-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
Mutations in the retinoblastoma tumor suppressor gene (rb1) cause both sporadic and familial forms of childhood retinoblastoma. Despite its clinical relevance, the roles of rb1 during normal retinotectal development and function are not well understood. We have identified mutations in the zebrafish space cadet locus that lead to a premature truncation of the rb1 gene, identical to known mutations in sporadic and familial forms of retinoblastoma. In wild-type embryos, axons of early born retinal ganglion cells (RGC) pioneer the retinotectal tract to guide later born RGC axons. In rb1 deficient embryos, these early born RGCs show a delay in cell cycle exit, causing a transient deficit of differentiated RGCs. As a result, later born mutant RGC axons initially fail to exit the retina, resulting in optic nerve hypoplasia. A significant fraction of mutant RGC axons eventually exit the retina, but then frequently project to the incorrect optic tectum. Although rb1 mutants eventually establish basic retinotectal connectivity, behavioral analysis reveals that mutants exhibit deficits in distinct, visually guided behaviors. Thus, our analysis of zebrafish rb1 mutants reveals a previously unknown yet critical role for rb1 during retinotectal tract development and visual function.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003106
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003106Summary
Mutations in the retinoblastoma tumor suppressor gene (rb1) cause both sporadic and familial forms of childhood retinoblastoma. Despite its clinical relevance, the roles of rb1 during normal retinotectal development and function are not well understood. We have identified mutations in the zebrafish space cadet locus that lead to a premature truncation of the rb1 gene, identical to known mutations in sporadic and familial forms of retinoblastoma. In wild-type embryos, axons of early born retinal ganglion cells (RGC) pioneer the retinotectal tract to guide later born RGC axons. In rb1 deficient embryos, these early born RGCs show a delay in cell cycle exit, causing a transient deficit of differentiated RGCs. As a result, later born mutant RGC axons initially fail to exit the retina, resulting in optic nerve hypoplasia. A significant fraction of mutant RGC axons eventually exit the retina, but then frequently project to the incorrect optic tectum. Although rb1 mutants eventually establish basic retinotectal connectivity, behavioral analysis reveals that mutants exhibit deficits in distinct, visually guided behaviors. Thus, our analysis of zebrafish rb1 mutants reveals a previously unknown yet critical role for rb1 during retinotectal tract development and visual function.
Introduction
Biallelic mutations in the retinoblastoma susceptibility gene rb1 are causal for intraocular childhood retinoblastomas. Rb1 is a member of a gene family that consists of three members, p105/Rb1, p107/Rb-like1, and p130/Rb-like2, collectively known as “pocket proteins” [1]. The activity of these proteins is controlled, in part, by cyclin/cyclin-dependant kinase complexes. Upon activation, Rb proteins bind to an array of proteins, including members of the E2F family of transcription factors to execute a range of cellular functions, including cell cycle exit, terminal differentiation, and cortical cell migration [2]. In humans, germline or somatic mutations occur throughout the 180 kb genomic region spanning the rb1 gene, including its promoter region, exons, and intronic essential splice sites, resulting in bilateral or unilateral retinoblastomas within the first 2 years of life [3], [4].
Given its clinical relevance, the role of Rb1 during embryonic development and during tumor suppression has been studied intensely, mostly using mouse models [5]. Rb1 is expressed ubiquitously during murine development, postnatally, and continues to be expressed in adults. Embryos harboring non-conditional Rb1 knockout alleles exhibit ectopic proliferation and apoptosis throughout the nervous system and die prenatally at embryonic day 14.5 [6], [7], [8]. Embryos with conditional loss of Rb1 in the retina display ectopic division and considerable apoptosis of retinal transition cells starting at E10 [9], [10], [11], [12]. Retinas in these animals contain reduced numbers of rods, bipolar cells, and RGCs, yielding a retina with a thin outer nuclear layer and a hypoplastic optic nerve. However, the etiology of optic nerve hypoplasia and if/how Rb1 functions in RGC axonal guidance has not been examined. Similarly, electroretinogram recordings from Rb1 deficient mouse retinas have revealed reduced photoreceptor to bipolar to amacrine signal transmission [10], yet the behavioral consequences have not been examined.
Here, we report that zebrafish space cadet mutants carry a rb1 mutation found in cases of sporadic and familial human retinoblastoma [13], [14], [15], [16]. In zebrafish rb1 mutants, RGC precursors show delayed exit from the cell cycle and hence a delay in the generation of early-born, postmitotic RGCs, whose axons are critical for pioneering the retinotectal tract. This delay leads to RGC intrinsic axon guidance defects, aberrant retinotectal connectivity, and deficits in phototactic behaviors. Together, this work describes a novel model for understanding the developmental role of rb1 and reveals a previously unknown role for rb1 in the formation of the retinotectal tract.
Results
space cadet harbors a disease causing mutation in rb1
We previously identified two mutant space cadet alleles, based on abnormal startle response behavior to acoustic or tactile stimuli [17], [18]. Using recombination mapping, we mapped the space cadette226a allele to a 1.1 cM interval on chromosome 21 between single nucleotide polymorphic markers in the myo5b locus (20 recombinants/2688 meioses), and in the ncam1 locus (12 recombinants/2688 meioses), respectively (Figure 1A). This genomic interval contains several annotated genes, including rb1, lpar6, and cystlr2, which have retained syntenic positional conservation between humans, mice and zebrafish (Figure 1B). Sequencing of rb1 cDNAs isolated from spcte226a larvae revealed the presence of 4 nucleotides inserted between exon 19 and exon 20. Subsequent sequencing of genomic DNA isolated from spcte226a larvae confirmed a single nucleotide change in the splice donor sequence of intron 19 (nt1912+1: G to A; Figure 1C). This generates a cryptic splice site donor, resulting in the 4 base pair insertion into the rb1 mRNA. This 4 base pair insertion causes a premature stop codon in exon 20, predicted to truncate the protein at amino acid 677, thereby severely truncating the B domain and cyclin domain essential for Rb1 function (Figure 1D) [15]. Interestingly, identical mutations have been reported in human patients with familial and sporadic forms of retinoblastoma [13], [14], [15], [16].
Fig. 1. spcte226a mutation induces a premature stop codon and truncation of Rb1. 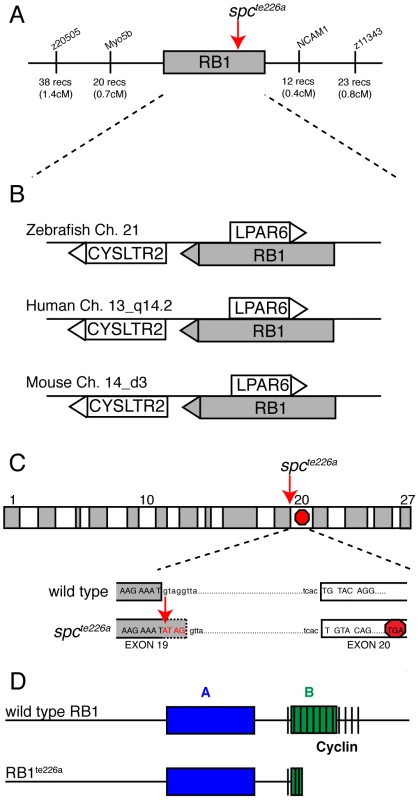
(A) Recombination mapping places the spcte226a mutant locus within a 1.1 cM interval on chromosome 21 that shows syntenic conservation (B) of rb1, lpar6, and cysltr2 transcripts with human chromosome 13_q14.2 and mouse chromosome 14_d3. (C) rb1 exons depicted as alternating gray/white blocks. spcte226a mutation at splice donor site of intron 19 (nt1912+1) causes frameshift and premature stop codon in exon 20. (D) Estimated Rb1te226a protein product with truncated B- and cyclin-domains. The zebrafish rb1 gene is 67% similar (52% identical-based on amino acid sequence) to the mouse and human rb1. The Rb1 protein consists of an A and B domain forming Rb1's binding “pocket”, and a cyclin binding domain (Figure 1D), and shows 81% amino acid similarity (66% identical) between zebrafish and mammalian rb1 in these critical domains. Thus, sequence homology, syntenic conservation, and cDNA sequence analysis provide compelling evidence that space cadet phenotypes are caused by an rb1 gene mutation known to cause retinoblastoma. Sequence analysis of the second mutant space cadetty85d allele did not reveal any changes in the coding sequence or in any of the splice donor and acceptor sites, suggesting that this allele is caused by a regulatory mutation in the rb1 locus. Importantly, analyses of spcte226a and spcty85d mutants revealed no significant differences with regards to the strength of the phenotypes examined below (Table 1). From here on, we will refer to the space cadet gene as rb1 and describe anatomical and behavioral defects in the rb1te226a allele.
