-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Broad Requirement for TLS Polymerases η and κ, and Interacting Sumoylation and Nuclear Pore Proteins, in Lesion Bypass during Embryogenesis
Translesion synthesis (TLS) polymerases are specialized DNA polymerases capable of inserting nucleotides opposite DNA lesions that escape removal by dedicated DNA repair pathways. TLS polymerases allow cells to complete DNA replication in the presence of damage, thereby preventing checkpoint activation, genome instability, and cell death. Here, we characterize functional knockouts for polh-1 and polk-1, encoding the Caenorhabditis elegans homologs of the Y-family TLS polymerases η and κ. POLH-1 acts at many different DNA lesions as it protects cells against a wide range of DNA damaging agents, including UV, γ-irradiation, cisplatin, and methyl methane sulphonate (MMS). POLK-1 acts specifically but redundantly with POLH-1 in protection against methylation damage. Importantly, both polymerases play a prominent role early in embryonic development to allow fast replication of damaged genomes. Contrary to observations in mammalian cells, we show that neither POLH-1 nor POLK-1 is required for homologous recombination (HR) repair of DNA double-strand breaks. A genome-wide RNAi screen for genes that protect the C. elegans genome against MMS–induced DNA damage identified novel components in DNA damage bypass in the early embryo. Our data suggest SUMO-mediated regulation of both POLH-1 and POLK-1, and point towards a previously unrecognized role of the nuclear pore in regulating TLS.
Published in the journal: . PLoS Genet 8(6): e32767. doi:10.1371/journal.pgen.1002800
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002800Summary
Translesion synthesis (TLS) polymerases are specialized DNA polymerases capable of inserting nucleotides opposite DNA lesions that escape removal by dedicated DNA repair pathways. TLS polymerases allow cells to complete DNA replication in the presence of damage, thereby preventing checkpoint activation, genome instability, and cell death. Here, we characterize functional knockouts for polh-1 and polk-1, encoding the Caenorhabditis elegans homologs of the Y-family TLS polymerases η and κ. POLH-1 acts at many different DNA lesions as it protects cells against a wide range of DNA damaging agents, including UV, γ-irradiation, cisplatin, and methyl methane sulphonate (MMS). POLK-1 acts specifically but redundantly with POLH-1 in protection against methylation damage. Importantly, both polymerases play a prominent role early in embryonic development to allow fast replication of damaged genomes. Contrary to observations in mammalian cells, we show that neither POLH-1 nor POLK-1 is required for homologous recombination (HR) repair of DNA double-strand breaks. A genome-wide RNAi screen for genes that protect the C. elegans genome against MMS–induced DNA damage identified novel components in DNA damage bypass in the early embryo. Our data suggest SUMO-mediated regulation of both POLH-1 and POLK-1, and point towards a previously unrecognized role of the nuclear pore in regulating TLS.
Introduction
DNA damaging agents from both endogenous and exogenous sources can induce replication-blocking DNA lesions that threaten cell cycle progression and, consequently, cell viability. To remove these DNA lesions cells are equipped with various specialized repair mechanisms [1], including nucleotide excision repair (NER) that deals with helix-distorting obstructions [2]. However, during embryogenesis, which entails phases of rapid cell division, only a limited time window is available for repair processes [3]. Consequently, unrepaired DNA damage may delay replication and cell cycle progression. In the nematode Caenorhabditis elegans a delay in replication is detrimental for the developmental program; timing of cell division is fixed and strictly regulated by the homologues of the checkpoint genes CHK1 and ATR [4]. Indeed, replication stress caused by depletion of nucleotide pools causes fatal errors in the correct timing of the first asynchronous divisions [5]. However, early embryos of C. elegans appear to be remarkably resistant to DNA damaging agents, suggesting an efficient way to prevent the induction of replication stress by DNA damage [6].
To be able to deal with replication obstructions, organisms evolved ways that allow bypass of the damaged template, thus ensuring continuity of the replication process [7]. Specialized TLS polymerases are capable of direct bypass of DNA lesions in an error free or error prone fashion, depending on their affinities for the specific lesion site. In eukaryotes, TLS is mediated by the DNA polymerases of the Y-family: Polη, Polκ, Polι and Rev1, and the B-family member Polζ. All members of the Y-family polymerases lack proofreading activity and share a conserved active site, which is different from the high-fidelity polymerases in its open and less sterically constrained structure. It allows for accommodation of a DNA lesion, but is also the basis for reduced fidelity [8]. The functional specificities of TLS polymerases are due to minor differences in the structural features of the active site.
The C. elegans genome encodes several Y-family TLS proteins, including POLH-1 and POLK-1, homologs of mammalian Polη and Polκ, respectively. Purified Polη of yeast and vertebrates is capable of replicating across a wide variety of DNA damages, including UV-induced cyclobutane pyrimidine dimers (CPDs), 7,8-dihydro-8-oxoguanine, O6-methylguanine, thymine glycol, cisplatin-induced intrastrand crosslinks, acetylaminofluorene-adducted guanine and benzo[a]pyrene-N2-guanine [9]. In humans, defective Polη has clinical implications: Polη is the product of the gene mutated in Xeroderma Pigmentosum complementation group “Variant” (XPV), a syndrome that is associated with a high predisposition towards developing skin cancers [10]. In addition to a role in damage bypass some studies have suggested a role for Polη in homologous recombination, as the polymerase responsible for extension of the invading strand in the D-loop recombination intermediate [11], [12]. Recently, it was reported that Polη plays a prominent role in early stages of nematode embryogenesis in C. elegans [6], [13]. Polκ displays structural similarity to Polη but is considered to be the most evolutionarily conserved member of the Y-family showing homology to prokaryotic DinB [8], [9]. Its substrate specificity in vitro is limited, although Polκ is an efficient extender of mispaired primer termini and some guanyl adducted lesion sites [14], [15]. Furthermore, Polκ has been suggested as one of the gap-filling polymerases in NER, explaining a moderate sensitivity of Polκ-deficient mammalian cells to UV [16], [17].
Here, we characterize the involvement of Polη and Polκ in various aspects of genome protection during animal development, using the model organism C. elegans. The advantages of this animal model are its spatial and temporal organization of gametogenesis and its rapid growth properties that allow monitoring DNA repair or lesion bypass during different developmental stages. We found that POLH-1 is involved in protection against a surprisingly wide range of DNA lesions, whereas the substrate specificity of POLK-1 is much more restricted. Both proteins can act redundantly on some lesions, since double mutants were extremely sensitive to the alkylating agent MMS, whereas both single mutants displayed profoundly less sensitivity to this carcinogen. In spite of their error proneness, POLH-1 and POLK-1 appear to be highly important in protection against DNA damage during embryonic development, while their role in later somatic development is limited. Finally, we used genome-wide RNAi to screen for factors that have a similar sensitivity profile leading to the identification of new factors that may play a role in the regulation of TLS.
Results
Isolation of C. elegans mutants for polh-1 and polk-1
To study the function of Y-family TLS polymerases in the DNA damage response at different stages of animal development, we set out to isolate mutants for the C. elegans homologs of the Polη and Polκ genes. Figure 1A illustrates a phylogenetic tree of the Y-family polymerase members from several species including C. elegans. The C. elegans genome encodes Polη, Polκ and Rev-1, but not Polι.
