-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Tissue-Specific Regulation of Chromatin Insulator Function
Chromatin insulators organize the genome into distinct transcriptional domains and contribute to cell type–specific chromatin organization. However, factors regulating tissue-specific insulator function have not yet been discovered. Here we identify the RNA recognition motif-containing protein Shep as a direct interactor of two individual components of the gypsy insulator complex in Drosophila. Mutation of shep improves gypsy-dependent enhancer blocking, indicating a role as a negative regulator of insulator activity. Unlike ubiquitously expressed core gypsy insulator proteins, Shep is highly expressed in the central nervous system (CNS) with lower expression in other tissues. We developed a novel, quantitative tissue-specific barrier assay to demonstrate that Shep functions as a negative regulator of insulator activity in the CNS but not in muscle tissue. Additionally, mutation of shep alters insulator complex nuclear localization in the CNS but has no effect in other tissues. Consistent with negative regulatory activity, ChIP–seq analysis of Shep in a CNS-derived cell line indicates substantial genome-wide colocalization with a single gypsy insulator component but limited overlap with intact insulator complexes. Taken together, these data reveal a novel, tissue-specific mode of regulation of a chromatin insulator.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003069
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003069Summary
Chromatin insulators organize the genome into distinct transcriptional domains and contribute to cell type–specific chromatin organization. However, factors regulating tissue-specific insulator function have not yet been discovered. Here we identify the RNA recognition motif-containing protein Shep as a direct interactor of two individual components of the gypsy insulator complex in Drosophila. Mutation of shep improves gypsy-dependent enhancer blocking, indicating a role as a negative regulator of insulator activity. Unlike ubiquitously expressed core gypsy insulator proteins, Shep is highly expressed in the central nervous system (CNS) with lower expression in other tissues. We developed a novel, quantitative tissue-specific barrier assay to demonstrate that Shep functions as a negative regulator of insulator activity in the CNS but not in muscle tissue. Additionally, mutation of shep alters insulator complex nuclear localization in the CNS but has no effect in other tissues. Consistent with negative regulatory activity, ChIP–seq analysis of Shep in a CNS-derived cell line indicates substantial genome-wide colocalization with a single gypsy insulator component but limited overlap with intact insulator complexes. Taken together, these data reveal a novel, tissue-specific mode of regulation of a chromatin insulator.
Introduction
Chromatin insulators are DNA-protein complexes that influence eukaryotic gene expression by organizing the genome into distinct transcriptional domains. Functionally conserved from Drosophila to humans, insulators regulate interactions between regulatory elements such as enhancers and promoters and demarcate silent and active chromatin regions (for review, see [1]). Chromatin insulators are thought to exert effects on gene expression by constraining the topology of chromatin and facilitating the formation of intra - and inter-chromosomal looping (for review, see [2]). These higher order interactions can vary between cell types, thereby facilitating tissue-specific transcriptional output.
Drosophila harbor several distinct classes of chromatin insulators, including the well studied gypsy insulator, also known as the Suppressor of Hairy wing (Su(Hw)) insulator. The zinc-finger DNA-binding protein, Su(Hw), recognizes a particular motif, imparting specificity to the gypsy insulator. In addition to Su(Hw), the core gypsy insulator complex contains Centrosomal protein 190 (CP190), which also harbors a zinc finger domain, and the non-DNA-binding protein, Modifier of mdg4 2.2 (Mod(mdg4)2.2). These core proteins are required for gypsy insulator activity [3]–[7]. Both CP190 and Mod(mdg4)2.2 contain broad complex, tramtrack, bric-a-brac (BTB) dimerization domains that have been suggested to mediate insulator-insulator interactions and facilitate the formation of long range insulator-mediated loops along the chromatin fiber [4], [8].
Specialized nuclear arrangement of gypsy insulator complexes correlates tightly with insulator function. The gypsy insulator proteins bind to thousands of sites throughout the genome with more than half of Su(Hw) binding sites occurring in intergenic regions and a large number of sites located within introns [9], [10]. Consistent with a role in boundary formation, Su(Hw) sites are positively correlated with both Lamin-associated domains and boundaries between transcriptionally active and silent chromatin [10], [11]. It has been shown that gypsy insulator proteins coalesce at a small number of foci in diploid nuclei, termed insulator bodies, which have been proposed to act either as hubs of higher order chromatin domains [8] or storage sites for insulator proteins [12]. Importantly, mutation of certain insulator components results in impaired insulator activity coincident with diffuse or smaller, more numerous insulator bodies [4], [8], [12]–[14]. However, formation of insulator bodies is not sufficient for gypsy insulator activity [15], [16], and a detailed mechanistic understanding of insulator bodies is still lacking. Nevertheless, the tight correlation between gypsy insulator function and insulator body localization suggests an important role for these structures. Finally, in addition to a variety of accessory proteins [17]–[19], a role for RNA in insulator function and insulator body organization was suggested based on RNA-dependent protein interaction with insulator complexes [20].
Genome-wide studies indicate that the locations of insulator protein binding sites are mainly consistent across different cell types but that insulator-dependent looping configurations may dictate differences in gene expression. In Drosophila, it has been shown that external stimuli can alter chromatin association of CP190, possibly leading to a change in chromatin looping [21]. Recent large-scale chromatin conformation capture (3C)-based studies have implicated insulator protein binding sites as key contact points mediating looping throughout the genome [22]–[25]. In several studies across species, specific chromatin conformations are observed in loci that produce tissue - or cell-type specific transcripts [26]–[32]. Whether insulators either establish tissue-specific chromatin organization or maintain configurations established via transcription is unclear. Furthermore, factors that control tissue-specific insulator-dependent chromatin organization remain unknown.
This study identifies a CNS enriched, RNA recognition motif (RRM) containing protein, Alan Shepard (Shep), as the first tissue-specific regulator of gypsy insulator activity and insulator body localization. We show that Shep interacts directly with Mod(mdg4)2.2 and Su(Hw) and also associates with gypsy insulator proteins in vivo. Using a novel quantitative, tissue-specific insulator assay, we find that Shep negatively regulates gypsy insulator activity in the CNS. In addition, mutation of Shep improves compromised insulator function and insulator body formation. Finally, genome-wide localization in the CNS-derived BG3 cell line reveals enrichment of overlap between Shep and Mod(mdg4)2.2 but less frequent than expected overlap among Shep, Su(Hw) and Mod(mdg4)2.2 together. These data suggest that gypsy chromatin insulator function can be regulated in a tissue-specific manner.
Results
Shep is a novel direct interactor of gypsy insulator complexes
The putative RNA-binding protein Shep was identified as a novel interaction partner of the gypsy insulator complex. Shep, encoded by the alan shepard locus, was found by yeast two-hybrid screening as a strong interactor of Mod(mdg4)2.2 [17; M. Capelson and V. Corces, personal communication]. The shep gene was named based on its identification in a gravitaxis screen [33] and is predicted computationally and suggested by EST data to produce four different protein isoforms with distinct N-terminal domains that share a mostly common C-terminal region bearing two highly conserved, tandemly arranged RNA recognition motifs (RRMs; Figure 1A). Isoforms B/D and E contain an additional 10 amino acid linker between the RRM domains, and all isoforms except B/D contain a 7 amino acid stretch at the C-terminus. Unlike core gypsy insulator proteins, Shep is conserved between flies and vertebrates (data not shown).
Fig. 1. Shep associates directly with gypsy insulator complexes. 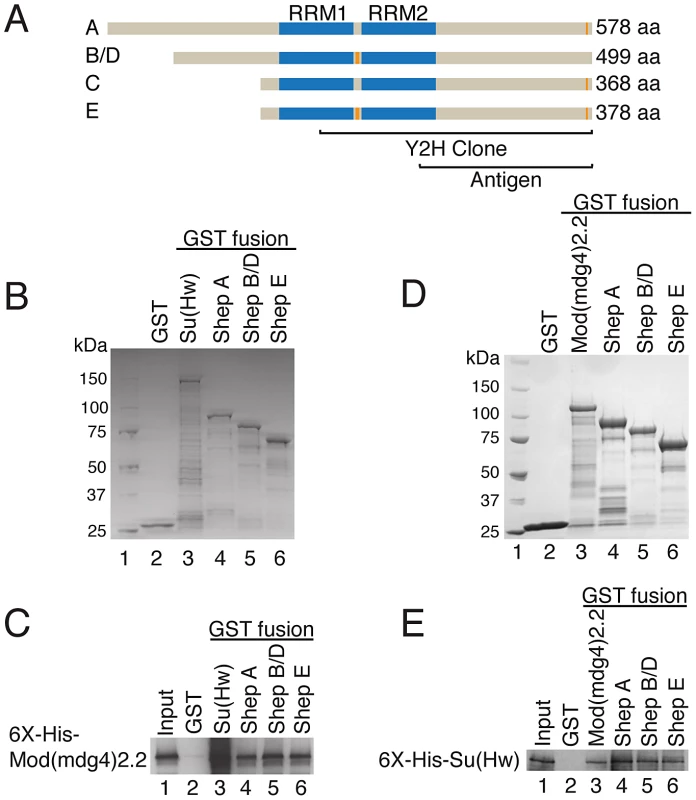
(A) Diagram of Shep protein isoforms. RRMs (blue) and alternative amino acid stretches (not to scale, orange) are shown. Regions of Shep utilized for antibody production or contained in the yeast two-hybrid clone, which corresponds to exons present in isoform E, are indicated. (B) Coomassie staining of recombinant GST fusion proteins used for binding reactions in (C). Protein marker is run in lane 1. (C) Interaction of purified, soluble His-Mod(mdg4)2.2 (lane 1, 4.5% input) with immobilized GST (lane 2), GST-Su(Hw) (lane 3) or GST-Shep isoforms (lanes 4–6). Binding of His-Mod(mdg4)2.2 to GST-fusion proteins was detected by Western blotting. (D) Coomassie staining of recombinant GST fusion proteins used for binding reactions in (E). (E) Interaction of purified, soluble His-Su(Hw) (lane 1, 6.3% input) with immobilized GST (lane 2), GST-Mod(mdg4)2.2 (lane 3) or GST-Shep isoforms (lanes 4–6). Binding of His-Su(Hw) to GST-fusion proteins was detected by Western blotting. We confirmed the Mod(mdg4)2.2-Shep physical interaction in vitro using recombinant proteins. GST-fusions of Shep isoforms A, B/D, and E (Figure 1B–1C, lanes 4–6) in comparison to GST-Su(Hw) as a positive control (lane 3) and GST alone as a negative control (lane 2) were isolated from bacterial extracts and tested for their ability to interact with purified recombinant His-Mod(mdg4)2.2. His-Mod(mdg4)2.2 is detected in the bound fraction in association with Su(Hw) and each Shep isoform but not GST alone, indicating a direct protein-protein interaction between Shep and Mod(mdg4)2.2.
