region with the elevated nonsynonymous to synonymous substitution rate, contributes an epistatic effect through interaction with the other coding region. The number of regions underlying sulfite resistance is likely dependent on how we identified FZF1. Tests of neutrality based on rate heterogeneity and dN/dS only indicate deviations from expected rates of divergence based on multiple substitutions. The accumulation of multiple changes at a single locus has also been found in other studies of interspecific differences [22]–[26], so it may not be an uncommon result when multiple regions are individually tested.
A limitation of our study is that we only quantified the effects of five regions and did not narrow their effects to individual substitutions. This limitation is in part due to the sensitivity of our sulfite resistance assay. As such, we did not determine whether the regions with the largest effect are caused by single or multiple substitutions, and whether there are epistatic effects between substitutions within a region. Further dissection of FZF1 divergence is needed to more accurately quantify the number, effect size and interactions among mutations affecting sulfite resistance.
Evolution of gene regulation
Transcription factors are often posited to be highly constrained during evolution due to their pleiotropic effects on the expression of other genes [47]. As such, many efforts to understand the evolution of gene regulation have focused on the evolution of cis-regulatory sequences rather than on trans-acting factors, e.g. [12], [48]. While changes in the expression of transcriptional regulators is hypothesized to be an important mode of evolutionary change [34], protein coding changes may also be important, e.g. [49]. We find that divergence in both the regulatory and coding sequence of FZF1 affects sulfite resistance and causes numerous downstream changes in gene expression. This raises the question of whether there have been any constraints on FZF1 divergence due to pleiotropy.
If FZF1 has been constrained by pleiotropy there must, at least under certain circumstances, be negative consequences to changes in FZF1 activity. Increased levels of FZF1 activity could reduce fitness in the absence of sulfites or after other exposures that activate FZF1, such as nitric oxide treatment [50]. The observation that the more potent S. paradoxus FZF1 allele is expressed at lower levels in the absence of sulfites provides some support for the idea that high levels of FZF1 activity may not always be advantageous. Assuming that there is some cost to constitutive increases in FZF1 activity, there are a number of ways in which this cost could be small enough to overcome or even eliminated.
One consideration is that SSU1 expression is likely the major determinant of sulfite resistance and so the benefit of increased SSU1 expression may outweigh any costs. In support of this possibility, SSU1 overexpression is able to rescue the effect of an FZF1 deletion (Figure S3) [38]. However, the expression of other genes may also be involved in sulfite resistance since divergence upstream of FZF1 affects sulfite resistance but only has a small, insignificant effect on SSU1 expression. Thus, coding changes in FZF1 that increase SSU1 expression may have outweighed any costs under other conditions, or may have been facilitated by lower levels of FZF1 expression in the absence of sulfites.
Another explanation for the lack of constraints on FZF1 divergence is compensatory changes in genes regulated by FZF1. In this scenario, slight changes in FZF1 activity may be compensated by cis-regulatory mutations in those FZF1 regulated genes where changes in gene expression are deleterious. A number of empirical studies have shown that transcription factors bind different targets between closely related species and even within species due in part to cis-regulatory sequence changes [11], [51]–[53]. Thus, it is also possible that cis-regulatory sequence evolution may have accommodated divergence in FZF1 activity.
A third explanation, suggested by the finding that transcription factors with few targets are less likely to be constrained by pleiotropy [54], is that FZF1 has few transcriptional targets and so is not greatly constrained by pleiotropy. In response to exogenously supplied nitric oxide, activation of only a small set of five genes, including SSU1, was found to specifically depend on the presence of FZF1 [50]. Another study found 21 upregulated and 37 downregulated genes two hours after sulfite treatment [55]. We found 1,096 FZF1-dependent expression changes, most of which showed the same direction of response to sulfite and only differed in magnitude. The observation that the sulfite-sensitive S. cerevisiae FZF1 allele caused more pronounced changes in gene expression relative to the S. paradoxus allele (Figure S2) is consistent with FZF1 causing indirect changes in gene expression mediated by its effects on sulfite resistance rather than by direct activation or repression of these genes. Furthermore, we found no enrichment of the FZF1 motif identified in the SSU1 promoter (TATCGTAT and CAACAA, [37]), defined by protein microarrays (CTGCTA, [56]), or by promoter bashing and response to nitrosative stress (YGSMNMCTATCAYTTYY, [50]) within the 271 genes showing a 2-fold significant increase in expression following sulfite treatment. Thus, most of the changes in gene expression that we observed may be an indirect consequence of a sulfite-induced stress response rather than a consequence of changes in direct targets of FZF1.
