-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
An Immunity-Triggering Effector from the Barley Smut Fungus Resides in an Ustilaginaceae-Specific Cluster Bearing Signs of Transposable Element-Assisted Evolution
Upon host infection, plant pathogens secrete suites of virulence effectors to suppress defense responses and support their own development. In certain cases, hosts evolve resistance genes that recognize such effectors or their actions to initiate defense responses. By deleting candidate genes, we identified the immune-triggering effector UhAvr1 from Ustilago hordei, a barley-infecting basidiomycete smut fungus. We show that this effector is expressed only when hyphae sense and infect barley coleoptile epidermal cells. Its presence in the fungus causes a necrotic reaction immediately upon penetration resulting in complete immunity in barley cultivars having resistance gene Ruh1. We show that fungal isolates that have mutated to change the expression of this non-crucial protein are avoiding recognition by the host, hence overcoming restriction by its immune response. In virulent isolates, transposable elements, known as genome modifiers, have separated the UhAvr1 coding region from its transcription signals. UhAvr1 is located in a larger cluster of ten effectors and is similar to clusters with more and further diversified effectors in the related maize pathogens U. maydis and Sporisorium reilianum. This study should lead us to discovering a mechanism by which this major cereal crop protects itself against this pathogen.
Published in the journal: . PLoS Pathog 10(7): e32767. doi:10.1371/journal.ppat.1004223
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004223Summary
Upon host infection, plant pathogens secrete suites of virulence effectors to suppress defense responses and support their own development. In certain cases, hosts evolve resistance genes that recognize such effectors or their actions to initiate defense responses. By deleting candidate genes, we identified the immune-triggering effector UhAvr1 from Ustilago hordei, a barley-infecting basidiomycete smut fungus. We show that this effector is expressed only when hyphae sense and infect barley coleoptile epidermal cells. Its presence in the fungus causes a necrotic reaction immediately upon penetration resulting in complete immunity in barley cultivars having resistance gene Ruh1. We show that fungal isolates that have mutated to change the expression of this non-crucial protein are avoiding recognition by the host, hence overcoming restriction by its immune response. In virulent isolates, transposable elements, known as genome modifiers, have separated the UhAvr1 coding region from its transcription signals. UhAvr1 is located in a larger cluster of ten effectors and is similar to clusters with more and further diversified effectors in the related maize pathogens U. maydis and Sporisorium reilianum. This study should lead us to discovering a mechanism by which this major cereal crop protects itself against this pathogen.
Introduction
Pathogenic microbes secrete hundreds of compounds and proteins into their host as part of the infection strategy. This arsenal of virulence factors, often small proteins with a predicted signal peptide (SP), effectors or candidate secreted effector proteins (CSEPs), functions to facilitate entry, to subdue defense responses that may be triggered through their recognition by the hosts' surveillance system, to divert nutrients and to ensure proliferation [1]–[4]. Plants use a variety of defense mechanisms to avoid pathogen invasion and subsequent disease, including physical barriers, preformed antimicrobial compounds, but also activation of defenses. In particular, defenses can be induced by the recognition of highly conserved pathogen molecules (Pathogen - Associated Molecular Patterns or PAMPs) resulting in a broad-based PAMP-triggered immunity (PTI). Certain pathogen effectors, whether secreted into the host apoplast or vessels and taken up, or delivered directly into cells to perform their function, are inadvertently recognized directly or through their action by a highly sophisticated system of which resistance (R) genes are a part, to elicit effector-triggered immunity or ETI [5]–[8]. Induced immunity includes cell wall strengthening, the generation of an environment toxic to the pathogen, encasement of the pathogen and localized programmed cell death (PCD) to arrest pathogen development [9]. Although the latter affects development of biotrophic pathogens, necrotrophic pathogens and hemibiotrophs at later stages of infection might have evolved to take advantage of triggering PCD [10], [11]. Earlier studies in several pathosystems demonstrated the presence of genetically dominant avirulence (Avr) genes in pathogens, the products of which have been shown more recently to often be effectors, interacting genetically with also often dominant host R genes. This concept was developed by Harold Flor using the flax-rust Melampsora lini pathosystem [12] and simultaneously by his contemporary, Arend Oort who studied the wheat-loose smut Ustilago tritici pathosystem but because of WWII could only publish his results in 1944 in Dutch [13].
In pathogen populations, there is strong selection to avoid recognition resulting in rapidly evolving effectors and, in response, evolving host R genes or effector targets [14], [15]. This natural arms race is accelerated in agricultural settings where invading pathogens necessitate the introduction of resistant host cultivars from breeding programs, thereby triggering boom-bust cycles. In many cases, Avr genes are present in genomic regions displaying high flexibility, such as telomeres [16], heterochromatic locations [17], [18], or are surrounded by transposable elements (TEs) [17], [19]–[22] which can facilitate effector gene mutation.
Basidiomycete smut fungi are important pathogens that cause disease world-wide and are of economic importance on many Poaceae [23]–[25]. Ustilago maydis, the maize smut fungus, has become the paradigm for molecular genetic studies on biotrophic basidiomycete plant pathogens [26], [27]. The barley covered smut fungus, U. hordei, is closely related but differs in important aspects: in U. hordei, race - and strain-specific virulence compatibility interactions exist whereas no dominant avirulence functions that genetically interact with dominant host resistance genes on a gene-for-gene basis have been identified in U. maydis. Moreover, U. hordei can infect only at the seed germination stage to develop quiescently in the meristematic region until sporulation occurs mainly in the seed heads [28] (Figure S1), a characteristic shared with many smut fungi, such as the maize-infecting Sporisorium reilianum [29]. In contrast, U. maydis can infect any above-ground part of the maize plant at any plant age, resulting in the proliferation and sporulation of the fungus in tumors it incites. In addition, U. hordei has differently organized mating-type loci affecting its biology [30], [31] and it has a larger genome due to a much higher content of repeats and TEs [32].
Six Avr genes have been genetically identified in U. hordei which in different combinations constitute 14 different reported races; six corresponding resistance genes have been proposed in barley [33]–[36]. UhAvr1 determines avirulence towards barley cultivar Hannchen which has matching resistance gene Ruh1 which we recently mapped to the short arm of barley chromosome 7H [37]. The UhAvr1 locus was located to an approximately 80-kb region contained on Bacterial Artificial Chromosome (BAC) clone 3-A2, using a marker-based approach in a mapping population of 54 progeny segregating for avirulence towards Hannchen, resulting from a cross between parental lines Uh362 (MAT-2 Uhavr1) and Uh364 (MAT-1 UhAvr1) [38]. We show here that this locus spans a cluster of predicted secreted protein genes on chromosome 18 and we identify through targeted deletions and complementation the gene with the UhAvr1 avirulence function, coding for a predicted secreted effector. The locus is syntenic to cluster 19A in both U. maydis and S. reilianum that also contains small proteins predicted to be secreted [26], [39], but has evolved differently. UhAvr1 is located in a transposon - and repeat-rich region and transposon activity seems responsible for breaking the avirulence towards Hannchen. In virulent isolates, it appears that insertion of transposable element sequences in the promoter of UhAvr1 has changed its expression.
Results
Sequence comparison of CSEP genes among Avr1 and vir1 phenotypes
BAC clone BAC3A-2, genetically harbouring U. hordei avirulence gene UhAvr1, was sequenced by GPS transposon insertion resulting in an assembled sequence of 117 kb. The presence of repeats and TE sequences made assembly challenging. On this BAC insert we identified 47 ORFs (Figure 1A, Table S1). Hybridization to DNA blots of separated chromosomes located this region to a 667-kb chromosome ([38], Figure S2), designated as U. hordei Chr 18, a homolog of U. maydis Chr19 in our recent comparative genome study [32]. In light of publications in which a number of avirulence gene products were CSEPs, we hypothesized that UhAVR1p could also be a secreted effector. On the sequenced BAC clone, ten predicted CSEPs were identified but only seven were likely candidates: gene 5 and gene 6 were located outside the genetic interval identified previously and gene 44 was very close to RFLP marker 2 which revealed three recombinants (Figure 1A, Table S1 [38]).
Fig. 1. Map of UhAvr1 and Uhavr1 locus regions. 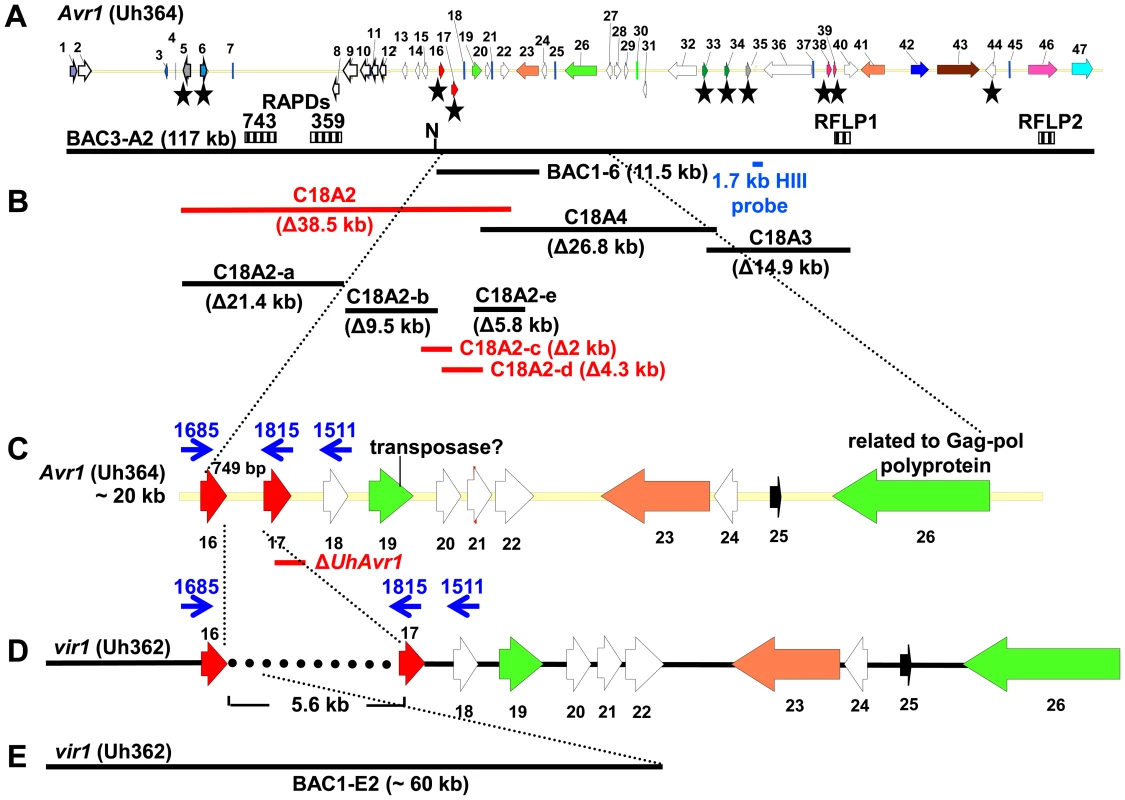
A. UhAvr1 locus region in U. hordei strain Uh364 with arrows representing all predicted ORFs with their direction of transcription (see Table S1 for gene calls and similarity to U. maydis cluster 19A homologs; [26]). Asterisks indicate the predicted secreted proteins encoding genes and N denotes the unique NotI site. BAC clone BAC3-A2 containing 117 kb of this locus is shown by a solid black bar. Complementing Xba1-fragment in subclone BAC1-6 is indicated underneath. Indicated by vertically striped boxes are RAPD markers 743 and 359 which identified and delimited the original region by revealing two and one recombinants in the population, respectively (∼2 and ∼1 cM distance; [38]) and, at the other genetic boundary, RFLP2 which revealed three different recombinants in the population or approximately 6 cM in distance; the 1.7 kb HindIII probe and RFLP marker 1 revealed no recombinants. B. Lines denote the regions in kb deleted in the respective Uh364 mutants; in red are the deletion mutants resulting in a virulent phenotype, whereas the others remained avirulent on Hannchen. C. Enlarged region containing gene 17 (UHOR_10022 as UhAvr1) and ten other ORFs. The red line below indicates the C-terminal deletion in gene 17 in mutant Uh1289 resulting in a virulent phenotype. The blue arrows and numbers refer to specific primers. D. Comparison to the syntenous region in the virulent parent Uh362 revealed the replacement of 634 bp by an insertion of a 5.5-kb, shown by the dotted line, part of which matches TE-related sequences. E. An overlapping BAC clone, BAC1E-2, containing the syntenous region and extending 1.2 kb past the end of gene 16 in the virulent parent Uh362, was used for sequencing. A change from avirulence to virulence would likely be caused by a mutation in the candidate gene such as a point mutation, leading to an amino acid change or a protein truncation, or a gene deletion or a change in transcription. To identify the UhAvr1 gene, we first checked the presence of the CSEP genes in the virulent parent Uh362. A PCR-amplification product for all ten genes was obtained from genomic DNA indicating their presence in the genome of the virulent parent. DNA sequence analysis of the seven candidate UhAvr1 genes did reveal point mutations in three of the alleles in Uh362 (Table 1). Since CSEP 35 displayed an amino acid difference that could have changed its charge between the virulent and avirulent form, gene 35 was deleted in parental avirulent strain Uh364. However, when crossed with virulent parent Uh362, it did not result in virulence on cultivar Hannchen harboring Ruh1. We therefore expanded the sequence comparisons to include a collection of field isolates from different parts of the world, four avirulent and six virulent on Hannchen (Table S2). Three of the six remaining likely candidate UhAvr1 genes were identical when comparing allelic sequences from virulent or avirulent isolates but in the other three, a few mutations were found (Table 1, Figure S3). Unfortunately, none of the revealed mutations could be correlated with the Avr1 or avr1 phenotypes. This indicated that there were other changes outside of the sequences we investigated, that were responsible for the change in phenotype, or that the avirulence function did not reside in the selected effector candidates.
