-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDegradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche
It is thought that the ability to degrade PDZ domain containing proteins is a hallmark of oncogenic papillomaviruses. However, since papillomaviruses did not evolve to be oncogenic, this hypothesis does not address the evolutionary importance of this phenotype. The present manuscript attempts to address whether HPV induced degradation of PDZ containing proteins is associated with oncogenic potential as determined by the clinical/epidemiological empirical cancer risk. Using Bayesian approaches to model trait evolution we show that it is highly unlikely for a virus to become oncogenic without first acquiring the ability to degrade PDZ proteins. Furthermore, the ability to degrade PDZ proteins allowed ancestral viruses to colonize a new cellular niche. However, in order to thrive in this new environment, these ancestral viruses had to acquire additional functions. We hypothesize that some of these additional phenotypes lead to oncogenicity. Importantly, our study illustrates the power of combining epidemiological, biochemical and evolutionary data with phylogenetic analysis in attempting to understand the relative role of specific pathogen phenotypes with host pathogenesis.
Published in the journal: . PLoS Pathog 11(6): e32767. doi:10.1371/journal.ppat.1004980
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004980Summary
It is thought that the ability to degrade PDZ domain containing proteins is a hallmark of oncogenic papillomaviruses. However, since papillomaviruses did not evolve to be oncogenic, this hypothesis does not address the evolutionary importance of this phenotype. The present manuscript attempts to address whether HPV induced degradation of PDZ containing proteins is associated with oncogenic potential as determined by the clinical/epidemiological empirical cancer risk. Using Bayesian approaches to model trait evolution we show that it is highly unlikely for a virus to become oncogenic without first acquiring the ability to degrade PDZ proteins. Furthermore, the ability to degrade PDZ proteins allowed ancestral viruses to colonize a new cellular niche. However, in order to thrive in this new environment, these ancestral viruses had to acquire additional functions. We hypothesize that some of these additional phenotypes lead to oncogenicity. Importantly, our study illustrates the power of combining epidemiological, biochemical and evolutionary data with phylogenetic analysis in attempting to understand the relative role of specific pathogen phenotypes with host pathogenesis.
Introduction
Papillomaviruses (PVs) are a diverse family of dsDNA viruses infecting most, if not all, amniotes. Based on nucleotide similarities, PVs are classified into genera identified by Greek letters. A genus is further divided into numbered species [1,2]. Persistent infection with specific human papillomaviruses (HPVs) has been shown to be necessary for the induction of cervical carcinoma [3,4]. All established oncogenic HPV types (OTs) belong to the genus Alphapapillomavirus [5]. Of note, phylogenetically, these oncogenic HPV types cluster into a so-called high-risk (HR) clade, indicating an evolutionary relationship between these viruses [5,6]. Importantly, available epidemiological data suggests that while some HPV types (e.g. HPV16) within this HR clade are strongly associated with cancer, others (e.g. HPV68) are only oncogenic in rare cases [7]. Throughout papillomavirus evolution PVs continuously adapted to new ecological niches on the host. This process selected for PVs with specific phenotypes needed to interact with changing cellular environments. It is highly unlikely that the ability to cause malignancy provided certain PVs with an evolutionary advantage. This suggests that the Alphapapillomaviruses acquired a particular combination of phenotypes while adapting to a specific ecological niche. In some viruses, the resulting cellular insult may inadvertently drive the infected host cell towards transformation. We previously postulated that use of phylogenetic, epidemiological and biochemical analyses would be essential for the identification of viral phenotypes specifically associated with oncogenicity [5,8,9,10].
Most papillomaviruses express at least 7 proteins, two of which—E6 and E7—have been demonstrated to be sufficient for oncogenesis. To date, the exact mechanisms by which these viral proteins cause cellular transformation is unknown. The viral E6 and E7 proteins interact with a diverse set of cellular pathways. Some of these interactions have been proposed to be unique to oncogenic viruses, while others appear to be shared by all investigated types [reviewed in 10,11,12,13,14,15,16].
It is well established that the E6 protein from specific HPVs targets PDZ containing proteins for degradation [17]. PDZ domains represent an abundant class of protein interaction modules that target specific motifs on partner proteins. PDZ containing proteins regulate multiple biological processes including differentiation and the maintenance of cellular polarity [17,18,19,20,21]. The interaction with PDZ containing proteins is dependent on a canonical class I PDZ binding motif (PBM) at the extreme C-terminal of the E6 protein [22]. Several E6 proteins interact with the PDZ-protein MAGI1 (a member of the membrane-associated guanylate kinase (MAGUK) protein family) [23], and MAGI1 is highly sensitive to degradation by E6 proteins [24,25]. However, only HPVs containing a type I PBM are able to degrade MAGI1 in vitro, and it has been stated that only oncogenic types contained such a motif [26], implying that interactions with PDZ containing proteins are critical for HPV-induced oncogenesis [27].
In the present paper, we provide evidence that all members of the HR-HPV clade (i.e., α-5, -6, -7, -9 and -11 species groups) indeed contain a class I PBM. Nevertheless, there does not appear to be a correlation between the presence of this motif and oncogenic classification. We also present data that all tested HR types, regardless of their oncogenic potential, degrade a human MAGI protein (hMAGI1d). Importantly, the design of our study (in which we tested E6 proteins from viruses representing all known species within the genus Alphapapillomavirus) allowed us to evaluate this phenotype from an evolutionary perspective. The derived evolutionary model demonstrates that the capability to degrade hMAGI1 entered the viral-host relationship prior to the oncogenic phenotype. In addition, while PDZ protein degradation is not sufficient for cancer development, the model suggests that it enabled the evolution of a phenotype associated with cell transformation. These data further support the notion that biological processes are best understood once their evolutionary origin is taken into account [28].
