-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
Cetaceans have a long history of commitment to a fully aquatic lifestyle that extends back to the Eocene. Extant species have evolved a spectacular array of adaptations in conjunction with their deployment into a diverse array of aquatic habitats. Sensory systems are among those that have experienced radical transformations in the evolutionary history of this clade. In the case of vision, previous studies have demonstrated important changes in the genes encoding rod opsin (RH1), short-wavelength sensitive opsin 1 (SWS1), and long-wavelength sensitive opsin (LWS) in selected cetaceans, but have not examined the full complement of opsin genes across the complete range of cetacean families. Here, we report protein-coding sequences for RH1 and both color opsin genes (SWS1, LWS) from representatives of all extant cetacean families. We examine competing hypotheses pertaining to the timing of blue shifts in RH1 relative to SWS1 inactivation in the early history of Cetacea, and we test the hypothesis that some cetaceans are rod monochomats. Molecular evolutionary analyses contradict the “coastal” hypothesis, wherein SWS1 was pseudogenized in the common ancestor of Cetacea, and instead suggest that RH1 was blue-shifted in the common ancestor of Cetacea before SWS1 was independently knocked out in baleen whales (Mysticeti) and in toothed whales (Odontoceti). Further, molecular evidence implies that LWS was inactivated convergently on at least five occasions in Cetacea: (1) Balaenidae (bowhead and right whales), (2) Balaenopteroidea (rorquals plus gray whale), (3) Mesoplodon bidens (Sowerby's beaked whale), (4) Physeter macrocephalus (giant sperm whale), and (5) Kogia breviceps (pygmy sperm whale). All of these cetaceans are known to dive to depths of at least 100 m where the underwater light field is dim and dominated by blue light. The knockout of both SWS1 and LWS in multiple cetacean lineages renders these taxa rod monochromats, a condition previously unknown among mammalian species.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003432
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003432Summary
Cetaceans have a long history of commitment to a fully aquatic lifestyle that extends back to the Eocene. Extant species have evolved a spectacular array of adaptations in conjunction with their deployment into a diverse array of aquatic habitats. Sensory systems are among those that have experienced radical transformations in the evolutionary history of this clade. In the case of vision, previous studies have demonstrated important changes in the genes encoding rod opsin (RH1), short-wavelength sensitive opsin 1 (SWS1), and long-wavelength sensitive opsin (LWS) in selected cetaceans, but have not examined the full complement of opsin genes across the complete range of cetacean families. Here, we report protein-coding sequences for RH1 and both color opsin genes (SWS1, LWS) from representatives of all extant cetacean families. We examine competing hypotheses pertaining to the timing of blue shifts in RH1 relative to SWS1 inactivation in the early history of Cetacea, and we test the hypothesis that some cetaceans are rod monochomats. Molecular evolutionary analyses contradict the “coastal” hypothesis, wherein SWS1 was pseudogenized in the common ancestor of Cetacea, and instead suggest that RH1 was blue-shifted in the common ancestor of Cetacea before SWS1 was independently knocked out in baleen whales (Mysticeti) and in toothed whales (Odontoceti). Further, molecular evidence implies that LWS was inactivated convergently on at least five occasions in Cetacea: (1) Balaenidae (bowhead and right whales), (2) Balaenopteroidea (rorquals plus gray whale), (3) Mesoplodon bidens (Sowerby's beaked whale), (4) Physeter macrocephalus (giant sperm whale), and (5) Kogia breviceps (pygmy sperm whale). All of these cetaceans are known to dive to depths of at least 100 m where the underwater light field is dim and dominated by blue light. The knockout of both SWS1 and LWS in multiple cetacean lineages renders these taxa rod monochromats, a condition previously unknown among mammalian species.
Introduction
Cetacea [dolphins, porpoises, and whales] represents a remarkable example of aquatic specialization within Mammalia [1]. With their return to river and marine environments, the ancestors of modern toothed cetaceans (Odontoceti) and baleen whales (Mysticeti) underwent extensive modifications that included the evolution of novel structures [e.g., baleen plates, tail flukes], major anatomical rearrangements (e.g., telescoping of the skull, development of fore-flippers), the loss or reduction of typical mammalian traits (e.g., olfactory structures, hair, hindlimbs), and associated behavioral changes (echolocation, filter-feeding, deep-diving) [2]–[4]. At the genetic level this restructuring includes evidence of positive selection in loci related to high-frequency audition [5]–[7], brain size [8], [9], and flipper development [10], as well as degradation of genes related to olfaction [11]–[13], taste [14], tooth enamel formation [15]–[17], and vomeronasal chemoreception [18].
In the case of vision, aquatic environments impose challenging constraints, and the cetacean eye exhibits both morphological and molecular specializations that enhance underwater sight [19]. Possible morphological adaptations include an extensive reflective tapetum lucidum, a spherical lens with high refractive power, a relatively large cornea, and a rod-dominated retina, all of which enhance visual capabilities under dim light conditions [20], [21]. At the molecular level, most mammals have dichromatic color vision based on presence of three visual pigments, each of which is a G protein-coupled receptor that consists of an opsin protein moiety linked via a Schiff base to a retinal chromophore [22]. The three opsins that characterize most mammals include a rod opsin (RH1) and two cone opsins, short wavelength-sensitive opsin (SWS1) and long wavelength-sensitive opsin (LWS). Rods mainly function in dim light conditions (scotopic/night vision) whereas cones require more light (photopic vision) and are necessary with color vision. By contrast with most other mammals, all cetaceans that have been investigated are thought to be L-cone monochromats that possess an inactivated copy of SWS1 and two functional opsins, RH1 and LWS, which are expressed in rod and L-cone cells of the retina, respectively [19], [23], [24].
Griebel and Peichl [19] and Peichl [24] suggested that retinal S-cones, which express SWS1 and are sensitive to blue wavelengths, were lost during an early, coastal period of cetacean evolution. Near-shore waters commonly have an underwater light spectrum that is red shifted owing to the absorption of blue light by organic and inorganic debris, and the loss of ‘jobless’ S-cones may have constituted an economical advantage in this environment by simplifying retinal and cortical visual information processing [19]. There are no inactivating frameshift mutations in SWS1 that are shared by all odontocetes and mysticetes [23], but Griebel and Peichl [19] suggested that an unidentified genetic change, possibly in the promoter region, thwarted expression of the SWS1 protein in the common ancestor of crown Cetacea. Following the knockout of SWS1, crown cetacean lineages that independently conquered the open ocean were forced to shift λmax [the wavelength of maximal absorption] of RH1 and LWS to bluer wavelengths because SWS1 had previously been inactivated [19], [24]. By contrast, Bischoff et al. [25] offered an alternative scenario in which RH1 was blue shifted in the common ancestor of Cetacea. Specifically, Bischoff et al. [25] speculated that the ancestral cetacean RH1 possessed 83Asn, 292Ser, and 299Ala at three key tuning sites, as in the deep-diving giant sperm whale [Physeter macrocephalus], but stopped short of using explicit methods to reconstruct the ancestral RH1 sequence of Cetacea. If RH1 was blue-shifted in the common ancestor of Cetacea, then SWS1 may have been inactivated independently in mysticetes and odontocetes, perhaps due to the inefficiency at S-cones at photon capture in dim light conditions [22].
Another intriguing hypothesis posits rod monochromacy, as opposed to L-cone monochromacy, in at least some cetaceans. McFarland [20] suggested that some cetaceans are probably rod monochromats in which S-cones, L-cones, and their associated opsin genes [SWS1 and LWS, respectively] are lacking, so that vision is based entirely on rods and the rod opsin gene RH1. Immunocytochemical studies have failed to support this hypothesis and instead demonstrated the presence of L-cones in representative odontocete species belonging to the families Delphinidae and Phocoenidae [26]. More recently, Fasick et al. [21] reported the first partial L-cone opsin sequence (LWS) of a mysticete, Eubalaena glacialis (Atlantic right whale), and suggested that the L-cone opsin in this taxon is blue shifted, as are the L-cone opsins of representative odontocetes [27]. A potential shortcoming of this study is that Fasick et al. [21] only sequenced exons 3 and 5 of the E. glacialis LWS gene. In addition, LWS sequences have not been characterized from several other cetacean families including the deep-diving Ziphiidae, Physeteridae, and Kogiidae. Thus, McFarland's [20] suggestion that some cetaceans are rod monochromats remains to be tested by a more complete sampling of opsin gene sequences from a broader array of species.
