-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genes That Bias Mendelian Segregation
Chromosome segregation during meiosis ensures that paternal and maternal chromosomes are equally transmitted to the progeny. Meiotic Drive Elements (MDs) are known to distort this 1∶1 ratio in many animal, plant, and fungal species by killing the gametes not carrying them. Most of the known MDs are complex genetic loci with separate genes for the killing activity and the resistance to said killing. Here, we report in a model fungus on two genes endowed with MD properties previously unreported. Both genes produce a single polypeptide and confer both killing and resistance. They exert their effect irrespective of their position in the genome. They can cross species barriers and promote bias in segregation in other species. As related genes are frequently observed in fungal genomes, we propose that they are representative of a novel kind of selfish genes that propagate by distorting the Mendel laws of segregation.
Published in the journal: . PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004387
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004387Summary
Chromosome segregation during meiosis ensures that paternal and maternal chromosomes are equally transmitted to the progeny. Meiotic Drive Elements (MDs) are known to distort this 1∶1 ratio in many animal, plant, and fungal species by killing the gametes not carrying them. Most of the known MDs are complex genetic loci with separate genes for the killing activity and the resistance to said killing. Here, we report in a model fungus on two genes endowed with MD properties previously unreported. Both genes produce a single polypeptide and confer both killing and resistance. They exert their effect irrespective of their position in the genome. They can cross species barriers and promote bias in segregation in other species. As related genes are frequently observed in fungal genomes, we propose that they are representative of a novel kind of selfish genes that propagate by distorting the Mendel laws of segregation.
Introduction
In many organisms, genetic factors, called Meiotic Drive Elements (MDs), have found ways to break Mendel's laws of heredity. MDs skew the expected 1∶1 ratio in their favor and are thus overrepresented in the progeny after meiosis. They have been observed in metazoans, plants and fungi [1]. They may play a critical role in population behavior, leading to sex ratio distortion and thus decreasing population size [2]. Additionally, fitness can also be altered by MD factors if they are genetically linked to alleles that confer deleterious traits. Investigation of “Segregation Distorter” in Drosophila [3], [4], “t-haplotypes” in mice [5], [6], [7] and the S5 locus in rice [8], [9] has showed that MDs are composed of at least two linked genes, the distorter that acts as a toxin by disrupting the formation of gametes, and the responder that acts as an antitoxin that protects from the deleterious distorter effects. These genes are generally embedded in large genomic regions devoid of recombination and containing numerous loci that affect positively or negatively meiotic distortion [3], [5]. In mouse and Drosophila, the distorters and responders originate from cellular genes that have acquired new functions [10], [11].
In fungi, MDs are known as Spore killers (Sks) [12]. In Neurospora, three Sks have been discovered [13]. They appear to follow rules similar to other known MDs, as they carry two genetically dissociable distorter and responder loci, embedded in a large region devoid of recombination [13]. The molecular basis for killing is unknown in Sk-2 and Sk-3, as the involved distorters have not yet been isolated. However, a resistance gene (responder) to Sk-2 and Sk-3 has been identified [14]. This gene, NCU09151, is of unknown function and restricted to species closely related to N. crassa (e.g., Sordaria macrospora). In the pseudo-homothallic fungus P. anserina, at least eight Sks have been observed [15]. One of them has been associated with deleterious effects during ascospore formation of the Het-s prion [16]. However, several additional Sks remain uncharacterized [15], including the first one discovered in fungi [17]. Here, we identify the distorters and responders of two P. anserina Sks. Unlike previously known MDs, both activities for these Sks are carried out by single genes acting autonomously irrespective of their position in the genome or of the fungal species and whose homologues are prevalent in many fungi.
Results
Identification of Spok1
In crosses between the S and T strains of P. anserina, only half the progeny reaches maturity in 90% of the asci (spore sacs; n>200), while all ascospores reach maturity in control S×S and T×T crosses (Fig. 1A). Back cross of the progeny retrieved from S×T crosses to the parental S and T strains showed 90% 2-spored and 100% 4-spored asci, respectively, indicating the presence of at least one Sk in strain T to which strain S is sensitive. It likely corresponds to the first described Sk in fungi [15], [17]. In our hands, this Sk triggers death in nine out of ten asci, and hence harbors a first division segregation (FDS) of 90%, suggesting a close linkage to a centromere (Fig. 1B). During P. anserina genome assembly verification by microsatellite genotyping of the progeny from a cross between the S and T strains [18], we observed a strong bias towards the transmission of the T centromere region of chromosome 5 in 50 progeny (Fig. S1) and this was not the case for the other chromosomes, pinpointing the Sk locus close to the centromere of chromosome 5.
Fig. 1. Structure of P. anserina asci. 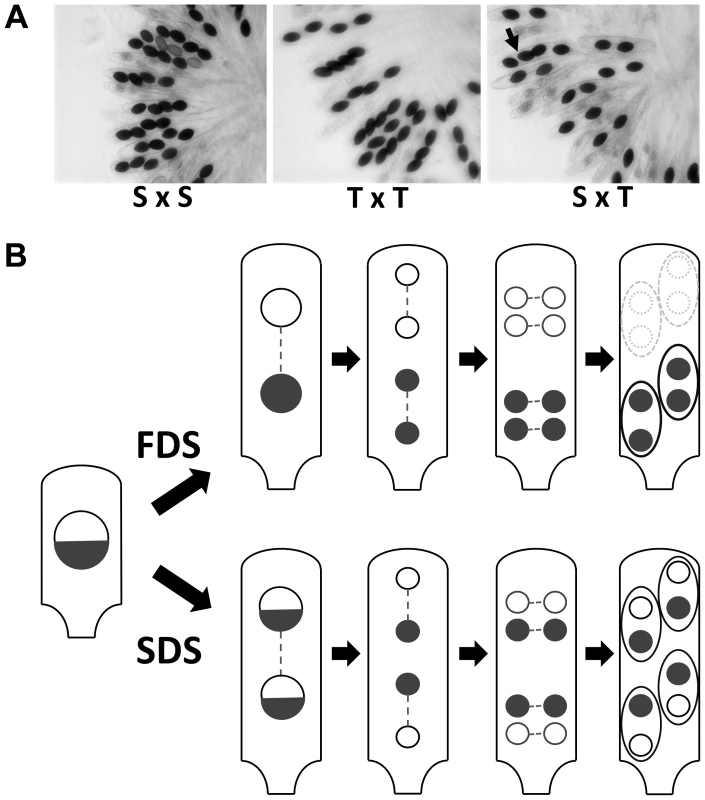
(A) S×S or T×T crosses yield 100% four binucleated ascospores per ascus. In S×T cross, 2-spored asci are indicative of spore killing. The Sk locus is linked to the centromere since only 10% of the asci have four ascospores (arrow in S×T). (B) Schematic representation of P. anserina FDS and SDS asci with Sk. FDS: no crossover between the Sk locus and the centromere results in First Division Segregation of Sk, triggering death of the two ascospores lacking the Sk locus. SDS: a crossover between the Sk locus and the centromere results in Second Division Segregation of Sk, generating four surviving heterokaryotic ascospores. Proportion of FDS and SDS asci depends upon the frequency of crossover and thus upon the genetic distance between Sk and the centromere. The 90% of 2-spored asci in S×T cross (A) is indicative of a close linkage of the Sk with the centromere. To narrow the region containing the Sk, we backcrossed a progeny (ST1) of the S×T cross twenty times to strain S, selecting each time for FDS asci. At each generation, we observed the Sk effect (i.e., 90% of 2-spored asci) and thus the final backcrossed strain (SKT20, Table S1) had a genome coming mostly from strain S, except for a small region containing the Sk locus from the T strain. Molecular analysis of polymorphic markers showed that SKT20 had its entire chromosome 5 coming from strain S except for a small region of 70 kb bordered by markers 5PGK and 5PGM (Table S2, Fig. 2). 5PGK differs between S and T for several SNPs that can be detected by sequence analysis. 5PGM differs by the presence of a 15 kb-region present in strain T and absent in strain S. The final backcrossed strain, SKT20, had the 5PGK and 5PGM markers of strain S. This strain displayed 90% 2-spored asci when crossed with S and 100% 4-spored asci when crossed with T, as expected if it contains the distorter and responder of the Sk from strain T (Table 1, Figure 3A). Final identification of the Sk was made by nested deletions in SKT20. A 6 kb-region located between coding sequences (CDS) Pa_5_4070 and Pa_5_4075 was found to be responsible for meiotic drive. This region encompassed a retroposon LTR and a single predicted gene, which we called Spok1 (Spore killer 1, Fig. 2, Fig. S2). Spok1 is absent in strain S, which has transposable elements at the same chromosomal location (i.e., between Pa_5_4070 and Pa_5_4075; Fig. 2, Fig. S2).
