-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
Non-autonomous cell-death is a cardinal feature of the disintegration of neural networks in neurodegenerative diseases, but the molecular bases of this process are poorly understood. The neural retina comprises a mosaic of rod and cone photoreceptors. Cone and rod photoreceptors degenerate upon rod-specific expression of heterogeneous mutations in functionally distinct genes, whereas cone-specific mutations are thought to cause only cone demise. Here we show that conditional ablation in cone photoreceptors of Ran-binding protein-2 (Ranbp2), a cell context-dependent pleiotropic protein linked to neuroprotection, familial necrotic encephalopathies, acute transverse myelitis and tumor-suppression, promotes early electrophysiological deficits, subcellular erosive destruction and non-apoptotic death of cones, whereas rod photoreceptors undergo cone-dependent non-autonomous apoptosis. Cone-specific Ranbp2 ablation causes the temporal activation of a cone-intrinsic molecular cascade highlighted by the early activation of metalloproteinase 11/stromelysin-3 and up-regulation of Crx and CoREST, followed by the down-modulation of cone-specific phototransduction genes, transient up-regulation of regulatory/survival genes and activation of caspase-7 without apoptosis. Conversely, PARP1+-apoptotic rods develop upon sequential activation of caspase-9 and caspase-3 and loss of membrane permeability. Rod photoreceptor demise ceases upon cone degeneration. These findings reveal novel roles of Ranbp2 in the modulation of intrinsic and extrinsic cell death mechanisms and pathways. They also unveil a novel spatiotemporal paradigm of progression of neurodegeneration upon cell-specific genetic damage whereby a cone to rod non-autonomous death pathway with intrinsically distinct cell-type death manifestations is triggered by cell-specific loss of Ranbp2. Finally, this study casts new light onto cell-death mechanisms that may be shared by human dystrophies with distinct retinal spatial signatures as well as with other etiologically distinct neurodegenerative disorders.
Published in the journal: . PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003555
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003555Summary
Non-autonomous cell-death is a cardinal feature of the disintegration of neural networks in neurodegenerative diseases, but the molecular bases of this process are poorly understood. The neural retina comprises a mosaic of rod and cone photoreceptors. Cone and rod photoreceptors degenerate upon rod-specific expression of heterogeneous mutations in functionally distinct genes, whereas cone-specific mutations are thought to cause only cone demise. Here we show that conditional ablation in cone photoreceptors of Ran-binding protein-2 (Ranbp2), a cell context-dependent pleiotropic protein linked to neuroprotection, familial necrotic encephalopathies, acute transverse myelitis and tumor-suppression, promotes early electrophysiological deficits, subcellular erosive destruction and non-apoptotic death of cones, whereas rod photoreceptors undergo cone-dependent non-autonomous apoptosis. Cone-specific Ranbp2 ablation causes the temporal activation of a cone-intrinsic molecular cascade highlighted by the early activation of metalloproteinase 11/stromelysin-3 and up-regulation of Crx and CoREST, followed by the down-modulation of cone-specific phototransduction genes, transient up-regulation of regulatory/survival genes and activation of caspase-7 without apoptosis. Conversely, PARP1+-apoptotic rods develop upon sequential activation of caspase-9 and caspase-3 and loss of membrane permeability. Rod photoreceptor demise ceases upon cone degeneration. These findings reveal novel roles of Ranbp2 in the modulation of intrinsic and extrinsic cell death mechanisms and pathways. They also unveil a novel spatiotemporal paradigm of progression of neurodegeneration upon cell-specific genetic damage whereby a cone to rod non-autonomous death pathway with intrinsically distinct cell-type death manifestations is triggered by cell-specific loss of Ranbp2. Finally, this study casts new light onto cell-death mechanisms that may be shared by human dystrophies with distinct retinal spatial signatures as well as with other etiologically distinct neurodegenerative disorders.
Introduction
The disintegration of neuronal networks owing to the non-autonomous death of neurons without primary damage is a hallmark manifestation of many neurodegenerative diseases and contributes determinately to their onset or progression [1]–[3]. Cone or rod photoreceptor neurons employ cell type-specific spectrally tuned and highly homologous phototransduction cascades. Neurodegenerative disorders affecting these neurons serve as excellent models to understand autonomous and non-autonomous cell death processes. First, early studies with chimeric mice with a mixture of healthy and unhealthy rod photoreceptors owing to the expression of rod-specific degenerative mutations by the latter showed that damaged rod photoreceptors promote the non-autonomous death of healthy rod photoreceptors [4]–[7], but the analogous event does not appear to occur between neighboring healthy and damaged cone photoreceptors [8]. Second, rod photoreceptor-specific mutations causing the death of rod photoreceptors promote ultimately the non-autonomous death of cone photoreceptors [3], [9]–[12]. This secondary loss of cone photoreceptors has the greatest impact on human vision, because cone photoreceptors mediate daylight and high acuity vision as well as color perception. For example, rod photoreceptor-specific mutations affecting phototransduction components of rod photoreceptors, such as the catalytic subunit of cGMP phosphodiesterase, rhodopsin and cyclic nucleotide-gated (CNG) channel subunits, lead to the degeneration of damaged rod and healthy cone photoreceptors [4], [9], [11]–[16]. By contrast, the cellular effects of cone photoreceptor-specific mutations causing cone degeneration are much less clear, but they are thought to spare the viability of rod photoreceptors. For example, cone-specific mutations impairing genes homologous to those of rod phototransduction cause the death of cone photoreceptors only [17]–[26].
A number of distinct models have been put forward to explain the secondary loss of healthy cone photoreceptors upon the primary degeneration of damaged rod photoreceptors. These include oxidative stress and metabolic imbalance caused by increased oxygen tension [27]–[29], loss of paracrine neurotrophic [30] or vasculotrophic support [31], microglia activation [32], [33] and release of rod-derived toxic byproducts [34]. Distinguishing what mechanisms trigger extrinsic-elicited cell death pathways is further complicated by our limited knowledge of the intrinsic and primary cell-death pathways affecting rod photoreceptor themselves, and whether these are consistent across rod degeneration models [35]–[45]. Likewise, the knowledge about the molecular and subcellular events underlying the primary demise of cone photoreceptors is very limited [19], [46]. Ascertaining the spatiotemporal processes causing autonomous and non-autonomous neural death is critical to our understanding of the pathogenesis of a variety of human photoreceptor dystrophies, such as retinitis pigmentosa (RP) and age-related macular degeneration (AMD), that harbor hallmark spatiotemporal manifestations presumably driven by distinct intrinsic and extrinsic factors.
The pleiotropic protein, Ran-binding protein-2 (RanBP2), is essential for organism viability and energy metabolism [47], [48]. Prior studies on RanBP2 indicate that it plays critical cell-type-dependent physiological roles in mediating gene-environment interactions. In this regard, distinct disease stressors, such as phototoxicity [49], [50], Parkinsonian toxic insults [51], and carcinogens [48], trigger a variety of cell-context-dependent clinical and pathophysiological manifestations upon partial deficits or mutations of Ranbp2. Further, semi-dominant mutations in human RANBP2 cause either acute necrotizing encephalopathies (ANE1) or acute transverse myelitis (ATM) upon exposure to a variety of infectious agents [52]–[54]. In this study we set out to determine the intrinsic and extrinsic effects of lack of Ranbp2 function in the survival of cone or rod photoreceptor neurons upon selective ablation of Ranbp2 in cone photoreceptors, where Ranbp2 is highly expressed [55]. We show that cone-specific ablation of Ranbp2 promotes the autonomous non-apoptotic death of cone photoreceptors and the cone-dependent apoptotic demise of rod photoreceptors by distinct cell-type death mechanisms. Hence, a primary impairment of cone photoreceptors can promote the secondary death of healthy rod photoreceptors, a paradigm-shift observation with implications to our understanding of human neurodegenerative diseases affecting distinct photoreceptor cell types with hallmark regional distributions in the retina and other neural networks of the central nervous system.
Results
Generation of mice with ablation of Ranbp2 selectively in cone photoreceptors
To determine the physiological role of Ranbp2 in cone photoreceptors and uncover the effects of its genetic ablation in autonomous and non-autonomous molecular and cellular events affecting targeted cones and healthy rod photoreceptors, Ranbp2 was selectively targeted in mouse cones by Cre-mediated recombination of floxed Ranbp2 [48], [56], [57] (Figure 1A). Hemizygous transgenic mice expressing Cre under control of the R/G cone opsin promoter (Tg-HRGP-cre) [56], [57] were crossed with Ranbp2Flox/+ mice [48] to produce Tg-HRGP-cre:Ranbp2Flox/+. These mice were then crossed to Ranbp2Flox/+ or Ranbp2Gt(pGT0pfs)630Wcs/+ , which harbors a constitutively disrupted allele of Ranbp2 [47] (Figure 1B), to generate the lines, Tg-HRGP-cre:Ranbp2Flox/Flox and Tg-HRGP-cre:Ranbp2Flox/Gt(pGT0pfs)630Wc. These lines were morphologically and functionally indistinguishable from each other and they are hereafter designated as HRGP-cre:Ranbp2−/−. An out-of-frame Ranbp2 transcript comprising the fusion of exons 1 and 3 is produced at P7 upon Cre expression at P6 (Figure 1C). Cre was specifically expressed in cell bodies of M - and S-cone photoreceptors (Figure 1D, Figure S1).
Fig. 1. Cre-mediated ablation of Ranbp2 selectively in cone photoreceptors. 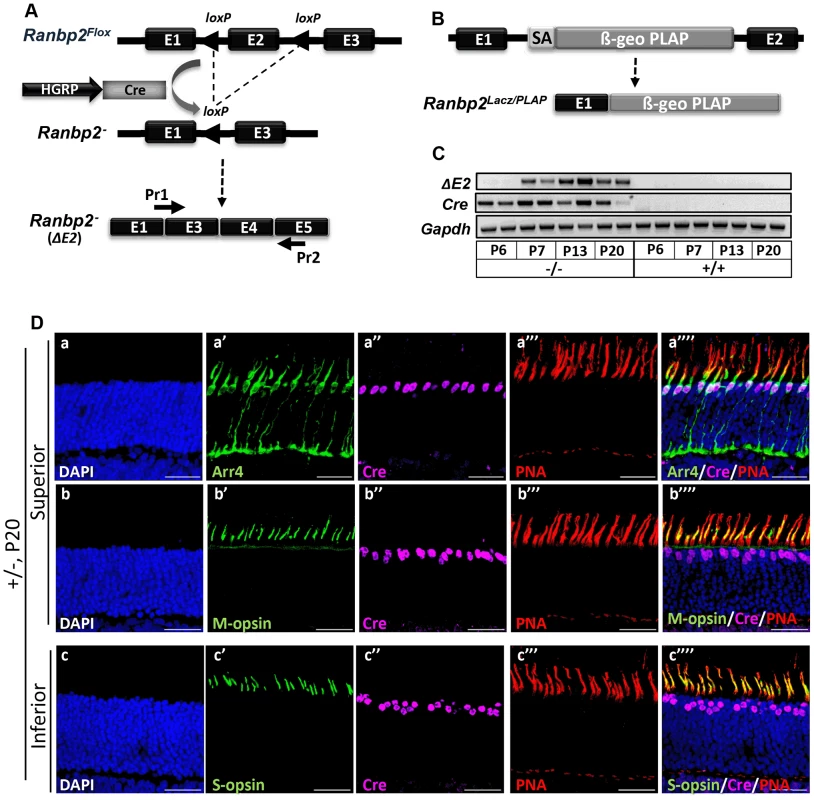
(A) Schematic diagram for targeted Ranbp2 allele upon HGRP-driven Cre excision of exon 2 and production of the recombinant mRNA of Ranbp2. Pr1 and Pr2 are specific to fused exons 1 and 3, and exon 5, respectively, and were used to monitor the functional excision of Ranbp2Flox allele. (B) Schematic diagram of a constitutively targeted Ranbp2 allele upon insertion of a promoterless bicistronic (β-geo-PLAP) cassette with a splicing acceptor site (SA) between exons 1 and 2. (C) RT-PCR of recombinant Ranbp2 mRNA without exon 2 using Pr1 and Pr2 primers. Excision of exon 2 (ΔE2) was detectable in HRGP-cre:Ranbp2−/− (−/−) but not wild-type (+/+) mice at P7 of age, a day after expression of Cre recombinase. (D) Co-expression (a″″–c″″) of Arr4 (a′), M-opsin (b′), S-opsin (c′), Cre (a″–c″) and PNA (a′″–c′″) in cone photoreceptor neurons of the superior (dorsal, a–b″″) or inferior (ventral, c–c″″) regions of the retina of HRGP-cre:Ranbp2+/− mice at P15. Sections were counterstained with DAPI (a–c). Legends: HGRP, L/M opsin promoter; Arr4, cone arrestin 4; PNA, Peanut Agglutinin. Scale bars = 25 µm. HRGP-cre:Ranbp2−/− mice present rampant degeneration of cone photoreceptors
We examined the temporal effects of loss of Ranbp2 expression in the morphology and survival of cones by comparing the immunostaining of retinal sections between HRGP-cre:Ranbp2−/− and HRGP-cre:Ranbp2+/− mice with anti-Cre and cone-specific anti-arrestin-4 (Arr4) antibodies [58] at P9, P13, P20 and P27 of age (Figure 2). In both genotypes, the cell bodies of Cre-expressing cone photoreceptors migrated to the distal (outer) region of the outer nuclear layer (ONL) by P13, where the majority of cone cell bodies are typically localized, and the outer segment (OS) and synaptic pedicles developed properly. However, few Cre+-cell bodies appeared displaced in the proximal (inner) ONL of HRGP-cre:Ranbp2−/− mice (Figure 2D″). By P20, cones of HRGP-cre:Ranbp2−/− mice presented prominent swelling of the synaptic pedicles (Figure 2F′) and retraction of some cell bodies to the proximal region of the ONL (Figure 2F″). By P27, only a very few surviving cones were present in HRGP-cre:Ranbp2−/− (Figure 2H′–2H″); by 6 weeks resilient cones lacking outer segments were rarely present (Figure S2), whereas no cones were present in 12-week old mice (data not shown).
