-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
article has not abstract
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003333
Category: Viewpoints
doi: https://doi.org/10.1371/journal.pgen.1003333Summary
article has not abstract
In our recent article [1], we reported an association between hypomethylation and genomic instability. A comment by Watson et al. [2] re-analyzes the data and claims that our findings may represent an artifact. We extend the methodological framework for analyzing copy number variants (CNVs) in the context of potential confounding factors to address the issues raised in the comment and to further research in this growing area of genomic science.
Watson et al. argue that the association we reported between hypomethylation of genomic DNA—determined from sperm methylomes [3]—and the density of CNVs can be explained by a combined confounding effect of known correlates of CNVs, namely repetitive elements and CpG islands. To support their argument, the authors eliminate many genomic regions providing a variety of justifications for why these regions create “spurious association”. Once the regions have been removed from the genome, Watson et al. claim the association between hypomethylation and genomic instability disappears.
We would first like to point out that the goal of our study was not to ignore potential relevance of these other factors—such as repetitive elements and CpG islands—but instead to broaden the scope of inquiry to examine possible additional explanatory power of hypomethylation. In deference to Watson et al., we initiated a re-analysis that systematically examined the purported confounding factors. Rather than pursuing the data exclusion approach used by Watson et al., we applied the standard multiple regression approach. The regression methods control for confounding without discarding data or otherwise biasing the inquiry to particular genomic sub-regions. Specifically, we first asked if the variables brought up by Watson et al. (LINE, SINE, LTR, Satellite, and CpG island content) individually or in combination explain the association between hypomethylation and CNV counts within 100-Kbp windows tiling the genome. In addition, using the Akaike information criterion (AIC) we measured the explanatory power of each of the six variables beyond the explanatory power of the other five.
We first applied the Negative Binomial regression model [4], because it is commonly used for overdispersed variables and is a well-accepted and robust method for count data such as CNV counts. We applied the same method to all five sample sets brought up by Watson et al.—HapMap270 [5], HapMap450 [6], WTCCC [7], Schizophrenia Cases, and Schizophrenia Controls [8]. As documented in Tables S1 and S2, in all five sample sets hypomethylation remained highly significantly associated with CNV density after correction for all of the “confounders” individually and in combination. As illustrated in Figure 1, as measured by AIC, methylation was more predictive of CNV counts per 100-Kbp window by an order of magnitude than any other factor.
