-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNatural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
A central problem when studying adaptation to a new environment is the interplay between genetic variation and phenotypic plasticity. Arabidopsis thaliana has colonized a wide range of habitats across the world and it is therefore an attractive model for studying the genetic mechanisms underlying environmental adaptation. Here, we study two collections of A. thaliana accessions from across Eurasia to identify loci associated with differences in climates at the sampling sites. A new genome-wide association analysis method was developed to detect adaptive loci where the alleles tolerate different climate ranges. Sixteen novel such loci were found including a strong association between Chromomethylase 2 (CMT2) and temperature seasonality. The reference allele dominated in areas with less seasonal variability in temperature, and the alternative allele existed in both stable and variable regions. Our results thus link natural variation in CMT2 and epigenetic changes to temperature adaptation. We showed experimentally that plants with a defective CMT2 gene tolerate heat-stress better than plants with a functional gene. Together this strongly suggests a role for genetic regulation of epigenetic modifications in natural adaptation to temperature and illustrates the importance of re-analyses of existing data using new analytical methods to obtain deeper insights into the underlying biology from available data.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004842
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004842Summary
A central problem when studying adaptation to a new environment is the interplay between genetic variation and phenotypic plasticity. Arabidopsis thaliana has colonized a wide range of habitats across the world and it is therefore an attractive model for studying the genetic mechanisms underlying environmental adaptation. Here, we study two collections of A. thaliana accessions from across Eurasia to identify loci associated with differences in climates at the sampling sites. A new genome-wide association analysis method was developed to detect adaptive loci where the alleles tolerate different climate ranges. Sixteen novel such loci were found including a strong association between Chromomethylase 2 (CMT2) and temperature seasonality. The reference allele dominated in areas with less seasonal variability in temperature, and the alternative allele existed in both stable and variable regions. Our results thus link natural variation in CMT2 and epigenetic changes to temperature adaptation. We showed experimentally that plants with a defective CMT2 gene tolerate heat-stress better than plants with a functional gene. Together this strongly suggests a role for genetic regulation of epigenetic modifications in natural adaptation to temperature and illustrates the importance of re-analyses of existing data using new analytical methods to obtain deeper insights into the underlying biology from available data.
Introduction
Arabidopsis thaliana has colonized a wide range of habitats across the world and it is therefore an attractive model for studying the genetic mechanisms underlying environmental adaptation [1]. Several large collections of A. thaliana accessions have either been whole-genome re-sequenced or high-density SNP genotyped [1]–[7]. The included accessions have adapted to a wide range of different climatic conditions and therefore loci involved in climate adaptation will display genotype by climate-at-sampling-site correlations in these populations. Genome-wide association or selective-sweep analyses can therefore potentially identify signals of natural selection involved in environmental adaptation, if those can be disentangled from the effects of other population genetic forces acting to change the allele frequencies. Selective-sweep studies are inherently sensitive to population-structure and, if present, the false-positive rates will be high as the available statistical methods are unable to handle this situation properly. Further experimental validation of inferred sweeps (e.g. [1], [8]) is hence necessary to suggest them as adaptive. In GWAS, kinship correction is now a standard approach to account for population structure that properly controls the false discovery rate. Unfortunately, correcting for genomic kinship often decreases the power to detect individual adaptive loci, which is likely the reason that no genome-wide significant associations to climate conditions were found in earlier GWAS analyses [1], [8]. Nevertheless, a number of candidate adaptive loci could despite this be identified using extensive experimental validation [1], [2], [8], showing how valuable these populations are as a resource for finding the genomic footprint of climate adaptation.
Genome-wide association (GWA) datasets based on natural collections of A. thaliana accessions, such as the RegMap collection, are often genetically stratified. This is primarily due to the close relationships between accessions sampled at nearby locations. Furthermore, as the climate measurements used as phenotypes for the accessions are values representative for the sampling locations of the individual accessions, these measurements will be confounded with the general genetic relationship [9]. Unless properly controlled for, this confounding might lead to excessive false-positive signals in the association analysis; this as the differences in allele-frequencies between loci in locations that differ in climate, and at the same time are geographically distant, will create an association between the genotype and the trait. However, this association could also be due to other forces than selection. In traditional GWA analyses, mixed-model based approaches are commonly used to control for population-stratification. The downside of this approach is that it, in practice, will remove many true genetic signals coming from local adaptation due to the inherent confounding between local genotype and adaptive phenotype. Instead, the primary signals from such analyses will be due to effects of alleles that exist in, and have similar effects across, the entire studied population. In general, studies into the contributions of genetic variance-heterogeneity to the phenotypic variability in complex traits is a novel and useful approach with great potential [10]. Here, we have developed and used a new approach that combines a linear mixed model and a variance-heterogeneity test, which addresses these initial concerns and shown that it is possible to infer statistically robust results of genetically regulated phenotypic variability in GWA data from natural populations.
This study describes the results from a re-analysis of data from the RegMap collection to find loci contributing to climate adaptation through an alternative mechanism: genetic control of plasticity. Such loci are unlikely to be detected with standard GWAS or selective-sweep analyses as they have a different genomic signature of selection and distribution across climate envelopes. The reason for this difference is that plastic alleles are less likely to be driven to fixation by directional selection, but rather that multiple alleles remain in the population under extended periods of time by balancing selection [11]. To facilitate the detection of such loci, we extend and utilize an approach [12], [13] that instead of mapping loci by differences in allele-frequencies between local environments, which is highly confounded by population structure, infer adaptive loci using a heterogeneity-of-variance test. This identifies loci where the minor allele is associated with a broader range of climate conditions than the major allele [12]. As such widely distributed alleles will be present across the entire population, they are less confounded with population structure and detectable in our GWAS analysis that utilizes kinship correction to account for population stratification.
Results
Genome-wide association analysis to detect loci with plastic response to climate
A genome-wide association analysis was performed for thirteen climate variables across ∼215,000 SNPs in 948 A. thaliana accessions from the RegMap collection, representing the native range of the species [1], [9]. In total, sixteen genome-wide significant loci were associated with eight climate variables (Table 1), none of which could be found using standard methods for GWAS analyses [1], [8], [14]–[16]. The effects were in general quite large, from 0.3 to 0.5 residual standard deviations (Table 1), meaning that the minor allele is associated with a climate that is between 21–35% more variable than that of the major allele. The detailed results from the association analysis for each of these climate variables are reported in S1 Figure–S13 Figure. As expected, there was low confounding between the alleles associated with a broader range of climate conditions and population structure. This is illustrated by the plots showing the distributions of these alleles across the population strata in relation to their geographic origin and the climate envelopes in S14 Figure–S35 Figure.
