-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
Chickens, pigs, and cattle are key reservoirs of Salmonella enterica, a foodborne pathogen of worldwide importance. Though a decade has elapsed since publication of the first Salmonella genome, thousands of genes remain of hypothetical or unknown function, and the basis of colonization of reservoir hosts is ill-defined. Moreover, previous surveys of the role of Salmonella genes in vivo have focused on systemic virulence in murine typhoid models, and the genetic basis of intestinal persistence and thus zoonotic transmission have received little study. We therefore screened pools of random insertion mutants of S. enterica serovar Typhimurium in chickens, pigs, and cattle by transposon-directed insertion-site sequencing (TraDIS). The identity and relative fitness in each host of 7,702 mutants was simultaneously assigned by massively parallel sequencing of transposon-flanking regions. Phenotypes were assigned to 2,715 different genes, providing a phenotype–genotype map of unprecedented resolution. The data are self-consistent in that multiple independent mutations in a given gene or pathway were observed to exert a similar fitness cost. Phenotypes were further validated by screening defined null mutants in chickens. Our data indicate that a core set of genes is required for infection of all three host species, and smaller sets of genes may mediate persistence in specific hosts. By assigning roles to thousands of Salmonella genes in key reservoir hosts, our data facilitate systems approaches to understand pathogenesis and the rational design of novel cross-protective vaccines and inhibitors. Moreover, by simultaneously assigning the genotype and phenotype of over 90% of mutants screened in complex pools, our data establish TraDIS as a powerful tool to apply rich functional annotation to microbial genomes with minimal animal use.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003456
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003456Summary
Chickens, pigs, and cattle are key reservoirs of Salmonella enterica, a foodborne pathogen of worldwide importance. Though a decade has elapsed since publication of the first Salmonella genome, thousands of genes remain of hypothetical or unknown function, and the basis of colonization of reservoir hosts is ill-defined. Moreover, previous surveys of the role of Salmonella genes in vivo have focused on systemic virulence in murine typhoid models, and the genetic basis of intestinal persistence and thus zoonotic transmission have received little study. We therefore screened pools of random insertion mutants of S. enterica serovar Typhimurium in chickens, pigs, and cattle by transposon-directed insertion-site sequencing (TraDIS). The identity and relative fitness in each host of 7,702 mutants was simultaneously assigned by massively parallel sequencing of transposon-flanking regions. Phenotypes were assigned to 2,715 different genes, providing a phenotype–genotype map of unprecedented resolution. The data are self-consistent in that multiple independent mutations in a given gene or pathway were observed to exert a similar fitness cost. Phenotypes were further validated by screening defined null mutants in chickens. Our data indicate that a core set of genes is required for infection of all three host species, and smaller sets of genes may mediate persistence in specific hosts. By assigning roles to thousands of Salmonella genes in key reservoir hosts, our data facilitate systems approaches to understand pathogenesis and the rational design of novel cross-protective vaccines and inhibitors. Moreover, by simultaneously assigning the genotype and phenotype of over 90% of mutants screened in complex pools, our data establish TraDIS as a powerful tool to apply rich functional annotation to microbial genomes with minimal animal use.
Introduction
Salmonella enterica is a facultative intracellular pathogen of worldwide importance, associated with c. 21.7 million cases of systemic typhoid fever and 93.8 million cases of non-typhoidal gastroenteritis in humans each year [1], [2]. Around 86% of human cases of non-typhoidal salmonellosis are the result of food-borne infections [2], and chickens, pigs and cattle are key reservoirs of infection [3]. The major S. enterica subspecies enterica encompasses a wide variety of serovars. Some of these, such as S. Typhimurium and S. Enteritidis, exhibit a wide host range, whereas others such as S. Typhi are largely restricted to a single host species. The molecular basis of the host - and tissue-tropism of S. enterica has long eluded researchers and there has been a disproportionate emphasis on the basis of Salmonella persistence and pathogenesis in murine models of colitis and typhoid fever. Comparative analyses of whole genome sequences have associated host-restriction of S. enterica serovars with gene decay. Interpretation of the impact of variation in the repertoire, sequence or expression of S. enterica genes requires an understanding of the roles of those genes in relevant hosts. Random transposon insertion mutants have been screened individually in chickens [4], but high-throughput simultaneous analysis of mutant phenotypes was made possible by the advent of signature-tagged mutagenesis (STM) [5]. STM allows the survival of individual mutants within a pool to be assessed qualitatively through hybridization of a probe to a unique tag sequence within the transposon. Comparison of hybridization signals obtained from “input pools” of mutants grown in vitro with the those obtained from the same set of mutants screened for survival in a model of infection (“output pool”) allows attenuated mutants to be identified. The insertion sites can then be identified by subcloning and sequencing. Analysis of pools of signature-tagged S. Typhimurium mutants in mice [5] led to the discovery of Salmonella Pathogenicity Island (SPI)-2 [6], a gene cluster that encodes a type III secretion system (T3SS-2) that acts as a molecular syringe for the secretion of effector molecules and influences systemic virulence and intracellular survival [7]. A distinct T3SS, encoded by SPI-1, was already known to be essential for infection of mice via the oral route [8]. Subsequently, STM libraries constructed in a range of Salmonella serovars were examined for their ability to colonize multiple hosts [9]–[14]. Comparative analysis of a library of 1045 mutants in chickens, pigs and calves suggested that S. Typhimurium deploys both conserved - and host-specific virulence factors [10], [12]. Notably, SPI-1 and -2 were vital in intestinal colonization of calves and pigs and to a lesser extent chickens [10], [12], yet a Type I protein secretion system encoded by SPI-4 appeared to influence infection of calves, but not chickens [12] or pigs [10].
Although STM has provided valuable insights into Salmonella pathogenesis, the technique is limited by the number of unique tags available, and the time and effort required to construct the library and identify attenuating mutations. Moreover, only negatively-selected mutants tend to be investigated and subjective judgments are used to compare signal intensities relative to the input and to other screened mutants. Transposon-directed insertion-site sequencing (TraDIS) [15], one of a new generation of STM-like techniques, addresses some of these limitations. TraDIS exploits Illumina sequencing [16] to obtain the sequence of the genomic region flanking each transposon. The massively parallel nature of the sequencing permits comparison of the number of specific reads derived from the input pools and the output pools after animal infection, providing a numerical measure of the extent to which mutants were negatively - or positively-selected during colonization (see Figure 1). TraDIS-like sequencing methods have been used to identify the essential gene complement of S. Typhi [15] and Streptococcus pneumoniae [17], genes involved in virulence of Haemophilus influenzae [18] and S. pneumoniae [19] in mice, and genes required for survival of the symbiont Bacteroides thetaiotaomicron in the murine gut [20]. Here we apply TraDIS to simultaneously assign the genotype and relative fitness of 7702 distinct S. Typhimurium mutants during intestinal colonization of chickens, pigs and cattle, providing highly relevant data for the control of zoonotic and animal salmonellosis.
