-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
Many pathogenic bacteria use cell–cell signaling systems involving the synthesis and perception of diffusible signal molecules to control virulence as a response to cell density or confinement to niches. Bacteria produce signals of diverse structural classes. Signal molecules of the diffusible signal factor (DSF) family are cis-2-unsaturated fatty acids. The paradigm is cis-11-methyl-2-dodecenoic acid from Xanthomonas campestris pv. campestris (Xcc), which controls virulence in this plant pathogen. Although DSF synthesis was thought to be restricted to the xanthomonads, it is now known that structurally related molecules are produced by the unrelated bacteria Burkholderia cenocepacia and Pseudomonas aeruginosa. Furthermore, signaling involving these DSF family members contributes to bacterial virulence, formation of biofilms and antibiotic tolerance in these important human pathogens. Here we review the recent advances in understanding DSF signaling and its regulatory role in different bacteria. These advances include the description of the pathway/mechanism of DSF biosynthesis, identification of novel DSF synthases and new members of the DSF family, the demonstration of a diversity of DSF sensors to include proteins with a Per-Arnt-Sim (PAS) domain and the description of some of the signal transduction mechanisms that impinge on virulence factor expression. In addition, we address the role of DSF family signals in interspecies signaling that modulates the behavior of other microorganisms. Finally, we consider a number of recently reported approaches for the control of bacterial virulence through the modulation of DSF signaling.
Published in the journal: . PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1004986
Category: Review
doi: https://doi.org/10.1371/journal.ppat.1004986Summary
Many pathogenic bacteria use cell–cell signaling systems involving the synthesis and perception of diffusible signal molecules to control virulence as a response to cell density or confinement to niches. Bacteria produce signals of diverse structural classes. Signal molecules of the diffusible signal factor (DSF) family are cis-2-unsaturated fatty acids. The paradigm is cis-11-methyl-2-dodecenoic acid from Xanthomonas campestris pv. campestris (Xcc), which controls virulence in this plant pathogen. Although DSF synthesis was thought to be restricted to the xanthomonads, it is now known that structurally related molecules are produced by the unrelated bacteria Burkholderia cenocepacia and Pseudomonas aeruginosa. Furthermore, signaling involving these DSF family members contributes to bacterial virulence, formation of biofilms and antibiotic tolerance in these important human pathogens. Here we review the recent advances in understanding DSF signaling and its regulatory role in different bacteria. These advances include the description of the pathway/mechanism of DSF biosynthesis, identification of novel DSF synthases and new members of the DSF family, the demonstration of a diversity of DSF sensors to include proteins with a Per-Arnt-Sim (PAS) domain and the description of some of the signal transduction mechanisms that impinge on virulence factor expression. In addition, we address the role of DSF family signals in interspecies signaling that modulates the behavior of other microorganisms. Finally, we consider a number of recently reported approaches for the control of bacterial virulence through the modulation of DSF signaling.
Introduction
The discovery of diffusible signal factor (DSF) signaling arose from a molecular genetic analysis of the regulation of the synthesis of virulence factors in the plant pathogen Xanthomonas campestris pv. campestris (Xcc) [1,2]. These studies identified a cluster of genes called rpf (standing for “regulation of pathogenicity factors”), mutation of which leads to coordinate down-regulation of the synthesis of a number of extracellular enzymes (including endoglucanase and protease) and the extracellular polysaccharide xanthan as well as reduced virulence to plants [1,2]. Subsequent work revealed that the products of several of these rpf genes were involved in a signaling system involving synthesis and perception of a diffusible molecule that was called DSF, for diffusible signal factor [2,3]. The synthesis of DSF in Xcc is totally dependent on RpfF, which has amino acid sequence relatedness to enoyl CoA hydratase and is partially dependent on RpfB, which is a long-chain fatty acyl CoA ligase. DSF sensing and signal transduction involves a two-component system comprising the sensor RpfC and regulator RpfG [3]. These proteins are encoded by the rpfGHC operon, which is adjacent to rpfF and convergently transcribed (Fig 1A). Perception of DSF by RpfC is linked to phosphorylation of the HD-GYP domain regulator RpfG and alteration in the cellular level of the second messenger cyclic di-GMP [3,4]. Different pathways then act to control different sub-sets of Rpf-regulated virulence functions. RpfC acts to positively regulate synthesis of virulence factors, but to negatively regulate DSF synthesis. The elevated level of DSF in rpfC mutants allowed characterization of the signal as the unsaturated fatty acid cis-11-methyl-dodecenoic acid (Fig 1B), representing the paradigm for a novel structural class of signal [3,5]. The cis unsaturated double bond at the 2-position is a key structural feature for activity; this structural motif is regarded as the signature for DSF family signals [5].
Fig. 1. DSF family signals and the organization of rpf gene clusters that direct signal synthesis and perception. 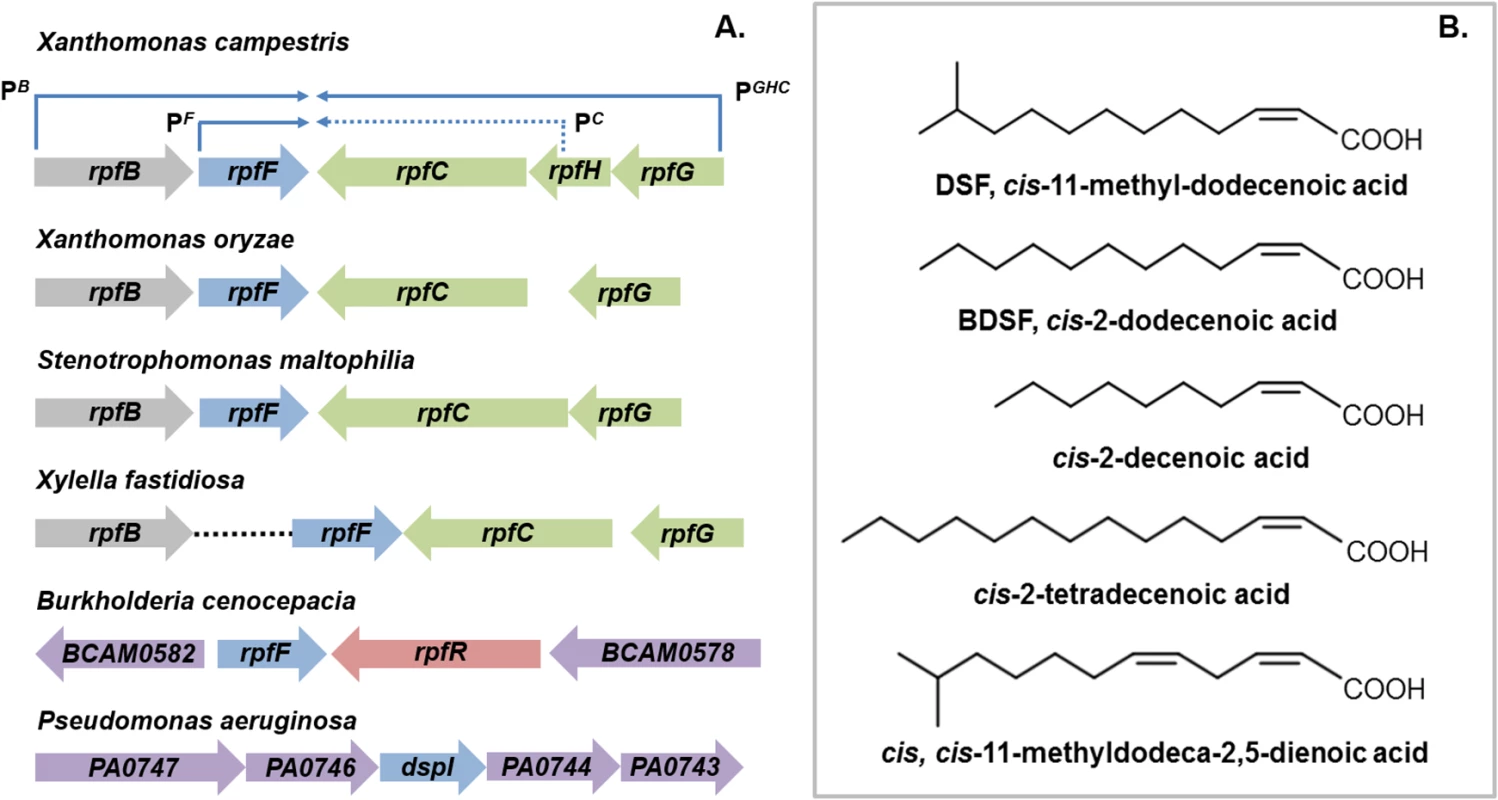