Tab. 1. Mutations found in 7 CSEPs among isolates. 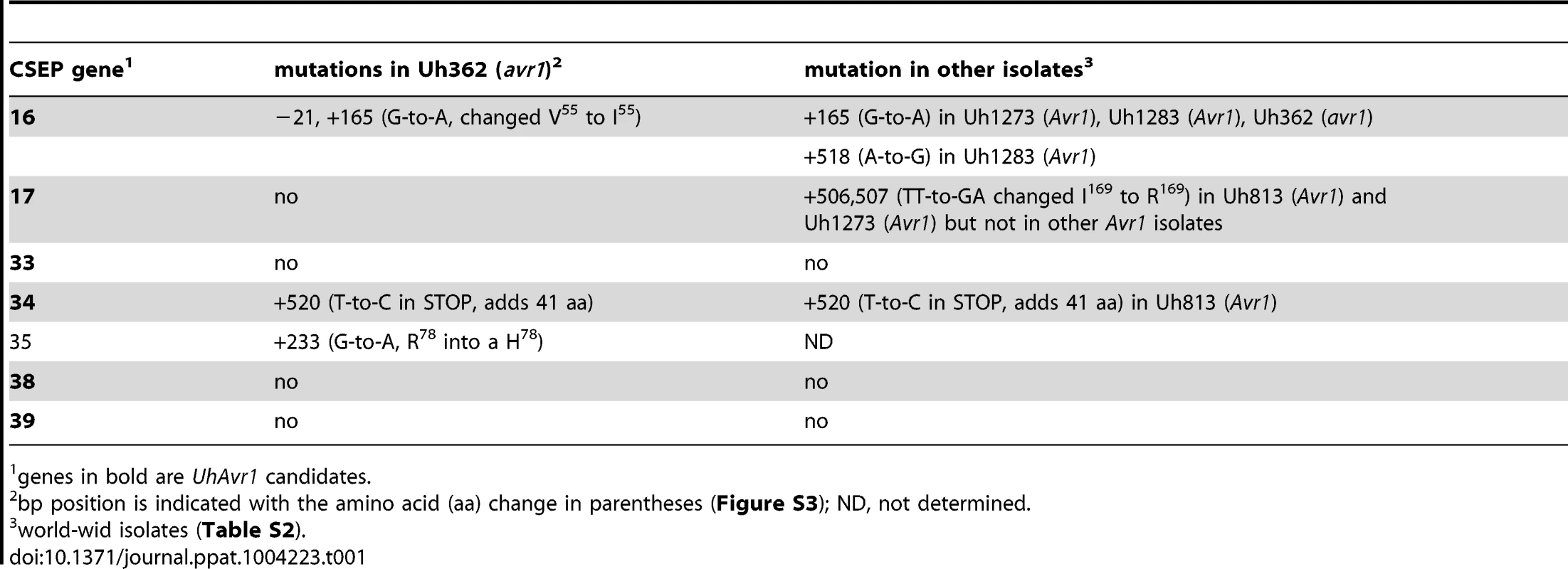
genes in bold are UhAvr1 candidates. Identification of UhAvr1 by deletion analysis
Since no likely candidate for UhAvr1 was found, a systematic deletion analysis of the 80-kb region delimited by the markers (Figure 1) was conducted using a marker-exchange method. In a first round, the region was divided into three sections, ranging from 15 to 38 kb in size, taking into account the location of the various predicted CSEP genes in the region (Figure 1B, Figure S4). No phenotypic differences or abnormal growth were observed for any of the haploid basidiospore deletion mutants and proper mating with compatible haploid basidiospores, such as virulent parental strain Uh362 necessary for pathogenicity tests, occurred. Mated strains were tested for pathogenicity by inoculating them on differential barley cultivars Hannchen (Ruh1) and Odessa (ruh1). Deletion of fragment C18A2 from avirulent parental strain Uh364 yielded strain Uh1041 (Uh364 Δ18A2) (Table S2) and resulted in disease on Hannchen after mating with compatible virulent wild-type strain Uh362 (Figure 2A), clearly indicating that the 38.5 kb fragment C18A2 contained avirulence gene UhAvr1. When Uh1041 was crossed with avirulent strain Uh365, a sibling to parental strain Uh364 but of opposite mating type, the resulting dikaryon caused disease on Odessa but not on Hannchen (Figure 2A), indicating complementation with the avirulence function and showing that no other functions in the recognition of the dominant UhAvr1 allele had been inadvertently compromised in the mutant.
Fig. 2. Pathogenicity test of deletion and complementation mutants. 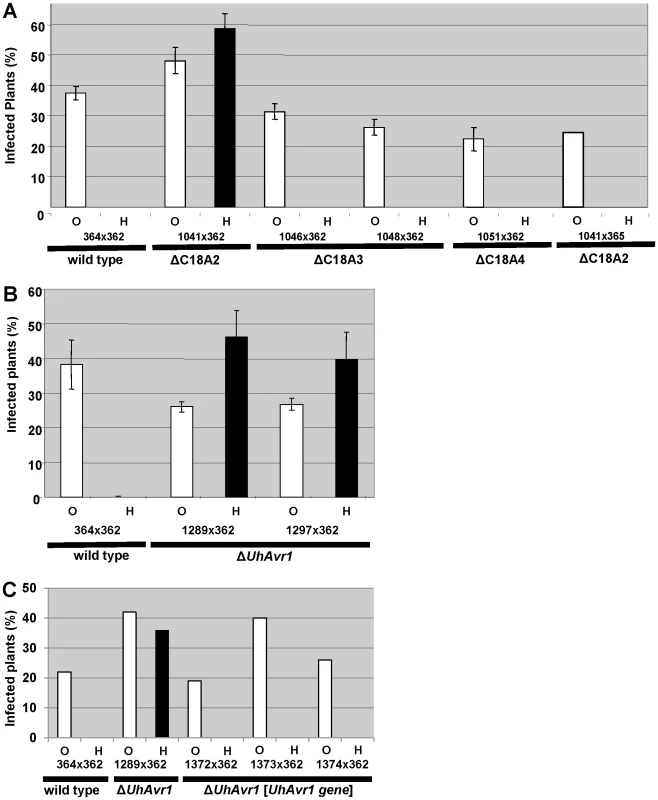
A. Fragment C18A2 contains the functional UhAvr1 as this deletion mutant (strain Uh1041, Figure 1B, Table S2) crossed with parental virulent strain Uh362 was virulent towards Hannchen (black bar). All other deletion mutants and wild type are avirulent towards Hannchen because of the presence of a functional UhAvr1, whereas all are virulent towards Odessa. Y-axis, percent infected plants out of the total number of inoculated plants. Average of three independent inoculation experiments with standard deviation is shown as error bars. Uh1041×avirulent Uh365, a Uh364 sibling of opposite mating type (Table S2), is a control cross. B. Deletion of the 3′-part of gene 17 (strains Uh1289 and Uh1297, Table S2) proves this gene represents UhAvr1. C. Random integration of gene 17 sequences including 5′- and 3′-flanking regions, complementing single ΔUhAvr1 deletion strain Uh1289, is sufficient to fully prevent disease on Hannchen (transformants Uh1372, Uh1373 and Uh1374, Tables S2 and S3). Fragment 18A2 harboring two CSEPs as the likely UhAvr1 allele, was further divided to make five additional deletion mutants (sub-sections C18A2-a to C18A2-e; Figure 1B). To generate the deletion constructs, primers were designed in such a way that the two CSEPs would be deleted in two different deletion constructs. Sixty-four PCR-positive deletion mutants were obtained for the five deletion constructs, which were further verified by DNA blot analysis (Figure S5). Nine deletion mutants were selected, two from each, except for C18A2-b for which only one expected deletion mutant was obtained. Among these, the two mutants for C18A2-c and the two for C18A2-d were virulent towards both barley cultivars Odessa and Hannchen in pathogenicity tests after mating with Uh362 (Figure S5G). The overlapping fragments C18A2-c and C18A2-d shared only gene 17 encoding a CSEP (Figure 1B) that was a strong candidate for UhAVR1p. To confirm this, another deletion mutant was produced in which the 3′ 319 bp of the ORF of this gene in parental strain Uh364 was deleted (Figure S6A and B). Two independent deletion mutants, Uh1289 and Uh1297, had this small deletion which resulted in virulence towards both Hannchen and Odessa when crossed with Uh362, producing disease in 40–50% of the plants (Figure 2B), and confirmed that gene 17 (UHOR_10022, GenBank CCF49778.1) is necessary for UhAvr1 avirulence function.
Complementation of the virulent Uh364 deletion mutants
A 11.5-kb XbaI fragment cloned in a modified BAC vector (pUSBAC5, converted for use in Ustilago species by introducing a specific hygromycin B resistance cassette [38]) yielded construct BAC1-6 which contained two predicted CSEPs: gene 16 and 17 (Figure 1B). This clone partially overlaps with fragment C18A2. C18A2 deletion mutant Uh1041, virulent towards Hannchen, was complemented with BAC1-6 and two stable transformants (Uh1205 and Uh1207; Table S2) were inoculated on barley cultivars Odessa and Hannchen after mating with compatible virulent strain Uh362. No abnormal growth or defect in mating behavior had been observed in these haploid complemented strains. After mating with Uh362, the complemented strains caused the same level of disease on Odessa as the wild-type cross and the deletion mutant, infecting from 30–40% of the plants. On Hannchen however, the level of disease was severely reduced compared to the deletion mutant and only ∼2.5% of the plants showed infected seed heads (Figure S7). Incomplete restoration of avirulence could have resulted from the integration of an incomplete fragment at random locations in the genome, affecting transcription. Similar results were obtained for Fusarium oxysporum f. sp lycopersici mutant strains complemented with the Six1 avirulence gene that did not restore complete avirulence towards tomato lines that contained the resistance gene I-3 [40].This suggested that BAC1-6 contained the functional UhAvr1 gene. To exclude any possible effects from other genes contained on the BAC clone or on the deleted C18A2 fragment, single UhAvr1 deletion mutant strain Uh1289 was complemented with complete wild-type gene 17 sequences, including 659 bp and 630 bp from the upstream and downstream regions, respectively. Three independent transformants, strains Uh1372, Uh1373 and Uh1374 (Table S2), completely prevented disease on Hannchen (Table S3), confirming that gene 17 is sufficient to restore avirulence and hence codes for UhAvr1 (Figure 2C).
UhAvr1 codes for a predicted full-length protein of 170 amino acids with a calculated Mw of 21 kDa. SignalP 4.1 predicts a 19 amino acids SP resulting in a processed mature protein of 18.9 kDa. Mature UhAVR1p has predicted coil, helix and extended beta structures but could not be modeled on any currently existing crystal structures (Figure S8) and no clear similarities could be found to known proteins or other domains.
Loss of avirulence is due to TE-activity upstream of the UhAvr1 ORF
From a BAC library constructed from genomic DNA from virulent strain Uh362, a BAC clone, BAC1-E2, was identified using gene 1 sequences as a probe (Figure 1E). We had found that all predicted CSEP ORFs could be amplified by PCR from genomic DNA of strain Uh362, but amplification of UhAvr1, gene 21 and subsequent genes further to the right could not be achieved from BAC1-E2; a probe representing gene 18 did not hybridize to a DNA blot from this BAC clone suggesting its insert did not cover this region. Sequencing and assembly of this BAC clone proved challenging due to the presence of many repeats and TEs, as it had been for BAC3-A2. Comparative analysis revealed indeed that one end of the BAC clone insert extended only 1187 bp past the stop codon of gene 16. Synteny between the virulent and avirulent parents however, was apparent only up to 115 bp upstream of the start codon of UhAvr1 (Figure 1C–E, Figure S3B). WUBLAST analysis found a 400-bp sequence after this break point, matching to two retrotransposon proteins (UHOR_14086 and UHOR_14170) in the U. hordei genome. To reveal the sequence upstream of the UhAvr1 ORF in the genome of Uh362, an inverse PCR was conducted with UhAvr1 ORF-specific primers on HindIII-digested and self-ligated genomic DNA. Sequence analysis confirmed the presence of intact UhAvr1 sequences including 115 bases upstream of its start codon. Further 5′, the sequence diverged revealing no other Uh364-derived sequences and after 166 bases matched sequences with high similarity to the common long-terminal repeat sequence LTR5 from U. hordei Tuh3, a copia-type retrotransposon also found in the mating-type region (Figure 3A, [32], [41]). A PCR product of 5.8 kb however, was amplified from gDNA from the virulent parent when using primer 1685 at the 3′-end of gene 16 and primer 1815 located at the 5′-end of UhAvr1 (Figure 1D, Figure 3B, Table S4). Several other primer combinations confirmed that an insertion of approximately 5.5 kb had occurred and that the genes to the right were preserved with respect to the organization in Uh364 (Figure 3B and not shown). Consistently, hybridization of three probes representing gene 16 (left of the breakpoint), UhAvr1 and gene 23 (approximately 12 kb right of the breakpoint) to separated chromosomes of parental strains Uh364 and Uh362, clearly revealed that all genes were located to the same Chr18 (Figure S2). Combined with sequence information from this insertion, the data suggested that in strain Uh362, TE activity had inserted a seemingly intact TE consisting of gag-pol sequences flanked by LTRs in the intergenic region between gene 16 and UhAvr1. We speculated that this event separated the UhAvr1 ORF from promoter elements thereby likely changing its expression and hence recognition in Hannchen, making this isolate virulent on this cultivar.
