-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
Faithful chromosome segregation during meiosis requires that homologous chromosomes associate and recombine. Chiasmata, the cytological manifestation of recombination, provide the physical link that holds the homologs together as a pair, facilitating their orientation on the spindle at meiosis I. Formation of most crossover (CO) events requires the assistance of a group of proteins collectively known as ZMM. HFM1/Mer3 is in this group of proteins and is required for normal progression of homologous recombination and proper synapsis between homologous chromosomes in a number of model organisms. Our work is the first study in mammals showing the in vivo function of mouse HFM1. Cytological observations suggest that initial steps of recombination are largely normal in a majority of Hfm1−/− spermatocytes. Intermediate and late stages of recombination appear aberrant, as chromosomal localization of MSH4 is altered and formation of MLH1foci is drastically reduced. In agreement, chiasma formation is reduced, and cells arrest with subsequent apoptosis at diakinesis. Our results indicate that deletion of Hfm1 leads to the elimination of a major fraction but not all COs. Formation of chromosome axial elements and homologous pairing is apparently normal, and Hfm1−/− spermatocytes progress to the end of prophase I without apparent developmental delay or apoptosis. However, synapsis is altered with components of the central region of the synaptonemal complex frequently failing to extend the full length of the chromosome axes. We propose that initial steps of recombination are sufficient to support homology recognition, pairing, and initial chromosome synapsis and that HFM1 is required to form normal numbers of COs and to complete synapsis.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003383
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003383Summary
Faithful chromosome segregation during meiosis requires that homologous chromosomes associate and recombine. Chiasmata, the cytological manifestation of recombination, provide the physical link that holds the homologs together as a pair, facilitating their orientation on the spindle at meiosis I. Formation of most crossover (CO) events requires the assistance of a group of proteins collectively known as ZMM. HFM1/Mer3 is in this group of proteins and is required for normal progression of homologous recombination and proper synapsis between homologous chromosomes in a number of model organisms. Our work is the first study in mammals showing the in vivo function of mouse HFM1. Cytological observations suggest that initial steps of recombination are largely normal in a majority of Hfm1−/− spermatocytes. Intermediate and late stages of recombination appear aberrant, as chromosomal localization of MSH4 is altered and formation of MLH1foci is drastically reduced. In agreement, chiasma formation is reduced, and cells arrest with subsequent apoptosis at diakinesis. Our results indicate that deletion of Hfm1 leads to the elimination of a major fraction but not all COs. Formation of chromosome axial elements and homologous pairing is apparently normal, and Hfm1−/− spermatocytes progress to the end of prophase I without apparent developmental delay or apoptosis. However, synapsis is altered with components of the central region of the synaptonemal complex frequently failing to extend the full length of the chromosome axes. We propose that initial steps of recombination are sufficient to support homology recognition, pairing, and initial chromosome synapsis and that HFM1 is required to form normal numbers of COs and to complete synapsis.
Introduction
Meiosis is a reductional type of cell division in which two rounds of chromosome segregation follow one round of DNA replication. This is a highly regulated and essential process to generate gametes containing the correct haploid chromosome complement. The first segregation, meiosis I, is reductional because homologous chromosomes, instead of sister chromatids, migrate to the opposite poles of the spindle. Recombination in meiosis I prophase prepares homologous chromosomes for segregation. Chiasmata, the cytological manifestation of recombination, provide physical links that hold the homologs in pairs and ensure the proper segregation of each chromosome of a pair to opposite poles of the spindle so that ultimately each gamete receives one copy of each chromosome [1], [2]. Mutations that reduce the level of recombination are invariably associated with increased errors in meiotic chromosome segregation [3], [4]. Examples of human diseases caused by aneuploidy include Down, Klinefelter, Edwards and Turner syndromes [5]. In order to explain the several causes of aneuploidy we need to identify and understand the mechanisms of homologous recombination that regulate proper number and positioning of COs in meiotic chromosomes.
The initial steps of homologous recombination involve introduction of double-strand breaks (DSBs) by the topoisomerase-like protein SPO11 [6], [7], [8], [9] followed by DNA end processing by exonucleases to generate 3′ single-stranded DNA tails [10], [11], [12]. Subsequently, RAD51 and DMC1 recombinases form filaments on the resected DNA tails and, together with ancillary proteins [13], [14], [15], catalyze the search for homologous sequences, resulting in repair of DSBs by promoting the invasion of intact double-stranded DNA by the single-stranded DNA ends (initial strand invasion) [15], [16], [17]. It is currently accepted that strand invasion intermediates proceed by one of two distinct pathways. They may dissociate after limited heteroduplex extension by DNA synthesis with subsequent re-joining of the broken chromosome by synthesis-dependent strand annealing to generate non-crossovers (NCO) [9], [18], [19]. Alternatively, initial strand invasion becomes stabilized by 3′ to 5′ unwinding of duplex DNA that promotes synthesis-mediated extension and gives rise to more stable single-end invasions [20], [21], [22] and double Holliday joins (dHJs) [23]. Intermediates in the second pathway complete repair to generate COs [18], [20], [21], [23], [24], [25]. Regulation of CO products is critical in meiosis but the players and mechanisms of control are poorly understood in higher eukaryotes. Two pathways have been proposed for the formation of COs in higher eukaryotes [26], [27], [28], [29], [30], [31], [32]. The majority of COs (90–95% in mouse) are processed by the ZMM group of proteins which include SYCP1 protein from the transverse filaments [33], [34], [35], [36], [37] of the synaptonemal complex, the ATP-dependent helicase Mer3 (HFM1 in mouse) [28], [38], [39], [40], the MSH4-MSH5 complex [34], [41], [42], [43], [44] and the MLH1-MLH3 heterodimer. This class of COs, termed class I, exhibits unique properties such as interference and homeostasis [45], [46], [47]. The second class of COs (Class II) depends on the structure specific endonuclease MUS81 that promotes the alternative MUS81-MMS4 pathway [48], [49].
Based on sequence homology with proteins known to be DNA helicases, mouse and human HFM1 and other Mer3 homologs are predicted to have seven motifs characteristic of the DExH box type of DNA/RNA helicases [40]. Mer3-deficient budding yeast strains carrying mutations in some of these motifs exhibit meiotic defects in DNA recombinational repair [50]. Biochemical evidence indicates that purified recombinant Mer3 from budding yeast works at an early step of recombination by stabilizing initial strand invasion catalyzed by RAD51 via DNA heteroduplex extension [51].
Mutants that inactivate HFM1/Mer3 orthologs in yeast, Arabidopsis, rice, Coprinus cinereus and Sordaria macrospora demonstrate the importance of Mer3 in the normal progression of several critical aspects of meiosis. Molecular analysis of intermediates of recombination in yeast Mer3 mutants reveals that DSBs are formed normally but that repair is delayed, single-end invasions are reduced (reaching a maximum of 15% of wild-type levels) and formation of meiotic recombination-dependent DNA synthesis is rare [21], [24]. Formation of later intermediates of recombination, dHJs, is delayed and severely reduced. This is accompanied by a 50–60% decrease in CO formation, and the remaining COs do not show interference [21], [24], [50], [52]. Similarly, genetic analysis of Arabidopsis Mer3 mutants shows that formation of COs is severely reduced (by 38%) but not eliminated, and that the remaining COs do not show interference [28], [38]. Notably, even though COs are severely affected in budding yeast, NCO intermediates are detected at normal levels in the absence of Mer3 [21], [24]. Cytological studies of Mer3 mutants suggest conserved function(s) of HFM1/Mer3 across all analyzed species. Hfm1/mer3 mutants display severe deficiencies in chiasma formation, high levels of chromosome missegregation at meiosis I and partial or total infertility [21], [24], [28], [38], [39], [50], [52], [53], [54]. For budding yeast, Arabidopsis, rice, C. Cinereus and S. macrospora Mer3 mutants, development of axial elements is apparent normal and, except for S. macrospora, where homolog alignment is inefficient, the onset of chromosome pairing shows no differences with respect to wild-type [21], [28], [38], [39], [50], [52], [53], [54]. Notably, later in prophase I, synaptic defects are manifested in all Hfm1/mer3 mutants. However, there is a wide variation in the extent of synapsis failure across species. Budding yeast and C. cinereus show the most severe synaptic phenotype, where the onset of synaptonemal complex formation is delayed [21], [24], [39]; in yeast, up to 90% of nuclei have separated homologous axes. Abnormal synapsis is also observed in S. macrospora where it persists among late pachytene cells and is accompanied by a high level of interwoven chromosomes, the latter being more severe after chromosome alignment [54]. Defects in synapsis are apparently moderate in Arabidopsis and rice, where EM and immunofluorescence analyses show only a partial asynapsis of chromosomes at pachytene [38], [53]. Results are controversial in Arabidopsis, however, in which one report describing the phenotype of four mutant alleles shows normal synaptonemal complex formation and no defect in synapsis [28]. These differences may reflect allelic differences. Nevertheless, failures in normal synaptonemal complex assembly across species indicate that the requirement for Mer3 in chromosome synapsis is evolutionarily conserved. Variability in the severity of synapsis defects and DNA repair abnormalities is reflected in the ability of Mer3 mutant meiocytes in different species to progress throughout prophase I. In yeast, arrest in meiotic progression results in dramatic reduction of spores and this strong arrest has been proposed to occur at late leptotene [21], [24], [50], [52]. In S. macrospora Mer3 mutants, the leptotene-pachytene transition is delayed, accompanied by defects in bouquet dynamics [54]. Initial stages of meiosis in both C. cinereus and Arabidopsis Mer3 mutants are apparently normal. However, in C. cinereus the majority of cells do not progress beyond meiosis I and became apoptotic [39]. In contrast, in Arabidopsis, one report shows that anomalous chromosome behavior first becomes apparent at diakinesis where all chromosomes are present as univalents which segregate randomly at metaphase I [28].
In this study we provide the first analysis of meiotic progression in HFM1/Mer3 null mice. We show that mutants of both sexes are infertile and that males show meiotic arrest at the end of prophase I. In a majority of Hfm1−/− spermatocytes the number of initial recombination events appears comparable to that observed in wild-type mice, but recombination is defective and the number of bivalents at diakinesis is reduced to ∼20%. These results are consistent with current models of recombination in which HFM1 participates in the major CO pathway. Although CO formation is severely defective in Hfm1−/− spermatocytes, chromosomes pair and initial synapsis is apparently normal. Our results suggest that early recombination transactions are sufficient for homologous recognition, pairing and initial synapsis. However, synapsis is incomplete for all but a small number of chromosomes in Hfm1−/− spermatocytes. We propose that completion of synapsis generally depends on an HFM1 activity that leads to the formation of COs.