Tab. 1. Loci with genome-significant, non-additive effects on climate adaptation and a functional analysis of nearby genes (r2>0.8) containing missense or nonsense mutations. 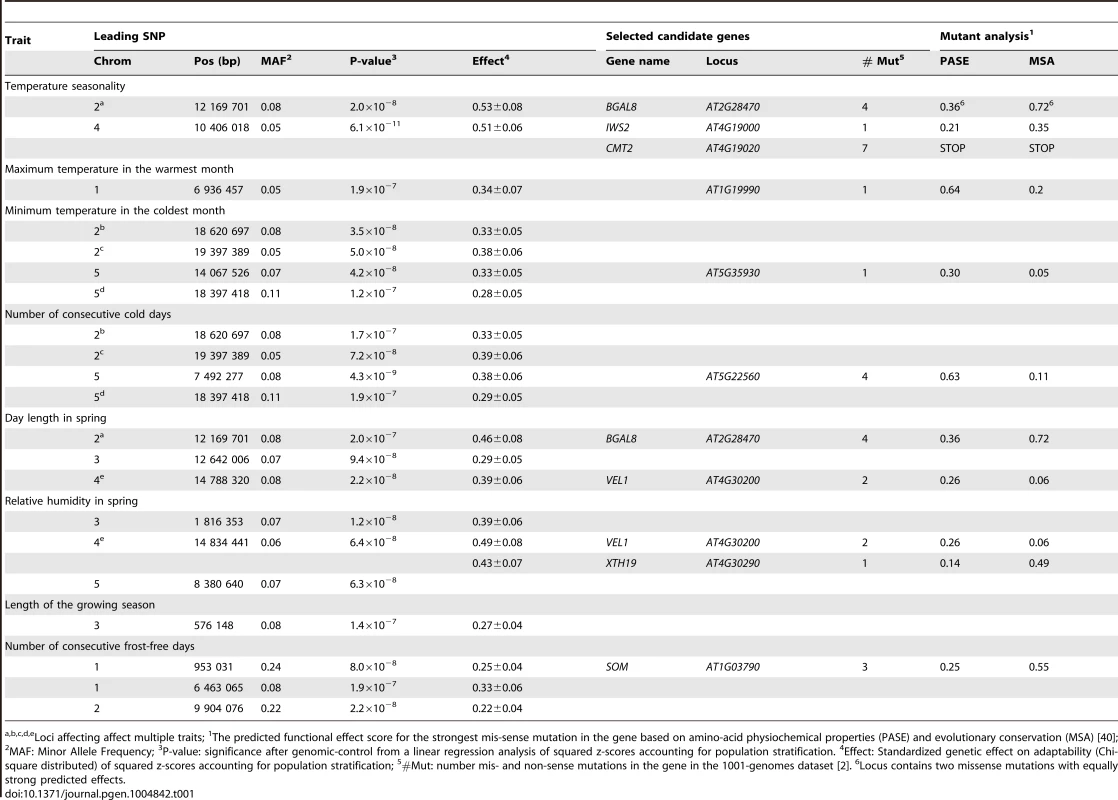
Loci affecting affect multiple traits; 1The predicted functional effect score for the strongest mis-sense mutation in the gene based on amino-acid physiochemical properties (PASE) and evolutionary conservation (MSA) [40]; 2MAF: Minor Allele Frequency; 3P-value: significance after genomic-control from a linear regression analysis of squared z-scores accounting for population stratification. 4Effect: Standardized genetic effect on adaptability (Chi-square distributed) of squared z-scores accounting for population stratification; 5#Mut: number mis- and non-sense mutations in the gene in the 1001-genomes dataset [2]. 6Locus contains two missense mutations with equally strong predicted effects. Identification of candidate mutations using re-sequencing data from the 1001-genomes project
Utilizing the publicly available whole-genome re-sequencing data from the 1001-genomes project [2]–[7] (http://1001genomes.org), we screened the loci with significant associations to the climate variables for candidate functional polymorphisms. Missense, nonsense or frameshift mutations in high linkage disequilibrium (LD; r2>0.8) with the leading SNPs were identified in five functional candidate genes associated with eight climate variables (for details on these see Table 1) and 11 less characterized genes (S1 Table). S2 Table provides 76 additional linked loci or genes without candidate mutations in their coding regions.
Several loci are associated with multiple climate variables
Interestingly, three out of the eight loci with missense mutations affected more than one climate variable, even though these were only marginally correlated. One such potentially pleiotropic adaptive effect for day length and relative humidity in the spring was associated with a locus containing the genes VEL1 and XTH19 (Table 1). The major allele at this locus was predominant in short-day regions, whereas the alternative allele was more plastic in relation to day-length. XTH19 has been implied as a regulator of shade avoidance [17], but information about its potential involvement in regulation of photoperiodic length is lacking. VEL1, is a Plant Homeo Domain (PHD) finger protein. PHD finger proteins are known to affect vernalization and flowering of A. thaliana, e.g. by silencing the key flowering locus FLC during vernalization, and is involved in photoperiod-mediated epigenetic regulation of MAF5 [18]–[20]. The finding that VEL1 is associated with day length and relative humidity is thus consistent with the role of previous reports on PHD finger proteins. It also makes this protein an interesting target for future studies into the genetics underlying simultaneous adaptation to day-length and humidity.
Another potentially pleiotropic adaptive effect was identified for two more highly correlated traits, minimum temperature and number of consecutive cold days (Pearson's r2 = 0.76). In total, 17 missense mutations were found at this locus. The top candidate gene containing a missense mutation is galactinol synthase 1 (GolS1). This gene has been reported to be involved in extreme temperature-induced synthesis [21], [22], making it an interesting target for further studies regarding the genetics of temperature adaptation.
Chromomethylase 2 (CMT2) is associated with temperature seasonality in the RegMap collection
A strong association to temperature seasonality, i.e. the ratio between the standard deviation and the mean of temperature records over a year, was identified near Chromomethylase 2 (CMT2; Table 1; Fig. 1). Stable areas are generally found near large bodies of water (e.g. London near the Atlantic 11±5°C; mean ± SD) and variable areas inland (e.g. Novosibirsk in Siberia 1±14°C). A premature CMT2 stop codon located on chromosome 4 at 10,414,556 bp (the 31st base pair of the first exon) segregated in the RegMap collection, with minor allele frequency of 0.05. This CMT2STOP allele had a genome-wide significant association with temperature seasonality (P = 1.1×10−7) and was in strong LD (r2 = 0.82) with the leading SNP (Fig. 1B). The geographic distributions of the wild-type (CMT2WT) and the alternative (CMT2STOP) alleles in the RegMap collection shows that the CMT2WT allele dominates in all major sub-populations sampled from areas with low or intermediate temperature seasonality. The plastic CMT2STOP allele is present, albeit at lower frequency, across all sub-populations in low - and intermediate temperature seasonality areas, and is more common in areas with high temperature seasonality (Fig. 2A; Fig. 3; S36 Figure). Such global distribution across the major population strata indicates that the allele has been around in the Eurasian population sufficiently long to spread across most of the native range and that the allele is not deleterious but rather maintained through balancing selection [11], perhaps by mediating an improved tolerance to variable temperatures.
