-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
In animal cell cytokinesis, constriction of an actomyosin ring at the equatorial cortex of dividing cells must be finely coordinated with plasma membrane remodeling and vesicle trafficking at the cleavage furrow. Accurate control of these events during cell cleavage is essential for maintaining ploidy and preventing neoplastic transformation. GOLPH3 has been recognized as a potent oncogene, involved in the development of several human tumors. However, the precise roles played by GOLPH3 in tumorigenesis are not yet understood. In this manuscript we demonstrate for the first time the requirement for GOLPH3 for cytokinesis. GOLPH3 protein localizes at the cleavage site of Drosophila dividing cells and is essential for cytokinesis in male meiotic cells and larval neuroblasts. We show that this protein acts as a key molecule in coupling plasma membrane remodeling with actomyosin ring assembly and stability during cytokinesis. Our studies indicate a novel connection between GOLPH3 and the molecular mechanisms of cytokinesis, opening new fields of investigation into the tumor cell biology of this oncogene.
Published in the journal: . PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004305
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004305Summary
In animal cell cytokinesis, constriction of an actomyosin ring at the equatorial cortex of dividing cells must be finely coordinated with plasma membrane remodeling and vesicle trafficking at the cleavage furrow. Accurate control of these events during cell cleavage is essential for maintaining ploidy and preventing neoplastic transformation. GOLPH3 has been recognized as a potent oncogene, involved in the development of several human tumors. However, the precise roles played by GOLPH3 in tumorigenesis are not yet understood. In this manuscript we demonstrate for the first time the requirement for GOLPH3 for cytokinesis. GOLPH3 protein localizes at the cleavage site of Drosophila dividing cells and is essential for cytokinesis in male meiotic cells and larval neuroblasts. We show that this protein acts as a key molecule in coupling plasma membrane remodeling with actomyosin ring assembly and stability during cytokinesis. Our studies indicate a novel connection between GOLPH3 and the molecular mechanisms of cytokinesis, opening new fields of investigation into the tumor cell biology of this oncogene.
Introduction
Cytokinesis in animal cells is achieved through the constriction of a plasma membrane-anchored actomyosin ring that drives cleavage furrow ingression from the equatorial cortex [1]. A network of cytoskeletal proteins, including Septins and Anillin, acts as a scaffold to anchor the actomyosin ring to the plasma membrane during furrowing [2], [3].
The central spindle (the prominent microtubule bundle that forms in ana-telophase between the segregating chromosomes) dictates the position of the cleavage furrow [4]–[6]. Central spindle assembly is regulated by the coordinated action of kinesin motor proteins, microtubule associated proteins (MAPs) and protein kinases [6]. A crucial signaling event that sets up the site of cleavage furrow formation for cytokinesis is the accumulation of active Rho GTPase at an equatorial position at the cortex. This localized active Rho GTPase drives both F-actin assembly at the plasma membrane and Myosin II activation [4], [5]. The central spindle transmits the spatial information required for cleavage furrow formation by delivering regulators of Rho to the equatorial cortex [4], [6].
Recent studies have shown that besides contractile ring constriction, cytokinesis also involves membrane trafficking from internal stores to the cleavage furrow [7], [8]. Work from several groups has implicated Golgi-derived vesicles in cytokinesis [7]–[9]. Endocytic traffic also contributes to this process. Clathrin and Dynamin, two proteins that promote endocytic vesicle budding from the plasma membrane and the endocytic recycling factors ARF6, Rab35 and Rab11 are required for completion of cytokinesis in several cell systems [7]–[9].
Successful cytokinesis also depends on a special plasma membrane lipid composition [9]–[11]. It has been suggested that special lipids at the cleavage site facilitate cell shape remodeling during furrow ingression, regulate membrane addition and/or provide signalling platforms controlling cytokinesis [8]. However the molecular mechanisms underlying membrane-cytoskeleton interactions during cleavage furrow ingression are not yet understood.
Several proteins involved in the phosphoinositide (PIP) cycle and specific types of PIPs have been implicated in cytokinesis. For example the Drosophila melanogaster phosphatidylinositol transfer protein Giotto/Vibrator (Gio/Vib) and the Golgi PI 4-kinase Four wheel drive (Fwd) are both required for furrow ingression in male meiotic cytokinesis [12]–[15]. Fwd is required to recruit Rab11 at the Golgi, for the synthesis of PI(4)P on Golgi membranes and for accumulation of Rab11 - and PI(4)P-containing organelles at the cell equator [15]. However Fwd does not accumulate at the cleavage furrow during cytokinesis [15].
GOLPH3, a highly conserved 34-kDa protein, functions as a PI(4)P effector at the Golgi [16], [17]. Recruitment of GOLPH3 and its yeast orthologue Vps74p to the Golgi depends on specific recognition of PI(4)P through a conserved binding pocket on the surface of the protein [16], [17]. Human GOLPH3 also interacts with MYO18A, mediating linkage with the F-actin cytoskeleton that has been proposed to facilitate flattening of the Golgi and vesicle formation [16]. GOLPH3 is also a potent oncogene, as it is commonly amplified in several solid tumours [18]–[27]. However, the molecular mechanisms through which this protein acts in malignant transformation are not yet understood. GOLPH3 has been implicated in cellular transformation via changes in the activity of the mammalian target of rapamycin (mTOR) [18]. GOLPH3 has been shown to activate mTOR signaling [18]. In addition GOLPH3 protein physically interacts with the VPS35 subunit of the retromer protein-recycling complex which has been linked to TOR signaling in yeast [18].
Here we present the first evidence to date implicating GOLPH3 in cytokinesis. We demonstrate that the Drosophila homologue of GOLPH3 controls both contractile ring formation and vesicle trafficking during cleavage furrow ingression in dividing cells. In addition, this protein accumulates at the midzone of telophase cells and interacts with components of both the cytokinetic apparatus and the membrane trafficking machinery. We propose that during cell cleavage, GOLPH3 acts as a key molecule to coordinate membrane remodeling and cytoskeletal dynamics.
Results
Isolation of mutants in Drosophila GOLPH3
The sauronz2217 (sauz2217) allele was identified in a screen for mutations that cause failure of cytokinesis in Drosophila spermatocytes [28]. The sauz2217 mutation failed to complement both Df(2L)Exel6007 and Df(2L)Exel7010 for the male sterility and male meiotic defects, defining a small chromosome region containing 21 genes. A P-element insertion in the annotated CG7085 gene, resulting in the lethal mutation l(2)s5379, failed to complement sauz2217 for both the male sterility and meiotic cytokinesis phenotype (Figure 1A and 1B). In addition, testes from males expressing double stranded RNA (dsRNA) against CG7085 in meiotic cells, contained frequent aberrant multi-nucleate spermatids, indicating cytokinesis defects comparable to those observed in sauz2217/l(2)s5379 or sauz2217/Df(2L)Exel7010 (sauz2217/Df) males (Figure 1A and 1B). CG7085 encodes a polypeptide of 293 amino acids that is 70% identical to human GOLPH3. Thus, hereafter we refer to CG7085 as Drosophila GOLPH3. DNA sequencing of sauz2217 allele (See Materials and Methods), revealed a single point mutation in the GOLPH3 gene, resulting in the replacement of the conserved Glutamic-acid 273 by Lysine at the C-terminus of the predicted polypeptide (Figure 1C). Based on these results, sauz2217 is a missense allele of the Drosophila homologue of GOLPH3 (Figure 1C). GFP-GOLPH3 protein expressed under the control of a tubulin promoter rescued the defects of sauz2217/Df and sauz2217/l(2)s5379 mutants, confirming that the cytokinesis phenotype is the consequence of alterations in the Drosophila GOLPH3 homologue.
Fig. 1. Drosophila GOLPH3 is required for spermatocyte cytokinesis. 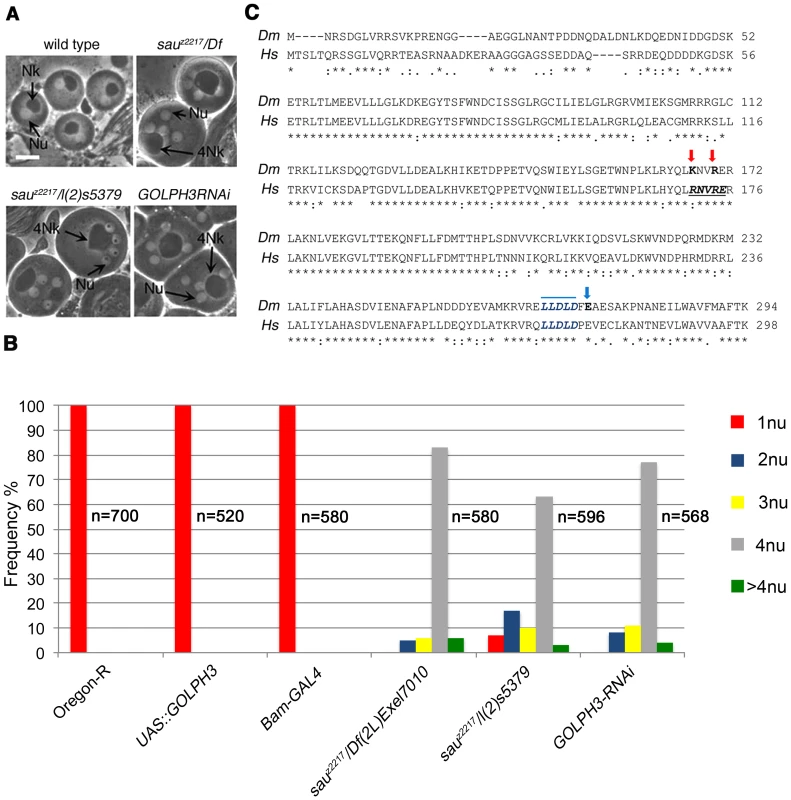
(A) Multinucleate onion stage spermatids due to cytokinesis failure during meiosis in GOLPH3 mutant males [sauz2217/Df(2L)Exel7010 and sauz2217/l(2)5379] and in males expressing dsRNA against CG7085 (GOLPH3RNAi), under control of the Bam-GAL4 driver [74]. (Nu) nuclei; (Nk) mitochondrial derivative; (4Nk) large mitochondrial derivative. Scale Bar; 10 µm. (B) Frequencies of spermatids containing 1, 2, 3, 4 or more than 4 nuclei per Nk, in sauz2217/Df(2L)Exel7010 and sauz2217/l(2)5379 mutant males and in males expressing dsRNA against CG7085 (GOLPH3RNAi) under control of Bam-GAL4. Oregon R males and males carrying either the Bam-GAL4 driver alone or the UAS::GOLPH3RNAi construct alone were used as control. (n); total number of spermatids (C) Alignment of Drosophila GOLPH3 (Dm) protein with human GOLPH3 (Hs). (Blue arrowhead) substitution of Glutamic acid by Lysine (E273K) in the sauz2217 male sterile allele; (red arrows) K167A and R170A mutations; (blue line) clathrin box sequence. (*) fully conserved residue; (:) conservation between groups of strongly similar properties; (.) conservation between groups of weakly similar properties. The Drosophila homologue of mammalian GOLPH3 localized to the cleavage site in dividing spermatocytes
Polyclonal antibodies raised against the entire Drosophila GOLPH3 protein, recognized a band of the predicted molecular weight in Western blots from extracts of adult testes (Figure 2A). This band appeared reduced in testis extracts from GOLPH3RNAi and sauz2217/Df males indicating that the antibodies specifically reacted with Drosophila GOLPH3 (Figure 2A).
