-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
Phospho - and sphingolipids are crucial cellular and intracellular compounds. These lipids are required for active transport, a number of enzymatic processes, membrane formation, and cell signalling. Disruption of their metabolism leads to several diseases, with diverse neurological, psychiatric, and metabolic consequences. A large number of phospholipid and sphingolipid species can be detected and measured in human plasma. We conducted a meta-analysis of five European family-based genome-wide association studies (N = 4034) on plasma levels of 24 sphingomyelins (SPM), 9 ceramides (CER), 57 phosphatidylcholines (PC), 20 lysophosphatidylcholines (LPC), 27 phosphatidylethanolamines (PE), and 16 PE-based plasmalogens (PLPE), as well as their proportions in each major class. This effort yielded 25 genome-wide significant loci for phospholipids (smallest P-value = 9.88×10−204) and 10 loci for sphingolipids (smallest P-value = 3.10×10−57). After a correction for multiple comparisons (P-value<2.2×10−9), we observed four novel loci significantly associated with phospholipids (PAQR9, AGPAT1, PKD2L1, PDXDC1) and two with sphingolipids (PLD2 and APOE) explaining up to 3.1% of the variance. Further analysis of the top findings with respect to within class molar proportions uncovered three additional loci for phospholipids (PNLIPRP2, PCDH20, and ABDH3) suggesting their involvement in either fatty acid elongation/saturation processes or fatty acid specific turnover mechanisms. Among those, 14 loci (KCNH7, AGPAT1, PNLIPRP2, SYT9, FADS1-2-3, DLG2, APOA1, ELOVL2, CDK17, LIPC, PDXDC1, PLD2, LASS4, and APOE) mapped into the glycerophospholipid and 12 loci (ILKAP, ITGA9, AGPAT1, FADS1-2-3, APOA1, PCDH20, LIPC, PDXDC1, SGPP1, APOE, LASS4, and PLD2) to the sphingolipid pathways. In large meta-analyses, associations between FADS1-2-3 and carotid intima media thickness, AGPAT1 and type 2 diabetes, and APOA1 and coronary artery disease were observed. In conclusion, our study identified nine novel phospho - and sphingolipid loci, substantially increasing our knowledge of the genetic basis for these traits.
Published in the journal: . PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002490
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002490Summary
Phospho - and sphingolipids are crucial cellular and intracellular compounds. These lipids are required for active transport, a number of enzymatic processes, membrane formation, and cell signalling. Disruption of their metabolism leads to several diseases, with diverse neurological, psychiatric, and metabolic consequences. A large number of phospholipid and sphingolipid species can be detected and measured in human plasma. We conducted a meta-analysis of five European family-based genome-wide association studies (N = 4034) on plasma levels of 24 sphingomyelins (SPM), 9 ceramides (CER), 57 phosphatidylcholines (PC), 20 lysophosphatidylcholines (LPC), 27 phosphatidylethanolamines (PE), and 16 PE-based plasmalogens (PLPE), as well as their proportions in each major class. This effort yielded 25 genome-wide significant loci for phospholipids (smallest P-value = 9.88×10−204) and 10 loci for sphingolipids (smallest P-value = 3.10×10−57). After a correction for multiple comparisons (P-value<2.2×10−9), we observed four novel loci significantly associated with phospholipids (PAQR9, AGPAT1, PKD2L1, PDXDC1) and two with sphingolipids (PLD2 and APOE) explaining up to 3.1% of the variance. Further analysis of the top findings with respect to within class molar proportions uncovered three additional loci for phospholipids (PNLIPRP2, PCDH20, and ABDH3) suggesting their involvement in either fatty acid elongation/saturation processes or fatty acid specific turnover mechanisms. Among those, 14 loci (KCNH7, AGPAT1, PNLIPRP2, SYT9, FADS1-2-3, DLG2, APOA1, ELOVL2, CDK17, LIPC, PDXDC1, PLD2, LASS4, and APOE) mapped into the glycerophospholipid and 12 loci (ILKAP, ITGA9, AGPAT1, FADS1-2-3, APOA1, PCDH20, LIPC, PDXDC1, SGPP1, APOE, LASS4, and PLD2) to the sphingolipid pathways. In large meta-analyses, associations between FADS1-2-3 and carotid intima media thickness, AGPAT1 and type 2 diabetes, and APOA1 and coronary artery disease were observed. In conclusion, our study identified nine novel phospho - and sphingolipid loci, substantially increasing our knowledge of the genetic basis for these traits.
Introduction
Phospho - and sphingolipids are present in all eukaryotic cell membranes and contribute to organelle structure and signalling events that influence cell behaviour and function [1]–[3]. Phosphatidylcholines (PC), phosphatidylethanolamines (PE), lysophosphatidylcholines (LPC) and PE-based plasmalogens (PLPE) are major classes of phospholipids that play an important role in several key processes such as cell survival and inflammation [4]–[6]. Sphingolipids are also essential components of plasma membranes and endosomes and are believed to play critical roles in cell surface protection, protein and lipid transport and sorting, and cellular signalling cascades [7]. In plasma, PC, PE and sphingomyelin (SPM) are included in the structure of lipoproteins; they constitute more than two-thirds of the total phospholipid content in HDL-C and LDL-C, as well as in platelets [8], [9]. Remarkable differences in plasma lipoprotein acceptor affinities for the phospholipids exist (LDL-C is the major acceptor for SPM, whereas HDL-C is the predominant acceptor for PC) [9]. Altered concentrations of circulating phospholipids have been implicated in the pathology of type 2 diabetes, dyslipidemia and cardiovascular disease [10]–[15], as well as a wide range of other common diseases including dementia and depression [16].
