-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
The seed maturation program only occurs during late embryogenesis, and repression of the program is pivotal for seedling development. However, the mechanism through which this repression is achieved in vegetative tissues is poorly understood. Here we report a microRNA (miRNA)–mediated repression mechanism operating in leaves. To understand the repression of the embryonic program in seedlings, we have conducted a genetic screen using a seed maturation gene reporter transgenic line in Arabidopsis (Arabidopsis thaliana) for the isolation of mutants that ectopically express seed maturation genes in leaves. One of the mutants identified from the screen is a weak allele of ARGONAUTE1 (AGO1) that encodes an effector protein for small RNAs. We first show that it is the defect in the accumulation of miRNAs rather than other small RNAs that causes the ectopic seed gene expression in ago1. We then demonstrate that overexpression of miR166 suppresses the derepression of the seed gene reporter in ago1 and that, conversely, the specific loss of miR166 causes ectopic expression of seed maturation genes. Further, we show that ectopic expression of miR166 targets, type III homeodomain-leucine zipper (HD-ZIPIII) genes PHABULOSA (PHB) and PHAVOLUTA (PHV), is sufficient to activate seed maturation genes in vegetative tissues. Lastly, we show that PHB binds the promoter of LEAFY COTYLEDON2 (LEC2), which encodes a master regulator of seed maturation. Therefore, this study establishes a core module composed of a miRNA, its target genes (PHB and PHV), and the direct target of PHB (LEC2) as an underlying mechanism that keeps the seed maturation program off during vegetative development.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003091
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003091Summary
The seed maturation program only occurs during late embryogenesis, and repression of the program is pivotal for seedling development. However, the mechanism through which this repression is achieved in vegetative tissues is poorly understood. Here we report a microRNA (miRNA)–mediated repression mechanism operating in leaves. To understand the repression of the embryonic program in seedlings, we have conducted a genetic screen using a seed maturation gene reporter transgenic line in Arabidopsis (Arabidopsis thaliana) for the isolation of mutants that ectopically express seed maturation genes in leaves. One of the mutants identified from the screen is a weak allele of ARGONAUTE1 (AGO1) that encodes an effector protein for small RNAs. We first show that it is the defect in the accumulation of miRNAs rather than other small RNAs that causes the ectopic seed gene expression in ago1. We then demonstrate that overexpression of miR166 suppresses the derepression of the seed gene reporter in ago1 and that, conversely, the specific loss of miR166 causes ectopic expression of seed maturation genes. Further, we show that ectopic expression of miR166 targets, type III homeodomain-leucine zipper (HD-ZIPIII) genes PHABULOSA (PHB) and PHAVOLUTA (PHV), is sufficient to activate seed maturation genes in vegetative tissues. Lastly, we show that PHB binds the promoter of LEAFY COTYLEDON2 (LEC2), which encodes a master regulator of seed maturation. Therefore, this study establishes a core module composed of a miRNA, its target genes (PHB and PHV), and the direct target of PHB (LEC2) as an underlying mechanism that keeps the seed maturation program off during vegetative development.
Introduction
Seed maturation is a highly coordinated developmental phase in which storage reserves, including seed storage proteins (SSPs), are synthesized and accumulated to high levels. The maturation genes need to be repressed, however, in order to allow seedling development to occur. Indeed, these genes are not expressed in vegetative organs of the plant [1]. Research in the past decade with the model plant Arabidopsis has led to the identification of repressors of seed maturation genes in vegetative organs (reviewed in [2]), including chromatin-remodelling ATPases [3]–[5], polycomb group (PcG) proteins [6]–[9], histone deacetylases [10], and DNA-binding transcription factors [11]–[13]. However, our understanding of the molecular mechanisms that repress the seed maturation program during vegetative development remains fragmented, and thus continued efforts are needed to identify additional factors involved and, more importantly, the molecular and functional links between the various components.
In Arabidopsis, ABA-INSENSITIVE3 (ABI3), FUSCA3 (FUS3), LEC1 and LEC2 are master regulators of seed maturation [14]–[17], and they regulate one another [18], [19]. ABI3, FUS3 and LEC2 are closely-related members of a plant-specific B3-domain transcription factor family. LEC1 encodes a novel homolog of the CCAAT-binding factor HAP3 subunit. Loss-of-function mutations in ABI3, FUS3, and LEC1 give rise to pleiotropic seed phenotypes including a strong reduction of SSPs. These regulatory genes are predominantly expressed in the seed. When misexpressed in vegetative tissues, they induce ectopic expression of the SSP genes and even the formation of somatic embryos [15], [17], [20]–[22]. It remains poorly understood, however, how the expression and activity of these master regulators are in turn regulated.
Small RNAs of 20–30 nucleotides (nt) have emerged as key sequence-specific regulators of gene expression that influence almost all aspects of plant biology (reviewed in [23]–[26]). There are two major types of small RNAs in plants, microRNA (miRNA) and small interfering RNA (siRNA). Plant miRNAs are generated from longer precursors arising from defined genomic loci – the MIRNA genes. The biogenesis of miRNAs involves several evolutionarily conserved families of proteins, including DICER-LIKE (DCL), ARGONAUTE (AGO), HUA ENHANCER 1 (HEN1), and HASTY (HST). Plant miRNAs regulate target mRNAs temporally and spatially through transcript cleavage and/or translational inhibition. Conserved miRNAs tend to target transcription factor genes that play crucial roles in almost all aspects of plant development. Plants are rich in endogenous siRNAs, which can be classified into several types, such as trans-acting siRNAs (ta-siRNAs), natural cis-antisense transcripts-associated siRNAs, and heterochromatic siRNAs.
Here, we show that mutations in AGO1 resulted in the ectopic expression of seed maturation genes in seedlings. Taking advantage of the weak ago1 allele identified in this work, we were able to identify the miRNA species (miR166) responsible for the repression of seed genes. We demonstrated that targets of miR166, the class III homeodomain leucine zipper (HD-ZIPIII) family of transcription factor genes, PHB and PHV, are positive regulators of seed genes. Further, we provided evidence to suggest that PHB acts directly at LEC2. This work thus uncovered an important role of miR166 in the repression of seed genes during seedling development.
Results
Identification of a Weak ago1 Allele That Causes Ectopic Expression of a Seed Gene Promoter Reporter
We have recently conducted a genetic screen in Arabidopsis to isolate mutants exhibiting ectopic expression of a soybean β-conglycinin gene promoter:GUS transgene (βCG:GUS), which is normally expressed only in the seed [5], [27], [28]. Here, we describe the characterization of one of the mutants identified from the screen, initially named essp5 (ectopic expression of seed storage proteins 5). The essp5 mutant plants exhibited strong ectopic GUS activity in leaves, but not in other organs (Figure 1A–1D). In addition, the mutant plants had pleiotropic developmental defects, such as late flowering, narrow and dark green leaves, shorter siliques and fewer seeds (Figure 1B and Figure S1).
