-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaControlling Pre-leukemic Thymocyte Self-Renewal
article has not abstract
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004881
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004881Summary
article has not abstract
T cell acute lymphoblastic leukemia (T-ALL) develops in a multistep process whereby thymic progenitor cells gradually accumulate genetic and epigenetic changes, eventually leading to fully transformed immature lymphoblasts. Aberrant activation of transcription factor oncogenes is considered a core component of the oncogenic program that drives malignant T cell transformation. For example, the TAL1 (SCL) and LYL1 basic Helix-Loop-Helix (bHLH) transcription factors, as well as the LIM domain–only proteins LMO1 or LMO2, are activated by chromosomal translocations or interstitial deletions in a large fraction of primary T-ALLs. Notably, these leukemias often present with activating NOTCH1 mutations, suggesting that enhanced NOTCH signaling and aberrant TAL1, LYL1, LMO1, and/or LMO2 expression are collaborative events in the multistep pathogenesis of T-ALL [1], [2]. Recent experimental evidence uncovered the existence of long-lived pre-leukemic stem cells (pre-LSCs) with self-renewal properties, allowing clonal expansion and subsequent acquisition of oncogenic mutations leading to cancer [3], [4]. For example, DNMT3A mutant pre-LSCs were shown to survive chemotherapy and represent a reservoir for leukemic progression and hematological relapse in acute myeloid leukemia (AML) [5], [6]. Although the concept of pre-leukemic stem cells has been previously proposed in the context of T-ALL [7], the actual molecular mechanisms by which T cell-specific oncogenes regulate pre-LSC activity of thymic precursors remains largely unexplored. Notably, these mechanistic insights could provide valuable input for the development of novel therapeutic strategies that can effectively eradicate quiescent and therapy-resistant clones [4], [6], [7].
Pre-leukemic Stem Cells: It's All in the DN3
In this issue of PLOS Genetics, Gerby et al. [8] used the SCLtgLMO1tg transgenic mouse model [9] to investigate the molecular pathways that mediate the transition of normal thymic precursors into pre-LSCs. Interestingly, combined SCL and LMO1 overexpression was sufficient to reprogram double negative 3 (DN3) T cell precursors with a finite lifespan into pre-LSCs with self-renewal activity and retained differentiation capacity (Fig. 1). Moreover, transcriptome analysis revealed that this transition towards pre-LSC was accompanied by a marked up-regulation of a stem cell signature, including Hhex, Nfe2, and Lyl1. Notably, Gerby et al. [8] also used LYL1tgLMO1tg transgenic mice to show that combined LYL1 and LMO1 overexpression results in similar reprogramming activities, as observed for SCL-LMO1. These results confirm recent work from McCormack and colleagues showing Lyl1 as an essential, but not sufficient, factor to reprogram Lmo2 overexpressing DN3 cells into pre-LSCs [10]. All together, these data convincingly show that combined activation of SCL or LYL1 together with LMO1 or LMO2 can drive oncogenic reprogramming of DN3 thymic precursors into self-renewing pre-LSCs.
