-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
In Drosophila, the MSL (Male Specific Lethal) complex up regulates transcription of active genes on the single male X-chromosome to equalize gene expression between sexes. One model argues that the MSL complex acts upon the elongation step of transcription rather than initiation. In an unbiased forward genetic screen for new factors required for dosage compensation, we found that mutations in the universally conserved transcription elongation factor Spt5 lower MSL complex dependent expression from the miniwhite reporter gene in vivo. We show that SPT5 interacts directly with MSL1 in vitro and is required downstream of MSL complex recruitment, providing the first mechanistic data corroborating the elongation model of dosage compensation.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003073
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003073Summary
In Drosophila, the MSL (Male Specific Lethal) complex up regulates transcription of active genes on the single male X-chromosome to equalize gene expression between sexes. One model argues that the MSL complex acts upon the elongation step of transcription rather than initiation. In an unbiased forward genetic screen for new factors required for dosage compensation, we found that mutations in the universally conserved transcription elongation factor Spt5 lower MSL complex dependent expression from the miniwhite reporter gene in vivo. We show that SPT5 interacts directly with MSL1 in vitro and is required downstream of MSL complex recruitment, providing the first mechanistic data corroborating the elongation model of dosage compensation.
Introduction
Drosophila dosage compensation is widely used as a model system to investigate how transcription is regulated by large scale chromatin modifications [1]. To equalize the expression of the X-linked genes between XY males and XX females, the single X-chromosome in males is hypertranscribed a modest, but essential ∼1.4–1.8 fold. This is accomplished by the MSL complex, which consists of at least five proteins and two noncoding roX (RNA on X) RNAs [2]. The complex contains the histone modifying enzymes MOF (H4K16ac) and MSL2 (H2BK34ub) [3]. MSL3 is a chromodomain protein implicated in MSL complex distribution to its target site [4]. MSL1 assembles the complex via discrete docking sites for MSL2, MSL3, and MOF. MLE is an ATPase/helicase with double stranded RNA binding motifs that associates with the complex in an RNA dependent manner.
A long-standing puzzle is the biochemical mechanism by which the MSL complex up regulates X-linked genes, each of which is controlled by different transcription factors. An elegant model that solves this problem posits that MSL complex does not act with diverse gene-specific transcription factors to alter initiation, but rather at the elongation step of transcription common to all genes [5]. This proposal is supported by the higher resolution mapping of MSL complex binding and H4K16 acetylation within the bodies of actively transcribed X-linked genes with a bias towards the 3′ end [6]–[8]. Global nuclear run on analysis showed that compared to autosomes, the male X-chromosome has higher levels of transcriptionally engaged RNAPII (RNA Polymerase II) within the distal portions of the genes [9]. In contrast to this, a recent study detected increased RNAPII occupancy at the promoters of X-linked genes in males leading to the alternate idea that dosage compensation operates at the level of transcription initiation [10]. It is not clear whether decondensation of the chromatin fiber by H4K16 acetylation aids passage or recruitment of RNAPII enough to explain dosage compensation [11], or if the MSL complex has additional interactions with the basal transcriptional machinery.
To search for new factors involved in dosage compensation we performed an unbiased forward genetic screen that relies on a sensitive eye pigmentation reporter of MSL complex activity. This approach was designed to recover heterozygous mutations in genes that are essential for general transcription in both sexes but play an additional role in male dosage compensation. We recovered multiple alleles of Spt5, a universally conserved transcription elongation factor. We found that SPT5 is required for dosage compensation in males and extensively colocalizes with the MSL complex on the X-chromosome. Moreover, we found that SPT5 and MSL1 directly interact with each other. We propose that SPT5 is required downstream of MSL complex recruitment to stimulate transcription elongation. The identification of SPT5 is strong mechanistic evidence supporting the elongation model of dosage compensation.
Results
A genetic screen for identifying new components of dosage compensation
The eye color of roX1 transgenic males is a sensitive reporter of MSL activity [12]. When roX1 transgenes occasionally land in repressive chromatin, the miniwhite marker is epigenetically silenced so that females have solid white eyes. Males have spotted eyes because the MSL complex binds the autosomal roX1 transgene and locally modifies the chromatin allowing miniwhite expression in a fraction of cells. We have previously described a strategy for isolating mutations that increased local MSL activity [13]. Here we use a similar method to isolate mutations that reduce MSL activity. This approach has two important advantages. First, it allows identification of factors that are instrumental in achieving dosage compensation of the male X-chromosome in vivo, but may associate with the MSL complex too weakly or transiently to copurify with MSL proteins. Second, our genetic strategy retains a wild type allele of the relevant gene allowing us to capture factors that have additional essential functions. Homozygous mutations in such factors would be lethal to both sexes and thus would have been missed in earlier genetic screens based on the male specific lethal phenotype.
We screened approximately 16,000 EMS mutagenized flies and identified 48 mutations that dramatically lowered the eye pigmentation in males (Figure 1A and 1B, Figure S1). It is difficult to estimate if the amount of pigmentation in any individual ommatidia changes. What the screen detects is a change in the fraction of ommatidia that do or do not derepress the miniwhite reporter linked to roX1. Mutants that were also recessive lethal to both sexes were placed in complementation groups (Figure S1). We tested the ability of the modifier mutants to suppress eye pigmentation in multiple mosaic roX1 transgenes inserted in distinct repressive locations reasoning that those were more likely to affect dosage compensation rather than the particular silencing factors acting on flanking chromatin (Figure S2). One uninteresting mechanism that might produce this phenotype would be mutations that globally strengthened the repressive chromatin environment responsible for silencing of the miniwhite gene in our roX1 reporters. We tested the effect of the new mutants on In(1)wm4 which displays classic position effect variegation in both sexes. Most of the candidate modifiers of dosage compensation did not affect pigmentation in In(1)wm4 (Figure 1C and 1D) arguing against a global increase in repressive chromatin.