Fig. 1. Y-family polymerases POLH-1 and POLK-1 of C. elegans. 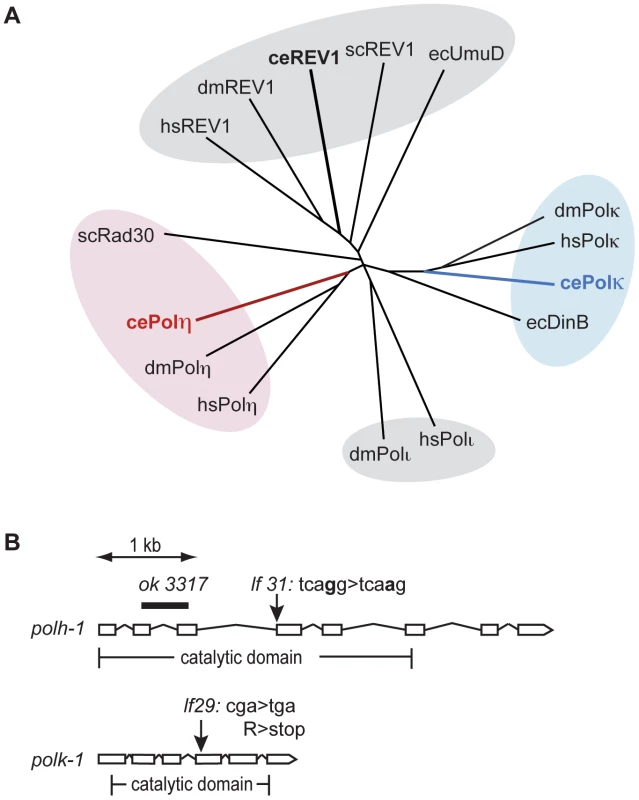
(A) Phylogenetic tree displaying Y-family polymerases from C. elegans, S. cerevisiae, D. melanogaster, E. coli and H. sapiens. Respectively red and blue branches show C. elegans POLH-1 (Polη) and POLK-1 (Polκ). (B) Gene structure of the C. elegans polh-1 and polk-1 genes and the molecular nature of the alleles used in this study. Full alignment of the C. elegans polh-1 and polk-1 gene products with their mammalian and yeast homologs reveals their well-conserved catalytic core (Figure S1). In addition, POLH-1 contains a C-terminal PIP box motif, which is essential for interaction with PCNA, and more recently, has also been shown to target the protein for degradation [13], [18]. The remaining part of the C-terminus is evolutionary less conserved. Human Polη/yeast Rad30 and human Polκ contain ubiquitin binding zinc finger (UBZ) domains, mediating their interaction with PCNA [19]. A UBZ domain was found in C. elegans POLK-1 but not in C. elegans POLH-1 (Figure S1). Furthermore, C. elegans and yeast Polη and Polκ lack previously identified mammalian motifs that mediate an interaction with the deoxycytidyl transferase Rev1 [20], [21].
Using a targeted mutagenesis approach [22] we isolated mutants for polh-1 and polk-1 (Figure 1B). polh-1 (lf31) has a single nucleotide substitution in the splice acceptor site of the fourth exon of the polh-1 gene; polk-1 (lf29) contains a premature stop in the fourth exon and encodes a severely truncated version of POLK-1, missing at least part of the catalytic domain. In the course of this study we obtained another polh-1 mutant from the Gene Knockout Consortium. This allele (ok3317) carries a deletion in polh-1 that results in a fusion of upstream sequences of exon 2 to downstream sequences of exon 3, removing 549 coding nucleotides (Figure 1B). These mutant strains were backcrossed to remove background mutations that resulted from the mutagenic treatment. No obvious abnormal phenotypes were observed for the mutant strains. Neither the number of progeny, embryonic survival rate nor post-embryonic development was affected by the absence of POLH-1 or POLK-1. However, double mutants of polh-1 (ok3317); polk-1 (lf29) and of polh-1 (lf31); polk-1 (lf29) show a minor but significant reduction in both brood size and embryonic survival (up to five percent of the progeny died, data not shown), suggesting some level of functional redundancy in promoting fecundity.
C. elegans POLH-1 in protection against UV and cisplatin
Because UV-induced CPDs are excellent substrates for Polη-mediated TLS in yeast and mammals [23], [24], we tested the sensitivity of polh-1 mutant animals to UV light by irradiating young adults and scoring progeny survival (Figure 2A and Figure S2A). In contrast to Polη-defective yeast and mammalian cells, that display only a mild hypersensitivity to UV [25], [26], POLH-1 deficiency leads to extreme sensitivity to UV irradiation. Both polh-1(lf31) and polh-1(ok3317) mutants are more sensitive to UV than animals carrying mutations in xpa-1, the worm homolog of NER gene XPA, which is essential for repair of UV damage [26], [27]. NER contributes to UV survival also in polh-1 compromised conditions as animals defective in both xpa-1 and polh-1 are more sensitive than either of the single mutants. (Figure S2B). In line with mammalian data, we observed that the protective role of POLH-1 is not restricted to UV-induced damage. polh-1 worms are severely sensitized to cisplatin treatment (Figure 2B and Figure S2C). This sensitivity was even more pronounced than for dog-1 mutant animals which are defective in the homolog of the Fanconi Anemia gene FANCJ, involved in crosslink repair [28].
Fig. 2. Germline sensitivity of polh-1 and polk-1 mutants to different sources of DNA damage. 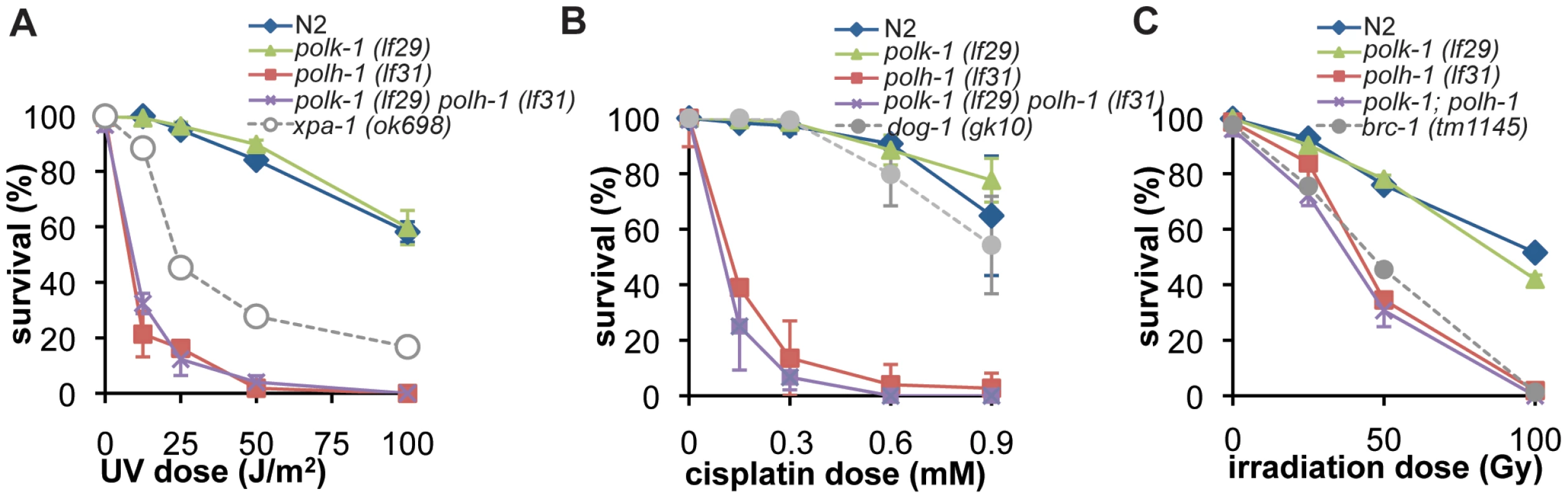
(A) Sensitivity to UV-irradiation. (B) Sensitivity to γ-irradiation. (C) Sensitivity to cisplatin. Adults were exposed to DNA damaging treatments and survival was quantified by counting dead embryos versus living progeny in the next generation. Each line represents the mean of minimal three independent experiments. Error bars denote the s.e.m. POLH-1 and XPA-1 protect against γ-irradiation–induced damage in the C. elegans germline
Vertebrate Polη has been implicated as the polymerase responsible for extension of HR intermediates [12], [29]. Since HR is the predominant repair pathway in C. elegans for γ-irradiation-induced breaks in germ cells [30], [31], we exposed L4 animals to γ-irradiation and scored survival of the progeny (Figure 2C and Figure S2D). We found that the sensitivity of polh-1 (lf31) and polh-1 (ok3317) mutants to irradiation was comparable to the sensitivity of animals that carry a mutation in brc-1, the worm homolog of the HR gene BRCA1 (Figure 2C). Worms defective for both polh-1 and brc-1 were more sensitive to γ-irradiation than either of the single mutants (Figure S2E–S2F), suggesting a brc-1-independent role for POLH-1 in protection against γ-irradiation. This conclusion is strengthened by data showing that POLH-1 and BRC-1 protect cells against radiation at very different developmental stages (see below).
To further test whether the sensitivity of the polh-1 mutants to γ-irradiation is due to a possible defect in HR of DSBs, we determined the role of Polη in response to DSBs endogenously produced by DNA transposition. Transposition is desilenced in the germline of rde-3 mutants [32] and the ensuing DSBs predominantly rely on HR for their repair [33]. However, embryonic lethality was not increased in polh-1; rde-3 double mutants, in contrast to increased lethality in brc-1; rde-3 doubles (Table S2). As an independent and a direct method to address a possible in vivo role of C. elegans Polη in HR, we measured repair of a site-specific DSB using a somatic HR reporter assay (Figure 3). In this assay, which will be described in more detail elsewhere, heat shock-induced expression of the yeast endonuclease I-SceI leads to a DSB in the coding sequence of a GFP transgene that is driven by the intestinal elt-2 promoter. This transgenic setup monitors intrastrand HR, specifically in E-lineage cells, which are still proficient to enter S-phase post embryonically (in contrast to many other post embryonic cells that arrest in G1 and rely on non-homologous end-joining to repair DSBs). A functional GFP transgene is generated following DSB induction only when repair uses a downstream GFP fragment as donor sequence (Figure 3A). This outcome will manifest as GFP expressing intestinal cells. While brc-1(tm1145) mutation resulted in a profound reduction in the number of cells that expressed GFP, polh-1(ok3317) mutant animals displayed similar numbers of cells expressing GFP with similar intensities as compared to wild type worms (Figure 3B and 3C). These data further support the notion that the observed sensitivity of polh-1 mutants to γ-irradiation is not caused by a defect in HR.