Fig. 3. Analysis of DNA sequences surrounding UhAvr1 in virulent and avirulent isolates. 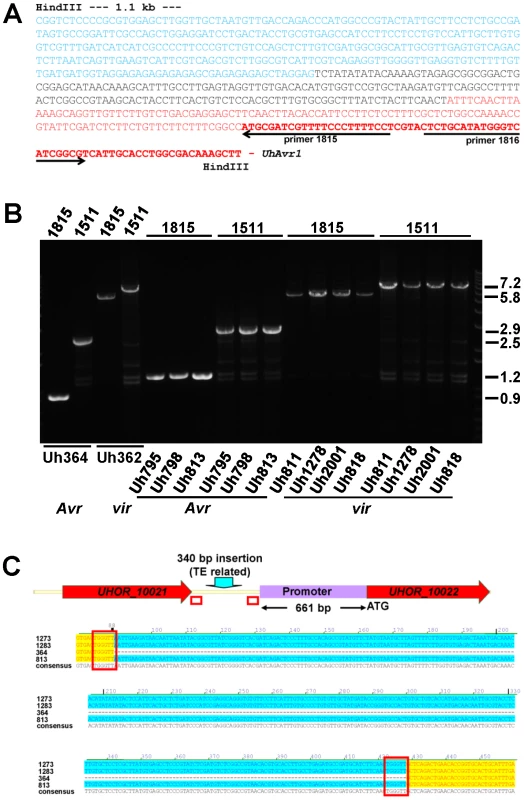
A. Inverse PCR of HindIII-digested, diluted and religated Uh362 gDNA, using outward-facing primers 1815 and 1816, generated a product of 1.8 kb. DNA sequencing from primer 1815 revealed the intact UhAvr1 ORF (red bold face) plus 115 bp of the 5′ upstream region (red) before the sequence started to diverge, indicating the insertion site. This unknown sequence consisted of 166 bases revealing no match to any U. hordei genome sequence or any sequences in public databases (black). Adjacent, the sequence (in blue) matched known TE sequences in the U. hordei genome sequence, in particular related to retrotransposon protein (E-value = 2.9e-36). Highly similar sequences were found on the same chromosome 18 in Uh364 (Avr1) at positions 18.6 and 27.2 kb from the UhAvr1 gene. B. EtBr-stained agarose gel showing length polymorphisms of PCR products among various U. hordei isolates avirulent (Avr) or virulent (vir) towards Hannchen, as indicated at the bottom (Table S2). On top, numbers refer to the primers used in combination with the “anchor” primer 1685 (Figure 1C and D). Sizes on the right are in Kb. C. Sequence comparison of the intergenic region between gene 16 and UhAvr1 suggests TE activity. The numbers indicate the base pair position within the 749 bp-intergenic region in the avirulent parent Uh364 and were compared to three different field isolates also avirulent on Hannchen (see Table S2). Note the insertion of 340 bp, matching TE sequences, upstream of the UhAvr1 promoter in the three other isolates (highlighted in blue) leaving a 661-bp promoter sequence apparently sufficient for avirulence function. This insert is flanked by two 6-bp direct repeats (TGGGTT, boxed), one of which is found in Uh364, possibly representing a “footprint”, i.e., a target site duplication, suggestive of (past) TE activity. See Figure S3C for details, also revealing other sequence variation in the remainder of the intergenic region among the field isolates. UhAvr1 is located in a region of the genome that is, with the mating-type region, among the richest in repeats and TEs, approaching 50% compared to an overall genome content of 8–10% [32]. This elevated presence/retention of TEs and repeats at this effector locus suggests that this region represents a more dynamic part of the genome, enabling evolutionary changes as proposed for other pathosystems [17], [19]–[22]. If this effector region is under selection pressure, e.g., to modify the expression of UhAvr1 to avoid triggering immune responses, it is conceivable that TE activity and insertions might have played a role. Since TE activity rates cannot easily be investigated, we assessed possible variation at the UhAvr1 locus in a number of field isolates. In Uh364, avirulent on Hannchen, 749 bp separate gene 16 and UhAvr1 from each other in the genome and amplification with primers 1685 and 1815 or 1511 (Figure 1C, Table S4) yields PCR products of 898 and 2462 bp respectively (Figure 3B). In five available avirulent field isolates, larger products were obtained for both primer combinations (results for three isolates are shown in Figure 3B) and upon sequencing revealed identical 340-bp insertions in the intergenic region in isolates Uh813, Uh1273 and Uh1283 (Figure 3C). This insertion also matched TE sequences in the U. hordei genome and was flanked by 6-bp repeats (TGGGTT), possibly a footprint of TE activity. This particular insertion, though apparently not affecting avirulence in these isolates, was not found in the virulent parent Uh362. The region was also analyzed from eight U. hordei field isolates virulent on Hannchen (Uh805, Uh811, Uh815, Uh818, Uh820, Uh822, Uh1278 and Uh2001-246; Table S2). Primer combination 1685 and 1815 or 1511produced PCR products of approximately 5.8 and 7.2 kb respectively, similar to the products obtained from Uh362 (Figure 3C). However, upon sequencing, variation was revealed among the TE sequences in the different virulent strains. One predominant mutation found in four virulent strains (Uh362, Uh805, Uh815, and Uh820) was a 10-bp insertion of a repeat (GAGAGAGAGC) that was however absent from three other virulent strains (Uh811, Uh818, and Uh822; Figure S3C). The 340-bp insertion discovered in three of the avirulent field isolates was not found in these eight virulent field isolates. Overall, the variation found in sequences surrounding UhAvr1 in field isolates both avirulent and virulent on Hannchen, and the similarity of those sequences to various U. hordei-specific repeats and TEs, suggest various transposition events have occurred in different isolates resulting in a variety of combinations upon which selection could act.
UhAvr1 causes programmed cell death and is expressed in hyphae during plant infection
Previous electronmicroscopy work revealed necrosis in cells immediately surrounding penetration sites early upon infection during an incompatible interaction on Hannchen [42]. We performed a microscopic analysis of the natural infection process by teliospores, previously produced on universal susceptible cultivar Odessa. Infection of coleoptiles of cultivar Hannchen by teliospores from crossed wild-type progenitor strains Uh364 (MAT-1 UhAvr1)×Uh362 (MAT-2 Uhavr1) caused extensive production of reactive oxygen species as visualized by DAB staining suggesting cell death could have been initiated (Figure 4A), and extensive callose deposition seemingly restricting pathogen development (Figure 4C and E). In stark contrast, teliospores produced from cross Uh1289 (Uh364 MAT-1, ΔUhAvr1)×Uh362 (MAT-2 Uhavr1) caused a natural infection of coleoptile epidermal cells of cultivar Hannchen, showing hyphal development, very little oxidative damage (Figure 4B) and limited, diffuse callose depositions (Figure 4D and E), illustrating a compatible interaction.
Fig. 4. Light microscopic analysis of compatible and incompatible infection types. 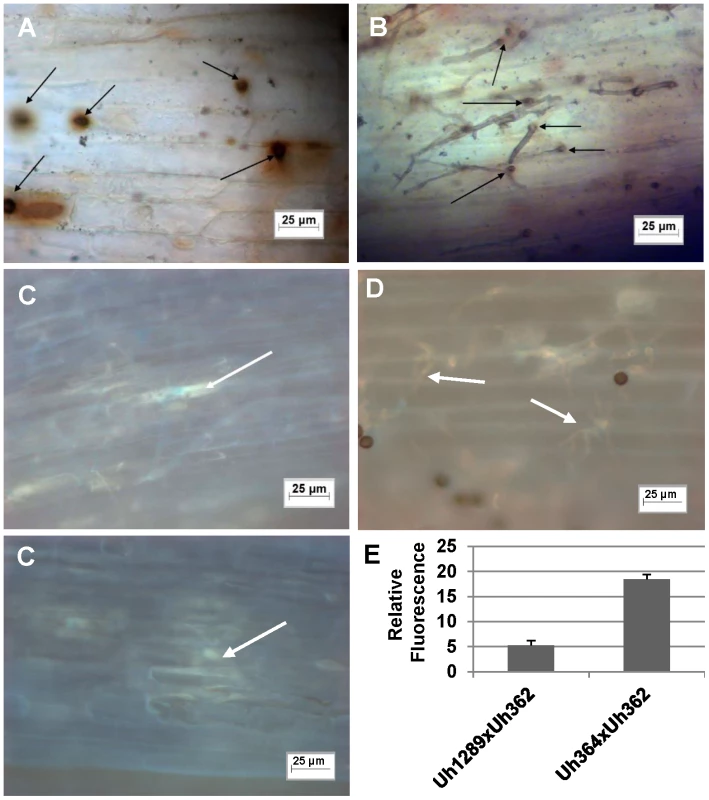
A. and C. Infection by teliospores from wild-type cross Uh364 (MAT-1, UhAvr1)×Uh362 (MAT-2, Uhavr1) on Hannchen (Ruh1) coleoptiles leads to restricted growth of infection hyphae after mating and penetration. Hypersensitive response-associated reactions include an extensive oxidative burst (DAB staining in A associated with infection sites (arrows) at 72 hrs after inoculation) and accumulation of callose and associated fluorescence around the restricted hyphae (arrows in C after 120 hrs; two panels showing different representative sites). B and D. Inoculation of U. hordei teliospores from cross Uh1289 (Uh364, ΔUhAvr1)×Uh362 (MAT-2, Uhavr1) showing compatibility on cv. Hannchen, leading to invasive growth where the oxidative burst is not extensively triggered at infection sites (arrows, DAB staining in B at 72 hrs after inoculation), and less callose and associated fluorescence around the spreading hyphae is observed at 120 hrs (arrows in D). E. Quantitation of fluorescence, representing callose, averaged over several penetration sites. Expression analysis of UhAvr1 by quantitative RT-PCR during infection proved challenging. No expression could be detected in haploid avirulent or virulent cells grown in liquid media, or during mating interactions on plates (data not shown). Weak and variable expression was observed in mated cells and teliospores applied to barley coleoptiles but always only when avirulent strain Uh364 was employed (Figure 5A); linear pre-amplification of cDNA to increase signal strength [43] corroborated these results but introduced variation (Figure 5B). This suggested that the expression of UhAvr1 might be induced only upon direct contact with, or actual infection of coleoptile epidermal cells. The low amount of transcript is likely due to the very few contact and penetration sites present resulting in a very small proportion of responding cells in the biological material (the inoculated coleoptiles) from which the RNA was isolated; this resulted in variable qRT-PCR results. From the combined data it was evident that in the virulent parental strain Uh362 (or its virulent sibling Uh359) the level of UhAvr1 mRNA bordered on the limit of detection, but was at most only 10% of the level seen in Uh364 after pre-amplification (Figure 5B).
