-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
Aurora B kinase (AURKB) is the catalytic subunit of the chromosomal passenger complex (CPC), an essential regulator of chromosome segregation. In mitosis, the CPC is required to regulate kinetochore microtubule (K-MT) attachments, the spindle assembly checkpoint, and cytokinesis. Germ cells express an AURKB homolog, AURKC, which can also function in the CPC. Separation of AURKB and AURKC function during meiosis in oocytes by conventional approaches has not been successful. Therefore, the meiotic function of AURKC is still not fully understood. Here, we describe an ATP-binding-pocket-AURKC mutant, that when expressed in mouse oocytes specifically perturbs AURKC-CPC and not AURKB-CPC function. Using this mutant we show for the first time that AURKC has functions that do not overlap with AURKB. These functions include regulating localized CPC activity and regulating chromosome alignment and K-MT attachments at metaphase of meiosis I (Met I). We find that AURKC-CPC is not the sole CPC complex that regulates the spindle assembly checkpoint in meiosis, and as a result most AURKC-perturbed oocytes arrest at Met I. A small subset of oocytes do proceed through cytokinesis normally, suggesting that AURKC-CPC is not the sole CPC complex during telophase I. But, the resulting eggs are aneuploid, indicating that AURKC is a critical regulator of meiotic chromosome segregation in female gametes. Taken together, these data suggest that mammalian oocytes contain AURKC to efficiently execute meiosis I and ensure high-quality eggs necessary for sexual reproduction.
Published in the journal: . PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004194
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004194Summary
Aurora B kinase (AURKB) is the catalytic subunit of the chromosomal passenger complex (CPC), an essential regulator of chromosome segregation. In mitosis, the CPC is required to regulate kinetochore microtubule (K-MT) attachments, the spindle assembly checkpoint, and cytokinesis. Germ cells express an AURKB homolog, AURKC, which can also function in the CPC. Separation of AURKB and AURKC function during meiosis in oocytes by conventional approaches has not been successful. Therefore, the meiotic function of AURKC is still not fully understood. Here, we describe an ATP-binding-pocket-AURKC mutant, that when expressed in mouse oocytes specifically perturbs AURKC-CPC and not AURKB-CPC function. Using this mutant we show for the first time that AURKC has functions that do not overlap with AURKB. These functions include regulating localized CPC activity and regulating chromosome alignment and K-MT attachments at metaphase of meiosis I (Met I). We find that AURKC-CPC is not the sole CPC complex that regulates the spindle assembly checkpoint in meiosis, and as a result most AURKC-perturbed oocytes arrest at Met I. A small subset of oocytes do proceed through cytokinesis normally, suggesting that AURKC-CPC is not the sole CPC complex during telophase I. But, the resulting eggs are aneuploid, indicating that AURKC is a critical regulator of meiotic chromosome segregation in female gametes. Taken together, these data suggest that mammalian oocytes contain AURKC to efficiently execute meiosis I and ensure high-quality eggs necessary for sexual reproduction.
Introduction
Haploid gametes are generated by meiosis, a unique cell division process that consists of a single round of DNA replication followed by two successive cell divisions. In the first division, meiosis I (MI), homologous chromosomes segregate. The second division, meiosis II (MII), is more similar to mitosis because sister chromatids segregate. An error in chromosome segregation can result in aneuploidy, the leading genetic cause of infertility and congenital birth defects in humans [1], [2]. It is now well appreciated that the incidence of aneuploidy is at least 10-fold higher in female gametes (oocytes) than it is in male gametes (sperm) [3]. Thus, understanding the underlying causes of oocyte aneuploidy could help address a majority of clinical aneuploidies in humans.
During meiosis there are a number of possible mistakes that could result in aneuploidy. These mistakes include, but are not limited to, defects in kinetochore-microtubule (K-MT) attachments, a faulty spindle assembly checkpoint (SAC), improper cytokinesis, or loss of sister chromatid cohesion [4]–[10]. In mitosis, the chromosomal passenger complex (CPC) is essential for steering the chromosomes through these obstacles [11]–[16]. The CPC does this through a sophisticated pattern of synchronized movements. At metaphase, the CPC localizes to kinetochores, and at anaphase, it relocates to the spindle midzone. This dynamic localization pattern ensures that the CPC phosphorylates the right substrates at the right time and place. Perturbing the CPC in oocytes often leads to errors in MI, thereby resulting in aneuploidy [6], [17].
In mitotically dividing cells, the CPC consists of a catalytic subunit, Aurora B kinase (AURKB), and regulatory subunits Inner Centromere Protein (INCENP), Survivin, and Borealin [18]–[20]. Meiotic cells, however, contain another enzymatic subunit, Aurora C kinase (AURKC), that can function in the CPC in place of AURKB [6], [21]–[25]. AURKB and AURKC are members of a conserved serine-threonine protein kinase family, and are highly similar in sequence within their catalytic domains. Both AURKs bind the IN box region of INCENP, but not at the same time [22]. This binding is essential to stimulate kinase activity and for subsequent phosphorylation of INCENP [26]. Because they are highly similar in sequence, AURKC can compensate for loss of AURKB when ectopically expressed in somatic cells and supports mitosis in preimplantation mouse embryos that lack AURKB [27]–[29]. Furthermore, AURKB compensates for the loss of AURKC in oocytes from Aurkc−/− mice [23].
The sequence similarities between AURKB and AURKC have hindered our understanding of their functions during meiosis. For example, small molecule inhibitors do not selectively inhibit the kinases [17], [22], [24], [25], [30], siRNA knockdown approaches are inefficient and lack specificity [17], [22], and, as mentioned, genetic knockout strategies allow for functional compensation [23], [29]. We hypothesized that expression of a dominant negative allele of AURKC (AURKC-DN) perturbed both AURKB and AURKC functions in oocytes [6]. To further understand the molecular mechanisms that lead to the high incidence of aneuploidy in mammalian oocytes, we sought to develop a tool to selectively disrupt AURKC function.
Mutation of the gatekeeper leucine residue in the ATP-binding pocket of AURKB inactivates the kinase in vivo and in vitro [31]. Here, we devised a similar strategy to inhibit AURKC activity and demonstrate that an AURKC-L93A gatekeeper mutant selectively disrupts AURKC, but not AURKB, function during oocyte meiosis. Using this strategy, we show that AURKC has non-overlapping functions with AURKB during MI. We find that loss of AURKC function results in misalignment of chromosomes and arrest at metaphase I (Met I), and, in oocytes that failed to arrest, aneuploidy at metaphase II (Met II). Oocytes expressing the AURKC mutant failed to correct erroneous K-MT attachments, which is the likely cause of the misaligned chromosomes and aneuploidy. We also find that AURKC-CPC is not uniquely required to maintain an active SAC or to execute cytokinesis. These events may be AURKB-CPC specific or require the activity of both AURKB and AURKC. This study is the first to ascribe non-overlapping functions of AURKC from AURKB during MI in mouse oocytes.
Results
Dominant negative AURKC disrupts both AURKB and AURKC function during oocyte meiosis
Expression of dominant negative AURKC (AURKC-T171A, T175A) (referred hereafter as AURKC-DN) in mouse oocytes causes cytokinesis failure and misaligned univalent chromosomes at MI ([6] and Figure S1). These phenotypes are identical to that of oocytes cultured in high concentrations of small molecule inhibitors of AURKB (ZM447439 and AZD1152) which likely also inhibit AURKC [17], [25], [32]. Two models could explain the similarity in phenotype between the two perturbations: 1) Either AURKB is not expressed in mouse oocytes or 2) AURKC-DN disrupts both AURKB and AURKC function. To investigate the first model, we assessed the protein expression of AURKB in oocytes undergoing meiosis via immunocytochemistry with an antibody previously validated to detect AURKB in mouse preimplantation embryos [29]. AURKB localized within the nucleus of prophase-arrested oocytes and with the meiotic spindle at Met I and II (Figure 1A). This localization pattern is different than that of AURKC, which localizes to kinetochores and the Met I inter-chromatid axis [23], [24]. To further confirm the specificity of the antibody, we examined AURKB in oocytes from Aurkc−/− mice. In oocytes from WT littermates, AURKB localized to the meiotic spindle, but in Aurkc−/− oocytes at Met II AURKB localized to kinetochores (Figure 1B). Exogenous AURKB-GFP also localized to kinetochores in Aurkb−/− and Aurkc−/− oocytes at Met II, as previously demonstrated [23] where it co-localized with Survivin, a CPC subunit (Figure 1C). In addition, this co-localization occurred at Met I (Figure S2). Similar to overexpression of AURKB-GFP in WT oocytes, we still detect AURKB-GFP in the spindle region in the single knockout oocytes (Figure S2). We also assessed the specificity of the antibody by immunoblot analysis. We microinjected wild-type oocytes with cRNAs encoding Aurka-Gfp, Aurkb-Gfp, or Aurkc-Gfp. Probing with anti-GFP antibody, confirmed expression in each group (Figure 1D, top panel). When we probed the membrane with the anti-AURKB antibody, it cross-reacted only with oocytes injected with Aurkb-Gfp (Figure 1D, middle panel). We, and others, previously demonstrated the presence of Aurkb mRNA in oocytes [6], [23]–[25], and another report documents AURKB protein in mouse oocytes [33]. Furthermore, when we probed pooled whole-cell lysates from 150 non-injected oocytes, we detected a band at ∼40 kDa, which is the expected size of endogenous AURKB (Figure 1D, middle panel, right-most lane). Taken together, these data support the model that AURKB is expressed in mouse oocytes. Furthermore, the localization of endogenous AURKB at kinetochores in oocytes from Aurkc−/− mice supports the hypotheses that either AURKB compensates for the loss of AURKC [23] and/or endogenous AURKC competes with AURKB for localization at kinetochores and the inter-chromatid axis.
