-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Potential for Enhancing the Power of Genetic Association Studies in African Americans through the Reuse of Existing Genotype Data
We consider the feasibility of reusing existing control data obtained in genetic association studies in order to reduce costs for new studies. We discuss controlling for the population differences between cases and controls that are implicit in studies utilizing external control data. We give theoretical calculations of the statistical power of a test due to Bourgain et al (Am J Human Genet 2003), applied to the problem of dealing with case-control differences in genetic ancestry related to population isolation or population admixture. Theoretical results show that there may exist bounds for the non-centrality parameter for a test of association that places limits on study power even if sample sizes can grow arbitrarily large. We apply this method to data from a multi-center, geographically-diverse, genome-wide association study of breast cancer in African-American women. Our analysis of these data shows that admixture proportions differ by center with the average fraction of European admixture ranging from approximately 20% for participants from study sites in the Eastern United States to 25% for participants from West Coast sites. However, these differences in average admixture fraction between sites are largely counterbalanced by considerable diversity in individual admixture proportion within each study site. Our results suggest that statistical correction for admixture differences is feasible for future studies of African-Americans, utilizing the existing controls from the African-American Breast Cancer study, even if case ascertainment for the future studies is not balanced over the same centers or regions that supplied the controls for the current study.
Published in the journal: . PLoS Genet 6(9): e32767. doi:10.1371/journal.pgen.1001096
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001096Summary
We consider the feasibility of reusing existing control data obtained in genetic association studies in order to reduce costs for new studies. We discuss controlling for the population differences between cases and controls that are implicit in studies utilizing external control data. We give theoretical calculations of the statistical power of a test due to Bourgain et al (Am J Human Genet 2003), applied to the problem of dealing with case-control differences in genetic ancestry related to population isolation or population admixture. Theoretical results show that there may exist bounds for the non-centrality parameter for a test of association that places limits on study power even if sample sizes can grow arbitrarily large. We apply this method to data from a multi-center, geographically-diverse, genome-wide association study of breast cancer in African-American women. Our analysis of these data shows that admixture proportions differ by center with the average fraction of European admixture ranging from approximately 20% for participants from study sites in the Eastern United States to 25% for participants from West Coast sites. However, these differences in average admixture fraction between sites are largely counterbalanced by considerable diversity in individual admixture proportion within each study site. Our results suggest that statistical correction for admixture differences is feasible for future studies of African-Americans, utilizing the existing controls from the African-American Breast Cancer study, even if case ascertainment for the future studies is not balanced over the same centers or regions that supplied the controls for the current study.
Introduction
A genetic association study estimating the main effects of single nucleotide polymorphisms (SNPs) or other genetic variants upon the risk of a rare or common disease in minority populations is a setting in which it is especially attractive to consider the use of existing genotype data as a supplementary or even a primary source of controls. DNA samples may be expensive and difficult to obtain, and response rates are often lower in minority populations [1]. Researchers might consider using an already available “stand in” population sample as controls, provided that genotype frequencies are equivalent to those in the population from which controls would be drawn. There are however two immediate concerns raised, one fundamental and the other technical in nature: The fundamental question is whether or not the controls are sampled from the same underlying population (or populations) as are the cases – or more generally the feasibility and “cost” (generally loss of statistical power) of controlling for case/control differences if they arise. The technical question is whether differences in genotyping, including differences in DNA preparation, and in the actual markers genotyped in cases and controls, i.e. when the platforms are not identical so that imputation is relied upon to make up the difference, may introduce false positive (or false negative) associations.
Consider the problem of conducting a genetic association study aimed at discovering genetic variants related to the risk of a disease, where there already exists extensive genotyping data, perhaps publicly available, for members of similar populations. If the disease under consideration is rare (so that genotype frequencies for controls may be expected to be the same as in the general population) then it is intuitively appealing to consider using existing control data from studies of other rare diseases (or population-based studies if they exist) to reduce the cost or increase the statistical power of an association study. From the classical case-control literature, a study that uses 1∶ matching of controls to each of cases will be equivalent in power to a 1∶1 matched study with cases (and an equal number of controls). Thus a study with a large number of controls, , for each case will have nearly twice the effective sample size of a 1∶1 matched study [2].
Note that 1∶m matched studies (with m>1) are only cost effective if it is more costly or difficult to obtain additional cases than it is to obtain additional suitable controls since the increment in effective sample by (for example) doubling the number of case-control pairs (in a 1∶1 matching) is twice that of adding two new controls to each existing pair (to achieve 1∶3 matching) to a study even though the total number of participants is the same. If however the cost of adding an additional control is far less than that of adding an additional case (because control data is already available) then adding the almost “free” controls is highly attractive although the returns diminish as more and more controls are added, with the increment in effective sample size governed by m/(m+1).
This paper considers these issues from both theoretical and empirical perspectives. We apply a recent generalization [3]–[6] of the testing procedure of Bourgain et al [7] to the situation where population substructure (in the broad sense), including admixture and relatedness between subjects, is estimated from marker data rather than being assumed to be known as the basis for our theoretical considerations of study power. We point out below the relationship between this procedure and that of the more widely known principal components technique [8]. Our empirical investigation of the use of existing controls utilizes data from a genome-wide association study (GWAS) of breast cancer in African American women, namely the African American Breast Cancer (AABC) study, in which cases and controls come from a total of 9 different studies widely distributed geographically throughout the United States. African Americans are a relatively understudied group (compared to European Americans) in studies of genetic susceptibility. African Americans are admixed with Europeans and (in some cases) with Hispanics (themselves an admixed group) and Native Americans [9], [10]. We examine empirically the false positive rates that occur when cases from one geographical location or study within the AABC study are combined with controls from other AABC locations or studies, as well as the success (and cost in terms of loss of effective sample size) of adjustment for the observed population differences in global genetic ancestry when analyzing such illustrative data sets derived from the AABC study.
Methods
Statistical Methods
We utilize an approach derived from that of Bourgain et al [7] which has been discussed extensively in recent papers [3]–[5]. This approach for accounting for relatedness between subjects in association tests adopts a “retrospective” approach towards the problem of testing for disease associations using marker data, in which (as in the Armitage test) the allele frequency of a variant is related to case-control status. In vector notation we have of observed values for a given SNP j. Here is the total number of subjects (cases+controls) in the study, and SNP values are coded as (0,1,2) for the number of copies of a specified allele, , carried by subject . (This coding of SNP genotype implies that we are interested in additive models for the relationship between disease risk and genotype but the approach can readily be extended to other codings). The retrospective approach models the mean of as a function of case-control status. If we define the design matrix C to have rows (1 ci) where ci is case-control status (0 or 1) for subject i then the mean, , of is written asRelatedness between subjects induces a covariance matrix for the number of copies of a given SNP of form(1)with specific to each SNP but with the same matrix for all SNPs. In fact, for known pedigree relationships, and unrelated founders, this matrix has diagonal elements equal to where is the inbreeding coefficient for subject and each off-diagonal element, , is twice the kinship coefficient for the relationship between subjects and [7].
