-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
Cyclin-dependent kinase 9 (Cdk9) promotes elongation by RNA polymerase II (RNAPII), mRNA processing, and co-transcriptional histone modification. Cdk9 phosphorylates multiple targets, including the conserved RNAPII elongation factor Spt5 and RNAPII itself, but how these different modifications mediate Cdk9 functions is not known. Here we describe two Cdk9-dependent pathways in the fission yeast Schizosaccharomyces pombe that involve distinct targets and elicit distinct biological outcomes. Phosphorylation of Spt5 by Cdk9 creates a direct binding site for Prf1/Rtf1, a transcription regulator with functional and physical links to the Polymerase Associated Factor (PAF) complex. PAF association with chromatin is also dependent on Cdk9 but involves alternate phosphoacceptor targets. Prf1 and PAF are biochemically separate in cell extracts, and genetic analyses show that Prf1 and PAF are functionally distinct and exert opposing effects on the RNAPII elongation complex. We propose that this opposition constitutes a Cdk9 auto-regulatory mechanism, such that a positive effect on elongation, driven by the PAF pathway, is kept in check by a negative effect of Prf1/Rtf1 and downstream mono-ubiquitylation of histone H2B. Thus, optimal RNAPII elongation may require balanced action of functionally distinct Cdk9 pathways.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004029
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004029Summary
Cyclin-dependent kinase 9 (Cdk9) promotes elongation by RNA polymerase II (RNAPII), mRNA processing, and co-transcriptional histone modification. Cdk9 phosphorylates multiple targets, including the conserved RNAPII elongation factor Spt5 and RNAPII itself, but how these different modifications mediate Cdk9 functions is not known. Here we describe two Cdk9-dependent pathways in the fission yeast Schizosaccharomyces pombe that involve distinct targets and elicit distinct biological outcomes. Phosphorylation of Spt5 by Cdk9 creates a direct binding site for Prf1/Rtf1, a transcription regulator with functional and physical links to the Polymerase Associated Factor (PAF) complex. PAF association with chromatin is also dependent on Cdk9 but involves alternate phosphoacceptor targets. Prf1 and PAF are biochemically separate in cell extracts, and genetic analyses show that Prf1 and PAF are functionally distinct and exert opposing effects on the RNAPII elongation complex. We propose that this opposition constitutes a Cdk9 auto-regulatory mechanism, such that a positive effect on elongation, driven by the PAF pathway, is kept in check by a negative effect of Prf1/Rtf1 and downstream mono-ubiquitylation of histone H2B. Thus, optimal RNAPII elongation may require balanced action of functionally distinct Cdk9 pathways.
Introduction
Regulation of RNA polymerase II (RNAPII) transcription elongation affects the expression of many eukaryotic genes [1]. A central regulator of metazoan elongation is positive transcription elongation factor b (P-TEFb), a complex comprising cyclin-dependent kinase 9 (CDK9) and a cyclin partner. P-TEFb was identified on the basis of its biochemical activity in overcoming promoter-proximal RNAPII pausing imposed by DRB-sensitivity inducing factor (DSIF; a complex of Spt4 and Spt5) and a negative elongation factor (NELF) [2]–[5]. Cdk9 orthologs are present and essential in Saccharomyces cerevisiae (Bur1) and Schizosaccharomyces pombe (Cdk9), where they have imputed positive roles in RNAPII elongation [6]–[12]. Cdk9 phosphorylates multiple proteins involved in elongation, including Spt5 and the Rpb1 subunit of RNAPII [13]. In addition to regulating elongation, these phosphorylations are implicated in the recruitment of chromatin-modifying enzymes and mRNA 3′-end processing factors [11], [14]–[16]. Cdk9 can also phosphorylate and activate the histone H2B ubiquitin-conjugating enzyme RAD6, indicating multiple links between Cdk9 and chromatin modification [17], [18].
Cdk9 phosphorylates both Rpb1 and Spt5 within repeated amino acid motifs at their C-termini. The Rpb1 C-terminal domain (CTD) repeat has consensus sequence Y1SPTSPS7 and is involved in coordinating co-transcriptional events [14]. Cdk9 can phosphorylate multiple sites within the CTD heptad, but its roles in RNAPII elongation have been commonly attributed to Ser2 phosphorylation [13]. Spt5 contains a C-terminal repeat domain (which varies in size and sequence among taxa) that is phosphorylated by Cdk9 on a conserved threonine residue within the repeated motif [4], [11], [16], [19]. The relative contributions of specific Cdk9 targets to its various functions in transcription are not known.
The Polymerase-Associated Factor (PAF) complex is a key mediator of Cdk9 function during elongation. PAF is minimally composed of conserved subunits Paf1, Leo1, Cdc73, and Ctr9 [20], and its functions broadly overlap with those of Cdk9. In vitro, PAF cooperates with the phosphorylated Spt5 CTD to relieve promoter-proximal RNAPII pausing, and stimulates RNAPII elongation on DNA and chromatin templates [21]–[24]. Like Cdk9, PAF also functions in mRNA 3′ end processing and co-transcriptional chromatin modification [25]–[28]. In budding yeast, PAF recruitment to transcribed genes requires Bur1-dependent Spt5 phosphorylation, which promotes a direct interaction between the Spt5 CTD and the Rtf1 subunit of PAF [15], [16], [18], [29]–[31].
Rtf1 is a stable component of PAF in budding yeast, but is biochemically separate from PAF in metazoans [22], [26], [32]–[37]. Both PAF and Rtf1 are present at transcribed genes and are required for co-transcriptional formation of mono-ubiquitylated histone H2B (H2Bub1) and methylation of histone H3 lysine 4 (H3K4me) [25], [26]. Mutation or knockdown of PAF1 and RTF1 revealed shared functions in telomeric gene silencing in S. cerevisiae, somite development and hematopoiesis in zebrafish, regulation of flowering time in A. thaliana, and mouse embryonic stem cell pluripotency [25], [38]–[41]. In contrast, zebrafish Rtf1 and PAF were found to have distinct functions in cardiac development [34]. Characterization of PAF mutants in S. cerevisiae revealed pleiotropic deficiencies for paf1Δ and ctr9Δ mutants but few phenotypes overall in an rtf1Δ strain [42]. Indeed, an rtf1Δ mutation was found to suppress the phenotypes of paf1Δ, suggesting that the relationship between Rtf1 and PAF could, in some circumstances, be antagonistic [37]. Lethality of bur1Δ in budding yeast is also suppressed by rtf1Δ, as are phenotypes caused by mutation of the associated cyclin Bur2 [43], [44]. Thus, Rtf1 may counteract an important shared function of Cdk9 and PAF.
Although both Rtf1 and PAF are required for formation of H2Bub1 and H3K4me, genetic analyses in S. cerevisiae indicate a particularly close functional connection between Rtf1 and these histone modifications [45], [46]. While the precise gene regulatory functions of H2Bub1 and H3K4me have not been elucidated, H2Bub1, like Rtf1, has been found to oppose Cdk9. In the fission yeast S. pombe, phenotypes associated with loss of H2Bub1 are suppressed by reduction of activity of Cdk9. These include a general 3′ shift of RNAPII density within genes, a pattern that could reflect enhanced elongation through coding regions caused by aberrant Cdk9 function [11]. H2Bub1 itself depends upon Cdk9 activity toward Spt5, and also contributes positively to Spt5 phosphorylation (Spt5-P) levels. Thus, H2Bub1 and Spt5-P may be part of an auto-regulatory mechanism that fine-tunes Cdk9 functions. The Cdk9 function antagonized by H2Bub1 involves a target other than Spt5, suggesting the involvement of a distinct Cdk9 pathway [11].
Using the fission yeast S. pombe as a model system, we report that PAF and Rtf1 (which we designate Prf1 in S. pombe) are involved in two distinct and opposing regulatory pathways, both of which depend on Cdk9 activity. We further show that these divergent functions involve different Cdk9 targets, and mirror the opposing functions of Cdk9 and H2Bub1 in elongation.
