-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaLong Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
Considerable evidence now supports the idea that the moderate telomere lengthening produced by recombinational telomere elongation (RTE) in a Kluyveromyces lactis telomerase deletion mutant occurs through a roll-and-spread mechanism. However, it is unclear whether this mechanism can account for other forms of RTE that produce much longer telomeres such as are seen in human alternative lengthening of telomere (ALT) cells or in the telomerase-resistant type IIR “runaway” RTE such as occurs in the K. lactis stn1-M1 mutant. In this study we have used mutationally tagged telomeres to examine the mechanism of RTE in an stn1-M1 mutant both with and without telomerase. Our results suggest that the establishment stage of the mutant state in newly generated stn1-M1 ter1-Δ mutants surprisingly involves a first stage of sudden telomere shortening. Our data also show that, as predicted by the roll-and-spread mechanism, all lengthened telomeres in a newly established mutant cell commonly emerge from a single telomere source. However, in sharp contrast to the RTE of telomerase deletion survivors, we show that the RTE of stn1-M1 ter1-Δ cells produces telomeres whose sequences undergo continuous intense scrambling via recombination. While telomerase was not necessary for the long telomeres in stn1-M1 cells, its presence during their establishment was seen to interfere with the amplification of repeats via recombination, a result consistent with telomerase retaining its ability to add repeats during active RTE. Finally, we observed that the presence of active mismatch repair or telomerase had important influences on telomeric amplification and/or instability.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003017
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003017Summary
Considerable evidence now supports the idea that the moderate telomere lengthening produced by recombinational telomere elongation (RTE) in a Kluyveromyces lactis telomerase deletion mutant occurs through a roll-and-spread mechanism. However, it is unclear whether this mechanism can account for other forms of RTE that produce much longer telomeres such as are seen in human alternative lengthening of telomere (ALT) cells or in the telomerase-resistant type IIR “runaway” RTE such as occurs in the K. lactis stn1-M1 mutant. In this study we have used mutationally tagged telomeres to examine the mechanism of RTE in an stn1-M1 mutant both with and without telomerase. Our results suggest that the establishment stage of the mutant state in newly generated stn1-M1 ter1-Δ mutants surprisingly involves a first stage of sudden telomere shortening. Our data also show that, as predicted by the roll-and-spread mechanism, all lengthened telomeres in a newly established mutant cell commonly emerge from a single telomere source. However, in sharp contrast to the RTE of telomerase deletion survivors, we show that the RTE of stn1-M1 ter1-Δ cells produces telomeres whose sequences undergo continuous intense scrambling via recombination. While telomerase was not necessary for the long telomeres in stn1-M1 cells, its presence during their establishment was seen to interfere with the amplification of repeats via recombination, a result consistent with telomerase retaining its ability to add repeats during active RTE. Finally, we observed that the presence of active mismatch repair or telomerase had important influences on telomeric amplification and/or instability.
Introduction
Recombination can maintain telomeres in many situations where telomerase is absent. Natural examples of this include the chromosomal telomeres in the mosquito Anopheles [1]–[2] and the mitochondrial telomeres in certain ciliates and yeasts [3]–[5]. Of particular importance are the 5–10% of human cancer cells where telomerase activity is undetectable and telomeres are maintained by a mechanism termed Alternative Lengthening of Telomeres (ALT) (for a review, see [6]). ALT cells are characterized by long and heterogeneous telomeres [7]–[10] and the presence of ALT-associated PML bodies (APB) that contain telomeric DNA as well as telomeric and recombinational proteins [8], [11]–[14].
Several lines of evidence suggest that recombination is involved in maintaining telomeres when the long and heterogeneous telomeres already exist in ALT cells. Plasmid tags introduced into a telomere can be duplicated to other telomeres or at the same telomere in ALT cells but not in telomerase positive cells [15]–[16]. Extrachromosomal telomeric circles (t-circles), likely products of intratelomeric recombination, are abundant in ALT cells [17]–[19]. Telomeric sister chromatid exchanges (t-SCE) occur at highly elevated rates in ALT cells [20]–[22]. However, the details of how recombination can establish these long and heterogeneous telomeres from normal-length telomeres in ALT cells are still unknown.
Recombinational telomere elongation (RTE) has been described in yeast mutants lacking telomerase in the species Sacchromyces cerevisiae [23], Kluyveromyces lactis [24], Candida albicans [25] and Schizosacchromyces pombe [26]. The recombination in these cases is thought to be caused by the shortening telomeres eventually losing part or all of their protective capping function. These mutants commonly display gradual growth senescence when telomeres are gradually shortening that is followed by the formation of better growing post-senescence survivors with longer telomeres [23]–[24], [27]. Two types of RTE were initially described in telomerase deletion mutants of S. cerevisiae. Both depend upon RAD52, suggesting that they require homologous recombination (HR). Type I RTE is characterized by amplification of subtelomeric Y′ elements and short tracts of telomeric repeats, and is dependent upon the canonical mitotic HR pathway involving RAD51, RAD54, RAD55, and RAD57. Type II RTE is characterized by lengthened tracts of telomeric repeats and is dependent upon a different pathway involving RAD50, RAD59, and SGS1 [28]–[33]. Only type II RTE normally occurs in K. lactis telomerase deletion mutants (ter1-Δ) [24]. Studies, particularly in K. lactis, have suggested that type II RTE occurs through a roll-and-spread mechanism, where a t-circle is used as a template to lengthen one short telomere which in turn can be used as a template to lengthen other telomeres via break-induced replication (BIR) events [34]–[37]. Consistent with this model, post-senescence survivors derived from cells with two kinds of telomeric repeats often contain repeating patterns in most or all lengthened telomeres [36]. Additionally, when a DNA circle containing telomeric repeats is transformed into a K. lactis telomerase deletion mutant, its sequence becomes efficiently amplified onto telomeric ends as long tandem arrays [36]. T-circles are also abundant in yeast mutants with telomere dysfunction [34], [38]–[39]. Furthermore, sequence from a single telomere is used as the source of all lengthened telomeres in K. lactis post-senescence survivors [37]. Type II RTE in S. cerevisiae has also been suggested to involve rolling circle copying of t-circles [39].
RTE can also be triggered by perturbation of telomeric capping proteins. For example, in S. cerevisiae, a cdc13-1 yku70 mutant can generate type II survivors without gradual growth senescence [40]. In K. lactis, telomerase deletion mutants containing telomeric repeats with defects in Rap1 binding develop much longer telomeres than equivalent mutants with only wild type repeats [37], [41]. Of particular interest is the stn1-M1 mutant of K. lactis [42]. Stn1 is a part of the Cdc13/Stn1/Ten1 (CST) complex that binds to the 3′ single-stranded telomeric overhang and protects the telomere termini from degradation and engagement in recombination (for a review see [43]). Stn1 also regulates telomerase recruitment and telomeric C-strand synthesis, the latter via its interaction with Polα/primase [44]–[45]. In many ways, the stn1-M1 mutant displays more similarity to ALT than do ter1-Δ mutants. It shares with ALT a steady state of very long and highly heterogeneous telomeres that are produced by recombination as well as the immediate presence of chronic but slight growth defects instead of the gradual growth senescence and survivor formation seen in ter1-Δ mutants [8], [42], [46]. Both ALT cells and stn1-M1 cells show high levels of telomere instability including elevated telomere recombination, rapid telomere deletions, and abundant extrachromosomal telomeric DNA including t-circles [10], [21], [38], [42], [47]–[48]. Finally, the phenotypes of stn1-M1 and of most ALT cells, in sharp contrast to that of ter1-Δ mutants, are largely not affected by whether telomerase is present or not, a property that we refer to as telomerase-resistant [8], [42]. While telomeric recombination in ter1-Δ cells appears repressed once telomeres are even moderately elongated, the telomere recombination in stn1-M1 cells is thought to occur at telomeres of all sizes. To distinguish the fundamental differences between telomere capping defects in the two mutants, the RTE in stn1-M1 mutant was termed type IIR for its “runaway” lengthening characteristics [42]. Given its similarities with ALT cells, the stn1-M1 mutant may therefore be a useful model system to obtain more clues about mechanisms that establish long telomeres in ALT cells.
In this work, we utilize mutationally tagged telomeric repeats to study the mechanism of type IIR RTE in stn1-M1 mutant during the establishment stage where long telomeres are generated from much shorter telomeres. Our results are consistent with predictions of the roll-and-spread model in demonstrating that sequence from one telomere is commonly spread to most or all telomeres of newly formed stn1-M1 mutants. Our results also suggest that rapid telomere truncations routinely precede the generation of long telomeres and that the presence of active mismatch repair or telomerase can impact the outcomes observed.
Results
Generating stn1-M1 mutants from precursors with mutationally tagged telomeric termini
To study the type IIR RTE that forms highly elongated telomeres in the stn1-M1 mutant of K. lactis, we generated stn1-M1 mutants from two kinds of precursors with mutationally tagged telomeric repeats. Previously, similar approaches were informative in studying the type II RTE that forms the more modestly elongated telomeres in K. lactis telomerase deletion (ter1-Δ) mutants [36]–[37]. The experimental setup for generating stn1-M1 cells from the first kind of precursor is diagramed in Figure 1A. An stn1-M1 ter1-Δ mutant was first transformed with a plasmid (pSTN1-TER1(ApaL)) containing both the STN1 and the TER1-20C(ApaL) genes. The TER1-20C(ApaL) gene forms a telomerase that adds mutated ApaL repeats onto all telomeric termini. ApaL repeats are phenotypically silent but contain a single base change that both forms an ApaLI site and eliminates the native RsaI site [49] (Figure 1A). A transformant was then serially passaged for ten streaks to allow telomeres to shorten to near normal length and to incorporate ApaL repeats at their termini. These passaged cells are referred to as the ApaL precursor cells.