Fig. 1. Hypothetical model describing pre-leukemic stem cells formation and T-ALL development in SCLtgLMO1tg mice. 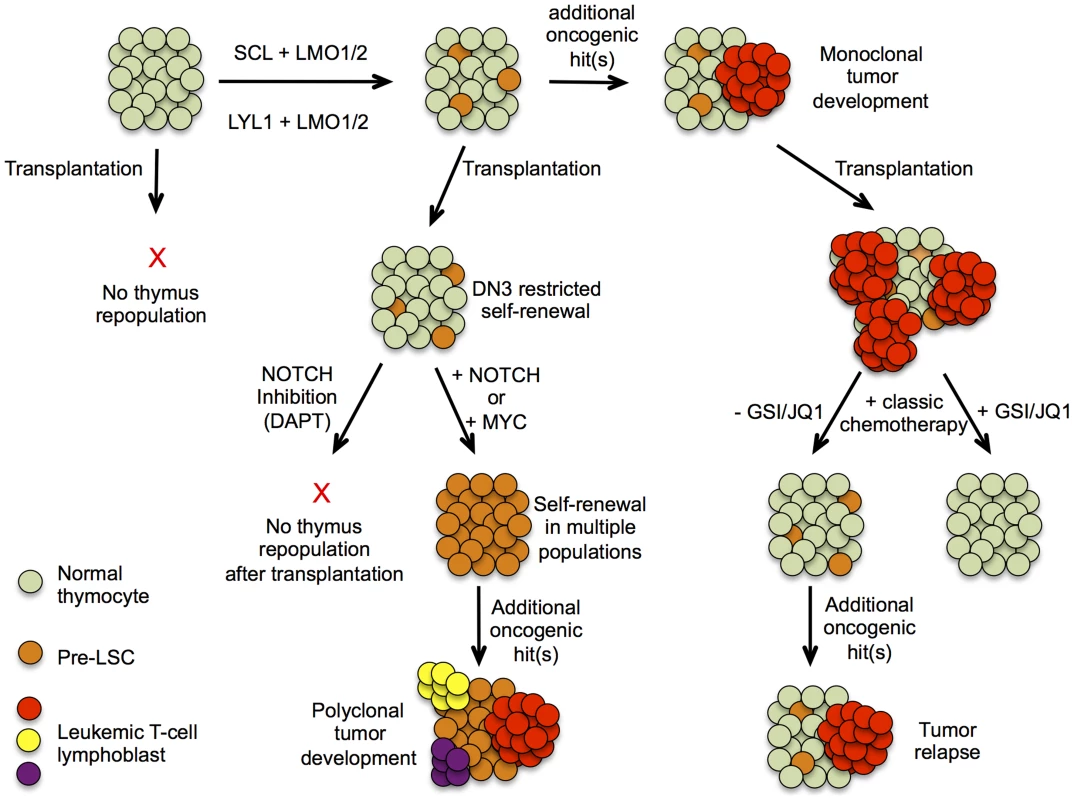
Schematic presentation of T-ALL progression based on experimental evidence presented by Gerby et al. Upper part represents the progression of T-ALL development in a thymus initiated by the formation of a pre-leukemic stem cell population, allowing clonal expansion and subsequent acquisition of oncogenic mutations, eventually leading to a mono/oligoclonal tumor. The lower part depicts results obtained after transplantation of thymocytes at different stages of tumor development and the effects of altering NOTCH1 signaling or its downstream component MYC on their repopulating capacity. NOTCHing up Pre-leukemic Stem Cells
High physiological levels of Notch1 signaling drive irreversible T-lineage commitment at the DN3 stage of T cell differentiation. Therefore, enhanced Notch1 activity could be critically involved in the reprogramming capacity of SCL-LMO1. Strikingly, Gerby and colleagues [8] showed that the self-renewal capacity of SCLtgLMO1tg DN3 cells was completely abrogated upon ablation of Notch1 activity by γ-secretase inhibitor treatment (Fig. 1). For the first time, these results unambiguously show that high physiological levels of Notch1 are critically required for SCL-LMO1–driven reprogramming of DN3 thymocytes into self-renewing pre-LSCs. Conversely, Gerby et al. [8] showed that supraphysiological levels of Notch1 significantly expanded the pool of pre-LSC; enhanced their engraftment and rendered them independent of the thymic microenvironment (Fig. 1). Remarkably, Notch1 activation by itself was not sufficient to trigger reprogramming activity, suggesting that the Notch1 oncogene is devoid of intrinsic self-renewal capacity, but confers a proliferative advantage to SCL-LMO1–primed pre-LSCs (Fig. 1). If the molecular mechanisms, as described above, turn out to be conserved in the human setting, inhibition of NOTCH signaling by γ-secretase inhibitors, small molecule inhibitors or antibodies targeting specific Notch receptors, could serve as an attractive approach to effectively target pre-LSCs for the treatment of human T-ALL. Most notably, in that scenario, eradication of the quiescent and chemo-resistant pre-LSCs by inactivation of NOTCH signaling might be applicable for a large fraction of primary human T-ALL patients, irrespective of their NOTCH1 mutational status at diagnosis. Furthermore, Gerby et al. [8] also showed that Myc acts as a critical mediator of pre-LSC activity downstream of Notch1, suggesting that combined NOTCH/MYC inhibition by γ-secretase and BET-bromodomain inhibitors [11] could further potentiate therapeutic targeting of the pre-LSC population in the context of human T-ALL (Fig. 1).