Here, we report complete or nearly complete protein-coding sequences for all three opsin genes (RH1, SWS1, LWS) from representatives of all extant families of Cetacea and the cetacean sister group, Hippopotamidae. Previous studies have characterized the evolutionary patterns of individual cetacean opsins in isolation, but have not yet integrated information from all three retinal opsin genes (RH1, LWS, SWS1) into a single, comprehensive analysis. We utilized selection intensity estimates, ancestral sequence reconstructions, shifts in spectral tuning, and shared missense/frameshift mutations to infer the complex history of opsin evolution in Cetacea. Our reconstructions suggest that RH1 was blue-shifted in the common ancestor of Cetacea prior to the independent inactivation of SWS1 on the stem mysticete and odontocete branches. LWS, in turn, was pseudogenized convergently in five different cetacean lineages [right whale plus bowhead, rorquals plus gray whale, Sowerby's beaked whale, giant sperm whale, pygmy sperm whale], all of which are deep divers that feed on bioluminescent organisms. The tandem inactivation of SWS1 and LWS in these taxa presumably renders them rod monochromats, a condition that was previously unknown within Mammalia.
Results
Phylogenetic Analyses
Maximum likelihood trees based on SWS1 exons plus introns, SWS1 exons, RH1 exons, and LWS exons are shown in Figures S1, S2, S3, S4. With a few exceptions, clades with high bootstrap support percentages (>90) on individual gene trees are in agreement with the species tree in Figure 1. All of the gene trees recovered Cetancodonta [Cetacea + Hippopotamidae], Cetacea, Mysticeti, Balaenidae, Balaenopteroidea, Physeteroidea, Ziphiidae, Iniidae + Pontoporiidae, Phocoenidae, Delphinidae, Delphinoidea, and Iniidae + Pontoporiidae + Delphinoidea [Delphinida]. Odontoceti was only recovered in the SWS1 analyses, but conflicting nodes in the RH1 and LWS trees had low bootstrap support values (≤53%).
Fig. 1. A hypothesis for the evolution of LWS, SWS1, and RH1 in Cetacea and outgroups. 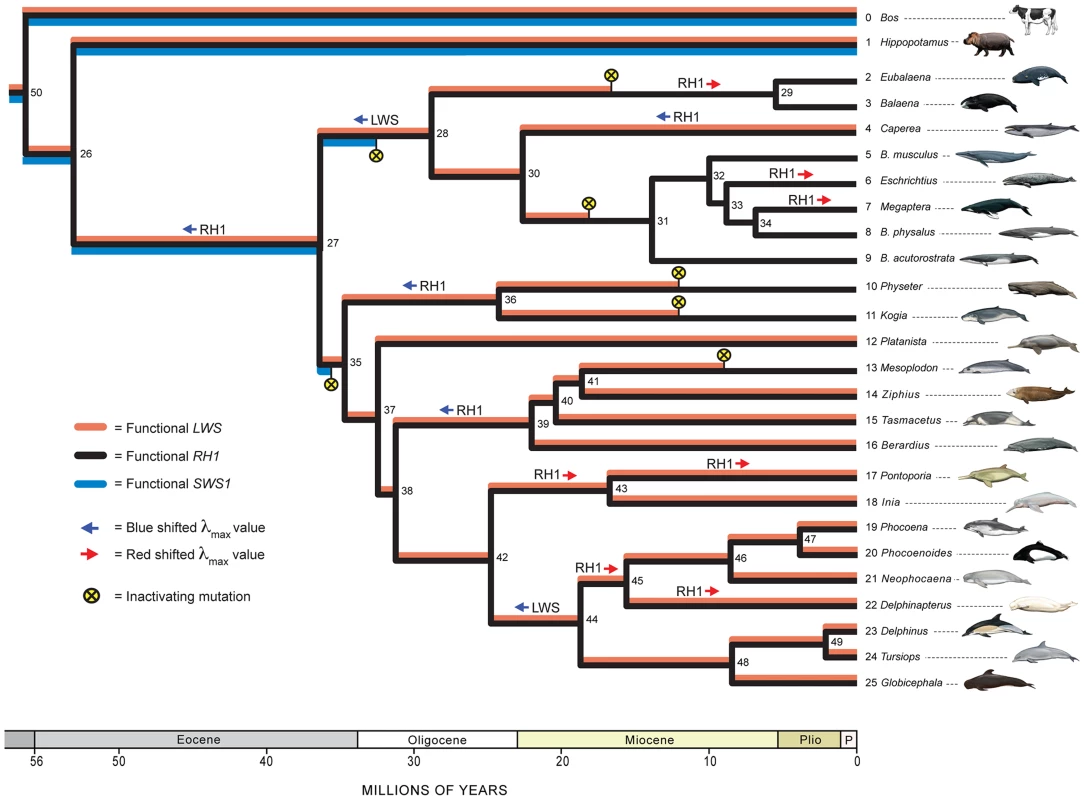
Phylogenetic relationships and divergence times for Cetacea follow McGowen et al. [42] and for outgroups are as in Meredith et al. [101]. Nodes in the tree are numbered (1–50) and are referenced to in the main text. The coloration of branches indicates inferred functional SWS1 (blue), functional LWS (red), and functional RH1 (black); for example, all three opsins are reconstructed as functional in the common ancestor of Cetacea (node 26 to node 27), but only RH1 is functional in balaenopteroid baleen whales (node 31 and its descendants). Inferred inactivation events (frameshifts, premature stop codons, etc.) are marked by yellow circles with Xs, and are arbitrarily placed at the midpoints of branches to which these events were optimized. Inferred red and blue shifts in λmax for LWS and RH1 are indicated by arrows at internodes. Geological time scale is shown at the bottom of the figure with time in millions of years (Plio = Pliocene; P = Pleistocene). For taxon names, the mysticete genus Balaenoptera is abbreviated as “B.”. Paintings are by Carl Buell. SWS1 Evolution
Inactivating mutations (frameshift indels, premature stop codons, disrupted intron splice sites, amino acid replacement at the Schiff's base counterion site) were apparent for all cetacean species in the SWS1 alignment (Table 1), but were lacking in SWS1 from the semiaquatic outgroup species, Hippopotamus amphibius (Figure 1). Although the SWS1 genes of all cetacean species show evidence of mutational decay, no inactivating mutations map to the last common ancestral branch of Cetacea (Figure 1, node 26 to node 27). Instead, different molecular lesions define various sublineages of Cetacea (Table 1). An amino acid replacement (E113G; bovine RH1 numbering) at the Schiff's base counterion site that is thought to disrupt opsin-chromophore binding [23] optimizes to the stem branch of Odontoceti (Figure 1; node 27 to node 35), and a four base-pair frameshift deletion was derived on the stem branch of Mysticeti (Figure 1; node 27 to node 28). These independent inactivating mutations imply that SWS1 was pseudogenized convergently in the two major subclades of Cetacea (Figure 1, Figure S5).