Fig. 2. Temporal and morphological profile of degeneration of cone photoreceptors in Ranbp2−/− mice. 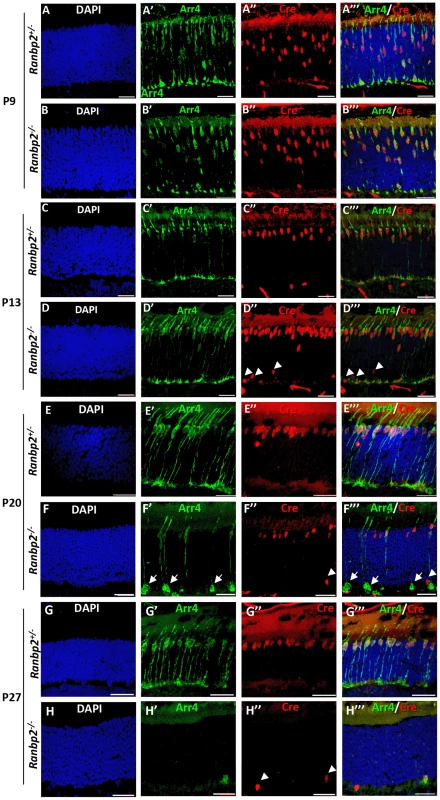
(A–H′″) Immunohistochemistry of retinal sections of HRGP-cre:Ranbp2+/− (Ranbp2+/−) and HRGP-cre:Ranbp2−/− (Ranbp2−/−) mice at P9 (A–B′″), P13 (C–D′″) P20 (E–F′″) and P27 (G–H′″) of age with the antibodies against Cre recombinase and cone arrestin (Arr4) showing rapid loss of cone photoreceptors in RanBP2−/−. Cre co-localized with Arr4 only in DAPI-stained cell bodies of cone cells throughout all ages regardless of genotype. Arrowheads in indicate Cre+ nuclei retracted to the proximal ONL (outer nuclear layer) in Ranbp2−/− at P13, P20 and P27 of age, respectively. Arrows point to prominent swellings of synaptic pedicles of cone photoreceptors in Ranbp2−/− at P20 of age. Scale bars = 25 µm. The degeneration of cone photoreceptors was quantitatively monitored in retinal flat mounts. We compared the number of M-cones and OS length between HRGP-cre:Ranbp2−/− and HRGP-cre:Ranbp2+/− mice (Figure 3). No differences were seen in the number of M-cones or in the length of their OS at P15. By P20, both measures were significantly decreased in HRGP-cre:Ranbp2−/− mice, and very few M-cones remained at P27 (Figure 3A–3C). Notably, the few surviving M-cones retained in P27 HRGP-cre:Ranbp2−/− mice still had OS which appeared of comparable length to those of HRGP-cre:Ranbp2+/− littermates (Figure 3A, 3C). At P20, the number of S-cones and the length of their OS were significantly decreased in HRGP-cre:Ranbp2−/− mice and prominent clumps of S-opsin were observed in the OS (Figure S3). We then examined the position of the Cre+-cell bodies within the proximal and distal ONL at peripheral and central regions of dorsal retinas (Figure 3D, 3E). Unlike HRGP-cre:Ranbp2+/− littermates, P13 HRGP-cre:Ranbp2−/− mice presented displaced Cre+-cell bodies in the peripheral and central regions of the proximal ONL. At P20, the number of Cre+-cell bodies had strongly decreased across the central and peripheral regions of the HRGP-cre:Ranbp2−/− retina, but this decrease was much more pronounced in the central retina (Figure 3E). By P27 and 3-months of age, respectively, very few and no Cre+ cells were observed in any regions of the retina (Figure 3E, data not shown).
Fig. 3. Topographic degeneration of M-cone photoreceptors and their outer segments in HRGP-cre:Ranbp2−/−. 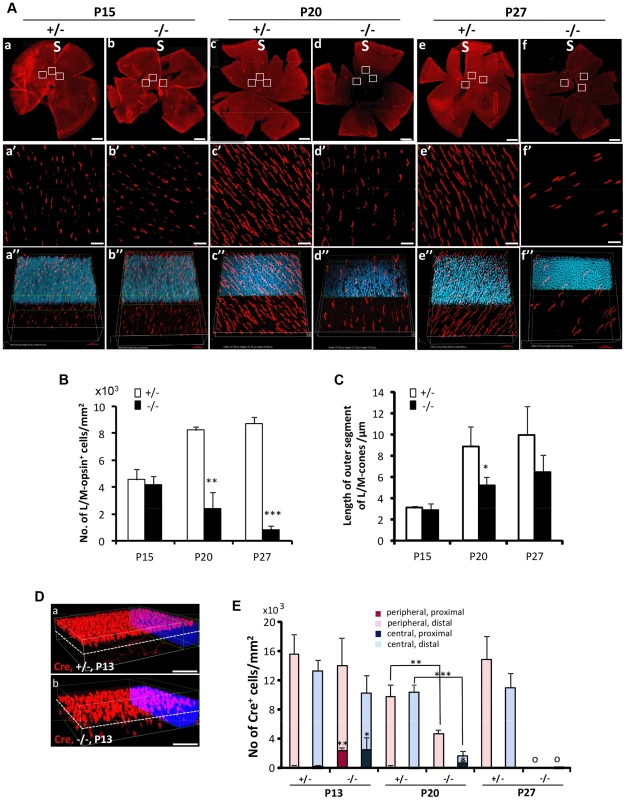
(A) Retinal flat mount images of HRGP-cre:Ranbp2−/− (−/−) and HRGP-cre:Ranbp2+/− (+/−) immunostained with an antibody against M-opsin showing progressive and severe M-cone cell loss in the retina of −/− mice. (a′–f′) Magnifications of the superior/central inset regions of a–f. (a″–f″) 3D-reconstruction images from a′–f′. (B–C) Quantitative and temporal analyses of the number (B) and length of outer segments (C) of M-cone photoreceptors in the superior (S)/central regions of retina of −/− and +/− mice. (D) Comparison of topographic distribution of Cre+ cells in the retina of −/− and +/− mice at P13 showing the proximal localization of a few displaced cone cells in −/− mice. White dashed line represents a virtual midline boundary between proximal and distal areas of the outer nuclear layer (ONL) used for tallying the topography of Cre+-cell bodies in (E). (E) Quantitative and morphometric analyses of the localization of Cre+ cells in the proximal versus distal ONL and central versus peripheral retina between −/− and +/− mice. About 20% of Cre+ cells are present in the proximal ONL region of −/− at P13 and there is greater number of Cre+-cell loss in the central retina of −/− mice at P20. Legend: Boxes in A (a–f) are ROIs used for quantitation of data shown in B and C. Data shown represent the mean ± SD, n = 3–5; **, p<0.01; ***, p<0.001. Scale bars = 150 µm (a–f), 20 µm (a′–f′), 50 µm (D). Non-autonomous death of rods upon demise of cone photoreceptors by distinct intrinsic mechanisms
OS shortening is thought to precede cell death of rod and cone photoreceptors [13], [28]. Hence, we employed multiple cell death markers to dissect out the molecular and subcellular processes underpinning the activation and progression of cell death between cone and rod photoreceptors of HRGP-cre:Ranbp2−/− mice. We used TUNEL staining to determine whether degenerating cone photoreceptors underwent apoptosis. As shown in Figures 4A and 4B, we did not identify any cell bodies that were TUNEL+ and Cre+. Instead, we found that all TUNEL+-cell bodies were Cre−, most likely rod photoreceptors, because these neurons comprise 97% of all photoreceptor cell types [59]. Morphometric analyses showed a drastic increase of TUNEL+-cell bodies at P20 (Figure 4B). Akin to the localization of Cre+ cells, this increase was significantly more pronounced also in central and peripheral regions of the distal ONL than in the counterpart proximal regions (Figure 4B).
Fig. 4. Spatiotemporal modalities of cell death between Cre+, Arr4+ and Arr4−-photoreceptor cell bodies in HRGP-cre:Ranbp2−/−. 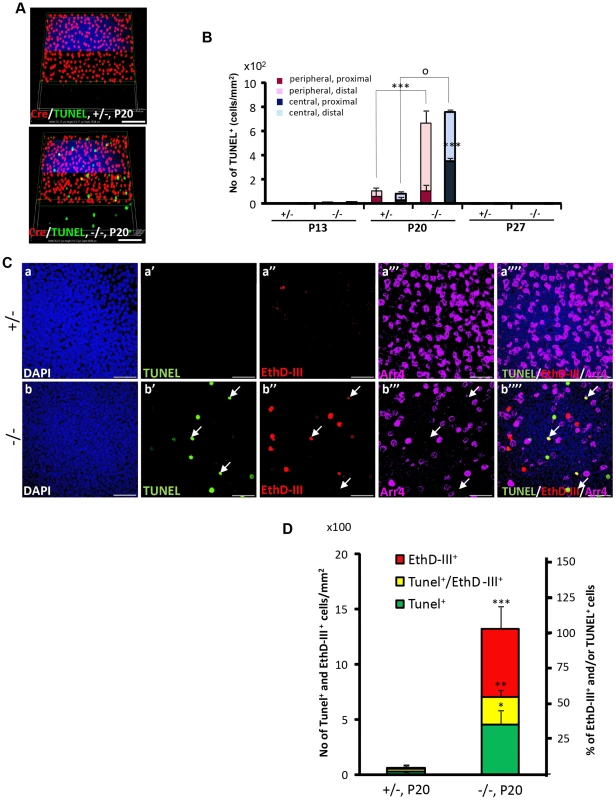
(A) Comparison of the relative 3-D distribution between Cre+-cone and TUNEL+ cells in the outer nuclear layer of retinas of HRGP-cre:Ranbp2+/− (+/−, top) and HRGP-cre:Ranbp2−/− mice (−/−, bottom) at P20 shows that all TUNEL+ cells are Cre−. (B) Morphometric and topographic distribution of TUNEL+ cells in outer nuclear layer (ONL, proximal vs distal) and regions of retina (peripheral vs central) between HRGP-cre:Ranbp2+/− (+/−) and HRGP-cre:Ranbp2−/− mice (−/−) at P13, P20 and P27. TUNEL+ cells become prominent at P20 with most localizing at the distal ONL of central and peripheral retina. No TUNEL+ cells were identified by P27. Data shown represent the mean ± SD, n = 4–5; ***, p<0.001; o, p<0.0001. (C) Identification and localization of TUNEL+ (apoptotic) and EthD-III+ (necrotic) in Arr4−-photoreceptor cell bodies of P20 retinas of HRGP-cre:Ranbp2+/− mice (+/−; a–a′″) and HRGP-cre:Ranbp2−/− mice (−/−; b–b′″). Representative images of TUNEL+ (a′–b′) or EthDIII+ (a″–b″) cell bodies of Arr4−-photoreceptors (a′″–b′″) and co-localization of these (a″″–b″″) are shown. No TUNEL+ and EthD-III+-cell bodies in Arr4+-photoreceptor cell bodies were identified. Sections were counterstained with DAPI (a–b). White arrows in b′–b′″ indicate the localization of TUNEL+EthD-III+ cell bodies. (D) Quantification analysis of C. Among the Arr4−-photoreceptor cell bodies tallied, approximately 50, 30 and 20% were TUNEL−EthD-III+, TUNEL+EthD-III− and TUNEL+EthD-III+, respectively, in −/− mice, whereas they were negligible in +/− mice. Data shown represent the mean ± SD, n = 3; ***, p<0.001; **, p<0.02; *, p<0.005; Scale bars = 50 µm (A), 20 µm (C). To further differentiate TUNEL+-apoptotic cell bodies from necrotic cells among cone and rod photoreceptors, we carried out morphometric analysis of co-localization of TUNEL+ and cone Arr4+-cell bodies of retinal explants incubated with the membrane-impermeable and DNA-binding fluorescent dye, ethidium homodimer III (EthD-III). There were no Arr4+Tunel+ or Arr4+EthD-III+ cells in either genotype (Figure 4C). Instead, we found that HRGP-cre:Ranbp2−/− at P20 had about 70 and 55% of EthD-III+ Arr4− and TUNEL+Arr4− cells, respectively, with 30% of these cells being EthD-III+TUNEL+Arr4− (Figure 4C, 4D). The presence of any of these apoptotic or necrotic cell bodies were negligible in HRGP-cre:Ranp2+/− (Figure 4C, 4D). These data support that the TUNEL+Arr4−, EthD-III+ Arr4− and EthD-III+TUNEL+Arr4− cells represent different cell death stages of rod photoreceptors.
To establish unequivocally that TUNEL+Cre−-apoptotic cell bodies are rod photoreceptors neurons, retinal sections were co-immunostained for Nr2E3, a transcription factor specifically expressed in cell bodies of rod photoreceptors [60]. As shown in Figure 5A (a–b″″), all Cre+-cell bodies of either genotype were Nr2E3−, whereas many TUNEL+-cell bodies were Nr2E3+ in HRGP-cre:Ranbp2−/−. We also identified a small subpopulation of TUNEL+-cell bodies with Nr2E3 aggregation at discrete perinuclear foci (Figure 5A, c–d″″), suggesting that Nr2E3 localization changes and its expression decreases during rod photoreceptor death. Quantitative morphometric analysis of triple-immunostained retinas showed that about 3.2% of the total cells in HRGP-cre:Ranbp2−/− were TUNEL+, while 52% and 44% of these TUNEL+ cell bodies were Cre−NR2E3+ or Cre−Nr2E3−, respectively; the remaining 4% were dying rods with discrete perinuclear aggregation of Nr2E3 (Figure 5B). Further examination of the TUNEL+Nr2E3− and TUNEL+Nr2E3+ cells indicated that they were TUNEL+EthD-III+ or TUNEL+EthD-III− (Figure S4). These data support that TUNEL+Nr2E3+ and TUNEL+Nr2E3− cells also represent different stages of cell death of rod photoreceptors.
