-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
Amphiphysin 2, encoded by BIN1, is a key factor for membrane sensing and remodelling in different cell types. Homozygous BIN1 mutations in ubiquitously expressed exons are associated with autosomal recessive centronuclear myopathy (CNM), a mildly progressive muscle disorder typically showing abnormal nuclear centralization on biopsies. In addition, misregulation of BIN1 splicing partially accounts for the muscle defects in myotonic dystrophy (DM). However, the muscle-specific function of amphiphysin 2 and its pathogenicity in both muscle disorders are not well understood. In this study we identified and characterized the first mutation affecting the splicing of the muscle-specific BIN1 exon 11 in a consanguineous family with rapidly progressive and ultimately fatal centronuclear myopathy. In parallel, we discovered a mutation in the same BIN1 exon 11 acceptor splice site as the genetic cause of the canine Inherited Myopathy of Great Danes (IMGD). Analysis of RNA from patient muscle demonstrated complete skipping of exon 11 and BIN1 constructs without exon 11 were unable to promote membrane tubulation in differentiated myotubes. Comparative immunofluorescence and ultrastructural analyses of patient and canine biopsies revealed common structural defects, emphasizing the importance of amphiphysin 2 in membrane remodelling and maintenance of the skeletal muscle triad. Our data demonstrate that the alteration of the muscle-specific function of amphiphysin 2 is a common pathomechanism for centronuclear myopathy, myotonic dystrophy, and IMGD. The IMGD dog is the first faithful model for human BIN1-related CNM and represents a mammalian model available for preclinical trials of potential therapies.
Published in the journal: . PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003430
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003430Summary
Amphiphysin 2, encoded by BIN1, is a key factor for membrane sensing and remodelling in different cell types. Homozygous BIN1 mutations in ubiquitously expressed exons are associated with autosomal recessive centronuclear myopathy (CNM), a mildly progressive muscle disorder typically showing abnormal nuclear centralization on biopsies. In addition, misregulation of BIN1 splicing partially accounts for the muscle defects in myotonic dystrophy (DM). However, the muscle-specific function of amphiphysin 2 and its pathogenicity in both muscle disorders are not well understood. In this study we identified and characterized the first mutation affecting the splicing of the muscle-specific BIN1 exon 11 in a consanguineous family with rapidly progressive and ultimately fatal centronuclear myopathy. In parallel, we discovered a mutation in the same BIN1 exon 11 acceptor splice site as the genetic cause of the canine Inherited Myopathy of Great Danes (IMGD). Analysis of RNA from patient muscle demonstrated complete skipping of exon 11 and BIN1 constructs without exon 11 were unable to promote membrane tubulation in differentiated myotubes. Comparative immunofluorescence and ultrastructural analyses of patient and canine biopsies revealed common structural defects, emphasizing the importance of amphiphysin 2 in membrane remodelling and maintenance of the skeletal muscle triad. Our data demonstrate that the alteration of the muscle-specific function of amphiphysin 2 is a common pathomechanism for centronuclear myopathy, myotonic dystrophy, and IMGD. The IMGD dog is the first faithful model for human BIN1-related CNM and represents a mammalian model available for preclinical trials of potential therapies.
Introduction
Amphiphysin 2 is one of the key factors in muscular membrane remodeling. The gene, BIN1, has recently been associated with two different muscle disorders: centronuclear myopathy (CNM, MIM #255200) [1] and myotonic dystrophy (DM, MIM #160900 and #602668) [2]. However, the muscle-specific role of the ubiquitous protein amphiphysin 2 and the pathological mechanisms underlying the muscle disorders are not well understood. This is mainly due to the lack of faithful animal models.
Centronuclear myopathies are characterized by a generalized muscle weakness, atrophy, predominance of type I fibers, and aberrant positioning of nuclei and mitochondria [3]. The different genetic forms are not or are only moderately progressive. The X-linked and dominant CNM forms result from mutations in the phosphoinositide phosphatase myotubularin (MTM1) and the large GTPase dynamin 2 (DNM2), respectively [4], [5]. The autosomal recessive form (ARCNM) is caused by mutations in BIN1, probably involving a partial loss-of-function as the protein level was found to be normal in previously described patients [1]. Amphiphysin 2, encoded by BIN1, contains a N-terminal amphipathic helix, a BAR (Bin/Amphiphysin/Rvs) domain, able to sense and maintain membrane curvature, a Myc-binding domain and a SH3 domain, both implicated in protein-protein interactions [6], [7], [8]. There are at least 12 different isoforms, mainly differing by the presence or absence of a phosphoinositide-binding domain and a clathrin-binding domain encoded by exon 11 and exons 13–16, respectively [9], [10]. The clathrin-binding domain is present in the brain isoforms, while the phosphoinositide-binding (PI) domain is found almost exclusively in skeletal muscle isoforms [10], [11], [12]. Sequencing of cDNA demonstrated that all BIN1 skeletal muscle isoforms contain exon 11 [12]. All ARCNM mutations described to date are in ubiquitously expressed exons [1], [13], [14], [15], raising the question about the molecular basis of the muscle-specificity of the disease. The BAR domain mutations strongly decreased the amphiphysin 2 membrane tubulating properties when expressed in cultured cells, while SH3 truncating mutations were shown to impair the binding and recruitment of dynamin 2 [1].
Mis-splicing of the BIN1 muscle-specific exon 11 was reported in different forms of myotonic dystrophy (DM) [2]. DM is one of the most common muscular dystrophies in neonates and adults, and results from the expression of mutant RNAs with expanded CUG or CCUG repeats leading to the sequestration of splicing factors and subsequent defects in RNA splicing. Splicing alterations of the muscle chloride channel CLCN1 are suggested to be responsible for the myotonia, whereas aberrant splicing of the insulin receptor INSR gene is thought to cause insulin resistance in DM patients. Complete or partial skipping of BIN1 exon 11 in congenital and adult DM was shown to involve structural T-tubule abnormalities and subsequently muscle weakness [2]. However, there are numerous splicing defects in DM. It is therefore challenging to assess the exact contribution of BIN1 exon 11 skipping to the DM phenotype, even though severe hypotonia, respiratory failure and histopathological features such as fiber hypotrophy and centrally located nuclei in the congenital forms of DM show intriguing similarities to CNM.
Amphiphysins are key regulators of membrane curvature and trafficking [16]. They can sense membrane curvature and presumably promote the curvature and fission of membranes [17]. Membrane binding occurs via BAR domain dimers, presenting a positively charged concave site that interacts with the negative membrane charges [17]. Amphiphysins also bind and recruit other regulators of endocytosis to sites of plasma membrane inward budding [18]. Amphiphysin 1 expression is restricted to neuronal tissues and the protein regulates synaptic vesicle recycling in the brain [19]. Amphiphysin 2 is highly expressed in adult striated muscle and its expression increases during muscle cell maturation [10], [11], [20], [21]. The polybasic residues encoded by BIN1 exon 11 are required for amphiphysin 2-induced membrane tubulation when exogenously expressed in cultured cells [1], [22]. In skeletal muscle, amphiphysin 2 is localized at the T-tubules, which are deep sarcolemmal invaginations enabling excitation-contraction coupling [11], i.e. the process converting an electrical stimulus into mechanical muscle work. This specific localization, together with the membrane tubulation properties of the muscle-specific isoform containing the PI domain, called iso8 or M-amphiphysin, has led to the suggestion that amphiphysin 2 is implicated in T-tubule biogenesis [22]. This is sustained by defects in the localization of nascent T-tubule markers such as caveolin 3 following BIN1 downregulation in cultured cells [23], and by the abnormal T-tubule structure seen in drosophila with null mutations in amph, the unique ortholog of mammalian amphiphysins 1 and 2 [24]. While faithful animal models were previously characterized for the MTM1 and DNM2 related CNM forms [25], the perinatal lethality of Bin1-null mice precludes the analysis of the role of amphiphysin 2 in skeletal muscle [26]. Therefore, critical questions concerning the muscle-specific function of amphiphysin 2 in mammals and the pathological mechanism of BIN1-related CNM remain unanswered. The lack of a faithful animal model for autosomal recessive centronuclear myopathy is a hurdle for a better comprehension of the pathological mechanisms and for the development of therapeutic approaches.
In this study, we identified and characterized the first human BIN1 mutation affecting the muscle-specific PI domain. We also identified a novel spontaneous canine model reproducing the human pathology and allowing investigations on the physiological role of amphiphysin 2 in skeletal muscle after birth. Characterization of the dog model revealed an important role for amphiphysin 2 in triad structure, and we provide the evidence for a physiological function of the membrane-deforming properties of amphiphysin 2 and its alternative splicing-dependent activity. Our data support the hypothesis that the alteration of the muscle-specific function of amphiphysin 2 on membrane remodeling is a common pathomechanism underlying several canine and human myopathies.
