-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
Hybrid sterility (HS) belongs to reproductive isolation barriers that safeguard the integrity of species in statu nascendi. Although hybrid sterility occurs almost universally among animal and plant species, most of our current knowledge comes from the classical genetic studies on Drosophila interspecific crosses or introgressions. With the house mouse subspecies Mus m. musculus and Mus m. domesticus as a model, new research tools have become available for studies of the molecular mechanisms and genetic networks underlying HS. Here we used QTL analysis and intersubspecific chromosome substitution strains to identify a 4.7 Mb critical region on Chromosome X (Chr X) harboring the Hstx2 HS locus, which causes asymmetrical spermatogenic arrest in reciprocal intersubspecific F1 hybrids. Subsequently, we mapped autosomal loci on Chrs 3, 9 and 13 that can abolish this asymmetry. Combination of immunofluorescent visualization of the proteins of synaptonemal complexes with whole-chromosome DNA FISH on pachytene spreads revealed that heterosubspecific, unlike consubspecific, homologous chromosomes are predisposed to asynapsis in F1 hybrid male and female meiosis. The asynapsis is under the trans - control of Hstx2 and Hst1/Prdm9 hybrid sterility genes in pachynemas of male but not female hybrids. The finding concurred with the fertility of intersubpecific F1 hybrid females homozygous for the Hstx2Mmm allele and resolved the apparent conflict with the dominance theory of Haldane's rule. We propose that meiotic asynapsis in intersubspecific hybrids is a consequence of cis-acting mismatch between homologous chromosomes modulated by the trans-acting Hstx2 and Prdm9 hybrid male sterility genes.
Published in the journal: . PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004088
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004088Summary
Hybrid sterility (HS) belongs to reproductive isolation barriers that safeguard the integrity of species in statu nascendi. Although hybrid sterility occurs almost universally among animal and plant species, most of our current knowledge comes from the classical genetic studies on Drosophila interspecific crosses or introgressions. With the house mouse subspecies Mus m. musculus and Mus m. domesticus as a model, new research tools have become available for studies of the molecular mechanisms and genetic networks underlying HS. Here we used QTL analysis and intersubspecific chromosome substitution strains to identify a 4.7 Mb critical region on Chromosome X (Chr X) harboring the Hstx2 HS locus, which causes asymmetrical spermatogenic arrest in reciprocal intersubspecific F1 hybrids. Subsequently, we mapped autosomal loci on Chrs 3, 9 and 13 that can abolish this asymmetry. Combination of immunofluorescent visualization of the proteins of synaptonemal complexes with whole-chromosome DNA FISH on pachytene spreads revealed that heterosubspecific, unlike consubspecific, homologous chromosomes are predisposed to asynapsis in F1 hybrid male and female meiosis. The asynapsis is under the trans - control of Hstx2 and Hst1/Prdm9 hybrid sterility genes in pachynemas of male but not female hybrids. The finding concurred with the fertility of intersubpecific F1 hybrid females homozygous for the Hstx2Mmm allele and resolved the apparent conflict with the dominance theory of Haldane's rule. We propose that meiotic asynapsis in intersubspecific hybrids is a consequence of cis-acting mismatch between homologous chromosomes modulated by the trans-acting Hstx2 and Prdm9 hybrid male sterility genes.
Introduction
Hybrid sterility (HS) is a postzygotic reproductive isolation barrier restricting gene flow between the related taxa during speciation. It is defined as a condition where two parental forms fertile inter se produce a hybrid that is sterile [1]. One of the most interesting findings coming from previous studies is a disproportionately large effect of Chr X on reproductive isolation, particularly on hybrid sterility and inviability. The large X-effect was described in diverse organisms, and evolutionary biologists designated it as one of the speciation rules [2]–[5]. Another speciation principle, called Haldane's rule [6], points to the empirical findings that hybrid inviability and sterility predominantly afflicts the heterogametic (XY or ZW) sex. The dominance theory originally proposed by Muller [7] explained the sex-dependent effect on hybrid fitness by the manifestation of recessive X-linked alleles in hemizygous XY males but not in XX females [8]–[10].
We have chosen M. m. musculus and M. m. domesticus subspecies (hereafter, Mmm and Mmd) as model organisms to study mammalian HS (for review see [11]–[13]). Both subspecies diverged from a common ancestor approximately 0.3 to 0.5 million years ago [14] and formed a hybrid zone across Europe after their secondary contact [15]. The repeated introgressions of Mmm genes into Mmd genome and vice versa across their hybrid zone indicate incomplete reproductive isolation between both young subspecies [16], [17]. Such early-stage model is superior in that it reduces the risk of analyzing HS genes that evolved as a consequence and not as the cause of speciation after full reproductive isolation of the related taxa [5], [18]. Numerous genetic and genomic tools are available for the mouse model, including the full genomic sequence of inbred strains representing both subspecies and additional 17 laboratory inbred strains [19] and a panel of 28 mouse intersubspecific chromosome substitution (consomic) strains carrying individual Mmm chromosomes or their parts on Mmd background [20]. A variety of commercially available antibodies detecting meiosis-specific proteins and histone modifications permit immunodetection of subnuclear structures important for meiotic chromosome synapsis and segregation [21], [22].
We identified the first hybrid sterility gene in mice, hybrid sterility 1 – Hst1 – as a polymorphic variant on Chr 17 between two laboratory strains, C57BL10/Sn and C3H/Di, both predominantly of Mmd origin (at that time still linkage group IX). When mated with Mmm wild mice trapped in Central Bohemia near Prague, these crosses produced sterile or fertile male hybrids, depending on their Hst1 alleles [23]. Recently, Hst1 was identified by the forward genetics approach as PR domain containing 9 (Prdm9) [24] and later was shown to control meiotic recombination hotspots [25], [26].
In a study of genetic architecture of F1 hybrid male sterility, the results of (Mmm×Mmd)×Mmd backcross predicted a minimum of four independently segregating HS loci. However, QTL analysis of the data revealed only two strong HS loci, Hst1/Prdm9 and a locus on Chr X [27]. This paradox could be explained either by the action of multiple minor HS loci undetected by relatively low-power QTL analysis or by different behavior of two major HS loci on the hybrid background. The latter alternative was supported in an experiment showing that these two HS loci are not sufficient to recapitulate the F1 HS phenotype on B6 (Mmd) genetic background [22]. Moreover, for the first time a compelling evidence was provided for the mechanism of HS, showing that aberrant meiotic pairing of heterosubspecific homologous chromosomes and meiotic arrest are the consequence of intersubspecific hybrid genetic background [22].
Here we report on the role of Chr X in male and female meiosis in mouse intersubspecific hybrids. We localized the Hstx2 locus controlling the asymmetry of HS in reciprocal intersubspecific F1 hybrid males to a 4.7 Mb interval on Chr X and mapped three autosomal loci that can abolish this asymmetry. We observed the predisposition of heterosubspecific homologs to asynapsis in male and female meiosis as the initial step of intrameiotic breakdown of the sterile hybrids. The effects of Hstx2 or Hst1/Prdm9 on the degree of asynapsis in pachynemas of male but not female intersubspecific hybrids concurred with the fertility of Mmm×Mmd F1 hybrid females homozygous for the Hstx2Mmm allele and resolved the apparent conflict with the dominance theory of Haldane's rule. Based on detailed meiotic analysis of the mouse model of intersubspecific F1 hybrids we propose that HS genes operate on “sensitized” genetic background resulting from the difficulties of proper meiotic synapsis of heterosubspecific autosomal homologs and consequent epigenetic dysregulation of X-Y chromosomes [22]. Accordingly, classical HS genes Prdm9 [24], [28] and Hstx2 [27] realize their HS-specific phenotypes by interacting, directly or indirectly, with the process of meiotic pairing and synapsis of heterospecific homologs.
Results
Fine mapping of Hstx2 and its role in HS asymmetry of reciprocal F1 hybrids
Dobzhansky-Muller incompatibilities (DMIs) between the nascent species often result in asymmetry of HS or inviability of reciprocal F1 hybrids [27], [29]–[31] although relatively little is known about their genetic control or mechanistic basis. Previously we have shown that asymmetry in male sterility of reciprocal hybrids between Mmm mouse subspecies represented by the PWD/Ph (hereafter PWD) inbred strain [32] and Mmd represented by the C57BL/6J inbred strain (hereafter B6) is controlled by the central region of Chr X (64.9 Mb–98.1 Mb, GRCm38). To localize the locus responsible for HS asymmetry we crossed consomic F1 females (B6.PWD-Chr X×B6) with PWD males. All 124 male offspring carried B6/PWD heterosubspecific autosomal pairs while Chr X loci were either PWD or B6, depending on the recombination breakpoints. Testes weight (TW, range 56–186 mg) and sperm count (SC, 0–13.5 million) were used as surrogate for QTL analysis of male fertility phenotypes. Their segregation localized a hybrid sterility locus to the 34.6–35.7 cM (1.5-LOD support interval) interval with the maximum LOD score 30 and 23 at 34.8 cM for TW and SC. As shown in Fig. 1A–D all males that received the PWD allele at the DXMit87 locus had small testes bellow 100 mg and little (below 106) sperm in ductus epididymis. This hybrid sterility locus causes complete sterility on F1 hybrid intersubspecific background, and we designate it hybrid sterility chromosome X 2, Hstx2. Earlier, we reported localization of the locus at lower resolution in the (PWD×B6)×B6 backcross (Dzur-Gejdosova 2012).
