-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
The cohesion of sister chromatids in the interval between chromosome replication and anaphase is important for preventing the precocious separation, and hence nondisjunction, of chromatids. Cohesion is accomplished by a ring-shaped protein complex, cohesin; and its release at anaphase occurs when separase cleaves the complex's α-kleisin subunit. Cohesin has additional roles in facilitating DNA damage repair from the sister chromatid and in regulating gene expression. We tested the universality of the present model of cohesion by studying cohesin in the evolutionarily distant protist Tetrahymena thermophila. Localization of tagged cohesin components Smc1p and Rec8p (the α-kleisin) showed that cohesin is abundant in mitotic and meiotic nuclei. RNAi knockdown experiments demonstrated that cohesin is crucial for normal chromosome segregation and meiotic DSB repair. Unexpectedly, cohesin does not detach from chromosome arms in anaphase, yet chromosome segregation depends on the activity of separase (Esp1p). When Esp1p is depleted by RNAi, chromosomes become polytenic as they undergo multiple rounds of replication, but fail to separate. The cohesion of such bundles of numerous chromatids suggests that chromatids may be connected by factors in addition to topological linkage by cohesin rings. Although cohesin is not detected in transcriptionally active somatic nuclei, its loss causes a slight defect in their amitotic division. Notably, Tetrahymena uses a single version of α-kleisin for both mitosis and meiosis. Therefore, we propose that the differentiation of mitotic and meiotic cohesins found in most other model systems is not due to the need of a specialized meiotic cohesin, but due to additional roles of mitotic cohesin.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003418
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003418Summary
The cohesion of sister chromatids in the interval between chromosome replication and anaphase is important for preventing the precocious separation, and hence nondisjunction, of chromatids. Cohesion is accomplished by a ring-shaped protein complex, cohesin; and its release at anaphase occurs when separase cleaves the complex's α-kleisin subunit. Cohesin has additional roles in facilitating DNA damage repair from the sister chromatid and in regulating gene expression. We tested the universality of the present model of cohesion by studying cohesin in the evolutionarily distant protist Tetrahymena thermophila. Localization of tagged cohesin components Smc1p and Rec8p (the α-kleisin) showed that cohesin is abundant in mitotic and meiotic nuclei. RNAi knockdown experiments demonstrated that cohesin is crucial for normal chromosome segregation and meiotic DSB repair. Unexpectedly, cohesin does not detach from chromosome arms in anaphase, yet chromosome segregation depends on the activity of separase (Esp1p). When Esp1p is depleted by RNAi, chromosomes become polytenic as they undergo multiple rounds of replication, but fail to separate. The cohesion of such bundles of numerous chromatids suggests that chromatids may be connected by factors in addition to topological linkage by cohesin rings. Although cohesin is not detected in transcriptionally active somatic nuclei, its loss causes a slight defect in their amitotic division. Notably, Tetrahymena uses a single version of α-kleisin for both mitosis and meiosis. Therefore, we propose that the differentiation of mitotic and meiotic cohesins found in most other model systems is not due to the need of a specialized meiotic cohesin, but due to additional roles of mitotic cohesin.
Introduction
Cohesin is a ring-shaped protein complex which holds sister chromatids together to prevent their untimely separation prior to anaphase (see [1]). It consists of four core components, Smc1 and Smc3, Scc3, and a member of the conserved α-kleisin family of proteins, Mdc1/Rad21/Scc1 in mitotic cells, or Rec8 in meiotic cells [1], [2].
In mitotic cells, newly synthesized sister chromatids are linked by cohesin. In some organisms, chromatids separate along their arms during prophase and at the centromeres during anaphase, while in other organisms separation occurs in a single step during anaphase. The resolution of cohesion during anaphase is initiated by the cleavage of the α-kleisin component of the cohesin, the opening of the ring structure, and the disappearance of the cohesin from the chromosomes. This allows the mitotic separation of the sister chromatids. The fact that ring opening releases the chromatids, together with other evidence (see [1]), led to the popular model that cohesin works by the enclosure of the two sisters by a single ring.
During meiosis, homologous chromosomes first pair and become connected by a protein structure, the synaptonemal complex (SC) (see [3]). At the same time, deliberate DNA double-strand breaks (DSBs) are formed and resolved in a way that leads to recombination [4]. Subsequent segregation of homologous chromosomes and sister chromatids is coordinated by the sequential loss of a specialized meiotic cohesin from the arms prior to the first meiotic division, and then from centromere regions prior to the second division. First, release of cohesin from regions distal to chiasmata allows the separation of homologous chromosomes, then release of centromeric or proximal regions separates sisters. Meiosis-specific regulation of cohesion works only when a specialized meiotic cohesin (containing the meiosis specific α-kleisin Rec8, and in some organisms also meiotic versions of other cohesin components) is loaded onto chromosomes during the premeiotic S-phase (for review see [5]). Kleisin cleavage in mitosis and meiosis is performed by a protein called separase, whose checkpoint-dependent activation relies upon the orderly association of chromosomes or bivalents with the division spindle, thus assuring their faithful disjunction (see [1], [6]). During meiotic prophase, cohesin is associated with the axial elements of SCs [7]–[11] and is involved in the repair and, in some organisms, the formation of meiotic DNA DSBs [12].
In addition to its well-established function in controlling chromatid and chromosome segregation, cohesin was found to play some additional roles, such as in DNA damage repair, where it promotes recombinational repair via sister DNA molecules [13], [14], in maintaining epigenetic inheritance states [15], in gene regulation [16] (and lit. cit. therein) and in chromosome conformation [17].
Tetrahymena thermophila is a ciliated protist. Like the other ciliates, Tetrahymena possesses its soma and its germline organized as two nuclei within one cell (see [18]). The germline micronucleus (MIC) carries a diploid number of 10 chromosomes that undergo closed mitosis and meiosis (Figure 1). The MIC genome is not expressed, and so the MIC is largely dispensable for vegetative proliferation of the cell; its only function is the propagation of heritable information during sexual reproduction. Transcription takes place only in the somatic macronucleus (MAC). The MAC contains ∼45-fold amplified copies of the genome which are distributed in ∼180 minichromosomes that lack centromeres and remain uncondensed. It does not divide by a mitotic process, but splits into roughly equal parts prior to cell division.
Fig. 1. Mitotic and meiotic stages. 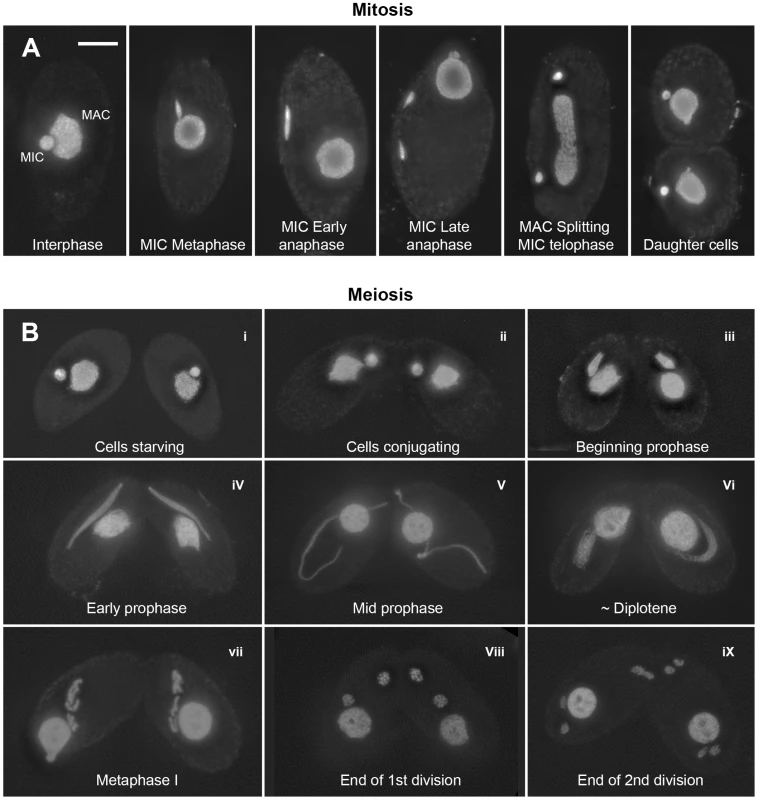
(A) Mitosis. In nondividing vegetative cells, the diploid micronucleus (MIC) rests in a pocket in the polyploid macronucleus (MAC). During mitosis, it becomes stretched and divides. The MAC splits (by an amitotic process) only after separate daughter MICs have formed. (B) Meiosis. Cells of complementing mating types fuse (“conjugate” – i–ii) and their diploid MICs perform synchronous meioses. During prophase (i–vi), the MIC elongates to >2× the cell length (iii–v), before it shortens again (vi). In contracting MICs, chromatin condensation culminates in the formation of condensed metaphase bivalents (vii). Subsequently, homologs separate in a first (viii) and sisters in a second meiotic division (ix). DNA stained by DAPI. Bar: 10 µm. Tetrahymena cells of complementing mating types join (“conjugate”) under starvation conditions and initiate synchronous meioses in their MICs (Figure 1). The MAC does not undergo meiosis but it degenerates, and a new MAC regenerates from the MIC in new sexual progeny. Meiosis is characterized by several unusual features: First, there is no dedicated premeiotic S-phase; cells at (micronuclear) G2 conjugate and enter the meiotic program. Moreover, Tetrahymena lacks an SC [19] and, most notably, the MIC undergoes an immense elongation, to up to 50 times its original diameter, during meiotic prophase [20]–[22]. Within the elongated MIC, chromosomes are arranged in parallel, which facilitates homologous pairing and recombination [23].
