-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
Danforth's short tail (Sd) is a semidominant mutation on mouse chromosome 2, characterized by spinal defects, urogenital defects, and anorectal malformations. However, the gene responsible for the Sd phenotype was unknown. In this study, we identified the molecular basis of the Sd mutation. By positional cloning, we identified the insertion of an early transposon in the Sd candidate locus approximately 12-kb upstream of Ptf1a. We found that insertion of the transposon caused overexpression of three neighboring genes, Gm13344, Gm13336, and Ptf1a, in Sd mutant embryos and that the Sd phenotype was not caused by disruption of an as-yet-unknown gene in the candidate locus. Using multiple knockout and knock-in mouse models, we demonstrated that misexpression of Ptf1a, but not of Gm13344 or Gm13336, in the notochord, hindgut, cloaca, and mesonephros was sufficient to replicate the Sd phenotype. The ectopic expression of Ptf1a in the caudal embryo resulted in attenuated expression of Cdx2 and its downstream target genes T, Wnt3a, and Cyp26a1; we conclude that this is the molecular basis of the Sd phenotype. Analysis of Sd mutant mice will provide insight into the development of the spinal column, anus, and kidney.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003204
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003204Summary
Danforth's short tail (Sd) is a semidominant mutation on mouse chromosome 2, characterized by spinal defects, urogenital defects, and anorectal malformations. However, the gene responsible for the Sd phenotype was unknown. In this study, we identified the molecular basis of the Sd mutation. By positional cloning, we identified the insertion of an early transposon in the Sd candidate locus approximately 12-kb upstream of Ptf1a. We found that insertion of the transposon caused overexpression of three neighboring genes, Gm13344, Gm13336, and Ptf1a, in Sd mutant embryos and that the Sd phenotype was not caused by disruption of an as-yet-unknown gene in the candidate locus. Using multiple knockout and knock-in mouse models, we demonstrated that misexpression of Ptf1a, but not of Gm13344 or Gm13336, in the notochord, hindgut, cloaca, and mesonephros was sufficient to replicate the Sd phenotype. The ectopic expression of Ptf1a in the caudal embryo resulted in attenuated expression of Cdx2 and its downstream target genes T, Wnt3a, and Cyp26a1; we conclude that this is the molecular basis of the Sd phenotype. Analysis of Sd mutant mice will provide insight into the development of the spinal column, anus, and kidney.
Introduction
Danforth's short tail (Sd) is a semidominant spontaneous mutant mouse characterized by severe spinal defects, urogenital defects, and anorectal malformations [1], [2], [3]. Heterozygous and homozygous Sd animals display a broad range of abnormalities in the vertebral column, including reduction or absence of the dens axis, reduction of all vertebral bodies in the dorsoventral axis, split vertebrae, and truncation of the caudal vertebral column [4], [5], [6]. The vertebral columns of Sd/Sd and Sd/+ mice are usually truncated at the seventh thoracic and the sixth caudal vertebral body, respectively [7]. The urogenital system in Sd heterozygotes may display malformations ranging from displaced to missing kidneys. Homozygotes invariably have missing or severely malformed and dislocated kidneys. The rectum and anal opening are missing, and the embryonic cloaca persists. Homozygous animals die within 24 h after birth [4].
Although Sd is known to map to mouse chromosome 2, little is known about the molecular nature of the mutation. Double mutants between the Sd and undulated (un) alleles showed reduced expression of Pax1 and enhancement of the vertebral malformations [8]. Pax1 expression is regulated by signals from the notochord [9], [10], thus providing a potential molecular link for the interaction between un and Sd. Zachgo et al. obtained a lacZ enhancer trap insertion called Etl4lacZ, which is tightly linked to Sd. If Etl4lacZ is present in trans (i.e., on the chromosome that is wild type (WT) for Sd), the Sd phenotype is enhanced [11]. In contrast, if Etl4lacZ is present in cis (i.e., on the same chromosome as Sd), the phenotype is attenuated, suggesting a direct interaction of the transgene insertion with the Sd gene at the DNA level. However, neither the Sd mutation nor the Sd gene is known [12], [13].
We previously obtained a mutant mouse line, SktGt, through gene-trap mutagenesis, and identified the Skt gene. We found that the SktGt locus was located 0.95 cM distal to the Sd locus, and that the Etl4lacZ site was located in the third intron of the Skt gene [14]. Because the Sd region had been shown to be located 0.15–0.3 cM distal to the marker D2Mit362 [13], [15] and 0.75 cM proximal to Etl4lacZ [11], it was clear that the Sd locus is located between D2Mit362 and the Skt gene. In this study, we identified the cause of the Sd mutation by generating an Sd-derived cosmid contig of approximately 0.5-Mb around the Sd locus and using it to guide the production of genetically engineered mice with particular genetic alterations.
Results
Phenotype of Sd mice
Although the Sd mouse was identified in 1940, detailed histological findings have not yet been fully described. In this study, we mainly analyzed three tissues—the vertebrae, urogenital tract, and kidney—because characteristic features are found in these tissues of Sd mice. Sd/Sd homozygotes had similar, but much more severe, abnormalities than Sd/+ heterozygotes in terms of truncation of the vertebrae at day 0 postpartum (Figure 1A, Text S1), defects of the nucleus pulposus in the intervertebral discs, anorectal malformations, and renal hypoplasia/agenesis (Figure 1B). We also revealed hypoplasia of the dens (Figure 1C) and sacral hypoplasia (Figure 1D, 1E) by high-resolution computed tomography (Text S1). Thus, Sd is considered a mouse model for caudal regression syndrome (CRS), characterized by vertebral, anorectal, and urogenital abnormalities. We also carried out a lung-floating test to analyze the cause of death. As shown in Figure 1F, the lungs of Sd/Sd neonates sank in water. Histological sections of the lung revealed atelectasis of the lung (Figure 1G), indicating that no breathing occurred after birth.
Fig. 1. Morphology of Sd mice. 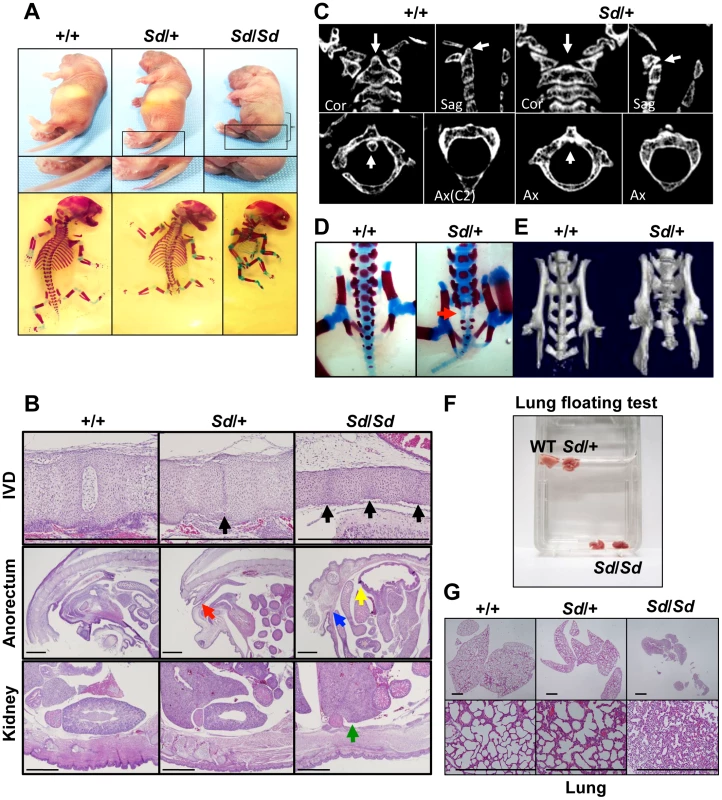
A. Truncation of tail and axial skeletal defects at day 0 postpartum. Sd homozygotes have similar, but much more severe, abnormalities than heterozygotes. The bracket indicates a short body trunk. B. Hematoxylin and eosin-stained histological sections of the thoracic intervertebral discs (IVDs), anorectum, and kidney of E18.5 embryos. Defects of the nucleus pulposus of the IVDs (black arrows) in hypoplastic intervertebral bodies were observed in both Sd/+ and Sd/Sd embryos. Sagittal sections of the anorectum showed blind-end-type anorectal malformations in Sd/Sd mice (yellow and blue arrows), and anal stenosis in Sd/+ (red arrow) mice. Sagittal sections of the kidney revealed renal agenesis (green arrow) in Sd/Sd mice. Bars: 1 mm. C. High-resolution computed tomography images of the cervical vertebral bodies. Upper panel: Coronal (Cor) and sagittal (Sag) images. Lower panel: Axial (Ax) images. The arrows in the wild-type (+/+) mice indicate the position of the dens in the cervical vertebral bodies. Sd/+ mice showed hypoplasia of the dens, as indicated by arrows. D. Alizarin red S and alcian blue skeletal staining of E18.5 embryos. Sd/+ embryos showed partial sacral defects that developed before birth (red arrow). E. High-resolution three-dimensional computed tomography images. Sd/+ mutant mice showed sacral hypoplasia, similar to that of Currarino syndrome. F. Lungs from Sd/Sd, but not wild-type (+/+) or Sd/+ mice, sank in water. G. Hematoxylin and eosin-stained histological sections showed atelectasis of Sd/Sd neonates' lungs, indicating no sign of breathing after birth. Bars: 500 µm. Insertion of a retrotransposon in the Sd mouse genome
The Sd locus had been shown to be located 0.15–0.3 cM distal to D2Mit362 and 0.75 cM proximal to Etl4lacZ [11], [13], [15]. We demonstrated that the insertion site of lacZ in the Etl4lacZ mutation was within the third intron of Skt [14]; thus, it was clear that Sd is located between D2Mit362 and Skt. Furthermore, we showed that the genetic distance between SktGt and Sd was 0.95 cM, and hence that Skt was genetically separated from Sd [14].
