-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Hooked and Cooked: A Fish Killer Genome Exposed
article has not abstract
Published in the journal: . PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003590
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003590Summary
article has not abstract
Few microorganisms match the impact that the oomycetes have had on mankind. This distinct lineage of eukaryotes is well-known for its most notorious member, Phytophthora infestans, the agent of the nineteenth century Irish potato famine, and several other devastating pathogens of cultivated and wild plants [1]. Indeed, more than 60% of oomycete species infect plants [2]. Less known is the fact that many oomycetes are parasitic on animals, from freshwater fish and crustaceans to mammals, such as livestock, pets, and humans [3]. Animal parasitic oomycetes have received much less attention than their plant pathogenic kin, and our understanding of their virulence mechanisms is rudimentary. However, research momentum is poised to accelerate with the first report of the genome of an animal parasitic oomycete. In this issue of PLOS Genetics, Jiang et al. [4] describe the 63 Mbp genome sequence of the fish pathogen Saprolegnia parasitica and highlight a distinct repertoire of candidate virulence genes.
Members of the genus Saprolegnia cause the disease saprolegniosis in both farmed and wild freshwater fish, such as the northern pike (Esox lucius) [3], [5], [6]. The disease is distinguished by mycelial growth on the fish skin and fins that can be followed by fatal invasion of the pathogen into muscles and blood vessels. Saprolegnia is particularly destructive in aquaculture, an industry of growing importance given the increased global consumption of fish and the decline of wild fish stocks. In high-value salmon farms, Saprolegnia causes significant damage with loss of about 10% of hatched fish [6]. The problem was exacerbated by the worldwide ban on the organic dye malachite green, which was widely used for chemical control but, as a toxic carcinogen, poses a health hazard to consumers [6], [7]. Consequently, saprolegniosis emerged as a significant problem for the aquaculture industry, and a proposed ban on another disinfectant, formalin, will further compound the problem.
The genome sequenced by Jiang et al. is of S. parasitica strain CBS223.65, which was isolated from infected pike fish (Esox lucius). The compact 63 Mbp genome encodes 17,065 predicted gene models. At one gene per 2.6 kb, it is the most gene-dense oomycete genome sequenced to date [8]. Extensive regions of the genome display loss of heterozygosity, which has also been observed in other oomycete genomes [9]. This may be a driver of genetic diversity. However, despite some similarities, the S. parasitica genome turns out to be quite different from other sequenced oomycetes with both loss and gain/expansion of gene classes that are consistent with the pathogen's lifestyle. The genome displays a large complement of kinases (kinome), larger than that of humans, and an expanded set of enzymes involved in chitin metabolism. However, it is the apparent adaptation to animal parasitism that is particularly exciting.
Pathogens and their hosts are engaged in a constant molecular arms race, with pathogens deploying virulence proteins as weapons to subvert hosts for their own benefit. Plant pathogenic oomycetes secrete a large array of cell wall–degrading enzymes that act to break down this protective physical barrier. These enzymes are essentially absent in S. parasitica as animal cells lack a cell wall. In contrast, the secretome of S. parasitica is dominated by proteases and lectins. One protease, which is highly expressed in mycelia (SPRG_14567), specifically degrades trout immunoglobulin M (IgM). This may represent a virulence activity designed to evade pathogen recognition by the fish immune system, suggesting secreted proteases can act to suppress host defences and are not necessarily deployed to attack host tissue.
It is well established that many plant parasitic oomycete genomes encode expanded families of putative host-translocated effectors characterised by amino acid sequence motifs such as RXLR, LFLAK, and CHXC [10]. Many of these effectors are expected to manipulate plant immunity. Interestingly, these types of effectors are completely absent in S. parasitica. Perhaps the lack of these classes of host-translocated effectors in the genome suggests they are not important for S. parasitica pathogenesis? Perhaps these types of effectors are not well suited for supporting infection of fish cells? However, one protein, SpHtp1, has previously been shown to translocate into fish cells [11]. The degree to which S. parasitica relies on other effectors in addition to SpHtp1 will be an interesting question to address in the future.