Identifying genetic variants that influence phospho - and sphingolipid concentrations will be an important step towards understanding pathways contributing to common human disease. Earlier studies of these metabolites identified a number of genetic loci associated with their levels in blood [17]–[19]. We conducted a meta-analysis of genome-wide association studies (GWAS) on plasma levels of 24 SPMs, 9 ceramides (CER), 57 PCs, 20 LPCs, 27 PEs and 16 PLPEs in five European populations: (1) the Erasmus Rucphen Family (ERF) study, conducted in the Netherlands, (2) the MICROS study from the Tyrol region in Italy, (3) the Northern Swedish Population Health Survey (NSPHS) in Norrbotten, Sweden, (4) the Orkney Complex Disease Study (ORCADES) in Scotland, and (5) the CROAS (CROATIA_Vis) study conducted on Vis Island, Croatia.
The top findings were further analysed by adjusting for plasma HDL-C, LDL-C, TG and TC levels. The influences of these top hits on within class lipid ratios were also assessed, to help elucidate potential mechanisms. Finally, the variants that were associated with plasma phospho - and sphingolipid levels were tested for association with carotid intima media thickness (IMT), type 2 diabetes (T2DM), and coronary-artery disease (CAD) using large consortia meta-analysis results.
Results
Table 1 provides an overview of the study populations. The mean age, gender ratio and mean values of major classes of phospho - and sphingolipids were comparable among the 5 populations. Means for the individual species are presented in Table S1. Figure 1A and 1B shows the combined Manhattan plot for the meta-analyses of the absolute values and proportions of all phospholipid traits, respectively; Figure 2A and 2B provides the same for the sphingolipids. Out