Fig. 1. Experimental strategy for TraDIS mutant screens. 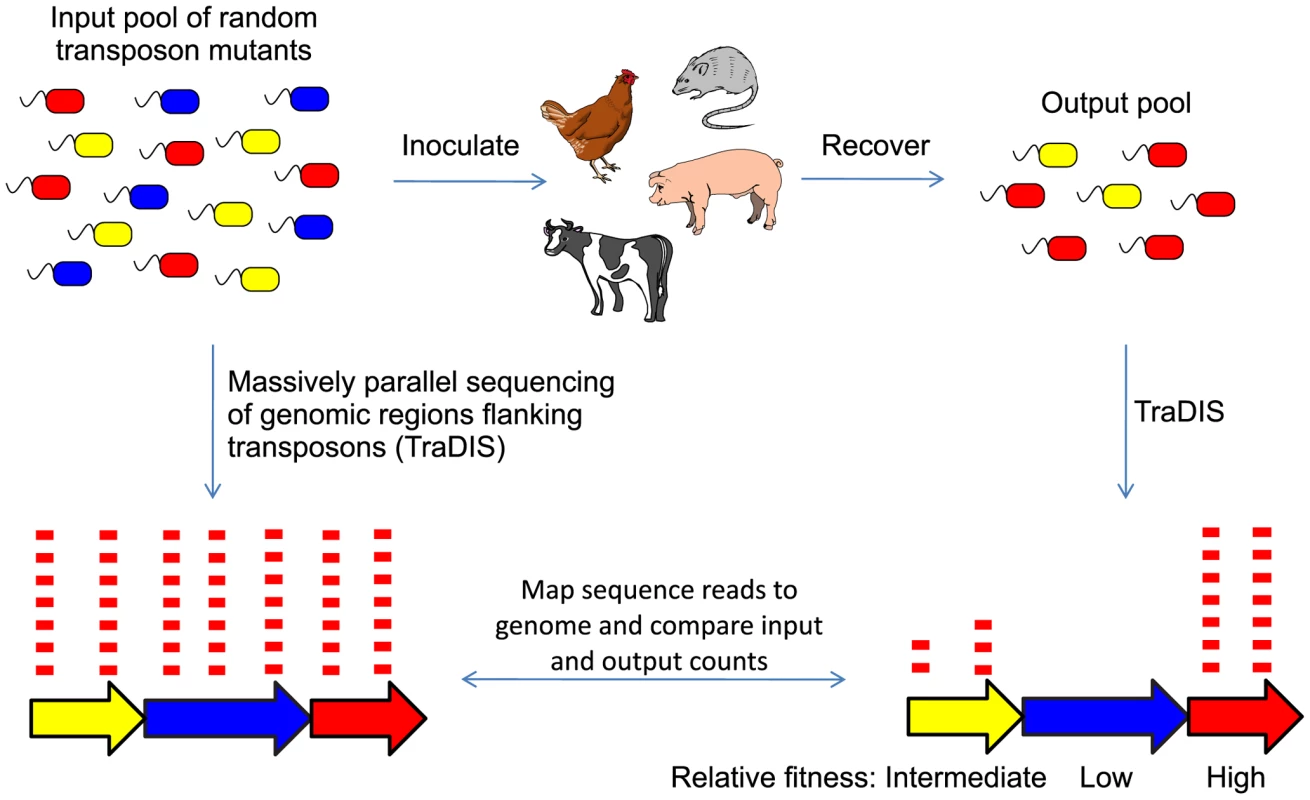
An input pool of random transposon insertion mutants is generated, and used to inoculate experimental animals. Output pools of bacteria that are capable of survival and growth in each host are isolated from an appropriate tissue. Massively parallel sequencing of the regions flanking each transposon allow the disrupted genes to be identified, and comparison of the sequence counts derived from the input and output pools allows the relative fitness of each mutant to be assessed. Results
Quantitative assessment of mutant fitness
To validate the quantitative nature of TraDIS, we applied it to investigate pools of Mu and mini-Tn5 mutants of S. Typhimurium strain SL1344 before and after intravenous infection of BALB/c mice (see Text S1). These mutant pools had been characterized previously using transposon-mediated differential hybridization (TMDH) [21], a microarray-based method that relies on hybridization of run-off transcripts arising from transposon-encoded T7 and SP6 promoters to high-density oligonucleotide arrays. In total, using TraDIS, 9792 distinct transposon insertions (4992 Mu and 4800 Tn5) were unambiguously mapped at the level of the single nucleotide to the SL1344 genome, providing relative fitness scores for 94.4% of the 10368 mutants screened, This is likely to be an underestimate of the performance of TraDIS, since it is likely that there were siblings of mutants with mapped insertions within the pool of 10368 mutants. The fitness scores assigned by TraDIS are defined as the log2-fold change in the number of sequence reads obtained across the boundaries of each transposon insertion between the input and output pools (see Materials and Methods), and were significantly correlated with the existing TMDH data (Figure S1; P<2.2×10−16), verifying the quantitative nature of TraDIS. TraDIS allowed identification of a number of mutants missed by TMDH (see Text S1), and provided finer mapping of insertion sites than can be achieved by hybridization of transposon-flanking sequences to tiling arrays, extending the conclusions of the earlier study and demonstrating the superiority of the TraDIS approach.
Assessment of mutant fitness in food animals
Though useful, previous attempts to assign comprehensively the role of S. Typhimurium genes relied on parenteral infection of atypically susceptible mice [21]–[23], and do not reflect the roles of Salmonella genes during intestinal colonization of food-producing animals infected via the natural oral route. To identify genes relevant to colonization of animal reservoirs and therefore zoonosis, we generated a library of 8550 mini-Tn5 mutants of S. Typhimurium strain ST4/74 and applied TraDIS to assess the survival of these mutants during oral infection of chickens, pigs and calves. Pools of 475 mutants were screened in individual pigs and calves and pools of 95 mutants were screened in duplicate chickens. Pilot studies in which selected pools had been repeatedly screened in each species indicated the reliable negative selection of the same mutants at these pool complexities in the absence of stochastic loss that may be due to population bottlenecks (data not shown). TraDIS mapped 7702 distinct insertions to the nucleotide level, representing at least 90.1% of the mutants screened, and demonstrated the random distribution of the transposon insertions around the genome (see Figure 2), which disrupt 2715 different genes. TraDIS analysis revealed evidence of random drop-out of mutants from the pools in one chicken, two pig and two calf experiments, likely owing to recovery of output pools of an inadequate size, and data from these animals were omitted from subsequent analysis (see Text S1).
