-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
X chromosome inactivation in eutherian mammals has been thought to be tightly controlled, as expected from a mechanism that compensates for the different dosage of X-borne genes in XX females and XY males. However, many X genes escape inactivation in humans, inactivation of the X in marsupials is partial, and the unrelated sex chromosomes of monotreme mammals have incomplete and gene-specific inactivation of X-linked genes. The bird ZW sex chromosome system represents a third independently evolved amniote sex chromosome system with dosage compensation, albeit partial and gene-specific, via an unknown mechanism (i.e. upregulation of the single Z in females, down regulation of one or both Zs in males, or a combination). We used RNA-fluorescent in situ hybridization (RNA-FISH) to demonstrate, on individual fibroblast cells, inactivation of 11 genes on the chicken Z and 28 genes on the X chromosomes of platypus. Each gene displayed a reproducible frequency of 1Z/1X-active and 2Z/2X-active cells in the homogametic sex. Our results indicate that the probability of inactivation is controlled on a gene-by-gene basis (or small domains) on the chicken Z and platypus X chromosomes. This regulatory mechanism must have been exapted independently to the non-homologous sex chromosomes in birds and mammals in response to an over-expressed Z or X in the homogametic sex, highlighting the universal importance that (at least partial) silencing plays in the evolution on amniote dosage compensation and, therefore, the differentiation of sex chromosomes.
Published in the journal: . PLoS Genet 9(7): e32767. doi:10.1371/journal.pgen.1003635
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003635Summary
X chromosome inactivation in eutherian mammals has been thought to be tightly controlled, as expected from a mechanism that compensates for the different dosage of X-borne genes in XX females and XY males. However, many X genes escape inactivation in humans, inactivation of the X in marsupials is partial, and the unrelated sex chromosomes of monotreme mammals have incomplete and gene-specific inactivation of X-linked genes. The bird ZW sex chromosome system represents a third independently evolved amniote sex chromosome system with dosage compensation, albeit partial and gene-specific, via an unknown mechanism (i.e. upregulation of the single Z in females, down regulation of one or both Zs in males, or a combination). We used RNA-fluorescent in situ hybridization (RNA-FISH) to demonstrate, on individual fibroblast cells, inactivation of 11 genes on the chicken Z and 28 genes on the X chromosomes of platypus. Each gene displayed a reproducible frequency of 1Z/1X-active and 2Z/2X-active cells in the homogametic sex. Our results indicate that the probability of inactivation is controlled on a gene-by-gene basis (or small domains) on the chicken Z and platypus X chromosomes. This regulatory mechanism must have been exapted independently to the non-homologous sex chromosomes in birds and mammals in response to an over-expressed Z or X in the homogametic sex, highlighting the universal importance that (at least partial) silencing plays in the evolution on amniote dosage compensation and, therefore, the differentiation of sex chromosomes.
Introduction
Vertebrates with heteromorphic sex chromosomes have either male heterogamety like humans (XX female and XY male), or female heterogamety like birds (ZZ male and ZW female). Degeneration of the non-recombining Y or W chromosome, central to the evolution of sex chromosomes, left genes on the X or Z as a single copy in the heterogametic sex. This resulted in an imbalance of X or Z gene dosage relative to the autosomes, and between the sexes.
Different dosage compensation systems have arisen independently in diverse organisms, suggesting that dosage compensation is critical for the survival of species with differentiated sex chromosomes. Ohno [1] hypothesized that degeneration of the Y/W chromosome would result in under expression from the X/Z in the heterogametic sex (equivalent to monosomy), which would result in pressure to up regulate the single X/Z to restore parity with the autosomes. Because the homogametic sex has two X/Z chromosomes, over expression would result in doubled normal expression (equivalent to tetrasomy), which would result in pressure for global down regulation of the X/Z to again resort parity with the autosome (reviewed in [2]). However, recent data has questioned global over regulation of the X/Z [3], [4], sparking considerable debate [5]–[7] and suggestion that dosage compensation evolved in response to a subset of dosage sensitive genes [8].
In eutherian mammals one whole X is transcriptionally silenced in the somatic cells of females, although many genes located on the evolutionarily more recent region escape silencing [9]. X inactivation (XCI) is established early in embryogenesis and is somatically heritable. In eutherian mammals the silenced X is chosen at random in a process governed by a large non-coding RNA transcribed from a locus called XIST (X-inactive-specific transcript) [10]–[12]. In all eutherian mammals, XIST is transcribed from the X to be inactivated, which it coats in cis during early development, although the timing and regulation of expression varies between species [13]. After one X is chosen for inactivation, a specific signature of epigenetic modifications is established [14], which appears to be conserved in even the most distantly related eutherians [15].
The increasing availability of genomic data now makes it possible to study dosage compensation in non-traditional model organisms such as marsupial and monotreme mammals. In marsupials, XCI is imprinted, with the paternal X always being the inactivated homologue [16]. X-inactivation in representative Australian marsupials is thought to be incomplete, tissue-specific and gene-specific [17]. RNA-fluorescent in situ hybridization (RNA-FISH) showed that within a population of fibroblasts cells derived from female tammar wallaby there was a mixture of 1X-active and 2X-active nuclei, the proportion of which was characteristic of each locus [18]. A similar profile was obtained for human and elephant X genes that escape inactivation [19]. However, it was shown (with RNA-FISH) in post-mortem tissue of the South American grey short-tailed opossum that X-inactivation is efficient [20], and RNA-sequencing revealed that X expression levels are similar between the sexes [21].
The egg-laying monotreme mammals (platypus and echidna species) have multiple sex chromosomes that share homology, not with the therian X, but the bird Z [22], [23]. Recent transcriptome sequencing in platypus showed that compensation is achieved via upregulation of X genes in males, and that global X-inactivation in females is likely unnecessary [21]. However, as for marsupials and escaper genes on the human X, RNA-FISH showed that within a population of platypus fibroblast cells, there was a mixture of 1X-active and 2X-active cells, each locus having a characteristic frequency of inactivation [24].
Data on dosage compensation in birds are fragmentary and contradictory. Real-time PCR showed equivalent expression of most Z-borne genes in ZZ male and ZW female chick embryos (that is, complete compensation) for most genes [25], but global microarray analysis in chicken, and a small cDNA microarray in zebra finch showed that male to female ratios were significantly higher for Z genes than for autosomal genes [26]. A male: female Z gene dosage of approximately 1.5 was demonstrated by microarray [27] and RNA-sequencing [21] analyses in chicken, and dosage ratios of 1.23 was observed in zebra finch [28] and 1.36 in crow [29]. The incomplete dosage compensation of Z-linked genes, at least in chicken, was reported to be regulated locally on a gene-by-gene basis, and is tissue and developmental stage specific [30].