Fig. 1. essp5 Results in Ectopic Expression of a Seed Marker Gene and Is Allelic to ARGONAUTE1. 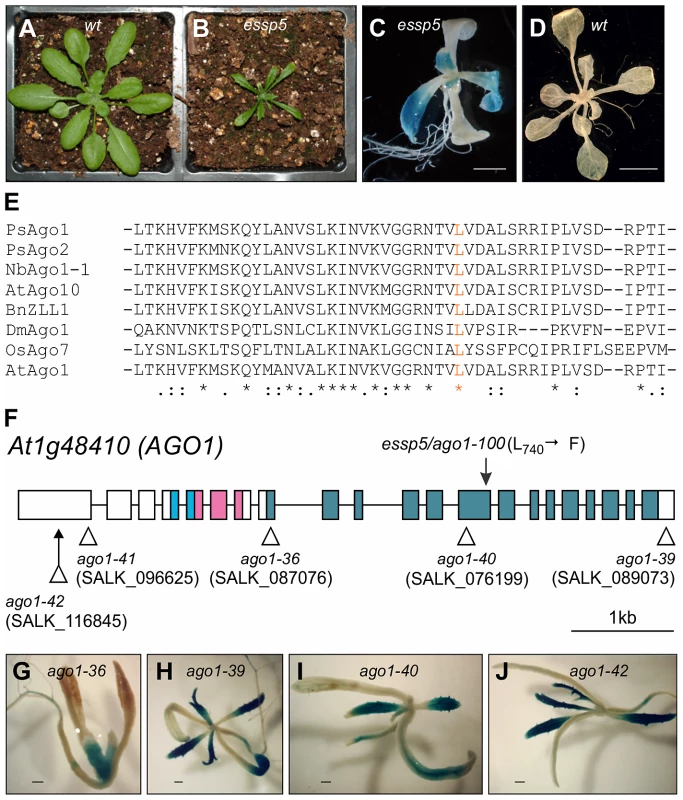
(A and B) Comparison of essp5 with wild type (wt) plants at two weeks. (C and D) GUS expression of the essp5 mutant (C) and wild type (wt, βCG:GUS) grown on agar at two weeks. Scale bars, 2 mm in (C) and 5 mm in (D). (E) Alignment of partial amino acid sequences (residues 707 to 755) of PIWI domains in AGO proteins from Arabidopsis (At), Rice (Os), Pea (Ps), Tobacco (Nb), Drosophila (Dm), Brassica (Bn). The essp5 mutation site (L740) is in orange. The asterisks indicate absolutely conserved residues, colons indicate high similarity, and dots indicate low similarity. (F) Structure of the AGO1 gene showing the location of the essp5 mutation and the T-DNA insertion sites of other ago1 alleles. Boxes and lines represent exons and introns, respectively. The colored boxes represent the conserved protein domains: light blue (PAZ), magenta (MID), and blue (PIWI). (G–J) GUS expression in four T-DNA insertion alleles of AGO1. Shown here is a representative F2 progeny from each of the crosses of the corresponding T-DNA allele with the βCG:GUS line. Scale bar, 500 µm. The mutation segregated as a recessive allele and was mapped to the AT1G48410 gene (Figure S2), which encodes AGO1, the major effector protein that associates with small RNAs [29]. A single missense mutation in this gene was identified that would lead to the conversion of a leucine residue at position 740 to a phenylalanine. The leucine 740 is a highly conserved residue in the PIWI domains of AGOs from diverse species (Figure 1E). A number of mutant ago1 alleles have been described previously and their pleiotropic morphological phenotypes have been documented [29]–[32]. The morphological phenotypes of essp5 resemble those documented for weak ago1 alleles. We obtained T-DNA insertion mutants of ago1, including ago1-36 (SALK_087076), ago1-39 (SALK_089073), ago1-40 (SALK_076199), ago1-41 (SALK_096625), and ago1-42 (SALK_116845) (Figure 1F), crossed them with βCG:GUS and examined GUS expression in F2 progeny seedlings. The T-DNA insertion lines, regardless of their insertion sites, all displayed similar morphological phenotypes: long rod-shape cotyledons, delayed emergence of true leaves, and premature death with only a couple of small true leaves. As shown in Figure 1G–1J, ectopic GUS activity was clearly observed in several T-DNA alleles with insertion sites located throughout the gene. Furthermore, we performed an allelism test to provide additional evidence that essp5 is a weak ago1 allele. A weak ago1 allele (ago1-25) that exhibits similar morphological phenotype [33] was crossed with essp5 and the F1 progeny were examined for GUS activity. As shown in Figure S3, the F1 seedlings displayed ectopic GUS expression, indicating that the essp5 GUS phenotype cannot be complemented by a weak ago1 allele. Together, these observations suggest that essp5 is allelic to AGO1 and thus designated as ago1-100.
Ectopic Expression of Seed Genes in ago1 Mutant Seedlings
To find out whether the endogenous seed maturation genes are indeed ectopically expressed in ago1 mutant seedlings, we performed northern blot analysis to profile the expression of both the 2S and 12S storage protein genes. As shown in Figure 2A, the transcripts of the storage protein genes are highly accumulated in the two strong alleles, ago1-41 and ago1-42, both with T-DNA insertion sites located in the 5′ end of the gene but barely detectable in ago1-100/essp5 and other weak alleles with insertion sites located in the middle and 3′ end of the AGO1 gene. We further examined the transcript levels of the four master regulators by quantitative real-time RT-PCR (qRT-PCR). In line with the ectopic expression of the storage protein genes, all the master regulators are expressed to varying levels in the mutants, especially ago1-42 (Figure 2B). In addition, we also profiled the temporal expression pattern of the maturation genes, using 2S2 as a marker. ago1-41 seedlings from 5 d to 19 d after germination were examined. As shown in Figure 2C, the 2S2 transcript peaked in abundance at around 13 d, but was clearly detected throughout the time course. These expression analyses clearly demonstrate the involvement of AGO1 in the repression of seed maturation genes.
Fig. 2. Expression Analysis of Seed Maturation Genes in ago1 Mutant Seedlings. 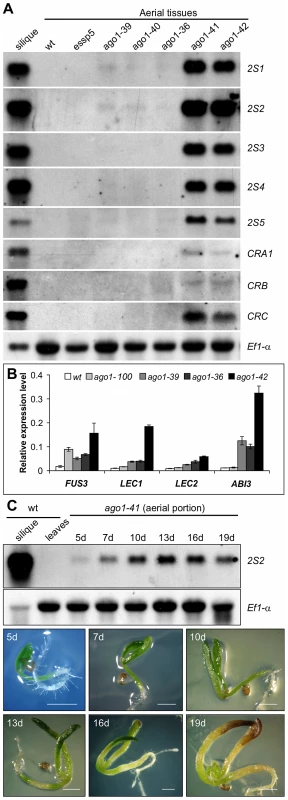
(A) RNA blot analysis of the expression of the five 2S genes and three 12S genes (CRA1, CRB, and CRC) in ago1 mutant seedlings grown for 14 days on MS agar. Wild type (Col) leaves and siliques were used as negative and positive controls, respectively. Same amount of RNA was used for each blot. Elongation factor 1α was used as loading control. (B) Real time quantitative RT-PCR (qRT-PCR) analysis of ABI3, FUS3, LEC1, and LEC2 genes in seedlings (aerial portion) of four ago1 mutants grown for 12 days on MS agar. Actin-8 was used as an internal control. The mean and standard error were determined from three biological replicates, for each of which the PCR was conducted in triplicated technical repeats. (C) RNA blot analysis of temporal expression of maturation genes (2S2 as a marker) in ago1-41 mutant seedlings grown on MS agar for 5, 7, 10, 13, 16, and 19 days. Wild type (Col) leaves and siliques were used as negative and positive controls, respectively. Same amount of RNA was used for each blot. Elongation factor 1α was used as a loading control. Shown at the bottom are images of the ago1-41 mutant on agar at various time points. Scale bar, 1 mm. Derepression of Seed Maturation Genes in Other miRNA Biogenesis Mutants
AGO1 associates with miRNAs and some endogenous siRNAs to mediate their activities [34]. AGO1 association also stabilizes the small RNAs such that ago1 mutants show a reduction in the steady-state levels of miRNAs and siRNAs [35]–[36]. To confirm that the ectopic expression of seed maturation genes in ago1 mutants is due to defects in small RNA biogenesis or activity, we examined seed gene expression in seedlings of loss-of-function alleles of genes commonly involved in small RNA biogenesis. For this purpose, we obtained mutant alleles of HEN1, hen1-5 (SALK_049197) and hen1-6 (SALK_090960), and of HST, hst-1 [37], hst-15 (SALK_079290) and hst-16 (SALK_056352). We introduced these mutations, individually, into the βCG:GUS background and examined GUS expression in the F2 generation. We were able to detect clear ectopic GUS activity in the hen1 backgrounds (hen1-5; Figure 3A and 3B), but not in the hst alleles. We then generated double mutants between ago1, hen1, and hst. As shown in Figure 3C–3J, both ago1-41 hst-16 and hen1-5 hst-16 double mutants exhibited a high level of expression of the storage protein genes and the master regulators. The ago1-41 hen1-5 plants were very small, which precluded seed gene expression analysis. These results indicate synergistic genetic interactions among AGO1, HEN1, and HST in repressing seed genes during seedling development and, more importantly, the involvement of a small RNA pathway(s) in this repression process.
