-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
Human blood groups are among the oldest known genetic polymorphisms. It has been proposed that blood group variation is a byproduct of pathogen-driven selection, including in the gastrointestinal tract where blood-group-related genes are often variably expressed. The B4galnt2 gene is responsible for the synthesis of the Sd(a)/Cad carbohydrate blood group antigen and displays variable tissue-specific expression patterns in wild mouse populations. Using an established model for Salmonella Typhimurium induced colitis, we found that loss of B4galnt2 expression in the intestinal epithelium decreases susceptibility to infection. These effects were strongly associated with the influence of B4galnt2 expression on the intestinal microbiota, whereby microbial diversity prior to infection was highly predictive of reduced inflammation and resistance to Salmonella Typhimurium infection. Additionally, B4galnt2 expression in blood vessels also distinctly influenced intestinal phenotypes and Salmonella susceptibility. These data lend new insights into bacterial community diversity as an “extended phenotype” that can be mediated by host genetic variation at blood-group-related genes. This work further provides strong experimental evidence in support of a scenario of complex selection on the B4galnt2 tissue-specific expression variants via host-microbe relationships and susceptibility to infectious disease.
Published in the journal: . PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1005008
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005008Summary
Human blood groups are among the oldest known genetic polymorphisms. It has been proposed that blood group variation is a byproduct of pathogen-driven selection, including in the gastrointestinal tract where blood-group-related genes are often variably expressed. The B4galnt2 gene is responsible for the synthesis of the Sd(a)/Cad carbohydrate blood group antigen and displays variable tissue-specific expression patterns in wild mouse populations. Using an established model for Salmonella Typhimurium induced colitis, we found that loss of B4galnt2 expression in the intestinal epithelium decreases susceptibility to infection. These effects were strongly associated with the influence of B4galnt2 expression on the intestinal microbiota, whereby microbial diversity prior to infection was highly predictive of reduced inflammation and resistance to Salmonella Typhimurium infection. Additionally, B4galnt2 expression in blood vessels also distinctly influenced intestinal phenotypes and Salmonella susceptibility. These data lend new insights into bacterial community diversity as an “extended phenotype” that can be mediated by host genetic variation at blood-group-related genes. This work further provides strong experimental evidence in support of a scenario of complex selection on the B4galnt2 tissue-specific expression variants via host-microbe relationships and susceptibility to infectious disease.
Introduction
The luminal surface of the intestinal mucosa is covered by distinct layers of highly glycosylated mucus that form a physical barrier between the intestinal microbial community and the host’s tissues. In addition to their important roles in host metabolism and signaling, glycans are known to contribute to the composition and physiology of the intestinal microbiota, thereby playing an important role in regulating microbe-host interactions [1]. Host glycans can contribute to a beneficial microenvironment for symbiotic microbes by providing carbohydrate sources or by serving as attachment sites [1–3], but glycans can in the same way also mediate pathogenic interactions [4, 5]. The patterns of intestinal carbohydrate structures, which vary along sites of the gastrointestinal tract, are the product of a combination of host glycosyltransferase expression programs as well as microbial influences [6, 7].
The genes responsible for synthesizing carbohydrate blood group antigens frequently display signatures of balancing selection and are implicated in the co-evolution of hosts and their pathogens [8]. A well-described example is the FUT2 gene, which encodes an α-1,2-fucosyltransferase that directs the expression of the H antigen in mucosal tissues and bodily secretions. Homozygosity for loss-of-function FUT2 mutations leads to loss of expression of ABO and H blood group glycans in secretions and is known as the “nonsecretor” phenotype, which is common in human populations [9]. Nonsecretor status has been implicated as a detrimental genetic risk factor for inflammatory disorders such as Crohn’s disease [10] and primary sclerosing cholangitis [11], while being positively associated with resistance to intestinal pathogens [12–14]. Glycosylation of the epithelium has recently been recognized as a direct immune cell mediated response to infection as a means to restore the protective functions of the microbial community and to ensure tissue homeostasis [15–17]. Glycans also mediate species specificity of pathogens. For example, the different associations of Helicobacter species to Lewis antigens in the canine gastric mucosa [18].
Gastrointestinal (GI) expression of the blood group glycosyltransferase β-1,4-N-acetylgalactosaminyltransferase 2 (B4galnt2), which directs biosynthesis of a carbohydrate antigen similar to blood group A termed the Sd(a) [19] is conserved across vertebrates [20]. However, in mice there is a common allele which confers a tissue specific switch in B4galnt2 expression from gut to blood vessels [21]. This allele is termed “Modifier of von Willebrand Factor-1” (Mvwf1) [22] because B4galnt2 vascular expression leads to aberrant glycosylation of the vascular-derived blood coagulation factor von Willebrand factor (VWF), resulting in accelerated VWF clearance from circulation [23]. Mvwf1 was first described in the RIIIS/J inbred mouse strain [22], and subsequent studies revealed RIIIS/J-like B4galnt2 alleles, which confer the B4galnt2 tissue-specific switch from gut (epithelial) to blood vessel (endothelial) expression, to be common in wild mouse populations [24]. Further, this variation appears to have been maintained in the mouse lineage for several million years despite the presumed detrimental effect of prolonged bleeding time, possibly due to a protective role in host-pathogen interactions [25]. A role for B4galnt2-glycans in intestinal host-microbe interactions is supported by the observation of significant alterations in the intestinal microbiota in B4galnt2-deficient mice [26]. Taken together, the prevalence of alleles conferring the tissue-specific switch in B4galnt2 expression in mice, the strong signatures of selection observed at the B4galnt2 locus in wild mouse populations and the altered resident microbiota found in B4galnt2-deficient mice support the hypothesis that variant tissue-specific B4galnt2 expression alters susceptibility to enteric infections in mice.
To investigate the role of variant host B4galnt2 expression in the context of intestinal infection, we challenged mice engineered to express B4galnt2 in various tissue-specific patterns with a mouse model of the intestinal pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium). Prior to - and during the course of infection, we examined histological and molecular markers of inflammation along with bacterial community profiles. We found that the composition of the intestinal microbiota was consistently influenced by the expression of B4galnt2-glycans, and that B4galnt2-associated intestinal microbial community profiles were predictive of - and responsible for susceptibility to S. Typhimurium infection. We demonstrate that mice deficient in intestinal B4galnt2 expression developed significantly less pathology after S. Typhimurium infection, in concert with attenuated induction of pro-inflammatory cytokines and infiltration of immune cells. Furthermore, we find that vascular B4galnt2 expression leads to decreased Salmonella colonization and increased inflammatory cytokine expression. Overall, our study elucidates a new role for this key host carbohydrate blood group antigen in the interplay between the host, commensals, and susceptibility to pathogen infections.
Results
B4galnt2 expression influences susceptibility to S. Typhimurium-induced colitis
To test the hypothesis that expression of intestinal B4galnt2 glycans influences host susceptibility to enteric pathogens, we used an established model for S. Typhimurium induced colitis [27]. Mice were bred to carry the desired combinations of alleles which express B4galnt2 in the intestinal epithelium (“B6”: referring to the endogenous C57BL6/J allele), vascular endothelium (“RIII”: referring to the RIIIS/J-derived Mvwf1 bacterial artificial chromosome transgene [21]), or lack a functional B4galnt2 gene due to a targeted knock-out allele (“B6 -/-”: referring to the B4galnt2 knock-out [23]). Twenty-four hours after streptomycin pre-treatment, mice were orally infected with S. Typhimurium SL1344 (“acute” infection, examined after 24 hours [28]) or the attenuated ΔaroA mutant (“chronic” infection, examined after 14 days [29]). None of the animals showed signs of inflammation or other pathology prior to infection. After infection in both the acute and chronic Salmonella models, mice expressing B4galnt2 in the intestinal epithelium (B6 +/- / RIII - and B6 +/- / RIII +) exhibited higher numbers of detached epithelial cells and neutrophils within the cecal lumen, increased inflammatory cell infiltration [29, 30] within the intestinal mucosa, and worsened submucosal edema in the ceca (Fig 1A). The dramatic reduction of cecum weight in infected B6 +/- mice compared to B6 -/- mice in acute Salmonella infection one day post infection (p.i.) indicated more severe disease [27] (Fig 1B). Accordingly, mice that did not express B4galnt2 in the intestinal epithelium (B6 -/-) developed significantly less cecal inflammation in both the acute and chronic infection model (Fig 1C).
Fig. 1. Tissue-specific expression of B4galnt2 glycans influence susceptibility to S. Typhimurium-induced colitis. 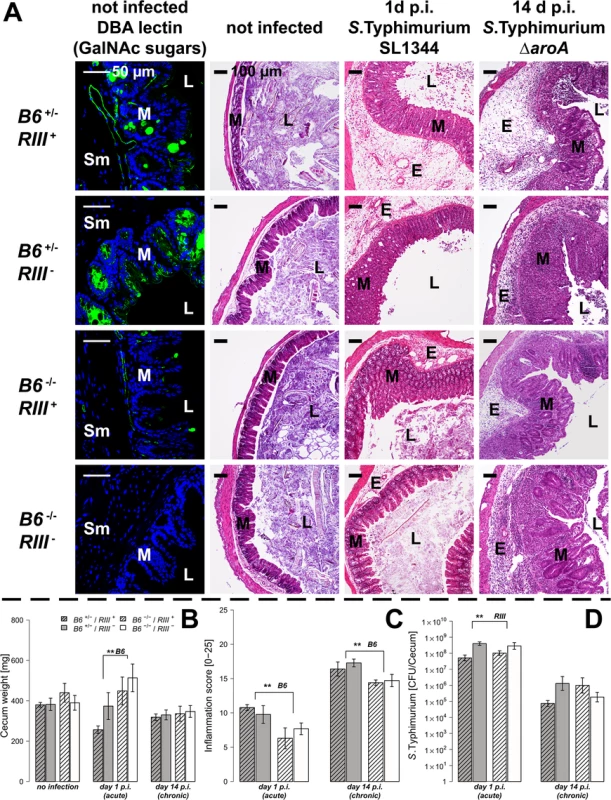
Mice were treated with streptomycin 24 h prior to infection with S. Typhimurium strain SL1344 for 24 h (acute) or with the attenuated strain S. Typhimurium ΔaroA for 14 days (chronic). (A) B4galnt2 expression phenotype is characterized by GalNAc residues, stained for by Dolichus biflorus agglutinin (DBA). H&E staining of cecal sections illustrated higher numbers of cells in the lumen (L), an increased influx of inflammatory cells to mucosa (M) and submucosa (Sm), epithelial cell desquamation and the formation of submucosal edema (E) upon infection with S. Typhimurium (bar = 100 μm). (B) Cecal weight indicated a significant influence of intestinal B4galnt2 glycans on S. Typhimurium induced colitis in the acute model (B6: F1,49 = 8.709, P = 0.0048; Linear model). (C) Histological scoring revealed higher inflammation in B6 +/- compared to B6 -/- mice (B6: F1,49 = 13.242, P = 0.0007; Linear model of X4 transformed inflammation scores). (D) Intestinal S. Typhimurium colonization was determined in tissue homogenates (RIII: F1,49 = 10.537, P = 0.0021; Linear model of log(CFU)). Data are presented as mean ± SEM, N = 9–19 per group in the acute model, N = 5–7 in the chronic model (# P<0.100, * P<0.050, ** P<0.010, *** P<0.001). In order to evaluate Salmonella colonization, colony forming units (CFUs) were quantified from homogenized ceca. While Salmonella burdens were comparable between different B4galnt2 intestinal epithelial-expressing genotypes (B6), RIII + (B4galnt2-endothelial expressing) animals exhibited lower Salmonella colonization in the acute Salmonella infections (Fig 1D). These results demonstrate a significant influence of intestinal epithelial B4galnt2 expression on susceptibility to Salmonella-induced colitis, and an independent effect of vessel-specific B4galnt2 expression on Salmonella burden. In contrast, infection of mice without prior streptomycin treatment resulted in equal bacterial organ colonization, organ weights, and elicited no intestinal inflammation regardless of the genotype of mice (S1 Fig). Due to the marked differences between mouse B4galnt2 genotypes in the acute infection model, we performed further studies only in this model.
B4galnt2-GalNAc residues have been shown to be detectable on the apical surface of intestinal epithelial cells [23, 26]. Immunohistochemical co-staining with Dolichos biflorus agglutinin (DBA) specifically detecting B4galnt2-derived β-1,4 linked GalNAc residues [21, 23] and MUCIN 2 (MUC2), the major secreted mucus protein in the large intestine, demonstrated a partial co-localization in goblet cells (Figs 2A and S2A). While MUC2 is considered to be glycosylated by B4GALNT2 [31], GalNAc residues were also detected in the intestinal mucosa of Muc2-deficient mice (S2B Fig), indicating the presence of other B4GALNT2-glycosylated substrates such as glycolipids [32, 33] and other glycoproteins [34–36]. To determine if B4galnt2-mediated glycosylation altered overall mucus thickness, which could make it easier for bacteria to cross the mucus layer and reach the epithelium, intestinal tissue of uninfected mice were fixed with Carnoy’s fixative, stained with alcian blue and the thickness of the dense inner mucus layer was determined. Although mucus thickness was not significantly affected by the lack of intestinal B4galnt2 expression (B6 -/-), it did show slight differences between RIII + and RIII - (Fig 2B and 2C). Furthermore, less DBA lectin staining was observed in the cecal mucosa of S. Typhimurium infected mice on day one p.i. compared to uninfected mice (Fig 2D). In contrast to the DBA staining (GalNAc), the detection of N-Acetylglucosamine (GlcNAc) residues recognized by Wheat Germ Agglutinin (WGA) showed no clear difference after infection, suggesting the alteration of mucosal DBA lectin-reactive carbohydrate profiles that occur in response to S. Typhimurium infection did not affect substrates glycosylated by WGA-reactive GlcNAc (Fig 2D). B4galnt2 gene expression was also down regulated upon infection (Fig 2E) which further corroborates the lectin staining results.
