-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
Escherichia coli pol V (UmuD′2C), the main translesion DNA polymerase, ensures continued nascent strand extension when the cellular replicase is blocked by unrepaired DNA lesions. Pol V is characterized by low sugar selectivity, which can be further reduced by a Y11A “steric-gate” substitution in UmuC that enables pol V to preferentially incorporate rNTPs over dNTPs in vitro. Despite efficient error-prone translesion synthesis catalyzed by UmuC_Y11A in vitro, strains expressing umuC_Y11A exhibit low UV mutability and UV resistance. Here, we show that these phenotypes result from the concomitant dual actions of Ribonuclease HII (RNase HII) initiating removal of rNMPs from the nascent DNA strand and nucleotide excision repair (NER) removing UV lesions from the parental strand. In the absence of either repair pathway, UV resistance and mutagenesis conferred by umuC_Y11A is significantly enhanced, suggesting that the combined actions of RNase HII and NER lead to double-strand breaks that result in reduced cell viability. We present evidence that the Y11A-specific UV phenotype is tempered by pol IV in vivo. At physiological ratios of the two polymerases, pol IV inhibits pol V–catalyzed translesion synthesis (TLS) past UV lesions and significantly reduces the number of Y11A-incorporated rNTPs by limiting the length of the pol V–dependent TLS tract generated during lesion bypass in vitro. In a recA730 lexA(Def) ΔumuDC ΔdinB strain, plasmid-encoded wild-type pol V promotes high levels of spontaneous mutagenesis. However, umuC_Y11A-dependent spontaneous mutagenesis is only ∼7% of that observed with wild-type pol V, but increases to ∼39% of wild-type levels in an isogenic ΔrnhB strain and ∼72% of wild-type levels in a ΔrnhA ΔrnhB double mutant. Our observations suggest that errant ribonucleotides incorporated by pol V can be tolerated in the E. coli genome, but at the cost of higher levels of cellular mutagenesis.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003030
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003030Summary
Escherichia coli pol V (UmuD′2C), the main translesion DNA polymerase, ensures continued nascent strand extension when the cellular replicase is blocked by unrepaired DNA lesions. Pol V is characterized by low sugar selectivity, which can be further reduced by a Y11A “steric-gate” substitution in UmuC that enables pol V to preferentially incorporate rNTPs over dNTPs in vitro. Despite efficient error-prone translesion synthesis catalyzed by UmuC_Y11A in vitro, strains expressing umuC_Y11A exhibit low UV mutability and UV resistance. Here, we show that these phenotypes result from the concomitant dual actions of Ribonuclease HII (RNase HII) initiating removal of rNMPs from the nascent DNA strand and nucleotide excision repair (NER) removing UV lesions from the parental strand. In the absence of either repair pathway, UV resistance and mutagenesis conferred by umuC_Y11A is significantly enhanced, suggesting that the combined actions of RNase HII and NER lead to double-strand breaks that result in reduced cell viability. We present evidence that the Y11A-specific UV phenotype is tempered by pol IV in vivo. At physiological ratios of the two polymerases, pol IV inhibits pol V–catalyzed translesion synthesis (TLS) past UV lesions and significantly reduces the number of Y11A-incorporated rNTPs by limiting the length of the pol V–dependent TLS tract generated during lesion bypass in vitro. In a recA730 lexA(Def) ΔumuDC ΔdinB strain, plasmid-encoded wild-type pol V promotes high levels of spontaneous mutagenesis. However, umuC_Y11A-dependent spontaneous mutagenesis is only ∼7% of that observed with wild-type pol V, but increases to ∼39% of wild-type levels in an isogenic ΔrnhB strain and ∼72% of wild-type levels in a ΔrnhA ΔrnhB double mutant. Our observations suggest that errant ribonucleotides incorporated by pol V can be tolerated in the E. coli genome, but at the cost of higher levels of cellular mutagenesis.
Introduction
Translesion synthesis (TLS) allows living organisms to tolerate DNA damage to their genome. The vast majority of TLS in Escherichia coli is catalyzed by the LexA-regulated damage-inducible polymerases II, IV and V, which alone, or in various combinations, are recruited to the sites of DNA damage [1]. The B-family pol II which is encoded by the polB gene, is a rare case of a specialized TLS polymerase possessing 3′-5′ exonuclease activity [2]. As a result, pol II-dependent replication of both undamaged and damaged DNA is quite accurate with the exception of an N2-acetylaminofluorene adducts, where it promotes −2 frameshifts [3]. Y-family polymerases, pol IV, encoded by the dinB gene [4], [5], and pol V, the product of the umuC and umuD genes [6], are devoid of exonucleolytic proofreading and are characterized by low-fidelity DNA synthesis on undamaged DNA [7], [8]. Nevertheless, pol IV is remarkably accurate when replicating past certain DNA lesions, such as N2-dG adducts [9]. While pol II and pol IV each appear to facilitate TLS of a narrow range of damaged substrates, pol V is able to accommodate a diverse spectrum of DNA lesions in its active site and bears the greatest burden of TLS in E. coli [1], [6], [10]. Pol V-dependent TLS is highly error-prone causing the majority of cellular mutagenesis after DNA damage [6], [11].
Pol V, a heterotrimeric UmuD′2C complex [12], requiring the presence of a RecA nucleoprotein filament (RecA*) for optimal activity [13]–[17], has intrinsically low base substitution fidelity [18], [19]. We have recently discovered that this polymerase is also characterized by substantially reduced sugar selectivity [20]. When the canonical Watson-Crick base pairing is preserved, purified pol V accompanied by accessory proteins readily incorporates all ribonucleotides (ribonucleoside monophosphates, rNMPs) except uracil and catalyzes efficient and highly processive RNA synthesis in vitro in the presence of all four rNTPs. The ability of pol V to incorporate ribonucleotides is dramatically enhanced by a Y11A substitution at the conserved steric gate residue of UmuC, and greatly reduced by an F10L substitution [20]. In contrast, a Y11F substitution affects sugar selectivity minimally [20]. All three alleles also have different effects on base substitution fidelity and TLS activity of the mutant polymerases. Because the Y11F mutant readily accommodates G∶T mispairs in the active site, it induces higher levels of mutagenesis than wild-type pol V [20], but the ability of the wild-type polymerase and Y11F mutant to replicate damaged DNA is similar. The F10L_UmuC variant is characterized by a significant increase in the accuracy of nucleotide incorporation and moderate decrease in TLS activity. Consistent with this observation, cells expressing the F10L mutant exhibit low levels of UV-induced mutagenesis [21]. In contrast, the in vivo phenotype of strains expressing pol V with the umuC_Y11A substitution contradicts its in vitro biochemical properties. UmuC_Y11A is highly inaccurate in vitro, yet exhibits low mutability in vivo [20], [21]. Furthermore, despite the observation that the UmuC_Y11A variant catalyzes TLS past a T-T cyclobutane pyrimidine dimer (CPD) in vitro at least as efficiently as the wild-type enzyme, it confers minimal UV-resistance to a ΔumuDC strain [21]. To explain these phenotypes, we suggest that the dramatic increase in rNMP incorporation promoted by UmuC_Y11A leads to the induction of downstream pathways involving rNMP processing. Presumably the rNMP-targeted repair pathways would not only reduce umuC_Y11A-dependent spontaneous and UV-induced mutagenesis, but also interfere with completion of TLS resulting in the observed decrease in UV resistance.
Recent studies have demonstrated that similar to pol V, various DNA polymerases are able to incorporate ribonucleotides into DNA although in most cases less efficiently (reviewed in [22]). Even replicative polymerases with much more rigorous steric exclusion mechanisms insert rNMPs in much higher amounts than it was previously assumed [23]. In addition to misinsertion during replication or repair, stable incorporation of rNMPs in the DNA backbone could result from the incomplete removal of RNA primers used during maturation of lagging-strand Okazaki fragments. Due to the presence of a reactive 2′ hydroxyl on the ribose ring, rNMPs embedded in genomic DNA could sensitize the DNA strand to spontaneous and enzymatic hydrolytic cleavage. They can also cause distortion to the structure of the double helix that disrupts the ability of DNA-binding proteins to recognize DNA, thereby interfering with subsequent replication and transcription processes. Therefore, efficient repair of RNA/DNA mismatches is a critical process for a living cell, so as to ensure maintenance of genome integrity and, ultimately, its viability. As a result, cells have evolved various pathways for recognizing and removing aberrant rNTP incorporated into DNA strands [24]–[27].