Fig. 1. Single QTL mapping of Hstx2 on Chr X. 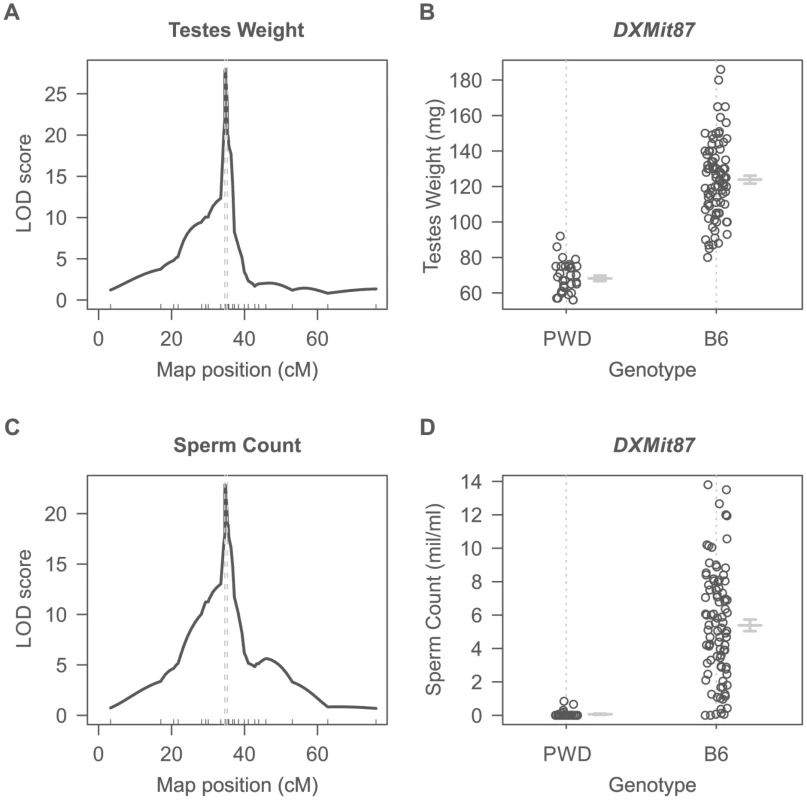
(A) QTL analysis of testes weight in the (B6.PWD-Chr X×B6)F1×PWD cross showed a 1.5-LOD support interval between 34.6 cM to 35.3 cM on Chr X. (B) Distribution of testes weight of males carrying PWD or B6 allele of DXMit87 marker with LOD score 30. (C) QTL analysis of sperm count in ductus epididymis shows the same 1.5 LOD support interval as for testes weight. (D) Distribution of sperm count of males carrying PWD or B6 allele of DXMit87 marker (Chr X: 66.65 Mb, GRCm38) with LOD score above 20. To further refine the position of Hstx2, a new partial consomic strain B6.PWD-Chr X.1s was created with extended proximal interval of the Chr XPWD sequence compared to B6.PWD-Chr X.1 (for strain description see ref [20]).
The borders of the PWD sequence of the introgressed Chr XPWD were compared to those of the existing proximal, central and distal partial consomic strains B6.PWD-Chr X.1, B6.PWD-Chr X.2 and B6.PWD-Chr X.3 [20] using high-resolution Mouse Universal Genotyping Array (MegaMUGA) (Table S1). To localize the region carrying Hstx2 on Chr XPWD, females of all four Chr X partial consomic strains were crossed with PWD males and the fertility of male offspring was examined (Fig. 2). The (B6.PWD-Chr X.1×PWD)F1 and (B6.PWD-Chr X.3×PWD)F1 hybrid males were semifertile with testes weight comparable to (B6×PWD)F1 males, while (B6.PWD-Chr X.1s×PWD)F1 and (B6.PWD-Chr X.2×PWD)F1 were fully sterile with small testes (p<0.0001, t-test) and no sperm in ductus epididymis (Fig. 2). Thus (B6.PWD-Chr.X.1s×PWD)F1 hybrids carried the Hstx2PWD allele and fully reconstructed the HS phenotype of (PWD×B6) males, showing meiotic arrest at epithelial stage IV and to lesser degree at late pachytene/diplotene stage (Table S2). The position of Hstx2 was localized to the 4.7 Mb interval delineated by UNC30904273 for the distal end of B6.PWD-Chr X.1 PWD sequence and UNC30934795 for the distal end of B6.PWD-Chr X.1s (X: 64,880,641–69,581,094, GRCm38).
Fig. 2. Fine mapping of Hstx1 and Hstx2 HS loci on Chr X using partial consomic strains. 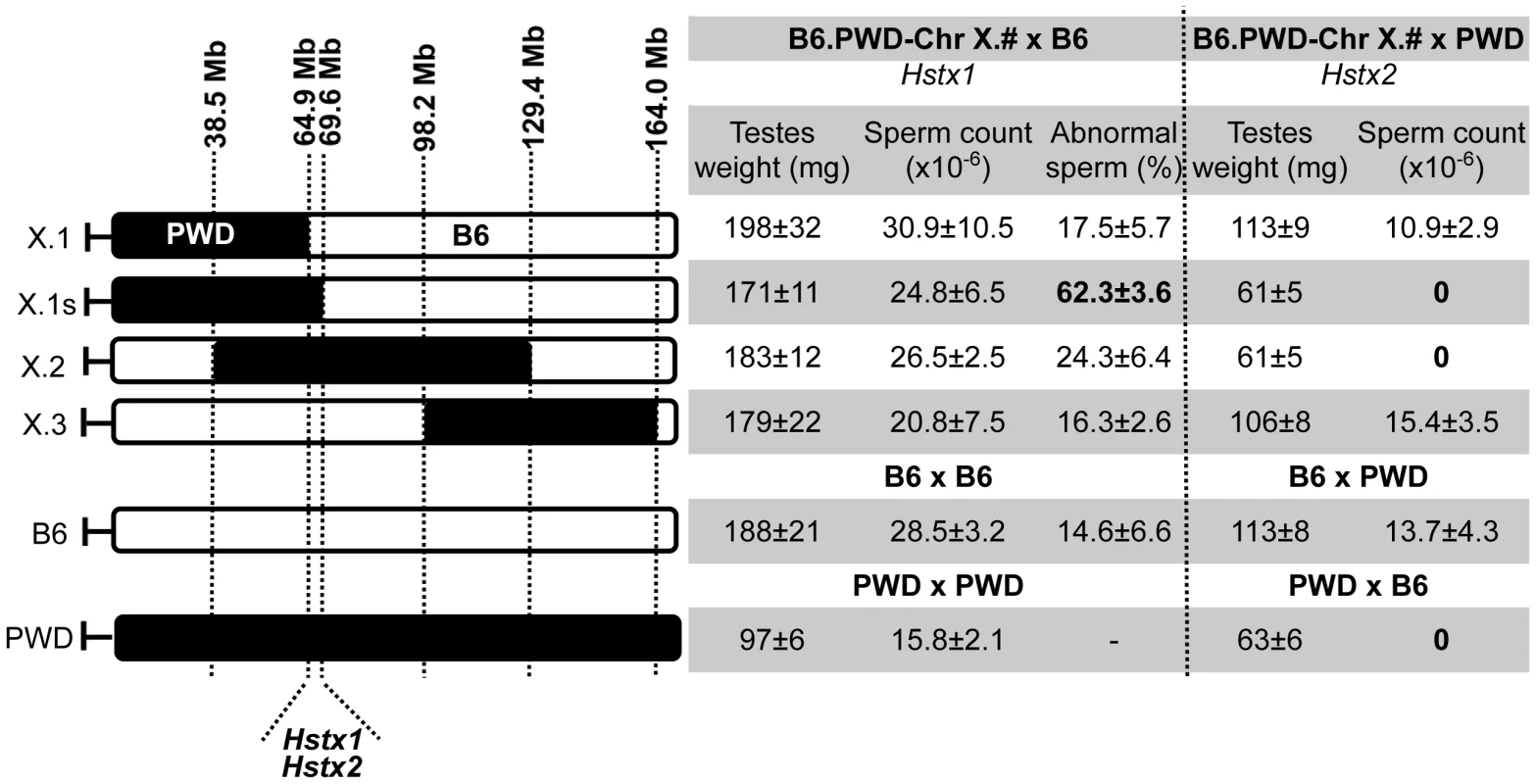
The partial consomic B6.PWD-Chr X.# females were crossed with B6 or PWD males for mapping Hstx1 and Hstx2, respectively. Testes weight and sperm count were used as fertility phenotypes. The borders of introgressed PWD sequence (black) were determined by MegaMUGA genotyping for B6.PWD-Chr X.1 (abbreviated here X.1), B6.PWD-Chr X.1s (X.1s) and B6.PWD-Chr X.2 (X.2). For B6.PWD-Chr X.3 (X.3) mapping see [20]. The map positions correspond to genome assembly GRCm38, megabase scale, for details see Table S1. Introgressed Hstx1PWD causes teratozoospermia in Mmd genome and maps to the same genomic region as Hstx2
The Hstx1 locus in the proximal part of Chr XPWD causes male sterility when introgressed onto the B6 background [33]. Contrary to the F1 hybrid meiotic arrest at late pachytene/diplotene stage controlled by Hstx2 [22], Hstx1PWD in the B6 genome causes postmeiotic breakdown and abnormal morphology of a fraction of non-functional sperm. Here we genotyped the male progeny of females heterozygous for PWD and B6 form of Chr X from the backcross generations 4–9 to B6 background and selected 71 Chr X single-recombinants with PWD centromeric end for fertility testing. The Hstx1 locus mapped within the interval spanned by DXMit76 and DXMit143 (Fig. S1).
To further localize Hstx1, we phenotyped all four B6.PWD-Chr X partial consomics and found a high percentage of abnormal sperm cells in B6.PWD-Chr X.1s compared to other three partial consomics and B6 males (p<0.05 t-test, Fig. 2). The analysis of partial consomic strains independently confirmed the Hstx1 localization to the same 4.7 Mb interval of Chr X that carries Hstx2. However, because B6.PWD-Chr X.2 males did not show high frequency of abnormal sperm in spite of their Hstx1PWD allele, we assumed that Hstx1 needs to interact with another genetic factor from the proximal region of Chr XPWD to manifest the abnormal sperm phenotype (Fig. 2, see also [33]).
Hstx1 and Hstx2 candidate genes
The 4.7 Mb Chr X candidate region of Hstx1 and Hstx2 loci carries 11 known protein-coding genes and 20 miRNA genes (Fig. S2). Of these, seven protein-coding genes, namely cancer/testis antigen 2 (Ctag2), RIKEN cDNA 4930447F04 gene (4930447F04Rik), SLIT and NTRK-like family, member 2 (Slitrk2), RIKEN cDNA 4933436I01 gene (4933436I01Rik), fragile X mental retardation syndrome 1 homolog (Fmr1), fragile X mental retardation 1 neighbor (Fmr1nb) and AF4/FMR2 family member 2 (Aff2) show high expression in adult testis. Of them, Aff2 is expressed pre-meiotically in spermatogonia, Fmr1 and Fmr1nb show expression in early prophase I, and Ctag2, 4930447F04Rik and Slitrk2 are expressed in meiotic and postmeiotic cells. Finally, 4933436I01Rik is expressed in haploid cells. Sorted populations of testicular cells from PWD and B6 strains and immature 14.5 dpp testes of their reciprocal hybrids did not show significant differences in relative mRNA expression levels of six meiotic and/or postmeiotic genes (Fig. S3). All 20 miRNAs in the candidate region are expressed in male germ cells but do not undergo meiotic sex chromosome inactivation (MSCI) [34]). Only the Mir465PWD cluster displayed a significant increase of expression in the first meiotic prophase of PWD and B6 sorted testicular cells (Figure 3A, B). The miRNA expression profiling of sterile (PWD×B6)F1 and fertile (B6×PWD)F1 14.5dpp testes showed 1.5 - to 2-fold upregulation of Mir883b-3p, Mir465a/b/c-3p and Mir465a/b -5p in sterile males, while Mir743a, Mir743-5p, Mir880 and Mir465c-5p showed 1.2 - to 4-fold down-regulation (Figure 3C, D).