It is difficult to compare dosage compensation between birds and mammals. However, recent comparative transcriptome sequencing [21] indicated that genes on the single Z in female chicken had equivalent expression levels to orthologous genes in outgroup species, where the Z is autosomal (i.e. expression from one Z equals expression from two proto-Zs), providing evidence for global upregulation of the single Z in females. In male chicken (with two Zs) the ZZ:proto-ZZ expression ratio was 1.13 to 1.56, indicating that Z gene upregulation was not specific to females. A similar pattern, although less clear, was observed for genes on platypus X5. Less efficient upregulation in the homogametic sex (i.e. ZZ/X5X5: ZW/X5Y5 expression ratio <2) indicated that upregulation was more efficient in the heterogametic sex, or that there was a mechanism to partially reduce expression of Z/X5 in the homogametic sex. Julien et al. [21] suggested that dosage compensation only mildly affects the homogametic sex in platypus and chicken and, as such, there was potentially no requirement for the evolution of Z/X5 inactivation.
Here we use RNA-FISH to examined chicken and platypus dosage compensation, which permits detection of transcription from one or both alleles at specific loci in individual nuclei. We found that genes on the chicken Z, as well as on the partially homologous platypus Xs, were expressed from one (or both) alleles in characteristic frequencies for different loci, just as on the independently evolved therian X. Our results indicate that silencing mechanisms were exapted multiple times in the homogametic sex (likely in response to the upregulation of genes on the Z/X in the heterogametic sex) resulting in transcriptional inactivation on non-homologous Zs and Xs in distantly related species.
Results
We examined transcription in individual fibroblasts, using RNA-FISH. As probes, we chose bacterial artificial chromosome (BAC) clones containing known genes on the Z chromosome in a representative bird (chicken), and on the X chromosomes in a representative monotreme (platypus). For both species there is a genome assembly available [31], [32].
Partial Inactivation of Z Genes in Chicken Cells
As controls, we chose ten BACs containing known autosomal genes, including a BAC containing GAPDH, which was used as a control in all experiments. In RNA-FISH experiments with each of the ten autosomal BACs, we observed two signals in at least 97% of nuclei in both male and female fibroblast cells (Table S1 and Figure S1). Thus, autosomal genes are generally transcribed from both alleles in cultured chicken fibroblasts.
We selected eleven Z-borne BACs, five of which contained a single gene, and six contained two or more genes (Table S1). We performed two-color RNA-FISH, with the autosomal control (GAPDH) and each test gene (Z-borne and autosomal), on male (ZZ) and female (ZW) fibroblasts (Table S1), and scored ≥100 cells for each hybridization. To control for polyploidy and accessibility of the probe into an individual nucleus, only nuclei with two signals from the control BAC were scored for the test BAC.
The frequency with which expression of the single Z in ZW females was detected as a single signal, controlled for the hybridization efficiency of each Z probe. All Z BAC probes displayed hybridization efficiencies of between 95–100% (Table S1). Even at the lowest hybridization efficiency of 95% (p = 0.95; q = 0.05), hybridization in ZZ male cells would produce few nuclei with 0 (q2 = 0.25%) or 1 (2pq = 9.5%) signal (see Materials and Methods).
For all probes hybridized to ZZ male nuclei, the cell population consisted of a mixture of nuclei that were 1Z-active (one signal) and 2Z-active (two signals) (Table S1). No locus was completely 1Z-active or completely 2Z-active. Instead, each Z locus was inactivated in a characteristic frequency of cells, ranging from 15% to 51% (Figure 1), that was significantly different (p<0.01 after Bonferroni correction and estimating experimental error; see Materials and Methods) from the number of 1-Z active nuclei expected due to inefficient hybridization. This frequency was reproducible between biological replicates for a subset of BACs, in which RNA-FISH was repeated on fibroblasts from a second individual (Table S1). The activity status of Z loci in a given nucleus did not appear to be clonally inherited (i.e. if the mother cell was 2Z-active both daughter cells should be 2Z-active; or conversely if the mother cell was 1Z-active both daughter cells should be 1Z-active); we observed daughter cells in our preparations in which one was 1Z-active and the other 2Z-active (Figure S2).
Fig. 1. RNA-FISH activity maps of the platypus Xs and chicken Z chromosomes. 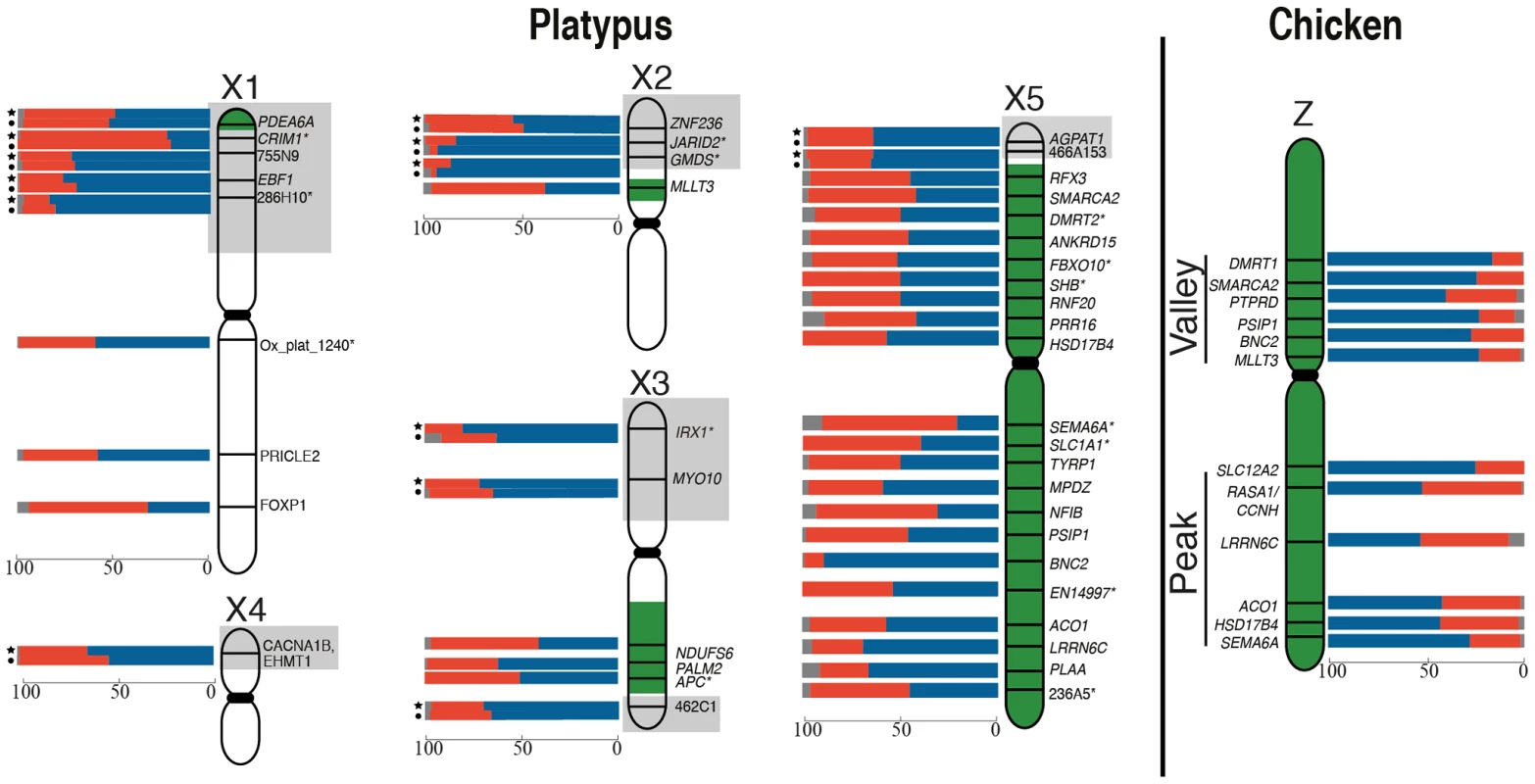
Bars represent the percentage of homogametic nuclei transcribing 2 (blue), 1 (red) or 0 (grey) alleles for each locus. Loci in pseudoautosomal regions (grey boxes) were tested in both male (indicated by a circle) and female (indicated by a star). Green coloring on platypus Xs represents homology to the chicken Z [23]. Platypus X chromosomes are not to scale (see Figure S5). Genes denoted by * were analysed in [24]. There was no obvious clustering of loci on the Z with particularly high or particularly low frequencies of 1Z-active cells, suggesting that the probability of transcriptional inhibition of a gene is independent of its physical location. The frequency of 1Z-active nuclei for the six BACs within the proposed dosage compensation ‘valley’ (129B9, 110A9, 164N4, 87K13, 57B13, and 89C2) was not lower than the five BACs in the ‘peak’ (65D18, 30H20, 73F14, 112C1, and 163I20) regions of the Z chromosome identified in chicken [28], [33]. However, there did appear to be a correlation of M∶F expression ratio (calculated from data in [34]; see Materials and Methods) of Z-genes, with the proportion of 1Z-active nuclei in males (R2 = 0.47, p = 0.03). If a gene was over expressed in males compared to females, there was a greater percentage of 1Z-active nuclei for that locus in males (Figure S3A).