Regardless of the mechanism, the concentration of multiple sequence changes in FZF1 suggests that it may have evolved without many genetic, functional or evolutionary constraints. However, the apparent absence of constraints could be a consequence of low basal levels of FZF1 expression. Under this scenario, changes in FZF1 regulation may have facilitated changes within its protein coding sequence.
Evolution of sulfite resistance
Even though FZF1 has diverged in its ability to confer resistance to sulfites, its impact on the evolution of sulfite resistance is hard to know. While there is substantial variation in sulfite resistance within and between species (Figure S4), divergence at other loci may be responsible for most differences in sulfite resistance and could compensate for any changes in FZF1. Within S. cerevisiae, variation in sulfite resistance is associated with a reciprocal translocation upstream of SSU1 that is more frequent in vineyard and wine strains than strains derived from other sources [39], [57]. The inferred loss of sulfite resistance conferred by changes in FZF1 along the lineage leading to S. cerevisiae, combined with the gain of sulfite resistance due to the translocation within some strains of S. cerevisiae, suggests that the evolution of sulfite resistance among species is not simple and compensatory changes may be involved.
Conclusions
In this study we find substantial divergence in function within the coding and upstream noncoding region of FZF1. Our finding that multiple regions underlie divergence in sulfite resistance is not unexpected given the patterns of non-neutral evolution, but differs from other studies that identify single changes of large effect based on genetic mapping or candidate gene approaches [14]. The contribution of both noncoding and coding regions to differences in sulfite resistance suggests that the distinction between evolution in noncoding and coding regions may be less important than the degree to which a gene has the capacity to evolve, unencumbered by constraints on its other functions [33]. In conclusion, our work supports a model whereby both gene expression and phenotypic divergence can be attributed to multiple mutations throughout the regulatory and protein-coding region of a single gene.
Materials and Methods
Screen for noncoding regions with substitution rate heterogeneity
S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus noncoding regions [58] were tested for substitution rate heterogeneity using a likelihood ratio test implemented using HyPhy [59]. The likelihood ratio test was used to compare a constrained model with a single substitution rate across lineages to an unconstrained model where each lineage was allowed to have a different substitution rate. For both models we used the HKY85 substitution model implemented in HyPhy, the known phylogenetic relationship among the species, and either a single parameter (constrained) or branch-specific parameters (unconstrained) for the ratio of the noncoding substitution rate at the locus of interest to the substitution rate at four-fold degenerate sites across the genome. Noncoding alignments were removed if the total length of insertion/deletions was more than 15% of the length of the entire alignment. While this filter eliminated the noncoding region upstream of FZF1, we had already initiated our functional analysis of FZF1 based on preliminary rate heterogeneity results and so retained it in our list of candidates. To examine whether substitution rate heterogeneity upstream of FZF1 depends on the alignment, we aligned the 5′ noncoding region using 6 alignment programs: Clustalw [60], MUSCLE [61], TCOFFEE [62], MAFFT [63], PRANK [64], and DCA [65]. The resulting alignments were tested for rate heterogeneity using the likelihood ratio test described above. For the coding sequence of FZF1, a sliding window analysis of dN/dS was performed for FZF1 using the K-estimator software [66] as described in Sawyer and Malik (2006). K-estimator uses Monte Carlo simulations to estimate the confidence intervals for estimates of dN/dS.
