-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
An important paradigm in evolutionary genetics is that of a delicate balance between genetic variants that favorably boost host control of infection but which may unfavorably increase susceptibility to autoimmune disease. Here, we investigated whether patients with psoriasis, a common immune-mediated disease of the skin, are enriched for genetic variants that limit the ability of HIV-1 virus to replicate after infection. We analyzed the HLA class I and class II alleles of 1,727 Caucasian psoriasis cases and 3,581 controls and found that psoriasis patients are significantly more likely than controls to have gene variants that are protective against HIV-1 disease. This includes several HLA class I alleles associated with HIV-1 control; amino acid residues at HLA-B positions 67, 70, and 97 that mediate HIV-1 peptide binding; and the deletion polymorphism rs67384697 associated with high surface expression of HLA-C. We also found that the compound genotype KIR3DS1 plus HLA-B Bw4-80I, which respectively encode a natural killer cell activating receptor and its putative ligand, significantly increased psoriasis susceptibility. This compound genotype has also been associated with delay of progression to AIDS. Together, our results suggest that genetic variants that contribute to anti-viral immunity may predispose to the development of psoriasis.
Published in the journal: . PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002514
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002514Summary
An important paradigm in evolutionary genetics is that of a delicate balance between genetic variants that favorably boost host control of infection but which may unfavorably increase susceptibility to autoimmune disease. Here, we investigated whether patients with psoriasis, a common immune-mediated disease of the skin, are enriched for genetic variants that limit the ability of HIV-1 virus to replicate after infection. We analyzed the HLA class I and class II alleles of 1,727 Caucasian psoriasis cases and 3,581 controls and found that psoriasis patients are significantly more likely than controls to have gene variants that are protective against HIV-1 disease. This includes several HLA class I alleles associated with HIV-1 control; amino acid residues at HLA-B positions 67, 70, and 97 that mediate HIV-1 peptide binding; and the deletion polymorphism rs67384697 associated with high surface expression of HLA-C. We also found that the compound genotype KIR3DS1 plus HLA-B Bw4-80I, which respectively encode a natural killer cell activating receptor and its putative ligand, significantly increased psoriasis susceptibility. This compound genotype has also been associated with delay of progression to AIDS. Together, our results suggest that genetic variants that contribute to anti-viral immunity may predispose to the development of psoriasis.
Introduction
Psoriasis is an immune-mediated, inflammatory skin disease that is associated with arthritis and other systemic co-morbidities [1]. Psoriasis is a highly heritable condition, with a monozygotic twin concordance rate of 70% [2] and an estimated sibling recurrence risk λs of 4–11 [3]. Linkage studies [4]–[6] and genome-wide association studies (GWAS) [7]–[11] have identified over 20 psoriasis susceptibilities alleles. However, the locus consistently displaying the strongest association signal, by many orders of magnitude, is the major histocompatibility complex (MHC).
We were intrigued by the observation that several of the most highly significant SNPs from psoriasis GWAS were identical to the top SNPs from GWAS of HIV-1 virologic control, a clinical phenotype whereby certain HIV-1 infected individuals, termed “HIV-1 controllers,” are able to maintain low levels of plasma HIV-1 RNA in the absence of antiretroviral therapy and who generally do not develop clinical symptoms [12]. For example, rs2395029 within the MHC gene HCP5 and a proxy for HLA-B*57, was identified in a psoriasis GWAS as the SNP with the largest odds ratio, OR = 4.1, p = 2.13×10−26 [8]. This same SNP has been shown to be the first or second most significant SNP in three GWAS of HIV-1 control [13]–[15]. The two most significant SNPs identified in multiple psoriasis GWAS are rs10484554 and rs12191877 near HLA-C (r2 = 1 with each other in Europeans) [8]–[10]. rs10484554 and rs12191877 were found to be associated with HIV-1 control [Supplementary Materials in [13], [15]] and both are in moderate linkage disequilibrium (r2 = 0.33) with rs9264942, another top SNP for HIV-1 control [14]–[16].
The relationship between psoriasis and HIV-1 is also interesting because of the clinical observation that HIV-1 infection can exacerbate existing psoriasis or trigger new-onset psoriasis [17]. As HIV-1 infection progresses and CD4+ T cell counts decrease, psoriasis can worsen [18], [19]. This has puzzled dermatologists and infectious disease clinicians because it has been convincingly established that psoriasis is an immune disorder that is mediated through activation of T cells. Several explanations for this “psoriasis HIV-1 paradox” have been proposed, including HIV-1 induced destruction of regulatory CD4+ T cells [20], an increase in number of memory CD8+ T cells late in disease [21], HIV-1 proteins acting as superantigens [22], or co-stimulation through traditional antigenic presentation [20].
Due to these genetic and clinical observations, we pursued a more in-depth analysis of the HLA region in psoriasis to determine whether patients with psoriasis are enriched for the major genetic determinants of HIV-1 control. The psoriasis data generated in this study were compared to the largest GWAS for HIV-1 control performed to date, involving 516 cases and 1,196 controls and for which detailed HLA allele information was available [15].
Results
Accurate Imputation of HLA Alleles
We imputed to four-digit resolution the HLA class I alleles (-A, -B, -C) and HLA class II alleles (-DQA1, -DQB1, -DRB1) of 1,727 psoriasis cases and 3,581 healthy controls which were obtained from 3 separate case-control cohorts of European ancestry (Table S1). Imputation was performed using the software HLA*IMP, which has been shown to have an accuracy of at least 96% for class I loci and 92% for Class II loci [23]. To further validate the accuracy of our imputation, we compared the imputed HLA alleles to empirically obtained HLA class I alleles for a subset of our samples (n = 98). The concordance was 566/581 alleles (97.4%), indicating that the imputation was of high accuracy. A sensitivity analysis examining the imputation accuracy of low frequency HLA alleles (allele frequency between 1% and 5%) demonstrated similar high accuracy (177/179 alleles = 98.9%). Only HLA alleles with a minor allele frequency greater than 1% in the control group were used for subsequent analyses.
HLA Association Testing Identifies a Similar Genetic Architecture between Psoriasis and HIV-1 Control
We tested all imputed HLA alleles for association with psoriasis using logistic regression, adjusting for gender, ancestry, and cohort. The top ten HLA associations for psoriasis are shown in Table 1 (Full four-digit and two-digit results in Tables S2 and S3, respectively). Overall, we observed a striking pattern in which the HLA alleles which are enriched in psoriasis patients are also enriched in HIV-1 controllers, and the HLA alleles which have decreased frequency in psoriasis patients are also decreased in HIV-1 controllers. We found that psoriasis patients are highly enriched for HLA-B*57 : 01 (12.5% in cases vs 3.9% in controls, p = 5.50×10−42, OR = 3.61), which in multiple studies has been shown to be the most significant predictor of both HIV-1 control and delayed progression time to AIDS [14], [15], [24]–[27]. Psoriasis patients also display a significant enrichment of the HIV-1 control allele B*13 : 02, whereas they display a relative paucity of B*07 : 02, B*40 : 01, and C*04 : 01 which are associated with lack of virologic control [15]. The HLA allele B*35, almost always seen with C*04 : 01, and the most significant HLA allele associated with rapid progression to AIDS [28], [29], was significantly protective against psoriasis in our dataset (p = 3.20×10−6, OR = 0.65 [0.54–0.78]). HLA-B*35 alleles can be segregated into B*35-Px and B*35-PY alleles, where Px alleles bind peptides with hydrophobic, non-tyrosine residues at position 9 and PY alleles bind peptides with tyrosine at position 9. It has been shown that the influence of HLA-B*35 in accelerating progression to AIDS is mostly attributable to HLA-B*35-Px alleles [30]. In our psoriasis dataset, the B*35-Px alleles B*35 : 02 and B*35 : 03 together demonstrated a stronger effect on psoriasis protection (p = 2.9×10−4, OR = 0.47 [0.31–0.71]) than the B*35-PY allele B*35 : 01 (p = 5.86×10−3, OR = 0.74 [0.60–0.92]).
Tab. 1. Top ten classical HLA alleles associated with psoriasis, and comparison to HIV-1 controllers as published in [15]. ![Top ten classical HLA alleles associated with psoriasis, and comparison to HIV-1 controllers as published in <em class="ref">[15]</em>.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/24d04e75476ae1ec90e4e86501c84bf2.png)
Alleles marked with † have an independent effect on HIV-1 control. A high degree of similarity is observed between psoriasis and HIV-1 control with respect to the magnitude and direction of the associated alleles. P values and ORs for psoriasis samples were adjusted by ancestry, gender, and cohort. ORs greater than 1.0 correspond to psoriasis susceptibility and control of HIV-1, whereas ORs less than 1.0 correspond to decreased psoriasis susceptibility and lack of virologic control. NS, not significant. Stepwise Regression Modeling Identifies HLA Alleles with Independent Effects on Psoriasis Susceptibility, Including the HIV-1 Control Alleles B*57 : 01 and B*27 : 05
To identify HLA alleles independently associated with psoriasis, we performed stepwise regression modeling, first conditioning the association results on the top allele HLA-C*06 : 02, and then adding alleles to the model in a stepwise manner. We identified HLA-C*06 : 02, B*38 : 01, A*02 : 01, B*39 : 01, B*27 : 05, B*08 : 01, B*14 : 02, B*55 : 01, and B*57 : 01 as HLA class I alleles independently associated with psoriasis (Table 2). In the multivariate regression model including all of these alleles, the HIV-1 viral control alleles B*57 : 01 and B*27 : 05 both had significant effect on psoriasis susceptibility (OR = 1.52 and 1.75, respectively). The contribution of B*27 : 05 was more apparent in the regression model than when B*27 : 05 was analyzed as a single allele (p = 0.016, OR = 1.32 [1.05–1.66]). The HIV-1 progression allele B*35 remained independently associated with psoriasis after conditioning on the top allele C*06 : 02 (p = 0.0064, OR = 0.77 [0.63–0.93]), but further conditioning on B*38 : 01 and A*02 : 01 resulted in a residual association signal for B*35 of p = 0.0168, OR = 0.78 [0.64–0.96]).
