-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate Receptors
GspB is a serine-rich repeat (SRR) adhesin of Streptococcus gordonii that mediates binding of this organism to human platelets via its interaction with sialyl-T antigen on the receptor GPIbα. This interaction appears to be a major virulence determinant in the pathogenesis of infective endocarditis. To address the mechanism by which GspB recognizes its carbohydrate ligand, we determined the high-resolution x-ray crystal structure of the GspB binding region (GspBBR), both alone and in complex with a disaccharide precursor to sialyl-T antigen. Analysis of the GspBBR structure revealed that it is comprised of three independently folded subdomains or modules: 1) an Ig-fold resembling a CnaA domain from prokaryotic pathogens; 2) a second Ig-fold resembling the binding region of mammalian Siglecs; 3) a subdomain of unique fold. The disaccharide was found to bind in a pocket within the Siglec subdomain, but at a site distinct from that observed in mammalian Siglecs. Confirming the biological relevance of this binding pocket, we produced three isogenic variants of S. gordonii, each containing a single point mutation of a residue lining this binding pocket. These variants have reduced binding to carbohydrates of GPIbα. Further examination of purified GspBBR-R484E showed reduced binding to sialyl-T antigen while S. gordonii harboring this mutation did not efficiently bind platelets and showed a significant reduction in virulence, as measured by an animal model of endocarditis. Analysis of other SRR proteins revealed that the predicted binding regions of these adhesins also had a modular organization, with those known to bind carbohydrate receptors having modules homologous to the Siglec and Unique subdomains of GspBBR. This suggests that the binding specificity of the SRR family of adhesins is determined by the type and organization of discrete modules within the binding domains, which may affect the tropism of organisms for different tissues.
Published in the journal: . PLoS Pathog 7(7): e32767. doi:10.1371/journal.ppat.1002112
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002112Summary
GspB is a serine-rich repeat (SRR) adhesin of Streptococcus gordonii that mediates binding of this organism to human platelets via its interaction with sialyl-T antigen on the receptor GPIbα. This interaction appears to be a major virulence determinant in the pathogenesis of infective endocarditis. To address the mechanism by which GspB recognizes its carbohydrate ligand, we determined the high-resolution x-ray crystal structure of the GspB binding region (GspBBR), both alone and in complex with a disaccharide precursor to sialyl-T antigen. Analysis of the GspBBR structure revealed that it is comprised of three independently folded subdomains or modules: 1) an Ig-fold resembling a CnaA domain from prokaryotic pathogens; 2) a second Ig-fold resembling the binding region of mammalian Siglecs; 3) a subdomain of unique fold. The disaccharide was found to bind in a pocket within the Siglec subdomain, but at a site distinct from that observed in mammalian Siglecs. Confirming the biological relevance of this binding pocket, we produced three isogenic variants of S. gordonii, each containing a single point mutation of a residue lining this binding pocket. These variants have reduced binding to carbohydrates of GPIbα. Further examination of purified GspBBR-R484E showed reduced binding to sialyl-T antigen while S. gordonii harboring this mutation did not efficiently bind platelets and showed a significant reduction in virulence, as measured by an animal model of endocarditis. Analysis of other SRR proteins revealed that the predicted binding regions of these adhesins also had a modular organization, with those known to bind carbohydrate receptors having modules homologous to the Siglec and Unique subdomains of GspBBR. This suggests that the binding specificity of the SRR family of adhesins is determined by the type and organization of discrete modules within the binding domains, which may affect the tropism of organisms for different tissues.
Introduction
The serine-rich repeat (SRR) glycoproteins of Gram-positive bacteria are an expanding family of microbial adhesins and virulence factors [1]–[6]. These proteins consist of a distinctive signal sequence and export-targeting region at the N-terminus, a short SRR region (∼50–170 amino acids), a ligand binding region, a second lengthy SRR region (∼400–4000 amino acids), and a cell wall anchoring motif at the C-terminus (Fig. 1) [1], [2], [4], [6]–[9]. The binding regions of the SRR glycoproteins contain significant sequence variation, which appears to account for their broad range of binding targets, including platelet membrane and salivary glycoproteins [1], [4], [9]–[11], endothelial cells [12], epithelial cells [13], erythrocytes [14], [15], and keratins [16], [17]. Expression of SRR proteins has been associated with increased virulence in several animal models of infection, including endocarditis [6], [18], meningitis [12], pneumonia [5], and bacteremia [7], [19].
Fig. 1. Schematic of the SRR adhesins. 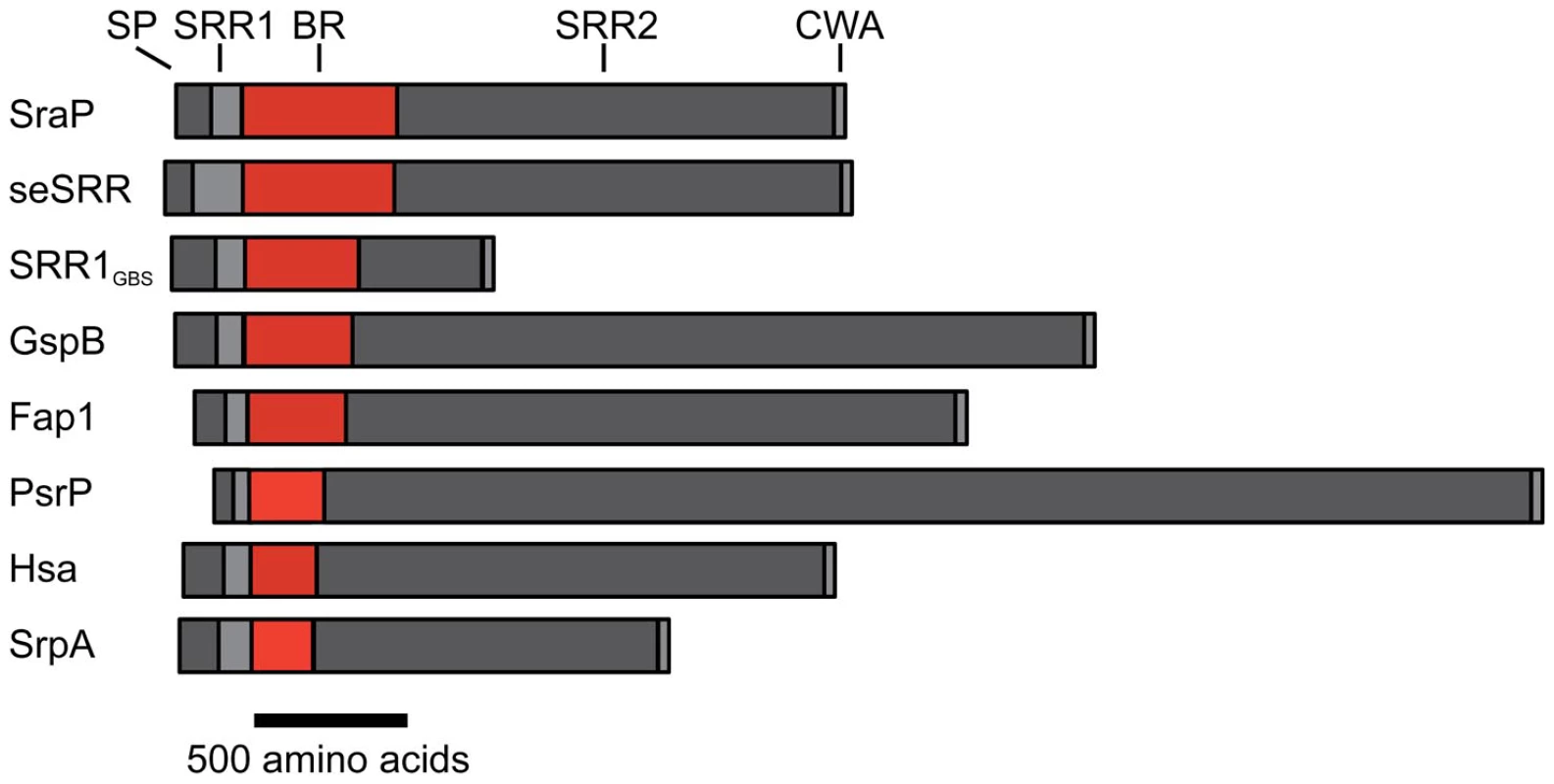
Selected SRR adhesins are aligned based on the N-terminus of their binding regions, with the largest binding region at the top and the smallest binding region at the bottom. The length of each domain and the entire protein is drawn to-scale. SP = signal peptide (dark gray), SRR1 = first serine rich repeat (light gray), BR = binding region (red), SRR2 = second serine rich repeat (dark gray), CWA = cell wall anchoring motif (light gray). The SRR adhesins used in this figure are S. aureus SraP (SraP); S. epidermidis SRR1 (seSRR), S. agalactiae SRR1 (SRR1GBS), S. gordonii strain M99 GspB (GspB), S. parasanguinis Fap1 (Fap1), S. pneumoniae PsrP (PsrP), S. gordonii strain Challis Hsa (Hsa), and S. sanguinis SrpA (SrpA). A number of bacterial surface components have been shown to mediate platelet binding in vitro, either by interacting directly with receptors on the platelet membrane, or via bridging molecules, such as fibrinogen [20]–[29]. The contribution of these interactions to virulence, however, has been assessed for relatively few of these adhesins. Previous studies have focused on the molecular basis for the SRR adhesin mediated binding of gram-positive bacteria to human platelets, and the role of this process in the pathogenesis of infective endocarditis. This interaction appears to be important for the attachment of blood-borne bacteria to platelets on the surface of damaged cardiac valves, thereby initiating infection. In addition, the subsequent deposition of platelets onto the infected endocardium may be due in part to bacterium-platelet binding, resulting in the formation macroscopic vegetations, which are the hallmark lesions of this disease [30]. Three of the SRR proteins (GspB of Streptococcus gordonii strain M99, Hsa of S. gordonii strain Challis, and SrpA of Streptococcus sanguinis strain SK36) bind human platelets through their interaction with glycocalicin, which is the carbohydrate-rich extracellular portion of the platelet membrane glycoprotein GPIbα [9], [11]. While the specific carbohydrate receptor for SrpA has not yet been identified, GspB and Hsa recognize sialylated trisaccharides [1], [11], [31], [32]. Dot blot assays using immobilized carbohydrates have demonstrated that GspB has high fidelity for sialyl-T antigen (i.e. NeuAcα(2–3)Galβ(1–3)GalNAc), one of the major carbohydrates on GPIbα [11], while Hsa binds to glycocalicin via either sialyl-T antigen or sialyllactose (Neu5Acα(2–6)Galβ(1–4)Glc) [31], [32]. Binding of these SRR adhesins to platelets is a high affinity process, with the interaction between GspB and glycocalicin having a KD of 2.38×10−8 M [11] and appears to be a major factor in the pathogenesis of infective endocarditis, since the loss of GspB or Hsa expression results in a marked reduction in virulence [33].
Structural information can enhance the understanding of the determinants of binding specificity. Here, we report the high-resolution crystal structure of GspBBR, both alone and in complex with the disaccharide α-2,3-sialyl (1-thioethyl)galactose, a precursor to synthetically-produced sialyl-T antigen. From these structures, we identified that a subdomain of GspBBR resembling mammalian sialic acid binding proteins binds to α-2,3-sialyl (1-thioethyl)galactose. Site directed mutagenesis and in vivo studies in a rat model of infective endocarditis verified that this carbohydrate binding site within GspBBR mediates binding of S. gordonii strain M99 to sialyl-T antigen, the host carbohydrates of the platelet membrane receptor GPIbα, and intact platelets, and that this interaction is important for virulence. Our analysis of the structure further identified that GspBBR contains an unusually modular fold, prompting us to re-analyze the sequences of the SRR family of adhesins. We determined that other structurally uncharacterized SRR adhesins also contain modules within their binding regions, suggesting that particular subdomains may be included, removed, or interchanged, manifesting in the broad range of binding partners observed in the family.
Results
Overall structure of GspBBR
We determined the structure of GspBBR to 1.4 Å resolution using the method of Multiwavelength Anomalous Dispersion from a single Dy3+ derivative (Fig. 2, Table 1, Table 2). GspBBR folds into an elongated rod, with dimensions of ∼130 Å×30 Å×30 Å. This rod is comprised of three apparently independently-folded subdomains arranged in a linear fashion, like beads on a string. The secondary structure of each of the three subdomains is predominated by β-strands. Interestingly, the first two subdomains are organized around core folds that resemble those found within the eukaryotic immunoglobulin (Ig) superfamily (Fig. 3). Ig-folds have previously been identified in prokaryotic proteins [34]–[41], and it has been noted that some of these bacterial proteins with Ig-folding topologies contain sequence similarity to their eukaryotic counterparts, while the others lack the residues conserved in the core of eukaryotic proteins with Ig-folds. GspBBR has homology with bacterial proteins that do not contain detectable sequence similarity to eukaryotic proteins with Ig-folds, lacking even the cysteines that normally form a disulfide bond.