Results
HFM1 is required for completion of meiosis
To understand the role of HFM1/Mer3 in mammals we generated Hfm1−/− mice. The gene trap vector utilized to disrupt Hfm1 contains a viral long terminal repeat (LTR) followed by a splice acceptor sequence (SA) and a promoter-less selectable marker neomycin (Neo) with a polyadenylation signal (pA). The vector also contains the PGK promoter followed by the first exon of the Bruton's Tyrosine Kinase (Btk) gene upstream of a splice donor (SD). The Btk exon contains termination codons in all reading frames to prevent translation of downstream fusion transcripts (Figure 1a). Hfm+/− and Flpe+ intercross results in the formation of Hfm−/− mice (Figure 1b–1g) in which no Hfm1 RNA transcript is detected (Figure 1c and 1d). Two pairs of oligonucleotides specific for RNA transcript in three different regions of Hfm1 were tested (exon boundaries 2–3, 15–16 and 36–37). RT-PCR analysis of Hfm1−/−/Flp+ mice detected no Hfm1 transcripts for any of the predicted alternative splicing forms of Hfm1(Figure 1c and 1d). Thus, we have generated mice that do not express any part of the HFM1 protein.
Fig. 1. Hfm1 gene target design and expression of Hfm1 in mutant mice. 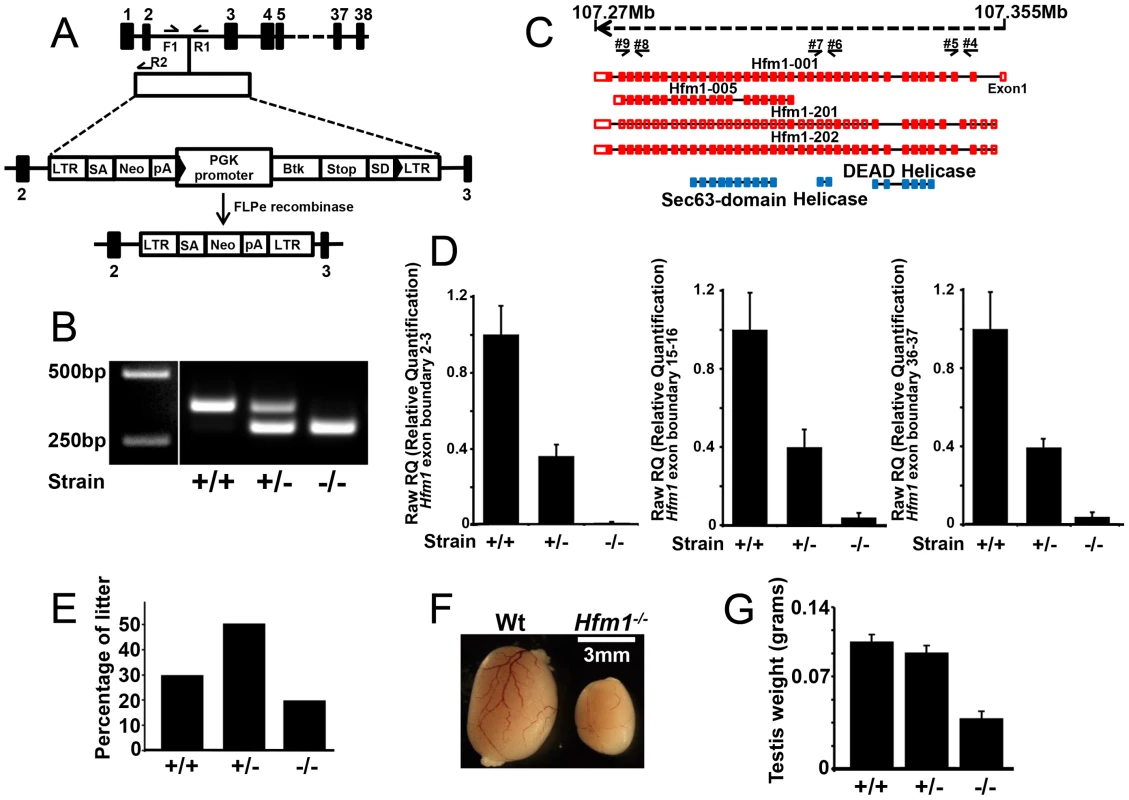
(a) The mouse Hfm1 gene-targeting construct. A trapping cassette was inserted into intron 2 of the Hfm1gene. F1/R1 and F1/R2 represent primers used for genotyping wild-type and knockout mice, respectively. Exons are shown as numbered black bars. (b) Genotyping strategy for identification of Hfm1−/− mice. (c) Predicted alternative splicing variants of Hfm1 (Hfm1 001, 005, 201 and 202). Exons are shown as red boxes and blue boxes represent portions of the HFM1 protein with recognized domains. Black arrows indicate the positions of primers used for RT-PCR analysis. (d) Transcript levels in testes of wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice measured by real time RT-PCR. (e) Percentage of litter obtained from Hfm1 heterozygous couples. (f) Hfm1−/− males display significantly smaller testes than wild-type littermates. (g) Quantification of testis weight for wild-type, heterozygous and homozygous mice. Hfm1−/− mice are viable. Out of the first 196 pups born, 39 (19.9%) are homozygous Hfm1−/−, a number that is close to a normal Mendelian distribution, and suggests that Hfm1 is not an essential gene (Figure 1e). Hfm1−/− adult mice appear normal in all aspects except for reproductive tissues. Hfm1−/− males (0.044 g±0.005, n = 10) had significantly smaller testes than wild-type (0.1 g±0.0063, n = 8; P≤0.0001, t test) littermates (Figure 1f and 1g). This substantial reduction in size is similar to those observed following deletion of other meiosis-specific homologous recombination genes, including Hop2, Dmc1 and Mlh1 [3], [55], [56].
Both male and female homozygous mutant mice were sterile due to severe blocks of spermatogenesis and oogenesis. Tissue analysis indicates that Hfm1−/− males develop testicular hypoplasia with hyperplasia of interstitial cells and lack spermatozoa. The number of seminiferous tubules is not reduced but the diameter is 30%–50% of normal. Primary spermatocytes represent the most advanced spermatogenic cells in the Hfm1−/− mice, indicating that spermatogenesis is blocked at diakinesis of meiosis I (Figure 2a, top row). TUNEL analysis for detection of apoptotic cells reveals that in contrast to wild-type, in which only occasional apoptotic cells are present, apoptosis of spermatocytes at diakinesis is common in seminiferous tubules of Hfm1−/− mice (Figure 2a, green cells after incorporation of Fluorescein-dUTP). We also analyzed hematoxylin-eosin histological sections of 45-days-old wild-type and Hfm1−/− ovaries. We note a significant reduction in ovary size, increase in stromal cells, reduced number of folicules and corpora lutea in the Hfm1−/− knockout mice. Nonetheless, we observed a small number of corpora lutea suggesting that ovulation could occur in the HFM1-deficient females, albeit infrequently (Figure 2b). Matings of heterozygous mice of both sexes produced litters of normal size but mating involving Hfm1−/− females were unsuccessful, suggesting that these females were infertile (results not shown).
Fig. 2. Hfm1−/− mice show profound defects in gametogenesis. 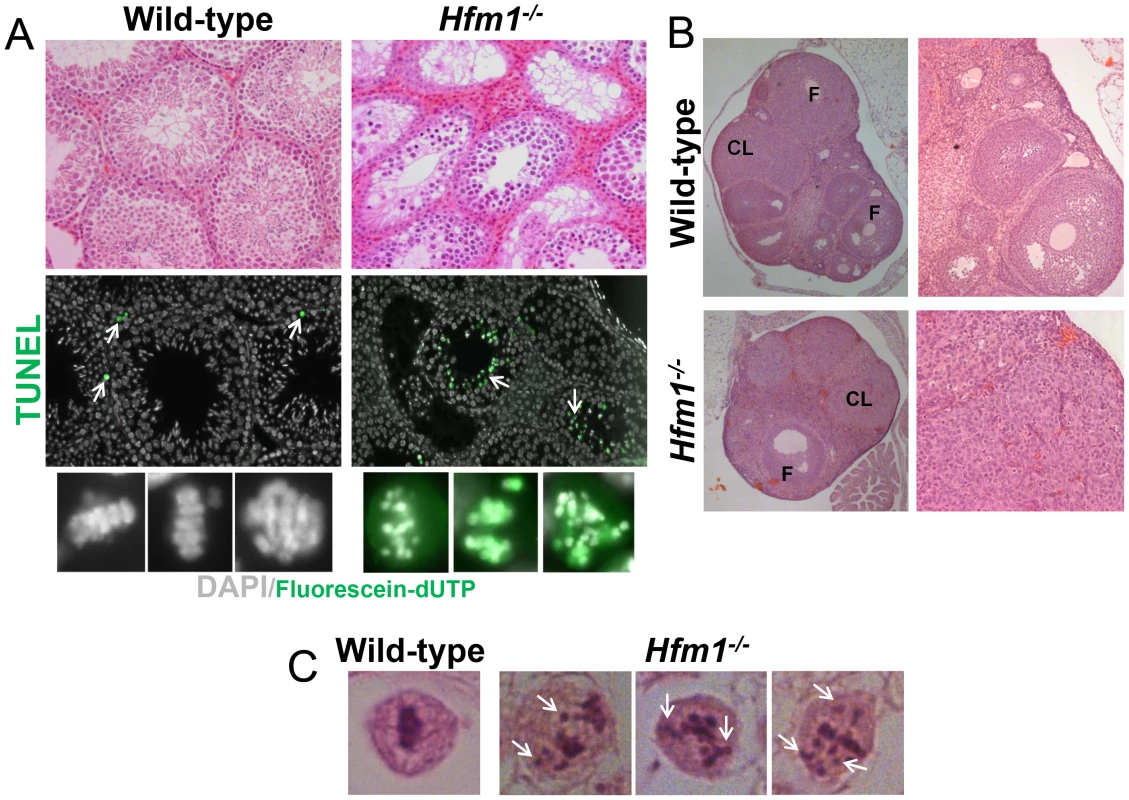
(a) Histological sections of wild-type and Hfm1−/− testes. TUNEL assay detecting apoptotic cells is also shown (middle and lower rows). Arrows mark examples of cells after incorporation of Fluorescein-dUTP. The bottom row shows higher magnification images of diakinesis spermatocytes. (b) Histological sections of wild-type and Hfm1−/− ovaries. CL and F identify corpus lutea and follicles respectively. (c) Single spermatocytes showing a typical metaphase I plate arrangement of chromosomes in wild-type but lagging chromosomes scattered throughout the nucleus in Hfm1−/−. In summary, tissue analysis suggests that spermatogenesis is apparently normal until the end of prophase I when spermatocytes arrest at diakinesis and indicates that prophase I checkpoints are not triggered in Hfm1−/− mice. Chromosomes appeared to be normally condensed at metaphase I in Hfm1−/− cells but, in contrast to wild-type cells, the chromosomes often failed to form a metaphase plate and remained scattered throughout the nucleus (Figure 2a (lower panel) and 2c).