Fig. 1. An LD block associated with temperature seasonality contains CMT2. 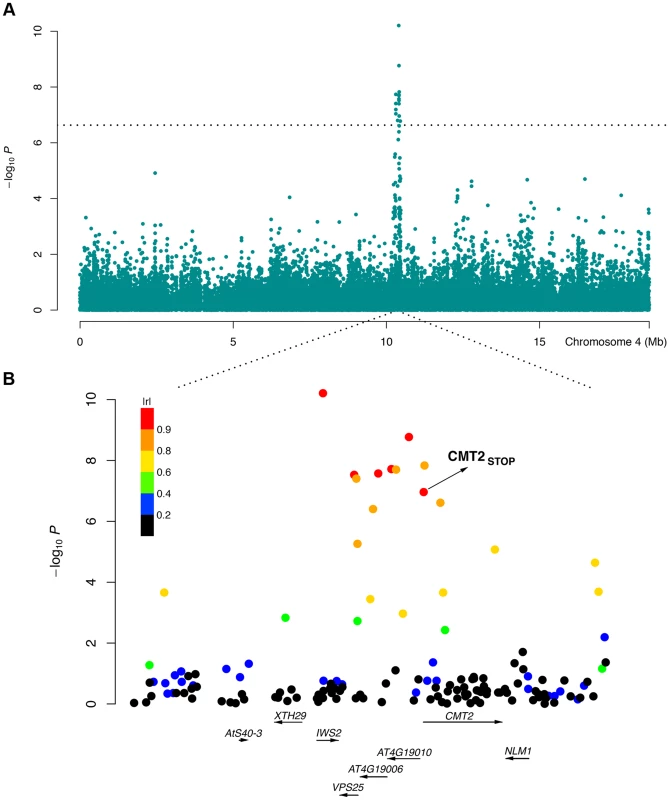
A genome-wide significant variance-heterogeneity association signal was identified for temperature seasonality in the RegMap collection of natural Arabidopsis thaliana accessions [1]. The peak on chromosome 4 around 10 Mb (A) mapped to a haplotype block (B) containing a nonsense mutation (CMT2STOP) early in the first exon of the Chromomethylase 2 (CMT2) gene. Color coding based on |r| (the absolute value of the correlation coefficient) as a measure of LD between each SNP in the region and the leading SNP in the association analysis. Fig. 2. Geographic distribution of, and heterogenous variance for, three CMT2 alleles in two collections of A. thaliana accessions. 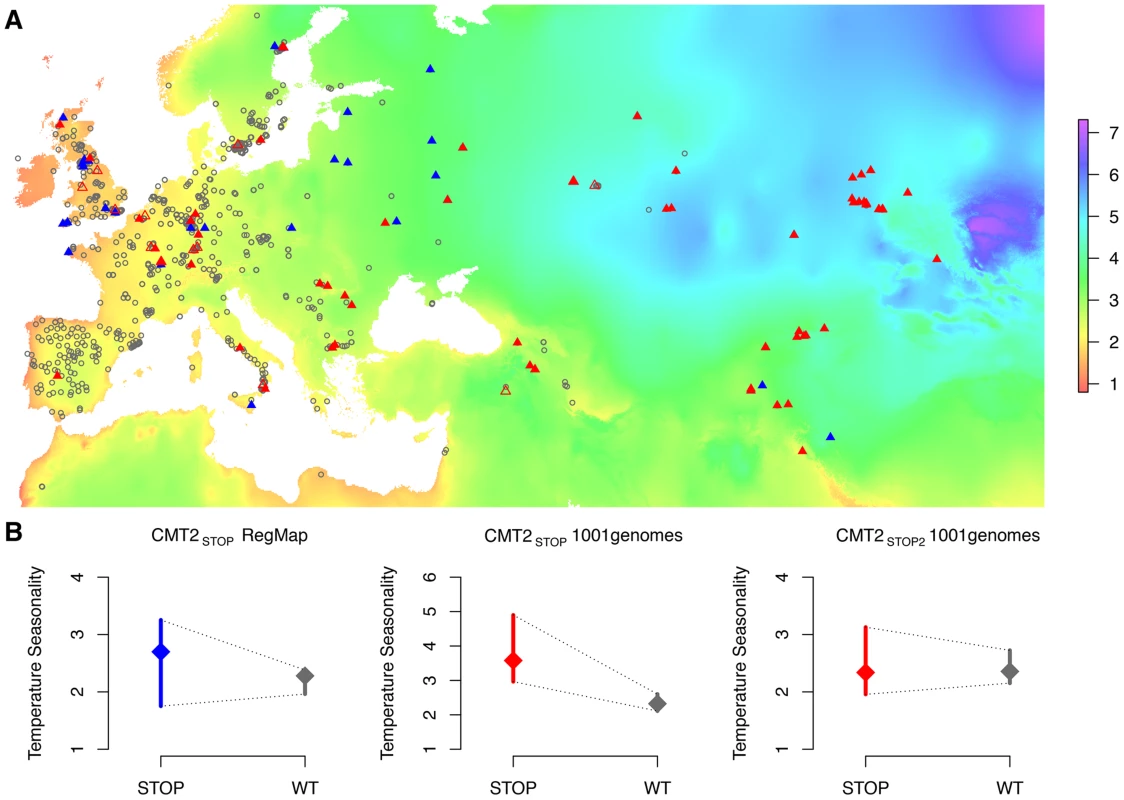
The geographic distributions (A) of the wild-type (CMT2WT; gray open circles) and two nonsense alleles (CMT2STOP/CMT2STOP2; filled/open triangles) in the CMT2 gene that illustrates a clustering of CMT2WT alleles in less variable regions and a greater dispersion of the nonsense alleles across different climates both in the RegMap [1] (blue) and the 1001-genomes [2](red) A. thaliana collections. The resulting variance-heterogeneity in temperature seasonality between genotypes is highly significant, as illustrated by the quantile plots in (B) where the median is indicated by a diamond and a bar representing the 25% to 75% quantile range. The color scale indicate the level of temperature seasonality across the map. The colorkey in (A) represent the temperature seasonality values, given as the standard-deviation in % of the mean temperature (K). Fig. 3. Principle components of the genomic kinship in the RegMap collection for the accessions carrying the alternative alleles at the Chromomethylase 2 locus (CMT2STOP and CMT2WT as filled and empty circles, respectively). 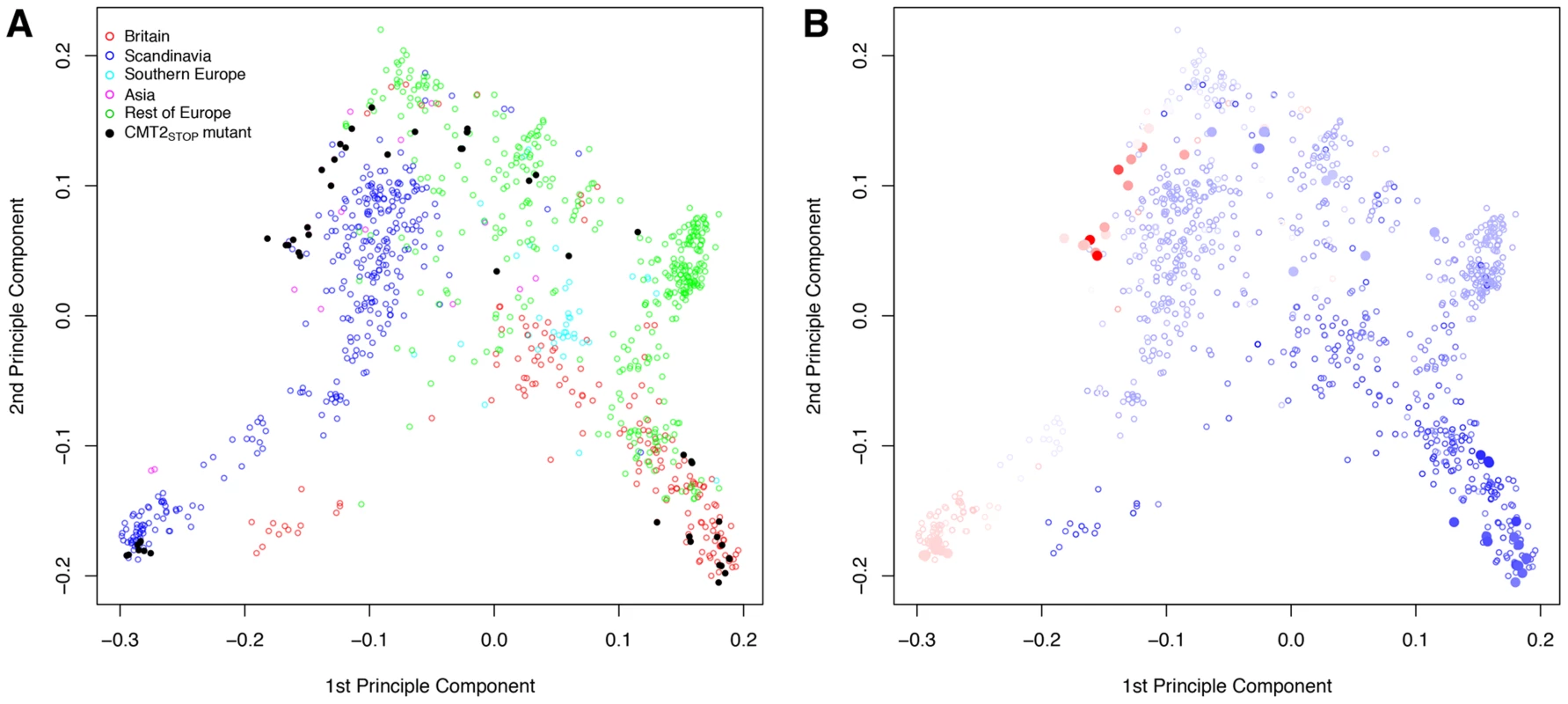
Coloring is based on (A) geographical regions (defined as in Figure S37) and (B) temperature seasonality, ranging from dark blue (least variable) to red (most variable). Broader geographic distribution of the CMT2STOP allele in the 1001-genomes collection
To confirm that the CMT2STOP association was not due to sampling bias in the RegMap collection, we also scored the CMT2 genotype and collected the geographical origins from 665 accessions that were part of the 1001-genomes project (http://1001genomes.org) [2], [3], [5]-[7]. In this more geographically diverse set (Fig. 2A), CMT2STOP was more common (MAF = 0.10) and had a similar allele distribution across Eurasia as in RegMap (Figure S36–S37). Two additional mutations were identified on unique haplotypes (r2 = 0.00) - one nonsense CMT2STOP2 at 10,416,213 bp (MAF = 0.02) and a frameshift mutation at 10,414,640 bp (two accessions). Both CMT2STOP and CMT2STOP2 had genotype-phenotype maps implying a plastic response to variable temperature (Fig. 2B) and the existence of multiple mutations disrupting CMT2 further suggest lack of CMT2 function as a potentially evolutionary beneficial event [23].