Fig. 2. B4galnt2 glycosylation in S. Typhimurium-induced colitis. 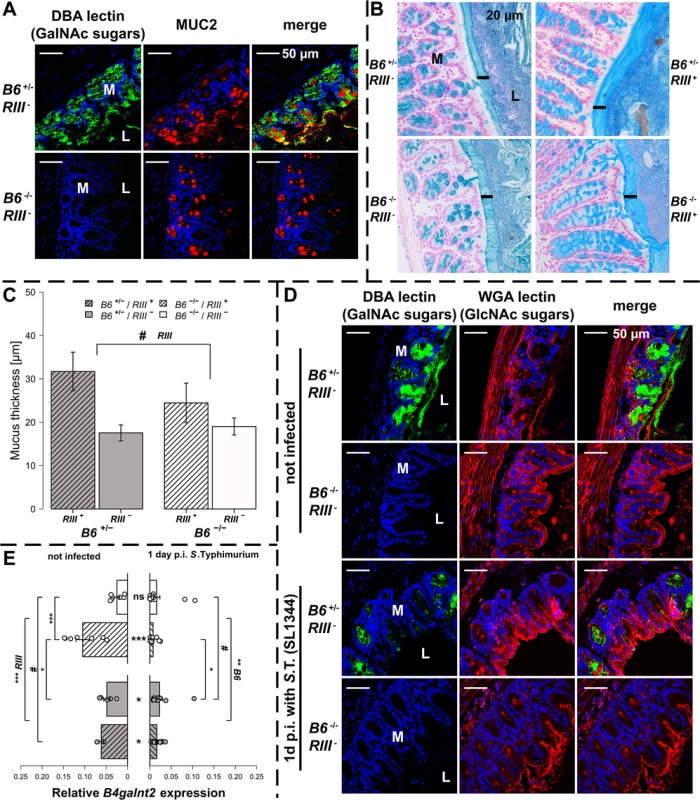
(A) MUC2 (red) and DBA lectin (green) staining in formalin fixed cecal tissue sections (B) Acidic mucus was stained with alcian blue in Carnoy’s-fixed tissue sections (bar = 20 μm). (C) Mucus thickness was determined at five different regions within one animal from which mean values were analysed (N = 3–5; Z = -1.807, P = 0.0816; Wilcoxon test via Monte-Carlo resampling). (D) B4galnt2 glycan residues (GalNAc) were stained with fluorescein labeled DBA (green) in formalin fixed cecal tissue sections before and 1 day p.i. with S. Typhimurium. GlcNAc residues were stained with Alexa633 labeled Wheat Germ Agglutinin (WGA) (red). (E) Relative expression of B4galnt2 before and after infection with S. Typhimurium showing significant differences between B6 and RIII genotypes before infection (B6: F1,17 = 0.216, P = 0.64779; RIII: F1,17 = 23.959, P = 0.00014; B6/RIII: F1,17 = 7.687, P = 0.01304 [pairwise comparisons- B6 -/-/RIII +|B6 -/-/RIII -: P = 0.00018, B6 +/-/RIII +|B6 -/-/RIII -: P = 0.08626, B6 -/-/RIII +|B6 +/-/RIII -: P = 0.02673]; Linear model and Tukey post-hoc test) and B6 genotype differences after infection (F1,53 = 11.787, P = 0.001165). Infection has additional influence on B4galnt2 expression (Z = 5.268, P<0.00001, Wilcoxon test via Monte-Carlo resampling), which is also genotype specific (B6 +/-/RIII +: Z = 2.6458, PBonferroni = 0.01192, B6 +/-/RIII -: Z = 2.6122, PBonferroni = 0.02832; B6 -/-/RIII +: Z = 3.5496, PBonferroni = 0.00016, B6 -/-/RIII -: Z = 2.0642, PBonferroni = 0.16132; Wilcoxon test via Monte-Carlo resampling; # P < 0.100, * P < 0.050, ** P < 0.010, *** P < 0.001; error bars indicate SEM). To test the direct effect of B4galnt2 expression on Salmonella’s interaction with the cecal epithelium, we performed both FISH staining of cecal sections 1 day p.i. as well as in vitro experiments with the intestinal epithelial Mode-K cell line and siRNA-mediated knockdown of B4galnt2 expression. Bacteria were stained by FISH using the Gam42a probe, which stains γ-Proteobacteria. In our experience virtually all Gam42a positive bacteria reaching the tissue in the streptomycin model at day 1 p.i. are Salmonella. Bacteria were counted if they were adherent to epithelial cells or invaded into the tissue in ten high power fields per cecal section. While adherent Salmonella were not significantly different in B6 +/- mice compared to B6 -/- mice, significantly more Salmonella were found to have invaded into the tissue of B6 +/- mice (Fig 3A). To further investigate whether B4galnt2 expression influences the interaction of Salmonella with epithelial cells, we used the intestinal epithelial Mode-K cell line and siRNA-mediated knockdown of B4galnt2 (knockdown efficiency: 96%; Fig 3B). Adhesion and invasion assays showed that knockdown of B4galnt2 expression does not significantly influence adhesion of Salmonella to epithelial cells (Fig 3C). However, invasion of S. Typhimurium into B4galnt2-expressing cells is slightly, but significantly increased relative to B4galnt2-knockdown cells (Fig 3C). This data shows that epithelial expression of B4galnt2 - both in vitro and in vivo - directly facilitates invasion by Salmonella.
Fig. 3. Epithelial B4galnt2-expression increases invasion by S. Typhimurium. 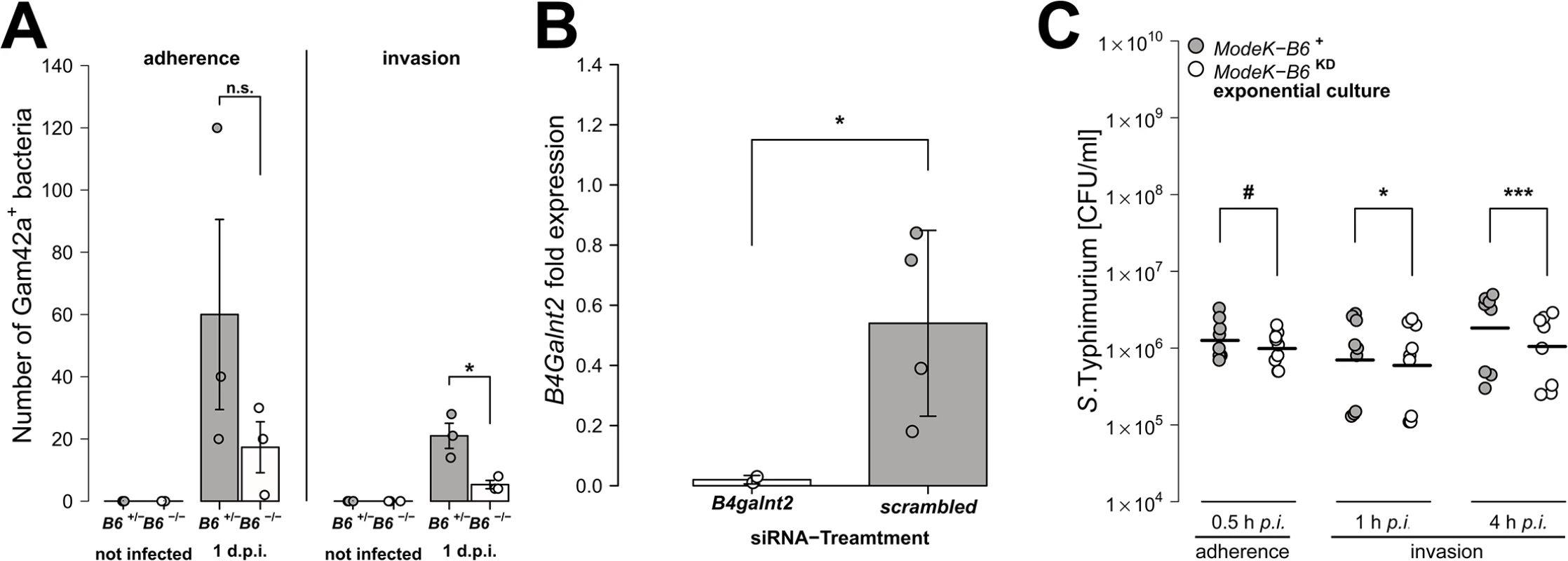
(A) Carnoy’s fixed cecal sections were stained by FISH (Gam42a probe) to visualize bacteria. Measurement of Salmonella adherence (t2.286 = -1.349, P = 0.2954, unpaired t-test) and mucosa invasion (t2.430 = -3.681, P = 0.0491; bacterial counts in 10 high power fields per individual, N = 3; unpaired t-test). (B) B4galnt2 expression in Mode-K cells after transfection with B4galnt2 specific siRNA and scrambled siRNA relative to untreated cells (t3.025 = -3.3601, P = 0.0432; unpaired t-test). (C) Salmonella invasion of Mode-K cell cultures transfected with B4galnt2 specific and scrambled siRNA, infected with S. Typhimurium. There is no significant effect of B4galnt2 expression for adhesion of the bacteria to Mode-K cells (0.5 h: F1,14 = 3.133, P = 0.0985; LMM with experiment as random factor), while invasion of S. Typhimurium into Mode-K cells expressing B4galnt2 was slightly better than into cells with B4galnt2 knockdown (1 h: F1,14 = 7.644, P = 0.0152, 4 h: F1,14 = 26.336, P = 0.0002; LMM with experiment as random factor; # P < 0.100, * P < 0.050, ** P < 0.010, *** P < 0.001; error bars indicate SEM). Intestinal epithelial B4galnt2 glycans are associated with elevated cytokine levels and higher numbers of inflammatory/immune cells after S. Typhimurium-induced colitis
We analyzed the transcript levels of pro-inflammatory cytokine genes in cecal tissues both prior to and after S. Typhimurium infection, focusing on those cytokines known to be induced early in Salmonella-triggered inflammation and associated with control of infection [37, 38]. The transcripts for the cytokines Tumor necrosis factor-α (Tnf-α), Interleukin-6 (Il-6), Interferon-γ (Ifn-γ) and Monocyte chemotactic protein-1 (Mcp-1) were elevated in all mice after infection, but to a significantly higher degree in B6 +/- mice compared to B6 -/- mice one day p.i. (Fig 4A–4D; Tnf-α: Z = -2.123, P = 0.0336; Il-6: Z = -2.458, P = 0.0138; Ifn-γ: Z = -2.417, P = 0.0147; Mcp-1: Z = -2.219, P = 0.0261; Wilcoxon test via Monte-Carlo resampling). Protein levels of Lipocalin-2 (LCN-2), a molecule implicated in antimicrobial defense and innate immunity [39, 40], were also increased in cecal tissue homogenates in B6 +/- mice compared to B6 -/- mice after infection (Fig 4E, S1 Table). Furthermore, vascular endothelial B4galnt2 expressing animals (RIII +) exhibited increased Il-6 expression (Z = -1.932, P = 0.0528), but decreased LCN-2 production (S1 Table), suggesting a role for vascular B4galnt2 expression in the host immune response to intestinal infection (Fig 3).
Fig. 4. B4galnt2-dependent immune response after S. Typhimurium infection. 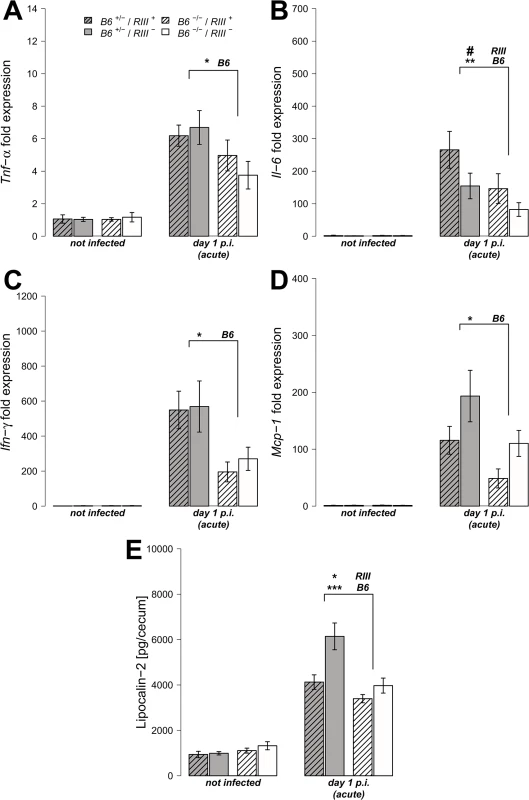
(A-D) Relative gene expression of Tnf-α, Il-6, Inf-γ and Mcp-1 was determined by RT-qPCR analysis. Values were normalized to Gapdh and Hprt and calculated as fold expression compared to the non-infected samples of each respective genotype. (E) Lipocalin-2 levels were measured by ELISA in supernatants of cecal homogenates (N = 3–11 per group) before- and one day p.i. with S. Typhimurium, showing a clear increase with infection (Z = -2.219, P = 0.0261; Wilcoxon test via Monte-Carlo resampling) and differences between B6 and RIII genotypes (S1 Table, # P < 0.100, * P < 0.050, ** P < 0.010, *** P < 0.001; error bars indicate SEM). We also analyzed cecal tissue sections for the presence of cells positive for CD68, which is strongly expressed by monocytes and macrophages, and CD3, which is expressed on mature T cells. Immunohistochemical staining and subsequent quantification of cell numbers revealed no difference in cell numbers according to endothelial B4galnt2 expression (RIII), but significantly fewer CD68 + and CD3 + cells were observed in the cecal tissues of B6 -/- mice (Figs 5A, 5B and S3A and S1 Table) after infection. The presence of neutrophils was further investigated by myeloperoxidase (MPO) staining. In line with our previous results, B6 -/- had fewer MPO positive cells in the intestinal mucosa (lumen and edema) compared to B6 +/- mice (Figs 4C and S3B) one day p.i., which was further quantified by the relative fluorescence signal intensity (P = 0.0001; Fig 4D, S1 Table). Overall, we detected significant differences in the abundance of CD68 + and CD3 + cells after infection with respect to the expression of B4galnt2 in the intestinal epithelium, but almost no differences with respect to vascular endothelial expression.