(A) RpfF, an enzyme of the crotonase superfamily, is involved in synthesis of DSF in all bacteria. Two core pathways of DSF perception have been described: (1) the RpfCG two-component system first identified in xanthomonads and (2) RpfR, a protein with PAS, GGDEF, and EAL domains, first identified in Burkholderia species. In Xanthomonas campestris, the genes encoding RpfC and RpfG are organized in an operon that is convergently transcribed to rpfF (top). This operon also contains rpfH, a gene encoding a protein similar to the input domain of RpfC but of no known function. The rpfF gene is found in an operon with rpfB, which encodes a fatty acyl CoA ligase but also has its own promoter. This organization of rpf genes occurs in all xanthomonads with the variations that rpfH is not widely conserved and rpfB can be located in a different genomic location, as is seen in Xylella fastidiosa. In Burkholderia species, rpfF is convergently transcribed with rpfR, which encodes the sensor protein. In Pseudomonas aeruginosa, the dspI gene that encodes an RpfF homolog is located in a cluster of genes encoding enzymes implicated in fatty acid metabolism. The identity of the sensor for the signal is not known. (B) The DSF family of signals comprises cis-2-unsaturated fatty acids of different chain lengths and branching. The paradigm cis-11-methyl–dodecenoic acid designated DSF, was first described in Xanthomonas campestris. Different signals were then described in Burkholderia cenocepacia (BDSF), Pseudomonas aeruginosa (cis-2-decenoic acid), Xylella fastidiosa (cis-2-tetradecenoic acid) and Xanthomonas oryzae (cis,cis-11 methyldodeca-2,5-dienoic acid). It is now established that each of these bacteria produces multiple DSF family signals, although each genus seems to be most responsive to the major signal that it produces. DSF family signals can be involved in bi-directional interspecies signaling. However unidirectional signaling is evident; Pseudomonas aeruginosa responds to DSF and BDSF produced by xanthomonads, but these latter organisms do not respond to cis-2-decenoic acid, the P. aeruginosa signal. Comparative genomic studies have indicated conservation of clustered rpfF, rpfG, and rpfC genes, and by extension DSF-mediated signaling, throughout the xanthomonads [6]. Furthermore, DSF synthesis and signaling is known to influence the virulence of several Xanthomonas spp. and Xylella fastidiosa, which are plant pathogens, and Stenotrophomonas maltophilia, some strains of which are opportunistic human pathogens [7,8]. In most of these bacteria, DSF acts to positively influence virulence, whereas in Xylella fastidiosa, DSF-deficient mutants show enhanced virulence [7,8]. Xylella fastidiosa is transmitted between plants exclusively by xylem sap-feeding insects. Intriguingly, DSF also influences bacterial interactions with these vectors. DSF-deficient mutants have a reduced capacity to colonize their insect vector and to form biofilms in the insect foregut [8]. This reduced retention leads to poor transmission to uninfected plants [8].
DSF synthesis was originally thought to be restricted to the xanthomonads and to unrelated bacteria with an rpfF-rpfG-rpfC gene cluster, such as Methylophaga, Leptospirillum, Thiobacillus, and Methylobacillus species [9]. Later findings, however, suggested a much broader significance for DSF signaling in the bacterial world. Structurally related molecules are produced by the unrelated bacteria Burkholderia cenocepacia and Pseudomonas aeruginosa: these molecules are cis-2-dodecenoic acid (BDSF) and cis-2-decenoic acid, respectively (Fig 1B) [10,11]. Signaling involving these DSF family members contributes to bacterial virulence, formation of biofilms, and antibiotic tolerance in these important human pathogens [11,12].
In addition to intraspecies signaling, DSF family signals have been implicated in interspecies and interkingdom signaling, in which they modulate the behavior of other microorganisms that do not produce the signal. For example, although Pseudomonas aeruginosa does not synthesize DSF or BDSF, it is capable of sensing these molecules, with consequences for bacterial behavior [9]. The dimorphic fungus Candida albicans can also respond to BDSF to modulate the yeast-hyphal transition [10].
Over the last five years, significant progress has been made in our understanding of DSF-family signaling. These advances have included (i) the discovery of the biosynthetic pathway/mechanism for DSF; (ii) the identification of new DSF family signals in a variety of additional bacterial pathogens; (iii) the discovery of alternate mechanisms for the sensing of DSF family signals that involved diverse proteins with PAS (Per-Arnt-Sim) domains; (iv) advances in the mechanistic understanding of the signal transduction pathways that follow DSF perception and their role in regulation of bacterial virulence; and (v) several seminal studies that demonstrate that modulation of DSF signaling may have potential in controlling bacterial disease. Here, we review these recent insights into the broad significance of DSF-family signaling systems in bacteria. We conclude by identifying some outstanding research questions concerning this fascinating family of signal molecules and their role in plant and animal pathogenic bacteria.
The Role of RpfF and RpfB in DSF Biosynthesis and Signaling
RpfF proteins have amino acid sequence similarity to enoyl CoA hydratases, which are members of the crotonase superfamily of enzymes [6,10]. Recent studies of BCAM0581, a homolog of RpfF, from B. cenocepacia have identified the immediate substrate(s) and some aspects of the enzymatic mechanism of these DSF synthases [13]. In vitro, BCAM0581 utilizes a 3-hydroxylated fatty acyl-ACP (acyl carrier protein) as substrate rather than a CoA derivative and through its desaturase and thioesterase activity generates the cis-2-unsaturated fatty acid BDSF (Fig 2). The current model for the action of BCAM0581 is that the enzyme first works as a dehydratase to convert 3-hydroxydodecanoyl-ACP to cis-2-dodecenoyl-ACP and then as a thioesterase to release free BDSF (cis-2-dodecenoic acid) [13]. The mechanistic details underpinning these two actions, which may be coupled in vivo, are unclear. RpfF is the only member of the crotonase superfamily with both desaturase and thioesterase activity. In vivo, the substrates for RpfF are presumably intermediates in fatty acid biosynthesis. Because RpfF has thioesterase activity, it can generate free saturated fatty acids from any fatty acyl ACP substrate. In the in vitro assay, an exogenous acyl-ACP synthetase is added to reverse this thioesterase reaction, promoting BDSF synthesis. Although RpfB was originally thought to be involved in DSF synthesis or processing [2,14], new work has established that it has a different role, acting in the mobilization of (saturated) free fatty acids generated by the thioesterase action of RpfF [15]. In Xcc, RpfB, which is a predicted fatty acid CoA ligase, activates these free fatty acid derivatives so that they can be used in phospholipid biosynthesis (Fig 2). In this way, RpfB counteracts the thioesterase activity of RpfF. RpfB has little activity against BDSF, however. Orthologs of RpfB occur widely in Burkholderia spp., although the encoding genes are not linked to rpfF, in contrast to what is seen in most xanthomonads (Fig 1A).