Fig. 1. Isolation of Spt5 mutations. 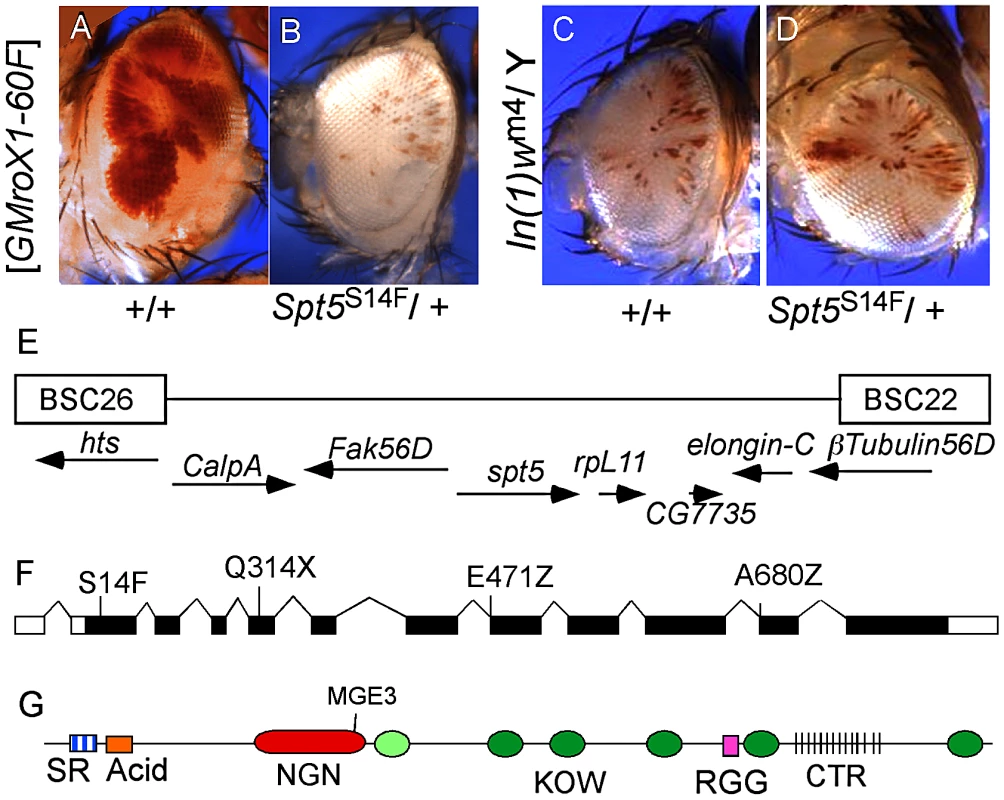
(A) Male flies carrying the [w+ GMroX1] transgene inserted at the 2R telomere (60F) have sectored pigmentation due to dosage compensation at the transgene. (B) Males heterozygous for mutations in Spt5 lose most red eye pigmentation due to reduced MSL complex activity. (C and D) Spt5 mutants have no effect on the PEV line In(1)wm4. (E) Genomic location of Spt5 and flanking deficiencies. (F) New Spt5 mutations failed to complement the previously reported Spt5MGE-3 allele [15]. (G) SPT5 domain features. SR, Serine/Arginine; NGN, N-terminal NusG; KOW, Kyrpides, Ouzounis, Woese light green oval indicates only partial match to consensus; RGG, arginine glycine glycine repeats; CTR, C-Terminal Repeats similar to RNAPII. Screen design and results are in Figure S1. SPT5 couples dosage compensation and transcription elongation
Complementation group C was chosen for detailed analysis. Meiotic recombination placed the locus near the polytene bands 56C-F but none of the available chromosome deficiencies uncovered the mutation [14]. Closer inspection revealed a gap in the deficiencies where elongation factors Spt5 and Elongin-C are located (Figure 1E). Available mutations in Elongin-C complemented all five group C alleles and we found no lesions in Elongin-C upon sequencing (data not shown). However, when we tested the Spt5MGE-3 mutation [15], it failed to complement all five group C alleles. Sequencing genomic DNA from these mutants identified one stop codon (Q314X), two splice junction mutations (E471Z and A680Z), and one missense mutation (S14F) (Figure 1F). The modular structure of SPT5 is summarized in Figure 1G.
Because SPT5 is such a critically important transcription elongation factor used by many genes, we were concerned that it appeared in our screen because reducing the level of any vital general transcription factor would lower expression of our eye color reporter. To address this concern, we screened the autosomal Bloomington Deficiency stock collection. We reasoned that if disrupting transcriptional efficiency in general affected our reporter, then many deficiencies would lower red pigmentation of the mosaic roX1 lines just like Spt5 mutations had. We crossed six different roX1 mosaic lines that carry roX1 transgenes in diverse chromatin environments to 190 deficiencies. The idea was that any deficiency that affected the eye coloration of multiple roX1 reporter lines was more likely to affect some aspect of dosage compensation rather than the particular repressive environment surrounding the different inserts. We found that only 10 intervals reduced MSL complex reporter activity (Table S1). Moreover, removing one copy of these 10 regions lowered MSL complex dependent red pigmentation across 4 or more of the mosaic roX1 lines supporting the notion that the relevant factors are somehow acting on dosage compensation. The deficiency screen shows that silencing the roX1 eye color reporter is an uncommon dominant haploinsufficient phenotype produced by only a few loci in the genome. Thus, the phenotype seen in Spt5 mutants is unlikely to be due to a general reduction of transcription.
We were still concerned that the white eye color gene used in our dosage compensation reporter might be particularly sensitive to SPT5 levels. We turned to strong hypomorphic alleles of white to test this possibility. The wa and we alleles each carry different transposon insertions that greatly reduce their expression resulting in orange eyes [17], [18]. On this background, small changes in white expression should be easily detectable by altered eye color. We crossed our Spt5 mutations into these two stocks and observed no difference in the eye pigmentation (Figure S3). We also crossed unrelated transgenes marked with miniwhite into our new Spt5 mutations and saw no change in eye pigmentation (data not shown). This shows that a 50% reduction in SPT5 levels does not alter the phenotype from hypomorphic white alleles or miniwhite. We conclude that Spt5 mutations dramatically affected the probability that males overcome silencing not because of global reduction of transcription across the genome or the white promoter itself, but rather because SPT5 plays some role in dosage compensation to which our roX1 reporter is responsive.
We used a sensitized genetic background to see if Spt5 affected dosage compensation of the X chromosome in addition to the roX1 eye color reporter transgene. Because SPT5 is essential for most transcription, homozygous null animals die early in development. The viability of Spt5/+ males demonstrates that dosage compensation must be adequate even with reduced SPT5 levels. The same is true for any of the msl/+ heterozygotes. However, males with limiting MSL complex might be more sensitive to reduced levels of SPT5. Males missing either roX1 or roX2 are alive but males missing both roX RNAs have greatly reduced male viability [19]. In our genetic background such roX1 roX2 double mutant males are completely lethal but can be rescued by an autosomal roX transgene [20]. Restoring male viability under these conditions depends on abundant MSL subunits. Males heterozygous for msl1 or mle showed reduced viability when roX1 RNA is also limiting [13]. Similarly, reducing SPT5 selectively lowered male viability to ∼15% when roX1 RNA was limiting consistent with a role in dosage compensation (Figure 2A). We assayed related factors to see how specific this phenotype was and found that lowering Elongin-C, another factor involved in elongation or Jil1, the histone H3S10 kinase that associates with the MSL complex had no effect on male viability (Figure 2A). However, Su(Tpl)S192, a mutation in the elongation factor ELL did reduce male viability. Others have reported that ELL RNAi lines display male specific lethality consistent with a role in dosage compensation [21]. These results are consistent with SPT5 playing a central role in dosage compensation that becomes more obvious when MSL activity is limited by low roX1 RNA levels.