Fig. 5. B4galnt2-dependent infiltration of immune cells after S. Typhimurium infection. 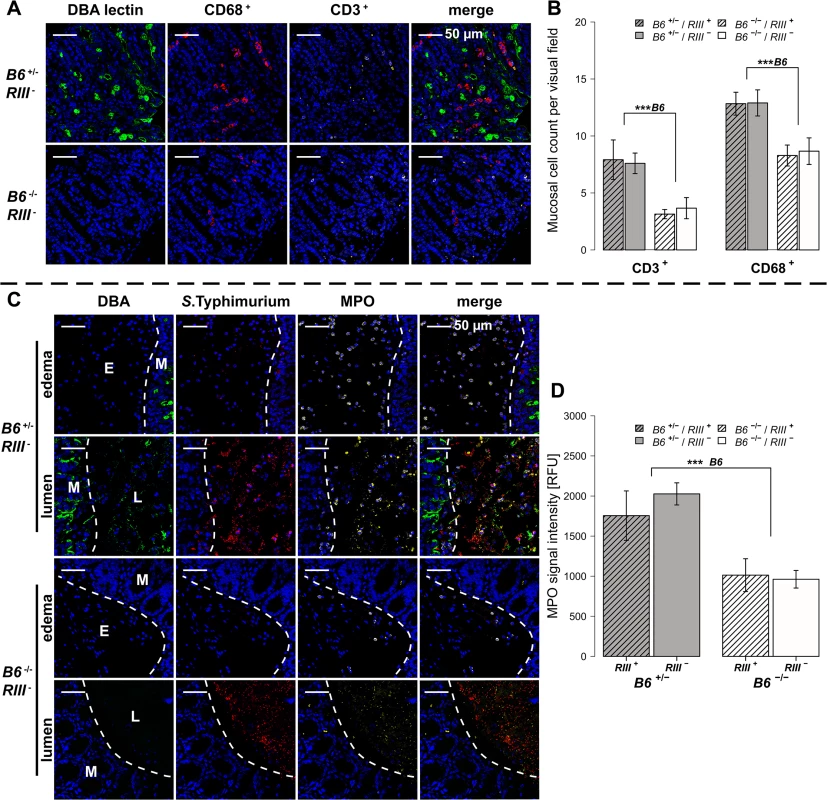
(A and B) Immunofluorescence staining and enumeration of positive cells per vision field showed that B6 +/- mice have higher numbers of CD68 (red) and CD3 (white) positive cells in the cecal mucosa 1d p.i. (N = 5–7). Nuclei were stained with DAPI (blue) and B4galnt2 glycans by using fluorescein labeled DBA (green). (C) Myeloperoxidase (MPO) positive cells (white) and S. Typhimurium (red) were determined by immunofluorescence staining. (D) MPO signal in lumen and edema was quantified and expressed as relative fluorescence units (RFU) (N = 7; Linear model; # P < 0.100, * P < 0.050, ** P < 0.010, *** P < 0.001, error bars indicate SEM). Bacterial diversity within and between mice is influenced by intestinal epithelial expression of B4galnt2
To examine the effect of B4galnt2 genotype on the intestinal microbiota in the context of infection, pyrosequencing of the 16S rRNA gene in fecal samples was performed for each individual before and after streptomycin treatment, and after S. Typhimurium infection. This resulted in a total of 122,818 sequences, with an average of 998.52 ± 13.49 SD reads per sample after normalization (Good’s coverage of OTUs: 92.46 ± 9.05% SD).
To obtain a detailed picture of the interaction of microbial communities with host factors, we first assessed within-sample (alpha) diversity at multiple complementary levels including species richness (Chao1), distribution (Shannon H), and two phylogenetic measures including Nearest Taxon Distance (NTI) and the Net Relatedness Index (NRI) [41]. Species diversity within and between bacterial communities was strongly influenced by the administration of streptomycin and S. Typhimurium (S4 Fig). Prior to streptomycin treatment and infection, the richness and evenness of operational taxonomic units (OTUs) show no significant differences according to B4galnt2 genotype (Fig 6A and 6B, and Table 1) in concordance with the results of Staubach et al. [26]. Phylogenetic clustering among close relatives (i.e. NTI) is significantly increased in animals with B4galnt2 expression in the endothelium (RIII +), while clustering of large phylogenetic groups (i.e. NRI) shows no discernable patterns (S5 Fig, Table 1).
Fig. 6. Analysis of microbial alpha diversity among genotypes and their association with intestinal inflammation. 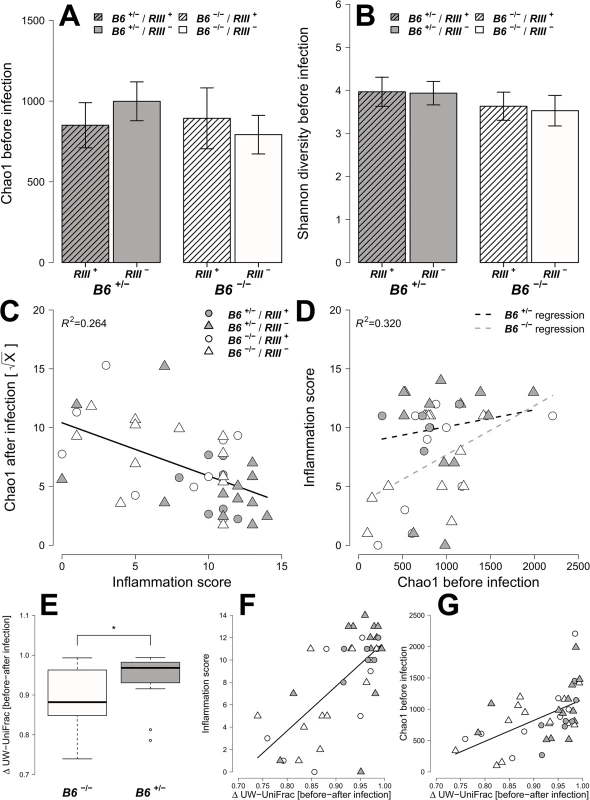
Microbial diversity was estimated from 97% species level OTUs and focused on the mean species richness (A; Chao1), and mean abundance based diversity (B; Shannon H), in the untreated animals. (C) The bacterial species richness is (i) decreasing with increasing inflammation (F1,22 = 14.2123, P = 0.0011; LMM), (ii) but highly predictive of inflammation with differences among B4galnt2 genotypes (D; Chao1 F1,21 = 9.8274, P = 0.005, B6: F1,21 = 9.2976, P = 0.0061, see also Table 2). The predictive power of alpha diversity for the outcome of infection is significantly improved by incorporating the B6 genotype (Chao1: R2LR = 0,320, ΔAIC = -5.936, LR = 7.9360, PLR-Test = 0.0048; Shannon H: R2LR = 0,271, ΔAIC = -6.1811, LR = 8.1811, PLR-Test = 0.0042; NTI: R2LR = 0.2625, ΔAIC = -8.8842, LR = 10.8842, PLR-Test = 0.001). The turnover of bacterial communities (Δ unweighted UniFrac) over the course of the experiment is strongest in animals expressing B4galnt2 in the epithelium (E; Z = -2.3213, P = 0.01978; Wilcoxon test via Monte-Carlo resampling), and is highest in animals with strong inflammation (F; ρ = 0.5894, P = 0.00005; Spearman rank correlation). The community disturbance is also highest in animals with a high species richness before treatment (G; ρ = 0.6040, P = 0.000042; Spearman rank correlation; # P < 0.100, * P < 0.050, ** P < 0.010, *** P < 0.001, error bars indicate SEM; only results of best models are shown and pairwise tests are indicated). Tab. 1. Results of the alpha diversity analyses before and after infection with S. Typhimurium (best models after REML fitting). 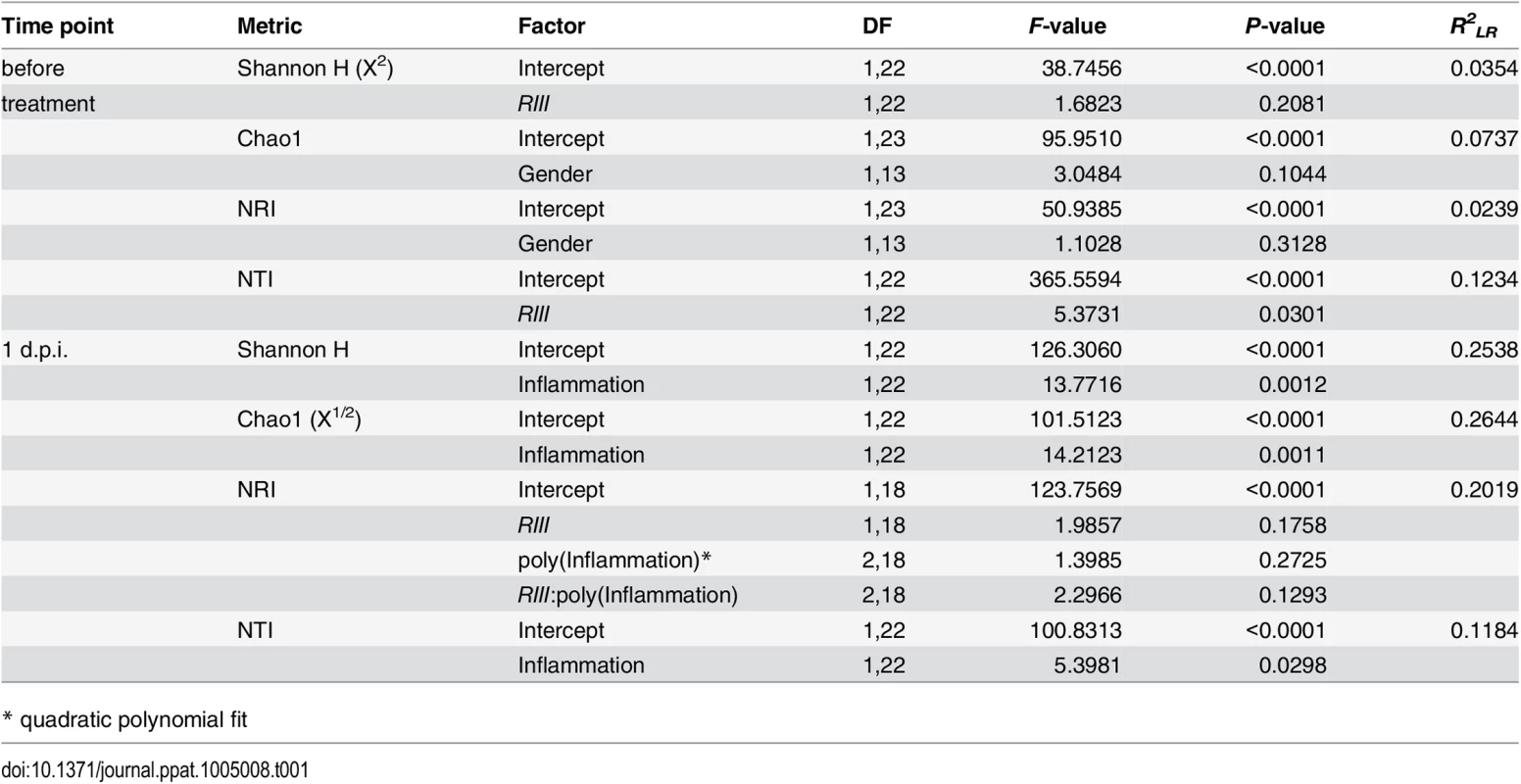
* quadratic polynomial fit After S. Typhimurium infection, the number of species and the evenness of their distribution showed a clear decrease with inflammation (Figs 6C and S5C). Phylogenetic clustering of deep branches, on the other hand, is only weakly influenced by genotype and inflammation after S. Typhimurium infection (S5E Fig, Table 1), while terminal phylogenetic clustering (NTI) shows a strong negative correlation to inflammation (S5 Fig, Table 1). In addition, the abundance of S. Typhimurium detected by 16S rRNA gene sequencing is influenced by B6- and RIII genotype, especially the low abundance observed in the RIII +/B6 -/- genotype (S6 Fig), which is consistent with the observations based on colony forming units (Fig 1D; see above).
Next, we attempted to determine which aspects of microbial communities may be associated with infection susceptibility by correlating diversity measurements prior to antibiotic treatment to the outcome of infection (inflammation score, S. Typhimurium load). Species richness, distribution, and the amount of phylogenetic clustering displayed a significant relationship to the severity of infection outcome, whereby pathology is predicted with relatively high power (Table 2). Furthermore, epithelial B4galnt2 expression (i.e. B6) significantly increases predictive power (Figs 6D and S7) and may therefore be an important factor modifying the involvement of the microbiota during pathogenesis. Specifically, species loss (ΔChao1) caused by the streptomycin and S. Typhimurium infection, which is higher in phylogenetically clustered and species rich communities (ΔChao1~NTI before infection, ρ = -0.4216, P = 0.006435, ΔChao1~Chao1 before infection, ρ = -0.9854, P < 2.2 × 10−16; Spearman rank correlation) may explain why high species diversity before treatment is correlated to a high inflammatory response (Table 2). Community resistance, measured here as the community turnover (Δunweighted UniFrac) between the pre - and post-infection time points, is higher in B6 -/- mice (i.e. lower Δunweighted UniFrac; Figs 6E, 6F, S8D, S8H and S8L) and shows a strong positive correlation with inflammation and species diversity (Figs 6F, 6G and S8 and S2 Table). Interestingly, the community turnover between the untreated and streptomycin treated communities (before infection) is not associated to the final Salmonella load or severity of inflammation. Thus, B4galnt2 expression in the gut epithelium influences the diversity and resistance of bacterial communities, which in turn is associated with the outcome of infection. Furthermore, these results also underscore the metastable character of highly diverse communities, as was already implied by May in 1972 [42].