Fig. 2. Structure of GspBBR. 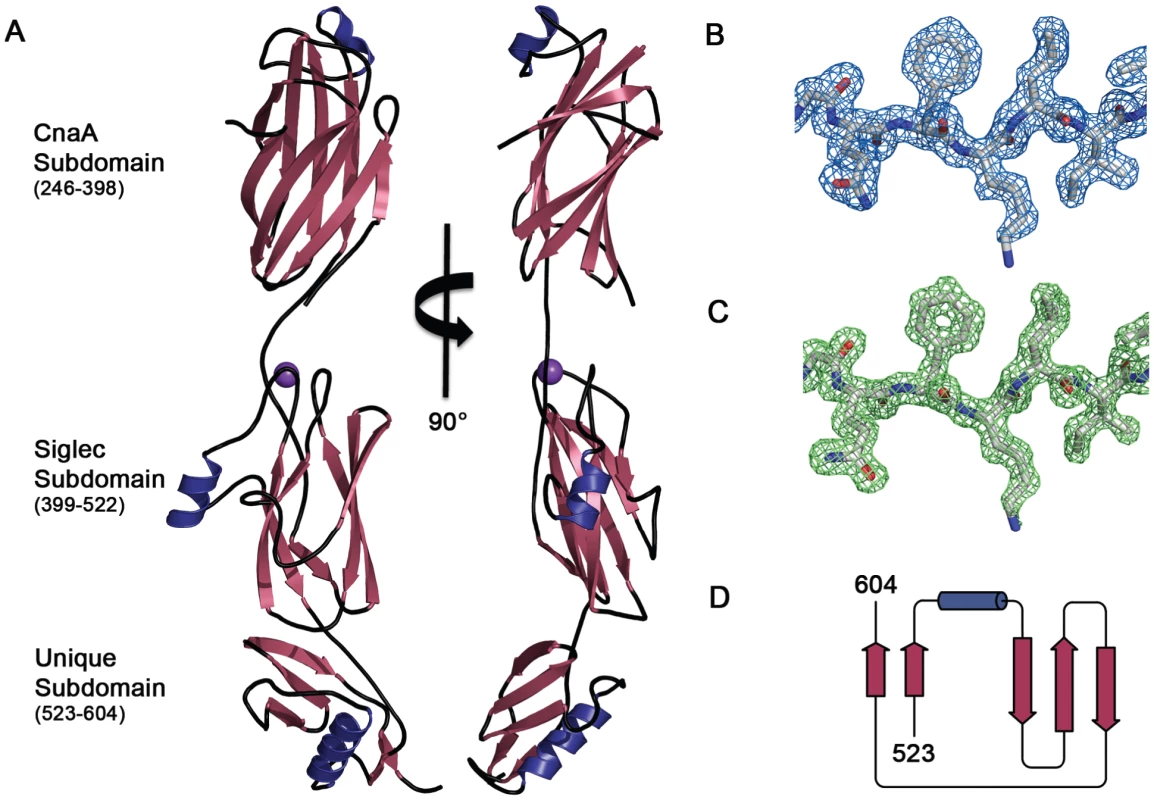
A. Two views of the structure of GspBBR separated by a 90° rotation. The N-terminus of the binding region is located at the top of the figure and the C-terminus is located at the bottom. β-Strands are colored magenta, α-helices are colored blue, and loops are colored black. A cation bound within the Siglec domain is shown as a purple sphere. B. Representative experimental electron density (blue mesh) at 2.0 Å resolution, contoured at 1.0 σ, and depicted superpositioned onto residues 515–520 of the final model. C. A m|Fo|−d|Fc| omit electron density map at 1.4 Å resolution (green mesh) calculated in REFMAC5 [65] after the removal of the model contoured at 3.0 σ and depicted superpositioned onto residues 515–520 of the final model. D. Folding diagram of the Unique subdomain. β-Strands are colored magenta and the α-helix is colored blue. Fig. 3. Ig-fold topologies. 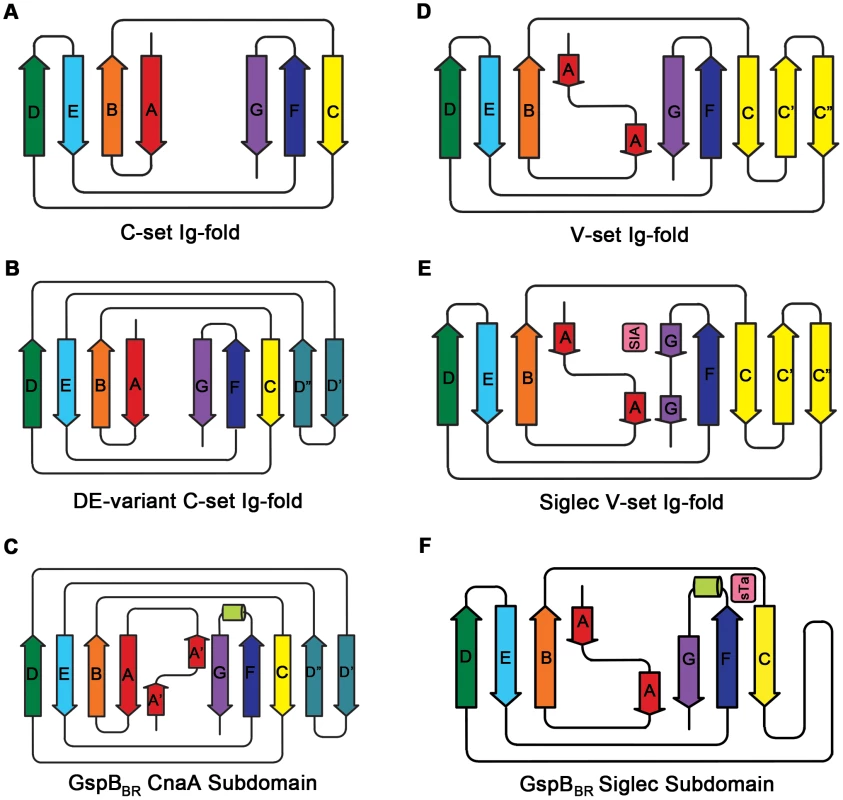
A. Topology diagram of a C-set Ig-fold. B. Topology diagram of the DE-variant of a C-set Ig-fold identified in MSCRAMMs [36]. C. Topology diagram of the GspBBR CnaA subdomain. D. Topology diagram of a V-set Ig-fold. E. Topology diagram of a Siglec [45] showing the binding location of the sialylated carbohydrate in pink. F. Topology diagram of the GspBBR Siglec subdomain showing the binding location of the sialyl-T antigen in pink. Tab. 1. X-ray data collection and refinement statistics. 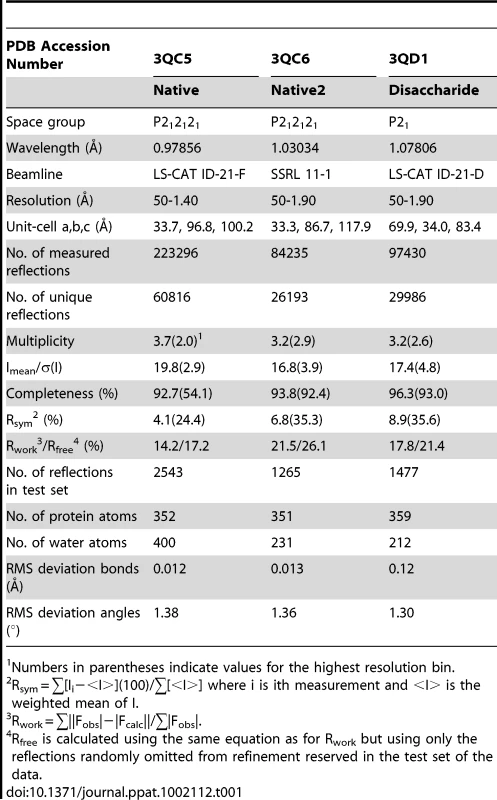
Numbers in parentheses indicate values for the highest resolution bin. Tab. 2. Derivative data collection and phasing statistics. 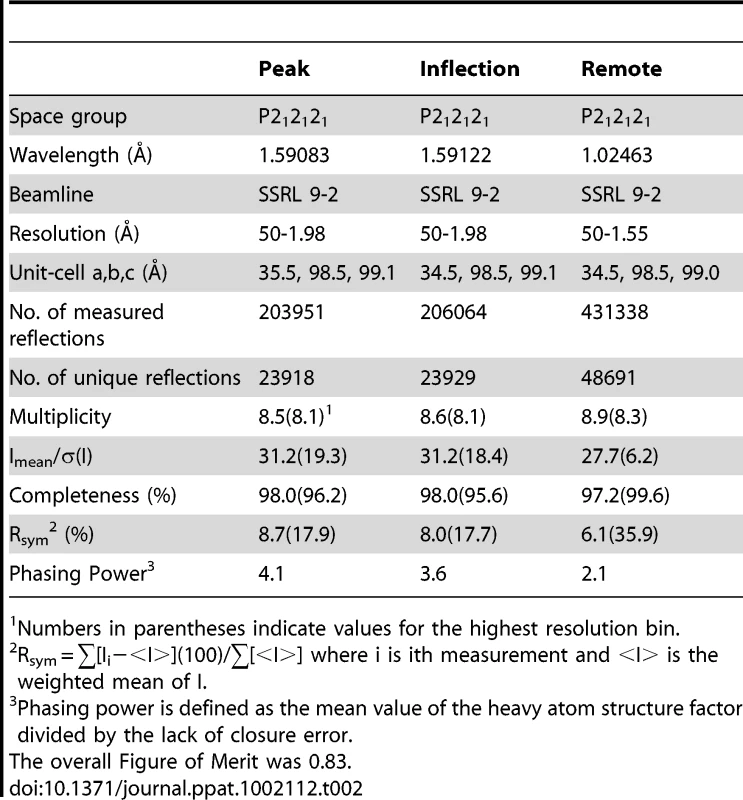
Numbers in parentheses indicate values for the highest resolution bin. Several topology variants of Ig-folds have been characterized. A structural homology search using the EMBL DaliLite server [42] identified that the N-terminal subdomain of GspBBR contains a folding topology, strand inserts, and inter-sheet angle reminiscent of the DE-variant of the Ig-fold [36] (Fig. 3A–C). This Ig-fold topology is found in prokaryotic proteins and was first identified within the A-region of the Staphylococcus aureus CNA protein [35]. CNA belongs to the family of microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) of Gram-positive pathogens [34]–[36]. Given its structural similarity, this N-terminal subdomain of GspBBR will be termed the CnaA subdomain. The highest structural similarity between the CnaA subdomain of GspBBR and any other structurally characterized protein is to the C-terminal subdomain of the binding region of SRR adhesin Fap1 from Streptococcus parasanguinis (Fap1NR-β) [43]. The RMS deviation of the structural alignment between the CnaA subdomain of GspBBR and the Fap1NR-β subdomain is 2.5 Å for 200 Cα atoms.
Surprisingly, a structural homology search using EMBL DaliLite [42] identified that the second subdomain contained a topology and strand inserts reminiscent of the V-set Ig fold adopted by eukaryotic sialic acid binding immunoglobulin-like lectins [44]–[46] (Siglecs; Fig. 3D–F). However, the C′ and C″ strands normally inserted into the V-set Ig-fold are instead replaced by a long loop inserted at the same location (Fig. 3D–F). The RMS deviation of the structural alignment between the second subdomain of GspBBR and Siglec-5 is 2.9 Å for 210 Cα atoms despite only 6% sequence identity. Like GspB, Siglecs bind carbohydrate receptors. Accordingly, the second domain is termed the Siglec subdomain. This subdomain of GspBBR contained electron density consistent with a cation-binding site (see Supporting Text S1, Fig. S1, Supporting Protocol S1). This was rather unexpected since Siglecs themselves do not bind cations in the carbohydrate-binding domain. The seven-coordinate number suggests that under physiological conditions, this should be a Ca2+ binding site. We explored the role of Ca2+ and other cations for the binding of S. gordonii strain M99 to glycocalicin. However, metal depletion, metal substitution, and site directed mutagenesis of the residues coordinating the ion were all consistent with the cation not being essential for carbohydrate receptor recognition (data not shown).
While still predominated by β-strands, the C-terminal subdomain of GspBBR does not contain an Ig-fold. In fact, a structural homology search using DaliLite [42] did not identify any proteins of known structure with significant similarity to this C-terminal subdomain (Fig. 2D). As a result, it will be termed the Unique subdomain.
Identification of the receptor binding site
We sought to identify the details of the interaction between GspBBR and the host receptor sialyl-T antigen. Free sialyl-T antigen is a rare reagent that is not commercially available and is challenging to synthesize. Therefore, we developed a 4-step synthesis for α-2,3-sialyl (1-thioethyl)galactose (NeuAcα(2–3)(1-CH3CH2S)Galβ) (see Supporting Protocol S1; Fig. S2) which is the disaccharide truncation of and a synthetic precursor to sialyl-T antigen (NeuAcα(2–3)Galβ(1–3)GalNAc). To confirm that α-2,3-sialyl (1-thioethyl)galactose binds to GspBBR, we assessed the ability of this disaccharide to inhibit the binding of S. gordonii to glycocalicin. In the presence of 44 mM α-2,3-sialyl (1-thioethyl)galactose, binding of S. gordonii to glycocalicin was reduced by 90% (Fig. 4A). Given the structural similarity to the native host receptor, this strongly suggests that α-2,3-sialyl (1-thioethyl)galactose competes directly for the sialyl-T antigen binding site.