Fig. 2. Circular diagrams of the S. Typhimurium SL1344 chromosome and plasmids (not to scale), showing the near random distribution and high density of transposon insertions mapped by TraDIS. 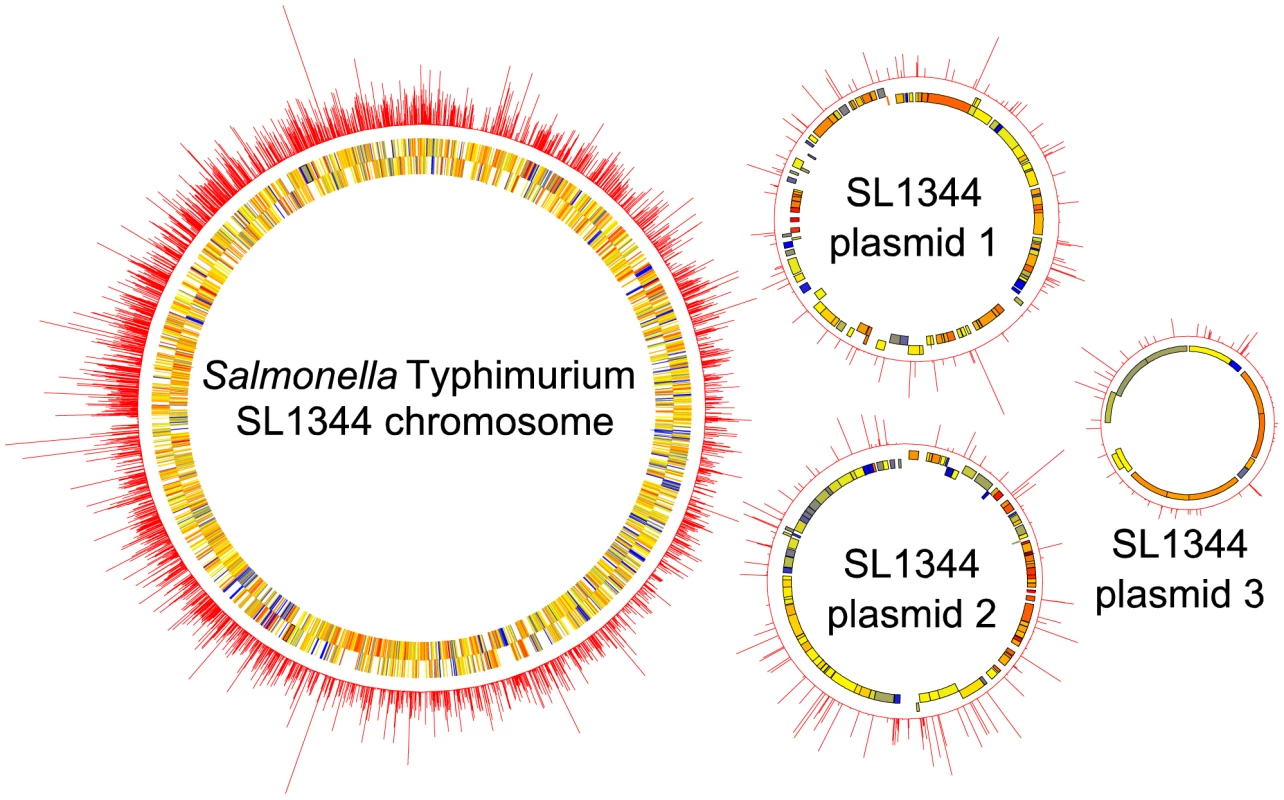
The inner two rings indicate the positions of annotated genes, coloured according to their GC content (blue = low, yellow = intermediate, red = high). The outer ring indicates the number of transposon-flanking sequence reads obtained at each position, with peaks corresponding to the presence of a transposon insertion. TraDIS assignments of the insertion sites and fitness scores of mutants are listed in Table S1 (BALB/c mice dosed intravenously) and Table S2 (chickens, pigs and calves dosed orally). The raw TraDIS sequence data are available from the NCBI Short Read Archive (accession numbers ERA000172 and ERP000286). To facilitate exploration of the TraDIS data, a user-friendly online genome browser was constructed with which the insertion site and fitness score can be viewed in the context of the linear genome, GC content, transcription start sites and existing annotation (http://www-tradis.vet.cam.ac.uk). Figure 3 shows the fitness scores obtained by TraDIS analysis of S. Typhimurium mutants screened in mice, chickens, pigs and calves, plotted against read coverage (equivalent to the “M/A” plots commonly used to display microarray data). The proportion of significantly attenuated mutants identified during intestinal colonization of food-producing animals was greater than in the murine typhoid model. In each host, a large proportion of mutations did not exert a strong negative or positive effect, indicating that a high number of accessory or redundant functions exist. P values were estimated using the available biological replicates (all pools were screened in duplicate in chickens, 2 pools were screened in duplicate calves and 3 pools were screened in triplicate pigs), and attenuated mutants were defined as those with a negative fitness score and P≤0.05.
Fig. 3. Relative fitness scores (defined as log2 fold changes in sequence read counts between input and output pools) for each transposon mutant, plotted against average read coverage (equivalent to an M/A plot, commonly used to display microarray data) for each of the four host species. 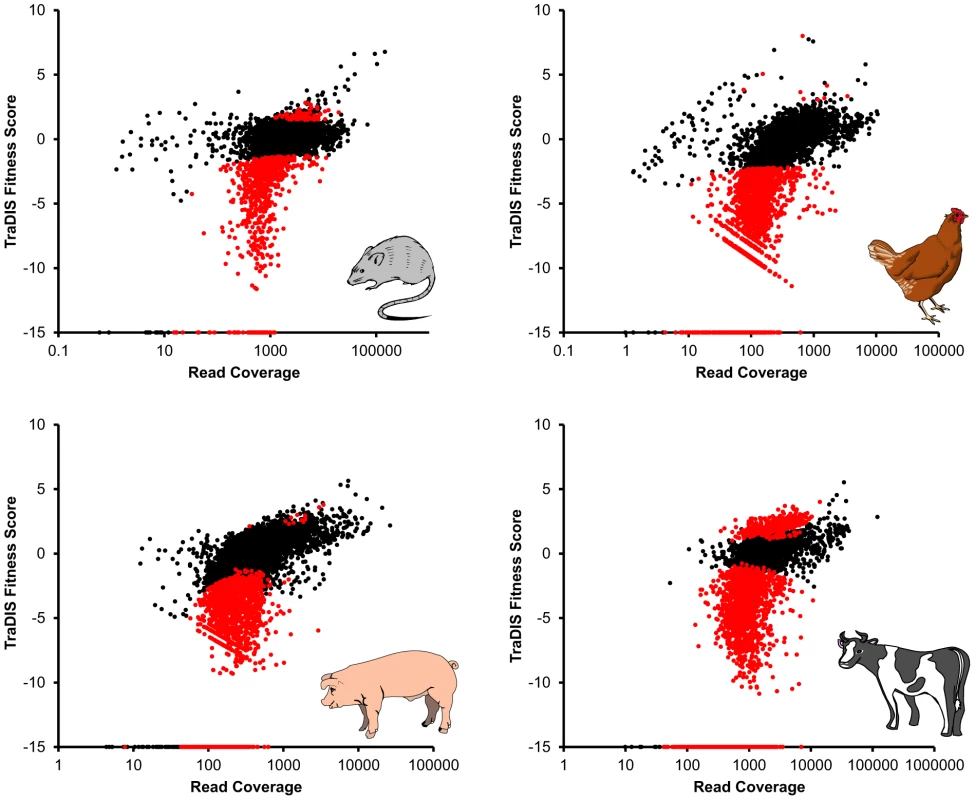
Mutants which showed a significant change in abundance in the output relative to the input are highlighted in red. Mutants which had no reads in the output pool are assigned an arbitrary fitness score of −15. To summarize the dataset further, genes were scored as potentially important in colonization if they were disrupted in at least one significantly attenuated mutant in any of the four host species. This enables comparisons between datasets derived from different transposon libraries (such as the mouse and chicken/pig/cattle datasets), and visualization of the data in comparison with other genome-wide datasets. For some genes, insertions at different subgenic locations can have divergent effects on the encoded protein resulting in contrasting fitness scores, so it is important to consider the genetic context of each individual transposon when interpreting the TraDIS data in detail. This is true for all transposon-based mutant screens, although earlier technologies such as STM and TMDH lacked sufficient resolution to permit such considerations. We recommend the use of the TraDIS browser (http://www-tradis.vet.cam.ac.uk) to assist interpretation of our data in the context of the genome annotation.