Fig. 2. Spok1 and Spok2 DNA regions. 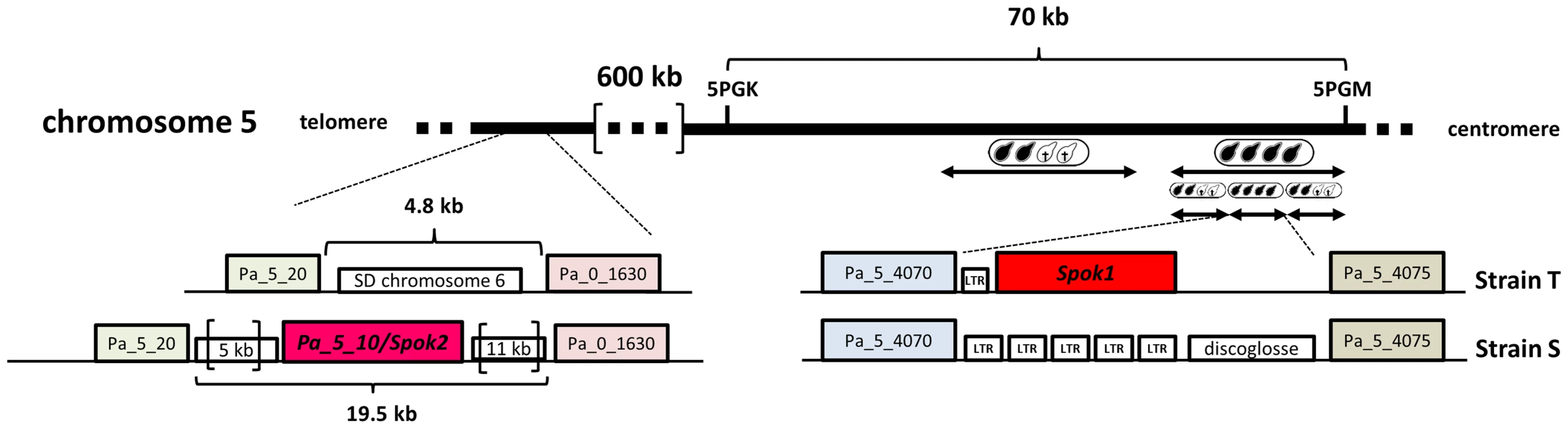
Double arrows define the sequences deleted to identify Spok1. 4-spored asci identify the deletions that abolish spore killing. LTR are Long Terminal Repeats of the crapaud retroposon and discoglosse is a DNA transposon [18]. SD: Segmental Duplication. 5 kb and 11 kb regions bordering Spok2 contain inactivated transposons. Pa_x_xxxx are P. anserina predicted CDS. Fig. 3. Rosettes of asci in indicated crosses. 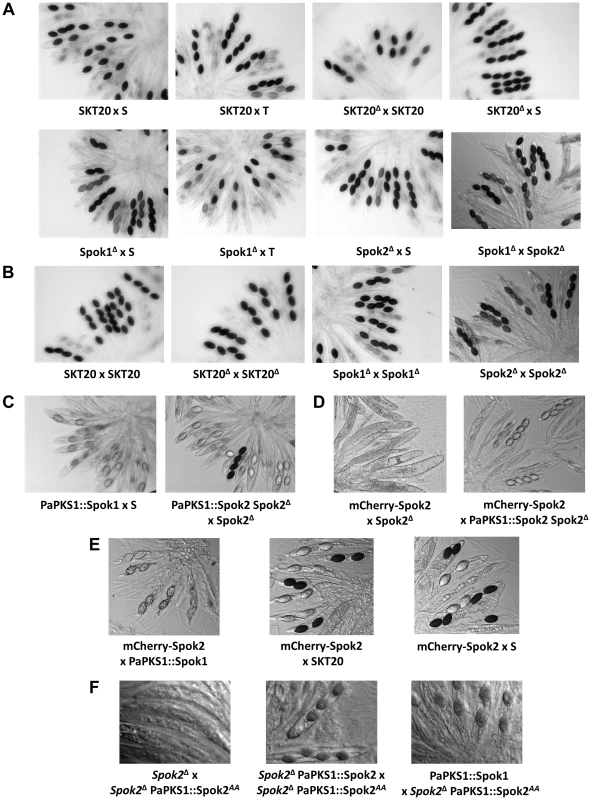
(A) Spok1 and Spok2 are Sks with 90% and 40% FDS, respectively; (B) Spok1 and Spok2 have no role in ascospore differentiation; (C) Spok1 and Spok2 are functional at the PaPKS1 locus; the asci with four dark spores results from SDS of the PaPKS1 locus; (D) complex phenotypes of mCherry-Spok2; (E) Spok1 and Spok2 confer resistance to mCherry-Spok2 spore killing; (F) complex phenotypes of Spok2AA. See text and table S1 for full genotype of strains. Tab. 1. Progeny analysis of Spok crosses. 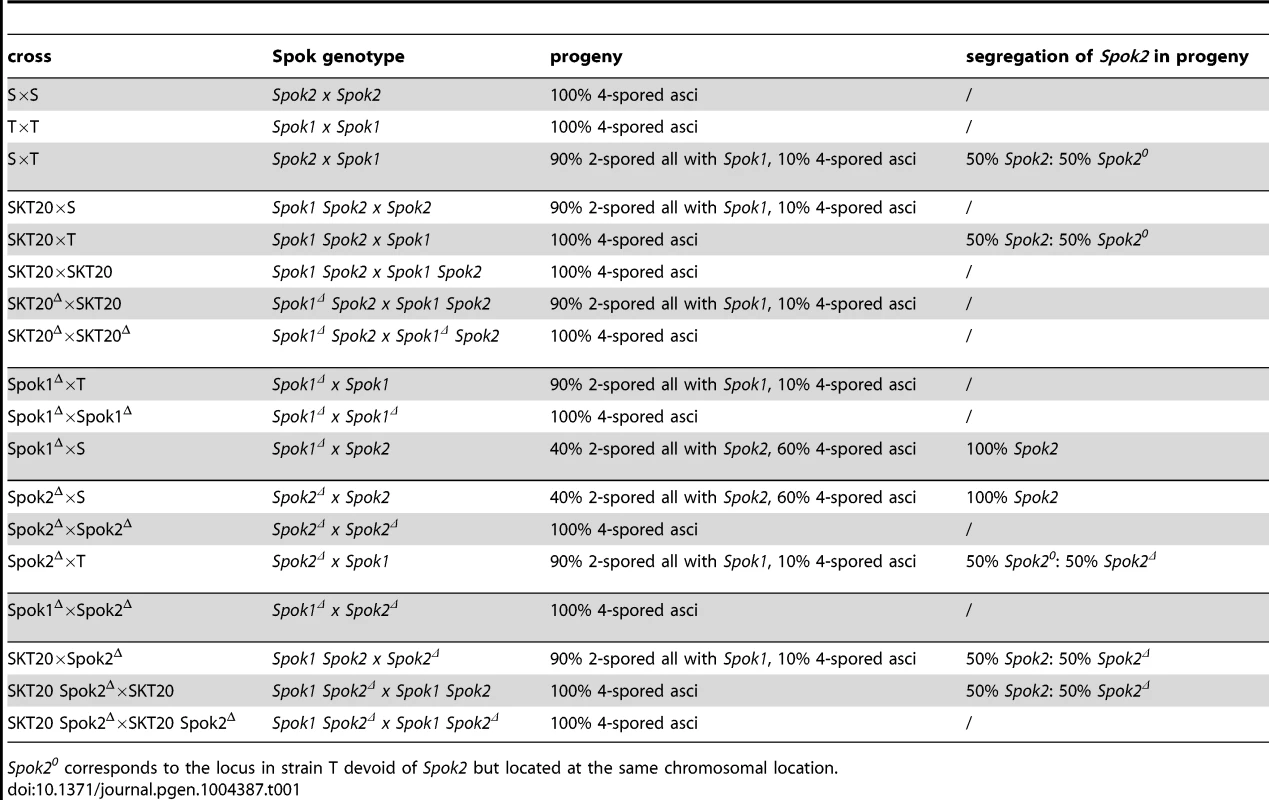
Spok20 corresponds to the locus in strain T devoid of Spok2 but located at the same chromosomal location. To validate that Spok1, and not an additional non-annotated gene present in the 6-kb region, was necessary and sufficient for spore killing, we first replaced solely its coding sequence with a hygromycin B-resistance marker (Fig. S2, see Materials and Methods for gene deletions and Fig. S3 for Southern Blot validations). The SKT20Δ strain had thus a genotype identical to SKT20, except that the Spok1 coding sequence was replaced. We observed the production of 100% 4-spored asci in crosses of SKT20Δ with S and 90% 2-spored asci in crosses of SKT20Δ with SKT20 (Fig. 3A), showing that Spok1 was responsible for both killing and resistance. Secondly, we inserted Spok1 in the PaPKS1 gene of strain S. PaPKS1 is located at the centromere of chromosome 2 and segregates with 99% FDS [19]. It encodes a polyketide synthase that controls the first step of melanin biosynthesis and PaPKS1 mutants are devoid of pigment at all stages of their life cycle [19]. This allows for easy screening of colorless recombinant transgenic strains carrying an insertion in the PaPKS1 gene. Additionally, ascospores carrying Spok1 should be devoid of pigment enabling their easy identification in crosses. Strains carrying Spok1 at PaPKS1 (PaPKS1::Spok1) yielded 99% of unpigmented 2-spored asci in crosses with the S strain (Fig. 3C). The 1% 4-spored asci recovered resulted from the expected second division segregation (SDS) of the PaPKS1::Spok1 locus. Thus, insertion of solely Spok1 into the PaPKS1 gene of strain S was sufficient to trigger both spore killing and resistance.
Phenotypic analysis of the whole life cycle of SKT20 and SKT20Δ (i.e., ascospore maturation and germination, mycelium growth, heterokaryon incompatibility and sexual reproduction including differentiation of fruiting body) showed no defects other than a lack of Sk activity in SKT20Δ, with SKT20×SKT20 and SKT20Δ×SKT20Δ homozygous crosses produced 100% 4-spored asci (Table 1, Fig. 3B). Sequence analysis of the 734 amino acid-long Spok1 did not reveal any functional domain (Fig. 4). However, PSORT [20] predicted a nuclear localization.
Fig. 4. Comparison of Spok1 and Spok2 protein sequences. 
The amino acids corresponding to the codons changed in the Spok2AA allele are underlined. Differences between Spok1 and Spok2 are shaded in grey. Strain S contains Spok2, a paralogue of Spok1, which also causes spore killing