Strain construction
FZF1 was deleted in YJF173 (S288c-background, Mat a, ho, ura3-52) using the KANMX deletion cassette [67]. FZF1 alleles were integrated into this strain at the ura3 locus by amplifying the entire FZF1 gene region, including the entire 5′ and 3′ noncoding regions along with 25 bases of ZRT1 and 45 bases of HXK2, using primers with homology to pRS306 and transforming the product along with the yeast integrative plasmid, pRS306 [68]. Integration of these constructs at the ura3 locus was achieved by selection on plates lacking uracil and each transformant was confirmed by PCR. Chimeras were generated using the same procedure but with FZF1 regions amplified from different species. The aligned ATG start site was used for all chimeras divided between the 5′ noncoding region and the coding region. A mutation of an alternate FZF1 start site in the S. paradoxus FZF1 allele did not significantly alter sulfite resistance compared to the non-mutated counterpart (data not shown). A subset of 2–5 transformants were sequenced to ensure that at least one transformant per construct contained no mutations.
Media and growth conditions
All experiments were conducted using YPD+TA (1% yeast extract, 2% peptone, 2% dextrose, 75 mM L-tartaric acid buffered to pH 3.5) [69]. Sulfite resistance was measured by comparing growth in the presence and absence of sodium sulfite. Strains were grown overnight in YPD+TA, diluted 1∶1000 in YPD+TA, grown for 3 hours, treated with either water or sodium sulfite (final concentration 0.7–0.9 mM sodium sulfite), and then grown for 20 hours in an iEMS plate reader at 30° with 1200 rpm shaking (model no. 1400; Thermo Lab Systems, Helsinki, Finland). For each strain, the sulfite-dependent delay in growth was determined by comparing the time at which maximum growth rate was observed for strains treated with sulfite relative to a water-treated control [70]. For each FZF1 construct, 4 to 8 independent transformants were phenotyped. To compare the sulfite-dependent delay in growth within and between yeast species, three replicate measurements were obtained for 6 S. cerevisiae strains: S288c (source: laboratory, obtained from: D. Botstein), YPS163 (source: oak exudate, United States, obtained from: P. Sniegowski), M8 and M33 (source: vineyard, Italy, obtained from R. Mortimer), YJM440 (source: clinical, United States, obtained from: J. McCusker), K9 (source: saké, Japan, obtained from: Nami Goto-Yamamoto), and five S. paradoxus strains: YPS138 (source: oak soil, United States), N17 (source: oak exudate, Russia), N44 (source: oak exudate, Russia), Y7 (source: oak bark, United Kingdom), and NRRL Y-17217 all obtained from G. Litti and E. Louis. Additional yeast species included: S. mikatae (IFO1815, obtained from: E. Louis), S. bayanus (NRRL Y-11845, obtained from: C. Kurtzman, ARS Culture Collection), Saccharomyces castellii (NRRL Y-12630, obtained from: M. Johnston), Saccharomyces kluyverii (NRRL Y-12651, obtained from: M. Johnston), and Kluyveromyces lactis (FM423, a haploid MAT á strain obtained from M. Johnston). All strains are diploid except as noted.
Analysis of sulfite resistance
Differences in sulfite resistance between species and species' chimeras were normalized for day effects and tested for significance using the nonparametric Kruskal-Wallis test. Pairwise differences between constructs were examined using the nonparametric Wilcoxon rank sum test with Bonferroni correction.
Differences in sulfite resistance among S. cerevisiae - S. paradoxus chimeric constructs of FZF1 were measured using linear mixed effect (lme) models to account for repeated measurements of the same construct. Sulfite resistance of each construct was measured three times and measurements on different days were standardized by a Z-score transformation. Sulfite resistance was fit to two models. The first model assumes each region from S. cerevisiae or S. paradoxus makes an additive contribution to differences in sulfite resistance: sulfite resistance = region1+region2+region3+region4+region5+(error | batch)+error, where each region has an effect that depends on the species the region came from and (error | batch) models random effects due to measurement of the same construct in different batches (96-well plates). The second model builds on the first model but adds in all pairwise interactions between regions: sulfite resistance = (region1+region2+region3+region4+region5)∧2+(error | batch)+error. The fit of the two models was compared using a likelihood ratio test with 10 degrees of freedom since the first and second models have 8 and 18 degrees of freedom, respectively. The percent variance explained by each model was calculated by R2 = 1−exp(−LR/n), where n is the sample size and LR is the likelihood ratio statistic defined by twice the difference in the log likelihood of the alternative relative to the null model [71]. The null model was fit using only an intercept: sulfite resistance = (error | batch)+error. For lme P-values, we tested whether the assumptions of the test were violated and resulted in inaccurate P-values by repeatedly permuting the data labels to obtain the distribution of P-values expected by chance. The permuted data showed no evidence for inaccurate P-values.