Fig. 5. Expression of UhAvr1 early in infection of coleoptiles. 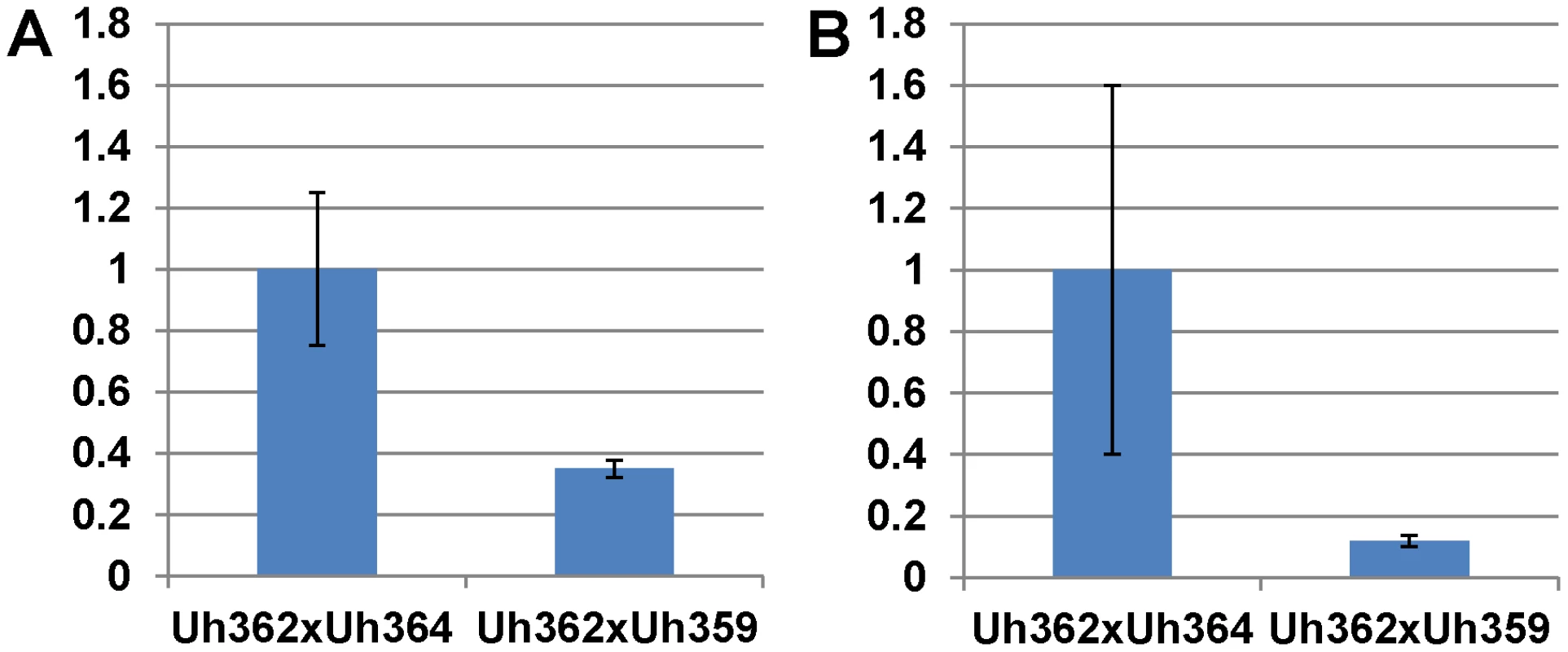
Quantitative Reverse Transcriptase PCR analysis measuring UhAvr1 gene transcript levels in total RNA isolated from barley cv. Hannchen (Ruh1) coleoptiles 48 hrs after inoculation with mated cell cultures of crosses as indicated: Uh362 (MAT-2, Uhavr1)×Uh364 (MAT-1, UhAvr1) versus Uh362×Uh359 (MAT-1, Uhavr1). A. Measurable UhAvr1 product was found when Uh364 was present but amplification from cross Uh362×Uh359 appeared not until cycle 38, at the limit of detection (P = 0.06, student t-test). B. cDNA from the UhAvr1 target and the U. hordei eIF-2B reference genes was pre-amplified for 10 cycles before quantitative RT-PCR with nested primers resulting in significant variation. Therefore, to substantiate the tentative expression results and to possibly localize UhAVR1p, a chimeric gene construct was made of UhAvr1 with its native promoter but linked to a green fluorescent protein (GFP) moiety at its C-terminal end. This was then used to replace by marker-exchange the ΔUhAvr1 deletion in strain Uh1289, thereby putting the chimer in its original expression site (Figure S6C). Confocal microscopy of this constructed strain Uh1353 (Table S2) clearly corroborated the qRT-PCR expression results since no fluorescence was observed in haploid or mated cells at the time of inoculation of barley coleoptiles (Figure 6A), whereas bright fluorescence comparable to GFP expressed from the strong U. maydis otef promoter [44] (Figure 6B) was apparent after 48 hrs in mated dikaryotic hyphae upon infection (Figure 6C) and while extending in the coleoptile later during the infection (Figure 6D). GFP fluorescence was seen in growing hyphal tips, possibly in vesicle-like structures (Figure 6D and E) and in older hyphae associated with the cell wall.
Fig. 6. Expression of UhAVR1:GFP chimers during infection. 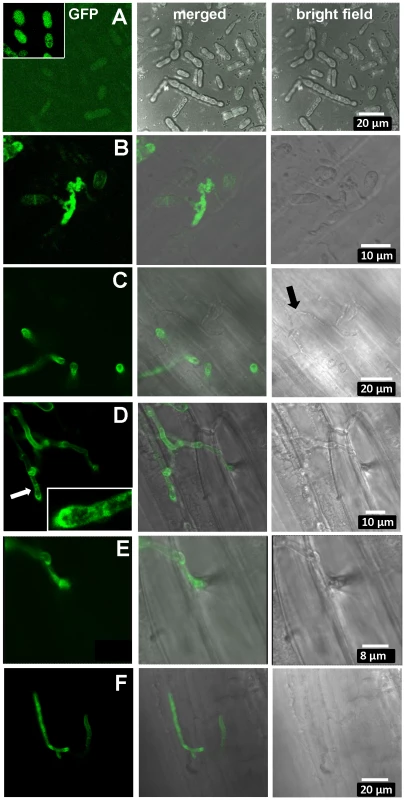
Confocal microscopy of mated U. hordei strains transformed with various GFP constructs. A. Free-floating and mated cells of cross Uh1353×Uh362 (MAT-2, Uhavr1) showing no green fluorescence, whereas GFP expressed from the strong constitutive otef promoter in strain Uh364 (Uh1351) shows bright fluorescence (insert); protein blot analysis verified no expression from Uh1353 and strong expression of just GFP from Uh1351 under these conditions (Figure S9B). B. As a control, Uh1357 (MAT-1 ΔUhAvr1 [otef:UhAvr1:GFP])×Uh362 on compatible Odessa coleoptiles at 48 hai shows strong GFP expression from the otef promoter in the same recipient strain. C. Uh1353×Uh362 on compatible Odessa coleoptiles at 48 hai shows septated hyphae on the surface devoid of cytoplasm and not fluorescing (arrow) whereas in invaded dikaryotic hyphae expression of the UhAVR1:GFP chimer is induced from its native promoter upon host “sensing” and penetration. D. As in C but at 100 hai, showing extended, septated hyphae. Fluorescence is visible in the hyphal cell wall but appears punctuated seemingly in vesicle-like structures in the growing points being concentrated at the tip (insert). E. Enlargement from D of penetration site on the right. F. Same cross as in C induces UhAvr1 expression in incompatible Hannchen at 48 hai, but no HR is seen. On cv. Hannchen, a very similar infection by fluorescent hyphae was observed but no obvious HR reaction, such as increased autofluorescence and/or cell collapse, was seen at 72 hrs (Figure 6F). This suggested that the chimeric protein seemed unable to trigger the R gene-based immunity. Indeed, pathogenicity tests with these complemented strains (Uh1353, Uh1354, Uh1355, Table S2) when mated with Uh362, were causing similar levels of disease on both Odessa and Hannchen (Table S3). In contrast, as reported above, strains complemented with wild-type UhAvr1 gene sequence including promoter and terminator elements did not cause any disease on Hannchen (Uh1372, Uh1373, Uh1374; Figure 2C, Table S3). These experiments suggested that the C-terminal GFP-moiety interfered with the process that led to resistance triggering. We have not been able to verify in these strains whether or not the intact chimer is produced when infecting and if so whether possibly proper translocation and targeting to the proper location is affected by the presence of a C-terminal moiety. Whether C-terminal extensions interfere with protein structure and/or obstruct proper recognition of the host target(s), R gene or R-gene complex, needs further study but has been shown to occur in the flax rust fungus [45].
When attempting to complement virulent deletion strain Uh1289 (ΔUhAvr1) with the UhAvr1 ORF under the control of the strong constitutive Ustilago Hsp70 promoter, the resulting transformants, when mated with compatible parental strain Uh362, did not trigger resistance in cultivar Hannchen and yielded levels of disease similar as on Odessa or from control crosses (Table S3, crosses 19–21, compare cross 6). Similar constructs with the Hsp70 or otef promoters driving the UhAvr1 ORF now linked at its C-terminal end to either the HA epitope tag or a GFP moiety, yielded transformants that similarly gave comparable levels of disease on both Odessa and Hannchen (Table S3, crosses 4, 5, 13–18). Protein blot analysis confirmed the production of the expected chimeric proteins in the transformants (Figure S9) and we assumed from these assays that the wild-type UhAVR1p effector is similarly expressed from the Hsp70 promoter in the transformants mentioned above. In many pathogens studied, cloned avirulence effectors have been shown to assert their avirulence function when reintroduced and expressed from non-native, strong promoters. In U. hordei, the expression of UhAvr1 is finely tuned (Figure 6) and it is possible that this regulation is essential for proper relocation and function, including R-triggered immunity.
UhAVR1p is not crucial for virulence
In several pathosystems, the deletion of avirulence effector genes was shown to affect virulence on host cultivars not harboring the cognate R gene. We tested in the ΔC18A2 deletion mutant whether or not genes 6 to 22, which included UhAvr1 and two other CSEP genes, have any virulence functions in U. hordei. To this end, an equivalent C18A2 deletion was generated in a MAT-2 mating partner by crossing Uh1041 (MAT-1 ΔC18A2) with virulent parent Uh362 (MAT-2 Uhavr1) on barley cultivar Hannchen. Carboxin-resistant basidiospores of mating type MAT-2 were collected by germinating teliospores from infected heads and lack of fragment C18A2 was verified by DNA blot analysis (Figure S10A). Each of three individual C18A2 deletion mutant progeny (Uh1116, Uh1117, Uh1118) was back-crossed with Uh1041, resulting in virulence towards Odessa that was similar to the wild-type cross (Figure S10B). One cross tested on Hannchen seemed also not affected in virulence compared to the single deletion mutant (Figure 2A). We concluded that genes 6 to 22 do not contribute significantly to virulence on barley. ΔUhAvr1 mutants crossed with Uh362 (Uhavr1) are always included in our pathogenicity tests and over many experiments, virulence, expressed as number of plants infected per total number of plants inoculated, has not differed significantly from wild-type crosses. This suggests that effector UhAVR1p is not contributing significantly to virulence. It is difficult to express virulence in a quantitative manner in this pathosystem and a subtle advantage of expressing UhAvr1 may play out at the population level over time.
The UhAvr1 locus resides in an evolving cluster of effectors in both U. hordei and U. maydis
The sequence analysis of clone BAC3-A2 revealed that the UhAvr1 locus is orthologous to a region on U. maydis chromosome Chr 19, spanning a cluster of 24 CSEPs, called cluster 19A, the largest of such clusters in the U. maydis genome [26] (Table S5). A similar cluster is found in S. reilianum, harboring 29 CSEPs [39]. In U. maydis, deletion of this cluster resulted in reduced disease on maize seedlings. SIMAP analysis [46] and two-directional BLASTp searches were used to find orthologs for the U. hordei predicted CSEPs at this region in the U. maydis genome (Table S1). There is synteny with conserved gene order between these U. hordei and U. maydis genomic regions flanking the predicted CSEPs (Figure 7A). However, the region containing the CSEPs is much diverged and rearrangements, including changes of gene orientation and several translocations of genes within the cluster, are apparent. For example, DigA (Uh gene 1 and Um gene 4) is conserved but a homolog of the adjacent oligosaccharyltransferase gene (Um gene 5) is found 52 kb away in an inverted orientation in U. hordei (Uh gene 23). On the other end, conserved homologs of U. maydis genes 35, 36 and 37 are found in a syntenous region in U. hordei (genes 42, 43 and 46, respectively), except that a CSEP gene (Uh gene 44) with homology to two Um CSEPs that are however located on a different Um Chr 10, and repeat sequences have inserted.