Similarly, we found that Shep also can interact directly with Su(Hw). GST-fusions of Shep isoforms A, B/D, and E (Figure 1D–1E, lanes 4–6) in comparison to GST alone (lane 2) and positive control, GST-Mod(mdg4)2.2 (lane 3), were tested for their ability to interact with purified recombinant His-Su(Hw). His-Su(Hw) is detected in the bound fraction in association with Mod(mdg4)2.2 and each Shep isoform but not GST alone. For both Su(Hw) and Mod(mdg4)2.2 binding assays, a near 1∶1 molar binding ratio between insulator proteins and Shep was observed, similar to the ratios observed between Mod(mdg4)2.2 and Su(Hw) in both binding experiments. These data provide evidence for direct protein interaction between Shep and Mod(mdg4)2.2 as well as between Shep and Su(Hw).
Coimmunoprecipitation of gypsy insulator proteins with Shep isoforms
Shep polyclonal antisera were generated using the common C-terminal region downstream of the RRMs allowing detection of all isoforms. Multiple bands are detected in larval extracts by Western blotting, and isoforms A, B/D, C and E were inferred by predicted molecular weights of 68, 60, 45, and 44 kDa, respectively (Figure 2A, lane 1). All bands are depleted upon shep RNAi hairpin knockdown, which targets all isoforms (lane 2), indicating antibody specificity. When the shepEY04794 allele, which contains a UAS sequence upstream of the shep C and E promoter, is induced ubiquitously using Gal4, the 45 kDa doublet is enriched over wildtype, identifying these two bands as isoforms C and E (lane 3). Finally, homozygous P-element insertion in shepKG10149 predicted to disrupt translation of isoform A causes specific loss of the largest band (lane 4). By process of elimination, isoform B/D corresponds to the apparent 60 kDa band.
Fig. 2. Coimmunoprecipitation of gypsy insulator proteins with Shep isoforms. 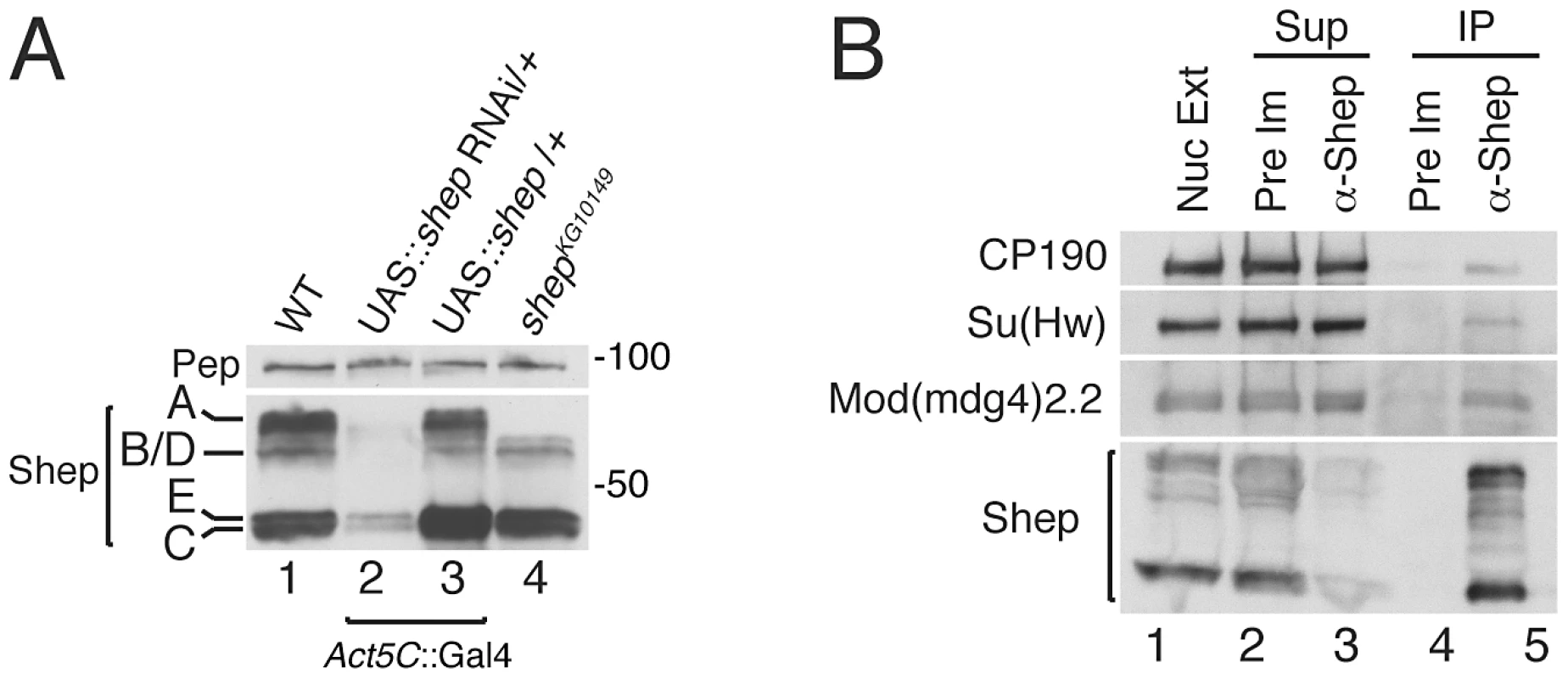
(A) Identification of Shep isoforms in vivo. Western blotting for Shep from larval extracts that are wildtype (lane 1), expressing Act5C::Gal4 driving single copy UAS-shep dsRNA (lane 2), expressing Act5C::Gal4 driving single copy UAS-shep C and E (lane 3), or containing a P-element insertion that disrupts the coding region of isoform A (lane 4). Pep is shown as a loading control. (B) Coimmunoprecipitation of gypsy insulator proteins with Shep. Embryo nuclear extracts (lane 1) were immunoprecipitated (IP) with either Pre-Immune (Pre Im; lanes 2 and 4) or α-Shep (lanes 3 and 5) serum. Shep, Mod(mdg4)2.2, Su(Hw), and CP190 were detected in nuclear extracts (Nuc Ext), supernatants (Sup) (lanes 2–3) and IPs (lanes 4–5) by Western blotting. Approximately 0.02% CP190, 0.02% Su(Hw), and 0.1% Mod(mdg4)2.2 of total were recovered in the IP. We used our specific Shep antisera to test whether gypsy insulator proteins associate with Shep in vivo by coimmunoprecipitation. When Shep complexes are immunoprecipitated from embryo nuclear extracts using Shep or control preimmune antisera, Shep is efficiently purified with the specific antibody (Figure 2B). Furthermore, a fraction of total gypsy insulator proteins CP190, Su(Hw) and Mod(mdg4)2.2 are detected in the bound fraction in association with Shep. The Polycomb Group (PcG) proteins, Pc and E(z) are not purified in the bound fraction, indicating specificity of the interaction between Shep and insulator proteins (Figure S1). Therefore, these data demonstrate that Shep interacts by direct protein interactions with Mod(mdg4)2.2 and Su(Hw) in vitro and associates with gypsy insulator proteins in vivo.
Identification of shep loss-of-function alleles
Direct physical interaction between Shep and gypsy insulator proteins prompted us to examine the functional relationship between shep and the gypsy insulator. We first obtained and characterized shep alleles bearing either P-element insertions or FRT-derived deletions independently generated from seven different genetic backgrounds [Figure 3A]; [ Table 1; 34,35]. To determine whether these alleles are loss-of-function, we performed quantitative RT-PCR for total and specific shep isoform transcript levels and observed decreases in larvae hemizygous for shep or containing homozygous shep P-element insertions (data not shown). Furthermore, four different homozygous P-element insertions result in loss of Shep protein, two greatly reducing all isoforms (Figure 3B, lanes 2–3) and two eliminating isoform A (lanes 7–8). No changes in Shep protein were observed when P-element alleles are heterozygous (data not shown), suggesting that these mutations are recessive. Additionally, Df(3L)Exel6104 transheterozygous deficiency combinations are viable and retain isoforms C and E, suggesting that isoforms A and B/D are not essential (Figure 3C, lanes 6–7). Other transheterozygous combinations of deficiencies or homozygous deficiencies cause lethality (Table 1), but due to deletion of neighboring essential genes, we cannot determine whether shep itself is essential for viability using these alleles. Importantly, no change in CP190, Su(Hw) or Mod(mdg4)2.2 protein levels is observed in shep mutants relative to wildtype levels (Figure 3B–3C). These data show that P-element insertions and deficiencies decrease Shep protein levels and likely constitute loss-of-function alleles.