Tab. 1. Summary of inactivating mutations in cetacean SWS1 and LWS genes. 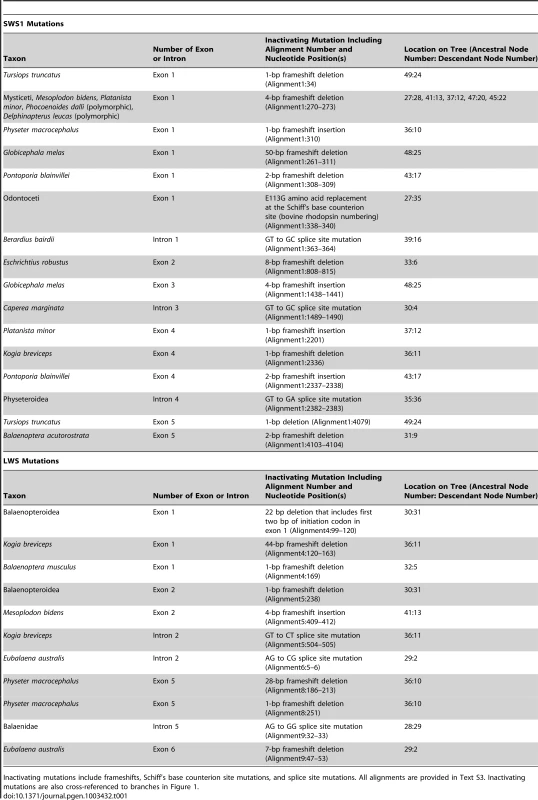
Inactivating mutations include frameshifts, Schiff's base counterion site mutations, and splice site mutations. All alignments are provided in Text S3. Inactivating mutations are also cross-referenced to branches in Figure 1. Estimates of ω (dN/dS) on different branches of the cetacean tree are consistent with parallel knockouts of SWS1 in Odontoceti and in Mysticeti. The ω estimate for SWS1 on the stem Cetacea branch (Figure 1, node 26 to node 27), just prior to the two inferred inactivating mutations, suggests a pattern of purifying selection based on analyses with two different codon frequency models (ω = 0.31, 0.35). Likewise, a signature of strong purifying selection (ω = 0.16, 0.17) was inferred on the stem Odontoceti branch (node 27 to node 35) (Table 2). Neutrality is predicted on the stem odontocete branch if SWS1 had previously been inactivated on the stem cetacean branch [19], but statistical tests rejected this hypothesis (Table 2). The ω estimate (0.75, 0.83) for the stem mysticete branch (node 27 to node 28), in turn, is only slightly lower than expected for complete neutrality (ω = 1.0) and suggests that pseudogenization occurred very early on this branch. Estimates of ω for crown odontocete + crown mysticete branches are in agreement with expectations for neutrality (Table 2), and in conjunction with numerous frameshift indels within these clades (Table 1) imply a release from selective constraints after the occurrence of inactivating mutations on the stem odontocete and stem mysticete branches (Figure 1).
Tab. 2. Results of χ2-tests for nonsynonymous and synonymous substitutions in SWS1. 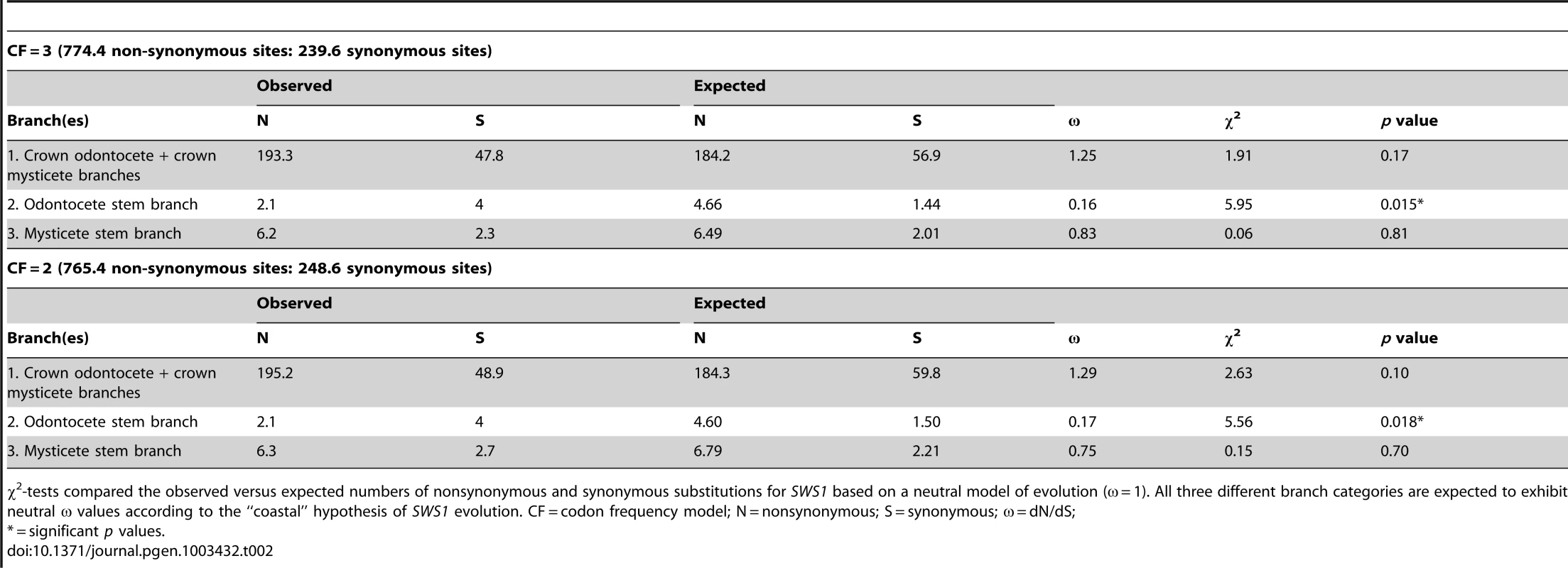
χ2-tests compared the observed versus expected numbers of nonsynonymous and synonymous substitutions for SWS1 based on a neutral model of evolution (ω = 1). All three different branch categories are expected to exhibit neutral ω values according to the “coastal” hypothesis of SWS1 evolution. CF = codon frequency model; N = nonsynonymous; S = synonymous; ω = dN/dS; RH1 Evolution
No inactivating mutations (frameshift indels, splice site mutations) were apparent in the RH1 alignment, implying that RH1 is functional in all of the species that were surveyed (Figure 1).
Ancestral amino acid sequences at key tuning sites (83, 292, 299) in Cetacea [21], [25] are shown in Table 3 for internal nodes with inferred blue or red shifts in λmax. Amino acid changes from DAS to NSS on the stem cetacean branch [node 26 to node 27] resulted in an inferred blue shift from 501 to 484 nm. Additional blue shifts (484 to 479 nm) are inferred in Caperea, in stem Physeteroidea (node 35 to node 36), and in stem Ziphiidae (node 38 to node 39) based on an amino acid changes at site 299 (NSS to NSA) that occurred independently in these three lineages. Seven red shifts were reconstructed in Cetacea, including three in Mysticeti (stem Balaenidae [node 28 to node 29], Megaptera, Eschrichtius) and four in Odontoceti (stem Iniidae + Pontoporiidae [node 42 to node 43], Pontoporia, stem Monodontoidea [node 44 to node 45], Delphinapterus) (Table 3).
Tab. 3. Amino acids at key tuning sites for RH1 [21], [25] and LWS [36]. ![Amino acids at key tuning sites for RH1 <em class="ref">[21]</em>, <em class="ref">[25]</em> and LWS <em class="ref">[36]</em>.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/82187dfc066de615fa2fcb42a4b82d34.png)
Node numbers correspond to Figure 1. RH1 λmax values are based on Fasick et al. [21] for DAS and Bischoff et al. [25] for all other amino acid combinations. LWS λmax values are based on Fasick et al. [21]. Among ten other amino acid sites that have been linked to spectral tuning in vertebrates [28], eight (sites 96, 102, 122, 183, 253, 261, 289, 317) are invariant among the cetaceans and the hippopotamid that were included in our taxon sampling, site 194 exhibits four amino acid replacements within Cetacea, and site 195 shows an amino acid replacement [L to P] on the stem Cetacea branch and four replacements within Cetacea.
Analyses with Codeml rejected site models 2 and 8, which add an extra category for positively selected sites, in favor of models 1 and 8a, respectively. By contrast, branch-site analyses with two different codon frequency models (CF) provided statistically significant support for a bin of five positively selected sites (7, 83, 123, 266, 292) on branches with λmax changes (CF2: P = 0.00036, ω = 5.53; CF3: P = 0.00018, ω = 6.43). Three of the five positively selected sites (83, 266, 292) have probabilities >0.95 of membership in this bin.