Fig. 2. The roles of RpfF and RpfB in DSF signaling. 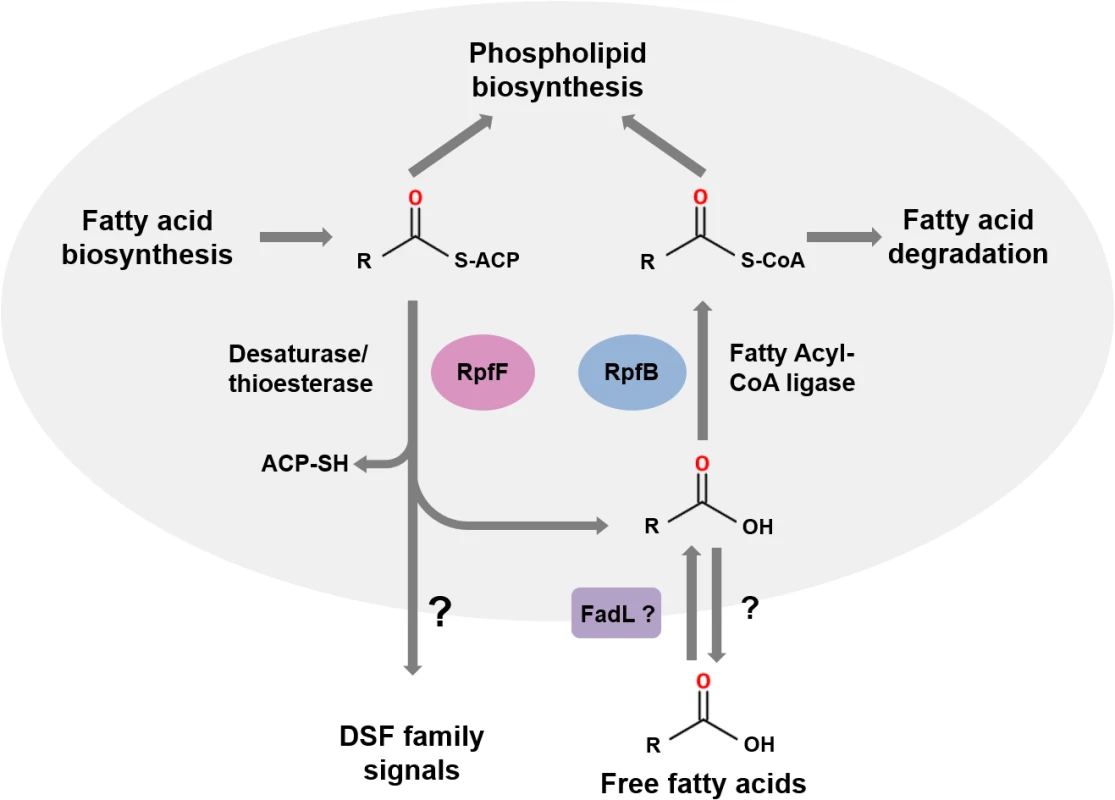
RpfF encodes an enzyme from the crotonase family that possesses both desaturase and thioesterase activities and uses fatty acyl ACP substrates derived from fatty acid biosynthesis as substrates. DSF family signals are generated from 3-hydroxy-fatty acyl ACPs by consecutive or coordinated desaturase and thioesterase action. In addition, the thioesterase activity of RpfF generates a range of free hydroxylated and non-hydroxylated fatty acids. DSF family signals leave the cell by as-yet-unknown efflux mechanisms (indicated by the question mark). Free fatty acids whose synthesis depends upon RpfF can also be found in the culture supernatant. The role of RpfB is in mobilization of these free fatty acids. Uptake, which may involve FadL, is followed by conversion to fatty acyl CoA derivatives that can be used in phospholipid biosynthesis or for degradation. This figure is a modified version of that published by Bi and colleagues [15]. The dual nature of BCAM0581 as a broad specificity thioesterase as well as a desaturase is also seen in vitro with the Xcc RpfF enzyme and is consistent with a number of previous observations on effects of rpfF mutation on fatty acid profiles in culture supernatants [16,17]. Loss by mutation of RpfF in S. maltophilia affects synthesis of not only DSF but also seven structurally related saturated and unsaturated fatty acids [17]. Culture supernatants of X. fastidiosa contain 12-methyl-tetradecanoic acid [18], as well as the authentic DSF-family signal cis-2-tetradecenoic acid, a recently described member of the DSF family (see Fig 1B) [19,20].
Individual bacteria can produce multiple DSF family signals. For example, bacteria within the Burkholderia cepacia complex (Bcc) as well as Xanthomonas spp. synthesize DSF, BDSF, and cis,cis-11-methyldodeca-2,5-dienoic acid (Fig 1B) [21–23]. These multiple DSF family signals are all dependent on RpfF for their synthesis. Swapping RpfF homologs between bacteria in the Bcc complex indicates that the pattern of DSF, BDSF, and cis,cis-11-methyldodeca-2,5-dienoic acid signals produced is regulated by the supply of different substrates, rather than differences in specificity of different RpfF synthases [22]. Consistent with this contention, in X. oryzae pv. oryzae, the rice pathogen, the three signals are present in different ratios depending on the culture medium and are produced with different time courses during growth [23]. The substrate for DSF synthesis in vivo must be 11-methyl-3-hydroxydodecanoyl-ACP, with the 11-methyl substitution presumably derived from leucine via the branched chain fatty acid synthetic pathway. Whether production of multiple DSF-family signals by one organism has any biological relevance is unclear. However the systems for sensing DSF family signals within a particular organism appear to be attuned to the major signal produced by that organism. For example, Xcc and X. fastidiosa generate cis-11-methyl-dodecenoic acid and cis-2-tetradecenoic acid respectively via highly related RpfF proteins, but each is more responsive to its own signal than the heterologous one [23].
The Interaction of RpfF and RpfC and Signal Transduction
As outlined above, RpfC in Xcc acts as a sensor for DSF but also in repression of DSF biosynthesis. DSF sensing and signal transduction leading to activation of the production of extracellular enzymes and EPS requires conserved amino acid residues of RpfC implicated in phosphorelay. In contrast, RpfC repression of DSF synthesis does not require phosphorelay but is mediated instead by protein–protein interactions between the REC domain of RpfC and RpfF, the DSF synthase [24]. Sequestration of RpfF in this manner may restrict synthesis of DSF; loss of RpfC by mutation results in a highly elevated level of DSF [24]. Release of RpfF as a result of conformational changes in RpfC that occur upon DSF binding has been invoked as a mechanism to allow rapid auto-induction of DSF synthesis [24,25]. However, subsequent experimental evidence indicates that DSF synthesis in Xcc is not auto-inductive, suggesting that this may not be the true role for this interaction [26]. Recent work in X. fastidiosa has provided potential further insight into the role for RpfF–RpfC interactions [26]. In Xcc, the synthesis of extracellular enzymes and EPS in an rpfF mutant can be restored to wild type by exogenous DSF. By contrast, addition of the Xylella DSF family signal does not restore the phenotype of the rpfF mutant to wild type. Instead DSF signal transduction in Xylella requires both RpfC and RpfF [26]. Enzymatically inactive variants of RpfF in which two conserved catalytic glutamate residues are replaced by alanine residues can also support DSF signal transduction. These findings reveal RpfF in Xylella to be a multifunctional protein. A plausible model for RpfF action is that it can interact with multiple REC domain proteins, and release from RpfC upon binding of DSF (or other signals) allows its interaction with these other regulators, thereby effecting signal transduction through “partner swapping.