Fig. 1. Predictive power of methylation and other genomic factors for CNV counts. 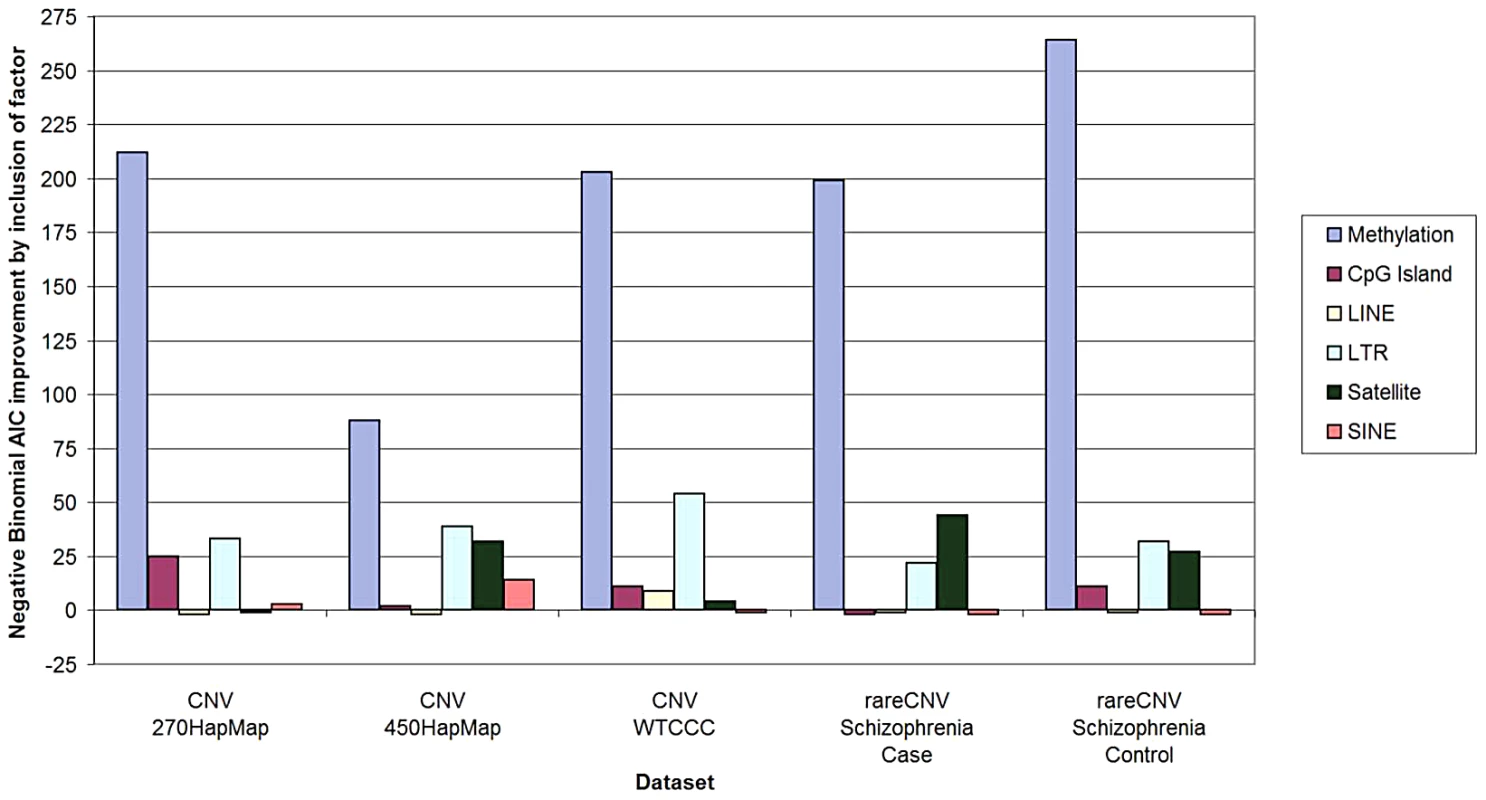
Predictive power of methylation, CpG island content, and repetitive element content (LINE, SINE, LTR, and Satellites) was measured using Akaike information criterion (AIC). For all five datasets, negative binomial regression was performed using all six factors and all six combinations of five factors (one factor being removed at a time). The y-axis represents the predictive power of a factor, as measured by the improvement of the AIC score based on all six factors relative to the AIC score without the factor. Note that this method measures predictive power of a factor after correction for any potential confounding due to other factors. (The detailed calculations and input data are in Supporting Information.) To examine robustness of our analyses with respect to modeling assumptions, we repeated the same analysis by applying the more widely used Poisson and linear regression models. All three models gave consistent results for all confounders in all five sample sets. (Table S2 describes regression models, output, and the input data extracted from our original paper sufficient to run a statistical program such as R to obtain the output.) We also employed zero-inflated negative binomial and Poisson regression models, and found completely concordant results. We therefore conclude that the assertions regarding “confounding” are not consistent with the data available. This is likely because Watson et al. resorted to an idiosyncratic selective data elimination procedure rather than pursuing a more standard statistical approach.
Second, Watson et al. bring up mappability of reads as a confounding factor while failing to mention that the original paper [1] considered and—using bisulfite sequencing data from embryonic stem cell H1 as a control—ruled out ascertainment biases due to read mappability: “We next examined the difference in methylation levels between sperm and H1. As illustrated in Figure S16, the difference shows even stronger association with structural mutability than the absolute methylation levels in sperm. This result rules out possible ascertainment biases due to low mappability of sequencing reads in potentially unstable and repetitive hypomethylated regions. It also suggests that structural mutability is associated with germline-specific hypomethylation.” Also quoting from our article: “We found significant negative correlation between the methylation scores in sperm and the heterozygosity rates (CNVs from 400 MGL samples: r≈−0.15, p≈10−9; CNVs from 270 HapMap samples: r≈−0.20, p≈10−10). In contrast, no significant correlation between the H1 methylation scores and the CNV heterozygosity rates was detected” [1].
