-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Stimulation of Host Immune Defenses by a Small Molecule Protects from Bacterial Infection
The nematode Caenorhabditis elegans offers currently untapped potential for carrying out high-throughput, live-animal screens of low molecular weight compound libraries to identify molecules that target a variety of cellular processes. We previously used a bacterial infection assay in C. elegans to identify 119 compounds that affect host-microbe interactions among 37,214 tested. Here we show that one of these small molecules, RPW-24, protects C. elegans from bacterial infection by stimulating the host immune response of the nematode. Using transcriptome profiling, epistasis pathway analyses with C. elegans mutants, and an RNAi screen, we show that RPW-24 promotes resistance to Pseudomonas aeruginosa infection by inducing the transcription of a remarkably small number of C. elegans genes (∼1.3% of all genes) in a manner that partially depends on the evolutionarily-conserved p38 MAP kinase pathway and the transcription factor ATF-7. These data show that the immunostimulatory activity of RPW-24 is required for its efficacy and define a novel C. elegans–based strategy to identify compounds with activity against antibiotic-resistant bacterial pathogens.
Published in the journal: . PLoS Genet 8(6): e32767. doi:10.1371/journal.pgen.1002733
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002733Summary
The nematode Caenorhabditis elegans offers currently untapped potential for carrying out high-throughput, live-animal screens of low molecular weight compound libraries to identify molecules that target a variety of cellular processes. We previously used a bacterial infection assay in C. elegans to identify 119 compounds that affect host-microbe interactions among 37,214 tested. Here we show that one of these small molecules, RPW-24, protects C. elegans from bacterial infection by stimulating the host immune response of the nematode. Using transcriptome profiling, epistasis pathway analyses with C. elegans mutants, and an RNAi screen, we show that RPW-24 promotes resistance to Pseudomonas aeruginosa infection by inducing the transcription of a remarkably small number of C. elegans genes (∼1.3% of all genes) in a manner that partially depends on the evolutionarily-conserved p38 MAP kinase pathway and the transcription factor ATF-7. These data show that the immunostimulatory activity of RPW-24 is required for its efficacy and define a novel C. elegans–based strategy to identify compounds with activity against antibiotic-resistant bacterial pathogens.
Introduction
Studies in the model nematode Caenorhabditis elegans have greatly expanded our understanding of development, neurobiology, host-pathogen interactions, and many other aspects of metazoan biology. Here we show that C. elegans-based assays enable the identification of immunostimulatory compounds, which can be employed together with genetic analyses to interrogate innate immune signaling pathways.
Previously, we demonstrated that C. elegans can be used in bacterial infection assays to identify novel antimicrobials [1]–[3]. Fifteen to twenty C. elegans animals fit comfortably in the wells of standard 384-well assay plates and the assay can be automated using image analysis software, which enables such studies to be conducted in high-throughput [1]. Using this system, we tested 37,214 compounds and identified 119 small molecules that prolonged the lifespan of nematodes infected with the Gram-positive human bacterial pathogen Enterococcus faecalis [1].
We hypothesized that the C. elegans-based screen for novel anti-infectives would identify small molecules that cure nematodes by stimulating the host innate immune response, in addition to compounds that block microbial virulence or directly inhibit bacterial growth [1]. In nature, nematodes consume bacteria for food and have evolved sophisticated innate immune mechanisms within their intestinal epithelium to defend against ingested pathogens [4]. C. elegans mount specific immune responses toward both bacterial and fungal pathogens using immune signaling mediators that are strongly conserved throughout evolution [5]–[10]. Principal among these regulators is the NSY-1/SEK-1/PMK-1 Mitogen Activated Protein (MAP) kinase pathway, orthologous to the ASK1 (MAP kinase kinase kinase)/MKK3/6 (MAP kinase kinase)/p38 (MAP kinase) pathway in mammals [10]. In C. elegans, the p38 MAP kinase pathway acts cell autonomously in the intestine [11] to coordinate the expression of immune effectors such as C-type lectins and genes that may encode antimicrobial peptides [8]. Recently, Shivers et al. found that the transcription factor ATF-7, an ortholog of mammalian ATF2/ATF7, is phosphorylated by the p38 MAP kinase PMK-1 and is also required for defense against ingested bacterial pathogens [12].
In this study, we describe a small molecule named RPW-24 that strongly stimulates the innate immune response of C. elegans in a manner that confers a survival advantage for nematodes during bacterial infection. We show that the activity of this compound is partially dependent on the C. elegans p38 MAP kinase cassette and ATF-7. These data demonstrate that C. elegans can be used in facile in vivo screens to identify compounds with desirable biological activities.
Results
A Compound with Curing Activity in a Live Animal Infection Model Is Not a Traditional Antibiotic
In a previous study, we screened 37,214 small molecules for those that prolonged the lifespan of C. elegans infected with the Gram-positive bacterial pathogen E. faecalis as a means to identify novel antimicrobials [1]. Of 119 compounds identified, 31 did not have any structural relationship to known antimicrobials and ten of these small molecules were effective against E. faecalis in the C. elegans infection model at doses that did not inhibit growth of the pathogen in an in vitro growth assay [1]. In contrast, all currently available antibiotics interfere with some aspect of bacterial growth or metabolism. In the C. elegans-based assay, traditional antibiotics, such as tetracycline, ciprofloxacin, ampicillin, and vancomycin, cured E. faecalis-infected nematodes only at doses several fold higher than the in vitro minimum inhibitory concentration (MIC) for bacterial growth [2]. We therefore hypothesized that a subset of these 31 small molecules conferred a survival advantage to infected worms by either stimulating the host immune response of C. elegans or by interfering with virulence factor production in the bacteria.
We reasoned that a small molecule, which demonstrated curing activity against diverse bacteria without directly affecting bacterial growth, would be a candidate immunostimulator. It would be less likely for such a compound to be a specific inhibitor of bacterial virulence determinants given the complex and presumably pathogen-specific nature of these factors. We therefore screened the 31 compounds described above that did not have any structural relationship to known antimicrobials for those that prolonged the lifespan of nematodes infected with Pseudomonas aeruginosa, a Gram-negative pathogen, at doses lower than the in vitro MIC for P. aeruginosa. We found that eight of the 31 small molecules demonstrated in vivo efficacy against nematodes infected with P. aeruginosa (Table 1, Figure 1A and Figure S1) at doses several fold lower than the in vitro MIC for these compounds against P. aeruginosa. Indeed, four of the tested compounds did not affect the growth of P. aeruginosa at any concentration tested (Table 1).
