-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
Ciliopathies, diseases arising from defects in the functions of primary cilia, have many different manifestations and vary dramatically in severity. How genetics influence ciliopathy phenotypes is poorly understood. Building off of our increasing knowledge of how different biochemical complexes contribute to ciliary function, we investigated how ciliopathy-associated genes interact to support ciliogenesis. Using a combination of nematode and mouse genetics, we found that genes encoding components of different biochemical complexes interact, whereas genes encoding different components within a single complex do not. These results revealed overlapping ciliary functions of biochemically distinct proteins complexes such as the BBSome, the transition zone MKS complex and the transition zone NPHP complex. This work indicates the genetic interactions that may alter the phenotypic consequences of human ciliopathy mutations.
Published in the journal: . PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005627
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005627Summary
Ciliopathies, diseases arising from defects in the functions of primary cilia, have many different manifestations and vary dramatically in severity. How genetics influence ciliopathy phenotypes is poorly understood. Building off of our increasing knowledge of how different biochemical complexes contribute to ciliary function, we investigated how ciliopathy-associated genes interact to support ciliogenesis. Using a combination of nematode and mouse genetics, we found that genes encoding components of different biochemical complexes interact, whereas genes encoding different components within a single complex do not. These results revealed overlapping ciliary functions of biochemically distinct proteins complexes such as the BBSome, the transition zone MKS complex and the transition zone NPHP complex. This work indicates the genetic interactions that may alter the phenotypic consequences of human ciliopathy mutations.
Introduction
Despite accelerating success in identifying genetic variations, the relationship between genotype and phenotype in humans often remains obscure. Even for many Mendelian diseases, the expressivity of disease alleles is not always predictable, indicating that additional genetic and epigenetic influences modify the disease-causing mutations. The genetic influences could include many common genetic variants with small effects, rare variants with large effects, and allele-specific gene-gene interactions. These genetic interactions can modify associated phenotypes through changes in gene expression, protein interactions, and effects on overlapping functions [1–4].
Ciliopathies, which arise from disrupted ciliary function, are prominent examples of disorders with complex inheritance. The clinical manifestations of ciliopathies are often variable and overlapping. For example, Meckel syndrome (MKS [MIM 249000]) is characterized by cystic kidney dysplasia, polydactyly, and occipital meningoencephalocoele. Nephronophthisis (NPHP [MIM 256100]), the most common genetic cause of renal failure in children, is characterized by cystic kidney dysplasia without limb or brain malformations. Bardet-Biedl syndrome (BBS [MIM 209900]), a disorder associated with at least nineteen loci, is characterized by cystic kidney dysplasia, polydactyly, retinal degeneration, obesity, and learning difficulties. Although BBS has been traditionally considered an autosomal recessive disorder affecting a single locus, in some instances three mutations in two loci are required to cause disease [5–9]. Thus, mutations in two loci can be required for ciliopathy penetrance.
As reflected by the broad spectrum of ciliopathy-associated phenotypes, primary cilia play numerous, often tissue-specific, roles in mammals. For example, cilia sense odorants and light, control growth in the kidney, and transduce the Hedgehog (Hh) signaling pathway, thereby playing integral functions in embryonic development and adult tissue homeostasis [10]. To transduce signals, cilia maintain compositions distinct from those of other cellular compartments. How cilia exclude some proteins while allowing others access remains a fundamental question [11]. Previous work has suggested that the transition zone, a region at the base of the cilium between the basal body and the axoneme proper, may be involved in this process [12–16]. The transition zone is defined ultrastructurally by the presence of Y-links that connect the microtubule core to the ciliary membrane [17, 18]. In mice, the proteins Tctn1, Tctn2, Tmem231, Tmem67, Mks1, B9d1, B9d2, Cep290 and Cc2d2a co-localize to the transition zone and form a large biochemical complex, the MKS complex [12, 16, 19]. A biochemically distinct complex that includes Nphp1 and Nphp4, termed the NPHP complex, also localizes to the transition zone [20, 21].
Mammals possess cilia on many cell types, whereas cilia in Caenorhabditis elegans are confined to the dendritic tips of 60 select sensory neurons in hermaphrodites. As in mammals, the C. elegans orthologs of the MKS complex genes, tmem-231 (MIM 614949), mks-1 (MIM 609883), mksr-1 (the ortholog of B9d1 [MEM 614144]), mksr-2 (the ortholog of B9d2 [MIM 611951]), mks-3 (the ortholog of Tmem67 [609884]), and mks-6 (the ortholog of Cc2d2a [MIM 612013]), as well as the orthologs of the NPHP complex genes nphp-1 (MIM 607100) and nphp-4 (MIM 607215), encode transition zone proteins [13, 22–26]. Individual loss of function of these genes does not dramatically compromise C. elegans ciliary structure. However, loss of function of one of several MKS complex genes when combined with loss of function of either nphp-1 or nphp-4 synergistically compromises ciliary structure in C. elegans [13, 22, 23].
Like MKS and NPHP complex proteins, many proteins associated with BBS form a large complex, the BBSome [27]. The BBSome functions as a vesicular coat that transports select G protein-coupled receptors (GPCRs) to the cilium through an association with intraflagellar transport (IFT) machinery [28, 29]. In contrast to MKS or NPHP components, abrogating the function of individual BBS components in C. elegans disrupts ciliary structure [30]. Whether the BBSome works together with the transition zone is unknown. However, mutation in MKS1 can cause BBS, and mutation in BBS4 (MIM 600374) can modify the ciliopathy phenotypes of CEP290 (MIM 610142), suggesting that the BBSome and transition zone may share some functions [31, 32].
We hypothesized that genetic interactions between MKS, NPHP, and BBS complex genes could contribute to the wide phenotypic spectrum observed in human ciliopathies. To test this hypothesis, we generated animals with intra - and inter-complex double mutations, with an emphasis on the ciliopathy gene Tectonic1 (Tctn1 [MIM 609863]). We previously found that Tctn1 is an essential component of the transition zone required for cilium-dependent Hh signaling in mice, and that human TCTN1 is mutated in another ciliopathy, Joubert syndrome [16, 33]. Here, we identify the C. elegans ortholog of Tctn1, tctn-1, and find that it also encodes a transition zone protein. Loss of TCTN-1 does not disrupt C. elegans ciliary structure, similar to other transition zone genes. tctn-1 interacts with nphp-4 and nphp-1, but not with the MKS complex genes mks-1, mksr-1, mksr-2 and mks-3, thereby genetically placing tctn-1 within the MKS module. These genetic interactions in nematodes predicted mammalian genetic interactions, as combining mouse Tctn1 and Nphp4 mutations also resulted in synthetic ciliary phenotypes. Additional double mutations between one gene of the MKS complex and one gene of the NPHP complex yielded similar synthetic effects, suggesting that the genetic interactions between MKS and NPHP complex components may pertain to many members. In contrast, double mouse mutations between genes within the same complex did not show such an interaction, consistent with findings in C. elegans. Furthermore, both C. elegans and mammals exhibited a genetic synergy between mutations affecting the transition zone and the BBSome, indicating that the MKS, NPHP, and BBS complexes share overlapping functions in ciliogenesis. Hence, mutations affecting distinct ciliopathy protein complexes synthetically interact to modify cilia-associated phenotypes.
Results
C. elegans Tectonic is a transition zone component
By sequence homology, we identified a single C. elegans ortholog of the three vertebrate Tectonic genes, E04A4.6, which we refer to as tctn-1. Mammalian Tectonics possess a signal peptide and a domain of unknown function (DUF1619) characteristic of the family. C. elegans tctn-1 also possesses a signal peptide and is most homologous to other Tectonics in the C-terminal region (S1 Fig). In nematodes, ciliary genes often contain a conserved X-box sequence that acts as a binding site for the ciliogenic RFX transcription factor, DAF-19 [34]. Consistent with a ciliary function, tctn-1 has a predicted X-box sequence (Fig 1A), is strongly downregulated in daf-19 mutants, and is preferentially expressed in ciliated sensory neurons [35–37]. For these reasons, we surmised that tctn-1 was likely to have a ciliary function in C. elegans.
Fig. 1. The C. elegans ortholog of Tectonic1 is a transition zone component. 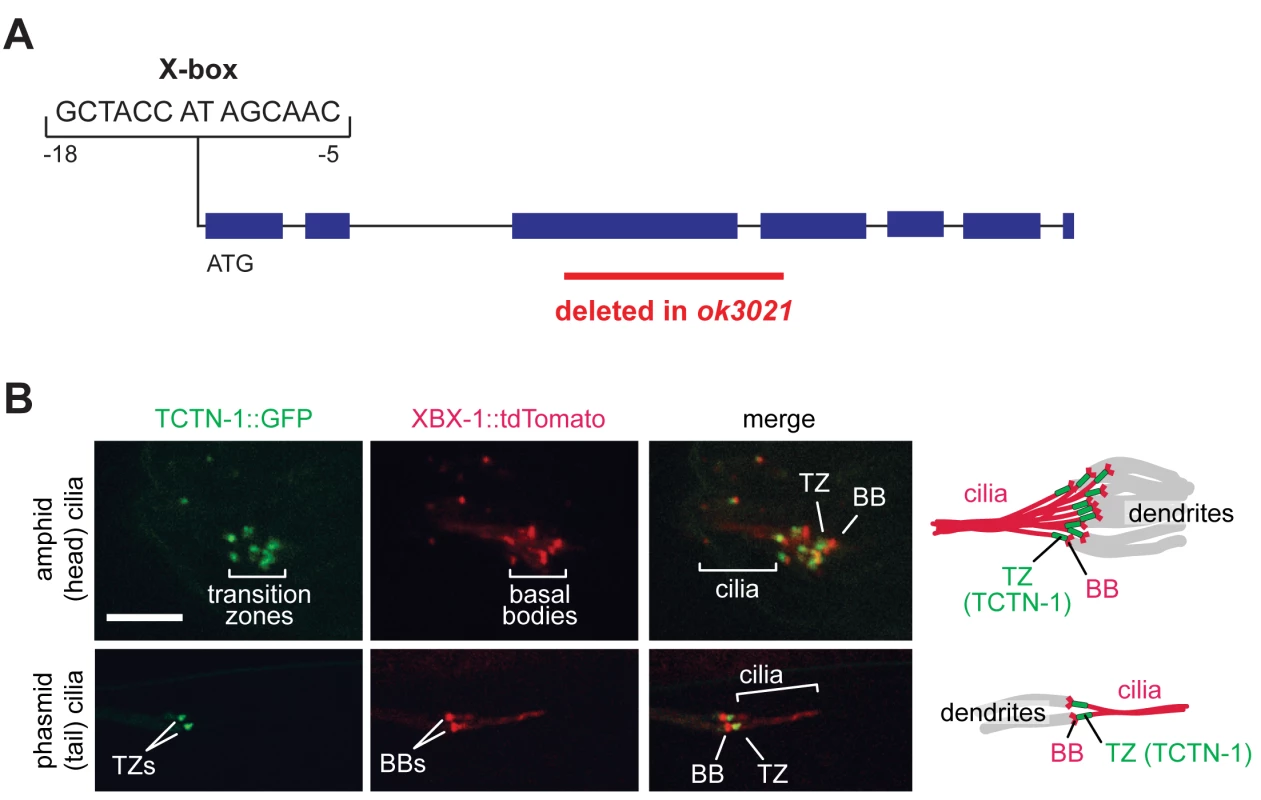
(A) Schematic of the X-box sequence and ok3021 allele of C. elegans tctn-1. (B) GFP-tagged TCTN-1 localizes specifically to the transition zones (TZs) in head (amphid) and tail (phasmid) cilia of C. elegans. Basal bodies (BBs) and ciliary axonemes are marked with tdTomato-tagged XBX-1, a component of the ciliary dynein. Schematics illustrate the position of TCTN-1 at the transition zone with respect to the basal body and axoneme. Scale bar, 5 μm. To investigate whether nematode TCTN-1 localizes to cilia, we generated a strain that expresses a carboxy-terminal GFP-tagged version of TCTN-1 under a bbs-8 promoter active in ciliated cells [38]. To visualize cilia, we marked basal bodies and axonemes with a tdTomato-tagged ciliary dynein light intermediate chain, XBX-1. TCTN-1-GFP was enriched specifically at the transition zone of cilia, immediately distal to the basal body and proximal to the axoneme (Fig 1B).
tctn-1 genetically interacts with NPHP complex genes, but not MKS complex genes
To investigate the function of TCTN-1, we obtained the E04A4.6(ok3021) mutant from the C. elegans Gene Knockout Consortium. The ok3021 allele contains a 515 base pair deletion spanning exons three and four of tctn-1, which generates a premature stop codon and is predicted to disrupt the function of tctn-1 (Fig 1A).