We created a cosmid library using embryonic day (E) 11.5 homozygous Sd/Sd embryos. We screened this cosmid library with 32 different DNA probes, obtaining 28 cosmid clones within the Sd region between D2Mit362 and SktGt. Based on physical mapping of 19 cosmid clones and 25 PCR products, the assembled contig spans a 542-kb region containing Sd (Figure S1). The region spanned by the contig contained three known genes, Ptf1a, Msrb2, and Skt, and five expressed sequence tags of unknown function: Gm13344, Gm13336, 4921504, E06Rik, and Otud1 (Figure S1, Text S1).
Interestingly, we found one cosmid clone for which one Not I digestion product was longer than that expected based on its end-sequence tags and wild-type genome informatics (C57BL/6 and Sv-129). We performed shotgun sequencing of the insert of this cosmid and determined it to be 36,440-bp long, which was considerably longer than the expected 27,936-bp (Figure S2A–S2C, Text S1). We submitted the 36,440-bp sequence to GenBank with accession number AB70168. Sequence analysis revealed that this cosmid contained a retrotransposon near the Gm13344, Gm13336, and Ptf1a genes. This retrotransposon was highly homologous to murine early transposon (ETn) endogenous retrovirus (ERV) 3 (ETnERV3)—Family: ERVK, Class: long-terminal repeat (98.6% identity)—whose size was 8,497 nucleotides (AB701682) (Figure 2A). This ETn was found to be tightly linked to the Sd phenotype, showing no recombination in 1,157 offspring. We performed Southern blot analysis using flanking DNA-specific genomic probes from the 5′-region to genotype Sd/Sd, Sd/+, and WT (+/+) mice. One 17,970-bp and one 9,461-bp band were detected in Sd/Sd E18.5 and +/+ embryos, respectively. Both were detected in Sd/+ E18.5 embryos (Figure 2B). In addition, using PCR primer pairs shown in Figure 2A, the genotype of offspring from the heterozygous intercross could be easily determined (Figure 2C). These data clearly suggest that the ETn insertion is associated with Sd phenotypes.
Fig. 2. Insertion of a retrotransposon in the Sd mouse. 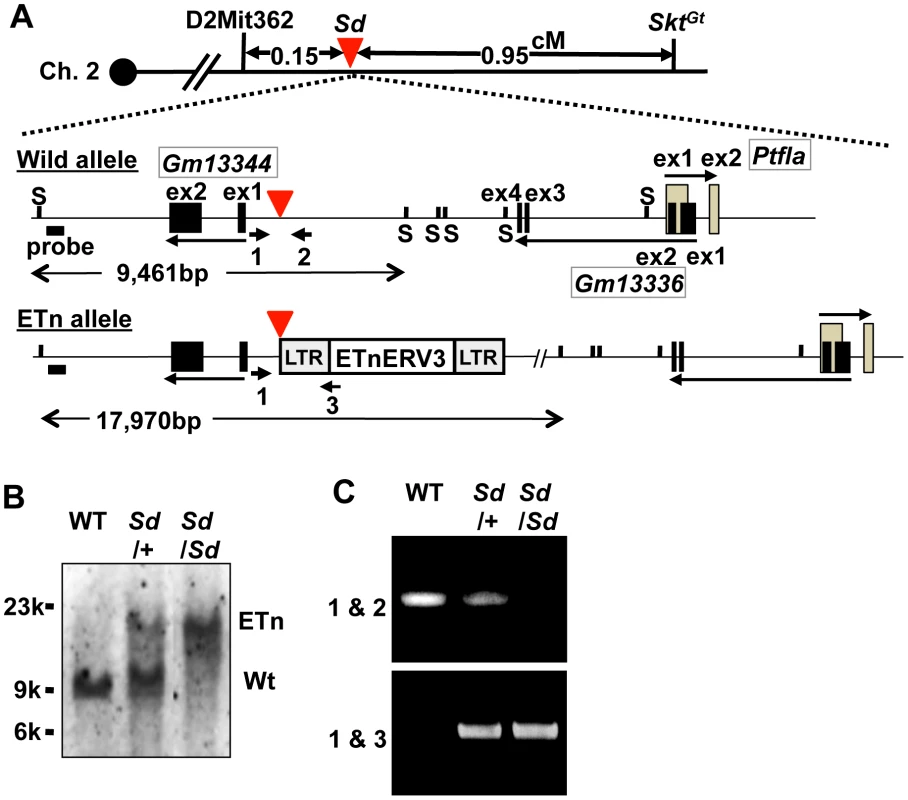
A. Structure of the wild-type and ETn alleles. The 8,497-bp of ETnERV3 was inserted 1,418-bp upstream of the first exon of Gm13344 and 8,114-bp downstream of the last exon of Gm13336. Each box indicates an exon. Arrows indicate the direction of transcription of the Gm13344, Gm13336, and Ptf1a genes. Black bold bars indicate genomic probes used for Southern blotting. “S” indicates Sph I restriction sites. Bold arrows with numbers indicate primers used for PCR analyses. The red arrowhead indicates the insertion point of the ETn. B. Genotyping by Southern blotting. An approximately 9.4-kb band or 18-kb band corresponded to the wild-type (WT) and ETn alleles, respectively. C. Genotyping by genomic PCR analysis. Primer pairs 1 and 2 or 1 and 3 were used to detect the WT or ETn alleles, respectively. Sd phenotypes are caused by the ETn insertion
The ETn insertion may disrupt an as-yet-unknown gene present in the insertion site or the presence of the ETn itself may cause the Sd phenotype. To distinguish these possibilities, we used a method that we developed for exchangeable gene targeting using Cre/lox [16], [17], [18]. Using this system, we were able to disrupt the target locus in the first step, and then produce an ETn knock-in allele (kiETn) by Cre-mediated site-specific recombination.
To examine whether the Sd phenotype was caused by disruption of an as-yet-unknown gene present at the insertion site of the ETn, we constructed a targeting vector that contained a 5′ homology region, the neomycin resistance gene (neo) flanked by loxP and lox2272, and a 3′ homology region. Using this targeting vector, we inserted neo flanked by loxP and lox2272 into embryonic stem (ES) cells at the site where the ETn is found in the Sd mouse (Figure 3A). We obtained neo knock-in mice through mating of germline chimeras (Figure S3A, S3B). The neo mice did not show any phenotype (Figure S3C). Thus, we could rule out the possibility that Sd is caused by the disruption of an as-yet-unidentified gene at the insertion site of the ETn.
Fig. 3. Sd phenotypes are caused by the ETn insertion. 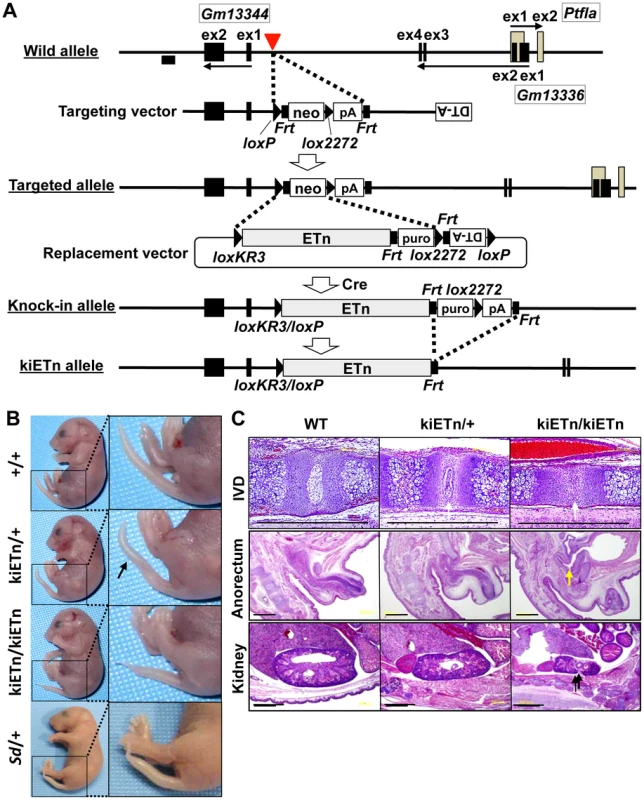
A. Strategy for creation of the knock-in (ki)ETn allele. The targeting vector was first used to create a targeted allele containing a neomycin resistance (neo) gene flanked by loxP and lox2272. Then, a replacement vector was used to replace neo with ETn by Cre-mediated recombination. Exons are shown as gray or black boxes. Black arrows indicate the direction of gene transcription. The red arrowhead indicates the insertion point of the ETn. The probe used for genotyping is shown as a black box. B. Morphology of the tails of E18.5 embryos. The kiETn/+ and the kiETn/kiETn embryos showed a kinked (arrow) and truncated tail, respectively. C. Hematoxylin and eosin staining of the intervertebral discs (IVDs), anorectum, and kidney of E18.5 embryos. The kiETn/+ embryos showed hypoplasia of the nucleus pulposus of the IVDs (white arrows). The kiETn/kiETn embryos showed a nucleus pulposus defect (white arrow) in the IVD, an anorectal malformation that was classified as fistulas of the urogenital tract (yellow arrow), and hypoplasia of the kidney (black arrow). Bars: 1 mm. We then examined whether the presence of ETn is required for the Sd phenotype. We first isolated an Xba I fragment containing the ETn with 287-bp upstream and 630-bp downstream regions from cosmid clone C3. Then, we prepared a replacement vector that contained the Xba I fragment flanked by loxKR3 and lox2272. By electroporating this replacement vector and a Cre expression vector, we created a knock-in allele in ES cells in which neo was replaced with ETn. The puromycin resistance (puro) gene was removed by expressing Flp, thus producing the kiETn allele (Figure 3A). The established kiETn heterozygous animals were fertile and showed a kinked tail phenotype. In this experiment, none of wild type mice (0/29) showed kinky tail, while 27% (6/22) kiETn/+ mice showed kinky tail (Figure 3B). There was no sex difference. Homozygotes showed characteristic Sd phenotypes, such as short tails with a defect of the nucleus pulposus of the intervertebral discs, anorectal malformations, and hypoplasia of the kidney, which were more severe than those in Sd heterozygotes (Figure 3B, 3C). All homozygous kiETn animals showed perinatal lethality. These data strongly supported the hypothesis that the ETn insertion is the cause of the Sd phenotype. However, the kiETn mutant phenotype was less severe than that observed in Sd mice. The kiETn allele was produced using an Xba I fragment containing the ETn with 287-bp 5′ and 630-bp 3′ flanking genomic sequences; these genomic flanking regions were duplicated in the kiETn allele. In addition, the loxKR3/loxP and Frt sequences were retained in the kiETn allele. Therefore, it is possible that the retention of these sequences attenuated the effect of the ETn on the phenotype.