Tab. 2. HLA class I alleles identified by stepwise logistic regression as independently associated with psoriasis. 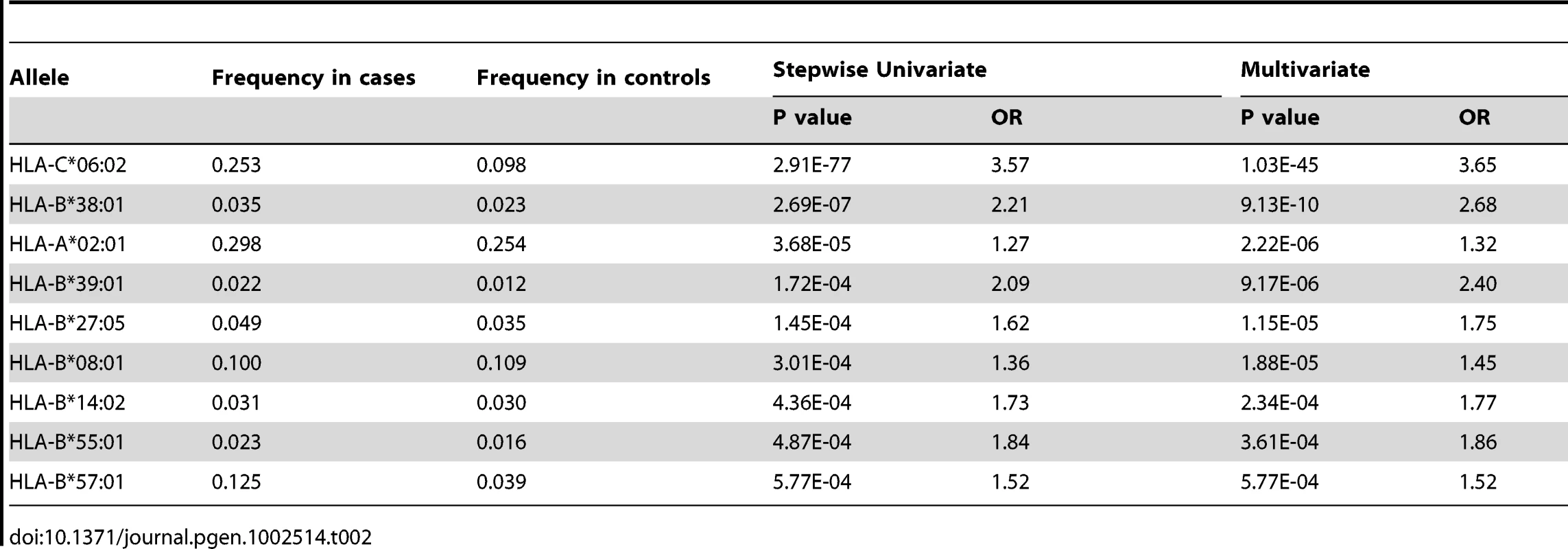
We also performed stepwise regression modeling combining class I and class II HLA alleles. At 4 digit resolution, C*06 : 02, B*38 : 01, DQB1*05 : 02, DQB1*06 : 04, and A*02 : 01 were found to be independent risk factors; however, at 2 digit resolution, the class II alleles DQB1*05 and DQB1*06 were no longer significant (data not shown).
The Extended Haplotype B*57 : 01–C*06 : 02–DQA1*02 : 01–DQB1*03 : 03–DRB1*07 : 01 Is Associated with Both Psoriasis Susceptibility and HIV-1 Control
We performed haplotype analysis in psoriasis patients and HIV-1 controllers to help understand how combinations of HLA alleles contribute to the observed association signals. We estimated the frequency of HLA haplotypes in our psoriasis case-control cohort as well as in 214 Caucasian HIV-1 infected individuals (52 HIV-1 controllers, 162 non-controllers) in the SCOPE cohort, whose HLA class I and II alleles had been previously genotyped. Our analysis revealed that both psoriasis patients and HIV-1 controllers are highly enriched for the B*57 : 01–C*06 : 02 haplotype as well as the extended haplotype B*57 : 01–C*06 : 02–DQA1*02 : 01–DQB1*03 : 03–DRB1*07 : 01, thus explaining why these individual alleles are associated with both phenotypes (Table 3 and Table 4). We found that the association of DQA1*02 : 01, DQB1*03 : 03, and DRB1*07 : 01 with psoriasis was nearly completely due to the effects of C*06 : 02 or B*57 : 01, since conditioning DQA1*0201 and DRB1*0701 on C*06 : 02 resulted in p = 0.017, OR = 1.21 and p = 0.199, OR = 1.11, respectively; and conditioning DQB1*0303 on B*57 : 01 resulted in p = 0.038, OR = 1.33. Thus, the primary genetic determinants of both psoriasis and HIV-1 control reside within the class I alleles.
Tab. 3. Association testing of HLA B–C haplotypes with psoriasis and HIV-1 control. 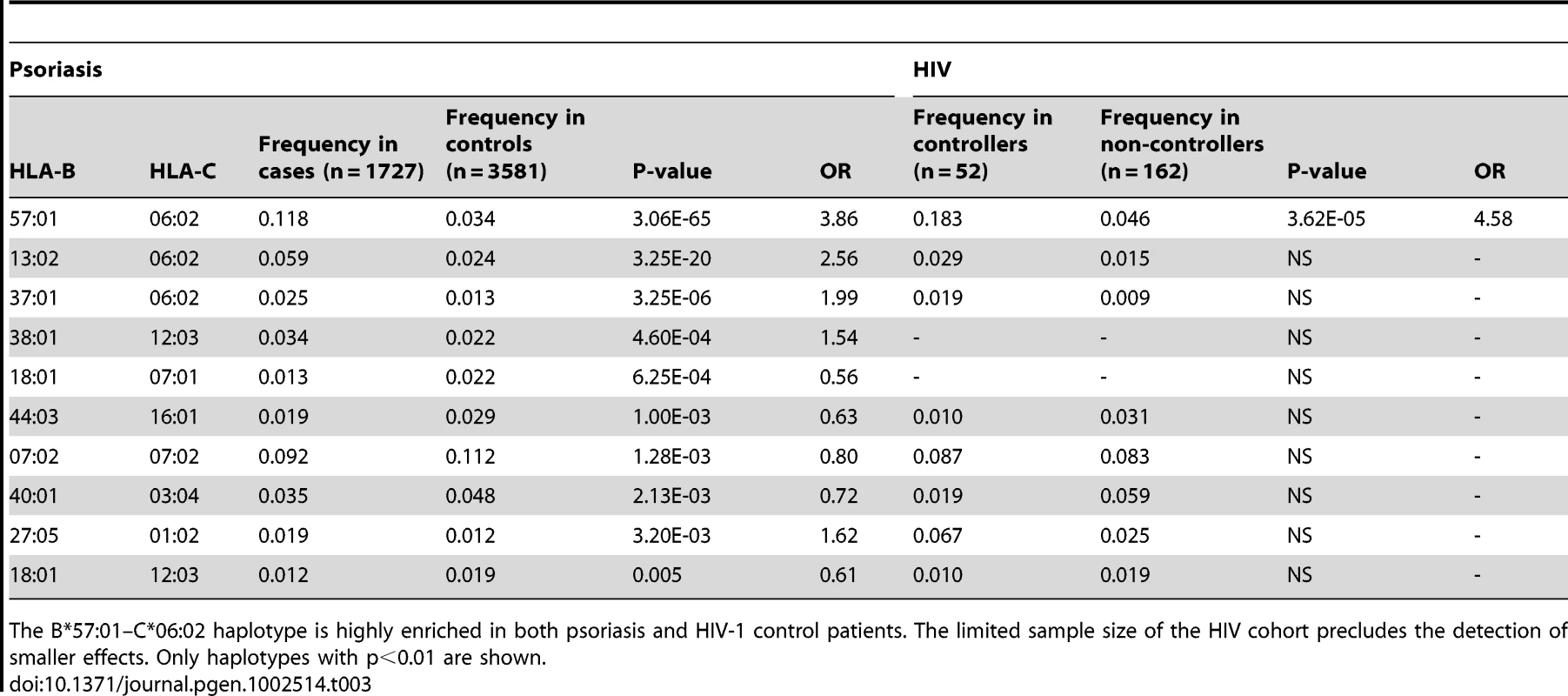
The B*57:01–C*06:02 haplotype is highly enriched in both psoriasis and HIV-1 control patients. The limited sample size of the HIV cohort precludes the detection of smaller effects. Only haplotypes with p<0.01 are shown. Tab. 4. Association testing of extended class I and II HLA haplotypes with psoriasis and HIV-1 control. 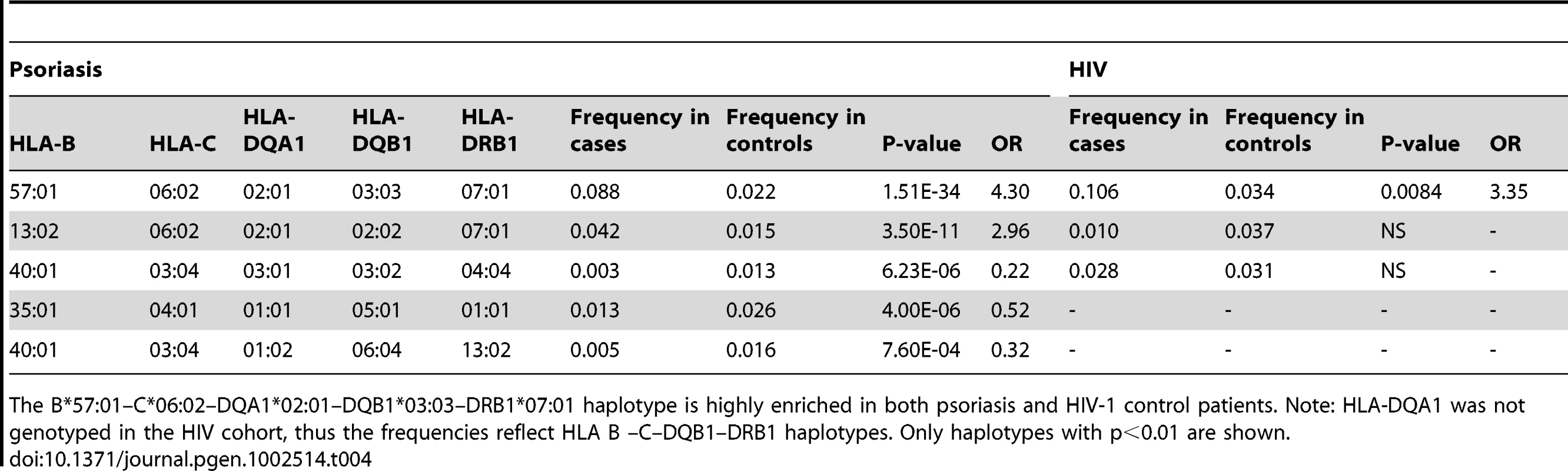
The B*57:01–C*06:02–DQA1*02:01–DQB1*03:03–DRB1*07:01 haplotype is highly enriched in both psoriasis and HIV-1 control patients. Note: HLA-DQA1 was not genotyped in the HIV cohort, thus the frequencies reflect HLA B –C–DQB1–DRB1 haplotypes. Only haplotypes with p<0.01 are shown. Amino Acids within HLA-B That Are Predictive of HIV-1 Viral Load Are Concordant between HIV-1 Controllers and Psoriasis Patients
Specific amino acid positions within the peptide binding groove of HLA class I molecules have been shown to serve as important mediators for the protective and risk effects of individual HLA alleles on HIV-1 control [15]. Namely, amino acid residues at positions 97, 67, and 70 within HLA-B were found to be more highly associated with HIV-1 control than HLA-B*57 : 01 and each of these amino acid positions was found to serve as a strong predictor of HIV-1 viral load levels in an independent cohort [15]. To determine whether psoriasis is associated with the groups of alleles that are marked by specific amino acids within HLA proteins, we used the official protein sequences [31] assigned to each four-digit HLA allele to perform association testing at each amino acid position within HLA-A, -B, -C, -DQA1, -DQB1, and -DRB1. As before, the association testing was adjusted for gender, ancestry, and cohort. We found that the five most significant amino acid positions for psoriasis occurred at 3 positions within HLA-B (residue 97 [p = 1.58×10−53], residue 67 [p = 4.00×10−45], and