Fig. 1. RPW-24 activates F35E12.5::GFP and prolongs the lifespan of C. elegans infected with P. aeruginosa without affect growth of the pathogen. 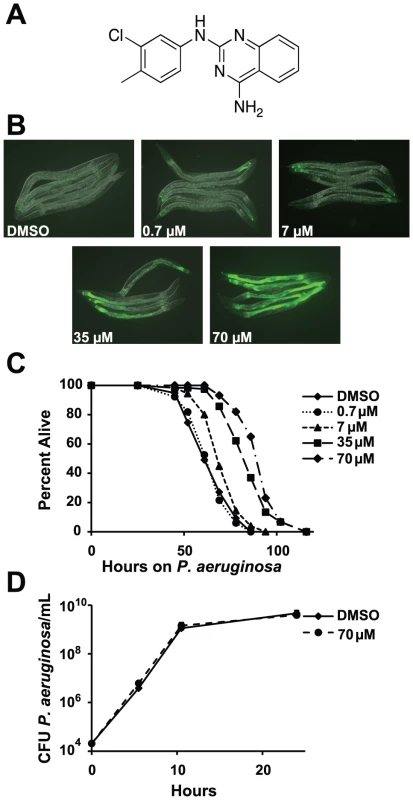
(A) The chemical structure of 2-N-(3-chloro-4-methylphenyl) quinazoline-2,4-diamine, which we have named RPW-24. (B) Fluorescence microscopy images of GFP expression from C. elegans acIs101 animals, which express a F35E12.5::GFP transgene, exposed to the indicated concentration of RPW-24 or DMSO, the solvent control, for 16 hours at 15°C. Green is GFP expression. (C) P. aeruginosa infection assay of wild-type nematodes exposed to different concentrations of RPW-24 compared to DMSO. The lifespan extension conferred by 70, 35 and 7 µM RPW-24 compared to DMSO treatment is significant in two biological replicates (P<0.0001). Data at each time point are the average of three plates per strain, each with approximately 50 animals per plate (sample sizes are given in Table S2). Data are representative of two independent experiments. (D) RPW-24 does not affect the growth of P. aeruginosa in the same media used for the C. elegans infection assay (slow-kill media). Data are the average of two biological replicates with error bars representing standard deviation. Tab. 1. Compounds effective against P. aeruginosa in the C. elegans assay at doses that do not inhibit bacterial growth. 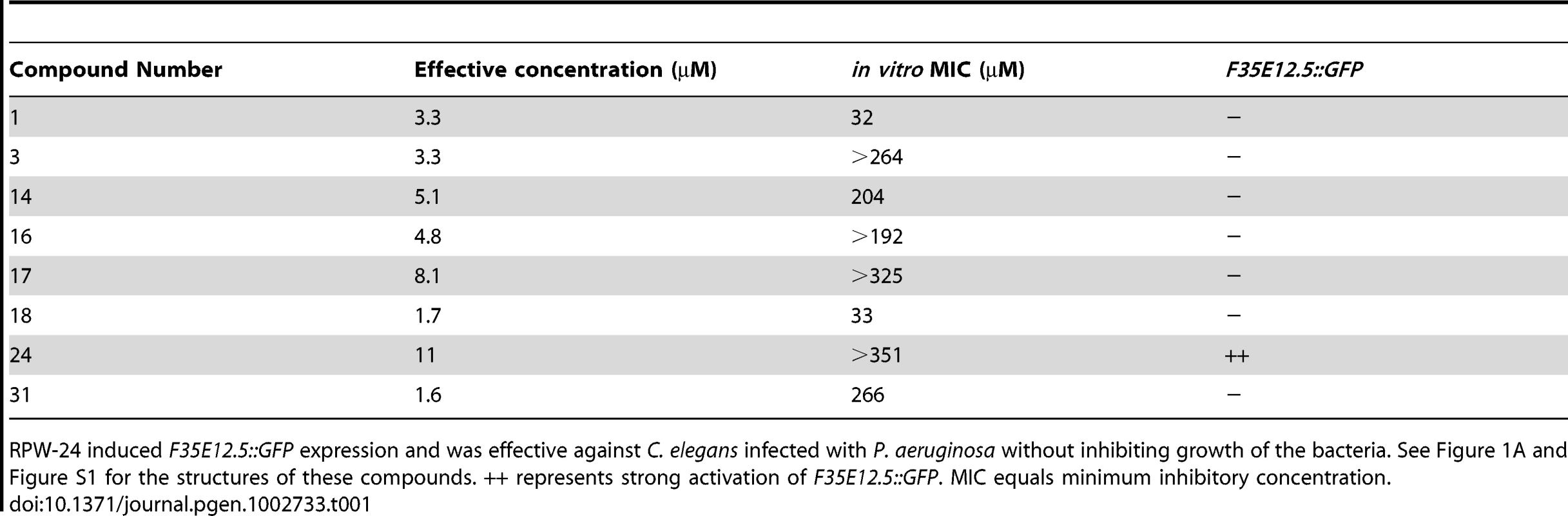
RPW-24 induced F35E12.5::GFP expression and was effective against C. elegans infected with P. aeruginosa without inhibiting growth of the bacteria. See Figure 1A and Figure S1 for the structures of these compounds. ++ represents strong activation of F35E12.5::GFP. MIC equals minimum inhibitory concentration. To ask if any of these eight compounds could activate the transcription of a putative immune effector, we used transgenic C. elegans animals carrying a transcriptional GFP reporter for the gene F35E12.5 [13]. F35E12.5 encodes a protein that contains a CUB-like domain and is involved in the transcriptional response towards several bacterial pathogens, including P. aeruginosa [8], [13], [14]. Interestingly, one of the four compounds that exhibited P. aeruginosa curing activity in the C. elegans infection assay without affecting growth of the pathogen (RPW-24, Figure 1A) activated the F35E12.5::GFP reporter in a dose dependent manner (Figure 1B). We therefore decided to focus on RPW-24 and explore its mode of action in more detail.
We found that RPW-24 promoted survival of P. aeruginosa-infected animals in a dose-dependent manner using an agar-based assay, the typical way that C. elegans infection assays are carried out in most laboratories (Figure 1C). C. elegans treated with 7, 35 and 70 µM of RPW-24 (but not 0.7 µM) were significantly resistant to P. aeruginosa infection compared to animals treated with the solvent control (DMSO)(Figure 1C). To determine if RPW-24 directly affects the growth of P. aeruginosa, we monitored in vitro bacterial growth in the presence 70 µM RPW-24 and DMSO. We conducted these experiments in the same media used for the nematode killing assay (Figure 1D) or in standard bacterial culture media (Luria broth, data not shown), but in the absence of C. elegans. In both cases, we found that 70 µM RPW-24 did not affect the growth rate of P. aeruginosa (Figure 1D and data not shown). In the C. elegans - P. aeruginosa pathogenicity assay, we observed that the intestines of the DMSO-treated animals were markedly distended and packed with P. aeruginosa cells 40 hours after infection (Figure S2), consistent with previous observations [9], [15]. In contrast, animals treated with RPW-24 had non-distended intestines, a morphology that was strikingly different from the DMSO controls (Figure S2).
In summary, RPW-24 is efficacious against both Gram-positive [1] and Gram-negative bacteria (Figure 1C) at concentrations that do not inhibit bacterial growth (Figure 1D, Table 1) and activates the transcription of a putative immune effector, F35E12.5 (Figure 1B). RPW-24 treatment also dramatically reduced the intestinal burden of P. aeruginosa 40 hours after infection compared to DMSO controls (Figure S2). These data suggest that RPW-24 affects the host immune response of C. elegans in a manner that confers a survival advantage against infection with diverse bacterial pathogens.
Exposure to RPW-24 Induces Immune Response Genes in C. elegans
To further study the effects of RPW-24 on C. elegans, we used Affymetrix whole genome GeneChips to generate transcriptome profiles of wild-type nematodes following exposure to either 70 µM RPW-24 or the solvent control DMSO in liquid culture media in the absence of bacterial pathogens for 16 hours at 15°C (Figure 2A). We found that RPW-24 induced a remarkably robust transcriptional response that involved only a small fraction (∼1.3%) of the genes of the C. elegans genome (Figure 2A). 269 genes were upregulated three-fold or greater (P<0.025), 125 of which were induced more than 50-fold during RPW-24 exposure (Table S1). The most highly upregulated gene was expressed more than 3500-fold higher in compound-exposed worms. For confirmation, we used qRT-PCR to analyze 10 genes that exhibited varying degrees of induction and expression levels in the microarray analysis (Figure 2A). We found that the transcriptional changes observed in the transcriptome profiling analysis directly correlated with the values obtained by qRT-PCR for all 10 genes tested (Figure S3). We also observed no difference in C. elegans gene induction whether the RPW-24 exposure occurred in liquid or on solid media (Figure S3). Taken together, these data suggest that RPW-24 strongly induces gene transcription in the absence of pathogen exposure.
Fig. 2. RPW-24 induces the transcription of putative immune effectors in C. elegans. 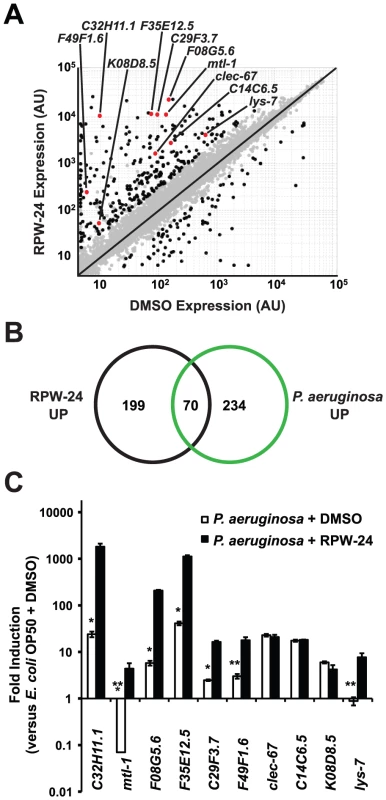
(A) A scatter plot compares gene expression levels of all 22,250 sequences on the Affymetrix GeneChip in wild-type C. elegans exposed to RPW-24 versus DMSO. Black dots highlight the 269 genes that were induced and the 62 that were repressed at least three-fold (P<0.025). Gray dots are genes whose expression levels did not significantly change in this study. The location on the scatter plot of the ten putative immune effectors whose expression was studied further by qRT-PCR are indicated and highlighted with red dots. Genes that fall on the black line are expressed at equal levels in both conditions. AU = arbitrary units. (B) Venn diagram gives the overlap of the C. elegans genes induced three-fold or greater by RPW-24 (P<0.025) in the microarray analysis (this study) with the genes upregulated during P. aeruginosa infection (greater than 2-fold, P<0.01) [8]. P<2.7×10−16 for the degree of overlap between these datasets versus the amount expected by chance alone. (C) Shown are qRT-PCR data of ten putative C. elegans immune effectors in wild-type animals infected with P. aeruginosa for eight hours and exposed to either 70 µM RPW-24 or DMSO, each plotted versus expression of the indicated genes in C. elegans exposed to E. coli OP50 and DMSO. The data are the average of two biological replicates each normalized to a control gene with error bars representing SEM. *P<0.05, **P = 0.06 and ***P = 0.08 for the comparison of fold change of the indicated gene in P. aeruginosa-infected animals exposed to RPW-24 versus DMSO. Examination of the 269 genes that were induced greater than three-fold revealed that RPW-24 causes induction of many genes previously shown to be involved in the C. elegans transcriptional response to pathogenic bacteria. We found that 70 of the 269 genes induced greater than 3-fold by RPW-24 were also activated during infection with P. aeruginosa (a gene set characterized by Troemel et al. [8])(Figure 2B), which is significantly more than the 3.3 gene overlap expected by chance alone (P<2.7×10−16). Among these induced genes are several gene classes that have been implicated in antimicrobial defenses, such as CUB-like domain-containing genes, ShK-like toxins, C-type lectins and small molecule kinases (Table S1) [8]. This result is particularly interesting considering that RPW-24 is active against P. aeruginosa in the worm infection model but does not affect growth of the pathogen in vitro.