The major enzymes initiating this pathway are ribonucleotide-specific endonucleases, Ribonucleases H (RNases H), which are present in organisms across all domains and are classified as types 1 and 2 based on sequence conservation and substrate preference [28]. Ribonucleases of both types are structurally related and have a similar mechanism of hydrolysis. However, while RNase HI cleaves the RNA moiety in the RNA/DNA hybrids with more than four sequential rNTPs embedded in a dsDNA strand, RNase HII enzymes can hydrolyze all kinds of hybrids, but prefer those which have a single rNTP embedded in DNA, rather than RNA/DNA duplexes with a stretch of riboses [29]. Although it is well established that RNase HI and HII are important for the release of rNMPs from a DNA duplex, the precise pathway initiated by these enzymes remains elusive. Based on in vitro studies, a general model describing the sequence of events that leads to the replacement of the ribose with deoxyribose has been developed for eukaryotic system. According to this model, after the phosphodiester bond of the nucleotide 5′ to the RNA-DNA junction is nicked by RNase H, an enzyme with 5′ to 3′ exonuclease activity makes a single cut 3′ to the rNTP, thus releasing the monoribonucleotide. After dissociation of the cleaved RNA, DNA polymerase fills the resulting gap and DNA ligase seals the nick [30], [31]. Previous studies suggest that the 3′ cut is made by FEN-1-like proteins [32] and that RNase HII nicking activity is promoted by binding to PCNA [33]. The importance of this pathway in repair of rNMPs incorporated by a DNA polymerase during replication has been emphasized by the observation that the lack of RNase HII in yeast strains expressing a mutant pol ε with relaxed sugar selectivity, leads to replicative stress and genome instability [34]. The main hallmark of this instability, deletion of 2–5 base pairs in short repetitive sequences, was also demonstrated in strains encoding wild-type pol ε and shown to require the endoribonuclease activity of Top1, a topoisomerase that relaxes supercoils by reversibly nicking duplex DNA [27]. The 2′-3′-cyclic phosphates formed after Top1-catalyzed cleavage between ribo - and deoxynucleotides, prohibit religation resulting in the generation of stable ssDNA breaks at the sites of incorporated rNMPs. Removal of all rNMPs is not a standard function of Top1, and it targets only some of the ribonucleotides in DNA/RNA hybrid when RNase H2 is defective [27].
Originally, it was hypothesized that the formation of deletions in cellular DNA in the absence of RNase HII occurs through a misalignment mechanism and involves mismatch repair (MMR) proteins, but it was subsequently shown to be independent of the status of the MMR machinery [35]. More recently it was revealed that the MMR system in both prokaryotes and eukaryotes competes with RNase H mechanisms to remove misincorporated ribonucleotides and restore DNA integrity when isolated rNMPs in chromosomal DNA also distort the Watson-Crick base-pairing [27].
The pathway of rNMP repair in prokaryotes is much less understood. Despite having multiple cellular functions, RNases HI and HII, encoded by the rnhA and rnhB genes respectively [29], are not essential for viability of bacteria, since the double mutants are viable, albeit temperature sensitive [36]. With respect to removal of rNMPs embedded in the genomic DNA, this means that other mechanisms can substitute for RNase H, or perhaps that prokaryotes can better tolerate DNA/RNA hybrid structures.
Taking advantage of the different capacities for ribonucleotide incorporation by pol V variants with substitutions at, or adjacent to the steric gate, we examined rNMP-processing pathways that cause phenotypic changes in strains expressing the pol V variants. We discovered that mutations in rnhB (encoding RNase HII), NER genes (uvrA and uvrC), and unexpectedly in dinB (encoding pol IV), play pivotal roles in modulating the extent of umuC_Y11A-dependent UV survival and mutagenesis. In addition, we show that in recA730 lexA(Def) ΔdinB strains lacking rnhB, rnhA helps to limit the extent of umuC_Y11A-dependent spontaneous mutagenesis imposed on the undamaged E.coli chromosome.
Results
Pol IV inhibits pol V–dependent incorporation of ribonucleotides during TLS in vivo
We have previously shown that the UV-resistance of recA730 lexA(Def) ΔumuDC cells expressing plasmid encoded umuC_Y11A is similar to that promoted by the vector plasmid alone [21]. This phenotype cannot be simply attributed to an inability to traverse the major UV-induced lesion because the highly purified pol V-Y11A enzyme bypassed the CPD efficiently in vitro [21]. Since UmuC_Y11A is characterized by low sugar discrimination fidelity [20], it seems plausible that the poor UV-survival of strains expressing the Y11A variant might be explained by large numbers of ribonucleotides incorporated during TLS that trigger repair pathways directed at rNMP removal. These pathways would excise pol V-dependent TLS tracts mimicking a pol V-deficient phenotype. To test this hypothesis, we measured the UV-sensitivity of strains expressing wild-type pol V and variants, but lacking an enzyme implicated in the repair of ribonucleotide-containing DNA. Based upon previous studies, the most logical choice was to assay UV-survival in strains lacking the RNase H proteins; RNase HI, the product of the rnhA gene [37] with the capacity to release RNA from RNA/DNA hybrids with multiple sequential rNMPs, and RNase HII encoded by the rnhB gene [38], which differs from the RNase HI by recognizing and cleaving a single ribonucleotide embedded in a DNA duplex. We therefore compared cell survival promoted by wild-type and mutant pol V variants in isogenic recA730 lexA(Def) ΔumuDC strains with ΔrnhA or ΔrnhB alleles alone, or in combination, after exposure to UV-light in a semi-quantitative “spot” assay. The plasmid-encoded pol V variants provide an excellent internal control for any effect of the RNase H proteins on cell survival, since the F10L mutant is essentially unable to incorporate ribonucleotides and should not exhibit any difference in the rnh+/ − strains, whereas Y11A efficiently incorporates ribonucleotides and any rNMP-mediated repair would be most evident by comparison of the rnh+/ − strains expressing this variant.
The first thing to note is that the ΔrnhA strain is more sensitive to UV-light than the isogenic rnh+ or ΔrnhB strains (Figure S1), In this strain background, chromosomal duplication is dysregulated and unlike normal genome duplication which is initiated at oriC, is likely to be initiated at D-loops formed at oriMs and R-loops formed at multiple oriKs [39]. Presumably the added load of DNA damage to cells with highly irregular modes of replication contributes to the observed increased UV-sensitivity. In all strains analyzed, wild-type pol V conferred considerable UV-resistance (Figure S1).
As previously reported [21], in a recA730 lexA(Def) ΔumuDC rnh+ background, umuC_Y11A confers minimal UV-resistance compared to either pGB2 vector, wild-type UmuC, Y11F, or F10L mutants (Figure S1A). In the ΔrnhA background, UV-survival of the Y11A variant was comparable to umuC_F10L and to the vector containing strain (Figure S1B). In contrast, while Y11A exhibited roughly the same overall UV-resistance as F10L in the ΔrnhB strain, it was considerably more UV-resistant than the vector containing strain (Figure S1C)
We were concerned, however, that these phenotypes were much less pronounced than we had anticipated, especially given the properties of the Y11A mutant in vitro [20], [21]. The strains employed here carry the lexA51(Def) allele which has a frameshift mutation in the C-terminus of LexA [40], that leads to constitutive expression of all LexA-regulated genes [41], including all three TLS polymerases, pol II, pol IV and pol V. Under these conditions, pol IV is the most abundant DNA polymerase in E.coli with an intracellular concentration of roughly 2500 molecules per cell [42]. Even though pol IV has not previously been implicated in the TLS of UV-induced lesions, we considered the possibility that the highly abundant enzyme could nevertheless compete with pol V to limit its access to stalled replication forks. To test this hypothesis, we generated isogenic recA730 lexA(Def) ΔumuDC ΔdinB ΔrnhA, ΔrnhB or ΔrnhA-ΔrnhB mutant strains and re-analyzed the effects of defects in RNase H on UV-survival of cells expressing wild-type pol V and its variants (Figure 1). As observed previously (Figure S1), the ΔrnhA allele rendered the strains more UV-sensitive than the isogenic rnh+ or ΔrnhB strains (Figure 1B and 1D, where cells were exposed to 20 J/m2 UV light compared to 40 J/m2 in Figure 1A and 1C). However, the ΔrnhA allele had no effect on the relative extent of UV-survival provided by the four pol V-expressing plasmids. In particular, the umuC_Y11A expressing plasmid conferred the least UV-resistance that was only marginally greater than the pGB2 vector containing strain.
Fig. 1. Effect of ΔrnhA and ΔrnhB on UV survival of recA730 lexA(Def) ΔumuDC ΔdinB strains expressing pol V variants. 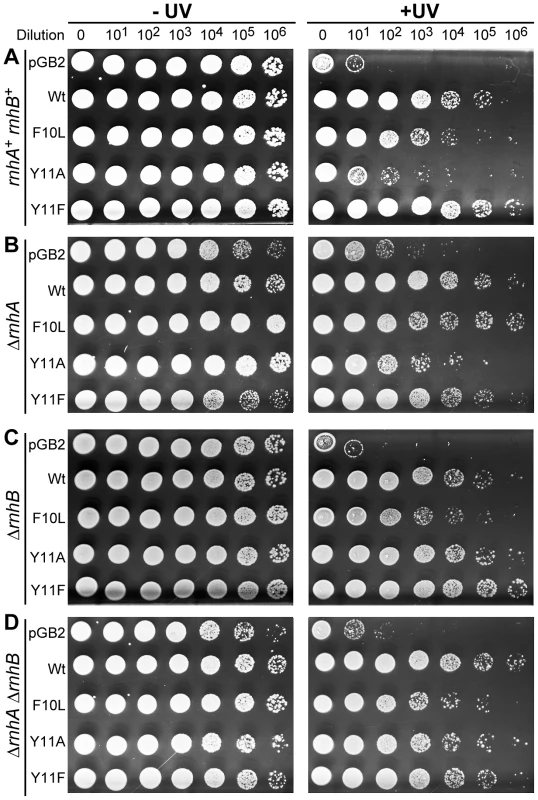
10 µl of 10-fold serial dilutions of overnight cultures were spotted onto the surface of rectangular LB agar plates and exposed to 40 J/m2 254 nM UV-light (panels A and C) and 20 J/m2 254 nM UV-light (panels B and D). Both unirradiated (−) and UV-irradiated (+) plates were incubated overnight at 37°C. In each panel, UV survival is shown for the recA730 lexA(Def) ΔumuDC ΔdinB strains either harboring pGB2 vector, or expressing pol V variants. The main observation of these experiments is that the UV-resistance of cells expressing umuC_Y11A increase dramatically in strains lacking rnhB, whereas survival of cells equipped with wild-type pol V, umuC_F10L, or umuC_Y11F is largely unaffected by the status of rnhB. A very different phenotype was observed in the ΔrnhB strain, where umuC_Y11A-dependent UV-survival was greatly enhanced (compare pGB2 and Y11A in Figure 1C). A similar enhancement of Y11A-dependent UV-survival was also observed in the more UV-sensitive ΔrnhA ΔrnhB double mutant strain (Figure 1D).