Results
BIN1 exon 11 splice mutation in patients with rapidly progressive centronuclear myopathy
To identify BIN1 mutations affecting its function in skeletal muscle, we sequenced the muscle-specific exon 11 and the adjacent splice-relevant intronic regions in a cohort of 84 patients with various forms of centronuclear myopathy and without mutations in MTM1, DNM2, or in the other BIN1 exons. We identified a homozygous BIN1 exon 11 splice acceptor mutation (IVS10-1G>A) in two affected members from a consanguineous family from Turkey (Figure 1A and 1B). DNA was not available for the third affected member, who is expected to carry the same homozygous BIN1 mutation as her monozygotic twin sister. The parents are healthy and do not present clinical signs of a muscle disorder. They are first-degree cousins and were found to be heterozygous for the BIN1 exon 11 splice acceptor mutation, confirming autosomal recessive inheritance of the disease. The mutation was not found in unaffected individuals from different origins, including 37 DNAs from an ethnically matched control population, and is not listed in the SNP databases as dbSNP, 1000 genomes, or the NHLBI exome variant server.
Fig. 1. Human BIN1 mutation of the exon 11 acceptor splice site and impact on splicing. 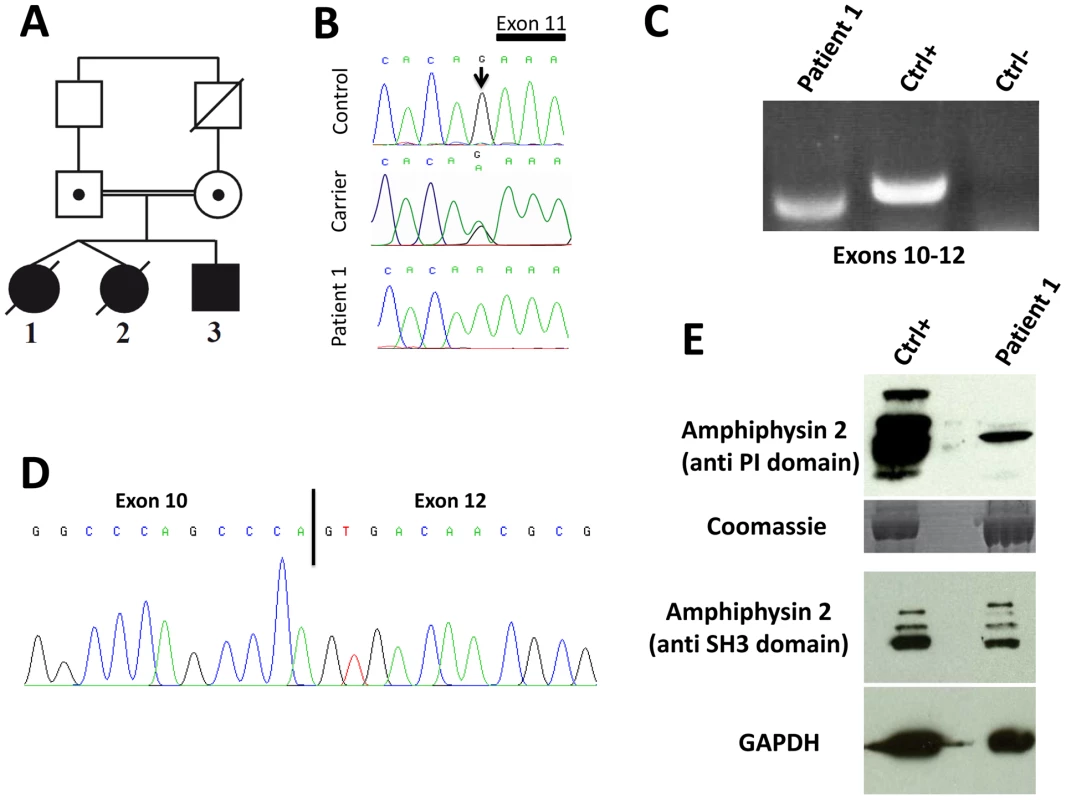
(A) Pedigree and (B) Chromatopherogram. Patients 1 and 3 are homozygous for the IVS10-1G>A mutation, while both parents are heterozygous carriers. DNA from patient 2 was not available. (C) RT-PCR on mRNA isolated from muscle using primers encompassing BIN1 exons 10–12 demonstrated amplification of a shorter product in patient 1 compared to a healthy control. For the negative control (Ctrl-) PCR was performed without cDNA. (D) Sequencing of the BIN1 cDNA from muscle demonstrated skipping of BIN1 exon 11 in patient 1. (E) Western blot analysis of patient muscle extracts detected a strong reduction of the amphiphysin 2 isoforms containing the exon 11 encoded PI-binding domain. The level of amphiphysin 2 detected with an anti-SH3 antibody was comparable between patient 1 and control. Patients 1 and 2 are dizygotic twins. Pregnancy and birth, as well as motor and speech development were normal. General muscle weakness was diagnosed at 3.5 years. Hypotonia, muscle weakness (predominantly of the lower limbs), respiratory distress (VC 50%) and loss of motor skills were rapidly progressive and the twins died from acute pneumonia involving cardiac failure at age 5 and 7, respectively. Patient 3 is the younger brother, and as for his sisters, pregnancy, birth, motor and speech development were normal. Age of onset was 3.5 years and the myopathy was highly progressive, contrasting the slow progression of muscle weakness in the reported CNM cases with BIN1 mutations in ubiquitous exons [1], [13], [14], [15]. Patient 3 presented with predominant proximal muscle weakness of the lower limbs requiring a wheelchair since the age of 5 years, facial weakness, but no respiratory distress. Eye movement defects, as seen in the majority of the MTM1, DNM2 and BIN1 patients, were not noted. Deep tendon reflexes were absent and the patient had progressive contractures in knees and ankles. Electrophysiological evaluation was normal or showed only unspecific myopathic changes, with normal nerve conduction velocity. Cardiac defects were not noted and CK levels were normal. Patient 3 is now 9 years old and presented at his last medical exam in April 2012 with low MRC grades for both upper and lower limbs.
Impact of the human BIN1 mutation on splicing
The BIN1 IVS10-1G>A variation changes the AG acceptor splice site into AA, and is predicted to impair exon 11 splicing by various algorithms. The wild-type acceptor site was recognized by NNSPLICE (score 0.84) and Human Splice Finder (88.5), while no acceptor splice site was predicted in the mutated sequence. To confirm an impact on exon 11 splicing, we performed RT-PCR after RNA isolation from a muscle biopsy of patient 1, amplified a fragment encompassing exons 10 to 12, and obtained a shorter product compared to the control (Figure 1C). To analyze the transcript(s), we cloned the PCR-products and sequenced the resulting clones. As we and others previously reported, the skeletal muscle BIN1 isoforms contain exon 11, but lack exons 7 and 13 to 16. Exon 17 can be either present or absent, corresponding to isoform 8 or M-amphiphysin [10], [11], [12]. Among the 30 analyzed clones, only a single clone contained exon 11. Twenty-nine clones did not contain exon 11 and directly combined exon 10 with exon 12, demonstrating a major skipping of the in-frame exon 11 in the patient muscle (Figure 1D). The impact of the mutation on the amphiphysin 2 protein level in skeletal muscle was investigated by Western blot (Figure 1E). Using an anti-PI domain antibody, we detected several bands in the control as previously reported [1], most probably reflecting post-translational modifications of the different isoforms containing exon 11. In the patient muscle, we found a significant decrease of the level of the amphiphysin 2 isoform containing the PI domain, confirming exon 11 skipping in most BIN1 muscle transcripts. The amphiphysin levels detected with the anti-SH3 domain antibody were similar in patient 1 and control. Together with the genetic data, we conclude that the rapidly progressive CNM form results from a splice mutation involving the skipping of the muscle-specific exon 11.
BIN1 exon 11 is required for membrane tubulation in muscle cells
Previous publications demonstrated the importance of the amphiphysin 2 PI domain in PtdIns(4,5)P2 binding and membrane tubulation [1], . We transfected C2C12 cells with BIN1 constructs including or excluding exon 11, and we induced the differentiation of the murine myoblasts into myotubes. Myotubes expressing the exon 11 containing isoform showed tubulation [22], [27], whereas the isoform without exon 11 did not induce this effect (Figure 2). Quantification revealed statistical significance. Immunolabelling of actin, caveolin-3 and RYR1 did not reveal obvious differences between the differentially transfected myotubes (data not shown), suggesting that the amphiphysin 2 PI domain is important for late muscle development or maintenance, rather than for early muscle development. This hypothesis is supported by the fact that the patients were unaffected at birth and during early childhood.