Fig. 2. Genetic interactions between Spt5 and dosage compensation. 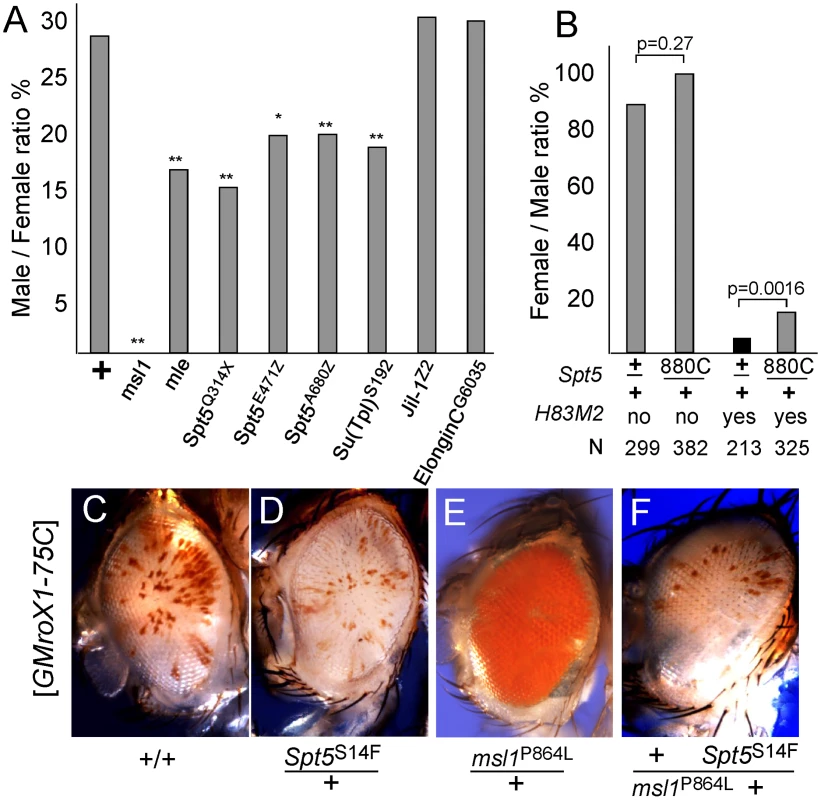
(A) Lowering SPT5 reduces male viability compared to sisters. All males were roX1 roX2 double mutants and partially rescued by one copy of the [GMroX1-75C] transgene. Males wild type for all other loci are rescued 29%, but flies missing one copy of the indicated dosage compensation genes have reduced male viability. ** p<0.01 * p<0.05 Fisher exact test. Detailed results in Table S2. (B) Reducing Spt5 rescues the sterility of [H83M2] females. y w; Spt5880C/CyO y+ females were mated to w/Y; [w+ H83M2]/+ males. Only the adults eclosing during the first two days are indicated to measure delayed development. [w+ H83M2] escaper females produced few eggs and were sterile (black bar). [w+ H83M2] females heterozygous for Spt5 regained fertility (gray bars). N = number of brothers recovered for each class. p calculated by Fisher exact test. (C) Males homozygous for the [GMroX1] transgene at 75C have a few pigmented sectors. (D) Singly, Spt5 slightly reduces and (E) msl1P864L dramatically increases local MSL activity. (F) When present together, Spt5S14F blocks the increased activity of the msl1P864L gain of function allele. See Figure S2 and Figure S3 for additional genetic analysis. Females normally lack dosage compensation because SXL blocks translation of msl2 mRNA. Ectopic dosage compensation can be induced in females by artificial expression of MSL2 by the [H83M2] transgene that escapes SXL regulation [22]. The inappropriate dosage compensation slows development resulting in delayed eclosion of adult females (Figure 2B). The resulting females produce very few eggs and are sterile. If reducing Spt5 weakens dosage compensation then that might reduce the toxic effects of inappropriate dosage compensation in [H83M2] overexpression females. When [H83M2] females also carried a mutation in Spt5, the female-specific developmental delay was modestly rescued (Figure 2B). However, the more striking result was that the Spt5/+ [H83M2] females produced abundant eggs that successfully developed into larvae. This argues that SPT5 is needed for MSL2 to drive inappropriate dosage compensation in females.
To further examine functional links between SPT5 and dosage compensation, we tested genetic interactions between the newly recovered Spt5 mutations and unusual gain of function msl1 alleles. We previously reported two missense alleles that partially disrupt the MSL1-MOF or MSL1-MSL3 interfaces [13]. Both mutations dominantly cause msl1*/+ males to produce solid red eyes (more MSL activity, Figure 2E) from the mosaic GMroX1-75C transgenic reporter whose basal pattern is mostly white with scattered small red sectors (Figure 2C). Spt5 mutations alone reduce sectoring slightly (Figure 2D). We constructed flies heterozygous for both the msl1P864L and Spt5S14F mutations that also carried the 75C dosage compensation eye color reporter. These males had white eyes (Figure 2F). This shows that even MSL complex containing the overly active P864L subunit can only act on the roX1 reporter when full SPT5 levels are present. Taken together, these in vivo results indicate a role for SPT5 in male X - dosage compensation beyond its general role in transcription of the entire genome in both sexes.
SPT5 and MSL complex colocalize on the X-chromosome
ChIP analysis found that SPT5 is enriched over the transcription start site (TSS) of most Drosophila genes, with additional binding across the transcribed regions [23]. At the level of polytene chromosomes, SPT5 binds many sites [16], [24] and colocalizes imperfectly with the MSL complex on the male X-chromosome [6]. To examine this issue in more detail, we raised new SPT5 antibodies. The serum recognized a single band around 135 kDa on SDS-PAGE which is larger than the predicted 119 kDa (Figure S4). Anomalous migration for SPT5 was reported earlier [16], [24]. The SPT5 serum recognized many bands on all polytene chromosomes in both sexes (Figure 3B control panel and Figure S5A). Most of the X-linked bands overlapped with MSL1 staining in males, but a few bands stain for only SPT5 or MSL1 (Figure 3B control panel and Figure S5A). The presence of SPT5 only bands is not surprising since several genes on the X escape dosage compensation [7]. A possible explanation for the MSL1 only bands will be presented below.