Accessions with the CMT2STOP allele has an altered genome-wide CHH-methylation pattern
CMT2 is a plant DNA methyltransferase that methylates mainly cytosines in CHH (H = any base but G) contexts, predominantly at transposable elements (TEs) [24], [25]. We tested the effect of CMT2STOP on genome-wide DNA methylation using 135 CMT2WT and 16 CMT2STOP accessions, for which high-quality MethylC-sequencing data was publicly available [7]. In earlier studies [24], [25], it has been shown that CMT2-mediated CHH methylation primarily affects TE-body methylation. In cmt2 knockouts in a Col-0 genetic background, this results in a near lack of CHH methylation at such sites. Here, we compared the levels of CHH-methylation across TEs between CMT2STOP and CMT2WT accessions. Our analyses revealed that the accessions carrying the CMT2STOP allele on average had a small (1%) average decrease in CHH-methylation across the TE-body compared to the CMT2WT accessions. A more detailed analysis showed that this difference was primarily due to two of 16 CMT2STOP accessions, Kz-9 and Neo-6, showing a TE-body CHH methylation pattern resembling that of the cmt2 knockouts in the data of [24]. Interestingly, none of the 135 CMT2WT accessions displayed such a decrease in TE-body CHH methylation, and hence there is a significant increase in the frequency of the cmt2 knockout TE-body CHH methylation pattern among the natural CMT2STOP accessions (P = 0.01; Fisher's exact test). Our analyses show that the methylation-pattern is more heterogeneous among the natural accessions than within the Col-0 accession, both for the CMT2STOP and CMT2WT accessions (both P = 0.01; Brown-Forsythe heterogeneity of variance test; Fig. 4). There is thus a significant association between the CMT2STOP polymorphism and decreased genome-wide TE-body CHH-methylation levels, and we show that this is apparently due to an increased frequency of the cmt2-mutant methylation phenotype. Further, the results also show a variable contribution of CMT2-independent CHH methylation pathways in the natural accessions. The reason why not all CMT2STOP accessions behave like null alleles is unclear, but the variability amongst in the level of CHH-methylation across the natural accessions suggest that it is possible that CMT2-independent pathways, such as the RNA-dependent DNA-methylation pathway, compensate for the lack of CMT2 due to segregating polymorphisms also at these loci. Alternatively, CMT2STOP alleles may not be null, maybe due to stop codon read-through, which is more common than previously thought [26]. Although our analyses of genome-wide methylation data have established that CMT2STOP allele has a quantitative effect on CHH methylation, further studies are needed to fully explore the link between the CMT2STOP allele, other pathways affecting genome-wide DNA-methylation and their joint contributions to the inferred association to temperature seasonality.