Materials and Methods
Ethics statement
The patient samples used in this study have been IRBB approved. (IRB number: 2009–274; Approval date: 07/08/2009; Title: Epigenetic Profiling of Cervical Cancer: A Pilot Study.)
Cell lines and constructs
The C-33A cell line is an HPV negative cervical cancer cell line expressing a mutant p53 protein [29,30] and was obtained from the ATCC (Manassas, VA, USA). The C-33A cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). Western blot analysis
C-33A cells were plated in 6-well plates (800,000 cells/well) and transfected with 1400 ng pQCXIN-HA-hMAGI1d, 600 ng of each pQCXIN based E6 expression vector and 100 ng pSUPER-GFP (transfection control) using lipofectamine LTX (Invitrogen) according to the manufacturer’s instructions. The cells were lysed in RIPA buffer (Millipore, Billerica, MA, USA), containing appropriate protease inhibitors. Equal amounts of protein lysates (10 ug) were separated by SDS-PAGE. Western blot membranes were probed with antibodies recognizing the HA-tag (Cell Signaling, Danvers, MA, USA), eGFP (Cell Signaling) or actin (Sigma).
Epidemiological classification and HPV type selection
Epidemiological classification of HPVs is based on a recent review of available world-wide reported data as conducted by an expert panel [7]. E6 proteins were selected to include at least one representative for each species within the genus Alphapapillomavirus. In addition, closely related viruses with differences in epidemiological classification were selected. The isolation and cloning of E6 ORFs has been described previously [8,9].
Phylogenetic tree construction
E6 sequences representing all viral types within the genus Alphapapillomavirus were downloaded from the PaVE database (http://pave.niaid.nih.gov/#home)[31]. The DNA sequences were translated to amino acids and aligned using MAFFT (the L-INS-I algorithm was used) [32], and the corresponding nucleotide coding regions were aligned within the Seaview program [33]. Bayesian tree reconstruction was performed using MrBayes [34,35] while implementing the GTR+I+G model (as selected by Jmodeltest2 [36,37]; S1 Table). To ensure the most efficient Bayesain analysis the convergence of the Markov chain Monte Carlo (MCMC) chains was explored graphically using AWTY [38]. The final posterior sample of trees is summarized as a majority rule consensus tree in Fig 1.
Fig. 1. Presence of C-terminal PDZ binding motif is correlated with phylogenetic classification. 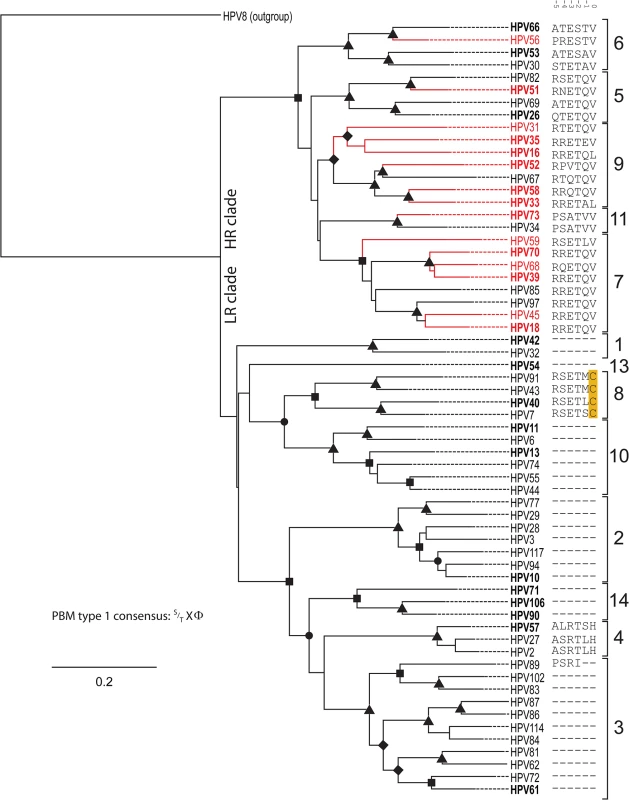
The Bayesian phylogenetic tree is constructed based on the E6 nucleotide sequences of all known human Alphapapillomaviruses. HPV8, a Betapapillomaviruses was used to root the tree. The solid branches indicate the optimized branch lengths. Dotted lines were added to the branches to facilitate visual inspection of the tree. Epidemiological classification divides the tree into two main clades (“high-risk” vs.”low-risk”). Oncogenic papillomavirus types are colored red (classification based on [7]). Posterior probability values are indicated using symbols at the nodes (triangle = 1; rectangle > 0.95; circle>0.90; diamond >0.80). The numbers to the right indicate the different viral species within the genus Alphapapillomaviruses. The sequence of the six C-terminal residues constituting a putative type 1 PBM is indicated following the virus name. A dash (“-“) indicates the absence of a residue at that position. Numbers above the sequences allow for easy identification of landmark residues as in [42]. Viruses in Bold were selected for the in vivo analyses. Analysis of trait evolution
The viral traits under study (MAGI1 degradation and oncogenic potential) were coded as discrete presence/absence variables. Clades for which no epidemiological and/or biochemical data were available were coded as missing, and were considered ambiguous by the program. The computer program BayesDiscrete [39] (available from http://www.evolution.rdg.ac.uk) was used to detect evidence of correlated evolution between discrete traits. In order to account for phylogenetic uncertainty during the tree building process, 500 trees were randomly sampled from the post-burn-in posterior sample of MrBayes trees. The reversible jump MCMC analysis was run for 1x109 iterations. After a burn-in of 1x106 iterations, the chains were sampled every 10,000th iteration. The alpha and beta parameters of a Gamma distribution were seeded from uniform distributions on the interval 0 to 2. To confirm the robustness of the results, the analysis was performed three independent times. Bayes factors were used to select between models [39]. Traditionally, a log Bayes Factor greater than 30 is considered as positive evidence for the model being tested.