Tab. 1. Comparison of the retinal phenotypes observed at 36 hpf in the two rb1 mutant alleles. 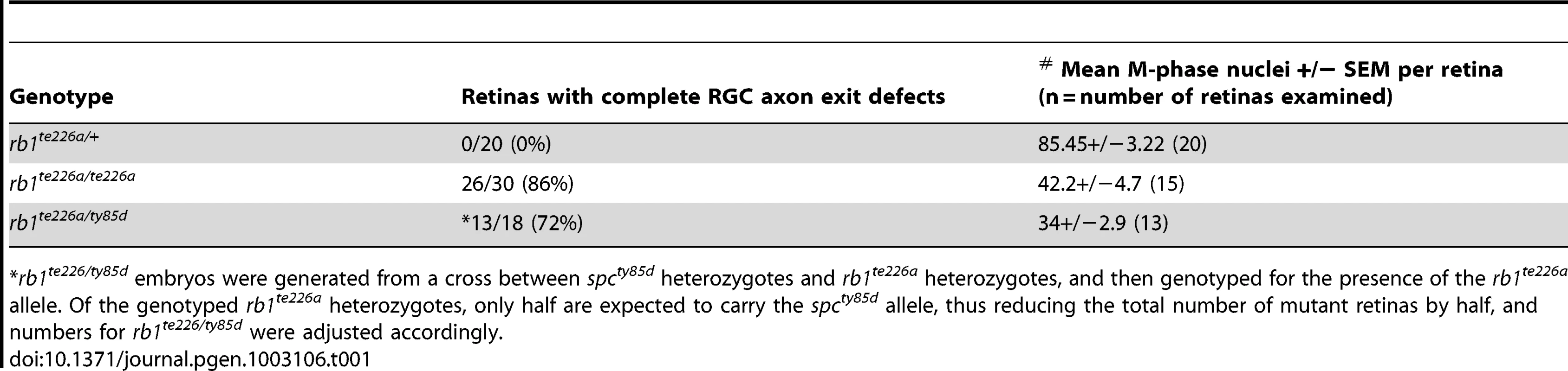
rb1te226/ty85d embryos were generated from a cross between spcty85d heterozygotes and rb1te226a heterozygotes, and then genotyped for the presence of the rb1te226a allele. Of the genotyped rb1te226a heterozygotes, only half are expected to carry the spcty85d allele, thus reducing the total number of mutant retinas by half, and numbers for rb1te226/ty85d were adjusted accordingly. rb1 mutant embryos show delayed RGC axon outgrowth and reduced tectal innervation
During zebrafish embryogenesis, the earliest born RGCs begin extending axons at 32 hpf, cross the ventral midline of the diencephalon to form the optic chiasm at 36 hpf, and project dorsally to the contralateral optic tectum by 48 h to form a retinotectal pathway critical for mediating visually guided behaviors by 120 hpf [19]. We previously reported that 120 hpf stage rb1 mutants display wild type like retinal lamination and expression of terminal RGC cell differentiation markers, but exhibit various RGC axonal pathfinding defects [18]. To determine the temporal onset and spatial site of RGC pathfinding errors in rb1 mutant embryos, we used the ath5:gfp transgenic line expressed in RGCs to examine the development of the retinotectal trajectory [20]. Importantly, between 28–96 hpf we did not detect a difference in the intensity of GFP fluorescence in the retinas of rb1 mutant compared to wild type retinas (Figure S1 and data not shown). At 36 hpf wild type RGCs have exited the retina and pioneered across the ventral midline to form an optic chiasm (n = 40, Figure 2A). In contrast, only 13% (n = 30) of rb1 mutant retinas had RGC axons that exited the retina (Figure 2B, 2C), suggesting that the loss of rb1 function causes a delay in the initial outgrowth of RGC axons from the retina. At 48 hpf the optic nerve in rb1 mutants was significantly thinner, with a mean diameter of 3.04 µm (n = 18), compared to the thicker optic nerves in wild type siblings (13.76 µm, n = 22; Figure 2D–2F). At 72 and 96 hpf, when wild type RGC axons have reached and innervated the optic tectum, rb1 mutant tecta show a significant reduction in RGC axon tectal innervation (Figure 2G–2L; see Methods for quantification details). Together, these results reveal that rb1 mutants exhibit a delay in RGC axonal outgrowth, leading to a delay in optic nerve development, and reduced innervation of the optic tectum.
Fig. 2. Rb1-deficient embryos possess delayed retinotectal projection. 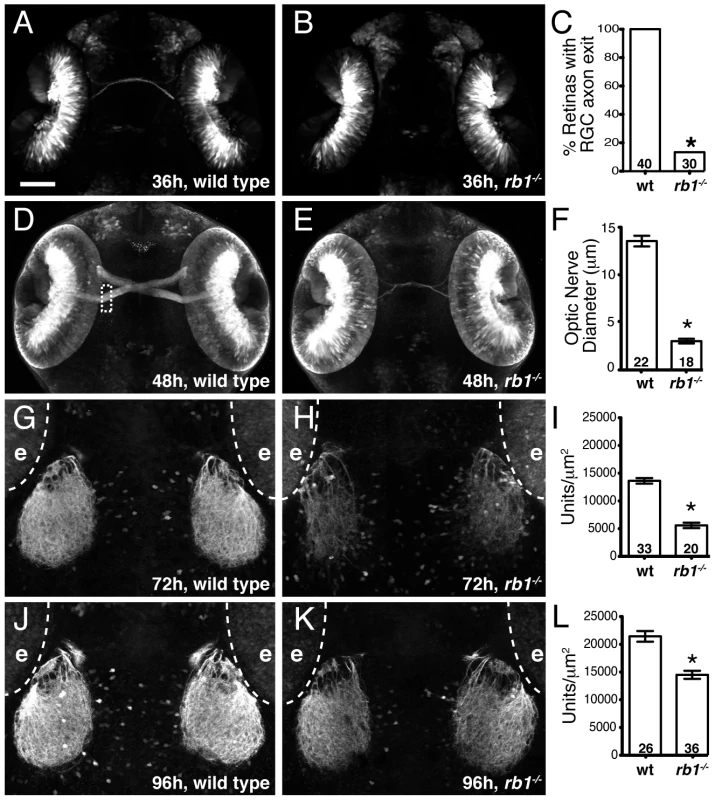
Dorsal view of maximum intensity projection of confocal z-stacks of RGC axons in wild type or rb1te226; ath5:gfp+/− embryos. (A–C) RGC axons fail to exit rb1te226 retina at 36 hpf. (D–F) rb1te226 optic nerves are hypoplastic. Diameter measured upon retinal exit, marked by dashed rectangle. (G–L) Tectal innervation is reduced in rb1te226 at 72 (G–I) and 96 hpf (J–L). Innervation measured as a function of pixel intensity units per µm2 from summation intensity projections of tectal area (not shown, see Experimental Procedures). Digitally excluded eyes (e) are outlined by dotted line. (C, F, I, L) Bar graphs represent the mean with error bars denoting SEM. *p<0.001; one-way ANOVA (F, I, L) or binomial z-test (C). N retinas, optic nerves, or tecta shown at base of bar graphs. Anterior to the top of panel, scale bar = 50 µm. Delayed retinotectal development is caused by a near complete loss of rb1 function in RGCs
To determine whether the identified mutation in rb1te226a is causative of the delay in retinotectal development, we injected wild type rb1 mRNA into one-cell stage rb1te226a mutants and examined optic nerve diameter at 48 hpf. Microinjection of wild type rb1 mRNA restored optic nerve diameter in rb1 deficient mutants in a dose dependent manner, demonstrating that mutations in zebrafish rb1 cause RGC outgrowth defects (Figure 3A–3C, 3E). To determine if and to which extent the mutant rb1te226a allele has retained biological activity, we examined the ability of rb1te226a mRNA to rescue retinotectal development in rb1te226a mutants (Figure 3D–3E). Injection of rb1te226a mRNA failed to significantly increase optic nerve diameter in rb1te226a mutants, suggesting that the rb1te226a protein product has very limited, if any, functionality. However, we cannot exclude the possibility that the mutant phenotype is ameliorated by maternal rb1 mRNA and/or protein deposition.