Fig. 4. Comparison of CHH methylation patterns inside TE-bodies, (A) between CMT2WT and CMT2STOP accessions using the data from [7], and (B) between four replicate Col-0 wild-type and cmt2 knock-outs from [24]. ![Comparison of CHH methylation patterns inside TE-bodies, (A) between <i>CMT2<sub>WT</sub></i> and <i>CMT2<sub>STOP</sub></i> accessions using the data from <em class="ref">[7]</em>, and (B) between four replicate Col-0 wild-type and <i>cmt2</i> knock-outs from <em class="ref">[24]</em>.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/6ef6977ac87cb6ac1cda11b64f678520.png)
For each accession, the curve is to illustrate the moving average methylation level in a sliding 100 bp window. On the x-axis, the two different strands of DNA are aligned in the middle, truncated at 5 kb from the edge of the TEs. Cmt2 mutant plants have an improved heat-stress tolerance
To functionally explore whether CMT2 is a likely contributor to the temperature-stress response, we have subjected cmt2 mutants to two types of heat-stress. First, we tested the reaction of Col-0 and the cmt2-5 null mutant (S45 Figure) to severe heat-stress (24 h at 37°C). This treatment was used because it can release transcriptional silencing of some TEs [27] and could thus be a good starting point to evaluate potential stress effects on cmt2. Under these conditions, the cmt2 mutant had significantly higher survival-rate (1.6-fold; P = 9.1×10−3; Fig. 5A) than Col-0. To evaluate whether a similar response could also be observed under less severe, non-lethal stress, we subjected the same genotypes to heat-stress of shorter duration (6 h at 37°C) and measured root growth after stress as a measure of the ability of plants to recover. Also under these conditions, the cmt2 mutant was found to be more tolerant to heat-stress, as its growth was less affected after being stressed (Fig. 5B; 1.9-fold higher in cmt2; P = 0.026, one-sided t-test). This striking improvement in tolerance to heat-stress of cmt2 plants suggests CMT2-dependent CHH methylation as an important alleviator of stress responses in A. thaliana and a candidate mechanism for temperature adaptation.
Fig. 5. cmt2 mutant plants display an increased tolerance to heat-stress. 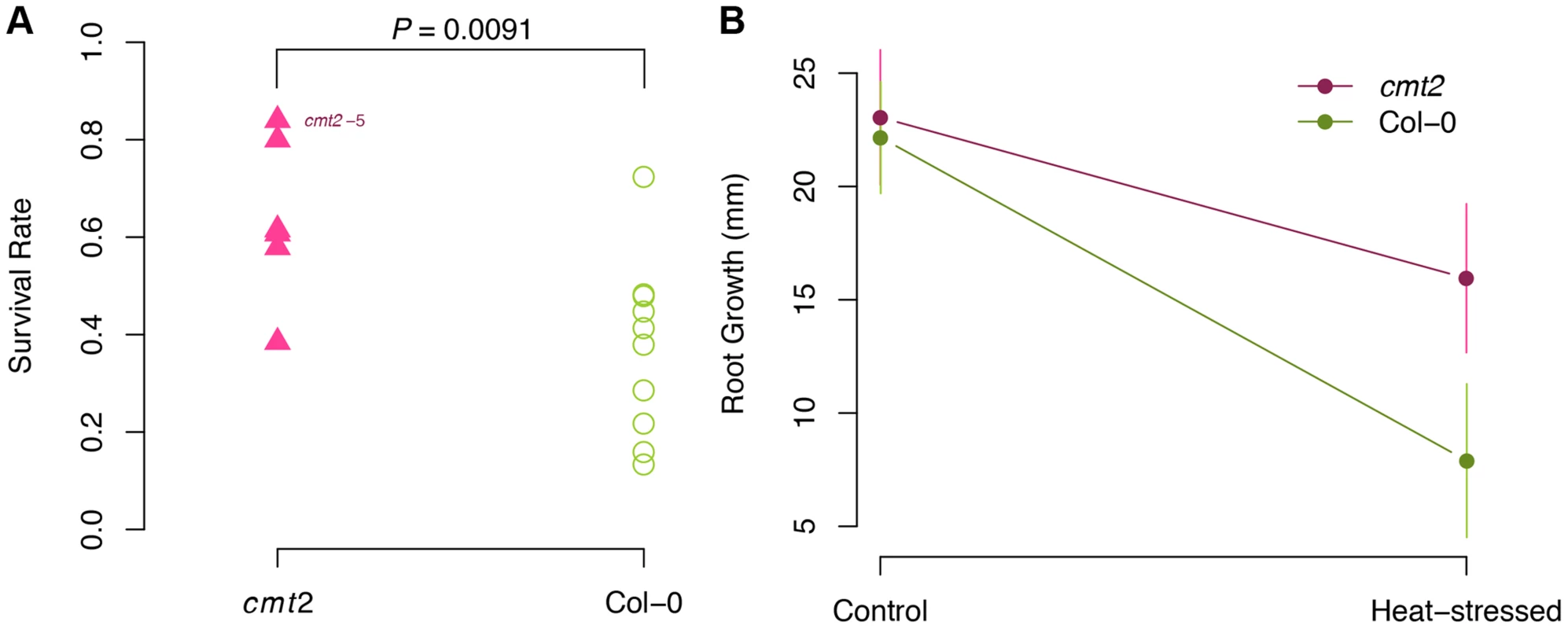
A. The survival rate is significantly higher for cmt2-5 mutant than for Col-0 plants under severe heat-stress (24 h at 37.5°C). P-values in A were obtained using a log-linear regression. B. The cmt2-5 mutant was also more tolerant to less severe heat-stress heat-stress (6 h at 37.5°C) than Col-0, here illustrated by its significantly faster growth of the root (P = 0.026; one-sided t-test) during the first 48 h following heat stress. The CMT2STOP allele is associated with increased leaf serration and higher disease presence after bacterial inoculation
To also explore the potential effects of the CMT2STOP allele on other phenotypes measured in collections of natural accessions, we tested for associations between this CMT2 polymorphism and the 107 phenotypes measured as part of a previous study [28]. Three phenotypes were found to be significantly associated with the genotype at this locus (S39 Figure).