of 357 meta-analyses performed, 202 outcomes yielded genome-wide significant findings, most of which were located around two genes, FADS and LIPC, which were identified previously [17], [19] as key lipid regulators and are associated with a large number of species (Table 2 and Table 3). Q-Q plots for the lipid GWAS that yielded significant associations are provided in Figure S1.
Fig. 1. Genome-wide association results for 115 phospholipid species. 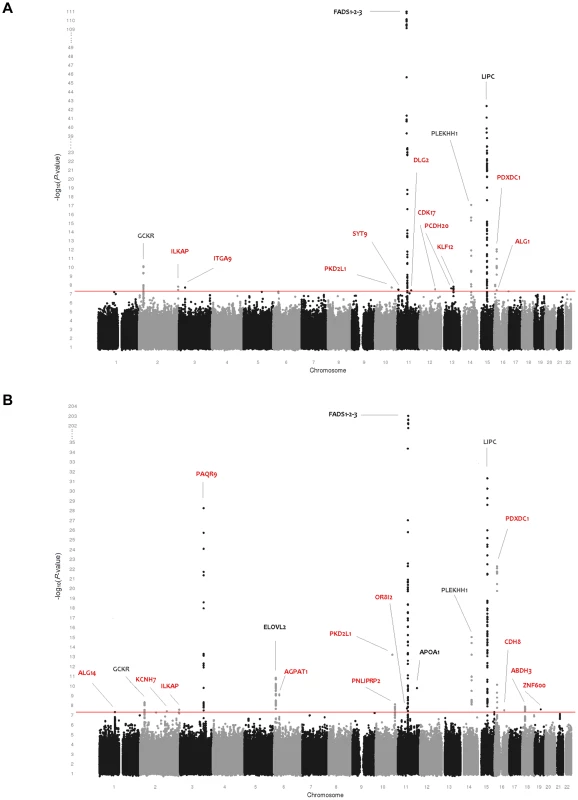
(A) Genome-wide association results for plasma levels of 115 phospholipid species. (B) Genome-wide association results for within-class proportions of 115 plasma phospholipid species. Manhattan plots show the combined association signals (−log10(P-value)) on the y-axis versus SNPs according to their position in the genome on the x-axis (NCBI build 36). Novel genes are represented in red, while previously known loci are represented in black. Fig. 2. Genome-wide association results for 33 sphingolipid species. 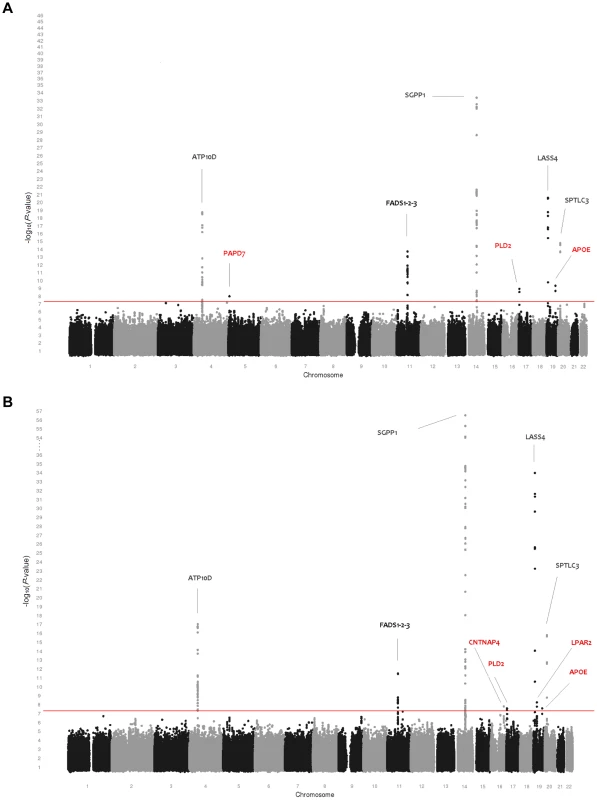
(A) Genome-wide association results for plasma levels of 33 sphingolipid species. (B) Genome-wide association results for within-class proportions of 33 sphingolipid species. Manhattan plots show the combined association signals (−log10(P-value)) on the y-axis versus SNPs according to their position in the genome on the x-axis (NCBI build 36). Novel genes are represented in red, while previously known loci are represented in black. Tab. 1. Study populations. 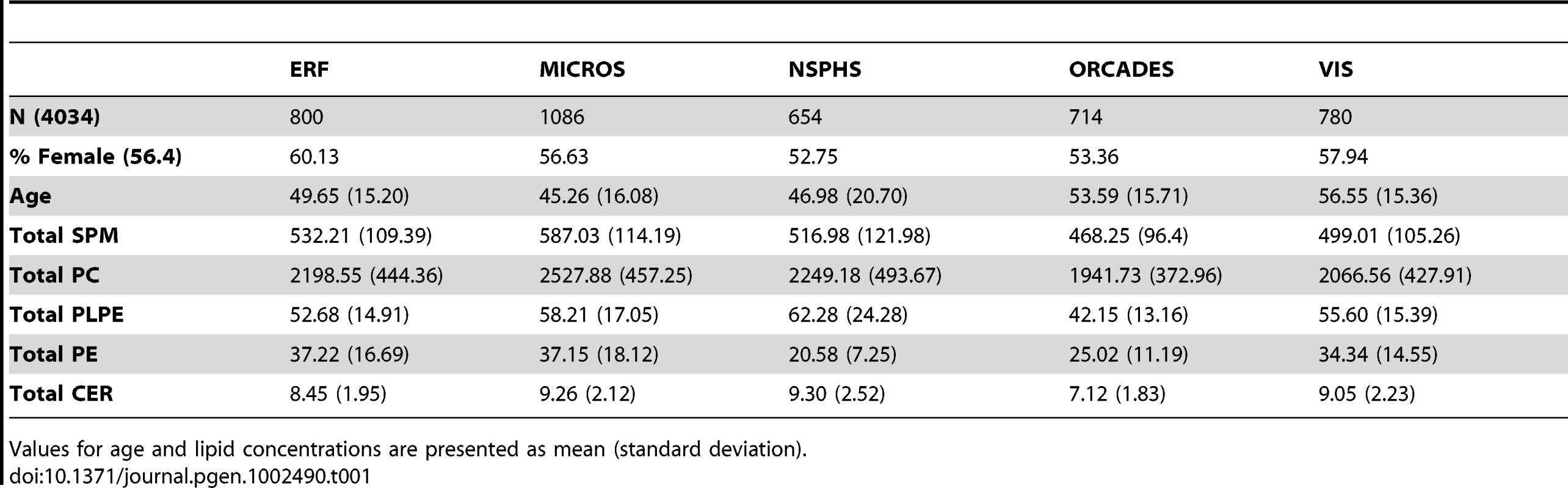
Values for age and lipid concentrations are presented as mean (standard deviation). Tab. 2. Variants significantly associated with circulating phospholipid levels and proportions. 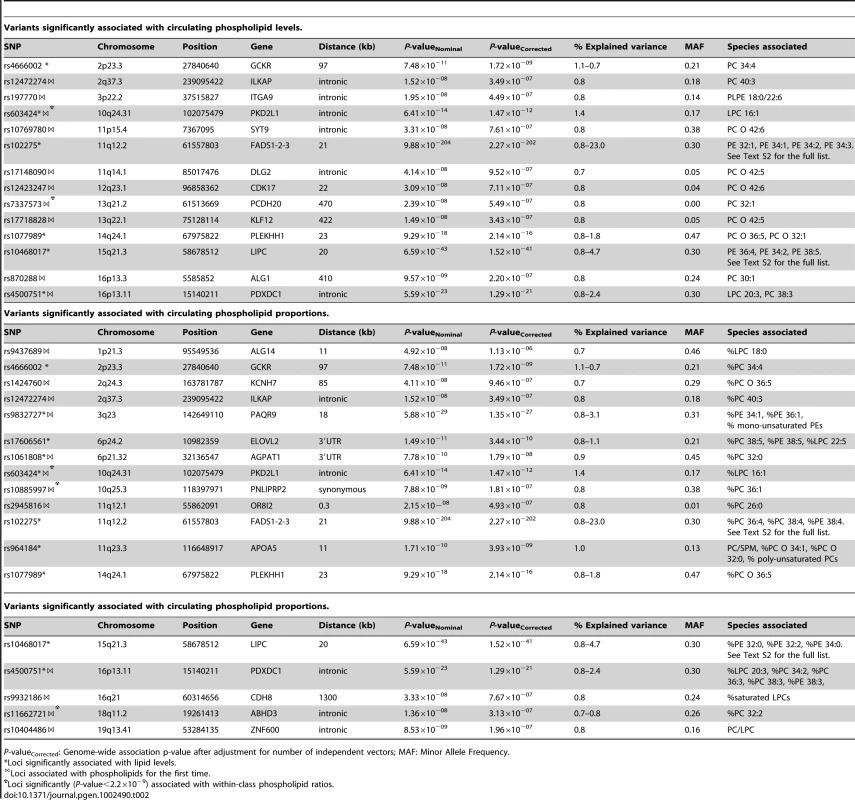