Overall, our data are consistent with the possibility that RNase H II activity actually promotes rNMP-dependent UV-induced cell killing. The fact that the increase in UV-resistance of ΔrnhB Y11A-expressing cells is much more dramatic in a ΔdinB background compared to the isogenic ΔrnhB dinB+ strain implies that pol IV interferes with pol-V-catalyzed replication during TLS of CPDs. Such inhibition is surprising, since the prevailing models for TLS suggest that the two polymerases may cooperate to ensure efficient TLS [1]. Furthermore, It has been proposed that the more processive and catalytically efficient pol IV replaces pol V at the replication fork in order to protect the primer terminus from proofreading by the exonuclease-proficient enzymes [43].
To test whether the inhibitory effect on pol V is pol IV-specific, we generated isogenic recA730 lexA(Def) ΔumuDC rnhB+/ − strains lacking pol II and determined UV-survival of cells expressing wild-type pol V and its variants (Figure S2). Deletion of pol II had very little effect on UV-resistance of the plasmid expressing strains. In general, in the rnh+ strains the relative UV resistance of Y11A was comparable to that promoted by the vector, pGB2, and less than that conferred by the F10L plasmid. There was a modest increase in UV-resistance in the ΔrnhB strains (Figure S2), but this was comparable in the ΔpolB and polB+ strains and certainly much less evident than observed with the ΔdinB/dinB+ ΔrnhB strains (c.f. Figure S1C and Figure 1C). Overall, our findings argue against a possible competition between pol II and pol V during the TLS of UV-induced lesions.
To characterize the effect of rNTP processing on UV-resistance and mutagenesis in strains expressing umuC_Y11A, we focused on the generally more UV-resistant recA730 lexA(Def) ΔumuDC ΔdinB rnhB+/ − strains, rather than the more sensitive ΔrnhA derivatives, where the interpretation of any results might be complicated due to more complex phenotypes involving constitutive and induced stable DNA replication [39]. As shown in Figure 2A, the UV-survival curves of the ΔdinB rnhB+ cells either lacking pol V, or expressing UmuC_Y11A are superimposable. Consistent with the semi-quantitative survival assay, the ΔdinB strain expressing UmuC_Y11A tolerates UV-damage much better in the absence of a functional RNase HII at all UV doses. In contrast, the absence of RNase HII has no effect on UV-resistance of strains either lacking pol V (pGB2), or expressing wild-type pol V.
Fig. 2. Quantitative UV survival and mutagenesis assays. 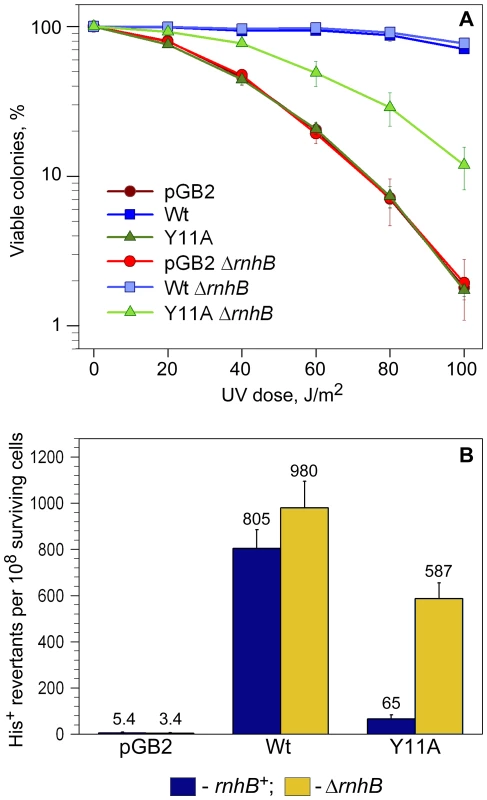
A: Survival. Exponentially growing cells were exposed to various doses of UV-light and serial dilutions spread on LB plates containing spectinomycin. The number of viable colonies was determined after overnight incubation at 37°C. Error bars indicate the standard error of the mean. Consistent with the semi-quantitative UV-survival assay shown in Figure 1, UV-resistance of strains expressing Y11A_UmuC increased significantly in the ΔrnhB background, while there was no change in UV-survival of the strain harboring vector, pGB2, or expressing wild-type pol V in the rnhB+/− strains. B: Mutagenesis. UV-induced mutagenesis was determined by exposing exponentially growing cells to 20 J/m2 UV light. Cell viability was in the range of 85–90% survival for wild-type pol V and ∼60–70% for vector control, pGB2, and the Y11A mutant. The average number of His+ revertants per 108 surviving cells ± standard error of the mean is indicated on the graph. The rnhB+ strains are indicated by navy-colored bars, while ΔrnhB strains are indicated by the gold-colored bars. As observed, the UmuC_Y11A-expressing cells exhibited an ∼9-fold increase in UV mutagenesis compared to the rnhB+ strain. Next, we compared the levels of UV-induced mutagenesis in the various strains by assaying reversion of the hisG4 ochre allele (Figure 2B). While there was a slight reduction in the level of UV-mutagenesis promoted by wild-type pol V in the ΔrnhB strain compared to the rnh+ strain, we observed an ∼9-fold increase in umuC_Y11A-dependent UV-induced mutagenesis in the ΔrnhB strain compared to the rnhB+ strain (Figure 2B). Our observations therefore indicate that RNase HII plays a major role in preventing Y11A-dependent ribonucleotide-driven mutagenesis in E.coli.
The rnhB gene encoding RNase HII is located in a multi-gene operon and is immediately upstream of the dnaE gene encoding the catalytic α-subunit of pol III [44]. To eliminate the possibility that the observed phenotypes of the ΔrnhB allele on Y11A-dependent mutagenesis might be non-specific, due to effects on expression of dnaE, we determined the levels of the α-subunit in isogenic dinB+/ − rnhB+/ − strains (Figure S3). In both the dinB+/ − cells, we observed an ∼25% reduction in the amount of α-subunit in the ΔrnhB strain compared to the rnhB+ strain. The reduced levels of the α-subunit do not, however, explain the Y11A-dependent increase in UV-resistance and UV-mutagenesis in the ΔrnhB strains, since if reduced levels of the α-subunit allows greater access of pol V to a primer terminus, then we would have also expected to observe a significant increase in wild-type pol V UV-mutagenesis, when in fact, we actually observed a small decrease (Figure 2B)
In vitro inhibition of pol V–dependent TLS by pol IV
Since pol IV appears to inhibit pol V-dependent TLS in vivo, we reconstituted TLS reactions in vitro using a circular vector with a unique T-T CPD [45], and a radiolabeled primer located five nucleotides 3′ from the CPD. To ensure maximal catalytic activity of both polymerases, the reaction conditions were optimized by including β-sliding processivity clamp, γ clamp-loading complex, and single-stranded DNA binding protein (SSB). The reactions also included a RecA nucleoprotein filament (RecA*), which has no noticeable effect on pol IV, but is required for pol V TLS in vivo [13]–[15] and in vitro [16], [17], [46]. The role of RecA* in pol V-catalyzed TLS is to transfer a molecule of RecA and ATP from its 3′-proximal tip to convert a barely active pol V to an activated UmuD′2C-RecA-ATP complex, termed pol V Mut [17], [46]. All reactions were carried out in a similar manner with some containing a single DNA polymerase, pol IV (Figure 3, lanes 2 and 7), wild-type pol V (lanes 3 and 8), or Y11A_UmuC pol V (lanes 5 and 10), while in other cases, pol V variants and pol IV were added simultaneously (lanes 4, 6, 9, and 11). DNA polymerases were used either at equi-molar concentrations (lanes 4 and 6), or with a 10-fold excess of pol IV over pol V (lanes 9 and 11).