Fig. 4. Identification of the receptor binding site. 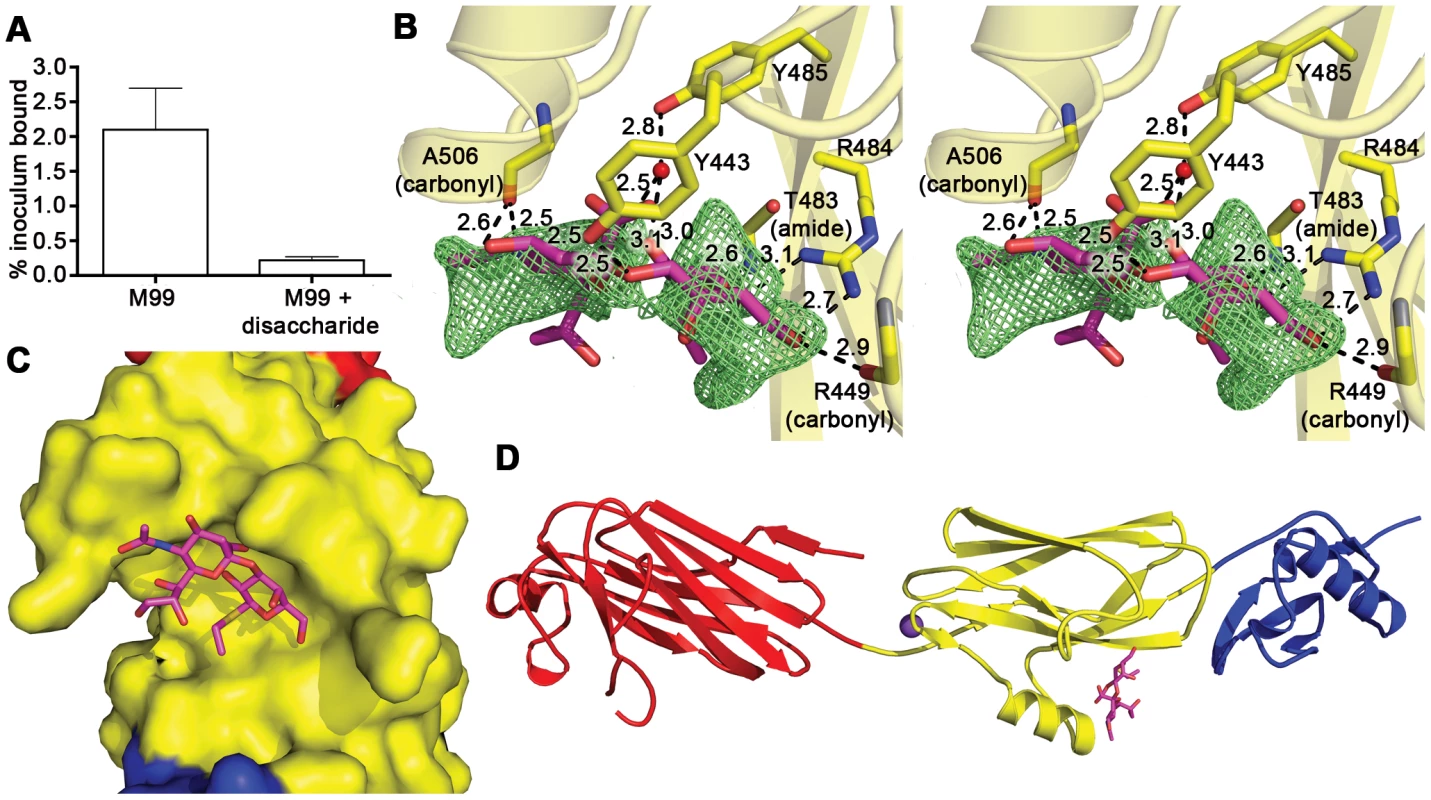
A. Inhibition of S. gordonii strain M99 binding to immobilized glycocalicin by the α-2,3-sialyl (1-thioethyl)galactose disaccharide. Binding was assessed in the absence or presence of 44 mM disaccharide, and is expressed as the percent of input bacteria that remained bound to glycocalicin after repeated washing of the microtiter wells (mean ± standard deviation). B. Stereo view of the disaccharide binding site superpositioned with m|Fo|−d|Fc| omit electron density (green mesh) calculated in REFMAC5 [65] and contoured to 2.5 σ after the removal of disaccharide from the model. C. A space filling model of GspBBR with bound disaccharide (stick model) demonstrates that the carbohydrate receptor binds within a pocket on the surface of the protein. The protein surface is colored by subdomain with the Siglec subdomain in yellow and the Unique subdomain in blue. The stick model of the disaccharide has carbon atoms colored magenta, oxygen atoms colored red, the nitrogen atom colored blue, and the sulfur atom colored yellow. The model is rotated 50° along the y-axis and 40° along the x-axis as compared to panel B. D. Overview of the GspBBR structure highlighting the location of the disaccharide binding site in the Siglec subdomain. The model is rotated 90° along the x-axis as compared to panel B. The disaccharide is shown with magenta bonds and the cation is highlighted as a purple sphere. We determined the co-crystal structure of α-2,3-sialyl (1-thioethyl)galactose in complex with GspBBR. New electron density consistent with bound disaccharide (Fig. 4B) was apparent within a defined pocket (Fig. 4C) located at the N-terminus of the C strand and C-terminus of the F strand in the Siglec domain (Fig. 3F) of the Siglec subdomain (Fig. 4D). While the similarity of the fold of the Siglec subdomain to mammalian Siglecs might predict a similar binding pocket, both the overall location of the α-2,3-sialyl (1-thioethyl)galactose binding site on the domain and the specific contacts between GspBBR to the carbohydrate differ from that of structurally characterized mammalian Siglecs [44]–[46] (Fig. 5). In fact, in GspBBR a helix is positioned in the location where carbohydrates bind to Siglecs, precluding the use of a structurally similar binding site. Instead, the local secondary structure surrounding the α-2,3-sialyl (1-thioethyl)galactose binding site resembles a β-grasp domain, which is a motif that commonly binds the α-2,3-linkage of sialic acid based multivalent carbohydrates [47]–[49]. In GspBBR, the backbone carbonyls of Ala506 and Arg449, the Nη1 and Nη2 of Arg484, the backbone amide nitrogen of Thr483 and the side chain hydroxyl of Tyr443 make direct contacts to the α-2,3-sialyl (1-thioethyl)galactose disaccharide, while the side chain hydroxyl of Tyr485 makes water-mediated contacts (Fig. 4B, Table 3).
Fig. 5. Comparison of carbohydrate binding in the Siglec subdomain of GspBBR and Siglec-5. 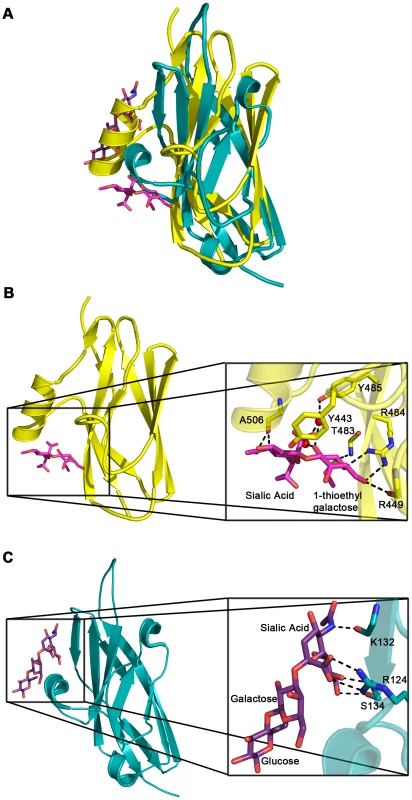
A. Overlay of the Siglec subdomain of GspBBR and Siglec-5. Siglec-5 is colored teal with the carbons of 3′ sialyllactose colored purple. The Siglec subdomain of GspBBR is colored yellow with the carbons of α-2,3-sialyl (1-thioethyl)galactose colored in magenta. In this panel, the Siglec carbohydrate binding site is occupied by a helix in the GspBBR Siglec subdomain therefore eliminating the possibility that this could be a second binding site for GspBBR. B & C. Comparison of the location and details of the carbohydrate binding site in (B) GspBBR with that of (C) Siglec-5 (PDB entry 2ZG3; [45]). The Siglec subdomain of GspBBR is colored yellow with the carbons of α-2,3-sialyl (1-thioethyl)galactose colored in magenta and Siglec-5 is colored teal with the carbons of 3′ sialyllactose colored purple. While Siglec-5 is used as an example, it is noted that for all structurally characterized Siglecs (Siglec-1 (aka sialoadhesin); PDB entry 1QFO; [44]), Siglec-5 (2ZG3; [45]), and Siglec-7 (PDB entry 2HRL; [74])), the sialic acid binding pocket is found at the same location. Tab. 3. Contacts at the receptor binding site. 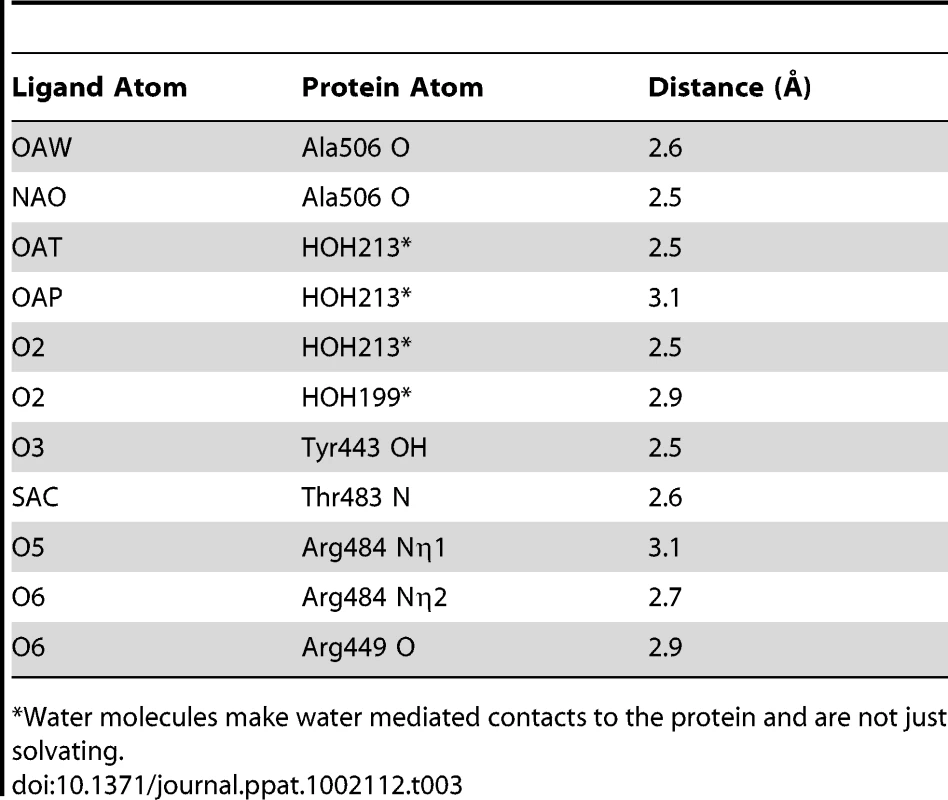
*Water molecules make water mediated contacts to the protein and are not just solvating. A comparison of the structure of GspBBR determined with and without the disaccharide did not show significant structural changes localized within the binding pocket, which suggests that it is pre-formed. Thus, while the physiologically-relevant receptor trisaccharide sialyl-T antigen is a rare reagent, we can qualitatively suggest its binding location by modeling the third carbohydrate onto α-2,3-sialyl (1-thioethyl)galactose. In our model, all three carbohydrates of the host receptor fit into a pre-formed binding pocket on the Siglec subdomain, with the third carbohydrate of sialyl-T antigen extending toward the Unique subdomain (Fig. S3).
While disaccharide binding to GspBBR did not appear to induce obvious changes in conformations of side chains within the binding pocket, the interdomain angle between the CnaA and Siglec subdomains unexpectedly straightened by 40° (Fig. 6; Video S1). The change in orientation between the CnaA and Siglec subdomains involves rotation around a single hinge consisting of residues Lys398-Asp399-Thr400, where the side chain Asp399 is one of the ion coordinating residues.