Fitness scores were obtained for 3194 distinct genes disrupted by transposon insertions in the mouse screen, and 2715 genes in the chicken, pig and calf screens, of which fitness score existed for 2435 genes in all three food-producing animals. Fitness scores were available from all four hosts for 1935 genes, of which 1069 had a significantly attenuated mutant in at least one host. Venn diagrams showing the numbers of significantly attenuated mutants, and the numbers of genes potentially important for colonization in chickens, pigs and cattle are shown in Figure 4. A further Venn diagram, combining the chicken/pig/cattle and mouse datasets, is available in Figure S2, and Figure S3 shows a comparison between the chicken, pig and cattle TraDIS data, and the data obtained from equivalent mutants in the earlier STM screens [10], [12]. A table of all genes disrupted by a transposon insertion, indicating if any of the mutants in each gene was significantly attenuated, is available in Table S3. To facilitate exploration of the TraDIS data, custom files have been prepared that allow proteins in KEGG metabolic pathway diagrams [24] to be coloured blue if an attenuated mutant was found in the encoding gene, or red if the gene was mutated but no significant attenuation was observed. The KEGG colour files are available from http://www-tradis.vet.cam.ac.uk. As an example, Figure S4 shows the effect of mutations affecting multiple steps in chorismate biosynthesis, which is known to influence persistence of S. Typhimurium in vivo. It is evident that mutations affecting sequential steps in the pathway are attenuating, with just two exceptions: the initial condensation of D-erythrose-4-phosphate and phosphoenolpyruvate into 3-deoxy-D-arabino-heptulosonate-7-phosphate, and the conversion of shikimate to shikimate-3-phosphate, both of which can be catalyzed by the products of multiple genes (aroFGH and aroKL, respectively). Thus, TraDIS identifies pathways in which defects exert a common effect, but also reveals steps at which functional redundancy exists. During the analysis of the TraDIS data it became clear that many regions of the genome with low GC content are important for intestinal colonization of chickens, pigs and cattle (see Text S1 and Figure S5).
Fig. 4. Venn diagrams showing the attenutation phenotypes observed in chickens, pigs, and cattle using TraDIS. 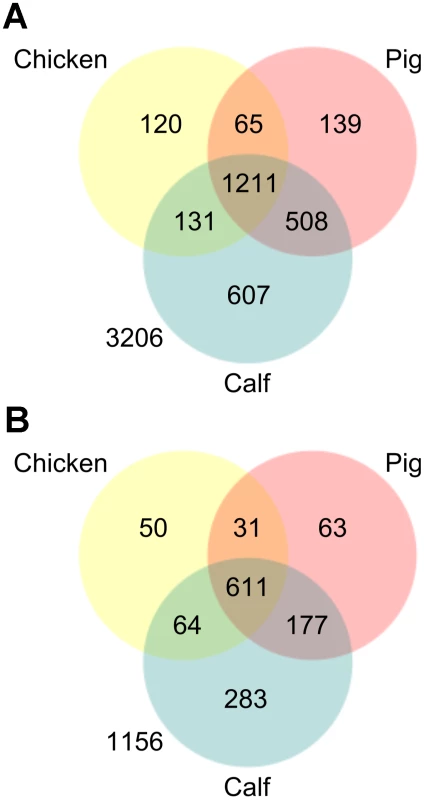
A) the numbers of transposon mutants which were significantly attenuated in each host B) the numbers of genes which were disrupted in the TraDIS mutant library, and which are potentially important for colonization (i.e. they had at least one significantly attenuated mutant) in each of the three reservoir hosts. Analysis of defined null mutants in chickens
Twelve genes were selected for further investigation based on the TraDIS data: carB, clpB, ilvC, mig-14, pagN, SL1344_0084 (STM0084), SL1344_4248 (STM4312), SL1344_3128 (STM3154), trxA, virK, ytfL and zirT (SL1344_1599). These targets were chosen based on their fitness scores (at least one mutant in each gene shows significant attenuation), and to include some genes with established roles in colonization in chickens (clpB [4]) or mice (trxA [25], mig-14 [26], virK [27]), genes with a postulated role in colonization (pagN [28], SL1344_0084 [12]), the mouse anti-virulence factor zirT [29], genes demonstrating variable fitness scores (SL1344_0084, SL1344_3128 and ytfL) and genes that demonstrate putative host-specific effects on colonization in the TraDIS data (clpB, ilvC and ytfL). Each gene was inactivated separately by λRed recombinase-mediated integration of linear PCR amplicons by homologous recombination [30]. Mutant phenotypes were evaluated in groups of 3 chickens per mutant per time interval. For each mutant, competitive indices (CIs) were derived 4, 6 and 10 days post-inoculation of age-matched chickens with the kanR-tagged mutant and ST4/74 nalR wild-type strain in a 1∶1 ratio (see Table 1).