Results
PAF and Prf1/Rtf1 are biochemically separate and functionally distinct
To further explore the relationship between Cdk9 activity and H2Bub1 in S. pombe, we assessed the biochemical and functional properties of orthologs of known PAF complex subunits. We first tagged the genes encoding homologs of Paf1 (systematic ID # SPAC664.03), Ctr9 (SPAC27D7.14c; named tpr1+ in S. pombe), and Rtf1 [SPBC651.09c; which we henceforth refer to as Prf1, for PAF-related factor 1, to avoid confusion with the unrelated gene SPAC22F8.07c currently annotated as rtf1+ [47]] with the tandem affinity purification (TAP) tag and purified the factors from whole-cell extracts [48]. Purified material was analyzed by SDS-PAGE alongside that from an untagged strain. Polypeptides uniquely present in the TAP purifications were excised from the SDS gel and analyzed by tandem mass spectrometry. Material purified from paf1-TAP and tpr1-TAP strains contained orthologs of the previously defined PAF complex components Paf1, Leo1 (SPBC13E7.08c), Tpr1, and Cdc73 (SPBC17G9.02c) (Figure 1A). Prf1 was not visible by SDS-PAGE in the Tpr1-TAP or Paf1-TAP preparations, nor were PAF components visible in the Prf1-TAP preparation (Figure 1A). Thus, Prf1 and PAF are predominantly separate in S. pombe cell extracts.
Fig. 1. Prf1 does not stably associate with the PAF complex. 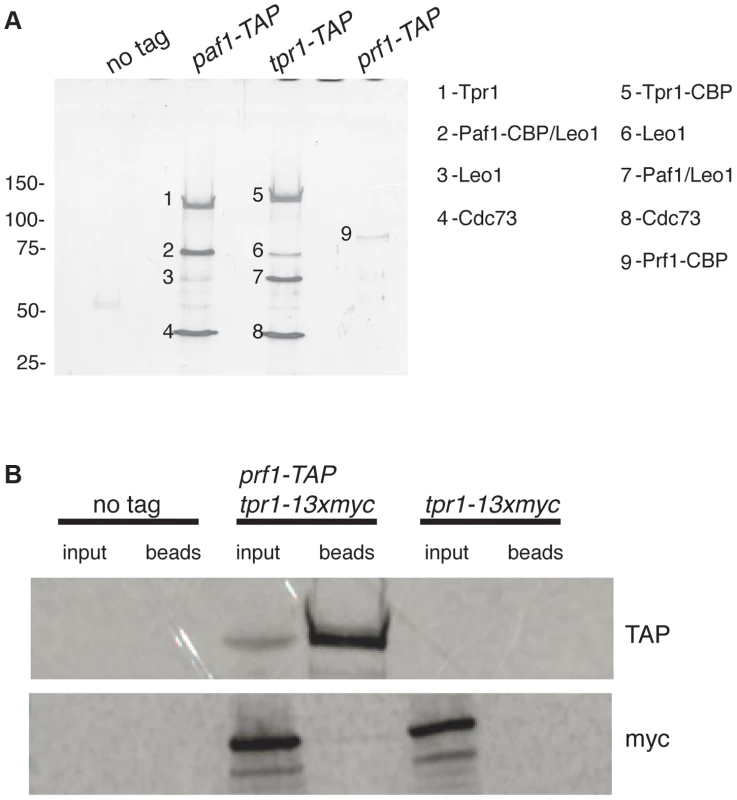
(A) TAP purification was carried out using whole cell extracts from the indicated strains and purified material was analyzed by SDS-PAGE and silver staining. The proteins contained in the labeled bands were identified by mass spectrometry (as indicated on the right). “CBP” denotes the residual fusion to the calmodulin binding peptide resulting from the TAP procedure. Leo1 was detected in two bands that also contained either Paf1-CBP (in the paf1-TAP lane) or Paf1 (in the tpr1-TAP lane). Molecular weight standards (in kD) are denoted on the left. (B) Single-step TAP purifications were performed using extracts from the indicated strains. 5% of input fractions (“input”) and 50% of bead-bound fractions (“beads”) were analyzed by SDS-PAGE and western blotting. To further confirm this finding we carried out in-solution mass spectrometry analysis on the Tpr1-TAP and Prf1-TAP preparations. Material derived from an untagged strain was analyzed in parallel as a control. This analysis identified the core PAF subunits Paf1, Leo1, Tpr1, and Cdc73 with high confidence in the Tpr1-TAP preparation, but not in the control or Prf1-TAP preparations (see Table S1). Conversely, Prf1 was identified with high confidence only in the Prf1-TAP purification.
We also investigated whether we could detect association between Prf1 and PAF in a single-step isolation procedure. We constructed a strain carrying both prf1-TAP and tpr1-13xmyc fusion genes. Whole-cell extracts from this strain, as well as from a tpr1-13xmyc strain, were incubated with IgG-sepharose beads to isolate Prf1-TAP and purified material was analyzed by western blotting. Whereas Prf1-TAP was efficiently purified, no Tpr1-13xmyc protein could be detected, further arguing that Prf1 and PAF do not stably associate in S. pombe cell extracts (Figure 1B).
We found that deletion of the S. pombe prf1+ gene, or of the tpr1+ and cdc73+ genes encoding PAF components, resulted in a total loss of H2Bub1 as gauged by immunoblotting, comparable to that observed in htb1-K119R (ubiquitin acceptor site) or brl2Δ (E3 ubiquitin ligase) mutants (Figure 2A). The presence of normal levels of H2Bub1 in the prf1-TAP and tpr1-TAP strains affirmed the functionality of the respective TAP-tagged proteins (Figure S1). As expected from the established dependence of H3K4me on H2Bub1 in yeast [49], [50], the prf1Δ, tpr1Δ, and cdc73Δ mutations also strongly diminished tri - and dimethyl forms of H3K4me, albeit to varying extents (Figure S2). Thus, Prf1 and PAF have a shared function in co-transcriptional histone modification. The paf1Δ and leo1Δ mutations caused only partial reduction in H2Bub1 and H3K4me levels, indicating divergent roles of PAF complex subunits in promoting these modifications (Figures 2A and S2). Because the H3K36me modification was maintained in these mutants, we infer a specific role for S. pombe PAF and Prf1 in the H2Bub1/H3K4me chromatin modification axis (Figure S2).
Fig. 2. Prf1 and PAF have shared roles in histone H2B ubiquitylation but are functionally distinct. 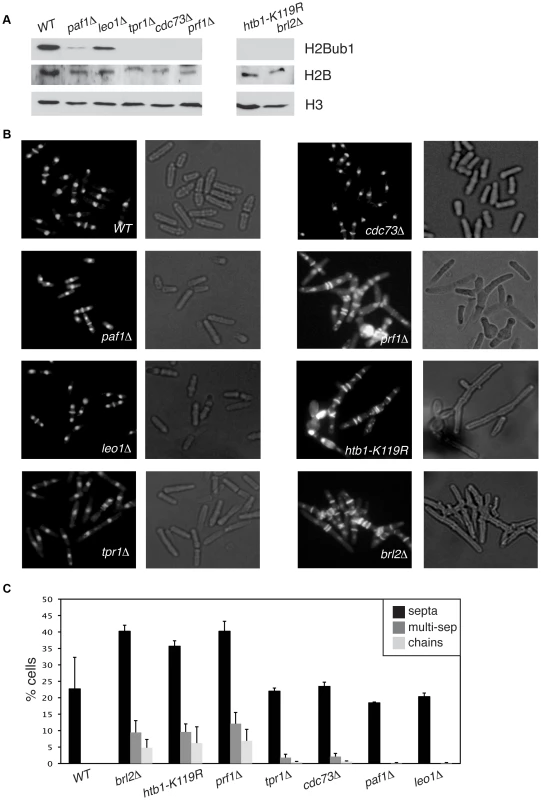
(A) Whole-cell extracts from strains of the indicated genotypes were analyzed by SDS-PAGE and western blotting with the indicated antibodies. (B) Cells of the indicated genotypes were fixed, stained with DAPI/calcofluor, and visualized by microscopy. For each strain fluorescent images (denoting DAPI/calcofluor staining) are shown on the left; bright-field images are shown on the right. (C) Quantification of the abnormal septation patterns in the indicated strains. “Septa” refers to cells with at least one visible division septum, “multi-sep” refers to cells with twinned septa or multiple septa between two nuclei, and “chains” refers to unseparated chains of cells. Error bars denote standard deviations from 3 independent experiments. Loss of H2Bub1 caused by htb1-K119R or brl2Δ mutations is associated with cell morphology and septation defects (including “twinned” septa and unseparated chains of cells) [11]. Microscopic examination of DAPI/calcofluor-stained cells revealed that prf1Δ caused qualitatively and quantitatively similar phenotypes (Figures 2B and 2C). To our surprise, the PAF complex mutations tpr1Δ and cdc73Δ, which eliminated H2Bub1 and reduced H3K4me levels, caused minimal cell morphology and septation defects. This indicates functional divergence between PAF and other factors promoting H2Bub1.