Fig. 1. Use of mutationally tagged repeats at telomeric termini to study RTE in stn1-M1 cells reveals evidence for repeating structures in telomeres of some newly generated stn1-M1 ter1-Δ mutants. 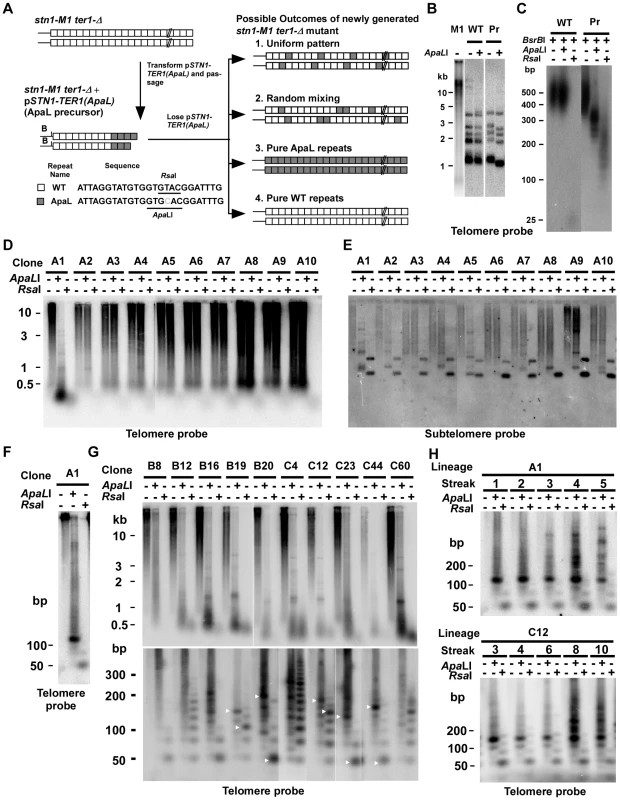
(A) Experiment outline. After transformation of pSTN1-TER1(ApaL) into stn1-M1 ter1-Δ strains, followed by serial passaging for 10 streaks, the ApaL precursor was generated that contains telomeres composed of terminal ApaL repeats (gray boxes) and internal WT repeats (white boxes). The sequences of a WT repeat and an ApaL repeat are shown. The positions of ApaLI and RsaI restriction sites in the repeats and the subtelomeric BsrBI(B) site are indicated. Upon loss of pSTN1-TER1(ApaL), newly generated stn1-M1 ter1-Δ mutants with long telomeres can be recovered. Four possible outcomes for the telomeres in these mutants are illustrated; 1. A uniformly repeating pattern of the two repeat types, 2. Randomly mixed repeats, 3. Pure ApaL repeats and 4. Pure WT repeats. (B) Southern blot, hybridized with a telomere probe, of EcoRI (indicated by “−”) and EcoRI+ApaLI (indicated by “+”) digested DNA from stn1-M1 (M1), WT and ApaL precursor (Pr). (C), BsrBI, BsrBI+ApaLI and BsrBI+RsaI-digested DNA from WT and ApaL precursor (Pr) separated on a 3% agarose gel electrophoresis and hybridized with the telomere probe. (D) Southern blot, hybridized with a telomere probe, of EcoRI (1st lane of every clone), EcoRI+ApaLI (2nd lane of every clone) and EcoRI+RsaI (3rd lane of every clone) digested DNA from 10 newly generated stn1-M1 ter1-Δ mutants generated from the ApaL precursor. (E) Same filter as in panel D after stripping and rehybridization with a subtelomeric probe. (F) EcoRI (1st lane), EcoRI+ApaLI (2nd lane) and EcoRI+RsaI (3rd lane) digested DNA from A1 clone in panel A separated on a 3% agarose gel and hybridized with a telomere probe. (G) Southern blot, hybridized with a telomere probe, of EcoRI (1st lane of every clone), EcoRI+ApaLI (2nd lane of every clone) and EcoRI+RsaI (3rd lane of every clone) digested DNA from 10 additional newly generated stn1-M1 ter1-Δ mutants separated by electrophoreses on 0.8% (Upper panel) and a 3% (Bottom panel) agarose gels. (H) Southern blot, hybridized with a telomere probe, of EcoRI (1st lane of every clone), EcoRI+ApaLI (2nd lane of every clone) and EcoRI+RsaI (3rd lane of every clone) digested DNA from A1 in panel D and C12 in panel G passaged for multiple streaks (as indicated) on YPD medium. All telomeres in the ApaL precursor cells had acquired ApaL repeats at termini as indicated by their EcoRI-digested telomeric fragments being shorter after ApaLI digestion (Figure 1B and data not shown). In contrast, telomeric fragments in a wild type control were digested away with RsaI but resistant to ApaLI (Figure 1B–1C). To estimate the number of ApaL repeats at telomeric termini of ApaL precursors, we digested their DNA by BsrBI+ApaLI and BsrBI+RsaI (Figure 1C and data not shown). BsrBI cleaves 3 bp away from 10 of 12 telomeres in K. lactis (Figure 1A), and ApaLI and RsaI specifically cleave ApaL and wild type (WT) repeats, respectively. The size ranges of the telomeric signal in BsrBI+ApaLI and BsrBI+RsaI digestions reflect the size ranges of internal WT repeats and terminal ApaL repeats, respectively. From this, we estimated that the terminal 100–300 bp of the 350–600 bp total telomere length was composed of ApaL repeats. This was verified by cloning and sequencing two telomeres from an ApaL precursor. One cloned telomere contained 11 basal WT repeats and three terminal ApaL repeats and the other contained ∼9 basal WT repeats and five terminal ApaL repeats (Figure S1A). To our surprise, clone 2 also contained a 13 bp repeat located between the WT and ApaL repeats (Figure S1A). The sequence of this 13 bp repeat suggested it arose when the terminal bases of a telomere annealed to the middle instead of to the 3′ end of the telomerase RNA template (Figure S1B). This half-sized repeat was not likely formed by the ApaL telomerase because it contained the native RsaI site of WT repeats rather than an ApaLI site.
We then selected for newly generated stn1-M1 ter1-Δ mutants by plating the ApaL precursors on medium containing 5-flouro-orotic acid (5-FOA) to select for loss of the pSTN1-TER1(ApaL) plasmid. As the loss of plasmid sequences simultaneously deletes both telomerase and the wild type STN1 gene, the generation of long telomeres in the newly formed stn1-M1 ter1-Δ cells should depend solely on recombination. Although the ter1-Δ mutation is, by itself capable of causing RTE, it is not likely to interfere with the type IIR RTE brought on by the stn1-M1 mutation. This is because the telomeric recombination induced by the ter1-Δ mutation is confined to occurring when telomeres become <∼150 bp in length [37] while telomeric recombination in stn1-M1 occurs even at very long telomeres. Consistent with this, the growth and telomere phenotypes of the stn1-M1 mutant are epistatic to those of the ter1-Δ mutant.
Because the pSTN1-TER1(ApaL) plasmid became integrated into a chromosome at the stn1-M1 locus in the ApaL precursor during passaging (data not shown), the rate of recovering stn1-M1 ter1-Δ mutants (generated by homologous recombination looping out the plasmid from the chromosome) was relatively low. A group of ten newly generated stn1-M1 ter1-Δ mutants was initially analyzed. All ten mutant clones showed the long and heterogeneous pattern of telomeres that is the characteristic of the stn1-M1 phenotype when EcoRI digests were observed in a Southern blot (Figure 1D). Telomeric signals in nine of these ten mutants (clones A2–A10, Figure 1D) were not obviously cleaved by ApaLI. Hybridization of the same filter to a subtelomeric probe (Figure 1E) showed that most signal from most clones was unchanged by ApaLI digestion. These results indicate that telomeres in these nine mutants were composed almost entirely of WT repeats. Consistent with this interpretation, telomeric signals from these nine mutants were virtually cleaved away by RsaI, which specifically cleaves WT repeats (Figure 1D). These results are very surprising, because the stn1-M1 phenotype forms rapidly without a period of growth senescence and should therefore not undergo any gradual sequence loss of terminal ApaL repeats before the formation of long telomeres by RTE [42]. Each of the A2–A10 clones did show a small percentage of the heterogeneously smeared subtelomeric signal shifted down to one or more bands, generally of ∼1 kb, from ApaLI digestion. This indicates that one or a small number of the telomeres in each clones retained at least one ApaL repeat near their base.
Strikingly, the telomeric signal of one stn1-M1 ter1-Δ mutant (clone A1) was cleaved into very small fragments by both ApaLI and RsaI (Figure 1D). When the same digests were run on a high-percentage agarose gel, the small fragments were observed to be composed largely of a ∼125 bp band in the ApaLI digests and a ∼50 bp band in the RsaI digests (Figure 1F). The former was predicted to contain four WT repeats and two half ApaL repeats and the latter was predicted to contain one ApaL repeat and two half WT repeats. These data suggest that telomeres in this stn1-M1 ter1-Δ mutant may contain repeating structures that consist of four WT repeats and one ApaL repeat as the repeating unit. To test this, we cloned and sequenced 38 telomeric fragments from this clone, which were produced by partial ApaLI digestion. Although mostly very small ApaLI fragments were recovered (Figure S2), the results showed that 19 of 51 (37%) blocks of WT repeats were ∼100–125 bp, of which 15 (79%) consisted of three WT repeats and one half WT repeat of the same sequence that was recovered from the ApaL precursor. Although these results rule out the presence of perfectly repeating patterns when DNA was isolated from the A1 mutant, the widespread presence of a particular pattern of repeats could suggest that telomeres with more perfect repeating patterns originally existed but was disrupted by numerous later recombination events.
We next analyzed 83 additional stn1-M1 ter1-Δ mutants generated from ApaL precursors. 73 of these clones had telomeric signals that were essentially uncleaved by ApaLI but were nearly fully cleaved by RsaI indicating that the lengthened telomeres were composed of virtually all WT repeats (data not shown). However, ten clones had telomeric signals that were cleaved partly or entirely into short fragments by both ApaLI and RsaI digestions (Figure 1G, upper panel). The same digests of these ten mutants were then run on a high-percentage agarose gel to resolve short DNA fragments (Figure 1G, lower panel). Several of these mutants, including B19, B20, C12, C23 and C44 showed favored fragment sizes in both ApaLI and RsaI digests (indicated by white arrows) which could be indicative of degraded repeating patterns. In each of these mutants, the most common fragment size of ApaL repeats was smaller than that of WT repeats. The other mutants examined, B8, B12, B16, C4 and C60, sometimes exhibited favored short fragments in RsaI digests but not obviously any in ApaLI digests. The average size of the telomeric signal in ApaLI digests of these clones tended to migrate at greater average size than that seen in the other clones. In some clones, most notably C4, the ApaLI digestion produced ladders of bands that included sizes consistent with the presence of the “half” telomeric repeats as was present in the A1 clone.
Two newly generated stn1-M1 ter1-Δ mutants that exhibited the best evidence of repeating patterns (clones A1 and C12) were serially passaged for 5–10 streaks on YPD plates and periodically examined for their telomeric DNA structure by Southern blots of ApaLI and RsaI digests run on high-percentage agarose gels (Figure 1H). The initial banding pattern of these mutants became more complicated after passaging. Specifically, the favored fragments of the two mutants in ApaLI digests, initially ∼110–150 bp, became more variable at later streaks and tended to produce new fragments of larger sizes. This observation supports the idea that telomeres in stn1-M1 mutants are extremely dynamic and are prone to high rates of recombination that can rapidly alter their structure.
The absence of mismatch repair leads to more heterogeneously structured telomeres in newly made stn1-M1 mutants
Base mismatches in DNA can reduce the rate of homologous recombination [50]. Therefore, the base mismatch between WT and ApaL repeats is likely to interfere with the recombination between telomeres that contain them. To test this, we disrupted the MSH2 gene (required for mismatch repair) in an ApaL precursor, and generated 17 stn1-M1 ter1-Δ msh2-Δ mutants by losing pSTN1-TER1(ApaL) as described above. Telomeric signals in 5 of these mutants showed little or no cleavage by ApaLI (data not shown). However, telomeric signals in the other 12 mutants were cleaved into broad ladders of bands with ∼25 bp steps by both ApaLI and RsaI (Figure 2A and data not shown). Unlike what was seen in mutants derived from a MSH2 background, these mutants showed no obvious favored fragments that might have indicated the presence of a degraded pattern. The appreciably higher percentage of stn1-M1 mutants made in the msh2-Δ background that had amplified ApaL repeats might also be related to mismatch repair. Alternatively, we cannot rule out the possibility that the increased incorporation of ApaL repeats occurring in the precursor during the additional cell divisions (equivalent to 2–3 streaks) needed to disrupt the MSH2 gene altered the result.