Results
PDZ domain binding motifs are evolutionary conserved in members of the high-risk clade within the genus Alphapapillomavirus
In an attempt to understand differences in oncogenic potential, researchers have traditionally compared viral phenotypes between the prototypical HR and LR types (HPV16/HPV18 vs. HPV6/HPV11) [13]. However, these viruses infect different anatomical niches on the human body and are separated by approximately 30 million years of evolutionary divergence, confounding simple comparison [10]. Phylogenetic analysis (Fig 1), clustered the members of the genus Alphapapillomvirus into two main clades [10]. Epidemiological evidence has shown that only a subset of the viruses in this HR clade (highlighted in red) is actually associated with cervical cancer [7,40,41]. The alignment of all known Alphapapillomavirus E6 proteins (see panel to the right of Fig 1; 6 C-terminal residues are shown) illustrates that the presence of a canonical type 1 PBM (S/TXΦ where Φ indicates any hydrophobic residue [42]) represents a synapomorphy of the high-risk papillomaviruses. In other words, all HR viruses, independent of associated oncogenic risk have a PDZ interacting domain. While most viruses in the LR clade do not contain comparable residues, HPV types belonging to the Alphapapillomavirus 8 species contain an E6 protein with a similar motif. The sequence analysis suggests that these alpha-8 E6 proteins may be able to target PDZ containing proteins for degradation. Thus, the E6 proteins from several non-oncogenic viruses contain putative PBMs, suggesting that the presence of a type 1 PBM does not allow for dichotomization between oncogenic and non-oncogenic HPV types. However, the presence of a sequence motif does not establish that these E6 proteins actually target PDZ proteins for degradation. Therefore, we examined the effects of E6 protein expression on steady state levels of a human PDZ-protein in vivo.
The E6 proteins from high-risk papillomaviruses induce degradation of a PDZ containing protein
In order to test whether different E6 proteins affect the steady state levels of a human PDZ protein, a novel isoform of MAGI was cloned from cervical cells (S1 Methods). We tested the effects of co-expressing HPV E6 proteins with the PDZ containing human protein hMAGI1d (Fig 2A). The figure shows a representative gel of the obtained results. These data demonstrate that all members of the HR-clade (highlighted in red) degrade hMAGI1d when compared to the empty vector control (pQCXIN). In addition, HPV40 E6, a representative of the alpha-8 species group, a viral type embedded in the LR-clade also degraded hMAGI1d in this system (Fig 2 and S1 Fig). Thus, the alpha-8 PBM (Fig 1) is associated with hMAGI1d degradation. It has been shown that the HPV E6 proteins target PDZ-proteins for proteasomal degradation [43,44]. We confirmed that the tested E6 proteins (including HPV40 E6) degrade human MAGI1d in a proteasome dependent manner (S1 Fig). HPV oncogenic potential and MAGI1d degradation is contrasted using mirrored trees in Fig 2B. The left tree is shaded according to epidemiological classification, while the tree on the right indicates the ability of HPV types to degrade MAGI1 as shown in Fig 2A. While this data indicates incoherence between both PDZ degradation and oncogenic activity, this hypothesis is formerly tested in the next paragraph.
Fig. 2. All members of the phylogenetic high-risk clade degrade human MAGI1d. 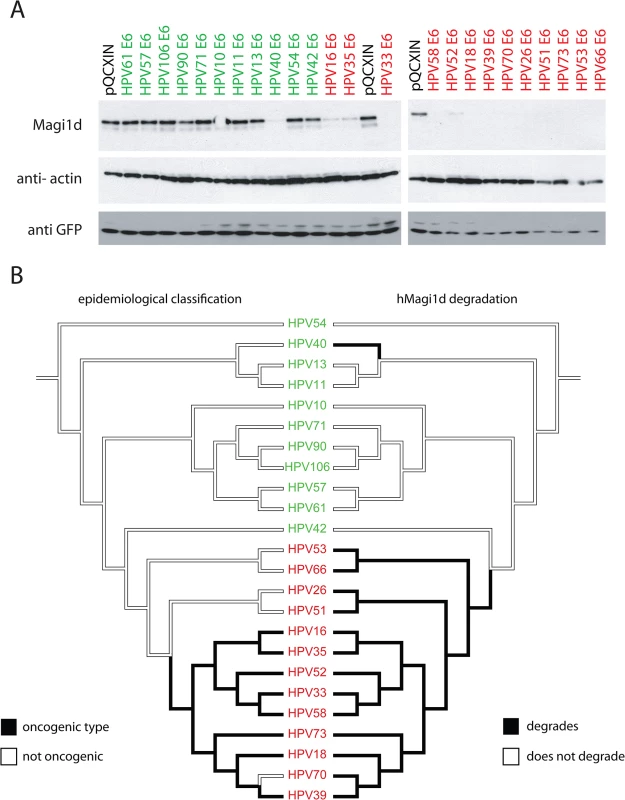
(A) C-33A cells were transfected with 24 different E6 proteins covering the known evolutionary spectrum within the Alphapapillomavirus genus. The western blot shows a representative experiment. GFP was probed as a transfection control indicating equal transfection. This figure shows that all High-Risk types (highlighted in red) target hMAGI1d for degradation. The pQCXIN vector was used as control. (B) Mirror trees comparing the epidemiological (left) and PDZ-protein degradation (right) phenotypes on the E6 based phylogeny. The viral names are colored according to phylogenetic classification. High-risk viruses are colored in red, while LR viruses are colored green. The branches of the tree are shaded according to the state of each character under investigation. hMAGI1d degradation by HPV E6 proteins represents an enabling phenotype towards oncogenicity
