-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
Unlike mammals, the identity of the master sex-determining gene varies among fish species, and it is not yet clear if there is a common molecular pathway regulating gonadal sex determination across teleosts. Here we show that a Y-linked duplicate of the anti-Mullerian hormone (amhy) is essential for male sex determination in tilapia. Mutation of amhy resulted in male to female sex reversal, while overexpression of it resulted in female to male sex reversal. A missense single nucleotide polymorphisms (SNP) (C/T) in the open reading frame (ORF) of amhy might contribute to male sex determination in tilapia. Knockout of the anti-Müllerian hormone receptor type II (amhrII) also resulted in male to female sex reversal. Taken the amhy in Patagonian pejerrey, amhrII in Takifugu rubripes, gsdfY in Oryzias luzonensis into consideration, these data highlight an important role for TGF-β signaling in teleost sex determination.
Published in the journal: . PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005678
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005678Summary
Unlike mammals, the identity of the master sex-determining gene varies among fish species, and it is not yet clear if there is a common molecular pathway regulating gonadal sex determination across teleosts. Here we show that a Y-linked duplicate of the anti-Mullerian hormone (amhy) is essential for male sex determination in tilapia. Mutation of amhy resulted in male to female sex reversal, while overexpression of it resulted in female to male sex reversal. A missense single nucleotide polymorphisms (SNP) (C/T) in the open reading frame (ORF) of amhy might contribute to male sex determination in tilapia. Knockout of the anti-Müllerian hormone receptor type II (amhrII) also resulted in male to female sex reversal. Taken the amhy in Patagonian pejerrey, amhrII in Takifugu rubripes, gsdfY in Oryzias luzonensis into consideration, these data highlight an important role for TGF-β signaling in teleost sex determination.
Introduction
Master sex-determining (SD) genes are the key genetic switches controlling the gonadal sex differentiation cascade leading to the development of either ovaries or testes. To date, master SD genes have been identified in only a few vertebrate species. SRY/Sry was the first sex determiner identified in mammals [1, 2]. With the recent discovery that sox3Y is the sex determiner in Oryzias dancena [3], Sox genes continue to figure prominently in discussions of vertebrate sex determination. Doublesex/mab (DM) related genes have been associated with sex determination in a wide range of species, including Dmrt1 in chicken and half-smooth tongue sole [4, 5], DM-W in African clawed frog [6], and dmy/dmrt1bY in Oryzias latipes [7, 8]. Other genes have been implicated as master sex determiners in particular lineages, including FOXL2 in goat [9], and sdY (irf9y) in rainbow trout [10]. Several recent studies have suggested that components of the transforming growth factor beta (TGF-β) signaling pathway are involved in sex determination in fishes. These include a Y-linked duplicate of the anti-Müllerian hormone (amhy) in the Patagonian pejerrey [11], a mutation in the amh receptor (amhrII) in Takifugu rubripes [12], and a Y-linked duplicate of a related ligand, gonadal soma derived growth factor (gsdfY) in Oryzias luzonensis [13]. These findings suggest a critical role for TGF-β signaling in gonadal sex determination in teleosts.
Studies of mammalian sex chromosomes have provided significant insights into the evolution of sex determination, but SD genes have not yet been identified in the vast majority of vertebrates. For example, teleost fishes make up nearly half of all living vertebrate species and show a wide variety of sex determination mechanisms [14], but only a handful of these sex determiners have been identified. Closely related species of fish frequently segregate different master sex determiners, suggesting that a delicately balanced network of gene interactions controls sex determination. For example, three different genes (dmy, gsdfY and sox3Y) have been identified as master sex determiners among closely related species of ricefish [3, 7, 8, 13]. Master sex determiners map to three different chromosomes among closely related species of stickleback [15]. Recent work has identified at least three sex determiners among strains of zebrafish [16, 17].
Numerous studies have investigated the mechanisms of sex determination in tilapia (Oreochromis niloticus), motivated in part by commercial interest in the higher growth rates of all-male progenies. Tilapia are gonochoristic teleosts in which sex is largely genetically determined [18], although environmental factors also play a role [19]. XX/XY sex determination systems have been described on both LG1 and LG23 in this species [20, 21]. All-XX and all-XY progenies can be obtained by crossing normal XX females to either experimentally sex-reversed XX pseudomales or YY supermales respectively [22].
A previous report identified several sex-linked markers near the amh gene on LG23 [21]. More recent studies identified a Y-linked duplication of amh on LG23, termed a male-specific amhy, differing from the sequence of amh by a 233 bp deletion in exonVII [23]. Our own analyses have identified five additional sex-linked markers on LG23 that map very close to amh [22]. amh is located in the center of this sex-linked region and shows sexually dimorphic expression in the gonads at 3 days post fertilization [24], making it an interesting gene for sex determination in this species.
Amh is responsible for the regression of Müllerian ducts in tetrapods [25]. It is also found in teleost fish despite the fact that they do not have Müllerian ducts [23, 26–28]. In mammals, Amh functions primarily through the type II receptor AmhrII [25]. Mutations of amhrII in medaka and Takifugu rubripes result in male to female sex reversal [12, 29]. These studies suggested that amh/amhrII signaling might play a role in fish sex determination.
Recent efforts have generated a number of important resources for tilapia research, including a genome sequence, a microarrayed fosmid library, and several gonadal transcriptomes [30–32]. TALEN and CRISPR/Cas9 gene knockout technologies have also been established in tilapia [33, 34]. The availability of these tools prompted us to try to isolate the SD gene in the Nile tilapia. In the present study, we isolated a Y-specific duplicate of the amh gene, designated as amhy, and confirmed its male-specific (XY and YY) expression by transcriptome analysis and Western blot. We used transgenic techniques to overexpress amhy in XX fish, and we used CRISPR/Cas9 mutagenesis to knockout amhy, and its putative receptor amhrII in XY fish. Our results suggest a conserved role for the TGF-βsignaling pathway in sex determination of vertebrates.
Results
Identification of a Y-linked duplication of amh
We used PCR to screen a microarrayed tilapia XY genomic library for a sex-specific marker. We identified X-specific and Y-specific fosmid clones (designated X278 and Y156) producing amplimers of 1422 and 982 bp, respectively (S1 Fig). The two fosmid clones were then sequenced using Illumina HiSeq2000 technology and carefully assembled using local Basic Local Alignment Search Tool (local BLAST) by hand based on the sequence differences of several genomic PCR products amplified from XX and YY individuals and the sequence differences between Y156 and X278 fosmid. Sequence analysis revealed that an amh gene, termed as amh, was present in the X278 fosmid clone. By gene prediction with GENSCAN and BLAST search in the tilapia genome using the assembled sequence of the Y 156 clone, only two genes, designated as amhy and amhΔ-y, were found in the Y156 fosmid clone. amhy is a tandem duplicate located immediately downstream of amhΔ-y (Fig 1a). The insertion of Y156 was about 40 kb, which was further confirmed by sequencing 25 fragments, each about 3 kb with partial overlapping ends.
Fig. 1. Schematic representation of amh/amhΔ-y/amhy gene structure on the Y and X chromosome. 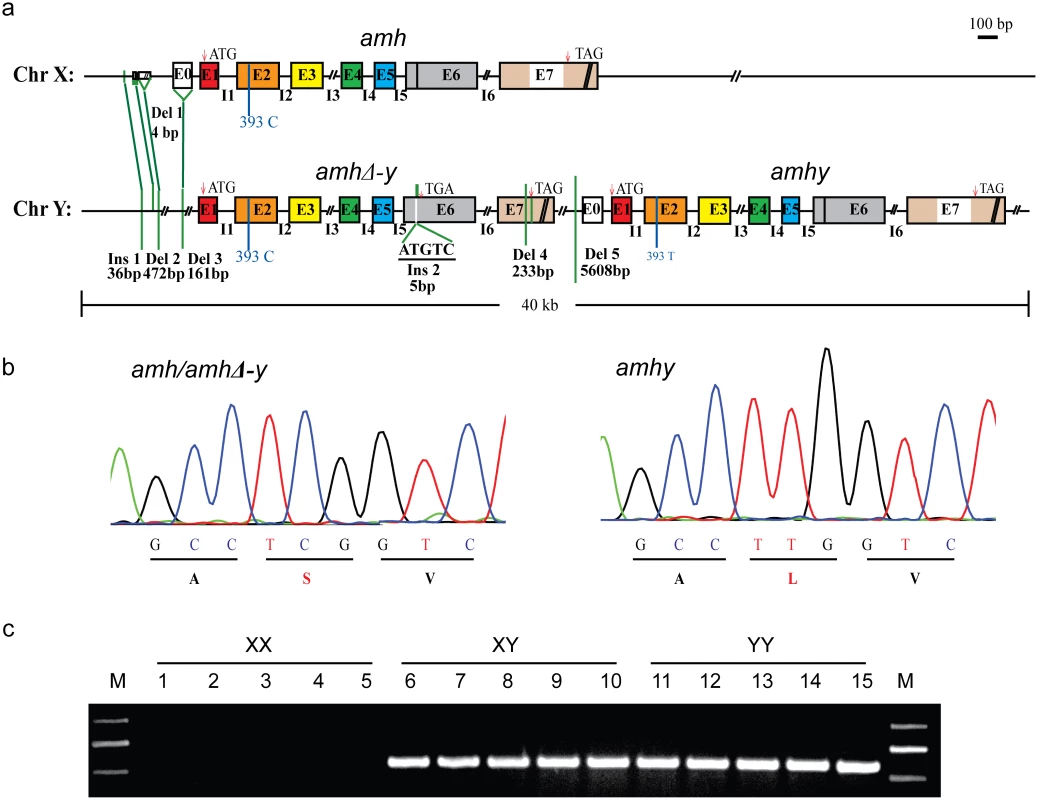
(a) Gene structure of amh, amhΔ-y and amhy. amhy was tandemly located downstream of amhΔ-y on the Y chromosome, with subsequent deletion of 5608 bp in the promoter. Compared with the X-linked amh, the transcribed region of amhΔ-y includes a 5 bp (ATGTC) insertion in exonVI and a deletion of 233 bp in exonVII. There are also three deletions (472, 161 and 4bp) and one insertion (36 bp) in the upstream of amhΔ-y start codon. (b) A missense SNP (C/T) was identified in the exonII of amh and amhy. This SNP converts amino acid from Serine to Leucine in the N-terminal region. (c) Genomic PCR amplification of the Y-specific fragments using an amhΔ-y specific primer F2 designed on the 5 bp insertion of amhΔ-y and a reverse primer R2 shared by amh, amhΔ-y and amhy (amhΔ-y-F2/R2). A 547 bp band was observed in XY and YY, but not XX genomic DNA. A comparison of the sequences of the two clones revealed numerous differences. The coding sequence of amhy was identical to the X-linked amh except a single nucleotide polymorphisms (SNP) (C/T) in exonII, which changes an amino acid (Ser/Leu92) in the N-terminal region. amhy has lost 5608 bp of promoter sequence that is found in the X-linked amh homolog (Fig 1a and 1b). amhΔ-y contains a 5 bp (ATGTC) insertion in exonVI, resulting in a frameshift mutation and a premature stop codon. The 5 bp insertion could be easily confirmed by Taqα I restriction enzyme digestion of the genomic PCR fragments spanning it, which resulted in two digested bands in the XY and YY fish, whereas no band was produced in XX fish (S2 Fig). amhΔ-y also has a deletion of 233 bp in exonVII that further precludes translation of the protein motif that binds to the TGF-βreceptor. There was one additional insertion (36 bp) and three other deletions (4, 472 and 161 bp, respectively) at -1972, -1756, -1664 and -625, respectively, of amhΔ-y start codon (ATG, A as 0 point), compared with amh on the X chromosome (Fig 1a). These differences between amhy, amh and amhΔ-y were further demonstrated by PCR amplification in XX, XY and YY genomic DNA pools (S3 Fig). The gene sequences of amhy, amhΔ-y and amh are shown in the S1 Sequence.