Homozygous tctn-1 mutants exhibited no alteration of growth, size, egg laying, brood size, or cilium-associated sensory behaviors (osmotic avoidance or chemotaxis to diacetyl or butanone) (S2 Fig). Additionally, the sensory neurons of tctn-1 mutants were indistinguishable from those of the wild type N2 reference strain in their ability to take up the hydrophobic dye, DiI. This dye filling assay is a simple method of indirectly testing the structural integrity of cilia of six head (amphid) neurons and two tail (phasmid) neurons [39], and suggests that ciliary structure is not grossly compromised in the absence of TCTN-1 (Fig 2A–2C). Other C. elegans transition zone gene mutants such as nphp-1, nphp-4, mks-1, mksr-1, mksr-2, and mks-3 mutants, similarly possess largely normal ciliary structure (Fig 2A–2C) [13, 22–24].
Fig. 2. C. elegans tctn-1 interacts with NPHP genes, but not MKS genes to affect ciliary structure. 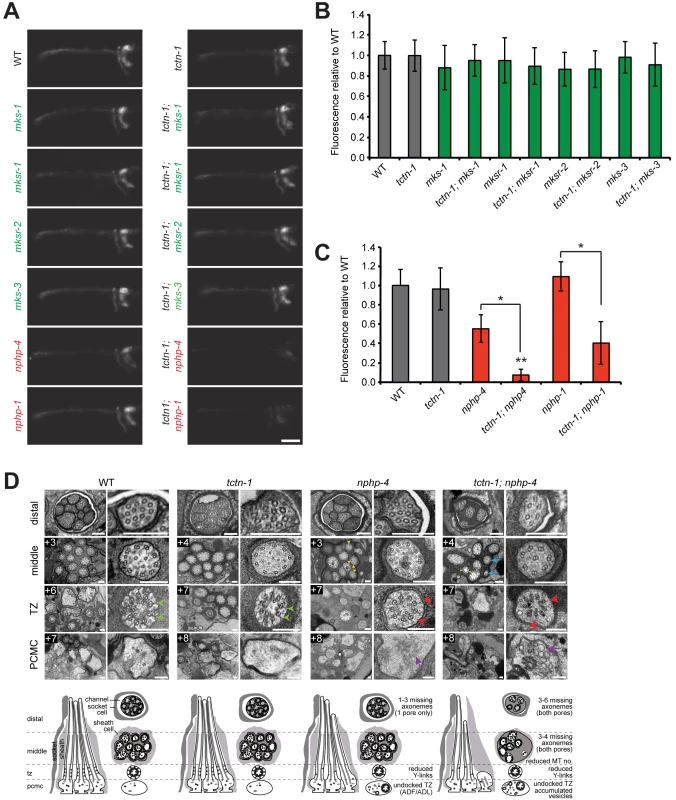
(A) Dye filling of amphid neurons in L4 nematodes in the indicated single transition zone mutants (left column) and tctn-1 double mutants (right column). Lateral views, with anterior to the left. Genotypes including an allele affecting a previously recognized MKS complex component are indicated in green. Genotypes including an allele affecting an NPHP complex component are indicated in red. Scale bar, 20 μm. (B and C) Fluorescence intensity of DiI filled amphid neurons in single mutants of the MKS complex or NPHP complex, and double mutants with tctn-1 relative to wild type. Error bars represent the standard deviation. Statistical significance according to unpaired Student’s t-tests (* p<0.001; ** p<0.001 compared to tctn-1). (D) Low and high magnification TEM cross-sections of the distal segment, middle segment, transition zone (TZ), and distal dendritic periciliary membrane compartment (PCMC) of amphid cilia with schematics below (lateral and transverse views). Green arrowheads indicate intact Y-links in wild type and tctn-1 transition zones whereas red arrowheads indicate reduced or missing Y-links, observed in nphp-4 and tctn-1; nphp-4 transition zones. Yellow arrowheads indicate open B-tubules and purple arrows indicate vesicle accumulation in the PCMCs of nphp-4 and tctn-1; nphp-4 mutants. tctn-1; nphp-4 mutants display several truncated axonemes and axonemes with fewer microtubule doublets (blue arrows) compared to wild type, tctn-1 and nphp-4 mutants. Boxed numbers indicate distances (μm) from the distal ciliary tips. Scale bars,100 nm. Previous studies have found that a mutation in either of the C. elegans homologs of the NPHP complex components nphp-1 or nphp-4, combined with a mutation in an MKS complex gene, synergistically disrupts ciliary structure and consequently causes a dye filling defect [13, 22, 23]. To test whether tctn-1 functions as an MKS complex or NPHP complex gene, we generated double mutants of tctn-1 and either MKS complex genes or NPHP complex genes. Double mutations affecting tctn-1 and any of four MKS complex genes (tctn-1; mks-1, tctn-1; mksr-1, tctn-1; mksr-2, and tctn-1; mks-3) did not alter dye filling, similar to tctn-1 single mutants (Fig 2A and 2B).
In contrast, dye filling in tctn-1; nphp-4 double mutants was dramatically disrupted compared to tctn-1 or nphp-4 single mutants. In tctn-1; nphp-4 double mutants, amphid neurons incorporated dye at low or undetectable levels, resulting in decreased fluorescence relative to either single mutant (Fig 2A and 2C). Similarly, tctn-1; nphp-1 double mutants were unable to incorporate dye compared to either single mutant (Fig 2A and 2C). Therefore, tctn-1 synergistically disrupts ciliary structure when combined with NPHP but not MKS complex mutations, genetically placing tctn-1 within the MKS complex.
TCTN-1 and NPHP-4 have overlapping functions in supporting ciliary structure
The inability of the sensory neurons of tctn-1; nphp-4 mutants to dye fill suggested that their ciliary structure or morphology was defective. tctn-1; nphp-4 phasmid cilia, visualized by expressing the fluorescently tagged ciliary protein XBX-1::tdTomato, were shorter than cilia of wild type or either single mutant (S3A and S3B Fig). Additionally, many tctn-1; nphp-4 cilia were mispositioned; for example, they were frequently abnormally close to the cell body at the ends of short dendrites, or not projecting posteriorly (S3C and S3D Fig). These perturbations in ciliary position are similar to those caused by concurrent loss of NPHP-4 and other MKS complex components [13, 22, 23].
To gain further insight into how TCTN-1 and NPHP-4 cooperatively support ciliogenesis, we examined the amphid channel cilia of tctn-1 and nphp-4 single mutants, and tctn-1; nphp-4 double mutants using transmission electron microscopy. In tctn-1 single mutants, the ten cilia of both amphid channel pores were indistinguishable from wild type. Notably, the ciliary transition zones of tctn-1 mutants contained intact Y-links and exhibited no overt abnormalities, suggesting that TCTN-1 is not essential for generating all structural components of the transition zone (Fig 2D).
In contrast, nphp-4 single mutants displayed cilia with moderate ultrastructural defects. In addition to the modest shortening of 1–3 axonemes in one amphid pore (the other pore is normal) and the occurrence of open B-tubules in the middle segment previously described [25], some nphp-4 mutant cilia exhibited a small accumulation of vesicles in the periciliary membrane compartment (PCMC) (Fig 2D). We also detected defects in the transition zone of nphp-4 mutants (Lambacher et al., manuscript in revision). Y-links, the structural hallmark of the transition zone, were frequently reduced in number and appeared less electron dense in nphp-4 mutants. Despite the reduction in Y-links, the ciliary membrane remained closely opposed to the axoneme in most nphp-4 transition zones. The only exceptions were the transition zones of the biciliated ADF and ADL neurons; in nphp-4 mutants, these transition zones were frequently fully disconnected from the ciliary membrane and positioned ectopically in the distal dendrite region (Fig 2D).
The ciliary structure of tctn-1; nphp-4 double mutants was more severely disrupted than that of nphp-4 single mutants. In tctn-1; nphp-4 mutants, at least 3–4 axonemes were missing in the middle and distal regions of both amphid pores, and remaining axonemes frequently possessed fewer microtubule doublets (Fig 2D). Additionally, tctn-1; nphp-4 mutant cilia displayed defects in Y-links at the transition zone similar to nphp-4 mutants: Y-links were either thinner than wild type or missing, and in some cases, transition zone microtubules were completely disconnected from the ciliary membrane (Fig 2D). An accumulation of membranes and vesicles was also observed in the PCMC of tctn-1; nphp-4 mutants. In contrast, tctn-1; mks-3 double mutants displayed no abnormalities in ciliary ultrastructure (S3E Fig), consistent with the lack of genetic interaction between tctn-1 and other MKS complex components. Thus, we conclude that ciliary structure is dependent on the overlapping functions of TCTN-1 and NPHP-4.
C. elegans TCTN-1 contributes to the ciliary gate function, but not to transition zone protein composition
We hypothesized that disruption of Y-links in nphp-4 single mutants and tctn-1; nphp-4 double mutants could reflect an abnormal composition of the transition zone. Using established markers of the ciliary base and axoneme, we assessed the localization of several fluorescently tagged transition zone proteins. MKSR-1, MKS-5, MKS-6 and NPHP-4 all localized to the transition zone of tctn-1 mutants in a manner indistinguishable from wild type (Fig 3A–3D). Similarly, despite the reduction in Y-links identified in nphp-4 and tctn-1; nphp-4 mutants, MKSR-1, MKS-6 and NPHP-4 localized to the transition zone of these mutants equivalently to wild type (Fig 3A, 3C and 3D). MKS-5 also localized to the transition zone of nphp-4 and tctn-1; nphp-4 mutants, although it partially mislocalized distally along the ciliary axoneme (Fig 3B).
Fig. 3. TCTN-1 contributes to C. elegans ciliary gate function. 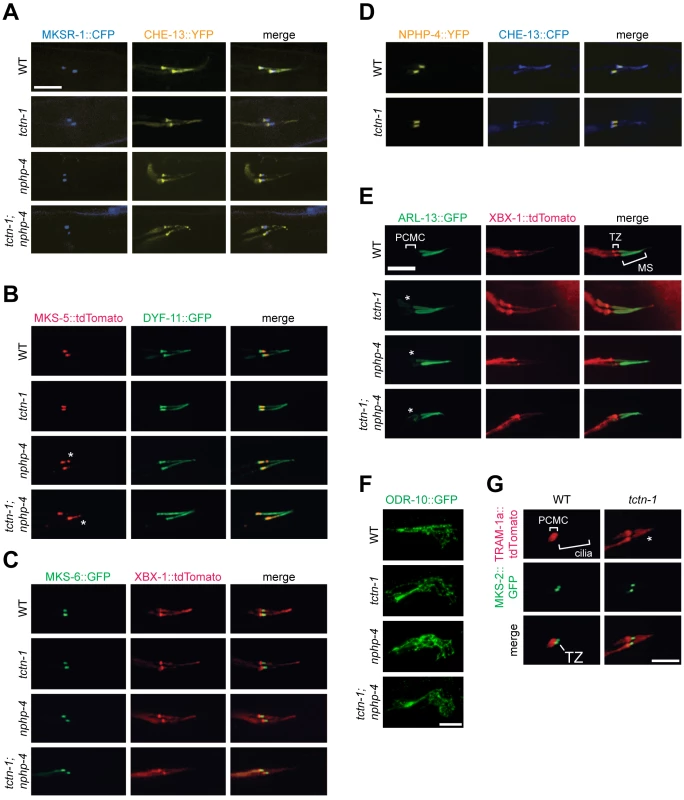
(A-D) Localization of different transition zone proteins in phasmid cilia of wild type, tctn-1, nphp-4, and tctn-1; nphp-4 mutants. Each GFP, CFP, YFP, or tdTomato-tagged transition zone protein is co-localized with a ciliary marker (CHE-13, DYF-11 or XBX-1), as indicated. Abnormal localization of MKS-5 in the nphp-4 single and tctn-1; nphp-4 double mutants is indicated by asterisks. (E) ARL-13::GFP localizes ectopically to the PCMC in tctn-1, nphp-4, and tctn-1; nphp-4 mutants as indicated by asterisks, but localizes normally to the middle segment (MS) of cilia. (F) ODR-10::GFP localizes to the cilia of AWA neurons in wild type, tctn-1, nphp-4, and tctn-1; nphp-4 mutants. Anterior is to the right. (G) TRAM-1a::tdTomato localizes only to the PCMC in wild type animals, but enters phasmid cilia in tctn-1 mutants, as indicated by asterisks. MKS-2::GFP marks the transition zone. Scale bars, 5 μm. The transition zone is proposed to function as a ciliary gate, controlling protein entry, retention, and exclusion [12–14, 16, 40]. The small GTPase, ARL-13 and its ortholog Arl13b localize to cilia in C. elegans and vertebrates, respectively [41, 42]. In C.elegans, transition zone genes including nphp-4 are required for ARL-13 to exclusively localize to cilia [43]. To test if C. elegans TCTN-1 regulates the localization of ARL-13, we examined a fusion of ARL-13 and GFP. As in wild type animals, in tctn-1 single and tctn-1; nphp-4 double mutants, ARL-13::GFP localized along the length of the axoneme. However, unlike wild type animals, tctn-1 and tctn-1; nphp-4 mutants displayed a modest accumulation of ARL-13::GFP in the PCMC (Fig 3E). Thus, similar to other C. elegans transition zone genes, TCTN-1 is not required for ARL-13::GFP to enter the cilium, but may be required for its retention within the cilium [43].