Fig. 6. Interdomain Angle. 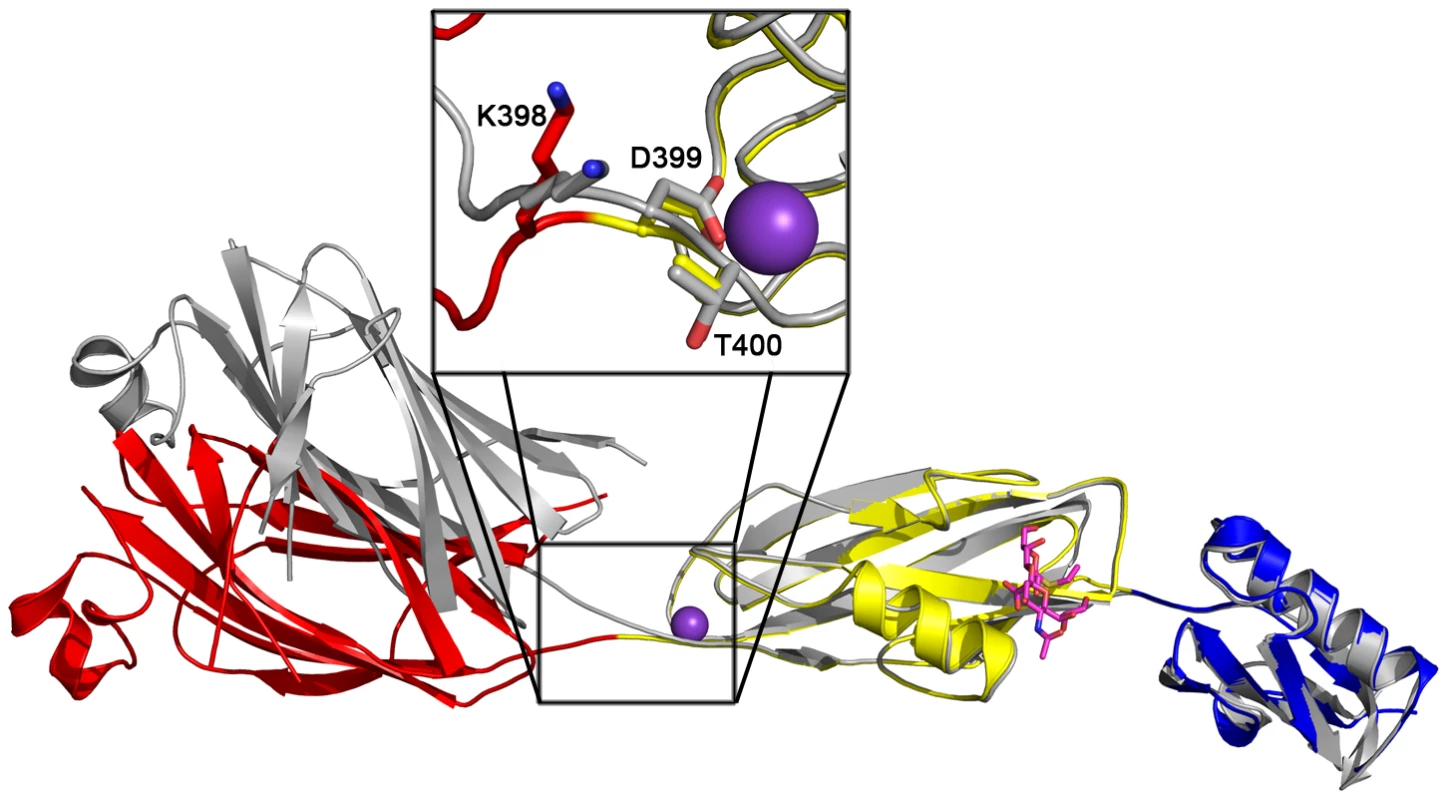
Differences in interdomain angles between structures of GspBBR. The structures of the Siglec and Unique subdomains of unbound and disaccharide-bound GspBBR were aligned using Dyndom [69]. The unbound structure is colored in silver, and the structure in complex with α-2,3-sialyl (1-thioethyl) galactose is colored by subdomain with the CnaA subdomain in red, the Siglec subdomain in yellow and the Unique subdomain colored in blue, and cation colored purple. The side chains of hinge residues Lys398, Asp399, and Thr400 are highlighted in the boxed inset. The difference in interdomain angle between the two structures is a 40°. A movie morphing these states is seen in Video S1. Functional relevance of the carbohydrate binding site
To confirm that this crystallographically-identified α-2,3-sialyl (1-thioethyl)galactose binding site is indeed important for GspB-mediated binding to GPIbα, we generated four isogenic variants of strain M99 with point mutations within the Siglec subdomain of GspB (Table 4). Three of these variants contained mutations within the crystallographically-identified binding site (Y443F, R484E and Y485F), while a control mutation (E401A) was located within the Siglec domain, but away from the receptor binding site. Importantly, none of the point mutations affected surface expression of GspB (Fig. 7A). Each of these isogenic variants had an 86% or greater decrease in binding to GPIbα in vitro, as compared with the parent strain (p<0.001) (Fig. 7B). This decrease is comparable to that of the gspB null mutant. In contrast, binding by the E401A variant was not significantly different from that of M99 (p = 0.44).
Fig. 7. Amino acid substitutions in the carbohydrate binding site of GspBBR. 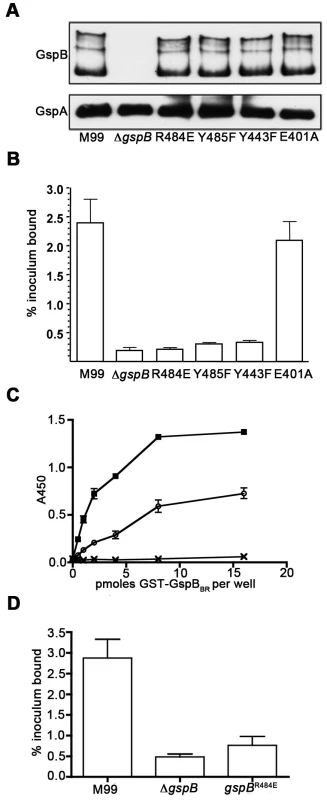
A. Expression of GspB and variant proteins on the S. gordonii cell surface. Peptidoglycan-linked proteins were extracted from the bacterial cell surface and analyzed by western blotting using a polyclonal anti-GspB serum (upper panel). The nitrocellulose membrane was then re-probed using a polyclonal anti-GspA serum (lower panel) in order to assess the relative efficiency of protein extraction and protein loading (GspA is another LPXTG-linked protein that is expressed by S. gordonii M99). B. Binding of S. gordonii strain M99 and derivative strains to glycocalicin immobilized in microtiter wells. Binding is expressed as the percent of input bacteria that remained bound to glycocalicin after repeated washing of the wells (mean ± standard deviation). The labels of each column indicate the sequence of the protein expressed in S. gordonii, with M99 indicating wild type, ΔgspB indicating a strain where the gene encoding GspB has been deleted, and remaining columns indicating the amino acid substitution present in the protein. E401 is a control residue not located near the binding pocket. C. Binding of sialyl-T antigen to purified GST-GspBBR. The indicated amounts of GST or GST-GspBBR wild-type (wt) and variant (R484E) proteins were immobilized in microtiter wells, and the binding of biotinylated sialyl-T antigen to each protein was detected by using peroxidase-conjugated streptavidin along with a chromogenic peroxidase substrate. Binding is expressed as the mean ± standard deviation (n = 3). The o represents purified GST-GspBBR-R484E, the x represents purified GST, and the represents purified GST-GspBBR. D. Binding of M99 and derivative strains to human platelets. Binding is expressed as the percent of input bacteria that remained bound to platelets after repeated washing of the wells (mean ± standard deviation). Tab. 4. Strain names with descriptions. 
We then selected the R484E mutation for more detailed study. The purified GST-GspBBR fusion protein harboring the R484E substitution exhibited a marked decrease in binding to biotinylated sialyl-T antigen (Fig. 7C). The R484E substitution also resulted in reduced binding to human platelets in vitro as assessed by quantifying the amount of input inoculum bound to fixed platelets (Fig. 7D). We next visualized platelets by Differential Interference Contrast (DIC) and fluorescence microscopy and quantified the number of platelets with surface-bound bacteria. In the presence of the DAPI-labeled PS846 (M99ΔgspB) or the PS2116 (M99 gspbR484E) strain, substantially fewer platelets had bacteria bound as compared to platelets in the presence of wild-type S. gordonii strain M99 (Fig. 8, Table 5). It should be noted that while the platelet numbers were normalized when placed onto the cover slip, during the experiment the wild type bacteria seemed to form microscopic aggregates with the platelets (data not shown) but this was not observed when platelets were mixed with strains PS2116 or PS846. This confirms the importance of this residue for carbohydrate binding.
Fig. 8. Binding of wild type S. gordonii strain M99, PS2116 (strain M99 with gspb harboring the R484E substitution), and PS846 (ΔgspB) to platelets. 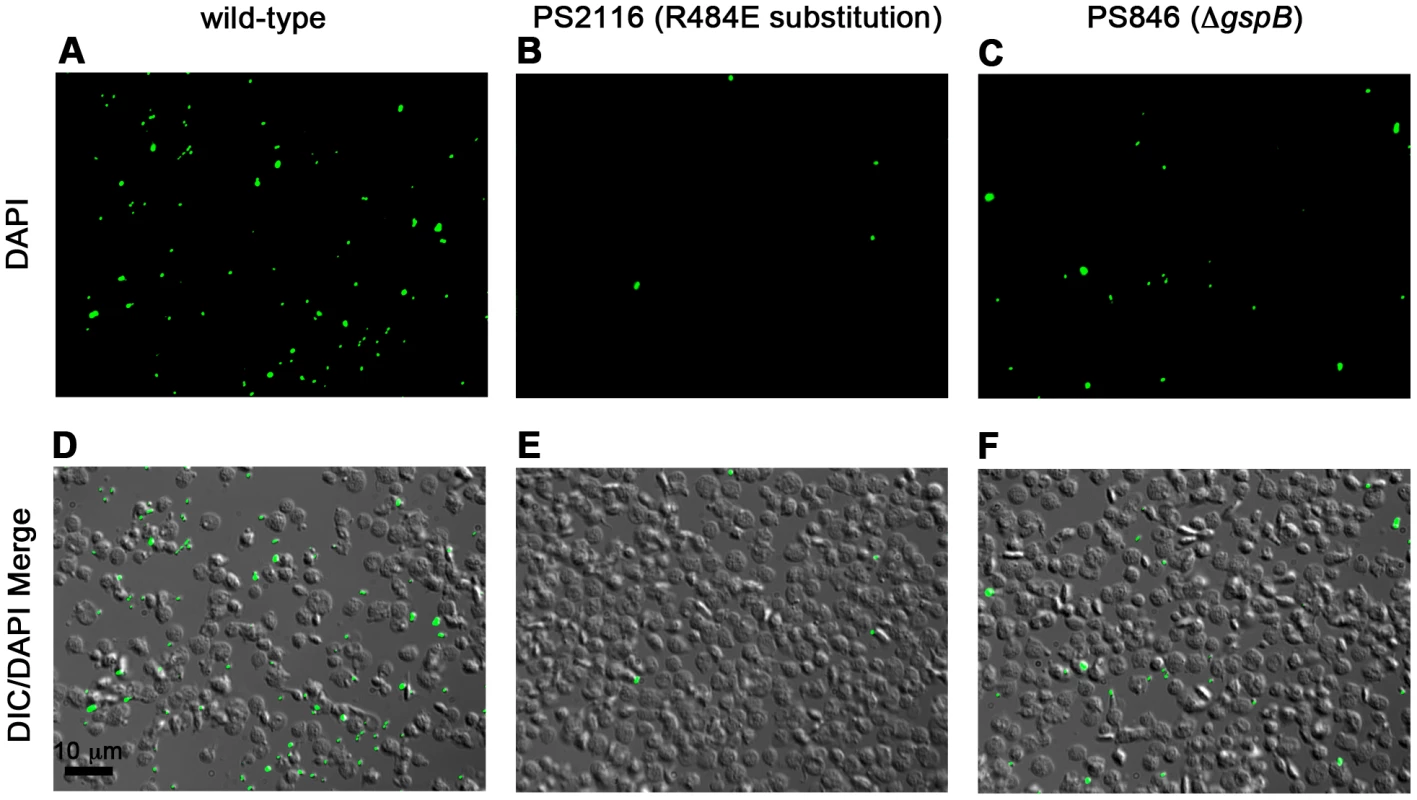
Fixed platelets were adhered to poly-L-lysine coated cover slips, quenched, and bound to live bacteria stained with DAPI. A–C. Fluorescent images of each strain with DAPI stain. D–F. Merge between the DIC image and the DAPI image to show relative number of platelets. Tab. 5. Binding of three variants of S. gordonii to human platelets. 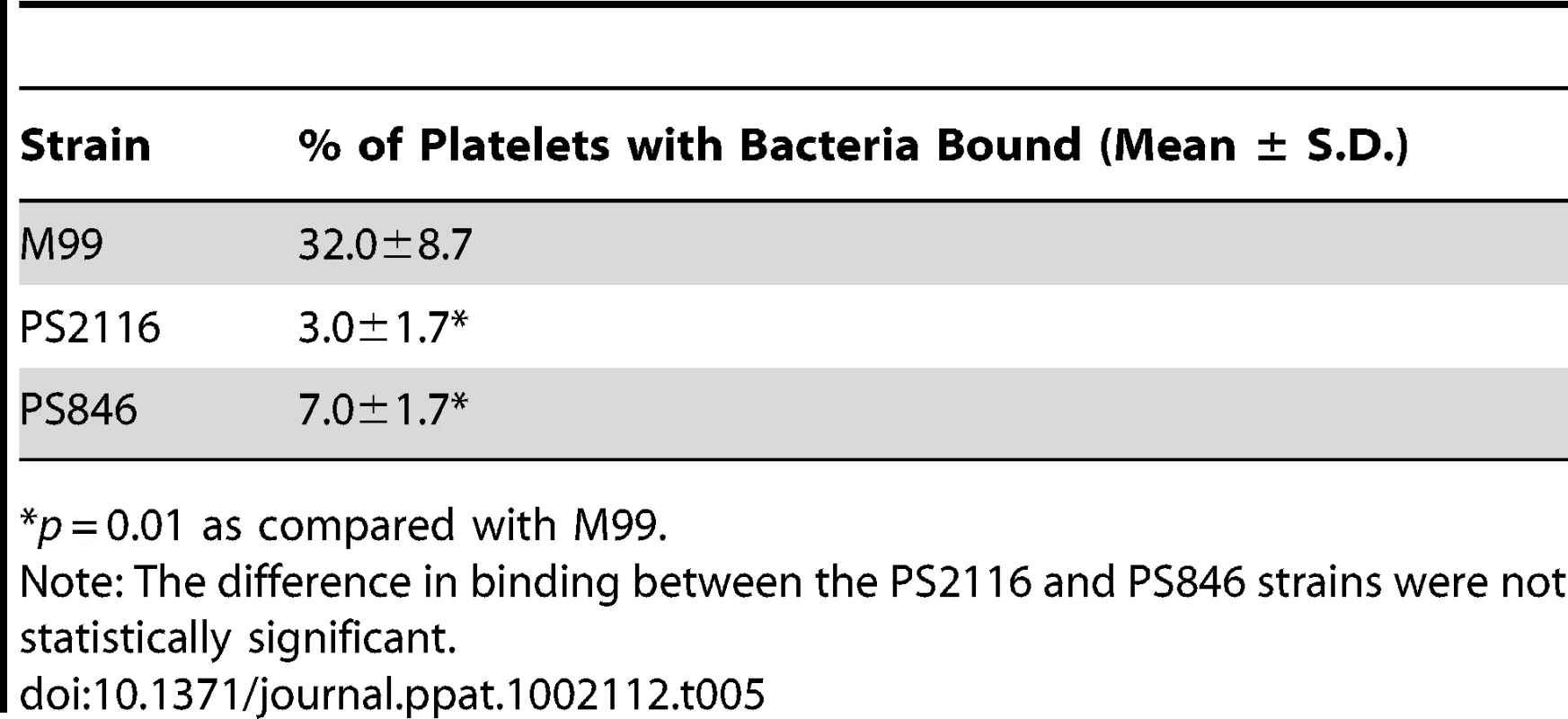
*p = 0.01 as compared with M99. Impact of Siglec-mediated binding on virulence
It has previously been demonstrated that loss of GspB expression results in a marked decrease in virulence, as measured by a rat model of infective endocarditis [18]. To assess whether binding via the Siglec subdomain to host carbohydrate receptors contributes to virulence, we examined the impact of the R484E substitution on the ability of S. gordonii strain M99 to produce endocardial infection. We first compared the relative virulence of M99 with strain PS2116, (M99 gspbR484E). Catheterized rats were simultaneously infected intravenously with 2×10−5 CFU of M99 and PS2116. After 72 h, the animals were sacrificed and the relative levels of bacteria within tissues determined. Animals co-infected with M99 and PS2161 (which carries the spec resistance marker just upstream of gspB) served as controls. When assessed at the above time-point, animals co-infected with M99 and PS2116 had significantly reduced densities of the mutant strain within vegetations (mean ± S.D. = 6.81±1.70 log10 CU/g. veg.) as compared with parental strain M99 (7.47±1.69 log10 CFU/g. veg; P<0.02). Loss of sialyl-T antigen binding was also associated with significantly reduced bacterial densities within kidneys and spleens (P<0.02 and P<0.001, respectively; Table 6). In parallel studies, no differences were observed between the M99 and the control strain (PS2161), as measured by CFU per gram of tissue within vegetations, kidneys, or spleens (data not shown). We also analyzed these findings by calculating a competition index, in which the ratio of M99 and PS2116 within tissues was normalized for the CFU of each strain within the inoculum. When analyzed by this approach, the densities of the GspB mutant strain PS2116 remained significantly reduced in all tissues, as compared with M99 (P<0.002), while the densities of the control strain were statistically similar.