Primers flanking differentiated sequences in the promoter and exon region were used for PCR-based sex genotyping. Y-specific primers amplified fragments in the XY and YY pooled genomic DNA respectively, but not in the pooled XX genomic DNA (Fig 1c). Genotypic sex based on these primers showed 100% correspondence with phenotypic sex based on gonadal histology in 300 individuals derived from four crosses (S1 Table).
Finally, there are five SNPs in the coding sequences of amhΔ-y and amhy. Three of them are non-synonymous and the amino acid changed (S4 Fig). amhΔ-y is close to neutral evolution in comparison to amhy (Ka/Ks<1) (S2 Table). The tilapia amhΔ-y cDNA isolated by rapid amplification of cDNA ends (RACE) is 2,065 bp long with a 46 bp 5' untranslated region (UTR), a 1,242 bp 3' UTR and an ORF of 777 bp encoding a putative protein of 258 aa (amino acid) without the TGF-β domain. The amh/amhy isolated by RACE has a 346 bp 5' UTR, a 782 bp 3' UTR and an ORF of 1,545 bp, encoding a putative protein of 514 aa with the TGF-β domain.
amh, amhy and amhrII are expressed in tilapia gonads
RT-PCR showed that among the twelve tissues examined, amh/amhy and amhrII were expressed exclusively in gonads, with greater expression in testis. amhΔ-y was expressed only in the XY testis, not in the XX ovary (S5 Fig). Transcriptome analysis revealed that amhy and amhΔ-y transcripts were only detected in XY gonads, with expression at 5 days after hatching (dah), the critical period for molecular sex determination in Nile tilapia, peaked at approximately 30 dah, and decreased at 90 and to very low level at 180 dah (Fig 2a and 2b). Similar expression profiles of amh and amhrII were observed in both XX and XY gonads, with significantly higher expression in XX than in XY gonads at 5 dah, while they showed higher expression in XY than in XX gonads at 30 dah onwards (Fig 2c and S6 Fig).
Fig. 2. Expression profiles of amhy, amhΔ-y and amh by transcriptome analysis. 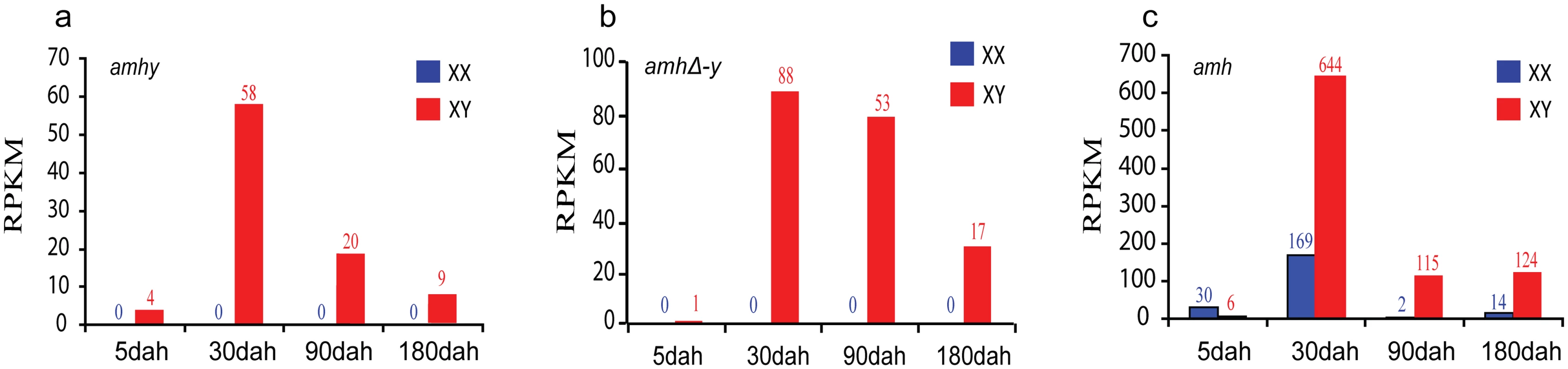
Transcriptome analysis revealed that amhy and amhΔ-y transcripts were only detected in XY gonads, with low expression at 5 days after hatching (dah), peaked at approximately 30 dah, and decreased expression at 90 dah (a, b). amh expression was observed in both XX and XY gonads with significant higher expression in XX than in XY gonads from 5 dah onwards, while it showed higher expression in XY than in XX gonads at 30 dah onwards (c). The expression of amh and amhy were analyzed by counting reads with the SNP (C/T) in exonII. The expression of amhΔ-y was analyzed by blast in the transcriptome data with the amhΔ-y specific 5 bp (ATGTC) insertion in exonVI. dah, days after hatching. A normalized measure of RPKM (reads per kb per million reads) was used to normalize the expression profiles of amhy, amhΔ-y and amh. The numbers over the bars indicate RPKM. Specificity of the Amh ployclonal antibody was characterized by Western blot analysis using recombinant protein (both purified and unpurified), and the proteins extracted from the XX ovary, XY testis, and YY testis. The Amh antibody can recognize the Amh, AmhΔ-y and Amhy protein. The blots revealed specific bands of ~54 kDa, corresponding to the calculated molecular weight of Amh/Amhy, in the protein samples extracted from the XX ovary, XY and YY testis. Another band of ~27 kDa, corresponding to AmhΔ-y, was detected exclusively in the protein samples extracted from the XY and YY testis. Amh/Amhy was detected in XY gonads of 5, 10, 30, 90, 180 and 300 dah tilapia, and also with expression of Amh in XX tilapia at these stages, while AmhΔ-y was only detected in 5, 10, 30 and 90 dah XY gonads. In addition, Amhy and AmhΔ-y were also detected in the proteins from YY gonads of 30 and 90 dah tilapia (S7 Fig). By immunohistochemistry, the Amh/AmhΔ-y/Amhy and AmhrII proteins were located in somatic cells surrounding germ cells in the XY gonads at 5 dah. At later stages, Amh/AmhΔ-y/Amhy is observed in myoid cells and Sertoli cells, while AmhrII was observed in spermatogonia and Sertoli cells of the testis at 30, 90 and 180 dah (S8 Fig).
Amhy knockout by CRISPR/Cas9 resulted in male to female sex reversal in XY fish
Two guide RNAs (gRNA) were designed, in exons II and III of amhy/amh/amhΔ-y, to increase the likelihood of successful targeting and also exclude the effects of off-target events on the phenotypes. The gRNAs contained BstN I or BsrB I sites adjacent to the protospacer adjacent motif (PAM) sequence for mutation analysis. Restriction enzyme digestion and Sanger sequencing were performed to confirm the insertions or deletions (indels), including both in-frame and frame-shift mutations, in pools of randomly selected embryos (Fig 3a). The screening results showed that 18% (8/45) of target 1 and 62% (28/45) of target 2 fish were mutated (S3 Table). Due to the high homology of amhy, amhΔ-y and amh, CRISPR/Cas9 disrupted three genes at both target sites. Restriction enzyme digestion showed that the CRISPR system disrupt amhy and amhΔ-y genes equally in F0 knockout XY fish (S9 Fig). RT-PCR with forward primers located in the targets using cDNA from 90 dah mutated gonads as templates demonstrated that the expression of amh/amhΔ-y/amhy mRNA in the F0 knockout group was much lower than that of control group (S10A Fig). Consistent with this, dramatically decreased Amh/Amhy protein levels and no AmhΔ-y were detected in the gonads of F0 knockout fish by Western blot (S10B Fig).