Fig. 1. AURKB is expressed in mouse oocytes. 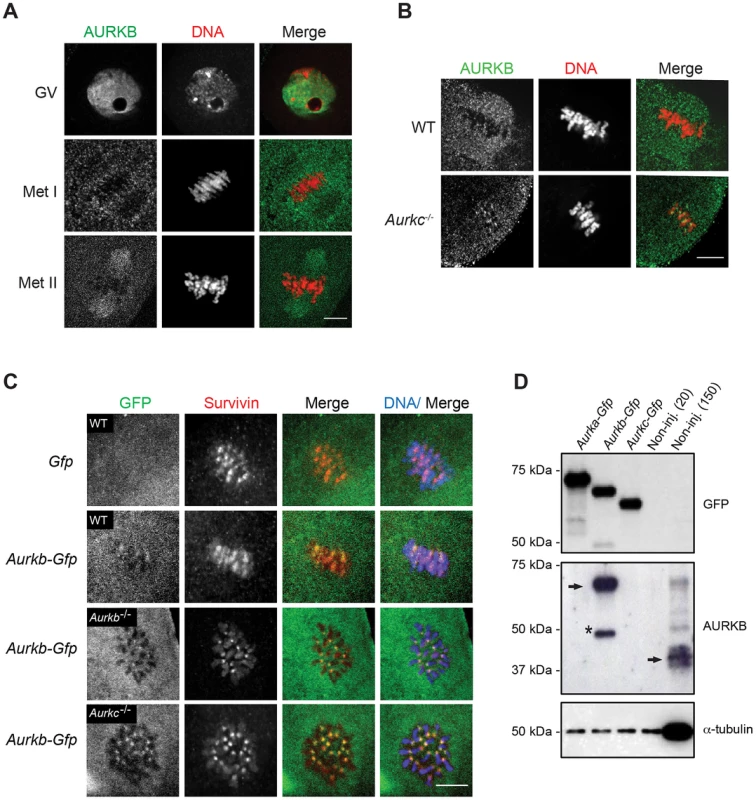
(A) GV-intact oocytes were collected from CF1 mice and matured in vitro for 8 h (Met I), or 16 h (Met II), prior to fixation and staining with an anti-AURKB antibody. (B) GV-intact oocytes were collected from WT and Aurkc−/− mice and matured in vitro for 16 h (Met II), prior to fixation and staining with an anti-AURKB antibody. Merged images show AURKB in green and DNA in red. (C) GV-intact oocytes were collected from WT, Aurkb−/−, and Aurkc−/− mice, microinjected with the indicated cRNA, and matured in vitro for 16 h (Met II), prior to fixation and staining with an anti-Survivin antibody. Merged images show AURKB-GFP in green, Survivin in red, and DNA in blue. These experiments were conducted with a minimum of 20 oocytes for each group. Shown are representative images (Scale bar, 10 µm). (D) 20 GV-intact oocytes were collected from CF1 mice and microinjected with the indicated cRNA. Two hours after injection, the oocytes were matured to Met II in vitro (16 h). The total numbers of non-injected control oocytes (Non-inj.) are indicated in parenthesis. Total cellular lysates were probed with the indicated antibody. The panels are images of the same membrane that was stripped and re-probed. The arrows indicate the specific AURKB protein band, and the asterisk indicates a presumed degradation product of AURKB-GFP. To investigate whether AURKC-DN (Figure 2A) disrupts both AURKB and AURKC function in oocyte meiosis, we eliminated issues with redundancies by using oocytes from Aurkc−/− mice (i.e. containing only AURKB) [23]. As anticipated, microinjection of WT or Aurkc−/− oocytes with Aurkc-DN cRNA resulted in the same phenotypes. These oocytes did not have detectable phosphorylated INCENP (pINCENP) (Figure 2B–C). They contained misaligned univalent chromosomes, and they failed to divide, as examined by polar body (PB) extrusion (Figure 2B, D). These data support the second model: AURKC-DN perturbs the function of both AURKB and AURKC, making it inadequate for assigning specific function to AURKC.
Fig. 2. Dominant-negative AURKC (AURKC-DN) disrupts both AURKB/C function in oocytes. 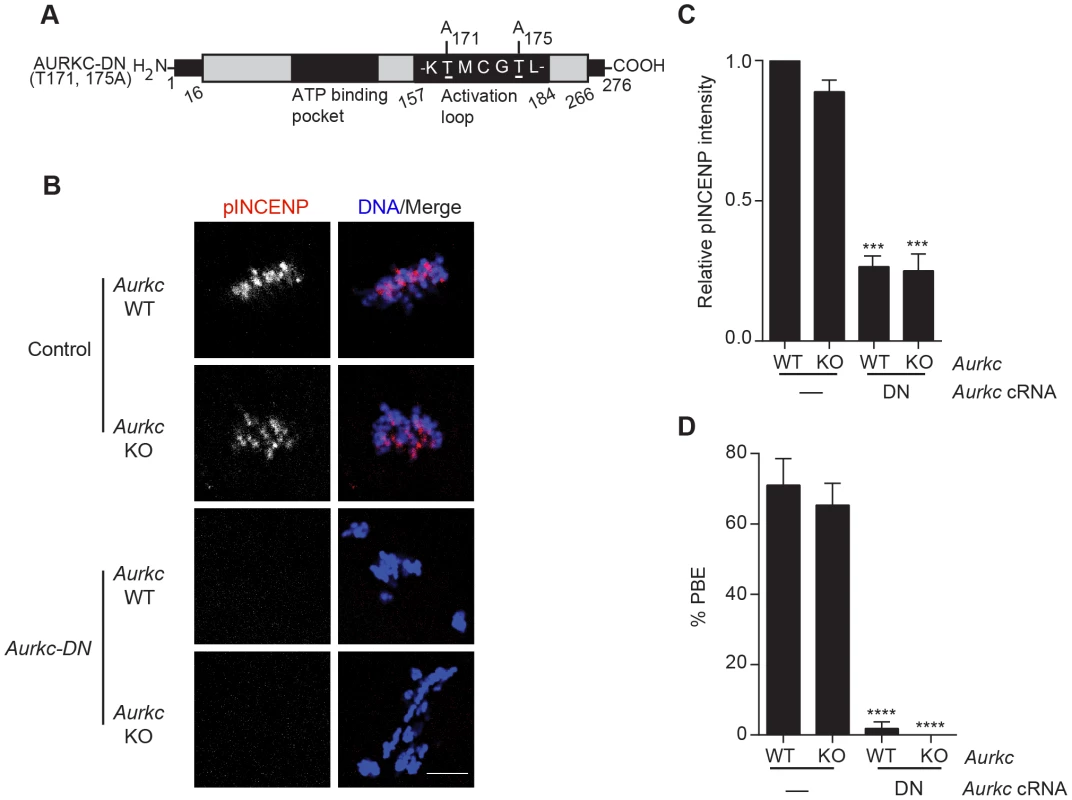
(A) Schematic representation of AURKC-DN. The mutated threonines (T) in the activation loop are underlined. (B–D) Full-grown WT (Aurkc WT) and Aurkc−/− (Aurkc KO) oocytes were injected with PBS or Gfp (Control) or Aurkc-DN cRNA. The injected oocytes were matured for 16 h, followed by fixation and immunostaining with a phospho-specific INCENP (pINCENP) antibody (red in merge). DNA was detected via DAPI staining (blue). Shown are representative confocal Z-projections (scale bar, 10 µm). (C) Corresponding quantification of pINCENP pixel intensities in B. This experiment was conducted 3 times with a minimum of 20 oocytes in each group. (D) Percentage of oocytes that extruded polar bodies (PBE). One-way ANOVA was used to analyze the data. *** P<0.001, **** P<0.0001. A gatekeeper AURKC mutant is catalytically inactive and selectively disrupts AURKC
Because AURKC-DN disrupts the function of both AURKB and AURKC, and because AURKB compensates for loss of AURKC [23], we sought to develop a tool to selectively perturb AURKC function. The dominant negative mutation involves changing two threonines in the activation loop to non-phosphorylatable alanine residues [6], [34] (Figure 2A). Therefore, this mutant, while not activated, can presumably bind ATP and substrates. Protein kinases are commonly bi-lobed in structure thereby generating a pocket for ATP binding and catalysis. Within the ATP binding pocket are conserved “gatekeeper” residues that restrict binding of other intracellular molecules [35], [36]. Mutation of a gatekeeper residue to an alanine enlarges the pocket, but generally does not perturb the function of most protein kinases. However, approximately 30% of kinases do not tolerate a mutation of gatekeeper residues [31], [35], [36]. Mutation of the gatekeeper residue (L159) in murine AURKB renders the kinase inactive, and it is unable to phosphorylate histone H3 in mitotic cell extracts [31]. Based on protein sequence alignment, we determined that the gatekeeper residue in AURKC is L93 (Figure 3A). Because mutation at this residue likely affects ATP binding instead of activation, we postulated that mutating L93 to A (hereafter referred to as AURKC-LA) might behave differently than the dominant negative and could selectively disrupt AURKC. To first confirm that AURKC-LA is not active, we microinjected WT oocytes with Aurkc-LA-Gfp cRNA. We assessed activity by immunostaining the injected oocytes with phospho-specific antibodies that recognize AURKB/C substrates. We found loss of auto-phosphorylated AURKC activation signal (pAURKC; pT171) and significantly decreased pINCENP (pS893/S894) compared to control injected oocytes (Figure 3B–E). Compared to controls, phosphorylated histone H3 (pH3S10) was reduced by ∼50% in LA-injected oocytes (Figure 3F–G). The levels of pAURKC and pINCENP were reduced almost to the same levels as they were in Aurkc-DN injected oocytes (Figure 3B, D). On the other hand, pH3S10 signals were completely inhibited only in the AURKC-DN oocytes, suggesting that H3S10 is a target of both AURKB and AURKC (Figure 3F–G). We note that phosphorylation of H3S10 in mitotic cells is less sensitive to localized AURKB activity compared to other substrates [37], and our data is consistent with this observation. These data suggest that AURKC-LA is catalytically inactive, and that it inhibits endogenous AURK/CPC activity.