Fig. 7. Comparison of the UhAvr1 loci in the parental strains and to the syntenous U. maydis cluster of effectors. 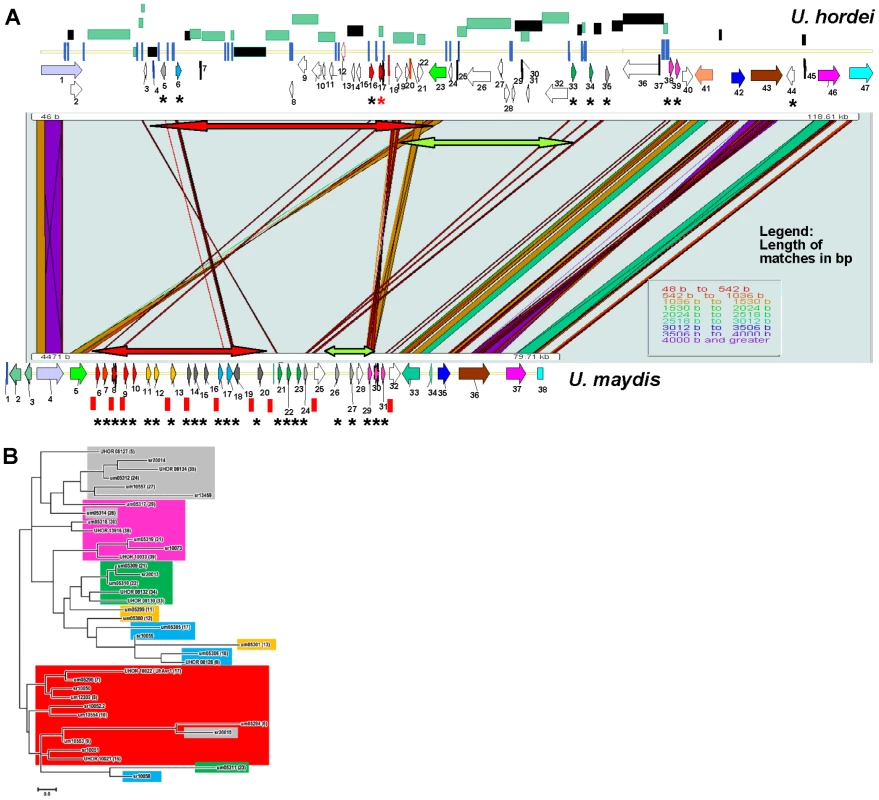
A. Comparison of the UhAvr1 locus (top) to the syntenic region in U. maydis harbouring cluster 19A (bottom) to illustrate the gross rearrangements present in the two genomes. Both loci are drawn to scale and PatternHunter output [97] was used to visualize synteny. Arrows indicate the position and direction of transcription of the genes and asterisks indicate predicted CSEPs. Light and dark green rectangles represent regions with LTRs and repeats, respectively. The red and green-colored two-sided arrows represent two regions that are inverted. Blue vertical lines represent small repeats and red vertical bars represent the 10-bp repeats scattered over the U. maydis genome and suggested to be TE footprints [32]. Gene numbering in U. hordei is as in Table S1; gene 17 is UhAvr1 (red asterisk in the top panel). Gene numbering in U. maydis is given in Table S5. B. Unrooted molecular phylogenetic tree of nine CSEPs found at the UhAvr1 locus of strain Uh364, 24 predicted CSEPs in the syntenous clusters 19A in U. maydis [26], and selected homologs in S. reilianum [39], revealed likely family members. The color code reflects the paralogous and homologous groups in the three species which are only depicted for U. hordei and U. maydis in panel B. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model [98] conducted in MEGA5 [99]. Overall, the U. hordei cluster region in between the syntenic blocks bordered by Uh genes 1 and 40, is 40.6 kb larger than in U. maydis, in part the result of the presence of TE and repetitive DNA sequences. Other important differences are in the complement of the predicted CSEP genes. In U. maydis, four families of CSEP genes that are arranged in tandem in clusters of several paralogs, were described [26] (Figure 7A). One U. maydis family (genes um05299, um05300 and um05301, genes 11, 12 and 13) is not represented in the U. hordei region or its genome [32] and seems species-specific. A molecular phylogenetic tree was generated and to reveal possible derived family members, we included several family members from the S. reilianum 19A CSEP cluster [39] (Figure 7B). UhAvr1 (UHOR_10022, gene 17) and its adjacent paralog UHOR_10021 (gene 16) are homologous to U. maydis CSEPs um05294 and um05295 (genes 6 and 7) residing in the tin1-1 to tin1-5 cluster, an expanded family of five adjacent, weakly related paralogous effectors [47]; S. reilianum has 3 homologs, sr10050, sr10051 and sr10052.2 (Figure 7A and B). Such an expansion in U. maydis is also seen for UHOR_10033 (gene 39) and UHOR_13916 (38) with three related paralogs in U. maydis (um05317, um05318 and um05319, genes 29, 30 and 31) and four in S. reilianum (sr10073, sr10075, sr10077 and sr10079), and for UHOR_08134 (gene 35) with various homologs in both U. maydis and S. reilianum. Overall, in U. hordei, the related families are more dispersed and separated from adjacent genes, sometimes in inverted orientation, by TE and repeat sequences. Virtually no such repeat sequences are present in the U. maydis cluster although U. maydis gene um05316 (gene 28) codes for a transposase indicating possible (past) TE activity (Figure 7A).
Discussion
Previously we showed that the UhAvr1 locus was located to an approximately 80-kb region contained on BAC clone 3-A2 [38]. In this study, we sequenced its insert to discover, among others, ten ORFs encoding small predicted secreted proteins at this genetic locus. RAPD and AFLP markers limited these to seven most-likely candidate avirulence-triggering effector genes. Sequence comparison of these ORFs from the virulent and avirulent parents used to generate the mapping population, as well as from ten additional virulent and avirulent field isolates from a world-wide collection, revealed that the change from avirulence (UhAvr1) to virulence (Uhavr1) is not due to mutations in the ORFs or the presence or absence of ORFs in the two parental strains. We subsequently identified UHOR_10022 through targeted deletions and complementation-based approaches as being U. hordei avirulence gene UhAvr1.
Quantitative RT-PCR analysis to verify expression of UhAvr1 during infection proved challenging because of the very low levels of fungal biomass at this stage relative to the tissue mass in the coleoptile. However, when microscopically investigating single cell events, the substantial fluorescence emanating from the UhAvr1:GFP fusion transcribed from its native promoter in its original genome location, only shortly after contact with barley coleoptiles (Figure 6), showed that this gene is induced during infection. In RNA samples isolated from immature and mature infected seed heads, no UhAvr1 mRNA could be detected by quantitative RT-PCR whereas high levels of expression were detected for U. hordei actin (UHOR_08813) and eIF-2B (UHOR_07772) genes that were used as references in the analysis (data not shown). This shows that UhAvr1 is expressed only during early infection, being highly regulated and suggests that UhAVR1p is needed only early during infection. In many plant pathogenic fungi and oomycetes, a subset of predicted effectors, some of which trigger avirulence, are expressed only upon infection and sometimes only in specific infection structures such as appressoria or haustoria [26], [45], [48]–[51].
Expression of UhAvr1 provides complete immunity in barley cultivars harboring Ruh1 and we show that UhAVR1p harbors an avirulence function which is somehow recognized by RUH1. Previously we had shown by electron microscopy that this interaction caused hyphal restriction, likely due to the deposition of electron-dense material, and necrosis in cells immediately surrounding penetration sites early upon infection [42]. This correlates well with the timing of expression of UhAvr1 and the accumulation of callose and associated fluorescence just around penetration sites and around restricted hyphae within 72 hours of infection (Figure 4C).
Sequence analysis among the limited collection of field isolates virulent and avirulent on Hannchen, suggest that UhAvr1 may encode a rather monomorphic protein; only two point mutations were identified in UhAvr1. In only one avirulent strain Uh813 that translated into a single amino acid substitution. Whether this points to indirect recognition of this avirulence effector by RUH1, more in line with the ‘guard model’ stipulating purifying selection as the guard recognizes modifications of the AVR protein on the guardee and imposes selection pressure against its function [52], [53] and which favours gene inactivation or deletion [54], whereas direct interaction according to the ‘receptor-ligand model’ tends to result in diversifying selection that generates highly divergent avirulence effector alleles in pathogen populations to escape this recognition by the R gene products [55]–[58], remains to be investigated. Future experiments are geared towards finding the target and mode of interaction of UhAVR1p.
In UhAVR1p, no clear similarities could be found to known proteins or domains. Interestingly, a RxLR tetrad is found in the paralogous U. maydis effectors um05295 (amino acid positions 99–102) and um10554 (125–128), and sr10052 (89–92) from Sporisorium reilianum. When compared, the RxLR motifs line up with a PDFR tetrad in UhAVR1 (Figure S8). The RxLR motif has been proposed to be involved in binding of specific plant and mammalian cell wall phospholipids (phosphatidylinositol 3-phosphate or PI3P), mediating effector uptake. However, among various fungal and oomycete effectors, this motif has been shown to allow for some variation [59] and its function in uptake has been controversial [60], [61]. Alternatively, PI3Ps are enriched in intracellular organelle membranes, specifically from early endosomes [62], [63] and we are investigating possible targeting of UhAVR1 to such locations. Intriguingly, if 20 amino acids were cleaved off, UhAVR1 is predicted to be myristoylated, suggesting a membrane association is involved. Several effectors have been shown to be myristoylated and that this was required for function [64], [65]. Moreover, amino acid K39 has a high probability of being a sumoylation site (Figure S8). Sumoylation is a reversible post-translational modification that affects an increasing number of biological processes by altering intracellular localization and protein-protein interactions.
We were not able to ascertain the virulence function(s) of UhAvr1 in this study. Examples exist of avirulence effectors with a clear role in virulence, such as AVR-a10 and AVR-k1 from Blumeria graminis that enhance fungal penetration in barley epidermal cells [66]. Similarly, AVR3a from P. infestans can suppress necrotic responses in Nicotiana benthamiana induced by INF1 elicitor [67]. Experiments expressing UhAvr1 in Nicotiana leaves did not support a role in the suppression of cell death initiated by several elicitors. This could be due to unavailable or too diverged targets of UhAVR1p in this heterologous system. A homologous system in barley would be needed, possibly in young coleoptiles if timing of expression is essential. In this context it is important to note that infection of barley by U. hordei only occurs early at seed germination, since the fungus needs to reach meristematic tissue; older plants or leaves cannot be successfully inoculated. In the related study by Brefort et al. [47], a U. maydis strain lacking the Tin1-1 to Tin1-5 effectors (genes 6–10 in Figure 7, with UhAVR1p closest related to Tin1-2) caused strong induction of endochitinases, SA-binding proteins and the apoplastic peroxidase POX12 in maize, indicative of enhanced defense responses and a possible role for these effectors in suppressing basal host immunity.
Whatever function UhAVR1p has, it does not seem to contribute significantly to virulence as shown in Figure 2B (and Figure S10B, where the paralogous gene 16 is also deleted). In U. maydis, deletion of the paralogous tin1-1 to tin1-5 effector family did not cause a statistically significant reduction in virulence [47]. However, it is very difficult to assess relative infection rates in this pathosystem for which no good quantitative measures exist and which relies on the number of infected plants out of a significant number of inoculated plants showing often considerable variation. Functional redundancy may exist in effectors located at other sites in the genome such as effector gene sr13459, a potential homolog of UHOR_08134 (gene 35), which is located on a different Chr 20 in S. reilianum. Although not easily measurable as reduced virulence in a few plant experiments, on a population level the UhAVR1 effector may contribute to overall fitness or virulence. It has also been argued that effectors (alleles) that contribute to virulence or fitness are maintained in a pathogen population [e.g. 68–70]. A TE insertion inactivated UhAvr1 but in the isolates we investigated, the genetic information of the ORF was still present. It is therefore possible that virulent isolates that have retained the (inactive) UhAvr1 ORF sequences may have a selective advantage because subsequent re-activation of the UhAvr1 ORF, i.e., by hooking it up again behind a promoter through transposition or gene conversion, will again bring about this population-level advantage if the selection pressure, i.e., plants with Ruh1, disappeared from the environment the fungal population occupies.
Our analysis of the U. hordei genome revealed many TEs and repeats with Repeat-Induced Point mutations, likely inactivating them [32]. However, complete TE (LTR-like) sequences with intact conserved predicted (gag/pol) proteins were also found indicating that these elements could be active transposons. In addition, comparison of the genomes of the three smuts, U. hordei, U. maydis and Sporisorium reilianum, suggested that a recent expansion had occurred of a few related TEs newly introduced in the U. hordei lineage after separation from a common ancestor, also indicating active elements (at least in its recent evolution). In our study, sequence comparisons between UhAvr1 loci from isolates avirulent and virulent on Hannchen revealed TE sequence variants upstream of UhAvr1 and that virulence towards Ruh1 was the result of TE activity and insertion of TE-derived sequences in the promoter region of UhAvr1 changing expression and likely recognition. TE activity and insertion at avirulence effector loci causing in situ mutations or changes in transcription leading to virulence phenotypes, have been described before in ascomycete pathogens [e.g. 19,21,71–73] but not in basidiomycetes.
At the locus, sequence variation involving TE sequences among various field isolates indicated transposition events, possibly of independent nature suggesting TE activity is an important mechanism to overcome resistance to Ruh1. In some field isolates, sequence variation was identical such as in virulent strains Uh362, Uh805, Uh815 and Uh820 (Figure S3C). Considering the geographic area the latter three were collected from (Kenya, Canary Island and Tunisia, respectively), this likely reflects a common ancestral event and regional spread; Uh362 was derived from a Canadian isolate backcrossed with an African isolate long ago to obtain homozygous material and likely acquired the virulent allele from this region. Similarly, the identical 340-bp insertion found in isolates Uh813, Uh1273 and Uh1283 (Figure 3C), respectively from Iran, Azerbaijan and Turkey, could have a regional ancestral origin. This illustrates the difficulty of sampling pathogens from a crop plant that is widely traded and grown in certain areas. In order to assess more-comprehensive variation, one would need to sample isolates from truly wild barley populations in remote locations.