Thymocyte Self-Renewal: In Which E Proteins Meet LMOs
From a mechanistic point of view, it has been previously postulated that the oncogenic potential of SCL and LYL1 is mediated by inhibition of E proteins [12]. Indeed, SCL and LYL1 can heterodimerise with E2A or HEB and form inactive transcriptional complexes that drive repression of E protein target genes, including critical regulators of T cell differentiation (Ptcra, Il7r, and Rag2) [12]. Alternatively, SCL and LYL1 can directly interact with LMO1 or LMO2 and recruit other co-activators to form active multiprotein transcriptional complexes that drive specific target gene expression [13]. Up until now, no concrete proof existed for whether the sequestration of E proteins is truly the major cause of SCL-driven leukemia development. Here, the authors generated a transgenic mouse model expressing a mutant form of SCL (SCLm13) that is defective in LMO1/2 binding, but still inhibits E proteins through heterodimerization [8]. Strikingly, the authors observed a strong delay in leukemia onset and a reduced tumor penetrance in the SCLm13tgLMO1tg versus SCLtgLMO1tg transgenic mice, suggesting that E protein inhibition is not the main oncogenic property of SCL in this model of murine T cell leukemogenesis. Furthermore, combined SCLm13 and LMO1 overexpression was unable to reprogram DN3 cells into pre-LSCs. All together, these data suggest that the SCL-LMO1 interaction is essential for the generation of pre-LSCs through reactivation of stem cell genes (Hhex, Lyl1) and serves as a critical mediator of malignant T cell transformation. Given this, targeting the SCL-LMO1/2 or LYL1-LMO1/2 [10] protein–protein interaction by small molecule inhibitors might represent an elegant new therapeutic approach to constrain self-renewing capacity of both leukemic and pre-leukemic cancer stem cells in T-ALL.
In conclusion, this study [8] nicely illustrates how cooperative transcription factor oncogenes can reprogram normal T cells into pre-LSCs during the earliest steps of malignant T cell transformation. Although these results provide novel insights in the multistep pathogenesis of T-ALL and have important therapeutic implications, it remains to be established whether gain of pre-leukemic self-renewal capacity is a true obligatory trait for human T-ALL development. Moreover, it would be interesting to know if other T-ALL–specific transcription factor oncogenes, including TLX1, TLX3, or HOXA, also possess the intrinsic capacity to induce self-renewal in T cell progenitors [7]. In regard to this notion, it is currently unclear if the results presented in this study are broadly relevant for the majority of primary human T-ALLs or if they specifically apply to a particular molecular genetic subtype of this disease. For example, early immature T-ALLs show high expression levels of SCL, LYL1, LMO1, and LMO2 as a reflection of their maturation arrest at the early double negative stages of T cell differentiation [14]. In contrast, chromosomal translocations or small genomic deletions targeting SCL, LYL1, and/or LMO2, are exclusively present in primary human T-ALLs with a late cortical tumor phenotype [1]. Therefore, the functional role of SCL, LYL1, and/or LMO2 in regulating self-renewal capacity might be influenced by the immunophenotypic background of the different molecular-genetic subtypes of human T-ALL, a notion that could be tested using transgenic models that overexpress the specific members of this bHLH transcription complex at later stages during T cell development.
Zdroje
1. Van VlierbergheP, FerrandoA (2012) The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest 122 : 3398–3406.
2. TremblayCS, HoangT, HoangT (2010) Early T cell differentiation lessons from T cell acute lymphoblastic leukemia. Prog Mol Biol Transl Sci 92 : 121–156.
3. PandolfiA, BarreyroL, SteidlU (2013) Concise review: preleukemic stem cells: molecular biology and clinical implications of the precursors to leukemia stem cells. Stem Cells Transl Med 2 : 143–150.
4. Corces-Zimmerman MR, Majeti R (2014) Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. E-pub ahead of print. doi:10.1038/leu.2014.211.
5. Corces-ZimmermanMR, HongWJ, WeissmanIL, MedeirosBC, MajetiR (2014) Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A 111 : 2548–2553.
6. ShlushLI, ZandiS, MitchellA, ChenWC, BrandweinJM, et al. (2014) Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506 : 328–333.
7. TremblayCS, CurtisDJ (2014) The clonal evolution of leukemic stem cells in T cell acute lymphoblastic leukemia. Curr Opin Hematol 21 : 320–325.
8. GerbyB, TremblayM, Rojas-SutterlinS, HébertJ, SauvageauG, et al. (2014) SCL, LMO1 and Notch1 reprogram thymocytes into self-renewing cells. PLoS Genet 10: e1004768 doi:10.1371/journal.pgen.1004768
9. TremblayM, TremblayCS, HerblotS, AplanPD, HebertJ, et al. (2010) Modeling T cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev 24 : 1093–1105.
10. McCormackMP, ShieldsBJ, JacksonJT, NasaC, ShiW, et al. (2013) Requirement for Lyl1 in a model of Lmo2-driven early T cell precursor ALL. Blood 122 : 2093–2103.
11. DelmoreJE, IssaGC, LemieuxME, RahlPB, ShiJ, et al. (2011) BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146 : 904–917.
12. HerblotS, SteffAM, HugoP, AplanPD, HoangT (2000) SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol 1 : 138–144.
13. MatthewsJM, LesterK, JosephS, CurtisDJ (2013) LIM-domain-only proteins in cancer. Nat Rev Cancer 13 : 111–122.
14. HayduJE, FerrandoAA (2013) Early T cell precursor acute lymphoblastic leukaemia. Curr Opin Hematol 20 : 369–373.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