Fig. 2. Drosophila GOLPH3 localizes to the cleavage furrow in dividing spermatocytes. 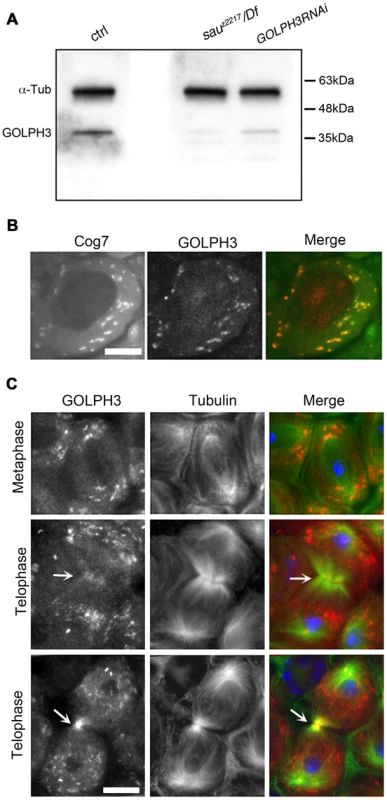
(A) Western blotting from adult testis extracts. Polyclonal antibodies raised against GOLPH3 protein, recognized a band of the predicted molecular weight. The band is reduced in testis extracts from sauz2217/Df(2L)Exel7010 and GOLPH3RNAi males. α-Tubulin was used as a loading control. (B) Colocalization of GOLPH3 (anti-GOLPH3, red) with the Golgi protein Cog7-GFP (green) in interphase spermatocytes. (C) In dividing spermatocytes GOLPH3 protein (red) was enriched in Golgi derived vesicles and accumulated at the cleavage furrow (arrows) during telophase (green) Tubulin; (blue) DNA. Scale Bar; 10 µm. Immunofluorescence analysis reveals that the GOLPH3 protein localized to Golgi structures in spermatocytes. In interphase spermatocytes at stage S5 (see Ref. [29] for spermatocyte stages), GOLPH3 was enriched at multiple round structures that also contained the Golgi marker Cog7 [30] (Figure S1 and 2B). In dividing spermatocytes at metaphase and anaphase, GOLPH3 was associated with vesicles in the polar regions of the cell (Figure 2C). Strikingly, during telophase GOLPH3 protein accumulated at the cleavage furrow (Figure S1 and Figure 2C) consistent with a role in cytokinesis.
Mutations in Drosophila GOLPH3 affect the Golgi stack structure in interphase primary spermatocytes
Consistent with the previous finding that depletion of human GOLPH3 affects Golgi architecture [16] GOLPH3 is essential for maintaining Golgi structure in Drosophila spermatocytes. In interphase wild type spermatocytes at stage S5, stained for the golgin Lava lamp (Lva, [31]), the average number of Golgi bodies per cell was 25 (Figure 3A and 3B). Conversely, spermatocytes from sauz2217/Df mutant males, stained for Lva at the same stage, exhibited a 1.9 - fold increase in the number of Golgi bodies (Figure 3A and 3B), with the average size decreased by 50% (Figure 3A and 3C). Previous work demonstrated that each Golgi in Drosophila has a paired structure consisting of two stacks held together through an actin-based mechanism [32]. The near-doubling of Golgi units observed in sauz2217/Df mutant spermatocytes raises the possibility that GOLPH3 may play a role in maintaining the paired Golgi organization in interphase spermatocytes.
Fig. 3. Mutations in GOLPH3 affect Golgi structure in primary spermatocytes. 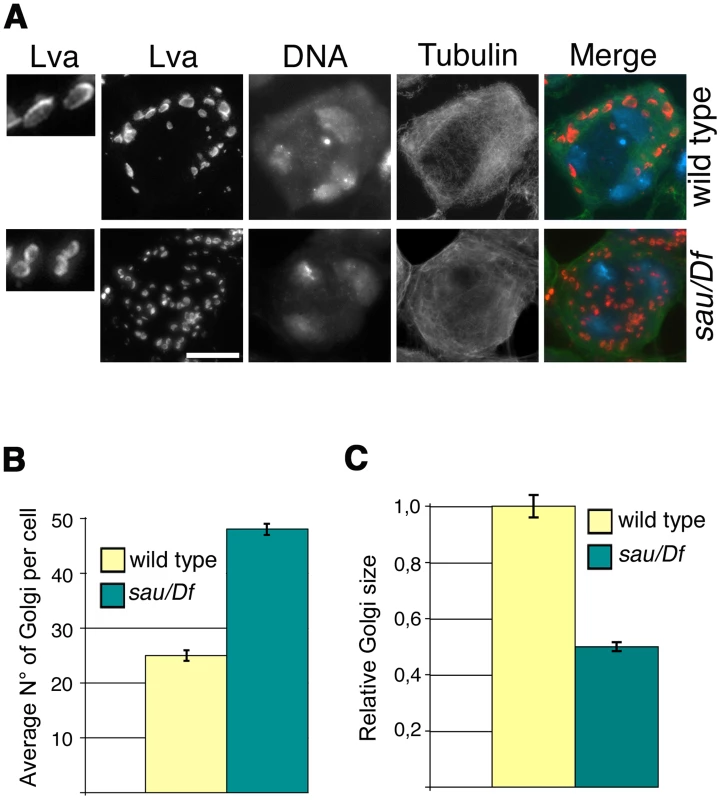
(A) G2 spermatocytes during the S5 stage, stained for Lva (red), Tubulin (green) and DNA (blue). Spermatocytes at this stage are characterized by three distinct chromatin clumps (corresponding to each major bivalents) and an interphase microtubule cytoskeleton (absence of asters). Left panels show enlargements of Golgi stacks in Lva images. Note that in wild type the size of Lva fluorescent units is approximately two fold when compared to sauz2217/Df(2L)Exel7010 (sau/Df) mutants. Scale Bar; 10 µm (B) Average number of Golgi bodies per cell (± SEM), visualized by Lva staining in spermatocytes at S5 stage from wild type and sauz2217/Df(2L)Exel7010 (sau/Df) mutant males. (C) Average size (± SEM), of Golgi stacks quantified by image J (Area in 2D images), in spermatocytes immunostained for Lva at S5 stage. Measures were normalized to control average size (set at 1). See also Materials and Methods. GOLPH3 is required for central spindle and contractile ring formation
Previous analysis of sauz2217/sauz2217 mutants, revealed defective central spindle in telophase spermatocytes [28]. Tubulin staining indicated defects in central spindle organization in sauz2217/Df hemizygous mutant males (Figure 4 and 5). Analysis of the dynamics of central spindle formation, performed in wild type (n = 10) and sauz2217/Df (n = 10) living spermatocytes expressing EGFP-β-tubulin [33], suggested a role for GOLPH3 in the initial steps of central spindle formation (Figure 4A). Localization of proteins associated with central spindle microtubules was also defective in sauz2217/Df mutant spermatocytes undergoing meiotic division. In both wild type and sauz2217/Df spermatocytes the microtubule bundling protein Fascetto/Prc1 (Feo, [34]) started to concentrate to microtubules at cell equator during anaphase (n = 44 mutant cells; n = 32 control cells; Figure 4B). However, in contrast to wild type, telophases from sauz2217/Df mutants did not exhibit a tight equatorial Feo band; instead Feo staining was either in puncta scattered between the two nuclei, or appeared diffuse (n = 50 mutant cells; n = 45 control cells; Figure 4B). Similar results were obtained in spermatocytes stained for the Polo kinase (Polo, [35]). Although Polo concentrated at the midzone of sauz2217/Df mutant spermatocytes during anaphase (n = 24 mutant cells; n = 24 control cells), it was dispersed in patches or diffuse during telophase (n = 34 mutant cells; n = 32 control cells; Figure 4C). Localization of the centralspindlin complex, which consists of the Rho GTPase activating protein (GAP) RacGAP50C [36] and the kinesin-like Pavarotti (Pav) [37], was also affected by loss of function of GOLPH3. sauz2217/Df mutant spermatocytes displayed faint concentration of both RacGAP50C (n = 32 mutant cells; n = 34 control cells) and Pav (n = 38 mutant cells; n = 40 control cells) at peripheral microtubules during anaphase (Figure 4D and 4E). In mutant spermatocytes at mid-late telophase these proteins failed to accumulate at both peripheral and interior microtubules (n = 50 mutant cells and n = 46 control cells examined for RacGAP50C; n = 44 mutant cells and n = 50 control cells examined for Pav; Figure 4D and 4E).
Fig. 4. GOLPH3 is required for central spindle assembly and localization of the centralspindlin components at the cell midzone. 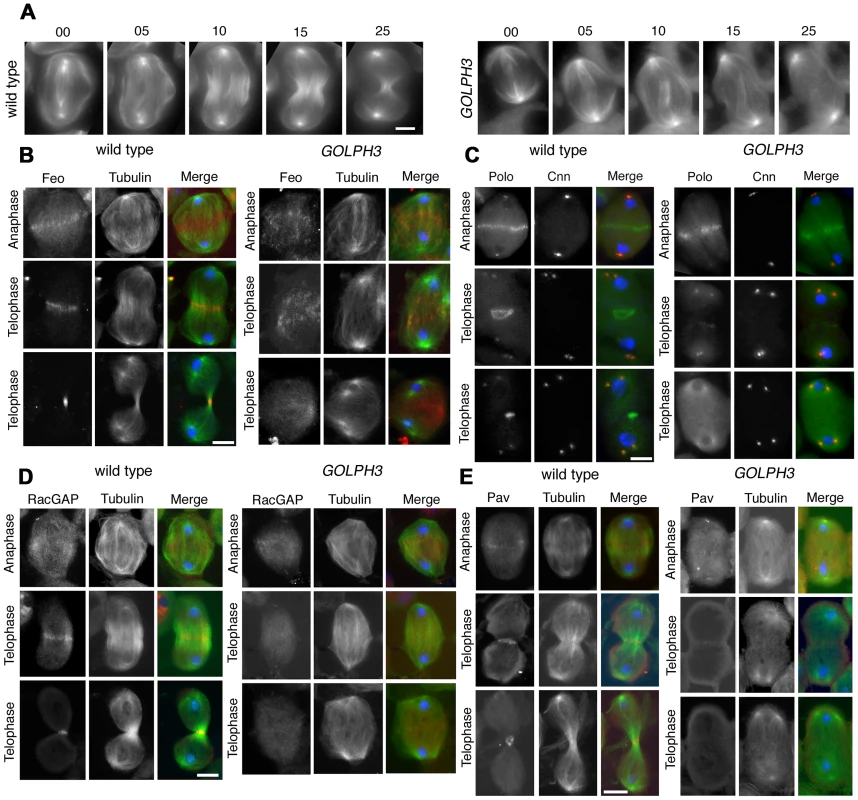
(A) Time lapse analysis of central spindle formation in spermatocytes from wild type and sauz2217/Df(2L)Exel7010 (GOLPH3) expressing EGFP-β-tubulin. In wild-type dividing spermatocytes at early anaphase, peripheral microtubules (MT) probed the equatorial cortex and started bundling (Frames 0, 5). Bundling of this population of MTs was accompanied by cell elongation and bundling of interior MTs (Frame 10). Furrow ingression compacted the MTs arrays (Frames 15) giving rise to the characteristic telophase central spindle that was pinched in the middle during cytokinesis (Frame 25). In sauz2217/Df(2L)Exel7010 (GOLPH3), peripheral MTs contacted the cortex at cell equator during early anaphase (Frames 0, 5) but failed to stably interdigitate (Frames 10, 15). In addition interior MTs failed to bundle at the cell midzone (Frame 15) and cells failed to constrict (Frames 15, 25). (B) Anaphase and telophase spermatocytes from wild type and sauz2217/Df(2L)Exel7010 (GOLPH3) stained for Feo (red), Tubulin (green), and DNA (blue). (C) Anaphase and telophase spermatocytes from wild type and sauz2217/Df(2L)Exel7010 (GOLPH3) stained for Polo (green), Centrosomin (Cnn, red) and DNA (blue). (D,E) Anaphase and telophase spermatocytes from wild type and sauz2217/Df(2L)Exel7010 (GOLPH3) stained for Tubulin (green), DNA (blue) and either RacGAP50C (D) or Pavarotti (Pav) (E) (red). Scale Bar; 10 µm. Fig. 5. GOLPH3 is required for maintaining Rho1 at the equatorial cortex and stabilizing Septin and Myosin rings. 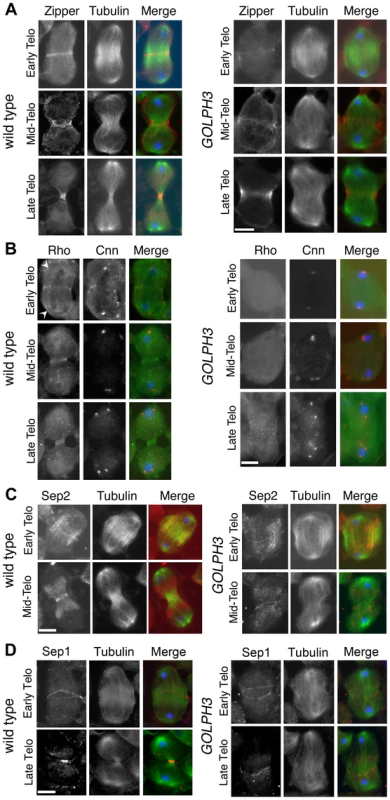
(A) Wild type and sauz2217/Df(2L)Exel7010 (GOLPH3) spermatocytes during late anaphase-early telophase and mid-late telophase stained for Tubulin (green), DNA (blue) and the Myo II heavy chain Zipper (red). (B) Wild type and sauz2217/Df(2L)Exel7010 (GOLPH3) spermatocytes during late anaphase-early telophase and mid-late telophase stained for Centrosomin (Cnn, red), DNA (blue) and Rho1 (green). Arrowheads indicate enrichment of Rho1 at the astral membranes. (C,D) Wild type and sauz2217/Df(2L)Exel7010 (GOLPH3) spermatocytes during telophase stained for Tubulin (green), DNA (blue) and either Sep2 (C) or Sep 1 (D) (red). Scale Bar; 10 µm. Previous analysis failed to detect F-actin rings in most telophase spermatocytes from sauz2217/sauz2217 homozygous males [28]. Staining for F-actin using an improved protocol (see Materials and Methods) revealed defects in F-actin ring assembly in sauz2217/Df mutant spermatocytes, with most telophases devoid of an F-actin ring (85%; n = 34 mutant cells; n = 44 control cells; Figure S2). sauz2217 mutation also impaired formation of regular Myosin II (Myo II) cortical rings. The Myo II heavy chain Zipper [38] formed a cortical ring at cell equator of wild type cells at late anaphase-early telophase (100% of cells, n = 48; Figure 5A). Zipper rings appeared tighter and constricted in mid - late telophase (100% of cells, n = 78; Figure 5A). Most sauz2217/Df mutant spermatocytes, fixed at late anaphase, displayed Zipper at cell equator. However in these cells Zipper was associated with thin rings at the equatorial cortex (n = 64; Figure 5A). In sauz2217/Df spermatocytes fixed at mid-late telophase, Zipper was associated with patches at cell equator (60% of cells) or formed unconstricted fragmented rings (30% of cells) (n = 120; Figure 5A). Contractile ring dynamics was examined in vivo in dividing spermatocytes expressing the GFP-tagged-regulatory light chain Spaghetti Squash-GFP (Sqh-GFP, [38], Figure S3). In wild type anaphase spermatocytes, Sqh-GFP concentrated into an equatorial ring that underwent constriction immediately after assembly (n = 10). In all mutant spermatocytes examined (n = 8), Sqh-GFP failed to concentrate into a well formed ring (Figure S3) and cells failed to constrict.