Intriguingly, analysis of a panel of clinical isolates of S. maltophilia revealed distribution of two sub-groups of rpf gene cluster that were associated with different levels of DSF synthesis and virulence regulation [27]. Two populations of RpfF called RpfF-1 and RpfF-2 were distinguished by differences in the N-terminal region of the protein. Each RpfF variant is associated with a specific RpfC variant (RpfF1 with RpfC1 and RpfF2 with RpfC2). RpfC1 and RpfC2 differ in the number of transmembrane helices in the sensory input domain [27]. Only RpfC-1-RpfF-1 variant strains display detectable DSF production; the observed DSF-deficient phenotype of RpfC-2-RpfF-2 variant strains is due to permanent repression of RpfF-2 by RpfC-2.
Expansion of Classes of Sensor Involved in DSF Signal Perception
The discovery of BDSF synthesis in B. cenocepacia through the action of the RpfF ortholog BCAM0581 raised the issue of the mechanisms by which the signal is perceived [10]. Two sensors for BDSF have now been described; BCAM0580, which is designated RpfR, and the sensor kinase BCAM0227 [28,29]. RpfR (see Fig 3) comprises an N-terminal PAS domain and GGDEF and EAL domains. GGDEF and EAL domains are implicated in the synthesis and degradation, respectively, of the second messenger cyclic di-GMP [29,30]. In vitro, RpfR exhibits cyclic di-GMP phosphodiesterase activity that is modulated by binding of BDSF to the N-terminal PAS domain [29]. Thus, in both B. cenocepacia and Xcc, sensing of a DSF family signal is linked to cyclic di-GMP turnover, but the mechanisms are completely different (Fig 3). Notably, this was the first description of the role of a PAS domain in sensing DSF-family signals. Bioinformatic analysis reveals that the RpfR-RpfF system is widely conserved not only in Burkholderia species but also in bacteria from related genera, such as Achromobacter, and unrelated Enterobacteriacaeae, including Yersinia, Serratia, Cronobacter, and Enterobacter.
Fig. 3. Signal transduction mechanisms for DSF family signals in different bacteria. 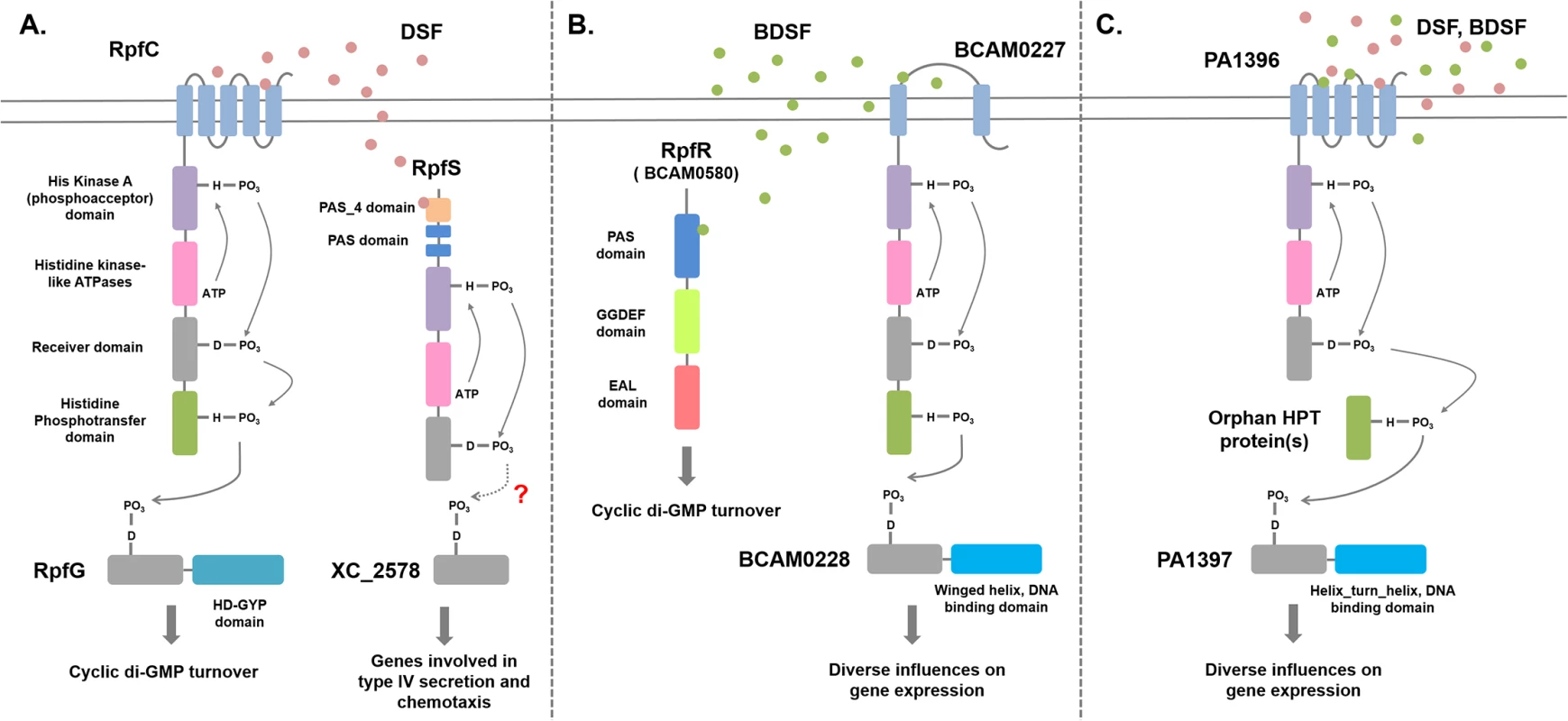
(A) Xanthomonas campestris. DSF perception involves the complex histidine kinase RpfC. Binding of DSF to the sensory input domain, which comprises five transmembrane helices, leads to autophosphorylation, phosphorelay via the receiver and histidine phosphotranfer domains, and phosphotransfer to the receiver domain of the response regulator RpfG. RpfG has an HD-GYP domain, which is a cyclic di-GMP phosphodiesterase; phosphorylation activates RpfG for cyclic di-GMP degradation. A second sensing system for DSF involves the soluble histidine kinase RpfS, which binds DSF through the N-terminal PAS_4 domain. RpfS influences the expression of a sub-set of DSF-regulated genes, in particular those involved in the epiphytic growth phase. This action is likely exerted through the response regulator XC_2578. The RpfGC system is “core,” being found in all xanthomonads, including Xylella fastidiosa, whereas RpfS is “accessory” and is not fully conserved. In Xylella, DSF signal transduction also involves RpfF (see text for details). (B) Burkholderia cenocepacia. BDSF perception involves the soluble PAS-GGDEF-EAL domain protein RpfR. Binding of BDSF to the N-terminal PAS domain activates RpfR for cyclic di-GMP degradation by the cyclic di-GMP phosphodiesterase EAL domain. A second sensing system for BDSF involves the histidine kinase BCAM0227, which differs from RpfC in having two transmembrane helices and a large periplasmic domain. Signal transduction from BCAM0227 involves autophosphorylation upon BDSF binding, phosphorelay, and phosphotransfer to the DNA-binding regulator BCAM0228. The RpfFR system is “core,” being found in widely in Burkholderia and other genera, whereas BCAM0227 is “accessory” and is only found in strains of B. cenocepacia. (C) Interspecies signaling in Pseudomonas aeruginosa. BDSF or DSF perception involves the membrane-associated histidine kinase PA1396, which resembles RpfC but does not have an HPt domain. Signal transduction from PA1396 involves autophosphorylation upon BDSF binding, phosphorelay, and phosphotransfer via an orphan HPt domain protein to the DNA-binding regulator PA1397. This system influences the expression of genes involved in polymyxin resistance and stress tolerance. BCAM0227 is a complex sensor kinase but is not a homolog of RpfC, the DSF sensor in Xcc [28]. BCAM0227 is predicted to have two transmembrane helices and a large periplasmic loop of 300 amino acids, whereas RpfC is predicted to have five transmembrane helices (Fig 3). Comparative transcriptome analysis showed that BCAM0227 is involved in regulation of a subset of functions that are controlled by BDSF in B. cenocapacia [28]. Unlike RpfR, BCAM0227 is restricted to B. cenocepacia [29]. This may suggest that RpfR-RpfF is a second “core” system for DSF signaling in addition to RpfF-RpfC-RpfG of the xanthomonads, whereas BCAM0227 is an accessory sensor.