Fig. 2. Essential role of BIN1 exon 11 in membrane tubulation in myotubes. 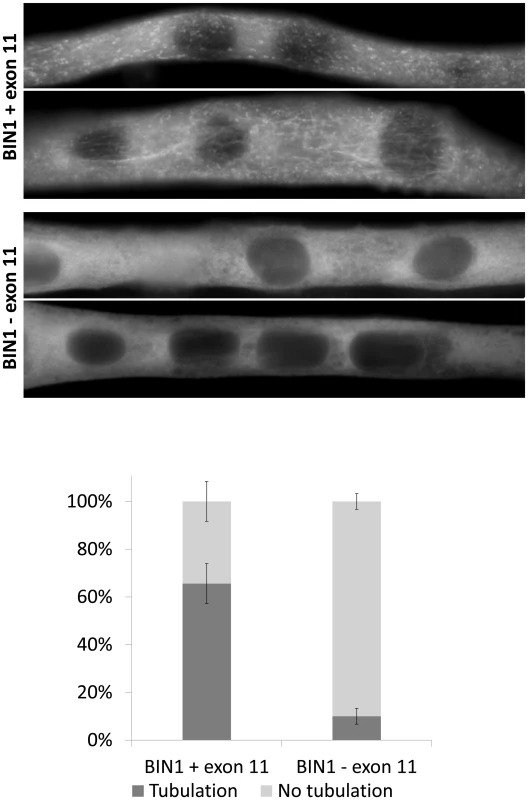
C2C12 myotubes overexpressing BIN1 isoform 8 (including exon 11) showed strong tubulation, whereas BIN1 isoform 9 (without exon 11) does not induce membrane tubulation 5 days post differentiation. Below: quantification of three independent experiments (>30 myotubes each) demonstrated that these findings were significant (p<0.01). BIN1 exon 11 splice mutation causes the canine Inherited Myopathy of Great Danes (IMGD)
The perinatal lethality of Bin1-null mice precludes investigations on the role of amphiphysin 2 in skeletal muscle maintenance [26]. To identify and characterize an animal model for BIN1-related CNM, we analyzed canine pedigrees with molecularly unsolved myopathies. The canine Inherited Myopathy of Great Danes (IMGD) is characterized by rapidly progressive muscle atrophy and exercise intolerance with an age of onset of about 6 months. Histological examinations of muscle biopsies from autosomal recessive cases from the UK, Canada and Australia revealed increased nuclear internalization and centralization [28], [29], [30], consistent with centronuclear myopathy. We excluded mutations in MTM1 [31] and PTPLA [32] before sequencing the coding regions and intron/exon boundaries of the canine BIN1 gene (XM_540990.3). We identified a homozygous AG to GG substitution of the BIN1 exon 11 acceptor splice site in five dogs from Canada, US and UK (IVS10-2A>G; Figures 3A and 3B). CK values for the dogs were normal or slightly elevated. Pedigree reconstruction revealed a distant relationship between the US and one UK dog (Figure 3C) and a previous publication reported a common ancestor for all IMGD dogs in the UK [29]. The BIN1 IVS10-2A>G mutation was not found in 112 healthy Great Danes and in 35 dogs from 12 other breeds, strongly suggesting its pathogenicity.
Fig. 3. The canine Inherited Myopathy of Great Danes results from a BIN1 mutation in the exon 11 acceptor splice site. 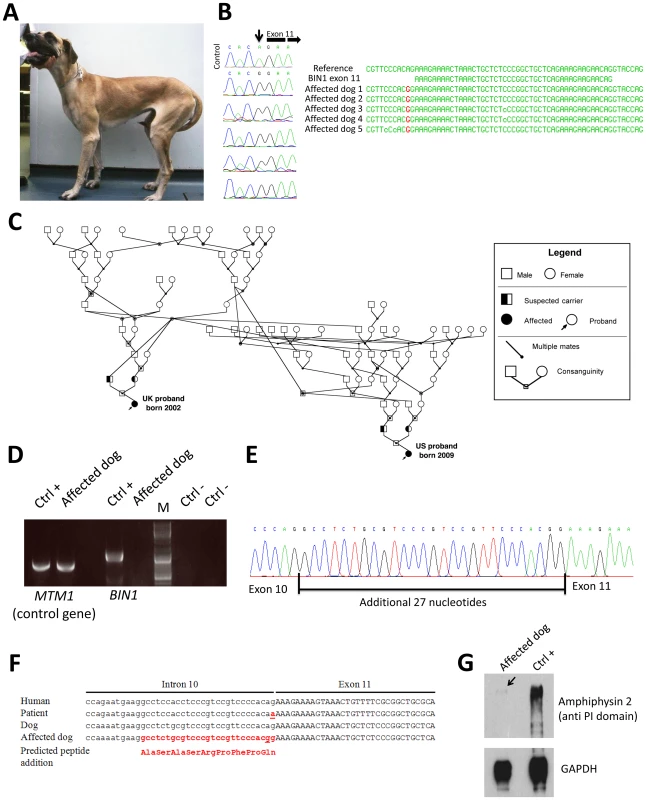
(A) Picture of an affected 3-year-old Great Dane dog. (B) Chromatopherograms and sequence alignment showing the BIN1 IVS10-2A>G mutation in 5 affected dogs. (C) Pedigree showing the distant relationship of two affected Great Dane dogs from the UK and US. (D) RT-PCR on skeletal muscle RNA showed a strong reduction of the BIN1 RNA level in the IMGD dog compared to the healthy canine control. Amplification of a control gene (MTM1) was normal. M = Marker (E) Sequencing of the residual cDNA revealed the presence of 27 additional nucleotides due to the use of a weak cryptic 5′ splice acceptor site. (F) Sequence alignment of human and canine BIN1 intron/exon boundary of exon 11. (G) Western blot using an anti-PI domain antibody showed a strong decrease of the amphiphysin 2 protein level. Impact of the canine BIN1 mutation on exon 11 splicing
Like the human BIN1 IVS10-1G>A mutation, the canine BIN1 IVS10-2A>G variation affects the exon 11 acceptor splice site. To assess its impact on splicing, we performed RT-PCR on RNA isolated from skeletal muscle biopsies and found a strong reduction of the BIN1 RNA level compared to healthy controls and compared to a control gene (MTM1, Figure 3D). We however detected a faint signal of expected size and cloned the amplicon. All three clones contained exon 11 with 27 additional upstream nucleotides, encoding the amino acid sequence ASASRPFPQ (Figure 3E). This in-frame extension results from the disposition of a weak cryptic 5′ acceptor site. The intronic sequence upstream of exon 11 slightly differs between human and dog, possibly explaining the cryptic splicing in dogs versus exon skipping in human patients (Figure 3F). To confirm the impact of the splice mutation on the amphiphysin 2 protein level, canine muscle extracts were probed with an anti-PI domain antibody on Western blot. Compared to the healthy control, amphiphysin 2 was significantly reduced in the affected dog (Figure 3G). Using an anti-SH3 antibody we detected a strong reduction of all skeletal muscle amphiphysin isoforms (Figure S1) in accordance with the RT-PCR data. We conclude that the canine Inherited Myopathy of Great Danes results from a BIN1 exon 11 splice mutation, provoking a strong reduction of the exon 11/PI domain-containing RNA and protein.
Similar histopathology in affected humans and dogs
Vastus lateralis muscle biopsies were performed for patient 1 as well as for patient 3 at the age of 3.5 years. H&E staining revealed prominent nuclear centralization (>60%, arrow), fiber atrophy and endomysial fibrosis (Figure 4), consistent with centronuclear myopathy. Similarly, H&E staining of biceps femoris muscle biopsies from affected dogs revealed nuclear internalization (>40%) and fiber atrophy. The central areas devoid of staining reflect perinuclear regions lacking myofibrils. Of note, the transverse muscle sections of patients and affected dogs showed an unusual lobulated appearance with indentations of the sarcolemma (arrowheads). NADH staining of human and canine sections revealed dense central areas in most fibers and “spoke of wheel” appearance in 5% of the fibers. ATPase staining showed no or only a slight predominance of type I muscle fibers as compared to the age matched controls. Gomori trichrome staining did not reveal any further abnormalities (data not shown). Taken together, human and canine histopathologies were comparable.