Tab. 6. Effect of Sialyl-T antigen binding on virulence in a rat model of infective endocarditis. 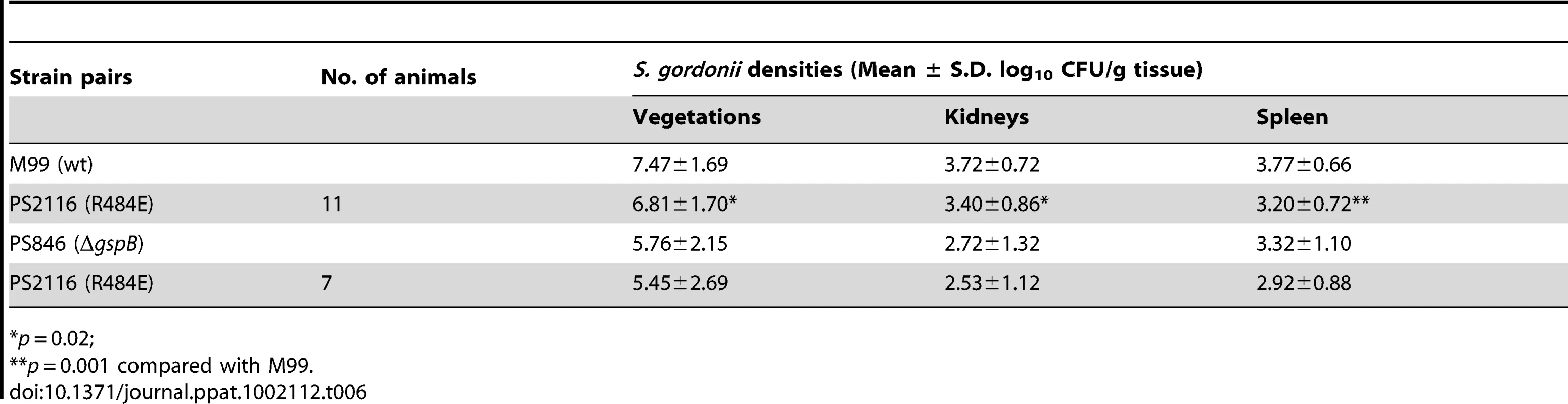
*p = 0.02; We then compared the relative virulence of PS2116 with PS846 (M99ΔgspB). Infective endocarditis was produced as above, using an inoculum containing the two strains in a 1∶1 ratio. When assessed at 72 h post-infection, PS2116 and PS846 had similar densities of organisms within all tissues (Table 6). Of note, the levels of both strains with vegetations were markedly lower than those achieved by wild-type M99 in the above studies. Thus, these two strains appeared comparably attenuated in the setting of endocarditis, indicating that platelet binding by the Siglec domain may be the predominant GspB interaction contributing to virulence in endocarditis.
The binding regions of SRR adhesins have a modular organization
While overall sequence analysis of members of the SRR family has identified unifying sequence trends (Fig. 1), sequence comparisons of the binding regions have shown little homology. A structural comparison between the three distinct subdomains of GspBBR (Fig. 2A) and the two distinct subdomains of the SRR adhesin Fap1 [43] immediately indicates that these family members adopt unrelated folds in their binding domains. Like GspBBR, Fap1NR appears to be composed of independently-folded subdomains; however its binding region only contains two modules whereas the binding region of GspBBR contains three. Intriguingly, Fap1NR contains a helical subdomain at its N-terminus (Fap1NR-α) that does not resemble any of the subdomains of GspBBR, and a CnaA subdomain at its C-terminus (Fap1NR-β) that resembles the N-terminal CnaA subdomain of GspBBR (GspBBR-C). This suggested to us that members of the SRR family could undergo reorganization of the modules within their host binding regions, with particular modules or combinations of modules conferring specific properties.
Accordingly, we re-analyzed the binding regions of selected members of the family using a new strategy, where we used BLAST [50] and ClustalW [51] to query the sequences of the binding regions of SRR adhesins with input sequences corresponding to subdomains of either GspBBR or Fap1NR, or of short regions (∼200 amino acids) of sequences of structurally uncharacterized binding regions of SRR adhesins. These modified sequence comparisons strongly suggest that the binding regions of members of the SRR family have indeed evolved to contain modules (Fig. 9), and show several distinct groupings. For example, all three assessed SRR adhesins with carbohydrate binding partners contain the Siglec and Unique subdomains in tandem, strongly suggesting that the inclusion of these modules within the binding region confers lectin-like binding characteristics.
Fig. 9. Modular organization of the binding regions within adhesins of the SRR superfamily. 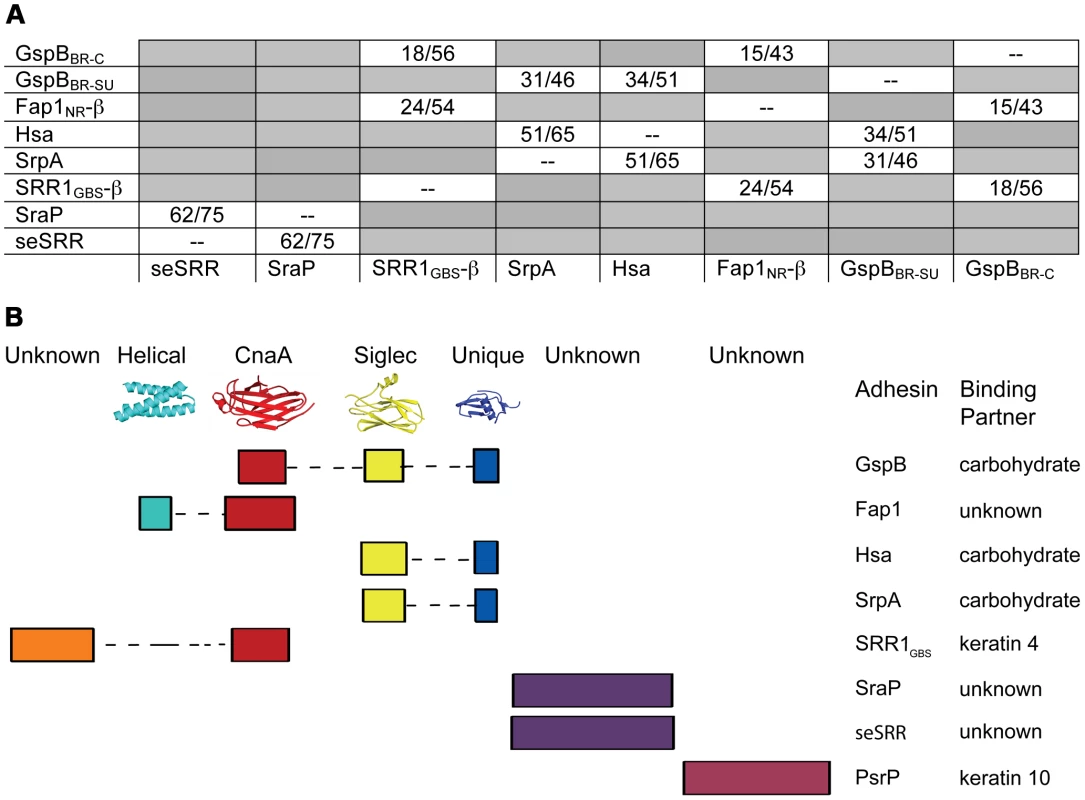
A. A summary of BLAST [50] or ClustalW [51] sequence alignments of the binding regions of SRR adhesins. The first number indicates sequence identity and the second indicates sequence similarity. GspBBR has been divided into its subdomains, with GspBBR-C indicating the CnaA subdomain and GspBBR-SU indicating both the Siglec and Unique subdomains. Only the C-terminal subdomain of SRR1GBS (SRR1GBS-β) and Fap1NR (Fap1NR-β) are used in this analysis since the N-terminal domains do not share detectable sequence identity with other sequences used in this analysis. Boxes in grey indicate SRR pairs that do not have detectable sequence identity within the binding region. B. A schematic summarizing (A) The colored rectangles represent similar regions of sequence, and are drawn to scale. If known, representative structures of each domain are illustrated along the top, and binding partners are indicated along the side. These focused sequence alignments additionally identify that a CnaA subdomain may be common in SRR adhesins, appearing in three of the eight binding regions that we assessed. As opposed to the combination of the Siglec and Unique subdomains, which are associated with carbohydrate binding, each adhesin containing a CnaA subdomain may have a different binding specificity (Fig. 9). Interestingly, in the SRR adhesins that were assessed, the CnaA subdomains were always found paired with sequence for another subdomain.
It is important to note that several of the binding regions of the SRR adhesins do not have significant sequence identity to any protein of known structure. For example, the binding region of S. agalactiae SRR1 (SRR1GBS) likely contains two subdomains, SRR1GBS-α at the N-terminus, which does not contain detectable sequence similarity to any protein of known fold, and SRR1GBS-β at the C-terminus, which has sequence similarity to MSCRAMMs and likely adopts a similar folding topology to CnaA. Likewise, no part of the binding regions of S. pneumoniae PsrP, S. aureus SraP, or S. epidermidis seSRRBR exhibits detectable sequence similarity to any known protein, but the latter two have high sequence similarity to each other.
Discussion
SRR adhesins have been identified in a variety of Gram-positive pathogens, and have been implicated as virulence factors in a wide spectrum of infections [5], [6], [12], [18]. The diversity of these infections (e.g., endocarditis, meningitis, pneumonia) and the broad scope of their anatomic locations are consistent with the binding regions of the SRR proteins differing considerably in their selectivity. Indeed, although only a few ligands for this family of proteins have been identified, they range from carbohydrates (such as sialyl-T antigen) [32] to proteins (such as keratins) [16], [17]. To date, however, the structural basis for this selectivity has been unknown.
In several members of the family (e.g. the SRR adhesins of S. gordonii and S. pneumoniae) the domain between the two SRR regions mediates binding. However, for many of these SRR adhesins (i.e. SraP of S. aureus), the host receptor has yet to be identified [6], [43], and the delineation of the binding region is assumed based upon sequence comparisons within the family. Among the best characterized of the SRR adhesins is GspB from S. gordonii, which has demonstrated binding affinity for the sialyl-T antigen carbohydrate decorating platelet glycoprotein GPIbα. Our crystal structure of the binding region, GspBBR, identified a modular organization with three subdomains (Fig. 2A), two of which are organized around Ig-folds. Proteins containing Ig-folds are commonly found within the mammalian immune system, where they exhibit a variety of functions; however, Ig-folds are not uncommon within pathogens, where they act exclusively as virulence factors. The first characterized bacterial protein to contain an Ig-fold was PapD, a chaperone for the assembly of pili in E. coli [37]. The Ig-fold has been observed in several other types of virulence factors, including components of pili (carbohydrate-binding fimbriae) [41], [52], and in adhesins of varying specificity for host receptors, including MSCRAMMs [34]–[36] and invasins [38]–[40].