Fig. 3. In vitro translesion synthesis past a TT-CPD lesion catalyzed by mixtures of pol IV and pol V. 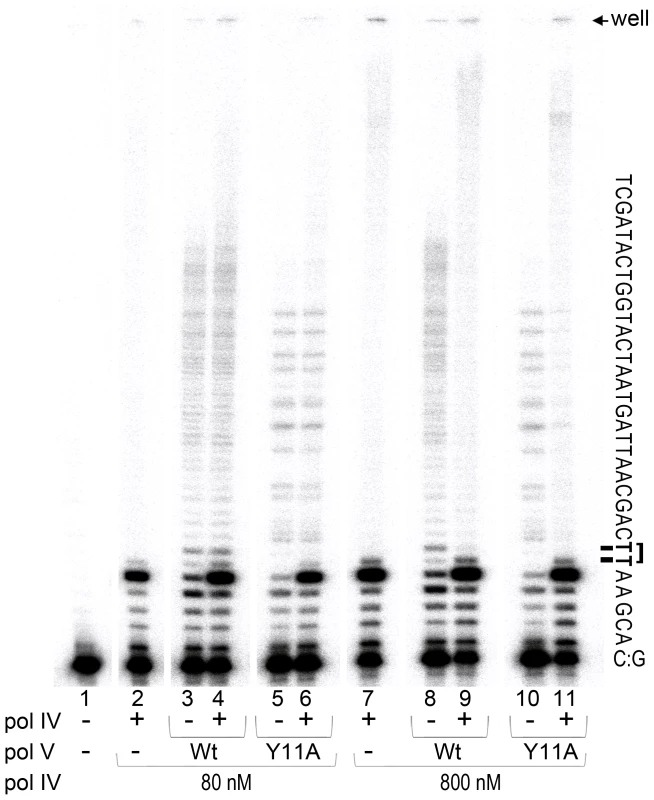
Translesion DNA synthesis was performed using a circular DNA template with a running-start primer with its 3′ end located 5 bases before the 3′T of the CPD. Primer extension reactions catalyzed by pol IV (80 nM, lane 1 or 800 nM, lane 6), wild-type pol V (80 nM, lanes 2 and 7), pol V (UmuC_Y11A) (60 nM, lanes 4 and 9), or a combination of pol IV (80 nM, lane 3 and 5 or 800 nM, lane 8 and 10) with either wild-type (80 nM, lanes 3 and 8) or polV (UmuC_Y11A) (60 nM, lanes 5 and 10) were performed for 30 sec as described in the Methods section. Part of the template sequence and position of the gel wells and a CPD lesion are indicated to the right of the gel panel. As clearly observed, when present in a 10-fold excess (similar to SOS induced conditions), pol IV inhibits TLS catalyzed by pol V. As expected, pol IV by itself was unable to bypass a CPD adduct [18] even when used at elevated levels (Figure 3, lanes 2 and 7). The major reaction product observed was located immediately adjacent to the 3′ base of the CPD, with a very small band corresponding to nucleotide incorporation opposite the 3′T of CPD, as previously reported [18]. Although the overall primer extension efficiency of pol V was lower than that of pol IV (total primer extension by pol V ranged between 15 and 20%, while pol IV, depending on the concentration used, extended 35 or 85% of primers), the ability of pol V to replicate past the lesion was substantially greater. For example, ∼70% of primers extended by pol V to the −1 position (relative to the CPD) were further extended past the CPD. In contrast, and independent of the polymerase concentration used, only 3% of the primers bypassed the CPD when they were extended by pol IV. TLS catalyzed by wild-type pol V and Y11A pol V was similarly efficient and processive, even though the distribution pattern of products differed (Figure 3, lanes 3, 5, 8 and 10, see also [21]). When pol IV and pol V were used at ∼ equi-molar concentrations (lanes 2–6), the extent of lesion bypass catalyzed by wild-type pol V and pol V UmuC_Y11A was unaffected by the presence of pol IV since the amount of reaction products extended past the lesion remained the same. The apparent increase in the proportion of replication products that accumulated opposite the template A immediately 3′ to CPD (in lanes 4 and 6 compared to lanes 3 and 5), is compensated by the increased proportion of elongated primers suggesting that pol V was unable to replace pol IV at the lesion site. When pol IV was present at ∼10-fold excess, which is roughly equivalent to the in vivo cellular ratio when maximally expressed during SOS-induction [42], [47] (lanes 7–11), significant inhibition of pol V-dependent TLS was observed (compare lanes 9 and 11 with lanes 8 and 10). In addition, the general distribution pattern of reaction products was similar to that observed in the reaction containing only pol IV (compare lanes 9 and 11 to lane 7, and the amount of primer elongated past the lesion expressed as a percent of primers elongated to the −1 position, was reduced to 3%, which is equivalent to the results observed in reactions in the presence of pol IV alone). The data suggests that under certain SOS-inducing conditions, pol IV may bind to the 3′-primer terminus of nascent DNA strand and thereby prevent access of pol V to the replicating fork thus serving as a cellular “competitive” inhibitor of pol V-dependent TLS at DNA lesions that pol IV itself is unable to bypass.
Effect of nucleotide excision repair on UV resistance of cells actively repairing misincorporated rNMPs
Our previous studies [21], and those described above (Figure 1 and Figure 2), indicate that in a strain actively repairing errantly-incorporated ribonucleotides, expression of umuC_Y11A confers minimal UV-resistance compared to wild-type pol V, or other pol V variants (umuC_F10L or umuC_Y11F). This phenotype, can, in part, be explained by the fact that the abundant ribonucleotides target the pol V-generated TLS tract for repair. As shown above, this process is initiated by RNase HII, which nicks the DNA backbone immediately 5′ of the misincorporated ribonucleotide, but the ribonucleotide must subsequently be physically replaced using other repair enzymes/polymerases with a limited ability to traverse UV-induced DNA lesions. To identify proteins involved in ribonucleotide removal, we constructed a series of isogenic recA730 lexA(Def) ΔumuDC ΔdinB strains with individual deletions of various DNA repair genes (unpublished data) and determined whether or not such an inactivation would lead to an increase in umuC_Y11A-specific UV-resistance, in a similar manner to that observed with defects in rnhB (Figure 1D and Figure 2). The control, or pol V-encoding plasmids were introduced into these repair deficient strains, and their sensitivity to UV-light was assayed in the semi-quantitative UV survival assay. In most cases, the ability of the particular pol V plasmid to confer UV resistance was the same as shown in Figure 4A, i.e., the relative UV-sensitivity of each plasmid-expressing strain remained the same. Wild-type pol V was similar to Y11F, and both were better than F10L at promoting UV-survival, while Y11A conferred minimal UV-resistance (unpublished data). However, a markedly different pattern of survival emerged in strains defective for nucleotide excision repair (NER), such as uvrA, which plays a critical role in damage recognition (Figure 4B), or uvrC, an endonuclease which cleaves the phosphodiester bond 3′ of the lesion, (Figure 4C). As expected because of their deficiency in NER, the ΔuvrA and ΔuvrC strains were much more sensitive to UV-light than the isogenic parental strains. As a consequence, while cells shown in Figure 4A were exposed to 40 J/m2 UV-light, the NER deficient cells shown in Figure 4B and 4C were only exposed to 1 J/m2 UV. However, the key observation is that in the NER-deficient strains, umuC_Y11A conferred considerable UV-resistance that was roughly similar to that observed for wild-type pol V and the umuC_Y11F variant (Figure 4B and 4C). Thus, the status of the NER machinery plays a critical role in the survival of cells exposed to DNA damage whilst concomitantly incorporating high levels of ribonucleotides.
Fig. 4. Role of NER in strains expressing pol V variants. 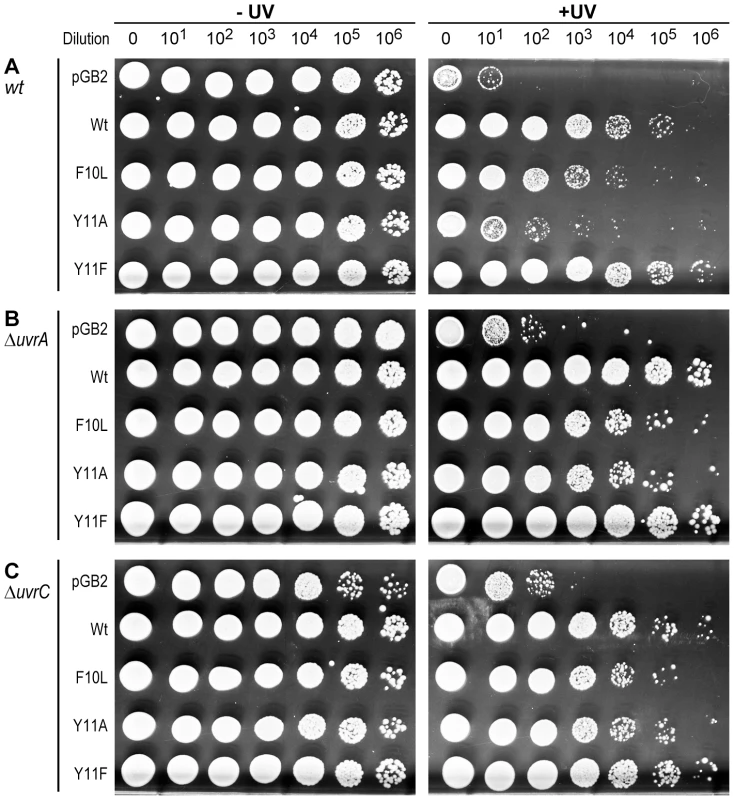
10 µl of 10-fold serial dilutions of overnight cultures were spotted onto the surface of rectangular LB agar plates and exposed to 40 J/m2 254 nM UV-light (Panel A), or 1 J/m2 254 nM UV-light (Panels B and C). Both unirradiated (−) and UV-irradiated (+) plates were incubated overnight at 37°C. In each panel, UV survival is shown for the recA730 lexA(Def) ΔumuDC ΔdinB strains either harboring pGB2 vector, or expressing pol V variants. Panel A (uvr+ strain) is reproduced from Figure 1A for direct comparison to the isogenic ΔuvrA (Panel B) and ΔuvrC (panel C) strains. The main observation of these experiments is that while the uvr− strains are considerably more UV-sensitive than the isogenic uvr+ strain, the relative sensitivity of the strains expressing pol V variants changes in the uvr− background, with UmuC_Y11A promoting an increase in UV-survival to a similar extent as wild-type pol V. Effect of RNase H on the levels of spontaneous mutagenesis in strains expressing pol V and variants
In addition to facilitating TLS, when expressed in a recA730 lexA(Def) background, pol V promotes high levels of spontaneous mutagenesis [15]. This mutagenesis is not the result of TLS of “cryptic” DNA lesions, but rather the ability of pol V to compete with E.coli's other DNA polymerases and gain access to undamaged genomic DNA where its low-fidelity synthesis is manifested as mutagenic events on the E.coli chromosome [48]. We have shown that despite its low sugar and base-substitution fidelity in vitro, when expressed in a recA730 lexA(Def) ΔumuDC strain, umuC_Y11A promotes low levels of spontaneous mutagenesis [20]. One obvious explanation, based upon our observations above, is that mutagenesis is limited via the actions of rnhB. However, the strain used in the earlier study also expresses pol IV and while it is believed that pol V and pol IV work together to promote spontaneous mutagenesis [43], we could not exclude the possibility that in a similar manner to its negative effect on the TLS of CPDs, pol IV might actually block access of the error-prone pol V_Y11A polymerase to undamaged chromosomal DNA. To test this hypothesis, we assayed spontaneous mutagenesis in isogenic recA730 lexA(Def) ΔumuDC ΔdinB rnh+/− strains (Figure 5). Expression of wild-type pol V in the rnhB+ cells resulted in a substantial increase in the number of spontaneously arising His+ revertants compared to the same strain lacking pol V. In the isogenic ΔrnhA strain, there was a considerable (3–5-fold) increase in the number of revertants promoted by pol V and variants, presumably because the increased number of R-loops in the ΔrnhA strain [39] help to hyperactivate the RecA730 protein [49] for its role in pol V-dependent mutagenesis [17]. While there was a 3.7-fold increase in the absolute number of Y11A-dependent mutations in the ΔrnhA strain compared to the rnh+ strain, when expressed as a percentage of wild-type pol V-dependent mutagenesis, umuC_Y11A mutagenesis actually decreased from 7 to 5% of the wild-type levels (Figure 5). In contrast, in the isogenic ΔrnhB strain, the number of umuC_Y11A-dependent revertants increased approximately 4.6-fold compared to the rnhB+ strain and reached ∼40% of the level of mutagenesis observed with wild-type pol V (Figure 5).