residue 70 [p = 1.35×10−40]) and 2 positions with HLA-C (residue 156 [p = 3.89×10−51] and residue 97 [p = 4.56×10−45]) (Table S4). Each of these 5 positions is located within the peptide-binding groove of the HLA molecule and directly contacts the bound peptide [32]. At each of these positions, we investigated whether the direction of the association signal was consistent between psoriasis patients and HIV-1 controllers. We confirmed that for each position examined, the amino acid residues associated with psoriasis susceptibility were associated with HIV-1 virologic control, and the amino acids associated with a protective effect on psoriasis risk were associated with HIV-1 progression (Figure 1). For example, alleles marked by Asn97, Thr97, and Val97 in HLA-B were associated with psoriasis susceptibility and HIV-1 control while those marked by Arg97 and Ser97 in HLA-B were associated with psoriasis protection and HIV-1 progression.
Fig. 1. Top 5 HLA amino acid positions associated with psoriasis and comparison with HIV-1 control and other autoimmune or inflammatory diseases. 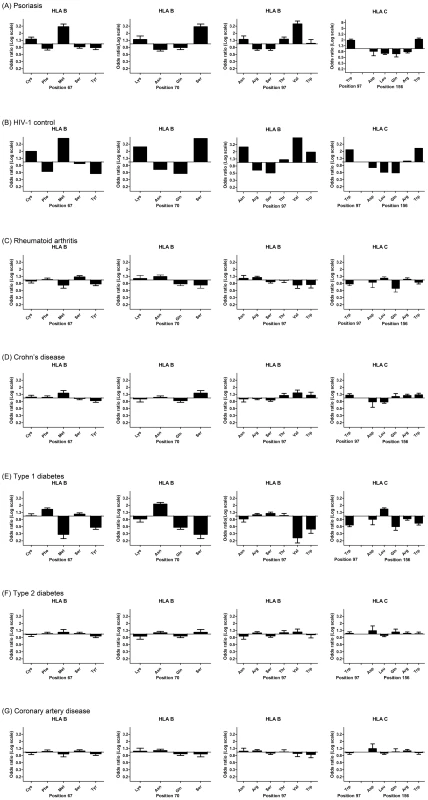
(A) Specific amino acid residues at positions 67, 70, and 97 within HLA-B and positions 97 and 156 within HLA-C are strongly associated with psoriasis susceptibility or protection, where the strength and direction of association are reflected by the odds ratio at each residue. All 5 positions occur in the peptide-binding groove of HLA-B or HLA-C. (B) Comparison of odds ratios to HIV-1 control, in which HLA-B positions 67, 70, and 97 are the top 3 three reported positions [15]. (C–G) Comparison of odds ratios to five other autoimmune or inflammatory diseases from the Wellcome Trust Case-Control Consortium. None of these demonstrate the same degree of similarity as between psoriasis and HIV-1 control. To demonstrate that this similarity cannot be entirely explained by the association of psoriasis with B*57:01 and C*06:02, we conditioned each of these residues on both B*57:01 and C*06:02 and found that Cys67, Ser67, Lys70, Asn97, Arg97 in HLA-B remained independently associated with psoriasis (all p<5×10−4) in the same direction as the association with HIV-1 control. This suggests that the repertoire of peptides bound at HLA-B may be similar between psoriasis patients and HIV-1 controllers. To rule out the possibility that the observed similarities between psoriasis and HIV-1 control were the result of systematic bias of the imputation process or general amino acid variability at those positions, we examined whether these 5 amino acids positions were associated with other autoimmune or inflammatory diseases. We examined GWAS data from 5 diseases studied by the Wellcome Trust Case Control Consortium [33]—rheumatoid arthritis, Crohn's disease, type 1 diabetes, type 2 diabetes, and coronary artery disease—and used the same imputation process as performed with psoriasis. We found that none of these diseases displayed the degree of similarity between psoriasis and HIV-1 control when considering the direction and magnitude of the association signal at these amino acid positions (Figure 1). Crohn's disease, which shares some pathophysiological features with psoriasis [34] and is also slightly enriched for HLA-C*06 : 02 (p = 4.2×10−5, OR 1.32) and HLA-B*57 : 01 (p = 3.68×10−4, OR 1.40), showed some similarity to psoriasis and HIV-1 control at these positions, but the magnitude of the association was smaller and several important residues such as Asn70 and Asn97 in HLA-B and Gln156 in HLA-C were not concordant. Interestingly, type 1 diabetics showed the opposite effect at many of these residues (i.e. patients with type 1 diabetes lack HIV-1 control alleles and have an excess of HIV-1 progression alleles), which could support the theory that type 1 diabetes is triggered by a viral infection.