Fig. 3. Identification of Shep loss-of-function alleles. 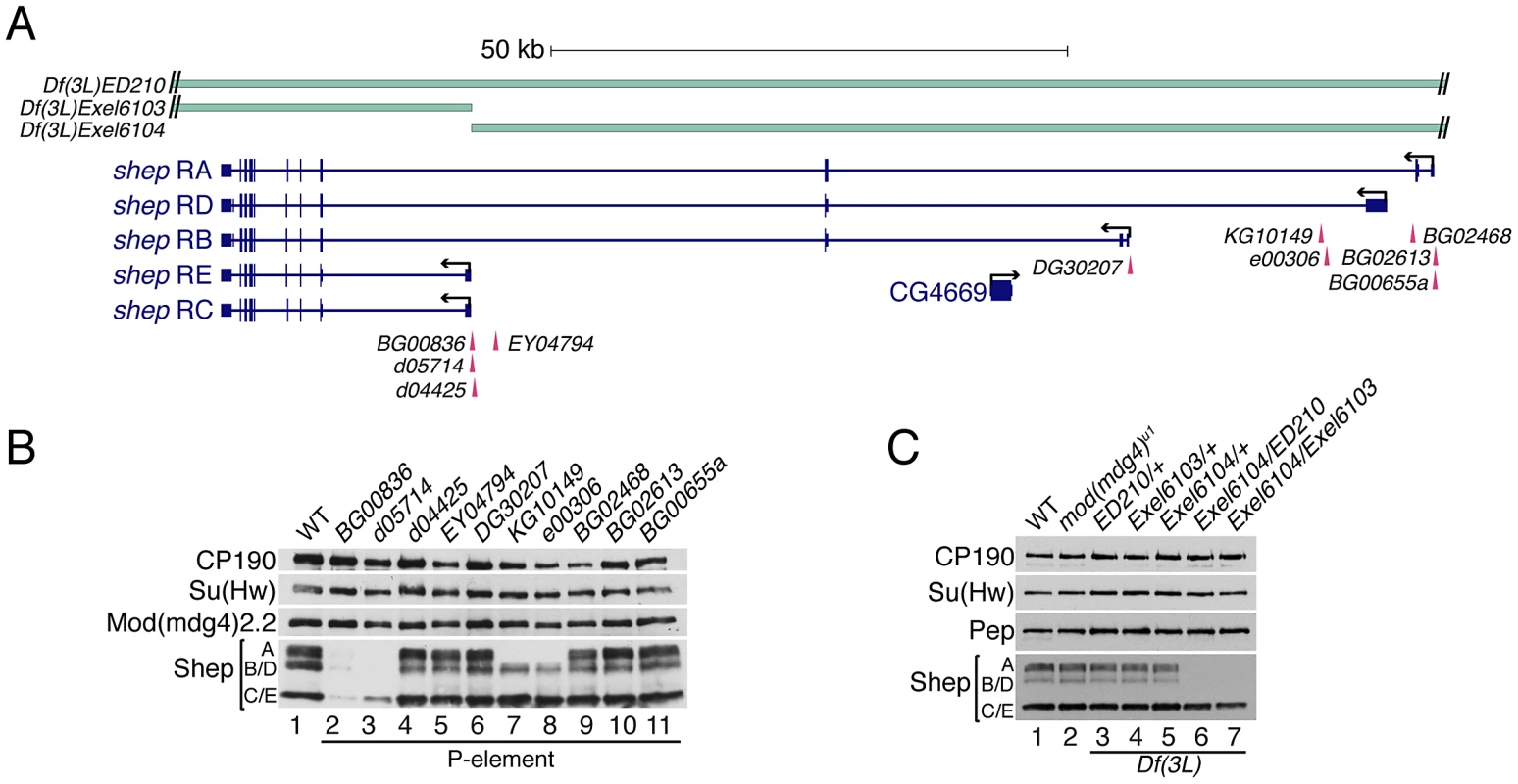
(A) Diagram of lesions in the shep locus. P-element insertion sites are denoted below the gene model, and genomic deficiencies are indicated above the gene model. Hatched lines indicate that deletions extend beyond the shep locus. See Table 1 for P-element details. (B) Western blotting of larval extracts of mod(mdg4)+ and homozygous shep P-element insertion larval extracts for Shep, Mod(mdg4)2.2, Su(Hw), and CP190 in the mod(mdg4)+ background. Lane numbers of gel are indicated. (C) Western blotting for CP190, Su(Hw), Pep, and Shep in larval extracts of mod(mdg4)+, mod(mdg4)u1, and heterozygous or transheterozygous shep deficiencies in the mod(mdg4)u1 background. Tab. 1. Summary of shep homozygous P element and heterozygous deficiency alleles. 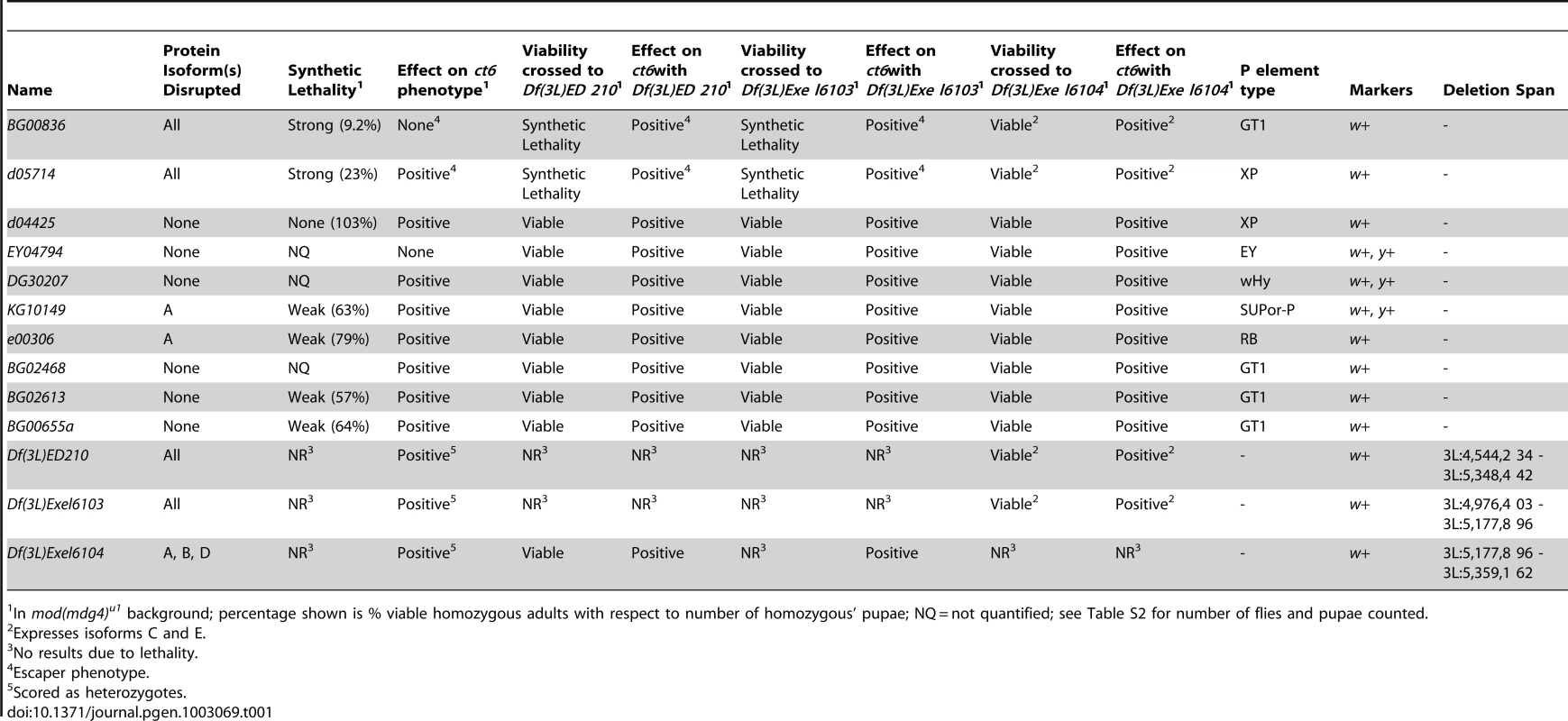
In mod(mdg4)u1 background; percentage shown is % viable homozygous adults with respect to number of homozygous' pupae; NQ = not quantified; see Table S2 for number of flies and pupae counted. Synthetic lethal relationships between mod(mdg4) and shep
We observed that mod(mdg4) mutants are particularly sensitive to shep expression levels. Homozygous shep P-element insertion alleles are viable in a wildtype background; however, in combination with mod(mdg4)u1, which is fully viable but null for the mod(mdg4)2.2 isoform, homozygous shep mutants displaying reduced Shep protein specifically exhibit strongly reduced viability (Table 1). We observed lethality in late pupal development and pharate adults; only 9.2% of shepBG00836 and 23% of shepd05714 mod(mdg4)u1 double mutant pupae survive to adulthood. Synthetic lethality was also observed for shep mutant alleles in combination with the mod(mdg4)T6 loss-of-function point mutation, confirming the genetic interaction. Moreover, overexpression of the shepEY04794 allele containing a UAS insertion or the Shep E isoform from a transgenic copy inserted on a different chromosome using the Act5C::Gal4 driver causes complete inviability of adult flies in the mod(mdg4)u1 background but not in wild type. In contrast, overexpression of the Shep E isoform harboring point mutations in the RRM domain designed to disrupt RNA-binding activity but not protein folding does not cause lethality in mod(mdg4)u1 flies despite both versions of Shep E protein being expressed at the same levels in wildtype flies (data not shown). The apparent sensitivity of mod(mdg4)u1 null mutants to alterations in Shep levels is consistent with direct physical interactions between Shep and insulator proteins and further suggests an antagonistic functional relationship between Mod(mdg4)2.2 and Shep, likely requiring Shep RNA-binding activity.