In addition to ARL-13, signaling proteins such as GPCRs can localize to cilia. To test if TCTN-1 regulates the ciliary localization of GPCRs, we examined whether it was required for the ciliary localization of ODR-10, a GPCR that senses the odorant diacetyl at the cilia of AWA neurons [44, 45]. ODR-10::GFP localized to AWA cilia equivalently in wild type and tctn-1, nphp-4, and tctn-1; nphp-4 mutants (Fig 3F), consistent with our finding that none of these genes is required for the behavioral responses to diacetyl (S2F Fig). Thus, C. elegans TCTN-1 is not essential for the ciliary localization of a GPCR.
In addition to promoting the localization of ciliary proteins, the transition zone is proposed to exclude non-ciliary proteins from entering the cilium. To test whether TCTN-1 participates in this aspect of transition zone function, we examined the localization of TRAM-1a, a transmembrane protein excluded from the cilium [13]. In wild type animals, TRAM-1a::tdTomato was confined to a ring of periciliary membrane at the distal end of the dendrite and was not observed in the ciliary axoneme. In contrast, TRAM-1a::tdTomato was present within the cilia of tctn-1 mutants (Fig 3G). Together, these findings indicate that C. elegans TCTN-1 is not essential for MKS or NPHP protein complex localization to the transition zone, nor for the ciliary localization of ARL-13::GFP or ODR-10::GFP, but is required to exclude a non-ciliary protein from the cilium. Therefore, TCTN-1 may be a key component of the ciliary barrier preventing the entry of non-ciliary proteins
The NPHP complex and BBS-5 have overlapping functions in C. elegans ciliogenesis
The genetic interaction between tctn-1 and NPHP complex genes strengthens previous indications that the MKS and NPHP complexes perform overlapping functions within the transition zone [13, 15, 22, 23]. We investigated whether a biochemical complex that does not specifically localize to the transition zone, the IFT-associated BBSome complex, could also have overlapping functions with transition zone complexes. Core BBSome components, BBS-1, BBS-2, BBS-4, BBS-5, BBS-7, BBS-8, and BBS-9 are conserved in C. elegans. Except for bbs-5 (MIM 603650) mutants, neurons of C. elegans bbs mutants do not incorporate dye, thus precluding the use of dye filling to analyze synthetic interactions with most BBS-associated genes [30, 46]. Although the bbs-5(gk507) allele contains a 680 bp deletion spanning the first two exons of the bbs-5 gene, neurons of homozygous bbs-5(gk507) mutants dye fill normally [46, 47], suggesting that this allele is hypomorphic or that BBS-5 is more dispensable for BBSome function than other components.
To test for genetic interactions between bbs-5 and transition zone genes, we examined dye filling in double mutants for bbs-5 and MKS complex genes or NPHP complex genes. bbs-5; tctn-1 and bbs-5; mksr-1 double mutants dye filled to the same extent as their single mutant counterparts, revealing no genetic interaction between bbs-5 and MKS complex genes (Fig 4A and 4B). In contrast, dye filling in bbs-5; nphp-4 and bbs-5; nphp-1 double mutants was dramatically decreased compared to their respective single mutants (Fig 4A and 4B). Therefore, BBS-5 and NPHP complex components have overlapping functions in supporting C. elegans ciliary integrity. The finding that bbs-5 synthetically interacts with NPHP complex genes but not MKS complex genes further suggests that the NPHP and MKS complexes perform distinct roles.
Fig. 4. C. elegans NPHP genes synthetically interact with bbs-5 to affect ciliary structure. 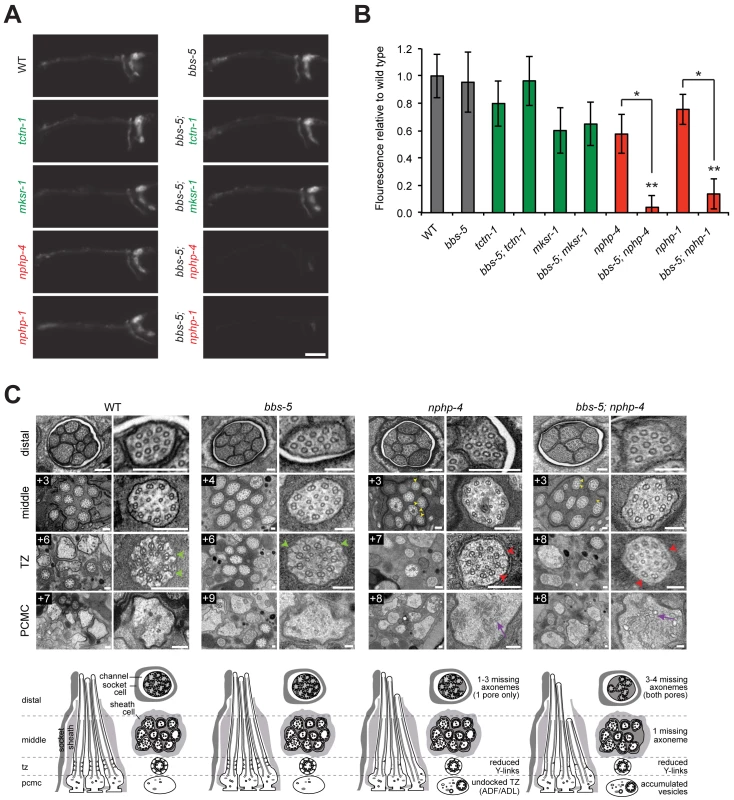
(A) Dye filling of amphid neurons in L4 nematodes in single transition zone mutants (left column) and bbs-5 double mutants (right column). Lateral views, with anterior to the left. Scale bar, 20 μm. (B) Fluorescence intensity of DiI filled amphid neurons relative to wild type. Genotypes including an allele affecting an MKS complex component are indicated in green. Genotypes including an allele affecting an NPHP complex component are indicated in red. Error bars represent the standard deviation. Statistical significance according to unpaired Student’s t-tests (* p<0.001; ** p<0.001 compared to bbs-5). (C) Low and high magnification TEM cross-sections of the distal segment, middle segment, transition zone (TZ), and PCMC of amphid cilia with schematics below (lateral and transverse views). bbs-5 mutants display normal ciliary structures, including intact Y-links (green arrowheads). bbs-5; nphp-4 mutant cilia display open B-tubules (yellow arrowheads), reduced Y-links (red arrowheads) and vesicle accumulation in the PCMC (purple arrows) similar to nphp-4 cilia, but have fewer axonemes that fully extend distally in both pores. Boxed numbers indicate distances (μm) from the distal ciliary tips. Scale bars,100 nm. To further investigate the interaction between bbs-5 and NPHP genes, we examined bbs-5; nphp-4 double mutant cilia by transmission electron microscopy. As predicted by the dye filling assay, bbs-5 mutants showed normal amphid channel cilium ultrastructure, with a full complement of ten axonemes extending through the length of each pore (Fig 4C). Moreover, the transition zones of bbs-5 mutants contained normal Y-links, and the ciliary membrane and microtubule core were indistinguishable from those of wild type animals (Fig 4C).
bbs-5; nphp-4 double mutant cilia displayed more pronounced ultrastructural defects than bbs-5 or nphp-4 single mutants. Similar to nphp-4 cilia, bbs-5; nphp-4 cilia possessed open B-tubules, a reduction in Y-links and vesicle accumulation in some PCMCs (Fig 4C). In contrast to the single mutants, bbs-5; nphp-4 double mutants were missing 3–4 axonemes in the distal region and one axoneme in the middle region of both amphid pores (Fig 4C). Thus, BBS-5 has overlapping functions with the NPHP complex in building or maintaining the ciliary axoneme.
Murine MKS and NPHP complexes have overlapping functions in limb development
Mutations in human NPHP4 cause nephronophthisis, either with or without retinal degeneration [48, 49]. To test whether the genetic interactions detected in C. elegans with MKS complex genes could contribute to the expressivity of NPHP4, we investigated whether the same genetic interactions occur in mammals. Nphp4nmf192/nmf192 mutant mice (hereinafter referred to as Nphp4n/n) are viable, but develop retinal degeneration and have reduced sperm motility [50]. Mouse Tctn1-/- mutants die during late gestation and exhibit heterotaxia, microphthalmia, and hindlimb polydactyly, phenotypes associated with ciliary malfunction and similar to those exhibited by human MKS affected individuals [16, 33]. Several other mouse MKS complex mutants, such as Tctn2-/- (MIM 613846) and Cc2d2a-/- (also known as Mks6), display highly similar phenotypes [16, 21].
To assess potential genetic interactions between the MKS and NPHP complexes in mammals, we generated mice doubly mutant for Tctn1 and Nphp4. Tctn1-/- mutants exhibited single digit polydactyly restricted to the hindlimbs and Nphp4n/n mutants had no limb abnormalities. In contrast, Tctn1-/- Nphp4n/n double mutants exhibited polydactyly in both the forelimb and hindlimb, and had an increased number of digits per limb compared to the polydactyly of Tctn1-/- mutants (Fig 5A–5C, S1 Table). In addition, Tctn1-/- Nphp4n/n double mutants exhibited partially penetrant exencephaly, which was not observed in Tctn1-/- single mutants or in Nphp4n/n single mutants (Fig 5D, S2 Table). Therefore, Tctn1 and Nphp4 interact synergistically in mammals, as they do in C. elegans.
Fig. 5. Mouse Tctn1 genetically interacts with NPHP complex genes, but not MKS complex genes. 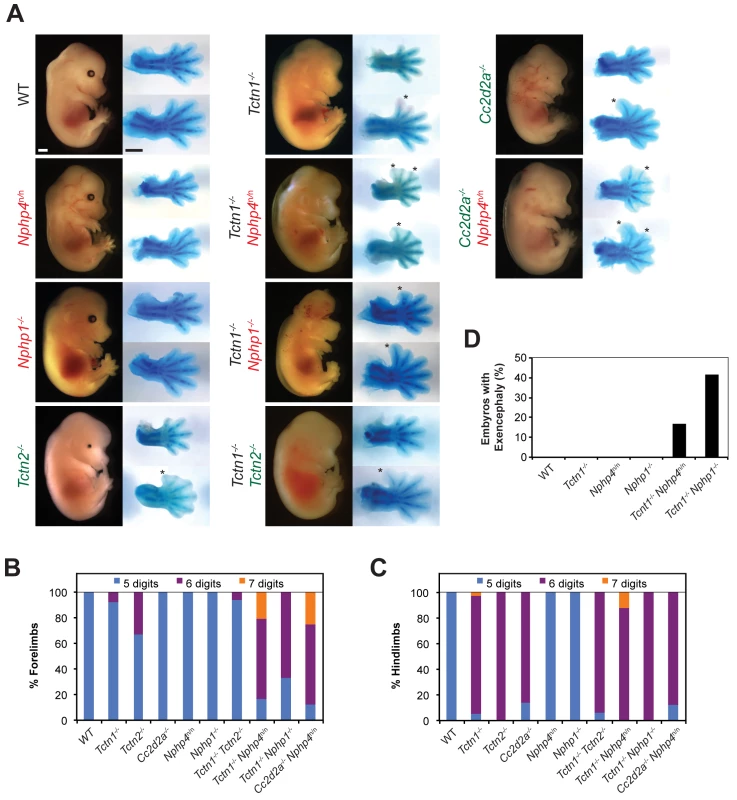
(A) Lateral views of wild type, single or double mutant mouse embryos of indicated genotype at E14.5. Exencephaly is apparent in the Tctn1-/- Nphp1-/- double mutant. Corresponding Alcian blue staining of the right forelimb (top) and hindlimb (bottom) are included, with asterisks indicating extra digits. Genes encoding components of the NPHP complex are indicated in red. Genes encoding components of the MKS complex are indicated in green. Scale bars, 1 mm. (B) Number of digits in the forelimbs and (C) hindlimbs of wild type, single or double mutant embryos of indicated genotypes. (D) Incidence of exencephaly in wild type, single or double mutants embryos of indicated genotypes. Numbers of animals analyzed for polydactyly and exencephaly are included in S1 and S2 Tables. Like NPHP4, human mutations in NPHP1 are associated with multiple ciliopathies, including nephronophthisis and Joubert syndrome, characterized by cerebellar vermis hypoplasia [51–53]. To determine if the genetic interaction between Tctn1 and Nphp4 was specific to Nphp4 or extended to another NPHP complex gene, we generated Tctn1-/- Nphp1-/- double mutant mice. As with Tctn1-/- Nphp4n/n double mutants, Tctn1-/- Nphp1-/- double mutants displayed more severe polydactyly and a higher penetrance of exencephaly compared to Tctn1-/- mutants (Fig 5A–5D, S1 and S2 Tables). These results reveal that in both nematodes and mice, Tctn1 displays a synthetic genetic interaction with both Nphp1 and Nphp4.