Fig. 5. Cell death in Nr2E3+-cell bodies of rod photoreceptors in HRGP-cre:Ranbp2−/−. 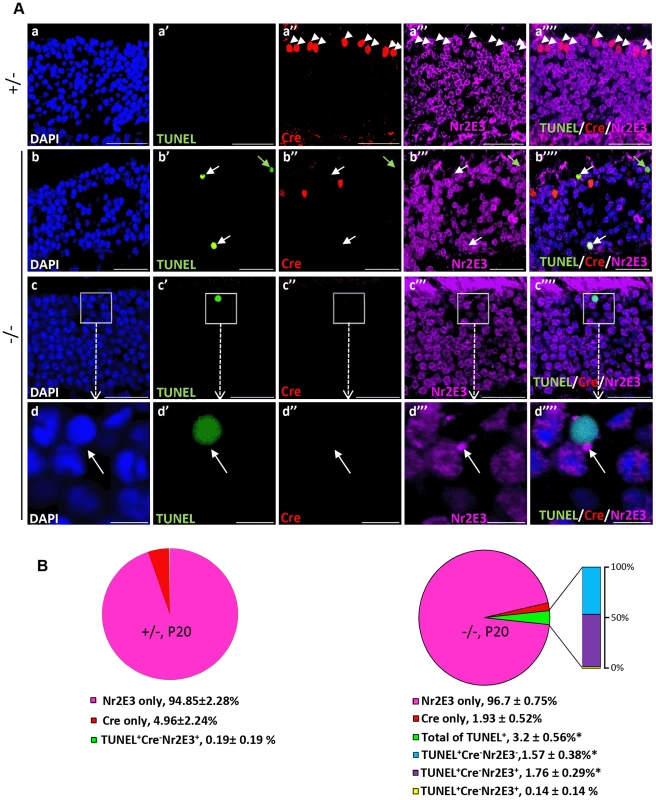
(A) Representative images of TUNEL+ (a′–d′), Cre+ (a″–d″) and Nr2E3+ (a′″–d′″) cell bodies and their colocalization (a″″–d″″) in rod photoreceptors of HRGP-cre:Ranbp2+/− (+/−; a–a″″) and HRGP-cre:Ranbp2−/− mice (−/−; b–d″″). d–d′″ are high magnifications of inset boxes of c–c″″. Sections were counterstained with DAPI (a–d). White arrowheads in a′–a″″ indicate Cre+Nr2E3− cells, white and green arrows in b′–b″″ point to TUNEL+Nr2E3+ and TUNEL+Nr2E3−, respectively. (B) Quantification analysis of A. Among the TUNEL+-cell bodies tallied, slightly over 50% were Nr2E3+ in −/− mice, whereas they were negligible in +/− mice. No TUNEL+Cre+-cell bodies were identified in −/− mice. % values are based on % of total DAPI+ cells (100%). Data shown represent the mean ± SD, n = 3; *; p<0.001. Scale bars = 20 µm (a–c″″), 5 µm (d–d″″). We next examined whether cone and rod photoreceptor degenerations were accompanied by the activation of caspases, a cardinal feature of cell death [61], [62]. We screened whole retinal extracts of HRGP-cre:Ranbp2−/− mice at P20 with substrates against specific caspases and found strong activation of caspases 3/7, mild activation of caspases 8 and 9, and no activation of caspases 1, 2 and 6, when compared to HRGP-cre:Ranbp2+/− mice (Figure S5). To define the spatiotemporal profile of caspase activation, we examined the activities of caspases 3 and 7 in retinal extracts and carried out morphometric analyses of retinal sections immunostained with antibodies against cleaved (activated) caspase 3, 7 or 9 of age-matched mice of both genotypes. In HRGP-cre:Ranbp2−/− mice, we found that the activities of caspase 3, caspase 7, or both, peaked at P13 (Figure 6A), well before the rise in TUNEL+-cell bodies in the ONL (Figure 4B), and that these activities were negligible by P27, when most cones had died (Figure 6A). To distinguish caspase 3 and 7 activities, we immunostained retinal sections of different ages for active caspase 3 and 7 and performed morphometric analysis (Figure 6B, 6C). These experiments showed that the majority of active caspase 3+-photoreceptor cell bodies were Cre−TUNEL− (Figure 6B), but there was also a small but significant fraction of active caspase-3+Cre+-cell bodies at P13 in HRGP-cre:Ranbp2−/− mice (Figure 6B, 6C). All types of active caspase 3+-photoreceptor cell bodies were drastically decreased in HRGP-cre:Ranbp2−/− mice and their presence were no different from HRGP-cre:Ranbp2+/− by P20 (Figure 6B, 6C a–a′″, d–d′″). By contrast, caspase 7+-cell bodies became prominent only at P20 and most were Cre+ (Figure 6C, b′–b′″, e′–e′″). Further, the temporal profiles of caspase 3 and 7 activities paralleled their transcriptional up-regulation (Figure S6). Collectively, the data support that activation of caspase 3 in rods and caspase 7 in cones contribute to the rise of activities of caspases 3 and 7 at P13 and P20, respectively.
Fig. 6. Spatiotemporal activation of caspases 3 and 7 and Parp1 in photoreceptors of HRGP-cre:Ranbp2−/− mice. 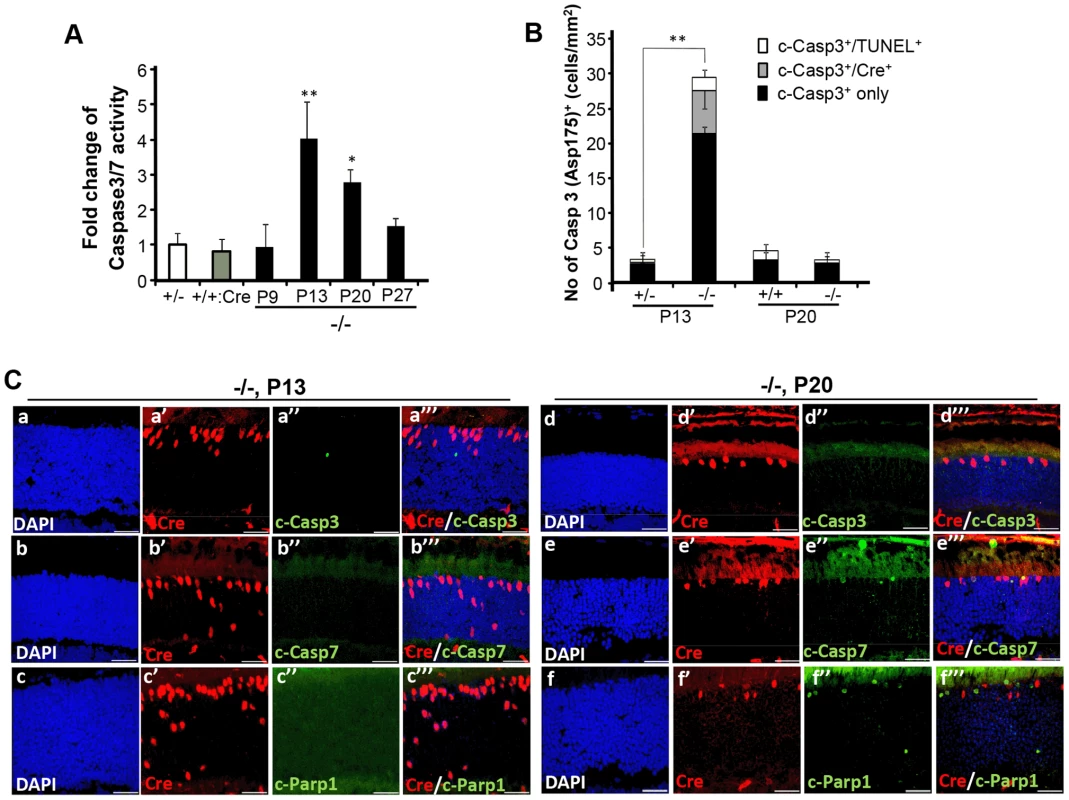
(A) Temporal profile of caspase 3/7 activation of retinal extracts of HRGP-cre:Ranbp2−/− mice (−/−) mice relative to age-matched +/− controls shows significant caspase3/7 activation at P13 and P20. The caspase 3/7 activities of +/− and +/+:Cre controls shown are at P20. (B) Quantitative, temporal and relative distributions between activated (cleaved) caspase 3 (c-casp3+), Cre+ and TUNEL+ in cell bodies of photoreceptors of HRGP-cre:Ranbp2+/− (+/−) and HRGP-cre:Ranbp2−/− mice (−/−) at P13 and P20. The majority of photoreceptor cell bodies are c-casp3+Cre−. The levels of any type of c-casp3+ cell bodies are negligible by P20. (C) Relative immunolocalizations of Cre+ and c-casp3+ (a–a′″, d–d′″), Cre+ and c-casp7+ (b–b′″, e–e′″), and Cre+ and activated Parp1 (c-Parp1+) (c–c′″, f–f′″) cell bodies of photoreceptors of HRGP-cre:Ranbp2−/− mice at P13 (a–c′″) and P20 (d–f′″). c-casp3+ develop at P13, c-casp7+ and c-Parp1+ at P20 and all c-Parp1+ cell bodies are Cre−, whereas c-casp7+ are Cre+. Legend: Data shown represent the mean ± SD, n = 4–5; *; p<0.05, **; p<0.01; scale bars = 25 µm; cleaved caspases 3 and 7, c-casp3 and c-casp7, respectively. Next, we examined the activation of Parp1 (Parp1+; Figure 6C, Figure S7), which is cleaved during apoptosis into 89 and 24 kDa fragments by caspase 3 or 7 [63], [64] and has been linked to rod photoreceptor degeneration [65]. We found that Parp1+cells were never Cre+ and that their appearance was biphasic with activity peaks at P9 and P20 (Figure 6C, Figure S7A, a–b″″). These temporal peaks of Parp1+-activities in rod photoreceptors coincided with the activation of caspase 9 in rods at P9 (Figure S7A, c–d″″) and caspase 7 in cones at P20 (Figure 6C). No Parp1+-cell bodies could be identified at P13 (Figure 6C). At P9, caspase 9+TUNEL+ and Parp1+TUNEL+ cell bodies were always cre− rods, while TUNEL+caspase 9−cre− and TUNEL+Parp1−cre− rods could also be observed (Figure S7A). These events indicate that caspase 9 and Parp1 activation in rod photoreceptors is short-lived. These manifestations were accompanied by transcriptional up-regulation of caspase 9 and Parp1 between P9 and P20, but not of caspase 8, which is typically activated by extrinsic cell-death mechanisms (Figure S7B) [61], [62].
The formation of rod apoptotic cell bodies prompted us to examine whether apoptosis-inducing factor (AIF) or cytochrome c was released from the mitochondria, as these are also hallmark events of the apoptotic cascade [66]–[68]. Subcellular fractionation of retinas showed no sign of AIF or cytochrome c in the cytosol fraction of either genotype at the peak of apoptosis (Figure S8A). Changes in AIF levels were also not observed in the nuclear-enriched fraction of either genotype (Figure S8B). In addition, the expression levels of the apoptotic protease activating factor-1 (Apaf-1) remained unchanged (data not shown). Finally, we examined markers for necroptosis, such as members of the receptor-interacting proteins, RIP1 and RIP3, and macroautophagy, such as the autophagosomal membrane marker, light chain 3B II (LC3B II), since they are thought to be induced upon photoreceptor degeneration [69]–[71]. Immunoassays of retinal extracts found no differences in these markers between HRGP-cre:Ranbp+/− and HRGP-cre:Ranbp2−/− mice at P13 (Figure S8C, S8D).
Selective activation of MMP11 in HRGP-cre:Ranbp2−/− mice
Histological examination of semi-thin retinal sections showed that retinas of HRGP-cre:Ranbp2−/− developed prominent interstitial spaces between photoreceptors cell bodies and across the ONL by P20, a phenotype not present in HRGP-cre:Ranbp2+/− mice (Figure 7A). The interstitial spaces could always be traced to prominent euchromatic nuclei typically localized at the distal (outer) edge of the ONL (Figure 7A), hallmark morphological and topographic features of cell bodies of cone photoreceptors. Detailed examination of the ultrastructure of the ONL showed that the interstitial spaces reflect degenerating lower fibers of cone photoreceptors that were dilated and very lucent (Figure 7B). This striking phenotype led us to hypothesize that ablation of Ranbp2 in cones promotes the activation of metalloproteinase(s) (MMPs) causing the weakening and degradation of the extracellular matrix, which normally organizes photoreceptor cell bodies within the ONL [72], and may contribute to the retraction of Cre+-cell bodies from the distal (outer) to the proximal (inner) region of the ONL (Figure 2). Hence, we screened retinal extracts for each the eleven MMP activities. In comparison to HRGP-cre:Ranbp2+/−, retinal extracts of HRGP-cre:Ranbp2−/− mice presented ∼3-fold higher activity of MMP11, but not of any other MMPs (Figure S9A). MMP11 activity was elevated at P13 and P20 and returned to control levels at P27 when most cones have degenerated (Figure 7C). The increase of MMP11 activity was also accompanied by a ∼3-fold increase of the active form of MMP11 (Figure 7D, 7E). The transcriptional up-regulation of Mmp11 in HRGP-cre:Ranbp2−/− mice as early as P9 followed its transient down-regulation at P7 and preceded the activation of MMP11 at P13, whereas the transcriptional levels of Timp3 remained largely unchanged across different ages (Figure S9B, 7C). To assess the cellular origin of MMP11 expression and activity, we performed immunohistochemistry of MMP-11 in retinal sections from P20 mice, and found that MMP-11 was localized prominently around cell bodies, inner segments and lower fibers of cone Arr4+-cells (Figure 7F). Collectively, these data confirm that ablation of Ranbp2 in cones promotes the up-regulation of MMP11 expression and activity in these neurons, an event which likely contributes to the development of interstitial spaces in the ONL and retraction of cones cell bodies to the proximal ONL in HRGP-cre:Ranbp2−/− mice.