The main goal of this study was to test whether degradation of PDZ containing proteins was associated with oncogenic potential as determined by the clinical/epidemiological empirical cancer risk [7]. The biochemical degradation of PDZ-proteins and viral oncogenic potential were coded as discrete variables, and tested to determine whether these pairs of discrete binary traits evolved together. Briefly, the test of correlated evolution, as implemented within the BayesTraits package [39], compares the fit of two models of evolution. In the independent model both traits are allowed to evolve independently on the tree. The dependent model requires that both traits evolve in a correlated fashion. In order to incorporate phylogenetic uncertainty we integrated the analysis over 500 trees randomly selected from the posterior distribution. We employed a reversible-jump (RJ) MCMC methodology [39]. The RJ-MCMC has the advantage that it travels through the posterior tree sample and tests all possible models of evolution in proportion to the likelihood of each individual model. This method provides a posterior sample (n = 100,000) of models with their associated transition rates. We obtain support for each model through the calculation of Bayes Factors based on the frequency with which each model occurs in the posterior sample compared to the prior expectation of observing that model (i.e., the ratio of posterior to prior odds) [45]. Bayes Factors are interpreted on a heuristic scale. Traditionally, Bayes Factors greater than 30 are considered very strong evidence in favor of one model. To ensure reproducibility of the RJ-MCMC analysis, the analysis was performed three independent times. Fig 3A shows the top 5 models selected by the RJ-MCMC approach. The selected model (“0ZZ0ZZ0Z”; Fig 3A) has a Bayes Factor of 130.41 (+/ - 1.99) demonstrating strong support for this model. We also reconstructed the ancestral combination of phenotypes at important nodes of the phylogenetic tree. This suggested that the most recent common ancestor (MRCA) of all high and low-risk Alphapapillomaviruses did not degrade PDZ proteins and was not oncogenic (probability P(0,0) = 0.79; Fig 3B, left panel). Likewise, the MRCA of the LR clade did not degrade PDZ proteins. However, the ancestor of the extant HR viruses acquired the ability to degrade PDZ proteins. Importantly, the analysis suggests that this ancestral HR virus would not have been oncogenic (P(0,1) = 0.74; Fig 3B, middle panel).
Fig. 3. Degradation of PDZ proteins is an enabling phenotype towards oncogenicity. 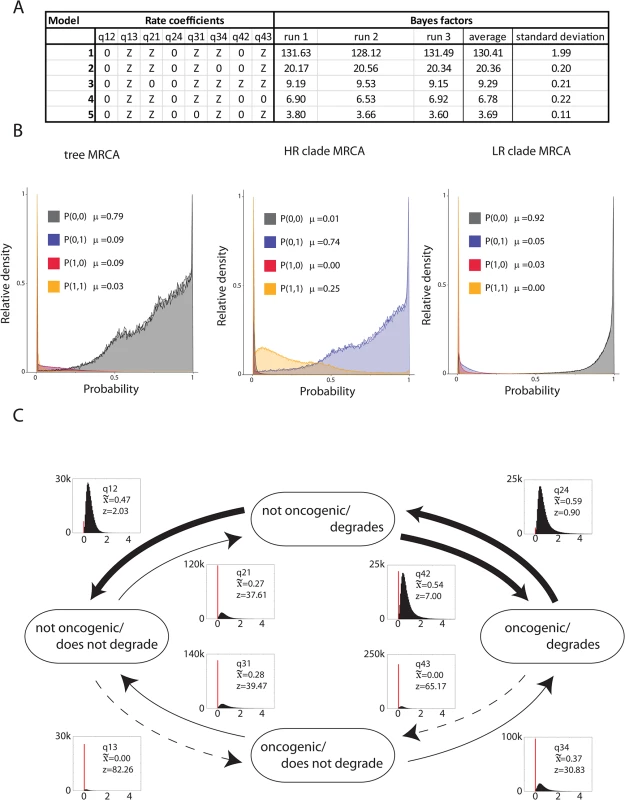
(A) Table of top five models as selected by the RJMCMC analysis. The RJMCMC was run three independent times to ensure that the reproducibility of the model analysis. Through comparing the ratio of posterior to prior odds (see materials and methods) we obtain Bayes Factors to support the choice of a specific model. The selected model has a Bayes Factor of 130.41 (st.dev. = 1.99) suggesting decisive evidence in favor of this model. (B) Ancestral phenotype reconstruction at three important nodes of the Alphapapillomavirus phylogeny. The graphs show the estimated marginal probability density plot for the common MRCA, the high-risk MRCA or the low-risk MRCA. The analysis suggests that the MRCA of the high-risk viruses acquired the ability to degrade PDZ-containing protein, but was likely not an oncogenic virus. (C) Estimated instantaneous rates of change between different combinations of viral phenotypes based on the RJMCMC analysis. Histograms show the posterior distribution of estimated values of the rate parameters. The red bars indicate the fraction of samples in which each rate is estimated to be zero. Arrow width is proportional to the median of these estimated rates. Z indicates the percentage of samples in which each rate parameter is estimated as zero. Consistent with the hypothesis that the ability to degrade PDZ containing proteins is an enabling phenotype, rates associated with oncogenic ability independent of PDZ protein interaction are often estimated as zero (i.e. they do not occur), and are thus represented as dotted line. Fig 3C shows the estimated instantaneous rates change between the four different phenotype combinations. The selected model suggests that the ancestor of the HR clade acquired the ability to degrade PDZ proteins, which is associated with a relatively low probability, likely reflecting the fact that a new interaction motif had to be created. This combination of phenotypes (able to degrade PDZ proteins while not oncogenic) appears to be highly unstable, and suggests that the ability to degrade certain PDZ proteins resulted in sub-optimal viral fitness. Under these assumptions, the model predicts two equally probable scenarios. In the first scenario, the progeny viruses would lose the capacity to degrade PDZ containing proteins. Since no extant members of the HR clade lost the ability to degrade PDZ proteins, we hypothesize that such viruses were unable to colonize the available niche and went extinct. The second scenario proposes that the ancestors of the extant HR viruses were able to colonize and thrive in a new ecological environment. It is likely that these viruses had to evolve ways to cope with the cellular milieu following the loss of PDZ containing proteins. Furthermore, the model predicts that it is highly unlikely to become oncogenic without degrading MAGI1. Taken together the evolutionary data suggests that the ability to degrade PDZ proteins represents an enabling phenotype that had to be acquired prior to further adaptation. Some of these additionally adopted phenotypes inadvertently resulted in cellular deregulation, transformation and cancer.