Fig. 3. Reporter system for homologous recombination in C. elegans. 
(A) Schematic representation of the reporter transgenes. Expression of the yeast endonuclease ISceI fused to Cherry is controlled by the heat shock promotor (pHS). The reporter transgene is placed behind the intestinal elt-2 promotor (pElt-2). Upon activation of ISceI following heat shock, the ISceI endonuclease cuts into the GFP coding sequence. Repair by gene conversion from an aborted copy of GFP results again in full length GFP. (B) Expression of GFP and Cherry in worms containing the reporter transgenes. Upon heat shock induction all intestinal cells express cherry::ISceI. Repair of the break site by HR from an aborted GFP template results in GFP expression in some intestinal cells. (C) Quantification of the fraction of GFP positive worms in different genetic backgrounds. Each bar represents the mean of three independent experiments. Error bars denote the s.e.m. (D) Germline sensitivity of polh-1 (lf31), xpa-1(ok698) and double mutants to γ-irradiation. Each line represents the mean of minimal three independent experiments. The percentage of surviving progeny is relative to the fraction of surviving progeny without any irradiation, since polh-1(lf31) and xpa-1(ok698) show about 30% synthetic lethality. Error bars denote s.e.m. We thus explored an alternative explanation, in which the increased cytotoxicity of polh-1 mutant animals towards γ-irradiation is the result of failed bypass of other (non-DSB) DNA lesions. Apart from DSBs, γ-irradiation induces single strand breaks (SSBs) as well as 8-Oxo-dG sites and thymine glycols [34]. We reasoned that base adducts in the DNA caused by γ-irradiation may resemble helix-distorting lesions that are substrates for NER and TLS. To address this hypothesis, we tested xpa-1 animals as well as animals defective for both xpa-1 and polh-1 for sensitivity to γ-irradiation (Figure 3D). Strikingly, a redundant effect of both factors was observed after exposure to γ-irradiation similar to the effect seen after UV-irradiation (Figure S2B). These results suggest that γ-irradiation of the germline causes replication-blocking lesions that are substrates for NER and can be bypassed by Polη. It also implies that genes previously found to be involved in γ-irradiation protection are not necessarily involved in the repair of DNA breaks [35].
Damage bypass by POLH-1 during early embryogenesis
C. elegans polh-1 mutants are far more sensitive to various DNA damaging agents as compared to vertebrate cells. We hypothesized that the dependence on POLH-1 for damage tolerance might be specific for early embryonic development, when TLS by POLH-1 is the predominant mechanism to avoid checkpoint activation by replication fork blocks on damaged DNA [6]. In differentiated cells, NER or other repair pathways may dominate the damage response. We therefore tested at which stage during development of C. elegans either POLH-1 mediated damage bypass or NER dominate the response to UV-irradiation. First, we exposed synchronized larvae of the L1 stage to UV light and quantified survival and growth (Figure 4A and Figure S3A). L1 larvae already contain 558 of the total 959 somatic cells that make up the adult animal, and thus mainly grow by cellular volume expansion as opposed to mitotic proliferation [36]. Although xpa-1 mutants completely arrest in L1 after a low dose of UV (Figure 4A, [27]), in polh-1 mutants L1 development is only slightly delayed (Figure S3A). Ultimately polh-1 mutants displayed similar survival as found for wildtype L1s following UV exposure (Figure 4A), indicating that in contrast to XPA, POLH-1 plays hardly any role in the UV damage response in L1. Second, we found that germ cell maturation in polh-1 mutants was comparable to wildtype following UV exposure (Figure 4B–4E), in contrast to xpa-1 mutants that (i) display an UV-induced expansion of the pachytene region and (ii) fail to generate normal-sized oocytes (Figure 4D), [27]. In addition we determined the apoptotic response in the germline after UV irradiation using a ced-1::GFP transgene that marks germ cells in the process of apoptosis [37]. In contrast to xpa-1 deficient animals [38], we found no reduction in the UV-dependent apoptotic response in polh-1 mutants as compared to wildtype animals (Figure S4). Together, these data indicate that NER is essential for normal gametogenesis and L1 development following UV exposure. Apparently, in polh-1 mutants there is sufficient time for repair of UV lesions in these developmental stages to prevent replication stress.
Fig. 4. C. elegans polh-1 and xpa-1 adults and embryos exposed to UV at different stages during development. 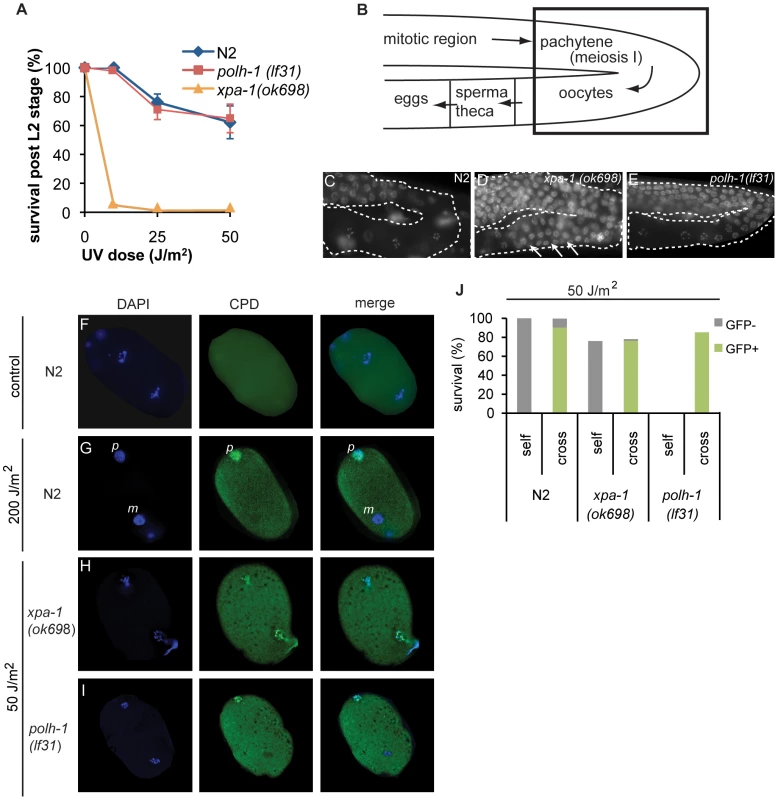
(A) Survival of larvae irradiated at L1 stage. Each line is the mean of three independent experiments; error bars denote s.e.m. (B) Schematic overview of the C. elegans germline. Boxed area shows the transition in the germline bend from the pachytene to maturating oocytes displayed in pictures C–D. (C–E) DAPI stainings of germlines of indicated genotype 16 hrs after exposure to 120 J/m2 UV. Morphology of the germline is completely disrupted in xpa-1 mutants (D) but not in wildtype (C) or polh-1 worms (E). Oocyte maturation in irradiated polh-1 mutants is normal (E), while most meiotic cells fail to progress into oocytes in xpa-1 worms after UV-irradiation (D), causing expansion of the pachytene region through the germline bend (arrows). (F–I) Presence of CPDs during the first embryonic divisions. Immunofluorescence on just fertilized N2 embryos 24 hrs after treatment with 200 J/m2 shows that only one of the two pronuclei carries CPDs (F and G). In UV irradiated xpa-1 embryos both pronuclei carry CPDs (H) while in polh-1 embryos, similar to wildtype (G), only one pronucleus contains CPDs (I). N.B. a lower UV dose was used in H and I to compare doses that induced similar levels of lethality. (J) UV-irradiated hermaphrodites were crossed with untreated males carrying a Pmyo-2::GFP transgene. UV-induced lethality is partly rescued in the cross progeny of polh-1, but not xpa-1 hermaphrodites. However, limited time for DNA repair is available immediately upon fertilization, when a C. elegans embryo goes through a 3-hour period of rapid divisions, according to a fixed and time-constrained lineage program [31]. Thus, in this developmental stage incomplete removal of DNA damage could account for the severe embryonic lethality of UV-exposed polh-1 mutants. To test this hypothesis, we studied the persistence of CPDs - the most abundant lesion type caused by UV – in pronuclei of oocytes, just after fertilization. We irradiated adults with 200 J/m2 and after 24 hours stained developing embryos for CPDs. Remarkably, in wildtype embryos CPDs were still present in the paternal pronucleus, while no CPDs were observed in the maternal pronucleus (Figure 4F–4G). We next assayed xpa-1 and polh-1 mutants after a dose of 50 J/m2 (leading to comparable levels of embryonic lethality). Mutants defective in xpa-1 displayed CPD staining in both pronuclei (Figure 4H), suggesting that in wildtype animals NER-dependent removal of CPDs has occurred during meiotic maturation of the germ cells. In contrast to xpa-1 mutants, but similar to wildtype animals, polh-1 mutants were proficient in removal of CPDs from the maternal pronucleus, whereas CPDs were clearly detectable in the paternal pronucleus (Figure 4I). Before migration and fusion with the maternal pronucleus, the paternal genome decondenses and is replicated in less than 12 minutes [39]. This time span is insufficient for NER to remove DNA damage. We hypothesize that the presence of unrepaired damage from the paternal DNA poses a problem on the first mitotic divisions in polh-1 early embryos. To address this hypothesis, we mated UV-irradiated wildtype or mutant hermaphrodites with untreated males, providing a source of undamaged sperm DNA (Figure 4J). To mark the progeny we used a transgenic line expressing Pmyo-2::GFP. Indeed, lethality in the progeny of irradiated polh-1 hermaphrodites is almost fully rescued by providing a source of undamaged sperm DNA. In contrast, mating of xpa-1 hermaphrodites with untreated males does not affect survival of the progeny. Together, these data indicate that correct progression of early embryonic cell divisions strongly relies on POLH-1 when the genome contains DNA damage. This dependency is not restricted to UV-induced damage but also extends to DNA damage induced by γ-irradiation. The increased sensitivity of polh-1 mutants to γ-irradiation can also be completely rescued by crossing irradiated hermaphrodites with untreated males, thus providing a non-damaged paternal genome (Figure S5). Importantly, this is in stark contrast to the sensitivity of brc-1 mutants, which cannot be rescued by providing non-damaged sperm. This developmental separation of the modes of action of these proteins further substantiates our findings that polh-1 and brc-1 act independently in protecting cells against γ-irradiation-induced DNA damage.