Fig. 3. Knockout of Amhy/AmhΔ-y/Amh by CRISPR/Cas9 resulted in male to female sex reversal in F0 XY fish. 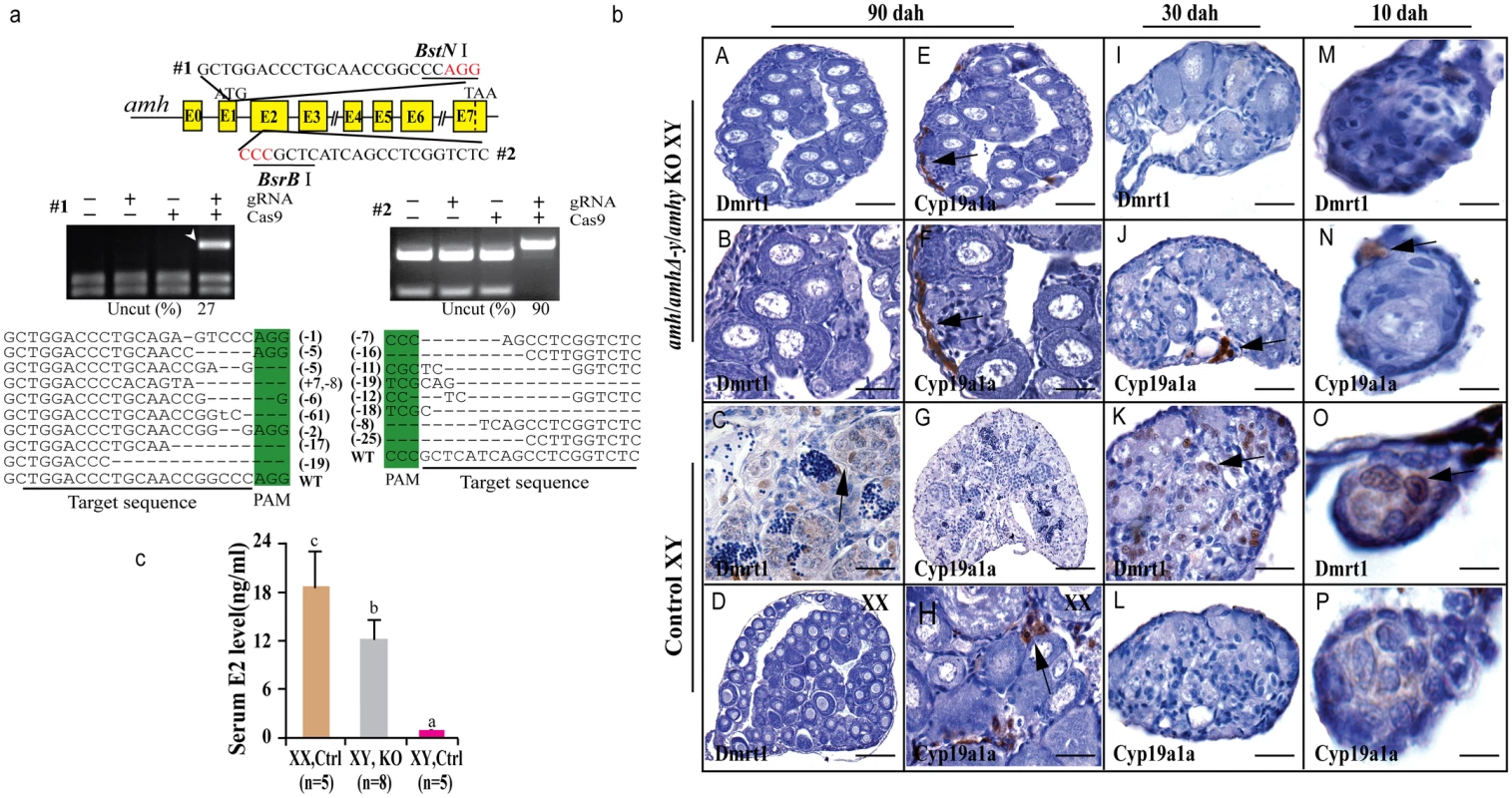
(a) Knockout of amhy/amhΔ-y/amh was achieved by CRISPR/Cas9. Two target sites were selected in the exon II and III of amhy/amhΔ-y/amh. Target site 1 and 2 were shared by amh, amhΔ-y and amhy. The two target sites included BstN I and BsrB I adjacent to the protospacer adjacent motif (PAM) for mutation analysis, respectively. For each target, digestion with the appropriate enzyme (BstN I, BsrB I) produced two cleavage bands in the control group, while an intact DNA fragment (indicated by white arrowheads) was observed in embryos injected with both Cas9 mRNA and target gRNA. Mutation sequences from the uncleaved bands were listed. The percentage of un-cleaved (i.e. mutant) DNA is shown for each target site. The indel frequency was calculated by dividing uncleaved band intensity to the total band intensity of the restriction enzyme digestion of pooled genomic DNA from up to 20 embryos. (b) The amh/amhΔ-y/amhy knockout F0 XY fish showed male to female sex reversal, as demonstrated by gonad histology and immunohistochemistry. Cyp19a1a, which was expressed in the control XX gonads but not XY gonads (G, L, P), was found to be expressed in the sex-reversed XY gonads at 10, 30 and 90 dah (E, F, H, J, N). In contrast, Dmrt1 disappeared in the sex-reversed XY gonads, which was different than the control testis at 10, 30 and 90 dah (A-D, I, M, K, O). (c) Higher serum E2 was observed in the sex-reversed XY fish compared with the XY control. Results are presented as the mean±SD. Different letters (a, b, c) indicate statistical differences at P<0.05 as determined by one-way ANOVA, followed by Tukey/Kramer post hoc test. Sample numbers are shown on the figure. The gonads of the mutagenized fish were subjected to both histological and immunohistochemical (IHC) analyses at 10, 30 and 90 dah. Macroscopic observation of the gonads of 3-month-old gRNA/Cas9 microinjected XY fish revealed that some of the mutated F0 fish showed male to female sex reversal. Gonadal sex differentiation in the sex-reversed XY fish was characterized by the formation of the ovarian cavity (OC) and the appearance of phase II oocytes. These gonads were topologically indistinguishable from the control XX ovary, but obviously different from the control XY testis (Fig 3b). IHC revealed that like the control ovary, male Sertoli cell marker Dmrt1 was not expressed in the sex-reversed XY gonads (Fig 3bA–3bD). Additionally, female specific marker Cyp19a1a was expressed in these sex-reversed XY gonads, like the control ovary (Fig 3bE–3bH). Consistent with the IHC results, higher serum estrogen (E2) was observed in the sex-reversed XY fish compared with the XY control (Fig 3c). The effects of amh/amhΔ-y/amhy deficiency on sex determination could be detected from a very early stage by IHC analysis of sexual dimorphic proteins such as Dmrt1 and Cyp19a1a. Tracing back, Cyp19a1a, which was not expressed in XY gonads at 10 and 30 dah, was detected in the sex-reversed XY gonads as early as 10 and 30 dah (Fig 3bJ, 3bN, 3bL and 3bP). In contrast, Dmrt1, which was detected in the control XY but not in XX gonad (Fig 3bK and 3bO), was not detected in sex-reversed XY gonads (Fig 3bI and 3bM).
Both target sites resulted in sex reversal. Of the 8 F0 fish with mutations in target site 1, 5 (62%) displayed complete male to female sex reversal. Of the 28 F0 fish with mutations in target site 2, 20 (71%) showed complete sex reversal (Table 1). The genetic sex of these sex-reversed XY fish with amh/amhΔ-y/amhy mutation was confirmed by amplification of a sex-linked marker (SPF/SPR) and all of these sex-reversed males were XY fish (S11 Fig). In addition, a number of oogonia, as demonstrated by Gsdf staining, and only a few primary and secondary oocytes, were observed in Amh deficient XX ovary at 90 dah (S12A Fig), compared with the control (S12B Fig).
Tab. 1. Phenotype of genetically modified fish produced in this study. 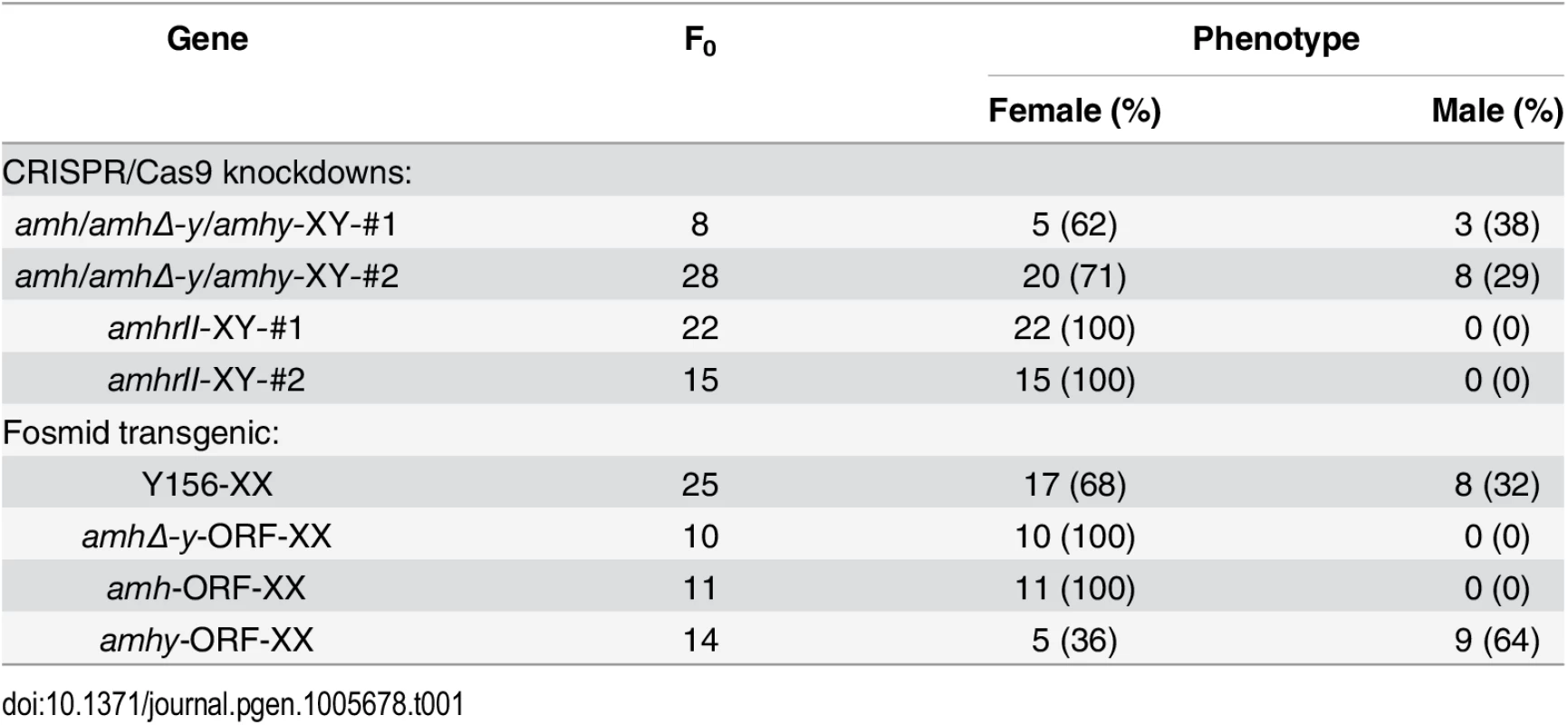
Sperm from the XY founders carrying different types of amh/amhΔ-y/amhy mutations (target 1 and 2) was mated with wild-type XX fish to produce F1 animals and independent mutant alleles having indels at the target of F1 XY were successfully obtained. These mutants caused frame shifts resulting in premature termination, predicted to yield truncated proteins (S13 Fig). The F1 XY fish with amhy mutant allele on the Y chromosome alone (n = 9+5) or double mutation of amhy and amhΔ-y (n = 14+10) displayed sex reversal with clear ovarian structure at 90 dah. In contrast, the F1 XY fish with mutation of amhΔ-y alone displayed no sex reversal with normal testis at 90 dah (n = 11+7) (Fig 4, Table 2).