The transcriptome profiling analysis demonstrated that RPW-24 induces the transcription of putative immune effectors while C. elegans animals are feeding on Escherichia coli OP50, a relatively non-pathogenic food source for nematodes. We wondered if RPW-24 would further enhance the induction of these genes when C. elegans is infected with a bacterial pathogen that activates expression of these effectors. We therefore used qRT-PCR to test the level of induction of ten putative immune effectors (Figure 2A) during P. aeruginosa infection in the presence and absence of RPW-24. This panel included five genes that contain a CUB-like domain (C32H11.1, F35E12.5, F08G5.6, C29F3.7, and K08D8.5), two ShK-like toxins (F49F1.6 and C14C6.5), one antibacterial lysozyme (lys-7), one C-type lectin (clec-67) and one metallothionein (mtl-1). Eight of these ten genes were activated by P. aeruginosa in the absence of RPW-24 (Figure 2C). Interestingly, the induction levels of five of these eight genes (C32H11.1, F08G5.6, F35E12.5, C29F3.7, and F49F1.6) were markedly increased in the presence of both RPW-24 and P. aeruginosa (Figure 2C). RPW-24 also caused the induction of one gene that was strongly repressed during P. aeruginosa infection (mtl-1) and another whose transcription was unaffected by P. aeruginosa exposure (lys-7). RPW-24 did not change the level of induction for clec-67, C14C6.5 or K08D8.5 in the presence of P. aeruginosa. Taken together, these data demonstrate that RPW-24 both enhances the expression of a subset of putative immune effectors that are normally activated during P. aeruginosa infection and causes the induction of others that are not activated by P. aeruginosa. We do not, however, think that the induction of any single gene is itself responsible for the efficacy of RPW-24 since others have shown that C. elegans immune effectors likely function redundantly to defend against P. aeruginosa infection [8]. The finding that putative immune effectors are robustly induced by RPW-24 during bacterial infection provides a potential explanation why this compound provides protection against P. aeruginosa-mediated killing.
Immunostimulatory Activity of RPW-24 Is Required for Its Efficacy and Is Partially Dependent on the p38 MAP Kinase Signaling Cassette
Because RPW-24 caused the induction of genes normally upregulated during bacterial infection, we reasoned that C. elegans immune gene activation by RPW-24 is important for its anti-infective activity and depends on conserved defense response pathways. Previous studies have identified a requirement for three C. elegans signaling pathways in the defense against P. aeruginosa infection: the p38 MAP kinase signaling cassette [10], the G-protein coupled receptor FSHR-1 [16], and the bZIP transcription factor ZIP-2 [17]. Using loss-of-function mutants, we asked whether the activity of any of these pathways is required for the efficacy of RPW-24 in treating C. elegans infected with P. aeruginosa. Indeed, we found that the magnitude of RPW-24-mediated lifespan prolongation of pmk-1(km25) null mutants infected with P. aeruginosa was significantly attenuated compared to the lifespan extension observed in compound-treated wild-type animals (Figure 3) (P<0.001 for the difference in the lifespan prolongation between pmk-1(km25) and wild-type animals in two biological replicates). In contrast to pmk-1(km25) mutants, fshr-1(ok778) loss-of-function mutants demonstrated significant lifespan extension by RPW-24 compared to pmk-1(km25) animals, despite the fact that fshr-1(ok778) animals are also hypersusceptible to P. aeruginosa infection (Figure 3) (P<0.001 for the difference in the lifespan prolongation between pmk-1(km25) and fshr-1(ok778) in two biological replicates). Likewise, the lifespan of zip-2(tm4067) animals infected with P. aeruginosa was also significantly prolonged by RPW-24, and to a greater degree than pmk-1(km25) (P<0.01 for the difference in two biological replicates). These data demonstrate that the activity of PMK-1, more so than FSHR-1 or ZIP-2, is important for RPW-24 to extend the lifespan of nematodes infected with P. aeruginosa. Morever, the observed reduction in the ability of RPW-24 to extend the lifespan of pmk-1(km25) animals is not likely to be secondary to decreased overall fitness of immunocompromised animals infected with a bacterial pathogen because RPW-24 prolongs the lifespan of fshr-1(ok778) animals, a strain that is also markedly hypersusceptible to P. aeruginosa infection (Figure 3).
Fig. 3. Full efficacy of RPW-24 in prolonging the lifespan of C. elegans infected with P. aeruginosa requires the p38 MAP kinase signaling cassette. 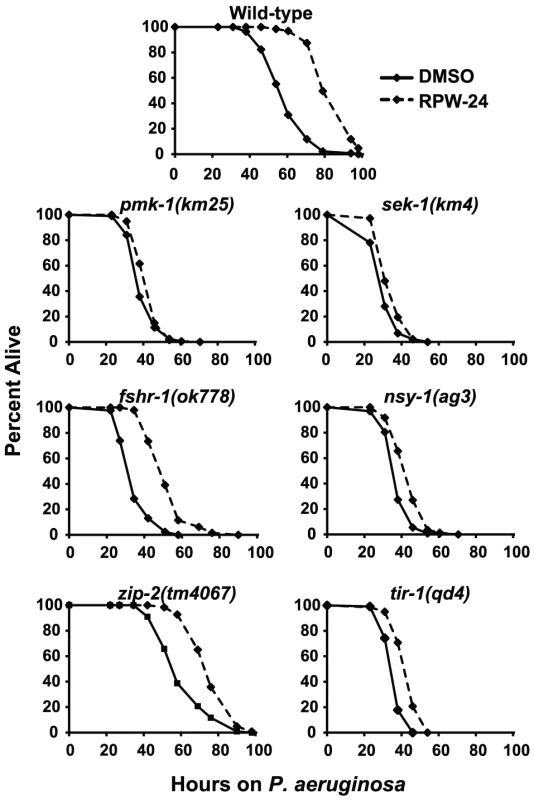
P. aeruginosa infection assays of C. elegans wild-type and mutant animals exposed to 70 µM RPW-24 compared to DMSO. Data at each time point are the average of three plates per strain, each with approximately 50 animals per plate (sample sizes are given in Table S2). Data are representative of two independent experiments. The p38 MAP kinase PMK-1 functions as part of a conserved signaling cassette to regulate host innate immune responses, which involves upstream activation by the MAP kinase kinase SEK-1, the MAP kinase kinase kinase NSY-1 and the Toll-Interleukin-1 receptor TIR-1. We tested the ability of RPW-24 to extend the lifespan of nematodes with mutations in each of these genes: sek-1(km4), nsy-1(ag3), and tir-1(qd4). As with the pmk-1(km25) mutants, we found that the curing activity of RPW-24 was attenuated in each of these mutant strains compared to the wild-type control (Figure 3), suggestive of a role for the entire TIR-1/NSY-1/SEK-1/PMK-1 signaling cassette in the activation of defense responses following exposure to RPW-24.