Discussion
In the present study we tested the ability of E6 proteins from different HPV types to degrade PDZ containing proteins. We used evolutionary hypothesis testing to reconstruct the plausible evolutionary events involved in E6-induced PDZ-protein degradation and HPV type cancer risk. These analyses indicated that the biochemical phenotype was not associated with the cancer risk, but rather with the evolution of extant viral types.
Due to their small genomic size, the complete genomic sequence of over 200 papillomaviruses has been characterized [31]. Importantly, the empirically derived contribution of most HPVs to (cervical) cancer has been well documented. While (cervical) carcinogenicity of the HR-HPV types likely varies in strength along a continuum without clear demarcation, from extremely strong (e.g. HPV16) to probably carcinogenic in rare situations (e.g. HPV 68), evidence suggests that only a small subset of viruses is responsible for virtually all cancers Of note, viruses closely related to these oncogenic HPV types are significantly less common in cancer, despite the fact that their overall prevalence is comparable. Suggesting that these related viruses are implicitly less carcinogenic. Taking into account some of the limitations associated with epidemiological classification, the present study only considered viruses as oncogenic if sufficient epidemiological evidence was available based on analysis by experts [41].
The analysis of biochemical assays in light of the available genomic information and epidemiological classification has allowed us to predict which viral phenotypes may be pathogenic and begin to explain why only a subset of these viruses is truly oncogenic. The use of evolutionary information has been valuable to interpret the relationship between HPV type specific phenotypes and HPV oncogenicity [8,9].
While developing the in vivo assay, we cloned and sequenced a novel isoform of the MAGI1 family, hMAGI1d. We provide evidence that this isoform is expressed in cervical cell lines and primary cervical tumors. We confirmed that, as was previously reported for other MAGI proteins, HPV16 E6 dramatically affects the steady state levels of exogenously expressed human MAGI1d [43] and that this reaction is dependent on the proteasome. Since hMAGI1d appears to be degraded by HPV E6 proteins in a manner similar to what has been reported for other PDZ containing proteins we used this human isoform as a representative member of the PDZ-protein family. Previous studies have shown that specific residues within the PBM and/or certain post-translational modifications may result in each E6 protein having a specific PDZ protein interactome [23,46,47,48]. However, the goal of this study was to relate the overall ability to degrade PDZ containing substrates to oncogenic potential. It was previously shown that most E6 proteins interact with MAGI1 with nano-molar affinity [49]. Therefore, by comparing the steady state levels of hMAGI1d, and not the kinetics of degradation, this approach allows for the representation of the degradation phenotype as a discrete variable.
The analysis of 24 tested viral E6 proteins showed that all members of the HR-clade, independent of oncogenic risk-classification were capable of degrading hMAGI1d. In addition, we provide evidence that the E6 protein of HPV40, a member of the alpha-8 species, contains a functional type 1 PBM. While HPV E6 proteins appear to prefer a Val or Leu residue, any hydrophobic residue can allow for interaction with PDZ containing proteins [42]. Indeed, the PBM found on the HBV core protein contains a terminal cysteine [50,51]. The observation that the members of the alpha-8 clade are the only low-risk viruses that encode E6 proteins with a functional PBM may be the result of convergent evolution. This is supported by the presence of a divergent cysteine (as opposed to the canonical valine or leucine). Importantly, this implies that (some) members of the low-risk clade have the inherent ability to degrade cellular targets. Indeed, we have previously shown that the E6 protein derived from the low-risk virus HPV71 is able to degrade p53 [8]. Furthermore, when the HPV18 PBM was grafted onto HPV11 E6, this chimeric protein was able to degrade cellular PDZ proteins [52]. This supports the notion that the ability to degrade cellular protein is inherently shared by all Alphapapillomaviruses [52].
We used evolutionary trait analysis to integrate biochemical, epidemiological and evolutionary data. This approach allowed us to model how two viral phenotypes (the ability to degrade PDZ containing proteins and viral oncogenic potential) evolved. Importantly, we were able to estimate the order of evolutionary events in the emergence of these traits in HPVs. Since all tested members of the High-risk clade are able to degrade hMAGI1d, the most parsimonious explanation suggests that the putative ancestor of this clade was likewise a degrader. Indeed, the trait analysis favors the hypothesis that the ancestor of the extant high-risk viruses gained the ability to degrade PDZ-containing proteins. Notably, this putative ancestral virus was likely not oncogenic. A key result indicates that the ability to degrade PDZ-proteins is a highly unstable phenotype. This suggests that even though the initial acquisition of a type I PBM lowered viral fitness, it may have allowed for the colonization of a new ecological niche. Furthermore, since the acquisition of PDZ degradation did not coincide with the ability of viruses to cause cancer, the oncogenic types must have acquired additional phenotype(s) that explain their association with human cancer. We have previously reported that the ability to increase cellular hTERT, the protein subunit of telomerase, shows strong association with epidemiological classification [9]. Importantly, since the oncogenic viruses do not form a monophyletic clade, oncogenicity may represent convergent evolution. Alternatively, the non-oncogenic types within the high-risk-clade may have reduced penetrance of the oncogenic phenotype(s), thereby making them less oncogenic.