Here, we take advantage of Tetrahymena's evolutionary distance from other commonly studied model systems to learn more about what features of cohesin are conserved, and what have been adapted to the needs of the particular organism. We also exploit the nuclear duality of Tetrahymena to separate functions of cohesin proteins that are important for chromosome division from functions related to gene expression and regulation. We demonstrate that Tetrahymena has evolved notable differences to the standard eukaryotic cohesion machinery. Investigation of these remarkable adaptations will lead to new insights into the flexibility of the chromosome cohesion and division processes.
Results
Tetrahymena possesses conserved components of the cohesion machinery, but only a single kleisin homolog
We performed a bioinformatic search of the T. thermophila proteome [24] for the presence of cohesin components (see Text S1). The protein encoded by TTHERM_00245660 was the top and significant hit identified in searches for homologs to the Scc1(Mcd1)/Rad21/Rec8 family of α-kleisin proteins [2]. It produced a significant match in region aa 579–607 of the 619 amino acid-protein to the conserved C-terminal winged helix domain [25]. An additional weaker similarity was found for the N-terminal aa 30–130 (Figure S1). Based on the alignment of the conserved N - and C-terminal regions, we generated a phylogenetic tree that shows a relatively close sequence relationship of the group of mitotic α-kleisins, whereas meiotic members and TTHERM_00245660p show higher sequence divergence (Figure 2).
Fig. 2. Phylogenetic relationships among eukaryotic α-kleisin family members. 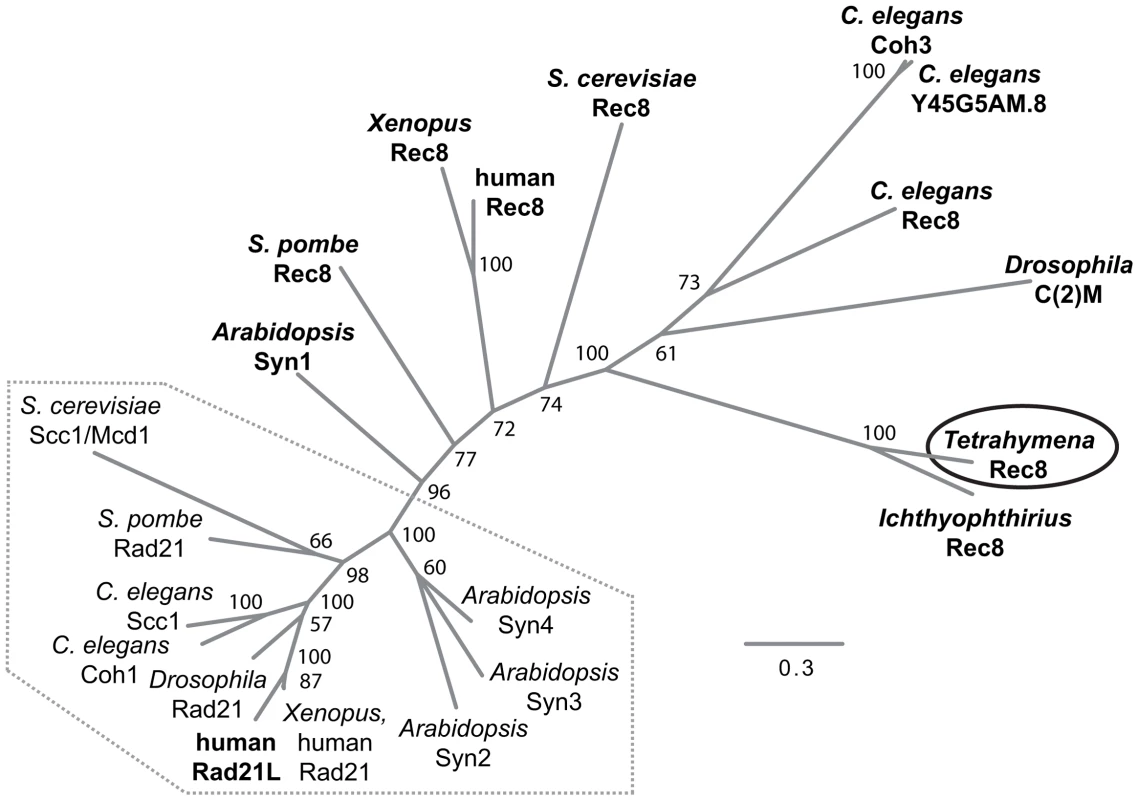
The unrooted phylogenetic tree was generated using MrBayes 3.2 [74] and visualized using FigTree. It shows experimentally confirmed meiotic (bold) and mitotic (regular) cohesins. Mitotic α-kleisins form a subgroup (dotted area) with closer sequence similarity, as indicated by the shorter branch lengths. Meiotic α-kleisins and Tetrahymena Rec8p show higher sequence diversity. The function of Drosophila C(2)M as the meiotic kleisin is still debated [62]. % Bayesian posterior probabilities are indicated. Bar: Number of amino acid substitutions per site. Another protein, TTHERM_00219160p, is characterized by a C-terminal winged helix domain with more distant similarity to kleisins. It was found as the second best and significant hit in profile searches with the winged helix domain that was used to identify TTHERM_00245660p. This similarity was also confirmed in reciprocal searches (see Text S1, Figure S1). However, sequence similarity beyond the C-terminal region could not be detected. TTHERM_00219160 mRNA has very low expression compared to the other cohesin gene homologs, and does not show significant up-regulation during conjugation, as the others do [26]. GFP tagging showed no detectable expression or localization of TTHERM_00219160p, even at high levels of replacement of the wildtype gene with the tagged gene (data not shown). All of this evidence suggests that this is not a true kleisin protein with cohesin function, leaving TTHERM_00245660p as the only likely candidate.
It is notable that Tetrahymena would have only a single α-kleisin homolog, because all eukaryotes studied to date in this respect have mitotic (Scc1/Rad21/Mcd1) and meiotic (Rec8) versions of this protein. Despite the lack of any closer relationship to the meiotic subgroup of α-kleisins (Figure 2), we will designate TTHERM_00245660p as Rec8p (and the gene as REC8) in the following, because of its function in mitosis and meiosis (see below). It shares this feature with the meiotic budding yeast Rec8 which, in principle, can take over the mitotic cohesion function [27]–[29].
Other core components of a putative cohesin complex show more conservation than Rec8. ORFs TTHERM_01048090 and TTHERM_00294810 encode clear Smc1 and Smc3 homologs, respectively [24], [30]. It is likely that the SMC3 ORF prediction is incorrect [24], because cDNA coverage for SMC3 in the Tetrahymena Functional Genomics Database (http://tfgd.ihb.ac.cn) suggests a protein of only 1187 amino acids. To confirm that putative cohesin proteins form a complex in Tetrahymena, an immunoprecipitation (IP) was performed using cells expressing Rec8-GFP. Wild type cells were used for a control IP. Mass spectrometry analysis of the precipitating proteins showed that Smc1 and Smc3 were enriched 13× and 9×, respectively, in the Rec8-GFP pulldown sample over the untagged Rec8 control sample. Of 1837 identified proteins, Smc1 and Smc3 ranked 1st and 4th with respect to the number of unique peptides identified in the pulldown sample but only at positions 521 and 703 in the control (Table S1). Sequence coverage was 56% vs. 3% for Smc1 and 47% vs. 8% for Smc3 in the pulldown and control samples (Figure S2). This is strong evidence for the involvement of Rec8, Smc1 and Smc3 in a protein complex and hence a confirmation of their correct identification as cohesin proteins.
An Scc3/Rec11 homolog candidate, encoded by TTHERM_00225630, was identified in profile searches versus the T. thermophila proteome with the region of best conservation from known Scc3/Rec11 orthologs, and was also confirmed in reciprocal searches (see Text S1). Although we did not find this homolog precipitating with Rec8-GFP, we will designate it as Scc3p. In addition to the core subunits of the cohesin complex, we were also able to identify the separase protein, another component of the cohesion/segregation machinery. TTHERM_00297160 is annotated in the Tetrahymena Genome database as the separase gene ESP1 [24]. The Esp1 protein sequence produced best reciprocal BLASTp hits with Saccharomyces cerevisiae Esp1, Schizosaccharomyces pombe Cut1, Caenorhabditis elegans SEP-1, Arabidopsis thaliana AESP and human ESPL1. We did not yet find obvious candidates for accessory cohesion proteins such as Eco1, Pds5, Wapl, sororin or securin.