The infection assays described above show that the p38 MAP kinase pathway is required for RPW-24 to prolong the lifespan of C. elegans infected with P. aeruginosa, suggesting that RPW-24 induces the expression of immune effectors regulated by this cascade. Previous work has shown that PMK-1 coordinates the transcription of CUB-like genes, ShK toxins and C-type lectins, gene classes which are also induced by RPW-24 [8]. Indeed, we found that 14 of the 86 genes whose basal expression depends on PMK-1 [8] were induced by exposure to RPW-24, which is 14.6 fold more than expected by chance alone (P = 0.002)(Table S1). To test whether RPW-24 activates immune effectors in a PMK-1-dependent manner, we used qRT-PCR to determine the induction levels of six putative immune effectors in pmk-1(km25) null mutants exposed to DMSO and RPW-24 in the absence of pathogen. Wild-type worms were used as the control. The set of six putative immune effectors (F35E12.5, F08G5.6, clec-67, C32H11.1, F49F1.6 and mtl-1) were chosen from the panel of ten genes described above on the criterion that they were robustly upregulated by RPW-24 (Figure 2A, Figure S3). Figure 4A shows that PMK-1 affects the basal expression levels of the RPW-24-induced genes F35E12.5, F08G5.6, clec-67 and F49F1.6 (defined as the relative expression of the gene in pmk-1(km25) mutants compared to its expression in wild-type animals)(Figure 4A). PMK-1 is also required for the RPW-24-mediated induction of clec-67 and C32H11.1 (defined as the fold difference in gene expression in the presence and absence of RPW-24 in wild-type or pmk-1(km25) mutant animals), and perhaps F49F1.6 and mtl-1, although the differences in induction of these later two genes did not reach statistical significance (Figure 4B). Importantly, the absolute expression levels of all six of these genes following RPW-24 exposure were significantly lower in pmk-1(km25) animals than wild-type controls (Figure 4A). Taken together with the P. aeruginosa infection assays in pmk-1(km25) mutants (Figure 3), these data suggest that PMK-1-dependent immune effectors mediate part of the protective effect of RPW-24 in P. aeruginosa-infected C. elegans.
Fig. 4. The expression levels of RPW-24–induced putative immune effectors are reduced in pmk-1(km25) mutants compared to wild-type animals. 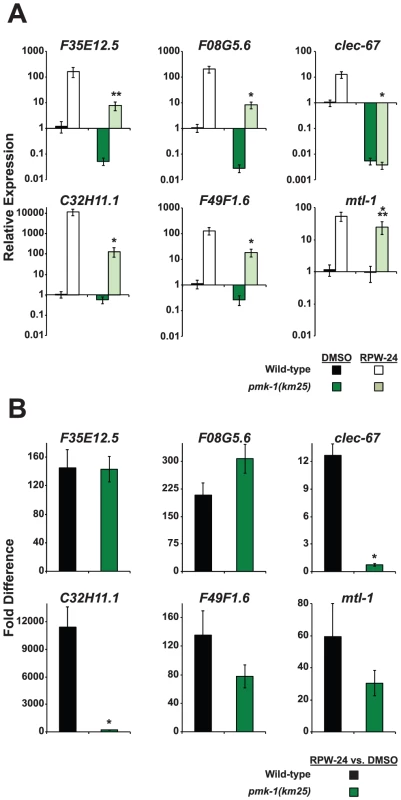
Six putative C. elegans immune effectors were analyzed by qRT-PCR in wild-type and pmk-1(km25) feeding on E. coli and exposed to either 70 µM RPW-24 or DMSO for 16 hours. Data are the average of two biological replicates each normalized to a control gene with error bars representing SEM. In (A), the data are relative to the average expression of the indicated gene in wild-type C. elegans exposed to DMSO. In (B), the fold change of the indicated gene above its expression in C. elegans exposed to DMSO is compared in wild-type and pmk-1(km25) mutant animals. *P<0.01, **P = 0.06, ***P = 0.09 for the comparison in (A) of relative expression levels in wild-type versus pmk-1(km25) animals, each exposed to RPW-24 and in (B) for the fold change in wild-type versus pmk-1(km25) mutant animals. RNAi Screen Reveals Role for the Transcription Factor ATF-7 in the Regulation of the RPW-24–Induced Immune Response
The phenotypic and genetic data presented above show that the p38 MAP kinase pathway is important for the RPW-24-induced modulation of C. elegans immune responses during P. aeruginosa infection. As described above, we found that 70 µM RPW-24 caused a striking increase in GFP production in the F35E12.5::GFP transcriptional reporter (Figure 1B). We thus reasoned that an RNAi screen could be used to find downstream regulator(s) of the RPW-24-induced immune response by identifying the genetic dependence of F35E12.5::GFP activation.
The basal regulation of F35E12.5 requires PMK-1, but its induction by RPW-24 occurs in a PMK-1-independent manner (Figure 4A and 4B). We therefore anticipated that a reverse genetic RNAi screen aimed at identifying transcription factors required for the RPW-24-mediated induction of F35E12.5::GFP would identify genetic regulators that act either downstream of or in parallel to the PMK-1 pathway. We used a feeding RNAi library containing bacterial clones that produce double stranded RNA (dsRNA) designed to individually knockdown the expression of 393 transcription factors in C. elegans, corresponding to 30–50% of the transcription factors in the C. elegans genome [18] and screened for RNAi clones that abrogated the RPW-24-mediated induction of F35E12.5::GFP. Among 393 screened, we found that a single clone, corresponding to the transcription factor ATF-7, caused a striking reduction of F35E12.5::GFP expression when nematodes were either growing on their normal laboratory food source (E. coli OP50) or infected with P. aeruginosa (Figure 5A). ATF-7 was previously shown to function downstream of PMK-1 in the regulation of immune response genes during P. aeruginosa infection [12].
Fig. 5. The C. elegans transcription factor ATF-7 regulates immune gene induction by RPW-24. 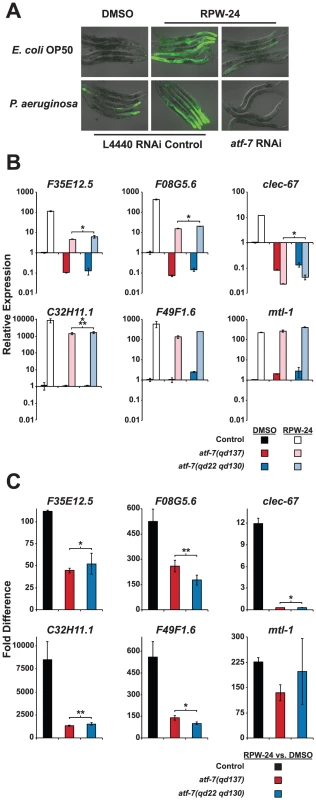
(A) Fluorescence microscopy images of C. elegans acIs101 animals, which express a F35E12.5::GFP transgene, exposed to the normal nematode food source E. coli OP50 or P. aeruginosa in the presence of DMSO or 70 µM RPW-24 for 16 hours at 25°C. C. elegans acIs101 animals were raised on L4440 RNAi control bacteria or an RNAi feeder strain designed to knockdown the expression of atf-7. Green is GFP expression. (B and C) Six putative C. elegans immune effectors were analyzed by qRT-PCR in wild-type, atf-7(qd137) and atf-7(qd22 qd130) animals feeding on E. coli and exposed to either 70 µM RPW-24 or DMSO for 16 hours. Data are presented as the average of two biological replicates each normalized to a control gene with error bars representing SEM. In (A), the data are relative to the average expression of the indicated gene in wild-type C. elegans exposed to DMSO. In (B), the fold change of the indicated gene above its expression in C. elegans exposed to DMSO is compared in wild-type, atf-7(qd137) and atf-7(qd22 qd130) animals. An N2-derived strain carrying the acIs219 transgene was used as the control strain because this transgene is also present in the atf-7(lof) strains [12]. *P<0.05, **P = 0.08, ***P = 0.09 for the comparison in (A) of the relative expression levels of both atf-7(lof) strains versus control animals, each exposed to RPW-24, and in (B) for the fold change in wild-type versus both atf-7(lof) mutants. qRT-PCR analysis confirmed that the reduction of F35E12.5::GFP expression was due to the knockdown of ATF-7 and not the consequence of non-specific transgene silencing. Specifically, the absolute level F35E12.5 expression following RPW-24 exposure was markedly reduced in two atf-7(lof) mutants [atf-7(qd137) and atf-7(qd22 qd130) [12]] compared to the control strain (Figure 5B), as were the levels of three other putative immune effectors (F08G5.6, clec-67 and C32H11.1). Moreover, the basal levels of F35E12.5, F08G5.6 and clec-67 were reduced to a similar degree in the atf-7(lof) mutants and the pmk-1(km25) animals, consistent with the previously described role for PMK-1 and ATF-7 in the basal regulation of immune effectors (compare Figure 4A and Figure 5B) [12]. Interestingly, we found that ATF-7 was also required for the full induction of five of six putative immune effectors (F35E12.5, F08G5.6, clec-67, C32H11.1, and F49F1.6)(Figure 5C), including two genes (F35E12.5 and F08G5.6) that were induced by RPW-24 independently of PMK-1 (Figure 4B).