Fig. 3. MSL complex binding to X-chromosome is not dependent on actively elongating RNA polymerase. 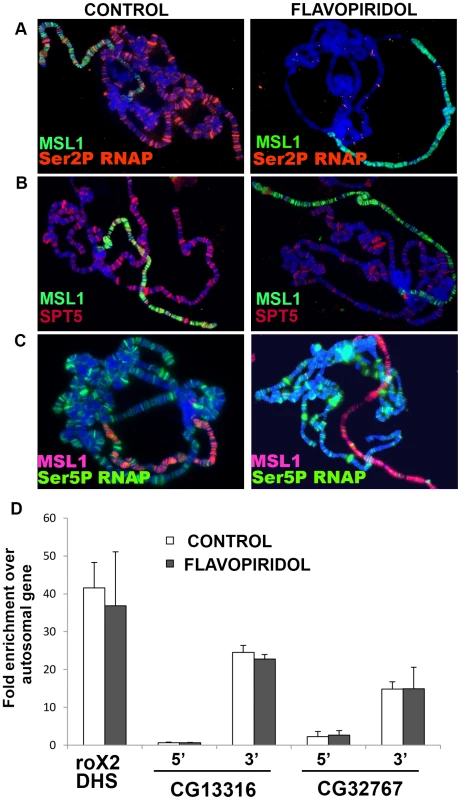
(A) Actively elongating RNA polymerase is found on all chromosomes and largely colocalizes with MSL complex on the male X. Flavopiridol treatment removes elongating polymerase but has no effect on MSL localization. (B) SPT5 decorates all chromosome arms and largely colocalizes with MSL complex on male X. The SPT5 pattern in B is shown more clearly in separate color channels in Figure S5. Flavopiridol treatment removes the bulk of SPT5 from all chromosomes. (C) Flavopiridol treatment has no effect on polymerase paused at the TSS. Chromosomes were stained with indicated antibodies against MSL1, Ser2 phopshorylated RNAP, Ser5 phosphorylated RNAP, and/or SPT5. (D) ChIP analysis of male S2 cells immunoprecipitated with anti-MSL1 antibodies. MSL1 is enriched near the 3′ ends of known MSL1 targets CG13316 and CG32767 with comparatively less MSL complex found near the 5′ TSS measured by quantitative real time PCR relative to the autosomal PKA gene. Blocking elongation with flavopiridol does not alter the MSL1 distribution. MSL1 binding to the roX2 DHS control region occurs by a sequence-dependent mechanism and does not require transcription [62]. MSL complex binding persists in the absence of elongation
In order to place SPT5 in the dosage compensation pathway, we focused on two of its most intensively studied roles. First, unphosphorylated SPT5 binds to and pauses RNA polymerase over the transcription start site (TSS). Release from this 5′ pause requires phosphorylation by P-TEFb at multiple sites at the C-termini of both RNA polymerase large subunit and SPT5 [25]–[27]. One way SPT5 might aid dosage compensation is if MSL complex stimulated release of the paused RNAPII/SPT5 complex at male X-linked genes. We refer to this as the Pause Release Model. Extra X-linked transcripts would result from clearing the 5′ end of genes freeing them for additional rounds of initiation. After phosphorylation by P-TEFb, SPT5 switches to a positive elongation factor that accompanies RNA polymerase down the gene. The MSL complex might instead enhance the processive action of SPT5 preventing pausing and/or premature termination as RNAPII moved across X-linked genes (Elongation Model).
The Pause Release model is less appealing because it calls for MSL action at the TSS, when MSL complex is instead predominantly located farther downstream [6]–[8]. One way to explain this discrepancy would be if the MSL complex only fleetingly interacts with P-TEFb or SPT5 at the 5′ end but then travels across the gene with the elongating RNAPII. If true, this model predicts that loss of SPT5 would lower MSL complex occupancy of the male X-chromosome because MSL complex could not enter the body of genes. Unfortunately, we cannot generate Spt5 null tissue. However, we can approximate that condition by using elongation inhibitors DRB and flavopiridol that block P-TEFb phosphorylation of RNAPII CTD Ser2 and SPT5 that are necessary for pause-release and entry into elongation [23], [28]. After exposure to these drugs, elongating RNAPII continues to the 3′ end of genes, but new RNAPII is trapped at the TSS, effectively stripping gene bodies of RNAPII and SPT5. Treating salivary glands with either inhibitor removed actively elongating RNAPII (Ser2P and Ser5P phosphorylated) from all chromosomes, but paused RNAPII that is Ser5P phosphorylated was unchanged consistent with previous reports [28] (Figure 3A and 3C). The banded SPT5 signal was strongly reduced on all chromosome arms after DRB or flavopiridol treatment (Figure 3B and data not shown). More importantly, the MSL complex staining pattern remained unchanged following inhibitor treatment (Figure 3 and data not shown). This shows that although MSL complex preferentially binds actively transcribed genes, binding persists for some time after the last polymerase has passed. This finding may explain the few loci bound by MSL complex but not SPT5 in untreated animals (Figure S5D and S5E). These may be dosage compensated genes whose developmentally controlled transcription ceased prior to fixation.
Modified histone H3K36me3 is found within active genes with a 3′ bias similar to the MSL complex. This modification may provide one component of MSL targeting specificity through the MSL3 chromodomain [29], but the issue is contentious [4]. We saw no difference in H3K36me3 staining between flavopiridol treated and mock treated tissue (Figure S6) consistent with earlier reports that these inhibitors only modestly lowered H3K36me3 [30].
To examine MSL binding at a higher resolution than is possible with polytenes, we turned to ChIP analysis of male S2 cells. We measured MSL1 binding to the 5′ and 3′ ends of two highly validated target genes after treatment with flavopiridol. If the scarcity of MSL complex at the 5′ ends of X-linked genes was caused by released RNAPII/SPT5 complex quickly carrying it into the body of genes, we might be able to trap MSL complex over the TSS by treatment with P-TEFb kinase inhibitors. The Pause Release Model predicts that flavopiridol treatment should cause the MSL signal to accumulate at the 5′ end of genes with a corresponding loss at the 3′ end. However, just as was seen with the polytene experiments, flavopiridol treatment did not alter MSL complex distribution as measured by ChIP (Figure 3D). These results argue against the Pause Release Model and instead favor the idea that MSL complex acts upon SPT5 during active elongation.
SPT5 interacts directly with MSL1 PEHE domain
We tested whether the genetic interactions observed between SPT5 and dosage compensation might arise from direct physical contacts. Early attempts to purify intact MSL complex did not recover SPT5 as a partner suggesting that if such interactions occur, they are transient [31], [32]. Dosage compensation in Drosophila is thought to have recruited an ancestral chromatin modifying complex found in most animals by evolving a new targeting strategy to the male X. If true, perhaps any SPT5-MSL interaction predates Drosophila dosage compensation and would be found in the most phylogenetically conserved regions of the complex. We tested the ancient PEHE domain of MSL1 that recruits MSL3 and MOF and forms a functionally critical core of the complex [13]. We asked whether purified subdomains of SPT5 (Figure 4A) could specifically pull down isolated MSL1 PEHE motif. We found specific binding between MSL1 PEHE and the N-terminal (N) and middle fragment (M) SPT5 fragments (Figure 4B). The N fragment contains the NusG-like domain that interacts with the RNAPII clamp domain to encircle the template DNA and makes RNAPII processive [33], [34] and one KOW motif (Figure 4A). The M segment contains additional KOW domains. KOW domains found in other proteins bind either protein or RNA partners [35]. We failed to detect any interactions with the C-terminal region that is phosphorylated at multiple sites by P-TEFb. Although this analysis does not exclude additional contacts between other MSL subunits or roX RNAs with regions of SPT5 in vivo, the data show that SPT5 and MSL complex have the ability to interact via MSL1 PEHE.