P-valueCorrected: Genome-wide association p-value after adjustment for number of independent vectors; MAF: Minor Allele Frequency. Tab. 3. Variants significantly associated with circulating sphingolipid levels and proportions. 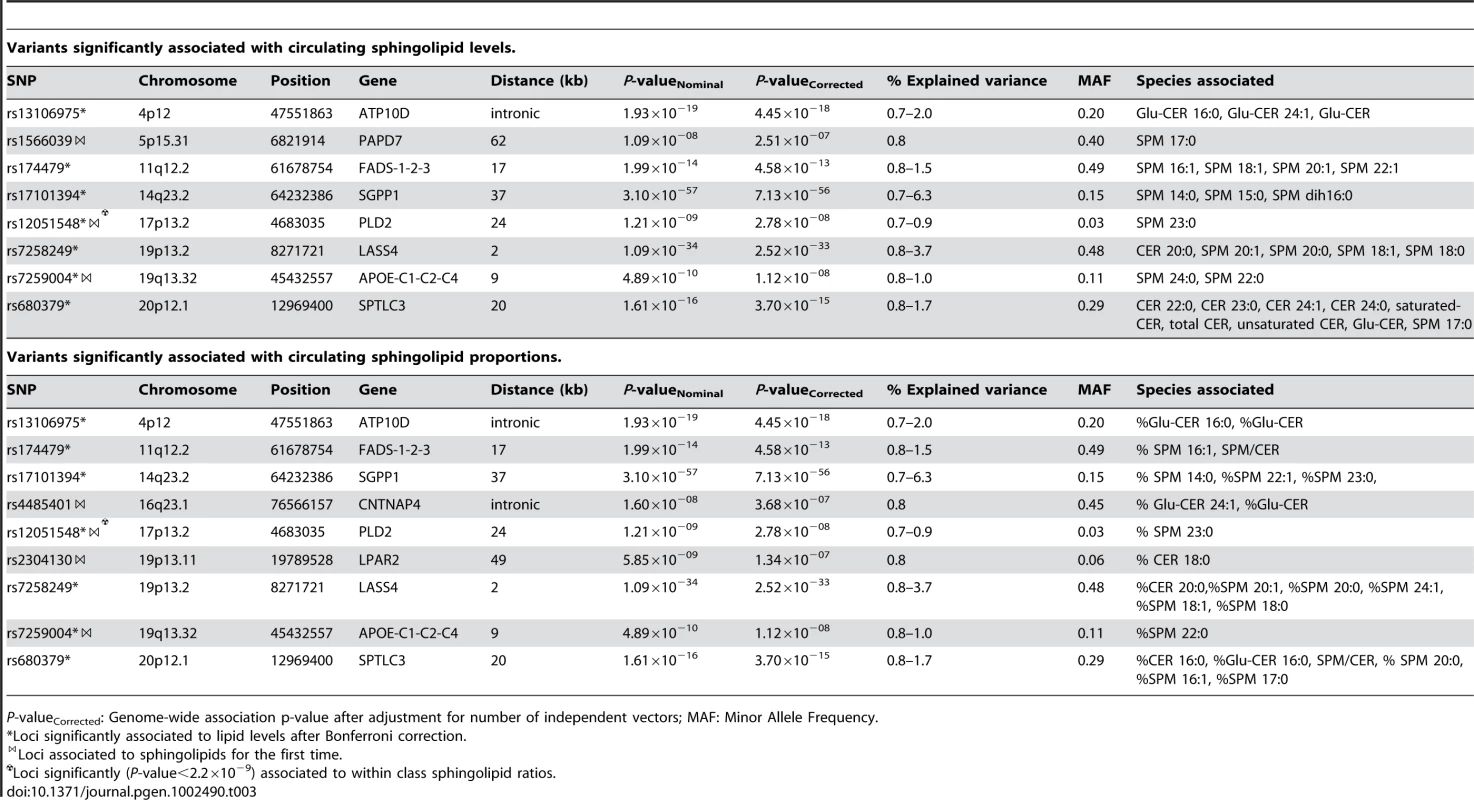
P-valueCorrected: Genome-wide association p-value after adjustment for number of independent vectors; MAF: Minor Allele Frequency. Phospholipids
As shown in Table 2, 25 loci were nominally associated (P-value<5×10−8) with absolute plasma levels and/or proportions of the phospholipid species. Among those loci, previously reported relationships between the FADS1, LIPC, PLEKHH1, GCKR, APOA1-5, and ELOVL2 loci and phospholipids were successfully replicated [17], [19]. Four novel genome-wide significant loci were also detected after a multiple testing correction to adjust for the approximate number of independent genotypes and phenotypes (n = 23) studied (P-value<2.2×10−9). These included PAQR9 on 3q23 (associated with %PE 34∶1 and %PE 36∶1), AGPAT1 on 6p21.32 (associated with PC 32∶0), PKD2L1 on 10q24.31 (LPC 16∶1), and PDXDC1 on 16p13.11 (LPC 20∶3, PC 34∶2, PC 36∶3 and PC 38∶3). Fifteen additional regions provided suggestive evidence of association (2.2×10−9<P-value<5×10−8) with phospholipids including the PNLIPRP2 locus, associated with %PC 36∶1; ZNF600 with PC/LPC ratio; ALG1 with PC 30∶1; ABHD3 with %PC 32∶2; KLF12 and DLG2, both associated with PC O 42∶5; ILKAP with PC 40∶3 and %PC 40∶3; ITGA9 with PLPE 18∶0/22∶6; OR8I2 with %PC 26∶0; PCDH20 with PC 32∶1; CDK17 and SYT9, both associated with PC O 42∶6; CDH8 with the proportion of saturated LPC; KCNH7 with %PC O 36∶5; and ALG14 with %LPC 18∶0. Regional association plots for all phospholipid loci are presented in Figure S2.