To assess whether the interaction of NPHP complex genes with Tctn1 extended to other MKS complex genes, we generated double mutants affecting Nphp4 and Cc2d2a. In addition to MKS, human CC2D2A mutations are associated with Joubert syndrome [54–56]. Like Tctn1-/- mutants, mouse Cc2d2a-/- mutants displayed single digit polydactyly restricted to the hindlimb [16]. Similar to Tctn1-/- Nphp4n/n double mutants, Cc2d2a-/- Nphp4n/n double mutants displayed polydactyly in the forelimbs unlike either single mutant (Fig 5A–5C, S1 Table). Therefore, the genetic interaction between the MKS and NPHP complexes extends to multiple genes of both complexes.
As mutations within the MKS complex or within the NPHP complex do not genetically interact when combined in C. elegans, we tested whether this principle held true in mice. We generated Tctn1-/- Tctn2-/- double mutant mice and found that they did not phenotypically differ from Tctn1-/- or Tctn2-/- single mutants (Fig 5A–5C, S1 Table). Similarly, we generated Nphp1-/- Nphp4n/n double mutant mice, which grow into adulthood at rates comparable to control littermates (S4A Fig). Thus, at a gross level in both nematodes and mammals, multiple mutations affecting different MKS complex components or multiple mutations affecting different NPHP complex components do not additively disrupt ciliary function. These results further suggest that the MKS complex and the NPHP complex do not possess residual function in the absence of individual components.
Murine Tctn1 and Nphp4 cooperatively support ciliogenesis
The genetic interaction of Tctn1 and Nphp4 suggested that the mammalian MKS and NPHP complexes have partially overlapping ciliary functions. To investigate how these two complexes participate in ciliary functions, we examined cilia in the forelimb, a tissue phenotypically affected in Tctn1-/- Nphp4n/n double mutants. Immunostaining revealed that ciliation in Tctn1-/- forelimbs was reduced compared to controls (Fig 6A). Tctn1-/- Nphp4n/n forelimbs were further depleted of cilia compared to Tctn1-/- forelimbs (Fig 6A). Transmission electron microscopy confirmed the depletion of cilia in the forelimb buds of Tctn1-/- Nphp4n/n double mutants, which possessed docked basal bodies, but lacked ciliary axonemes (Fig 6A and 6B). The greater ciliogenesis defect in Tctn1-/- Nphp4n/n forelimbs likely accounts for the increased polydactyly, as cilia are required for Shh-dependent patterning of the limb buds [57–59].
Fig. 6. Mouse Tctn1 and Nphp4 have distinct roles in transition zone composition and overlapping roles in ciliogenesis. 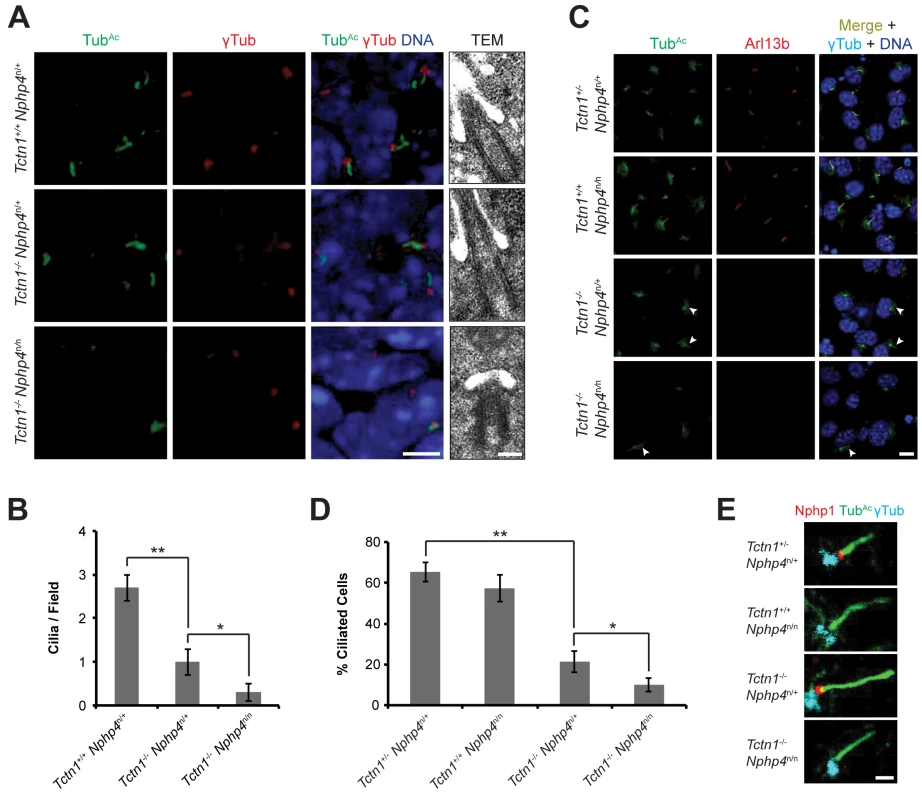
(A) Limb bud sections (left) from E11.5 embryos were stained for acetylated tubulin (TubAc, green) to mark the ciliary axonemes, γ-tubulin (red) to mark the basal bodies and DAPI (blue) to mark nuclei. Scale bar, 5 μm. Transmission electron microscopy (TEM, right) for each genotype. Scale bar, 200 nm. (B) Quantification of limb bud cilia in control, Tctn1-/- Nphp4n/+, and Tctn1-/- Nphp4n/n mutants from TEM fields of view. Error bars represent the standard error of the mean. Statistical significance according to unpaired Student’s t-tests (* p<0.05; ** p<0.001). (C) Fibroblasts derived from E13.5 limb buds of the indicated genotypes stained for TubAc (green), Arl13b (red), γ-tubulin (cyan), and DAPI (blue). White arrowheads indicate cilia in Tctn1-/- Nphp4n/+ and Tctn1-/- Nphp4n/n mutants. Scale bar, 10 μm. (D) Quantification of proportion of ciliated cultured limb bud fibroblasts. Error bars represent the standard deviation. Statistical significance according to unpaired Student’s t-tests (* p<0.05; ** p<0.0001). (E) Immunostaining of limb bud fibroblasts for Nphp1 (red), TubAc (green), and γ-tubulin (cyan). Scale bar, 1 μm. To further investigate how Tctn1 and Nphp4 participate in ciliary functions, we cultured fibroblasts derived from mutant and control limb buds. Limb bud fibroblasts showed similar degrees of ciliation in vitro as they did in vivo (Fig 6C and 6D). Although the majority of Tctn1-/- Nphp4n/n mutant limb fibroblasts lacked cilia, a small percentage of cells remained ciliated. In the ciliated Tctn1-/- Nphp4n/n mutant cells, we investigated whether ciliary or transition zone protein composition was disturbed. The small GTPase Arl13b localized to cilia in control and Nphp4n/n mutant fibroblasts, but was absent in Tctn1-/- and Tctn1-/- Nphp4n/n mutant cilia (Fig 6C). Interestingly, C. elegans TCTN-1 was dispensable for much of the cellular ARL-13:GFP to localize to cilia (Fig 3E), demonstrating a difference in the function of C. elegans and mouse Tctn1.
We examined the transition zone localization of Nphp1, a member of the NPHP complex. In control and Tctn1-/- mutant fibroblasts, Nphp1 localized to the transition zone, but in Nphp4n/n single and Tctn1-/- Nphp4n/n double mutant fibroblasts, Nphp1 was absent from the transition zone (Fig 6E). Hence, mouse Nphp4 is required for the localization of Nphp1, as it is in C. elegans [24]. Therefore, in mice, ciliary localization of Arl13b depends specifically on Tctn1, and not on Nphp4, whereas transition zone localization of Nphp1 depends specifically on Nphp4, and not on Tctn1. Thus, Tctn1 and Nphp4 have overlapping functions in promoting ciliogenesis, and distinct roles in controlling ciliary protein localization.
As cilia are required for vertebrate Hh signaling, we examined whether Tctn1-/- and Tctn1-/- Nphp4n/n mutant cells could respond to Hh pathway stimulation. Upon stimulation with Smo agonist (SAG), a Hh pathway agonist [60, 61], control and Nphp4n/n limb bud fibroblasts induced Hh target genes Gli1 (MIM 165220) and Ptch1 (MIM 601309) (Fig 7A and 7B). In contrast, Tctn1-/- and Tctn1-/- Nphp4n/n mutants did not increase Gli1 and Ptch1 transcription in response to SAG, indicating defective Hh signal transduction (Fig 7A and 7B).
Fig. 7. Roles for mouse Tctn1 and Nphp4 in Hh signaling and ciliary localization of Hh pathway components. 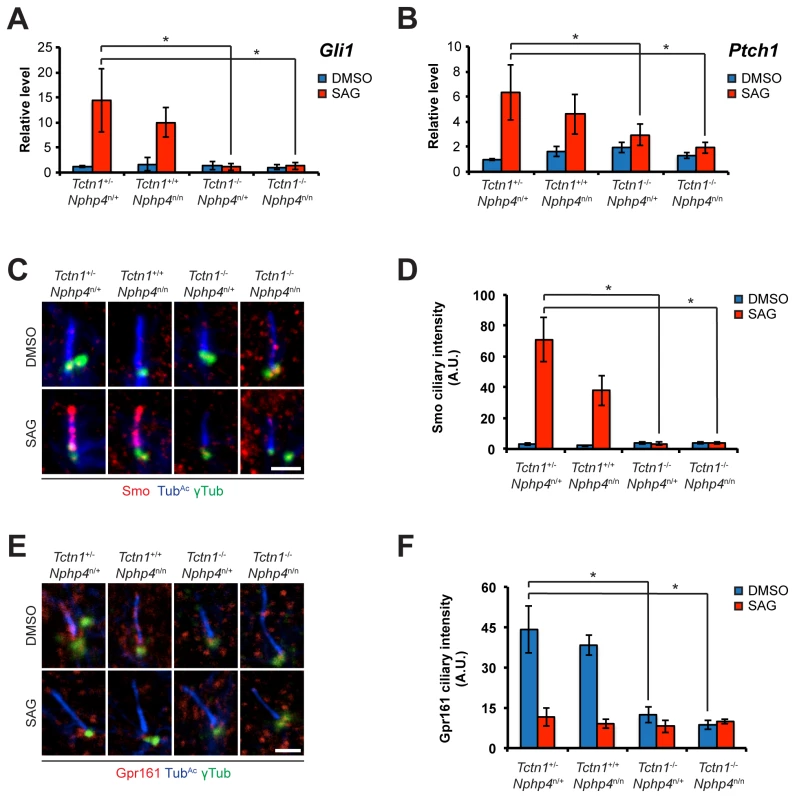
(A and B). mRNA levels of Gli1 and Ptch1 normalized to β-actin in forelimb bud cells treated with DMSO or SAG. Error bars represent standard deviations. Statistical significance according to unpaired Student’s t-tests (* p<0.05). (C) Limb bud fibroblasts treated with DMSO or SAG, then immunostained for Smo (red), TubAc (blue), and γ-tubulin (green). (D) Quantitation of Smo ciliary intensity in DMSO or SAG treated limb bud fibroblasts. (E) Limb bud fibroblasts treated with DMSO or SAG, then immunostained for Gpr161 (red), TubAc (blue), and γ-tubulin (green). (F) Quantitation of Gpr161 ciliary intensity in DMSO or SAG treated limb bud fibroblasts. Error bars represent standard error of the mean. Statistical significance according to unpaired Student’s t-tests (* p<0.05). Scale bars, 2 μm. To further assess the role of Tctn1 and Nphp4 in Hh signaling, we examined the localization of Hh pathway components in those mutant cells that were ciliated. Upon SAG treatment, Smo translocated into the cilia of control and Nphp4n/n cells, but failed to do so in Tctn1-/- and Tctn1-/- Nphp4n/n cells (Fig 7C and 7D). Tctn1-/- and Tctn1-/- Nphp4n/n cilia also failed to localize Gpr161 (Fig 7E and 7F), a negative regulator of the Hh pathway that exits cilia upon pathway activation [62]. Tctn1-/- and Tctn1-/- Nphp4n/n cells therefore have defects in Hh signal transduction, and disrupted ciliary localization of some Hh pathway components.