What other virulence proteins are deployed by S. parasitica? Remarkably, Jiang et al. discovered a large number of genes that are typical of animal parasites and are missing in plant pathogenic oomycetes. These include toxins (Haemolysin E-like), lectins and disintegrins (which may mediate host cell binding), CHAPs (cysteine, histidine-dependent amidohydrolases/peptidases), and secreted nucleases (Figure 1A). Even more remarkably, Jiang et al. show that these genes may have been acquired by horizontal gene transfer (HGT), possibly from bacteria. Further, the most abundant transposon in the S. parasitica genome belongs to the LINE repeat group. This repetitive element is prevalent in animal genomes and may have been acquired from an animal host. The role of the virulence factors acquired by HGT in progression of saprolegniosis will surely be a target for further study.
Fig. 1. Multiple virulence factors are deployed by Saprolegnia parasitica and an overview of oomycete phylogeny. 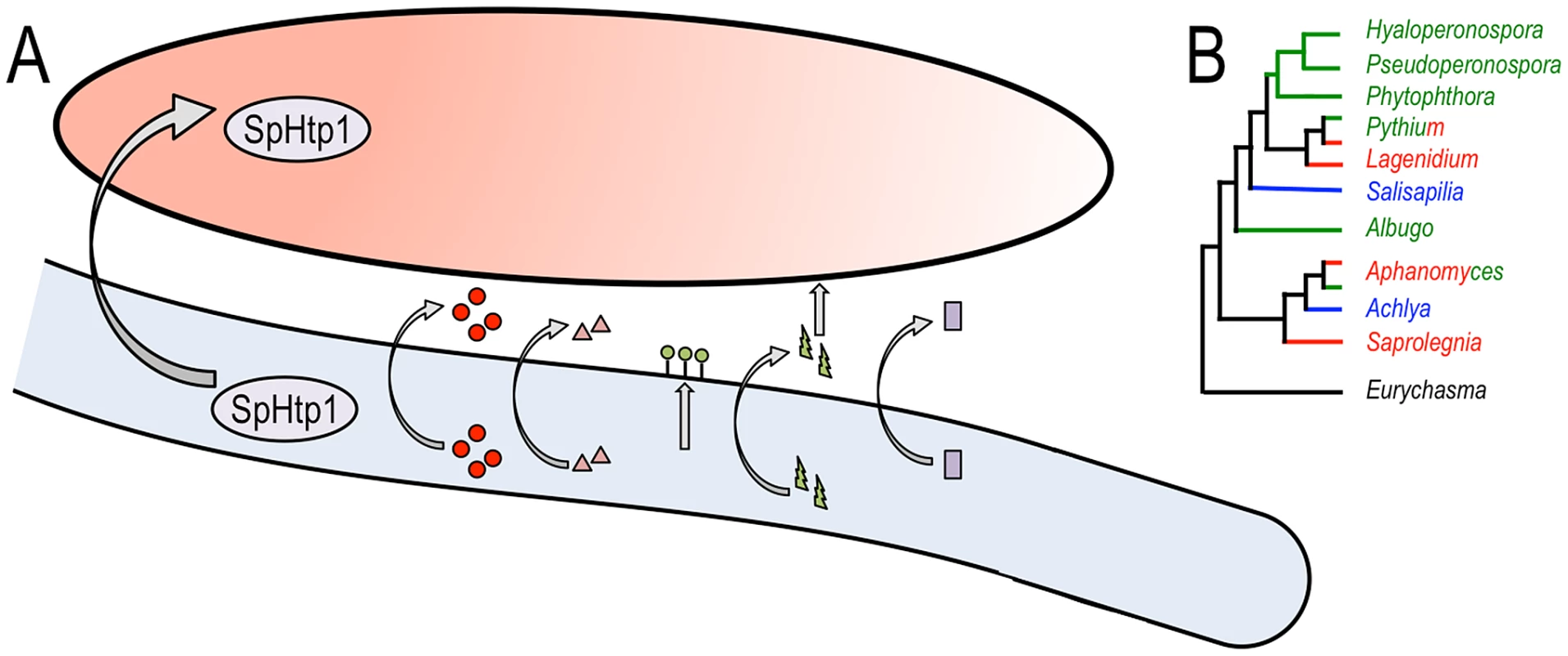
(A) Schematic representation of a Saprolegnia parasitica hypha (light blue) deploying virulence factors against a fish cell (salmon color). SpHtp1 is translocated inside the host cell, and other factors are secreted to the cell surface (lectins [green circles]) or the extracellular space (proteases [red circles], CHAPs [pink triangles], toxins [HlyE, which presumably targets the host membrane, green bolts] and nucleases [purple squares]). B. An overview of oomycete phylogeny. The main genera are displayed with the plant pathogenic lineages in green, animal parasites in red, and saprophytes in blue. Some genera, such as Pythium and Aphanomyces, include both plant and animal parasitic species. The early branching Eurychasma is an obligate pathogen of marine brown algae. As highlighted by Jiang et al., the genetic makeup of the fish parasite S. parasitica turned out to be markedly different from those of plant pathogenic oomycetes. Thus, it is important to sample across a diverse range of parasitic and saprophytic lifestyles to gain a thorough understanding of the structure and function of oomycete genomes (an overview of oomycete phylogeny is given in Figure 1B). To date, genome sequence analyses of plant pathogenic oomycetes of the genera Albugo, Hyaloperonsopora, Phytophthora, Pseudoperonospora, and Pythium have been published [9], [12]–[19]. Yet we still await the complete genome sequence of saprophytic species and additional animal parasites such as Aphanomyces spp. [20]. With these genome sequences in hand, comparative analyses will shed further light on how these enigmatic eukaryotes have adapted to a variety of ecological niches and host species.