Fig. 3. Expression profiling of a cluster of MiRNA genes within the Hstx1/Hstx2 critical region. 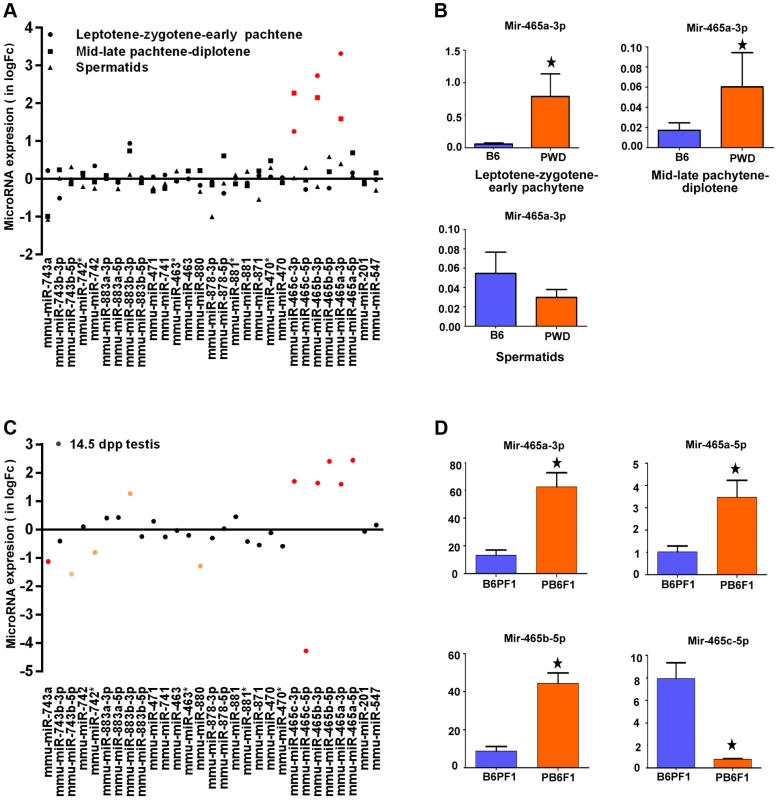
(A) Log fold-change of expression ratio of PWD versus B6 MiRNA genes in flow-sorted testicular cells. Significant overexpression of Mir465 cluster in PWD germ cells is shown in red. (B) Validation of Mir465 overexpression in PWD primary spermatocytes by qRT PCR. Data normalized to U6 non-coding RNA (C) Log fold-change of expression ratio of MiRNA genes in (PWD×B6)F1 versus (B6×PWD)F1 (abbreviated PB6F1 and B6PF1) 14.5 d old testes. Significant upregulation in red (P<0.05). (D) qRT PCR validation of differences in Mir465 expression. Data normalized to Mir152. Re-sequencing candidate genes from PWD genomic DNA and BAC clones [35] and inspection of the PWD exome sequence revealed seven non-synonymous substitutions of the 4933436I01Rik PWD allele compared to B6 (see also [36]). Of the remaining genes, Aff2 carries five, Fmr1nb and Slitrk2 carry two, 4930447F04Rik and Ctag2 carry one, and Fmr1 does not carry any non-synonymous substitution. Inspection of Sanger Institute Mouse Genome Project (http://www.sanger.ac.uk/resources/mouse/genomes/) confirmed the same SNPs for PWK, a closely related Mmm inbred strain. Search of miRNA sequences revealed one SNP in the seed sequence of Mir743a, changing AAAGACA in B6 to AAAGACG in PWD (Table 1).
Tab. 1. Missense SNPs of candidate genes in the Hstx1/Hstx2 critical region on Chr X. 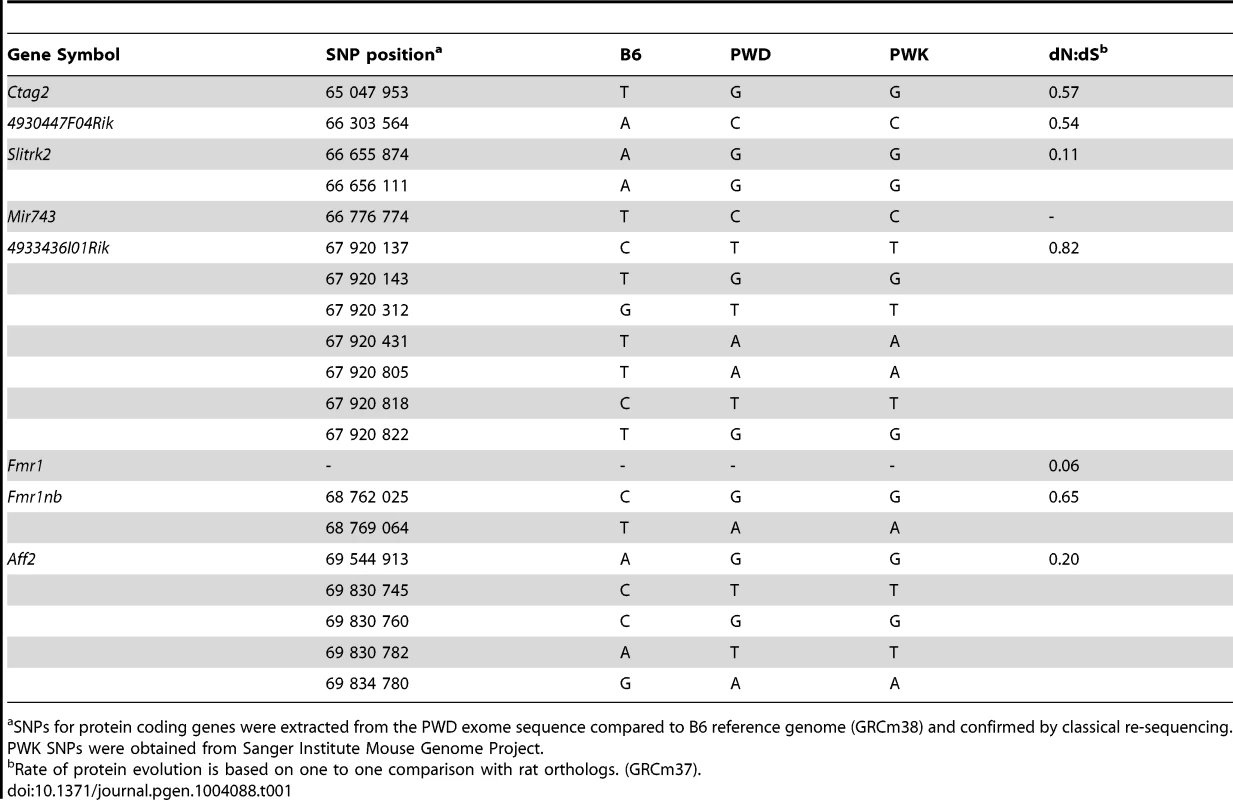
SNPs for protein coding genes were extracted from the PWD exome sequence compared to B6 reference genome (GRCm38) and confirmed by classical re-sequencing. PWK SNPs were obtained from Sanger Institute Mouse Genome Project. Reproductive isolation genes in Drosophila and in mouse have been shown to evolve rapidly and to undergo positive selection [37]–[39]. Three of the candidates for the Hstx1/2 locus, Ctag2, 4933436I01Rik and Fmr1nb, displayed an elevated rate of protein evolution (Table 1). In particular, 4933436I01Rik is among the most rapidly evolving genes on Chr X [40], [41], showing weak but significant expression in primary spermatocytes and strong post-meiotic expression [42], https://www.genevestigator.com/gv/). Fmr1nb, another possible candidate, shows high expression in pre-pachytene spermatocytes [43].
Intrasubpecific autosomal polymorphisms suppress asymmetry of HS
The ability of Hstx2B6 to rescue the meiotic arrest of Mmm×Mmd F1 hybrids is subject to intrasubspecific Mmm polymorphisms. While asymmetric male sterility of (PWD×B6)F1 hybrids depends on the presence of the Hstx2PWD allele, another inbred strain derived from Mmm, known as STUS produces fully sterile F1 hybrid males with B6 mice regardless of the direction of the cross [44]. To map the STUS/PWD autosomal allelic variants that ensure full intrameiotic arrest in males carrying Mmm Chr XB6, we genotyped 84 test-cross males from crosses of B6 females with (PWD×STUS)F1 or (STUS×PWD)F1 males. QTL analysis of the sperm count (binomial, sperm cells present or absent) revealed QTLs on Chr 3, Chr 9 and Chr 13, while QTL for the testes weight mapped on Chr 3 and Chr 13 (Fig. 4). The reciprocal cross of (STUS×PWD) females with B6 males yielded only sterile male offspring without sperm in ductus epididymis (Table S3), strongly indicating that these autosomal QTLs interact with Mmm Chr XB6. Several interesting candidate genes with meiotic functions have been found in these QTL regions, including Hormad1, Sycp1, H2afx or Msh3 (Table S4). Admittedly, 1.5-LOD support intervals of the QTLs proved quite large and none of the selected candidates displayed a dN/dS ratio indicative of their rapid evolution.