Tctn1 and Bbs1 have overlapping functions in mammalian ciliogenesis
As the genetic interactions between MKS and NPHP complex genes in C. elegans were conserved in mouse, we examined genetic interactions between mouse MKS and NPHP complex genes and BBS-associated genes to determine if these genetic interactions were also conserved. As described above, we observed a synthetic interaction between C. elegans NPHP complex genes and bbs-5 (Fig 4A and 4B). Because a Bbs5 mouse model has not been described, we investigated the function of another BBS-associated gene, Bbs1 (MIM 209901). Bbs1-/- mutant mice are obese, lack sperm flagella, and develop retinal degeneration, but do not exhibit either the polydactyly or renal abnormalities observed in human BBS-affected individuals [63]. Mouse Nphp4n/n Bbs1-/- double mutant embryos survived through the end of embryogenesis with no gross abnormalities, similar to each single mutant (S4B Fig), indicating that, in contrast to the genetic interaction between nphp-4 and a BBS-associated gene in C. elegans, there is no strong genetic interaction between mouse Nphp4 and Bbs1.
Unlike C. elegans, in which tctn-1 and bbs-5 did not interact, mouse Tctn1 and Bbs1 did genetically interact. Tctn1-/- Bbs1-/- double mutant embryos displayed dramatically increased polydactyly and exencephaly compared to either single mutant embryos (Fig 8A–8D, S3 and S4 Tables). Thus, a BBS-associated gene interacts with NPHP complex genes in C. elegans, but not in mouse, whereas a BBS-associated gene interacts with an MKS complex gene in mouse, but not in C. elegans. BBSome and transition zone complex genes interact in both organisms, but the specific genetic interactions differ between nematodes and mammals, suggesting that some ciliopathy gene interactions are species and/or cell-type specific, and may reflect evolutionary differences in the function of the nematode and mammalian MKS and NPHP complexes.
Fig. 8. Mouse Tctn1 genetically interacts with Bbs1. 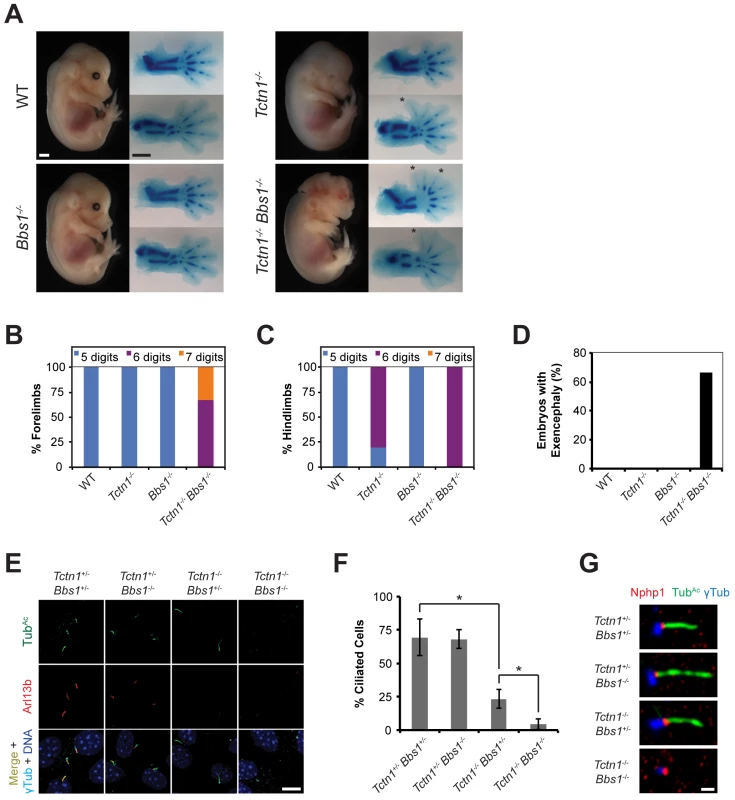
(A) Lateral views of wild type, Tctn1-/-, Bbs1-/- and Tctn1-/- Bbs1-/- embryos at E14.5. Exencephaly is apparent in the Tctn1-/- Bbs1-/- double mutant. Alcian blue staining of the corresponding genotype’s forelimb (top) and hindlimb (bottom) are included, with asterisks indicating extra digits. Scale bars, 1 mm. (B-D) Number of digits in the (B) forelimbs and (C) hindlimbs, and (D) the incidence of exencephaly for the indicated genotypes. Numbers of animals analyzed for polydactyly and exencephaly are shown in S3 and S4 Tables. (E) Limb bud fibroblasts of the indicated genotypes immunostained for TubAc (green), Arl13b (red), γ-tubulin (cyan), and DAPI (blue). Scale bar, 10 μm. (F) Quantification of proportion of ciliated limb bud fibroblasts. Error bars represent the standard deviation. Statistical significance according to unpaired Student’s t-tests (* p<0.0001). (G) Limb bud fibroblasts immunostained for Nphp1 (red), TubAc (green), and γ-tubulin (blue). Scale bar, 1 μm. To analyze the cell biological basis of the genetic interaction between Tctn1 and Bbs1, we examined the cilia of fibroblasts derived from developing forelimb buds. Tctn1-/- Bbs1-/- double mutant cells failed to form cilia, as detected by immunostaining for acetylated tubulin and Arl13b (Fig 8E and 8F). However, Tctn1-/- Bbs1-/- cells still possessed basal bodies, as identified by γ-tubulin staining (Fig 8E). Even when lacking axonemes, Tctn1-/- Bbs1-/- cells still localized Nphp1 appropriately to the transition zone (Fig 8G), unlike Nphp4n/n or Tctn1-/- Nphp4n/n cells (Fig 6E), suggesting that Tctn1 and Bbs1 have overlapping roles in mammalian ciliogenesis, but not in basal body or transition zone formation.
We hypothesized that the dramatic loss of cilia in Tctn1-/- Bbs1-/- double mutants compared to Tctn1-/- single mutants could account for the increased polydactyly of Tctn1-/- Bbs1-/- embryos, as decreased ciliary function and consequent decreased Gli3 processing results in polydactyly [10]. To test this possibility, we measured the mRNA expression levels of the Shh target genes Gli1 and Ptch1 in limb bud fibroblasts stimulated with SAG. Tctn1-/- mutant fibroblasts showed a reduction in Gli1 and Ptch1 expression compared to controls, and Tctn1-/- Bbs1-/- double mutants showed a further reduction compared to Tctn1-/- mutants (Fig 9A and 9B). Thus Tctn1-/- Bbs1-/- double mutants have abrogated Hh pathway activation, likely resulting in the exacerbation of ciliopathy phenotypes compared to either Tctn1 or Bbs1 single mutants.
Fig. 9. Mouse Tctn1 and Bbs1 have overlapping roles in Hh signaling and ciliary localization of Hh pathway components. 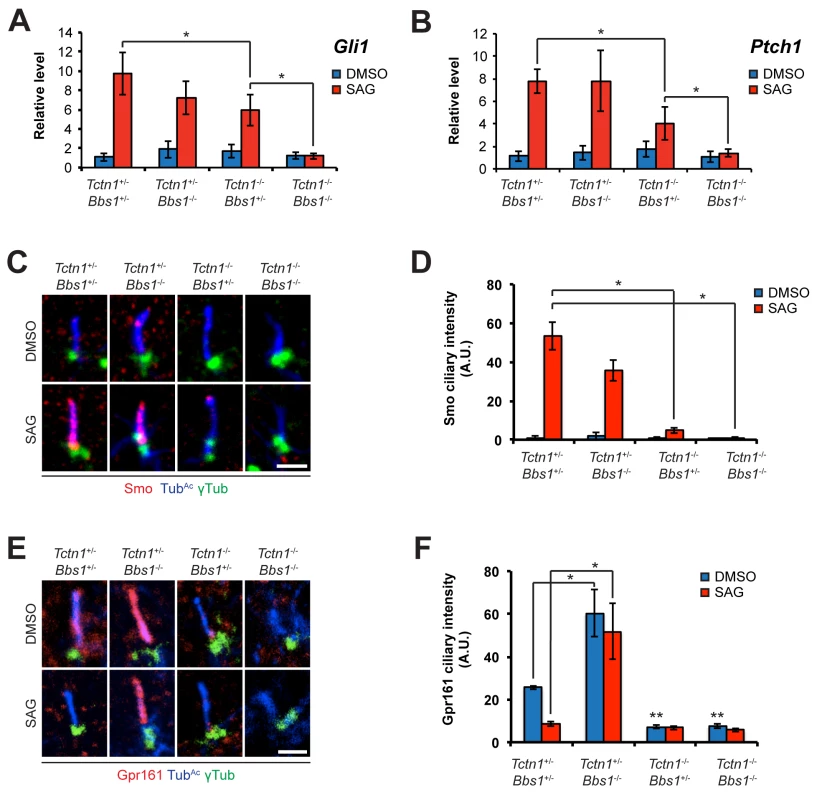
(A and B) mRNA levels of Gli1 and Ptch1 normalized to β-actin in forelimb bud cells treated with DMSO or SAG. Error bars represent the standard deviations. Statistical significance according to unpaired Student’s t-tests (* p<0.05). (C) Limb bud fibroblasts treated with DMSO or SAG, then immunostained for Smo (red), TubAc (blue), and γ-tubulin (green). (D) Quantitation of Smo ciliary intensity in DMSO or SAG treated limb bud fibroblasts. *p<0.05 in unpaired Student’s t-tests. (E) Limb bud fibroblasts treated with DMSO or SAG, then immunostained for Gpr161 (red), TubAc (blue), and γ-tubulin (green). (F) Quantitation of Gpr161 ciliary intensity in DMSO or SAG treated limb bud fibroblasts. *p<0.01 and **p<0.001 in unpaired Student’s t-tests with respect to Tctn1+/- Bbs1+/- mutant fibroblasts treated with DMSO. Error bars represent standard error of the mean. Scale bars, 2 μm. To further probe the role of Tctn1 and Bbs1 in Hh signal transduction, we examined the localization of the Hh pathway components Smo and Gpr161. Consistent with reduced Shh signaling, and in contrast to control and Bbs1-/- fibroblasts, Smo failed to translocate to cilia upon SAG treatment in Tctn1-/- and Tctn1-/- Bbs1-/- fibroblasts (Fig 9C and 9D). Gpr161, a negative regulator of Hh signaling, was present in cilia of control cells and was removed from cilia upon SAG treatment. Unexpectedly, Bbs1-/- mutant cells displayed increased levels of Gpr161 in cilia. Moreover, Gpr161 remained in Bbs1-/- mutant cilia upon SAG treatment, demonstrating that Bbs1 is required for the removal of Gpr161 from cilia (Fig 9E and 9F). Tctn1-/- cilia lacked Gpr161 even in the absence of SAG, as did Tctn1-/- Bbs1-/- double mutant cilia (Fig 9E and 9F). Thus, Bbs1 promotes the exit of Gpr161 from cilia and Tctn1 promotes the localization of Gpr161 to cilia.