Gene expression analysis
Gene expression was measured using four independent transformants of each FZF1 construct. Strains were resuspended in YPD+TA at an OD600 of 0.25 from an overnight YPD+TA culture and grown in 100 mL cultures at 30°C, 200 rpm. After 3 hours, each culture was sampled at 0, 15, 30 and 60 minutes after addition of sodium sulfite to a final concentration of 1 mM. Cells were centrifuged, washed and frozen in a dry ice/ethanol bath and stored at −80°C. RNA was isolated and cDNA prepared using Qiagen's RNaeasy Mini Kit and Omniscript RT Kit, respectively (Valencia, CA).
Quantitative PCR was used to measure expression of FZF1 and SSU1. A 20-fold dilution of cDNA reactions was used for the real-time PCR assays with gene specific primers and Strategene's Brilliant II SYBR Green QPCR Master Mix (Santa Clara, CA). Expression was assayed on Stratagene's MX3000P QPCR machine. For FZF1, species-specific primers were used and a plate specific correction factor, estimated for each plate from quantitative PCR measurements of DNA extracted from a heterozygous strain containing both the S. cerevisiae and S. paradoxus FZF1 alleles, was used to account for the difference in PCR efficiency between the S. cerevisiae and S. paradoxus primers. Data were mean normalized for day and batch effects and expression levels were measured relative to ACT1. The Wilcoxon rank sum test with Bonferroni correction was used to identify significant differences in expression due to FZF1 alleles.
Genome-wide measurements of gene expression were obtained using Agilent Technologies (Santa Clara, CA) yeast (V2) gene expression microarrays (8×15K, Catalog number: G4813A-016322) following the manufacturers protocols. Sample labeling, hybridization and microarray scanning was conducted by the Expression and Genotype Core at Washington University's Genome Center. Gene expression was measured for three independent replicates at the 0 and 15 minute time-points. Each sample was compared to a reference made up of a pool of all RNA samples. Expression data was deposited in the GEO database under accession GSE35308. After median normalization of each microarray, differences in gene expression were tested using an analysis of variance (ANOVA) with the model: expression = allele*time+technical replicate+error, where allele measures the effect of the different FZF1 alleles, time measures the effect of each time-point, and technical replicate accounts for differences between replicated features on the microarray. The rate of false positives was estimated by permuting the sample labels 100 times and repeating the analysis. For each gene showing a significant difference in expression, a second ANOVA was performed to identify expression changes that could be attributed to the coding or 5′ noncoding region or an interaction between the two regions. For genes showing expression differences that depended on the FZF1 construct we used the model: expression = noncoding*coding+error, and for genes showing differences that depended on an interaction between the FZF1 construct and time we used the model: expression = noncoding*coding*time+error. Gene sets enriched for gene ontology (GO) categories were identified using DAVID [72].