Fig. 4. Single QTL scan for autosomal loci supporting Hstx2 independent intrameiotic arrest of (Mmd×Mmm)F1 intersubspecific hybrids. 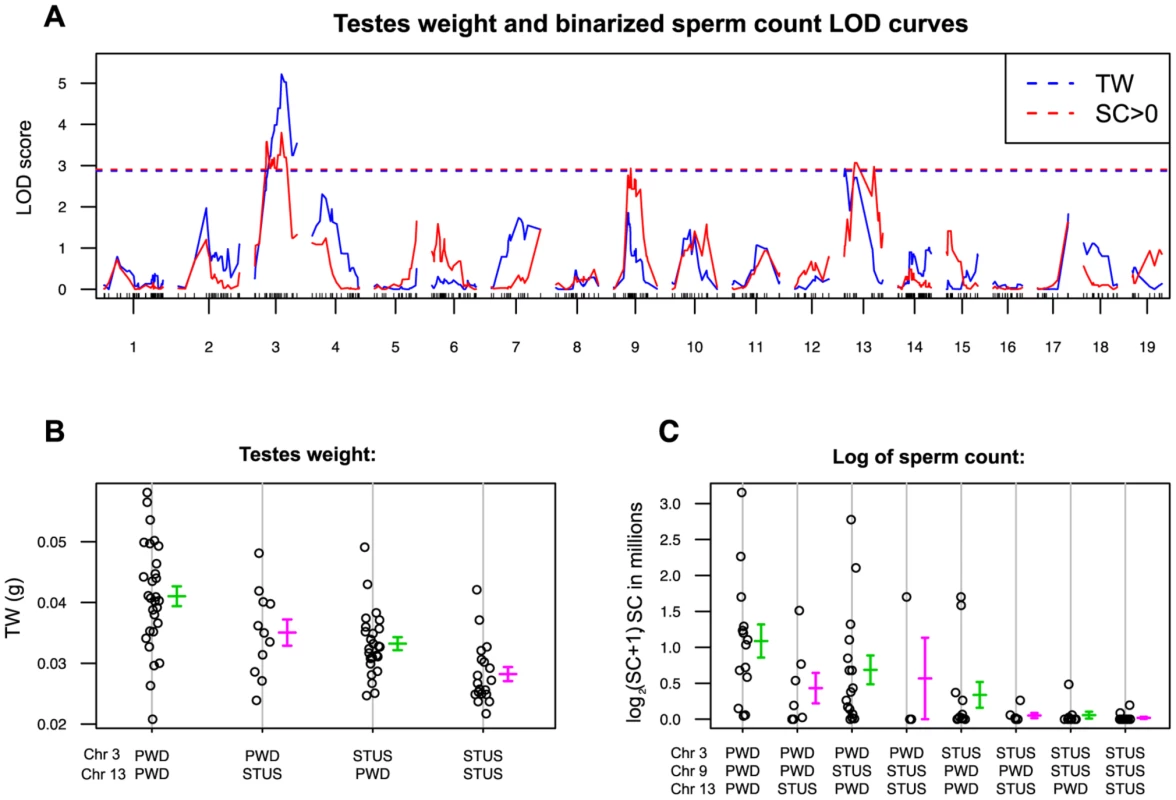
(A) Testes weight QTLs (red) reached significance on Chrs 3 (marker UNC030163295, Chr3: 104,320,699) and 13 (JAX00357337 Chr 13:47,975,634) and QTLs for sperm count on Chrs 3, 9 (JAX00171568, Chr 9:56,491,601) and 13. Sperm count was evaluated as a binary trait (SC = 0, SC>0). (B) and (C) Additive effect of QTLs on testes weight and sperm count. For map positions and possible candidate genes see Table S4. Hstx2 affects meiotic pairing and spermatogenic differentiation
The meiotic arrest of (PWD×B6)F1 hybrid males is associated with failure of proper synapsis of homologous heterosubspecific autosomes, delay of DNA double-strand break (DSB) repair on unsynapsed autosomes and dysregulation of meiotic sex chromosome inactivation (MSCI) at the first meiotic prophase. However, full fertility and complete autosomal synapsis is restored when Chr 17 is PWD/PWD consubspecific on otherwise PWD/B6 F1 background [22]. Here we focused on the fertility parameters and pachytene chromosome synapsis in F1 hybrid males differing at the Hstx2 locus (Fig. S4A–E). The Hstx2PWD allele in (B6.PWD-Chr X.1s×PWD) hybrid males ensured full sterility, meiotic arrest at mid-late pachynemas, almost absent diplotene spermatocytes and a lack of sperm. Immunostaining of SYCP3 and SYCP1 components of lateral and central elements of synaptonemal complexes or HORMAD2 protein revealed unsynapsed autosomes in >90% of pachynemas of both, (PWD×B6) and (B6.PWD-Chr X1s×PWD) F1 hybrid males. The super-resolution structured illumination microscopy documented irregular spots of SYCP1 on some univalents and nonhomologous synapsis and/or translocations (Fig. 5), resembling “tangles” observed in pachynemas with reduced frequency of DSBs [45]. In contrast, (B6.PWD-Chr X.1×PWD) males carrying Hstx2B6 were semifertile, with partial meiotic arrest at late pachytene stage. Only 34% of pachynemas showed asynapsis of one or two pairs of autosomes (Fig. S4A–E). It can be concluded that in F1 hybrid males the Hstx2B6 allele partially restores fertility and significantly reduces the frequency of pachynemas with asynapsis and the number of unsynapsed autosomes per cell.
Fig. 5. Super-resolution microscopy of synaptonemal complexes on spreads of (B6.PWD-Chr X.1s×PWD) pachytene spermatocytes. 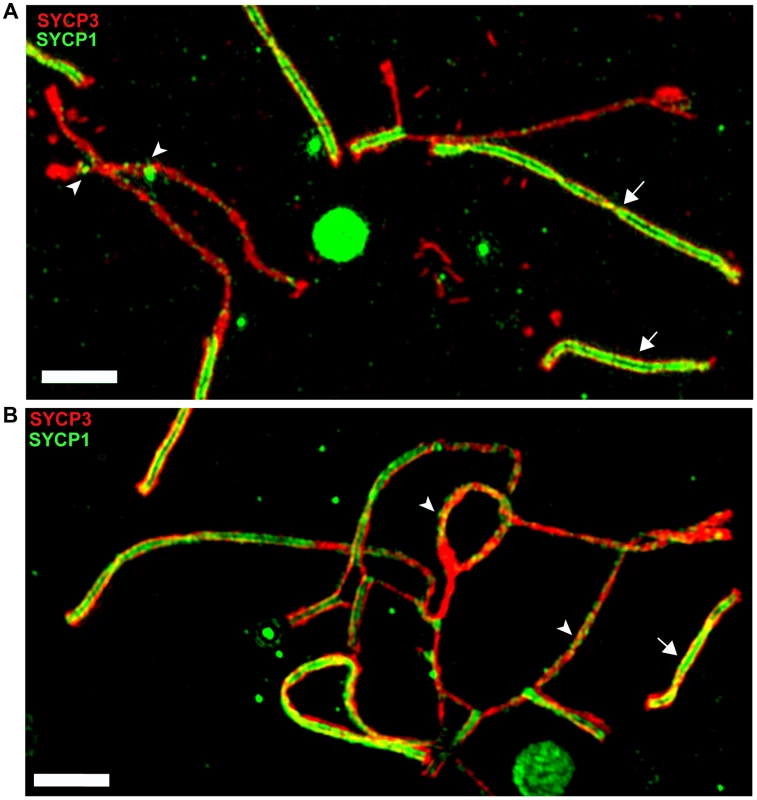
(A) Detail of a pachytene spermatocyte of a sterile male immunostained by SYCP3 (red) and SYCP1 (green) antibodies. Properly synapsed bivalents (arrows) show two parallel threads of transverse filaments decorated by SYCP1 antibody which form the central region embedded in SYCP3 lateral elements. Unsynapsed chromosomes lack transverse filaments but display some irregular SYCP1 spots (arrowheads). (B) Example of a nonhomologous pairing and/or translocations, and asynapsis in pachynema of (B6.Chr X.1s×PWD)F1 sterile male. Bar 2000 nM. To test whether the occurrence of meiotic asynapsis is not limited to the (PWD×B6) strain combination, we checked chromosome synapsis in pachytene spermatocytes of (STUS×B6)F1 and (PWD×SCHEST)F1 hybrids. SCHEST is a wild-derived strain of Mmd (J.P., unpublished). Both F1 hybrids were sterile, showing no sperm (STUS×B6) or few sperm (PWD×SCHEST, <0.9 mil.) in ductus epididymis. In both cases >90% of pachytene spermatocytes revealed multiple pairs of unsynapsed autosomes (Fig. S5A, B). Thus, asynapsis in Mmm×Mmd intersubspecific hybrids is a more general phenomenon, not confined to the incompatibilities between B6 and PWD genome.
Hstx2 and Hst1/Prdm9 regulate male but not female asynapsis of heterosubspecific homologs
Asynapsis preferentially affects autosomal pairs with heterosubspecific homologs, and their pairing failure is strongly influenced by the Prdm9 and Hstx2 genes in sterile hybrid males ([22] and above). We asked whether the genetic control of meiotic asynapsis differs between male and female gametogenesis of intersubspecific hybrids. The pachytene chromosome asynapsis was not observed in PWD and B6 spermatocytes, but occurred in 14% and 29% of pachytene oocytes of the same genotype. In (PWD×B6)F1 hybrid females, 47.5% of pachynemas showed asynapsis, but contrary to the F1 hybrid males the frequency of asynaptic oocytes was not dependent on the Prdm9 and Hstx2 genotypes (Fig. 6A, B). The conclusion was reached from the comparison of male and female meiosis in hybrids between particular consomics and PWD. Thus, 46% of (PWD×B6.PWD-Chr 17)F1 pachytene oocytes displayed asynapsis that was completely absent in spermatocytes of the same genotype. Moreover 46.5% and 44% of oocytes of (B6.PWD-Chr X.1×PWD)F1 and (B6.PWD-Chr X.1s×PWD)F1 hybrids showed asynaptic autosomes (Fig. 6B), compared to 34.1% and 96.4% of pachynemas of the corresponding male genotypes. It can be concluded that contrary to male meiosis, Chr 17 and Hstx2 do not change the overall frequency of asynaptic pachynemas in female meiosis of intersubspecific hybrids. However, detailed analysis of female hybrids consubspecific for Chr 17PWD showed a lower number of unsynapsed autosomes per cell (p<0.01) when compared with the other intersubspecific F1 hybrid genotypes (Fig. 6C). Thus, Prdm9/Hst1 and/or some other genes on Chr 17 exert a limited effect on asynapsis in female hybrids as well. The elevated incidence of asynaptic oocytes predetermined to elimination in intersubspecific ovaries was reflected by a reduced ratio of diplotene/pachytene oocytes in ovarian cell spreads (Fig. 6D).