TEs play important roles in shaping genomes, causing rearrangements such as deletions, inversions, duplications, translocations, but also neo-functionalizations. Recently, genome analyses of several fungal and oomycete pathogens revealed that many effector genes reside in TE and repeat-rich regions (including at telomeres), a feature that may have evolved to allow for variations necessary for parasites under high host selection pressures to quickly adapt when their virulence effectors are triggering defenses [e.g.], [18], [22], [68], [74]–[77]. The UhAvr1 gene is located in a region of the genome that sports ten CSEP genes and is, with the mating-type region, among the richest in repeats and TEs, approaching 50% (Figure 7A [32], [41]). Incidentally, the UhAvr1 locus revealed conserved synteny in regions flanking cluster 19A, the largest cluster of CSEP genes in U. maydis, and to some extend among its coded effectors (Figure 7). Transcription of the U. maydis CSEP genes is induced after infection of maize and deletion of this whole cluster severely reduces disease [26]. It appears that these species, including related S. reilianum, share some of these likely ancestral genes but that possibly because of their obligate biotrophic interaction with diverse hosts, these effectors have evolved differently. Phylogeny revealed expanded CSEP gene families in U. maydis and S. reilianum. Interestingly, in the U. maydis-maize pathosystem, no effector-R gene interactions involving avirulence and resistance genes have been genetically identified to date [78]–[80]. It is possible that the higher number of paralogs in the U. maydis (and S. reilianum) effector gene families represent past diversifying selection acting on these effectors to avoid host recognition and making U. maydis better adapted to host populations. This could have resulted from adaptation to changed effector target molecules or the defeat of major resistance genes over time.
While in U. hordei the mechanism to avoid host recognition involves the activity of TEs, U. maydis and S. reilianum have more streamlined genomes with few deleterious repeats and TEs [26], [32], [39]. The question arises how the latter organisms have created the needed variation. One scenario could be past TE activity, followed by purging of TEs and repeats brought about by a highly active homologous recombination system known to exist in U. maydis. The numerous small (10 bp) repeats in the U. maydis genome have been suggested to be footprints of past TE activities [32], [39] and 26 are found exactly in between the effector genes in the U. maydis cluster 19A (Figure 7A). Alternatively, if TE activity did not play role in these organisms, highly active recombination followed by genetic drift may have caused sufficient variability. However, the evolution of these pathogens is more complex and involves sex [81]; U. hordei with its bipolar mating system which promotes inbreeding, may select for the use of TEs as genome modifiers whereas U. maydis and S. reilianum with their tetrapolar mating systems which cause reduced inbreeding potential, can create variation through recombining with outside partners [77]. Undoubtedly, the selection pressure imposed by the host has had a major impact on maintaining the variability among populations, as has been shown for the U. maydis-maize interaction [82].
Materials and Methods
Plant and fungal strains
Two barley cultivars, ‘Odessa’ (ruh1, universal susceptible) and a differential, ‘Hannchen’ (Ruh1) were used for pathogenicity assays. Fungal strains and mutants generated are listed in Table S2. U. hordei haploid parental strains Uh364 (alias Uh4857-4, MAT-1 UhAvr1) and Uh362 (alias Uh4854-10, MAT-2 Uhavr1) were described previously [38].
Fungal growth conditions and U. hordei transformation
Haploid U. hordei strains were grown in liquid Potato Dextrose Broth (PDB), complete medium (CM [83]) or YEPS (1% yeast extract, 2% peptone, 2% sucrose), while 2.5 µg/ml carboxin (Sigma-Aldrich), 100 µg/ml Hygromycin B (Calbiochem, La Jolla, CA, USA) or 40 µg/ml Zeocin (Invitrogen, Valencia, CA, USA) were added when appropriate. Strains were grown at 22°C. For genetic transformation of U. hordei, protoplasts were prepared according to a modified protocol [84], instead using 384 mg/ml Vinoflow FCE (Gusmer Enterprises) as enzyme mix for digesting the fungal cell wall [85]. Protoplasts were transformed with 5 µg DNA mixed with 1 µl of a 15 mg/ml heparin (Sigma) in STC (10 mM Tris-HCl pH 7.5, 100 mM CaCl2, 1M sorbitol) solution and selected on double-complete medium plate (DCM) supplemented with 1 M sorbitol and appropriate antibiotic. After 5–7 days incubation at 22°C, colonies from DCM-S were transferred to CM medium and incubated for two days at 22°C before transferring to liquid CM medium for further analysis.
Pathogenicity assays
Two haploid cultures of opposite mating type (OD600 of ∼1, tested in mating assays as described in [86]) were mixed 1∶1 v/v before inoculation of barley seeds. Seeds were dehulled, surface sterilized for 3 min with 70% EtOH, followed by 10 min with 1% bleach, and rinsed several times with sterile ddH2O. Surface-sterilized seeds were dipped in mated cultures and a vacuum of 20 psi was applied for 20 min. Subsequently, excess inoculum was drained and seeds were kept for 6 hrs at room temperature before sowing in potting mix (Pro-Mix BX) at a density of 3 seeds per 3×3″ pot of which 18 were placed in a tray. Plants were grown in controlled-environment chambers with an 18 hour light-6 hour dark cycle at 22°C. Disease ratings were scored at heading, approximately 2 months after planting, by counting infected plants among all inoculated plants. The same inoculum was always applied to both barley cultivars Hannchen (Ruh1) and Odessa (ruh1) simultaneously to verify effectiveness.
Sequencing and analysis of BAC clones and ORFs
BAC3-A2 containing the UhAvr1 locus from the avirulent parent Uh364 was sequenced using the GPS-Mutagenesis System (New England Biolabs) with a few modifications. In the donor vector, the kanamycin resistance cassette within the transprimer was replaced with a phleomycin resistance cassette driven by both the Em7 bacterial promoter and the U. maydis glyceraldehydes-3-phosphate dehydrogenase (GAPDH) promoter and terminator [87]. This generated an insertion that could be used directly as a marker-exchange construct to generate deletions within U. hordei through homologous recombination. After in vitro recombination and transformation in E. coli, BAC clones from 6×96 random bacterial colonies were sequenced using primers N and S, yielding paired sequence reads from the ends outwards of the randomly inserted transprimers. These DNA sequences and several BAC end-sequences covering this region from clones of the source BAC genomic library [41] were entered in the PCAP.REP genome assembly program [88]. To place certain sequences and to verify their location, physical mapping was performed by using the unique Not1 restriction enzyme sites in the transprimer and BAC insert and measuring generated fragment sizes on CHEF gels (data not shown). BAC clone 1-E2 covering the Uhavr1 locus in the virulent parental strain Uh362, was recovered from a BAC library via hybridization. This BAC clone, as well as BAC3-A2 for confirmation, were sequenced using the 454 technology at the Plant Biotechnology Institute (Saskatoon, SK). The resulting reads were assembled using the Newbler program (Roche Applied Science). Alignment of the BAC sequences from the virulent parent along the avirulent backbone was facilitated by a custom Perl script. The order of contigs was confirmed by PCR and gaps were corrected through manual sequencing. Genes were predicted using FGENESH [89] and VectorNTI (Invitrogen). Predicted proteins were searched for secretion signals using the SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/), by TargetP v1.1 [90] to identify and remove proteins that were predicted to be mitochondrial, and by ProtComP 9.0 (http://linux1.softberry.com/berry.phtml) which compares them to proteins in the LocDB and PotLocDB databases which hold proteins with known or reliably predicted localization. The sequence of clone BAC3-A2 was contributed to the Uh364 genome sequencing effort ([32] http://www.helmholtz-muenchen.de/en/ibis/institute/groups/fungal-microbial-genomics/resources/muhdb/index.html) and is part of UHOR_scaffold_5.00017, NCBI #CAGI01000148.1 with UHOR_10022, protein ID CCF49778.1, at position 159450–160022; the sequence of the region containing the breakpoint in virulent parent Uh362 on BAC clone1-E2 is accessible under NCBI #KF640593. To sequence ORFs in Uh362 and field isolates, predicted CSEP genes and intergenic regions were amplified by PCR. Primers were designed 100 bp upstream and 100 bp downstream of the ORFs (Table S4) using the Primer3 software (http://sourceforge.net/projects/primer3/files/). Sequencing of the purified products was carried out using the Big Dye terminator v3 chemistry (Applied Biosystems). Large PCR products were generated using LongAmp DNA Polymerase (New England Biolabs, M0323S).
Deletion analysis of the UhAvr1-containing region
One gene, UHOR_08134, was deleted using a double-jointed PCR method [91] and the hygromycin resistance cassette to generate a marker-exchange construct. All other deletion mutants involving individual target genes or clusters of genes were constructed using marker-exchange plasmids generated by the DelsGate method [92]. Briefly, primers were designed separately for each construct to amplify by PCR 1.5 to 2 kb of 5′ - and 3′ - sequences flanking the target region (Table S4), using Uh364 genomic DNA as template. Primers 5L and 5R were then used for the amplification of a 5′-flanking fragment adding an I-SceI recognition sequence tail upstream and an attB1 sequence tail downstream of the flank sequence. Primers 3L and 3R were used to amplify the 3′-flanking fragment, adding the attB2 sequence tail upstream and the I-SceI sequence tail downstream. The two PCR-amplified fragments were then gel-purified using the QIAquick Gel extraction kit (Qiagen) and subsequently recombined into the pDnorCbx vector (NCBI accession # EU360889 [92]) using the Gateway BP Clonase II enzyme Mix (Invitrogen). To assess the resulting marker-exchange plasmids, two PCR reactions were performed using 5′ - gene-specific primer 5R in combination with the SceIF primer, and 3′ - gene-specific primer 3L in combination with primer SceIR primer (Table S4). SceIF and SceIR primers were designed for the I-SceI enzyme recognition site in the forward and reverse orientation, respectively. The deletion constructs were verified by sequencing and were then linearized with I-SceI enzyme (New England, Biolabs) and used directly for U. hordei transformation.
Carboxin-resistant mutants were analyzed for proper gene deletion by PCR reactions on purified gDNA. Sixty to as many as 300 carboxin-resistant colonies sometimes needed to be screened (depending on the region targeted) to get at least four PCR positive transformants for each construct which were then verified by DNA blot analysis. For DNA blot hybridization, 10 µg of gDNA was digested with selected restriction enzymes and run out in 0.8% agarose gels in 1xTAE buffer (40 mM Tris-acetate, 1 mM EDTA). Blotting to nylon membranes (Amersham Biosciences, Buckinghamshire, UK) and hybridization were carried out following standard procedures [93]. DNA probes for either the 5′ - or 3′-flanks were amplified using PCR and labeled with [α-32P] dCTP using the random primer labeling system kit (Amersham Biosciences) according to manufacturer's recommendations. The efficiency of homologous recombination was different for different constructs and seemed dependent on the size of the deletion fragment; the efficiency was higher for small fragments.
Plasmid constructs
Gene expressing constructs were designed to make use of the GateWay technology (Invitrogen). U. hordei ORFs, either with or without the sequence coding for the SP, but without their stop codon, were amplified by PCR with a CACC tetranucleotide sequence at the 5′-end to allow for directional cloning into Gateway entry vector pENTR/D-TOPO (Invitrogen; Table S4). Cloned inserts were sequenced and were subsequently transferred to a designed GateWay destination vector, pUBleX1Int:GateWay:HA (a derivative of Ustilago-specific integrative expression vector pUBleX1Int [94]), using LR recombineering. For the transient assays and microscopy after bombardment, the above-mentioned pENTR clones (UHOR_10022-SP-STOP) were recombined into a modified pMCG161 vector (ChromDB at http://www.chromdb.org; NCBI accession no. AY572837) to create N - or C-terminal GFP-expressing chimers from the maize ubiquitin promoter (Ubi:GFP:UhAvr1-SP and Ubi:UhAvr1-SP:GFP). A control construct expressed just GFP. Details on the constructs and destination vectors can be obtained from the authors.
Quantitative RT-PCR analysis
Barley cv. Hannchen coleoptiles were inoculated with mated cell cultures as described above and infection was allowed to proceed for 48 hrs on sterile filter paper in petri dishes in the dark at 22°C. Coleoptiles from 3 biological replicates were dissected from the seed and roots and total RNA from 100 mg of sample was isolated using Trizol Reagent (Invitrogen, Cat. No. 15596-018). Ten µg of total RNA was then treated with TURBO DNase (Applied Biosystems, Cat. No. AM22380. After quantitation, cDNA synthesis was carried using SuperScript III Reverse Transcriptase (Invitrogen, Cat. No. 18080-093). Quantitative RT-PCR assays were carried out on a CFX96 Real-Time System (Bio-Rad) with the following cycling conditions: (1) 2 min 95°C incubation, (2) cycling at 95°C 10 sec, 55°C 30 sec for 40 cycles, (3) melt curve from 65°C to 95°C at 0.5 degree increments. Analyses and statistics were carried out with the Bio-Rad CFX Manage Software. To overcome the very low expression levels observed, nested real time PCR was carried out as per [43]. An initial 10 cycle pre-amplification with flanking primers 1689+1249 for UhAvr1 and 1804+1805 for reference gene UheIF-2B ( = UHOR_07772; Table S4) was carried out on a Bio-Rad MyCycler (conditions: (1) 95°C 2 min., (2) 10 cycles of 95°C 30 sec, 55°C 30 sec, 72°C 60 sec), followed by the qRT-PCR process above performed with nested internal primers 1798+1799 for UhAvr1 and 1811+1812 for reference gene UheIF-2B (Table S4).