Another important amino acid within HLA-B that may be relevant for HIV-1 progression is position 116, which not only interacts with the carboxy-terminal residues of peptides in the F pocket, but also strongly influences the interaction of HLA class I molecules with the peptide-loading complex [35], [36]. Studies of HLA-B*44 : 05 and HLA-B*44 : 02, which only differ at position 116, have shown that B*44 : 05 (containing tyrosine at position 116, “116Y”) utilizes a tapasin-independent pathway that leads to a less optimal peptide repertoire compared to the tapasin-dependent HLA-B*44 : 02 (116D) [36]. 116Y is strongly associated with lack of HIV-1 control, p = 1.6×10−10, OR = 0.57 [15]. Our data show that 116Y is strongly associated with decreased susceptibility to psoriasis, p = 7.96×10−17, OR = 0.66 [0.60–0.73] (Table S4). In our dataset, all HLA-B alleles containing 116Y had an odds ratio less than 1.0, including B*07 : 02 (p = 3.4×10−6, OR = 0.71), B*08 : 01 (p = 0.092, OR = 0.88), B*35 : 02 (p = 0.038, OR = 0.54), B*40 : 01 (p = 7.24×10−6, OR = 0.60), B*40 : 02 (p = 0.013, OR = 0.53), and B*51 : 01 (p = 0.084, OR = 0.82). Thus, protection against psoriasis may be associated with presentation of a less-optimized peptide repertoire.
Together, our data indicate that the genetic similarity between psoriasis patients and HIV-1 controllers extends to specific amino acid residues within class I molecules that mediate peptide binding, influence peptide loading, and which mark viral control or progression. We additionally performed stepwise regression to identify amino acid residues that were independently associated with psoriasis and found Trp156 and Ala24 in HLA-C; Val97, Leu145, Cys67, and Tyr99 in HLA-B; and Gly107 in HLA-A to be markers independently associated with psoriasis (Table S5). Similar to HIV-1 control [15], we found that HLA-B positions 97 and 67 remained in the model, while position 70 dropped out due to linkage disequilibrium with positions 97 and 67. However, we caution against an interpretation that the amino acids identified here as independent are necessarily the functional ones. Due to the complex LD patterns between the amino acids, the final output of the stepwise regression model is affected by the starting variable, and potentially functionally significant amino acids can be lost because they are tagged by other residues. For example, HLA-B Gly62, part of the α1 helix located in the B-pocket of the peptide binding groove, shows strong independent association with HIV-1 control (p = 4.6×10−27, OR = 5.03) [15]. Gly62 is also strongly associated with susceptibility to psoriasis (p = 2.03×10−39, OR = 3.20) but is in high LD with HLA-B Val97 and thus drops out of the final psoriasis model.
The HIV-1 Protective HLA-C 3′UTR Deletion Polymorphism rs67384697 Is Enriched in Psoriasis
Expression levels of HLA-C are modulated by the G/ - polymorphism rs67384697 located within the 3′ UTR of HLA-C, where the presence of the deletion inhibits the binding of the microRNA hsa-miR-148 to the 3′UTR and results in higher HLA-C surface expression [37]. In a multivariate model of HIV-1 control, the deletion allele of rs67384697 has a strong effect on viral control independent of the classical alleles HLA-B*57 : 01 and HLA-B*27 : 05, although the high LD of rs67384697 with other HLA-B and HLA-C alleles makes it difficult to determine whether rs67384697 (high HLA-C expression) is directly mediating this effect, or whether the HLA alleles themselves are causal [37]. Nevertheless, rs67384697 has been proposed to be the functional variant that explains the previously identified protective effect of rs9264942 on HIV-1 control, where rs9264942 is located -35 kb upstream of HLA-C and is in moderately high LD with rs67384697 (r2 = 0.74). We investigated whether rs67384697 was associated with psoriasis by imputing the deletion genotype of all psoriasis cases and controls. This was made possible by the near perfect LD between HLA-C four digit classical alleles and presence or absence of the deletion [Supplementary Table 2 in [37]]. To confirm the validity of our imputation, we sequenced the region of the HLA-C 3′UTR containing rs67384697 in a subset of our samples (n = 70) and found a concordance rate of 138/140 alleles (98.6%), indicating the imputation was robust. Using logistic regression and adjusting for sex, ancestry, and cohort, we found that deletion allele of rs67384697 was significantly associated with psoriasis (p = 1.02×10−29, OR = 1.72) (Table 5), again confirming the similarity between psoriasis patients and HIV-1 controllers. We found that the association of rs67384697 with psoriasis was largely driven by HLA-C*06 : 02, since conditioning on HLA-C*06 : 02 resulted in only a marginally significant p-value for rs67384697 (p = 0.044, OR = 1.12). We note, however, that among all HLA-C allotypes, HLA-C*06 : 02 shows the highest level of cell surface expression, which could explain, to some extent, its strong association with psoriasis.
Tab. 5. Association of rs67384697 with psoriasis and conditional analysis on HLA-C*06:02 and B*57:01. 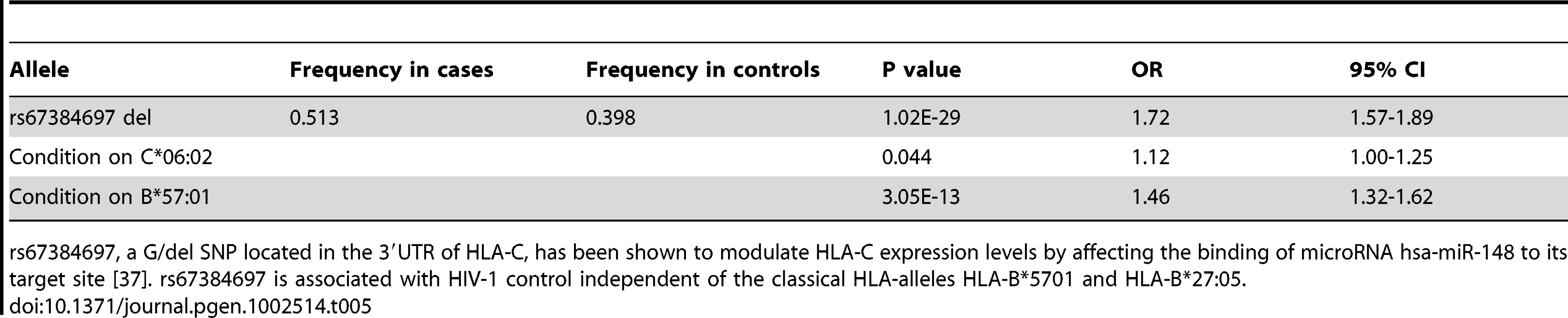
rs67384697, a G/del SNP located in the 3′UTR of HLA-C, has been shown to modulate HLA-C expression levels by affecting the binding of microRNA hsa-miR-148 to its target site [37]. rs67384697 is associated with HIV-1 control independent of the classical HLA-alleles HLA-B*5701 and HLA-B*27:05. KIR3DS1 Plus HLA-B Bw4-80I Is a Risk Factor for Psoriasis
Natural killer (NK) cells, a major component of the innate immune system, respond in the early stages of viral infection by producing cytokines and killing infected cells. NK-cell responses are regulated in part by activating and inhibitory killer immunologlubulin-like receptors (KIRs) on NK cells which engage HLA class I molecules on target cells. The activating KIR allele KIR3DS1 on chromosome 19, alone or in combination with its putative HLA-B ligand Bw4, has been associated with delayed progression to AIDS and improved HIV-1 outcomes [38]–[43]. The HLA-B Bw4 epitope can be identified by the presence of isoleucine or threonine at amino acid position 80, whereas the Bw6 epitope contains asparagine at position 80.
Our HLA data revealed that psoriasis is associated with HLA-B alleles carrying the Bw4 epitope (p = 1.28×10−25, OR = 1.66, Table S4). The association was stronger for Bw4-80I [isoleucine] (p = 8.28×10−22, OR = 1.80) than for Bw4-80T [threonine] (p = 3.41×10−4, OR = 1.22), which is interesting because Bw4-80I is thought to have a higher binding affinity for its KIR receptor than Bw4-80T [44]. We therefore hypothesized that psoriasis susceptibility might be mediated through activation of NK cells through KIR3DS1 and its putative partner HLA-B Bw4-80I. We genotyped KIR3DS1 in a subset of our psoriasis samples (n = 397) and compared the results to a healthy control cohort with available KIR3DS1 and HLA genotypes (n = 282). We found that the presence of the compound genotype KIR3DS1+Bw4-80I was a strong risk factor for psoriasis (frequency 22.7% in cases vs 6.9% in controls, p = 1.54×10−7, OR = 3.92, Table 6). Individuals positive for KIR3DS1 but lacking Bw4-80I had no increased risk for psoriasis (p = 0.63, OR = 0.91), and individuals positive for Bw4-80I but lacking KIR3DS1 had only a borderline increased risk of psoriasis (p = 0.058, OR = 1.53). To our knowledge, this is the first report that the compound genotype KIR3DS1+Bw4-80I is a strong risk factor for psoriasis susceptibility. This finding is again consistent with our observation that there is significant genetic similarity between psoriasis patients and HIV-1 controllers; however, replication of the KIR3DS1+Bw4-80I association in additional psoriasis cohorts is warranted.