Supporting Information
Zdroje
1. BergmannSIhmelsJBarkaiN 2004 Similarities and differences in genome-wide expression data of six organisms. PLoS Biol 2 e9 doi:10.1371/journal.pbio.0020009
2. LelandaisGTantyVGeneixCEtchebestCJacqC 2008 Genome adaptation to chemical stress: clues from comparative transcriptomics in Saccharomyces cerevisiae and Candida glabrata. Genome Biol 9 R164
3. ChanETQuonGTChuaGBabakTTrochessetM 2009 Conservation of core gene expression in vertebrate tissues. J Biol 8 33
4. WhiteheadACrawfordD 2006 Variation within and among species in gene expression: raw material for evolution. Mol Ecol 15 1197 1211
5. FayJCMcCulloughHLSniegowskiPDEisenMB 2004 Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol 5 R26
6. KhaitovichPWeissGLachmannMHellmannIEnardW 2004 A neutral model of transcriptome evolution. PLoS Biol 2 e132 doi:10.1371/journal.pbio.0020132
7. LemosBMeiklejohnCCaceresMHartlD 2005 Rates of divergence in gene expression profiles of primates, mice, and flies: stabilizing selection and variability among functional categories. Evolution 59 126 137
8. FayJCWittkoppPJ 2008 Evaluating the role of natural selection in the evolution of gene regulation. Heredity 100 191 199
9. WittkoppPJHaerumBKClarkAG 2008 Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40 346 350
10. EmersonJJHsiehLSungHWangTHuangC 2010 Natural selection on cis and trans regulation in yeasts. Genome Res 20 826 836
11. IhmelsJBergmannSGerami-NejadMYanaiIMcClellanM 2005 Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309 938 940
12. DonigerSWFayJC 2007 Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol 3 e99 doi:10.1371/journal.pcbi.0030099
13. TsankovAMThompsonDASochaARegevARandoOJ 2010 The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol 8 e1000414 doi:10.1371/journal.pbio.1000414
14. SternDLOrgogozoV 2008 The loci of evolution: how predictable is genetic evolution? Evolution 62 2155 2177
15. YvertGBremRWhittleJAkeyJFossE 2003 Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet 35 57 64
16. BrownKMLandryCRHartlDLCavalieriD 2008 Cascading transcriptional effects of a naturally occurring frameshift mutation in Saccharomyces cerevisiae. Mol Ecol 17 2985 2997
17. KimHSHuhJFayJC 2009 Dissecting the pleiotropic consequences of a quantitative trait nucleotide. FEMS Yeast Res 9 713 722
18. RockmanMV 2012 The QTN program and the alleles that matter for evolution: all that's gold does not glitter. Evolution 66 1 17
19. BullardJHMostovoyYDudoitSBremRB 2010 Polygenic and directional regulatory evolution across pathways in Saccharomyces. Proc Natl Acad Sci U S A 107 5058 5063
20. FraserHBMosesAMSchadtEE 2010 Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc Natl Acad Sci U S A 107 2977 2982
21. FraserHBBabakTTsangJZhouYZhangB 2011 Systematic detection of polygenic cis-regulatory evolution. PLoS Genet 7 e1002023 doi:10.1371/journal.pgen.1002023
22. WangXChamberlinHM 2002 Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-48 ovo gene. Genes Dev 16 2345 2349
23. McGregorAPOrgogozoVDelonIZanetJSrinivasanDG 2007 Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448 587 590
24. JeongSRebeizMAndolfattoPWernerTTrueJ 2008 The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132 783 793
25. RebeizMPoolJEKassnerVAAquadroCFCarrollSB 2009 Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326 1663 1667
26. FrankelNErezyilmazDFMcGregorAPWangSPayreF 2011 Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474 598 603
27. LiYLiangHGuZLinZGuanW 2009 Detecting positive selection in the budding yeast genome. J Evol Biol 22 2430 2437
28. BirdCPStrangerBELiuMThomasDJIngleCE 2007 Fast-evolving noncoding sequences in the human genome. Genome Biol 8 R118
29. PollardKSSalamaSRKingBKernADDreszerT 2006 Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2 e168 doi:10.1371/journal.pgen.0020168
30. PrabhakarSNoonanJPPääboSRubinEM 2006 Accelerated evolution of conserved noncoding sequences in humans. Science 314 786