Many of the genome-wide significant and suggestive loci in Table 2 were associated with the percentage of each lipid molecule within its own class (mol%) rather than to absolute values. Single SNP analysis of ratios showed that rs4500751 (PDXDC1) was strongly associated with PC 36∶3/PC 34∶2 (P-value = 4.37×10−25) and LPC 20∶3/LPC 16∶1 (P-value = 6.84×10−23) (Table S2). Further, rs11662721 (ABHD3) was associated with the ratio of PC 32∶2 to PC 36∶2 (P-value = 9.35×10−10), but also to PC 36∶3 (P-value = 1.80×10−9) and PC 38∶3 (P-value = 6.71×10−9). rs9437689 (ALG14) and rs603424 (PKD2L1) were associated with the ratios of LPC 16∶0 to LPC 18∶0 (P-value = 2.70×10−8) and LPC 16∶1 (P-value = 2.25×10−15), respectively. SNP rs10885997 (PNLIPRP2) was associated with PC 36∶1/PC 34∶1 (P-value = 3.28×10−10) and PC 36∶1/PC 34∶3 (P-value = 1.15×10−9). SNP rs7337585 (PCDH20) was associated with the ratio of PC 32∶1 to several ether-bound PC species (the strongest association was with PC 32∶1/PC O 32∶0; P-value = 1.82×10−18) and, finally, rs2945816 (OR8I2) was associated with the ratio of PC 26∶0 to several long chain PCs (the strongest association was with PC 26∶0/PC 36∶1; P-value = 2.93×10−9).
Sphingolipids
Table 3 shows the 10 loci that were associated with either absolute plasma levels (panel A) or percentages (panel B) of sphingomyelin species or ceramides. Among those loci, 5 (ATP10D, FADS1-3, SGPP1, SPTLC3, LASS4) were previously described in genome-wide analyses [18], [19]. These loci retained significance after adjustment for the number of genotypes and phenotypes tested. In addition, five novel loci were identified at a nominal P-value of 5×10−8 (PAPD7, CNTNAP4, PLD2, LPAR2, and APOE). Two of these, APOE on 19q13.32 (associated with SPM 24∶0 and SPM 22∶0) and PLD2 on 17p13.2 (associated with SPM 23∶0), remained significant after correction for the number of phenotypes tested. The other three showed suggestive evidence of association (2.2×10−9<P-value<5×10−8) to either sphingomyelins or ceramides: PAPD7 on 5p15.31 (SPM 17∶0), the CNTNAP4 region on 16q23.1 (% Glu-CER 24∶1, %Glu-CER) and LPAR2 on 19p13.11 (%C 18∶0). Regional association plots for the sphingolipid loci are presented in Figure S3.
When studying the ratios of the index lipid to the other lipids within the same class, the strongest association for rs12051548 (PLD2) was found with the SPM 23∶0/SPM 16∶1 ratio (P-value = 2.43×10−10). SNP rs7259004 in the APOE locus was strongly associated with the ratio of SPM 24∶0 to SPM 24∶2 (P-value = 5.11×10−9) and SPM 16∶1 (P-value = 4.79×10−8) but also with the ratio of SPM 22∶0 to the same lipids (SPM 24∶2: P-value = 2.91×10−8 and SPM 16∶1: P-value = 1.98×10−8).
HDL-C, LDL-C, TG, and TC
As a point of reference, the genome-wide significant findings (P-value<5×10−8) from the GWAS of TC, LDL-C, HDL-C, and TG in these samples are provided in Table S3. CETP was associated with HDL-C levels (P-value = 8.5×10−20), APOE was associated with LDL-C (P-value = 9.2×10−26) and TC levels (P-value = 4.6×10−11). APOA1-5 (P-value = 1.6×10−8) and PDCD11 (P-value = 2.7×10−10) were associated with TG levels. Except for the PDCD11 locus, these associations have all been previously reported [20].
To determine if the associations of the phospho - and sphingolipid loci were mediated by these major classes of plasma lipoproteins, conditional analyses were performed. Table S4 shows the effect size, standard error, and P-values for the genome-wide significant loci when adjusted for HDL-C, LDL-C, TG and TC. Only the association of the APOE locus (rs7259004) with SPMs was greatly affected by the incorporation of LDL-C and TC. No other major differences were observed in effect size or P-value.
Pathway analyses
Additionally, we investigated whether the genes from the GWAS fit into previously known sphingolipid and glycerophospholipid pathways, which are available among the canonical pathways from various data bases provided by ConsensusPathDB [21]. By testing for enrichment of known pathways, glycerolipid metabolism (P-value = 0.002; KEGG), chylomicron-mediated lipid transport (P-value = 0.003; Reactome), triglyceride biosynthesis (P-value = 0.006; Reactome), metabolism of lipids and lipoproteins (P-value = 0.002; Reactome) and biosynthesis of the N-glycan precursor (P-value = 0.005; Reactome) were found to be significantly enriched among the phospholipid related loci. Considering the sphingolipid associated loci, the same analysis implicated the sphingolipid metabolism (P-value = 1.0×10−5; Reactome), metabolism of lipids and lipoproteins (P-value = 1.0×10−5; Reactome), and LPA receptor mediated events (P-value = 0.002; PID) pathways. These analyses suggested that, among genes from the same locus, SRD5A1 is a more likely candidate than PAPD7 and LPAR2 is a more likely candidate than neighbouring ZNF101 and ATP13A1 (Tables S5 and S6).
Figure S4 places all of the nearest, or most likely, genes from genome-wide significant and suggestive loci in the Ingenuity glycerophospholipid metabolism pathway [22]. Of the 25 loci associated with phospholipids at a nominal P-value<5×10−8, 13 genes (KCNH7, AGPAT1, PNLIPRP2, SYT9, FADS2, DAGLA, DLG2, APOA1, APOC3, ELOVL2, CDK17, LIPC and PLA2G10) from 11 loci can be mapped to the glycerophospholipid metabolism pathway; among the 10 loci associated with sphingomyelins or ceramides, 6 genes (FADS2, DAGLA, PLD2, LASS4, APOE, APOC2) from 4 loci can be mapped to the same pathway (Figure S4). Figure S5 maps the same genes onto the Ingenuity sphingolipid metabolism pathway. Of the 10 sphingomyelin or ceramide loci, 9 genes from 5 loci (FADS1, FADS2, C11orf10, SGPP1, APOE, APOC1, APOC2, ,LASS4, and PLD2) can be placed in this pathway, as was the case for 12 genes from 8 loci implicated in phospholipids (ILKAP, ITGA9, AGPAT1, FADS1, FADS2, C11orf10, APOA1, APOA5, APOC3, PCDH20, LIPC, and PDXDC1).