Fig. 5. Effect of ΔrnhA and ΔrnhB on spontaneous mutagenesis in recA730 lexA(Def) ΔumuDC ΔdinB strains expressing pol V variants. 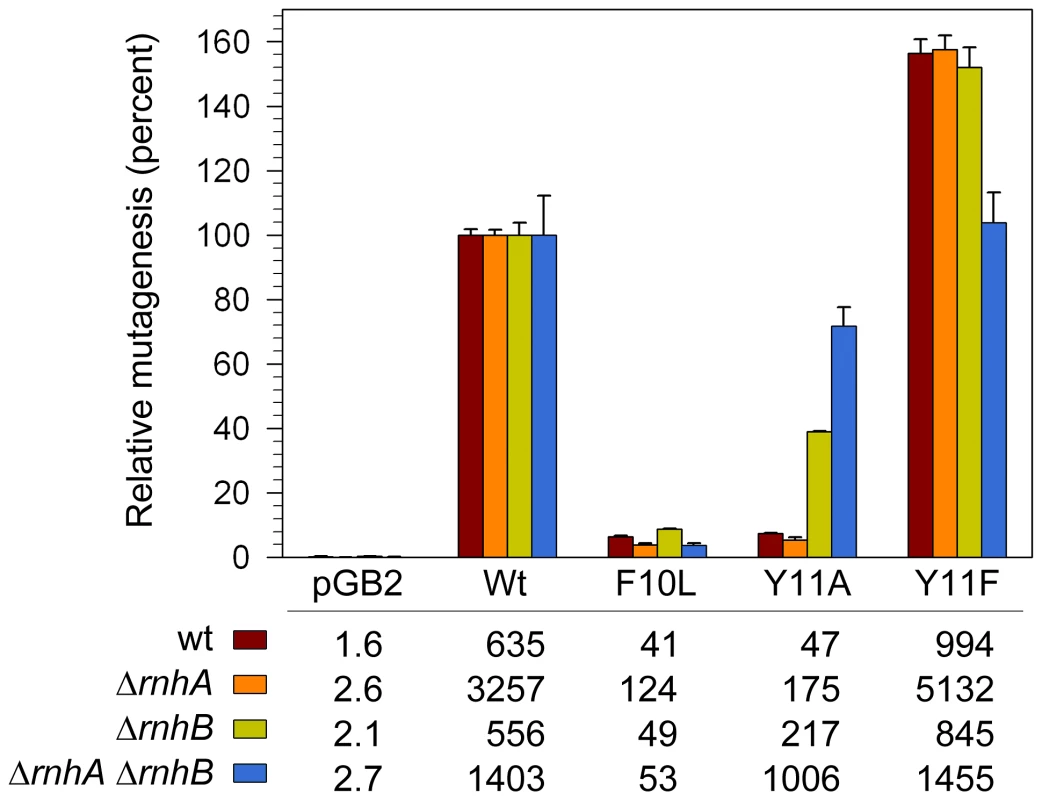
Spontaneous mutagenesis was measured by assaying reversion of the hisG4 ochre allele (leading to histidine prototophy) as described in Materials and Methods. The average number of His+ revertants per plate ± standard error of the mean is indicated in the table. Since the extent of mutagenesis promoted by wild-type pol V differed in the various strains, we have expressed the level of mutagenesis promoted by the variants as a percentage of wild-type mutagenesis. As clearly observed, umuC_Y11A-dependent mutagenesis increased in the ΔrnhB strain and was further elevated in the ΔrnhA ΔrnhB double mutant. In contrast, umuC_F10L gave consistently low levels of mutagenesis in all strains, and umuC_Y11F higher than wild-type levels in all strains. Our studies therefore show that RNase HII clearly participates in a repair pathway that reduces the accumulation of rNMPs, as well as incorrect dNMPs incorporated into undamaged and damaged DNA by UmuC_Y11A. Based upon its in vitro properties [21], we expected pol V umuC_Y11A to be as mutagenic, if not more so, than the wild-type pol V, but even in the ΔrnhB strain, Y11A-dependent mutagenesis was less than half of that observed with wild-type pol V (Figure 5), suggesting that perhaps additional repair pathways act to reduce the mutagenic consequences of rNMPs incorporated by the highly error-prone umuC_Y11A. Indeed, in the isogenic ΔrnhA ΔrnhB strain umuC_Y11A spontaneous mutagenesis increased significantly to ∼72% of the level observed with wild-type pol V (Figure 5). Thus, although Rnase HI alone does not appear to participate in the removal of ribonucleotides incorporated by umuC_Y11A, in the absence of Rnase HII, where there is likely to be a significant accumulation of ribonucleotides into DNA, Rnase HI helps reduce the mutagenic burden of errant ribonucleotide incorporation into the E.coli genome.
Discussion
In order to maintain its genomic integrity a cell must protect its DNA from constant assaults coming from different sources. Among these is the attempt to replace the sugar moiety of a nucleotide, which appears to be one of the most persistent potential sources of “damage”. Incorporation of rNMP into the DNA backbone most frequently occurs during DNA replication and repair due to mistakes made by DNA polymerases. Seemingly a harmless event, assuming that the base of the ribonucleotide being incorporated is a correct Watson-Crick pair, it nevertheless can threaten the cell's well-being because the presence of rNMP with a reactive 2′ hydroxyl on the ribose ring makes the DNA strand more susceptible to spontaneous or enzymatic cleavage. It can also lead to a B- to A-form conformational DNA transition and disrupt interactions of DNA-binding proteins thereby compromising various DNA processing pathways. E.coli pol V appears to be one of the least discriminate DNA polymerases. The sugar selectivity of pol V can be significantly improved by an F10L substitution in the catalytic subunit UmuC and vice versa, a Y11A substitution in UmuC significantly reduces the ability of pol V to select a nucleotide with the correct sugar [20].
To prevent the deleterious effects of ribonucleotides incorporated into DNA, E.coli is equipped with enzymes capable of hydrolyzing the phosphodiester bond between ribo - and deoxyribonucleotides, thereby triggering repair pathways leading to removal of rNMPs. In the present study, we show that ribonucleotide-specific endonuclease RNase HII plays an important role in the correction of mistakes made by error-prone pol V. The basic mechanism of ribonucleotide repair appears to be evolutionary conserved as Nick McElhinny et al., recently reported that RNase H2-dependent repair is necessary for the prevention of replicative stress and genome instability in yeast strains expressing a pol ε variant with compromised sugar selectivity [23]. However, in contrast to other studies demonstrating that deletion of RNase H2 increases spontaneous mutagenesis in yeast strains with wild-type DNA polymerases [25], [50], [51], no increase in spontaneous or UV-induced mutagenesis was observed upon in a ΔrnhB strain expressing wild-type pol V (Figure 2B and Figure 5). Nevertheless, these data do not imply that RNase HII is not important for correction of pol V-dependent mistakes, but rather suggest that sugar selectivity of the polymerase must be significantly reduced for endonuclease function to be readily detectable. Indeed, the lack of RNase HII caused a significant increase in mutagenesis in strains expressing UmuC_Y11A for which ribonucleotide processing is most important. Therefore, the pathway initiated by RNase HII not only leads to removal of nucleotides with an incorrect sugar, but also to the repair of base substitutions, explaining the low mutability of the rnhB+ strain expressing highly error-prone UmuC_Y11A.
While we observed a minimal effect of ΔrnhA alone on the level of umuC_Y11A-dependent spontaneous mutability, there was a dramatic increase in spontaneous mutagenesis in combination with the ΔrnhB allele (Figure 5). Presumably this is due to the accumulation of ribonucleotides in the absence of Rnase HII, and the propensity of the Y11A variant to catalyze synthesis of polynucleotide chains containing multiple sequential rNMPs [20].
