-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAnalysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
In plants, naturally evolving disease resistance (R) genes can cause autoimmunity when combined with different genetic backgrounds. This phenomenon, called immune-related hybrid incompatibility (HI), leads to growth inhibition and fitness loss due to inappropriate activation of defense. HI likely reflects different evolutionary paths of immune-related genes in nature. We have examined the genetic architecture of a complex R locus present in a Central European accession (Ler) which underlies HI with Central Asian accessions of Arabidopsis. We show that expression of one gene (R3) within the Ler cluster of eight tandem R genes (R1–R8) controls the balance between growth and defense but that R3 needs at least one other co-acting member within the R locus to condition HI. We traced the R1–R8 haplotype to a local population of Ler relatives in Poland where it also underlies HI with Central Asian accessions. Occurrence of the incompatible haplotype in ∼30% of genetically diverse local individuals, suggests that it has not arisen recently and has been maintained through selection or drift. Co-occurrence in the same population of individuals containing different R genes that do not cause HI provides a basis for determining genetic and environmental forces influencing how plant immunity genes evolve and diversify.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004848
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004848Summary
In plants, naturally evolving disease resistance (R) genes can cause autoimmunity when combined with different genetic backgrounds. This phenomenon, called immune-related hybrid incompatibility (HI), leads to growth inhibition and fitness loss due to inappropriate activation of defense. HI likely reflects different evolutionary paths of immune-related genes in nature. We have examined the genetic architecture of a complex R locus present in a Central European accession (Ler) which underlies HI with Central Asian accessions of Arabidopsis. We show that expression of one gene (R3) within the Ler cluster of eight tandem R genes (R1–R8) controls the balance between growth and defense but that R3 needs at least one other co-acting member within the R locus to condition HI. We traced the R1–R8 haplotype to a local population of Ler relatives in Poland where it also underlies HI with Central Asian accessions. Occurrence of the incompatible haplotype in ∼30% of genetically diverse local individuals, suggests that it has not arisen recently and has been maintained through selection or drift. Co-occurrence in the same population of individuals containing different R genes that do not cause HI provides a basis for determining genetic and environmental forces influencing how plant immunity genes evolve and diversify.
Introduction
Understanding the processes by which new species arise is an important evolutionary question [1]. In plants, polyploidy is one of the best known mechanisms in speciation, together with other pre-zygotic and post-zygotic barriers [2]. However, more discrete and often cumulative changes in plant genomes can lead to reproductive barriers and, potentially, isolation [3]. Intrinsic to speciation is the divergence of populations, which allows accumulation of genetic differences as a result of drift, local adaptation or coevolution. Such evolutionary processes may create novel alleles or genes that, when combined with other forms from divergent populations, cause hybrid failure to various degrees [3], [4]. These alleles and the resulting hybrids are referred to as ‘incompatible’ and they have been documented in plant breeding programs e.g. [5], although incompatibilities are not limited to crops, as demonstrated by their occurrence in Arabidopsis thaliana [6] and Mimulus guttatus [7] populations.
In some genetically recessive hybrid incompatible (HI) interactions, parental lineages may have experienced alternate loss-of-function of duplicated genes required for viability as a result of relaxed purifying selection [8], [9]. Other more complex dominant and recessive hybrid incompatibilities in plants involve allelic mismatches between immune-related genes that trigger constitutive activation of defenses in the absence of pathogen challenge. Hybrid necrosis is often a symptom of resistance deregulation and its cost on growth and reproduction [10].
Plants are frequently attacked by microbial pathogens which cause disease by deploying virulence factors (effectors) that interfere with plant host defenses [11]. Pathogen effectors are in turn recognized, directly or indirectly, by intracellular nucleotide binding-leucine rich repeat (NLR) receptors to induce effector-triggered immunity, which is a rapid host cellular resistance response often associated with localized programmed cell death [11]. NLRs broadly fall into two structural sub-classes carrying either an N-terminal Toll/Interleukin1-receptor domain (known as TNLs) or a coiled-coil domain (CNLs). Consistent with their role as sensors at the molecular interface with rapidly evolving pathogen effector arsenals, there has been massive expansion and diversification of NLR gene families across plant lineages [12], [13]. Nevertheless, the rate of NLR variation is unlikely to keep pace with microbial change, and therefore maintenance of diverse NLR alleles within a population might be an important determinant of host-pathogen coevolution [14]. Accordingly, some TNL genes exhibit molecular signatures consistent with patterns of balancing selection [14]–[16] which would contribute to the standing genetic variation present in nature. Further evidence suggests that plants also extend their resistance spectrum by intercepting actions of multiple effectors on a limited set of cellular targets [17]. There are examples of NLRs monitoring or ‘guarding’ the status of an effector target (the guardee) [11]. Maintaining appropriate ‘guard-guardee’ associations while tolerating NLR variation presents a challenge because even small molecular rearrangements might disturb NLR homeostasis, leading to autoimmunity and impaired growth. Such mismatches would be particularly exposed in interactions in which protein pairs have diverged independently in populations and natural variants arisen that mimic effector modifications [18].
TNLs are highly polymorphic genes in Arabidopsis and many reside in clusters [12], [19]. Their characteristic three-domain composition and within-gene sequence repetition can further promote high levels of polymorphism through non-allelic homologous recombination [12], [20], [21] and TNLs are among plant genes with the highest naturally occurring sequence variation, closely followed by receptor-like kinase (RLK) genes [9], [12], [22].
Several cases have been reported in which autoimmunity results from allelic interactions involving TNL or RLK genes. Intraspecific incompatibilities in Arabidopsis involve interactions between TNL genes [6], TNLs with RLKs [23], or TNLs with a gene encoding a cysteine biosynthetic enzyme [24]. Interspecific HI between crossable species has been reported to involve the CNL receptor-guarded effector target RIN4 in lettuce [25] and NLR/RLK combinations in rice [26], [27]. The nature of the epistatic interaction is therefore not entirely predictable, although the frequency with which a polymorphic cluster of NLR and/or RLK genes is involved likely reflects the occurrence of high genetic variation at these loci, or lower phenotypic buffering capacity for NLR and/or RLK variation [12], [28]. In Arabidopsis accession Ws-0, TNL genes within the Recognition of Peronospora parasitica1 (RPP1) resistance locus confer specific resistance to infectious downy mildew Hyaloperonospora arabidopsidis (Hpa) isolates (formerly Peronospora parasitica) [29], [30]. Interestingly, an Arabidopsis RPP1-like locus with no known pathogen recognition specificity underlies three independent autoimmune interactions [6], [24], [31], two of which involve the RPP1-like Landsberg erecta (Ler) haplotype [24], [31]. This suggests that the RPP1-like locus is predisposed to immune-related hybrid incompatibility in this species.
We reported the occurrence of HI between Ler and individuals from Central Asian populations (Kas-2 and Kond). Incompatible Ler/Kas-2 and Ler/Kond hybrids exhibit dwarfism and sterility at moderately low temperature (14–16°C) typical for ambient temperatures during the natural growing season of many A. thaliana accessions, and these phenotypes are suppressed at higher temperature (20–22°C) [23], [31]. Ler/Kas-2 and Ler/Kond incompatibilities are caused by a common recessive genetic interaction between the RPP1-like Ler locus and Kas-2 or Kond alleles of RLK Strubbelig Receptor Family 3 (SRF3). The RPP1-like locus in Ler contains eight tandemly arranged TNL genes (R1–R8, including R6 which is a truncated form). Col-0 has only two RPP1-like genes at this locus (At3g44630 and At3g44670), similar to its close relative Arabidopsis lyrata [32], suggesting that Ler carries a derived RPP1-like haplotype. An RPP1-like structural variant in accession Uk-1 likely triggers incompatibility with Uk-3, an accession from the same local population [6]. Strikingly, different genetic determinants underlie the RPP1-like Uk-1 and Ler incompatibilities [31]. The Uk-1 RPP1-like locus is incompatible with a Uk-3 allelic form of the TNL gene, SSI4 (suppressor of salicylic acid insensitivity of npr1-4) [6], but not with Kas-2 or Kond alleles of SRF3 [31]. Hence, independent epistatic networks underlie these incompatibilities.
Incompatible SRF3 alleles are frequently found in Central Asia and exhibit molecular patterns consistent with signatures of a recent selective sweep, suggesting that incompatibility with Ler has arisen as a by-product of selection [23]. By contrast, little is known about the natural distribution of the RPP1-like Ler haplotype in Central Europe or the potential benefit or cost of carrying it. Here, we have examined the genetic basis of the RPP1-like Ler incompatibility and the occurrence of the RPP1-like Ler haplotype worldwide. We find that it has been maintained in a local Central European natural population over at least seven decades. We tested for involvement of individual RPP1-like Ler genes in HI between Ler and Kas-2 using artificial microRNA (amiRNA) silencing and analysis of RPP1-like Ler transgenes in neutral or incompatible Arabidopsis backgrounds. We establish that individual RPP1-like Ler genes contribute differently to the trade-off between growth and disease resistance. Incompatibility between Ler and Kas-2 is associated with higher expression of the RPP1-like R3 gene and engineered R3 over expression causes autoimmunity. However, individual RPP1-like Ler gene members are insufficient to condition HI with Kas-2 and Kond. We conclude that a minimum expression threshold of two or more RPP1-like Ler genes in combination with Kas-2 or Kond SRF3 allelic forms is required for autoimmunity and HI. Finally, we show that the incompatible RPP1-like Ler haplotype is frequent in a local Central European population where it co-occurs with other RPP1-like genes not triggering incompatibility with Kas-2 or Kond. Our study reveals the complex nature and local genetic diversity of the RPP1-like Ler locus underlying incompatibility with Central Asian populations of Arabidopsis.