Consistent with the defects in F-actin ring assembly at the plasma membrane, cortical accumulation of Rho GTPase at an equatorial position was defective in mutant dividing spermatocytes. In wild type spermatocytes at late anaphase-early telophase, Rho1 was enriched at the astral membranes and the parafusorial membranes and started to concentrate into a narrow ring at the equatorial cortex (n = 32; Figure 5B). The Rho1 ring appeared thicker in mid-telophase spermatocytes and fully constricted in late telophase (n = 36; Figure 5B). In contrast, in most dividing spermatocytes from sauz2217/Df, Rho1 was diffuse in the cytoplasm and failed to concentrate into clear cortical rings at the cell equator during either late anaphase/early telophase (93%, n = 27; Figure 5B) or mid-late telophase (93%, n = 30; Figure 5B). Localization of Septins was also affected in GOLPH3 mutants. These cytoskeletal scaffolding proteins, bind to phosphoinositides and form a membrane-associated filament system required to anchor the actomyosin ring to the plasma membrane [39]. In wild type dividing spermatocytes both Septin1 (Sep1, [40]; n = 25) and Septin2 (Sep2, [41]; n = 45) concentrated into thin equatorial rings during early telophase (Figure 5C and 5D). At later telophase stages, rings of both Sep2 (n = 63) and Sep1 (n = 44) appeared thicker as constriction progressed cells (Figure 5C and 5D). In sauz2217/Df telophases Septins concentrated at the cell midzone (n = 42 cells examined for Sep2; n = 24 cells examined for Sep1; Figure 5C and 5D). However in mutant spermatocytes fixed at mid-late telophase Septin rings appeared thin and fragmented (n = 62 for Sep 2, n = 33 for Sep1; Figure 5C and 5D). Thus wild type function of GOLPH3 protein is required to maintain active Rho to the equatorial cortex and stabilization of Myo II and Septin-containing rings.
PI(4)P binding is essential for GOLPH3 function in cytokinesis
To test whether binding to PI(4)P is essential for GOLPH3 function in cytokinesis, we constructed a mutant version of GFP-GOLPH3 (GFP-GOLPH3K167A/R170L) carrying two substitutions (red arrows in Figure 1C) that, on the basis of crystal structure of human GOLPH3 [16], [17] are predicted to impair GOLPH3 binding to PI(4)P. GFP-GOLPH3K167A/R170L was unable to rescue the cytokinetic phenotype of sauz2217 (Figure 6A) and failed to localize to Golgi membranes and to the cleavage site in both wild type and sauz2217/Df (Figure 6B and 6C). Localization of wild type GOLPH3 protein at Golgi stacks was abolished in spermatocytes from mutants in fwd (Figure 6B), further supporting the requirement for PI(4)P for GOLPH3 recruitment to this organelle. The Glu-Lys (E273K) mutation in our original sauz2217 allele also affected the ability of GOLPH3 to bind to phosphoinositides. Drosophila wild type GOLPH3, and GOLPH3K167A/R170L, GOLPH3E273K mutant proteins were expressed as GST-tagged proteins and their ability to bind phosphoinositides was assessed through protein-lipid overlay assays (Figure 6D). Wild type GOLPH3 protein interacted not only with PI(4)P but also with PI(3)P, PI(5)P and phosphatidic acid (PA, a glycerophospholipid) (Figure 6D). Remarkably, the pattern of lipid interaction for GOLPH3E273K was comparable to that of GOLPH3K167A/R170L indicating defective binding to both PI(4)P and PI(5)P (Figure 6D). Consistent with the defective PI(4)P lipid interaction of GOLPH3E273K protein, GOLPH3 protein was not detected at the Golgi stacks of sauz2217 mutant spermatocytes immunostained with anti-GOLPH3 antibodies (Figure S1). Moreover GOLPH3 failed to concentrate at the equatorial cortex of sauz2217 dividing spermatocytes (Figure S1). To assess whether E273K and K167A/R170L mutations affect the stability of GOLPH3 protein the expression level of each GFP-tagged GOLPH3 protein was assessed by Western blot analysis. As shown in Figure 6E, in testes GFP-GOLPH3E273K protein was decreased by 52% and GFP-GOLPH3K167A/R170L protein by 54% relative to wild type GFP-GOLPH3. Together our data indicate that E273K and K167A/R170L mutations impair binding to PI(4)P and disrupt both the subcellular localization and the stability of GOLPH3 protein.
Fig. 6. Binding to PI(4)P is essential for GOLPH3 function during cytokinesis. 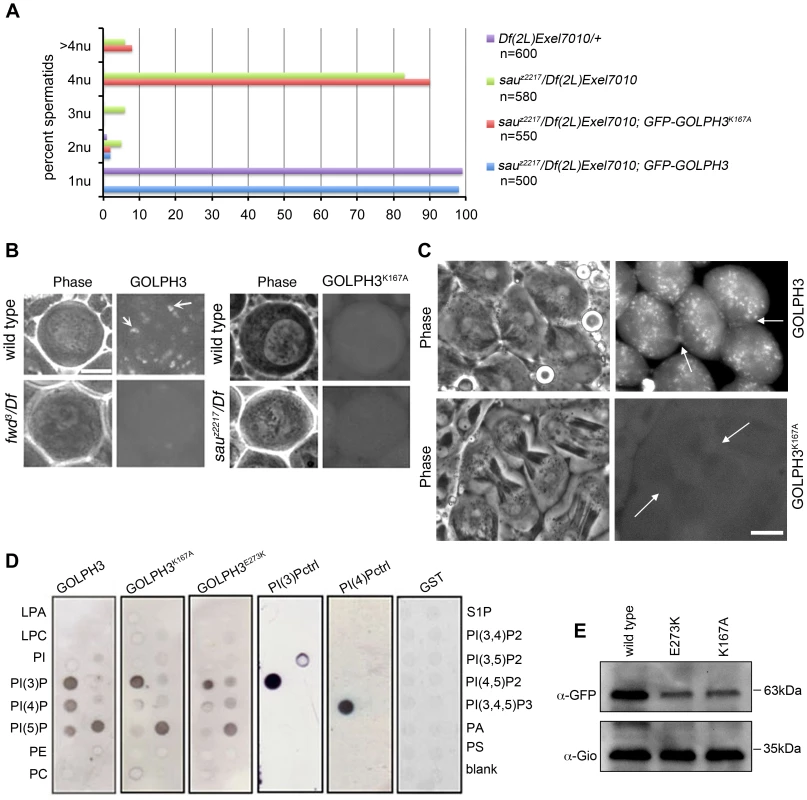
(A) Frequencies of spermatids (percentage) containing 1, 2, 3, 4 or more than 4 nuclei (nu) per Nk in testes from males of the indicated genotypes. (n); total number of spermatids counted per each genotype. Scale Bar; 10 µm. (B) Live spermatocytes of the indicated genotypes, expressing either GFP-GOLPH3 or a mutant version of GFP-GOLPH3 carrying K167A/R170L substitutions (GFP-GOLPH3K167A) were imaged at the same exposure time. GFP-GOLPH3 failed to localize to Golgi stacks (fluorescent round bodies indicated by arrows) in fwd3/Df(3L)7C [fwd3/Df] mutant spermatocytes. GFP-GOLPH3K167A failed to localize at the Golgi membranes in both wild type and sauz2217/Df(2L)Exel7010 (sauz2217/Df) mutant spermatocytes. (C) Phase-contrast and corresponding fluorescence images of live telophase spermatocytes expressing either GFP-GOLPH3 (GOLPH3) or GFP-GOLPH3 carrying K167A/R170L substitutions (GOLPH3K167A). Arrows indicate cleavage furrows. Scale Bar; 10 µm. (D) PIP strips (Echelon) were incubated with the indicated GST-tagged protein or GST, followed by anti-GST antibody for detection of lipid binding. PI(3)P ctrl and PI(4)P ctrl indicate GST-tagged control proteins that bind to either PI(3)P or PI(4)P. (E) Immunoblotting analysis of wild type testes expressing either GFP-GOLPH3 (wild type), GFP-GOLPH3E273K (E273K) or GFP-GOLPH3K167A/R170L (K167A). Giotto (Gio) was used as a loading control. Reciprocally the localization of PI(4)P was disrupted in telophase cells of sauz2217/Df spermatocytes. An RFP fusion to the PI(4)P marker PH-FAPP localized on Golgi organelles in both premeiotic and dividing spermatocytes (Figure 7A and data not shown) and was enriched at the midzone during telophase (Figure 7A and 7B) consistent with data of Polevoy and coauthors [15]. In premeiotic spermatocytes from sauz2217/Df, RFP-PH-FAPP signals at the Golgi membranes, were as intense as in wild type (data not shown). However in dividing spermatocytes RFP-PH-FAPP signals were delocalized and failed to accumulate at the midzone during telophase (Figure 7A and 7B). Visualization of PI(4,5)P2, the phosphorylated derivative of PI(4)P, using PLCδ-PH-GFP [42] showed this lipid at the cleavage furrow plasma membrane in wild type but not in GOLPH3 mutants (Figure 7C). Consistent with the idea that Fwd and GOLPH3 proteins contribute to the same process, double mutants carrying both fwd and GOLPH3 mutations were synthetic lethal. Animals that were heterozygous for GOLPH3 and hemizygous for fwd [sauz2217/+; fwd3/Df(3L)7C] and animals that were heterozygous for fwd and homozygous/hemizygous for GOLPH3 [sauz2217/sauz2217; fwd3/+ or sauz2217/Df(2L)Exel7010; fwd3/+] were both viable. However, sauz2217 was fully lethal in combination with fwd3/Df(3L)7C; individuals of genotypes sauz2217/Df(2L)Exel7010; fwd3/Df(3L)7C or sauz2217/sauz2217; fwd3/Df(3L)7C died during larval stages.