Fig. 4. Histopathological comparison of muscles from human patient and IMGD dog. 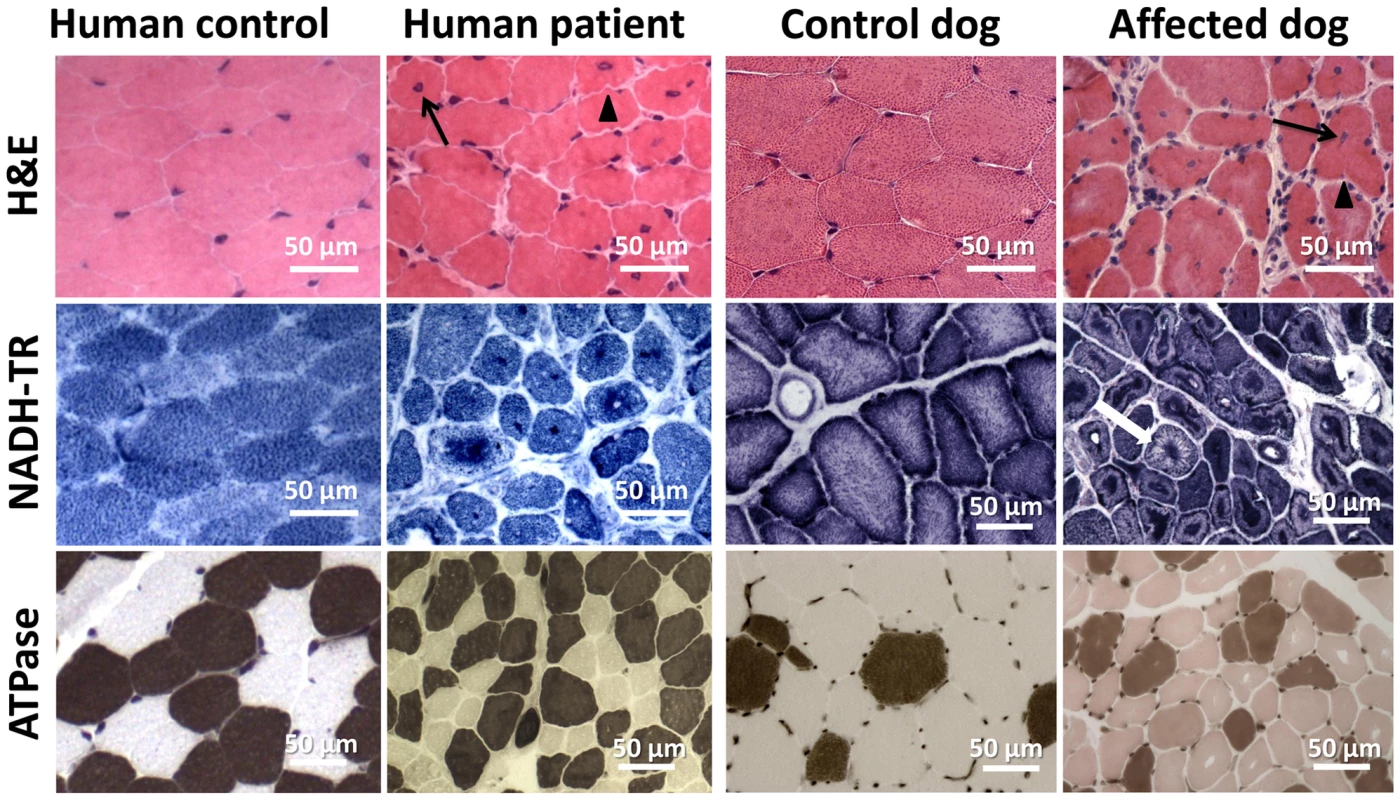
Human and canine muscle biopsy sections revealed nuclear centralization (arrows), fiber atrophy and lobulation as well as sarcolemmal invaginations (arrowheads) on H&E staining. NADH-TR staining demonstrated central dense areas in many fibers and “spoke of wheel” appearance in a few fibers (white arrow). ATPase staining (pH 4.3) revealed no or only a slight predominance of type I fibers compared to the age-matched controls. Common ultrastructural and membrane defects in affected patients and dogs
To uncover the pathological defects underlying this highly progressive form of centronuclear myopathy and to validate the canine model, we analyzed human and dog muscle biopsies by electron microscopy. Ultrastructural analysis of the human muscle biopsy revealed centralized nuclei surrounded by an area devoid of myofibrils and containing glycogen granules and other organelles (Figure 5A, Figure S2), as commonly seen in MTM1, DNM2 and BIN1-related CNM. Myofibrillar disintegration with occasional Z-band streaming (arrow, Figure 5A) was seen in the adjacent sarcomeres. Triad structures were found to be aberrant and we observed frequent enlarged structures, most probably originating from the sarcoplasmic reticulum (arrow, Figure 5D). We also noted other membrane alterations, including accumulations of membranes and tubules, vacuoles containing whorled membranes (arrow, Figure 5B), as well as a high number of myelin-like membranous structures suggestive of autophagosomes (arrow, Figure 5C). Likewise, ultrastructural analysis of muscle biopsies from an affected Great Dane dog showed nuclear internalization, mitochondrial accumulations around the internalized nuclei and myofibrillar disarray (Figure 5E, Figure S3). We furthermore found membranous whorls (arrow, Figure 5F) as reported for the X-linked CNM Labrador retriever model with MTM1 mutation [31], deep membrane invaginations (arrowhead, Figure 5F), lipofuscin granules (arrow, Figure 5G), and abnormal triads in almost all fibers (arrow, Figure 5H). Sarcolemmal invaginations contained basement membranes and often pointed towards centralized nuclei. Taken together and considering the histological analysis described above, histopathology of IMGD dogs and human patients appear strikingly similar, emphasizing common alterations of membrane structures.
Fig. 5. Common ultrastructural and membrane defects in affected human and dog. 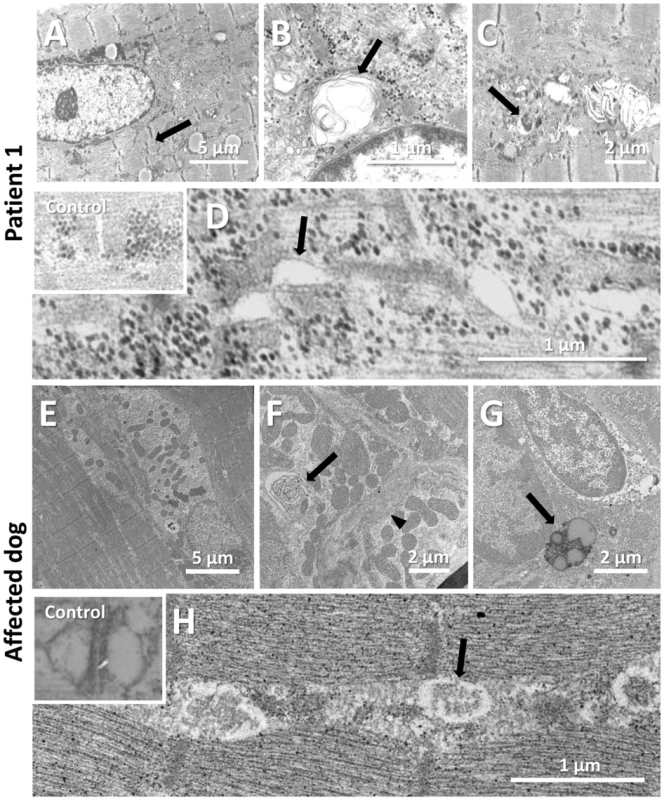
(A–D) Electron microscopic analysis of a patient biopsy showing a centralized nucleus surrounded by organelles and mild sarcomeric disarray (arrow, A), accumulations of membranes and vacuoles containing whorled membranes (arrow, B), autophagic vacuoles containing myelin-like material (arrow, C), and widened tubules at the triads (arrow, D). The inset shows normal triads in an age-matched biopsy. (E–H) Ultrastructural analysis of a IMGD dog biopsy revealed central nuclei surrounded by mitochondrial accumulations (E), membranous whorls (arrow, F), deep membrane invaginations (arrowhead, F), lipofuscin granules (arrow, G), and abnormal triads (arrow, H). The inserted picture shows a normal triad in an age-matched canine control. Amphiphysin 2 is present but altered in affected human and dog muscles
To further characterize the pathophysiology of the rapidly progressive human CNM and canine IMGD, we performed immunolocalization experiments on muscle biopsies. Using the R3062 antibody recognizing most amphiphysin isoforms or the PI-domain specific R2405 antibody, signals were detected as an intracellular network in transverse sections of human and canine controls (Figure 6). Signals were also detected in sections of muscles from patient and affected dog, reflecting the presence of different amphiphysin 2 isoforms as shown by Western blot. Despite the decrease of BIN1 RNA in affected dogs, the remaining mis-spliced in-frame transcripts can explain the detection of amphiphysin 2 on muscle sections, especially because immunohistochemistry is not quantitative. The amphiphysin 2 network appeared however abnormal in patient and IMGD sections. In some fibers we noted central areas without any signal, while in other fibers accumulations around centralized nuclei were observed (arrows). To determine whether these anomalies were specific for the BIN1 exon 11 splice mutation or rather a general BIN1-related CNM feature, we analyzed a muscle biopsy from a patient with the previously reported BIN1 p.Asp151Asn mutation and a classical ARCNM phenotype [1]. We observed similar accumulations of amphiphysin 2 (Figure 6A), suggesting that different BIN1 mutations in humans and dogs lead to similar amphiphysin 2 mis-localization in muscle.