Associations were found to two phenotypes related to disease presence following inoculation with Pseudomonas viridiflava (strains PNA3.3a and ME3.1b; P = 4.8×10−3 and P = 1.3×10−4, respectively). Scoring of disease was done by eye four days after inoculation in 6 replicates per strain × accession using a scale from 0 (no visible symptom) to 10 (leaves collapse and turn yellow) with an increment of 1 [28]. The connection between an increased susceptibility (0.6 and 0.7 units for PNA3.3a and ME3.1b, respectively) to disease and an increased tolerance to temperature seasonality is not obvious. However, recent work by [29] has shown that widespread dynamic CHH-methylation is important for the response to Pseudomonas syringae infection. In light of this finding, it is therefore not unlikely that these phenotypes are functionally related via an altered CMT2-mediated CHH-methylation in response to abiotic and biotic stress.
An association was also found for the level of leaf serration (increase by 0.23 units for the CMT2STOP allele; P = 3.3×10−3), determined after growth for 8 weeks at 10°C (level from 0: entire lamina, to 1.5: sharp/jagged serration), across 4 plants per accession [28]. Measures of leaf serration were also available at 16 and 22°C, and interestingly there was a significant CMT2 genotype × temperature interaction (P = 0.048). The CMT2STOP accessions have the same level of serration across the three measured temperatures, whereas the level of serration decreases with temperature for the CMT2WT accessions (S38 Figure). Although we are not aware of any earlier results connecting leaf serration to the CMT2 locus or the level of CHH-methylation in the plant, this result further indicates that the effects of the CMT2STOP and the CMT2WT alleles depend on temperature.
Discussion
A major challenge in attempts to identify individual loci involved in climate adaptation is the strong confounding between geographic location, climate and population structure in the natural A. thaliana population. Earlier genome-wide association analyses in large collections of natural accessions experienced a lack of statistical power when correcting for population-structure [1], [8]. We used an alternative GWAS approach [12] to test for a variance-heterogeneity, instead of a mean difference, between genotypes. This analysis identifies loci where the minor allele is more plastic (i.e. exist across a broader climatic range) than the major allele. As it has low power to detect cases where the minor allele is associated with a lower variance (here with local environments), it will not map private alleles in local environments in a genome-wide analysis [12], [30]. In contrast, a standard GWAS map loci where the allele-frequencies follow the climatic cline. Although plastic alleles might be less frequent in the genome, they are easier to detect in this data due to their lower confounding with population-structure. This overall increase in power is also apparent when comparing the signals that reach a lower, sub-GWAS significance level (S40 Figure–S44 Figure).
Several novel genome-wide significant associations were found to the tested climate variables, and a locus containing VEL1 was associated to both day length and relative humidity in the spring. A thaliana is a facultative photoperiodic flowering plant and hence non-inductive photoperiods will delay, but not abolish, flowering. A genetic control of this phenotypic plasticity is thus potentially an adaptive mechanism. VEL1 regulates the epigenetic silencing of genes in the FLC-pathway in response to vernalization [19] and photoperiod length [20] resulting in an acceleration of flowering under non-inductive photoperiods. Our results suggest that genetically plastic regulation of flowering, via the high-variance VEL1 allele, might be beneficial under short-day conditions where both accelerated and delayed flowering is allowed. In long-daytime areas, accelerated flowering is potentially detrimental hence the wild-type allele has the highest adaptive value. It can be speculated whether this is connected to the fact that day-length follows a latitudinal cline, where early flowering might be detrimental in northern areas where accelerated flowering, when the day-length is short, could lead to excessive exposure to cold temperatures in the early spring and hence a lower fitness.
A particularly interesting finding in our vGWAS was the strong association between the CMT2-locus and temperature seasonality. Here the allele associated with higher temperature seasonality (i.e the plastic allele) had an altered genome-wide CHH methylation pattern where some accessions displayed a TE-body CHH methylation pattern similar to that of cmt2 mutant plants. Interestingly, a recent study by Dubin et al. [31] in a collection of Swedish A. thaliana accessions report that CHH methylation is temperature sensitive, and that the CMT2-locus is a major trans-acting controller of the observed variation in genome-wide CHH-methylation between the accessions. These findings, together with our experimental work showing that cmt2 mutants were more tolerant to both mild and severe heat-stress, strongly implicate CMT2 as an adaptive locus and clearly illustrate the potential of our method as a useful approach to identify novel associations of functional importance.
It is not clear via which mechanism CMT2-dependent CHH methylation might affect plant heat tolerance. Although our results show that the CMT2STOP allele is present across regions with both low and high temperature seasonality, it remains to be shown whether this is due to this allele being generally more adaptable across all environments, or whether the CMT2WT allele is beneficial in environments with stable temperature and the CMT2STOP in high temperature seasonality areas. Regardless, we consider it most likely that the effect will be mediated by TEs in the immediate neighborhood of protein-coding genes. Heterochromatic states at TEs can affect activity of nearby genes and thus potentially plant fitness [32]. Consistent with a repressive role of CMT2 on heat stress responses, CMT2 expression is reduced by several abiotic stresses including heat [33]. Because global depletion of methylation has been shown to enhance resistance to biotic stress [29], it is possible that DNA-methylation has a broader function in shaping stress responses than currently thought.
Our results show that CMT2STOP accessions have more heterogeneous CHH methylation patterns than CMT2WT accessions. The CMT2STOP polymorphism is predicted to lead to a non-functional CMT2 protein, and hence a genome-wide CHH-methylation profile resembling that of a complete cmt2 mutant [24]. Although some of the accessions carrying the CMT2STOP allele displayed this pattern with a lower CHH-methylation inside TE-bodies, most of these accessions did not have any major loss of genome-wide CHH methylation. Such heterogeneity might indicate the presence of compensatory mechanisms and hence that the effects of altered CMT2 function could be dependent on the genetic-background. This is an interesting finding that deserves further investigation, although such work is beyond the scope of the current study. Our interpretation of the available results is that our findings reflect the genetic heterogeneity among the natural accessions studied. In light of the recent report by [25], who showed a role also of CMT3 in TE-body CHH methylation, it is not unlikely that the regulation of CHH methylation may result from the action and interaction of several genes.
We identified several alleles associated with a broader range of climates across the native range of A. thaliana, suggesting that a genetically mediated plastic response might of important for climate adaptation. Using publicly available data from several earlier studies, we were able to show that an allele at the CMT2 locus displays an altered genome-wide CHH-methylation pattern was strongly associated with temperature seasonality. Using additional experiments, we also found that cmt2 mutant plants tolerated heat-stress better than wild-type plants. Together, these findings suggest this genetically determined epigenetic variability as a likely mechanism contributing to a plastic response to the environment that has been of adaptive advantage in natural environments.
Materials and Methods
Climate data and genotyped Arabidopsis thaliana accessions
Climate phenotypes and genotype data for a subset of the A. thaliana RegMap collection were previously analyzed by [1]. We downloaded data on 13 climate variables and genotypes of 214,553 single nucleotide polymorphisms (SNPs) for 948 accessions from: http://bergelson.uchicago.edu/regmap-data/climate-genome-scan. The climate variables used in the analyses were: aridity, number of consecutive cold days (below 4 degrees Celsius), number of consecutive frost-free days, day-length in the spring, growing-season length, maximum temperature in the warmest month, minimum temperature in the coldest month, temperature-seasonality, photosynthetically active radiation, precipitation in the wettest month, precipitation in the driest month, precipitation-seasonality, and relative humidity in the spring. More information on these variables is provided by [1]. No squared pairwise Pearson's correlation coefficients between the phenotypes were greater than 0.8 (S7 Figure of [1]).
