-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Wide World of Ribosomally Encoded Bacterial Peptides
article has not abstract
Published in the journal: . PLoS Pathog 10(7): e32767. doi:10.1371/journal.ppat.1004221
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004221Summary
article has not abstract
Bacterial Peptides: Enormous Diversity
Peptides are defined as short chains of amino acids that are linked by peptide bonds. In eukaryotes, peptides encompass an enormous range of structure and function, from signaling hormones, to anti-pathogen molecules, to powerful toxins. In bacteria, ribosomally produced peptides known as bacteriocins have been historically investigated for their potential antimicrobial activities [1]. However, recent efforts in genomics and natural product discovery have led to a tremendous “explosion” in the sheer number and diversity of ribosomally produced peptides from the prokaryotic domain. It has been estimated that all bacteria and archaea produce at least one, but more likely multiple, bacteriocin-like peptides that have a wide range of functions including antimicrobial toxins, virulence factors, and bacterial “hormones” that allow bacterial communities to organize multicellular behaviors such as biofilm formation. This article provides an overview of ribosomally produced bacterial peptides and their diverse roles in bacterial lifestyles, along with future prospects and recent computational and bioinformatic approaches aimed at decoding the overall “language” of these bacterially produced peptides.
Structure and Classification of Small Bacterial Peptides
Ribosomally produced bacterial peptides are a large class of compounds that encompass an extraordinary amount of chemical, structural, and functional diversity (Figure 1) [2], [3]. These small peptides can range from unmodified linear forms to highly modified, and sometimes circularized, structures. These modifications serve to confer specific chemical properties that could not be obtained via peptide synthesis alone, further increasing the number and complexity of these bacterial peptide families. Furthermore, certain modifications are thought to serve as an important safety mechanism to regulate the toxic activities of the bacterial peptide, thereby providing a level of control and self-immunity [2], [4]. Some of the major chemical classifications of ribosomally produced bacterial peptides include Lantibiotics such as Nisin, Linear azo(line)-containing peptides such as Microcin B17, Lasso peptides such as the Escherichia coli antibacterial peptide Microcin J25, and many others that continue to be discovered at a rapid pace [5]. Approaches to systematically classify all known ribosomally produced bacterial peptides have involved dividing groups based on: (1) particular prolific producers such as lactic acid bacteria, (2) particular modifications of the bacterial peptide, or (3) specific peptide activities. Indeed, given the sheer number and diversity of these bacterially produced compounds, there is tremendous potential in the discovery and development of these natural products as therapeutics. It has been noted that with respect to bacteriocins, bacteria have, in essence, “already designed what clinicians and pharmaceutical industries are once again struggling to obtain” [2].