Discussion
We investigated the genetic interactions between distinct biochemical complexes involved in ciliary function in both nematodes and mammals. We found that TCTN-1, the C. elegans ortholog of the mammalian Tectonics, localized to the ciliary transition zone. Loss of tctn-1 did not abrogate ciliogenesis on its own or in combination with mutations affecting other MKS complex components. However, loss of tctn-1 in combination with loss of components of the NPHP complex, a biochemically distinct transition zone complex, synergistically abrogated ciliogenesis (Fig 10). This interaction between MKS and NPHP complex genes also occurred in mice, suggesting that the overlapping functions of the MKS and NPHP complexes in supporting ciliary structure are evolutionarily conserved. To extend our understanding of how different ciliopathy protein complexes work together, we analyzed how BBS-associated proteins function with transition zone complexes. Components of the BBSome cooperate with MKS and NPHP complexes, but the specific genetic interactions that support ciliogenesis in C. elegans and mice differ (Fig 10). Thus, we have uncovered evolutionarily conserved and non-conserved functional interactions between three biochemically distinct ciliopathy complexes.
Fig. 10. Genetic interactions between transition zone complexes and the BBSome in C. elegans and mice. 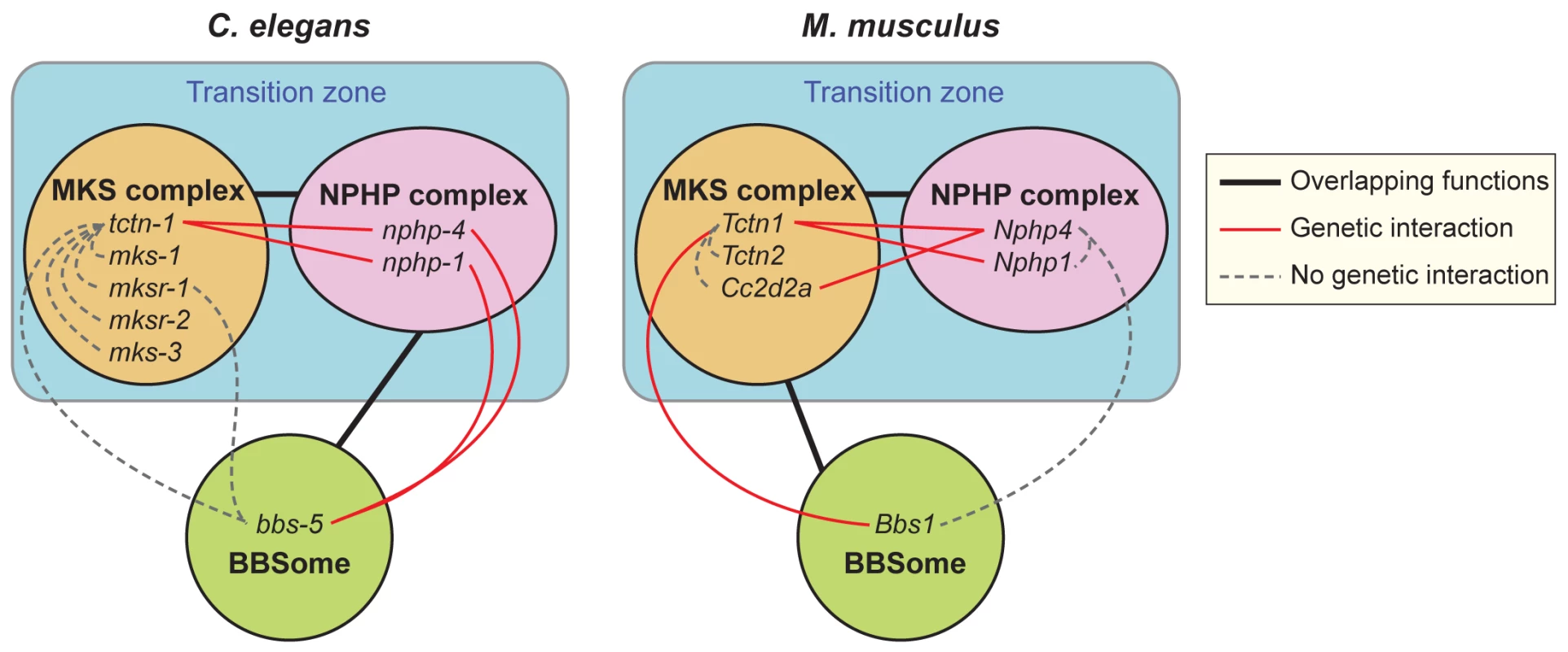
In C. elegans, MKS complex genes interact with NPHP complex genes, and a BBS-associated gene interacts with NPHP complex genes. In M. musculus, MKS complex genes interact with NPHP complex genes, and a BBS-associated gene interacts with at least one MKS complex gene. Tctn1 behaves as an MKS component in both organisms. Protein complexes that share overlapping functions in ciliogenesis are connected by black lines. Within protein complexes, specific genes that display a genetic interaction are connected by red lines. Genes that show no genetic interaction are connected by dashed grey lines. In both mammals and C. elegans, MKS and NPHP complex proteins localize to the transition zone at the ciliary base. Both the mouse and C. elegans orthologs of Tctn1 possess signal peptides, and are thus predicted to localize to the extracellular side of the transition zone. Where Tctn1 is relative to the remainder of the MKS complex, the NPHP complex, and the electron-micrographically defined components of the transition zone, such as the Y-links, remains to be determined. How the transition zone regulates the protein composition of cilia is not well understood, though the Y-links themselves have been implicated [13, 14, 40]. We found that C. elegans nphp-4 and tctn-1; nphp-4 mutant cilia had missing or reduced Y-links, but still localized ARL-13 and ODR-10, suggesting that the presence of intact Y-links is not critical for all ciliary protein localization. Additional analyses will be required to clarify the composition of Y-links and the relationship of Y-link structure to the control of ciliary composition.
Strikingly, Tctn1 was required for the ciliary localization of Arl13b in mouse, but not for the ciliary entry of ARL-13 in C. elegans, a difference that suggests that Tctn1 or the transition zone function distinctly in mammals and nematodes. In addition to Arl13b, which associates with membranes through palmitoyl anchors [42], mouse Tctn1 is required for the ciliary localization of other membrane-associated proteins, such as Smo, Pkd2 and Inpp5e [16, 26, 64]. We found that C. elegans TCTN-1 controls ciliary membrane composition similarly to other transition zone proteins, by excluding the non-ciliary membrane protein TRAM-1a from the cilium [13]. One possibility is that the mammalian MKS complex functions primarily to promote the localization of ciliary membrane-associated proteins, whereas the nematode MKS complex functions primarily to prevent the entry of non-ciliary membrane-associated proteins.
We found a genetic interaction between transition zone genes and BBS-associated genes. Intriguingly, the NPHP complex synergizes with a BBS-associated gene in C. elegans whereas an MKS complex gene synergizes with a BBS-associated gene in mice (Fig 10). No interactions were found between the MKS complex genes and a BBS-associated gene in C. elegans or between an NPHP complex gene and a BBS-associated gene in mice. Functional differences between bbs-5 and Bbs1 may account for the different genetic interactions observed in nematodes and mice. Despite these limitations, the specificity of the synergistic interactions between Bbs1 and Tctn1 in mice, and between bbs-5 and nphp-4 or nphp-1 in C. elegans suggests that the transition zone has altered its relationship to the BBSome through evolution. Intriguingly, the NPHP complex in C. elegans also localizes to the basal body-transition fibers, where the BBSome and IFT proteins dock [13, 40]. It will be interesting to determine whether the precise locations or functions of the MKS and NPHP complexes in the transition zone are different in nematodes and mammals, and whether these differences account for the distinct interactions with the BBSome.
How compromising the MKS and NPHP complexes, MKS and BBS complexes in mammals, or NPHP and BBS complexes in nematodes results in synthetic ciliary defects remains unclear. In mammals, Tctn1 is required for the localization of select transition zone proteins and a subset of ciliary membrane proteins [16]. Given that the BBSome facilitates the ciliary localization of other membrane proteins [29], compromising both the BBSome and MKS complex function may abrogate transport sufficiently to disrupt ciliary function and ciliogenesis. Physical interaction between Tctn1 and the BBSome has not been detected, but intriguingly, the MKS complex interactor, Cep290, binds to Bbs4 [16, 32]. Furthermore, loss of Bbs4 exacerbates the mislocalization of Rhodopsin in Cep290 mutant retinas [32], indicating that MKS complex interactors and the BBSome can cooperate to transport proteins to cilia.
BBS-associated genes have also been implicated in the export of proteins from cilia [65–68]. Our finding that Bbs1 is required for the ciliary exclusion of Gpr161, whereas Tctn1 is required for the ciliary localization of Gpr161, suggests that the BBSome and the transition zone may also act antagonistically to dynamically localize some ciliary proteins. Arl6, a BBSome-associated GTPase, is similarly required to remove Gpr161 from cilia [69]. This shared requirement suggests that removing Gpr161 from cilia may involve most or all BBS-associated proteins.
As the MKS and NPHP complexes also have overlapping functions in supporting ciliogenesis in mice, the NPHP complex may, like the MKS complex, regulate protein trafficking to cilia. Likewise, in C. elegans, in which the NPHP complex genes interact with both MKS and BBS genes, the NPHP complex may have overlapping functions in regulating ciliary protein composition. In support of this possibility, C. elegans NPHP-4 is required for proper ciliary localization of certain IFT proteins and Chlamydomonas Nphp4 regulates the entry of a subset of membrane proteins into flagella [25, 70].
As phenotypes reflect gene function, new phenotypes generated by the interaction of multiple mutations can reveal previously unknown functions of the genes involved. On their own, Nphp4 and Nphp1 do not play critical roles in limb bud or neural tube development. However, mutation of Nphp4 or Nphp1 modifies the Tctn1-/- limb and neural tube phenotypes, thereby demonstrating their involvement in the affected tissues. Similarly, human BBS patients have extra digits, yet mouse models of BBS have thus far not displayed polydactyly. Our results demonstrate that mouse Bbs1 does indeed affect limb patterning, exposed in the context of a Tctn1-/- mutation. Therefore, studying genes in combination can reveal roles for those genes that might otherwise be masked by the overlapping function of other genes. By identifying overlapping functions for genes in model organisms, particularly those from distinct biochemical complexes, we may be able to predict which genes interact and which do not in humans.
Thus far, understanding how human genotypes predict phenotypes is insufficient except for the most straightforward traits. For ciliopathy-affected individuals, even when the principal disease-causing mutation is identified, how mutations in that gene manifest as a range of phenotypes in different populations remains unknown. Genome-wide association studies (GWAS) have identified polymorphisms associated with many diseases and traits. However, these only account for a small percentage of the estimated heritability, provoking searches for sources of “missing heritability” [71, 72]. Work in simple organisms such as yeast has suggested that more extensive accounting for the effects of allelic variation can explain the phenotypic variation in disease and common traits [73]. Our work suggests that in heterogeneous, oligogenic disorders such as ciliopathies, missing heritability could arise not from the effects of polymorphisms by themselves but from gene specific interactions between polymorphisms, which GWAS fail to detect. Our double mutant analyses demonstrate that the phenotypes that arise for a given mutation depend on the genetic context, such that an innocuous mutation in one genetic background may alter a disease phenotype in another. In support of this possibility in the specific case of ciliopathies, human genetic studies have found NPHP and BBS patients with multiple genetic lesions that contribute to their phenotypes [9, 74, 75]. As common or rare variants may be shared within a family, they may be a source of heritability and significant modifiers of disease phenotypes. Detecting variants that affect expressivity only in specific genetic contexts will require power and computation not currently present in GWAS. Therefore, as a complement to GWAS, animal genetic models provide a valuable means to identify the oliogenic interactions that define inherited disease phenotypes.
Materials and Methods
Ethics statement
Mouse protocols were approved by the Institutional Animal Care and Use Committee at the University of California, San Francisco (approval AN091380). Euthanasia was performed by cervical dislocation, and all animal studies were conducted in accordance with internationally-accepted standards.
C. elegans strains
All strains (S5 Table) were generated and maintained under standard conditions [76]. The tctn-1 mutant strain, E04A4.6(ok3021) IV (WormBase ID: WBVar00094107), was obtained from the C. elegans Gene Knockout Consortium via the Caenorhabditis Genetics Center and outcrossed to wild type (N2) six times. Standard mating procedures were used to generate double mutants and to introduce fluorescently tagged protein constructs into the specified genetic backgrounds.