We calculated the temperature seasonality for at sampling locations of a selection of 1001-genomes (http://1001genomes.org) accessions. Raw climate data was downloaded from http://www.worldclim.org/, re-formatted and thereafter processed by the raster package in R. The R code for generating this data is provided in S1 Text. The genotype for the CMT2STOP polymorphism was obtained by extracting the corresponding SNP data for the 1001-genomes accessions.
Statistical modeling in genome-wide scans for adaptability
The climate data at the geographical origins of the A. thaliana accessions were treated as phenotypic responses. Each climate phenotype vector for all the accessions was normalized via an inverse-Gaussian transformation. The squared normalized measurement of accession is modeled by the following linear mixed model to test for an association with climate adaptability (i.e. a greater plasticity to the range of the environmental condition):
where is an intercept, the SNP genotype for accession , the genetic SNP effect, the polygenic effects and the residuals. is coded 0 and 2 for the two homozygotes (inbred lines). The genomic kinship matrix is constructed via the whole-genome generalized ridge regression method HEM (heteroscedastic effects model) [13] as , where is a number of individuals by number of SNPs matrix of genotypes standardized by the allele frequencies. is a diagonal matrix with element for the j-th SNP, where is the SNP-BLUP (SNP Best Linear Unbiased Prediction) effect estimate for the j-th SNP from a whole-genome ridge regression, and is the hat-value for the j-th SNP. Quantities in can be directly calculated using the bigRR package [13] in R. An example R source code for performing the analysis is provided in S1 Text.The advantage of using the HEM genomic kinship matrix , rather than an ordinary genomic kinship matrix , is that HEM is a significant improvement of the ridge regression (SNP-BLUP) in terms of the estimation of genetic effects [13], [34]. Due to this, the updated genomic kinship matrix better represents the relatedness between accessions and also accounts for the genetic effects of the SNPs on the phenotype.
Testing and quality control for association with climate adaptability
The test statistic for the SNP effect is constructed as the score statistic [35]:
implemented in the GenABEL package [36], where are the centered genotypic values and the centered phenotypic measurements. The statistic has an asymptotic distribution with 1 degree of freedom. Subsequent genomic control (GC) [37] of the genome-wide association results was performed under the null hypothesis that no SNP has an effect on the climate phenotype. SNPs with minor allele frequency (MAF) less than 0.05 were excluded from the analysis. A 5% Bonferroni-corrected significance threshold was applied. As suggested by [30], the significant SNPs were also analyzed using a Gamma generalized linear model to exclude positive findings that might be due to low allele frequencies of the high-variance SNP.Statistical testing for associations between the CMT2STOP polymorphism and phenotypes measured in a collection of natural accessions
The CMT2STOP genotype was extracted from the publicly available genome-wide genotype data with 107 phenotype measured from [28]. The association between the CMT2STOP genotype and each phenotype was tested by fitting a normal linear mixed model to account for population stratification, where the genomic kinship matrix was calculated by the ibs(, weight = 'freq') procedure in the GenABEL package [36], and the linear mixed model was fitted using the hglm package [38].
Functional analysis of polymorphisms in loci with significant genome-wide associations to climate
All the loci that showed genome-wide significance in the association study was further characterized using the genome sequences of 728 accessions sequenced as part of the 1001-genomes project (http://1001genomes.org). Mutations within a ±100Kb interval of each leading SNP and that are in LD with the leading SNP (r2>0.8) were reported (S1 Table). The consequences of the identified polymorphisms were predicted using the Ensembl variant effect predictor [39] and their putative effects on the resulting protein estimated using the PASE (Prediction of Amino acid Substitution Effects) tool [40].
Evaluation of TE-body methylation of CMT2STOP and CMT2WT natural accessions
In a previous study, the methylation levels were scored at 43,182,344 sites across the genome using MethylC-sequencing in 152 natural A. thaliana accessions (data available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43857) [7]. 135 of these accessions carried the CMT2WT and 17 the CMT2STOP alleles. Upon further inspection, the accession Rd-0 was excluded as it did not have sufficient sequence coverage to be used in the analyses. For each accession, across all TEs, moving averages of the CHH methylation level were calculated using a 100 bp sliding window from the borders of the TEs. The same analysis was also performed for four wild-type and four cmt2 knockout accessions (data available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41302) [24]. The results showing the TE-body CHH methylation patterns are visualized in Fig. 4.
Heat-stress treatments on cmt2 knockouts and natural CMT2STOP accessions
A CMT2 T-DNA insertion line (SAIL_906_G03, cmt2-5 [24], [41]) was ordered from NASC. Seeds of Col-0 wild-type and cmt2-5 was then used for heat stress experiments based on a previously described protocol [27]. This treatment was used because it was shown to interfere with epigenetic gene silencing as evident from transcription of some TE [27]. Seeds were plated on ½ MS medium (0.8% agar, 1% sucrose), stratified for two days at 4°C in the dark and transferred to a growth chamber with 16 h light (110 µmol m−2 s−1, 22°C) and 8 h dark (20°C) periods. Ten-day-old seedlings were transferred to 4°C for one hour and subsequently placed for 6 h or 24 h at 37.5°C in the dark. Plant survival was scored two days after 24 h of heat stress with complete bleaching of shoot apices as lethality criterion (S46 Figure). Experiments were repeated six times, each with ∼30 plants per genotype. Root length was measured immediately before the 6 h heat stress and two days after heat stress.
A log-linear regression was conducted to test for the difference in survival rate between Col-0 and cmt2-5 knockout, i.e.
where is the number of surviving plants of accession , the corresponding total number of plants, the experiment effect, the accession effect, and an intercept. The model fitting procedure was implemented using the glm() procedure in R, with option family = gaussian(link = log), as response, as offset, and , , as fixed effects.Supporting Information
Zdroje
1. HancockAM, BrachiB, FaureN, HortonMW, JarymowyczLB, et al. (2011) Adaptation to Climate Across the Arabidopsis thaliana Genome. Science 334 : 83–86.