Fig. 3. Ectopic Expression of Seed Maturation Genes in miRNA Biogenesis Mutants. 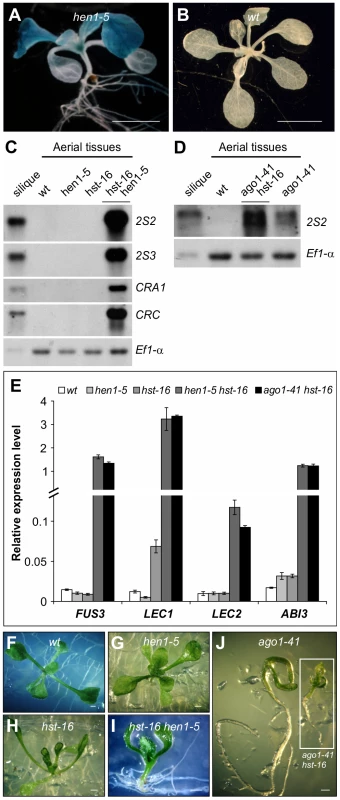
(A and B) GUS expression in hen1 mutant (A) and wild type (wt, βCG:GUS) grown on agar for two weeks. Shown here are representative F2 progeny from the crosses of hen1-5 with the βCG:GUS line. Scale bars, 3 mm in (A) and 5 mm in (B). (C and D) RNA blot analysis of the 2S and 12S genes in seedlings (aerial tissues) in various miRNA biogenesis mutant backgrounds grown for 14 days on MS agar. Wild type (Col) leaves and siliques were used as negative and positive controls, respectively. Same amount of RNA was used for each blot. Elongation factor 1α was used as a loading control. (E) qRT-PCR analysis of ABI3, FUS3, LEC1, and LEC2 genes in seedlings (aerial portion) of various miRNA biogenesis mutants grown for 15 days on MS agar. Actin-8 was used as an internal control. The mean and standard error were determined from three biological replicates. (F–J) Morphological phenotypes of 13-day seedlings of various miRNA biogenesis mutants. Scale bar, 1 mm. Since AGO1, HEN1 and HST are essential players in small RNA biogenesis and are involved in several small RNA pathways [38], it was necessary to determine which pathway underlies the mutant phenotype. To this end, we took advantage of pathway-specific components to define the specific pathway responsible for the ago1 mutant phenotype. Specifically, RDR2 is an essential component of the heterochromatin pathway and RDR6 is required for the biogenesis of trans-acting siRNAs. We obtained and introduced the rdr2-1 (SAIL_1277_H08) and rdr6-11 [39] mutations into the βCG:GUS background by genetic crosses and examined GUS expression in the F2 progeny. A large number of F2 seedlings were stained for GUS and no ectopic GUS activity was observed in either population. This genetic evidence suggests that it is unlikely that the trans-acting siRNA or the hc-siRNA pathway is involved in the repression of seed genes in seedlings. Since AGO1, HEN1, and HST all act in miRNA biogenesis, a miRNA(s) is thought to be a strong candidate for the repression of seed genes during vegetative development.
Reduced Accumulation of Conserved miRNAs in ago1 and Other miRNA Biogenesis Mutants
To provide evidence that a miRNA pathway is indeed underlying the mutant phenotype, we examined the steady-state levels of a number of conserved miRNAs in ago1 and other mutant backgrounds. In ago1 mutants, it was documented previously that the accumulations of a number of conserved miRNAs decline markedly and their target gene transcripts are concomitantly elevated [35]–[36]. Here, we performed a miRNA northern blot analysis to examine and compare the accumulation of conserved miRNA species in ago1, hen1, hst, and the two double mutants, ago1 hst and hen1 hst. As shown in Figure 4, we confirmed the published observation for ago1 in that all the miRNAs examined were clearly reduced. More importantly, we observed further reduced accumulation of most examined miRNAs in ago1 hst and hen1 hst double mutants compared to the single mutants (Figure 4). These findings are consistent with a genetic model for explaining the ectopic seed gene expression in ago1and other mutants: the steady-state level of a specific miRNA was reduced below a threshold to lead to the ectopic expression of its target gene, which encodes a positive regulator of seed maturation genes leading to the ectopic expression of seed genes in leaves.
Fig. 4. Reduced Accumulation of miRNAs in Various miRNA Biogenesis Mutant Backgrounds as Determined by Northern Blotting. 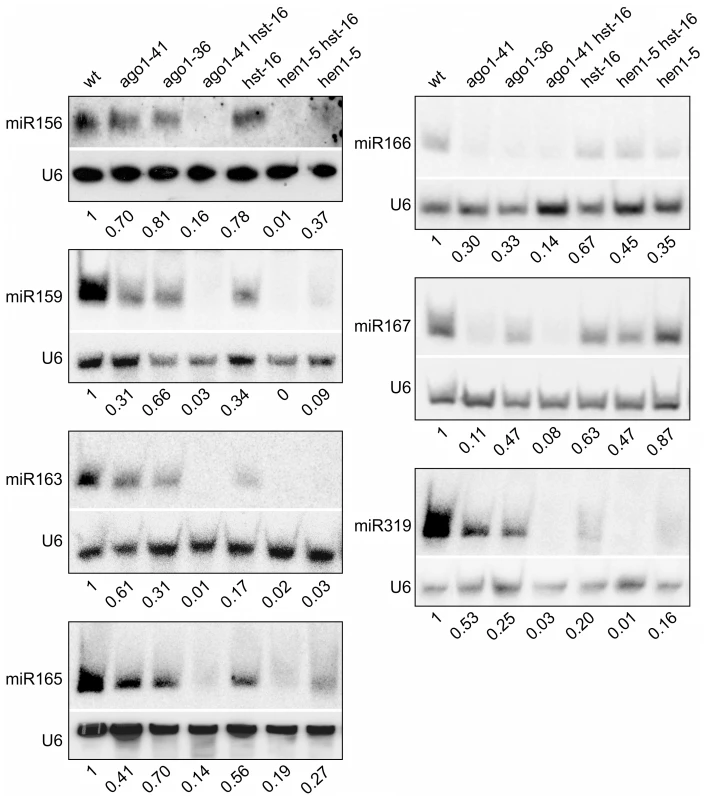
The genotypes of the mutants are indicated at the top. Total RNAs were extracted from two-week old seedlings (aerial portion). The levels of each small RNA were normalized to those of U6 and compared with wild type (Col). The numbers below the gel images indicate the relative abundance of the small RNAs. Overexpression of miRNA166 Rescues the essp5/ago1-100 Mutant Phenotype and Loss of miR166 Causes Ectopic Expression of Seed Maturation Genes
Post-germination repression of seed genes is critical in order for the seedling to develop normally. We thus reasoned that such a fundamental developmental program should be controlled by a conserved miRNA(s). Therefore, to find out which miRNA was involved in conferring the essp5/ago1-100 GUS phenotype, we over-expressed each of the 15 conserved miRNA species, as listed in [23] and Table S1, in the essp5/ago1-100 background and examined GUS expression of the resulting transgenic plants. The transgenic plants overexpressing different miRNAs displayed unique morphological phenotypes, which are consistent with previously published observations (reviewed in [23]).