Zdroje
1. Lamour K, Kamoun S (2009) Oomycete Genetics and Genomics. Diversity, Interactions and Research Tools: Wiley-Blackwell.
2. ThinesM, KamounS (2010) Oomycete-plant coevolution: recent advances and future prospects. Curr Opin Plant Biol 13 : 427–433.
3. PhillipsAJ, AndersonVL, RobertsonEJ, SecombesCJ, van WestP (2008) New insights into animal pathogenic oomycetes. Trends Microbiol 16 : 13–19.
4. JiangRHY, de BruijnI, HaasBJ, BelmonteR, LöbachL, et al. (2013) Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLOS Genet 9: e1003272 doi:10.1371/journal.pgen.1003272
5. Bruno DW, van West P, Beakes GW (2009) Fish Diseases and Disorders, Volume 3: Viral, Bacterial and Fungal Infections, 2nd edition. Bruno DW, Woo PTK, editors. CABI.
6. van WestP (2006) Saprolegnia parasitica, an oomycete pathogen with a fishy appetite: new challenges for an old problem. Mycologist 20 : 99–104.
7. CulpSJ, BelandFA (1996) Malachite green: A toxicological review. J Am Coll Toxicol 15.
8. RaffaeleS, KamounS (2012) Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol 10 : 417–430.
9. LamourKH, MudgeJ, GobenaD, Hurtado-GonzalesOP, SchmutzJ, et al. (2012) Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol Plant Microbe Interact 25 : 1350–1360.
10. BozkurtTO, SchornackS, BanfieldMJ, KamounS (2012) Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol 15 : 483–492.
11. WawraS, BainJ, DurwardE, de BruijnI, MinorKL, et al. (2012) Host-targeting protein 1 (SpHtp1) from the oomycete Saprolegnia parasitica translocates specifically into fish cells in a tyrosine-O-sulphate-dependent manner. Proc Natl Acad Scie U S A 109 : 2096–2101.
12. BaxterL, TripathyS, IshaqueN, BootN, CabralA, et al. (2010) Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330 : 1549–1551.
13. HaasBJ, KamounS, ZodyMC, JiangRH, HandsakerRE, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461 : 393–398.
14. KemenE, GardinerA, Schultz-LarsenT, KemenAC, BalmuthAL, et al. (2011) Gene Gain and Loss during Evolution of Obligate Parasitism in the White Rust Pathogen of Arabidopsis thaliana. PLoS Biol 9: e1001094.
15. RaffaeleS, FarrerRA, CanoLM, StudholmeDJ, MacLeanD, et al. (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330 : 1540–1543.
16. TylerBM, TripathyS, ZhangX, DehalP, JiangRH, et al. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313 : 1261–1266.
17. TianM, WinJ, SavoryE, BurkhardtA, HeldM, et al. (2011) 454 Genome sequencing of Pseudoperonospora cubensis reveals effector proteins with a QXLR translocation motif. Mol Plant Microbe Interact 24 : 543–553.
18. LevesqueCA, BrouwerH, CanoL, HamiltonJP, HoltC, et al. (2010) Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol 11: R73.
19. CookeDE, CanoLM, RaffaeleS, BainRA, CookeLR, et al. (2012) Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog 8: e1002940.
20. GaulinE, MadouiMA, BottinA, JacquetC, MatheC, et al. (2008) Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS ONE 3: e1723.
Štítky
Genetika Reprodukční medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene ClusterČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 6- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Růst a vývoj dětí narozených pomocí IVF
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