Co-crystallization of GspBBR with α-2,3-sialyl (1-thioethyl)galactose identified a specific binding site for this disaccharide within the Siglec subdomain (Fig. 4B–D), and the introduction of single point mutations within this region significantly reduced levels of binding of S. gordonii to glycocalicin. Moreover, the R484E substitution in GspB reduced platelet binding by M99 (Fig. 7D, Fig. 8, Table 5), and had a marked reduction in binding of the GST-GspBBR fusion protein to sialyl-T antigen (Fig. 7C), confirming the importance of the Siglec subdomain for carbohydrate binding. These results indicate that this binding pocket within the Siglec subdomain is required for binding to sialyl-T antigen, and that the interaction of this GspB subdomain with sialyl-T antigen is important for binding to platelets.
When examined in a co-infection model of infective endocarditis, the isogenic variant of M99 expressing GspB-R484E (strain PS2116) was also significantly reduced in virulence (most notably a 78% reduction in bacterial levels within vegetations as compared with the parent strain). When PS2116 was tested with PS846 (M99ΔgspB) in this model, the strains had comparable but reduced densities within target tissues (as compared with WT M99), indicating that they were similarly reduced in virulence. These findings indicate that the predominant property of GspB contributing to the pathogenesis of endocarditis is its interaction with sialyl-T antigen. In previous in vivo studies, co-infection with M99 and PS846 yielded more pronounced differences between the strains, as measured by densities of organisms within target tissues [18]. This may reflect subtle experimental differences, such as inoculum size, but could also be due to residual binding activity of the mutated GspB. Of note, PS2116 had low but detectable levels of platelet binding in vitro. Similarly, the GST-GspBBR-R484E had measurable levels of binding to sialyl-T antigen, as compared to GST alone. This residual binding seen in vitro, presumably due to the other key residues identified by crystallography, may account for the residual virulence of the PS2116 strain. Alternatively, although ligands for GspB other than sialyl-T antigen have not been identified, it is conceivable that GspB has other interactions in vivo that contribute to virulence in this setting, beyond those mediating platelet binding.
Upon binding of the α-2,3-sialyl (1-thioethyl)galactose to GspBBR, a 40° interdomain angle straightening is observed between the CnaA and Siglec subdomains (Fig. 6, Video S1). The interdomain hinge is centered at the ion binding site, making the interdomain angle change reminiscent of calcium dependent interdomain angle changes observed in C-cadherin, which also contains modules of Ig-fold topology [53]. There are several possibilities for the origin and role of this interdomain reorganization in GspBBR. It is first possible that GspBBR exhibits natural flexibility between these two domains, and that the difference in crystal contacts artifactually resulted in the straighter form of the protein being trapped in the co-structure of GspBBR with α-2,3-sialyl (1-thioethyl)galactose. Intriguingly, however, an interdomain straightening is also observed in small angle x-ray scattering (SAXS) of the binding region of SRR protein Fap1NR upon lowering the pH to 5.0, where Fap1NR has highest affinity for its host receptor [43]. Those experiments, performed in solution, are not limited by crystal contacts and suggest that interdomain straightening upon ligand binding could be a conserved feature of members of the SRR family. Physiological roles of interdomain straightening upon ligand binding include improved hydrodynamics in the cardiovascular system or oral cavity. The elongation upon ligand binding is perhaps more compellingly reminiscent of the interdomain straightening that occurs upon the binding of P-selectin and β3-integrin to their respective ligands [54]. These proteins have been suggested to form “catch bonds” in order to have increased affinity to their ligand upon the application of tensile force. Each of these possibilities for the observed straightening of GspBBR upon ligand binding is currently under investigation.
While both Siglecs and GspBBR bind to carbohydrate receptors, the structural details of disaccharide binding to this novel bacterial Siglec bear little resemblance to the binding of sialic acid moieties to characterized mammalian Siglecs [44]–[46] (Fig. 5). Indeed, comparison of the binding sites reveals that the secondary structure of GspBBR has a helix that is found at the same location where 3′ sialyllactose binds to Siglec-5 (Fig. 5A), thus eliminating the possibility of analogous carbohydrate binding. Instead, GspBBR appears to use a β-grasp domain to form the specific host receptor binding site, a domain found in another group of sialic acid binding proteins, staphylococcal superantigen-like proteins (SSLs) that forms a V-like cleft for carbohydrate binding [47]–[49]. Nevertheless, both mammalian Siglecs and GspBBR have binding pockets predominated by tyrosines and arginines. With a closer look at each binding site, in Siglecs, the sialic acid moiety of the binding partner makes a salt bridge with an arginine residue. This salt bridge is not present in the binding site of GspBBR despite the presence of R484 and its key role in binding; this arginine instead binds to the C6 hydroxyl and the pyranose oxygen of the 1-thioethylgalactose (Table 3).
Importantly, the structure of GspBBR provides insight into the binding repertoire of other SRR proteins, and specifically highlights which modules mediate binding to host carbohydrate receptors. Homologues of GspB containing regions with high sequence similarity to the Siglec and Unique subdomains (such as S. gordonii strain Challis Hsa and S. sanguinis SrpA) are also demonstrated lectins. Supporting this sequence analysis, our co-crystal structure of GspBBR with α-2,3-sialyl (1-thioethyl)galactose demonstrates that the first two carbohydrates of host receptor bind within a pre-formed pocket on the Siglec subdomain. Using this experimental co-structure as a starting point, we could model binding of the sialyl-T antigen. This model is consistent with the third carbohydrate of this trisaccharide binding to an extended lobe on this pocket. This region of the binding pocket is still within the Siglec subdomain but extends toward the Unique subdomain (Fig. S3).
The structures of both GspBBR and Fap1NR identified a CnaA-like subdomain within the binding region, and our sequence analysis (Fig. 9) additionally predicts a CnaA subdomain at the C-terminus of the binding region of S. agalactiae SRR1 (SRR1GBS-β), an adhesin for keratin 4 [16]. As opposed to the Siglec subdomain, which contains an identifiable receptor binding site, the precise function of the CnaA subdomain is less clear. MSCRAMMs, such as CnaA from S. aureus, recognize peptides through the formation of a binding site between two Ig-like domains (Fig. S4). The bound peptide forms an additional strand that becomes a part of the Ig-fold. In contrast, GspBBR, Fap1NR, and SRR1GBS each apparently contain a single CnaA subdomain amongst the modules in the binding region (Fig. 9). Our modeling suggests that binding of a peptide between two domains in a manner analogous to binding of a peptide to other MSCRAMMs is not feasible. Indeed, an additional strand found in the subdomain of GspBBR occupies the peptide binding site of MSCRAMMs. (Fig. S4, Fig. 3C strand A′). Nevertheless, in SRR adhesins, the CnaA subdomain is always found together with at least one other subdomain, suggesting that the function may either require or be tuned by the presence of a second subdomain. Indeed, recent studies on the binding region of Fap1 support this hypothesis. The structures of Fap1NR-α (helical subdomain) and Fap1NR-β (CnaA subdomain) identified a region of surface-exposed hydrophobic residues on each domain that is predicted to be contiguous based upon SAXS. Fap1 normally mediates binding of this bacterium to the oral cavity, and mutagenesis of these hydrophobic residues abrogated binding to an in vitro tooth model consisting of saliva-coated hydroxylapatite [43].
Our sequence analysis additionally predicts that the binding regions of other SRR adhesins contain modules that are structurally distinct from those identified in either GspBBR or Fap1NR. Two of these, S. aureus SraPBR and S. epidermidis seSRRBR, have high sequence identity to each other, but no detectable sequence similarity to any other SRR adhesin (Fig. 9). This strongly suggests that they bind a common, as yet unidentified, host receptor that is distinct from the carbohydrates and keratins recognized by other SRR adhesin family members. By comparison, neither S. pneumoniae PsrPBR, which has been demonstrated to bind keratin 10, nor the N-terminal subdomain of S. agalactiae SRR1BR, which binds keratin 4, have detectable sequence similarity to any currently available sequence (Fig. 9). Given this analysis, it is clear that a more detailed understanding of the binding characteristics of the SRR adhesin family will require that for each module type, the binding partner should be identified, and the structure of the binding region should be determined both alone and in complex with the appropriate host receptor.
Materials and Methods
Ethics statement
All procedures involving rats were approved by the Los Angeles Biomedical Research Institute animal use and care committee, following the National Institutes of Health guidelines for animal housing and care. Platelets were collected using a protocol approved by the UCSF Committee on Human Research (H1193-25513-07) and by the VUMC Human Research Protection Program IRB Committee (110364).
Protein expression and purification
The DNA encoding residues 233–617 of GspB (GspBBR) was cloned into the pGEX vector containing an N-terminal glutathione S-transferase (GST) fusion tag as described [11]. This clone contains a single base pair change as compared to the deposited NCBI sequence that results in a serine at position 444 instead of an asparagine. The protein was expressed with E. coli BL21 Gold (Stratagene) in Luria Broth Medium as described [55] and purified using a GST affinity column (GE Healthcare). The GST tag was removed from GspBBR using Factor Xa, and GspBBR was further purified using size exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare) with buffer containing 20 mM Tris pH 7.4 [55].
Crystallization
All crystals of GspBBR were grown using the hanging drop vapor diffusion technique at 23°C [55] using 1 µl protein solution and 1 µl of reservoir solution equilibrated against 1 ml of the reservoir solution. Crystals of native GspBBR grew from two chemically distinct sets of conditions. The first set of conditions included GspBBR at a concentration of 6 mg/ml buffered in 20 mM Tris pH 7.4 and equilibrated against a reservoir solution containing 33% Jeffamine ED-2001, 0.1 M HEPES pH 7.5 and 0.15 M KCl at 23°C. These crystals belonged to the primitive orthorhombic space group P212121 with unit cell dimensions a = 33.7 Å, b = 96.8 Å, c = 100.2 Å with α = β = γ = 90°. Prior to data collection, crystals were cryo-protected in a solution containing all of the chemical components of each reservoir solution and 15% glycerol then flash-cooled in liquid nitrogen. The dataset used for refinement merged to 1.4 Å resolution.
The second type of native GspBBR crystals grew when the protein was equilibrated against a reservoir solution containing 25% polyethylene glycol 3350, 0.1 M HEPES pH 7.5, 0.15 M NH4CH3COO, and 10 mM spermidine and induced crystal growth from 10 mg/ml GspBBR buffered in 20 mM HEPES pH 7.5 at 18°C. These crystals also belonged to the orthorhombic space group P212121, but had altered unit cell dimensions of a = 33.4 Å, b = 86.7 Å, c = 117.9 Å, α = β = γ = 90°. Prior to data collection, these crystals were flash cooled in liquid nitrogen without additional cryo-protectant. The best dataset from this crystal form merged to 2.0 Å resolution.
Crystals of GspBBR in complex with the α-2,3-sialyl (1-thioethyl)galactose disaccharide were grown using the hanging drop vapor diffusion method with 6 mg/mL GspBBR in buffer containing 1 mM α-2,3-sialyl (1-thioethyl)galactose and 20 mM Tris pH 7.4. The reservoir solution contained 8% PEG 3350, 7.5 mM CoCl2, 7.5 mM NiCl2, 7.5 mM CdCl2, 7.5 mM MgCl2, and 0.1 M HEPES pH 7.5. Crystals grew within two days and were cryo-protected in reservoir solution supplemented with 20% glycerol and flash-cooled in liquid nitrogen. The disaccharide co-crystals formed in the primitive monoclinic space group P21 with unit cell dimensions a = 69.9 Å, b = 34.0 Å, c = 83.4 Å with α = γ = 90° and β = 99.2°. The best data from this crystal form merged to 1.9 Å resolution.
Data collection and processing
Crystals were assessed for diffraction quality at the Stanford Synchrotron Radiation Lightsource (SSRL) beamlines 9-2, 11-1, and 12-2 and the Life Sciences Collaborative Access Team (LS-CAT) beamlines ID-21-D/F/G. Datasets were collected using the beamlines, temperatures, wavelengths, and detectors listed in Table 1 and Table 2. All data were processed using the HKL2000 [56] and CCP4 [57] suites of programs.
Preparation of heavy atom derivatives and structure determination
A Dy3+ derivative was prepared by soaking pre-formed crystals of GspBBR in 1 mM DyCl3 for three days. Data were collected at three wavelengths near the Dy3+ L3 edge (Table 2). Dy3+ bound to a single site in the protein as determined using the SHELXD [58] subroutine in the program SHARP [59]. While HoCl3 also successfully derivatized GspBBR, non-isomorphism between crystals prevented the use of this second derivative in a traditional MIR calculation. The non-isomorphism was so severe that phases calculated in SHARP [59] only used data from a single DyCl3 soaked crystal and did not include a native dataset for reference. This process resulted in reasonable phasing statistics and an overall figure of merit of 0.83 at 2.0 Å resolution (Table 2). Phases were improved by solvent flattening in DM [60] which produced electron density maps of high quality (Fig. 2B). Automated chain tracing was performed using PHENIX [61], which was able to trace residues 251–316 and 327–601, representing 94.7% of the model. This resulted in an initial Rcryst of 23.8% and Rfree of 25.5%.