Fig. 7. Localization of PI(4)P is disrupted in telophase cells from GOLPH3 mutants. 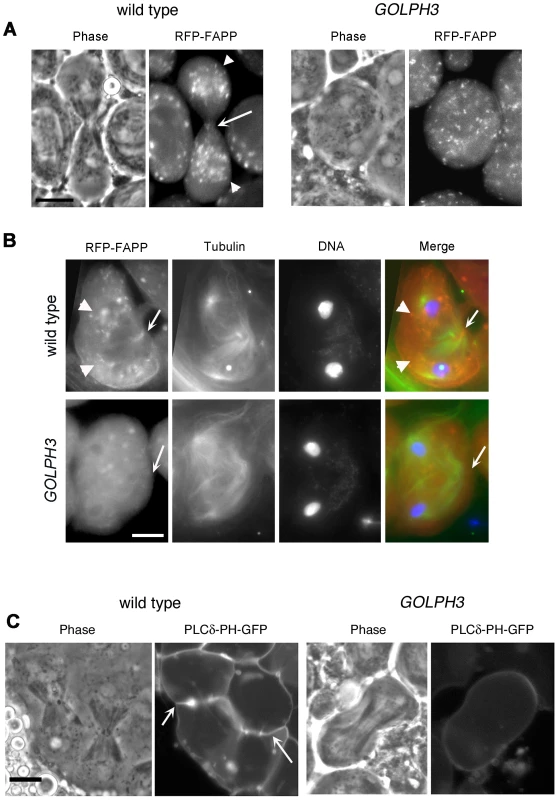
(A,C) Live spermatocytes expressing either RFP-PH-FAPP (RFP-FAPP) or PLCδ-PH-GFP were imaged at the same exposure time during telophase. Note that the distribution of both RFP-PH-FAPP and PLCδ-PH-GFP is affected in GOLPH3 mutant spermatocytes indicating a defective PI(4)P and PI(4,5)P2 subcellular localization and/or synthesis. Arrows indicate accumulation of RFP-PH-FAPP (A) and PLCδ-PH-GFP (C) at the cleavage furrows of wild type cells. (B) Fixed telophase spermatocyes expressing RFP-PH-FAPP (RFP-FAPP) stained for RFP (red), tubulin (green) and DNA (blue). Arrows indicate the cell midzone. Arrowheads indicate the enrichment of RFP signals at the poles of the wild type cell. Scale Bar; 10 µm. Like the requirement for Fwd for localization of both PI(4)P-associated secretory organelles and Rab11 at the cleavage furrow [15], [43], loss of GOLPH3 also affected concentration of Rab11 at cell equator. In contrast to wild type, Rab11 was dispersed during cytokinesis in sauz2217/Df spermatocytes (Figure 8A). GOLPH3 protein function was also required to localize other membrane trafficking markers at the cleavage furrow. In both wild type and sauz2217/Df interphase spermatocytes at stage S5, a GFP fusion to clathrin light chain (Clc-GFP, [44]) was enriched in clusters of vesicular structures (Figure S4A). However, although Clc-GFP accumulated to cell equator of wild type telophase spermatocytes (Figure S4A) in sauz2217/Df fluorescent signals were scattered and failed to accumulate at the cleavage furrow in cytokinesis (Figure S4A). Likewise analysis of a GFP-fusion to Rab5 [45], indicated that this protein, similarly to Rab11 and Clc-GFP, accumulated to the cleavage furrow in wild type but not in sauz2217/Df (Figure S4B). The small GTPase proteins Rab5 controls early endocytic events such as clathrin-coated-vesicle-mediated transport from the plasma membrane to the early endosomes and homotypic early endosome fusion [46].
Fig. 8. Rab11 forms a complex with GOLPH3 and depends on GOLPH3 for localization at the cleavage site. 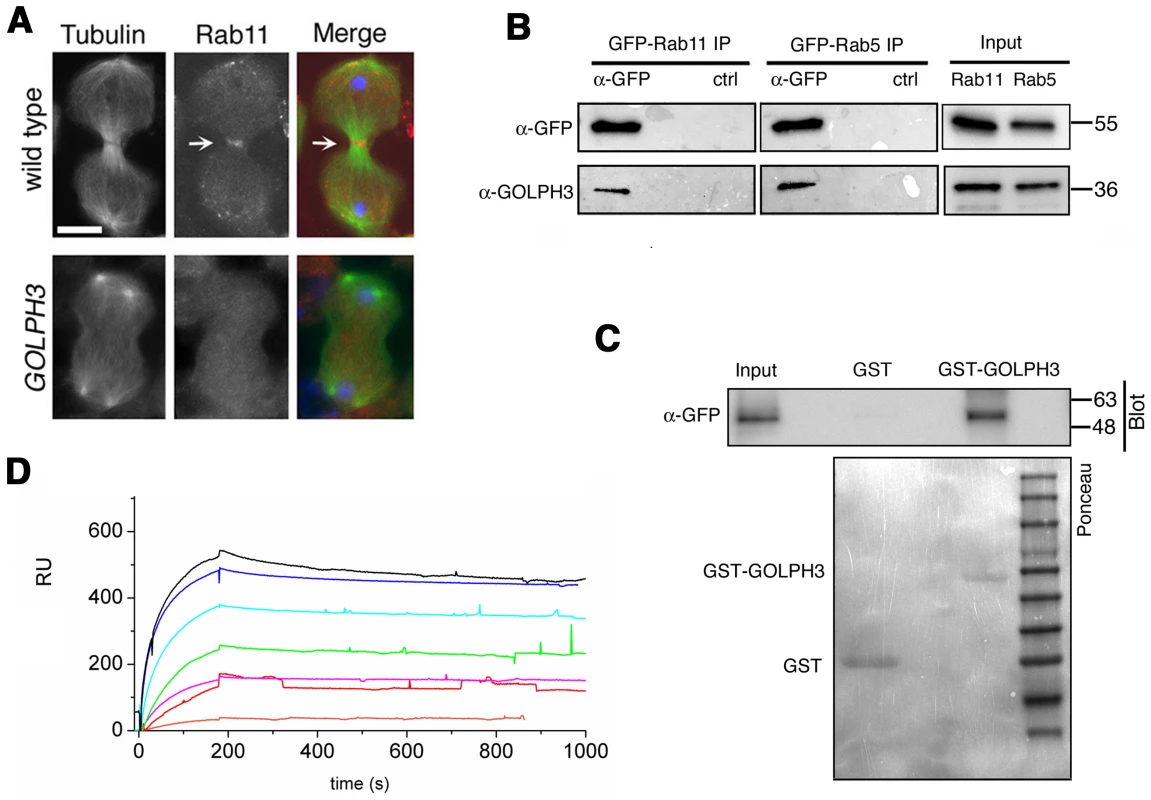
(A) Telophase spermatocytes from wild type and sauz2217/Df(2L)Exel7010 (GOLPH3) stained for Tubulin (green), Rab11 (red) and DNA (blue). Arrows indicate accumulation of Rab11 at the cleavage furrow of wild type spermatocyte. Scale Bar; 10 µm. (B) GOLPH3 protein coprecipitated with Rab11 and Rab5 in testis extracts. Protein extracts from testes expressing either GFP-Rab5 or GFP-Rab11 were immunoprecipitated with GFP-trap beads (α-GFP) and blotted for either GFP or GOLPH3. Control binding beads (ctrl) were used in control experiments. Input is 4% of lysates. Molecular masses are indicated in kilodaltons. (C) Bacterially expressed GST-GOLPH3 was purified by gluthatione-sepharose beads and incubated with testis lysates expressing GFP-Rab11. GST bound to gluthatione-sepharose beads was used as a negative control. GST-GOLPH3 precipitated GFP-Rab11 from testis protein extracts. Ponceau staining (Ponceau) is shown as loading control. Input is 4% of lysates. Molecular masses are indicated in kilodaltons. (D) Interaction of GOLPH3 immobilized on a CM5 sensorchip with Rab11, measured via Surface Plasmon Resonance experiments. Sensorgrams were obtained using GOLPH3 as ligand and Rab11 as analyte. Rab11 concentrations were as follows: 50 nM (orange), 200 nM (red), 400 nM (magenta), 1.0 µM (green), 2.0 µM (cyan), 4.0 µM (blue), 10.0 µM (black). The interaction of immobilized GOLPH3 ligand with the analyte was detected through mass concentration-dependent changes in the refractive index on the sensor chip surface, expressed as resonance units (RU). The increase in RU relative to baseline (0–180 s) indicates complex formation, whereas the decrease in RU represents dissociation of Rab11 from immobilized GOLPH3 upon injection of buffer. KD = 180 nM indicates that a high affinity complex is formed; the dissociation kinetics are very slow (kd about 1×10−4 s−1). We next tested whether GOLPH3 interacts with central spindle and contractile ring components. Co-immunoprecipitation (Co-IP) and GST-pulldown experiments with testis extracts revealed that GOLPH3 interacts with the cytokinesis proteins Pav, Zipper and Septins (Figure 9A–9C and Figure S5B). We also explored whether GOLPH3 interacts with membrane trafficking proteins. Consistent with data from mammalian cells [18], we found that GOLPH3 and the retromer Vps35 protein form a complex also in Drosophila (Figure S5A and S5B). In addition we found that GOLPH3 protein interacts with Rab11 (Figure 8B and 8C) in Drosophila testes. Surface Plasmon Resonance (SPR) analysis indicated that Rab11 and GOLPH3 proteins form a high affinity, stable complex (Figure 8D), since the measured KD was 180 nM, and the dissociation kinetics were very slow (kd about 2×10−4 s−1). Finally Co-IP and GST-pulldown experiments indicated interactions of GOLPH3 protein with Rab5 and Clathrin in male germ cells (Figure 8B, S4C and S4D).