Rec8p and Smc1p localize to both mitotic and meiotic nuclei and remain associated with chromosome arms during divisions
In order to observe the localization of cohesin in Tetrahymena, we used strains expressing tagged fusion proteins Rec8-GFP and Smc1-HA. Immunostaining against GFP or HA revealed a micronuclear localization of both proteins, in both vegetatively growing and sexually reproducing cells. This localization is maintained throughout all stages of mitosis and meiosis (Figure 3A–3D, Figure S3A, Figure S4). In addition, we tagged the putative Scc3p with mCherry and found its localization to be the same (Figure 3E). In elongated meiotic nuclei, which correspond to the prophase stage of meiosis, Rec8p, Smc1p and Scc3p can be seen along the entire length of the chromosomes (Figure 3C, 3D, 3E, Figure S4). When only one of two conjugating meiotic cells carried the tagged proteins (as shown for Smc1-HA and Scc3-mCherry in Figure 3D, 3E and Figure S4), the protein was found retained in this cell. This is in contrast to the previously observed exchange of proteins between conjugating partners [31] and indicates a low turnover of cohesin proteins.
Fig. 3. Localization of Rec8p, Smc1p, and Scc3p to mitotic and meiotic chromosomes. 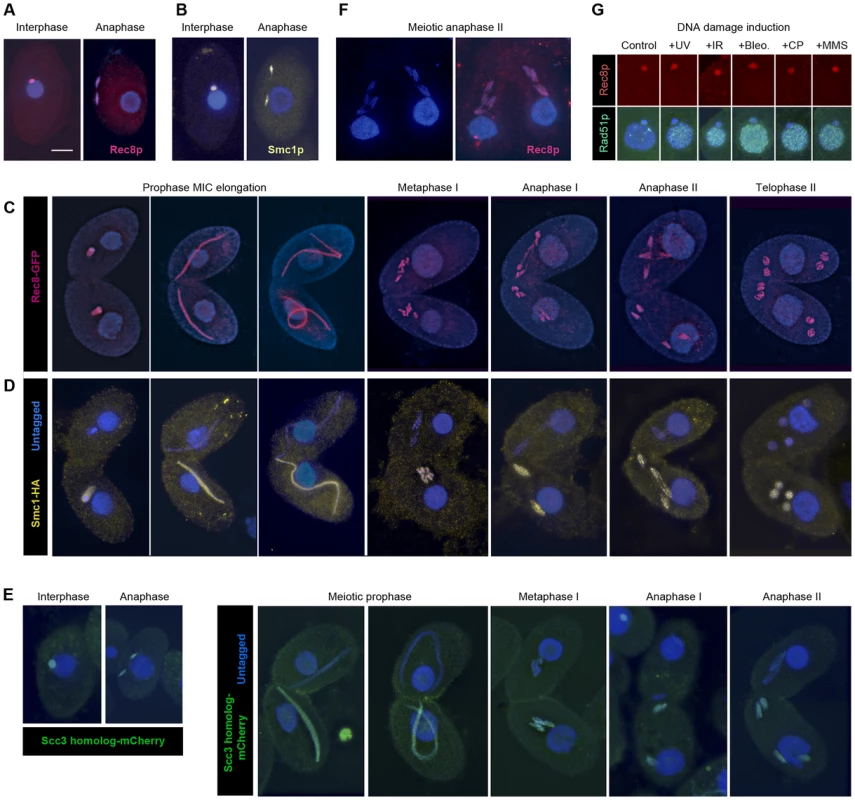
(A, B) Rec8-GFP (red) and Smc1-HA (yellow) localize to interphase and mitotic MICs. (C) Rec8p localizes to MIC chromosomes during all stages of meiosis (both mating partners express GFP-tagged Rec8). (D) Smc1p localizes to MIC chromosomes during all stages of meiosis (only one partner expresses HA-tagged Smc1). Note the absence of both cohesion proteins from the MACs. (E) The Scc3p homolog TTHERM_00225630p (green) localizes to mitotic and meiotic MICs of all stages. (F) Paired cells in meiotic anaphase II prepared by a spreading method to remove free nuclear proteins. Persistance of Rec8 staining (red) demonstrates that it remains attached to the arms of separating chromatids. (G) MAC localization of Rec8p is not increased, whereas repair protein Rad51 is strongly expressed in the MAC upon DNA damage. Bar: 10 µm in A–F. Notably, in mitotic anaphase as well as in anaphase I and II of meiosis, the proteins could be seen along the full length of the stretched chromosome arms (Figure 3A–3E). This is in contrast to the expected behavior shown by other organisms [7], [32]–[36], where cohesin disappears from chromosome arms during anaphase, but is similar to fission yeast mitosis [37], where the separation of sister chromatids is not accompanied by the removal of cohesin. It is possible that the anaphase signal could result from cleaved cohesin rings that remain in the nucleoplasm or loosely associated with chromosomes during closed divisions. In order to address this possibility, detergent-spread fixations were performed in which free proteins are washed out of the MIC [38]. Immunofluorescent stainings of these fixations still showed the continuous localization of Smc1-HA and Rec8-GFP on chromosome arms throughout anaphase (Figure 3F). This indicates that cohesin remains attached to chromatin during divisions.
It was shown by Feulgen microspectrophotometry that in the Tetrahymena vegetative cell cycle, MIC DNA replication starts during division, as early as late anaphase or telophase [39]. We confirmed this staging by BrdU incorporation (Figure S3B). This early replication may provide an explanation for the presence of cohesin along mitotic chromatids. Cohesin may be present in a non-cohesive form in order to establish cohesion as soon as replication starts.
Cohesin is not detected in the amitotic macronucleus
Tetrahymena allows separate observation of different cohesin functions because mitosis and transcription are carried out in separate nuclei within one and the same cell. We did not observe Rec8p nor Smc1p in the MAC of vegetatively growing or meiotic cells (Figure 3A–3D, 3F). In principle, there should be no strict need for cohesin in the MAC, because the polyploid macronuclear chromosomes separate randomly by an amitotic process, and therefore it is not necessary to hold sister chromatids together after replication. However, an increasing number of non-cohesive functions of cohesin have been reported in studies of other model organisms (for reviews see [1], [6], [40]). If cohesin performs these functions in Tetrahymena, it could conceivably be needed in the MAC as well as the MIC. Because cohesin has previously been shown to localize to sites of broken DNA [41], [42], we hypothesized that macronuclear cohesin might be induced by DNA damage. It has been shown in Tetrahymena that DNA damage induces expression of Rad51p in the MAC [43]. This suggests that a recombinational repair pathway is triggered that might rely on the physical linkage of sister DNA molecules by cohesin. Cells expressing Rec8-GFP were treated with UV-C, γ-radiation, MMS, cisplatin, or bleomycin (see Materials and Methods), all of which are known to induce DNA damage in Tetrahymena [23], [44]. Cells were fixed 0.5 or 1.5 h after treatment. Immunostaining against GFP showed no macronuclear localization while the DNA repair protein Rad51 was abundant in the MAC in response to the DNA damage (Figure 3G). 200 cells with Rad51-positive MACs were evaluated per experiment. In all cases, Rec8p was detected in the MIC but never in the MAC. A similar experiment did not detect damage-induced Smc1-HA in the MAC, either (data not shown). These results seem to indicate that cohesin does not localize to the MAC. Therefore, if cohesin functions in the MAC, it must be at concentrations too low to observe cytologically.
Loss of cohesin impairs vegetative propagation and causes MIC chromosome mis-segregation
To learn more about the function of cohesin in Tetrahymena, depletion of Rec8p or Smc1p was performed by RNA interference (RNAi). For this, cells were transformed with constructs to express hairpin (hp) RNA molecules from a cadmium-inducible promoter, and RNAi was elicited in SMC1hp and REC8hp cells by growth in medium with CdCl2.
Induction of rec8RNAi (rec8i) in growing cells resulted in a very slow depletion of Rec8p, as can be seen by Western blotting in cells expressing both the Rec8-GFP and the REC8hp construct (Figure 4A). After 4 h of RNAi induction (approximately 1–2 cell divisions), large amounts of Rec8p remained in the cells, whereas REC8 mRNA was substantially reduced even after 2 h, as seen by RT-PCR (Figure 4B). Only after 24 hours of RNAi, was the protein almost completely depleted (Figure 4A). This data also confirms that Rec8p has a very low turnover in the cell (see above), in contrast to RNAi targets studied previously, which needed only short periods of RNAi induction to produce phenotypes [38], [45].