The lack of isomorphism between crystals prevented transfer of these initial coordinates to other data sets using a simple rigid body refinement. As a result, the model from the Dy3+ data set was transferred to the remaining data sets using the program PHASER [62] followed by rigid body refinement in CNS [63]. Each structure was subjected to alternate rounds of model building using the program COOT [64] and refinement using CNS [63] and REFMAC [65].
The coordinates for the α-2,3-sialyl (1-thioethyl)galactose were built using CCP4i Sketcher [66] and PRODRG [67]. Refinement statistics for all final models are listed in Table 1. Figures were created using PyMOL [68], and inter-domain rotations were determined using DynDom [69].
Synthesis of the α-2,3-sialyl (1-thioethyl)galactose disaccharide
Based upon work by Danifshesky and coworkers [70], we developed a four-step synthesis for the sialyl-T antigen precursor, α-2,3-sialyl (1-thioethyl)galactose (see Supporting Protocol S1). The correct synthesis of the disaccharide was verified by NMR (Fig. S2). The α-2,3-sialyl (1-thioethyl)galactose was resuspended in water for all applications.
Site-directed Mutagenesis
Point mutations of gspB were introduced into the S. gordonii chromosome via a strategy that involved recombination by double cross-over between gspB codons 487–602 and a gene approximately 300 bp upstream. This approach ensured incorporation of only the intended mutation of gspB codons ranging from 399 to 485, and avoided possible imprecise recombination within the SRR regions. As a first step, the 5′ end of gspB (codons 1 to 486 along with 200 nts from a non-coding region upstream) was replaced with a chloramphenicol resistance cassette as follows. A 0.5 kb segment of a gene of unknown function upstream of gspB was amplified using PCR. The product was digested with XhoI and ClaI and then cloned upstream of the cat gene in pC326 [71]. A segment spanning gspB codons 487 to 602 was then amplified using primers B487F and B602R, digested with SpeI and NotI, and cloned downstream of the cat gene. The resulting plasmid, pC326Δ5′B, was used to transform S. gordonii strain M99 as described [4]. One of the chloramphenicol-resistant transformants, (M99 Δ5′gspB::cat), was selected for subsequent gene replacement.
A series of plasmids was then constructed to facilitate the replacement of the 5′ end of gspB in M99 Δ5′gspB::cat. The XhoI-ClaI fragment from pC326Δ5′B was cloned upstream of the spec gene in pS326 [72], and a 1 kb NsiI-SpeI fragment of gspB (spanning codons 1 to 296), was cloned downstream. A SpeI-NotI fragment spanning codons 296 to 602 was then cloned downstream of the NsiI-SpeI fragment. Point mutations in the resulting plasmid, pS326B602, were generated by a two-stage PCR procedure. In the first stage, primer 25F along with a reverse gspB primer, or the corresponding gspB forward primer along with primer B602R, were used to amplify the upstream or downstream segments, respectively. The two PCR products were combined for the second stage and then amplified using primers 25F and B602R. The PCR product was digested with SpeI and NotI and then used to replace the corresponding segment of pS326B602. The incorporation of only the intended change in any segment generated by PCR was confirmed by DNA sequence analysis. Plasmids were then used to transform M99 Δ5′gspB::cat, resulting in a replacement of Δ5′gspB::cat with a 5′gspB::spec variant. As a control, the wild-type gspB sequence along with the spec cassette (pS326B602) was also crossed into the M99 Δ5′gspB::cat chromosome (generating strain PS2161). Transformants were selected on spectinomycin and scored for the loss of chloramphenicol resistance. Expression of the variant GspB proteins on the bacterial cell surface was verified by western blotting as described [72].
GspBBR binding to biotinylated sialyl-T antigen
GST, GST-GspBBR and GST-GspBBR-R484E were purified from E. coli as described [55]. The purified proteins were diluted to 320 µM into DPBS, serial two-fold dilutions were made, and 50 µl of each dilution was added to wells of a 96-well microtiter plate. After incubating the plate overnight at 4°C, unbound proteins were removed by aspiration and wells were rinsed with 100 µl DPBS. Biotinylated sialyl-T antigen (sialyl-T antigen conjugated to biotin via a polyacrylamide linker; GlycoTech Corporation) was diluted to 50 µg/ml in DPBS containing 1× Blocking Reagent (Roche), 50 µl was added to each well, and the plate was incubated for 2 h at RT with vigorous rocking. After removing unbound biotin-sialyl-T antigen, wells were rinsed three times with 100 µl DPBS, 50 µl of streptavidin-conjugated horseradish peroxidase (0.1 µg/ml in DPBS) was added to each well and the plate was incubated for 1 h at 23°C. The wells were washed twice with 100 µl DPBS, and then 200 µl of a solution of OPD (0.4 mg/ml citrate-phosphate buffer) was added to each well. The contents of the wells were mixed by gently vortexing the plate, and the absorbance at 450 nm was measured 20 min after the addition of the OPD substrate. Data were plotted as the means ± standard deviations, with n = 3.
Binding of S. gordonii to glycocalicin and platelet monolayers
The binding of S. gordonii to immobilized glycocalicin was performed as described previously [32]. In brief, strains were grown for 18 hr, washed twice with DPBS, sonicated briefly to disrupt aggregated cells, and then diluted to approximately 2×107 per ml. To determine whether α-2,3 sialyl (1-thioethyl)galactose inhibited binding to glycocalicin, the washed and sonicated bacteria were diluted into DPBS or DPBS containing 44 mM α-2,3-sialyl (1-thioethyl)galactose, pH 7.5. The bacterial suspensions were then applied to wells of a microtiter plate that had been coated with glycocalicin (1.25 µg/well). After 2 h at room temperature, the unbound bacteria were removed by aspiration. Wells were washed three times with DPBS, and the bound bacteria were released by trypsinization. The number of input and bound bacteria were determined by plating serial dilutions of the bacterial suspensions on sheep blood agar plates, and binding was expressed as the percent of the input bound to glycocalicin. The binding of S. gordonii to immobilized human platelets was assessed as described previously [4]. Results of both assays are reported as the means ± standard deviations, with n = 6. Differences in binding were compared by the unpaired t-test.
DIC and fluorescence microscopy
S. gordonii strains M99 wild type, PS2116, and PS846 were grown in 5 mL of Todd Hewitt Broth at 37°C without shaking for 18 hours. Cells were then vortexed to resuspend bacteria and spun down at 4000× g for ten minutes. The supernatant was removed and the bacteria washed twice with 5 mL of DPBS containing MgCl2 and CaCl2. The bacteria were then resuspended in 5 mL of DPBS and sonicated briefly to disrupt aggregated clumps. To 1 mL of each cell suspension, 500 nM 4′,6-diamidino-2-phenylindole (DAPI) was added.
Platelets were fixed using 1.6% paraformaldehyde. 500 µL of platelets were mounted on poly-L-lysine coated cover slips (in a 6-well tray) which was spun at 400× g for ten minutes in order to promote platelet adherence to the cover slips. Excess platelets were removed by washing with Tris Buffered Saline (TBS) and 1 mL of TBS was added to each well. 500 µL of the bacterial suspension was added to the platelets and the samples were rocked vigorously for 30 minutes at 23°C. Excess bacteria were then removed by washing three times with TBS. Each sample was mounted onto a slide for microscopy. Slides were imaged using Nikon TiE Inverted light microscope equipped with a Photometrics CoolSnap HQ CCD camera. Platelets were imaged with a 100×1.49NA objective using DIC optics and a DAPI filter cube. Image J software was used to create the contrast and composite images.
Rat model of infective endocarditis
A competition model of infective endocarditis was produced in Sprague–Dawley female rats (250–300 g) as described previously [18]. In brief, the animals were anesthetized with ketamine (35 mg/kg) and xylazine (10 mg/kg). A sterile polyethylene catheter was surgically placed across the aortic valve of each animal, such that the tip was positioned in the left ventricle. Catheters were left in place throughout the study. Catheterized animals were then infected intravenously (IV) with an inoculum containing 2×105 CFU of both strains (i.e., a 1∶1 mixture of i) S. gordonii strain M99 and strain PS2116 (5′gspBR484E::spec), ii) M99 and strain PS2161 (5′gspB::spec control strain), or iii) PS2116 and strain PS846 (M99 ΔgspB::pEVP3; CmR) [18], [73]. At 72 h post-infection, animals were sacrificed with thiopental (100 mg, intraperitoneally). Animals were included in the final analysis only if the catheters were correctly positioned across the aortic valve at the time of sacrifice, and if macroscopic vegetations were seen. All cardiac vegetations, as well as samples of the kidneys and spleens were harvested, weighed, homogenized in saline, serially diluted, and plated onto Todd Hewitt agar (THA) (for the parental S. gordonii strain M99) and THA containing 100 µg/ml of spectinomycin (for strains PS2116 and PS2161) or chloramphenicol 5 mg/ml (for strain PS846) for quantitative culture, to determine the number of CFU/g of S. gordonii strains within tissues. After 48 h of incubation at 37°C, bacterial colonies were counted. The number of bacteria within tissues was expressed as the log10 CFU per gram of tissue. Differences between means were compared for statistical significance by the paired t-test, using p≤0.05 as the threshold for significance. The data were also analyzed by calculating a “competition index,” which was defined as the ratio of S. gordonii strain M99 and strain PS2116 or PS2161, as well as PS2116 and PS846, within tissues for each animal, normalized by the ratio of organisms in the inoculum. The mean of the log10 normalized ratios was tested against the hypothesized ‘no effect’ mean value of 0, as described previously, using a paired t-test.
Accession numbers
The coordinates and structure factors for S. gordonii strain M99 GspBBR have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank with accession codes 3QC5 (native 1), 3QC6 (native 2, crystal form 2), and 3QD1 (α-2,3-sialyl (1-thioethyl)galactose bound).