Analysis of leaf GUS expression was conducted in the T2 generation. For each miRNA transgene, multiple independent transgenic lines were analyzed (in most cases 10 lines); and for each line, at least 20 T2 progeny homozygous for essp5 were stained for GUS activity. We only observed loss of leaf GUS activity in miR166 and miR156 overexpressing lines. In this study, we have focused on the characterization of miR166. In total, we only obtained four miR166 transgenic lines, miR166ox-1-4, of which two showed clear loss of leaf GUS activity (miR166ox-1-2) while the other two (miR166ox-3-4) did not show as obvious a change compared with essp5/ago1-100 seedlings (Figure 5A–5D). The extremely low rate of positive transgenic plants for miR166 is likely due to the fact that some transgenic seedlings failed to develop the shoot apical meristem and could not survive in soil, as observed by others [40]. To confirm that the loss of leaf GUS activity in the transgenic lines was indeed due to the elevated accumulation of miR166, a northern blot analysis was performed. As shown in Figure 5E, there were clearly higher levels of miR166 in lines miR166ox-1-2 than lines miR166ox-3-4. In addition, we observed the formation of aberrant structures on leaves of miRNA166ox-1-2 (Figure 5F). Similar aberrant structures were observed by Zhou et al in miRNA166 overexpressors [40]. These observations suggest that the reduction of miRNA166 and the concomitant accumulation of its target gene transcripts are likely the cause underlying the ectopic GUS phenotype of essp5/ago1-100 seedlings.
Fig. 5. Over-Expression of miRNA166 Rescues the essp5 GUS Phenotype, and Loss of miR166 Causes Ectopic Expression of Seed Maturation Genes. 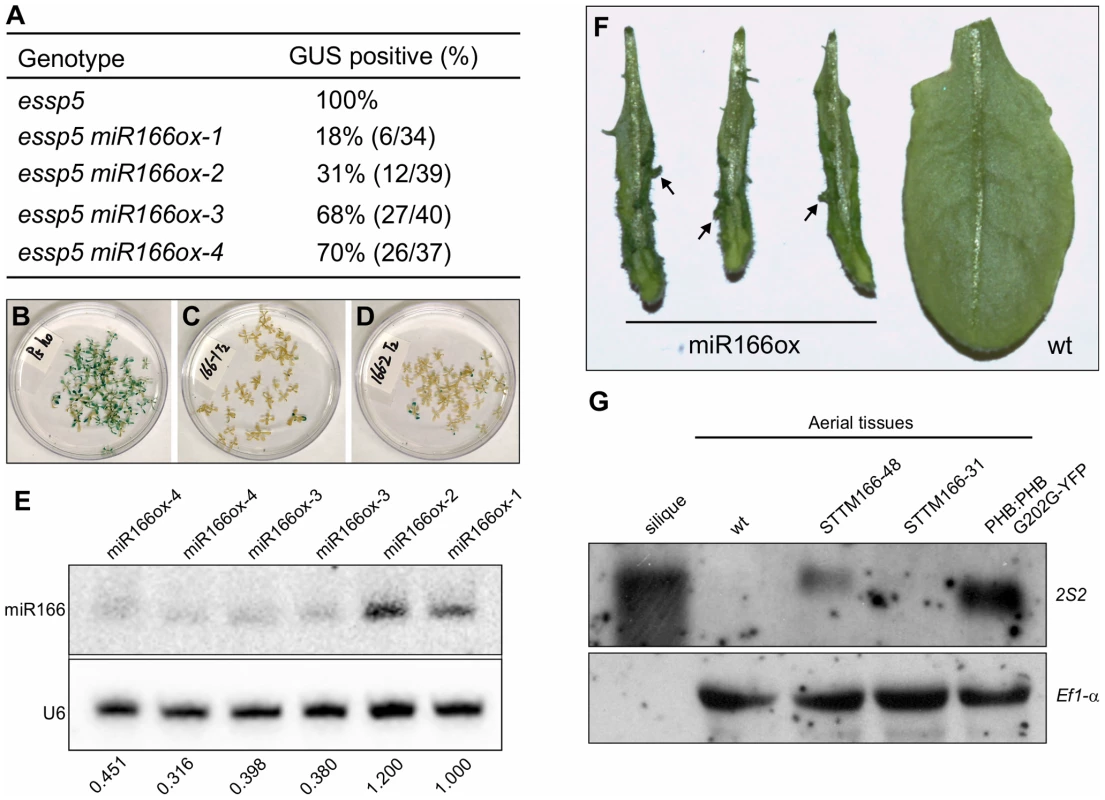
(A) Loss of ectopic GUS activity in essp5 plants overexpressing miR166. The miR166 transgene driven by the CaMV 35S promoter was introduced into essp5/+ background. Four T1 transgenic lines were identified, designated essp5 miR166ox-1–4. T2 seeds were germinated on selective MS agar plates such that only seedlings transgenic for miR166 were selected. After 13 days of growth, essp5-like seedlings were harvested and stained for GUS activity. (B–D) Photos showing the GUS phenotypes of essp5 (B), essp5 miR166ox-1 (C), and essp5 miR166ox-2 (D), respectively, as listed in (A). (E) Accumulation of miR166 in the four transgenic lines as listed in (A), i.e., essp5 miR166ox-1–4, as determined by northern blotting. The genotypes of the plants are indicated at the top. Total RNAs were extracted from two-week old seedlings (aerial portion). The numbers below the gel images indicate the relative abundance of the small RNAs. (F) Comparison of leaf morphology of miRNA166 overexpressors with that of wild type. Note the aberrant structures formed on the transgenic leaves (arrows). (G) RNA blot analysis of the 2S2 gene in seedlings (aerial tissues) in the miR166 silencing lines STTM165/166-48 and STTM165/166-31 grown for 14 days on MS agar. Wild type (Col) leaves and siliques were used as negative and positive controls, respectively. The PHB:PHB G202G-YFP transgenic plants were also included as a positive control. Elongation factor 1α was used as a loading control. To demonstrate that the specific loss of miR166 can cause the ectopic expression of seed maturation genes, we obtained the recently developed transgenic lines that exhibit a dramatic reduction in miR165/166 accumulation achieved by the expression of a short tandem target mimic (STTM165/166) [41]. RNA blot analysis was performed to examine the expression of seed storage protein genes in these transgenic lines, using 2S2 as a probe. As shown in Figure 5G, the 2S2 gene is clearly expressed in the strongest line (STTM165/166-48), but not detectable in a weaker line (STTM165/166-31). This observation indicates that miR166 plays an important role in repressing seed genes in seedlings.
Ectopic Expression of the HD-ZIPIII Genes PHB or PHV Is Sufficient for Seed Gene Expression in Seedlings
It has been well established that the miR165/166 family miRNAs target the transcripts of the HD-ZIPIII genes, controlling their expression level and domain, to fulfill their roles in plant development including leaf polarity determination [42]–[45]. The HD-ZIPIII family consists of five transcription factors (REV, PHB, PHV, AtHB8, and AtHB15), and they play both redundant and unique roles in diverse plant developmental processes [46]. In this context, it is worth noting that the transcript level of PHB was found to be decreased in miR166 overexpressors (Figure S4). To investigate whether the HD-ZIPIII proteins are responsible for conferring the ectopic GUS phenotype of essp5/ago1-100, we introduced loss-of-function mutations in PHB and PHV genes into essp5/ago1-100 by genetic crosses and examined leaf GUS expression in F2 and F3 seedlings. A large number of F2/F3 seedlings were examined and no clear loss of leaf GUS activity was observed in phb essp5 or phv essp5. We further introduced phb phv double mutations into the essp5/ago1-100 background, but still saw no detectable loss of leaf GUS activity. Obviously, the potential redundancy among the five HD-ZIPIII genes could be confounding the genetic analyses above.