Results
amiRNA silencing of RPP1-like Ler genes
Previously, we mapped the Ler locus causing incompatibility with Kas-2 and Kond Central Asian accessions (SRF3 forms) to a large ∼87 kb cluster of RPP1-like genes on chromosome 3 [31]. We also established that HI was suppressed by loss-of-function mutations of the TNL immunity regulator EDS1 or by depletion of the defense signaling hormone salicylic acid (SA), consistent with TNL genes driving HI [31]. To ascertain whether HI is due to one or more RPP1-like Ler genes within the locus, we used artificial microRNA (amiRNA) silencing of an incompatible Ler/Kas-2 near isogenic line (NIL) which contains a Ler introgression spanning the RPP1-like locus in a Kas-2 background [31]. Incompatible NIL plants were transformed with amiRNAs KB209, KB212 and KB228 originally designed against RPP1-like genes in Uk-1 (S1 Table). Of the three amiRNAs used, only KB209 and KB212 have predicted complementarity with RPP1-like Ler genes. Multiple independent NIL lines transformed with amiRNAs KB209, KB212 and KB228 (the latter used as a negative control) were tested for suppression of incompatible phenotypes at 14–16°C. We observed suppression of incompatibility in all NIL plants transformed with amiRNA KB209, and in most KB212 transformants (Fig. 1A). As expected, KB228 did not rescue the incompatible NIL phenotype (Fig. 1A).
Fig. 1. Growth phenotype of amiRNA lines at 14–16°C and RPP1-like gene expression. 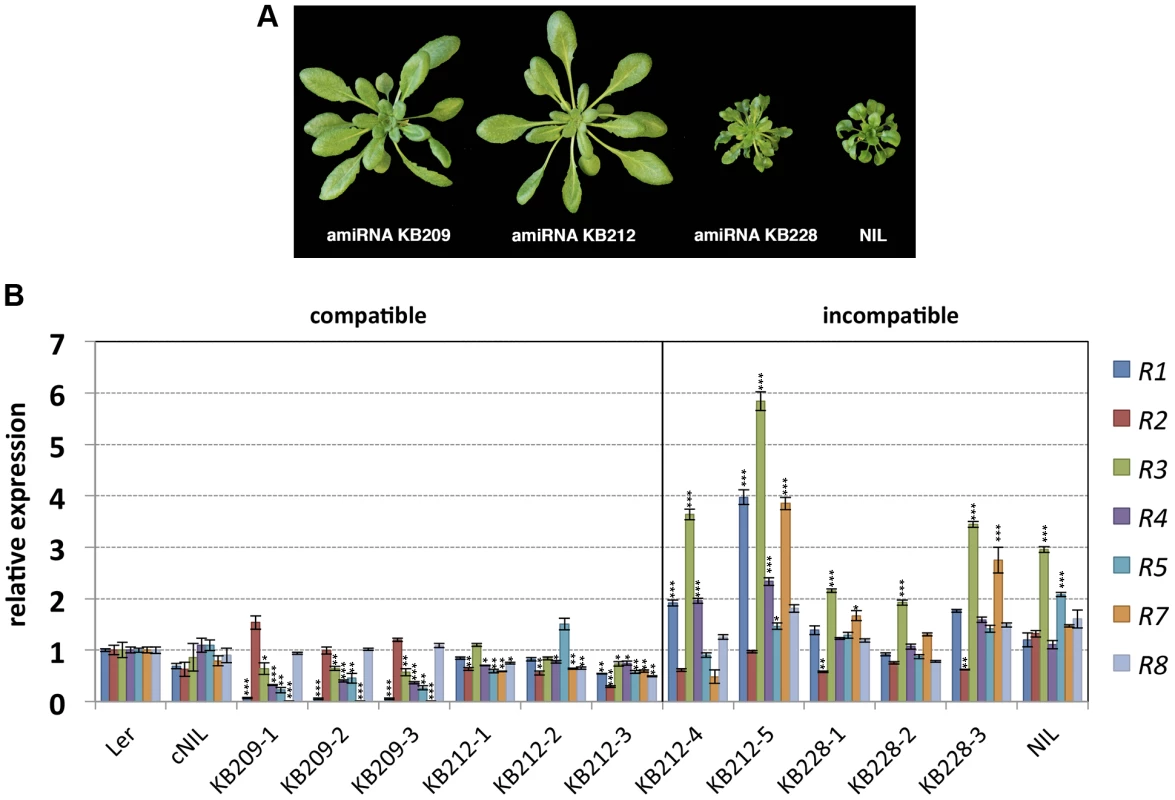
(A) Growth phenotype of 5-week old Ler/Kas-2 NIL plants transformed with amiRNAs effective (KB209 and KB212) or not effective (KB228) suppressing incompatibility. (B) Expression of individual RPP1-like Ler genes determined by qRT-PCR in suppressed (compatible) amiRNA KB209 lines (KB209-1, KB209-2, KB209-3), KB212 (KB212-1, KB212-2, KB212-3) and non-suppressed (incompatible) amiRNA lines KB212 (KB212-4, KB212-5), KB228 (KB228-1, KB228-2, KB228-3). Values are relative to Ler and the mean ± SD of three biological replicates each using three technical replicates. cNIL (complemented NIL), NIL (incompatible Ler/Kas-2 near-isogenic line) [23], [31]. Significant differences in gene expression between Ler and other genotypes using Student's t-test are indicated by asterisks: *P<0.05, **P<0.01, ***P<0.005. Using quantitative RT-PCR (qRT-PCR) with primer pairs that discriminated between individual RPP1-like Ler genes (S2 Table), we determined expression of each gene within the RPP1-like Ler cluster in the amiRNA lines with and without HI suppression, in the original NIL background, a complemented NIL line (cNIL; transformed with the compatible SRF3 Ler allele [23]), and the Ler parental accession (Fig. 1B). Quantitative expression analyses indicated silencing of most RPP1-like Ler genes by amiRNAs KB209 and KB212 in suppressed lines (compatible), except for R2 and R8, which were expressed at wild-type (Ler) levels in KB209 lines (Fig. 1B). These results narrowed the potential RPP1-like incompatibility determinants down to R1, R3, R4, R5 and R7. In non-suppressed incompatible lines (KB228, KB212-4 and KB212-5) and the NIL, we consistently detected higher RPP1-like R3 expression levels (≥2-fold) compared to Ler, the cNIL and compatible amiRNA lines (Fig. 1B). These results show that loss of HI correlates with reduced expression of multiple RPP1-like Ler genes, whereas maintenance of HI is associated with enhanced expression of R3. Thus, R3 might be a key factor in the incompatibility with Kas-2.
Contribution of RPP1-like Ler genes to the trade-off between growth and disease resistance
Prolonged activation of defenses bears a fitness cost for the plant [33], [34] and this might shape the genetic composition of Resistance (R) genes in natural populations [13]. We measured the contribution of individual RPP1-like Ler genes to the trade-off between growth and disease resistance by transforming a neutral (compatible) background, accession Col-0, with genomic constructs of each RPP1-like Ler gene under control of its native 5′ and 3′ sequences. These lines are referred to as ColRPP1Ler. We also included RPP1-like Ler R1 and R5 genes, which contain stop codons in their coding sequences (S1 Figure). Expression of the RPP1-like Ler transgenes in Col-0 was detectable and ranged from 0.5 to 5.5-fold their native expression levels in Ler (Fig. 2). Interestingly, ColRPP1Ler R3 lines with higher expression (lines 12, 13 and 27) exhibited dwarfism and sterility at 14–16°C, which were suppressed at 20–22°C (S2 Figure). By contrast, ColRPP1Ler R1, R2, R4, R5, R7 and R8 lines did not show obvious growth defects at 14–16°C regardless of the transgene expression level (Fig. 2 and S3 Figure). Expression of the defense marker gene PR-1 was used to monitor defense activation in the different transgenic lines (Fig. 2). ColRPP1Ler R3 lines with higher transgene expression also exhibited high PR-1 expression. Expression of PR-1 remained low in ColRPP1-like Ler R1, R2, R4, R5, R7 and R8 lines and variation in PR1 transcript levels between lines did not correlate with transgene expression (Fig. 2). Cell death lesions were detected in leaves of the stunted ColRPP1Ler R3 lines at 14–16°C, as observed previously for Ler/Kas-2 incompatible lines [31], but were absent in all other ColRPP1-like Ler R1, R2, R4, R5, R7 and R8 lines grown under the same conditions (S4 Figure).