Fig. 6. Amphiphysin 2 is present but altered in muscles from affected humans and dogs. 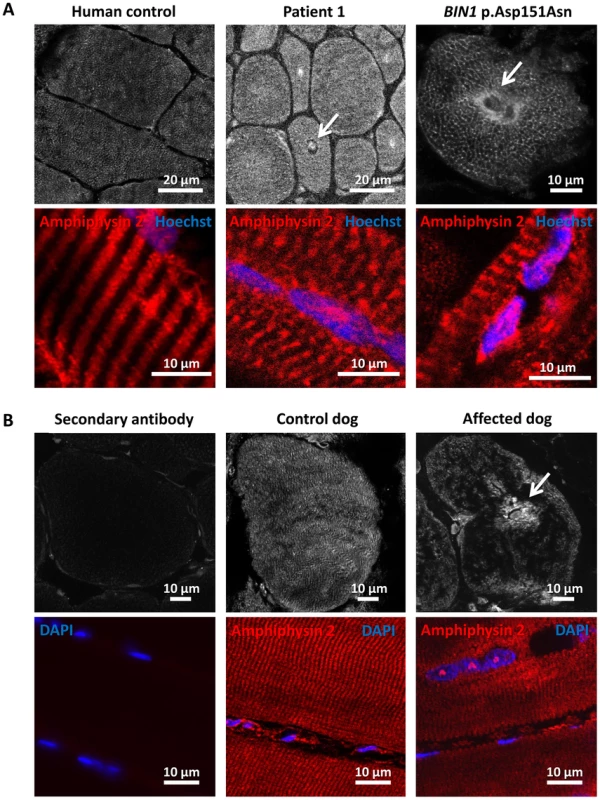
(A) Amphiphysin 2 localization in control (left), patient 1 (middle) and a CNM patient with the p.Asp151Asn mutation (right). Arrows indicate abnormal accumulations of amphiphysin 2 around centralized nuclei in both patient muscles. (B) Abnormal localization of amphiphysin 2 on transversal and longitudinal muscle sections from an IMGD dog compared to a control. The secondary without the primary antibody was applied on control canine sections to withdraw non-specific background staining. Alteration of triad and membrane trafficking regulators
Amphiphysin 2 has been proposed to be implicated in T-tubule biogenesis, but the exact link has barely been documented in mammalian skeletal muscle [22]. We therefore examined the skeletal muscle triad using antibodies against the junctional sarcoplasmic calcium channel RYR1 and the T-tubule marker DHPR in human and dog (Figure 7). Both proteins were profoundly altered, showing focal accumulations or central areas without signal in the fibers. Compared to the control longitudinal sections, the transversal orientation of RYR1-labeled triads was lost in patient and canine muscle. Similarly, the longitudinal sarcoplasmic calcium pump SERCA1 was mislocalized in sections from affected dogs.
Fig. 7. Alteration of triad components and proteins regulating membrane trafficking. 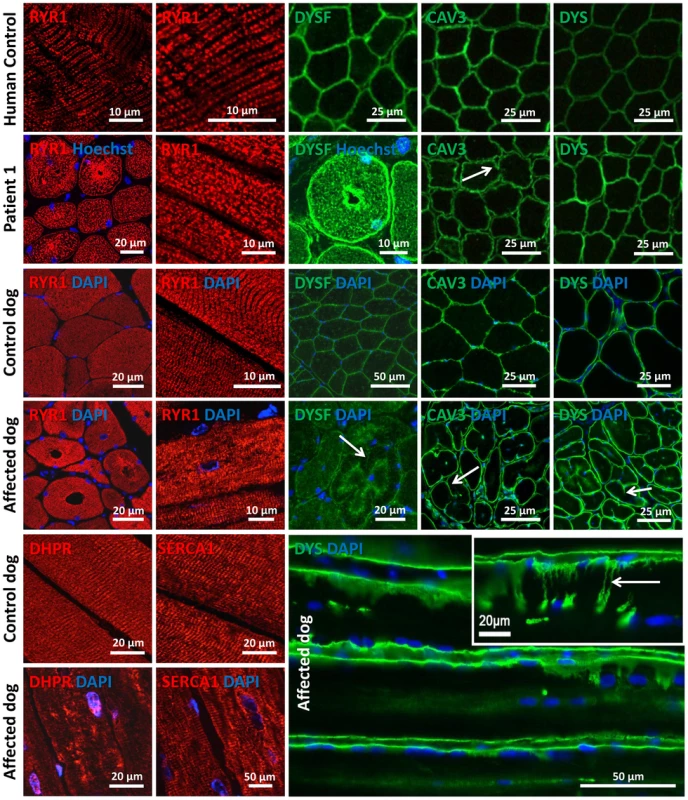
Immunolocalization on longitudinal and transversal sections from patients and affected dogs revealed an abnormal pattern of the junctional sarcoplasmic calcium channel RYR1, the T-tubule marker DHPR and the longitudinal sarcoplasmic calcium pump SERCA1. Especially RYR1 was found to accumulate around internalized nuclei. Intracellular dysferlin signals were detected in patients and affected dogs, but not in the age-matched controls. Labeling of the sarcolemmal markers caveolin 3 and dystrophin demonstrated prominent lobulation and deep indentations of the plasma membrane in patients and affected dogs on transversal and longitudinal sections (arrows). We next wanted to know whether the aberrant triad structure was concurrent with more generalized membrane defects. Dysferlin and caveolin 3, key regulators of membrane repair and trafficking [33], [34], were found to be mainly located at the sarcolemma in control muscle sections. In contrast, transverse sections of patient 1 and of an affected Great Dane dog revealed striking intracellular accumulations of dysferlin, mainly around central nuclei (Figure 7). Labeling of the sarcolemmal markers dysferlin, caveolin 3 and dystrophin confirmed the presence of numerous fibers with unusual lobulated and indented sarcolemma, representing deep sarcolemmal invaginations pointing towards the center of the fibers (arrows, Figure 7). Taken together, our data correlate the highly progressive human CNM and canine IMGD with general membrane alterations at the triad, the sarcolemma and within the fibers. However, these defects did not reflect a general disorganization of the sarcomere, as alpha-actinin labeling appeared largely normal (not shown). Staining of developmental myosin revealed no difference between affected and control dogs, indicating that there is no excessive fiber regeneration in IMGD dogs (Figure S3).
Altered myotubularin localization in BIN1-mutated canine muscles
As MTM1 is mutated in X-linked human and canine CNM, we investigated the localization of myotubularin in muscle sections of IMGD dogs. Myotubularin formed an intracellular network in control sections and the signal was stronger in type II fibers labeled with the SERCA1 antibody (Figure 8). In both analyzed IMGD muscles, myotubularin was mainly located as concentric strands pointing to the center in both type I and type II fibers. We conclude that altered splicing of BIN1 has a strong impact on myotubularin localization in muscle, revealing a potential link between IMGD and X-linked CNM.
Fig. 8. Myotubularin is mis-localized in IMGD muscle. 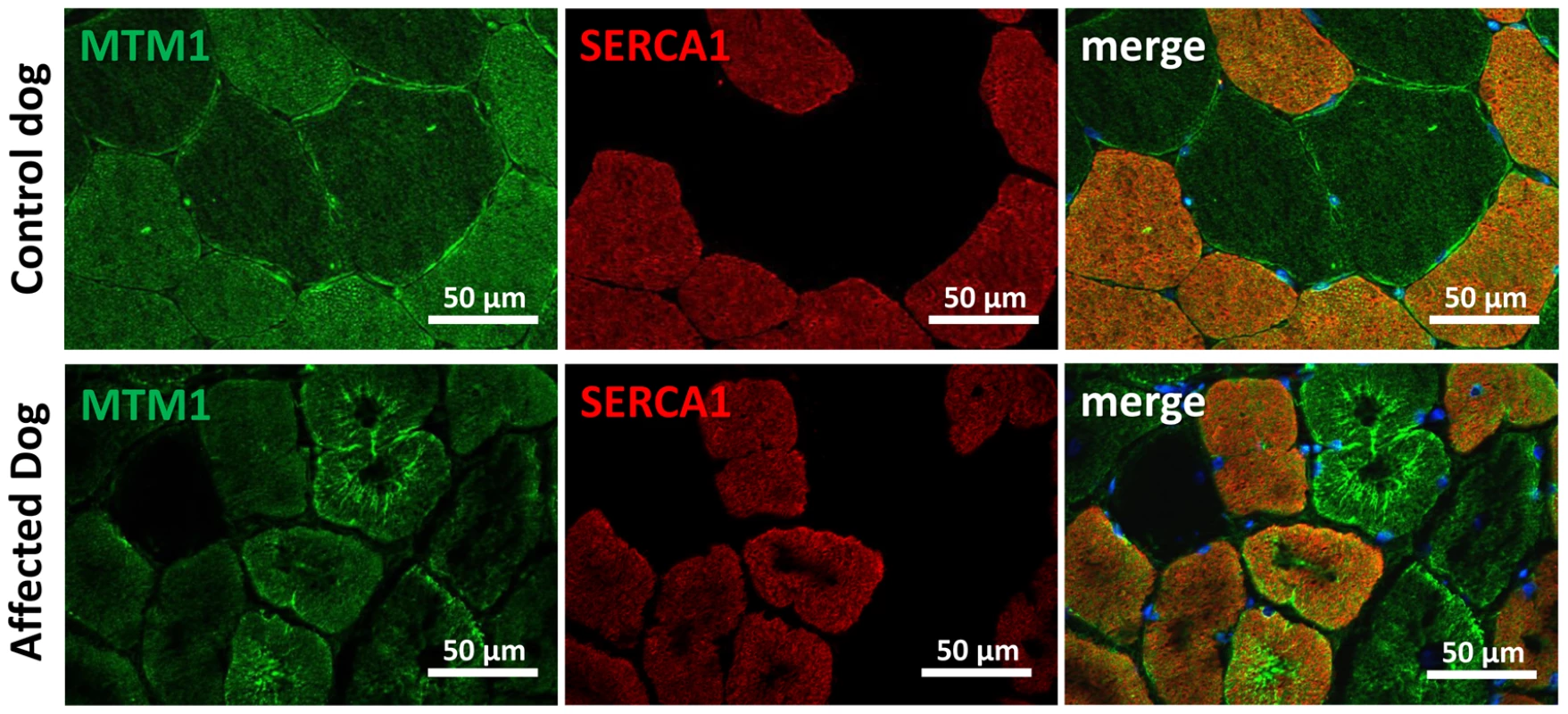
In muscle sections from control dogs, myotubularin is predominantly expressed in type II fibers expressing SERCA1. In affected IMGD dogs, massive myotubularin accumulations formed a concentric network around the fiber center. Discussion
In this study we identified and characterized BIN1 mutations affecting the splicing of the muscle-specific exon 11, resulting in a rapidly progressing myopathy in humans and dogs. The IMGD dog is the first faithful mammalian model for BIN1-related centronuclear myopathy and particularly for the highly progressive form, and is the only characterized mammalian model available for preclinical trials of potential therapies for this severe congenital myopathy. Our data provide strong evidence for muscle-specific functions of amphiphysin 2 in membrane structural organization and remodelling and allow novel insights into the overlapping pathogenesis of centronuclear myopathy and myotonic dystrophy. A schematic representation of the amphiphysin 2 protein domains and of the position of the mutations and splicing alterations causing classical autosomal recessive centronuclear myopathy, rapidly progressive human CNM and canine IMGD as well as myotonic dystrophy is shown in Figure 9.