Fig. 4. Mutation of Amhy resulted in male to female sex reversal in F1 XY fish. 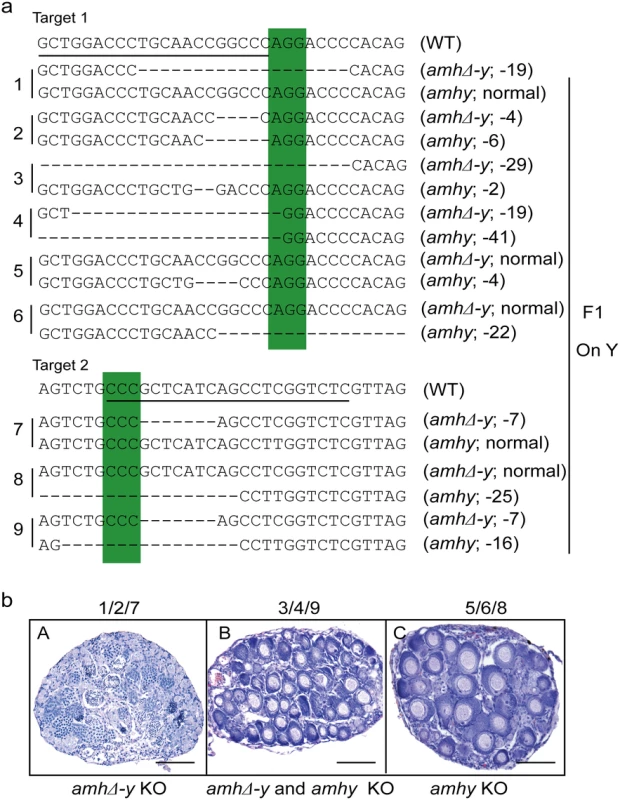
(a) Nine independent mutations at the target site 1 and 2 were detected in F1 fish: three types of amhy mutations, three types of amhΔ-y mutation and three types of amhy and amhΔ-y double mutations. (b) The XY fish with amhy mutant allele (n = 9+5) or with double mutation of amhy and amhΔ-y (n = 14+10) on the Y chromosome displayed sex reversal with typical ovary characterized by the presence of ovarian cavity (OC) and the appearance of oocytes. The XY fish with amhΔ-y (n = 11+7) mutant allele on the Y chromosome displayed no sex reversal with typical testis characterized by different stages of spermatogenic cells. Scale bars, 25 μm. Tab. 2. Phenotype of F<sub>1</sub> XY fish with <i>amhy</i>/<i>amhΔ-y</i> mutations. 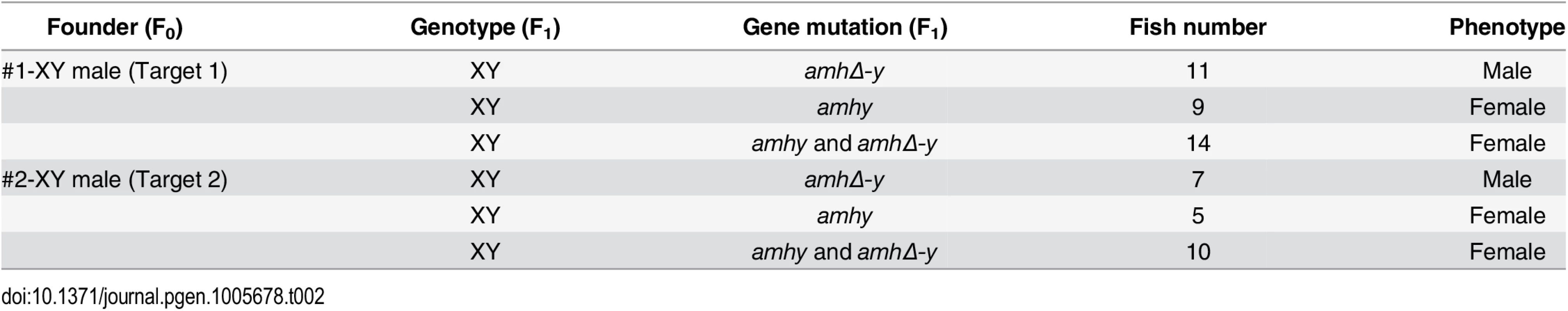
Overexpression of Y156 fosmid or Amhy ORF causes female to male sex reversal in F0 XX fish
Gonad histological analysis revealed that 8 (32%) of 25 Y156 fosmid transgenic F0 XX fish displayed complete female to male sex reversal at 90 dah (Fig 5a, Table 1). The sex-reversed gonads exhibited a clear testicular structure. IHC showed that they expressed the testicular specific gene Dmrt1, and did not express the ovarian specific Cyp19a1a (Fig 5aA–5aF). The integration and mRNA expression of the transgene in the XX fry were examined and confirmed by genomic PCR and RT-PCR, using amhΔ-y specific primers, at 3 and 5 dah (Fig 5b).
Fig. 5. Transgenic overexpression of Y156 fosmid and Amhy ORF resulted in sex reversal in F0 XX fish. 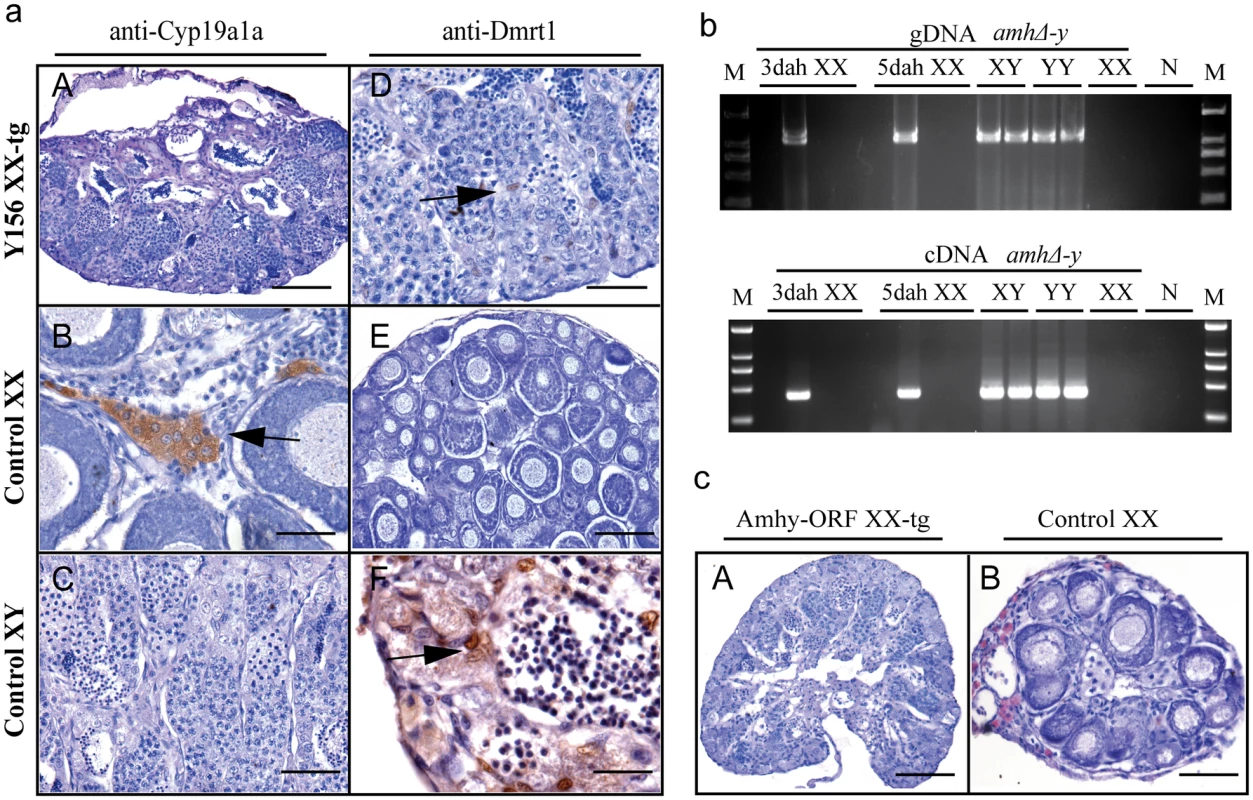
(a) The overexpression of fosmid Y156 containing amhy in F0 XX fish resulted in sex reversal (8 (31.6%) of 25), which was histologically indistinguishable from the control testis. Immunohistochemistry analysis showed that Cyp19a1a was absent from the sex-reversed XX fish gonads, like the control testis, but unlike the control ovary (A-C). In contrast, like the control testis but different from the control ovary, Dmrt1 was expressed in the sex-reversed XX fish gonads (D-F). Scale bars, 50μm (A, E); 15μm (B, D); 25μm (C); 10μm (F). (b) Integration of the transgene and mRNA expression in the XX fry confirmed by genomic PCR and RT-PCR using amhΔ-y specific primers at 3 and 5 dah. Successful transgene of amhy was indirectly reflected by amhΔ-y mRNA detection. gDNA, genomic DNA; dah, days after hatching; N, negative control. M, DNA molecular standard. (c) Transgenic overexpression of amhy ORF in F0 XX fish resulted in sex reversal. H.E. staining gonads from the 3-month-old XX transgenic fish displayed a clear testicular structure (n = 9), while gonads from the negative transgene fish showed typical ovarian structure (n = 21). Scale bars, 50 μm. To investigate the ability of the amhy ORF to induce sex reversal, we constructed overexpression vector pIRES-hrGFP-1a in which CMV controlled the amhy cDNA. Morphologically, the amhy-transgene XX F0 fish also displayed sex reversal with a clear testicular structure (n = 9), while no sex reversal was found in the control XX group (n = 21) at 90 dah (Fig 5c). In contrast, the amh or amhΔ-y-transgene XX F0 fish displayed no sex reversal with a clear ovarian structure at 90 dah (amh, n = 11; amhΔ-y, n = 10) (S14 Fig, Table 1).
AmhrII knockout causes male to female sex reversal in XY fish
Two gRNAs were designed in the exon II and III of amhrII (Fig 6a), an autosomal gene located on LG20. In total, 73% (22/30) of target 1 and 50% (15/30) of target 2 were mutated by CRISPR/Cas9. The mutation frequencies within individuals ranged from 22% to 68% (S3 Table). All fish with amhrII mutations showed complete male to female sex reversal, as visualized by the formation of the ovarian cavity (OC) and the appearance of pre-vitellogenic oocytes when checked by gonad histology at 90 dah (Fig 6b). IHC analysis showed that Cyp19a1a was expressed like the control ovary, while Dmrt1 was not expressed in these sex-reversed XY gonads (Fig 6bA–6bD). The effect of AmhrII deficiency on gonadal differentiation could be detected from a very early stage. Cyp19a1a was expressed, while Dmrt1 was not expressed in AmhrII deficient XY gonads even at 10 and 30 dah (Fig 6bE–6bL). Consistently, knockout of amhrII in the XY fish resulted in increased serum E2 level compared with the control fish (Fig 6c). The genetic sex (XY) of these sex-reversed fish was further confirmed by PCR of a sex-linked marker (SPF/SPR) (Fig 6d).