Fig. 4. Phenotypes of cohesin-depleted cells. 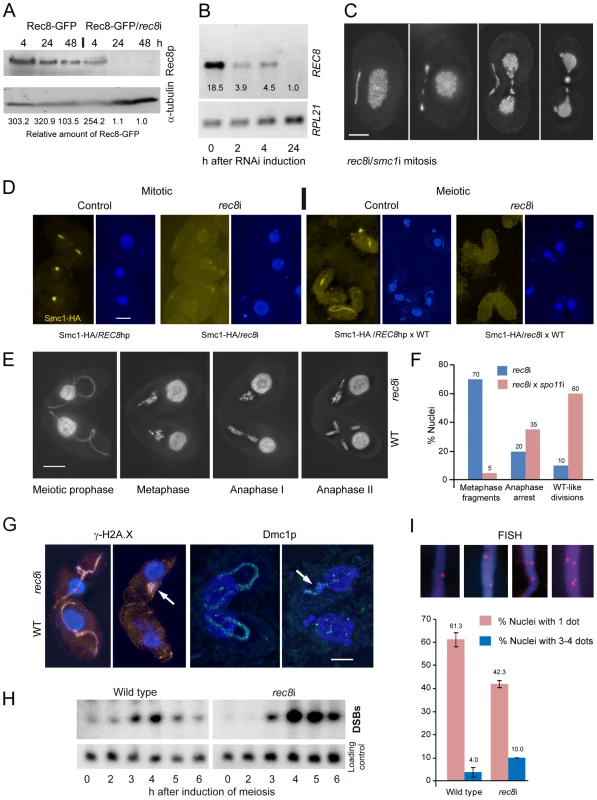
(A) Rec8p is depleted by cadmium (Cd)-induced expression of hairpin RNA and monitored by the detection of GFP-tagged Rec8p on a Western gel. RNAi for 24 h leads to virtually complete loss of Rec8p. GFP band intensities were determined using the “Analyze” tool of ImageJ (Wayne Rasband, N.I.H.; http://rsb.info.nih.gov/ij/), normalized to the α-tubulin loading control, and used to calculate relative protein amounts. (B) RT-PCR shows that REC8 RNA is greatly reduced after 2 h of RNAi induction, and nearly absent after 24 h. (C) Mitotic division of the MIC shows delayed segregation and lagging chromosomes, and the MAC also splits abnormally in cells depleted of Rec8p and Smc1p. Bar: 10 µm. (D) Depletion of Rec8p causes the loss of Smc1p in mitotic and meiotic MICs. In the presence of Rec8p, Smc1-HA localizes to MICs in vegetatively propagating cells, but disappears when rec8 RNAi is induced. When REC8hp/Smc1-HA cells are mated to a wild type (WT) partner, the former show micronuclear Smc1-HA localization, which disappears upon rec8 RNAi. Bar: 10 µm. (E) rec8i cells (top) arrest at an abnormal metaphase-anaphase I stage, whereas the WT mating partners (bottom) show the normal progression of meiosis. In WT metaphase, 5 distinct bivalents can be seen (arrow points to one well-separated bivalent), whereas the rec8i partner shows fragmented chromosomes. Bar: 10 µm. (F) Quantitation of rec8i meiosis arrest 4,5 h after meiosis induction. Eliminating DSBs by mating rec8i cells with spo11i partners rescues the rec8i phenotype. Stages of rec8i cells were scored in cell pairs where their respective WT or spo11i mating partners had progressed beyond the stage indicated (n = 100 cells in both experiments). (G) DSB markers g-H2AX (orange) and Dmc1p (green) highlight elongated meiotic prophase MICs (left) in both rec8i and WT cells of a mating pair. Later in meiosis (right), when they have disappeared from the WT partner, they are still present in the arrested MICs of the rec8i partner (arrows). (H) Detection of DSBs by PFGE in WT and rec8i meiosis. The lower panel shows a control hybridization to the same membrane as a test for equal DNA loading and Southern transfer. (I) Evaluation of pairing (scored as the presence of a single FISH signal) and loss of cohesion (scored as the presence of 3 or 4 signals) in elongated meiotic prophase MICs, displayed by different genotypes. Examples of MICs with different numbers of FISH signals are shown on top. Values are means of three repeats with 50 nuclei evaluated, each. Error bars indicate standard deviation. Depletion of Rec8p and/or Smc1p impaired the growth of cells. Viability was tested by isolating single cells of wild type (WT), REC8hp, SMC1hp or REC8hp/SMC1hp strains (n = 47 each) in drops of medium containing 0.1 µg/ml CdCl2 to induce RNAi. After 48 h of growth, 100% of WT clones had more than 100 cells, whereas 0% of rec8i, 4.2% of smc1i and 6.4% of rec8i/smc1i clones had grown to the same density.
Cells depleted of cohesins for at least 24 hours often showed lagging chromosomes in mitotic anaphase and, unlike in wild-type controls (Figure 1A), the MACs began to elongate before mitosis was complete (Figure 4C). This suggests that a lack of cohesion causes abnormal spindle attachments that result in a delay in chromatid segregation. rec8i/smc1i cells displayed 45% (n = 100) abnormal anaphases whereas the phenotype was less prevalent in the smc1i line (13% (n = 100) of anaphases were defective) and absent in the rec8i line. Thus, the additive effect of the double RNAi is necessary to strongly affect segregation. Notably, telophase MICs appeared normal, presumably because lagging or misoriented chromatids eventually manage to migrate to the poles. MACs also had problems in splitting (Figure 4C, Figure S5A), presumably as a consequence of delayed MIC mitosis. Cells often contained large amounts of DNA left between newly divided MACs, which is a less prevalent phenomenon in the wild type [46].
We next tested if depletion of Rec8p influences Smc1p localization. We constructed a strain that carries both the REC8hp and Smc1-HA. This strain showed the normal micronuclear localization of HA-tagged Smc1p (in 200 of 200 evaluted nuclei) when REC8 RNAi was not induced (Figure 4D). However, when RNAi was induced (rec8i), Smc1p staining was lost (Figure 4D). In quantitative terms, in 96% of nuclei, the Smc1 signal was completely lost, and in 4% it was strongly reduced (n = 200 nuclei). When this strain was mated to wild type, Smc1-HA localized to meiotic MICs (in 200 of 200 evaluted nuclei), but mostly failed to do so upon RNAi depletion of Rec8 (Figure 4D). The Smc1 signal was reduced or completely lost, in 8% and 85% of nuclei, respectively (n = 200 nuclei). The dependency of Smc1 localization on Rec8 confirms a strong interdependence of Rec8 and Smc1 protein expression or stability, as would be expected for members of a protein complex such as cohesin. Moreover, it indicates that Rec8p is the predominant, if not only, kleisin partner of Smc1p, otherwise more Smc1p, which was forming a complex with another kleisin, would persist.
Meiotic divisions are arrested upon the depletion of cohesin components
To evaluate the meiotic phenotype of rec8i and smc1i cells, RNAi was performed on growing cells for 24 hours to deplete cohesin prior to inducing meiosis. The cohesin-depleted cells were then mated with a WT strain expressing Rec8-GFP. Due to the low turnover rate of the cohesion proteins (see above), RNAi affected only the hairpin-carrying partner of mating pairs and allowed direct comparison of defective stages with the corresponding meiotic stages in the WT partner. DAPI staining of these mating cells showed that rec8i and smc1i cells begin meiosis normally and elongate the MIC, as in WT. However, whereas in WT cells, the MIC contracts and chromosomes condense to form bivalents in metaphase, in rec8i or smc1i cells, the chromosomes never fully condense (Figure 4E, Figure S5). In these cells, meiosis arrests at an abnormal metaphase-anaphase-like state, with clumps of fragmented and stretched chromatin, instead of distinct chromosomes, while the WT partner progresses normally through the meiotic divisions (Figure 4E). Quantitation of the rec8i meiotic arrest showed that in pairs where the WT partner had progressed to metaphase or beyond, 70% of the rec8i partners were arrested in the fragmented metaphase state, 20% attempted to undergo an abnormal anaphase, and 10% showed a normal looking division (Figure 4F). To determine if the rec8i arrest phenotype was dependent on meiotic DSBs, we depleted the Spo11 DSB nuclease by RNAi. spo11 RNAi of one cell inhibits or reduces DSB formation in the partner cell as well (data not shown). When we mated rec8i cells with spo11i partners, the rec8i arrest phenotype was partially rescued. 60% of cells completed anaphase I, 35% showed abnormal divisions, and only 5% arrested with fragmented metaphase chromosomes (Figure 4F). This suggests that rec8i cells have a defect in DSB repair.
Depletion of cohesin impairs DNA repair in MICs
To further test if meiotic DSB repair was affected, matings of rec8i and WT cells were stained with antibodies against γ-H2A.X or the recombination protein Dmc1. These DSB markers are normally seen primarily in the elongated pachytene MIC, and they disappear in later stages after DSBs are repaired [38], as can be seen in the WT partner (Figure 4G). However, γ-H2A.X and Dmc1 staining can still be seen in the rec8i cells, after their WT partners have completed DSB repair. This suggests that DSBs are not completely repaired in the absence of cohesin. The persistence of DSBs was also confirmed by electrophoretic detection of DSB-dependent chromosome fragments in mating rec8i cells (Figure 4H). Whole cell DNA preparations of mating cells were separated by pulsed field gel electrophoresis, blotted, and probed with a repetitive sequence found only in the MIC [44]. Under the conditions used, unbroken MIC chromosomes do not enter the gel, but chromosomes fragmented by DSBs migrate as a mass, creating a distinct band. In WT cells, DSBs appeared at 3 hours after induction of meiosis, and disappeared at 5–6 h as breaks were repaired and meiosis was completed (Figure 4H). In rec8i matings, however, the band representing meiotic DSBs appeared normally at 3 h, but seemed to accumulate to a higher level at 4 and 5 h, and did not disappear by 6 h after induction of meiosis. Therefore, we can conclude that cohesin is required for the repair of meiotic DSBs.