Fig. 2. The effect of mismatch repair deficiency on telomeric structure in stn1-M1 mutants derived from the ApaL precursor. 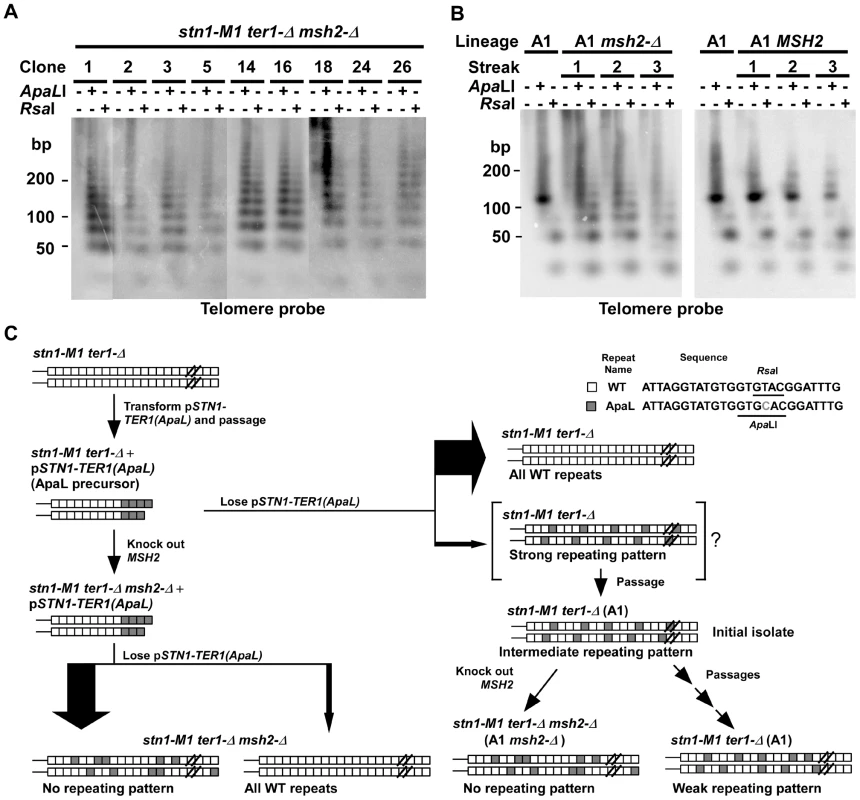
(A) Southern blot, hybridized with a telomere probe, of EcoRI (1st lane of every clone), EcoRI+ApaLI (2nd lane of every clone) and EcoRI+RsaI (3rd lane of every clone) digested DNA from nine newly generated stn1-M1 ter1-Δ mutants in a msh2-Δ background separated on a 3% agarose gel. (B) Southern blot, hybridized with a telomere probe, of EcoRI (1st lane of every clone), EcoRI+ApaLI (2nd lane of every clone) and EcoRI+RsaI (3rd lane of every clone) digested DNA from the A1 clone (in Figure 2A) with (A1 msh2-Δ) or without (A1 MSH2) successful deletion of MSH2. Each clone is shown at each of 3 serial streaks. The DNA was separated on a 3% agarose gel. The A1 clone from Figure 2A is also shown. (C) Diagram summarizing the experimental outline and results from generating stn1-M1 ter1-Δ MSH2 and stn1-M1 ter1-Δ msh2-Δ mutants from ApaL precursors. Only a minority of newly generated mutants retain both wild type telomeric repeats (white boxes) and ApaL repeats (grey boxes) in their lengthened telomeres. The stn1-M1 ter1-Δ MSH2 clones such as the A1 clone are shown as hypothetically being derived from a newly established mutant cell with long telomeres with a strongly repeating pattern of wild type and ApaL repeats (“strong repeating pattern”). By the time enough cells are available for DNA analysis, recombination is likely to have partly degraded the initial pattern (“intermediate repeating pattern”). Subsequent passaging of these cells leads to further pattern degradation (“weak repeating pattern”). In contrast, an equivalent generation of stn1-M1 ter1-Δ msh2-Δ mutants (left side of figure) leads to no sign of repeating patterns in cells retaining both ApaL and WT repeats, even immediately after their isolation. We speculated that stn1-M1 ter1-Δ mutants made in a msh2-Δ background had sufficiently high levels of telomeric recombination to rapidly break down any repeating structure that might have been formed initially. To test this idea, we attempted to knock out the MSH2 gene in the A1 clone of stn1-M1 ter1-Δ mutant that had highly favored small telomeric fragments in both ApaLI and RsaI digests (Figure 1D). Among 72 clones transformed with the knockout cassette, only one had the MSH2 gene successfully disrupted. As homologous gene disruption rate, even with the fragment used in this experiment, is generally 10–40% in STN1 cells, it is conceivable that very high levels of telomeric recombination might interfere with homologous recombination elsewhere in the genome of stn1-M1 cells. The one stn1-M1 msh2-Δ clone we did recover was serially passaged on YPD plates for three streaks and its telomeres were examined after each streak by Southern blot (Figure 2B). The same procedure was carried out with a control transformant that still had the intact MSH2 gene. Upon disruption of MSH2, the simple banding patterns of the A1 clone, particularly the larger fragment in ApaLI digests, become distinctly heterogeneous as soon as the samples could be analyzed (Figure 2B). This heterogeneous pattern is similar to those from stn1-M1 ter1-Δ mutants generated directly in a msh2-Δ background shown in Figure 2A. In contrast, the control transformant having the intact MSH2 exhibited a banding pattern that remained distinctly more stable. This result demonstrates that the elevated telomeric recombination in a msh2-Δ background is sufficient enough to rapidly break down a repeating structure that might have been formed initially in telomeres of a newly generated stn1-M1 ter1-Δ mutant. A diagram summarizing our experiments with stn1-M1 ter1-Δ mutants generated from ApaL precursors is shown in Figure 2C.
The sequence from a single telomere can be spread to all other telomeres during the establishment of the stn1-M1 ter1-Δ state
RTE in ter1-Δ mutants of K. lactis regularly involves the spreading of sequence from a single telomere to all other telomeres to generate modest elongated telomeres [37]. To test whether the stn1-M1 mutant can also use this mechanism to generate highly elongated telomeres, we constructed telomeric DNA fragments consisting of the subtelomeric sequence, a HIS3 marker and Bcl telomeric repeats (Figure 3A). The subtelomeric sequence is shared by 11 of 12 K. lactis telomeres allowing the transformed fragments to replace a native telomere through homologous recombination. The Bcl repeats each contain a phenotypically silent base change that generates a BclI site [51]–[53]. DNA fragments containing either ∼11 or ∼40 Bcl repeats were transformed into stn1-M1 ter1-Δ cells containing pSTN1-TER1 integrated at the stn1-M1 locus (Figure 3A). While the shorter telomeric fragment generated a new telomere of normal length (Figure 3B), the long telomeric fragment generated a telomere substantially longer than those of wild type cells (Figure 3F). The resulting transformants are hereafter referred to as “normal length Bcl precursors” and “long Bcl precursors”, respectively.
Fig. 3. Spreading of a telomeric sequence from a single telomere source during RTE in stn1-M1 ter1-Δ cells. 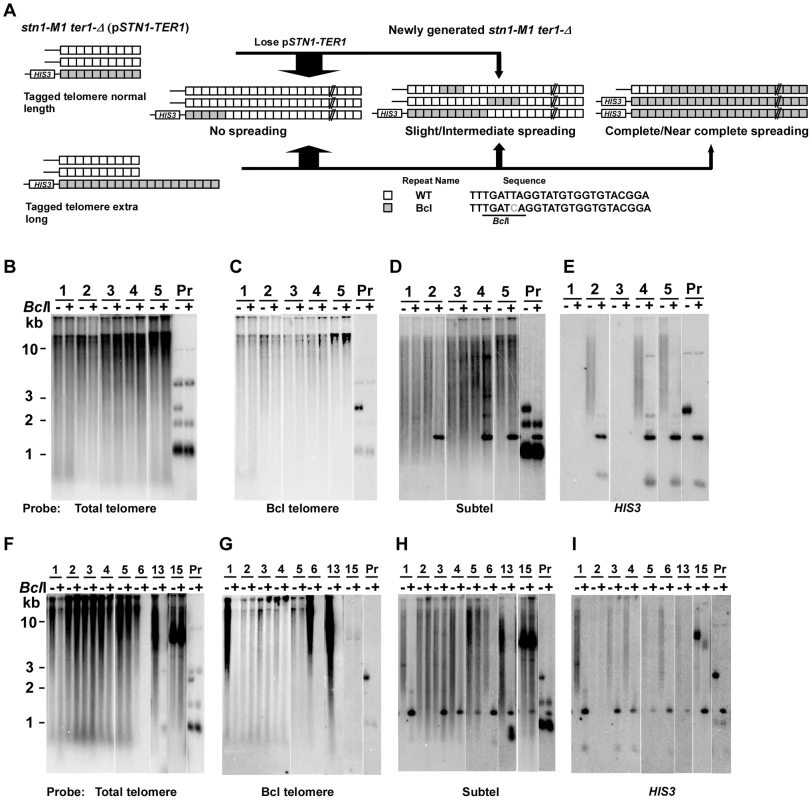
(A) Experimental outline showing generation of stn1-M1 ter1-Δ mutants from STN1 TER1 precursors containing a single telomere of either normal length or ∼2× longer than normal length that are composed of Bcl repeats. The sequence of the Bcl repeat is indicated. (B) Southern blot, hybridized with a telomere probe, of EcoRI (indicated by “−”) and EcoRI+BclI (indicated by “+”) digested DNA from 5 newly generated stn1-M1 ter1-Δ clones from normal length Bcl precursors as well as from one normal length Bcl precursor STN1 TER1 control. (C–E) Same filter as in panel B after stripping and rehybridization with a Bcl telomere probe, subtelomeric probe and HIS3 probe, respectively. (F) Southern blot, hybridized with a telomere probe, of EcoRI (indicated by “−”) and EcoRI+BclI (indicated by “+”) digested DNA from 8 newly generated stn1-M1 ter1-Δ clones from long Bcl precursors as well as from one long Bcl precursor STN1 TER1 control. (G–I) Same filter as in panel F after stripping and rehybridization with a Bcl telomere probe, subtelomeric probe and HIS3 probe, respectively. Next, we generated 55 clones of stn1-M1 ter1-Δ mutants from four normal length Bcl precursors by plating the precursors on 5-FOA-containing medium that selected for the loss of the pSTN1-TER1 plasmid. Telomeric signals in 49 of these clones appear essentially identical in both EcoRI and EcoRI+BclI digests (Clones 1, 3, 4 and 5 in Figure 3B and data not shown) which indicated that telomeres in these clones were composed mostly or entirely of WT repeats with very few or no Bcl repeats. Other data were also consistent with this view. The weak signal observed in these clones with a probe specific to Bcl telomeric repeats was not sensitive to BclI digestion, suggesting that it was due to background hybridization to the heavily amplified wild type repeats (Figure 3C). Additionally, subtelomeric signals in these clones were largely resistant to BclI digestion, except for ∼1.5 kb fragments generated in some clones (Clones 2, 4 and 5 in Figure 3D–3E) that also hybridized to HIS3 probe, consistent with them being derived from the original integrated telomere. We classified these 49 clones as having no amplification or spreading of Bcl repeats (Figure 4, column 1). Our results suggest that Bcl repeats were actively avoided as a source of sequence to be amplified during establishing the long telomeres in the stn1-M1 ter1-Δ mutant.