Fig. 6. Meiotic asynapsis in female hybrids. 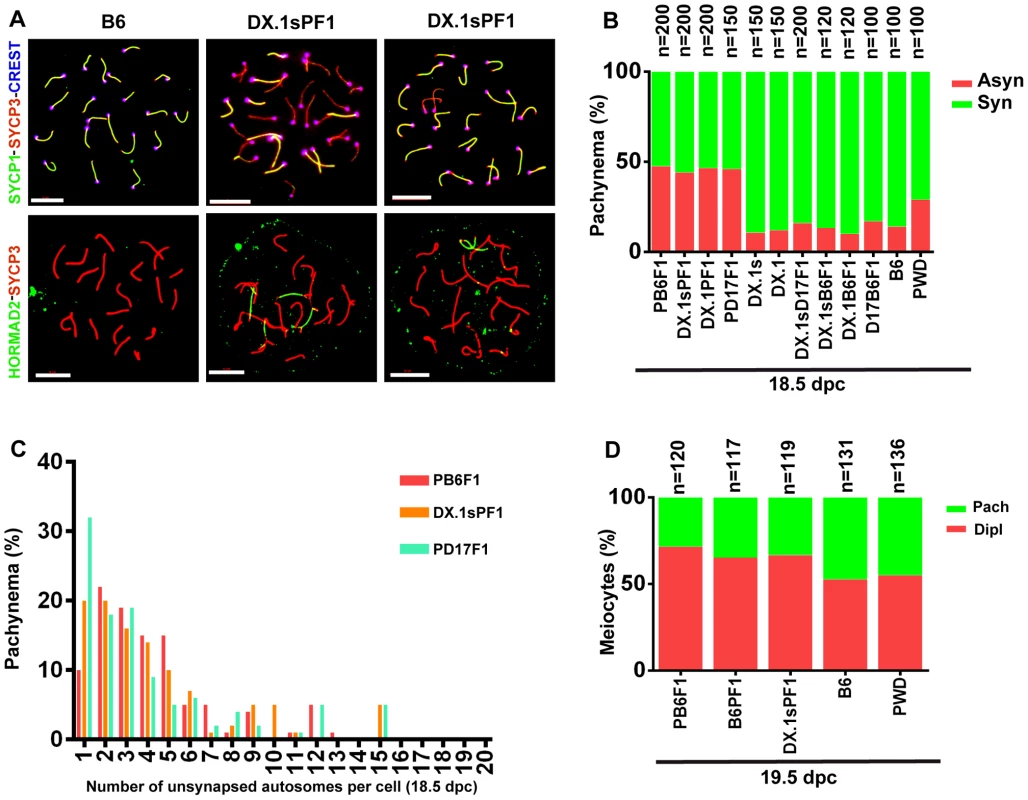
Abbreviations of consomic strains and their hybrids: DX.1 – B6.PWD-Chr X.1; DX.1PF1 – (B6.PWD-Chr X.1×PWD)F1; DX.1sPF1 – (B6.PWD-Chr X.1s×PWD)F1; DX.1B6F1 – (B6.PWD-Chr X.1×B6)F1; DX.1sB6F1 – (B6.PWD-Chr X.1s×B6)F1; DX.1sD17F1 – (B6.PWD-Chr X.1s×B6.PWD-Chr 17)F1; D17B6F1 – (B6.PWD-Chr 17×B6)F1; B6PF1 – (B6×PWD)F1, PB6F1 – (PWD×B6)F1; PD17F1 – (PWD×B6.PWD-Chr 17)F1. (A) Chromosome synapsis in pachytene oocytes of B6 and (B6.PWD-Chr X.1s×PWD)F1 18.5–19.5 dpc female fetuses was analyzed by combination of SYCP1, SYCP3 and CREST (centromeric heterochromatin) immunostaining or by HORMAD2 and SYCP3 to detect unsynapsed chromosomes. Bar, 10 µm. (B) The frequency of oocytes showing one or more asynaptic chromosomes is similar (>40%) irrespective of Hstx2 and Prdm9/Hst1 genotype. (C) Although the (PWD×B6), (B6.PWD-Chr X.1s×PWD), (B6.PWD-Chr X.1×PWD) and (B6.PWD-Chr 17×PWD)F1 hybrid females do not differ in percentage of pachytene oocytes with asynapsis, the (B6.PWD-Chr 17×PWD)F1 females, conspecific for Chr 17PWD, carry significantly less asynapsed chromosomes per cell. (D) The frequency of diplonemas in spread oocyte preparations was significantly lower (p<0.01, χ2 test) in intersubspecific hybrids than in parental inbred strains.. Heterosubspecific but not consubspecific homologs are sensitized to asynapsis in female intersubspecific hybrids as well
We have shown that consubspecific (PWD/PWD) homologous autosomes evade asynapsis in otherwise heterosubspecific (PWD/B6) genomic background of hybrid males. The finding indicated a cis-type of asynapsis control based on some kind of mismatch between orthologous chromosomes of Mmm and Mmd origin [22]. Considering the difference between male and female hybrids in the overall frequency of asynapsis and the male-limited effect of HS genes we asked whether the mismatch of heterosubspecific homologs lowering their synapsis efficiency also operates in female meiosis.
For this purpose we analyzed primary oocytes of female hybrids between PWD and chromosome substitution strains carrying Chr 17PWD, Chr X.1PWD or Chr X.1sPWD, respectively. We compared the efficacy of meiotic synapsis of consubspecific (PWD/PWD) Chr 17 and Chr X homologs with matching heterosubspecific pairs in (PWD×B6)F1 pachytene oocytes and oocytes from the parental controls. Using the whole-chromosome DNA FISH we also visualized Chrs 2, 16, 18, 19 and X. In (PWD×B6)F1 pachynemas Chr 2 showed the lowest incidence of asynapsis. The frequency of univalents of small autosomes 16, 17, 18 and 19 varied between 18% and 49% in asynaptic pachytene oocytes. Strikingly, Chr X displayed the highest frequency (64%) of asynapsis (Table 2).
Tab. 2. Asynapsis of individual chromosomes in pachytene oocytes of intersubspecific F1 hybrids and parental controls. 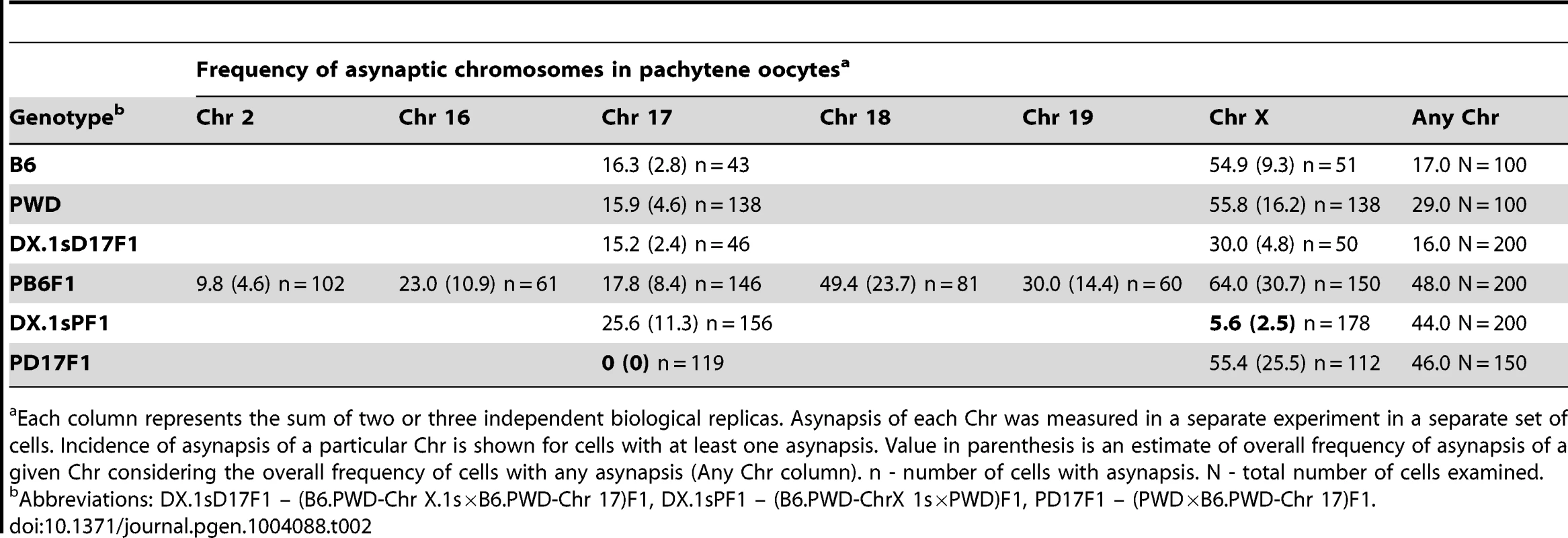
Each column represents the sum of two or three independent biological replicas. Asynapsis of each Chr was measured in a separate experiment in a separate set of cells. Incidence of asynapsis of a particular Chr is shown for cells with at least one asynapsis. Value in parenthesis is an estimate of overall frequency of asynapsis of a given Chr considering the overall frequency of cells with any asynapsis (Any Chr column). n - number of cells with asynapsis. N - total number of cells examined. The asynapsis of consubspecific Chr 17PWD/PWD homologs dropped to zero in (PWD×B6.PWD-Chr 17)F1 oocytes, although the total frequency of pachynemas with asynapsis was the same as in (PWD×B6) hybrids. In (PWD×B6.PWD-Chr X.1s)F1 oocytes, 69.9 Mb of the centromeric part of Chr X was consubspecific for the PWD sequence, while the end of the chromosome, 101.4 Mb in length, was PWD/B6 heterosubspecific. Nevertheless, the partial PWD homozygosity was sufficient to reduce Chr X asynapsis from 64% down to 5.6% of pachytene oocytes (Table 2). It can be concluded that asynapsis in intersubspecific female and male hybrids follows the same rule, depending on yet unspecified sequence incompatibility between individual homologs of Mmm and Mmd origin.
Pursuing the dominance theory of Haldane's rule
To explain Haldane's rule of hybrid sterility, the dominance theory posits the recessive nature of X-linked variants that disrupt gametogenesis in hemizygous (XY) but not in homozygous (XX) sex [9]. In its most straightforward interpretation the F1 hybrid females should be sterile in the same way as their hemizygous male sibs if their genotype were made homozygous for an incompatible Chr X variant. We constructed such genotype by crossing consomic females B6.PWD-Chr.X.1s with PWD males. The resulting female hybrids were PWD/B6 heterosubspecific for the whole autosomal genome but consubspecific for proximal 69.9 Mb of Chr XPWD, encompassing the Hstx1/2 hybrid sterility locus. Contradicting the simple interpretation of Muller's dominance hypothesis the (B6.PWD-Chr.X.1s×PWD)F1 hybrid females were fully fertile, as were the parental controls (Table S5). However, the results were concordant with the testis-specific function of Hstx2PWD. Thus, to follow the dominance theory, the preponderance of male-limited HS in species with heterogametic sex could be explained by a predominance of recessive, compared to dominant, mutations of HS genes and their male-limited expression. Admittedly, the latter premise is in conflict with HS obeying the Haldane's rule in birds and Lepidoptera.
Discussion
The role of Hstx2 in F1 hybrid sterility
Disproportionate involvement of Chr X in HS has been well documented in classical studies of Drosophila hybrids (for review see [46], [47]) and repeatedly reported in hybrids of the house mouse subsp [27], [33], [36], [48]. The introgression of Chr XPWD of Mmm into Mmd B6 genetic background resulted in abnormal sperm morphology and inability to fertilize eggs. This phenotype is controlled mainly by the Hstx1 locus supported by additional loci on Chr X [33]. With the aim to positionally clone the Hstx2 gene we narrowed down the critical region to a 4.7 Mb interval (Chr X: 64.88 Mb–69.58 Mb) and showed that it also carries the Hstx1 locus. In contrast to Hstx1 phenotype, the intrameiotic arrest is controlled from a single Hstx2 locus on Chr X. The Hstx2/Hstx1 critical interval is overlapped by and may be identical with the 8.4 Mb QTL responsible for the sterilizing effect of Mmm Chr XPWK introgressed into the genetic background of Mmd LEWES inbred strain [30]. Using the position on Chr X, spermatogenic expression, and dN∶dS ratio Good and coworkers [41] predicted nine candidate genes for X-linked hybrid sterility, three of which (Ctag2, 4933436I01Rik and Fmr1nb) also occur in the present list of Hstx1/2 candidates. Moreover, the Sha2 locus of the Japanese house mouse Mus m. molossinus [49], potentially identical with Hstx1, maps to the same interval. Sha2 is responsible for spermiogenic arrest in B6 males carrying Chr XM.m.molossinus introgression [49]. Among the candidate genes in the region, Fmr1nb and 4933436I01Rik are expressed in the appropriate cell type during germ cell differentiation and display two and seven non-synonymous substitutions, respectively. The critical region also contains the Mir465 cluster of miRNA genes, which show a significant difference in expression between reciprocal F1 hybrids. Moreover, Mir743a carries a single SNP in its seed sequence. Admittedly, the Hstx1/Hstx2 candidate region is still too large to finalize the list of Hstx2/Hstx1 candidate genes. Recently, many ampliconic genes on the mouse and human Chr X were shown to be unique for a given species and expressed predominantly in testicular germ cells [50]. These features make them potential candidates for reproductive isolation genes. The Hstx1/Hstx2 critical region is flanked by amplicons 4930527E24Rik and Xlr (amplicons 7 and 9 in [51]). However, none of the candidate protein-coding genes or Mir genes is located within a known amplicon and all protein-coding candidates have an ortholog in other mammalian species.