Fig. 1. Functional diversity of ribosomally produced bacterial peptides. 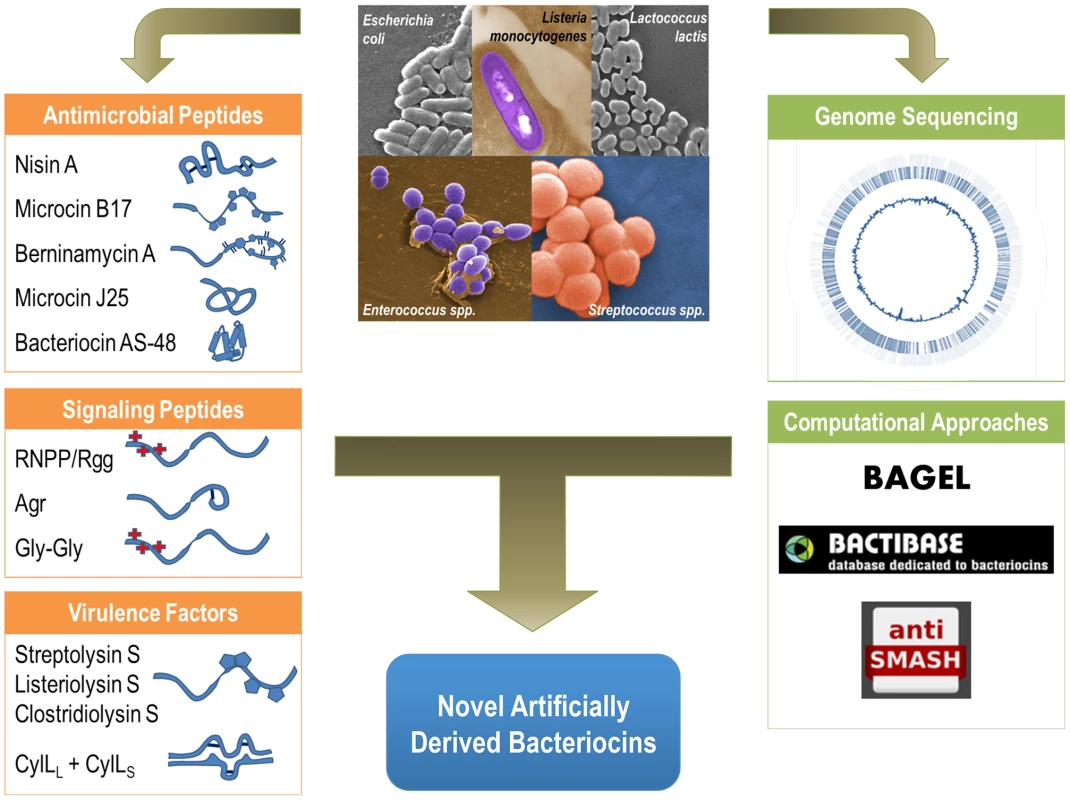
Bacterial peptides produced by both gram-positive and gram-negative bacteria include antimicrobial peptides such as Nisin and Microcin B17, known host virulence factors such as the Streptolysin S-like cytolysins, and the peptide cytolysin from E. faecalis. Bacterial peptides that structurally resemble bacteriocins are also utilized as signaling molecules. Computational and genomic approaches, such as BAGEL, BACTIBASE, and Anti-SMASH, can be combined with genomic data to catalog and discover new ribosomally produced bacterial peptides. Combining computational approaches with experimental data can guide the development of novel antimicrobials and artificially derived peptides with specific functions and targets. Bacterial Peptides as Antimicrobial Compounds
Many bacterial peptides have been known to exhibit potent antimicrobial activity against competitor species of the producing microorganism. Nisin, whose activity was first reported in 1928 [6], has been used widely in the food industry to prevent food-borne pathogens [7]. The mechanism of Nisin's action is to bind to an intermediate in bacterial cell wall synthesis, resulting in bacterial killing through pore formation [8]. Nisin belongs to a larger group of bacteriocins known as lantibiotics; these compounds have been highly studied, given their ability to specifically target clinically important pathogens such as Streptococcus pneumoniae, Methicillin-resistant strains of Staphylococcus aureus (MRSA), and Vancomycin-resistant enterococci (VRE) [9]. Microcin B17, a linear peptide produced by particular strains of E. coli, is one of many bacteriocins that are enzymatically modified by the addition of heterocyclic residues [10]. Microcin B17 targets susceptible bacteria by inhibiting bacterial DNA gyrase [11]. In contrast to Nisin, whose antimicrobial activity is most active against gram-positive bacteria, Microcin B17 has been shown to be effective against a wide range of gram-negative pathogens, such as E. coli, Salmonella, Shigella, and Yersinia species. A widely studied example of an unmodified bacterial peptide is the enterococcal bacteriocin AS-48, which has antimicrobial activity against gram-positive pathogens such as Listeria monocytogenes [12]. AS-48 bacteriocin belongs to a larger class of unmodified peptides that adopt a particular native structure that is critical for their activity.