Surprisingly, when we inactivated Spok1 in parental strain T (Spok1Δ strain), this did not result in the expected absence of 2-spored asci in Spok1Δ×S crosses, as 40% of the asci were 2-spored (Table 1, Fig. 3A). However, the strain yielded 100% 4-spored asci when crossed with itself and 90% 2-spored asci when crossed with strain T, as expected (Table 1, Fig. 3A and 3B). Strain S, but not strain T, carries Pa_5_10, a CDS with 87% amino-acid identity to Spok1 (Fig. 4) and bordered by two large regions composed of Repeat Induced Point mutation (RIP)-inactivated transposons [21] (Fig. 2 and Fig. S2). Pa_5_10 is 600 kb away from Spok1 on the same chromosome arm, in a region with an expected FDS of 40% (Fig. 2). In strain T, this position is occupied by a segmental duplication of chromosome 6 (Fig. S2). The Pa_5_10 gene (hereafter named Spok2 for Spore killer 2) was thus a good candidate for the killing of ascospores in 40% of the asci of the Spok1Δ×S crosses. Spok2 was deleted by replacing its coding sequence with a hygromycin B-resistance marker to yield strain Spok2Δ (Fig. S3). When Spok2Δ was crossed with the parental strain S, 40% 2-spored asci were observed (Fig. 3A). Analysis of the homokaryotic ascospores recovered from a Spok2Δ×S cross showed that they were all sensitive to hygromycin B, indicative of a specific killing of the ascospores carrying Spok2Δ (hygromycin B-resistant) by those carrying wild-type Spok2. As expected from its chromosomal location, Spok2 causes ascospore death in only 40% of asci (n>200; Fig. 3A). Moreover, when the Spok2 coding sequence was inserted in the PaPKS1 gene of the Spok2Δ strain, using the same strategy as for Spok1 (PaPKS1::Spok2), it caused ascospore death in 99% of the asci when crossed to the Spok2Δ strain (Fig. 3C). This showed that Spok2 can also be responsible for spore killing. In crosses between Spok1Δ (strain T) and Spok2Δ (strain S), 100% 4-spored asci were observed (Table 1, Fig. 3A), showing that Spok2 was responsible for the 40% 2-spored asci present in the Spok1Δ×S crosses.
Like Spok1, Spok2 does not appear to be involved in any aspect of the physiology and development of P. anserina, as we could not detect any defect in the mycelium, fruiting body and ascospores of the Spok2Δ strain, with the homozygous cross of this strain yielding 100% 4-spored asci (Table 1, Fig. 3B). Sequence analysis of the Spok2 protein predicted with low probability an ATP binding site of a kinase domain acting on low molecular weight molecules. However, this domain is not predicted for Spok1 despite the great sequence identity (87%) between the two proteins (Fig. 4). Like Spok1, Spok2 was predicted by PSORT to be in nuclei.
Spok1 is a resistance factor to Spok2
In S×T crosses, we did not detect any obvious meiotic drive created by Spok2, i.e., excess transmission of the S genotype in the region surrounding Spok2 (Fig. S1). This was surprising since in this cross both Spok1 and Spok2 are in heterozygous configuration, which should enable killing by both Spok1 and Spok2 (Table 1). Possibly, Spok1 could act as a resistance factor to Spok2. To directly test this hypothesis, the 2-spored-asci progeny of Spok1Δ x S crosses was analyzed. Data showed that all the recovered ascospores had two nuclei containing both the Spok2 gene (11 asci analyzed), suggesting that Spok2 exerted spore killing only when the cross was devoid of Spok1 (Table 1). Homokaryotic “SKT20 Spok2Δ” strains, carrying a functional Spok1 recombined with a deleted Spok2, were successfully isolated in the progeny of a cross between SKT20 (which contain functional Spok1 and Spok2; Table 1) and Spok2Δ. Upon crossing these SKT20 Spok2Δ strains with SKT20, approximately 40% of homokaryotic descendants (9 of 23) carried the Spok2Δ deletion. Altogether, this showed that Spok2 triggered spore killing only in the absence of Spok1. On the contrary, Spok2 did not confer any resistance to Spok1, as we never obtained homokaryotic progeny that did not carry Spok1, in S×T crosses.