Cytological markers suggest that later stages of recombination are defective in Hfm1−/−
As described in the Introduction, deletion of HFM1/Mer3 in a number of species results in abnormal progression of homologous recombination. We tested progression of recombination in mouse Hfm1−/− spermatocytes. DSB repair can be assayed indirectly by staining chromosomes for γH2AX [57], [58], which appears on chromatin near DSBs [59] then disappears with repair. Wild-type and Hfm1−/− pachytene spermatocytes were identified by typical morphological characteristics. Chromosomes look very compact and relatively stiff, at least 95% of the axial/lateral elements (SYCP3 signal) of homologous chromosomes are closely coaligned (synapsed), and sex chromosome axes appear in a typical pachytene conformation as previously defined by Dresser, et. al. [60]. In the case of γH2AX staining, for quantitation purposes, pachytene-like Hfm1−/− spermatocytes were subdivided into two categories by the state of X and Y chromosome pairing (XY = tightly associated and X-Y = loosely associated). We note that in wild-type cells at pachytene and diplotene, immunostaining with γH2AX marks only the sex body (n = 100). For a fraction of Hfm1−/− spermatocytes, however, some γH2AX persists not only in the sex body but also in one or several patches along autosomes in pachytene-like spermatocytes with either tightly (30.8%±7, n = 210) or loosely associated (43.2%±5.1, n = 120) X and Y chromosomes. Remnants of γH2AX can also be observed in a fraction of diplotene Hfm1−/− spermatocytes (27.4%±6.2, n = 240) (Figure 3a and 3b).
Fig. 3. Abnormal accumulation of γH2AX, RAD51, and MSH4 foci in meiotic chromosomes of Hfm1−/− spermatocytes. 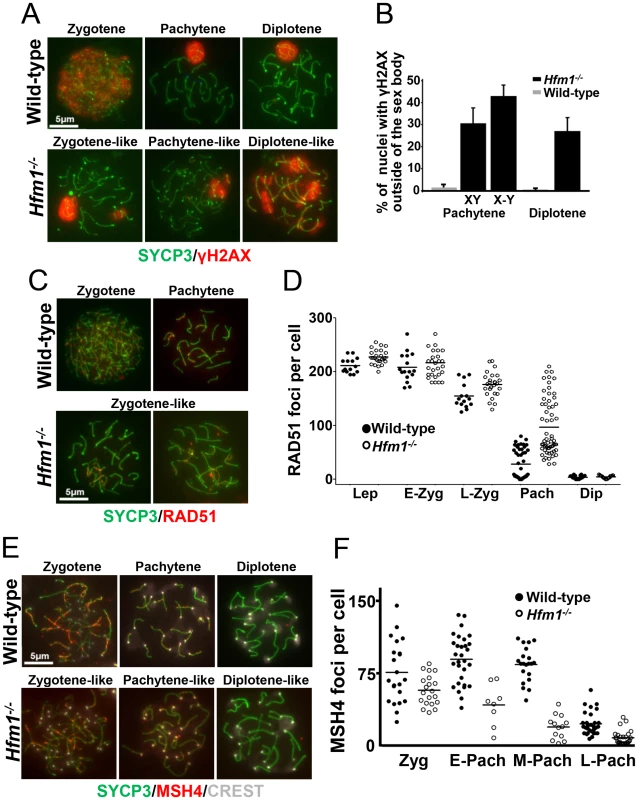
(a) Chromosome spreads of wild-type and Hfm1−/− spermatocytes immunostained for SYCP3 and γH2AX. Magnification bar represents 5 µm. (b) Quantification of wild-type and Hfm1−/− spermatocytes showing additional γH2AX signal. (c) Representative spermatocytes of wild-type and Hfm1−/− mice at different stages of prophase I immunostained for SYCP3 and RAD51. Magnification bar represents 5 µm. (d) Quantification of the number of RAD51 foci per cell at the indicated stages. Lep, leptotene; E-Zyg, early zygotene; L-Zyg, late zygotene; Pach, pachytene and Dip, diplotene. Horizontal lines denote means. See Table 1 for summary of means, standard deviations, and results of statistical tests. (e) Spread nuclei of wild-type and Hfm1−/− spermatocytes immunostained for SYCP3 and MSH4. (f) Quantification of MSH4 foci per cell at the indicated stages. Horizontal lines denote means. See Table 1 for summary of means, standard deviations, and results of statistical tests. RAD51 forms cytologically-detectable complexes at sites of ongoing recombination [61], [62]. We determined the kinetics of formation and disapeareance of RAD51 foci on meiotic chromosomes of wild-type and Hfm1−/− mice. RAD51 foci decrease from late zygotene (154.6±21.6 foci per cell, n = 15) to pachytene (27.9±27.1 foci per cell, n = 55) in wild-type spermatocytes but were elevated in Hfm1−/− spermatocytes in late zygotene (176.1±22.2 foci per cell, n = 28, P≤0.001, t test) and persisted at particularly high levels in 31% of pachytene-like nuclei (96.9±41.2 foci per cell, n = 80, P≤0.001, t test) (Figure 3c and 3d and Table 1). Persistent RAD51 foci at pachytene, as previously described for yeast and S. macrospora Mer3 mutants [52], [54], may indicate delayed and/or inefficient DSB repair. However, it is possible that persistent RAD51 foci reflect the failure to remove RAD51 from dsDNA even though repair is complete.
Tab. 1. Number of recombination-associated foci in spermatocytes. 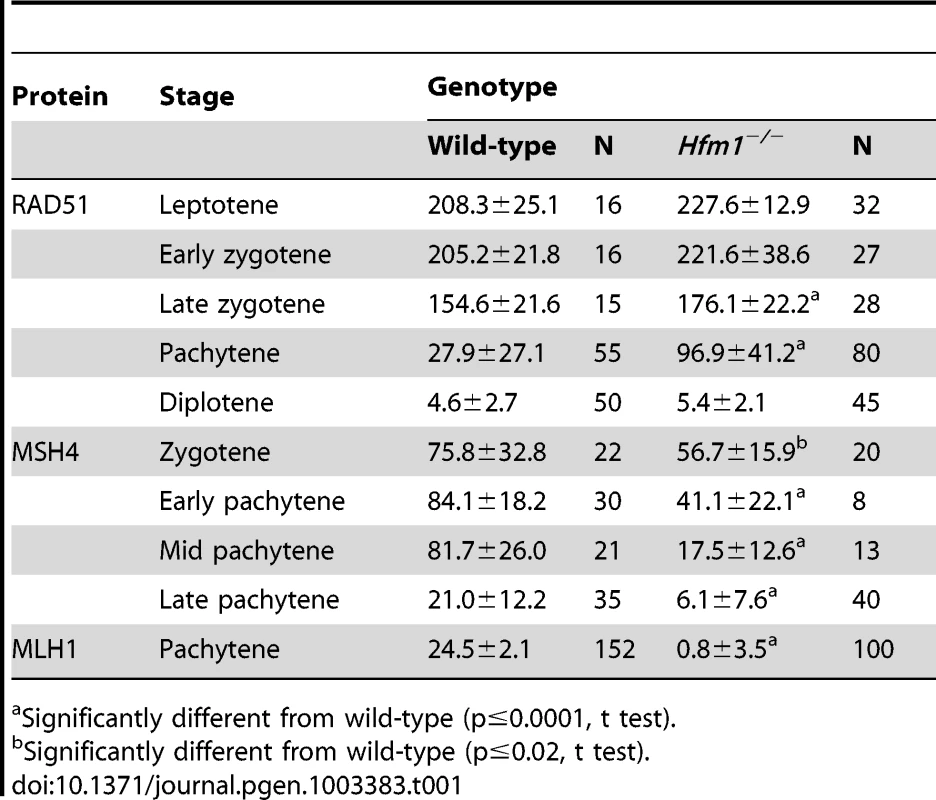
Significantly different from wild-type (p≤0.0001, t test). In sum, our results suggest that DSBs are formed essentially at wild-type levels in Hfm1−/− spermatocytes, and that for a major fraction of Hfm1−/− cells the numbers and kinetics of γH2AX and RAD51 immunosignals are similar to wild-type.
MSH4 is a member of the mammalian mismatch repair gene family whose members are involved in the control of meiotic recombination. Previous studies have shown that MSH4 is present in the nuclei of spermatocytes early in prophase I and that it forms discrete foci along meiotic chromosomes during the zygotene and pachytene stages of meiosis [43]. Compared to wild-type cells, Hfm1−/− spermatocytes showed a reduced number of MSH4 foci at zygotene (74.9% P≤0.041, t-test), early pachytene (48.8%, P≤0.0007, t test), mid pachytene (21.4%±, P≤0.0001, t test) and late pachytene (29%, P≤0.0001, t test) (Figure 3e and 3f and Table 1). Thus, HFM1 is required for accumulation of normal numbers of MSH4 foci.
MLH1 mismatch repair protein is required for CO formation in mice. The accumulation of MLH1 foci along chromosome axes in early to mid pachytene is thought to occur at sites where COs are established [26], [63]. In agreement with published results, in wild-type cells we observed an average of 24.5±2.1 MLH1 foci per cell (n = 152) [63], [64], [65]. However, we observed only background signal in pachytene Hfm1−/− spermatocytes (0.8±3.5, n = 100) (Figure 4a and 4b and Table 1), suggesting a defect in the class I crossover pathway.