Fig. 7. Activation of matrix metalloproteinase 11 (MMP11) upon ablation of Ranbp2 in cone photoreceptors. 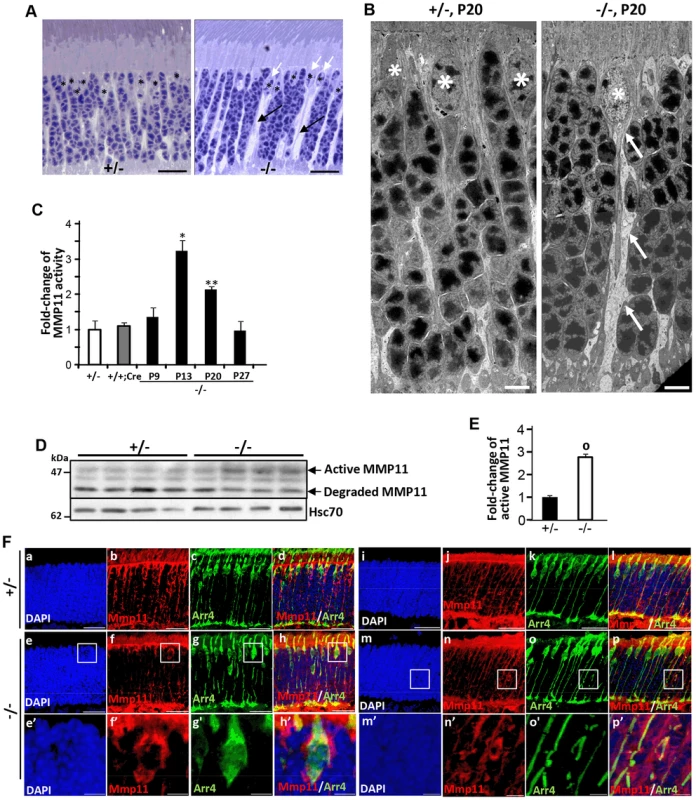
(A) Light microscopy images of methylene blue stained semi-thin retinal sections showing the presence of prominent interstitial spaces (black arrows) originating from cone cell bodies (white arrows) and between cell bodies of rod photoreceptors in HRGP-cre:Ranbp2−/− (−/−) compared to HRGP-cre:Ranbp2+/− (+/−) mice. * denote euchromatic nuclei of cones. (B) Electron micrographs depicting ultrastructural changes in the lower fiber of a −/− cone photoreceptor. Note the swelling of the lower fiber (arrows) traced to the cell body of a cone nucleus characterized by its prominent euchromatin (*). *, cone nuclei. (C) Temporal profile of MMP11 activity of retinal extracts of HRGP-cre:Ranbp2−/− mice (−/−) mice relative to age-matched +/− controls shows significant MMP11 activity at P13 and P20. The MMP11 activity of +/− and +/+:Cre controls shown are at P20. (D) Immunoblots of MMP11 from retinal homogenates of HRGP-cre:Ranbp2−/− (−/−) compared to +/− mice at P20 showing an increase of the active form of MMP11 in −/−. Hsc70 is cytosolic heat shock protein 70 used as loading control. (E) Quantitation analysis of the levels of active MMP11 in (D) of HRGP-cre:Ranbp2−/− (−/−) relative to age-matched +/− mice at P20. (F) Retinal sections immunostained with antibodies against cone arrestin (Arr4) and MMP11. (a–d) central retinal region of HRGP-cre:Ranbp2+/− (+/−); (e–f) central retinal region of HRGP-cre:Ranbp2−/− (−/−); (e′–f′) magnification of boxed regions shown in e–f; (i–l) peripheral retinal region of HRGP-cre:Ranbp2+/− (+/−); (m–p) peripheral retinal region of HRGP-cre:Ranbp2−/− (−/−); (m′–p′) magnification of lower fibers of cones of boxed regions shown in m–p. MMP11 predominantly localizes to cone photoreceptors, such as interstitial space around cell bodies, lower fibers and inner segments. Legend: Data shown represent the mean ± SD, n = 4; *, p<0.05; **, p<0.01; o, p<0.0001; scale bars = 25 µm (A, F), 5 µm (B). Erosive ultrastructural destruction of cones and damage of rod photoreceptors
To ascertain in greater detail the morphological changes of degenerating cone photoreceptors, we examined the ultrastructure of HRGP-cre:Ranbp2+/− and HRGP-cre:Ranbp2−/− retinal sections at P20. The subcellular subcompartments of cone and rod photoreceptors of HRGP-cre:Ranbp2+/− mice had normal morphologies (Figure 8A, 8A′, 8F, 8F′, 8I), whereas cone photoreceptors of HRGP-cre:Ranbp2−/− mice exhibited distinct morphological changes across multiple subcellular compartments. Across different photoreceptor cells of HRGP-cre:Ranbp2−/−, we found that cone OS membranes were extended, disorganized and collapsed into large, amorphous and lucent areas (Figure 8B, 8B′, 8C, 8C′), that electrodense material accumulated at the connecting cilium (Figure 8D, 8D′), and that prominent electron lucent areas were present in the inner segments (Figure 8E, 8E′). Additional abnormalities were seen at the synaptic pedicles including accumulation of multilamellar bodies (Figure 8G, 8G′), mitochondria with widespread electron lucent matrix areas without cristae and formation of cytosolic lucent areas without a limiting membrane around subcellular debris (Figure 8H, 8H′). Albeit less extensive, cristae of some mitochondria in the rod synaptic spherules were also disrupted resulting in the formation of lucent areas within the matrix (Figure 8J).
Fig. 8. Ultrastructural changes of cone and rod photoreceptors upon ablation of Ranbp2 in cones at P20. 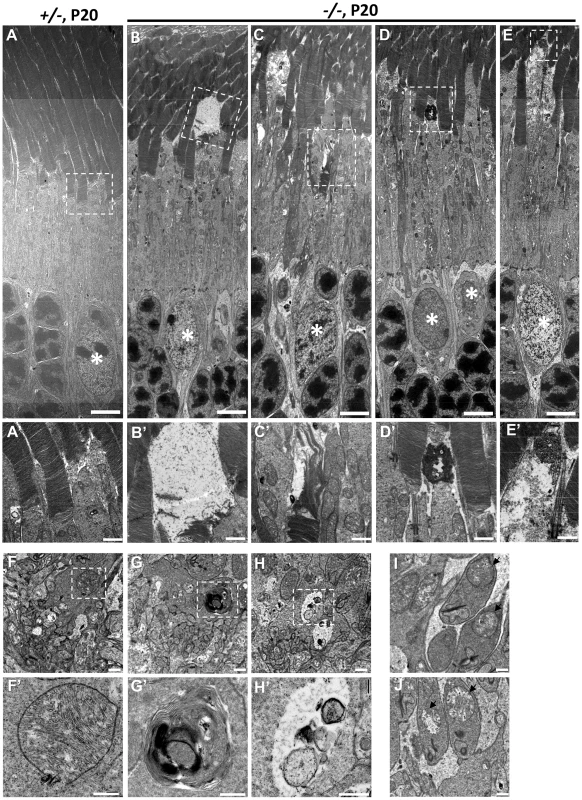
(A) Ultrastructural image of rod and cone photoreceptors outer and inner segments of HRGP-cre:Ranbp2+/− (+/−). All structures look unremarkable. (B–E) Representative ultrastructural images of multiple features rod and cone photoreceptors outer and inner segments of HRGP-cre:Ranbp2−/− (−/−). (B) shows the collapse of the cone outer segment and formation of a large electron lucent area, (C) shows extended discs and partial erosion of cone outer segment, (D) shows the accumulation of amorphous electron dense material at the connecting cilium of a cone photoreceptor, (E) shows the formation of electron lucent areas in the inner segment of a cone photoreceptor. White stars, euchromatic nuclei of cone photoreceptors. (A′–E′) are magnifications of boxed areas in A–E showing the aforementioned pathological features. (F) Synaptic pedicles of cones with normal morphology of HRGP-cre:Ranbp2+/− (+/−). (G) Multi-lamellar body with electrodense material in a cone synaptic pedicle of HRGP-cre:Ranbp2−/− (−/−). (H) Electron lucent area surrounding subcellular debris in a cone synaptic pedicle of HRGP-cre:Ranbp2−/− (−/−). F′–H′ are magnified images of boxed areas in F–H. (I) Synaptic spherule of a rod photoreceptor containing unremarkable mitochondria (black arrows) in HRGP-cre:Ranbp2+/− (+/−). (J) Synaptic spherule of a rod photoreceptor with abnormal mitochondria (black arrows) containing disorganized cristae and electron lucent areas in the matrix in HRGP-cre:Ranbp2−/− (−/−). Scale bars = 5 µm (A–E), 1 µm (A′–E′), 500 nm (F–H) and 200 nm (F′–H′). Multiphasic and temporal changes in gene expression and pathways upon Ranbp2 ablation
We examined the effect of Ranbp2 ablation selectively in cones on the expression of cone photoreceptor-specific and other pertinent genes by quantitative real time-PCR (qRT-PCR) (Figure 9, Table S1). We found that Ranbp2 ablation led to rapid declines of M-opsin (Opn1mw) and S-opsin (Opn1sw) mRNAs as early as P13 (Figure 9A), when there are yet no prominent cellular changes in M-opsin and Cre+-neurons between genotypes (Figure 3B, 3E), and such declines occurred without concomitant changes in rhodopsin (Rho) mRNA (Figure 9A). Other cone-specific genes showed a similar pattern of down-regulation, including Pde6h, Pde6c and Gnat3 (Figure 9B). By P27, when cones have degenerated, the expression of all cone-specific genes was at or below the detection limit in HRGP-cre:Ranbp2−/− retinas (Figure 9A, 9B). By contrast, the expression of the pan-photoreceptor markers, Rcvn and Osgep, increased transiently at P13 (Figure 9C).
Fig. 9. Temporal profiles of changes in gene expression by ablation of Ranbp2 in cone photoreceptors. 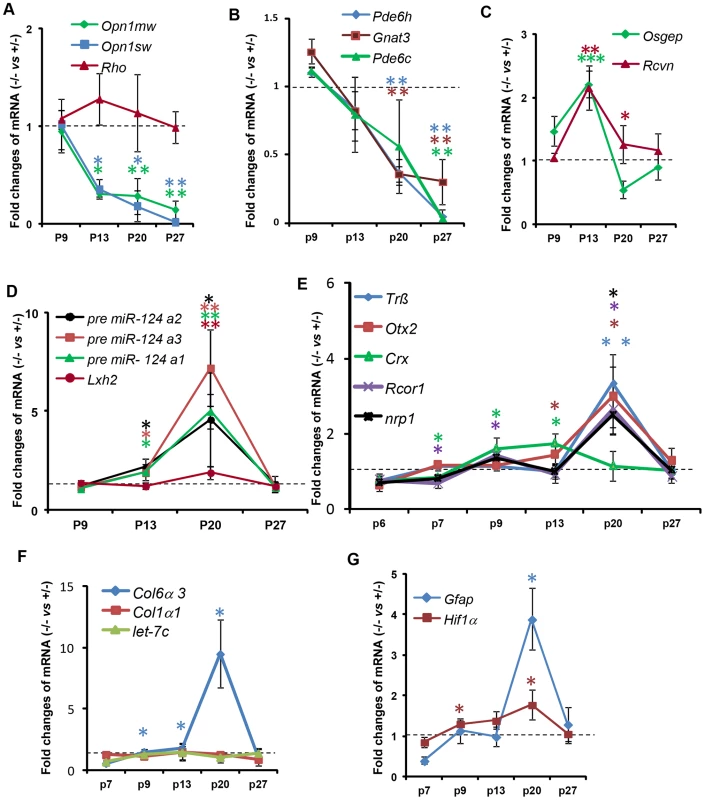
(A–G) qRT-PCR of cone-specific (A, B), pan-photoreceptor (C), cone survival (D) and cone transcription factors (E), col6α3 and col1α1 (F), and Gfap and Hif1α transcripts (G). There is a decrease of cone-specific transcripts beginning at P13 (A, B), whereas pan-photoreceptor (C) and cone survival genes (D) begin transient up-regulations at the same age. The expression of cone survival genes peaks at P20 (D). (E) The transcription factors, Crx and CoREST (Rcor1), are the first nuclear factors whose changes of their transcriptional levels coincides with the genetic ablation of Ranbp2 at P7. These events are followed by the up-regulations at P20 of the transcription factors, Nrp1, Trß2 and Otx2 (E). The up-regulations of transcripts encoding the substrate of MMP11, col6α3, begins at P9 and peaks at P20 (F), whereas those for the hypoxia marker, Hif1α, and inflammatory marker, Gfap, begin at P9 and P20, respectively (G). Legends: Data shown represent the mean ± SD, n = 3–4; *, p<0.05; **, p<0.01; ***, p<0.001; refer to table S1 for gene designations/symbols. We also examined the genetic variants of miR-124a (miR-124a1/Rncr3, miR-124a2, miR-124a3), which mediate cone survival, and its downstream target transcript, Lhx2, whose translation is suppressed by miR-124a [73]. We found strong up-regulation of miR-124a in the HRGP-cre:Ranbp2−/− retina, which was accompanied by increased levels of Lhx2, albeit of a lesser magnitude (Figure 9D). The time course of these changes was similar, beginning at P13, peaking at P20 and returning to HRGP-cre:Ranbp2+/− levels at P27 when cones have degenerated (Figure 9D). We also assessed the expression of Trß2, Otx2 and Crx, encoding transcription factors critical to the maturation and maintenance of cone photoreceptors (Figure 9E) [74]–[78]. Crx expression was decreased at P7, when Cre-mediated excision of Ranbp2 occurs (Figure 1C), and then rose steadily up to P13, and then decreased to HRGP-cre:Ranbp2+/− levels at P27. In comparison, expression of Trß2 and Otx2 increased later, with a sharp peak at P20, and then declined to basal levels at P27 (Figure 9E). Like Crx, CoREST (also known as Rcor1), a cofactor of REST (repressor element 1 silencing transcription factor), was transiently down-regulated at P7 and then rose at P9 until P20, when there was a transient up-regulation of neuropilin-1 (nrp1), a receptor whose transcriptional expression is modulated by miR-124 and CoREST (Figure 9E) [79]. The expressions of CoREST and nrp1 also returned to basal levels at P27 (Figure 9E).
Because Ranbp2 ablation induced the expression and activation of MMP11, a metalloproteinase which is known to cleave selectively the α3 chain of collagen VI (Col6α3) [80], we examined the time course changes of Col6α3 expression in the HRGP-cre:Ranbp2−/− retina. A selective rise in Col6α3 expression, but not α1 chain of collagen I (Col1α1), was detected as early as P9, shortly after a transcriptional increase in Mmp11 was also detected (Figure 9F, S9B). Cola6α3 expression peaked at P20 and then became indistinguishable from HRGP-cre:Ranbp2+/− mice at P27. We did not observe any transcriptional changes in let-7c, a miRNA reported to promote the down-regulation of Mmp11 (Figure 9F) [81]. Finally, we examined transcriptional changes in markers associated with neurodegenerative mechanisms including with autophagy (e.g. cathepsin S, lysozyme and culsterin), glycolysis (6-Pfk), hypoxia (Hif1α) and inflammation (Gfap). Among these, we found changes only in Hif1α and Gfap with Hif1α rising from P9 until P20 and Gfap transiently spiking at P20 when most cones are undergoing degeneration (Figure 9G).