To further probe the functional relationships between Cdk9, PAF, Prf1, and H2Bub1, we assessed growth of the relevant mutant strains under conditions previously shown to make requirement for Cdk9 activity more stringent, including minimal medium (EMM) and medium containing the nucleoside biosynthesis inhibitor mycophenolic acid (MPA) [51]. We found that tpr1Δ and cdc73Δ phenocopied the hypomorphic cdk9-T212A mutant (which lacks the phosphorylation site for the CDK-activating enzyme Csk1; [7]) under these conditions, arguing for a functional link between these factors and Cdk9 activity (Figure 3). The paf1Δ and leo1Δ mutants shared the dramatic EMM sensitivity of tpr1Δ and cdc73Δ but were only mildly sensitive to MPA. Importantly, htb1-K119R and brl2Δ mutants lacked the phenotypic signature associated with reduced Cdk9 activity and loss of PAF, as did prf1Δ (Figure 3). The brl2Δ and prf1Δ mutations did cause moderate growth defects on EMM, perhaps due to H2Bub1-independent functions of Brl2 and Prf1 proteins. Thus, whereas PAF is functionally aligned with Cdk9, Prf1 is intimately connected to factors directly involved in H2Bub1 formation.
Fig. 3. PAF mutant phenotypes resemble cdk9 loss-of-function. 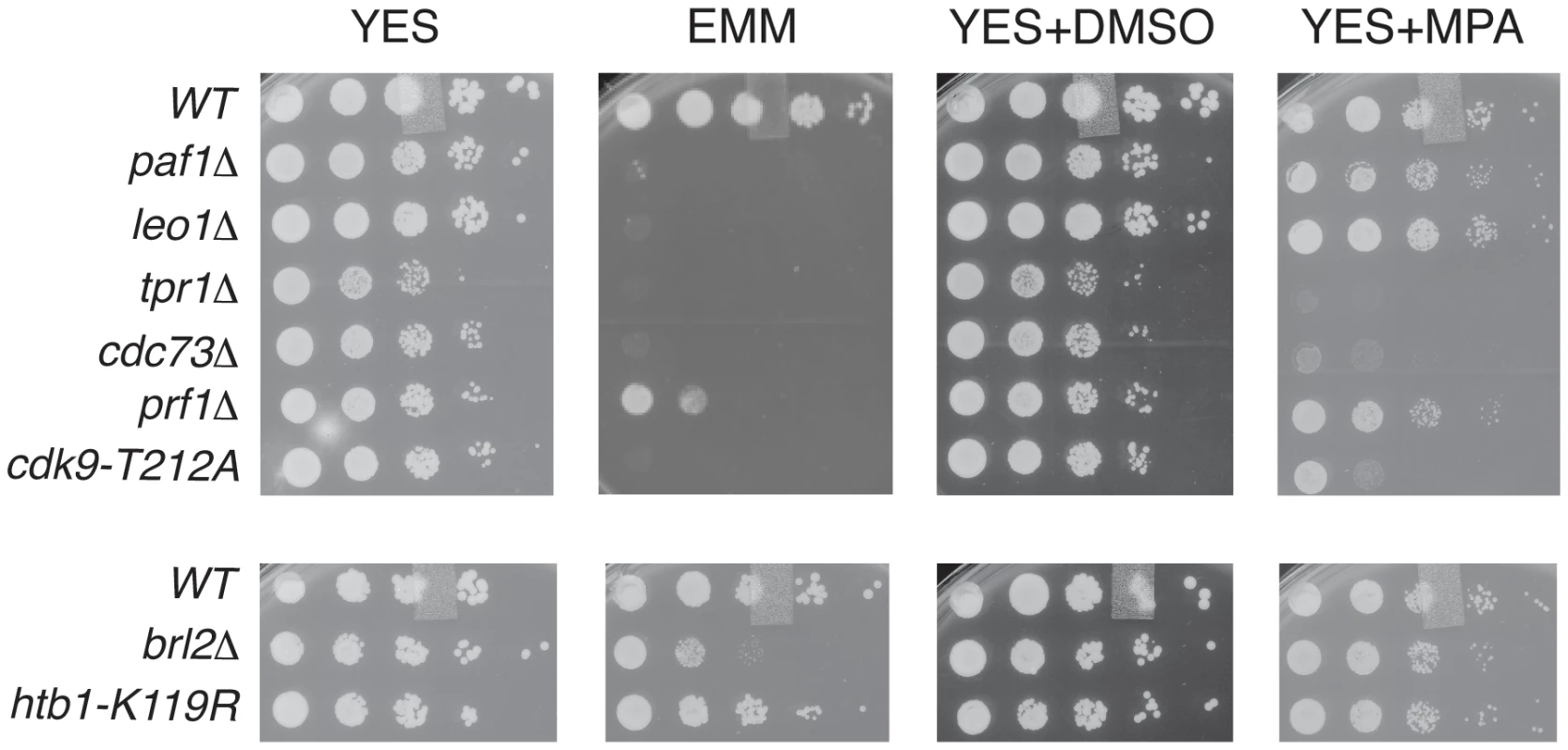
Five-fold serial dilutions of the indicated strains were spotted on agar plates containing rich media (YES), minimal media (EMM), rich media with dimethyl sulfoxide (YES+DMSO), or rich media with mycophenolic acid (YES+MPA). Recruitment of PAF and Prf1 to transcribed chromatin involves distinct Cdk9 targets
To further define the distinct functions of PAF and Prf1 we investigated mechanisms that target these factors to transcribed genes. Prompted by findings in budding yeast linking PAF chromatin association to Bur1 activity [15], [16], [30], we performed chromatin immunoprecipitation (ChIP) in prf1-TAP, tpr1-TAP, and paf1-TAP strains that also carried a Cdk9 variant (cdk9as) engineered to be sensitive to inhibition by bulky ATP analogues [12]. We found that chromatin recruitment of all three proteins to the constitutive act1+ and adh1+ genes depended on Cdk9 activity (Figure 4A). Acute inhibition of Cdk9as in these strains had no effect on overall levels of the respective TAP-tagged proteins (Figure S3A) and did not greatly impair RNAPII occupancy at act1+ or adh1+ (Figure S3B). This argues that chromatin association of both PAF and Prf1 depends on Cdk9 activity.
Fig. 4. Prf1 and PAF are recruited to chromatin via alternate mechanisms. 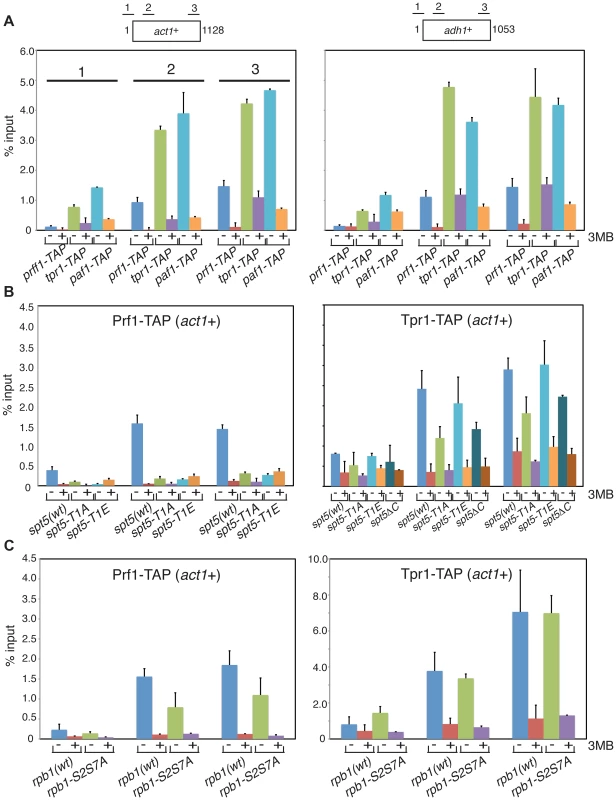
(A) Strains used for ChIP with IgG resin (recognizing the TAP tag) are indicated at the bottom. All strains also harbored the cdk9as allele and were treated with either DMSO (−) or 20 µM 3-MB-PP1 (+) for two hours before crosslinking for ChIP (also indicated by “3MB” at the far right). Assays were quantified by qPCR using primers specific for the act1+ (left) or adh1+ (right) genes. Lengths of the gene coding regions (in base pairs) and positions of PCR amplicons are indicated at the top. Numbering of the datasets for the corresponding PCR amplicons shown for act1+ (left) is used throughout the figure. Error bars denote standard deviations from 2–3 independent experiments. (B and C) Strains of genotypes indicated at the bottom were used for ChIP of either Prf1-TAP (left) or Tpr1-TAP (right). Cultures were treated with DMSO or 3-MB-PP1 as in (A). We reasoned that different Cdk9 targets might mediate the distinct functions of PAF and Prf1. The Spt5 CTD is a specific target of Cdk9 in S. pombe; the Spt5 CTD consists of tandem copies of a nonapeptide motif of consensus sequence T1PAWNSGSK9. We reported previously that changing the Thr1 phosphoacceptor site to alanine reduced H2Bub1 levels [11]. We analyzed Prf1 and PAF recruitment to transcribed chromatin by ChIP in strains harboring Spt5 CTD mutations. The strains were constructed by replacing the endogenous spt5+ CTD domain (which contains 18 nonapeptide repeats) with a truncated (but fully functional) CTD domain containing eight consensus repeats or otherwise identical repeat arrays with T1A or T1E mutations [19]. The strains also carried the cdk9as allele, which allowed assessment of the contribution of specific loss of Spt5-P to the effect of general Cdk9as inhibition.