From these initial RNA-FISH results it could not be determine if transcriptional inhibition of different loci were on the same Z chromosome, or on different Z chromosomes. Therefore we used two-color RNA-FISH to examine transcription of two pairs of neighbouring Z genes in female and male chicken nuclei: SMARCA2/PTPRD (∼2 Mb apart), and BNC2/MLLT3 (∼1.5 Mb apart) (Table 1) (Figure 2). In ZW female nuclei, we observed co-location (close proximity) of signals in 96% of nuclei that expressed both genes. In male nuclei, for each cell in which both loci were 1Z-active, we observed co-location of signals in 96% of nuclei (Figure 2A), indicating transcription from the same Z chromosome. This is consistent with the presence of an active Z (Za), and an inactive Z (Zi) on which genes are prone to silencing.
Fig. 2. Transcriptional activity of neighbouring chicken Z loci and platypus X5 loci in fibroblasts. 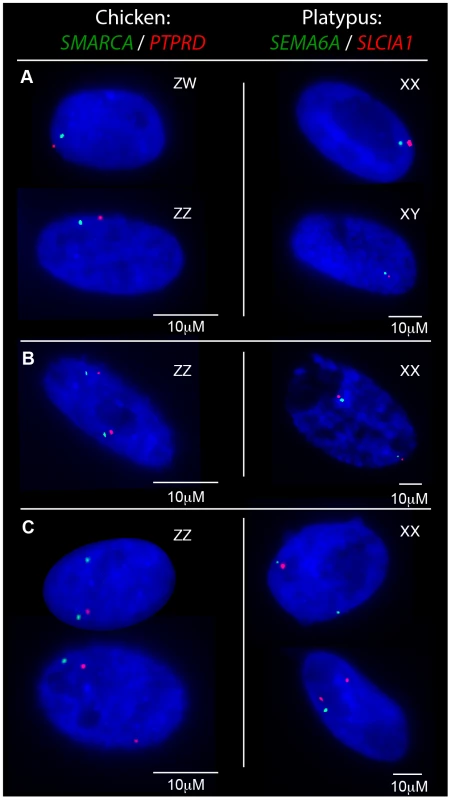
Gene names are color coded to correspond to signal color. A) In nuclei with only one allele active for both genes the signals co-locate in both sexes. B) Nuclei from the homogametic sex in which both genes are 2Z/2X-active. C) Nuclei from the homogametic sex in which the active Z/X expresses both genes and the other (inactivatable) Z/X expresses only one gene. Tab. 1. RNA-FISH analysis of transcription from neighbouring loci in homogametic nuclei. 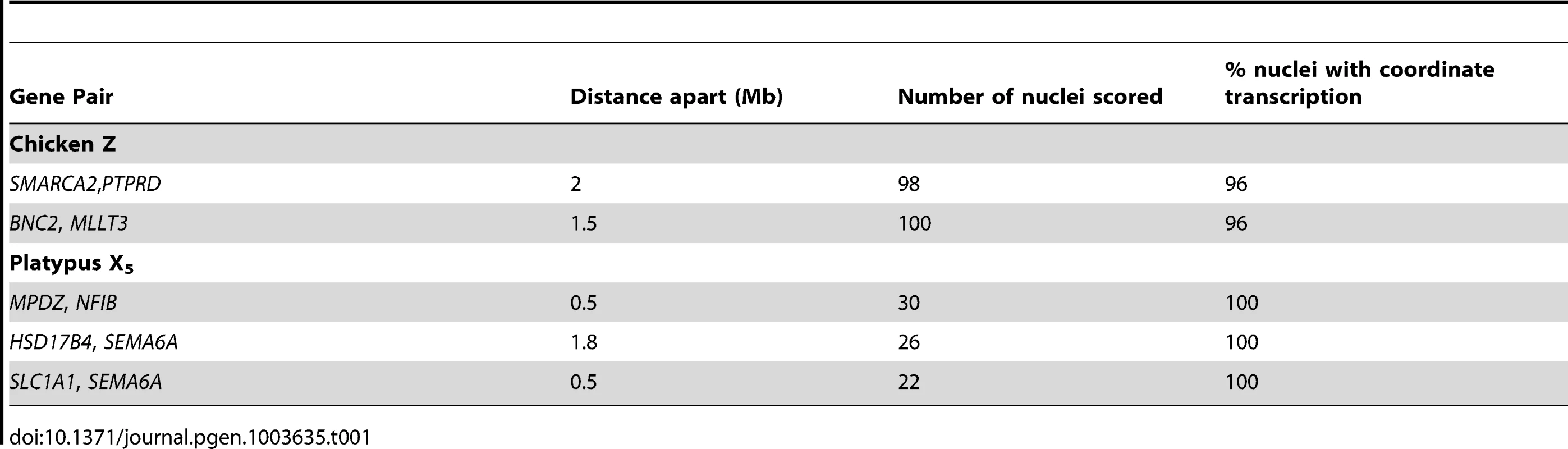
Two-color RNA-FISH experiments with the same gene pairs also provided the opportunity to determine whether the silencing of neighbouring genes on the Zi chromosome was coordinated. In cells in which at least one of the neighbouring gene pairs was 2Z-active, we observed (for both Z gene pairs) that the second locus was simultaneously transcribed (i.e. both loci were 2Z-active in the same nucleus; Figure 2B–C) at a frequency no greater than expected by chance (see Materials and Methods; Table 2), consistent with independent silencing of tightly linked genes on Zi. The frequency of nuclei, in which both loci in a gene pair are expected to be 2Z - active, was the product of 2Z-active frequencies of each gene (from initial RNA-FISH results; Table S1).