Fig. 2. Transgene and PR-1 expression in ColRPP1 lines. 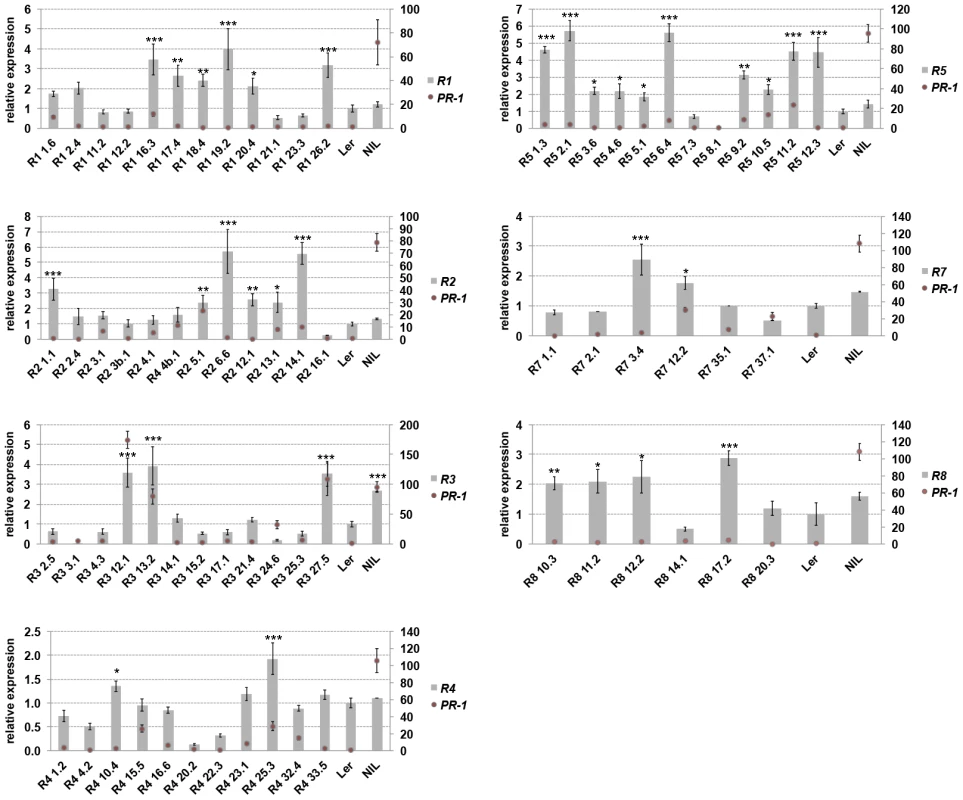
Expression of RPP1-like Ler R1, R2, R3, R4, R5, R7 and R8 transgenes (left axis) and PR-1 (right axis) determined in individual homozygous ColRPP1 lines, Ler and NIL plants grown at 14–16°C by qRT-PCR. Values are relative to Ler and the mean ± SD of three biological replicates each using three technical replicates. NIL (incompatible Ler/Kas-2 near-isogenic line [31]). Significant differences in gene expression between Ler and different ColRPP1 lines using Student's t-test are indicated by asterisks: *P<0.05, **P<0.01, ***P<0.005. We tested the different ColRPP1 lines for basal immunity to the virulent Hpa isolate Noco2 (S5 Figure) [23], and for TNL (RPP2)-mediated immunity to avirulent Hpa isolate Cala2 (S6 Figure). Effector-triggered TNL immunity is often accompanied by a hypersensitive response (HR) involving localized plant cell death at infection sites [35]. These pathology assays showed that RPP1-Ler R1, R2, R4, R5, R7 or R8 genes did not alter Col basal disease resistance against Hpa Noco2 (S5 Figure) or TNL resistance against Hpa Cala2 (S6 Figure). The results argue against a contribution of individual RPP1-like Ler R1, R2, R4, R5, R7 or R8 to defense, as measured by their Hpa disease resistance phenotypes. However, overexpression of RPP1-like Ler R3 in Col-0 caused increased cell death in response to virulent Hpa isolate Noco2 (S5 Figure) and extended HR-like lesioning in response to avirulent Hpa Cala2 (S6 Figure). These infection phenotypes are similar to those observed in incompatible Ler/Kas-2 lines [31]. Basal and TNL triggered resistance phenotypes were not altered in transgenic lines with lower R3 expression (S7 Figure). In summary, the results show that overexpression of RPP1-like Ler R3 causes HI-like autoimmunity and suggest that RPP1-like Ler R3 expression affects the trade-off between plant growth and disease resistance.
Effects of RPP1-like Ler R3 expression with different allelic combinations at SRF3 and QTL5
The RPP1-like Ler locus is incompatible with homozygous Kas-2 alleles at SRF3 and a third locus (QTL5) involved in HI with Kas-2 [31]. Because RPP1-like Ler R3 overexpression causes enhanced immunity in a neutral background, we asked whether allelic variation at the SRF3 and QTL5 interacting loci affect expression of genes within the RPP1-like Ler locus. For this, we measured expression of individual RPP1-like Ler genes in Ler/Kas-2 recombinant inbred lines (RILs) [36] carrying incompatible (RPP1-like/SRF3/QTL5: Ler/Kas-2/Kas-2) or compatible combinations of alleles at RPP1, SRF3 and QTL5 (Ler/Ler/Kas-2 and Ler/Kas-2/Ler) (Fig. 3). Expression analyses showed that RPP1-like Ler R3 transcript levels were significantly (≥2.0-fold) higher in RILs carrying incompatible alleles (Fig. 3). Also, R3 transcriptional up-regulation did not occur in an incompatible Ler/Kas-2 line carrying the eds1-2 mutation (Fig. 3). These results underscore the relationship between incompatibility and RPP1-like Ler R3 expression inferred from the amiRNA analysis (Fig. 1B), and suggest that incompatibility involves EDS1-dependent upregulation of RPP1-like Ler R3.
Fig. 3. Effects of R3 expression with different allelic combinations at SRF3 and QTL5. 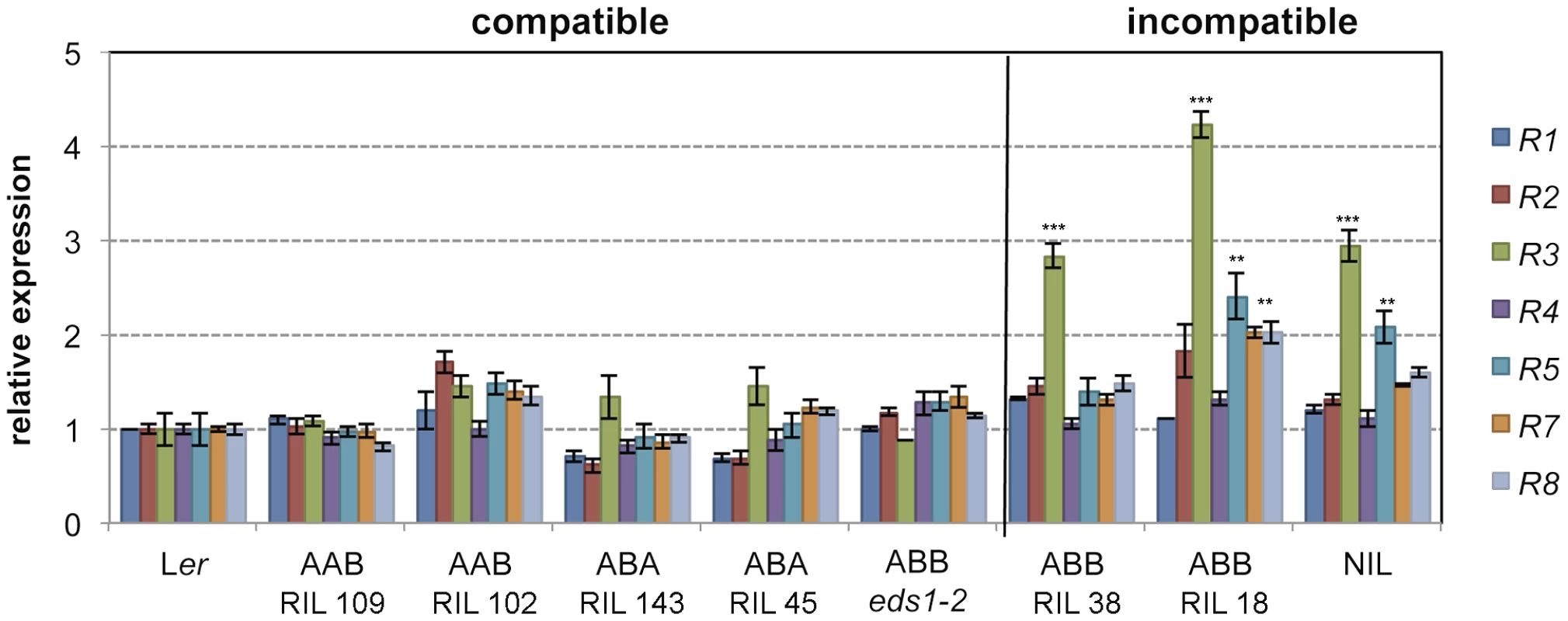
Expression of RPP1-like Ler genes determined by qRT-PCR in different Ler/Kas-2 recombinant inbred lines (RIL) carrying homozygous allelic combinations at RPP1-like/SRF3/QTL5 loci: Ler/Ler/Kas-2 (AAB), Ler/Kas-2/Ler (ABA), Ler/Kas-2/Kas-2 (ABB). AAB and ABA represent compatible allele combinations, whereas ABB triggers incompatibility. The ABB eds1-2 line carrying an EDS1 loss-of-function mutation is described in [31]. Values are relative to Ler and the mean ± SD of three biological replicates each using three technical replicates. Significant differences in the gene expression between Ler and different genotypes using Student's t-test are indicated by asterisks: *P<0.05, **P<0.01, ***P<0.005. Contribution of individual RPP1-like Ler genes to incompatibility
We then examined the contribution of individual RPP1-like Ler genes to incompatibility with Kas-2 and Kond, by crossing these accessions with multiple independent ColRPP1 transgenic lines differing in RPP1-like Ler transgene expression (S3 Table). We generated and scored an average of 220 F2 plants per population for the segregation of HI phenotypes at 14–16°C. As expected, progeny derived from the cross of ColRPP1 R3 dwarf lines (12 and 27) with Kas-2 and Kond segregated for incompatible phenotypes in the F2 (S3 Table). Incompatible phenotypes were not detected in crosses of any of the other ColRPP1 lines with Kas-2 and Kond (S3 Table). F2 populations derived from ColRPP1 R3 lines with lower transgene expression (14, 21, 24 and 25) also did not display incompatibility (S3 Table). These results suggest that individual members of the RPP1-like Ler locus, including R3 under native expression conditions, are insufficient to condition HI with Kas-2 and Kond.