Subcellular protein localization
Strains bearing TCTN-1 C-terminal GFP translational fusion constructs were generated by stitch-PCR, essentially as previously described [77]. In brief, the bbs-8 promoter region, followed by the complete genomic coding region of the tctn-1 gene, was fused in-frame to EGFP, and used to create transgenic lines. Live animals were anaesthetized using 10 mM levamisole, mounted on 5% agarose pads, and observed by spinning-disc confocal microscopy. The subcellular localization patterns of the fluorescent marker-tagged proteins were assessed in either wild type (N2) animals or in the indicated mutant backgrounds. Mislocalization phenotypes were confirmed by analyzing at least 50 animals for each strain. For visualization of ODR-10::GFP, animals were fixed and immunostained with anti-GFP (600-101-215; Rockland) following the Finney-Ruvkun protocol [78], and imaged on a Leica TCS SPE confocal microscope.
C. elegans assays
To assess sensory neuron dye filling, 300 synchronized L4 worms were washed with S-basal and immersed in 10 mg/ml DiI in S-basal for two hours. Worms were rinsed with S-basal, and plated onto an OP50 seeded plate. After one hour, the animals were anesthetized with 0.2 M sodium azide, mounted on a 2% agarose pad, and imaged on a Zeiss Axioplan microscope. Assays were performed on at least three separate occasions for each genotype. Statistical significance was assessed using the unpaired Student’s t test.
To measure growth, 150 synchronized L1 larvae were plated on OP50 and incubated at 20°C. After 45 hours, the animals were counted and staged. Assay was repeated three times. To measure size, DIC images of synchronized day 1 gravid adults were captured on a Zeiss Axioplan microscope. Perimeter and area were measured using Fiji software for 20–35 animals of each genotype. Egg laying was measured by picking synchronized day 1 gravid adults onto individual plates. One hour later, the number of eggs on the plate was counted. Brood size was measured by plating synchronized L4 animals, and transferring the animals onto fresh plates daily until the end of the reproductive period. Progeny from each plate was counted for at least ten animals. Standard osmotic avoidance and chemotaxis assays were performed as previously described using 8M glycerol to create high osmolarity, and diacytyl (1 : 1000 dilution) and butanone (1 : 1000 dilution) as chemoattractants [79, 80].
Mouse alleles and mutant analysis
Tctn1−(Tctn1Gt(KST296)Byg), Tctn2−(Tctn2tm1.1Reit), Nphp4n(Nphp4nmf192), Nphp1−(Nphp1tm1Jgg) Cc2d2a−(Cc2d2aGt(AA0274)Wtsi), and Bbs1−(Bbs1tm1Vcs) alleles have been previously described [16, 21, 33, 50, 63, 81]. Single and double mutant embryos were generated by timed matings of trans-heterozygous animals. For gross phenotypic examination, embryos were harvested at E14.5, fixed in 4% paraformaldehyde, and imaged on a Zeiss Discovery V12 steREO microscope. At least three embryos of each double mutant combination were analyzed. Limb bud digits were visualized by fixing E14.5 embryos in ethanol and staining in Alcian blue and Alizarin red, as previously described [82].
Limb fibroblast derivation
Limb fibroblast cell lines were generated from E13.5 mouse embryos that were harvested and transferred into Dulbecco's PBS with penicillin, streptomycin and fungizone (P/S/F) at 37°C. For each embryo, both forelimb buds were extracted and incubated for 2 minutes at room temperature in 0.05% Trypsin-EDTA solution, after which they were disaggregated by pipetting. The resulting cell suspensions were immediately transferred into DMEM medium supplemented with 20% fetal bovine serum (FBS) and P/S/F, and incubated overnight with gentle shaking at 37°C and 5% CO2. The next day cells were confluent and were passaged using the trypsinization procedure described above. Cells were maintained in DMEM+15%FBS+P/S/F and were not diluted more than five times when passaging. All analyses were performed in cells passaged less than ten times.
Immunofluorescence
For immunostaining, confluent limb bud fibroblasts were starved for 48 hours in OptiMEM medium to induce ciliation. For immunostaining of Smo or Gpr161, confluent limb bud fibroblasts were starved for 24 hours in Optimem, and treated DMSO or SAG (1 μM) for 6 hours. Cells were fixed by incubating them first in DPBS+4% paraformaldehyde (10 min, RT), followed by cold methanol (3 min, -20°C). Cells were blocked and permeabilized for at least 15 min at RT in blocking solution (PBS+2% donkey serum+0.1% Triton-X100+0.02% sodium azide). Coverslips were then incubated with primary antibodies diluted in blocking solution for 3 hours at RT. After two rinses in PBS, coverslips were incubated with fluorophore (Alexa Fluor)-conjugated donkey secondary antibodies and DAPI for 1 hour at RT in darkness. This was followed by two rinses in milliQ water and the mounting of coverslips on slides using gelvatol mounting medium. Slides were imaged on a Leica TCS SPE confocal microscope. Antibodies used were: acetylated tubulin (clone 6-11B-1; Sigma), γ-tubulin (C-20; Santa Cruz), Arl13b (17711-1-AP; Proteintech), Nphp1 (gift from Dr. Greg Pazour), Smoothened (ab38686; Abcam) and Gpr161 (13398-1-AP; Proteintech). For quantitation of cilia numbers, fields of cells were imaged at seven positions spanning the whole sample. Acetylated tubulin-positive cilia associated with a γ-tubulin-positive basal body were counted in each field and normalized to the number of nuclei in the field. Statistical significance was assessed using the unpaired Student’s t test. Quantification of Smo and Gpr161 ciliary intensity and immunostaining of limb bud cryosections were performed as previously described [16, 64].
Shh signaling assay
To measure Shh signaling, confluent limb bud fibroblasts were starved in OptiMEM for 24 hours, then treated with DMSO or 200nM SAG (Cayman Chemical) for 24 hours. RNA was extracted from these cells using the RNeasy Mini Kit (Qiagen) and cDNA was synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR was performed using EXPRESS SYBR GreenER qPCR Supermix, with premixed ROX (Invitrogen) on a 7300 Real-Time PCR machine (Applied Biosystems). Transcript levels of Gli1 and Ptch1 were normalized to levels of β-actin. The primers used were Gli1F, 5’-GGTGCTGCCTATAGCCAGTGTCCTC-3’; Gli1R, 5’-GTGCCAATCCGGTGGAGTCAGACCC-3’; Ptch1F, 5’-CTCTGGAGCAGATTTCCAAGG-3’; Ptch1R, 5’-TGCCGCAGTTCTTTTGAATG-3’; β-actinF, 5’-CACAGCTTCTTTGCAGCTCCTT-3’; and β-actinR, 5’-CGTCATCCATGGCGAACTG-3’. Experiments were performed in triplicate and repeated at least three times. Statistical significance was assessed using the unpaired Student’s t test.
Electron microscopy
Transmission electron microscopy of C. elegans amphid cilia was performed as previously described [13, 42]. Images depicting nphp-4 single mutant cilia were of strain PT709: nphp-4(tm925) him-5(e1490) V. No differences in ciliary structure were detected between nphp-4 and nphp-4 him-5 genotypes (Lambacher et al., manuscript in revision). For TEM analysis of limb cilia in mice, we isolated forelimb buds from E11.5 embryos, washed with Dulbecco's PBS and incubated in fixative solution containing 0.1M sodium cacodylate (pH 7.2), 4% formaldehyde, 4% glutaraldehyde, 4% tannic acid and 5% sucrose, where they remained for two days. The limb buds were processed for TEM as previously described [16]. Images were acquired using a JEOL JEM-1400 electron microscope. For quantification of limb bud cilia, axonemes with docked basal bodies were counted per field in at least seven fields. Statistical significance was assessed using the unpaired Student’s t test.
Supporting Information
Zdroje
1. Morell RJ, Brewer CC, Ge D, Snieder H, Zalewski CK, King KA, et al. A twin study of auditory processing indicates that dichotic listening ability is a strongly heritable trait. Hum Genet. 2007;122(1):103–11. doi: 10.1007/s00439-007-0384-5 17533509.
2. Adato A, Kalinski H, Weil D, Chaib H, Korostishevsky M, Bonne-Tamir B. Possible interaction between USH1B and USH3 gene products as implied by apparent digenic deafness inheritance. Am J Hum Genet. 1999;65(1):261–5. doi: 10.1086/302438 10364543; PubMed Central PMCID: PMCPMC1378101.
3. Floeth M, Bruckner-Tuderman L. Digenic junctional epidermolysis bullosa: mutations in COL17A1 and LAMB3 genes. Am J Hum Genet. 1999;65(6):1530–7. doi: 10.1086/302672 10577906; PubMed Central PMCID: PMCPMC1288363.
4. Vincent S, Planells R, Defoort C, Bernard MC, Gerber M, Prudhomme J, et al. Genetic polymorphisms and lipoprotein responses to diets. Proc Nutr Soc. 2002;61(4):427–34. 12691171.
5. Hichri H, Stoetzel C, Laurier V, Caron S, Sigaudy S, Sarda P, et al. Testing for triallelism: analysis of six BBS genes in a Bardet-Biedl syndrome family cohort. Eur J Hum Genet. 2005;13(5):607–16. doi: 10.1038/sj.ejhg.5201372 15770229.
6. Abu-Safieh L, Al-Anazi S, Al-Abdi L, Hashem M, Alkuraya H, Alamr M, et al. In search of triallelism in Bardet-Biedl syndrome. Eur J Hum Genet. 2012;20(4):420–7. doi: 10.1038/ejhg.2011.205 22353939; PubMed Central PMCID: PMCPMC3306854.
7. Nakane T, Biesecker LG. No evidence for triallelic inheritance of MKKS/BBS loci in Amish Mckusick-Kaufman syndrome. Am J Med Genet A. 2005;138(1):32–4. doi: 10.1002/ajmg.a.30593 16104012.
8. Katsanis N, Eichers ER, Ansley SJ, Lewis RA, Kayserili H, Hoskins BE, et al. BBS4 is a minor contributor to Bardet-Biedl syndrome and may also participate in triallelic inheritance. Am J Hum Genet. 2002;71(1):22–9. doi: 10.1086/341031 12016587; PubMed Central PMCID: PMCPMC384990.
9. Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, et al. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293(5538):2256–9. doi: 10.1126/science.1063525 11567139.
10. Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–44. doi: 10.1038/nrg2774 20395968; PubMed Central PMCID: PMCPMC3121168.
11. Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J Cell Biol. 2012;197(6):697–709. doi: 10.1083/jcb.201111146 22689651; PubMed Central PMCID: PMCPMC3373398.
12. Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, et al. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2012;14(1):61–72. doi: 10.1038/ncb2410 22179047.
13. Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192(6):1023–41. doi: 10.1083/jcb.201012116 21422230; PubMed Central PMCID: PMCPMC3063147.
14. Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, et al. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190(5):927–40. doi: 10.1083/jcb.201006105 20819941; PubMed Central PMCID: PMCPMC2935561.
15. Huang L, Szymanska K, Jensen VL, Janecke AR, Innes AM, Davis EE, et al. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am J Hum Genet. 2011;89(6):713–30. doi: 10.1016/j.ajhg.2011.11.005 22152675; PubMed Central PMCID: PMCPMC3234373.
16. Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43(8):776–84. doi: 10.1038/ng.891 21725307; PubMed Central PMCID: PMCPMC3145011.
17. Gilula NB, Satir P. The ciliary necklace. A ciliary membrane specialization. J Cell Biol. 1972;53(2):494–509. 4554367; PubMed Central PMCID: PMCPMC2108734.
18. Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117(2):456–87. 2428682.
19. Dowdle WE, Robinson JF, Kneist A, Sirerol-Piquer MS, Frints SG, Corbit KC, et al. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet. 2011;89(1):94–110. doi: 10.1016/j.ajhg.2011.06.003 21763481; PubMed Central PMCID: PMCPMC3135817.
20. Fliegauf M, Horvath J, von Schnakenburg C, Olbrich H, Müller D, Thumfart J, et al. Nephrocystin specifically localizes to the transition zone of renal and respiratory cilia and photoreceptor connecting cilia. J Am Soc Nephrol. 2006;17(9):2424–33. doi: 10.1681/ASN.2005121351 16885411.
21. Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145(4):513–28. doi: 10.1016/j.cell.2011.04.019 21565611; PubMed Central PMCID: PMCPMC3383065.
22. Williams CL, Winkelbauer ME, Schafer JC, Michaud EJ, Yoder BK. Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditis elegans ciliogenesis. Mol Biol Cell. 2008;19(5):2154–68. doi: 10.1091/mbc.E07-10-1070 18337471; PubMed Central PMCID: PMCPMC2366840.