Fig. 9. GOLPH3 protein interacts with both cytokinetic proteins in testis extracts. 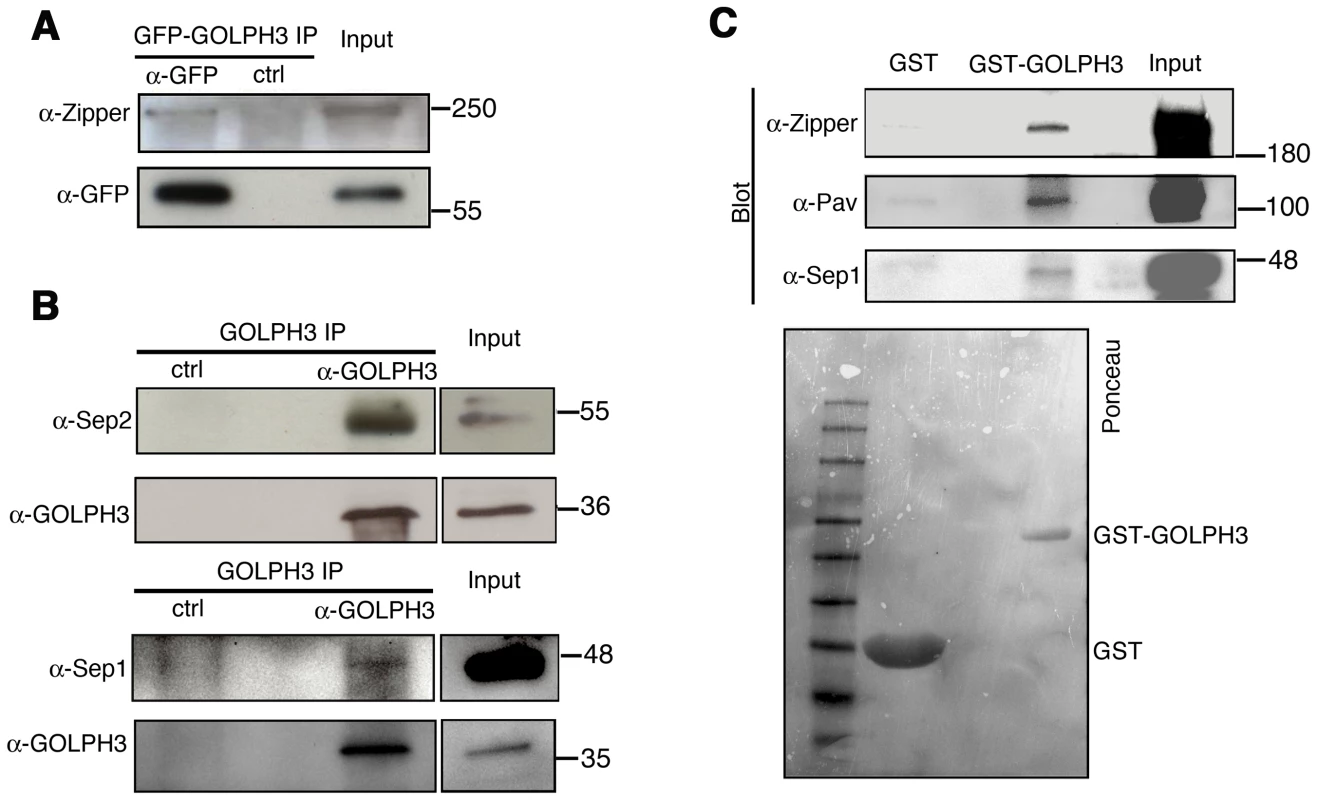
(A,B) Co-IP of GOLPH3 with Zipper and Septins. (A) Equal fractions of protein extracts from testes expressing GFP-GOLPH3 were immunoprecipitated with either mouse anti-GFP (α-GFP) or non-specific mouse IgG (ctrl) and blotted for either Zipper or GFP. (B) Protein extracts from wild type testes were immunoprecipitated with antibodies against Drosophila GOLPH3 (α-GOLPH3) and blotted for either Sep2, Sep1 or GOLPH3. Preimmune serum was used as control (ctrl). (C) Bacterially expressed GST-GOLPH3 was purified by gluthatione-sepharose beads and incubated with testis lysate. GST bound to gluthatione-sepharose beads was used as a negative control. GST-GOLPH3 precipitated Zipper, Pav and Sep1 from testis protein extracts. Ponceau staining (Ponceau) is shown as a loading control. In all the experiments input is 4% of lysates. Molecular masses are indicated in kilodaltons. Drosophila GOLPH3 is required for cytokinesis in larval brain neuroblasts
The gene GOLPH3 is essential for normal development and viability in Drosophila; l(2)s5379 homozygotes or l(2)s5379/Df(2L)Exel7010 hemizygotes die in early larval stages and individuals of genotype sauZ2217/Df have a reduced life span when compared with control siblings. Moreover we found an early lethal phenotype in individuals carrying the UAS::GOLPH3-RNAi construct in combination with drivers for ubiquitous RNAi (such as Actin-GAL4 or tubulin-GAL4). We thus wondered whether GOLPH3 is required for cytokinesis also in somatic cells. In dividing neuroblasts of larval central nervous system (CNS) GOLPH3 was enriched at the cleavage furrow consistent with a requirement for cytokinesis (Figure 10A). To knockdown the gene GOLPH3 in larval neuroblasts, UAS::GOLPH3-RNAi was expressed in larval CNS using the GAL4 system (tubulin-GAL4) with a temperature sensitive GAL80 (GAL80ts, [47]) (Figure 10B–10F). GOLPH3 depletion in larval CNS resulted in a significant frequency of tetraploidy: 6% of metaphases were tetraploid in larval brains expressing dsRNA against GOLPH3 (n = 1588 metaphases scored, Figure 10C) versus 0% in larval brains carrying the UAS::GOLPH3-RNAi transgene alone (n = 1800 metaphases scored). Immunostaining of larval brains for tubulin and Zipper also indicated cytokinesis defects in larval neuroblasts expressing UAS::GOLPH3-RNAi with 30% of mid-telophases failing to form a compact, fully constricted ring versus 0% in control cells (n = 150 mutant mid-telophases; n = 160 control mid-telophases; Figure 10D). Immunofluorescence analysis of mid-telophase neuroblasts depleted of GOLPH3 also revealed failure to maintain Septin proteins at the cleavage furrow and to form a compact central spindle microtubule array (Figure 10E–10F).
Fig. 10. GOLPH3 is required for cytokinesis in larval neuroblasts. 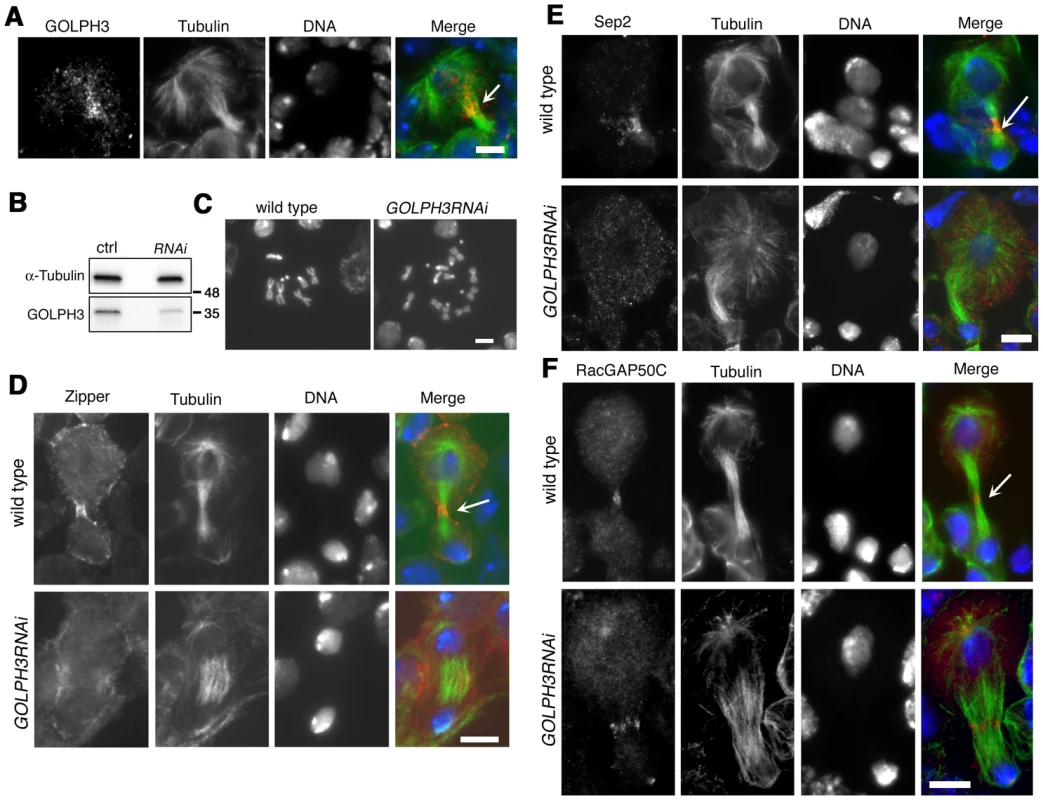
(A) GOLPH3 accumulates at the cleavage furrow of larval dividing neuroblasts. Larval neuroblast stained for Tubulin (green), GOLPH3 (red) and DNA (blue). Arrow indicates the midzone. Scale Bar, 5 µm. (B) Western blotting from larval brain extracts. Polyclonal antibodies raised against GOLPH3 protein, recognized a band of the predicted molecular weight. The band is reduced in larval brain extracts from animals expressing UAS::GOLPH3RNAi under control of Tub-GAL 4 Tub-GAL80ts. α-Tubulin was used as a loading control. Molecular masses are indicated in kilodaltons. (C) Normal male metaphase from wild type and tetraploid male metaphase from animals animals expressing UAS::GOLPH3RNAi under control of Tub-GAL4 Tub-GAL80ts. Scale Bar, 5 µm. (D,E) Knockdown of GOLPH3 in larval neuroblasts affects assembly of a stable, fully constricted contractile ring and impairs formation of a compact central spindle. Localization of Zipper (D), Sep2 (E) and RacGAP50C (F) in late telophase neuroblasts from wild type animals and in animals expressing UAS::GOLPH3RNAi under control of Tub-GAL 4 Tub-GAL80ts, [GOLPH3RNAi]. Larval brains were stained for Tubulin (green), DNA (blue) and either Zipper, Sep 2 or RacGAP50C (red). Telophase stage is based on chromatin condensation. Arrows point to the midzone. Scale Bar, 5 µm. Discussion
In this paper we have provided the first compelling demonstration of the requirement for GOLPH3 protein function for cytokinesis. Our data demonstrate that the Drosophila homologue of human GOLPH3, plays an essential role in both contractile ring formation and vesicle trafficking. In most GOLPH3 mutant dividing spermatocytes Rho1 appeared diffuse (this study) and F-actin rings failed to assemble (this study and [28]). GOLPH3 mutation also prevented formation of robust Myosin II/Septin rings and disrupted central spindle assembly. GOLPH3 protein localized to the cleavage site during cytokinesis and biochemical analyses suggested interactions with Septins, Myosin and the centralspindlin component Pav.
Our observations of an enrichment of Sqh and Zipper at the cleavage site in the absence of a clear Rho1 ring are in agreement with a previous study on male germ cells depleted for anillin [48] and suggest that a pathway independent of Rho1 might be involved in the initial recruitment of Myosin II components in spermatocytes. Interestingly besides accumulating at the cleavage site, Rho1 is also enriched at the astral membranes in wild type dividing spermatocytes (arrowheads Figure 5B). It is then possible that a mechanism based on vesicle-mediated transport is implicated in maintaining Rho1 at the cleavage site during cytokinesis.
The requirement for GOLPH3 in spermatocyte cytokinesis is intimately connected to the ability of this protein to bind PI(4)P. A mutant version of GOLPH3 that was unable to associate with PI(4)P, failed to rescue the cytokinesis defects of sauz2217/Df mutants. GOLPH3 depended on PI(4)P for its recruitment to both the Golgi and the cleavage furrow. Moreover PI(4)P concentration at the cleavage furrow required wild type function of GOLPH3.
Human GOLPH3 was shown to bind to PI(4)P and recruit MYO18A to the Golgi [16]. It has been proposed that PI(4)P, GOLPH3 and MYO18A are required for tethering Golgi membranes to the actin cytoskeleton to stretch the Golgi and produce a tensile force that facilitates vesicle budding [16]. We showed that GOLPH3 is required to maintain the integrity of paired Golgi stacks in Drosophila interphase spermatocytes. Remarkably a proteomic analysis of PI(4)P - containing liposomes demonstrated that these organelles are enriched in Rab11, PI 4-kinase IIIβ and the actin regulatory factors Rac1 and Scar/WAVE [49]. It is then likely that GOLPH3 participates in a PI(4)P dependent recruitment of these actin regulatory factors that contribute to regulate pairing of the Golgi stack structure. We speculate that a module containing PI(4)P-GOLPH3-MyoII is involved in producing a tensile force at the cleavage furrow required to shape the actomyosin ring architecture during cleavage furrow constriction.
A possible model to illustrate how GOLPH3 function might be involved in cytokinesis is depicted in Figure 11. Because GOLPH3 protein binds to both PI(4)P and Rab11 and both PI(4)P-and Rab11 were delocalized in sauz2217 mutant spermatocytes, GOLPH3 protein might have a role in targeting PI(4)P - and Rab11-secretory vesicles to the cleavage furrow. PI(4)P is the substrate for PI(4)P 5-kinase for the synthesis of the PI(4,5)P2 lipid that is generated in the cleavage furrow and plays crucial roles during cytokinesis [10], [11]. Indeed imaging of dividing spermatocytes expressing PLCδ-PH-GFP, revealed enrichment of PI(4,5)P2 at the cleavage furrow in wild type but not in GOLPH3 mutants. PI(4,5)P2 is known to enhance actin polimerization by modulating activity of proteins that regulate F-actin dynamics including profilin and cofilin [50], [51]. Thus, failure to assemble F-actin rings in GOLPH3 mutants might be the consequence of a defective PI(4,5)P2 pool at the cleavage site. Recent data demonstrate that several cytokinesis players required in early stages including Rho, RhoGEF and centralspindlin subunit MgcRacGAP, also interact with phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] and/or PI(4)P and have plasma membrane interactions regulated by those phosphoinositides [52]–[55]. Defective PI(4,5)P2/PI(4)P concentration also affects Septin filament formation in the cleavage furrow. Septins form a gauze-like mesh tightly associated with plasma membranes [56]. Work in both mammalian and yeast cells demonstrated that Septins are able to interact in vitro with PIPs including P(4,5)P2, PI(3,4,5)P3 and PI(4)P through a polybasic domain [57]–[59]. Moreover Septin polymerization into filaments is enhanced by association with lipid bilayers [59], [60]. It has also been suggested that interaction of Septin filaments with phosphoinositides facilitates bending of plasma membranes and stabilizes the cleavage furrow [10]. In addition one of the mammalian septins (SEPT2) directly binds Nonmuscle Myo II and this interaction is required for Myo II full activation during cytokinesis [61].