31. PrabhakarSViselAAkiyamaJAShoukryMLewisKD 2008 Human-specific gain of function in a developmental enhancer. Science 321 1346 1350
32. PollardKSSalamaSRLambertNLambotMCoppensS 2006 An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443 167 172
33. SternDLOrgogozoV 2009 Is genetic evolution predictable? Science 323 746 751
34. CarrollSB 2008 Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134 25 36
35. SawyerSLMalikHS 2006 Positive selection of yeast nonhomologous end-joining genes and a retrotransposon conflict hypothesis. Proc Natl Acad Sci U S A 103 17614 17619
36. CasaloneEColellaCMDalySFontanaSTorricelliI 1994 Cloning and characterization of a sulphite-resistance gene of Saccharomyces cerevisiae. Yeast 10 1101 1110
37. AvramDLeidMBakalinskyAT 1999 Fzf1p of Saccharomyces cerevisiae is a positive regulator of SSU1 transcription and its first zinc finger region is required for DNA binding. Yeast 15 473 480
38. ParkHBakalinskyAT 2000 SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast 16 881 888
39. Pérez-OrtínJEQuerolAPuigSBarrioE 2002 Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res 12 1533 1539
40. Markova-RainaPPetrovD 2011 High sensitivity to aligner and high rate of false positives in the estimates of positive selection in the 12 Drosophila genomes. Genome Res 21 863 874
41. WongKMSuchardMAHuelsenbeckJP 2008 Alignment uncertainty and genomic analysis. Science 319 473 476
42. SchmidKYangZ 2008 The trouble with sliding windows and the selective pressure in BRCA1. PLoS ONE 3 e3746 doi:10.1371/journal.pone.0003746
43. RonaldJBremRWhittleJKruglyakL 2005 Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet 1 e25 doi:10.1371/journal.pgen.0010025
44. GaschASpellmanPKaoCCarmel-HarelOEisenM 2000 Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11 4241 4257
45. GaschAWerner-WashburneM 2002 The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics 2 181 192
46. PontingCP 2011 What are the genomic drivers of the rapid evolution of PRDM9? Trends Genet 27 165 171
47. WrayGHahnMAbouheifEBalhoffJPizerM 2003 The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20 1377 1419
48. VenkataramSFayJC 2010 Is transcription factor binding site turnover a sufficient explanation for cis-regulatory sequence divergence? Genome Biol Evol 2 851 858
49. LynchVJMayGWagnerGP 2011 Regulatory evolution through divergence of a phosphoswitch in the transcription factor CEBPB. Nature 480 383 386
50. SarverADeRisiJ 2005 Fzf1p regulates an inducible response to nitrosative stress in Saccharomyces cerevisiae. Mol Biol Cell 16 4781 4791
51. OdomDTDowellRDJacobsenESGordonWDanfordTW 2007 Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39 730 732
52. BornemanARGianoulisTAZhangZDYuHRozowskyJ 2007 Divergence of transcription factor binding sites across related yeast species. Science 317 815 819
53. SchmidtDWilsonMDBallesterBSchwaliePCBrownGD 2010 Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328 1036 1040
54. ZhouLMaXArbeitmanMNSunF 2009 Chromatin regulation and gene centrality are essential for controlling fitness pleiotropy in yeast. PLoS ONE 4 e8086 doi:10.1371/journal.pone.0008086
55. ParkHHwangY 2008 Genome-wide transcriptional responses to sulfite in Saccharomyces cerevisiae. J Microbiol 46 542 548
56. BadisGChanETvan BakelHPena-CastilloLTilloD 2008 A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32 878 887
57. YuasaNNakagawaYHayakawaMIimuraY 2004 Distribution of the sulfite resistance gene SSU1-R and the variation in its promoter region in wine yeasts. J Biosci Bioeng 98 394 397
58. KellisMPattersonNEndrizziMBirrenBLanderE 2003 Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423 241 254
59. PondSFrostSMuseS 2005 HyPhy: hypothesis testing using phylogenies. Bioinformatics 21 676 679
60. ThompsonJDGibsonTJPlewniakFJeanmouginFHigginsDG 1997 The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876 4882
61. EdgarRC 2004 MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 1792 1797
62. NotredameCHigginsDGHeringaJ 2000 T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302 205 217