Fig. 6. Knockout of amhrII by CRISPR/Cas9 resulted in male to female sex reversal in XY fish. 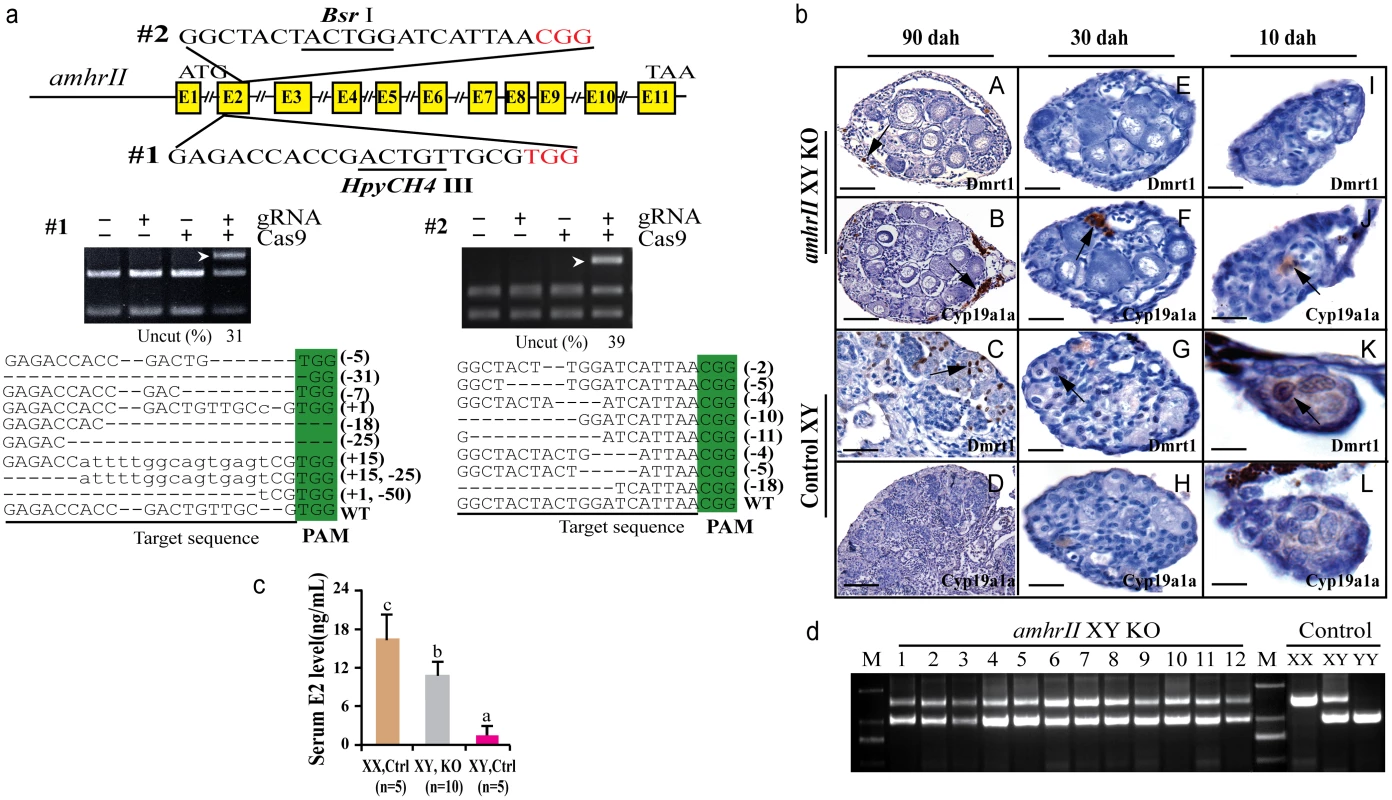
(a) Two target sites were selected in the exonII and III of amhrII. The target sites contain Bsr I and HpyCH4 III adjacent to PAM for mutation analysis. The un-cleaved bands were observed in the knockout group after Bsr I and HpyCH4 III digestion, while the DNA of the control groups were completely digested. The percentage of uncleaved band is shown below the gel images. Mutation sequences from the un-cleaved bands are listed. (b) The amhrII knockout XY fish exhibited male to female sex reversal as demonstrated by gonad histology and immunohistochemistry (IHC). IHC analysis showed that, like the control XX gonads but unlike the control XY gonads, Cyp19a1a was expressed in the sex-reversed XY gonads at 10, 30 and 90 dah (B, D, F, J, H, L). In contrast, Dmrt1 was absent from the sex-reversed XY gonads, which was distinct from the control XY gonads at 10, 30 and 90 dah (A, C, E, I, G, K). (c) Knockout of amhrII in the XY fish resulted in increased serum E2 level compared with the control fish. Results are presented as the mean±SD. Different letters (a, b, c) indicate statistical differences at P<0.05 as determined by one-way ANOVA followed by Tukey/Kramer post hoc test. Sample numbers are shown. (d) Genotype of the sex-reversed XY fish was confirmed with a sex specific marker (SPF/SPR). 1–12, sex-reversed fish with amhrII knockout. M, DNA molecular standard. Scale bars, 50μm (A, B); 15μm (C, D, E-H); 10μm (I-L). Discussion
In recent years, several different master SD genes have been identified in various fish species, giving the impression that the molecular mechanism underlying sex determination is different in each group. However, the limited data available for teleost SD genes suggest that members of the TGF-β superfamily (gsdfY, amhy and amhrII) could be part of a common pathway for sex determination in fish [11–13]. Tilapia sex determination has been widely studied with the goal of producing all-male progenies that have an enhanced growth rate in aquaculture. In Nile tilapia, some studies using fish from Egypt and Ghana have indicated that the main sex determining locus is on LG1 (with unresolved evidence regarding LG3) [35–37]. Recently, a sex-determining locus was mapped adjacent to the amh on LG23 in the Swansea strain of this species, using Simple Sequence Repeats (SSR) and sex specific markers [21, 24]. Thereafter, amhy, a Y-specific duplicate of the amh gene, was identified and suggested to be the candidate sex determiner [23]. However, this gene, named as amhΔ-y by us, is not responsible for sex determination in our strain. Another duplicated copy of amh with a missense SNP and a large fragment of promoter loss, which is located immediately downstream of amhΔ-y and designated as amhy, was found and demonstrated to be critical for male sex determination in our strain. Therefore, it is possible that different strain possesses different sex chromosome and different SD genes. Judging from the assembled sequence, it is reasonable to conclude that amhy arose from duplication of amh gene followed by 5608 bp promoter loss. amhΔ-y is an allele of the X-linked amh as most of the sequences in the upstream promoter of amhΔ-y were identical to the upstream of amh on the X chromosome.
amhy could be considered a candidate SD gene of the Nile tilapia because of its Y specific expression profile. Due to the high homology of three amh, four pairs of gonadal transcriptomes from different development stages were used to analyze their expression. It was found that amhy expression was restricted to XY testis, with expression at the beginning of critical sex determination period 5 dah, clearly preceding the first signs of morphological differentiation of ovaries and testes (~25 dah) [38]. According to the transcriptome data, both amhy and amh mRNA were detected in the XY gonads at 5 dah. Therefore, Western blot results indirectly suggested the expression of Amhy protein in XY gonads at 5 dah. In contrast, amh expression in XY gonads was much lower than that of XX gonads at 5 dah. Although the amhΔ-y was also found to be restricted to XY testis, we found there is a 5 bp insertion in exonVI of the amhΔ-y in addition to the previously reported TGF-β domain 233 bp deletion in exonVII [23]. It is this frameshift mutation that generates a truncated Amh lacking the TGF-β domain, which is important for binding to AmhrII. The truncated Amh could not directly bind to AmhrII even if the 233 bp sequence in exonVII was not deleted. Therefore, amhΔ-y might be a degenerated gene in tilapia.
The best way to understand the function of a gene in sex determination is to perform gain/loss of function studies and to characterize the resulting biological effects. For instance, overexpression of a 117 kb genomic DNA fragment that carries dmy in XX medaka, or the presence of a genomic fragment that included gsdfY, converts XX individuals into sex-reversed XX males [13, 39]. Knockdown of the Patagonian pejerrey amhy in XY fish led to an up-regulation of female factors and the development of ovaries [11]. In the present study, knockout of amhy or both amhy and amhΔ-y using CRISPR/Cas9 resulted in ovarian development in XY fish, while mutation of amhΔ-y alone could not. In contrast, overexpression from the amhy genomic region or its ORF under the control of CMV promoter induced testicular differentiation in XX fish, while overexpression of amhΔ-y alone in XX fish resulted in no sex reversal. Therefore, we demonstrated that amhy is necessary and sufficient to induce testicular differentiation in tilapia. A detailed analysis showed that the missense SNP and the large fragment promoter loss in amhy might contribute to sex determination in tilapia. Recent studies reported that a few regulatory or coding sequence mutations in other pre-existing and duplicated genes can generate new SD genes in fishes. For example, a few differences in the cis-regulatory region of gsdfY and sox3Y contribute to male sex determination in two medaka species. A missense SNP in AmhrII is the only difference associated with phenotypic sex in Takifugu rubripes. In this study, overexpression of amhy ORF in XX fish led to sex reversal, while overexpression of amh in XX fish resulted in no sex reversal, implying that the missense SNP (C/T) in the coding sequence might contribute to male sex determination in tilapia. In another Nile tilapia strain, a missense SNP in exonVI of amh was also associated with autosomal and temperature dependent sex reversal [40]. These studies indicated that small variations in the coding sequence of amh might have taken over a critical role in tilapia sex determination. The important characteristic of a master SD gene is its tight linkage with the non-recombinant part of the heterochromosome. Right now, we do not know the size of the male-specific non-recombining region and how many genes are located in the region in tilapia. Therefore, amhy is considered as the candidate sex determining gene in this strain of the Nile tilapia.