Alternative sensors for DSF family signals have also been proposed for Xanthomonas spp and Xylella [31–33]. These suggestions were based on comparative transcriptome analysis of wild type and rpfF, rpfC, and rpfG mutants [31–33]. The effects of these mutations were not consistent with the existence of a single linear pathway, and pointed to the occurrence of alternative sensors for DSF and alternative regulators that interact with RpfC [31–34]. A genetic screen identified XC_2579 (RpfS) as a second sensor for DSF in Xcc [35]. RpfS is a histidine sensor kinase with an N-terminal PAS domain (PAS_4 domain) that binds DSF and is required for regulation (Fig 3). RpfS controls expression of a specific subset of genes controlled by DSF and has a role in the epiphytic phase of the disease cycle [35]. Homologs of RpfS occur in many, but not all, Xanthomonas and Stenotrophomonas species, suggesting that it is an accessory element in DSF signaling.
PAS domains are known to be highly divergent at the primary sequence level, but nevertheless have a conserved three-dimensional architecture within which structural clades can be discerned [36]. This structural division of PAS domains may reflect, in part, the class of small molecule that they bind. The PAS domains of XC_2579 and RpfR lack significant sequence similarity, and further work will be needed to show if they are nonetheless highly related, structurally.
DSF Signaling and the Regulation of Bacterial Virulence, Biofilm Formation, and Antibiotic Resistance
In many bacteria, DSF family-mediated signaling contributes to bacterial virulence, formation of biofilms and antibiotic tolerance; examples are given in Table 1. Perception of DSF-family signals by the two “core” pathways is linked to cyclic di-GMP turnover. This second messenger interacts with a range of effectors to exert a regulatory action at transcriptional, post-transcriptional, and post-translational levels [30]. Hence, identifying factors under DSF control has required transcriptomic and proteomic approaches as well as assessment of phenotypes [31–34,37–39].
Tab. 1. Virulence-related and other factors that are regulated by DSF family signals in diverse pathogens. 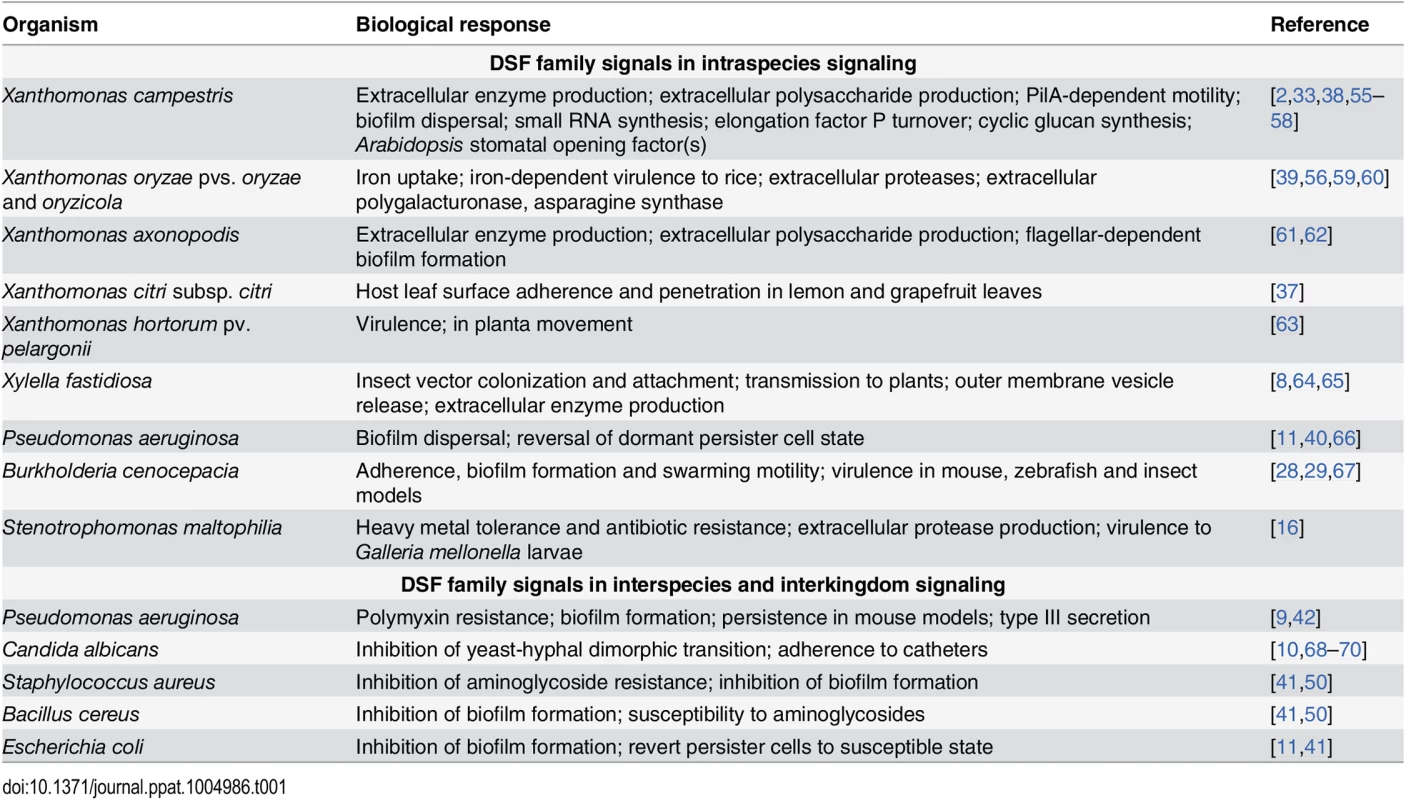
DSF Family Signals and Intra - and Inter-species Signaling in Pseudomonas aeruginosa
Studies in P. aeruginosa have provided an insight into signaling involving the DSF family through identification of new members of the family (cis-2-decenoic acid; Fig 1) [11], and the role that DSF has in interspecies communication (Fig 4) [9,40]. Cis-2-decenoic acid was identified as a factor produced by P. aeruginosa that can induce dispersion of biofilms produced by P. aeruginosa as well as other bacteria, and inhibit the growth of Staphylococcus aureus (Fig 4) [11,41]. An RpfF homolog called DspI (PA0745) is required for production of cis-2-decenoic acid [40]. The action of the purified DspI protein has not been tested, but is inferred from the effects of mutation of the dspI gene on the production of cis-2-decenoic acid in the culture medium [40]. DspI contains 15 out of the 29 conserved amino acids of the predicted ligand-binding site of RpfF from Xanthomonas oryzae [40] and has the two conserved glutamates known to be essential for DSF production by RpfF from Xanthomonas campestris and Xylella fastidiosa [25,26], suggesting that it acts by the same mechanism. Intriguingly, PA0745 is located in a putative operon with genes encoding other fatty acid–handling enzymes including an acyl-CoA dehydrogenase, a 3-hydroxyisobutyrate dehydrogenase and a putative enoyl-CoA hydratase/isomerase. Homologs of DspI occur in over ten Pseudomonas species, suggesting that production of DSF family signal molecules may be widespread among members of the genus [40]. Cis-2-decenoic acid does not effectively activate the DSF signaling pathway in Xcc, but conversely, DSF and BDSF can activate changes in gene expression, alter biofilm formation, and increase antibiotic tolerance in P. aeruginosa [9,30]. This action is exerted via PA1396, a sensor kinase with an input domain very similar to that of RpfC of Xcc [9]. Such interspecies signaling may occur in multispecies infections, such as those associated with cystic fibrosis (CF) in which P. aeruginosa is present together with S. maltophilia and Burkholderia species (Fig 4) [42]. The detection of DSF and BDSF signals at physiologically relevant levels in the sputum of CF patients supports this contention [42]. PA1396 does not respond to cis-2-decenoic acid, however, so the identity of the sensor for this intra-species signal remains obscure.