Fig. 4. Frequency of spreading of sequence from a single tagged telomere to other telomeres in K. lactis stn1-M1 mutants with different genetic backgrounds. 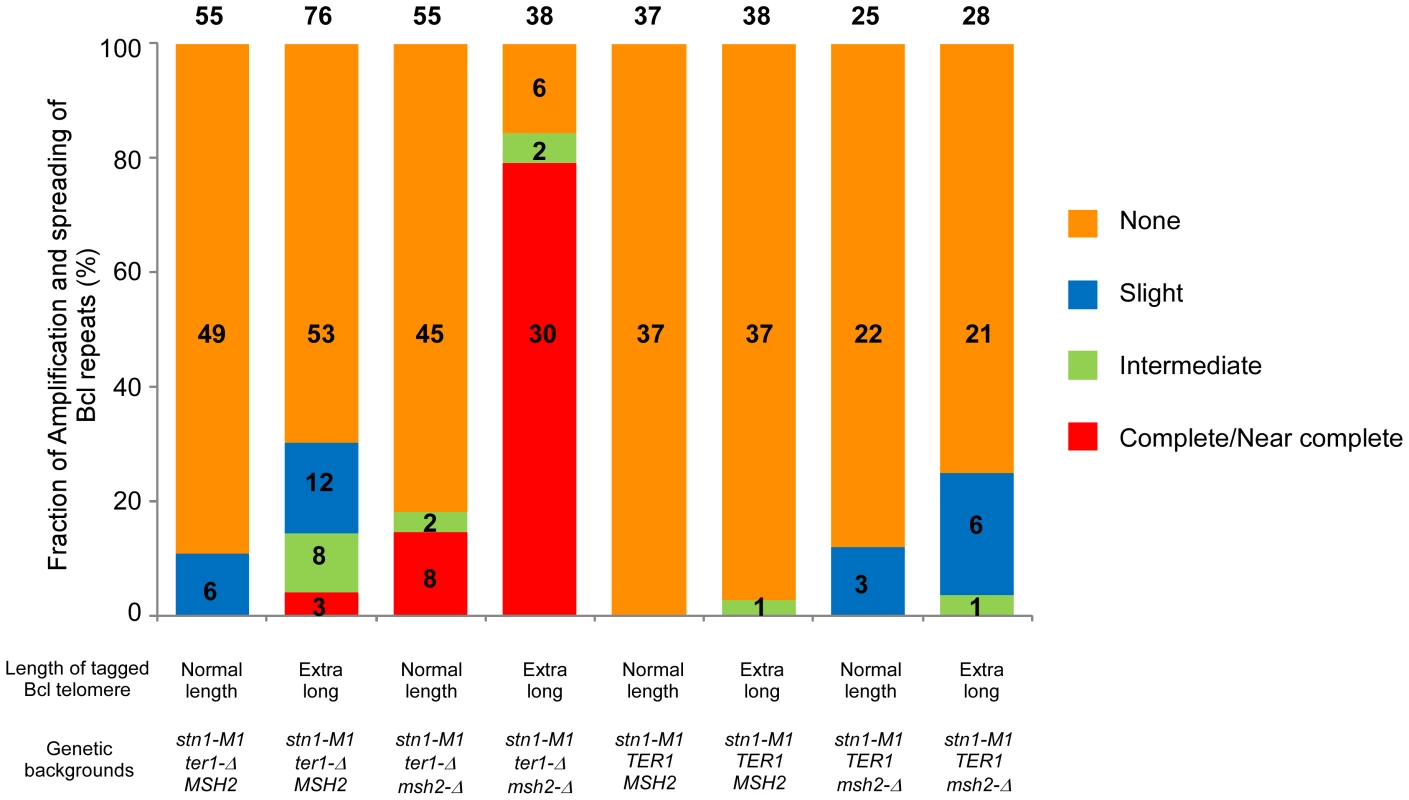
Columns 1–8 show histograms of the frequencies of four different types of outcomes (none, slight, intermediate and complete/near complete) with regard to the extent and spreading of ApaL repeats in newly generated stn1-M1 mutants. Numbers above each column indicate the total number of clones examined. Numbers within histograms indicate the number of clones of each type when those numbers were above zero. Definitions of the outcome types are as follows. None: defined by each of the following being true: 1) total EcoRI-digested telomeric signal is reduced no more than 1.5 fold by additional BclI digestion; 2) Bcl specific telomeric signal is less than 2.5 fold more than that of “precursor”; 3) EcoRI-digested subtelomeric signal larger than 2 kb is reduced no more than 1.5 fold by BclI digestion. Slight: defined by each of the following being true: 1) total EcoRI-digested telomeric signal is reduced no more than 3 fold by additional BclI digestion; 2) Bcl specific telomeric signal is less than 5 fold more than that of “precursor”; 3) EcoRI-digested subtelomeric signal larger than 2 kb is reduced more than 1.5 fold by BclI digestion. Intermediate: defined by each of the following being true: 1) total EcoRI-digested telomeric signal is reduced no more than 3 fold by additional BclI digestion; 2) Bcl specific telomeric signal is 5 fold or more higher than that of “precursor”; 3) EcoRI-digested subtelomeric signal larger than 2 kb is reduced more than 1.5 fold by BclI digestion. Complete/Near Complete: defined by each of the following being true: 1) total EcoRI- digested telomeric signal is reduced at least 3 fold by additional BclI digestion; 2) Bcl specific telomeric signal is 8 fold or more than that of “precursors”; 3) EcoRI-digested subtelomeric signal larger than 2 kb is reduced more than 1.5 fold by BclI digestion. The remaining 6 of 55 clones, including clone 2 in Figure 3B, showed amplification of Bcl repeats estimated to be 3–5 times that present in the single telomere of the precursor strain (Figure 4 and data not shown). At least two of these clones showed extra copies of the HIS3 gene (data not shown), suggesting that the modest amplification of Bcl repeats occurred primarily by subtelomeric break-induced replication (BIR) events that produced extra copies of the tagged telomere but without the Bcl repeats otherwise becoming amplified.
Of the 55 total clones, 41 (including 38 of 49 clones with no amplification and spreading of Bcl repeats) showed no HIS3 signal (Figure 3E and data not shown). This indicates that these mutants lost the HIS3-tagged telomere and probably all the Bcl repeats originally attached to that telomere. Such frequent loss of the subtelomeric HIS3 was not entirely surprising as that stn1-M1 cells showed very high rates of loss of a URA3 gene inserted at the same subtelomeric location [42]. These deletions are likely BIR events that replace one telomere with sequence from another telomere. The same mechanism is likely responsible for occasions in some mutants where the HIS3-tagged telomere became duplicated.
Analysis of 76 newly generated clones of stn1-M1 ter1-Δ mutants from long Bcl precursors showed somewhat different results (Figure 4, column 2). While 53 clones (70%) showed no amplification or spreading of Bcl repeats, the other 23 clones (30% of the total) did show some degree of amplification and spreading of Bcl repeats. Most notably, telomeric signals in three of these clones, including clones 6 and 13 in Figure 3F, can be completely or nearly completely cleaved away by BclI digestion, suggesting that telomeres in these clones were composed mostly or entirely of Bcl repeats (Figure 4). Consistent with this interpretation, the Bcl repeat-specific signals of these three clones were intense in EcoRI digests but were eliminated by BclI digestion (Figure 3G). Furthermore, the long smeared subtelomeric signal of these clones in EcoRI digests was largely or entirely cleaved by BclI into fragments that were generally <2 kb (Figure 3H). Interestingly, these three clones showed no amplification of the subtelomeric HIS3 gene (Figure 3I and data not shown), suggesting that the Bcl repeats were amplified by inter-telomeric recombination rather than by subtelomeric duplication.
As indicated in Figure 4, 8 of the 76 clones derived from the long Bcl precursor (including clone 1 in Figure 3F–3I) were classified as having intermediate amplification and spreading of Bcl repeats based on: 1) total telomeric signal that was partially cleaved by BclI (Figure 3F); 2) Bcl-specific telomeric signal that was at least 5 times that of the precursor (Figure 3G and data not shown); and 3) subtelomeric signal that was substantially cleaved into one or more short fragments by BclI digestion (Figure 3H). These results suggest that both Bcl and WT repeats were amplified and copied onto multiple telomeres in these mutants. This group of clones had no obvious telomeric fragments of 50–500 bp in EcoRI+BclI digests (Figure 3F and data not shown), suggesting that the amplified Bcl and WT repeats were not generally interspersed closely together. Indeed, the signal from WT repeats that remained after BclI digestion was generally very long, suggesting that WT repeats in these clones were often in long continuous arrays (Figure 3F and data not shown). Twelve other clones (including clone 4 in Figure 3F–3I) were classified as having slight amplification and spreading of Bcl repeats based on lesser degrees of both amplification of Bcl repeats and subtelomeric signal that was cleaved by BclI (Figure 4, column 2).
One clone of the stn1-M1 ter1-Δ mutant, clone 15 in Figure 3F–3I, showed a unique outcome. It displayed a ∼9–10 kb band that hybridized intensely with both telomeric and subtelomeric probes but was resistant to BclI digestion (Figure 3F and 3H). These results are consistent with the possibility that this clone contained tandem arrays composed of both WT telomeric repeats and subtelomeric sequences, but not Bcl repeats. HIS3 was detected in the clone but apparently was not within the amplified fragment (Figure 3I). Conceivably, the putative tandem arrays originated from a native telomere rather than the Bcl telomere. This clone is reminiscent of type I post-senescent survivors in Sacchromyces cerevisiae, which are characterized by amplified subtelomeric Y′ elements and short tracts of telomeric repeats [31], [54]. Although type I-like survivors with alternating telomere and non-telomeric sequences can occur in K. lactis cells that are transformed with a circle containing telomeric repeats and the URA3 gene [35], [48], to our knowledge, this is the first report of RTE amplifying subtelomeric sequences in K. lactis.
23 of 76 stn1-M1 ter1-Δ mutants (30%) derived from long Bcl precursors had lost HIS3 signal (Figure 3I and data not shown). All 23 of these clones showed no amplification and spreading of Bcl repeats. This loss rate of HIS3 was significantly less (p<0.001, in Fisher exact test) than that seen in mutants derived from normal Bcl precursors where 41 of 55 clones (75%) showed no HIS3 signal. This result suggests that a long telomere stabilizes the adjacent subtelomeric sequences from being lost during the establishment of an stn1-M1 ter1-Δ mutant.
The absence of mismatch repair facilitates the spreading of sequence from a single tagged telomere to other telomeres
To test whether the low frequency of spreading Bcl repeats to other telomeres in stn1-M1 ter1-Δ mutants was affected by mismatch repair, we constructed normal length Bcl precursor and long Bcl precursor strains containing pSTN1-TER1 and a msh2-Δ mutation (Figure 5A). After screening for loss of pSTN1-TER1, we identified 55 stn1-M1 ter1-Δ msh2-Δ mutants from three normal length Bcl precursors and 38 mutants from two long Bcl precursors. The results showed that the msh2-Δ background permitted a substantially higher frequency of amplification and spreading of Bcl repeats than a MSH2 background did. Eight of 55 stn1-M1 ter1-Δ msh2-Δ mutants (15%) derived from normal length Bcl precursors (including clones 1–2 in Figure 5B–5E) showed complete or near complete spreading of Bcl repeats (Figure 4, column 3). Two mutants (4%), including clone 6 in Figure 5B–5E, showed intermediate amplification and spreading of Bcl repeats and the remaining mutants (45 of 55; 81%), including clones 5 and 11–13 in Figure 5B–5E, showed no amplification (Figure 4, column 3).