Spok2 but not Spok1 is prevalent in P. anserina strains
Because Spok1 and Spok2 behaved as selfish genetic elements propagating through meiotic drive, we evaluated their presence by PCR amplification of a 630 bp product using primers hybridizing in regions conserved in both Spok1 and Spok2 (Table S2) in various strains of P. anserina and its sibling species, P. comata (Table 2). PCR amplification products were obtained for 19 out of the 22 tested strains. Among the three remaining ones, two (X and CBS411.78) behaved as expected if they lacked both Spok1 and Spok2 in crosses with Spok1 - or Spok2 - containing strains. The third one (A406) exhibited a surprising behavior, as it was non-killing but resistant to both Spok1 and Spok2. Similar strains with non-killing activities but resistant to all Sks have been found in N. crassa [22].
Tab. 2. Spok Sk in P. anserina geographic races. 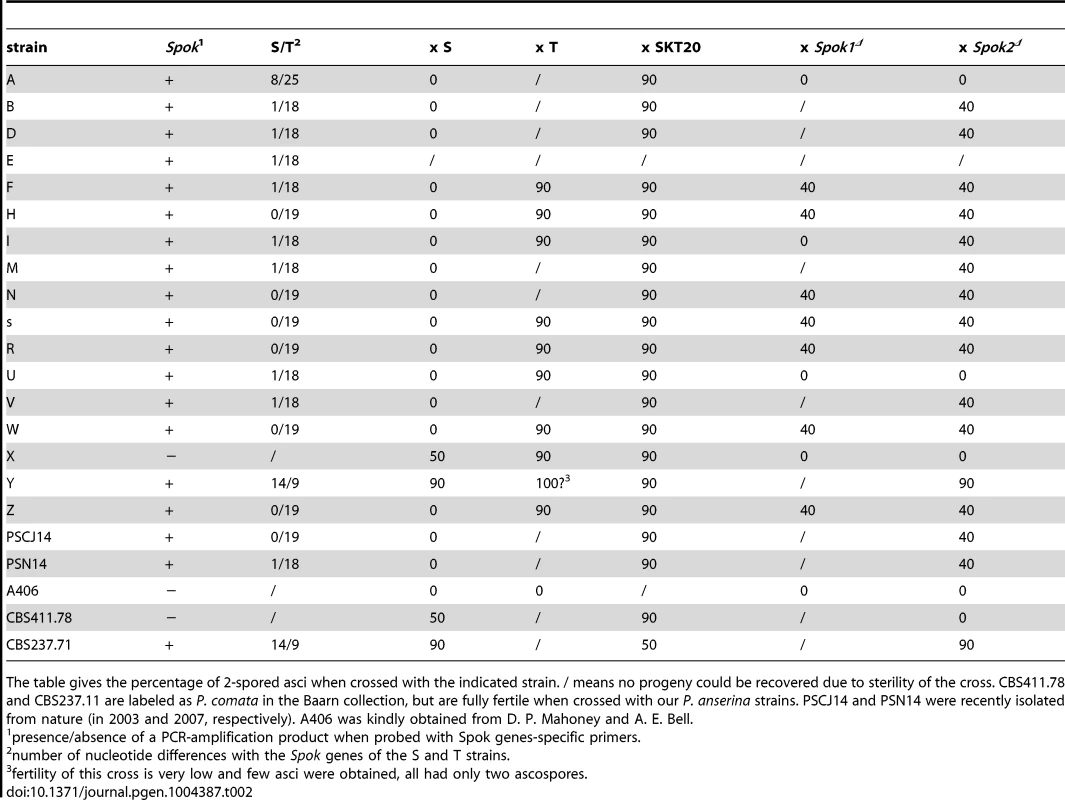
The table gives the percentage of 2-spored asci when crossed with the indicated strain. / means no progeny could be recovered due to sterility of the cross. CBS411.78 and CBS237.11 are labeled as P. comata in the Baarn collection, but are fully fertile when crossed with our P. anserina strains. PSCJ14 and PSN14 were recently isolated from nature (in 2003 and 2007, respectively). A406 was kindly obtained from D. P. Mahoney and A. E. Bell. The 630 pb amplification products were sequenced to assess whether they originated from Spok1 or Spok2. Spok1 was not found in any of the other strains tested here and is thus so far only present in strain T, while analysis of the remaining strains showed that all but two (Y and CBS237.71) contained Spok2 or a variant of it. Strains B, D, E, F, I, M, U and PSN14 carried a Spok2 variant with a silent nucleotide polymorphism (A to G at nucleotide N° 1194) and these strains behaved like strain S. Strain A carried another Spok2 variant with the silent A1194 to G substitution, an A to G substitution at position 1029, resulting in a Tyr343 to Cys polymorphism, and a GCCGGT insertion at position 1041 resulting in an insertion of two amino acids (Arg-Cys) after amino acid n°346. This Spok2 allele was active for resistance to Spok2 as shown by the recovery of 100% 4-spored asci from A×S crosses. The A×SKT20 crosses yielded 90% of 2-spored asci, showing that Spok1 was still able to act as a Sk in presence of the Spok2 allele from strain A. Interestingly, A×Spok1Δ and A×Spok2Δ crosses produced 100% 4-spored asci, showing that this allele was inactive for killing in such crosses (Table 2). Inoperativeness for killing was confirmed by analysis of A×Spok2Δ progeny, in which 50% homokaryotic hygromycin B-resistant ascospores carrying the Spok2 deletion were present.
P. comata CBS237.71 and P. anserina strain Y contained the same Spok-related gene having 14 and 9 differences with S and T, respectively, in the sequenced region. This gene may be another distinct functional Sk. Indeed, strain Y was previously reported as containing a Sk in crosses with strain S [15]. We confirm this (Table 2), as Y×S crosses presented 90% 2-spored asci. Unfortunately, fertility of strain Y was very low when crossed with strain T and SKT20. However, the few recovered asci suggested a complex interaction between the Sk of strain T and Y, a phenomenon previously seen in crosses with strain O and Us5 [23]. Therefore, it is highly probable that a third Spok gene (Spok3) endowed with spore killing activity and segregating with 90% SDS is present in Y. Spok3 could also be present in P. comata CBS237.11, which also displays 90% 2-spored asci when crossed with strain S (data not shown). Unfortunately, this strain cannot be crossed with strain T.
GFP and mCherry tagging alter distorter or responder activities of Spok proteins
To gain some insight into the molecular mechanisms of Spok1 and Spok2 action, we tagged the two proteins at the carboxy - and amino-termini with GFP (Spok1) and mCherry (Spok2). Spok1-GFP and Spok2-mCherry proteins tagged at their C-termini were obtained by introducing the GFP or mCherry CDS upstream of the stop codon of Spok1 and Spok2. GFP-Spok1 and mCherry-Spok2 proteins tagged at their N-termini were obtained by inserting in vitro the GFP or mCherry CDS downstream of the Spok genes start codons. The chimaeric constructs were then inserted at the PaPKS1 locus. When crossed with the S, T, SKT20, SKT20Δ, Spok1Δ and Spok2Δ strains, the strains carrying the GFP and mCherry constructs exhibited unexpected patterns. Crosses of Spok1-GFP with all strains yielded 100% asci with four spores, indicating that, while unable to promote killing, the transgene enabled resistance to Spok1 and Spok2 killing. Similarly, crosses with Spok2-mCherry showed that this allele was unable to kill Spok2Δ, yet was resistant to Spok2. N-terminally tagged mCherry-Spok2 produced empty asci when crossed with Spok2Δ (Fig. 3D), as if the transgene conserved the killing activity but lost the resistance one. However, in crosses with the strain having Spok2 at the PaPKS1 locus, only 4-spored asci were obtained, as if the responder activity was restored in the presence of a wild-type copy of Spok2. Both Spok1 and Spok2 enabled resistance to mCherry-Spok2 killing, as crosses with S, SKT20 and PaPKS1::Spok1 yielded ascospores with the expected segregation if full resistance occurred (Fig. 3E). On the contrary, GFP-Spok1 exhibited a pattern of asci expected for a protein endowed with both distorter and responder activities (data not shown). These data indicated that it was possible to independently inactivate either the responder or the distorter activities of Spok proteins, as previously gathered from the variant present in strain A.