Fig. 9. Schematic representation of the amphiphysin 2 domains and BIN1 alterations in different myopathies. 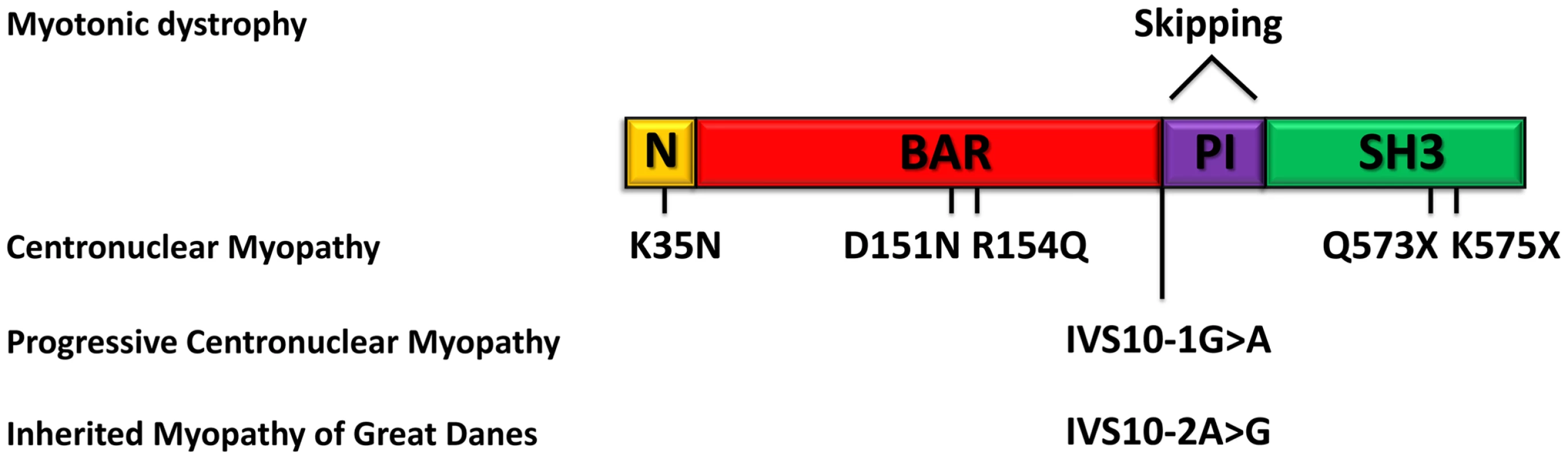
Schematic representation of the amphiphysin 2 protein domains and position of the known mutations causing autosomal recessive centronuclear myopathy, the new splice mutations resulting in rapidly progressive centronuclear myopathy and canine Inherited Myopathy of Great Danes. Myotonic dystrophy induces mis-splicing of BIN1 exon 11. Nomenclature is based on isoform 1 (NM_139343). BIN1 mutations in classical and highly progressive centronuclear myopathies
Classical BIN1-related ARCNM has been described with neonatal or childhood onset, hypotonia and ptosis and all mutations were found in ubiquitously expressed exons [1], [13], [14], [15]. The muscle weakness was mildly to moderately progressive, and some patients could still walk at older age. In contrast, our patients with a splice mutation affecting the muscle-specific exon 11 showed a rapid disease progression involving strong care-dependence and leading to death within a few years, despite normal motor development and disease-onset not before 3.5 years. The histopathological findings of our patients and of the previously reported ARCNM cases partially overlap, including atrophy, prominent nuclear internalization and central dense areas upon NADH-TR staining of muscle sections. However, there is no evidence for type I fiber predominance in the muscle biopsies of our patients. Previous RT-PCR experiments demonstrated a progressive integration of exon 11 in BIN1 mRNA during human skeletal muscle development [2]. We therefore hypothesize that the muscle-specific exon 11 plays a major role in muscle maintenance, rather than in early muscle development. This is in accordance with the highly progressive phenotype of humans and dogs with a disease onset several months or years after birth. Consistently, we detected amphiphysin 2 in muscle tissue, but RNA analysis revealed major skipping of BIN1 exon 11. This suggests that the patients mainly express an embryonic BIN1 isoform, which might not be able to assume the function of the adult BIN1 isoform, possibly explaining the more progressive phenotype compared to patients with BIN1 mutations in the ubiquitously expressed exons.
The canine Inherited Myopathy of Great Danes is a faithful model for BIN1-related centronuclear myopathy
The characterization of the pathological mechanisms leading to BIN1-related CNM and the development of potential therapeutic approaches is obviated by the lack of a faithful animal model. Bin1-null mice are perinatally lethal [26], so that a comprehensive analysis of skeletal muscle alterations during disease development is not possible. We sought for dog breeds with molecularly unsolved congenital myopathies and we identified the canine Inherited Myopathy of Great Danes as a disease model reproducing the histological and physiological defects observed in BIN1-related CNM patients. IMGD has been reported for cases in Canada, Australia and UK and is characterized by generalized muscle atrophy, exercise intolerance, exercise-induced tremor and muscle wasting [29]. The disease typically starts before 10 months of age, is highly progressive, and most of the affected dogs are euthanized before 18 months of age due to severe debilitating muscle weakness. Histological examinations revealed internalized or central nuclei without evidence of inflammation, disruption of the sarcomeric architecture with central fiber areas devoid of myofibrils, and central accumulations of mitochondria and glycogen granules ([28], [29], [30] and our data). In addition, type I fiber predominance in combination with an increased expression of genes implicated in the slow-oxidative metabolism was described [35]. In this study we demonstrate that IMGD and progressive CNM have a comparable etiopathology and both conditions result from mutations of the AG acceptor splice site of the BIN1 muscle-specific exon 11. The histopathology and the cellular organization defects of the human and canine muscle disorders are almost identical, we therefore consider IMGD as a faithful mammalian model for BIN1-related centronuclear myopathy.
Veterinary implications
Some dogs of our IMGD cohort were found to be negative for BIN1 mutations, suggesting that IMGD encompasses several disorders with similar clinical and overlapping histopathological features. The proven relationship of two affected Great Dane dogs demonstrates a common origin of the BIN1 exon 11 splice mutation, and it is likely that all five affected dogs described here can be traced back to a common ancestor. As the muscle disorder is inherited as a recessive trait, and as canine pedigrees are generally highly inbred, it is likely that the mutation can be found in Great Dane dog populations from all over the world, as recently demonstrated for another autosomal recessive CNM form in Labrador retrievers [36]. It is therefore of veterinary medical interest to sequence BIN1 exon 11 in Great Dane dogs. Also, veterinarians and veterinary pathologists should consider BIN1 mutations as a possible cause of any unexplained progressive myopathy in dogs, especially when the biopsy displays internal nuclei and lobulated or indented sarcolemma.