Supporting Information
Zdroje
1. TakahashiYKonishiKCisarJOYoshikawaM 2002 Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect Immun 70 1209 1218
2. HandleyPSCorreiaFFRussellKRosanBDiRienzoJM 2005 Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol Immunol 20 131 140
3. WuHMintzKPLadhaMFives-TaylorPM 1998 Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol 28 487 500
4. BensingBASullamPM 2002 An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol 44 1081 1094
5. SanchezCJShivshankarPStolKTrakhtenbroitSSullamPM 2010 The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog 6 8 e1001044 doi:10.1371/journal.ppat.1001044
6. SibooIRChambersHFSullamPM 2005 Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus, in binding to human platelets. Infect Immun 73 2273 2280
7. SeifertKNAddersonEEWhitingAABohnsackJFCrowleyPJ 2006 A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152 1029 1040
8. WuHFives-TaylorPM 1999 Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis. Mol Microbiol 34 1070 1081
9. PlummerCWuHKerriganSWMeadeGCoxD 2005 A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br J Haematol 129 101 109
10. TakamatsuDBensingBAPrakobpholAFisherSJSullamPM 2006 Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun 74 1933 1940
11. TakamatsuDBensingBAChengHJarvisGASibooIR 2005 Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ib alpha. Mol Microbiol 58 380 392
12. van SorgeNMQuachDGurneyMASullamPMNizetV 2009 The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J Infect Dis 199 1479 1487
13. MistouMYDramsiSBregaSPoyartCTrieu-CuotP 2009 Molecular dissection of the secA2 locus of group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J Bacteriol 191 4195 4206
14. YajimaAUrano-TashiroYShimazuKTakashimaETakahashiY 2008 Hsa, an adhesin of Streptococcus gordonii DL1, binds to alpha2-3-linked sialic acid on glycophorin A of the erythrocyte membrane. Microbiol Immunol 52 69 77
15. TakahashiYSandbergALRuhlSMullerJCisarJO 1997 A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to alpha2-3-linked sialic acid-containing receptors. Infect Immun 65 5042 5051
16. SamenUEikmannsBJReinscheidDJBorgesF 2007 The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect Immun 75 5405 5414
17. ShivshankarPSanchezCRoseLFOrihuelaCJ 2009 The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol 73 663 679
18. XiongYQBensingBABayerASChambersHFSullamPM 2008 Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb Pathog 45 297 301
19. ObertCSublettJKaushalDHinojosaEBartonT 2006 Identification of a Candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun 74 4766 4777
20. PetersenHJKeaneCJenkinsonHFVickermanMMJesionowskiA 2010 Human platelets recognize a novel surface protein, PadA, on Streptococcus gordonii through a unique interaction involving fibrinogen receptor GPIIbIIIa. Infect Immun 78 413 422
21. KerriganSWJakubovicsNSKeaneCMaguirePWynneK 2007 Role of Streptococcus gordonii surface proteins SspA/SspB and Hsa in platelet function. Infect Immun 75 5740 5747
22. JakubovicsNSKerriganSWNobbsAHStrombergNvan DolleweerdCJ 2005 Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect Immun 73 6629 6638
23. GaneshVKRiveraJJSmedsEKoYPBowdenMG 2008 A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog 4 e1000226
24. McDevittDNanavatyTHouse-PompeoKBellETurnerN 1997 Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem 247 416 424
25. MiajlovicHLoughmanABrennanMCoxDFosterTJ 2007 Both complement - and fibrinogen-dependent mechanisms contribute to platelet aggregation mediated by Staphylococcus aureus clumping factor B. Infect Immun 75 3335 3343
26. LoughmanAFitzgeraldJRBrennanMPHigginsJDownerR 2005 Roles for fibrinogen, immunoglobulin and complement in platelet activation promoted by Staphylococcus aureus clumping factor A. Mol Microbiol 57 804 818
27. O'BrienLKerriganSWKawGHoganMPenadesJ 2002 Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol Microbiol 44 1033 1044
28. FitzgeraldJRFosterTJCoxD 2006 The interaction of bacterial pathogens with platelets. Nat Rev Microbiol 4 445 457
29. SeoHSXiongYQMitchellJSeepersaudRBayerAS 2010 Bacteriophage lysin mediates the binding of streptococcus mitis to human platelets through interaction with fibrinogen. PLoS Pathog 6 8 e1001047 doi:1001010.1001371/journal.ppat.1001047
30. DurackDT 1975 Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J Pathol 115 81 89
31. YajimaATakahashiYKonishiK 2005 Identification of platelet receptors for the Streptococcus gordonii DL1 sialic acid-binding adhesin. Microboil Immunol 49 795 800
32. BensingBALopezJASullamPA 2004 The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ib alpha. Infect Immun 72 6528 6537
33. TakahashiYTakashimaEShimazuKYagishitaHAobaT 2006 Contribution of sialic acid-binding adhesin to pathogenesis of experimental endocarditis caused by Streptococcus gordonii DL1. Infect Immun 74 740 743
34. PonnurajKBowdenMGDavisSGurusiddappaSMooreD 2003 A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115 217 228
35. SymerskyJPattiJMCarsonMHouse-PompeoKTealeM 1997 Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat Struct Biol 4 833 838
36. DeivanayagamCCWannERChenWCarsonMRajashankarKR 2002 A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J 21 6660 6672
37. HolmgrenABrandenCI 1989 Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature 342 248 251
38. CotaEJonesCSimpsonPAltroffHAndersonKL 2006 The solution structure of the invasive tip complex from Afa/Dr fibrils. Mol Microbiol 62 356 366
39. HamburgerZABrownMSIsbergRRBjorkmanPJ 1999 Crystal structure of invasin: a bacterial integrin-binding protein. Science 286 291 295
40. JedrzejczakRDauterZDauterMPiatekRZalewskaB 2006 Structure of DraD invasin from uropathogenic Escherichia coli: a dimer with swapped beta-tails. Acta Crystallogr D Biol Crystallogr 62 157 164
41. VengadesanKMaXDwivediPTon-ThatHNarayanaSV 2011 A Model for Group B Streptococcus Pilus Type 1: The Structure of a 35-kDa C-Terminal Fragment of the Major Pilin GBS80. J Mol Biol 407 731 743
42. HolmLRosenstromP 2010 Dali server: conservation mapping in 3D. Nucleic Acids Res 38 W545 549
43. RamboarinaSGarnettJAZhouMLiYPengZ 2010 Structural insights into serine-rich fimbriae from gram-positive bacteria. J Biol Chem 285 32446 32457
44. MayAPRobinsonRCVinsonMCrockerPRJonesEY 1998 Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ sialyllactose at 1.85 A resolution. Mol Cell 1 719 728
45. ZhuravlevaMATrandemKSunPD 2008 Structural implications of Siglec-5-mediated sialoglycan recognition. J Mol Biol 375 437 447
46. AttrillHImamuraASharmaRSKisoMCrockerPR 2006 Siglec-7 undergoes a major conformational change when complexed with the alpha(2,8)-disialylganglioside GT1b. J Biol Chem 281 32774 32783
47. ChungMCWinesBDBakerHLangleyRJBakerEN 2007 The crystal structure of staphylococcal superantigen-like protein 11 in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition. Mol Microbiol 66 1342 1355
48. BakerHMBasuIChungMCCaradoc-DaviesTFraserJD 2007 Crystal structures of the staphylococcal toxin SSL5 in complex with sialyl Lewis X reveal a conserved binding site that shares common features with viral and bacterial sialic acid binding proteins. J Mol Biol 374 1298 1308
49. ImbertyAVarrotA 2008 Microbial recognition of human cell surface glycoconjugates. Curr Opin Struct Biol 18 567 576
50. AltschulSFGishWMillerWMyersEWLipmanDJ 1990 Basic local alignment search tool. J Mol Biol 215 403 410
51. ChennaRSugawaraHKoikeTLopezRGibsonTJ 2003 Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 3497 3500
52. De GreveHWynsLBouckaertJ 2007 Combining sites of bacterial fimbriae. Curr Opin Struct Biol 17 506 512
53. SotomayorMSchultenK 2008 The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys J 94 4621 4633
54. SokurenkoEVVogelVThomasWE 2008 Catch-bond mechanism of force-enhanced adhesion: counterintuitive, elusive, but … widespread? Cell Host Microbe 4 314 323
55. PyburnTMYankovskayaVBensingBACecchiniGSullamPM 2010 Purification, crystallization and preliminary X-ray diffraction analysis of the carbohydrate-binding region of the Streptococcus gordonii adhesin GspB. Acta Crystallogr F Struct Biol Cryst Commun 66 1503 1507
56. OtwinowskiZMinorW 1997 Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A 276 307 326
57. BaileyS 1994 The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D Biol Crystallogr 50 760 763
58. SheldrickGM 2008 A short history of SHELX. Acta Crystallogr A 64 112 122
59. de La FortelleEBricogneG 1997 Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Macromolecular Crystallography, Pt A 276 472 494
60. BaileyS 1994 The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D Biol Crystallogr 50 760 763
61. AdamsPDAfoninePVBunkocziGChenVBDavisIW 2010 PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66 213 221
62. MccoyAJGrosse-KunstleveRWAdamsPDWinnMDStoroniLC 2007 Phaser crystallographic software. J Appl Crystallogr 40 658 674
63. BrungerAT 2007 Version 1.2 of the Crystallography and NMR system. Nat Protoc 2 2728 2733
64. EmsleyPCowtanK 2004 Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 2126 2132
65. MurshudovGNVaginAADodsonEJ 1997 Refinement of macromolecular structures by the maximum-likelihood method. A Acta Crystallogr D Biol Crystallogr 53 240 255
66. PottertonEBriggsPTurkenburgMDodsonE 2003 A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr 59 1131 1137
67. SchuttelkopfAWvan AaltenDM 2004 PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60 1355 1363
68. DeLanoWL 2002 The PyMOL Molecular Graphics System
69. HaywardSBerendsenHJ 1998 Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30 144 154
70. SchwarzJBKudukSDChenXTSamesDGlunzPW 1999 A broadly applicable method for the efficient synthesis of alpha-O-linked glycopeptides and clustered sialic acid residues. J Am Chem Soc 121 2662 2673
71. MitchellJSibooIRTakamatsuDChambersHFSullamPM 2007 Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol Microbiol 64 844 857
72. TakamatsuDBensingBASullamPM 2004 Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol Microbiol 52 189 203
73. BensingBATakamatsuDSullamPM 2005 Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol Microbiol 58 1468 1481
74. AlpheyMSAttrillHCrockerPRvan AaltenDM 2003 High resolution crystal structures of Siglec-7. Insights into ligand specificity in the Siglec family. J Biol Chem 278 3372 3377
75. KleywegtGJ 1996 Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D Biol Crystallogr 52 842 857
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek A Typhimurium-Typhi Genomic Chimera: A Model to Study Vi Polysaccharide Capsule Function In VivoČlánek SUMO Pathway Dependent Recruitment of Cellular Repressors to Herpes Simplex Virus Type 1 GenomesČlánek Dynamic Evolution of Pathogenicity Revealed by Sequencing and Comparative Genomics of 19 IsolatesČlánek Widespread Endogenization of Genome Sequences of Non-Retroviral RNA Viruses into Plant GenomesČlánek The Cost of Virulence: Retarded Growth of Typhimurium Cells Expressing Type III Secretion System 1Článek A Role for the Chemokine RANTES in Regulating CD8 T Cell Responses during Chronic Viral Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- What Do We Really Know about How CD4 T Cells Control ?
- “Persisters”: Survival at the Cellular Level
- E6 and E7 from Beta Hpv38 Cooperate with Ultraviolet Light in the Development of Actinic Keratosis-Like Lesions and Squamous Cell Carcinoma in Mice
- Selection of Resistant Bacteria at Very Low Antibiotic Concentrations
- The Extracytoplasmic Domain of the Ser/Thr Kinase PknB Binds Specific Muropeptides and Is Required for PknB Localization
- CD39/Adenosine Pathway Is Involved in AIDS Progression
- Hypoxia and a Fungal Alcohol Dehydrogenase Influence the Pathogenesis of Invasive Pulmonary Aspergillosis
- One Is Enough: Effective Population Size Is Dose-Dependent for a Plant RNA Virus
- Effects of Interferon-α/β on HBV Replication Determined by Viral Load
- A Typhimurium-Typhi Genomic Chimera: A Model to Study Vi Polysaccharide Capsule Function In Vivo
- Dual Chaperone Role of the C-Terminal Propeptide in Folding and Oligomerization of the Pore-Forming Toxin Aerolysin
- Rotavirus Stimulates Release of Serotonin (5-HT) from Human Enterochromaffin Cells and Activates Brain Structures Involved in Nausea and Vomiting
- Dissociation of Infectivity from Seeding Ability in Prions with Alternate Docking Mechanism
- The Impact of Recombination on dN/dS within Recently Emerged Bacterial Clones
- The Regulation of Sulfur Metabolism in
- Illumination of Parainfluenza Virus Infection and Transmission in Living Animals Reveals a Tissue-Specific Dichotomy
- A Permeable Cuticle Is Associated with the Release of Reactive Oxygen Species and Induction of Innate Immunity
- A Concerted Action of Hepatitis C Virus P7 and Nonstructural Protein 2 Regulates Core Localization at the Endoplasmic Reticulum and Virus Assembly
- SUMO Pathway Dependent Recruitment of Cellular Repressors to Herpes Simplex Virus Type 1 Genomes
- Re-localization of Cellular Protein SRp20 during Poliovirus Infection: Bridging a Viral IRES to the Host Cell Translation Apparatus
- Divergent Effects of Human Cytomegalovirus and Herpes Simplex Virus-1 on Cellular Metabolism
- A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate Receptors
- Transformation of Natural Genetic Variation into Genomes
- EBV Latency Types Adopt Alternative Chromatin Conformations
- Global mRNA Degradation during Lytic Gammaherpesvirus Infection Contributes to Establishment of Viral Latency
- Dynamic Evolution of Pathogenicity Revealed by Sequencing and Comparative Genomics of 19 Isolates
- Microbial Virulence as an Emergent Property: Consequences and Opportunities
- Widespread Endogenization of Genome Sequences of Non-Retroviral RNA Viruses into Plant Genomes
- Structural Basis of Chemokine Sequestration by CrmD, a Poxvirus-Encoded Tumor Necrosis Factor Receptor
- Cross-Species Transmission of a Novel Adenovirus Associated with a Fulminant Pneumonia Outbreak in a New World Monkey Colony
- An Interaction between KSHV ORF57 and UIF Provides mRNA-Adaptor Redundancy in Herpesvirus Intronless mRNA Export
- Elevated 17β-Estradiol Protects Females from Influenza A Virus Pathogenesis by Suppressing Inflammatory Responses
- The Role of IL-15 Deficiency in the Pathogenesis of Virus-Induced Asthma Exacerbations
- Fluorescence Lifetime Imaging Unravels Metabolism and Its Crosstalk with the Host Cell
- Programmed Death (PD)-1-Deficient Mice Are Extremely Sensitive to Murine Hepatitis Virus Strain-3 (MHV-3) Infection
- Hemoglobin Promotes Nasal Colonization
- Crystallography of a Lewis-Binding Norovirus, Elucidation of Strain-Specificity to the Polymorphic Human Histo-Blood Group Antigens
- The Cost of Virulence: Retarded Growth of Typhimurium Cells Expressing Type III Secretion System 1
- A Genome-Wide Approach to Discovery of Small RNAs Involved in Regulation of Virulence in
- Requires Glycerol for Maximum Fitness During The Tick Phase of the Enzootic Cycle
- C Metabolic Flux Analysis Identifies an Unusual Route for Pyruvate Dissimilation in Mycobacteria which Requires Isocitrate Lyase and Carbon Dioxide Fixation
- A Role for the Chemokine RANTES in Regulating CD8 T Cell Responses during Chronic Viral Infection
- Glycosaminoglycans and Sialylated Glycans Sequentially Facilitate Merkel Cell Polyomavirus Infectious Entry
- Regulation of Stomatal Tropism and Infection by Light in : Evidence for Coordinated Host/Pathogen Responses to Photoperiod?
- Multiple Translocation of the Effector Gene among Chromosomes of the Rice Blast Fungus and Related Species
- Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens
- Unique Cell Adhesion and Invasion Properties of O:3, the Most Frequent Cause of Human Yersiniosis
- C-Terminal Region of EBNA-2 Determines the Superior Transforming Ability of Type 1 Epstein-Barr Virus by Enhanced Gene Regulation of LMP-1 and CXCR7
- Novel Chikungunya Vaccine Candidate with an IRES-Based Attenuation and Host Range Alteration Mechanism
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Requires Glycerol for Maximum Fitness During The Tick Phase of the Enzootic Cycle
- Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens
- The Role of IL-15 Deficiency in the Pathogenesis of Virus-Induced Asthma Exacerbations
- “Persisters”: Survival at the Cellular Level
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