Bacterial Peptides as Virulence Factors
Recent discoveries have uncovered the fact that long-known potent bacterial toxins such as Streptolysin S (SLS) are actually small, ribosomally produced peptides, whose enzymatic modifications are highly similar to those of known bacteriocins, such as Microcin B17. In addition to their well-described role in microbial warfare against competing bacterial species, these peptide toxins are beginning to be recognized as key contributors to initiating host disease. Streptolysin S has been identified as a major contributing factor in successful translocation of Streptococcus pyogenes across the epithelial barrier through a mechanism involving the disruption of intracellular junctions via cleavage of occludin and E-cadherin [13]. The ability of peptide toxins such as SLS to prevent phagocytic clearance can also be mediated through direct killing of immune cells. A series of simple in vitro experiments exploring the effects of SLS on mouse peritoneal macrophages in the early 1970s provided the first indication that bacteriocin-like toxins can exhibit leukotoxic effects [14]. Like S. pyogenes, Enterococcus faecalis also produces a peptide cytolysin (encoded by the cyIL gene cluster) that is capable of lysing neutrophils and macrophages to avoid immune clearance [15]. Interestingly, several microbial peptide toxins have also been shown to have synergistic activity with other bacterial virulence factors, suggesting that, in fact, these bacterial peptides may serve the dual role of causing direct damage to the host while also increasing the overall virulence output. For example, Hung et al. utilized a murine infection model to demonstrate that the peptide toxin SLS synergizes with the unrelated streptococcal pyrogenic exotoxin B (SpeB) during infection to enhance several features of pathogenesis, including inhibition of phagocytic clearance and the induction of macrophage apoptosis [16]. In commensal bacteria such as Lactobacillus plantarum, it has been shown that production of antimicrobial bacteriocins can modulate the immune response of dendritic and peripheral blood mononuclear cells as well as alter host cytokine profiles versus nonbacteriocin producing mutants [17].
Bacterial Peptides as Communication Signals
Many gram-positive bacteria use small peptides to communicate within a multicellular community to regulate processes such as cellular density, biofilm formation, competence for mating, and coordinated control of virulence [18]. Quorum sensing, the act of bacterial communication via extracellular diffusible molecules, allows bacteria in many cases to synchronize group behavior and facilitate coordinated events. In gram-negative bacteria, N-acyl homoserine lactones have been extensively studied for their role as communication molecules; however, in gram-positive bacteria, recent studies have revealed that small peptides function as the predominant signaling molecule of choice. One of the earliest discoveries of peptide pheromone signaling is the Agr system in Staphylococcus aureus, which uses a small cyclic peptide, known as autoinducing peptide (AIP), to communicate in a multicellular setting to control the expression of virulence genes for a coordinated effect on host pathogenesis [19]. Additionally, three other major groups of bacterial peptides have been studied for their roles in intercellular communication, which are the Gly-Gly processed peptides, the RNPP systems peptides, and the Rgg motif signaling peptides [20]. In some cases, reports are emerging that these peptides control multiple behaviors in bacteria. In Streptococcus intermedius, the quorum signal peptide known as competence stimulating peptide (CSP) was found not only to be involved in regulating competence for natural transformation but also to regulate and promote biofilm formation [21]. Multifunctional roles for bacteriocins are becoming a wider and more accepted phenomenon. For example, reports have suggested that Nisin can act both as an antimicrobial molecule and through an autocrine signaling mechanism to potentiate Nisin production in a cell density–dependent manner [22].
Understanding the “Grammar” of Bacterial Peptides
Genome mining has been an important technological resource in the discovery of novel natural products, such as bacteriocins. Bacteriocin-like peptides are highly attractive candidates for genome mining, as these natural products are genetically encoded with nearby genes encoding their corresponding modifying enzymes. Proximity to genes encoding known modifying enzymes can aid in the identification of peptide biosynthesis gene clusters [23]. In many cases, several metabolites have been identified from “cryptic” or “orphan gene clusters” [24]. These cryptic gene clusters have demonstrated that new, as-yet-uncharacterized enzymology is likely to be involved in the assembly of the final natural product, likely leading to greater diversities of bacterial peptides than have previously been appreciated.
Many web-based Bacteriocin gene mining and annotation tools have been developed to aid in the identification, characterization, and classification of novel bacteriocins. These include mining tools such as BAGEL (http://bagel.molgenrug.nl) and bacteriocin repositories such as BACTIBASE (http://bactibase.pfba-lab-tun.org/main.php). Anti-SMASH is a recently developed website that expands genome mining to not only bacteriocins but a host of other genetically identifiable antibiotics and other secondary metabolites [25]. Although these tools are rapidly expanding the repertoire of bacterially produced peptides, one elusive goal has been to deduce the function of a given bacterial peptide from the structural and genetic information alone. Recent computing approaches, however, have begun to address the goal of using purely in silico approaches in order to predict the specific activity of peptides and proteins. Loose et al. have postulated a “linguistic” model for the design of antimicrobial peptides. The repeated usage of particular amino acid sequences that are common to antimicrobial peptides led these investigators to propose a certain grammar that governs the “language” of antimicrobial peptides [26]. In a recent study, Gupta et al. used a database of toxic and nontoxic peptides coupled with machine learning and a quantitative matrix approach to predict whether a given peptide will have toxic properties [27]. Their program, Toxipred, functions to essentially predict the relative toxicity of a given peptide based on sequence alone. Although still relatively new, programs such as these and others, combined with the rapid pace of genomic discovery, will rapidly accelerate the pace of drug discovery in these bacterial peptide families.