At least two homozygous copies of RPP1-like Ler genes are required for incompatibility
We isolated F2 individuals carrying each of the RPP1-like Ler transgenes and Kas-2 incompatible allele combinations at SRF3 and QTL5 (Fig. 4A). These lines did not exhibit HI phenotypes at 14–16°C. We then studied the effect of adding one copy of the entire RPP1-like Ler cluster to these lines, maintaining homozygous Kas-2 incompatible alleles at SRF3 and QTL5 (Fig. 4B), or compatible heterozygous alleles at SRF3 (S8 Figure). These materials were generated by crossing the above F2 plants and their controls with the NIL (see Material and Methods). Growing the RPP1-like Ler hemizygous lines at 14–16°C did not reveal incompatibility (Fig. 4B). Similar growth phenotypes to the respective controls carrying compatible heterozygous SRF3 alleles were observed (S8 Figure). We concluded that immune-related HI between Ler and Kas-2 requires a minimum expression threshold of two or more RPP1-like Ler genes in combination with the Kas-2 allelic forms.
Fig. 4. Growth phenotype at 14–16°C of ColRPP1/Kas-2 plants and RPP1-like Ler hemizygous lines. 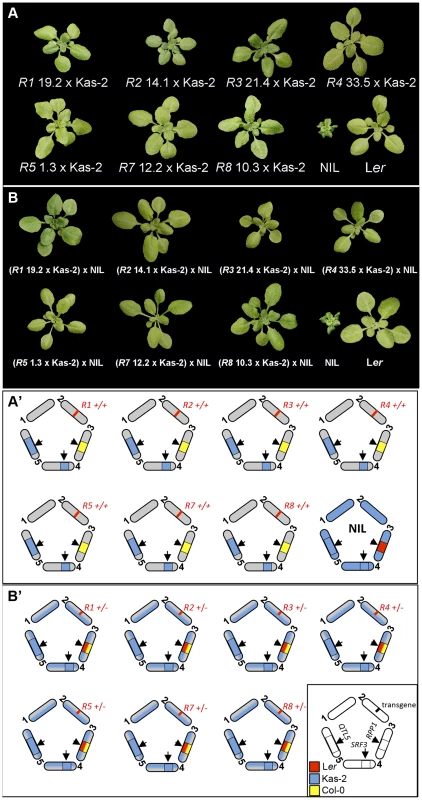
(A) Growth phenotype of representative F3 plants derived from the cross of ColRPP1 R1, R2, R3, R4, R5, R7 and R8 transgenic lines with Kas-2, which carry their respective transgenes in combination with incompatible alleles at interacting loci. (B) Growth phenotype of the F1 progeny derived from the cross of genotypes in (A) with the incompatible Ler/Kas-2 NIL [31]. Genotypes are shown in the lower panel (A′ and B′). Distribution of the RPP1-like Ler haplotype in an Arabidopsis local population
We reported that the RPP1-like Ler incompatible haplotype is rare in Europe based on analysis of thirty F2 populations derived from a cross of Kas-2 with different Central European accessions [23]. This contrasts with the high frequency of SRF3 incompatible alleles found in Central Asia [23]. We also reported that the Landsberg ERECTA (La-0) accession is incompatible with Kas-2, indicating that the incompatible RPP1-like locus was already present in the parental Landsberg La-0 background [31]. Through an optimized PCR screen based on amplification of full-length RPP1-like Ler R1-R8 genes, we searched for the presence of a conserved RPP1-like Ler haplotype in 346 A. thaliana accessions representing a diverse global sample (S4 Table). None of these shared the RPP1-like haplotype with Ler. Absence of a broad distribution led us to hypothesize that the RPP1-like Ler haplotype, if present in the wild, is geographically restricted.
In May 2011, we traced the origin of Ler (Landsberg an der Warthe, Germany 1939) [37] to the area of Gorzów Wielkopolski (Poland) (S9 Figure). There we collected 167 individuals (named Gorzów, Gw), which were genotyped using 149 genome-wide SNPs [38]. With these markers, we could distinguish at least 44 different multi-locus haplotypes (Figs. 5 and 6A) which shared 58–72% SNPs with Ler. This is well above the mode of ∼44% seen for arbitrary pairs of worldwide accessions [39]. Three Gw individuals (1.8% of the entire sample) carried heterozygous alleles at various markers across the genome including the RPP1-like cluster, suggesting that outcrossing occurs between local accessions, as previously observed [40]. Structure [41] and PCA analyses of Gw individuals and other accessions originally from neighboring countries (Austria, Czech Republic and Germany), as well as more geographically distant accessions (The Netherlands, Russia and former Soviet Union, and Central Asia), confirmed that the Gw population is most closely related to other Central European A. thaliana accessions and that, with K = 3 and above, forms a distinct group (S10 Figure).