Fig. 4. Signals of the DSF family in interspecies and inter-kingdom signaling. 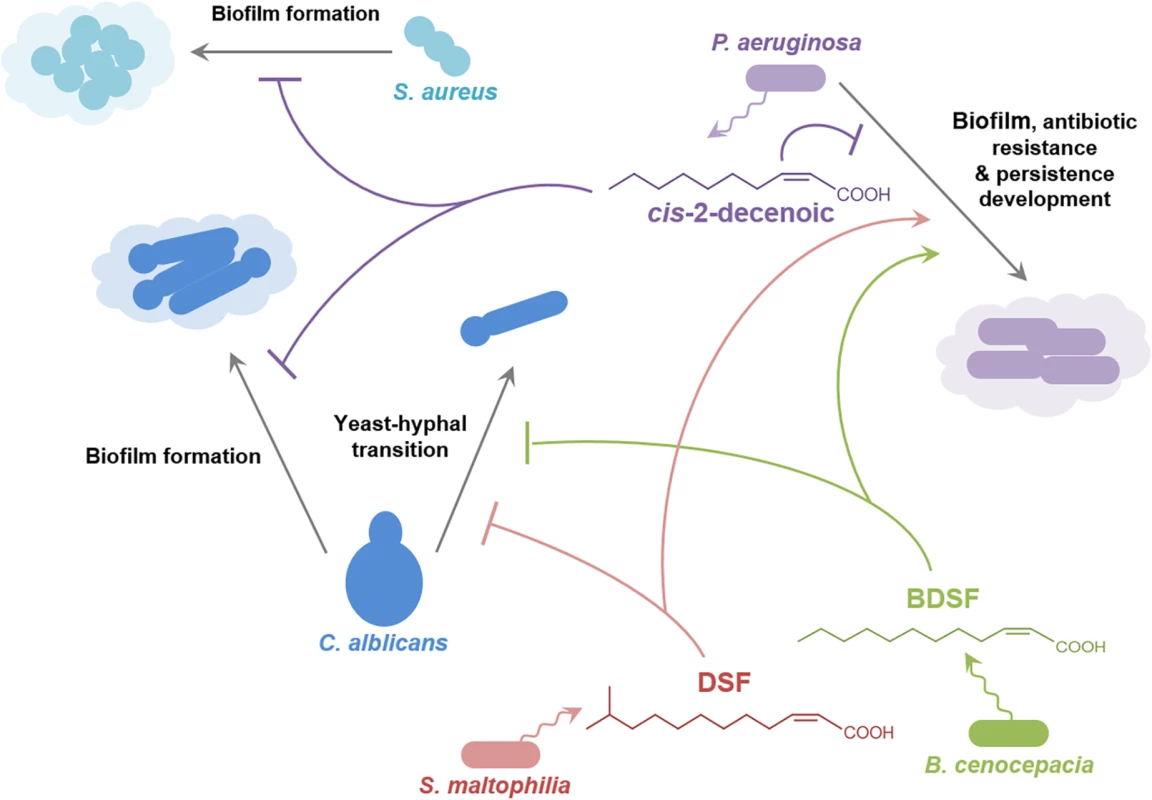
DSF family signals produced by Stenotrophomonas maltophilia (principally DSF) and Burkholderia cenocepacia (principally BDSF) influence the behavior of Pseudomonas aeruginosa and the yeast–hyphal transition in Candida albicans. The P. aeruginosa signal cis-2-decenoic acid prevents biofilm by Candida albicans and growth and biofilm formation by Staphylococcus aureus. The effects of DSF and BDSF on Pseudomonas aeruginosa are exerted via the sensor kinase PA1396, which does not respond to cis-2-decenoic. The production of DSF and BDSF may allow interspecies communication between Stenotrophomonas maltophilia and Burkholderia cenocepacia. Other signal molecules are also involved in interaction between some of these organisms, but are not indicated here for clarity. For example, 3-oxo-dodecanoyl-homoserine lactone produced by P. aeruginosa can influence C. albicans and B. cenocepacia. The Role of DSF Family Signals in Interkingdom Signaling
The DSF family of signal molecules has been implicated in inter-kingdom signaling between bacteria and the dimorphic fungal pathogen C. albicans. DSF, BDSF, and CDSF can all act to modulate the morphological yeast-hyphal transition of C. albicans (Fig 4) [10,43]. Such interactions may be important in the CF airway, in which C. albicans can be part of the polymicrobial community. A range of other compounds able to inhibit the Candida yeast–hyphal transition include the fungal signals farnesol and farnesoic acid, dodecanol, the bacterial quorum sensing signal 3-oxo-C12-homoserine lactone and trans-2-decenoic acid produced by Streptococcus mutans [44]. It is not known if all these molecules exert their action through the same mechanism.
There is an expanding literature on the influence that bacterial signal molecules such as N-acyl homoserine lactones and the Pseudomonas quinolone signal (PQS) can have on mammalian host cells [44,45]. In contrast, there is, as yet, almost no information on the action of DSF family molecules on plant or mammalian hosts. Examination of the effects of DSF on the salicylic acid and abscisic acid signaling pathways in rice revealed little impact on marker genes for the former and only a minor suppressive effect on one marker gene for the latter [46].
Establishment of Methods for Interference with DSF Family Signaling As a Route to Control Bacterial Disease
The role of DSF signaling in regulation of virulence factors has indicated that it may be a potential target for interference to allow control of disease. Work on the behavior of X. fastidiosa and Xcc has indicated that DSF signaling is normally finely balanced during the disease process and that such a fine balance might therefore be readily disrupted by either degradation or over-production of the signal, with consequences for disease control [31]. One approach has been to engineer plants to express RpfF and hence elevate DSF levels [47,48]. Plant expression of RpfF from X. fastidiosa in citrus and grape reduces the virulence of the important plant pathogens X. citri and X. fastidiosa respectively [47,48]. Although the synthesis of virulence factors in X. citri within transgenic citrus is reduced, the underlying mechanisms are not clear. It is plausible that synthesis of DSF activates plant defense responses that impair bacterial growth and gene expression. Alternatively, expression of RpfF may generate DSF and/or structural analogs that directly influence cell-cell signaling. Expression of the Xylella RpfF in grape conferred the production of not only cis-2-tetradecenoic acid (the Xylella signal) but also DSF and cis-2-hexadecenoic acid, two compounds that have not been found in cultures of X. fastidiosa [19].
Inoculation of bacteria able to degrade DSF can reduce virulence and symptom production by X. fastidiosa in grape and Xcc in brassica [49]. This suggests that it should be possible to select or engineer strains for use as biocontrol agents for plant diseases. The mechanisms by which bacteria degrade or inactivate DSF are unknown, although rapid degradation or inactivation of DSF by a Pseudomonas spp strain G requires carAB, which encode enzymes responsible for the synthesis of carbamoylphosphate, a precursor in biosynthesis of pyrimidines and arginine [49].