Association with IMT, T2DM, and CAD risk
The top 35 SNPs were assessed for association with IMT, T2DM, and CAD using the GWAS results from the CHARGE [23], DIAGRAM [24] and CARDIoGRAM [25] consortia, respectively. For IMT, we observed a significant association (P-value = 7×10−4) with the FADS1-2-3 locus SNP rs102275 (Table S7). rs1061808, located in the HLA region on chromosome 6, and two SNPs from the FADS1-2-3 region (rs174479 and rs102275) were associated with T2DM risk (nominal P-value<0.05) (Table S8). rs964184 from the APOA1-5 region was previously reported to be associated with CAD risk (P-value = 8.02×10−10) by the CARDIoGRAM meta-analysis study (Table S9). For all three outcomes, the observed P-value distribution differed significantly from that expected under the null hypothesis (Kolmogorov Smirnov P-value≤3.3×10−16; Figure S6).
Discussion
This genome-wide association study of more than 350 phospho - and sphingolipid measurements in five European populations yielded 25 loci associated with phospholipids and 10 loci associated with sphingolipids using a nominal P-value of 5×10−8. After correction for the number of independent phenotypes, the novel genome-wide significant loci included: PAQR9, AGPAT1, PKD2L1, PDXDC1, APOE and PLD2. In addition, further analysis of suggestive SNPs with lipid ratios showed significant association for an additional 3 loci (ABDH3, PNLIPRP2, and PCDH20).
The strongest association in the PAQR9 locus was observed between rs9832727 and the proportion of mono-unsaturated PEs, especially with the ratios PE 34∶1/PE 34∶2 and PE 36∶1/PE 36∶2. The protein coded byPAQR9 is an integral membrane receptor and functions as receptor for the hormone adiponectin, suggesting a molecular link with obesity and T2DM [26]. However, we did not observe an association between T2DM risk and this variant.
In the AGPAT1 locus, rs1061808 was associated with the proportion of PC 32∶0, and, especially, with the ratio of PC 32∶0/PC 34∶1. AGPAT1 is directly connected to phospholipid metabolism (Figures S4 and S5), as the product of this gene converts lysophosphatidic acid (LPA) into phosphatidic acid (PA) [27]. The locus lies 400 kb distant from the HLA-DRB1 gene which was previously associated with insulin secretion [28]. A suggestive association between rs1061808 and increased T2DM risk was observed in the DIAGRAM consortium meta-analysis results.
We found two loci that strongly influence plasma LPC levels: PKD2L1 and PDXDC1. An intronic variant, rs603424 in the PKD2L1 gene, was strongly associated with LPC 16∶1. Pathway analyses suggest that another gene in the same region, SCD (FADS-5), 25 kb away, may be a better candidate since it encodes the stearoyl-CoA desaturase (delta-9-desaturase) enzyme which is involved in fatty acid desaturation. Other members of the FADS family are the strongest genetic regulators of phospholipid metabolism identified to date. In the PDXDC1 locus, the strongest association was observed for intronic SNP rs4500751. This variant is 300 kb distant from PLA2G10, a gene that plays a major role in releasing arachidonic acid from cell membrane phospholipids [29] and the protein can be mapped to both the glycerophospholipid and the sphingolipid metabolism pathways by Ingenuity (Figures S4 and S5). In our study, the variant was strongly associated with the ratios of 20∶3 fatty acid carrying LPCs, as well as PEs, and PCs, but not with the others, suggesting a fatty-acid specific mechanism for this enzyme.
Another index SNP (rs7259004), associated with SPMs, maps to the well known APOE locus, which also includes three other lipid genes (APOC1, APOC2 and APOC4). Results from the conditional analyses (Table S4) suggest that the effect of this variant on SPM 22∶0 levels is dependent on plasma LDL-C levels and that SPM 22∶0 and SPM 24∶0 are likely be abundant in LDL-C particles, which can also be inferred from their high phenotypic correlations with LDL-C (r = 0.6, P-value = 2.8×10−68 for SPM 22∶0 and r = 0.6, P-value = 2.8×10−66 for SPM 24∶0).
A second locus associated with the SPMs is PLD2 (phospholipase D2). PLD2 catalyzes the hydrolysis of PC to produce phosphatidic acid and choline and the PLD2 signalling pathway is involved in the destabilization of ABCA1 and, therefore, plays a role in the generation of plasma HDL-C particles [30]. PLD2-related processes may be responsible, in part, for determining the SPM content of HDL-C. Unexpectedly, we did not observe an association between PC levels and the PLD2 locus.
The analysis of the ratios of the phospholipids uncovered three additional associations significant at the adjusted genome-wide threshold (P-value<2.2×10−9): ABDH3, PNLIPRP2 and PCDH20. The exact function of the ABDH3 and PCDH20 proteins, and how they relate to phospholipid metabolism, has not been determined. PNLIPRP2 (pancreatic lipase-related protein 2) fulfils a key function in dietary fat absorption by hydrolyzing triglycerides into diglycerides and, subsequently, into monoglycerides and free fatty acids (Figure S4) [31]. We found that a synonymous coding SNP (rs10885997) in PNLIPRP2 was associated with the ratios PC 36∶1/PC 34∶1 and PC 36∶1/PC 34∶3, suggesting a fatty-acid specific turnover between these lipids.