Spok1 distorter and responder domains cannot be easily dissociated
In view of the above results, we tried to determine whether two separate domains carrying either the distorter or the responder activities could be identified in the Spok1 protein. In frame deletions in Spok1 were made in vitro and the truncated genes were reintroduced in the PaPKS1 gene, as done with full length Spok1 (Fig. S4). None of the construct carried any functional killing and resistance activity (n>30), showing that the two functions could not easily be separated on two independent DNA fragments.
Aspartate707/Glutamate708 are important for Spok2 activity
As mentioned above, a putative kinase domain was predicted at the C-terminus of Spok2, but not in Spok1. Pfam analysis [24] identified aspartate707 as a potential catalytic residue in Spok2. Because the next residue (n°708) was a glutamate, which may substitute to aspartate707 in the catalytic center, we mutated both the aspartate707 and glutamate708 to alanines. The recovered mutant, Spok2AA, was inserted in the PaPKS1 gene in the Spok2Δ strain. When crossed with Spok2Δ, the strain carrying Spok2AA yielded empty asci, as if the distorter activity was active and the responder one was inactive (Fig. 3F). However, cross of Spok2AA with the strain carrying a wild-type Spok2 allele at the PaPKS1 locus produced asci with four colorless ascospores (Fig. 3F). In such cross, both the Spok2 and Spok2AA ascospores survive, as if the Spok2AA responder activity was active. The behavior of the Spok2AA allele was thus identical to the one of N-terminally tagged mCherry-Spok2. Finally, when crossed with a strain carrying Spok1 at the PaPKS1 locus, asci containing two colorless ascospores were obtained in 99% of the asci, as if Spok1 acted alone.
Spok proteins accumulate in nuclei
For the four constructs, GFP and mCherry fluorescence was detected in the mycelium and in the fruiting bodies. During ascosporogenesis, similar patterns were observed for all constructs up to the post-meiotic mitosis: a diffuse cytoplasmic presence and an accumulation inside nuclei, as predicted by PSORT (Fig. 5). After this mitosis, at the stage at which two nuclei are present in each ascospore [25] (Fig. 1B), fluorescence was clearly discernible in nuclei of all ascospores in the asci were spore killing occurred (mCherry-Spok2 x S and GFP-Spok1 x S crosses). At the beginning of ascospore development, fluorescence was detected in the nuclei of all spores (Fig. 5C and 5D), including those undergoing death. At later stages, fluorescence persisted only in the surviving two ascospores, while the two others degenerated (Fig. 5E and 5F). In asci of crosses where no death occurs (Spok2-mCherry x Spok2Δ and Spok1-GFP x S), we could not detect fluorescence in two out of the four ascospores as early as after post-meiotic mitosis in asci undergoing FDS for Spok genes (Fig. 5H, 5I, 5K), suggesting that lack of death in these crosses was due to reduced levels of Spok1 or Spok2 in sensitive ascospores.
Fig. 5. Localization of Spok protein in developing asci. 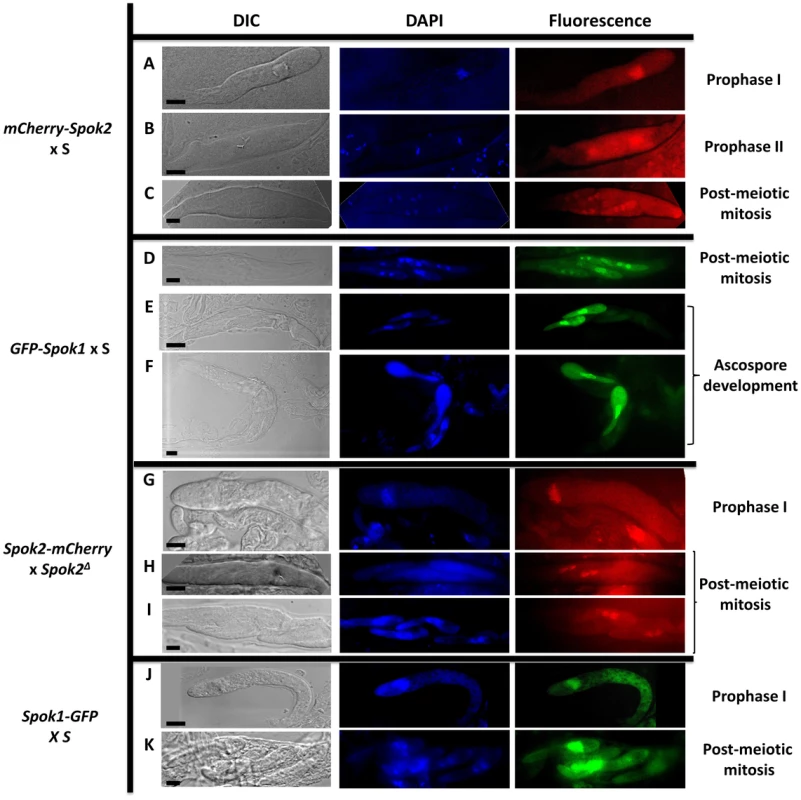
A to F: Localization of N-terminus tagged proteins with maintained spore killing activity. During prophase I, Spok proteins are located mainly in the nucleus and to a lesser extent in the cytoplasm (A & B). After the post-meiotic mitosis, Spok proteins are present in all nuclei (C & D). During ascospore development, sensitive spores are killed and the Spok proteins localize mainly in the nucleus of the resistant spores (E & F). G to K: Localization of the C-terminus tagged proteins without spore killing activity. Fluorescence pattern in prophase I is identical to the one with N-terminus-tagged proteins (G & J). After the post-meiotic mitosis, fluorescence is observed only in the nuclei of the two surviving spores (H, I & K). bar = 5 µm. The genomes of P. anserina and other fungi contain genes related to Spok1 and Spok2
Mining available databases of complete genome sequences showed that homologues of Spok genes are present and prevalent in the genomes of many filamentous ascomycetes (spore sac fungi; Fig. 6). They are present in all major classes of Pezizomycotina except in the basal classes Orbiliomycetes and Pezizomycetes, but in a patchy distribution with closely related species having or lacking Spok-related genes. Numbers can go up to 9 and 11 Spok-like genes in the genomes of Fusarium oxysporum and Microsporum canis, respectively. Interestingly, tree construction with selected species showed that the Spok-like gene phylogeny did not follow the known evolution of fungi, indicative of possible horizontal transfers (Fig. 6). Moreover, they were often present as pseudogenes, identified by the presence of mutations interrupting the coding sequence or by truncation. P. anserina contains three more Spok-like genes (Pa_7_3950, Pa_4_4000 and Pa_1_5015), all with transposable elements in their vicinity. They are all present in S and T, each occupying the same locus in both strains.