Conclusions
Ribosomally produced bacterial peptides have seen a recent surge in interest due to new structural, biochemical, and microbiological tools, along with advances in genome sequencing and bioinformatics. Computational algorithms and bacterial peptide databases are rapidly growing as more of these compounds are discovered and deposited. Although traditionally bacteriocins have been classified as having antimicrobial properties, recent findings suggest that bacteria utilize bacteriocin-like peptides to perform a myriad of roles, including intercellular communication, host colonization and manipulation, as well as the exciting finding that these small peptides can have multiple functions in a given microorganism. Computational-based approaches, coupled with experimental data, will allow investigators to decipher the overall “language” of these ribosomally produced bacterial peptides such that novel antimicrobials and artificially derived peptides with specific functions and targets can be soon developed—provided that we will be able to “speak its language.”
Zdroje
1. CotterPD, RossRP, HillC (2013) Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol 11 : 95–105.
2. BabaT, SchneewindO (1998) Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol 6 : 66–71.
3. SitCS, VederasJC (2008) Approaches to the discovery of new antibacterial agents based on bacteriocins. Biochem Cell Biol 86 : 116–123.
4. GarridoMC, HerreroM, KolterR, MorenoF (1988) The export of the DNA replication inhibitor Microcin B17 provides immunity for the host cell. Embo J 7 : 1853–1862.
5. ArnisonPG, BibbMJ, BierbaumG, BowersAA, BugniTS, et al. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30 : 108–160.
6. RogersLA (1928) The Inhibiting Effect of Streptococcus Lactis on Lactobacillus Bulgaricus. J Bacteriol 16 : 321–325.
7. LubelskiJ, RinkR, KhusainovR, MollGN, KuipersOP (2008) Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci 65 : 455–476.
8. BierbaumG, SahlHG (2009) Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10 : 2–18.
9. PiperC, CotterPD, RossRP, HillC (2009) Discovery of medically significant lantibiotics. Curr Drug Discov Technol 6 : 1–18.
10. WalshCT, NolanEM (2008) Morphing peptide backbones into heterocycles. Proc Natl Acad Sci U S A 105 : 5655–5656.
11. YorgeyP, LeeJ, KordelJ, VivasE, WarnerP, et al. (1994) Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc Natl Acad Sci U S A 91 : 4519–4523.
12. MendozaF, MaquedaM, GalvezA, Martinez-BuenoM, ValdiviaE (1999) Antilisterial activity of peptide AS-48 and study of changes induced in the cell envelope properties of an AS-48-adapted strain of Listeria monocytogenes. Appl Environ Microbiol 65 : 618–625.
13. SumitomoT, NakataM, HigashinoM, JinY, TeraoY, et al. (2011) Streptolysin S contributes to group A streptococcal translocation across an epithelial barrier. J Biol Chem 286 : 2750–2761.
14. OfekI, Bergner-RabinowitzS, GinsburgI (1972) Oxygen-stable hemolysins of group A streptococci. 8. Leukotoxic and antiphagocytic effects of streptolysins S and O. Infect Immun 6 : 459–464.
15. MiyazakiS, OhnoA, KobayashiI, UjiT, YamaguchiK, et al. (1993) Cytotoxic effect of hemolytic culture supernatant from Enterococcus faecalis on mouse polymorphonuclear neutrophils and macrophages. Microbiol Immunol 37 : 265–270.
16. HungCH, TsaoN, ZengYF, LuSL, ChuanCN, et al. (2012) Synergistic effects of streptolysin S and streptococcal pyrogenic exotoxin B on the mouse model of group A streptococcal infection. Med Microbiol Immunol 201 : 357–369.