To influence disease in animals or humans, a rational approach would be the identification of molecules that block key signal sensing or transduction steps. Structural analogues of DSF may have a role in control of virulence factor synthesis in different pathogens. Such molecules may represent lead compounds for new drugs. The signal molecules themselves may also be of use. BDSF can act to enhance the antimicrobial efficacy of antibiotics against some bacterial pathogens and inhibit C. albicans adherence to catheters [50,51]. Cis-2-decenoic acid can inhibit growth and biofilm formation by S. aureus [41], in combination with antibiotics can eradicate pre-established biofilms of a number of bacteria [52,53], and can revert antimicrobial-insensitive persister cells of P. aeruginosa and Escherichia coli to a susceptible state [54]. Although such approaches discussed above are still in their infancy, the examples outlined in combination with other control measures could facilitate improved approaches to prevent and treat bacterial infections associated with a range of plant, animal, and human diseases.
Concluding Remarks
It is now evident that signal molecules of the DSF family play a significant role in regulation of virulence and disease progression in a wide range of plant, animal, and human bacterial pathogens. However, the recent advances outlined above pose a new set of outstanding research questions. Will the determination of the structure of RpfF from both Xanthomonas and Stenotrophomonas species aid in defining the mechanistic details of DSF synthesis and allow the rational development of inhibitors? Will analysis of PAS domains identify structural features specifying DSF binding? The expansion of the classes of DSF receptor suggests that further DSF-family–mediated intra-species or inter-species signaling remains to be predicted and identified. How does the P. aeruginosa signal-response network involve cis-2-decenoic function? Will this represent a third class of widely conserved DSF-family signaling system? What are the actions of DSF family signals on fungi other than C. albicans and on eukaryotic hosts of plant and animal pathogens? Unquestionably, future research to address these issues will most likely uncover further fascinating aspects of this family of signal molecules.
Zdroje
1. Tang JL, Liu YN, Barber CE, Dow JM, Wootton JC, et al. (1991) Genetic and Molecular analsysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol Gen Genet 226 : 409–417.
2. Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJG, et al. (1997) A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol 24 : 555–566.
3. Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM (2000) A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol 38 : 986–1003.
4. Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, et al. (2006) Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA 103 : 6712–6717.
5. Wang LH, He YW, Gao YF, Wu JE, Dong YH, et al. (2004) A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol 51 : 903–912.
6. Fouhy Y, Lucey JF, Ryan RP, Dow JM (2006) Cell-cell signaling, cyclic di-GMP turnover and regulation of virulence in Xanthomonas campestris. Research in Microbiology 157 : 899–904. 17008065
7. Chatterjee S, Newman KL, Lindow SE (2008) Cell-to-cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol Plant Microbe Interact 21 : 1309–1315.
8. Newman KL, Almeida RP, Purcell AH, Lindow SE (2004) Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc Natl Acad Sci U S A 101 : 1737–1742.
9. Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, et al. (2008) Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol 68 : 75–86.
10. Boon C, Deng Y, Wang L-H, He Y, Xu J-L, et al. (2008) A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2 : 27–36.
11. Davies DG, Marques CNH (2009) A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191 : 1393–1403.
12. Ryan RP, McCarthy Y, Watt SA, Niehaus K, Dow JM. (2009) Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J Bacteriol. 191 : 5013–5019.
13. Bi H, Christensen QH, Feng Y, Wang H, Cronan JE (2012) The Burkholderia cenocepacia BDSF quorum sensing fatty acid is synthesized by a bifunctional crotonase homologue having both dehydratase and thioesterase activities. Mol Microbiol 83 : 840–855.
14. Almeida RPP, Killiny N, Newman KL, Chatterjee S, Ionescu M, et al. (2012) Contribution of rpfB to cell-to-cell signal synthesis, virulence, and vector transmission of Xylella fastidiosa. Mol Plant Microbe Interact 25 : 453–462.
15. Bi H, Yu Y, Dong H, Wang H, Cronan JE (2014) Xanthomonas campestris RpfB is a fatty Acyl-CoA ligase required to counteract the thioesterase activity of the RpfF diffusible signal factor (DSF) synthase. Mol Microbiol 93 : 262–275.
16. Fouhy Y, Scanlon K, Schouest K, Spillane C, Crossman L, et al. (2007) Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J Bacteriol 189 : 4964–4968.
17. Huang T-P, Lee Wong AC (2007) A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl Environ Microbiol 73 : 5034–5040.
18. Colnaghi Simionato AV, da Silva DS, Lambais MR, Carrilho E (2007) Characterization of a putative Xylella fastidiosa diffusible signal factor by HRGC-EI-MS. J Mass Spectrom 42 : 1375–1381.
19. Lindow S, Newman K, Chatterjee S, Baccari C, Lavarone AT, et al. (2014) Production of Xylella fastidiosa diffusible signal factor in transgenic grape causes pathogen confusion and reduction in severity of Pierce's disease. Mol Plant Microbe Interact 27 : 244–254.
20. Beaulieu ED, Ionescu M, Chatterjee S, Yokota K, Trauner D, et al. (2013) Characterization of a diffusible signaling factor from Xylella fastidiosa. mBio 4: e00539–12.
21. Deng Y, Liu X, Wu J, Lee J, Chen S, Cheng Y, Zhang C, Zhang LH. (2015) The host plant metabolite glucose is the precursor of diffusible signal factor (DSF) family signals in Xanthomonas campestris. Appl Environ Microbiol 81 : 2861–2868.
22. Deng Y, Wu Je, Eberl L, Zhang L-H (2010) Structural and Functional Characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl Environ Microbiol 76 : 4675–4683.
23. He Y-W, Wu Je, Cha J-S, Zhang L-H (2010) Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF-family signals in regulation of virulence factor production. BMC Microbiol 10.
24. He Y-W, Wang C, Zhou L, Song H, Dow JM, et al. (2006) Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain-protein interaction. J Biol Chem 281 : 33414–33421.
25. Cheng Z, He Y-W, Lim SC, Qamra R, Walsh MA, et al. (2010) Structural basis of the sensor-synthase interaction in autoinduction of the quorum sensing signal DSF biosynthesis. Structure 18 : 1199–1209.
26. Ionescu M, Baccari C, Da Silva AM, Garcia A, Yokota K, et al. (2013) Diffusible Signal Factor (DSF) Synthase RpfF of Xylella fastidiosa is a multifunction protein also required for response to DSF. J Bacteriol 195 : 5273–5284.
27. Huedo P, Yero D, Martinez-Servat S, Estibariz I, Planell R, et al. (2014) Two different rpf clusters distributed among a population of Stenotrophomonas maltophilia clinical strains display differential diffusible signal factor production and virulence regulation. J Bacteriol 196 : 2431–2442.
28. McCarthy Y, Yang L, Twomey KB, Sass A, Tolker-Nielsen T, et al. (2010) A sensor kinase recognizing the cell-cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia. Mol Microbiol 77 : 1220–1236.
29. Deng Y, Schmid N, Wang C, Wang J, Pessi G, et al. (2012) Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc Natl Acad Sci USA 109 : 15479–15484.
30. Römling U, Galperin MY, Gomelsky M. (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 77 : 1–52.
31. Chatterjee S, Wistrom C, Lindow SE (2008) A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc Natl Acad Sci USA 105 : 2670–2675.
32. Dow M (2008) Diversification of the function of cell-to-cell signaling in regulation of virulence within plant pathogenic Xanthomonads. Sci Signaling 1: pe23.
33. An S-Q, Febrer M, McCarthy Y, Tang D-J, Clissold L, et al. (2013) High-resolution transcriptional analysis of the regulatory influence of cell-to-cell signalling reveals novel genes that contribute to Xanthomonas phytopathogenesis. Mol Microbiol 88 : 1058–1069.
34. Vorhoelter F-J (2013) RNA-Seq facilitates a new perspective on signal transduction and gene regulation in important plant pathogens. Mol Microbiol 88 : 1041–1046.
35. An S-Q, Allan JH, McCarthy Y, Febrer M, Dow JM, et al. (2014) The PAS domain-containing histidine kinase RpfS is a second sensor for the diffusible signal factor of Xanthomonas campestris. Mol Microbiol 92 : 586–597.