Tab. 6. Association Testing of KIR3DS1 with Psoriasis. 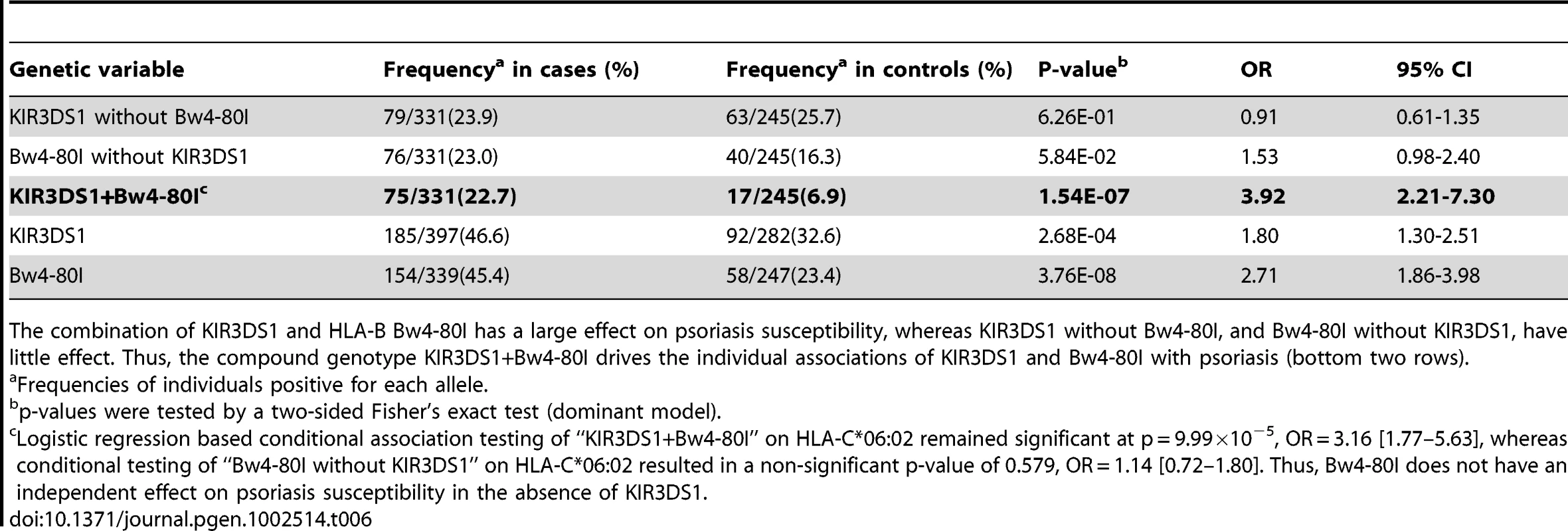
The combination of KIR3DS1 and HLA-B Bw4-80I has a large effect on psoriasis susceptibility, whereas KIR3DS1 without Bw4-80I, and Bw4-80I without KIR3DS1, have little effect. Thus, the compound genotype KIR3DS1+Bw4-80I drives the individual associations of KIR3DS1 and Bw4-80I with psoriasis (bottom two rows). Discussion
In this study, we followed up on the observation that several of the top SNPs from genome-wide association studies of psoriasis were identical to the top SNPs from genome wide association studies of HIV-1 control. Using imputation of HLA alleles, we found that psoriasis patients are enriched for several of the most significant known genetic variants associated with HIV-1 control: HLA-B*57 and HLA-B*27, which are associated with decreased viral load and increased time to AIDS [14], [15], [24]–[27]; specific amino acid residues at HLA-B positions 97, 67, and 70 that are strong markers of HIV-1 controller status and viral load [15]; the deletion SNP rs67384697 which is associated with decreased viral load independent of HLA-B*57 and HLA-B*27 [37]; and the activating KIR3DS1 allele in combination with HLA-B Bw4-80I [42]. Psoriasis patients also demonstrate a significant paucity of HLA alleles and variants associated with HIV-1 disease progression [15], [28], including HLA-B*35 (especially B*35-Px), B*07 : 02, B*40, C*04 : 01, C*07, and tyrosine 116 in HLA-B associated with sub-optimal peptide loading (Table S2, Table S4). These effects were consistent between the 3 psoriasis cohorts examined in this study, demonstrating that the effects observed were real (Table S6).
An important question to address, however, is whether the structural similarity between HLA alleles in psoriasis and HIV-1 control reflects the same underlying causal variants, or merely a coincidental association due to linkage disequilibrium. Our data suggest that some, but not all, of the observed similarity can be attributed to linkage disequilibrium. Our haplotype analysis shows that both psoriasis patients and HIV-1 controllers are enriched for the same extended haplotype, B*57 : 01–C*06 : 02–DQA1*02 : 01–DQB1*03 : 03–DRB1*07 : 01. In the HIV-1 controllers, this haplotype is likely primarily driven by selection for B*57 : 01, since previous studies have shown that the association of C*06 : 02 with HIV-1 control is dependent on B*57 : 01 in Europeans [15] and B*5801 in Africans [45], although one indirect benefit of C*06 : 02 for HIV-1 control is its high LD (D′ = 1) with the rs67384697 deletion polymorphism. In psoriasis, the haplotype association appears to be driven more by C*06 : 02 than B*57 : 01, since C*06 : 02 remains significant after conditioning on B*57 : 01 (p = 6.86×10−39, OR = 3.04) and a number of C*06 : 02 haplotypes that do not contain B*57 : 01 still remain associated with psoriasis (Table 3). In addition, the association of the deletion allele of rs67384697 with psoriasis appears to be largely driven by LD with C*06 : 02. However, it should be noted that variants in high LD with C*0602 may be contributing to the observed association signal for C*0602. Interestingly, one can take the association signal for C*06 : 02 in psoriasis and perform stepwise conditioning on all coding amino acids within C*06 : 02 to demonstrate that the coding residues do not account for the entire association signal (Table S7). Therefore, the association of C*06 : 02 with psoriasis reflects, in part, other variants in high LD with C*06 : 02.
Despite the effects of linkage disequilibrium, our data suggest that some HIV-1 control variants indeed contribute independently to psoriasis susceptibility. First, both HLA-B*57 : 01 and HLA-B*27 : 05 remain associated with psoriasis after conditioning on C*06 : 02 (B*57 : 01 p = 1.43×10−3, OR = 1.45; B*27 : 05 p = 4.83×10−4, OR = 1.52) and both remain independently associated with psoriasis in our stepwise regression model (Table 2). A previous analysis of the HLA region in psoriasis by Feng et al. [46] also found that B*57 : 01 was independent of C*06 : 02 in Caucasians; moreover, in this study B*57 : 01 was also found to be independent of C*06 : 02 in a Chinese psoriasis cohort, which is notable because the LD between C*06 : 02 and B*57 : 01 is lower in Asians (D′ = 0.41) compared to Europeans (D′ = 0.90) [47]. Prior studies have also shown that B*27 is a strong risk factor for psoriasis in the subset of patients with psoriatic arthritis, especially those with axial disease [48]–[51]. Second, linkage disequilibrium with C*06 : 02 does not explain the lower frequency of the HIV-1 progression allele B*35 in psoriasis, nor can it account for the concordance of amino acid residues at HLA-B positions 67, 70, and 97 whose association with psoriasis was shown to be independent of HLA-B*57 : 01 and HLA-C*06 : 02 (Cys67, Ser67, Lys70, Asn97, Arg97, see Figure 1. legend). Finally, the provisional association of KIR3DS1+HLA-B Bw4-80I with psoriasis cannot be due to linkage disequilibrium, because KIR3DS1 is located on chromosome 19 which segregates independently of chromosome 6.
Although B*57 : 01, B*27 : 05, and possibly B*35 may have independent effects in psoriasis, additional studies are needed to clarify the precise mechanism(s) by which these and other psoriasis-associated HLA alleles contribute to psoriasis susceptibility or protection. The observation that psoriasis patients and HIV-1 controllers display concordant amino acids within the peptide binding groove of HLA-B suggests the possibility that an unknown psoriasis antigen shares homology with HIV-1 epitopes. An alternative possibility is that B*57 : 01, B*27 : 05, and B*35 do not restrict antigen presentation in psoriasis, but primarily function through their ability or inability to activate NK cells. We have provisionally shown a strong effect of KIR3DS1+Bw4-80I on psoriasis susceptibility, and B*57 : 01 contains the Bw4-80I epitope. The second strongest HLA allele in our stepwise regression model, B*38 : 01, also contains the Bw4-80I epitope. B*27 : 05 contains the Bw4-80T epitope, while protective alleles B*35 and B*40 contain the Bw6 epitope, which do not serve as ligands for KIR. Previous studies have shown that the activating KIR allele KIR2DS1 also contributes to psoriasis or psoriatic arthritis susceptibility [52]–[55], supporting the notion that NK cells may play a role in the pathogenesis of psoriasis. Finally, we have discussed the potential role of peptide processing on susceptibility to psoriasis, with the presence of tyrosine at HLA-B position 116 associated with protection against psoriasis, where position 116 is located near the C terminus of the bound peptide. A role for peptide processing influencing psoriasis risk has been previously identified for the gene ERAP1, an amino peptidase which regulates the quality of peptides bound to MHC class I molecules through trimming the peptide N terminus [10].
The genetic similarity between psoriasis patients and HIV-1 controllers has interesting implications. On a population level, the data would predict that Caucasian individuals with psoriasis are more likely than Caucasian individuals without psoriasis to be HIV-1 controllers, and HIV-1 controllers are more likely than non-controllers to develop psoriasis. This does not imply that every individual with psoriasis will be an HIV-1 controller, since only a fraction of psoriasis patients will harbor, for example, B*57, and even the presence of B*57 does not guarantee HIV-1 control, as this allele is present in some HIV-1 progressors. Nevertheless, one would expect an enrichment of HIV-1 controllers in the psoriasis population relative to a non-psoriatic population.