Fig. 4. Reduced number of COs in HFM1-deficient spermatocytes. 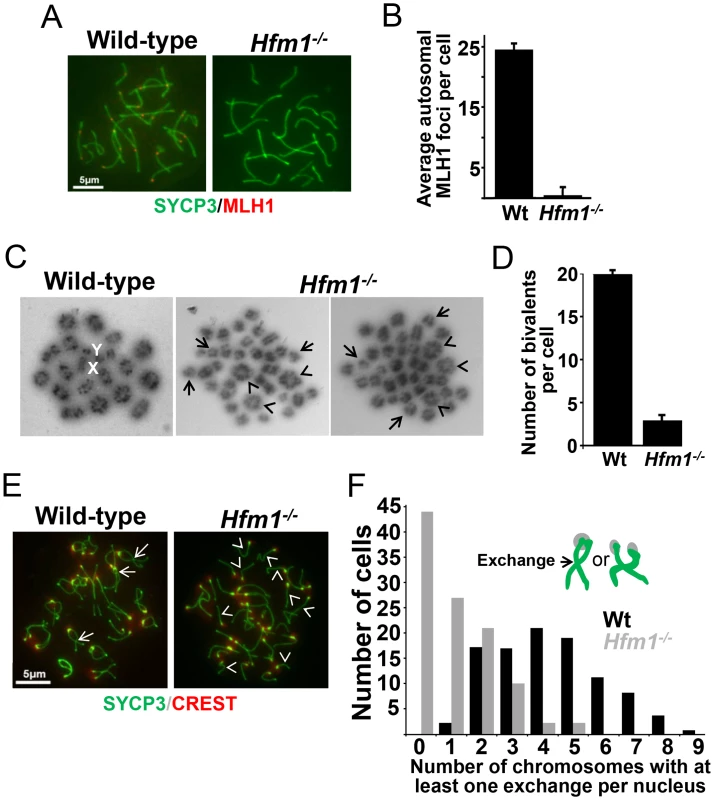
(a) Spread nuclei of wild-type and Hfm1−/− pachytene spermatocytes immunostained for SYCP3 and MLH1. Magnification bar represents 5 µm. (b) Quantification of autosomal MLH1 foci per pachytene cells. See Table 1 for summary of means, standard deviations, and results of statistical tests. (c) Metaphase spreads of wild-type and Hfm1−/− spermatocytes. Note the reduced number of bivalents (arrowheads) and increased number of univalents (tailed arrows) in Hfm1−/− cells. X and Y indicate the sex chromosomes. (d) Quantification (mean ± standard deviation) of metaphase bivalents per cell. (e) Representative diplotene wild-type and Hfm1−/− spermatocytes stained with anti-SYCP3 and CREST (a centromere marker) antibody. Note the increase in univalent chromosomes of knockout cells (arrowheads). Tailed arrows indicate examples of forming chiasmata in wild-type spermatocytes. Magnification bar represents 5 µm. (f) Quantification of cells in (e). Chiasma frequency is substantially reduced in Hfm1−/− spermatocytes
To confirm that deletion of HFM1 reduces CO formation, we analysed chiasma frequency in wild-type and Hfm1−/− spermatocytes. Chiasmata join homologs to make stable bivalents, so we first scored the number of bivalent chromosomes per cell in air-dried diakinesis spreads. We found an average of 20±0.5 (n = 100) bivalents per wild-type spermatocyte and only 3.7±2.2 (n = 100) bivalents per Hfm1−/− spermatocyte (Figure 4c and 4d). The dramatic reduction (81.3%, t test, P≤0.001) in the number of bivalents observed in Hfm1−/− cells strongly suggests a requirement for HFM1 in the formation of COs. As a second test, we measured the number of chromosomes at mid-diplotene with apparent mid-arm chiasmata (Figure 4e and 4f). We again observed a significant difference in the distribution of chiasmata per cell for wild-type (mode of 5 chiasmata per cell) compared to Hfm1−/− (mode of 0 chiasmata per cell) (P = 0.03, Wilcoxon rank-sum test). The reduction in the number of chiasmata in knockout spermatocytes indicates a requirement for HFM1 in the formation of the major fraction of COs in mice.
HFM1 is required for normal levels of XY synapsis but not for sex body formation
In early pachytene spermatocytes, X and Y chromosome pairs normally form a short stretch of synaptonemal complex encompassing the small region of homology in the pseudo-autosomal region (PAR). Synapsis at PAR is very sensitive to reduced rates of recombination and perturbations in synapsis, and thus can be used to detect defects in these processes [65]. The sex body formed apparently normally in all pachytene and diplotene Hfm1−/− spermatocytes. However, the X and Y chromosome PAR regions are unsynapsed and separated in 43.5% of Hfm1−/− cells (n = 161) versus only 5% of wild-type cells (n = 110, P≤0.001 Fisher's exact test) (Figure 5a and 5b). It is possible that XY synapsis fails to initiate at wild-type frequencies. However, a later defect in synapsis (see below) suggests the alternative that XY association is lost prematurely.
Fig. 5. Effect of Hfm1 deletion on sex chromosome association. 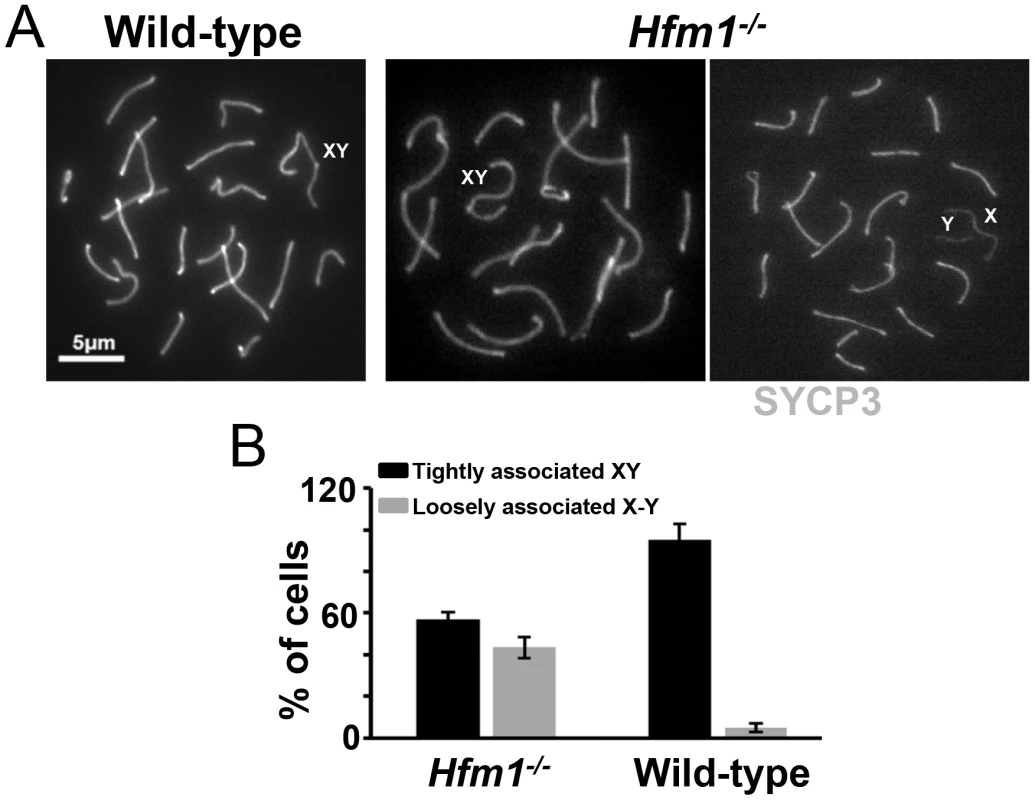
(a) Representative pachytene wild-type and Hfm1−/− spermatocytes immunostained for SYCP3 showing tightly or loosely associated X and Y chromosomes. Magnification bar represents 5 µm. (b) Quantification of cells in (a). Bars represent standard deviation obtained from 4 wild-type and 3 Hfm1−/− mice. Homologous chromosomes pair but synapsis is frequently defective in Hfm1−/− spermatocytes
Given that meiotic recombination and synapsis (i. e., synaptonemal complex formation) are codependent processes [66], we tested the requirement for HFM1 in synapsis by localizing synaptonemal complex proteins SYCP3 of the axial/lateral element and SYCP1 of the central element. Autosomal axes of similar lengths are found in coaligned pairs, indicating that development of the axial element of the synaptonemal complex and homologous chromosome pairing are normal in all Hfm1−/− spermatocytes. In addition, absence of splitting of the axes indicates that sister chromatid cohesion is not affected by the absence of HFM1 (21±0.7 centromeres for wild-type (n = 100) and 21±1.2 centromeres for Hfm1−/− (n = 100) spermatocytes at pachytene) (Figure 6a). Spermatocytes progress through meiotic prophase I in Hfm1−/− mice (Figure 6b and 6c). When individual stages of asynchronous populations of wild-type and Hfm1−/− spermatocytes from adult mice were compared, we observed an increase in the percentage of diplotene (wild-type, 40.1±2, n = 672; Hfm1−/−, 49.96±3, n = 686; P≤0.008, t test) and diakinesis (wild-type, 1.7±0.2, n = 20; Hfm1−/−, 4.81±0.5, n = 66; P≤0.0001 t test) and a decrease in pachytene (wild-type, 52.17±4.2, n = 614; Hfm1−/−, 36.27±3.2, n = 620; P≤0.0008, t test) in Hfm1−/− spermatocytes relative to wild-type. However, no significant differences were observed when the distribution of spermatocytes for the entire population of spermatocytes (from leptotene to diakinesis) from wild-type and Hfm1−/− were compared (Wilcoxan rank P = 0.3, one sided).