Fig. 4. MSL1 PEHE domain physically interacts with SPT5. 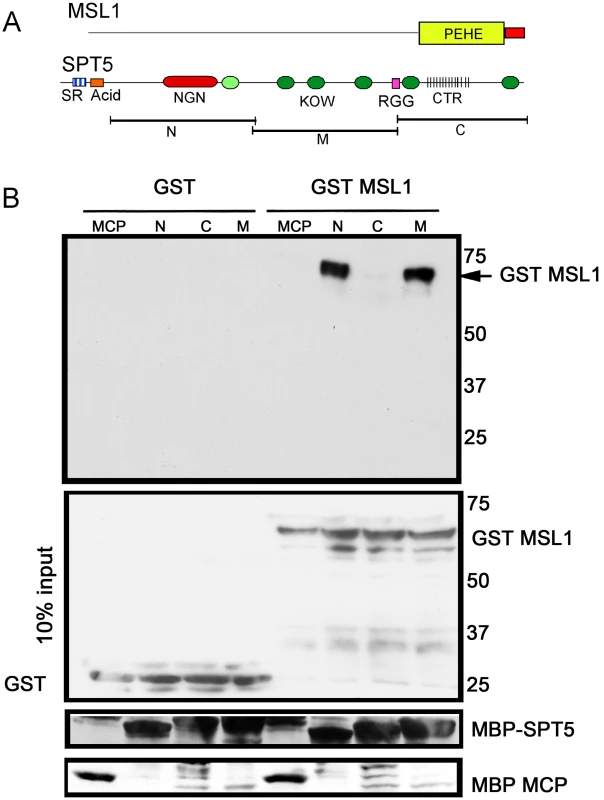
(A) MSL1 PEHE domain (aa 751–1039) was expressed as GST (Glutathione S-transferase) fusion protein. SPT5 fragments N, M, and C were expressed as MBP (Maltose binding Protein) fusion proteins [16]. (B) Purified SPT5-MBP fragments immobilized on amylose beads were allowed to interact with either GST-MSL1PEHE or GST. After washing, the recovered proteins were analyzed anti-GST Westerns. The membrane was stripped and reprobed to visualize the MBP fusion proteins. The middle input panel corresponds to 10% of the input GST proteins visualized with anti-GST antibodies. Other factors found in the screen
Although we focused our analysis on Spt5, we wondered if the other modifier mutations found in the screen might identify a new class of factors needed for dosage compensation. We were able to map a few modifiers to previously characterized genes. In the case of deficiencies, we tested whether point mutations of candidate genes could recapitulate the effect of deficiency. That approach showed that Chromator was the relevant gene that dominantly suppresses the MSL complex dependent reporter expression in Df(3L)BSC21. CHRO is a chromodomain protein that localizes specifically to the interband regions and is implicated in maintaining chromosome structure [36], [37]. Importantly, it copurifies with the MSL complex [31] underscoring the validity of our genetic approach to search for factors involved in dosage compensation. Additionally, we found that a complementation group from the EMS mutagenesis screen fell within the Df(2R)vg-C (Figure S1B). This interval contained three strong candidates, Spt4, iswi, and Sin3A. Complementation tests eliminated iswi. In eukaryotes, SPT5 usually acts in a complex with SPT4. No point mutations in Spt4 have been reported in flies, but the gene is not essential in yeast [38]. All the new EMS mutations instead failed to complement a known lesion in Sin3A [39], the first indication that the SINA/RPD3 histone deacetylase complex may play a role in dosage compensation. No deficiency removes Spt5 so this region was not covered in our deficiency screen.
Discussion
It is possible that dosage compensation in Drosophila is entirely a consequence of the known histone modifications carried out by its subunits, H4K16ac (MOF) and H2BK34ub (MSL2). However, if additional factors are required, new approaches may be needed to identify them. Biochemical purification is challenging due to the very large size of the MSL complex, the presence of the noncoding roX RNAs, and the fact that active MSL complex is tightly associated with transcribed chromatin. Extraction methods strong enough to release soluble MSL complex from chromatin may destroy critical contacts with key partners. Genetic approaches also face limitations. If an important partner performs additional functions beyond dosage compensation, mutations would likely be lethal to both sexes masking its interaction with the MSL complex.
We developed an unbiased forward genetic screen able to detect subtle changes in MSL activity that are not large enough to prevent dosage compensation of the male X, but sufficient to alter a sensitive eye pigmentation reporter. This screen implicated Spt5, a universally conserved transcription processivity factor for RNAPs, in the MSL pathway [27], [33], [40], [41]. The validity of our approach is illustrated by the identification of mutations in known components involved in the process such as msl1, mle and Chro [13]. The value of a genetic approach to detect protein interactions that may only be stable on actively transcribed chromatin is evident. The Drosophila protein interaction map (DPiM) identified dozens of proteins that are candidate interactors with SPT5 but surprisingly found no stable contacts with subunits of either RNA polymerase II or P-TEFb, the most highly validated partners known from other studies. This search also found no contacts with MSL subunit [42], [43]. The technical difficulty most likely rests with the problem of isolating an enormous complex of many megadaltons tightly tethered to DNA.
Multiple lines of evidence support a role for SPT5 in dosage compensation. The effect of Spt5 mutations on the white eye color reporter was entirely dependent upon the adjacent roX1 locus that can recruit soluble MSL complex to any location in the genome. Spt5 mutations had no effect on white or miniwhite gene expression when not linked to roX1. Spt5 mutations acted on all mosaic roX1 reporter transgenes regardless of the chromatin environment surrounding the inserts. Interactions of Spt5 mutants and gain of function msl1 alleles suggest that SPT5 acts between MSL complex and RNA polymerase. Mutations in Spt5 selectively reduced male viability under limiting roX RNA conditions in a manner comparable to the effect of mle mutations. Additionally mutations in Spt5 partially suppressed the toxic effects of ectopic dosage compensation in females. An independent screen of the Drosophila deficiency collection showed that the Spt5 phenotype is rare. Removing one allele of almost any transcription related factor had no effect on the eye pigmentation levels of mosaic roX1 reporters arguing that dosage compensation is particularly sensitive to SPT5 protein levels. Finally, we found that the most ancient and conserved segment of the MSL1 protein physically binds to two different regions of the SPT5 protein consistent with the largely overlapping patterns of chromatin occupancy across the body of X-linked genes.
In eukaryotes SPT5 along with SPT4 forms the DSIF (DRB: 5, 6-dichloro-1-β-D-ribofuranosylbenzimidazole Sensitivity Inducing Factor) [27]. The highly conserved NusG like domain (NGN) docks to RNAP through its interaction with the RNAP clamp domain and closes the cleft where the tightly bent melted DNA template resides preventing RNAP from falling off the template [33] (Figure 1G). The multiple KOW domains may contact either the emerging nascent transcript or other transcription factors. Although some studies indicated that SPT5 acts on a restricted set of genes [44], genome wide ChIP analysis showed that SPT5 and RNAPII colocalize throughout the genome [23], [28]. SPT5 arrests RNAPII near the transcription start site as the short nascent transcript emerges from the enzyme [25]–[27]. The highly regulated release from pause is controlled by the P-TEFb kinase phosphorylating multiple sites near the C-terminus of SPT5 and RNAPII CTD [25], [27], [45].
While regulated release from pause was originally described using the highly inducible hsp70 gene from Drosophila [46], it is now recognized as a widespread step in transcriptional regulation [23], [28], [47]. Although other factors, such as cMyc, stimulate transcription through pause-release of SPT5 [28], our results argue against a similar mechanism operating in Drosophila dosage compensation. MSL complex occupancy is lowest around the TSS and does not depend on continuous association with the elongating RNAPII/SPT5 to be enriched within the gene bodies. Instead we propose that the effect of SPT5 on dosage compensation is downstream of MSL complex recruitment (Figure 5).