The spt5-T1A mutation reduced chromatin association of Prf1 at act1+ and adh1+ to a level close to that observed upon inhibition of Cdk9as (5 - to 10-fold; Figures 4B and S4A). Thus, Prf1 recruitment to transcribed chromatin strongly depends on phosphorylation of the Spt5 CTD by Cdk9. The phosphomimetic spt5-T1E mutation also impaired ChIP of Prf1, but rendered the residual crosslinking of Prf1 Cdk9-independent. Thus, although a negative charge at all Thr1 positions within a truncated CTD is not sufficient for normal Prf1 recruitment it does allow some Prf1 binding through an altered, Cdk9-independent mechanism.
In contrast, ChIP of Tpr1-TAP revealed Cdk9-dependent chromatin association in the presence of spt5-T1A and spt5-T1E mutations, as well as in a strain in which the Spt5 CTD is deleted altogether (spt5ΔC)(Figures 4B and S4A). This argues that PAF recruitment to chromatin involves a Cdk9 target other than the Spt5 CTD. The modest (∼2-fold) decrease in ChIP of Tpr1-TAP in the spt5-T1A and spt5ΔC mutants (that was not apparent in the phosphomimetic spt5-T1E mutant) suggests that Spt5-P does make some contribution to PAF recruitment. These data point to the existence of two functionally distinct Cdk9 pathways (which we will refer to as the PAF pathway and the Prf1 pathway) that involve distinct phosphoacceptor targets.
We note that global levels of Spt5-P were reduced by prf1Δ, as was found previously for htb1-K119R and brl2Δ mutations (Figure S5A). This implicates Prf1 as part of the positive feedback loop linking Spt5-P and H2Bub1. Mutations in PAF components either did not affect (tpr1Δ and cdc73Δ) or increased (paf1Δ and leo1Δ) global levels of Spt5-P, further highlighting the specific functional connection between Spt5-P and the Prf1 pathway.
The Rpb1 CTD is a target of Cdk9. In S. pombe, Cdk9 contributes to global levels of Rpb1-Ser2 phosphorylation and promotes the formation of multiply phosphorylated Rpb1 CTD repeats [12], [51]. Budding yeast PAF components Cdc73 and Ctr9 both interact with phosphorylated Rpb1 CTD peptides in vitro and enhance global levels of Rpb1-Ser2 phosphorylation in vivo [30], [32], [52]. This latter function is shared by the corresponding PAF components in S. pombe (but not by Prf1; see Figure S5B). To investigate the role of Rpb1 CTD phosphorylation in Prf1 and PAF recruitment, we carried out ChIP assays in strains harboring phosphoacceptor site mutations in all the Rpb1 CTD repeats. The “wild-type” control in these experiments carried a truncated but fully functional Rpb1 CTD domain [53]. ChIP assays using an rpb1-S2AS7A double mutant (in which both serines 2 and 7 are changed to alanine in every CTD repeat) showed very little effect on chromatin association of either Prf1-TAP or Tpr1-TAP at the active genes analyzed (although a ∼2-fold, gene-specific reduction of Prf1-TAP association was observed at act1+) (Figures 4C and S4B).
We also tested the effect of an rpb1-S5A mutation, the lethality of which was bypassed by fusion of rpb1-S5A to the MCE1 gene encoding the mammalian mRNA capping enzyme [53]. As compared to the wild-type rpb1-MCE1 fusion, the rpb1-S5A-MCE1 fusion had little impact on crosslinking of either Prf1 or Tpr1 to the two loci tested (Figure S6). These data suggest that Rpb1 CTD phosphorylation on serines 2, 5, or 7 is not required for Prf1 or PAF association with transcribed chromatin in vivo.
The marked dependence of Prf1 chromatin association on Spt5-P led us to test whether this could reflect a direct interaction between the Spt5 CTD and Prf1. Biotinylated peptides corresponding to the unmodified or the Thr1-P form of the Spt5 CTD were coupled to streptavidin beads and incubated with TAP-purified, native Prf1 or PAF. We detected binding of Prf1 (∼10% bound) to the unmodified Spt5 CTD peptide that was enhanced by phosphorylation (∼35% bound) (Figures 5A and 5B). This interaction was specific, as we detected no binding of Prf1 to Rpb1 CTD phospho-peptides modified at Ser2, Ser5, Ser7, or Ser5 and Ser7. Given that TAP-purified Prf1 is not visibly associated with other proteins (Figure 1), this argues that there is a direct interaction between Prf1 and Spt5-P.
Fig. 5. Direct and specific association of Prf1/Rtf1 with the phosphorylated Spt5 CTD. 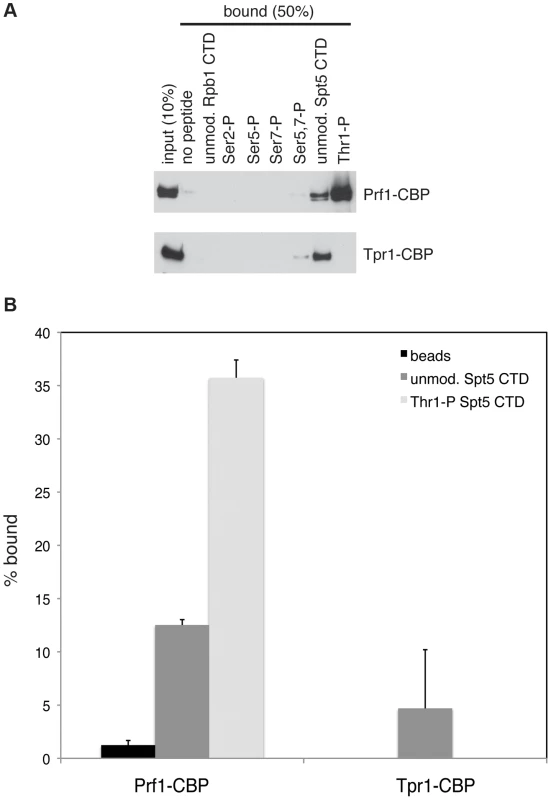
(A) The indicated biotinylated peptides were immobilized on streptavidin magnetic beads and incubated with purified Prf1-CBP (top) or Tpr1-CBP (bottom). Input and bound material was analyzed by SDS-PAGE and western blotting with a TAP tag antibody as shown. (B) Quantification of binding experiments represented in (A). Error bars denote standard deviations from three independent experiments. PAF (purified from the tpr1-TAP strain) also bound (∼5% of total) to the unmodified Spt5 CTD peptide but failed to bind detectably to the Spt5-P peptide, suggesting that, in vitro, phosphorylation inhibits PAF association with the Spt5 CTD (Figures 5A and 5B). Similar results were obtained with PAF purified from the paf1-TAP strain (Figure S7). Thus, we presume that the modest negative effect of the spt5-T1A mutation on Tpr1-TAP crosslinking to active genes was due to another Spt5 CTD-binding factor. Consistent with our ChIP data, we did not detect PAF association with unmodified or singly modified Rpb1 CTD peptides, and detected weak association with the peptide that was doubly phosphorylated at Ser5 and Ser7.
Evidence that PAF and Prf1 have opposing effects on RNAPII elongation
We previously argued that the relationship between Cdk9 and H2Bub1 is mediated by two Cdk9 pathways, one involving Spt5-P and another involving Spt5-P and an additional target [11]. Our data suggested the two pathways were functionally opposed. We wished to test whether the Prf1 and PAF pathways described here might mediate these opposing functions.
We first assessed genetic interactions between prf1Δ and reduction of Cdk9 activity. We introduced the cdk9as allele into the prf1Δ background and analyzed double mutants by DAPI/calcofluor staining after inhibition of Cdk9as with 3-MB-PP1. Cdk9as inhibition strongly suppressed the cell morphology and septation phenotypes caused by prf1Δ (as it did for brl2Δ; Figure 6A). A hypomorphic cdk9-T212A mutation similarly rescued these phenotypes. These results indicate that the cell morphology and septation defects in prf1Δ, brl2Δ, and htb1-K119R strains depend upon Cdk9 activity, and suggest that Prf1, Brl2, and H2Bub1 itself act to limit some aspect of Cdk9 function.