To determine if the activity of ATF-7 is important for the efficacy of RPW-24, we tested the ability of RPW-24 to prolong the lifespan of atf-7(lof) mutants exposed to P. aeruginosa. The magnitude of lifespan extension conferred by RPW-24 was reduced in both the atf-7(qd137) and the atf-7(qd22 qd130)(Figure S4A and S4B, respectively) mutants compared to the control strain. We therefore conclude that RPW-24 stimulates the C. elegans immune response genes in a manner that involves both the p38 MAP kinase cassette PMK-1 and the conserved transcription factor ATF-7, consistent with the placement of ATF-7 downstream of PMK-1 [12]. Expression analysis of RPW-24-induced genes (Figure 5C) suggests that in addition to functioning downstream of PMK-1, ATF-7 receives inputs from a PMK-1 independent pathway to coordinate the induction of putative immune effectors (such as F35E12.5 and F08G5.6) and the RPW-24-mediated resistance to P. aeruginosa infection. This conclusion is based on the finding that the RPW-24-mediated activation of F35E12.5 and F08G5.6 is PMK-1 independent (as opposed to their basal level of expression), but is at least partially dependent on ATF-7 (compare Figure 4B with Figure 5C). That is, the basal levels of expression of F35E12.5 and F08G5.6 are both PMK-1 and ATF-7 dependent, whereas the fold induction of these genes following RPW-24 is not affected in the pmk-1(km25) mutant, but is reduced by at least half in the atf-7(lof) mutants. The biological significance of this PMK-1-independent transcriptional activator is not known.
It is also interesting to note that RPW-24 exhibited an attenuated, but significant (P<0.001) and reproducible ability to rescue the atf-7(lof) mutants. Therefore, the anti-infective activity of RPW-24 may involve another immune signaling pathway, independent of both the p38 MAP kinase pathway and ATF-7. This conclusion is consistent with the observation that RPW-24 also modestly, but significantly (P<0.001), extends the lifespan of pmk-1(km25), sek-1(km4), nsy-1(ag3), and tir-1(qd4) mutant animals infected with P. aeruginosa (Figure 3), and the fact that the induction of F35E12.5 and F08G5.6 is not completely abrogated in the atf-7(lof) mutants. An alternate explanation is that RPW-24 affects virulence factor production by P. aeruginosa or exerts a subtle effect on growth of the pathogen.
RPW-24 Is Toxic to C. elegans
In addition to inducing a preponderance of genes involved in the transcriptional response to pathogenic bacteria, RPW-24 caused the upregulation of genes involved in the detoxification of small molecules (Table S1). The microarray analysis revealed that 58 of the 269 genes activated 3 fold or more and 31 of the 57 genes activated 50 fold or more were UDP-glucuronosyltransferases (UDPs), cytochrome P450s (CYPs), glutathione-s-transferases (GSTs) or short-chain dehydrogenases (SDRs). These gene classes play integral roles in the Phase I and II detoxification of both endobiotic and xenobiotic toxins in both nematodes and mammals [19], [20]. Indeed, seven of ten CYPs, two of three UDPs, one of four GSTs, three of three carboxylesterases, and three of six C-type lectins were induced both by RPW-24 and exposure to five xenobiotic toxins [21]. Taken together, these data suggest that RPW-24 induces xenobiotic detoxification pathways in C. elegans.
To determine if RPW-24 adversely affects wild-type nematodes growing in the absence of pathogen, we first used a behavioral assay designed to study the aversion response of C. elegans to xenobiotic toxins [22]. The addition of some poisons to the center of small lawns of non-pathogenic E. coli causes C. elegans animals to leave the lawn, presumably to minimize toxin exposure. Interestingly, we observed a significant aversion response to 70 µM RPW-24 (Figure 6A and 6B). After 16 hours, 51% of the nematodes had left the lawn containing RPW-24, whereas only 7% of animals left a control lawn (P = 0.002; Figure 6B). Next, we conducted a lifespan assay on nematode growth media supplemented with either DMSO or varying concentrations of RPW-24 and found that RPW-24 shortened C. elegans lifespan in a dose-dependent manner (Figure 6C), with 70 µM RPW-24 resulting in a 24% reduction in median lifespan. Interestingly, we observed lifespan shortening only at compound concentrations that rescued C. elegans from P. aeruginosa infection [7, 35 and 70 µM, but not 0.7 µM]. We also found that 70 µM RPW-24 slowed the development of animals when they were exposed at the first larval stage (L1)(Figure 6D).
Fig. 6. RPW-24 is toxic to C. elegans. 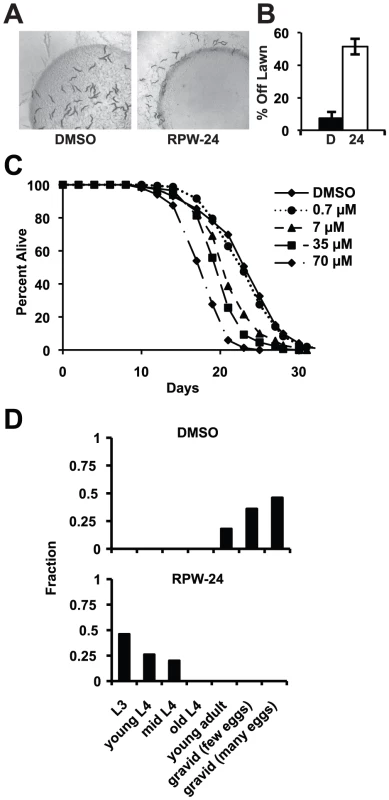
(A) Wild-type C. elegans leave a lawn of E. coli containing 70 µM RPW-24 more so than DMSO after 16 hours of exposure. (B) The average percentage of wild-type C. elegans that were off a lawn of E. coli supplemented with either DMSO or 70 µM RPW-24 after 16 hours of exposure from three plates per condition [as in (A)] is presented with error bars representing SEM. P = 0.002 for the comparison of the two conditions. (C) RPW-24 shortens the lifespan of wild-type C. elegans in a dose-dependent manner. Data at each time point are the average of three plates per strain, each with approximately 50 animals per plate (sample sizes are given in Table S2). (D) RPW-24 slows development of wild-type C. elegans. Developmental stage was accessed in 50 animals per treatment group and presented as the percentage of the population that was at the indicated development stage after 65 hours of incubation at 20°C. In C. elegans, oxidative stress is a potent inducer of phase II detoxification genes. Moreover, the p38 MAP kinase PMK-1 regulates the cellular response to oxidative and arsenite stress, but through the transcription factor SKN-1, not ATF-7 [12], [23]–[25]. We therefore wondered whether RPW-24 might confer protection against arsenite stress. We found, however, that 70 µM RPW-24 did not protect nematodes exposed to 5 mM sodium arsenite for 16 hours at 20°C (Figure S5). In fact, the toxicity of RPW-24 and sodium arsenite were synergistic in this assay, resulting in nearly 100% mortality of wild-type nematodes exposed to both compounds (Figure S5).
While RPW-24 confers a survival advantage for nematodes infected with pathogenic bacteria, these data demonstrate that it is toxic to C. elegans growing under standard laboratory conditions. Whether this toxicity is a consequence of direct effects of the compound on C. elegans or hyper-activation of the immune system by RPW-24 is unknown.
Discussion
We hypothesized that a C. elegans-based screen for novel anti-infectives would identify small molecules that act by stimulating the host immune response. Four lines of evidence allow us to conclude that RPW-24 protects C. elegans from bacterial infection by inducing the production of putative immune effectors via an evolutionarily conserved immune pathway. First, RPW-24 confers a survival advantage for nematodes infected with the Gram-positive bacteria E. faecalis and the Gram-negative bacteria P. aeruginosa at doses that do not inhibit in vitro growth of these pathogens. Second, whole genome microarray analysis demonstrates that RPW-24 induces a transcriptional response in C. elegans in the absence of pathogen exposure. We observed that 26% of the genes induced 3-fold or greater are also induced by infection with P. aeruginosa. Moreover, we showed that the RPW-24-mediated induction of several putative immune effectors is enhanced in the context of P. aeruginosa infection. Third, using C. elegans animals deficient in immune pathway signaling in pathogenesis assays and gene expression analyses, we demonstrated that the TIR-1/NSY-1/SEK-1/PMK-1 cascade in C. elegans is required for RPW-24 to exert its full effect. Lastly, an RNAi screen indicated that the transcription factor ATF-7 controls the RPW-24-mediated induction of a putative immune effector gene. The ability of RPW-24 to promote survival of P. aeruginosa-infected nematodes is partially dependent on this transcription factor. ATF-7 has been shown to function downstream of the MAPK PMK-1 to activate the expression of immune effectors [12]. Here we show that ATF-7 also activates the expression of genes independently of the PMK-1 p38 MAPK pathway. In summary, these data strongly suggest that immune gene activation by RPW-24 is required for its protective effects. While it is theoretically possible that RPW-24 confers protection for C. elegans by also inhibiting virulence factor production in both E. faecalis and P. aeruginosa, we feel this is less likely given that clear effects of RPW-24 on the nematode and the evolutionary diversity between these bacterial pathogens.