Tab. 2. Frequency of nuclei transcribing both alleles of neighbouring loci. 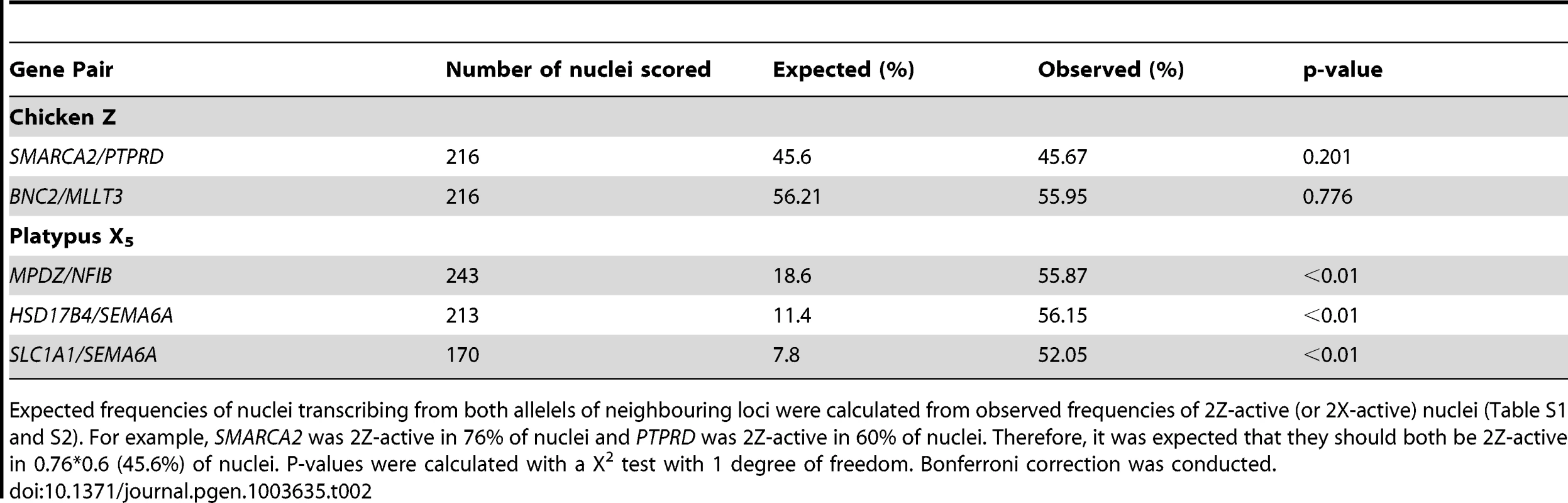
Expected frequencies of nuclei transcribing from both allelels of neighbouring loci were calculated from observed frequencies of 2Z-active (or 2X-active) nuclei (Table S1 and S2). For example, SMARCA2 was 2Z-active in 76% of nuclei and PTPRD was 2Z-active in 60% of nuclei. Therefore, it was expected that they should both be 2Z-active in 0.76*0.6 (45.6%) of nuclei. P-values were calculated with a X2 test with 1 degree of freedom. Bonferroni correction was conducted. Partial Inactivation of X Genes in Platypus Cells
A total of 40 platypus BACs were analyzed: 19 in X-specific regions (two on X1, one on X2, two on X3, 14 on X5), nine in pseudoautosomal regions (PAR), and 12 on autosomes. The autosomal BAC bearing HPRT (on chromosome 6) was used as the autosomal control in all experiments. Hybridization efficiencies for each probe were assessed in male cells. These ranged from 94% to 100% (Table S2). At least 100 cells were scored for each hybridization.
In RNA-FISH experiments with each of the 12 autosomal BACs, we observed two signals in at least 95% of nuclei in both male and female fibroblast cells (Table S2 and Figure S4). Thus autosomal genes are generally transcribed from both alleles in cultured platypus fibroblasts.
For all X specific loci we observed 1X-active female nuclei at a significantly greater frequency (p<0.01 after Bonferroni correction and estimating experimental error; see Materials and Methods) than expected from inefficient hybridization. No loci were completely 1X-active, or 2X-active in every nucleus, and frequencies of 1X-active nuclei ranged from 25% to 62% for different loci. There was no obvious clustering of genes on any X with particularly high, or particularly low frequencies of 1X-active nuclei (Figure 1). However, for loci that were expressed at a higher level in females compared to males, there did appear to be a correlation of F∶M expression ratio (in cultured fibroblast calculated from data in [21]; see Materials and Methods) of X-genes with the proportion of 1X-active nuclei in females (R2 = 0.54, p = 0.004). If a gene was over expressed in females compared to males, there was a greater percentage of 1X-active nuclei for that locus in females (Figure S3B).
The activity status of X loci in a given nucleus was not strictly clonally inherited (i.e. if the mother cell is 2× active both daughter cell should be 2× active; or conversely if the mother cell is 1× active both daughter cell should be 1× active); we observed daughter cells in which one was 1X-active and the other 2X-active (Figure S2). We therefore conclude that genes on the platypus X chromosomes are subject to inactivation in a proportion of nuclei that is characteristic for each gene.
To determine if transcriptional inhibition of adjacent loci was coordinated, we used two-color RNA-FISH to examine transcription of three pairs of neighboring genes on platypus X5: MPDZ/NFIB (∼500 kb apart), HSD17B4/SEMA6A (∼1.8 Mb apart) and SEMA6A/SLC1A1 (∼500 kb apart) (Table 1) (Figure 2A–C). As expected, in male controls we observed co-location of signals in 100% of nuclei for each gene pair. In female nuclei in which both loci were 1X-active, we also observed co-location of signals in 100% of nuclei for each gene pair tested (Figure 2A), consistent with the presence of a single active X5 (X5a), and an X5 (X5i) on which genes are prone to silencing.
Subsequently, we tested whether inactivation of adjacent loci was synchronized on X5i for these same neighbouring gene pairs by observing, in cells with one gene 2X-active, the frequency at which the second gene was 2X-active. We observed 2X-active nuclei at frequencies that were significantly (p<0.01) greater than expected by chance (Table 2, Figure 2; see Materials and Methods), indicating at least regional coordination of transcription on X5i. It was not possible to assess coordinated transcription over extended regions because FISH signals derived from X5a could not be distinguished from signals derived from X5i (see Materials and Methods).
Using RNA-FISH we also examined, in both male and female fibroblasts, transcription from genes located in the PAR of platypus sex chromosomes. We studied one locus in each of PARs X2/Y2, X3/Y2, X3/Y3 and X4/Y3, two loci in X5/Y4, and three loci in X1/Y1 (Figure S5). We expected to see nearly 100% 2X - active nuclei in both males and females, as for loci in the human PAR1, and the autosomal controls. However, all nine pseudoautosomal BACs tested were 1× active in 27–42% of male nuclei, and 16–47% of female nuclei, indicating significant (p<0.01 after Bonferroni correction) PAR gene inactivation in both sexes for seven of the nine loci tested (Table S2, Figure 1).