POLH-1 and POLK-1 act in a redundant fashion in protection against the methylating agent MMS
We next wondered whether a similar developmentally restrained function could be attributed to TLS polymerase POLK-1. To address this question, we exposed polk-1(lf29) mutant worms to different doses of UV, cisplatin or γ-irradiation (Figure 2A–2C), but found no difference in sensitivity as compared to wildtype animals, indicating that POLK-1 is not involved in protection against these sources of DNA damage in C. elegans. However, akin to the outcome of published RNAi experiments [6], both polk-1 and polh-1 mutants are sensitive to chronic exposure to the alkylating agent MMS, albeit that the sensitivity in polh-1 mutants was much more pronounced (Figure 5A, Figure S6), indicating that both POLH-1 and POLK-1 play a role in bypass of MMS-induced DNA damage. We next assayed polh-1 (lf31); polk-1 (lf29) double mutants and polh-1 (ok3317) polk-1(RNAi) animals for MMS sensitivity (Figure 5A, Figure S6A). Interestingly, double mutants were extremely sensitive to MMS, and complete lethality was observed at a dose that was 100 times lower than the effective dose for any of the single mutants (Figure 5A, Figure S6A). We did not observe any synergistic effect for any of the other types of lesions we tested (Figure 2).
Fig. 5. polh-1 and polk-1 protect in a redundant fashion against the methylating agent MMS. 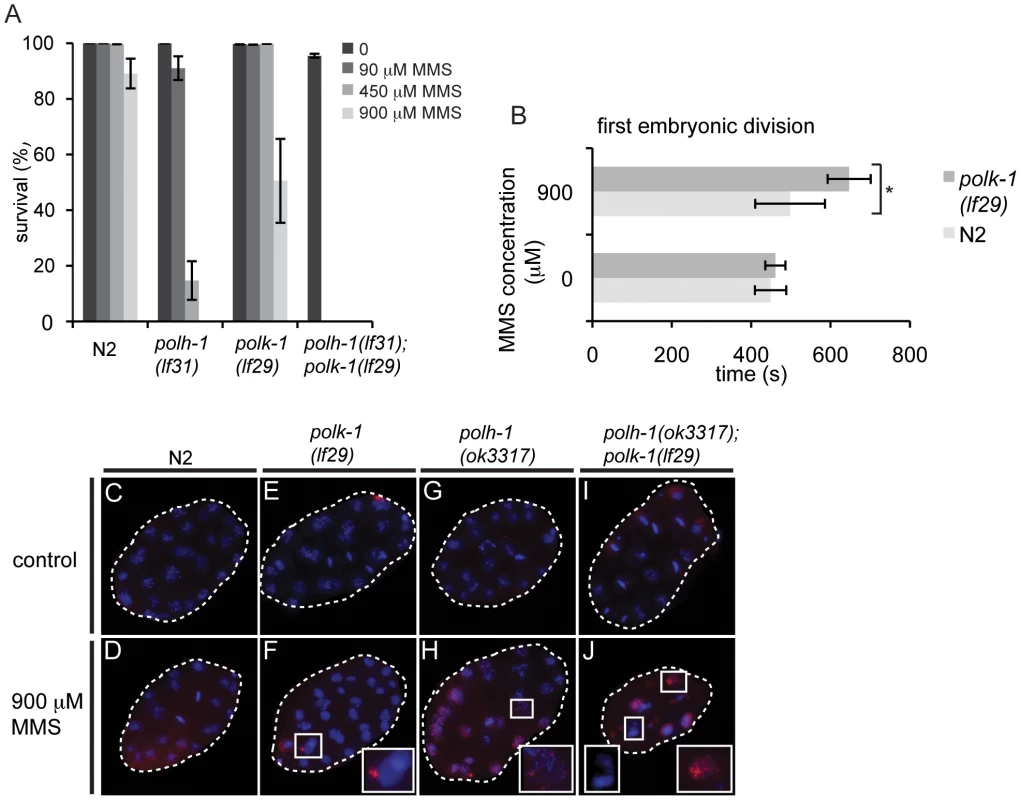
(A) Double mutants of polh-1 and polk-1 are severely sensitized to MMS exposure. Results of a representative experiment are shown. Error bars denote SD. (B) Delayed progression through the first embryonic division after MMS exposure in polk-1 mutants. The interval between passing of the paternal pronucleus over the midline till the start of cytokinesis is timed for at least 5 embryos per datapoint. Statistical significance for the difference in delay between N2 and polk-1 embryos treated with MMS was calculated with a student's t-test (p = 0.012). (C–J) RAD51 immunostainings of early embryos treated with MMS. Morphology of polh-1; polk-1 double mutant embryos is abnormal after MMS exposure, displaying chromatin bridges and abundant RAD51 staining (J). polh-1(ok3317) and polk-1(lf29) single mutants show incidental RAD51 foci in embryos (F and H), while such foci were never observed in untreated controls (C, E, G and I). POLH-1 has previously been shown to be involved in avoiding DNA damage-induced checkpoint activation [6]. In C. elegans embryogenesis, checkpoints - mediated by the C. elegans homologs of the checkpoint genes ATR and CHK-1 - are used to time the first asynchronous cell divisions that are essential for embryonic patterning and thus embryonic viability [5]. Checkpoint activation due to DNA damage interferes with the developmental role of the checkpoint, causing patterning defects and embryonic lethality. Our results with null mutants for polh-1 and polk-1 suggest that both POLH-1 and POLK-1 can act to avoid checkpoint activation. To test the involvement of POLK-1 in checkpoint avoidance directly, we timed the first embryonic division of polk-1 embryos after exposure to MMS. Figure 5B illustrates a delayed first embryonic division in polk-1 mutants when compared to wildtype embryos. Importantly, we also observed examples of polk-1 embryos that after MMS treatment fully arrested at the 1-cell stage (Videos S1 and S2), while we never observed such cases for MMS-treated wildtype embryos. Two other phenotypes are also indicative of replication stress during early embryonic divisions of MMS treated polh-1 and polk-1 mutant animals. First, polh-1; polk-1 double mutant embryos displayed foci of the DSB repair marker RAD51 (Figure 5C–5J), indicative of DSBs resulting from trying to replicate damaged genomes [40]. Second, DAPI staining revealed chromatin bridges and a disrupted nuclear morphology in the early embryo (Figure 5J), suggesting division of disentangled or incompletely replicated genomes. These phenotypes were less profound, but noticeable, in both single mutants, while never observed in wild type embryos exposed to similar MMS concentrations (Figure 5C–5H).