Fig. 6. Prf1 and PAF pathways have opposing biological effects. 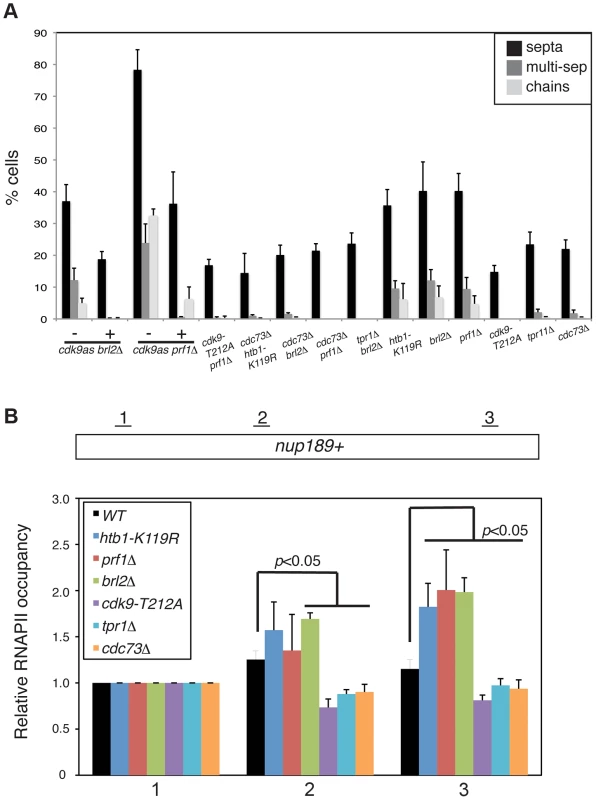
(A) Quantification of the abnormal septation patterns in the indicated strains. Strains carrying the cdk9as allele were cultured in DMSO (−) or 2 µM 3-MB-PP1 (+) for 15 hours prior to fixation for microscopy. Error bars denote standard deviations from 3 independent experiments. (B) ChIP of RNAPII was carried out in the indicated strains and quantified by qPCR using primers specific to the nup189+ gene. Values were normalized to that for primer pair 1. Error bars denote standard deviations from three independent experiments. Significant differences from wild-type values (unpaired t-test) are indicated. We next determined whether PAF mutations, which phenocopy cdk9-T212A, could also suppress the cell morphology and septation phenotypes of htb1-K119R, brl2Δ, and prf1Δ mutants. We found that cdc73Δ, when combined with htb1-K119R, brl2Δ, or prf1Δ, suppressed aberrant septation. Similarly, brl2Δ septation phenotypes were effectively suppressed by tpr1Δ (Figure 6A). The dependence of these phenotypes on both Cdk9 activity and PAF is consistent with the Prf1 pathway acting in opposition to the PAF pathway.
We also tested whether htb1-K119R, brl2Δ, and prf1Δ could reciprocally suppress cdk9 and PAF mutant phenotypes. None of these mutations affected the growth of cdk9 or PAF mutants on minimal media. Whereas htb1-K119R suppressed the MPA sensitivity of cdk9-T212A, prf1Δ did not, and brl2Δ enhanced this phenotype (Figure S8). These results indicate that the Prf1 pathway does not generally limit PAF pathway function.
In order to ascertain how these two pathways regulate RNAPII elongation, we carried out RNAPII ChIP experiments in strains harboring mutations in either the Prf1 or PAF pathways. We quantified the ChIP by qPCR with primers in nup189+ and SPBC354.10+, genes that exemplify the 3′ shift in RNAPII distribution that we documented previously [11]. Consistent with our previous data, the htb1-K119R mutation led to an increase in RNAPII occupancy near the 3′ end of the nup189+ gene relative to the 5′ end (Figures 6B and S9A). A similar effect was also observed for brl2Δ and prf1Δ mutations, implying that the 3′ RNAPII shift is characteristic of the Prf1 pathway. In contrast, cdk9-T212A, cdc73Δ, and tpr1Δ mutations led to a relative decrease in RNAPII density toward the 3′ end of the gene, consistent with defective elongation (Figure 6B). The effects on RNAPII distribution were generally less significant at SPBC354.10+, but we nonetheless observed a similar pattern in which RNAPII occupancy increased toward the 3′ end of the gene in Prf1 pathway mutants and decreased or was unaffected in PAF pathway mutants (Figure S9A).
We also found that the Prf1 and PAF pathway mutations differentially affected the steady-state levels of mRNA produced from these genes. PAF pathway mutants exhibited modest decreases in mRNA levels, whereas no change or modest increases were observed in the Prf1 pathway mutant strains (Figure S9B). These changes are likely to reflect altered RNAPII elongation, since the absolute levels of RNAPII occupancy near the 5′ ends of these genes did not differ between Prf1 and PAF pathway mutant strains (Figure S9C). Overall, our data suggest that the PAF and Prf1 pathways have differential effects on behavior of the RNAPII elongation complex (Figure 7).
Fig. 7. Model depicting the roles of the Prf1/Rtf1 and PAF pathways in RNAPII elongation. 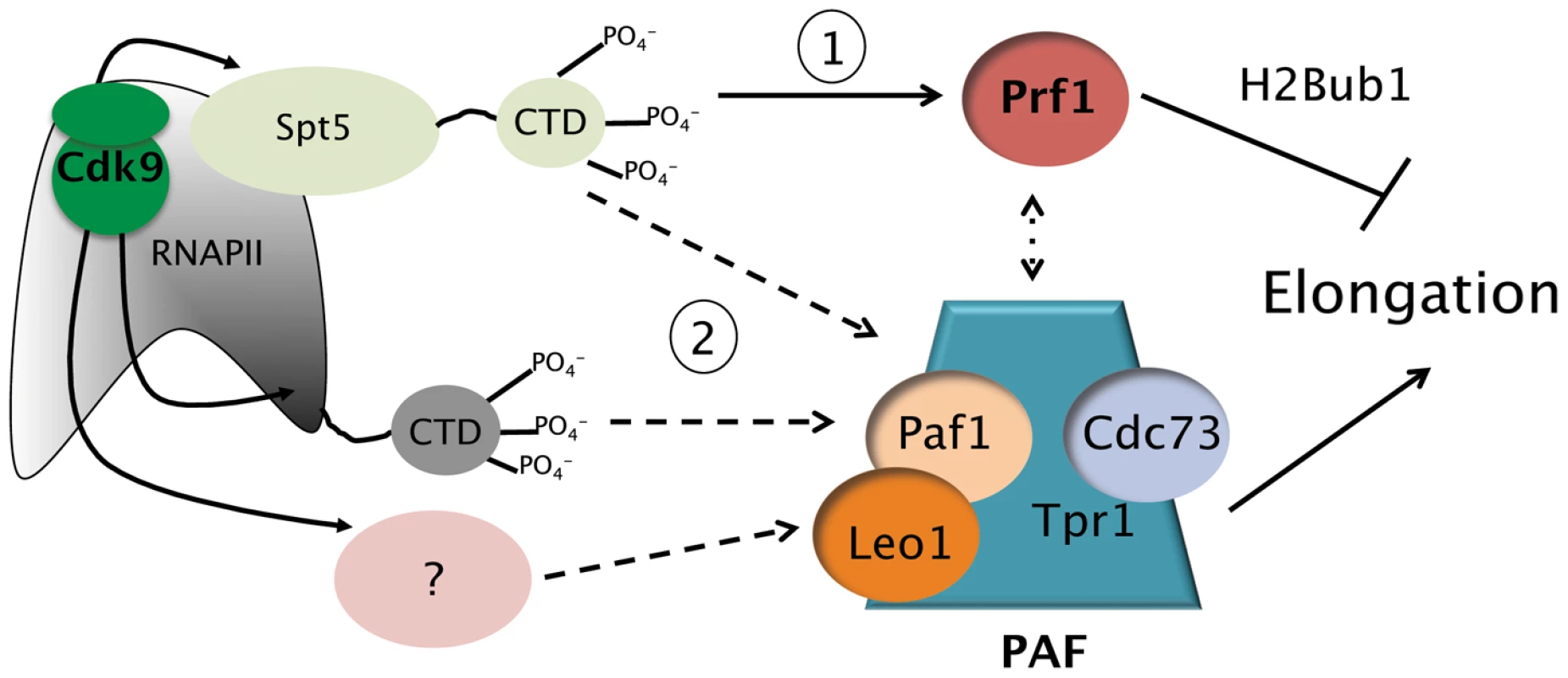
The Prf1 pathway, involving direct association of Prf1 with phosphorylated Spt5 CTD, is labeled “1.” The PAF pathway, involving multiple Cdk9 targets, is labeled “2.” Potential crosstalk between the two pathways is indicated by the broken double arrow. See text for details. Discussion
Cdk9 influences multiple aspects of RNAPII transcription. Our results here reveal that phosphorylation of the Spt5 CTD by Cdk9 directly and specifically recruits the conserved transcriptional regulator Prf1/Rtf1 to transcribed chromatin. We further define a second Cdk9 pathway, involving phosphorylation of at least one additional target, which recruits the PAF complex. We show that these two pathways have distinct and opposing functions, suggesting that balanced action of Cdk9 pathways is a feature of RNAPII elongation.