Fig. 3. rb1 mRNA overexpression rescues optic nerve hypoplasia in rb1te226 mutants. 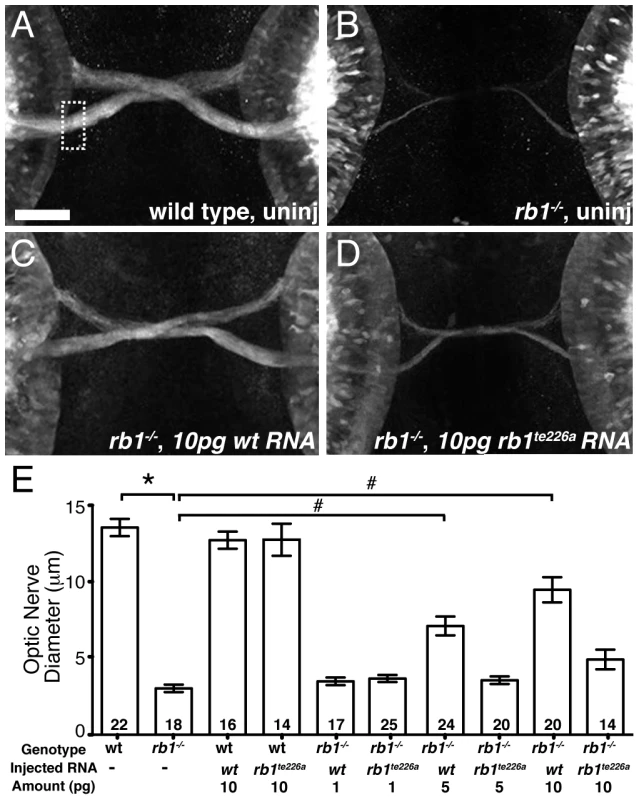
(A–B) Optic nerves of uninjected wild type (A) and rb1te226a; ath5:gfp+/− embryos (B) at 48 hpf. (C–D) Optic nerves of rb1te226a; ath5:gfp+/− embryos injected with 10 pg of wild type rb1 (C) or rb1te226a (D) mRNA. Dorsal view of maximum intensity projection of confocal z-stacks. (E) Mean optic nerve diameter, measured at dashed rectangle in (A). Error bars denote SEM. *p<0.001, #p<0.01; one-way ANOVA with brackets indicating compared groups. N optic nerves shown at base of bar graphs. Anterior to the top of panel, scale bar = 50 µm. Finally, we asked whether Rb1 functions within RGCs for their axons to exit from the retina and enter the retinotectal path. Because zebrafish rb1 is expressed ubiquitously throughout development (Figure 4A–4B), we generated chimeric embryos by transplanting cells at the blastula stage between rb1 mutant and wild type embryos, and then examined their ability to exit from the retina (Figure 4C). A significant fraction of axons from genotypically mutant rb1 RGCs transplanted into wild type hosts failed to exit from the retina (19% of retinas showed failure of transplanted rb1 mutant RGC axons to exit, n = 31, Figure 4E–4F), consistent with the low but significant frequency of rb1 mutant retinas in which we observed a complete failure of RGCs to exit from the eye (11%; see below). Conversely, 100% of rb1 mutant retinas showed exit of axons from transplanted wild type RGCs (n = 69, Figure 4D). Thus, during zebrafish development rb1 acts RGC autonomously for axons to exit the retina and to form the optic nerve.
Fig. 4. rb1 is required in RGC axons to regulate retinal exit. 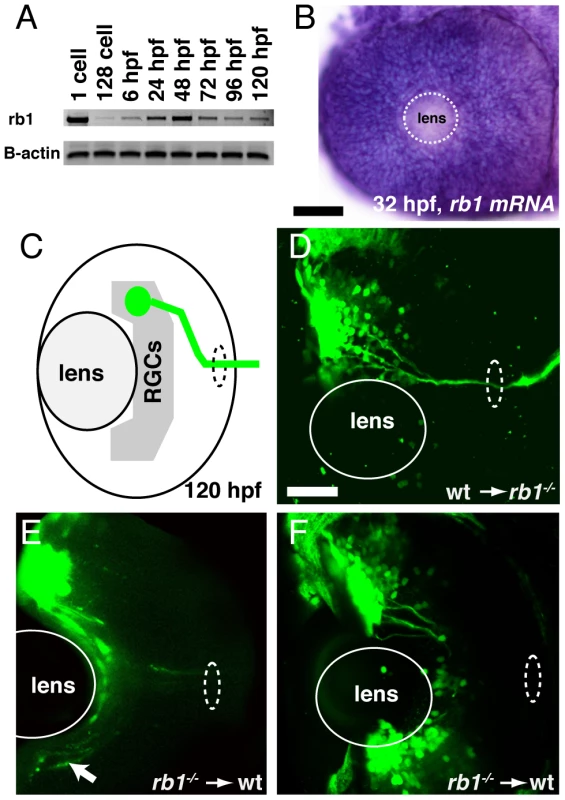
(A–B) rb1 expression by RT-PCR from the 1-cell to 120 hpf larval stage and in the retina by in situ hybridization at 32 hpf. B-actin shown as control. White dashed circle marks lens position. (C) Schematic representation of retinal exit by ath5:gfp labeled transplanted RGC clones. (D–F) Transplanted ath5:gfp labeled clones from wild type donors to rb1te226 mutant retinas (D) and vice versa (E–F). White dashed oval marks retinal exit point. Arrow indicates intraretinally misrouted axons (E). Scale bar = 50 µm. Delayed cell cycle exit of rb1 mutant RGC precursors leads to reduction of “early born” RGCs
Given the RGC intrinsic defects observed in rb1 mutants, we next wanted to determine the primary defect leading to the delay of RGC axons to exit from the retina. Rb1 canonically functions to regulate cell cycle checkpoints, promoting cell cycle exit and differentiation of progenitors and suppressing cell cycle re-entry of differentiated cells [2]. In the retina, rb1 has been shown to promote the exit of retinal progenitor cells from the cell cycle into the various postmitotic cell types that populate the retinal lamina [9], [10], [11], [12]. To examine rb1 deficient retinas for cell cycle defects, we labeled wild type and rb1 mutant retinas for M-phase positive nuclei with an anti-phosphohistone-H3 antibody (anti-pH3) during the initial phase of RGC birth and axon outgrowth, between 28 and 36 hpf. During this time window, premitotic ath5 positive retinal progenitors divide, with one daughter becoming a postmitotic RGC and the other maintaining its progenitor potency to give rise to other retinal cell types that become postmitotic at later stages of development [21]. Although the total number of M-phase positive increased with time between 28 and 36 hpf in rb1 mutant and wild type retinas, we observed fewer M-phase positive nuclei in rb1 deficient retinas, compared to wild type retinas, at each time point examined (Figure 5).
Fig. 5. rb1te226a retinas show delayed cell cycle exit during early retinogenesis. 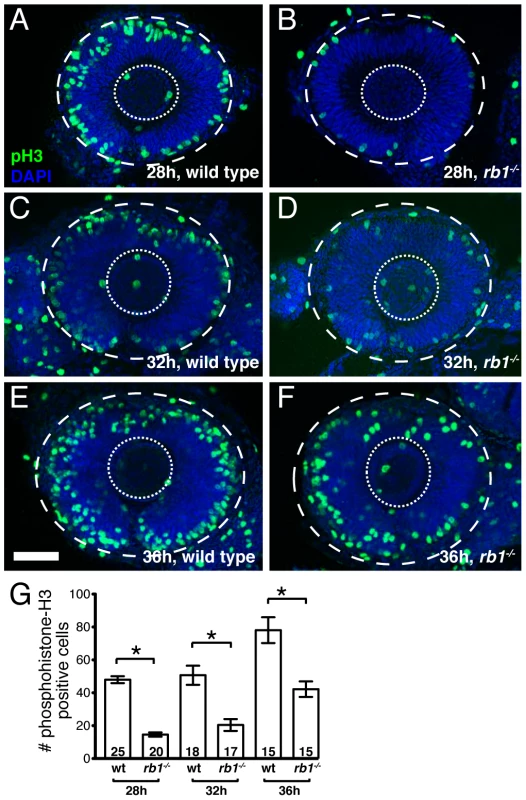
Retinas removed from wild type (A, C, E) or rb1te226a embryos (B, D, F) at 28 (A–B), 32 (C–D), or 36 hpf (E–F). Retinas labeled with anti-phosphohistone H3 antibody (green) to label M-phase nuclei and counterstained with DAPI (blue). Lateral view of maximum intensity projection of confocal z-stacks. Inner dashed circle outlines lens and outer dashed circle outlines retina. (G) Mean number of anti-phosphohistone-H3 labeled cells per retina. Error bars denote SEM. *p<0.01; one-way ANOVA with brackets indicating compared groups. N retinas shown at base of bar graphs. Anterior to the left, dorsal to the top of each panel. Scale bar = 50 µm. One possibility is that the reduction of M-phase retinal precursors in rb1 deficient retinas is due to increased cell death. Indeed, compared to wild type retinas, rb1 deficient retinas showed a slight, but significant increase of TUNEL positive nuclei between 28 and 36 hpf (Figure S1). Importantly though, comparing the increased number of TUNEL positive nuclei to the decreased number of pH3 positive nuclei in rb1 mutants at 28, 32, and 36 hpf revealed that apoptosis accounts for only 18–26% of the observed reduction in M-phase positive retinal precursors in rb1 mutant retinas at each time point examined. This suggests that in rb1 mutant retinas cell death contributes only partially to the deficiency of M-phase positive nuclei (Figure S1G). Thus, the reduction in M-phase retinal precursors in rb1 mutant retinas suggests a prolonged terminal cell cycle for the retinal precursors, which need to exit their final cycle to become the earliest population of postmitotic RGCs.