Insights into amphiphysin 2 muscle-specific functions and pathological mechanisms of centronuclear myopathy
Detailed immunohistochemical and ultrastructural analyses of muscles from patients and affected Great Dane dogs revealed common membrane alterations and abnormal accumulations of proteins regulating membrane trafficking. Similar findings were observed on biopsies from patients with DNM2 or MTM1 mutations [12], suggesting that mislocalization of triad proteins reflects common aberrations in CNM and that the amphiphysin 2 muscle-specific isoform plays an important role in triad formation and/or maintenance. This is in accordance with the known biochemical function of amphiphysin 2 and other N-BAR domain proteins to sense membrane curvature and to potentially induce curvature through the insertion of an amphipathic helix into the membrane bilayer. In vitro and cell culture experiments have led to the suggestion that the exon 11 encoded PI-binding motif is essential for membrane tubulation in cultured muscle cells [22]. Indeed, Drosophila mutated for amphiphysin, the ortholog of both amphiphysin 1 and amphiphysin 2, display an abnormal T-tubule system [24]. T-tubule alterations and muscle weakness were reproduced in murine Tibialis anterior injected with a U7 small nuclear RNA construct harboring an antisense sequence promoting BIN1 exon 11 skipping [2]. However, nuclear centralization and atrophy were not observed, contrasting with the IMGD model. This difference might be species-related, is possibly due to a low efficacy of the AAV-U7 method or alternatively to the examination time point 4 months post injection. As the triad is the membrane structure controlling excitation-contraction coupling, this suggests that impaired excitation-contraction coupling and subsequent calcium homeostasis defects are a primary cause of the myopathy. Of note, abnormal intracellular calcium release was observed in isolated murine muscle fibers after BIN1 shRNA-mediated knock-down [37]. Together with the present characterization of the IMGD model, these data indicate that amphiphysin 2 has an important muscle-specific role in triad structural maintenance, and provide additional evidence that triad modifications are a common defect in centronuclear myopathies, IMGD and myotonic dystrophies [2], [12].
Triads are not the only membrane compartment affected in patients and dogs harboring BIN1 exon 11 mutations. We also noted central accumulations of caveolin 3 and dysferlin, two key regulators of membrane trafficking in skeletal muscle, numerous membranous whorls, and a peculiar remodeling of the sarcolemma, manifesting an indented fiber perimeter and invaginations towards the center of the fibers. Caveolin 3 regulates membrane tension at the sarcolemma and dysferlin controls membrane exocytosis in sarcolemmal membrane repair [33], [34]. As both proteins are also present on regenerating T-tubules [38], their mislocalization resulting from a BIN1 mutation would be in accordance with defective T-tubule regeneration. Moreover, data mainly obtained in cultured cells support a key role of amphiphysins in the formation of endocytic vesicles [16], and a study in Caenorhabditis elegans suggested a role of amphiphysin in vesicle recycling [39]. Defects in intracellular signaling resulting from calcium defects and impaired transport of ion channels and growth factor might explain the muscle weakness and atrophy in BIN1-related CNM.
Amphiphysin 2 links several forms of centronuclear myopathies and myotonic dystrophy
Our findings on the IMGD model uncovered possible links between BIN1-related and other forms of CNM. Altered triads and the presence of membranous whorls were reported for MTM1 dog, mouse and zebrafish models as well as for patients with MTM1 mutations involving protein loss [12], [31], [40], [41], [42]. Abnormal triad markers were also reported for MTM1-related and DNM2-related CNM [12], [43]. Dysferlin localization was not extensively studied in MTM1-CNM but was increased in the cytoplasm of a mouse model and in patients with DNM2-CNM [44]. Moreover, we found myotubularin localization was strongly impaired in IMGD muscles. These findings suggest that myotubularin and amphiphysin 2 are in the same pathway regulating membrane remodeling in skeletal muscle and strengthen the hypothesis of a common pathological mechanism of the X-linked and the autosomal recessive CNM forms.
Alternative splicing of BIN1 exon 11 is mis-regulated in patients with myotonic dystrophy [2]. The parallel inclusion of exon 7 was noted, but its impact has not been assessed yet. Here we report the first mutation affecting the muscle-specific exon 11 of BIN1 and having an impact on splicing. The major clinical and histological aspects of the patients and IMGD dogs include general muscle weakness, atrophy and nuclear centralization, consistent with the muscle phenotype in DM patients. Our data therefore support the hypothesis that mis-splicing of BIN1 exon 11 partially accounts for the muscle-specific signs in myotonic dystrophy.
Materials and Methods
Ethics statement
Human sample collection was performed with informed consent from the patients according to the declaration of Helsinki and experimentation was performed as part of routine diagnosis. All dogs were examined with the consent of their owners. Blood and biopsies were obtained as part of routine clinical procedures for diagnostic purposes. Cheek cells were collected by owners or veterinarians using non-invasive swabs. As the data were from client-owned dogs undergoing normal veterinary exams, there was no “animal experiment” according to the legal definitions in Europe and the US. All local regulations related to clinical procedures were observed. Cryopreserved muscle specimens were processed and stored at the University of California, San Diego, under a tissue transfer approval from the institutional Animal Care and Use Committee.
Molecular genetics
Human Genomic DNA was prepared from peripheral blood by routine procedures and sequenced for all coding exons and intron/exon boundaries of MTM1, DNM2, and BIN1 as described elsewhere [1], [4], [5]. Patient 1 had a normal CTG repeat length at the DMPK locus (7 and 13 repeats) and was therefore excluded for myotonic dystrophy. Control DNAs were from healthy individuals of Turkish origin.
Dog DNA samples were extracted from cheek cells, venous blood or muscle biopsy specimens (cryosections or paraffin embedded tissue) by routine procedures and sequenced for all coding exons and intron/exon boundaries of canine MTM1 [31], PTPLA [32] and BIN1 (primer sequences in Table S1). Control samples were from a world-wide collection of healthy Great Danes as well as from healthy individuals of 13 other breeds.
RNA studies
RNA was extracted from muscle biopsies by routine procedures and reverse transcribed using the SuperScript III kit (Invitrogen, Carlsbad, USA). Human and dog amplicons were cloned into the pGEM-T Easy vector (Promega, Madison, USA) and transfected into E.coli DH5α cells. Blue/white selection, repeated twice, resulted in 30 clones for the human cDNA and 3 clones for the canine cDNA. Control dog was an unaffected Drahthaar (German Wirehaired Pointer). Primer sequences are listed in Table S1.
Protein studies
Western blot and immunofluorescence were performed using routine protocols. Biceps femoris and tibialis anterior biopsies from two affected dogs (14 months and 22 months, respectively) and from healthy age-matched Golden Retrievers or Belgian Shepherds as controls have been used for the analysis. Following antibodies were used for the study: R2406 (home-made rabbit anti-BIN1 PI binding domain), R2444 (home-made rabbit anti-BIN1 SH3 domain), R3062 (home-made rabbit anti-BIN1 exon 12 epitope), R2867 and R2868 (home-made rabbit anti-MTM1), mouse anti-GAPDH (Merck Millipore, Darmstadt, Germany), mouse anti-ryanodine receptor 1 (Affinity BioReagents, Golden, USA), mouse anti-SERCA 1 (Affinity BioReagents, Golden, USA), rabbit anti-dysferlin (Euromedex, Souffelweyersheim, France), goat anti-caveolin-3 (Tebu-BIO, Le-Perray-en-Yvelines, France), rabbit anti-caveolin-3 (Affinity BioReagents, Golden, USA), mouse anti-DHPR (Affinity BioReagents, Golden, USA), and mouse anti-dystrophin (Leica Microsystems, Germany). For immunohistofluorescence, transverse cryosections were prepared, fixed and stained by routine methods. Nuclei were stained with Hoechst or DAPI (Sigma-Aldrich, St. Louis, USA). Sections were mounted with slowfade antifade reagent (Invitrogen, Carlsbad, USA) and viewed using a laser scanning confocal microscope (TCS SP2; Leica Microsystems, Wetzlar, Germany) or a a Zeiss Axio Observer Z.1 microscope equipped with a 20×, 40× or 63× lens and Axioplan imaging with structured illumination (Carl Zeiss, Jena, Germany).
Muscle histology
For histochemical analyses, transverse sections of muscle cryosections (8 µm) of vastus lateralis and biceps femoris muscle biopsies were stained with hematoxylin-eosin, modified Gomori trichrome, NADH-TR and myofibrillar ATPase and then assessed for centralized nuclei, fiber morphology, fiber type distribution, cores, protein accumulation and cellular infiltrations.
Electron microscopy
Muscle biopsies were processed for electron microscopy as described previously [45]. Briefly, the tissue was fixed either in 6% phosphate-buffered glutaraldehyde (human patient) or in 2.5% paraformaldehyde, 2.5% glutaraldehyde, and 50 mM CaCl2 in 0.1 M cacodylate buffer at pH 7.4 (dog), and post-fixed with 2% OsO4, 0.8% K3Fe(CN)6 in 0.1 M cacodylate buffer (pH 7.4) for 2 h at 4°C and incubated with 5% uranyl acetate for 2 h at 4°C. Samples were dehydrated in graded series of ethanol and embedded in epoxy resin 812. Ultrathin sections (70 nm) were contrasted with uranyl acetate and lead citrate.
Membrane tubulation assay
Murine C2C12 myoblasts were seeded on coverslips and transfected at 50–60% confluency using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) either with GFP-BIN1 isoform 8 (including exon 11) or isoform 9 (excluding exon 11, both were a kind gift from Pietro de Camilli, Howard Hughes Medical Institute, USA). Cells were differentiated after 24 h by changing to medium containing 2% horse serum instead of FCS and fixed and stained after 5 days of differentiation by routine methods. Nuclei were stained with Hoechst/DAPI (Sigma-Aldrich, St. Louis, USA) and sections were mounted with slowfade antifade reagent and viewed using a laser scanning confocal microscope (TCS SP2; Leica Microsystems, Wetzlar, Germany).