Prf1/Rtf1 and PAF are biochemically separate in S. pombe
Prf1/Rtf1 and PAF do not form a stable complex in S. pombe whole-cell extracts, a finding consistent with the properties of these proteins in metazoans. Biochemical studies of human Rtf1 have shown that it can be assembled into a complex with PAF in vitro, and that a C-terminal fragment of Rtf1 can be detected in association with PAF purified from nuclear extracts [22]. Although the physiological significance of these findings has not been explored, they suggest the possibility of a regulated interaction between full-length Rtf1 and PAF. Modulation of this interaction could underlie the shared and distinct functions of Rtf1 and PAF in different biological contexts.
The similarities we have found between the biochemical properties of Prf1 and PAF in fission yeast and those of their metazoan orthologs underscore the utility of the S. pombe system as a model for study of these factors. S. pombe Prf1 and PAF also show functional resemblance to their human counterparts in that they promote the H2Bub1/H3K4me histone modification axis but not H3K36me, a modification that is PAF-dependent in S. cerevisiae [26], [43].
We found that individual subunits within S. pombe PAF were functionally distinct: loss of either Tpr1 or Cdc73 was associated with more severe phenotypic consequences in our assays than loss of Paf1 or Leo1. This contrasts with PAF functional organization in S. cerevisiae, where paf1Δ causes pleiotropic phenotypes [42]. Further investigation into physical interactions between individual PAF subunits and other factors will help to clarify these functional differences.
Prf1/Rtf1 is a direct downstream effector of phosphorylated Spt5 CTD
Our data show that Prf1/Rtf1 directly and specifically associates with the phosphorylated Spt5 CTD, a finding that is consistent with recent reports on budding yeast and human orthologs of this protein [30], [31], [54]. In vitro, budding yeast Rtf1 was found to bind Spt5-P and the phosphorylated Rpb1 CTD with comparable affinities, and Rtf1 mutations that abrogate its interaction with Spt5-P prevented the association of multiple PAF subunits with transcribed chromatin in vivo [30], [54]. The budding yeast Ctr9 and Cdc73 proteins could also interact with phosphorylated peptides corresponding to either the Spt5 CTD or the Rpb1 CTD [30]. The apparently overlapping mechanisms involved in recruitment of Rtf1 and other PAF components to chromatin in budding yeast are in line with the robust association of Rtf1 with PAF found in this organism. Our findings in S. pombe point to a specific and primary role for the Prf1/Spt5-P interaction in Prf1 recruitment to transcribed chromatin, and suggest an alternate recruitment mechanism for PAF.
The direct interaction between Prf1/Rtf1 and Spt5-P helps explain the critical role of Spt5-P in establishment of H2Bub1 levels. This interaction also raises mechanistic questions regarding the reciprocal dependence of Spt5-P on H2Bub1. Loss of Spt5-P in the htb1-K119R mutant is accompanied by a reduction in crosslinking of Cdk9 to transcribed chromatin [11]. In the prf1Δ strain, it is possible that loss of Spt5-P is also attributable to the absence of the Prf1/Spt5-P interaction, which could provoke dephosphorylation of Spt5-P.
The spt5-T1A mutation did not result in cell morphology and septation defects observed in prf1Δ, brl2Δ, or htb1-K119R mutant strains, suggesting that some Prf1 function is maintained in the absence of Spt5 CTD phosphorylation [11]. This is consistent with our ChIP results, which demonstrated a substantial but incomplete loss of Prf1 chromatin recruitment in spt5-T1A mutant strains. Since residual Prf1 recruitment was further reduced by Cdk9as inhibition, other Cdk9 targets are likely to play some role in this process, although the bypass of Cdk9 dependence in the spt5-T1E mutant suggests that the Spt5 CTD is the primary Cdk9 target involved. Multiple domains of S. cerevisiae Rtf1 can be independently recruited to chromatin, indicating that mechanisms other than Spt5 phosphorylation are also important for Rtf1 chromatin association [31], [46], [55]. It is also possible that Prf1 may retain some function even when it is not stably bound to chromatin, as has been observed for the S. cerevisiae PAF complex [56].
PAF association with transcribed chromatin requires phosphorylation of multiple targets by Cdk9
We found that PAF recruitment to chromatin in vivo was only modestly sensitive to Spt5 CTD phosphorylation at both transcribed loci tested. Since we found no interaction between the purified PAF complex and Spt5-P peptide in vitro, we presume that this reflects an indirect effect on PAF association with chromatin mediated by another factor that binds to Spt5-P. This is unlikely to be Prf1, as PAF chromatin association is not affected by the spt5-T1E mutation (unlike that of Prf1).
Because crosslinking of Tpr1-TAP and Paf1-TAP to transcribed loci was dramatically reduced by Cdk9as inhibition even in an spt5-T1A mutant, PAF recruitment must require at least one other Cdk9 target in addition to Spt5. Given that the Rpb1 CTD is an established target of Cdk9 activity, we were surprised to find that simultaneous elimination of Ser2 and Ser7 in the Rpb1 CTD repeat, or removal of Ser5, had no substantial effect on PAF recruitment in vivo. This could reflect PAF association with forms of the Rpb1 CTD repeat phosphorylated on both Ser5 and another residue, as has been suggested by recent biochemical experiments with budding yeast PAF components [30]. Alternatively, the role of the Rpb1 CTD may be redundant with the Spt5 CTD [19], [57]. Pleiotropic phenotypes associated with mutant combinations that include Rpb1-Ser5 have precluded thorough genetic tests of these possibilities. Recruitment of the PAF complex to chromatin may also involve another, as yet unidentified, Cdk9 target.
Balanced action of Prf1/Rtf1 and PAF pathways in RNAPII elongation
Septation and cell morphology phenotypes of Prf1 pathway mutants were reduced by cdk9-T212A, cdc73Δ, and tpr1Δ mutations, indicating that these components act aberrantly in the absence of the Prf1 pathway. Prf1 pathway mutations did not generally suppress cdk9 or PAF mutant phenotypes, suggesting that the Prf1 pathway has a specialized role in limiting certain aspects of PAF pathway function.
ChIP of RNAPII reveals differential effects of the two pathways on RNAPII distribution within the nup189+ and SPBC354.10+ gene-coding regions, with the PAF pathway promoting RNAPII occupancy at promoter-distal sequences and the Prf1 pathway limiting occupancy at these locations. This observation supports our earlier studies on the opposing effects of Cdk9 activity and H2Bub1 on intragenic RNAPII distribution, and implies that these findings also characterize the two Cdk9 pathways we have described.
We propose that Cdk9-dependent recruitment of PAF, through phosphorylation of the Spt5 CTD and other targets, promotes RNAPII elongation through gene coding regions, as supported by our RNAPII ChIP and mRNA expression data. Spt5 CTD phosphorylation also recruits Prf1/Rtf1 and promotes formation of H2Bub1, which may act as an auto-regulatory mechanism to balance the positive effects of the PAF pathway. Previous studies documenting a positive role for PAF in elongation have shown Rtf1 to be dispensable for this function, consistent with the distinctive behavior of Prf1 in S. pombe [21]–[23]. Although we did not observe significant effects of the Prf1 pathway mutations on expression of nup189+ and SPBC354.10+ (with the exception of a slight increase for SPBC354.10+ in the htb1-K119R mutant), it is possible that loss of the putative Cdk9 auto-regulatory function in these mutants would be associated with defective mRNA processing or export, rather than a change in mRNA levels per se. Further elucidation of the mechanisms through which these pathways affect gene expression will be an important goal of future research.
A negative role for the Prf1 pathway in elongation could be related to the functions of H2Bub1 in directing nucleosome assembly and histone deacetylation within gene coding regions [58]–[61]. Indeed, H2Bub1 and the Hos2 deacetylase complex have been linked to reduced RNAPII processivity in budding yeast [61].