Increased expression of Gm13344, Gm13336, and Ptf1a transcripts
To examine the effects of the ETn on transcription of the Gm13344, Gm13336, and Ptf1a genes, we first obtained full-length sequences for the Gm13344, Gm13336, and Ptf1a transcripts by rapid amplification of cDNA ends (RACE) and reverse transcription PCR (RT-PCR) (Text S1). The full-length sequences of the Gm13344, Gm13336, and Ptf1a transcripts were determined by compiling the sequences of the 5′ RACE and 3′ RACE products with those of expressed sequence tags that showed 100% homology to the RACE products. The final Gm13344 and Gm13336 cDNA sequences were 1,557-bp (AB701678) and 917-bp (AB701680) long, respectively (Figure 4A). In the case of Gm13344, we identified two alternative splicing products, 1,557-bp and 1,403-bp (AB701679) long; splicing in the first exon produced the latter transcript. Unexpectedly, we found a 1,105-bp fusion transcript (AB701681) containing the first and second exons of Gm13336 and 456-bp of the ETn sequence in Sd mutant embryos, which we termed the mutant Gm13336 transcript (mGm13336) (Figure 4A). Our sequencing data suggested that the Gm13344 and Gm13336 genes do not contain a significant open reading frame (ORF). Moreover, Gm13344 partially overlaps with Gm13336, and Gm13336 partially overlaps with Ptf1a. Interestingly, quantitative RT-PCR analyses revealed increased expression of all four transcripts in the Sd mutant at E9.0 and E9.5 (Figure 4B, 4C), but not at E10.0.
Fig. 4. Increased expression of transcripts from the Gm13344 and Gm13336-Ptf1a loci. 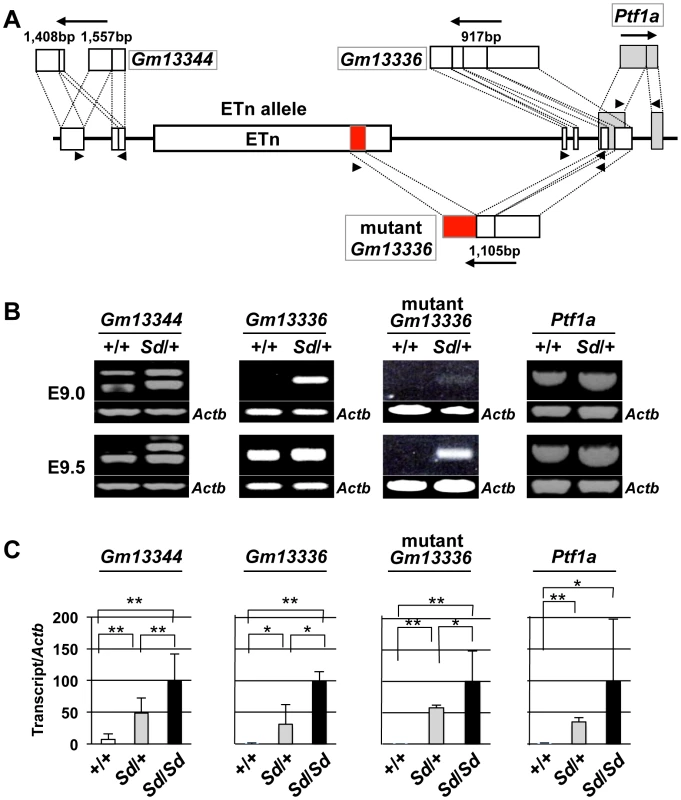
A. Schematic representation of transcripts. Two alternatively spliced transcripts were produced from the Gm13344 locus. One transcript was produced from each of the Gm13336 locus and the Ptf1a locus. In addition, one fusion transcript was produced from Gm13336 and the ETn. The part of the ETn sequence contained in the fusion transcript is shown as a red box. Each exon of Gm13344, Gm13336, and Ptf1a is shown as a white or gray box. The black arrows indicate the direction of transcription. Triangles indicate PCR primers. B. RT-PCR analyses. Increased expression of Gm13344, normal Gm13336, mutant Gm13336, and Ptf1a was observed in E9.0 and E9.5 Sd/+ embryos. C. Quantitative RT-PCR analyses. Expression of Gm13344, Gm13336, mutant Gm13336, and Ptf1a was increased in E9.5 embryos of Sd/+ and Sd/Sd littermates. The data represent the mean ± SD of independent whole embryos (Gm13344, +/+: n = 4, Sd/+: n = 6, Sd/Sd: n = 6; Gm13336, +/+: n = 5, Sd/+: n = 5, Sd/Sd: n = 3; mutant Gm13336, +/+: n = 5, Sd/+: n = 5, Sd/Sd: n = 3; Ptf1a, +/+: n = 8, Sd/+: n = 16, Sd/Sd: n = 10). *p<0.05; ** p<0.01. Tail phenotype in ETn-Gm13336-Ptf1a transgenic mice
To determine whether any of these four transcripts is responsible for Sd, we first generated two lines of transgenic mice carrying either a proximal genomic fragment including Gm13344 and the ETn (Gm13344-ETn) or a distal genomic fragment including the ETn, Gm13336, and Ptf1a (ETn-Gm13336-Ptf1a) (Figure 5A). Although all four transcripts were expressed in transgenic mice at E10.5 or neonatally (Figure 5B), Gm13344-ETn transgenic embryos showed no phenotype (Figure 5C), while ETn-Gm13336-Ptf1a transgenic mice showed a short tail similar to that of the Sd mutant (Figure 5C). These results strongly suggested that the increased expression of Gm13336, mGm13336, and/or Ptf1a is the cause of Sd.
Fig. 5. Tail phenotype in Gm13336-Ptf1a transgenic mice. 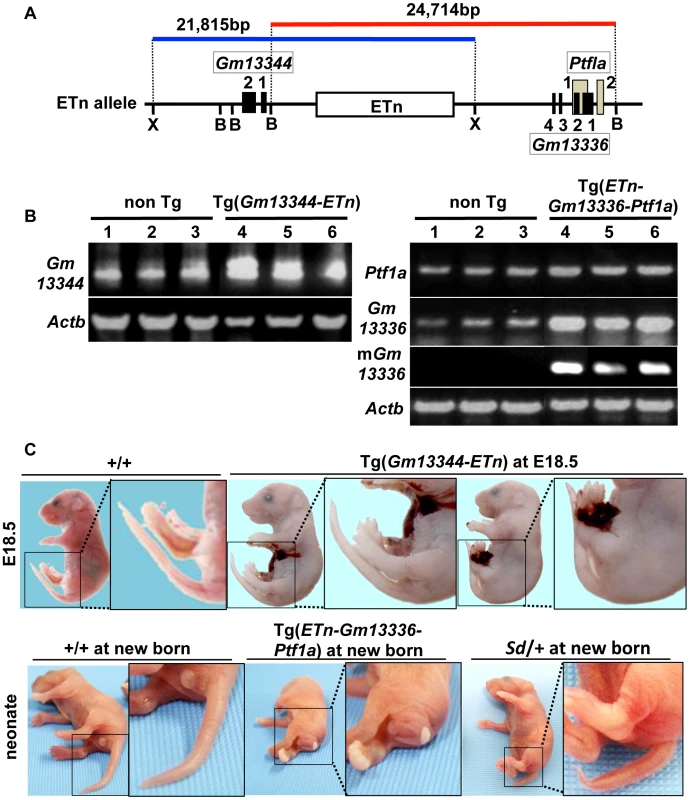
A. Structure of the transgene containing the ETn. Schematic map of the ETn insertion locus. The proximal Xho I (X) fragment spanning 21,815-bp (blue bar) contains Gm13344 and the ETn. The distal Bsm BI fragment spanning 24,714-bp (red bar) contains the ETn and Gm13336-Ptf1a. Exons of the Gm13344, Gm13336, and Ptf1a genes are shown as black or gray boxes. Numbers indicate corresponding exon numbers. B. RT-PCR analyses. Increased expression of Gm13344 in transgenic (Tg)(ETn-Gm13344) and of Gm13336, mGm13336, and Ptf1a in Tg(ETn-Gm13336-Ptf1a) embryos was observed at E10.5. C. Morphology of the tail in E18.5 embryos and neonates. Tg(ETn-Gm13344) E18.5 embryos showed a normal tail. Tg(ETn-Gm13336-Ptf1a) and Sd/+ neonates showed short tails. Phenotypic rescue by disruption of the Gm13336 and Ptf1a genes
Because Gm13336 and Ptf1a overlap each other, the strategy we employed to identify the Sd gene was as follows. First, we created a null allele for all three transcripts—Gm13336, mGm13336, and Ptf1a. Then, we expressed each gene by returning it into this locus by the exchangeable gene targeting method, to determine whether the Sd phenotype could be reproduced. For this purpose, we successfully obtained germline-competent ES cell lines using blastocysts obtained by mating Sd/+ mice and +/+ mice. Four of 15 ES cell lines carried the Sd allele and three were positive for the Sry gene (Figure S4A, Text S1). All the chimeric mice showed a short tail (Figure S4B). We used this germline-competent Sd/+ ES cell line for disruption of the Gm13336-Ptf1a allele. The vector used for homologous recombination in Sd/+ ES cells is shown in Figure 6A. The neo cassette was inserted between the first and second exons of Gm13336, resulting in deletion of the first exon of Ptf1a. Nine targeted ES clones lacking Gm13336-Ptf1a were identified by Southern blot analysis with both a 5′ probe and a 3′ probe (Figure 6A) and were used to generate chimeric mice. We obtained eight chimeric mice and four of them were germline chimeras. The Gm13336-Ptf1a+/neo mice were healthy and fertile, and indistinguishable from their negative littermates. Then, we examined whether the Gm13336-Ptf1aneo allele and the ETn segregated in the next generation. We obtained 20 offspring from three germline chimeras. None showed segregation of the Gm13336-Ptf1aneo and ETn alleles (Figure S5A), suggesting that these offspring carry the Gm13336-Ptf1aneo and ETn alleles on the same chromosome [ETn-Gm13336-Ptf1aneo/+-+; cis configuration]. These mice were intercrossed to produce homozygous ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo mice. The ratio of +/+, ETn-Gm13336-Ptf1aneo/+-+, and ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo mice at E18.5 was 13∶25∶15 (n = 53), which is accordance with the expected Mendelian ratio. In ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo embryos, no Gm13336 or Ptf1a transcripts were detected (Figure 6B), indicating the creation of a null allele. Interestingly, ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo embryos exhibited no abnormalities in vertebral, urogenital, or anorectal development (Figure 6C, 6D), despite the presence of the ETn allele. However, the pancreas was missing in ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo E18.5 embryos, as expected (Figure S5B). Using the targeted ES clone, we obtained one germline chimera, which transmitted the ETn and Gm13336-Ptf1aneo alleles independently into its offspring. In this case, F1 mice carried the ETn and the Gm13336-Ptf1aneo alleles on different chromosomes [ETn-+/+-Gm13336-Ptf1aneo; trans configuration] and thus showed a similar phenotype to that of the heterozygous Sd mutant. These findings clearly indicated that the cis configuration of ETn-Gm13336-Ptf1a causes the Sd phenotype.