Fig. 11. Diagram illustrating how GOLPH3 function might be involved in cytokinesis. 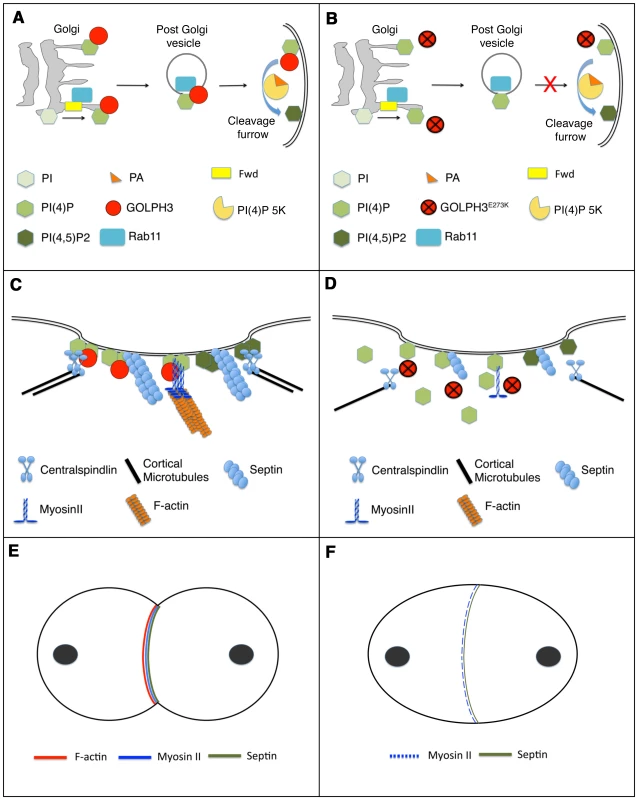
(A,C,E), wild type; (B, D, F) sauz2217/Df mutants. (A) Golgi membranes are depicted in grey. Synthesis of PI(4)P at the Golgi requires function of the PI-4 kinase Fwd. Fwd also recruits Rab11 protein to Golgi where it becomes associated with secretory vesicles containing PI(4)P. GOLPH3 localization to the Golgi depends on the PI(4)P phosphoinositide. Wild type function of GOLPH3 is required for recruitment of PI(4)P-and Rab11-containing vesicles to the cleavage furrow. PI(4)P is the immediate precursor of PI(4,5)P2 that is generated in the cleavage furrow by PI(4)P 5-kinase. Binding to phosphatidic acid (PA), GOLPH3 might also contribute to PI(4)P 5-kinase activation and membrane curvature during cytokinesis. (B) In sauz2217/Df mutant cells GOLPH3 protein fails to concentrate to Golgi membranes and to the cleavage furrow. PI(4)P- and Rab11-containing vesicles fail to accumulate at the cleavage furrow. PI(4)P- 5 kinase [PI(4)P 5K] activation might be also impaired in sauz2217/Df mutants. (C) PI(4)P-GOLPH3 and PI(4,5)P2 regulate interaction of centralspindlin, septins and actomyosin with plasma membrane during cytokinesis. For the sake of simplicity, each cytokinesis protein is depicted separately. (D) In sauz2217/Df mutant cells PI(4)P-GOLPH3 does not localize to the cleavage site. As a result, localization of centralspindlin at the equatorial cortex is not maintained, centralspindlin-associated microtubules fail to stably bundle at the midzone and F-actin ring assembly is impaired. sauz2217 mutations also affect stability of Myosin II and Septin filaments at the cleavage furrow. (E) In wild type telophase cells, rings containing F-actin, Myosin II and Septins constrict at the cleavage furrow during cytokinesis. (F) In sauz2217/Df mutant cells F-actin ring assembly fails, Myosin II and Septin rings are patchy or thin and fail to constrict. The defects in cytokinesis in GOLPH3 mutant spermatocytes also revealed malfunctioning of membrane trafficking pathways. Several studies have suggested an intimate connection between vesicle trafficking and actomyosin ring assembly/stability during cytokinesis. Syntaxin 1 is required for proper F-actin ring assembly in Drosophila S2 cells [62] and F-actin is targeted to the cleavage furrow of Drosophila postcellularized embryonic cells on endosomal vesicles [63]. In Dyctiostelium most clathrin null cells fail to undergo furrow ingression; in these cells MyoII is frequently associated with patches instead of forming a ring, a phenotype that is comparable to that of GOLPH3 [64], [65]. Our data demonstrate that GOLPH3 protein is required for proper localization of Clathrin and Rab5 in dividing spermatocytes and might function as a molecular partner for both proteins in Drosophila male meiotic cells. A potential molecular interaction between Clathrin and GOLPH3 is further suggested by the presence in the GOLPH3 amino acid sequence of a putative clathrin binding motif [66], “LLDLD” indicated by a blue line in Figure 1C.
Because GOLPH3 not only binds to PI(4)P but also to PI(3)P and PI(5)P in lipid binding assays, we believe GOLPH3 protein may be also involved in membrane trafficking pathways that depend on phosphoinositides other than PI(4)P during cytokinesis. Indeed recent studies have involved PI(3)P in cytokinesis; PI(3)P positive endosomes localize to the midzone of both fungal and mammalian cells [67], [68] and inhibition of PI(3)P production results in cytokinesis failures in HeLa cells [69]. Strikingly, we have found that GOLPH3 protein also binds to the glycerophospholipid phosphatidic acid (PA) in lipid binding assays. It has been shown that PA is important for plasma membrane curvature [70] and is also an activator of PI(4)P 5-kinase [71]. Thus we speculate that may also contribute to PI(4)P 5-kinase activation and membrane remodelling during cytokinesis, binding to PA (Figure 11).
Our study demonstrates that GOLPH3 is essential for normal development and viability in Drosophila. In addition the presence of tetraploid metaphases and defective contractile rings in larval neuroblasts depleted of GOLPH3 indicates a requirement for this protein for cytokinesis of somatic cells. Because cytokinesis failures have been associated with premalignant disease and cancer [72], [73] our findings suggest novel insight into molecular circuits that involve the oncogene GOLPH3 in cytokinesis.
Materials and Methods
Fly stocks and transgenes
The sauz2217 allele was from C. Zuker collection of ethylmethane sulfonate-induced lines that were screened for defects in spermatocyte cytokinesis [28]. The chromosomal deficiencies Df(2L)Exel6007, Df(2L)Exel7010, Df(3L)7C and the P element l(2)s5379 were obtained from the Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN). Flies carrying the fwd3 mutant allele and flies expressing GFP-Cog7 were previously described [12], [30]. UAS::GOLPH3-RNAi flies were from the Vienna Drosophila RNAi Collection (VDRC line 46150). Bam-GAL4 [74] was used to inactivate GOLPH3 in spermatocytes and to drive the expression of Chc-RFP from the UAS::Chc-RFP transgene. P{tubP-GAL4}LL7 (tub-GAL4) flies were from the Bloomington Drosophila Stock Center. Flies carrying both tub-GAL4 and tubGAL80ts transgenes (on third chromosome) were kindly provided by Timothy Megraw (University of Florida).
The following GFP/RFP transgenic lines were a gift from H.C. Chang (Pardue University): β2t::GFP-Rab5, β2t::GFP-Rab11, β2t::Clc-GFP, UAS::Chc-RFP [44], [45]. The β-tubulin-EGFP stock [33] was kindly provided by D. Glover (University of Cambridge, UK). The Sqh-GFP stock [38] was a gift from R. Karess (University of Paris, Paris France). The RFP-PH-FAPP and PLCδ-PH-GFP strains [15], [42] were kindly provided from J. A. Brill (University of Toronto).
Molecular cloning
To identify the mutation in the EMS-induced sauz2217 allele, genomic DNA corrisponding to CG7085/GOLPH3 gene was amplified by PCR and sequenced on both strands (Bio-Fab research service). DNA sequences from sauz2217/sauz2217 individuals were compared to sequences of the original Zuker-background chromosome to confirm the missense mutation. To generate the GFP-GOLPH3 fusion construct, the EGFP CDS was fused in frame to the aminoterminus of the full length cDNA corrisponding to CG7085 gene and cloned into the pCaSpeR4 under the control of α-tubulin promoter (pCaSpeR4-tubulin, see Ref. [75] for details on this vector). To generate the Vps35-mRFP transgene, DNA encoding Vps35-mRFP was PCR amplified from a Vps35-mRFP expression plasmid (gift from Viktor Korolchuk, [76]) and cloned into pCaSper4-tubulin. To generate the Vps35-FLAG transgene, the cDNA of Vps35 was cloned into pCaSper4-tubulin in frame with C-terminal 3XFLAG sequence. Transgenic flies were generated by P-element mediated germline transformation, performed by Bestgene Inc. (Chino Hills, CA). GFP-GOLPH3 was crossed into the sauz2217 mutant background to test for phenotypic rescue of male sterility and meiotic cytokinesis failure. S2 cultures were transfected with Vps35-FLAG and either GFP or GFP-GOLPH3 plasmids using Cellfectin II Reagent (Invitrogen) as recommended by the manufacturer. Site-directed mutagenesis, used to generate K167A/R170L substitutions, was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). DNA constructs obtained after mutagenesis were confirmed by DNA sequencing on both strands. To obtain gluthatione S-transferase (GST) fusion proteins, full-length cDNAs corrisponding to either Drosophila wild type GOLPH3, GOLPH3K167A/R170L or GOLPH3E273K, were cloned into a pGEX-6p-2 (GE Healthcare) at the GST C-terminus.
Protein expression and purification, antibody generation
GST-full-length Drosophila GOLPH3 (dGOLPH3) proteins and GST-full-length Drosophila Rab11 were expressed in BL21-CodonPlus [DE3] cells (Invitrogen) and purified using HiTrap affinity columns (GTtrap FF, and GSTtrap HP columns, GE Healthcare) operated with AKTA 900 Fast Protein Liquid Chromatography. Polyclonal antisera were raised against the purified GST-dGOLPH3 protein. Immunization was carried out at Agro-Bio Services (www.agro-bio.com); two rabbits and two mice were injected using standard procedures. The anti-GST-GOLPH3 antisera were first depleted of anti-GST antibodies and then affinity-purified against GST-GOLPH3.