It is notable that an interaction between PAF and the elongation factor TFIIS that stimulates elongation in mammalian cells is antagonized by the H2Bub1 ubiquitin ligase RNF20 [62]. Although we find no role for TFIIS in opposing H2Bub1 functions in S. pombe (data not shown), a role for the Prf1 pathway and H2Bub1 in altering PAF function in elongation may be conserved.
In metazoans, Cdk9 activity has an essential role in release of RNAPII from promoter-proximal pause sites. Phosphorylation of multiple targets, including the Rpb1 CTD, Spt5, and NELF, is thought to drive RNAPII from the pause, implying coordinate action of Cdk9 phosphorylation events in pause release [4], [63]. Interestingly, the experiments defining the role of PAF in P-TEFb-dependent pause release in vitro used PAF complex devoid of Rtf1 [21]. We suggest that the P-TEFb pause release function in metazoans, which involves multiple Cdk9 targets and PAF, is analogous to the PAF pathway we have documented in S. pombe. The metazoan analog of the Prf1 pathway might involve Rtf1 recruitment via Spt5-P subsequent to pause release, as part of a mechanism to keep the stimulatory effects of PAF on elongation in check. Thus, the roles of Cdk9 targets in regulating the RNAPII elongation complex may diverge during the course of elongation.
Materials and Methods
Strains and media
S. pombe strains used in this study are listed in Table S2. Liquid cultures used standard YES media (yeast extract 5 g/L, D-glucose 30 g/L, supplemented with 250 mg/L each of histidine, leucine, adenine and uracil). Minimal media (EMM) was purchased from MP Biomedicals. EMM complete was supplemented with 250 mg/L each of histidine, leucine, adenine, and uracil. Mycophenolic acid (MPA; Bioshop) was dissolved in dimethyl sulfoxide (DMSO) and used at a final concentration of 25 µg/mL. The TAP and 13xmyc tags were introduced at the relevant chromosomal loci as described previously [64]. All other genetic manipulations were carried out using standard methods [65].
Tandem affinity purification and mass spectrometry
Paf1-TAP, Prf1-TAP and Tpr1-TAP were purified from whole cell extracts as described [48]. For mass spectrometry analysis of polypeptides excised from polyacrylamide gels, purified material was concentrated by addition of 0.02% sodium deoxycholate, incubation on ice for 30 min, and precipitation with trichloroacetic acid (15% final concentration) overnight. The precipitate was collected by centrifugation, washed twice with ice-cold ethanol and then resuspended with 10 µl of SDS-PAGE sample buffer. Proteins were resolved by SDS-PAGE on a 4–20% gradient acrylamide gel and visualized by Coomassie (Bio-rad) or silver staining before mass spectrometry analysis.
To analyze the purified material in solution, the TAP purification procedure was modified as follows: Glycerol and detergent were omitted from the final wash and elution steps, and EGTA was replaced with EDTA in the elution buffer. The eluted material was subjected to mass spectrometry analysis, details of which are provided in the Supplemental Material (see Text S1).
For single-step TAP purification, whole cell extracts were incubated with IgG-sepharose beads (GE Healthcare) and the beads were collected and washed as described [48]. Beads were suspended in SDS sample buffer and bound material was analyzed by SDS-PAGE and western blotting.
Immunological methods
Immunoblotting was performed as previously described [11]. The following commercial antibodies were used: H2Bub1 (Active Motif #39623), H3K4me1 (Abcam #ab8895), H3K4me2 (Abcam #ab32356), H3K4me3 (Abcam #ab8580), H3K36me3 (Abcam #ab9050), histone H3 (Abcam #ab1791), histone H2B (Millipore #07-371), TAP tag (Fisher Scientific #PICAB1001), Rpb1 (8WG16; Covance #MMS-126R-200), Rpb1-Ser2-P (H5; Covance #MMS-129R), myc (9E10; Covance #MMS-150P). Antibodies against S. pombe Spt5 and Spt5-P were described previously [11].
Phenotypic analyses
DAPI/calcofluor staining was carried out as described previously [11]. For spot tests, 4×105 cells were harvested from exponentially growing cultures, and resuspended in YES media. Ten-fold serial dilutions of the culture were spotted onto the appropriate plates and incubated at 30°C for 3–6 days.
Chromatin immunoprecipitation
ChIP was carried out as previously described [48] with minor modifications. Briefly, 1.5×107 cells grown in YES media were fixed with 1% formaldehyde for 30 min. Glycine (pH 2.5) was added at a final concentration of 125 mM to stop the crosslinking. Cells were washed twice with cold 1XTBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Tween-20) and snap frozen in liquid nitrogen. Cell pellets were resuspended in 0.5 mL of lysis buffer [50 mM HEPES, pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM PMSF, protease inhibitor cocktail (Roche)], 200 µl of sterile glass beads were added, and cells were lysed at 4°C using a mini-beat beater (4×30 s with 1 min rests on ice). The lysates were transferred to 1.5 mL tubes and centrifuged at top speed for 15 min to collect the chromatin. Chromatin pellets were resuspended in 1 mL lysis buffer, sonicated for 20 minutes using a Bioruptor waterbath sonicator (Diagenode) (30 s ON/OFF with 2 min rest on ice after 10 min) and centrifuged at top speed for 5 minutes. The supernatant (700–800 µl) was incubated for 2–3 h with either 50 µl of IgG sepharose beads (GE Healthcare)(for TAP immunoprecipitation) or with 15 µl of protein G Dynabeads (Invitrogen) coupled to 8WG16 antibody (for RNAPII immunoprecipitation) at 4°C with rocking. The beads were washed successively with 0.5 mL of each of the following buffers: lysis buffer +0.1% SDS, lysis buffer +0.1% SDS+500 mM NaCl, LiCl buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 250 mM LiCl, 0.5% sodium deoxycholate, 0.5% NP-40) and TE (10 mM Tris-HCl, pH 7.5, 1 mM EDTA). Chromatin was eluted with 100 µl of elution buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 1% SDS) at 65°C for 30 min. Reversal of crosslinks, DNA purification, and quantitative PCR were as described [48]. Primers specific for the act1+, adh1+, nup189+, and SPBC354.10+ genes have been described previously [11], [66].
Peptide-binding assays
15 µl of streptavidin Dynabeads (Invitrogen) were washed three times with 400 µl of 1xPBS/0.1% Triton-X100. Beads were coupled to 50 µg biotinylated peptide in 380 µl of 1xPBS. Incubation was at room temperature for 3 h with rocking. Spt5 CTD peptides were synthesized at the Keck Foundation Research Laboratory (Yale University) and had the sequence [biotin]-GSKTPAWNSGSKTPAWNS. In the phospho-peptide both Thr residues were phosphorylated. Rpb1 CTD peptides were a generous gift from Francois Robert (IRCM, Montreal) and had the following sequences: [biotin]-YSPTSPSYSPTSPSYSPTSPS, [biotin]-YSPTSPSY(pSer)PTSPSYSPTSPS, [biotin]-SPTSPSYSPT(pSer)PSYSPTSPSY, [biotin]-TSPSYSPTSP(pSer)YSPTSPSYSP, [biotin]-PTSPSYSPT(pSer)P(pSer)YSPTSPSYS. Beads were washed twice with washing buffer (20 mM HEPES pH 7.6, 20% v/v glycerol, 1 mM EDTA, 0.1% Triton X-100, 250 mM KOAc, 10 mM β-glycero-3-phosphate, 1 mM PMSF) and resuspended with 150 µl of the binding buffer (20 mM HEPES pH 7.6, 0.1% Triton X-100, 1 mM PMSF, 10 mM β-glycero-3-phosphate). Binding reactions contained 20 mM Hepes pH 7.6, 0.075% Triton X-100, 0.75 mM PMSF, 7.5 mM β-glycero-3-phosphate, 5% glycerol, 37.5 mM KOAc, 2.5 mM MgOAc, 0.25 mM EDTA, 0.25 mM DTT and 50–200 ng of purified factors (∼50 ng for Prf1-TAP or 200 ng for Paf1-TAP and Tpr1-TAP) in a volume of 200 µl. The mixtures were incubated at 4°C for 1 h with shaking. Beads were washed 4 times with 1 mL of washing buffer, resuspended with 20 µl of SDS-PAGE sample buffer, boiled for 5 minutes at 95°C, and subjected to SDS-PAGE and immunoblotting with a TAP antibody. Signal intensities were quantified using ImageJ software and used to calculate percent bound.