Meiotic pairing and sister chromatid cohesion are reduced in cells depleted of Rec8p
Because defective DSB repair causes reduced meiotic pairing [38], we wondered if pairing was affected by the absence of Rec8. In meiosis of Tetrahymena, close pairing of homologous chromosomes is established in elongated MICs. Fluorescence in situ hybridization (FISH) can be used to monitor meiotic pairing of a chromosomal locus, which is indicated by the presence of a single FISH signal instead of two (Figure 4I). While a single signal could also result from mitotic nondisjunction (see above), rec8i did not show a mitotic phenotype. Therefore we were able to use rec8i cells to score pairing, using a probe to an intercalary chromosomal locus. In WT cells, 34.7% of meiotic prophase nuclei showed two FISH signals, corresponding to the unpaired homologous loci, and 61.3% showed only one signal, indicating pairing of that locus had occurred. In rec8i cells, pairing was reduced to 42.3%. (Figure 4I). At the same time, FISH was also used to evaluate cohesion. 10.0% of rec8i elongated nuclei showed 3 or 4 signals, as compared to 4.0% in WT (Figure 4I). Altogether, the depletion of cohesin results in a reduction of meiotic sister chromatid cohesion and homologous pairing.
Separase (Esp1p) knockdown prevents chromosome segregation and causes polyploidization
The continuous association of cohesin with mitotic and meiotic chromosomes during and after anaphase led us to ask if the separase cleavage mechanism used in other organisms was conserved in Tetrahymena. We therefore designed an RNAi construct to target Tetrahymena's separase homolog, ESP1. After induction of the esp1i construct in vegetative growth, cells immediately showed problems dividing the MICs. Normally the MIC divides before the MAC (Figure 1A and Figure S3), whereas in esp1i cells, MICs arrested in an anaphase-like state by the time the MAC divided (Figure 5A). As a result, the cleavage furrow formed while the MIC was still attempting division, leaving one daughter cell containing the undivided MIC, and one cell without a MIC (Figure 5A). Staining for phosphorylated (Ser10) histone H3, which is a marker for chromosome condensation [47], showed that the MICs stay in a mitosis-like condensation state (Figure 5A). After 24 h of esp1i induction, only 10.5% of cells contained MICs, as opposed to 98.5% of uninduced cells. This decreased to 7% after 48 h, and 3% after 72 h of esp1i induction (n = 200 cells per genotype and timepoint). After 48 h of esp1i induction, the remaining MICs were very large, presumably as a result of polyploidization by numerous rounds of replication and failed separation. Many of these large MICs had a ropy appearance, suggesting a bundled, polytenic state of chromosomes (Figure 5B). FISH staining of a unique chromosomal locus showed signal clusters whose strength indicated that they contained multiple copies, again suggesting a polytenic organization of unseparated chromatids (Figure 5C). To obtain a rough estimate of the degree of polyploidization, FACScan was performed of cultures 48 h after esp1i induction. While nuclei from WT cells sorted into two clear peaks corresponding to MICs and MACs, esp1i cells showed only one peak for both MICs and MACs (Figure 5D). This indicates that aberrant MICs may reach roughly the same DNA content as MACs and suggests that they may become 16–32-ploid.
Fig. 5. RNAi depletion of separase Esp1p prevents mitotic and meiotic division and causes MIC polyploidization. 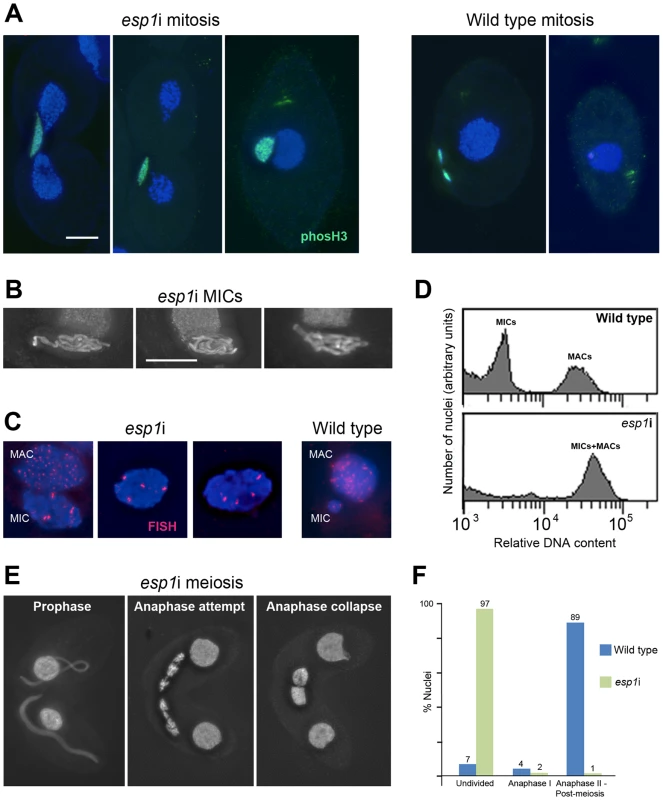
(A) Chromosomes fail to segregate during mitosis. Whereas in the WT MIC, division precedes the splitting of the MAC, the esp1i MIC unsuccessfully attempts division and eventually remains as a single unreduced MIC in one of the two daughter cells. Staining for phosH3(Ser10) (green) highlights only condensed mitotic chromosomes in the WT, while undivided esp1i nuclei remain in a permanently condensed state. (B) After 48 hours of esp1i induction, MICs become very large and contain ropy structures, suggesting bundles of unseparated chromatids. (C) FISH with a probe against a unique chromosomal locus shows that, whereas the WT MIC contains two copies of the locus, esp1i MICs are polyploid. Unlike in the MAC, multiple copies of the locus are not dispersed, but form clusters. This suggests that replicated chromatids do not fall apart but remain in bundles resembling polytenic chromosomes. (D) FACScan analysis after 48 hours of esp1i induction confirms that MICs become polyploid due to failing segregation. While MICs and MACs form peaks of different DNA contents in the WT, in the esp1i sample they coalesce into a single peak roughly equivalent to the WT MAC DNA content. (E) Meiotic prophase stages are normal but bivalents fail to separate at anaphase I and coalesce back to a single MIC in the absence of Esp1p (compare with the WT situation in Figure 1B). (F) Quantitation of the esp1i meiotic arrest 5,5 h after meiosis induction. Bar in (A) represents 10 µm in (A) and (E), bar in (B) represents 10 µm in (B) and (C). When two ESP1hp cells were mated and RNAi was induced during meiosis, MICs elongated normally and chromosomes condensed at metaphase I. However, most cells arrested in anaphase I, and chromosomes coalesced back to enlarged undivided MICs (Figure 5E, 5F). Together, these observations suggest that cohesin cleavage does not occur in the absence of Esp1p, which prevents sister chromatid separation in mitosis and the separation of homologs in meiosis I.
Discussion
The role of cohesin in chromosome organization
Efficient depletion of cohesin proteins caused problems with MIC chromosome segregation in mitosis, which is suggestive of loss of cohesion. Loss of Rec8p also led to a reduction of cohesion in meiosis. It has been shown in other organisms that cohesion is relatively insensitive to reduced amounts of cohesin [48], [49] whereas even small decreases in the levels of cohesin drastically affect meiotic DNA repair [7], [48], [50], [51]. This explains why meiosis is more affected than mitosis in the rec8i and smc1i single knockdown strains. Also, cohesin-independent mechanisms, such as relational coiling or catenation of the sister chromatids, protein-protein interactions, or locally delayed replication could provide additional forces to hold sister chromatids together and contribute to proper segregation [52]–[54]. After long periods of cohesin depletion, mitotic cultures deteriorated. However, this was not due to MIC aneuploidy caused by segregation problems, because Tetrahymena can propagate with a defective or severely aneuploid MIC for numerous generations [55].
In contrast to MICs, MACs did not display cytologically detectable cohesin. Not even upon artificial DNA damage could we induce notable cohesin loading, although the Rad51 DNA damage response was robust. Nevertheless, depletion of cohesin results in a defect in MAC division. Dividing MACs in cohesin-depleted cells leave behind substantial amounts of DNA in the cleavage furrow, and this DNA remains in the cells as extra-nuclear bodies. This defect, along with the growth defect observed in cohesin depleted cells, may indicate a role of cohesin in normal cell division, either through a direct function in the MAC, or through cross-talk between the MIC and the MAC [56].