It should also be noted that the role of RNase HII-initiated repair in determining UV-sensitivity of Y11A-expressing cells was most pronounced in ΔdinB strains. We assume that in cells expressing dinB (Pol IV), there is competition for a primer-terminus between pol IV and pol V that limits the extent of the pol V-dependent TLS tract, which in the case of UmuC_Y11A will concomitantly reduce the number of incorporated rNMPs into the genome, and the need for RNase HII-mediated repair (Figure S1C). In a similar vein, since the number of rNMPs incorporated by the wild-type pol V, UmuC_Y11F, and especially UmuC_F10L pol V, is significantly lower than that of UmuC_Y11A, the effect of RNase HII and pol IV on UV-resistance is negligible in the strains expressing these polymerases (Figure 1).
RNase HII-initiated rNMP repair, which at the same time leads to the correction of base substitutions, readily explains the low mutability of strains expressing a pol V variant with an impaired steric gate. In the current study, we found that the low UV-resistance, which is somewhat unexpected for cells equipped with an efficient TLS polymerase such as UmuC_Y11A [20], is also connected to ribonucleotide incorporation and repair. For example, we show that the sensitivity to UV-light in cells expressing umuC_Y11A is reduced by RNase HII to the level detected in the strain completely lacking pol V. The observation that removal of rNMPs actually diminishes cells viability implies that unlike yeast, in which unrepaired rNMPs lead to genome instability, bacterial cells can tolerate the presence of ribonucleotides in genomic DNA quite well. This assumption is supported by the findings that all the strains defective in rNMPs repair have similar colony size and growth rates independent of the sugar discrimination properties of a pol V variant. However, even assuming that E.coli is able to tolerate the presence of ribonucleotides in its genome, it nevertheless seems counterintuitive that activation of a pathway directed at the removal of rNMPs would reduce cell viability after UV-treatment. In order to explain these observations, we propose the following model: simultaneous attempts of the NER machinery to repair UV-induced lesions on the parental template strand and of the RNase HII-initiated pathway to remove numerous rNMPs from the TLS-tract in the nascent strand, lead to the formation of multiple and persistent DNA double-strand breaks that cause cell death (Figure 6). Pulse Field Gel Electrophoresis (PFGE) and bacterial COMET assays are currently underway to test this hypothesis. When either of these repair pathways is inactivated, UmuC_Y11A confers significant UV resistance (Figure 1D, Figure 2A, and Figure 4) presumably because of its ability to facilitate TLS and in spite of the fact that ribonucleotides are concomitantly incorporated into the E.coli genome. However, the increased survival comes at the steep cost of increased cellular mutagenesis (Figure 2B, Figure 5).
Fig. 6. Model for the effect of RNase HII and NER proteins on UV sensitivity of strains proficient for ribonucleotide incorporation. 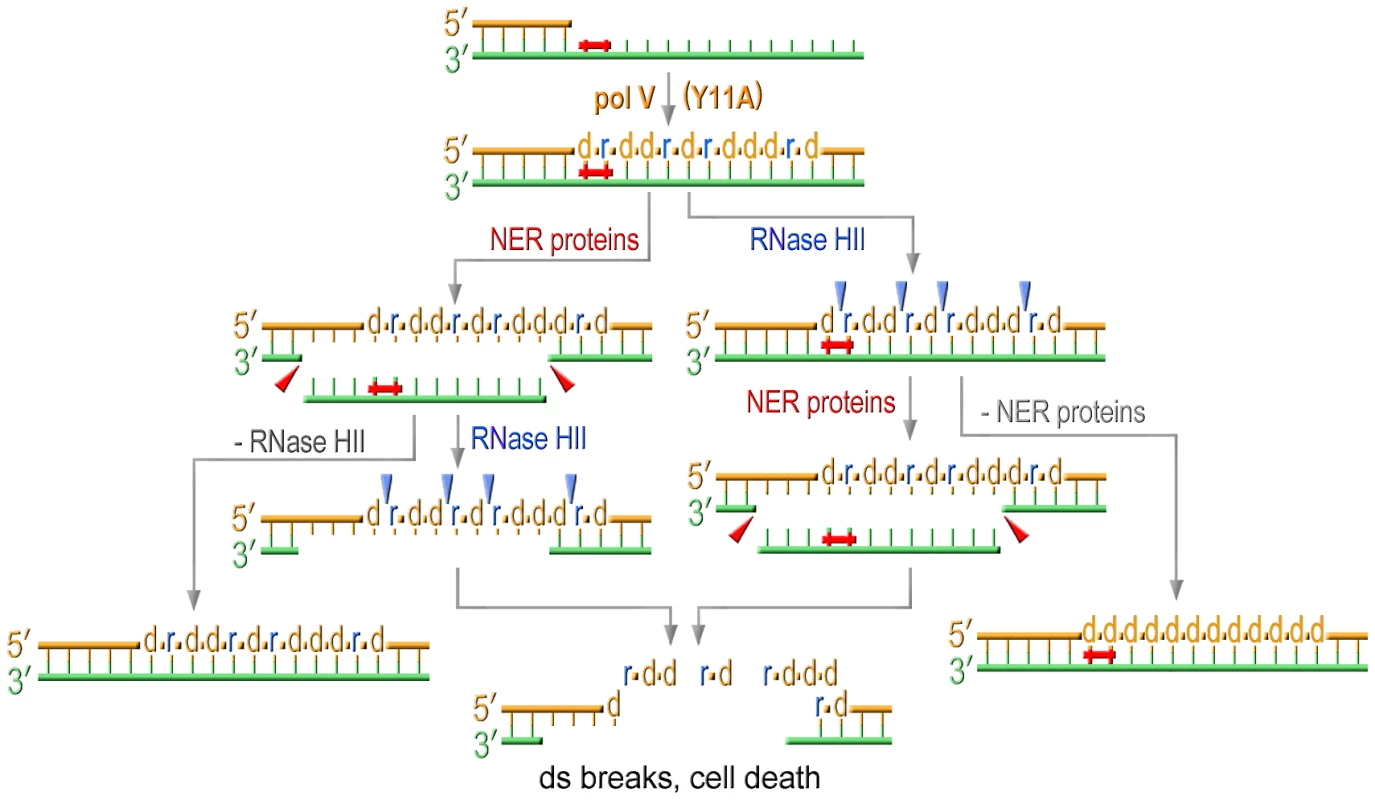
Translesion replication catalyzed by Y11A mutant produces a TLS tract containing multiple ribonucleotides. NER excises UV-induced lesions and produces gaps on the template strand. RNase HII initiating removal of multiple rNMPs incorporated during TLS produces nicks on the daughter strand. The concerted action of both these repair pathways results in formation of persistent double strand breaks ultimately leading to cell death. Inactivation of either repair pathway selectively improves UV-resistance of cells expressing UmuC_Y11A. In summary, E.coli utilizes a variety of mechanisms to minimize pol V-dependent ribonucleotide incorporation into its genome. The first line of defense is a competition between pol IV and pol V during TLS that limits the access of the errant pol V to a stalled primer terminus. In the absence of pol IV, pol V can incorporate ribonucleotides, but these are rapidly removed from the genome in an RNase HII-initiated repair pathway. Concurrent with rNTP removal, NER of the UV-lesion results in double-strand breaks leading to cell death. The Rnase HII-mediated repair pathway minimizes both UV-induced and spontaneous mutagenesis to the bacterial chromosome and in its absence, and in the presence of a mutant pol V with a propensity to incorporate polyribonucleotides (UmuC_Y11A), RNase HI serves as a backup to RNase HII to limit the mutagenic consequences of excessive ribonucleotide accumulation into the E.coli genome.
Materials and Methods
Bacterial strains
Most of the E. coli K-12 strains used in this study are derivatives of RW584 (full genotype: recA730 lexA51(Def) ΔumuDC596::ermGT thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 glnV44 galK2 hisG4 rpsL31 xyl-5 mtl-1 argE3 thi-1 sulA211 [52]. All derivatives were made by standard methods of P1 transduction using P1vir [53] (Table 1). The various alleles were selected by conferring resistance to spectinomycin (20 µg/ml), zeocin (25 µg/ml), chloramphenicol (20 µg/ml) and kanamycin (50 µg/ml) respectively and subsequently confirmed by PCR [54]–[56].