In this report, an XY specific up-regulation was detected in the expression of amhrII in the gonads from 5 dah onwards, coincident with sex determination in tilapia. Higher levels of amhrII expression in testis have been consistently observed in four tilapia species and several other fishes [12, 28, 41]. Importantly, mutations in amhrII in medaka and Takifugu rubripes lead to male to female sex reversal [12, 29]. In the present study, knockout of amhrII in XY fish resulted in 100% male to female sex reversal in tilapia. However, as in medaka, but unlike in Takifugu rubripes, amhrII is an autosomal gene located on LG20 in Nile tilapia. It is well documented that AmhrII is the receptor for Amh in mammals [25]. However, the receptors for amhy and amh in fish have not been identified. Notably, the expression profiles of amh and amhy was similar to that of amhrII during tilapia gonadal development and these factors were found to be co-localized in cells surrounding the germ cells at 5 dah. Both Amhy and Amh have the identical TGF-β domain, which is responsible for binding to its receptor. According to these results, Amhy might be the ligand of AmhrII. Therefore, Amhy signal functions through AmhrII to determine sex determination in tilapia. Further investigations may reveal the mechanism by which Amhy/AmhrII signal pathway determines tilapia male sex. In addition, knockout of amhrII resulted in 100% sex reversal, while knockout of amhy in F0 fish XY only resulted in about 60% sex reversal. As the F0 fish were mosaic, the mutation rate varies individually. Therefore, only some of the F0 knockout fish displayed sex reversal. A thorough analysis of gene mutation of F1 knockout fish indicated that the ratio for fish bearing amhy frameshift mutation versus fish bearing frameshift mutation is approximately 60% (23/34), which is exactly the ratio of sex reversed fish to F1 positive fish. The probability for amhy and amhΔ-y mutation in different cell types of F0 fish should be the same as that of the germ cells. This explains why only 60% of the F0 positive fish displayed male to female sex reversal. In contrast, knockout of amhrII is equal to disrupt whole signal pathway, and therefore, resulted in 100% sex reversal.
amhy was first reported as the sex determiner in Patagonian pejerrey [11]. Existing evidence support the notion that the Y-specific duplication of Amh arose independently in tilapia and Patagonian pejerrey. This study provides a new example of convergent evolution for the formation of SD gene. Although there are reports showing sex determination roles for Amh and AmhrII in several species, our results are the first that demonstrate that both genes are critical for sex determination in a single species (Table 3). Even though Amh and AmhrII are not the master sex determination genes in mammals, chicken, and other fish species, they are also essential for testicular differentiation [3, 25, 28, 42, 43]. For example, loss-of-function mutants of AMH in the male mouse lead to partial hermaphroditism, with the uterus and oviduct present along with the testis, but no ovaries [25]. Our study highlights the significance of TGF-β signaling pathway in fish sex determination. Its role in sex determination and differentiation in other vertebrates deserves further investigation.
Tab. 3. Overview of the <i>amh</i>, <i>amhy</i>, <i>amhΔ-y</i> and <i>amhrII</i> in tilapia sex determination. 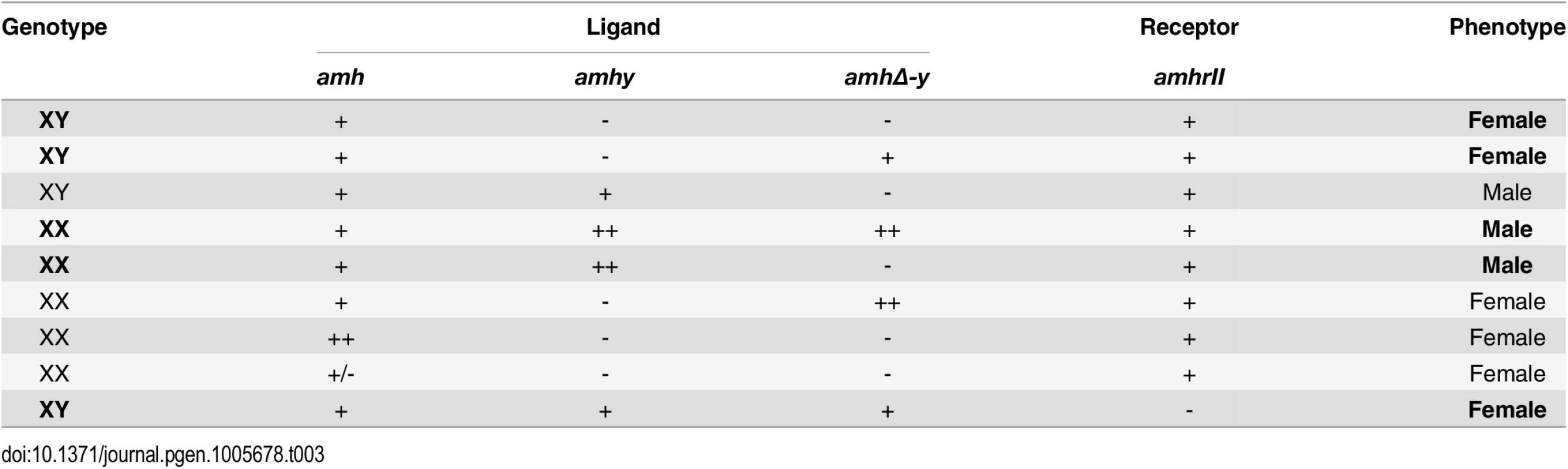
Estrogen plays a critical role in ovarian differentiation and maintenance in a variety of vertebrates [44–48]. For instance, administration of estrogens can reverse phenotypic males to females in marsupials [49], birds [50], reptiles [51], and teleosts [52, 53]. Foxl2 is a key factor involved in female sex determination in vertebrates, including fishes and mammals [9, 34, 46, 54]. It is worth noting that Foxl2 directly activates the expression of cyp19a1a, encoding aromatase, a key enzyme responsible for estrogen production in tilapia [44], goat [55], mouse [54], and human cells [56]. A possible mechanism of Amh/AmhrII action in fish is through suppression of aromatase expression, decreasing estrogen levels so as to promote testis formation, as has been described in mammals and birds [57, 58]. Knockdown of Patagonian pejerrey amhy in XY fish resulted in up-regulation of foxl2 and cyp19a1a expression [11]. Similar results were also obtained in hotei mutant medaka [29]. Consistent with these reports, knockout of amhy and amhrII in XY tilapia resulted in increased Cyp19a1a expression and serum E2 levels during sex reversal. On the other hand, amhy/amh transgenic overexpression of amhy in XX fish displayed no Cyp19a1a expression. Therefore, Amh/AmhrII signaling might play a critical role in male sex determination via regulation of the Foxl2-aromatase pathway in teleosts.
In conclusion, our results suggest that the tandem duplicated amhy is essential for male sex determination in the Nile tilapia. Mutation of amhy in XY fish resulted in male to female sex reversal and mutation of amhΔ-y could not, while overexpression of amhy in XX fish resulted in female to male sex reversal. Further, knockout of the amh type II receptor (amhrII) in XY fish also resulted in male to female sex reversal. Our findings highlight the significance of TGF-β signaling pathway in fish sex determination. Amhy/AmhrII play a critical role in the regulation of sex determination, probably via regulation of aromatase expression in teleosts. The role of this pathway in sex determination of other vertebrates deserves further investigation.
Materials and Methods
Ethics statement
Animal experiments were conducted in accordance with the regulations of the Guide for Care and Use of Laboratory Animals and were approved by the Committee of Laboratory Animal Experimentation at Southwest University.
Fish
The founder strain of the Nile tilapia, which was first introduced from Egypt in Africa, was obtained from Prof. Nagahama (Laboratory of Reproductive Biology, National Institute for Basic Biology, Okazaki, Japan) and reared in large tanks with a circulating aerated freshwater system. All-XX and all-XY progenies were obtained by crossing sex-reversed XX pseudomales or YY supermales with normal females (XX).
Screening of tilapia's fosmid library and sequence assembling
The microarrayed fosmid library of XY tilapia genomic DNA was constructed using the pCC2FOS vector (Epicentre, USA) according to the manufacturer's protocol [30]. X - and Y - specific clones were isolated by PCR screening of the library using a pair of sex specific primers (SPF: ATGGCTCCGAGACCTTGACTG; SPR: CAGAAATGTAGACGCCCAGGTAT) from marker-5 which amplified a 1422 bp fragment from the X chromosome and a 982 bp fragment from the Y chromosome [22]. DNA sequencing was carried out using Illumina HiSeq2000 technology by Invitrogen Corporation (Shanghai, China). The sequence was assembled by hand together with the local Blast software based on the following rationale: 1) the number of reads from the duplicated regions of Y156 were approximately twice of the un-duplicated region by Local Blast analysis with the sequence of X278 which is identical to the released genome sequences; 2) some reads from the Y156 fosmid displayed differential sequences (deletions and insertions) and SNPs; 3) some primers designed spanning the differential region can produce two fragments with the Y156 fosmid and YY genomic DNA while one band with X278 fosmid and XX genomic DNA, which can be used to help scaffold together the Illumina reads. To confirm the assembled sequences, twenty five PCR fragments were amplified and sequenced using 25 pair primers which were designed in the differential regions of amhy and amhΔ-y, as indicated in S15 Fig. The differences between the Y and X fosmid were further confirmed by PCR using XX, XY and YY fish genomic DNA as templates. Primers were listed in S4 Table.
DNA, RNA extraction and cDNA synthesis
Tail fin was clipped from XX, XY and YY fish and incubated for 3 hrs at 55°C in lysis buffer containing 0.5% sodium dodecyl sulfate (SDS), 25 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 10 mM Tris-HCl (pH 8.0), and 200 μg/ml of proteinase K. The lysate was extracted with phenol/chloroform and precipitated with isopropanol. The extracted DNA was dissolved, quantified and then used as template for subsequent analyses.
Total RNA was extracted from various tissues (brain, pituitary, gill, heart, liver, spleen, intestine, ovary, testis, kidney, Muscle and head kidney) of pooled three XX and three XY fish at 180 dah using a column-based RNA extraction kit (Qiagen, Germany). After DNase I (RNase free) treatment, total RNA (500 ng) from each sample was reverse transcribed into first-strand cDNA using PrimeScript RT Master Mix Perfect Real Time Kit (Takara, Japan) according to the manufacturer's instructions. For each sample, in order to confirm that DNase I (RNase free) treatment of the RNA was complete, a negative control cDNA synthesis reaction without reverse transcriptase was performed.
Tissue distribution analysis by RT-PCR
RT-PCR was performed to reveal the tissue distribution expression patterns of amh, amhy, amhΔ-y, and amhrII. Positive and negative controls were set up with plasmid DNA and negative control cDNA, respectively. β-actin was used as an internal control. Primer sequences used for RT-PCR were listed in S4 Table. The PCR conditions were as follows: after an initial denaturation at 94°C for 3 min, a 33-cycle reaction was carried out at 94°C for 30 s, 58°C for 30 s and 72°C for 30 s. For β-actin amplification, a 25-cycle reaction was used with an extension time of 45 s, other conditions being identical.