Next, taking advantage of the previously identified gain-of-function mutations in HD-ZIPIII family genes, we investigated whether these proteins are sufficient to cause the ectopic expression of seed genes. These gain-of-function alleles have mutations in the miR166 target regions to cause a mismatch between the miRNA and the target mRNA and thus render the transcripts resistant to miRNA-mediated degradation and consequently the ectopic accumulation of HD-ZIPIII transcripts. First, the gain-of-function mutations phb-1d [47] and phv-1d [44] were introduced into the βCG:GUS background by genetic crosses and ectopic GUS activity was examined in F2 seedlings (Figure 6A–6I). Meanwhile, another gain-of-function phb allele driven by the CaMV 35S promoter, 35S:PHB G202G [43], was also introduced into the βCG:GUS background by Agrobacterium-mediated transformation and GUS activity was examined for each independent T1 plant (Figure 6A and 6J–6M). As shown in Figure 6A–6M, ectopic GUS activity was clearly observed for phb-1d, phv-1d, and 35S:PHB G202G. Further, we performed northern blot and qRT-PCR analyses to examine the ectopic expression of endogenous seed maturation genes in the gain-of-function mutant plants (Figure 6N and 6O). Two representative maturation genes 2S2 and 2S3 were clearly expressed in the mutant seedlings (Figure 6N). Similarly, the four master regulators were all elevated to varying levels (Figure 6O). In addition, a gain-of-function REV mutant, rev-10d [42], was also analyzed in the βCG:GUS background but no ectopic GUS activity was detected. In summary, our gain-of-function genetic evidence indicates that the HD-ZIPIII proteins PHB and PHV are each sufficient for ectopic expression of seed genes.
Fig. 6. HD-ZIPIII Genes PHB/PHV Are Sufficient for Activating Ectopic Expression of Seed Genes in Seedlings. 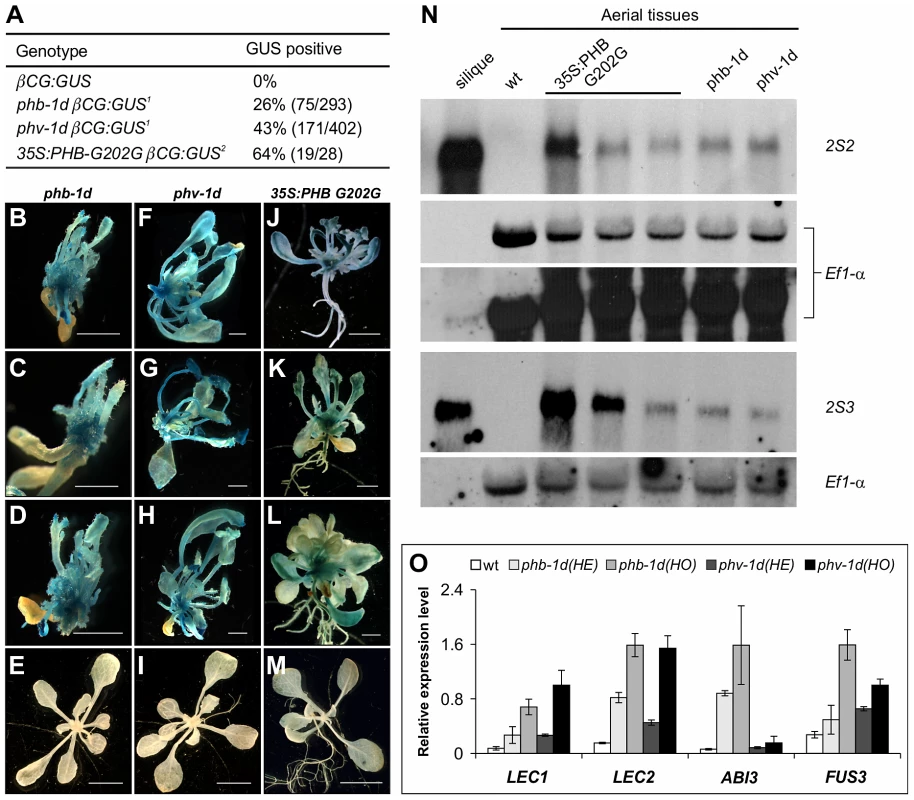
(A) GUS phenotypes of gain-of-function mutations in PHB and PHV genes. Note1: phb-1d or phv-1d was crossed with βCG:GUS. In the F2 progeny grown on agar for three weeks, phb-1d- or phv-1d-like plants were selected and stained for GUS activity. Note2: The 35S:PHB G202G transgene was introduced into the βCG:GUS background by agrobacterium-mediated transformation. Independent T1 transgenic seedlings were selected and stained for GUS activity. (B–M) GUS phenotypes of gain-of-function phb and phv mutants grown on agar for three weeks. Shown here are representative F2 progeny from the cross of phb-1d or phv-1d with βCG:GUS, as well as the T1 transgenic lines containing the 35S:PHB G202G in βCG:GUS background. Wild type (wt, βCG:GUS) seedlings (two weeks old) were also stained as negative controls (E, I, and M). Scale bars, 2 mm in B–D, F–H and J–K; 5 mm in E, I and M. (N) RNA blot analysis of the expression of the maturation genes (2S2 and 2S3 as markers) in the gain-of-function phb and phv mutant seedlings grown for 22 days on MS agar. For the 35S:PHB G202G transgenic plants, independent T1 plants were selected and grouped into three groups according to the severity of their morphological phenotypes. Note the difference in the hybridization intensities. Wild type (Col) leaves and siliques were used as negative and positive controls, respectively. Same amount of RNA was used for each blot. Elongation factor 1α was used as a loading control. In the top panel, two exposures of the EF1- α loading control were shown. (O) qRT-PCR analysis of ABI3, FUS3, LEC1, and LEC2 expression in seedlings (aerial portion) of phb-1d and phv-1d mutants grown for 20 days on MS agar. For each mutant allele, heterozygous (HE) and homozygous (HO) mutant seedlings were measured separately for comparison. Actin-8 was used as an internal control. The mean and standard error were determined from three biological replicates. Physical Occupancy of PHB at the LEC2 Gene Promoter
HD-ZIP proteins are plant-specific transcription factors and named for the combination of homeodomain and leucine zipper domains at their N termini [48]. They bind a palindromic DNA sequence in vitro as dimers [49]. To determine whether PHB acts directly at maturation gene loci, we performed chromatin immunoprecipation (ChIP) experiments to examine PHB occupancy at the promoters of these genes.
For the ChIP assays, we generated Arabidopsis plants transgenic for a YFP-tagged gain-of-function allele of PHB under the control of the PHB native promoter (PHB:PHB G202G-YFP). Morphologically, the transgenic plants resemble the phb-1d mutant (Figure 7A–7D). When the transgene was introduced into the βCG:GUS background, it resulted in ectopic GUS activity (Figure 7E–7H). Expression of the master regulators of seed maturation in the transgenic seedlings was also examined by qRT-PCR. As shown in Figure 7I, these genes were ectopically expressed to similar levels compared to those of phb-1d, and the expression levels in homozygous seedlings were clearly higher than those in the hemizygous siblings. These observations demonstrate that the PHB:PHB G202G-YFP plants resemble phb-1d. In addition, we observed, at a low frequency, disorganized growth and/or formation of somatic embryo-like structures in the transgenic plants (Figure 7J–7L). Some parts of these plants could be stained by the neutral lipid dye fat red (Figure 7M–7O), indicating a high level accumulation of seed storage-specific triacylglycerols in these plants (fat red/sudan red stains only seed storage-specific lipids).