Fig. 6. Phylogenetic tree of Spok and Spok-like genes in representative fungal species. 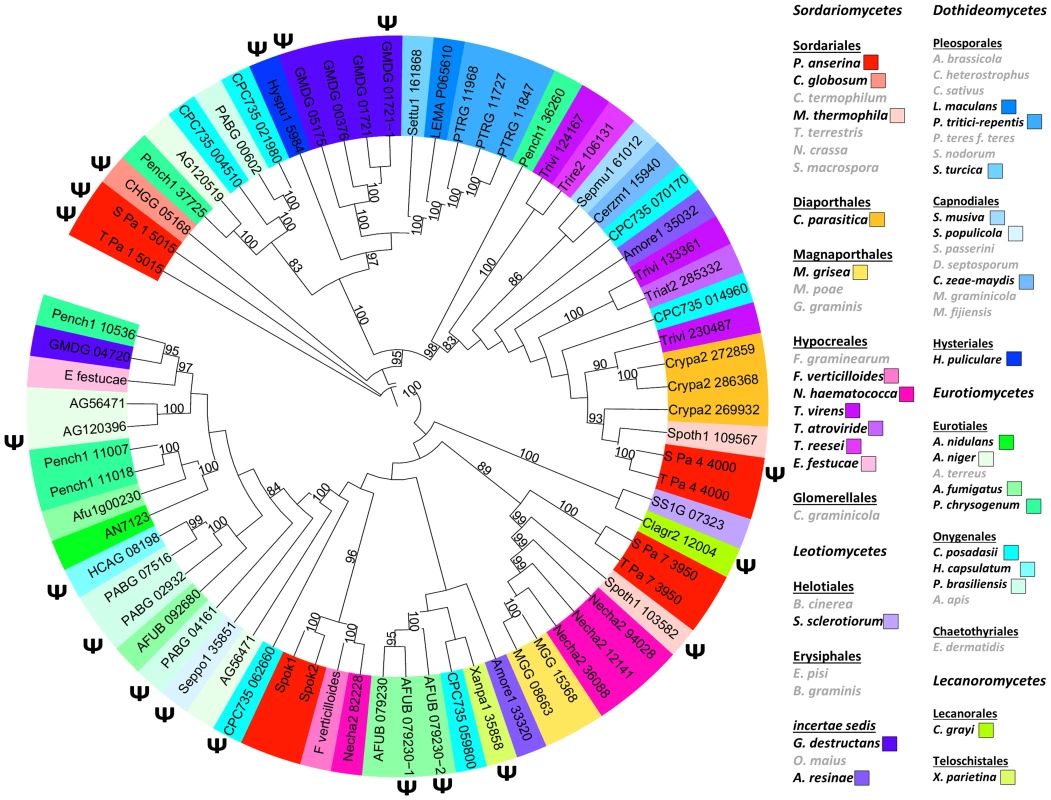
ψ: putative pseudogenes. Right: species in grey: no detected Spok or Spok-like gene and pseudogene. The other species were color coded according to the known phylogeny. Spok triggers spore killing in other fungi
To determine whether Spok1 was able to trigger ascospore death in another species, it was introduced under the expression of its own promoter in Sordaria macrospora along with a hygromycin B-resistance marker. This fungus is related to P. anserina, even if the genetic distance (i.e., average percentage identity between orthologous proteins) between Laesiophaeriaceae to which P. anserina belongs and Sordariaceae to which S. macrospora belongs is equivalent to that between mammals and fishes [18]. It is homothallic and ascospore morphogenesis is different, as eight ascospores are differentiated around single nuclei [26]. Genome sequence analysis indicates that S. macrospora is devoid of Spok genes (Fig. 6). Eight transformants carrying Spok1 were recovered and crossed to a strain devoid of Spok1. Resulting asci contained four wild-type-looking darkly-pigmented spores and four smaller often-abnormal unpigmented spores (Fig. 7). Wild-type-looking and abnormal ascospores were germinated and tested for resistance to hygromycin B and spore killing activity. 45 out of 66 wild-type-looking ascospores germinated, all were resistant to hygromycin B. 17 were successfully crossed to the strain devoid of Spok1. All showed a segregation of four normal and four abnormal ascospores. Two white ascospores out of 66 germinated. Both were resistant to hygromycin B. One was successfully crossed to the strain devoid of Spok1. Progeny was composed of asci with four wild-type-looking spores and four abnormal spores. The two germinated spores contained Spok1 and likely corresponded to unripe ascospores devoid of pigments, as sometime seen in crosses. Therefore, Spok1 is able to create meiotic drive in S. macrospora.
Fig. 7. Heterologous expression of Spok genes. 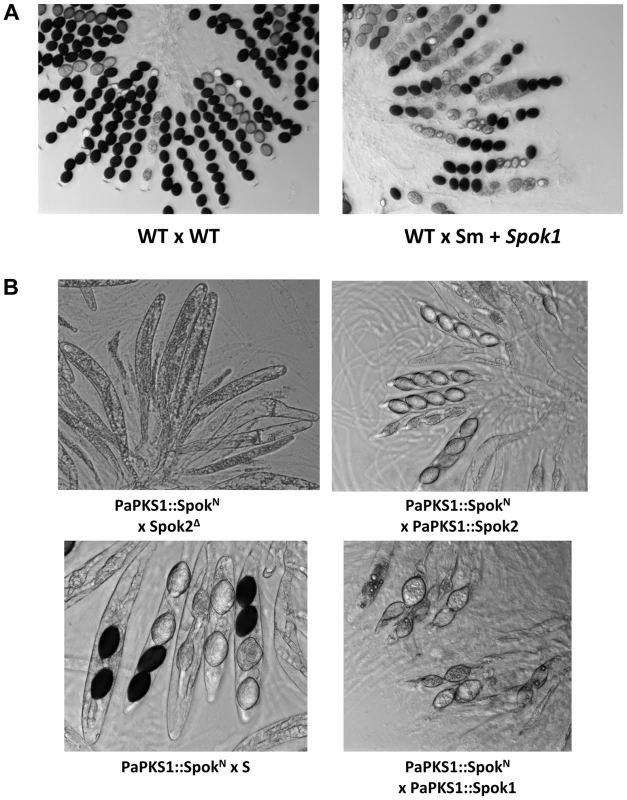
(A) S. macrospora crosses showing Sk activity of Spok1 in another species. (B) Expression of SpokN at the PaPKS1 locus of the Spok2Δ strain results in empty asci, showing that heterologous expression of SpokN results in efficient killing and lack of resistance to said killing. As for mCherry-Spok2 and Spok2AA, crosses with PaPKS1::Spok2 result in 4-spored asci; PaPKS1::SpokN x PaPKS1::Spok1 and PaPKS1::SpokN x S crosses showed that Spok1 and Spok2 promote resistance to SpokN, respectively. Finally, we introduced the Necha2_82228 gene from Nectria haematococca (Fig. 6) into the PaPKS1 gene of strain Spok2Δ, with its own promoter and terminator sequences. Despite being the closest relative of Spok1/Spok2 on the tree of Fig. 6, the SpokN protein is only 34% and 33% identical to Spok1 and Spok2, respectively. Note that the evolutionary gap between P. anserina and N. haematococca, is much larger than that between S. macrospora and P. anserina, arguing for strong differences in modality of ascospore differentiation [27]. PaPKS1::SpokN x Spok2Δ crosses were barren, as all asci were empty, indicating that the SpokN protein had a non-autonomous killing action, reminiscent of the Spok2AA and mCherry-Spok2 proteins that kills but does not allow for resistance. As for these two proteins, PaPKS1::SpokN x PaPKS1::Spok2 crosses produced 4-spored asci. Likewise, PaPKS1::SpokN x PaPKS1::Spok1 and PaPKS1::SpokN x S crosses showed that Spok1 and Spok2 promote resistance to SpokN, respectively. Therefore, despite the great divergence between SpokN and Spok1/Spok2, killing activity is retained and meiotic drive could be promoted by linking genetically SpokN with a resistance factor such as Spok1 or Spok2.