Shep negatively regulates gypsy enhancer blocking activity
In order to assess whether shep loss-of-function affects insulator activity in vivo, we examined the phenotypes of two well-characterized gypsy-dependent alleles, y2 and ct6. These alleles result from gypsy retrotransposon insertion between the upstream body enhancer and promoter of y or between the upstream distal wing margin enhancer and promoter of ct [36]. These insertions block enhancer function, resulting in loss of abdominal cuticle pigmentation or misshapen wing margin, respectively. In an otherwise wildtype background, shep P-element alleles and deficiencies produce no decrease in enhancer blocking activity at y2 or ct6 (data not shown), and since y2 and ct6 are fully active for enhancer blocking, an increase in insulator activity cannot be assessed. In order to sensitize the assay, y2 and ct6 were examined in the presence of the mod(mdg4)u1 mutation. This mutation disrupts insulator function and allows partial restoration of enhancer-promoter communication. The gypsy-dependent phenotypes in homozygous shep P-element alleles in the mod(mdg4)u1 background were scored for ct6 on a scale of 0–4 with increasing severity of phenotype. Approximately half of male mod(mdg4)u1 wings display a score of zero (Figure 4A). In contrast, for eight of ten homozygous P-element and all heterozygous deficiency alleles of shep, we observed positive effects on enhancer blocking activity at ct6 in the mod(mdg4)u1 background (Figure 4A, Table 1, Table S1), indicating increased gypsy insulator activity. For shepBG00836 and shepd05714 mod(mdg4)u1 double homozygous mutants, only escapers could be scored due to synthetic lethality. Similar changes in insulator phenotypes were observed for shep hemizygous mutations (Figure 4B) but not heterozygous mutations in the mod(mdg4)u1 background (data not shown), indicating that these shep mutations are recessive with respect to insulator activity.
Fig. 4. Loss-of-function shep alleles disrupt gypsy insulator activity at ct6. 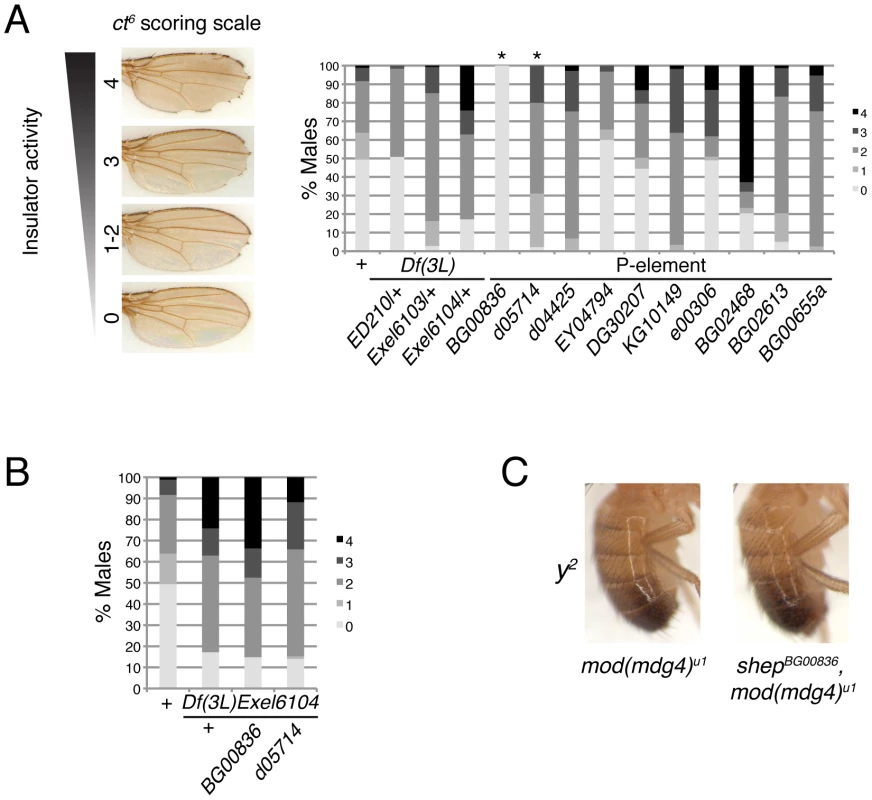
(A) Effects of shep mutations on the ct6 phenotype. All flies are homozygous for mod(mdg4)u1. At the shep locus, flies are wildtype (shep+), harbor a heterozygous deficiency, or contain a homozygous P-element insertion as indicated. Percent of population scored on a scale of 0–4 is indicated for each genotype. 0, no notching; 1, slight notching in one wing; 2, slight notching in both wings; 3, pronounced notching in hinge distal wing margin; 4, severe notching in both hinge proximal and distal margins. Asterisks denote P-element insertions showing extensive synthetic lethal interaction with mod(mdg4)u1 for which rare escapers were scored (49≤n≤180 for all genotypes). (B) Hemizygous alleles of shep affect ct6. Phenotypes of ct6 of shepBG00836 and shepd05714 mutations transheterozygous with Df(3L)Exel6104. All flies are homozygous for mod(mdg4)u1. Flies were scored in parallel with those in (A) (85≤n≤180). (C) Male abdominal pigmentation due to y2 expression is unchanged in mod(mdg4)u1 compared to shepBG00836, mod(mdg4)u1 flies. To verify that the P-element insertion alleles are loss-of-function for enhancer blocking activity, the insulator phenotypes of each shep P-element allele crossed to each deficiency were examined. We found that insulator phenotypes and synthetic lethality remained the same or insulator function was slightly increased compared to homozygous P-elements, except when shepBG00836 and shepd05714 are transheterozygous with Df(3L)Exel6104 (Table 1). In these cases, synthetic lethality is rescued, corresponding to elevated isoform C and E transcript and protein levels likely due to artificial juxtaposition of the C and E promoter to a cis-regulatory element from a partially deleted upstream gene or mini-w+of the original P-element remaining after FRT excision (data not shown). Nevertheless, insulator activity of these shepBG00836 and shepd05714 transheterozygous mutants is improved compared to mod(mdg4)u1, confirming that shepBG00836 and shepd05714 are loss-of-function alleles (Figure 4B).
We determined that shep P-element mutants in the mod(mdg4)u1 background do not affect the phenotype of ctn, caused by insertion of a roo transposable element (data not shown). This result suggests that the effect of shep on ct6 is due to changes in gypsy insulator activity and not direct regulation of ct expression. Importantly, since shep mutants affect insulator activity in mod(mdg4)u1 null mutants, it likely that, in vivo, Shep can interact with Su(Hw) in the absence of Mod(mdg4)2.2. Overall, these data indicate that the wildtype function of Shep is to negatively regulate gypsy insulator activity.
In contrast to positive effects on ct6, shep mutations in the mod(mdg4)u1 background do not affect y2. The phenotype of y2 remained unchanged by mutation or deletion of shep in the mod(mdg4)u1 background (Figure 4C, data not shown). The specific effect at ct6 but not y2 in shep mutants raises the possibility that shep negatively regulates a subset of gypsy insulators.
Shep alters gypsy insulator localization in a tissue-specific manner
In order to determine how Shep regulates insulator function and in what contexts, we examined the distribution of Shep in late stage wildtype embryos. We find that Shep protein is enriched in the embryonic CNS including the brain and ventral nerve cord, areas that are also positive for the neuron-specific protein Elav (Figure 5A). The overlap between Shep and Elav is partial in that Shep is also expressed in glial cells. Shep levels are low but detectable in non-CNS tissues; likewise, microarray expression data from various developmental stages are consistent with our results [37]. In the third instar larval stage, higher overall protein levels are detected in the brain compared to eye, leg, or wing imaginal discs or salivary glands by Western blotting (Figure 5B) as well as immunofluorescence (data not shown). These data demonstrate that Shep is a CNS-enriched protein at both embryonic and larval stages.