Fig. 5. Spreading of a telomeric sequence from a single telomere source during RTE in stn1-M1 ter1-Δ msh2-Δ cells. 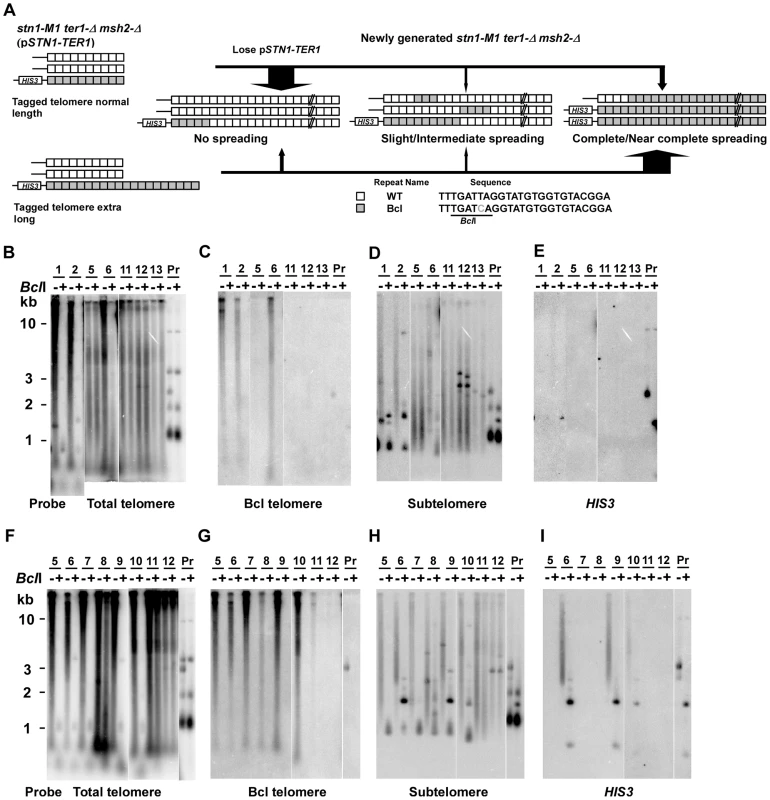
(A) Experimental outline showing generation of stn1-M1 ter1-Δ msh2-Δ mutants from STN1 TER1 msh2-Δ precursors containing a single telomere of either normal length or ∼2× longer than normal length that are composed of Bcl repeats. The sequences of a WT repeat and a Bcl repeat are indicated. (B) Southern blot, hybridized with a telomere probe, of EcoRI (indicated by “−”) and EcoRI+BclI (indicated by “+”) digested DNA from 7 newly generated stn1-M1 ter1-Δ msh2-Δ clones from normal length Bcl precursors in a msh2-Δ background as well as one normal length Bcl STN1 TER1 msh2-Δ precursor control. (C–E) Same filter as in panel B after stripping and rehybridization with a Bcl telomere probe, subtelomeric probe and HIS3 probe, respectively. (F) Southern blot, hybridized with a telomere probe, of EcoRI (indicated by “−”) and EcoRI+BclI (indicated by “+”) digested DNA from 8 newly generated stn1-M1 ter1-Δ msh2-Δ clones from long Bcl precursors in a msh2-Δ background as well as one long Bcl STN1 TER1msh2-Δ control. (G–I) Same filter as in panel F after stripping and rehybridization with a Bcl telomere probe, subtelomeric probe and HIS3 probe, respectively. Remarkably, 30 of 38 mutants (78%) derived from long Bcl precursors (including clones 5–7, 9, and 10 in Figure 5F–5I) showed complete or near complete spreading of Bcl repeats (Figure 4, column 4). At least 25 of these clones appeared to contain a small percentage of WT repeats interspersed among the Bcl repeats, as indicated by telomeric signal of <500 bp present in EcoRI+BclI digestion (e.g., clones 5–7, 9, and 10 in Figure 5F). However, as these telomeric signals were not intense, it is likely that the WT repeats were not interspersed throughout the telomeres. Only two clones (6%), including clone 8 and 11 in Figure 5F–5I, showed intermediate amplification and spreading of Bcl repeats (Figure 4, column 4). As the disruption of mismatch repair presumably permits Bcl repeats to recombine with WT repeats in an unperturbed fashion, we conclude that the long telomeres formed during the establishment of an stn1-M1 ter1-Δ mutant are regularly derived primarily from a single telomere source.
45 of 55 (82%) stn1-M1 ter1-Δ msh2-Δ mutants derived from normal Bcl precursors (Figure 5E and data not shown) and 21 of 38 (55%) mutants derived from long Bcl precursors (Figure 5I and data not shown) had lost the HIS3 genes. This difference was significant (p = 0.0005 in Fisher exact test). Strikingly, many clones that had complete or nearly complete spreading of Bcl repeats (4 of 8 mutants from the normal length Bcl precursor and 14 of 30 from the long Bcl precursor) had lost the HIS3 genes (clone 5 and 7 in Figure 5I and data not shown). On the other hand, 4 of 18 clones that had complete or near complete spreading of Bcl repeats contained estimated 5–10 copies of HIS3 genes (data not shown). These results showed that the spreading of Bcl repeats to all other telomeres could occur either with or without concurrent spreading of the subtelomeric HIS3 to other telomeres.
The presence of telomerase inhibits the spreading of sequence from a single tagged-telomere to other telomeres
To test the effect of telomerase on amplification and spreading of Bcl repeats, we constructed normal length Bcl precursors and long Bcl precursors in both stn1-M1 TER1 MSH2 and stn1-M1 TER1 msh2-Δ backgrounds that were complemented by plasmid pSTN1. These precursors were similar to those used above except that the subtelomeric marker gene was URA3 and the complementing plasmid carried STN1 and HIS3 (Figure 6A). We generated stn1-M1 mutants from all four precursor types by screening for loss of pSTN1 after streaking onto YPD plates and identifying clones with the rough colony phenotype characteristic of stn1-M1 mutants. Results from this showed that telomerase significantly inhibited the amplification and spread of Bcl repeats (Figure 6; Figure S3; and Figure 4, columns 5–6). None of 37 stn1-M1 TER1 MSH2 clones derived from normal-length precursors showed amplification and spreading of Bcl repeats (Figure S3A–S3D and Figure 4, column 5), and only 1 of 38 stn1-M1 TER1 MSH2 clones from the long precursors showed detectable amplification and spreading (clone 9 in Figure S3E–S3H and Figure 4, column 6). The one mutant that did show amplification (an intermediate level) was estimated to contain three copies of URA3 (data not shown). This may suggest that amplification and spreading of Bcl repeats in this clone occurred primarily through subtelomeric BIR events that produced extra copies of the tagged telomere rather than by telomere-telomere recombination.
Fig. 6. Spreading of a telomeric sequence form a single telomere source during RTE in stn1-M1 TER1 msh2-Δ cells. 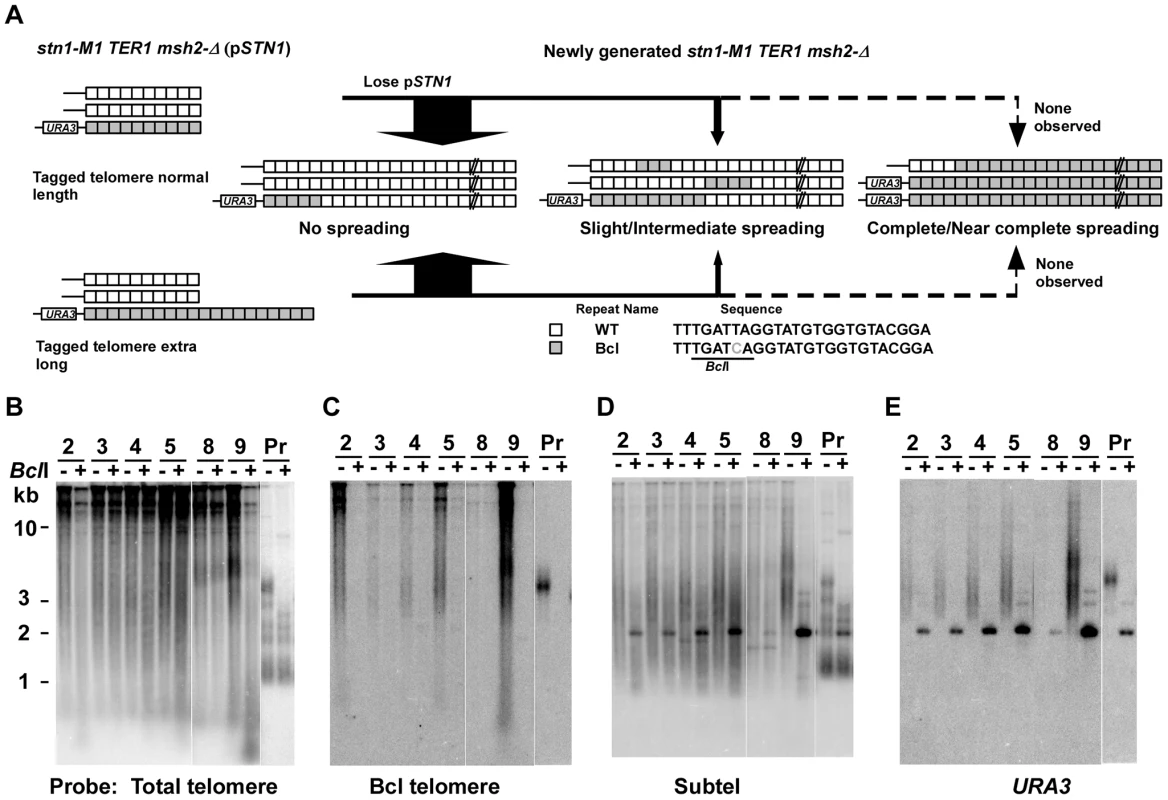
(A) Experimental outline showing generation of stn1-M1 TER1 msh2-Δ mutants from STN1 TER1 msh2-Δ precursors containing a single telomere composed of Bcl repeats that is either normal length or ∼2× longer than normal length. The sequences of a WT repeat and a Bcl repeat are indicated. (B) Southern blot, hybridized with a telomere probe, of EcoRI (indicated by “−”) and EcoRI+BclI (indicated by “+”) digested DNA from six newly generated stn1-M1 TER1 msh2-Δ clones from long Bcl precursors in a TER1 msh2-Δ background as well as one normal length Bcl STN1 TER1 msh2-Δ precursor control. (C–E) Same filter as in panel B after stripping and rehybridization with a Bcl telomere probe, subtelomeric probe and URA3 probe, respectively. Similar outcomes were seen with stn1-M1 TER1 msh2-Δ mutants. There, the frequency of amplification and spreading of Bcl repeats was markedly lower than that seen in stn1-M1 ter1-Δ msh2-Δ mutants (Figure 4, columns 7–8). None of 28 stn1-M1 TER1 msh2-Δ mutants obtained from the long precursors showed complete amplification and spreading of Bcl repeats, and only 7 of 28 mutants showed intermediate or slight amplification. Clone 9 in Figure 6B–6E is the sole example of intermediate amplification. Among stn1-M1 TER1 msh2-Δ mutants derived from a normal length Bcl precursor, only 3 of 25 mutants, including clones 6 and 7 in Figure S3I–S3L, showed slight Bcl amplification (Figure 4, column 7).
The subtelomeric URA3 marker was comparatively stable in the stn1-M1 TER1 mutants. Only 7 of 67 stn1-M1 TER1 msh2-Δ mutants and no stn1-M1 TER1 MSH2 mutants (0 of 39) had lost URA3 (Figure S3D, S3H, S3L; Figure 6E; and data not shown). These results suggest that an active telomerase in stn1-M1 mutants helps to stabilize the adjacent subtelomeric sequences against loss or amplification.
Discussion
Accumulated previous evidence suggests that a roll-and-spread mechanism is involved in generating the moderately elongated telomeres formed in K. lactis ter1-Δ post-senescence survivors (reviewed in [27]). Our studies here suggest that this mechanism, involving amplification of sequence from a single t-circle is also involved in establishing the more ALT-like highly elongated telomeres in the K. lactis stn1-M1 mutant. Some support for the importance of t-circles in stn1-M1 cells already existed. Previous data had shown that t-circles are abundant in stn1-M1 cells [38]. Additionally, a recent study showed that introducing t-circles into stn1-M1 cells leads to tandem arrays of the circle's sequence becoming incorporated at multiple telomeres [48].
The use of mutationally tagged repeats was critical to earlier studies on how RTE occurs in a ter1-Δ mutant [35]–[37]. However, the type II RTE in this mutant is episodic, occurring when telomeres drop below a critical length and essentially shutting off once telomeres are moderately lengthened [24]. The transient stability of the lengthened telomeres allowed Southern blots that examined the structure of telomeric DNA from populations of cells to be very informative. We anticipated that the telomerase-resistant type IIR RTE of the stn1-M1 mutant, with its apparently continuous high rate of telomeric recombination, would be more problematic to study by this method as any long telomere initially generated would be expected to be unstable and would become altered by further recombination events in many if not most cells of any culture large enough to be studied. The first tagging approach used in this study involved generating stn1-M1 MSH2 mutants from precursors with ApaL repeats at all telomeric termini. Among the mutant clones that had both amplified WT and ApaL repeats, roughly half had telomeric blocks of both types of repeats that had single favored sizes. Although other interpretations cannot be ruled out, this result is consistent with a roll-and-spread mechanism that derived all amplified telomeres from the sequence of a single small t-circle if the uniformly repeating patterns predicted from rolling circle copying of a t-circle containing both WT and ApaL repeats had been extensively disrupted by later recombination events.