The incompatible alleles of major HS loci are not fixed to homozygosity within Mmm and Mmd subspecies, in agreement with the idea of the early stage of their speciation. Hst1/Prdm9 is polymorphic for “sterility” and “fertility” alleles in natural populations and in inbred strains [23], [44], [52] and the same is true for the X-linked HS QTLs [44], [53]. The asymmetric contribution of Mmm and Mmd genomes to HS of reciprocal hybrids reveals polymorphic control as well. Chr XB6 causes meiotic arrest depending on PWD/STUS polymorphic modifiers on Chrs 3, 9 and 13.
The proximal part of Chr X carries loci for major incompatibilities also outside the Mus musculus group of mouse subpecies. Introgression of Chr X of Mus spretus into B6 inbred strain reduced testes weight and fertility [54], with the largest LOD score mapping approximately 10 Mb proximal to the Hstx1/Hstx2 region. The males were semifertile, with many tubules containing normally developing spermatozoa but some tubules completely devoid of germ cells. The testes of (Mus macedonicus×B6) F1 hybrid males displayed premeiotic block with Sertoli cells and dividing spermatogonia. The QTL analysis of a backcross population revealed two major loci, again on Chr 17 and Chr X, but with positions distal to Prdm9 and to Hstx1/Hstx2 loci, respectively [55]. These crosses confirmed the large X-effect in mice but did not reveal common HS genes beyond the group of house mouse subspecies.
Laboratory crosses of wild-derived inbred strains continue to reveal new details about the genetic control and mechanistic basis of hybrid sterility, but their direct role in speciation is less clear. In an alternative approach, evolutionary biologists collect wild mice from the house mouse hybrid zone to analyze the introgression of (sub)species-specific DNA markers. Because this approach can reflect all kinds of incompatibilities restricting the gene flow, the results are usually complex, uncovering multiple regions on autosomes and Chr X. Nonetheless, at least two common themes have emerged from the field data and laboratory crosses, namely the large X-effect and a specific role of 60 Mb–80 Mb interval of Chr X in reproductive isolation of Mmm/M. m. molossinus and Mmd [16], [17], [56], [57]
The role of meiotic chromosome asynapsis in intersubspecific reproduction barrier
More than 90% of primary spermatocytes of sterile Mmm×Mmd F1 hybrids fail to synapse properly their chromosomes at the pachytene stage of meiosis. Unsynapsed autosomes carry DMC1/RAD51 foci on unrepaired DSBs and are decorated by the phosphorylated form of histone H2AFX. The sex body containing X and Y chromosomes is often malformed or disappears, and transcriptional inactivation of sex chromosomes (MSCI) is disrupted [22], [58]. Such failure of chromosomes to synapse can be under trans - or cis-control. Mutations of various meiotic genes involved directly or indirectly in meiotic chromosome pairing and synapsis cause asynapsis of multiple autosomes in trans [59]. A null allele of the Prdm9 gene on Mmd background causes male and female sterility associated with asynapsis and failure to form the sex body [60], the phenotype similar to sterile Mmm×Mmd hybrids [22], [24]. Null mutations of Sycp1, Hormad1 or Mei4, the candidate genes regulating the Hstx2-controlled asymmetry of HS, also cause asynapsis. Contrary to genes acting in trans, certain structural mutations of chromosomes such as translocations or inversions can cause local, cis-acting asynapsis, followed by meiotic arrest and sterility [43], [61]–[63]. To distinguish between the trans - and cis - control of asynapsis in sterile Mmm×Mmd males, we modified the F1 hybrids using the intersubspecific chromosome substitution strains B6.PWD-Chr# [20]. In (B6.PWD-Chr#×PWD)F1 hybrids we compared the ability of consubspecific and heterosubspecific (PWD/PWD vs PWD/B6) chromosomes to synapse on otherwise F1 hybrid background. These experiments clearly showed that asynapsis is regulated primarily in cis, because heterosubspecific chromosome pairs in male as well as in female meiosis were more prone to failure to synapse. The sensitivity of heterosubspecific chromosome pairs to asynapsis was strongly modified in trans by the Prdm9 and Hstx2 HS genes because only Prdm9PWD/B6 and Hstx2PWD allelic combinations on F1 hybrid background resulted in asynapsis of multiple autosomes in >90% of pachytene spermatocytes and full meiotic arrest. A few proteins required for meiotic chromosome alignment and pairing have been described [59]; however, the cis-acting signals which ensure pairing and synapsis of homologous chromosomes are still unknown. Our assay of meiotic synapsis, comparing the consubspecific versus heterosubspecific pairing homologs, offers a genetic approach to solving the problem. Such experiments are in progress.
Haldane's rule and male-specific effect of Hstx2PWD in mouse hybrids
Hybrid sterility affects preferentially heterogametic sex, males in Drosphila or mammals, and females in birds and Lepidoptera. Several hypotheses have been proposed to explain Haldane's rule, including the dominance theory, the faster-male theory, the faster X theory and meiotic drive [47]. According to Muller's dominance theory, Haldane's rule can be explained by hemizygosity of the X (Z) chromosome in the heterogametic sex. Both, recessive and dominant X (Z)-linked HS genes could control HS in heterogametic sex, but only dominant HS genes could sterilize the homogametic sex. Using attached-X chromosome, Coyne and others tested the prediction by making Drosophila hybrid females homozygous for the recessive-acting X chromosome. In D. simulans×D. mauritiana or D. sechellia hybrids the female hybrids carrying two D. simulans Chr X were fertile, contrary to the prediction (for review see [47] and references therein). The authors argued that because HS genes are male specific, suggesting extra developmental sensitivity of spermatogenesis relative to oogenesis, their recessive forms manifest DMI in male but not in female gametogenesis. Indeed, the mouse Hst1/Prdm9 and Hstx2 are also male-specific HS loci, therefore not contradicting the fertility of female Mmm×Mmd F1 hybrids homozygous for the Hstx2Mmm allele. To accommodate the dominance theory with X-linked, male-specific HS genes, their recessive nature could be tested by a dominant transgene from the Mmm subspecies.
A mechanistic model of F1 hybrid sterility
We propose a mechanistic model of HS based on the assumption that cis-controlled pachytene asynapsis is the primary cause of apoptosis of primary spermatocytes and sterility in intersubspecific hybrids of the house mouse (Fig. 7). We have found that the number of unsynapsed autosomes per cell varies, indicating that the same type of cis-acting mechanism operates on individual autosomes. Although the molecular mechanism of homologous chromosome recognition during meiotic pairing and synapsis is unknown in mammals, one possibility is that fast evolving noncoding DNA and/or RNA sequences interfere with homology search of single-strand 3′ends on heterosubspecific homologs during DSB repair, thus interfering with their synapsis during the first meiotic prophase (see also [45]). Alternatively, the process of homolog recognition can be affected earlier, before meiotic recombination begins.
Fig. 7. The proposed sequence of events leading to male limited sterility of intersubspecific hybrids of house mouse. 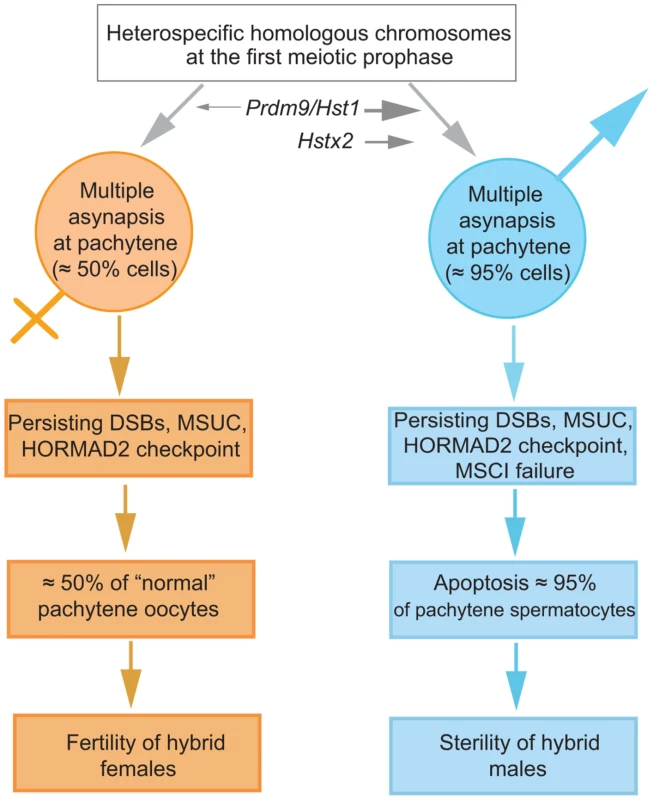
Susceptibility of heterosubspecific homologs to asynapsis is common to both sexes. Prdm9/Hst1 and Hstx2 hybrid sterility genes can modulate this sensitivity from 0% to >95% in spermatogenesis but not in oogenesis, depending on allelic combinations of epistatic DMIs. Multiple asynaptic autosomes provoke MSCI, contributing to hybrid male sterility. Approximately one half of unaffected oocytes ensure fertility of hybrid females. It remains to be established what is the cause of asynapsis of heterosubspecific homologs. The male-specific action of the Hstx2 or Prdm9/Hst1 HS genes could explain why the same perturbation of sequence homology acts differently in male and female meiosis. While in female pachynemas the percentage of meiocytes with asynaptic chromosomes ranges between 40% and 45% and is ultimately unaffected by the Hstx2 or Prdm9/Hst1 genes, meiotic asynapsis of F1 males depends considerably on their genotype. Full sterility and complete meiotic block is associated with the Prdm9PWD/B6 Hstx2PWD genotype only, while the substitution of Hstx2PWD for Hstx2B6 on otherwise F1 hybrid background reduces the incidence of cells with asynapsis to the level found in female meiosis. Moreover, full recovery of fertility and meiotic pairing can be achieved in F1 hybrid males consubspecific for Chr 17PWD (homozygous Prdm9PWD/PWD). We expect that analysis of recombinant heterosubspecific/consubspecific chromosome pairs could provide the genetic approach to identifying the cis-acting sites important for meiotic pairing and synapsis.