Fig. 5. Shep negatively regulates gypsy activity in the CNS. 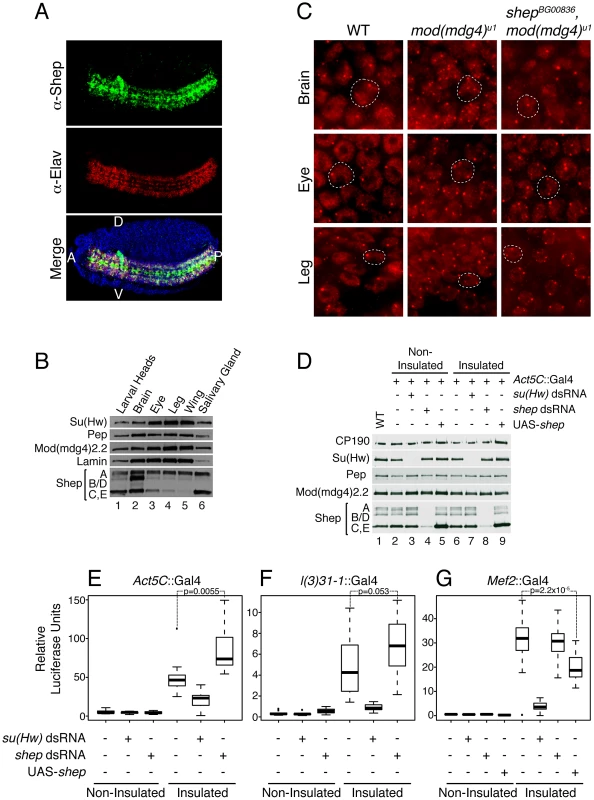
(A) Confocal imaging of Shep distribution in stage 14 wildtype Oregon R embryo by indirect immunofluorescence using guinea pig α-Shep (green) and mouse α-Elav (red) antibodies detected by α-guinea pig Alexa-488 and α-mouse Alexa-594 secondary antibodies. DAPI staining (blue) is also shown in the merged image. A, anterior; P, posterior; D, dorsal; V, ventral. (B) Western blotting of anterior third instar larval extracts (lane 1), brains (lane 2), eye discs (lane 3), leg discs (lane 4), wing discs (lane 5), and salivary glands (lane 6) for Shep, Su(Hw), Mod(mdg4)2.2, Pep, and Lamin. (C) Epifluorescence imaging of insulator body localization by indirect immunofluorescence using rabbit α-CP190 and α-rabbit Alexa-594 in whole mount brain, leg imaginal disc, or eye imaginal disc tissues in wild type; mod(mdg4)u1; or shepBG00836, mod(mdg4)u1 larvae. White dotted lines outline one example nucleus in each image. (D) Western blotting of larval extracts for Shep, Su(Hw), CP190, Mod(mdg4)2.2 and Pep in wildtype (lane 1), non-insulated (lanes 2–5), and insulated (lanes 6–9) luciferase lines. Act5c::Gal4 was used to drive single copy UAS-su(Hw) dsRNA (lanes 3 and 7), UAS-shep dsRNA (lanes 4 and 8) or Shep overexpression (UAS-shep, lanes 5 and 9). (E–G) Relative luciferase units were quantified in individual larvae expressing Act5C::Gal4 (E), l(3)31-1::Gal4 (F) Mef2::Gal4 (G), dsRNA hairpin, and/or UAS-shep as indicated. Luciferase values across the population are plotted as box and whisker plots where boxes represent upper and lower quartiles proximal to the median, and whiskers represent the range excluding outliers. Populations were compared by 1-way ANOVA, and pairwise p values were calculated by Tukey HSD post hoc tests. Outliers falling outside a normal distribution are shown (dots) but were not used to calculate p values. For each genotype, n≥12 larvae. For (F), non-insulated control vs. non-insulated shep RNAi, p = 0.18; for (G), insulated control vs. insulated shep RNAi, p = 0.99. In order to examine whether Shep affects insulator complexes in a tissue-specific manner, we examined the localization of insulator bodies in the presence and absence of Shep in larval brain compared to non-CNS cell types. Wild type, mod(mdg4)u1 and double mutant shepBG00836, mod(mdg4)u1 whole mount larval brain and imaginal disc tissues were stained using antibodies directed against CP190. Because the brain contains heterogeneous cell types, we focused on peripheral cells in the medulla of the brain lobe in which 1–2 insulator bodies are visible in the nucleus per focal plane. In mod(mdg4)u1 mutants, insulator bodies are disrupted in all tissues including the brain, resulting in an increased number of foci compared to wild type (Figure 5C). In shepBG00836, mod(mdg4)u1 double mutants, insulator body localization in the brain reverts to a wildtype appearance (observed in 8 of 9 experiments). The same effect is also observed in perineurial glia of the outer cell layer surrounding the brain hemispheres (data not shown). In contrast, peripheral cells of the eye and leg imaginal discs, which display low Shep expression, insulator bodies are indistinguishable in shepBG00836, mod(mdg4)u1 compared to mod(mdg4)u1 mutants. We also did not observe differences in CP190 localization in peripheral cells of the wing imaginal disc; however, insulator bodies in all genotypes are less prominent in this tissue type (data not shown). Additional shep mutants examined, shepKG10149, shepe00306, shepBG00655a, and shepBG02613, display similar effects (data not shown). Restoration of mislocalized insulator bodies when shep levels are reduced in the brain but not non-CNS tissue suggests a tissue-specific role for Shep in disrupting insulator activity.
Shep represses insulator barrier activity in CNS tissue
In order to determine whether Shep affects insulator activity in the CNS, we developed a versatile barrier assay that allows quantification of gypsy insulator activity using identical reporters in essentially any tissue of interest. This assay relies on three transgenes: the transcriptional reporter UAS-luciferase inserted into a defined attP landing site, either insulated by flanking Su(Hw) binding sites or non-insulated [38]; a Gal4-inducible dsRNA hairpin construct for knockdown of a gene of interest [39]; and a tissue-specific Gal4 driver. This system allows for directly comparable quantification of luciferase activity in the insulated or non-insulated context in the presence or absence of a protein of interest. Use of the Gal4 system allows interrogation of a specific subset of cells for both the reporter as well as the hairpin knockdown within an otherwise wildtype organism, which is not easily achieved using standard genetic manipulation of existing mutants. We used luciferase reporter constructs inserted into attP3 on the X chromosome [40], which display extremely low basal expression unless insulated (Figure 5E–5G) relative to other attP insertion sites tested [38]. Insulator-dependent expression at attP3 is likely due to its positioning within a PcG repressed region (Figure S2). Addition of insulators flanking the UAS-luciferase reporter likely stops the spread of repressive chromatin, allowing for measurable activity. Due to high variability of expression among individuals, luciferase levels were measured in individual whole third instar larvae, and values for each population (n≥12) were compared by one-way ANOVA. As proof of principle, ubiquitously expressed Act5C::Gal4 induces high luciferase activity in insulated compared to non-insulated lines (Figure 5E). As expected, su(Hw) knockdown causes a drastic reduction in both Su(Hw) protein (Figure 5D, lanes 3 and 7) and luciferase activity in insulated but not non-insulated lines (Figure 5E), indicating that luciferase expression directly reports Su(Hw)-mediated insulation. In contrast, upon shep knockdown (Figure 5D, lanes 4 and 8) an increase in luciferase activity is observed for the insulated line (p = 0.0055, Tukey's HSD post hoc test), indicating an increase in insulator activity (Figure 5E). Therefore, Shep negatively influences both gypsy-dependent barrier and enhancer blocking activities.
Since ubiquitous knockdown of shep could report an increase in insulator activity in any or all tissues, CNS-specific Gal4 expression was utilized to quantitatively address whether shep affects gypsy insulator activity in the CNS. Localized Gal4 expression in the CNS with l(3)31-1::Gal4 induces luciferase to a lower level than ubiquitous Gal4 due to its restricted expression pattern (Figure 5F). Upon su(Hw) knockdown in the CNS, luciferase expression returns to non-insulated levels. In contrast, when shep is knocked down, a marginally significant increase in luciferase levels is observed (p = 0.053), demonstrating that shep negatively affects insulator activity in the CNS.
Finally, we tested whether Shep affects barrier activity in muscle cells, a tissue type that expresses low levels of Shep. Muscle-specific Mef2::Gal4 induces high levels of luciferase activity; accordingly, su(Hw) knockdown results in a dramatic decrease in luciferase activity (Figure 5G). In contrast, shep knockdown in muscle tissue has no significant effect compared to Mef2::Gal4 alone (p = 0.99), demonstrating that shep does not play a substantial role in insulator activity in muscle tissue. However, ectopic overexpression of Shep C and E using shepEY04794 in muscle tissue is sufficient to result in decreased insulator activity (p = 2.2×10−5). Therefore, in muscle cells, artificially reaching a certain threshold of Shep protein expression reduces insulator activity. This quantitative and tissue-specific insulator assay further supports a role for Shep as a negative regulator of gypsy insulator activity.
Comparison of Shep and gypsy insulator protein genome-wide localization
In order to determine the extent to which Shep colocalizes with insulator proteins, we mapped the genome-wide chromatin association profiles of Su(Hw), Mod(mdg4)2.2, and Shep by ChIP-seq in the BG3 larval CNS-derived cell line. Using previously characterized Su(Hw) and Mod(mdg4)2.2 antibodies [16], [41], [42] as well as our specific Shep antisera (see methods), we observe sharp peaks of Su(Hw), Mod(mdg4)2.2, and Shep, as well as broader peaks of Shep signal (Figure 6A–6B). Using the SPP algorithm [43] at a 1% false discovery rate (FDR), we detected 4099 Su(Hw) peaks, 1575 Mod(mdg4)2.2 peaks, and 4443 Shep peaks (Figure 6C), numbers in agreement with previous studies of Su(Hw) and Mod(mdg4)2.2 binding profiles in various cell types [9], [10], [42]. Similar to previous studies [10], [42], [44], the majority of Mod(mdg4)2.2 sites overlap with Su(Hw), and strong enrichment of overlap is observed compared to random expectation (Figure 6D). As expected, Su(Hw) is found mostly in inter - and intragenic regions [9], [10], [44] (Figure 6C). In contrast, Shep binding is mainly observed over genes, with 65% of Shep peaks falling in transcription start sites (TSSs). An intermediate distribution pattern is observed for Mod(mdg4)2.2.