Transcriptome analysis
Four pairs of XX and XY gonads from tilapia at 5, 30, 90 and 180 dah were sequenced using Illumina2000 HiSeq technology in our previous study [32]. The sequence with 5 bp (ATGTC) insertion in exonVI or the missense SNP (C/T) in exonII was used as query sequence (60 bp in length) to blast against the transcriptome clean reads using local BLAST software. amh and amhy were analyzed by counting reads with the SNP (C/T) in exonII. The expression of amhΔ-y was analyzed by blast in the transcriptome data with the amhΔ-y specific 5 bp (ATGTC) insertion in exonVI. A normalized measure of RPKM (reads per kb per million reads) was used to normalize the expression profiles of amhy, amhΔ-y, amh and amhrII. In addition, to get more precise expression data for amhy and amh in the 5 dah XY and XX gonads, two more pairs of gonadal transcriptomes from 5 dah fish (totally six gonadal transcriptomes, 3 from XX fish and 3 from XY fish), were sequenced to analyze amhy and amh expression.
Production, characterization of antibody and western blot analysis
The production of Amh polyclonal antibody was performed as follows: The recombinant constructs of amh and amhΔ-y were prepared by cloning the ORFs of these genes into the pCold I vector (Takara, Japan). Recombinant Amh and AmhΔ-y were expressed and purified. The Amh was used as the antigen used to immunize rabbits for the production of polyclonal antibody. Ten days after the last immunization, rabbit serum was collected and recombinant protein purified by affinity chromatography on Sepharose 4B Fast Flow Resin (Sigma, Germany). Subsequently, the purified ployclonal antibody was evaluated by Western blotting. Briefly, total proteins extracted from XX and XY gonads from 5, 10, 30, 90, 180, 300 dah, YY gonads from 30 and 90 dah tilapia and the recombinant proteins (both purified and unpurified) were separated using 12% SDS-PAGE under reducing condition. Notably, the proteins used for 5 and 10 dah Western blot was extracted from the fish after removing head, tail, viscera and muscle. Separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes and then blocked with 5% fat milk and incubated with primary antibody of Amhy at 1 : 500 dilution, and then with a second antibody conjugated with horseradish peroxidase (Bio-Rad, USA) at 1 : 2000. Finally, the immunoreactive signals were detected with BeyoECL Plus Kit (Beyotime, China) and visualized on Fusion FX7 (Vilber Lourmat, France).
Immunohistochemistry (IHC)
Gonads of 5-, 30-, 90 - and 180 - dah fish were dissected, fixed in Bouin's solution for 24 hrs at room temperature, and subsequently dehydrated, embedded in paraffin and serially sectioned at 5 μm thickness. IHC was performed to determine the cellular localization of Amh/AmhΔ-y/Amhy and AmhrII in gonads using the Amh and AmhrII antibodies at 1 : 500, 1 : 1000 dilution respectively. IHC staining was carried out as follows: After washing with PBS, the sections were treated in blocking solution, incubated with the primary rabbit polyclonal antibodies overnight at 4°C. Subsequently, the sections were incubated with a second antibody conjugated with horseradish peroxidase (Bio-Rad, USA) at 1 : 2000. Immunoreactive signals were visualized using diaminobenzidine (Sigma, USA) as substrate. Sections were counterstained with hematoxylin.
gRNA design and Cas9 mRNA in vitro transcription
Two gRNA target sites were selected for each gene on the sense or antisense strand of amh/amhy, amhrII and gsdf with ZIFIT Targeter. BLAST with the tilapia genome was performed to avoid off-targets according to the principles reported previously [59]. In addition, a restriction enzyme cutting site adjacent to the NGG (PAM) was included for convenient mutagenesis analysis. The DNA template preparation, PCR conditions, gRNA in vitro transcription, gRNA purification, Cas9 mRNA in vitro transcription and purification were carried out as described previously [34]. In vitro transcription was performed with the Megascript T7 Kit (Ambion, USA) for 4 hrs at 37°C using 500 ng purified DNA as templates. The Cas9 plasmids were linearized with Xba I and purified by ethanol precipitation as templates. Cas9 mRNA was produced by in vitro transcription of 1 μg linearized template DNA with a T7 mMESSAGE mMACHINE Kit (Ambion, USA) according to the manufacturer’s instructions.
Knockout of amhy and amhrII in tilapia
About 500 pg of the gRNA (150 ng/μl) and Cas9 mRNA (500 ng/μl), mixed at a molar ratio of 1 : 1, was microinjected directly into XX or XY fertilized eggs. Mutated fish were identified by loss of the restriction enzyme site. Mutation efficiencies and sequences of the mutated targets were evaluated by restriction enzyme digestion and Sanger sequencing as follows: the DNA fragments spanning the target for each fish were amplified. The recovered PCR products were purified and digested by restriction enzyme within the target. The uncleaved bands were recovered, sequenced and then aligned with the wild type. In addition, the percentage of uncleaved band (i.e., potential mutations in target site) was measured by quantifying the band intensity of the restriction enzyme digestion with Quantity One Software (Bio-Rad, USA). The indel frequency was calculated by dividing uncleaved band intensity to the total band intensity.
All the procedures for RNA and protein extraction from 90 dah control (n = 5) and F0 fish gonads (n = 5), cDNA synthesis, RT-PCR and Western blot were performed as described above. Two forward primers were designed in the target sites, together with reverse primers in another exon, for detection of amh/amhΔ-y/amhy mRNA expression in mutated gonads by RT-PCR. The primers were listed in S4 Table.
Transgenic overexpression of Y156 fosmid, Amhy or AmhΔ-y ORF in XX fish
Not I linearized fosmid clone containing amhy (Y156) was injected into all XX fertilized eggs of tilapia. To detect amhΔ-y and amhy mRNA expression in transgenic fish, total RNA was extracted from the whole bodies of XX fry after removal of the yolk at 3 and 5 dah. All the procedures for RNA extraction, cDNA synthesis, and RT-PCR using Y specific primers (amhΔ-y-F1/R1) were carried out as described above. The detection of amhΔ-y expression was indirectly stated the successful overexpression of amhy. PCR was performed with Y specific primers (amhΔ-y-F1/R1) to detect the transgene in extracted genomic DNA (S4 Table).
Overexpression of Amhy or AmhΔ-y alone in XX fish was performed as follows: the amhy or amhΔ-y ORF was subcloned into the multiple cloning sites downstream of the cytomegalovirus (CMV) promoter of the pIRES-hrGFP-1a vector. Transgenic overexpression was carried out by injection of these constructs into the blastodisc of fertilized eggs of XX population. Genomic DNA extraction and positive fish screening were performed as described previous study [44]. Additionally, the transgenic fish gonads were subjected to histological and IHC assays to examine the effects of Amhy overexpression on sex determination.
Phenotype, histological and IHC analyses of F0 knockout and overexpression fish
Gonads of amh/amhΔ-y/amhy, or amhrII knockout and control fish were dissected at 10, 30 and 90 dah. After fixation in Bouin's solution for 24 hours at room temperature, they were dehydrated and embedded in paraffin. Tissue blocks were sectioned at 5 μm and stained with hematoxylin and eosin for histological analysis or used for IHC. IHC using Dmrt1 and Cyp19a1a antibodies, which were diluted at 1 : 100, and 1 : 2500 respectively, was performed. Additionally, the gonads of fish overexpressing Y156 fosmid or amhy were subjected to histological and IHC assays at 90 dah after injection. The genotypes of all sex reversed fish were confirmed by a pair of sex specific primer (SPF/SPR) (S4 Table).
Estrogen assay
Blood samples were collected from the caudal veins of the 3-month-old knockout (F0) as well as control fish. Serum estradiol-17β (E2) levels were measured using the E2 enzyme immunoassay kits (Cayman, USA). Sample purification and assays were performed according to the manufacturer's instructions.
Production and gonadal phenotype analysis of the F1 XY fish
The amh/amhΔ-y/amhy mutant XY fish (target 1 and 2) with the moderate indel frequency were randomly selected as F0 founders. They were raised to sexual maturity and mated with wild-type XX tilapia to produce F1 fish. At 90 dah, the genomic DNA from F1 fish was extracted individually for genotyping and mutation assays. The genotype of all F1 fish were determined by a pair of sex specific primer (SPF/SPR). For F1 XY individual mutation assays, one pair of gene specific primers was designed to ensure specific amplification of amhy and amhΔ-y respectively in the first round of PCR. Then, the first round PCR products were diluted and used as template for the second round of PCR using the knockout fish screening primers (S4 Table). Restriction enzyme BstN I digestion of the amplified fragments from second PCR and Sanger sequencing were performed to confirm mutation types of F1 fish. The SNPs near the two targets were used to distinguish amhΔ-y and amhy. The fish were processed for histological analysis to analyze their gonadal phenotype.
Supporting Information
Zdroje
1. Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, et al. (1990) A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346 : 240–244. 1695712
2. Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R (1991) Male development of chromosomally female mice transgenic for Sry. Nature 351 : 117–21. 2030730
3. Takehana Y, Matsuda M, Myosho T, Suster ML, Kawakami K, et al. (2014) Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat Commun 5 : 4157. doi: 10.1038/ncomms5157 24948391
4. Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, et al. (2009) The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461 : 267–271. doi: 10.1038/nature08298 19710650
5. Chen S, Zhang G, Shao C, Huang Q, Liu G, et al. (2014) Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46 : 253–260. doi: 10.1038/ng.2890 24487278
6. Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, et al. (2008) A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci USA 105 : 2469–2474. doi: 10.1073/pnas.0712244105 18268317
7. Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, et al. (2002) DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417 : 559–563. 12037570
8. Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, et al. (2002) A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci USA 99 : 11778–11783. 12193652
9. Boulanger L, Pannetier M, Gall L, Allais-Bonnet A, Elzaiat M, et al. (2014) FOXL2 is a female sex-determining gene in the goat. Curr Biol 24 : 404–408. doi: 10.1016/j.cub.2013.12.039 24485832
10. Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, et al. (2012) An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr Biol 22 : 1423–1428. doi: 10.1016/j.cub.2012.05.045 22727696
11. Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, et al. (2012) A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci USA 109 : 2955–2959. doi: 10.1073/pnas.1018392109 22323585
12. Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, et al. (2012) A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet 8: e1002798. doi: 10.1371/journal.pgen.1002798 22807687
13. Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, et al. (2012) Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191 : 163–170. doi: 10.1534/genetics.111.137497 22367037
14. Mank JE, Avise JC (2009) Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev 3 : 60–67. doi: 10.1159/000223071 19684451
15. Ross JA, Urton JR, Boland J, Shapirom MD, Peichel CL (2009) Turnover of sex chromosomes in the stickleback fishes (gasterosteidae). PLoS Genet 5: e1000391. doi: 10.1371/journal.pgen.1000391 19229325
16. Bradley KM, Breyer JP, Melville DB, Broman KW, Knapik EW, et al. (2011) An SNP-Based Linkage Map for Zebrafish Reveals Sex Determination Loci. G3 (Bethesda) 1 : 3–9.