Fig. 5. Gorzów population structure. 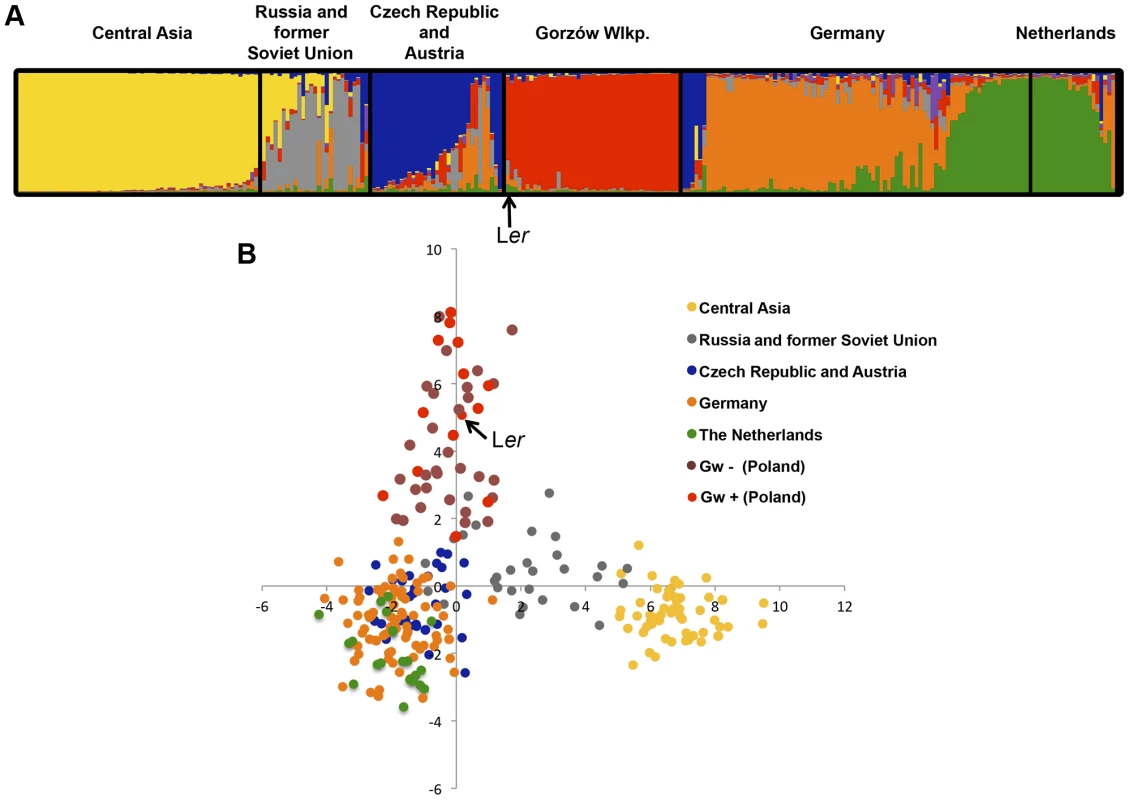
Estimated population structure (A) and principal component analysis, PCA (B), derived from the analysis of 149 genome-wide SNP in genetically distinct individuals in the Gorzów population (n = 44) and accessions from neighboring countries (Czech Republic and Austria, n = 33; Germany, n = 88) or more distant regions (Netherlands, n = 21; Russia and former Soviet Union, n = 27; Central Asia, n = 61). Each accession is colored in segments depicting individual's estimated membership fractions in six main clusters (optimal K = 6, for lower K values see S10 Figure). Gw individuals carrying (Gw+) or not (Gw−) the RPP1-like Ler haplotype are represented in different colors in the PCA plot. Fig. 6. Genetic diversity of the RPP1-like locus in the Gorzów population. 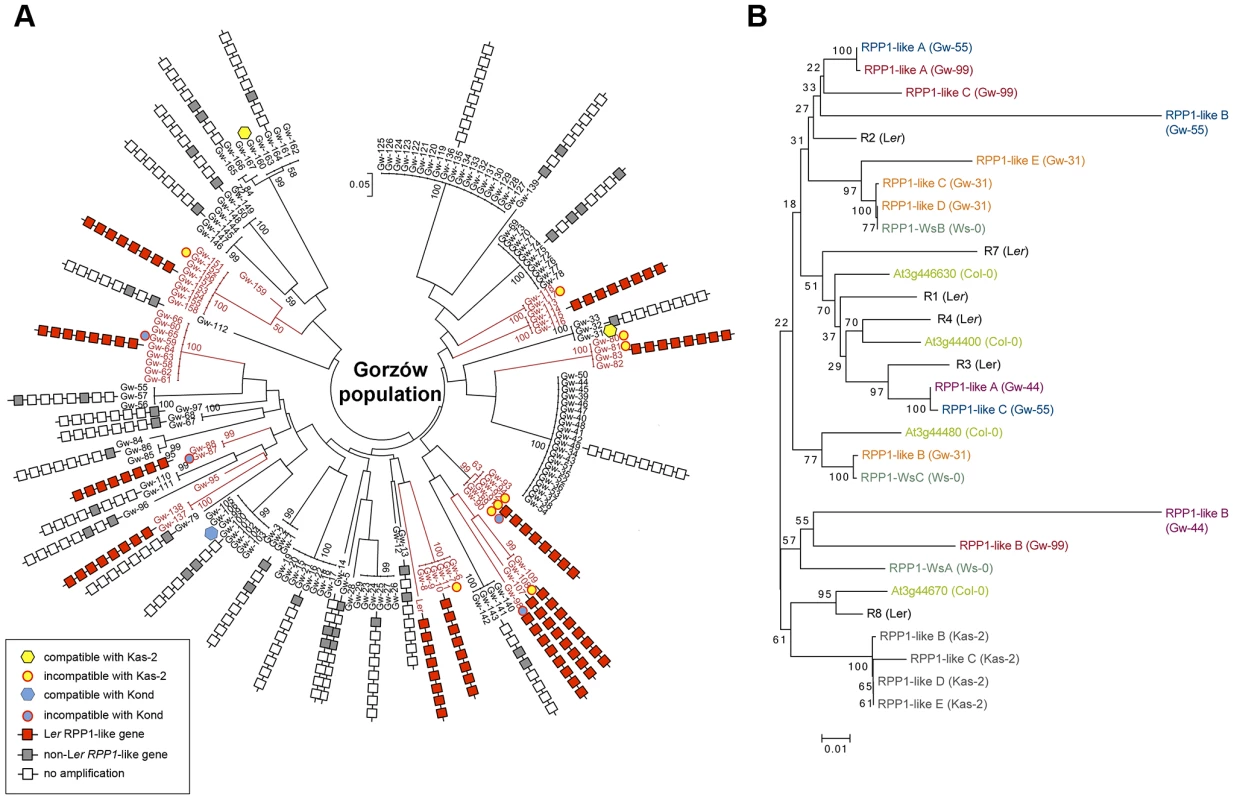
(A) Neighbor-joining tree showing the genome-wide genetic diversity among Gw accessions, estimated from a set of 149 genome-wide SNPs. Boxes represent genes within the RPP1-like haplotype (R1 to R8, with R8 the closest to the tree) conserved with Ler (red) differing from Ler (gray), or absent (white), based on amplification and sequencing of RPP1-like genes with specific primers designed for the RPP1-like Ler cluster. Accessions carrying the RPP1-like Ler haplotype are highlighted in red. Accessions used for crosses with Kas-2 or Kond and their compatibility/incompatibility outcome are indicated. (B) Neighbor-joining tree of RPP1-like genes in Gw− and Kas-2 showing extensive allelic variation. Phylogeny is based in sequencing RPP1-like genes in Gw− individuals (Gw-31, Gw-44, Gw-55 and Gw-99) and Kas-2. The Ler accession derived from plants collected in the same region in 1939 showed a close genetic relationship with the modern Gorzów population (Fig. 5 and S10 Figure). About a third of Gw individuals, representing 14 different multilocus-haplotypes, shared the RPP1-like incompatibility genes found in Ler (Fig. 5B and S11 Figure). This finding suggests that the derived RPP1-like Ler haplotype has been maintained in the original location over at least 72 generations (assuming a generation time of one year) since the first sampling of the population in 1939 [37]. However, the RPP1-like Ler haplotype has not spread in Central Europe (S4 Table).
For simplicity, we will refer to the accessions with RPP1-like Ler alleles as Gw+ and those lacking it as Gw−. Both types of individuals were not differentiated from each other in PCA and neighbor-joining tree analyses based on genome-wide SNP data (Figs. 5B and 6A). We also analyzed the genetic diversity of RPP1-like genes within the Gw− genetic groups (Fig. 6B). Using the same primer combinations designed to amplify RPP1-like Ler genes, we observed variation in the sequence and composition of the RPP1-like cluster in Gw− individuals (Fig. 6A). Due to the variable nature of the cluster, we designed primers annealing to conserved RPP1-like sequences for cloning RPP1-like genes in Gw− individuals. In this way, we isolated RPP1-like genes from four genetically different Gw− accessions sharing 61%–70% SNP with Ler (Gw-31, Gw-44, Gw-55 and Gw-99) and from Kas-2. Phylogenetic analyses, based on sequencing RPP1-like genes of isolated clones, showed a high degree of RPP1-like gene variation between Gw− accessions (Fig. 6B). RPP1-like Kas-2 genes clustered together within the same branch of the tree. However, most branches were formed by RPP1-like genes from different accessions without an obvious relationship (Fig. 6B). Neighbor-Net analysis of RPP1-like genes from Gw− and Gw+ accessions produced evidence for parallelograms in the network, mostly between RPP1-like genes from Gw− accessions, which is suggestive of recombination (S12 Figure). Notably, nucleotide diversity flanking the RPP1-like locus was higher in Gw− than Gw+ haplotypes, extending from ∼−50 kb to +150 kb from the locus (S13 Figure). These results suggest that recombination at the RPP1-like locus is suppressed in Gw+ accessions but not in Gw−, in agreement with observations made during the fine-mapping of the Ler QTL in Ler/Kas-2 heterozygous inbred families [31]. Based on these observations, we hypothesize that the derived RPP1-like Ler haplotype has increased in frequency by introgression to different genetic backgrounds in the Gw population, and that low recombination has contributed to maintaining the RPP1-like Ler cluster as an extended haplotype. The increase in frequency of the derived RPP1-like Ler haplotype is perhaps the result of a selective advantage, drift or complex demographic events, but less likely due to a recent bottleneck, since it was found in diverse genetic backgrounds. The co-occurrence of different RPP1-like allelic forms in Gw− individuals would be expected for Resistance genes in natural populations that are maintained by negative-frequency dependent selection [14]. We conclude that there is extensive diversity of RPP1-like genes co-occurring in a local population.
Incompatibility between Gw and Kas-2 or Kond accessions
Having found that the RPP1-like haplotype is maintained within genetically different individuals from a local Gorzów population, we determined whether it also conditions incompatibility with Kas-2 and Kond. Therefore 13 Gw+ individuals belonging to 10 mutlilocus-haplotype groups (Gw-7, Gw-59, Gw-80/81, Gw-87, Gw-89, Gw-91/92, Gw-98, Gw-108, Gw-117 and Gw-152) and three Gw− controls (Gw-31, Gw-99 and Gw-160) were crossed to Kas-2 and Kond and equal numbers of F2 populations generated (S5 Table). Approximately 250 F2 plants per population were then scored for the segregation of incompatible phenotypes at 14–16°C. We detected the incompatibility in all crosses derived from Gw+ individuals with Kas-2 and Kond (S5 Table; Fig. 7). Incompatible F2 individuals always carried Gw (Ler) homozygous alleles at the RPP1-like locus and Kas-2 or Kond homozygous alleles at SRF3 (S5 Table). The data are consistent with involvement of at least two recessive loci in the Gw+/Kas-2 and Gw+/Kond incompatibilities. By contrast, F2 plants derived from crosses of Gw− with Kas-2 did not display HI at 14–16°C (Fig. 7 and S5 Table). These results show that Gw+ individuals carrying the RPP1-like Ler haplotype are incompatible with Kas-2 and Kond. Therefore, the incompatibility reported between Ler/Kas-2 and Ler/Kond can be extended to a population scale.