Supporting Information
Zdroje
1. ZhouQ, LiT, PriceDH (2012) RNA polymerase II elongation control. Annu Rev Biochem 81 : 119–143.
2. MarshallNF, PriceDH (1995) Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 270 : 12335–12338.
3. WadaT, TakagiT, YamaguchiY, WatanabeD, HandaH (1998) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. Embo J 17 : 7395–7403.
4. YamadaT, YamaguchiY, InukaiN, OkamotoS, MuraT, et al. (2006) P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21 : 227–237.
5. YamaguchiY, TakagiT, WadaT, YanoK, FuruyaA, et al. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97 : 41–51.
6. MurrayS, UdupaR, YaoS, HartzogG, PrelichG (2001) Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol Cell Biol 21 : 4089–4096.
7. PeiY, DuH, SingerJ, St AmourC, GranittoS, et al. (2006) Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo. Mol Cell Biol 26 : 777–788.
8. PeiY, SchwerB, ShumanS (2003) Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J Biol Chem 278 : 7180–7188.
9. PeiY, ShumanS (2003) Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J Biol Chem 278 : 43346–43356.
10. KeoghMC, PodolnyV, BuratowskiS (2003) Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol 23 : 7005–7018.
11. SansoM, LeeKM, ViladevallL, JacquesPE, PageV, et al. (2012) A Positive Feedback Loop Links Opposing Functions of P-TEFb/Cdk9 and Histone H2B Ubiquitylation to Regulate Transcript Elongation in Fission Yeast. PLoS Genet 8: e1002822.
12. ViladevallL, St AmourCV, RosebrockA, SchneiderS, ZhangC, et al. (2009) TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell 33 : 738–751.
13. PeterlinBM, PriceDH (2006) Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23 : 297–305.
14. HsinJP, ManleyJL (2012) The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26 : 2119–2137.
15. ZhouK, KuoWH, FillinghamJ, GreenblattJF (2009) Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A 106 : 6956–6961.
16. LiuY, WarfieldL, ZhangC, LuoJ, AllenJ, et al. (2009) Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 29 : 4852–4863.
17. ShchebetA, KarpiukO, KremmerE, EickD, JohnsenSA (2012) Phosphorylation by cyclin-dependent kinase-9 controls ubiquitin-conjugating enzyme-2A function. Cell Cycle 11 : 2122–2127.
18. WoodA, SchneiderJ, DoverJ, JohnstonM, ShilatifardA (2005) The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell 20 : 589–599.
19. SchneiderS, PeiY, ShumanS, SchwerB (2010) Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol Cell Biol 30 : 2353–2364.
20. TomsonBN, ArndtKM (2013) The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta 1829 : 116–126.
21. ChenY, YamaguchiY, TsugenoY, YamamotoJ, YamadaT, et al. (2009) DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev 23 : 2765–2777.
22. KimJ, GuermahM, RoederRG (2010) The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140 : 491–503.
23. RondonAG, GallardoM, Garcia-RubioM, AguileraA (2004) Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep 5 : 47–53.
24. PavriR, ZhuB, LiG, TrojerP, MandalS, et al. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125 : 703–717.
25. KroganNJ, DoverJ, WoodA, SchneiderJ, HeidtJ, et al. (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11 : 721–729.
26. ZhuB, MandalSS, PhamAD, ZhengY, Erdjument-BromageH, et al. (2005) The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev 19 : 1668–1673.
27. PenheiterKL, WashburnTM, PorterSE, HoffmanMG, JaehningJA (2005) A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20 : 213–223.
28. Rozenblatt-RosenO, NagaikeT, FrancisJM, KanekoS, GlattKA, et al. (2009) The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci U S A 106 : 755–760.
29. LaribeeRN, KroganNJ, XiaoT, ShibataY, HughesTR, et al. (2005) BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol 15 : 1487–1493.
30. QiuH, HuC, GaurNA, HinnebuschAG (2012) Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. Embo J 31 : 3494–3505.
31. MayekarMK, GardnerRG, ArndtKM (2013) The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex. Mol Cell Biol 33 : 3259–3273.
32. NordickK, HoffmanMG, BetzJL, JaehningJA (2008) Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell 7 : 1158–1167.
33. AdelmanK, WeiW, ArdehaliMB, WernerJ, ZhuB, et al. (2006) Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol 26 : 250–260.
34. LangenbacherAD, NguyenCT, CavanaughAM, HuangJ, LuF, et al. (2011) The PAF1 complex differentially regulates cardiomyocyte specification. Dev Biol 353 : 19–28.
35. Rozenblatt-RosenO, HughesCM, NannepagaSJ, ShanmugamKS, CopelandTD, et al. (2005) The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol 25 : 612–620.
36. KroganNJ, KimM, AhnSH, ZhongG, KoborMS, et al. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22 : 6979–6992.
37. MuellerCL, JaehningJA (2002) Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol 22 : 1971–1980.
38. AkanumaT, KoshidaS, KawamuraA, KishimotoY, TakadaS (2007) Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep 8 : 858–863.
39. BaiX, KimJ, YangZ, JurynecMJ, AkieTE, et al. (2010) TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142 : 133–143.
40. HeY, DoyleMR, AmasinoRM (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18 : 2774–2784.
41. DingL, Paszkowski-RogaczM, NitzscheA, SlabickiMM, HeningerAK, et al. (2009) A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4 : 403–415.
42. BetzJL, ChangM, WashburnTM, PorterSE, MuellerCL, et al. (2002) Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol Genet Genomics 268 : 272–285.
43. ChuY, SimicR, WarnerMH, ArndtKM, PrelichG (2007) Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. Embo J 26 : 4646–4656.
44. KeoghMC, KurdistaniSK, MorrisSA, AhnSH, PodolnyV, et al. (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123 : 593–605.
45. LenstraTL, BenschopJJ, KimT, SchulzeJM, BrabersNA, et al. (2011) The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 42 : 536–549.
46. PiroAS, MayekarMK, WarnerMH, DavisCP, ArndtKM (2012) Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc Natl Acad Sci U S A 109 : 10837–10842.
47. EydmannT, SommarivaE, InagawaT, MianS, KlarAJ, et al. (2008) Rtf1-mediated eukaryotic site-specific replication termination. Genetics 180 : 27–39.
48. RacineA, PageV, NagyS, GrabowskiD, TannyJC (2012) Histone H2B ubiquitylation promotes activity of the intact Set1 histone methyltransferase complex in fission yeast. J Biol Chem 287 : 19040–19047.
49. TannyJC, Erdjument-BromageH, TempstP, AllisCD (2007) Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev 21 : 835–847.
50. ChandrasekharanMB, HuangF, SunZW (2010) Histone H2B ubiquitination and beyond: Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics 5 : 460–468.
51. St AmourCV, SansoM, BoskenCA, LeeKM, LarochelleS, et al. (2012) Separate domains of fission yeast Cdk9 (P-TEFb) are required for capping enzyme recruitment and primed (Ser7-phosphorylated) CTD substrate recognition. Mol Cell Biol 32 : 2372–2383.
52. PhatnaniHP, JonesJC, GreenleafAL (2004) Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43 : 15702–15719.
53. SchwerB, ShumanS (2011) Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell 43 : 311–318.
54. WierAD, MayekarMK, HerouxA, ArndtKM, VandemarkAP (2013) Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci U S A 110 : 17290–17295.
55. WarnerMH, RoinickKL, ArndtKM (2007) Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol 27 : 6103–6115.
56. MuellerCL, PorterSE, HoffmanMG, JaehningJA (2004) The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 14 : 447–456.
57. LindstromDL, HartzogGA (2001) Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159 : 487–497.
58. FlemingAB, KaoCF, HillyerC, PikaartM, OsleyMA (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31 : 57–66.
59. ChandrasekharanMB, HuangF, SunZW (2009) Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A 106 : 16686–16691.
60. BattaK, ZhangZ, YenK, GoffmanDB, PughBF (2011) Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev 25 : 2254–2265.
61. WeinbergerL, VoichekY, TiroshI, HornungG, AmitI, et al. (2012) Expression Noise and Acetylation Profiles Distinguish HDAC Functions. Mol Cell 47 : 193–202.
62. ShemaE, KimJ, RoederRG, OrenM (2011) RNF20 Inhibits TFIIS-Facilitated Transcriptional Elongation to Suppress Pro-oncogenic Gene Expression. Mol Cell 42 : 477–488.
63. FujinagaK, IrwinD, HuangY, TaubeR, KurosuT, et al. (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24 : 787–795.
64. TastoJJ, CarnahanRH, McDonaldWH, GouldKL (2001) Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18 : 657–662.
65. MorenoS, KlarA, NurseP (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194 : 795–823.
66. GuiguenA, SoutourinaJ, DewezM, TafforeauL, DieuM, et al. (2007) Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J 26 : 1552–1559.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