Fig. 6. Phenotypic rescue by disruption of the Gm13336-Ptf1a gene. 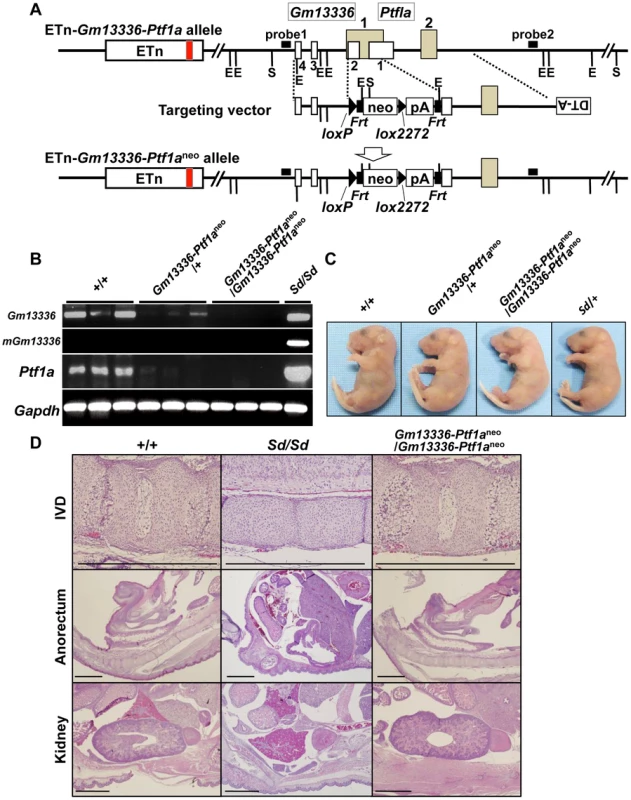
A. Strategy for disruption of the Gm13336-Ptf1a gene. The Gm13336-Ptf1a gene was disrupted using a targeting vector that contained a neomycin resistance (neo) gene flanked by loxP and lox2272. Exons of the Gm13336 and Ptf1a genes are shown as white and gray boxes, respectively. B. RT–PCR analyses of E9.5 embryos. Expression of Gm13336, mutant Gm13336, and Ptf1a was not detected in ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo embryos. C. Tail phenotype of E18.5 embryos. Both ETn-Gm13336-Ptf1aneo/+-+ and ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo embryos showed normal tails, although these embryos have the ETn. D. Hematoxylin and eosin-stained sections from E18.5 embryos. The Sd phenotype observed in the intervertebral discs (IVDs), anorectum, and kidneys was not present in ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo mice. Bars: 1 mm. Identification of Ptf1a as a cause of Sd
To identify the gene causing Sd, we prepared two replacement vectors. One contained the Ptf1a ORF flanked by loxKR3 and lox2272 [16], [18]. Using Cre-mediated recombination, we replaced the phosphoglycerate kinase 1 promoter (PGK)-neo gene with the Ptf1a ORF in ETn-Gm13336-Ptf1aneo ES cells (Figure 7A). The other replacement vector contained exons 1 and 2 of the Gm13336 gene with a CAG promoter flanked by loxKR3 and lox2272. Using Cre-mediated recombination, we replaced the PGK-neo gene with the CAG-exons 1 and 2 of Gm13336 (CAG-Gm13336(1–2)) in ETn-Gm13336-Ptf1aneo ES cells (Figure 7A). Although we confirmed the expression of Gm13336 and mGm13336 in ETn-Gm13336-Ptf1aGm13336(1–2)/+-+ and ETn-Gm13336-Ptf1aGm13336(1–2)/ETn-Gm13336-Ptf1aGm13336(1–2) mice, we observed no abnormal phenotypes (Figure 7B). In contrast, the ETn-Gm13336-Ptf1aPtf1a/+-+ mice showed a short tail similar to that in Sd/+ mice, while ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a neonates showed no tail and a short trunk (Figure 7B and Figure S6A). Histological examination revealed defects of the nucleus pulposus of the intervertebral discs and anorectal malformations similar to the defects in Sd/Sd mice, although the kidney defect was less severe (Figure 7C). As expected, the pancreas was restored to normal in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a mice (Figure 7C). The vertebral columns of ETn-Gm13336-Ptf1aPtf1a/+-+ and ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a mice were usually truncated at the eighth caudal and the tenth thoracic vertebrae, respectively (Figure S6B).
Fig. 7. Identification of Ptf1a as a cause of Sd. 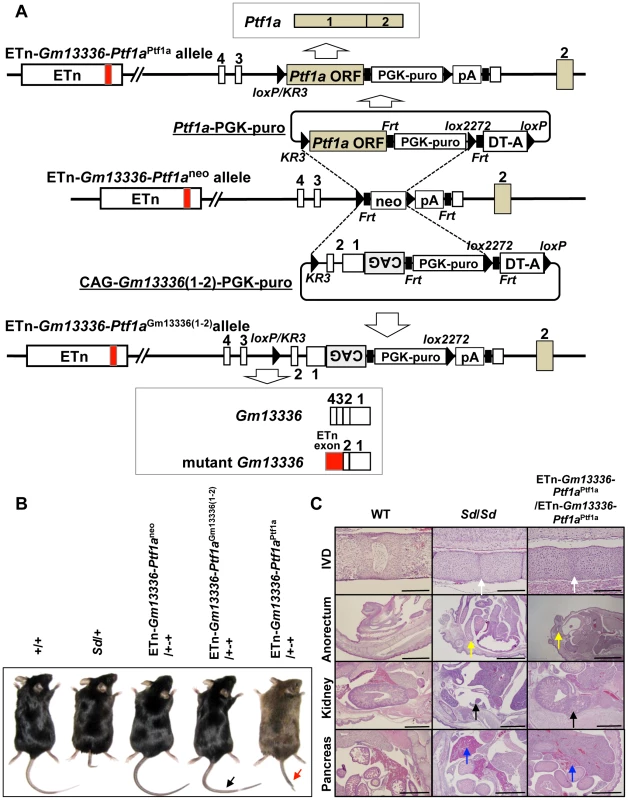
A. Strategy for creation of a knock-in allele. Using the ES cell line carrying the ETn-Gm13336-Ptf1aneo allele, the neomycin resistance (neo) gene was replaced with the Ptf1a ORF or CAG-Gm13336(1–2) using the replacement vector Ptf1a ORF-PGK-puro or CAG-Gm13336(1–2)-PGK-puro, respectively. Exons are shown as white, gray or red boxes. B. Tail phenotype in adult mice. ETn-Gm13336-Ptf1aCAG-Gm13336(1–2)/+-+ mice showed a normal tail (black arrow), despite the rescue of Gm13336 and mGm13336 expression. However, ETn-Gm13336-Ptf1aPtf1a/+-+ mice showed a short tail (red arrow). C. Hematoxylin and eosin-stained sections of neonates. Defects of the nucleus pulposus of the intervertebral discs (IVDs) (white arrows) and anorectal malformations (yellow arrows) in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a mice were similar to those in Sd homozygous mutants, although the kidney defect (black arrows) was less severe. The defect of the pancreas was restored to normal in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a mice (blue arrows). Bars: 1 mm. To exclude the possibility that Gm13336 or mGm13336 is responsible for Sd, we introduced CAG-Gm13336 or CAG-mGm13336 into the gene-trap ES cell line 21-B137 (Figure S7A, Text S1). We confirmed the expression of these transcripts and observed no abnormalities (Figure S7B).