Ranbp2 has multifaceted roles in biology and pathology across tissues, cell types and cell-stages. This complexity reflects the interaction of the diverse structural modules of Ranbp2 with multifunctional partners. Hence, we employed the Ingenuity pathway analysis (IPA) to identify and delineate connectivity maps linking Ranbp2 with the regulation of expression of genes/proteins identified by this study and genetic, protein and metabolic networks. The top network hit generated by IPA (score 22) was associated to “Cellular Development, Nervous System Development and Function and Carbohydrate Metabolism” (Figure 10). This network comprised thirty-two gene products and three endogenous chemicals, D-glucose, sn-glycero-3-phosphocholine and tretinoin (all-trans retinoic acid) (Figure 10). Remarkably, this connectivity map revealed links between transcription factors, many of which belong to the orphan nuclear receptor family and are known to be modulated by Ranbp2 levels, and metabolites, whose levels are also affected by Ranbp2 and regulate transcriptional activities of nuclear factors [47], [49], [51], [82], [83]. Among other novel points of interest, the IPA revealed central roles of i) huntingtin cross-talk with nuclear factors modulated by Ranbp2, a mechanism which is thought to be disrupted in Huntington's disease (HD) [84], and ii) two secreted and extracellular signaling proteins, wingless-type MMTV integration site family, member 2 (WNT2B) and brain-derived neurotrophic factor (BDNF), intersecting multiple nodes modulated by Ranbp2 and other factors. WNT2B and BDNF are known to play important roles in regulation of cell growth, differentiation, tumorigenesis, and to support the survival of existing neurons, respectively [85]–[87].
Fig. 10. Connectivity map of a gene network linked to Ranbp2 functions. 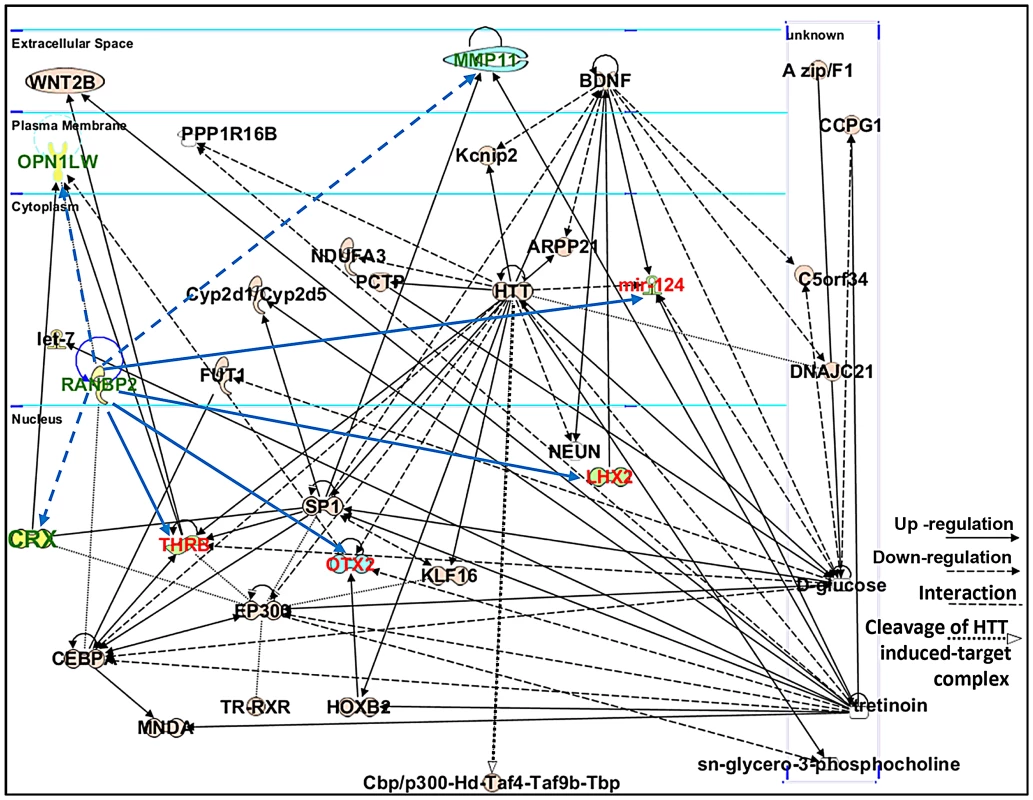
Ingenuity pathway analysis (IPA) of factors modulated by loss of Ranbp2 identified a top network implicated in “Cellular and Nervous System Development and Function, and Carbohydrate Metabolism” (score of 22). Dashed and solid blue arrows were added manually to the network to reflect the down-regulation and up-regulation of genes (shown in green and red), respectively, observed first upon ablation of Ranbp2 in cone photoreceptors as identified by this work. Note that Crx and Mpp11 expressions underwent first transient down-modulation at P7 followed by their sustained up-regulation. Legend: ARPP21, cAMP-regulated phosphoprotein, 21 kDa; BDNF, Brain derived neurotrophic factor; C5orf34, chromosome 5 open reading frame 34; cbp, CREB-binding protein; CCPG1, cell cycle progression 1; CEBPA, CCAAT/enhancer-binding protein alpha; CRX, Cone-rod homeobox protein; Cyp2d1/Cyp2d5, cytochrome P450, family 2, subfamily d, polypeptide 1/cytochrome P450, family 2, subfamily d, polypeptide 5; DNAJC21, DnaJ homolog subfamily C member 21; EP300, also called p300, E1A binding protein p300; FUT1, fucosyltransferase 1; HOXB2, homeobox B2; HTT or Hd, huntingtin; Kcnip2, Kv channel-interacting protein 2; KLF16, Kruppel-like factor 16; let-7, Let-7 microRNA precursor; LHX2, LIM/homeobox protein2; mir-124, microRNA 124; MMP11, Matrix Metalloproteinase 11; MNDA, myeloid cell nuclear differentiation antigen; NDUFA3, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3; NEUN, Feminizing Locus on X-3, Fox-3, or Hexaribonucleotide Binding Protein-3; OPN1LW, long-wave-sensitive opsin (cone pigment); OTX2, orthodenticle homeobox 2; PCTP, Phosphatidylcholine transfer protein; PPP1R16B, protein phosphatase 1, regulatory subunit 16B; sn-glycero-3-phosphocholine, 2,3-dihydroxypropyl 2-(trimethylazaniumyl)ethyl phosphate; SP1, Sp1 transcription factor; THRB, thyroid hormone receptor, beta; Taf4, TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor; Taf9b, TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor; Tbp, TATA box binding protein; TR, Thyroid Hormone Receptor; RXR, Retinoic acid receptor; tretinoin, all-trans retinoic acid or ATRA; WNT2B, wingless-type MMTV integration site (WNT) family2B. Retinal physiological deficits upon impairment and loss of cone photoreceptors
Genetic excision of Ranbp2 is detectable by P7, which coincides with immediate transcriptional changes in the levels of Mmp11, Crx and CoREST (Figure 9E, S9B). Reduced levels of cone-specific transcripts, such as those encoding phototransduction proteins, were not detectable until P13 and changes in cone morphology were not observed until P20. These observations, and the non-autonomous molecular and cellular effects of cones on rod photoreceptors, prompted us to determine the onset and progression of cone and rod physiological dysfunction caused by ablation of Ranbp2 in cones. We measured cone and rod function by light - and dark-adapted electroretinograms (ERGs), respectively. Figure 11 summarizes the ERG data obtained from mice between P13 and 150 days. At P13, dark-adapted ERGs of HRGP-cre:Ranbp+/− and HRGP-cre:Ranbp2−/− mice were comparable (Figure 11A, left; Figure 11B), indicating equivalent retention of rod-mediated outer retinal activity at this age. In comparison, light-adapted ERGs, reflecting cone activity [88], [89] were already reduced significantly at P13 (Figure 11A, right; Figure 11C). By P22, the amplitude of the light-adapted ERGs were markedly reduced in HRGP-cre:Ranbp2−/− mice (Figures 11D), and cone ERGs were extinguished in HRGP-cre:Ranbp2−/− mice at P29 (Figure 11A, 11E). The amplitude of the dark-adapted ERG a-wave, which reflects phototransduction in the outer segments of rod photoreceptors, was not significantly different between HRGP-cre:Ranbp2+/− and HRGP-cre:Ranbp2−/− mice at any age examined (Figure 11A, 11B, 11F, 11G). In mice aged P29 and P150, the dark-adapted ERG b-wave was reduced, but in a luminance-dependent fashion (Figure 11F, 11G). At low stimulus luminances, where the b-wave reflects synaptic transmission from rod photoreceptors to rod bipolar cells, there was no significant reduction in amplitude. At higher stimulus luminances, where both rod - and cone-mediated synaptic activity contribute to the b-wave, a significant amplitude reduction was observed (Figure 11F). The synaptic localization of post-synaptic density 95 (PSD95) protein was comparable between genotypes at P20 (Figure S10), indicating that the b-wave reductions noted at high flash luminances reflects a loss of the cone pathway contribution to the ERG b-wave at these stimulus levels, and not to an alteration in synaptic transmission between rod photoreceptors and bipolar cells. Figure 11G plots the amplitude of HRGP-cre:Ranbp2−/− light - and dark-adapted ERG components relative to those of control littermates (HRGP-cre:Ranbp2+/−) ranging from P13 to 21-weeks in age. HRGP-cre:Ranbp2−/− mice have a selective reduction in the light-adapted ERG indicating that the dark-adapted ERG is not sensitive to the limited cone-induced rod loss seen by other cell biological measures previously described in this study. In agreement with the electrophysiological observations, the complete loss of cone photoreceptors and the limited cone-induced loss of rod photoreceptors did not cause significant changes in ONL thickness and cell body density when analyzed by light microscopy in mice as old as 12-weeks of age (Figure S11).
Fig. 11. Electrophysiological responses of cones and rod photoreceptors upon ablation of Ranbp2 in cones. 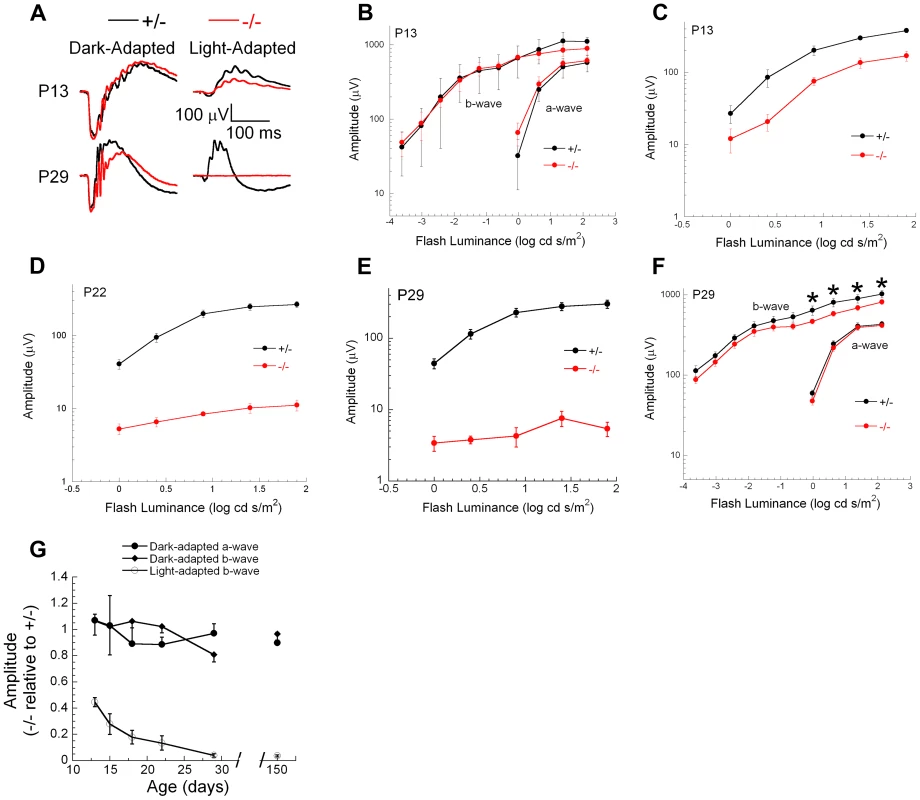
(A) Representative electroretinograms (ERGs) of control HRGP-cre:Ranbp2+/− (+/−, black) and HRGP-cre:Ranbp2−/− mice (−/−, red) mice at P13 and P29 to a 1.4 log cd s/m2 stimulus flash presented in darkness (left) or superimposed upon a steady rod-desensitizing adapting field (right). (B–F) Luminance-response functions for the major components of the dark-adapted rod (B, F) or light-adapted cone ERGs (C–E) at P13 (B, C), P22 (D) or P29 (E, F). Repeated measures analysis of variance was used to compare luminance-response functions of +/− and −/− mice for the amplitude of the major ERG components. Cone ERG amplitudes were significantly reduced in −/− mice at all ages examined (P13, P18, P22, P29, P150). Dark-adapted a-wave amplitudes were not different between +/− and −/− mice at any age examined. Dark-adapted b-wave luminance-response functions were also not different between +/− and −/− mice at any age examined. At P29 (F) and P150, dark-adapted ERG b-waves were significantly reduced (p<0.05, t-test, denoted by *) for stimuli above 0.0 log cd s/m2, but not for lower luminance stimuli. (G) Amplitude of −/− responses expressed relative to those of +/− littermates for the major ERG response components. Each measure reflects a single stimulus condition: ‘Light-adapted b-wave’ represents cone ERGs obtained to a 1.4 log cd s/m2 stimulus flash superimposed upon a steady adapting field; ‘Dark-adapted a-wave’ represents rod ERG a-waves obtained to a 1.4 log cd s/m2 stimulus flash presented in darkness; ‘Dark-adapted b-wave’ represents rod ERG b-waves obtained to a −1.2 log cd s/m2 stimulus flash presented in darkness. Data points indicate the average ± s.e.m. for n = 5–10 mice. Discussion
The results of this study demonstrate: i) cone-specific ablation of Ranbp2 triggers the demise of cone photoreceptors with concomitant non-autonomous cell death of rod photoreceptors, ii) the death of rod photoreceptors is contingent on the presence of cone photoreceptors, and iii) the cell death mechanisms of cone and rod photoreceptors are intrinsically distinct with the former and latter undergoing atypical features of necrosis and apoptosis, respectively [61], [62]. We found that cone photoreceptors undergo necrotic changes including massive erosive destruction of their intracellular contents and late caspase 7 activation, but without apparent changes in loss of membrane permeability. In comparison, rods undergo apoptosis, caspase 3 and Parp1 activations, and loss of membrane permeability.