63. KatohKKumaKTohHMiyataT 2005 MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33 511 518
64. LöytynojaAGoldmanN 2010 webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 11 579
65. StoyeJ 1998 Multiple sequence alignment with the Divide-and-Conquer method. Gene 211 GC45 56
66. ComeronJ 1999 K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15 763 764
67. WachABrachatAPöhlmannRPhilippsenP 1994 New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793 1808
68. SikorskiRSHieterP 1989 A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19 27
69. ParkHLopezNIBakalinskyAT 1999 Use of sulfite resistance in Saccharomyces cerevisiae as a dominant selectable marker. Curr Genet 36 339 344
70. KimHSFayJC 2007 Genetic variation in the cysteine biosynthesis pathway causes sensitivity to pharmacological compounds. Proc Natl Acad Sci U S A 104 19387 19391
71. MageeL 1990 R2 measures based on Wald and likelihood ratio joint significance tests. American Statistician 44 250 253
72. HuangDWShermanBTLempickiRA 2009 Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 44 57
73. BussereauFLafayJBolotin-FukuharaM 2004 Zinc finger transcriptional activators of yeasts. FEMS Yeast Res 4 445 458
74. Goto-YamamotoNKitanokShikiKYoshidaYSuzukiT 1998 SSU1-R, a sulfite resistance gene of wine yeast, is an allele of SSU1 with a different upstream sequence. J Ferment Bioeng 86 427 433
Štítky
Genetika Reprodukční medicína
Článek Uracil-Containing DNA in : Stability, Stage-Specific Accumulation, and Developmental InvolvementČlánek Fuzzy Tandem Repeats Containing p53 Response Elements May Define Species-Specific p53 Target GenesČlánek Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early EmbryosČlánek Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2α Kinase GCN-2Článek Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human CellsČlánek TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome EndsČlánek Attenuation of Notch and Hedgehog Signaling Is Required for Fate Specification in the Spinal CordČlánek Genome-Wide Functional Profiling Identifies Genes and Processes Important for Zinc-Limited Growth ofČlánek MicroRNA93 Regulates Proliferation and Differentiation of Normal and Malignant Breast Stem Cells
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 6- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Transfer zmraženého embrya zlepšuje výsledky IVF
- Velké děti po kryoembryotransferu
-
Všechny články tohoto čísla
- Rumors of Its Disassembly Have Been Greatly Exaggerated: The Secret Life of the Synaptonemal Complex at the Centromeres
- Mimetic Butterflies Introgress to Impress
- Tipping the Balance in the Powerhouse of the Cell to “Protect” Colorectal Cancer
- Selection-Driven Gene Loss in Bacteria
- Decreased Mitochondrial DNA Mutagenesis in Human Colorectal Cancer
- Parallel Evolution of Auditory Genes for Echolocation in Bats and Toothed Whales
- Diverse CRISPRs Evolving in Human Microbiomes
- The Rad4 ATR-Activation Domain Functions in G1/S Phase in a Chromatin-Dependent Manner
- Stretching the Rules: Monocentric Chromosomes with Multiple Centromere Domains
- Uracil-Containing DNA in : Stability, Stage-Specific Accumulation, and Developmental Involvement
- Fuzzy Tandem Repeats Containing p53 Response Elements May Define Species-Specific p53 Target Genes
- Adaptive Introgression across Species Boundaries in Butterflies
- G Protein Activation without a GEF in the Plant Kingdom
- Synaptonemal Complex Components Persist at Centromeres and Are Required for Homologous Centromere Pairing in Mouse Spermatocytes
- An Engineering Approach to Extending Lifespan in
- Incompatibility and Competitive Exclusion of Genomic Segments between Sibling Species
- Effects of Histone H3 Depletion on Nucleosome Occupancy and Position in
- Patterns of Evolutionary Conservation of Essential Genes Correlate with Their Compensability
- Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis
- Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early Embryos