Ectopic expression of Ptf1a from the ETn-Gm13336-Ptf1a allele
If the Ptf1a gene is responsible for Sd, Ptf1a should be expressed in tissues where the various phenotypes are observed. To examine the expression pattern of Ptf1a during embryonic development, we first tried to detect Ptf1a mRNA by in situ hybridization or PTF1a protein by immunohistochemistry. However, we failed to detect the expression of Ptf1a by either method, probably because of low-level expression and the low sensitivity of these methods. Thus, we used Cre-mediated recombination to replace the PGK-neo cassette in ETn-Gm13336-Ptf1aneo ES cells with the lacZ gene (Figure 8A). These mice were examined for lacZ expression by whole mount X-gal staining during embryonic development at E8.5, E9.5, E10.5, and E11.5 (Figure 8B). Interestingly, lacZ expression was detected in the notochord and hindgut at E8.5 and E9.5. LacZ expression extended to the cloaca and mesonephros at E10.5 and to the pancreatic bud at E10.5 and E11.5. LacZ expression was strongly detected in the notochord, mesonephros, and cloaca at E9.5 (Figure 8C).
Fig. 8. Ectopic expression of Ptf1a and downregulation of Cdx2 and its downstream targets. 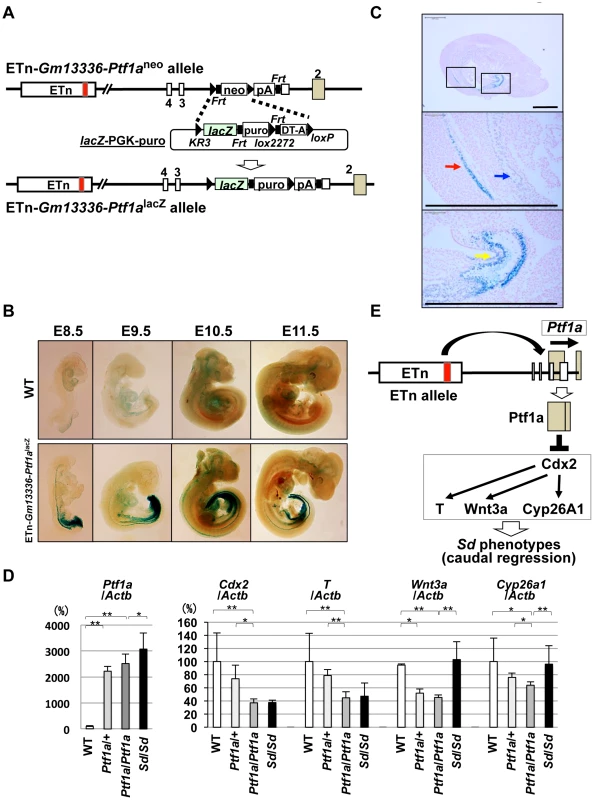
A. Strategy for generating ETn-Gm13336-Ptf1alacZ mice. The PGK-neo gene was replaced with the lacZ gene in the ETn-Gm13336-Ptf1aneo allele using Cre-mediated recombination. B. Whole-mount X-gal staining of ETn-Gm13336-Ptf1alacZ embryos. Upper panel: wild-type (WT) littermates. Lower panel: ETn-Gm13336Ptf1alacZ embryos. LacZ expression was detected in the notochord and hindgut at E8.5 to E9.5. Then, lacZ expression extended to the cloaca and mesonephros at E10.5 and to the pancreatic bud at E10.5 and E11.5. C. Ectopic lacZ expression. LacZ expression was strongly detected in the notochord (red arrow), mesonephros (blue arrow), and cloaca (yellow arrow) by histological analysis with X-gal staining at E9.5. The areas shown at higher magnification in the lower panels are indicated by boxes. Bars: 500 µm. D. Quantitative RT-PCR analyses in E9.5+/+, Ptf1a/+, and Ptf1a/Ptf1a littermates and in Sd/Sd embryos with a C57BL/6 genetic background. Although the expression of Ptf1a in ETn-Gm13336-Ptf1aPtf1a/+-+ and homozygous ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a mice was slightly lower than that in Sd/Sd mice, it was much higher than that in WT mice. The expression levels of Cdx2 and T in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a mice were similar to those in Sd/Sd mice, but significantly lower than those in WT mice. The expression of Wnt3a and Cyp26a1 was decreased in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a mice, but not in Sd/Sd mice. The data represent the mean ± SD of independent whole embryos (+/+: n = 8, Ptf1a/+: n = 4, Ptf1a/Ptf1a: n = 6, Sd/Sd: n = 3–10). *p<0.05; ** p<0.01. E. Model of Ptf1a action. The ETn insertion induces ectopic expression of Ptf1a, resulting in downregulation of Cdx2 and its downstream targets T, Wnt3a, and Cyp26a1. This combined deficiency of gene expression causes the Sd phenotype. Kawaguchi et al. previously reported that expression of lacZ in Ptf1aCre;ROSA26R mice was restricted to the pancreas during development in Ptf1a-Cre transgenic mice in which the Cre gene was inserted into the Ptf1a locus [19]. Therefore, this ectopic expression in ETn-Gm13336-Ptf1alacZ embryos was considered to be induced by ETn insertion. Taken together, the ectopic expression pattern of Ptf1a is consistent with the phenotypes observed in Sd.
Effect of ectopic Ptf1a expression
To examine the effects of ectopic Ptf1a expression, we carried out transcriptional profiling using E10.0 Sd/Sd and WT whole embryos (Tables S1 and S2) (Text S1). As expected, Ptf1a was upregulated 3.7-fold in Sd/Sd embryos. Among the significantly downregulated genes, we focused on the Cdx2 and T genes, because Cdx2 is known to regulate the expression of genes such as T, Wnt3a, and Cyp26a1, which are essential for development of the posterior embryo [20]. In fact, mice mutant for these genes show tail phenotypes similar to those in Sd mice [21], [22], [23], [24], [25], [26], [27]. Thus, the expression of these genes may be suppressed by the ectopic expression of Ptf1a in tissues such as the notochord, gut, and mesonephros. We analyzed the mRNA expression of Ptf1a, Cdx2, T, Wnt3a, and Cyp26a1 in ETn-Gm13336-Ptf1aPtf1a/+-+ and ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a embryos as well as in Sd E9.5 embryos. The level of Ptf1a expression in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a embryos was about 80% of that in Sd embryos, but was 20-fold higher than that in WT embryos (Figure 8D). As mentioned above, RNA profiling showed a 3.7-fold increase, but this discrepancy could be caused by differences in sensitivity and specificity. Both Cdx2 and T expression in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a and Sd embryos decreased to about 40% of that in WT embryos at E9.5. Wnt3a and Cyp26a1 expression was decreased to 42% and 62% of WT levels, respectively, in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a embryos, while their expression in Sd/Sd embryos was similar to that in WT embryos. However, only two genes, Cdx2 and T, were downregulated at E10.0 and E11.5 in ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a mice (Figure S8).
We further analyzed whether expression of Ptf1a can suppress the expression of Cdx2, T, Wnt3a, and Cyp26a1 in ES cells. We inserted the CAG-Ptf1a gene into the 21-B137 allele using the same method (see Figure S9A). As expected, the mRNA expression of Cdx2, T, Wnt3a, and Cyp26a1 was significantly decreased (Figure S9A). Furthermore, we transiently expressed Ptf1a in WT ES cells by electroporating in the expression vector CAG-Ptf1a. As shown in Figure S9B, the expression of Cdx2, T, Wnt3a, and Cyp26a1 was decreased compared with that in ES cells transfected with the control CAG-enhanced green fluorescent protein (EGFP) (Text S1).
As suggested by our ES cell data shown in Figure S9, Ptf1a expression can downregulate the expression of four genes: Cdx2, T, Wnt3a, and Cyp26a1. Accordingly, all four genes were downregulated in E9.5 ETn-Gm13336-Ptf1aPtf1a/ETn-Gm13336-Ptf1aPtf1a embryos (Figure 8D). However, only two genes, Cdx2 and T, were downregulated at E10.0 and E11.5 (Figure S8), similar to the result observed at E9.5 in Sd embryos. The precise spatial or temporal regulation of Ptf1a gene expression in Sd mice could differ from that in ETn-Gm13336-Ptf1aPtf1a mice because of the difference in allele structure.
Taken together, our results strongly suggest that ectopic expression of Ptf1a induced by ETn insertion suppresses the Cdx2 gene and its downstream targets such as T, Wnt3a, and Cyp26a1, resulting in the characteristic phenotypes observed in Sd mice (Figure 8E).
Discussion
In this study, we have revealed the nature of the Sd mutation to be the insertion of an ETn causing ectopic expression of Ptf1a in the caudal region of the embryo, resulting in suppression of Cdx2 and its downstream target genes.
Endogenous retroviruses are present in the genomes of all vertebrates [28], [29]. Retrotransposons are genetic elements that can amplify themselves in a genome and are ubiquitous components of the DNA of many eukaryotic organisms. They are responsible for the majority of ERV-induced de novo germline mutations [30]. Most commonly, germline mutations caused by retrotransposon insertions occur in an intron, disrupting gene expression by causing premature polyadenylation, aberrant splicing, or ectopic transcription driven by the long terminal repeat. For ETn insertions, the most commonly reported defect is premature polyadenylation within the ETn, coupled with aberrant splicing because of a few commonly used cryptic splice signals [31]. However, ETn-promoted ectopic gene expression has not previously been observed [31]. Kano et al. reported that dactylaplasia (Dac) is a LTR retroptransposon insertion caused by the type D mouse endogenous provirus element (MusD), and that the ectopic MusD expression at the apical ectodermal ridge of limb buds correlates with the dactylaplasia phenotype [32]. However, in this Dac mutation, ETn-promoted ectopic expression of any endogenous genes has not been observed. In this study, we revealed the insertion of an ETn approximately 12 kb upstream of the transcription initiation site of the Ptf1a gene, resulting in the misexpression of Ptf1a. As the expression of Gm13344 and Gm13336 was also increased by this insertion, ETn acts as an enhancer, instead of as a transcription initiator. To the best of our knowledge, this is the first report of an ETn insertion causing ectopic gene expression.