An unexpected observation in our transcriptome profiling analysis was that RPW-24 caused the dramatic induction of genes involved in the detoxification of small molecules, including UDP-glucuronosyltransferases (UGTs), cytochrome P450s (CYPs), glutathione-s-transferases (GSTs) and short-chain dehydrogenases (SDRs). In mammals, these gene classes act together to detoxify xenobiotic small molecules via two successive reactions [19]. Phase I reactions involve the addition of chemically reactive functional groups to the toxins and are predominantly mediated by CYPs and SDRs. In Phase II, UGTs and GSTs add side groups that increase the solubility of small toxic molecules, which aides in their excretion. These mediators of detoxification are highly conserved throughout evolution and are present in C. elegans [20], [26], [27]. Their marked induction by RPW-24 suggests that this compound may be recognized as a toxin. Consistent with this hypothesis, RPW-24 caused a strong behavioral avoidance phenotype, shortened nematode lifespan, and delayed the development of nematodes growing on non-pathogenic bacteria.
We have identified a low molecular weight molecule that can potently activate the innate immune response of C. elegans. The target of this compound is not known, nor are the mechanisms that act upstream of the p38 MAP kinase cassette to trigger the RPW-24-mediated immune activation in C. elegans. Indeed, it also unclear how any of the C. elegans immune pathways, including the p38 MAP kinase cassette, are activated during bacterial infection. Recently, several investigators have shown that the nematode monitors the integrity of cellular processes as a means to detect pathogen invasion, and to trigger defense responses and behavioral avoidance phenotypes. McEwan et al. [28] and Dunbar et al. [29] each found that the inhibition of translation by a bacterial toxin induced a protective immune response in C. elegans that was dependent on the p38 MAP kinase and ZIP-2 pathways. Similarly, Melo et al. showed that disruption of core cellular processes, such as translation, mitochondrial respiration and proteasome function, by bacterial toxins induced a behavioral avoidance phenotype [22]. Thus, it is possible that the toxic effects of RPW-24 trigger immune response pathways and a behavioral aversion response in an analogous manner. Alternatively, RPW-24 itself could directly activate immune pathways in C. elegans.
In this study, we have demonstrated the utility of using small molecules in conjunction with classical epistasis analysis and RNAi screens to dissect immune signaling pathways in C. elegans. We therefore hypothesize that RPW-24 can be used as a tool in additional genetic analyses both to identify the target(s) of this small molecule and to determine mechanism by which the p38 MAP kinase pathway is activated by RPW-24, which may offer insights into how C. elegans detects bacterial pathogens.
The World Health Organization has declared that antimicrobial-resistant pathogens are one of the three greatest threats to human health. Exacerbating this problem is the striking absence of novel antibiotics in the development pipeline. In 2008, there were only 16 antimicrobial compounds in late stage clinical trials and only one of these agents had a novel mechanism of action [30]. Furthermore, all of these agents target some aspect of bacterial replication or metabolism. Indeed, it has been suggested that the obvious bacterial targets amenable for antimicrobial drug design have been exhausted [31]. Identifying host-acting small molecules that modulate innate immune responses is a promising approach to identify novel antimicrobial therapies [32]. In theory, such agents should place minimal selection pressure on bacteria to acquire resistance determinants and could have broad-spectrum antimicrobial activity. Agonists of the mammalian Toll-like and NOD-like receptors are among the most promising compounds that are being explored for this purpose [32], [33]. We propose that C. elegans-based compound screening assays can be used to mine large chemical libraries for additional small molecule anti-infectives that act by stimulating host immune defenses. Whether or not these host-acting small molecules will be effective for the treatment of bacterial infections in mammals, genetic dissection of the C. elegans signaling pathways activated by such compounds may suggest strategies for the development of new classes of antimicrobial therapies.
Materials and Methods
C. elegans Strains
C. elegans were maintained and propagated on E. coli OP50 as described [34]. The C. elegans strains used in this study were: N2 Bristol [34], pmk-1(km25) [10], sek-1(km4) [10], nsy-1(ag3) [10], tir-1(qd4) [11], atf-7(qd137) [12], atf-7(qd22 qd130) [12], fshr-1(ok778) [16], zip-2(tm4067) [17] glp-4(bn-2) [35], AU78 [agIs219 (pT24B8.5::GFP::unc-54-3′UTR pttx-3::GFP::unc-54-3′utr)] [11] and AY101 [acIs101[pDB09.1(pF35E12.5::GFP); pRF4(rol-6(su1006))] [13].
C. elegans Bacterial Infection and Other Assays
Slow-killing P. aeruginosa solid media infection assays were performed as previously described [15] with some modifications. A single colony of P. aeruginosa PA14 was innoculated into 3-mL of Luria-Bertani (LB) media and allowed to incubate at 37°C for 14 to 15 hours. 10 µL of this culture was added to 35-mm tissue culture plates containing 4 mL of slow kill agar supplemented with 1% DMSO and the indicated conentration of RPW-24. Others have shown that this concentration of DMSO has little effect on growth or development of C. elegans [36], [37]. Plates were incubated for 24 hours at 37°C and 24 hours at 25°C. C. elegans lifespan assays were conducted on nematode growth media (NGM) supplemented with the indicated concentraion of RPW-24 and seeded with OP50. The sensitivity of RPW-24-treated wild-type animals to oxidative stress was determined using 5 mM sodium arsenite following a previously described protocol [12]. For the infection, lifespan and sodium arsenite assays, 0.1 mg/mL 5-fluorodeoxyuridine (FUDR) was added to the media 1 to 2 hours before the start of the assay to prevent progeny from hatching. Approximately 50 L4 staged nematodes were picked to each of three or four assay plates per experimental condition. Animals were scored as live or dead on a daily basis by gently touching them with a platinum wire. Worms that crawled onto the wall of the tissue culture plate were eliminated from the analysis. The P. aeruginosa killing assays were conducted at 25°C. The lifespan and sodium arsenite assays were performed at 20°C. The sample sizes for each of these experiments are given in Table S2. For the experiments with the atf-7(lof) mutants, we used an N2-derived strain carrying the acIs219 transgene (AU78) as the control strain because this transgene is also present in the atf-7 mutant strains [12]. The C. elegans liquid media infection assay used to screen the 31 compounds for those with activity against P. aeruginosa-infected nematodes was developed in our laboratory and was conducted in either 384 or 96 well plates using glp-4(bn-2) animals (Kirienko, NV and Ausubel FM, unpublished data). Growth of P. aeruginosa in the presence of RPW-24 was determined by inoculating 1.0×104 bacteria in liquid slow-kill media containing either 70 µM RPW-24 or DMSO and allowing the culture to grow at 37°C in a roller drum. At the indicated time points, 10 µL of the culture was removed and CFUs were determined by plating serial dilutions.
The propensity of wild-type C. elegans to leave a lawn of bacteria supplemented with RPW-24 was assayed using a previously described protocol [22]. Briefly, 6-well tissue culture plates containing NGM were seeded with concentrated E. coli OP50. 70 µM RPW-24 or an equal volume of DMSO was added to the center of the E. coli lawn and allowed to dry. 70 to 90 young L4 animals were added to the center of the lawns and animals were scored as either on or off the lawn after 16 hours incubation at room temperature. To determine if RPW-24 slowed the development of wild-type animals, 70 µM RPW-24 or DMSO was added to NGM plates and seeded with OP50. L1 staged animals, synchronized by hypochlorite treatment, were added to these plates and allowed to incubate at 20°C for 65 hours. Developmental stages of 50 animals per treatment group were determined by microscopic examination of the gonad.
C. elegans Microarray Analysis
N2 animals were synchronized by hypochlorite treatment. Arrested L1s were plated on 10 cm NGM plates seeded with E. coli OP50 and grown at 20°C until the late L4 larval stage. Animals were incubated for 15 hours at 15°C in 2 mL of liquid S-basal complete medium [38] containing 70 µM RPW-24 or DMSO and supplemented with E. coli OP50. The final concentration of DMSO in both samples was 1%. RNA was extracted from three biological replicates using TRI Reagent (Molecular Research Center) according to the manufacturer's instructions and purified using an RNeasy column (Qiagen). RNA samples were prepared and hybridized to Affymetrix full-genome GeneChips for C. elegans at the Harvard Medical School Biopolymer Facility (Boston, MA) following previously described protocols [5], [8] and instructions from Affymetrix. Data were analyzed using GenePattern version 2.0 software using GC-RMA and quantile normalization [39]. Conditions were compared using GenePattern to determine the fold change between conditions for each probe set and to generate a P value using a modified t-test. Probe sets were considered differentially expressed if the fold change was 3-fold or greater (P<0.025).