Materials and Methods
Animals and ethics statement
The animals were maintained at the Institute of Molecular Genetics in Prague and Institute of Vertebrate Biology in Studenec (Academy of Sciences), Czech Republic. The project was approved by the Institutional Animal Care and Use Committee of the Institute of Molecular Genetics, AS CR, protocol No. 141/2012. The principles of laboratory animal care (NIH Publication No. 85-23, revised 1985) as well as specific Czech Law No. 246/1992 Sb. compatible with the corresponding EU regulations and standards, namely Council Directive 86/609/EEC and Appendix A of the Council of Europe Convention ETS123, were observed.
C57BL/6J (B6, mostly M. m. domesticus) originated from The Jackson Laboratory (Bar Harbor, ME). The PWD/Ph (PWD) and STUS strains were derived from wild M. m. musculus [32], [53]. The chromosome substitution (consomic) strains C57BL/6J-Chr #PWD [20], abbreviated here B6.PWD-Chr #, were maintained in a pathogen-free barrier facility with a 12 h light/12 h dark cycle. The mice had ad libitum access to a standard rodent diet (VELAZ, ST-1, 3.4% fat) and acidified water. All males were sacrificed at the age of 60 to 70 days.
Genotyping, phenotyping and histology
All 124 male mice from (B6.PWD-Chr X×B6)F1×PWD cross and 71 male progeny from (B6.PWD-Chr X×B6)F1x B6 cross were genotyped using SSLP Chr X markers listed in Table S6. Genomic DNA from mouse tails and spleens was prepared by the HotSHOT method [64] followed by phenol-chloroform cleaning [65]. For genotyping B6.PWD-Chr X# partial consomics and 84 males of B6×(PWD×STUS) test-cross we used MegaMUGA high-density genotyping array carrying 77,000 markers on Illumina Infinium Platform (http://csbio.unc.edu/CCstatus/index.py).
Eight weeks old male progeny of the crosses were phenotyped for testes weight (TW), sperm count (SC) and percentage of abnormal spermatozoa as described [33]. The B6×(PWD×STUS) males were phenotyped at 60 days of age as described [44]. For histological analysis the paraffin-embedded testicular sections were stained with periodic acid Schiff and hematoxylin-eosin and observed using a Nikon Eclipse 200 microscope. The Penguin 150CL CCD color camera (Pixera) was used to capture photographs, which were processed using Adobe Photoshop (Adobe Systems).
QTL analysis
QTL mapping of recombinant males from (B6.PWD-Chr X×B6)F1×PWD and B6×(PWD×STUS) crosses was performed using the R 12.1 and its R/qtl package [66] Marker positions were taken from MGI mouse genetic map [67]. Standard interval mapping was implemented using scanone function. TW and logSC were modeled as continuous variables, fertility/sterility as a binary variable. Genotype probabilities between markers were calculated at a grid size of 5 cM and with genotyping error rate of 0.01%. Genome-wide significance was calculated by 1000 permutations and compared to α = 5% threshold.
Fluorescence-activated cell sorting of spermatogenic populations
Three spermatogenic populations (leptotene+zygotene+early pachytene, mid-late pachytene+diplotene, and spermatids) were isolated using fluorescence-activated cell sorting (FACS) as described earlier [43], [68] from PWD and B6 testes. The cells were directly sorted into QIAzol lysis reagent of the miRNeasy Mini isolation kit (QIAGEN). Small aliquots of cells were sorted in Krebs-Ringer bicarbonate medium for indirect immunofluorescence analysis. The population composition was estimated by staining with anti-SYCP3, anti-SYCP1, anti-γH2AFX antibodies (details below) and DAPI (Vectashield). All sorted populations showed 85–90% purity of the desired cell type.
Microarray miRNA and protein-coding gene transcription analysis and qRT-PCR validation
Total RNA was isolated from sorted cells and 14.5 dpc testis using the miRNeasy Mini isolation kit (QIAGEN) as recommended. RNA concentration was determined by NanoDrop (NanoDrop Technologies) and its integrity checked in Agilent 2100 bioanalyzer, RNA Lab-On-a-Chip (Agilent Technologies). The total RNA (20–30 ng for gene expression and 120 ng for miRNA expression) was converted to cRNA using the Affymetrix Two-Cycle Target Labeling kit according to the manufacturer's instructions or using the Affymetrix 3′ IVT Express Kit. Affymetrix GeneChip Mouse Genome 430.2.0 array, and Affymetrix GeneChip miRNA 1.0 Array was hybridized with cRNA. The data obtained from the experiments were analyzed using Bioconductor [69] (http://www.bioconductor.org/) and the R project for statistical computing (version 2.12; http://www.r-project.org/). The probes were annotated to Ensembl gene identifiers using the custom chip description file, which was based on NCBI build 37. The data were normalized using RMA (Affymetrix GeneChip Mouse Gene 1.0ST Array) and quantile normalization (Affymetrix GeneChip miRNA 1.0). We used Linear Models for Microarray Data Package, limma version 3.6 [70] for statistical evaluations of expression differences as described [22]. The microarray dataset is deposited in the NCBI Gene Expression Omnibus (GEO) with series accession number GSE41707 [22] GSE49442 and GSE49443. Expression of different X-linked protein-coding genes on spermatogenic populations were derived from NCBI GEO profiles or NCBI GEO database GSE7306 [43] and GSE49444.
For qRT-PCR of protein-coding genes, reverse transcription of isolated RNA samples was carried out using Applied Biosystems (ABI) high-capacity cDNA reverse transcription kit. The quantification of mRNAs was performed using FastStart DNA Master SYBR Green I kit (Roche) and amplified in LightCycler 2000 (Roche). Reactions without reverse transcriptase were utilized as negative control. The assays were done in biological and technical triplicates. The data were analyzed using LightCycler Software version 3.5.3 (Roche). For validation of miRNA expression we used ABI TaqMan MicroRNA assays and followed the manufacturer's instructions. The reactions were cycled in Applied Biosystems 7300 Real-time PCR system, and associated software was used for data analysis. The reactions were also carried out using biological and technical triplicates and proper negative controls. The highest and stably expressed miRNAs, U6 non-coding RNA for sorted cells and Mir152 for 14.5 dpc testis, were used as the reference for data normalization. The primers were designed using Primer 3 software (http://frodo.wi.mit.edu/). Sequences of primers are given in Table S6.
Sequencing, SNP and PWD exome analysis
Exome sequence analysis was carried out for PWD/Ph mice at BGI Europe using Illumina HiSeq 2000 sequencers. The PWD exome sequence was aligned to NCBIm37 genome (BAM format, http://samtools.sourceforge.net/SAM1.pdf) and deposited at the Sequence Read Archive (SRA) accession SRR942524.
All the non-synonymous mutations between PWD and B6 for 4.7 Mb Hstx2 locus were tabulated. Sequence validations on PWD cDNA (for protein-coding genes) and PWD BACs (for miRNAs) [35] were carried out as described [24] using sequencing capillary machine ABI310 (Applied Biosystems). The sequences of primers are listed in Table S3. Some of the SNPs were also confirmed using the Mouse Phenome database (http://phenome.jax.org/). The dN∶dS ratio (an indicator of evolutionary selective pressure on genetic processes) of different X-linked protein-coding genes between rat and mouse was calculated using Ensemble Biomart.
Immunostaining and DNA FISH of spread meiocytes
The meiocyte spreads were prepared by using the hypotonic protocol as described earlier [21], [71]. The nuclei were immunostained using following primary antibodies; rat polyclonal anti-SYCP3 (Abcam, #15092), mouse monoclonal anti-SYCP1 (Abcam, #15087), guinea pig anti-histone linker H1t [72], human autoimmune anti-centromere (AB-Incorporated, #15-235), mouse monoclonal anti-γH2AFX (Upstate, #05-636), mouse monoclonal antibody anti-SYCP3(D-1) (Santa Cruz #74569), rabbit polyclonal antibody HORMAD2(C-18) (Santa Cruz #82192) and the secondary antibodies: goat anti-Rabbit IgG-AlexaFluor488 (Molecular Probes, A -11034), goat anti-Mouse IgG-Alexa Fluor 568 (Molecular Probes, A-11031), goat anti-Rabbit IgG-Alexa Fluor 568 (Molecular Probes, A-11036), goat anti-Mouse IgG-Alexa Fluor 350 (Molecular Probes, A - 21049), goat anti-Mouse IgG-Alexa Fluor 647 (Molecular Probes, A-21236), goat anti-Rabbit IgG-Alexa Fluor 647 (Molecular Probes, A-21245) and goat anti-Guinea pig IgG-Cy3 (Chemicon, #AP108C). The immunolabeled meiocytes were subjected to DNA FISH using Metasystems XMP XCyting Mouse Chromosome N Whole Painting Probes for Chrs 2, 16, 17, 18, 19 and X as described [73]. The images were acquired using a Nikon Eclipse 400 (Tokyo, Japan) microscope with motorized stage control using a Plan Fluor objective, 60× (Nikon, MRH00601) and captured using a DS-QiMc monochrome CCD camera (Nikon) and NIS elements program. The fluorescent intensity of images was adjusted using Adobe Photoshop CS software (Adobe Systems). For super-resolution microscopy the meiotic spreads were examined with the Zeiss Axioimager Z.1 platform equipped with the Elyra PS.1 super-resolution system for SR SIM and the LSM780 module for CLSM, using Alpha Pln Apo 63×/1.40 oil Zeiss objective (total magnification 1008×) with appropriate oil (Immersol 518F, by Zeiss). SR-SIM setup was 5 rotations and 5 phases for each image layer. Up to 7 (usually 3) 110 nm Z-stacks were acquired per image. Staging of meiotic prophase I in males and females were done as described earlier [22].
Statistics
Multiple biological replicates of each genotype were analyzed for cellular phenotypes and RNA expression. The significance of body weight, testes weight, sperm morphology and breeding phenotypes was computed using Welsch's t-test. Sizes of chromatin areas covered by the hybridization signal in synapsed and unsynapsed autosomes were compared by Analysis of Variance (ANOVA) with Tukey's correction for multiple testing. Differences between cellular phenotypes were determined with χ2 test. All computations were done using R 2.15.0 or Graphpad Prism (http://www.graphpad.com/scientific-software/prism/#1).