17. Wilson CA, High SK, McCluskey BM, Amores A, Yan YL, et al. (2014) Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics 198 : 1291–1308. doi: 10.1534/genetics.114.169284 25233988
18. Mair GC, Scott AG, Penman DJ, Beardmorem JA, Skibinski DO (1991) Sex determination in the genus Oreochromis: 1. Sex reversal, gynogenesis and triploidy in o. niloticus (L.). Theor Appl Genet 82 : 144–152. doi: 10.1007/BF00226205 24213058
19. Baroiller JF, D'Cotta H, Bezault E, Wessels S, Hoerstgen-Schwark G (2009) Tilapia sex determination: Where temperature and genetics meet. Comp Biochem Physiol A Mol Integr Physiol 153 : 30–38. doi: 10.1016/j.cbpa.2008.11.018 19101647
20. Lee BY, Penman DJ, Kocher TD (2003) Identification of a sex-determining region in Nile tilapia (Oreochromis niloticus) using bulked segregant analysis. Anim Genet 34 : 379–383. 14510676
21. Eshel O, Shirak A, Weller JI, Slossman T, Hulata G, et al. (2011) Fine-mapping of a locus on linkage group 23 for sex determination in Nile tilapia (Oreochromis niloticus). Anim Genet 42 : 222–224. doi: 10.1111/j.1365-2052.2010.02128.x 24725231
22. Sun YL, Jiang DN, Zeng S, Hu CJ, Ye K, et al. (2014) Screening and characterization of sex-linked DNA markers and marker-assisted selection in the Nile tilapia (Oreochromis niloticus). Aquaculture 433 : 19–27.
23. Eshel O, Shirak A, Dor L, Band M, Zak T, et al. (2014) Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics 15 : 774. doi: 10.1186/1471-2164-15-774 25199625
24. Eshel O, Shirak A, Weller JI, Hulata G, Ron M (2012) Linkage and Physical Mapping of Sex Region on LG23 of Nile Tilapia (Oreochromis niloticus). G3. 2 : 35–42. doi: 10.1534/g3.111.001545 22384380
25. Josso N, di Clemente N, Gouédard L (2001) Anti-Müllerian hormone and its receptors. Mol Cell Endocrinol 179 : 25–32. 11420127
26. Miura T, Miura C, Konda Y, Yamauchi K (2002) Spermatogenesis-preventing substance in Japanese eel. Development 129 : 2689–2697. 12015296
27. Yoshinaga N, Shiraishi E, Yamamoto T, Iguchi T, Abe S, et al. (2004) Sexually dimorphic expression of a teleost homologue of Müllerian inhibiting substance during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem Biophys Res Commun 322 : 508–513. 15325259
28. Wu GC, Chiu PC, Lyu YS, Chang CF (2010) The expression of amh and amhr2 is associated with the development of gonadal tissue and sex change in the protandrous black porgy, Acanthopagrus schlegeli. Biol Reprod 83 : 443–453. doi: 10.1095/biolreprod.110.084681 20505169
29. Morinaga C, Saito D, Nakamura S, Sasaki T, Asakawa S, et al. (2007) The hotei mutation of medaka in the anti-Müllerian hormone receptor causes the dysregulation of germ cell and sexual development. Proc Natl Acad Sci USA 104 : 9691–9696. 17535919
30. Li MH, Wu FR, Xiong CQ, Zeng S, Yang SJ, et al. (2011) Construction of microarray fosmid library and its application in gene isolation in Nile tilapia, Oreochromis niloticus [in Chinese]. Journal of Fisheries of China 35 : 28–34.
31. Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, et al. (2014) The genomic substrate for adaptive radiation in African cichlid fish. Nature 513 : 375–381. doi: 10.1038/nature13726 25186727
32. Tao W, Yuan J, Zhou L, Sun L, Sun Y, et al. (2013) Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS One 8: e63604. doi: 10.1371/journal.pone.0063604 23658843
33. Li MH, Yang HH, Li MR, Sun YL, Jiang XL, et al. (2013) Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology 154 : 4814–4825. doi: 10.1210/en.2013-1451 24105480
34. Li M, Yang H, Zhao J, Fang L, Shi H, et al. (2014) Efficient and Heritable Gene Targeting in Tilapia by CRISPR/Cas9. Genetics 197 : 591–599. doi: 10.1534/genetics.114.163667 24709635
35. Ezaz MT, Harvey SC, Boonphakdee C, Teale AJ, McAndrew BJ, et al. (2004) Isolation and physical mapping of sex-linked AFLP markers in Nile tilapia (Oreochromis niloticus L.). Mar Biotechnol 6 : 435–445. 15791488
36. Cnaani A, Kocher TD. (2008) Sex-linked markers and microsatellite locus duplication in the cichlid species Oreochromis tanganicae. Biol Lett. 4 : 700–3. doi: 10.1098/rsbl.2008.0286 18700198
37. Gammerdinger WJ, Conte MA, Acquah EA, Roberts RB, Kocher TD. (2014) Structure and decay of a proto-Y region in Tilapia, Oreochromis niloticus. BMC Genomics. 15 : 975. doi: 10.1186/1471-2164-15-975 25404257
38. Ijiri S, Kaneko H, Kobayashi T, Wang DS, Sakai F, et al. (2008) Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol Reprod 78 : 333–341. 17942796
39. Matsuda M, Shinomiya A, Kinoshita M, Suzuki A, Kobayashi T, et al. (2007) DMY gene induces male development in genetically female (XX) medaka fish. Proc Natl Acad Sci USA 104 : 3865–3870. 17360444
40. Wessels S, Sharifi RA, Luehmann LM, Rueangsri S, Krause I, et al. (2014) Allelic variant in the anti-Müllerian hormone gene leads to autosomal and temperature-dependent sex reversal in a selected Nile tilapia line. PLoS One. 9:e104795. doi: 10.1371/journal.pone.0104795 25157978
41. Böhne A, Sengstag T, Salzburger W (2014) Comparative transcriptomics in East African cichlids reveals sex and specied specific expression and new candidated for sex differentiation in fishes. Genome Biol Evol 6 : 2567–2585. 25364805
42. Cutting AD, Ayers K, Davidson N, Oshlack A, Doran T, et al. (2014) Identification, expression, and regulation of anti-Müllerian hormone type-II receptor in the embryonic chicken gonad. Biol Reprod 90 : 106. doi: 10.1095/biolreprod.113.116491 24621923
43. Horiguchi R, Nozu R, Hirai T, Kobayashi Y, Nagahama Y, et al. (2013) Characterization of gonadal soma-derived factor expression during sex change in the protogynous wrasse, Halichoeres trimaculatus. Dev Dyn 242 : 388–399. doi: 10.1002/dvdy.23929 23335393
44. Wang DS, Kobayashi T, Zhou LY, Paul-Prasanth B, Ijiri S, et al. (2007) Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol Endocrinol 21 : 712–725. 17192407
45. Guiguen Y, Fostier A, Piferrer F, Chang CF (2010) Ovarian aromatase and estrogens: a pivotal role for gonadal sex differentiation and sex change in fish. Gen Comp Endocrinol 165 : 352–366. doi: 10.1016/j.ygcen.2009.03.002 19289125
46. Okada E, Yoshimoto S, Ikeda N, Kanda H, Tamura K, et al. (2009) Xenopus W-linked DM-W induces Foxl2 and Cyp19 expression during ovary formation. Sex Dev 3 : 38–42. doi: 10.1159/000200080 19339816
47. Matsumoto Y, Buemio A, Chu R, Vafaee M, Crews D (2013) Epigenetic control of gonadal aromatase (cyp19a1) in temperature-dependent sex determination of red-eared slider turtles. PLoS One 8: e63599. doi: 10.1371/journal.pone.0063599 23762231
48. Lambeth LS, Cummins DM, Doran TJ, Sinclair AH, Smith CA (2013) Overexpression of aromatase alone is sufficient for ovarian development in genetically male chicken embryos. PLoS One 8: e68362. doi: 10.1371/journal.pone.0068362 23840850
49. Coveney D, Shaw G, Renfree MB (2001) Estrogen-induced gonadal sex reversal in the tammar wallaby. Biol Reprod 65 : 613–621. 11466233
50. Scheib D (1983) Effects and role of estrogens in avian gonadal differentiation. Differentiation 23: S87–S92. 6444180
51. Merchant-Larios H, Ruiz-Ramirez S, Moreno-Mendoza N, Marmolejo-Valencia A (1997) Correlation among thermo sensitive period, estradiol response, and gonad differentiation in the sea turtle Lepidochelys olivacea. Gen Comp Endocrinol 107 : 373–385. 9268618
52. Piferrer F (2001) Endocrine sex control strategies for the feminization of teleost fish. Aquaculture 197 : 229–28.
53. Kobayashi T, Kajiura-Kobayashi H, Nagahama Y (2003) Induction of XY sex reversal by estrogen involves altered gene expression in a teleost, tilapia. Cytogenet Genome Res 101 : 289–294. 14684997
54. Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, et al. (2009) Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139 : 1130–1142. doi: 10.1016/j.cell.2009.11.021 20005806
55. Pannetier M, Fabre S, Batista F, Kocer A, Renault L, et al. (2006) FOXL2 activates P450 aromatase gene transcription: towards a better characterization of the early steps of mammalian ovarian development. J Mol Endocrino. 36 : 399–413.
56. Fleming NI, Knower KC, Lazarus KA, Fuller PJ, Simpson ER, et al. (2010) Aromatase is a direct target of FOXL2: C134W in granulosa cell tumors via a single highly conserved binding site in the ovarian specific promoter. PLoS One 5: e14389. doi: 10.1371/journal.pone.0014389 21188138
57. di Clemente N, Ghaffari S, Pepinsky RB, Pieau C, Josso N, et al. (1992) A quantitative and interspecific test for biological activity of anti-müllerian hormone: the fetal ovary aromatase assay. Development 114 : 721–727. 1319894
58. Nishikimi H, Kansaku N, Saito N, Usami M, Ohno Y, et al. (2000) Sex differentiation and mRNA expression of P450c17, P450arom and AMH in gonads of the chicken. Mol Reprod Dev 55 : 20–30. 10602270
59. Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339 : 819–823. doi: 10.1126/science.1231143 23287718
Štítky
Genetika Reprodukční medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis inČlánek Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 11- Délka menstruačního cyklu jako marker ženské plodnosti
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
-
Všechny články tohoto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