The present study provides evidence that all members of the High-Risk clade are able to target PDZ containing proteins for degradation. Phosphorylation of the PBM by cellular kinases modulates the ability of viral E6 proteins to recognize PDZ containing substrates. The observation that evolutionarily related E6 proteins are substrates of divergent kinases [48], suggests that the acquisition of a PBM predates the (convergent) evolution of regulation through post-translational modifications.
By analyzing extant E6 proteins, a previous study did not find evidence for a strict correlation between in vitro PDZ protein degradation and oncogenicity [26]. However, in order to fully understand the importance of E6/PDZ-protein interaction and degradation in the malignant process, it is important to analyze this phenotype in relationship to the evolutionary history of these viruses. By taking evolutionary relationships into account we propose a model in which the ability to degrade PDZ proteins allowed for the colonization of a new cellular niche (e.g the cervical transformation zone).
A role for PDZ protein degradation during the viral lifecycle is supported by the observation that mutants of HPV31 unable to interact with PDZ proteins are less fit compared to wild type viruses. Specifically, cellular proliferation, viral copy number control and other early viral functions were affected [20]. A PBM is required for long-term replication of the viral genome. Interestingly, this requirement is alleviated when p53 is removed from the cell using shRNA [53]. While, all tested members of the HR-clade were capable of degrading both p53 and hMAGI1d [8], low-risk viruses do not show clear correlation between both phenotypes. HPV71, a virus previously shown to degrade p53 [8], did not affect the steady state levels of MAGI1d. Inversely, HPV40 does not degrade p53. Therefore, the association between the ability to degrade p53 and PDZ-proteins may be less clear than previously suggested [43].
In conclusion, the ability of E6 proteins to interact with PDZ proteins allowed papillomaviruses to colonize a new ecosystem in the host. However, in order to thrive within this new environment the virus evolved additional ways to usurp the host cells’ machinery. Through interfering with normal differentiation and cell cycle control pathways, long term persistent infection may prime the cell for malignant transformation. The acquisition of hTERT promoter activation (and/or other interactions) by the oncogenic viruses might begin to explain why these viruses are associated with significantly higher cancer rates compared to non-oncogenic types [9].
Finally, this study highlights the importance of combining epidemiological, biochemical and evolutionary data with phylogenetic analysis in attempting to understand the relative role of specific viral phenotypes with host pathogenesis.
Supporting Information
Zdroje
1. de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H (2004) Classification of papillomaviruses. Virology 324 : 17–27. 15183049
2. Bernard HU, Burk RD, Chen Z, van Doorslaer K, Zur Hausen H, et al. (2010) Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401 : 70–79. doi: 10.1016/j.virol.2010.02.002 20206957
3. Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S (2007) Human papillomavirus and cervical cancer. Lancet 370 : 890–907. 17826171
4. Bodily J, Laimins LA (2011) Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol 19 : 33–39. doi: 10.1016/j.tim.2010.10.002 21050765
5. Burk RD, Chen Z, Van Doorslaer K (2009) Human papillomaviruses: genetic basis of carcinogenicity. Public Health Genomics 12 : 281–290. doi: 10.1159/000214919 19684441
6. Schiffman M, Herrero R, DeSalle R, Hildesheim A, Wacholder S, et al. (2005) The carcinogenicity of human papillomavirus types reflects viral evolution. Virology 337 : 76–84. 15914222
7. IARC (2012) IARC Monographs on the Evaluation of Carcinogenic Risks to Humans Volume 100B: A Review of Human Carcinogens: Biological Agents.
8. Fu L, Van Doorslaer K, Chen Z, Ristriani T, Masson M, et al. (2010) Degradation of p53 by Human Alphapapillomavirus E6 Proteins Shows a Stronger Correlation with Phylogeny than Oncogenicity. PLoS ONE 5: e12816. doi: 10.1371/journal.pone.0012816 20862247
9. Van Doorslaer K, Burk RD (2012) Association between hTERT activation by HPV E6 proteins and oncogenic risk. Virology 433 : 216–219. doi: 10.1016/j.virol.2012.08.006 22925336
10. Van Doorslaer K, Burk RD (2010) Evolution of Human Papillomavirus Carcinogenicity. Advances in Virus Research 77 : 41–62. doi: 10.1016/B978-0-12-385034-8.00002-8 20951869
11. Moody CA, Laimins LA (2010) Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 10 : 550–560. doi: 10.1038/nrc2886 20592731
12. McLaughlin-Drubin ME, Munger K (2009) Oncogenic activities of human papillomaviruses. Virus Res 143 : 195–208. doi: 10.1016/j.virusres.2009.06.008 19540281
13. Klingelhutz AJ, Roman A (2012) Cellular transformation by human papillomaviruses: Lessons learned by comparing high - and low-risk viruses. Virology 424 : 77–98. doi: 10.1016/j.virol.2011.12.018 22284986
14. White EA, Kramer RE, Tan MJ, Hayes SD, Harper JW, et al. (2012) Comprehensive Analysis of Host Cellular Interactions with Human Papillomavirus E6 Proteins Identifies New E6 Binding Partners and Reflects Viral Diversity. J Virol.