Fig. 3. AURKC-LA and AURKC-DN are catalytically inactive. 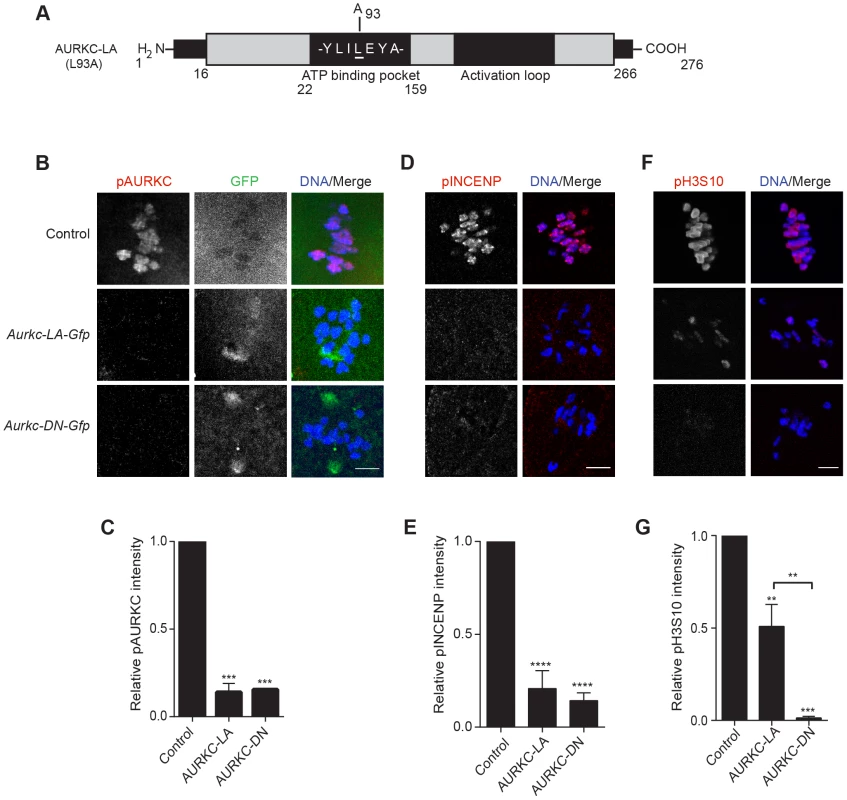
(A) Schematic representation of AURKC-LA. The mutated leucine (L) residue in the ATP binding pocket is underlined. (B–G) Full-grown WT oocytes were injected with the indicated cRNA; controls were injected with PBS or Gfp cRNA. Met I oocytes were fixed and examined for phosphorylated AURKC (pAURKC) (red in merge) and GFP expression (green in merge) (B), phosphorylated INCENP (pINCENP) (red in merge) (D) and phosphorylated H3S10 (pH3S10) (red in merge) (F). DNA was detected via DAPI staining (blue). Shown are representative Z-projections from confocal microscopy (scale bars, 10 µm). (C, E, and G) Corresponding quantification of fluorescence intensity of B, D, and F, respectively. The experiment was conducted at least 2 times with a minimum of 20 oocytes in each group. Shown are representative images. One-way ANOVA was used to analyze the data. **P<0.01; *** P<0.001; **** P<0.0001. To test our hypothesis that the gatekeeper mutant specifically inhibits AURKC, we first microinjected Aurkc-LA in Aurkb−/− oocytes (which express only AURKC; Figure 4A–C) and in Aurkc−/− oocytes (which express only AURKB, and that compensates for AURKC [23]; Figure 4D–F). Control mice were WT littermates from each genetic background. AURKC-LA significantly reduced INCENP phosphorylation and PB emission in Aurkb−/− oocytes suggesting that the catalytically inactive AURKC-LA efficiently disrupts endogenous AURKC function (Figure 4A–C). Importantly, Aurkc−/− oocytes expressing AURKC-LA extruded PBs and had normal levels of phosphorylated INCENP, similar to WT and KO injected controls (Figure 4D–F). These data confirm that AURKC-LA selectively disrupts AURKC function without disrupting endogenous AURKB function.
Fig. 4. AURKC-L93A (AURKC-LA) is catalytically inactive, and selectively disrupts AURKC function. 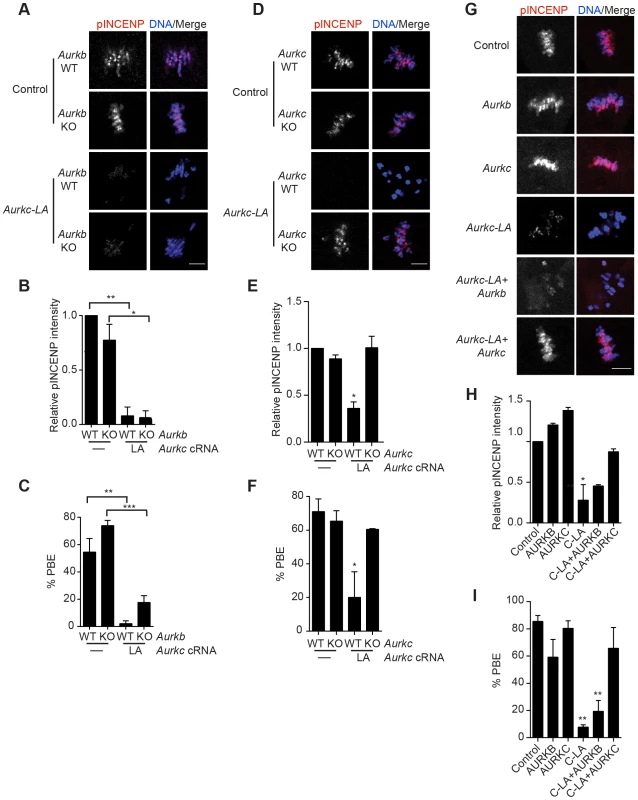
(A–I) Full-grown WT or Aurkb−/− (A–C), WT or Aurkc−/− oocytes (D–F) or WT CF1 oocytes (G–I) were injected with the indicated cRNA; controls were injected with either PBS or Gfp cRNA. The microinjected oocytes were matured in vitro to Met II (16 h) followed by pINCENP detection (red in merge) via confocal microscopy. DNA was detected by DAPI staining (blue). Shown are representative Z-projections (scale bar, 10 µm). (B, E, H) Corresponding quantification of pINCENP intensities. (C, F, I) Percentage of oocytes that extruded polar bodies (PBE). The experiments were conducted 3 times with a minimum of 15 oocytes in each group. One-way ANOVA was used to analyze the data. * P<0.05, ** P<0.01, *** P<0.001. To further test the specificity of AURKC-LA, we conducted a rescue experiment in WT oocytes. Co-expression of WT AURKC rescued the AURKC-LA phenotypes. INCENP was phosphorylated to near control levels and PBs were extruded. Co-expression of WT AURKB did not rescue the phenotypes (Figure 4G–I). These data further confirm that AURKC-LA does not perturb AURKB function and suggests that the defect in INCENP phosphorylation and block in meiotic maturation are specific for loss of AURKC function.
AURKC is required to retain CPC localization during MI of mouse oocytes
In budding yeast, aurora kinase activity is required for proper CPC localization and prevents premature localization of the CPC to the spindle [38]. In mitotically dividing tissue culture cell lines, inactive AURKB mutants fail to localize normally at centromeres [31], [39]. To investigate if AURKC-LA behaves similar to WT AURKC, we analyzed its subcellular localization in oocytes at Met I. WT AURKC localized to kinetochores and inter-chromatid axes of Met I oocytes (Figure 5A) as previously reported [23], [24]. On the other hand, AURKC-LA and AURKC-DN failed to localize normally. Both mutants localized predominantly with the spindle. Therefore, AURKC activity may be required to regulate CPC localization.
Fig. 5. AURKC is required to retain CPC localization at Met I. 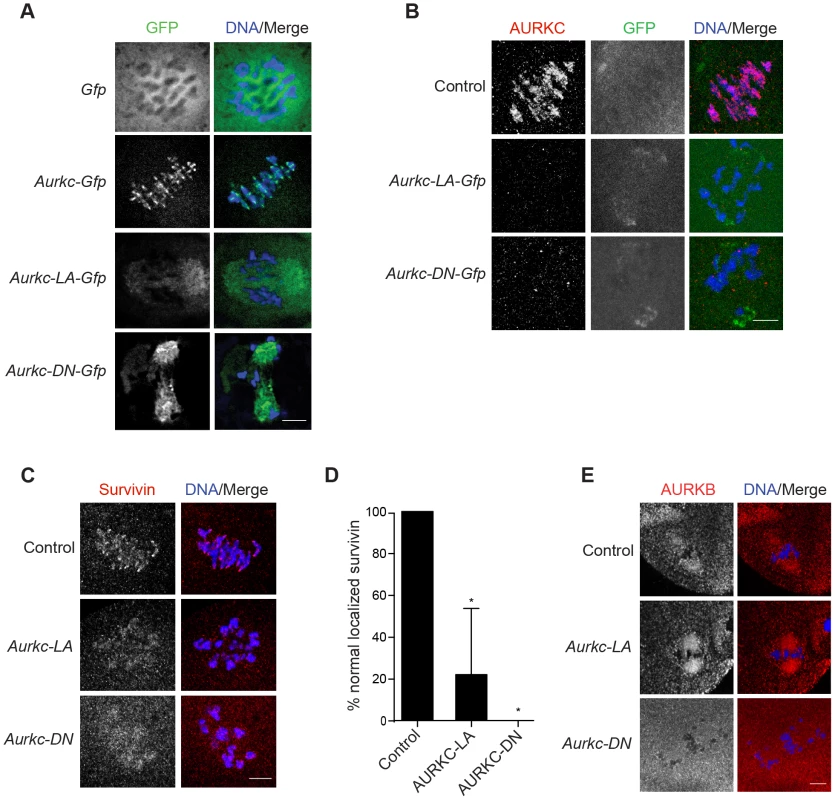
Full-grown oocytes were injected with the indicated cRNA; controls were injected with PBS or Gfp cRNA. After 8 h of in vitro maturation, Met I oocytes were fixed and examined for localization of the GFP-tagged mutant protein (green in merge) (A), endogenous AURKC (red in merge) (B), endogenous Survivin (red in merge) (C), and endogenous AURKB (red in merge) (E). DNA was detected via DAPI staining (blue). Shown are representative confocal Z-projections (scale bars, 10 µm). (D) Corresponding quantification of oocytes with properly localized Survivin in B. The experiments were conducted at least 2 times with a minimum of 20 oocytes in each group. One-way ANOVA was used to analyze the data. *P<0.05. To examine the changes in CPC localization, we first assessed the localization of endogenous AURKC. When oocytes expressed either AURKC-LA or AURKC-DN we could not detect endogenous AURKC on the chromosomes (Figure 5B). For reasons not determined, we note that the antibody used to detect AURKC on chromosomes is not compatible with detecting de-localized AURKC-LA. Survivin is also a member of the CPC, and is expressed during mouse oocyte meiosis [40], [41]. Similar to AURKC-DN, oocytes expressing AURKC-LA resulted in displacement of the CPC at Met I as evidenced by the loss of kinetochore and inter-chromatid axis localization of endogenous Survivin (Figure 5C–D). Importantly, AURKC-LA did not alter the spindle localization of AURKB as compared to AURKC-DN, further supporting our evidence that AURKC-LA selectively perturbs AURKC (Figure 5E). These findings are consistent with previous observations that loss of AURKB/C kinase activity by using small molecule inhibitors results in displacement and atypical localization of the CPC in mitosis [31], [42] and oocyte meiosis (our unpublished observations).