Tab. 2. Prediction of inflammatory response by different aspects of alpha diversity (best models after REML fitting). 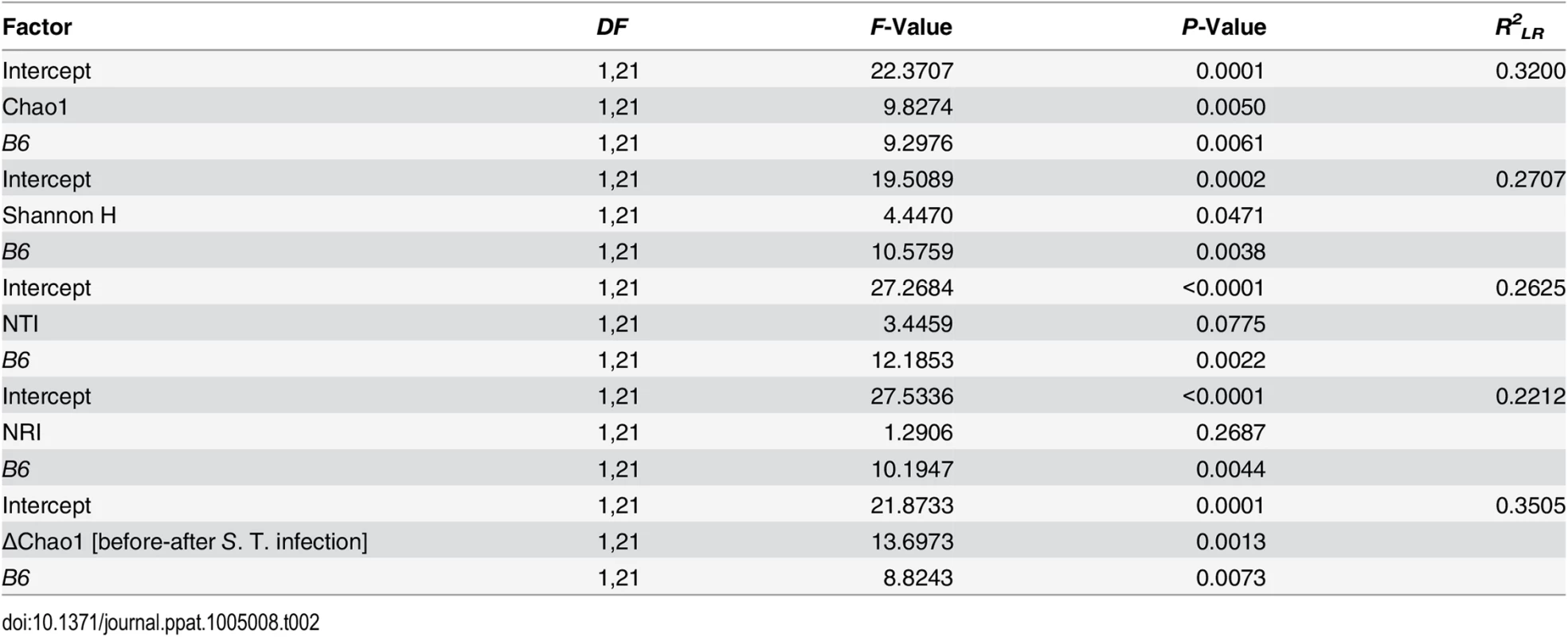
To infer whether differences between the bacterial communities of mice with different B4galnt2 expression patterns may contribute to susceptibility, we performed beta diversity analyses. Accordingly, diversity between communities was measured based on different characteristics in untreated animals, including OTU - presence/absence (Jaccard/JA),-abundance (Bray-Curtis/BC) and-distribution (Redundancy Analysis/RDA), in addition to the presence/absence - (unweighted UniFrac/UW-UF) and abundance of phylogenetic branches (weighted UniFrac/W-UF). This yielded similar community differences with respect to B6 genotype in nearly all measures (Figs 7A and S9 and S3 Table) and importantly, confirms the previous findings of Staubach et al. 2012 [26] with the current cohort of mice, which were re-derived and housed in a different animal facility. In addition, the bacterial communities among B6 +/- animals displayed far less inter-individual variation in their community composition than B6 -/- animals (S9 and S10 Figs).
Fig. 7. Treatment wise Principle Coordinate Analysis (unweighted UniFrac) of untreated- and S. Typhimurium inoculated mice and distribution of indicator bacteria among mice. 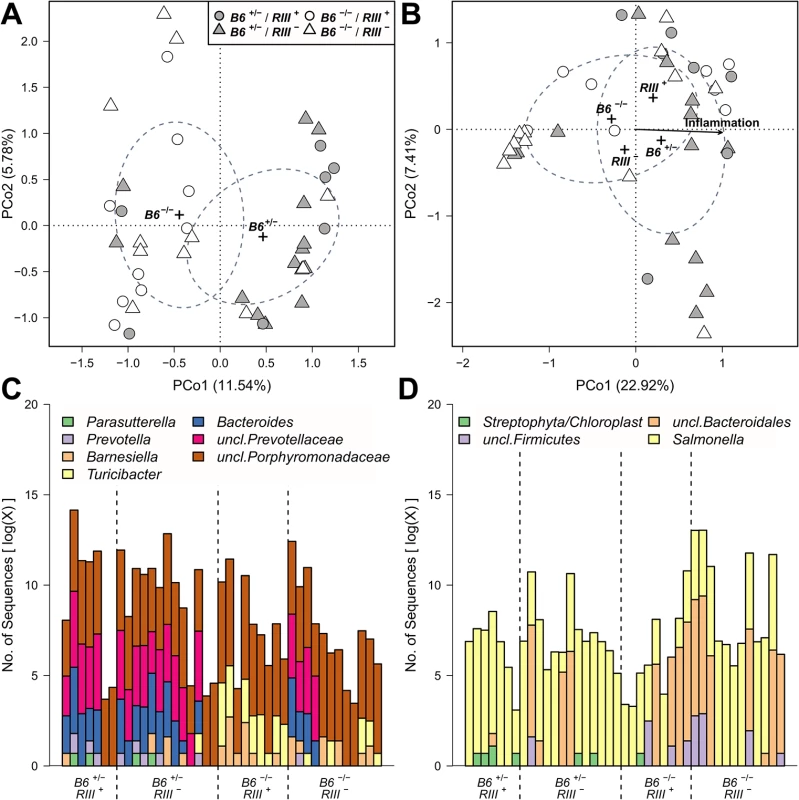
The significant sample clusters and correlations are shown, displaying a strong influence of epithelial B4galnt2 expression on the microbial community composition (no treatment (A): R2 = 0.1480, P = 0.0019; Salmonella treatment (B): B6: R2 = 0.0607, P = 0.08669, RIII: R2 = 0.0781, P = 0.040, inflammation: R2 = 0.5531, P<0.0001). Abundance distribution of indicator genera before (C) and after S. Typhimurium infection (D) for B4galnt2 gut expression among mice. Differences in community structure after S. Typhimurium infection were also evaluated and correlated with inflammation score as an additional variable. This showed that differences in communities with respect to B4galnt2 genotype are also present after infection. Furthermore, the communities changed their species composition with increasing inflammation, which appeared to be most prominent in the microbiota of B6 +/- animals (RDA: B6-F1,38 = 3.4908, P = 0.0022, inflammation - F1,38 = 5.0547, P = 0.0002, adjusted R2 = 0.1406; Figs 7B and S8 and S3 Table). Lastly, the inter-individual distance among B6 -/- also remained higher after S. Typhimurium infection (S9 and S10 Figs).
Indicator species and genera characterize the bacterial communities according to intestinal epithelial expression of B4galnt2
To investigate the drivers of community differentiation between B4galnt2 genotypes, we employed indicator species analysis. Before treatment and subsequent infection, several genera and species were associated with B4galnt2 expression (B6 +/-) in the gut, including members of the Bacteroidales (Bacteroides, Prevotella, Prevotellaceae) and Parasutterella (Proteobacteria), while Turicibacter (Firmicutes) and other members of the Bacteroidales (Barnesiella, Porphyromonas, Porphyromonadaceae) were indicative of mice lacking B4galnt2 expression in the gut (B6 -/-; Fig 7C and 7D and S4 and S5 Tables). In addition, Turicibacter, Erysipelotrichaceae, and Marvinbryantia (Firmicutes) are associated to endothelial expression of B4galnt2 glycans (RIII +; Fig 7C and 7D and S4 Table). To further understand the nature of potential interactions among indicator taxa, we performed a targeted correlation network analysis using Spearman rank correlations of the indicator genera to the remaining community members. Interestingly, the genera displaying differential preferences with respect to B4galnt2 genotype were also negatively correlated with one another, suggesting competitive exclusion mediated by the presence/absence of B4galnt2 glycans (Turicibacter-Bacteroides: ρ = -0.485, P = 0.0013; Turicibacter-uncl.Prevotellaceae: ρ = -0.447, P = 0.0034). Further, only Turicibacter, which is an indicator for the lack of B4galnt2 expression in the gut, is directly correlated to the indicators of B6 +/- genotype while uncl. Porphyromonadaceae (B6 -/- indicator) are only associated to Turicibacter abundance (Fig 8A). Through this analysis we additionally found Parabacteroides as negatively associated to Bacteroides and Prevotellaceae, suggesting either competition for B4galnt2 glycans or a secondary indicator for their absence (Fig 8A and S6 Table). Furthermore, we detected associations of taxa post infection, such as an overabundance of Salmonella and Cyanobacteria in B6 +/-, and uncl. Bacteroidales and uncl. Firmicutes in B6 -/- mice. Interestingly, we found taxa associated to B4galnt2 expression in the gut overlapping with a previous study by Staubach et al. (2012), such as Barnesiella and Porphyromonadaceae (S4 and S5 Tables) [26], which further strengthens the evidence for interactions with B4galnt2 given the independence of these cohorts of mice (see above). Lastly, we explored the dataset for individual taxon associations with inflammation, revealing Turicibacter and Salmonella to be positively associated to inflammation, potentially benefiting from the inflammatory reactions at the epithelial barrier. Other indicators for the absence of B4galnt2 glycans like Parabacteroides or Prophyromonadaceae, however, decline with increasing inflammation (S7 Table). Only the unclassified Erysipelotrichaceae, which are secondary indicators for the absence of B4galnt2 glycans in the epithelium (see Fig 8A and S6 and S7 Tables), are potential probiotic bacteria whose abundance prior to treatment decreases with inflammation (ρ = -0.320, P = 0.0417). The analysis of the complete co-occurrence network revealed strong dependencies among community members before treatment (Fig 8B). Specifically, we found a higher incidence of weak negative interactions (competition), and a low number of very strong positive interactions (Fig 8B). The co-occurrence network after S. Typhimurium infection shows a comparable distribution of positive and negative interactions, as observed before infection (S11A Fig). Further, it reveals the widespread impact of Salmonella (indicator of B6 +/-) on the microbial community, as its position is highly central and strongly influences several other highly integrated parts of the community (S11B Fig).
Fig. 8. Targeted co-occurrence network analysis of indicator genera and overall network analysis. 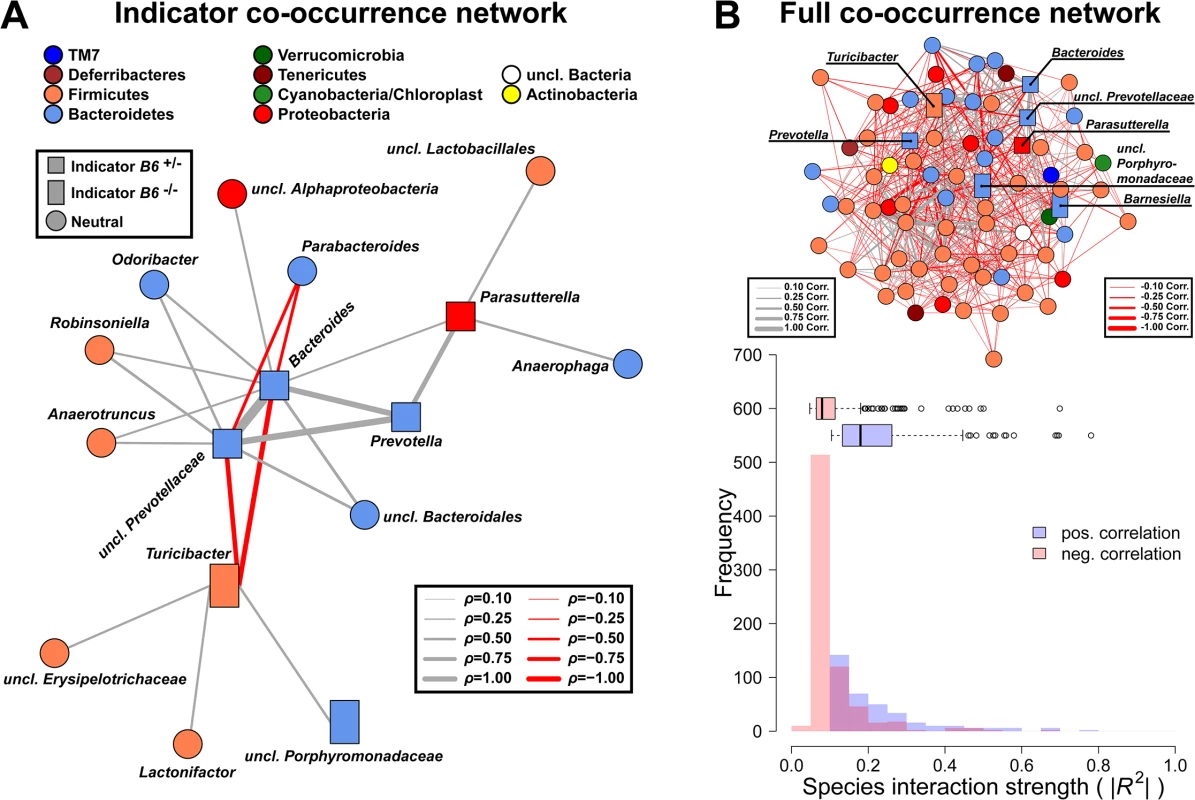
(A) Indicator genera for B6 genotypes were correlated to abundances of the remaining community members to investigate proximate interactions among indicator genera and the surrounding community (interactions are Spearman correlations see S6 Table; square - B6 +/- indicator, rectangle—B6 -/- indicator, circle—no indicator/neutral). (B) Microbial co-occurrence network based on genera abundances (only significant associations shown), with indicator species highlighted. Microbial communities show significant higher interaction strength among positive interactions (i.e. potential mutualistic; SPF: W = 489396, P < 2.20 × 10−16; Wilcoxon test). However, the higher frequency of negative weak interactions overall has a stabilizing effect preventing the communities from collapsing (positive/negative interactions = 0.482; # P < 0.100, * P < 0.050, ** P < 0.010, *** P < 0.001). Increased susceptibility of B6 +/- mice to S. Typhimurium triggered inflammation is dependent on microbiota composition
In order to determine whether the microbiota composition contributes to the elevated susceptibility of B6 +/- mice to inflammation, we transplanted feces from B6 +/- and B6 -/- donor mice into germfree C57BL/6J (B6 +/+) recipient mice. 21 days post fecal transplantation, mice were treated with streptomycin and 24 hours later infected with S. Typhimurium. Cecum weight and S. Typhimurium colonization (CFU count) do not differ significantly between the fecal donor genotypes (Fig 9A, 9B and 9C). However, the extent of tissue inflammation caused by S. Typhimurium infection was significantly lower in mice transplanted with microbiota from B6 -/- mice due to decreased mucosal damage and decreased submucosal edema (Fig 9A and 9D). These results demonstrate that the differences in microbiota composition from B6 +/- and B6 -/- mice are responsible for the lower susceptibility of B6 -/- mice to Salmonella induced inflammation.