Fig. 7. PHB Occupancy at LEC2 Promoter. 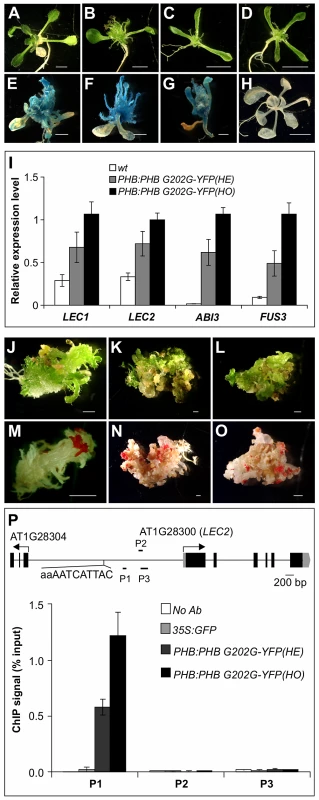
(A–D) Morphological phenotype of the 35S:GFP (A) and PHB:PHB G202G-YFP (B–D) transgenic plants grown on agar for three weeks. (B) a hemizygous plant. (C–D) homozygous plants. Note that multiple cotyledons (3 or 4) were found in the majority (∼80%) of PHB:PHB G202G-YFP homozygous plants. Scale bar, 3 mm. (E–H) GUS phenotype of PHB:PHB G202G-YFP transgenic seedlings (in βCG:GUS background) grown on MS agar for three weeks (E–G) and wild type (wt, βCG:GUS) grown on MS agar for two weeks. Scale bars, 1 mm in E–G and 5 mm in H. (I). qRT-PCR analysis of ABI3, FUS3, LEC1, and LEC2 expression in seedlings (aerial portion) of hemizygous (HE) and homozygous (HO) PHB:PHB G202G-YFP seedlings grown for 20 days on MS agar. Actin-8 was used as an internal control. The mean and standard error were determined from three biological replicates. (J–O) Fat red staining of PHB:PHB G202G-YFP plants that exhibited disorganized growth. Such plants were found at a rate of approximately 10% among the homozygous transgenic plants. Shown in J (20 d), K (35 d), and L (35 d) are plants before staining that correspond to those in (M), (N), and (O), respectively, after staining. Scale bar, 1 mm. (P) PHB occupancy at LEC2 promoter as determined by ChIP using anti-GFP antibody in PHB:PHB G202G-YFP plants. The 35S:GFP plants served as negative control. ChIP DNAs were analyzed by qPCR. The results were reproduced in two biological replicates. Standard deviations were calculated from three technical repeats. Shown at the top is a schematic representation of the LEC2 gene. Boxes and lines represent exons and introns, respectively. Transcription start sites are indicated by arrows. Black bars labeled P1–P3 represent the regions examined by ChIP-qPCR. The potential PHB binding sequence is also indicated. ChIP was performed with anti-GFP antibodies and an Arabidopsis line transgenic for GFP driven by the CaMV 35S promoter (35S:GFP) was used as a negative control. The ChIP DNAs were analyzed by qPCR to examine the enrichment of promoter region genomic DNAs of the four master regulator genes. One region in the LEC2 promoter was highly enriched relative to the 35S:GFP and no antibody controls (Figure 7P), but no enrichment was found for the promoter regions of other maturation genes examined (Figure S5). The enrichment level in homozygous plants was about double that in the hemizygous siblings, consistent with the ectopic expression level of seed genes in these plants (Figure 7I). Interestingly, a partial palindromic sequence, aaAATCATTAC, was found in the vicinity of the enriched genomic region in the LEC2 promoter, but not at other maturation loci. This sequence is very similar to the HD-ZIPIII binding consensus sequence, GTAAT(G/C)ATTAC, derived from an in vitro binding site selection experiment [49]. These observations suggest that PHB, when ectopically expressed, binds to the LEC2 promoter and activates the expression of the gene. LEC2 can, in turn, activate a network of maturation-related genes including ABI3, FUS3, LEC1, and the SSP genes (Figure 8).
Fig. 8. A Model of MicroRNA–Mediated Repression of Seed Genes in Leaves. 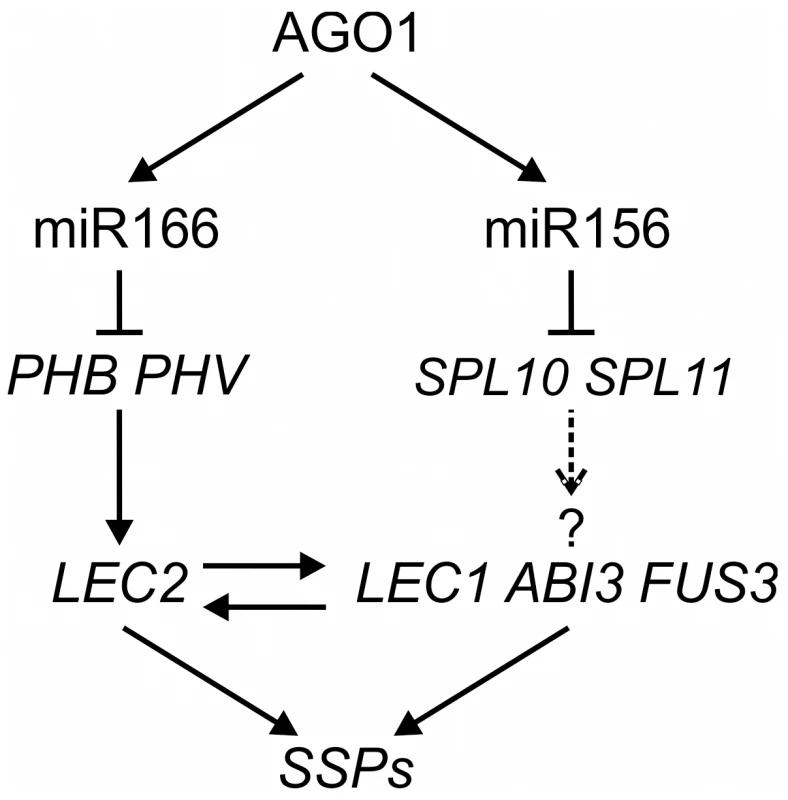
In wild type seedlings, AGO1 and miR166 repress the expression of PHB/PHV, which promotes the seed maturation program by directly activating LEC2 expression and indirectly that of other positive regulators of the seed maturation program. miR156 may also play a role in the process, possibly by repressing SPL10 and SPL11, based on this work and that of Nodine and Bartel [53]. It is not known, however, whether SPLs are sufficient for inducing the expression of seed genes or whether they directly activate the seed genes. Discussion
In this work, we first identified a weak EMS ago1 allele, which exhibited ectopic expression of a GUS reporter driven by a seed gene promoter. Taking advantage of the weak ago1 allele and its GUS phenotype, we then performed a series of transgenic and genetic analyses to search for the molecular mechanisms underlying the mutant phenotype. We first demonstrated that miR166 reduction is a major cause of the mutant phenotype and further showed that the targets of miR166, PHB and PHV, are sufficient for derepressing seed maturation genes in seedlings. Finally, our ChIP assay using a tagged PHB transgenic line suggests that PHB may act directly at the LEC2 promoter (summarized in Figure 8). However, in addition to LEC2, PHB may also regulate other factors that in turn regulate seed maturation genes directly or indirectly. Future studies, such as ChIP-seq, are needed to address this issue. Therefore, this work has added miR165/166 to the documented repertoire of postgermination repressors of the embryonic program (reviewed in [2]), and more importantly, established PHB, and possibly PHV, as direct positive regulators of the master regulator of seed maturation LEC2. A major future challenge in the field is to find out the genetic and molecular relationships amongst the various players, including transcription factors, chromatin remodelers and modifiers, and the newly added miRNA, and build an integrated genetic network.