Fig. 6. Comparison of Su(Hw), Mod(mdg4)2.2, and Shep ChIP–seq profiles in BG3 cells. 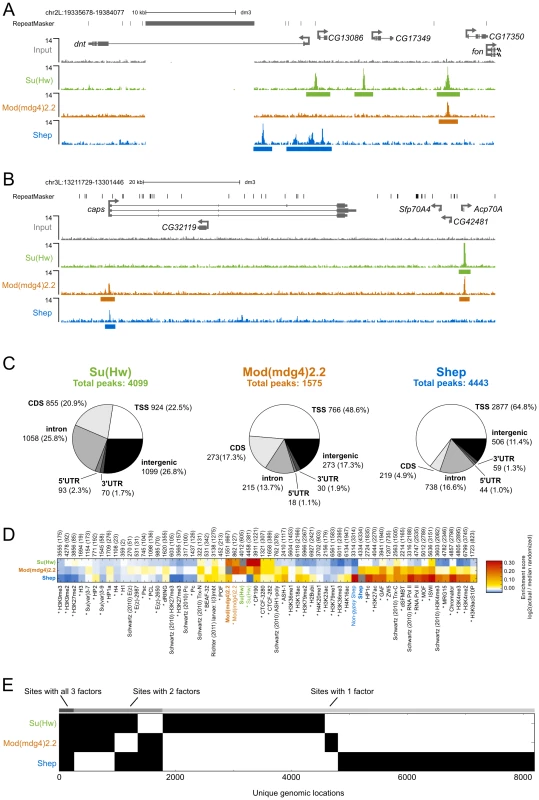
(A) Screenshot of Su(Hw), Mod(mdg4)2.2, and Shep ChIP-seq signals at the dnt neuronal-expressed locus. The large gap in ChIP signal corresponds to a highly repetitive region to which sequence reads could not be aligned with high confidence. (B) Screenshot of the caps neuronal-expressed locus. (C) Classification of Su(Hw), Mod(mdg4)2.2, and Shep ChIP-seq peaks in BG3 cells. Number of sites and percentage of total in parentheses corresponding to TSS, transcription start site; CDS, coding sequence; 5′ UTR, 5′ untranslated region; 3′ UTR, 3′ untranslated region. See methods for classification hierarchy of overlapping categories. (D) Heat map of log2 enrichment scores for pairwise comparisons of binding sites for Su(Hw), Mod(mdg4)2.2, Shep, and additional data sets. Color scale corresponding to enrichment value is indicated (right). Positive values indicate significant enrichment while negative values indicate significant negative correlation of enrichment. Self-self comparisons are indicated in grey, and pairwise comparisons that are not statistically significant (p>0.001) are indicated in white. Numbers along top of each column indicate the total number of features in each data set, and the number of sites overlapping with Shep are indicated in parentheses. Data from Richter (2011) were derived from larval brains and imaginal discs; all other datasets are derived from BG3 cells. Data from modENCODE are indicated by an asterisk. Full heat map with hierarchical clustering is shown in Figure S4. (E) Binary heat map of Su(Hw), Mod(mdg4)2.2, and Shep binding sites in BG3 cells ordered by supervised hierarchical clustering. Each row represents a single genomic location, and a mark in a column represents the presence of a particular factor. Given that Shep can interact directly with either Su(Hw) or Mod(mdg4)2.2 and copurifies with a fraction of total gypsy insulator core proteins, we expected a substantial degree of overlap between Shep and either Su(Hw) or Mod(mdg4)2.2. Indeed, nearly half of Mod(mdg4)2.2 sites overlap with Shep, and 16% of Shep sites overlap with Mod(mdg4)2.2 (Figure 6E). The observed overlap between Shep and Mod(mdg4)2.2 is greater than random expectation (Figure 6D). In contrast, no enrichment is observed for colocalization between Shep and Su(Hw). Nevertheless, nearly one quarter of Shep binding sites overlap with either Su(Hw) or Mod(mdg4)2.2 (Figure 6E), supporting the notion that a substantial fraction of chromatin-associated Shep harbors insulator-related activity. Although expressed at low levels in salivary glands, Shep localization in polytene chromosomes also shows partial overlap between Shep and gypsy insulator proteins (Figure S3). Chromatin association of Shep at non-gypsy insulator sites could reflect alternate unknown functions of Shep or a gypsy insulator-independent means of recruitment.
We next compared Shep genome-wide localization with that of a variety of chromatin-associated factors and histone modification marks in BG3 cells. Enrichment scores for two-way overlaps between all factors were calculated, and unsupervised hierarchical clustering was performed (Figure S4). This analysis reveals high similarity of binding profiles of the insulator proteins Su(Hw), Mod(mdg4)2.2, CP190, and CTCF (Figure 6D). In contrast, Shep genome-wide localization most closely resembles factors associated with active transcription such as RNA polymerase II. Analysis of Shep sites not overlapping with either Su(Hw) or Mod(mdg4)2.2 also overlap significantly with active transcription marks. Consistent with our comparative analysis, Shep localization is likewise observed at highly transcribed puff regions of polytene chromosomes (Figure S3). Interestingly, Shep genome-wide localization also displays similarity to that of Chromator, a protein recently implicated as a boundary factor potentially capable of organizing physical chromatin domains [25] and also overlaps significantly with CP190 and BEAF (Figure 6D).
Consistent with Shep functioning as a negative regulator of gypsy insulator activity, we noted a significantly lower than expected frequency of three-way overlap among Shep, Su(Hw) and Mod(mdg4)2.2. In fact, the three factors are only observed together at 271 sites (Figure 6E). Considering the 1403 Mod(mdg4)2.2 sites that colocalize with either Su(Hw) or Shep, this degree of three-way overlap is lower than expected by chance (p<1×10−4, permutation test; p = 2.2×10−16, hypergeometric test). The same results are obtained when this analysis is performed on Su(Hw) sites that overlap with either Mod(mdg4)2.2 and Shep as well as the Shep sites that overlap with either Su(Hw) or Mod(mdg4)2.2 (see methods). Taken together, these results indicate substantial colocalization of Shep with Mod(mdg4)2.2 but limited three-way overlap among Shep and both gypsy insulator proteins.
Discussion
Here we have demonstrated a role for the CNS-enriched RRM protein, Shep, in the tissue-specific, negative regulation of gypsy chromatin insulator activity. Shep interacts directly with either Su(Hw) or Mod(mdg4)2.2 in vitro and associates physically with gypsy insulator complexes in vivo. Mutations in shep improve enhancer blocking activity and cause synthetic lethality with mod(mdg4)2.2 mutations. Two lines of evidence indicate that Shep affects insulator activity in a tissue-specific manner. First, insulator body localization is altered in CNS but not other tissues of shep mutants. Second, barrier activity is improved in CNS but not muscle tissue when Shep levels are reduced. Finally, genome-wide mapping of Shep and gypsy insulator proteins in BG3 cells reveals substantial overlap with individual insulator proteins but lack of three-way overlap, further supporting a role for Shep in negative regulation of insulator activity in certain tissues.
Shep negatively regulates gypsy insulator activity in a tissue-specific manner
Shep acts as a tissue-specific negative regulator of gypsy insulator function and insulator body localization. Shep localization is most enriched in the CNS at both embryonic and larval stages; however, it is also expressed at lower levels in additional tissues. Although we have demonstrated that Shep functions in the CNS, Shep can also repress enhancer blocking activity in the wing and could possibly affect insulator activity in other tissues. For example, ubiquitous reduction of Shep levels strongly improves overall barrier activity, suggesting that tissues outside of the CNS may also harbor Shep activity. Nonetheless, Shep does not appear to function in all tissues; knockdown of Shep does not affect barrier activity in muscle tissue, no changes in insulator body localization are observed in eye or leg tissue of shep mutants, and no effect is observed for y2 enhancer blocking in pigment cells of shep mutants. Interestingly, when Shep is overexpressed in muscle tissue, reduction of barrier activity is observed, suggesting that a certain threshold of Shep protein is needed to repress insulator activity. Since Shep protein can be detected at least at low levels in all tissues tested thus far, it is unlikely that the mere presence of Shep protein is sufficient to disrupt gypsy insulator activity. It remains to be determined what other cofactors, such as proteins or RNAs, may contribute to Shep activity.
Shep may negatively regulate insulator activity by interfering with insulator protein interactions required for their activity. ChIP-seq analyses shows that the genome-wide binding profile of Shep in CNS-derived BG3 cells overlaps substantially with that of Mod(mdg4)2.2 but not extensively with both Su(Hw) and Mod(mdg4)2.2 combined. Lack of three-way overlap is not entirely unexpected given that Shep is a negative regulator of gypsy insulator activities. Shep coimmunoprecipitation experiments copurify only a small fraction of total insulator proteins present in nuclear extracts, suggesting that Shep-insulator complexes are not abundant or not stable in vivo. Since Shep can bind either Mod(mdg4)2.2 or Su(Hw) in vitro at a 1∶1 ratio, Shep binding could compete with direct interaction between Mod(mdg4)2.2 and Su(Hw) or their interactions with other factors such as CP190. Moreover, our finding that mod(mdg4) mutants are highly sensitive to Shep dosage suggests an antagonistic functional relationship between Mod(mdg4)2.2 and Shep. Specifically, Shep may negatively regulate higher order insulator-insulator complex interactions, which appear to be mediated by direct interaction between Mod(mdg4)2.2 and CP190 [4]. Insulator body localization in larval brains of shep, mod(mdg4)u1 mutants reverts back to a wildtype pattern compared to compromised mod(mdg4)u1 mutants, perhaps indicating that the normal function of Shep may be to prevent larger insulator complexes from forming in these cell types.
Functional consequences of Shep activity in the CNS
Our results are consistent with the possibility that Shep promotes tissue-specific chromatin configurations by modulating insulator complexes. While differential occupancy of insulator proteins at their respective binding sites may play a role in regulating certain loci [21], occupancy throughout the genome does not differ extensively between cell types [9], [45]. Therefore, alternate mechanisms to control insulator activity likely exist. Shep activity could prevent insulator-insulator contacts otherwise present in tissues that do not express shep, resulting in relief of enhancer blocking or repression by silencers. Interestingly, shep was identified as a regulator of complex behavioral traits in screens for altered sensory-motor responsiveness to gravity [33] and aggressive behavior [46], suggesting the possibility that regulation of an insulator-based mechanism could exist to effect changes in neurological function.
Potential involvement of RNA in Shep and chromatin insulator function
Given that Shep is an RRM-containing protein, RNA-binding may contribute to the ability of Shep to associate with insulator complexes in vivo. Shep RRMs are highly conserved, and lethality caused by Shep overexpression in the mod(mdg4) mutant background is not observed when the RRMs are mutated. This result suggests that Shep RRMs may be functional with respect to insulator activity. One possibility is that the specific RNA bound by Shep could affect targeting of Shep to insulator sites. Another not mutually exclusive prospect is that Shep is recruited to chromatin cotranscriptionally by binding nascent transcripts. It will be important to determine in future studies if Shep binds RNA while in complex with gypsy insulator proteins as well as the identities of Shep and insulator-associated RNA. Our results point to a novel role for Shep and possibly RNA to regulate insulator activity in a tissue-specific manner.
Materials and Methods
Drosophila strains
Stocks were raised at 25°C on standard cornmeal medium. Shep P-element insertion alleles, shep deficiencies, Act5C::Gal4, Mef2::Gal4, and l(3)31-1::Gal4 were obtained from the Bloomington and Exelixis Stock Centers. Lines expressing su(Hw) (10724 GD) or shep dsRNA (37863 GD) were obtained from the Vienna Drosophila RNAi Center. UAS::luciferase constructs were inserted into the attP3 landing site [38]. The ct6 phenotype was scored in flies on the first day after eclosion. For all genotypes, males show a more severe wing notching phenotype than females. The y2 phenotype was scored in flies aged for 1 d at 25°C. Larvae for luciferase insulator assays and whole mount immunofluorescence were raised at 25°C. Larvae for polytene chromosome staining were raised at 18°C. Embryos aged 0–24 h for nuclear extracts and immunofluorescence were collected from a population cage as described [20]. Anterior thirds of larvae were used for Western blotting.