Efficient meiotic progression and chromosome alignment requires AURKC activity during MI
Similar to AURKC-DN expressing oocytes, oocytes expressing AURKC-LA were defective in meiotic progression. The majority of oocytes expressing AURKC-LA (∼60%) failed to extrude PBs. The kinetics with which those that did extrude PBs were ∼1 h delayed compared to controls. For AURKC-DN expressing oocytes, this failure was more pronounced (∼95%); these oocytes initially extruded PBs, but then retracted them, as previously reported (Figure 6A) [6]. The discrepancy suggests that AURKB carries out meiotic functions during MI that do not require AURKC activity.
Fig. 6. Meiotic progression to Met II and chromosome alignment at Met I requires AURKC. 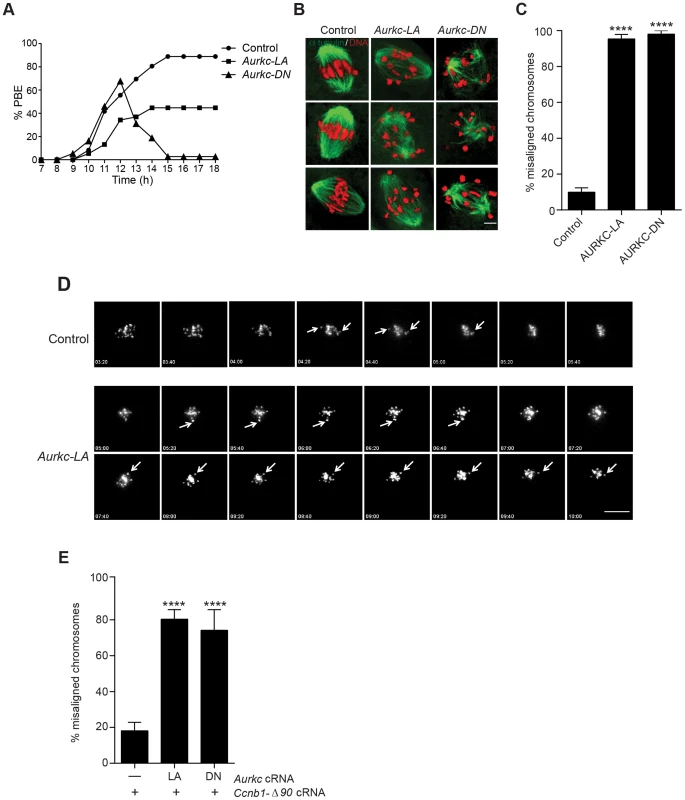
(A) Full-grown WT oocytes from CF1 mice were injected with the indicated cRNA, followed by in vitro maturation (16 h) and analysis of the timing of polar body extrusion (PBE) by live cell imaging. The experiment was carried out 2 times with a minimum of 30 oocytes in each group. (B) Representative confocal Z-projections of DNA (red) and spindle configurations (green) from oocytes at Met I (7 h after milrinone washout) that were injected with the indicated cRNA. The experiment was conducted 3 times with a minimum of 30 oocytes in each group (Scale bar, 10 µm). (C) Quantification of the number of oocytes with misaligned chromosomes analyzed in B. (D) Representative H2B-mCherry fluorescence images of oocytes coinjected with the indicated cRNA and H2B-mCherry cRNA; the white arrows indicate non-aligned bivalent chromosomes (Scale bar, 50 µm) (E) Met I exit was blocked by microinjection of non-degradable cyclin B (150 ng/µl) mixed with the indicated cRNA, and examined for chromosome alignment by immunocytochemistry. Controls were injected with either PBS or Gfp cRNA. The experiment was conducted 2 times with a minimum of 20 oocytes in each group. One-way ANOVA was used to analyze the data. **** P<0.0001. To understand the biological significance of this different phenotype, we first focused on Met I. Both AURKC-LA and AURKC-DN expressing oocytes have nearly the same chromosome misalignment phenotype at Met I (Figure 6B–C), suggesting that AURKC-CPC is the main CPC complex from prophase of MI through Met I, and that AURKC is essential for chromosome alignment. To examine the chromosome alignment phenotype in more detail, we imaged control-injected and AURKC-LA-injected oocytes live. Both groups expressed H2B-GFP to mark chromosomes. Unlike in controls, chromosomes in AURKC-LA oocytes oscillated between nearly aligned and misaligned for the duration of the imaging (Figure 6D, Movies S1, S2).
The presence of misaligned chromosomes in oocytes expressing AURKC-LA could be due to a chromosome alignment problem, or may reflect a cell-cycle delay in oocyte progression to Met I. To discriminate between these possibilities, we blocked Met I exit by injecting oocytes with non-degradable cyclin B1 (Ccnb1-Δ90) [23], [43] to allow oocytes more time to align their chromosomes. Strikingly, unlike control oocytes, the majority of the oocytes expressing AURKC-LA still had misaligned chromosomes even after spending 8 hours at Met I (Figure 6E). These data indicate that AURKC activity is indispensable for chromosome alignment in mouse oocyte meiosis.
AURKC-CPC is not the sole CPC complex that regulates the SAC
The majority of oocytes expressing AURKC-LA arrested at Met I with bivalent chromosomes (Figure S1). Given the severe chromosome misalignment at Met I, it was expected that AURK-DN expressing oocytes would also arrest at Met I [44]. As previously reported, all oocytes expressing AURKC-DN contain univalent chromosomes (Figure S1) [6]. The presence of univalents suggests an active Anaphase Promoting Complex/Cyclosome (APC/C) and separation of homologous chromosomes. The regulatory mechanism responsible for controlling APC/C is called the Spindle Assembly Checkpoint (SAC). The SAC signals the delay of anaphase onset until all chromosomes acquire the correct kinetochore-microtubule attachment either in mitosis [45], [46] or oocyte meiosis [9], [10], [47], [48].
We investigated the ability of AURKC-LA and AURKC-DN to maintain the SAC. To conduct these studies, we incubated control, Aurkc-LA or Aurkc-DN injected oocytes in nocodazole, a microtubule-depolymerizing drug that keeps the SAC active in WT cells because of an absence of K-MT attachments. As expected only oocytes expressing AURKC-DN extruded PBs in the presence of nocodazole (Figure 7A,B). We obtained similar results when nocodazole was used at a lower dose that does not completely depolymerize the spindle (Figure S3). These results indicate that AURKB has a role in maintaining an active SAC signal.
Fig. 7. AURKC does not maintain SAC activation by itself. 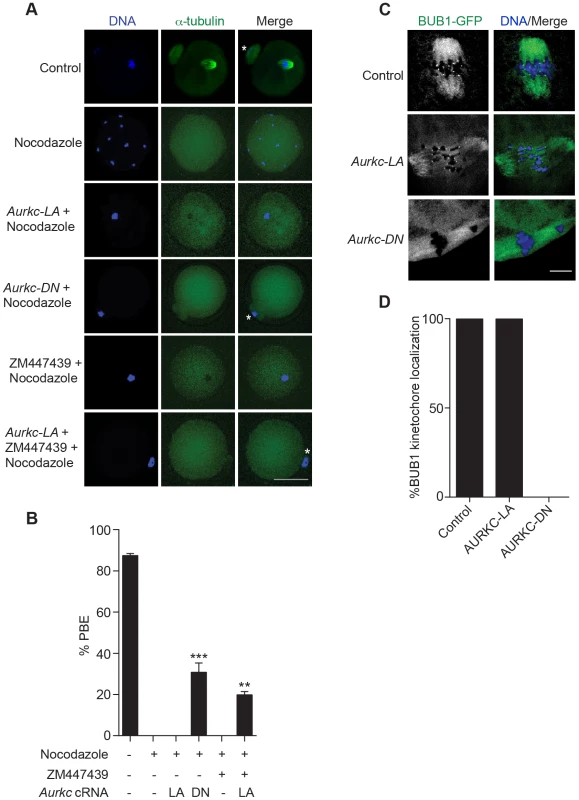
(A) Full-grown oocytes were injected with the indicated cRNA; controls were injected with PBS or Gfp cRNA. Nocodazole and ZM447439 were added to the maturation medium as indicated to a final concentration of 5 µM and 2 µM, respectively. After maturation for 16 h, the oocytes were examined for extrusion of the first polar body (PBE) and spindle formation (green) via fluorescence microscopy (scale bar, 50 µm). DNA was detected via DAPI staining (blue). The experiment was conducted 3 times with a minimum of 30 oocytes in each group. Shown are representative images; the white asterisks mark PBs. (B) Quantification of the percentage of oocytes that extrude a polar body (PBE) in A. One-way ANOVA was used to analyze the data. ** P<0.01; *** P<0.001. (C) Bub1-Gfp cRNA (300 ng/µl) was co-injected with the indicated cRNA and in vitro matured oocytes were then examined by confocal microscopy to detect GFP (green in merge). DNA was detected via DAPI staining (blue). Shown are representative Z-projections (scale bar, 10 µm). The experiment was conducted 2 times with a minimum of 20 oocytes in each group. (D) Quantification of the percentage of oocytes in C that contained BUB1-GFP at kinetochores. ZM447439 is a pan-Aurora kinase inhibitor with higher specificity for AURKB than AURKC and AURKA [49]. Oocytes incubated in a high concentration of ZM447439 (10 µM) bypass the SAC [30]. This dose likely inhibits both AURKB and AURKC. Our data indicate that AURKC-CPC is not the sole CPC involved in SAC signaling, but it possible that its function overlaps with AURKB. To investigate this possibility, we incubated oocytes expressing AURKC-LA with a low dose of ZM447439 (2 µM) that does not normally bypass the SAC (Figure 7A–B), in the presence of nocodazole. This is a dose that likely only inhibits AURKB. When AURKB was inhibited in oocytes expressing AURKC-LA, they bypassed the SAC and extruded PBs (Figure 7A,B). These data suggest that the SAC is controlled by both AURKB and AURKC.