Web resources
1000 genomes - A Deep Catalog of Human Genetic Variation (URL: http://www.1000genomes.org/)
Database of Single Nucleotide Polymorphisms (dbSNP). Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine. (dbSNP Build ID: 134).
(URL: http://www.ncbi.nlm.nih.gov/SNP/)
Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/)
Online Mendelian Inheritance in Man (OMIM) (URL: http://www.omim.org/)
NNsplice: prediction of splice mutations (URL: http://www.fruitfly.org/seq_tools/splice.html)
Human Splicing finder (URL: http://www.umd.be/HSF/)
Supporting Information
Zdroje
1. NicotAS, ToussaintA, ToschV, KretzC, Wallgren-PetterssonC, et al. (2007) Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat Genet 39 : 1134–1139.
2. FugierC, KleinAF, HammerC, VassilopoulosS, IvarssonY, et al. (2011) Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med 17 : 720–725.
3. JungbluthH, Wallgren-PetterssonC, LaporteJ (2008) Centronuclear (myotubular) myopathy. Orphanet J Rare Dis 3 : 26.
4. BitounM, MaugenreS, JeannetPY, LaceneE, FerrerX, et al. (2005) Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet 37 : 1207–1209.
5. LaporteJ, HuLJ, KretzC, MandelJL, KioschisP, et al. (1996) A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet 13 : 175–182.
6. GrabsD, SlepnevVI, SongyangZ, DavidC, LynchM, et al. (1997) The SH3 domain of amphiphysin binds the proline-rich domain of dynamin at a single site that defines a new SH3 binding consensus sequence. J Biol Chem 272 : 13419–13425.
7. ItohT, De CamilliP (2006) BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta 1761 : 897–912.
8. SakamuroD, ElliottKJ, Wechsler-ReyaR, PrendergastGC (1996) BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Genet 14 : 69–77.
9. RenG, VajjhalaP, LeeJS, WinsorB, MunnAL (2006) The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol Mol Biol Rev 70 : 37–120.
10. Wechsler-ReyaR, SakamuroD, ZhangJ, DuhadawayJ, PrendergastGC (1997) Structural analysis of the human BIN1 gene. Evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J Biol Chem 272 : 31453–31458.
11. ButlerMH, DavidC, OchoaGC, FreybergZ, DaniellL, et al. (1997) Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of Ranvier in brain and around T tubules in skeletal muscle. J Cell Biol 137 : 1355–1367.
12. ToussaintA, CowlingBS, HniaK, MohrM, OldforsA, et al. (2011) Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta Neuropathol 121 : 253–266.
13. BohmJ, YisU, OrtacR, CakmakciH, KurulSH, et al. (2010) Case report of intrafamilial variability in autosomal recessive centronuclear myopathy associated to a novel BIN1 stop mutation. Orphanet J Rare Dis 5 : 35.
14. ClaeysKG, MaisonobeT, BohmJ, LaporteJ, HezodeM, et al. (2010) Phenotype of a patient with recessive centronuclear myopathy and a novel BIN1 mutation. Neurology 74 : 519–521.
15. MejaddamAY, NennesmoI, SejersenT (2009) Severe phenotype of a patient with autosomal recessive centronuclear myopathy due to a BIN1 mutation. Acta Myol 28 : 91–93.
16. QualmannB, KochD, KesselsMM (2011) Let's go bananas: revisiting the endocytic BAR code. EMBO J 30 : 3501–3515.
17. PeterBJ, KentHM, MillsIG, VallisY, ButlerPJ, et al. (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303 : 495–499.
18. OwenDJ, WiggeP, VallisY, MooreJD, EvansPR, et al. (1998) Crystal structure of the amphiphysin-2 SH3 domain and its role in the prevention of dynamin ring formation. EMBO J 17 : 5273–5285.
19. Di PaoloG, SankaranarayananS, WenkMR, DaniellL, PeruccoE, et al. (2002) Decreased synaptic vesicle recycling efficiency and cognitive deficits in amphiphysin 1 knockout mice. Neuron 33 : 789–804.
20. MaoNC, SteingrimssonE, DuHadawayJ, WassermanW, RuizJC, et al. (1999) The murine Bin1 gene functions early in myogenesis and defines a new region of synteny between mouse chromosome 18 and human chromosome 2. Genomics 56 : 51–58.
21. Wechsler-ReyaRJ, ElliottKJ, PrendergastGC (1998) A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol Cell Biol 18 : 566–575.
22. LeeE, MarcucciM, DaniellL, PypaertM, WeiszOA, et al. (2002) Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 297 : 1193–1196.
23. KojimaC, HashimotoA, YabutaI, HiroseM, HashimotoS, et al. (2004) Regulation of Bin1 SH3 domain binding by phosphoinositides. Embo J 23 : 4413–4422.
24. RazzaqA, RobinsonIM, McMahonHT, SkepperJN, SuY, et al. (2001) Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev 15 : 2967–2979.
25. CowlingBS, ToussaintA, MullerJ, LaporteJ (2012) Defective membrane remodeling in neuromuscular diseases: insights from animal models. PLoS Genet 8: e1002595.
26. MullerAJ, BakerJF, DuHadawayJB, GeK, FarmerG, et al. (2003) Targeted disruption of the murine Bin1/Amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Mol Cell Biol 23 : 4295–4306.
27. SpiegelhalterC, ToschV, HentschD, KochM, KesslerP, et al. (2010) From dynamic live cell imaging to 3D ultrastructure: novel integrated methods for high pressure freezing and correlative light-electron microscopy. PLoS One 5: e9014.
28. DaviesSE, DaviesDR, RichardsRB, BruceWJ (2008) Inherited myopathy in a Great Dane. Aust Vet J 86 : 43–45.
29. Lujan Feliu-PascualA, SheltonGD, TargettMP, LongSN, ComerfordEJ, et al. (2006) Inherited myopathy of great Danes. J Small Anim Pract 47 : 249–254.
30. McMillanCJ, TaylorSM, SheltonGD (2006) Inherited myopathy in a young Great Dane. Can Vet J 47 : 891–893.
31. BeggsAH, BohmJ, SneadE, KozlowskiM, MaurerM, et al. (2010) MTM1 mutation associated with X-linked myotubular myopathy in Labrador Retrievers. Proc Natl Acad Sci U S A 107 : 14697–14702.
32. PeleM, TiretL, KesslerJL, BlotS, PanthierJJ (2005) SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet 14 : 1417–1427.
33. BansalD, MiyakeK, VogelSS, GrohS, ChenCC, et al. (2003) Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 423 : 168–172.
34. SinhaB, KosterD, RuezR, GonnordP, BastianiM, et al. (2011) Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 144 : 402–413.
35. ChangKC, McCullochML, AndersonTJ (2010) Molecular and cellular insights into a distinct myopathy of Great Dane dogs. Vet J 183 : 322–327.
36. MaurerM, MaryJ, GuillaudL, FenderM, PeleM, et al. (2012) Centronuclear Myopathy in Labrador Retrievers: A Recent Founder Mutation in the PTPLA Gene Has Rapidly Disseminated Worldwide. PLoS One 7: e46408.
37. TjondrokoesoemoA, ParkKH, FerranteC, KomazakiS, LesniakS, et al. (2011) Disrupted membrane structure and intracellular Ca(2)(+) signaling in adult skeletal muscle with acute knockdown of Bin1. PLoS One 6: e25740.
38. KlingeL, HarrisJ, SewryC, CharltonR, AndersonL, et al. (2010) Dysferlin associates with the developing T-tubule system in rodent and human skeletal muscle. Muscle Nerve 41 : 166–173.
39. PantS, SharmaM, PatelK, CaplanS, CarrCM, et al. (2009) AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol 11 : 1399–1410.
40. Buj-BelloA, LaugelV, MessaddeqN, ZahreddineH, LaporteJ, et al. (2002) The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc Natl Acad Sci U S A 99 : 15060–15065.
41. DowlingJJ, VreedeAP, LowSE, GibbsEM, KuwadaJY, et al. (2009) Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet 5: e1000372.
42. PiersonCR, Dulin-SmithAN, DurbanAN, MarshallML, MarshallJT, et al. (2012) Modeling the human MTM1 p.R69C mutation in murine Mtm1 results in exon 4 skipping and a less severe myotubular myopathy phenotype. Hum Mol Genet 21 : 811–825.
43. RomeroNB, BitounM (2011) Centronuclear myopathies. Semin Pediatr Neurol 18 : 250–256.
44. DurieuxAC, VignaudA, PrudhonB, ViouMT, BeuvinM, et al. (2010) A centronuclear myopathy-dynamin 2 mutation impairs skeletal muscle structure and function in mice. Hum Mol Genet 19 : 4820–4836.
45. WeisJ, SchroderJM (1988) Adult polyglucosan body myopathy with subclinical peripheral neuropathy: case report and review of diseases associated with polyglucosan body accumulation. Clin Neuropathol 7 : 271–279.
Štítky
Genetika Reprodukční medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene ClusterČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 6- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