Tab. 1. Competitive indices of defined null mutants of S. Typhimurium lacking candidate virulence-associated loci identified by TraDIS. 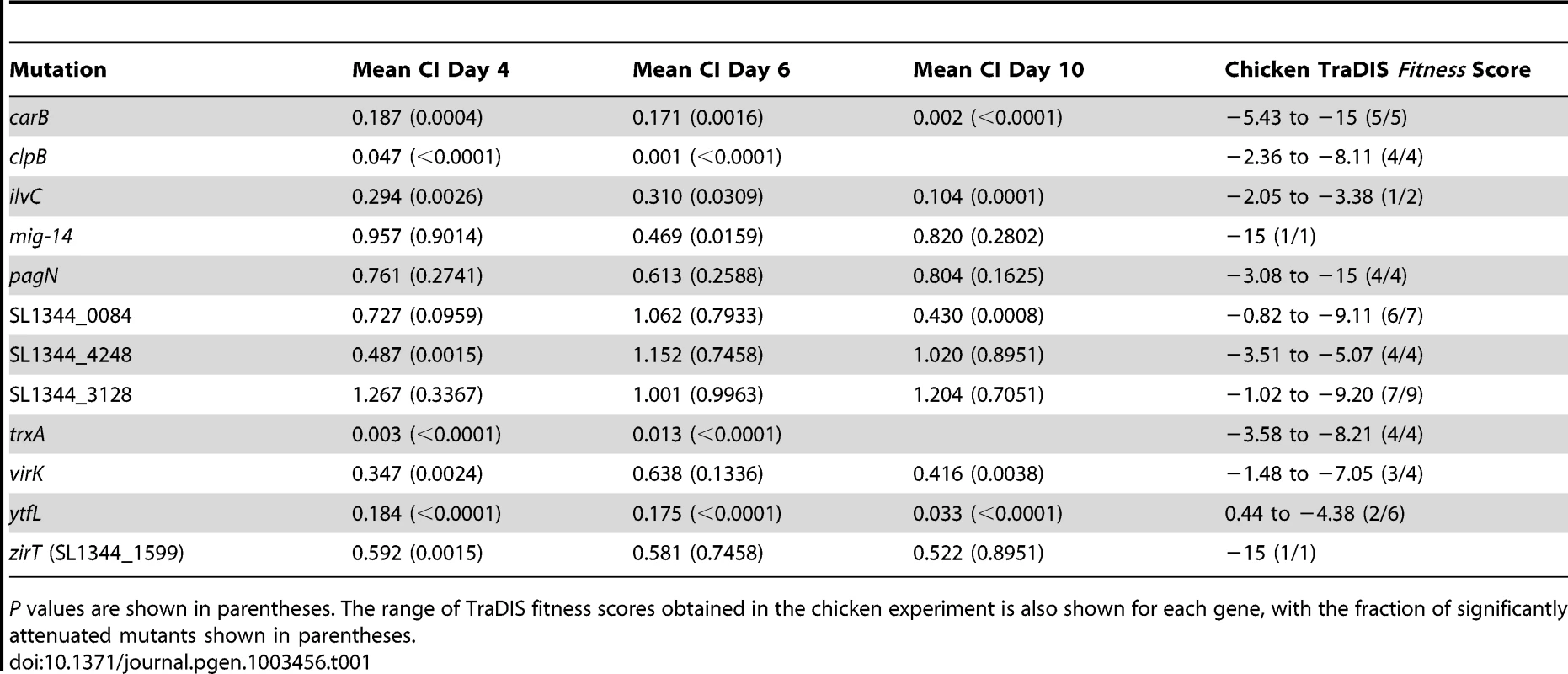
P values are shown in parentheses. The range of TraDIS fitness scores obtained in the chicken experiment is also shown for each gene, with the fraction of significantly attenuated mutants shown in parentheses. With a single exception (SL1344_3128) all mutants were negatively-selected relative to the parent strain at day 4 post-inoculation, which corresponds to the time at which mutants were recovered for the TraDIS analysis. The difference in the mutant∶wild-type ratio was significantly different from the ratio in the inocula for 8 of the mutants at day 4. For a further 2 mutants, significant differences were detected at later time points. Taken together with comparisons to existing datasets for signature-tagged mutants of the same strain in the same animal models [10], [12] (see Text S1), the data strongly support evidence of attenuation detected by TraDIS. Variance from TraDIS fitness scores is likely to reflect differences in competition dynamics for a given mutant relative to co-screened wild-type or mutant bacteria. For one gene (SL1344_3128 ), no evidence was found of any attenuation of the defined mutant, which performed comparably to the wild-type at all time intervals. This gene was chosen for further investigation because its mutants exhibited a wide range of TraDIS fitness scores (−1.02 to −9.20). Interestingly, mutants in the gene cluster SL1344_3128-30 are predicted to be deficient in swarming motility [31], suggesting the possibility that such motility may be an occasional but not universal requirement for colonization.
Discussion
The genetic basis of intestinal colonization
The TraDIS dataset is a powerful resource for understanding intestinal colonization of a range of highly relevant hosts by Salmonella, and thus zoonotic transmission and animal disease. The data suggest that the definition of what constitutes a colonization gene is not straightforward, encompassing genes involved in metabolism, stress responses and transcriptional regulation, together with genes with well-established roles in virulence. The T3SSs encoded by SPI-1 and SPI-2 are both essential for infection in chickens, pigs and cattle, although there are some mutants within both regions that are not attenuated or exhibit a less pronounced phenotype in chickens. T3SSs allow the secretion of effector molecules into the host cytoplasm, these effectors being encoded both within SPI-1 and 2 and distally. Most of the known effector genes, including sopA, sopB, sopE2, sipA, avrA, sipC, sseG, sseI, sifA, sseK1, pipB2 and sopD2, were identified by TraDIS as being important for infection of all three food-producing animals, although as with the T3SS structural genes, the phenotype was often less pronounced in chickens. Other T3SS effectors, including sptP, slrP, gogB and sspH2, could be disrupted without affecting colonization. Of these, slrP has been implicated as a host-specificity factor, essential for oral infection of mice but not required for calf infection [14]. Null mutants of sptP are not impaired in their interactions with cultured macrophages or epithelial cells [32]–[34], and sspH2 mutants do not show any defect in vacuole-associated actin polymerization [35]. For both sptP and sspH2, the lack of phenotype was suggested to be due to functional redundancy amongst T3SS effectors.
There are also attenuated mutants that harbour insertions within the other recognized Salmonella pathogenicity islands [36]. In SPI-3, mutants in some genes (mgtC, marT, SL1344_3717 and SL1344_3721) were attenuated, with others (misL, sugR, slsA and mgtB) showing no attenuation. Interestingly, marT, which encodes a transcriptional regulator, is a pseudogene in S. Typhi, and restoring it reduces survival during infection of a human cell culture [37]. A role for marT in infection of chickens, pigs and cattle suggests a selection pressure for its retention in the S. Typhimurium genome. SPI-4 was previously thought to play a role in infection of cattle but not chickens or pigs, based on STM screens [10], [12]. TraDIS suggests a role for SPI-4 in all three species, although the phenotypes in chickens and pigs are more subtle than in calves, highlighting the increased sensitivity of TraDIS relative to STM. Some transposon insertions within the central highly repetitive region of siiE are tolerated in chickens and pigs, but are attenuated in cattle. Interestingly, siiE is split into two ORFs in both S. Typhi genome sequences, and part of the repetitive central region is absent from the genome of S. Paratyphi A. SiiE is secreted [38], indicating that in trans complementation by co-screened mutants does not obscure the identification of secreted colonization factors by TraDIS. All of the genes of the enteritis-associated SPI-5 that were disrupted by a transposon (pipACD, sopB and orfX) were required in all three species, although often with a milder phenotype in chickens. Several clusters of attenuated mutants were also found in the Salmonella chromosomal island (SCI, also known as SPI-6 in S. Typhi), including mutants in the hypothetical genes sciJ, sciQ, SL1344_0286A and sciX, the fimbrial subunit safA and its chaperone safB, the regulator sinR, SL1344_0301 (STM0305) which encodes a putative cytoplasmic protein, the pagN adhesin (STM0306) and sciZ (STM0307), a homologue of Shigella virG. Deletion of SCI affects invasion and virulence in a mouse intraperitoneal infection model [39], and the phenotype of a defined safA mutant has been confirmed in pigs [10].