Fig. 9. B4galnt2-dependent microbiota composition is responsible for enhanced susceptibility to inflammation. 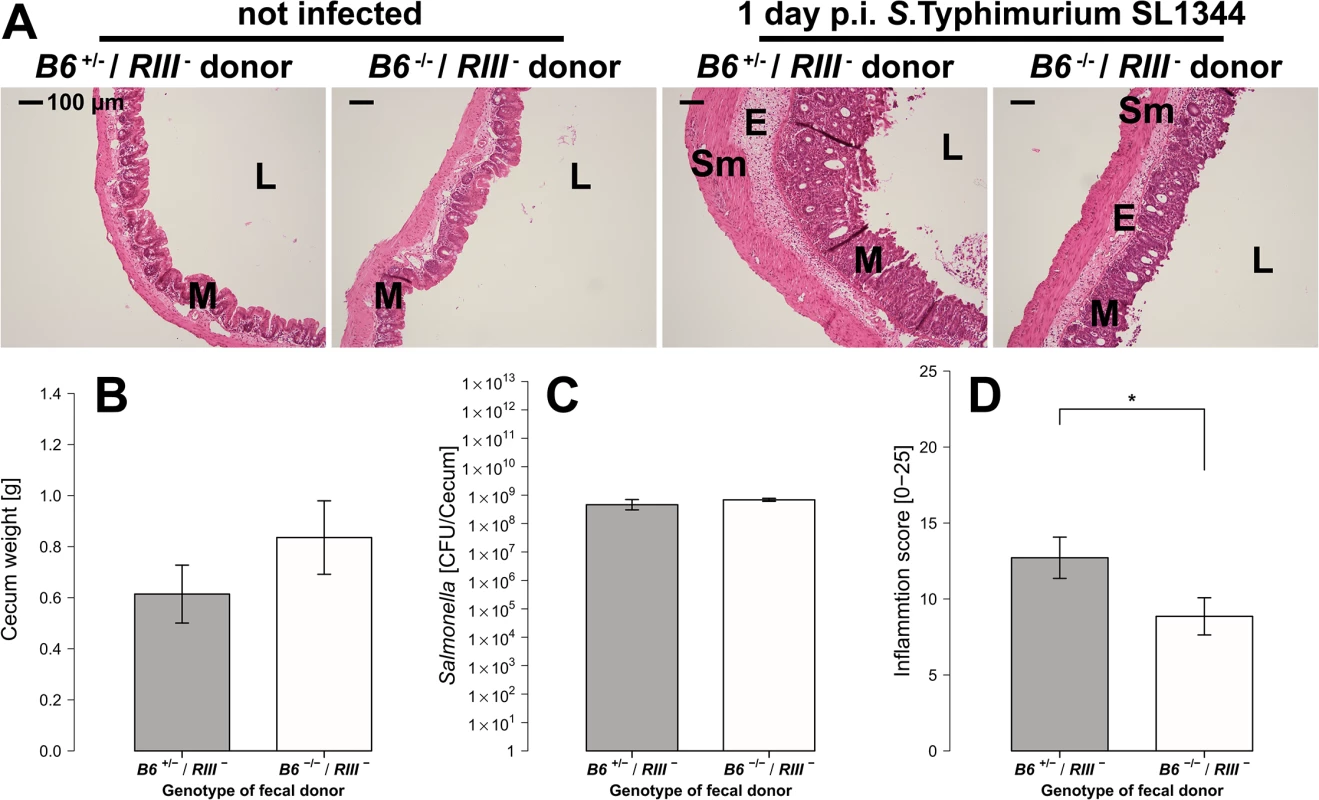
(A) Representative H&E staining of cecal sections with higher number of luminal cells (L), increased influx of inflammatory cell populations into the mucosa (M) and epithelial cell desquamation and submucosal edema (E) upon infection with S. Typhimurium (bar = 100 μm). (B) Cecum weight (Z = 1.087, P = 0.3013, (C) and Salmonella abundance in the cecum (Z = 0.447, P = 0.7098) do not differ between donor genotypes (N = 7 infected and N = 3 uninfected controls per donor genotype). (D) Histological inflammation is significantly reduced in mice that received a B6 -/- microbiome (Z = -2.074, P = 0.0459; Wilcoxon test via Monte-Carlo resampling, # P < 0.100, * P < 0.050, ** P < 0.010, *** P < 0.001, error bars indicate SEM). Discussion
Infectious diseases are one of the strongest selective forces on many levels of biological complexity. Over time, a steady cycle of adaptation and counter-adaptation has left molecular traces in the genomes of many organisms including humans [43]. The most prominently affected members are genes associated with the immune system, e.g. MHC [44], however, others including blood-group-related genes display similar signatures of selection [3, 8, 45–47]. In this study, we investigated intestinal infection as a potential driver of selection at B4galnt2 observed in the wild by studying the effect of variant tissue-specific expression of B4galnt2 on host-microbiota interactions and susceptibility to intestinal infection with Salmonella. This revealed strong evidence for the influence of B4galnt2-specific host glycosylation on microbial community composition and a role in pathogen resistance.
Our experiments revealed less intestinal pathology, lower inflammatory responses, and changes in microbial community structure and composition in animals lacking B4galnt2 expression in their intestinal epithelium. Host mucosal glycans can directly interact with the microbiota by serving as specific attachment sites or as nutrient sources for some microorganisms. Thus, host mucosal carbohydrates can influence, directly and indirectly, the establishment of overlapping competitive niches, which serve as a barrier against potential pathogens (i.e. “colonization resistance”) [48, 49]. We found B4galnt2-expression-dependent characteristics of the intestinal microbiota, such as species and phylogenetic diversity, which predict the colonization success of S. Typhimurium and the severity of the accompanying intestinal inflammation. In our experiment, species-rich and phylogenetically clustered microbial communities appear to be more vulnerable to Salmonella infection, and ultimately inflammation. Before the seminal works of May and others [42, 50, 51], high diversity habitats were synonymous with high stability and productivity [52, 53]. However, the diversity-stability debate remains unresolved [54–56]. High diversity only has a stabilizing effect if reactions of community members are asynchronous, which balances the reduction of one species by the complementary increase of other community members [57–59]. This “portfolio” - [60] or “insurance” effect [61] dampens perturbations by a release of inter-species competition, or by differential susceptibility to the environmental stressors [54]. Diverse communities also exhibit an intrinsically higher tendency of community change, as a large number of species (especially rare species) are prone to becoming lost through environmental perturbation and stochastic events due to their limited relative abundance [62]. This has been observed in grassland communities, where compositional instability increases with community diversity [63]. Thus, the comparably high number of strong positive interactions in the bacterial communities of this study (see Fig 8B) may therefore explain the tendency of exacerbated species loss and inflammation after disturbance, as the stabilizing effects of competitive release are lower [57, 60, 64, 65]. Furthermore, evolutionary relatedness among community members has a strong influence on community reactions and productivity. Closely related species (e.g. phylogenetically clustered) presumably overlap in their niches and functional capacities [66, 67] and react in similar ways to environmental stressors, which dampens the insurance effect (i.e. “negative insurance effect”) as observed in the investigated microbial communities [68, 69].
Antibiotic treatment usually has long lasting effects, but previous studies show that a certain degree of resilience occurs through short-term repopulation of dormant bacteria [49, 70]. The disturbance in microbial communities appears to be buffered in mice not expressing B4galnt2 glycans in the epithelium, possibly by conferring “colonization resistance” via a higher potential to compete with invading Salmonella and by dampening the effects of community disturbance [67, 71, 72]. Thus, in the context of a diminished and disturbed microbial community after streptomycin treatment [73], it is likely that the more resilient/resistant communities in B6 -/- mice maintain a greater potential for rapid recovery [48, 70, 74]. We further postulate that B4galnt2 genotype-dependent host-microbe interactions modulate the host’s immune response, contributing to less severe pathology and increased pathogen clearance in mice lacking intestinal epithelial B4galnt2 expression.
Commensal gut bacteria benefit from the intestinal mucus and its diverse glycan residues, as they offer a complex repertoire of binding sites and carbohydrate sources independent of the host diet [1, 3, 75, 76]. The indicator species identified for mucosal B4galnt2 expression, Prevotella and Bacteroides, are known to digest and bind a large spectrum of glycans [77]. These bacteria of high metabolic potential show signs of niche competition with the genus Turicibacter, an indicator for B4galnt2-deficient mice. Turicibacter, e.g. Turicibacter sanguinis, is a known member of the human and murine gut microbiome, but can only utilize a narrow range of carbohydrates [78]. As suggested by Dimitriu et al. (2013) [79], the trade-off between low metabolic capacity and competitive abilities [78, 80] with the potential for fast colonization might explain the association of Turicibacter with B6 -/- mice and the co-increase with S. Typhimurium [81–83]. It was also suggested that Turicibacter possesses immune modulatory characteristics (increasing iNK T cell, and marginal zone B cell abundance [84]), and may thus help to lower the susceptibility to gut inflammation in B6 -/- compared to B6 +/- mice in the face of equivalent Salmonella burdens [79]. However, Turicibacter could also benefit from existing tissue inflammation, as several genomic features such as laminin, internalin, or a collagen binding pilus allow this genus to act as an opportunistic pathogen, and thus explain its association with tissue inflammation [78, 80]. Similarly, Barnesiella shows repeated association to the absence of B4galnt2 glycans [26]. This genus has the potential to counteract inflammatory responses and thus appears to play a central role in the gut microbiome [85].
Co-staining of MUC2 and DBA lectin demonstrated a partial co-localization in goblet cells, suggesting that MUC2 is glycosylated by B4galnt2 in agreement with previously published data [31]. However, B4galnt2 glycans were also detectable in the cecal mucosa of Muc2-deficient mice (Figs 2A and S2A), indicating additional intestinal targets of B4GALNT2 glycosylation. Other glycosylation targets for B4galnt2 are Sd(a)/Cad antigens, which have been shown to be present in colonic mucins [34, 36], glycolipids and glycoproteins [32, 33, 35, 86]. Intestinal mucin glycans, including blood group α-1,2 fucosylated receptors, have been proposed as attachment sites for Salmonella [87, 88], but Salmonella does not appear to directly bind B4galnt2-GalNAc residues in vitro [4]. The glycan profile may also change in animals not expressing B4galnt2 in addition to the lack of β1,4-GalNac residues/Sd(a), whereby the increase or decrease of other residues may offer new nutrient sources or attachment sites for bacteria or immune cells [35, 89]. Nevertheless, we found slightly increased invasion into epithelial cells in vivo and in vitro when B4galnt2 is expressed. However, our fecal transfer experiments demonstrate that the altered bacterial community of B6 -/- mice confers resistance towards Salmonella induced inflammation. Thus, it is likely that indirect mechanisms, such as the microbial community and its capability of glycan liberation, subsequent changes in nutrient or microbe abundances [90] and the type of interactions [72], are responsible for the higher susceptibility of mice expressing B4galnt2 in the intestinal epithelium to S. Typhimurium infection.
Our study reveals an increased production of pro-inflammatory mediators, higher numbers of immune/inflammatory cells, and more severe colitis after S. Typhimurium infection in the ceca of mice expressing B4galnt2 in the intestinal epithelium. Although endothelial B4galnt2 expression did not impact the development of colitis as judged by histology, RIII + mice had lower pathogen burden in the cecum and lower levels of Mcp-1 and LCN-2 compared to RIII - mice, supporting a role for vascular B4galnt2 in host immune defense in the face of intestinal pathogens. Functionally, carbohydrate differentiation antigens play an important role in the homing and differentiation of intraepithelial lymphocytes in the small intestine, indicating a plausible phenotype that may result from the expression of B4galnt2 in endothelial cells [91–94]. The recruitment of neutrophils and CD3 + cells [35], as well as leukocyte infiltration, were reported to be influenced through the glycosylation of selectin receptors [95] and could be associated with the elimination of carbohydrate ligands for selectins. B4galnt2 expression in gastrointestinal cancers has been shown to reduce metastatic dissemination, adding to the role of the Sd(a) antigen in cell motility [96, 97]. Further studies focusing on the role of endothelial B4galnt2 expression are needed to understand the impact of B4galnt2-GalNAc residues in host immune responses and its potential role for homing of immune cells to the intestine.