Given the well-established expression patterns and roles of miR166 and its targets in leaf polarity determination (reviewed in [50], [51]), an obvious outstanding question is why the normal expression of the PHB and PHV genes in the adaxial domain of leaf primordia in wild type plants is not sufficient to cause the ectopic expression of seed maturation genes. miR165/166 is concentrated in the abaxial domain to restrict the expression of the HD-ZIPIII transcription factor genes to the adaxial domain in the lateral organs in Arabidopsis [42]–[44] and maize [45]. In phb and phv gain-of-function mutants, the expression of PHB and PHV is not restricted to the adaxial domain but extends into the entire primordium. We observed ectopic expression of seed maturation genes only in these gain-of-function mutants, indicating that the normal, adaxial expression of the HD-ZIP III genes is not sufficient to activate the seed maturation program. There could be at least two underlying reasons. First, the ectopic expression of the seed maturation genes in the phb and phv gain-of-function mutants only occurs in the abaxial domain. In this scenario, the lack of necessary co-factors or the presence of negative factors in the adaxial domain may prevent the HD-ZIPIII genes from activating the seed maturation genes. Alternatively, it might be a matter of thresholds – the adaxial domain normally does not have sufficient levels of HD-ZIPIII expression to trigger the seed maturation program, but when the miRNA is compromised, the expression level is high enough to trigger the program. Our preliminary observation is in support of the first scenario. GUS expression along the adaxial-abaxial axis in essp5/ago1-100 was examined and GUS activity was found only on the abaxial side (Figure S6). In addition, interestingly, GUS was also observed in both the upper and lower epidermal cells (Figure S6).
The seed maturation program is a tightly regulated developmental process. Mechanisms are in place to not only ensure its repression during seedling development but also prevent its precocious induction during early embryogenesis [2], [52]. The induction of seed maturation is also referred to as the morphogenesis-to-maturation phase transition of embryogenesis. While our studies have established miR165/166 and implicated miR156 as players in the repression of the seed maturation program in vegetative development, two recent studies have also revealed important roles of miRNAs in regulating the morphogenesis-to-maturation phase transition [53], [54]. Of these, the work of Nodine and Bartel [53] demonstrated that miR156 and two of its target genes SPL10 and SPL11 play a major role in early embryo patterning and in preventing the precocious expression of maturation genes. An obvious question is whether miR165/166 also acts similarly in early embryogenesis to control the morphogenesis-to-maturation phase transition. Previous studies have shown that PHB and PHV promote embryonic development, and that the expression of these genes must be repressed by miR165/166 for embryonic development to proceed normally. For example, Grigg et al showed that serrate (se) mutants cause ectopic expression of PHB and PHV in the root pole of embryos, and that the embryonic lethal phenotype of se mutants can be rescued by loss-of-function mutations in PHB and PHV [55]. Smith and Long also showed that PHB and PHV promote shoot development during embryogenesis [56]. These studies focused on the roles of the miR165/166-PHB/PHV module in early embryo patterning. Our finding that this module plays an important role in repressing seed maturation genes during seedling development prompted us to test its role in the morphogenesis-to-maturation phase transition. We performed a ChIP analysis using a transgenic line expressing a tagged PHB driven by its endogenous promoter (PHB:PHB-YFP). Preliminary data suggests that PHB acts directly at LEC2 during embryogenesis (Figure S7). Future investigations are needed to sort out the contributions of each miRNA to the repression of the seed maturation program during the pre - and post-maturation stages.
Materials and Methods
Plant Material, Growth Conditions, and Genotype Analysis
Seeds of mutants including the T-DNA insertion mutants were obtained from the ABRC, unless otherwise indicated. Seeds were stratified at 4°C for 3-d. Then the seeds were sowed on soil or on agar plates containing 4.3 g/L Murashige and Skoog nutrient mix (Sigma-Aldrich), 1.5% sucrose, 0.5 g/L MES (pH 5.7), and 0.8% agar. Plants were grown under 16 h-light (22°C)/8 h-dark (20°C) cycles; except that the phb-1d/+ and phv-1d/+ mutants were grown at 17°C during reproductive development as described [47]. Homozygous T-DNA insertion mutants were identified by genotyping.
Map-Based Cloning of essp5
The mutant essp5 was isolated from the same genetic screen as essp1 and essp3 [5], [27]. For genetic mapping of the essp5 mutation, mutant plants were crossed with wild type plants of the Ler ecotype. A total of 644 homozygous essp5 mutants were collected from the F2 segregating population. Genomic DNA extracted from these seedlings was used for PCR-based mapping with simple sequence polymorphism markers, and the essp5 locus was mapped to a ∼127 kb genomic interval on BACs F11A17, T1N15 and F9P7 on chromosome one (17,852–17,979 kb). Sequencing of the genomic region revealed a mutation in At1g48410.
Histochemical GUS and Fat Red Staining
The modified GUS staining solution (0.5 mg/mL 5-bromo-4-chloro-3-indolyl-glucuronide, 20% methanol, 0.01 M Tris-HCl, pH 7.0) was used [5]. Seedlings immersed in GUS staining solution were placed under vacuum for 15 min, and then incubated at 37°C overnight. The staining solution was removed and samples were cleared by sequential incubation in 75% and 95% ethanol. Fat red staining was performed by incubating samples in a saturated solution of Sudan red 7B (Sigma) in 70% ethanol for 1 h at room temperature. Samples were then rinsed with 70% ethanol [57].
Gene Expression Analysis by qRT–PCR and Northern Blotting
Plants grown on MS media were used for gene expression analyses. RT-PCR and RNA blot analyses were preformed as described previously [5]. Probes for detecting transcripts of the CRA1, CRB, and CRC genes were designed based on Pang et al [58]. Real-time PCR was conducted using the Bio-Rad CFX96 real-time PCR detection system and the SsoFast™ EvaGreen® Supermix kit (Bio-Rad Laboratories, Inc.). Data from three biological replicates were analyzed by the software Bio-Rad CFX96 Managertm V1.6.541.1028, using Actin8 as the internal reference. DNA oligonucleotides used as probes or in real-time PCR are listed in Table S1.
miRNA Northern Blot Analysis
RNA isolation and hybridization for miRNA detection were performed as described [59], [60]. 5′-end-labeled 32P antisense DNAs or an LNA oligonucleotide (for miRNA166) were used to detect miRNAs from total RNAs (10 µg each sample). Oligonucleotide probes used are listed in Table S1.
miRNA Transgene Constructs
Genome sequences surrounding the selected MIRNA genes (listed in Table S1) were amplified by PCR from genomic DNA isolated from wild-type Arabidopsis (Col). The amplified DNA was first cloned into the pDNR221 vector (Invitrogen), confirmed by sequencing, and then recombined into the pEarlyGate100 Gateway-compatible destination vector [61] where the MIRNA genes are under the control of the CaMV 35S promoter. The constructs were introduced into essp5 homozygous or heterozygous plants (essp5/+). PCR primers used for amplifying the MIRNA genes are listed in Table S1. Transgenic plants were selected on Basta, allowed to grow to maturity and seeds were collected, and GUS expression was analyzed in the next generation.
ChIP
For the construction of the PHB:PHB G202G-YFP transgene plasmid, the PHB promoter was PCR amplified from Arabidopsis (Col-0) genomic DNA by Fusion DNA Polymerase (NEB, M0530) using primers EcoRI-PHBpr and PHBpr-NcoI, and inserted into the pBluscript SK vector. The plasmid was then fully digested by SpeI and partially digested by EcoRI. The full-length promoter fragment was purified and ligated with the pEARLEYGATE 104 vector [61] to generate the plasmid pEG104-PHBpro. The PHB G202G coding sequence was amplified from cDNAs made from 35S:PHB G202G transgenic plants [43] with primers PHBf and PHBr, cloned into the pENTR-D-topo vector (invitrogen), and subsequently cloned into the destination vector pEARLEYGATE104 by LR reaction. The generated plasmid pEG104-PHB G202G were digested by NcoI and SpeI, and the PHB G202G-YFP fragment was recovered and ligated with pEG104-PHBpro to obtain the pEG104-PHB:PHB G202G-YFP plasmid. The PHB:PHB-YFP transgene plasmid was constructed using a similar strategy. Primers are listed in Table S1.