Ptf1a, which encodes a basic helix-loop-helix transcription factor, was originally reported to be a pancreatic determiner that drives undifferentiated cells in the foregut endoderm to differentiate into a pancreatic lineage [19], [33]. In humans, Ptf1a was identified as responsible for the human permanent neonatal diabetes mellitus associated with cerebellar ataxia, and was reported to be involved in cerebellar development [34]. In fact, ectopic expression of Ptf1a conferred inhibitory gamma aminobutyric acid characteristics to neural progenitors, which were normally fated to become glutamatergic excitatory neurons [35]. These loss - and gain-of-function experiments suggested that Ptf1a itself acts as a cell fate determinant. However, in our case, Ptf1a overexpression and/or ectopic expression resulted in downregulation of Cdx2 and its downstream targets T, Wnt3a, and Cyp26a1. The shortened tail phenotype of the Sd mutant is similar to that of the Cdx2 mutant, although heterozygous Cdx2 mutants show an anterior homeotic shift of the cervical and thoracic spine and rib abnormalities that are not observed in Sd mice [23]. The shortened tail phenotype of the T mutant [23], [36] is also similar to that of the Sd mutant. A mutation in the Wnt3a gene results in a lack of caudal somites, a disrupted notochord, and failure to form a tail bud [37], [38]. Cyp26a1-null mutants die during mid–late gestation and show a number of major morphogenetic defects, such as truncation of the tail, deficiencies of the external genitalia, anal atresia, and horseshoe kidneys [25], [26]. Interestingly, expression of T and Wnt3a in the tail bud was downregulated in Cyp26a1-deficient mice. Taken together, these results suggest that the phenotypes observed in Sd mice are caused by the combination of partial deficiency of Cdx2 and its downstream target genes.
We demonstrated that the mRNA expression of Cdx2, T, Wnt3a, and Cyp26a1 was significantly decreased in ES cells overexpressing Ptf1a. Ptf1 is a trimeric transcription factor comprising Ptf1a, an E protein (such as Tcf4 or Tcf12 [39]), and a third protein such as the mammalian Suppressor of Hairless (RBP-J) or its paralog RBP-L [40]. However, the binding site for Ptf1 has not been reported in the Cdx2 gene, nor in the T, Wnt3a, or Cyp26a1 genes. Instead, there are two Tcf-binding elements in the promoters of both the Cdx2 and T genes in the mouse [41], [42]. It is possible that overexpressed Ptf1a can bind to Tcf4, preventing the binding of Tcf4 to the promoter, leading to attenuation of the expression of Cdx2 and T. Decreased expression of Cdx2 may cause activation of β-catenin-mediated transcriptional activity, because Cdx2 directly binds β-catenin and disrupts the β-catenin–Tcf protein complex [42], [43], [44]. However, nuclear β-catenin interacts with the Tcf/lymphoid enhancer factor (Lef) family of DNA-binding proteins to regulate the expression of numerous Wnt target genes [45], [46], [47]. Thus, the removal of Tcf by Ptf1a may result in the degradation of β-catenin and downregulation of the Wnt target genes.
CRS is a congenital heterogeneous constellation of caudal anomalies that includes varying degrees of agenesis of the spinal column, anorectal malformations, and genitourinary anomalies. Its pathogenesis is unclear, but it could be the result of excessive physiologic regression of the embryonic tail. As described above, the various mouse mutants have shown that caudal agenesis occurs a result of hypodevelopment of the anterior–posterior axis. The Sd mouse is considered a model for human CRS based on phenotypic similarity in the spine, hindgut, and urogenital system. The exact etiology in humans is unknown, except in some cases of Currarino syndrome. Currarino syndrome is a form of CRS with hemisacrum, anorectal malformations, and presacral mass, such as teratoma [48]. Previous reports have shown that HLXB9 is a major causative gene for Currarino syndrome [49], [50]. HLXB9 is a homeobox protein that contains a glycine-and-alanine-rich region and a strongly acidic region next to the amino-terminal and carboxy-terminal side of the homeodomain, respectively [51]. However, a null mutation of the Hlxb9 gene in mice showed agenesis of the dorsal pancreas but no skeletal truncation [52], [53]. It is of interest that both Ptf1a and Hlxb9 are expressed in the pancreas, and that Ptf1a is required for Hlxb9 expression [54]. Although it is not clear whether PTF1A is involved in human CRS, the evidence from this study implicates PTF1A and possibly HLXB9 in the caudal abnormalities. Further studies using Sd mice will provide further insight into the development of human CRS.
Materials and Methods
Danforth's short tail (Sd) mice
Sd mice purchased from the Jackson Laboratory (Bar Harbor, ME) were backcrossed to C57BL/6 mice for at least ten generations. All experiments were performed in accordance with the Declaration of Helsinki and were approved by the Kumamoto University Ethics Committee for Animal Experiments (authorization number in Kumamoto University: C23-262, C24-278).
Identification of Sd mice by PCR and Southern blotting
PCR was used to genotype Sd alleles. For the WT allele, the 5′ primer 5′Sd-S1 (5′-GAAAGCAAAGGGCTGCTTAC-3′) and the 3′ primer 3′Sd-A1 (5′-TATTCTTGCAGGGAGAGTTG-3′) were used to amplify a 283-bp fragment. To detect the Sd allele, the 5′ primer 5′Sd-S1 (5′-GAAAGCAAAGGGCTGCTTAC-3′) and the 3′ primer 5′Tn-A1 (5′-TCTCGTGTGATCTGTCTGTC-3′), located in the ETn, were used to amplify a 228-bp fragment. The PCR conditions were as follows: denaturation at 94°C; followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. PCR products were visualized on a 1% agarose gel using ethidium bromide. For Southern blotting, genomic DNA was digested overnight with Sph I and subjected to electrophoresis on a 1% agarose gel. DNA was transferred onto a positively charged nylon membrane (Roche, Indianapolis, IN). After baking at 80°C for 1 h, the membrane was hybridized with a flanking genomic DNA-specific probe (Figure 2A).
ES cell culture
ES cells were cultured in KSR-GMEM medium consisting of Glasgow Minimum Essential Medium (Sigma, St Louis, MO) with 1×nonessential amino acids (Gibco Invitrogen, Grand Island, NY), 0.1 mM β-mercaptoethanol, 1 mM sodium pyruvate, 1% fetal bovine serum (HyClone; Thermo Fisher Scientific Inc., Waltham, MA), 14% Knockout Serum Replacement (Gibco Invitrogen), and 1100 U/ml leukemia inhibitory factor (ESGRO; Chemicon, Temecula, CA).
Generation of ETn knock-in mice
The knock-in method used in this study was developed by us and has been described [16], [17], [18]. In the first step, the targeting vector—containing a 5′ homology region, loxP, Frt, PGK-neo cassette, lox2272, polyadenylation signal (pA), Frt, 3′ homology region, and an MC1 promoter-diphtheria toxin A fragment with a pA (MC1-DT-A)—was constructed using pBluescript II containing the PGK-neo cassette (p03) [55]. Targeting vectors were electroporated into feeder-free KTPU8 ES cell lines derived from the TT2 ES cell line, according to previously described methods [56], [57]. Three targeted ES clones were obtained from 288 G418-resistant clones. ES cells were aggregated with ICR morulas to produce chimeric mice. Germline transmission was confirmed in all three lines. In the second step, the replacement vector—containing loxKR3 (KR3), ETn, Frt, puromycin N-acetyl-transferase (puro), lox2272, Frt, MC1-DT-A, and loxP—was electroporated into the targeted ES cell clones to establish the kiETn allele [18].
The PGK-puro cassette was removed by transient Flp expression. One replaced ES clone was obtained from 28 puromycin-resistant clones. These kiETn ES cells were used to produce germline chimeras. Heterozygous kiETn mice were backcrossed to C57BL/6 for at least five generations. Then, heterozygous kiETn mice were intercrossed to produce homozygous kiETn mice.
Identification of kiETn mice
Founder kiETn mice were identified by PCR and Southern blotting. Genomic DNA was extracted from an ear clip. The ETn fragment digested by Xba I contained both 5′ and 3′ flanking genomic sequences; partial tandem duplication (5′: 287-bp; 3′: 630-bp) of flanking genomic sequences was confirmed by PCR analyses. To detect the kiETn allele, the 5′ primer 5′Sd-S1 (5′-GAAAGCAAAGGGCTGCTTAC-3′), located in the 5′ flanking genomic region, and the 3′ primer 5′Tn-A1 (5′-TCTCGTGTGATCTGTCTGTC-3′), located in the ETn, generated 228-bp and 650-bp fragments, respectively. To detect the WT allele, the 5′ primer 5′Sd-S1 (5′-GAAAGCAAAGGGCTGCTTAC-3′), located in the 5′ flanking genomic region, and the 3′ primer 3′Sd-A1 (5′-TATTCTTGCAGGGAGAGTTG-3′), located in the 3′ flanking genomic region, generated a 283-bp fragment. To detect the targeted allele, the 5′ and 3′ primers neo-F (5′-AGAGGCTATTCGGCTATGAC-3′) and neo-R (5′-CACCATGATATTCGGCAAGC-3′), respectively, both located in the targeting vector, generated a 545-bp fragment. For Southern blotting, genomic DNA was digested overnight with Sph I and subjected to electrophoresis on a 1.0% agarose gel. DNA was transferred onto a positively charged nylon membrane (Roche). After baking at 80°C for 1 h, the membrane was prehybridized and then hybridized using flanking genomic DNA-specific probes (Figure 2A) prepared using a Digoxigenin DNA Labeling and Detection Kit (Roche).