In summary, we demonstrate that different patterns of tissue-specific B4galnt2 expression not only influence intestinal microbial communities, but also change host susceptibility and immunological responses to S. Typhimurium infection [45, 98]. Thus, a complex scenario including B4galnt2-dependent changes in microbial communities, vascular immune phenotypes, bleeding tendencies and susceptibility to intestinal infections likely contributes to the maintenance of variation at B4galnt2 in wild mouse populations.
Material and Methods
Animal models of variant B4galnt2 tissue-specific expression
All genetically engineered mouse lines used in the study were backcrossed >20 generations to a C57BL/6J background prior to breeding of the experimental animals. C57BL/6J (B6 +/+) mice were purchased from The Jackson Laboratory. Mice heterozygous for the B4galnt2 knock-out allele (B6 +/-) [23] and RIIIS/J-B4galnt2 BAC transgenic (RIII +) mice which exhibit the Mvwf1 phenotype [21] were re-derived at the University Clinic Eppendorf, Hamburg, Germany. Intercross of B6 +/- × B6 +/-RIII + generated heterozygous B6 +/-/RIII +, B6 -/-/RIII +, B6 +/-/RIII - and B6 -/-/RIII - offspring, which were raised and housed together as littermates under specific pathogen-free conditions in individually ventilated cages at the animal facility of the University of Kiel, Germany. Standard chow (ssniff, Soest, Germany) and water were provided ad libitum. Germ-free C57BL/6J mice were produced at the gnotobiotic facility of the Hannover Medical School. Experiments were conducted in the animal facility of the Leibniz Research Center Borstel, Germany and at the animal facility of University Hospital Schleswig-Holstein Kiel.
Ethics statement
All experiments were conducted consistent with the ethical requirements of the Animal Care Committee of the Ministry of Energy, Agriculture, the Environment and Rural Areas of Schleswig-Holstein, Germany and in direct accordance with the German Animal Protection Law. The protocols were approved by the Ministry of Energy, Agriculture, the Environment and Rural Areas of Schleswig-Holstein, Germany (Protocol: V312-72241.123–3 and V312-7224.123–3).
Salmonella infection of mice
Streptomycin (20mg per mouse) (Sigma-Aldrich, Hamburg, Germany) was given by oral gavage to mice aged 10–14 weeks. 24 hours after antibiotic administration, mice were infected with either S. Typhimurium SL1344 (acute infection; [28]) or the attenuated S. Typhimurium ΔaroA (chronic infection; [29]) at a dose of 3 × 106 bacteria in 100 μL HEPES buffer (100 mM, pH 8.0; PAA, Cölbe, Germany). Control mice (mock-infection) were given 100 μL HEPES buffer. Bacterial loads were determined by plating serial dilutions of homogenized organs on Luria Bertani agar (Roth, Karlsruhe, Germany) containing streptomycin (100 μg/mL).
siRNA knockdown and tissue culture infections
Mouse intestinal epithelial Mode-K cells were grown in DMEM supplemented with 5% fetal bovine serum (Biochrom, Berlin, Germany) and 1% HEPES (GE Healthcare, Frankfurt, Germany). For the siRNA knockdown of B4galnt2 1 × 105 cells per well were seeded in a 24 well plate containing 10nM siRNA and lipofectamine (Life Technologies, Darmstadt, Germany) according to manufacturer’s instructions for reverse transfection. As a negative control cells were treated with scrambled siRNA. 24h post transfection cells were infected with an MOI 50 of wildtype S. Typhimurium grown to late-logarithmic phase. 30 min p.i., cells were washed and extracellular bacteria were killed by addition of medium containing gentamicin (100 μg/ml). Cells were lysed at various timepoints (30 min, 1 h and 4 h) and the number of adherent and invaded bacteria was determined by plating serial dilutions.
Fluorescence in situ hybridization (FISH) staining
Cecal tissues were fixed in Carnoy’s fixative overnight, embedded in paraffin, and then cut in 5 μm sections on glass slides. Sections were deparaffinized and incubated with a Texas red-conjugated EUB338 general bacterial probe (5’-GCTGCCTCCCGTAGGAGT-3’) and an AlexaFluor 488 conjugated Gam42a probe (5’-GCCTTCCCACATCGTTT-3’) that recognizes bacteria that belong to the γ-Proteobacteria class (37°C, O/N, dark). Tissue samples were washed with hybridization buffer (0.9 M NaCl, 0.1 M Tris pH 7.2, 0.1% SDS). This step was repeated with FISH Washing Buffer (0.9 M NaCl, 0.1 M Tris pH 7.2) with gentle shaking for 15 minutes. Sections were washed with water and mounted using Prolong GOLD with DAPI (Molecular Probes) and imaged using an AxioImager microscope equipped with an AxioCam HRm camera operating through AxioVision software. High power field (HPF) (630X) was used for enumerating intracellular and extracellular S. Typhimurium.
Staining of acidic mucus and mucus thickness
Carnoy’s-fixed paraffin-embedded tissues were sectioned (5 μm), deparaffinized, and stained with 1% Alcian Blue (Sigma-Aldrich, Hamburg, Germany) solution (in 1% acetic acid) for 10 min, counterstained in nuclear fast red solution (1%), dehydrated, and mounted for examination. Photographs were taken at an original magnification of 100x and mucus thickness was measured at six random locations per section using NIS-Element Software (Nikon, Dusseldorf, Germany).
Fecal transplantation experiments
Fresh feces from B6 +/- or B6 -/- mice was sampled and immediately homogenized (1 : 10 w/v) in transfer buffer (sterile phosphate buffered saline containing 0.05% cysteine HCl (Sigma-Aldrich)). After centrifugation, the supernatant was collected and 200 μL were orally gavaged into germ-free adult C57BL/6J recipient mice. 21 days post transplantation mice were treated with streptomycin and 24 hours later infected with S. Typhimurium.
Histopathological analysis
Tissues were fixed in 10% neutral buffered formalin overnight and embedded in paraffin. 5 μm sections were deparaffinized and stained with haematoxylin and eosine (H&E). Histological scores in the ceca of infected mice were determined as previously described [30]. Briefly, pathological changes were assessed by evaluating various parameters such as presence of luminal cells, infiltrating immune cells, crypt abscesses and the formation of edema in the respective layer of the intestinal bowel wall including the surface epithelium, mucosa and submucosa.
Immunohistochemistry
Formalin fixed tissue sections (5 μm) were deparaffinized and rehydrated. After antigen retrieval with 10 mM sodium citrate buffer (pH 6.0) and blocking with 2% normal goat serum, specimens were incubated with antibodies specific for S. Typhimurium (Clone B395M, Dunn Laboratories, Asbach, Germany), CD3 (Abcam, Cambridge, UK), CD68 (Abcam, Cambridge, UK), myeloperoxidase (MPO) (Thermo Fisher Scientific, Schwerte, Germany), and MUC2 (Santa Cruz, Dallas, TX, USA) followed by fluorescently labeled secondary antibodies (Molecular Probes, Invitrogen, Carlsbad, CA, USA) or with fluorescently labelled DBA (dolichus biflorus agglutinin) and WGA (wheat germ agglutinin) lectins (Vector laboratories, Burlingame, CA, USA). Counterstaining of nuclei was performed using 4,6-Diamidin-2-phenylindol (DAPI) (Invitrogen, Carlsbad, CA, USA). Images were obtained using a Leica SP5 confocal microscope (Leica, Wetzlar, Germany).
Lipocalin-2 enzyme-linked immunosorbent assay (ELISA)
Lipocalin-2 concentrations in the supernatant of tissue homogenates were determined with a mouse specific ELISA Development Kit by R&D Systems (R&D Systems, Wiesbaden, Germany) according to the manufacturer’s instructions.
Real-time quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from cecal tips by using the High Pure RNA Tissue Kit (Roche Diagnostics, Mannheim, Germany) and reverse transcription was conducted with the Transcriptor High Fidelity cDNA Synthesis Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. RT-qPCR was performed with Quantitect SYBR-Green Mastermix (QIAGEN, Hilden, Germany) for the following genes: Ifn-γ, fw TCAAGTGGCATAGATGTGGAAGAA, rev TGGCTCTGCAGGATTTTCATG; Tnf-α,fw CCACCACGCTCTTCTGTCTAC, rev AGGGTCTGGGCCATAGAACT; Il-6, fw GAGGATACCACTCCCAACAGACC, rev AAGTGCATCATCGTTGTTCATACA; Mcp-1, fw CCTGCTGTTCACAGTTGCC, rev ATTGGGATCATCTTGCTGGT; B4galnt2, fw TGGCAAGTCCTACCATGAGG, rev GTCTGCAGAAGTGGCTGGA; Gapdh, fw ATTGTCAGCAATGCATCCTG, rev ATGGACTGTGGTCATGAGCC; Hprt, fw AGTGTTGGATACAGGCCAGAC, rev CGTGATTCAAATCCCTGAAGT. Relative gene expression was calculated using geNORM and the 2-∆∆Ct method, with Gapdh and Hprt as housekeeping genes [99].
DNA extraction and 16S rRNA gene sequencing
DNA was extracted from fecal samples (stored at -80°C) using the PowerSoil DNA Isolation Kit (MO Bio Laboratories, Carlsbad, CA) following the manufacturer’s protocol. The 16S rRNA gene was amplified using barcoded primers flanking the V1 and V2 hypervariable regions (27F-338R) and were sequenced following the methods describe in Rausch et al. 2011 [100].
Sequence processing and quality control
Raw sequences were trimmed by mothur 1.31.2 requiring no ambiguous bases, a mean quality score within a window of 50 base pairs of ≥ 35 and a minimum length of 200 nucleotides for the coupled V1-V2 region [101]. Chimeric sequences were determined using USEARCH 4.25 (database informed UCHIME algorithm) [102]. Sequences were confirmed as bacterial using the RDP classifier with ≥ 60% bootstrap threshold [103]. For all downstream analyses of diversity and habitat association, we took a random subset of 1000 sequences per sample to normalize the read distribution (Good’s Coverage; no treatment: 85.67 ± 6.61% SD; Streptomycin: 97.38 ± 3.13% SD; S. Typhimurium infection: 98.36 ± 1.74% SD). These sequences were aligned to the curated SILVA seed database using the NAST alignment procedure as implemented in mothur and subsequently OTU binning was carried out via average distance clustering [104]. Phylogenetic tree construction on representative OTU sequences (average distant sequence of the OTU) was done by FastTree 2.1 using the CAT substitution model with gamma correction [105]. Raw sequence data can be accessed online under the accession number PRJEB5269 at the European Nucleotide Archive.
Statistical analysis
Species diversity indices (Chao1 species richness, Shannon-Weaver index), as well as the phylogenetic distance at the tips of the phylogenetic tree (Nearest Taxon Index, NTI) and its deep branches (Net Relatedness Index, NRI) were calculated in R [106–108]. The phylogenetic measures of beta diversity (unweighted - and weighted UniFrac) and metrics based on shared OTU presence (Jaccard) or abundance (Bray-Curtis) were calculated in “vegan” [109–111]. Statistical analysis of community composition based on different beta diversity metrics was performed with Principal Coordinate Analysis (PCoA) and non-parametric multivariate analysis of variance and multivariate dispersion as implemented in the “vegan” package for R with 105 permutations. For constrained ordination (Redundancy Analysis) the OTU table was Hellinger-transformed and RDA was carried out following Legendre and Legendre [112]. Significance of factors and axes was ascertained using a permutative ANOVA approach (5000 permutations). Linear mixed models (LMM, cage as random factor) were applied to alpha diversity measures and optimized with model selection by AIC criterion, normality of model residuals and refitting of the final model under Restricted Maximum Likelihood (REML) [113]. The R2LR values of the final mixed model were calculated using the MuMIN package for R [114, 115]. Lipocalin-2 levels, fluorescence signals, inflammation scores, CFU counts, and cecum weights were analyzed in a linear model framework with parameter selection to minimize the AIC value and no significant reduction of fit. For the comparison of expression values among genotypes we employed a Wilcoxon test with Monte-Carlo resampling [116]. Salmonella counts (Gam24a + cells) in Mode-K cell cultures were analyzed using an LMM with the independent rounds of experiments as random factor to incorporate experimental variation. Indicator species analysis was based on 105 permutations using the indicator value to assess the association for each taxon [117]. All P-values of the genera and OTU associations were adjusted by the Benjamini-Hochberg procedure. Taxon co-occurence networks were calculated by SPARCC based on 105 permutations and significant associations (P < 0.05) were included in the network construction [118].
Supporting Information
Zdroje
1. Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Micro. 2012;10(5):323–35. Epub 2012/04/12. doi: 10.1038/Nrmicro2746 ISI:000302938700010; PubMed Central PMCID: PMC4005082.
2. Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–9. 15790854
3. Varki A, Freeze HH, Gagneux P. Evolution of Glycan Diversity. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. 2010/03/20 ed. Cold Spring Harbor NY: The Consortium of Glycobiology Editors, La Jolla, California; 2009.
4. Giannasca KT, Giannasca PJ, Neutra MR. Adherence of Salmonella typhimurium to Caco-2 cells: identification of a glycoconjugate receptor. Infection and immunity. 1996;64(1):135–45. Epub 1996/01/01. 8557331; PubMed Central PMCID: PMC173738.
5. Kobayashi M, Lee H, Nakayama J, Fukuda M. Roles of gastric mucin-type O-glycans in the pathogenesis of Helicobacter pylori infection. Glycobiology. 2009;19(5):453–61. Epub 2009/01/20. doi: 10.1093/glycob/cwp004 WOS:000265096000001; PubMed Central PMCID: PMC2667159.
6. Henry SM. Molecular diversity in the biosynthesis of GI tract glycoconjugates. A blood-group-related chart of microorganism receptors. Transfusion clinique et biologique: journal de la Societe francaise de transfusion sanguine. 2001;8(3):226–30. Epub 2001/08/14. 11499965.
7. Moran AP, Gupta A, Joshi L. Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut. 2011;60(10):1412–25. Epub 2011/01/14. doi: 10.1136/gut.2010.212704 21228430.