Supporting Information
Zdroje
1. Dobzhansky T (1951). Genetics and the origin of Species: Columbia University, New York.
2. Coyne JA, Orr HA (2004) Speciation: Sinauer Associates, Inc., Sunderland, Massachusetts U.S.A.
3. MoehringAJ, LlopartA, ElwynS, CoyneJA, MackayTF (2006) The genetic basis of postzygotic reproductive isolation between Drosophila santomea and D. yakuba due to hybrid male sterility. Genetics 173 : 225–233.
4. SlotmanM, Della TorreA, PowellJR (2004) The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics 167 : 275–287.
5. MaheshwariS, BarbashDA (2011) The Genetics of Hybrid Incompatibilities. Annu Rev Genet 45 : 331–55.
6. HaldaneJ (1922) Sex ration and unisexual sterility in animal hybrids. J Genet 12 : 101–109.
7. MullerH, PontecorvoG (1942) Recessive genes causing interspecific sterility and other disharmonies between Drosophila melanogaster and simulans. Genetics 27 : 157.
8. CoyneJA (1989) Genetics of sexual isolation between two sibling species, Drosophila simulans and Drosophila mauritiana. Proc Natl Acad Sci U S A 86 : 5464–5468.
9. TurelliM, OrrHA (1995) The dominance theory of Haldane's rule. Genetics 140 : 389–402.
10. TurelliM, OrrHA (2000) Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154 : 1663–1679.
11. ForejtJ (1996) Hybrid sterility in the mouse. Trends Genet 12 : 412–417.
12. ForejtJ (1985) Chromosomal and genic sterility of hybrid type in mice and men. Experimental and Clinical Immunogenetics 2 : 106–119.
13. Forejt J, Pialek J, Trachtulec Z (2012) Hybrid male sterility genes in the mouse subspecific crosses. In: Macholan M, Baird SJE, Muclinger P, Pialek J, editors. Evolution of the House Mouse. Cambridge: Cambridge University Press. pp 482–503.
14. DinW, AnandR, BoursotP, DarvicheD, DodB, et al. (1996) Origin and radiation of the house mouse: Clues from nuclear genes. Journal of Evolutionary Biology 9 : 519–539.
15. Baird SJE, Macholan M (2012) What can the Mus musculus musculus/M. m. domesticus hybrid zone tell us about speciation? In: Macholan M, Baird SJ, Muclinger P, Pialek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press. pp. 334–372.
16. PayseurBA, KrenzJG, NachmanMW (2004) Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58 : 2064–2078.
17. MacholanM, BairdSJ, DufkovaP, MunclingerP, BimovaBV, et al. (2011) Assessing multilocus introgression patterns: a case study on the mouse X chromosome in central Europe. Evolution 65 : 1428–1446.
18. Lewontin R (1974) The genetic basis of evolutionary change. New York: Columbia University Press.
19. KeaneTM, GoodstadtL, DanecekP, WhiteMA, WongK, et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477 : 289–294.
20. GregorovaS, DivinaP, StorchovaR, TrachtulecZ, FotopulosovaV, et al. (2008) Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res 18 : 509–515.
21. AndersonLK, ReevesA, WebbLM, AshleyT (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151 : 1569–1579.
22. BhattacharyyaT, GregorovaS, MiholaO, AngerM, SebestovaJ, et al. (2013) Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Natl Acad Sci U S A 110: E468–477.
23. ForejtJ, IvanyiP (1974) Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet Res 24 : 189–206.
24. MiholaO, TrachtulecZ, VlcekC, SchimentiJC, ForejtJ (2009) A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323 : 373–375.
25. BaudatF, BuardJ, GreyC, Fledel-AlonA, OberC, et al. (2010) PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327 : 836–840.
26. ParvanovED, PetkovPM, PaigenK (2010) Prdm9 controls activation of mammalian recombination hotspots. Science 327 : 835.
27. Dzur-GejdosovaM, SimecekP, GregorovaS, BhattacharyyaT, ForejtJ (2012) Dissecting the genetic architecture of F1 hybrid sterility in house mice. Evolution 66 : 3321–3335.
28. FlachsP, MiholaO, SimecekP, GregorovaS, SchimentiJ, et al. (2012) Interallelic and intergenic incompatibilities of the Prdm9 (Hst1) gene in mouse hybrid sterility. Plos Genet 8: e1003044.
29. TurelliM, MoyleLC (2007) Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics 176 : 1059–1088.
30. GoodJM, HandelMA, NachmanMW (2008) Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution Int J Org Evolution 62 : 50–65.
31. BrideauNJ, BarbashDA (2011) Functional conservation of the Drosophila hybrid incompatibility gene Lhr. BMC Evol Biol 11 : 57.
32. GregorovaS, ForejtJ (2000) PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies–a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol (Praha) 46 : 31–41.
33. StorchovaR, GregorovaS, BuckiovaD, KyselovaV, DivinaP, et al. (2004) Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome 15 : 515–524.
34. SongR, RoS, MichaelsJD, ParkC, McCarreyJR, et al. (2009) Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet 41 : 488–493.
35. JansaP, DivinaP, ForejtJ (2005) Construction and characterization of a genomic BAC library for the Mus m. musculus mouse subspecies (PWD/Ph inbred strain). BMC Genomics 6 : 161.
36. GoodJM, VanderpoolD, SmithKL, NachmanMW (2011) Extraordinary sequence divergence at Tsga8, an X-linked gene involved in mouse spermiogenesis. Mol Biol Evol 28 : 1675–1686.
37. TingC, TsaurS, WuM, WuC (1998) A rapidly evolving homeobox at the site of a hybrid sterility gene [see comments]. Science 282 : 1501–1504.
38. BarbashDA, SiinoDF, TaroneAM, RooteJ (2003) A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci U S A 100 : 5302–5307.
39. PresgravesDC, BalagopalanL, AbmayrSM, OrrHA (2003) Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423 : 715–719.
40. BonoH, YagiK, KasukawaT, NikaidoI, TominagaN, et al. (2003) Systematic expression profiling of the mouse transcriptome using RIKEN cDNA microarrays. Genome Res 13 : 1318–1323.
41. GoodJM, DeanMD, NachmanMW (2008) A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179 : 2213–2228.
42. HruzT, LauleO, SzaboG, WessendorpF, BleulerS, et al. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008 : 420747.
43. HomolkaD, IvanekR, CapkovaJ, JansaP, ForejtJ (2007) Chromosomal rearrangement interferes with meiotic X chromosome inactivation. Genome Res 17 : 1431–1437.
44. VyskocilovaM, PrazanovaG, PialekJ (2009) Polymorphism in hybrid male sterility in wild-derived Mus musculus musculus strains on proximal chromosome 17. Mamm Genome 20 : 83–91.
45. KauppiL, BarchiM, LangeJ, BaudatF, JasinM, et al. (2013) Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 27 : 873–886.
46. Coyne JA, Orr HA (1989) Two rules of speciation. In: Otte D, Endler J, editors. Speciation and Its Consequences. Sunderland, Massachusetts: Sinauer. pp. 180–207.
47. Coyne JA, Orr HA (2004) Speciation. SunderlandMassachusetts: Sinauer Associates. 545 p.
48. GoodJM, GigerT, DeanMD, NachmanMW (2010) Widespread over-expression of the X chromosome in sterile F hybrid mice. PLoS Genet 6: e1001148.
49. OkaA, MitaA, Sakurai-YamataniN, YamamotoH, TakagiN, et al. (2004) Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166 : 913–924.
50. MuellerJL, SkaletskyH, BrownLG, ZaghlulS, RockS, et al. (2013) Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet 45 : 1083–1087.
51. MuellerJL, MahadevaiahSK, ParkPJ, WarburtonPE, PageDC, et al. (2008) The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 40 : 794–799.
52. TrachtulecZ, VlcekC, MiholaO, GregorovaS, FotopulosovaV, et al. (2008) Fine haplotype structure of a chromosome 17 region in the laboratory and wild mouse. Genetics 178 : 1777–1784.
53. PialekJ, VyskocilovaM, BimovaB, HavelkovaD, PialkovaJ, et al. (2008) Development of unique house mouse resources suitable for evolutionary studies of speciation. J Hered 99 : 34–44.
54. ElliottR, MillerD, PearsallR, HohmanC, ZhangY, et al. (2001) Genetic analysis of testis weight and fertility in an interspecies hybrid congenic strain for Chromosome X. Mamm Genome 12 : 45–51.
55. ElliottRW, PoslinskiD, TabaczynskiD, HohmanC, PazikJ (2004) Loci affecting male fertility in hybrids between Mus macedonicus and C57BL/6. Mamm Genome 15 : 704–710.
56. TeeterKC, PayseurBA, HarrisLW, BakewellMA, ThibodeauLM, et al. (2008) Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res 18 : 67–76.
57. JanousekV, WangL, LuzynskiK, DufkovaP, VyskocilovaMM, et al. (2012) Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol Ecol 21 : 3032–3047.
58. CampbellP, GoodJM, NachmanMW (2013) Meiotic sex chromosome inactivation is disrupted in sterile hybrid male house mice. Genetics 193 : 819–828.
59. Bolcun-FilasE, SchimentiJC (2012) Genetics of meiosis and recombination in mice. Int Rev Cell Mol Biol 298 : 179–227.
60. HayashiK, YoshidaK, MatsuiY (2005) A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438 : 374–378.
61. BurgoynePS, MahadevaiahSK, TurnerJM (2009) The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 10 : 207–216.
62. BurgoynePS, MahadevaiahS, MittwochU (1985) A reciprocal autosomal translocation which causes male sterility in the mouse also impairs oogenesis. J Reprod Fertil 75 : 647–652.
63. SetterfieldLA, MahadevaiahS, MittwochU (1988) Chromosome pairing and germ cell loss in male and female mice carrying a reciprocal translocation. J Reprod Fertil 82 : 369–379.
64. TruettGE, HeegerP, MynattRL, TruettAA, WalkerJA, et al. (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29 : 52–54.
65. HomolkaD, IvanekR, ForejtJ, JansaP (2011) Differential expression of non-coding RNAs and continuous evolution of the X chromosome in testicular transcriptome of two mouse species. PLoS One 6: e17198.
66. Broman KW, Sen S (2009) A guide to QTL mapping with R/qtl. \New York: Springer.
67. BultCJ, EppigJT, KadinJA, RichardsonJE, BlakeJA (2008) The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res 36: D724–728.
68. BastosH, LassalleB, ChicheporticheA, RiouL, TestartJ, et al. (2005) Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65 : 40–49.
69. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
70. SmythGK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 : 3.
71. TurnerJM, MahadevaiahSK, Fernandez-CapetilloO, NussenzweigA, XuX, et al. (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37 : 41–47.
72. InselmanA, EakerS, HandelMA (2003) Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet Genome Res 103 : 277–284.
73. KauppiL, BarchiM, BaudatF, RomanienkoPJ, KeeneyS, et al. (2011) Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331 : 916–920.
Štítky
Genetika Reprodukční medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 2- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