It is worth noting that in general the topic of relatedness includes what is often considered to be population substructure. For example consider two large but isolated populations (freely mixing within each population) that have been separated for many generations. While a random sample of people from the same population (with sample size small relative to the population size) might be considered unrelated to each other when considering that population separately, when considering the two populations together people from one isolated population are considered to be related to each other, relative to those in the other population. In particular, genetic markers will, through a process of random drift and other factors, be able to distinguish members from the two populations, and this will be detectable when calculation of the matrix is performed. A standard method of simulating genetic markers for divergent populations stemming from the same ancestral population (e.g. the Balding-Nichols model [11]) can readily be shown to produce covariance matrices of the form of expression (1).
If and are both known then the best linear unbiased estimate (BLUE) of the regression vector is of weighted least squares formand the variance covariance matrix of the estimates is in the form ofThus inference on the significance of the allele frequency difference between cases and controls may be based upon the Wald test statistic(2)with the (2,2) element of
In general of course, and are not known, except in the case of known pedigrees and unrelated founders, where can be computed from first principles. The estimation of using marker data has been considered by a number of authors and both method of moments [4], [5] and maximum likelihood methods [3] have been considered. A method of moments estimator of can be concisely written [5] as(3)and the estimate of asOne value of this approach, which is exploited here, is that it is relatively easy to compute the power of the Wald test if we can hypothesize a form of the relatedness matrix . For a given form for (below we consider several forms for both isolated population models and more complex admixed populations) then for a given sample size, , a given allele frequency for a causal SNP, and a hypothesized difference in allele frequencies between cases and controls (which can then be related to odds ratios in typical case/control analysis) we can compute the non-centrality parameter of (and hence the power of the test) as(4)We illustrate the computation of this non-centrality parameter for a number of important special cases in the results section below. It is worth noting now, however, that the Bourgain test appears to be reasonably powerful compared to other procedures, and can sometimes be considered as a compromise between the principal components method [8] and genomic control [12]. We attempt to justify this last statement in the results section below.
In addition to the Bourgain test we used several well known tools for addressing population structure in the AABC data. For example we computed eigenvectors through the use of the program EIGENSTRAT [8]. Briefly, each eigenvector explains a proportion of the genetic variation among samples in the analysis so that the leading eigenvector explains the greatest variation, followed by the second eigenvector, and so forth. The full set of eigenvectors form an orthonormal basis so that each eigenvector is scaled on the unit interval and linearly independent from all other eigenvectors. Note that the EIGENSTRAT procedure is operating on the same estimated matrix, , that we have described above.
To assess ancestry within the AABC study in relation to reference populations from HapMap, we performed a principal components analysis based on ancestry informative markers that were genotyped in both the AABC study and the HapMap Phase 3 populations. The 2,546 ancestry informative markers (contained within the Illumina 1M genotyping array which was used in the AABC scan) were selected based on low inter-marker correlation and high correlation to a previously determined eigenvector that explained African and European ancestry.
We quantified percent African ancestry for each of the nine AABC study populations by running the program STRUCTURE for each study population. The program implements a Markov Chain Monte Carlo algorithm that provides the posterior estimates of the proportion of ancestry from each of k clusters for each individual, where k is specified by the investigator. For each AABC study population, we assigned k = 3, including genotypes from the same ancestry informative markers used in PCA genotyped in YRI, CEU, and JPT from HapMap Phase 3.
Studies in AABC
AABC included 9 epidemiological studies of breast cancer among African American women, which comprise a total of 3,153 cases and 2,831 controls. Below is a brief description of these studies.
The Multiethnic Cohort Study (MEC)
The MEC is a prospective cohort study of 215,000 men and women in Hawaii and Los Angeles [1] between the ages of 45 and 75 years at baseline (1993–1996). Through December, 31 2007, a nested breast cancer case-control study in the MEC included 556 African American cases (554 invasive and 12 in situ) and 1,003 African American controls. An additional 178 African American breast cancer cases (ages: 50–84) diagnosed between June 1, 2006 and December 31, 2007 in Los Angeles County (but outside of the MEC) were combined with the MEC samples in the analysis.
The Los Angeles component of The Women's Contraceptive and Reproductive Experiences (CARE) Study
The NICHD Women's CARE Study is a large multi-center population-based case-control study that was designed to examine the effects of oral contraceptive (OC) use on invasive breast cancer risk among African American women and white women ages 35–64 years in five U.S. locations [13]. Cases in Los Angeles County were diagnosed from July 1, 1994 through April 30, 1998, and controls were sampled by random-digit dialing (RDD) from the same population and time period; 380 African American cases and 224 African American controls were included in stage 1 of the scan.
The Women's Circle of Health Study (WCHS)
The WCHS is an ongoing case-control study of breast cancer among women of European and African descent residing in the New York City boroughs (Manhattan, the Bronx, Brooklyn and Queens) and in seven counties in New Jersey (Bergen, Essex, Hudson, Mercer, Middlesex, Passaic, and Union) [14]. Eligible cases included women diagnosed with invasive breast cancer between 20 and 74 years of age; controls were identified through RDD. The WCHS contributed 272 invasive African American cases and 240 African American controls to stage 1 of the GWAS.
The San Francisco Bay Area Breast Cancer Study (SFBC)
The SFBC is a population-based case-control study of invasive breast cancer in Hispanic, African American and non-Hispanic White women conducted between 1995 and 2003 in the San Francisco Bay Area [15]. African American cases, ages 35–79 years, were diagnosed between April 1, 1995 and April 30, 1999, with controls identified through RDD. Stage 1 included 172 invasive African American cases and 231 African American controls from SFBC.
The Northern California Breast Cancer Family Registry (NC–BCFR)
The NC-BCFR is an on-going population-based family study conducted in the Greater San Francisco Bay Area, and is one of 6 sites collaborating in the Breast Cancer Family Registry (BCFR), an international consortium funded by NCI [16]. African American breast cancer cases in NC-BCFR were diagnosed after January 1, 1995 and between the ages of 18 and 64 years; population controls were identified through random digit dialing (RDD). Stage 1 genotyping was conducted for 440 invasive African American cases and 53 African American controls.
The Carolina Breast Cancer Study (CBCS)
The CBCS is a population-based case-control study conducted between 1993 and 2001 in 24 counties of central and eastern North Carolina [17]. Cases were identified by rapid case ascertainment system in cooperation with the North Carolina Central Cancer Registry and controls were selected from the North Carolina Division of Motor Vehicle and United States Health Care Financing Administration beneficiary lists. Participants' ages ranged from 20 to 74 years. For stage 1, DNA samples were provided from 656 African American cases with invasive breast cancer and 608 African American controls.
The Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) Cohort
PLCO, coordinated by the U.S. National Cancer Institute (NCI) in 10 U.S. centers, enrolled during 1993–2001 approximately 155,000 men and women, aged 55–74 years, in a randomized, two-arm trial to evaluate the efficacy of screening for these four cancers [18]. A total of 64 African American invasive breast cancer cases and 133 African American controls contributed to stage 1 of the GWAS.