To investigate whether the dependency on POLH-1 and POLK-1 for tolerance to MMS was restricted to embryogenesis - similar to the requirement of POLH-1 in UV tolerance - we followed the outgrowth of L1 animals exposed to increasing concentrations of MMS (Figure S6B). The development of polh-1 larvae was mildly affected, while no delay was observed for polk-1 animals. As for UV, NER deficient xpa-1 larvae were profoundly more sensitive to MMS than either polk-1 or polh-1 deficient larvae (Figure S6B), while the opposite is true for embryonic stages: xpa-1 embryos are less sensitive to MMS than polh-1 embryos [6]. This again argues that TLS is more important than DNA repair at developmental stages that are characterized by fast replication cycles.
A genetic approach for identifying new factors in TLS regulation in the early embryo
Since POLH-1 and POLK-1 together appear to be extremely important in protecting the developing embryo against MMS, we wondered whether there might be a general pathway underlying the regulation of the two TLS enzymes. To identify new factors regulating TLS in the early embryo, we performed a genome-wide RNAi screen for genes sensitizing embryos to MMS. Out of 16,757 genes tested (covering ∼86% of all predicted C. elegans genes), we found 87 genes that resulted in sensitivity to MMS upon knockdown, including polk-1. polh-1 was not identified in this screen, probably due to insufficient knockdown by the RNAi clone targeting this gene. We next inspected these RNAi knockdowns for phenotypes reminiscent of polh-1;polk-1 double mutants. All 87 hits were analysed by DAPI for altered nuclear morphology after exposure to MMS (Figure 5A and Figure S7). Four clones were selected for follow-up analysis based on perturbed embryonic divisions as indicated by chromatin bridges and malformed nuclei. These clones targeted the genes gei-17, ulp-1, npp-2 and npp-22 (Figure 6). gei-17 encodes a SUMO-protease that was previously shown to interact with POLH-1 after DNA damage [13]. ulp-1 encodes a ubiquitin-like protease (ULP) that deconjugates SUMO moieties from their target proteins [41]. npp-2 and npp-22 encode two components of the C. elegans nuclear pore complex (NPC)[42]. Null alleles of gei-17, npp-2 and npp-22 are embryonic lethal. Knockdowns of the four genes reduced tolerance to MMS to a similar extent as mutations in polh-1 and polk-1 (Figure 6K). In line with published data [6] we found that gei-17 knockdown led to abundant RAD51 foci in embryos treated on MMS, indicative of replication stress. Also knockdown of ulp-1, npp-2 and npp-22 lead to MMS-induced RAD51 foci, although to a lesser extend than gei-17 knockdown. This is consistent with the observation that these knockdowns also display less dramatic effects on progeny survival. Foci formation was never observed in mock-treated knockdowns or wild type controls (Figure 6A–6J).
Fig. 6. New factors in damage tolerance in the early embryo. 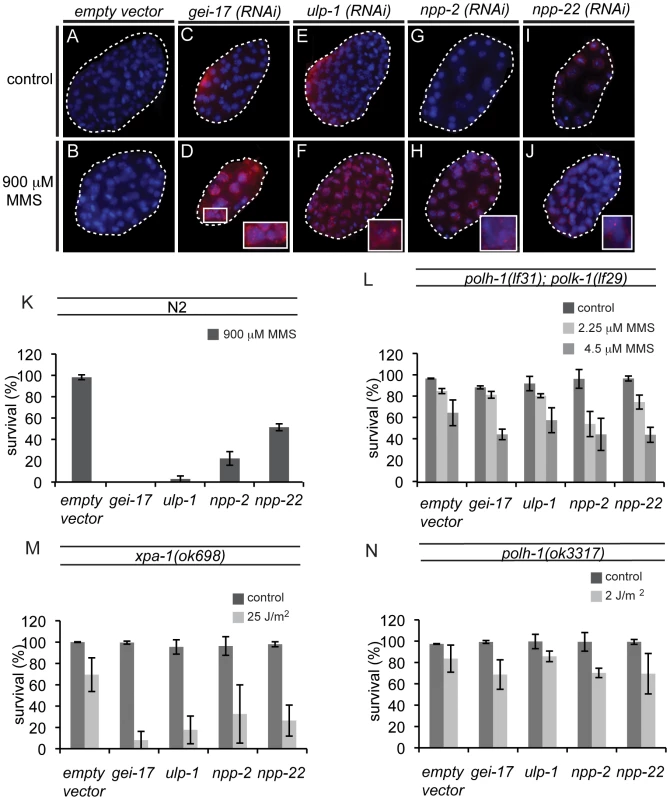
(A–J) RAD51 stainings of early embryos treated with MMS. gei-17 RNAi knockdown embryos show abundant RAD51 staining after treatment with MMS (D). Incidental RAD51 foci are observed in ulp-1, npp-2 and npp-22 knockdown embryos (F, H, J) but not in wildtype controls (B). (K) MMS sensitivity of gei-17, ulp-1, npp-2 and npp-22 knockdowns. (L) Sensitivity to MMS is not further reduced in polh-1; polk-1 mutants by any of the tested RNAi clones. (M) Sensitivity to UV is further reduced by indicated RNAi food against gei-17, ulp-1, npp-2 and npp-22 in a xpa-1 mutant background. (N) Sensitivity to a low dose of UV is not further reduced in a polh-1 mutant background after knockdown by indicated RNAi foods. We hypothesized that if these genes were in a common pathway with POLH-1 and POLK-1, then knockdown of these factors would not further increase sensitivity of polh-1;polk-1 worms to a low dose of MMS. Indeed, MMS sensitivity was not further increased when ulp-1, gei-17, npp-2 and npp-22 were knocked down in polh-1;polk-1 double mutant animals (Figure 6L), placing all four factors in an epistatic relation to the TLS genes in the response to alkylating damage. To substantiate this epistatic relationship we also studied another source of DNA damage infliction, by exposing young adults to UV light. We previously showed that polh-1 is important for embryonic development in the presence of UV damage, and that an additional mutation in the NER factor xpa-1 renders the animals even more sensitive to low doses of UV (Figure 2A and Figure S2B). We argued that if these factors act in a common pathway with Polη in the response to UV, we would expect their knockdowns to be epistatic with a polh-1 mutation, but increase the sensitivity of xpa-1 defective animals. Indeed, knockdowns of gei-17, ulp-1, npp-2 or npp-22 all further increased the sensitivity of xpa-1 mutant animals to a low dose of UV (Figure 6M), but did not change sensitivity of polh-1 mutants (Figure 6N). Together these data indicate that ulp-1, npp-2 and npp-22 are novel factors in TLS mediated by POLH-1 and POLK-1 in response to DNA damage during early embryogenesis in C. elegans. The SUMO protease gene gei-17 has previously been shown to promote damage tolerance by sumoylating POLH-1 [13]. Our results suggest that GEI-17 is implicated in TLS mediated by both POLH-1 and POLK-1.
Discussion
Here we demonstrate that there is modulation of the choice between repair and bypass of damaged template DNA in a developing organism. A priori one would expect error-free repair by NER to be the favoured option in germ cells to prevent the accumulation of mutations in subsequent generations. Indeed, we and others found that both for germ cell maturation and post-embryonic somatic development, NER is indispensable in response to specific DNA damages [26], [27], [38]. However, and in line with previously published data [6], we found that immediately after fertilization of the oocyte, during stages of rapid cell divisions in the early embryo, survival is determined by TLS factors and not by NER. The need for efficient TLS must be viewed in light of the strict timing of the developmental program, which likely does not allow time to “wait” for repair processes to be completed. Our observation that wild type animals can easily survive UV doses up to 50 J/m2 without substantially repairing CPDs from their sperm or decondensed paternal pronucleus indicates that TLS-proficient zygotes can replicate a damaged genome containing 10–50.000 UV lesions in less than 12 minutes - the time it takes for the paternal nucleus to double its genome content - without delaying cell division [39].
We found that C. elegans POLH-1 has a broader substrate specificity than POLK-1; POLH-1 is involved in bypass of damage induced upon exposure to UV light, γ-irradiation, cisplatin and MMS. We considered the possibility that all treatments may lead to a common substrate that causes the observed cytotoxicity, such as DSBs brought about by replication fork obstruction and collapse. This notion has been supported by studies in vertebrates, in which Polη was suggested to act in HR repair of DSBs by extending the D-loop intermediate structure [11], [12]. However, we observed a wild type response to either transposon-mediated or ISceI-induced DSBs, thus arguing against a role for POLH-1 in DSB repair, in either germline or somatic tissue of C. elegans. We ascribe the sensitivity of polh-1 mutants towards γ-irradiation to the induction of other non-DSB lesions, which may be NER substrates. Consistent with this interpretation we observed a synergistic relationship between xpa-1 and polh-1 with respect to IR sensitivity. The induction of free radicals by ionizing radiation causes a plethora of lesions in the DNA, such as 8-oxo-dG sites, which may require Polη-mediated bypass to prevent checkpoint activation [43].