Fimbriae play a well-established role in Salmonella attachment and intestinal colonization [40]. All twelve fimbrial operons were disrupted by multiple transposons in the TraDIS screen. No obvious host-specific phenotypes were seen, with a common pattern that mutants of fimbrial subunit genes were attenuated, whereas assembly genes were often dispensable, suggesting cross-talk in the assembly pathways. Stress responses are also important in the infection process, as Salmonella is subjected to a range of stresses including low pH, oxidative stress and heat shock [41]. The genetic components of these stress responses overlap [42], and many of these genes harboured transposons that resulted in attenuation. These included the sigma factor gene rpoE and its anti-sigma factor resA, the heat shock chaperone genes dnaK and dnaJ and the heat shock protease gene degP (htrA). Interestingly, several stress response genes are variably attenuated in the different hosts, suggesting species-specific stresses. These include the two-component regulatory system genes envZ and ompR, and the oxidative stress response genes dps, katE and proV which are all attenuated in pigs and cattle but show little or no attenuation in chickens. Conversely, transposon mutants in clpB, clpP and clpX, which encode proteases and are involved in the regulation of rpoS, are attenuated in chickens but not pigs or cattle.
Many S. Typhimurium genes beyond the classical virulence factors and stress response genes were revealed to be important for oral infection of livestock species. These include genes involved in nucleotide metabolism (pyrCD, purADGH, dgt, dcd, guaA, pyrCD and carAB), aromatic amino acid biosynthesis (aroABCDE), inorganic ion transport (trkAH, znuABC, fepCDG), protein synthesis (tufAB, fusA, efp, rplI, rpsK), protein export (tatABC, yajC) and many genes involved in carbohydrate metabolism. Additionally, numerous low GC clusters of genes with putative metabolic functions and multiple attenuating mutations were identified. Several global regulators, including crp, smpB and dam, result in attenuation in all three hosts, whereas another, fnr, appeared only to be important for infection in chickens. On occasion, TraDIS revealed functional data at a sub-genic level. For example, most of the insertions that disrupt the SPI-1 gene sptP result in attenuation, but insertions close to the 3′ end of the gene are tolerated. The gene rpoC, which encodes the β′ subunit of RNA polymerase, is essential in S. Typhimurium [43]. However, one transposon insertion in the chicken, pig and cattle dataset, and two in the mouse dataset, were identified close to the 3′ end of rpoC. These insertions would disrupt the extreme C-terminal end of the encoded protein, and were found to reduce the fitness of the mutants in the animal screens. Similarly, an insertion was found at the 3′ end of the essential polA gene which encodes DNA polymerase I, and this mutant was significantly attenuated in chickens, pigs and cattle.
The recent RNAseq-based analysis of the S. Typhimurium SL1344 transcriptome [44] identified a number of small-regulatory RNAs. As indicated in Table S4 several of these were implicated in colonization by TraDIS. For many it is difficult to demonstrate conclusively a colonization-associated phenotype from the TraDIS data alone, since we cannot preclude the potential for polar effects on adjacent genes. This is the case for InvR, which is encoded within SPI-1. Table S4 details only the sRNA genes annotated in the SL1344 genome (which differs from ST4/74 by just 8 SNPs [45]), but in the TraDIS data there are a number of attenuating transposons within large intergenic regions that could reveal the presence of novel sRNA genes.
Putative host-specificity determinants of S. Typhimurium
The chicken, pig and cattle TraDIS data presented in Figure 4 indicate that a shared core set of 611 genes is required for efficient colonization of all three species, with a smaller set of species-specific colonization factors. The core set comprises approximately two thirds of the genetic requirements for infection of each individual species, and 48% of the total set of colonization-associated genes. There are 259 genes which are required for systemic infection of mice for which comparable data are available from the food-producing animals (Figure 4); of these, 140 also contribute to oral infection of chickens, pigs and cattle, and only 43 are putative mouse-specific factors. Many of the differences between the mouse and chicken/pig/cattle datasets may arise from the additional genetic requirements for infection via the oral route.
Although most colonization factors were necessary for infection of chickens, pigs and cattle, there were some patterns amongst the colonization factors that appeared to function in a host-specific manner that may reflect underlying differences in host biology. There are many genes associated with flagellar motility that are essential for infection of pigs but not required for chicken or calf infection, including fliY, flgK, fliN, flgN, fliB and fliZ. Several other flagellum-associated genes (flgB, flgL, fliL) are required for infection of cattle but not chickens or pigs. In chickens, many genes that are involved in anaerobic growth are required; these include genes involved in the production of group I hydrogenase (hypOBF, hybABCDF), fumarate reductase (frdAD), pflB, pfkA, rNTP reductase (nrdDG) and the global regulator Fnr. Differences in oxygen tension proximal to villus tips have been detected that modulate the regulation of Shigella virulence genes [46], therefore the requirement for distinct respiratory pathways by S. Typhimurium in food animals may reflect differences in the niches occupied. Also required in chickens, but apparently not calves or pigs, are the virK homologue ybjX, ilvGE, clpB and the his operon. The observation that many of the host-specific phenotypes were observed independently in multiple genes affecting the same pathways strongly suggests that these effects are due to differences in the within-host environment. For example, in relation to fumarate reductase there were nine independent frdA mutants that were all significantly attenuated in chickens; of these only one showed significant attenuation in pigs, and none in calves. Similarly, in relation to group I hydrogenase, from a total of 15 mutations affecting the hybABCDF genes, 12 showed significant attenuation in chickens, but none in pigs or calves (Table S2).