Shep cloning
Coding regions of FlyBase annotated Shep isoforms RA, RB/RD, and RE were amplified by PCR from Trizol (Invitrogen) extracted, Oregon R embryonic cRNA that was oligo-dT primed and reverse transcribed by Superscript III (Invitrogen). The isoform RA clone obtained differs from FlyBase annotations in 2 locations where either a downstream splice site was used, as in isoforms B, D, and E, resulting in 12 extra amino acids and an additional unannotated exon was included resulting in an additional 11 amino acids; RRM domains remain intact in this isoform A variant. The shep, su(Hw), and mod(mdg4)2.2 cDNAs were inserted into pENTR/D-TOPO and recombined into pDEST 15 (Invitrogen) to generate N-terminal GST fusion constructs. All plasmids were sequenced for verification.
Recombinant protein and GST-pulldown
Expression of GST, GST-Shep, GST-Su(Hw), GST-Mod(mdg4)2.2, His-Su(Hw), and His-Mod(mdg4)2.2 was induced in E. coli BL21 cells by 1 mM Isopropyl β-D-1-thiogalactopyranoside at 37° C. Proteins were purified under native conditions by affinity using Glutathione-Agarose (Pierce) or Ni-NTA-Agarose (Qiagen). 2 µg immobilized GST or GST fusion proteins were incubated with 35 µg soluble His-Mod(mdg4)2.2 in PBSMT (137 mM NaCl, 2.7 mM KCl, 10 mM NaH2PO4, 1.8 mM KH2PO4, 250 mM MgCl2, 0.3% Triton X-100 supplemented with Complete protease inhibitors (Roche), 1 mM PMSF, 1 mM DTT, and 100 mg/mL BSA) in a volume of 350 µL. Binding reactions were carried out at 4°C rotating for 2 h. Unbound protein was removed, and beads were washed 5 times in PBSMT. Bound His-Mod(mdg4)2.2 was eluted in sample buffer, separated by SDS-PAGE, and detected by Western blotting. His-Su(Hw) binding reactions were carried out in the same manner except in HBSM (50 mM HEPES, pH 6.7; 150 mM NaCl; 5 mM KCl; 2.5 mM MgCl2) supplemented with 0.3% Triton-X 100, 0.2 M KCl, protease inhibitors, 1 mM PMSF, 1 mM DTT, and 100 mg/mL BSA.
Antibodies and antibody production
6X-His-Shep isoform E (AA169–368), CP190 antigen [4], Su(Hw) antigen [41] and Mod(mdg4)2.2 antigen [47] were expressed in BL21 cells, affinity purified by Ni-NTA-agarose (Qiagen) according to the manufacturer's protocol under denaturing conditions and used to immunize rabbits and guinea pigs using standard procedures (Covance Research Products). For Western blotting, guinea pig α-Shep serum was used at 1∶2000, guinea pig α-CP190 was used at 1∶10,000, guinea pig α-Mod(mdg4)2.2 [41] was used at 1∶1000, guinea pig α-Su(Hw) [41] was used at 1∶7500, α-Pc [32] was used at 1∶1000, α-E(z) [48] was used at 1∶1000, and α-Pep [49] was used at 1∶1000. For insulator body staining, rabbit α-CP190 [4] was used at 1∶30,000. The monoclonal α-Elav9AF89 was obtained from the Developmental Studies Hybridoma Bank and used at 1∶1000, and guinea pig α-Shep serum was used at 1∶200 for IF.
Coimmunoprecipitation
Nuclei from 20 g of embryos were prepared as described [20]. Nuclei were lysed in 4 mL HBSM supplemented with 0.3% TritonX-100 (HBSMT), complete protease inhibitors and 1 mM PMSF by dounce homogenization with the B pestle. Extracts were cleared of insoluble material by centrifugation, and half of the supernatant was incubated with pre-immune serum and half with α-Shep serum pre-conjugated to protein A sepharose. IPs were carried out for 1 h at 4°C, rotating. Unbound protein was removed and beads were washed 4 X in HBSMT and 1 X in HBSM. Bound protein was eluted in sample buffer, separated by SDS-PAGE, and detected by Western blotting.
Immunofluorescence
Polytene chromosome spreads were prepared as described previously [8]. Brains and imaginal discs were dissected from at least 5 larvae of each genotype per experiment and stained as described previously [32]. Chromosomes and discs were imaged using a Leica DM5000B epifluorescent microscope and captured using OpenLab software.
Indirect immunofluorescence of mixed stage Oregon R embryos was carried out as described [50], [51]. Blocked embryos were incubated rotating with primary antibodies overnight at 4°C and secondary antibodies for 2 h at 37°C. After washing, embryos were incubated in DAPI and mounted in 2.5% DABCO (Sigma) in 70% glycerol. Embryos were imaged on a Zeiss 510 confocal microscope.
Luciferase insulator assay
Females homozygous for attP3::UAS-luciferase transgenes were crossed to Gal4 expressing males; luciferase in individual F1 male larvae was quantified. Any homozygous lethal Gal4 lines were selected against GFP expressing balancer chromosomes. Larvae were collected on dry ice and stored at −80°C until use, at which time they were homogenized in 30 µL Glo Lysis buffer (Promega) and incubated at room temperature for 10 min. Debris was cleared from extracts by centrifugation, and 20 µL soluble material was dispensed into opaque 96-well plates; the same volume of luciferase reagent (Promega) was added to each well, and plates were incubated in the dark for 10 min. Light emission was quantified using a Spectramax II Gemini EM plate reader (Molecular Devices). Luciferase values were normalized to total protein determined by Bradford assay carried out in parallel. Luciferase values between genotype populations were log transformed to obtain a normal distribution and compared by one-way ANOVA. Tukey HSD post hoc tests were used to determine pairwise p values between genotypes. For further information including additional Gal4 lines tested, see Text S1.
Cell culture
BG3-c2 cells were grown in S2 medium (Sigma) supplemented with 10% fetal calf serum and 10 µg/mL insulin. Cells were maintained in monolayer at 25°C.
Chromatin immunoprecipitation and ChIP–seq library construction
Cells were fixed in 1% formaldehyde added directly to cells in culture medium for 10 min at RT with gentle agitation; formaldehyde was quenched by addition of glycine to 0.125 M with gentle agitation for 5 min at RT. 5×106 to 107 cells were used per IP. Cells were pelleted at 400 rcf and washed twice in ice cold PBS. Cells were resuspended in 1 mL ice cold cell lysis buffer (5 mM PIPES, pH 8, 85 mM KCl, 0.5% NP-40) supplemented with protease inhibitors, and nuclei were released by Dounce homogenization with the B pestle and pelleted by centrifugation at 9190 rcf for 5 min at 4°C. Nuclei and chromatin were further processed as described [41]. Chromatin was fragmented to an average size of 300 bp by sonication and validated by agarose gel electrophoresis. Sequencing libraries were prepared according to the standard Illumina ChIP-seq protocol. Highly similar profiles were obtained with two independent α-Shep antibodies; therefore, the antibody (guinea pig) displaying the highest signal to noise ratio was utilized for subsequent analyses. Rabbit α-Su(Hw) [16] and rabbit α-Mod(mdg4)2.2 [42] were used for ChIP-seq. Highly similar profiles were obtained with two independent α-Mod(mdg4)2.2 antibodies [41]; therefore, the antibody displaying the highest signal to noise ratio was utilized for subsequent analyses. Libraries were constructed with TruSeq adapters and sequenced on an Illumina HiSeq multiplexed in a single lane. For directed ChIP, quantitative PCR was performed as previously reported [32].
Computational analyses
ChIP–seq
36 bp reads from the Illumina HiSeq 2000 sequencer were mapped to dm3 chromosomes except chrUextra, using Bowtie v0.12.7 with parameters “–best –strata -m1 -n2 –tryhard”. Reads from repetitive regions were removed, and duplicates were removed with MarkDuplicates from Picard 1.49. Peak-calling was performed with SPP [43] using default parameters (e.g., FDR = 0.01, z-threshold = 3) with the exception of “srange = c(50, 200)” when calculating binding site characteristics to improve symmetry of the auto-correlation curve. Broad peak regions were added to binding site point positions, and final peaks were merged. Sequence data are deposited in the Gene Expression Omnibus under accession number GSE40797.
Downstream analyses
Intersections, classification, and randomizations in the below analyses were performed with pybedtools v0.6 [52], gffutils v0.8, and BEDTools v2.16.2 [53].
Pie charts
Feature classes [TSSs (1 bp transcript start position), CDSs, introns, 5′UTRs, and 3′UTRs] were extracted from all annotated isoforms of all annotated genes in FlyBase release 5.33. Intergenic regions were defined as the remainder of dm3. Since a ChIP-seq peak can fall in more than one class, we classified a peak by its highest priority annotation class, where the priorities from highest to lowest are TSS, CDS, intron, 5′UTR, 3′UTR, and intergenic.
Heat maps
Data files containing called peaks were downloaded from GEO and modENCODE [54]–[56] and converted to BED files. Shep peaks that overlapped either a Su(Hw) peak or a Mod(mdg4)2.2 peak by at least one base were filtered out to create a set of non-gypsy Shep peaks. Enrichment scores were calculated as follows: For each pairwise comparison between files A and B, the Jaccard statistic (intersection of bp divided by union of bp; as described previously [57]) was computed to obtain the “actual” statistic. Then, features in file A were shuffled to a random position on the same chromosome, and the Jaccard statistic was again calculated. After 1000 such shufflings, the actual statistic was divided by the median of the empirical distribution to get an enrichment score, (actual+1)/(median randomized+1), for the comparison. The full enrichment matrix was hierarchically clustered using correlation as a distance metric and complete linkage clustering as implemented in SciPy, with rows clustered identically as columns. Selected rows from the full clustered matrix in Figure S4 are shown in Figure 6D.