Other results clearly demonstrated that ongoing recombination in stn1-M1 mutants can disrupt existing signs of repeating patterns. This was most strikingly seen when a stn1-M1 ter1-Δ MSH2 mutant with a very non-random WT/ApaL repeat pattern had its MSH2 gene disrupted. As soon as this msh2-Δ derivative could be examined, its telomeric repeats had acquired a much more randomized pattern similar to those seen in stn1-M1 ter1-Δ mutants established directly in a msh2-Δ background. As discussed further below, loss of mismatch repair is expected to elevate recombination rates between WT and ApaL repeats to levels that would occur if no mismatches were present. Thus, even the complete absence of preferred sizes of blocks of WT and ApaL repeats seen in stn1-M1 ter1-Δ msh2-Δ clones is not inconsistent with a single t-circle having been the original source of the amplified telomeres.
A result of particular importance in our study was that Bcl repeats present at a single telomere in a precursor cell were sometimes the source of virtually all of the amplified telomeric DNA sequences in newly generated stn1-M1 mutants. This effect was most prominent in a strain lacking both telomerase and MSH2. This mutant combination is probably the most relevant of those examined that used Bcl repeats to study telomeric recombination. This is because the absence of telomerase assures that all telomeric lengthening is due to recombination while the absence of mismatch repair presumably permits recombination between wild type and Bcl telomeres to occur at the same frequency as recombination between two wild type telomeres in otherwise equivalent circumstances. In the ter1-Δ msh2-Δ strain, the percentage of stn1-M1 mutants that displayed complete or near complete amplification and spreading of Bcl repeats was 14% in mutants derived from the normal length Bcl precursors and 78% in mutants derived from the long Bcl precursors (Figure 4). These results are very similar to those seen in an earlier study with K. lactis ter1-Δ mutants, where total spreading of Bcl repeats to all telomeres from a single normal length Bcl telomere and a single long Bcl telomere was measured at 10% and 94%, respectively [37]. As was proposed with ter1-Δ mutants, we suggest that a roll-and-spread mechanism, where rolling circle amplification of a single t-circle followed by spreading of that sequence to all telomeres by BIR events, can account for these observations. When all twelve telomeres are the same length and each has the same chance to be amplified and spread, the predicted frequency of clones where Bcl repeats have been amplified and spread would be roughly one twelfth. The much greater frequency of spreading of Bcl repeats that is observed from mutants derived from the long Bcl precursors indicates that a longer telomere is much better able than a shorter telomere at promoting the spread of its sequence to other telomeres. The roll-and-spread model predicts that this could occur because the long Bcl telomere is used directly as a template for lengthening other telomeres and/or is better at forming t-circles. Although t-circles are common in established stn1-M1 cells, the fact that all amplified telomeric sequences can be derived from a single telomere may suggest that t-circle formation is limiting during the initial establishment of the mutant state. Additionally, the copying of sequence from a lengthened telomere onto other telomeric ends might be facilitated by the physical clustering of uncapped telomeres. In favor of this, it has been reported that multiple double-strand DNA breaks (DSBs) are often recruited to a single Rad52-containing focus for DNA repair in S. cerevisiae [55].
The amplification and spreading of Bcl repeats in newly generated stn1-M1 mutants was strongly inhibited by the mismatch repair system. This is not surprising as mismatch repair can inhibit recombination involving sequences with imperfect homology [50]. For example, in S. cerevisiae, 1% and 6% sequence divergence can reduce mitotic recombination ∼20 and ∼140 fold, respectively [56]. Therefore, the 4% sequence divergence between Bcl repeats and WT repeats in our study may significantly reduce the recombination between the telomeres containing them. Mismatch repair has been suggested to inhibit recombination by rejecting or processing the heteroduplex formed by strand invasion occurring with homeologous sequences [50]. Loss of MSH2 was previously shown to facilitate the recombination that generates post-senescence survivors in telomerase deletion mutants of S. cerevisiae and K. lactis [57]. This was attributed to increased recombination between the homeologous telomeric repeats in S. cerevisiae and between the homeologous subtelomeric sequences in K. lactis. Deficiency in mismatch repair can also facilitate ALT-like telomerase-independent telomere elongation in human colon cancer cell lines and in gastric carcinomas [20], [58]. Conceivably, this effect stems from the degenerate telomeric repeats that are common in the basal regions of human telomeres [59]–[60].
In stn1-M1 mutants formed in a ter1-Δ MSH2 background, there was not only a substantial reduction in the frequency of clones exhibiting total spreading of Bcl repeats but also, when spreading was observed, it was far more likely to be partial, accompanied by substantial amplification of WT repeats as well. We suggest that in these clones, formation of a t-circle from the Bcl telomere will occur efficiently (as no mismatches would be present during intratelomeric recombination) but copying sequence from a t-circle composed purely of Bcl repeats onto any of the 11 wild type telomeres would be impeded. When copying of a Bcl repeat t-circle did occur, the impeded ability of the Bcl repeats to recombine with the resident wild type telomeres could allow time for wild type t-circles to form and have their sequences amplified.
Interestingly, mismatch repair was apparently not an appreciable barrier to recombination between Bcl and WT repeats in STN1 during the formation of post-senescence survivors in ter1-Δ mutants [37]. One possibility to account for this is that the extremely short telomeres in senescing cells of these mutants recombine in a different way than the somewhat longer telomeres in newly forming stn1-M1 mutants. This is supported by the fact that the type II RTE in S. cerevisiae depends on a pathway involving Rad50 when occurring at very short telomeres in a senescing telomerase deletion mutant but depends on the more standard Rad51-dependent pathway when occurring at longer telomeres uncapped by defects in telomere capping proteins [30], [40]. Also, Rad51-dependent recombination is inhibited by mismatch repair 13-fold more than Rad50-dependent recombination [61]. The formation of post-senescence survivors in K. lactis STN1 ter1-Δ mutants has recently been shown to require both the Rad50 - and the Rad51 - pathways (Basenko and McEachern, unpublished data).
Telomerase is active in stn1-M1 cells, but its presence does not grossly affect the phenotype of the mutant [42]. This indicates that the recombination by itself is capable of generating and maintaining the very long and unstable telomeres of stn1-M1 cells and that the recombinational processes of type IIR RTE in stn1-M1 cells are not suppressed by the presence of telomerase. Our results here demonstrate that the presence of telomerase at the establishment of the stn1-M1 state can largely inhibit the amplification and spreading of Bcl repeats. While we cannot rule out the possibility that telomerase fundamentally alters the mechanism by which telomeres are maintained in stn1-M1 cells, we believe this is unlikely. Rather, we suggest that sequence addition by telomerase masks the effects of recombination. In particular, we suggest that the stn1-M1 mutation, like certain telomeric repeat mutations shown to produce type IIR RTE, is disrupted not only in telomeres' ability to block recombination but also in their ability to negatively regulate sequence addition by telomerase [37], [41]. Consistent with this possibility is our observation that telomerase inhibits the spreading of Bcl repeats not only from a normal length telomere, but also from long Bcl telomere in stn1-M1 cells. The latter is resistant to telomerase addition in wild type K. lactis cells [37]. With multiple telomerase-synthesized WT telomeric repeats added onto the ends of both long and normal length Bcl telomeres, t-circles formed from terminal deletions of these telomeres would likely often be composed only of WT repeats. This would of course interfere with the ability of the Bcl repeats to amplify and spread to other telomeres. The relative contribution of recombination and telomerase to new telomeric repeat synthesis in stn1-M1 TER1 cells is not fully known. In experiments we performed where TER1-20C(ApaL) was present during the establishment of newly generated stn1-M1 mutants, we found that ApaL repeats were present but only as a minority of the telomeric repeats in each of multiple clones examined (data not shown). This argues that recombination is the predominant mechanism for telomeric repeat amplification in stn1-M1 cells. It is reasonable to believe, however, that the contribution of telomerase might be greatest at the earliest stages of formation of the stn1-M1 state, before recombination has abundant long telomeric sequences available for it to copy.
One unexpected result from our work was that precursor cells with ApaL repeats present at the termini of all telomeric ends produced stn1-M1 mutants where the ApaL repeats were generally completely absent. This occurred in both msh2-Δ and MSH2 backgrounds and therefore does not appear to require mismatch repair. As the ApaL telomeric base change does not appear to alter telomere function [49], this effect is also not likely to be due to selection against amplification of ApaL repeats. The simplest explanation for this outcome would be that a significant fraction of telomeric termini, at least a third of the ∼350–600 bp telomeres by our estimates, is routinely deleted prior to the initiation of recombination events that elongate telomeres when the stn1-M1 mutant state is established. Telomere shortening also precedes RTE in yeast telomerase deletion mutants. In that case, however, the shortening occurs very gradually over many tens of cell divisions from replicative sequence loss [54], [62]–[63]. Such gradual telomere shortening cannot explain the terminal sequences loss we see in stn1-M1 cells. Most newly generated spores of the stn1-M1 mutant die within a few cell divisions [42]. This indicates that they experience a severe growth problem immediately after their generation and suggests that the terminal telomeric loss occurs very rapidly. Indeed, the rapid telomeric deletion might help account for the poor viability of stn1-M1 spores.
A number of mechanisms have been proposed for generating telomeric deletions (for a review see [64]). One well studied mechanism is telomere rapid deletion (TRD) which is thought to be an intratelomeric recombination event that occurs after a telomeric 3′ overhang strand invades into the double-stranded region of the same telomere (for a review see [65]). TRD can lead to telomere deletion in wild type yeast cells that contain artificially elongated telomeres [66]–[68]. Processes similar to TRD, requiring the recombination protein XRCC3, can lead to shortening of both normal and dysfunctional mammalian telomeres [18], [69]. TRD has been proposed to be a mechanism that can generate t-circles [34], [68].
An obvious question that arises from our data is why telomeric truncations would predominate at the earliest stage of the formation of a mutant that ultimately generates and maintains highly elongated telomeres. At least two possibilities exist. One is that the physiological conditions at the earliest stage of stn1-M1 mutants, when telomere deletions occur, are different from those at later stages, when telomere elongation predominates. Conceivably, later stages might be influenced by the presence of chronic DNA damage and react differently to dysfunctional telomeres compared to cells at the earliest stage. This idea is supported by the finding that S. cerevisiae telomerase deletion mutants showed expression changes in hundreds of genes during the senescence caused by shorting telomeres [70]. Another possibility is that telomeric deletions are always more frequent than recombination events that lengthen telomeres in stn1-M1 cells. In this scenario, net lengthening might only predominate over shortening once long telomeres or t-circles are present and abundant enough to serve as efficient templates for elongation events that can generate long extensions. Some observations support this possibility as well. In wild type K. lactis cells, deletions from TRD are approximately an order of magnitude more frequent than telomere elongation by recombination [67]. Also, telomeric deletions are very frequent in stn1-M1 cells and in other mutants undergoing type IIR RTE [41], [48].