Importantly, two chicken autosomal BACs, orthologous to the platypus PAR genes that are subject to partial silencing, were 2-allele-active. Additionally, we demonstrated that four of the partially inactivated chicken Z/platypus X5 loci tested here are always 2 allele-active in human fibroblasts (Table S3), where they are autosomal. Finally, two of the biallelically expressed autosomal chicken BACs (Table S1), and four of the biallelically expressed autosomal platypus BACs (Table S2), tested here contained genes orthologous to X genes in human where they are subject to silencing. These experiments demonstrate that inactivation was dependent on the type of chromosome a locus was located on (i.e. sex chromosome or autosome), rather than it being a phenomenon unique to the loci examined here.
Discussion
Partial Sex Chromosome Inactivation
Using RNA-FISH, we demonstrate for the first time that Z inactivation plays a role in chicken dosage compensation, and confirmed that inactivation is also a feature of orthologous platypus X loci, as was previously shown by Deakin et al. [24].
Our observations conflict with those of a previous study of five chicken Z loci using RNA-FISH. Kuroda et al. [35] reported that most male nuclei expressed both Z alleles in five different tissue types (two Z loci were tested in liver, and one each in kidney, spleen and retina), and suggested that there was no inactivation on the chicken Z chromosome. The inconsistency in findings with this study may be due to the different tissue types used, and different inactivation profiles of chicken Z genes in other tissue types would not be surprising. We used fibroblast cells because they are easily collected and cultured from species that are difficult to sample, and to be consistent with our previous studies of sex chromosome silencing in mammals [18], [19], [24].
Additionally, Kuroda et al. [35] concluded that inactivation does not occur on the chicken Z because the frequency of detecting two signals of hybridization was similar for the autosomal probe and the Z probes (70–80%). However, the detection frequencies (1 or 2 signals observed) in male nuclei for the autosomal and Z probes were low (which were 66–77%, and were not co-hybridized). In this study we co-hybridized our autosomal control and test genes using one cell type, and only cells with two autosomal signals were scored for the test gene, controlling for ploidy and probe accessibility into individual nuclei. As a result we obtained much higher hybridization efficiencies, providing for much simpler interpretation.
An exhaustive study of male-female expression ratios using microarrays showed that different loci on the chicken Z are compensated to different extents [26], [30], and Julien et al. [21] eloquently demonstrated that the Z/X5 is upregulated in the heterogametic sex in chicken/platypus. These studies assessed transcription across whole cell populations, so could not distinguish between partial expression in all nuclei, and heterogeneity of expression of different cells. In male chicken fibroblast nuclei, we found for all loci a mixture of 1Z-active and 2Z active cells at reproducible frequencies (15% to 51% 1Z-active, depending on the locus). Therefore, we demonstrate that in the homogametic sex there is partial inactivation of one Z allele.
As such, these results demonstrate that an integral step of the partial bird dosage compensation system must include the silencing of one Z allele in males, analogous to the inactivation we observed here (and previously [24]) on the orthologous (although independently evolved) platypus X5, and the independently evolved therian mammal X chromosome. We also observe that loci over expressed in the homogametic sex were more likely to be 1Z/X-active, indicating that there is pressure to reduce expression of these genes in the homogametic sex. We conclude that partial inactivation on the Z in birds, and at least on four of the five X chromosomes in platypus, is likely in response to Z/X upregulation (perhaps of only several dosage sensitive genes) carrying through to the homogametic sex. Because higher transcript levels in the homogametic sex appear to correlate with a greater percentage of 1Z/X-active nuclei, evolution to down regulate the chicken Z (and platypus Xs) is conceivably an ongoing process. Were everything in equilibrium 1Z/1X-activity should correlate with a 1∶1 transcript ratio between the sexes.
Distribution of Inactivation over the Sex Chromosome
We found no correlation between the extent of inactivation and the location of the gene on the chicken Z (Figure 1), and did not observe stronger inactivation of loci in the dosage compensation valley, which was detected on the chicken Z chromosome using microarrays [26], [28], [33], and is differentially methylated on the two Z chromosomes in males [36]. The functional significance of this dosage compensation valley is unclear because it is not conserved in zebra finch [28]. Other studies of chicken concluded that the dosage compensated valley region, concentrated in genes with low male to female expression ratios, simply contains genes with a strong female bias [30], [37], although this interpretation has also been challenged [38], [39]. It is important to note that our data does not necessarily conflict with original reports on the dosage compensation valley region [33], in which it was suggested that females up-regulate genes, rather than males down-regulating genes.
We also observed no clustering of platypus X-borne genes with very low or very high frequencies of 1X-active nuclei (Figure 1). This implies that the probability of X inactivation is not correlated with gene location, and provides no evidence for an X chromosome inactivation center from which inactivation might spread.
Coordinate Control of Sex Chromosome Activity
We observed that adjacent genes were always expressed from the same Za chromosome in male chicken cells. This is consistent with a hypothesis that all loci on one Z chromosome are active (Za), and loci on the other Z (Zi) are prone to inactivation in a proportion of cells. We found that the probabilities of expression of adjacent loci on the Zi were not correlated, suggesting that escape from inactivation of each locus on Zi is independently regulated.
In female platypus cells, our analysis of three platypus X5 pairs showed co-location of signals for neighbouring loci (Table 2; Figure 2A), consistent with coordinate activity on one X5 (X5a) and inactivation of loci on the other X5 (X5i) in a proportion of cells in females. Partial activity of genes on Xi is consistent with the previous demonstration that both alleles were transcribed [24]. Unlike chicken, neighbouring genes on platypus X5 showed concordant 2X-activity (Figure 2B), implying that the regulation of transcription from X5i is at least under regional control.
We suggest, therefore, that in cells from male chickens there is one active Za, and one partially inactive (“inactivatable”) Zi on which loci have specific probabilities of being inactivated. Likewise, we propose that in cells from female platypus, there is one active Xa and one partially inactive (“inactivatable”) Xi on which loci have specific probabilities of being inactivated. Without allelic markers that would distinguish paternal and maternal Z/X chromosomes, we cannot determine whether silencing is random or imprinted. Allelic differences were used to demonstrate that both alleles are expressed, apparently at equivalent levels, in female platypus heterozygous for FBX010, GLIS3, and SHB [24]. These loci are all expressed from Xi in about half the cells, so the approximately equal representation of the two alleles might favor a random inactivation hypothesis; however peak intensity on sequencing trace files is at best semi-quantitative, so this question remains open.
Ancient Gene Silencing and the Evolution of XCI
Because platypus sex chromosomes share homology with the chicken Z chromosome, our observation of inactivation in a proportion of cells for genes on the sex chromosomes in the homogametic sex of a representative bird (ZZ male chicken) and monotreme (XX female platypus) could be interpreted as the evolution of a silencing mechanism on an ancient bird-like Z in a common reptile-mammal ancestor. However, it is uncertain whether the bird Z/platypus X homology represents identity by descent (reviewed in [40]–[42]), but under either scenario the sex chromosome inactivation observed here is likely independently evolved [43].