LWS Evolution
Inactivating mutations are apparent in LWS sequences from ten cetacean species (Figure 1, Figure S6, Table 1). Reconstructions of ancestral sequences imply eight frameshift indels and three splice site disruptions within Cetacea, with convergent inactivation of LWS on the following five branches (Figure 1): Physeter macrocephalus, Kogia breviceps, Mesoplodon bidens, stem Balaenidae (node 28 to node 29), and stem Balaenopteroidea (node 30 to node 31). All of these separate knockouts of LWS postdate prior inactivations of SWS1 and therefore result in rod monochromacy (Figure 1).
Estimates of ω throughout the species tree generally are consistent with multiple, independent knockouts of LWS within Cetacea. Branches reconstructed as functional for LWS exhibit a strong signature of purifying selection (ω = 0.09, 0.10). By contrast, ω estimates on “transitional” branches [16], where inactivating mutations in LWS were reconstructed, generally show elevated rates of nonsynonymous substitution (Physeter: ω = 0.38, 0.41, Kogia: ω = 0.73, 0.78, stem balaenopteroid branch: ω = 0.34, 0.37). Exceptions are the short transitional branches for stem Balaenidae (1.5 to 1.7 inferred substitutions, ω = 0.0001) and Mesoplodon (2.4 to 3.1 inferred substitutions, ω = 0.13, 0.19). Branches within crown Balaenopteroidea (node 31 and descendant branches) plus crown Balaenidae (node 29 and descendant branches), which are interpreted as pseudogenic based on the prior occurrence of inactivating mutations, have an ω value based on two codon models (0.69, 0.70) that does not deviate significantly from neutral expectations (ω = 1.00) based on χ2-tests.
Reconstructions of ancestral amino acid sequences at five key tuning sites (amino acids 180, 197, 277, 285, 308) [21], [25] are shown in Table 3 for branches with inferred shifts in λmax. For the five tuning sites, AHYTA (λmax = 552 nm) is the inferred ancestral condition for Cetancodonta (node 26) and for the last common ancestor of extant cetaceans (node 27). Three changes at LWS tuning sites were reconstructed within Cetacea. Parallel changes from AHYTA to AHYTS on the stem Mysticeti branch [node 27 to node 28] and on the stem Delphinoidea branch (node 42 to node 44) imply blue shifts from 552 nm to 522–531 nm. A change from AHYTA to AHYTP was reconstructed on the terminal Inia branch, but the functional effect of A308P is unknown (Table 3).
Analyses with Codeml rejected site models 2 and 8, which add an extra category for positively selected sites, in favor of models 1 and 8a, respectively. Similarly, positively selected sites were not identified in branch-site analyses.
Discussion
Opsin Evolution in Cetacea
Here, we assembled complete or nearly complete protein-coding sequences for RH1, SWS1, and LWS for representatives of all extant cetacean families. These sequences, in combination with molecular evolutionary analyses, permit a detailed, synthetic reconstruction of opsin evolution in Cetacea (Figure 1).
Recent phylogenetic hypotheses imply that the aquatic ancestry of Cetacea extends back to its last common ancestor with the semi-aquatic Hippopotamidae in the early Eocene, >50 Ma [4], [29], [30]. Whales and hippos share a variety of “aquatic” specializations including sparse hair, loss of sebaceous glands, and the ability to birth and nurse underwater [4], [31], [32], but these features traditionally have been interpreted as parallel evolutionary derivations in these two lineages. Given the hypothesis that the common ancestor of cetaceans and hippos was aquatic/semi-aquatic (Figure 1, node 50 to node 26), shared mutations in opsin genes that enhance vision in aquatic environments might be expected in whales and hippos. ML reconstructions of ancestral opsin sequences imply only two replacement substitutions (LWS: E41D; RH1: L216M) on the stem lineage. These replacements are not at key tuning sites and fail to provide compelling evidence for an aquatic shift in opsin properties in the common ancestor of hippos and whales.
Following divergence from Hippopotamidae, the unique evolutionary history of Cetacea began on the stem cetacean branch (Figure 1, node 26 to node 27). The fossil record indicates that the stem cetacean lineage was marked by a profound transition in anatomy from primitive semi-aquatic forms to obligately aquatic taxa with vestigial hindlimbs [3], [33]–[35]. Ancestral reconstructions imply that stem cetaceans retained dichromatic color vision with functional SWS1, LWS, and RH1 as in Hippopotamus and more distantly related artiodactyls; a blue shift in RH1 also occurred on the stem cetacean branch (Figure 1). Specifically, the amino acid array at three key tuning sites (83, 292, 299) [21], [25] changed from DAS to NSS, with an inferred λmax shift from 501 to 484 nm. Our ML reconstruction supports Bischoff et al.'s [25] hypothesis that RH1 was blue shifted on the stem cetacean branch, but contradicts their assertion that the ancestral cetacean expressed the amino acids NSA as in deep-diving physeteroids.
In addition to replacements at sites 83 and 292, a change at tuning site 195 (K to T) of RH1 occurred on the stem cetacean branch. This change from a polar amino acid to a positively charged residue has been retained in the deep-diving physeteroids (giant sperm whale, pygmy sperm whale). The inferred shift in λmax that results from a K to T replacement at this site, if any, remains to be investigated with mutagenesis studies. Unlike tuning sites 83, 292, and 299, that are situated in transmembrane regions of RH1 and are in close proximity to the chromophore, site 195 is positioned in the luminal face of RH1 [36]. The nature of long distance interactions between this amino acid site and the chromophore are unknown [36].
The basal split in Cetacea defines the separation of Odontoceti from Mysticeti, and also marks the evolution of profound changes in anatomy/feeding strategy in both clades [4], [37]. Echolocation capabilities and degradation of olfactory structures were derived on the stem odontocete branch (Figure 1, node 27 to node 35), whereas the transition to bulk filter feeding with a keratinous baleen sieve evolved on the stem mysticete branch (Figure 1, node 27 to node 28). These divergent specializations represent changes in feeding style that would be expected to impact demands on visual systems.
Following the blue shift in RH1 on the stem cetacean branch, SWS1 was inactivated independently in stem odontocetes and in stem mysticetes, coincident with the evolution of divergent specializations in these two clades (Figure 1). Two lines of evidence support this reconstruction and argue against an earlier knockout of SWS1 in the common ancestor of Cetacea. First, comprehensive sequencing of SWS1 exons and introns revealed no shared inactivating mutations common to all extant cetaceans. Odontocetes have a common missense mutation at the Schiff's base counterion site (E113G) that disrupts opsin-chromophore binding [23]. Mysticetes, in turn, share a 4-bp frameshift mutation in exon 1 of SWS1 that results in a premature stop codon. Frameshift indels in the same position occur in several odontocetes, but these deletions are most parsimoniously reconstructed as convergent between Mysticeti and multiple odontocete subclades (Text S1). Several mutations that disrupt intron boundaries were identified, but in all cases these substitutions map to branches within Odontoceti or within Mysticeti. Second, dN/dS values on the stem odontocete and stem mysticete branches should indicate an absence of selective constraints if SWS1 was inactivated earlier in the common ancestor of Cetacea. Estimates of dN/dS (0.75, 0.83) for the stem mysticete branch are consistent with neutral evolution (dN/dS = 1.00), but neutrality was rejected given the low dN/dS estimates (0.16, 0.17) for the stem odontocete branch, indicative of purifying selection and thus functionality after the split between Odontoceti and Mysticeti (Figure 1, node 27; Table 2).
Together, our reconstructions for the evolution of RH1 and SWS1 contradict the coastal knockout hypothesis [19], [24]. This scenario postulates that SWS1 was inactivated during an early amphibious phase of cetacean history when semi-aquatic whales occupied coastal waters that absorbed blue light, and that RH1 was subsequently blue shifted in crown cetaceans that moved to open ocean environments dominated by blue light. The coastal knockout hypothesis requires an as yet undiscovered inactivating mutation in SWS1, perhaps in the promoter region of this gene or at splice sites [23]. Instead, our results fit the hypothesis that RH1 was blue shifted in the common ancestor of Cetacea, and that SWS1 was convergently knocked out in Odontoceti and in Mysticeti after cetaceans had invaded open ocean habitats. This is perhaps surprising given that SWS1 is well suited to detect the blue light that dominates the open ocean. However, the relative scarcity of S cones in the mammalian retina, which diminishes the efficiency of photon capture under dim light conditions, may have predisposed S-cones to eventual loss through relaxed selection [22]. By contrast, rods are much more efficient at photon capture under dim light conditions because of their higher density in the mammalian retina and their integration with large, sparsely distributed ganglion cells that sum photon detection over huge receptive fields [38]. The preferential retention of a functional copy of LWS rather than SWS1 in some cetaceans may reflect the higher density of L-cones and their greater impact on visual acuity [22].