To determine if loss of rb1 function indeed causes an initial delay in the presence of postmitoic, differentiated RGCs, we examined expression of isl2b-gfp, one of the earliest transgenic markers indicative for postmitotic RGCs [22]. We found that in wild type retinas postmitotic, differentiating RGCs marked by isl2b-gfp expression emerged first at 32 hpf, increased significantly in their abundance by 36 hpf, and by 48 hpf isl2b-gfp positive RGCs were densely packed throughout the ganglion cell layer (Figure 6A, 6C, 6E, n = 25, 16, and 29, respectively). In contrast, isl2b-gfp positive RGCs were present in only 13% of rb1 deficient retinas at 32 hpf (n = 23). Because of the cytoplasmic localization of the GFP signal and the density at which RGCs normally populate the ganglion cell layer, it is difficult to determine the total number of isl2b-gfp positive RGCs. Nonetheless, semi-quantitative analysis revealed that by 36 hpf, isl2b-gfp positive RGCs were present in 90% of rb1 mutant retinas (n = 29); however, their distribution with the retina was more similar to that of younger wild type retinas at 32 hpf (Figure 6B, 6D). By 48 hpf, all rb1 mutant retinas harbored isl2b-gfp neurons (n = 32), although differentiation still appeared to lag in 81% (n = 32) of the rb1 mutant retinas compared to the more densely packed ganglion cell layer in wild type retinas (n = 29, Figure 6E–6F). Despite the reduced number of RGCs present at 48 hpf, rb1 mutant isl2b-gfp positive RGCs express DM-GRASP, a late marker of RGC differentiation, demonstrating that mutant RGCs were fully differentiated once becoming postmitotic (Figure S2). Importantly, the number of ath5-gfp positive RGC precursors was unaffected in rb1 mutants (Figure S1 and data not shown). Thus, the rb1 deficiency causes a delay in the transition of RGC precursors to postmitotic RGCs, but not in the specification of RGC precursors.
Fig. 6. rb1te226a retinas possess fewer postmitotic RGCs. 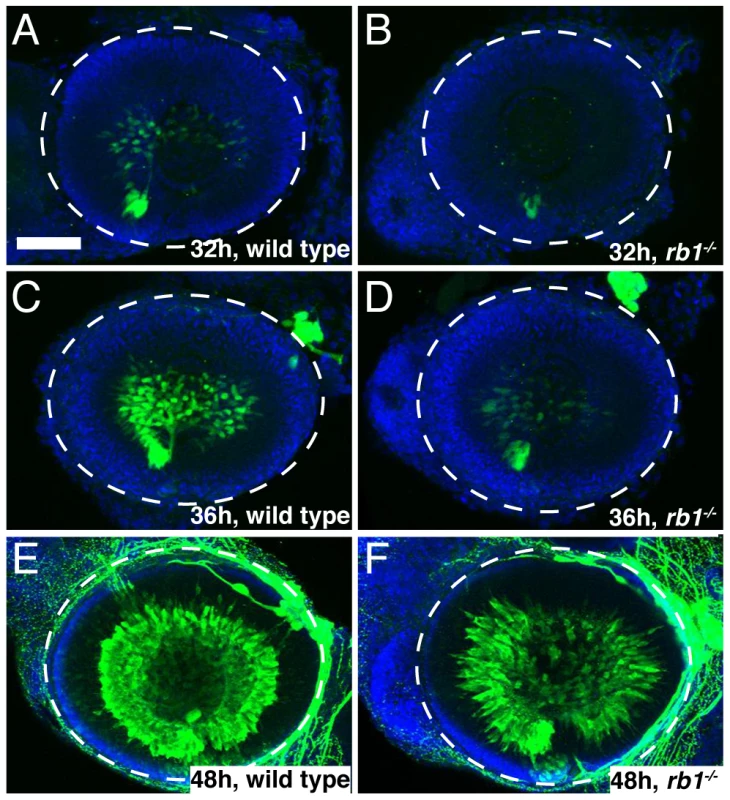
Retinas removed from wild type (A, C, E) or rb1te226a; isl2b:gfp embryos (B, D, F) at 32 (A–B), 36 (C–D), or 48 hpf (E–F). Retinas labeled with anti-GFP (green) to label postmitotic, isl2b expressing RGCs and counterstained with DAPI (blue). Lateral view of maximum intensity projection of confocal z-stacks. White circle outlines retina. Anterior to the left, dorsal to the top of each panel. Scale bar = 50 µm. Aside from the reduced population of early born RGCs, rb1 mutant retinas appear grossly normal, and at 120 hpf, show proper lamination by each retinal cell type [18]. Although we did not determine whether birth dating of other retinal cell types is affected in rb1 mutant retinas, netrin-positive exit glial cells and Muller glia cells are present in appropriate numbers and location, indistinguishable from wild-type retinas (Figure S2). Taken together, these results suggest that a delay in cell cycle exit by rb1 deficient RGC precursors leads to a transient reduction in the early born postmitotic RGCs without consequence to the gross morphology and overall cellular landscape of the rb1 mutant retina.
rb1-deficient RGC axons exhibit intraretinal and midline pathfinding errors
The early born RGCs are located within the central retina and pioneer the retinotectal tract to the contralateral optic tectum [22]. In the absence of the early pioneering RGC axons, the axons of later born, more peripherally located RGCs fail to exit the eye and project aberrantly within the retina [22]. Given the reduced number of these early born, central RGCs in rb1 mutants, we sought to determine whether peripheral RGC axon trajectories were affected. For this, we labeled small groups of RGCs in the anterior peripheral retina with DiO (green) and in the posterior peripheral retina with DiI (red, Figure 7A–7F). In 120 hpf larvae wild type larvae, all labeled axons from anterior and posterior RGCs fasciculated shortly after sprouting from their soma and extended as an axon bundle, forming a path directly toward the retinal exit point (n = 83, Figure 7A–7B, 7D–7E). In contrast, 91% (n = 65) of rb1 deficient retinas harbored a significant subset of axons that had extended aberrantly throughout the retina and failed to exit (Figure 7C, 7F). These results demonstrate that the delayed differentiation of the early born RGCs in rb1 mutants impairs the ability of later born RGC axons to exit the retina.