Taken together, our results suggest that the establishment of long and heterogeneous telomeres in stn1-M1 via type IIR RTE may involve the following events as summarized in Figure 7. First, upon initiating the stn1-M1 state, the uncapped telomeres rapidly undergo net deletion to generally lose at least one third of the repeats from telomeric termini. Next, an occasional telomeric recombination event results in the production of a small t-circle that is used as a template to lengthen one or more telomeres. These t-circles are likely derived from the more basal repeats of a telomere as indicated by the absence of amplification of ApaL repeats in most clones in our experiments. Finally, the initial elongated telomere(s) serve as the templates for lengthening most or all remaining telomeres, generally before other t-circles can form and compete for being copied. Another study of ours [48] that examined the stability of tandem arrays at telomeres in stn1-M1 cells, suggests that once the long telomere state is established, the maintenance of it likely includes secondary formation and copying of t-circles. However, the spreading of sequence from a single source to all telomeres in these secondary amplifications was rare or absent, presumably because cells already contained many potential templates (both t-circles and linear telomeric tracts) that could be copied to generate long telomeric arrays.
Fig. 7. Modified roll-and-spread model of recombinational telomere elongation during the establishment of stn1-M1 ter1-Δ cells. 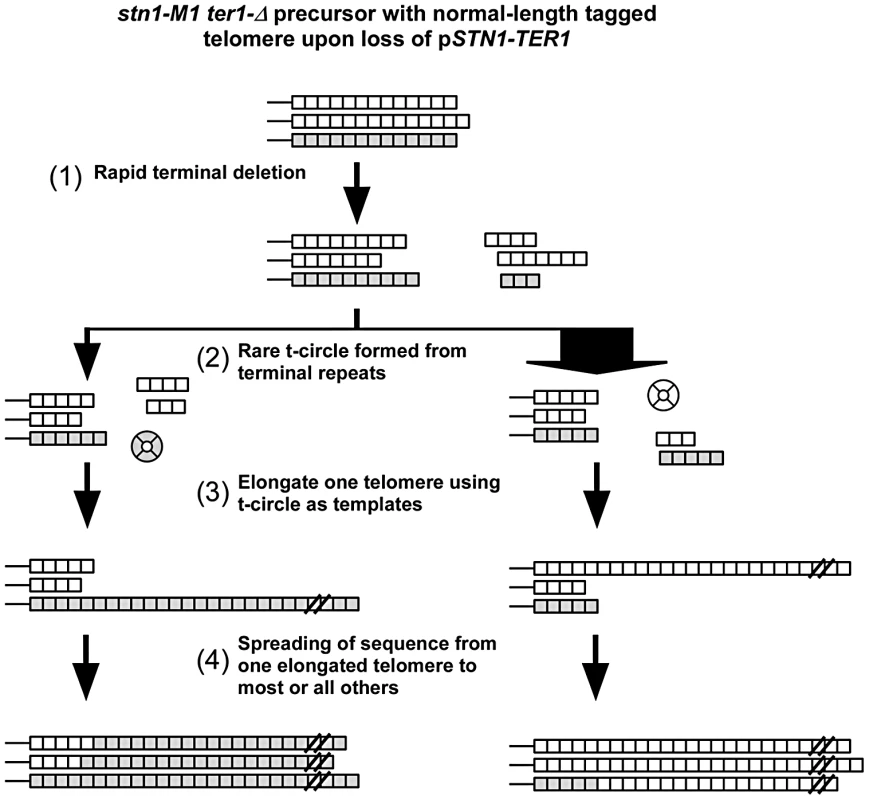
(1) Upon loss of the complemented wild type STN1 and TER1 alleles, the terminal telomeric repeats are subject to rapid deletion; (2) A rare formation of a t-circle, likely as a deletion product of a single telomere. As 11 of 12 telomeres are composed of wild type repeats, most t-circles formed would have wild type repeats, as indicated by the width of the arrows; (3) At least one telomere is elongated through rolling circle synthesis using the t-circle as the template; (4) the sequence of the elongated telomere is spread to most or all other telomeres. The extent to which our results with stn1-M1 cells bear on human ALT cells remains to be determined. T-circles are known to be abundant in ALT cells [17], however, there are conflicting data regarding their importance. While Ku mutations inhibit formation of t-circles and block proliferation of ALT cells [71], mutations in Xrcc3 and Nbs1 have been reported to eliminate t-circles without blocking ALT cell growth [72]. Our study suggests that it will be important to examine not only the maintenance of long telomeres in established ALT cells but also the role of t-circles during the initial establishment of the ALT cell state. Our data also suggest that a single telomeric template DNA might commonly be the source of elongation of multiple, even all, other telomeres. Interestingly, multiple telomeres in ALT cells have been shown to cluster into single foci which co-localize with PML bodies in a manner that could potentially facilitate recombination between telomeres [73]. However, there are other reasons to believe that a single telomere is unlikely to be the source of all telomere elongation during the establishment of ALT. These include that there far more telomeres in a mammalian cell and that the establishment of the ALT state might not be nearly as abrupt as the establishment of the stn1-M1 state. Our finding that telomerase can have some effects on telomeric recombination in stn1-M1 cells may have parallels in ALT. Indeed, expression of telomerase was shown to reduce the number of telomeres clustered in single foci in the ALT cells and could therefore decrease the frequency of recombination between telomeres [73]. Finally we would note that ALT does not arise in human cells from a simple lack of telomerase activity. This suggests that ALT likely requires mutations that allow telomeric recombination to occur at elevated frequencies. Mutations in yeast such as the stn1-M1 mutation may suggest that alterations in human telomere binding proteins might be a possible cause or contributor to the ALT phenotype. Much further work will be needed to fully understand the mechanisms involved in the establishment and maintenance of ALT.
Materials and Methods
Strains and plasmid
All K. lactis strains used are derivatives of wild type (WT) 7B520 [74]. K. lactis stn1-M1, stn1-M1 ter1-Δ strains were described previously [42]. The precursors of K. lactis stn1-M1 msh2-Δ and stn1-M1 ter1-Δ msh2-Δ were constructed as follows. The MSH2 gene was first amplified as a 4.4 kb fragment from genomic DNA of 7B520 cells by PCR (forward primer: 5′AGGGATCCGGGAGGCTCCAATAACAACA3′; reverse primer: 5′ACCTCGAGTTGCGAGTGATTCGTTCAAG3′) and cloned into the BamHI and XhoI sites of pBluescript IIKS(−) resulting in pMSH2. Then, pMSH2 was digested by BglII and EcoRI to delete an 891 bp fragment of the ORF of MSH2, and a 1.4 kb PCR-amplified (forward primer: 5′AGGCAGATCTGGATGGCGGCGTTAGTATCG3′; reverse primer: 5′AGGAATTCCCAGCGACATGGAGGCCCAG3′) fragment of KANMX gene from the genomic DNA of SAY557 [75] was inserted into the BglII+EcoRI − digested pMSH2 to produce the pMSH2::KANMX. The 3.2 kb disruption cassette containing MSH2::KANMX was then amplified from pMSH2::KANMX by PCR (forward primer: 5′ATATTGCAGAGGAGCGAGGA3′; reverse primer: 5′CTTGTACGGACGGGTCATCT3′) and was transformed into either stn1-M1 cells complemented with pSTN1 or stn1-M1 ter1-Δ cells complemented with pSTN1-TER1 or pSTN1-TER1(ApaL). The knockout of the MSH2 gene was confirmed by Southern blotting and hybridization to a MSH2 probe.
The plasmid pSTN1 was constructed in following two steps. First, a 3.4 kb fragment containing the 1.3 kb ORF of the STN1 gene and 1.6 kb upstream and 0.5 kb downstream sequences was obtained by PCR (forward primer: 5′ACGAGCTCTGGCAACCCACTTGTGACTA3′; reverse primer: 5′ACCTCGAGTGCTCAGCCAATTTCTGTTG3′) using the genomic DNA of the 7B520 strain as the template. Second, the PCR fragment, which contains flanking SacI and XhoI sites, was inserted into the polylinker SacI and XhoI sites of pKL313(HIS3) [51] to generate the pSTN1. Plasmids pSTN1-TER1 and pSTN1-TER1(ApaL) were constructed as follows. First, the STN1 gene was cloned as a 3.4 kb fragment from genomic DNA of 7B520 cells by PCR (forward primer: 5′ACGAGCTCTGGCAACCCACTTGTGACTA3′; reverse primer: 5′ACGGTACCTGCTCAGCCAATTTCTGTTG3′) into the SacI and KpnI sites of pCXJ18 [76] to result in plasmid pCXJ18-STN1. The 2.6 kb TER1 or TER1(ApaL) gene fragments flanked by XbaI and KpnI sites were obtained from pJR31 [37], and pJR31 derivative that contained a mutation in the template of TER1 changing T20 to C to create an ApaLI restriction site by oligonucleotide mediated site-directed mutagenesis as described elsewhere [77]. Subsequently, the 2.6 kb TER1 or TER1(ApaL) fragment was cloned into the pCXJ18-STN1 to generate pSTN1-TER1 or pSTN1-TER1(ApaL).
The URA3-tagged single Bcl telomeres containing ∼11 and ∼40 Bcl-telomeric repeats were described before [37]. The HIS3-tagged single Bcl telomeres were based on the URA3-tagged single Bcl telomeres, which were cleaved by PstI and NruI to excise the URA3 gene and replace it with a 1.2 kb HIS3 fragment amplified from pKL313 [51] by PCR (forward primer: 5′ACAGTGCTGCAGCGGCATCAGAGCAGATTGTA3′; reverse primer: 5′ACTGAGTCGCGATCTGTGCGGTATTTCACACC3′). Either URA3-tagged or HIS3-tagged single Bcl telomeres were transformed into stn1-M1 cells with a MSH2 or a msh2-Δ genetic background complemented by pSTN1, or stn1-M1 ter1-Δ cells with a MSH2 or a msh2-Δ genetic background complemented by pSTN1-TER1 respectively. Ura+ or His+ colonies were then examined by Southern blotting to confirm that URA3 or HIS3-tagged single Bcl telomeres had replaced a single native telomere by subtelomeric recombination.
K. lactis transformation was done by electroporation as described for S. cerevisiae [78]. Passaging of complemented cells was carried out by serial streaking of single colonies on rich medium (YPD plates) at 30°C. Strains were streaked every 3 days down to single cells that grew into colonies. Each streak was estimated to be 20–25 cell divisions.
Telomere cloning and sequencing
The yeast genomic DNA sample from clone A1 in Figure 2A was partially digested with ApaLI. This was terminated by an equal volume of 12.5 mM EDTA added to the digestion reaction. The ApaLI partially digested DNA was ligated with the ApaLI-digested pACYC177 plasmid, and transformed into DH5α cells. The clones with telomeric fragments were confirmed by a Southern blot hybridized to telomeric probe. Positive clones were then sequenced.
Southern hybridizations
Yeast genomic DNA samples digested with restriction enzymes were run on 0.8% or 3% agarose gels and then transferred onto Hybond N+ membrane in 0.4 M NaOH. All hybridization were carried out in Na2HPO4 and SDS as described [79]. The γ-32P-labeled telomeric probe is Klac 1–25 [63] The temperature of hybridization and washing for this probe was between 45–50°C. The γ-32P-labeled Bcl telomeric probe was KTelBcl (GATCAGGTATGTGG) [51] The temperature of hybridization and washing was 40°C and 34–36°C respectively. The subtelomeric probe was generated from pKL11-B (Insert of ∼1 kb telomeric EcoRI-SmaI fragment into pBluescript SK−), which was digested with XbaI and ligated back together to excise all the telomeric sequence and was then digested by EcoRI and XbaI to generate a ∼600 bp subtelomeric fragment for probe. The URA3 probe was described before [80]. The RAD50 gene probe was the purified PCR product from K. lactis genomic DNA (Forward primer: 5′GATAGGTCTACCGCGACCAA3′; Reverse primer: 5′GCGTAAGAGGACGCATTCAT3′). Subtelomeric, URA3, and RAD50 probes were prepared using a random priming kit (NEB). Temperature of hybridization and washing for these probes was 65°C. The membranes were autoradiographed and visualized using a Molecular Dynamics Storm PhosphorImager.