- A Mouse Model of Acrodermatitis Enteropathica: Loss of Intestine Zinc Transporter ZIP4 (Slc39a4) Disrupts the Stem Cell Niche and Intestine Integrity
- Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2α Kinase GCN-2
- Geographic Differences in Genetic Susceptibility to IgA Nephropathy: GWAS Replication Study and Geospatial Risk Analysis
- Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human Cells
- TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome Ends
- Stimulation of Host Immune Defenses by a Small Molecule Protects from Bacterial Infection
- A Broad Requirement for TLS Polymerases η and κ, and Interacting Sumoylation and Nuclear Pore Proteins, in Lesion Bypass during Embryogenesis
- Genome-Wide Identification of Ampicillin Resistance Determinants in
- The CCR4-NOT Complex Is Implicated in the Viability of Aneuploid Yeasts
- Gustatory Perception and Fat Body Energy Metabolism Are Jointly Affected by Vitellogenin and Juvenile Hormone in Honey Bees
- Genetic Variants on Chromosome 1q41 Influence Ocular Axial Length and High Myopia
- Is a Key Regulator of Pancreaticobiliary Ductal System Development
- The NSL Complex Regulates Housekeeping Genes in
- Attenuation of Notch and Hedgehog Signaling Is Required for Fate Specification in the Spinal Cord
- Dual-Level Regulation of ACC Synthase Activity by MPK3/MPK6 Cascade and Its Downstream WRKY Transcription Factor during Ethylene Induction in Arabidopsis
- Genome-Wide Functional Profiling Identifies Genes and Processes Important for Zinc-Limited Growth of
- Base-Pair Resolution DNA Methylation Sequencing Reveals Profoundly Divergent Epigenetic Landscapes in Acute Myeloid Leukemia
- MicroRNA93 Regulates Proliferation and Differentiation of Normal and Malignant Breast Stem Cells
- Phylogenomic Analysis Reveals Dynamic Evolutionary History of the Drosophila Heterochromatin Protein 1 (HP1) Gene Family
- Found: The Elusive ANTAR Transcription Antiterminator
- The Mutation in Chickens Constitutes a Structural Rearrangement Causing Both Altered Comb Morphology and Defective Sperm Motility
- Control of CpG and Non-CpG DNA Methylation by DNA Methyltransferases
- Polymorphisms in the Mitochondrial Ribosome Recycling Factor Compromise Cell Respiratory Function and Increase Atorvastatin Toxicity
- Gene Expression Profiles in Parkinson Disease Prefrontal Cortex Implicate and Genes under Its Transcriptional Regulation
- Global Regulatory Functions of the Endoribonuclease III in Gene Expression
- Extensive Evolutionary Changes in Regulatory Element Activity during Human Origins Are Associated with Altered Gene Expression and Positive Selection
- The Regulatory Network of Natural Competence and Transformation of
- Brain Expression Genome-Wide Association Study (eGWAS) Identifies Human Disease-Associated Variants
- Quantifying the Adaptive Potential of an Antibiotic Resistance Enzyme
- Divergence of the Yeast Transcription Factor Affects Sulfite Resistance
- The Histone Demethylase Jhdm1a Regulates Hepatic Gluconeogenesis
- RNA Methylation by the MIS Complex Regulates a Cell Fate Decision in Yeast
- Rare Copy Number Variants Observed in Hereditary Breast Cancer Cases Disrupt Genes in Estrogen Signaling and Tumor Suppression Network
- Genome-Wide Location Analysis Reveals Distinct Transcriptional Circuitry by Paralogous Regulators Foxa1 and Foxa2
- A Mutation Links a Canine Progressive Early-Onset Cerebellar Ataxia to the Endoplasmic Reticulum–Associated Protein Degradation (ERAD) Machinery
- The Mechanism for RNA Recognition by ANTAR Regulators of Gene Expression
- Limits to the Rate of Adaptive Substitution in Sexual Populations
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Rumors of Its Disassembly Have Been Greatly Exaggerated: The Secret Life of the Synaptonemal Complex at the Centromeres
- The NSL Complex Regulates Housekeeping Genes in
- Tipping the Balance in the Powerhouse of the Cell to “Protect” Colorectal Cancer
- Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