15. Vande Pol SB, Klingelhutz AJ (2013) Papillomavirus E6 oncoproteins. Virology 445 : 115–137. doi: 10.1016/j.virol.2013.04.026 23711382
16. Roman A, Munger K (2013) The papillomavirus E7 proteins. Virology 445 : 138–168. doi: 10.1016/j.virol.2013.04.013 23731972
17. Pim D, Bergant M, Boon SS, Ganti K, Kranjec C, et al. (2012) Human papillomaviruses and the specificity of PDZ domain targeting. FEBS J 279 : 3530–3537. doi: 10.1111/j.1742-4658.2012.08709.x 22805590
18. Doorbar J (2006) Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 110 : 525–541. 16597322
19. James MA, Lee JH, Klingelhutz AJ (2006) Human papillomavirus type 16 E6 activates NF-kappaB, induces cIAP-2 expression, and protects against apoptosis in a PDZ binding motif-dependent manner. J Virol 80 : 5301–5307. 16699010
20. Lee C, Laimins LA (2004) Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus type 31. J Virol 78 : 12366–12377. 15507623
21. Massimi P, Gammoh N, Thomas M, Banks L (2004) HPV E6 specifically targets different cellular pools of its PDZ domain-containing tumour suppressor substrates for proteasome-mediated degradation. Oncogene 23 : 8033–8039. 15378012
22. Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, et al. (1997) Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275 : 73–77. 8974395
23. Fournane S, Charbonnier S, Chapelle A, Kieffer B, Orfanoudakis G, et al. (2011) Surface plasmon resonance analysis of the binding of high-risk mucosal HPV E6 oncoproteins to the PDZ1 domain of the tight junction protein MAGI-1. J Mol Recognit 24 : 511–523. doi: 10.1002/jmr.1056 20842623
24. Kranjec C, Massimi P, Banks L (2014) Restoration of MAGI-1 Expression in Human Papillomavirus-Positive Tumor Cells Induces Cell Growth Arrest and Apoptosis. J Virol 88 : 7155–7169. doi: 10.1128/JVI.03247-13 24696483
25. Thomas M, Glaunsinger B, Pim D, Javier R, Banks L (2001) HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Oncogene 20 : 5431–5439. 11571640
26. Muench P, Hiller T, Probst S, Florea AM, Stubenrauch F, et al. (2009) Binding of PDZ proteins to HPV E6 proteins does neither correlate with epidemiological risk classification nor with the immortalization of foreskin keratinocytes. Virology 387 : 380–387. doi: 10.1016/j.virol.2009.02.018 19285702
27. Thomas M, Narayan N, Pim D, Tomaic V, Massimi P, et al. (2008) Human papillomaviruses, cervical cancer and cell polarity. Oncogene 27 : 7018–7030. doi: 10.1038/onc.2008.351 19029942
28. Dobzhansky T (1973) Nothing in biology makes sense except in the light of evolution. The American Biology Teacher 35 : 125–129.
29. Auersperg N (1964) Long-Term Cultivation of Hypodiploid Human Tumor Cells. J Natl Cancer Inst 32 : 135–163. 14114965
30. Yee C, Krishnan-Hewlett I, Baker CC, Schlegel R, Howley PM (1985) Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol 119 : 361–366. 2990217
31. Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, et al. (2012) The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res.
32. Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30 : 3059–3066. 12136088
33. Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27 : 221–224. doi: 10.1093/molbev/msp259 19854763
34. Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294 : 2310–2314. 11743192
35. Huelsenbeck JP, Bollback JP (2001) Empirical and hierarchical Bayesian estimation of ancestral states. Syst Biol 50 : 351–366. 12116580
36. Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25 : 1253–1256. doi: 10.1093/molbev/msn083 18397919
37. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52 : 696–704. 14530136
38. Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL (2008) AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24 : 581–583. 17766271
39. Pagel M, Meade A (2006) Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat 167 : 808–825. doi: 10.1086/503444 16685633
40. Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, et al. (2009) A review of human carcinogens—Part B: biological agents. Lancet Oncol 10 : 321–322. 19350698
41. Schiffman M, Clifford G, Buonaguro FM (2009) Classification of weakly carcinogenic human papillomavirus types: addressing the limits of epidemiology at the borderline. Infect Agent Cancer 4 : 8. doi: 10.1186/1750-9378-4-8 19486508
42. Beuming T, Skrabanek L, Niv MY, Mukherjee P, Weinstein H (2005) PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics 21 : 827–828. 15513994
43. Ainsworth J, Thomas M, Banks L, Coutlee F, Matlashewski G (2008) Comparison of p53 and the PDZ domain containing protein MAGI-3 regulation by the E6 protein from high-risk human papillomaviruses. Virol J 5 : 67. doi: 10.1186/1743-422X-5-67 18518978
44. Massimi P, Shai A, Lambert P, Banks L (2008) HPV E6 degradation of p53 and PDZ containing substrates in an E6AP null background. Oncogene 27 : 1800–1804. 17934525
45. Currie TE, Greenhill SJ, Gray RD, Hasegawa T, Mace R (2010) Rise and fall of political complexity in island South-East Asia and the Pacific. Nature 467 : 801–804. doi: 10.1038/nature09461 20944739
46. Chi CN, Bach A, Engstrom A, Stromgaard K, Lundstrom P, et al. (2011) Biophysical characterization of the complex between human papillomavirus E6 protein and synapse-associated protein 97. J Biol Chem 286 : 3597–3606. doi: 10.1074/jbc.M110.190264 21113079
47. Thomas M, Massimi P, Navarro C, Borg JP, Banks L (2005) The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24 : 6222–6230. 16103886
48. Boon SS, Tomaic V, Thomas M, Roberts S, Banks L (2015) Cancer-causing human papillomavirus E6 proteins display major differences in the phospho-regulation of their PDZ interactions. J Virol 89 : 1579–1586. doi: 10.1128/JVI.01961-14 25410862
49. Fournane S, Charbonnier S, Chapelle A, Kieffer B, Orfanoudakis G, et al. (2010) Surface plasmon resonance analysis of the binding of high-risk mucosal HPV E6 oncoproteins to the PDZ1 domain of the tight junction protein MAGI-1. J Mol Recognit.