Our data also suggest a hypothesis that the existence of psoriasis may represent aberrant activation of evolutionarily-derived viral control alleles [56]. Barreiro et al. have shown that several of the top MHC SNPs associated with both psoriasis and HIV-1 control reside on haplotypes which show strong evidence of recent positive selection in the genome, as evidenced by long haplotypes indicative of rapid expansion of an advantageous allele in the population [57]. Psoriasis could subsequently result from the activation of viral control alleles due to the presence of a psoriasis antigen with sequence homology to HIV-1, or due to other environmental triggers. Although this study has focused on HLA and KIR alleles, other non-MHC psoriasis genes are plausibly associated with host response to viral infection. ERAP1 is involved in class I peptide processing and demonstrates epistasis with C*06 : 02 [10] and IFIH1 encodes a cytoplasmic helicase that mediates induction of interferon response to viral RNA [10], [58]. TNIP1, TNFAIP3, TRAF3IP2, NFKBIA, and REL are associated with the TNF-α pathway and activation of NF-κB; while IL23R, IL12B, IL23A, and TYK2 are associated with activation of the Th17 pathway. Psoriasis is characterized by the upregulation of the cytokines IFN-α, IFN-γ, TNF-α, IL-17, IL-22, and IL-23 [59], while TNF-α, IFN-γ, and Th17+ T cells have been shown to be important in HIV-1 controllers [60]–[62].
The enrichment of viral control alleles in psoriasis patients may also help explain the psoriasis HIV-1 paradox (Figure 2). Psoriasis patients are more likely to harbor alleles such as HLA-B*57, HLA-B*27, HLA-C*06 (in high LD with the HLA-C 3′UTR deletion polymorphism), and KIR3DS1+Bw4-80I, theoretically resulting in vigorous cytotoxic T cell and NK cell responses upon infection with HIV-1 virus. The pro-inflammatory environment created by these anti-viral responses, resulting in the production of cytokines such as TNF-α and IFN-γ, would worsen the psoriasis. In addition, if the psoriasis antigen had sequence homology to HIV-1, then antigen specific immune responses directed against HIV-1 might cross-react with the psoriasis antigen and also flare the psoriasis. In either case, reduction of viral load and removal of the antigenic trigger through treatment with anti-retroviral therapies would improve the psoriasis, which is indeed seen clinically [17]. This explanatory model is consistent with several observational studies that patients with severe psoriasis and HIV-1 infection tend to carry the HLA-C*06 and HLA-B*27 alleles [63], [64], because such alleles would trigger the vigorous immune response associated with exacerbation of psoriasis.
Fig. 2. Proposed model of relationship between psoriasis and HIV-1 control. 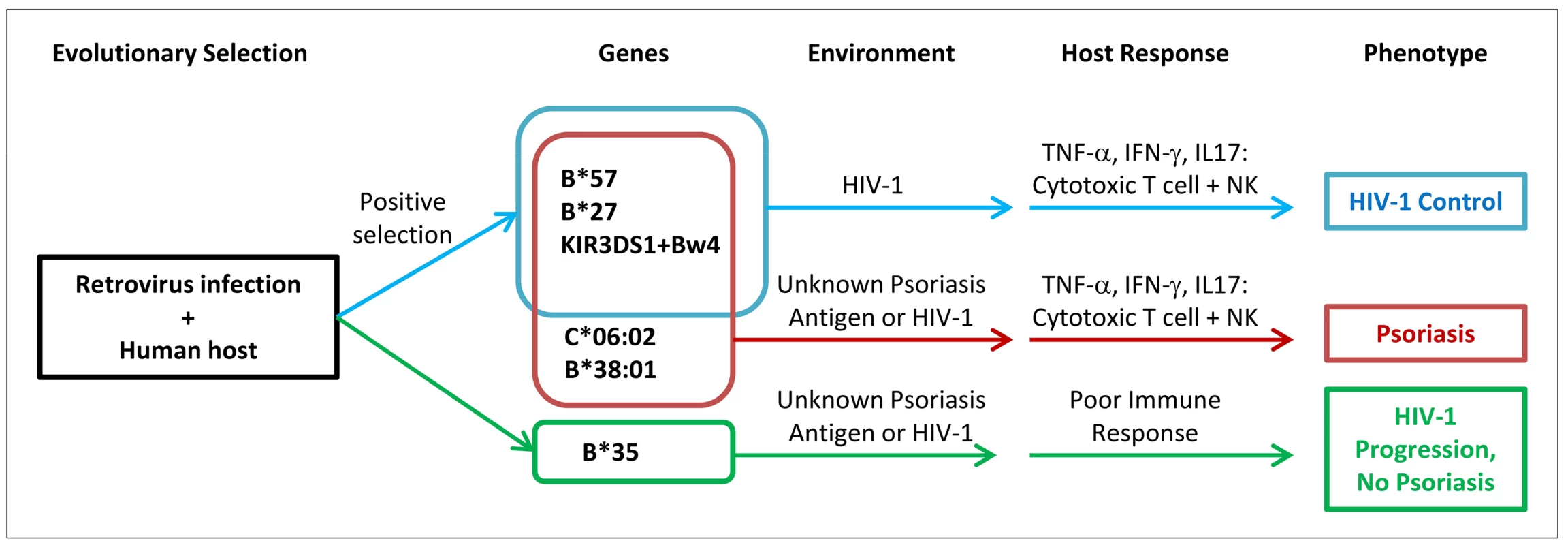
Human ancestors encountered retroviruses similar to HIV-1, leading to positive selection for viral control alleles such as B*57. Individuals who develop psoriasis are enriched for viral control alleles that are aberrantly activated by environmental triggers or unknown skin antigens (possibly sharing homology to HIV-1 epitopes). When individuals with psoriasis become infected with HIV-1, they mount vigorous cytotoxic T cell and natural killer cell immune responses leading to secretion of pro-inflammatory cytokines which worsens the psoriasis. The genetic determinants for psoriasis and HIV-1 control are overlapping, but not identical. The data presented here with psoriasis and HIV-1 control illustrate the delicate balance of the human immune response, in which processes that safeguard the body against pathogens may also engage deleterious inflammatory responses. A similar example occurs with a genetic variant in the SH2B3 gene which may be protective against bacterial infection but which increases susceptibility to celiac disease, an autoimmune disease of the gut resulting from gluten intolerance [65]. Another example can be seen with the identification of genetic variants in immune function genes that increase the risk of sepsis, a systemic inflammatory response to infection which can lead to death [66], [67].
In summary, using a large dataset of psoriasis cases and controls, we have shown that psoriasis patients and HIV-1 controllers share a high degree of similarity at their HLA loci. While some of this similarity is attributable to linkage disequilibrium, we present evidence that much of the similarity may be attributable to shared biological mechanisms including activation of natural killer cells, specificity of antigen presentation, and use of optimal MHC class I peptide processing. The genetic similarity between psoriasis and HIV-1 control suggests the possibility that psoriasis represents aberrant activation of pathways associated with anti-viral immunity. If this hypothesis is true, then the study of the biological pathways active in psoriasis may provide new therapeutic insights for the treatment of HIV-1.
Materials and Methods
Study Subjects
The study population and source are shown in Table S1. Two independent genome-wide association scan datasets were used as cohort 1 and cohort 2 in the present study. All cases and controls were of European descent. More details on subject characteristics and recruitment can be found in Liu et al. [8] and Nair et al. [9]. Only the subjects whose HLA alleles were successfully imputed (see below) were included in our analysis. In cohort 3, 169 psoriasis cases recruited from Washington University, St. Louis were directly typed for the class I HLA alleles by combining locus-specific amplification with hybridization of sequence-specific oligonucleotide probes as described in [68]; 1,711 control samples of European ancestry were obtained from studies 66 and 67 of illumina iControlDB. There was no overlap between the subjects among the three cohorts. Informed consent was obtained from each participant.
HIV Cohorts
Data generated by this study were primarily compared against a published genome-wide association study of HIV-1 control involving 516 HIV controllers of European ancestry (viral load <2,000 RNA copies/ml by three measurements over at least 12 months without antiviral therapy) and 1,196 controls (treatment-naïve chronically infected individuals with advanced disease, median viral load 61,698 copies/ml) [15]. HLA haplotype analysis was performed on 214 Caucasian HIV-1 infected individuals (52 HIV-1 controllers, 162 non-controllers) from the SCOPE cohort (Study of the Consequences of the Protease Inhibitor Era), whose HLA class I and II alleles had been previously directly genotyped. SCOPE HIV-1 controllers were antiretroviral therapy-naïve subjects who had at least one year duration of documented plasma HIV RNA below 2,000 copies/ml, while SCOPE non-controllers were subjects who had at least one documented viral load above 10,000 copies/ml.
WTCCC Data
The Wellcome Trust Case-Control Consortium data were obtained from the WTCCC official website (http://www.wtccc.org.uk/). In this study, we used Affymetrix 500 K genotyping data from approximately 2,000 samples from each of five diseases (rheumatoid arthritis, Crohn's disease, type 1 diabetes, type 2 diabetes, and coronary artery disease) and 3,000 shared control samples from the 1958 birth cohort (58C) and the National Blood Service (NBS). More details about these samples are described elsewhere [33].