Fig. 7. Incompatible phenotypes derived from the cross of Gw+/Kas-2 and Gw+/Kond accessions. 
Gw+ individuals were crossed to Kas-2 and Kond and their F2 progenies screened for the occurrence of incompatible phenotypes at 14–16°C (see S5 Table). Dwarf plants on the left of each cross carry homozygous Gw (Ler) alleles at RPP1-like locus and homozygous SRF3 Kas-2 or Kond alleles, which are not present in normal sized sister F2 plants from the same cross (right). Incompatibility is absent in the cross of Gw− accessions with Kas-2 or Kond (Gw-160 as representative; see S5 Table). Discussion
The arrangement of NLR genes in clusters is the result of tandem, ectopic or large-scale segmental duplications that can be followed by local rearrangements. The clusters may be homogeneous, containing structurally similar NLR genes, or complex when formed by a heterogeneous class of TNL and CNL (Coiled Coil-NLR) genes [12], [42], [43]. The RPP1-like Ler cluster is homogeneous and formed by a sequence of highly related TNL genes [31]. Different repertoires of RPP genes are known to recognize effectors delivered by genetically diverse strains of the Arabidopsis oomycete pathogen Hpa [44], consistent with there being coevolution between naturally evolving Hpa isolates and local Arabidopsis populations. Compared to Col-0 and Arabidopsis lyrata [32], accession Ler contains six additional RPP1-like genes at this locus, one of which (R6) is a truncated form [31]. Nonreciprocal crossover is a source of copy number variation that can generate clusters of different sizes which would limit recombination [45]. Given the larger number of genes within the cluster, the RPP1-like Ler haplotype appears to be a derived one. Sequence alignment and exon-intron organization suggests that RPP1-like genes R3 and R4 are the result of ectopic recombination and duplication of the At3g44400 TNL (S1 Figure). R2 and R8 RPP1-like Ler are the most closely related genes to At3g44630 and At3g44670 in Col-0 respectively, suggesting a common ancestry (S1 Figure).
Activation of defense incurs a cost for the plant, which is often translated into reduced growth and reproductive fitness [18]. However, naturally occurring alleles that lower the thresholds for activation of defense have been found at the ACD6 locus together with wild-type alleles in Arabidopsis natural populations [46]. We found that the majority of RPP1-like Ler genes, except R3, do not confer an obvious fitness cost in neutral backgrounds (Col-0) regardless of their expression levels, nor any clear advantage for disease resistance to the virulent Hpa isolate Noco2 (S3, S4, S5 Figures). R3 overexpression in ColRPP1 lines enhances resistance to Hpa Noco2 (S5 Figure) and reduces fitness at 14–16°C (S2 Figure), thus phenocopying Ler/Kas-2 HI [31]. R3 overexpression in Ler has the same deleterious effect as in Col-0, and is therefore a background-independent phenotype. Incompatible and autoimmune phenotypes were not evident in ColRPP1 lines with wild-type R3 expression levels (S7 Figure). The fitness cost of R3 overexpression suggests a need for tight R3 transcriptional regulation to provide balanced growth and defense. Indeed, a large fraction of the methylation variability in Arabidopsis is detected in regions containing NLR genes [47], [48] and restricting NLR expression is important for normal plant development [49], [50].
Crossing R3 lines exhibiting Ler wild-type expression levels with Kas-2 and Kond failed to reconstitute HI in the F2 generation, nor did crossing individual ColRPP1 R1, R2, R4, R5, R7 and R8 transgenic lines with the same accessions, or adding one supporting copy of the RPP1-like Ler cluster to the above lines (Fig 4; S3 Table). These results suggest that minimum expression of at least two RPP1-like Ler genes is required for incompatibility with Kas-2. However, non-suppressed NIL amiRNA lines and incompatible Ler/Kas-2 RILs exhibited higher R3 transcript levels than suppressed NIL amiRNA lines, Ler/Kas-2 RILs containing compatible allele combinations or incompatible RILs in an immunity suppressed eds1-2 background (Fig. 1B and Fig. 3). Therefore, a strong positive correlation exists between R3 expression and HI. Moreover, R3 overexpression is sufficient to induce plant stunting (S2 Figure). We conclude that HI between Ler and Kas-2 likely involves an amplification of RPP1-like Ler R3 expression in an EDS1-dependent manner. Therefore, both RPP1-like expression and protein differences contribute to HI. The manner in which specific Kas-2 and Kond SRF3 allelic forms contribute to this, and a basis for co-action between R3 and at least one other RPP1-like Ler gene within the locus need to be clarified.
The multigenic nature of an incompatible locus was recently reported in an interspecific cross in rice that requires the presence of two tandem RLK genes for immune-related hybrid weakness [27]. Due to their close proximity and likely reduced recombination rate, genes within the RPP1-like Ler cluster will tend to co-segregate, thus maintaining the locus. Underscoring this, the incompatible RPP1-like Ler haplotype is maintained in the wild (Gorzów), and accessions carrying it are incompatible with Kas-2 and Kond (Fig. 7 and S5 Table). An arms race based on selective sweeps of plant R and pathogen effector genes has been proposed to drive the coevolution between plants and pathogens. However, such dramatic ‘boom and bust’ cycles do not explain all observations made for the genetic composition and infection outcomes in wild populations [51]. Current evidence points to negative-frequency dependent selection, in which rare R alleles can gain a selective advantage and mitigate fitness costs, thereby promoting R gene cyclic dynamics and diversity [51], [52]. In addition, the possibility of neutral evolution when selection is weak, and other non-selective processes such as isolation-by-distance, might shape the genetic diversity of wild populations [53].
Here we have studied the natural distribution of the RPP1-like Ler haplotype in the wild. Our screen in 346 global accessions shows that the RPP1-like Ler cluster is not geographically widespread (S4 Table). Rather, we found it in a population of genetically related Ler individuals in Gorzów Wielkopolski (Poland) (Figs. 5 and 6). This pattern contrasts with the wide distribution of SRF3 Kas-2 and Kond alleles in Central Asian populations [23]. However, the genetic variation among Gorzów individuals (Fig. 5B) suggests that it is not a recent population. Indeed, the original La-0 and La-1 accessions were collected in 1939 [37] and the modern Gorzów population reported here in 2011. Strikingly, Gw individuals carrying the RPP1-like Ler haplotype (Gw+) are not more genetically related to each other than to Gw− individuals not carrying it (Figs. 5B and 6A). Therefore, it is unlikely that this haplotype increased in frequency as result of a recent bottleneck. We favor an evolutionary scenario in which the derived RPP1-like Ler haplotype has increased in frequency (∼30%) by introgression to different Gw genetic backgrounds (Figs. 5B and 6A), and low recombination has helped to maintain RPP1-like Ler genes in genetically different Gw+ individuals (S11 and S13 Figures).
The observed high diversity of RPP1-like genes in Gw− individuals (Fig. 6B) is a pattern expected for wild populations in which multiple (rare) alleles are maintained by negative-frequency dependent selection [14], [54]. Phylogenetic network analysis of RPP1-like genes from Gw+ and Gw− accessions revealed patterns suggestive of recombination between RPP1-like genes from Gw− accessions (S12 Figure). Therefore, recombination between RPP1-like Gw− genes might be a source of RPP1-like gene diversity in the local Gw population (Fig. 6B). However, such patterns were not evident between Gw+ and Gw− RPP1-like genes (S12 Figure). This argues against the high diversity of Gw− RPP1-like genes observed in the population being derived from multiple independent recombination/deletion events acting on the RPP1-like Gw+ (Ler) cluster.
Increased expression of RPP1-Ler R3 might confer a selective advantage in disease resistance or a fitness cost in terms of growth and reproduction at moderately low temperature (14–16°C), highlighting the potential importance of trade-offs and genotype-by-environment interactions in the evolutionary dynamics of RPP1-like genes in wild populations. Whether the increase in frequency of the RPP1-Ler haplotype in Gorzów is due to drift, demography or selection requires further investigation. Such an evolutionary study will entail determining the genetic diversity of R genes and local pathogen populations as well as fitness effects modulated by local environment. These complex interactions, involving also beneficial or commensal microbial communities that might compete with pathogens for resources [55], are likely to shape how plants and pathogens coevolve in nature.