In addition to the blue shift of RH1 in the ancestral cetacean branch, several amino acid replacements in RH1 imply further adjustments in λmax (Figure 1). These shifts are generally consistent with the photic environments that are occupied by different cetacean species [20], [25]. Among these changes are blue shifts in deep-diving physeteroids [sperm whales] and in ziphiids [beaked whales], a red shift in the common ancestor of Inia and Pontoporia, both of which are found in shallow water environments, and red shifts in several mysticetes [e.g., Eschrichtius, Megaptera]. Bischoff et al. [25] suggested that the red-shifted pigments that occur in some mysticetes are better adapted to relatively shallower foraging environments than the ancestral mysticete pigment. The blue shifts in Physeteroidea and Ziphiidae occured in parallel and in both cases involve amino acid replacements [serine to alanine] at tuning site 299 [21], [25]. Sperm whales and beaked whales rank among the deepest diving mammals and specialize on a cephalopod-rich diet [39]. Several phylogenetic studies of anatomical evidence grouped these suction feeding species, presumably based on convergent character states related to their deep diving habits [40], [41], but most recent work indicates that ziphiids are more closely related to dolphins and porpoises than to physeteroids [37], [42], [43].
Nozawa et al. [44] suggested that Yang's [45] codeml program is not useful for identifying adaptive sites in visual pigments. Our results support Nozawa et al.'s [44] finding that site analyses fail to identify adaptive changes in visual pigments. However, branch-site tests identified five codons in RH1 that have evolved under positive selection on branches with inferred changes in λmax. The ω value for the five positively selected sites is well above one (5.53–6.43), and supports the hypothesis that changes affecting λmax in cetacean RH1 proteins are adaptive. The failure of site analyses to detect positively selected sites in RH1 may be a consequence of mixing positive selection on foreground branches with purifying selection on background branches. Nozawa et al. [44] criticized branch-site tests [45]–[47] for their proclivity to generate false positive results based on simulations, but Yang et al. [48] correctly noted that false positives only occurred in 32/14,000 cases, which is much lower than the nominal significance level (5%) and demonstrates that the branch-site test is conservative. Among the positively selected sites, two (83, 292) are known tuning sites that in part are the basis for inferring changes in λmax (Figure 1). Changes at site 83 may also be important in dim-light conditions because the amino acid at this position affects the rate at which photoreceptor cells generate electrical signals [49]. The other three sites (7, 123, 266) have not been predicted to affect λmax. Site 7 occurs in the extracellular domain, site 123 occurs in transmembrane helix III, and site 266 occurs in transmembrane helix 6 [50]. The functional consequences of mutations at these amino acid positions in cetacean RH1 sequences remain unknown, although conformational changes associated with transmembrane domains III and VI of G protein-coupled receptors may be important in receptor activation [51].
Changes in LWS spectral sensitivity coincide with deployment of cetaceans to diverse aquatic habitats (Figure 1). A blue shift in LWS in stem mysticetes co-occurs with an SWS1 frameshift mutation on the same branch, although the sequence of these events is unclear. An additional LWS blue shift in λmax maps to the common ancestor of Delphinoidea [dolphins, porpoises, beluga], but the most striking feature of LWS evolution in Cetacea is the convergent knockout of this gene in five different lineages: Balaenopteroidea (rorquals and gray whale), Balaenidae (bowhead and right whale), Mesoplodon bidens (Sowerby's beaked whale), Physeter macrocephalus (giant sperm whale), and Kogia breviceps (pygmy sperm whale) (Figure 1). Given that SWS1 is also debilitated in each of these species (Figure 1), the genetic data imply that these taxa are rod monochromats. This iterated degeneration of cetacean LWS was not apparent in earlier studies because complete protein-coding LWS sequences had been generated for only a few cetacean species [21].
Rod Monochromacy in Cetacea
Historically, the pure rod retina has been proposed as the “extreme” adaptation to low light levels [52]. Walls [52] and McFarland [20] suggested the possibility of rod monochromacy in at least some cetaceans. More generally, early studies on retinal anatomy hinted at this condition in a variety of nocturnal and aquatic mammalian species with rod dominated retinas, including night monkeys, lemurs, tarsiers, chinchillas, seals, and bats [52]–[57]. Recent work has shown that representative cetaceans are instead L-cone monochromats and retain a functional copy of LWS [21], [26]. Similarly, primates, rodents, pinnipeds, and bats that were previously hypothesized to be rod monochromats are now known to be L-cone monochromats with functional LWS or even cone dichromats with functional LWS and SWS1 [22], [58]–[61]. The present survey of cetacean opsins, which documents pseudogenization of both SWS1 and LWS in multiple cetacean lineages, vindicates McFarland's [20] hypothesis that some cetaceans are rod monochromats (Figure 1). To our knowledge these are the only known examples of rod monochromacy in Mammalia or even Amniota. The observation that five independent derivations of mammalian rod monochromacy are all clustered within Cetacea is striking, and suggests that one or more features of cetacean biology have been pivotal in driving the degenerative pattern of opsin evolution in this aquatic clade.
The naked mole rat (Heterocephalus glaber) is the only other mammal, aside from the cetacean species characterized here, that is known to lack a functional copy of LWS, but H. glaber retains an intact SWS1 and is interpreted as a cone monochromat [62]. This condition contrasts with other mammalian cone monochromats [pinnipeds, dolphins, porpoises, some procyonids, some rodents, some bats] that combine a pseudogenic SWS1 with a functional copy of LWS [22], [23], [28], [60], [63]–[71]. It has been suggested that cetacean cone monochromats [e.g. bottlenose dolphin, Tursiops truncatus] can distinguish colors, possibly via interactions between LWS and RH1 [38], [72], but any vestiges of color vision presumably have been lost in the various rod monochromatic cetacean species documented here (Figure 1).
Among other vertebrates, rod monochromatic taxa are rare and to our knowledge have only been documented in bony and cartilaginous fishes [73]–[81], caecilians [22], [82], and the cave salamander Proteus anguinus [83], although presumed rod monochromacy based entirely on immunocytochemistry, microscopy or spectral analysis does not preclude the possibility that other minor visual pigment classes exist [77], [81], [83]. Most of the rod monochromatic fish species inhabit the deep sea or are nocturnal; caecilians are generally fossorial and/or nocturnal with poorly developed eyes; and the cave salamander Proteus lives in a virtually light-free environment.
The phylogenetic evidence for multiple, independent knockouts of both SWS1 and LWS within Cetacea raises the question of why convergent pseudogenization and rod monochromacy evolved in this clade but not in other mammalian groups. All rod monochromatic cetacean species that were genetically characterized in our survey are capable of diving to depths that exceed 100 m, with sperm and beaked whales ranking among the deepest diving mammals [39], [84]–[88]. The selective pressures on mammalian retinal opsins in deep-water habitats are drastically different from those on land. In the open marine environment, the electromagnetic radiation of visible light is weakened with depth due to absorption and scattering [89]. In the mesopelagic zone (150–1000 m), down-welling sunlight becomes more monochromatic and the spectrum shifts towards shorter, bluer wavelengths with depth [81], [82]. Below 1000 m (bathypelagic zone), there is no down-welling sunlight and localized bluish bioluminescence becomes the predominant source of light [90].