KIR Analysis Cohorts
KIR3DS1 typing was performed on 397 psoriasis subjects from cohorts 2 and 3 described above. Control HLA and KIR3DS1 data were obtained from 282 healthy Caucasian blood donors from the Carrington laboratory.
Imputation and Validation of Classical HLA Alleles
The program HLA*IMP [23] was used to impute HLA loci -A, -B, -C, -DQA1, -DQB1 and -DRB1 to 4-digit resolution in our genome-wide SNP datasets. Individuals or SNPs with a missing data frequency above 0.20 were excluded as recommended in the software manual. A call threshold of 0.7 on the modes of the posterior HLA type distributions was employed, which represents a good compromise between accuracy and call rate as suggested by the author. Imputation accuracy was assessed by comparing the imputed HLA alleles to directly typed HLA class I alleles in a subset of our samples (n = 98), comprising 42 samples imputed from Illumina 300 K SNP data and 56 samples imputed from Perlegen SNP data. We found that HLA*IMP produces highly accurate HLA type imputations at HLA class I loci at the 4-digit level. The concordance for the Illumina 300 K platform was 244/252 alleles (96.8%) and the concordance for the Perlegen platform was 322/329 alleles (97.9%), for an overall concordance rate of 566/581 alleles (97.4%). To examine the imputation accuracy of infrequent/rare HLA alleles, we identified all HLA class I alleles with a population frequency of less than 5% in individuals of European descent, according to the online database: http://www.allelefrequencies.net. We then examined the accuracy of imputation at the 4 digit level for these infrequent/rare HLA alleles in our subjects for whom we had both directly genotyped HLA alleles and imputed HLA alleles. The concordance of HLA alleles with frequency <5% was 95/102 alleles (93.1%) for the Illumina 300 K platform and 116/122 alleles (95.1%) for the Perlegen platform, for an overall concordance rate of 211/224 alleles (94.2%). However, our manuscript excludes HLA alleles with frequency less than 1%. For HLA alleles with frequency greater than 1% but less than 5%, the concordance was 78/79 alleles (98.7%) for Illumina 300 K and 99/100 alleles (99.0%) for Perlegen, for an overall concordance rate of 177/179 alleles (98.9%).
Association Testing and Adjustment for Covariates
Additive logistic regression models in PLINK [69] were used for most of the association tests, except for the HLA haplotype association tests. To account for potential population stratification or admixture in these samples, principal component analyses (PCA) was performed using the EIGENSTRAT [70]. Seven PCs in cohort 1 and ten PCs in cohort 2 were used for ancestry adjustment, based on leveling off of the PCA scree plot. The principal component score for each individual was included as a covariate in all models along with cohort and gender in logistic regression models. Multivariate logistic regression was performed in R software package (http://www.r-project.org/). To examine the consistency of association signals seen in the 3 cohorts used, a heterogeneity index was calculated using the meta-analysis module in PLINK.
Stepwise Regression Modeling of Independent HLA Alleles
Conditional and stepwise logistic regression was performed using the ‘condition’ function in PLINK to determine whether independent effects existed. The method begins with an empty model to which variables are added in an iterative process as described by Barcellos et al [71]. Briefly, starting with HLA-C*06 : 02 which exhibits the strongest association with psoriasis, we conditioned candidate HLA alleles on C*06 : 02 to determine the next most significant independent effect. For the model including both class I and class II alleles, the iterative process completes when no candidate allele demonstrates p<0.0006, which corresponds to the Bonferroni correction for the 88 HLA candidate alleles with MAF>1% tested. For the model including only class I alleles, the iterative process completes when no candidate allele demonstrates p<0.00096, which corresponds to the Bonferroni correction for the 52 class I candidate alleles with MAF>1% tested.
Association Testing of Amino Acid Positions (Including WTCCC Diseases)
The amino acid sequence of all HLA alleles is completely determined by the HLA type at four-digit resolution. We used the official amino acids sequences defined for known HLA alleles [31] and our imputed HLA allele data to determine the frequency of amino acid residues in cases and controls. HLA amino acids residues were tested for association using a logistic regression model that corrects for population substructure, gender and cohort using PLINK. For amino acid positions with >2 alleles, the omnibus test in the conditional haplotype analysis module in PLINK was used to determine a single p-value for all alleles at that position.
Stepwise Regression Modeling of Amino Acid Residues
We performed stepwise logistic regression in PLINK to determine the amino acid residues that were independently associated with psoriasis, using the same approach as done with the HLA alleles. For the amino acid analysis, the algorithm completes when no remaining candidate residue has p<0.0001, which corresponds to the Bonferroni correction for the 480 HLA amino acid residues with MAF>1% tested. For the stepwise regression modeling of HLA-C 06 : 02 association signal (Table S7), the algorithm completes when no candidate residue has p<0.0006(0.05/87).
Imputation and Validation of rs67384697 Deletion Polymorphism
Since there is strong linkage disequilibrium between specific HLA-C alleles and rs67384697 [Supplementary Table 2 in [37]], we were able to determine the rs67384697 genotype of all subjects using their HLA-C four digit classical alleles. To ensure the accuracy of our imputation, we directly sequenced (ABI 3730 DNA analyzer, Quintara Biosciences, Berkeley, CA) the region of the HLA-C 3′UTR containing rs67384697 in a subset of our samples (n = 70) and determined a high concordance rate of 138/140 alleles (98.6%). The following primers were used for sequencing of genomic DNA samples: forward 5′-gtgagattctggggagctga and reverse 5′-gaacagcaactaggcacagg as specified in [37].
Haplotype Analysis
Arlequin V3.5, based on the EM algorithm, was used to estimate the frequency of HLA allele haplotypes in our psoriasis cohorts and the SCOPE HIV cohort. To ensure the accuracy of haplotype construction, we compared the haplotype frequencies generated by Arlequin to the frequencies obtained by direct counting of the phased HLA alleles output from HLA*IMP, and found the two methods to yield nearly identical results. Haplotype frequencies were tested for statistically significant differences between case and control groups using the Chi Square test or Fisher's exact test in the R software package.
KIR Genotyping
KIR3DS1 genotyping was performed by using multiplex PCR-SSP (sequence-specific priming) according to Kulkarni et al with other minor modifications [72]. Briefly, each reaction contained 15 ng of DNA, 200 µM dNTP, 1.5 mM MgCl2, 0.5 µl 10× PCR buffer, 1 µM of each primer for KIR3DS1 and KIR3DL1 and 0.8 µM of each primer for HLA-DRB1, and 0.025 µl of Platinum Taq polymerase (Invitrogen, Carlsbad, CA) in a 5 µL final volume. The polymerase chain reaction (PCR) conditions were: 3 min at 94°C; 5 cycles of 15 s at 94°C, 15 s at 65°C, 30 s at 72°C; 25 cycles of 15 s at 94°C, 15 s at 60°C, 30 s at 72°C; 4 cycles of 15 s at 94°C, 1 min at 55°C, 2 min at 72°C followed by a final 7 min extension step at 72°C. To confirm the accuracy of the results, samples were replicated using a second set of KIR3DS1 and KIR3DL1 primers from [73]. Phenotype frequencies for the presence of each gene were estimated by direct counting. Frequency differences between psoriasis and control groups were tested for significance by two-sided Fisher's exact test.