50. Hsu EC, Lin YC, Hung CS, Huang CJ, Lee MY, et al. (2007) Suppression of hepatitis B viral gene expression by protein-tyrosine phosphatase PTPN3. J Biomed Sci 14 : 731–744. 17588219
51. Banks L, Pim D, Thomas M (2012) Human tumour viruses and the deregulation of cell polarity in cancer. Nat Rev Cancer 12 : 877–886. doi: 10.1038/nrc3400 23175122
52. Brimer N, Lyons C, Vande Pol SB (2007) Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 358 : 303–310. 17023019
53. Brimer N, Vande Pol SB (2014) Papillomavirus E6 PDZ interactions can be replaced by repression of p53 to promote episomal human papillomavirus genome maintenance. J Virol 88 : 3027–3030. doi: 10.1128/JVI.02360-13 24352452
54. Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95 : 5857–5864. 9600884
55. Letunic I, Doerks T, Bork P (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37: D229–232. doi: 10.1093/nar/gkn808 18978020
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage TraffickingČlánek An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived NeuronsČlánek Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPsČlánek Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African TrypanosomiasisČlánek Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin ReleaseČlánek HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T CellsČlánek Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 6- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Diagnostický algoritmus při podezření na syndrom periodické horečky
- Autoinflamatorní onemocnění: prognózu zlepšuje včasná diagnostika a protizánětlivá terapie
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Introducing “Research Matters”
- Exploring Host–Pathogen Interactions through Biological Control
- Analysis of Bottlenecks in Experimental Models of Infection
- Expected and Unexpected Features of the Newly Discovered Bat Influenza A-like Viruses
- Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
- Recombinant Murine Gamma Herpesvirus 68 Carrying KSHV G Protein-Coupled Receptor Induces Angiogenic Lesions in Mice
- TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING
- Targeting Human Transmission Biology for Malaria Elimination
- Two Cdc2 Kinase Genes with Distinct Functions in Vegetative and Infectious Hyphae in
- An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived Neurons
- Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs
- Virulence Factors of Induce Both the Unfolded Protein and Integrated Stress Responses in Airway Epithelial Cells
- Peptide-MHC-I from Endogenous Antigen Outnumber Those from Exogenous Antigen, Irrespective of APC Phenotype or Activation
- Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African Trypanosomiasis
- An Ultrasensitive Mechanism Regulates Influenza Virus-Induced Inflammation
- The Role of Human Transportation Networks in Mediating the Genetic Structure of Seasonal Influenza in the United States
- Host Delivery of Favorite Meals for Intracellular Pathogens
- Complement-Opsonized HIV-1 Overcomes Restriction in Dendritic Cells
- Inter-Seasonal Influenza is Characterized by Extended Virus Transmission and Persistence
- A Critical Role for CLSP2 in the Modulation of Antifungal Immune Response in Mosquitoes
- Twilight, a Novel Circadian-Regulated Gene, Integrates Phototropism with Nutrient and Redox Homeostasis during Fungal Development
- Surface-Associated Lipoproteins Link Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels
- Latent Membrane Protein LMP2A Impairs Recognition of EBV-Infected Cells by CD8+ T Cells
- Bank Vole Prion Protein As an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains
- Neuronal Subtype and Satellite Cell Tropism Are Determinants of Varicella-Zoster Virus Virulence in Human Dorsal Root Ganglia Xenografts
- Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2'-Fluoro-4'-Chloromethyl-Cytidine Triphosphate
- Structure of the Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome
- Activated Brain Endothelial Cells Cross-Present Malaria Antigen
- Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in
- Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin Release
- A Macrophage Subversion Factor Is Shared by Intracellular and Extracellular Pathogens
- A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT
- Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor
- The Role of ExoS in Dissemination of during Pneumonia
- IRF-5-Mediated Inflammation Limits CD8 T Cell Expansion by Inducing HIF-1α and Impairing Dendritic Cell Functions during Infection
- Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
- Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7
- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- HPV16 Down-Regulates the Insulin-Like Growth Factor Binding Protein 2 to Promote Epithelial Invasion in Organotypic Cultures
- The νSaα Specific Lipoprotein Like Cluster () of . USA300 Contributes to Immune Stimulation and Invasion in Human Cells
- RSV-Induced H3K4 Demethylase KDM5B Leads to Regulation of Dendritic Cell-Derived Innate Cytokines and Exacerbates Pathogenesis
- Leukocidin A/B (LukAB) Kills Human Monocytes via Host NLRP3 and ASC when Extracellular, but Not Intracellular
- Border Patrol Gone Awry: Lung NKT Cell Activation by Exacerbates Tularemia-Like Disease
- The Curious Road from Basic Pathogen Research to Clinical Translation
- From Cell and Organismal Biology to Drugs
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
- A 21st Century Perspective of Poliovirus Replication
- Is Development of a Vaccine against Feasible?
- Waterborne Viruses: A Barrier to Safe Drinking Water
- Battling Phages: How Bacteria Defend against Viral Attack
- Archaea in and on the Human Body: Health Implications and Future Directions
- Degradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche
- Natural Variants of the KPC-2 Carbapenemase have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability
- Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers
- The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly
- Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites
- The Proteome of the Isolated Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components
- NK-, NKT- and CD8-Derived IFNγ Drives Myeloid Cell Activation and Erythrophagocytosis, Resulting in Trypanosomosis-Associated Acute Anemia
- Successes and Challenges on the Road to Cure Hepatitis C
- BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses
- A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers
- Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria
- Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration
- Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
- EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival
- Single-Cell and Single-Cycle Analysis of HIV-1 Replication
- TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase
- The Herpes Simplex Virus Protein pUL31 Escorts Nucleocapsids to Sites of Nuclear Egress, a Process Coordinated by Its N-Terminal Domain
- Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections – A Prospective Cohort Study
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- Battling Phages: How Bacteria Defend against Viral Attack
- A 21st Century Perspective of Poliovirus Replication
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Revma Focus: Spondyloartritidy
nový kurz
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