A rod-dominated retina is advantageous in dim light conditions [38]. Therefore, the convergent pseudogenization of SWS1 and LWS in multiple cetacean lineages may be an adaptation to deep-water habitats and/or feeding at night on bioluminescent invertebrate prey. Cone opsins have a higher rate of thermal activation [i.e., dark noise] than RH1 [91] and may interfere with rod sensitivity under scotopic conditions. Thus, combined SWS1 and LWS pseudogenization may have increased RH1 sensitivity in physeteroids and ziphiids that feed in the mesopelagic and bathypelagic zones. Echolocation is a key specialization that has enabled odontocete taxa such as these to forage at night and at great depths on individual prey items, in particular cephalopods [92]; rod monochromatism may be an additional adaptive feature that has enabled predation at depth. Balaenopteroid and balaenid mysticetes are not known to feed in the bathypelagic zone, do not echolocate, and instead batch filter aggregations of small prey items. However, baleen whales do feed at night and much of their diet is composed of bioluminescent prey including krill [93], [94]. The ability to take advantage of this huge resource offers a compelling selective driver on the evolution of visual systems in Mysticeti, and the detection of schools of tiny prey at night would seem to be problematic without echolocation. The reliance of various mysticete species on RH1 might represent one solution for improved night vision given that rods are more useful than cones for contrast detection and hence picking out schools of prey from the background. Along these lines, the parallel pseudogenization of both cone opsins in Cetacea (Figure 1) could be the result of natural selection favoring an all-rod retina, in which case cone opsins were either selected against because of interference with RH1, or were rendered ‘jobless’ by the elimination of cones and released from selective constraints on color vision in this aquatic clade [22].
Conclusions
The emergence of Cetacea represents a profound macroevolutionary transition that entails comprehensive remodeling at both the genetic and morphological levels [4]. Our results elucidate key events in the evolutionary history of cetacean opsins, including an initial blue shift of RH1 in stem Cetacea, parallel knockouts of SWS1 in Odontoceti and Mysticeti, and five independent inactivations of LWS in deep-diving cetacean lineages. As correctly surmised by McFarland [20], some cetaceans are rod monochromats and have evolved eyes that are highly specialized for dim-light vision.
Materials and Methods
Taxon and Gene Sampling
Previously published RH1, SWS1, and LWS sequences for Cetacea were combined with new sequences that were generated through PCR and dideoxy sequencing. We targeted complete coding regions of all three opsin genes for representatives of Hippopotamidae and all extant cetacean families (Text S2). RH1, LWS, and SWS1 sequences for additional cetartiodactyl families [Bovidae, Cervidae, Suidae, Camelidae] were assembled from Ensembl, Pre-Ensembl, and NCBI based on availability with minor augmentation by new sequences (Table S1, Text S2).
PCR and Sequencing
Aligned sequences for Bos taurus, Sus scrofa, Tursiops truncatus, Vicugna pacos, and available GenBank sequences (Table S1) were used to design PCR primers for SWS1, RH1, and LWS. SWS1 (exons 1–4; partial exon 5; introns 1–4) was amplified in five overlapping segments. PCR primers for RH1 (exons 1–5) and LWS (exons 1–6) were positioned in the flanking intronic regions of each exon (see Text S2 for additional details on PCR reactions). Accession numbers for new cetartiodactyl sequences are KC676796–KC677023 (Table S1). Primer sequences are provided in Table S2.
Alignments and Phylogenetic Analyses
Sequences were aligned manually using Se-Al [95]. The virtual mRNA alignment lengths were 1014 base pairs (bp) for SWS1, 1092 bp for LWS, and 1044 bp for RH1. The complete alignment for SWS1, including exons and introns, was 4163 bp. All alignments for phylogenetic and PAML analyses, along with alignments for non-overlapping PCR amplicons (exons plus partial introns for LWS and RH1), are provided in Text S3 in nexus format. Phylogenetic analyses were performed with RAxML 7.2.7 [96] and the GTR + Γ model of sequence evolution. Additional details are provided in Text S2.
Inactivating Mutations
Opsin alignments were manually inspected for putative inactivating mutations, including substitutions that result in stop codons, changes at intron splice donor/acceptor sites, and frameshift indels. We also examined SWS1 sequences for a missense mutation at Schiff's counterion site (E113G; bovine RH1 numbering) that disrupts opsin-chromophore binding [23].
Ancestral Sequence Reconstructions and Character State Mapping
Ancestral DNA sequences for SWS1, LWS, and RH1 were reconstructed with the Baseml program implemented in PAML 4.4b [45]. We used the REV model and a composite species tree based on McGowen et al. [42] for cetaceans and Gatesy [97] for all other cetartiodactyls. Frameshift mutations and other indels were optimized with Fitch parsimony, as implemented in Mesquite [98].
λmax Estimation
Spectral tuning in RH1 is influenced by at least 13 amino acid sites [28], although replacements at only three of these sites (83, 292, 299) fully explain the absorbance difference between cow RH1 (Bos taurus, λmax = 500 nm) and bottlenose dolphin RH1 (Tursiops truncatus, λmax = 488 nm) [99]. These replacements are D83N, A292S, and A299S. Different combinations of ancestral and derived amino acids at these three sites have been tested in mutagenic studies of Bos RH1 to explain the various λmax values that occur in other cetaceans [21], [25]. For LWS, Yokoyama [36] suggested a “five-sites” rule whereby λmax values between 510 and 560 in vertebrates can be fully explained by amino acid changes S180A, H197Y, Y277F, T285A, A308S and their interactions. Here, we follow Fasick et al. [21] and Bischoff et al. [25] and provide λmax estimates for newly determined RH1 and LWS sequences based on directly determined λmax values from expressed RH1 and LWS pigments that possess identical amino acids at the same key sites for each of these opsins. It will be important in future studies to perform direct measurements of λmax on reconstructions of ancestral RH1 sequences. Even without these experiments, empirical measurements on a diverse array of opsins from cetacean species and Bos taurus (wild type and mutagenesis variants) provide a strong foundation for inferring λmax values in ancestral cetacean sequences [21], [25].
dN/dS Analyses
The Codeml program in PAML 4.4b [45] was used to estimate the ratio (ω) of the non-synonymous substitution rate [dN] to the synonymous substitution rate (dS) at individual sites (RH1, LWS) and on branches (SWS1, LWS). We also performed branch-site analyses [45]–[47], [100] on RH1 and LWS sequences. In both cases, branches with predicted changes in λmax (Figure 1) of the relevant opsin were assigned to the foreground, and all other branches were assigned to the background. We used a composite species tree for all cetartiodactyl taxa as detailed above. Statistical tests of neutrality [complete absence of functional constraints] for branches and sets of branches were executed as in Meredith et al. [16]. See Text S2 for details.
Supporting Information
Zdroje
1. ThewissenJGM, BajpaiS (2001) Whale origins as a poster child for macroevolution. BioScience 51 : 1037–1049.
2. ThewissenJGM, CooperLN, GeorgeJC, BajpaiS (2009) From land to water: the origin of whales, dolphins, and porpoises. Evo Edu Outreach 2 : 272–288.
3. UhenMD (2010) The origin(s) of whales. Annu Rev Earth Planet Sci 38 : 189–219.
4. GatesyJ, GeislerJH, ChangJ, BuellC, BertaA, et al. (2013) A phylogenetic blueprint for a modern whale. Mol Phylogenet Evol 66 : 479–506.
5. LiY, LiuZ, ShiP, ZhangJ (2010) The hearing gene Prestin unites echolocating bats and whales. Curr Biol 20: R55–R56.
6. LiuY, RossiterS, HanX, CottonJ, ZhangS (2010) Cetaceans on a molecular fast track to ultrasonic hearing. Curr Biol 20 : 1834–1839.
7. DaviesKTJ, CottonJA, KirwanJD, TeelingEC, RossiterSJ (2012) Parallel signatures of sequence evolution among hearing genes in echolocating mammals: an emerging model of genetic convergence. Heredity 108 : 480–489.
8. McGowenMR, MontgomerySH, ClarkC, GatesyJ (2011) Phylogeny and adaptive evolution of the brain-development gene microcephalin (MCPH1) in cetaceans. BMC Evol Biol 11 : 98.
9. XuS, ChenY, ChengY, YangD, ZhouX, et al. (2012) Positive selection at the ASPM gene coincides with brain size enlargements in cetaceans. Proc R Soc B 279 : 4433–4440.