The Nashville Breast Health Study (NBHS)
The NBHS is a population-based case-control study of incident breast cancer conducted in the Tennessee [19]. The study was initiated in 2001 to recruit patients with invasive breast cancer or ductal carcinoma in situ, and controls, recruited through RDD between the ages of 25 and 75 years. NBHS contributed 310 African American invasive cases (57 in situ), and 186 African American controls to the stage 1 analysis.
Wake Forest University Breast Cancer Study (WFBC)
African American breast cancer cases and controls in WFBC were recruited at Wake Forest University Health Sciences from November 1998 through December 2008 [20]. Controls were recruited from the patient population receiving routine mammography at the Breast Screening and Diagnostic Center. Age range of participants was 30–86 years. WFBC contributed 125 cases (116 invasive and 9 in situ) and 153 controls to the stage 1 analysis.
Genotyping
Genotyping in stage 1 was conducted using the Illumina Human1M-Duo BeadChip. Of the 5,984 samples from these studies (3,153 cases and 2,831 controls), we attempted genotyping of 5,932, removing samples (n = 52) with DNA concentrations <20 ng/ul by pico green assay. After clustering the genotype data we removed samples based on the following exclusion criteria: 1) unknown replicates (≥98.9% genetically identical) that we were able to confirm (only one of each duplicate was removed, n = 15); 2) unknown replicates that we were not able to confirm through discussions with study investigators (pair or triplicate removed, n = 14); 3) samples with call rates <95% after a second attempt (n = 100); 4) samples with ≤5% African ancestry (n = 36) (discussed below); and, 5) samples with <15% mean heterozygosity of SNPs in the X chromosome and/or similar mean allele intensities of SNPs on the X and Y chromosomes (n = 6) (these are likely to be males).
In the analysis, we removed SNPs with <95% call rate or minor allele frequencies (MAFs) <1%. To assess genotyping reproducibility we included 138 replicate samples; the average concordance rate was 99.95% (>99.93% for all pairs). We also eliminated SNPs with genotyping concordance rates <98% based on the replicates. The final analysis dataset included 3,016 cases and 2,745 controls, with an average SNP call rate of 99.7% and average sample call rate of 99.8%. Hardy-Weinberg equilibrium (HWE) was not used as a criterion for removing SNPs for this analysis.
Results
Non-Centrality Parameter for the Bourgain Test in the Case of Isolated Populations
We use the Balding Nichols model [11] for allele frequency differences between isolated populations. In this model allele frequencies for a SNP in modern data populations are distributed according to a beta distribution with the ancestral allele frequency of that SNP. In this model the variance of the modern day allele frequency is , thus is a parameter specifying the degree of separation between the modern day and ancestral population. As described in Rakovski and Stram 2009 [4] if genotypes are obtained for randomly sampled individual from two modern day isolated populations using this model and the separation of each modern day population from the ancestral population equals (for ) statistic then the covariance matrix, between subjects for the jth SNP will have diagonal terms equal to for members of the first population, diagonal terms equal to for members of the second, off diagonal terms of , , or zero for pairs of individuals who are either both from the first population, both from the second population or from different populations respectively. Here is the frequency in the ancestral population of SNP . Consider now a study in which all cases come from one isolated population, and all controls from another. Assume for simplicity that (both populations have the same degree of separation from their ancestral source), and that the number of cases and controls are both equal to so that total sample size is (the calculations below can be readily altered for different matching fractions if necessary). Thus we can write the variance of the estimator of the case-control difference for SNP aswith the first column of C being a vector of 1's and the second column of C a vector of 1's and 0's indicating case-control (and population) status. Using a readily derived formula for the inverse of an matrix of compound symmetric formwe can easily write(5)Thus the non-centrality parameter, = of a test of association does not increase linearly in N, but rather is bounded above by the value . This can impose severe limitations on the power of any study in which there are such differences. To put this in perspective, consider two isolated populations which are each separated from their ancestral population with an F value of 0.0005, and consider an allele that exhibits 40 percent frequency in the ancestral population. The variance of the difference between the two isolated modern day populations in the frequency of this allele is equal to so that we would expect by chance that there is a about a 1.5 percent difference in allele frequencies between cases and controls for such an allele. Consider now the detection, in a study of 5,000 cases and 5,000 controls, of a disease-causing allele of the same frequency associated with a 10 percent difference in allele frequencies between cases and controls ( = .2). The difference in allele frequencies is approximately 6 times larger than expected due to population differences, and can be seen to correspond to an odds ratio for disease, under a multiplicative risk model, of 1.5 per copy of the risk allele. From equation (2) the non-centrality parameter will be equal to 34.73 in this case; on the other hand if between cases and controls is 0 the non-centrality parameter will equal 208.33. Thus a study that would, given no differences between cases and controls in population of origin, have overwhelming power (>.9999) to reject the null hypothesis at a genome wide level significance (p<) is, under this alternative, reduced to having power of only 56 percent after correcting for the differences in origins of cases and controls. The survey of European populations by Nelis et al [21] estimates fixation indices, , (which can be equated to under the Balding Nichols model) between populations in SNP allele frequencies which range from less than .001 between neighboring populations to 0.023 for Southern Italy versus parts of Finland. Because our values as defined above are between present day and ancestral populations the fixation indices calculated between present day populations by Nelis et al need to be multiplied by ½ to be consistent with our definition of . Thus the example we have given () corresponds only to the nearest neighbor populations in Europe and would appear to throw into doubt any thought of using control data not perfectly matched in ancestry to cases.
Admixed Populations
While the calculations given above appear to be pessimistic regarding the usefulness of shared control data it is important to note that the completely isolated population model is naïve and makes assumptions not applicable to the study subjects for the AABC study or indeed for most modern populations. Therefore we broaden our discussion to admixed populations, specifically, when the DNA from both cases and controls come from groups that are admixed from the same two ancestral populations. We consider this in two parts, first deriving results for comparisons between “completely” admixed populations, i.e. where the two populations have different levels of admixture between the ancestral populations, but when there is no within-population heterogeneity in ancestry. Next we focus on the much more realistic setting of incompletely admixed populations serving as cases and controls.
Completely admixed populations
Here we consider each of two populations of interest (one supplying cases and the other controls) as consisting of randomly mating populations derived by admixture from the same two ancestral populations. In the first modern day population we assume that 100 percent of the ancestors are from the first ancestral population and the remaining 100 percent are from the other ancestral population. In the second population the fractions are and respectively. We further assume that the two ancestral populations are themselves derived from a single earlier ancestral population and that the values of these populations compared to the earlier populations are both equal to the same value. Under these simplifying assumptions (which can be readily relaxed if need be) we can easily derive the covariance matrix for a SNP vector between subjects to have elements
Var() = for subject in the first population (i.e. cases)
Cov() = for both subjects and in the first population
Var() = for subject in the second population (i.e. controls)
Cov() = for both subjects and in the second population and finally
Cov() = for subjects and in different populations
With sampling of N cases from the first modern day admixed population and N controls from the second after repeated use of the matrix formulae for the inverse of a compound symmetric matrix we can compute the variance term for as the [2,2] element of the variance covariance matrix, ,(6)asThis expression is bounded from below by the quantity .