Similar patterns of 1X gene expression in a proportion of cells have been observed in marsupials (tammar wallaby) [18] and escaper genes on the X in eutherians (elephant, mouse and human) [19]. The marsupial and eutherian X shares a large conserved region, which is completely non-homologous with the sex chromosomes of birds and monotremes. Thus, the probabilistic inactivation we observed in birds, monotremes and therian mammals was independently exapted from an ancient toolkit of mechanisms to turn off transcription of one allele. Indeed, this partial silencing system shares characteristics with monoallelic expression of some autosomal mammalian genes, such as interleukins (IL2, IL4, IL5, IL10, IL13) [44]–[48].
We propose that probabilistic silencing of the Z or X chromosome in the homogametic sex is an early step in the evolution of sex chromosome inactivation. A major difference in the inactivation systems is the extent of coordination. This is most evident for X inactivation in eutherian mammals, which is coordinately controlled by XIST, a locus that is absent outside eutherians [49]. In both chicken and platypus, the co-expression of neighbouring loci on the Za/X5a implies that the choice of which Zi/Xi to inactivate is controlled at least at the regional level, and possibly at the level of the whole chromosome, and in platypus (but not chicken) the probability of expression of neighbouring loci on X5i is at least regionally coordinated.
In eutherian mammals silencing was augmented by additional molecular changes (including histone modification and DNA methylation), into the stable and complex inactivation system typical of most genes on the conserved region of the eutherian X (XCR), which shows tight 1X-active expression observed at the single cell, as well as the population level [9], [19]. In contrast, the evolutionarily younger X added region of the human X (XAR) contains many escaper genes that show a probabilistic expression pattern similar to that of birds and monotremes. The marsupial X, though homologous to the eutherian XCR, displays a pattern of expression similar to the eutherian XAR, monotreme Xs and bird Z, suggesting that changes to the regulation of the eutherian XCR occurred after the divergence of marsupials from eutherian mammals 150 million years ago. This is consistent with the different epigenetic profiles displayed by the marsupial X and the eutherian XCR [15], [20], [50].
Inactivation of Pseudoautosomal Genes
PAR inactivation has intriguing implications for sex chromosome evolution. The accepted hypothesis for the evolution of sex chromosomes and dosage compensation is that Y degeneration resulted in loss of Y gene function, which in turn drove the evolution of dosage compensation (resulting in XCI in therian mammals) [1], [51]. However, an alternate hypothesis is that the spread of the XCI signal into undifferentiated X regions preceded, and drove, degradation of homologous Y regions [52].
Genes in the human and mouse PAR are exempt from X-inactivation because the Y copy complements the X [53]. We therefore expected X and Y alleles of platypus PAR genes to be active in all nuclei in both sexes, as they are for genes in the human PAR1. Surprisingly, we observed a significantly lower frequency of 2X-active cells in both sexes for seven of the nine PAR loci. This confirms the observation [24] that other PAR loci were 1-allele active in platypus, and implies that inactivation of PAR genes regularly occurs in platypus. This contrasts with the expression of all autosome loci from both alleles in nearly all cells.
In females, inactivation of genes on the X PAR might be achieved via spreading of inactivation from the X-specific regions, as has been observed in the recently evolved human PAR 2 [54], [55]. However, PAR inactivation in males cannot be the result of XCI spreading, instead Y PARs could be inactivated by their proximity to heterochromatin on the male-specific region of the Y. It is possible that upregulation of the active X chromosomes in platypus, which maintained parity with autosomal genes, also resulted in upregulation of genes in the PARs. To mitigate this there was selection for partial inactivation of Xi PAR genes in females, and Y PAR genes in males.
In conclusion, our studies of dosage compensation on independently evolved amniote sex chromosomes reveals common patterns of inactivation. This suggests that repressive molecular mechanisms were independently exapted to reduce the probability of transcription from one Z or X chromosome in the homogametic sex, in response to upregulation (perhaps of only a subset genes) in the heterogametic sex.
Materials and Methods
Ethics Statement
The study was approved, and all samples were collected and held under The Australian National University Animal Experimentation Ethics Committee proposal numbers R.CG.11.06 and R.CG.14.08.
Identification of BAC Gene Content
Genes were chosen based on three criteria: 1) Widespread expression in other species to maximize the chance they were expressed in platypus and chicken fibroblast cells. 2) A BAC was preferentially chosen if it contained a single gene. 3) Loci were selected that were distributed at roughly even intervals along chicken Z and platypus X5. On some of the platypus X chromosomes loci selection was limited by the paucity of anchored BACs. BACs containing chicken and platypus genes were ordered from CHORI BACPAC Resources Centre (http://bacpac.chori.org/). Chicken BACs bearing genes of interest were chosen using the UCSC genome browser BAC track (http://genome.ucsc.edu/cgi-bin/hgGateway). Platypus BACs were from Veyrunes et al. [23]. Additional platypus BACs were identified by blasting the BAC end trace archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi).
Cell Culture and RNA-FISH on Chicken and Platypus Interphase Nuclei
Chicken fibroblast cell lines were established from 8-day-old chicks; metaphase spreads were karyotypically normal. Platypus fibroblast cell lines were established from adult wild animals that were karyotypically normal.
Platypus and chicken fibroblast cells were cultured at 32°C and 37°C respectively in 5.0% CO2 on gelatin-coated coverslips in 1∶1 AmnioMax C100 medium (Invitrogen)/DMEM 10% FCS to a density of 60 to 80%. RNA-FISH was carried out as previously described [18]. Hybridization of the probe to homologous DNA will not occur because there is no DNA denaturation step in the RNA-FISH protocol.
RNA-FISH Scoring and Statistical Analysis
For chicken RNA-FISH experiments a BAC from chromosome 1, which contained the gene GAPDH (CH261-14L1), was used as the autosomal control. For platypus RNA-FISH experiments a BAC from chromosome 6, which contained HPRT1 (OaBb_405M2; GenBank Accession No. AC148426), was the autosomal control. Only diploid nuclei (two signals from the autosomal BAC) were scored for the test gene (X/Z or pseudoautosomal).
For each test gene, 1X - or 1Z-active nuclei were observed as one signal, and 2X - or 2Z-active nuclei were observed as two signals. Hybridization efficiencies (p) were obtained from results in the heterogametic sex (in which one signal is expected in all nuclei). These were used to calculate the expected frequency of nuclei with two signals, one signal, and no signals in the homogametic sex using the formula p2+2pq+q2 = 1; where p2 is the expected frequency of nuclei with two signals, 2pq (q = 1–p) is the frequency of nuclei with one signal, and q2 is the frequency with no signal. P-values were calculated using a χ2 test with two degrees of freedom and Bonferroni correction was conducted.
For a more rigorous statistical analysis we removed no signal cells from the dataset, and calculated experimental error using missed hybridization events in our autosomal RNA-FISH experiments. In chicken we conducted 16 autosomal RNA-FISH experiments and scored a total of 1756 nuclei, each of which should produce 2 signals, for a total of 3512 signals. We observed a total of 12 nuclei with no signals and 16 nuclei with one signal, which is equal to 40 missed hybridisations out of 3512 (1.14%). Following a Poisson distribution, 95% confidence limits for 40 events gives a minimum of 28.58 and a maximum of 54.47. The upper value of the 95% confidence limits represents 1.55% experimental error (i.e. 54.47 out of 3512). For platypus, using exactly the same approach (18 autosomal RNA-FISH experiments, 1884 nuclei scored in total, 3768 expected signals, 69 missed hybridisations, upper 95% confidence interval of 87.32), maximum experimental error was estimated at 2.32%. All observed values were adjusted towards the expected values by 1.55% and 2.32% in chicken and platypus respectively. P-values were recalculated using a χ2 test with one degree of freedom. Significance remained (after Bonferroni correction) for all BACs. Taking into account an arbitrary conservative experimental error of 10% (±5%), and adjusting all the observed values by 5% towards the expected values still results in significant p-values.