10. WangZ, YuanL, RossiterSJ, ZuoX, RuB, et al. (2009) Adaptive evolution of 5′HoxD genes in the origin and diversification of the cetacean flipper. Mol Biol Evol 26 : 613–622.
11. KishidaT, KubotaS, ShirayamaY, FukamiH (2007) The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: evidence for reduction of the functional proportions in cetaceans. Biol Lett 3 : 428–430.
12. McGowenMR, ClarkC, GatesyJ (2008) The vestigial olfactory receptor subgenome of odontocete whales: phylogenetic congruence between gene-tree reconciliation and supermatrix methods. Syst Biol 57 : 574–590.
13. HaydenS, BekaertM, CriderTA, MarianiS, MurphyWJ, et al. (2010) Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res 20 : 1–9.
14. JiangP, JosueJ, LiX, GlaserD, LiW, et al. (2012) Major taste loss in carnivorous mammals. Proc Natl Acad Sci USA 109 : 4956–4961.
15. DeméréTA, McGowenMR, BertaA, GatesyJ (2008) Morphological and molecular evidence for a stepwise evolutionary transition from teeth to baleen in mysticete whales. Syst Biol 57 : 15–37.
16. MeredithRW, GatesyJ, MurphyWJ, RyderOA, SpringerMS (2009) Molecular decay of the tooth gene enamelin (ENAM) mirrors the loss of enamel in the fossil record of placental mammals. PLoS Genet 5: e1000634 doi:10.1371/journal.pgen.1000634.
17. MeredithRW, GatesJ, ChengJ, SpringerMS (2011) Pseudogenization of the tooth gene enamelysin (MMP20) in the common ancestor of extant baleen whales. Proc Roy Soc Lond B 278 : 993–1002.
18. YuL, JinW, WangJ, ZhangX, ChenM, et al. (2010) Characterization of TRPC2, an essential genetic component of VNS chemoreception, provides insights into the evolution of pheromonal olfaction in secondary-adapted marine mammals. Mol Biol Evol 27 : 1467–1477.
19. GriebelU, PeichlL (2003) Colour vision in aquatic mammals - facts and open questions. Aquat Mamm 29 : 18–30.
20. McFarlandWN (1971) Cetacean visual pigments. Vision Res 11 : 1065–1076.
21. FasickJI, BischoffN, BrennanS, VelasquezS, AndradeG (2011) Estimated absorbance spectra of the visual pigments of the North Atlantic right whale (Eubalaena glacialis). Mar Mamm Sci 27: E321–E331.
22. DaviesWIL, CollinSP, HuntDM (2012) Molecular ecology and adaptation of visual photopigments in craniates. Mol Ecol 21 : 3121–3158.
23. LevensonDH, DizonA (2003) Genetic evidence for the ancestral loss of SWS cone pigments in mysticete and odontocete cetaceans. Proc Roy Soc London B 270 : 673–679.
24. PeichlL (2005) Diversity of mammalian photoreceptor properties: adaptations to habitat and lifestyle. Anat Rec 287A: 1001–1012.
25. BischoffN, NickleB, CroninTW, VelasquezS, FasickJI (2012) Deep-sea and pelagic rod visual pigments identified in the mysticete whales. Vis Neurosci 29 : 95–103.
26. PeichlL, BehrmannG, KrogerRHH (2001) For whales and seals the ocean is not blue: A visual pigment loss in marine mammals. Eur J Neurosci 13 : 1520–1528.
27. NewmanLA, RobinsonPR (2005) Cone visual pigments of aquatic mammals. Vis Neurosci 22 : 873–879.
28. ZhaoH, RuB, TeelingEC, FaulkesCG, ZhangS, et al. (2009) Rhodopsin molecular evolution in mammals inhabiting low light environments. PLoS ONE 4: e8326 doi:10.1371/journal.pone.0008326.
29. GeislerJH, TheodorJM (2009) Hippopotamus and whale phylogeny. Nature 458: E1–E4.
30. SpauldingM, O'LearyMA, GatesyJ (2009) Relationships of Cetacea (Artiodactyla) among mammals: increased taxon sampling alters interpretations of key fossils and character evolution. PLoS ONE 4: e7062 doi:10.1371/journal.pone.0007062.
31. GatesyJ, HayashiC, CroninA, ArctanderP (1996) Evidence from milk casein genes that cetaceans are close relatives of hippopotamid artiodactyls. Mol Biol Evol 13 : 954–963.
32. GatesyJ (1997) More DNA support for a Cetacea/Hippopotamidae clade: the blood-clotting protein gene gamma-fibrinogen. Mol Biol Evol 14 : 537–543.
33. GingerichPD, HaqMU, ZalmoutIS, KhanIH, MalakaniMS (2001) Origin of whales from early artiodactyls: hands and feet of Eocene Protocetidae from Pakistan. Science 293 : 2239–2242.
34. ThewissenJGM, WilliamsEM, RoeLJ, HussaiST (2001) Skeletons of terrestrial cetaceans and the relationship of whales to artiodactyls. Nature 413 : 277–281.
35. ThewissenJGM, CooperLN, ClementzMT, BajpaiS, TiwariBN (2007) Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 450 : 1190–1194.
36. YokoyamaS (2008) Evolution of dim-light and color vision pigments. Annu Rev Genomics Hum Genet 9 : 259–282.
37. GeislerJH, McGowenRM, YangG, GatesyJ (2011) A supermatrix analysis of genomic, morphological, and paleontological data from crown Cetacea. BMC Evol Biol 11 : 112.
38. Reuter T, Peichl L (2008) Structure and function of the retina in aquatic tetrapods. In: Thewissen JGM, Nummela S, eds. Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates. Berkeley and Los Angeles, California: Univ California Press. pp 149–174.
39. BloodworthBE, OdellDK (2008) Kogia breviceps (Cetacea: Kogiidae). Mamm Species 819 : 1–12.
40. FordyceRE (1994) Waipatia maerewhenua, new genus and new species (Waipatiidae, new family), an archaic late Oligocene dolphin (Cetacea: Odontoceti: Platanistoidea) from New Zealand. Proc San Diego Soc Nat Hist 29 : 147–176.
41. GeislerJH, SandersAE (2003) Morphological evidence for the phylogeny of Cetacea. J Mammal Evol 10 : 23–129.
42. McGowenMR, SpauldingM, GatesyJ (2009) Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol Phylogenet Evol 53 : 891–906.
43. SteemanME, HebsgaardMB, FordyceRE, HoSYW, RaboskyDL, et al. (2009) Radiation of extant cetaceans driven by restructuring of the oceans. Syst Biol 58 : 573–585.
44. NozawaM, SuzukiY, NeiM (2009) Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci USA 106 : 6700–6705.
45. YangZ (2007) PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol 24 : 1586–1591.
46. YangZ, WongWSW, NielsenR (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22 : 1107–1118.
47. ZhangJ, NielsenR, YangZ (2005) Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22 : 2472–2479.
48. YangZ, NielsenR, GoldmanN (2009) In defense of statistical methods for detecting positive selection. Proc Natl Acad Sci USA 106: E95.
49. SugawaraT, ImaiH, NikaidoM, ImamotoY, OkadaN (2010) Vertebrate rhodopsin adaptation to dim light via rapid meta-II intermediate formation. Mol Biol Evol 27 : 506–519.
50. RaderAJ, AndersonG, IsinB, KhoranaHG, BaharI, et al. (2004) Identification of core amino acids stabilizing rhodopsin. Proc Natl Acad Sci USA 101 : 7246–7251.
51. GetherU, LinS, GhanouniP, BallesterosJA, WeinsteinH, et al. (1997) Agonists induce conformational changes in transmembrane domains III an VI of the β2 adrenoceptor. EMBO J 16 : 6737–6747.
52. Walls GL (1942) The vertebrate eye and its adaptive radiation. Bloomfield Hills, Michigan: Cranbrook Institute of Science.