A closer examination of the findings published by Illig et al., supports the association signals within 100 kb of loci PDXDC1 (same SNP, P-value = 2.8×10−7), AGPAT1 (P-value = 4.9×10−7), PNLIPRP2 (P-value = 2.7×10−7), KLF12 (P-value = 5.9×10−7), ALG1 (P-value = 4.7×10−3), CDH8 (P-value = 7.6×10−7), PLD2 (P-value = 9.4×10−4) and ZNF600 (P-value = 3.3×10−7) for various phospho - and sphingolipid outcomes. SNP rs603424 in PKD2L1 was previously associated with acylcarnitine C 16∶1, although this result was not replicated [19].
The significant hits from the current study were further studied for potential associations with IMT, T2DM, and CAD. For all three outcomes, the P-value distributions differed significantly from the expected null distribution even after exclusion of nominally significant SNPs, suggesting that some of these variants contribute to these outcomes even when they do not achieve statistical significance.
Among our top hits, rs102275 from the FADS cluster was associated with IMT in the CHARGE meta-analysis results [23]. This finding demonstrates the involvement of the FADS locus in the development of atherosclerosis.
In addition, the top SNP from the APOA1-5 locus was implicated in CAD risk in the CARDIoGRAM study [25]. This locus, previously associated with TG levels [20], influenced two ether-bound PCs and the PC/SPM ratio in our study. APOA1 and APOA2 are the predominant proteins in HDL-C particles, which also transport TG. The association between the phospholipids and rs964184 remained significant after adjustment for TG levels, suggesting that this signal is not due solely to TG mediated effects. APOA1 is also a cofactor for lecithin cholesterol acyltransferase (LCAT) which converts cholesterol and PC to cholesteryl esters and LPC on the surface of HDL-C [32] and it is possible that the association we observe here is due to LCAT mediated phospholipid cleavage.
Mapping the findings into the glycerophospholipid and sphingolipid metabolism pathways uncovered several enzymes, kinases, peptidases and G-protein coupled receptors that may also be relevant for phospho - and sphingolipid metabolism. Among those involved in sphingolipid metabolism (Figure S5), HNF4A (hepatocyte nuclear factor-4) appears to be a common interacting factor for several genes (PCDH20, APOC1, AGPAT1, ITGA9, PLD2, C11ORF10, APOC2, GCKR, APOE, APOC3 and LIPC) from our GWAS. It is already known that the extinction of many hepatic functions and their expression are correlated with expression of HNF4A which is a candidate transcription factor for further research on lipidomics [33].
In conclusion, we identified 15 previously undescribed loci that were suggestively associated (2.2×10−9<P-value<5×10−8) with phospho - and sphingolipid levels. These included interesting candidate genes such as LPAR2. These loci will require follow-up to definitively establish their relationship with these phenotypes. We also identified nine novel loci below the corrected genome-wide significance threshold (P-value<2.2×10−9). These loci considerably expand our knowledge of genes/regions involved in the determination of phospho - and sphingolipid concentrations and provide interesting avenues for future research into this important topic.
Materials and Methods
All studies were approved by the local ethical committees. Detailed descriptions of the study populations that contributed to the meta-analysis, as well as detailed information on ethical statements, genotyping, lipid measurements and pathway analysis, are presented in Text S1. Briefly, lipid species were quantified by electrospray ionization tandem mass spectrometry (ESIMS/MS) using methods validated and described previously [34], [35]. For each lipid molecule, we adopted the naming system where lipid side chain composition is abbreviated as Cx∶y, where x denotes the number of carbons in the side chain and y the number of double bonds. For example, PC 34∶4 denotes an acyl-acyl phosphatidylcholine with 34 carbons in the two fatty acid side chains and 4 double bonds in one of them. Lipid traits were analysed individually as well as aggregated into groups of species with similar characteristics (e.g. unsaturated ceramides). These were then analyzed as both absolute concentrations (µM) and as molar percentages within lipid sub-classes (mol%) (calculated as the proportion of each lipid molecule among its own class (e.g. PC, PE, PLPE, LPC). The additive value of the analyses of molar proportions is that it may bring to light genes involved in the transition of one species to another, such as through fatty acid chain elongation or (de)saturation. We also performed single SNP association analyses for each novel locus and the ratio of the index lipid (for example, PC 34∶1) to the other lipids in the same class (in the example, PC 34∶1/PC 36∶1, PC 34∶1/PC 38∶1) so that we could determine whether the SNP might be involved in elongation or (de)saturation.
DNA samples were genotyped according to the manufacturer's instructions on Illumina Infinium HumanHap300v2, HumanHap300v1 or HumanCNV370v1 SNP bead microarrays. Genotype data for these five populations were imputed using MACH 1.0 (v1.0.16) [36], [37] using the HapMap CEU population (release 22, build 36).
As all of the studies included related individuals, testing for association between lipid and allele dosage were performed using a mixed model approach as implemented with the ‘mmscore’ option in the GenABEL software [38]. Results from the five populations were combined using inverse variance weighted fixed-effects model meta-analyses using the METAL software [39]. To correct for multiple testing, we adopted a Bonferroni correction for the number of phenotypes studied. Since most of the lipid values are correlated with each other, we used the number of principal components (n = 23) that accounted for 79% of the phenotypic variance for this correction and applied it to the classical genome-wide significance threshold (5×10−8).