Fig. 6. Meiotic prophase I progress in wild-type and Hfm1−/− mice. 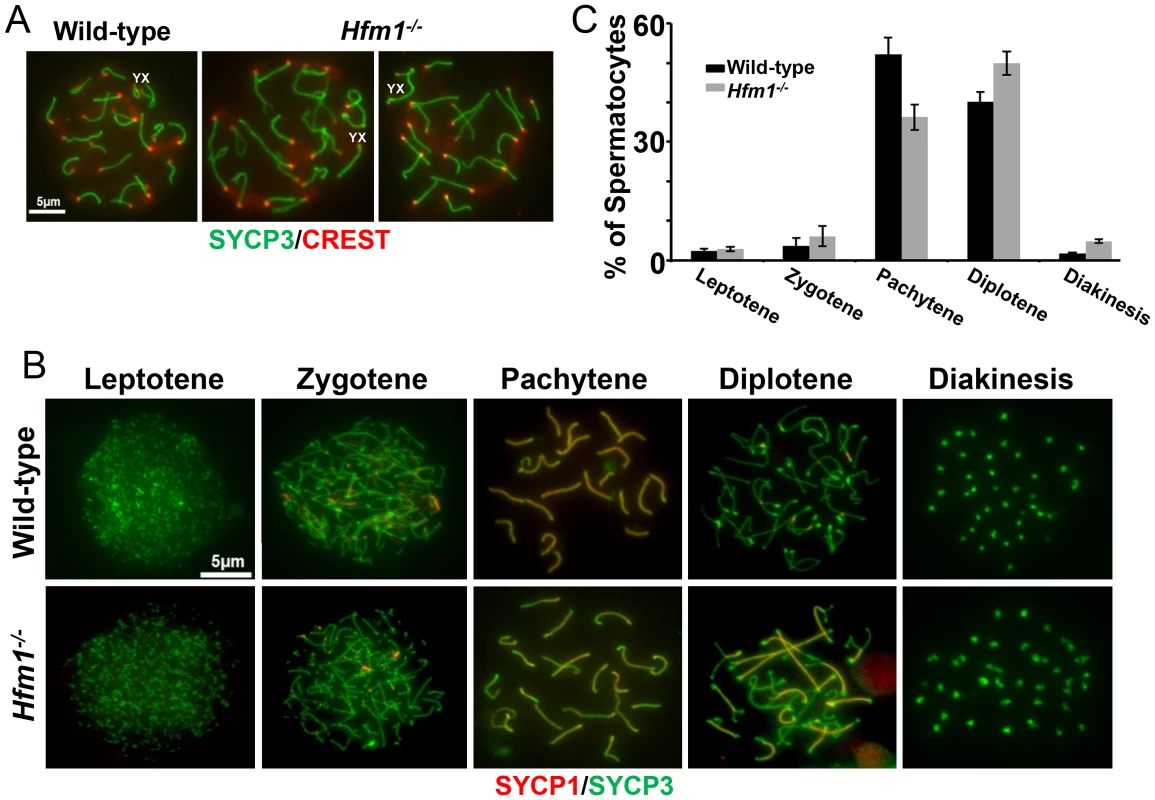
(a) Representative pachytene wild-type and Hfm1−/− spermatocytes stained with SYCP3 and CREST. Magnification bar represents 5 µm. (b) Co-immunostaining of SYCP3 and SYCP1 in wild-type and Hfm1−/− spermatocytes with no apparent synaptic defects at different stages of prophase I. Note that diakinesis is the last stage of spermatogenesis detected for Hfm1−/− cells. (c) Composition of spermatocyte population in wild-type and Hfm1−/− spermatocytes. Notably, synaptic defects are evident early in pachytene spermatocytes (Figure 7a–7d). We analyzed the numbers and types of synaptic anomalies in pachytene where at least 95% of the axial/lateral elements (SYCP3 signal) of homologous chromosomes are closely coaligned (synapsed) and sex chromosome axes show the typical early pachytene configuration [60]. Pachytene-like spermatocytes with at least one abnormal synaptic conformation constituted 46.2% of all scored cells (n = 91) and 25.7% of total scored chromosomes showed some type of synaptic anomaly. Incompletely synapsed bivalents had one end unsynapsed, generally the centromere distal end (62% of the total chromosomes exhibiting anomalies) (Figure 7c). Thus, chromosome homologous recognition and pairing appears normal in Hfm1−/− spermatocytes but synapsis is frequently defective.
Fig. 7. Homologous chromosomes pair normally and achieve initial synapsis in Hfm1−/− spermatocytes. 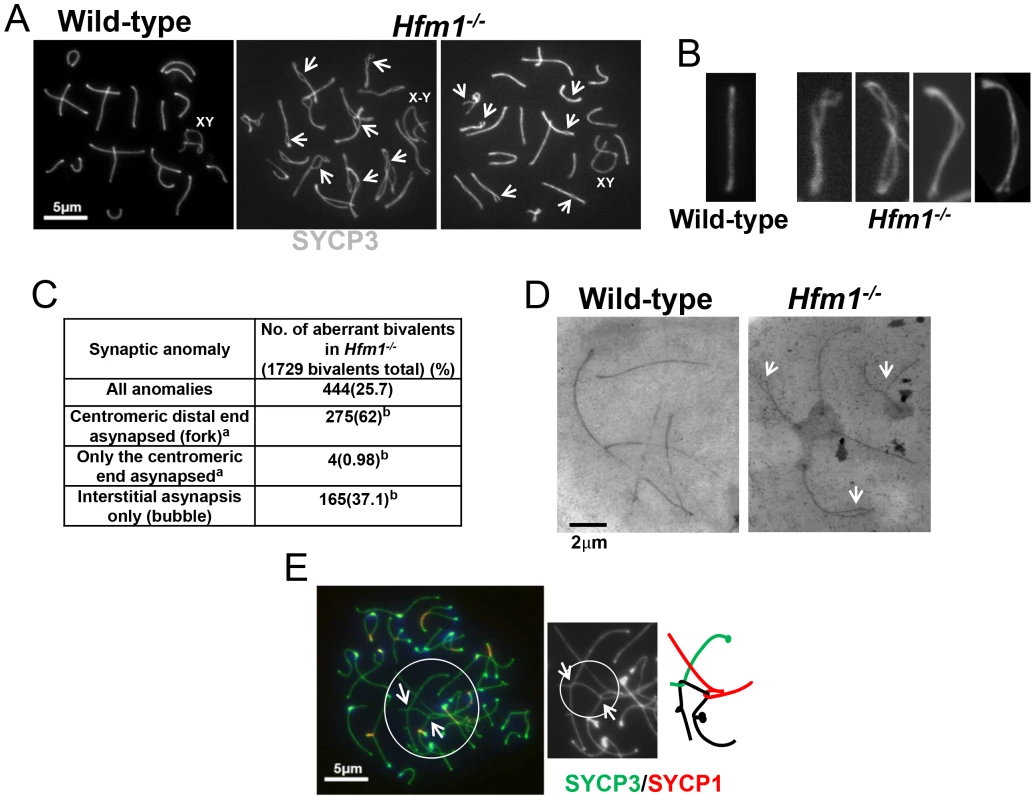
(a) Immunostaining of SYCP3 in wild-type and Hfm1−/− spermatocytes. Examples of pachytene-like spermatocytes are shown. Arrows mark sites of synaptic defects. XY and X-Y indicate positions of tightly and loosely associated sex chromosomes, respectively. (b) Magnified chromosomes show details of synaptic defects. (c) Quantification of synaptic anomalies in meiotic chromosomes of Hfm1−/− spermatocytes. a - Irrespective of whether interstitial asynapsis was also present. b - % calculated with respect to total chromosomes displaying anomalies. (d) Electron microscopy of silver stained pachytene-like spermatocytes. Arrows mark sites of synaptic defects. (e) Example of tangled chromosomes in diplotene Hfm1−/− spermatocytes. Arrows indicate sites of entangled chromosome axes. Magnification bar represents 5 µm. Chromosome interlocks, as visualized in the mer3 mutant of S. macrospora [54], were not seen in Hfm1−/− spermatocytes but entangled chromosomes appeared in a fraction of Hfm1−/− diplotene nuclei (3.6%, n = 190; none were detected in wild-type, n = 320; P≤0.001) (Figure 7e). Whether this entangling occurs earlier but only becomes visible at diplotene, or develops only as chromosomes desynapse in diplotene, is not clear. It is possible that defects in resolving partial interlocks (e. g., those involving chromatin loops but not chromosome axes, which would be invisible in our assay) originated earlier in prophase and resulted in the synaptic discontinuities and entangling that we observed later in prophase.
A novel characteristic of chromosomes in Hfm1−/− spermatocytes is the substantially reduced immunostaining for SYCP1 (82.9%, n = 380) and components of the synaptonemal complex central element TEX12 (80%, n = 120) and SYCE1 (70%, n = 97) compared with wild-type pachytene cells, in which all chromosomes show strong, uninterrupted immunolabeling (scored chromosomes, n = 608) (Figure 8). In Hfm1−/− spermatocytes, SYCP1, SYCE1 and TEX12 localize in patches that in some areas are remarkably evenly spaced (Figure 8a and 8b; see [54]). Intriguingly, many Hfm1−/− spermatocytes contained a small number of bivalents with wild-type levels of SYCP1 staining and uninterrupted synapsis, even though the remainder of bivalents had uniformly reduced levels of SYCP1 and gaps in synapsis (60% Hfm1−/− spermatocytes with synapsed XY, n = 59, and 76% Hfm1−/− spermatocytes with unsynapsed XY, n = 67) (Figure 9a). The average intensity of fluorescent signal for SYCP1 from wild-type (18.24±0.70, n = 50 chromosomes) and from Hfm1−/− bivalents with reduced SYCP1 signal (Hfm1 I, 5.65±0.39, n = 50) are significantly different (P≤0.0001, t test). However, no significant difference was observed when wild-type was compared with the fraction of Hfm1−/− bivalents with higher levels of SYCP1 staining (Hfm1 II; 17.60±0.47, n = 50; P≤0.45, t test) (Figure 9b).