This study unveils early molecular events triggered by and concomitant with the ablation of Ranbp2 in cone photoreceptors, including changes in Mmp11, Crx and CoREST expressions at P7 and of MMP11 activity at P13, followed by the activation of caspase 7 and a compensatory burst in the expression of the MMP11 substrate, Cola6α3, at P20, when the degeneration of cones is well underway. These events are accompanied by the up-regulation of cone survival genes, such as genetic variants of miR-124a and its downstream target transcript, Lhx2, at P13, when the down-regulation of cone-specific genes, such as M - and S-opsins, becomes significant. miR-124 promotes the translational suppression of Lhx2, a process which has been linked to the suppression of apoptosis during cone photoreceptor differentiation and aberrant sprouting of hippocampal neurons [73]. In contrast to these observations, up-regulation of miR-124a does not prevent the demise of mature cone photoreceptors and occurs concomitantly with a rise of Lhx2 levels. Further, changes in CoREST levels, a target of miR-124 [79], preceded those of all miR-124 variants. These observations suggest that the miR-124 variants may act on other substrates, such as Foxa2 [90], and that may modulate the survival of cones (or rods) independently of Lhx2 and CoREST suppression [79]. Finally, even though cones account for only ∼3% of all photoreceptor types of the mouse retina [59], the return of cone-derived caspase 7 and MMP11 activities and rod-derived caspase 3 activity to control levels after cones have degenerated strongly support these events are triggered solely by the Ranbp2-dependent dysfunction of cones.
The early and selective activation of MMP11 followed by the development of prominent interstitial spaces between photoreceptor cell bodies, swelling of the lower fibers of cones, and the loss of membrane permeability of rods supports that MMP11 plays an important role in the development of autonomous and non-autonomous photoreceptor degeneration. These roles may include autocrine, paracrine and even intracrine actions, whereby intracrine and paracrine functions contribute to intracellular erosion of cones and non-autonomous death of rods, respectively. In comparison to other MMP proenzymes, MMP11 shows important functional differences. While most MMPs are activated extracellularly, MMP11 is secreted in the active form [91]. This exposes intracellular substrates to active MMP11, a potential intracrine signal, and its extracellular proteolytic functions may stimulate autocrine and/or paracrine signaling. Regardless of its molecular mechanisms of action(s), increased levels of MMP11 expression is reported to modulate cell survival [92]–[96], to act as a negative prognostic of cancer patient survival [97]–[99], and to promote oncogenic homing, tumorigenesis and metastasis [92], [96], cardinal manifestations triggered also by haploinsufficiency and hypomorphism of Ranbp2 [48].
MMP11 up-regulation has been reported to promote the suppression of apoptotic and necrotic cell death, rather than stimulation of cell proliferation [95], [96]. These cellular manifestations appear at odds with the physiological phenotypes of our study. It is possible that the distinct pathological outcomes produced by the induction of MMP11 upon loss of Ranbp2 reflect intrinsically distinct tissue/cell-type-dependent signaling cues produced by MMP11 substrates. Hence, etiological distinct disorders, such as cancer and neurodegeneration, may share pathomechanisms with distinct clinical outcomes. The identification of pathophysiological substrates of MMP11 will be critical to uncover the scope of its pathobiological roles and aid toward the design of tissue-selective MMP11 inhibitors. It will be interesting to explore whether paracrine factors and players within the network uncovered by the IPA presented in this work, such as Wnt2b and BDNF, their receptors or nuclear transcriptional factors, are substrates of MMP11 or exert regulatory effects on MMP11 expression or activity. Although a specific pharmacological inhibitor of MMP11 is not available [100], [101], the genetic interactions between Mmp11 and Ranbp2 may be revealed by assessing the effect of genetic ablation of Mmp11 on the pathophysiological manifestations observed in HRGP-cre:Ranbp2−/− (e.g. loss of membrane permeability of rods and rod apoptosis) and to define a potential role for MMP11 in neuroprotection against autonomous and non-autonomous cell death mechanisms affecting cone or rod photoreceptor survival.
Another critical outcome of this work is the finding of non-autonomous apoptotic death of rod (Nr2E3+) photoreceptors upon cone dysfunction and death. TUNEL+ cones were never identified at any age, while dying rods comprised a mixed population of TUNEL+, EthD-III+ or both. Thus, rods undergo a programmed cell death that is triggered by the demise of cones. Important, rod cell death ceases at P27, when cone degeneration is complete based on the lack of apoptotic bodies, the absence of Parp1+ and caspase3/7+ cells, and the preservation of the ONL and rod photoreceptor function at P27 and later ages.
Our results also indicate the presence of atypical mechanisms of cell death. While classical cell death features were identified during cone or rod degeneration, the following observations do not match previously recognized canonical paradigms. First, caspase 7 activation, typically observed in apoptosis, was found in dying cones lacking cleaved Parp1, a substrate of caspase 7. Instead, cone death was characterized by the rampant disintegration of critical subcellular structures, such as outer segments and mitochondria, swelling of lower fibers and cone pedicles, features that are consistent with necrosis. However, EthD-III+ cones were not identified as would be expected from the typical loss of membrane permeability caused by necrosis. Second, although rod photoreceptor death is consistent with apoptosis as supported by the presence of caspase 3+, Parp1+ and TUNEL+-cell bodies, classic necrotic features were also present, including a loss of plasma membrane permeability (EthD-III+ cell bodies) and limited formation of electron lucent areas in the matrix of mitochondria at synaptic spherules. These observations indicate that activation of caspase 3 and caspase 7 does not determine the mode of rod or cone cell death, respectively. This conclusion is supported by comparable rates of rod death in rd1 mice on a wild-type or caspase3−/− background [43]. Third, we did not observe a release of cytochrome C and AIF to the cytoplasm, as typified in apoptosis [66] and observed in other photoreceptor degeneration models [35], [45], an induction of RIP1, RIP3, or caspase 8 activation as observed in necroptosis [61], [69] and lipidation of microtubule-associated protein 1 light chain 3 (LC3/Atg8) that is typical of autophagy [70], [71]. Altogether these data support the existence of complex, shared, unique and thus atypical cell death mechanisms between rod and cone photoreceptors. These manifestations are likely determined by the cell-type dependent pleiotropic molecular and metabolic activities of Ranbp2 [47]–[54]. Emerging mouse models of Ranbp2 harboring losses in selective domains and functional activities of Ranbp2 will aid in parsing the contribution of such activities to cellular functions and intrinsic or extrinsic cell death modalities unique or shared by several diseases.
Finally, our data show that the non-autonomous death of rods upon loss of Ranbp2 in cones is not promoted by the absence of cones or loss of rod-cone cell contacts. Instead, a likely mechanism is the release of a diffusible factor by cone photoreceptors upon their dysfunction that is deleterious to rod photoreceptors. MMP11 activation is an excellent candidate to play a critical role in such paracrine and death signaling. Our data predict that the topographic density of cone and rod photoreceptors in the retina will play a determinant role in the level of rod photoreceptor degeneration. This issue is of crucial significance to human retinal dystrophies, because the unique and heterogeneous topographic distributions of photoreceptor types across the human retina often mirror retinal pathologies with regional tissue and clinical hallmarks, such as RP, cone-rod dystrophies and age-related macular degeneration (AMD). It is likely that loss of biological activities regulated by RANBP2 will exert prominent pathological outcomes in regions of the retina, such as the macula, where the topographic density of cone and rod photoreceptors is similar.
Materials and Methods
Animals
HRGP-Cre mice (kindly provided by Yun-Zheng Le, University of Oklahoma Health Sciences Center) [56], [57] were crossed to Ranbp2+/flox mice (kindly provided by Jan M. van Deursen, Mayo Clinic College of Medicine) [48] to produce HRGP-cre:Ranbp2+/flox. HRGP-cre:Ranbp2+/flox were then crossed to Ranbp2+/flox or Ranbp2Gt(pGT0pfs)630Wcs/+ [47] to generate HRGP-cre:Ranbp2+/− (+/−) and HRGP-cre:Ranbp2−/− (−/−). Mice were in the following mixed genetic background: 129 SvJ,C57BL/6J,FVB/N,129olaHsd. Mice were screened also for rd1 and rd8 alleles. Mice were raised in a pathogen-free transgenic barrier facility at <70 lux and given ad libitum access to water and chow diet 5LJ5 (Purina, Saint Louis, MO). Animal protocols were approved by the Institutional Animal Care and Use Committees at Duke University and Cleveland Clinic, and all procedures adhered to the ARVO guidelines for the Use of Animals in Vision Research.
Antibodies
Primary antibodies used this study were: mouse anti-Cre (Covance, Princeton, NJ), rabbit anti-Cre (Novagen, Gibbstown, NJ), rabbit anti-Arr4 (Millipore/Pel-Freez Biologicals , Billerica, MA), rabbit anti-L/M opsin (#Ab 21069) [55], rabbit monoclonal anti-cleaved caspase3 Asp175 (Cell Signaling, Danvers, MA), rabbit polyclonal anti-cleaved-caspase 7 Asp353 (Cell Signaling), rabbit anti-cleaved caspase 8 Asp391 (Cell Signaling), rabbit anti-cleaved caspase 9 Asp 330 (Cell Signaling), rabbit anti-cleaved Parp1 Asp214 (Cell Signaling), mouse anti-full-length Parp1 (BD Bioscience, San Jose, CA), mouse anti-LC3 B (Cell Signaling, Danvers, MA), mouse anti-cytochrome C (BD Bioscience, San Jose, CA), mouse anti-mitochondrial heat shock protein 70 (Affinity Bioreagent, Golden, CO), rabbit-cytosolic heat shock protein 70 (Assay Design, Farmingdale, NY), rabbit anti-GFAP (DAKO, Carpinteria, CA), mouse anti-glutamine synthase (Sigma Aldrich, Saint Louis, MO), mouse anti-RIP1 (BD Bioscience, San Jose, CA), rabbit anti-RIP3 (Sigma Aldrich, Saint Louis), rabbit anti-GAPDH and goat anti-AIF1 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Apaf-1 (LSBio, Seattle, WA), mouse anti PSD-95 (Affinity BioReagent), mouse anti-MMP11(Thermo Scientific, Waltham, MA), rabbit anti-S-opsin (Millipore/Pel-Freez Biologicals), rabbit anti-Nr2E3 (Proteintech, Hayward, CA) and Peanut Agglutinin TRTIC conjugate (Sigma-Aldrich). Alexa-conjugated secondary antibodies (408, 488, 568 and Cy5) were from Invitrogen (Carlsbad, CA).
RT-PCR and qRT-PCR
For total RNA and pre-miRNA isolation, retinas were homogenized with the TRIZOL Regent (Invitrogen) using Bullet Blender BBX24 (Next Advance Inc., Averill Park, NY) in the presence of 0.5 mm zirconium oxide beads (Next Advance Inc., Averill Park, NY) for 3 min at 8,000 rpm. RNA was reverse transcribed into cDNA using SuperScript II reverse transcriptase (Invitrogen). For RT-PCR, the ∼380 bp amplicon encompassing fused exons 1 and 3 of recombinant Ranbp2 mRNA was amplified from 250 ng of retinal cDNA. PCR was performed using primer 1 (Pr1:CGCCCCGAGAGTACATTTCTA) and primer 2 (Pr2:AAGTTTATTCCATCCATCTTCA) with GoTaq Green Master Mix (Promega, Madison, WI) under the following cycling conditions: 5 min/95°C of initial denaturation, 35 cycles/94°C (30 s), 55°C(30 s) and 72°C(30 s) with final elongation step at 72°C for 3 mins. Same cycling conditions were applied for Cre (CTAATCGCCATCTTCCAGCAGG, AGGTGTAGAGAAGGCACTTAGC) and Gapdh (GCAGTGGCAAAGTGGAGATT, GAATTTGCCGTGAGTGGAGT). qRT-PCR reactions were carried out with 8 ng of cDNA, 800 nM forward and reverse primers, 10 µl of 2×SYBR® Green PCR Master Mix (Applied Bioscience, Warrington, MA) in a 20 µl final volume in 48-well plates using the ECO™ Real-Time PCR system (Illumina, San Diego, CA). The relative amount of transcripts was calculated by the ΔΔ CT method using Gapdh as reference (n = 3–4). Primer sequences and designations are provided in Table S1.
Immunohistochemistry
The superior region of mouse cornea was burned using Low Temperature Cautery (Bovie Medical Corporation, St. Peterburg, FL) immediately after mice were killed. For immunohistochemistry, eyeballs were removed and fixed with 2% paraformaldehyde/phosphate-buffer saline (PBS), pH 7.4 for 4 hr after small incisions were made in the anterior portion. Upon removal of the lens, eyecups were infiltrated with 5% sucrose/100 mM PBS, pH 7.4, for 5 hr followed by 30% sucrose/100 mM PBS for 12 hours, embedded in Tissue-Tek O.C.T. compound (Sakura, Torrance) and stored at −80°C. 12 µm thick retinal cryosections along the vertical meridian of the eyecup were mounted on glass slides. For flat mounts, retinas were removed from fixed eyeballs, cut in a four-quadrant cloverleaf pattern using the caruncle as an orientation landmark and fixed for an additional 15 min in a 24-well plate. Specimens were incubated in blocking buffer (PBS, pH 7.4, containing 0.1% Triton X-100, 10% normal goat serum) for 1 hr at RT (for sections) or 12 hr at 4°C (flat mounts), followed by incubation with primary antibodies in incubation buffer (PBS, pH 7.4, containing 0.1% Triton X-100, 5% normal goat serum) overnight at RT (for sections) or for 3 days at 4°C (flat mounts). Specimens were washed thrice with washing buffer (PBS/0.1% Triton X-100) for 10 min, and incubated in incubation buffer for 2 hr with Alexa-conjugated secondary antibodies (1∶1,000; Invitrogen). For Nr2E3 immunostaining, eyeballs were fixed instead with 1% paraformaldehyde for 1 hr at room temperature. 6 µm-thick retinal cryosections were incubated in blocking buffer for 1 hr at room temperature followed by treatment with proteinase K (20 µg/ml, Promega, Madison, WI) for 9 min and standard immunostaining protocols as described earlier. Specimens were washed again thrice and mounted on glass slides for visualization and image acquisition. Images were acquired with a Nikon C1+-laser scanning confocal microscope coupled with a LU4A4 launching base of 4-solid state diode lasers (407 nm/100 mW, 488 nm/50 mW, 561 nm/50 mW, 640 nm/40 mW) and controlled by the Nikon EZC1.3.10 software (v6.4).
TUNEL assay
TUNEL assays were performed with the DeadEnd Fluorometric TUNEL System (Promega, Medison) with the following modifications from the manufacturer's instructions. Briefly, specimens were incubated with 20 µg/ml proteinase K for 15 min at RT followed by fixation with 4% paraformaldehyde for 15 min, and incubations with primary and secondary antibodies along with DAPI (Invitrogen, Carlsbad, CA). Specimens were equilibrated for 5 min in manufacturer's buffer before undergoing TdT reactions for 60 min at 37°C. Reactions were stopped with 2×SSC, washed three times with PBS and mounted.