Supporting Information
Zdroje
1. HolthuisJCPomorskiTRaggersRJSprongHVan MeerG 2001 The organizing potential of sphingolipids in intracellular membrane transport. Physiol Rev 81 1689 1723
2. LajoiePGoetzJGDennisJWNabiIR 2009 Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J Cell Biol 185 381 385
3. MerrillASK 2002 Sphingolipids: metabolism and cell signaling. VanceDVJE Biochemistry of Lipids, Lipoproteins and Membranes 373 407
4. BakovicMFullertonMDMichelV 2007 Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of CTP:phosphoethanolamine cytidylyltransferase (Pcyt2). Biochem Cell Biol 85 283 300
5. PoliGLeonarduzziGBiasiFChiarpottoE 2004 Oxidative stress and cell signalling. Curr Med Chem 11 1163 1182
6. van MeerGVoelkerDRFeigensonGW 2008 Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9 112 124
7. ZhengWKollmeyerJSymolonHMominAMunterE 2006 Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta 1758 1864 1884
8. BroekmanMJHandinRIDerksenACohenP 1976 Distribution of phospholipids, fatty acids, and platelet factor 3 activity among subcellular fractions of human platelets. Blood 47 963 971
9. EngelmannBKoglCKulscharRSchaippB 1996 Transfer of phosphatidylcholine, phosphatidylethanolamine and sphingomyelin from low - and high-density lipoprotein to human platelets. Biochem J 315 Pt 3 781 789
10. BruggerBErbenGSandhoffRWielandFTLehmannWD 1997 Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci U S A 94 2339 2344
11. HodgeAMEnglishDRO'DeaKSinclairAJMakridesM 2007 Plasma phospholipid and dietary fatty acids as predictors of type 2 diabetes: interpreting the role of linoleic acid. Am J Clin Nutr 86 189 197
12. MalerbaGSchaefferLXumerleLKloppNTrabettiE 2008 SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids 43 289 299
13. MannheimDHerrmannJVersariDGosslMMeyerFB 2008 Enhanced expression of Lp-PLA2 and lysophosphatidylcholine in symptomatic carotid atherosclerotic plaques. Stroke 39 1448 1455
14. MatsumotoTKobayashiTKamataK 2007 Role of lysophosphatidylcholine (LPC) in atherosclerosis. Curr Med Chem 14 3209 3220
15. WangLFolsomARZhengZJPankowJSEckfeldtJH 2003 Plasma fatty acid composition and incidence of diabetes in middle-aged adults: the Atherosclerosis Risk in Communities (ARIC) Study. Am J Clin Nutr 78 91 98
16. FarooquiAAHorrocksLAFarooquiT 2000 Glycerophospholipids in brain: their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem Phys Lipids 106 1 29
17. GiegerCGeistlingerLAltmaierEHrabe de AngelisMKronenbergF 2008 Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet 4 e1000282
18. HicksAAPramstallerPPJohanssonAVitartVRudanI 2009 Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet 5 e1000672
19. IlligTGiegerCZhaiGRomisch-MarglWWang-SattlerR 2009 A genome-wide perspective of genetic variation in human metabolism. Nat Genet 42 137 141
20. TeslovichTMMusunuruKSmithAVEdmondsonACStylianouIM 2010 Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466 707 713
21. KamburovAWierlingCLehrachHHerwigR 2009 ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res 37 D623 628
22. Jimenez-MarinACollado-RomeroMRamirez-BooMArceCGarridoJJ 2009 Biological pathway analysis by ArrayUnlock and Ingenuity Pathway Analysis. BMC Proc 3 Suppl 4 S6
23. BisJCKavousiMFranceschiniNIsaacsAAbecasisGR 2011 Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nat Genet 43 940 947
24. VoightBFScottLJSteinthorsdottirVMorrisAPDinaC 2010 Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42 579 589
25. SchunkertHKonigIRKathiresanSReillyMPAssimesTL 2011 Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43 333 338
26. TangYTHuTArterburnMBoyleBBrightJM 2005 PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J Mol Evol 61 372 380
27. AguadoBCampbellRD 1998 Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J Biol Chem 273 4096 4105
28. HowsonJMRosingerSSmythDJBoehmBOToddJA 2011 Genetic Analysis of Adult-Onset Autoimmune Diabetes. Diabetes 60 2645 2653
29. SinghDKSubbaiahPV 2007 Modulation of the activity and arachidonic acid selectivity of group X secretory phospholipase A2 by sphingolipids. J Lipid Res 48 683 692
30. WangYOramJF 2005 Unsaturated fatty acids phosphorylate and destabilize ABCA1 through a phospholipase D2 pathway. J Biol Chem 280 35896 35903
31. GillerTBuchwaldPBlum-KaelinDHunzikerW 1992 Two novel human pancreatic lipase related proteins, hPLRP1 and hPLRP2. Differences in colipase dependence and in lipase activity. J Biol Chem 267 16509 16516
32. BreslowJLRossDMcPhersonJWilliamsHKurnitD 1982 Isolation and characterization of cDNA clones for human apolipoprotein A-I. Proc Natl Acad Sci U S A 79 6861 6865
33. ChartierFLBossuJPLaudetVFruchartJCLaineB 1994 Cloning and sequencing of cDNAs encoding the human hepatocyte nuclear factor 4 indicate the presence of two isoforms in human liver. Gene 147 269 272
34. LiebischGDrobnikWLieserBSchmitzG 2002 High-throughput quantification of lysophosphatidylcholine by electrospray ionization tandem mass spectrometry. Clin Chem 48 2217 2224
35. LiebischGLieserBRathenbergJDrobnikWSchmitzG 2004 High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim Biophys Acta 1686 108 117
36. NothnagelMEllinghausDSchreiberSKrawczakMFrankeA 2009 A comprehensive evaluation of SNP genotype imputation. Hum Genet 125 163 171
37. LiYWillerCJDingJScheetPAbecasisGR 2010 MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34 816 834
38. AulchenkoYSRipkeSIsaacsAvan DuijnCM 2007 GenABEL: an R library for genome-wide association analysis. Bioinformatics 23 1294 1296
39. WillerCLiYAbecasisG 2010 METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26 2190 2191
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Prenatální expozice ftalátům a anogenitální vzdálenost u novorozenců
- Vztah užívání alkoholu a mužské fertility
-
Všechny články tohoto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