In somatic cells, the CPC kinase (AURKB) promotes the kinetochore recruitment of key SAC components including BUB1 (Budding uninhibited by benzimidazoles 1) [50]. To further validate our findings, we microinjected Bub1-Gfp cRNA [48] along with Aurkc-LA or Aurkc-DN cRNAs into oocytes. Again, loss of AURKC function alone did not perturb BUB1 kinetochore localization (Figure 7C–D). But when AURKB was also inhibited, BUB1 failed to localize to the kinetochores (Figure 7C–D). These data confirm that AURKC is not the sole CPC kinase involved in SAC signaling.
AURKC-CPC is not the sole CPC for cytokinesis
Arrest at Met I is the predominant phenotype observed in oocytes expressing AURKC-LA, but there is small percentage of oocytes which do extrude PBs (Figure 6A). Consistent with Yang et al., AURKC-DN expressing oocytes began to extrude PBs, but failed to complete cytokinesis and subsequently retracted the PBs [6] (Figure 8A, Movie S4). This phenotype is reminiscent of oocytes cultured in pan Aurora kinase inhibitors ZM447439 and AZD1152 [17], [25], [32]. Unlike AURKC-DN, AURKC-LA expressing oocytes that progressed through Met I extruded PBs normally without any evidence of cytokinesis failure suggesting that AURK-CPC is not the sole CPC controlling cytokinesis, and that AURKB may be important for this function (Figure 8A–C; Movie S3). Progression to Met II did not depend on expression level of the mutant protein. In a zoomed out image of supplemental movie 3 (Movie S5), the oocyte expressing less AURKC-LA arrested at Met I, and the oocyte expressing more AURKC-LA extruded a polar body. To further confirm our hypothesis, we investigated pINCENP as a marker of CPC activity at telophase I (Telo I). Similar to controls, oocytes expressing AURKC-LA contained phosphorylated INCENP at the mid-body (Figure 8D). These data further support our observations that AURKC activity is dispensable for cytokinesis in oocytes. Oocytes lacking AURKB contain pINCENP at the midbody, but when we microinjected Aurkc-LA cRNA in Aurkb−/− oocytes (contain only AURKC) we did not detect phosphorylated INCENP. These data suggest overlapping AURKB-CPC and AURKC-CPC activities control cytokinesis (Figure 8D).
Fig. 8. AURKC alone does not regulate cytokinesis and loss of its function leads to aneuploid eggs. 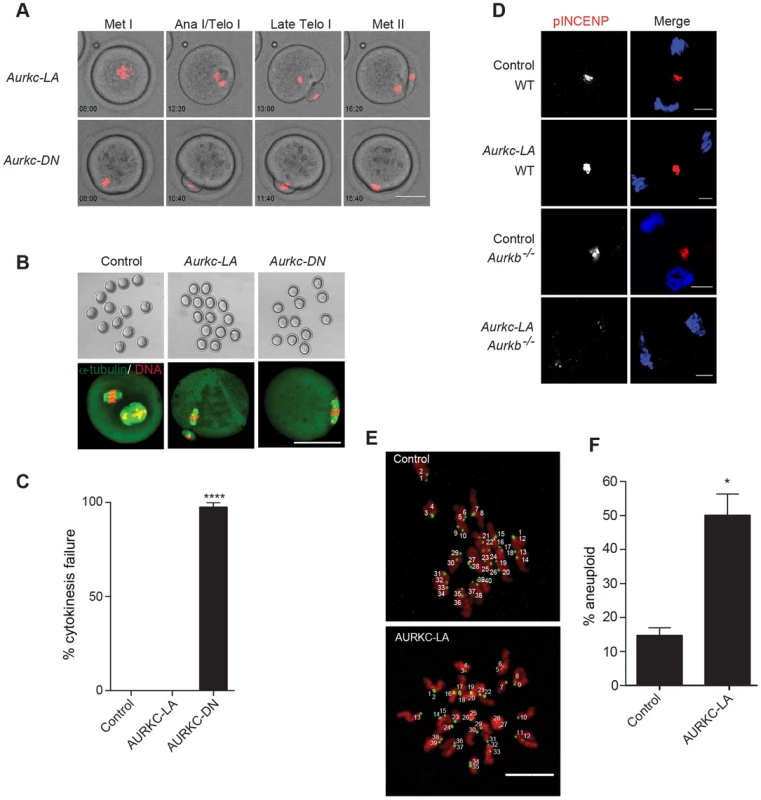
(A) Snapshots from a time-lapse series showing chromatin (H2B-mCherry; red) and bright field images from oocytes co-injected with H2B-mCherry and the indicated GFP-tagged cRNA (Scale bar, 50 µm). (B) Representative Z-projections obtained by confocal microscopy of spindle (green) and DNA (red) configurations of Met II eggs (scale bar, 50 µm). (C) Percentage of oocytes that failed cytokinesis. The experiment was conducted 3 times and at least 20 oocytes were examined in each group. (D) WT and Aurkb−/− oocytes were microinjected with Aurkc-LA cRNA followed by maturation to telophase I and examination of phosphorylated INCENP (pINCENP) (red in merge) (scale bar, 10 µm). DNA was detected by DAPI (blue). Shown are representative examples. (E) Met II eggs from the indicated groups were treated with monastrol followed by detection of DNA (red) and kinetochores with Crest anti-sera (green) (scale bar, 10 µm). The number of kinetochores was counted in each egg, and an aberration of 40 was scored as aneuploid. The experiment was conducted 3 times with a minimum of 20 oocytes in each experiment. Shown are representative Z-projections. (F) Quantification of D. One-way ANOVA was used to analyze the data in B and Student's t-test was used to analyze the data in E. Controls were injected with either PBS or Gfp cRNA. Values with asterisks vary significantly, *P<0.05; **** P<0.0001. Disruption of AURKC function leads to aneuploid eggs
To investigate the biological significance of selectively perturbing AURKC during MI, AURKC-LA expressing oocytes were examined for aneuploidy using an in situ chromosome spread method [7], [51]. The percentage of aneuploid eggs was significantly higher in Aurkc-LA-injected oocytes (that did not arrest at Met I) compared to controls (Figure 8E–F). We did not assess ploidy when both AURKB and AURKC kinases were perturbed because no PBs were extruded, and therefore resulted in 100% polyploidy, as previously described [6]. Thus, AURKC function is critical for faithful chromosome segregation in oocyte meiosis.
AURKC-CPC is the predominant CPC that corrects erroneous K-MT attachments
In mouse oocytes lateral interactions between microtubules and chromosomes drive the early stages of pro-Met I, but the final and sharp alignment of chromosomes at the Met I plate requires end-on K-MT attachments [52]. Brunet and colleagues performed nocodazole washout experiments to examine spindle recovery in mouse oocytes. They found that some chromosomes moved towards the spindle poles (where K-MT end-on attachment is established) before congressing to the metaphase plate. In agreement with this observation, in tissue culture cell lines the mal-oriented, but not bi-oriented, chromosomes move to the mitotic spindle pole until correct attachments are made, and then alignment at the metaphase plate is achieved [53]. We therefore hypothesized that failure to correct erroneous K-MT attachments leads to the misaligned chromosomes that are adjacent to the spindle poles (Figure 6B), in Aurkc-LA-injected oocytes.
When K-MT attachments are correct at Met I, the bivalent chromosomes are bi-oriented with monotelic attachment of each sister pair to opposite poles. This type of attachment generates tension leading to greater separation between the two sister-kinetochore pairs of each homologous chromosome. Incorrect attachment (merotelic and syntelic) leads to decreased tension and reduced separation between the two sister-kinetochore pairs of each homologous chromosome (Figure 9A–B) [10], [54]. Similar to AURKC-DN-expressing oocytes, oocytes expressing AURKC-LA showed significantly shorter inter-kinetochore distance (detected by CREST anti-serum) compared to control oocytes. These data imply that the error correction mechanism is impaired in these oocytes. Moreover, the majority of the misaligned bivalents had incorrect attachments as evidenced by the decrease of the inter-kinetochore distance (Figure 9C). These data suggest that the chromosome misalignment phenotype after disruption of AURKC function might be, at least in part, due to a defect in correcting improper K-MT attachments. This result is consistent with the conclusion that mitotic cells lacking AURKB activity fail to align chromosomes due to inability to correct abnormal attachment [54].
Fig. 9. AURKC is the primary CPC kinase that corrects erroneous K-MT attachments. 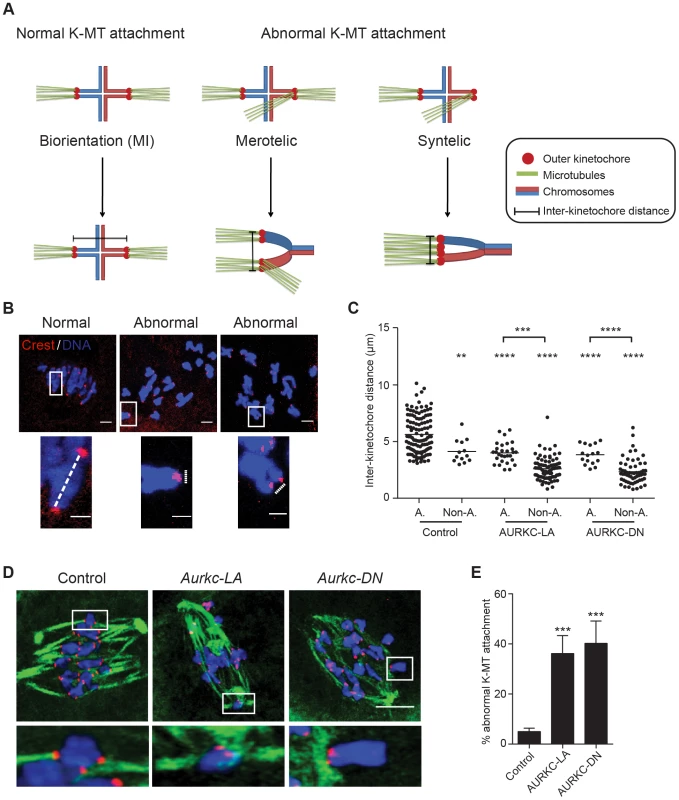
(A) Schematic representation of normal and abnormal K-MT attachments. Sister chromatids are indicated in the same color. Note that chiasmata linking the homologous chromosomes were omitted for simplicity purposes. (B–E) GV oocytes were microinjected with the indicated cRNA and matured to Met I. Kinetochores were labeled with CREST (red) and inter-kinetochore distance was measured using Image J (dotted line). DNA was counterstained with DAPI (blue). The scale bar represents 10 µm for the original images and 2 µm for the magnified images. (C) Quantification of the inter-kinetochore distance from aligned (A.) and misaligned (Non-A.) chromosomes. Each data point is the distance between two sister kinetochore pairs within a bivalent chromosome in an oocyte. The experiment was conducted 3 times with a minimum of 20 oocytes in each group. (D) Representative images of K-MT attachments. Oocytes were incubated in ice-cold medium to depolymerize non-kinetochore attached tubulin prior to fixation and detection of kinetochores (red), tubulin (green) and DNA (blue) (Scale bar, 10 µm). The experiment was conducted 3 times with a minimum of 15 oocytes in each group. (E) Quantification of abnormal K-MT attachments. One-way ANOVA was used to analyze the data. ** P<0.01, *** P<0.001, **** P<0.0001. To further confirm that correcting erroneous K-MT attachments depends upon AURKC, we conducted an assay to determine the presence of stable end-on attachments of K-MTs to kinetochores. Microtubules that form stable kinetochore attachment are cold stable, whereas microtubules that do not form stable attachments with kinetochores are cold labile [55]. We exposed Met I oocytes to a pulse of cold medium, prior to fixation and immunocytochemistry to detect kinetochores and microtubules. Aurkc-LA-injected oocytes had a significantly greater percentage of abnormal (merotelic and syntelic) attachments than mCherry-injected controls (Figure 9D–E). The percentage of abnormal K-MT attachments in Aurkc-LA-injected oocytes was similar to that of Aurkc-DN-injected oocytes (Figure 9E and [6]). We suggest that AURKC-CPC is the predominant form of the CPC that corrects erroneous K-MT attachments and for chromosome alignment in mouse oocyte meiosis.
Discussion
Distinguishing the roles of AURKB and C has been complicated by many factors. The two kinases are highly similar in sequence and appear to compensate for one another. One logical interpretation is that AURKB is the predominant CPC kinase in mitosis while AURKC is the predominant CPC kinase in meiosis. This model is supported by a report that detected no AURKB protein in mouse oocytes by immunoblotting [6]. But a collection of observations suggests that AURKB is found in oocytes. For example, oocytes express Aurkb mRNA [24], [25] and overexpression of AURKB, but not AURKC, rescues defects induced by a low dose of ZM447439, a pan Aurora kinase inhibitor with highest affinity for AURKB [24], [33], [49]. In this report, using a different antibody and mouse strains than in the previous report [6] we detected AURKB protein in mouse oocytes by immunoblot, and showed that it localized to centromeres in Aurkc−/− oocytes (Figure 1). These data provide evidence that AURKB is expressed in mouse oocytes and support our previous report [23]. Dominant negative alleles of AURKB and AURKC perturb themselves and one another when expressed in mitosis [22]. It is therefore not surprising that when we expressed AURKC-DN in Aurkc−/− oocytes, endogenous AURKB was also perturbed (Figure 2B). A second model to consider is the notion that the mouse genome may contain multiple copies of Aurkc [56] and that the knockout is not completely void of AURKC protein. Another group has revisited the updated mouse genome sequence and found that coding regions of Aurkc are not duplicated [57]. Moreover, when we probed oocytes from Aurkc−/− mice, we did not detect any Aurkc transcript or protein [23]. Importantly, until this study, whether AURKB and AURKC have any non-overlapping functions was not known.
By selectively disrupting AURKC function in oocytes, we have shown for the first time that AURKC has distinct functions from AURKB in mouse oocytes. We find that AURKC corrects erroneous K-MT attachments, a likely cause of chromosome misalignment at Met I (Figures 6, 9). This failure to align chromosomes caused a Met I arrest, as one would expect given an intact SAC (Figure 7). But the small percentage of oocytes that presumably had mild chromosome misalignment, likely below the threshold of maintaining SAC activation, extruded PBs without any evidence of cytokinesis failure (Figure 8). We found that these oocytes were aneuploid (Figure 8). In our experiments, AURKC-LA is expressed from the GV stage though out meiosis. We note that, this prolonged duration of expression could make analysis of the Met II phenotype more challenging. These later roles of AURKB and AURKC will be important to address in future studies. Thus, AURKC-CPC appears to be the predominant CPC that corrects improper K-MT attachments, a function essential for preventing aneuploidy.
We are interested in understanding why meiosis might require two CPC kinases whereas most mitotic cells have only one. In mitosis, AURKB directly maintains SAC activation by recruiting components such as BUB1 to kinetochores [39], [50], [58], [59] and indirectly participates in the SAC by destabilizing K-MTs. Given the presence of two forms of the CPC in oocyte meiosis, we propose a separation of function model: AURKB-CPC recruits BUB1 to kinetochores, while AURKC-CPC destabilizes improper K-MT attachments. In agreement with this hypothesis, we observed bypass of SAC-inducing conditions only when we inhibited both AURKB/C (Figure 6) and arrest at Met I when we inhibited only AURKC (Figure 6). This strategy to use complementary AURKB and AURKC functions to control the SAC may be critical to provide an insurance mechanism to prevent aneuploidy in a transcriptionally quiescent cell type where AURKB protein is not stable [23].
Loss of AURKC function did not affect pINCENP at the mid-body or induce cytokinesis failure (Figure 8). These data suggest that the CPC containing AURKC as the catalytic subunit is not the predominant form of the CPC that regulates cytokinesis. AURKB-CPC plays an important role in mitotic cytokinesis by phosphorylating many substrates, including INCENP, at the midbody [15], [16], [60], [61]. However, INCENP is phosphorylated in oocytes that lack AURKB (Figure 8). It is possible that AURKC compensates for loss of AURKB in the knockout oocytes. Interestingly, mitotic cytokinesis requires an increased amount of AURKB activity, as compared to its metaphase functions [62]. AURKB and AURKC both localize to the midbody in oocytes [24], [33]. Therefore, it is possible that oocytes satisfy the need for elevated AURK activity at the midbody by having overlapping functions of 2 forms of the CPC available, differing only in the catalytic subunit, and further examination is needed.
We have not yet determined why the gatekeeper mutant displays specificity for affecting only AURKC. We have eliminated that possibility that these mutants were expressed at different levels in our system or in the different genetic backgrounds (Figure S4). We have also ruled out possible differences in catalytic activity because oocytes expressing AURKC-LA also showed complete loss of AURKC and INCENP phosphorylation (Figure 3). In both mutants the activation loop is not phosphorylated but the proteins are different. In the DN protein, the threonines are mutated to alanines, whereas in the LA protein the threonines are present but do not contain phosphate. The activation loop of protein kinases is important not only for catalytic activity but also for conformation stabilization, the ability to bind substrates, and for substrate specificity [63]. The conformation of the ATP binding pocket is also critical for protein structure [22]. Similar to our observations with AURKC-DN, mutation of the activation loop threonines to alanines in protein kinase C (PKC) alpha loosens its specificity and the mutant inhibits the other PKC isoforms [64]. Although we are not certain as to the mechanism of inhibition of the LA protein, one model to investigate is that AURKC functions as a dimer within the CPC, and that the gatekeeper mutant functions as a dominant negative only in the context of an AURKC dimer. To our knowledge there is no evidence that AURKB dimerizes, and it would be interesting if this mechanism were AURKC-specific. Alternatively, AURKC-LA may function as a pseudokinase. Pseudokinases have high sequence homology to kinases but do not have detectable catalytic activity [65], [66]. Some of these proteins contain amino acid substitutions in gatekeeper residues of their ATP binding pockets that would ablate ATP binding or efficient catalysis [65]. If AURKC-LA were acting as a pseudokinase it could be preventing WT AURKC from binding the CPC. Most significantly, in Aurkc−/− oocytes, where AURKB is the only CPC kinase, expression of AURKC-LA but not AURKC-DN resulted in normal meiotic progression and CPC kinase activity. Therefore it is clear that the main difference between these mutants is the inability of AURKC-LA to compete with endogenous AURKB function.
To our knowledge, this is the first report to separate AURKB and AURKC meiotic functions, and is consistent with some of the proposed models [17], [23]. AURKC is expressed in other cell types, including testes, neuronal tissue and some cancer cells [21], [29], [67], [68]. The biological significance of AURKC expression in cancer cells is also of clinical interest, but not well understood. Because these cells also express AURKB, studying the functions of AURKC in cancer cell division poses the same specficitiy difficulties as in oocytes. With our validation of AURKC-LA being specific for disrupting AURKC function, we propose that this gatekeeper mutant will be helpful tool for answering questions relevant to the reproductive and cancer fields
Materials and Methods
Generation and genotyping of Aurkc−/− mice and Aurkbfl/fl ZP3-Cre mice
Details for generating and genotyping Aurkc−/− mice were described previously [23], [67]. The Aurkbfl/fl mice were a generous gift from M. Malumbres (CNIO, Spain) [29]. For generating Aurkbfl/fl ZP3-Cre mice, female mice carrying the Aurkb floxed alleles were crossed with ZP3-Cre males (Jackson laboratories) [69], and genotyping for the LoxP sites was carried out as previously described [29]. Cre genotyping was carried out as described by Jackson Laboratories. A detailed phenotypic description will be described elsewhere. All animals were in a mixed background of C57BL/6J, 129/Sv, and CD1 and maintained following Institutional Animal Use and Care Committee and National Institutes of Health (NIH) guidelines.
Cloning, mutagenesis and in vitro cRNA synthesis
Generation of non-degradable cyclin B, Aurka, Aurkb, and Aurkc-Gfp were described previously [24], [43]. To generate Bub1-Gfp, murine Bub, sequence was amplified via PCR from a cDNA clone, (Open Biosystems, #3671932) and ligated into pIVT-GFP [70]. Aurkc-LA and Aurkc-DN mutants were generated by site-directed mutagenesis using the QuikChange Multi-site Mutagenesis kit (Agilent Technologies) following manufacturer's instructions. To generate Aurkc-DN T171 and 175 were changed to an A (ACA and ACT to GCC; Figure 2A). To generate Aurkc-L93 was changed to an A (CTG to GCC; Figure 3A).
DNA linearization of all Gfp - and mCherry - containing constructs was carried out using Nde I (New England BioLabs). After DNA linearization, the digests were purified (Qiagen, QIAquick PCR Purification) and in vitro transcription was carried out using an mMessage mMachine T7 kit (Ambion) according to the manufacturer's instructions. Finally, the cRNA was purified using an RNAEasy kit (Qiagen).
Oocyte collection, microinjection and culture
Full-grown, GV-intact oocytes were obtained from pregnant mare serum gonadotropin - (PMSG) (Calbiochem #367222) primed (44–48 h before collection), 6-wk-old female mice as previously described [71]. The collection and injection medium for oocytes was bicarbonate-free minimal essential medium (MEM) containing, 25 mM Hepes, pH 7.3, 3 mg/ml polyvinylpyrollidone (MEM/PVP) and 2.5 µM milrinone (Sigma #M4659) to prevent meiotic resumption [72].
Denuded GV oocytes were microinjected with ∼10 pl of 0.8–1 µg/µl of the indicated cRNA, unless otherwise noted. Following microinjection, the oocytes were cultured in Chatot, Ziomek, and Bavister (CZB) medium containing 2.5 µM milrinone. All culture and in vitro meiotic maturation occurred in a humidified incubator with 5% CO2 in air at 37°C. For the oocytes that were examined at Met II, we incubated the injected oocytes for 1–3 h prior to meiotic maturation, and for the oocytes that were examined at Met I, we incubated the injected oocytes overnight (14 h) prior to meiotic maturation. In vitro meiotic maturation was conducted in milrinone-free CZB medium for periods of 6–7 h (Met I), 9 h (Telo I) or 16 h (Met II).
Nocodazole (Sigma #M1404) and ZM447439 (Tocris #2458) were dissolved in dimethyl sulfoxide (DMSO). Nocodazole and ZM447439 were added to CZB culture medium to a final concentration of 5 µM and 2 µM, respectively, and in vitro maturation was performed in a humidified chamber (Becton Dickinson #353037).
Immunocytochemistry and confocal microscopy
For analysis of cold-stable microtubules, oocytes were incubated for 5 minutes on ice in MEM/PVP, and then fixed for 25 minutes at 37°C in 3.7% formaldehyde in 100 mM Pipes, pH 6.8, containing 10 mM EGTA, 1 mM MgCl2 and 0.2% Triton X-100 [73]. AURKC-GFP was detected after fixation in 3.7% paraformaldehyde in phosphate-buffered saline (PBS) for 1 hour; survivin was detected by similar fixation conditions plus 0.1% Triton X-100. In all other experiments, oocytes were fixed in 2–2.5% paraformaldehyde in PBS for 20 minutes at room temperature. After fixation, the cells were permeabilized with 0.1% Triton X-100 in PBS for 15 minutes and transferred to blocking buffer (PBS+0.3% BSA+0.01% Tween-20) for 15 minutes. Immunostaining was performed by incubating the fixed oocytes with the primary antibody for 1 hour. After washing in blocking solution, the oocytes were incubated in secondary antibodies for 1 hour; omission of the primary antibody served as negative control. DNA was stained and mounted with 4′, 6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI; Life Technologies #D1306; 1∶170) diluted in VectaShield (Vector Laboratories) under a coverslip with gentle compression. Fluorescence was detected on Zeiss 510 Meta laser-scanning confocal microscope under a 63× objective.
All oocytes in the same experiment were processed at the same time. The laser power was adjusted to a level where signal intensity was just below saturation for the group that displayed the highest intensity and all images were then scanned at that pre-determined laser power. The intensity of fluorescence was quantified with NIH image J software keeping the processing parameters identical when experimental analysis required intensity measurements.
Live cell imaging
Oocytes microinjected with the indicated cRNAs and histone H2B-mCherry cRNA were transferred into separate drops of CZB medium covered with mineral oil in a 96 well dish (Greiner Bio One, #655892). Bright field, GFP and mCherry image acquisition was started at the GV stage using an EVOS FL Auto Imaging System (Life Technologies) with a 20× objective. The microscope stage was heated to 37°C and 5% CO2 was maintained using the EVOS Onstage Incubator. Images of individual cells were acquired every 20 min and processed using NIH image J software.
Antibodies
The following primary antibodies were used in immunofluorescence: CREST autoimmune serum (Antibodies Incorporated; #15-234; 1∶30), AURKB (Abcam #AB2254; 1∶50), AURKC (Bethyl #A400-023A - BL1217; 1∶30), pAURKC (kind gift of T. Tang, Institute of Biomedical Science, Taiwan [6]; 1∶500), phospho-specific Ser893/Ser894 INCENP (kind gift of M. Lampson, UPenn [74]; 1∶1,000), survivin (Cell Signaling Technology #2808S; 1∶500), α-tubulin-Alexa Fluor 488 conjugate (Life Technologies #322588; 1∶100), phospho-specific H3S10 (Millipore; #05-806;1∶100).
In situ chromosome counting
Monastrol treatment, immunocytochemical detection of kinetochores and chromosome counting were performed as previously described [75]. Briefly, eggs were cultured for 2 hours in CZB containing 100 µM monastrol (Sigma) to disperse the chromosomes by collapsing the bipolar spindle to a monopolar spindle. Eggs were fixed in freshly prepared 2% paraformaldehyde and stained with CREST anti-serum to detect kinetochores and DAPI to detect DNA. Images were collected at 0.6-µm Z-intervals to capture the entire region of the MII spindle (16–20 µm total). To obtain a chromosome count for each egg, serial confocal sections were analyzed to determine the total number of kinetochores and calculated using NIH image J software.
Immunoblotting
Oocytes were lysed in 1% SDS, 1% β-mercaptoethanol, 20% glycerol, and 50 mM Tris–HCl (pH 6.8), and denatured at 95°C for 5 min. Proteins separated by electrophoresis in 10% SDS polyacrylamide precast gel. Stained proteins of known molecular mass (range: 14–200 kDa) were run simultaneously as standards. The electrophoretically separated polypeptides were transferred to nitrocellulose membranes using a Trans-Blot Turbo Transfer System (Bio-Rad) then blocked by incubation in 2% blocking (ECL blocking; Amersham) solution in TBS-T (Tris-buffered saline with 0.1% Tween 20) for 1 h. The membranes were then incubated with primary antibodies at 4°C overnight (GFP (Sigma #G6539; 1∶1,000), β actin (Abcam #ab20272; 1∶10,000), AURKB (Abcam #ab2254; 1∶500), α-tubulin (Sigma #T-6074; 1∶10,000). After washing with TBS-T five times, the membranes were incubated with a secondary antibody labeled with horseradish peroxidase for 1 h followed with washing with TBS-T five times. The signals were detected using the ECL Select Western blotting detection reagents (Amersham) following the manufacturer's protocol.
Statistical analysis
One-way ANOVA and Student's t-test, as indicated in figure legends, were used to evaluate the differences between groups using GraphPad Prism. The differences of p<0.05 were considered significant.
Supporting Information
Zdroje
1. HassoldT, ChiuD (1985) Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum Genet 70 : 11–17.
2. HassoldT, HallH, HuntP (2007) The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet 16 Spec No. 2: R203–208.
3. HassoldT, HuntP (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2 : 280–291.
4. LelandS, NagarajanP, PolyzosA, ThomasS, SamaanG, et al. (2009) Heterozygosity for a Bub1 mutation causes female-specific germ cell aneuploidy in mice. Proc Natl Acad Sci U S A 106 : 12776–12781.
5. JonesKT (2008) Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum Reprod Update 14 : 143–158.
6. YangKT, LiSK, ChangCC, TangCJ, LinYN, et al. (2010) Aurora-C kinase deficiency causes cytokinesis failure in meiosis I and production of large polyploid oocytes in mice. Mol Biol Cell 21 : 2371–2383.
7. ChiangT, DuncanFE, SchindlerK, SchultzRM, LampsonMA (2010) Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol 20 : 1522–1528.
8. DavydenkoO, SchultzRM, LampsonMA (2013) Increased CDK1 activity determines the timing of kinetochore-microtubule attachments in meiosis I. J Cell Biol 202 : 221–229.
9. HomerHA, McDougallA, LevasseurM, MurdochAP, HerbertM (2005) Mad2 is required for inhibiting securin and cyclin B degradation following spindle depolymerisation in meiosis I mouse oocytes. Reproduction 130 : 829–843.
10. LaneSI, YunY, JonesKT (2012) Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development 139 : 1947–1955.
11. CarmenaM, WheelockM, FunabikiH, EarnshawWC (2012) The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 13 : 789–803.
12. NicklasRB (1997) How cells get the right chromosomes. Science 275 : 632–637.
13. MaldonadoM, KapoorTM (2011) Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol 13 : 475–482.
14. MatsonDR, DemirelPB, StukenbergPT, BurkeDJ (2012) A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes Dev 26 : 542–547.
15. Murata-HoriM, FumotoK, FukutaY, IwasakiT, KikuchiA, et al. (2000) Myosin II regulatory light chain as a novel substrate for AIM-1, an aurora/Ipl1p-related kinase from rat. J Biochem 128 : 903–907.