Discussion
The Spok MDs that we report here are constituted of single genes that carry both distorter and responder activities, unlike those of animals, plants and Neurospora, which are large and complex loci with two major genes, the distorter and the responder, and several additional ones that quantitatively modify the effects of either the distorter or the responder [3], [5], [6], [8], [14]. Unlike other MDs, Spok MDs may not have originated from cellular genes that have acquired additional functions disrupting normal gamete/spore formation, as they do not appear to be endowed with any physiological function. In P. anserina, another MD has been linked to the Het-s prion [16], in which spore killing results from an incompatibility reaction triggered by the co-presence of both the het-s protein in a prion-aggregated form and the soluble Het-S protein. A similar reaction is observed in vegetative hyphae when the two proteins are mixed together. From a genetic point of view, both the Het-s and Het-S alleles need to be present in crosses to see the MD effects. Spok genes act differently from the Het-s/Het-S MD, since their mere presence at a locus is sufficient to trigger the preferential transmission of this locus during meiosis. Spok genes thus define a new class of selfish elements that propagate vertically via meiotic drive and possibly horizontally in association with mobile elements. Indeed, at least in P. anserina, Spok-like genes are always in the vicinity of transposons and they do not seem to play any role in normal development.
Bioinformatic analysis did not provide many clues regarding the potential mode of action of Spok genes. An ATP-binding site of a kinase domain was predicted at the C-terminus of Spok2 with low probability, but not in Spok1, questioning its validity. A first model based on the presence of such a domain to explain the dual activity of the Spok genes can be put forward as follows. Spok proteins could be bifunctional enzymes that catalyze both the formation of a toxin from cellular metabolites and its inactivation. The toxin could diffuse in the ascus, while the enzyme and hence the detoxifying activity could not. This should result in the death of the ascospores not expressing the Spok proteins. If the kinase domain is involved in the formation of the toxin, its mutation should result in an allele inactive for killing but active for resistance. On the contrary, if the kinase domain is involved in detoxification, the mutation should produce an allele active for killing and inactive for resistance. However, our data show that this model is unlikely since the mutation of the putative catalytic residues, Aspartate707/Glutatmante708, has a more complex effect as it creates a Spok2AA allele unable to resist the toxin it produces, but which is fully resistant in the presence of a wild-type Spok2 allele in the cross. Therefore, it is as if wild-type Spok2 could activate the responder activity in Spok2AA, while Spok2AA could not. A second model for the dual mode of action of Spok proteins could be inspired from the yeast killer toxins for which the preprotoxin is a precursor of the toxin but confers resistance by complex formation with the toxin and subsequent degradation [28]. In yeasts, the preprotoxin genes are carried by double-stranded RNA viruses and are not known to trigger meiotic drive. Although, Spok1 and Spok2 do not present any obvious sequence similarity with the yeast killer toxin genes, we could propose that they would operate in a similar manner in which the preprotoxin (responder) is involved in resistance to the toxin (distorter). In such model, killing and resistance will depend on a subtle balance between toxin production from preprotoxin processing and preprotoxin/toxin complexes degradation. This may account for all the features presented by Spok Sks, including cross-resistance triggered by Spok1 to the Spok2 Sk (Spok1 could inactivate the Spok2 toxin, while Spok2 could not remove the Spok1 toxin), the inability to separate two domains by deletion analysis as well as the phenotype of the mCherry-Spok2, Spok2AA, SpokN alleles. Indeed, interactions between the Spok2 preprotoxin or toxin and the mCherry-Spok2, Spok2AA, SpokN proteins could result in their rapid degradation leading to inability to produce sufficient amounts of toxins. Cytological observations are also compatible with this model as GFP - and mCherry-tagged proteins that do not promote killing disappear very early during ascus maturation. A last model would posit that the Spok proteins may be the toxins and the Spok genes the responders that would inactivate the toxins by sequestering them at a defined place inside the nucleus. This would require protein/DNA as well as protein/protein interaction to allow the binding of many toxin molecules on few DNA sequences. In this model, the putative ATP binding site could thus be involved in DNA (nucleotide) binding rather than in a kinase activity. In this model, the Spok2 protein could bind the Spok1 gene, while the converse would not be possible explaining the resistance of Spok1 over Spok2 effects. The mCherry-Spok2, Spok2AA, SpokN proteins would be unable either to enter the nucleus or bind directly the Spok2/SpokN gene, but could do so in the presence of the wild-type Spok2 protein through protein/protein interactions. It is also compatible with the nuclear localization of the Spok proteins after delimitation of the ascospores.
Spok1 and Spok2 have many similar homologues in a wide array of filamentous ascomycetes, including in P. anserina itself. These are present in a patchy phylogenetic distribution, even in the P. anserina populations, do not follow the known fungal evolution and are often present as pseudogenes. The hypothesis that meiotic drive elements are invasive, can result in fewer progeny and can transport bad hitchhikers may explain both the unusual phylogeny and pseudogenes. The fact that a Spok-like gene can be a resistance factor to other Spok-like Sks also complicates the evolution of this family of genes. As Spok1 alone is able to trigger meiotic drive in S. macrospora and because SpokN may do so in P. anserina when associated with a Spok resistance factor(s), we surmise that Spok-like genes may account, in part, for the additional Sks detected in many other fungi, including P. anserina itself.
Materials and Methods
All protocols for cultivation, genetic and molecular analysis with P. anserina are available at http://podospora.igmors.u-psud.fr. Similar culture techniques were used for S. macrospora. Crosses were performed on M2 minimal medium using the S-derived strains as females and T-derived strains as males (supplementary Table S1). Sk was detected by the presence of 2-spored asci in the F1 progeny and assignments to Spok1 or Spok2 were made by measuring the ratio of 2-spored versus 4-spored asci and by backcrossing the F1 progeny to the S, T, Spok1Δ and Spok2Δ strains and observing the F2 progeny. The sequence of Spok1 has been deposited in GenBank with accession n° JX560967.
Polymorphic marker analysis
Genomic DNA was extracted from 50 progenies of an S×T cross (ST1 to ST50). Markers were amplified by PCR with 5 min denaturation at 94°C followed by 30 cycles [30 sec 94°C, 30 sec 55°C, 1 min 72°C] and finished by 10 min elongation at 72°C. The primer pairs used are given in Supplementary Table S2. DNA was separated on 2% agarose gels. DNA extracted from the parental S and T strains was used as a control.
Deletion
Spok1 and Spok2 deletions were made on strains TΔmus51 and SΔmus51, respectively, as described for PaTLK2 in [29] using Hygromycin B resistance as a selection marker. Table S2 gives the primers used for deletions. The deletions were verified by Southern blotting as in [30].
Insertion of Spok alleles at the centromere of chromosome 2
Insertion of Spok alleles was made with a strategy involving integration of a plasmid with a single crossover into the PaPKS1 gene resulting in its inactivation. A 1395 bp DNA fragment from PaPKS1 was amplified by PCR using S genomic DNA and primers 193SkFSII and 193SkRSI. A 4344 bp DNA fragment surrounding Spok1 was amplified by PCR with the Pfu DNA polymerase from Promega (Madison, WI, USA) using T genomic DNA and primers 510FSI and 510RNI. The PaPKS1 fragment was digested with SalI and SacII enzymes, the Spok1 fragment with SalI and NotI and both were ligated into pBC-phleo vector [31] cut with SacII and NotI to yield pEnterprise1. pEnterprise1 was introduced by transformation into the SΔmus51::su8-1 strain and one transformant devoid of pigment, was selected for further analysis. Spok2 and SpokN, including their own promoters and terminators, were fused by PCR with the 1395 bp PaPKS1 DNA fragment. The fused PCR fragments was digested with SacII and NotI and cloned into pBC-phleo and pBC-Genet vectors to yield pEnterprise2 and pEnterpriseNectria, respectively. Both plasmids were then introduced into P. anserina as for pEnterprise1. The same strategy was used to introduce GFP-Spok1 and mCherry-Spok2 and the truncated alleles (used primers in Table S2).