RT–PCR
PCR amplification of each DNA fragment and gene was performed using TaKaRa EX or LA Taq (Takara, Kyoto, Japan), according to the manufacturer's protocol. The primer sequences were as follows: Gm13344 (accession number AB701678 and AB701679): 5′-ACGAATGGGGTGTTCAGACG-3′ (sense) and 5′-CGACTGCCAGACCCAGGAAG-3′ (antisense), generating two alternative splicing products, 446-bp and 297-bp fragments; Gm13336 (accession number AB701680): 5′-TGACGCTTTGTGAGTGATCC-3′ (sense) and 5′-AACACTCCTGTGATGTGTAG-3′ (antisense), generating a 226-bp fragment; mGm13336: 5′-TGACGCTTTGTGAGTGATCC-3′ (sense) and 5′-GAACAATACGATTTCTTTTTACCTG-3′ (antisense), generating a 488-bp fragment; Ptf1a: 5′-TGAGGGACCTACCCGAATTG-3′ (sense) and 5′-ACAATATGCACAAAGACGCG-3′ (antisense), generating a 1,105-bp fragment; Actb (β-actin; control): 5′-ATGTACGTAGCCATCCAGGC-3′ (sense) and 5′-AAGAAGGAAGGCTGGAAAAG-3′ (antisense), generating a 407-bp fragment; Gapdh (glyceraldehyde 3-phosphate dehydrogenase; control): 5′-GGAAAGCTGTGGCGTGATG-3′ (sense) and 5′-CTGTTGCTGTAGCCGTATTC-3′ (antisense), generating a 392-bp fragment. Thermal cycling was carried out with denaturation at 94°C; followed by 30, 35, or 40 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. PCR products were visualized on 1% agarose gels using ethidium bromide.
Quantitative real-time RT–PCR
Reverse transcribed products were used for quantitative real-time PCR using an Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA). TaqMan Gene Expression Master Mix and TaqMan Gene Expression Assays for Gm13336 (a custom-made Gm13336 TaqMan probe), mGm13336 (a custom-made mGm13336 TaqMan probe), Ptf1a (Mm 00479622), Cdx2 (Mm 01212280), T (Mm 01318252), Wnt3a (Mm 00437337), Cyp26a1 (Mm 00514486), and Actb (Mm 00607939) were purchased from Applied Biosystems. Reactions were carried out under the following conditions: 2 min at 50°C and 10 min at 95°C; followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Calibration was conducted using the relative standard curve method. To construct a standard curve, a standard sample cDNA was prepared from E10.5 embryos for Cdx2, T, Wnt3a, Cyp26a1, and Actb, or Ptf1a cDNA for Ptf1a, or Gm13336 cDNA for Gm13336, or mGm13336 cDNA for mGm13336. For each PCR assay, the standard curve was generated using the same standard sample. The relative concentration of the target gene in each sample was calculated from the constructed standard curve, and the ratio of the relative concentration of the target gene to Actb in each sample was calculated. This ratio represented the relative expression of the target gene normalized to Actb compared with the standard sample. Actb is recommended by Applied Biosystems as a suitable endogenous internal control for TaqMan RT-PCR analyses. Real-time PCR for Gm13344 transcripts was performed using the THUNDERBIRD SYBR qPCR mix (Toyobo, Osaka, Japan). The following primer pair was used for the shorter splice variant Gm13344 transcripts: 5′-TGTGCTGGACCCAAACATAGCCAAAG-3′ (sense) and 5′-CGACTGCCAGACCCAGGAAG-3′ (antisense), generating a 293-bp fragment. The relative concentration of Gm13344 in each sample was calculated from the constructed standard curve, and the ratio of the relative concentration of Gm13344 to Actb was calculated. This ratio represented the relative expression of the target gene normalized to Actb compared with the standard sample.
Generation of transgenic mice
To isolate the 21,815-bp DNA fragment containing Gm13344-ETn and the 24,714-bp DNA fragment containing ETn-Gm13336 from a cosmid clone, cosmid DNA was digested with Xho I and Bsm BI, respectively. Isolated DNA fragments were microinjected into fertilized eggs obtained from C57BL/6 mice, at a final concentration of 1 µg/ml in Tris-EDTA buffer.
Targeting vector construction and generation of ETn-Gm13336-Ptf1aneo mice
Genomic DNA containing the coding region of the Ptf1a gene was isolated from the cosmid clone containing the ETn. The pBluescript II construct containing the PGK-neo cassette (p03) and the MC1-DT-A fragment was used as a backbone to construct the targeting vector. The targeting vector comprised a 5′ homology region, loxP, Frt, PGK-neo, lox2272, pA, Frt, 3′ homology region, and MC1-DT-A (Figure 6A). The 5′ homology region contained the first exon, first intron, second exon, and part of the second intron of Gm13336; part of the first exon of Ptf1a was deleted by insertion of the neo cassette. The targeting vector was electroporated into Sd/+ ES cells as described above. Nine targeted ES clones were obtained from 192 G418-resistant clones. ES cells were aggregated with ICR morulas as described above. Germline chimeras were obtained from four ES lines. This strain of mouse was designated ETn-Gm13336-Ptf1aneo. ETn-Gm13336-Ptf1aneo/+-+ mice were backcrossed to C57BL/6 mice for at least three generations. Then, ETn-Gm13336-Ptf1aneo/+-+ mice were intercrossed to produce ETn-Gm13336-Ptf1aneo/ETn-Gm13336-Ptf1aneo mice.
Identification of ETn-Gm13336-Ptf1aneo mice
Founder ETn-Gm13336-Ptf1aneo mice were identified by PCR and Southern blotting. Genomic DNA was extracted from an ear clip. To detect the targeted allele, the 5′ primer neo-F (5′-AGAGGCTATTCGGCTATGAC-3′) and the 3′ primer neo-R (5′-CACCATGATATTCGGCAAGC-3′), located in the neo cassette, generated a 545-bp fragment. To detect the WT allele, the 5′ primer 5′AKKO-S1 (5′-ATTGCTCAGAACCCCTAGGG-3′), located in the 5′ flanking genomic region of the second exon of Ptf1a, and the 3′ primer 3′AKKO-A1 (5′-GATTCCCTGAGCTGTGAAGC-3′), located in the 3′ flanking genomic region of the second exon of Gm13336, generated a 1,777-bp fragment. For Southern blotting, genomic DNA was digested overnight with Eco RI and Spe I and electrophoresed on 1.0% agarose gels. DNA was transferred onto a positively charged nylon membrane (Roche). After baking at 80°C for 1 h, the membrane was prehybridized and then hybridized using 5′ and 3′ flanking genomic DNA-specific probes (Figure 6A) prepared using a Digoxigenin DNA Labeling and Detection Kit (Roche).
Construction of the Ptf1a ORF replacement vector and establishment of ETn-Gm13336-Ptf1aPtf1a ES cells and mouse lines
To insert the Ptf1a ORF into the ETn-Gm13336-Ptf1a allele, ETn-Gm13336-Ptf1aneo ES cells were used. The ES cell clones were electroporated with the Cre expression vector and a replacement vector assembled from pKR3-Frt-del.pA-puro-2272 with the cloned Ptf1a ORF to establish ETn-Gm13336-Ptf1aPtf1a ES cell and mouse lines (Figure 7A). ETn-Gm13336-Ptf1aPtf1a mice were backcrossed to C57BL/6 mice for at least three generations. Gm13336-Ptf1aPtf1a/+-+ mice were intercrossed to produce Gm13336-Ptf1aPtf1a/Gm13336-Ptf1aPtf1a mice.
Construction of CAG-Gm13336(1–2) and establishment of ETn-Gm13336-Ptf1aCAG-Gm13336(1–2) ES cells and mouse lines
To insert the CAG-Gm13336(1–2) into the ETn-Gm13336-Ptf1aneo allele, ETn-Gm13336-Ptf1aneo ES cells were used. The ES cell clones were electroporated with the Cre expression vector and a replacement vector assembled from pKR3-Frt-del.pA-puro-2272 with the cloned Gm13336(1–2) to establish ETn-Gm13336-Ptf1aCAG-Gm13336(1–2) ES cell and mouse lines (Figure 7A). ETn-Gm13336-Ptf1aCAG-Gm13336(1–2) mice were backcrossed to C57BL/6 mice for at least three generations. Then, Gm13336-Ptf1aCAG-Gm13336(1–2)/+-+ mice were intercrossed to produce Gm13336-Ptf1aCAG-Gm13336(1–2)/Gm13336-Ptf1aCAG-Gm13336(1–2) mice.
Construction of the lacZ replacement vector and establishment of the ETn-Gm13336-Ptf1alacZ mouse line
ETn-Gm13336-Ptf1aneo ES cells were used to insert lacZ into the ETn-Gm13336-Ptf1a allele. The ES cell clones were electroporated with a replacement vector assembled from pKR3-Frt-del.pA-puro-2272 and cloned lacZ to establish ETn-Gm13336-Ptf1alacZ mouse lines (Figure 8A). ETn-Gm13336-Ptf1alacZ mice were backcrossed to C57BL/6 mice for at least two generations.
Histological analysis
Embryos and neonates were fixed in phosphate-buffered 15% formaldehyde overnight, rinsed twice for 1 h in phosphate-buffered saline (PBS), dehydrated through increasing concentrations of ethanol, equilibrated with xylene, embedded in paraffin wax, and sectioned at 4 µm. Sagittal sections were stained with hematoxylin and eosin and examined by light microscopy.
Detection of β-galactosidase (lacZ) activity
Samples were fixed for 30 min at room temperature in fix solution [1% formaldehyde, 0.2% glutaraldehyde, and 0.02% NP-40 in PBS]. Fixed samples were washed twice with PBS and incubated overnight at 30°C in staining solution (5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 2 mm MgCl2, 0.5% X-gal in PBS). Samples were rinsed twice in PBS and then post-fixed in 10% formaldehyde. For observation of whole-mount X-gal staining, samples were made transparent using benzylalcohol/benzylbenzoate (1∶2), after dehydration with a series of ethanol steps (25%, 50%, 70%, 100%, and 100%, 1 h each). For histological analysis, samples were sectioned at 8 µm and counterstained with Nuclear Fast red (Funakoshi, Tokyo, Japan) after X-gal staining.
Statistical analysis
The results are presented as the mean ± standard deviation (SD) of independent experiments as detailed separately in each corresponding figure legend. Data were compared using the Student's t-test and were considered significantly different at p<0.05.