Third, Watson et al. argue that the higher mode with zero scores for the Methylation Index (MI = 0) is likely an artifact due to small SNP and CpG counts. In this context it is surprising that Watson et al. fail to mention that our article considered, examined, and ruled out this possibility: “One could expect that if the windows with MI = 0 were due to low probing density, the windows within the higher mode would have fewer SNPs or CpGs. However, we examined potential biases in MI estimation due to variations in the number of SNPs, CpGs, read coverage (Figure S6CD), or sampling events (Figure S7BD) and found no significant difference between the two modes, ruling out the possibility that the two modes may be explained by variation in mappability or shallow sampling. In addition, a simulation experiment showed that the statistical variance of methylation estimates due to CpG sampling of windows with MI = 0 was a relatively small fraction of biological variance in methylation observed between the two sperm methylomes (Figure S8). We therefore hypothesize that the higher mode may either indicate hypomethylation specific to the female germline, given that male and female germline methylation patterns are highly dimorphic [47], or may be due to other germline hypomethylation detected by MI that is absent from sperm.”
Fourth, the “confounders” brought up by Watson et al. do not influence genomic instability independently of the methylation state and therefore do not meet the common definition of confounding [9]. In the specific case of CpG islands, the striking pattern where hypomethylated CpG islands are enriched in unstable regions (Figure 2A and 2B in Watson et al. [2]) would in fact be expected to occur if hypomethylation were mechanistically linked to genomic instability. Watson et al. ignore this possibility without sound justification while claiming that this pattern somehow provides evidence against any connection of hypomethylation and genomic instability.
Fifth, contrary to what Watson et al. claim, our article does not state that hypomethylation plays a causative role in genomic instability. Specifically, in the discussion section of our article we state three possible mechanistic explanations for the observed association: DNA break–inducing germline-specific demethylation during embryogenesis; mutagenic effects of germline-specific gene expression in hypomethylated loci; and mutagenic effects of transcription factor binding to hypomethylated loci.
In summary, we thank Watson et al. for their efforts and further examination of our reported observations. Nevertheless, we find that the arguments put forward in the comment do not diminish the strength of our reported findings. Specifically, our analyses of the confounding factors suggested by Watson et al. do not diminish the contention that genomic correlates may provide only a partial explanation for the hotspots of genomic instability. Thus, broadening inquiry to also include the epigenome may be warranted.
Supporting Information
Zdroje
1. LiJ, HarrisRA, CheungSW, CoarfaC, JeongM, et al. (2012) Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet 8 (5) e1002692 doi:10.1371/journal.pgen.1002692
2. WatsonCT, GargP, SharpAJ (2013) Comment on “Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome”. PLoS Genet 9: e1003332 doi:10.1371/journal.pgen.1003332
3. MolaroA, HodgesE, FangF, SongQ, McCombieWR, et al. (2011) Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146 (6) 1029–1041.
4. Hilbe J. (2011) Negative binomial regression. Cambridge, UK: Cambridge University Press. p. 553.
5. McCarrollSA, KuruvillaFG, KornJM, CawleyS, NemeshJ, et al. (2008) Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 40 (10) 1166–1174.
6. ConradDF, PintoD, RedonR, FeukL, GokcumenO, et al. (2010) Origins and functional impact of copy number variation in the human genome. Nature 464 (7289) 704–712.
7. The Wellcome Trust Case Control Consortium (2010) Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464 (7289) 713–720.
8. The International Schizophrenia Consortium (2008) Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455 (7210) 237–241.
9. Pearl J (2000) Causality: models, reasoning, and inference. Cambridge, UK: Cambridge University Press. 384 p.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Transfer zmraženého embrya zlepšuje výsledky IVF
- Velké děti po kryoembryotransferu
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