Materials and Methods
Plant materials and growth conditions
The identity and stock numbers of the Arabidopsis thaliana accessions used in this study are listed in S4 Table. Seeds were obtained from the Nottingham Arabidopsis Stock Center (NASC) or collected by the authors. Seeds from the Gorzów population were harvested in Gorzów Wielkopolski (Poland) in May 2011, and amplified between June–October 2011 as single seed descent at the greenhouse facilities of the Max Planck Institute for Plant Breeding Research (Cologne, Germany). Progeny of Gw individuals was directly used for SNP genotyping (see below). The genotype of the incompatible Ler/Kas-2 NIL used in this study has been described before [31]. For growth of plants, seeds were stratified on wet filter paper at 4°C in the dark for 2 to 4 days, and transferred to soil for germination. Plants were germinated and grown under 12 h dark/12 h light cycles, 14°C/16°C and 70% relative humidity in growth chambers (Percival Scientific, USA).
amiRNA lines
amiRNA constructs in binary vectors targeting RPP1-like genes used in this study, KB209 (21mer: TGACACATAAACTCCATCGGT), KB212 (21mer: TACATTTCAACTGCGAGCGTC) and KB228 (21mer: TATATCCGTAATGATTGCGGC) designed using WMD3 [56], were transformed into Ler/Kas-2 NIL plants by floral dip [57]. For the selection of transgenic lines, seeds were surface-sterilized and selected on MS ½ solid media containing 50 µg/ml kanamycin (Sigma-Aldrich) and 200 µg/ml ticarcillin/clavulanic acid 15∶1 (Duchefa). T1 plants were transferred to soil at 23°C and T2 homozygous lines isolated by segregation analyses of T3 seeds on selective media.
Gene expression analyses
Total RNA isolated from 5-week old plants was extracted using TRIzol reagent (Invitrogen). Two micrograms of RNA was treated with DNAse I (Invitrogen) and first strand cDNA synthesized using Superscript II (Invitrogen) and oligo dT. Quantitative real-time PCR using SYBR Green I dye method was performed on Roche LightCycler 480 II detector system following the PCR conditions: 95°C 2 min, 40 cycles (95°C, 15 s; 60°C, 30 s; 68°C, 20 s). Standard curves were performed for quantification. Primers used for gene expression analyses are listed in S2 Table. The specificity of RPP1-like Ler oligonucleotides was determined by comparison of the amplification efficiency using a series of equimolecular premixes of RPP1-like Ler R1-R8 plasmid DNAs in which the target genes were absent or present. qRT-PCR analyses were always performed on at least three biological replicates with three technical replicates each using UBQ10 (At4g05320) as housekeeping gene.
Generation of Col-0 RPP1-like Ler transgenic lines
Genomic regions of RPP1-like Ler genes were amplified from 200 ng of freshly extracted genomic DNA (BioSprint Workstation, Qiagen) of the Ler accession (code number N20), using primer combinations listed in Table S2 and LA Taq DNA polymerase (Takara). PCR conditions were: 94°C 5 min, followed by 30 cycles (94°C, 15 s; 55°C, 30 s; 68°C, 4 min), 68°C 10 min. PCR products were separated on 1% agarose gel stained with ethidium bromide, and purified by gel scission (Gel extraction Kit, Qiagen). Purified fragments were cloned into pGEM T-easy (Promega), and the clones sequenced using T7, SP6 and primers listed in Table S2. Sanger sequencing was performed at the Max Planck Genome Center Cologne (Cologne, Germany). The RPP1-like Ler genes were released from pGEMT-easy by digestion with Not I and cloned into the pCambia1300 binary vector (www.cambia.org) modified to contain the PspOMI site in the MCS. The resulting clones were transformed into Agrobacterium tumefaciens GV3101 pMP90 strain [58] for transformation of Col-0 plants [57]. T0 seeds were selected on MS ½ media supplemented with hygromycin 15 µg/ml and 200 µg/ml ticarcillin/clavulanic acid 15∶1 (Duchefa). Homozygous lines with single T-DNA insertions were determined by segregation analyses and selected for further analyses.
Generation of RPP1-like Ler hemizygous lines
To generate RPP1-like hemizygous lines, we isolated multiple F2 plants from populations described in Table S3 with the genotypes: RPP1-like Ler transgene (+/+), RPP1-like locus (Col-0/Col-0), SRF3: Kas-2/Kas-2 or Col-0/Col-0, QTL5: Kas-2/Kas-2. F3 plants were then crossed to the NIL (RPP1-like: Ler/Ler; SRF3: Kas-2/Kas-2; QTL5: Kas-2/Kas-2) and isolated F1 plants carrying one copy of RPP1-like Ler transgene (+/−), the RPP1-like Ler cluster in hemizygosity (Ler/Col-0) and incompatible (Kas-2/Kas-2) or compatible (Kas-2/Col-0) alleles at SRF3, and Kas-2/Kas-2 at QTL5, as confirmed by genotyping using markers previously reported [23], [31].
Histochemical analyses and pathogen infection assays
Cell death was determined by staining with lactophenol trypan blue [31] and visualization under light microscope (Axioplan, Carl Zeiss). Images were captured in a Leica DFC490 digital camera. Infection with Hpa Cala2 and Noco2 isolates was performed as described [23], [31]. Plant cell death and Hpa infection structures were visualized under light microscope after 4 days of infection.
Screen of RPP1-like Ler haplotype in Arabidopsis accessions
Genomic DNA from 5-week-old Arabidopsis accessions listed in Table S4 was extracted using DNA BioSprint Workstation (Qiagen) and arrayed in 96-well plates including Ler controls on every plate. 200 ng of freshly extracted DNA was used for amplification of full-length RPP1-like R1-R8 Ler genes using primer combinations listed in Table S2 and LA Taq DNA polymerase (Takara). Amplification of UBQ10 was used as control. PCR conditions were as described above for cloning RPP1-like Ler genes. Products of the PCR reaction were separated on 1% agarose gels stained with EthBr. Amplicons and sizes were documented using the Gel Doc XR (BioRad) system.
Genotyping, structure, PCA and phylogenetic analyses
Genomic DNA from Gw accessions was extracted from leaves of 5-week old plants grown in the greenhouse using the BioSprint Workstation (Qiagen) platform by following manufacturer's instructions. SNP multilocus genotypes [38] were determined using the genotyping facility services of the University of Chicago (Chicago, USA). Presence/absence of RPP1-like Ler haplotype in the Gw population was determined by PCR amplification and sequencing of RPP1-like genes using specific primers (Table S2). The population structure of the Gw population was inferred using the software STRUCTURE [41] and previously described settings [23]. To adjudicate the correct number of genetic clusters K, we applied the ΔK method [59] in combination with the absolute value of ln P(X|K). Principal component analysis was performed using R. Neighbor joining-tree was performed from aligned sequences using Mega6.06 and 5000 bootstrap repetitions. Neighbor-net was constructed using SplitsTree4 [60]. Nucleotide diversity was computed using DnaSP (DNA Sequence Polymorphism version 5.10) [61].
Cloning of RPP1-like genes in Gw- accessions
Genomic DNA was extracted from leaves of Gw accessions using DNeasy Plant Mini Kit (Qiagen) according to manufacturer's instructions. RPP1-like genes were amplified by PCR using RA+RC and RB+RC primer combinations and Phusion High-Fidelity DNA Polymerase (Thermo Scientific) and the same PCR conditions as above (Table S2). Amplified fragments were purified by gel scission (Gel Extraction Kit, Qiagen), ligated into the vector pSPARKII (Canvax) and transformed into E. coli DH10B (Clontech). The plasmid DNA from at least 16 independent clones was extracted using Plasmid MiniPrep Kit (Qiagen) and used for Sanger sequencing (Table S2) to identify unique clones. Sequences obtained were assembled using MacVector 12.7.5 and single contigs aligned to the A. thaliana genome using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm their identity based on similarity to known RPP1 and RPP1-like genes. Sequences are deposited in GenBank under accession numbers KM575915–KM575930.