An explanation for the broad substrate specificity of POLH-1 may reside in the flexible active site of POLH-1, which may allow for bypass of lesions that are structurally very different. Indeed, studies in chicken DT40 cells indicate that Polη is a much more versatile polymerase than the phenotype of XPV cells would suggest [44]. Alternatively, Polη could have an indirect role by recruiting other TLS proteins to the damage site. In human cells Rev1 is recruited to UV damages via an interaction with Polη [45]. Interestingly, Polκ has been shown to serve as a ‘backup’ polymerase in XPV cells in the bypass of both UV-induced CPDs as well as cisplatin adducts [46], [47]. We here show that in nematodes this genetic interaction is restricted to damage induced by the Sn1 methylating agent MMS. The molecular effects of MMS include the formation of N-7 methylguanine (which by spontaneous depurination can lead to an abasic site), N3 - methyladenine, N3-cytosine and O6-methylguanine [48]. Although we cannot deduce from our in vivo analysis which of these damages underlies the cytotoxicity observed in nematodes, all of these base damages have residual coding capacity, and are less structurally perturbing than some of the DNA lesions induced by cisplatin or IR treatment. This notion may explain the redundant role of the functionally more restricted POLK-1 on MMS, while no contribution was seen following UV, IR or cisplatin treatment.
In order to find novel factors that are directly or indirectly involved in TLS, we screened for RNAi knockdowns that rendered cells sensitive to MMS and UV, only in the context of TLS functionality. Out of ∼17,000 clones we identified four genes whose knockdown sensitized wildtype but not TLS-deficient animals to MMS treatment. One of these genes, gei-17, was previously reported to regulate Polη; GEI-17 was shown to sumoylate POLH-1 near its PIP-box motif resulting in protection of the protein from degradation [13]. The profound effects of gei-17 RNAi on cellular tolerance to MMS suggest that this SUMO-ligase most likely acts on both POLH-1 and POLK-1 (Figure 5A and 5I); we note that C. elegans POLK-1 may also contain a PIP-box motif (Figure S1). In addition to GEI-17 we also identified the SUMO protease ULP-1 as a factor in TLS-mediated MMS - and UV-sensitivity. This result suggests that, apart from sumoylation, also desumoylation may play a role in the regulation of TLS proteins. Additional studies that identify targets of ULP-1 needs to establish whether its role is direct, by desumoylation of the TLS polymerases, or indirect. Ubiquitin-like proteases (ULPs) deconjugate SUMO from their target proteins and therefore the damage sensitivity of ULP-1 knockdown may also be explained by disturbed regulation because of a shortage of SUMO. SUMO proteases and ligases may anchor to the NPCs in order to sumoylate or desumoylate their targets [49]. Here we show that, similar to gei-17 and ulp-1, RNAi against nuclear pore components npp-2 and npp-22 is compatible with viability but results in sensitivity to UV lesions and MMS. However, sensitivity was not further increased in the absence of POLH-1 and POLK-1. This finding suggests a role for the NPC (or NPC subunits) in TLS mediated damage tolerance, possibly in the localization of SUMO-regulation. npp-2 encodes the C. elegans homolog of yeast Nup85, which is one of the proteins of the scNup84 scaffolding complex. In yeast, mutants in the Nup84 and Nup60/Mlp1-2 complexes have similar phenotypes in the response to DNA damage as Ulp1 mutants [50]. A direct link of NPCs to the DNA damage response in yeast was also suggested by Nagai et al., who showed relocation of damaged DNA to nuclear pores [51] and recently by Bermejo et al. who showed involvement of inner basket proteins in replication fork stability [52]. npp-22 encodes the C. elegans homolog of yeast and mammalian NDC1, which is crucial for nuclear pore assembly [53]. Future work on gei-17, ulp-1 and the nuclear pore components npp-2 and npp-22 is needed to substantiate the role of sumoylation and desumoylation processes and a possible link to the NPC (subunits) in regulating TLS.
Materials and Methods
C. elegans genetics
All strains were cultured according to standard methods [54]. Wildtype N2 (Bristol) worms were used in all control experiments. The polh-1 (lf31) and polk-1 (lf29) mutants were isolated in our own laboratory. polh-1 (ok3317) worms, that were kindly provided by Joel Meyer (Duke University, Durham NC, USA), have been generated by the C. elegans knock-out consortium. BCN2081, carrying a single copy integrated Pmyo::GFP transgene, was a gift from Ben Lehner (EMBL Centre for Genomic Regulation, Barcelona, Spain) [55]. All other alleles (xpa-1 (ok698); rde-3 (ne298); brc-1 (tm1145); dog-1 (gk10)) and the transgenic line MD701 (bcIs39[P(lim-7)ced-1::GFP+lin-15(+)]) were obtained from the C. elegans Genetics Center (St Paul, MN, USA). All mutant strains were backcrossed six times before performing experiments. Newly generated strains are listed in Table S1 in the supplementary information.
Survival assays
Staged animals were exposed to different doses of various DNA damaging agents. To assess germline sensitivity three plates with three worms were allowed to lay eggs for 24–48 hrs per experimental condition. 24 hrs later, the number of unhatched eggs and the number of surviving progeny was determined. All experiments were performed in triplicate. To measure germline sensitivity to UV, staged young adults one day post L4 were transferred to empty NGM plates and exposed to different doses of UV-C (predominantly 254 nm, Philips). Animals were placed on fresh OP50 plates and allowed to lay eggs for 32 hrs.
To determine whether lethality could be rescued by the supply of undamaged sperm, UV irradiated hermaphrodites were mated with untreated BCN2081 worms, which have Pmyo-2::GFP transgenes integrated in their genomes. After 24 hrs of male contact, the hermaphrodites were transferred to individual plates and allowed to lay eggs for 24 hrs. The mother was subsequently removed and 24 hrs later the number of non-hatched eggs and the number of GFP+ and GFP - progeny was determined.
The sensitivity of L1 larval stage animals to UV was measured as described previously [27]. L1s were synchronized by bleaching, and exposed to UV-C on empty NGM plates. Per plate, at least 100 L1 animals were counted. For three subsequent days the development of L1-treated animals was monitored.
To measure germline sensitivity to γ-irradiation, different doses were delivered by an X-ray generator (dose rate 7 Gy/min; YXLON International) to L4 animals. Animals were allowed to lay eggs for 48 hrs, and scored 24 hrs later for hatching.
Sensitivity to cisplatin was determined by incubating staged L4 animals for 3 hrs in M9 containing different concentrations of cisplatin (Sigma-Aldrich). After 1 hr recovery on OP50 plates, animals were placed on fresh OP50 plates and allowed to lay eggs for 48 hrs. The mother was removed and the survival of the progeny was scored 24 hrs later.
To measure sensitivity to chronic exposure to MMS, staged L4 animals were placed for 24 hrs on NGM plates containing different concentrations of MMS (Sigma-Aldrich). After 24 hrs, the number of non-hatched eggs and surviving progeny was determined.
Homologous recombination (HR) assay
A HR reporter plasmid was constructed consisting of a GFP/LacZ fusion under the control of the intestinal specific elt-2 promotor [56]. An ISceI recognition sequence was inserted that disrupted the GFP ORF. To provide a template for homologous recombination, part of the GFP coding region was PCR amplified and inserted downstream of the disrupted GFP/LacZ locus. The ISceI expressing plasmid pRP3001 (hsp-16.41::ISceI ORF) [57], was modified to include the mCherry ORF leading to a functional ISceI/mCherry protein to visualize and monitor the expression of the ISceI endonuclease. A detailed description of the reporter system and its validation will be published elsewhere.
For reading out HR, synchronized L4 animals were transferred and incubated for 1.5 hrs at 34°C. After 24 hrs, GFP expression in the intestine was analyzed on a Leica DM6000 microscope.
Microscopy
Nuclear stainings on germlines and embryos were performed by incubation of staged young adults for 10 minutes in ethanol containing 10 µg/mL 4′,6-diamidino-2-phenylindole (DAPI). After two washes with PBS, worms were mounted on object slides in 30% glycerol.
To detect CPDs, eggs were liberated from UV-irradiated worms and fixed with 3% paraformaldehyde. Fixed eggs were permeabilized by freeze cracking and subsequently washed with 1% Triton and methanol (−20°C). CPDs were visualized by subsequent staining with an anti-CPD mouse monoclonal antibody and an Alexa488-labelled goat-anti-mouse secondary antibody (Molecular Probes Inc) combined with 10 µg/mL DAPI. Dissected worms and eggs were mounted on object slides in Vectashield.