Protein blot analysis
Total protein was isolated from frozen ground cells, as described [95]. Protein samples were boiled for 5 min and spun briefly for 30 sec before being separated by 12.5% SDS-PAGE on a Bio-Rad Mini-Protean III apparatus. Protein was transferred from the gel to Sequi-Blot PVDF Western blotting membrane (Bio-Rad) using a Bio-Rad liquid transfer apparatus following the manufacturer's protocols. Membranes were probed with 200 ng/ml rat anti-HA (hemagglutenin) high affinity monoclonal antibody (Roche Applied Science) or anti-GFP (Clontech Living Colors JL-8 anti-GFP monoclonal). For detection of primary bound antibody, membranes were incubated with peroxidase-conjugated AffiniPure Goat Anti-Rat-Ig (H+L) secondary antibody according to supplier's instruction. For visualization of bound antibody, the Enhanced Chemiluminescence system (ECL) plus Western Blotting Detection Reagents (Amersham Biosciences/GE Healthcare) were used.
Microscopic analyses
To inoculate barley coleoptiles with teliospores, seed hulls were removed by hand to expose the embryo. Seeds were surface sterilized as above and germinated for 48 hrs in the dark at 18°C on sterile filter paper. Emerged coleoptiles were then dusted gently with a paintbrush with teliospores previously released from an infected seed head by gentle grinding. Alternatively, seeds germinated for 24 hrs with emerged coleoptiles, were immersed in cell cultures of OD600 ∼1, mated for 24 hrs after mixing MAT-1 and MAT-2 strains in a 1∶1 ratio, under a vacuum of 20 psi for 20 min, after which the inoculum was drained. After inoculation, seedlings were kept moist and were further incubated in the dark at 18°C. Observation of GFP-expressing fungal infection was done on a Leica SP2-AOBS laser scanning confocal microscope at 488 nm excitation and detection at 499–552 nm.
For light microscopy, seedlings were sampled at 72, 96, 120 and 144 hrs following inoculation. Plants were gently washed and crown tissues consisting of a 1 to 2 cm section of the coleoptile surrounding the crown region were excised, split longitudinally in half and both halves were mounted in lactophenol-cotton (aniline) blue to stain for callose [96]. Sections were viewed with a Zeiss Universal microscope using the 330–385 nm and 460–490 nm excitation and emission filters, respectively, and a HBO103W/2 light source. For detection of the oxidative burst, hydrogen peroxide was detected by vacuum infiltrating dissected coleoptiles for 10 min with 1 mg/ml 3,3′ - diaminobenzidine tetrahydrochloride (DAB, Sigma) in 10 mM Na2HPO4, pH 7 and 0.05% v/v Tween 20, incubation for 6 hrs, and subsequent bleaching in a 3∶1∶1 ethanol ∶ acetic acid ∶ glycerol solution. The numbers of DAB stained sites and their relative size on both halves of 1 cm coleoptile sections were counted from a minimum of 5 seedlings per replication. Three replications were employed and the study was repeated two times. For quantitation of callose, average fluorescence associated with penetration sites was measured on 5 (compatible interaction) to 11 (incompatible interaction) TIF images imported into ImageJ software (National Institutes of Health, Bethesda, Maryland) and the average background fluorescence was subtracted. Data were analyzed using PROC GLM with SAS software (SAS Institute, Cary, NC, USA) and means were separated using Duncan's multiple range test (P≤0.05).
Supporting Information
Zdroje
1. StergiopoulosI, De WitPJ (2009) Fungal effector proteins. Annu Rev Phytopathol 47 : 233–263.
2. KoeckM, HardhamAR, DoddsPN (2011) The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cell Microbiol 13 : 1849–1857.
3. AliS, BakkerenG (2011) Fungal and oomycete effectors – strategies to subdue a host. Can J Plant Pathol 33 : 425–446.
4. GiraldoMC, ValentB (2013) Filamentous plant pathogen effectors in action. Nat Rev Micro 11 : 800–814.
5. BollerT, HeSY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324 : 742–744.
6. HückelhovenR, SchweizerP (2011) Quantitative disease resistance and fungal pathogenicity in Triticeae. J Plant Physiol 168 : 1–2.
7. GassmannW, BhattacharjeeS (2012) Effector-Triggered Immunity signaling: from gene-for-gene pathways to protein-protein interaction networks. Mol Plant Microbe Int 25 : 862–868.
8. BoydLA, RidoutC, O'SullivanDM, LeachJE, LeungH (2013) Plant–pathogen interactions: disease resistance in modern agriculture. Trends Genet 29 : 233–240.
9. DoddsPN, RathjenJP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nature Rev Genet 11 : 539–548.
10. KleemannJ, Rincon-RiveraLJ, TakaharaH, NeumannU, van ThemaatEVL, et al. (2012) Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog 8: e1002643.
11. LiuZ, ZhangZ, FarisJD, OliverRP, SymeR, et al. (2012) The cysteine-rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathogens 8: e1002467.
12. FlorHH (1942) Inheritance of pathogenicity in Melampsora lini. Phytopathology 32 : 653–669.
13. OortAJP (1944) Onderzoekingen over stuifbrand II. Overgevoeligheid van tarwe voor stuifbrand, Ustilago tritici. Hypersensitiviness of wheat to loose smut. Tijdschrift over plantenziekten 50 : 73–106.
14. van der HoornRA, De WitPJ, JoostenMH (2002) Balancing selection favors guarding resistance proteins. Trends Plant Sci 7 : 67–71.
15. RavensdaleM, NemriA, ThrallPH, EllisJG, DoddsPN (2011) Co-evolutionary interactions between host resistance and pathogen effector genes in flax rust disease. Mol Plant Pathol 12 : 93–102.
16. OrbachMJ, FarrallL, SweigardJA, ChumleyFG, ValentB (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12 : 2019–2032.
17. FudalI, RossS, GoutL, BlaiseF, KuhnM-L, et al. (2007) Heterochromatin-like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map-based cloning of AvrLm6. Mol Plant-Microbe Interact 20 : 459–470.
18. ParlangeF, DaverdinG, FudalI, KuhnML, BalesdentMH, et al. (2009) Leptosphaeria maculans avirulence gene AvrLm4-7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4-mediated recognition through a single amino acid change. Mol Microbiol 71 : 851–863.
19. KangS, LebrunMH, FarrallL, ValentB (2001) Gain of virulence caused by insertion of a Pot3 transposon in a Magnaporthe grisea avirulence gene. Mol Plant-Microbe Interact 14 : 671–674.
20. GoutL, FudalI, KuhnM-L, BlaiseF, EckertM, et al. (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol 60 : 67–80.
21. ZhouE, JiaY, SinghP, CorrellJC, LeeFN (2007) Instability of the Magnaporthe oryzae avirulence gene AVR-Pita alters virulence. Fungal Genet Biol 44 : 1024–1034.
22. HaasBJ, KamounS, ZodyMC, JiangRHY, HandsakerRE, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461 : 393–398.
23. Fisher GW, Holton CS (1957) Biology and control of the smut fungi. New York: Ronald Press.
24. HoltonCS, HoffmannJA, DuranR (1968) Variation in the smut fungi. Ann Rev Phytopathol 6 : 213–242.
25. Vanky K (2012) Smut fungi of the world. St. Paul, MN, USA: APS Press. 1480 p.
26. KamperJ, KahmannR, BolkerM, MaL-J, BrefortT, et al. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444 : 97–101.
27. BrefortT, DoehlemannG, Mendoza-MendozaA, ReissmannS, DjameiA, et al. (2009) Ustilago maydis as a pathogen. Ann Rev Phytopathol 47 : 423–445.
28. HuGG, LinningR, BakkerenG (2002) Sporidial mating and infection process of the smut fungus, Ustilago hordei, in susceptible barley. Can J Bot 80 : 1103–1114.
29. GhareebH, BeckerA, IvenT, FeussnerI, SchirawskiJ (2011) Sporisorium reilianum infection changes inflorescence and branching architectures of maize. Plant Physiology 156 : 2037–2052.
30. BakkerenG, KronstadJW (1994) Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc Natl Acad Sci USA 91 : 7085–7089.
31. BakkerenG, KämperJ, SchirawskiJ (2008) Sex in smut fungi: Structure, function and evolution of mating-type complexes. Fungal Genet Biol 45: S15–S21.
32. LaurieJD, AliS, LinningR, MannhauptG, WongP, et al. (2012) Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 24 : 1733–1745.
33. TapkeVF (1945) New physiologic races of Ustilago hordei. Phytopathology 35 : 970–976.
34. SidhuG, PersonC (1972) Genetic control of virulence in Ustilago hordei. II. Identification of genes for host resistance and demonstration of gene-for-gene relations. Can J Genet Cytol 14 : 209–213.
35. EbbaT, PersonC (1975) Genetic control of virulence in Ustilago hordei. IV. Duplicate genes for virulence and genetic and environmental modification of a gene-for-gene relationship. Can J Genet Cytol 17 : 631–636.
36. ThomasPL (1976) Interaction of virulence genes in Ustilago hordei. Can J Genet Cytol 18 : 141–149.
37. GrewalTS, RossnagelBG, BakkerenG, ScolesGJ (2008) Identification of resistance genes to barley covered smut and mapping of the Ruh1 gene using Ustilago hordei strains with defined avirulence genes. Can J Plant Pathol 30 : 277–284.
38. LinningR, LinD, LeeN, AbdennadherM, GaudetD, et al. (2004) Marker-based cloning of the region containing the UhAvr1 avirulence gene from the basidiomycete barley pathogen Ustilago hordei. Genetics 166 : 99–111.
39. SchirawskiJ, MannhauptG, MünchK, BrefortT, SchipperK, et al. (2010) Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330 : 1546–1548.
40. RepM, van der DoesHC, MeijerM, van WijkR, HoutermanPM, et al. (2004) A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol 53 : 1373–1383.
41. BakkerenG, JiangG, WarrenRL, ButterfieldY, ShinH, et al. (2006) Mating factor linkage and genome evolution in basidiomycetous pathogens of cereals. Fungal Genet Biol 43 : 655–666.
42. HuG-G, LinningR, BakkerenG (2003) Ultrastructural comparison of a compatible and incompatible interaction triggered by the presence of an avirulence gene during early infection of the smut fungus, Ustilago hordei, in barley. Physiol Mol Plant Pathol 62 : 155–166.
43. Hernandez-ArteagaS, Lopez-RevillaR (2010) Ultrasensitive quantitation of human papillomavirus type 16 E6 oncogene sequences by nested real time PCR. Infect Agent Cancer 5 : 9.
44. SpelligT, BottinA, KahmannR (1996) Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet 252 : 503–509.
45. CatanzaritiAM, DoddsPN, LawrenceGJ, AyliffeMA, EllisJG (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 : 243–256.
46. RatteiT, ArnoldR, TischlerP, LindnerD, StumpflenV, et al. (2006) SIMAP: the similarity matrix of proteins. Nucleic Acids Res 34: D252–256.
47. BrefortT, TanakaS, NeidigN, DoehlemannG, VinconV, et al. (2014) Characterization of the largest effector gene cluster of Ustilago maydis. PLoS Pathog 10: e1003866.
48. EllisJG, DoddsPN, LawrenceGJ (2007) The role of secreted proteins in diseases of plants caused by rust, powdery mildew and smut fungi. Curr Opin Microbiol 10 : 326–331.
49. de WitPJGM, MehrabiR, BurgHAVd, StergiopoulosI (2009) Fungal effector proteins: past, present and future. Mol Plant Pathol 10 : 735–747.
50. DoehlemannG, van der LindeK, AssmannD, SchwammbachD, HofA, et al. (2009) Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog 5: e1000290.
51. HacquardS, JolyDL, LinY-C, TisserantE, FeauN, et al. (2012) A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Mol Plant-Microbe Interact 25 : 279–293.
52. DanglJL, JonesJD (2001) Plant pathogens and integrated defence responses to infection. Nature 411 : 826–833.
53. Schulze-LefertP, PanstrugaR (2011) A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci 16 : 117–125.
54. BentAF, MackeyD (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45 : 399–436.
55. EllisJG, DoddsPN, LawrenceGJ (2007) Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol 45 : 289–306.
56. RehmanyAP, GordonA, RoseLE, AllenRL, ArmstrongMR, et al. (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two arabidopsis lines. Plant Cell 17 : 1839–1850.
57. HallSA, AllenRL, BaumierRE, BaxterLA, FisherK, et al. (2009) Maintenance of genetic variation in plants and pathogens involves complex networks of gene-for-gene interactions. Mol Plant Pathol 10 : 449–457.
58. ChouS, KrasilevaKV, HoltonJM, SteinbrennerAD, AlberT, et al. (2011) Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci USA 108 : 13323–13328.
59. KaleSD, TylerBM (2011) Entry of oomycete and fungal effectors into plant and animal host cells. Cell Microbiol 13 : 1839–1848.
60. WawraS, DjameiA, AlbertI, NürnbergerT, KahmannR, et al. (2013) In vitro translocation experiments with RxLR-reporter fusion proteins of Avr1b from Phytophthora sojae and AVR3a from Phytophthora infestans fail to demonstrate specific autonomous uptake in plant and animal cells. Mol Plant-Microbe Interact 26 : 528–536.
61. TylerBM, KaleSD, WangQ, TaoK, ClarkHR, et al. (2013) Microbe-independent entry of oomycete RxLR effectors and fungal RxLR-like effectors into plant and animal cells is specific and reproducible. Mol Plant Microbe Int 26 : 611–616.