Fig. 8. Synaptic defects in Hfm1−/− spermatocytes. 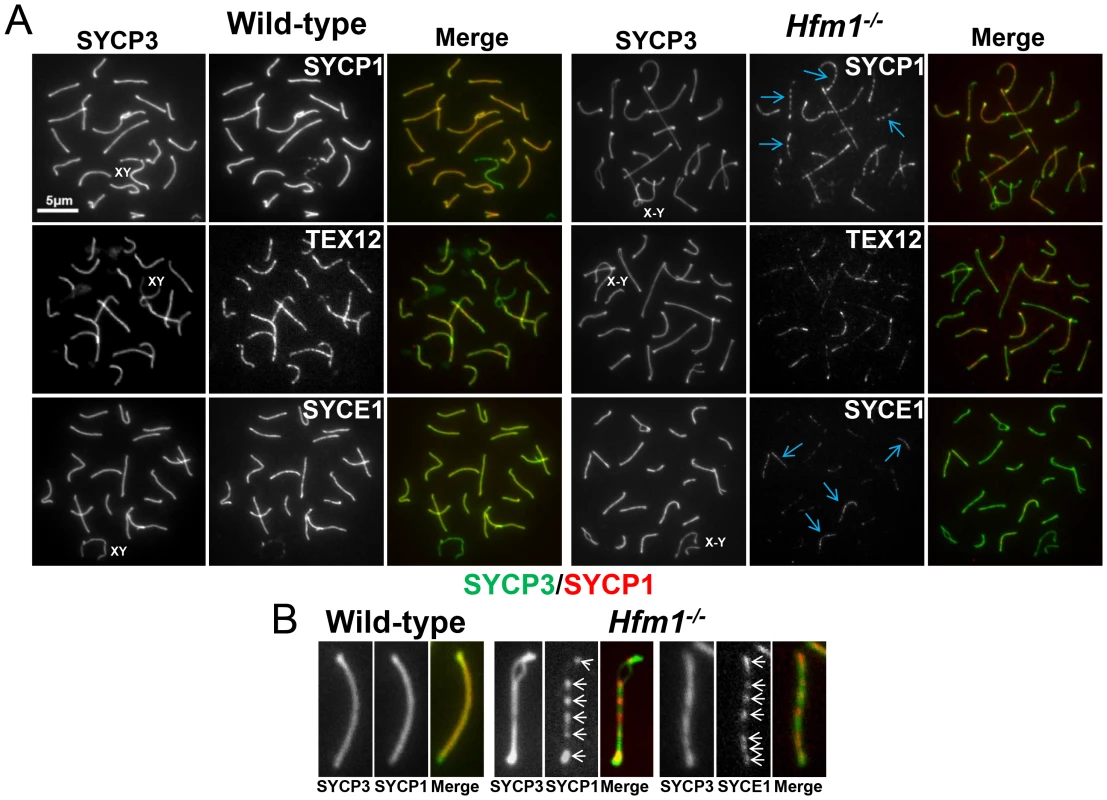
(a) Defects in SYCP1, TEX12 and SYCE1 deposition on synaptonemal complex of Hfm1−/− pachytene-like spermatocytes. Wild-type is shown for comparison. XY and X-Y represents the sex chromosomes. Arrows indicate examples of chromosomes with patches of central element and transverse filament components of the synaptonemal complex. (b) Magnified chromosomes display details of patchy staining for SYCP1 and SYCE1 in Hfm1−/− spermatocytes. Arrows indicate SYCP1 and SYCE1. Fig. 9. Hfm1−/− spermatocytes contain a small number of bivalents with wild-type levels of SYCP1 staining and uninterrupted synapsis. 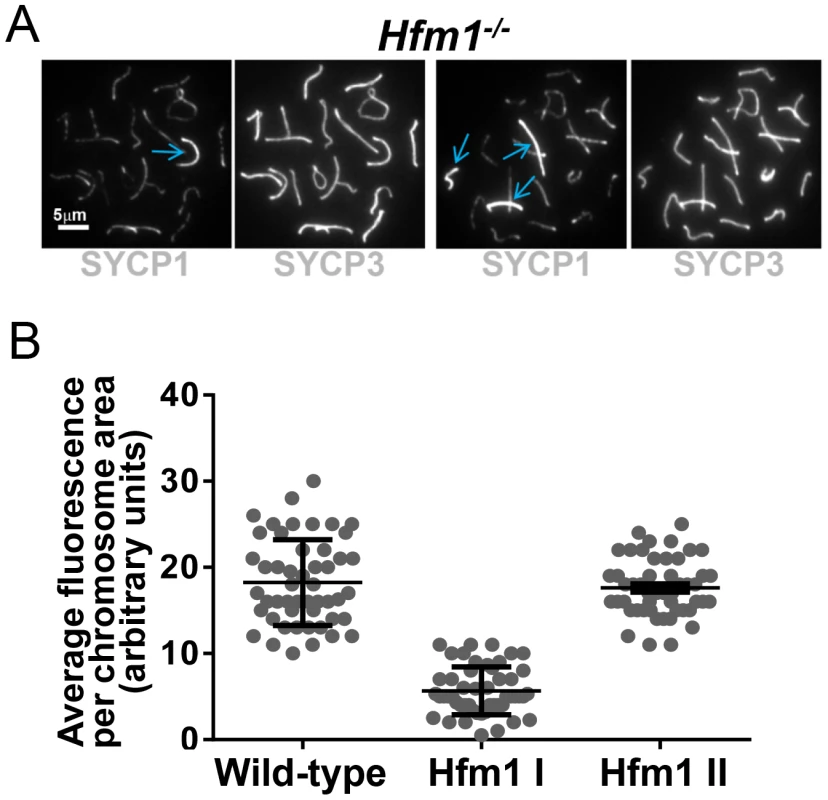
(a) Hfm1−/− nuclei with 1 (left) and 3 (right) bivalents displaying wild-type levels of SYCP1 staining. Arrows indicate chromosomes with wild-type like distribution and signal intensity for SYCP1. (b) Quantification of SYCP1 immunosignal for wild-type and Hfm1−/− spermatocytes. 50 chromosomes were randomly picked for wild-type and 100 chromosomes for Hfm1−/− spermatocytes. Hfm1 I and Hfm1 II represent chromosome populations from Hfm1−/− spermatocytes with and without reduced immunofluorescence with respect to wild-type. Plotted values were obtained using the Metamorph program to determine average intensity per chromosomal area, which was corrected for background fluorescence and normalized by the length of the chromosome. In summary, efficient homologous recognition and synaptic initiation occur in the absence of HFM1. However, in Hfm1−/− spermatocytes, (1) for all but a small fraction of chromosomes, synapsis is incomplete and characterized by subnormal amounts of central region components of the synaptonemal complex and by regions of complete asynapsis, (2) the X and Y chromosomes are frequently unsynapsed even though they colocalze in the sex body, suggesting premature desynapsis, though failure to initiate synapsis cannot be ruled out, and (3) chromosome entanglement becomes visible at diplotene.
Discussion
Recombination defects in HFM1/Mer3-deficient meiocytes
We observed that knocking out HFM1 in mouse spermatocytes results in a dramatic reduction of MLH1 foci and chiasma formation. These results indicate a recombination defect that inactivates the major pathway for CO formation. Similar observations have been made in mutants that inactivate Mer3 orthologs in budding yeast, Arabidopsis, rice, C. cinereus and S. macrospora [21], [24], [28], [38], [39], [50], [52], [53], [54]. Moreover, the physical analysis of intermediates of recombination performed in yeast Mer3 mutants revealed a dramatic decrease of single-end invasion and dHJ intermediates [21], which correlates with 50–60% disappearance of COs [21], [24], [50], [52]. Genetic analysis of Arabidopsis Mer3 mutants alleles also shows that formation of COs is severely reduced (by 38%) but not eliminated. In agreement, cytological analysis of Mer3 mutants in all studied species confirmed the severe deficiency in CO formation by showing a dramatic decrease in chiasma formation. This phenotype is in general correlated with cell cycle arrest or chromosome missegregation at meiosis I with partial or total infertility. CO formation in mouse Hfm1−/− spermatocytes is severely reduced but not eliminated (Figure 4). Interestingly, genetic analysis in budding yeast and Arabidopsis demonstrate that remnant COs do not show interference, therefore deletion of Mer3 led only to the elimination of interference-sensitive CO Class I [24], [28], [38], [50], [52]. By extension of these results and as previously proposed [26], [29], we speculate that at least two pathways for CO formation exist in mammals, the HFM1-dependent pathway which gives rise to interference-sensitive COs and another, presumably MUS81-dependent, pathway which is expected to produce a small number of randomly distributed COs [26] (Figure 10a).
Fig. 10. The meiotic role of mouse HFM1. 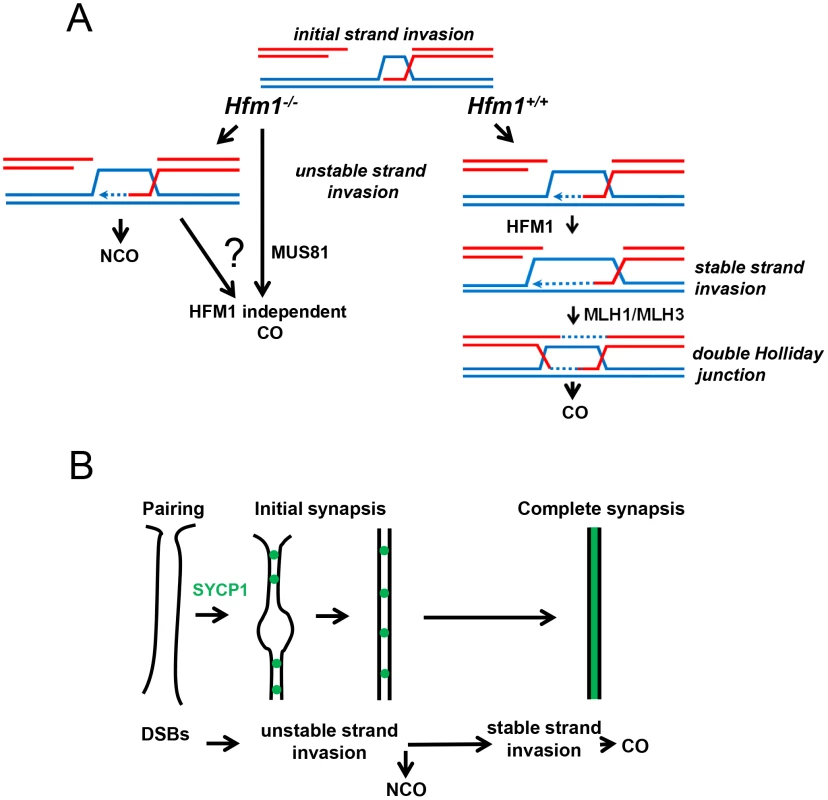
The proposed model summarizes observations of this and previous studies. (a) HFM1 is required for normal progression of meiotic recombination. Deletion of HFM1 in spermatocytes leads to the absence of most but not all COs. (b) Initial stages of recombination are sufficient to support homologous chromosomes pairing and initial synapsis. However, in the absence of CO-specific intermediates synapsis cannot be completed along the full lengths of bivalents. An important observation from the molecular analysis of recombination intermediates performed in yeast is that while formation of COs is severely reduced, NCO intermediates are detected at normal levels in the absence of Mer3 [21], [24]. In this scenario it is attractive to think that in the major fraction of Hfm1−/− spermatocytes, DSBs are repaired as NCOs because most cells progress to diakinesis and show no immunosignal for markers of incompleted DNA repair such as γH2AX and RAD51. However, it is also possible that some DSBs are repaired as COs by using the sister chromatid as template.
An intriguing characteristic of the Hfm1−/− mutant spermatocytes is that the number of MSH4 foci in Hfm1−/− pachytene spermatocytes is substantially reduced with respect to wild-type, suggesting a defect in the middle stages of recombination. One possibility is that the deficiency of MSH4 may be explained by inefficient recruitment of this protein to chromosomes because HFM1 and/or other ZMM components are not loaded at recombination sites. However, other scenarios are possible. We also speculate that the less prominent decrease in MSH4 foci in zygotene may reflect an earlier HFM1-independent chromosome localization and perhaps function of MSH4. This may explain the more severe synaptic phenotype observed in MSH4 than in HFM1 mutants. Indeed, immunolocalization of Mer3 and Msh4 in yeast shows that only 50–70% of Mer3 foci co-localize with Mhs4 [67], and Msh4−/− and Msh5−/− [43] mice have a more severe meiotic phenotype than Hfm1−/−. However, further studies such as the analysis of Hfm1/Msh4 double mutants will be needed to test this possibility.