To study RAD51 foci formation, a similar procedure as described for CPD staining was followed. Fixed eggs were permeabilized by freeze cracking and subsequently washed with 1% Triton and methanol (−20°C). RAD51 was visualized by subsequent staining with an anti-RAD51 rabbit monoclonal antibody and an Alexa488-labelled goat-anti-rabbit secondary antibody (Molecular Probes Inc) combined with 10 µg/mL DAPI. Dissected worms and eggs were mounted on object slides in Vectashield.
For the analysis of apoptosis, transgenic MD701 animals, expressing a ced1::GFP fusion behind a lim-7 promotor, were used to visualize sheath cells surrounding apoptotic germ cells [37]. All microscopy was performed with a Leica DM6000 microscope.
RNAi screen and RNAi experiments
Using the Ahringer Lab RNAi feeding library a genome-wide screen was performed for clones sensitizing animals to MMS. The procedure is an adaptation from a genome-wide RNAi screen for radiation sensitivity by Van Haaften et al, described in detail in their supplementary data [35]. Briefly, L1 worms were grown to L4s in liquid on RNAi food. At the L4 stage MMS was added to a concentration of 0.01%. After three days survival of the progeny was scored by visual inspection. For knockdown of polk-1, gei-17, ulp-1, npp-2 and npp-22 genes, individual Ahringer clones were grown on IPTG containing NGM plates. Staged L4s were transferred to RNAi plates; analysis was performed on the progeny of these animals.
Supporting Information
Zdroje
1. CicciaAElledgeSJ 2010 The DNA damage response: making it safe to play with knives. Mol Cell 40 179 204 doi:10.1016/j.molcel.2010.09.019
2. NouspikelT 2009 DNA repair in mammalian cells : Nucleotide excision repair: variations on versatility. Cell Mol Life Sci 66 994 1009 doi:10.1007/s00018-009-8737-y
3. O'FarrellPHStumpffJSuTT 2004 Embryonic cleavage cycles: how is a mouse like a fly? Curr Biol 14 R35 R45
4. EncaladaSEMartinPRPhillipsJBLyczakRHamillDR 2000 DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Developmental Biology 228 225 238 doi:10.1006/dbio.2000.9965
5. BrauchleMBaumerKGönczyP 2003 Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr Biol 13 819 827
6. HolwayAHKimS-HLa VolpeAMichaelWM 2006 Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. The Journal of Cell Biology 172 999 1008 doi:10.1083/jcb.200512136
7. AndersenPLXuFXiaoW 2008 Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res 18 162 173 doi:10.1038/cr.2007.114
8. PrakashSJohnsonREPrakashL 2005 Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74 317 353 doi:10.1146/annurev.biochem.74.082803.133250
9. WatersLSMinesingerBKWiltroutMED'SouzaSWoodruffRV 2009 Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73 134 154 doi:10.1128/MMBR.00034-08
10. JohnsonREKondratickCMPrakashSPrakashL 1999 hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285 263 265
11. KawamotoTArakiKSonodaEYamashitaYMHaradaK 2005 Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell 20 793 799 doi:10.1016/j.molcel.2005.10.016
12. McIlwraithMJMcllwraithMJVaismanALiuYFanningE 2005 Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell 20 783 792 doi:10.1016/j.molcel.2005.10.001
13. KimS-HMichaelWM 2008 Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol Cell 32 757 766 doi:10.1016/j.molcel.2008.11.016
14. HaracskaLPrakashLPrakashS 2002 Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc Natl Acad Sci USA 99 16000 16005 doi:10.1073/pnas.252524999
15. CarlsonKDJohnsonREPrakashLPrakashSWashingtonMT 2006 Human DNA polymerase kappa forms nonproductive complexes with matched primer termini but not with mismatched primer termini. Proc Natl Acad Sci USA 103 15776 15781 doi:10.1073/pnas.0605785103
16. OgiTLehmannAR 2006 The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol 8 640 642 doi:10.1038/ncb1417
17. OgiTLimsirichaikulSOvermeerRMVolkerMTakenakaK 2010 Three DNA Polymerases, Recruited by Different Mechanisms, Carry Out NER Repair Synthesis in Human Cells. Mol Cell 37 714 727 doi:10.1016/j.molcel.2010.02.009
18. HaracskaLKondratickCMUnkIPrakashSPrakashL 2001 Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol Cell 8 407 415
19. BienkoMGreenCMCrosettoNRudolfFZapartG 2005 Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310 1821 1824 doi:10.1126/science.1120615
20. KosarekJNWoodruffRVRivera-BegemanAGuoCD'SouzaS 2008 Comparative analysis of in vivo interactions between Rev1 protein and other Y-family DNA polymerases in animals and yeasts. DNA Repair 7 439 451 doi:10.1016/j.dnarep.2007.11.016
21. OhashiEHanafusaTKameiKSongITomidaJ 2009 Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells 14 101 111 doi:10.1111/j.1365-2443.2008.01255.x
22. CuppenEGortEHazendonkEMuddeJvan de BeltJ 2007 Efficient target-selected mutagenesis in Caenorhabditis elegans: toward a knockout for every gene. Genome Res 17 649 658 doi:10.1101/gr.6080607
23. JohnsonREPrakashSPrakashL 1999 Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science 283 1001 1004
24. MasutaniCArakiMYamadaAKusumotoRNogimoriT 1999 Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J 18 3491 3501 doi:10.1093/emboj/18.12.3491
25. McDonaldJPLevineASWoodgateR 1997 The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147 1557 1568
26. AstinJWO'NeilNJKuwabaraPE 2008 Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues, RAD-3 and WWP-1. DNA Repair 7 267 280 doi:10.1016/j.dnarep.2007.10.004
27. LansHMarteijnJASchumacherBHoeijmakersJHJJansenG 2010 Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet 6 e1000941 doi:10.1371/journal.pgen.1000941
28. YoudsJLBarberLJWardJDCollisSJO'NeilNJ 2008 DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol 28 1470 1479 doi:10.1128/MCB.01641-07
29. RattrayAJStrathernJN 2005 Homologous recombination is promoted by translesion polymerase poleta. Mol Cell 20 658 659 doi:10.1016/j.molcel.2005.11.018
30. BoultonSJMartinJSPolanowskaJHillDEGartnerA 2004 BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol 14 33 39
31. ClejanIBoerckelJAhmedS 2006 Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics 173 1301 1317 doi:10.1534/genetics.106.058628
32. ChenC-CGSimardMJTabaraHBrownellDRMcColloughJA 2005 A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol 15 378 383 doi:10.1016/j.cub.2005.01.009
33. PlasterkRH 1991 The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J 10 1919 1925
34. RoosWPKainaB 2012 DNA damage-induced apoptosis: From specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett doi:10.1016/j.canlet.2012.01.007
35. van HaaftenGRomeijnRPothofJKooleWMullendersLHF 2006 Identification of conserved pathways of DNA-damage response and radiation protection by genome-wide RNAi. Curr Biol 16 1344 1350 doi:10.1016/j.cub.2006.05.047
36. AltunZFHallDH 2009 Introduction to C. elegans anatomy. WormAtlas
37. SchumacherBHanazawaMLeeM-HNayakSVolkmannK 2005 Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120 357 368 doi:10.1016/j.cell.2004.12.009
38. StergiouLDoukoumetzidisKSendoelAHengartnerMO 2007 The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ 14 1129 1138 doi:10.1038/sj.cdd.4402115
39. EdgarLGMcGheeJD 1988 DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53 589 599
40. HolwayAHHungCMichaelWM 2005 Systematic, RNA-interference-mediated identification of mus-101 modifier genes in Caenorhabditis elegans. Genetics 169 1451 1460 doi:10.1534/genetics.104.036137
41. ZhangHSmolenGAPalmerRChristoforouAvan den HeuvelS 2004 SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat Genet 36 507 511 doi:10.1038/ng1336
42. GalyVMattajIWAskjaerP 2003 Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell 14 5104 5115 doi:10.1091/mbc.E03-04-0237
43. LeeD-HPfeiferGP 2008 Translesion synthesis of 7,8-dihydro-8-oxo-2′-deoxyguanosine by DNA polymerase eta in vivo. Mutat Res 641 19 26 doi:10.1016/j.mrfmmm.2008.02.006
44. HirotaKSonodaEKawamotoTMotegiAMasutaniC 2010 Simultaneous disruption of two DNA polymerases, Polη and Polζ, in Avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet 6 doi:10.1371/journal.pgen.1001151
45. AkagiJ-IMasutaniCKataokaYKanTOhashiE 2009 Interaction with DNA polymerase eta is required for nuclear accumulation of REV1 and suppression of spontaneous mutations in human cells. DNA Repair 8 585 599 doi:10.1016/j.dnarep.2008.12.006
46. ShacharSZivOAvkinSAdarSWittschiebenJ 2009 Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 28 383 393 doi:10.1038/emboj.2008.281