Rec8p and Smc1p are required for meiotic DSB repair and pairing
We have shown that meiotic cells depleted of either Rec8p or Smc1p arrest prior to anaphase I. Similar to the situation in budding yeast [7], [57], [58] and C. elegans [8], meiotic DSBs are formed normally in the absence of Rec8p. However, these breaks are not repaired, which likely causes the prophase arrest.
Cohesion has been previously found to be crucial for meiotic DSB repair, to the extent that even a partial loss of cohesin function can result in unrepaired breaks [7], [48], [50], [51]. Exactly how cohesin participates in DNA repair is not known. It is thought that cohesin facilitates repair directed by the sister chromatid by keeping sisters tightly bound together (see [59]). However, one could speculate that close access to the sister chromatid would be less crucial for meiotic DSB repair, which can, and often prefers, to use the homologous chromosome as a template. Therefore, it seems likely that cohesin plays a more direct role in DNA repair than simply providing cohesion, because the repair function is much more sensitive to slight perturbations in cohesin dosage [48]; see [14].
We have also shown that depletion of cohesin in Tetrahymena results in a moderate reduction in meiotic pairing. In other organisms, cohesin forms the backbone of the SC, a structure that is central to meiotic recombination (see Introduction). Although Tetrahymena does not form an SC, it is quite likely that cohesin plays a role in defining chromosome axes and providing the necessary architecture to support homologous recombination. Precise pairing of homologs in meiosis of Tetrahymena is dependent on homologous recombination [23], and so the reduction of pairing in the absence of cohesin may be a result of the defect in DSB repair.
Cohesin persists on anaphase chromosomes
The persistence of cohesin on anaphase chromosome arms might suggest that dissolution of cohesion works by a mechanism different from kleisin cleavage. However, we have shown that depletion of Esp1p prevents the segregation of chromosomes in both mitosis and meiosis. This phenotype is consistent with the canonical separase-dependent loss of cohesion. There are several possible explanations for the maintenance of Rec8p and Smc1p on chromosome arms: First, it may be related to the absence of an apparent G1 interval in Tetrahymena and the early onset of MIC DNA replication during late anaphase or telophase [39] (see also [60]). Thus, the permanent presence of cohesins could be due to the temporal overlap of their removal from separating sisters in anaphase and loading to chromatin in preparation for cohesion establishment during replication, shortly after. This explanation may seem less applicable to the association of cohesins with anaphase I chromosome arms, since this division is not followed by DNA replication. However, the two meiotic divisions occur in rapid succession and are followed quickly by post-meiotic replication [60]. Therefore, carrying non-cohesive cohesin along on the chromosomes may allow a more rapid establishment of cohesion. To maintain this “dormant” population of cohesin on chromosomes, there could be a mechanism in which all cohesin rings are opened by separase cleavage of Rec8p and yet continue to bind to chromatin. Because this would require replacement of the cleaved Rec8, it seems more likely that only a small subset of cohesin complexes connect sisters, and only the intersister subpopulation is removed by cleavage. This interpretation was preferred by Tomonaga et al. (2000) [37] for their similar observation in fission yeast mitosis.
Polytene MIC chromosomes suggest cohesion between multiple sisters
Prolonged depletion of Esp1p in vegetative cells results in many cells without MICs and a few cells with grossly enlarged MICs, presumably as a result of multiple rounds of replication without division. Polyploidization was also found after inactivation of separase in several mouse tissues, in D. melanogaster and in non-yeast fungi, whereas budding yeast chromosomes break during attempted separation and daughter cells die as a result [61] (and lit. cit. therein). In Tetrahymena, the chromosomes of polyploid MICs appear to be polytenic, because FISH against one chromosomal arm locus shows clustered or banded signals. The embrace model of cohesion limits the number of 10-nm chromatin fibers that can be encircled by a cohesin ring of an estimated diameter of ca. 40–45 nm to about four to six. This is hardly consistent with our observation of ∼4 chromatid bundles and the estimation of 16–32-ploid chromatid content. Thus, chromatid bundles may be formed by cohesin rings randomly encircling sisters as they are replicated within the bundle, so that a large, networked, multi-sister bunch is created, instead of individual pairs of sisters. Alternatively, some form of non-embrace-type of cohesion could be employed (see [1]).
Tetrahymena possesses a variant of the canonical cohesion-segregation machinery, with a single kleisin
We have found that Tetrahymena possesses components of the canonical cohesion-segregation machinery characterized in other model systems such as Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis and mice. Interestingly, only one α-kleisin homolog was found in Tetrahymena, which functions in both mitosis and meiosis. In contrast, most other organisms (with the possible exception of Drosophila [62]) have two paralogs, e.g. the mitotic Scc1/Mcd1 and meiotic Rec8 of yeast. Mammalian systems even have three homologs: Rad21, Rec8, and Rad21L, with the latter two being meiosis specific [63]–[65]. One reason for having only one multipurpose α-kleisin in Tetrahymena is its lack of a dedicated pre-meiotic S phase [66], [67]. Cells go from G2 alternatively into mitosis or meiosis, and therefore there is no opportunity to load a specific cohesin.
Special features in the meiotic kleisin have been considered important for maintaining cohesion of sister centromeres in meiosis I so that they segregate together. This is in contrast to the mitotic cohesin, which ensures sisters segregate to opposite poles (for review see [5]). It will be interesting to learn how Tetrahymena Rec8p performs both functions. On the other hand, meiotic Rec8 can take over most mitotic cohesin function in budding yeast [27]–[29]. Thus, the primary question may not be how Tetrahymena can make with a single α-kleisin, but rather why budding yeast and the others require a mitotic and a meiotic version. It was found that Scc1, but not Rec8, can induce DSB-dependent cohesion in mitosis [68], and it is conceivable that Scc1 also performs better in non-canonical cohesin roles such as gene regulation. Thus, a specialized kleisin may have diverged during evolution to be optimized for functions that may be of subordinate importance in Tetrahymena, due to its allocation of gene expression and propagation to different nuclei.
Clearly, the function of cohesin in Tetrahymena has many parallels with previously studied organisms. However, future studies are needed to explore the numerous differences that we have found, including Tetrahymena's use of a single α-kleisin and the continuous association of cohesin with chromatin throughout anaphase.
Materials and Methods
Strains and cell culture
Tetrahymena thermophila strains B2086 and Cu428 served as wild types and as the source strains for transformation with RNAi constructs. Cells were propagated vegetatively at 30°C according to standard methods (see [69]). For meiosis experiments, cells were grown to a density of ∼2×105 cells/ml and made competent for conjugation by starvation in 10 mM Tris–Cl (pH 7.4) for at least 16 h. Conjugation and meiosis were induced by mixing starved cultures of different mating types.
RNAi gene knockdown
The ESP1hp construct was created by amplifying a ∼500 bp region of the ESP1 ORF and cloning it into the rDNA based RNAi vector to create a hairpin expression cassette [45] (see Table S2 for primer sequences). Transformation and selection was performed as previously described [38].
For REC8 RNAi, the transformation strategy was changed slightly. A new vector was created using pBS-CHX, a vector targeting knock-ins to the RPL29 gene which also confers cycloheximide resistance (gift of Chad Pearson). The hairpin expression cassette was released from the rDNA vector using NotI digestion and cloned into the NotI site of the pBS-CHX multiple cloning site (MCS). Then the XmaI site in the MCS was destroyed by blunting and religation in order to allow direct cutting and pasting of hairpin fragments into the PmeI/XmaI and ApaI/XhoI halves of the expression cassette. An ∼500 bp fragment of the REC8 gene was then amplified from genomic DNA using primers that added the appropriate restriction sites, and these fragments were cloned into the two sides of the hairpin cassette. For a map of vector REC8hpCYH see Figure S6. This construct was digested with BlpI and introduced into vegetatively growing Tetrahymena by biolistic transformation. Cells were grown overnight, then selected in 7.5 µg/ml cycloheximide. Transformants were successively grown in higher concentrations of cycloheximide up to 20 µg/ml, to increase the macronuclear copy number of the hairpin containing chromosome.
In order to create a strain for double RNAi of both REC8 and SMC1, it was necessary to use a different selection marker for the SMC1hp, therefore another RNAi vector was created using pMNMM3 (gift of Kazufumi Mochizuki), which carries the NEO5 (paromomycin resistance) gene and targets knock-ins to the MTT1 locus, utilizing the MTT1 cadmium-inducible metallothionein promoter for expression. To facilitate cloning into this vector, a new hairpin linker was amplified from genomic DNA using primers that introduced BamHI and PmeI restriction sites on one end, and XmaI and PstI sites on the other end. This was cloned into the MCS of pMNMM3, and then the ∼500 base pair fragment of SMC1 was amplified and cloned into either end of the linker as in previous constructs. For a map of vector SMC1hpNEO see Figure S6. The construct was digested with NotI and XhoI and introduced into vegetatively growing Tetrahymena (either WT strains or REC8hp strains) by biolistic transformation [70]. Transformants were selected in 120 µg/ml paromomycin, then grown in successively higher concentrations of drug, up to 2 mg/ml.