Fig. 7. Intraretinal and midline RGC axon pathfinding errors in rb1te226a mutants. 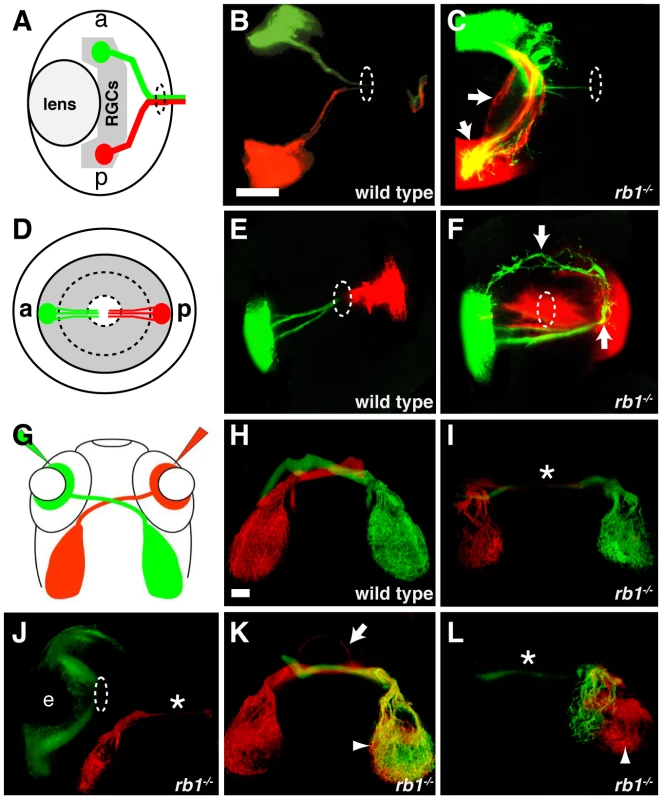
(A–C) Dorsal, intraretinal views of DiO (green) labeled anterior and DiI (red) labeled posterior RGCs. (D–F) Lateral, intraretinal views of DiO/DiI labeled RGCs. (G–L) Dorsal views of retinotectal projection labeled by whole retina fills with DiI/DiO. Eyes removed in H–I, K–L. Dashed circles indicate retinal exit point. Arrows mark misprojecting axons. Asterisk shows optic nerve hypoplasia. Arrowheads mark ipsilateral tectal innervation. Abbreviations: (a) anterior, (p) posterior, (e) eye. Scale bar = 50 µm. The delayed cell cycle exit and differentiation of pioneering RGCs lacking rb1 may also affect axon navigation by later born RGC axons at key choice points: the ventral midline of the diencephalon and/or the optic tectum. To examine these possibilities, we filled the RGC layer of the left and right eyes of wild type and rb1 mutant larvae with either DiI or DiO, respectively (Figure 7G). In wild type siblings, 99% of dye filled optic nerves projected to their appropriate contralateral tectum (n = 946, Figure 7H). In contrast, rb1 mutant optic nerves displayed a variety of phenotypes. The majority of rb1 deficient optic nerves were significantly thinner than their wild type counterparts (37%, n = 663, Figure 7I–7J, 7L), consistent with what we observed in with ath5:gfp (Figure 2). In a significant portion of rb1 deficient optic nerves, 17%, RGCs projected to both the contralateral but also to the ipsilateral tectum, indicative of midline pathfinding defects (n = 663, Figure 7K–7L). Focal DiI/DiO labeling of RGC axons arising from the anterior and posterior retina revealed that retinotopic mapping, a function of retinal cell body location [23], remains intact in rb1 mutants despite the aberrant pathfinding en route to the optic tectum (Figure S3). Finally, in 11% of rb1 mutant retinas, there was a complete failure of RGCs to exit from the eye, even at 120 hpf (Figure 7J). Taken together, these results suggest that rb1 deficient RGC axons make intraretinal and midline pathfinding errors, leading to reduced and incorrect tectal innervation.
rb1 mutants display deficits in visually guided behaviors
By 120 hpf, zebrafish larvae perform an array of sensorimotor behaviors, including responses to visual stimulation. For example, changes in visual field illumination, such as the sudden absence of light or a shift from uniform to focal illumination, elicit specific, stereotyped turning behaviors [24], [25]. We first examined the ability of rb1 mutant larvae to perform positive phototaxis, defined as navigating toward a target light source that is presented after extinguishing the pre-adapted uniform light field [25]. Positive phototactic navigation is characterized by larvae first turning towards the target light source and then swimming forward towards the target. As previously reported, when presented with a target light source wild type larvae facing away from the light target show significant initiation of turns, which are preferentially biased towards the light target (Figure 8A–8B). Once facing the target, wild type larvae initiate forward scoot swims (Figure 8C). In contrast, turn initiation in rb1 mutant larvae facing away from the light target was dramatically reduced (Figure 8A). On the few occasions when they initiated a turn, turning direction was unbiased with respect to the light target (Figure 8B). Moreover, rb1 mutants facing the light target did not show an increase in forward scoot swim initiation above baseline (Figure 8C). To further determine whether rb1 mutants respond to more extreme changes in illumination, we examined their ability to perform an O-bend response to a visual dark flash stimulus, a sudden extinction of light [24]. Again, compared to their wild type siblings, rb1 mutants displayed a minimal O-bend response to dark flash stimulation (Figure 8D). Despite their impaired visual responses, rb1 mutants showed no difference in the spontaneous initiation of turning or swimming behaviors compared to wild type siblings (Figure 8D). Importantly, the kinematic parameters of spontaneously occurring turning and swimming movements were indistinguishable between rb1 mutants and their wild type siblings, demonstrating that the neural circuits required for initiation and execution of turning behaviors are largely intact in rb1 mutants. Together, these results demonstrate that rb1 mutants exhibit visual deficits.
Fig. 8. <i>rb1<sup>te226a</sup></i> larvae fail to execute visually guided behaviors. 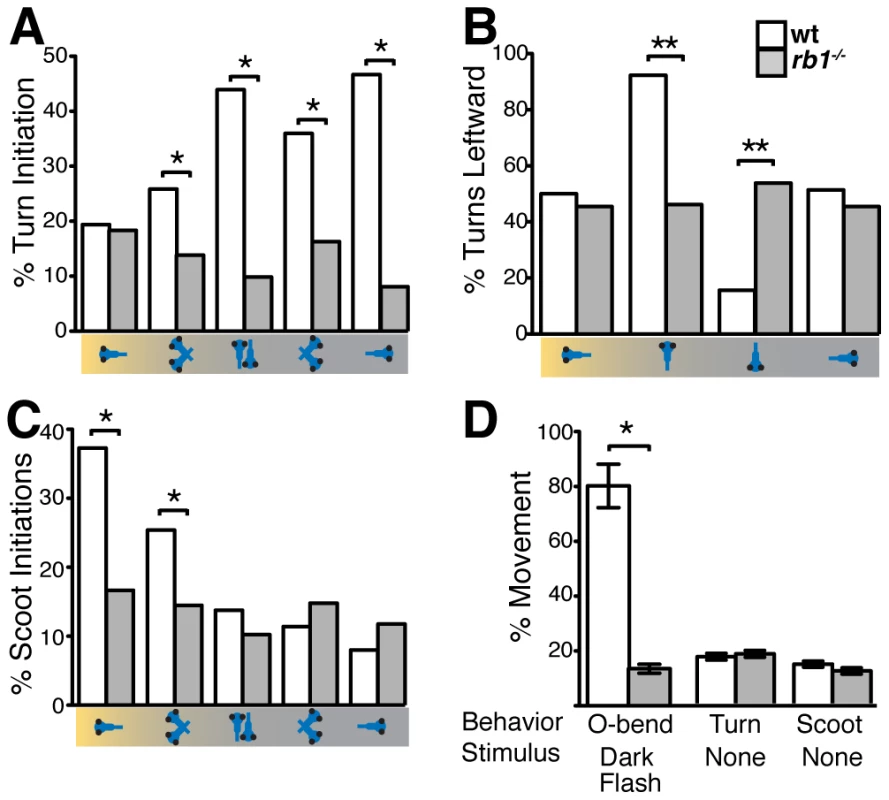
Discussion
Children with biallelic germline or sporadic inactivation of rb1 are likely to form ocular tumors during early childhood. Initially, the retinas of affected individual show an otherwise grossly normal morphology. In contrast, even conditional rb1 knockout mouse models exhibit ectopic proliferation and cell death leading to significant morphological defects throughout affected retinas. We find that inactivation of the zebrafish rb1 gene through a rb1 causing mutation results in mutant retinas that display very limited signs of cell death, with differentiated retinal cell types that are properly laminated, similarly to childhood retinas lacking rb1. Thus, the fairly ‘normal’ retinal landscape of zebrafish rb1te226a mutants provided us with a unique opportunity to investigate if and how rb1 is required to establish the retinotectal projection. Our analysis reveals a RGC autonomous requirement for rb1 in regulating RGC axon pathfinding within the retina and at presumptive choice points en route to the optic tectum. Moreover, we demonstrate that zebrafish rb1te226a mutants exhibit deficits in visually guided behaviors, suggesting that the retinotectal path defects in rb1 mutants may be sufficient to impair vision. Together, this work reveals a novel role for rb1 in the establishment of RGC axon projections during development and establishes a unique model for understanding the developmental and tumor suppressor roles of the rb1 gene.