47. ZivOGeacintovNNakajimaSYasuiALivnehZ 2009 DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci USA 106 11552 11557 doi:10.1073/pnas.0812548106
48. FuDCalvoJASamsonLD 2012 Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 12 104 120 doi:10.1038/nrc3185
49. PalancadeBDoyeV 2008 Sumoylating and desumoylating enzymes at nuclear pores: underpinning their unexpected duties? Trends Cell Biol 18 174 183 doi:10.1016/j.tcb.2008.02.001
50. PalancadeBLiuXGarcia-RubioMAguileraAZhaoX 2007 Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol Biol Cell 18 2912 2923 doi:10.1091/mbc.E07-02-0123
51. NagaiSDubranaKTsai-PflugfelderMDavidsonMBRobertsTM 2008 Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322 597 602 doi:10.1126/science.1162790
52. BermejoRCapraTJossenRColosioAFrattiniC 2011 The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146 233 246 doi:10.1016/j.cell.2011.06.033
53. StavruFHülsmannBBSpangAHartmannECordesVC 2006 NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. The Journal of Cell Biology 173 509 519 doi:10.1083/jcb.200601001
54. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
55. SempleJIGarcia-VerdugoRLehnerB 2010 Rapid selection of transgenic C. elegans using antibiotic resistance. Nat Methods 7 725 727 doi:10.1038/nmeth.1495
56. FukushigeTHendzelMJBazett-JonesDPMcGheeJD 1999 Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc Natl Acad Sci USA 96 11883 11888
57. PontierDBTijstermanM 2009 A robust network of double-strand break repair pathways governs genome integrity during C. elegans development. Curr Biol 19 1384 1388 doi:10.1016/j.cub.2009.06.045
Štítky
Genetika Reprodukční medicína
Článek Uracil-Containing DNA in : Stability, Stage-Specific Accumulation, and Developmental InvolvementČlánek Fuzzy Tandem Repeats Containing p53 Response Elements May Define Species-Specific p53 Target GenesČlánek Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early EmbryosČlánek Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2α Kinase GCN-2Článek Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human CellsČlánek TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome EndsČlánek Attenuation of Notch and Hedgehog Signaling Is Required for Fate Specification in the Spinal CordČlánek Genome-Wide Functional Profiling Identifies Genes and Processes Important for Zinc-Limited Growth ofČlánek MicroRNA93 Regulates Proliferation and Differentiation of Normal and Malignant Breast Stem Cells
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 6- Intrauterinní inseminace a její úspěšnost
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
- Růst a vývoj dětí narozených pomocí IVF
- Akutní intermitentní porfyrie
- Příjem alkoholu a menstruační cyklus
-
Všechny články tohoto čísla
- Rumors of Its Disassembly Have Been Greatly Exaggerated: The Secret Life of the Synaptonemal Complex at the Centromeres
- Mimetic Butterflies Introgress to Impress
- Tipping the Balance in the Powerhouse of the Cell to “Protect” Colorectal Cancer
- Selection-Driven Gene Loss in Bacteria
- Decreased Mitochondrial DNA Mutagenesis in Human Colorectal Cancer
- Parallel Evolution of Auditory Genes for Echolocation in Bats and Toothed Whales
- Diverse CRISPRs Evolving in Human Microbiomes
- The Rad4 ATR-Activation Domain Functions in G1/S Phase in a Chromatin-Dependent Manner
- Stretching the Rules: Monocentric Chromosomes with Multiple Centromere Domains
- Uracil-Containing DNA in : Stability, Stage-Specific Accumulation, and Developmental Involvement
- Fuzzy Tandem Repeats Containing p53 Response Elements May Define Species-Specific p53 Target Genes
- Adaptive Introgression across Species Boundaries in Butterflies
- G Protein Activation without a GEF in the Plant Kingdom
- Synaptonemal Complex Components Persist at Centromeres and Are Required for Homologous Centromere Pairing in Mouse Spermatocytes
- An Engineering Approach to Extending Lifespan in
- Incompatibility and Competitive Exclusion of Genomic Segments between Sibling Species
- Effects of Histone H3 Depletion on Nucleosome Occupancy and Position in
- Patterns of Evolutionary Conservation of Essential Genes Correlate with Their Compensability
- Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis
- Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early Embryos
- A Mouse Model of Acrodermatitis Enteropathica: Loss of Intestine Zinc Transporter ZIP4 (Slc39a4) Disrupts the Stem Cell Niche and Intestine Integrity
- Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2α Kinase GCN-2
- Geographic Differences in Genetic Susceptibility to IgA Nephropathy: GWAS Replication Study and Geospatial Risk Analysis
- Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human Cells
- TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome Ends
- Stimulation of Host Immune Defenses by a Small Molecule Protects from Bacterial Infection
- A Broad Requirement for TLS Polymerases η and κ, and Interacting Sumoylation and Nuclear Pore Proteins, in Lesion Bypass during Embryogenesis
- Genome-Wide Identification of Ampicillin Resistance Determinants in
- The CCR4-NOT Complex Is Implicated in the Viability of Aneuploid Yeasts
- Gustatory Perception and Fat Body Energy Metabolism Are Jointly Affected by Vitellogenin and Juvenile Hormone in Honey Bees
- Genetic Variants on Chromosome 1q41 Influence Ocular Axial Length and High Myopia
- Is a Key Regulator of Pancreaticobiliary Ductal System Development
- The NSL Complex Regulates Housekeeping Genes in
- Attenuation of Notch and Hedgehog Signaling Is Required for Fate Specification in the Spinal Cord
- Dual-Level Regulation of ACC Synthase Activity by MPK3/MPK6 Cascade and Its Downstream WRKY Transcription Factor during Ethylene Induction in Arabidopsis
- Genome-Wide Functional Profiling Identifies Genes and Processes Important for Zinc-Limited Growth of
- Base-Pair Resolution DNA Methylation Sequencing Reveals Profoundly Divergent Epigenetic Landscapes in Acute Myeloid Leukemia
- MicroRNA93 Regulates Proliferation and Differentiation of Normal and Malignant Breast Stem Cells
- Phylogenomic Analysis Reveals Dynamic Evolutionary History of the Drosophila Heterochromatin Protein 1 (HP1) Gene Family
- Found: The Elusive ANTAR Transcription Antiterminator
- The Mutation in Chickens Constitutes a Structural Rearrangement Causing Both Altered Comb Morphology and Defective Sperm Motility
- Control of CpG and Non-CpG DNA Methylation by DNA Methyltransferases
- Polymorphisms in the Mitochondrial Ribosome Recycling Factor Compromise Cell Respiratory Function and Increase Atorvastatin Toxicity
- Gene Expression Profiles in Parkinson Disease Prefrontal Cortex Implicate and Genes under Its Transcriptional Regulation
- Global Regulatory Functions of the Endoribonuclease III in Gene Expression
- Extensive Evolutionary Changes in Regulatory Element Activity during Human Origins Are Associated with Altered Gene Expression and Positive Selection
- The Regulatory Network of Natural Competence and Transformation of
- Brain Expression Genome-Wide Association Study (eGWAS) Identifies Human Disease-Associated Variants
- Quantifying the Adaptive Potential of an Antibiotic Resistance Enzyme
- Divergence of the Yeast Transcription Factor Affects Sulfite Resistance
- The Histone Demethylase Jhdm1a Regulates Hepatic Gluconeogenesis
- RNA Methylation by the MIS Complex Regulates a Cell Fate Decision in Yeast
- Rare Copy Number Variants Observed in Hereditary Breast Cancer Cases Disrupt Genes in Estrogen Signaling and Tumor Suppression Network
- Genome-Wide Location Analysis Reveals Distinct Transcriptional Circuitry by Paralogous Regulators Foxa1 and Foxa2
- A Mutation Links a Canine Progressive Early-Onset Cerebellar Ataxia to the Endoplasmic Reticulum–Associated Protein Degradation (ERAD) Machinery
- The Mechanism for RNA Recognition by ANTAR Regulators of Gene Expression
- Limits to the Rate of Adaptive Substitution in Sexual Populations
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Rumors of Its Disassembly Have Been Greatly Exaggerated: The Secret Life of the Synaptonemal Complex at the Centromeres
- The NSL Complex Regulates Housekeeping Genes in
- Tipping the Balance in the Powerhouse of the Cell to “Protect” Colorectal Cancer
- Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