Supporting Information
Zdroje
1. RothCW, KobeskiF, WalterMF, BiessmannH (1997) Chromosome end elongation by recombination in the mosquito Anopheles gambiae. Mol Cell Biol 17 : 5176–5183.
2. BiessmannH, DonathJ, WalterMF (1996) Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol Biol 5 : 11–20.
3. MorinGB, CechTR (1988) Mitochondrial telomeres: surprising diversity of repeated telomeric DNA sequences among six species of Tetrahymena. Cell 52 : 367–374.
4. TomaskaL, NosekJ, MakhovAM, PastorakovaA, GriffithJD (2000) Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res 28 : 4479–4487.
5. NosekJ, RycovskaA, MakhovAM, GriffithJD, TomaskaL (2005) Amplification of telomeric arrays via rolling-circle mechanism. J Biol Chem 280 : 10840–10845.
6. CesareAJ, ReddelRR (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11 : 319–330.
7. MurnaneJP, SabatierL, MarderBA, MorganWF (1994) Telomere dynamics in an immortal human cell line. EMBO J 13 : 4953–4962.
8. HensonJD, NeumannAA, YeagerTR, ReddelRR (2002) Alternative lengthening of telomeres in mammalian cells. Oncogene 21 : 598–610.
9. JeyapalanJN, Mendez-BermudezA, ZaffaroniN, DubrovaYE, RoyleNJ (2008) Evidence for alternative lengthening of telomeres in liposarcomas in the absence of ALT-associated PML bodies. Int J Cancer 122 : 2414–2421.
10. PerremK, ColginLM, NeumannAA, YeagerTR, ReddelRR (2001) Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol Cell Biol 21 : 3862–3875.
11. YeagerTR, NeumannAA, EnglezouA, HuschtschaLI, NobleJR, et al. (1999) Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59 : 4175–4179.
12. JiangWQ, ZhongZH, HensonJD, NeumannAA, ChangAC, et al. (2005) Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol Cell Biol 25 : 2708–2721.
13. JiangWQ, ZhongZH, HensonJD, ReddelRR (2007) Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene 26 : 4635–4647.
14. PottsPR, YuH (2007) The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol 14 : 581–590.
15. DunhamMA, NeumannAA, FaschingCL, ReddelRR (2000) Telomere maintenance by recombination in human cells. Nat Genet 26 : 447–450.
16. MuntoniA, NeumannAA, HillsM, ReddelRR (2009) Telomere elongation involves intra-molecular DNA replication in cells utilizing alternative lengthening of telomeres. Hum Mol Genet 18 : 1017–1027.
17. CesareAJ, GriffithJD (2004) Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol 24 : 9948–9957.
18. WangRC, SmogorzewskaA, de LangeT (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119 : 355–368.
19. ZengS, XiangT, PanditaTK, Gonzalez-SuarezI, GonzaloS, et al. (2009) Telomere recombination requires the MUS81 endonuclease. Nat Cell Biol 11 : 616–623.
20. BechterOE, ZouY, WalkerW, WrightWE, ShayJW (2004) Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res 64 : 3444–3451.
21. Londono-VallejoJA, Der-SarkissianH, CazesL, BacchettiS, ReddelRR (2004) Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res 64 : 2324–2327.
22. BaileySM, BrennemanMA, GoodwinEH (2004) Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res 32 : 3743–3751.
23. LundbladV, BlackburnEH (1993) An alternative pathway for yeast telomere maintenance rescues est1 - senescence. Cell 73 : 347–360.
24. McEachernMJ, BlackburnEH (1996) Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev 10 : 1822–1834.
25. CiudadT, AndaluzE, Steinberg-NeifachO, LueNF, GowNA, et al. (2004) Homologous recombination in Candida albicans: role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol Microbiol 53 : 1177–1194.
26. NakamuraTM, CooperJP, CechTR (1998) Two modes of survival of fission yeast without telomerase. Science 282 : 493–496.
27. McEachernMJ, HaberJE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75 : 111–135.
28. TsukamotoY, TaggartAK, ZakianVA (2001) The role of the Mre11-Rad50-Xrs2 complex in telomerase - mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol 11 : 1328–1335.
29. ChenQ, IjpmaA, GreiderCW (2001) Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol 21 : 1819–1827.
30. TengSC, ChangJ, McCowanB, ZakianVA (2000) Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell 6 : 947–952.
31. TengSC, ZakianVA (1999) Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19 : 8083–8093.
32. LeS, MooreJK, HaberJE, GreiderCW (1999) RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152 : 143–152.
33. CohenH, SinclairDA (2001) Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc Natl Acad Sci USA 98 : 3174–3179.
34. Groff-vindmanC, CesareAJ, NatarajanS, GriffithJD, McEachernMJ (2005) Recombination at long mutant telomeres produces tiny single - and double-stranded telomeric circles. Mol Cell Biol 25 : 4406–4412.
35. NatarajanS, Groff-vindmanC, McEachernMJ (2003) Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot Cell 2 : 1115–1127.
36. NatarajanS, McEachernMJ (2002) Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol 22 : 4512–4521.
37. TopcuZ, NicklesK, DavisC, McEachernMJ (2005) Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc Natl Acad Sci USA 102 : 3348–3353.
38. BasenkoEY, CesareAJ, IyerS, GriffithJD, McEachernMJ (2010) Telomeric circles are abundant in the stn1-M1 mutant that maintains its telomeres through recombination. Nucleic Acids Res 38 : 182–189.
39. LinCY, ChangHH, WuKJ, TsengSF, LinCC, et al. (2005) Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot Cell 4 : 327–336.
40. GrandinN, CharbonneauM (2003) The Rad51 pathway of telomerase-independent maintenance of telomeres can amplify TG1–3 sequences in yku and cdc13 mutants of Saccharomyces cerevisiae. Mol Cell Biol 23 : 3721–3734.
41. BechardLH, ButunerBD, PetersonGJ, TopcuZ, McEachernMJ (2009) Mutant relomeric repeats in yeast can disrupt the negative regulation of recombination-mediated telomere maintenance and create an alternative lengthening of telomeres-like phenotype. Mol Cell Biol 29 : 626–639.
42. IyerS, ChadhaAD, McEachernMJ (2005) A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol Cell Biol 25 : 8064–8073.
43. PriceCM, BoltzKA, ChaikenMF, StewartJA, BeilsteinMA, et al. (2010) Evolution of CST function in telomere maintenance. Cell Cycle 9 : 3157–3165.
44. GrandinN, DamonC, CharbonneauM (2000) Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol 20 : 8397–8408.
45. PuglisiA, BianchiA, LemmensL, DamayP, ShoreD (2008) Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J 27 : 2328–2339.
46. RoganEM, BryanTM, HukkuB, MacleanK, ChangAC, et al. (1995) Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol Cell Biol 15 : 4745–4753.
47. OganesianL, KarlsederJ (2011) Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell 42 : 224–236.
48. XuJ, McEachernMJ (2012) Maintenance of very long telomeres by recombination in the Kluyveromyces lactis stn1-M1 mutant involves extreme telomeric turnover, telomeric circles and concerted telomeric amplification. Mol Cell Biol 32 : 2992–3008.
49. UnderwoodDH, McEachernMJ (2004) Genetic dissection of the Kluyveromyces lactis telomere and evidence for telomere capping defects in TER1 mutants with long telomeres. Eukaryot Cell 3 : 369–384.
50. JiricnyJ (2006) The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 7 : 335–346.
51. RoyJ, FultonTB, BlackburnEH (1998) Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev 12 : 3286–3300.
52. UnderwoodDH, McEachernMJ (2001) Totally mutant telomeres: single-step mutagenesis of tandem repeat DNA sequences. Biotechniques 30 : 934–935, 938.
53. McEachernMJ, UnderwoodDH, BlackburnEH (2002) Dynamics of telomeric DNA turnover in yeast. Genetics 160 : 63–73.
54. LundbladV, SzostakJW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57 : 633–643.
55. LisbyM, MortensenUH, RothsteinR (2003) Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 5 : 572–577.
56. ChenW, Jinks-RobertsonS (1999) The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics 151 : 1299–1313.
57. RizkiA, LundbladV (2001) Defects in mismatch repair promote telomerase-independent proliferation. Nature 411 : 713–716.
58. OmoriY, NakayamaF, LiD, KanemitsuK, SembaS, et al. (2009) Alternative lengthening of telomeres frequently occurs in mismatch repair system-deficient gastric carcinoma. Cancer Sci 100 : 413–418.
59. BairdDM, JeffreysAJ, RoyleNJ (1995) Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J 14 : 5433–5443.
60. AllshireRC, DempsterM, HastieND (1989) Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res 17 : 4611–4627.
61. SpellRM, Jinks-RobertsonS (2003) Role of mismatch repair in the fidelity of RAD51 - and RAD59-dependent recombination in Saccharomyces cerevisiae. Genetics 165 : 1733–1744.
62. SingerMS, GottschlingDE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266 : 404–409.
63. McEachernMJ, BlackburnEH (1995) Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376 : 403–409.
64. BairdDM (2008) Mechanisms of telomeric instability. Cytogenet Genome Res 122 : 308–314.
65. LustigAJ (2003) Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet 4 : 916–923.
66. LiB, LustigAJ (1996) A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev 10 : 1310–1326.
67. BechardLH, JamiesonN, McEachernMJ (2011) Recombination can cause telomere elongations as well as truncations deep within telomeres in wild-type Kluyveromyces lactis cells. Eukaryot Cell 10 : 226–236.
68. BucholcM, ParkY, LustigAJ (2001) Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol Cell Biol 21 : 6559–6573.
69. PickettHA, HensonJD, AuAY, NeumannAA, ReddelRR (2011) Normal mammalian cells negatively regulate telomere length by telomere trimming. Hum Mol Genet
70. NautiyalS, DeRisiJL, BlackburnEH (2002) The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99 : 9316–9321.
71. LiB, ReddyS, ComaiL (2011) Depletion of Ku70/80 reduces the levels of extrachromosomal telomeric circles and inhibits proliferation of ALT cells. Aging (Albany NY) 3 : 395–406.
72. ComptonSA, ChoiJH, CesareAJ, OzgurS, GriffithJD (2007) Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res 67 : 1513–1519.
73. DraskovicI, ArnoultN, SteinerV, BacchettiS, LomonteP, et al. (2009) Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci USA 106 : 15726–15731.
74. WrayLVJr, WitteMM, DicksonRC, RileyMI (1987) Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol 7 : 1111–1121.
75. KegelA, MartinezP, CarterSD, AstromSU (2006) Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res 34 : 1633–1645.
76. ChenXJ (1996) Low - and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene 172 : 131–136.
77. KunkelTA, RobertsJD, ZakourRA (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol 154 : 367–382.
78. BeckerDM, FikesJD, GuarenteL (1991) A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci USA 88 : 1968–1972.
79. ChurchGM, GilbertW (1984) Genomic sequencing. Proc Natl Acad Sci USA 81 : 1991–1995.
80. McEachernMJ, IyerS (2001) Short telomeres in yeast are highly recombinogenic. Mol Cell 7 : 695–704.
81. WangZR, GuoL, ChenL, McEachernMJ (2009) Evidence for an additional base-pairing element between the telomeric repeat and the telomerase RNA template in Kluyveromyces lactis and other yeasts. Mol Cell Biol 29 : 5389–5398.
82. FultonTB, BlackburnEH (1998) Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol Cell Biol 18 : 4961–4970.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Intrauterinní inseminace a její úspěšnost
- Příjem alkoholu a menstruační cyklus
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
- Růst a vývoj dětí narozených pomocí IVF
- Akutní intermitentní porfyrie
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