Colocalization of Su(Hw), Mod(mdg4)2.2, and Shep
To assess the possibility of Shep and Mod(mdg4)2.2 binding mutually exclusively to Su(Hw) sites, we created an N×M binary matrix of binding sites where N = 8194 is the number of binding sites containing any of Su(Hw), Shep, or Mod(mdg4)2.2 (using the pybedtools.contrib.plotting.binary_heatmap() function) and M = 3 for the three factors. We then took the set of 1356 Su(Hw) sites with Shep, Mod(mdg4)2.2, or both, and extracted the Shep and Mod(mdg4)2.2 vectors for these sites representing a total of 663 Shep+Su(Hw) and 964 Mod(mdg4)2.2+Su(Hw) sites. There were 271 Shep+Mod(mdg4)2.2 colocalization events in these vectors. We then randomly shuffled the vectors 10,000 times, computing colocalization each time, and obtained a mean of 472 colocalization events with no iteration giving less than 438 colocalization events. Therefore, of the Su(Hw) sites containing either Shep or Mod(mdg4)2.2, the actual Shep+Mod(mdg4)2.2 overlap of 271 suggests Shep, Mod(mdg4)2.2, and Su(Hw) colocalize significantly less often than expected (empirical p<1×10−4). These results are also consistent with a hypergeometric test using n = 1356, n1 = 663, n2 = 964, and m = 271 (p = 2.2×10−16).
The same analysis was performed for 1403 Mod(mdg4)2.2 sites with Su(Hw), Shep, or both as well as 964 Mod(mdg4)2.2+Su(Hw) and 710 Mod(mdg4)2.2+Shep extracted sites. The mean of randomized iterations was 488 and none had less than 454 colocalization events (empirical p<1×10−4) and (p = 2.2×10−16, hypergeometric test).
For 1102 Shep sites with Su(Hw), Mod(mdg4)2.2, or both as well as 710 Shep+Mod(mdg4)2.2 and 663 Shep+Su(Hw) extracted sites. The mean of randomized iterations was 427 and none had less than 393 colocalization events (empirical p<1×10−4) and (p = 2.2×10−16, hypergeometric test).
Supporting Information
Zdroje
1. GasznerM, FelsenfeldG (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7 : 703–713.
2. PhillipsJE, CorcesVG (2009) CTCF: master weaver of the genome. Cell 137 : 1194–1211.
3. KimJ, ShenB, RosenC, DorsettD (1996) The DNA-binding and enhancer-blocking domains of the Drosophila suppressor of Hairy-wing protein. Mol Cell Biol 16 : 3381–3392.
4. PaiCY, LeiEP, GhoshD, CorcesVG (2004) The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16 : 737–748.
5. GerasimovaTI, GdulaDA, GerasimovDV, SimonovaO, CorcesVG (1995) A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82 : 587–597.
6. CaiHN, LevineM (1997) The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J 16 : 1732–1741.
7. GeyerPK, CorcesVG (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6 : 1865–1873.
8. GerasimovaTI, ByrdK, CorcesVG (2000) A chromatin insulator determines the nuclear localization of DNA. Mol Cell 6 : 1025–1035.
9. BusheyAM, RamosE, CorcesVG (2009) Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23 : 1338–1350.
10. NegreN, BrownCD, ShahPK, KheradpourP, MorrisonCA, et al. (2010) A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6: e1000814 doi:10.1371/journal.pgen.1000814.
11. van BemmelJG, PagieL, BraunschweigU, BrugmanW, MeulemanW, et al. (2010) The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE 5: e15013 doi:10.1371/journal.pone.0015013.
12. GolovninA, VolkovI, GeorgievP (2012) SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J Cell Sci 125 : 2064–2074.
13. GerasimovaTI, CorcesVG (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92 : 511–521.
14. GhoshD, GerasimovaTI, CorcesVG (2001) Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J 20 : 2518–2527.
15. GolovninA, MelnikovaL, VolkovI, KostuchenkoM, GalkinAV, et al. (2008) ‘Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep 9 : 440–445.
16. GerasimovaTI, LeiEP, BusheyAM, CorcesVG (2007) Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell 28 : 761–772.
17. CapelsonM, CorcesVG (2005) The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell 20 : 105–116.
18. RamosE, TorreEA, BusheyAM, GurudattaBV, CorcesVG (2011) DNA topoisomerase II modulates insulator function in Drosophila. PLoS ONE 6: e16562 doi:10.1371/journal.pone.0016562.
19. CapelsonM, CorcesVG (2006) SUMO conjugation attenuates the activity of the gypsy chromatin insulator. EMBO J 25 : 1906–1914.
20. LeiEP, CorcesVG (2006) RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38 : 936–941.
21. WoodAM, Van BortleK, RamosE, TakenakaN, RohrbaughM, et al. (2011) Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell 44 : 29–38.
22. Lieberman-AidenE, van BerkumNL, WilliamsL, ImakaevM, RagoczyT, et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326 : 289–293.
23. DixonJR, SelvarajS, YueF, KimA, LiY, et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485 : 376–380.
24. HandokoL, XuH, LiG, NganCY, ChewE, et al. (2011) CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43 : 630–638.
25. SextonT, YaffeE, KenigsbergE, BantigniesF, LeblancB, et al. (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148 : 458–472.
26. SplinterE, HeathH, KoorenJ, PalstraRJ, KlousP, et al. (2006) CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 20 : 2349–2354.
27. XuZ, WeiG, ChepelevI, ZhaoK, FelsenfeldG (2011) Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat Struct Mol Biol 18 : 372–378.
28. SimonisM, KlousP, SplinterE, MoshkinY, WillemsenR, et al. (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38 : 1348–1354.
29. HouC, DaleR, DeanA (2010) Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A 107 : 3651–3656.
30. LiHB, MullerM, BahecharIA, KyrchanovaO, OhnoK, et al. (2011) Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol 31 : 616–625.
31. WangKC, YangYW, LiuB, SanyalA, Corces-ZimmermanR, et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472 : 120–124.
32. MoshkovichN, NishaP, BoylePJ, ThompsonBA, DaleRK, et al. (2011) RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev 25 : 1686–1701.
33. ArmstrongJD, TexadaMJ, MunjaalR, BakerDA, BeckinghamKM (2006) Gravitaxis in Drosophila melanogaster: a forward genetic screen. Genes Brain Behav 5 : 222–239.
34. BellenHJ, LevisRW, LiaoG, HeY, CarlsonJW, et al. (2004) The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 : 761–781.
35. RyderE, BlowsF, AshburnerM, Bautista-LlacerR, CoulsonD, et al. (2004) The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167 : 797–813.
36. GdulaDA, GerasimovaTI, CorcesVG (1996) Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci U S A 93 : 9378–9383.
37. ChintapalliVR, WangJ, DowJA (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39 : 715–720.
38. MarksteinM, PitsouliC, VillaltaC, CelnikerSE, PerrimonN (2008) Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40 : 476–483.
39. DietzlG, ChenD, SchnorrerF, SuKC, BarinovaY, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 : 151–156.
40. GrothAC, FishM, NusseR, CalosMP (2004) Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166 : 1775–1782.
41. MoshkovichN, LeiEP (2010) HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet 6: e1000880 doi:10.1371/journal.pgen.1000880.
42. Van BortleK, RamosE, TakenakaN, YangJ, WahiJE, et al. (2012) Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res In press.
43. KharchenkoPV, TolstorukovMY, ParkPJ (2008) Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26 : 1351–1359.
44. SchwartzYB, Linder-BassoD, KharchenkoPV, TolstorukovMY, KimM, et al. (2012) Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res In press.
45. KimTH, AbdullaevZK, SmithAD, ChingKA, LoukinovDI, et al. (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128 : 1231–1245.
46. EdwardsAC, ZwartsL, YamamotoA, CallaertsP, MackayTF (2009) Mutations in many genes affect aggressive behavior in Drosophila melanogaster. BMC Biol 7 : 29.
47. MongelardF, LabradorM, BaxterEM, GerasimovaTI, CorcesVG (2002) Trans-splicing as a novel mechanism to explain interallelic complementation in Drosophila. Genetics 160 : 1481–1487.
48. KahnTG, SchwartzYB, DellinoGI, PirrottaV (2006) Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem 281 : 29064–29075.
49. AmeroSA, ElginSC, BeyerAL (1991) A unique zinc finger protein is associated preferentially with active ecdysone-responsive loci in Drosophila. Genes Dev 5 : 188–200.
50. DeFalcoTJ, VerneyG, JenkinsAB, McCafferyJM, RussellS, et al. (2003) Sex-specific apoptosis regulates sexual dimorphism in the Drosophila embryonic gonad. Dev Cell 5 : 205–216.
51. PatelNH (1994) Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol 44 : 445–487.
52. DaleRK, PedersenBS, QuinlanAR (2011) Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 27 : 3423–3424.
53. QuinlanAR, HallIM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 : 841–842.
54. CelnikerSE, DillonLA, GersteinMB, GunsalusKC, HenikoffS, et al. (2009) Unlocking the secrets of the genome. Nature 459 : 927–930.
55. SchwartzYB, KahnTG, StenbergP, OhnoK, BourgonR, et al. (2010) Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 6: e1000805 doi:10.1371/journal.pgen.1000805.
56. RichterC, OktabaK, SteinmannJ, MullerJ, KnoblichJA (2011) The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat Cell Biol 13 : 1029–1039.
57. FavorovA, MularoniL, CopeLM, MedvedevaY, MironovAA, et al. (2012) Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput Biol 8: e1002529 doi:10.1371/journal.pcbi.1002529.
58. SchuettengruberB, GanapathiM, LeblancB, PortosoM, JaschekR, et al. (2009) Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13 doi:10.1371/journal.pbio.1000013.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