The serovar Typhimurium strain ST4/74 investigated here is a natural bovine isolate that elicits pathology typical of clinical salmonellosis in all the host species used in this study. However, some atypical S. Typhimurium strains exist that have lost the capability to colonize a broad range of hosts. For example, the laboratory-adapted strain LT2 and its derivatives tend to be avirulent or less virulent in mice relative to natural isolates of serovar Typhimurium [47] and ST4/74-based strains. It is noteworthy that the rpoS gene encoding the sigma factor σS, which is defective in LT2 and associated with the relative avirulence of this strain [48], was found to be required in all four species tested by screening of the S. Typhimurium libraries we describe (Table S3). TraDIS identified additional regions of the ST4/74 genome that are required for infection, but which are absent from the LT2 genome. These include several genes that are encoded within the same phage element: SL1344_1965, which is required for infection of mice, and SL1344_1929/30 and SL1344_1976, which are required for infection of chickens, pigs and cattle. Moreover, some Typhimurium strains have become adapted to a particular host, and the genome sequence of a human-adapted variant [49] reveals the decay of a number of genes important in colonization of food animals (e.g. allP, sseI, pipD, ydeE), but also other pseudogenes for which no role in food animals for the intact gene could be detected (e.g. ratB, ygbE, yhjU). Integration of genome sequences with high-resolution functional data of the kind we describe will provide further clues to explain the differential virulence of host-adapted or laboratory strains relative to natural isolates.
Our study represents the first comprehensive genome-wide survey of the role of thousands of Salmonella genes during colonization of the primary reservoirs of human non-typhoidal salmonellosis. TraDIS simultaneously assigned the genotype and relative fitness of 7702 distinct S. Typhimurium insertion mutants in chickens, pigs and cattle, representing over 90% of the mutants screened in pools of up to 475 mutants per animal. TraDIS therefore represents a significant advance in the reduction, refinement and replacement of animal models relative to STM, where only negatively-selected mutants tend to be interrogated and the vast majority of insertion sites and phenotypes are unreported [50]. Multiple lines of evidence suggest that the TraDIS data are robust and reliably reflect the fitness of the screened mutants in each host animal. Many of the attenuated mutants were found to harbour transposons in genes known to be involved in colonization. The TraDIS fitness scores correlated well with established datasets obtained using STM [10], [12] and TMDH [21]. Multiple mutations within the same gene or pathway usually gave comparable phenotypes, and most of the attenuated mutants demonstrated the same phenotype independently in the three different food-producing animal hosts. The examples of putatively host-specific attenuation tended to be restricted to particular pathways with multiple independent mutations. Finally, analysis of defined knockout mutants of targets chosen based on the TraDIS data reproduced attenuated phenotypes in all but one case.
Many novel colonization-associated genes were identified within the S. Typhimurium genome and the data provide an invaluable resource for the community to mine and extend. Moreover, TraDIS indicated that thousands of mutations exerted little or no effect in vivo, implying functional redundancy that may limit and refine the selection of targets for novel inhibitors, as previously suggested [51]. Unlike library screens conducted in murine typhoid models to date, we provide highly relevant data for control of intestinal S. enterica infections in food-producing animals, and thus zoonosis. Attenuating mutations may be suitable for selection and refinement of live vaccines for food-animals, and these in turn may express heterologous antigens. Further, the data will guide the interpretation of existing and fast emerging datasets on the repertoire, sequence and expression of Salmonella genes and aid the modelling of virulence in a wider evolutionary and ecological context. Our data reflect mutant phenotypes at a specific time and site, and further studies on the temporal and spatial role of Salmonella genes are likely to be informative. This study also establishes TraDIS as a quantitative technology in functional genomics, which has potential for widespread application beyond the realm of microbial pathogenicity.
Methods
Ethics statement
Animal experiments were conducted according to the requirements of the Animals (Scientific Procedures) Act 1986 (project license number 30/2485) with the approval of the local Ethical Review Committee.
For full details of experimental animals, bacterial strains, materials, molecular biological techniques and statistical methods see Text S1. Briefly, a library of 8550 mini-Tn5 mutants was generated in a spontaneous nalidixic acid resistant variant of S. Typhimurium ST4/74. The mutants were combined into pools of 95 for chickens, and 475 for pigs and calves. Animals were inoculated orally and killed humanely 4 days (chickens and calves) or 3 days (pigs) after infection, or earlier if the clinical endpoint was reached. A section of an appropriate tissue (whole caeca for chickens, spiral colonic mucosa for pigs and distal ileal mucosa for calves) was homogenized and grown overnight on MacConkey agar plates to isolate the output bacteria.
Genomic DNA was prepared from the inocula and output samples, and fragmented to ∼300 bp. An Illumina adapter was ligated to the fragments, and PCRs were performed using an adapter-specific primer in conjunction with primers homologous to each end of the transposon. The sequences of all oligonucleotide primers used in this study are detailed in Table S5. The resultant products were sequenced on single end Illumina flowcells using a sequencing primer designed to read a 10 bp tag of transposon-derived sequence, plus 27 bp of flanking genomic DNA. Sequences containing the tag were mapped to the S. Typhimurium SL1344 genome sequence. A transposon was inferred to be present if there were corresponding reads derived from each end of the transposon in the input pool.
The number of reads corresponding to each transposon in the input pool, and the number of reads mapping to the equivalent position in the output pool data, were compared using DESeq [52]. The ratio of input∶output read counts was determined, after normalisation to account for variations in the total number of reads obtained for each sample, and expressed as log2(fold change), referred to as the fitness score. A negative fitness score indicates an attenuated mutant, a positive score indicates a mutant which was more abundant in the output pool than in the input. For strongly attenuated mutants, no reads were obtained in the output pool, so it was not possible to calculate a finite log2(fold change); such mutants were assigned an arbitrary fitness score of −15. For each individual mutant, the hypothesis that the fitness score was equal to zero (i.e. that the mutant was present at equivalent levels in the input and output pools) was tested using the negative binomial distribution as implemented in DESeq. DESeq models variance under the assumption that mutants with comparable levels of sequence coverage exhibit similar levels of dispersion. We exploited this model to estimate P values for all mutants whilst minimising the number of biological replicates by fitting using only those mutants for which replicate data points were available, and applying the resultant model to the data derived from all mutants.
Defined null mutants were obtained for twelve genes identified as attenuated in the TraDIS screen, and assessed in competition with wild-type ST4/74 during oral infection of chickens. The ratios of mutant∶wild-type bacteria from caecal isolates at day 4, 6 and 10 were compared with those in the inoculum, and the significance of any differences was tested using Student's t-test.
Supporting Information
Zdroje
1. CrumpJA, LubySP, MintzED (2004) The global burden of typhoid fever. Bull World Health Organ 82 : 346–353.
2. MajowiczSE, MustoJ, ScallanE, AnguloFJ, KirkM, et al. (2010) The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50 : 882–889.
3. StevensMP, HumphreyTJ, MaskellDJ (2009) Molecular insights into farm animal and zoonotic Salmonella infections. Philos Trans R Soc Lond B Biol Sci 364 : 2709–2723.
4. TurnerAK, LovellMA, HulmeSD, Zhang-BarberL, BarrowPA (1998) Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect Immun 66 : 2099–2106.
5. HenselM, SheaJE, GleesonC, JonesMD, DaltonE, et al. (1995) Simultaneous identification of bacterial virulence genes by negative selection. Science 269 : 400–403.