Tab. 1. E. coli strains used in this study. 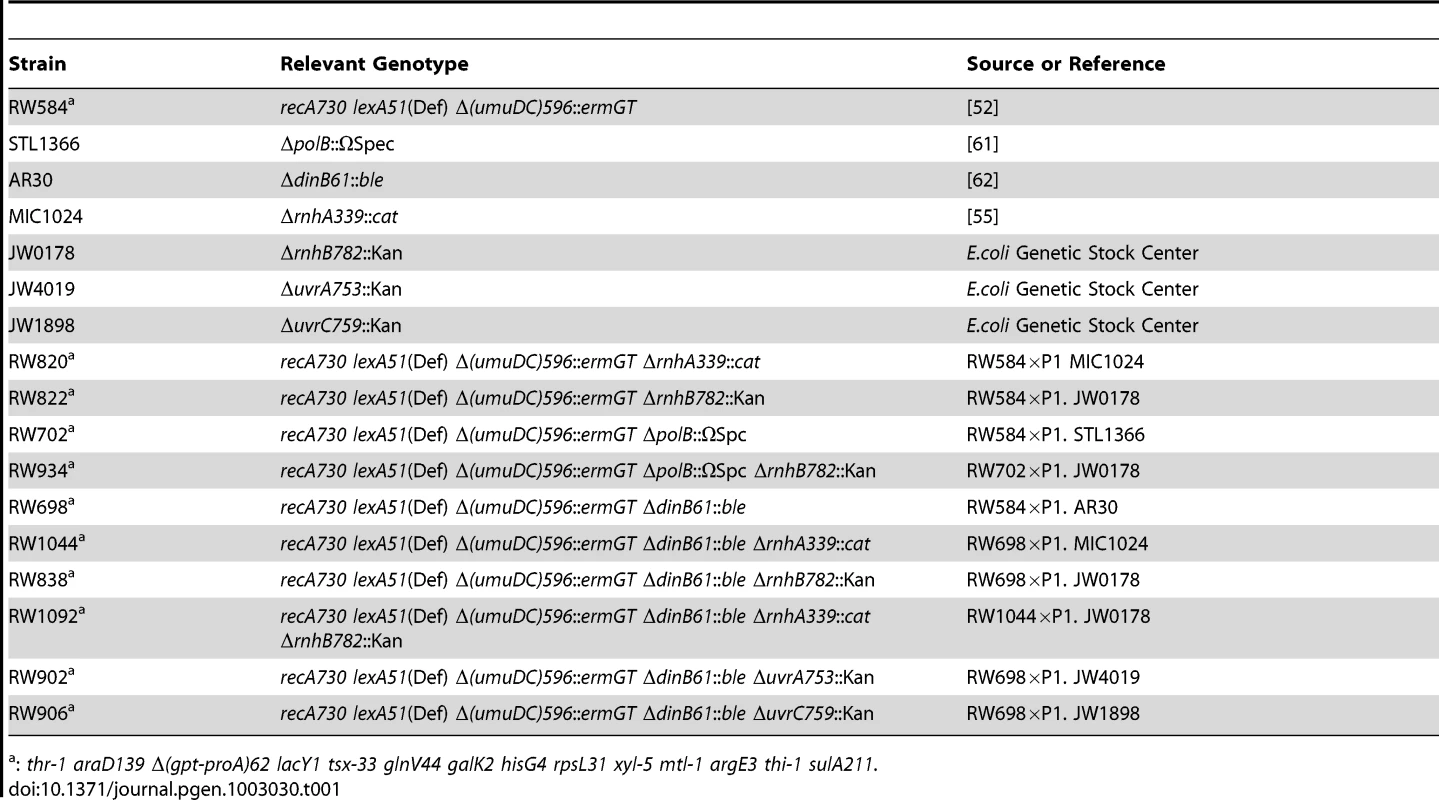
: thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 glnV44 galK2 hisG4 rpsL31 xyl-5 mtl-1 argE3 thi-1 sulA211. Plasmids
The low-copy-number plasmids used for expression of UmuC variants are derived from pGB2 [57]. Spectinomycin resistant plasmids pRW134, pJM964, pJM963, and pJM952 encode E.coli UmuD′ along with wild-type UmuC or F10L, Y11A, and Y11F variants, respectively and the proteins are expressed from the native umu promoter [20]. Ampicillin resistant derivatives were generated by replacing the BspHI-BspHI vector fragment encoding resistance to spectinomycin with a BspHI-BspHI fragment from pET22b+ encoding resistance to ampicillin. Bacteria harboring plasmids were grown in LB media containing appropriate antibiotics (50 µg/ml spectinomycin, or 100 µg/ml ampicillin).
Semi-quantitative UV survival assays
Cells were grown overnight at 37°C in Luria–Bertani (LB) plus spectinomycin. The next morning, the cultures were sequentially diluted 10-fold in eppendorf tubes containing SM buffer [58]. 10 µl of each serial dilution was then spotted on the surface of a 12×8 cm rectangular LB agar plate (Nunc, ThermoFisher). The plates were irradiated with UV light (254 nm) and incubated overnight at 37°C. Images of the irradiated plates/cultures were captured with a FluorChem HD2 imaging system (Alpha Innotec).
Quantitative spontaneous mutagenesis, UV-induced mutagenesis, and UV survival assays
Cells transformed with the vector plasmid, pGB2, or one of the low-copy number plasmids expressing wild-type pol V or UmuC variants were grown overnight at 37°C in LB media plus spectinomycin. The next day, cultures were diluted 100-fold into 10 ml fresh media and grown at 37°C until they reach an OD600 of 0.1 (roughly 3 hrs). Cells were centrifuged and resuspended in an equal volume of SM buffer [58] and transferred to a petri dish. Aliquots were removed and saved as the unirradiated control for the experiments. The culture was irradiated at a UV fluence of ∼2 J/m2 per second and aliquots removed at 20 J/m2 increments. At least three independent cultures were assayed for each strain and all experiments were performed under yellow light to avoid unwanted photoreactivation.
For UV survival assays, appropriate serial dilutions (based upon trial assays) were plated on LB agar plates containing spectinomycin and incubated overnight at 37°C. The surviving fraction was determined by dividing the number of viable cells exposed to UV by the number of viable cells in the unirradiated culture. Error bars represent the standard error of the mean (SEM).
To determine the number of spontaneously arising histidine mutants on the plate, as well as UV-induced mutants, the unirradiated cell culture was seeded on the Davis and Mingioli minimal agar plates [59] plus glucose (0.4% wt/vol); agar (1.0% wt/vol); proline, threonine, valine, leucine, and isoleucine (all at 100 µg/ml); thiamine (0.25 µg/ml); and either no histidine, or histidine (1 µg/ml). On the plates containing no histidine, only pre-existing His+ mutants grew to form colonies. However, on the plates containing 1 µg/ml histidine, 100–200 His− cells are able to grow on the limiting amount of histidine, so that a viable cell count can be obtained under the exact same conditions where His+ mutant arise. When ∼4×107 bacteria were seeded, they grew to form a lawn, concomitantly exhausting the low level of histidine. Spontaneously arising His+ mutants grew up through the lawn and were counted after 4 days incubation at 37°C.
To determine the extent of UV-induced mutagenesis, cells that had been irradiated with 20 J/m2 UV were used for analysis. This UV dose was chosen since even the UV-sensitive strains exhibited minimal cell killing at this exposure. These conditions therefore provide a window to observe UV-induced mutagenesis without the complications associated with differential levels of cell killing in the various strains. The UV-induced mutation frequencies were calculated as previously described [60]. This equation not only takes into account the number of mutants spontaneously arising on the low histidine plates, but also any effect of reduced cell viability on the number of pre-existing His+ mutants in the culture. The data reported in Figure 4 represent the average number of His+ mutants from 3 separate experiments (± standard error of the mean [SEM]).
Western blots
Cells were grown overnight at 37°C in LB plus appropriate antibiotics. The next morning, cultures were diluted 1∶100 in fresh LB, plus antibiotics and grown with aeration at 37°C until they reached an OD600 of ∼0.5. Cultures were harvested by centrifugation, resuspended in 1× SDS sample buffer (50 mM Tris-HCl [pH 6.8], 10% glycerol, 2.3% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, 10 mM dithiothreitol), and immediately frozen in dry ice. Cells were lysed by multiple freeze-thaw cycles and boiled for 5 mins at 95–100°C. Extracts were immediately applied to a 15% SDS-PAGE gel. After separation, proteins were transferred to an Immobilon-P membrane (Millipore) using standard Western blot protocols. The membrane was incubated overnight with a 1∶1000 dilution of mouse monoclonal antibodies raised against the α-subunit of pol III (kindly provided by Charles McHenry, University of Colorado). The membrane was then incubated with secondary anti-mouse alkaline phosphatase conjugated antibodies and visualized using the CSPD-Western light assay (Applied Biosystems). Pictures were captured on a FluorChem HD2 imaging system (Alpha Innotec).
In vitro replication assays
Wild-type pol V, the UmuC_Y11A variant and pol IV, β-clamp and γ-complex were purified as previously described [46]. pSOcpd plasmid, containing a unique CPD adduct, was also constructed as previously described [45]. All oligonucleotides were synthesized by Lofstrand Laboratories (Gaithersburg, MD) and gel purified prior to use. 5′-32P labeled M13-TT (5′ – GAT-CGA-TGG-TAC-GGA-CG) primer was annealed to pSOcpd ssDNA templates at a 1.5∶1 molar ratio by heating in an annealing buffer (50 mM Tris-HCl (pH 8), 5 mM MgCl2, 50 µg/ml BSA, 1.42 mM 2-mercaptoethanol) for 10 min at 100°C followed by slow cooling to room temperature. 4 mM RecA (New England Biolabs, Ipswich, MA) was incubated with 0.25 µM 48-mer single-stranded oligonucleotide in the presence of 1 mM adenosine 5′[γ-thio]triphosphate (ATPγS, Biolog Life Science Institute, Bremen, Germany) in the 1× reaction buffer [20 mM Tris-HCl pH 7.5, 8 mM MgCl2, 8 mM DTT, 80 µg/ml BSA, 4% glycerol] at 37°C for 5 min to form RecA nucleoprotein filament on ssDNA (RecA*). Reaction mixture containing 1 mM ATP, 50 µM dNTPs, 2 nM DNA templates, 100 nM SSB (Epicentre Biotechnologies, Madison WI), 50 nM β clamp, and 5 nM γ complex in the 1× reaction buffer was preincubated for 3 min at 37°C. Purified pol V variants (80 nM) were first combined with RecA*(0.25 µM) to form pol V Mut and then added to the reaction mixture. When indicated, Pol V was either substituted, or mixed with indicated amounts of purified pol IV. Reactions were incubated at 37°C for 20 mins and terminated by adding 10 ml of 2× loading buffer [97% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue]. The products were heat-denatured and resolved by denaturing PAGE (8 M urea, 15% acrylamide), followed by visualization on a Fuji image analyzer FLA-5100.