Zebrafish rb1te226a mutants harbor a human retinoblastoma causing rb1 gene mutation. The mutant protein is truncated in the B-domain and lacks the cyclin-binding domain, reducing Rb1's capacity to form a ‘pocket’, and reducing its capacity for phosphorylation by cyclin dependent kinases [2], [26]. Consistent with the notion that the rb1te226a mutant allele is largely non-functional, mRNA over-expression in rb1 mutants does not ameliorate the rb1 mutant phenotype. Despite the absence of biological activity of the truncated rb1te226a protein, mutant zebrafish show a significantly milder retinal phenotype compared to conditional or even germline rb1 mouse knockouts [6], [7], [8], [9], [10], [11], [12]. One possible explanation is the strong maternal contribution of rb1 in zebrafish (Figure 4A), which may suppress phenotypic expressivity at early stages of development. Consistent with this idea, formation of the initial scaffold of axon tracts during the first day of development appears unaffected in rb1te226a mutants, yet visual and hindbrain pathways that develop after the first day of development show defects [18].
In humans the rb1 nt1960+1 mutation, which is identical to the zebrafish rb1te226a mutation, causes ocular tumors [13], [14], [15], [16], raising the possibility that zebrafish rb1 mutants might also develop tumors as juveniles. However, rb1 mutants fail to inflate a functional swim bladder, and die ∼7 days post fertilization, precluding the analysis of ocular tumors in juveniles. Although wild type rb1 mRNA injection rescues the early RGC and retinotectal defects through 48 hpf, these transiently rescued rb1te226a mutants do not survive beyond 7 days of development, indicating that rb1 plays a critical role after the injected mRNA has been degraded. Establishing stable, inducible rb1 transgenic lines to rescue developmental deficits will therefore be required to monitor juvenile and adult zebrafish for retinal tumors.
In rb1te226a mutants, a significant subset of RGC axons fail to exit the retina, and many of the exiting axons then project incorrectly to the ipsilateral tectum, revealing a previously unrecognized requirement for rb1 in regulating axon pathfinding. One possible explanation for the RGC guidance defects is that in rb1te226a mutant RGCs the expression of guidance factors might be disrupted. Interestingly, cortical cell migration was shown to be dependent on rb1 regulated neogenin expression [27], suggesting that rb1 deficient RGC axons might lack guidance factors required to navigate towards the retinal exit point and properly cross the ventral midline. To investigate this possibility, we performed microarray gene expression analysis of the retina and brains of 32 hpf rb1te226a mutants (MAW and MG, unpublished). However, this approach did not reveal a significant change in the expression levels of neogenin or other known axon guidance genes. Although it remains possible that rb1 regulates expression of un-identified guidance factors, it is more likely that rb1 regulates axon pathfinding indirectly by ensuring the timely exit of RGC precursors from the cell cycle and hence the appropriate temporal appearance of differentiated RGCs. In fact, genetic ablation of the earliest born RGCs prevents the formation of the retinotectal tract [22]. This suggests that RGC birth order imprints a critical hierarchical pathfinding role on RGC axons, such that axons from the earliest born RGCs pioneer the retinotectal tract that later born RGC axons will follow [22]. The delayed onset of RGC birth in rb1te226a mutants may therefore reduce the population of pioneering RGCs present during a restricted window of environmentally expressed guidance factors.
Zebrafish rb1te226a mutants display deficits in the acoustic startle response [17], [18] and in visually guided behaviors, reflecting the importance of rb1 function for the development of neural circuitry underlying behavior. The deficits in startle behavior are due to defects in a small subset of hindbrain neurons, the spiral fiber neurons [18], and giving the results presented here, it is tempting to speculate that rb1 plays a similar role for the transition of these neurons from precursors to postmitotic neurons. Unfortunately, markers that follow the development of spiral fiber neurons are not available, precluding such analysis. Therefore, we focused on the well-characterized development of RGCs. Although we demonstrate a defect in the early development of these cells and their axonal connectivity, we cannot exclude the possibility that zebrafish rb1 mutants exhibit defects in the development and/or function of other retinal cell types, and that these defects contribute to the deficits in visual behaviors we observe. Future analysis of transgenic lines expressing the wild type Rb1 gene in individual retinal cell types will reveal which cell type(s) and connections are causative of the visual deficit.
In summary, we report a zebrafish mutant carrying a human disease causing rb1 mutation, which reveals novel roles of rb1 in regulating RGC axon pathfinding and visually guided motor behavior. Furthermore, these mutants provide a non-murine vertebrate model of rb1 and offer new potential for identifying the elusive retinoblastoma cell of origin and further insight into the developmental role of rb1.
Materials and Methods
Ethics statement
All experiments were conducted according to an Animal Protocol fully approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC) on January 27, 2011, protocol number 803446. Veterinary care is under the supervision of the University Laboratory Animal Resources (ULAR) of the University of Pennsylvania.
Animals and fish maintenance
The zebrafish (Danio rerio) strain used in this study was the spcte226a allele (now referred to as rb1te226a) of space cadet [17], [18], maintained on a mixed TLF and Tubingen background. The rb1te226a allele was also crossed into the ath5:gfp and isl2b:gfp transgenic backgrounds for RGC analysis [20], [22]. rb1te226a+/−;ath5:gfp+/− or rb1te226a+/−;isl2b:gfp+/− adults were always crossed with rb1te226a+/−;TLF adults to ensure that rb1te226a embryos analyzed for GFP-expressing RGCs were hemizygous for GFP. Throughout the manuscript, rb1−/−, “rb1 deficient”, and rb1 mutant refers to rb1te226a homozygotes. The other space cadet allele spcty85d [17] was only used where mentioned. Embryos were collected in the morning, maintained on a 14/10 hour light/dark cycle at 28°C, and staged as described previously [28]. Larvae were raised in 6 cm plastic Petri dishes at a density of 20–30 per 7 mL in E3 medium (5 mM NaCl, 0.17 m mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) with medium changes at 48 hpf (hours post fertilization) and 96 hpf. Behavioral experiments were conducted on 120 hpf larvae.
Recombination mapping and molecular cloning of rb1
A three generation mapping cross between rb1te226a heterozygous and WIK fish was generated, and pools of 25 F2 mutant and F2 sibling 5 dpf larvae were collected in the F2 generation and used for bulk segregant mapping (see Table 2 for simple sequence length and single nucleotide polymorphic markers). Mutant larvae were identified by performing successive, unilateral C-bends to acoustic or tactile stimulation [17], [18]. To identify the mutation, cDNA was prepared following total mRNA extraction from 5 dpf larvae as previously described [29]. rb1 cDNA was amplified with primers (rb1:1–6, Table 2) designed against overlapping regions of the rb1 reference sequence (Ensembl) with the following RT-PCR conditions: 94°C for 3 min and then 40 cycles of 94°C for 45 sec, 57°C for 1 min, and 70°C for 1 min. Products were gel purified and cloned into the pCR2.1-TOPO-TA vector for sequencing. After detecting a frameshift and 4 nucleotide addition to the end of exon 19 in rb1te226a cDNA clones, gDNA was isolated from 5 dpf larvae, and intron 19 was amplified with the rb1:8 primers, using identical PCR conditions to those described above.