We see that this bound is zero only if either or if . The non-centrality parameter will (as in the previous example) then again be bounded from above. Consider for example two groups of “completely” admixed modern-day subpopulations one with 25 percent of ancestry from one ancestral population (and 75 percent from the other population) and the other with 20 percent ancestry from the first ancestral population (and 80 percent from the second). If we take the fixation index, value of 0.153 of Nelis et al for (applicable for HapMap Yorubans versus HapMap Europeans) and the allele frequency and odds ratios as above (40 percent and 1.5 per copy respectively) then the non-centrality parameter will be equal to 72.77 compared to 208.33 in the non-mixed case, leading again to loss of power although not as extreme as in the previous example.
Incompletely admixed groups
The above example is still unrealistic because we have assumed that all members within each of the two study populations (cases and controls) have exactly the same fraction of ancestry coming from the two differing ancestral groups: from population 1 for the cases and from population 1 for the controls. This is not what is seen in most complex population groups today, and is not what we see in the AABC data considered below. Instead in modern admixed populations there is a relatively wide range of within-population admixture proportions. If there is overlap due to ancestry for cases and controls then tests like the Bourgain test, principal components, multidimensional scaling [22], and genomic matching [23], [24] have a much greater likelihood of success than in the previous examples. Again we use the Bourgain test to quantify this. For example we can assume a distribution of values of the admixture fraction in the case and control samples while keeping a difference “on average” in ancestry between the two groups. We then can calculate “appropriate” values of the matrix K and use this in additional calculations. For our purposes the beta distribution provides a reasonably flexible model for the distribution of individual values of the admixture proportion in a modern day admixed population. Figure 1 gives a plot of the density function for a number of beta distributions. The beta distribution is usually described in terms of two parameters, and with the mean of the beta distribution equal to and the variance equal to . For convenience we re-parameterize this distribution in terms of its mean, , and a heterogeneity parameter, , with , so that . Thus samples from this distribution have mean and variance . This parameterization is useful when comparing the overlap of two beta distributions with different means but similar within group variances as in Figure 1.
Fig. 1. Plot of the density function of beta distributions parameterized by mean and heterogeneity factor . 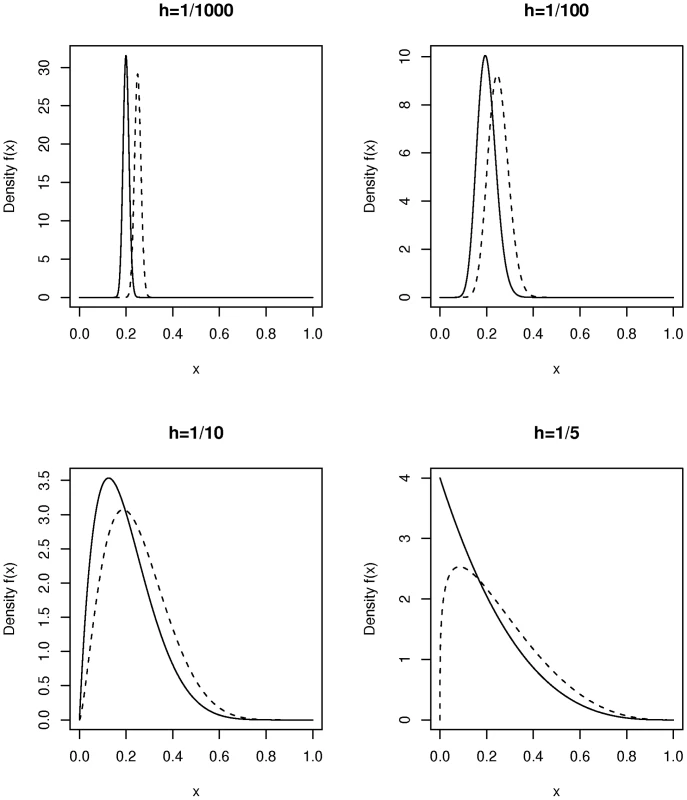
On each subplot the density is shown for two choices of namely = 0.2 (solid line) and = 0.25 (dotted lines). We consider now the non-centrality parameter for the Bourgain test when two incompletely admixed populations are used as cases and controls where there is an overall difference in admixture fraction in the two populations but where each population is heterogeneous with respect to the fraction of ancestry from the two mixing populations. Figure 2 plots the non-centrality parameter (NCP) for the Bourgain test against the number, , of cases and controls (1∶1 case-control ratio for pairs) for the example used above (a risk variant of frequency 40 percent overall, and a OR for disease equal to 1.5 per copy) it is clear from the figure that the non-centrality parameter is very much determined by the degree of heterogeneity in admixture percentage within each population (cases versus controls) as parameterized here by .
Fig. 2. Plot of non-centrality parameter for the Bourgain test for a case-control study using two incompletely admixed populations as sources of cases and controls respectively. 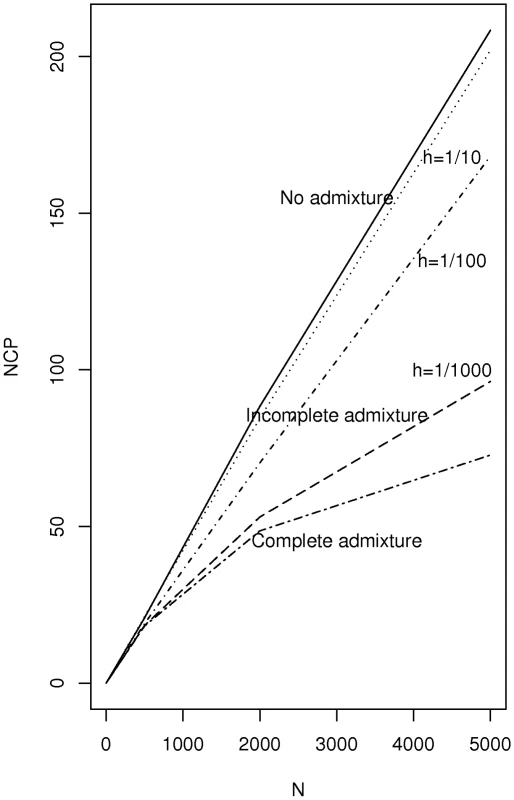
The parameters chosen refer to a test of a variant associated with disease which has 40 percent overall allele frequency and which is associated with a 10 percent difference in frequency between cases and controls (OR = 1.5 per copy). Cases are assumed to have average admixture percentage of 20 percent and controls 25 percent. The within population heterogeneity is specified by a single common heterogeneity parameter as used in Figure 1. AABC Study Results
Our analysis focuses upon (1) estimating a more appropriate model for the distribution of ancestry in the data from the AABC data than the homogeneous “complete” admixture described above, (2) checking the adequacy of this model, enriching it if necessary, and (3) describing the implications of the model for the likely power to detect associations in studies in which all cases come from outside the AABC study populations, and controls are chosen from within the AABC. We can partly mimic such studies by making up “pseudo” case-control studies using the data from the different study sites within the AABC study.