8. Fumagalli M, Cagliani R, Pozzoli U, Riva S, Comi GP, Menozzi G, et al. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome Res. 2009;19(2):199–212. Epub 2008/11/11. doi: 10.1101/gr.082768.108 WOS:000263132600004; PubMed Central PMCID: PMC2652214.
9. Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, Blancher A, et al. A Natural History of FUT2 Polymorphism in Humans. Molecular Biology and Evolution. 2009;26(9):1993–2003. Epub 2009/06/03. doi: 10.1093/molbev/msp108 WOS:000269001500006.
10. McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Human molecular genetics. 2010;19(17):3468–76. Epub 2010/06/24. doi: 10.1093/hmg/ddq248 20570966; PubMed Central PMCID: PMC2916706.
11. Folseraas T, Melum E, Rausch P, Juran BD, Ellinghaus E, Shiryaev A, et al. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. Journal of hepatology. 2012;57(2):366–75. Epub 2012/04/24. doi: 10.1016/j.jhep.2012.03.031 22521342; PubMed Central PMCID: PMC3399030.
12. Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindbland L, et al. Human susceptibility and resistance to Norwalk virus infection. Nature Medicine. 2003;9(5):548–53. Epub 2003/04/15. doi: 10.1038/nm860 WOS:000182610600035.
13. Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni Binds Intestinal H(O) Antigen (Fucα1, 2Galβ1, 4GlcNAc), and Fucosyloligosaccharides of Human Milk Inhibit Its Binding and Infection. Journal of Biological Chemistry. 2003;278(16):14112–20. Epub 2003/02/04. doi: 10.1074/jbc.M207744200 12562767.
14. Magalhaes A, Gomes J, David L, Haas R, Boren T, Reis C. FUT2-Null Mice Show Impaired BabA-Mediated Adhesion of H. pylori to Gastric Mucosa. Helicobacter. 2009;14(4):371-. WOS:000268269300177.
15. Pham Tu Anh N, Clare S, Goulding D, Arasteh Julia M, Stares Mark D, Browne Hilary P, et al. Epithelial IL-22RA1-Mediated Fucosylation Promotes Intestinal Colonization Resistance to an Opportunistic Pathogen. Cell Host & Microbe. 2014;16(4):504–16. Epub 2014/09/30. doi: 10.1016/j.chom.2014.08.017 PMC4190086; PubMed Central PMCID: PMC4190086.
16. Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345(6202). doi: 10.1126/science.1254009
17. Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;advance online publication. doi: 10.1038/nature13823 http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature13823.html#supplementary-information.
18. Amorim I, Freitas DP, Magalhães A, Faria F, Lopes C, Faustino AM, et al. A comparison of Helicobacter pylori and non-Helicobacter pylori Helicobacter spp. Binding to Canine Gastric Mucosa with Defined Gastric Glycophenotype. Helicobacter. 2014;19(4):249–59. doi: 10.1111/hel.12125 24689986
19. Lo Presti L, Cabuy E, Chiricolo M, Dall'Olio F. Molecular cloning of the human beta1,4 N-acetylgalactosaminyltransferase responsible for the biosynthesis of the Sd(a) histo-blood group antigen: the sequence predicts a very long cytoplasmic domain. Journal of biochemistry. 2003;134(5):675–82. Epub 2003/12/23. 14688233.
20. Stuckenholz C, Lu L, Thakur P, Kaminski N, Bahary N. FACS-Assisted Microarray Profiling Implicates Novel Genes and Pathways in Zebrafish Gastrointestinal Tract Development. Gastroenterology. 2009;137(4):1321–32. Epub 2009/07/01. doi: http://dx.doi.org/10.1053/j.gastro.2009.06.050. 19563808; PubMed Central PMCID: PMC2785077. doi: 10.1053/j.gastro.2009.06.050
21. Johnsen JM, Levy GG, Westrick RJ, Tucker PK, Ginsburg D. The endothelial-specific regulatory mutation, Mvwf1, is a common mouse founder allele. Mammalian Genome. 2008;19(1):32–40. doi: 10.1007/s00335-007-9079-4 WOS:000252483800005.
22. Mohlke KL, Nichols WC, Westrick RJ, Novak EK, Cooney KA, Swank RT, et al. A novel modifier gene for plasma von Willebrand factor level maps to distal mouse chromosome 11. Proceedings of the National Academy of Sciences. 1996;93(26):15352–7. Epub 1996/12/24. 8986815; PubMed Central PMCID: PMC26408.
23. Mohlke KL, Purkayastha AA, Westrick RJ, Smith PL, Petryniak B, Lowe JB, et al. Mvwf, a Dominant Modifier of Murine von Willebrand Factor, Results from Altered Lineage-Specific Expression of a Glycosyltransferase. Cell. 1999;96(1):111–20. Epub 1999/02/16. doi: http://dx.doi.org/10.1016/S0092-8674(00)80964-2. 9989502.
24. Johnsen JM, Teschke M, Pavlidis P, McGee BM, Tautz D, Ginsburg D, et al. Selection on cis-Regulatory Variation at B4galnt2 and Its Influence on von Willebrand Factor in House Mice. Molecular Biology and Evolution. 2009;26(3):567–78. Epub 2008/12/18. doi: 10.1093/molbev/msn284 ISI:000263420900009; PubMed Central PMCID: PMC2727395.
25. Linnenbrink M, Johnsen JM, Montero I, Brzezinski CR, Harr B, Baines JF. Long-term balancing selection at the blood group-related gene B4galnt2 in the genus Mus (Rodentia; Muridae). Molecular Biology and Evolution. 2011;28(11):2999–3003. Epub 2011/06/10. doi: 10.1093/molbev/msr150 21652612.
26. Staubach F, Künzel S, Baines AC, Yee A, McGee BM, Bäckhed F, et al. Expression of the blood-group-related glycosyltransferase B4galnt2 influences the intestinal microbiota in mice. ISME J. 2012;6(7):1345–55. Epub 2012/01/27. doi: http://www.nature.com/ismej/journal/vaop/ncurrent/suppinfo/ismej2011204s1.html. 22278669; PubMed Central PMCID: PMC3379640. doi: 10.1038/ismej.2011.204
27. Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, et al. Pretreatment of Mice with Streptomycin Provides a Salmonella enterica Serovar Typhimurium Colitis Model That Allows Analysis of Both Pathogen and Host. Infection and Immunity. 2003;71(5):2839–58. doi: 10.1128/iai.71.5.2839–2858.2003 ISI:000182501500061.
28. Hoiseth SK, Stocker BA. Genes aroA and serC of Salmonella typhimurium constitute an operon. Journal of Bacteriology. 1985;163(1):355–61. Epub 1985/07/01. 2989248; PubMed Central PMCID: PMC219121.
29. Grassl GA, Valdez Y, Bergstrom KSB, Vallance BA, Finlay BB. Chronic Enteric Salmonella Infection in Mice Leads to Severe and Persistent Intestinal Fibrosis. Gastroenterology. 2008;134(3):768–80.e2. Epub 2008/03/08. doi: http://dx.doi.org/10.1053/j.gastro.2007.12.043. 18325390. doi: 10.1053/j.gastro.2007.12.043
30. Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infection and immunity. 2005;73(6):3219–27. Epub 2005/05/24. doi: 10.1128/iai.73.6.3219–3227.2005 15908346; PubMed Central PMCID: PMC1111876.
31. Wei X, Yang Z, Rey Federico E, Ridaura Vanessa K, Davidson Nicholas O, Gordon Jeffrey I, et al. Fatty Acid Synthase Modulates Intestinal Barrier Function through Palmitoylation of Mucin 2. Cell Host & Microbe. 2012;11(2):140–52. Epub 2012/02/22. doi: http://dx.doi.org/10.1016/j.chom.2011.12.006. 22341463; PubMed Central PMCID: PMC3285413.
32. Blanchard D, Piller F, Gillard B, Marcus D, Cartron JP. Identification of a novel ganglioside on erythrocytes with blood group Cad specificty. Journal of Biological Chemistry. 1985;260(13):7813–6. 4008478
33. Piller F, Blanchard D, Huet M, Cartron JP. Identification of a α-NeuAc-(2→3)-β-d-galactopyranosyl N-acetyl-β-d-galactosaminyltransferase in human kidney. Carbohydrate Research. 1986;149(1):171–84. 2425965
34. Dohi T, Yuyama Y, Natori Y, Smith PL, Lowe JB, Oshima M. Detection of N-acetylgalactosaminyltransferase mRNA which determines expression of Sda blood group carbohydrate structure in human gastrointestinal mucosa and cancer. International Journal of Cancer. 1996;67(5):626–31. doi: 10.1002/(sici)1097-0215(19960904)67 : 5<626::aid-ijc6>3.0.co;2-w
35. Dall'Olio F, Malagolini N, Chiricolo M, Trinchera M, Harduin-Lepers A. The expanding roles of the Sda/Cad carbohydrate antigen and its cognate glycosyltransferase B4GALNT2. Biochimica et Biophysica Acta (BBA)—General Subjects. 2014;1840(1):443–53. Epub 2013/10/12. doi: http://dx.doi.org/10.1016/j.bbagen.2013.09.036. 24112972.
36. Capon C, Maes E, Michalski JC, Leffler H, Kim YS. Sd(a)-antigen-like structures carried on core 3 are prominent features of glycans from the mucin of normal human descending colon. Biochem J. 2001;358(3):657–64. Epub 2001/10/02. 11577689; PubMed Central PMCID: PMC1222115.
37. Godinez I, Haneda T, Raffatellu M, George MD, Paixao TA, Rolan HG, et al. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun. 2008;76(5):2008–17. Epub 2008/03/19. IAI.01691-07 [pii] doi: 10.1128/IAI.01691-07 18347048; PubMed Central PMCID: PMC2346712.
38. de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. Host-pathogen interaction in invasive Salmonellosis. PLoS Pathog. 2012;8(10):e1002933. Epub 2012/10/12. doi: 10.1371/journal.ppat.1002933 PPATHOGENS-D-12-00748 [pii]. 23055923; PubMed Central PMCID: PMC3464234.
39. Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, Vijay-Kumar M. Fecal Lipocalin 2, a Sensitive and Broadly Dynamic Non-Invasive Biomarker for Intestinal Inflammation. PLoS One. 2012;7(9):e44328. Epub 2012/09/08. doi: 10.1371/journal.pone.0044328 22957064; PubMed Central PMCID: PMC3434182.
40. Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5(5):476–86. Epub 2009/05/21. doi: S1931-3128(09)00108-5 [pii]doi: 10.1016/j.chom.2009.03.011 19454351; PubMed Central PMCID: PMC2768556.
41. Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33(1):475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448 ISI:000180007000018.
42. May RM. Will a Large Complex System be Stable? Nature. 1972;238(5364):413–4. 4559589
43. Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admettla A, Pattini L, Nielsen R. Signatures of Environmental Genetic Adaptation Pinpoint Pathogens as the Main Selective Pressure through Human Evolution. PLoS Genet. 2011;7(11):e1002355. Epub 2011/11/11. doi: 10.1371/journal.pgen.1002355 22072984; PubMed Central PMCID: PMC3207877.
44. Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 1997;17(2):179–224. Epub 1997/01/01. ISI:A1997WR04600004.
45. Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9(8):747–55. Epub 1999/07/16. 10406840.
46. Varki A. Nothing in Glycobiology Makes Sense, except in the Light of Evolution. Cell. 2006;126(5):841–5. Epub 2006/09/09. doi: 10.1016/j.cell.2006.08.022 16959563.
47. Andres AM, Hubisz MJ, Indap A, Torgerson DG, Degenhardt JD, Boyko AR, et al. Targets of balancing selection in the human genome. Mol Biol Evol. 2009;26(12):2755–64. Epub 2009/08/29. doi: 10.1093/molbev/msp190 MEDLINE:19713326; PubMed Central PMCID: PMC2782326.
48. Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota. PLoS Biol. 2007;5(10):e244. Epub 2007/09/01. doi: 10.1371/journal.pbio.0050244 17760501; PubMed Central PMCID: PMC1951780.
49. Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, Benecke A, et al. The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Salmonella Diarrhea. PLoS Pathog. 2010;6(9):e1001097. doi: 10.1371/journal.ppat.1001097 20844578
50. Levins R. COMPLEX SYSTEMS1970. 73–88 p.
51. Gardner MR, Ashby WR. CONNECTANCE OF LARGE DYNAMIC (CYBERNETIC) SYSTEMS—CRITICAL VALUES FOR STABILITY. Nature. 1970;228(5273):784-&. doi: 10.1038/228784a0 WOS:A1970H798400060.
52. Macarthur R. FLUCTUATIONS OF ANIMAL POPULATIONS, AND A MEASURE OF COMMUNITY STABILITY. Ecology. 1955;36(3):533–6. doi: 10.2307/1929601 WOS:A1955WR61800032.
53. Odum EP. Fundamentals of ecology1953. v+384p. Illus.-v+p. Illus. p.
54. Loreau M. Linking biodiversity and ecosystems: towards a unifying ecological theory2010 2010-01-12 00 : 00 : 00. 49–60 p.
55. Holling CS. Resilience and Stability of Ecological Systems. Annu Rev Ecol Syst. 1973;4(ArticleType: research-article / Full publication date: 1973 / Copyright 1973 Annual Reviews):1–23. doi: 10.2307/2096802
56. Ives AR, Carpenter SR. Stability and Diversity of Ecosystems. Science. 2007;317(5834):58–62. doi: 10.1126/science.1133258 17615333
57. Tilman D. Biodiversity: Population Versus Ecosystem Stability. Ecology. 1996;77(2):350–63. doi: 10.2307/2265614
58. Pfisterer AB, Schmid B. Diversity-dependent production can decrease the stability of ecosystem functioning. Nature. 2002;416(6876):84–6. 11882897
59. Isbell F, Calcagno V, Hector A, Connolly J, Harpole WS, Reich PB, et al. High plant diversity is needed to maintain ecosystem services. Nature. 2011;advance online publication. doi: http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature10282.html#supplementary-information.