36. Henry JT, Crosson S. (2011) Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 65 : 261–286.
37. Guo Y, Zhang Y, Li J-L, Wang N (2012) Diffusible signal factor-mediated quorum sensing plays a central role in coordinating gene expression of Xanthomonas citri subsp citri. Mol Plant Microbe Interact 25 : 165–179.
38. O'Connell A, An S-Q, McCarthy Y, Schulte F, Niehaus K, et al. (2013) Proteomics analysis of the regulatory role of Rpf/DSF cell-to-cell signaling system in the virulence of Xanthomonas campestris. Mol Plant Microbe Interact 26 : 1131–1137.
39. Qian G, Zhou Y, Zhao Y, Song Z, Wang S, et al. (2013) Proteomic analysis reveals novel extracellular virulence-associated proteins and functions regulated by the diffusible signal factor (DSF) in Xanthomonas oryzae pv. oryzicola. J Proteome Res 12 : 3327–3341.
40. Amari DT, Marques CNH, Davies DG (2013) The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic Acid. J Bacteriol 195 : 4600–4610.
41. Jennings JA, Courtney HS, Haggard WO (2012) Cis-2-decenoic Acid Inhibits S. aureus Growth and Biofilm In Vitro: A Pilot Study. Clin Orthop Relat Res 470 : 2663–2670.
42. Twomey KB, O'Connell OJ, McCarthy Y, Dow JM, O'Toole GA, et al. (2012) Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J 6 : 939–950.
43. Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. (2008) A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2 : 27–36.
44. Vilchez R, Lemme A, Ballhausen B, Thiel V, Schulz S, et al. (2010) Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signaling molecule trans-2-decenoic acid (SDSF). Chembiochem 11 : 1552–1562.
45. Freestone PPE, Sandrini SM, Haigh RD, Lyte M (2008) Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol 16 : 55–64.
46. Short FL, Murdoch SL, Ryan RP (2014) Polybacterial human disease: the ills of social networking. Trends Microbiol 22 : 508–516.
47. Xu J, Zhou L, Venturi V, He YW, Kojima M, Sakakibari H, Höfte M, De Vleesschauwer D. (2015) Phytohormone-mediated interkingdom signaling shapes the outcome of rice - Xanthomonas oryzae pv. oryzae interactions. BMC Plant Biol 21 : 10.
48. Caserta R, Picchi SC, Takita MA, Tomaz JP, Pereira WEL, et al. (2014) Expression of Xylella fastidiosa RpfF in citrus Disrupts signaling in Xanthomonas citri subsp citri and thereby its virulence. Mol Plant Microbe Interact 27 : 1241–1252.
49. Newman KL, Chatterjee S, Ho KA, Lindow SE (2008) Virulence of plant pathogenic bacteria attenuated by degradation of fatty acid cell-to-cell signaling factors. Mol Plant Microbe Interact 21 : 326–334.
50. Deng Y, Lim A, Lee J, Chen S, An S, et al. (2014) Diffusible signal factor (DSF) quorum sensing signal and structurally related molecules enhance the antimicrobial efficacy of antibiotics against some bacterial pathogens. BMC Microbiol 14 : 51.
51. Tian J, Weng LX, Zhang YQ, Wang LH. (2013) BDSF inhibits Candida albicans adherence to urinary catheters. Microb Pathog. 64 : 33–38.
52. Rahmani-Badi A, Sepehr S, Mohammadi P, Soudi MR, Babaie-Naiej H, et al. (2014) A combination of cis-2-decenoic acid and antibiotics eradicates pre-established catheter-associated biofilms. J Med Microbiol 63 : 1509–1516.
53. Sepehr S, Rahmani-Badi A, Babaie-Naiej H (2014) Unsaturated fatty acid, cis-2-decenoic acid, in combination with disinfectants or antibiotics removes pre-established biofilms formed by food-related bacteria. PLoS One 9: e101677.
54. Marques CN, Morozov A, Planzos P, Zelaya HM. (2010) The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl Environ Microbiol 80 : 6976–6991.
55. Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL. 2003 Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci U S A.100 : 10995–11000.
56. Rai R, Javvadi S, Chatterjee S. (2015) Cell-cell signaling promotes ferric iron uptake in Xanthomonas oryzae pv. oryzicola that contribute to its virulence and growth inside rice. Mol Microbiol 96 : 708–727.
57. Gudesblat GE, Torres PS, Vojnov AA. (2009) Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiol 149 : 1017–1027.
58. Rigano LA, Payette C, Brouillard G, Marano MR, Abramowicz L, Torres PS, Yun M, Castagnaro AP, Oirdi ME, Dufour V, Malamud F, Dow JM, Bouarab K, Vojnov AA. (2007) Bacterial cyclic beta-(1,2)-glucan acts in systemic suppression of plant immune responses. Plant Cell 19 : 2077–2089.
59. Chatterjee S. & Sonti R. V. (2002) rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol Plant Microbe Interact 15 : 463–471.
60. Qian G, Liu C, Wu G, Yin F, Zhao Y, Zhou Y, Zhang Y, Song Z, Fan J, Hu B, Liu F. (2013) AsnB, regulated by diffusible signal factor and global regulator Clp, is involved in aspartate metabolism, resistance to oxidative stress and virulence in Xanthomonas oryzae pv. oryzicola. Mol Plant Pathol 14 : 145–157.
61. Thowthampitak J, Shaffer BT, Prathuangwong S. Loper JE. (2008) Role of rpfF in virulence and exoenzyme production of Xanthomonas axonopodis pv. glycines, the causal agent of bacterial pustule of soybean. Phytopathology 98 : 1252–1260.
62. Malamud F. Torres PS, Roeschlin R, Rigano LA, Enrique R, Bonomi HR, Castagnaro AP, Marano MR, Vojnov AA. (2011) The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology 157 : 819–829.
63. Barel V, Chalupowicz L, Barash I, Sharabani G, Reuven M, Dror O, Burdman S, Manulis-Sasson S. (2014) Virulence and in planta movement of Xanthomonas hortorum pv. pelargonii are affected by the diffusible signal factor (DSF)-dependent quorum sensing system. Mol Plant Pathol. E-pub ahead of print. doi: 10.1111/mpp.12230
64. Ionescu M, Zaini PA, Baccari C, Tran S, da Silva AM, Lindow SE. 2014 Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc Natl Acad Sci U S A. 111 : 3910–3918.
65. Baccari C, Killiny N, Ionescu M, Almeida RP, Lindow SE. (2014) Diffusible signal factor-repressed extracellular traits enable attachment of Xylella fastidiosa to insect vectors and transmission. Phytopathology. 104 : 27–33.
66. Deng Y, Boon C, Chen S, Lim A, Zhang LH. (2013) Cis-2-dodecenoic acid signal modulates virulence of Pseudomonas aeruginosa through interference with quorum sensing systems and T3SS. BMC Microbiol. 13 : 231.
67. Ryan RP, McCarthy Y, Watt SA, Niehaus K, Dow JM. (2009) Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J Bacteriol. 191 : 5013–5019.
68. de Rossi B. P., Garcia C., Alcaraz E. & Franco M. (2014) Stenotrophomonas maltophilia interferes via the DSF-mediated quorum sensing system with Candida albicans filamentation and its planktonic and biofilm modes of growth. Revista Argentina Microbiol 46 : 288–297.
69. Kim S., Kim E., Shin D. S., Kang H. & Oh K. B. (2002). Evaluation of morphogenic regulatory activity of farnesoic acid and its derivatives against Candida albicans dimorphism. Bioorg Med Chem Lett 12, 895–898.
70. Tian J, Weng LX, Zhang YQ, Wang LH. (2013) BDSF inhibits Candida albicans adherence to urinary catheters. Microb Pathog. 64 : 33–38.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 CellsČlánek Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