Creation of the Spok2AA allele
To create the Spok2AA allele, the plasmid pEnterprise2 was amplified by PCR using primers Spok2MutF and Spok2MutR (Table S2, in bold red are the nucleotides used to change the aspartate and glutamate codons to alanine ones). The PCR product was transformed into Escherichia coli and candidates were selected by sequencing the mutated region. One candidate was completely sequenced, found devoid of mutations and introduced in a Spok2Δ Δmus51 strain. Two transformants were selected based on their lack of pigments and used for further analysis.
Insertion of Spok1 in S. macrospora
The Spok1 fragment was excised from pEnterprise1 and cloned into the pBC-Hygro plasmid cut with SalI and NotI. The pBC-Hygro containing Spok1 plasmid was transformed into the spo11 mutant of S. macrospora. The wild-type strain and 27 hygromycin B-resistant transformants were crossed with the spo55 mutant to force outcrossing.
Microscopy analysis
Perithecium contents for fluorescence analysis were prepared as in [26]. Pictures were taken with a Leica DMIRE 2 microscope coupled with a 10-MHz Cool SNAPHQ charge-coupled device camera (Roper Instruments). They were analyzed with ImageJ.
Phylogenetic analysis
The phylogenetic analysis of Supplementary Fig. S2 was carried out by aligning the sequences with MAFFT [32] and trimming them with Jalview to retain informative positions [33]. The tree was constructed using PhyML [34] with the default parameters [35] and 100 bootstrapped data sets. The tree was visualized with the iTOL server [36]. Pseudogenes were defined by the presence of several mutations (deletions, frameshifts or read-through) inactivating the coding sequences.
Supporting Information
Zdroje
1. PennisiE (2003) Meiotic drive. Bickering genes shape evolution. Science 301 : 1837–1839.
2. SaupeSJ (2012) A fungal gene reinforces Mendel's laws by counteracting genetic cheating. Proc Natl Acad Sci U S A 109 : 11900–11901.
3. LarracuenteAM, PresgravesDC (2012) The Selfish Segregation Distorter Gene Complex of Drosophila melanogaster. Genetics 192 : 33–53.
4. SandlerL, HiraizumiY, SandlerI (1959) Meiotic Drive in Natural Populations of Drosophila Melanogaster. I. the Cytogenetic Basis of Segregation-Distortion. Genetics 44 : 233–250.
5. LyonMF (2003) Transmission ratio distortion in mice. Annu Rev Genet 37 : 393–408.
6. BauerH, SchindlerS, CharronY, WillertJ, KusecekB, et al. (2012) The nucleoside diphosphate kinase gene Nme3 acts as quantitative trait locus promoting non-Mendelian inheritance. PLoS Genet 8: e1002567.
7. Dobrovolskaia-ZavadskaiaN (1927) Sur la mortification spontanee de la queue chez la souris nouveau-nee et sur l'existence d'un caractere (facteur) hereditaire “non-viable”. C R Seances Soc Biol Fil 97 : 114–116.
8. YangJ, ZhaoX, ChengK, DuH, OuyangY, et al. (2012) A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337 : 1336–1340.
9. ChenJ, DingJ, OuyangY, DuH, YangJ, et al. (2008) A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci U S A 105 : 11436–11441.
10. KusanoA, StaberC, GanetzkyB (2002) Segregation distortion induced by wild-type RanGAP in Drosophila. Proc Natl Acad Sci U S A 99 : 6866–6870.
11. HerrmannBG, KoschorzB, WertzK, McLaughlinKJ, KispertA (1999) A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature 402 : 141–146.
12. RajuN (1994) Ascomycetes Spore killers: Chromosomal elements that distort genetic ratios among the products of meiosis. Mycologia 86 : 461–473.
13. TurnerBC, PerkinsDD (1979) Spore killer, a chromosomal factor in Neurospora that kills meiotic products not containing it. Genetics 93 : 587–606.
14. HammondTM, RehardDG, XiaoH, ShiuPK (2012) Molecular dissection of Neurospora Spore killer meiotic drive elements. Proc Natl Acad Sci U S A 109 : 12093–12098.
15. van der GaagM, DebetsAJ, OosterhofJ, SlakhorstM, ThijssenJA, et al. (2000) Spore-killing meiotic drive factors in a natural population of the fungus Podospora anserina. Genetics 156 : 593–605.
16. DalstraHJP, SwartK, DebetsAJM, SaupeSJ, HoekstraRF (2003) Sexual transmission of the [Het-s] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci U S A 100 : 6616–6621.
17. PadieuE, BernetJ (1967) Mode d'action des gènes responsables de l'avortement de certains produits de la méiose chez l'Ascomycète Podospora anserina. C R Acad Sci Paris 264 : 2300–2303.
18. EspagneE, LespinetO, MalagnacF, Da SilvaC, JaillonO, et al. (2008) The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol 9: R77.
19. CoppinE, SilarP (2007) Identification of PaPKS1, a polyketide synthase involved in melanin formation and its use as a genetic tool in Podospora anserina. Mycol Res 111 : 901–908.
20. HortonP, ParkKJ, ObayashiT, FujitaN, HaradaH, et al. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–587.
21. GalaganJE, SelkerEU (2004) RIP: the evolutionary cost of genome defense. Trends Genet 20 : 417–423.
22. TurnerBC (2001) Geographic distribution of Neurospora spore killer strains and strains resistant to killing. Fungal Genet Biol 32 : 93–104.
23. HamannA, OsiewaczHD (2004) Genetic analysis of spore killing in the filamentous ascomycete Podospora anserina. Fungal Genetics and Biology 41 : 1088–1098.
24. PuntaM, CoggillPC, EberhardtRY, MistryJ, TateJ, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–D301.
25. BeckettA, WilsonIM (1968) Ascus cytology of Podospora anserina. J Gen Microbiol 53 : 81–87.
26. Thompson-CoffeC, ZicklerD (1994) How the cytoskeleton recognizes and sorts nuclei of opposite mating type during the sexual cycle in filamentous ascomycetes. Dev Biol 165 : 257–271.
27. HanlinRT (1971) Morphology of Nectria haematococca. Am J Bot 58 : 105–116.
28. BreinigF, SendzikT, EisfeldK, SchmittMJ (2006) Dissecting toxin immunity in virus-infected killer yeast uncovers an intrinsic strategy of self-protection. Proc Natl Acad Sci U S A 103 : 3810–3815.
29. LalucqueH, MalagnacF, BrunS, KickaS, SilarP (2012) A Non-Mendelian MAPK-Generated Hereditary Unit Controlled by a Second MAPK Pathway in Podospora anserina. Genetics 191 : 419–433.
30. GrognetP, LalucqueH, SilarP (2012) The PaAlr1 magnesium transporter is required for ascospore development in Podospora anserina. Fung Biol 116 : 1111–1118.
31. SilarP (1995) Two new easy-to-use vectors for transformations. Fungal Genet Newsl 42 : 73.
32. KatohK, TohH (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9 : 286–298.
33. WaterhouseAM, ProcterJB, MartinDMA, ClampM, BartonGJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 : 1189–1191.
34. GuindonS, GascuelO (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52 : 696–704.
35. DereeperA, GuignonV, BlancG, AudicS, BuffetS, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–469.
36. LetunicI, BorkP (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23 : 127–128.
37. CarverTJ, RutherfordKM, BerrimanM, RajandreamMA, BarrellBG, et al. (2005) ACT: the Artemis Comparison Tool. Bioinformatics 21 : 3422–3423.
Štítky
Genetika Reprodukční medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