Supporting Information
Zdroje
1. Vaisman A, McDonald JP, Woodgate R (2012) Translesion DNA Synthesis. In: Böck A, Curtiss R, Kaper JB, Karp PD, Neidhardt FC et al.., editors. EcoSal: Escherichia coli and Salmonella: cellular and molecular miology. Washington, DC: American Society for Biology.
2. CaiH, YuH, McEnteeK, KunkelTA, GoodmanMF (1995) Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from Escherichia coli. J Biol Chem 270 : 15327–15335.
3. BecherelOJ, FuchsRP (2001) Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc Natl Acad Sci U S A 98 : 8566–8571.
4. OhmoriH, HatadaE, QiaoY, TsujiM, FukudaR (1995) dinP, a new gene in Escherichia coli, whose product shows similarities to UmuC and its homologues. Mut Res 347 : 1–7.
5. WagnerJ, GruzP, KimSR, YamadaM, MatsuiK, et al. (1999) The dinB gene encodes an novel Escherichia coli DNA polymerase (DNA pol IV) involved in mutagenesis. Mol Cell 4 : 281–286.
6. KatoT, ShinouraY (1977) Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet 156 : 121–131.
7. JaroszDF, BeuningPJ, CohenSE, WalkerGC (2007) Y-family DNA polymerases in Escherichia coli. Trends Microbiol 15 : 70–77.
8. SchlacherK, GoodmanMF (2007) Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol 8 : 587–594.
9. JaroszDF, GodoyVG, DelaneyJC, EssigmannJM, WalkerGC (2006) A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439 : 225–228.
10. Friedberg EC, Walker GC, Siede W, Wood R, Schultz RA, et al.. (2006) DNA Repair and Mutagenesis. Washington, DC: ASM Press.
11. SteinbornG (1978) Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet 165 : 87–93.
12. WoodgateR, RajagopalanM, LuC, EcholsH (1989) UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc Natl Acad Sci U S A 86 : 7301–7305.
13. NohmiT, BattistaJR, DodsonLA, WalkerGC (1988) RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A 85 : 1816–1820.
14. DutreixM, MoreauPL, BailoneA, GalibertF, BattistaJR, et al. (1989) New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol 171 : 2415–2423.
15. SweasyJB, WitkinEM, SinhaN, Roegner-ManiscalcoV (1990) RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol 172 : 3030–3036.
16. SchlacherK, CoxMM, WoodgateR, GoodmanMF (2006) RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442 : 883–887.
17. JiangQ, KarataK, WoodgateR, CoxMM, GoodmanMF (2009) The active form of DNA polymerase V is UmuD′2C-RecA-ATP. Nature 460 : 359–363.
18. TangM, PhamP, ShenX, TaylorJ-S, O′DonnellM, et al. (2000) Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404 : 1014–1018.
19. PhamP, RangarajanS, WoodgateR, GoodmanMF (2001) Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli. Proc Natl Acad Sci U S A 98 : 8350–8354.
20. VaismanA, KubanW, McDonaldJP, KarataK, YangW, et al. (2012) Critical amino acids in Escherichis coli responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res 40 : 6144–6157.
21. KubanW, VaismanA, McDonaldJP, KarataK, YangW, et al. (2012) Escherichia coli UmuC active site mutants: effects on translesion DNA synthesis, mutagenesis and cell survival. DNA Repair 11 : 726–732.
22. BrownJA, SuoZ (2011) Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry 50 : 1135–1142.
23. Nick McElhinnySA, WattsBE, KumarD, WattDL, LundstromEB, et al. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A 107 : 4949–4954.
24. ShenY, KohKD, WeissB, StoriciF (2012) Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nat Struc Mol Biol 19 : 98–104.
25. Nick McElhinnySA, KumarD, ClarkAB, WattDL, WattsBE, et al. (2010) Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol 6 : 774–781.
26. LazzaroF, NovarinaD, AmaraF, WattDL, StoneJE, et al. (2012) RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell 45 : 99–110.
27. KimN, HuangSN, WilliamsJS, LiYC, ClarkAB, et al. (2011) Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332 : 1561–1564.
28. KanayaS (2009) Ribonuclease H. The FEBS journal 276 : 1481.
29. Hollis T, Shaban NM (2011) Structure and functions of RNase H enzymes. In: Nicholson AW, editor. Ribonucleases: Springer. pp. 299–318.
30. TurchiJJ, HuangL, MuranteRS, KimY, BambaraRA (1994) Enzymatic completion of mammalian lagging-strand DNA replication. Proc Natl Acad Sci U S A 91 : 9803–9807.
31. GoulianM, RichardsSH, HeardCJ, BigsbyBM (1990) Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem 265 : 18461–18471.
32. RydbergB, GameJ (2002) Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci USA 99 : 16654–16659.
33. BubeckD, ReijnsMA, GrahamSC, AstellKR, JonesEY, et al. (2011) PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res 39 : 3652–3666.
34. Nick McElhinnySA, KisslingGE, KunkelTA (2010) Differential correction of lagging-strand replication errors made by DNA polymerases α and δ. Proc Natl Acad Sci U S A 107 : 21070–21075.
35. ClarkAB, LujanSA, KisslingGE, KunkelTA (2011) Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase ε. DNA Repair 10 : 476–482.
36. ItayaM, OmoriA, KanayaS, CrouchRJ, TanakaT, et al. (1999) Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J Bacteriol 181 : 2118–2123.
37. CarlPL, BloomL, CrouchRJ (1980) Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J Bacteriol 144 : 28–35.
38. ItayaM (1990) Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc Natl Acad Sci U S A 87 : 8587–8591.
39. AsaiT, KogomaT (1994) D-loops and R-loops: alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J Bacteriol 176 : 1807–1812.
40. Fernández de HenestrosaAR, OgiT, AoyagiS, ChafinD, HayesJJ, et al. (2000) Identification of additional genes belonging to the LexA-regulon in Escherichia coli. Mol Microbiol 35 : 1560–1572.
41. MountDW (1977) A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc Natl Acad Sci U S A 74 : 300–304.
42. KimSR, MatsuiK, YamadaM, GruzP, NohmiT (2001) Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Gen Genomics 266 : 207–215.
43. KubanW, Banach-OrlowskaM, SchaaperRM, JonczykP, FijalkowskaIJ (2006) Role of DNA polymerase IV in Escherichia coli SOS mutator activity. J Bacteriol 188 : 7977–7980.
44. WelchMM, McHenryCS (1982) Cloning and identification of the product of the dnaE gene of Escherichia coli. J Bacteriol 152 : 351–356.
45. KarataK, VidalAE, WoodgateR (2009) Construction of a circular single-stranded DNA template containing a defined lesion. DNA Repair 8 : 852–856.
46. KarataK, VaismanA, GoodmanMF, WoodgateR (2012) Simple and efficient purification of E.coli DNA polymerase V: cofactor requirements for optimal activity and processivity in vitro. DNA Repair 11 : 431–440.
47. WoodgateR, EnnisDG (1991) Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet 229 : 10–16.
48. FijalkowskaIJ, DunnRL, SchaaperRM (1997) Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J Bacteriol 179 : 7435–7445.
49. VlašićI, ŠimatovićA, Brčić-KostićK (2011) Genetic requirements for high constitutive SOS expression in recA730 mutants of Escherichia coli. J Bacteriol 193 : 4643–4651.
50. QiuJ, QianY, FrankP, WintersbergerU, ShenB (1999) Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol 19 : 8361–8371.
51. ChenJZ, QiuJ, ShenB, HolmquistGP (2000) Mutational spectrum analysis of RNase H(35) deficient Saccharomyces cerevisiae using fluorescence-based directed termination PCR. Nucleic Acids Res 28 : 3649–3656.
52. MeadS, VaismanA, Valjavec-GratianM, KarataK, VandewieleD, et al. (2007) Characterization of polVR391: a Y-family polymerase encoded by rumA′B from the IncJ conjugative transposon, R391. Mol Microbiol 63 : 797–810.
53. Miller JH (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press.
54. CurtiE, McDonaldJP, MeadS, WoodgateR (2009) DNA polymerase switching: effects on spontaneous mutagenesis in Escherichia coli. Mol Microbiol 71 : 315–331.
55. ItayaM, CrouchRJ (1991) A combination of RNase H (rnh) and recBCD or sbcB mutations in Escherichia coli K12 adversely affects growth. Mol Gen Genet 227 : 424–432.
56. BabaT, AraT, HasegawaM, TakaiY, OkumuraY, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2 : 2006.0008.
57. ChurchwardG, BelinD, NagamineY (1984) A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31 : 165–171.
58. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory.
59. DavisBD, MingioliES (1950) Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol 60 : 17–28.
60. SedgwickSG, BridgesBA (1972) Survival, mutation and capacity to repair single strand DNA breaks after gamma irradiation in different exr − strains of Escherichia coli. Mol Gen Genet 119 : 93–102.
61. RangarajanS, WoodgateR, GoodmanMF (1999) A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci U S A 96 : 9224–9229.
62. BordenA, O'GradyPI, VandewieleD, Fernández de HenestrosaAR, LawrenceCW, et al. (2002) Escherichia coli DNA polymerase III can replicate efficiently past a T-T cis-syn dimer if DNA polymerase V and the 3′ to 5′ exonuclease proofreading function encoded by dnaQ are inactivated. J Bacteriol 184 : 2674–2681.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Příjem alkoholu a menstruační cyklus
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
- Akutní intermitentní porfyrie
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