Principal components analysis of the AABC study
We used the EIGENSTRAT program to calculate the eigenvectors and eigenvalues of the matrix given in expression (3) using the set of 2,457 AIMs described above. We first noted that the first eigenvalue of was much larger than the remaining eigenvalues (504.85 compared to 29.71 for the next largest). We strongly suspected that the eigenvector associated with this eigenvalue corresponds to the fraction of European ancestry based on CEU of HapMap. To clarify this we ran the program STRUCTURE on AABC study data and included HapMap genotype data (CEU, YRI, and CBT+YRI) for these same AIMs. The first eigenvector of the AABC data was highly correlated (r = 0.991) with the estimate of STRUCTURE for the percentage of European ancestry. For illustrative purposes we added the HapMap subject's data to the EIGENSTRAT analysis and plotted the first 4 eigenvectors in Figure 3.
Fig. 3. Principal components plots of AABC and selected HapMap samples. 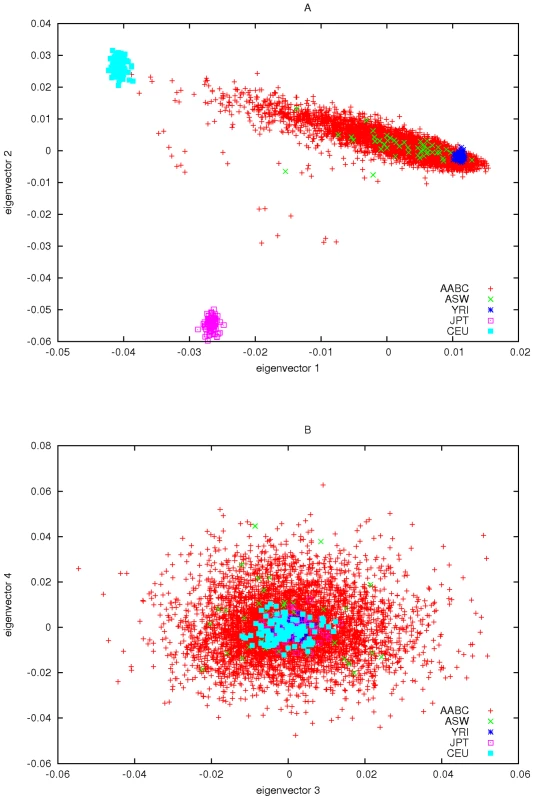
Variation among AABC study sites in admixture percentage
Figure 4 shows box plots of the STRUCTURE estimates of the fraction of European ancestry according to AABC study site. For each study site, we estimated the mean and variance of the distribution and related those by the method of moments to the two parameters ( and ) in our parameterization described above. The estimate of average European ancestry varied significantly by study, from approximately 0.25 for the MEC cohort and other studies in California to approximately 0.19 for the CBCS and other studies in the East and South-east United States. The heterogeneity parameter also varied by site but was generally estimated to be close to 1/7 for all sites, which indicated a large degree of overlap between the admixture fractions even between the most different studies.
Fig. 4. Plot of estimate of proportion of African ancestry from STRUCTURE by participating AABC study. 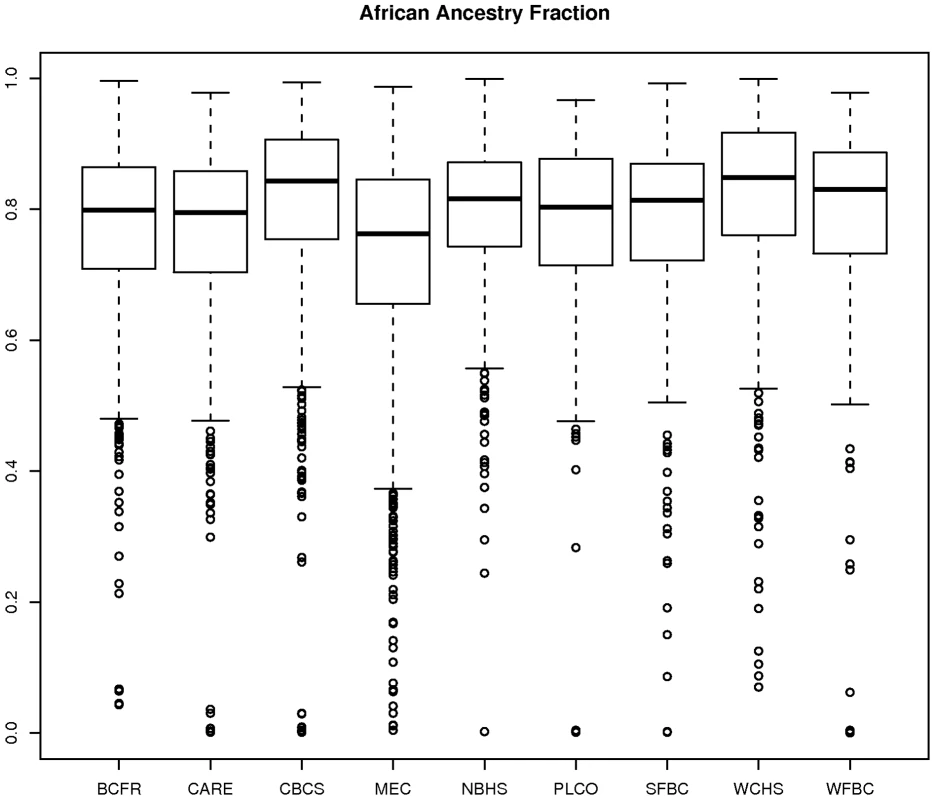
Other population structure in the AABC study
If the only ancestry differences among the control populations sampled by the AABC study relates to fraction of European versus African ancestry then our previous analysis would indicate that very little loss of power, compared to using perfectly matched controls, would result from a case-control study that uses African American cases, and one (or all) of the AABC control samples as convenience controls, after adjustment for differences in European versus African ancestry. Figure 3 gives evidence for some degree of admixture with an Asian as well as European group, which we assume [10] is largely due to admixture with Native American or Hispanic populations. The requirement of controlling for this additional admixture will also affect the power of using the AABC study as a source of shared controls, as would other types of admixture or population stratification. In order to quantify the total effect of the significant eigenvectors in the AABC data we did the following: Using the Tracy-Widom statistic as incorporated into EIGENSTRAT we estimated the number of “significant” eigenvalues (i.e. those significantly larger than the remaining eigenvalues) and hence the “significant” eigenvectors. Using these significant eigenvectors we formed a “smoothed” version of the matrix as(7)where . Here is the number of significant eigenvectors of , the , are the eigenvectors and is the associated eigenvalue. Formula (7) gives a full rank estimate which has the same first eigenvectors and eigenvalues as as well as the same trace (sum of all N eigenvalues). We use this to estimate non-centrality parameters for several example studies, based on the AABC data. We found that approximately the first 200 eigenvalues were nominally significant (at a 5 percent type I error rate) using the Tracy-Widom statistic and considered the effect of adjusting for a range of 1 to all 200 eigenvectors in our calculations. Fewer than all 200 eigenvectors likely to needed in a realistic analysis, here we are examining an extreme case. The fact that this many eigenvectors are nominally significant may be due in part to not controlling for multiple comparisons or may also be reflective of unsuspected close or distant relatedness between certain participants.