Transcript Abundance
Expression values were obtained for known genes on each BAC in chicken and platypus (from [21], [34]), and expression ratios calculated. If a BAC carried more than one gene, expression data from the gene that spanned the largest proportion of the BAC was used (DMRT1/3 expression data was not included due to its involvement in sex determination). For chicken expression data was available for brain, cerebellum, heart, kidney and liver. Because no data were available for cultured fibroblasts, an average M∶F expression ratio was taken across all tissues to best control for tissue specific expression changes. For platypus, data were available for cultured fibroblast cells, which was used to calculate F∶M expression ratios because the present study was conducted in cultured fibroblasts.
These expression ratios were plotted against the percentage of 1Z/1X-active nuclei observed for the relevant BAC. If an RNA-FISH experiment was repeated for a BAC, an average observed percentage of 1Z/X activity was used. Scatterplots were drawn (Figure S3), and R2 and p-values were calculated in Microsoft Excel.
Scoring Coordinated Transcription/Transcription Inhibition of Adjacent Genes
RNA-FISH signals co-locate in the nucleus when closely linked and transcribed from the same chromosome, whereas signals from genes transcribed from different chromosomes (or distantly linked on the same chromosome) are further apart. We only used pairs of genes physically close enough to each other to give unmistakable results.
To determine if the there was a single active Z (or X for platypus), all nuclei that were 1Z (or 1X) active for both loci in a pair were scored. Co-location of the two signals was interpreted as a single active Z (or X) in that nucleus. The expected frequency of nuclei that were 2Z (or 2X) active for both loci in a gene pair was the product of individual 2Z (or 2X) frequencies of each gene (from initial RNA-FISH results). To determine if inactivation of neighbouring gene pairs was coordinated, this frequency was compared to the observed frequency of nuclei that were 2Z (or 2X) active for both loci. P-values were calculated using a χ2 test with one degree of freedom and Bonferroni correction was conducted.
Supporting Information
Zdroje
1. Ohno S (1967) Sex chromosomes and sex-linked genes. New York: Springer-Verlag.
2. HeardE (2004) Recent advances in X-chromosome inactivation. Curr Opin Cell Biol 16 : 247–255.
3. XiongY, ChenX, ChenZ, WangX, ShiS, et al. (2010) RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet 42 : 1043–1047.
4. LinF, XingK, ZhangJ, HeX (2012) Expression reduction in mammalian X chromosome evolution refutes Ohno's hypothesis of dosage compensation. Proc Natl Acad Sci U S A 109 : 11752–11757.
5. DengX, HiattJB, NguyenDK, ErcanS, SturgillD, et al. (2011) Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet 43 : 1179–1185.
6. LinH, HalsallJA, AntczakP, O'NeillLP, FalcianiF, et al. (2011) Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno's hypothesis. Nat Genet 43 : 1169–1170; author reply 1171–1162.
7. KharchenkoPV, XiR, ParkPJ (2011) Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet 43 : 1167–1169; author reply 1171–1162.
8. PessiaE, MakinoT, Bailly-BechetM, McLysaghtA, MaraisGA (2012) Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A 109 : 5346–5351.
9. CarrelL, WillardHF (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434 : 400–404.
10. BrownCJ, BallabioA, RupertJL, LafreniereRG, GrompeM, et al. (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349 : 38–44.
11. BallabioA, WillardHF (1992) Mammalian X-chromosome inactivation and the XIST gene. Curr Opin Genet Dev 2 : 439–447.
12. BrockdorffN, AshworthA, KayGF, McCabeVM, NorrisDP, et al. (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71 : 515–526.
13. OkamotoI, PatratC, ThepotD, PeynotN, FauqueP, et al. (2011) Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472 : 370–374.
14. HeardE (2005) Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr Opin Genet Dev 15 : 482–489.
15. ChaumeilJ, WatersPD, KoinaE, GilbertC, RobinsonTJ, et al. (2011) Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: epigenetic modifications in distantly related mammals. PLoS One 6: e19040.
16. SharmanGB (1971) Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature 230 : 231–232.
17. CooperD, JohnstonPG, GravesJAM (1993) X-inactivation in marsupials and monotremes. Semin Cell Dev Biol 4 : 117–128.
18. Al NadafS, WatersPD, KoinaE, DeakinJE, JordanKS, et al. (2010) Activity map of the tammar X chrmosome shos that marsupial S inactivation is incomplete and escape is stochastic. Genome Biology 11: R122.
19. Al NadafS, DeakinJE, GilbertC, RobinsonTJ, GravesJA, et al. (2012) A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosoma 121 : 71–78.
20. MahadevaiahSK, RoyoH, VandeBergJL, McCarreyJR, MackayS, et al. (2009) Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol 19 : 1478–1484.
21. JulienP, BrawandD, SoumillonM, NecsuleaA, LiechtiA, et al. (2012) Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol 10: e1001328.
22. RensW, O'BrienPC, GrutznerF, ClarkeO, GraphodatskayaD, et al. (2007) The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol 8: R243.
23. VeyrunesF, WatersPD, MiethkeP, RensW, McMillanD, et al. (2008) Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res 18 : 965–973.
24. DeakinJE, HoreTA, KoinaE, GravesJA (2008) The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet 4: e1000140.
25. McQueenHA, McBrideD, MieleG, BirdAP, ClintonM (2001) Dosage compensation in birds. Curr Biol 11 : 253–257.
26. ItohY, MelamedE, YangX, KampfK, WangS, et al. (2007) Dosage compensation is less effective in birds than in mammals. J Biol 6 : 2.
27. EllegrenH, Hultin-RosenbergL, BrunstromB, DenckerL, KultimaK, et al. (2007) Faced with inequality: chickens do not have a general dosage compensation of sex-linked genes. BMC Biol 5 : 40.
28. ItohY, ReplogleK, KimYH, WadeJ, ClaytonDF, et al. (2010) Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res 20 : 512–518.
29. WolfJB, BrykJ (2011) General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics 12 : 91.
30. MankJE, EllegrenH (2009) All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity 102 : 312–320.
31. WallisJW, AertsJ, GroenenMA, CrooijmansRP, LaymanD, et al. (2004) A physical map of the chicken genome. Nature 432 : 761–764.
32. WarrenWC, HillierLW, Marshall GravesJAM, BirneyE, PontingCP, et al. (2008) Genome analysis of the platypus reveals unique signatures of evolution. Nature 453 : 175–183.
33. MelamedE, ArnoldAP (2007) Regional differences in dosage compensation on the chicken Z chromosome. Genome Biol 8: R202.
34. BrawandD, SoumillonM, NecsuleaA, JulienP, CsardiG, et al. (2011) The evolution of gene expression levels in mammalian organs. Nature 478 : 343–348.