Fig. 5. A model for dosage compensation. 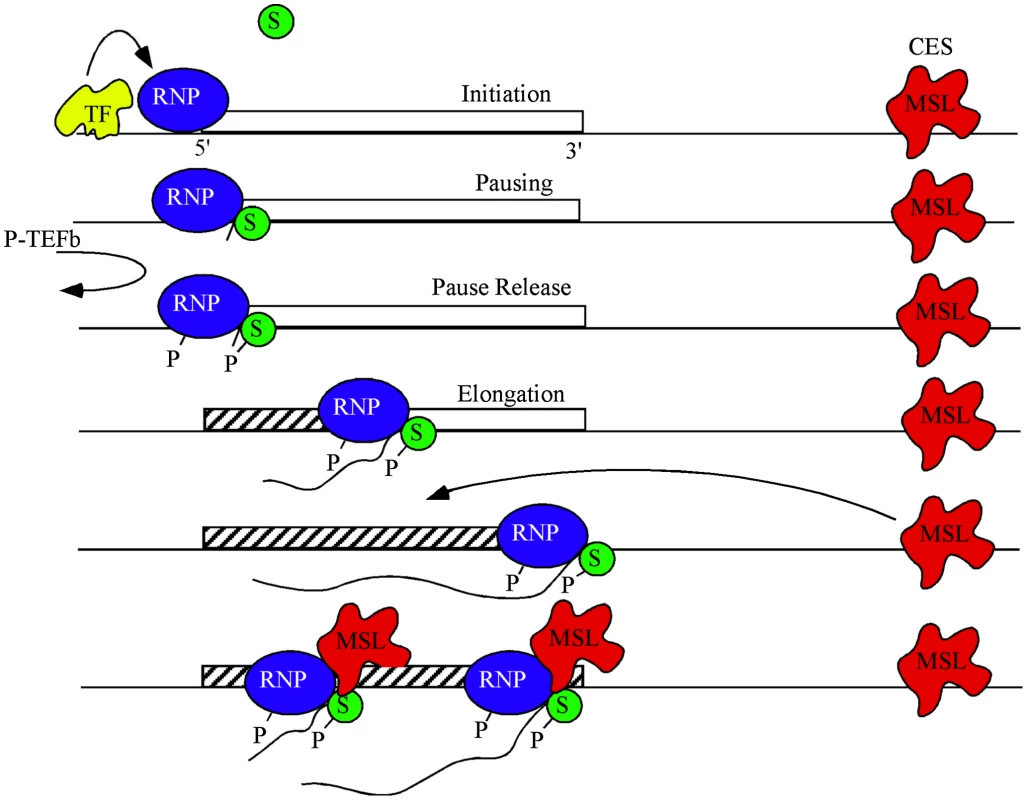
A highly simplified view separates transcription into phases of initiation controlled by gene-specific transcription factors (yellow), pausing of RNA polymerase II (blue) near the TSS by SPT4/5 (green), and release of pausing when P-TEFb phosphorylates both the CTD of RNAP and SPT5 leading to productive elongation. MSL complex (red) is attracted to the X chromosome by high affinity or chromatin entry sites (CES) scattered along the chromosome. The pioneer RNP may lay down new chromatin marks (hatch) characteristic of active genes. Some feature of active chromatin recruits MSL complex from local CES. During subsequent rounds of transcription MSL complex interacts with SPT5 to promote processivity. MSL complex mediated H4K16ac is enriched within the body of genes and drives decondensation of chromatin possibly facilitating easier passage of RNAPII [5], [48]. It is plausible that within this chromatin domain, SPT5 impacts dosage compensation via its known interactions with SPT6, which eases RNAPII passage by nucleosomal removal [16], [24], [40], [49], [50] and thereby improves the elongation rate of RNAPII. Alternatively, the interaction between MSL complex and SPT5 may increase elongation rates of dosage compensated genes on the X by enhancing RNAPII processivity [33]. We hypothesize that passage of a pioneer RNAPII generates certain transcription-specific epigenetic modifications such as H3K36me3 across a gene. These modifications recruit MSL complex from nearby X-linked sequence specific binding sites called Chromatin Entry Sites (CES) or High Affinity Sites (HAS) [1]. Once recruited, MSL complex is stably bound and stimulates elongation via its transient interaction with the transcribing SPT5/RNAPII (Figure 5). At least one point of this interaction is via the MSL1 PEHE domain and SPT5 NusG like and KOW domains.
While we have focused on the analysis of Spt5 in this report, our genetic approach also yielded additional candidates. So far, we have mapped two of these to Chro and Sin3A. CHRO, a chromodomain protein copurifies with the MSL complex [31]. Interestingly, CHRO recruits and localizes with JIL-1, a histone kinase that has also been implicated in dosage compensation [36], [37], [51], [52]. The CHRO/JIL-1 kinase complex is thought to maintain chromosomal integrity [36], [37]. It is conceivable that this complex plays a similar role in maintaining the specialized X-chromatin architecture in male flies. SIN3A, part of the SIN3A/RPD3 histone deacetylase complex is attracted by phosphorylated SPT5 and Ser2 phosphorylated CTD of RNAPII to deacetylate histones in the wake of transcribing RNAPII within the H3K36me3 chromatin domain [53], [54]. Therefore, phosphorylated SPT5, in addition to modulating processivity may also play a role in erasing transcription dependent acetylation via recruiting the SIN3A/RPD3 complex. This serves to suppress spurious transcription initiation from cryptic promoters within the coding region [53], [54]. Alternatively, the SIN3A/RPD3 complex may play a role in MSL complex recruitment to the GAGA element rich MSL recognition elements (MRE) sequences via its interaction with GAGA factor [55]. Further enquiry into the specific role played by these newly identified factors will result in an improved understanding of the mechanism of dosage compensation.
An independent RNAi screen using an MSL complex dependent luciferase expression as a reporter in S2 cells also identified a role for CHRO and SIN3A in dosage compensation [56]. Recovering overlapping cofactors from rather different genetic screens increases confidence that these strategies are identifying authentic components of the dosage compensation pathway. However, SPT5 was not found using the RNAi screen. This is not surprising since a general transcription factor such as SPT5 probably affects the expression of the normalizing control used in luciferase reporter assays. This again highlights the usefulness of an in vivo genetic strategy.
Our results provide direct in vivo support for the elongation model of dosage compensation by linking the SPT5 elongation factor to the MSL complex [5]. The finding that the Drosophila males have ∼1.4 fold more transcriptionally engaged RNAPII at the distal ends of X-linked genes as compared to autosomes also supports the idea of increased elongation [9]. Conversely, a recent report that compared global RNAPII occupancy in males and females found an increase in RNAPII levels across the entire body of the gene including the promoters on male X-linked genes [10]. This observation raises the possibility that dosage compensation may operate at the level of transcription initiation. A caveat of this study is that only a subset of X-linked genes (n = 242) had detectable RNAPII within the body of genes, possibly due to technical difficulties in the ability to detect elongating RNAPII. An alternate explanation for the results is that lowered RNAPII pausing [9], [10] and increased elongation improves RNAPII recycling from the 3′ to 5′ end of genes possibly via gene looping interactions and may be reflected in ChIP seq studies as an increase in RNAPII levels at the promoter. Moreover, Conrad et al postulate that H4K16ac at promoters is the key to dosage compensation. However, H4K16ac at promoters occurs both on male autosomes and all chromosomes in females and is not specific to the male X [48], [57]. On the other hand, H4K16ac within gene bodies is a unique feature of transcribed genes on the male X-chromosome and is therefore an attractive candidate to drive dosage compensation by improving RNAPII passage across the chromatin fibre during elongation.