53. WoollardHH (1927) The differentiation of the retina in the primates. Proc Zool Soc Lond 97 : 1–18.
54. WoollardHH, BeattieJ (1927) The comparative anatomy of the lateral geniculate body. J Anat 61 : 414–423.
55. DetwilerSR (1941a) The eye of the owl monkey (Nyctipithecus). Anat Rec 80 : 233–241.
56. DetwilerSR (1941b) Some biological aspects of vision. Sigma Xi Quart 29 : 112–129.
57. DetwilerSR (1956) The eye and its structural adaptations. Am Sci 44 : 45–72.
58. NagaoK, TakenakaN, HiraiM, KawamuraS (2005) Coupling and decoupling of evolutionary mode between X - and Y-Chromosomal red-green opsin genes in owl monkeys. Gene 352 : 32–91.
59. TanY, YoderAD, YamashitaN, LiW-H (2005) Evidence from opsin genes rejects nocturnality in ancestral primates. Proc Natl Acad Sci USA 102 : 14712–14716.
60. MüllerB, GoodmanSM, PeichlL (2007) Cone photoreceptor diversity in the retinas of fruit bats (Megachiroptera). Brain Behav Evol 70 : 90–104.
61. BowmakerJK (2008) Evolution of vertebrate visual pigments. Vision Res 48 : 2022–2041.
62. KimEB, FangX, FushanAA, HuangZ, LobanovAB, et al. (2011) Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479 : 223–227.
63. JacobsGH, DeeganDFII (1992) Cone photopigments in nocturnal and diurnal procyonids. J Comp Physiol A 171 : 351–358.
64. JacobsGH, DeeganDFII, NeitzJ, CrognaleMA, NeitzM (1993) Photopigments and color vision in the nocturnal monkey, Aotus. Vision Res 33 : 1773–1783.
65. JacobsGH, NeitzM, NeitzJ (1996) Mutations in S-cone pigment genes and the absence of colour vision in two species of nocturnal primate. Proc Royal Soc London B 263 : 705–710.
66. DeeganDFII, JacobsGH (1996) Spectral sensitivity and photopigments of a nocturnal prosimian, the bushbaby (Otolemur crassicaudatus). Am J Primatol 40 : 55–66.
67. PeichlL, MoutairouK (1998) Absence of short-wavelength sensitive cones in the retinae of seals (Carnivora) and African giant rats (Rodentia). Eur J Neurosci 10 : 2586–2594.
68. CrognaleMA, LevensonDH, PonganisPJ, DeeganJF, JacobsGH (1998) Cone spectral sensitivity in the harbor seal (Phoca vitulina) and implications for color vision. Can J Zool 76 : 2114–2118.
69. KawamuraS, KuboteraN (2004) Ancestral loss of short wave-sensitive cone visual pigment in Lorisiform prosimians, contrasting with its strict conservation in other prosimians. J Mol Evol 58 : 314–321.
70. CarvalhoLPS, CowinJA, WilkieSE, BowmakerJK, HuntDM (2006) Shortwave visual sensitivity in tree and flying squirrels reflects changes in life style. Curr Biol 16: R81–R83.
71. JacobsGH (2009) Evolution of colour vision in mammals. Phil Trans R Soc B 364 : 2957–2967.
72. GriebelU, SchmidA (2002) Spectral sensitivity and color vision in the bottlenose dolphin (Tursiops truncatus). Mar Fresh Behav Physiol 35 : 129–137.
73. PartridgeJC, ShandJ, ArcherSN, LythgoeJN, van Groningen-LuybenWAHM (1989) Interspecific variation in the visual pigments of deep-sea fishes. J Comp Physiol A 164 : 513–529.
74. RippsH, DowlingJE (1991) Structural features and adaptive properties of photoreceptors in the skate retina. J Exp Zool Suppl 5 : 46–54.
75. DouglasRH, PartridgeJC, HopeAJ (1995) Visual and lenticular pigments in the eyes of demersal deep-sea fishes. J Comp Physiol A 177 : 111–122.
76. O'BrienJ, RippsH, Al-UbaidiMR (1997) Molecular cloning of a rod opsin cDNA from the skate retina. Gene 193 : 141–150.
77. BozzanoA, MurgiaR, Vallerga, HiranoJ, ArcherS (2001) The photoreceptor system in the retinae of two dogfishes, Scyliorhinus canicula and Galeus melastomus: possible relationship with depth distribution and predatory lifestyle. J Fish Biol 59 : 1258–1278.
78. BozzanoA (2004) Retinal specialisations in the dogfish Centroscymnus coelolepis from the Mediterranean deep-sea. Scientia Marina 68 (Suppl 3) 185–195.
79. HartNS, TheissSM, HarahushBK, CollinSP (2011) Microspectrophotometric evidence for cone monochromacy in sharks. Naturwissenschaften 98 : 193–201.
80. LisneyTJ, TheissSM, CollinsSP, HartNS (2012) Vision in elasmobranchs and their relatives: 21st century advances. J Fish Biol 80 : 2024–2054.
81. SchieberNL, CollinS, HartNS (2012) Comparative retinal anatomy in four species of elasmobranch. J Morphol 273 : 423–440.
82. MohunSM, DaviesWL, BowmakerJK, PisaniD, HimstedtW, et al. (2010) Identification and characterization of visual pigments in caecilians (Amphibia: Gymnophiona), an order of limbless vertebrates with rudimentary eyes. J Exp Biol 213 : 3586–3592.
83. KosM, BulogB, SzélA, RöhlichP (2001) Immunocytochemical demonstration of visual pigments in the degenerate retinal and pineal photoreceptors of the blind cave salamander (Proteus anguinus). Cell Tissue Res 303 : 15–25.
84. SchreerJF, KovacsKM (1997) Allometry of diving capacity in air-breathing vertebrates. Can J Zool 75 : 339–358.
85. PanigadaS, ZanardelliM, CaneseS, JahodaM (1999) How deep can baleen whales dive? Mar Ecol Prog Ser 187 : 309–311.
86. AmanoM, YoshiokaM (2003) Sperm whale diving behavior monitored using a suction-cup-attached TDR tag. Mar Ecol Prog Ser 258 : 291–295.
87. CrollDA, MarinovicB, BensonS, ChavezFP, BlackN, et al. (2005) From wind to whales: trophic links in a coastal upwelling system. Mar Ecol Prog Ser 289 : 117–130.
88. WatwoodSL, MillerPJO, JohnsonM, MadsenPT, TyackPL (2006) Deep-diving foraging behavior of sperm whales (Physeter macrocephalus). J Anim Ecol 75 : 814–825.
89. Jerlov NG (1968) Optical oceanography. Amsterdam, The Netherlands: Elsevier Sci Publ Co.
90. WarrantEJ, LocketNA (2004) Vision in the deep sea. Biol Rev 79 : 671–712.
91. FuY, KefalovV, LuoD-G, XueT, YauK-W (2008) Quantal noise from human red cone pigment. Nature Neurosci 11 : 565–571.
92. LindbergDR, PyensonND (2007) Things that go bump in the night: evolutionary interactions between cephalopods and cetaceans in the tertiary. Lethaia 40 : 335–343.
93. WareC, FriedlaenderAS, NowacekDP (2010) Shallow and deep lunge feeding of humpback whales in fjords of the West Antarctic Peninsula. Mar Mamm Sci 27 : 587–605.
94. GoldbogenJA (2011) Mechanics, hydrodynamics and energetics of blue whale lunge feeding: efficiency dependence on krill density. J Exp Biol 214 : 131–146.
95. Rambaut A (1996) Se-Al: sequence alignment editor, version 2.0a11. Available: http://tree.bio.ed.ac.uk/software/seal/.
96. StamatakisA (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 : 2688–2690.
97. Gatesy J (2009) Whales and even-toed ungulates (Cetartiodactyla). In: Hedges SB, Kumar S, eds. The timetree of life. Oxford, UK: Oxford Univ Press. pp 511–515.
98. Maddison WP, Maddison DR (2010) Mesquite: a modular system for evolutionary analysis, version 2.73. Available: http://mesquiteproject.org.
99. FasickJI, RobinsonPR (1998) Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry 37 : 433–438.
100. YangZ, dos ReisM (2011) Statistical properties of the branch-site test of positive selection. Mol Biol Evol 28 : 1217–1228.
101. MeredithRW, JanečkaJE, GatesyJ, RyderOA, FisherCA, et al. (2011) Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334 : 521–524.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