Quantitative RT–PCR (qRT–PCR) Analyses
Animals of the indicated genotype were treated and RNA was extracted as described for the microarray analysis. For the experiments with the atf-7(lof) mutants, we used strain AU78 as as the control strain [12]. For gene expression analysis of nematodes on solid media, 70 µM RPW-24 or DMSO was added to 20 mL nematode growth media in 10 cm petri dishes seeded with E. coli OP50. For qRT-PCR studies of nematodes infected with P. aeruginosa, 20 mL of slow killing media was added to 10 cm petri dishes containing either DMSO or 70 µM RPW-24. Plates were seeded with either 250 µL of E. coli OP50 or 50 µL P. aeruginosa diluted in 200 µL LB, each from overnight cultures. The plates were incubated for 24 hours at 37°C and 24 hours at 25°C. Old L4/young adult animals were added to the assay plates and incubated at 25°C for eight hours. RNA was reverse transcribed to cDNA using the Retroscript kit (Ambion). cDNA was analyzed by qRT-PCR using a CFX1000 machine (Bio-Rad) and previously published primers [8]. All values were normalized against the control gene snb-1, which has been used previously in qRT-PCR studies of C. elegans innate immunity [5], [8], [9], [12], [40]. Analysis of the microarray expression data revealed that the expression of snb-1 did not vary under the conditions tested in the experiment. Fold change was calculated using the Pfaffl method [41].
Feeding RNAi Screen
The RNAi screen of 393 transcription factors was conducted using RNAi clones from the Ahringer and Vidal RNAi libraries and an established protocol [42]. The RNAi clone for atf-7 was developed by Shivers et al [12]. Briefly, overnight cultures of feeding RNAi clones were added to each well of a 96-well RNAi plate and allowed to grow at room temperature overnight. 40–60 L1 staged acIs101 animals, which express a F35E12.5::GFP transgene, were added to each well and allowed to grow for two days at 20°C until they were at the L4 or young adult stage. Animals were then washed from the RNAi plates into S-basal complete media [38] containing 70 µM RPW-24. All experiments with feeding RNAi used a gfp RNAi as the negative control, which resulted in no visible GFP expression in acIs101 transgenic animals. acIs101 animals treated with the empty vector L4440 and exposed to 70 µM RPW-24 was used as the positive control. Animals were scored for GFP expression following photograph of each well using an Image Xpress Micro microscope (Molecular Devices Corporation, Sunnyvale, CA).
Microscopy
Nematodes were mounted onto agar pads, paralyzed with 10 mM levamisole (Sigma) and photographed using a Zeiss AXIO Imager Z1 microscope with a Zeiss AxioCam HRm camera and Axiovision 4.6 (Zeiss) software.
Statistical Analyses
Differences in survival of C. elegans animals infected with P. aeruginosa on slow-killing assay plates were determined with the log-rank test. To determine if the increase in survival conferred by RPW-24 treatment was different in one population compared to another [for example, in wild-type versus pmk-1(km25) animals], we examined the difference in the effect of RPW-24 treatment on the hazard in each group using a Cox proportional hazard model (Stata11, Stata, College Station, TX). Fold changes in the qRT-PCR analyses and the differences in survival in the arsenite assays were compared using unpaired, two-tailed student t-tests. When comparing microarray datasets, the overlap expected by chance alone was determined in 50 groups of randomly selected C. elegans genes using Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/), a technique that has been used for similar analyses [43]. P values were determined using chi-square tests.
Accession Numbers
Accession numbers for the genes and gene products mentioned in this paper are given for Wormbase, a publicly available database that can be accessed at http://www.wormbase.org. These accession numbers are pmk-1 (B0218.3), nsy-1 (F59A6.1), sek-1 (R03G5.2), atf-7 (C07G2.2), zip-2 (K02F3.4), fshr-1 (C50H2.1), skn-1 (T19E7.2), C32H11.1, F35E12.5, F08G5.6, C29F3.7, K08D8.5, F49F1.6, C14C6.5, lys-7 (C02A12.4), clec-67 (F56D6.2) and mtl-1 (K11G9.6). The microarray dataset can be downloaded from the National Center for Biotechnology Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo). The accession number for these data is GSE37266.
Supporting Information
Zdroje
1. MoyTIConeryALLarkins-FordJWuGMazitschekR 2009 High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem Biol 4 527 533
2. MoyTIBallARAnklesariaZCasadeiGLewisK 2006 Identification of novel antimicrobials using a live-animal infection model. Proc Natl Acad Sci USA 103 10414 10419
3. Pukkila-WorleyRHolsonEWagnerFMylonakisE 2009 Antifungal Drug Discovery through the Study of Invertebrate Model Hosts. Curr Med Chem 16 1588 1595
4. Pukkila-WorleyRAusubelFM 2012 Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Opin Immunol 24 3 9
5. Pukkila-WorleyRAusubelFMMylonakisE 2011 Candida albicans Infection of Caenorhabditis elegans Induces Antifungal Immune Defenses. PLoS Pathog 7 e1002074 doi:10.1371/journal.ppat.1002074
6. Pukkila-WorleyRPelegAYTampakakisEMylonakisE 2009 Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryotic Cell 8 1750 1758
7. Pukkila-WorleyRMylonakisE 2010 From the outside in and the inside out: Antifungal immune responses in Caenorhabditis elegans. Virulence 1 111 112
8. TroemelERChuSWReinkeVLeeSSAusubelFM 2006 p38 MAPK Regulates Expression of Immune Response Genes and Contributes to Longevity in C. elegans PLoS Genet 2 e183 doi:10.1371/journal.pgen.0020183
9. IrazoquiJETroemelERFeinbaumRLLuhachackLGCezairliyanBO 2010 Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus PLoS Pathog e1000982 6 doi:10.1371/journal.ppat.1000982
10. KimDHFeinbaumRAlloingGEmersonFEGarsinDA 2002 A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297 623 626
11. ShiversRPKooistraTChuSWPaganoDJKimDH 2009 Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe 6 321 330
12. ShiversRPPaganoDJKooistraTRichardsonCEReddyKC 2010 Phosphorylation of the Conserved Transcription Factor ATF-7 by PMK-1 p38 MAPK Regulates Innate Immunity in Caenorhabditis elegans PLoS Genet e1000892 6 doi:10.1371/journal.pgen.1000892
13. BolzDDTenorJLAballayA 2010 A conserved PMK-1/p38 MAPK is required in Caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J Biol Chem 285 10832 10840
14. O'RourkeDBabanDDemidovaMMottRHodgkinJ 2006 Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res 16 1005 1016
15. TanMWMahajan-MiklosSAusubelFM 1999 Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA 96 715 720
16. PowellJRKimDHAusubelFM 2009 The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc Natl Acad Sci USA 106 2782 2787
17. EstesKADunbarTLPowellJRAusubelFMTroemelER 2010 bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc Natl Acad Sci USA 107 2153 2158
18. Reece-HoyesJSDeplanckeBShinglesJGroveCAHopeIA 2005 A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol 6 R110
19. AmsdenGWBallowCHBertinoJSJKashubaAD 2005 Pharmacokinetics and Pharmacodynamics of Anti-infective Agents. MandellGLBennettJEDolinR 271 281 Principals and Practice of Infectious Diseases. Philadelphia, PA: Elsevier.