6. SheaJE, HenselM, GleesonC, HoldenDW (1996) Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A 93 : 2593–2597.
7. HenselM (2000) Salmonella pathogenicity island 2. Mol Microbiol 36 : 1015–1023.
8. GalanJE, CurtissR3rd (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86 : 6383–6387.
9. BisphamJ, TripathiBN, WatsonPR, WallisTS (2001) Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect Immun 69 : 367–377.
10. CarnellSC, BowenA, MorganE, MaskellDJ, WallisTS, et al. (2007) Role in virulence and protective efficacy in pigs of Salmonella enterica serovar Typhimurium secreted components identified by signature-tagged mutagenesis. Microbiology 153 : 1940–1952.
11. LichtensteigerCA, VimrER (2003) Systemic and enteric colonization of pigs by a hilA signature-tagged mutant of Salmonella choleraesuis. Microb Pathog 34 : 149–154.
12. MorganE, CampbellJD, RoweSC, BisphamJ, StevensMP, et al. (2004) Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol 54 : 994–1010.
13. ShahDH, LeeMJ, ParkJH, LeeJH, EoSK, et al. (2005) Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 151 : 3957–3968.
14. TsolisRM, TownsendSM, MiaoEA, MillerSI, FichtTA, et al. (1999) Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun 67 : 6385–6393.
15. LangridgeGC, PhanM-D, TurnerDJ, PerkinsTT, PartsL, et al. (2009) Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Research 19 : 2308–2316.
16. BentleyDR, BalasubramanianS, SwerdlowHP, SmithGP, MiltonJ, et al. (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456 : 53–59.
17. van OpijnenT, BodiKL, CamilliA (2009) Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6 : 767–772.
18. GawronskiJD, WongSMS, GiannoukosG, WardDV, AkerleyBJ (2009) Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proceedings of the National Academy of Sciences 106 : 16422–16427.
19. van OpijnenT, CamilliA (2012) A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Research 22 : 2541–2551.
20. GoodmanAL, McNultyNP, ZhaoY, LeipD, MitraRD, et al. (2009) Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6 : 279–289.
21. ChaudhuriRR, PetersSE, PleasanceSJ, NorthenH, WillersC, et al. (2009) Comprehensive identification of Salmonella enterica serovar Typhimurium genes required for infection of BALB/c mice. PLoS Pathog 5: e1000529 doi:10.1371/journal.ppat.1000529.
22. LawleyTD, ChanK, ThompsonLJ, KimCC, GovoniGR, et al. (2006) Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog 2: e11 doi:10.1371/journal.ppat.0020011.
23. SantiviagoCA, ReynoldsMM, PorwollikS, ChoiSH, LongF, et al. (2009) Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog 5: e1000477 doi:10.1371/journal.ppat.1000477.
24. KanehisaM, GotoS, HattoriM, Aoki-KinoshitaKF, ItohM, et al. (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–357.
25. BjurE, Eriksson-YgbergS, AslundF, RhenM (2006) Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect Immun 74 : 5140–5151.
26. ValdiviaRH, CirilloDM, LeeAK, BouleyDM, FalkowS (2000) mig-14 is a horizontally acquired, host-induced gene required for salmonella enterica lethal infection in the murine model of typhoid fever. Infect Immun 68 : 7126–7131.
27. DetweilerCS, MonackDM, BrodskyIE, MathewH, FalkowS (2003) virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol Microbiol 48 : 385–400.
28. LambertMA, SmithSG (2009) The PagN protein mediates invasion via interaction with proteoglycan. FEMS Microbiol Lett 297 : 209–216.
29. Gal-MorO, GibsonDL, BalutaD, VallanceBA, FinlayBB (2008) A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis. PLoS Pathog 4: e1000036 doi:10.1371/journal.ppat.1000036.
30. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 : 6640–6645.
31. WangQ, MaricondaS, SuzukiA, McClellandM, HarsheyRM (2006) Uncovering a large set of genes that affect surface motility in Salmonella enterica serovar Typhimurium. J Bacteriol 188 : 7981–7984.
32. ChenLM, KanigaK, GalanJE (1996) Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol 21 : 1101–1115.
33. FuY, GalanJE (1998) The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol 27 : 359–368.
34. KanigaK, UralilJ, BliskaJB, GalanJE (1996) A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol 21 : 633–641.
35. MiaoEA, BrittnacherM, HaragaA, JengRL, WelchMD, et al. (2003) Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol Microbiol 48 : 401–415.
36. HenselM (2004) Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol 294 : 95–102.
37. RetamalP, Castillo-RuizM, VillagraNA, MorgadoJ, MoraGC (2010) Modified intracellular-associated phenotypes in a recombinant Salmonella Typhi expressing S. Typhimurium SPI-3 sequences. PLoS ONE 5: e9394 doi:10.1371/journal.pone.0009394.
38. MorganE, BowenAJ, CarnellSC, WallisTS, StevensMP (2007) SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect Immun 75 : 1524–1533.
39. FolkessonA, LofdahlS, NormarkS (2002) The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res Microbiol 153 : 537–545.
40. EdwardsRA, PuenteJL (1998) Fimbrial expression in enteric bacteria: a critical step in intestinal pathogenesis. Trends Microbiol 6 : 282–287.
41. RychlikI, BarrowPA (2005) Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol Rev 29 : 1021–1040.
42. MorganRW, ChristmanMF, JacobsonFS, StorzG, AmesBN (1986) Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci U S A 83 : 8059–8063.
43. KnuthK, NiesallaH, HueckCJ, FuchsTM (2004) Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol Microbiol 51 : 1729–1744.
44. KrogerC, DillonSC, CameronAD, PapenfortK, SivasankaranSK, et al. (2012) The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proceedings of the National Academy of Sciences of the United States of America 109: E1277–1286.
45. RichardsonEJ, LimayeB, InamdarH, DattaA, ManjariKS, et al. (2011) Genome sequences of Salmonella enterica serovar typhimurium, Choleraesuis, Dublin, and Gallinarum strains of well - defined virulence in food-producing animals. J Bacteriol 193 : 3162–3163.
46. MarteynB, WestNP, BrowningDF, ColeJA, ShawJG, et al. (2010) Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465 : 355–358.
47. Sanderson KE, Stocker BAD (1987) Salmonella typhimurium strains used in genetic analysis. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M et al.., editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington D.C.: ASM Press. pp. 1220–1224.
48. SwordsWE, CannonBM, BenjaminWHJr (1997) Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infection and Immunity 65 : 2451–2453.
49. KingsleyRA, MsefulaCL, ThomsonNR, KariukiS, HoltKE, et al. (2009) Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res 19 : 2279–2287.
50. EckertS, DzivaF, ChaudhuriRR, LangridgeGC, TurnerDJ, et al. (2011) Retrospective application of transposon-directed insertion-site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J Bacteriol 193 : 1771–1776.
51. BeckerD, SelbachM, RollenhagenC, BallmaierM, MeyerTF, et al. (2006) Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440 : 303–307.
52. AndersS, HuberW (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