62. TakenawaT, ItohT (2006) Membrane targeting and remodeling through phosphoinositide-binding domains. IUBMB Life 58 : 296–303.
63. KutateladzeTG (2010) Translation of the phosphoinositide code by PI effectors. Nat Chem Biol 6 : 507–513.
64. NimchukZ, MaroisE, KjemtrupS, LeisterRT, KatagiriF, et al. (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101 : 353–363.
65. LewisJD, AbadaW, MaW, GuttmanDS, DesveauxD (2008) The HopZ family of Pseudomonas syringae Type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana. J Bacteriol 190 : 2880–2891.
66. RidoutCJ, SkamniotiP, PorrittO, SacristanS, JonesJDG, et al. (2006) Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell 18 : 2402–2414.
67. BosJIB, KannegantiT-D, YoungC, CakirC, HuitemaE, et al. (2006) The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J 48 : 165–176.
68. SacristanS, VigourouxM, PedersenC, SkamniotiP, Thordal-ChristensenH, et al. (2009) Coevolution between a family of parasite virulence effectors and a class of LINE-1 retrotransposons. PLoS ONE 4: e7463.
69. BrownJK, TellierA (2011) Plant-parasite coevolution: bridging the gap between genetics and ecology. Annu Rev Phytopathol 49 : 345–367.
70. SteinbrennerAD, GoritschnigS, KrasilevaKV, SchreiberKJ, StaskawiczBJ (2012) Effector recognition and activation of the Arabidopsis thaliana NLR innate immune receptors. Cold Spring Harbor Symp Quant Biol 77 : 249–257.
71. FarmanML, EtoY, NakaoT, TosaY, NakayashikiH, et al. (2002) Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol Plant Microbe Interact 15 : 6–16.
72. LudererR, TakkenFL, de WitPJ, JoostenMH (2002) Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol Microbiol 45 : 875–884.
73. CuiL, YinW, DongS, WangY (2012) Analysis of polymorphism and transcription of the effector gene Avr1b in Phytophthora sojae isolates from China virulent to Rps1b. Mol Plant Pathol 13 : 114–122.
74. RouxelT, GrandaubertJ, HaneJK, HoedeC, van de WouwAP, et al. (2011) Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nature Comm 2 : 202.
75. SpanuPD (2012) The genomics of obligate (and nonobligate) biotrophs. Annu Rev Phytopathol 50 : 91–109.
76. SchmidtS, HoutermanP, SchreiverI, MaL, AmyotteS, et al. (2013) MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genomics 14 : 119.
77. LaurieJ, D., LinningR, WongP, BakkerenG (2013) Do TE activity and counteracting genome defenses, RNAi and methylation, shape the sex lives of smut fungi? Plant Signal Behav 8: e23853.
78. LubberstedtT, KleinD, MelchingerAE (1998) Comparative QTL mapping of resistance to Ustilago maydis across four populations of European flint-maize. Theor Appl Genet 97 : 1321–1330.
79. ParisseauxB, BernardoR (2004) In silico mapping of quantitative trait loci in maize. Theor Appl Genet 109 : 508–514.
80. BaumgartenAM, SureshJ, MayG, PhillipsRL (2007) Mapping QTLs contributing to Ustilago maydis resistance in specific plant tissues of maize. Theor Appl Genet 114 : 1229–1238.
81. WhittleCA, NygrenK, JohannessonH (2011) Consequences of reproductive mode on genome evolution in fungi. Fungal Genet Biol 48 : 661–667.
82. MunkacsiAB, StoxenS, MayG (2008) Ustilago maydis populations tracked maize through domestication and cultivation in the Americas. Proc Biol Sci 275 : 1037–1046.
83. HollidayR (1961) The genetics of Ustilago maydis. Genet Res Camb 2 : 204–230.
84. TsukudaT, CarletonS, FotheringhamS, HollomanWK (1988) Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol Cell Biol 8 : 3703–3709.
85. SzewczykE, NayakT, OakleyCE, EdgertonH, XiongY, et al. (2006) Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc 1 : 3111–3120.
86. BakkerenG, KronstadJW (1993) Conservation of the b mating-type gene complex among bipolar and tetrapolar smut fungi. Plant Cell 5 : 123–136.
87. KinalH, TaoJS, BruennJA (1991) An expression vector for the phytopathogenic fungus, Ustilago maydis. Gene 98 : 129–134.
88. HuangX, YangSP, ChinwallaAT, HillierLW, MinxP, et al. (2006) Application of a superword array in genome assembly. Nucleic Acids Res 34 : 201–205.
89. SalamovAA, SolovyevVV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10 : 516–522.
90. EmanuelssonO, BrunakS, von HeijneG, NielsenH (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2 : 953–971.
91. YuJH, HamariZ, HanKH, SeoJA, Reyes-DominguezY, et al. (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41 : 973–981.
92. García-PedrajasMD, NadalM, DennyT, Baeza-MontañezL, PazZ, et al. (2010) DelsGate: a robust and rapid method for gene deletion. Methods Mol Biol 638 : 55–76.
93. Sambrook J, Russell DW, editors (1999) Molecular Cloning: a laboratory manual. 3rd Edition, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
94. HuG, KampA, LinningR, NaikS, BakkerenG (2007) Complementation of Ustilago maydis MAPK mutants by a wheat leaf rust, Puccinia triticina homolog: potential for functional analyses of rust genes. Mol Plant-Microbe Interact 20 : 637–647.
95. LaurieJ, LinningR, BakkerenG (2008) Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis. Curr Genet 53 : 49–58.
96. HoodME, ShewHD (1996) Applications of KOH-aniline blue fluorescence in the study of plant-fungal interactions. Phytopathol 86 : 704–708.
97. MaB, TrompJ, LiM (2002) PatternHunter: faster and more sensitive homology search. Bioinformatics 18 : 440–445.
98. JonesDT, TaylorWR, ThorntonJM (1992) The rapid generation of mutation data matrices from protein sequences. Comp Appl Biosciences 8 : 275–282.
99. TamuraK, PetersonD, PetersonN, StecherG, NeiM, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28 : 2731–2739.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDSČlánek The Semen Microbiome and Its Relationship with Local Immunology and Viral Load in HIV InfectionČlánek Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
-
Všechny články tohoto čísla
- Bacteriophages as Vehicles for Antibiotic Resistance Genes in the Environment
- Helminth Infections, Type-2 Immune Response, and Metabolic Syndrome
- Defensins and Viral Infection: Dispelling Common Misconceptions
- Holobiont–Holobiont Interactions: Redefining Host–Parasite Interactions
- The Wide World of Ribosomally Encoded Bacterial Peptides
- Microbial Egress: A Hitchhiker's Guide to Freedom
- Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDS
- HIV-1 Capture and Transmission by Dendritic Cells: The Role of Viral Glycolipids and the Cellular Receptor Siglec-1
- Tetherin Can Restrict Cell-Free and Cell-Cell Transmission of HIV from Primary Macrophages to T Cells
- The Frustrated Host Response to Is Bypassed by MyD88-Dependent Translation of Pro-inflammatory Cytokines
- Larger Mammalian Body Size Leads to Lower Retroviral Activity
- The Semen Microbiome and Its Relationship with Local Immunology and Viral Load in HIV Infection
- Lytic Gene Expression Is Frequent in HSV-1 Latent Infection and Correlates with the Engagement of a Cell-Intrinsic Transcriptional Response
- Phase Variation of Poly-N-Acetylglucosamine Expression in
- A Screen of Mutants Reveals Important Roles for Dot/Icm Effectors and Host Autophagy in Vacuole Biogenesis
- Structure of the Trehalose-6-phosphate Phosphatase from Reveals Key Design Principles for Anthelmintic Drugs
- The Impact of Juvenile Coxsackievirus Infection on Cardiac Progenitor Cells and Postnatal Heart Development
- Vertical Transmission Selects for Reduced Virulence in a Plant Virus and for Increased Resistance in the Host
- Characterization of the Largest Effector Gene Cluster of
- Novel Drosophila Viruses Encode Host-Specific Suppressors of RNAi
- Pto Kinase Binds Two Domains of AvrPtoB and Its Proximity to the Effector E3 Ligase Determines if It Evades Degradation and Activates Plant Immunity
- Genetic Analysis of Tropism Using a Naturally Attenuated Cutaneous Strain
- Plasmacytoid Dendritic Cells Suppress HIV-1 Replication but Contribute to HIV-1 Induced Immunopathogenesis in Humanized Mice
- A Novel Mouse Model of Gastroenteritis Reveals Key Pro-inflammatory and Tissue Protective Roles for Toll-like Receptor Signaling during Infection
- Pathogenicity of Is Expressed by Regulating Metabolic Thresholds of the Host Macrophage
- BCKDH: The Missing Link in Apicomplexan Mitochondrial Metabolism Is Required for Full Virulence of and
- Independent Bottlenecks Characterize Colonization of Systemic Compartments and Gut Lymphoid Tissue by
- Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress
- G3BP1, G3BP2 and CAPRIN1 Are Required for Translation of Interferon Stimulated mRNAs and Are Targeted by a Dengue Virus Non-coding RNA
- Cytolethal Distending Toxins Require Components of the ER-Associated Degradation Pathway for Host Cell Entry
- The Machinery at Endoplasmic Reticulum-Plasma Membrane Contact Sites Contributes to Spatial Regulation of Multiple Effector Proteins
- Arabidopsis LIP5, a Positive Regulator of Multivesicular Body Biogenesis, Is a Critical Target of Pathogen-Responsive MAPK Cascade in Plant Basal Defense
- Plant Surface Cues Prime for Biotrophic Development
- Real-Time Imaging Reveals the Dynamics of Leukocyte Behaviour during Experimental Cerebral Malaria Pathogenesis
- The CD27L and CTP1L Endolysins Targeting Contain a Built-in Trigger and Release Factor
- cGMP and NHR Signaling Co-regulate Expression of Insulin-Like Peptides and Developmental Activation of Infective Larvae in
- Systemic Hematogenous Maintenance of Memory Inflation by MCMV Infection
- Strain-Specific Variation of the Decorin-Binding Adhesin DbpA Influences the Tissue Tropism of the Lyme Disease Spirochete
- Distinct Lipid A Moieties Contribute to Pathogen-Induced Site-Specific Vascular Inflammation
- Serovar Typhi Conceals the Invasion-Associated Type Three Secretion System from the Innate Immune System by Gene Regulation
- LANA Binds to Multiple Active Viral and Cellular Promoters and Associates with the H3K4Methyltransferase hSET1 Complex
- A Molecularly Cloned, Live-Attenuated Japanese Encephalitis Vaccine SA-14-2 Virus: A Conserved Single Amino Acid in the Hairpin of the Viral E Glycoprotein Determines Neurovirulence in Mice
- Illuminating Fungal Infections with Bioluminescence
- Comparative Genomics of Plant Fungal Pathogens: The - Paradigm
- Motility and Chemotaxis Mediate the Preferential Colonization of Gastric Injury Sites by
- Widespread Sequence Variations in VAMP1 across Vertebrates Suggest a Potential Selective Pressure from Botulinum Neurotoxins
- An Immunity-Triggering Effector from the Barley Smut Fungus Resides in an Ustilaginaceae-Specific Cluster Bearing Signs of Transposable Element-Assisted Evolution
- Establishment of Murine Gammaherpesvirus Latency in B Cells Is Not a Stochastic Event
- Oncogenic Herpesvirus KSHV Hijacks BMP-Smad1-Id Signaling to Promote Tumorigenesis
- Human APOBEC3 Induced Mutation of Human Immunodeficiency Virus Type-1 Contributes to Adaptation and Evolution in Natural Infection
- Innate Immune Responses and Rapid Control of Inflammation in African Green Monkeys Treated or Not with Interferon-Alpha during Primary SIVagm Infection
- Chitin-Degrading Protein CBP49 Is a Key Virulence Factor in American Foulbrood of Honey Bees
- Influenza A Virus Host Shutoff Disables Antiviral Stress-Induced Translation Arrest
- Nsp9 and Nsp10 Contribute to the Fatal Virulence of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus Emerging in China
- Pulmonary Infection with Hypervirulent Mycobacteria Reveals a Crucial Role for the P2X7 Receptor in Aggressive Forms of Tuberculosis
- Syk Signaling in Dendritic Cells Orchestrates Innate Resistance to Systemic Fungal Infection
- A Repetitive DNA Element Regulates Expression of the Sialic Acid Binding Adhesin by a Rheostat-like Mechanism
- T-bet and Eomes Are Differentially Linked to the Exhausted Phenotype of CD8+ T Cells in HIV Infection
- Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health
- Influence of ND10 Components on Epigenetic Determinants of Early KSHV Latency Establishment
- Antibody to gp41 MPER Alters Functional Properties of HIV-1 Env without Complete Neutralization
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDS
- Holobiont–Holobiont Interactions: Redefining Host–Parasite Interactions
- BCKDH: The Missing Link in Apicomplexan Mitochondrial Metabolism Is Required for Full Virulence of and
- Helminth Infections, Type-2 Immune Response, and Metabolic Syndrome
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