20. McElweeJJSchusterEBlancEThomasJHGemsD 2004 Shared transcriptional signature in Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem 279 44533 44543
21. ReichertKMenzelR 2005 Expression profiling of five different xenobiotics using a Caenorhabditis elegans whole genome microarray. Chemosphere 61 229 237
22. MeloJARuvkunG 2012 Inactivation of Conserved C. elegans Genes Engages Pathogen - and Xenobiotic-Associated Defenses. Cell 149 452 466
23. AnJHBlackwellTK 2003 SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17 1882 1893
24. InoueHHisamotoNAnJHOliveiraRPNishidaE 2005 The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19 2278 2283
25. AnJHVranasKLuckeMInoueHHisamotoN 2005 Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci USA 102 16275 16280
26. GemsDMcElweeJJ 2005 Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech Ageing Dev 126 381 387
27. ChakrapaniBPSKumarSSubramaniamJR 2008 Development and evaluation of an in vivo assay in Caenorhabditis elegans for screening of compounds for their effect on cytochrome P450 expression. J Biosci 33 269 277
28. McEwanDLKirienkoNVAusubelFM 2012 Host Translational Inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an Immune Response in Caenorhabditis elegans. Cell Host Microbe 11 364 374
29. DunbarTLYanZBallaKMSmelkinsonMGTroemelER 2012 C. elegans Detects Pathogen-Induced Translational Inhibition to Activate Immune Signaling. Cell Host Microbe 11 375 386
30. BoucherHWTalbotGHBradleyJSEdwardsJEGilbertD 2009 Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48 1 12
31. BumannD 2008 Has nature already identified all useful antibacterial targets? Curr Opin Microbiol 11 387 392
32. HancockREWNijnikAPhilpottDJ 2012 Modulating immunity as a therapy for bacterial infections. Nat Rev Microbiol 10 243 254
33. HennessyEJParkerAEO'NeillLAJ 2010 Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov 9 293 307
34. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
35. BeananMJStromeS 1992 Characterization of a germ-line proliferation mutation in C. elegans. Development 116 755 766
36. GoldsteinPMagnanoL 1988 Effects of dimethyl sulphoxide on early gametogenesis in Caenorhabditis elegans: ultrastructural aberrations and loss of synaptonemal complexes from pachytene nuclei. Cytobios 56 45 57
37. RandJBJohnsonCD 1995 Genetic pharmacology: interactions between drugs and gene products in Caenorhabditis elegans. EpsteinHFShakesDC 187 204 Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego: Academic Press Inc., Vol. 48.
38. StiernagleT 1999 Maintenance of C. elegans. HopeIA 59 60 C. elegans: a practical approach. Oxford University Press, USA.
39. ReichMLiefeldTGouldJLernerJTamayoP 2006 GenePattern 2.0. Nat Genet 38 500 501
40. RichardsonCEKooistraTKimDH 2010 An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463 1092 1095
41. PfafflMW 2001 A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45
42. O'RourkeEJConeryALMoyTI 2009 Whole-animal high-throughput screens: the C. elegans model. Methods Mol Biol 486 57 75
43. KirienkoNVMcEnerneyJDKFayDS 2008 Coordinated regulation of intestinal functions in C. elegans by LIN-35/Rb and SLR-2 PLoS Genet 4 e1000059 doi:10.1371/journal.pgen.1000059
Štítky
Genetika Reprodukční medicína
Článek Uracil-Containing DNA in : Stability, Stage-Specific Accumulation, and Developmental InvolvementČlánek Fuzzy Tandem Repeats Containing p53 Response Elements May Define Species-Specific p53 Target GenesČlánek Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early EmbryosČlánek Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2α Kinase GCN-2Článek Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human CellsČlánek TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome EndsČlánek Attenuation of Notch and Hedgehog Signaling Is Required for Fate Specification in the Spinal CordČlánek Genome-Wide Functional Profiling Identifies Genes and Processes Important for Zinc-Limited Growth ofČlánek MicroRNA93 Regulates Proliferation and Differentiation of Normal and Malignant Breast Stem Cells
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 6- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- Rumors of Its Disassembly Have Been Greatly Exaggerated: The Secret Life of the Synaptonemal Complex at the Centromeres
- Mimetic Butterflies Introgress to Impress
- Tipping the Balance in the Powerhouse of the Cell to “Protect” Colorectal Cancer
- Selection-Driven Gene Loss in Bacteria
- Decreased Mitochondrial DNA Mutagenesis in Human Colorectal Cancer
- Parallel Evolution of Auditory Genes for Echolocation in Bats and Toothed Whales
- Diverse CRISPRs Evolving in Human Microbiomes
- The Rad4 ATR-Activation Domain Functions in G1/S Phase in a Chromatin-Dependent Manner
- Stretching the Rules: Monocentric Chromosomes with Multiple Centromere Domains
- Uracil-Containing DNA in : Stability, Stage-Specific Accumulation, and Developmental Involvement
- Fuzzy Tandem Repeats Containing p53 Response Elements May Define Species-Specific p53 Target Genes
- Adaptive Introgression across Species Boundaries in Butterflies
- G Protein Activation without a GEF in the Plant Kingdom
- Synaptonemal Complex Components Persist at Centromeres and Are Required for Homologous Centromere Pairing in Mouse Spermatocytes
- An Engineering Approach to Extending Lifespan in
- Incompatibility and Competitive Exclusion of Genomic Segments between Sibling Species
- Effects of Histone H3 Depletion on Nucleosome Occupancy and Position in
- Patterns of Evolutionary Conservation of Essential Genes Correlate with Their Compensability
- Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis
- Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early Embryos
- A Mouse Model of Acrodermatitis Enteropathica: Loss of Intestine Zinc Transporter ZIP4 (Slc39a4) Disrupts the Stem Cell Niche and Intestine Integrity
- Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2α Kinase GCN-2
- Geographic Differences in Genetic Susceptibility to IgA Nephropathy: GWAS Replication Study and Geospatial Risk Analysis
- Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human Cells
- TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome Ends
- Stimulation of Host Immune Defenses by a Small Molecule Protects from Bacterial Infection
- A Broad Requirement for TLS Polymerases η and κ, and Interacting Sumoylation and Nuclear Pore Proteins, in Lesion Bypass during Embryogenesis
- Genome-Wide Identification of Ampicillin Resistance Determinants in
- The CCR4-NOT Complex Is Implicated in the Viability of Aneuploid Yeasts
- Gustatory Perception and Fat Body Energy Metabolism Are Jointly Affected by Vitellogenin and Juvenile Hormone in Honey Bees
- Genetic Variants on Chromosome 1q41 Influence Ocular Axial Length and High Myopia
- Is a Key Regulator of Pancreaticobiliary Ductal System Development
- The NSL Complex Regulates Housekeeping Genes in
- Attenuation of Notch and Hedgehog Signaling Is Required for Fate Specification in the Spinal Cord
- Dual-Level Regulation of ACC Synthase Activity by MPK3/MPK6 Cascade and Its Downstream WRKY Transcription Factor during Ethylene Induction in Arabidopsis
- Genome-Wide Functional Profiling Identifies Genes and Processes Important for Zinc-Limited Growth of
- Base-Pair Resolution DNA Methylation Sequencing Reveals Profoundly Divergent Epigenetic Landscapes in Acute Myeloid Leukemia
- MicroRNA93 Regulates Proliferation and Differentiation of Normal and Malignant Breast Stem Cells
- Phylogenomic Analysis Reveals Dynamic Evolutionary History of the Drosophila Heterochromatin Protein 1 (HP1) Gene Family
- Found: The Elusive ANTAR Transcription Antiterminator
- The Mutation in Chickens Constitutes a Structural Rearrangement Causing Both Altered Comb Morphology and Defective Sperm Motility
- Control of CpG and Non-CpG DNA Methylation by DNA Methyltransferases
- Polymorphisms in the Mitochondrial Ribosome Recycling Factor Compromise Cell Respiratory Function and Increase Atorvastatin Toxicity
- Gene Expression Profiles in Parkinson Disease Prefrontal Cortex Implicate and Genes under Its Transcriptional Regulation
- Global Regulatory Functions of the Endoribonuclease III in Gene Expression
- Extensive Evolutionary Changes in Regulatory Element Activity during Human Origins Are Associated with Altered Gene Expression and Positive Selection
- The Regulatory Network of Natural Competence and Transformation of
- Brain Expression Genome-Wide Association Study (eGWAS) Identifies Human Disease-Associated Variants
- Quantifying the Adaptive Potential of an Antibiotic Resistance Enzyme
- Divergence of the Yeast Transcription Factor Affects Sulfite Resistance
- The Histone Demethylase Jhdm1a Regulates Hepatic Gluconeogenesis
- RNA Methylation by the MIS Complex Regulates a Cell Fate Decision in Yeast
- Rare Copy Number Variants Observed in Hereditary Breast Cancer Cases Disrupt Genes in Estrogen Signaling and Tumor Suppression Network
- Genome-Wide Location Analysis Reveals Distinct Transcriptional Circuitry by Paralogous Regulators Foxa1 and Foxa2
- A Mutation Links a Canine Progressive Early-Onset Cerebellar Ataxia to the Endoplasmic Reticulum–Associated Protein Degradation (ERAD) Machinery
- The Mechanism for RNA Recognition by ANTAR Regulators of Gene Expression
- Limits to the Rate of Adaptive Substitution in Sexual Populations
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Rumors of Its Disassembly Have Been Greatly Exaggerated: The Secret Life of the Synaptonemal Complex at the Centromeres
- The NSL Complex Regulates Housekeeping Genes in
- Tipping the Balance in the Powerhouse of the Cell to “Protect” Colorectal Cancer
- Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