Supporting Information
Zdroje
1. LososJB, ArnoldSJ, BejeranoG, BrodieED, HibbettD, et al. (2013) Evolutionary biology for the 21st century. PLoS Biol 11: e1001466 doi:10.1371/journal.pbio.1001466
2. RiesebergLH, WillisJH (2007) Plant speciation. Science 317 : 910–914 doi:10.1126/science.1137729
3. BombliesK (2010) Doomed lovers: mechanisms of isolation and incompatibility in plants. Annu Rev Plant Biol 61 : 109–124 doi:10.1146/annurev-arplant-042809-112146
4. AbbottR, AlbachD, AnsellS, ArntzenJW, BairdSJE, et al. (2013) Hybridization and speciation. J Evol Biol 26 : 229–246 doi:10.1111/j.1420-9101.2012.02599.x
5. OuyangY, LiuY-G, ZhangQ (2010) Hybrid sterility in plant: stories from rice. Curr Opin Plant Biol 13 : 186–192 doi:10.1016/j.pbi.2010.01.002
6. BombliesK, LempeJ, EppleP, WarthmannN, LanzC, et al. (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236 doi:10.1371/journal.pbio.0050236
7. MacNairMR, ChristieP (1983) Reproductive isolation as a pleiotropic effect of copper tolerance in Mimulus guttatus? Heredity (Edinb) 50 : 295–302 doi:10.1038/hdy.1983.31
8. BikardD, PatelD, Le MettéC, GiorgiV, CamilleriC, et al. (2009) Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323 : 623–626 doi:10.1126/science.1165917
9. ClarkRM, SchweikertG, ToomajianC, OssowskiS, ZellerG, et al. (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 : 338–342 doi:10.1126/science.1138632
10. BombliesK, WeigelD (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8 : 382–393 doi:10.1038/nrg2082
11. DoddsPN, RathjenJP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11 : 539–548 doi:10.1038/nrg2812
12. GuoY-L, FitzJ, SchneebergerK, OssowskiS, CaoJ, et al. (2011) Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol 157 : 757–769 doi:10.1104/pp.111.181990
13. JonesJDG, DanglJL (2006) The plant immune system. Nature 444 : 323–329 doi:10.1038/nature05286
14. KarasovTL, HortonMW, BergelsonJ (2014) Genomic variability as a driver of plant-pathogen coevolution? Curr Opin Plant Biol 18 : 24–30 doi:10.1016/j.pbi.2013.12.003
15. SalvaudonL, GiraudT, ShykoffJA (2008) Genetic diversity in natural populations: a fundamental component of plant-microbe interactions. Curr Opin Plant Biol 11 : 135–143 doi:10.1016/j.pbi.2008.02.002
16. StahlEA, DwyerG, MauricioR, KreitmanM, BergelsonJ (1999) Dynamics of disease resistance polymorphism at the RPM1 locus of Arabidopsis. Nature 400 : 667–671 doi:10.1038/23260
17. MukhtarMS, CarvunisA-R, DrezeM, EppleP, SteinbrennerJ, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333 : 596–601 doi:10.1126/science.1203659
18. AlcázarR, ParkerJE (2011) The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci 16 : 666–675 doi:10.1016/j.tplants.2011.09.001
19. MeyersBC, KaushikS, NandetyRS (2005) Evolving disease resistance genes. Curr Opin Plant Biol 8 : 129–134 doi:10.1016/j.pbi.2005.01.002
20. WickerT, YahiaouiN, KellerB (2007) Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J 51 : 631–641 doi:10.1111/j.1365-313X.2007.03164.x
21. JacobF, VernaldiS, MaekawaT (2013) Evolution and conservation of plant NLR functions. Front Immunol 4 : 297 doi:10.3389/fimmu.2013.00297
22. CaoJ, SchneebergerK, OssowskiS, GüntherT, BenderS, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43 : 956–963 doi:10.1038/ng.911
23. AlcázarR, GarcíaAV, KronholmI, de MeauxJ, KoornneefM, et al. (2010) Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat Genet 42 : 1135–1139 doi:10.1038/ng.704
24. TahirJ, WatanabeM, JingH-C, HunterDA, TohgeT, et al. (2012) Activation of R-mediated innate immunity and disease susceptibility is affected by mutations in a cytosolic O-acetylserine (thiol) lyase in Arabidopsis. Plant J doi:10.1111/tpj.12021
25. JeukenMJW, ZhangNW, McHaleLK, PelgromK, den BoerE, et al. (2009) RIN4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21 : 3368–3378 doi:10.1105/tpc.109.070334
26. YamamotoE, TakashiT, MorinakaY, LinS, WuJ, et al. (2010) Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol Genet Genomics 283 : 305–315 doi:10.1007/s00438-010-0514-y
27. ChenC, ChenH, LinY-S, ShenJ-B, ShanJ-X, et al. (2014) A two-locus interaction causes interspecific hybrid weakness in rice. Nat Commun 5 : 3357 doi:10.1038/ncomms4357
28. SangsterTA, SalathiaN, LeeHN, WatanabeE, SchellenbergK, et al. (2008) HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105 : 2969–2974 doi:10.1073/pnas.0712210105
29. BotellaMA, ParkerJE, FrostLN, Bittner-EddyPD, BeynonJL, et al. (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10 : 1847–1860.
30. RehmanyAP, GordonA, RoseLE, AllenRL, ArmstrongMR, et al. (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17 : 1839–1850 doi:10.1105/tpc.105.031807
31. AlcázarR, GarcíaAV, ParkerJE, ReymondM (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci U S A 106 : 334–339 doi:10.1073/pnas.0811734106
32. HuTT, PattynP, BakkerEG, CaoJ, ChengJ-F, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43 : 476–481 doi:10.1038/ng.807
33. TianD, TrawMB, ChenJQ, KreitmanM, BergelsonJ (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423 : 74–77 doi:10.1038/nature01588
34. Van HultenM, PelserM, van LoonLC, PieterseCMJ, TonJ (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci U S A 103 : 5602–5607 doi:10.1073/pnas.0510213103
35. SinapidouE, WilliamsK, NottL, BahktS, TörM, et al. (2004) Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J 38 : 898–909 doi:10.1111/j.1365-313X.2004.02099.x
36. el-LithyME, BentsinkL, HanhartCJ, RuysGJ, RovitoD, et al. (2006) New Arabidopsis recombinant inbred line populations genotyped using SNPWave and their use for mapping flowering-time quantitative trait loci. Genetics 172 : 1867–1876 doi:10.1534/genetics.105.050617
37. KranzA, KirchheimB (1987) Genetic resources in Arabidopsis. Arab Inf Serv 24 : 160–161.
38. WarthmannN, FitzJ, WeigelD (2007) MSQT for choosing SNP assays from multiple DNA alignments. Bioinformatics 23 : 2784–2787 doi:10.1093/bioinformatics/btm428
39. PlattA, HortonM, HuangYS, LiY, AnastasioAE, et al. (2010) The scale of population structure in Arabidopsis thaliana. PLoS Genet 6: e1000843 doi:10.1371/journal.pgen.1000843
40. BombliesK, YantL, LaitinenRA, KimS-T, HollisterJD, et al. (2010) Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet 6: e1000890 doi:10.1371/journal.pgen.1000890
41. FalushD, StephensM, PritchardJK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164 : 1567–1587.
42. LeisterD (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20 : 116–122.
43. McDowellJM, SimonSA (2006) Recent insights into R gene evolution. Mol Plant Pathol 7 : 437–448 doi:10.1111/j.1364-3703.2006.00342.x
44. CoatesME, BeynonJL (2010) Hyaloperonospora arabidopsidis as a pathogen model. Annu Rev Phytopathol 48 : 329–345 doi:10.1146/annurev-phyto-080508-094422
45. ChinDB, Arroyo-GarciaR, OchoaOE, KesseliRV, LavelleDO, et al. (2001) Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa). Genetics 157 : 831–849.
46. TodescoM, BalasubramanianS, HuTT, TrawMB, HortonM, et al. (2010) Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465 : 632–636 doi:10.1038/nature09083
47. SchmitzRJ, SchultzMD, UrichMA, NeryJR, PelizzolaM, et al. (2013) Patterns of population epigenomic diversity. Nature 495 : 193–198 doi:10.1038/nature11968
48. YuA, LepèreG, JayF, WangJ, BapaumeL, et al. (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci U S A 110 : 2389–2394 doi:10.1073/pnas.1211757110
49. BoccaraM, SarazinA, ThiébeauldO, JayF, VoinnetO, et al. (2014) The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP - and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog 10: e1003883 doi:10.1371/journal.ppat.1003883
50. GloggnitzerJ, AkimchevaS, SrinivasanA, KusendaB, RiehsN, et al. (2014) Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16 : 376–390 doi:10.1016/j.chom.2014.08.010
51. TackAJM, ThrallPH, BarrettLG, BurdonJJ, LaineA-L (2012) Variation in infectivity and aggressiveness in space and time in wild host-pathogen systems: causes and consequences. J Evol Biol 25 : 1918–1936 doi:10.1111/j.1420-9101.2012.02588.x
52. ThrallPH, LaineA-L, RavensdaleM, NemriA, DoddsPN, et al. (2012) Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecol Lett 15 : 425–435 doi:10.1111/j.1461-0248.2012.01749.x
53. GandonS (2002) Local adaptation and the geometry of host-parasite coevolution. Ecol Lett 5 : 246–256 doi:10.1046/j.1461-0248.2002.00305.x
54. McDowellJM, DhandaydhamM, LongTA, AartsMG, GoffS, et al. (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10 : 1861–1874.
55. BulgarelliD, RottM, SchlaeppiK, Ver Loren van ThemaatE, AhmadinejadN, et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488 : 91–95 doi:10.1038/nature11336
56. OssowskiS, SchwabR, WeigelD (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53 : 674–690 doi:10.1111/j.1365-313X.2007.03328.x
57. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743 doi:10.1046/j.1365-313x.1998.00343.x
58. KonczC, SchellJ (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. MGG Mol Gen Genet 204 : 383–396 doi:10.1007/BF00331014
59. EvannoG, RegnautS, GoudetJ (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14 : 2611–2620 doi:10.1111/j.1365-294X.2005.02553.x
60. HusonDH, BryantD (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23 : 254–267 doi:10.1093/molbev/msj030
61. LibradoP, RozasJ (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 : 1451–1452 doi:10.1093/bioinformatics/btp187
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