Supporting Information
Zdroje
1. KimNThrashBMenterA 2010 Comorbidities in psoriasis patients. Semin Cutan Med Surg 29 10 15
2. FarberEMNallMLWatsonW 1974 Natural history of psoriasis in 61 twin pairs. Arch Dermatol 109 207 211
3. BhaleraoJBowcockAM 1998 The genetics of psoriasis: a complex disorder of the skin and immune system. Hum Mol Genet 7 1537 1545
4. NairRPStuartPENistorIHiremagaloreRChiaNV 2006 Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet 78 827 851
5. TomfohrdeJSilvermanABarnesRFernandez-VinaMAYoungM 1994 Gene for familial psoriasis susceptibility mapped to the distal end of human chromosome 17q. Science 264 1141 1145
6. VealCDCaponFAllenMHHeathEKEvansJC 2002 Family-based analysis using a dense single-nucleotide polymorphism-based map defines genetic variation at PSORS1, the major psoriasis-susceptibility locus. Am J Hum Genet 71 554 564
7. HuffmeierUUebeSEkiciABBowesJGiardinaE 2010 Common variants at TRAF3IP2 are associated with susceptibility to psoriatic arthritis and psoriasis. Nat Genet 42 996 999
8. LiuYHelmsCLiaoWZabaLCDuanS 2008 A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet 4 e1000041 doi:10.1371/journal.pgen.1000041
9. NairRPDuffinKCHelmsCDingJStuartPE 2009 Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet 41 199 204
10. StrangeACaponFSpencerCCKnightJWealeME 2010 A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet 42 985 990
11. StuartPENairRPEllinghausEDingJTejasviT 2010 Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet 42 1000 1004
12. DeeksSGWalkerBD 2007 Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27 406 416
13. FellayJGeDShiannaKVColomboSLedergerberB 2009 Common genetic variation and the control of HIV-1 in humans. PLoS Genet 5 e1000791 doi:10.1371/journal.pgen.1000791
14. FellayJShiannaKVGeDColomboSLedergerberB 2007 A whole-genome association study of major determinants for host control of HIV-1. Science 317 944 947
15. PereyraFJiaXMcLarenPJTelentiAde BakkerPI 2010 The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330 1551 1557
16. ThomasRAppsRQiYGaoXMaleV 2009 HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet 41 1290 1294
17. MorarNWillis-OwenSAMaurerTBunkerCB 2010 HIV-associated psoriasis: pathogenesis, clinical features, and management. Lancet Infect Dis 10 470 478
18. ObuchMLMaurerTABeckerBBergerTG 1992 Psoriasis and human immunodeficiency virus infection. J Am Acad Dermatol 27 667 673
19. SadickNSMcNuttNSKaplanMH 1990 Papulosquamous dermatoses of AIDS. J Am Acad Dermatol 22 1270 1277
20. FifeDJWallerJMJeffesEWKooJY 2007 Unraveling the paradoxes of HIV-associated psoriasis: a review of T-cell subsets and cytokine profiles. Dermatol Online J 13 4
21. RoedererMDubsJGAndersonMTRajuPAHerzenbergLA 1995 CD8 naive T cell counts decrease progressively in HIV-infected adults. J Clin Invest 95 2061 2066
22. TorresBAJohnsonHM 1994 Identification of an HIV-1 Nef peptide that binds to HLA class II antigens. Biochem Biophys Res Commun 200 1059 1065
23. DiltheyATMoutsianasLLeslieSMcVeanG 2011 HLA*IMP–an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 27 968 972
24. AltfeldMAddoMMRosenbergESHechtFMLeePK 2003 Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17 2581 2591
25. CatanoGKulkarniHHeWMarconiVCAganBK 2008 HIV-1 disease-influencing effects associated with ZNRD1, HCP5 and HLA-C alleles are attributable mainly to either HLA-A10 or HLA-B*57 alleles. PLoS ONE 3 e3636 doi:10.1371/journal.pone.0003636
26. KaslowRACarringtonMAppleRParkLMunozA 1996 Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med 2 405 411
27. MiguelesSASabbaghianMSShupertWLBettinottiMPMarincolaFM 2000 HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A 97 2709 2714
28. CarringtonMNelsonGWMartinMPKissnerTVlahovD 1999 HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283 1748 1752
29. CarringtonMO'BrienSJ 2003 The influence of HLA genotype on AIDS. Annu Rev Med 54 535 551
30. GaoXNelsonGWKarackiPMartinMPPhairJ 2001 Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med 344 1668 1675
31. HLA Nomenclature. Accessed 2012 Jan 06. http://hla.alleles.org/alleles/index.html
32. RechePAReinherzEL 2003 Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol 331 623 641
33. Wellcome Trust Case Control C 2007 Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447 661 678
34. ChoJHBrantSR 2011 Recent insights into the genetics of inflammatory bowel disease. Gastroenterology 140 1704 1712
35. ElliottTWilliamsA 2005 The optimization of peptide cargo bound to MHC class I molecules by the peptide-loading complex. Immunol Rev 207 89 99
36. WilliamsAPPehCAPurcellAWMcCluskeyJElliottT 2002 Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity 16 509 520
37. KulkarniSSavanRQiYGaoXYukiY 2011 Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature 472 495 498
38. BarbourJDSriramUCaillierSJLevyJAHechtFM 2007 Synergy or independence? Deciphering the interaction of HLA Class I and NK cell KIR alleles in early HIV-1 disease progression. PLoS Pathog 3 e43 doi:10.1371/journal.ppat.0030043
39. BouletSSharafiSSimicNBruneauJRoutyJP 2008 Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS 22 595 599
40. JennesWVerheydenSDemanetCAdje-ToureCAVuylstekeB 2006 Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol 177 6588 6592
41. LongBRNdhlovuLCOksenbergJRLanierLLHechtFM 2008 Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol 82 4785 4792
42. MartinMPGaoXLeeJHNelsonGWDetelsR 2002 Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31 429 434
43. RavetSScott-AlgaraDBonnetETranHKTranT 2007 Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood 109 4296 4305
44. CellaMLongoAFerraraGBStromingerJLColonnaM 1994 NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med 180 1235 1242
45. KiepielaPLeslieAJHoneyborneIRamduthDThobakgaleC 2004 Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432 769 775
46. FengBJSunLDSoltani-ArabshahiRBowcockAMNairRP 2009 Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet 5 e1000606 doi:10.1371/journal.pgen.1000606
47. CaoKHollenbachJShiXShiWChopekM 2001 Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol 62 1009 1030
48. GladmanDDAnhornKASchachterRKMervartH 1986 HLA antigens in psoriatic arthritis. J Rheumatol 13 586 592
49. MarsalSArmadans-GilLMartinezMGallardoDRiberaA 1999 Clinical, radiographic and HLA associations as markers for different patterns of psoriatic arthritis. Rheumatology (Oxford) 38 332 337
50. RahmanPRoslinNMPellettFJLemireMGreenwoodCM 2011 High resolution mapping in the major histocompatibility complex region identifies multiple independent novel loci for psoriatic arthritis. Ann Rheum Dis 70 690 694
51. Torre AlonsoJCRodriguez PerezAArribas CastrilloJMBallina GarciaJRiestra NoriegaJL 1991 Psoriatic arthritis (PA): a clinical, immunological and radiological study of 180 patients. Br J Rheumatol 30 245 250
52. HolmSJSakurabaKMallbrisLWolkKStahleM 2005 Distinct HLA-C/KIR genotype profile associates with guttate psoriasis. J Invest Dermatol 125 721 730
53. LuszczekWManczakMCisloMNockowskiPWisniewskiA 2004 Gene for the activating natural killer cell receptor, KIR2DS1, is associated with susceptibility to psoriasis vulgaris. Hum Immunol 65 758 766
54. MartinMPNelsonGLeeJHPellettFGaoX 2002 Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J Immunol 169 2818 2822
55. WilliamsFMeenaghASleatorCCookDFernandez-VinaM 2005 Activating killer cell immunoglobulin-like receptor gene KIR2DS1 is associated with psoriatic arthritis. Hum Immunol 66 836 841
56. NogralesKEBrasingtonRDBowcockAM 2009 New insights into the pathogenesis and genetics of psoriatic arthritis. Nat Clin Pract Rheumatol 5 83 91
57. BarreiroLBQuintana-MurciL 2010 From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet 11 17 30
58. LiYLiaoWCargillMChangMMatsunamiN 2010 Carriers of rare missense variants in IFIH1 are protected from psoriasis. J Invest Dermatol 130 2768 2772
59. NestleFOKaplanDHBarkerJ 2009 Psoriasis. N Engl J Med 361 496 509
60. CicconeEJGreenwaldJHLeePIBiancottoAReadSW 2011 CD4+ T Cells, Including Th17 and Cycling Subsets, Are Intact in the Gut Mucosa of HIV-1-Infected Long-Term Nonprogressors. J Virol 85 5880 5888
61. OwenREHeitmanJWHirschkornDFLanteriMCBiswasHH 2010 HIV+ elite controllers have low HIV-specific T-cell activation yet maintain strong, polyfunctional T-cell responses. AIDS 24 1095 1105
62. SalgadoMRallonNIRodesBLopezMSorianoV 2011 Long-term non-progressors display a greater number of Th17 cells than HIV-infected typical progressors. Clin Immunol 139 110 114
63. MallonEYoungDBunceMGotchFMEasterbrookPJ 1998 HLA-Cw*0602 and HIV-associated psoriasis. Br J Dermatol 139 527 533
64. ReveilleJDConantMADuvicM 1990 Human immunodeficiency virus-associated psoriasis, psoriatic arthritis, and Reiter's syndrome: a disease continuum? Arthritis Rheum 33 1574 1578
65. ZhernakovaAElbersCCFerwerdaBRomanosJTrynkaG 2010 Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet 86 970 977
66. ArcaroliJFesslerMBAbrahamE 2005 Genetic polymorphisms and sepsis. Shock 24 300 312
67. NamathAPattersonAJ 2009 Genetic polymorphisms in sepsis. Crit Care Clin 25 835 856, x
68. HelmsCSacconeNLCaoLDawJACaoK 2005 Localization of PSORS1 to a haplotype block harboring HLA-C and distinct from corneodesmosin and HCR. Hum Genet 118 466 476
69. PurcellSNealeBTodd-BrownKThomasLFerreiraMA 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
70. PriceALPattersonNJPlengeRMWeinblattMEShadickNA 2006 Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38 904 909
71. BarcellosLFMaySLRamsayPPQuachHLLaneJA 2009 High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet 5 e1000696 doi:10.1371/journal.pgen.1000696
72. KulkarniSMartinMPCarringtonM 2009 KIR genotyping by multiplex PCR-SSP. Methods Mol Biol 612 365 375
73. HsuKCLiuXRSelvakumarAMickelsonEO'ReillyRJ 2002 Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol 169 5118 5129
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 2- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Příjem alkoholu a menstruační cyklus
- Doporučení pro diagnostiku a léčbu akutních jaterních porfyrií
- Doc. Miloš Kubánek: Nemocní se srdeční amyloidózou jsou často skryti a sledováni pod jinými diagnózami
-
Všechny články tohoto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