Supporting Information
Zdroje
1. DunnLC (1940) A new mutation in the mouse affecting spinal column and urogenital system. Journal of heredity 31 : 343–348.
2. GrünebergH (1958) Genetical studies on the skeleton of the mouse. XXII. The development of Danforth's short-tail. J Embryol Exp Morphol 6 : 124–148.
3. FavreA, BrianoS, MazzolaC, BrizzolaraA, TorreM, et al. (1999) Anorectal malformations associated with enteric dysganglionosis in Danforth's short tail (Sd) mice. J Pediatr Surg 34 : 1818–1821.
4. Gluecksohn-SchoenheimerS (1943) The morphological manifestations of a dominant mutation in mice affecting tail and urogenital system. Genetics 28 : 341–348.
5. Gluecksohn-SchoenheimerS (1945) The Embryonic development of mutants of the Sd-strain in mice. Genetics 30 : 29–38.
6. GrünebergH (1953) Genetical studies on the skeleton of the mouse. VI. Danforth's short-tail. J Genet 51 : 317–326.
7. AndoT, SembaK, SudaH, SeiA, MizutaH, et al. (2011) The floor plate is sufficient for development of the sclerotome and spine without the notochord. Mech Dev 128 : 129–140.
8. KosekiH, WallinJ, WiltingJ, MizutaniY, KispertA, et al. (1993) A role for Pax-1 as a mediator of notochordal signals during the dorsoventral specification of vertebrae. Development 119 : 649–660.
9. Brand-SaberiB, EbenspergerC, WiltingJ, BallingR, ChristB (1993) The ventralizing effect of the notochord on somite differentiation in chick embryos. Anat Embryol (Berl) 188 : 239–245.
10. EbenspergerC, WiltingJ, Brand-SaberiB, MizutaniY, ChristB, et al. (1995) Pax-1, a regulator of sclerotome development is induced by notochord and floor plate signals in avian embryos. Anat Embryol (Berl) 191 : 297–310.
11. ZachgoJ, KornR, GosslerA (1998) Genetic interactions suggest that Danforth's short tail (Sd) is a gain-of-function mutation. Dev Genet 23 : 86–96.
12. LanePW (1993) BirkenmeierCS (1993) Urogenital syndrome (us): a developmental mutation on chromosome 2 of the mouse. Mamm Genome 4 : 481–484.
13. AlfredJB, RanceK, TaylorBA, PhillipsSJ, AbbottCM, et al. (1997) Mapping in the region of Danforth's short tail and the localization of tail length modifiers. Genome Res 7 : 108–117.
14. SembaK, ArakiK, LiZ, MatsumotoK, SuzukiM, et al. (2006) A novel murine gene, Sickle tail, linked to the Danforth's short tail locus, is required for normal development of the intervertebral disc. Genetics 172 : 445–456.
15. MaatmanR, ZachgoJ, GosslerA (1997) The Danforth's short tail mutation acts cell autonomously in notochord cells and ventral hindgut endoderm. Development 124 : 4019–4028.
16. ArakiK, ArakiM, YamamuraK (2002) Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res 30: e103.
17. ZhaoG, LiZ, ArakiK, HarunaK, YamaguchiK, et al. (2008) Inconsistency between hepatic expression and serum concentration of transthyretin in mice humanized at the transthyretin locus. Genes Cells 13 : 1257–1268.
18. ArakiK, OkadaY, ArakiM, YamamuraK (2010) Comparative analysis of right element mutant lox sites on recombination efficiency in embryonic stem cells. BMC Biotechnol 10 : 29.
19. KawaguchiY, CooperB, GannonM, RayM, MacDonaldRJ, et al. (2002) The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 32 : 128–134.
20. SavoryJGBN, PierreV, RijliFM, De RepentignyY, KotharyR, LohnesD (2009) Cdx2 regulation of posterior development through non-Hox targets. Development 136 : 4099–4110.
21. TakadaS, StarkKL, SheaMJ, VassilevaG, McMahonJA, et al. (1994) Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev 8 : 174–189.
22. GrecoTL, TakadaS, NewhouseMM, McMahonJA, McMahonAP, et al. (1996) Analysis of the vestigial tail mutation demonstrates that Wnt-3a gene dosage regulates mouse axial development. Genes Dev 10 : 313–324.
23. ChawengsaksophakK, JamesR, HammondVE, KontgenF, BeckF (1997) Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386 : 84–87.
24. MeislerMH (1997) Mutation watch: mouse brachyury (T), the T-box gene family, and human disease. Mamm Genome 8 : 799–800.
25. Abu-AbedS, DolleP, MetzgerD, BeckettB, ChambonP, et al. (2001) The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev 15 : 226–240.
26. SakaiY, MenoC, FujiiH, NishinoJ, ShiratoriH, et al. (2001) The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev 15 : 213–225.
27. GaoN, WhiteP, KaestnerKH (2009) Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell 16 : 588–599.
28. BaillieGJ, van de LagemaatLN, BaustC, MagerDL (2004) Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J Virol 78 : 5784–5798.
29. PennisiE (2007) Evolution. Jumping genes hop into the evolutionary limelight. Science 317 : 894–895.
30. KazazianHHJr (2004) Mobile elements: drivers of genome evolution. Science 303 : 1626–1632.
31. MaksakovaIA, RomanishMT, GagnierL, DunnCA, van de LagemaatLN, et al. (2006) Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2: e2 doi:10.1371/journal.pgen.0020002
32. KanoH, KurahashiH, TodaT (2007) Genetically regulated epigenetic transcriptional activation of retrotransposon insertion confers mouse dactylaplasia phenotype. Proc Natl Acad Sci U S A 104 : 19034–19039.
33. KrappA, KnoflerM, LedermannB, BurkiK, BerneyC, et al. (1998) The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev 12 : 3752–3763.
34. SellickGS, BarkerKT, Stolte-DijkstraI, FleischmannC, ColemanRJ, et al. (2004) Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet 36 : 1301–1305.
35. HoshinoM, NakamuraS, MoriK, KawauchiT, TeraoM, et al. (2005) Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47 : 201–213.
36. ChesleyP (1935) Development of the short-tailed mutant in the house mouse. J Exp Zool 70 : 429–459.
37. GrecoTL, TakadaS, NewhouseMM, McMahonJA, McMahonAP, et al. (1996) Analysis of the vestigial tail mutation demonstrates that Wnt-3a gene dosage regulates mouse axial development. Genes Dev 10 : 313–324.
38. YoshikawaY, FujimoriT, McMahonAP, TakadaS (1997) Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev Biol 183 : 234–242.
39. HoriK, Cholewa-WaclawJ, NakadaY, GlasgowSM, MasuiT, et al. (2008) A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev 22 : 166–178.
40. BeresTM, MasuiT, SwiftGH, ShiL, HenkeRM, et al. (2006) PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol 26 : 117–130.
41. ArnoldSJ, StappertJ, BauerA, KispertA, HerrmannBG, et al. (2000) Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech Dev 91 : 249–258.
42. SaegusaM, HashimuraM, KuwataT, HamanoM, WaniY, et al. (2007) A functional role of Cdx2 in beta-catenin signaling during transdifferentiation in endometrial carcinomas. Carcinogenesis 28 : 1885–1892.
43. GuoRJ, HuangE, EzakiT, PatelN, SinclairK, et al. (2004) Cdx1 inhibits human colon cancer cell proliferation by reducing beta-catenin/T-cell factor transcriptional activity. J Biol Chem 279 : 36865–36875.
44. GuoRJ, FunakoshiS, LeeHH, KongJ, LynchJP (2010) The intestine-specific transcription factor Cdx2 inhibits beta-catenin/TCF transcriptional activity by disrupting the beta-catenin-TCF protein complex. Carcinogenesis 31 : 159–166.
45. KorinekV, BarkerN, MorinPJ, van WichenD, de WegerR, et al. (1997) Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/ − colon carcinoma. Science 275 : 1784–1787.
46. MorinPJ, SparksAB, KorinekV, BarkerN, CleversH, et al. (1997) Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275 : 1787–1790.
47. RubinfeldB, RobbinsP, El-GamilM, AlbertI, PorfiriE, et al. (1997) Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science 275 : 1790–1792.
48. CurrarinoG, ColnD, VottelerT (1981) Triad of anorectal, sacral, and presacral anomalies. AJR Am J Roentgenol 137 : 395–398.
49. RossAJ, Ruiz-PerezV, WangY, HaganDM, SchererS, et al. (1998) A homeobox gene, HLXB9, is the major locus for dominantly inherited sacral agenesis. Nat Genet 20 : 358–361.
50. BelloniE, MartuccielloG, VerderioD, PontiE, SeriM, et al. (2000) Involvement of the HLXB9 homeobox gene in Currarino syndrome. Am J Hum Genet 66 : 312–319.
51. HarrisonKA, DrueyKM, DeguchiY, TuscanoJM, KehrlJH (1994) A novel human homeobox gene distantly related to proboscipedia is expressed in lymphoid and pancreatic tissues. J Biol Chem 269 : 19968–19975.
52. HarrisonKA, ThalerJ, PfaffSL, GuH, KehrlJH (1999) Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet 23 : 71–75.
53. LiH, ArberS, JessellTM, EdlundH (1999) Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat Genet 23 : 67–70.
54. ThompsonN, GesinaE, ScheinertP, BucherP, Grapin-BottonA (2012) RNA profiling and chromatin immunoprecipitation-sequencing reveal that PTF1a stabilizes pancreas progenitor identity via the control of MNX1/HLXB9 and a network of other transcription factors. Mol Cell Biol 32 : 1189–1199.
55. ThomasKR, CapecchiMR (1987) Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51 : 503–512.
56. YagiT, TokunagaT, FurutaY, NadaS, YoshidaM, et al. (1993) A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem 214 : 70–76.
57. TaniwakiT, HarunaK, NakamuraH, SekimotoT, OikeY, et al. (2005) Characterization of an exchangeable gene trap using pU-17 carrying a stop codon-beta geo cassette. Dev Growth Differ 47 : 163–172.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