EthD-III staining
Fresh P20 retinal explants were incubated with 5 µM EthD-III (Biotium, Hayward, CA) in the Neurobasal−A Medium/B-27 Supplement (Invitrogen) for 4 hr at 37°C in a humidified 5% CO2 atmosphere, washed vigorously 3 times with 100 mM PBS for 10 min and fixed with 2% paraformaldehyde/100 mM PBS for 20 min at room temperature. Specimens were then processed as described for immunostaining and TUNEL procedures.
Morphometric analyses
Morphometric analyses of M-cone photoreceptors were performed from 127×127 µm image fields captured with a Nikon C1+-laser scanning confocal microscope. Optical slices were 3D-reconstructed for the whole length of outer segments (∼25 µm, step size of 0.5 µm) from retinal flat mounts immunostained with an L/M-opsin antibody [55]. M-cone photoreceptors and the length of their outer segments were then tallied and measured from three image fields for each retina with the post-acquisition Nikon Elements AR (ver3.2) software. Cre+, c-casp3+ and TUNEL+-cells bodies were imaged from flat mounts and three image fields of 127×127 µm and collapsed for the whole depth of ONL (∼60 µm, step size of 3 µm) from the central or peripheral retinal regions. For TUNEL+, EthD-III+ and Arr4+-cells bodies, three image fields of 127×127×5 µm from retinal flat mounts were randomly selected and quantitatively analyzed. For tallying of DAPI+, TUNEL+,−Nr2E3+or Cre+-cell bodies, whole vertical meridian sections were counted and averaged to generate pie graphs (Excel, Microsoft, Seattle, WA). The distal region of the ONL was arbitrarily defined as the 15 µm distal segment of the ONL up to the external limiting membrane and the remaining segment was defined as proximal. 3D-reconstruction of collapsed images and morphometric analysis was performed with Nikon Elements AR software (ver. 3.2). Two-tailed equal or unequal variance t-test statistical analysis was performed. p≤0.05 was defined as significant.
SDS-PAGE and immunoblots
Retinas were homogenized at 4°C with Bullet Blender BBX24 (Next Advance Inc., Averill Park, NY) in the presence of 0.5 mm zirconium oxide beads (Next Advance Inc.) and RIPA buffer containing complete protease inhibitors (Roche Applied Bioscience, Penzberg, Germany) and 10 mM iodoacetamide (Sigma Aldrich). Protein concentration was measured by the BCA method using BSA as a standard. Samples (55 µg) were resolved on 11% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotted and developed using the SuperSignal Pico West (Thermo Scientific) as described previously [50]. Blots were probed with mouse anti-MMP-11 antibody (Thermo Scientific). Densitometry analysis of immunoblots was performed with Metamorph v7.0 (Molecular Devices, Sunnyvale, CA). Two-tail t-test statistical analysis was performed for validation of significant change (p<0.05).
Caspases' assays
Retinas were homogenized using BulletBlender BBX24 (Next Advance Inc.) with 0.5 mm zirconium oxide beads (Next Advance Inc.) and RIPA buffer. Retinal homogenates were centrifuged at 10,000 g for 15 min at 4°C. Supernatants were collected and protein concentrations determined by the BCA method using BSA as standard. Retinal homogenates were diluted in caspase assay buffer and caspase profiling assays were performed with the Sensolyte AFC Caspase sampler kit according to company's protocol (AnaSpec, Fremont, CA). The following caspase substrates were used for screening: Ac-YVAD-AFC (SB1) and Ac-WEHD-FAC (SB2) for caspase 1, Ac-VDVAD-FAC (SB3) for caspase 2, Ac-IETD-FAC, (SB4) for caspase 8, Ac-DEVD-FAC (SB5) and Z-DEVD-AFC (SB6) for caspases 3/7, Ac-LEHD-FAC (SB7) for caspase 9 and Ac-VEID-AFC (SB8) for caspase 6. Assays were first optimized for substrate dilution, extract concentration and reaction time. Substrate screenings were carried with three different protein concentrations. Analytical assays of caspase 3/7 (SB5) were performed with diluted 50 µl of retinal homogenate (280 ng) and 50 µl of substrate (1∶200 dilution in caspase assay buffer), mixed in the 96 well-plate, incubated with shaking for 1 hr under dark at room temperature. Measurements of fluorescence were performed at excitation/emission/cutoff = 380/500/495 nm with SpectraMax M5 (Molecular Devices, Sunnyvale, CA). Control reactions without extracts were subtracted from the samples' readings.
Matrix metalloproteinases' assays
MMPs' screening assays were performed with Sensolyte 520 generic MMP assay kit per company's protocol (AnaSpec, Fremont, CA). Retinas extracts were prepared as described previously with the exception that RIPA buffer was replaced with MMP assay buffer (AnaSpec). Briefly, MMP assays are based on the dequenching of fluorescence intensity of 5-FAM upon proteolytic cleavage of 5-FRAM/QXL520 peptide substrate by MMPs. MMPs screenings, except MMP11, were performed upon activation with 1 mM 4-aminophenylmercuric acetate (APMA) at different time intervals (MMP2, 7, 8 and 13 : 1 hr activation; MMP9, 12 and 14 : 2 hr activation; MMP1 : 3 hr activation; MMP3 and 10 : 24 hr activation) at 37°C followed by 1 hr incubation with 5-FRAM/QXL520 peptide substrate. For MMP11, retinal extract was directly mixed with substrate and incubated for 1 hr before fluorescence reading. 50 µl of activated/non-activated protein extract at 56, 112 or 224 ng and 50 µl of substrate (final substrate dilution 1∶200) were mixed in a 96 well-plate, incubated with shaking for 1 h at room temperature at dark. Measurements of fluorescence were performed at excitation/emission/cutoff = 490/520/495 nm with SpectraMax M5 (Molecular Devices, Sunnyvale, CA). Control measurements without retinal extracts under the same conditions were subtracted from the sample readings. Analytical assays with MMP11 were performed with 56 ng of retinal extract.
Subcellular fractionation
Mitochondrial and cytosolic fractions of the retina were isolated with the Mitochondria Isolation Kit for Tissue (Abcam, Cambridge, MA) per manufacturer's instruction. Briefly, retinas were washed with washing buffer, homogenized with a Kontes Microtube Pellet Pestle Rod with motor in isolation buffer, centrifuged at 1,000× g for 10 mins, the pellet was saved (nuclear-enriched fraction) and supernatant was centrifuged again at 12,000× g for 15 min. The supernatants (cytosolic fraction) were saved, the pellets (mitochondrial fraction) washed with Isolation buffer twice and re-suspended with isolation buffer with complete protein inhibitor cocktail (Roche Applied Bioscience, Penzberg, Germany).
Semi-thin sections and transmission electron microscopy
Eyeballs were removed and fixed with 2% glutaraldehyde:paraformaldehyde/0.1% cacodylate buffer, pH 7.2, overnight at 4°C. For semi-thin histological sections, 0.5 µm sections along the vertical meridian were mounted on glass slides and stained with 1% methylene blue. Light images of the retina sections were acquired with a Axiopan-2 light microscope controlled by Axovision Rel 4.6 and coupled to a AxioCam HRc digital camera (Carl Zeiss, Germany). For electron microscopy, specimens were post-fixed in 2% osmium tetraoxide in 0.1% cacodylate buffer and embedded in Spurr resin. 60 nm-thick sections were cut with Leica Ultracut S (Leica Microsystems, Waltzer, Germany), stained with 2% uranyl acetate/4% lead citrate and imaged with JEM-1400 transmission electron microscope (JEOL, Tokyo, Japan) coupled with an ORIUS 1000CCD camera.
Ingenuity pathway analysis
The Ingenuity pathway analysis (IPA, Ingenuity Systems, Redwood City, CA) was used to examine the gene dataset modulated by loss of Ranbp2 and to define connectivity maps and networks. Network of genes are algorithmically produced based on their connectivity. Significant network scores reflect the negative logarithm of a P value associated with the likelihood of connectivity of a set of genes in a network. The network with highest score (score of 22) was chosen for further analysis.
Electroretinography
After overnight dark adaptation, mice were anesthetized (ketamine: 80 mg/kg; xylazine: 16 mg/kg) and eyedrops were used for pupil dilation (1% tropicamide; 2.5% phenylephrine HCl; 1% cyclopentolate HCl) and corneal anesthesia (1% proparacaine HCl). The active electrode was a stainless-steel wire active electrode that contacted the corneal surface through 1% methylcellulose; needle electrodes placed in the cheek and tail served as reference and ground leads, respectively. Responses were differentially amplified (0.3–1,500 Hz), averaged, and stored using a UTAS E-3000 signal averaging system (LKC Technologies, Gaithersburg, MD). Strobe flash stimuli were initially presented in darkness within a ganzfeld bowl. Flash luminance ranged from −3.6 to 2.1 log cd s/m2 and stimuli were presented in order of increasing luminance. A steady adapting field (20 cd/m2) was then presented in the ganzfeld. After a 7 min light adaptation period cone ERGs were evoked by strobe flash stimuli superimposed upon the adapting field. Flash luminance ranged from −0.8 to 1.9 log cd s/m2 and stimuli were presented at 2 Hz in order of increasing luminance. The amplitude of the a-wave was measured 8 ms after flash onset from the prestimulus baseline. The amplitude of the b-wave was measured from the a-wave trough to the peak of the b-wave or, if no a-wave was present, from the pre-stimulus baseline.
Supporting Information
Zdroje
1. PalopJJ, ChinJ, MuckeL (2006) A network dysfunction perspective on neurodegenerative diseases. Nature 443 : 768–773.
2. IlievaH, PolymenidouM, ClevelandDW (2009) Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol 187 : 761–772.
3. BramallAN, WrightAF, JacobsonSG, McInnesRR (2010) The genomic, biochemical, and cellular responses of the retina in inherited photoreceptor degenerations and prospects for the treatment of these disorders. Annu Rev Neurosci 33 : 441–472.
4. Portera-CailliauC, SungCH, NathansJ, AdlerR (1994) Apoptotic photoreceptor cell death in mouse models of retinitis pigmentosa. Proc Natl Acad Sci USA 91 : 974–978.
5. KedzierskiW, BokD, TravisGH (1998) Non-cell-autonomous photoreceptor degeneration in rds mutant mice mosaic for expression of a rescue transgene. J Neurosci 18 : 4076–4082.
6. Mohand-SaidS, HicksD, LeveillardT, PicaudS, PortoF, et al. (2001) Rod-cone interactions: developmental and clinical significance. Prog Retin Eye Res 20 : 451–467.
7. HuangPC, GaitanAE, HaoY, PettersRM, WongF (1993) Cellular interactions implicated in the mechanism of photoreceptor degeneration in transgenic mice expressing a mutant rhodopsin gene. Proc Natl Acad Sci USA 90 : 8484–8488.
8. LewisA, WilliamsP, LawrenceO, WongRO, BrockerhoffSE (2010) Wild-type cone photoreceptors persist despite neighboring mutant cone degeneration. J Neurosci 30 : 382–389.
9. Carter-DawsonLD, LaVailMM, SidmanRL (1978) Differential effect of the rd mutation on rods and cones in the mouse retina. Invest Ophthalmol Vis Sci 17 : 489–498.
10. HartongDT, BersonEL, DryjaTP (2006) Retinitis pigmentosa. Lancet 368 : 1795–1809.
11. JaissleGB, MayCA, ReinhardJ, KohlerK, FauserS, et al. (2001) Evaluation of the rhodopsin knockout mouse as a model of pure cone function. Invest Ophthalmol Vis Sci 42 : 506–513.
12. CideciyanAV, HoodDC, HuangY, BaninE, LiZY, et al. (1998) Disease sequence from mutant rhodopsin allele to rod and cone photoreceptor degeneration in man. Proc Natl Acad Sci U S A 95 : 7103–7108.
13. JohnSK, SmithJE, AguirreGD, MilamAH (2000) Loss of cone molecular markers in rhodopsin-mutant human retinas with retinitis pigmentosa. Mol Vis 6 : 204–215.
14. HumphriesMM, RancourtD, FarrarGJ, KennaP, HazelM, et al. (1997) Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat Genet 15 : 216–219.
15. DryjaTP, FinnJT, PengYW, McGeeTL, BersonEL, et al. (1995) Mutations in the gene encoding the alpha subunit of the rod cGMP-gated channel in autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci USA 92 : 10177–10181.
16. BareilC, HamelCP, DelagueV, ArnaudB, DemailleJ, et al. (2001) Segregation of a mutation in CNGB1 encoding the beta-subunit of the rod cGMP-gated channel in a family with autosomal recessive retinitis pigmentosa. Hum Genet 108 : 328–334.
17. ChangB, GrauT, DangelS, HurdR, JurkliesB, et al. (2009) A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc Natl Acad Sci USA 106 : 19581–19586.
18. ThiadensAA, den HollanderAI, RoosingS, NabuursSB, Zekveld-VroonRC, et al. (2009) Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am J Hum Genet 85 : 240–247.
19. TrifunovicD, DenglerK, MichalakisS, ZrennerE, WissingerB, et al. (2010) cGMP-dependent cone photoreceptor degeneration in the cpfl1 mouse retina. J Comp Neurol 518 : 3604–3617.
20. ChangB, HawesNL, HurdRE, DavissonMT, NusinowitzS, et al. (2002) Retinal degeneration mutants in the mouse. Vision Res 42 : 517–525.
21. GardnerJC, WebbTR, KanugaN, RobsonAG, HolderGE, et al. (2010) X-linked cone dystrophy caused by mutation of the red and green cone opsins. Am J Hum Genet 87 : 26–39.
22. GlicksteinM, HeathGG (1975) Receptors in the monochromat eye. Vision Res 15 : 633–636.
23. WissingerB, GamerD, JagleH, GiordaR, MarxT, et al. (2001) CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet 69 : 722–737.
24. NishiguchiKM, SandbergMA, GorjiN, BersonEL, DryjaTP (2005) Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum Mutat 25 : 248–258.
25. BielM, SeeligerM, PfeiferA, KohlerK, GerstnerA, et al. (1999) Selective loss of cone function in mice lacking the cyclic nucleotide-gated channel CNG3. Proc Natl Acad Sci USA 96 : 7553–7557.
26. DingXQ, HarryCS, UminoY, MatveevAV, FlieslerSJ, et al. (2009) Impaired cone function and cone degeneration resulting from CNGB3 deficiency: down-regulation of CNGA3 biosynthesis as a potential mechanism. Hum Mol Genet 18 : 4770–4780.
27. ShenJ, YangX, DongA, PettersRM, PengYW, et al. (2005) Oxidative damage is a potential cause of cone cell death in retinitis pigmentosa. J Cell Physiol 203 : 457–464.