Specifically we considered a breast cancer study in which all African-American cases ( = 635) came from the CBCS and all controls ( = 990) came from the Los Angeles component of the MEC. Accordingly we extracted the 1625 by 1625 submatrix (corresponding to these selected cases and controls) of for use in the calculation of noncentrality parameters. The basic idea is to calculate using the estimate of for this study, compared to a study of the same size but where cases and controls are sampled from the same non-admixed population. Because the appropriate and is not known, but is estimated from the data, our analysis of power becomes slightly more complicated than above. For a given SNP, , we estimated the variance of by the scalar(8)The expected value of this variance estimate can be written as(9)The expected value depends upon the details of admixture and other types of population structure in this subset of AABC participants and can be written as(10)In a study of homogeneous non-admixed case and control populations, and under the null hypothesis that is zero, this reduces to where is the allele frequency of marker . Substituting expression (10) into expression (9) with either = (for a homogeneous study) or for (for our example) allows us to compare the variance of the estimators when a total of eigenvectors are adjusted for in our hypothesized case-control study, compared to the homogeneous study. Figure 5 plots the values of expression (9) for varying values of for a marker with frequency 20 percent. It appears that correcting for increasing numbers of eigenvectors (at least beyond the first two components) has relatively little effect on and hence on study effective sample size and power. Even using all 200 “significant” eigenvectors in the calculation of the smoothed estimator only increases the variance of the estimator by about 7 percent relative to a study that needs no correction. Of course we also need to confirm that the false positive rate for the Bourgain test is properly controlled for by using a particular number, , of eigenvectors. Figure 6 gives a quantile-quantile (QQ) plot of the () p-values from tests of association of each of the AIMs used to estimate K with case-control status in our hypothesized study, while using either 0 (uncorrected), 1 (the CEU – YRI component), 10, or 200 eigenvectors to form . Here the uncorrected analysis is very highly over-dispersed with a very large number of associations globally significant (using the Bonferonni test). However even using just 1 eigenvector appears to give a reasonably adequate control of the type I error while correcting for 200 gives nearly perfect control, with little loss of additional power (as indicated in Figure 5).
Fig. 5. Plot of variance. 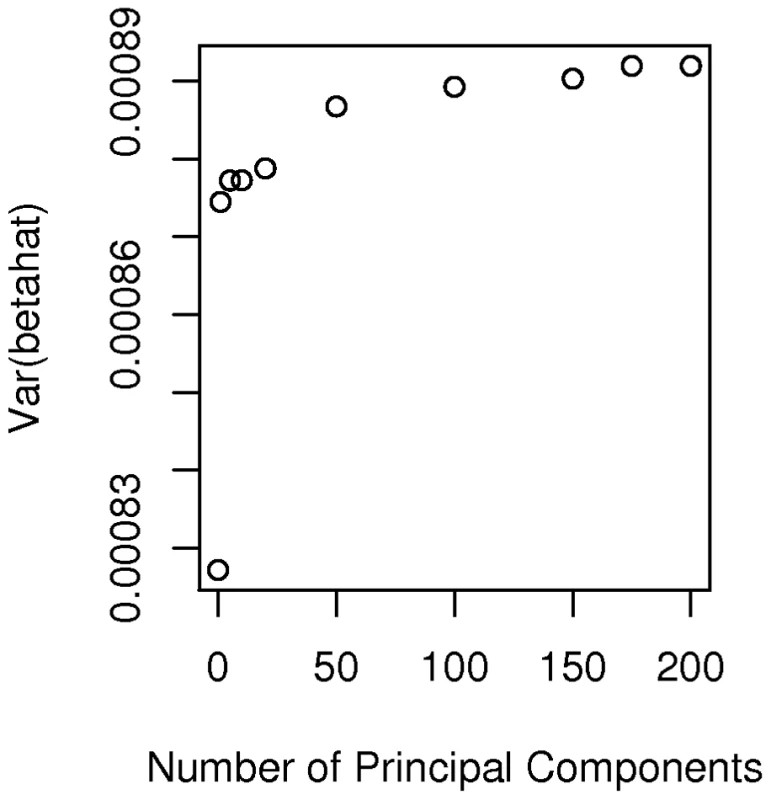
Plot of the variance of according to the number, , of eigenvectors adjusted for in the Bourgain test of association. Fig. 6. Quantile-quantile plot of p-values from association tests in the hypothesized case-control study in which cases from the CBCS and controls from the MEC are used. 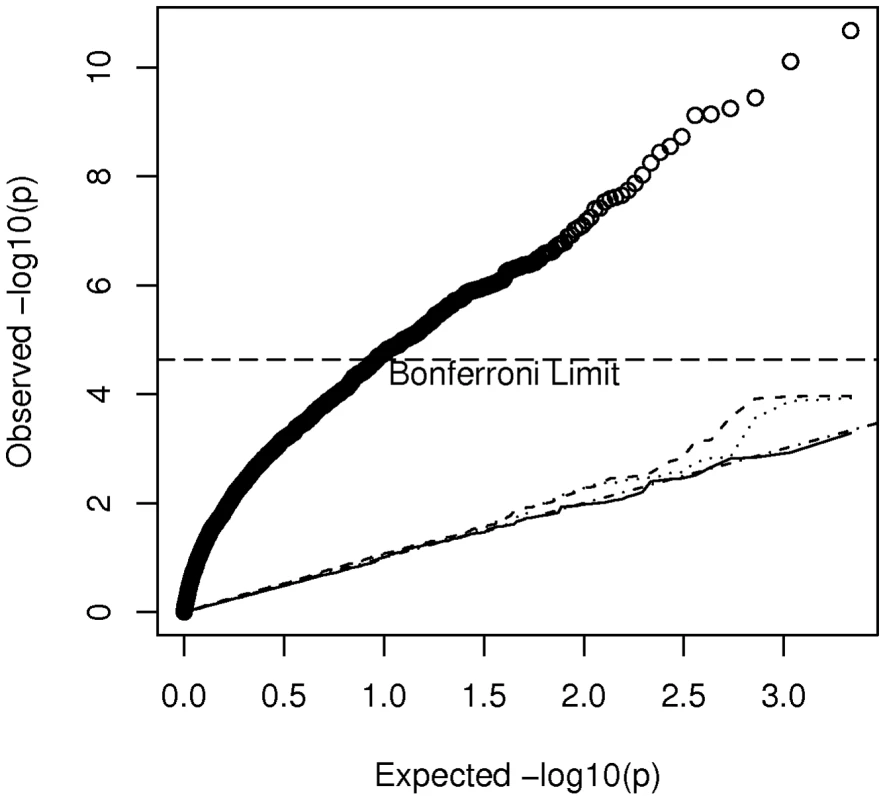
The plotted values indicate adjustment for 0 (uncorrected heavy solid line), 1 (dashed line), 10 (dotted line) and 200 (thin solid line) eigenvectors, by using these components in the calculation of . Discussion
We have adopted a somewhat non-standard approach in relying upon the Bourgain test rather than principal components [8] or related methods [22]–[24] to control for population structure in a GWAS of a minority population with cases/controls drawn from multiple studies with different designs and recruitment approaches. We have done this mainly because we can give certain theoretical results for the Bourgain test when assuming specific forms for the true kinship matrix using this procedure. It is worth noting that the Bourgain test can be regarded as a random effects version of the usual principal components method. In particular the Bourgain model can be alternatively described as a model for the mean of conditional upon all eigenvectors of as(10)Now consider the coefficients as being independent random effects with mean zero and variances equal to times the associated eigenvalues, of . Averaging over all the random will yield the unconditional mean and variance covariance matrix . Our “smoothed” estimate, , of is motivated by expression (10), and choosing a value of is analogous to choosing the number of eigenvectors to be used as fixed effects by EIGENSTRAT. The random effects framework also highlights the relationship between the Bourgain procedure and the genomic control method of Devlin and Roeder [25]. In genomic control one additional parameter that governs the dispersion of the test statistic is used to assess the association between and case-control status. In the smoothed version of the Bourgain test introduced here, we choose a total of such parameters.