Chromatin immunoprecipitation (ChIP) was carried out as described previously [62]. One gram of twenty-day-old Arabidopsis plants grown on MS agar was used for each ChIP. The sonicated chromatin was immunoprecipitated with 5 µL of anti-GFP antibody (ab290, Abcam). Quantitative ChIP PCR was performed with three technical replicates, and results were presented as percentage of input. ChIP experiments were performed at least two times. See Table S1 for primer sequences used for ChIP-PCR and construction of the PHB:PHB G202G-YFP transgene.
Supporting Information
Zdroje
1. Vicente-CarbajosaJ, CarboneroP (2005) Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49 : 645–651.
2. ZhangH, OgasJ (2009) An epigenetic perspective on developmental regulation of seed genes. Mol Plant 2 : 610–627.
3. HendersonJT, LiHC, RiderSD, MordhorstAP, Romero-SeversonJ, et al. (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134 : 995–1005.
4. LiHC, ChuangK, HendersonJT, RiderSDJr, BaiY, et al. (2005) PICKLE acts during germination to repress expression of embryonic traits. Plant J 44 : 1010–1022.
5. TangX, HouA, BabuM, NguyenV, HurtadoL, et al. (2008) The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol 147 : 1143–1157.
6. MoonYH, ChenL, PanRL, ChangHS, ZhuT, et al. (2003) EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15 : 681–693.
7. ChanvivattanaY, BishoppA, SchubertD, StockC, MoonYH, et al. (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131 : 5263–5276.
8. SchubertD, ClarenzO, GoodrichJ (2005) Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol 8 : 553–561.
9. MakarevichG, LeroyO, AkinciU, SchubertD, ClarenzO, et al. (2006) Different Polycomb group complexes regulate common target genes in Arabidopsis. Embo Rep 7 : 947–952.
10. TanakaM, KikuchiA, KamadaH (2008) The Arabidopsis Histone Deacetylases HDA6 and HDA19 Contribute to the Repression of Embryonic Properties after Germination. Plant Physiol 146 : 149–161.
11. TsukagoshiH, MorikamiA, NakamuraK (2007) Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci USA 104 : 2543–2547.
12. SuzukiM, WangHH, McCartyDR (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143 : 902–911.
13. GaoMJ, LydiateDJ, LiX, LuiH, GjetvajB, et al. (2009) Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21 : 54–71.
14. GiraudatJ, HaugeBM, ValonC, SmalleJ, ParcyF, et al. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 : 1251–1261.
15. LotanT, OhtoM, YeeKM, WestMA, LoR, et al. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 : 1195–1205.
16. LuerssenH, KirikV, HerrmannP, MiseraS (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15 : 755–764.
17. StoneSL, KwongLW, YeeKM, PelletierJ, LepiniecL, et al. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98 : 11806–11811.
18. KagayaY, ToyoshimaR, OkudaR, UsuiH, YamamotoA, et al. (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46 : 399–406.
19. ToA, ValonC, SavinoG, GuilleminotJ, DevicM, et al. (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 : 1642–1651.
20. ParcyF, ValonC, RaynalM, Gaubier-ComellaP, DelsenyM, et al. (1994) Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 : 1567–1582.
21. GazzarriniS, TsuchiyaY, LumbaS, OkamotoM, McCourtP (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7 : 373–385.
22. Santos MendozaM, DubreucqB, MiquelM, CabocheM, LepiniecL (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579 : 4666–4670.
23. Jones-RhoadesMW, BartelDP, BartelB (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57 : 19–53.
24. RamachandranV, ChenX (2008) Small RNA metabolism in Arabidopsis. Trends Plant Sci 13 : 368–374.
25. ChenX (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25 : 21–44.
26. VoinnetO (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136 : 669–687.
27. LuQ, TangX, TianG, WangF, LiuK, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61 : 259–270.
28. TangX, LimM-H, PelletierJ, TangM, NguyenV, KellerWA, TsangEWT, WangA, RothsteinSJ, HaradaJJ, CuiY (2012) Synergistic Repression of the Embryonic Program by SET DOMAIN GROUP 8 and EMBRYONIC FLOWER 2 in Arabidopsis Seedlings. J Exp Bot 63 : 1391–1404.
29. VaucheretH (2008) Plant ARGONAUTES. Trends Plant Sci 13 : 350–358.
30. BohmertK, CamusI, BelliniC, BouchezD, CabocheM, et al. (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. Embo J 17 : 170–180.
31. KidnerCA, MartienssenRA (2004) Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 : 81–84.
32. YangL, HuangW, WangH, CaiR, XuY, et al. (2006) Characterizations of a hypomorphic argonaute1 mutant reveal novel AGO1 functions in Arabidopsis lateral organ development. Plant Mol Biol 61 : 63–78.
33. MorelJB, GodonC, MourrainP, BéclinC, BoutetS, et al. (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14 : 629–39.
34. BaumbergerN, BaulcombeDC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A 102 : 11928–11933.
35. VaucheretH, VazquezF, CrétéP, BartelDP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18 : 1187–1197.
36. VaucheretH, MalloryAC, BartelDP (2006) AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22 : 129–136.
37. TelferA, PoethigRS (1998) HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125 : 1889–1898.
38. VaucheretH (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20 : 759–771.
39. PeragineA, YoshikawaM, WuG, AlbrechtHL, PoethigRS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18 : 2368–2379.
40. ZhouGK, KuboM, ZhongR, DemuraT, YeZH (2007) Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol 48 : 391–404.
41. YanJ, GuY, JiaX, KangW, PanS, et al. (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24 : 415–427.
42. EmeryJF, FloydSK, AlvarezJ, EshedY, HawkerNP, et al. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13 : 1768–1774.
43. MalloryAC, ReinhartBJ, Jones-RhoadesMW, TangG, ZamorePD, et al. (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. Embo J 23 : 3356–3364.
44. McConnellJR, EmeryJ, EshedY, BaoN, BowmanJ, et al. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 : 709–713.
45. JuarezMT, KuiJS, ThomasJ, HellerBA, TimmermansMC (2004) microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 : 84–88.
46. PriggeMJ, OtsugaD, AlonsoJM, EckerJR, DrewsGN, et al. (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 : 61–76.
47. McConnellJR, BartonMK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125 : 2935–2942.
48. SchenaM, DavisRW (1992) HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci USA 89 : 3894–3898.
49. SessaG, SteindlerC, MorelliG, RubertiI (1998) The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 38 : 609–622.
50. KidnerCA, TimmermansMC (2007) Mixing and matching pathways in leaf polarity. Curr Opin Plant Biol 10 : 13–20.
51. PulidoA, LaufsP (2010) Co-ordination of developmental processes by small RNAs during leaf development. J Exp Bot 61 : 1277–1291.
52. BraybrookSA, HaradaJJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13 : 624–630.
53. NodineMD, BartelDP (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev 24 : 2678–2692.
54. WillmannMR, MehalickAJ, PackerRL, JenikPD (2011) MicroRNAs Regulate the Timing of Embryo Maturation in Arabidopsis. Plant Physiol 155 : 1871–1884.
55. GriggSP, GalinhaC, KornetN, CanalesC, ScheresB, TsiantisM (2009) Repression of apical homeobox genes is required for embryonic root development in Arabidopsis. Curr Biol 19 : 1485–1490.
56. SmithZR, LongJA (2010) Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464 : 423–426.
57. BratzelF, López-TorrejónG, KochM, Del PozoJC, CalonjeM (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20 : 1853–1859.
58. PangPP, PruittRE, MeyerowitzEM (1988) Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopis thaliana. Plant Mol Biol 11 : 805–820.
59. ParkW, LiJ, SongR, MessingJ, ChenX (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 : 1484–1495.
60. PallGS, Codony-ServatC, ByrneJ, RitchieL, HamiltonA (2007) Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res 35: e60.
61. EarleyKW, HaagJR, PontesO, OpperK, JuehneT, et al. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 : 616–629.
62. GendrelAV, LippmanZ, MartienssenR, ColotV (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2 : 213–218.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Transfer zmraženého embrya zlepšuje výsledky IVF
- Velké děti po kryoembryotransferu
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