2. CaoJ, SchneebergerK, OssowskiS, GüntherT, BenderS, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43 : 956–963 doi:10.1038/ng.911
3. OssowskiS, SchneebergerK, ClarkRM, LanzC, WarthmannN, et al. (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Research 18 : 2024–2033 doi:10.1101/gr.080200.108
4. SchneebergerK, HagmannJ, OssowskiS, WarthmannN, GesingS, et al. (2009) Simultaneous alignment of short reads against multiple genomes. Genome Biol 10: R98 doi:10.1186/gb-2009-10-9-r98
5. SchneebergerK, OssowskiS, OttF, KleinJD, WangX, et al. (2011) Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proceedings of the National Academy of Sciences 108 : 10249–10254 doi:10.1073/pnas.1107739108
6. LongQ, RabanalFA, MengD, HuberCD, FarlowA, et al. (2013) Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45 : 884–890 doi:10.1038/ng.2678
7. SchmitzRJ, SchultzMD, UrichMA, NeryJR, PelizzolaM, et al. (2013) Patterns of population epigenomic diversity. Nature 495 : 193–198 doi:10.1038/nature11968
8. Fournier-LevelA, KorteA, CooperMD, NordborgM, SchmittJ, et al. (2011) A Map of Local Adaptation in Arabidopsis thaliana. Science 334 : 86–89.
9. HortonMW, HancockAM, HuangYS, ToomajianC, AtwellS, et al. (2012) Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44 : 212–216 doi:10.1038/ng.1042
10. Geiler-SamerotteK, BauerC, LiS, ZivN, GreshamD, et al. (2013) The details in the distributions: why and how to study phenotypic variability. Current Opinion in Biotechnology: 1–8. doi:10.1016/j.copbio.2013.03.010
11. PetterssonME, NelsonRM, CarlborgÖ (2012) Selection on variance-controlling genes: adaptability or stability. Evolution 66 : 3945–3949 doi:10.1111/j.1558-5646.2012.01753.x
12. ShenX, PetterssonM, RönnegårdL, CarlborgÖ (2012) Inheritance beyond plain heritability: variance-controlling genes in Arabidopsis thaliana. PLoS Genet 8: e1002839 doi:10.1371/journal.pgen.1002839
13. ShenX, AlamM, FikseF, RönnegårdL (2013) A novel generalized ridge regression method for quantitative genetics. Genetics 193 : 1255–1268 doi:10.1534/genetics.112.146720
14. BaxterI, BrazeltonJN, YuD, HuangYS, LahnerB, et al. (2010) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet 6: e1001193 doi:10.1371/journal.pgen.1001193
15. TrontinC, TisnéS, BachL, LoudetO (2011) What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr Opin Plant Biol 14 : 225–231 doi:10.1016/j.pbi.2011.03.024
16. WeigelD (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiology 158 : 2–22 doi:10.1104/pp.111.189845
17. SasidharanR, ChinnappaCC, StaalM, ElzengaJTM, YokoyamaR, et al. (2010) Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiology 154 : 978–990 doi:10.1104/pp.110.162057
18. SungS, SchmitzRJ, AmasinoRM (2006) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes & Development 20 : 3244–3248 doi:10.1101/gad.1493306
19. De LuciaF, CrevillenP, JonesAME, GrebT, DeanC (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proceedings of the National Academy of Sciences 105 : 16831–16836 doi:10.1073/pnas.0808687105
20. KimD-H, SungS (2010) The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proceedings of the National Academy of Sciences 107 : 17029–17034 doi:10.1073/pnas.1010834107
21. PanikulangaraTJ, Eggers-SchumacherG, WunderlichM, StranskyH, SchöfflF (2004) Galactinol synthase1. A novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiology 136 : 3148–3158 doi:10.1104/pp.104.042606
22. TajiT, OhsumiC, IuchiS, SekiM, KasugaM, et al. (2002) Important roles of drought - and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29 : 417–426.
23. BarrickJE, LenskiRE (2013) Genome dynamics during experimental evolution. Nat Rev Genet 14 : 827–839 doi:10.1038/nrg3564
24. ZemachA, KimMY, HsiehP-H, Coleman-DerrD, Eshed-WilliamsL, et al. (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153 : 193–205 doi:10.1016/j.cell.2013.02.033
25. StroudH, DoT, DuJ, ZhongX, FengS, et al. (2013) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21 : 64–72 doi:10.1038/nsmb.2735
26. Joshua G Dunn CKFNGBERGJSW (2014) Correction: Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 3. doi:10.7554/eLife.03178.
27. ItoH, GaubertH, BucherE, MirouzeM, VaillantI, et al. (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472 : 115–119 doi:10.1038/nature09861
28. AtwellS, HuangYS, VilhjálmssonBJ, WillemsG, HortonM, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465 : 627–631 doi:10.1038/nature08800
29. DowenRH, PelizzolaM, SchmitzRJ, ListerR, DowenJM, et al. (2012) Widespread dynamic DNA methylation in response to biotic stress. Proceedings of the National Academy of Sciences 109: E2183–E2191 doi:10.1073/pnas.1209329109
30. ShenX, CarlborgÖ (2013) Beware of risk for increased false positive rates in genome-wide association studies for phenotypic variability. Front Genet 4 : 93 doi:10.3389/fgene.2013.00093
31. Dubin MJ, Zhang P, Meng D, Remigereau M-S, Osborne EJ, et al. (2014) DNA methylation variation in Arabidopsis has a genetic basis and shows evidence of local adaptation. arXiv: 1410.5723 [q-bio.GN].
32. KöhlerC, WolffP, SpillaneC (2012) Epigenetic mechanisms underlying genomic imprinting in plants. Annu Rev Plant Biol 63 : 331–352 doi:10.1146/annurev-arplant-042811-105514
33. KilianJ, WhiteheadD, HorakJ, WankeD, WeinlS, et al. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50 : 347–363 Available: http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2007.03052.x/full.
34. Shen X, Li Y, Rönnegård L, Uden P, Carlborg Ö (2014) Application of a genomic model for high-dimensional chemometric analysis. Journal of Chemometrics: n/a–n/a.
35. ChenW-M, AbecasisGR (2007) Family-based association tests for genomewide association scans. Am J Hum Genet 81 : 913–926 doi:10.1086/521580
36. AulchenkoYS, RipkeS, IsaacsA, van DuijnCM (2007) GenABEL: an R package for genome-wide association analysis. Bioinformatics 23 : 1294–1296.
37. DevlinB, RoederK (1999) Genomic control for association studies. Biometrics 55 : 997–1004.
38. RönnegårdL, ShenX, AlamM (2010) hglm: A package for fitting hierarchical generalized linear models. The R Journal 2 : 20–28.
39. McLarenW, PritchardB, RiosD, ChenY, FlicekP, et al. (2010) Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26 : 2069–2070 doi:10.1093/bioinformatics/btq330
40. LiX, KierczakM, ShenX, AhsanM, CarlborgÖ, et al. (2013) PASE: a novel method for functional prediction of amino acid substitutions based on physicochemical properties. Front Genet 4 : 21 doi:10.3389/fgene.2013.00021
41. AlonsoJM (2003) Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science 301 : 653–657 doi:10.1126/science.1086391
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání






