-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
The Elongator complex, including the histone acetyl transferase Sin3/Elp3, was isolated as an RNA polymerase II-interacting complex, and cells deficient in Elongator subunits display transcriptional defects. However, it has also been shown that Elongator mediates the modification of some tRNAs, modulating translation efficiency. We show here that the fission yeast Sin3/Elp3 is important for oxidative stress survival. The stress transcriptional program, governed by the Sty1-Atf1-Pcr1 pathway, is affected in mutant cells, but not severely. On the contrary, cells lacking Sin3/Elp3 cannot modify the uridine wobble nucleoside of certain tRNAs, and other tRNA modifying activities such as Ctu1-Ctu2 are also essential for normal tolerance to H2O2. In particular, a plasmid over-expressing the tRNALysUUU complements the stress-related phenotypes of Sin3/Elp3 mutant cells. We have determined that the main H2O2-dependent genes, including those coding for the transcription factors Atf1 and Pcr1, are highly expressed mRNAs containing a biased number of lysine-coding codons AAA versus AAG. Thus, their mRNAs are poorly translated after stress in cells lacking Sin3/Elp3 or Ctu2, whereas a mutated atf1 transcript with AAA-to-AAG lysine codons is efficiently translated in all strain backgrounds. Our study demonstrates that the lack of a functional Elongator complex results in stress phenotypes due to its contribution to tRNA modification and subsequent translation inefficiency of certain stress-induced, highly expressed mRNAs. These results suggest that the transcriptional defects of these strain backgrounds may be a secondary consequence of the deficient expression of a transcription factor, Atf1-Pcr1, and other components of the transcriptional machinery.
Published in the journal: . PLoS Genet 9(7): e32767. doi:10.1371/journal.pgen.1003647
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003647Summary
The Elongator complex, including the histone acetyl transferase Sin3/Elp3, was isolated as an RNA polymerase II-interacting complex, and cells deficient in Elongator subunits display transcriptional defects. However, it has also been shown that Elongator mediates the modification of some tRNAs, modulating translation efficiency. We show here that the fission yeast Sin3/Elp3 is important for oxidative stress survival. The stress transcriptional program, governed by the Sty1-Atf1-Pcr1 pathway, is affected in mutant cells, but not severely. On the contrary, cells lacking Sin3/Elp3 cannot modify the uridine wobble nucleoside of certain tRNAs, and other tRNA modifying activities such as Ctu1-Ctu2 are also essential for normal tolerance to H2O2. In particular, a plasmid over-expressing the tRNALysUUU complements the stress-related phenotypes of Sin3/Elp3 mutant cells. We have determined that the main H2O2-dependent genes, including those coding for the transcription factors Atf1 and Pcr1, are highly expressed mRNAs containing a biased number of lysine-coding codons AAA versus AAG. Thus, their mRNAs are poorly translated after stress in cells lacking Sin3/Elp3 or Ctu2, whereas a mutated atf1 transcript with AAA-to-AAG lysine codons is efficiently translated in all strain backgrounds. Our study demonstrates that the lack of a functional Elongator complex results in stress phenotypes due to its contribution to tRNA modification and subsequent translation inefficiency of certain stress-induced, highly expressed mRNAs. These results suggest that the transcriptional defects of these strain backgrounds may be a secondary consequence of the deficient expression of a transcription factor, Atf1-Pcr1, and other components of the transcriptional machinery.
Introduction
Unicellular organisms are particularly exposed to the environment, and the major changes in microbial gene expression programs arise as a consequence of extracellular stresses. Most regulation is achieved by transcriptional events, with a shift of the transcriptional machinery from growth - to stress-related genes. Therefore, the classical large complexes which contribute to a strong and efficient RNA polymerase II (Pol II) gene transcription, such as Mediator and SAGA, do contribute to stress survival, and genetic defects in non-essential components of these complexes can render phenotypes of sensitivity to stress.
In fission yeast, the MAP kinase Sty1 pathway is essential to induce massive changes in the gene expression programs in response to environment insults (for reviews, see [1], [2]). Upon different types of life-threatening insults such as osmotic or oxidative stress, heat shock or nutrient deprivation, a cascade of phosphorylations results in the activation of Sty1, which then accumulates in the nucleus and triggers a broad transcriptional change of up to 5–10% of the genome. Thus, hundreds of genes become repressed, while hundreds of others are activated, to promote survival. These genes, positively or negatively controlled by different stresses in a Sty1-dependent manner, were called CESR (core environmental stress response) genes [3]. The main effector of such transcriptional events, or at least the activation ones, is the heterodimeric transcription factor Atf1-Pcr1 [3], [4]. Several activities modulating chromatin accessibility and compactness modulate the Sty1-dependent transcription program (for a review, see [5]). For instance, the absence of the histone acetyl transferase (HAT) and SAGA component Gcn5 renders cells sensitive to several stresses due to defective chromatin remodelling along the stress genes [6], [7].
Thus, regarding the fission yeast stress response it is plausible to hypothesize that strains defective in chromatin remodeling activities and/or in components of the large complexes which contribute to an efficient Pol II transcription may be sensitive to stress. That would be the case of the transcription complex named Elongator, isolated in Saccharomyces cerevisiae as essential to trigger chromatin remodeling [8]. This multi-component complex includes Elp3, a HAT that regulates the levels of histone H3 lysine (Lys) 14 and H4 Lys8 acetylation [9]. A number of reports described the participation of Elongator in chromatin modulation, and strains devoid of some of its six components exhibit a pleiotropic phenotype, including transcriptional elongation defects and problems with polarized exocytosis (for a review, see [10]).
In 1985, an S. pombe strain named sin3-193 was reported to have defects in transfer RNA (tRNA) modification, since digestion of tRNAs from this mutant strain to nucleosides and subsequent nucleoside analysis demonstrated the absence of one particular modification in uridine (U) [11]. The laboratory of Bystrom identified in 2005 the sin3/elp3 gene product as one of the Elongator components, and isolated an equivalent tRNA modifying regulatory activity in S. cerevisiae's Elp3 [12]. Later the same was found for Elongator from Arabidopsis thaliana [13] and Caenorhabditis elegans [14]. Importantly enough, most defects initially associated with a role of Elongator in transcription and exocytosis were bypassed by elevated levels of specific tRNAs, those normally modified by the complex (see below) [15], [16]. Thus, the diverse roles of Elongator are a matter of debate (for a review, see [17]).
The genetic code is degenerated, so that most amino acids are encoded by more than one triplet, some of which are more common than others and define the codon usage of a given organism. Up to 75–100 different post-transcriptional nucleoside modifications have been reported in eukaryotic tRNAs [18], many of which occur at the anticodon loop. In particular, a dual modification of a U (U34) at the 5′ wobble position of the anticodon of several tRNAs [those coding for glutamine (Gln), Lys and glutamic acid (Glu), having a UUB (‘B’ being G, C or U) anticodon] has been suggested to have a role in either translation fidelity [19]–[21] or efficiency [22]–[25], and to even be required for viability in yeast [26]. In S. cerevisiae, these modifications consist in the addition of a methoxycarbonylmethyl at carbon 5 of U by the Elongator complex (mcm5U34), and in a thiolation at carbon 2 by the Nfs1-Uba4-Urm1-Ncs6-Ncs2 network (s2U34) [15], [26]–[32]. As indicated above, Sin3/Elp3 of S. pombe has been reported to be required to generate the mcm5s2U modification in tRNAs [11], [12], and Ctu1-Ctu2 are the sequence and functional homologs of Ncs6-Ncs2 [33]. An S. pombe strain lacking both tRNA-modifying activities has recently been shown to display cell cycle defects [34].
In a genetic search for deletion mutants with altered sensitivity to H2O2, we have isolated the putative histone H3 HAT Sin3/Elp3, a component of the Elongator complex. Our initial assumption was that mutations in a chromatin-modifying activity such as Elongator should result in cells displaying stress sensitivity due to defects in transcriptional efficiency. However, our results indicate that Sin3/Elp3 mutant does not display enough alterations in transcriptional events as to explain the substantial sensitivity to peroxides. In fact, the levels of acetylated H3 (total or associated to stress genes) are not significantly affected in Δsin3/elp3 cells. Instead, the wobble U of tRNAs for Lys, Gln and Glu is not modified in cells lacking Sin3/Elp3. This defect in tRNA modification seems to be sufficient to cause the oxidative stress phenotype, since cells devoid of the second modification pathway, required for the formation of s2U34, such as Ctu1 or Ctu2, are also sensitive to stress. Furthermore, over expression of one of these tRNA species, tRNALysUUU, is sufficient to complement all the stress defects of cells lacking Sin3/Elp3. Importantly, we show here that the mRNAs for the Atf1 and Pcr1 transcription factors, which are critical for CESR gene expression and are enriched in the AAA codon for Lys, are not greatly affected in the knock-out strain (Δsin3/elp3). However, Atf1 and Pcr1 protein levels are severely decreased in Δsin3/elp3 and Δctu2 cells. Furthermore, a mutated atf1 transcript with AAA-to-AAG lysine codons is efficiently translated in all strain backgrounds.
Results
Cells lacking Sin3/Elp3 are sensitive to H2O2, but only display minor transcriptional defects
Defects in activities required to mediate massive changes in gene expression should result in cells displaying stress sensitivity. On this basis, we screened a collection of S. pombe deletion strains searching for mutants with impaired survival against H2O2 on solid plates. We isolated several strains with defects in chromatin modifying activities, such as the HAT Gcn5 [7]. Another strain displaying even more severe growth sensitivity to peroxides is that lacking Sin3/Elp3, a HAT and component of the Elongator complex (Figure 1A). We analyzed mutants in other components of Elongator present in our deletion collection (Table S1), and at least subunits Elp4, Iki3/Elp1 and SPAC30.02c [the homolog to KTI12, an S. cerevisiae protein associated to Elongator [35]] are required for wild-type tolerance to H2O2 (Figure 1A).
Fig. 1. Cells lacking Sin3/Elp3 are sensitive to H2O2, but do not display major transcriptional defects. 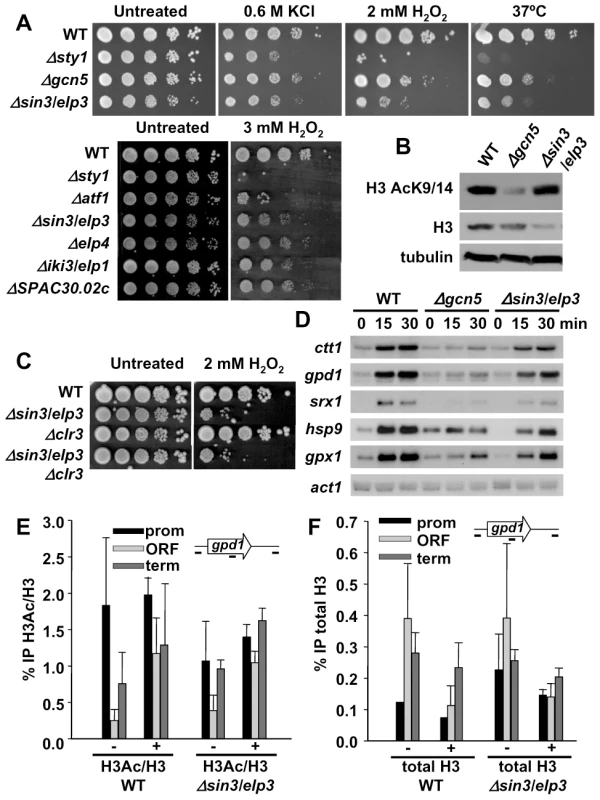
(A) Δsin3/elp3 strain and other mutants of Elongator complex are sensitive to oxidative stress. Serial dilutions from cultures of strains 972 (WT), AV18 (Δsty1), MS161 (Δgcn5), IV16 (Δsin3/elp3), MS98 (Δatf1), IV72 (Δelp4), IV66 (Δiki3/elp1) and IV68 (ΔSPAC30.02c; S. pombe ortholog of kti12) were spotted onto rich plates without (Untreated) or with the indicated concentrations of H2O2 or KCl, and grown at 30°C unless indicated (37°C). (B) Total levels of histone H3 acetylation at lysines 9 and 14 are not affected in a Δsin3/elp3 strain. Protein extracts from strains 972 (WT), MS161 (Δgcn5) and IV16 (Δsin3/elp3) were analyzed by Western blot with antibodies against acetylated Lys9 and Lys14 of histone H3 (H3K9K14-Ac) or total H3, as a loading control. (C) Deletion of clr3 does not rescue Δsin3/elp3 sensitivity to oxidative stress. Serial dilutions from cultures of strains 972 (WT), IV16 (Δsin3/elp3), SPK19 (Δclr3) and IV95 (Δsin3/elp3 Δclr3) were spotted onto rich plates without (Untreated) or with 2 mM H2O2. (D) Stress-dependent transcriptional analysis of wild-type and Δsin3/elp3 cells. Culture of strains 972 (WT), MS161 (Δgcn5) and IV16 (Δsin3/elp3) were treated with 1 mM H2O2 for the indicated times. Total RNA was analyzed by Northern blot with probes for ctt1, gpd1, hsp9, srx1, and gpx1. act1 is shown as a loading control. (E) Stress-dependent H3 acetylation at CESR genes does not require Sin3/Elp3. Cultures of strains 972 (WT) and IV16 (Δsin3/elp3) were treated (+) or not (−) with 1 mM H2O2 for 5 min. Chromatin immunoprecipitation (ChIP) assays were performed using antibodies specific for acetylated Lys9 and Lys14 of histone H3 (H3Ac) or against unmodified C-terminal domain of H3 (H3). The percentage of immunoprecipitation of acetylated H3 versus total H3 is indicated (% IP H3Ac/H3). ChIP experiments were performed using primers covering promoter (prom), coding (ORF) and termination (term) sequences of the gpd1 gene. (F) Stress-dependent nucleosome eviction at CESR genes does not require Sin3/Elp3. The same experiment as in E is represented here as the percentage of immunoprecipitation of total H3 (%IP total H3). Error bars (SEM) for all ChIP experiments were calculated from biological triplicates. Recently, it had been shown that Gcn5 is the major contributor to H3 acetylation in fission yeast, with only a very modest decrease in H3 acetylation at Lys9 and Lys14 in cells lacking Sin3/Elp3 [36] (Figure 1B). Furthermore, we could not suppress the Δsin3/elp3 defects to H2O2 stress by further deletion of the stress-related histone H3 deacetylase Clr3 (Figure 1C). Finally, the expression levels and/or the induction kinetics of several H2O2–inducible genes are affected in Δsin3/elp3 cells, but to a lesser extent than cells lacking the HAT Gcn5 (Figure 1D), even though the sensitivity to peroxides of Δsin3/elp3 cells is more severe than that of Δgcn5 cells (Figure 1A).
Next, we performed chromatin immunoprecipitation (ChIP) experiments in an attempt to detect Sin3/Elp3 at or close to stress genes, as previously found for Gcn5 [7], but we were unable to find Sin3/Elp3 associated with CESR genes (data not shown). Furthermore, we performed ChIP analysis of total and acetylated histone H3 to detect a localized effect of the lack of Sin3/Elp3 on the nucleosomes of the stress genes. However, the levels of histone acetylation, as determined by the ratio of acetylated H3 per total histone H3, were not significantly altered upon stress in cells lacking Sin3/Elp3 compared to wild-type cells (Figure 1E for gpd1 and Figure S1A for ctt1). Also, a similar decrease in total histone H3 levels at CESR genes, as an indicator of nucleosome eviction, was detected upon stress imposition in both wild-type and Δsin3/elp3 cells (Figure 1F for gpd1 and Figure S1B for ctt1). Thus, we conclude that stress-dependent histone acetylation and nucleosome eviction at stress genes does not significantly rely on Sin3/Elp3.
Sin3/Elp3 is required to modify the tRNAs of Lys, Glu and Gln at their wobble U
As detailed in the Introduction, some cytoplasmic tRNAs (those with a UUB anticodon, ‘B’ being U, C or G in the tRNAs for Lys, Glu of Gln, respectively), are subjected to diverse modifications at their U34 (5′ position) of the anticodon to yield mcm5s2U34 (Figure 2A). Thus, Elongator mutants in S. cerevisiae have been shown to fully lack the mcm5U modification [12], and present a 50% decrease in the s2U modification at these specific tRNAs [27], [30]. In an attempt to confirm the requirement of Sin3/Elp3 in the dual modification of these tRNAs, we purified tRNA and used an electrophoretic mobility shift assay which allows the detection of thio-containing tRNA molecules [37]. As a control, we used strains lacking Ctu1 and Ctu2, which had recently been demonstrated to be required for the thiolation step [33] (Figure 2A). As we show in Figure 2B, Sin3/Elp3 seems to be also required for thiolation of U34 at tRNALysUUU, tRNAGlnUUG and tRNAGluUUC. The presence of a contaminant inhibitor in the tRNA samples of the different mutants was discarded by mixing those with wild-type RNA and performing the same band shift assay: the thio-contaning tRNA molecules of wild-type RNA were perfectly detected (Figure S2). These findings suggest that introduction of the mcm5 at U34 is the first step in the dual modification at these tRNA anticodon residues in S. pombe. This is consistent with the observation in budding yeast that defects in Elongator not only abolish the mcm5 modification, but also partially compromise the thiolation of position 2 of U [27], [30], whereas the absence of the Ctu2 homologue, Ncs2, does not affect the formation of the mcm5 chain [15].
Fig. 2. Sin3/Elp3 is required for modification of uridine-34 (U34) at the anticodon of some cytoplasmic tRNAs. 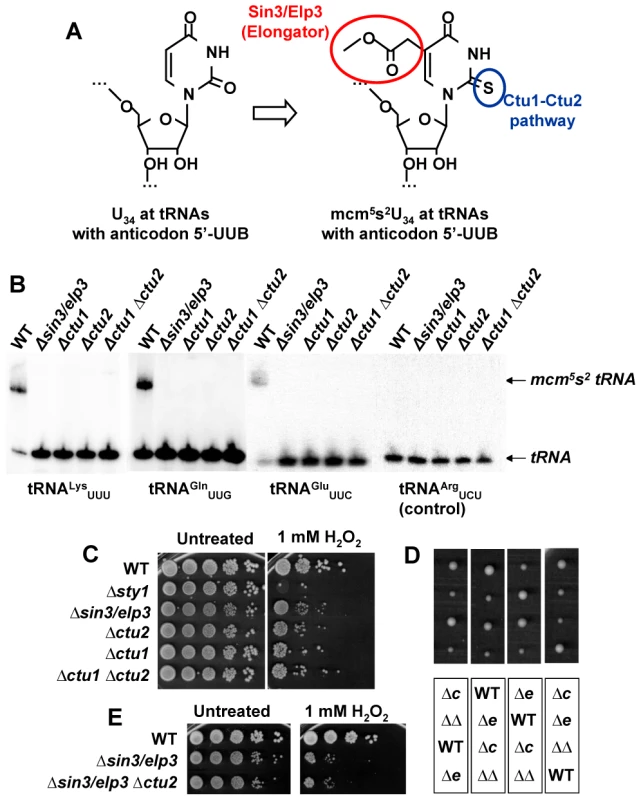
(A) Sin3/Elp3 and the Ctu1-Ctu2 pathways are required for the mcm5 and s2 modifications, respectively, at U34 of some anticodons. (B) Northern blot analysis of bulk tRNA isolated from WT (972), IV16 (Δsin3/elp3), YDH 644 (Δctu1), IV86 (Δctu2), and YDH 254 (Δctu1 Δctu2) using specific probes against tRNALysUUU, tRNAGlnUUG, tRNAGluUUC and tRNAArgUCU (negative control) by the APM-gel retardation method. The position of the unmodified (tRNA) or modified (mcm5s2 tRNA) tRNAs are indicated with arrows. (C) Δctu1 and Δctu2 strains are sensitive to oxidative stress. Serial dilutions from cultures of strains 972 (WT), AV18 (Δsty1), IV16 (Δsin3/elp3), IV86 (Δctu2), YDH 644 (Δctu1), YDH 254 (Δctu1 Δctu2) were spotted onto rich plates without (Untreated) or with 1 mM H2O2. (D) Double mutant Δsin3/elp3 Δctu2 colonies are similar in size to single mutant Δsin3/elp3. Tetrad analysis of a cross between IV16 (Δsin3/elp3) and IV86 (Δctu2) strains. Each vertical box corresponds to the four spores of a tetrad. The genotypes of each colony of the top panel are indicated in the corresponding positions of the bottom panel: WT, Δctu2 (Δc), Δsin3/elp3 (Δe) or Δsin3/elp3 Δctu2 (ΔΔ). (E) Single mutant Δsin3/elp3 and double mutant Δsin3/elp3 Δctu2 show similar sensitivity to oxidative stress. Serial dilutions from cultures of strains 972 (WT), IV16 (Δsin3/elp3) and JF73 (Δsin3/elp3 Δctu2) were spotted onto rich plates without (Untreated) or with 1 mM H2O2. Defects in U34 thiolation by Ctu1-Ctu2 are also rendering cells sensitive to oxidative stress
According to our results, Sin3/Elp3 is required for the mcm5s2U34 modification of tRNAs but does not affect histone H3 acetylation. This suggests that the absence of the mcm5s2U34 modification could be the cause of stress sensitivity of cells lacking Sin3/Elp3. If this hypothesis is correct, then Δctu1 or Δctu2 strains should share the same phenotypes as Elongator mutants, as shown before in budding yeast [15]. As shown in Figure 2C, the tolerance to peroxides of strains lacking Sin3/Elp3, Ctu1 or Ctu2 is very similar and lower than that of wild-type cells. Furthermore, cells lacking Sin3/Elp3 and Ctu2 are not synthetic lethal (Figure 2D) and the double mutant displays similar H2O2 sensitivity than the Δsin3/elp3 mutant (Figure 2E), suggesting that the catalytic activities of the Elongator and Ctu1/2 complexes are sequential (Figure 2B), and that the absence of only one of them is sufficient to avoid full modification of the wobble U34 nucleosides and disturb the function of the target tRNAs.
The stress phenotypes of cells lacking Sin3/Elp3 or Ctu1-Ctu2 are suppressed by over expression of tRNALysUUU
Cells lacking Sin3/Elp3, Ctu1 or Ctu2 cannot generate the mcm5s2U34 modification at some specific tRNAs, and are sensitive to oxidative stress. These modifications have been proposed to contribute to either translation efficiency [22]–[25] or fidelity [19]–[21] of mRNAs containing the complementary codons. If the stress sensitivity is a consequence of defects in codon specific translation, then over-expression of the target tRNAs may alleviate the phenotypes, as shown before in S. cerevisiae [15]. We constructed episomal plasmids and used them to over-express different tRNAs in Δsin3/elp3 cells (Figure 3A). As shown in Figure 3B, the sensitivity to peroxides of cells lacking Sin3/Elp3 was largely rescued with over-expression of tRNALysUUU, but not with plasmids containing other tRNAs which are also modified by Elongator and by Ctu1-Ctu2, such as those for tRNAGlnUUG or tRNAGluUUC. As a negative control, complementation was not observed over-expressing the Elongator-independent tRNALysCUU. Sensitivity of cells lacking Ctu2 was also suppressed with tRNALysUUU over-expression (Figure S3). We conclude that this complementation provides genetic evidence for the role of tRNA modification in the Δsin3/elp3 phenotype, and in particular for the role of modified tRNALysUUU in the stress response.
Fig. 3. Over-expression of tRNALysUUU supresses the growth defects of Δsin3/elp3 upon oxidative stress. 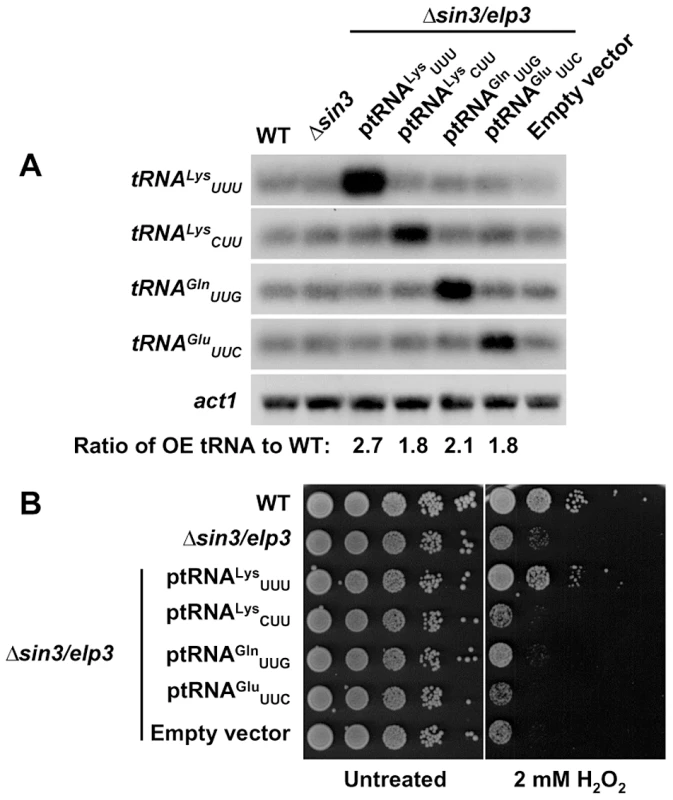
(A) Relative levels of tRNA over-expression by Northern blot. Total RNA from strains 972 (WT), IV16 (Δsin3/elp3), or JF77 (Δsin3/elp3) transformed with episomal plasmids p465 (ptRNALysUUU), p466 (ptRNALysCUU), p467 (ptRNAGlnUUG), p468 (ptRNAGluUUC) or the empty vector pREP.42x, was analyzed by Northern blot with probes of dsDNA of the indicated tRNAs labeled at their antisense strand. act1 is shown as a loading control. The numbers below last panel indicate the relative levels of the corresponding over-expressed tRNA normalized to basal levels in wild-type strain. (B) Serial dilutions from cultures of strains as in A were spotted onto synthetic rich media plates without (Untreated) or with 2 mM H2O2. The codon usage for Lys at stress genes is not optimized for highly expressed genes
Modifications of U34 at the tRNAs for Lys, Gln and Glu (with UUB anticodons) are important for proper cell growth, and S. cerevisiae cells lacking one or several genes required for these modifications display various phenotypes [15], [26]. However, the function of these modifications in the process of translation is far from being understood. In recent years, mcm5s2U34 tRNA modifications have been proposed to modulate translation through a number of distinct mechanisms: increasing the efficiency of codon recognition [25] or aminoacyl-tRNA synthetase interaction [22]–[24]; or increasing translation fidelity by helping to bind only to the correct codon [19], by avoiding frame shifts [20], or both [21]. Lys, Gln and Glu can be coded by two nucleotide triplets, each of which can be recognized by a specific tRNA; only one of the two tRNAs for each of the three amino acids carries a U at the 5′ position of the anticodon which is modified by the Elongator and Ctu1-Ctu2 complexes (see Table 1). The intracellular concentration of each tRNA is assumed to be proportional to the number of copies of the tRNA coding genes [38]–[41]. Regarding the cytoplasmic tRNAs decoding for Lys, Gln and Glu, the S. pombe genome displays the largest disequilibrium in gene copy number for the tRNAs for Lys: there are 3 copies of tRNALysUUU gene but 9 copies of the tRNALysCUU gene (Table 1). However, the overall codon usage for Lys in fission yeast indicates that 62% of the Lys codons require the less abundant tRNALysUUU [42] (Table 1), and this codon usage dramatically changes for highly expressed genes, where only 10% of the codons are read by the Sin3/Elp3 modifiable tRNALysUUU. This bias of codon usage seems to be a result of optimizing translation during evolution, since the abundant tRNAs should be used for the translation of highly expressed mRNAs, which may require an efficient and fast translation machinery [42].
Tab. 1. Codon usage and tRNA gene copy number for Lys, Gln and Glu. 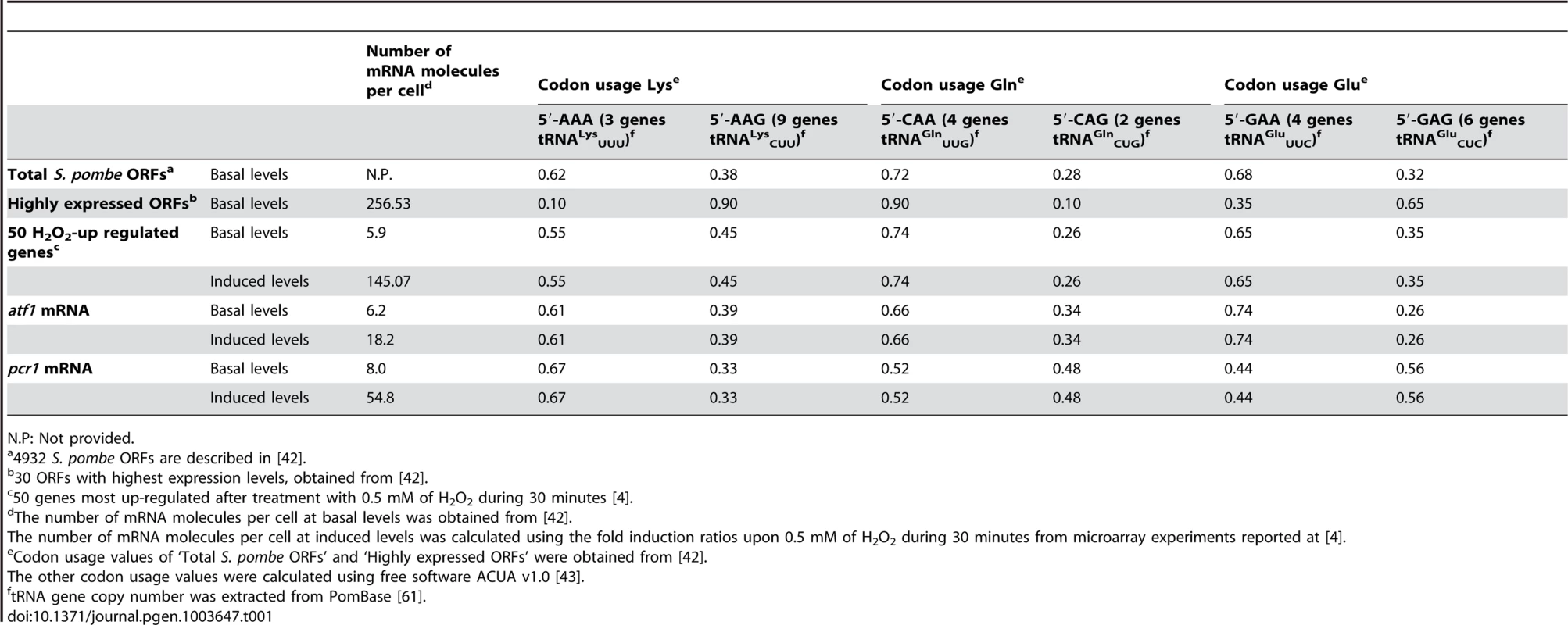
N.P: Not provided. The CESR genes are highly expressed upon stress conditions. We wanted to test whether upon induction these stress genes reach mRNA levels which are comparable to those of highly expressed genes. Using microarray expression data from basal [42] and H2O2 conditions [4], we calculated the mean number of mRNA molecules per cell for the 50 most induced genes upon peroxide exposure (Table 1). On average, we calculated 145 mRNA molecules per cell, which is comparable to the mean 257 mRNA molecules per cell for the group of ‘Highly expressed ORFs’ (open reading frames) [42]. We further calculated the codon usage for these subset of S. pombe genes using the ACUA software [43], and found it similar to the mean of total fission yeast genes (Table 1). This indicates that there has not been an evolutionary adaptation to change the codon usage of the stress genes, which are only highly expressed under some stress conditions. Instead, optimization of the recognition of the Lys codon AAA by the less abundant tRNALysUUU at H2O2-up regulated mRNAs seems to be achieved by modifying the wobble nucleoside U34. That explains why the U34-modifying activities are specifically critical especially for stress survival.
The stress-dependent protein levels, but not the mRNA levels, of the transcription regulators Atf1 and Pcr1 are severely decreased in cells lacking Sin3/Elp3
If lack of Sin3/Elp3 is causing defects due to its tRNA-modifying activity, then translation of stress mRNAs may be affected. The transcriptional response to stress is driven by the MAP kinase Sty1 and the heterodimeric transcription factor Atf1-Pcr1, which trigger massive transcriptional changes. Based on our analysis of codon usage we speculated that induction of CESR gene expression should be accompanied with efficient translation of the newly synthesized mRNAs (Figure 4A). We used act1 mRNA and tubulin as loading and quantification controls in our Northern and Western blots, respectively. Whereas the gene coding for actin is constitutively expressed, the atf1 and pcr1 genes are induced upon H2O2 exposure (Figure 4B; WT), and proper over-expression of Atf1 upon stress is required to fully achieve a complete transcriptional cellular response. The codon usage of the genes coding for these transcriptional regulators has not been evolutionary adapted towards highly expressed genes (Table 1). We therefore speculated that Atf1 and Pcr1 protein synthesis, rather than their mRNA accumulation, would be defective in cells missing the tRNALysUUU-modifying activities Sin3/Elp3 or Ctu2, as would occur with the expression of all the other CESR proteins. As shown in Figure 4B, H2O2-dependent expression of atf1 and pcr1 mRNAs is not dramatically impaired in cells lacking Sin3/Elp3 or Ctu2. However, the amount of translated Atf1 and Pcr1 proteins are clearly diminished in the mutant strains (Figure 4C). Thus, the defective accumulation upon stress of the Atf1-Pcr1 transcription factor and other CESR proteins in the Δsin3/elp3 or Δctu2 strains (Figure 4D) could explain the sensitivity to stress of cells lacking any of these two tRNA-modifying complexes. We also determined that moderate levels of over-expression of tRNALysUUU (Figure 3A) were not able to fully recover the wild-type levels of Atf1 or Pcr1 proteins in the mutant strains, as determined by Western blot (data not shown): we suspect that since up to 500 stress genes are expressed more than 2-fold upon 0.5 mM H2O2 stress [4], most of the corresponding mRNAs may need excess tRNALysUUU for proper translation in Δsin3/elp3 or Δctu2 strains.
Fig. 4. Protein levels of the stress transcription factors Atf1 and Pcr1 depend on the U34 modifying activities Sin3/Elp3 and Ctu2. 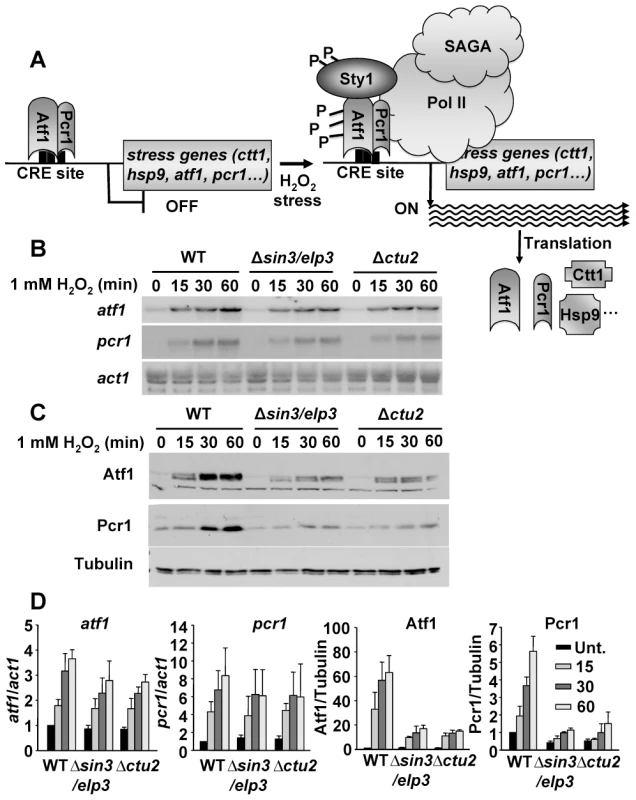
(A) Scheme illustrating the activation of the stress gene expression program by Atf1 and Pcr1 (see text for details). (B and C) Absence of Sin3/Elp3 or Ctu2 barely affects transcription of the atf1 and pcr1 genes but largely affects Atf1 and Pcr1 protein levels. Rich media cultures of strains 972 (WT), IV16 (Δsin3/elp3) and IV86 (Δctu2) treated with 1 mM H2O2 at the indicated time points were analyzed to determine transcription levels of atf1 and pcr1 genes by Northern blot (B), or Atf1 and Pcr1 protein levels by Western blot using polyclonal antibodies (C). act1 mRNA or tubulin were used as loading controls for B and C, respectively. (D) Quantification of the relative mRNA and protein levels for atf1, pcr1, Atf1 and Pcr1 in wild-type and mutant strains. The Northern or Western blot panels of experiments as in B and C, respectively, were quantified and represented here relative to untreated wild-type levels (with an assigned value of 1). The atf1 and pcr1 mRNA levels normalized to act1 are shown in the left two panels, whereas the Atf1 and Pcr1 protein levels normalized to tubulin are shown in the two right panels. Error bars (SEM) were calculated from biological duplicates. Further confirmation of the role of Sin3/Elp3 on mRNA translation came from the fact that expression of a synthetic atf1 gene, in which all the AAA codons of its ORF had been changed by the synonymous AAG, rendered Atf1 expression not sensitive to the absence of Sin3/Elp3 or Ctu2 (Figure 5 and Figure S4 for wild-type and AAA-to-AAG HA-Atf1 expressed from an heterologous and constitutive promoter; Figure S5 for wild-type and AAA-to-AAG Atf1 expressed from its own promoter). The shift in electrophoretic mobility of the H2O2 - and Sty1-dependent phosphorylated Atf1 has been widely reported [44], [45]. Importantly enough, cells lacking Sin3/Elp3 or Ctu2 expressing the AAG-only Atf1 and therefore reaching wild-type levels of Atf1 still displayed sensitivity to grow on H2O2 plates (Figure 5C for Δsin3/elp3 and Figure S4C for Δctu2), since the Lys codon usage of the hundreds of genes over-expressed upon oxidative stress remain rich in the AAA codon (Table 1).
Fig. 5. Expression of a synthetic AAA-to-AAG atf1 gene rendered wild-type Atf1 protein levels in Elongator mutants. 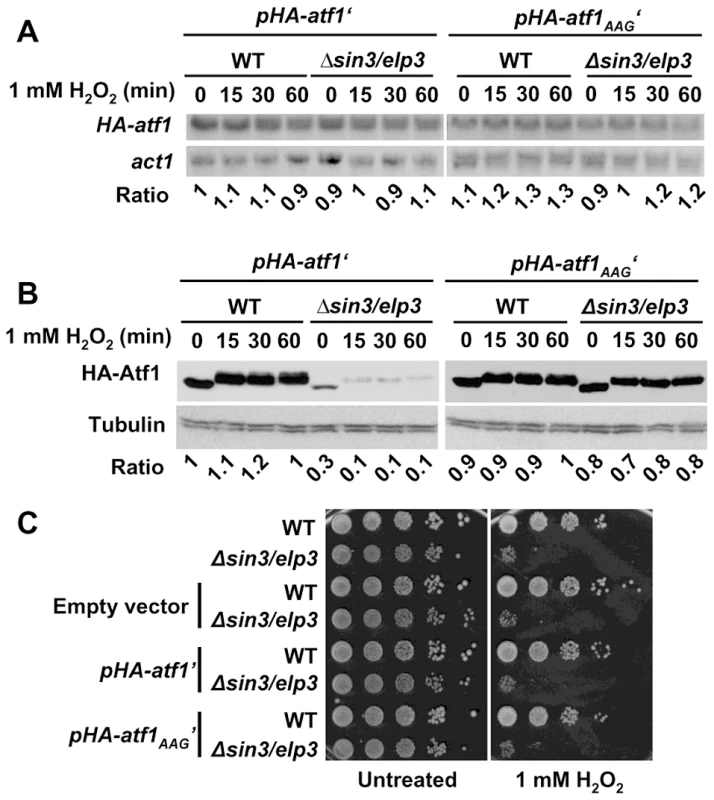
(A and B) Vectors carrying a constitutively expressed wild-type (pHA-atf1′) or mutated atf1 gene (pHA-atf1AAG′) were integrated in the chromosomes of wild-type or Δsin3/elp3 strains. Rich media cultures of strains JF91 (WT+pHA-atf1′), JF92 (Δsin3/elp3+pHA-atf1′), JF94 (WT+pHA-atf1AAG′) and JF95 (Δsin3/elp3+pHA-atf1AAG′), either untreated (0) or treated with 1 mM H2O2 for the indicated times, were analyzed to determine HA-atf1 mRNA levels by Northern blot using an anti-HA probe (A) or HA-Atf1 protein levels by Western blot using monoclonal antibody against HA (B). The numbers below the Northern or Western blot panels indicate the relative levels of HA-atf1/act1 mRNAs (panel A) or HA-Atf1/tubulin protein levels (panel B). (C) Expression of the mutant AAA-to-AAG Atf1 protein does not suppress the growth defects of Δsin3/elp3 cells upon oxidative stress. Empty vector or plasmids carrying a wild-type (pHA-atf1′) or a mutated atf1 gene (pHA-atf1AAG′) were integrated in the chromosomes of wild-type or Δsin3/elp3 strains. Cultures from the resulting strains JF88 (WT+empty vector), JF89 (Δsin3/elp3+empty vector), JF91 (WT+pHA-atf1′), JF92 (Δsin3/elp3+pHA-atf1′), JF94 (WT+pHA-atf1AAG′) and JF95 (Δsin3/elp3+pHA-atf1AAG′) were serially diluted and spotted onto rich media plates without (Untreated) or with 1 mM H2O2. Discussion
Optimal performance of a biological process such as cellular adaptation to environmental changes requires that a complex phenomenon like protein expression to be carried out with high efficiency and fidelity. Thus, not only transcription but also mRNA homeostasis and translation have to be performed with maximum efficiency, or survival would be hampered. Our study not only provides new insights into the way cells respond to oxidative stress, but also provides important clues regarding the role of the Elongator complex in translation efficiency. Furthermore, our study reveals how the absence of the mcm5s2U34 modification at tRNALysUUU in cells lacking Sin3/Elp3 or Ctu1/Ctu2 contributes to the observed phenotype of sensitivity to peroxides of these mutant strains.
One of the singularities of the redundancy of the genetic code is that it allows to choose between alternative codons for the same amino acid, and that may exert important consequences on the efficiency of translation depending mainly on the concentrations of each one of the corresponding tRNAs. As intracellular concentrations of different tRNAs are not easily measured, the amount of each species in cells are often considered to be proportional to the copy number of the tRNA-coding genes in the genome [38], [41]. Thus, codon usage may be biased towards the use of abundant tRNAs when strong and efficient translation is required, i.e. highly expressed genes. Out of the three tRNAs species modified at their 5′-uridine of the anticodon to mcm5s2U34, tRNAs for Lys display the largest imbalance according the gene copy number (only one tRNALysUUU for 3 tRNALysCUU; Table 1). Accordingly, the codon usage for highly expressed ORFs suffers the major deviation from the average S. pombe ORFs for the Lys codons (only 10% of the Lys codons are AAA in highly expressed ORFs, whereas the average is 62%; Table 1). This adaptation of the Lys codon usage in highly expressed genes, which has not been adopted by the CESR genes, would justify the need for the mcm5s2U34 modification at the tRNALysUUU to set up a strong stress response, which should enhance the efficiency of translation of the atf1 and pcr1 mRNAs.
Recently, a mass spectrometry-based analysis by the groups of Dedon and Begley has revealed that the spectrum of nucleoside modifications of S. cerevisiae changes upon cell exposure to H2O2 and other stressors [46]. In some cases, but not all, increases of a specific modified ribonucleoside correlate with stress sensitivity of mutant cells lacking an enzyme involved in its biosynthesis [46]. In particular, the mcm5s2U34 is not enhanced by peroxides in budding yeast, while other modifications such as m5C are [47]. Similarly, cells lacking Trm9, required to introduce the mcm5s2U34 in yeast, are not sensitive to H2O2 [46], while we have shown here that cells lacking Elongator components, or Ctu1 or Ctu2, display impaired survival when exposed to peroxides. How the changes in modified nucleosides are triggered upon stress in S. cerevisiae is still to be determined. Whether the mcm5s2U34 modification accumulates upon H2O2 in S. pombe will have to be elucidated, but at least the activities required for this modification (Elongator and Ctu1-2) are not up-regulated at the transcriptional level upon peroxide exposure [4].
We have shown here that generation of the mcm5U34 by Elongator is required for wild-type tolerance to oxidative stress. In the absence of Elongator, the thiolation at carbon 2 of U34 is not generated by Ctu1-Ctu2 (Figure 2A), since cells lacking Sin3/Elp3 do not incorporate any modification (mcm5 or S2) at U34. However, cells lacking Ctu2 or Ctu1 accumulate mcm5U34 [33]; unfortunately, this modification does not seem to be sufficient to allow proper translation of atf1 and pcr1 mRNAs (Figure 4B), and therefore Δctu2 cells show sensitivity to peroxides (Figure 2C).
Cells lacking Sin3/Elp3 already display some defects in the absence of stress, since translation of some growth-related mRNAs is probably dampened as well. Thus, the size of colonies derived from Δsin3/elp3 spores is significantly smaller than that of wild-type or Δctu2 ones (Figure 2D). Also, both the duplication time and maximum OD600 of cultures are defective in cells lacking Sin3/Elp3 (Figure S6A). Importantly enough, the growth-related Δsin3/elp3 defects are totally suppressed by over-expression of tRNALysUUU (Figure S6B). It has recently been reported that fission yeast cells devoid of Elongator result in three phenotypes unrelated to stress: thermosensitivity, cell elongation and multiple and misplaced septa, common to defects in cell-cycle progression [11], [34]. In this report, the authors performed a screen of the fission yeast proteome for translational defects which pointed towards the kinase Cdr2, a central regulator of mitosis, as the target of translational control by Elongator due to an unusual Lys codon usage bias of its mRNA. Analysis of their proteome-wide data indicates that Atf1 and Pcr1 are also down-regulated even under basal, unstressed conditions, supporting the conclusions of our manuscript [34]. It is worth pointing out that cells lacking Ctu2 display defects in atf1 and pcr1 translation and subsequent sensitivity to stress, whereas they do not seem to have growth defects (Figure 2D and Figure S6A). This suggests that the non-thiolated mcm5U34 tRNAs of Δctu2 cells may have some intermediate activity, indicating that some mRNAs could be efficiently translated but not others.
In conclusion, our study points to the fact that the biologically essential functions of Elongator in S. pombe are related to tRNA modification. This function is significantly relevant for the efficient translation of mRNAs with a biased high AAA to AAG ratio of codons for Lys, especially when these mRNAs are expressed at high copy number. Highly expressed house-keeping genes have evolutionary circumvented this problem by enhancing the AAG to AAA ratio. The wobble position U34 of tRNAs with an UUB anticodon is almost universally modified [48]–[50], and our report suggests that specific highly expressed mRNAs with a high VAA content (‘V’ being A, C or G) should be inefficiently translated in the absence of Elongator, what would determine a particular phenotype for each cell type.
Materials and Methods
Yeast strains, plasmids and growth conditions
We used the wild-type S. pombe 972 (h−) and mutants thereof. The origins and genotypes of strains used in this study are outlined in Table S2. To construct episomal plasmids containing tRNA coding genes, the tRNA genomic sequences were PCR-amplified from S. pombe genomic DNA using primers specific for the tRNAs' upstream and downstream sequences, and cloned into the fission yeast episomal plasmid pREP.42x [51], with a ura4-selectable marker, previously digested with PstI and SacI to eliminate the nmt promoter and the transcription terminator sequences, so that each tRNA is expressed from its endogenous sequences (500 bp genomic flanking sequences on each side of the tRNAs). The replication origin ars1 contained in the pREP.42x vector allows an average plasmid copy number of approximately 8 copies/cell [52]. We obtained plasmids p465 [expressing tRNALysUUU (SPBTRNALYS.06)], p466 [expressing tRNALysCUU (SPCTRNALYS.11)], p467 [expressing tRNAGlnUUG (SPBTRNAGLN.02)] and p468 [expressing tRNAGluUUC (SPATRNAGLU.02)]. All the PCR-amplified DNA fragments cloned in these plasmids were confirmed by sequencing. To generate a mutated version of atf1 where all eleven AAA codons were replaced by AAG, full length gene synthesis was performed (GeneScript). The inducible nmt promoter of plasmid p123.41x [53] was replaced by the constitutive sty1 promoter (0.8 kb from ATG). The resulting plasmid was digested to release the sty1 promoter fused to the HA coding sequence, and cloned into pAY025 [54] to obtain the plasmid p386′ (empty vector). Then, the atf1 and atf1AAG ORFs were cloned into p386′ to obtain p428′ and p428′AAG respectively. These plasmids, allowing the constitutive expression of HA-Atf1 and HA-Atf1AAG, were integrated at the leu1 loci of wild-type and mutant strains, yielding strains JF91 to JF96 (see Table S2). Both wild-type and mutated atf1AAG alleles were also introduced at the endogenous atf1 locus of strain JF85, JF86 (Δsin3/elp3) and JF87 (Δctu2) by standard recombination techniques, yielding strains JF106, JF107 and JF108 (wild-type atf1) and JF109, JF110 and JF111 (atf1AAG) (see Table S2). Cells were grown in rich medium (YE) in most of the experiments, or in synthetic minimal medium when indicated [55].
H2O2 sensitivity assay
For survival on solid plates, S. pombe strains were grown, diluted and spotted (105 to 10 cells per spot) in YE medium or synthetic minimal medium agar plates as described previously [54], containing or not H2O2 at the indicated concentrations.
RNA analysis
Total RNA from S. pombe rich medium cultures was obtained, processed and transferred to membranes as described previously [56]. Membranes were hybridized with the [α-32P]dCTP-labelled ctt1, gpd1, hsp9, srx1, gpx1, atf1, pcr1 and act1 probes, containing the complete ORFs. To determine the levels of tRNA over-expression in strains carrying episomal tRNA plasmids, we PCR-amplified from genomic DNA 300-bp products corresponding to each specific tRNA, and then performed a primer extension of each antisense strand with Klenow polymerase, dNTPs and [α-32P]dCTP, following standard molecular biology techniques [57]. A similar strategy was used to label the HA-coding DNA, which was used in Figures 5A and S4A. Membranes were exposed to a phosphorimager plate (GE Healthcare) and scanned on a Typhoon 8600 (GE Healthcare). Relative quantifications were performed using the ImageQuant 5.2 program (GE healthcare), using act1 mRNA as loading control.
Preparation of S. pombe TCA extracts and immuno blot analysis
Modified trichloroacetic acid (TCA) extracts were prepared as previously described [54]. Immunoblotting to analyze the in vivo acetylation state of total histone H3 was performed as described previously [7]. Pcr1 and Atf1 were immunodetected with polyclonal anti-Pcr1 and polyclonal anti-Atf1 antiserums, as described previously [54]. HA-Atf1 was detected with monoclonal anti-HA antibody. Monoclonal anti-tubulin (Sigma) was used as a loading control. Relative quantifications of protein levels in Western blots was performed using the free Image J software.
Chromatin immunoprecipitation
To test histone H3 acetylation and total H3 upon stress imposition, the indicated strains were grown in rich media, and chromatin isolation and immunoprecipitation was performed as described previously [7]. The error bars (SEM) were calculated from biological triplicates.
tRNA isolation
Cells were grown at 30°C in 100 ml rich media and harvested at OD600 of 0.5. The cell pellet was resuspended in 4 ml 0.9% NaCl. The cells suspension was vortexed at room temperature for 5 min in the presence of 4 ml of acidic phenol and 3 ml of glass beads. Subsequently 0.4 ml chloroform were added and the suspension vortexed for another 30 sec. The suspension was cleared by centrifugation at 3000 rpm for 20 min at room temperature. The water phase was collected and re-extracted with 2 ml of acidic phenol and 0.2 ml of chloroform until the interphase was clean. The final water phase was collected, mixed with 2.5 vol of 100% ethanol and 0.1 vol of 20% potassium acetate to precipitate tRNA. Precipitated tRNA was purified by column purification as described previously [58].
tRNA modification analysis
0.5 µg of bulk tRNA per lane (or mixed 0.5 µg+0.5 µg of two different types of bulk tRNA per lane when indicated; Figure S2) were analyzed on 10% acrylamide gels, 0.5× TBE; 7 M urea. (N-Acryloylamino) phenyl mercuric chloride (APM) was added to a final concentration of 50 µg per ml. Northern blot analysis was performed essentially as described previously [59], using probes CTCCCACTGCGAGATTCGAACTCGC to detect tRNALysUUU, GGTCGTACTGGGAATCGAACCCAGG to detect tRNAGlnUUG, CTCCGTTGCGGGGAGTCGAA to detect tRNAGluUUC and CTCCCGGCGGGACTCGAA to detect the negative control tRNAArgUCU. Membranes were exposed to a phosphorimager plate (GE Healthcare) and scanned on a Typhoon FLA 7000 (GE Healthcare). In the absence of APM in the gels, the corresponding shifts of thiolated tRNAs were not observed (data not shown).
Tetrad analysis
Tetrad analysis was performed essentially as described [60]. Briefly, asci and spores was separated using the MSM 400 Yeast Dissection Microscope (Singer Instruments) and germinated in YE medium agar plates. To determine the genotype of each spore, colonies generated were replicated into the same media containing or not kanamycin (KAN) and/or nourseothricin (NAT).
Supporting Information
Zdroje
1. VivancosAP, JaraM, ZuinA, SansoM, HidalgoE (2006) Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol Genet Genomics 276 : 495–502.
2. VealEA, DayAM, MorganBA (2007) Hydrogen peroxide sensing and signaling. Mol Cell 26 : 1–14.
3. ChenD, TooneWM, MataJ, LyneR, BurnsG, et al. (2003) Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell 14 : 214–229.
4. ChenD, WilkinsonCR, WattS, PenkettCJ, TooneWM, et al. (2008) Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol Biol Cell 19 : 308–317.
5. SansoM, Vargas-PerezI, GarciaP, AyteJ, HidalgoE (2011) Nuclear roles and regulation of chromatin structure by the stress-dependent MAP kinase Sty1 of Schizosaccharomyces pombe. Mol Microbiol 82 : 542–554.
6. JohnssonA, Xue-FranzenY, LundinM, WrightAP (2006) Stress-specific role of fission yeast Gcn5 histone acetyltransferase in programming a subset of stress response genes. Eukaryot Cell 5 : 1337–1346.
7. SansoM, Vargas-PerezI, QuintalesL, AntequeraF, AyteJ, et al. (2011) Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res 39 : 6369–6379.
8. WittschiebenBO, FellowsJ, DuW, StillmanDJ, SvejstrupJQ (2000) Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. Embo J 19 : 3060–3068.
9. WinklerGS, KristjuhanA, Erdjument-BromageH, TempstP, SvejstrupJQ (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci U S A 99 : 3517–3522.
10. SvejstrupJQ (2007) Elongator complex: how many roles does it play? Curr Opin Cell Biol 19 : 331–336.
11. HeyerWD, ThuriauxP, KohliJ, EbertP, KerstenH, et al. (1984) An antisuppressor mutation of Schizosaccharomyces pombe affects the post-transcriptional modification of the “wobble” base in the anticodon of tRNAs. J Biol Chem 259 : 2856–2862.
12. HuangB, JohanssonMJ, BystromAS (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. Rna 11 : 424–436.
13. MehlgartenC, JablonowskiD, WrackmeyerU, TschitschmannS, SondermannD, et al. (2010) Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 76 : 1082–1094.
14. ChenC, TuckS, BystromAS (2009) Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 5: e1000561.
15. EsbergA, HuangB, JohanssonMJ, BystromAS (2006) Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 24 : 139–148.
16. ChenC, HuangB, EliassonM, RydenP, BystromAS (2011) Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet 7: e1002258.
17. VerseesW, De GroeveS, Van LijsebettensM (2010) Elongator, a conserved multitasking complex? Mol Microbiol 76 : 1065–1069.
18. AgrisPF, VendeixFA, GrahamWD (2007) tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol 366 : 1–13.
19. KrugerMK, PedersenS, HagervallTG, SorensenMA (1998) The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J Mol Biol 284 : 621–631.
20. UrbonaviciusJ, QianQ, DurandJM, HagervallTG, BjorkGR (2001) Improvement of reading frame maintenance is a common function for several tRNA modifications. Embo J 20 : 4863–4873.
21. PatilA, ChanCT, DyavaiahM, RooneyJP, DedonPC, et al. (2012) Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol 9 : 990–1001.
22. AgrisPF, SollD, SenoT (1973) Biological function of 2-thiouridine in Escherichia coli glutamic acid transfer ribonucleic acid. Biochemistry 12 : 4331–4337.
23. SylversLA, RogersKC, ShimizuM, OhtsukaE, SollD (1993) A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry 32 : 3836–3841.
24. CusackS, YaremchukA, TukaloM (1996) The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. Embo J 15 : 6321–6334.
25. AshrafSS, SochackaE, CainR, GuentherR, MalkiewiczA, et al. (1999) Single atom modification (O–>S) of tRNA confers ribosome binding. Rna 5 : 188–194.
26. BjorkGR, HuangB, PerssonOP, BystromAS (2007) A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. Rna 13 : 1245–1255.
27. NakaiY, NakaiM, HayashiH (2008) Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem 283 : 27469–27476.
28. HuangB, LuJ, BystromAS (2008) A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. Rna 14 : 2183–2194.
29. SchliekerCD, Van der VeenAG, DamonJR, SpoonerE, PloeghHL (2008) A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A 105 : 18255–18260.
30. NomaA, SakaguchiY, SuzukiT (2009) Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 37 : 1335–1352.
31. LeidelS, PedrioliPG, BucherT, BrostR, CostanzoM, et al. (2009) Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458 : 228–232.
32. PedrioliPG, LeidelS, HofmannK (2008) Urm1 at the crossroad of modifications. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep 9 : 1196–1202.
33. DewezM, BauerF, DieuM, RaesM, VandenhauteJ, et al. (2008) The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci U S A 105 : 5459–5464.
34. BauerF, MatsuyamaA, CandiracciJ, DieuM, ScheligaJ, et al. (2012) Translational control of cell division by Elongator. Cell Reports 1 : 1–10.
35. FichtnerL, FrohloffF, BurknerK, LarsenM, BreunigKD, et al. (2002) Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol Microbiol 43 : 783–791.
36. NugentRL, JohnssonA, FlehartyB, GogolM, Xue-FranzenY, et al. (2010) Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation. BMC Genomics 11 : 59.
37. IgloiGL (1988) Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27 : 3842–3849.
38. DongH, NilssonL, KurlandCG (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260 : 649–663.
39. PercudaniR, PavesiA, OttonelloS (1997) Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J Mol Biol 268 : 322–330.
40. KanayaS, YamadaY, KudoY, IkemuraT (1999) Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238 : 143–155.
41. TullerT, CarmiA, VestsigianK, NavonS, DorfanY, et al. (2010) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141 : 344–354.
42. HiraokaY, KawamataK, HaraguchiT, ChikashigeY (2009) Codon usage bias is correlated with gene expression levels in the fission yeast Schizosaccharomyces pombe. Genes Cells 14 : 499–509.
43. VetrivelU, ArunkumarV, DorairajS (2007) ACUA: a software tool for automated codon usage analysis. Bioinformation 2 : 62–63.
44. KanohJ, WatanabeY, OhsugiM, IinoY, YamamotoM (1996) Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1 : 391–408.
45. ShiozakiK, RussellP (1996) Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev 10 : 2276–2288.
46. ChanCT, DyavaiahM, DeMottMS, TaghizadehK, DedonPC, et al. (2010) A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247.
47. ChanCT, PangYL, DengW, BabuIR, DyavaiahM, et al. (2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3 : 937.
48. GustiloEM, VendeixFA, AgrisPF (2008) tRNA's modifications bring order to gene expression. Curr Opin Microbiol 11 : 134–140.
49. GingoldH, PilpelY (2011) Determinants of translation efficiency and accuracy. Mol Syst Biol 7 : 481.
50. GrosjeanH, de Crecy-LagardV, MarckC (2010) Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett 584 : 252–264.
51. MaundrellK (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123 : 127–130.
52. BrunC, DubeyDD, HubermanJA (1995) pDblet, a stable autonomously replicating shuttle vector for Schizosaccharomyces pombe. Gene 164 : 173–177.
53. VivancosAP, CastilloEA, BiteauB, NicotC, AyteJ, et al. (2005) A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc Natl Acad Sci U S A 102 : 8875–8880.
54. SansoM, GogolM, AyteJ, SeidelC, HidalgoE (2008) Transcription factors Pcr1 and Atf1 have distinct roles in stress - and Sty1-dependent gene regulation. Eukaryot Cell 7 : 826–835.
55. MorenoS, KlarA, NurseP (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194 : 795–823.
56. CastilloEA, AyteJ, ChivaC, MoldonA, CarrascalM, et al. (2002) Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol Microbiol 45 : 243–254.
57. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
58. BjorkGR, JacobssonK, NilssonK, JohanssonMJ, BystromAS, et al. (2001) A primordial tRNA modification required for the evolution of life? Embo J 20 : 231–239.
59. LauNC, LimLP, WeinsteinEG, BartelDP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 : 858–862.
60. SmithGR (2009) Genetic analysis of meiotic recombination in Schizosaccharomyces pombe. Methods Mol Biol 557 : 65–76.
61. WoodV, HarrisMA, McDowallMD, RutherfordK, VaughanBW, et al. (2012) PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40: D695–699.
Štítky
Genetika Reprodukční medicína
Článek Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and MammalsČlánek The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and DevelopmentČlánek Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative ElementsČlánek Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine ExposureČlánek Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 7- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- An Solution for Crossover Formation
- Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, , as a Genetic Component of Neural Tube Defects in Humans
- Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
- Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
- Genomic Analysis of Natural Selection and Phenotypic Variation in High-Altitude Mongolians
- Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
- Role of CTCF Protein in Regulating Locus Transcription
- Gene Set Signature of Reversal Reaction Type I in Leprosy Patients
- Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups
- Is Required for Formation of the Genital Ridge in Mice
- Monopolin Subunit Csm1 Associates with MIND Complex to Establish Monopolar Attachment of Sister Kinetochores at Meiosis I
- Recombination Dynamics of a Human Y-Chromosomal Palindrome: Rapid GC-Biased Gene Conversion, Multi-kilobase Conversion Tracts, and Rare Inversions
- Mechanisms of Protein Sequence Divergence and Incompatibility
- Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack
- Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies
- Combinatorial Regulation of Meiotic Holliday Junction Resolution in by HIM-6 (BLM) Helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 Nucleases
- The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
- The Role of Interruptions in polyQ in the Pathology of SCA1
- Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in
- Fine Time Course Expression Analysis Identifies Cascades of Activation and Repression and Maps a Putative Regulator of Mammalian Sex Determination
- Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90
- Oxidative Stress and Replication-Independent DNA Breakage Induced by Arsenic in
- A Moonlighting Enzyme Links Cell Size with Central Metabolism
- Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
- The Conserved Intronic Cleavage and Polyadenylation Site of CstF-77 Gene Imparts Control of 3′ End Processing Activity through Feedback Autoregulation and by U1 snRNP
- The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in through Maintaining a Progenitor-like Cell State
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- The RNA-binding Proteins FMR1, Rasputin and Caprin Act Together with the UBA Protein Lingerer to Restrict Tissue Growth in
- Pattern Dynamics in Adaxial-Abaxial Specific Gene Expression Are Modulated by a Plastid Retrograde Signal during Leaf Development
- A Network of HMG-box Transcription Factors Regulates Sexual Cycle in the Fungus
- Bacterial Adaptation through Loss of Function
- ENU-induced Mutation in the DNA-binding Domain of KLF3 Reveals Important Roles for KLF3 in Cardiovascular Development and Function in Mice
- Interplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
- FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription
- The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression
- Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative Elements
- The Conserved ADAMTS-like Protein Lonely heart Mediates Matrix Formation and Cardiac Tissue Integrity
- The cGMP-Dependent Protein Kinase EGL-4 Regulates Nociceptive Behavioral Sensitivity
- RBM5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility
- Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of Expression at the 6q23.2 Coronary Heart Disease Locus
- Yeast Pol4 Promotes Tel1-Regulated Chromosomal Translocations
- A Dual Role for SOX10 in the Maintenance of the Postnatal Melanocyte Lineage and the Differentiation of Melanocyte Stem Cell Progenitors
- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle,
- Prediction of Complex Human Traits Using the Genomic Best Linear Unbiased Predictor
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
- Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine Exposure
- Exquisite Light Sensitivity of Cryptochrome
- miR-133a Regulates Adipocyte Browning In Vivo
- Strabismus Promotes Recruitment and Degradation of Farnesylated Prickle in Planar Polarity Specification
- Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
- Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
- Joint Molecule Resolution Requires the Redundant Activities of MUS-81 and XPF-1 during Meiosis
- The Mating Competence of Geographically Diverse Strains in Their Natural and Unnatural Sand Fly Vectors
- Defective Repair of Oxidative Base Lesions by the DNA Glycosylase Nth1 Associates with Multiple Telomere Defects
- Effective Blocking of the Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function
- Trans-Ancestral Studies Fine Map the SLE-Susceptibility Locus
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Bacterial Adaptation through Loss of Function
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