35. KurodaY, AraiN, AritaM, TeranishiM, HoriT, et al. (2001) Absence of Z-chromosome inactivation for five genes in male chickens. Chromosome Res 9 : 457–468.
36. ItohY, KampfK, ArnoldAP (2011) Possible differences in the two Z chromosomes in male chickens and evolution of MHM sequences in Galliformes. Chromosoma 120 : 587–598.
37. MankJE, EllegrenH (2009) Sex bias in gene expression is not the same as dosage compensation. Heredity (Edinb) 103 : 434.
38. MelamedE, ElashoffD, ArnoldAP (2009) Evaluating dosage compensation on the chicken Z chromosome: should effective dosage compensation eliminate sexual bias? Heredity 103 : 357–359.
39. ZhangSO, MathurS, HattemG, TassyO, PourquieO (2010) Sex-dimorphic gene expression and ineffective dosage compensation of Z-linked genes in gastrulating chicken embrios. BMC Genomics 11 : 13.
40. Marshall GravesJA, PeichelCL (2010) Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol 11 : 205.
41. WatersPD, Marshall GravesJA (2009) Monotreme sex chromosomes–implications for the evolution of amniote sex chromosomes. Reprod Fertil Dev 21 : 943–951.
42. O'MeallyD, EzazT, GeorgesA, SarreSD, GravesJA (2012) Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res 20 : 7–19.
43. LivernoisAM, GravesJA, WatersPD (2012) The origin and evolution of vertebrate sex chromosomes and dosage compensation. Heredity (Edinb) 108 : 50–58.
44. BixM, LocksleyRM (1998) Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science 281 : 1352–1354.
45. CaladoDP, PaixaoT, HolmbergD, HauryM (2006) Stochastic monoallelic expression of IL-10 in T cells. J Immunol 177 : 5358–5364.
46. HollanderGA, ZuklysS, MorelC, MizoguchiE, MobissonK, et al. (1998) Monoallelic expression of the interleukin-2 locus. Science 279 : 2118–2121.
47. KellyBL, LocksleyRM (2000) Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J Immunol 165 : 2982–2986.
48. RiviereI, SunshineMJ, LittmanDR (1998) Regulation of IL-4 expression by activation of individual alleles. Immunity 9 : 217–228.
49. DuretL, ChureauC, SamainS, WeissenbachJ, AvnerP (2006) The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312 : 1653–1655.
50. RensW, WallduckMS, LovellFL, Ferguson-SmithMA, Ferguson-SmithAC (2010) Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci U S A 107 : 17657–17662.
51. MarinI, SiegalML, BakerBS (2000) The evolution of dosage-compensation mechanisms. Bioessays 22 : 1106–1114.
52. GravesJA, SchmidtMM (1992) Mammalian sex chromosomes: design or accident? Curr Opin Genet Dev 2 : 890–901.
53. MoreyC, AvnerP (2011) The demoiselle of X-inactivation: 50 years old and as trendy and mesmerising as ever. PLoS Genet 7: e1002212.
54. D'EspositoM, CiccodicolaA, GianfrancescoF, EspositoT, FlagielloL, et al. (1996) A synaptobrevin-like gene in the Xq28 pseudoautosomal region undergoes X inactivation. Nat Genet 13 : 227–229.
55. CiccodicolaA, D'EspositoM, EspositoT, GianfrancescoF, MigliaccioC, et al. (2000) Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet 9 : 395–401.
Štítky
Genetika Reprodukční medicína
Článek The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and DevelopmentČlánek Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative ElementsČlánek Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine ExposureČlánek Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 7- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- An Solution for Crossover Formation
- Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, , as a Genetic Component of Neural Tube Defects in Humans
- Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
- Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
- Genomic Analysis of Natural Selection and Phenotypic Variation in High-Altitude Mongolians
- Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
- Role of CTCF Protein in Regulating Locus Transcription
- Gene Set Signature of Reversal Reaction Type I in Leprosy Patients
- Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups
- Is Required for Formation of the Genital Ridge in Mice
- Monopolin Subunit Csm1 Associates with MIND Complex to Establish Monopolar Attachment of Sister Kinetochores at Meiosis I
- Recombination Dynamics of a Human Y-Chromosomal Palindrome: Rapid GC-Biased Gene Conversion, Multi-kilobase Conversion Tracts, and Rare Inversions
- Mechanisms of Protein Sequence Divergence and Incompatibility
- Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack
- Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies
- Combinatorial Regulation of Meiotic Holliday Junction Resolution in by HIM-6 (BLM) Helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 Nucleases
- The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
- The Role of Interruptions in polyQ in the Pathology of SCA1
- Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in
- Fine Time Course Expression Analysis Identifies Cascades of Activation and Repression and Maps a Putative Regulator of Mammalian Sex Determination
- Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90
- Oxidative Stress and Replication-Independent DNA Breakage Induced by Arsenic in
- A Moonlighting Enzyme Links Cell Size with Central Metabolism
- Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
- The Conserved Intronic Cleavage and Polyadenylation Site of CstF-77 Gene Imparts Control of 3′ End Processing Activity through Feedback Autoregulation and by U1 snRNP
- The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in through Maintaining a Progenitor-like Cell State
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- The RNA-binding Proteins FMR1, Rasputin and Caprin Act Together with the UBA Protein Lingerer to Restrict Tissue Growth in
- Pattern Dynamics in Adaxial-Abaxial Specific Gene Expression Are Modulated by a Plastid Retrograde Signal during Leaf Development
- A Network of HMG-box Transcription Factors Regulates Sexual Cycle in the Fungus
- Bacterial Adaptation through Loss of Function
- ENU-induced Mutation in the DNA-binding Domain of KLF3 Reveals Important Roles for KLF3 in Cardiovascular Development and Function in Mice
- Interplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
- FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription
- The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression
- Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative Elements
- The Conserved ADAMTS-like Protein Lonely heart Mediates Matrix Formation and Cardiac Tissue Integrity
- The cGMP-Dependent Protein Kinase EGL-4 Regulates Nociceptive Behavioral Sensitivity
- RBM5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility
- Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of Expression at the 6q23.2 Coronary Heart Disease Locus
- Yeast Pol4 Promotes Tel1-Regulated Chromosomal Translocations
- A Dual Role for SOX10 in the Maintenance of the Postnatal Melanocyte Lineage and the Differentiation of Melanocyte Stem Cell Progenitors
- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle,
- Prediction of Complex Human Traits Using the Genomic Best Linear Unbiased Predictor
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
- Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine Exposure
- Exquisite Light Sensitivity of Cryptochrome
- miR-133a Regulates Adipocyte Browning In Vivo
- Strabismus Promotes Recruitment and Degradation of Farnesylated Prickle in Planar Polarity Specification
- Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
- Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
- Joint Molecule Resolution Requires the Redundant Activities of MUS-81 and XPF-1 during Meiosis
- The Mating Competence of Geographically Diverse Strains in Their Natural and Unnatural Sand Fly Vectors
- Defective Repair of Oxidative Base Lesions by the DNA Glycosylase Nth1 Associates with Multiple Telomere Defects
- Effective Blocking of the Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function
- Trans-Ancestral Studies Fine Map the SLE-Susceptibility Locus
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Bacterial Adaptation through Loss of Function
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