Lipid binding assay
PIP Strips were purchased from Echelon (Echelon Biosciences) and were used following the manufacturer's instructions. Briefly, GST and GST fusion proteins, expressed and purified following the protocol described above, were incubated with PIP Strips at the concentration recommended by Echelon. Control GST-fusion proteins, for testing binding to a specific phosphoinositide, were purchased from Echelon and used at the recommended concentration. To detect the bound protein to PIP Strips, the following reagents were used: mouse anti-GST antibody, (1∶2000, Sigma #G-1160), anti mouse IgG conjugated with Horseradish peroxidase (HRP), (1∶5000, Sigma #A-9917); ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Western blotting and immunoprecipitation
For immunoblotting analysis of GOLPH3 protein, 40 adult testes from males of each genotype, were homogenized in 100 µl of Lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40, 1 mM PMSF, 1× Protease inhibitor Cocktail) using a Dounce homogenizer, for 40 minutes on ice. Cell lysates were cleared by centrifugation and protein concentration of supernatants was determined using the Bradford protein assay (Bio-Rad, Hercules CA). Equal amounts of protein extracts were analysed by SDS-PAGE and Western blotting. Samples were separated on Mini-protean TGX precast gels (Bio-Rad) and blotted to PVDF membranes (Bio-Rad). Membranes were blocked in Tris-buffered saline (Sigma-Aldrich) with 0,05% Tween-20 (TBST) containing 5% nonfat dry milk (Bio-Rad; Blotting GradeBlocker) for 3–4 hours at room temperature followed by incubation with primary and secondary antibodies diluited in TBST. For the coimmunoprecipitation (Co-IP) experiment shown in Figure 9A, 400 adult testes expressing GFP-GOLPH3 were homogenized in 500 µl of Lysis buffer (see above) for 40 minutes on ice. Lysate was clarified by centrifugation and protein concentration was quantified using the Bradford protein assay. 4% of lysate was retained as the “input”, the remainder was precleared with Sepharose-Protein G (Sigma) and divided into two. Fractions were incubated with either 2 µg of mouse monoclonal anti-GFP antibody (Roche) or 2 µg of non-specific mouse IgG (Sigma) for 2 hours at 4°C, followed by the addition of 40 µl of protein G-sepharose beads for further two hours. The beads were washed extensively with Lysis buffer, boiled in SDS sample buffer and separated by SDS-PAGE. To immunoprecipitate Drosophila GOLPH3 (dGOLPH3), testis lysate from 400 adult testes was precleared and divided into two. Fractions were incubated with either 5 µg of mouse anti-dGOLPH3 antibody S11047/1/56 or 5 µg of mouse pre immune serum (S11047/1), from the same animal before the immunization). After antibody incubation, immunoprecipitation was performed using the immunoprecipitation kit-Protein G (Roche) following the manufacturer's instructions. Other Co-IP experiments from testes expressing GFP and/or RFP-tagged proteins, were performed using the GFP/RFP trap-A kits and control binding beads purchased from ChromoTek (Planegg-Martinsried), following the protocol that was previously described [30]. Primary antibodies, used for immunoblotting were as follows: Rabbit anti-dGOLPH3, (1∶2500; G49139/77 this study), mouse monoclonal anti-α-Tubulin (1∶5000) (Sigma-Aldrich T6199), Rabbit anti-Zipper (1∶2000), gift from R.E. Karess; Rabbit anti-Pav (1∶2000), gift from D. Glover (University of Cambridge); Rabbit anti-Sep1 (1∶300) and Rabbit anti-Sep2 (1∶500) gift from J. Pringle (Stanford University, CA); Rabbit anti-Gio (1∶2000); Rat monoclonal anti-RFP (1∶1000), (ChromoTek, 5F8); mouse HRP anti-GFP (1∶1000), (Vector-Lab); mouse HRP anti-Flag (1∶1000), (clone M2 Sigma). HRP-conjugated secondary antibodies (GE Healthcare) were used at 1∶5000. After incubation with the antibodies, blots were washed in TBST and imaged using ECL detection kit (GE Healthcare).
GST pulldown assays
GST and GST-GOLPH3 proteins were expressed in bacteria and purified using Glutathione-Sepharose 4B beads (GE Healthcare) following the manufacturer's instructions. To obtain testis protein extracts, at least 600 adult testes were homogenized for 40 minutes on ice in 500 µl of Lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 0,5% NP-40, 1 mM EDTA) with the addition of Protease and phosphatase inhibitors cocktails (Roche), using a Dounce homogenizer. Procedures to obtain lysates from S2 cells, were previously described [75]. Lysates from either testes or S2 cells were cleared by centrifugation and protein concentration of the supernatants was determined by Bradford Assay (Bio-Rad). GST-pulldown was performed by incubating lysates with either GST or GST-GOLPH3 (at the appropriate concentration) bound to Glutathione-Sepharose 4B beads, at 4°C for two hours, with gentle rotation. The beads were washed three times in “wash buffer” (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, Protease and phosphatase inhibitors), boiled in SDS sample buffer, and separated by SDS-PAGE. The bound proteins were analysed by Western Blotting (see above). Before immunoblotting PVDF membranes were stained with Ponceau S (Sigma-Aldrich) as loading control.
Surface Plasmon Resonance experiments
For Surface Plasmon Resonance (SPR) experiments, following elution of the GST fusion proteins from Glutathione Sepharose, the eluate was dialysed extensively against Cleavage Buffer (20 mM TRIS pH 7.0, 150 mM NaCl, 1 mM DTT). Digestion was performed with 2 units PreScission Protease for each 100 µg of fusion protein in the eluate, for 4 hours at 4°C. Once digestion was complete, the sample was applied to Glutathione Sepharose pre-equilibrated in Cleavage Buffer, to remove the GST portion of the fusion protein and the PreScission Protease from the protein of interest. Cleaved proteins were recovered from the flowthrough, and concentrated using 10 K Amicon Ultra-15 Centrifugal Filter Units. SPR experiments were carried out using a BIACORE X system (BIAcore AB, Uppsala, Sweden). The sensor chip (CM5, Biacore AB) was activated chemically by a 35 µl injection of a 1∶1 mixture of N-ethyl-N′-(3-(diethylaminopropyl) carbodiimide (200 mM) and N-hydroxysuccinimide (50 mM) at a flow rate of 5 µl/min. GOLPH3 was immobilized on the activated sensor chip via amine coupling. The immobilization was carried out in 20 mM sodium acetate at pH 5.2; the remaining ester groups were blocked by injecting 1 M ethanolamine hydrochloride (35 µl). This procedure ensures immobilization of GOLPH3 principally via the N-terminus. As a control, the sensor chip was treated as described above in the absence of GOLPH3. The level of immobilization of GOLPH3 was about 1200 RU. Rab11 (in 10 mM Hepes pH 7.4, 150 mM NaCl +0,005% surfactant P20) was injected on the sensor chip at a constant flow (30 µl/min). The interaction of immobilized GOLPH3 ligand with the analyte was detected through mass concentration-dependent changes in the refractive index on the sensor chip surface, expressed as resonance units (RU). The increase in RU relative to baseline indicates complex formation; the plateau region represents the steady-state phase of the interaction, whereas the decrease in RU represents dissociation of Rab11 from immobilized GOLPH3 after injection of buffer. A response change of 1000 RU typically corresponds to a change in the protein concentration on the sensor chip of 1 ng/mm2. The sensorgrams were analyzed by BIAevaluation software.
Microscopy image acquisition and immunofluorescence
Cytological preparations were made with brains from third instar larvae and testes from either third instar larvae or adults. sauz2217/Df(2L)Exel7010 mutants were used in all the immunofluorescence experiments for GOLPH3 testes. To deplete GOLPH3 in larval neuroblasts UAS::GOLPH3-RNAi flies were crossed to tub-GAL4 tubGAL80ts flies. Progeny was kept at 18°C until the third instar stage and subsequently transferred to 29°C for 18 h before the analysis of the cytokinetic phenotype.
To visualize α Tubulin with either Clc-GFP or RFP-PH-FAPP, Lva with GFP-Cog7 or to stain F-actin with Rhodamine-phalloidin (Invitrogen), larval testes were fixed in 4% formaldehyde as previously described [30]. To visualize α Tubulin and GOLPH3, larval testes or brains were fixed and stained following the protocol described by Starr and coauthors [77]. Briefly testes and brains were dissected in 0.7% NaCl and transferred into a drop of PHEMT (60 mM Pipes, 25 mM HEPES, pH 7.0, 10 mM EGTA, 4 mM MgSO4, 0.5% Triton X-100) for two minutes. Testes and brains were then transferred to 4 µl of PHEMT containing 3.7% formaldehyde on a coverslip, gently squashed on an inverted slide and fixed for 10 minutes before immersing in liquid nitrogen. After removing the coverslip, preparations were immersed in methanol for 20 minutes (−20°C) and rehydrated in PBS containing 0.1% Triton for 20 minutes at room temperature. For immunostaining with other antibodies, preparations were fixed using 3.7% formaldehyde in PBS and then squashed in 60% acetic acid as previously described [43]. Monoclonal antibodies were used to stain α-Tubulin (1∶300; Sigma-Aldrich, T6199), RFP (1∶200; ChromoTek, 5F8); Rho1 (1∶100, p1D9 [78]; Developmental Studies Hybridoma Bank, University of Iowa), and Polo (1∶20; Mouse anti-Polo M294; [35]) gift from D. Glover. Polyclonal antibodies were as follows: Rabbit anti-dGOLPH3, (1∶2500; G49139/77 this study), rabbit anti-Zipper (1∶400; [38]), gift from R.E. Karess, rabbit anti-Lva (1∶500; [31]), gift from O. Papoulas (University of Texas); rabbit anti-Rab11 (1∶30; [79]), gift from M. Gonzalez-Gaitan (Max Planck Institute of Molecular cell Biology, Dresden) anti-Pav (1∶200; [37]) and anti-RacGAP50C (1∶200; [36]) kindly provided by D. Glover; anti-Feo (1∶200; [34]), gift from F. Vernì (Università Sapienza, Roma); anti-Sep1 and anti Sep2 (1∶30; [40], ). Alexa 555-conjugated anti rabbit IgG (Molecular Probes) and rhodamine/FITC-conjugated anti-mouse IgG (Jackson Immunoresearch), were used as secondary antibodies (1∶250 and 1∶20 respectively). Preparations fixed to visualize Clc-GFP together with tubulin, were first incubated with monoclonal anti-α–Tubulin (see above) followed by incubation with GFP-Booster (ChromoTek) diluited 1∶100 in PBS containing rhodamine-conjugated anti-mouse IgG. To visualize α-Tubulin together with F-actin, preparations were first incubated with anti α-Tubulin followed by incubation with Rhodamine-phalloidin (Invitrogen) diluited 20 U/ml in PBS containing FITC-conjugated anti-mouse IgG. In all cases preparations were mounted in Vectashield medium with DAPI (Vector labs). Images were captured with a charged-coupled device (CCD camera, Photometrics Coolsnap HQ), connected to a Zeiss Axioplan, epifluorescence microscope, equipped with an HBO 100-W mercury lamp, and 40× or 100× objectives as previously described [13], [43]. The number of Golgi stacks per cell was counted manually, by analyzing images of G2 spermatocytes at S5 stage, stained for tubulin, Lva and DNA. In total, 32 cells were examined for both wild type and sauz2217/Df(2L)Exel7010, in images captured in 4 duplicated experiments (n = 8 representative cells per each experiment). The size of Golgi stacks in spermatocytes was analysed using image J software (NIH; http://rsbweb.nih.gov/ij/). In total, 110 randomly selected Golgi were measured for both wild type and sauz2217/Df(2L)Exel7010 using Image J by manual demarcation with a limiting polygon and calculation of its area (n = 4 duplicated experiments). Each measure was normalized to control average size (set at 1). To visualize mitotic chromosomes, larval brains were dissected in NaCl 0.7%, treated with hypotonic solution for 7 minutes and fixed in 45% acetic acid. Preparations were then immersed in liquid nitrogen, processed as per [28] and mounted in Vectashield medium with DAPI (Vector Laboratories, Burlingame, CA).
Live imaging
Larval testes were prepared and imaged for time lapse as previously described [43]. Spermatocytes were examined with a Zeiss Axiovert 20 microscope equipped with a 63×, 1. 25 NA and a 63×, 1.4 NA objectives and a filter wheel combination (Chroma Technology Corp.). Images were collected at 1 minute time intervals with a CoolSnap HQ camera (Photometrics) controlled through a Metamorph software (Universal imaging); nine to eleven fluorescent optical sections were captured at 1-µm z steps and maximally projected using the Metamorph software.
Supporting Information
Zdroje
1. GreenRA, PaluchE, OegemaK (2012) Cytokinesis in animal cells. Annu Rev Cell Dev Biol 28 : 29–58.
2. D'AvinoPP (2009) How to scaffold the contractile ring for a safe cytokinesis - lessons from Anillin-related proteins. J Cell Sci 122 : 1071–1079.
3. MostowyS, CossartP (2012) Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol 13 : 183–194.
4. D'AvinoPP, SavoianMS, GloverDM (2005) Cleavage furrow formation and ingression during animal cytokinesis: a microtubule legacy. J Cell Sci 118 : 1549–1558.
5. PieknyA, WernerM, GlotzerM (2005) Cytokinesis: welcome to the Rho zone. Trends Cell Biol 15 : 651–658.
6. GlotzerM (2009) The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol 10 : 9–20.
7. McKayHF, BurgessDR (2011) ‘Life is a highway’: membrane trafficking during cytokinesis. Traffic 12 : 247–251.
8. NetoH, CollinsLL, GouldGW (2011) Vesicle trafficking and membrane remodelling in cytokinesis. Biochem J 437 : 13–24.
9. GiansantiMG, FullerMT (2012) What spermatocytes tell us about the mechanims underlying cytokinesis. Cytoskeleton (Hoboken) 69 : 869–881.