28. PunzoC, KornackerK, CepkoCL (2009) Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci 12 : 44–52.
29. KomeimaK, RogersBS, LuL, CampochiaroPA (2006) Antioxidants reduce cone cell death in a model of retinitis pigmentosa. Proc Natl Acad Sci USA 103 : 11300–11305.
30. LeveillardT, Mohand-SaidS, LorentzO, HicksD, FintzAC, et al. (2004) Identification and characterization of rod-derived cone viability factor. Nat Genet 36 : 755–759.
31. OtaniA, DorrellMI, KinderK, MorenoSK, NusinowitzS, et al. (2004) Rescue of retinal degeneration by intravitreally injected adult bone marrow-derived lineage-negative hematopoietic stem cells. J Clin Invest 114 : 765–774.
32. ZengHY, ZhuXA, ZhangC, YangLP, WuLM, et al. (2005) Identification of sequential events and factors associated with microglial activation, migration, and cytotoxicity in retinal degeneration in rd mice. Invest Ophthalmol Vis Sci 46 : 2992–2999.
33. ZeissCJ, JohnsonEA (2004) Proliferation of microglia, but not photoreceptors, in the outer nuclear layer of the rd-1 mouse. Invest Ophthalmol Vis Sci 45 : 971–976.
34. RippsH (2002) Cell death in retinitis pigmentosa: gap junctions and the ‘bystander’ effect. Exp Eye Res 74 : 327–336.
35. DoonanF, DonovanM, CotterTG (2003) Caspase-independent photoreceptor apoptosis in mouse models of retinal degeneration. J Neurosci 23 : 5723–5731.
36. Sancho-PelluzJ, Arango-GonzalezB, KustermannS, RomeroFJ, van VeenT, et al. (2008) Photoreceptor cell death mechanisms in inherited retinal degeneration. Mol Neurobiol 38 : 253–269.
37. FrassonM, SahelJA, FabreM, SimonuttiM, DreyfusH, et al. (1999) Retinitis pigmentosa: rod photoreceptor rescue by a calcium-channel blocker in the rd mouse. Nat Med 5 : 1183–1187.
38. PawlykBS, LiT, ScimecaMS, SandbergMA, BersonEL (2002) Absence of photoreceptor rescue with D-cis-diltiazem in the rd mouse. Invest Ophthalmol Vis Sci 43 : 1912–1915.
39. Pearce-KellingSE, AlemanTS, NickleA, LatiesAM, AguirreGD, et al. (2001) Calcium channel blocker D-cis-diltiazem does not slow retinal degeneration in the PDE6B mutant rcd1 canine model of retinitis pigmentosa. Mol Vis 7 : 42–47.
40. TakeuchiK, NakazawaM, MizukoshiS (2008) Systemic administration of nilvadipine delays photoreceptor degeneration of heterozygous retinal degeneration slow (rds) mouse. Exp Eye Res 86 : 60–69.
41. JosephRM, LiT (1996) Overexpression of Bcl-2 or Bcl-XL transgenes and photoreceptor degeneration. Invest Ophthalmol Vis Sci 37 : 2434–2446.
42. ChenJ, FlanneryJG, LaVailMM, SteinbergRH, XuJ, et al. (1996) bcl-2 overexpression reduces apoptotic photoreceptor cell death in three different retinal degenerations. Proc Natl Acad Sci U S A 93 : 7042–7047.
43. ZeissCJ, NealJ, JohnsonEA (2004) Caspase-3 in postnatal retinal development and degeneration. Invest Ophthalmol Vis Sci 45 : 964–970.
44. JomaryC, NealMJ, JonesSE (2001) Characterization of cell death pathways in murine retinal neurodegeneration implicates cytochrome c release, caspase activation, and bid cleavage. Mol Cell Neurosci 18 : 335–346.
45. SangesD, ComitatoA, TammaroR, MarigoV (2006) Apoptosis in retinal degeneration involves cross-talk between apoptosis-inducing factor (AIF) and caspase-12 and is blocked by calpain inhibitors. Proc Natl Acad Sci USA 103 : 17366–17371.
46. SchaeferhoffK, MichalakisS, TanimotoN, FischerMD, BecirovicE, et al. (2010) Induction of STAT3-related genes in fast degenerating cone photoreceptors of cpfl1 mice. Cell Mol Life Sci 67 : 3173–3186.
47. AslanukovA, BhowmickR, GurujuM, OswaldJ, RazD, et al. (2006) RanBP2 Modulates Cox11 and Hexokinase I Activities and Haploinsufficiency of RanBP2 Causes Deficits in Glucose Metabolism. PLoS Genet 2: e177.
48. DawlatyMM, MalureanuL, JeganathanKB, KaoE, SustmannC, et al. (2008) Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 133 : 103–115.
49. ChoKI, YiH, TserentsoodolN, SearleK, FerreiraPA (2010) Neuroprotection resulting from insufficiency of RANBP2 is associated with the modulation of protein and lipid homeostasis of functionally diverse but linked pathways in response to oxidative stress. Dis Model Mech 3 : 595–604.
50. ChoKI, YiH, YehA, TserentsoodolN, CuadradoL, et al. (2009) Haploinsufficiency of RanBP2 is neuroprotective against light-elicited and age-dependent degeneration of photoreceptor neurons. Cell Death Differ 16 : 287–297.
51. ChoKI, SearleK, WebbM, YiH, FerreiraPA (2012) Ranbp2 haploinsufficiency mediates distinct cellular and biochemical phenotypes in brain and retinal dopaminergic and glia cells elicited by the Parkinsonian neurotoxin, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP). Cell Mol Life Sci 69 : 3511–3527.
52. NeilsonDE, AdamsMD, OrrCM, SchellingDK, EibenRM, et al. (2009) Infection-Triggered Familial or Recurrent Cases of Acute Necrotizing Encephalopathy Caused by Mutations in a Component of the Nuclear Pore, RANBP2. Am J Hum Genet 84 : 44–51.
53. WolfK, Schmitt-MechelkeT, KolliasS, CurtA (2013) Acute necrotizing encephalopathy (ANE1): rare autosomal-dominant disorder presenting as acute transverse myelitis. J Neurol [Epub ahead of print]
54. LonnqvistT, IsohanniP, ValanneL, Olli-LahdesmakiT, SuomalainenA, et al. (2011) Dominant encephalopathy mimicking mitochondrial disease. Neurology 76 : 101–103.
55. MavlyutovTA, CaiY, FerreiraPA (2002) Identification of RanBP2 - and Kinesin-Mediated Transport Pathways with Restricted Neuronal and Subcellular Localization. Traffic 3 : 630–640.
56. LeYZ, AshJD, Al-UbaidiMR, ChenY, MaJX, et al. (2004) Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol Vis 10 : 1011–1018.
57. LeYZ, AshJD, Al-UbaidiMR, ChenY, MaJX, et al. (2006) Conditional gene knockout system in cone photoreceptors. Adv Exp Med Biol 572 : 173–178.
58. NikonovSS, BrownBM, DavisJA, ZunigaFI, BraginA, et al. (2008) Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron 59 : 462–474.
59. JeonCJ, StrettoiE, MaslandRH (1998) The major cell populations of the mouse retina. J Neurosci 18 : 8936–8946.
60. ChenJ, RattnerA, NathansJ (2005) The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci 25 : 118–129.
61. GalluzziL, VitaleI, AbramsJM, AlnemriES, BaehreckeEH, et al. (2012) Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 19 : 107–120.
62. KroemerG, GalluzziL, VandenabeeleP, AbramsJ, AlnemriES, et al. (2009) Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ 16 : 3–11.
63. KaufmannSH, DesnoyersS, OttavianoY, DavidsonNE, PoirierGG (1993) Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res 53 : 3976–3985.
64. GermainM, AffarEB, D'AmoursD, DixitVM, SalvesenGS, et al. (1999) Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J Biol Chem 274 : 28379–28384.
65. Paquet-DurandF, SilvaJ, TalukdarT, JohnsonLE, AzadiS, et al. (2007) Excessive activation of poly(ADP-ribose) polymerase contributes to inherited photoreceptor degeneration in the retinal degeneration 1 mouse. J Neurosci 27 : 10311–10319.
66. SleeEA, HarteMT, KluckRM, WolfBB, CasianoCA, et al. (1999) Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol 144 : 281–292.
67. KleinJA, Longo-GuessCM, RossmannMP, SeburnKL, HurdRE, et al. (2002) The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature 419 : 367–374.
68. LiptonSA, Bossy-WetzelE (2002) Dueling activities of AIF in cell death versus survival: DNA binding and redox activity. Cell 111 : 147–150.
69. MurakamiY, MatsumotoH, RohM, SuzukiJ, HisatomiT, et al. (2012) Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc Natl Acad Sci U S A 109 : 14598–14603.
70. KunchithapauthamK, RohrerB (2007) Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy 3 : 433–441.
71. KunchithapauthamK, RohrerB (2007) Autophagy is one of the multiple mechanisms active in photoreceptor degeneration. Autophagy 3 : 65–66.
72. Page-McCawA, EwaldAJ, WerbZ (2007) Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol 8 : 221–233.
73. SanukiR, OnishiA, KoikeC, MuramatsuR, WatanabeS, et al. (2011) miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci 14 : 1125–1134.
74. HennigAK, PengGH, ChenS (2008) Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res 1192 : 114–133.
75. NgL, HurleyJB, DierksB, SrinivasM, SaltoC, et al. (2001) A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet 27 : 94–98.
76. RobertsMR, SrinivasM, ForrestD, Morreale de EscobarG, RehTA (2006) Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci USA 103 : 6218–6223.
77. NishidaA, FurukawaA, KoikeC, TanoY, AizawaS, et al. (2003) Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci 6 : 1255–1263.
78. KoikeC, NishidaA, UenoS, SaitoH, SanukiR, et al. (2007) Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol 27 : 8318–8329.
79. BaudetML, ZivrajKH, Abreu-GoodgerC, MuldalA, ArmisenJ, et al. (2012) miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat Neurosci 15 : 29–38.
80. MotrescuER, BlaiseS, EtiqueN, MessaddeqN, ChenardMP, et al. (2008) Matrix metalloproteinase-11/stromelysin-3 exhibits collagenolytic function against collagen VI under normal and malignant conditions. Oncogene 27 : 6347–6355.
81. HanHB, GuJ, ZuoHJ, ChenZG, ZhaoW, et al. (2012) Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J Pathol 226 : 544–555.
82. Stehlin-GaonC, WillmannD, ZeyerD, SanglierS, Van DorsselaerA, et al. (2003) All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nat Struct Biol 10 : 820–825.
83. FujiedaH, BremnerR, MearsAJ, SasakiH (2009) Retinoic acid receptor-related orphan receptor alpha regulates a subset of cone genes during mouse retinal development. J Neurochem 108 : 91–101.
84. HelmlingerD, YvertG, PicaudS, MerienneK, SahelJ, et al. (2002) Progressive retinal degeneration and dysfunction in R6 Huntington's disease mice. Hum Mol Genet 11 : 3351–3359.
85. HaradaC, GuoX, NamekataK, KimuraA, NakamuraK, et al. (2011) Glia - and neuron-specific functions of TrkB signalling during retinal degeneration and regeneration. Nat Commun 2 : 189.
86. WilsonRB, KunchithapauthamK, RohrerB (2007) Paradoxical role of BDNF: BDNF+/ − retinas are protected against light damage-mediated stress. Invest Ophthalmol Vis Sci 48 : 2877–2886.
87. van AmerongenR, MikelsA, NusseR (2008) Alternative wnt signaling is initiated by distinct receptors. Sci Signal 1: re9.
88. SharmaS, BallSL, PeacheyNS (2005) Pharmacological studies of the mouse cone electroretinogram. Vis Neurosci 22 : 631–636.
89. XuX, QuiambaoAB, RoveriL, PardueMT, MarxJL, et al. (2000) Degeneration of cone photoreceptors induced by expression of the Mas1 protooncogene. Exp Neurol 163 : 207–219.
90. BaroukhN, RavierMA, LoderMK, HillEV, BounacerA, et al. (2007) MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem 282 : 19575–19588.
91. PeiD, WeissSJ (1995) Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature 375 : 244–247.
92. MassonR, LefebvreO, NoelA, FahimeME, ChenardMP, et al. (1998) In vivo evidence that the stromelysin-3 metalloproteinase contributes in a paracrine manner to epithelial cell malignancy. J Cell Biol 140 : 1535–1541.
93. AndarawewaKL, MotrescuER, ChenardMP, GansmullerA, StollI, et al. (2005) Stromelysin-3 is a potent negative regulator of adipogenesis participating to cancer cell-adipocyte interaction/crosstalk at the tumor invasive front. Cancer Res 65 : 10862–10871.
94. ManesS, MiraE, BarbacidMM, CipresA, Fernandez-ResaP, et al. (1997) Identification of insulin-like growth factor-binding protein-1 as a potential physiological substrate for human stromelysin-3. J Biol Chem 272 : 25706–25712.
95. WuE, MariBP, WangF, AndersonIC, SundayME, et al. (2001) Stromelysin-3 suppresses tumor cell apoptosis in a murine model. J Cell Biochem 82 : 549–555.
96. BoulayA, MassonR, ChenardMP, El FahimeM, CassardL, et al. (2001) High cancer cell death in syngeneic tumors developed in host mice deficient for the stromelysin-3 matrix metalloproteinase. Cancer Res 61 : 2189–2193.
97. JonesLE, HumphreysMJ, CampbellF, NeoptolemosJP, BoydMT (2004) Comprehensive analysis of matrix metalloproteinase and tissue inhibitor expression in pancreatic cancer: increased expression of matrix metalloproteinase-7 predicts poor survival. Clin Cancer Res 10 : 2832–2845.
98. AsanoT, TadaM, ChengS, TakemotoN, KuramaeT, et al. (2008) Prognostic values of matrix metalloproteinase family expression in human colorectal carcinoma. J Surg Res 146 : 32–42.
99. YangYH, DengH, LiWM, ZhangQY, HuXT, et al. (2008) Identification of matrix metalloproteinase 11 as a predictive tumor marker in serum based on gene expression profiling. Clin Cancer Res 14 : 74–81.
100. FisherJF, MobasheryS (2006) Recent advances in MMP inhibitor design. Cancer metastasis reviews 25 : 115–136.
101. MatziariM, DiveV, YiotakisA (2007) Matrix metalloproteinase 11 (MMP-11; stromelysin-3) and synthetic inhibitors. Med Res Rev 27 : 528–552.
Štítky
Genetika Reprodukční medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene ClusterČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 6- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