Tab. 2. PCR primers for recombination mapping, molecular cloning, and genotyping of <i>rb1<sup>te226a</sup></i>. 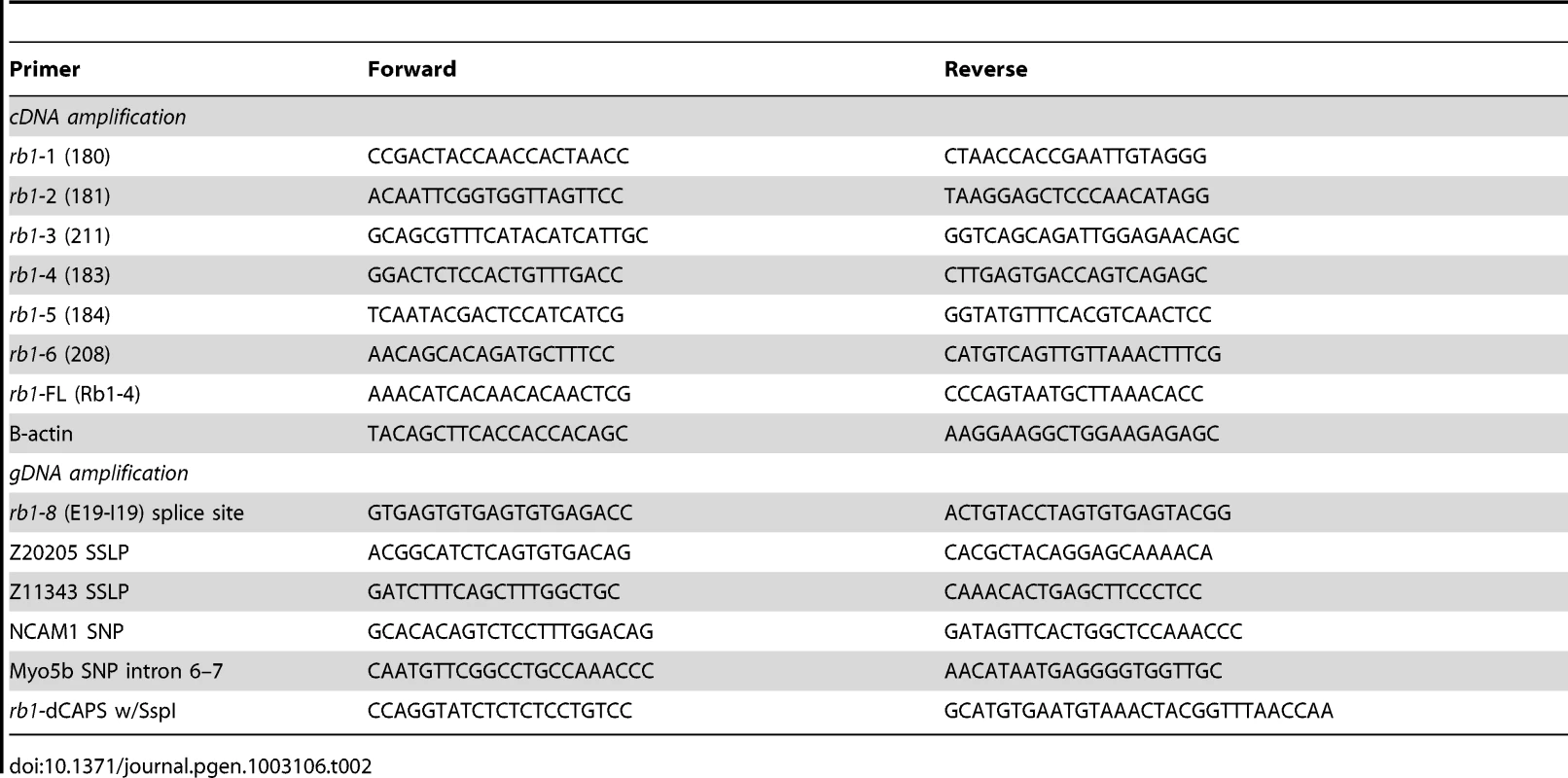
For rb1 RNA injection, cDNA was prepared from genotyped homozygous wild type or rb1te226a mutant 5 dpf larvae (dCAPS protocol, see below) and amplified with the rb1:FL primers (similar conditions as above, except extension time increased to 3 min), which includes the coding region of rb1, and cloned into the pCS2+ vector. Wild type rb1 and rb1te226a mRNA was prepared using the mMessage mMachine kit (Ambion, NY) and injected at the 1-cell stage at doses ranging from 1–100 picograms. Embryos injected with 20 or greater picograms of rb1 mRNA showed gross morphological abnormalities and necrosis, whereas embryos injected with 10 picograms or less appeared morphologically normal.
PCR genotyping rb1te226a
To genotype rb1te226a embryos, we developed a dCAPS assay [30] using the dCAPS program (http://helix.wustl.edu/dcaps/dcaps.html) to design appropriate primers (Table 2). After gDNA isolation, PCR was performed as described above. The PCR product is then digested with SspI (New England Biolabs, Ipswich, MA), cleaving the rb1te226a allele and producing a 120 bp fragment that can be distinguished from the 150 bp wild type allele on a 3% agarose gel containing 1.5% Metaphor agarose (Lonza, Rockland, ME). All genotyping, except for BrdU labeled embryos, was performed following immunolabeling experiments.
Immunohistochemistry and in situ hybridization
For immunostaining, embryos were fixed in 4% paraformaldehyde (PFA) overnight at 4°C, permeabilized with 1 mg/mL collagenase, and blocked for 1 hour with 5% normal goat serum in 0.1 M phosphate buffer. Embryos were then incubated in the primary antibodies anti-GFP (1∶200 mouse JL8, Clontech, Mountain View, CA or 1∶500 rabbit, Invitrogen, Carlsbad, CA), anti-phosphohistone-H3 (Millipore, Charlottesville, VA), 1∶100 anti-BrdU (Roche, Branchburg, NJ), and/or 1∶50 A2-J-22 polyclonal antisera (recognizes carbonic anhydrase II, kindly provided by Dr. P. Linser) overnight at 4°C in blocking solution, washed out, and then detected by the addition of AlexaFluor488 or AlexaFluor594 conjugated secondary antibodies (1∶500, Invitrogen, Carlsbad, CA). TUNEL assay was performed as previously described [31] using Apoptag Peroxidase In Situ Apoptosis Detection Kit (Chemicon, Temecula, CA). After staining, samples were mounted in DAPI containing Vectashield (Vector Labs, Burlingame, CA). Images were acquired with a Zeiss 710 confocal laser scanning microscope (LSM 710) using ZEN2010 software.
For in situ hybridization, digoxygenin-UTP labeled antisense riboprobes for rb1 were synthesized and hydrolyzed from the full length rb1 cDNA construct [32]. Whole-mount in situ hybridization was performed as described previously [33]. Images were acquired with a Zeiss Axioskop compound microscope. For RT-PCR based expression analysis, the rb1:FL and B-actin primers (Table 2) were run against cDNA prepared from total mRNA extracted from 25 embryos/larva at each stage.
Lipophilic dye labeling of retinal ganglion cells
120 hpf larvae were anesthetized (0.01% Tricaine) and fixed in 4% paraformaldehyde at 4°C overnight. Larvae were removed from fix, washed briefly in phosphate buffered saline (PBS), and mounted dorsal side up for whole retinal injection or laterally for discreet RGC labeling on glass microscope slides in a bed of 1.5% agarose. To label all RGCs, the vitreal space of each eye was filled with either of the fluorescent lipophilic dyes DiI (red) or DiO (green) (Molecular Probes, Eugene, OR) dissolved in 1% chloroform, using a WPI PV820 picopump injector fitted with a glass micropipette. For discreet labeling, a small region of the exposed eye was labeled with pulses of DiI/DiO dissolved in 0.5% dimethylformamide. Injected larvae were kept moist with PBS and incubated overnight at room temperature in a humidity chamber in darkness. Larvae were then examined for phenotype analysis using a Zeiss Axioplan compound fluorescent microscope. Eyes were carefully removed from selected representative larvae, which were then remounted on coverslips in agarose for imaging. Images were recorded using a Zeiss 510 confocal laser scanning microscope (LSM510) and Zeiss LSM510 analytic software.
Cell transplantation
For transplant direction wild type donor into space cadet host, wild type transgenic Tg(ath5:gfp) and rb1te226a heterozygous fish were used to generate wild type GFP expressing donor embryos and non-GFP expressing rb1te226a mutant embryos, respectively. For transplant direction space cadet donor into wild type host, rb1te226a; Tg(ath5:gfp) double heterozygotes and either TU or TLF strain wild type mating pairs were used to generate rb1te226 GFP expressing donor embryos and non-GFP expressing wild type embryos, respectively. Once the appropriate donor-host embryos were collected, embryos were immediately placed in E3 medium and kept at room temperature. Donor embryos were pressure injected into the yolk sac at the 1–2 cell stage with the lineage tracer tetramethylrhodamine dextran, 3 Kd, 5% w/v (Molecular Probes, Eugene, OR) dissolved in 0.2 M KCL and filter sterilized. Donor and host embryos were then incubated at 28.5°C in E3 medium in darkness to grow synchronously to the 1000 cell stage. Embryos were then transferred into room temperature complete E2 medium (E2) to retard growth, and dechorionated using Pronase (1∶50 in E2 of 30 mg/ml stock, Roche) in glass 60 mm petri dishes. Dechorionated embryos were washed extensively with E2, transferred using a fire polished glass Pasteur pipette into individual wells in a transplantation dish containing E2, and properly oriented. Transplantation needles were made using #1BBL No Fil borosilicate glass pipettes (WPI), pulled to produce fine tips in a P87 pipette puller (Sutter Instruments, Novato, CA), broken at various diameter openings, and polished using a microforge. Needles were then inserted into a standard pipette holder connected to a modified manual injection apparatus, and mounted in a micromanipulator arm for precision control. Thirty to fifty blastomeres were carefully removed from the donor embryo using the transplantation pipette/manual injector apparatus, and transferred into the adjacent host embryo at the apex of the animal pole (eye/nose region). Operated embryos were maintained in the transplantation dish wells in E2 at 28.5°C in darkness following transplantation, and were allowed to develop undisturbed until epiboly completed. Embryos were then transferred from the transplantation wells/dish into either separate 1.5% agarose coated 60 mm plastic Petri dishes for donors and hosts, or 1.5% agarose coated wells in 12-well tissue culture plates as host-donor pairs, depending on the direction of the transplant, and incubated at 28.5°C for five days. The later was necessary in order to correctly identify rb1te226a donors from each donor-host pair, as the motility phenotype does not manifest itself until 120 hpf. 120 hpf larvae were screened for the presence of GFP expressing RGC clones using a Leica MZFIII fluorescence stereomicroscope, and further analyzed for misprojecting RGC axons using a Zeiss axioplan compound fluorescence microscope. Host larvae suspected of containing misprojecting RGC axons (ie, not exiting the eye, or midline defects) were then fixed and stained with anti-GFP antibody as described above, and imaged using a Zeiss LSM510 microscope and Zeiss LSM510 analytic software. Confocal z-stacks were of sufficient depth (150–220 µm) to insure optic nerves were not inadvertently missed.