10. BrillJA, WongR, WildeA (2011) Phosphoinositide function in cytokinesis. Curr Biol 21: R930–R934.
11. EchardA (2012) Phosphoinositides and cytokinesis: the “PIP” of the iceberg. Cytoskeleton (Hoboken) 69 : 893–912.
12. BrillJA, HimeGR, Scharer-SchukszM, FullerMT (2000) A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development 127 : 3855–3864.
13. GiansantiMG, BonaccorsiS, KurekR, FarkasRM, DimitriP, et al. (2006) The class I PITP giotto is required for Drosophila cytokinesis. Curr Biol 16 : 195–201.
14. GattMK, GloverDM (2006) The Drosophila phosphatidylinositol transfer protein encoded by vibrator is essential to maintain cleavage-furrow ingression in cytokinesis. J Cell Sci 119 : 2225–2235.
15. PolevoyG, WeiHC, WongR, SzentpeteryZ, KimYJ, et al. (2009) Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J Cell Biol 187 : 847–858.
16. DippoldHC, NgMN, Farber-KatzSE, LeeSK, KerrML, et al. (2009) GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell 139 : 337–351.
17. WoodCS, SchmitzKR, BessmanNJ, SettyTG, FergusonKM, et al. (2009) PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J Cell Biol 87 : 967–975.
18. ScottKL, KabbarahO, LiangMC, IvanovaE, AnagnostouV, et al. (2009) GOLPH3 modulates mTOR signaling and rapamycin sensitivity in cancer. Nature 459 : 1085–1090.
19. LiXY, LiuW, ChenSF, ZhangLQ, LiXG, et al. (2011) Expression of the Golgi phosphoprotein-3 gene in human gliomas: a pilot study. J Neurooncol 105 : 159–163.
20. LiH, GuoL, ChenSW, ZhaoXH, ZhuangSM, et al. (2012) GOLPH3 overexpression correlates with tumor progression and poor prognosis in patients with clinically N0 oral tongue cancer. J Transl Med 10 : 168 doi:10.1186/1479-5876-10-168
21. KunigouO, NagaoH, KawabataN, IshidouY, NaganoS, et al. (2012) Role of GOLPH3 and GOLPH3L in the proliferation of human rhabdomyosarcoma. Oncol Rep 26 : 1337–1342.
22. HuBS, HuH, ZhuCY, GuYL, LiJP (2012) Overexpression of GOLPH3 is associated with poor clinical outcome in gastric cancer. Tumour Biol 34 : 515–520.
23. HuaX, YuL, PanW, HuangX, LiaoZ, et al. (2012) Increased expression of Golgi phosphoprotein-3 is associated with tumor aggressiveness and poor prognosis of prostate cancer. Diagn Pathol 7 : 127 doi:10.1186/1746-1596-7-127
24. WangJH, ChenXT, WenZS, ZhengM, DengJM, et al. (2012) High expression of GOLPH3 in esophageal squamous cell carcinoma correlates with poor prognosis. PLos One 7: e45622 doi:10.1371/journal.pone.0045622
25. ZengZ, LinH, ZhaoX, LiuG, WangX, et al. (2012) Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin Cancer Res 18 : 4059–4069.
26. ZhouJ, XuT, QinR, YanY, ChenC, et al. (2012) Overexpression of Golgi phosphoprotein-3 (GOLPH3) in glioblastoma multiforme is associated with worse prognosis. J Neurooncol 110 : 195–203.
27. ZhouX, ZhanW, BianW, HuaL, ShiQ, et al. (2013) GOLPH3 regulates the migration and invasion of glioma cells though RhoA. Biochem Biophys Res Commun 433 : 338–344.
28. GiansantiMG, FarkasRM, BonaccorsiS, LindsleyDL, WakimotoBT, et al. (2004) Genetic Dissection of Meiotic Cytokinesis in Drosophila. Mol Biol Cell 15 : 2509–2522.
29. CenciG, BonaccorsiS, PisanoC, VerniF, GattiM (1994) Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatognesis. J Cell Sci 107 : 3521–3534.
30. BelloniG, SechiS, RiparbelliMG, FullerMT, CallainiG, et al. (2012) Mutations in Cog7 affect Golgi structure, meiotic cytokinesis and sperm development during Drosophila spermatogenesis. J Cell Sci 125 : 5441–5452.
31. SissonJC, FieldC, VenturaR, RoyouA, SullivanW (2000) Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J Cell Biol 151 : 905–918.
32. KondylisV, SpoorendonkKM, HerpersB, Friggi-GrelinF, RabouilleC (2007) The golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev Cell 12 : 901–915.
33. InoueYH, SavoianMS, SuzukiT, MáthéE, YamamotoMT, et al. (2004) Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J Cell Biol 166 : 49–60.
34. VernìF, SommaMP, GunsalusKC, BonaccorsiS, BelloniG, et al. (2004) Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr Biol 14 : 1569–1575.
35. CarmenaM, RiparbelliMG, MinestriniG, TavaresAM, AdamsR, et al. (1998) Drosophila polo kinase is required for cytokinesis. J Cell Biol 143 : 659–671.
36. D'AvinoPP, SavoianMS, CapalboL, GloverDM (2006) RacGAP50C is sufficient to signal cleavage furrow formation during cytokinesis. J Cell Sci 119 : 4402–4408.
37. AdamsRR, TavaresAA, SalzbergA, BellenHJ, GloverDM (1998) Pavarotti encodes a kinesin–like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev 12 : 1483–1494.
38. RoyouA, SullivanW, KaressR (2002) Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J Cell Biol 158 : 127–137.
39. MostowyS, CossartP (2012) Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol 13 : 183–194.
40. FaresH, PeiferM, PringleJR (1995) Localization and possible function of Drosophila Septins. Mol Biol Cell 6 : 1843–1859.
41. FieldCM, al-AwarO, RosenblattJ, WongML, AlbertsB, et al. (1996) A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol 133 : 605–616.
42. WongR, HadjiyanniI, WeiHC, PolevoyG, McBrideR, et al. (2005) PIP2 hydrolysis and calcium release are required for cytokinesis in Drosophila spermatocytes. Curr Biol 15 : 1401–1406.
43. GiansantiMG, BelloniG, GattiM (2007) Rab11 is required for membrane trafficking and actomyosin ring constriction in meiotic cytokinesis of Drosophila males. Mol Biol Cell 18 : 5034–5047.
44. ZhouX, FabianL, BayraktarJL, DingHM, BrillJA, et al. (2011) Auxilin is required for formation of Golgi-derived clathrin-coated vesicles during Drosophila spermatogenesis. Development 138 : 1111–1120.
45. AbdallahAM, ZhouX, KimC, ShahKK, HodgenC, et al. (2013) Activated Cdc42 kinase regulates Dock localization in male germ cells during Drosophila spermatogenesis. Dev Biol 378 : 141–153.
46. ZerialM, McBrideH (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2 : 107–117.
47. McGuireSE, PhuongPT, OsbornAJ, MatsumotoK, DavisRL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302 : 1765–1768.
48. GoldbachP, WongR, BeiseN, SarpalR, TrimbleWS, et al. (2010) Stabilization of the actomyosin ring enables spermatocyte cytokinesis in Drosophila. Mol Biol Cell 21 : 1482–1493.
49. BaustT, CzupallaC, CrauseE, Bourel-BonnetL, HoflackB (2006) Proteomic analysis of adaptor protein 1A coats selectively assembled on liposomes. Proc Natl Acad Sci U S A 103 : 3159–3164.
50. LoganMR, MandatoCA (2006) Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol Cell 98 : 377–388.
51. SaarikangasJ, ZhaoH, LappalainenP (2010) Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev 90 : 259–289.
52. YoshidaS, BartoliniS, PellmanD (2009) Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev 23 : 810–823.
53. SuKC, TakakiT, PetronczkiM (2011) Targeting of the RhoGEF Ect2 to the equatorial membrane controls clavage furrow formation during cytokinesis. Dev Cell 21 : 1104–1115.
54. FrenetteP, HainesE, LoloyanM, KinalM, PakarianP, et al. (2012) An anillin-Ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PloS One 7: e34888 doi:10.1371/journal.pone.0034888
55. LekomtsevS, SuKC, PyeVE, BlightK, SundaramoorthyS, et al. (2012) Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature 492 : 276–279.
56. OhY, BiE (2011) Septin structure and function in yeast and beyond. Trends Cell Biol 21 : 141–148.
57. ZhangJ, KongC, XieH, McPhersonPS, GrinsteinS, et al. (1999) Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol 9 : 1458–1467.
58. CasamayorA, SnyderM (2003) Molecular dissectin of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol Cell Biol 23 : 2762–2777.
59. BertinA, McMurrayMA, ThaiL, GarciaG3rd, VotinV, et al. (2010) Phosphatidylinositol-4,5-biphospate promotes budding yeast septin filament assembly and organization. J Mol Biol 404 : 711–731.
60. Tanaka-TakiguchiY, KinoshitaM, TakiguchiK (2009) Septin-mediated uniform bracing of phospholipid membranes. Curr Biol 19 : 140–145.
61. JooE, SurkaMC, TrimbleWS (2007) Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev Cell 13 : 677–690.
62. SommaMP, FasuloB, CenciG, CundariE, GattiM (2002) Molecular dissection of cytokinesis by RNA interference in Drosophila cutured cells. Mol Biol Cell 13 : 2448–2460.
63. AlbertsonR, CaoJ, HsiehTS, SullivanW (2008) Vesicles and actin are targeted to the cleavage furrow via furrow microtubules and the central spindle. J Cell Biol 181 : 777–790.
64. NiswongerML, O'HalloranTJ (1997) A novel role for clathrin in cytokinesis. Proc Natl Acad Sci U S A 94 : 8575–8578.
65. GeraldNJ, DamerCK, O'HalloranTJ, De LozanneA (2001) Cytokinesis failure in clathrin-minus cells is caused by cleavage furrow instability. Cell Motil Cytoskeleton 48 : 213–223.
66. ter HaarET, HarrisonSC, KirchhausenT (2000) Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci U S A 97 : 1096–1100.
67. SagonaAP, NezisIP, PedersenNM, LiestølK, PoultonJ, et al. (2010) PtdIns(3)P controls cytokinesis through KIF13A-mediated recrutment of FYVE-CENT to the midbody. Nat Cell Biol 12 : 362–371.
68. SchinkKO, BolkerM (2009) Coordination of cytokinesis and cell separation by endosomal targeting of a Cdc42-specific guanine nucleotide exchange factor in Ustilago maydis. Mol Biol Cell 20 : 1081–1088.
69. MontagnacG, EchardA, ChavrierP (2008) Endocytic traffic in animal cell cytokinesis. Curr Opin Cell Biol 20 : 454–461.
70. KooljmanEE, KingKE, GangodaM, GerickeA (2005) Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry 44 : 2097–2102.
71. MoritzA, De GraanPN, GispenWH, WitzKW (1992) Phosphatidic acid is a specific activator of phosphatidylinositol-4-phosphate kinase. J Biol Chem 267 : 7207–7210.
72. FujiwaraT, BandiM, NittaM, IvanovaEV, BronsonRT, et al. (2005) Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437 : 1043–1047.
73. StorchovaZ, KufferC (2008) The consequences of tetraploidy and aneuploidy. J Cell Sci 121 : 3859–3866.
74. ChenD, McKearinDM (2003) A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130 : 1159–1170.
75. RaffaGD, RaimondoD, SorinoC, CugusiS, CenciG, et al. (2010) Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev 24 : 1596–1601.
76. KorolchukVI, SchützMM, Gómez-LlorenteC, RochaJ, LansuNR, et al. (2007) Drosophila Vps35 function is necessary for normal endocytic trafficking and actin cytoskeleton organization. J Cell Sci 120 : 4367–4376.
77. StarrDA, WilliamsBC, HaysTS, GoldbergML (1998) Zw10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol 142 : 763–774.
78. MagieCR, Pinto-SantiniD, ParkhurstSM (2002) Rho1 interacts with p120ctn and alpha-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development 129 : 3771–3782.
79. EmeryG, HuttererA, BerdnikD, MayerB, Wirtz-PeitzF, et al. (2005) Asymmetryc Rab11 endosomes regulate delata recycling and specify cell fate in the Drosophila nervous system. Cell 122 : 763–773.
Štítky
Genetika Reprodukční medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