60. Doak DF, Bigger D, Harding EK, Marvier MA, O'Malley RE, Thomson D. The statistical inevitability of stability-diversity relationships in community ecology. American Naturalist. 1998;151(3):264–76. doi: 10.1086/286117 WOS:000072128600006.
61. Yachi S, Loreau M. Biodiversity and ecosystem productivity in a fluctuating environment: The insurance hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(4):1463–8. doi: 10.1073/pnas.96.4.1463 WOS:000078698400054.
62. Hubbell SP. The unified neutral theory of biodiversity and biogeography. Monographs in Population Biology. 2001;32:i–xiv, 1–375. ZOOREC:ZOOR13700052777.
63. Sankaran M, McNaughton SJ. Determinants of biodiversity regulate compositional stability of communities. Nature. 1999;401(6754):691–3.
64. McNaughton SJ. STABILITY AND DIVERSITY OF ECOLOGICAL COMMUNITIES. Nature. 1978;274(5668):251–3. doi: 10.1038/274251a0 WOS:A1978FG65000039.
65. McCann K, Hastings A, Huxel GR. Weak trophic interactions and the balance of nature. Nature. 1998;395(6704):794–8.
66. Cadotte MW, Cardinale BJ, Oakley TH. Evolutionary history and the effect of biodiversity on plant productivity. Proceedings of the National Academy of Sciences. 2008;105(44):17012–7. doi: 10.1073/pnas.0805962105
67. Cadotte MW, Dinnage R, Tilman D. Phylogenetic diversity promotes ecosystem stability. Ecology. 2012;93(sp8):S223–S33. doi: 10.1890/11-0426.1 ISI:000307302400018.
68. Petchey OL, Casey T, Jiang L, McPhearson PT, Price J. Species richness, environmental fluctuations, and temporal change in total community biomass. Oikos. 2002;99(2):231–40. doi: 10.1034/j.1600-0706.2002.990203.x WOS:000179715200003.
69. Zhang Q-G, Zhang D-Y. Species richness destabilizes ecosystem functioning in experimental aquatic microcosms. Oikos. 2006;112(1):218–26. doi: 10.1111/j.0030-1299.2006.14220.x
70. Bohnhoff M, Drake BL, Miller CP. Effect of Streptomycin on Susceptibility of Intestinal Tract to Experimental Salmonella Infection. Experimental Biology and Medicine. 1954;86(1):132–7. Epub 1954/05/01. doi: 10.3181/00379727-86-21030 13177610.
71. Srivastava DS, Cadotte MW, MacDonald AAM, Marushia RG, Mirotchnick N. Phylogenetic diversity and the functioning of ecosystems. Ecology Letters. 2012;15(7):637–48. Epub 2012/05/16. doi: 10.1111/j.1461-0248.2012.01795.x 22583836.
72. Jones EI, Nuismer SL, Gomulkiewicz R. Revisiting Darwin's conundrum reveals a twist on the relationship between phylogenetic distance and invasibility. Proceedings of the National Academy of Sciences. 2013;110(51):20627–32. doi: 10.1073/pnas.1310247110
73. Dethlefsen L, Huse S, Sogin ML, Relman DA. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. Plos Biology. 2008;6(11):e280. Epub 2008/11/21. doi: 10.1371/journal.pbio.0060280 BIOSIS:PREV200900045692; PubMed Central PMCID: PMC2586385.
74. Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, et al. Like Will to Like: Abundances of Closely Related Species Can Predict Susceptibility to Intestinal Colonization by Pathogenic and Commensal Bacteria. PLoS Pathog. 2010;6(1):e1000711. Epub 2010/01/12. doi: 10.1371/journal.ppat.1000711 20062525; PubMed Central PMCID: PMC2796170.
75. Sonnenburg JL, Xu J, Leip DD, Chen C-H, Westover BP, Weatherford J, et al. Glycan Foraging in Vivo by an Intestine-Adapted Bacterial Symbiont. Science. 2005;307(5717):1955–9. Epub 2005/03/26. doi: 10.1126/science.1109051 15790854.
76. Holmén Larsson JM, Karlsson H, Sjövall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19(7):756–66. doi: 10.1093/glycob/cwp048 19321523
77. Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Micro. 2013;11(7):497–504. Epub 2013/06/12. doi: 10.1038/nrmicro3050 http://www.nature.com/nrmicro/journal/v11/n7/abs/nrmicro3050.html#supplementary-information. 23748339.
78. Bosshard PP, Zbinden R, Altwegg M. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium. International Journal of Systematic and Evolutionary Microbiology. 2002;52(4):1263–6. Epub 2002/08/01. doi: 10.1099/ijs.0.02056–0 12148638.
79. Dimitriu PA, Boyce G, Samarakoon A, Hartmann M, Johnson P, Mohn WW. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environmental Microbiology Reports. 2013;5(2):200–10. Epub 2013/04/16. doi: 10.1111/j.1758-2229.2012.00393.x 23584963.
80. Cuív PÓ, Klaassens ES, Durkin AS, Harkins DM, Foster L, McCorrison J, et al. Draft Genome Sequence of Turicibacter sanguinis PC909, Isolated from Human Feces. Journal of Bacteriology. 2011;193(5):1288–9. Epub 2010/12/25. doi: 10.1128/jb.01328-10 21183674; PubMed Central PMCID: PMC3067595.
81. Wang GH. Plant traits and soil chemical variables during a secondary vegetation succession in abandoned fields on the Loess Plateau. Acta Botanica Sinica. 2002;44(8):990–8. WOS:000177681900019.
82. Suter M, Edwards PJ. Convergent succession of plant communities is linked to species’ functional traits. Perspectives in Plant Ecology, Evolution and Systematics. 2013;15(4):217–25. doi: http://dx.doi.org/10.1016/j.ppees.2013.05.001.
83. Lohbeck M, Poorter L, Martínez-Ramos M, Rodriguez-Velázquez J, van Breugel M, Bongers F. Changing drivers of species dominance during tropical forest succession. Functional Ecology. 2014;28(4):n/a-n/a. doi: 10.1111/1365-2435.12240 ISI:000340673900028.
84. Presley LL, Wei B, Braun J, Borneman J. Bacteria Associated with Immunoregulatory Cells in Mice. Applied and Environmental Microbiology. 2010;76(3):936–41. Epub 2009/12/17. doi: 10.1128/aem.01561-09 20008175; PubMed Central PMCID: PMC2813032.
85. Weiss GA, Chassard C, Hennet T. Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. Br J Nutr. 2014 : 1–9. Epub 2014/01/15. doi: S0007114513004200 [pii]doi: 10.1017/S0007114513004200 24411010.
86. Drouilhet L, Mansanet C, Sarry J, Tabet K, Bardou P, Woloszyn F, et al. The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the B4GALNT2 Gene within the Ovary. PLoS Genet. 2013;9(9):e1003809. Epub 2013/10/03. doi: 10.1371/journal.pgen.1003809 24086150; PubMed Central PMCID: PMC3784507.
87. Vimal DB, Khullar M, Gupta S, Ganguly NK. Intestinal mucins: the binding sites for Salmonella typhimurium. Molecular and cellular biochemistry. 2000;204(1–2):107–17. Epub 2000/03/16. 10718631.
88. Chessa D, Winter MG, Jakomin M, Baumler AJ. Salmonella enterica serotype Typhimurium Std fimbriae bind terminal alpha(1,2)fucose residues in the cecal mucosa. Molecular microbiology. 2009;71(4):864–75. Epub 2009/02/03. doi: 10.1111/j.1365-2958.2008.06566.x 19183274.
89. Groux-Degroote S, Wavelet C, Krzewinski-Recchi M-A, Portier L, Mortuaire M, Mihalache A, et al. B4GALNT2 gene expression controls the biosynthesis of Sda and sialyl Lewis X antigens in healthy and cancer human gastrointestinal tract. The International Journal of Biochemistry & Cell Biology. 2014;53(0):442–9. doi: http://dx.doi.org/10.1016/j.biocel.2014.06.009.
90. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;advance online publication(7469):96–9. Epub 2013/09/03. doi: 10.1038/nature12503http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature12503.html#supplementary-information. 23995682; PubMed Central PMCID: PMC3825626. 23995682
91. Lefrancois L. Carbohydrate differentiation antigens of murine T cells: expression on intestinal lymphocytes and intestinal epithelium. Journal of immunology (Baltimore, Md: 1950). 1987;138(10):3375–84. Epub 1987/05/15. 2437191.
92. Zarbock A, Ley K, McEver RP, Hidalgo A. Leukocyte ligands for endothelial selectins: specialized glycoconjugates that mediate rolling and signaling under flow. Blood. 2011;118(26):6743–51. Epub 2011/10/25. doi: 10.1182/blood-2011-07-343566 22021370; PubMed Central PMCID: PMC3245201.
93. Kobayashi M, Fukuda M, Nakayama J. Role of Sulfated O-Glycans Expressed by High Endothelial Venule-Like Vessels in Pathogenesis of Chronic Inflammatory Gastrointestinal Diseases. Biological & Pharmaceutical Bulletin. 2009;32(5):774–9. Epub 2009/05/08. WOS:000266047300003; PubMed Central PMCID: PMC2718737.
94. Gauguet J-M, Rosen SD, Marth JD, von Andrian UH. Core 2 branching β1,6-N-acetylglucosaminyltransferase and high endothelial cell N-acetylglucosamine-6-sulfotransferase exert differential control over B - and T-lymphocyte homing to peripheral lymph nodes. Blood. 2004;104(13):4104–12. Epub 2004/08/21. doi: 10.1182/blood-2004-05-1986 15319280.
95. Lowe JB. Glycan-dependent leukocyte adhesion and recruitment in inflammation. Current opinion in cell biology. 2003;15(5):531–8. Epub 2003/10/02. 14519387.
96. Kawamura YI, Kawashima R, Fukunaga R, Hirai K, Toyama-Sorimachi N, Tokuhara M, et al. Introduction of Sd(a) carbohydrate antigen in gastrointestinal cancer cells eliminates selectin ligands and inhibits metastasis. Cancer research. 2005;65(14):6220–7. Epub 2005/07/19. doi: 10.1158/0008-5472.can-05-0639 16024623.
97. Kawamura YI, Adachi Y, Curiel DT, Kawashima R, Kannagi R, Nishimoto N, et al. Therapeutic adenoviral gene transfer of a glycosyltransferase for prevention of peritoneal dissemination and metastasis of gastric cancer. Cancer Gene Ther. 2014;21(10):427–33. Epub 2014/09/13. doi: 10.1038/cgt.2014.46 25213663.
98. Littman Dan R, Pamer Eric G. Role of the Commensal Microbiota in Normal and Pathogenic Host Immune Responses. Cell Host & Microbe. 2011;10(4):311–23. Epub 2011/10/25. doi: 10.1016/j.chom.2011.10.004 22018232; PubMed Central PMCID: PMC3202012.
99. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7):research0034.1—research.11. Epub 2002/08/20. doi: 10.1186/gb-2002-3-7-research0034; PubMed Central PMCID: PMC126239.
100. Rausch P, Rehman A, Künzel S, Häsler R, Ott SJ, Schreiber S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proceedings of the National Academy of Sciences. 2011;108(47):19030–5. Epub 2011/11/10. doi: 10.1073/pnas.1106408108 22068912; PubMed Central PMCID: PMC3223430.
101. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open Source, Platform-independent, Community-supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009;75(23):7537–41. Epub October 2, 2009. doi: 10.1128/aem.01541-09 19801464; PubMed Central PMCID: PMC2786419.
102. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. Epub 2010/08/17. doi: 10.1093/bioinformatics/btq461 20709691.
103. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. Epub 2007/06/26. doi: 10.1128/aem.00062-07 17586664; PubMed Central PMCID: PMC1950982.
104. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res. 2007;35(21):7188–96. doi: 10.1093/nar/gkm864 17947321
105. Price MN, Dehal PS, Arkin AP. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One. 2010;5(3):e9490. Epub 2010/03/13. doi: 10.1371/journal.pone.0009490 20224823; PubMed Central PMCID: PMC2835736.
106. Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26(11):1463–4. Epub 2010/04/17. doi: 10.1093/bioinformatics/btq166 20395285.
107. Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RB, Simpson GL, et al. vegan: Community Ecology Package. 1.17–6 ed: http://CRAN.R-project.org; 2011.
108. Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2012.
109. McArdle BH, Anderson MJ. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology. 2001;82(1):290–7. doi: 10.1890/0012-9658(2001)082[0290:Fmmtcd]2.0.Co;2 ISI:000166488200024.
110. Anderson MJ. Distance-Based Tests for Homogeneity of Multivariate Dispersions. Biometrics. 2006;62(1):245–53. Epub 2006/03/18. doi: 10.1111/j.1541-0420.2005.00440.x 16542252.
111. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and Environmental Microbiology. 2005;71(12):8228–35. doi: 10.1128/aem.71.12.8228–8235.2005 WOS:000234417600073.
112. Legendre P, Legendre L. Numerical ecology. Second English edition. Developments in Environmental Modelling. 1998;20:i–xv, 1–853. ZOOREC:ZOOR13500057538.
113. Pinheiro J, Bates D, DebRoy S, Sarkar D, Team RDC. nlme: Linear and Nonlinear Mixed Effects Models. http://CRAN.R-project.org; 2011.
114. Bartoń K. MuMIn: multi-model inference, R package version 1.9.13. 2013.
115. Magee L. R2 Measures Based on Wald and Likelihood Ratio Joint Significance Tests. The American Statistician. 1990;44(3):250–3. doi: 10.2307/2685352
116. Hothorn T, Hornik K, Van de Wiel MA, Zeileis A. A Lego system for conditional inference. American Statistician. 2006;60(3):257–63. doi: 10.1198/000313006x118430 WOS:000239411800006.
117. De Cáceres M, Legendre P, Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119(10):1674–84. doi: 10.1111/j.1600-0706.2010.18334.x
118. Friedman J, Alm EJ. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput Biol. 2012;8(9):e1002687. doi: 10.1371/journal.pcbi.1002687 23028285
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 CellsČlánek Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