16. ToureA, MzaliR, LiotC, SeguinL, MorinL, et al. (2008) Phosphoregulation of MgcRacGAP in mitosis involves Aurora B and Cdk1 protein kinases and the PP2A phosphatase. FEBS Lett 582 : 1182–1188.
17. SharifB, NaJ, Lykke-HartmannK, McLaughlinSH, LaueE, et al. (2010) The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci 123 : 4292–4300.
18. RuchaudS, CarmenaM, EarnshawWC (2007) Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 8 : 798–812.
19. KellyAE, FunabikiH (2009) Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol 21 : 51–58.
20. van der WaalMS, HengeveldRC, van der HorstA, LensSM (2012) Cell division control by the Chromosomal Passenger Complex. Exp Cell Res 318 : 1407–1420.
21. TsengTC, ChenSH, HsuYP, TangTK (1998) Protein kinase profile of sperm and eggs: cloning and characterization of two novel testis-specific protein kinases (AIE1, AIE2) related to yeast and fly chromosome segregation regulators. DNA Cell Biol 17 : 823–833.
22. ChenHL, TangCJ, ChenCY, TangTK (2005) Overexpression of an Aurora-C kinase-deficient mutant disrupts the Aurora-B/INCENP complex and induces polyploidy. J Biomed Sci 12 : 297–310.
23. SchindlerK, DavydenkoO, FramB, LampsonMA, SchultzRM (2012) Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci U S A 109: E2215–2222.
24. ShudaK, SchindlerK, MaJ, SchultzRM, DonovanPJ (2009) Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev 76 : 1094–1105.
25. SwainJE, DingJ, WuJ, SmithGD (2008) Regulation of spindle and chromatin dynamics during early and late stages of oocyte maturation by aurora kinases. Mol Hum Reprod 14 : 291–299.
26. HondaR, KornerR, NiggEA (2003) Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell 14 : 3325–3341.
27. SasaiK, KatayamaH, StenoienDL, FujiiS, HondaR, et al. (2004) Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil Cytoskeleton 59 : 249–263.
28. LiX, SakashitaG, MatsuzakiH, SugimotoK, KimuraK, et al. (2004) Direct association with inner centromere protein (INCENP) activates the novel chromosomal passenger protein, Aurora-C. J Biol Chem 279 : 47201–47211.
29. Fernandez-MirandaG, TrakalaM, MartinJ, EscobarB, GonzalezA, et al. (2011) Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development 138 : 2661–2672.
30. LaneSI, ChangHY, JenningsPC, JonesKT (2010) The Aurora kinase inhibitor ZM447439 accelerates first meiosis in mouse oocytes by overriding the spindle assembly checkpoint. Reproduction 140 : 521–530.
31. HengeveldRC, HertzNT, VromansMJ, ZhangC, BurlingameAL, et al. (2012) Development of a chemical genetic approach for human aurora B kinase identifies novel substrates of the chromosomal passenger complex. Mol Cell Proteomics 11 : 47–59.
32. MortlockAA, FooteKM, HeronNM, JungFH, PasquetG, et al. (2007) Discovery, synthesis, and in vivo activity of a new class of pyrazoloquinazolines as selective inhibitors of aurora B kinase. J Med Chem 50 : 2213–2224.
33. VogtE, KippA, Eichenlaub-RitterU (2009) Aurora kinase B, epigenetic state of centromeric heterochromatin and chiasma resolution in oocytes. Reprod Biomed Online 19 : 352–368.
34. ChenSH, TangTK (2002) Mutational analysis of the phosphorylation sites of the Aie1 (Aurora-C) kinase in vitro. DNA Cell Biol 21 : 41–46.
35. GarskeAL, PetersU, CortesiAT, PerezJL, ShokatKM (2011) Chemical genetic strategy for targeting protein kinases based on covalent complementarity. Proc Natl Acad Sci U S A 108 : 15046–15052.
36. ZhangC, KenskiDM, PaulsonJL, BonshtienA, SessaG, et al. (2005) A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat Methods 2 : 435–441.
37. WangF, UlyanovaNP, DaumJR, PatnaikD, KatenevaAV, et al. (2012) Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. J Cell Biol 199 : 251–268.
38. NakajimaY, CormierA, TyersRG, PigulaA, PengY, et al. (2011) Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics. J Cell Biol 194 : 137–153.
39. DitchfieldC, JohnsonVL, TigheA, EllstonR, HaworthC, et al. (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 161 : 267–280.
40. SunSC, WeiL, LiM, LinSL, XuBZ, et al. (2009) Perturbation of survivin expression affects chromosome alignment and spindle checkpoint in mouse oocyte meiotic maturation. Cell Cycle 8 : 3365–3372.
41. SunSC, LiuHL, SunQY (2012) Survivin regulates Plk1 localization to kinetochore in mouse oocyte meiosis. Biochem Biophys Res Commun 421 : 797–800.
42. WangF, UlyanovaNP, van der WaalMS, PatnaikD, LensSM, et al. (2011) A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr Biol 21 : 1061–1069.
43. SchindlerK, SchultzRM (2009) CDC14B acts through FZR1 (CDH1) to prevent meiotic maturation of mouse oocytes. Biol Reprod 80 : 795–803.
44. GuiL, HomerH (2012) Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development 139 : 1941–1946.
45. KhodjakovA, PinesJ (2010) Centromere tension: a divisive issue. Nat Cell Biol 12 : 919–923.
46. MusacchioA, SalmonED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8 : 379–393.
47. BrunetS, PahlavanG, TaylorS, MaroB (2003) Functionality of the spindle checkpoint during the first meiotic division of mammalian oocytes. Reproduction 126 : 443–450.
48. McGuinnessBE, AngerM, KouznetsovaA, Gil-BernabeAM, HelmhartW, et al. (2009) Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr Biol 19 : 369–380.
49. GirdlerF, GascoigneKE, EyersPA, HartmuthS, CrafterC, et al. (2006) Validating Aurora B as an anti-cancer drug target. J Cell Sci 119 : 3664–3675.
50. VigneronS, PrietoS, BernisC, LabbeJC, CastroA, et al. (2004) Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell 15 : 4584–4596.
51. SteinP, SchindlerK (2011) Mouse oocyte microinjection, maturation and ploidy assessment. J Vis Exp 53: pii: 2851.
52. BrunetS, MariaAS, GuillaudP, DujardinD, KubiakJZ, et al. (1999) Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J Cell Biol 146 : 1–12.
53. LampsonMA, RenduchitalaK, KhodjakovA, KapoorTM (2004) Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 6 : 232–237.
54. JellumaN, BrenkmanAB, van den BroekNJ, CruijsenCW, van OschMH, et al. (2008) Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell 132 : 233–246.
55. RiederCL (1981) The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 84 : 145–158.
56. HuHM, ChuangCK, LeeMJ, TsengTC, TangTK (2000) Genomic organization, expression, and chromosome localization of a third aurora-related kinase gene, Aie1. DNA Cell Biol 19 : 679–688.
57. Ben KhelifaM, ZouariR, HarbuzR, HalouaniL, ArnoultC, et al. (2011) A new AURKC mutation causing macrozoospermia: implications for human spermatogenesis and clinical diagnosis. Mol Hum Reprod 17 : 762–768.
58. SantaguidaS, VernieriC, VillaF, CilibertoA, MusacchioA (2011) Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J 30 : 1508–1519.
59. FamulskiJK, ChanGK (2007) Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr Biol 17 : 2143–2149.
60. BirkenfeldJ, NalbantP, BohlBP, PertzO, HahnKM, et al. (2007) GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev Cell 12 : 699–712.
61. QiM, YuW, LiuS, JiaH, TangL, et al. (2005) Septin1, a new interaction partner for human serine/threonine kinase aurora-B. Biochem Biophys Res Commun 336 : 994–1000.
62. XuZ, VagnarelliP, OgawaH, SamejimaK, EarnshawWC (2010) Gradient of increasing Aurora B kinase activity is required for cells to execute mitosis. J Biol Chem 285 : 40163–40170.
63. LahamLE, MukhopadhyayN, RobertsTM (2000) The activation loop in Lck regulates oncogenic potential by inhibiting basal kinase activity and restricting substrate specificity. Oncogene 19 : 3961–3970.
64. Garcia-ParamioP, CabrerizoY, BornancinF, ParkerPJ (1998) The broad specificity of dominant inhibitory protein kinase C mutants infers a common step in phosphorylation. Biochem J 333(Pt 3): 631–636.
65. HuJ, YuH, KornevAP, ZhaoJ, FilbertEL, et al. (2011) Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proc Natl Acad Sci U S A 108 : 6067–6072.
66. BoudeauJ, Miranda-SaavedraD, BartonGJ, AlessiDR (2006) Emerging roles of pseudokinases. Trends Cell Biol 16 : 443–452.
67. KimminsS, CrosioC, KotajaN, HirayamaJ, MonacoL, et al. (2007) Differential functions of the Aurora-B and Aurora-C kinases in mammalian spermatogenesis. Mol Endocrinol 21 : 726–739.
68. KimuraM, MatsudaY, YoshiokaT, OkanoY (1999) Cell cycle-dependent expression and centrosome localization of a third human aurora/Ipl1-related protein kinase, AIK3. J Biol Chem 274 : 7334–7340.
69. LewandoskiM, WassarmanKM, MartinGR (1997) Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol 7 : 148–151.
70. IgarashiH, KnottJG, SchultzRM, WilliamsCJ (2007) Alterations of PLCbeta1 in mouse eggs change calcium oscillatory behavior following fertilization. Dev Biol 312 : 321–330.
71. SchultzRM, MontgomeryRR, BelanoffJR (1983) Regulation of mouse oocyte meiotic maturation: implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev Biol 97 : 264–273.
72. TsafririA, ChunSY, ZhangR, HsuehAJ, ContiM (1996) Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev Biol 178 : 393–402.
73. LampsonMA, KapoorTM (2005) The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol 7 : 93–98.
74. SalimianKJ, BallisterER, SmoakEM, WoodS, PanchenkoT, et al. (2011) Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol 21 : 1158–1165.
75. DuncanFE, ChiangT, SchultzRM, LampsonMA (2009) Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol Reprod 81 : 768–776.
Štítky
Genetika Reprodukční medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
-
Všechny články tohoto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