Mammals also contain a version of the MSL complex composed of MSL1, 2, 3 and MOF, but apparently lacking a large noncoding RNA component and RNA helicase [58], [59]. Like flies, the human MSL complex, is bound within the bodies of genes with a distinct 3′ bias, acetylates histone H4K16 in the body of genes and increases transcription by approximately two fold [58]–[60]. Our results linking the most conserved domain of MSL1 with the conserved transcription elongation factor SPT5 in flies indicate that mammalian MSL complex is likely to also act upon transcription elongation.
Materials and Methods
Fly stocks
Mutagenesis was performed as described [13]. Detailed mutagenesis scheme and protocol is included in Text S1. For the deficiency screen the transgenic lines [w+ GMroX1-58D], [w+ GMroX1-60F], [w+ GMroX1-69C], [w+ GMroX1-75C], [w+ GMroX1-99F] and [w+ GMroX1-102C] were used. The full genotype of ΔroX1,roX2 stock is y w roX1ex6 Df(1)roX252 [w+ cos4Δ4.3] [19].
Antibody generation and affinity purification
Plasmids for bacterial expression of MBP fusion SPT5 protein fragments, SPT5-N (aa 112–393), M (aa 389–733) and C (aa 732–1054) were a kind gift from Dr. John Lis [16]. Antibodies were raised by Cocalico Biologicals, Pennsylvania.
Polytene squashes
Polytene squashes were prepared as described in [61]. Primary antibodies were rabbit anti-MSL1 antibodies (1∶50), guinea pig anti-SPT5 antibodies (1∶100), mouse H5 monoclonal anti-Ser2P RNAP (Covance, 1∶30), mouse H14 monoclonal anti-Ser5P RNAP (Covance, 1∶50) and rabbit anti-H3K36me3 (Invitrogen, 1∶50). Appropriate secondary antibodies were used in combinations that allowed for dual protein localization.
Chromatin immunoprecipitation
Detailed protocol can be found in Text S1. For inhibitor treatment 500 nM flavopiridol or 100 µM DRB was used. Briefly S2 cells were crosslinked with 1% formaldehyde and nuclei extracted in 15 mM HEPES, 5 mM MgCl2, 0.2 mM EDTA, 0.5 mM EGTA, 10 mM KCl, 350 mM Sucrose, 0.1% Tween 20, 0.5 mM PMSF, 1 mM DTT. Chromatin was sheared to 300–700 bp fragments and pulled down with anti MSL1 antibodies (Gift from M.Kuroda). After several washes eluted chromatin was used. For real time PCR (ABI 7900 qPCR model), SYBR green master mix (ABI), 1 µM primers and 1 µl of input and ChIP DNA was used. Primer sequences are provided in Text S1.
Protein interaction
MBP-SPT5N, MBP-SPT5M, MBP-SPT5C, the unrelated protein MBP-MCP (MS2 phage coat protein), GST-MSL1 C-terminal domain fusion protein and GST were expressed and isolated from bacteria. Equivalent molar concentrations of MBP proteins bound to amylose beads were incubated with GST proteins. After three washes with 150 mM NaCl, 0.1% NP40 and 20 mM Tris for 10 mins at 4°C, proteins were eluted by boiling in SDS-loading buffer and separated on 8% SDS-polyacrylamide gels. Westerns were performed as described [13]. We used anti-GST antibodies (Sigma) to detect GST and affinity purified guinea pig anti-SPT5 sera to detect MBP-bound SPT5. The proteins were visualized by using appropriate HRP conjugated secondary antibodies (Jackson Immuno) and lunimol reagent (Santa Cruz).
Supporting Information
Zdroje
1. ConradT, AkhtarA (2012) Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet 13 : 123–134 doi:10.1038/nrg3124.
2. GelbartM, KurodaM (2009) Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136 : 1399–1410.
3. WuL, ZeeB, WangY, GarciaB, DouY (2011) The RING Finger Protein MSL2 in the MOF Complex Is an E3 Ubiquitin Ligase for H2B K34 and Is Involved in Crosstalk with H3 K4 and K79 Methylation. Mol Cell 43 : 132–144 doi:10.1016/j.molcel.2011.05.015.
4. KimD, BlusB, ChandraV, HuangP, RastinejadF, KhorasanizadehS (2010) Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat Struct Mol Biol 17 : 1027–1029.
5. SmithE, AllisC, LucchesiJ (2001) Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem 276 : 31483–31486 doi:10.1074/jbc.C100351200.
6. LegubeG, McWeeneyS, LercherM, AkhtarA (2006) X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev 20 : 871–883 doi:10.1101/gad.377506.
7. AlekseyenkoAA, LarschanE, LaiW, ParkP, KurodaM (2006) High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20 : 848–857 doi:10.1101/gad.1400206.
8. GilfillanGD, StraubT, de WitE, GreilF, LammR, et al. (2006) Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 20 : 858–870 doi:10.1101/gad.1399406.
9. LarschanE, BishopE, KharchenkoP, CoreL, LisJ, et al. (2011) X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471 : 115–118 doi:10.1038/nature09757.
10. ConradT, CavalliF, VaquerizasJ, LuscombeN, AkhtarA (2012) Drosophila dosage compensation involves enhanced Pol II recruitment to male X-linked promoters. Science 337 : 742–746.
11. Shogren-KnaakM, IshiiH, SunJ-M, PazinMJ, DavieJR, et al. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311 : 844–847 doi:10.1126/science.1124000.
12. KelleyR, KurodaM (2003) The Drosophila roX1 RNA gene can overcome silent chromatin by recruiting the male-specific lethal dosage compensation complex. Genetics 164 : 565–574.
13. PrabhakaranM, KelleyR (2010) A new strategy for isolating genes controlling dosage compensation in Drosophila using a simple epigenetic mosaic eye phenotype. BMC Biol 8 : 80 doi:10.1186/1741-7007-8-80.
14. ZhaiR, HiesingerP, KohT, VerstrekenP, SchulzeK, et al. (2003) Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci U S A 100 : 10860–10865.
15. MahoneyM, ParksA, RuddyD, TiongS, EsengilH, et al. (2006) Presenilin-based genetic screens in Drosophila melanogaster identify novel Notch pathway modifiers. Genetics 172 : 2309–2324.
16. AndrulisED, GuzmaE, DoP, WernerJ, LisJT (2000) High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev 2635–2649 doi:10.1101/gad.844200.Krumm.
17. O'HareK, AlleyMRK, CullingfordTE, DriverA, SandersonMJ (1991) DNA sequence of the Doc retroposon in the white-one mutant of Drosophila melanogaster and of secondary insertions in the phenotypically altered derivatives white-honey and white-eosin. Mol Gen Genet 225 : 17–24.
18. LevisR, BinghamP, RubinG (1982) Physical map of the white locus of Drosophila melanogaster. Proc Natl Acad Sci U S A 79 : 564–568.
19. MellerVH, RattnerBP (2002) The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J 21 : 1084–1091 doi:10.1093/emboj/21.5.1084.
20. KelleyR, LeeO-K, ShimY-K (2008) Transcription rate of noncoding roX1 RNA controls local spreading of the Drosophila MSL chromatin remodeling complex. Mech Dev 125 : 1009–1019 doi:10.1016/j.mod.2008.08.003.