HFM1/Mer3 is required for complete synapsis
Initial stages of meiotic chromosome restructuring are apparently not affected by HFM1/Mer3 deletion as in all tested species assembly of axial elements appears normal [21], [24], [28], [38], [39], [53], [54]. In addition, except for the case of S. macrospora, in which the onset of chromosome alignment is delayed [54], in all Mer3 mutants homologous recognition and chromosome pairing apparently proceed as in wild-type strains. Notably, another evolutionarily conserved characteristic of Mer3 is its requirement for normal synapsis. However, there is a wide variation in the extent of synapsis failure in Mer3 mutants in different species. Budding yeast and C. cinereus show the most severe phenotypes [21], [24], [39]. For example, Mer3 yeast cells stained with antibodies against Zip1 and Red1 show that up to 90% of cells have separated homologous axes [24], and EM analysis in C. cinereus mutants shows no formation of synaptonemal complex [39]. Abnormal synapsis is also observed in S. macrospora where asynapsed chromosomes persist among late pachytene cells and is accompanied by other chromosomal defects such as high levels of interwoven chromosomes [54]. We observed that in mouse Hfm1−/− spermatocytes, similar to rice and Arabidopsis Mer3 mutants, the defect in synapsis is apparently moderate. We found that a fraction of bivalents in pachytene have short lengths of widely separated axes and that components of the central region of the synaptonemal complex fail to extend along the entire lengths of most chromosome axes (Figure 7 and Figure 8).Thus, the requirement for HFM1/Mer3 to promote completed synapsis is conserved in mice.
A simple explanation for the largely normal pairing and early synapsis in mouse Hfm1−/− spermatocytes is that initial recombination transactions, those occurring before HFM1 activity, are sufficient to establish recognition of homology between chromosome pairs (Figure 10b). The idea that earlier DNA transactions leading to NCOs as well as to COs promote initial stages of homologous chromosome interactions has been proposed previously [21], [54], and also finds support in double mutants of the budding yeast gene sgs1 (a homolog of mammalian BLM) with zip1, zip3 or mer3. In these mutants, interhomolog associations occur at many more “axial association” sites than the ones giving rise to COs and generate such uniform and close alignment of homologous axes that they appear “pseudosynapsed” [24], [68].
Although initial steps of recombination and chromosome pairing apparently are not affected by the absence of HFM1 and the concomitant reduction in the numbers of COs, incomplete synaptonemal complex formation is observed in Hfm1−/− spermatocytes. This suggests that completion of synapsis is dependent on later steps of recombination that lead to CO formation or to CO formation per se. Alternatively, the HFM1 contribution to completion of synapsis may be unrelated to its function in recombination.
The synaptic phenotypes of mouse mutants for proteins that apparently act before HFM1 (DMC1 and HOP2) and after HFM1 (MLH1 and MLH3) are consistent with the idea that HFM1 provides an “intermediate activity” in recombination that is required for completion of synapsis. In Dmc1−/− and Hop2−/− spermatocytes, homologous pairing and synapsis are absent [55], [56]. In contrast, in Mlh1−/− and Mlh3−/− spermatocytes, mature, complete synaptonemal complexes form [3], [69]. Our explanation of the synaptic phenotype in Hfm1−/− spermatocytes and the link to recombination may be extended to other model organisms with similar HFM1/Mer3-dependencies, as in Arabidopsis, rice and, to a lesser extent, S. macrospora. The more extensive synapsis defects observed in yeast and C. cinereus Mer3 mutants presumably indicate different dependencies in these organisms in the mechanisms directing and/or coordinating synaptonemal complex formation with recombination.
Defects in Hfm1−/− spermatocytes do not cause developmental delay or cell apoptosis during prophase I
To avoid deleterious outcomes in recombination and synaptic defects, checkpoints survey meiotic errors and ultimately eliminate cells containing unresolved defects. In budding yeast and mice, among other species, meiocytes with defects in DSB repair and/or chromosome synapsis trigger meiotic arrest during prophase I [2], [70], [71], [72]. However, because recombination is interdependent with synapsis it has remained uncertain whether there is a distinct checkpoint that responds to defects in meiotic recombination or synapsis alone [73]. An intriguing characteristic of the HFM1 mutation in mouse spermatocytes is that despite abnormalities in recombination and synapsis, Hfm1−/− spermatocytes progress through prophase I with no apparent sign of checkpoint activation. For a major fraction of Hfm1−/− spermatocytes, we assume that most DSBs are repaired so that no intermediates remain that could trigger a DNA repair checkpoint. In this case, despite incomplete synaptonemal complex and partial defects, mutant spermatocytes are still able to satisfy the surveillance mechanisms. In this scenario, however, it is not clear to us why the fraction of Hfm1−/− spermatocytes that exhibit persistent γH2AX and RAD51 immunosignals do not show apparent prophase I delay or a cell response to an overly long delay/arrest such as apoptosis. A possible explanation is that persistent γH2AX and RAD51 foci are a consequence of inefficient removal of these proteins following abnormal (but completed) DNA repair. Alternatively, a low level of apoptosis during prophase I might not be detected by our assay. It also is possible that HFM1/Mer3 itself is required for a checkpoint, so that in the absence of Mer3 the checkpoint is inactivated. Regardless of the details, the requirements for HFM1/Mer3 for cell-cycle progression in mouse are different from those in budding yeast or C.cinereus, where Mer3 deletion results in mid prophase I arrest, or in S. macrospora, where Mer3 mutants show a delay in the leptotene-zygotene transition. These differences among species indicate that the biological functions that require HFM1/Mer3 and/or the responses to its absence have diverged along with other features of meiotic chromosome metabolism in these organisms.
Materials and Methods
Mice
Mice used in this study were wild-type (C57BL/6), transgenic Flpe B6.Cg-Tg (ACT FLPe) 9205Dym and HFM1 knockout. Experiments conformed to relevant regulatory standards and were approved by the IACUC (Institutional Animal Care and Use Committee).
Generation of Hfm1-deficient mice
The mouse embryonic stem (ES) cell line Hfm1Gt(ost347241)Lex containing a gene trap insertion in the second intron of the Hfm1 gene (Figure 1a) was obtained from the Texas A&M Institute for Genomic Medicine (informatics.jax.org). The gene-trapping vector used to create this line, VICTR48, was designed to prevent translation of downstream fusion transcripts while maintaining the presence of the Hfm1 transcript. To identify the exact insertion site within Hfm1 intron 2, PCR reactions were performed using one primer hybridizing at the 5′ end of the gene trap vector and the complementary primer obtained by 3′ RACE PCR from the mouse embryonic stem cell clone Hfm1Gt(ost347241)Lex. The PCR product was sequenced revealing that the insertion site was 1510 bp into intron 2. The Hfm1Gt (ost347241) Lex embryonic stem cells were injected into C57BL/6 blastocysts to create chimeric mice, which were bred with C57BL/6 to generate the first progeny of Hfm1+/− heterozygous mice genotyped by PCR analysis (Figure 1b). Elimination of Hfm1 transcript by removing the PGK/Btk/SD cassette was accomplished by crossing heterozygous Hfm1+/− mice with a mouse line expressing FLPe recombinase (B6.Cg-Tg (ACT FLPe) 9205Dym/J) (Figure 1a).
Genotyping of mice by PCR
The genotyping was carried out by PCR using oligonucleotide #1 (F1) and #2 (R1) to amplify the wild-type allele (a 340 bp fragment) and primers #1(F1) and #3 (R2) (Table S1) to amplify the knockout allele (a 270 bp fragment). The cycling conditions were: 94°C 5 min; 94°C 30 sec, 58°C 45 sec, 72°C 60 sec for 35 cycles; 72°C 5 min. The presence of Flpe was assessed by genotyping using the primers (#10 and #11) under the same PCR conditions.
Real-time RT–PCR
Total RNA was isolated from adult testis with the RNeasy Mini Kit (Qiagen). 2.0 µg of RNA was oligo dT primed and reverse-transcribed with the High capacity RNA-to-cDNA kit (Applied Biosystems). Three different exon boundaries of Hfm1 were amplified using Power Syber Green PCR Master Mix (Applied biosystems) with the following primers: #4 and #5, #6 and #7 and #8 and #9, respectively. The cycling conditions were: 95°C 10 min; 95°C 10 sec, 60°C 30 sec, for 40 cycles. The melt curve was of 95°C 10 sec and 65°C to 95°C performed in increments of 0.5°C.
Histological analysis
Testes and ovaries for histological examination were removed and fixed overnight in 10% neutral-buffered formalin (Sigma). Serial sections from either testes or ovaries were positioned on microscope slides and analyzed using either hematoxylin and eosin staining or a TUNEL assay (Roche).
Cytology
We employed established experimental approaches for the visualization of chromosomes in chromosome surface spreads [74]. Incubations with primary antibodies were carried out for 12 h at 4°C in 1× PBS plus BSA 2%. To detect SYCP1 and SYCP3 we used polyclonal rabbit antibody raised against mouse SYCP1 at 1∶200 dilution (Novus Biologicals) and polyclonal mouse antibody raised against mouse SYCP3 at 1∶300 dilution (Abcam). Centromeres were detected using the human centromere protein antibody (CREST, Antibody Incorporated) at 1∶50 dilution. Other primary antibodies used in this study were as follows: monoclonal mouse antibody raised against mouse γH2AX at 1∶500 dilution (Millipore), polyclonal rabbit antibody raised against mouse RAD51 at 1∶200 dilution (Santa Cruz Biotechnology), polyclonal rabbit antibody raised against mouse MSH4 at 1∶200 dilution (AbCam), monoclonal mouse antibody raised against mouse MLH1 at 1∶50 dilution (BD pharmingen), polyclonal rabbit antibody raised against SYCE1 at 1∶200 dilution (kindly provided by Ch. Hoog) and polyclonal rabbit antibody raised against TEX12 at 1∶200 dilution (kindly provided by Ch. Hoog). Following three washes in 1× PBS, slides were incubated for 1 h at room temperature with secondary antibodies. A combination of Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Jackson laboratories) with Rhodamine-conjugated goat anti-mouse IgG and Cy5-conjugated goat anti-human IgG each diluted 1∶450 were used for simultaneous triple immunolabeling. Slides were subsequently counterstained for 3 min with 2 µg/ml DAPI containing Vectashield mounting solution (Vector Laboratories) and sealed with nail varnish. For RAD51, MSH4 and MLH1 focus counts, nuclei were staged using the extent of SYCP3 immunolabeling and synapsis as markers for meiotic prophase progression. Only foci that co-localized with the chromosome axes were counted. We use Axiovision SE 64 (Carl Zeiss, inc.) for imaging acquisition and processing. Statistical tests were as described in the text and table legends.