Image analysis and quantification
Confocal stacks were processed into maximum and/or summation intensity projections using ImageJ for quantification. We used the full width half maximum algorithm to calculate optic nerve diameter from maximum intensity projections of GFP-labeled retinal ganglion cell axons. Tectal innervation was determined by making 20 µm summation projections of GFP labeled tecta, tracing the area of the labeled tectum to determine the Raw Integrated Density (RID) per µm2, and subtracting the RID/µm2 of an unlabeled, background region. TUNEL and anti-pH3 labeled nuclei were counted from 30 µm stacks using Volocity (PerkinElmer, Waltham, MA), with individual cells distinguished by fluorescent intensity and size. Statistical analysis was performed on all data using the Graphpad prism software (www.graphpad.com).
Behavioral analysis
Behavioral experiments were performed on 120 hpf larvae and analyzed with the FLOTE software package as previously described [25], [34], [35]. rb1te226a and wild type siblings were identified based on acoustic startle behavior [17], [18] and then grouped by phenotype for visual behavior testing in 6 cM petri dishes at a density of 12 fish per dish. For all behavioral experiments, N = 48 rb1te226a and 48 wild type sibling larvae. For phototaxis experiments, video recordings were triggered every 500 msec, with each recording covering a 400 msec time window, for a total duration of 4 sec of recorded behavior. Each group of 12 larva were subjected to 3 rounds of phototaxis testing, with 3 min between trials. Orientation of larvae to target light was determined at the beginning of each 400 ms recording as previously described [25], such that the behavior of each larva was tested multiple times and in different orientations with respect to the target light. Therefore, the N for Figure 8A and 8C ranged from 75 to 547 for wild type siblings and 136 to 311 for rb1te226a larvae. In Figure 8B, the N ranged from 39 to 106 for wild type siblings and 13–33 for rb1te226a larvae. For dark flash response experiments, N = 4 groups of 12 larvae. Spontaneous behavior was analyzed on individually housed larvae on a 4×4 grid array.
Supporting Information
Zdroje
1. CobrinikD (2005) Pocket proteins and cell cycle control. Oncogene 24 : 2796–2809.
2. BurkhartDL, SageJ (2008) Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8 : 671–682.
3. LohmannDR (1999) RB1 gene mutations in retinoblastoma. Hum Mutat 14 : 283–288.
4. ValverdeJR, AlonsoJ, PalaciosI, PestanaA (2005) RB1 gene mutation up-date, a meta-analysis based on 932 reported mutations available in a searchable database. BMC Genet 6 : 53.
5. MacphersonD (2008) Insights from mouse models into human retinoblastoma. Cell Div 3 : 9.
6. ClarkeAR, MaandagER, van RoonM, van der LugtNM, van der ValkM, et al. (1992) Requirement for a functional Rb-1 gene in murine development. Nature 359 : 328–330.
7. JacksT, FazeliA, SchmittEM, BronsonRT, GoodellMA, et al. (1992) Effects of an Rb mutation in the mouse. Nature 359 : 295–300.
8. LeeEY, ChangCY, HuN, WangYC, LaiCC, et al. (1992) Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359 : 288–294.
9. ChenD, Livne-barI, VanderluitJL, SlackRS, AgochiyaM, et al. (2004) Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5 : 539–551.
10. ChenD, OpavskyR, PacalM, TanimotoN, WenzelP, et al. (2007) Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol 5: e179 doi:10.1371/journal.pbio.0050179.
11. MacPhersonD, SageJ, KimT, HoD, McLaughlinME, et al. (2004) Cell type-specific effects of Rb deletion in the murine retina. Genes Dev 18 : 1681–1694.
12. ZhangJ, GrayJ, WuL, LeoneG, RowanS, et al. (2004) Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet 36 : 351–360.
13. LohmannDR, GerickM, BrandtB, OelschlagerU, LorenzB, et al. (1997) Constitutional RB1-gene mutations in patients with isolated unilateral retinoblastoma. Am J Hum Genet 61 : 282–294.
14. TaylorM, DehainaultC, DesjardinsL, DozF, LevyC, et al. (2007) Genotype-phenotype correlations in hereditary familial retinoblastoma. Hum Mutat 28 : 284–293.
15. AbouzeidH, MunierFL, ThonneyF, SchorderetDF (2007) Ten novel RB1 gene mutations in patients with retinoblastoma. Mol Vis 13 : 1740–1745.
16. HoudayerC, DehainaultC, MattlerC, MichauxD, Caux-MoncoutierV, et al. (2008) Evaluation of in silico splice tools for decision-making in molecular diagnosis. Hum Mutat 29 : 975–982.
17. GranatoM, van EedenFJ, SchachU, TroweT, BrandM, et al. (1996) Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 123 : 399–413.
18. LorentK, LiuKS, FetchoJR, GranatoM (2001) The zebrafish space cadet gene controls axonal pathfinding of neurons that modulate fast turning movements. Development 128 : 2131–2142.
19. HutsonLD, ChienCB (2002) Wiring the zebrafish: axon guidance and synaptogenesis. Curr Opin Neurobiol 12 : 87–92.
20. MasaiI, LeleZ, YamaguchiM, KomoriA, NakataA, et al. (2003) N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development 130 : 2479–2494.
21. PoggiL, VitorinoM, MasaiI, HarrisWA (2005) Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol 171 : 991–999.
22. PittmanAJ, LawMY, ChienCB (2008) Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development 135 : 2865–2871.
23. McLaughlinT, O'LearyDD (2005) Molecular gradients and development of retinotopic maps. Annu Rev Neurosci 28 : 327–355.
24. BurgessHA, GranatoM (2007) Modulation of locomotor activity in larval zebrafish during light adaptation. J Exp Biol 210 : 2526–2539.
25. BurgessHA, SchochH, GranatoM (2010) Distinct retinal pathways drive spatial orientation behaviors in zebrafish navigation. Curr Biol 20 : 381–386.
26. WeinbergRA (1995) The retinoblastoma protein and cell cycle control. Cell 81 : 323–330.
27. AndrusiakMG, McClellanKA, Dugal-TessierD, JulianLM, RodriguesSP, et al. (2011) Rb/E2F regulates expression of neogenin during neuronal migration. Mol Cell Biol 31 : 238–247.
28. KimmelCB, BallardWW, KimmelSR, UllmannB, SchillingTF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203 : 253–310.
29. PetersonSM, FreemanJL (2009) RNA isolation from embryonic zebrafish and cDNA synthesis for gene expression analysis. J Vis Exp
30. NeffMM, TurkE, KalishmanM (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18 : 613–615.
31. LoweryLA, SiveH (2005) Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development 132 : 2057–2067.
32. CoxKH, DeLeonDV, AngererLM, AngererRC (1984) Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol 101 : 485–502.
33. HalloranMC, SeveranceSM, YeeCS, GemzaDL, RaperJA, et al. (1999) Analysis of a Zebrafish semaphorin reveals potential functions in vivo. Dev Dyn 214 : 13–25.
34. BurgessHA, GranatoM (2007) Sensorimotor gating in larval zebrafish. J Neurosci 27 : 4984–4994.
35. BurgessHA, JohnsonSL, GranatoM (2009) Unidirectional startle responses and disrupted left-right co-ordination of motor behaviors in robo3 mutant zebrafish. Genes Brain Behav 8 : 500–511.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Intrauterinní inseminace a její úspěšnost
- Příjem alkoholu a menstruační cyklus
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
- Růst a vývoj dětí narozených pomocí IVF
- Akutní intermitentní porfyrie
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