17. van HemertS, MeijerinkM, MolenaarD, BronPA, de VosP, et al. (2010) Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol 10 : 293.
18. LyonGJ, NovickRP (2004) Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25 : 1389–1403.
19. KongKF, VuongC, OttoM (2006) Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol 296 : 133–139.
20. CookLC, FederleMJ (2013) Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38 : 473–492.
21. PetersenFC, FimlandG, ScheieAA (2006) Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol Microbiol 61 : 1322–1334.
22. KleerebezemM (2004) Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25 : 1405–1414.
23. ChallisGL (2008) Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology 154 : 1555–1569.
24. GrossH (2007) Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Appl Microbiol Biotechnol 75 : 267–277.
25. BlinK, MedemaMH, KazempourD, FischbachMA, BreitlingR, et al. (2013) antiSMASH 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41: W204–212.
26. LooseC, JensenK, RigoutsosI, StephanopoulosG (2006) A linguistic model for the rational design of antimicrobial peptides. Nature 443 : 867–869.
27. GuptaS, KapoorP, ChaudharyK, GautamA, KumarR, et al. (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 8: e73957.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDSČlánek The Semen Microbiome and Its Relationship with Local Immunology and Viral Load in HIV InfectionČlánek Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Bacteriophages as Vehicles for Antibiotic Resistance Genes in the Environment
- Helminth Infections, Type-2 Immune Response, and Metabolic Syndrome
- Defensins and Viral Infection: Dispelling Common Misconceptions
- Holobiont–Holobiont Interactions: Redefining Host–Parasite Interactions
- The Wide World of Ribosomally Encoded Bacterial Peptides
- Microbial Egress: A Hitchhiker's Guide to Freedom
- Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDS
- HIV-1 Capture and Transmission by Dendritic Cells: The Role of Viral Glycolipids and the Cellular Receptor Siglec-1
- Tetherin Can Restrict Cell-Free and Cell-Cell Transmission of HIV from Primary Macrophages to T Cells
- The Frustrated Host Response to Is Bypassed by MyD88-Dependent Translation of Pro-inflammatory Cytokines
- Larger Mammalian Body Size Leads to Lower Retroviral Activity
- The Semen Microbiome and Its Relationship with Local Immunology and Viral Load in HIV Infection
- Lytic Gene Expression Is Frequent in HSV-1 Latent Infection and Correlates with the Engagement of a Cell-Intrinsic Transcriptional Response
- Phase Variation of Poly-N-Acetylglucosamine Expression in
- A Screen of Mutants Reveals Important Roles for Dot/Icm Effectors and Host Autophagy in Vacuole Biogenesis
- Structure of the Trehalose-6-phosphate Phosphatase from Reveals Key Design Principles for Anthelmintic Drugs
- The Impact of Juvenile Coxsackievirus Infection on Cardiac Progenitor Cells and Postnatal Heart Development
- Vertical Transmission Selects for Reduced Virulence in a Plant Virus and for Increased Resistance in the Host
- Characterization of the Largest Effector Gene Cluster of
- Novel Drosophila Viruses Encode Host-Specific Suppressors of RNAi
- Pto Kinase Binds Two Domains of AvrPtoB and Its Proximity to the Effector E3 Ligase Determines if It Evades Degradation and Activates Plant Immunity
- Genetic Analysis of Tropism Using a Naturally Attenuated Cutaneous Strain
- Plasmacytoid Dendritic Cells Suppress HIV-1 Replication but Contribute to HIV-1 Induced Immunopathogenesis in Humanized Mice
- A Novel Mouse Model of Gastroenteritis Reveals Key Pro-inflammatory and Tissue Protective Roles for Toll-like Receptor Signaling during Infection
- Pathogenicity of Is Expressed by Regulating Metabolic Thresholds of the Host Macrophage
- BCKDH: The Missing Link in Apicomplexan Mitochondrial Metabolism Is Required for Full Virulence of and
- Independent Bottlenecks Characterize Colonization of Systemic Compartments and Gut Lymphoid Tissue by
- Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress
- G3BP1, G3BP2 and CAPRIN1 Are Required for Translation of Interferon Stimulated mRNAs and Are Targeted by a Dengue Virus Non-coding RNA
- Cytolethal Distending Toxins Require Components of the ER-Associated Degradation Pathway for Host Cell Entry
- The Machinery at Endoplasmic Reticulum-Plasma Membrane Contact Sites Contributes to Spatial Regulation of Multiple Effector Proteins
- Arabidopsis LIP5, a Positive Regulator of Multivesicular Body Biogenesis, Is a Critical Target of Pathogen-Responsive MAPK Cascade in Plant Basal Defense
- Plant Surface Cues Prime for Biotrophic Development
- Real-Time Imaging Reveals the Dynamics of Leukocyte Behaviour during Experimental Cerebral Malaria Pathogenesis
- The CD27L and CTP1L Endolysins Targeting Contain a Built-in Trigger and Release Factor
- cGMP and NHR Signaling Co-regulate Expression of Insulin-Like Peptides and Developmental Activation of Infective Larvae in
- Systemic Hematogenous Maintenance of Memory Inflation by MCMV Infection
- Strain-Specific Variation of the Decorin-Binding Adhesin DbpA Influences the Tissue Tropism of the Lyme Disease Spirochete
- Distinct Lipid A Moieties Contribute to Pathogen-Induced Site-Specific Vascular Inflammation
- Serovar Typhi Conceals the Invasion-Associated Type Three Secretion System from the Innate Immune System by Gene Regulation
- LANA Binds to Multiple Active Viral and Cellular Promoters and Associates with the H3K4Methyltransferase hSET1 Complex
- A Molecularly Cloned, Live-Attenuated Japanese Encephalitis Vaccine SA-14-2 Virus: A Conserved Single Amino Acid in the Hairpin of the Viral E Glycoprotein Determines Neurovirulence in Mice
- Illuminating Fungal Infections with Bioluminescence
- Comparative Genomics of Plant Fungal Pathogens: The - Paradigm
- Motility and Chemotaxis Mediate the Preferential Colonization of Gastric Injury Sites by
- Widespread Sequence Variations in VAMP1 across Vertebrates Suggest a Potential Selective Pressure from Botulinum Neurotoxins
- An Immunity-Triggering Effector from the Barley Smut Fungus Resides in an Ustilaginaceae-Specific Cluster Bearing Signs of Transposable Element-Assisted Evolution
- Establishment of Murine Gammaherpesvirus Latency in B Cells Is Not a Stochastic Event
- Oncogenic Herpesvirus KSHV Hijacks BMP-Smad1-Id Signaling to Promote Tumorigenesis
- Human APOBEC3 Induced Mutation of Human Immunodeficiency Virus Type-1 Contributes to Adaptation and Evolution in Natural Infection
- Innate Immune Responses and Rapid Control of Inflammation in African Green Monkeys Treated or Not with Interferon-Alpha during Primary SIVagm Infection
- Chitin-Degrading Protein CBP49 Is a Key Virulence Factor in American Foulbrood of Honey Bees
- Influenza A Virus Host Shutoff Disables Antiviral Stress-Induced Translation Arrest
- Nsp9 and Nsp10 Contribute to the Fatal Virulence of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus Emerging in China
- Pulmonary Infection with Hypervirulent Mycobacteria Reveals a Crucial Role for the P2X7 Receptor in Aggressive Forms of Tuberculosis
- Syk Signaling in Dendritic Cells Orchestrates Innate Resistance to Systemic Fungal Infection
- A Repetitive DNA Element Regulates Expression of the Sialic Acid Binding Adhesin by a Rheostat-like Mechanism
- T-bet and Eomes Are Differentially Linked to the Exhausted Phenotype of CD8+ T Cells in HIV Infection
- Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health
- Influence of ND10 Components on Epigenetic Determinants of Early KSHV Latency Establishment
- Antibody to gp41 MPER Alters Functional Properties of HIV-1 Env without Complete Neutralization
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDS
- Holobiont–Holobiont Interactions: Redefining Host–Parasite Interactions
- BCKDH: The Missing Link in Apicomplexan Mitochondrial Metabolism Is Required for Full Virulence of and
- Helminth Infections, Type-2 Immune Response, and Metabolic Syndrome
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