We have shown that if cases and controls come from genetically distinct populations but ones that have only recently diverged (so that the parameter is very small) then some limited power remains to detect true marker associations so long as the true value of is very large compared to the “typical” differences between cases and controls seen with the other markers. This is also analogous in interpretation to the genomic control method. However our explicit description of the upper bound on the noncentrality parameter of the association test for such a study clearly shows the limits of this design, and by implication, the limits of the genomic control procedure as well. Basically neither of these two methods behaves “properly” from a statistical point of view as sample size increases, i.e. the non-centrality parameter under an alternative hypothesis (and hence power) does not increase correspondingly. In genomic control the overdispersion parameter that the procedure corrects for increases with sample size, while for the Bourgain test the noncentrality parameter is bounded from above.
For case-control studies involving two or more similarly admixed populations that differ in admixture fraction, the key issue in assessing the power of a study using cases from one population and controls from another is in determining the within-population heterogeneity of the admixture fractions, relative to the between population differences in average admixture. If the within-population heterogeneity is small then the situation is equivalent to the case of isolated populations, i.e., there will be a bound on the power of a study to detect an effect with the bound determined by the upper limit on the noncentrality parameter as a function of as computed above.
Despite the concerns raised in our theoretical considerations, in our assessment of the observed marker data from the AABC study we tentatively conclude that reuse of controls data from this study in future work may be statistically feasible. While there are clear differences in average admixture fraction between studies these are dwarfed by the within-study heterogeneity. Other signs of hidden structure in the AABC studies (as evidenced by additional eigenvalues which are significant by the Tracy-Widom test) do not appear to have a very large impact (Figure 5) on the power of our hypothetical study using the CBCS and MEC cases and controls respectively. Control for the first few (1–200 in our case) eigenvectors appears to dramatically reduce false positive associations with very little power loss (about a 7 percent reduction in effective sample size) relative to studies of homogeneous sets of cases and controls. We have used a specific set of ancestry informative markers in our analysis but the existence of genome-wide data for the AABC allows for considerable latitude in selecting SNPs to control for admixture, and even randomly selected SNPs, if enough are considered, can be used for admixture correction. Our use of the Bourgain test when considering the feasibility of a particular study design allows us to consider noncentrality parameters (and hence power) in particularly simple and helpful ways. While we have focused on the Bourgain method to correct for admixture differences in the AABC study our specific finding (that little loss in power is anticipated when re-using control data from this study) is likely to apply also to fixed-effects methods such as treating principal components or STRUCTURE estimates of percentage ancestry from ancestral populations as covariates. Our reasoning is based upon the close relationship between the principal components methods and the random effects rationale for the Bourgain test as given in equation (10) and also on the high correlation seen between STRUCTURE estimates of African ancestry in the AABC study and the first eigenvector from principal components.
Zdroje
1. KolonelLN
HendersonBE
HankinJH
NomuraAM
WilkensLR
2000 A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol 151 346 357
2. RothmanKJ
GreenlandS
1998 Modern Epidemiology Philadelphia Lippincott-Raven
3. ChoiY
WijsmanEM
WeirBS
2009 Case-control association testing in the presence of unknown relationships. Genet Epidemiol 33 668 678
4. RakovskiCS
StramDO
2009 A kinship-based modification of the Armitage trend test to address hidden population structure and small differential genotyping errors. PLoS One 4 e5825 doi:10.1371/journal.pone.0005825
5. AstleW
BaldingDJ
2010 Population structure and cryptic relatedness in genetic association studies. Statistical Science In press
6. ThorntonT
McPeekMS
2010 ROADTRIPS: Case-Control Association Testing with Partially or Completely Unknown Population and Pedigree Structure. Am J Hum Genet 86 172 184
7. BourgainC
HoffjanS
NicolaeR
NewmanD
SteinerL
2003 Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am J Hum Genet 73 612 626
8. PriceAL
PattersonNJ
PlengeRM
WeinblattME
ShadickNA
2006 Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38 904 909
9. CooperRS
TayoB
ZhuX
2008 Genome-wide association studies: implications for multiethnic samples. Hum Mol Genet 17 R151 155
10. CampbellMC
TishkoffSA
2008 African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet 9 403 433
11. BaldingD
NicholsR
1995 A method for quantifying differentiation between populations at multi-allelic locus and its implications for investigating identify and paternity. Genetica 3 3 12
12. DevlinB
RoederK
WassermanL
2001 Genomic control, a new approach to genetic-based association studies. Theoretical Population Biology 60 155 166
13. MarchbanksPA
McDonaldJA
WilsonHG
BurnettNM
DalingJR
2002 The NICHD Women's Contraceptive and Reproductive Experiences Study: methods and operational results. Ann Epidemiol 12 213 221
14. AmbrosoneCB
CiupakGL
BanderaEV
JandorfL
BovbjergDH
2009 Conducting Molecular Epidemiological Research in the Age of HIPAA: A Multi-Institutional Case-Control Study of Breast Cancer in African-American and European-American Women. J Oncol 2009 871250
15. JohnEM
SchwartzGG
KooJ
WangW
InglesSA
2007 Sun exposure, vitamin D receptor gene polymorphisms, and breast cancer risk in a multiethnic population. Am J Epidemiol 166 1409 1419
16. JohnEM
HopperJL
BeckJC
KnightJA
NeuhausenSL
2004 The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res 6 R375 389
17. NewmanB
MoormanPG
MillikanR
QaqishBF
GeradtsJ
1995 The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat 35 51 60
18. ProrokPC
AndrioleGL
BresalierRS
BuysSS
ChiaD
2000 Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials 21 273S 309S
19. ZhengW
CaiQ
SignorelloLB
LongJ
HargreavesMK
2009 Evaluation of 11 breast cancer susceptibility loci in African-American women. Cancer Epidemiol Biomarkers Prev 18 2761 2764
20. SmithTR
LevineEA
FreimanisRI
AkmanSA
AllenGO
2008 Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 29 2132 2138
21. NelisM
EskoT
MagiR
ZimprichF
ZimprichA
2009 Genetic structure of Europeans: a view from the North-East. PLoS One 4 e5472 doi:10.1371/journal.pone.0005472
22. LeeAB
LucaD
KleiL
DevlinB
RoederK
2010 Discovering genetic ancestry using spectral graph theory. Genet Epidemiol 34 51 59
23. LucaD
RingquistS
KleiL
LeeAB
GiegerC
2008 On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am J Hum Genet 82 453 463
24. GuanW
LiangL
BoehnkeM
AbecasisGR
2009 Genotype-based matching to correct for population stratification in large-scale case-control genetic association studies. Genet Epidemiol 33 508 517
25. DevlinB
RoederK
1999 Genomic control for association studies. Biometrics 55 997 1004
Štítky
Genetika Reprodukční medicína
Článek Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator ofČlánek Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline AnalysisČlánek Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoAČlánek Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.Článek Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 9- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Příjem alkoholu a menstruační cyklus
- Hysteroskopická resekce děložního septa zlepšuje šanci na graviditu žen s jinak nevysvětlenou infertilitou
-
Všechny články tohoto čísla
- Optimal Strategy for Competence Differentiation in Bacteria
- Mutational Patterns Cannot Explain Genome Composition: Are There Any Neutral Sites in the Genomes of Bacteria?