The SPO11hp construct was prepared in the same way as the REC8hp, using the primers listed (Table S2). Transformation and selection was also performed as for rec8i. In all cases, RNAi was induced by expression of dsRNA from the MTT1 promoter by the addition of 0.2 µg/ml of CdCl2 (final concentration:) to cells carrying the hairpin construct.
Protein tagging
Rec8-GFP, Smc1-HA and Scc3-mCherry expressing cells were created using a knock-in approach to fuse the tag to the C-terminus of each gene at its native genomic locus. Tagging constructs were created by amplifying the last ∼500 bp of the ORF as well as a ∼500 bp region downstream of the gene, and fusing these two products with the tagging cassette using overlapping PCR. (See Table S2 for primer sequences). The tagging cassettes were amplified from pHA-Neo4 or pEGFP-Neo4 (gifts of Kazufumi Mochizuki). Tagging constructs were introduced into the MAC of vegetatively growing B2086 and Cu428 cells by biolistic transformation. Transformants were selected in media containing paromomycin in increasing concentrations up to 50 mg/ml. For detecting Rec8-GFP by Western blotting, protein extracts were prepared by trichloroacetic acid precipitation, run on 10% SDS-PAGE gels and blotted. Membranes were incubated with anti-GFP antiserum and with appropriate HRP-conjugated secondary antibody, and the protein bands were detected by chemiluminescence.
Protein immunoprecipitation and mass spectrometry
Immunoprecipitation against GFP tag was performed using magnetic GFP-trap beads (ChromoTec, Martinsried, Ger), according to the manufacturer's protocol. Lysates were created by sonicating 5×107 mating cells (5 h after mixing) in 2 ml of lysis buffer +1 mM PMSF and protease inhibitors (1× cOmplete mini, Roche, Indianapolis, IN), 4×25 sec at 37% power, duty cycle 5. After 2 h incubation with lysate, the beads were washed 7×5 min with wash buffer (10 mM Tris/Cl pH7.5, 150 mM NaCl, 0.5 mM EDTA), and 2×5 min with 150 mM NaCl. The bead bound proteins were trypsinized, and peptides were loaded on a Dionex UltiMate 3000 HPLC system (Thermo Scientific, San Jose, CA). Peptides separated in a 0.1% formic acid/0% acetonitrile – 0.08% formic acid/80% acetonitrile gradient in water were injected into the mass spectrometer via an electrospray-interface. MS/MS analysis was carried out with a Q Exactive mass spectrometer (Thermo Scientific), and peptide spectra were recorded over a mass range of 350–2000 m/z (for details see [71]).
For peptide identification, the .RAW-files were loaded into Proteome Discoverer 1.4.0.282 (Thermo Scientific). MS/MS spectra created were searched using Mascot 2.2.07 (Matrix Science, London, UK) against the NCBI non-redundant protein sequence database, using the taxonomy group Alveolata.
DNA damage assay
For induction of DNA damage by UV irradiation, 5 ml aliquots of cells were placed in a 90 mm plastic Petri dish. Open dishes were placed in a Stratalinker crosslinker and treated with 254 nm UV (UV-C) at a dosage of 150 Joules/m2. Treatment with ionizing radiation was performed by exposure to 5000 rads of γ-radiation from a 137Cs source. For chemical induction of DNA damage, cells were treated with 100 µg/ml cisplatin (from a 2 mg/ml stock solution in 10 mM Tris-HCl), 50 µg/ml bleomycin (from a 10 mg/ml stock solution in 10 mM Tris-HCl) or 4 mM methyl methane sulfonate (MMS, from a 100 mM stock solution in 10 mM Tris-HCl). Cells were fixed and prepared for immunostaining (see below). Immunostaining of DNA repair protein Rad51 allowed cytological detection of DNA damage in the MAC.
Cell preparation for FACScan analysis
Cell suspensions were centrifuged and Carnoy's fixative (methanol, chloroform, acetic acid 6∶3∶2) was quickly added to the pellet. This disrupts the cells and separates MICs and MACs. Carnoy's fixative was replaced by 70% ethanol after 1 h at room temperature, and the fixations were stored in the freezer. Shortly before measuring, the nuclei were pelleted by centrifugation, resuspended in 1× PBS and stained by the addition of DAPI (4′,6-diamidino-2-phenylindole; final concentration: 0.2 µg/ml).
Cytological preparation, immunostaining, and fluorescence in situ hybridization (FISH)
Different preparation methods were applied for subsequent immunostaining and FISH. A combined formaldehyde fixation and detergent permeabilization treatment [72] was used for subsequent chromosome staining with DAPI or for immunostaining of Dmc1p/Rad51p, PhosH3, and GFP and HA tags. An enforced detergent spreading method for the removal of free nuclear proteins [44] was applied for probing chromatin associated proteins. For immunostaining of γ-H2A.X, cells were fixed with Schaudinn's fixative, washed and resuspended in methanol [73]. For subsequent FISH, cells were fixed with Carnoy's fixative (methanol, chloroform, acetic acid 6∶3∶2). Drops of fixed cell suspensions were dried down on slides.
For immunostaining, slides were washed with 1×PBS and 1×PBS+0.05% Triton, incubated with primary antibodies over night at 4°C, washed again, incubated with appropriate FITC - or Cy3-labeled secondary antibodies for 2 h, and washed and mounted with anti-fading buffer supplemented with 0.5 ìg/ml DAPI. The following primary antibodies were applied: Mouse monoclonal antibody against the related DNA repair proteins Dmc1 and Rad51 (1∶50, Clone 51RAD01, NeoMarkers, Fremont, CA), mouse anti-γ-H2A.X antibody (1∶200, BioLegend, San Diego, CA), rabbit anti-phosphorylated H3Ser10 (1∶500, Upstate Biotechnology, Charlottesville, VA), rabbit anti-GFP (1∶100, Molecular Probes, Eugene, OR), monoclonal mouse anti-HA (1∶50, Roche Diagnostics GmbH, Mannheim, GER) and rabbit anti-HA (1∶100, Sigma St. Louis, MO).
For FISH, a probe against an intercalary chromosomal region, scaffold scf_8254686 (http://ciliate.org), the same as described in ref. [23], was used. DNA on slides and the Cy3-labeled hybridization probe were denatured and hybridized for 36–48 h at 37°C [72].
In all cases, chromatin was counterstained with DAPI, and z stacks of pictures were taken under a fluorescence microscope equipped with the appropriate filters. Picture stacks were deconvolved, projected, assigned false colors, and multicolor images were merged.
Detection of DSBs by pulsed-field gel electrophoresis (PFGE)
For details of the detection of DSB-generated fragments by PFGE, see [44]. In short, chromosome-sized DNA was prepared in agarose plugs. The run was performed in 1% agarose with 0.5× TBE buffer at 200 V, 6°C for 14 h with 60-s pulses, 10 h with 90-s pulses, and 1 h with 120-s pulses in a Bio Rad Chef-DR III system. Under these conditions, intact MIC chromosomes do not enter the gel, whereas fragments migrate as a single band. Since numerous small MAC chromosomes are distributed along the entire gel and cover the DSB-generated signal, MIC-borne DNA fragments were highlighted by Southern detection of a MIC-specific DNA [44]. The membrane was stripped and re-hybridized with a probe against a 121-kb MAC chromosome as a marker to test equal DNA loading and Southern transfer for different time points.
Supporting Information
Zdroje
1. NasmythK, HaeringCH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43 : 525–558.
2. SchleifferA, KaitnaS, Maurer-StrohS, GlotzerM, NasmythK, et al. (2003) Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell 11 : 571–575.
3. ZicklerD, KlecknerN (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33 : 603–754.
4. San FilippoJ, SungP, KleinH (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77 : 229–257.
5. PetronczkiM, SiomosMF, NasmythK (2003) Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell 112 : 423–440.
6. PetersJ-M, TedeschiA, SchmitzJ (2008) The cohesin complex and its roles in chromosome biology. Genes Dev 22 : 3089–3114.
7. KleinF, MahrP, GalovaM, BuonomoSBC, MichaelisC, et al. (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 : 91–103.
8. PasierbekP, JantschM, MelcherM, SchleifferA, SchweizerD, et al. (2001) A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 15 : 1349–1360.
9. SeversonAF, LingL, van ZuylenV, MeyerBJ (2009) The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev 23 : 1763–1778.
10. LiuZ, MakaroffCA (2006) Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis. Plant Cell 18 : 1213–1225.
11. EijpeM, OffenbergH, JessbergerR, RevenkovaE, HeytingC (2003) Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1ß and SMC3. J Cell Biol 160 : 657–670.
12. EllermeierC, SmithGR (2005) Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci USA 102 : 10952–10957.
13. WatrinE, PetersJ-M (2006) Cohesin and DNA damage repair. Exp Cell Res 312 : 2687–2693.
14. DorsettD, StrömL (2012) The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol 22: R240–R250.
15. LandeiraD, BartJ-M, Van TyneD, NavarroM (2009) Cohesin regulates VSG monoallelic expression in trypanosomes. J Cell Biol 186 : 243–254.