Spermatocyte chromosome spreads for electron microscopy analysis was performed as previously described [60].
Supporting Information
Zdroje
1. KlecknerN (1996) Meiosis: how could it work? Proc Natl Acad Sci U S A 93 : 8167–8174.
2. RoederGS (1997) Meiotic chromosomes: it takes two to tango. Genes Dev 11 : 2600–2621.
3. EdelmannW, CohenPE, KaneM, LauK, MorrowB, et al. (1996) Meiotic pachytene arrest in MLH1-deficient mice. Cell 85 : 1125–1134.
4. HassoldT, HuntP (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2 : 280–291.
5. HassoldTJ, JacobsPA (1984) Trisomy in man. Annu Rev Genet 18 : 69–97.
6. RomanienkoPJ, Camerini-OteroRD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6 : 975–987.
7. KeeneyS, GirouxCN, KlecknerN (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 : 375–384.
8. KeeneyS, BaudatF, AngelesM, ZhouZH, CopelandNG, et al. (1999) A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics 61 : 170–182.
9. PâquesF, HaberJE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews 63 : 349–404.
10. MimitouEP, SymingtonLS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455 : 770–774.
11. ZhuZ, ChungWH, ShimEY, LeeSE, IraG (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134 : 981–994.
12. GravelS, ChapmanJR, MagillC, JacksonSP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22 : 2767–2772.
13. PezzaRJ, VoloshinON, VanevskiF, Camerini-OteroRD (2007) Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes & Development 21 : 1758–1766.
14. ChiP, San FilippoJ, SehornMG, PetukhovaGV, SungP (2007) Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev 21 : 1747–1757.
15. NealeMJ, KeeneyS (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442 : 153–158.
16. BiancoPR, TracyRB, KowalczykowskiSC (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci 3: D570–603.
17. SungP, KrejciL, Van KomenS, SehornMG (2003) Rad51 recombinase and recombination mediators. J Biol Chem 278 : 42729–42732.
18. AllersT, LichtenM (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106 : 47–57.
19. NassifN, PenneyJ, PalS, EngelsWR, GloorGB (1994) Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Molecular and cellular biology 14 : 1613–1625.
20. HunterN, KlecknerN (2001) The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106 : 59–70.
21. BornerGV, KlecknerN, HunterN (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 : 29–45.
22. BishopDK, ZicklerD (2004) Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117 : 9–15.
23. SchwachaA, KlecknerN (1995) Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83 : 783–791.
24. JessopL, RockmillB, RoederGS, LichtenM (2006) Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet 2: e155 doi:10.1371/journal.pgen.0020155
25. SzostakJW, Orr-WeaverTL, RothsteinRJ, StahlFW (1983) The double-strand break repair model for recombination. Cell 33 : 25–35.
26. HollowayJK, BoothJ, EdelmannW, McGowanCH, CohenPE (2008) MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet 4: e1000186 doi:10.1371/journal.pgen.1000186
27. LynnA, SoucekR, BornerGV (2007) ZMM proteins during meiosis: crossover artists at work. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 15 : 591–605.
28. MercierR, JolivetS, VezonD, HuppeE, ChelyshevaL, et al. (2005) Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3,whereas the other one is not. Current biology : CB 15 : 692–701.
29. GuillonH, BaudatF, GreyC, LiskayRM, de MassyB (2005) Crossover and noncrossover pathways in mouse meiosis. Molecular Cell 20 : 563–573.
30. BerchowitzLE, FrancisKE, BeyAL, CopenhaverGP (2007) The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3: e132 doi:10.1371/journal.pgen.0030132
31. HigginsJD, BucklingEF, FranklinFC, JonesGH (2008) Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. The Plant journal : for cell and molecular biology 54 : 152–162.
32. CopenhaverGP (2005) Plant genetics: when not to interfere. Current biology : CB 15: R290–291.
33. SymM, EngebrechtJA, RoederGS (1993) ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72 : 365–378.
34. de VriesFA, de BoerE, van den BoschM, BaarendsWM, OomsM, et al. (2005) Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19 : 1376–1389.
35. HigginsJD, Sanchez-MoranE, ArmstrongSJ, JonesGH, FranklinFC (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes & Development 19 : 2488–2500.
36. ColaiacovoMP, MacQueenAJ, Martinez-PerezE, McDonaldK, AdamoA, et al. (2003) Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Developmental cell 5 : 463–474.
37. PageSL, HawleyRS (2004) The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 20 : 525–558.
38. ChenC, ZhangW, TimofejevaL, GerardinY, MaH (2005) The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. The Plant journal : for cell and molecular biology 43 : 321–334.
39. SugawaraH, IwabataK, KoshiyamaA, YanaiT, DaikuharaY, et al. (2009) Coprinus cinereus Mer3 is required for synaptonemal complex formation during meiosis. Chromosoma 118 : 127–139.
40. TanakaK, MiyamotoN, Shouguchi-MiyataJ, IkedaJE (2006) HFM1, the human homologue of yeast Mer3, encodes a putative DNA helicase expressed specifically in germ-line cells. DNA sequence : the journal of DNA sequencing and mapping 17 : 242–246.
41. ZalevskyJ, MacQueenAJ, DuffyJB, KemphuesKJ, VilleneuveAM (1999) Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153 : 1271–1283.
42. KellyKO, DernburgAF, StanfieldGM, VilleneuveAM (2000) Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156 : 617–630.
43. KneitzB, CohenPE, AvdievichE, ZhuL, KaneMF, et al. (2000) MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev 14 : 1085–1097.
44. HigginsJD, ArmstrongSJ, FranklinFC, JonesGH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes & Development 18 : 2557–2570.
45. Martinez-PerezE, ColaiacovoMP (2009) Distribution of meiotic recombination events: talking to your neighbors. Current opinion in genetics & development 19 : 105–112.
46. HillersKJ (2004) Crossover interference. Current biology : CB 14: R1036–1037.
47. KlecknerN, ZicklerD, JonesGH, DekkerJ, PadmoreR, et al. (2004) A mechanical basis for chromosome function. Proceedings of the National Academy of Sciences of the United States of America 101 : 12592–12597.
48. de los SantosT, HunterN, LeeC, LarkinB, LoidlJ, et al. (2003) The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164 : 81–94.
49. CopenhaverGP, HousworthEA, StahlFW (2002) Crossover interference in Arabidopsis. Genetics 160 : 1631–1639.
50. NakagawaT, KolodnerRD (2002) Saccharomyces cerevisiae Mer3 Is a DNA Helicase Involved in Meiotic Crossing Over. Mol Cell Biol 22 : 3281–3291.
51. MazinaOM, MazinAV, NakagawaT, KolodnerRD, KowalczykowskiSC (2004) Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117 : 47–56.
52. NakagawaT, OgawaH (1999) The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J 18 : 5714–5723.
53. WangK, TangD, WangM, LuJ, YuH, et al. (2009) MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. Journal of cell science 122 : 2055–2063.
54. StorlazziA, GarganoS, Ruprich-RobertG, FalqueM, DavidM, et al. (2010) Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141 : 94–106.
55. PetukhovaGV, RomanienkoPJ, Camerini-OteroRD (2003) The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell 5 : 927–936.
56. PittmanDL, CobbJ, SchimentiKJ, WilsonLA, CooperDM, et al. (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1 : 697–705.
57. HunterN, Valentin BornerG, LichtenM, KlecknerN (2001) Gamma-H2AX illuminates meiosis. Nat Genet 27 : 236–238.
58. BannisterLA, PezzaRJ, DonaldsonJR, de RooijDG, SchimentiKJ, et al. (2007) A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol 5: e105 doi:10.1371/journal.pbio.0050105
59. MahadevaiahSK, TurnerJM, BaudatF, RogakouEP, de BoerP, et al. (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27 : 271–276.
60. DresserM, PisetskyD, WarrenR, McCartyG, MosesM (1987) A new method for the cytological analysis of autoantibody specificities using whole-mount, surface-spread meiotic nuclei. J Immunol Methods 104 : 111–121.
61. AshleyT, PlugAW, XuJ, SolariAJ, ReddyG, et al. (1995) Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma 104 : 19–28.
62. MoensPB, ChenDJ, ShenZ, KolasN, TarsounasM, et al. (1997) Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106 : 207–215.
63. AndersonLK, ReevesA, WebbLM, AshleyT (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151 : 1569–1579.
64. de BoerE, StamP, DietrichAJ, PastinkA, HeytingC (2006) Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci U S A 103 : 9607–9612.
65. RoigI, DowdleJA, TothA, de RooijDG, JasinM, et al. (2010) Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet 6: e1001062 doi:10.1371/journal.pgen.1001062
66. PetronczkiM, SiomosMF, NasmythK (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112 : 423–440.
67. TerasawaM, OgawaH, TsukamotoY, ShinoharaM, ShirahigeK, et al. (2007) Meiotic recombination-related DNA synthesis and its implications for cross-over and non-cross-over recombinant formation. Proceedings of the National Academy of Sciences of the United States of America 104 : 5965–5970.
68. RockmillB, FungJC, BrandaSS, RoederGS (2003) The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr Biol 13 : 1954–1962.
69. LipkinSM, MoensPB, WangV, LenziM, ShanmugarajahD, et al. (2002) Meiotic arrest and aneuploidy in MLH3-deficient mice. Nature Genetics 31 : 385–390.
70. GhabrialA, SchupbachT (1999) Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nature cell biology 1 : 354–357.
71. BhallaN, DernburgAF (2005) A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310 : 1683–1686.
72. AshleyT, GaethAP, CreemersLB, HackAM, de RooijDG (2004) Correlation of meiotic events in testis sections and microspreads of mouse spermatocytes relative to the mid-pachytene checkpoint. Chromosoma 113 : 126–136.
73. BurgoynePS, MahadevaiahSK, TurnerJM (2009) The consequences of asynapsis for mammalian meiosis. Nature reviews Genetics 10 : 207–216.
74. PetersAH, PlugAW, van VugtMJ, de BoerP (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome research 5 : 66–68.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