- Frail Hypotheses in Evolutionary Biology
- Genetic Architecture of Complex Traits and Accuracy of Genomic Prediction: Coat Colour, Milk-Fat Percentage, and Type in Holstein Cattle as Contrasting Model Traits
- Allelic Variation at the 8q23.3 Colorectal Cancer Risk Locus Functions as a Cis-Acting Regulator of
- Allelic Selection of Amplicons in Glioblastoma Revealed by Combining Somatic and Germline Analysis
- Germline Variation Controls the Architecture of Somatic Alterations in Tumors
- Mice Doubly-Deficient in Lysosomal Hexosaminidase A and Neuraminidase 4 Show Epileptic Crises and Rapid Neuronal Loss
- Analysis of Population Structure: A Unifying Framework and Novel Methods Based on Sparse Factor Analysis
- FliO Regulation of FliP in the Formation of the Flagellum
- Cdc20 Is Critical for Meiosis I and Fertility of Female Mice
- dMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells
- DCAF26, an Adaptor Protein of Cul4-Based E3, Is Essential for DNA Methylation in
- Genome-Wide Double-Stranded RNA Sequencing Reveals the Functional Significance of Base-Paired RNAs in
- An Immune Response Network Associated with Blood Lipid Levels
- Genetic Variants and Their Interactions in the Prediction of Increased Pre-Clinical Carotid Atherosclerosis: The Cardiovascular Risk in Young Finns Study
- The Histone H3K36 Methyltransferase MES-4 Acts Epigenetically to Transmit the Memory of Germline Gene Expression to Progeny
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Lactic Acidosis Triggers Starvation Response with Paradoxical Induction of TXNIP through MondoA
- Identification of Early Requirements for Preplacodal Ectoderm and Sensory Organ Development
- Orphan CpG Islands Identify Numerous Conserved Promoters in the Mammalian Genome
- Analysis of the Basidiomycete Reveals Conservation of the Core Meiotic Expression Program over Half a Billion Years of Evolution
- ETS-4 Is a Transcriptional Regulator of Life Span in
- The SR Protein B52/SRp55 Is Required for DNA Topoisomerase I Recruitment to Chromatin, mRNA Release and Transcription Shutdown
- The Baker's Yeast Diploid Genome Is Remarkably Stable in Vegetative Growth and Meiosis
- Chromatin Landscape Dictates HSF Binding to Target DNA Elements
- The APETALA-2-Like Transcription Factor OsAP2-39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice
- Accurately Assessing the Risk of Schizophrenia Conferred by Rare Copy-Number Variation Affecting Genes with Brain Function
- Widespread Over-Expression of the X Chromosome in Sterile F Hybrid Mice
- The Characterization of Twenty Sequenced Human Genomes
- The Genome of a Pathogenic : Cooptive Virulence Underpinned by Key Gene Acquisitions
- A Single Element Maintains Repression of the Key Developmental Regulator
- Identification of New Genetic Risk Variants for Type 2 Diabetes
- Effect of Correlated tRNA Abundances on Translation Errors and Evolution of Codon Usage Bias
- Evidence of Selection upon Genomic GC-Content in Bacteria
- Proteomic Changes Resulting from Gene Copy Number Variations in Cancer Cells
- Rice a Cinnamoyl-CoA Reductase-Like Gene Family Member, Is Required for NH1-Mediated Immunity to pv.
- Longitudinal Genome-Wide Association of Cardiovascular Disease Risk Factors in the Bogalusa Heart Study
- Response to Mechanical Stress Is Mediated by the TRPA Channel Painless in the Heart
- DNMT3L Modulates Significant and Distinct Flanking Sequence Preference for DNA Methylation by DNMT3A and DNMT3B
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
- Incremental Genetic Perturbations to MCM2-7 Expression and Subcellular Distribution Reveal Exquisite Sensitivity of Mice to DNA Replication Stress
- Loss of Maternal ATRX Results in Centromere Instability and Aneuploidy in the Mammalian Oocyte and Pre-Implantation Embryo
- Comparative Genomic Hybridization (CGH) Reveals a Neo-X Chromosome and Biased Gene Movement in Stalk-Eyed Flies (Genus )
- Differentiation of Zebrafish Melanophores Depends on Transcription Factors AP2 Alpha and AP2 Epsilon
- Gene–Environment Interactions at Nucleotide Resolution
- Dementia Revealed: Novel Chromosome 6 Locus for Late-Onset Alzheimer Disease Provides Genetic Evidence for Folate-Pathway Abnormalities
- Critical Functions of Rpa3/Ssb3 in S-Phase DNA Damage Responses in Fission Yeast
- Preferential Re-Replication of Heterochromatin in the Absence of Geminin
- The Potential for Enhancing the Power of Genetic Association Studies in African Americans through the Reuse of Existing Genotype Data
- Evidence That Mutation Is Universally Biased towards AT in Bacteria
- Perturbation Analysis of Heterochromatin-Mediated Gene Silencing and Somatic Inheritance
- Diversity of Eukaryotic DNA Replication Origins Revealed by Genome-Wide Analysis of Chromatin Structure
- Genetic Deletion of the Desmosomal Component Promotes Tumor Microinvasion in a Mouse Model of Pancreatic Neuroendocrine Carcinogenesis
- The Metabolic Enzyme ManA Reveals a Link between Cell Wall Integrity and Chromosome Morphology
- SNPs Associated with Cerebrospinal Fluid Phospho-Tau Levels Influence Rate of Decline in Alzheimer's Disease
- Synthesizing and Salvaging NAD: Lessons Learned from
- A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis
- An Insect Herbivore Microbiome with High Plant Biomass-Degrading Capacity
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Synthesizing and Salvaging NAD: Lessons Learned from
- Optimal Strategy for Competence Differentiation in Bacteria
- Long- and Short-Term Selective Forces on Malaria Parasite Genomes
- Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