16. UhlmannF (2008) Molecular biology: cohesin branches out. Nature 451 : 777–778.
17. HadjurS, WilliamsLM, RyanNK, CobbBS, SextonT, et al. (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460 : 410–413.
18. OriasE, CervantesMD, HamiltonE (2011) Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res Microbiol 162 : 578–586.
19. WolfeJ, HunterB, AdairWS (1976) A cytological study of micronuclear elongation during conjugation in Tetrahymena. Chromosoma 55 : 289–308.
20. RayCJr (1956) Meiosis and nuclear behaviour in Tetrahymena pyriformis. J Protozool 3 : 88–96.
21. SugaiT, HiwatashiK (1974) Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J Protozool 21 : 542–548.
22. MartindaleDW, AllisCD, BrunsPJ (1982) Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp Cell Res 140 : 227–236.
23. LoidlJ, MochizukiK (2009) Tetrahymena meiotic nuclear reorganization is induced by a checkpoint kinase-dependent response to DNA damage. Mol Biol Cell 20 : 2428–2437.
24. EisenJA, CoyneRS, WuM, WuD, ThiagarajanM, et al. (2006) Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol 4: e286 doi:10.1371/journal.pbio.0040286.
25. HaeringCH, SchoffneggerD, NishinoT, HelmhartW, NasmythK, et al. (2004) Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell 15 : 951–964.
26. MiaoW, XiongJ, BowenJ, WangW, LiuY, et al. (2009) Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE 4: e4429 doi:10.1371/journal.pone.0004429.
27. BuonomoSBC, ClyneRK, FuchsJ, LoidlJ, UhlmannF, et al. (2000) Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8p by separin. Cell 103 : 387–398.
28. KitajimaTS, KawashimaSA, WatanabeY (2004) The konserved kinetochore protein Shugoshin protects centromeric cohesion during meiosis. Nature 427 : 510–517.
29. ZenvirthD, LoidlJ, KleinS, ArbelA, ShemeshR, et al. (1997) Switching yeast from meiosis to mitosis: double-strand break repair, recombination and synaptonemal complex. Genes Cells 2 : 487–498.
30. CervantesMD, CoyneRS, XiXH, YaoMC (2006) The condensin complex is essential for amitotic segregation of bulk chromosomes, but not nucleoli, in the ciliate Tetrahymena thermophila. Mol Cell Biol 26 : 4690–4700.
31. McDonaldBB (1966) The exchange of RNA and protein during conjugation in Tetrahymena. J Protozool 13 : 277–285.
32. WaizeneggerIC, HaufS, MeinkeA, PetersJ-M (2000) Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103 : 399–410.
33. CaiX, DongFG, EdelmannRE, MakaroffCA (2003) The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J Cell Sci 116 : 2999–3007.
34. KitajimaTS, YokobayashiS, YamamotoM, WatanabeY (2003) Distinct cohesin complexes organize meiotic chromosome domains. Science 300 : 1152–1155.
35. LosadaA, HiranoM, HiranoT (2002) Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev 16 : 3004–3016.
36. KudoNR, WassmannK, AngerM, SchuhM, WirthKG, et al. (2006) Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell 126 : 135–146.
37. TomonagaT, NagaoK, KawasakiY, FuruyaK, MurakamiA, et al. (2000) Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev 14 : 2757–2770.
38. Howard-TillRA, LukaszewiczA, LoidlJ (2011) The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genet 7: e1001359 doi:10.1371/journal.pgen.1001359.
39. WoodardJ, KaneshiroE, GorovskyMA (1972) Cytochemical studies on the problem of macronuclear subnuclei in Tetrahymena. Genetics 70 : 251–260.
40. JessbergerR (2009) Cohesin's dual role in the DNA damage response: repair and checkpoint activation. EMBO J 28 : 2491–2493.
41. StrömL, LindroosHB, ShirahigeK, SjögrenC (2004) Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16 : 1003–1015.
42. ÜnalE, Arbel-EdenA, SattlerU, ShroffR, LichtenM, et al. (2004) DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16 : 991–1002.
43. CampbellC, RomeroDP (1998) Identification and characterization of the RAD51 gene from the ciliate Tetrahymena thermophila. Nucleic Acids Res 26 : 3165–3172.
44. LukaszewiczA, Howard-TillRA, NovatchkovaM, MochizukiK, LoidlJ (2010) MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma 119 : 505–518.
45. Howard-TillRA, YaoMC (2006) Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol Cell Biol 26 : 8731–8742.
46. CleffmanG (1980) Chromatin elimination and the genetic organisation of the macronucleus in Tetrahymena thermophila. Chromosoma 78 : 313–325.
47. WeiY, MizzenCA, CookRG, GorovskyMA, AllisCD (1998) Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci USA 95 : 7480–7484.
48. BaudrimontA, PenknerA, WoglarA, MamnunYM, HulekM, et al. (2011) A new thermosensitive smc-3 allele reveals involvement of cohesin in homologous recombination in C. elegans. PLoS ONE 6: e24799 doi:10.1371/journal.pone.0024799.
49. Heidinger-PauliJM, MertO, DavenportC, GuacciV, KoshlandD (2010) Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol 20 : 957–963.
50. BrarGA, HochwagenA, EeLS, AmonA (2009) The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol Biol Cell 20 : 1030–1047.
51. LightfootJ, TestoriS, BarrosoC, Martinez-PerezE (2011) Loading of meiotic cohesin by SCC-2 is required for early processing of DSBs and for the DNA damage checkpoint. Curr Biol 21 : 1421–1430.
52. FarcasA-M, UluocakP, HelmhartW, NasmythK (2011) Cohesin's concatenation of sister DNAs maintains their intertwining. Mol Cell 44 : 97–107.
53. ShimadaK, GasserSM (2007) The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell 128 : 85–99.
54. GuacciV, KoshlandD (2012) Cohesin-independent segregation of sister chromatids in budding yeast. Mol Biol Cell 23 : 729–739.
55. BrunsPJ, BrussardTEB (1981) Nullisomic Tetrahymena: eliminating germinal chromosomes. Science 213 : 549–551.
56. Frankel J (2000) Cell biology of Tetrahymena thermophila. In: Asai DJ, Forney JD, editors. Tetrahymena thermophila. San Diego: Academic Press. pp. 27–125.
57. KimKP, WeinerBM, ZhangL, JordanA, DekkerJ, et al. (2010) Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143 : 924–937.
58. KugouK, FukudaT, YamadaS, ItoM, SasanumaH, et al. (2009) Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell 20 : 3064–3076.
59. HiranoT (2005) Cell biology: holding sisters for repair. Nature 433 : 467–468.
60. Cole E, Sugai T (2012) Developmental progression of Tetrahymena through the cell cycle and conjugation. In: Collins K, editors. Tetrahymena thermophila. San Diego: Academic Press. pp. 177–236.
61. WirthKG, WutzG, KudoNR, DesdouetsC, ZetterbergA, et al. (2006) Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol 172 : 847–860.
62. KhetaniRS, BickelSE (2007) Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J Cell Sci 120 : 3123–3137.
63. LeeJ, OgushiS, SaitouM, HiranoT (2011) Condensins I and II are essential for construction of bivalent chromosomes in mouse oocytes. Mol Biol Cell 22 : 3465–3477.
64. HerranY, Gutierrez-CaballeroC, Sanchez-MartinM, HernandezT, VieraA, et al. (2011) The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J 30 : 3091–3105.
65. IshiguroK, KimJ, Fujiyama-NakamuraS, KatoS, WatanabeY (2011) A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep 12 : 267–275.
66. McDonaldBB (1962) Synthesis of deoxyribonucleic acid by micro - and macronuclei of Tetrahymena pyriformis. J Cell Biol 13 : 193–203.
67. DoerderFP, De BaultLE (1975) Cytofluorometric analysis of nuclear DNA during meiosis, fertilization and macronuclear development in the ciliate Tetrahymena pyriformis, syngen 1. J Cell Sci 17 : 471–493.
68. Heidinger-PauliJM, ÜnalE, GuacciV, KoshlandD (2008) The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell 31 : 47–56.
69. Orias E, Hamilton EP, Orias JD (2000) Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. In: Asai DJ, Forney JD, editors. Tetrahymena thermophila. San Diego: Academic Press. pp. 189–211.
70. Cassidy-HanleyD, BowenJ, LeeJH, ColeE, VerPlankLA, et al. (1997) Germline and somatic transformation of mating Tetrahymena termophila by particle bombardment. Genetics 146 : 135–147.
71. KöcherT, PichlerP, SwartR, MechtlerK (2012) Analysis of protein mixtures from whole-cell extracts by single-run nanoLC-MS/MS using ultralong gradients. Nature Protoc 7 : 882–890.
72. LoidlJ, ScherthanH (2004) Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci 117 : 5791–5801.
73. MochizukiK, NovatchkovaM, LoidlJ (2008) DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J Cell Sci 121 : 2148–2158.
74. RonquistF, TeslenkoM, van der MarkP, AyresDL, DarlingA, et al. (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol Epub ahead of print.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