23. Williams CL, Masyukova SV, Yoder BK. Normal ciliogenesis requires synergy between the cystic kidney disease genes MKS-3 and NPHP-4. J Am Soc Nephrol. 2010;21(5):782–93. doi: 10.1681/ASN.2009060597 20150540; PubMed Central PMCID: PMCPMC2865747.
24. Winkelbauer ME, Schafer JC, Haycraft CJ, Swoboda P, Yoder BK. The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J Cell Sci. 2005;118(Pt 23):5575–87. doi: 10.1242/jcs.02665 16291722.
25. Jauregui AR, Nguyen KC, Hall DH, Barr MM. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J Cell Biol. 2008;180(5):973–88. doi: 10.1083/jcb.200707090 18316409; PubMed Central PMCID: PMCPMC2265406.
26. Roberson EC, Dowdle WE, Ozanturk A, Garcia-Gonzalo FR, Li C, Halbritter J, et al. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J Cell Biol. 2015;209(1):129–42. doi: 10.1083/jcb.201411087 25869670; PubMed Central PMCID: PMCPMC4395494.
27. Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129(6):1201–13. doi: 10.1016/j.cell.2007.03.053 17574030.
28. Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci U S A. 2008;105(11):4242–6. doi: 10.1073/pnas.0711027105 18334641; PubMed Central PMCID: PMCPMC2393805.
29. Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141(7):1208–19. doi: 10.1016/j.cell.2010.05.015 20603001; PubMed Central PMCID: PMCPMC2898735.
30. Blacque OE, Reardon MJ, Li C, McCarthy J, Mahjoub MR, Ansley SJ, et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18(13):1630–42. doi: 10.1101/gad.1194004 15231740; PubMed Central PMCID: PMCPMC443524.
31. Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, Rix S, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40(4):443–8. doi: 10.1038/ng.97 18327255.
32. Zhang Y, Seo S, Bhattarai S, Bugge K, Searby CC, Zhang Q, et al. BBS mutations modify phenotypic expression of CEP290-related ciliopathies. Hum Mol Genet. 2014;23(1):40–51. doi: 10.1093/hmg/ddt394 23943788; PubMed Central PMCID: PMCPMC3857943.
33. Reiter JF, Skarnes WC. Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev. 2006;20(1):22–7. doi: 10.1101/gad.1363606 16357211; PubMed Central PMCID: PMCPMC1356097.
34. Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5(3):411–21. 10882127.
35. Chen N, Mah A, Blacque OE, Chu J, Phgora K, Bakhoum MW, et al. Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 2006;7(12):R126. doi: 10.1186/gb-2006-7-12-r126 17187676; PubMed Central PMCID: PMCPMC1794439.
36. Phirke P, Efimenko E, Mohan S, Burghoorn J, Crona F, Bakhoum MW, et al. Transcriptional profiling of C. elegans DAF-19 uncovers a ciliary base-associated protein and a CDK/CCRK/LF2p-related kinase required for intraflagellar transport. Dev Biol. 2011;357(1):235–47. doi: 10.1016/j.ydbio.2011.06.028 21740898; PubMed Central PMCID: PMCPMC3888451.
37. Colosimo ME, Brown A, Mukhopadhyay S, Gabel C, Lanjuin AE, Samuel AD, et al. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr Biol. 2004;14(24):2245–51. doi: 10.1016/j.cub.2004.12.030 15620651.
38. Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425(6958):628–33. doi: 10.1038/nature02030 14520415.
39. Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, et al. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139(1):171–88. 7705621; PubMed Central PMCID: PMCPMC1206316.
40. Jensen VL, Li C, Bowie RV, Clarke L, Mohan S, Blacque OE, et al. Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J. 2015. doi: 10.15252/embj.201488044 26392567.
41. Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12(5):767–78. doi: 10.1016/j.devcel.2007.03.004 17488627.
42. Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, Foley-Fisher C, et al. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol. 2010;188(6):953–69. doi: 10.1083/jcb.200908133 20231383; PubMed Central PMCID: PMCPMC2845074.
43. Cevik S, Sanders AA, Van Wijk E, Boldt K, Clarke L, van Reeuwijk J, et al. Active transport and diffusion barriers restrict Joubert Syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet. 2013;9(12):e1003977. doi: 10.1371/journal.pgen.1003977 24339792; PubMed Central PMCID: PMCPMC3854969.
44. Sengupta P, Chou JH, Bargmann CI. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84(6):899–909. 8601313.
45. Dwyer ND, Troemel ER, Sengupta P, Bargmann CI. Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell. 1998;93(3):455–66. 9590179.
46. Lee BH, Liu J, Wong D, Srinivasan S, Ashrafi K. Hyperactive neuroendocrine secretion causes size, feeding, and metabolic defects of C. elegans Bardet-Biedl syndrome mutants. PLoS Biol. 2011;9(12):e1001219. doi: 10.1371/journal.pbio.1001219 22180729; PubMed Central PMCID: PMCPMC3236739.
47. Xu Q, Zhang Y, Wei Q, Huang Y, Li Y, Ling K, et al. BBS4 and BBS5 show functional redundancy in the BBSome to regulate the degradative sorting of ciliary sensory receptors. Sci Rep. 2015;5 : 11855. doi: 10.1038/srep11855 26150102; PubMed Central PMCID: PMCPMC4493597.
48. Mollet G, Salomon R, Gribouval O, Silbermann F, Bacq D, Landthaler G, et al. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat Genet. 2002;32(2):300–5. doi: 10.1038/ng996 12244321.
49. Otto E, Hoefele J, Ruf R, Mueller AM, Hiller KS, Wolf MT, et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am J Hum Genet. 2002;71(5):1161–7. doi: 10.1086/344395 12205563; PubMed Central PMCID: PMCPMC385091.
50. Won J, Marín de Evsikova C, Smith RS, Hicks WL, Edwards MM, Longo-Guess C, et al. NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum Mol Genet. 2011;20(3):482–96. doi: 10.1093/hmg/ddq494 21078623; PubMed Central PMCID: PMCPMC3016909.
51. Hildebrandt F, Otto E, Rensing C, Nothwang HG, Vollmer M, Adolphs J, et al. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet. 1997;17(2):149–53. doi: 10.1038/ng1097-149 9326933.
52. Konrad M, Saunier S, Heidet L, Silbermann F, Benessy F, Calado J, et al. Large homozygous deletions of the 2q13 region are a major cause of juvenile nephronophthisis. Hum Mol Genet. 1996;5(3):367–71. 8852662.
53. Parisi MA, Bennett CL, Eckert ML, Dobyns WB, Gleeson JG, Shaw DW, et al. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am J Hum Genet. 2004;75(1):82–91. doi: 10.1086/421846 15138899; PubMed Central PMCID: PMCPMC1182011.
54. Gorden NT, Arts HH, Parisi MA, Coene KL, Letteboer SJ, van Beersum SE, et al. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am J Hum Genet. 2008;83(5):559–71. doi: 10.1016/j.ajhg.2008.10.002 18950740; PubMed Central PMCID: PMCPMC2668034.
55. Noor A, Windpassinger C, Patel M, Stachowiak B, Mikhailov A, Azam M, et al. CC2D2A, encoding a coiled-coil and C2 domain protein, causes autosomal-recessive mental retardation with retinitis pigmentosa. Am J Hum Genet. 2008;82(4):1011–8. doi: 10.1016/j.ajhg.2008.01.021 18387594; PubMed Central PMCID: PMCPMC2427291.
56. Tallila J, Jakkula E, Peltonen L, Salonen R, Kestilä M. Identification of CC2D2A as a Meckel syndrome gene adds an important piece to the ciliopathy puzzle. Am J Hum Genet. 2008;82(6):1361–7. doi: 10.1016/j.ajhg.2008.05.004 18513680; PubMed Central PMCID: PMCPMC2427307.
57. Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1(4):e53. doi: 10.1371/journal.pgen.0010053 16254602; PubMed Central PMCID: PMCPMC1270009.
58. Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132(13):3103–11. doi: 10.1242/dev.01894 15930098.
59. May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287(2):378–89. doi: 10.1016/j.ydbio.2005.08.050 16229832.
60. Frank-Kamenetsky M, Zhang XM, Bottega S, Guicherit O, Wichterle H, Dudek H, et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1(2):10. 12437772; PubMed Central PMCID: PMCPMC137065.
61. Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99(22):14071–6. doi: 10.1073/pnas.182542899 12391318; PubMed Central PMCID: PMCPMC137838.
62. Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152(1–2):210–23. doi: 10.1016/j.cell.2012.12.026 23332756.
63. Davis RE, Swiderski RE, Rahmouni K, Nishimura DY, Mullins RF, Agassandian K, et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci U S A. 2007;104(49):19422–7. doi: 10.1073/pnas.0708571104 18032602; PubMed Central PMCID: PMCPMC2148305.
64. Garcia-Gonzalo FR, Phua SC, Roberson EC, Garcia G, Abedin M, Schurmans S, et al. Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling. Dev Cell. 2015;34(4):400–9. doi: 10.1016/j.devcel.2015.08.001 26305592; PubMed Central PMCID: PMCPMC4557815.
65. Domire JS, Green JA, Lee KG, Johnson AD, Askwith CC, Mykytyn K. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell Mol Life Sci. 2011;68(17):2951–60. doi: 10.1007/s00018-010-0603-4 21152952; PubMed Central PMCID: PMCPMC3368249.
66. Zhang Q, Nishimura D, Seo S, Vogel T, Morgan DA, Searby C, et al. Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc Natl Acad Sci U S A. 2011;108(51):20678–83. doi: 10.1073/pnas.1113220108 22139371; PubMed Central PMCID: PMCPMC3251145.
67. Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187(7):1117–32. doi: 10.1083/jcb.200909183 20038682; PubMed Central PMCID: PMCPMC2806276.
68. Lechtreck KF, Brown JM, Sampaio JL, Craft JM, Shevchenko A, Evans JE, et al. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol. 2013;201(2):249–61. doi: 10.1083/jcb.201207139 23589493; PubMed Central PMCID: PMCPMC3628507.
69. Liew GM, Ye F, Nager AR, Murphy JP, Lee JS, Aguiar M, et al. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev Cell. 2014;31(3):265–78. doi: 10.1016/j.devcel.2014.09.004 25443296; PubMed Central PMCID: PMCPMC4255629.
70. Awata J, Takada S, Standley C, Lechtreck KF, Bellvé KD, Pazour GJ, et al. NPHP4 controls ciliary trafficking of membrane proteins and large soluble proteins at the transition zone. J Cell Sci. 2014;127(21):4714–27. doi: 10.1242/jcs.155275 25150219; PubMed Central PMCID: PMCPMC4215714.
71. Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–51. doi: 10.1038/nrg2554 19293820.
72. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494 19812666; PubMed Central PMCID: PMCPMC2831613.
73. Bloom JS, Ehrenreich IM, Loo WT, Lite TL, Kruglyak L. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494(7436):234–7. doi: 10.1038/nature11867 23376951; PubMed Central PMCID: PMCPMC4001867.
74. Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet. 2003;72(5):1187–99. doi: 10.1086/375178 12677556; PubMed Central PMCID: PMCPMC1180271.
75. Hoefele J, Wolf MT, O'Toole JF, Otto EA, Schultheiss U, Dêschenes G, et al. Evidence of oligogenic inheritance in nephronophthisis. J Am Soc Nephrol. 2007;18(10):2789–95. doi: 10.1681/ASN.2007020243 17855640.
76. Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. 4366476; PubMed Central PMCID: PMCPMC1213120.
77. Inglis PN, Blacque OE, Leroux MR. Functional genomics of intraflagellar transport-associated proteins in C. elegans. Methods Cell Biol. 2009;93 : 267–304. doi: 10.1016/S0091-679X(08)93014-4 20409822.
78. Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63(5):895–905. 2257628.
79. Culotti JG, Russell RL. Osmotic avoidance defective mutants of the nematode Caenorhabditis elegans. Genetics. 1978;90(2):243–56. 730048; PubMed Central PMCID: PMCPMC1213887.
80. Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74(3):515–27. 8348618.
81. Louie CM, Caridi G, Lopes VS, Brancati F, Kispert A, Lancaster MA, et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet. 2010;42(2):175–80. doi: 10.1038/ng.519 20081859; PubMed Central PMCID: PMCPMC2884967.
82. Nagy A, Gertsensten M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Press; 2003.
Štítky
Genetika Reprodukční medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis inČlánek Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 11- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