21. SmithE, WinterB, EissenbergJ, ShilatifardA (2008) Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci U S A 105 : 8575–8579.
22. KelleyRL, SolovyevaI, LymanLM, RichmanR, SolovyevV, et al. (1995) Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81 : 867–877.
23. GilchristDA, Dos SantosG, FargoDC, XieB, GaoY, et al. (2010) Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143 : 540–551 doi:10.1016/j.cell.2010.10.004.
24. KaplanCD, MorrisJR, WuC, WinstonF (2000) Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev 14 : 2623–2634 doi:10.1101/gad.831900.tion.
25. WadaT, TakagiT, YamaguchiY, WatanabeD, HandaH (1998) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J 15 : 7395–7403.
26. MissraA, GilmourDS (2010) Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A 107 : 11301–11306 doi:10.1073/pnas.1000681107.
27. WadaT, TakagiT, YamaguchiY, FerdousA, ImaiT, et al. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12 : 343–356.
28. RahlP, LinC, SeilaA, FlynnR, McCuineS, et al. (2010) c-Myc Regulates Transcriptional Pause Release. Cell 141 : 432–445.
29. LarschanE, AlekseyenkoA, GortchakovA, PengS, LiB, et al. (2007) MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28 : 121–133 doi:10.1016/j.molcel.2007.08.011.
30. VakocC, SachdevaM, WangH, BlobelG (2006) Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26 : 9185–9195.
31. MendjanS, TaipaleM, KindJ, HolzH, GebhardtP, et al. (2006) Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21 : 811–823 doi:10.1016/j.molcel.2006.02.007.
32. PrestelM, FellerC, StraubTHM, BeckerP (2010) The Activation Potential of MOF Is Constrained for Dosage Compensation. Mol Cell 38 : 815–826.
33. Martinez-RucoboFW, SainsburyS, CheungACM, CramerP (2011) Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J 30 : 1302–1310 doi:10.1038/emboj.2011.64.
34. HirtreiterA, DamsmaGE, CheungACM, KloseD, GrohmannD, et al. (2010) Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res 38 : 4040–4051 doi:10.1093/nar/gkq135.
35. SteinerT, KaiserJ, MarinkoviçS, HuberR, WahlM (2002) Crystal structures of transcription factor NusG in light of its nucleic acid - and protein-binding activities. EMBO J 21 : 4641–4653.
36. GanM, MoebusS, EggertH, SaumweberH (2011) The Chriz–Z4 complex recruits JIL-1 to polytene chromosomes,a requirement for interband-specific phosphorylation of H3S10. J Biosci 36 : 425–438 doi:10.1007/s12038-011-9089-y.
37. RathU, DingY, DengH, QiH, BaoX, et al. (2006) The chromodomain protein, Chromator, interacts with JIL-1 kinase and regulates the structure of Drosophila polytene chromosomes. J Cell Sci 119 : 2332–2341 doi:10.1242/jcs.02960.
38. MaloneE, FasslerJ, WinstonF (1993) Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol Gen Genet 237 : 449–459.
39. SpradlingA, SternaD, BeatonbA, RehmbJ, LavertyT, et al. (1999) The Berkeley Drosophila Genome Project Gene Disruption Project: Single P-Element Insertions Mutating 25% of Vital Drosophila Genes. Genetics 153 : 135–177.
40. HartzogG, WadaT, HandaH, WinstonF (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 12 : 357–369.
41. KimD, InukaiN, YamadaT, FuruyaA, SatoH, et al. (2003) Structure-function analysis of human Spt4: evidence that hSpt4 and hSpt5 exert their roles in transcriptional elongation as parts of the DSIF complex. Genes Cells 8 : 371–378.
42. flybase (n.d.).
43. GuruharshaK, RualJ, ZhaiB, MintserisJ, VaidyaP, et al. (2011) A protein complex network of Drosophila melanogaster. Cell 147 : 690–703.
44. KomoriT, InukaiN, YamadaT, YamaguchiY, HandaH (2009) Role of human transcription elongation factor DSIF in the suppression of senescence and apoptosis. Genes Cells 14 : 343–354.
45. IvanovD, KwakYT, GuoJ, GaynorRB (2000) Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol 20 : 2970–2983.
46. O'BrienT, LisJ (1991) RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol 11 : 5285–5290.
47. NechaevS, FargoD, dos SantosG, LiuL, GaoY, et al. (2010) Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327 : 335–338.
48. GelbartME, LarschanE, PengS, ParkPJ, KurodaMI (2009) Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol 16 : 825–832 doi:10.1038/nsmb.1644.
49. ArdehaliMB, YaoJ, AdelmanK, FudaNJ, PeteschSJ, et al. (2009) Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J 28 : 1067–1077 doi:10.1038/emboj.2009.56.
50. BortvinA, WinstonF (1996) Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272 : 1473–1476.
51. JinY, WangY, JohansenJ, JohansenKM (2000) JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J Cell Biol 149 : 1005–1010.
52. RegnardC, StraubT, MitterwegerA, DahlsveenI, FabianV, et al. (2011) Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet 7: e1001327 doi:10.1371/journal.pgen.1001327.
53. DrouinS, LaraméeL, JacquesP-É, ForestA, BergeronM, et al. (2010) DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet 6: e1001173 doi:10.1371/journal.pgen.1001173.
54. CarrozzaMJ, LiB, FlorensL, SuganumaT, SwansonSK, et al. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 : 581–592 doi:10.1016/j.cell.2005.10.023.
55. EspinásM, CanudasS, FantiL, PimpinelliS, CasanovaJ, et al. (2000) The GAGA factor of Drosophila interacts with SAP18, a Sin3-associated polypeptide. EMBO Rep 1 : 253–259.
56. LarschanE, SorucoM, LeeO, PengS, BishopE, et al. (2012) Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Drosophila Dosage Compensation. PLoS Genet 8: e1002830 doi:10.1371/journal.pgen.1002830.
57. KindJ, VaquerizasJM, GebhardtP, GentzelM, LuscombeNM, et al. (2008) Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 133 : 813–828 doi:10.1016/j.cell.2008.04.036.
58. TaipaleM, ReaS, RichterK, VilarA, LichterP, et al. (2005) hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 25 : 6798–6810 doi:10.1128/MCB.25.15.6798.
59. SmithE, CayrouC, HuangR, LaneW, CoJ, et al. (2005) A Human Protein Complex Homologous to the Drosophila MSL Complex Is Responsible for the Majority of Histone H4 Acetylation at Lysine 16. Mol Cell Biol 25 : 9175–9188 doi:10.1128/MCB.25.21.9175.
60. LiX, WuL, CorsaC, KunkelS, DouY (2009) Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell 36 : 290–301.
61. KelleyRL, MellerVH, GordadzePR, RomanG, DavisRL, et al. (1999) Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98 : 513–522.
62. BaiX, AlekseyenkoAa, KurodaMI (2004) Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J 23 : 2853–2861 doi:10.1038/sj.emboj.7600299.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Intrauterinní inseminace a její úspěšnost
- Příjem alkoholu a menstruační cyklus
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
- Růst a vývoj dětí narozených pomocí IVF
- Akutní intermitentní porfyrie
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



